Abstract
Cancer is a leading cause of morbidity and mortality worldwide. Over the past decades, the concept of precision cancer medicine has emerged as a novel approach in the field of oncology that aims to tailor the most effective treatment options to each individual cancer patient based on the genetic profile of the tumor of each individual patient. Recently, tissue biopsy has become an essential part of cancer care and is widely used to characterize the tumor. However, tissue biopsy techniques face different challenges due to their invasiveness, cost, time, and adversity in potential sampling due to tissue heterogeneity. To overcome these issues, a non-invasive approach has developed, which is known as liquid biopsy. It is a simple, fast, and worthwhile technique based on the analysis of circulating tumor DNA (which is a fraction of cfDNA), circulating tumor cells (CTCs), and other tumor-derived material in blood plasma. This review provides an overview of the concept of liquid biopsy and briefly discusses the role of ctDNA and CTC analysis as tools for early diagnosis and prognosis of cancer. In this review, we also speculate on the advantages of liquid biopsy as opposed to tissue biopsy and postulate that liquid biopsy may be a comprehensive approach to overcome the current limitations associated with costly, invasive, and time-consuming tissue biopsy.
Keywords: Cancer, liquid biopsy, tissue biopsy, circulating tumor DNA (ctDNA), circulating tumor cells (CTCs)
Introduction
Cancer is a term for a distinctive disease characterized by uncontrolled abnormal cell proliferation, which can invade nearby tissues and metastasize to distant organs through the blood and lymph systems.1,2 Metastatic spread of cancer to distant sites remains the main cause of death for cancer patients in ~90% of cases. 3 For cancer treatment, conventional cancer therapies practiced in the clinic include systemic chemotherapy (including hormonal therapy and molecular targeted drugs), surgery and radiotherapy, 4 and the choice of therapy depends on the type and stage of the tumor, as well as the general state and age of the patient.4,5 These conventional therapies are generally effective against early-stage localized tumors, whereas their effectiveness is not the same against a later stage of cancer and there are chances of frequent relapse.4,6
Drug resistance appears to be a serious problem in treatment of cancer; currently, 90% of chemotherapy failures occur during the invasion and metastasis of cancers related to drug resistance. In chemotherapy, by following the administration of a certain drug, a large number of patient tumor cells become resistant to the drug. 7
Although many types of cancer are initially susceptible to chemotherapy, over time they can develop resistance to traditional therapies through different mechanisms, such as drug inactivation, epigenetics, drug target alteration, drug efflux, DNA damage repair, cell death inhibition, and epithelial-mesenchymal transition (EMT). These mechanisms promote direct or indirect drug resistance in human cancer cells, acting independently or in combination and through various signal transduction pathways. 8
One study showed that mutations in self-controlling negative feedback in signal transmission can produce resistance for drugs targeting such signaling pathways. Furthermore, tumor cells may also lack antigenic mutations, or some key molecules of the immune system are perhaps absent, such as HLA, and these cells can become unsusceptible to immunotherapy drugs. 9
In addition, various agents used in conventional therapies are harmful both to normal cells and cancer cells, which can lead to prominent side effects. These conventional cytotoxic treatments also lack specificity for tumor cells, making them ineffective in providing lifelong protection against cancer.4,6
Therefore, there was an urgent need for new therapeutic strategies that allow more effective monitoring of cancer status, thereby improving the efficacy of treatment by specifically eliminating cancer cells and sparing normal cells from toxicity to develop long-lasting protection. 10
As a result of recent advancements in medical and pharmacogenetic research, the field of oncology has introduced a profound innovative approach that overcomes the old one-size-fits-all strategy; this new paradigm is known as precision medicine, which is designed to customize therapies for patients in relation to the personalized molecular profile of the tumor.11,12 So far, the path of survival of cancer patients is tissue dependent, known as tissue biopsy, which has several advantages over conventional cancer treatments.
Tissue Biopsy
Tissue biopsy involves the examination of tumor tissue by obtaining tissue through a fine needle or open surgery, 13 these tissue samples are genetically analyzed to define the type of tumor and then the best personalized treatment regimen is designed.
Drawbacks of tissue biopsy
Despite the informative nature of tissue biopsy, it is painful, invasive, costly, time consuming, and, most importantly, sometimes inappropriate to capture tumor heterogeneity. 14
Apart from other issues related to tissue biopsy, intra-tumoral heterogeneity is always a potential issue in the precision of this procedure, which induces significant challenges in selecting an effective treatment strategy based only on tissue biopsy. 15
Tissue biopsies typically obtain a sample of only a part of the tumor and therefore encompass only a fraction of tumor heterogeneity, as serial tumor sampling is not clinically practical with invasive tissue biopsy; therefore, complete information on the levels of genetic and epigenetic variability of a patient’s cancer is not achieved.16,17
Invasive tissue biopsy has also been studied to be impossible after surgical resection and if there is no detectable metastatic mass. 18
Moreover, unsatisfactory detection of early-stage tumor or residual lesions, also causing limitations of this therapy. 19
Therefore, a worthwhile and noninvasive approach has been developed that could proficiently capture the genomic content of tumors in fluids and detect tumor heterogeneity in the course of cancer treatments; this approach is known as liquid biopsy that provides noninvasive multifunctional biomarkers for early diagnosis, accurate prognosis, selection of therapeutic targets, monitoring of metastases, as well as monitoring response and resistance to treatment. 20
Liquid Biopsy
The term liquid biopsy is basically introduced for the analysis of CTCs 21 but is now also associated with ctDNA and other biomarkers such as miRNAs. 22
In addition to blood, several body fluids such as blood, plasma, serum, saliva, urine, and gastric juice could be used to perform liquid biopsy for non-invasive clinical evaluation.23-25
The clinical use of liquid biopsy has increased significantly since 2004, when Guardant Health released the first commercially available liquid biopsy test. 26 Liquid biopsy (restricted to ctDNA analysis) was officially adopted as a diagnostic tool in the main reference center laboratories during the course of 2016. 17
Currently, there are several commercially available assays that are FDA approved; some are considered proficient in treatment capability by insurance companies. For example, in 2016, the FDA approved cobas® EGFR Mutation Test v2 to determine the eligibility of patients with non-small cell lung cancer to receive certain EGFR tyrosine kinase inhibitors.27-29
Liquid biopsy: A comprehensive approach to overcome the current limitations associated with tissue biopsy
Liquid biopsy represents a contemporary diagnostic and prognostic tool in precision oncology that has received considerable recognition over the past years to overcome the current limitations associated with tissue biopsies. 30
For example, cancer is often found in organs or tissues of the body that are difficult to access, such as in lung cancer, tissue-resident biomarker measurements are clinically often not possible due to significant clinical risk, such as bleeding, injury, or infection as a consequence of an invasive tissue biopsy; therefore, liquid biopsy has the advantages of being a noninvasive and sustainable technique to solve this problem. 31
In the perspective of tumor heterogeneity, serial liquid biopsy can be a better representative of the entire population of tumor cells than a tissue biopsy sample, can monitor tumor heterogeneity longitudinally, and allows successive monitoring of the tumor genetic profile.15,18,32-34
Furthermore, unlike tissue biopsies, liquid biopsy enables the detection of tumor at an early stage; early detection of tumor not only prevents cancer mortality, but also reduces morbidity and costs. 35
Approaches to liquid biopsy analysis
Liquid biopsy comprises the noninvasive analysis of ctDNA and CTCs. In addition to ctDNA and CTC biomarkers, other blood components recently discovered as sources of blood-based biomarkers, such as exosomes or platelets, 36 but in this review we will focus on circulating tumor DNA (ctDNA) and circulating tumor cell (CTC).
Cell-free DNA (cfDNA)/circulating tumor DNA (ctDNA)
Circulating cell-free DNA (cfDNA) and circulating tumor DNA are significant non-invasive blood biomarkers used to assess tumor diagnosis and prognosis. 37
Discovery of cfDNA and ctDNA
Mandel and Metais discovered the presence of cfDNA in the blood of healthy individuals in 1948. 38 Decades later, many researchers have extended Mandel and Metais’ work to the identification of tumor-derived cfDNA (also known as circulating tumor DNA-ctDNA) in the blood of cancer patients.38,39
According to previous reports, ctDNA possesses many cancer-associated molecular characteristics, such as single nucleotide mutations, methylation changes, cancer-derived viral sequences, rearrangements, amplifications, MSI and LOH,40-43 therefore, it was considered derived from tumor tissue.
Presence of cfDNA and ctDNA in the body
It is mainly found in the bloodstream; now it can be derived from cerebrospinal fluid, saliva, and can be excreted through urine with extremely low content. Recently, ctDNA has been shown to be present in the CSF of patients with brain tumors, and a high percentage of tumor DNA has also been observed in the saliva of patients with oral cavity cancer.44-46
Release of cfDNA and ctDNA in the blood stream
Two mechanisms for the release of DNA into the blood circulation have been postulated. The first is the passive release of DNA through cell death either by apoptosis or necrosis. 47
Another mechanism of ctDNA release is active secretion by viable tumor cells,47,48 secretion occurs by the release of extracellular vesicles, such as exosomes and prostasomes, which contain DNA pieces of around 150 to 250 bp. 49
Elevated level of cfDNA and ctDNA in cancer patient
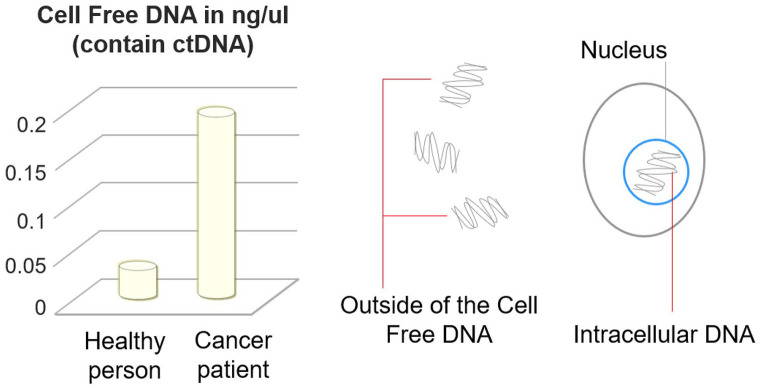
In general, cancer patients have an elevated level of circulating free DNA (Figure 1) than healthy individuals.50-52
Figure 1.
Cell-free DNA (cfDNA) concentration in healthy and cancer patients and its relation to circulating tumor DNA (ctDNA). ctDNA originates mainly from necrosis, apoptosis, and/or circulating tumor cells (CTCs). Fragments of this tumor-derived DNA circulate in the blood with cfDNA(derived from all types of cells, cancerous and noncancerous). A portion of this cfDNA can also come from live cells when they spontaneously release DNA.
In 1977, scientists made the dynamic observation of abnormally high levels of cfDNA in plasma and serum from cancer patients compared to healthy individuals, and this fraction of cfDNA originated from cancer patients was assumed to represent mainly circulating tumor DNA (ctDNA).51,53
Concentration and size of cfDNA and ctDNA fragments in cancer patient
Initial studies showed that the concentration of cfDNA in cancer patients increased significantly compared to a healthy individual that ranges from 0 to 1000 ng/mL of blood, with an average of 180 ng/mL. In contrast, cfDNA in healthy individuals ranges from 0 to 100 ng/mL of blood, with an average of 30 ng/mL.49,51
Another amazing feature of ctDNA is the wide variability in the fragment size. This variability can be attributed to the variety of mechanisms that underlie dead cancer cells. In healthy individuals, cfDNA is released predominantly through apoptosis with small and uniform fragments of around 185 to 200 bp.54,55
In cancer patients, due to incomplete and random digestion of genomic DNA by the nuclease enzyme, necrosis creates a spectrum of DNA fragments of various sizes, interestingly; shorter fragments of ctDNA have been reported in some tumor types (eg, hepatocellular carcinomas) as well as large fragments of cfDNA of thousands of base pairs.50,54-58
These studies show that the concentration and length of the ctDNA fragment can be used for the prediction, early diagnosis, and prognosis of cancer. 59
Clinical application of cfDNA and ctDNA
Conventionally, protein biomarkers are used in cancer diagnosis and in the evaluation of therapeutic responses, such as carcinoembryonic antigen (CEA), prostate specific antigen (PSA), cancer antigen (CA) 19-9 and CA-125, but the specificity and reliability of these protein biomarkers are not satisfactory.60,61 Recently, ctDNA has been the most commonly used type of blood-based biomarker candidate in clinical practice. Specifically, ctDNA analysis has been used primarily to monitor response to therapy, detect minimal residual disease throughout treatment, and evaluate the development of resistance to therapy.62-67
Of many potential clinical benefits of ctDNA, early detection in cancer diagnosis can be an important approach to reduce cancer mortality, 68 and several studies have also reported the detection of genetic alterations in patients with early stage cancer that support the role of ctDNA in early detection of cancer.69-73
Many studies have also shown that ctDNA can be a powerful prognostic factor. For example, in the case of triple-negative breast cancer patients, the detection of ctDNA during or after neoadjuvant chemotherapy, and prior to surgery, has been shown to have a strong prognostic value. 74
Another study in pancreatic cancer showed that higher levels of total percentage of ctDNA were an independent prognostic factor for overall survival. 75
In addition, several studies have compared cfDNA levels in cancer patients with healthy individuals and those with benign conditions, showing promising diagnostic and prognostic applications of cfDNA. For example, plasma cfDNA levels have been reported to be elevated in metastatic RCC relative to localized disease, 76 and in the case of bladder cancer, urine cfDNA levels were found to be significantly elevated compared to controls. 77
Plasma levels of cfDNA have also been reported to be elevated compared to benign prostatic hyperplasia (BPH). 78 Similar to some studies in melanoma, breast, ovarian, and colon cancers,79-82 elevated levels of cfDNA can serve as a noninvasive predictor and prognostic biomarker of cancer.
Furthermore, ctDNA can potentially help detect cancer recurrence much earlier than current conventional procedures. 83 For example, a study of breast cancer patients shows that ctDNA monitoring is a precise method for the early detection of asymptomatic metastatic recurrence in patients diagnosed with primary breast cancer. 84
According to another study, patient-specific ctDNA analysis can be a more sensitive and specific approach to detecting recurrence in breast cancer compared to standard clinical and radiological surveillance, and provides evidence that ctDNA can be detected in most patients with breast cancer several months before clinical relapse. 85
Postoperative detection of ctDNA has also been shown to remain strongly predictive of recurrence among patients with both lower risk (pathological complete response—pCR) and higher risk (pathologic node-positive—pN+) disease. 86
Another potential role for analyzing ctDNA levels is to assess treatment response or resistance; which involves quantitative measurement of the level of ctDNA over time in response to cancer treatments. 87
Monitoring of treatment response is essential to avoid continuing ineffective therapies, to prevent unnecessary side effects, and to determine the benefit of new therapies. Treatment response is generally assessed using serial imaging, but radiographic measurements often do not detect changes in tumor burden. Dawson et al 62 reported that circulating tumor DNA levels showed a greater correlation with changes in tumor burden, and provided the earliest assessment of treatment response in 10 of 19 women (53%) measuring tumor burden with high sensitivity and specificity.
Circulating tumor cells (CTCs)
CTCs discovered by Ashworth in 1869 during an autopsy of a patient with metastatic cancer. 88 These intact tumor cells are released into the bloodstream from primary and secondary tumor sites,89-91 and their migration to the circulatory system results in the development of distant metastases.90-92
Morphology of CTCs
CTCs are a highly effective cell population, characterized by high heterogeneity at the genetic, transcriptomic, proteomic, and metabolomic levels. 93 They do not have well-defined morphological aspects and may vary according to cancer type and stage, 92 and are present as single cell or clusters of cells. 94 They may cluster with parental tumor cells or with fibroblasts, leukocytes, endothelial cells, or platelets and form aggregates with a higher propensity to develop distant metastases.95,96
Numbers of CTCs present in the blood stream
They are rarely found in clinically healthy people or in people with non-malignant tumors. A study showed that patients with metastases have 1 to 10 CTCs per mL of blood,89,91 with even lower numbers in early-stage diseases. 97
Basically, the low number of CTCs isolated from a patient, especially at the beginning of the disease, is the main challenge in using CTCs for diagnostic purposes. 98
However, with the development of technologies capable of dealing with minimal cell numbers, such as microfluidics and NGS, 99 the CTC analysis is now in a position to address highly relevant questions.
Isolation and detection of CTCs
CTCs are observed in different types of cancer, such as melanoma, breast, ovarian, prostate, lung, colorectal, pancreatic, head, neck, and bladder cancer.100,101
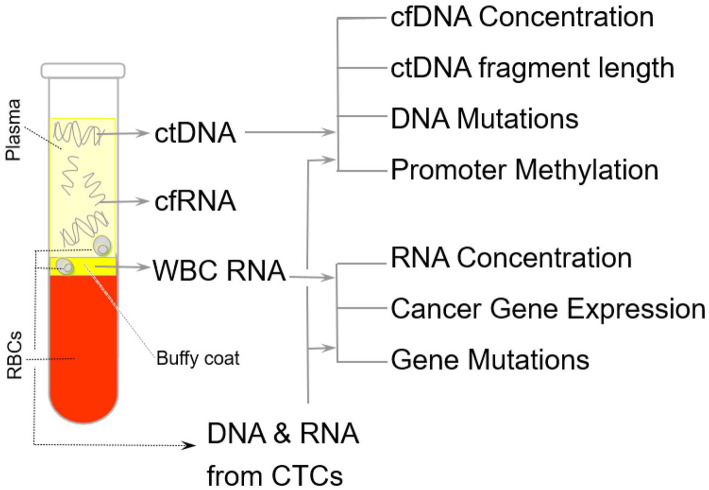
They are present in various biological fluids of cancer patients, such as peripheral blood (Figure 2), urine, pleural effusion, ascites, and cerebrospinal fluid. 47
Figure 2.
This is a schematic diagram showing the collection resource for genetic material from various gravitational layers of blood. The genetic material for liquid biopsy can be collected from various body fluids, including blood compartments, ctDNA from plasma, and CTC from buffy coat. Different types of change can be detected as tumor markers from CTC-derived RNA, such as oncogene expression and mutation. ctDNA can be used for the detection of mutations, the promotor methylation of tumor suppressor genes, and the length of DNA fragments.
Techniques for the detection and isolation of these biomarkers were first described in 1960 and gradually improved over the next 40 years.102,103
Detection of CTC has been improved through the development of specialized technologies based on various principles, including antibody capture,104-106 size exclusion, 107 red and white blood cell depletion, 108 or dielectrophoresis. 109
The CellSearch system is reported as a validated method for CTC detection that has been approved by the US Food and Drug Administration. The CellSearch system, designed for the CTC enumeration in 7.5 mL of blood, was first introduced in 2004 where the analytical accuracy, reproducibility, and linearity of the system were demonstrated. 110
Clinical application of CTCs
Analysis of CTCs provides information on cellular components, including DNA, RNA, and protein, 7 and can be used as an independent prognostic factor for tumors such as prostate cancer, breast cancer, and colon cancer.111-114
CTC analysis also has the potential to be used as a biomarker for the screening and early detection of cancer. 115 Early detection and characterization of CTCs is an important tool for monitoring and preventing the development of overt metastatic disease.116,117
In patients with metastatic breast cancer, the detection of CTC has been reported to have a great prognostic value. 118
Another role of circulating tumor cell (CTC) analysis has been reported to monitor response to therapy in patients with advanced non-small cell lung cancer (NSCLC). Changes in CTCs number during treatment have been proposed as a predictive biomarker of response to both chemotherapy and targeted therapies 119 .
CTCs have also been reported to replace tissue biopsies to predict tumor recurrence 120 and in cases of inaccessible neoplastic sites or unsuccessful sampling; CTCs would be a perceptible approach over tissue biopsy. 121
Conclusion
In summary, liquid biopsy is a significant innovation in the field of precision medicine and has made a competent potential in the treatment of cancer; this revolutionary non-invasive technique consists of the detection and isolation of circulating tumor DNA, circulating tumor cells, and exosomes, as a source of genomic and proteomic information in cancer patients. 121 Analysis of these biomarkers offers a valuable range of information that allows non-invasive diagnosis, prognosis, and treatment of cancer.
Both ctDNA and CTCs are interesting complementary biomarkers of liquid biopsy that can be used in parallel in various aspects of cancer management. Analysis of each biomarker has its own advantages and disadvantages; specifically, ctDNA analysis is an attractive approach because it is simple to isolate and analyze ctDNA, but it is limited to the analysis of DNA-related abnormalities; in contrast, CTC analysis provides profiling of the entire cell; however, it is difficult to isolate a population of rare cells10,17 and is not easily adopted in most laboratories. Thus, its clinical applicability will require further research and verification. 35
Clearly, liquid biopsy opens up a new possibility of non-invasive, safe, and easily repeatable practice in cancer treatment.
Despite the advantages of liquid biopsy, more studies are required to address the challenges and limitations of this technique, which restricted its implementation in clinical practice.
However, the multifaceted advantages of liquid biopsy demonstrate its application as a promising prognosis and diagnostic tool in precision oncology, and it may be a comprehensive approach to overcome the current limitations associated with invasive tissue biopsy and can identify multiple tumor genes present in the bloodstream at multiple time points.
Footnotes
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the authorship, and/or publication of this article.
Authors Contributions: R.Q. coceptualized, researched and wrote this manuscript. All authors have read and agreed to the published version of the final manuscript.
Ethics Approval and Consent to Participate: This manuscript does not involve the use of animal or human data or tissue, so ethical approval is not applicable in this section.
ORCID iDs: Tetsuyuki Hirahata  https://orcid.org/0000-0003-1260-754X
https://orcid.org/0000-0003-1260-754X
Reeshan ul Quraish  https://orcid.org/0000-0002-3609-7583
https://orcid.org/0000-0002-3609-7583
References
- 1. Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7-30. [DOI] [PubMed] [Google Scholar]
- 3. Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559-1564. [DOI] [PubMed] [Google Scholar]
- 4. Damyanov CA, Maslev IK, Pavlov VS, Avramov L. Conventional treatment of cancer realities and problems. Ann Complement Altern Med. 2018;1:1002. [Google Scholar]
- 5. Glatzer M, Panje C, Sirén C, Cihoric N, Putora P. Decision making criteria in oncology. Oncology. 2020;98:370-378. [DOI] [PubMed] [Google Scholar]
- 6. Chakraborty S, Rahman T. The difficulties in cancer treatment. Ecancermedicalscience. 2012;6:ed16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goodman LS, Wintrobe MM, Dameshek W, Goodman MJ, Gilman A, McLennan MT. Nitrogen mustard therapy: use of methyl-bis (beta-chloroethyl) amine hydrochloride and tris (beta-chloroethyl) amine hydrochloride for Hodgkin’s disease, lymphosarcoma, leukemia and certain allied and miscellaneous disorders. JAMA. 1946;132:126-132. [DOI] [PubMed] [Google Scholar]
- 8. Housman G, Byler S, Heerboth S, et al. Drug resistance in cancer: an overview. Cancers. 2014;6:1769-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168:707-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vu BT, Tan Le D, Van Pham P. Liquid biopsies: tumour diagnosis and treatment monitoring. Biomed Res Ther. 2016;3:745-756. [Google Scholar]
- 11. Hodson R. Precision medicine. Nature. 2016;537:S49. [DOI] [PubMed] [Google Scholar]
- 12. Savonarola A, Palmirotta R, Guadagni F, Silvestris F. Pharmacogenetics and pharmacogenomics: role of mutational analysis in anti-cancer targeted therapy. Pharmacogenomics J. 2012;12:277-286. [DOI] [PubMed] [Google Scholar]
- 13. Kasraeian S, Allison DC, Ahlmann ER, Fedenko AN, Menendez LR. A comparison of fine-needle aspiration, core biopsy, and surgical biopsy in the diagnosis of extremity soft tissue masses. Clin Orthop Relat Res. 2010;468:2992-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mclarty JL, Yeh CH. Circulating cell-free DNA: the blood biopsy in cancer management. MOJ Cell Sci Rep. 2015;2:27-29. [Google Scholar]
- 15. Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qin Z, Ljubimov VA, Zhou C, Tong Y, Liang J. Cell-free circulating tumor DNA in cancer. Chin J Cancer. 2016;35:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Castro-Giner F, Gkountela S, Donato C, et al. Cancer diagnosis using a liquid biopsy: challenges and expectations. Diagnostics. 2018;8:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boeddinghaus I, Johnson SR. Serial biopsies/fine-needle aspirates and their assessment. Methods Mol Med. 2006;120:29-41. [DOI] [PubMed] [Google Scholar]
- 19. Yong E. Cancer biomarkers: written in blood. Nature. 2014;511:524-526. [DOI] [PubMed] [Google Scholar]
- 20. Tan CR, Zhou L, El-Deiry WS. Circulating tumor cells versus circulating tumor DNA in colorectal cancer: pros and Cons. Curr Colorectal Cancer Rep. 2016;12:151-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem. 2013;59:110-118. [DOI] [PubMed] [Google Scholar]
- 22. Ding Y, Li W, Wang K, Xu C, Hao M, Ding L. Perspectives of the application of liquid biopsy in colorectal cancer. Biomed Res Int. 2020;2020:6843180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reimers N, Pantel K. Liquid biopsy: novel technologies and clinical applications. Clin Chem Lab Med. 2019;57:312-316. [DOI] [PubMed] [Google Scholar]
- 24. Aro K, Wei F, Wong DT, Tu M. Saliva liquid biopsy for point-of-care applications. Front Public Health. 2017;5:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jain S, Lin SY, Song W, Su YH. Urine-Based liquid biopsy for nonurological cancers. Genet Test Mol Biomarkers. 2019;23:277-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Coverage of liquid biopsies for cancer care increases greatly over 5 years. HemOnctoday. July 9, 2020. [Google Scholar]
- 27. U.S. Food and Drug Administration. “cobas® EGFR Mutation Test v2 - P150047.” Medical Devices. U.S. Food and Drug Administration. September 9, 2016. World Wide Web. Accessed September 20, 2017. https://r.search.yahoo.com/_ylt=Awr9DtmAjPNhM98AoZ1XNyoA;_ylu=Y29sbwNncTEEcG9zAzEEdnRpZANCQVNFTElORUNfMQRzZWMDc3I-/RV=2/RE=1643379969/RO=10/RU=https%3a%2f%2fwww.fda.gov%2fmedia%2f130429%2fdownload/RK=2/RS=tGWD2E0y4MeIT7xK.g1gCO4JaoU
- 28. Brown P. The cobas® EGFR mutation test v2 assay. Future Oncol. 2016;12:451-452. [DOI] [PubMed] [Google Scholar]
- 29. Greig SL. Osimertinib: first global approval. Drugs. 2016;76:263-273. [DOI] [PubMed] [Google Scholar]
- 30. De Rubis G, Rajeev Krishnan S, Bebawy M. Liquid biopsies in cancer diagnosis, monitoring, and prognosis. Trends Pharmacol Sci. 2019;40:172-186. [DOI] [PubMed] [Google Scholar]
- 31. Sholl LM, Aisner DL, Allen TC, et al. Liquid biopsy in lung cancer: a perspective from members of the pulmonary pathology society. Arch Pathol Lab Med. 2016;140:825-829. [DOI] [PubMed] [Google Scholar]
- 32. Bidard FC, Weigelt B, Reis-Filho JS. Going with the flow: from circulating tumor cells to DNA. Sci Transl Med. 2013;5:207ps14. [DOI] [PubMed] [Google Scholar]
- 33. Cheung AH, Chow C, To KF. Latest development of liquid biopsy. J Thorac Dis. 2018;10:S1645-S1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bedard PL, Hansen AR, Ratain MJ, Siu LL. Tumour heterogeneity in the clinic. Nature. 2013;501:355-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Etzioni R, Urban N, Ramsey S, et al. The case for early detection. Nat Rev Cancer. 2003;3:243-252. [DOI] [PubMed] [Google Scholar]
- 36. Neumann MHD, Bender S, Krahn T, Schlange T. ctDNA and CTCs in liquid biopsy – current status and where we need to progress. Comput Struct Biotechnol J. 2018;16:190-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Alix-Panabières C, Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6:479-491. [DOI] [PubMed] [Google Scholar]
- 38. Mandel P, Metais P. Les acidesnucleiques du plasma sanguin chez l’homme. C R Seances Soc Biol Fil. 1948;142:241-243. [PubMed] [Google Scholar]
- 39. Leung F, Kulasingam V, Diamandis EP, et al. Circulating tumor DNA as a cancer biomarker: fact or fiction? Clin Chem. 2016;62:1054-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chan KC, Jiang P, Zheng YW, et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem. 2013;59:211-224. [DOI] [PubMed] [Google Scholar]
- 41. Balgkouranidou I, Chimonidou M, Milaki G, et al. Breast cancer metastasis suppressor-1 promoter methylation in cell-free DNA provides prognostic information in non-small cell lung cancer. Br J Cancer. 2014;110:2054-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chan KCA, Hung ECW, Woo JKS, et al. Early detection of nasopharyngeal carcinoma by plasma Epstein-Barr virus DNA analysis in a surveillance program. Cancer. 2013;119:1838-1844. [DOI] [PubMed] [Google Scholar]
- 43. Elshimali Y, Khaddour H, Sarkissyan M, Wu Y, Vadgama J. The clinical utilization of circulating cell free DNA (CCFDNA) in blood of cancer patients. Int J Mol Sci. 2013;14:18925-18958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. De Mattos-Arruda L, Mayor R, Ng CKY, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015;6:8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang Y, Springer S, Mulvey CL, et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med. 2015;7:293ra104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Melkonyan HS, Feaver WJ, Meyer E, et al. Transrenal nucleic acids: from proof of principle to clinical tests. Ann N Y Acad Sci. 2008;1137:73-81. [DOI] [PubMed] [Google Scholar]
- 47. Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223-238. [DOI] [PubMed] [Google Scholar]
- 48. Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P. About the possible origin and mechanism of circulating DNA: apoptosis and active DNA release. Clin Chim Acta. 2001;313:139-142. [DOI] [PubMed] [Google Scholar]
- 49. Minciacchi VR, Zijlstra A, Rubin MA, Di Vizio D. Extracellular vesicles for liquid biopsy in prostate cancer: where are we and where are we headed? Prostate Cancer Prostatic Dis. 2017;20:251-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Delgado PO, Alves BCA, de Sousa Gehrke F, et al. Characterization of cell-free circulating DNA in plasma in patients with prostate cancer. Tumour Biol. 2013;34:983-986. [DOI] [PubMed] [Google Scholar]
- 51. Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646-650. [PubMed] [Google Scholar]
- 52. Hashad D, Sorour A, Ghazal A, Talaat I. Free circulating tumor DNA as a diagnostic marker for breast cancer. J Clin Lab Anal. 2012;26:467-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chakravadhanula M, Tembe W, Legendre C, et al. Detection of an atypical teratoid rhabdoid brain tumor gene deletion in circulating blood using next-generation sequencing. J Child Neurol. 2014;29:n81-n85. [DOI] [PubMed] [Google Scholar]
- 54. Jiang P, Lo YMD. The long and short of circulating cell-free DNA and the Ins and outs of molecular diagnostics. Trends Genet. 2016;32:360-371. [DOI] [PubMed] [Google Scholar]
- 55. Giacona MB, Ruben GC, Iczkowski KA, Roos TB, Porter DM, Sorenson GD. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas. 1998;17:89-97. [DOI] [PubMed] [Google Scholar]
- 56. Wang BG, Huang HY, Chen YC, et al. Increased plasma DNA integrity in cancer patients. Cancer Res. 2003;63:3966-3968. [PubMed] [Google Scholar]
- 57. Krishnamurthy N, Spencer E, Torkamani A, Nicholson L. Liquid biopsies for cancer: coming to a patient near you. J Clin Med. 2017;6:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Thierry AR, Mouliere F, Gongora C, et al. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res. 2010;38:6159-6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shukla D, Kale AD, Hallikerimath S, Yerramalla V, Subbiah V. Can quantifying free-circulating DNA be a diagnostic and prognostic marker in oral epithelial dysplasia and oral squamous cell carcinoma? J Oral Maxillofac Surg. 2013;71:414-418. [DOI] [PubMed] [Google Scholar]
- 60. Yoshimasu T, Maebeya S, Suzuma T, et al. Disappearance curves for tumor markers after resection of intrathoracic malignancies. Int J Biol Markers. 1999;14:99-105. [DOI] [PubMed] [Google Scholar]
- 61. Ito K, Hibi K, Ando H, et al. Usefulness of analytical CEA doubling time and half-life time for overlooked synchronous metastases in colorectal carcinoma. Jpn J Clin Oncol. 2002;32:54-58. [DOI] [PubMed] [Google Scholar]
- 62. Dawson SJ, Tsui DWY, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199-1209. [DOI] [PubMed] [Google Scholar]
- 63. Diaz LA, Jr, Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7:302ra133. [DOI] [PubMed] [Google Scholar]
- 65. Schiavon G, Hrebien S, Garcia-Murillas I, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med. 2015;7:313ra182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Siravegna G, Mussolin B, Buscarino M, et al. Erratum: clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21:827-801. [DOI] [PubMed] [Google Scholar]
- 67. Roschewski M, Dunleavy K, Pittaluga S, et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: a correlative biomarker study. Lancet Oncol. 2015;16:541-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Aravanis AM, Lee M, Klausner RD. Next-Generation sequencing of circulating tumor DNA for early cancer detection. Cell. 2017;168:571-574. [DOI] [PubMed] [Google Scholar]
- 69. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, et al. Detection of circulating tumor DNA in early- and late stage human malignancies. Sci Transl Med. 2014;6:224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545:446-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cohen JD, Javed AA, Thoburn C, et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc Natl Acad Sci U S A. 2017;114:10202-10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017;9:eaan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cavallone L, Aguilar A, Aldamry M, et al. Circulating tumor DNA (ctDNA) during and after neoadjuvant chemotherapy and prior to surgery is a powerful prognostic factor in triple-negative breast cancer (TNBC). J Clin Oncol. 2019;37:594-594. [Google Scholar]
- 75. Patel H, Okamura R, Fanta P, et al. Clinical correlates of blood-derived circulating tumor DNA in pancreatic cancer. J Hematol Oncol. 2019;12:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wan J, Zhu L, Jiang Z, Cheng K. Monitoring of plasma cell-free DNA in predicting postoperative recurrence of clear cell renal cell carcinoma. Urol Int. 2013;91:273-278. [DOI] [PubMed] [Google Scholar]
- 77. Brisuda A, Pazourkova E, Soukup V, et al. Urinary cell-free DNA quantification as non-invasive biomarker in patients with bladder cancer. Urol Int. 2016;96:25-31. [DOI] [PubMed] [Google Scholar]
- 78. Chun FK, Müller I, Lange I, et al. Circulating tumour-associated plasma DNA represents an independent and informative predictor of prostate cancer. BJU Int. 2006;98:544-548. [DOI] [PubMed] [Google Scholar]
- 79. Shinozaki M, O’Day SJ, Kitago M, et al. Utility of circulating B-RAF DNA mutation in serum for monitoring melanoma patients receiving biochemotherapy. Clin Cancer Res. 2007;13:2068-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bidard FC, Madic J, Mariani P, et al. Detection rate and prognostic value of circulating tumor cells and circulating tumor DNA in metastatic uveal melanoma. Int J Cancer. 2014;134:1207-1213. [DOI] [PubMed] [Google Scholar]
- 81. Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4:136ra68. [DOI] [PubMed] [Google Scholar]
- 82. Tie J, Kinde I, Wang Y, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol. 2015;26:1715-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chakrabarti S, Xie H, Urrutia R, Mahipal A. The promise of circulating tumor DNA (ctDNA) in the management of early-stage colon cancer: aa critical review. Cancers. 2020;12:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Olsson E, Winter C, George A, et al. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol Med. 2015;7:1034-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Coombes RC, Page K, Salari R, et al. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clin Cancer Res. 2019;25:4255-4263. [DOI] [PubMed] [Google Scholar]
- 86. Tie J, Cohen J, Wang Y, et al. The potential of circulating tumor DNA (ctDNA) to guide adjuvant chemotherapy decision making in locally advanced rectal cancer (LARC). J Clin Oncol. 2017;35:3521-3521. [Google Scholar]
- 87. Merker JD, Oxnard GR, Compton C, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J Clin Oncol. 2018;36:1631-1641. [DOI] [PubMed] [Google Scholar]
- 88. Ashworth TR. A case of cancer in which cells similar to those in the tumors were seen in the blood after death. Med J Aust. 1869;14:146-149. [Google Scholar]
- 89. Lianidou ES, Strati A, Markou A. Circulating tumor cells as promising novel biomarkers in solid cancers. Crit Rev Clin Lab Sci. 2014;51:160-171. [DOI] [PubMed] [Google Scholar]
- 90. Masuda T, Hayashi N, Iguchi T, Ito S, Eguchi H, Mimori K. Clinical and biological significance of circulating tumor cells in cancer. Mol Oncol. 2016;10:408-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hardingham JE, Grover P, Winter M, Hewett PJ, Price TJ, Thierry B. Detection and clinical significance of circulating tumor cells in colorectal cancer–20 years of progress. Mol Med. 2015;21:S25-S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Parkinson DR, Dracopoli N, Petty BG, et al. Considerations in the development of circulating tumor cell technology for clinical use. J Transl Med. 2012;10:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Micalizzi DS, Maheswaran S, Haber DA. A conduit to metastasis: circulating tumor cell biology. Genes Dev. 2017;31:1827-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Aceto N, Bardia A, Miyamoto D, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Krebs MG, Metcalf RL, Carter L, Brady G, Blackhall FH, Dive C. Molecular analysis of circulating tumour cells-biology and biomarkers. Nat Rev Clin Oncol. 2014;11:129-144. [DOI] [PubMed] [Google Scholar]
- 96. Labelle M, Begum S, Hynes RO. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell. 2011;20:576-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Young R, Pailler E, Billiot F, et al. Circulating tumor cells in lung cancer. Acta Cytol. 2012;56:655-660. [DOI] [PubMed] [Google Scholar]
- 98. Yap TA, Lorente D, Omlin A, Olmos D, de Bono JS. Circulating tumor cells: a multifunctional biomarker. Clin Cancer Res. 2014;20:2553-2568. [DOI] [PubMed] [Google Scholar]
- 99. Ma S, Murphy TW, Lu C. Microfluidics for genome-wide studies involving next generation sequencing. Biomicrofluidics. 2017;11:021501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Mavroudis D. Circulating cancer cells. Ann Oncol. 2010;21:vii95-100. [DOI] [PubMed] [Google Scholar]
- 101. Pantel K, Alix-Panabières C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med. 2010;16:398-406. [DOI] [PubMed] [Google Scholar]
- 102. Salgado I, Hopkirk JF, Long RC, Ritchie AC, Ritchie S, Webster DR. Tumour cells in the blood. Can Med Assoc J. 1959;81:619-622. [PMC free article] [PubMed] [Google Scholar]
- 103. Alexander RF, Spriggs AI. The differential diagnosis of tumour cells in circulating blood. J Clin Pathol. 1960;13:414-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Nagrath S, Sequist LV, Maheswaran S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Stott SL, Hsu CH, Tsukrov DI, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci USA. 2010;107:18392-18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Stott SL, Lee RJ, Nagrath S, et al. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Sci Transl Med. 2010;2:25ra23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Xu L, Mao X, Imrali A, et al. Optimization and evaluation of a novel size based circulating tumor cell isolation system. PLoS One. 2015;10:e0138032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ozkumur E, Shah AM, Ciciliano JC, et al. Inertial focusing for tumor antigen–dependent and – independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5:179ra47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Chiu TK, Chou WP, Huang SB, et al. Application of optically-induced-dielectrophoresis in microfluidic system for purification of circulating tumour cells for gene expression analysis - cancer cell line model. Sci Rep. 2016;6:32851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Allard WJ, Matera J, Miller MC, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897-6904. [DOI] [PubMed] [Google Scholar]
- 111. Lack J, Gillard M, Cam M, Paner GP, VanderWeele DJ. Circulating tumor cells capture disease evolution in advanced prostate cancer. J Transl Med. 2017;15:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Bidard FC, Michiels S, Riethdorf S, et al. Circulating tumor cells in breast cancer patients treated by neoadjuvant chemotherapy: a meta-analysis. J Natl Cancer Inst. 2018;110:560-567. [DOI] [PubMed] [Google Scholar]
- 113. Wang L, Zhou S, Zhang W, et al. Circulating tumor cells as an independent prognostic factor in advanced colorectal cancer: a retrospective study in 121 patients. Int J Colorectal Dis. 2019;34:589-597. [DOI] [PubMed] [Google Scholar]
- 114. Rahbari NN, Aigner M, Thorlund K, et al. Meta-analysis shows that detection of circulating tumor cells indicates poor prognosis in patients with colorectal cancer. Gastroenterology. 2010;138:1714-1726. [DOI] [PubMed] [Google Scholar]
- 115. Tsai WS, Nimgaonkar A, Segurado O, et al. Prospective clinical study of circulating tumor cells for colorectal cancer screening. J Clin Oncol. 2018;36:556-556. [Google Scholar]
- 116. Hong B, Zu Y. Detecting circulating tumor cells: current challenges and new trends. Theranostics. 2013;3:377-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Coumans FA, Siesling S, Terstappen LW. Detection of cancer before distant metastasis. BMC Cancer. 2013;13:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781-791. [DOI] [PubMed] [Google Scholar]
- 119. Gallo M, De Luca A, Frezzetti D, Passaro V, Maiello MR, Normanno N. The potential of monitoring treatment response in non-small cell lung cancer using circulating tumour cells. Expert Rev Mol Diagn. 2019;19:683-694. [DOI] [PubMed] [Google Scholar]
- 120. Cai LL, Ye HM, Zheng LM, Ruan RS, Tzeng CM. Circulating tumor cells (CTCs) as a liquid biopsy material and drug target. Curr Drug Targets. 2014;15:965-972. [PubMed] [Google Scholar]
- 121. Palmirotta R, Lovero D, Cafforio P, et al. Liquid biopsy of cancer: a multimodal diagnostic tool in clinical oncology. Ther Adv Med Oncol. 2018;10:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]