Abstract
Coronavirus disease 2019 (COVID-19) represents a systemic disease that may cause severe metabolic complications in multiple tissues including liver, kidney, and cardiovascular system. However, the underlying mechanisms and optimal treatment remain elusive. Our study shows that impairment of ACE2 pathway is a key factor linking virus infection to its secondary metabolic sequelae. By using structure-based high-throughput virtual screening and connectivity map database, followed with experimental validations, we identify imatinib, methazolamide, and harpagoside as direct enzymatic activators of ACE2. Imatinib and methazolamide remarkably improve metabolic perturbations in vivo in an ACE2-dependent manner under the insulin-resistant state and SARS-CoV-2-infected state. Moreover, viral entry is directly inhibited by these three compounds due to allosteric inhibition of ACE2 binding to spike protein on SARS-CoV-2. Taken together, our study shows that enzymatic activation of ACE2 via imatinib, methazolamide, or harpagoside may be a conceptually new strategy to treat metabolic sequelae of COVID-19.
Keywords: COVID-19, SARS-CoV-2, ACE2, metabolic complication, enzymatic activator, imatinib, methazolamide, harpagoside
Graphical abstract
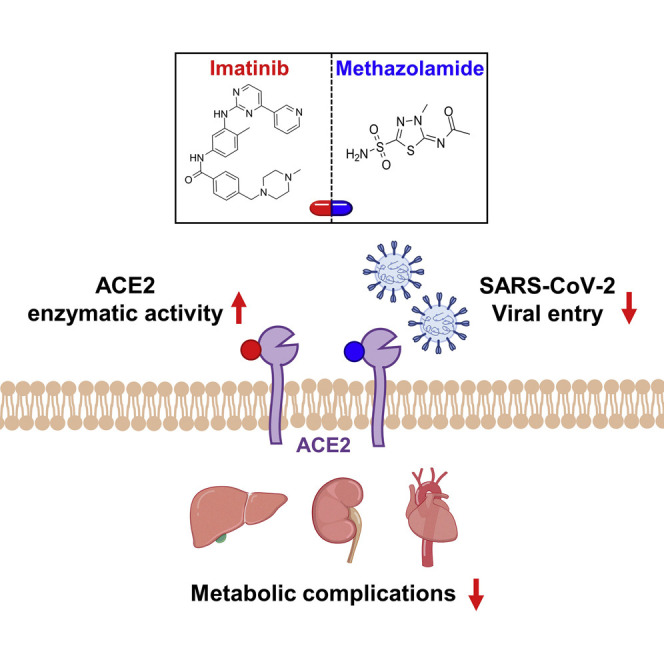
Li et al. identify imatinib and methazolamide as enzymatic activators of ACE2, which improve metabolic complications under SARS-CoV-2 infection and reduce viral entry via allosteric inhibition of binding between ACE2 and spike protein, suggesting that activation of ACE2 might be a conceptually new strategy to treat metabolic sequelae of COVID-19.
Introduction
Coronavirus disease 2019 (COVID-19) represents a systemic disease that significantly disturbs multiple tissues and organs throughout the body (Guo et al., 2021; Nunes Duarte-Neto et al., 2020). Notably, emerging evidence has demonstrated that SARS-CoV-2 infection promotes metabolic complications at both system and organ levels in patients without pre-existing metabolic disorders. Systemic metabolic defects including hyperglycemia, high blood pressure, and low high-density lipoprotein cholesterol (HDL-C) have been extensively reported (Gupta et al., 2020; Wei et al., 2020). “New-onset” diabetes has been reported in 5%–29% of the patients with COVID-19 and is associated with worse outcomes (He et al., 2021; Singh and Singh, 2020). Known COVID-19-related metabolic defects at the organ level include hepatic steatosis, glomerulopathy, endothelial dysfunction, and thrombosis (Díaz et al., 2020; Ramos-Casals et al., 2021). Supporting this, a proteomic analysis of autopsy samples reveals a dysregulation of glucose and fatty acid metabolism in multiple organs including the liver, kidney, and heart (Nie et al., 2021). Collectively, SARS-CoV-2 may cause substantial metabolic defects as severe and/or long-term complications.
The pivotal mechanisms mediating the detrimental metabolic effects of SARS-CoV-2, however, remain largely unknown. Here, we conducted omics analysis to identify differentially expressed genes between COVID-19 and metabolic diseases including diabetes, hypertension, diabetic nephropathy, and atherosclerosis. Surprisingly, we identified downregulation of ACE2 as a top candidate hub potentially mediating SARS-CoV-2-induced metabolic defects. We combined connectivity map (CMAP), supercomputer-based docking, and experimental approaches to identify potential strategies to restore the impaired ACE2 pathway. We identified three new ACE2 enzymatic activators: imatinib, methazolamide, and harpagoside. Two repurposed drugs, imatinib and methazolamide, could act as ACE2 enzymatic activators to potently improve glucose and lipid metabolisms after SARS-CoV-2 infection in vivo. Moreover, imatinib and methazolamide displayed direct antiviral effect via inhibition of ACE2 binding to spike protein. Given the very limited understanding of SARS-CoV-2 pathology and the scarcity of effective therapy for COVID-19 and its metabolic complications, such repurposed drugs may serve as promising therapeutic candidates for rapid clinical application during this urgent situation.
Results
ACE2 is a key molecule potentially linking COVID-19 to associated metabolic defects
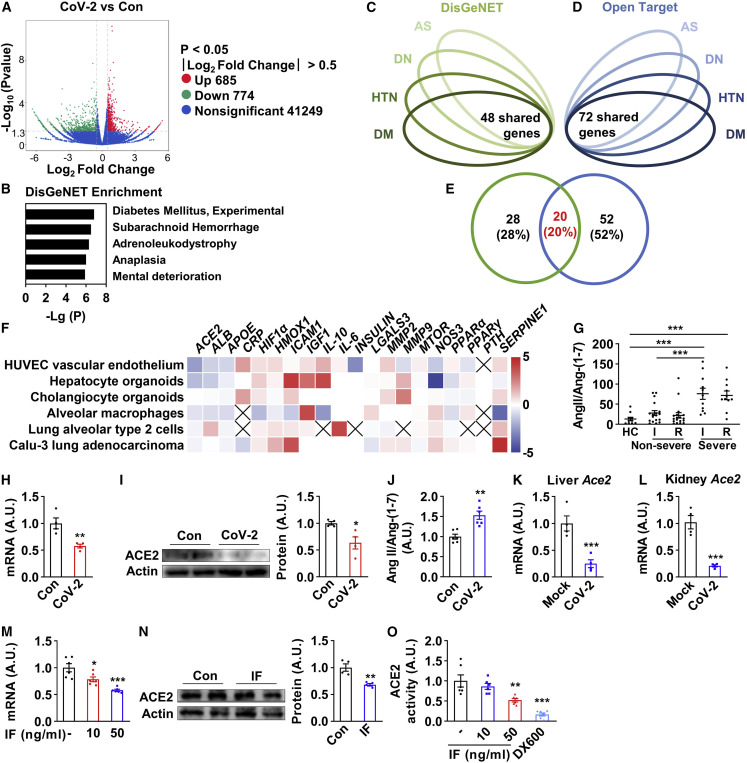
Clinical studies reveal remarkable metabolic defects in patients with SARS-CoV-2 infection. However, the molecular mechanisms remain elusive. Our in vitro transcriptomic data from human umbilical vein endothelial cells (HUVECs) infected by SARS-CoV-2 showed that the differentially expressed genes after viral infection were enriched in metabolism-related diseases/pathways including “diabetes mellitus” and “glycolysis/gluconeogenesis,” alongside virus-infection-related pathways including “human papillomavirus infection” (Figures 1A, 1B, S1A, and S1B). Based on this finding, we hypothesized that SARS-CoV-2 may disturb the expression of the selected genes critical for the development of COVID-19-related metabolic complications. Given that hyperglycemia, high blood pressure, low HDL-C, glomerulopathy, endothelial dysfunction, and thrombosis represent the main metabolic defects in COVID-19 patients and have similar symptoms as diabetes, hypertension, diabetic nephropathy, and atherosclerosis (Beckman et al., 2002; Sowers and Epstein, 1995), we speculated that common genes shared by these four diseases may be central to the pathogenic process of COVID-19-related metabolic disorders. Thus, the top 400 markedly altered genes of four metabolic diseases were selected from DisGeNET and Open Target database (Tables S1 and S2) (Carvalho-Silva et al., 2019; Piñero et al., 2020), and 48 (Figure 1C) and 72 (Figure 1D) genes were shared among these four metabolic diseases chosen from two different databases (Tables S1 and S2). Furthermore, 20 genes were shared between the above two lists (Figure 1E). We postulated that these 20 genes may play a central role in the pathogenic progress of these COVID-19-related metabolic diseases. Next, we compared the expression patterns of these 20 genes in our transcriptome and SARS-CoV-2-infected organoids and cells described previously (Blanco-Melo et al., 2020; Grant et al., 2021; Huang et al., 2020). Notably, the virus receptor ACE2 was one of the most consistently suppressed genes after virus infection in different transcriptomic datasets (Figure 1F), suggesting that ACE2 may serve as an essential hub linking SARS-CoV-2 infection to its secondary metabolic defects. Moreover, a direct quantification of ACE2 enzymatic substrate Ang II and its product Ang-(1–7) in plasma of 18 non-severe COVID-19 patients, 12 severe patients, and 10 healthy controls demonstrated reductions of ACE2 enzymatic activity in both non-severe and severe patients, and the ACE2 enzymatic activity remained depressed in the recovery phase (Figure 1G).
Figure 1.
ACE2 is a key molecule potentially linking COVID-19 to associated metabolic defects
(A) Volcano plot of differentially expressed genes after in vitro infection of SARS-CoV-2 (MOI = 0.005) in HUVECs for 24 h.
(B) DisGeNET pathway enrichment of differentially expressed genes after infection (combined 685 upregulated and 774 downregulated genes from A).
(C–E) Bioinformatics analysis of the mutual target disease-associated genes of diabetes mellitus (DM), hypertension (HTN), diabetic nephropathy (DN), and atherosclerosis (AS) that were queried from DisGeNET and Open Target database. The overlapping 48 shared genes from DisGeNET (C), the 72 shared genes from Open Target (D), and 20 genes identified on both DisGeNET (left) and Open Target database (right) as mutual target disease-associated genes of four diseases (E) were shown.
(F) The heatmap to represent the alterations of the above 20 genes upon SARS-CoV-2 infection. Data were shown as log2 fold change. The cross mark represents the undetectable gene from the transcriptomic study.
(G) ELISA quantification of plasma angiotensin II (Ang II) and angiotensin-(1–7) (Ang-(1–7)) in COVID-19 patients, shown in Ang II against Ang-(1–7) ratio. HC, healthy control; I, infection phase; R, recovery phase. Health control group, n = 10, mean age ± SD = 42.30 ± 7.35, male/female = 5/5; non-severe group, n = 18, mean age ± SD = 51.61 ± 10.13, male/female = 9/9; severe group, n = 12, mean age ± SD = 57.75 ± 14.55, male/female = 6/6.
(H and I) HUVECs were infected with SARS-CoV-2 (MOI = 0.005) for 24 h and were subjected to real-time PCR of ACE2 (H) and immunoblotting (I, blot shown on the left, quantification on the right, n = 4).
(J) HUVECs were infected with SARS-CoV-2 (MOI = 0.005) for 48 h, and then the medium was removed and 100 nM Ang II in 1 mL HEPES solution was applied for the cells. After 1 h treatment in the incubator at 37°C, the supernatant containing Ang II and cleaved Ang-(1–7) was examined (n = 6).
(K and L) Sixteen-week-old human ACE2 transgenic mice were intranasally challenged with 4 × 104 FFU SARS-CoV-2 after 10-week high-fat-diet treatment. After 7 days post-infection, all mice were fasted for 6 h and sacrificed. Livers (K) and kidneys (L) were subjected to real-time PCR (n = 3–4).
(M–O) HUVECs were treated with a “cocktail” of different inflammatory factors (IFs), namely the combination of 10 or 50 ng/mL of TNF-α, IL-4, IL-6, and IFN-γ or 1 μM DX600 for 48 h and were subjected to real-time PCR of ACE2 (M) (n = 6), immunoblotting (N, blot shown on the left, quantification on the right, n = 4), and enzymatic activity assay (O, data were shown as the area under the kinetic activity curves, n = 6). DX600 was used as a negative control. Error bars represent SEM; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. HUVECs, human umbilical vein endothelial cells; AU, arbitrary unit; CoV-2, SARS-CoV-2.
See also Figure S1.
To further investigate the effects and mechanism of virus infection on the regulation of ACE2, we infected HUVECs with SARS-CoV-2. A significant reduction of ACE2 mRNA and protein upon infection was observed (Figures 1H and 1I), indicating the impairment of ACE2 pathway. Moreover, the ACE2 enzymatic activity was detected with infected cells incubated with Ang II in HEPES buffer. The ratio of Ang II/Ang-(1–7) was increased, indicating that ACE2 enzymatic activity was impaired after the virus infection (Figure 1J). More importantly, Ace2 mRNA was also significantly reduced in the liver and the kidney of diet-induced obesity hACE2 transgenic mice 7 days post-infection of SARS-CoV-2 (Figures 1K and 1L). To test whether inflammatory response following SARS-CoV-2 infection could regulate ACE2 expression, we also treated HUVECs with the “cocktail” of TNF-α, IL-4, IL-6, and IFN-γ, the most representative inflammatory factors that appeared in COVID-19-induced cytokine storm, and found a significant reduction of ACE2 mRNA and protein, as well as the enzymatic activity (Figures 1M–1O and S1C). Likewise, a significant decrease of ACE2 protein in kidney and heart was also observed in a COVID-19 autopsy study (Nie et al., 2021). Taken together, we conclude that ACE2 level and enzymatic activity are reduced during COVID-19, presumably due to the direct virus infection and secondary inflammatory cytokine challenge.
ACE2 plays an important role in maintaining metabolic homeostasis
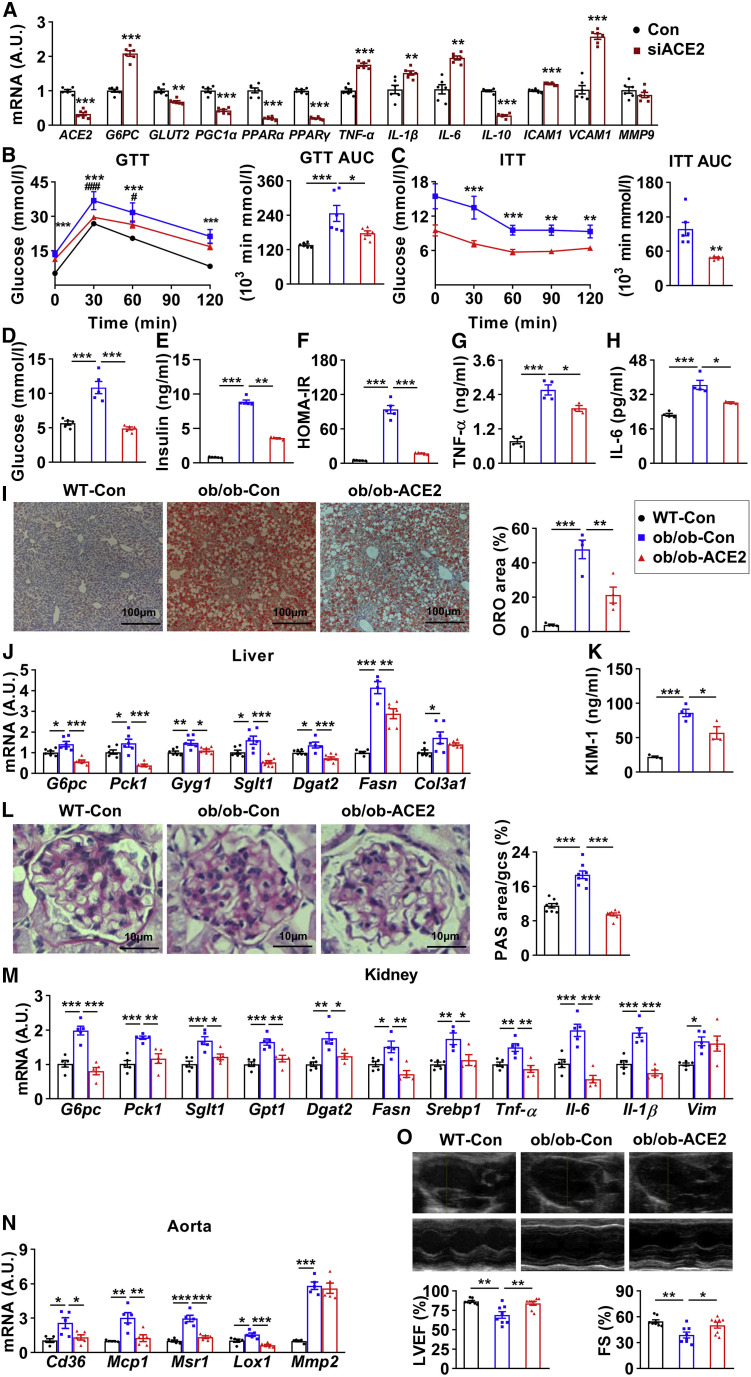
ACE2 is a key member of the renin-angiotensin system (RAS). The RAS consists of two arms that counteract each other to maintain homeostasis: a pro-inflammatory and pro-oxidative axis mainly formed by Ang II and angiotensin type 1 (AT1) receptor and an anti-inflammatory and antioxidative axis composed of Ang-(1–7) and Mas receptor (MasR). ACE2 plays a vital role in the balance as it converts Ang II into Ang-(1–7) (Santos et al., 2018). Although a few findings have indicated that ACE2 regulates glucose and lipid metabolism (Dong et al., 2012; Nadarajah et al., 2012; Xuan et al., 2018), the role of ACE2 in systemic metabolism remains elusive. We first measured the expression of genes involved in metabolic regulation upon manipulation of ACE2 expression. Interestingly, we found that following ACE2 knockdown in HUVECs, (1) regarding the genes related to glucose metabolism, G6PC was increased and GLUT2 was reduced; (2) regarding the genes related to lipid metabolism, PGC1α, PPARα, and PPARγ were reduced; (3) regarding the genes related to inflammation, TNF-α, IL-1β, and IL-6 were increased and IL-10 was reduced; and (4) regarding the genes related to vascular injury, ICAM1 and VCAM1 were increased and MMP9 was unaltered (Figure 2 A). Conversely, the expressions of PPARα and PPARγ were increased, whereas TNF-α and IL-6 were reduced upon ACE2 overexpression (Figures S2A–S2D).
Figure 2.
ACE2 plays an important role in maintaining metabolic homeostasis
(A) HUVECs were infected with 10 nM control siRNA (Con) or ACE2 siRNA (siACE2) for 24 h and subjected to real-time PCR (n = 6).
(B and C) Eight-week-old male ob/ob mice were treated with intravenous injection of AAV9-CAG-hACE2-EGFP (ob/ob-ACE2) and corresponding control virus (ob/ob-Con) and their wild-type littermates with control virus (WT-Con). Glucose tolerance testing (GTT) was performed at 10 weeks (B), and insulin tolerance testing (ITT) was performed at 11 weeks (C) (n = 6). The significance of ob/ob-Con versus WT-Con was shown as ∗; ob/ob-ACE2 versus ob/ob-Con as #.
(D–F) At 12 weeks, all mice were fasted for 6 h and sacrificed. Quantification of fasting blood glucose (D), plasma insulin (E), and homeostatic model assessment of insulin resistance (HOMA-IR) (F) were shown (n = 5).
(G and H) Plasma TNF-α (G) and IL-6 (H) were measured (n = 4).
(I and J) Livers were subjected to oil red O (ORO) staining (I, images shown on the left, quantification on the right; n = 4) and real-time PCR (n = 6) (J).
(K) Plasma KIM-1 (n = 4).
(L and M) Kidneys were subjected to periodic acid-Schiff (PAS) staining (L, images shown on the left, quantification on the right; n = 8) and real-time PCR (M) (n = 5).
(N) Aortas were subjected to real-time PCR (n = 5).
(O) Hearts were subjected to echocardiography (representative M-mode echocardiography images shown on the top, quantification of LVEF (%) and FS (%) on the bottom; n = 8).
Error bars represent SEM, ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001; #p < 0.05, ##p < 0.01, and ###p < 0.001. AUC, area under the curve; KIM-1, kidney injury molecule-1; LVEF, left ventricular ejection fraction; FS, fractional shortening. See also Figure S2.
We next investigated the role of ACE2 on systemic metabolism by overexpressing ACE2 in ob/ob mice. An intravenous injection of AAV9-hACE2 resulted in a 19-fold increase of ACE2 protein in murine liver (Figure S2E), as well as 42.2% reduction in the ratio between plasma Ang II and Ang-(1–7) (Figure S2F). Overexpression of ACE2 modestly reduced the body weight gain, epididymal fat index, and food and water intake (Figures S2G–S2J). Importantly, the glucose tolerance (Figure 2B), insulin sensitivity (Figure 2C), fasting glucose (Figure 2D), and insulin (Figure 2E), as well as the homeostatic model assessment of insulin resistance (HOMA-IR) (Figure 2F) in obese mice were significantly improved and almost normalized to the level of lean mice. Furthermore, the plasma triglyceride (TG) level showed a trend toward reduction, while total cholesterol (TC) was significantly improved (Figures S2K and S2L). The inflammatory factors of TNF-α and IL-6 in plasma were significantly reduced in ACE2-overexpressed mice (Figures 2G and 2H), suggesting an amelioration of systemic inflammation. Given that liver, kidney, and cardiovascular system are the major metabolic tissues impaired upon COVID-19 and also express a fair amount of ACE2, we analyzed the metabolic alterations of these tissues in obese mice upon ACE2 overexpression. The fat accumulation in the liver was dramatically ameliorated as determined by the oil red O (ORO) and hematoxylin and eosin (H&E) staining (Figures 2I and S2M), suggesting an improvement in hepatic steatosis. Not surprisingly, the hepatic TC was significantly improved (Figure S2N). Moreover, the elevated genes of G6pc, Pck1, Gyg1, Sglt1, Dgat2, and Fasn were significantly decreased, while the expression of Col3a1 was not altered (Figure 2J), suggesting improved glucose and lipid metabolism in the liver. Regarding kidney, the level of renal injury marker kidney injury molecule-1 (KIM-1) was significantly reduced (Figure 2K), whereas the glomerular damage was also ameliorated as determined by periodic acid-Schiff (PAS) staining (Figure 2L). Similarly in liver, elevated genes related to glucose metabolism (G6pc, Pck1, Sglt1, and Gpt1), lipid metabolism (Dgat2, Fasn, and Srebp1), and inflammation (Tnf-α, Il-6, and Il-1β) were all significantly suppressed, while Vim was not altered (Figure 2M), suggesting improvements of glucose and lipid metabolic disorders, inflammation, as well as cell integrity in kidney. Regarding the cardiovascular system, the expressions of Cd36, Mcp1, Msr1, and Lox1 in aorta were significantly reduced, and the expression of Mmp2 was not altered (Figure 2N). The impaired left ventricular ejection fractions (LVEFs) and fractional shortening (FS) were normalized to the control levels as determined by echocardiography (Figure 2O), suggesting improved metabolism and function in the cardiovascular system. Taking these data together, we conclude that ACE2 is beneficial for maintaining systemic metabolic homeostasis.
Imatinib, harpagoside, and methazolamide are identified as ACE2 activators
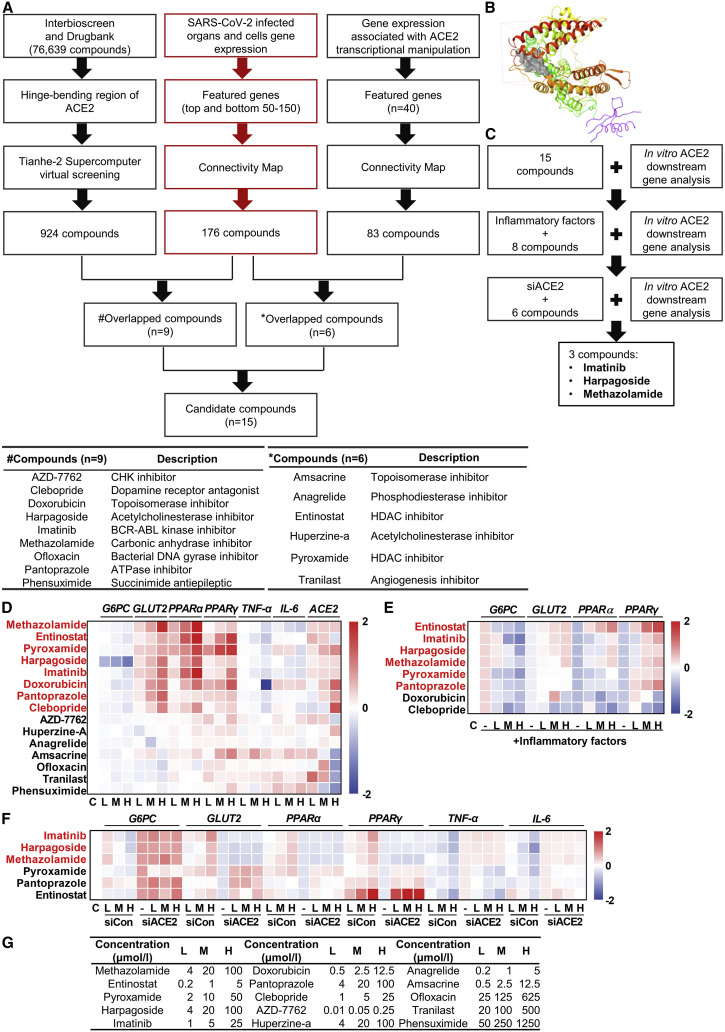
We postulated that the impaired ACE2 pathway may be a key factor causing SARS-CoV-2-associated metabolic defects. Therefore, we speculated that restoring ACE2 expression or activity may be a new and effective strategy in the treatment of COVID-19-associated metabolic disorders. Since specific ACE2 activators have not yet been validated (Haber et al., 2014), we aimed to identify pharmacological compounds to activate ACE2. Figure 3 A illustrates the workflow of high-throughput compound screening. We used two unbiased approaches to identify the potential activators: (1) enzymatic activators, compounds directly binding to ACE2 to enhance its enzymatic activity, and (2) transcriptional activators, compounds transcriptionally increasing ACE2 expression. InterBioScreen and DrugBank database (76,639 compounds in total) were applied for in silico virtual docking screening via the Tianhe-2 supercomputer. The crystal structure of the apo form of human ACE2 was used for molecular docking (Towler et al., 2004). We targeted a specific structural pocket in the hinge-bending region of ACE2, as binding in this region would potentially stabilize the “open” conformation of ACE2 from converting to “closed” conformation (Figure S3A). Using in silico virtual docking screening method, from the above-mentioned 76,639 compounds, we first selected 924 compounds with the potential to directly bind to and activate ACE2 (Figures 3A, left branch, and 3B; Table S3). Next, using CMAP and 40 featured genes correlated with ACE2 transcriptional activation (Table S3), we determined 83 compounds as the potential transcriptional activator of ACE2 (Figure 3A, right branch; Table S3). Furthermore, using CMAP and featured genes of SARS-CoV-2 infection, we identified 176 compounds that can potentially reverse COVID-19-altered gene expression patterns (Figure 3A, middle branch; Table S3). Next, we overlapped the 176 and 924 compounds and identified 9 shared compounds as potential ACE2 enzymatic activators counteracting COVID-19, whereas overlapping the 176 and 83 compounds revealed 6 shared compounds as ACE2 transcriptional activator counteracting COVID-19. Thus, we identified 15 compounds, including nine potential ACE2 enzymatic activators and six potential ACE2 transcriptional activators, that may be capable of reversing COVID-19-induced gene alteration (Figure 3A). The names and descriptions of these 15 compounds were listed in the bottom of Figure 3A. The virtual docking site of ACE2 for the compound screening was shown in Figure 3B.
Figure 3.
Imatinib, harpagoside, and methazolamide are identified as ACE2 activators
(A) Flow chart depicting the process of in silico identification of potential ACE2 activators with Tianhe-2 supercomputer virtual screening and CMAP bioinformatic analysis (top) and detailed information of the overlapped compounds (bottom, # and ∗ indicated different overlapping strategy).
(B) The docking site (gray) of ACE2 protein (PDB: 1R42).
(C) Schematic of the in vitro experimental workflow to identify the final three compounds.
(D–F) Real-time PCR of gene expression in HUVECs. Log10 fold change was calculated based on treated samples against control samples. HUVECs were treated with 15 compounds for 16 h (D), inflammatory factors for 32 h (50 ng/mL TNF-α, IL-4, IL-6, and IFN-γ) following eight compounds for 16 h (E), or 10 nM control siRNA (siCon) and ACE2 siRNA (siACE2) for 8 h following six compounds for 16 h (F) (n = 3). The selected compounds based on the scoring system were highlighted in red and applied for the following screening.
(G) The low (L), medium (M), and high (H) concentrations of 15 compounds applied in the above three experiments.
See also Figure S3.
We next sought to validate these potential ACE2 activators via a series of in vitro experiments (Figure 3C). We treated HUVECs with these 15 compounds and measured the mRNA levels of genes downstream of ACE2, namely G6PC, GLUT2, PPARα, PPARγ, TNF-α, and IL-6. Eight compounds were most capable of regulating the expression of these genes (Figure 3D) in a similar pattern with activation of ACE2 (Figures 2A and S2A–S2D). We next pretreated HUVECs with the “cocktail” of inflammatory factors and then with the above eight compounds. Interestingly, six compounds most robustly reversed the change of gene expression upon ACE2 reduction (Figure 3E). Finally, we treated HUVECs with these six compounds after ACE2 knockdown (Figure S3B), and we found that three compounds (imatinib, harpagoside, and methazolamide) were capable of regulating genes in an ACE2-dependent manner (Figure 3F). The concentrations of these 15 compounds were listed in Figure 3G, and the cell viability after the treatment with each compound at the highest concentrations was determined by lactate dehydrogenase (LDH) assay, which demonstrated no significant toxic effects of these compounds except for amsacrine (Figure S3C). The expressions of ACE2 downstream genes after these three compounds treatment were shown in Figures S3D–S3S. Notably, imatinib, harpagoside, and methazolamide showed much higher potency to activate ACE2 in comparison with diminazene aceturate (DIZE), a previously described ACE2 activator (Figures S3D–S3I) (Goru et al., 2017; Shenoy et al., 2013). Furthermore, these three compounds tended to modestly increase the ACE2 protein (1.2- to 1.9-fold) while significantly elevating its mRNA (1.9- to 2.5-fold) (Figures S3T–S3W). Taken together, our data identified imatinib, harpagoside, and methazolamide as new ACE2 activators.
Imatinib, harpagoside, and methazolamide directly bind to and activate ACE2
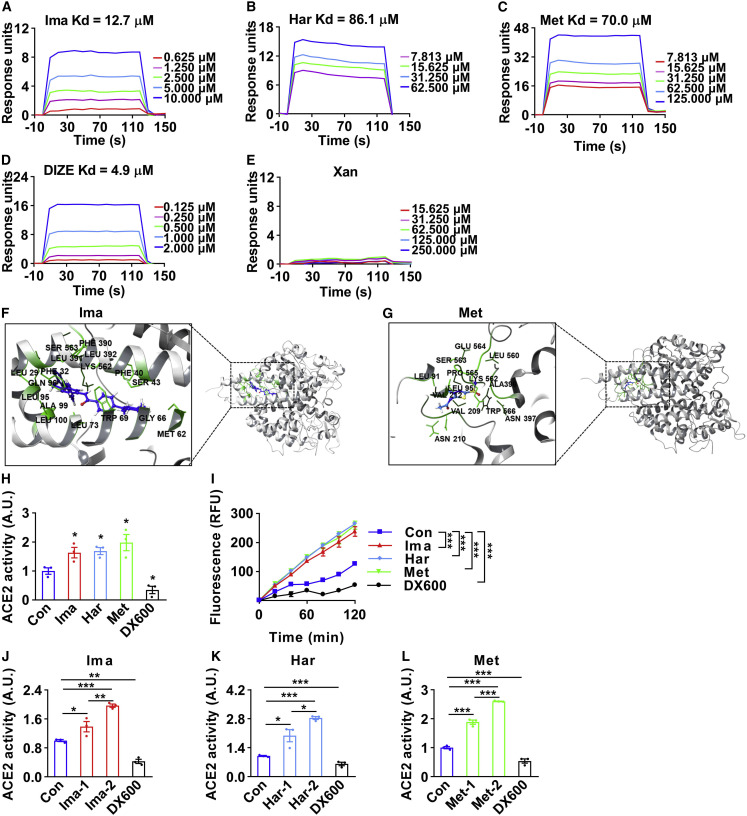
Given that all these three compounds were screened initially as a direct enzymatic activator of ACE2, we next determined whether these compounds can directly bind to ACE2 using the Biacore assay. Imatinib, harpagoside, and methazolamide bound to ACE2 protein with Kd values (equilibrium dissociation constant) at 12.7, 86.1, and 70.0 μM, respectively, whereas the corresponding Kd value for the previously proven binder DIZE was 4.9 μM. As a negative control, Xanthone (Xan) was not able to bind to ACE2 (Figures 4A–4E). The molecular dynamics simulation of the structural binding between ACE2 protein/amino acids and imatinib or methazolamide was shown in Figures 4F and 4G. Harpagoside was not available for simulation due to the lack of structure information of 1,5,6,7a-tetrahydrocyclopenta[c]pyran in GROMACS. Next, we measured the enzymatic activity of ACE2 after the treatment of these compounds. We first treated HUVECs with imatinib, harpagoside, or methazolamide and found that the enzymatic activity was significantly elevated, whereas the enzymatic inhibitor of ACE2 (DX600) (Huang et al., 2003) showed an opposite effect (Figure 4H). ACE2 enzymatic activity was also significantly elevated in HUVEC lysates after exposure to these compounds in time- and dose-dependent manners (Figures 4I–4L). Importantly, human and mouse ACE2 proteins shared 82.1% in sequence homology (Figure S4A), suggesting highly conserved ACE2 function across different species. Supporting this, knockdown of ACE2 in mouse AML12 cells resulted in increased expressions of G6pc, Tnf-α, and Il-6 and decreased expressions of Glut2, Pparα, and Pparγ (Figure S4B). Finally, imatinib, harpagoside, and methazolamide effectively increased the mRNA levels of Pparα, Pparγ, and Ace2 in AML12 cells (Figures S4C–S4E), indicating that these compounds serve as direct activators of ACE2 in both human and mouse cells.
Figure 4.
Imatinib, harpagoside, and methazolamide directly bind to and activate ACE2
(A–E) The direct binding was illustrated by surface plasmon resonance (SPR) assay of purified ACE2 protein with imatinib (Ima) (A), harpagoside (Har) (B), methazolamide (Met) (C), diminazene aceturate (DIZE) (D), or xanthone (Xan) (E). Xanthone was used as a negative control. The Kd values (equilibrium dissociation constant) of compounds binding to ACE2 protein were calculated based on the fitted curves.
(F and G) The detailed binding between ACE2 protein (gray) and imatinib (F, blue) and methazolamide (G, blue) was simulated with molecular dynamics simulation by GROMACS. Residues involved in binding were marked in green.
(H) The HUVECs were treated with 25 μM imatinib, 100 μM harpagoside, 100 μM methazolamide, or 1 μM DX600 for 16 h, then subjected to ACE2 enzymatic activity assay. Data shown as the area under the kinetic activity curves. The ACE2 enzymatic inhibitor DX600 was used as a negative control (n = 3).
(I–L) The HUVEC lysates were prepared and treated with 1.5 × 10−6 M imatinib, 1.5 × 10−6 M harpagoside, 1.5 × 10−6 M methazolamide, or 1.0 × 10−6 M DX600 for 30 min, then subjected to ACE2 enzymatic activity assay in a kinetics model. The kinetic activity curves were shown (I) (n = 3). The effects of imatinib (J) at concentrations of 1.5 × 10−7 M (Ima-1) and 1.5 × 10−6 M (Ima-2), harpagoside (K) at concentrations of 0.5 × 10−7 M (Har-1) and 1.5 × 10−6 M (Har-2), or methazolamide (L) at concentrations of 1.5 × 10−7 M (Met-1) and 1.5 × 10−6 M (Met-2) on the enzymatic activity of ACE2 in HUVEC lysates were shown as the area under the kinetic activity curves (n = 3).
Error bars represent SEM, ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. See also Figure S4.
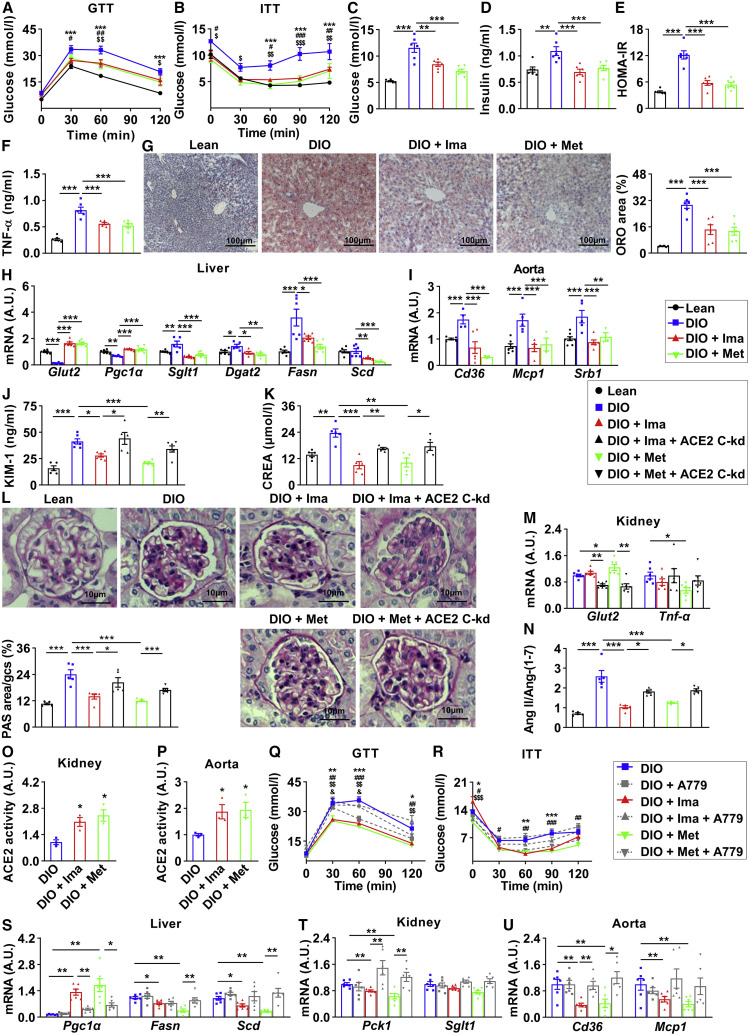
Imatinib and methazolamide ameliorate metabolic defects in insulin-resistant mice via ACE2
Since imatinib and methazolamide have been commercially available for clinical therapy, it is more tempting to repurpose these two drugs for COVID-19-associated metabolic diseases due to the urgent pandemic. We next investigated the role of imatinib and methazolamide in metabolic regulation in vivo. We treated the high-fat-diet-induced insulin-resistant mice with imatinib or methazolamide and monitored the systemic metabolic outcomes. The body weight, epididymal fat index, as well as food and water intake were reduced in mice treated with imatinib or methazolamide (Figures S5A–S5D). The glucose tolerance (Figure 5 A), insulin sensitivity (Figure 5B), fasting glucose and insulin (Figures 5C and 5D), and HOMA-IR (Figure 5E) were significantly improved after imatinib or methazolamide intervention, whereas plasma TG and TC were also reduced (Figures S5E and S5F). The inflammatory factor TNF-α in plasma was decreased (Figure 5F). Importantly, the hepatic steatosis was dramatically improved (Figure 5G), and the ameliorations of plasma alanine transaminase (ALT) and aspartate transaminase (AST), and hepatic TG and TC were observed as well (Figures S5G–S5J). Moreover, the downregulated genes of Glut2 and Pgc1α and the upregulated genes of Sglt1, Dgat2, Fasn, and Scd were significantly improved to lean mice levels (Figure 5H). Additionally, the protein expression of FoxO1 in liver was significantly reduced (Figure S5K), whereas the hepatic ACE2 was modestly upregulated only upon methazolamide intervention (Figure S5L). For aorta, the upregulated genes of Cd36, Mcp1, and Srb1 were significantly improved (Figure 5I). These data indicate systemic improvements of glucose and lipid metabolism after imatinib or methazolamide intervention.
Figure 5.
Imatinib and methazolamide ameliorate metabolic defects in insulin-resistant mice via ACE2
(A–I) Twenty-eight-week-old male mice with 23-week high-fat-diet treatment (DIO) and controlled lean mice (Lean) were treated with vehicle, 250 mg/kg imatinib (DIO + Ima), or 100 mg/kg methazolamide (DIO + Met) through gavage once each day for 4 weeks.
(A and B) Glucose tolerance testing (GTT) was performed at 30 weeks (A); insulin tolerance testing (ITT) was performed at 31 weeks (B) (n = 6). The significance of lean versus DIO was shown as ∗, DIO versus DIO + Ima as #, and DIO versus DIO + Met as $. ∗,#,$p < 0.05, ∗∗,##,$$p < 0.01, and ∗∗∗,###,$$$p < 0.001. At 32 weeks, all mice were fasted for 6 h and sacrificed.
(C–F) Fasting blood glucose (C), plasma insulin (D), homeostatic model assessment of insulin resistance (HOMA-IR) (E), and plasma TNF-α (F) (n = 6).
(G and H) Livers were subjected to oil red O (ORO) staining (G, images shown on the left, quantification on the right) and real-time PCR (H) (n = 6).
(I) Aortas were subjected to real-time PCR (n = 6).
(J–P) For kidney conditional knockdown of ACE2 (ACE2 C-kd), 26-week-old male mice with 21-week high-fat-diet treatment (DIO) and controlled lean mice (Lean) were treated with transparenchymal renal pelvis injection of AAV9-CAG-mACE2shRNA-EGFP or control virus. After 2-week recovery, mice were given vehicle, 250 mg/kg imatinib (DIO + Ima and DIO + Ima + ACE2 C-kd), or 100 mg/kg methazolamide (DIO + Met and DIO + Met + ACE2 C-kd) through gavage once each day for 4 weeks. At 32 weeks, all mice were fasted for 6 h and sacrificed.
(J and K) Plasma KIM-1 (J) and CREA (K) (n = 6).
(L and M) Kidneys were subjected to periodic acid-Schiff (PAS) staining (L, images and quantification were shown; n = 5) and real-time PCR (M) (n = 6).
(N) Quantification of plasma ratio of Ang II against Ang-(1–7) (n = 5).
(O and P) The lysates of kidneys (O) and aortas (P) were subjected to ACE2 enzymatic activity assay, and data were shown as the area under the kinetic activity curves (n = 3).
(Q–U) For A779 (MasR inhibitor) intervention, 28-week-old male mice with 23-week high-fat-diet treatment were given vehicle (DIO and DIO + A779), 250 mg/kg imatinib (DIO + Ima and DIO + Ima + A779), or 100 mg/kg methazolamide (DIO + Met and DIO + Met + A779) with or without 3 mg/kg A779 once each day for 4 weeks.
(Q and R) Glucose tolerance testing (GTT) was performed at 30 weeks (Q), and insulin tolerance testing (ITT) was performed at 31 weeks (R) (n = 6). The significance of DIO versus DIO + Ima was shown as ∗, DIO versus DIO + Met as #, DIO + Ima versus DIO + Ima + A779 as $ and DIO + Met versus DIO + Met + A779 as &. ∗,#,$,&p < 0.05, ∗∗,##,$$,&&p < 0.01, and ∗∗∗,###,$$$,&&&p < 0.001. At 32 weeks, all mice were fasted for 6 h and sacrificed.
(S–U) Livers (S), kidneys (T), and aortas (U) were subjected to real-time PCR.
Error bars represent SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. CREA, creatinine. See also Figure S5.
To confirm the necessity of ACE2 in mediating these effects, we injected the AAV-shACE2 via tail vein (ACE2 kd) to knock down ACE2 in high-fat-diet-induced insulin-resistant mice. Such AAV-shACE2 injection led to approximately 60% reduction of Ace2 mRNA in liver, without significant alteration in kidney and aorta (Figure S5M). Only modest improvements of glucose tolerance and insulin sensitivity were observed in an ACE2-dependent manner for imatinib or methazolamide intervention (Figures S5N and S5O), probably due to the insufficient knockdown efficiency. Given that the acute kidney injury represents one of the most common metabolic damages upon COVID-19, and ACE2 is abundantly expressed in kidney, we aimed to specifically focus on the metabolic outcomes upon ACE2 knockdown in kidney. We injected AAV-shACE2 in kidney via transparenchymal renal pelvis puncture (ACE2 conditional knockdown, ACE2 C-kd). As a result, the mRNA level of Ace2 in kidney was reduced by approximately 50% compared with those in control (Figure S5P). Notably, after imatinib or methazolamide intervention, KIM-1 and creatinine (CREA) levels in plasma were significantly reduced (Figures 5J and 5K), accompanied by the amelioration of the glomerular damage as determined by PAS staining (Figure 5L). Moreover, the gene expression of Glut2 was significantly increased, whereas Tnf-α was reduced (Figure 5M). Importantly, knockdown of ACE2 in kidney partially or even remarkably reversed all of these effects (Figures 5J–5M), indicating that imatinib and methazolamide improve metabolic defects in an ACE2-dependent manner at least in kidney.
Finally, we tested the role of ACE2 enzymatic activity in mediating these effects. Imatinib or methazolamide with or without A779 (MasR inhibitor) were given to insulin-resistant mice, our data first confirmed a drug-induced elevation of ACE2 enzymatic activity based on the following observations: (1) drug treatments reduced the Ang II/Ang-(1–7) ratio in an ACE2-dependent manner (Figure 5N) and (2) the enzymatic activity of ACE2 in kidney and aorta was significantly increased after drug treatments (Figures 5O and 5P) as well as the Ace2 mRNA level (Figures S5Q and S5R). Importantly, antagonizing ACE2-MasR axis by A779 blocked the improvement of the glucose tolerance (Figure 5Q), insulin sensitivity (Figure 5R), and the altered genes of Pgc1α, Fasn, and Scd in liver (Figure 5S), Pck1 and Sglt1 in kidney (Figure 5T), and Cd36 and Mcp1 in aorta (Figure 5U) induced by imatinib or methazolamide, suggesting the requirement of ACE2 enzymatic product Ang-(1–7) to mediate the regulatory effects of imatinib and methazolamide.
ACE2 enzymatic activators improve metabolic defects and inhibit virus entry upon SARS-CoV-2 infection
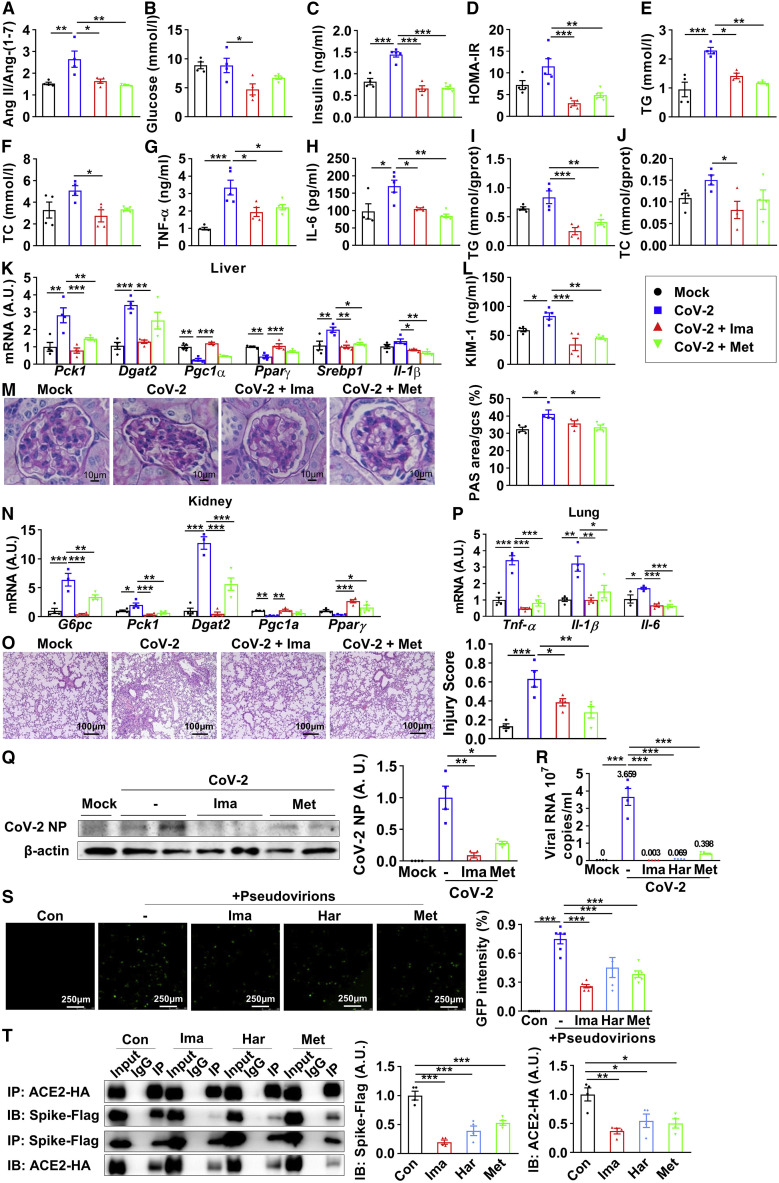
To further clarify the regulatory effects of imatinib and methazolamide upon SARS-CoV-2 infection, hACE2 transgenic mice (with replacement of endogenous mouse ACE2) were pretreated with imatinib or methazolamide for 4 weeks after diet-induced obesity and then infected with SARS-CoV-2. Of note, the Ang II/Ang-(1–7) ratio of mice plasma was significantly upregulated after infection and reversed to control level after compounds treatment (Figure 6 A), suggesting inhibition of ACE2 enzymatic activity upon infection and the activation upon compound treatment. While the fasting glucose was not significantly altered upon infection (Figure 6B), the fasting insulin (Figure 6C) and HOMA-IR (Figure 6D) were significantly upregulated after virus infection in mice without any treatment, suggesting an early stage of insulin resistance. Imatinib or methazolamide intervention, however, significantly ameliorated SARS-CoV-2-induced insulin resistance (Figures 6B–6D). In parallel, the TG, TC, TNF-α, and IL-6 in plasma were elevated upon infection and reversed after the treatment of ACE2 activators (Figures 6E–6H). In liver, hepatic TG and TC levels showed a strong increasing trend following infection, which was dramatically ameliorated after interventions (Figures 6I and 6J). The disrupted metabolic genes of Pck1, Dgat2, Pgc1α, Pparγ, Srebp1, and Il-1β were significantly improved after imatinib or methazolamide treatment (Figure 6K). Regarding the kidney, after compound treatment, the elevated KIM-1 upon infection was significantly blunted (Figure 6L), whereas the glomerular damage was also ameliorated as determined by PAS staining (Figure 6M), indicating protective effects of the ACE2 activators on renal function. Moreover, the disrupted metabolic genes of G6pc, Pck1, Dgat2, Pgc1α, and Pparγ were significantly improved with the treatment of imatinib or methazolamide (Figure 6N). Taken together, these data showed that imatinib and methazolamide restored the ACE2 enzymatic activity and largely improved the metabolic defects caused by SARS-CoV-2 infection in insulin-resistant mice.
Figure 6.
ACE2 enzymatic activators improve metabolic defects and inhibit virus entry upon SARS-CoV-2 infection
(A–Q) Twelve-week-old human ACE2 transgenic mice were given vehicle (Mock, CoV-2), 250 mg/kg imatinib (CoV-2 + Ima), and 100 mg/kg methazolamide (CoV-2 + Met) through gavage once each day for 4 weeks after 6-week high-fat-diet treatment and were intranasally challenged with 4 × 104 FFU SARS-CoV-2. After 7 days of infection, all mice were fasted for 6 h and sacrificed.
(A–H) Quantification of the ratio of Ang II against Ang-(1–7) in plasma (A), fasting blood glucose (B), plasma insulin (C), homeostatic model assessment of insulin resistance (HOMA-IR) (D), plasma triglyceride (E), plasma total cholesterol (F), plasma TNF-α (G), and plasma IL-6 (H) (n = 4).
(I–K) Livers were subjected to lipid assay (I and J) and real-time PCR (K) (n = 3–4).
(L) Plasma KIM-1 (n = 4–5).
(M and N) Kidneys were subjected to periodic acid-Schiff (PAS) staining (M, images and quantification were shown; n = 4) and real-time PCR (N) (n = 4).
(O–Q) Lungs were subjected to H&E staining (O, images and quantification were shown; n = 4), real-time PCR (P) (n = 4), and immunoblotting of viral nucleocapsid protein (NP) (Q, blot shown on the left, quantification on the right; n = 4).
(R) Vero E6 cells were pretreated with 25 μM imatinib, 100 μM harpagoside, or 100 μM methazolamide for 6 h, followed by SARS-CoV-2 (MOI = 0.005) infection for 42 h; the supernatant was subjected to real-time PCR (n = 4).
(S) HEK293T cells expressing hACE2 were pretreated with 25 μM imatinib, 100 μM harpagoside, or 100 μM methazolamide for 6 h, followed by pseudovirion treatment for 66 h, and were observed with fluorescence microscopy (images shown on the left, quantification on the right; n = 5).
(T) The HEK293T cells overexpressing spike-FLAG and ACE2-HA were treated with 25 μM imatinib, 100 μM harpagoside, or 100 μM methazolamide for 48 h. Precipitated proteins were subjected to immunoblotting (blot shown on the left, quantification on the right; n = 4).
Error bars represent SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. See also Figure S6.
We next investigated the regulation of ACE2 enzymatic activators on virus infection. First, we found that imatinib or methazolamide intervention significantly improved virus-induced lung damage including septal thickening and alveolar infiltration of inflammatory cells as determined by H&E staining (Figure 6O). Consistently, viral-infection-induced elevation of Tnf-α, Il-1β, and Il-6 expression was significantly reversed by imatinib or methazolamide treatment (Figure 6P). Importantly, the expression level of viral nucleocapsid protein (NP) in the lung, as determined by immunoblotting (Figure 6Q) and immunohistochemical staining (Figure S6A), was significantly reduced after imatinib or methazolamide intervention, suggesting a direct antiviral effect of both imatinib and methazolamide. We further validated the effect of all three ACE2 activators on viral infection in Vero E6 cells. Pretreatment of imatinib, harpagoside, or methazolamide dramatically reduced the viral RNA in the cell culture medium by up to 89.1%–99.9% upon SARS-CoV-2 infection (Figure 6R). In line with that, the titer of virus in the supernatant of infected Vero E6 cells after treatment with each of the three compounds, as determined by focus formation assay, was also reduced largely (Figure S6B), even though slight upregulations of ACE2 mRNA level in Vero E6 cells were observed (Figure S6C).
To further investigate the underlying mechanism, we first treated cells with the “false virus” fused with spike protein to mimic SARS-CoV-2 entry. HEK293T cells expressing hACE2 were pretreated with imatinib, harpagoside, or methazolamide, followed by pseudovirion treatment. The virus entry was significantly reduced following imatinib, harpagoside, or methazolamide treatment (Figure 6S), suggesting a potential competition of the binding between these three compounds and spike protein to ACE2 receptor. The cell toxicity of pseudovirions was not observed upon interventions (Figure S6D). Moreover, our virtual computation analysis further supported these important observations. We simulated the binding kinetics between the compounds, spike protein, and ACE2 protein via the Tianhe-2 supercomputer and confirmed a potential competition between these molecules. As shown in Figure S6E, when imatinib or methazolamide was added, the free energy binding between ACE2 and spike protein increased from −264.784 to −249.744 kcal/mol for imatinib and from −264.784 to −258.103 kcal/mol for methazolamide. Interestingly, the conformational alterations of ACE2 and spike protein structure were also observed after adding imatinib or methazolamide, as determined by molecular dynamics simulation by supercomputer (Figures S6F and S6G). To validate our in silico results, co-immunoprecipitation (coIP) assay was performed to assess the association between ACE2 and spike protein in presence of these three compounds. As shown in Figure 6T, imatinib, harpagoside, and methazolamide significantly reduced the spike protein level within the ACE2 immunocomplex and vice versa. These data suggest that the antiviral effect of these three compounds was at least partially due to the inhibition of spike protein binding to ACE2. Finally, we performed a meta-analysis and showed that the treatment for hypertension with ACEi or ARB in COVID-19 patients did not exacerbate the risk of severity but significantly decreased the risk of mortality of COVID-19 (Figure S6H), despite a potential modest elevation of ACE2 level upon using these antihypertension drugs (Lebek et al., 2020; Reynolds et al., 2020), indicating the safety of activating ACE2 in drug design for treating COVID-19.
Discussion
Our data show that ACE2 serves as a key link bridging COVID-19 and its associated metabolic dysfunctions. Furthermore, we have screened and identified three compounds (imatinib, methazolamide, and harpagoside) as direct enzymatic activators of ACE2. The repurposed drugs of imatinib and methazolamide show considerable improvements in metabolic dysfunctions in both diet-induced insulin-resistant mice and SARS-CoV-2-infected mice and surprisingly decrease the infectivity of SARS-CoV-2 in vivo and in vitro.
It remains highly controversial what roles ACE2 plays during the progression of COVID-19 and its complications, although it is well established that ACE2 serves as the receptor of the spike protein on SARS-CoV-2 (Wan et al., 2020). Overexpression of ACE2 enhances viral entry, whereas blockage with ACE2 antibody reduces SARS-CoV-2 infection (Hoffmann et al., 2020). Therefore, inhibition of ACE2 expression has been proposed as a strategy to treat COVID-19 (Ahmad et al., 2021; Ivanov et al., 2021). In contrast, ACE2, as an important component of RAS axis, exerts anti-inflammatory and antioxidative functions and maintains metabolic homeostasis as supported by our current findings. Our data show that SARS-CoV-2 infection results in a significant reduction of ACE2 expression, consistent with a recent finding that ACE2 is downregulated in both kidney and heart of COVID-19 patients (Nie et al., 2021). Notably, impaired ACE2 function appears to be a dominant driver for the induction of metabolic defects upon COVID-19. Our data show that administration of ACE2 enzymatic activators, identified by our unbiased screening, dramatically improves metabolic abnormalities after virus exposure in vivo with the restoration of impaired ACE2. Moreover, such ACE2 activators reduce virus infection via allosteric inhibition of ACE2 binding to spike protein. Taken together, our findings strongly suggest a new pharmacologic strategy to combat COVID-19 and its metabolic complications by restoring ACE2 enzymatic activity upon SARS-CoV-2 infection, instead of further inhibiting ACE2 level.
Several approaches are currently available for activation of ACE2 pathway, including ACE pathway antagonist, soluble ACE2, Ang-(1–7) receptor agonist, etc. However, the potential consequence of hypotension with ACE inhibition may limit its clinical application as an indirect ACE2 activator in COVID-19 patients. The large molecular size of soluble ACE2 may impede its penetrance on specific organs, and the expensive cost also limits its clinical application. No clinical or translational studies have been reported on Ang-(1–7) receptor agonists, such as AVE 0991 (Suski et al., 2013). More importantly, none of the above-mentioned approaches exert functions as direct enzymatic activators of ACE2. In this study, we aim to identify potent ACE2 activators. By utilizing CMAP and virtual docking computation, we have identified three compounds (imatinib, methazolamide, and harpagoside) as ACE2 enzymatic activators. We show that these compounds are very effective to improve the metabolic abnormalities. Imatinib and methazolamide dramatically improve hyperglycemia, dyslipidemia, inflammation, hepatic steatosis, and vascular disorders in vivo. They also ameliorate kidney dysfunctions in an ACE2-dependent manner. Notably, the enzymatic activity of ACE2 plays an important role given that MasR inhibitor A779 can partially block these effects.
The repurposed drugs are of great advantage for clinical application due to the validated side effects. Imatinib is a potent inhibitor of the BCR-ABL tyrosine kinase associated with chronic myeloid leukemia (Druker et al., 2001). Methazolamide, a potent carbonic anhydrase (CA) inhibitor, has been extensively used in the treatment of glaucoma and the prevention of altitude sickness (Fossati et al., 2016). Besides these traditional targets and effects, very few reports have investigated their regulatory roles on metabolism. A recent study has shown that imatinib exhibits beneficial effects on insulin sensitivity (Pichavaram et al., 2021). Similarly, methazolamide has also been identified as an insulin sensitizer (Konstantopoulos et al., 2012). Our current data show that imatinib and methazolamide serve as ACE2 enzymatic activators and dramatically ameliorate systemic metabolic defects in liver, kidney, and cardiovascular system of insulin-resistant mice. Furthermore, our in vitro data show that imatinib and methazolamide enhance ACE2 enzymatic activity and reduce Ang II/Ang-(1–7) ratio. Furthermore, the virtual in silico simulation and in vitro binding assay confirm the direct binding of these two drugs to ACE2. Despite that several previous reports have claimed that chemical compounds xanthenone (XNT) and DIZE could bind to and activate ACE2 to improve hypertension (Hernández Prada et al., 2008; Kulemina and Ostrov, 2011), controversial results from others suggest that XNT and DIZE are not the direct enzymatic activator of ACE2, since the beneficial effect of XNT on blood pressure still exists upon ACE2 deficiency, and the direct stimulatory effects of XNT and DIZE on ACE2 are not observed in ex vivo kidneys of mice and rats (Haber et al., 2014). Therefore, the identification of new ACE2 enzymatic activators is of great significance in the potential treatment of COVID-19-associated diseases given the versatile role of ACE2 in metabolic regulation.
One unexpected observation of this study is that these ACE2 enzymatic activators could slightly elevate the expression level of ACE2, which may raise the concern of enhancing SARS-CoV-2 entry. However, previous clinical findings have shown that patients receiving antihypertensive treatment with an ACEi are not more susceptible to SARS-CoV-2 infection (Reynolds et al., 2020) despite the fact that ACE2 levels are partially elevated upon administration of ACEi (Reynolds et al., 2020). Supporting this, our meta-analysis shows that ACEi/ARB does not change the risk of severity but rather reduces the mortality in patients with COVID-19. Based on these studies, several guidelines recommend not to discontinue ACEi/ARB for COVID-19 patients with hypertension (Alexandre et al., 2020; Mehta et al., 2020).
Interestingly, a recent screening reveals that imatinib may bind to ACE2 and inhibit virus infection, the mechanism of which remains elusive (Han et al., 2021). Due to its antiviral effect, imatinib has been reported in case reports and clinical trials on COVID-19. More stable pulmonary opacities have been observed in a COVID-19 patient 3 days after application of imatinib (Morales-Ortega et al., 2020), and a large-scale clinical trial has reported that imatinib intervention reduces the hazard ratio on mortality (Aman et al., 2021). Our data indicate that imatinib, along with another identified ACE2 activator, methazolamide, can inhibit virus infection both in vivo and in vitro, at least partially via the direct interaction with ACE2 protein. This consequently reduces ACE2’s binding to spike protein, therefore inhibiting the virus entry. These observations implicate that the application of imatinib and methazolamide in the treatment of COVID-19 would benefit from the ACE2-dependent metabolic improvement, as well as the direct reduction of SARS-CoV-2 load within the cells.
In summary, COVID-19 is a systemic disease that may induce severe metabolic defects in multiple tissues, and impairment of ACE2 pathway is a key factor linking virus infection to its metabolic sequelae. Here, we identify imatinib, methazolamide, and harpagoside as the direct enzymatic activators of ACE2 and show that imatinib and methazolamide are able to improve SARS-CoV-2-induced metabolic disorders by enzymatic activation of ACE2 and inhibit viral entry via allosteric inhibition of ACE2 binding to spike protein; therefore, they can be repurposed for the potential treatment of COVID-19 and its metabolic complications.
Limitations of study
Given that a high-throughput virtual screening workflow via Tianhe-2 supercomputer, instead of traditional high-throughput cell-culture-based in vitro screening, was applied for initial ACE2 activator identification, some potential activators besides imatinib, methazolamide, and harpagoside might be neglected. Moreover, the potential of imatinib and methazolamide to improve metabolism in COVID-19 patients remains unclear and requires clinical investigation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-FoxO1 | Cell Signaling Technology | Cat. #2880; RRID: AB_2106495 |
| Rabbit polyclonal anti-ACE2 | Abcam | Cat. ab15348; RRID: AB_301861 |
| Rabbit monoclonal anti-β-Actin | Cell Signaling Technology | Cat. #4970; RRID: AB_2223172 |
| Rabbit monoclonal anti-SARS-CoV-2 Nucleocapsid | Sino Biological Company | Cat. # 40143-R001; RRID: AB_2827974 |
| Rabbit monoclonal anti-HA | Cell Signaling Technology | Cat. #3724; RRID: AB_1549585 |
| Mouse monoclonal anti-Flag | Cell Signaling Technology | Cat. #8146; RRID: AB_10950495 |
| Anti-rabbit IgG, HRP-linked Antibody | Cell Signaling Technology | Cat. #7074; RRID: AB_2099233 |
| Bacterial and virus strains | ||
| SARS-CoV-2 (hCoV-19/CHN/SYSU-IHV/2020 strain) | Sun Yat-sen University | GISAID: EPI_ISL_444969 |
| AAV9-CAG-humanACE2-EGFP | PackGene Biotech | N/A |
| AAV9-CAG-mACE2shRNA-EGFP | PackGene Biotech | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| A779 | MedChenExpress | Cat. HY-P0216 |
| Amsacrine | MedChenExpress | Cat. HY-13551 |
| Anagrelide | MedChenExpress | Cat. HY-B0523A |
| AZD-7762 | MedChenExpress | Cat. HY-10992 |
| Clebopride | MedChenExpress | Cat. HY-B1613A |
| Diminazene aceturate (DIZE) | MedChenExpress | Cat. HY-12404 |
| Doxorubicin | MedChenExpress | Cat. HY-15142 |
| DX600 | MedChenExpress | Cat. HY-P2222 |
| Entinostat | MedChenExpress | Cat. HY-12163 |
| Harpagoside | MedChenExpress | Cat. HY-N0396 |
| Huperzine-A | MedChenExpress | Cat. HY-17387 |
| Imatinib | MedChenExpress | Cat. HY-15463 |
| Methazolamide | MedChenExpress | Cat. HY-B0553 |
| Ofloxacin | MedChenExpress | Cat. HY-B0125 |
| Pantoprazole | MedChenExpress | Cat. HY-17507 |
| Phensuximide | MedChenExpress | Cat. HY-B(1730) |
| Pyroxamide | MedChenExpress | Cat. HY-13216 |
| Tranilast | MedChenExpress | Cat. HY-B0195 |
| Xanthone | MedChenExpress | Cat. HY-N0126 |
| Human angiotensin-I converting enzyme 2 (ACE2) | Sino Biological Company | Cat. 10108-H05H |
| Human IL-4 | Sino Biological Company | Cat. 11846-HNAE |
| Human IL-6 | Sino Biological Company | Cat. 10395-HNAE |
| Human IFN-γ | Sino Biological Company | Cat. 11725-HNAS |
| Human TNF-α | Sino Biological Company | Cat. 10602-HNAE |
| Critical commercial assays | ||
| Human Angiotensin II ELISA kit | CUSABIO | Cat. CSB-E04493h |
| Human Angiotensin-(1-7) ELISA kit | CUSABIO | Cat. CSB-E14242h |
| Mouse TNF-α ELISA kit | CUSABIO | Cat. CSB-E04741m |
| Mouse IL-6 ELISA kit | CUSABIO | Cat. CSB-E04639m |
| Mouse Angiotensin II ELISA kit | CUSABIO | Cat. CSB-E04495m |
| Mouse Angiotensin-(1-7) ELISA kit | CUSABIO | Cat. CSB-E13763m |
| Mouse kidney injury molecule 1 (KIM-1) ELISA kit | CUSABIO | Cat. CSB-E08809m |
| Mouse insulin ELISA kit | CUSABIO | Cat. CSB-E05071m |
| RNA extraction reagent | Accurate Biology Company | Cat. AG21102 |
| Evo M-MLV Reverse Transcription kit | Accurate Biology Company | Cat. AG11711 |
| SYBR Green Premix Pro Taq HS qPCR kit | Accurate Biology Company | Cat. AG11701 |
| ACE2 activity assay Kit (Fluorometric) | Abcam | Cat. ab273297 |
| Triglyceride assay kits | Nanjing Jiancheng Bioengineering Institute | Cat. A110-1-1 |
| Total cholesterol assay kits | Nanjing Jiancheng Bioengineering Institute | Cat. A111-1-1 |
| HDL-C assay kits | Nanjing Jiancheng Bioengineering Institute | Cat. A112-1-1 |
| LDL-C assay kits | Nanjing Jiancheng Bioengineering Institute | Cat. A113-1-1 |
| Series S Sensor Chip CM7 | GE Healthcare Life Sciences | Cat. 10281415 |
| Deposited data | ||
| Raw RNA-seq data | This paper | GSA: HRA001795 |
| Experimental models: Cell lines | ||
| HUVEC | ATCC | CRL-1730; RRID: CVCL_2959 |
| HEK293T | NICR, China | N/A |
| AML12 | ATCC | CRL-2254; RRID: CVCL_0140 |
| Vero E6 | ATCC | CRL-1586; RRID: CVCL_0574 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6JGpt | GemPharmatech | Cat. N000013; RRID: IMSR_GPT:N000013 |
| Mouse: C57BL/6JGpt ob/ob | GemPharmatech | Cat. T001461; RRID: IMSR_GPT:T001461 |
| Mouse: B6/JGpt-Ace2em1Cin(hACE2)/Gpt | GemPharmatech | Cat. T037659; RRID: IMSR_GPT:T037659 |
| Oligonucleotides | ||
| See Table S4 for oligos used in this study | N/A | N/A |
| Software and algorithms | ||
| Graphpad Prism 8 | GraphPad Software | N/A |
| Review Manager 5.4 | Cochrane Collaboration | N/A |
| HISAT2 | http://daehwankimlab.github.io/hisat2/ | N/A |
| GENCODE | https://www.gencodegenes.org/ | GENCODE 37 |
| DESeq2 package | https://bioconductor.org/packages/release/bioc/html/DESeq2.html | N/A |
| Metascape | https://metascape.org/ | N/A |
| ImageJ | NIH | N/A |
| Other | ||
| 60% High fat diet | Trophic Animal Feed High-Tech Co.,Ltd | Cat. TP23300 |
Resource availability
Lead contact
Additional information and requests for resources or reagents can be addressed to the Lead Contact, Sifan Chen (chensf26@mail.sysu.edu.cn).
Materials availability
This study did not generate new unique materials or reagents.
Experimental model and subject details
Animal studies
General
Male C57BL/6J mice (age, 4 weeks), male ob/ob mice (age, 5-6 weeks) and their wild-type littermates (age, 5-6 weeks), and male human ACE2 transgenic mice (age, 5-6 weeks) were purchased from GemPharmatech, China. Animals were housed under 12 h of light/dark cycles with unrestricted access to food and water in specific pathogen free (SPF) conditions. The body weight and food intake were measured every other day. All conventional animal studies were approved by the Institutional Animal Care and Use Committees of the First Affiliated Hospital of Sun Yat-sen University. Authentic SARS-CoV-2 challenge studies were carried out in strict accordance with the guidelines and regulations of Laboratory Monitoring Committee of Guangdong Province of China and were approved by Ethics Committee of Zhongshan School of Medicine of Sun Yat-sen University on Laboratory Animal Care.
Dietary, AAV, virus and Compounds Treatments
To establish the diet-induced obese (DIO) mouse model, wild-type C57BL/6J mice were fed with 60% high fat diet (HFD; TP23300, TROPHIC, China) at five-week-old, and maintained on the same diet for 21-23 weeks before experiments. Mice in ob/ob-ACE2 and DIO + ACE2 knockdown group were treated with an intravenous injection of AAV9-CAG-humanACE2-EGFP and AAV9-CAG-mACE2shRNA-EGFP at a dose of 1.0×1013 genome copies (GC), respectively. The control group was treated with corresponding control viruses at a dose of 1.0×1013 GC. For knockdown of ACE2 in kidney, antegrade renal pelvis injection was performed via transparenchymal renal pelvis puncture with 0.5×1013 GC viruses in each kidney. Two weeks after virus injection, 250 mg/kg of imatinib or 100 mg/kg of methazolamide was given daily by gavage for 4 weeks, and 3 mg/kg of A779, an antagonist of Mas receptor, was given daily by intraperitoneal injection for 4 weeks. The same treatments (dose and duration) of imatinib or methazolamide were applied in DIO mice to study the direct effects of these two drugs on metabolic regulation and in vivo infection. For the infection of SARS-CoV-2, twelve-week-old human ACE2 transgenic mice were given vehicle, 250 mg/kg of imatinib and 100 mg/kg methazolamide through gavage one time each day for 4 weeks after 6 weeks diet-induced obesity and were intranasally challenged with 4×104 FFU SARS-CoV-2 (hCoV-19/CHN/SYSU-IHV/2020 strain; Accession ID on GISAID: EPI_ISL_444969). After 7 days infection, all mice were fasted for 6 h and sacrificed under anesthetized condition.
Cell Culture
Human umbilical vein cells (HUVECs) were cultured in F-12K medium (Hyclone) supplemented with 10% fetal bovine serum (FBS, Gibco), 0.1 mg/mL heparin and 30 μg/mL Endothelial Cell Growth Supplement and containing 100 IU/ml penicillin and 100 μg/ml streptomycin. HEK293T and Vero E6 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Hyclone) supplemented with 10% FBS and 100 IU/ml penicillin and 100 μg/ml streptomycin. AML12 cells were cultured in DMEM/F12 medium (Hyclone) supplemented with insulin-transferrin-selenium, penicillin-streptomycin, 10% FBS and 100 nM dexamethasone. All cell lines were maintained at 37°C in a 5% CO2 cell culture incubator. Lipofectamine 2000 (Invitrogen, USA) was used for plasmid and siRNA transfections according to the manufacturers’ instructions. HUVECs cultured in 12-well plate were transfected with 2 μg ACE2 plasmid for 72 h for overexpression experiments. The concentration of the siRNA was 10 nM for each well in 12-well plate, treated with compounds in 8 h, and the cells were collected after 24 h for the ACE2 knockdown experiments. HUVECs were plated in 12-well plates at 1×104 cells per well, treated with imatinib, harpagoside, or methazolamide for 16 h, harvesting for RNA analysis. For cytokines treatment experiment, cells were treated with recombinant human TNF-α, IFN-γ, IL-4 and IL-6 (Sino Biological, China) at each concentration of 50 ng/ml for 32 h, and then treated with compounds for another 16 h. For the infection of SARS-CoV-2, 5×105 HUVECs or Vero E6 cells were incubated with SARS-CoV-2 (MOI = 0.005) for 24-48 h at 37°C, and then were harvested for Western blotting or RNA analysis. SARS-CoV-2 (hCoV-19/CHN/SYSU-IHV/2020 strain; Accession ID on GISAID: EPI_ISL_444969) was isolated from the sputum of a female infected individual, and propagated in Vero E6 cells, the virus stocks were obtained from the supernatant of Vero E6 after inoculation for 48 h, and the titers were determined by FRNT assay targeting nucleocapsid protein.
Method details
Glucose Tolerance Test (GTT) and Insulin Tolerance Test (ITT)
For GTT, animals were fasted from the beginning of dark cycle (7 PM). After 15 h fasting, blood glucose levels were measured from the tail vein with a glucometer. Subsequently, saline or dextrose solution (2 g/kg for lean and DIO mice, 1 g/kg for ob/ob mice) was administered intraperitoneally. Blood glucose levels were measured from the tail vein at 30, 60 and 120 min following glucose injection.
For ITT, mice were fasted for 6 h during the light cycle. Saline or insulin (1 IU/kg for lean and DIO mice, 2 IU/kg for ob/ob mice) were administered intraperitoneally. Blood glucose levels were measured at 30, 60, 90 and 120 min following insulin injection from the tail vein.
For fasting glucose, mice were fasted for 6 h during the light cycle, and blood glucose levels were measured from the tail vein.
Biochemical measurements
Blood samples were centrifuged at 3,000 g for 10 min, and plasma was pipetted and stored at –80°C. Biochemical parameters including alanine aminotransferase (ALT), aspartate aminotransferase (AST), total cholesterol (TC), triglycerides (TG) and plasma creatinine levels were measured according to the manufacturer’s instructions (Servicebio, China).
Lipid profile analysis
A commercial assay kits (Nanjing Jiancheng Bioengineering Institute, China) was used to measure the levels of TC and TG in the liver of ob/ob or DIO mice. The samples were weighed and homogenized with 9 times volume of ethyl alcohol, then centrifuged at 12,000 rpm for 5 min at 4°C. The supernatants were applied to lipid profile analysis according to the manufacturer’s instructions.
Echocardiography
Echocardiography was performed using Vevo3100 (VisualSonics, Canada) equipped with a 30 MHz linear transducer (Visual Sonics, Canada). Fractional shortening was calculated as follows: %FS = [(LVEDD − LVESD)/LVEDD] × 100, LVEDD represents left ventricular end-diastolic diameter, and LVESD represents left ventricular end-systolic diameter. M-mode images were obtained for measurement of wall thickness and chamber dimensions with the use of the leading-edge convention adapted by the American Society of Echocardiography.
Histology
Tissues were fixed with 4% formalin and embedded in paraffin, 7 μm-thick sections were prepared for hematoxylin and eosin (H/E), Oil red O, periodic acid-Schiff (PAS) and immunohistochemistry staining. The images were obtained using a microscope DM 2500B (Leica, Germany). Liver Oil red O staining (Huang et al., 2021), kidney PAS staining (Liu et al., 2019) and lung injury score (based on five histological findings of neutrophils in the alveolar space, neutrophils in the interstitial space, hyaline membranes, proteinaceous debris filling the airspaces and alveolar septal thickening) (Matute-Bello et al., 2011) were quantified using ImageJ software (National Institutes of Health, USA), according to previously defined methods.
RNA Sequencing of Viral Infected Cells
Approximately 5×105 HUVECs per sample were incubated with SARS-CoV-2 (MOI = 0.005) for 24 h at 37°C, then were harvested. Total RNA from infected and mock infected cells was lysed in AG RNAex Pro Reagent (Accurate Biotechnology (Hunan), China) and extracted. mRNA was purified from total RNA using poly-T oligo-attached magnetic beads and fragmentated using divalent cations under elevated temperature in First Strand Synthesis Reaction Buffer (5×). Double-stranded cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase for first strand, following DNA Polymerase I and dNTP for second strand synthesis. PCR was performed with Phusion High-Fidelity DNA polymerase (Thermo Scientific, USA) according to the manufacturer’s instructions. At last, PCR products were purified (AMPure XP system), following libraries sequencing on an Illumina Novaseq platform.
Focus formation assay
Focus formation assay was performed as reported previously (Sun et al., 2020). Vero E6 cells were pre-treated with 25 μM imatinib, 100 μM harpagoside or 100 μM methazolamide for 6 h, followed by SARS-CoV-2 (MOI = 0.005) infection for 42 h, the supernatant was collected and serially diluted to obtain dilutions in the range of 1:10−1 to 1:10−6 with DMEM medium for the focus formation assay. Vero E6 cells were cultured in 96-well plates (20,000 cells/well), after full confluency, growth media was removed from the Vero E6 cells and 50 μL of virus dilutions was added. Virus was incubated for 1 h at room temperature. After incubation, 125 μl of 1.6% carboxymethyl cellulose (CMC) was added to each well of the 96-well plate and then incubated at 37°C with 5% CO2 for 24 h. 200 μl 4% paraformaldehyde per well were added and incubated at room temperature for 1 h. After removal of paraformaldehyde, monolayers were permeabilized with 50 μl 0.2% Triton X-100 supplemented with 1% BSA per well at temperature for 30 min. Monolayers were then blocked for 1 h in PBS with 1% BSA. After blocking, cells were incubated with SARS-CoV NP primary antibody (SinoBiologicals, China; 40143-R001, 1:2000 dilution) for 1 h at 37°C in PBS with 1% BSA. Monolayers were washed with PBS and incubated with an HRP-conjugated secondary antibody for 1 h at 37°C in PBS with 1% BSA. Then the secondary antibody was removed, monolayers were washed with PBS, and then developed using TrueBlue substrate (KPL) for 5-10 min. Plates were imaged on an EliSpot reader and foci were detected.
Pseudovirus Entry Assays
HEK293T/ACE2 stable cell line (PackGene Biotech, China), which is constructed by transduction of human ACE2 (hACE2) into HEK293T cells, were pretreated with imatinib, harpagoside, or methazolamide for 6 h, respectively. Subsequently, SARS-CoV-2 pseudovirions (PackGene Biotech, China) were added to the cells and incubated at 37°C for another 66 h. The SARS-CoV-2 pseudovirus-infected GFP-expressing HEK293T/ACE2 cells were observed by fluorescence microscopy (Leica DMI8, Germany) and the quantification of GFP density was performed via ImageJ software (National Institutes of Health, USA).
Real-time PCR (RT-PCR)
Total RNA was extracted by using AG RNAex Pro Reagent (Accurate Biotechnology (Hunan), China). With 1 μg of total RNA for reverse transcription, cDNA was synthesized using Evo M-MLVRT kit (Accurate Biotechnology (Hunan), China). qRT-PCR was performed with SYBR Green Premix Pro Taq HS qPCR Kit (Accurate Biotechnology (Hunan), China) and LightCycle480 II thermal cycler (Roche, Switzerland). Relative expression of each gene was normalized to GAPDH. The value of the control group was set to 1. The primer sequences are listed in the Table S4.
Western Blotting
For Western blotting, cells and tissues were lysed in RIPA buffer (Tris-HCl 25 mM, pH 7.6; NaCl 150 mM; NP-40 1%; sodium deoxycholate 1%; SDS 0.1%) with protease inhibitors and phosphatase inhibitors. Protein concentration was measured using bicinchoninic acid (BCA) (ComWin Biotech, China). Equal amounts of protein were separated by SDS-PAGE and transferred to a PVDF membrane which was incubated in 5% milk in Tris-buffered saline for 1 h at room temperature, followed by incubation with primary antibodies against ACE2 (Cell Signaling Technology, USA, 1:1000 dilution), FoxO1 (Cell Signaling Technology, USA, 1:1000 dilution), SARS-CoV-2 Nucleocapsid (NP) (SinoBiologicals, China, 1:1000 dilution), HA (Cell Signaling Technology, USA, 1:1000 dilution) and Flag (Cell Signaling Technology, USA, 1:1000 dilution) overnight at 4°C, and a secondary antibody (1:10000 dilution) conjugated with horseradish peroxidase (Cell Signaling Technology, USA) for 1 h at room temperature. Membranes were developed with chemiluminescent ECL reagents (Millipore, USA). The relative expression of target protein to the control β-actin were determined by densitometric analysis using ImageJ software (National Institutes of Health, USA).
Co-immunoprecipitation
The HEK293T cells were overexpressed with each 10 μg plasmid of Spike-Flag and ACE2-HA, and treated with 25 μM imatinib, 100 μM harpagoside, or 100 μM methazolamide for 48 h. Cells were lysed in 20 mM Tris-HCl PH8.0, 0.2 mM EDTA PH8.0, 50 mM MgCl2, 10% Glycerol, 300 mM KCl, 0.1% NP40 and protease inhibitors PMSF. The lysates were centrifuged at 14,000 g for 30 min. An aliquot of supernatants was used as input, and the rest of the supernatants were incubated with the respective antibodies (anti-HA and anti-Flag, Cell Signaling Technology, USA) or control IgG and mixed with Dynabeads Protein G beads (Invitrogen, USA) overnight at 4°C. Precipitated proteins were eluted with the corresponding epitope 3×Flag peptides or with elution buffer (50 mM glycine, pH 2.8) and analyzed by western blotting.
Enzyme-linked Immunosorbent Assay (ELISA)
For the quantitative determination of plasma Ang II and Ang-(1-7) in COVID-19 patients, a serial of fasting blood samples from 30 patients with laboratory confirmed SARS-CoV-2 infection were collected. The blood samples of infection phase were collected on the following day of admission with overnight fasting in SARS-CoV-2 positive patients, while the blood samples of recovery phase were collected within 3 days before patients discharge after overnight fasting. For comparison, the blood samples from 10 healthy volunteers were collected. All blood samples were inactivated under the temperature of 56°C for 30 min. This experiment was approved by the Institutional Review Board of Guangzhou Eighth People's Hospital. For the quantitative determination of Ang II and Ang-(1-7) in the supernatant from infected HUVECs, HUVECs were infected with SARS-CoV-2 (MOI = 0.005) for 48 h, then the medium was washed away and 100 nM Ang II in 1 ml HEPES solution was applied for the cells (Kopf and Campbell, 2013), after 1 h treatment in the incubator at 37°C, the supernatant was collected and inactivated under the temperature of 56°C for 30 min, then subjected to ELISA assay. For the quantitative determination of plasma Ang II, Ang-(1-7), TNF-α, IL-6, insulin and KIM-1 concentrations in mice, plasma samples were diluted and added to ELISA assay plates (CUSABIO Biotech, China) pre-coated with specific antibodies. Samples were bound by the immobilized antibodies, reacted with avidin-conjugated Horseradish Peroxidase (HRP), and developed with 3,3’,5,5’-tetramethylbenzidine (TMB) substrate. Reactions were terminated, and the absorbance was measured at 450 nm using a microplate reader (Tecan, Swiss) within 5 min.
ACE2 Enzymatic Activity Assay
Activity of ACE2 in cell lines, cell lysates and tissues were measured by ACE2 activity assay kit (Fluorometric) (Abcam, UK), which utilizes the ability of an active ACE2 to cleave a synthetic MCA based peptide substrate to release a free fluorophore. The measurements were performed in black plates (Corning, USA) with a total volume of 100 μl lysates and ACE2 substrate. HUVECs were treated with 25 μM imatinib, 100 μM harpagoside and 100 μM methazolamide respectively for 16 h, then lysed and followed by measurement according to the kit. Similarly, tissues from the mice treated with imatinib or methazolamide were applied for ACE2 enzymatic activity measurement. HUVEC lysates and imatinib (1.5×10-7 M, 1.5×10-6 M), harpagoside (0.5×10-7 M, 1.5×10-6 M), or methazolamide (1.5×10-7 M, 1.5×10-6 M) were incubated respectively for 30 min at room temperature to assess the time dependency of the fluorescence signal. Fluorescence kinetics was measured by use of a multiplate reader (SpectraMax 5, USA) at an excitation wavelength of 320 nm and an emission wavelength of 420 nm. ACE2 inhibitor DX600 was used as a negative control.
Surface Plasmon Resonance (SPR)
The SPR assays were performed to analyze the interactions between the compounds and ACE2 protein (Sino Biological, China) by using a Biacore T100 machine with Sensor Chip CM7 (GE Healthcare, USA) at 25°C. ACE2 was immobilized onto CM7 chips, sensorgrams were recorded by injecting various concentrations of compounds. The binding kinetics (Kd) was analyzed with the software BIA evaluation Version 4.1.
Bioinformatics
An initial list of target-disease associated genes of diabetes, diabetic nephropathy, hypertension and atherosclerosis were identified by querying data available in the DisGeNET 7.0 database (Piñero et al., 2020) and Open Targets Platform (Carvalho-Silva et al., 2019). DisGeNET (http://www.disgenet.org) and Open Targets Platform (https://www.opentargets.org/) were accessed in September 2020. For DisGeNET, all Unified Medical Language System Concept Unique Identifiers (CUIs) that relate to these four diseases were selected from DisGeNET, and further data frames of genes with evidence for a relationship with these four diseases were created. CUIs queried in DisGeNET were as follows: diabetes Mellitus [C0011849], hypertensive disease [C0020538], diabetic nephropathy [C0011881] and atherosclerosis [C0004153]. For Open Targets Platform, the term “diabetes mellitus”, “hypertension”, “diabetic nephropathy” and “atherosclerosis” were searched, and further data frames of genes with evidence for a relationship with these four diseases were generated from six perspectives of disease genetics, transcriptomics, approved drugs and clinical candidates, animal models, biochemical pathways and text mining from the medical literature. Next, the top 400 target-disease associated genes in four diseases in each platform were overlapped to search for mutual genes of these four diseases.
In all, 20 genes were found in this process and further expression levels were searched from the transcriptome data in COVID-19 literature. Namely, GSE151803 (Yang et al., 2020), GSE147507 (Blanco-Melo et al., 2020), GSE155249 (Grant et al., 2021) and GSE153277 (Huang et al., 2020) were searched and the expression levels of these 20 genes were shown in alphabetical order.
CMAP
The CMAP (https://clue.io/), an online platform which conducts pattern-matching algorithms to compare input gene signatures with gene signatures in diverse contexts of over 450,000 chemical perturbagens (Chen et al., 2021; Subramanian et al., 2017), was utilized to identify compounds that create gene expression patterns opposite to SARS-CoV-2 infection. Six articles reporting transcriptomics of SARS-CoV-2-infected organs or cells (Appelberg et al., 2020; Liao et al., 2020; Liu et al., 2020; Shen et al., 2020; Wang and Xu, 2020; Xiong et al., 2020) were searched out in May 18, 2020, top and bottom 50–150 featured genes were selected from each article and submitted online for CMAP query, and then one ranked list of compounds was generated for each article. Compounds with negative “connectivity” and “enrichment” scores, which indicate a potential capability of inhibiting SARS-CoV-2 virus infection or secondary injuries, were subsequently filtered to a new list of compounds for each article. A total of 176 compounds that presented in more than one list were included for further study.
A total of 40 potential downstream featured genes correlated with ACE2 transcriptional manipulation were searched out from literature (Cao et al., 2016, 2019a, 2019b; Kawabe et al., 2019; Patel et al., 2016; Takeshita et al., 2018) and submitted for CMAP query, a ranked list of compounds creating gene expression patterns similar to ACE2 transcriptional activation was generated. In this list, a total of 83 compounds with positive “connectivity” and “enrichment” scores were included for further investigation.
Compounds Scoring System of in vitro Experiments for Validation of ACE2 Activators
The selected 15 compounds (Figure 3A) were further validated with in vitro experiments. In brief, for the first step as shown in Figure 3D, the ranking value of each compound was calculated with the sum of each gene expression level against control (log10) after treatment with low-, medium- and high-concentration, respectively. The top 8 compounds from the first step were selected. For the second step shown in Figure 3E, the ranking value of each compound was calculated with the sum of each gene expression level after treatments of each compound minus the expression level after treatment of inflammatory factors. The top 6 compounds from the second step were selected. For the final step shown in Figure 3F, compounds with significant alteration of each gene after ACE2 knockdown comparing with control siRNA were considered as ACE2-dependent candidates, and imatinib, harpagoside and methazolamide were selected.
Structure-based High Throughput Virtual Screening
The crystal structure of the apo form of human ACE2 in the open conformation (PDB: 1R42) was used for molecular docking. The Glide in SP mode of Schrodinger suite 2019 version was used for in silico screening of 76,639 compounds, of which 10,275 compounds were obtained from the DurgBank database (https://www.drugbank.com), and the remaining 66,364 compounds were obtained from the InterBioScreen database (https://www.ibscreen.com). We targeted a specific structural pocket in the hinge-bending region of ACE2 as shown in Figure 3B. Related research has shown that this region is implicated in conformational shuttling between the two status of the enzyme, as binding in this region would potentially stabilize ACE2 active conformation from shuffling to inactive conformation (Kulemina and Ostrov, 2011). We showed the diagram of conformational changes of the two types in Figure S3A. The grid sites were created using Glide receptor grid generator with docking length of 20 Å. Grids centers were determined from active resides on target protein as shown in Figure 3B.
Detailed Modeling of Binding Structure
Imatinib and methazolamide were selected for further molecular dynamics simulation for detailed docking binding structure. The GROMACS package (Abraham et al., 2015) was used to carry out molecular dynamics simulation. The general AMBER force field (GAFF) (Wang et al., 2004) was used for imatinib and methazolamide and the AMBER force field ff99SB (Hornak et al., 2006) for the protein receptor. Parameters for imatinib and methazolamide were assigned using antechamber, and the charges were calculated using Gaussian09 at the HF/6–13G∗ level and the RESP method (Bayly et al., 1993) was used to derive atomic charges. The SPC/E water model (Berendsen et al., 1987) was used to solve and sodium cations were used to neutralize the binding systems. A 1 μs molecular dynamics simulation was performed and the last 100 ns trajectories were used for analysis. The binding free energy was calculated using MM-GBSA approach (Baker et al., 2001; Rashmi Kumari et al., 2014).
Meta-analysis
The guideline for meta-analyses and systematic reviews (preferred reporting items for systematic reviews and meta-analysis) was followed (Moher et al., 2009). Search strategy: a systematic article search was performed using the PubMed, Embase, Cochrane, bioRxiv and medRxiv databases until March 1, 2021. The terms “SARS-CoV-2”, “angiotensin-converting enzyme inhibitors” and “angiotensin receptor blockers” and their synonyms were used to identify all potentially relevant studies. No filters or restrictions were applied. References from these articles were further screened for additional relevant studies. The inclusion criteria were: (1) patients with confirmed or clinically suspected COVID-19 taking ACE inhibitor (ACEi) or angiotensin receptor blocker (ARB), and (2) a primary end point reported. The exclusion criteria were: (1) publication not in English, (2) full-text version not available, (3) hypertension was not clarified, and (4) number of patients less than four. Duplicates were removed manually. All citations were independently screened by two of co-authors (D.F. and C.L.) using the inclusion and exclusion criteria (Table S5). When discrepancies occurred, a decision was reached through further discussion. Data extraction: patients were divided into ACEi/ARB group and non-ACEi/ARB group by the method of treatment. The patients’ demographics, basic information, laboratory tests (i.e., blood glucose, triglyceride, total cholesterol), blood pressure, and follow-up data were extracted. The primary end point was death, and the secondary end point was disease severity.
Quantification and statistical analysis
All non-RNA-seq statistical data analysis was performed using Graphpad Prism 8 (GraphPad Software, USA). ImageJ was used for quantification of western blots and histology. Two-tailed unpaired Student’s t test was used to compare two groups of data, while one-way ANOVA was used to compare multiple groups of data. All data were shown as mean ± SEM. All results shown were representative of at least three independent experiments. For RNA-seq, raw reads were aligned to the human genome (hg38) using HISAT2 (v2.0.5). Genes were annotated and counted by featureCounts (v1.5.0) with GENCODE gene annotation (v37). Differentially expressed genes were called by using DESeq2 package (v1.20.0) with the following criteria: p value < 0.05 and |Log2 Fold Change| > 0.5 unless specifically defined. Metascape (Zhou et al., 2019) was used for enrichment of differentially expressed genes in Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al., 2019), Gene Ontology (GO) (Carbon et al., 2009) and DisGeNET pathways (Piñero et al., 2020). For meta-analysis, the Fisher exact test was used for categorical variables in accordance with the principles outlined in the Cochrane Handbook (Higgins et al., 2021). The outcomes of ACEi/ARB were analyzed using random effects models with Review Manager 5.4 (Cochrane Collaboration, Oxford, UK). A p value less than 0.05 is considered as statistically significant.
Acknowledgments
We thank Prof. Hui Zhang’s lab (Institute of Human Virology, Key Laboratory of Tropical Disease Control of Ministry of Education, Zhongshan School of Medicine, Sun Yat-sen University) for providing the SARS-CoV-2 virus strain. This study was supported by the Guangdong Science and Technology Department (2020B1212060018, 2020B1212030004) and the special COVID-19 grant from the Project of Educational Commission of Guangdong Province of China (no. 2020KZDXZ1215). The funders had no role in study design, data collection and analysis, or preparation of the manuscript.
Author contributions
Z.L., M.P., C.L., A.H., Yixin Zhang., J.P., H.C., J. Li, D.F., K.H., and S.C. performed the experiments; P.C., Y. Lu, and M.P. performed compound screening; J. Liu, Y. Li, W.L., W.Z., Yang Zhang, F.L., Z.Z., Z.G., and H.L. provided technical support; D.G., X.H., Y. Zhu, L.L., B.H., W.C., L.G., and L.Y. provided materials; D.G., B.H., W.C., L.G., and L.Y. discussed the project; and Z.L., M.P., P.C., C.L., K.D., L.Y., and S.C. wrote and edited the manuscript. S.C. conceived of the project idea.
Declaration of interests
The authors declare no competing interests.
Published: February 11, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cmet.2022.01.008.
Supplemental information
Data and code availability
All raw sequencing data were uploaded to the GSA (Genome Sequence Archive) under the accession number GSA: HRA001795 in the National Genomics Data Center (NGDC, https://bigd.big.ac.cn/).
References
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- Ahmad I., Pawara R., Surana S., Patel H. The repurposed ACE2 inhibitors: SARS-CoV-2 entry blockers of Covid-19. Top. Curr. Chem. (Cham) 2021;379:40. doi: 10.1007/s41061-021-00353-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandre J., Cracowski J.L., Richard V., Bouhanick B., ‘Drugs, COVID-19’ working group of the French Society of Pharmacology, Therapeutics Renin-angiotensin-aldosterone system and COVID-19 infection. Ann. Endocrinol. (Paris) 2020;81:63–67. doi: 10.1016/j.ando.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman J., Duijvelaar E., Botros L., Kianzad A., Schippers J.R., Smeele P.J., Azhang S., Bartelink I.H., Bayoumy A.A., Bet P.M., et al. Imatinib in patients with severe COVID-19: a randomised, double-blind, placebo-controlled, clinical trial. Lancet Respir. Med. 2021;9:957–968. doi: 10.1016/S2213-2600(21)00237-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelberg S., Gupta S., Svensson Akusjärvi S., Ambikan A.T., Mikaeloff F., Saccon E., Végvári Á., Benfeitas R., Sperk M., Ståhlberg M., et al. Dysregulation in Akt/mTOR/HIF-1 signaling identified by proteo-transcriptomics of SARS-CoV-2 infected cells. Emerg. Microbes Infect. 2020;9:1748–1760. doi: 10.1080/22221751.2020.1799723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly C.I., Cieplak P., Cornell W., Kollman P.A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- Beckman J.A., Creager M.A., Libby P. Diabetes and atherosclerosis: epidemiology, pathophysiology, and management. JAMA. 2002;287:2570–2581. doi: 10.1001/jama.287.19.2570. [DOI] [PubMed] [Google Scholar]
- Berendsen H.J.C., Grigera J.R., Straatsma T.P. The missing term in effective pair potentials. J. Phys. Chem. 1987;91:6269–6271. [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Yang F., Shi T., Yuan M., Xin Z., Xie R., Li S., Li H., Yang J.K. Angiotensin-converting enzyme 2/angiotensin-(1–7)/Mas axis activates Akt signaling to ameliorate hepatic steatosis. Sci. Rep. 2016;6:21592. doi: 10.1038/srep21592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Lu X.M., Tuo X., Liu J.Y., Zhang Y.C., Song L.N., Cheng Z.Q., Yang J.K., Xin Z. Angiotensin-converting enzyme 2 regulates endoplasmic reticulum stress and mitochondrial function to preserve skeletal muscle lipid metabolism. Lipids Health Dis. 2019;18:207. doi: 10.1186/s12944-019-1145-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Song L.N., Zhang Y.C., Li Q., Shi T.T., Yang F.Y., Yuan M.X., Xin Z., Yang J.K. Angiotensin-converting enzyme 2 inhibits endoplasmic reticulum stress-associated pathway to preserve nonalcoholic fatty liver disease. Diabetes Metab. Res. Rev. 2019;35:e3123. doi: 10.1002/dmrr.3123. [DOI] [PubMed] [Google Scholar]
- Carbon S., Ireland A., Mungall C.J., Shu S., Marshall B., Lewis S., AmiGO Hub. Web Presence Working Group AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Silva D., Pierleoni A., Pignatelli M., Ong C., Fumis L., Karamanis N., Carmona M., Faulconbridge A., Hercules A., McAuley E., et al. Open targets platform: new developments and updates two years on. Nucleic Acids Res. 2019;47:D1056–D1065. doi: 10.1093/nar/gky1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Liu X., Peng C., Tan C., Sun H., Liu H., Zhang Y., Wu P., Cui C., Liu C., et al. The phytochemical hyperforin triggers thermogenesis in adipose tissue via a Dlat-AMPK signaling axis to curb obesity. Cell Metab. 2021;33:565–580.e7. doi: 10.1016/j.cmet.2021.02.007. [DOI] [PubMed] [Google Scholar]
- Díaz L.A., Idalsoaga F., Cannistra M., Candia R., Cabrera D., Barrera F., Soza A., Graham R., Riquelme A., Arrese M., et al. High prevalence of hepatic steatosis and vascular thrombosis in COVID-19: a systematic review and meta-analysis of autopsy data. World J. Gastroenterol. 2020;26:7693–7706. doi: 10.3748/wjg.v26.i48.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B., Yu Q.T., Dai H.Y., Gao Y.Y., Zhou Z.L., Zhang L., Jiang H., Gao F., Li S.Y., Zhang Y.H., et al. Angiotensin-converting enzyme-2 overexpression improves left ventricular remodeling and function in a rat model of diabetic cardiomyopathy. J. Am. Coll. Cardiol. 2012;59:739–747. doi: 10.1016/j.jacc.2011.09.071. [DOI] [PubMed] [Google Scholar]
- Druker B.J., Talpaz M., Resta D.J., Peng B., Buchdunger E., Ford J.M., Lydon N.B., Kantarjian H., Capdeville R., Ohno-Jones S., et al. Efficacy and safety of a specific inhibitor of the bcr-abl tyrosine kinase in chronic myeloid leukemia. N. Engl. J. Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- Fossati S., Giannoni P., Solesio M.E., Cocklin S.L., Cabrera E., Ghiso J., Rostagno A. The carbonic anhydrase inhibitor methazolamide prevents amyloid beta-induced mitochondrial dysfunction and caspase activation protecting neuronal and glial cells in vitro and in the mouse brain. Neurobiol. Dis. 2016;86:29–40. doi: 10.1016/j.nbd.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goru S.K., Kadakol A., Malek V., Pandey A., Sharma N., Gaikwad A.B. Diminazene aceturate prevents nephropathy by increasing glomerular ACE2 and AT receptor expression in a rat model of type1 diabetes. Br. J. Pharmacol. 2017;174:3118–3130. doi: 10.1111/bph.13946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant R.A., Morales-Nebreda L., Markov N.S., Swaminathan S., Querrey M., Guzman E.R., Abbott D.A., Donnelly H.K., Donayre A., Goldberg I.A., et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature. 2021;590:635–641. doi: 10.1038/s41586-020-03148-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Jiang S., Li Z., Chen S. Metabolic syndrome “interacts” with COVID-19. Bio Integr. 2021;1:168–177. [Google Scholar]
- Gupta A., Madhavan M.V., Sehgal K., Nair N., Mahajan S., Sehrawat T.S., Bikdeli B., Ahluwalia N., Ausiello J.C., Wan E.Y., et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PubMed] [Google Scholar]
- Haber P.K., Ye M., Wysocki J., Maier C., Haque S.K., Batlle D. Angiotensin-converting enzyme 2-independent action of presumed angiotensin-converting enzyme 2 activators: studies in vivo, ex vivo, and in vitro. Hypertension. 2014;63:774–782. doi: 10.1161/HYPERTENSIONAHA.113.02856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Duan X., Yang L., Nilsson-Payant B.E., Wang P., Duan F., Tang X., Yaron T.M., Zhang T., Uhl S., et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589:270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Liu C., Peng J., Li Z., Li F., Wang J., Hu A., Peng M., Huang K., Fan D., et al. COVID-19 induces new-onset insulin resistance and lipid metabolic dysregulation via regulation of secreted metabolic factors. Signal Transduct. Target. Ther. 2021;6:427. doi: 10.1038/s41392-021-00822-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández Prada J.A., Ferreira A.J., Katovich M.J., Shenoy V., Qi Y., Santos R.A.S., Castellano R.K., Lampkins A.J., Gubala V., Ostrov D.A., et al. Structure-based identification of small-molecule angiotensin-converting enzyme 2 activators as novel antihypertensive agents. Hypertension. 2008;51:1312–1317. doi: 10.1161/HYPERTENSIONAHA.107.108944. [DOI] [PubMed] [Google Scholar]
- Higgins J.P.T., Thomas J., Chandler J., Cumpston M., Li T., Page M.J., Welch V. Cochrane handbook for systematic reviews of interventions version 6.2 (updated February 2021) (Cochrane) 2021. https://training.cochrane.org/handbook
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Sexton D.J., Skogerson K., Devlin M., Smith R., Sanyal I., Parry T., Kent R., Enright J., Wu Q.L., et al. Novel peptide inhibitors of angiotensin-converting enzyme 2. J. Biol. Chem. 2003;278:15532–15540. doi: 10.1074/jbc.M212934200. [DOI] [PubMed] [Google Scholar]
- Huang J., Hume A.J., Abo K.M., Werder R.B., Villacorta-Martin C., Alysandratos K.D., Beermann M.L., Simone-Roach C., Lindstrom-Vautrin J., Olejnik J., et al. SARS-CoV-2 infection of pluripotent stem cell-derived human lung alveolar type 2 cells elicits a rapid epithelial-intrinsic inflammatory response. Cell Stem Cell. 2020;27:962–973.e7. doi: 10.1016/j.stem.2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Liu C., Peng M., Su Q., Liu R., Guo Z., Chen S., Li Z., Chang G. Glycoursodeoxycholic acid ameliorates atherosclerosis and alters gut microbiota in apolipoprotein E-deficient mice. J. Am. Heart Assoc. 2021;10 doi: 10.1161/JAHA.120.019820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov V., Goc A., Ivanova S., Niedzwiecki A., Rath M. Inhibition of ACE2 expression by ascorbic acid alone and its combinations with other natural compounds. Infect. Dis. (Auckl) 2021;14 doi: 10.1177/1178633721994605. 1178633721994605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Furumichi M., Morishima K., Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47:D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe Y., Mori J., Morimoto H., Yamaguchi M., Miyagaki S., Ota T., Tsuma Y., Fukuhara S., Nakajima H., Oudit G.Y., et al. ACE2 exerts anti-obesity effect via stimulating brown adipose tissue and induction of browning in white adipose tissue. Am. J. Physiol. Endocrinol. Metab. 2019;317:E1140–E1149. doi: 10.1152/ajpendo.00311.2019. [DOI] [PubMed] [Google Scholar]
- Konstantopoulos N., Molero J.C., McGee S.L., Spolding B., Connor T., de Vries M., Wanyonyi S., Fahey R., Morrison S., Swinton C., et al. Methazolamide is a new hepatic insulin sensitizer that lowers blood glucose in vivo. Diabetes. 2012;61:2146–2154. doi: 10.2337/db11-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf P.G., Campbell W.B. Endothelial metabolism of angiotensin II to angiotensin III, not angiotensin (1–7), augments the vasorelaxation response in adrenal cortical arteries. Endocrinology. 2013;154:4768–4776. doi: 10.1210/en.2013-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulemina L.V., Ostrov D.A. Prediction of off-target effects on angiotensin-converting enzyme 2. J. Biomol. Screen. 2011;16:878–885. doi: 10.1177/1087057111413919. [DOI] [PubMed] [Google Scholar]
- Kumari R., Kumar R., Open Source Drug Discovery Consortium, Lynn A. g_mmpbsa—a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- Lebek S., Tafelmeier M., Messmann R., Provaznik Z., Schmid C., Maier L.S., Birner C., Arzt M., Wagner S. Angiotensin-converting enzyme inhibitor/angiotensin II receptor blocker treatment and haemodynamic factors are associated with increased cardiac mRNA expression of angiotensin-converting enzyme 2 in patients with cardiovascular disease. Eur. J. Heart Fail. 2020;22:2248–2257. doi: 10.1002/ejhf.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Liu W., Chen X., Wang Y., Chen Y., Chen S., Gong W., Chen T., Sun L., Zheng C., Yin B., et al. Micheliolide ameliorates diabetic kidney disease by inhibiting Mtdh-mediated renal inflammation in type 2 diabetic db/db mice. Pharmacol. Res. 2019;150:104506. doi: 10.1016/j.phrs.2019.104506. [DOI] [PubMed] [Google Scholar]
- Liu X., Chen Y., Tang W., Zhang L., Chen W., Yan Z., Yuan P., Yang M., Kong S., Yan L., et al. Single-cell transcriptome analysis of the novel coronavirus (SARS-CoV-2) associated gene ACE2 expression in normal and non-obstructive azoospermia (NOA) human male testes. Sci. China Life Sci. 2020;63:1006–1015. doi: 10.1007/s11427-020-1705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute-Bello G., Downey G., Moore B.B., Groshong S.D., Matthay M.A., Slutsky A.S., Kuebler W.M., Acute Lung Injury in Animals Study Group An official American Thoracic Society workshop report: features and measurements of experimental acute lung injury in animals. Am. J. Respir. Cell Mol. Biol. 2011;44:725–738. doi: 10.1165/rcmb.2009-0210ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta N., Kalra A., Nowacki A.S., Anjewierden S., Han Z., Bhat P., Carmona-Rubio A.E., Jacob M., Procop G.W., Harrington S., et al. Association of use of angiotensin-converting enzyme inhibitors and angiotensin II receptor blockers with testing positive for coronavirus disease 2019 (COVID-19) JAMA Cardiol. 2020;5:1020–1026. doi: 10.1001/jamacardio.2020.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D.G., PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Ortega A., Bernal-Bello D., Llarena-Barroso C., Frutos-Pérez B., Duarte-Millán M.Á., García de Viedma-García V., Farfán-Sedano A.I., Canalejo-Castrillero E., Ruiz-Giardín J.M., Ruiz-Ruiz J., et al. Imatinib for COVID-19: a case report. Clin. Immunol. 2020;218:108518. doi: 10.1016/j.clim.2020.108518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajah R., Milagres R., Dilauro M., Gutsol A., Xiao F., Zimpelmann J., Kennedy C., Wysocki J., Batlle D., Burns K.D. Podocyte-specific overexpression of human angiotensin-converting enzyme 2 attenuates diabetic nephropathy in mice. Kidney Int. 2012;82:292–303. doi: 10.1038/ki.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie X., Qian L., Sun R., Huang B., Dong X., Xiao Q., Zhang Q., Lu T., Yue L., Chen S., et al. Multi-organ proteomic landscape of COVID-19 autopsies. Cell. 2021;184:775–791.e14. doi: 10.1016/j.cell.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes Duarte-Neto A., de Almeida Monteiro R.A., da Silva L.F.F., Malheiros D., de Oliveira E.P., Theodoro Filho J., Pinho J.R.R., Soares Gomes-Gouvêa M., Salles A.P.M., de Oliveira I.R.S., et al. Pulmonary and systemic involvement of COVID-19 assessed by ultrasound-guided minimally invasive autopsy. Histopathology. 2020;77:186–197. doi: 10.1111/his.14160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel V.B., Mori J., McLean B.A., Basu R., Das S.K., Ramprasath T., Parajuli N., Penninger J.M., Grant M.B., Lopaschuk G.D., et al. ACE2 deficiency worsens epicardial adipose tissue inflammation and cardiac dysfunction in response to diet-induced obesity. Diabetes. 2016;65:85–95. doi: 10.2337/db15-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichavaram P., Shawky N.M., Hartney T.J., Jun J.Y., Segar L. Imatinib improves insulin resistance and inhibits injury-induced neointimal hyperplasia in high fat diet-fed mice. Eur. J. Pharmacol. 2021;890:173666. doi: 10.1016/j.ejphar.2020.173666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñero J., Ramírez-Anguita J.M., Saüch-Pitarch J., Ronzano F., Centeno E., Sanz F., Furlong L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48:D845–D855. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Casals M., Brito-Zerón P., Mariette X. Systemic and organ-specific immune-related manifestations of COVID-19. Nat. Rev. Rheumatol. 2021;17:315–332. doi: 10.1038/s41584-021-00608-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds H.R., Adhikari S., Pulgarin C., Troxel A.B., Iturrate E., Johnson S.B., Hausvater A., Newman J.D., Berger J.S., Bangalore S., et al. Renin-angiotensin-aldosterone system inhibitors and risk of Covid-19. N. Engl. J. Med. 2020;382:2441–2448. doi: 10.1056/NEJMoa2008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos R.A.S., Sampaio W.O., Alzamora A.C., Motta-Santos D., Alenina N., Bader M., Campagnole-Santos M.J. The ACE2/angiotensin-(1–7)/MAS axis of the renin-angiotensin system: focus on angiotensin-(1–7) Physiol. Rev. 2018;98:505–553. doi: 10.1152/physrev.00023.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., Quan S., Zhang F., Sun R., Qian L., et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182:59–72.e15. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy V., Gjymishka A., Jarajapu Y.P., Qi Y., Afzal A., Rigatto K., Ferreira A.J., Fraga-Silva R.A., Kearns P., Douglas J.Y., et al. Diminazene attenuates pulmonary hypertension and improves angiogenic progenitor cell functions in experimental models. Am. J. Respir. Crit. Care Med. 2013;187:648–657. doi: 10.1164/rccm.201205-0880OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.K., Singh R. Hyperglycemia without diabetes and new-onset diabetes are both associated with poorer outcomes in COVID-19. Diabetes Res. Clin. Pract. 2020;167:108382. doi: 10.1016/j.diabres.2020.108382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers J.R., Epstein M. Diabetes mellitus and associated hypertension, vascular disease, and nephropathy. An update. Hypertension. 1995;26:869–879. doi: 10.1161/01.hyp.26.6.869. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Narayan R., Corsello S.M., Peck D.D., Natoli T.E., Lu X., Gould J., Davis J.F., Tubelli A.A., Asiedu J.K., et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171:1437–1452.e17. doi: 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Zhuang Z., Zheng J., Li K., Wong R.L., Liu D., Huang J., He J., Zhu A., Zhao J., et al. Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell. 2020;182:734–743.e5. doi: 10.1016/j.cell.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suski M., Olszanecki R., Stachowicz A., Madej J., Bujak-Giżycka B., Okoń K., Korbut R. The influence of angiotensin-(1–7) Mas receptor agonist (AVE 0991) on mitochondrial proteome in kidneys of apoE knockout mice. Biochim. Biophys. Acta. 2013;1834:2463–2469. doi: 10.1016/j.bbapap.2013.08.008. [DOI] [PubMed] [Google Scholar]
- Takeshita H., Yamamoto K., Nozato S., Takeda M., Fukada S.I., Inagaki T., Tsuchimochi H., Shirai M., Nozato Y., Fujimoto T., et al. Angiotensin-converting enzyme 2 deficiency accelerates and angiotensin 1–7 restores age-related muscle weakness in mice. J. Cachexia Sarcopenia Muscle. 2018;9:975–986. doi: 10.1002/jcsm.12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler P., Staker B., Prasad S.G., Menon S., Tang J., Parsons T., Ryan D., Fisher M., Williams D., Dales N.A., et al. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004;279:17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94 doi: 10.1128/JVI.00127-20. e00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Xu X. scRNA-seq profiling of human testes reveals the presence of the ACE2 receptor, a target for SARS-CoV-2 infection in spermatogonia, Leydig and Sertoli cells. Cells. 2020;9:920. doi: 10.3390/cells9040920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- Wei X., Zeng W., Su J., Wan H., Yu X., Cao X., Tan W., Wang H. Hypolipidemia is associated with the severity of COVID-19. J. Clin. Lipidol. 2020;14:297–304. doi: 10.1016/j.jacl.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z., et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan X., Gao F., Ma X., Huang C., Wang Y., Deng H., Wang S., Li W., Yuan L. Activation of ACE2/angiotensin (1–7) attenuates pancreatic β cell dedifferentiation in a high-fat-diet mouse model. Metabolism. 2018;81:83–96. doi: 10.1016/j.metabol.2017.12.003. [DOI] [PubMed] [Google Scholar]
- Yang L., Han Y., Nilsson-Payant B.E., Gupta V., Wang P., Duan X., Tang X., Zhu J., Zhao Z., Jaffré F., et al. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27:125–136.e7. doi: 10.1016/j.stem.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequencing data were uploaded to the GSA (Genome Sequence Archive) under the accession number GSA: HRA001795 in the National Genomics Data Center (NGDC, https://bigd.big.ac.cn/).