Abstract
Genetically engineered microbes that secrete therapeutics, sense and respond to external environments, and/or target specific sites in the gut fall under an emergent class of therapeutics, called live biotherapeutic products (LBPs). As live organisms that require symbiotic host-interactions, LBPs offer unique therapeutic opportunities, but also face distinct challenges in the gut microenvironment. In this review, we describe recent approaches (often demonstrated using traditional probiotic microorganisms) to discover LBP chassis and genetic parts utilizing omics-based methods and highlight LBP delivery strategies, with a focus on addressing physiological challenges that LBPs encounter after oral administration. Finally, we share our perspective on the opportunity to apply an integrated approach, wherein discovery and delivery strategies are utilized synergistically, towards tailoring and optimizing LBP efficacy.
Keywords: live biotherapeutic products, multi-omics, drug discovery, drug delivery, gastrointestinal physiology
Tailoring the Development of Live Biotherapeutic Products for Human Application
Live biotherapeutic products (LBPs) are an emerging therapeutic modality that encompass living microbes (e.g., bacteria, yeast), are not vaccines, and are used to prevent, treat, or cure human diseases [1]. LBPs may include a single microbial strain or a consortium of multiple microbial strains. The functions of LBPs may be conferred through processes innate to the microbe(s) or enabled through genetic engineering. In all cases, LBPs perform specific therapeutic functions, thus distinguishing them from probiotic supplements [2]. One of the first examples of a genetically engineered LBP was a Lactococcus lactis was engineered to secrete interleukin-10 to locally treat intestinal inflammation through in vivo optimization in small animals, introduction of biocontainment strategies, and evaluation in human clinical trials [3–5]. While there are currently no FDA approved LBPs, many current clinical trials utilize either consortia-based or single strain-based LBPs for the treatment of recurrent Clostridium difficile infection [6, 7] amongst a variety of other indications including cancer, inflammation, metabolic disorders, and rare diseases. Over 50% of current clinical trials using LBPs for cancer treatment are in combination with immunomodulatory antibodies, exemplifying their importance in immune regulation [8]. This validates the potential for LBPs to target a wide variety of indications and substantiates the need to optimize the discovery and delivery process to rapidly expand LBP translation to the clinic [9].
In this review, we focus on genetically engineered LBPs that are administered via the oral route, due to their rapid emergence in clinical trials and because of their unique potential to be tailored for specific therapeutic functions using omics-based methodologies and formulation strategies. We primarily focus on studies performed in several traditional probiotic chassis (see Glossary) organisms while highlighting rising LBP chassis candidates in the field. We close by providing a perspective on the challenges and opportunities in the clinical translation of novel LBPs.
LBPs provide a wide range of beneficial functions such as regulation of the mucosal immune system, in situ production of therapeutics, nutrient/toxin metabolism, or even act as living diagnostics [10, 11]. Unlike other therapeutic modalities (e.g., small molecules, biologics, gene therapies) [12], LBPs are living, growing, and dynamically interacting with their hosts. As such, design of LBPs presents unique challenges such as their required survival within the host, their potential interactions with the host immune system, and their close competition with existing members of the gut microbiome [13]; these challenges stem from the unique physiology of the gut and the need for LBPs to navigate and synergistically interact with the host (Figure 1, Key Figure).
Figure 1, Key Figure. Physiological Challenges in the Human Gut.
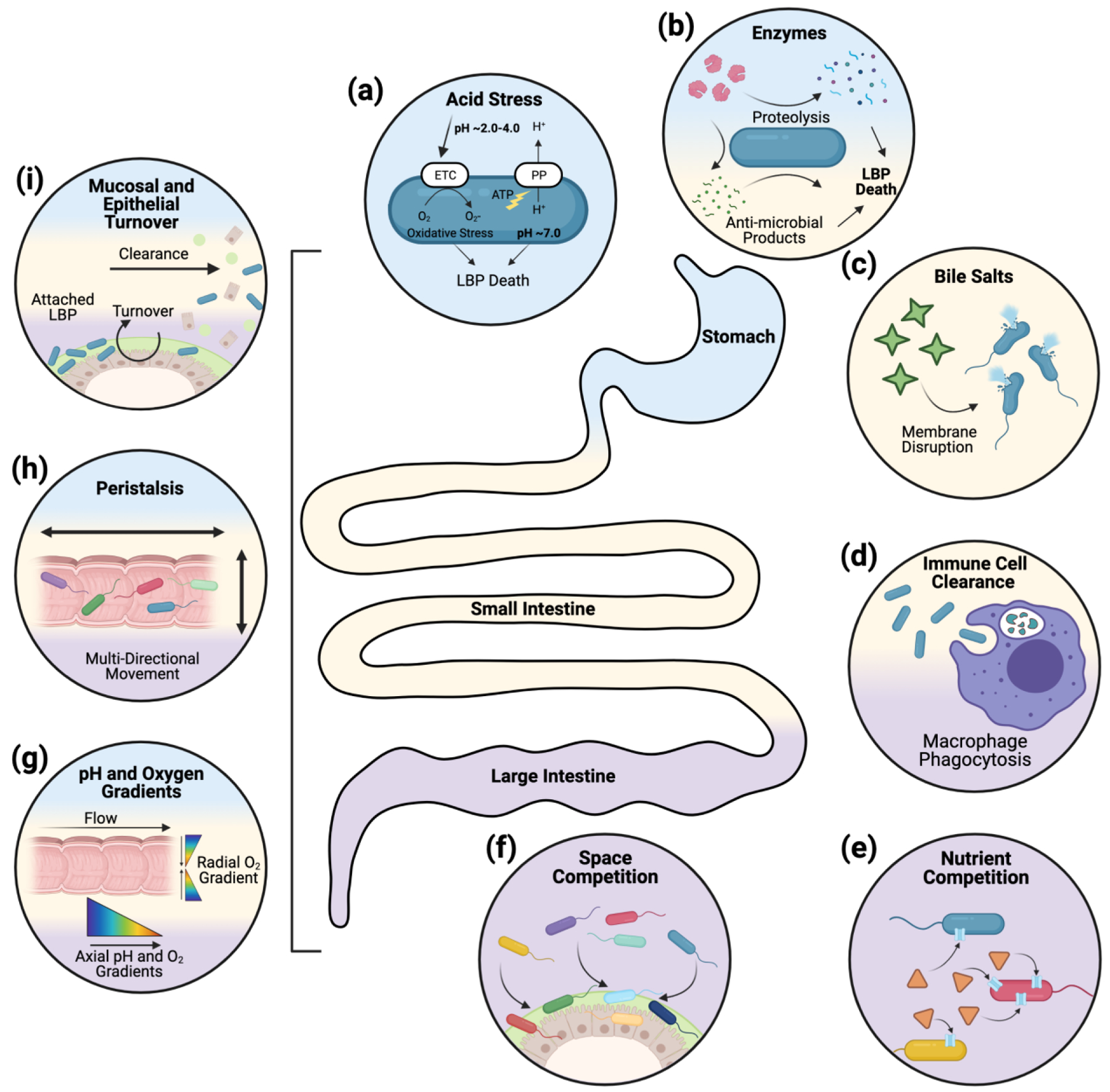
Upon oral administration, LBPs will encounter various physiological challenges during passage from the stomach (blue), through the small intestine (yellow), and to the colon/large intestine (purple). There are a variety of chemical challenges secreted in the gut, such as (a) acid (stomach), (b) digestive enzymes (stomach and small intestine), and (c) bile salts (small intestine) which either disrupt essential LBP components (e.g., cell wall) or cause internal stresses that lead to LBP death. (d) Immune cells in the gut (small intestine and colon) can actively sense, interact with, and clear exogeneous LBPs. Competition, arising from the existing microbiota (large intestine), can limit LBPs ability to access sufficient (e) nutrients, for growth and metabolism, or (f) space, for adherence, growth, and colonization. Physiological challenges can also be ubiquitously encountered through the gut such as chemical gradients (g) (pH and oxygen) or physical phenomena including (h) peristalsis and (i) epithelial/mucosal turnover. These physiological aspects, and how they serve as challenges to LBPs upon oral administration, are discussed in more detail in Box 1.
A major limitation in the translation of LBPs is the lack of quantitative and high-throughput approaches to enable discovery of LBP chassis or components (e.g., promoters, colonization factors) that can navigate host physiology and efficiently perform therapeutic functions. Another, distinct limitation, is the lack of strategies that enable controlled LBP delivery (e.g., target site localization, controlled transit times) to the gut to provide therapeutically optimal LBP concentrations. In this review, we consider gut physiology (Box 1) to identify major challenges (Figure 1) that limit LBP efficacy. We then highlight how these physiological challenges can be circumvented, overcome, or even exploited via: (i) discovery of LBP chassis or genetic components using quantitative omics methodologies, and (ii) novel delivery strategies including pharmaceutical formulations or genetic engineering approaches. Finally, we provide our perspective as to how discovery and delivery can be integrated to enable the rational design, development, and implementation of next-generation LBPs.
Box 1. Host-LBP Interactions: Physiological Challenges.
The dynamic interactions between LBPs and the host microenvironment govern LBP fate and function. Following oral administration, LBPs encounter various physiological challenges (Figure 1) that decrease survival, prevent colonization, and alter therapeutic function of LBPs. In the stomach, acids induce protein denaturation [96] and enzymes cause proteolysis of the LBP [97]. In the small intestine, enzymes persist and bile salts, which solubilize cell wall lipids and proteins, are secreted [98]. As LBPs enter the colon (large intestine), LBP clearance via immune cells (macrophages) occurs [99]. Since the colon is home to the largest number of microbes (~1014 CFU) in the body, it is a highly competitive (for both space and nutrients) environment for LBPs [100]. Furthermore, since many LBPs are fermented in optimal in vitro conditions prior to administration, processed during formulation, and stored at various temperatures, LBPs must rapidly adapt to new nutritional sources and chemical environments (oxygen or pH differences) while competing with microbiota members that have evolved to thrive in that specific environment. A separate set of challenges such as peristalsis, mucus/epithelium turnover, and both pH and oxygen gradients are dynamic and persist through the stomach to the colon [101]. The physical contractions associated with peristalsis breakdown and transport food through the gut, which leads lead to rapid transit times (2h in the stomach, 2h in the small intestine, and 6h in the colon [101]); this limits LBP interactions with target tissues or sites. Mucus and epithelium turnover are active processes that occur dynamically and at different rates in the stomach, small intestine, and colon; mucus turnover contributes to gut transit of LBPs and influences residence time [102]. Oxygen gradients in the gut (~21% in the stomach with gradual reduction to almost 0% in the colon), can affect LBP performance since LBP metabolism and thus therapeutic function can be influenced by oxygen content [103]. Similarly, gut pH (generally 1.0–2.5 in stomach, 7–7.4 in small intestine, and to 6–6.7 in colon [104]), can affect LBP growth and metabolism. Collectively, chemical challenges (e.g., stomach acids, enzymes, bile salts) can lead to the rapid reduction of viable LBPs, physical challenges (e.g., peristalsis, mucus turnover) can limit the ability of viable LBPs to colonize or persist in specific parts of the gut, and competitive challenges (e.g., nutrient competition, space competition) can affect LBP metabolism and their therapeutic functions.
Overcoming Physiological Challenges in LBP Discovery
In this section, we discuss omics-based strategies (Table 1) for evaluating and engineering the interactions between LBPs and both the indigenous microbiota (e.g., competition for nutrients or niches, nutritional cross-feeding) and the human host (e.g., binding to gut surfaces, immune recognition, metabolic crosstalk) (Figure 2).
Table 1.
Omics Strategies for Discovering Novel LBP Design Elements.
| Omics Science | Characterizes/Quantifies | Organisms investigated | Application to LBPs | Refs |
|---|---|---|---|---|
| Genomics | DNA | Bifidobacterium bifidum, 36 Bifidobacterium spp, Bacillus coagulans, Bacteroides thetaiotaomicron, Bacteroides ovatus, Bacteroides uniformis, Bacteroides vulgatus, Blautia hydrogenotrophica, Collinsella aerofaciens, Clostridium hiranonis, Desulfovibrio piger, Eggerthella lenta, Eubacterium rectale, Faecalibacterium prausnitzii, Prevotella copri | Colonization factors and in vivo adaptation mechanisms via comparative genomics | [15, 18] |
| Inter-microbe interactions via profiling compositional changes in native and synthetic communities | [21, 22] | |||
| Transcriptomics | RNA | Saccharomyces cerevisiae, Lactobacillus plantarum, Lactococcus lactis, Lactobacillus rhamnosus GG, Lactobacillus crispatus, Bacteroides fragilis, Akkermansia mucinipila, Ruminococcus gnavus, Bacteroides thetaiotaomicron | Inter-microbe interactions via differential expression analysis in synthetic communities | [34, 35] |
| In vivo adaptation mechanisms by differential expression analysis in gut-like conditions | [30, 33, 105, 106] | |||
| Genetic parts for engineered LBP design via differential expression analysis in response to defined gut cues | [31, 37, 38] | |||
| Colonization factors via spatially-resolved differential expression analysis | [107] | |||
| Proteomics | Proteins | Lactobacillus acidophilus, Lactobacillus salivarius | In vivo adaptation mechanisms via differential proteome analysis in gut-like conditions | [40–43, 108] |
| Genetic parts for engineered LBP design via proteomics of engineered strains | [109, 110] | |||
| Metabolomics | Metabolites (Excluding DNA, RNA, or protein) | Lactobacillus acidophilus, Bacteroides thetaiotaomicron, Bacteroides ovatus, Bacteroides uniformis, Bacteroides vulgatus, Blautia hydrogenotrophica, Collinsella aerofaciens, Clostridium hiranonis, Desulfovibrio piger, Eggerthella lenta, Eubacterium rectale, Faecalibacterium prausnitzii, Prevotella copri, Ruminococcus gnavus, Ruminococcus bromii | In vivo adaptation mechanisms via metabolomics of gut-adapted strains | [48, 49, 51] |
| Inter-microbe interactions via metabolic profiling of synthetic communities | [22, 29] | |||
| Functional genomics | Engineered gene function | Escherichia coli Nissle, Bacteroides thetaiotaomicron, Saccharomyces cerevisiae, Saccharomyces boulardii, Bacteroides fragilis | Promoter design | [34, 55, 59] |
| Pathway optimization | [56, 57] | |||
| In situ genetic tractability | [53, 54] | |||
| In vivo adaptation mechanisms | [62, 63] |
Figure 2. Discovery Approaches to Overcome Physiological Challenges and Control Microenvironment Interactions.
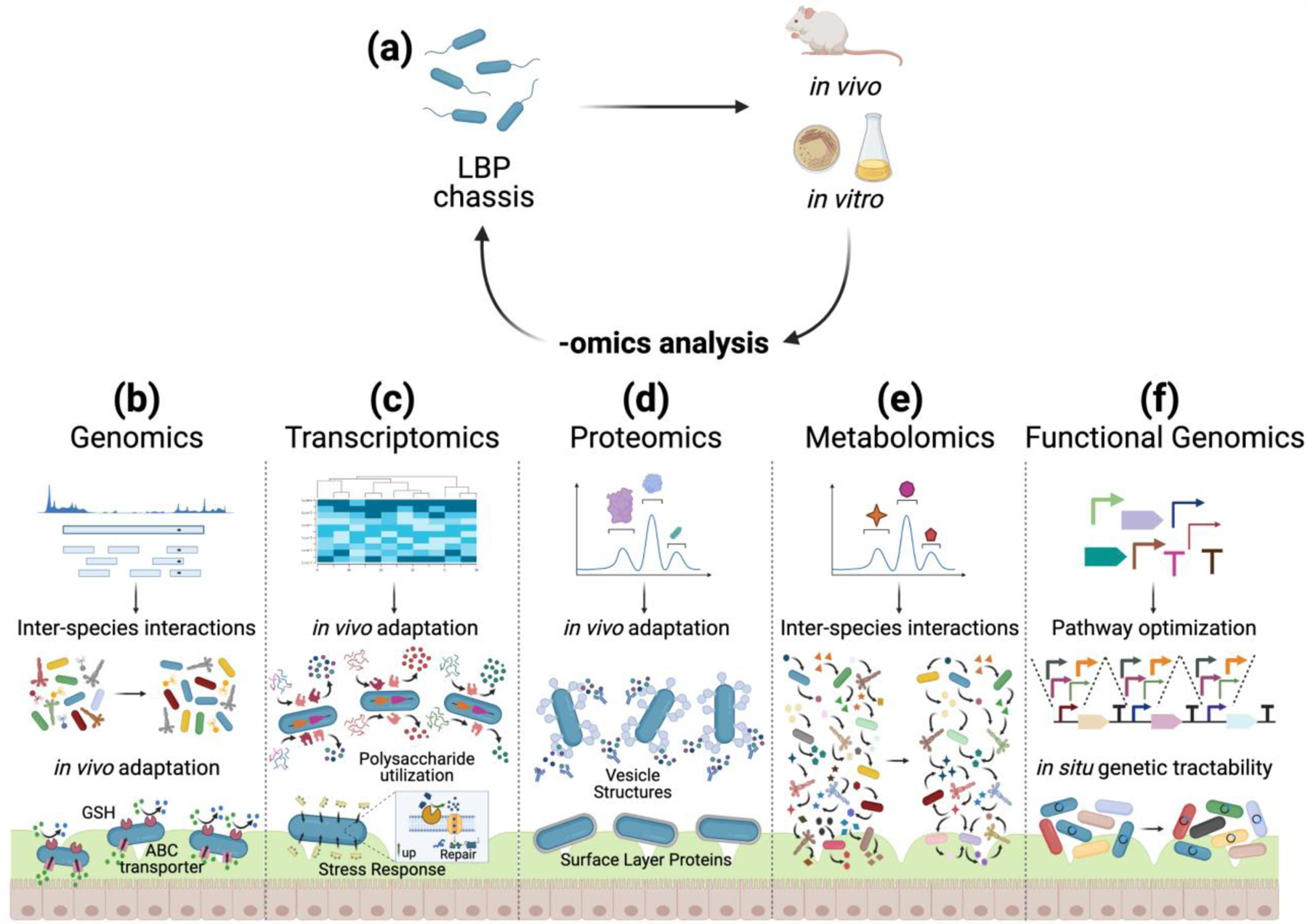
(a) Discovery of LBPs can be guided by investigating chassis in in vitro and in vivo settings, using a variety of ‘omics pipelines. (b) Genomics measures DNA composition at both the organism and community levels and can be used to determine the presence and abundance of LBPs in various locations in the gut and under various microenvironment conditions. These insights identify factors that drive the LBP’s adaptation to the gut and control interactions with other microbiota members. (c) Transcriptomics can identify genes that are differentially regulated in response to physiological challenges in the gut. These differentially regulated genes illuminate in vivo adaptation mechanisms of LBPs. (d) Proteomics can identify proteins in LBPs (as well as their subcellular localization) that facilitate in vivo adaptation. Since proteins on LBP surfaces mediate extracellular interactions, proteomics can provide insight into how LBPs interact and communicate with the dynamic gut microenvironment. (e) Metabolomics can identify the metabolic activity of LBPs in a multi-species community through identification of metabolites and their fluxes, which in turn illuminates how an LBP chassis adapts to gut microenvironments. (f) Functional genomics can be used to achieve pathway optimization and high-throughput strain screening for engineered LBP applications. Pathway optimization enables high activity of the engineered function even under the burden of physiological challenges. High-throughput strain screening can identify candidate chassis that are already gut colonizers and that can receive genetic payloads in situ through horizontal gene transfer.
Genomics
Genomics is the study of the structure, function, and evolution of genomes within a given organism or community of organisms [14]. Genomics analyses can be used to identify LBP chassis that contain colonization-promoting gene functions [15, 16], and identify interactions between the LBP and gut symbionts [17] (Figure 2b). For instance, in silico analysis of the Bifidobacterium bifidum genome revealed genes encoding for glycosyl hydrolases and ABC-type transporters that are inherent features of mucus-colonizing bacteria [18]. These genes enable degradation and transportation of mucin-derived host glycans, thus ensuring ensure nutrient availability in the gut for this LBP chassis [19]. To translate these predictions to function (e.g., colonization) in competitive gut environments, in vivo studies can validate these in silico hypotheses; approaches such as shotgun metagenomics can describe compositional shifts in the microbiota in vivo and therefore identify microbe-microbe interactions. For example, oral administration of Lactobacillus plantarum shifted the composition of the microbiota towards an increase in Bacteroides and decrease in Firmicutes in a mouse model of inflammatory bowel disease [20]. Further mechanistic understanding of how microbe-microbe interactions challenge LBPs can be revealed through genomics-assisted computational modeling of the microbial community [21] as well as experimental approaches. Pairwise combinations of a 12-member synthetic human gut microbiota were cultured to parameterize the generalized Lotka-Volterra equations, revealing how specific species drive community stability [22]. Looking forward, similar mathematical models can be constructed for LBPs, enabling discovery of inter-species interactions and prediction of LBP colonization. Separately, in studies involving human fecal microbiota transplants, metagenomic analyses have enabled the identification of colonization factors which govern the occupation of specific niches in the human gut microbiome [23–25], providing clinically translatable data which can be considered in future engineered LBP design.
Transcriptomics
Transcriptomics is the study of the structure, function, and evolution of the transcriptome (i.e., the entirety of RNA transcripts produced by the genome) of a given organism or community of organisms under a variety of conditions [26]. Here, we focus on transcriptomics analyses performed on microbes placed under conditions relevant to human gut physiology to uncover gene regulation pathways relevant to overcoming physiological challenges (Figure 2c). Transcriptomic analysis of Akkermansia muciniphila demonstrated that several systems involved in bile acid resistance (e.g. ABC transporters, RND transporters, hopanoid synthesis, exopolysaccharide synthesis) were differentially regulated while under a bile acid challenge [27]. Further, a transcriptomic study of Akkermansia muciniphila cultured in the presence of mucin showed upregulation of fructosidase, β-galactosidase, and hexosaminidase, which convert mucin to various oligosaccharides, which are then converted into monosaccharides to be used in glycolysis [28]. A similar transcriptomic study has also been conducted on a smaller community consisting of Faecalibacterium prausnitzii, Blautia hydrogenotrophica, and Roseburia intestinalis revealing that each species exhibited different metabolic activities when cultured individually or together and showing that these strains competed for fructose and cross-fed formate [29]. Other transcriptomic analyses of mucin utilization have also been conducted on Bacteroides thetaiotaomicron, Ruminococcus gnavus, and Bacteroides fragilis [30–32] to understand how different chassis adapt and utilize mucin structures in the gut. Collectively, presence and/or activation of certain cellular processes can provide competitive nutritional advantage to LBP chassis over other microbes, and these patterns can be identified via transcriptomics.
Transcriptomic analysis can be performed for other physiological stresses such as acid [33], oxygen [34], and other microbes [35]. Lactobacillus acidophilus profiling in germ free mice revealed that carbohydrate, nucleotide, and amino acid metabolism genes, as well as genes encoding for mucus-binding proteins, surface adhesins, and surface-layer (S-layer) proteins, are differentially regulated in the gut [36]. These genes exhibited differential spatial expression patterns, indicating that Lactobacillus acidophilus alters gene expression depending on niche association. Once identified by transcriptomics, these regulatory networks can be synthetically optimized to further increase chassis fitness or can be recombinantly introduced in other less-fit LBPs to resist physiological challenges [37, 38].
Proteomics
Proteomics is the study of the structure, function, and evolution of the proteome (i.e., the entire set of proteins expressed) by an organism or community of organisms at a given time and under a variety of conditions [39]. Here, we primarily focus on proteomic studies which evaluate the protein expression profiles of single microbial strains in response to physiological challenges relevant to the human gut (Figure 2d). Previously, proteome responses of several LBPs in the presence of bile [40], nutrient availability [41], and oxygen gradients [42] have been captured. A proteomics study conducted on the surface proteins of different Lactobacillus strains revealed strain-specific adaptation to physiological challenges such as bile, immune responses, and other microbes [43]. Identifying and characterizing surface proteins with immunomodulatory activities can provide insight into circumventing immune challenges in the gut. Recent proteomic studies conducted on Lactobacillus acidophilus surface proteins identified several S-layer proteins and S-layer associated proteins with immunomodulatory characteristics [44]. LBP surface protein content (both quantity and type) may drive LBP chassis’ long-term adaptation in the gut, allowing colonization to reach “steady-state”. Proteomics enables high-throughput analysis of the proteins involved in overcoming physiological challenges specific to the gut environment which can assist in selecting a chassis for LBP design or incorporating engineered elements utilizing these proteins.
Metabolomics
Metabolomics analyses may be categorized into two distinct approaches: 1) untargeted metabolomics wherein all molecules (e.g., sugars, lipids, fatty acids, phenolic compounds) other than DNA, RNA, or proteins in an organism or community are evaluated, and 2) targeted metabolomics wherein specific and known molecules are evaluated in a given organism or community under a variety of experimental conditions [45]. Here, we primarily discuss targeted metabolomic approaches because it can be used to distinguish which mechanisms microbes use to overcome specific physiological challenges in the gut (Figure 2e). Metabolomics can be used to reveal the ability of the LBP to tune its metabolic functions to transform host-derived, microbiota-derived, diet-derived, and xenobiotic compounds to facilitate survival in the gut [46]. For example, a metabolomic study of 22 Lactobacillus plantarum strains revealed the extent of bile acid deconjugation in a strain- and substrate-specific manner. In addition to mitigating bile stress, deconjugated bile acids restrict proliferation of certain microbial species, including opportunistic pathogens, therefore reducing LBP competition for nutrients and space [47]. Similarly, a metabolomics study on a mucin-degrading Ruminococcus gnavus strain and the resistant-starch-degrader Ruminococcus bromii investigated their cross-feeding dynamics in presence of host-derived (mucin) and diet-derived (resistant starch) sugars, revealing that these strains compete for malto-oligosaccharides [48]. Additionally, the metabolic capacity of the LBP can simultaneously benefit the host and enable LBP energy acquisition in the gut. Metabolomic profiling of Lactobacillus acidophilus and Lactobacillus gasseri characterized the metabolic ability and extent of degradation of dietary oxalate, which is a toxic compound involved in kidney disorders including primary hyperoxaluria [49]. Similarly, metabolomic studies on Bacteroides thetaiotaomicron revealed that sphingolipid biosynthesis and outer membrane vesicles were essential for maintenance and development of immune system and gut symbiosis [50, 51]. Collectively, metabolomic analysis of LBPs reveals mechanisms to overcome physiological challenges at the molecular scale.
Functional genomics
Functional genomics uses genetic engineering for high-throughput investigation of gene functions, regulatory parts, and transformability [52] (Figure 2f). Engineering functions into gut-adapted bacteria can minimize the burden of physiological challenges[53]. For example, a technique called MAGIC (metagenomic alteration of gut microbiome by in situ conjugation) demonstrated genetic engineering of gut bacteria through horizontal gene transfer; afterwards, identities of modifiable gut bacteria were elucidated by metagenomic sequencing [54]. Although this approach may enable high-throughput identification of genetically modifiable bacteria in the gut environment, it may not be feasible for therapeutic purposes without additional safeguards in place due to biocontainment concerns. Another approach for ensuring activity of therapeutic functions in presence of physiological challenges is to characterize genetic regulatory parts under gut-mimicking conditions. For instance, using transcript barcoding, activity levels of 30 promoters in Escherichia coli Nissle 1917 (EcN) were characterized in vitro and in vivo [55]; this approach can enable better predictions of engineered function in vivo to aid in regulatory part selection [56]. A similar approach can be applied to LBPs engineered to express complex metabolic pathways. For example, 9 promoters were combinatorically assembled in vivo in Saccharomyces boulardii to achieve rapid optimization of β-carotene and violacein productivities [57]. Broadly, high-throughput characterizations of regulatory parts can identify functions that “turn on” in response to defined gut microenvironments which can enable control over LBP function in hosts. Functional genomic screens, either loss-of-function (i.e., transposon insertion sequencing) or gain-of-function (i.e., genomic fragment library), enable identification of colonization factors in LBPs [58]. Considerable efforts have also recently focused on the engineering of Bacteroides; broadly, these efforts have focused on developing a genetic toolkit including ribosome binding sites, promoters, CRIPSR-Cas systems and recombinases to achieve tunable expression of innate and heterologous genes, and demonstrated a range of gene expression, up to 10,000-fold with constitutive promoters and up to 100-fold with inducible promoters [59–61]. High-throughput screening of 2100 Bacteroides vulgatus clones containing fragments of the genome of a natural colonizer, Bacteroides fragilis, revealed a unique class of polysaccharide utilization loci that enabled colonization of Bacteroides vulgatus by allowing it to better utilize gut mucins [62]. In another study, a metagenomic library generated from healthy infants and their mothers was expressed in EcN. Upon competition of these strains in vivo, it was revealed that expression of genes involved in polysaccharide utilization, acid tolerance, and mucin utilization enabled increased colonization of EcN in germ-free and gnotobiotic mice [63]. Altogether, multi-omic analyses provide a powerful platform for identifying synthetic “parts” that enable an engineered LBP to change its behavior in response to the host’s immune system, disease state, microbiota composition, and metabolic signaling pathways.
Overcoming Physiological Challenges in LBP Delivery
Here, we discuss pharmaceutical formulations and genetic engineering approaches (Figure 3) that can modulate LBP interactions with physiological surfaces, overcome physiological challenges, and address biocontainment issues as they relate to delivery of LBPs.
Figure 3. Formulation and Genetic Engineering Strategies to Improve LBP Delivery.
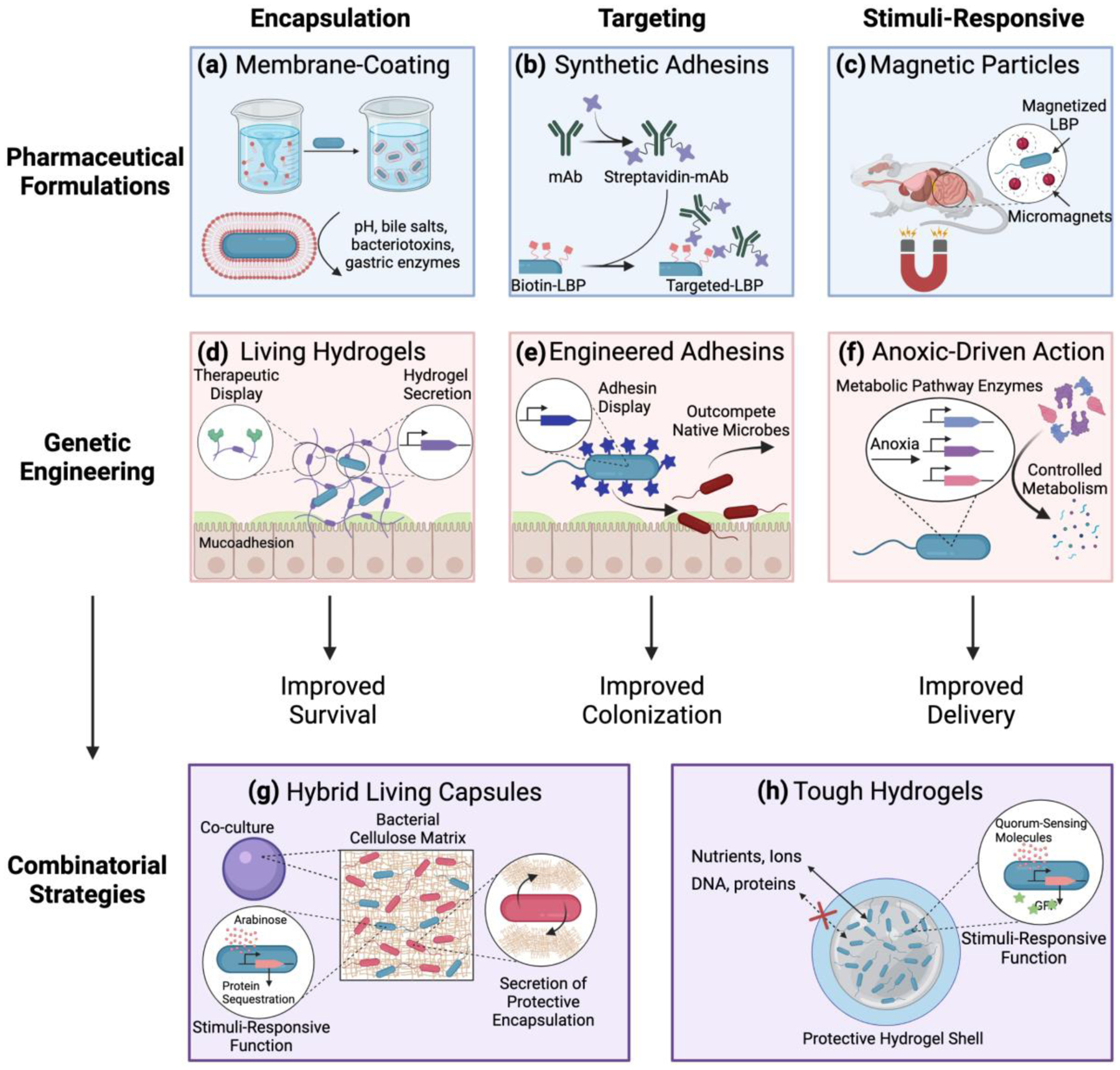
Pharmaceutical formulations and genetic engineering strategies can be leveraged to both overcome physiological challenges and utilize physiological microenvironments to improve LBP delivery and LBP function. Examples of pharmaceutical formulation approaches include (a) encapsulation via phospholipid bi-layer membrane LBP coatings, (b) targeting via synthetic adhesin-surface modification of the LBP, and (c) endowing stimuli-responsive functions by using external magnets to direct the movement of magnetized LBPs. Examples of genetic engineering approaches include (d) engineered LBPs which secrete an encapsulating protective and mucoadhesive hydrogel with conjugated therapeutic modalities, (e) LBPs engineered to express adhesins with high binding affinities on the microbial surface for targeting, and (f) the expression of several enzymes for controlled metabolism in response to anoxic conditions for stimuli-responsive therapeutic function. Recently, pharmaceutical formulations and genetic engineering strategies have been combined to (g) encapsulate genetically engineered LBPs in a hydrogel bead which allows diffusion of nutrients into the bead to maintain the engineered function while achieving biocontainment, and (h) enable co-culture of a bacterial cellulose-secreting probiotic organism with engineered LBPs to enable protective encapsulation of the LBP while maintaining its engineered function.
Formulation Strategies
Pharmaceutical formulations are used to control interactions between a therapeutic and its physiological environment [12]. For LBPs, pharmaceutical formulations can be used to mitigate chemical challenges, interface with physiological tissues, and target specific areas that provide competitive or therapeutic advantages. In recent work, coating of probiotics via self-assembly of biocompatible lipids in a 15-minute vortex step (Figure 3a) enhanced probiotic protection against enzymes, low/high pH, antibiotics, and ethanol. The lipid coating increased LBP residence time in mice and improved therapeutic efficacy in two models of murine colitis [64]. This coating approach has also been used with biocompatible mucoadhesive polymers to protect and improve in vivo survival of coated-microbes [65]. Some LBP formulations are inspired by the innate abilities and functions of gut symbionts. Natural biofilm formation was induced in Bacillus subtilis as a self-encapsulation approach. The biofilm-coated LBP had improved resistance against low pH and gut enzymes in vitro and enhanced LBP mucoadhesion in vivo, leading to 17-fold greater intestinal colonization [66]. Other biofilm-inspired formulations have also been described; alginate-based microparticles were used to simultaneously encapsulate probiotic microbes, enable secretion of small molecule therapeutics and probiotic by-products, and limit diffusion of chemical challenges into the microparticle [67]. Inspired by adhesins, a surface modification approach was used to decorate the exterior of EcN with anti-MUC2 antibodies (Figure 3b). This involved the chemical conjugation of biotin to primary amines on the microbe surface and subsequent introduction of streptavidin conjugated antibodies. This approach enabled molecular targeting to mucus and accelerated EcN colonization [68]. Stimuli-responsive materials can also be used to direct LBP function. Recently, externally applied magnetic fields were used to manipulate the gut transit of EcN that was orally co-administered with micromagnets (Figure 3c) [69]. Importantly, this approach facilitated targeted microbe localization and enabled stable colonization in mice, without the use of antibiotics. The encapsulation approaches, bio-inspired strategies, and stimuli-responsive functionalities described here, and in other recent work [70, 71], could improve delivery of LBPs by increasing survival following oral administration, prolonging residence time, accelerating colonization speed, or specifying the location of colonization.
Genetic Engineering Strategies
Genetic engineering strategies can be applied towards improving delivery by overcoming physiological challenges, facilitating site specific targeting, enabling increased survival and persistence in the gut, and controlling drug release. EcN was genetically engineered to form a living hydrogel through secretion of curli fibers which encapsulate and protect the LBP in situ, promote mucoadhesion, and mimic biofilm formation, thereby mitigating LBP clearance due to mucosal turnover, peristalsis, space competition, and acids/enzymes [72]. This platform was also developed to display therapeutic trefoil factors conjugated to the curli fibers to promote mucosal healing (Figure 3d) [73]. Similar to the surface modified adhesins described above, LBPs can be genetically engineered to display ligands on their surface to enable specific molecular interactions within the host; recent work described the genetic engineering of L. lactis and EcN to express a variety of mucus-binding proteins on the microbial surface [74]. Being naturally resistant to stomach acids and gastric enzymes, spore-forming bacteria (e.g. Clostridia, Firmicutes) are also being studied as potential chassis [75] or for identifying proteins involved in the process of colonizing the late gastrointestinal tract [76]. Further, lactic acid bacteria were engineered to surface-express a fragment of the Clostridioides difficile adhesin, SlpA (Figure 3e); this enabled the LBP to outcompete and prevent colonization of C. difficile in hamster and piglet in vivo models of infection [77].
LBPs can also be genetically engineered to sense and respond to the dynamic environment of the gut. For example, recent work described a genetically engineered lactic acid bacteria that release both antimicrobial and anti-biofilm proteins in response to the P. aeruginosa-specific quorum-sensing molecule, 3-oxo-C12-homoserine lactone [78]. Recently, the dynamic oxygen gradients in the gut were leveraged to initiate therapeutic function by engineering EcN to express phenylalanine-degrading enzymes in response to anoxic environments for the management of phenylketonuria (Figure 3f) [9]. Additionally, the LBP was genetically engineered to be auxotrophic to enable biocontainment. Auxotrophy, or the inability to produce an essential metabolite, is a key element often utilized in genetically engineered microbes for human application to enable biocontainment without the need for plasmid-based, antibiotic resistance markers. In other work, two genetically engineered functions, molecular targeting, and stimuli-responsive drug secretion, were combined in a single LBP. Here, EcN was genetically engineered to: (i) selectively bind to the colorectal surface antigen, heparan sulfate proteoglycan, and (ii) secrete the enzyme myrosinase which converted dietary glucosinolate into the chemotherapeutic sulforaphane. This approach resulted in a 7-fold reduction in tumor occurrence in a murine model of colorectal cancer [79]. Collectively, these and other recent examples [80, 81] highlight the potential of using genetic engineering approaches to control LBP interactions with the host on a molecular scale, or secrete therapeutics in response to external cues.
Combination Approaches: Pharmaceutical Formulations to Deliver Genetically Engineered LBPs
Synergistic strategies that combine both pharmaceutical formulations and genetic engineering strategies to improve LBP delivery have begun to emerge. Recent work utilized the material properties of bacterial cellulose (e.g., biodegradability, barrier properties), secreted by Gluconacetobacter hansenii and the genetic tractability of EcN to generate hybrid living capsules which combined both strains in a single delivery system. EcN, genetically engineered to perform a variety of sense-and-respond functions (e.g., biomolecule sequestration, enzymatic catalysis) was co-cultured with Gluconacetobacter hansenii, which naturally produced a cellulose-based capsule that enabled the encapsulation and protection of the genetically engineered EcN (Figure 3g). Importantly, EcN maintained its genetically engineered functions while encapsulated within the Gluconacetobacter hansenii bacterial cellulose-based capsule [82]. Further, genetically engineered E. coli was encapsulated into crosslinked sodium-alginate beads containing polyacrylamide on the terminal exterior; this created a “tough” hydrogel-like shell on the exterior of the bead (Figure 3h). This formulation approach protected the encapsulated microbes from external challenges (e.g., antibiotic, low pH) while ensuring: (i) biocontainment of the genetically modified LBP via prevention of release/escape of the encapsulated LBP, and (ii) maintenance of genetically engineered functions (e.g., response to external chemical stimuli, secretion of signaling molecules, sensing of heavy-metal contaminants) [83]. This example, which utilizes biomaterial encapsulation of genetically engineered microbes points towards possible next-generation LBP delivery systems wherein biomaterial-mediated biocontainment can be achieved and genetically engineered functions can be maintained. Biocontainment is a critical component in designing genetically engineered LBPs with several other approaches including nutritional auxotrophy, kill switches, transcriptional regulation pathways, and devices to remove genetically engineered constructs, which have been extensively reviewed elsewhere [10, 84]. Further, although stable gut colonization may be desirable in some cases, deriving a therapeutic effect from an engineered LBP is not necessarily dependent on colonization and several genetically engineered LBPs studied in clinical trials were intentionally designed not to colonize the gut for the purpose of improving LBP biocontainment [3, 85]. We envision that both genetically engineered approaches and formulation approaches will continue to be developed and potentially combined in single LBP systems, ideally leading to the establishment of a biocontainment toolbox that can be used for a variety of specific strains, applications, and end-functions.
Concluding Remarks and Future Perspectives
The challenges encountered by LBPs upon oral administration and during gut transit are multifactorial (Figure 1) and can affect LBP functions (e.g., survival, site-specific targeting, in situ drug production, transit time, therapeutic action). These complexities present challenges to both LBP design and delivery, as no single set of criteria is sufficient to design or deliver LBPs that are appropriate for all use cases. On the other hand, the myriad of distinct biomolecular environments in the gut are advantageous, as they enable LBPs to be tailored to reside or perform functions at specific sites.
Multi-omics analyses play a major role in discovery of novel microbial chassis, colonization factors, and regulatory parts that enable LBPs to overcome gut physiological challenges (Figure 2, Table 1). Multi-omics-based LBP discovery has primarily been accomplished in two ways. First, analyses are performed on wild-type, non-engineered symbionts, or probiotic strains. This process illuminates the natural mechanisms microbes use to overcome physiological challenges. Second, and more recently, candidate gene edits are delivered at high throughput to an LBP chassis, and gene edits with beneficial effects are recovered by applying omics-based analyses and innovative selection conditions. It is expected that the recent application of machine learning techniques towards analysis of microbiome data [86] will extend to LBP design. Furthermore, advances in single-cell [87] and spatially-resolved [88] omics analyses can provide greater insights into the heterogeneity of microbial behaviors in vivo. Additionally, while most LBP discovery efforts to date have focused on bacteria, other kingdoms of life (e.g., fungi, viruses, and archaea) may provide other therapeutic opportunities.
As LBPs have been discovered, developed, and evaluated in humans, two distinct strategies to improve their delivery have emerged. The use of pharmaceutical formulations and genetic engineering strategies (Figure 3) are actively being developed and employed to control LBP location (target site), duration (residence time), and concentration (dose) to improve safety and efficacy. Pharmaceutical formulation strategies (e.g., encapsulation, target-functionalization, stimuli-responsive control) can potentially be applied to all LBPs since they rely on using physical and chemical modifications to the chassis. Moreover, formulation approaches are modular, and can be modified for specific LBPs (e.g., material-microbe compatibility), delivery functions (e.g., release), or host tissues (e.g., disease sites). On the other hand, genetic engineering approaches can be uniquely applied to LBPs since unlike other therapeutics, LBPs are living and actively sense, respond, and perform therapeutic functions dynamically within the host. Together, the use of pharmaceutical formulations or genetic engineering strategies offer advantages that can synergistically cooperate to provide unmatched control over LBP delivery and function; indeed, recent examples (Figure 3c) highlight the potential of combining these approaches to address unmet needs in LBP delivery.
Distinct challenges in the clinical translation of LBPs from academic labs to generally available therapies include the translation from small animal models to humans and the ability for model systems to predict in vivo utility. Each type of model system (e.g. in vitro, in vivo, in silico) has both unique benefits and limitations as it relates to generating multi-omics data. In vitro culture and adhesion models allow for identification of specific parameters which may influence colonization and/or survivability in the gut such as metabolite consumption or microbial surface binding proteins [89–92]. In contrast in vivo models enable the generation of physiologically relevant multi-omics data; however, the identification of specific parameters influencing LBP performance can be more challenging. In silico models allow for the rapid generation of diverse sets of data however can be limited by assumptions in the computational model [93, 94]. Further, the colonization profiles observed in mice or other animal models do not always reflect colonization profiles observed in humans, necessitating translational studies and predictive modeling to scale to humans.
Traditionally, LBP discovery (Figure 4a) and delivery (Figure 4b) have occurred sequentially, with delivery considerations arising after the discovery process. However, we posit that these operations can synergize with each other if performed simultaneously (Figure 4c) (see Outstanding Questions). For example, multi-omics analyses described above can be applied to LBPs residing in the delivery vehicle to describe their effects on microbial physiology, and design LBPs that are highly compatible with delivery formulations. Also, the impact of LBP formulations on the gut microbiota and host can be queried, enabling researchers to select LBPs that minimize detrimental effects. Moving forward, more sophisticated quantitative approaches are needed to accelerate the process of designing and formulating engineered LBPs for therapeutic application; the utilization of predictive modeling may enable this (Figure 4d). Specifically, modeling approaches that integrate gut physiology (commonly used in pharmacokinetic modeling) with microbial metabolism and growth (commonly used by microbial ecologists) will enable rapid evaluation of different engineering and delivery strategies in silico before experimental testing [95]. By performing LBP discovery with delivery strategies in mind, and vice versa, we expect that engineered LBPs will be able to treat diseases with much tighter therapeutic windows, and with more strict requirements for site specificity.
Figure 4. Integrating Discovery and Delivery for Future LBP Development.
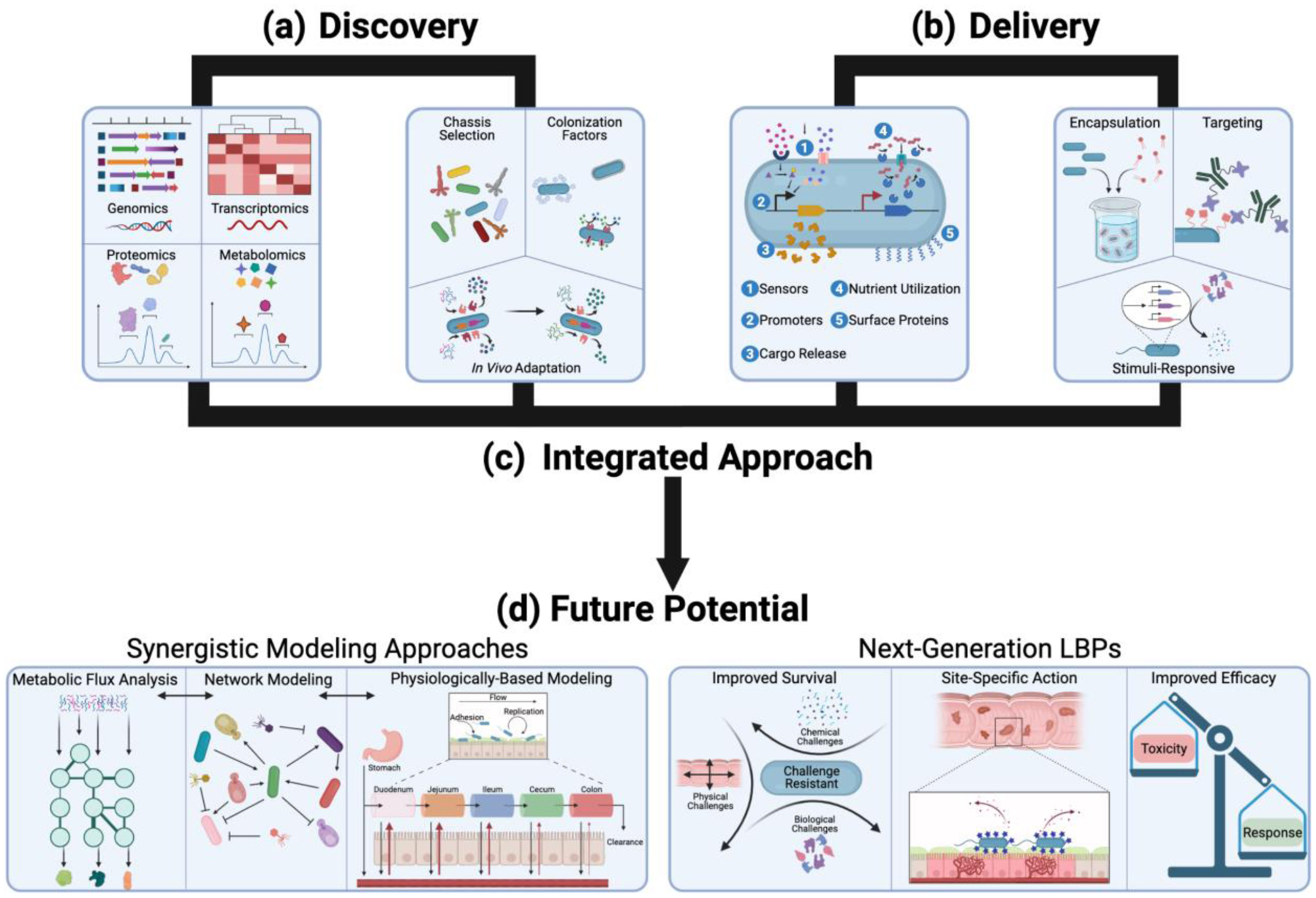
(a) Recent trends in the discovery of LBPs utilize multi-omics (e.g., genomics, transcriptomics, proteomics, metabolomics) strategies to uncover potential new microbial chassis, colonization factors, and adaptation mechanisms to overcome the physiological challenges of the gut. (b) Recent trends in the delivery of LBPs utilize both pharmaceutical formulation and genetic engineering strategies to encapsulate, target, and/or generate stimuli-responsive functions to overcome physiological challenges. (c) The current strategies can be integrated wherein discovery and delivery occur simultaneously for synergistic LBP design (e.g., chassis selection, genetic engineering, formulation approaches). (d) Synergistic LBP discovery and delivery has the potential to inform more sophisticated modeling approaches wherein micro-scale approaches (e.g., metabolic flux analysis, network modeling) are combined with macro-scale approaches (e.g., physiologically-based pharmacokinetic modeling) to enable better prediction for LBP efficacy. Synergistic LBP discovery and delivery also has the potential to pave the way for next-generation LBPs which have both improved survival and site-specific action through genetic engineering and pharmaceutical formulations approaches, ultimately leading to more efficacious LBPs with increased therapeutic response and decreased off-target toxicities.
Outstanding Questions Box.
How can discovery approaches and delivery strategies be synergistically combined to facilitate rational development of LBPs?
How can genome-scale, ecological, and physiologically-based pharmacokinetic modeling be integrated to achieve predictive design of LBPs?
What are the optimum in vitro conditions that can predict and represent in vivo activity of engineered functions using ‘omics analysis?
How can predictive models and ‘omics data be combined to minimize the metabolic burden created by the engineered function?
How can functional genomics selections be applied to select for LBPs that improve host health?
How can multi-omics approaches be used for optimization of LBP formulations?
How can host- and microbiota-response to delivery formulations be quantified?
For LBPs that integrate discovery approaches and delivery strategies, how can the most important and efficacy-driving LBP features be identified?
How can pharmaceutical formulations be integrated into existing manufacturing processes for LBPs?
Highlights.
The physiological microenvironment of the gut influences the efficacy of LBPs and both discovery and delivery strategies can be used to overcome physiological challenges in the gut
Multi-omics illuminates colonization mechanisms of non-engineered LBPs to inspire engineering strategies
Functional genomics generates and tests engineered LBPs in high throughput manner to provide improved strains
Pharmaceutical formulations can be used to control the interactions between LBPs and their physiological microenvironment, creating modular technologies and approaches that can be applied to all LBPs
Genetic engineering approaches can improve LBP delivery through overcoming physiological challenges, enabling molecular interactions with host surfaces, controlling therapeutic functions in response to local physiological cues
ACKNOWLEDGEMENTS
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R35GM137898 (to Aaron C. Anselmo), the National Science Foundation under Award Number CBET-1934284 (to Nathan Crook), and the Novo Nordisk Foundation under Award Number NNF19SA0035474 (to Nathan Crook). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. Illustrations created with BioRender.com.
Glossary
- adhesin
components or appendages of bacteria that facilitate adhesion or adherence to other cells or to surfaces.
- auxotrophy
the inability of an organism to synthesize a particular compound required for its own growth, requiring an external supply of that compound for survival.
- biocontainment
the prevention of engineered microbes from entering, being metabolically active in, or growing in environments outside of the host.
- biofilm
a protective film primarily composed of polysaccharides, proteins, nucleic acids, and lipids that is secreted by a microbe and enables microbial adherence to surfaces.
- chassis
an organism that contains and supports the genetic components encoding for a desired engineered function.
- colonization
when microbes continuously grow and maintain metabolic activity in/on a host.
- colonization factor
a gene, or set of genes, that enable a microbe to colonize a host.
- cross-feeding
intra- and inter-species exchange of nutrients.
- genetic tractability
the amenability of a microbe for genetic manipulation.
- hydrogel
a water-containing gel composed of a network of crosslinked polymer chains.
- Lotka-Volterra equations
a pair of first-order nonlinear differential equations, frequently used to describe the dynamics of biological systems in which multiple species interact.
- metabolic crosstalk
interaction between the host and the microbiota which can determine fate and function of the microbe and the disease state of the host.
- microparticle
a particle between 1 and 1000 μm in size often composed of biocompatible lipids and/or polymers which can encapsulate drug molecules or microbes.
- niche
the position of a species within an ecosystem encompassing both the physical and environmental factors required for survival and the interactions with other species.
- pharmacokinetics
the time course of a drug moving through the distinct compartments of the body.
- promoter
a sequence of DNA that controls the expression level of downstream coding regions.
- quorum-sensing molecule
a molecule that signals the presence of related microbes nearby; often induces the expression of virulence-related genes.
- regulatory networks
sets of macromolecules (e.g., proteins, RNA) that interact to control the level of expression of various genes in an organism.
- regulatory part
a DNA sequence that influences the rate of transcription of nearby genes.
- symbiont
microbe associated with the host without implication of benefit or harm.
- transformability
capacity for a microbe to be transformed with foreign genetic material.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.FDA (2016) Early Clinical Trials with Live Biotherapeutic Products: Chemistry, Manufacturing, and Control Information.
- 2.O’Toole PW, et al. (2017) Next-generation probiotics: the spectrum from probiotics to live biotherapeutics. Nat Microbiol 2, 17057. [DOI] [PubMed] [Google Scholar]
- 3.Steidler L, et al. (2003) Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat Biotechnol 21, 785–789 [DOI] [PubMed] [Google Scholar]
- 4.Steidler L, et al. (2000) Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 289, 1352–1355 [DOI] [PubMed] [Google Scholar]
- 5.Braat H, et al. (2006) A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn’s disease. Clin Gastroenterol Hepatol 4, 754–759 [DOI] [PubMed] [Google Scholar]
- 6.Garber K (2020) First microbiome-based drug clears phase III, in clinical trial turnaround. Nat Rev Drug Discov 19, 655–656 [DOI] [PubMed] [Google Scholar]
- 7.Blount KF, et al. (2019) Restoration of Bacterial Microbiome Composition and Diversity Among Treatment Responders in a Phase 2 Trial of RBX2660: An Investigational Microbiome Restoration Therapeutic. Open Forum Infectious Diseases 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang LL-W, et al. (2021) Cell therapies in the clinic. Bioengineering & Translational Medicine n/a, e10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Isabella VM, et al. (2018) Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nature Biotechnology 36, 857–864 [DOI] [PubMed] [Google Scholar]
- 10.Riglar DT and Silver PA (2018) Engineering bacteria for diagnostic and therapeutic applications. Nat Rev Microbiol 16, 214–225 [DOI] [PubMed] [Google Scholar]
- 11.Charbonneau MR, et al. (2020) Developing a new class of engineered live bacterial therapeutics to treat human diseases. Nat Commun 11, 1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vargason AM, et al. (2021) The evolution of commercial drug delivery technologies. Nat Biomed Eng [DOI] [PubMed] [Google Scholar]
- 13.Mimee M, et al. (2016) Microbiome therapeutics - Advances and challenges. Adv Drug Deliv Rev 105, 44–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koonin EV, et al. (2021) Evolution of Microbial Genomics: Conceptual Shifts over a Quarter Century. Trends Microbiol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapse NG, et al. (2019) Functional annotation of the genome unravels probiotic potential of Bacillus coagulans HS243. Genomics 111, 921–929 [DOI] [PubMed] [Google Scholar]
- 16.Alayande KA, et al. (2020) Integrated genome-based probiotic relevance and safety evaluation of Lactobacillus reuteri PNW1. PLoS One 15, e0235873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta S, et al. (2020) Investigating the dynamics of microbial consortia in spatially structured environments. Nature communications 11, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turroni F, et al. (2010) Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci U S A 107, 19514–19519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tailford LE, et al. (2015) Mucin glycan foraging in the human gut microbiome. Front Genet 6, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, et al. (2017) Lactobacillus plantarum LPOnlly alters the gut flora and attenuates colitis by inducing microbiome alteration in interleukin10 knockout mice. Mol Med Rep 16, 5979–5985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devika NT and Raman K (2019) Deciphering the metabolic capabilities of Bifidobacteria using genome-scale metabolic models. Scientific Reports 9, 18222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venturelli OS, et al. (2018) Deciphering microbial interactions in synthetic human gut microbiome communities. Mol Syst Biol 14, e8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jouhten H, et al. (2020) Cultivation and Genomics Prove Long-Term Colonization of Donor’s Bifidobacteria in Recurrent Clostridioides difficile Patients Treated With Fecal Microbiota Transplantation. Frontiers in Microbiology 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li SS, et al. (2016) Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science 352, 586. [DOI] [PubMed] [Google Scholar]
- 25.Maldonado-Gómez MX, et al. (2016) Stable Engraftment of Bifidobacterium longum AH1206 in the Human Gut Depends on Individualized Features of the Resident Microbiome. Cell Host Microbe 20, 515–526 [DOI] [PubMed] [Google Scholar]
- 26.Heintz-Buschart A and Wilmes P (2018) Human Gut Microbiome: Function Matters. Trends Microbiol 26, 563–574 [DOI] [PubMed] [Google Scholar]
- 27.Hagi T, et al. (2020) The effect of bile acids on the growth and global gene expression profiles in Akkermansia muciniphila. Applied Microbiology and Biotechnology 104, 10641–10653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, et al. (2021) Transcriptomics and metabolomics reveal the adaption of Akkermansia muciniphila to high mucin by regulating energy homeostasis. Scientific Reports 11, 9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D’Hoe K, et al. (2018) Integrated culturing, modeling and transcriptomics uncovers complex interactions and emergent behavior in a three-species synthetic gut community. Elife 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crost EH, et al. (2016) The mucin-degradation strategy of Ruminococcus gnavus: The importance of intramolecular trans-sialidases. Gut microbes 7, 302–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryan D, et al. (2020) A high-resolution transcriptome map identifies small RNA regulation of metabolism in the gut microbe Bacteroides thetaiotaomicron. Nature Communications 11, 3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao Y, et al. (2016) cis-Encoded Small RNAs, a Conserved Mechanism for Repression of Polysaccharide Utilization in Bacteroides. Journal of Bacteriology 198, 2410–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koskenniemi K, et al. (2011) Proteomics and transcriptomics characterization of bile stress response in probiotic Lactobacillus rhamnosus GG. Mol Cell Proteomics 10, M110 002741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ponomarova O, et al. (2017) Yeast Creates a Niche for Symbiotic Lactic Acid Bacteria through Nitrogen Overflow. Cell Syst 5, 345–357 e346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maligoy M, et al. (2008) Transcriptome analysis of Lactococcus lactis in coculture with Saccharomyces cerevisiae. Appl Environ Microbiol 74, 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goh YJ, et al. (2021) In Vivo Transcriptome of Lactobacillus acidophilus and Colonization Impact on Murine Host Intestinal Gene Expression. mBio 12, e03399–03320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Avall-Jaaskelainen S, et al. (2003) Surface display of the receptor-binding region of the Lactobacillus brevis S-layer protein in Lactococcus lactis provides nonadhesive lactococci with the ability to adhere to intestinal epithelial cells. Appl Environ Microbiol 69, 2230–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Antikainen J, et al. (2002) Domains in the S-layer protein CbsA of Lactobacillus crispatus involved in adherence to collagens, laminin and lipoteichoic acids and in self-assembly. Mol Microbiol 46, 381–394 [DOI] [PubMed] [Google Scholar]
- 39.Kleiner M (2019) Metaproteomics: Much More than Measuring Gene Expression in Microbial Communities. mSystems 4, e00115–00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lv LX, et al. (2017) Integrated transcriptomic and proteomic analysis of the bile stress response in probiotic Lactobacillus salivarius LI01. J Proteomics 150, 216–229 [DOI] [PubMed] [Google Scholar]
- 41.Celebioglu HU, et al. (2017) Mucin- and carbohydrate-stimulated adhesion and subproteome changes of the probiotic bacterium Lactobacillus acidophilus NCFM. J Proteomics 163, 102–110 [DOI] [PubMed] [Google Scholar]
- 42.Calderini E, et al. (2017) Comparative proteomics of oxidative stress response of Lactobacillus acidophilus NCFM reveals effects on DNA repair and cysteine de novo synthesis. Proteomics 17 [DOI] [PubMed] [Google Scholar]
- 43.Celebioglu HU and Svensson B (2017) Exo- and surface proteomes of the probiotic bacterium Lactobacillus acidophilus NCFM. PROTEOMICS 17, 1700019. [DOI] [PubMed] [Google Scholar]
- 44.do Carmo FLR, et al. (2018) Extractable Bacterial Surface Proteins in Probiotic-Host Interaction. Front Microbiol 9, 645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Treuren W and Dodd D (2020) Microbial Contribution to the Human Metabolome: Implications for Health and Disease. Annu Rev Pathol 15, 345–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng L, et al. (2020) Identifying determinants of bacterial fitness in a model of human gut microbial succession. Proc Natl Acad Sci U S A 117, 2622–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Theriot CM, et al. (2016) Antibiotic-Induced Alterations of the Gut Microbiota Alter Secondary Bile Acid Production and Allow for Clostridium difficile Spore Germination and Outgrowth in the Large Intestine. mSphere 1, e00045–00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crost EH, et al. (2018) Mechanistic Insights Into the Cross-Feeding of Ruminococcus gnavus and Ruminococcus bromii on Host and Dietary Carbohydrates. Frontiers in Microbiology 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chamberlain CA, et al. (2019) Metabolomic profiling of oxalate-degrading probiotic Lactobacillus acidophilus and Lactobacillus gasseri. PLoS One 14, e0222393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bryant WA, et al. (2017) In Silico Analysis of the Small Molecule Content of Outer Membrane Vesicles Produced by Bacteroides thetaiotaomicron Indicates an Extensive Metabolic Link between Microbe and Host. Frontiers in Microbiology 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown EM, et al. (2019) Bacteroides-Derived Sphingolipids Are Critical for Maintaining Intestinal Homeostasis and Symbiosis. Cell Host & Microbe 25, 668–680.e667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Helm E, et al. (2018) The evolving interface between synthetic biology and functional metagenomics. Nat Chem Biol 14, 752–759 [DOI] [PubMed] [Google Scholar]
- 53.Neil K, et al. (2020) Highly efficient gene transfer in the mouse gut microbiota is enabled by the Incl2 conjugative plasmid TP114. Communications Biology 3, 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ronda C, et al. (2019) Metagenomic engineering of the mammalian gut microbiome in situ. Nature Methods 16, 167–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crook N, et al. (2020) Transcript Barcoding Illuminates the Expression Level of Synthetic Constructs in E. coli Nissle Residing in the Mammalian Gut. ACS Synth Biol 9, 1010–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mitchell LA, et al. (2015) Versatile genetic assembly system (VEGAS) to assemble pathways for expression in S. cerevisiae. Nucleic Acids Res 43, 6620–6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Durmusoglu D, et al. (2021) In Situ Biomanufacturing of Small Molecules in the Mammalian Gut by Probiotic Saccharomyces boulardii. ACS Synth Biol [DOI] [PubMed] [Google Scholar]
- 58.Dantas G, et al. (2013) Experimental approaches for defining functional roles of microbes in the human gut. Annu Rev Microbiol 67, 459–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mimee M, et al. (2015) Programming a Human Commensal Bacterium, Bacteroides thetaiotaomicron, to Sense and Respond to Stimuli in the Murine Gut Microbiota. Cell Systems 1, 62–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Horn N, et al. (2016) A Novel Tightly Regulated Gene Expression System for the Human Intestinal Symbiont Bacteroides thetaiotaomicron. Frontiers in Microbiology 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jones DR, et al. (2019) Engineering dual-glycan responsive expression systems for tunable production of heterologous proteins in Bacteroides thetaiotaomicron. Scientific Reports 9, 17400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee SM, et al. (2013) Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501, 426–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crook N, et al. (2019) Adaptive Strategies of the Candidate Probiotic E. coli Nissle in the Mammalian Gut. Cell Host Microbe 25, 499–512 e498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cao Z, et al. (2019) Biointerfacial self-assembly generates lipid membrane coated bacteria for enhanced oral delivery and treatment. Nat Commun 10, 5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anselmo AC, et al. (2016) Layer-by-Layer Encapsulation of Probiotics for Delivery to the Microbiome. Adv Mater 28, 9486–9490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang X, et al. (2020) Bioinspired oral delivery of gut microbiota by self-coating with biofilms. Sci Adv 6, eabb1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Z, et al. (2018) Biofilm-Inspired Encapsulation of Probiotics for the Treatment of Complex Infections. Adv Mater 30, e1803925. [DOI] [PubMed] [Google Scholar]
- 68.Vargason AM, et al. (2020) Surface Modifications for Improved Delivery and Function of Therapeutic Bacteria. Small 16, e2001705. [DOI] [PubMed] [Google Scholar]
- 69.Buss MT, et al. (2021) Spatial Control of Probiotic Bacteria in the Gastrointestinal Tract Assisted by Magnetic Particles. Adv Mater 33, e2007473. [DOI] [PubMed] [Google Scholar]
- 70.Park BW, et al. (2017) Multifunctional Bacteria-Driven Microswimmers for Targeted Active Drug Delivery. ACS Nano 11, 8910–8923 [DOI] [PubMed] [Google Scholar]
- 71.Zhang H, et al. (2018) A pH-Responsive Gel Macrosphere Based on Sodium Alginate and Cellulose Nanofiber for Potential Intestinal Delivery of Probiotics. ACS Sustainable Chemistry & Engineering 6, 13924–13931 [Google Scholar]
- 72.Duraj-Thatte AM, et al. (2019) Genetically Programmable Self-Regenerating Bacterial Hydrogels. Adv Mater 31, e1901826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Praveschotinunt P, et al. (2019) Engineered E. coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut. Nat Commun 10, 5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mays ZJS, et al. (2020) Quantifying and Engineering Mucus Adhesion of Probiotics. ACS Synth Biol 9, 356–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Henn MR, et al. (2021) A Phase 1b Safety Study of SER-287, a Spore-Based Microbiome Therapeutic, for Active Mild to Moderate Ulcerative Colitis. Gastroenterology 160, 115–127.e130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Green E (2021) US & EU Patents awarded to UK Biotech. https://chainbiotech.com/2021/05/07/us-eu-patents-awarded-to-uk-biotech/
- 77.Vedantam G, et al. (2018) An Engineered Synthetic Biologic Protects Against Clostridium difficile Infection. Front Microbiol 9, 2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chappell TC and Nair NU (2020) Engineered lactobacilli display anti-biofilm and growth suppressing activities against Pseudomonas aeruginosa. NPJ Biofilms Microbiomes 6, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ho CL, et al. (2018) Engineered commensal microbes for diet-mediated colorectal-cancer chemoprevention. Nat Biomed Eng 2, 27–37 [DOI] [PubMed] [Google Scholar]
- 80.Swofford CA, et al. (2015) Quorum-sensing Salmonella selectively trigger protein expression within tumors. Proc Natl Acad Sci U S A 112, 3457–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mimee M, et al. (2015) Programming a Human Commensal Bacterium, Bacteroides thetaiotaomicron, to Sense and Respond to Stimuli in the Murine Gut Microbiota. Cell Syst 1, 62–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Birnbaum DP, et al. (2021) Hybrid Living Capsules Autonomously Produced by Engineered Bacteria. Advanced Science n/a, 2004699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tang TC, et al. (2021) Hydrogel-based biocontainment of bacteria for continuous sensing and computation. Nat Chem Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pedrolli DB, et al. (2019) Engineering Microbial Living Therapeutics: The Synthetic Biology Toolbox. Trends in Biotechnology 37, 100–115 [DOI] [PubMed] [Google Scholar]
- 85.Kurtz CB, et al. (2019) An engineered E. coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Science Translational Medicine 11, eaau7975. [DOI] [PubMed] [Google Scholar]
- 86.Camacho DM, et al. (2018) Next-Generation Machine Learning for Biological Networks. Cell 173, 1581–1592 [DOI] [PubMed] [Google Scholar]
- 87.Woyke T, et al. (2017) The trajectory of microbial single-cell sequencing. Nat Methods 14, 1045–1054 [DOI] [PubMed] [Google Scholar]
- 88.Tropini C, et al. (2017) The Gut Microbiome: Connecting Spatial Organization to Function. Cell Host Microbe 21, 433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mays ZJS, et al. (2020) Quantifying and Engineering Mucus Adhesion of Probiotics. ACS synthetic biology 9, 356–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nelson MT, et al. (2021) Characterization of an engineered live bacterial therapeutic for the treatment of phenylketonuria in a human gut-on-a-chip. Nature Communications 12, 2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Williamson IA, et al. (2018) A High-Throughput Organoid Microinjection Platform to Study Gastrointestinal Microbiota and Luminal Physiology. Cell Mol Gastroenterol Hepatol 6, 301–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Harimoto T, et al. (2019) Rapid screening of engineered microbial therapies in a 3D multicellular model. Proceedings of the National Academy of Sciences 116, 9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Diener C, et al. (2020) MICOM: Metagenome-Scale Modeling To Infer Metabolic Interactions in the Gut Microbiota. mSystems 5, e00606–00619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Charbonneau MR, et al. (2021) Development of a mechanistic model to predict synthetic biotic activity in healthy volunteers and patients with phenylketonuria. Communications Biology 4, 898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thiele I, et al. (2020) Personalized whole-body models integrate metabolism, physiology, and the gut microbiome. Mol Syst Biol 16, e8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lund P, et al. (2014) Coping with low pH: molecular strategies in neutralophilic bacteria. FEMS Microbiol Rev 38, 1091–1125 [DOI] [PubMed] [Google Scholar]
- 97.Zhu H, et al. (2006) Bacterial killing in gastric juice--effect of pH and pepsin on Escherichia coli and Helicobacter pylori. J Med Microbiol 55, 1265–1270 [DOI] [PubMed] [Google Scholar]
- 98.Begley M, et al. (2005) The interaction between bacteria and bile. FEMS Microbiol Rev 29, 625–651 [DOI] [PubMed] [Google Scholar]
- 99.Hirayama D, et al. (2017) The Phagocytic Function of Macrophage-Enforcing Innate Immunity and Tissue Homeostasis. Int J Mol Sci 19, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sender R, et al. (2016) Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol 14, e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hua S, et al. (2015) Advances in oral nano-delivery systems for colon targeted drug delivery in inflammatory bowel disease: selective targeting to diseased versus healthy tissue. Nanomedicine 11, 1117–1132 [DOI] [PubMed] [Google Scholar]
- 102.Arike L, et al. (2020) Protein Turnover in Epithelial Cells and Mucus along the Gastrointestinal Tract Is Coordinated by the Spatial Location and Microbiota. Cell Rep 30, 1077–1087 e1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zheng L, et al. (2015) Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A Review in the Theme: Cellular Responses to Hypoxia. American journal of physiology. Cell physiology 309, C350–C360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Koziolek M, et al. (2015) Investigation of pH and Temperature Profiles in the GI Tract of Fasted Human Subjects Using the Intellicap((R)) System. J Pharm Sci 104, 2855–2863 [DOI] [PubMed] [Google Scholar]
- 105.Bang M, et al. (2018) Transcriptional Response and Enhanced Intestinal Adhesion Ability of Lactobacillus rhamnosus GG after Acid Stress. J Microbiol Biotechnol 28, 1604–1613 [DOI] [PubMed] [Google Scholar]
- 106.Koskenniemi K, et al. (2011) Proteomics and transcriptomics characterization of bile stress response in probiotic Lactobacillus rhamnosus GG. Molecular & cellular proteomics : MCP 10, M110.002741–M002110.002741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Donaldson GP, et al. (2020) Spatially distinct physiology of Bacteroides fragilis within the proximal colon of gnotobiotic mice. Nat Microbiol 5, 746–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dean SN, et al. (2019) Isolation and characterization of Lactobacillus-derived membrane vesicles. Scientific Reports 9, 877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Klotz C, et al. (2017) Investigating the Effect of Growth Phase on the Surface-Layer Associated Proteome of Lactobacillus acidophilus Using Quantitative Proteomics. Frontiers in Microbiology 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.O’Flaherty S and Klaenhammer TR (2016) Multivalent Chromosomal Expression of the Clostridium botulinum Serotype A Neurotoxin Heavy-Chain Antigen and the Bacillus anthracis Protective Antigen in Lactobacillus acidophilus. Appl Environ Microbiol 82, 6091–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]


