Abstract
Simple Summary
Thymine DNA Glycosylase (TDG) is a DNA repair protein that plays an important role in gene regulation. Recent studies have shown that TDG interacts with various transcription factors to activate target genes. TDG also functions in a pathway known as active DNA demethylation, which removes 5-mC from DNA and replaces it with unmethylated cytosine. In this review, we summarize the various functions of TDG in gene regulation as well as the physiological relevance of TDG in cancer.
Abstract
DNA methylation is an essential covalent modification that is required for growth and development. Once considered to be a relatively stable epigenetic mark, many studies have established that DNA methylation is dynamic. The 5-methylcytosine (5-mC) mark can be removed through active DNA demethylation in which 5-mC is converted to an unmodified cytosine through an oxidative pathway coupled to base excision repair (BER). The BER enzyme Thymine DNA Glycosylase (TDG) plays a key role in active DNA demethylation by excising intermediates of 5-mC generated by this process. TDG acts as a key player in transcriptional regulation through its interactions with various nuclear receptors and transcription factors, in addition to its involvement in classical BER and active DNA demethylation, which serve to protect the stability of the genome and epigenome, respectively. Recent animal studies have identified a connection between the loss of Tdg and the onset of tumorigenesis. In this review, we summarize the recent findings on TDG’s function as a transcriptional regulator as well as the physiological relevance of TDG and active DNA demethylation in cancer.
Keywords: Thymine DNA Glycosylase, transcription, coactivator, active DNA demethylation, chromatin reorganization, tumor suppressor, cancer
1. Introduction
The methylation of cytosine at the 5th carbon (5-mC) is a prevalent form of DNA modification in mammals that is essential for various biological processes, such as X-chromosome inactivation, genomic imprinting, cell differentiation, and the suppression of mobile genetic elements [1]. In addition, aberrant DNA methylation is a common molecular lesion in cancer, usually causing global hypomethylation and locus-specific hypermethylation of tumor suppressor genes. 5-mC occurs predominantly in the context of CpG islands, which are short, interspersed, CG-rich regions of DNA that were originally identified as an unmethylated fraction of mouse genomic DNA [2]. CpG islands are typically 500–3000 base pairs in length and are found within or proximal to nearly half of the promoters of mammalian genes [3]. For example, all housekeeping genes are associated with promoter CpG islands that are enriched in permissive histone marks, contain multiple transcription factor-binding sites, and are typically unmethylated [4]. CpG islands can also be found within enhancer, intergenic, and intronic regions and are often associated with transcriptional regulation of associated genes.
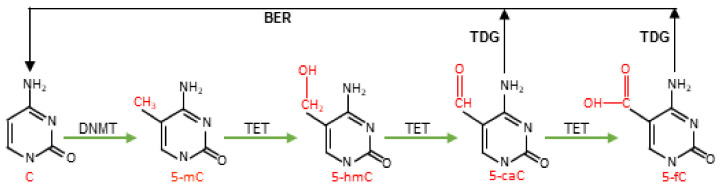
5-mC marks are established during embryonic development by the de novo DNA methyltransferases 3a/3b (DNMT3a/3b), which catalyze the transfer of a methyl group from S-adenosylmethionine (SAM) to the C5 carbon of cytosine [5]. The 5-mC mark is faithfully reproduced over successive replication cycles by the maintenance DNA methyltransferase 1 (DNMT1) in association with the co-factor Ubiquitin-like containing PHD and RING finger domains 1 (UHRF1) [6]. Although 5-mC has long been considered a relatively stable epigenetic mark, several studies have established that DNA methylation, like histone modifications, is dynamic [7,8,9]. In mammals, there are two basic mechanisms involved in the removal of the 5-mC mark: passive and active DNA demethylation. In passive DNA demethylation, the methylated DNA is diluted over successive replication cycles by the deactivation or nuclear exclusion of maintenance DNMT1 or its associated co-factor UHRF1. This is evident during embryonic development, where the maternal genome undergoes passive DNA demethylation by the nuclear exclusion of oocyte specific UHRF1 [10]. Alternatively, 5-mC can be removed in a process called active DNA demethylation that is independent of the cell cycle. Rather, this process is dependent on the oxidative function of the ten eleven translocation (TET) family of proteins, of which there are three functional paralogs (TET1, TET2, and TET3) that exist in mammals due to a gene triplication event that occurred in jawed vertebrates [11,12]. In this process, TETs recognize and successively oxidize 5-mC to 5-hydroxymethylcytosine (5-hmC), and then to 5-formylcytosine (5-fC) and 5-carboxylcytosine (5-caC). 5-fC and 5-caC are then specifically recognized and excised by the base excision repair (BER) protein Thymine DNA Glycosylase (TDG). This generates an apurinic/apyrimidinic (AP) site [13], which is repaired by the combined actions of AP endonuclease 1, DNA Polymerase β and XRCC1-DNA Ligase IIIα complex [14]. This cycle of methylation and active DNA demethylation involving de novo DNMTs, TETs, and BER proteins affords a powerful route to transcriptional manipulation in a replication-independent manner (Figure 1).
Figure 1.
Active DNA demethylation pathway. DNA methyltransferases (DNMT) methylate unmodified C to generate 5-methylcytosine (5-mC), which can be successively oxidized by ten eleven translocation (TET) enzymes to generate 5-hydroxymethylcytosine (5-hmC), 5-formylcytosine (5-fC), and 5-carboxylcytosine (5-caC). Highly oxidized cytosine derivatives, 5-fC and 5-caC, are excised by Thymine DNA Glycosylase (TDG) and repaired through base excision repair (BER) to regenerate unmodified C.
In this review, we will first discuss the role of TDG in transcriptional regulation and in active DNA demethylation. We also discuss the recent work using animal models, which establishes a connection between TDG and tumorigenesis in vivo. A large body of evidence has corroborated the enzymatic function of TDG as well as its scaffolding activities in gene regulation. In addition, the physiological relevance of TDG and active DNA demethylation are becoming clearer since a disruption in these processes leads to symptoms of insulin resistance, loss of bile acid homeostasis, and ultimately cancer. The goal of this review is to provide the reader with an overview of the mechanistic work that has been documented on the role of TDG in gene regulation as a framework for understanding its physiological relevance in cancer (a list of all the abbreviations used in this review can be found at the end of the main text).
2. The Role of TDG in Transcriptional Regulation
The human Tdg gene consists of 10 exons spanning a 23 Kbp region and encodes for a 410 amino acid protein consisting of a centrally positioned catalytic domain flanked by an amino and carboxy terminus, which are lysine-rich and confers important regulatory functions. TDG belongs to the mammalian uracil DNA glycosylase (UDG) superfamily, which all share a common α/β fold [15]. TDG was originally discovered in HeLa cell extracts as a BER enzyme that catalyzes the excision of U:G and T:G mismatches [16]. TDG was then found to glycosylate a variety of mismatched pyrimidine bases, as well as oxidized/halogenated bases such as thymine glycol (Tg), 5-formyluracil (fU), 5-fluorouracil (5-FU), 5-chlorouracil (ClU), 5-hydroxyuracil (5-OHU), 5-hydroxymethyluracil (5-hmU), 3,N4-ethenocytosine (εC), 5-hydroxycytosine (5-OHC), 7,8-dihydro-8-oxoadenine (8oxoA), and 5-bromocytosine (BrC) [17,18,19,20,21,22,23,24]. Recent developments have tied TDG to transcriptional regulation by participating in co-activator complex assembly and active DNA demethylation [25]. Much of the evidence supporting a role for TDG in transcriptional regulation is derived from studies demonstrating its interaction with nuclear hormone receptors. Early reports demonstrated a direct interaction between TDG and the retinoic acid receptor α (RARα) and retinoid X receptor α (RXRα) [26,27]. TDG interacts with RARα/RXRα via its catalytic domain in a ligand-dependent manner, thereby enhancing the affinity of RARα for its response element at target genes [27,28]. Ligand binding was also found to trigger the recruitment of additional co-activators, such as CREB-binding protein (CBP) or its related family member p300, that form a ternary complex with TDG and the RARα [29,30]. Importantly, loss of Tdg was found to be embryonic lethal at E11.5 that resulted partly from a dysregulation in retinoic acid signaling [31,32]. Furthermore, the differentiation of mouse embryonic stem cells (mESCs) in response to retinoic acid was inhibited in Tdg-null mESCs [31]. Mechanistically, TDG was shown to maintain DNA methylation homeostasis and facilitate the recruitment of CBP/p300 and other co-activators at retinoid-dependent target genes. Surprisingly, Tdg knockout also prevented the reprogramming of mouse embryonic fibroblasts to induced pluripotent stem cells, demonstrating the requirement of TDG and active DNA demethylation in cell lineage conversion [33].
TDG has also been shown to interact with other members of the nuclear receptor superfamily, including the androgen receptor (AR), glucocorticoid receptor (GR), progesterone receptor (PR), vitamin D3 receptor (VDR), peroxisome proliferator-activated receptor (PPAR), thyroid hormone receptor (TR), and the estrogen receptor (ER) [27,28,34,35,36]. Utilizing a combination of functional and genomic analysis, the involvement of TDG in ER signaling has recently been documented. ER is a ligand-dependent nuclear receptor overexpressed in many breast cancers and is the target of endocrine-based cancer therapies. TDG was shown to localize at a subset of enhancers of ER target genes in an estrogen-dependent manner. Importantly, approximately half of the TDG binding sites characterized were found to overlap with E2-mediated ER binding [37]. Many of the enhancers occupied by TDG and ER were found to actively transcribe enhancer RNAs (eRNAs) and facilitate a 3-dimensional chromatin reorganization to bring promoter and enhancer element in proximity at target genes. Surprisingly, TDG-dependent eRNA transcription and chromatin reorganization were found to be essential for gene expression of some ER target genes.
To corroborate the effects of TDG on chromatin reorganization, a recent study has shown that TDG has the capacity to alter chromatin structure directly through its physical interactions with DNA [38]. Nucleosome array experiments demonstrated that TDG can decondense or open individual chromatin fibers through its interactions with linker DNA. Remarkably, TDG also promotes condensation through long-range interactions between fibers resulting in oligomerization into higher-order chromatin structures. The terminal domains of TDG are critical for this function and appear to have opposing roles during chromatin condensation. TDG mediates chromatin condensation through its amino terminal domain, whereas its carboxyl terminal domain has an antagonizing effect in the process. Moreover, the authors showed that TDG-mediated chromatin condensation can be reversed by growth arrest and DNA damage-inducible protein alpha (GADD45a), providing evidence that TDG’s interactions with GADD45a and other interacting proteins influences its ability to dynamically control chromatin architecture. Altogether, this comprehensive interaction network depicts TDG as a potential scaffold protein and an important player in protein complex stability at target genes.
3. TDG in Active DNA Demethylation
Numerous studies have shown that TDG efficiently excises 5-fC and 5-caC oxidation products of 5-mC generated by TET enzymes, and it is TDG’s role in active DNA demethylation, which most likely accounts for the embryonic lethality of Tdg knockout mice [39,40,41,42]. Biochemical reconstitution studies using purified recombinant proteins have demonstrated a direct interaction between TET1 and TDG [43]. Furthermore, the TET/TDG complex was highly active and capable of initiating active DNA demethylation in vitro. In the presence of additional BER factors, active DNA demethylation is then completed to correctly re-establish unmodified cytosine on both strands in a sequential manner. TET1 and TDG have also been found to interact physically and are targeted to chromatin by GADD45a [43,44]. GADD45a is a multi-faceted nuclear protein, which has been implicated in DNA demethylation, DNA repair, and genomic stability. Overexpression of GADD45a leads to global DNA demethylation in the presence of TDG and TET proteins, demonstrating that GADD45a enhances DNA demethylation by TDG [45].
The oxidized 5-mC derivatives generated by TETs act as intermediaries for active DNA demethylation and can also accumulate at specific regions throughout the genome. In normal tissues/cell-types, 5-hmC is more abundant than 5-fC/5-caC (>10-fold), as TETs convert 10% of 5-mC to 5-hmC, and only a subset (1–10%) of 5-hmC is converted to 5-fC/5-caC [7]. Although 5mC, 5hmc, and 5fC are all substrates for TET mediated oxidation, they appear to exhibit different substrate and/or catalytic activities. Enzyme kinetic studies suggest that conversion of 5mC to 5hmC is faster than 5hmC to 5fC and 5fC to 5caC. This implies that TET/TDG-mediated oxidation may stall at the 5-hmC step [7].
Genome-wide mapping experiments have shown that 5-hmC is enriched at: (1) promoters that have low CpG density and/or associated with bivalent domains, which are regions that contain both activating and repressive histone marks, typically found in developmental genes that are repressed in ESCs but activated during differentiation, (2) gene bodies of actively transcribed genes, and (3) distal regulatory elements including enhancers, insulators, and regions flanking transcription factor binding sites [46]. The 5-fC and 5-caC metabolites also become enriched within active enhancers, exons, as well as active promoters containing the H3K4me3 chromatin mark, suggestive of an active DNA demethylation process associated with actively transcribing genes [47,48]. Notably, in ESCs, 5-fC/5-caC enrichment increases with the level of promoter accessibility and coincides with the genomic localization of TDG and TET proteins [47].
At a molecular level, 5-fC/5-caC have been shown to impart changes to the physical properties of DNA such as increased DNA flexibility. This can affect supercoiling and packaging of DNA, which can influence gene expression by establishing distinct regulatory regions that directly control the recruitment of specific proteins [49,50]. Interacting proteins specific for each oxidized derivative have been identified that function in DNA repair, transcription, and chromatin modification, suggesting that each metabolite may have a unique biological function [51]. Collectively, these results suggest that 5-mC oxidized derivatives function as stable modifications to modulate biological activity independent of their role as demethylation intermediates.
4. TDG in Cancer
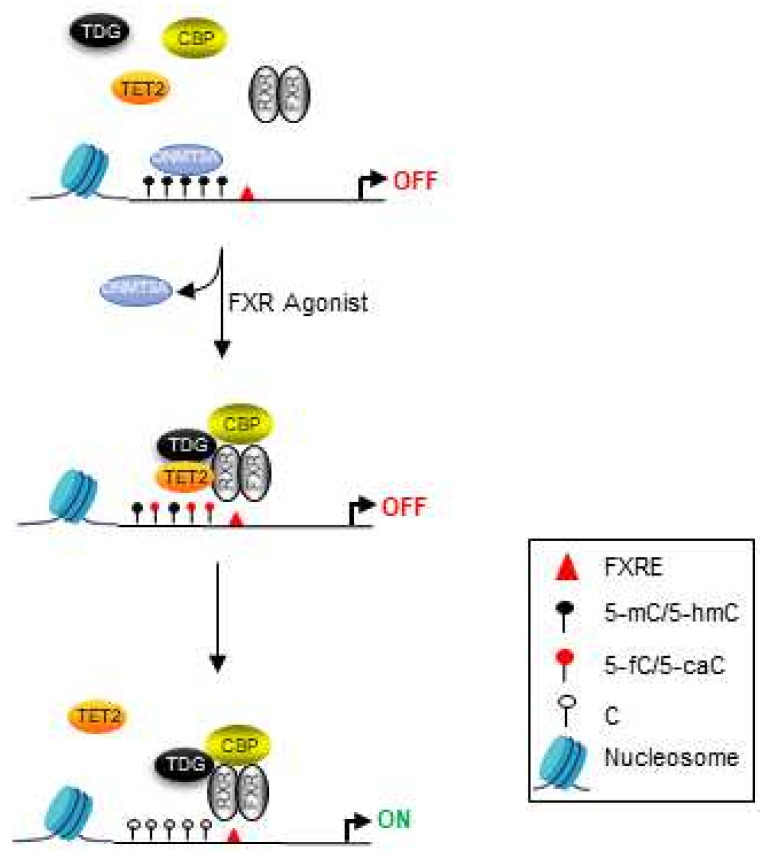
TDG’s involvement in p53 signaling was one of the earliest indications of TDG as a potential tumor suppressor. TDG was found to potentiate p53 signaling, which in turn regulates its own expression [52]. TDG was also found to be essential for the expression of several tumor suppressors genes in vitro, such as p15ink4b, Hic1, Rarβ, and Nr0b2 [29,53,54]. Several conditional Tdg knockout studies performed in mice have since provided support for TDG as a tumor suppressor in vivo [53,55]. One study demonstrated that intestinal-specific loss of Tdg in ApcMin mice, a well-characterized model of tumor disposition, resulted in a two-fold increase in small intestinal adenomas [55]. This phenotype was observed predominantly in female mice, suggesting that sexual dimorphism may contribute to cancer incidence in response to Tdg loss. Utilizing a novel conditional Tdg knockout mouse model containing the tamoxifen-inducible Cre-ERT2 to excise Tdg in all tissues in a temporal fashion [53], our lab demonstrated that the conditional deletion of Tdg in adult mice (TdgcKO) resulted in the development of late-onset hepatocellular carcinoma (HCC) and hepatoblastoma (HB). Interestingly, a sex bias in HCC incidence in the TdgcKO mice was observed, with male mice displaying an approximately 2-fold increase in HCC incidence compared to females. Moreover, male TdgcKO mice displayed increased body weight and glucose intolerance, which are common symptoms associated with obesity and type 2 diabetes, which are major risk factors for HCC [56]. Etiologically, loss of BA homeostasis is a major driver for HCC development in mice and humans [57]. Accordingly, male TdgcKO mice display increased hepatic and serum bile acids (BAs) with age. Immunohistochemistry of TdgcKO livers found that 5-caC staining was more intense in a subpopulation of cells in the liver. This provides evidence that the deletion of Tdg may block active DNA demethylation, leading to an accumulation of 5-caC in the liver in addition to loss of co-activator recruitment and associated transcriptional consequences. The observed phenotypes in TdgcKO mice have been recapitulated using a liver-specific Tdg knockout mouse model (unpublished observations). Through high-throughput transcriptomic analysis of male TdgcKO livers followed by gene-set enrichment analysis, metabolism was identified as the most dysregulated pathway in TdgcKO mice. Considering that the Farnesoid X Receptor (FXR) is the master regulator of diverse metabolic processes, including hepatic BA and glucose metabolism, it is likely that TDG’s coactivating role in FXR signaling plays a considerable role in the maintenance of hepatic homeostasis. Similar to TdgcKO mice, Fxr knockout mice also develop a late-onset HCC and display symptoms associated with obesity and type 2 diabetes, including glucose intolerance and the accumulation of primary bile acids with age [58,59]. Importantly, intraperitoneal injection of mice with an FXR agonist GW4064 caused rapid recruitment of an FXR complex consisting of FXR, TDG, the lysine acetyltransferase CBP, and TET2 to a subset of FXR target genes (Figure 2). Collectively, these findings demonstrate that a loss of Tdg leads to a dysregulation in the FXR-SHP axis in the liver and that TdgcKO mice exhibit an increased prevalence of HCC in a background of elevated serum and intrahepatic BAs. The onset of liver cancer in TdgcKO mice was surprising considering that expression of Tdg is ubiquitous and is in stark contrast to Tet knockouts, which result in predominantly hematopoietic abnormalities and malignancies [60]. Unlike the hematopoietic system, hepatocytes, under normal physiological conditions, are mitotically dormant and mostly found in the quiescent state (G0). This may provide a more favorable environment for the accumulation of 5-fC/5-caC in TdgcKO livers during active DNA demethylation, which is a replication-independent process.
Figure 2.
Transcriptional regulation of FXR-target genes by TET/TDG-mediated active DNA demethylation. In the absence of ligand, DNMT3A is bound to methylated DNA in a transcriptionally inactive state. In the presence of FXR agonist, the FXR/RXR heterodimer recruits TDG, CBP, and TET2 to form a ternary complex at target genes. DNMT3A is displaced and 5-mC undergoes oxidation to 5-fC/5-caC in a TET2/TDG dependent manner. TDG excises 5-fC and 5-caC leading to restoration of the unmethylated cytosine and transcriptional activation.
In contrast to its tumor suppressive properties, two studies have shown that TDG can promote tumourigenesis and may be a potential target for cancer therapy. The first study showed that Tdg is overexpressed in a subset of human colorectal cancer (CRC) patients [61]. TDG acts as a positive regulator of WNT signalling by functioning as an adaptor protein for the transcription factor TCF4 and recruiting CBP/p300. Moreover, stable transfection of TDG shRNA into several CRC cell lines inhibited cell growth. Importantly, stable knockdown of Tdg reduces the ability of CRC cells to form tumors in xenograft assays suggesting that TDG is required for CRC cell proliferation in vivo. More recently, utilizing melanoma cell line models it was shown that inactivation of TDG causes cell cycle arrest and senescence along with increased DNA methylation at a subset of CpG sites [62]. Furthermore, Tdg knockdown was shown to supress tumor formation of melanoma cell lines in xenograft models suggesting that TDG activity is critical for tumor induction and/or progression. Using a high throughput screening assay dependent on TDG catalytic activity, the authors identified first generation TDG inhibitors that decreased viability and clonogenic capacity of melanoma lines.
To date no homozygous mutations have been identified in cancer patients. A heterozygous missense mutation in Tdg that is associated with reduced TDG protein levels has been identified in rectal cancer [63]. In humans, several single nucleotide polymorphisms (SNPs) in the Tdg gene have been associated with an increased risk of cancer development. For example, the SNP rs4135054 is associated with esophageal squamous cell carcinoma (ESCC) [64]. Additionally, a non-synonymous coding SNP rs2888805 (V367M mutation) and an intronic SNP rs4135150 are associated with an increased risk of developing non-melanoma skin cancer (NMSC) and other cancers [65]. A more recent study determined that two other SNPs (rs4135113 and rs1866074) are associated with an increased risk of colorectal cancer [66]. The AA genotype of the SNP rs4135113 increased the risk of colon cancer development by more than 3.6-fold, whereas the minor allele A increased the risk by 1.6-fold. Collectively, these findings suggest that TDG possesses both tumor suppressive properties as well as oncogenic properties depending on the type of cancer involved (Table 1).
Table 1.
Differing roles of TDG in various cancers.
| Cancer | Species | Role of TDG | Phenotype/Effect | References |
|---|---|---|---|---|
| HCC/HB | Mouse | Tumor suppressor | Loss of Tdg results in increased HCC/HB incidence predominantly in male mice | [53] |
| Intestinal adenoma | Mouse | Tumor suppressor | Loss of Tdg results in a two-fold increase in small-intestinal adenomas predominantly in female mice | [55] |
| CRC | Human | Oncogene | Tdg expression is upregulated in human CRC | [61] |
| Human | - | Two SNPs (rs4135113/rs1866074) are associated with increased risk of CRC | [66] | |
| Mouse | Oncogene | Tdg knockdown inhibits xenografted CRC growth in nude mice | [61] | |
| Melanoma | Human | Oncogene | Tdg knockdown/inhibition reduces viability of melanoma cell lines | [62] |
| Rectal cancer | Human | Tumor suppressor | Decreased TDG expression due to D284Y mutation is associated with increased risk of rectal cancer | [63] |
| ESCC | Human | - | SNP (rs4135054) is associated with increased risk of ESCC | [64] |
| NMSC | Human | - | Two SNPs (rs288805/rs4135150) are associated with increased risk of NMSC | [65] |
HCC—hepatocellular carcinoma; HB—hepatoblastoma; CRC—colorectal cancer; ESCC—esophageal squamous-cell carcinoma; NMSC—nonmelanoma skin cancer.
5. Deregulated Active DNA Demethylation and Cancer
Not surprisingly, the genome-wide distribution of 5-mC derivatives is dramatically altered in most cancers compared to normal tissues, which raises the possibility that dysregulation of the DNA demethylation machinery may lead to a reprogramming of the epigenomic landscape in cancer. 5-hmC levels are dramatically reduced in several cancer types including breast, liver, lung, gastric, prostrate, pancreatic, renal, as well as glioblastoma and melanoma [67,68,69,70,71,72,73]. This reduction in 5-hmC largely occurs within gene bodies and regulatory regions such as enhancers and transcription factor binding sites, which may account for promoter hypermethylation observed in most cancers [74]. Low 5-hmC levels serve as a predictive marker for poor prognosis and survival in several cancers and is also correlated with decreased expression of TETs [75]. Furthermore, TET mutations are common in several types of cancers, and the DNA methylation patterns found in TET-deficient cells are similar to those of cancer genomes showing promoter hypermethylation in combination with widespread hypomethylation within heterochromatin [76]. Measurement of 5-fC in ES cells using a chemical pulldown approach has shown that upon Tdg knockdown, the gene promoters that showed the largest increase in 5-fC tended to gain methylation during differentiation of ES cells suggesting that normal 5-fC excision may be critical for the establishment of correct methylation patterns [77]. We have shown that the Hypermethylated in cancer 1 (Hic1) gene undergoes active DNA demethylation in response to retinoic acid and that the loss of active DNA demethylation precedes hypermethylation and silencing of Hic1 in the same tissue [29].
6. Future Perspectives
The recent observation that TDG can alter chromatin structure through direct interaction with DNA, as well as functioning in long range chromatin fiber interactions, adds an additional layer of complexity to the multi-faceted nature of this protein. However, the chromatin remodeling activity has been identified largely using in vitro assays. Additional studies are necessary using genomic approaches based on chromosome capture technology to examine the dependency of the spatial organization of the genome on TDG. The N- and C-terminal regions of TDG are essential for interactions with other proteins and contain important sites for post-translational modifications such as acetylation, phosphorylation, and SUMO conjugation [78]. While it has been suggested that SUMO conjugation causes the release of TDG from abasic sites [79,80], the exact role for these covalent modifications in active DNA demethylation remains unclear and should be explored. Moreover, further studies should investigate potential mechanisms contributing to the sexual dimorphism in cancer development observed in both conditional Tdg knockout mouse models. TDG’s coactivating role in various sex hormone-related pathways (i.e., androgen signaling) may likely have a contributing effect to these sex differences. The discovery of first-generation inhibitors of TDG suggests that TDG is a druggable target in cancer. These inhibitors should be tested on other in vivo models to determine their efficacy for treating other types of cancers. Alternatively, developing molecular tools to introduce site-specific DNA demethylation at hypermethylated, cancer-associated loci may prove to be beneficial as a potential cancer therapy [81]. Further research should be conducted regarding TDG’s role in FXR signaling and hepatic homeostasis, as well as the role of 5-fC and 5-caC in cancer regarding their use as potential biomarkers for various cancers.
7. Conclusions
TDG has key functions in DNA repair, DNA demethylation, and as a transcriptional co-activator. These functions play overlapping roles in gene regulation through associations with various interacting partners. It is becoming evident that active DNA demethylation is a critical aspect of gene regulation that has important ramifications in cancer research. The recent conditional Tdg knockout studies have demonstrated TDG’s role as a tumor suppressor in vivo and as a tumor promoter in some contexts, highlighting the importance of TDG’s role for maintaining normal cellular homeostasis.
Abbreviations
| 5-caC | 5-carboxylcytosine |
| 5-fC | 5-formylcytosine |
| 5-FU | 5-fluorouracil |
| 5-hmC | 5-hydroxymethylcytosine |
| 5-hmU | 5-hydroxymethyluracil |
| 5-mC | 5-methylcytosine |
| 5-OHC | 5-hydroxycytosine |
| 5-OHU | 5-hydroxyuracil |
| 8oxoA | 7,8-dihydro-8-oxoadenine |
| AP | Apurinic/Apyrimidinic |
| AR | Androgen Receptor |
| BAs | Bile Acids |
| BER | Base Excision Repair |
| BrC | 5-bromocytosine |
| CBP | CREB-binding Protein |
| ClU | 5-chlorouracil |
| CRC | Colorectal cancer |
| DNMT | DNA methyltransferase |
| ER | Estrogen Receptor |
| eRNAs | enhancer RNAs |
| ESCC | Esophageal Squamous Cell Carcinoma |
| ɛC | 3,N4-ethenocytosine |
| fU | 5-formyluracil |
| FXR | Farnesoid X Receptor |
| GADD45a | Growth Arrest and DNA damage-inducible Protein Alpha |
| GR | Glucocorticoid Receptor |
| HB | Hepatoblastoma |
| HCC | Hepatocellular Carcinoma |
| Hic1 | Hypermethylated in Cancer 1 |
| mESCs | Mouse Embryonic Stem Cells |
| NMSC | Nonmelanoma Skin Cancer |
| PPAR | Peroxisome Proliferator-Activated Receptor |
| PR | Progesterone Receptor |
| RARα | Retinoic Acid Receptor α |
| RXRα | Retinoid X Receptor α |
| SAM | S-adenosylmethionine |
| SNPs | Single Nucleotide Polymorphisms |
| TDG | Thymine DNA Glycosylase |
| Tdg cKO | Conditional TDG Knockout |
| TET | Ten Eleven Translocation |
| tg | thymine glycol |
| TR | Thyroid Hormone Receptor |
| UDG | Uracil DNA Glycosylase |
| UHRF1 | Ubiquitin-like containing PHD and RING finger domains 1 |
| VDR | Vitamin D3 Receptor |
Author Contributions
Conceptualization, J.T., O.O. and H.M.H.; funding acquisition, J.T.; writing—original draft preparation, O.O., J.T. and H.M.H.; writing—review and editing, J.T., O.O., H.M.H. and M.I. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the CIHR grant PJT155982 and NSERC grant 06542-2018RGPIN to J.T. and O.O. was funded by an NSERC grant 06542-2018RGPIN; HMH was funded by an operating grant from the Cancer Research Society #R2854A12.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bird A. DNA Methylation Patterns and Epigenetic Memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Bird A., Taggart M., Frommer M., Miller O.J., Macleod D. A Fraction of the Mouse Genome That Is Derived from Islands of Nonmethylated, CpG-Rich DNA. Cell. 1985;40:91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- 3.Akan P., Deloukas P. DNA Sequence and Structural Properties as Predictors of Human and Mouse Promoters. Gene. 2008;410:165–176. doi: 10.1016/j.gene.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deaton A.M., Bird A. CpG Islands and the Regulation of Transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okano M., Bell D.W., Haber D.A., Li E. DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for De Novo Methylation and Mammalian Development. Cell. 1999;99:247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 6.Bostick M., Kim J.K., Estève P.-O., Clark A., Pradhan S., Jacobsen S.E. UHRF1 Plays a Role in Maintaining DNA Methylation in Mammalian Cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 7.An J., Rao A., Ko M. TET Family Dioxygenases and DNA Demethylation in Stem Cells and Cancers. Exp. Mol. Med. 2017;49:e323. doi: 10.1038/emm.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhutani N., Burns D.M., Blau H.M. DNA Demethylation Dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohli R.M., Zhang Y. TET Enzymes, TDG and the Dynamics of DNA Demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Messerschmidt D.M., Knowles B.B., Solter D. DNA Methylation Dynamics during Epigenetic Reprogramming in the Germline and Preimplantation Embryos. Genes Dev. 2014;28:812–828. doi: 10.1101/gad.234294.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iyer L.M., Tahiliani M., Rao A., Aravind L. Prediction of Novel Families of Enzymes Involved in Oxidative and Other Complex Modifications of Bases in Nucleic Acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pastor W.A., Aravind L., Rao A. TETonic Shift: Biological Roles of TET Proteins in DNA Demethylation and Transcription. Nat. Rev. Mol. Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu H., Zhang Y. Reversing DNA Methylation: Mechanisms, Genomics, and Biological Functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fromme J.C., Verdine G.L. Base Excision Repair. Adv. Protein Chem. 2004;69:1–41. doi: 10.1016/S0065-3233(04)69001-2. [DOI] [PubMed] [Google Scholar]
- 15.Aravind L., Koonin E.V. The Alpha/Beta Fold Uracil DNA Glycosylases: A Common Origin with Diverse Fates. Genome Biol. 2000;1:research0007.1. doi: 10.1186/gb-2000-1-4-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiebauer K., Jiricny J. In Vitro Correction of G.T Mispairs to G.C Pairs in Nuclear Extracts from Human Cells. Nature. 1989;339:234–236. doi: 10.1038/339234a0. [DOI] [PubMed] [Google Scholar]
- 17.Bennett M.T., Rodgers M.T., Hebert A.S., Ruslander L.E., Eisele L., Drohat A.C. Specificity of Human Thymine DNA Glycosylase Depends on N-Glycosidic Bond Stability. J. Am. Chem. Soc. 2006;128:12510–12519. doi: 10.1021/ja0634829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hang B., Medina M., Fraenkel-Conrat H., Singer B. A 55-KDa Protein Isolated from Human Cells Shows DNA Glycosylase Activity toward 3,N4-Ethenocytosine and the G/T Mismatch. Proc. Natl. Acad. Sci. USA. 1998;95:13561–13566. doi: 10.1073/pnas.95.23.13561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardeland U., Bentele M., Jiricny J., Schär P. Separating Substrate Recognition from Base Hydrolysis in Human Thymine DNA Glycosylase by Mutational Analysis. J. Biol. Chem. 2000;275:33449–33456. doi: 10.1074/jbc.M005095200. [DOI] [PubMed] [Google Scholar]
- 20.Madabushi A., Hwang B.-J., Jin J., Lu A.-L. Histone Deacetylase SIRT1 Modulates and Deacetylates DNA Base Excision Repair Enzyme Thymine DNA Glycosylase. Biochem. J. 2013;456:89–98. doi: 10.1042/BJ20130670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saparbaev M., Laval J. 3,N4-Ethenocytosine, a Highly Mutagenic Adduct, Is a Primary Substrate for Escherichia Coli Double-Stranded Uracil-DNA Glycosylase and Human Mismatch-Specific Thymine-DNA Glycosylase. Proc. Natl. Acad. Sci. USA. 1998;95:8508–8513. doi: 10.1073/pnas.95.15.8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talhaoui I., Couve S., Gros L., Ishchenko A.A., Matkarimov B., Saparbaev M.K. Aberrant Repair Initiated by Mismatch-Specific Thymine-DNA Glycosylases Provides a Mechanism for the Mutational Bias Observed in CpG Islands. Nucleic Acids Res. 2014;42:6300–6313. doi: 10.1093/nar/gku246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon J.-H., Iwai S., O’Connor T.R., Pfeifer G.P. Human Thymine DNA Glycosylase (TDG) and Methyl-CpG-Binding Protein 4 (MBD4) Excise Thymine Glycol (Tg) from a Tg:G Mispair. Nucleic Acids Res. 2003;31:5399–5404. doi: 10.1093/nar/gkg730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Talhaoui I., Couvé S., Ishchenko A.A., Kunz C., Schär P., Saparbaev M. 7,8-Dihydro-8-Oxoadenine, a Highly Mutagenic Adduct, Is Repaired by Escherichia Coli and Human Mismatch-Specific Uracil/Thymine-DNA Glycosylases. Nucleic Acids Res. 2013;41:912–923. doi: 10.1093/nar/gks1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cortázar D., Kunz C., Saito Y., Steinacher R., Schär P. The Enigmatic Thymine DNA Glycosylase. DNA Repair. 2007;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 26.Chevray P.M., Nathans D. Protein Interaction Cloning in Yeast: Identification of Mammalian Proteins That React with the Leucine Zipper of Jun. Proc. Natl. Acad. Sci. USA. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Um S., Harbers M., Benecke A., Pierrat B., Losson R., Chambon P. Retinoic Acid Receptors Interact Physically and Functionally with the T:G Mismatch-Specific Thymine-DNA Glycosylase. J. Biol. Chem. 1998;273:20728–20736. doi: 10.1074/jbc.273.33.20728. [DOI] [PubMed] [Google Scholar]
- 28.Léger H., Smet-Nocca C., Attmane-Elakeb A., Morley-Fletcher S., Benecke A.G., Eilebrecht S. A TDG/CBP/RARα Ternary Complex Mediates the Retinoic Acid-Dependent Expression of DNA Methylation-Sensitive Genes. Genom. Proteom. Bioinform. 2014;12:8–18. doi: 10.1016/j.gpb.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hassan H.M., Kolendowski B., Isovic M., Bose K., Dranse H.J., Sampaio A.V., Underhill T.M., Torchia J. Regulation of Active DNA Demethylation through RAR-Mediated Recruitment of a TET/TDG Complex. Cell Rep. 2017;19:1685–1697. doi: 10.1016/j.celrep.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 30.Tini M., Benecke A., Um S.-J., Torchia J., Evans R.M., Chambon P. Association of CBP/P300 Acetylase and Thymine DNA Glycosylase Links DNA Repair and Transcription. Mol. Cell. 2002;9:265–277. doi: 10.1016/S1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- 31.Cortázar D., Kunz C., Selfridge J., Lettieri T., Saito Y., MacDougall E., Wirz A., Schuermann D., Jacobs A.L., Siegrist F., et al. Embryonic Lethal Phenotype Reveals a Function of TDG in Maintaining Epigenetic Stability. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 32.Cortellino S., Xu J., Sannai M., Moore R., Caretti E., Cigliano A., Le Coz M., Devarajan K., Wessels A., Soprano D., et al. Thymine DNA Glycosylase Is Essential for Active DNA Demethylation by Linked Deamination-Base Excision Repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu X., Zhang L., Mao S.-Q., Li Z., Chen J., Zhang R.-R., Wu H.-P., Gao J., Guo F., Liu W., et al. Tet and TDG Mediate DNA Demethylation Essential for Mesenchymal-to-Epithelial Transition in Somatic Cell Reprogramming. Cell Stem Cell. 2014;14:512–522. doi: 10.1016/j.stem.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Chen D., Lucey M.J., Phoenix F., Lopez-Garcia J., Hart S.M., Losson R., Buluwela L., Coombes R.C., Chambon P., Schär P., et al. T:G Mismatch-Specific Thymine-DNA Glycosylase Potentiates Transcription of Estrogen-Regulated Genes through Direct Interaction with Estrogen Receptor Alpha. J. Biol. Chem. 2003;278:38586–38592. doi: 10.1074/jbc.M304286200. [DOI] [PubMed] [Google Scholar]
- 35.Chiang S., Burch T., Van Domselaar G., Dick K., Radziwon A., Brusnyk C., Edwards M.R., Piper J., Cutts T., Cao J., et al. The Interaction between Thymine DNA Glycosylase and Nuclear Receptor Coactivator 3 Is Required for the Transcriptional Activation of Nuclear Hormone Receptors. Mol. Cell. Biochem. 2010;333:221–232. doi: 10.1007/s11010-009-0223-1. [DOI] [PubMed] [Google Scholar]
- 36.Lucey M.J., Chen D., Lopez-Garcia J., Hart S.M., Phoenix F., Al-Jehani R., Alao J.P., White R., Kindle K.B., Losson R., et al. T:G Mismatch-Specific Thymine-DNA Glycosylase (TDG) as a Coregulator of Transcription Interacts with SRC1 Family Members through a Novel Tyrosine Repeat Motif. Nucleic Acids Res. 2005;33:6393–6404. doi: 10.1093/nar/gki940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolendowski B., Hassan H., Krstic M., Isovic M., Thillainadesan G., Chambers A.F., Tuck A.B., Torchia J. Genome-Wide Analysis Reveals a Role for TDG in Estrogen Receptor-Mediated Enhancer RNA Transcription and 3-Dimensional Reorganization. Epigenet. Chromatin. 2018;11:5. doi: 10.1186/s13072-018-0176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deckard C.E., Sczepanski J.T. Reversible Chromatin Condensation by the DNA Repair and Demethylation Factor Thymine DNA Glycosylase. Nucleic Acids Res. 2021;49:2450–2459. doi: 10.1093/nar/gkab040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He Y.-F., Li B.-Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., et al. Tet-Mediated Formation of 5-Carboxylcytosine and Its Excision by TDG in Mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L., Lu X., Lu J., Liang H., Dai Q., Xu G.-L., Luo C., Jiang H., He C. Thymine DNA Glycosylase Specifically Recognizes 5-Carboxylcytosine-Modified DNA. Nat. Chem. Biol. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pidugu L.S., Flowers J.W., Coey C.T., Pozharski E., Greenberg M.M., Drohat A.C. Structural Basis for Excision of 5-Formylcytosine by Thymine DNA Glycosylase. Biochemistry. 2016;55:6205–6208. doi: 10.1021/acs.biochem.6b00982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maiti A., Drohat A.C. Thymine DNA Glycosylase Can Rapidly Excise 5-Formylcytosine and 5-Carboxylcytosine: Potential Implications for Active Demethylation of CpG Sites. J. Biol. Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weber A.R., Krawczyk C., Robertson A.B., Kuśnierczyk A., Vågbø C.B., Schuermann D., Klungland A., Schär P. Biochemical Reconstitution of TET1–TDG–BER-Dependent Active DNA Demethylation Reveals a Highly Coordinated Mechanism. Nat. Commun. 2016;7:10806. doi: 10.1038/ncomms10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Z., Gu T.-P., Weber A.R., Shen J.-Z., Li B.-Z., Xie Z.-G., Yin R., Guo F., Liu X., Tang F., et al. Gadd45a Promotes DNA Demethylation through TDG. Nucleic Acids Res. 2015;43:3986–3997. doi: 10.1093/nar/gkv283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barreto G., Schäfer A., Marhold J., Stach D., Swaminathan S.K., Handa V., Döderlein G., Maltry N., Wu W., Lyko F., et al. Gadd45a Promotes Epigenetic Gene Activation by Repair-Mediated DNA Demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 46.Wu X., Zhang Y. TET-Mediated Active DNA Demethylation: Mechanism, Function and Beyond. Nat. Rev. Genet. 2017;18:517–534. doi: 10.1038/nrg.2017.33. [DOI] [PubMed] [Google Scholar]
- 47.Neri F., Incarnato D., Krepelova A., Rapelli S., Anselmi F., Parlato C., Medana C., Dal Bello F., Oliviero S. Single-Base Resolution Analysis of 5-Formyl and 5-Carboxyl Cytosine Reveals Promoter DNA Methylation Dynamics. Cell Rep. 2015;10:674–683. doi: 10.1016/j.celrep.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 48.Neri F., Incarnato D., Krepelova A., Parlato C., Oliviero S. Methylation-Assisted Bisulfite Sequencing to Simultaneously Map 5fC and 5caC on a Genome-Wide Scale for DNA Demethylation Analysis. Nat. Protoc. 2016;11:1191–1205. doi: 10.1038/nprot.2016.063. [DOI] [PubMed] [Google Scholar]
- 49.Raiber E.-A., Portella G., Martínez Cuesta S., Hardisty R., Murat P., Li Z., Iurlaro M., Dean W., Spindel J., Beraldi D., et al. 5-Formylcytosine Organizes Nucleosomes and Forms Schiff Base Interactions with Histones in Mouse Embryonic Stem Cells. Nat. Chem. 2018;10:1258–1266. doi: 10.1038/s41557-018-0149-x. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y., Zhou C. Formation and Biological Consequences of 5-Formylcytosine in Genomic DNA. DNA Repair. 2019;81:102649. doi: 10.1016/j.dnarep.2019.102649. [DOI] [PubMed] [Google Scholar]
- 51.Spruijt C.G., Gnerlich F., Smits A.H., Pfaffeneder T., Jansen P.W.T.C., Bauer C., Münzel M., Wagner M., Müller M., Khan F., et al. Dynamic Readers for 5-(Hydroxy)Methylcytosine and Its Oxidized Derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 52.Kim E.-J., Um S.-J. Thymine–DNA Glycosylase Interacts with and Functions as a Coactivator of P53 Family Proteins. Biochem. Biophys. Res. Commun. 2008;377:838–842. doi: 10.1016/j.bbrc.2008.10.058. [DOI] [PubMed] [Google Scholar]
- 53.Hassan H.M., Isovic M., Kolendowski B., Bauer-Maison N., Onabote O., Cecchini M., Haig A., Maleki Vareki S., Underhill T.M., Torchia J. Loss of Thymine DNA Glycosylase Causes Dysregulation of Bile Acid Homeostasis and Hepatocellular Carcinoma. Cell Rep. 2020;31:107475. doi: 10.1016/j.celrep.2020.03.039. [DOI] [PubMed] [Google Scholar]
- 54.Thillainadesan G., Chitilian J.M., Isovic M., Ablack J.N.G., Mymryk J.S., Tini M., Torchia J. TGF-β-Dependent Active Demethylation and Expression of the P15ink4b Tumor Suppressor Are Impaired by the ZNF217/CoREST Complex. Mol. Cell. 2012;46:636–649. doi: 10.1016/j.molcel.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 55.Xu J., Cortellino S., Tricarico R., Chang W.-C., Scher G., Devarajan K., Slifker M., Moore R., Bassi M.R., Caretti E., et al. Thymine DNA Glycosylase (TDG) Is Involved in the Pathogenesis of Intestinal Tumors with Reduced APC Expression. Oncotarget. 2017;8:89988–89997. doi: 10.18632/oncotarget.21219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanyal A.J., Yoon S.K., Lencioni R. The Etiology of Hepatocellular Carcinoma and Consequences for Treatment. Oncologist. 2010;15:14–22. doi: 10.1634/theoncologist.2010-S4-14. [DOI] [PubMed] [Google Scholar]
- 57.Wang X., Fu X., Van Ness C., Meng Z., Ma X., Huang W. Bile Acid Receptors and Liver Cancer. Curr. Pathobiol. Rep. 2013;1:29–35. doi: 10.1007/s40139-012-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim I., Morimura K., Shah Y., Yang Q., Ward J.M., Gonzalez F.J. Spontaneous Hepatocarcinogenesis in Farnesoid X Receptor-Null Mice. Carcinogenesis. 2007;28:940–946. doi: 10.1093/carcin/bgl249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang F., Huang X., Yi T., Yen Y., Moore D.D., Huang W. Spontaneous Development of Liver Tumors in the Absence of the Bile Acid Receptor Farnesoid X Receptor. Cancer Res. 2007;67:863–867. doi: 10.1158/0008-5472.CAN-06-1078. [DOI] [PubMed] [Google Scholar]
- 60.Ko M., An J., Pastor W.A., Koralov S.B., Rajewsky K., Rao A. TET Proteins and 5-Methylcytosine Oxidation in Hematological Cancers. Immunol. Rev. 2015;263:6–21. doi: 10.1111/imr.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu X., Yu T., Shi J., Chen X., Zhang W., Lin T., Liu Z., Wang Y., Zeng Z., Wang C., et al. Thymine DNA Glycosylase Is a Positive Regulator of Wnt Signaling in Colorectal Cancer. J. Biol. Chem. 2014;289:8881–8890. doi: 10.1074/jbc.M113.538835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mancuso P., Tricarico R., Bhattacharjee V., Cosentino L., Kadariya Y., Jelinek J., Nicolas E., Einarson M., Beeharry N., Devarajan K., et al. Thymine DNA Glycosylase as a Novel Target for Melanoma. Oncogene. 2019;38:3710–3728. doi: 10.1038/s41388-018-0640-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vasovcak P., Krepelova A., Menigatti M., Puchmajerova A., Skapa P., Augustinakova A., Amann G., Wernstedt A., Jiricny J., Marra G., et al. Unique Mutational Profile Associated with a Loss of TDG Expression in the Rectal Cancer of a Patient with a Constitutional PMS2 Deficiency. DNA Repair. 2012;11:616–623. doi: 10.1016/j.dnarep.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li W.-Q., Hu N., Hyland P.L., Gao Y., Wang Z.-M., Yu K., Su H., Wang C.-Y., Wang L.-M., Chanock S.J., et al. Genetic Variants in DNA Repair Pathway Genes and Risk of Esophageal Squamous Cell Carcinoma and Gastric Adenocarcinoma in a Chinese Population. Carcinogenesis. 2013;34:1536–1542. doi: 10.1093/carcin/bgt094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ruczinski I., Jorgensen T.J., Shugart Y.Y., Schaad Y.B., Kessing B., Hoffman-Bolton J., Helzlsouer K.J., Kao W.H.L., Wheless L., Francis L., et al. A Population-Based Study of DNA Repair Gene Variants in Relation to Non-Melanoma Skin Cancer as a Marker of a Cancer-Prone Phenotype. Carcinogenesis. 2012;33:1692–1698. doi: 10.1093/carcin/bgs170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reddy Parine N., Alanazi I.O., Shaik J.P., Aldhaian S., Aljebreen A.M., Alharbi O., Almadi M.A., Azzam N.A., Alanazi M. TDG Gene Polymorphisms and Their Possible Association with Colorectal Cancer: A Case Control Study. J. Oncol. 2019;2019:7091815. doi: 10.1155/2019/7091815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bhattacharyya S., Yu Y., Suzuki M., Campbell N., Mazdo J., Vasanthakumar A., Bhagat T.D., Nischal S., Christopeit M., Parekh S., et al. Genome-Wide Hydroxymethylation Tested Using the HELP-GT Assay Shows Redistribution in Cancer. Nucleic Acids Res. 2013;41:e157. doi: 10.1093/nar/gkt601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen K., Zhang J., Guo Z., Ma Q., Xu Z., Zhou Y., Xu Z., Li Z., Liu Y., Ye X., et al. Loss of 5-Hydroxymethylcytosine Is Linked to Gene Body Hypermethylation in Kidney Cancer. Cell Res. 2016;26:103–118. doi: 10.1038/cr.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kudo Y., Tateishi K., Yamamoto K., Yamamoto S., Asaoka Y., Ijichi H., Nagae G., Yoshida H., Aburatani H., Koike K. Loss of 5-Hydroxymethylcytosine Is Accompanied with Malignant Cellular Transformation. Cancer Sci. 2012;103:670–676. doi: 10.1111/j.1349-7006.2012.02213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lian C.G., Xu Y., Ceol C., Wu F., Larson A., Dresser K., Xu W., Tan L., Hu Y., Zhan Q., et al. Loss of 5-Hydroxymethylcytosine Is an Epigenetic Hallmark of Melanoma. Cell. 2012;150:1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu C., Liu L., Chen X., Shen J., Shan J., Xu Y., Yang Z., Wu L., Xia F., Bie P., et al. Decrease of 5-Hydroxymethylcytosine Is Associated with Progression of Hepatocellular Carcinoma through Downregulation of TET1. PLoS ONE. 2013;8:e62828. doi: 10.1371/journal.pone.0062828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Turcan S., Rohle D., Goenka A., Walsh L.A., Fang F., Yilmaz E., Campos C., Fabius A.W.M., Lu C., Ward P.S., et al. IDH1 Mutation Is Sufficient to Establish the Glioma Hypermethylator Phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang H., Liu Y., Bai F., Zhang J.-Y., Ma S.-H., Liu J., Xu Z.-D., Zhu H.-G., Ling Z.-Q., Ye D., et al. Tumor Development Is Associated with Decrease of TET Gene Expression and 5-Methylcytosine Hydroxylation. Oncogene. 2013;32:663–669. doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Azizgolshani N., Petersen C.L., Chen Y., Levy J.J., Salas L.A., Perreard L., Nguyen L.N., Christensen B.C. DNA 5-Hydroxymethylcytosine in Pediatric Central Nervous System Tumors May Impact Tumor Classification and Is a Positive Prognostic Marker. Clin. Epigenet. 2021;13:176. doi: 10.1186/s13148-021-01156-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rasmussen K.D., Helin K. Role of TET Enzymes in DNA Methylation, Development, and Cancer. Genes Dev. 2016;30:733–750. doi: 10.1101/gad.276568.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lio C.-W.J., Yue X., Lopez-Moyado I.F., Tahiliani M., Aravind L., Rao A. TET Methylcytosine Oxidases: New Insights from a Decade of Research. J. Biosci. 2020;45:21. doi: 10.1007/s12038-019-9973-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Raiber E.-A., Beraldi D., Ficz G., Burgess H.E., Branco M.R., Murat P., Oxley D., Booth M.J., Reik W., Balasubramanian S. Genome-Wide Distribution of 5-Formylcytosine in Embryonic Stem Cells Is Associated with Transcription and Depends on Thymine DNA Glycosylase. Genome Biol. 2012;13:R69. doi: 10.1186/gb-2012-13-8-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mohan R.D., Litchfield D.W., Torchia J., Tini M. Opposing Regulatory Roles of Phosphorylation and Acetylation in DNA Mispair Processing by Thymine DNA Glycosylase. Nucleic Acids Res. 2010;38:1135–1148. doi: 10.1093/nar/gkp1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mohan R.D., Rao A., Gagliardi J., Tini M. SUMO-1-Dependent Allosteric Regulation of Thymine DNA Glycosylase Alters Subnuclear Localization and CBP/P300 Recruitment. Mol. Cell. Biol. 2007;27:229–243. doi: 10.1128/MCB.00323-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Steinacher R., Schär P. Functionality of Human Thymine DNA Glycosylase Requires SUMO-Regulated Changes in Protein Conformation. Curr. Biol. 2005;15:616–623. doi: 10.1016/j.cub.2005.02.054. [DOI] [PubMed] [Google Scholar]
- 81.Neja S.A. Site-Specific DNA Demethylation as a Potential Target for Cancer Epigenetic Therapy. Epigenetics Insights. 2020;13:2516865720964808. doi: 10.1177/2516865720964808. [DOI] [PMC free article] [PubMed] [Google Scholar]