Significance
Toxin–antitoxin systems are enigmatic and diverse elements of bacterial and bacteriophage genomes. We have uncovered remarkable versatility in an antitoxin protein domain that has evolved to neutralize dozens of different toxin domains. We find that antitoxins carrying this domain—Panacea—form complexes with their cognate toxins, indicating a direct neutralization mechanism, and that Panacea can be evolved to neutralize a noncognate and nonhomologous toxin with just two amino acid substitutions. This raises the possibility that this domain could be an adaptable universal or semi-universal protein neutralizer with significant biotechnological and medical potential.
Keywords: toxin, antitoxin, bacteriophage, evolution, panacea
Abstract
Toxin–antitoxin (TA) gene pairs are ubiquitous in microbial chromosomal genomes and plasmids as well as temperate bacteriophages. They act as regulatory switches, with the toxin limiting the growth of bacteria and archaea by compromising diverse essential cellular targets and the antitoxin counteracting the toxic effect. To uncover previously uncharted TA diversity across microbes and bacteriophages, we analyzed the conservation of genomic neighborhoods using our computational tool FlaGs (for flanking genes), which allows high-throughput detection of TA-like operons. Focusing on the widespread but poorly experimentally characterized antitoxin domain DUF4065, our in silico analyses indicated that DUF4065-containing proteins serve as broadly distributed antitoxin components in putative TA-like operons with dozens of different toxic domains with multiple different folds. Given the versatility of DUF4065, we have named the domain Panacea (and proteins containing the domain, PanA) after the Greek goddess of universal remedy. We have experimentally validated nine PanA-neutralized TA pairs. While the majority of validated PanA-neutralized toxins act as translation inhibitors or membrane disruptors, a putative nucleotide cyclase toxin from a Burkholderia prophage compromises transcription and translation as well as inducing RelA-dependent accumulation of the nucleotide alarmone (p)ppGpp. We find that Panacea-containing antitoxins form a complex with their diverse cognate toxins, characteristic of the direct neutralization mechanisms employed by Type II TA systems. Finally, through directed evolution, we have selected PanA variants that can neutralize noncognate TA toxins, thus experimentally demonstrating the evolutionary plasticity of this hyperpromiscuous antitoxin domain.
Toxin–antitoxin systems (TAs) are diverse two-gene elements that are widespread in plasmids and chromosomes of bacteria and archaea (1, 2) as well as in genomes of temperate bacteriophages that prey on these microbes (3–6). The various protein toxins target different core processes of the encoding cell to dramatically inhibit growth while their cognate antitoxins efficiently neutralize the toxicity. Known TA toxins exert their toxicity in a variety of ways (1), often targeting translation through modification or cleavage of the ribosome, translation factors, transfer RNAs (tRNAs), or messenger RNAs (mRNAs). Similarly, antitoxins counteract the toxins through diverse mechanisms (1, 7). The means of neutralization are often classified into four or more subtypes, the main four being base pairing of the antitoxin RNA with the toxin mRNA (Type I TA systems), direct protein–protein binding and inhibition (Type II), inhibition of the protein toxin by the antitoxin RNA (Type III), or indirect nullification of toxicity (Type IV). Plasmid-encoded TA systems have long been known to function as addiction modules that promote plasmid maintenance and stability (8). The biological function of chromosomal TAs has been harder to pin down, with different TAs being implicated—with varying levels of confidence—in modulation of bacterial physiology in response to the environment, stabilization of genomic elements, and bacteriophage defense (1, 2, 9, 10).
We have recently discovered a class of TA systems that employs RelA/SpoT homologue (RSH) enzymes—so-called toxic Small Alarmone Synthetases (toxSASs)—as toxic enzymes to abrogate bacterial growth (4). Housekeeping RSH enzymes such as the Escherichia coli ribosome-associated amino acid starvation sensor RelA synthesize the nucleotide alarmone (p)ppGpp, a pyrophosphorylated derivative of GDP/GTP (11, 12). The toxicity of Cellulomonas marina toxSAS FaRel relies on the production of the related toxic alarmone (pp)pApp from housekeeping adenosine nucleotides AMP, ADP, and ATP (4). The accumulation of (pp)pApp results in dramatic depletion of ATP, which, in turn, leads to the cessation of transcription followed by the inhibition of translation and replication (4, 13). The synthesis of (pp)pApp is not the only mechanism of toxicity employed by toxSAS enzymes: we have found that the majority of experimentally explored toxSASs, such as PhRel2 from Bacillus subtilis strain Ia1a, act as specific protein synthesis inhibitors that pyrophosphorylate the 3′ CCA end of tRNA to abrogate aminoacylation (12). In the case of Mycobacterial phage Phrann protein Gp29, a translation-inhibiting toxSAS in the PhRel subfamily that likely pyrophosphorylates tRNA (12), the biological function appears to be defense against phage superinfection (5).
ToxSASs are neutralized by several different antitoxins that act via Type II and Type IV mechanisms. The cognate antitoxin of B. subtilis Ia1a PhRel2 (a tRNA-modifying toxSAS) belongs to a widespread domain family of unknown function designated by the Pfam database as DUF4065, in which DUF stands for domain of unknown function (14). Clues about the roles of DUF4065 are limited; however, it is found in so-called genetic element protein A, previously associated with TA loci (15, 16), and is also present in the proteolysis-promoting SocA antitoxin of the replication-inhibiting SocB toxin (17). This unusual mechanism of neutralization by an antitoxin is referred to as Type VI. We have earlier identified the DUF4065 domain in a putative alternative antitoxin to the ribonuclease (RNase) MqsR, but this was not tested experimentally (15).
We asked whether, given the broad distribution of DUF4065 across multiple phyla of bacteria and archaea, the analysis of the genomic neighborhood of DUF4065 can enable the prediction of novel TA systems. Using our tool FlaGs (for flanking genes) (18) to analyze diverse genomes across the tree of life, we find that DUF4065 is the predicted antitoxin counterpart of at least 1,268 different putative TA system families corresponding to at least 88 distinct putative toxin–DUF4065 domain combinations, found in diverse bacteria, archaea, and bacteriophages. While many of the toxins of these systems are related to classical TA toxins such as various mRNA interferases (19, 20), Fic/Doc-type protein modification enzymes (21), and toxSASs (4), others have little similarity to known domains or proteins with solved structures. We have experimentally verified nine DUF4065-containing antitoxins as neutralizers of their cognate toxin partners. These toxins include translation inhibitors, membrane disruptors, and a putative nucleotide cyclase that pleiotropically affects metabolism, compromising transcription and translation, as well as inducing RelA-dependent accumulation of the guanosine tetraphosphate alarmone nucleotide (p)ppGpp. Complex formation indicates DUF4065-containing antitoxins neutralize toxins via direct protein–protein interaction [that is, act as Type II TA systems (1, 2)], and we have identified substitutions that confer the ability of one antitoxin to neutralize a noncognate toxin. Given the versatility of the antitoxin function of DUF4065, we have named the domain Panacea after the Greek goddess of universal remedy.
Results
The Domain DUF4065 Is Found in Diverse TA-Like Loci across Bacteria, Archaea, and Bacteriophages.
As DUF4065 has previously been associated with TA systems (15–17), we asked whether it may constitute a widespread antitoxin domain paired in operons with novel toxin domains. To answer this, we used sensitive sequence searching combined with an analysis of gene neighborhoods using our tool FlaGs (18) (see SI Appendix, Fig. S1 for a graphical overview of the procedure). Using the hidden Markov model (HMM) of the DUF4065 domain (14) to scan 20,209 genomes across cellular life and viruses, we identified 2,281 hits (Dataset S1) in prokaryotes and bacteriophages comprising 27 phyla of bacteria, 3 phyla of archaea, and 17 different bacteriophages (Dataset S1). Of those 2,281, 76 are present in complete prokaryotic genomes, allowing the determination of whether they are chromosome or plasmid encoded according to the genome annotations. All but two of our identified DUF4065 homologs in complete genomes are chromosome localized. The two exceptions annotated as plasmid encoded (but may be minichromosomes) are archaeal, found in Haloarchaea (protein accessions WP_050049451.1 and WP_049938427.1). Most DUF4065-carrying taxa only carry a single homolog; 217 taxa have two, 45 have three, 14 have four, 12 have five, and 5 have more than five. Of these five taxa, the taxon with the most DUF4065 homologs is the Mollicute bacterium “strawberry lethal yellows phytoplasma” strain NZSb11. This genome contains 25 DUF4065 homologs, of which three are predicted as being encoded in TA-like loci by our in silico analysis pipeline.
Adapting FlaGs for analyzing gene neighborhood conservation (SI Appendix, Fig. S1), we find that around half of the identified DUF4065-containing proteins can be detected as being encoded in two-gene loci that are conserved across multiple species, reminiscent of TA systems (Dataset S1, representatives in Fig. 1, Dataset S2, and SI Appendix, Fig. S2). In total, we predicted 1,313 preliminarily TA (pTA)-like loci using the criteria 1) that there should be a maximum distance of 100 nucleotides between the two genes, 2) that this architecture is conserved in two or more species, and 3) the conservation of the gene neighborhood does not suggest longer operons than three genes (SI Appendix, Fig. S1). We allowed three-gene architectures into our analysis, as TAs can sometimes be found with a conserved third gene, such as mazG in the case of mazEF (22), chaperones in the case of tripartite TA–chaperone modules (23), or transcriptional regulators in the case of the paaR-paaA-parE system (3). By allowing three-part clusters, we have identified 25 clusters that are conserved as a third gene in a subset of genomes that encode a particular predicted TA pair (Dataset S1). We call these accessory proteins, annotations of which include DNA/nucleotide and protein/amino acid modification enzymes, helicases, proteases, and nucleases. Each detected accessory third gene was only present in a small fraction of the genomes in which the main TA pair was identified, suggesting that whatever the role of these third genes, they probably do not play a general role in toxicity and neutralization.
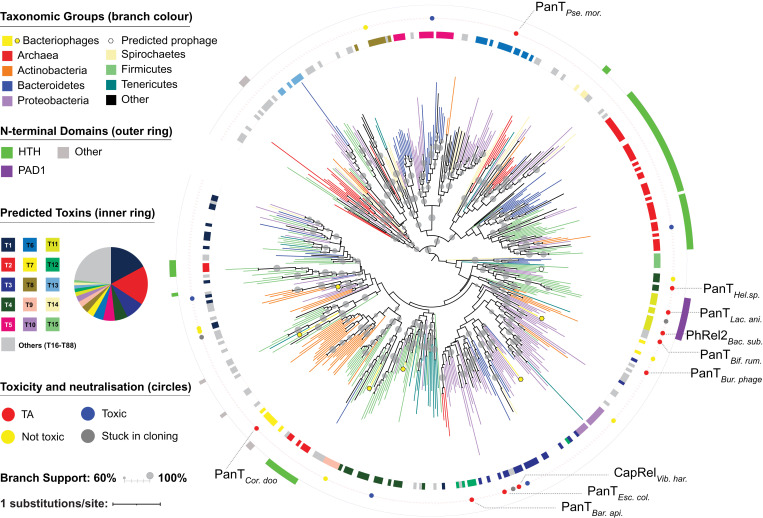
Fig. 1.
The domain DUF4065/Panacea is found in a wide variety of TA-like loci across bacteria, archaea, and bacteriophages. Branches of the IQTree maximum likelihood phylogenetic tree of representative PanA sequences are colored by major taxonomic groupings as per the upper left key with an additional symbol to highlight bacteriophages. Rectangles in the outer and inner rings indicate the presence and absence of N-terminal domains in the PanA sequences and predicted associated toxin groups, respectively, according to the left-hand keys. Colored circles between the rings indicate putative TA pairs that have been tested in toxicity neutralization assays and the results of those assays. “TA” means the expression of the toxin compromises E. coli growth, and coexpression of the antitoxin either fully or partially counteracts the toxicity. “Toxic” means toxicity is confirmed, but the cognate PanA sequence does not rescue in this E. coli system. “Stuck in cloning” refers to cases in which the putative toxin genes could not be successfully chemically synthesized and plasmid subcloned, potentially because of the toxicity being too severe. Gray circles on the branches indicate branch support from IQTree ultrafast bootstrapping (53). Tree annotation was carried out with iTOL (54).
It is conceivable that some homologous genes are found adjacent to DUF4065-encoding genes in multiple genomes purely by chance and are not part of genuine TA systems. Therefore, we used a BlastP-based (24) reciprocity test to filter out putative “toxins” that are at risk as being spurious hits (SI Appendix, Fig. S1C). From the 1,313 pTA-like loci, we determined that 67 proteins (of which 39 are predicted toxins and 28 are accessory proteins) are likely spurious hits (Dataset S1). Major classes of these spurious hits are transposases/integrases that are commonly found in TA-encoding neighborhoods and various ATPases that are captured into homologous clusters because of their well-conserved ATP-binding motifs (Dataset S1).
The remaining 1,268 putative TA loci that we predict to be relatively reliable correspond to 88 clusters of potential toxins. We number these clusters with a T prefix; for example, SocB is in cluster T10. The vast majority of these are annotated as “hypothetical protein,” as they share only weak similarity to proteins of known function. Therefore, we searched the putative toxin protein sequences against the National Center for Biotechnology Information Conserved Domains Database (NCBI CDD) to detect the presence of known domains (Dataset S1). Of the 1,268 putative toxins, 938 sequences (belonging to 41 clusters) had no hit to a domain, and of the others, the most predominant domains were MqsR like (n = 90), Fic/Doc like (n = 32), and toxSAS like (domain names NT_Pol-beta-like, RelA_SpoT, and NT_Rel-Spo-like; n = 31). Other known toxin domains that were represented in the CDD results were PemK (mRNAse) and ParE (DNA gyrase inhibitor). For clusters that failed to find a hit in the CDD database, HHPred (25) was run with one to two representative sequences per cluster, revealing additional potential homology to proteins of known structures for 30 clusters (Dataset S1; see the following sections of Results for examples among our verified TAs).
The variety in the potential toxin domains suggests that the DUF4065 domain may be a universal or semi-universal antitoxin domain capable of neutralizing various different toxic proteins. In light of this, we suggest renaming DUF4065 to Panacea and abbreviate each Panacea-containing putative antitoxin and putative toxin protein as PanA and PanT, respectively. We refer to the two-gene system with the handle PanAT. In each PanAT system, the order of two genes can differ: either antitoxin first or toxin first. However, antitoxin first is the more common arrangement (943 versus 325) as is typical for Type II TA systems (1, 2).
Maximum likelihood phylogenetic analysis shows the PanA tree largely does not follow taxonomic relationships, reflecting a high degree of mobility (Fig. 1, SI Appendix, Fig. S2, and Dataset S2). While the deepest branches are poorly supported (not surprising for a small protein), there are a number of groups with medium to strong (over 60 to 100%) bootstrap support that include different bacterial—and sometimes archaeal—phyla. While Panacea is present broadly across prokaryotes, it does not appear to be present in eukaryotes. The only PanA we discovered in eukaryotes was in the Pharoah ant (Monomorium pharaonic; XP_028045404.1), and this appears to be a case of contamination, as an identical sequence is found in the bacterium Stenotrophomonas maltophilia. Surprisingly, a strongly supported clade of PanA sequences does not necessarily mean they all share the same PanT as shown by the inner ring in Fig. 1 and the toxin partner swapping in focus in Fig. 2A and SI Appendix, Fig. S2. Indeed, the exchange of toxin partners within a clade appears to be frequent. We refer to this kind of domain-level partner swapping as hyperpromiscuity, to distinguish from the promiscuity that can be seen when one single antitoxin sequence can nullify multiple con-cognate but homologous toxins (26–28).
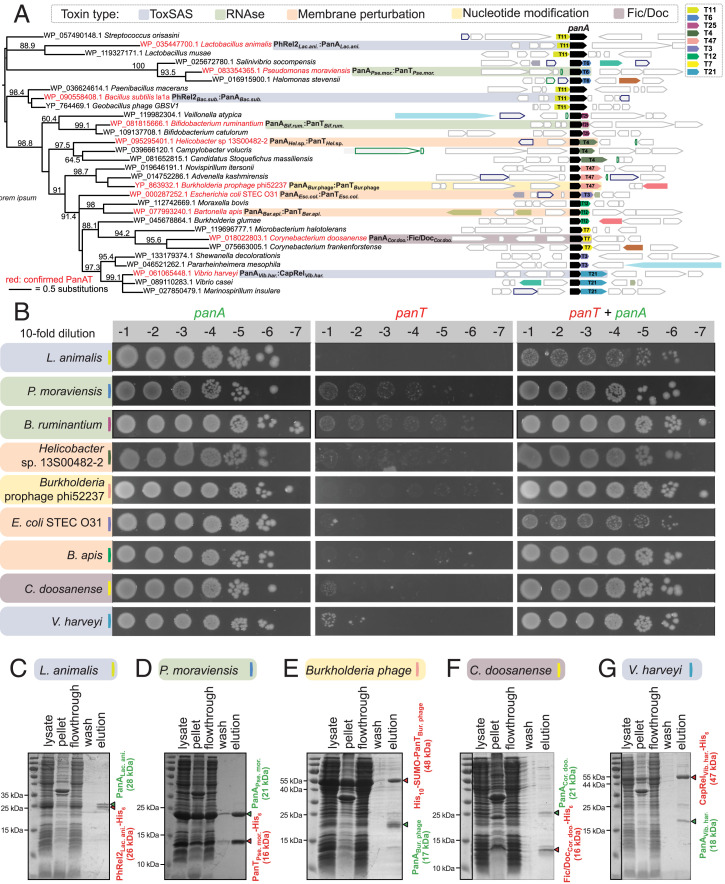
Fig. 2.
PanA antitoxins form stable complexes with evolutionarily diverse TA toxins. (A) The maximum likelihood tree of PanA sequences, annotated with conserved gene neighborhoods generated with FlaGs (18). Numbers on branches show IQTree ultrafast bootstrap support (53). Genes belonging to homologous clusters are colored the same; the PanA antitoxin is universally shown in black. Numbers on genes preceded by a T indicate toxin clusters. (B) Validation of panAT TA pairs by toxicity neutralization assays. Overnight cultures of E. coli strains transformed with pBAD33 and pKK223-3 vectors or derivatives expressing putative panT toxins and panA antitoxins, correspondingly, were adjusted to OD600 1.0, serially diluted, and spotted on LB medium supplemented with appropriate antibiotics and inducers (0.2% arabinose for panT induction and 1 mM IPTG for panA induction). (C–G) A pull-down assay demonstrates complex formation between PanA antitoxins and PanT toxins. Untagged PanA representatives were coexpressed in the E. coli BL21 DE3 strain together with N-terminally affinity-tagged (His10-SUMO in the case of Burkholderia prophage phi52237 PanT; His6 in all other cases) cognate PanT toxin. Filtered lysate was incubated with buffer-equilibrated Ni-beads, and PanAT complexes were eluted with 300 mM imidazole and resolved on 15% SDS-PAGE. The theoretical molecular weights for tagged toxins and antitoxins are indicated in red and green, respectively.
Some—but not all—PanAs carry additional N-terminal domain regions (Fig. 1). Often, these match a known helix-turn-helix (HTH) domain, of which a number of variations exist in the NCBI CDD. We aligned all the identified regions with hits to HTH models to make our own updated HTH model. From this, we identified HTH domains in the N-terminal regions of 343 PanA sequences (Dataset S1). HTH domains are often DNA binding, are frequently found in transcription factors, and are common in Type II antitoxins (1, 2). This suggests that, like many other Type II systems, Panacea domain–containing antitoxins can in some cases also regulate the TA function at the level of transcription. Apart from HTH domains, the only widely conserved N-terminal extension appears to correspond to a new domain, which we refer to as PanA-associated domain 1 (PAD1) (SI Appendix, Fig. S3). All but two of the TA-predicted PAD1 containing PanAs are paired with toxSAS-like toxins (the exception being putative ATPases from Clostridia [PanT group T62; Dataset S1]). The position at the N terminus and the presence of conserved histidines may indicate that PAD1 is a new DNA-binding domain, although it has no detectable homology with any known domain. PAD1 is also present in nine Panacea-containing proteins that do not meet the criteria for TA-like loci (Dataset S1). In all cases in which PanA contains the PAD1 domain and is in a TA-like locus, the toxin is encoded upstream of the antitoxin, the less common arrangement in the data set as a whole and in TA systems in general (1, 2).
PanA Is a Hyperpromiscuous Antitoxin Domain.
Sampling broadly across PanA diversity, we selected 25 of the putative novel TAs for experimental validation in toxicity neutralization assays (Figs. 1 and 2A, Table 1, SI Appendix, Table S1, and Dataset S2). Putative toxins and antitoxins were expressed in E. coli strain BW25113 under the control of arabinose- and isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible promoters, respectively (4). For a gene pair to classify as a bona fide TA, two criteria need to be fulfilled: 1) the expression of the toxin should compromise E. coli growth, and 2) coexpression with the antitoxin should—either fully or partially—rescue from growth inhibition by the toxin. In addition to the PanA-neutralized PhRel2Bac. sub. toxSAS from B. subtilis Ia1a that we have validated earlier (4), we have verified here nine PanAT pairs as being genuine TA loci (Table 1 and Fig. 2B).
Table 1.
Summary of experimentally characterized PanAT pairs
| Organism | Description | Toxin MOA/toxin family | Toxin accession | Antitoxin accession |
| Escherichia coli STEC O31 | PanAEsc. col.-PanTEsc. col. | Membrane | WP_000019185.1 | WP_000287252.1 |
| Helicobacter sp. 13S00482-2 | PanAHel.sp.-PanTHel.sp. | Membrane | WP_095295403.1 | WP_095295401.1 |
| Bartonella apis | PanABar. api.-PanTBar. api. | Membrane | WP_077993242.1 | WP_077993240.1 |
| Burkholderia prophage phi52237 | PanABur. phage-PanTBur. phage | Nucleotide cyclase | YP_293707.1 | YP_863932.1 |
| Bifidobacterium ruminantium | PanABif. rum.-PanTBif. rum. | RNase | WP_026646888.1 | WP_081815666.1 |
| Pseudomonas moraviensis | PanAPse. mor.-PanTPse. mor. | RNase | WP_083201923.1 | WP_083354365.1† |
| Vibrio harveyi | PanAVib. har.-CapRelVib. har. | toxSAS | WP_061065447.1 | WP_061065448.1 |
| Bacillus subtilis Ia1a | PanABac. sub.-PhRel2Bac. sub. | toxSAS | WP_090558406.1 | WP_090558408.1 |
| Lactobacillus animalis | PanALac. ani.-PanTLac. ani. | toxSAS | WP_052006344.1 | WP_035447700.1 |
| Corynebacterium doosanense | PanACor. doo.-PanTCor. doo. | Fic/Doc | WP_018022804.1 | WP_018022803.1 |
†The NBCI sequence of PanA from P. moraviensis appeared to be truncated at the N terminus relative to its homologues, and therefore we took an upstream start codon, equivalent to adding ten amino acids, MIFSEQKVAQ, to the N terminus.
PanA-neutralized toxins from Lactobacillus animalis (PhRel2Lac. ani.) and Vibrio harveyi (CapRelVib. har.) belong to two different toxSAS subfamilies and, as we have shown recently, the majority of toxSAS target translation by inhibiting tRNA aminoacylation through the pyrophosphorylation of the 3′ CCA end of tRNA (12). Toxins from Pseudomonas moraviensis strain LMG 24280 (PanTPse. mor.) and Bifidobacterium ruminantium strain DSM 6489 (PanTBif. rum.) have no hits against the NCBI CDD but are predicted to be structurally similar to EndoA/PemK/MazF family RNases with HHPred (25) and thus may act as translational inhibitors similarly to the archetypal TA toxin MazF that cleaves mRNA at ACA nucleotide sequences (29). The Corynebacterium doosanense toxin (PanTCor. doo.) is predicted to be a member of the Fic/Doc protein family, which includes the Doc TA toxin that inhibits protein synthesis by phosphorylating the essential translation elongation factor EF-Tu (21). Burkholderia prophage phi52237 (PanTBur. phage) has no detectable homology to any protein domain in the NCBI CDD. However, HHpred predicts similarity to adenylate and guanylate cyclase with 97% probability, suggesting that its toxicity could be via the production of a toxic cyclic nucleotide species. Finally, many of the predicted toxin genes encode putative small peptides with predicted transmembrane helices (SI Appendix, Fig. S4). Of the verified TAs, the toxins with putative membrane spanning segments are those originating from E. coli strain STEC O31 (PanTEsc. col.), Helicobacter sp. 13S00482-2 (PanTHel. sp.), and Bartonella apis strain BBC0122 (PanTBar. api.) (Table 1 and SI Appendix, Fig. S4). The clusters containing PanTEsc. col. (T3) and PanTBar. api. (T12) have similar sequence compositions consisting of a charged N-terminal region followed by a hydrophilic C-terminal region where the transmembrane regions are predicted (SI Appendix, Figs. S2A and S4). It is possible that T3 and T12 are homologous, although they are dissimilar enough that they are not clustered together by FlaGs (SI Appendix, Fig. S2A). The transmembrane helices of PanTHel. sp. are found at its C terminus, while its N-terminal region is similar to coiled-coil regions found in the synaptonemal complex protein 1 superfamily (30) and a Salmonella phage tail needle protein (25). For additional confirmation that PanTs vary substantially on the protein fold level, we de novo–predicted the structures of representative validated PanTs. using trRosetta, a deep learning–based method (31) (SI Appendix, Fig. S5). In agreement with the HHpred results, the PanTs are predicted to adopt different, structurally unrelated folds.
Of the potential TA pairs that were selected and could not be verified, three of the putative toxin genes could not be successfully plasmid subcloned by the commercial provider (SI Appendix, Table S1). While we cannot be sure of the reason for this, it is likely that their toxicity was too severe to allow cloning in E. coli. Five PanTs were toxic but were not rescued by their cognate PanA, and in three of these cases, PanA itself was toxic (SI Appendix, Table S1 and Fig. S6 A–D). For example, while the PanA-associated mRNAse MqsR from Herbaspirillum frisingense GSF30 was—as we predicted earlier (15)—toxic, its toxicity was not countered by its cognate PanA when coexpressed in E. coli (SI Appendix, Fig. S6C). Finally, eight PanTs were not toxic when tested in E. coli—but this does not rule out the possibility of toxicity in the original host (SI Appendix, Table S1).
PanAT Pairs Are Type II TA Systems.
The Panacea domain–containing SocA antitoxin of Caulobacter crescentus acts as a proteolytic adaptor, bringing the toxin SocB into contact with the protease ClpXP (17). To test whether other PanAs act as such adapters, we repeated our neutralization assays in E. coli strains lacking ClpXP and Lon proteases. These proteases are not necessary for neutralization by PanA (SI Appendix, Fig. S7). We therefore hypothesized that the general neutralization mechanism of PanA is through direct binding and inhibition typical of classical Type II systems. To test this, we carried out pull-down assays using coexpressed cognate native PanA antitoxins together with N-terminally affinity-tagged PanT toxins (with either His6 or His10-SUMO tags). We validated stable complex formation for five PanAT pairs: L. animalis PhRel2Lac. ani.:PanALac. ani. (Fig. 2C), P. moraviensis PanTPse. mor.:PanAPse. mor. (Fig. 2D), Burkholderia prophage phi52237 PanTBur. phage: PanABur. phage (Fig. 2E), C. doosanense Fic/DocCor. doo.: PanACor. doo. (Fig. 2F), and V. harveyi CapRelVib. har.:PanAVib. har. (Fig. 2G).
Protein Synthesis Is a Major Target of PanT Toxins.
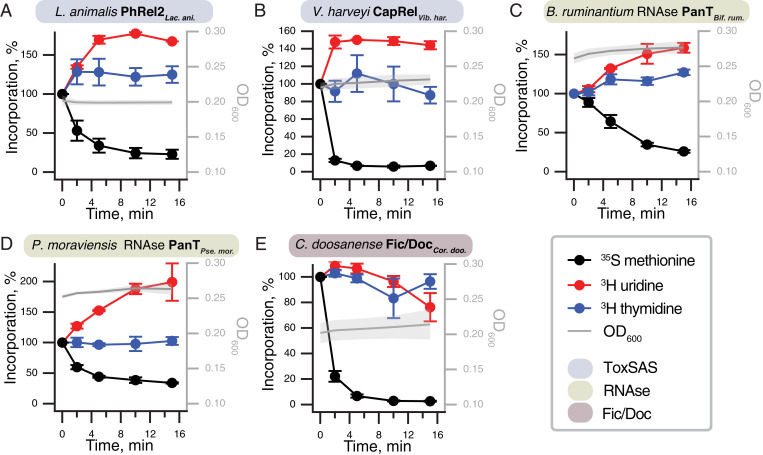
To address the molecular mechanisms of PanT toxicity, we assayed the effects of PanT expression on macromolecular synthesis by following the incorporation of 35S methionine in proteins, 3H uridine in RNA, and 3H thymidine in DNA, comparing to the effects of E. coli MazF RNase as a positive control (SI Appendix, Fig. S8A). As predicted, five of the identified PanT—L. animalis PhRel2Lac. ani. and V. harveyi CapRelVib. har. toxSAS, putative RNases PanTPse. mor. and PanTBif. rum. and C. doosanense Fic/Doc toxin, Fic/DocCor. doo.—specifically inhibit protein synthesis (Fig. 3 A–E). The mechanism of action of all the protein synthesis–inhibiting toxins can be predicted by homology. ToxSASs L. animalis PhRel2 and V. harveyi CapRel are closely related to other representatives we have characterized earlier (12) and almost certainly pyrophosphorylate the CCA end of tRNA. The C. doosanense Fic/Doc toxin Fic/DocCor. doo. presumably modifies EF-Tu as observed for other Doc enzymes (32). Predicted RNases PanTBif. rum. and PanTPse. mor. likely inhibit translation by cleaving mRNA or tRNA as do their—albeit distant—relatives (33).
Fig. 3.
Protein synthesis is a major target of PanT toxins. Metabolic-labeling assays following the incorporation of 35S methionine (black traces), 3H uridine (red), and 3H thymidine (blue) upon expression of translation-inhibiting PanT representatives: (A) L. animalis PhRel2 and (B) V. harveyi CapRel toxSAS; putative RNases (C) PanTBif. rum. and (D) PanTPse. mor.; and (E) C. doosanense Fic/Doc toxin, Fic/DocCor. doo. Expression of PanTs in E. coli BW25113 was induced with 0.2% L-arabinose.
Burkholderia Prophage phi52237 PanT Is a Pleiotropic Toxin that Induces the RelA-Mediated Stringent Response.
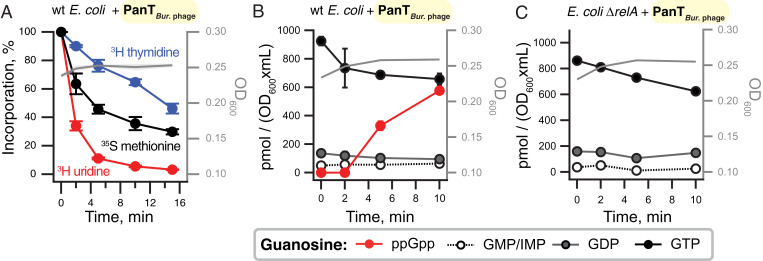
The Burkholderia prophage PanTBur. phage toxin is unique among our verified toxins in that it predominantly inhibits transcription, with weaker effects on translation and even weaker on replication (Fig. 4A). The mode of inhibition is reminiscent of that of C. marina FaRel toxSAS (4) and P. aeruginosa Type VI secretion system RSH effector Tas1 (12, 13) that act though production of the toxic nucleotide alarmone (pp)pApp, leading to dramatic depletion of ATP and GTP. Therefore, we used our high performance liquid chromatography (HPLC)-based approach to study the effects of PanTBur. phage toxin expression on E. coli nucleotide pools (34). In contrast to the drastic drop in GTP and ATP seen upon expression of C. marina FaRel toxSAS (4), expression of PanTBur. phage results in only a slight decrease in GTP (Fig. 4B) without affecting the ATP levels (SI Appendix, Fig. S9A). Surprisingly, despite having no detectable sequence or structural homology with RSH protein family members, PanTBur. phage expression causes an accumulation of the alarmone nucleotide ppGpp (Fig. 4B). This suggests that either 1) the toxin activates cellular RSH enzymes—given the strength of the effect, likely the most potent E. coli (p)ppGpp synthetase RelA—or 2) the PanTBur. phage toxin itself is capable of producing the alarmone. To distinguish between the two scenarios, we analyzed nucleotide levels upon toxin expression in an E. coli strain lacking relA. No accumulation of ppGpp is detected upon PanTBur. phage expression in the relA-deficient strain (Fig. 4C), and just as in the case of wild type, there is no effect on ATP levels (SI Appendix, Fig. S9B). Therefore, we conclude that expression of this toxin directly or indirectly induces ppGpp production by RelA. To deconvolute the direct effects of Burkholderia prophage PanTBur. phage toxin on 35S methionine, 3H uridine, and 3H thymidine incorporation from the secondary effects caused by RelA-dependent ppGpp accumulation, we performed metabolic labeling in the ΔrelA E. coli strain (SI Appendix, Fig. S9C). Just as in the wild-type strain, the main target is transcription, closely followed by translation. Thus, the growth inhibition and metabolic-labeling effects observed upon PanTBur. phage expression are not related to ppGpp accumulation.
Fig. 4.
PanTBur. phage toxin from Burkholderia prophage phi52237 compromises transcription and translation as well as inducing the RelA-mediated stringent response. (A) Metabolic-labeling assay using wild-type E. coli BW25113 expressing PanTBur. phage toxin. (B and C) Guanosine nucleotide pools in either wild-type (B) or ΔrelA (C) E. coli BW25113 expressing PanTBur. phage toxin. Cell cultures were grown in defined minimal MOPS medium supplemented with 0.5% glycerol at 37 °C with vigorous aeration. Expression of PanTBur. phage toxin was induced with 0.2% L-arabinose at the OD600 0.2. Intracellular nucleotides are expressed in pmol per OD600 × mL as per inset. Error bars indicate the SE of the arithmetic mean of three biological replicates.
The Cell Membrane Is Another Major Target of PanT Toxins.
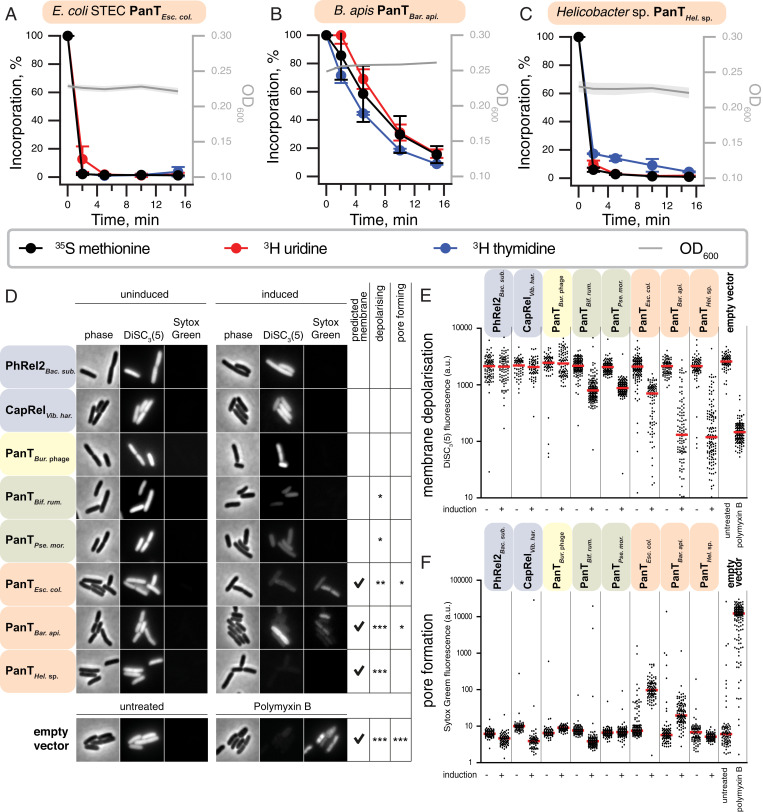
Next, we performed 35S methionine, 3H uridine, and 3H thymidine metabolic labeling experiments with the predicted transmembrane domain harboring toxins PanTEsc. col. (Fig. 5A), PanTBar. api. (Fig. 5B), and PanTHel. sp. (Fig. 5C). Unlike the toxins of the previous two sections of Results that predominantly target translation or transcription, expression of these toxins indiscriminately inhibited transcription, translation, and DNA replication, consistent with a more general shutdown of metabolic activities caused by membrane disruption. Indeed, a comparable response was observed with the induction of membrane-depolarizing E. coli HokB TA toxin (35) (SI Appendix, Fig. S8B) and treatment with the membrane-targeting inhibitor of oxidative phosphorylation carbonyl cyanide 3-chlorophenylhydrazone (SI Appendix, Fig. S8C).
Fig. 5.
Membrane integrity is a major target of PanT toxins. (A–C) Metabolic-labeling assays with wild-type E. coli BW25113 expressing (A) PanTEsc. col., (B) PanTBar. api., or (C) PanTHel. sp. toxins. (D) Phase-contrast (Left) and fluorescence images (Middle and Right) of E. coli cells costained with membrane potential–sensitive dye DiSC3(5) and membrane permeability indicator SYTOX Green. Depicted are representative cells carrying either an empty or PanT-expressing vector under uninducing (no L-arabinose) or inducing (30-min induction with 0.2% L-arabinose) conditions. As a positive control, cells containing empty vector (in MOPS-glucose medium) were incubated for 15 min with membrane-depolarizing and pore-forming antibiotic Polymyxin B. High DiSC3(5) fluorescence levels indicate high membrane potential levels found in well-energized, metabolically active cells. High SYTOX Green levels, in contrast, indicate formation of pores in the inner membrane. (E and F) Quantification of (E) DiSC3(5) and (F) SYTOX Green fluorescence for individual cells from the same imaging dataset (n = 92 to 165 cells). Median fluorescence intensity is indicated with a red line. Shading indicates toxin type as per Fig. 2A.
To directly test this hypothesis, we analyzed the integrity of cell membranes upon toxin induction using a combination of the membrane potential–sensitive dye “DiSC3(5)” (36) and inner membrane permeability indicator SYTOX Green (37). A strong membrane depolarization combined with an increased SYTOX Green permeability was observed for PanTBar. api. and PanTEsc. col. (Fig. 5 D–F). Expression of PanTHel. sp., in contrast, triggered strong depolarization without an increase in SYTOX Green permeability. Thus, we conclude PanTEsc. col., PanTHel. sp., and PanTBar. api. exert their toxic activity through membrane depolarization which, in the case of PanTEsc. col. and PanTBar. api., is caused by large pore formation. Finally, weak membrane depolarization was also observed for PanTBif. rum. and PanTPse. mor., although these are not predicted to contain transmembrane helices and are instead predicted to be RNases. Therefore, the effect of these toxins on cell membranes is more likely to be indirect through disturbances in respiration or central carbon metabolism. A potential membrane-spanning region is predicted for PanATBur. phage, albeit with relatively weak support (55%) (SI Appendix, Fig. S4D). As this protein does not appear to affect membrane integrity, its toxicity that is particularly striking in its effect on transcription as described above is more likely to result from its enzymatic activity, putatively cyclic nucleotide synthesis.
While PanAs Are Naturally Specific for their Cognate PanT Toxins, Their PanT Neutralization Spectrum Can Be Expanded through Directed Evolution.
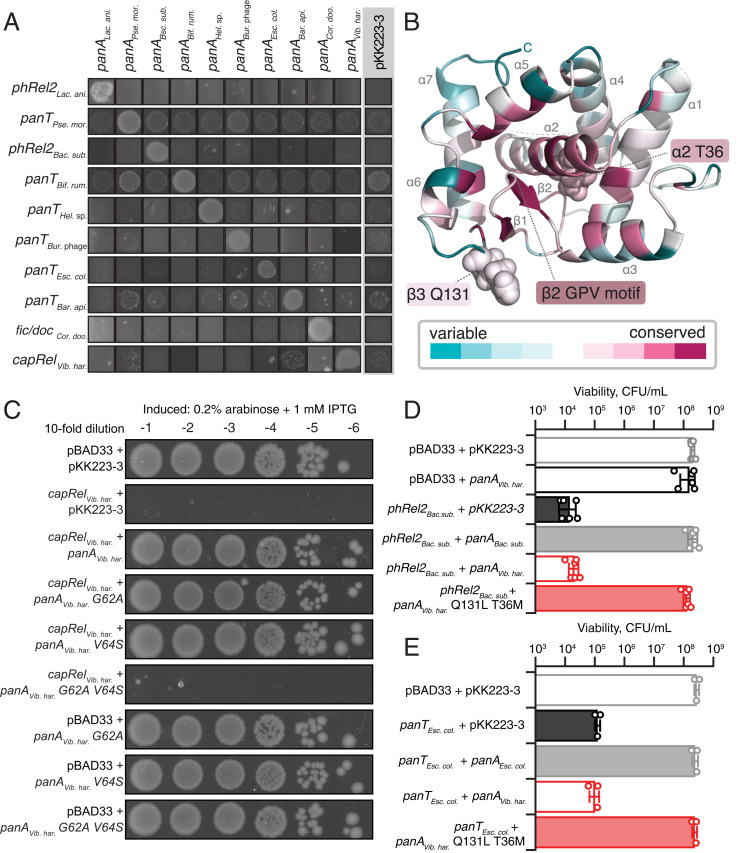
We have earlier shown that Type II antitoxins neutralizing toxSAS toxins—such as B. subtilis Ia1a PanABac. sub. neutralizing PhRel2Bac. sub.—are specific for their cognate toxins (4). PanA is clearly a versatile domain that can evolve to neutralize—and become specific for—a range of different toxin domains. Therefore, we performed exhaustive cross-inhibition testing, resulting in a 10 × 10 cross-neutralization matrix (Fig. 6A and SI Appendix, Fig. S10). A clear diagonal signal is indicative of PanA antitoxins naturally efficiently protecting only from cognate toxins—even within groups of evolutionary related toxic effectors such as toxSAS CapRelVib. har., PhRel2Lac. ani., and PhRel2Bac. sub. Conversely, on the evolutionary timescale, Panacea changes its toxin specificity and swaps partners, which raises the questions of what the structurally important regions for neutralization are and how a new specificity profile can be evolved.
Fig. 6.
PanA specificity can be readily evolved though directed evolution. (A) Exhaustive cross-neutralization testing establishes the strict specificity of PanA antitoxins toward their cognate toxins. The overnight cultures of E. coli strains transformed with pBAD33 and pKK223-3 vectors or derivatives thereof expressing toxin and PanA antitoxins was adjusted to 1.0, cultures serially diluted from 101- to 106-fold and spotted on LB agar medium supplemented with appropriate antibiotics as well as inducers (0.2% arabinose for toxin induction and 1 mM IPTG for induction PanA variants); 101-fold dilution is shown. (B) trRosetta-predicted structure (TM score 0.70, “very high confidence”) of the PanAVib. har. antitoxin colored by degree of conservation as per SI Appendix, Fig. S11. (C) Effect of amino acid substitutions in conserved GPV motif on of PanAVib. har. antitoxin functionality. (D) Neutralization of PhRel2Bac. sub. toxin by evolved noncognate PanAVib. har. T36M Q131L antitoxin. To quantify the effects of PanA/PanT coexpression on bacterial viability, the overnight cultures were subdiluted and spread on the LB agar, and individual colonies were counted. Analogous experiments with single-substituted T36M and Q131L PanAVib. har. variants are shown in SI Appendix, Fig. S11. (E) Neutralization of PanTEsc. col. toxin by wild-type and T36M Q131L PanAVib. har. variants.
The Panacea domain is not identifiably homologous to any protein with a known structure. Therefore, we have de novo predicted the structure of PanAVib. har. using trRosetta, a deep learning–based method (31) (Fig. 6B). The model has a confidence categorized as “very high,” with an estimated template modeling (TM) score of 0.704. Independent structural prediction with AlphaFold2 (10) indicates the same overall fold, with an RMSD of 1.05 Å as calculated by PyMol (SI Appendix, Fig. S11). The structure is comprised of a central helix (ɑ2) surrounded by five further helices and a small three-strand β-sheet that contains a strongly conserved GPV amino acid sequence motif in the β2 strand proximal to the central helix ɑ2 (Fig. 6B and SI Appendix, Fig. S12). The β3 and ɑ2 elements are particularly well conserved in the sequence alignment (SI Appendix, Fig. S12). We probed the functional importance of the GPV motif in toxicity neutralization assays. While individual G62A and V64S substitutions did not affect the ability of panAVib. har. to neutralize its cognate toxin capRelVib. har., the G62A V64S double substitution resulted in the loss of neutralization activity (Fig. 6C), supporting that the GPV motif is, indeed, functionally important.
Next, we subjected a pair of toxSAS:PanA TA systems with effectors belonging to two distinct toxSAS subfamilies—PhRel2 and CapRel—to directed evolution experiments and screened for mutant variants of PanAVib. har. that are able to neutralize B. subtilis PhRel2Bac. sub. Even though the amino acid identity between PanAVib. har. and PanABac. sub. proteins is only 30 to 40%, just two substitutions—T36M and Q131L—were sufficient for cell viability as judged by colony-counting experiments (Fig. 6D and SI Appendix, Fig. S13A). Individual T36M and Q131L substitutions are not sufficient to elicit cross-reactivity (SI Appendix, Fig. S13A). T36 is part of the well-conserved central helix ɑ2, while Q131 is located in a small, variable β3 strand. The β2 strand containing the conserved GPV motif is sandwiched between these structural elements (Fig. 6B). Notably, the T36M Q131L PanAVib. har. variant is still capable of protecting from the cognate CapRelVib. har. toxin. However, the protection from PhRel2Bac. sub. toxicity is less efficient than that conferred by the cognate PanABac. sub. antitoxin: the bacterial colonies are smaller, indicative of incomplete detoxification (SI Appendix, Fig. S13A). Therefore, we hypothesized that the T36M Q131L double substitution does not result in specificity switching in a strict sense but rather relaxes the specificity, thus allowing the neutralization of noncognate toxins. To probe this hypothesis, we tested if T36M Q131L PanAVib. har. could protect from other noncognate PanTs (Fig. 6E and SI Appendix, Fig. S13B). We found that T36M Q131L PanAVib. har. can protect from the noncognate cell membrane–targeting PanTEsc. col. (Fig. 6E), although incompletely, as evident from the smaller colony size (SI Appendix, Fig. S13B); no increased protection from other noncognate PanTs was detected (SI Appendix, Fig. S13C).
Discussion
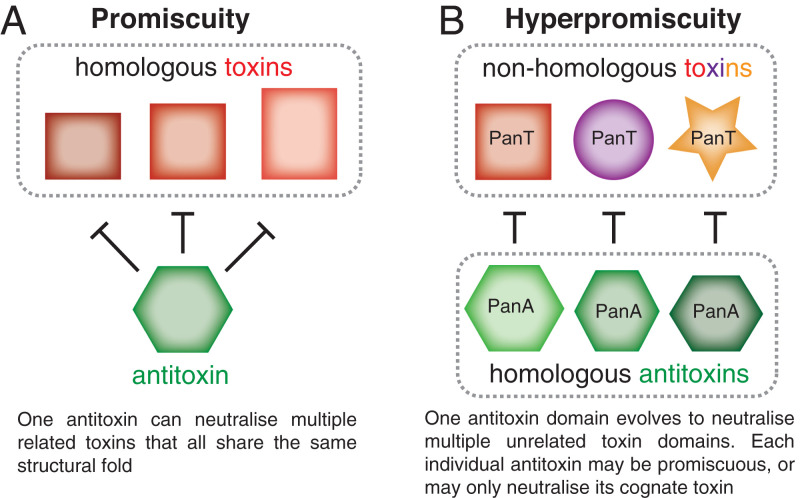
Type II TAs are highly specific at the sequence level; however, small changes can result in promiscuous intermediates allowing the neutralization of additional homologous but noncognate toxins (28, 38, 39). Through selection experiments, we have demonstrated that via just two amino acid substitutions, Panacea-containing antitoxins can be made to neutralize not just noncognate but nonhomologous noncognate toxins that have different cellular targets and mechanisms of action. This reveals a remarkable versatility of the Panacea domain. We suggest describing the ability of an antitoxin domain to evolve to neutralize different toxin domains as hyperpromiscuity, distinguishing from promiscuity, in which one individual antitoxin can neutralize noncognate but homologous toxins sharing the same structural fold (Fig. 7). A naturally occurring example of the latter can be seen in the bacteriophage T4 antitoxin Dmd that neutralizes the homologous mRNase toxins RnlA and LsoA (26, 27).
Fig. 7.
Antitoxin promiscuity versus hyperpromiscuity. (A) A promiscuous antitoxin has relaxed neutralization specificity toward its target toxin and can neutralize a range of related toxins which all share the same structural fold. Examples include cross regulation of RelBE-like modules in Mycobacterium tuberculosis (55) and promiscuous ParD antitoxins generated through directed evolution that neutralize noncognate ParE toxins (28). (B) A hyperpromiscuous antitoxin domain, as exemplified by Panacea, can evolve to neutralize unrelated toxins that share neither structural fold nor mechanism of action.
Other versatile antitoxin domains have also previously been observed in computational analyses to be associated with multiple toxin-like domains (16, 40, 41), indicating potentially similar plasticity and hyperpromiscuity. One example is the Phd-related antitoxin domain found in proteins that can neutralize RelE-like mRNases, in addition to those that neutralize the EF-Tu phosphorylating toxin Doc (40). DUF4065/Panacea has previously avoided identification as a widespread antitoxin domain, despite its broad distribution in prokaryotic chromosomes. Further bioinformatic investigations of TA systems are required to understand just how unique Panacea is in its hyperpromiscuity and how many more hyperpromiscuous TAs are waiting for discovery in the vast wealth of microbial genomes.
A number of other outstanding questions about PanA remain. Firstly, how is one single domain able to neutralize so many different toxins while insulating itself against noncognate interactions? The answer to this will come from structural analyses of multiple PanAs—both alone and in complex with cognate toxins—combined with additional directed evolution experiments using different PanA pairs and an analysis of sequence coevolution. A structural analysis of complexes will also reveal the molecular function of the conserved GPV motif that is a signature of the Panacea domain and is critical for neutralization. The second question is just how much of a role proteases play in the function of PanA in some species—given the previously observed function of the Panacea domain–containing antitoxin SocA in the proteolytic degradation of toxin SocB in Caulobacter (17). While our results are most consistent with a Type II direct mechanism of inactivation rather than the indirect Type VI–like mechanism observed for SocA, the two means of neutralization may not be mutually exclusive. Dual antitoxin functions have previously been observed; for example, DarG of the DarTG system both removes a toxic modification from DNA (Type IV) and directly binds to and inhibits the modifying enzyme DarT (Type II) (42). Finally, the evolutionary forces that drive and enable such ready partner swapping of PanAT pairs are unclear. One answer to this is hinted at in the kinds of proteins that are encoded near PanATs (Dataset S1) and in the analysis of TA (although not PanAT) gene locations near recombination sites of Tn3 transposases (43). We have found that many PanATs are encoded in close enough vicinity to transposase genes for the latter to be predicted as third component TA system genes or even false positive potential toxins that were filtered out by our pipeline (Dataset S1). It is not surprising, nor is it a new observation, that TAs can be associated with transposons; they can potentially act as addiction modules, similar to their role on plasmids (2). It is tempting to speculate that the presence of PanATs near hotspots of genomic rearrangements involving transposons and prophages—combined with an inbuilt versatility of the Panacea domain—could be driving the recombining of pairs that we observe.
Materials and Methods
Identification of PanA in Proteomes across the Tree of Life.
From the NCBI genomes index (http://ftp.ncbi.nlm.nih.gov/genomes), we downloaded 20,209 predicted proteomes, selecting all viruses and one representative proteome per species for archaea, bacteria, and eukaryotes. The full taxonomy was also retrieved from NCBI. To detect the presence of PanA across the tree of life, we used the HMM of the DUF4065 domain from the Pfam database (14). We used HMMer v3.1b2 (44) to scan our database of proteomes with the DUF4065 HMM using thresholds set to the HMM profile’s gathering cutoffs. We found that the DUF4065 domain was present in 2,281 identified sequences. We stored the sequences, taxonomy of the source organism, and domain composition in a MySQL database. We used this dataset and subsets of it for further phylogenetic analysis (SI Appendix, Methods: Representative sequence dataset assembly and Phylogenetic analysis).
Prediction of Sequence Features and Structure.
Structural modeling was carried out with the trRosetta server (31). This prediction is based on de novo folding, guided by deep learning restraints. The model was colored by conservation using the Consurf server and an alignment of the sequences shown in Fig. 2 (45). Additional structural prediction was carried out for PanAVib. har. with the AlphaFold2 (46) Colab notebook with default settings (“advanced” version; https://github.com/sokrypton/ColabFold). RSMD was calculated using structural alignment in PyMol v2.4.2 (pymol.org). Transmembrane regions were predicted with the TMHMM 2.0 sever (default settings). See SI Appendix, Methods: Prediction of sequence features and structure for details of sequence analyses for the prediction of protein domains and the identification of prophage-like genomic regions.
Prediction of TA Loci.
Our Python tool FlaGs (18), which takes advantage of the sensitive sequence search method Jackhmmer (44), was adapted to identify conserved two- or three-gene conserved architectures that are typical of TA loci. Full details of the method are described in SI Appendix, Methods: Prediction of TA loci, with a schematic of the workflow shown in SI Appendix, Fig. S1. All scripts and datasets are available at https://github.com/GCA-VH-lab/Panacea.
Metabolic Labeling with 35S Methionine, 3H Uridine, or 3H Thymidine.
Metabolic-labeling assays were performed as described previously (4). For details, see SI Appendix, Methods: Metabolic labeling.
Construction of Plasmids.
All bacterial strains and plasmids used in the study are listed in Dataset S3, and details can be found in SI Appendix, Methods: Construction of plasmids.
HPLC-Based Nucleotide Quantification.
E. coli strain BW2511324 and E. coli BW25113 ΔrelA were transformed with PanTBur. phage–expressing plasmid (pBAD33—Burkholderia prophage phi52237) as well as empty pKK223-3 vector. The starter cultures were pregrown overnight at 37 °C with vigorous shaking (200 rpm) in Neidhardt MOPS minimal media supplemented with 1 µg/mL thiamine, 1% glucose, 0.1% caa, 100 µg/mL carbenicillin, and 20 µg/mL chloramphenicol. The overnight cultures were diluted to optical density (OD)600 0.05 in 115 mL prewarmed medium MOPS supplemented with 0.5% glycerol as carbon source and grown until OD600 ∼ 0.2 at 37 °C, 200 rpm. At this point, 0.2% arabinose was added to induce the expression of the toxin. A total of 26-mL samples were collected for HPLC analyses at 0, 2, 5, and 10 min after the addition of arabinose and 1 mM IPTG. Nucleotide extraction and HPLC analyses were performed as described previously (34). The OD600 measurements were performed in parallel with a collection of the samples for HPLC analyses.
Toxicity Neutralization Assays.
Toxicity-neutralization assays were performed on Lysogeny broth (LB) medium (Lennox) plates (VWR). E. coli BW25113 strains transformed with pBAD33 derivative plasmids encoding toxins [medium copy number, p15A origin of replication, CmlR, toxins are expressed under the control of a PBAD promoter (47)] and pKK223-3 derivatives encoding antitoxins [medium copy number, ColE1 origin of replication, AmpR, antitoxins are expressed under the control of a PTac promoter (48)] were grown in liquid LB medium (BD) supplemented with 100 µg/mL carbenicillin (AppliChem) and 20 µg/mL chloramphenicol (AppliChem) as well as 1% glucose (repression conditions). Serial 10-fold dilutions were spotted (5 µL per spot) on solid LB plates containing carbenicillin and chloramphenicol in addition to either 1% glucose (repressive conditions) or 0.2% arabinose combined with 1 mM IPTG (induction conditions). Plates were scored after an overnight incubation at 37 °C.
To quantify bacterial viability (colony forming units, CFU), overnight cultures were diluted to OD600 either in the range from 0.1 to 0.01 (for the strains expressing PhRel2Bac. sub., with and without coexpression of wild-type PanAVib. har.) or OD600 ranging from 1.0 × 10−4 to 1.0 × 10−5 (all other strains) and spread on the LB agar medium as described above for the spot-test toxicity neutralization assay. The final CFU/mL estimates were normalized to OD600 1.0.
PanAT Complex Formation.
The plasmids were transformed into the E. coli BL21 DE3 strain. Fresh transformants were washed from an LB (BD Difco-Fisher Scientific) agar plate and used to inoculate a 1-L culture LB supplemented with kanamycin (50 µg/mL). The cells were grown on 37 °C until OD600 reached 0.4 to 0.5 and induced with 0.5 mM IPTG. The cells were harvested after overnight cultivation on 18 °C, 220 rpm. The cells were opened with sonication in binding buffer (BB: 25 mM Hepes, pH 7.5; 300 mM NaCl; 10 mM imidazole; 2 mM CaCl2; 2 mM β-ME). Filtered lysate was incubated with 1 mL previously buffer-equilibrated Ni-beads (His60 Ni Superflow Resin, TaKaRa) for 30 min. Bound protein was washed with BB on a gravity column and eluted with 300 mM imidazole. Fractions were resolved on a 15% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) gel and stained with SimplyBlue SafeStain.
Fluorescence Microscopy.
Overnight cultures were grown at 37 °C in MOPS minimal medium supplemented with 1% glucose. Next morning, the cells were washed, resuspended, and diluted in MOPS medium supplemented with 0.5% glycerol in order to remove glucose followed by incubation at 37 °C until an OD600 of 0.3. For maintaining the plasmids, all cultures were grown in the presence of 34 µg/mL chloramphenicol. Toxin production was induced by the addition of arabinose (0.2%) for 30 min followed by staining with 200 nM of the membrane permeability indicator SYTOX Green (37) alongside the induction and 250 nM membrane potential–sensitive dye DiSC3(5) (49) for the last 5 min (36, 50). The samples were immobilized on microscope slides covered with a thin layer of H2O/1.2% agarose and imaged immediately. As a positive control for pore formation, BW25113 E. coli cells transformed with the empty pBAD33 vector were incubated with 10 µg/mL polymyxin B for 15 min (51). Microscopy was performed using a Nikon Eclipse Ti equipped with a Nikon Plan Apo 100×/1.40 Oil Ph3 objective, CoolLED pE-4000 light source, Photometrics BSI sCMOS camera, and Chroma 49002 (excitation [EX] 470/40, dichroic mirror [DM] 495 lpxr, and emission [EM] 525/50) and Semrock Cy5-4040C (EX 628/40, DM 660 lp, EM 692/40) filter sets. The images were acquired with Metamorph 7.7 (MolecularDevices) and analyzed with Fiji (52).
Selection of Cross-Inhibiting PanA Mutants.
An error-prone PCR mutant library of Vibrio Harveyii PanA antitoxins was created as described in SI Appendix, Methods: Selection of cross-neutralizing PanAs: preparation of the antitoxin mutant library. A total of 5 µL (around 1 µg) antitoxin mutant library was transformed into the BW25113 E. coli strain carrying a noncognate toxin expression plasmid PhRel2Bac. sub. toxSAS toxin from B. subtilis Ia1a (VHp303). The transformants were let to recover for 1 h in 1 mL SOC media at 37 °C and added to 20 mL LB media supplemented with ampicillin (100 µg/mL), chloramphenicol (25 µg/mL), 0.2% L-arabinose, and 1 mM IPTG. The bacteria were grown overnight at 37 °C while expressing both toxin and antitoxin. Next day, the plasmid was extracted from 3 mL culture using a Favorprep Plasmid Extraction Mini Kit (Favorgen Biotech Corp.). A total of 500 ng plasmid mix was again transformed into BW25113 carrying a toxin expression plasmid and let to recover as before. A total of 100 µL recovery culture was spread on LB agar plates containing corresponding antibiotics as well as 0.2% glucose (control of transformation efficiency), and the rest of the culture was collected by centrifugation and spread on an LB agar plate containing corresponding antibiotics as well as 0.2% L-arabinose and 1 mM IPTG.
Overnight cultures were started from selected colonies for further testing of cross-inhibition. The plasmids were extracted with Favorprep Plasmid Extraction Mini Kit and cleaved with FastDigest SacI restriction enzyme (Thermo Scientific) to eliminate the toxin plasmids. To ensure the purity of the antitoxin mutant mix, it was transformed into the E. coli DH5α strain, and the plasmids were extracted from the offspring of a single colony. The E. coli BW25113 strain expressing the cognate or noncognate toxin was then transformed with 500 ng mutated plasmid. Again, 100 µL recovery culture was spread onto LB supplemented with corresponding antibiotics as well as 0.2% glucose agar plates, and the rest of the bacteria was collected and spread on LB agar plates supplemented with corresponding antibiotics, 0.2% arabinose and 1 mM IPTG. Phusion High-Fidelity DNA Polymerase (Thermo Scientific) was used to amplify the panA mutant (pK223_fwd_CPEC and pK223_rev_CPEC primers) and toxin genes (pBAD_fwd and pBAD_rev primers) with colony PCR and sequenced using pK223_rev_CPEC or pBAD_fwd primer correspondingly. The plasmid mixes and bacterial colonies were tested for possible contamination at various steps using FIREPol DNA Polymerase (Solis BioDyne): antitoxins were tested with the combination of pK223_rev_CPEC and STEC_panA_ctrl2, VH_panA_ctrl1, or Bsup_panA_ctrl1 primers and toxins with the combination of pBAD_fwd and STEC_TOX_ctrl1, VH_TOX_ctrl1, and Bsup_TOX_ctrl1 (Dataset S3).
Supplementary Material
Acknowledgments
We are thankful to Tsigereda Ghebretnsae Ghebrelul and Constantine Stavropoulos for their help with microbiological assays, Niilo Kaldalu and Villu Kasari for sharing an E. coli strain lacking Lon and ClpXP, Michael Laub for sharing SocA/B-encoding plasmids, and to Bhanu Chouhan for structural prediction with the AlphaFold2 Colab notebook. We are also grateful to Mikael Lindberg and the Umeå University Protein Expression Platform. This research was supported by grants from the Knut and Alice Wallenberg Foundation (2020.0037 to G.C.A.), Umeå University Medical Faculty (Biotechnology Grant to G.C.A.), Umeå Center for Microbial Research (Gender Policy Grant to G.C.A.), Kempestiftelserna (SMK-1858.3 to G.C.A. and Scholarship Fall 2020 to C.K.S.), Carl Tryggers Stiftelse för Vetenskaplig Forskning (CTS19:24 to G.C.A.), the Swedish Research Council (Vetenskapsrådet) Grants (2017-03783 to V.H. and 2019-01085 to G.C.A.), Ragnar Söderbergs Stiftelse (to V.H.), the European Union from the European Regional Development Fund through the Center of Excellence in Molecular Cell Engineering (2014 to 2020.4.01.15-0013 to T.T. and V.H.), Molecular Infection Biology Estonia—Research Capacity Building Project (H2020-WIDESPREAD-2018-2020/GA: 857518 to T.T. and V.H.), and the Estonian Research Council (PRG335 to T.T. and V.H.). The V.H. group was also supported by the Swedish Research Council (2018-00956 to V.H.) within the RIBOTARGET consortium under the framework of The Joint Programming Initiative on Antimicrobial Resistance (JPIAMR). H.S. was supported by UKRI (UK Research and Innovation) Biotechnology and Biological Sciences Research Council Grant (BB/S00257X/1) and J.A.B. by UKRI Medical Research Council Grant MR/N013840/1.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. G.v.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102212119/-/DCSupplemental.
Data Availability
Python code and text files of alignments, trees, and HMMs have been deposited in GitHub (https://github.com/GCA-VH-lab/Panacea) (56). All other study data are included in the article and/or supporting information.
References
- 1.Harms A., Brodersen D. E., Mitarai N., Gerdes K., Toxins, targets, and triggers: An overview of toxin-antitoxin biology. Mol. Cell 70, 768–784 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Fraikin N., Goormaghtigh F., Van Melderen L., Type II toxin-antitoxin systems: Evolution and revolutions. J. Bacteriol. 202, e00763-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hallez R., et al. , New toxins homologous to ParE belonging to three-component toxin-antitoxin systems in Escherichia coli O157:H7. Mol. Microbiol. 76, 719–732 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Jimmy S., et al. , A widespread toxin-antitoxin system exploiting growth control via alarmone signaling. Proc. Natl. Acad. Sci. U.S.A. 117, 10500–10510 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dedrick R. M., et al. , Prophage-mediated defence against viral attack and viral counter-defence. Nat. Microbiol. 2, 16251 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo Y., et al. , RalR (a DNase) and RalA (a small RNA) form a type I toxin-antitoxin system in Escherichia coli. Nucleic Acids Res. 42, 6448–6462 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Bruyn P., Girardin Y., Loris R., Prokaryote toxin-antitoxin modules: Complex regulation of an unclear function. Protein Sci. 30, 1103–1113 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogura T., Hiraga S., Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. U.S.A. 80, 4784–4788 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopatina A., Tal N., Sorek R., Abortive infection: Bacterial suicide as an antiviral immune strategy. Annu. Rev. Virol. 7, 371–384 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Song S., Wood T. K., A primary physiological role of toxin/antitoxin systems is phage inhibition. Front. Microbiol. 11, 1895 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haseltine W. A., Block R., Gilbert W., Weber K., MSI and MSII made on ribosome in idling step of protein synthesis. Nature 238, 381–384 (1972). [DOI] [PubMed] [Google Scholar]
- 12.Kurata T., et al. , RelA-SpoT Homolog toxins pyrophosphorylate the CCA end of tRNA to inhibit protein synthesis. Mol. Cell 81, 3160–3170.e9 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Ahmad S., et al. , An interbacterial toxin inhibits target cell growth by synthesizing (p)ppApp. Nature 575, 674–678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Gebali S., et al. , The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaldalu N., Kasari V., Atkinson G., Tenson T., “Type II toxin-antitoxin loci: The unusual mqsRA locus” in Prokaryotic Toxin-Antitoxins, Gerdes K., Ed. (Springer Berlin Heidelberg, Berlin, Heidelberg, 2013), pp. 93–105. [Google Scholar]
- 16.Makarova K. S., Wolf Y. I., Koonin E. V., Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct 4, 19 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aakre C. D., Phung T. N., Huang D., Laub M. T., A bacterial toxin inhibits DNA replication elongation through a direct interaction with the β sliding clamp. Mol. Cell 52, 617–628 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saha C. K., Sanches Pires R., Brolin H., Delannoy M., Atkinson G. C., FlaGs and webFlaGs: Discovering novel biology through the analysis of gene neighbourhood conservation. Bioinformatics 37, 1312–1314 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamaguchi Y., Park J. H., Inouye M., MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J. Biol. Chem. 284, 28746–28753 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christensen S. K., Pedersen K., Hansen F. G., Gerdes K., Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332, 809–819 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Castro-Roa D., et al. , The Fic protein Doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat. Chem. Biol. 9, 811–817 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gross M., Marianovsky I., Glaser G., MazG – A regulator of programmed cell death in Escherichia coli. Mol. Microbiol. 59, 590–601 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Bordes P., et al. , SecB-like chaperone controls a toxin-antitoxin stress-responsive system in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 108, 8438–8443 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camacho C., et al. , BLAST+: Architecture and applications. BMC Bioinformatics 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimmermann L., et al. , A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Short F. L., Blower T. R., Salmond G. P., A promiscuous antitoxin of bacteriophage T4 ensures successful viral replication. Mol. Microbiol. 83, 665–668 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Otsuka Y., Yonesaki T., Dmd of bacteriophage T4 functions as an antitoxin against Escherichia coli LsoA and RnlA toxins. Mol. Microbiol. 83, 669–681 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Aakre C. D., et al. , Evolving new protein-protein interaction specificity through promiscuous intermediates. Cell 163, 594–606 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y., et al. , MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12, 913–923 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Marchler-Bauer A., et al. , CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J., et al. , Improved protein structure prediction using predicted interresidue orientations. Proc. Natl. Acad. Sci. U.S.A. 117, 1496–1503 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veyron S., Peyroche G., Cherfils J., FIC proteins: From bacteria to humans and back again. Pathog. Dis. 76, fty012 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Masuda H., Inouye M., Toxins of prokaryotic toxin-antitoxin systems with sequence-specific endoribonuclease activity. Toxins (Basel) 9, 140 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varik V., Oliveira S. R. A., Hauryliuk V., Tenson T., HPLC-based quantification of bacterial housekeeping nucleotides and alarmone messengers ppGpp and pppGpp. Sci. Rep. 7, 11022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerdes K., et al. , The hok killer gene family in gram-negative bacteria. New Biol. 2, 946–956 (1990). [PubMed] [Google Scholar]
- 36.Te Winkel J. D., Gray D. A., Seistrup K. H., Hamoen L. W., Strahl H., Analysis of antimicrobial-triggered membrane depolarization using voltage sensitive dyes. Front. Cell Dev. Biol. 4, 29 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roth B. L., Poot M., Yue S. T., Millard P. J., Bacterial viability and antibiotic susceptibility testing with SYTOX green nucleic acid stain. Appl. Environ. Microbiol. 63, 2421–2431 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walling L. R., Butler J. S., Structural determinants for antitoxin identity and insulation of cross talk between homologous toxin-antitoxin systems. J. Bacteriol. 198, 3287–3295 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lite T. V., et al. , Uncovering the basis of protein-protein interaction specificity with a combinatorially complete library. eLife 9, e60924 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leplae R., et al. , Diversity of bacterial type II toxin-antitoxin systems: A comprehensive search and functional analysis of novel families. Nucleic Acids Res. 39, 5513–5525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akarsu H., et al. , TASmania: A bacterial toxin-antitoxin systems database. PLoS Comput. Biol. 15, e1006946 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jankevicius G., Ariza A., Ahel M., Ahel I., The toxin-antitoxin system DarTG catalyzes reversible ADP-ribosylation of DNA. Mol. Cell 64, 1109–1116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lima-Mendez G., et al. , Toxin-antitoxin gene pairs found in Tn3 family transposons appear to be an integral part of the transposition module. MBio 11, e00452-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eddy S. R., Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashkenazy H., et al. , ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guzman L. M., Belin D., Carson M. J., Beckwith J., Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brosius J., Holy A., Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc. Natl. Acad. Sci. U.S.A. 81, 6929–6933 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waggoner A., Optical probes of membrane potential. J. Membr. Biol. 27, 317–334 (1976). [DOI] [PubMed] [Google Scholar]
- 50.Kepplinger B., et al. , Mode of action and heterologous expression of the natural product antibiotic vancoresmycin. ACS Chem. Biol. 13, 207–214 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Vaara M., Agents that increase the permeability of the outer membrane. Microbiol. Rev. 56, 395–411 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Letunic I., Bork P., Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang M., Gao C., Wang Y., Zhang H., He Z. G., Characterization of the interaction and cross-regulation of three Mycobacterium tuberculosis RelBE modules. PLoS One 5, e10672 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.C. K. Saha, G. C. Atkinson, GCA-VH-lab/Panacea. Panacea data and scripts. https://github.com/GCA-VH-lab/Panacea. Deposited 15 November 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Python code and text files of alignments, trees, and HMMs have been deposited in GitHub (https://github.com/GCA-VH-lab/Panacea) (56). All other study data are included in the article and/or supporting information.