Significance
We found that BRD4 interacts with the LSD1/NuRD complex and colocalizes with this repressive complex on super-enhancers and that decommissioning BRD4-directed super-enhancer organization of transcription repression programs inflicts widespread drug resistance in breast cancer. The study uncovers a functional duality of BRD4 in super-enhancer organization of transcription activation and repression linking to oncogenesis and chemoresistance, respectively, supporting the pursuit of a combined targeting of BRD4 and PELI1 in effective treatment of breast cancer.
Keywords: BRD4, super-enhancer, LSD1/NuRD complex, JQ1 resistance
Abstract
BRD4 is well known for its role in super-enhancer organization and transcription activation of several prominent oncogenes including c-MYC and BCL2. As such, BRD4 inhibitors are being pursued as promising therapeutics for cancer treatment. However, drug resistance also occurs for BRD4-targeted therapies. Here, we report that BRD4 unexpectedly interacts with the LSD1/NuRD complex and colocalizes with this repressive complex on super-enhancers. Integrative genomic and epigenomic analyses indicate that the BRD4/LSD1/NuRD complex restricts the hyperactivation of a cluster of genes that are functionally linked to drug resistance. Intriguingly, treatment of breast cancer cells with a small-molecule inhibitor of BRD4, JQ1, results in no immediate activation of the drug-resistant genes, but long-time treatment or destabilization of LSD1 by PELI1 decommissions the BRD4/LSD1/NuRD complex, leading to resistance to JQ1 as well as to a broad spectrum of therapeutic compounds. Consistently, PELI1 is up-regulated in breast carcinomas, its level is negatively correlated with that of LSD1, and the expression level of the BRD4/LSD1/NuRD complex–restricted genes is strongly correlated with a worse overall survival of breast cancer patients. Together, our study uncovers a functional duality of BRD4 in super-enhancer organization of transcription activation and repression linking to oncogenesis and chemoresistance, respectively, supporting the pursuit of a combined targeting of BRD4 and PELI1 in effective treatment of breast cancer.
Bromodomain-containing protein 4 (BRD4), along with BRD2, BRD3, and testes/oocyte-specific BRDT, constitutes the bromodomain and extraterminal (BET) protein family in mammals (1). At the molecular level, these proteins act as epigenetic readers to specifically recognize acetylated lysine on histones (2), and biologically, the BET family is implicated in a broad spectrum of cellular processes such as cell proliferation, differentiation, metabolism, and DNA repair (3–6).
BRD4 is the best-characterized member of the BET family. Initially identified in the mammalian Mediator complex (7), a multiprotein assembly that links transcription factors to RNA polymerase II (Pol II), BRD4 is subsequently described as a general regulator for Pol II–dependent transcription through interaction with P-TEFb (8). Consistently, analysis of the genomic landscape of BRD4 found that BRD4 is associated with essentially all active promoters and a significant proportion of active enhancers in the genome of various normal and transformed cell types (9, 10). In concordance with its general role in transcription regulation, BRD4 has also been implicated in multiple pathological states, particularly inflammation, obesity, and tumorigenesis including midline carcinoma (11), acute myeloid leukemia (12), gastric cancer (6), and breast cancer (13). The tumorigenic potential of BRD4 has been largely attributed to its role in enhancing transcription elongation of genes such as c-MYC (14) and BCL2 (15) that are essential for the cell cycle and apoptosis. As such, BET proteins are being pursued as exciting novel targets for cancer treatment. Potent BET inhibitors with promising antitumor efficacy in a number of preclinical cancer models have been developed in recent years, and encouraging signs of efficacy in suppressing tumor growth by repressing the expression of oncogenes have already been reported (14). One of the BET inhibitors is JQ1 (16), a small molecule that occupies the bromodomain pocket of BET proteins with a high affinity in a manner that is competitive with acetylated histones. However, drug resistance also occurs for JQ1 (17, 18). Considering a wide range of sensitivity to JQ1, the intrinsic nature of JQ1 resistance needs to be explored to gain an optimal efficacy in epigenetic therapies.
Super-enhancers are clusters of enhancers that are occupied by master regulators such as Oct4, Sox2, BRD4, and Mediator (19, 20) in close genomic proximity. These enhancers are demarcated by certain epigenetic signatures, such as high enrichments of histone 3 lysine 4 monomethylation (H3K4me1) and H3K27 acetylation (H3K27ac) (21, 22). It is believed that super-enhancers govern lineage-specific gene expression and regulate oncogene activation to drive tumorigenesis (23). Accordingly, inhibition of super-enhancer–dependent transcription also represents a promising therapeutic strategy for cancer intervention (24, 25). Interestingly, further genome-wide analyses indicated that BRD4 is preferentially bound to super-enhancers (9, 10). Indeed, it was shown that super-enhancer–associated genes are highly sensitive to JQ1 (24). Nevertheless, the functional link between super-enhancers and JQ1 resistance is poorly understood.
The vertebrate Mi-2/nucleosome–remodeling and deacetylase (NuRD) complex is a multisubunit protein assembly that possesses both chromatin-remodeling ATPase and histone deacetylase and functions in transcription repression (26, 27). The NuRD complex contains several subunits whose pattern of expression is heterogeneous in different cell and tissue types, conferring this assembly with additional regulatory capacity and unique property (28). We reported previously that lysine-specific demethylase 1 (LSD1) is enlisted to the NuRD complex, expanding the enzymatic repertoire of this complex to include a histone demethylase (29). Owing to its importance in the regulation of histone methylation dynamics, LSD1 has also been implicated in various disease states (30, 31) and is also being pursued as a therapeutic target for cancers (32). However, despite the role of LSD1 in transcription regulation and the significance of its catalyzed demethylation of H3K4me1/2, the influence of LSD1 on the function of super-enhancers has been largely unexplored.
Pellino (PELI) proteins are signal-responsive ubiquitin ligases that are characterized by a cryptic fork head–associated domain for substrate recognition and an atypical RING domain catalyzing K63- or K48-linked polyubiquitination (33). Recent studies indicate that these proteins play important roles in innate immunity, DNA repair, and tumorigenesis (34, 35). Specifically, loss of PELI1 leads to hyperactivation and nuclear accumulation of c-Rel that contribute to the development of autoimmune disease (33); in diffuse large B-cell lymphoma, PELI1 expression is positively correlated with the expression of MYC, BCL6, BCL2, and MUM1 (34, 36), and high expression of PELI1 is associated with a frequent bone-marrow dissemination and shorter relapse-free survival (36). However, the potential role for PELI1 in breast carcinogenesis is currently unknown.
In the current study, we report an unexpected interaction of BRD4 with the LSD1/NuRD repression complex. We show that the BRD4/LSD1/NuRD complex physically occupies super-enhancers and functionally restricts the activation of a panel of genes including those that have well-established roles in drug resistance. We find that long-time treatment of breast cancer cells with BRD4 inhibitor JQ1 or destabilizing LSD1 by PELI1 decommissions the BRD4/LSD1/NuRD complex, leading to resistance to JQ1 as well as to a broad spectrum of antitumor compounds. We explore the clinicopathological significance of the PELI1-LSD1-BRD4/LSD1/NuRD axis in breast carcinogenesis.
Results
BRD4 Is Physically Associated with the LSD1/NuRD Complex.
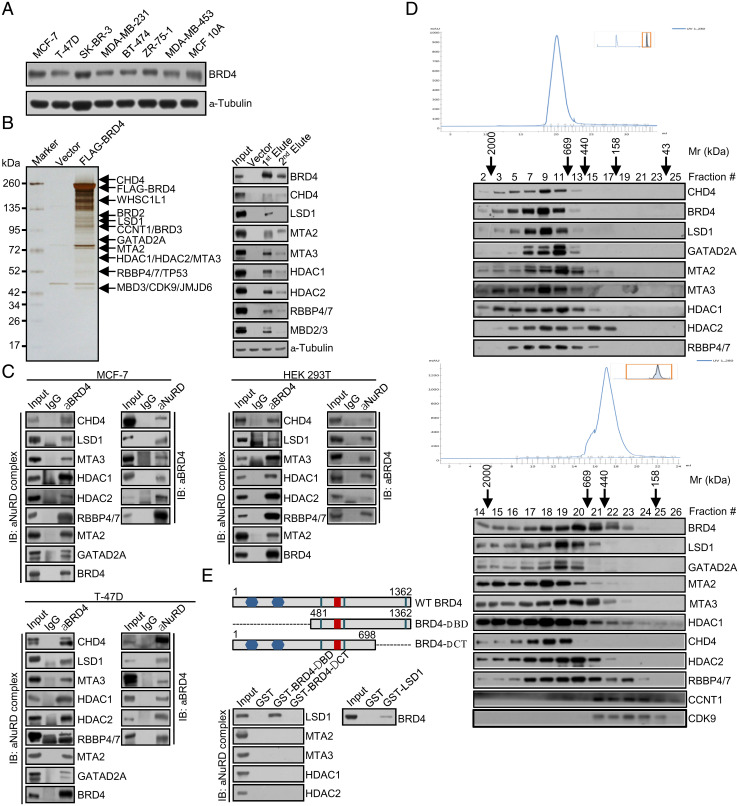
The molecular mechanism underlying breast carcinogenesis has long been the focus in our laboratory (29, 37–40). In order to further explore the role of BRD4 in the development and progression of breast cancer and to gain mechanistic insights into the drug resistance to JQ1, we first profiled the expression pattern of BRD4 in different breast cancer cell lines. Cellular proteins were extracted and analyzed by Western blotting with a monoclonal antibody against BRD4. The results showed that endogenous BRD4 is a protein with a molecular weight of ∼172 kDa and that BRD4 is expressed at variable levels in different cell lines (Fig. 1A).
Fig. 1.
BRD4 is physically associated with the LSD1/NuRD complex. (A) BRD4 expression in breast cancer cell lines. Cellular proteins were extracted from the indicated cell lines for Western blotting analysis using a monoclonal antibody against BRD4. (B) Immunopurification and mass spectrometry analysis of BRD4-associated proteins. Cellular extracts from FLAG-BRD4–expressing MCF-7 cells were subjected to affinity purification with anti-FLAG affinity columns and eluted with excess FLAG peptides. The elutes were resolved by SDS-PAGE and silver stained (Left). The protein bands were retrieved and analyzed by mass spectrometry. Column-bound proteins were analyzed by Western blotting using antibodies against the indicated proteins (Right). (C) Coimmunoprecipitation assays in MCF-7 cells, T-47D cells, or HEK 293T cells with anti-BRD4 followed by immunoblotting (IB) with antibodies against the indicated proteins or with antibodies against the indicated proteins followed by IB with anti-BRD4. (D) Cofractionation of BRD4 and the LSD1/NuRD complex by FPLC in MCF-7 cells or FLAG-BRD4–expressing MCF-7 cells. Chromatographic elution profiles and IB of the chromatographic fractions are shown. Equal volume from each fraction was analyzed and the elution positions of the calibration proteins with known molecular masses (Kilodaltons, kDa) are indicated. (E) Schematic diagrams of BRD4 deletion mutants (Upper). GST pull-down assays with bacterially expressed GST or GST-fused BRD4 deletion mutants and in vitro–transcribed/translated individual components of the LSD1/NuRD complex as indicated (Lower Left). Reciprocal GST pull-down experiments with GST-fused LSD1 and in vitro–transcribed/translated BRD4 as indicated (Lower Right).
We then employed immunoaffinity purification-coupled mass spectrometry to interrogate the BRD4 interactome in vivo. To this end, FLAG-tagged BRD4 (FLAG-BRD4, NM_058243.3) was stably expressed in human breast adenocarcinoma MCF-7 cells. Cellular extracts were prepared and subjected to immunoaffinity purification using anti-FLAG M2 magnetic beads. The bound proteins were eluted, resolved, and visualized by silver staining on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Mass spectrometric analysis revealed that BRD4 was copurified with TP53, JMJD6, WHSC1L1, and two components of the P-TEFb complex, CDK9 and CCNT1 (Fig. 1B), all known to interact with BRD4 (8, 41–43). Unexpectedly, LSD1, CHD4, MTA2, MTA3, HDAC1, HDAC2, and RBBP4/7, all the components of the LSD1/NuRD complex (29), were also detected in the BRD4 interactome (Fig. 1B). Copurification of the components of the LSD1/NuRD complex with BRD4 was verified by Western blotting of column-bound proteins with antibodies against the corresponding proteins (Fig. 1B). The detailed result of the mass spectrometry is provided in SI Appendix, Table S1.
To confirm the interaction of BRD4 with the LSD1/NuRD complex, coimmunoprecipitation experiments were carried out in MCF-7 cells with antibodies detecting endogenous proteins. Immunoprecipitation with antibodies against BRD4 followed by immunoblotting with antibodies against LSD1, CHD4, MTA2, MTA3, HDAC1, HDAC2, or RBBP4/7 demonstrated that all these proteins were efficiently coimmunoprecipitated with BRD4 (Fig. 1C). Reciprocally, immunoprecipitation with antibodies against LSD1, CHD4, or MTA3 followed by immunoblotting with antibodies against BRD4 also showed that BRD4 was efficiently coimmunoprecipitated with these components of the LSD1/NuRD complex (Fig. 1C). Similar results were also obtained in human breast carcinoma T-47D cells as well as in human embryonic kidney (HEK) 293T cells (Fig. 1C).
To further support the interaction between BRD4 and the LSD1/NuRD complex, nuclear proteins from MCF-7 cells were fractionated by fast protein liquid chromatography (FPLC) with Superose 6 columns and a high salt extraction and size-exclusion approach. Notably, native BRD4 in MCF-7 cells was eluted with an apparent molecular mass much greater than that of the monomeric protein; BRD4 immunoreactivity was detected in chromatographic fractions with a relative peak centered between 669 and 2,000 kDa (Fig. 1D). Importantly, the chromatographic pattern of BRD4 largely overlapped with that of the components of the LSD1/NuRD complex (Fig. 1D). In addition, analysis of FLAG-BRD4 affinity elutes from FPLC after Superose 6 gel filtration showed that the majority of the purified FLAG-BRD4 existed in a multiprotein complex, which peaked in fractions 18 and 20 and contained the subunits of the LSD1/NuRD complex (Fig. 1D). The BRD4 elution pattern also overlapped with that of the P-TEFb complex between 158 and 440 kDa (Fig. 1D). Together, these results support the existence of the BRD4/LSD1/NuRD complex in vivo.
To substantiate the interaction of BRD4 with the LSD1/NuRD complex and to investigate the molecular detail involved in the interaction of these proteins, glutathione S-transferase (GST)-fused BRD4 deletion mutants were generated in which the N-terminal bromodomains (ΔBD) or the carboxyl terminus (ΔCT) were deleted (Fig. 1E). GST pull-down assays with bacterially purified GST-fused BRD4 deletion mutants and in vitro–transcribed/translated components of the LSD1/NuRD complex including LSD1, MTA2, MTA3, HDAC1, and HDAC2 showed that BRD4 was capable of interacting with LSD1 but not with the other components of the LSD1/NuRD complex that we tested and that the carboxyl-terminal region of BRD4 was responsible for mediating the interaction of BRD4 with LSD1 (Fig. 1E). GST pull-down experiments with GST-fused LSD1 and in vitro–transcribed/translated BRD4 corroborated these observations (Fig. 1E). Collectively, the results in GST pull-down assays further indicate that BRD4 interacts with the LSD1/NuRD complex in vivo.
BRD4 and the LSD1/NuRD Complex Occupy Super-Enhancers.
Given that BRD4 is preferentially bound to super-enhancers (9, 10) and our observation that BRD4 is associated with the LSD1/NuRD complex, we thus asked the question whether BRD4 and the LSD1/NuRD complex are colocalized at super-enhancers. To this end, we first performed the proximity ligation assay (PLA) in HeLa cells with antibodies against BRD4, LSD1, and MTA3 and found that BRD4 was in proximity with the components of the LSD1/NuRD complex but not with HDAC3, which was included as a negative control (Fig. 2A).
Fig. 2.
BRD4 and the LSD1/NuRD complex occupy super-enhancers. (A) Confocal microscopic analysis for the subcellular localization of BRD4 and the LSD1/NuRD complex. HeLa cells were fixed and immunostained with antibodies against BRD4, LSD1, or MTA3. PLA signals were detected predominantly in cell nuclei (green), and DAPI staining was included to visualize the nucleus (blue). (Scale bars: 50 μm.) (B) The interactome of BRD4, LSD1, and MTA3 was analyzed by RIME assays in MCF-7 cells with the indicated antibodies followed by mass spectrometry. The data are represented as word clouds, in which the size of the protein names represents the strength and confidence of the interactions based on the unique peptides. (C) Venn diagram of chromatin-bound BRD4/LSD1/NuRD and H3K4me1. The STRING analysis of BRD4, LSD1, MTA3, and H3K4me1 interactomes in MCF-7 cells is shown. (D) Schematic representation of the interaction between BRD4 and the LSD1/NuRD complex at enhancers.
Rapid immunoprecipitation-coupled mass spectrometry (RIME) of endogenous proteins was then performed in MCF-7 cells in which cross-linked complexes were immunoprecipitated with antibodies against BRD4, LSD1, MTA3, or H3K4me1. Mass spectrometric analysis of digested immunoprecipitates revealed that BRD4 was copurified with almost all the components of the LSD1/NuRD complex, including LSD1, MTA2, MTA3, HDAC1, HDAC2, GATAD2A, GATAD2B, and MBD3 (Fig. 2B), consistent with the observations in Fig. 1 and further reinforcing the physical interaction of BRD4 with the LSD1/NuRD complex. Importantly, the LSD1 interactome contained, in addition to the subunits of the NuRD complex, including MTA1, MTA2, GATAD2B, HDAC1, HDAC2, RBBP4/7, several proteins, including BRD4 and MED1, that are known to reside at super-enhancers (20) (Fig. 2B). Likewise, interrogation of the MTA3 interactome found, apart from the components of the NuRD complex, a number of proteins, including BRD4, MED1, ZMYND8, and KDM5C, that are known to localize at super-enhancers (20, 44) (Fig. 2B), and, remarkably, a total of 327 proteins from the BRD4/LSD1/NuRD complex interactome were shared by the interactome of H3K4me1 (Fig. 2C). These observations strongly support the notion that the BRD4/LSD1/NuRD complex occupies super-enhancers (Fig. 2D), consistent with the role of BRD4 in super-enhancer organization (20, 24).
Genomic Landscape of the BRD4/LSD1/NuRD Complex in Breast Cancer Cells.
As mentioned earlier, BRD4 is a transcriptional activator (8). The identification of the physical association of BRD4 with the LSD1/NuRD complex was unexpected, as the LSD1/NuRD complex is known to act in transcription repression (29, 45). To explore the functional significance of the physical association between BRD4 and the LSD1/NuRD complex at super-enhancers, chromatin immunoprecipitation-coupled massive parallel DNA sequencing (ChIP-seq) experiments were performed in MCF-7 cells using antibodies against BRD4, LSD1, MTA3, H3K4me1, and H3K27ac. With Model-based Analysis of ChIP-seq version 2 (MACS2) (46) and a q value cutoff of 0.05, 43,300 BRD4-binding peaks, 65,574 LSD1-binding summits, 40,264 MTA3-binding sites, 149,985 H3K4me1-enriched peaks, and 94,907 H3K27ac-enriched peaks were called. These peaks were then annotated by specific genic features using the ChIPseeker R Bioconductor package (47) with promoter specification centered on ±3,000 bp of the transcription start site (SI Appendix, Fig. S1A). The targeted sequences by BRD4, LSD1, and MTA3 were then cross-analyzed and stitched if there was at least 1-bp overlap among them, and these sequences were considered to be the targets of the BRD4/LSD1/NuRD complex. Venn diagram and Genome Browser tracking showed a significant overlap between LSD1-, MTA3-, and BRD4-binding events, yielding a total of 22,577 targets that were cobound by BRD4, LSD1, and MTA3 (SI Appendix, Fig. S1B). Heatmap analysis of the binding patterns revealed that LSD1 and MTA3 were indeed significantly enriched in regions surrounding BRD4 summits (SI Appendix, Fig. S1C). Significantly, HOMER (Hypergeometric Optimization of Motif EnRichment) (http://homer.ucsd.edu/homer/) analysis of the BRD4, LSD1, and MTA3 peaks also revealed that the binding summits of BRD4, LSD1, and MTA3 indeed contained similar sequence motifs (SI Appendix, Fig. S1D), strongly supporting the physical interaction and functional connection between BRD4 and the LSD1/NuRD complex. Importantly, comparing the characteristic genomic landscapes of BRD4, LSD1, and MTA3 indicated that these proteins were indeed significantly enriched in regions surrounding 3,000 bp of the H3K4me1 or H3K27ac ChIP-seq peaks, which had at least 1-bp overlap with BRD4 binding sites (SI Appendix, Fig. S1E), consolidating the notion that the BRD4/LSD1/NuRD complex occupies super-enhancers.
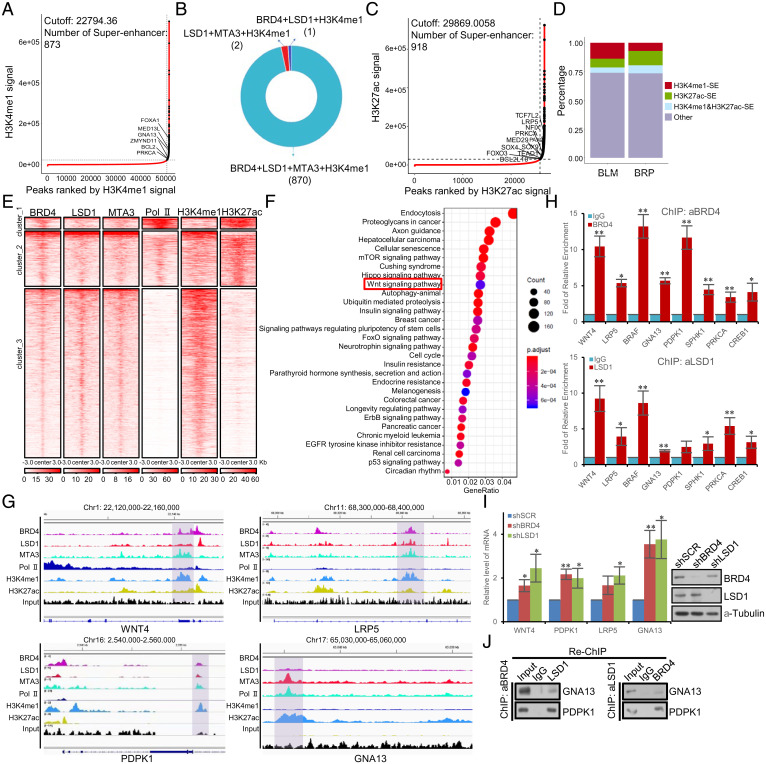
To substantiate the argument that the BRD4/LSD1/NuRD complex is localized at super-enhancers, Rank Ordering of Super-Enhancers (ROSE) was utilized to identify super-enhancers (19). With a ROSE-estimated optimal cutoff of the H3K4me1 signal as 22,794.36, a total of 873 super-enhancers were identified (Fig. 3A). Strikingly, among these 873 H3K4me1–defined super-enhancers, 870 were coenriched with BRD4, LSD1, MTA3, and H3K4me1, only two were coenriched with LSD1, MTA3, and H3K4me1 and only one was coenriched for BRD4, LSD1, and H3K4me1 (Fig. 3B). With a ROSE-estimated optimal cutoff of the H3K27ac signal as 29,869.0058, a total of 918 super-enhancers were identified (Fig. 3C). Combined with previous studies that BRD4 interacts with the P-TEFb complex and stimulates Pol II–dependent transcriptional elongation, we further analyze the functional roles of the BRD4/Pol II complex and the BRD4/LSD1/NuRD complex at super-enhancers. With a q value cutoff of 0.05, 77,101 Pol II–binding peaks were called. The peaks of BRD4 were stitched if there was an at least 1-bp overlap with LSD1 and MTA3 peaks or with Pol II peaks and then cross-analyzed with super-enhancers defined by H3K4me1 or H3K27ac. Stacked plots showed that the BRD4/LSD1/NuRD complex largely occupied H3K4me1-defined super-enhancers, whereas the BRD4/Pol II complex occupied H3K27ac-defined super-enhancers (Fig. 3D). Strikingly, tag density of BRD4, LSD1, MTA3, H3K4me1, H3K27ac, and Pol II was remarkably different across distinct groups of BRD4 peaks, which had at least 1-bp overlap with super-enhancers defined by H3K4me1 or H3K27ac peaks (Fig. 3E), further supporting the notion that BRD4 executes different functions at different super-enhancers through interacting with different complexes. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis using the R Bioconductor clusterProfiler package (48) with a Benjamini–Hochberg adjusted P value cutoff of 0.05 for the genes that were cobound by BRD4, LSD1, and MTA3 revealed that the BRD4/LSD1/NuRD complex–directed super-enhancers influence several prominent cellular signaling pathways, including autophagy, Hippo, and WNT pathways that are critically involved in cell proliferation, survival, or homeostasis (Fig. 3F). The detailed results are provided in SI Appendix, Table S2. Interestingly, a collection of the genes that are implicated in these signaling pathways, including WNT4 (49), LRP5 (50), BRAF (51), GNA13 (52), PDPK1 (53), SPHK1 (54), PRKCA (55), and CREB1 (56), have well-recognized roles in drug resistance. Further genomic profiling of the components of the BRD4/LSD1/NuRD complex showed a significant enrichment of these components at the locus of WNT4, PDPK1, LRP5, and GNA13 (Fig. 3G). In addition, quantitative ChIP (qChIP) analysis in MCF-7 cells using specific antibodies against BRD4 or LSD1 on the enhancer of a panel of selected genes including WNT4, LRP5, BRAF, GNA13, PDPK1, SPHK1, PRKCA, and CREB1 showed a strong enrichment of BRD4 and LSD1 on the enhancer of these genes (Fig. 3H). Moreover, real-time RT-PCR analysis of the expression of the representative genes, including WNT4, PDPK1, LRP5, and GNA13, in BRD4- or LSD1-depleted MCF-7 cells showed that knockdown of either BRD4 or LSD1 indeed resulted in an increase in the expression of these genes (Fig. 3I).
Fig. 3.
Genome-wide identification of the transcriptional targets of the BRD4/LSD1/NuRD complex at super-enhancers. (A) Super-enhancers based on normalized H3K4me1 occupancy in MCF-7 cells using the ROSE algorithm. (B) Pie chart of H3K4me1-marked super-enhancers and the LSD1/NuRD complex– and/or BRD4-binding summits. (C) Super-enhancers based on normalized H3K27ac occupancy in MCF-7 cells using the ROSE algorithm. (D) Stacked plots of the distribution of BRD4 ChIP-seq peaks, which had at least 1-bp overlap with LSD1/MTA3 (BLM) or Pol II (BRP) on H3K4me1- or H3K27ac-defined super-enhancers. (E) Tag density of BRD4, LSD1, MTA3, H3K4me1, H3K27ac, and Pol II against BRD4 peaks, which had at least 1-bp overlap with super-enhancers defined by H3K4me1 or H3K27ac peaks. Peaks were clustered into three groups according to the ChIP-seq signals based on the k-means method. (F) Dot plot of the top 30 most significantly enriched KEGG pathways derived from the overlapping genes of BRD4, LSD1, and MTA3 obtained through the clusterProfiler R Bioconductor package. The details of the ChIP-seq experiments are provided in Materials and Methods, and results from the pathway analysis are provided in SI Appendix, Table S2. (G) Snapshot of BRD4, LSD1, MTA3, H3K4me1, and H3K27ac signals at the locus of WNT4, PDPK1, LRP5, and GNA13 by Integrative Genomics Viewer (IGV). (H) qChIP verification of ChIP-seq results on the enhancer of the indicated genes with antibodies against the indicated proteins in MCF-7 cells. Results are presented as fold of change over control. Each bar represents the mean ± SD for triplicate experiments (*P < 0.05, and **P < 0.01). (I) qPCR analysis of the expression of the indicated genes selected from ChIP-seq results in MCF-7 cells under depletion of BRD4 or LSD1 using lentivirus-delivered shRNA. Each bar represents the mean ± SD for triplicate experiments (*P < 0.05, and **P < 0.01). The knockdown efficiency was validated by Western blotting. (J) ChIP/Re-ChIP experiments on the enhancer of the indicated genes with antibodies against the indicated proteins in MCF-7 cells.
To verify that BRD4 and the LSD1/NuRD complex occupy the enhancer of their target genes as one protein complex, sequential ChIP or ChIP/Re-ChIP (sequential chromatin immunoprecipitation) experiments were performed on the enhancer of two representative target genes, GNA13 and PDPK1. In these experiments, soluble chromatins were first immunoprecipitated with antibodies against BRD4, and the immunoprecipitates were subsequently reimmunoprecipitated with antibodies against LSD1. The results showed that, in precipitates, the enhancer of GNA13 and PDPK1 that was immunoprecipitated with antibodies against BRD4 could be reimmunoprecipitated with antibodies against LSD1 (Fig. 3J). Similar results were obtained when the initial ChIP was done with antibodies against LSD1 (Fig. 3J). Collectively, these observations support the notion that BRD4 and the LSD1/NuRD complex are physically and functionally associated with super-enhancers to restrict the activation of a panel of genes including those that are well recognized for drug resistance.
Decommissioning the BRD4/LSD1/NuRD Complex Leads to JQ1 Resistance.
As stated earlier, BRD4 inhibitors are being pursued as novel targets for cancer treatment, and effective BRD4 inhibitors with promising antitumor efficacy have been developed in several preclinical cancer models with encouraging signs of efficacy in suppressing tumor growth (14). However, probably not surprisingly, drug resistance also occurs for BRD4 inhibitors such as JQ1 (17, 18). Given our observations that the BRD4/LSD1/NuRD complex acts to restrict the activation of drug-resistant genes, we next investigated the functional relationship between JQ1 and the BRD4/LSD1/NuRD complex in the context of antitumor effect and drug resistance. To this end, MCF-7 cells were treated with JQ1 for different periods of time, and the proliferation of these cells was then analyzed using cell counting kit 8 (CCK8). Notably, JQ1 treatment was associated with an inhibited proliferation of MCF-7 cells, an effect that lasted for about 3 wk (SI Appendix, Fig. S2A). Nonetheless, drug resistance emerged in MCF-7 cells after about 4 wk of JQ1 treatment; the proliferation of MCF-7 cells was no longer inhibited by JQ1 treatment (SI Appendix, Fig. S2A). Analysis by qChIP indicated that JQ1 treatment led to a decreased occupancy of BRD4 on the enhancer of GNA13 and PDPK1 genes, even at an early stage of JQ1 treatment (SI Appendix, Fig. S2B), which is expected. However, intriguingly, the enrichment of LSD1 and MTA3 persisted for up to 3 wk of JQ1 treatment; the occupancy of LSD1 and MTA3 on the enhancer of GNA13 and PDPK1 genes also diminished after 4 wk of JQ1 treatment (SI Appendix, Fig. S2B). In agreement, qPCR analysis showed that the expression of GNA13 and PDPK1 increased significantly in MCF-7 cells only after 4 wk of JQ1 treatment (SI Appendix, Fig. S2C). Thus, it appears that a short treatment of JQ1 only affects the chromatin binding of BRD4, whereas a longer treatment leads to a disappearance of the chromatin binding of the LSD1/NuRD complex, an event corresponding to JQ1 resistance. This scenario is consistent with the repression of the drug-resistant genes by the LSD1/NuRD complex. Based on the role of BRD4 as a general regulator for super-enhancer organization (20, 24), if our interpretation is correct, it means that the function of BRD4 in the BRD4/LSD1/NuRD complex on chromatin is to organize super-enhancers in a way that fosters a proper chromatin environment conducive to the recruitment of the LSD1/NuRD complex. Moreover, the observations in SI Appendix, Fig. S2 B and C also point to a scheme that once it is recruited to chromatin, the LSD1/NuRD complex can bind to thus repress the target genes for a longer time. However, a sustained chromatin binding of the LSD1/NuRD complex, and thus the repression of target genes, requires a sustained super-enhancer organization by BRD4. To support this notion, MCF-7 cell clones with BRD4 stably depleted with lentivirus-delivered small hairpin RNA (shRNA) were generated, and the recruitment of the LSD1/NuRD complex on the enhancer of GNA13 and PDPK1 genes in these cells was measured by qChIP. The results showed that without BRD4, the binding of the LSD1/NuRD complex on target genes was not detected (SI Appendix, Fig. S2D). Remarkably, however, reconstitution of BRD4 expression by ectopic expression of an shRNA-resistant BRD4 construct in these cells resulted in the detection of the LSD1/NuRD complex on the enhancer of GNA13 and PDPK1 genes (SI Appendix, Fig. S2D). Additionally, MCF-7 cell clones with LSD1 stably depleted with lentivirus-delivered shRNA were collected, and the recruitment of BRD4 on the enhancer of GNA13 and PDPK1 in these cells was measured by qChIP. The results showed that depletion of LSD1 had no effect on the recruitment of BRD4 to the enhancers (SI Appendix, Fig. S2D), strongly supporting the argument that BRD4 is required for the recruitment of the LSD1/NuRD complex to super-enhancers to restrict the activation of these enhancers.
To exclude the possibility that the diminished occupancy of the BRD4/LSD1/NuRD complex on chromatin and decommissioning this assembly was a result of diminished expression of the proteins in the BRD4/LSD1/NuRD complex upon JQ1 treatment, Western blotting analysis was performed in MCF-7 cells under the treatment of JQ1. We found that the level of all the protein components of the BRD4/LSD1/NuRD complex had no significant changes during JQ1 treatment (SI Appendix, Fig. S2E), except for LSD1, whose level decreased with a longer treatment of JQ1.
To gain mechanistic insights into the decrease in the level of LSD1 during JQ1 treatment and to investigate the significance of LSD1 down-regulation in JQ1 resistance, we further measured the messenger RNA (mRNA) level of LSD1 by qPCR in MCF-7 cells under the treatment of JQ1. We found that while the protein level of LSD1 declined under the treatment of JQ1, the level of LSD1 mRNA did not change (SI Appendix, Fig. S2F). Thus, the decreased protein level of LSD1 was probably a result of proteasome-mediated protein degradation. In agreement, the effect could be effectively blocked by the proteasome-specific inhibitor MG132 (SI Appendix, Fig. S2G).
The E3 Ligase PELI1 Specifically Targets LSD1 for Degradation.
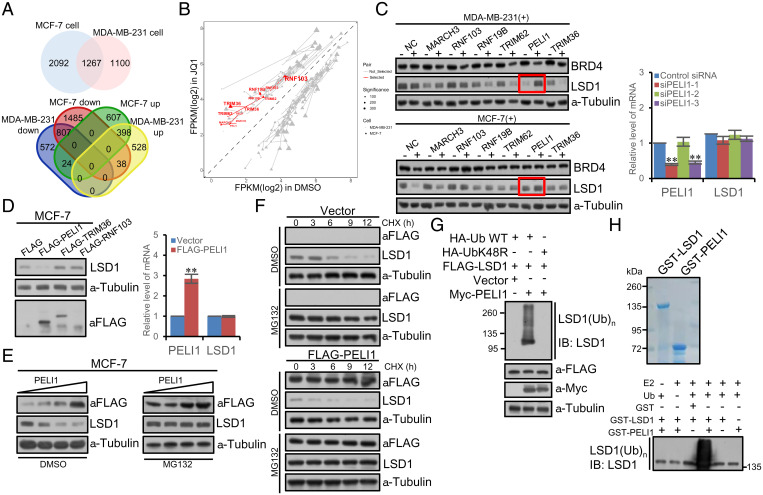
To search for the E3 ligase that is responsible for LSD1 down-regulation under JQ1 treatment, we first profiled the differentially expressed genes in breast cancer cells with or without JQ1 treatment by RNA-based deep sequencing (RNA-seq) to identify the genes that are functionally linked to protein degradation. To this end, total RNAs were extracted from MCF-7 and MDA-MB-231 cells that were untreated or treated with JQ1 for 4 wk, followed by complementary DNA synthesis, library construction, and sequencing using BGIseq500 with single-end 50-bp reads. The quality of raw data was examined by FastQC, and sequencing adapter and low-quality reads, including those with more than five “N” bases and mean Phred quality score less than 15, were removed through fastp. Spliced Transcripts Alignment to a Reference software (57) was utilized to align clean reads to the unmasked human reference genome (GRCh38 and hg38). Raw counts of the reads mapped to genes were selected to extract differentially expressed genes using the DESeq2 Bioconductor package (58) with P value <0.05 and absolute log 2 (fold change) >1 as the threshold. A total of 3,359 differentially expressed genes were identified in MCF-7 cells, including 1,029 up-regulated and 2,330 down-regulated, and 2,367 differentially expressed genes were identified in MDA-MB-231 cells, including 964 up-regulated and 1,403 down-regulated. Cross-analysis yielded a total of 1,267 differentially expressed genes, of which 436 were up-regulated and 831 were down-regulated under the treatment of JQ1 (Fig. 4A). We reasoned that the E3 ligase that targets for LSD1 degradation must be the one that was up-regulated under JQ1 treatment. Based on this rationale, we identified seven E3 ligases that were among the up-regulated group under JQ1 treatment in MCF-7 and MDA-MB-231 cells. Since LSD1 is localized and functions in the nucleus, we thus focused our search on the E3 ligases that have been reported or are projected to localize in the nucleus, and this categorized six E3 ligases including RNF103, MARCH3, RNF19B, TRIM62, PELI1, and TRIM36 (Fig. 4B).
Fig. 4.
The E3 ligase PELI1 specifically targets LSD1 for degradation. (A) RNA-seq assays in MCF-7 or MDA-MB-231 cells that were untreated or treated with JQ1 for 4 wk. Venn diagram of the differentially expressed genes analyzed with a cutoff of P value <0.05 and absolute log 2 (fold change) >1. (B) Scatter plot of differentially expressed genes in MCF-7 (triangle) or MDA-MB-231 cells (dot) that were treated with JQ1 (y-axis) or dimethyl sulfoxide (DMSO) (x-axis). The lines connecting the triangles or dots indicate the same genes. Red indicates the up-regulated nuclear E3 ligases. (C) Analysis of the protein level of LSD1 by Western blotting in MCF-7 or MDA-MB-231 cells upon knocking down the E3 ligases using corresponding siRNAs. The mRNA expression of LSD1 was also analyzed by qPCR. Each bar represents the mean ± SD for triplicate experiments (**P < 0.01). (D) Western blotting analysis of the LSD1 protein in MCF-7 cells upon overexpression of PELI1, TRIM36, or RNF103 (Left). The mRNA expression of LSD1 was also analyzed by qPCR (Right). Each bar represents the mean ± SD for triplicate experiments (**P < 0.01). (E) Western blotting analysis of PELI1 dose-dependent decay of LSD1 in MCF-7 cells under MG132 treatment. The level of LSD1 decreased upon overexpression of PELI1; an effect could be effectively blocked by MG132. (F) CHX chase assays in MCF-7 cells overexpressing FLAG-PELI1. (G) In vivo ubiquitination assays in MCF-7 cells overexpressing FLAG-LSD1. Cells were cotransfected with Myc-PELI1 and HA-Ub or ubiquitin mutant UbK48R and treated with MG132 for 10 h prior to immunoprecipitation. (H) In vitro ubiquitination assays with GST-LSD1 or GST-PELI1 in the presence or absence of ubiquitin and E2. The reaction mixture was analyzed by Western blotting with anti-LSD1.
Next, functional screening using small interfering RNA (siRNA)-directed loss-of-function of the six nuclear E3 ligases was performed in MCF-7 and MDA-MB-231 cells that were under the treatment of JQ1 for at least 3 wk. In these experiments, the expression of each individual of the six E3 ligases was knocked down using pooled three different sets of corresponding siRNAs, and the level of LSD1 was measured by Western blotting. We found that while loss-of-function of RNF103, MARCH3, RNF19B, TRIM62, and TRIM36 had only marginal effects on the level of LSD1 protein, knockdown of PELI1 resulted in a dramatic increase in LSD1 level (Fig. 4C), which was not due to transcriptional up-regulation of LSD1, as measured by qPCR under the experimental condition (Fig. 4C), suggesting that PELI1 is the E3 ligase that targets LSD1 for proteasome-dependent degradation.
To further support this, MCF-7 cells were transfected with FLAG-tagged PELI1. Western blotting analysis of cellular lysates revealed that the steady-state level of LSD1 markedly decreased upon PELI1 overexpression (Fig. 4D), whereas overexpression of TRIM36 or RNF103, two other E3 ligases identified in RNA-seq, did not result in detectable changes in LSD1 steady-state level. The decreased LSD1 protein expression under PELI1 overexpression was not due to transcriptional regulation of LSD1, as qPCR measurements indicated that PELI1 overexpression did not affect LSD1 mRNA level (Fig. 4D). Meanwhile, Western blotting analysis showed that PELI1 dose-dependent decay of LSD1 in MCF-7 cells could be effectively blocked by MG132 (Fig. 4E). Moreover, cycloheximide (CHX) chase assays in MCF-7 cells that were transfected with FLAG-PELI1 revealed that PELI1 overexpression was associated with a decrease in the half-life of LSD1, effects that only occurred in the absence of MG132 (Fig. 4F).
To gain further support that PELI1 targets LSD1 for proteasome-dependent degradation, we next determined whether PELI1-promoted LSD1 destabilization is a consequence of LSD1 ubiquitination. To this end, MCF-7 cells were cotransfected with FLAG-LSD1 together with Myc-PELI1 and HA-tagged ubiquitin (HA-Ub) in the presence of MG132. Immunoprecipitation with the anti-FLAG M2 magnetic beads followed by immunoblotting with an antibody against LSD1 indicated that overexpression of PELI1 was associated with an increase in LSD1 polyubiquitination (Fig. 4G). However, when wild-type ubiquitin used in ubiquitination assays was replaced with UbK48R, a ubiquitin mutant defective for polyubiquitin chain assembly (59), PELI1-promoted LSD1 polyubiquitination was no longer detected (Fig. 4G). Moreover, in vitro ubiquitination assays with bacterially expressed GST-PELI1 and in vitro–transcribed/translated LSD1 showed that PELI1 promotes LSD1 polyubiquitination but only in the presence of ubiquitin-conjugating enzyme UbcH6 (E2) and ubiquitin (Fig. 4H). Collectively, these observations indicate that PELI1 is a bona fide E3 ubiquitin ligase for LSD1.
Elimination of PELI1 Improves the Therapeutic Efficacy of JQ1 in Breast Cancer Cells.
Given our observation that the BRD4/LSD1/NuRD complex acts to repress a panel of genes that are functionally associated with drug resistance, we next investigated the effect of destabilization of LSD1 by PELI1 on the function of the BRD4/LSD1/NuRD complex and on JQ1 resistance in breast cancer. To this end, MCF-7 cell clones stably expressing PELI1 were generated and treated with JQ1 for different periods of time. CCK8 cell-proliferation assays indicated that PELI1 overexpression accelerated the emergence of JQ1 resistance; PELI1-overexpressing MCF7 cells were still inhibited at the first week of JQ1 treatment but became resistant to JQ1 treatment around 2 wk of JQ1 administration as corresponding cells grew faster than untreated controls (SI Appendix, Fig. S3 A, Upper), corresponding to a diminished enrichment of both LSD1 and MTA3 on the enhancer of GNA13 and PDPK1 genes, although the level of MTA3 was not affected by PELI1 overexpression in MCF-7 cells treated with JQ1 for 2 wk (SI Appendix, Fig. S3 B, Upper). Consistently, the expression of GNA13 and PDPK1 increased in PELI1-overexpressing MCF-7 cells, as measured by qPCR (SI Appendix, Fig. S3 C, Upper). These results indicate that short treatments of PELI1-overexpressing MCF-7 cells with JQ1 affected the chromatin binding of the BRD4/LSD1/NuRD complex, an event corresponding to JQ1 resistance.
We also generated MCF-7 cell clones with PELI1 stably depleted by lentivirally delivered PELI1 shRNA. CCK8 cell-proliferation assays indicated that knockdown of PELI1 indeed sensitized MCF-7 cells to JQ1; the proliferation of PELI1-depleted MCF-7 cells continued to decrease after 4 wk of JQ1 treatment, and almost no viable cells were detected after 5 wk of JQ1 treatment (SI Appendix, Fig. S3 A, Lower). In agreement, qChIP experiments still detected the occupancy of LSD1 on the enhancer of GNA13 and PDPK1 genes at 4 wk of JQ1 treatment in PELI1-depleted MCF-7 cells (SI Appendix, Fig. S3 B, Lower), and qPCR measurement indicated that the expression of GNA13 and PDPK1 remained low at 4 wk of JQ1 treatment (SI Appendix, Fig. S3 C, Lower). These results indicate that long treatments of PELI1-depleted MCF-7 cells only affect the chromatin binding of BRD4. In addition, colony-formation assays showed that PELI1 knockdown was associated with a marked reduction in colony number of MCF-7 cells, an effect that could be rescued, at least partially, by overexpression of an shRNA-resistant PELI1 plasmid (SI Appendix, Fig. S3D). Collectively, these results support a notion that inhibition of PELI1 improves the therapeutic efficacy of JQ1 in breast cancer cells.
Given the scope and variety of the drug-resistant genes that are influenced by the BRD4/LSD1/NuRD complex, we next investigated the impact of decommissioning the BRD4/LSD1/NuRD assembly by inhibiting BRD4 or/and destabilizing LSD1 on the efficacy of other antitumor drugs. To this end, MCF-7 cells were treated for 4 to 5 wk with JQ1 along with the treatment with a panel of antitumor compounds including DNA synthesis inhibitors bleomycin and mitomycin C (60, 61), topoisomerase inhibitors topotecan and doxorubicin (62), microtubule depolymerizing agents paclitaxel and nocodazole (63), DNA cross-linking agents cisplatin and carboplatin (64), DNA base alkylating agents thiotepa and mechlorethamine (65), and antimetabolites for depleting deoxynucleotide pool in DNA replication gemcitabine and 5-fluorouracil (66). CCK8 cell-proliferation assays indicated that JQ1-resistant MCF-7 cells were also insensitive to mitomycin C, doxorubicin, and gemcitabine, as the IC50s (half-maximal inhibitory concentration) of these compounds were significantly higher in JQ1-resistant MCF-7 cells (SI Appendix, Fig. S3E). In corroboration, analysis of the IC50s of a series of therapeutic compounds in breast cancer cell lines in the Genomics of Drug Sensitivity in Cancer (GDSC) database (https://www.cancerrxgene.org/) and the mRNA level of PELI1 in corresponding cell lines in the Cancer Cell Line Encyclopedia (CCLE) database (https://sites.broadinstitute.org/ccle/) also indicated that the sensitivity of bleomycin, topotecan, doxorubicin, cisplatin, or gemcitabine is negatively correlated with the mRNA level of PELI1 (SI Appendix, Fig. S3F). These results suggest that decommissioning the BRD4/LSD1/NuRD complex also enables MCF-7 cells to evade the selective pressure by these antitumor drugs.
The Clinicopathological Significance of the PELI1-LSD1-BRD4/LSD1/NuRD Axis in Breast Cancer.
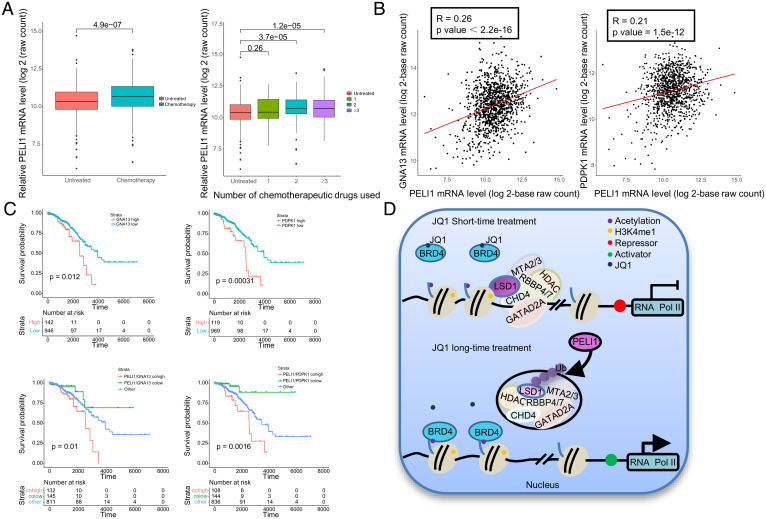
In order to gain further support of the role of the PELI1-LSD1-BRD4/LSD1/NuRD axis in drug resistance in breast cancer and to extend our observations to a clinicopathologically relevant context, we first interrogated the mRNA expression of PELI1 in 1,091 breast cancer samples collected from the Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/), of which 579 were treated with at least one chemotherapeutic drug and 512 were untreated or received other types of treatment such as hormonal treatment or immunotherapy. We found that PELI1 is up-regulated in breast cancer samples receiving at least two chemotherapeutic drugs (two-sided t test), and, strikingly, the level of its expression is positively correlated with the number of chemotherapeutic drugs used in patients (Spearman correlation coefficient: 0.145, P value: 1.551 × 10−6) (Fig. 5A), implying that PELI1 function may be particularly relevant in drug-resistant conditions. Moreover, consistent with our identification that GNA13 and PDPK1 are among the downstream targets of the BRD4/LSD1/NuRD complex, when the relative level of GNA13 or PDPK1 was plotted against that of PELI1 in the 1,091 breast-carcinoma samples, statistically significant positive correlations were observed (Fig. 5B). In addition, analysis of the correlation in the expression between LSD1 and PELI1 using a breast cancer landscape dataset (https://www.breastcancerlandscape.org/index.html) found a statistically significant negative correlation in the protein levels of PELI1 and LSD1 in 45 breast cancer samples (P = 0.021, R = −0.34) (SI Appendix, Fig. S4A).
Fig. 5.
The clinicopathological significance of the PELI1-LSD1-BRD4/LSD1/NuRD axis in breast cancer. (A) The mRNA expression of PELI1 in 1,091 breast cancer samples collected from TCGA and stratified by their therapeutic regimens. (B) The correlation of mRNA expression between PELI1, GNA13, and PDPK1. The relative level of PELI1 was plotted against that of GNA13 or PDPK1. (C) Survival analysis for the relationship between survival time and PELI1/ GNA13 or PELI1/PDPK1 signatures in breast cancer patients based on the 1,091 breast carcinoma samples from the TCGA dataset. (D) The proposed model for the PELI1-LSD1-BRD4/LSD1/NuRD axis in breast cancer.
Finally, to substantiate the clinicopathological significance of our observations, we analyzed the expression levels of PELI1 and LSD1 in breast cancer and their correlations with clinical behaviors of breast cancer patients. Kaplan–Meier survival analysis of all of the Gene Expression Omnibus (GEO) datasets included in the online tool (http://kmplot.com/analysis/) showed that either high LSD1 expression (P = 1.4×10−7) or low PELI1 expression (P = 0.012) was associated with a better relapse-free survival of breast cancer patients (SI Appendix, Fig. S4B). Prognostic analysis based on the 1,091 breast carcinomas samples revealed that high expression of either GNA13 (P = 0.012) or PDPK1 (P = 0.00031) is strongly correlated with an inferior overall survival of breast cancer patients and that concurrently high expressions of PELI1 and GNA13 (P = 0.01) or PELI1 and PDPK1 (P = 0.0016) are correlated with worse overall survival of breast cancer patients (Fig. 5C). These data are consistent with a role for PELI1 to destabilize LSD1 and thus to decommission the BRD4/LSD1/NuRD complex, leading to the derepression of drug-resistant genes and JQ1 resistance in breast cancer therapy (Fig. 5D).
Discussion
Combating cancer remains a daunting task, not only because of the difficulty in identifying and approaching the genuine oncogenic driver but also owing to the apparently inevitable and notorious drug resistance. Accordingly, discerning the driving force behind carcinogenesis and developing the strategy to avoid drug resistance remain the keys for therapeutic intervention of cancer.
Most of malignancies are associated with complex abnormalities in both genetics and epigenetics. Epigenetics not only influences information transfer through generations but also regulates various pathophysiological processes (67). In recent years, the field of epigenetic therapy is also flourishing and novel treatments for numerous diseases, including cancer, derived from epigenetic system are becoming reality.
BRD4 is an epigenetic reader (2) and a general regulator of transcription (8). More recent studies indicate that this epigenetic factor is associated with active promoters and preferentially binds to super-enhancers (9, 10). It is a current belief that, at the molecular level, BRD4 functions in super-enhancer organization and transcription activation. Biologically, the BET family, including BRD4, has been implicated in a variety of cellular processes such as cell proliferation, differentiation, metabolism, and DNA repair (3–6). As such, BRD4 has also been implicated in a broad range of cancer types including breast cancer (13), and the oncogenic potential of BRD4 is attributed to its role in transcription activation of genes including c-MYC (14) and BCL2 (15) that are critically involved in cell proliferation and survival. Unexpectedly, we found in the current study that BRD4 is also physically associated with the LSD1/NuRD complex. Since this complex has been well characterized as a transcriptionally repressive assembly (29, 45), our observation means that BRD4 also has a role in transcription repression. Indeed, we demonstrated that BRD4 and the LSD1/NuRD complex colocalize at super-enhancers and that the BRD4/LSD1/NuRD complex transcriptionally represses a cohort of genes, including GNA13 and PDPK1, that are well documented to play roles in drug resistance (52, 53). Comparing our results to the results from previous studies found that ∼10% of the 436 up-regulated genes in our study were significantly up-regulated under JQ1 treatment in K562 and MV4-11 cells (68) and MOLT4 cells (69), while eight up-regulated genes in our study were also up-regulated upon BRD4 degradation in MOLT4 cells (69). It is becoming increasingly clear that BRD4 is a chromatin organizer that executes transcription activation or repression in a context-dependent manner (70).
The finding that BRD4 is involved in transcription repression is intriguing. However, literature indicates that the transcriptionally repressive activity of BRD4 was also reported in previous studies in the regulation of autophagy and lysosomal function (71, 72). Considering its genic signature in super-enhancers and its role in transcription regulation, it is possible that super-enhancer organization is the genuine function of BRD4 and that its transcriptional regulatory function is context-dependent. In line with this, a dual function in transcription regulation has also been reported for CDK9, SOX2, and Mediator (73–75), which, together with BRD4, belongs to the so-called master regulator of super-enhancers.
Super-enhancers are clusters of cis-elements that are bound in trans by the master regulators (19). It is believed that super-enhancers contribute to tissue specification by dictating lineage-specific gene expression (23), a mission that is likely accomplished by coordinated activation or/and repression of structurally and functionally related clusters of genes through influencing long-range high-order chromatin configuration. Such a scheme has apparent evolutionary advantages in terms of the spatiotemporal pattern and precisely controlled manner of gene expression in eukaryotic cells. Although the physical and structural features of the clustered genes by super-enhancers need further delineation, the functional relevance of the grouped genes is not only expected but also evidenced (24, 76, 77). Our observations that the BRD4/LSD1/NuRD complex represses a cohort of genes that are known to play roles in drug resistance are consistent with this notion.
Drug resistance is almost becoming a general feature of cancer chemotherapies, and understanding of the molecular mechanism underlying drug resistance is of paramount importance for optimizing therapeutic efficacy. Owing to its transcription activation of several well-established oncogenes, such as c-MYC (14) and BCL2 (15), BRD4 and the other BET proteins are being pursued as promising novel targets for cancer treatment (14). The current drugging strategy for the BET proteins is to design small-molecule inhibitors, such as JQ1 (16), that bind to the bromodomain pocket of BET proteins to compete with acetylated histones. However, disappointingly, as stated in previous studies, drug resistance also occurs for JQ1 (17, 18). We found that, intriguingly, treatment with JQ1 results in no immediate activation of the drug-resistant genes on super-enhancers but that long treatment or destabilization of LSD1 by the E3 ligase PELI1 leads to decommissioning of the BRD4/LSD1/NuRD complex and JQ1 resistance in breast cancer cells. These observations are consistent with a role for BRD4 in the regulation of chromatin dynamics in which BRD4 orchestrates super-enhancers to bring these enhancer elements into close proximity, facilitating the recruitment of the LSD1/NuRD complex to chromatin. Our model suggests a scheme in which once the LSD1/NuRD complex is loaded to chromatin, it stays there and thus represses the transcription of drug-resistant genes for a period of time without the presence of BRD4. However, sustained recruitment of the LSD1/NuRD complex, and thus transcription repression of drug-resistant genes, still requires BRD4 for the maintenance of super-enhancer organization. Accordingly, it has also been reported that super-enhancer–associated genes are highly sensitive to JQ1 (24). These observations highlight the importance of super-enhancers in shaping the chromatin structure and in dictating the pattern of gene expression. Indeed, inhibition of super-enhancer–dependent transcription also represents a promising therapeutic strategy for cancer intervention (24, 25), as mentioned earlier. Ultimately, super-enhancer–targeted therapy will rely on trans-acting factors like BRD4 to be druggable.
We found that the LSD1 protein level is down-regulated in breast cancer during JQ1 treatment, and we identified that PELI1 is responsible for LSD1 decay through its E3 ligase activity and via the ubiquitin-proteasomal pathway. The reason for LSD1 destabilization during JQ1 treatment is currently unclear. Based on our observation that the expression of PELI1 is up-regulated under JQ1 treatment and that BRD4 knockdown was not associated with changes in PELI1 expression (SI Appendix, Fig. S5), it is conceivable that the up-regulation of PELI1 represents a survival strategy for breast cancer cells to evade the selective pressure of JQ1 treatment, as elevated PELI1 will destabilize LSD1, leading to the dysfunction of the BRD4/LSD1/NuRD complex and derepression of drug-resistant genes. Whether this feedback regulation also exists in JQ1 treatment of other malignancies is currently unknown. Perhaps more importantly, whether a similar system is also implemented in normal physiology and what is its biological significance need further investigation. Nevertheless, our observations are consistent with a previous report that PELI1 is positively correlated with the expression of MYC, BCL6, BCL2, and MUM1 in diffuse large B-cell lymphoma (36).
The physiological significance of the BRD4/LSD1/NuRD complex and its targeted genes remain to be investigated, as mentioned in this study. We showed that the BRD4/LSD1/NuRD complex–mediated restriction of super-enhancer activation influences various genes, including WNT4, LRP5, BRAF, GNA13, PDPK1, SPHK1, PRKCA, and CREB1, and impacts several cellular signaling pathways including the PI3K-AKT and WNT pathways. Although these genes have been documented to play important roles in drug resistance (49–56), the PI3K-AKT and WNT pathways are important cellular signaling mechanisms profoundly affecting biological activities such as cell growth and differentiation, transcription, replication, apoptosis, and aging (78, 79). It will be interesting in future studies to investigate the regulation of these cellular processes by the BRD4/LSD1/NuRD complex and the biological readouts stemmed from the BRD4/LSD1/NuRD complex–mediated restriction of super-enhancer activation. It is equally important to investigate whether the functional link of the LSD1/NuRD complex to super-enhancers is a general phenomenon in terms of the repression function of this complex. Nevertheless, we report in the current study that BRD4 is physically associated with the LSD1/NuRD complex and functionally coordinates with this repressive assembly to restrict the hyperactivation of super-enhancers in breast cancer cells. We found that the BRD4/LSD1/NuRD complex represses a panel of genes including those that are known to be involved in drug resistance. We demonstrated that long JQ1 treatment or destabilization of LSD1 by PELI1 leads to decommissioning the BRD4/LSD1/NuRD complex and resistance to JQ1 as well as to other antitumor therapeutics in breast cancer cells. We found that PELI1 is up-regulated during breast cancer chemotherapies, and the level of its expression is negatively correlated with that of LSD1. We showed that the levels of the BRD4/LSD1/NuRD complex–restricted genes, such as GNA13 and PDPK1, are strongly correlated with an inferior overall survival of breast cancer patients. Our study uncovers a functional duality for BRD4 in transcription regulations, which link to tumorigenesis and chemoresistance, supporting a functional link of the LSD1/NuRD complex to super-enhancers and the pursuit of a combined therapeutic targeting for BRD4 and PELI1 in effective treatment of breast cancer. In this regard, it is exciting to note that BBT-401, a potent first-in-class PELI1 inhibitor for treatment of ulcerative colitis, underwent Phase I study in May 2020.
Materials and Methods
Colony-Formation Assay.
MCF-7 cells were maintained in culture media for 14 d, fixed with 4% paraformaldehyde, stained with crystal violet for colony observation, and counted using a light microscope. Each experiment was performed in triplicate and repeated at least three times.
Statistical Analysis.
Results from biological triplicate experiments are presented with error bars as mean ± SD. A P value of less than 0.05 was considered statistically significant. Survival was conducted using the Kaplan–Meier method, and the difference between the survival curves was analyzed with the log-rank test. Detailed information about the materials and methods can be found in the SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by a Grant (2021YFA1300603 to Y.S.) from the Ministry of Science and Technology of China, and Grants (82188102, 81730079, and 31991164 to Y.S.) from the National Natural Science Foundation of China.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2109133119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information. The raw data of ChIP-seq and RNA-seq are deposited at the GEO database (https://www.ncbi.nlm.nih.gov/geo/) with accession number GSE171908.
References
- 1.Belkina A. C., Denis G. V., BET domain co-regulators in obesity, inflammation and cancer. Nat. Rev. Cancer 12, 465–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filippakopoulos P., Knapp S., Targeting bromodomains: Epigenetic readers of lysine acetylation. Nat. Rev. Drug Discov. 13, 337–356 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Cheung K. L., et al. , Distinct roles of Brd2 and Brd4 in potentiating the transcriptional program for Th17 cell differentiation. Mol. Cell 65, 1068–1080.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sdelci S., et al. , MTHFD1 interaction with BRD4 links folate metabolism to transcriptional regulation. Nat. Genet. 51, 990–998 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stanlie A., Yousif A. S., Akiyama H., Honjo T., Begum N. A., Chromatin reader Brd4 functions in Ig class switching as a repair complex adaptor of nonhomologous end-joining. Mol. Cell 55, 97–110 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Ba M., et al. , BRD4 promotes gastric cancer progression through the transcriptional and epigenetic regulation of c-MYC. J. Cell. Biochem. 119, 973–982 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Jiang Y. W., et al. , Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc. Natl. Acad. Sci. U.S.A. 95, 8538–8543 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jang M. K., et al. , The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19, 523–534 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Chapuy B., et al. , Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell 24, 777–790 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stratton M. S., et al. , Signal-dependent recruitment of BRD4 to cardiomyocyte super-enhancers is suppressed by a microRNA. Cell Rep. 16, 1366–1378 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.French C. A., et al. , BRD4 bromodomain gene rearrangement in aggressive carcinoma with translocation t(15;19). Am. J. Pathol. 159, 1987–1992 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuber J., et al. , RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–528 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crawford N. P., et al. , Bromodomain 4 activation predicts breast cancer survival. Proc. Natl. Acad. Sci. U.S.A. 105, 6380–6385 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delmore J. E., et al. , BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peirs S., et al. , Targeting BET proteins improves the therapeutic efficacy of BCL-2 inhibition in T-cell acute lymphoblastic leukemia. Leukemia 31, 2037–2047 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Filippakopoulos P., et al. , Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shu S., et al. , Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 529, 413–417 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rathert P., et al. , Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature 525, 543–547 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whyte W. A., et al. , Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sabari B. R., et al. , Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heintzman N. D., et al. , Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Rothschild G., Basu U., Lingering questions about enhancer RNA and enhancer transcription-coupled genomic instability. Trends Genet. 33, 143–154 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X., Cairns M. J., Yan J., Super-enhancers in transcriptional regulation and genome organization. Nucleic Acids Res. 47, 11481–11496 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lovén J., et al. , Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peeters J. G., et al. , Inhibition of super-enhancer activity in autoinflammatory site-derived T cells reduces disease-associated gene expression. Cell Rep. 12, 1986–1996 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Humphrey G. W., et al. , Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J. Biol. Chem. 276, 6817–6824 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Tong J. K., Hassig C. A., Schnitzler G. R., Kingston R. E., Schreiber S. L., Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395, 917–921 (1998). [DOI] [PubMed] [Google Scholar]
- 28.Bowen N. J., Fujita N., Kajita M., Wade P. A., Mi-2/NuRD: Multiple complexes for many purposes. Biochim. Biophys. Acta 1677, 52–57 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Wang Y., et al. , LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 138, 660–672 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Shi Y., Histone lysine demethylases: Emerging roles in development, physiology and disease. Nat. Rev. Genet. 8, 829–833 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Han X., et al. , Destabilizing LSD1 by Jade-2 promotes neurogenesis: An antibraking system in neural development. Mol. Cell 55, 482–494 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Maiques-Diaz A., Somervaille T. C., LSD1: Biologic roles and therapeutic targeting. Epigenomics 8, 1103–1116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moynagh P. N., The roles of Pellino E3 ubiquitin ligases in immunity. Nat. Rev. Immunol. 14, 122–131 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Park H. Y., et al. , Pellino 1 promotes lymphomagenesis by deregulating BCL6 polyubiquitination. J. Clin. Invest. 124, 4976–4988 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ha G. H., et al. , Pellino1 regulates reversible ATM activation via NBS1 ubiquitination at DNA double-strand breaks. Nat. Commun. 10, 1577 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choe J. Y., et al. , PELI1 expression is correlated with MYC and BCL6 expression and associated with poor prognosis in diffuse large B-cell lymphoma. Mod. Pathol. 29, 1313–1323 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Shan L., et al. , FOXK2 elicits massive transcription repression and suppresses the hypoxic response and breast cancer carcinogenesis. Cancer Cell 30, 708–722 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Si W., et al. , Dysfunction of the reciprocal feedback loop between GATA3- and ZEB2-nucleated repression programs contributes to breast cancer metastasis. Cancer Cell 27, 822–836 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Yan R., et al. , SCF(JFK) is a bona fide E3 ligase for ING4 and a potent promoter of the angiogenesis and metastasis of breast cancer. Genes Dev. 29, 672–685 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y., et al. , Nucleation of DNA repair factors by FOXA1 links DNA demethylation to transcriptional pioneering. Nat. Genet. 48, 1003–1013 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Liu W., et al. , Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell 155, 1581–1595 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen C., et al. , NSD3-short is an adaptor protein that couples BRD4 to the CHD8 chromatin remodeler. Mol. Cell 60, 847–859 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu S. Y., Lee A. Y., Lai H. T., Zhang H., Chiang C. M., Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol. Cell 49, 843–857 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen H., et al. , Suppression of enhancer overactivation by a RACK7-histone demethylase complex. Cell 165, 331–342 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Q., et al. , Binding of the JmjC demethylase JARID1B to LSD1/NuRD suppresses angiogenesis and metastasis in breast cancer cells by repressing chemokine CCL14. Cancer Res. 71, 6899–6908 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y., et al. , Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu G., Wang L. G., He Q. Y., ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Yu G., Wang L. G., Han Y., He Q. Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shackleford M. T., et al. , Estrogen regulation of mTOR signaling and mitochondrial function in invasive lobular carcinoma cell lines requires WNT4. Cancers (Basel) 12, 2931 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller-Kleinhenz J., et al. , Dual-targeting Wnt and uPA receptors using peptide conjugated ultra-small nanoparticle drug carriers inhibited cancer stem-cell phenotype in chemo-resistant breast cancer. Biomaterials 152, 47–62 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J., et al. , A secondary mutation in BRAF confers resistance to RAF inhibition in a BRAFV600E-mutant brain tumor. Cancer Discov. 8, 1130–1141 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rasheed S. A. K., et al. , GNA13 expression promotes drug resistance and tumor-initiating phenotypes in squamous cell cancers. Oncogene 37, 1340–1353 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan J., et al. , PDK1 signaling toward PLK1-MYC activation confers oncogenic transformation, tumor-initiating cell activation, and resistance to mTOR-targeted therapy. Cancer Discov. 3, 1156–1171 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Imbert C., et al. , Resistance of melanoma to immune checkpoint inhibitors is overcome by targeting the sphingosine kinase-1. Nat. Commun. 11, 437 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coppé J. P., et al. , Mapping phospho-catalytic dependencies of therapy-resistant tumours reveals actionable vulnerabilities. Nat. Cell Biol. 21, 778–790 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pan W., et al. , Abiraterone acetate induces CREB1 phosphorylation and enhances the function of the CBP-p300 complex, leading to resistance in prostate cancer cells. Clin. Cancer Res. 27, 2087–2099 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M., Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ben-Yehoyada M., et al. , Checkpoint signaling from a single DNA interstrand crosslink. Mol. Cell 35, 704–715 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hecht S. M., Bleomycin: New perspectives on the mechanism of action. J. Nat. Prod. 63, 158–168 (2000). [DOI] [PubMed] [Google Scholar]
- 62.Chen A. Y., Liu L. F., DNA topoisomerases: Essential enzymes and lethal targets. Annu. Rev. Pharmacol. Toxicol. 34, 191–218 (1994). [DOI] [PubMed] [Google Scholar]
- 63.Čermák V., et al. , Microtubule-targeting agents and their impact on cancer treatment. Eur. J. Cell Biol. 99, 151075 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Bruno P. M., et al. , A subset of platinum-containing chemotherapeutic agents kills cells by inducing ribosome biogenesis stress. Nat. Med. 23, 461–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fu D., Calvo J. A., Samson L. D., Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer 12, 104–120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaye S. B., New antimetabolites in cancer chemotherapy and their clinical impact. Br. J. Cancer 78 (suppl. 3), 1–7 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Allis C. D., Jenuwein T., The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Muhar M., et al. , SLAM-seq defines direct gene-regulatory functions of the BRD4-MYC axis. Science 360, 800–805 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winter G. E., et al. , BET bromodomain proteins function as master transcription elongation factors independent of CDK9 recruitment. Mol. Cell 67, 5–18.e19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Linares-Saldana R., et al. , BRD4 orchestrates genome folding to promote neural crest differentiation. Nat. Genet. 53, 1480–1492 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sakamaki J. I., et al. , Bromodomain protein BRD4 is a transcriptional repressor of autophagy and lysosomal function. Mol. Cell 66, 517–532.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sakamaki J. I., Ryan K. M., Transcriptional regulation of autophagy and lysosomal function by bromodomain protein BRD4. Autophagy 13, 2006–2007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Balciunas D., Gälman C., Ronne H., Björklund S., The Med1 subunit of the yeast mediator complex is involved in both transcriptional activation and repression. Proc. Natl. Acad. Sci. U.S.A. 96, 376–381 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang H., et al. , Targeting CDK9 reactivates epigenetically silenced genes in cancer. Cell 175, 1244–1258.e26 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Otsubo T., Akiyama Y., Yanagihara K., Yuasa Y., SOX2 is frequently downregulated in gastric cancers and inhibits cell growth through cell-cycle arrest and apoptosis. Br. J. Cancer 98, 824–831 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chandra S., Ehrlich K. C., Lacey M., Baribault C., Ehrlich M., Epigenetics and expression of key genes associated with cardiac fibrosis: NLRP3, MMP2, MMP9, CCN2/CTGF and AGT. Epigenomics 13, 219–234 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hnisz D., et al. , Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fresno Vara J. A., et al. , PI3K/Akt signalling pathway and cancer. Cancer Treat. Rev. 30, 193–204 (2004). [DOI] [PubMed] [Google Scholar]
- 79.Zhan T., Rindtorff N., Boutros M., Wnt signaling in cancer. Oncogene 36, 1461–1473 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information. The raw data of ChIP-seq and RNA-seq are deposited at the GEO database (https://www.ncbi.nlm.nih.gov/geo/) with accession number GSE171908.