Abstract
Pathogenesis-related 1 (PR-1) proteins, which are defense proteins in plant–pathogen interactions, play an important role in the resistance and defense of plants against diseases. Blister blight disease is caused by Exobasidium vexans Massee and a major leaf disease of tea plants (Camellia sinensis (L.) O. Kuntze). However, the systematic characterization and analysis of the PR-1 gene family in tea plants is still lacking, and the defense mechanism of this family remains unknown. In this study, 17 CsPR-1 genes were identified from the tea plant genome and classified into five groups based on their signal peptide, isoelectric point, and C-terminus extension. Most of the CsPR-1 proteins contained an N-terminal signal peptide and a conserved PR-1 like domain. CsPR-1 genes comprised multiple cis-acting elements and were closely related to the signal-transduction pathways involving TCA, NPR1, EDS16, BGL2, PR4, and HCHIB. These characteristics imply an important role of the genes in the defense of the tea plant. In addition, the RNA-seq data and real-time PCR analysis demonstrated that the CsPR-1-2, -4, -6, -7, -8, -9, -10, -14, -15, and -17 genes were significantly upregulated under tea blister-blight stress. This study could help to increase understanding of CsPR-1 genes and their defense mechanism in response to tea blister blight.
Keywords: PR-1, blister blight, Camellia sinensis, defense mechanism
1. Introduction
The plant immune system has multiple layers of defense responses to intercept the infection and damage caused by pathogenic microorganisms [1]. In the first barrier of the immune system, plants recognize the conserved pathogen-associated molecular patterns (PAMPs) of slowly evolving microorganisms or pathogens through the pattern-recognition receptors (PRRS) located on the surface of the cell membrane to activate pattern-triggered immunity (PTI) [2]. However, compatible pathogens can overcome this first layer of defense. Plants specifically recognize effectors via polymorphic nucleotide-binding-domain and leucine-rich repeat (NB-LRR) proteins encoded by most resistance genes (R genes) directly or indirectly to activate effector-triggered immunity (ETI) [3]. According to the products encoded by defense genes and their functions, these genes can be divided into secondary metabolite-synthesis genes, cell-wall-modification -related genes, protease-inhibitor genes, and plant-pathogenesis-related (PR) genes [4].
Numerous studies have shown that PR genes play important roles in the resistance and defense of plants against diseases [5,6,7]. PR proteins are induced and accumulated in host plants during attacks by oomycetes, fungi, bacteria, viruses, or insects. Seventeen families of PR proteins have been classified and characterized according to the homology of amino-acid sequences, serological relationships, and enzyme activities [8]. PR-1 proteins are related to cysteine-rich secretory proteins, the core regions of which are relatively conservative in various species [9]. The existence of a PR-1-like domain helps to deal with the stress of an adverse environment and improves the stability of PR-1-like proteins [10]. The CAP-derived peptide 1 (CAPE1, consensus motif PxGNxxxxxPY) has been found in the PR-1-like domain, which is related to stress signaling and conserved between the monocots and dicots [11]. PR-1 proteins occur in multigene families within plant genomes and can be further classified as acidic or basic, depending on their theoretical isoelectric point (pI) [12]. Numerous studies have demonstrated that PR-1 proteins abundantly accumulate during plant-defense responses and are ubiquitous across plant species [8,13]. In grapevine (‘BN5-4’), the basic-type VvPR1b1 gene confers high resistance against Pseudomonas syringae [14]. The WjAMP-2 (PR-1) protein has been purified from Wasabia japonica and strongly inhibits activity against phytopathogenic fungi, and the overexpression of WjAMP-2 in tobacco could enhance resistance to Botrytis cinerea [15]. In addition, PR-1 gene expression has been considered a reliable marker for the salicylic-acid (SA)-mediated defense pathway and the activation of systemic acquired resistance (SAR) in various plant species [16]. Recently, with the development of gene-sequencing technology and the improvement of bioinformatics prediction systems, a growing number of gene families has been identified [17,18,19,20,21,22]. At present, the PR-1 gene family has been identified, as well expression profiles in response to biotic and abiotic stresses in many species—for instance, 11 genes in Piper nigrum [13], 13 in Solanum lycopersicum [23], 21 in Vitis vinifera [14], 22 in Arabidopsis [8], 23 in Triticum aestivum [24], 24 in Glycine max [25], and 32 in Oryza sativa [8]. Clarifying the classification and composition of gene-family members in the genome is the first step to explore species-related characteristics and biological problems and can lay a solid foundation for subsequent gene-function characterization and genetic manipulation.
Tea, which is the most consumed nonalcoholic beverage worldwide, is made of the apical buds and tender leaves of tea plants (Camellia sinensis (L.) O. Kuntze) [26]. During the growth of these plants, tea leaves are susceptible to various fungal pathogens, severely reducing the yield and quality of tea. Tea blister blight disease, caused by Exobasidium vexans Massee, is one of the serious diseases of tea plants [27]. E. vexans mainly damages the tender leaves and stems of tea plants, causing the young, diseased tissues to be covered with white blisters [28]. As the disease develops, the fungal infection leads to necrosis and withering of a large number of leaf tissues, and the tea leaves gradually turn brown and curl [29]. Nevertheless, the resistance mechanism of the response of tea plants to blister blight disease has not yet been explained in detail. Transcriptome analysis has identified 149 putative key defense-related transcripts/genes involving defense-related enzymes, resistance genes, multidrug-resistant transporters, and transcription factors [30]. Foliar application of 0.01% chitosan could induce the expression of different defense-related enzymes via NO signaling to reduce the incidence of blister blight [31]. Although an important gene family in the resistance and defense of plants against diseases, PR-1 genes have not been revealed in tea plants, and the biological functions of the members of this family remain unknown. In this study, CsPR-1 genes were identified based on transcriptome data analysis and the tea genome database. The genes were characterized, and expression patterns were detected and analyzed. The results could provide an important foundation for the exploration of the biological functions of the CsPR-1 gene family in tea plants.
2. Results
2.1. Identification and Characterization of CsPR-1 Gene-Family Members
Using the Tea Plant Genome Database and bioinformatics analysis, 17 CsPR-1 gene members and their corresponding amino-acid sequences were confirmed and listed in Table S1. These CsPR-1 genes were named CsPR-1-1 to CsPR-1-17 according to the sequence of their genomic output. The CsPR-1 proteins ranged from 156 aa (CsPR-1-10) to 340 aa (CsPR-1-2), and the molecular weights (MWs) were between 17.37 kDa (CsPR-1-10) and 38.28 kDa (CsPR-1-2). The pI values varied from 4.84 (CsPR-1-10) to 10.07 (CsPR-1-2), indicating the presence of PR-1 protein-encoding genes with both acidic and basic amino acids in the tea plant genome. Hydropathicity (GRAVY) analysis showed that the GRAVY values of all analyzed CsPR-1 genes were negative, indicating their high hydrophilicity. More detailed information, including the sequence ID, chromosomes, and subcellular localization, is listed in Table 1.
Table 1.
Identified PR1 genes in the genome of the tea plant.
| Sequence ID | Gene | Chromosome | Protein | |||||
|---|---|---|---|---|---|---|---|---|
| Length (aa) | MW (kD) | pI | Gravy | SP | Subcellular Location | |||
| TEA022692.1 | CsPR1-1 | Chr04 | 163 | 18.07 | 7.08 | −0.353 | 24 | extr: 7, chlo: 6, cyto: 1 |
| TEA002521.1 | CsPR1-2 | Chr06 | 340 | 38.28 | 10.07 | −1.089 | 27 | vacu: 5, nucl: 3, cyto: 2, plas: 2, chlo: 1, E.R.: 1 |
| TEA017943.1 | CsPR1-3 | Chr06 | 162 | 17.93 | 8.74 | −0.213 | 24 | extr: 6, vacu: 5, chlo: 2, golg: 1 |
| TEA021774.1 | CsPR1-4 | Chr13 | 162 | 17.93 | 8.74 | −0.213 | 24 | extr: 6, vacu: 5, chlo: 2, golg: 1 |
| TEA022146.1 | CsPR1-5 | Chr01 | 173 | 18.61 | 6.81 | −0.227 | 21 | extr: 4, chlo: 3, vacu: 3, cyto: 2, plas: 2 |
| TEA004542.1 | CsPR1-6 | Chr06 | 160 | 17.59 | 4.96 | −0.235 | 24 | extr: 12, vacu: 2 |
| TEA022585.1 | CsPR1-7 | Chr08 | 170 | 19.25 | 9.22 | −0.375 | 24 | chlo: 12, extr: 1, vacu: 1 |
| TEA028234.1 | CsPR1-8 | Chr01 | 162 | 18.26 | 9.16 | −0.399 | 24 | chlo: 4, vacu: 3, extr: 2, E.R.: 2, nucl: 1, mito: 1, plas: 1 |
| TEA004551.1 | CsPR1-9 | Chr06 | 159 | 17.52 | 8.47 | −0.27 | 22 | chlo: 14 |
| TEA028218.1 | CsPR1-10 | Chr01 | 156 | 17.37 | 4.84 | −0.406 | 28 | chlo: 11, cyto: 2, nucl: 1 |
| TEA022150.1 | CsPR1-11 | Chr01 | 191 | 21.53 | 9.16 | −0.34 | \ | vacu: 6, chlo: 4, extr: 2, nucl: 1, plas: 1 |
| TEA030748.1 | CsPR1-12 | unkown | 204 | 22.60 | 5.66 | −0.257 | \ | E.R.: 5.5, cyto: 5, E.R._plas: 3.5, nucl: 1, mito: 1, vacu: 1 |
| TEA025681.1 | CsPR1-13 | Chr03 | 196 | 22.61 | 6.09 | −0.479 | 23 | extr: 7, chlo: 4, E.R.: 2, vacu: 1 |
| TEA022240.1 | CsPR1-14 | unkown | 162 | 17.88 | 6.86 | −0.297 | 24 | chlo: 13, vacu: 1 |
| TEA021361.1 | CsPR1-15 | Chr03 | 160 | 17.62 | 5.38 | −0.269 | 24 | extr: 11, vacu: 2, cyto: 1 |
| TEA011597.1 | CsPR1-16 | unkown | 168 | 17.94 | 8.57 | −0.213 | 24 | chlo: 6, vacu: 5, extr: 2, golg: 1 |
| TEA004541.1 | CsPR1-17 | Chr06 | 162 | 17.87 | 8.62 | −0.345 | 24 | extr: 7, vacu: 4, chlo: 2, mito: 1 |
2.2. Analysis of the Genomic and Domain Structures of CsPR-1 Genes
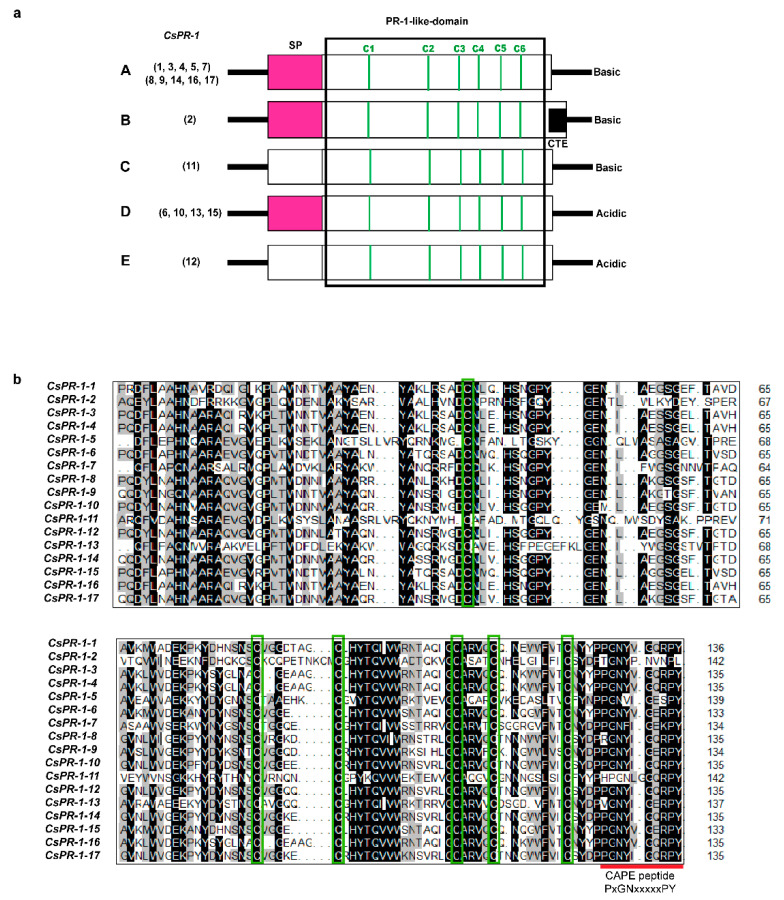
According to the calculated pI values, the predicted signal peptide (SP), and the C-terminal extension (CTE) of the deduced proteins, the 17 CsPR-1 genes were classified into five major groups (Figure 1a). Group A included 10 genes (CsPR-1-1, -3, -4, -5, -7, -8, -9, -14, -16, and -17), which all encoded basic (pI = 7.08-9.17) PR-1 proteins containing an N-terminal SP and a PR-1 like domain of 134 aa to 139 aa. Group B included only CsPR-1-2, which encoded basic (pI = 10.64) protein containing an SP and a PR-1-like domain of 142 aa, followed by a CTE. Group C was comprised of CsPR-1-11, encoded basic (pI = 9.13) protein with a PR-1-like domain of 141 aa. Group D included four genes (CsPR-1-6, -10, -13, -14, and -15), which all encoded basic (pI = 4.75–6.49) proteins containing an SP, followed by a conserved PR-1like domain of 133 aa to 137 aa. Group E (CsPR-1-12) encoded a basic (pI = 5.07) protein containing a PR-1 like domain of 135 aa. The amino-acid sequences of the PR-1-like domain of the 17 deduced CsPR-1 proteins were aligned. Most CsPR-1 proteins have three strictly conserved cysteine-residue pairs and a CAPE 1 peptide (Figure 1b).
Figure 1.
Genomic and domain structures of the CsPR-1 genes. (a) Diagram of the genomic structures of the CsPR-1 genes. Open boxes signify the open reading frames (ORFs). Signal-peptide (SP) regions are shaded in pinkish red. Vertical green solid bars represent the positions of the six conserved cysteine residues (C1-C6). The box drawn with black solid lines indicates the conserved PR-1-like domains (pfam cd05381). The interior black box indicates the C-terminal extension (CTE). (b) Amino-acid alignment of the PR-1-like domain of the deduced CsPR-1 proteins. DNAMAN 7.0 was used to mark the amino-acid residues with an identity of more than 50% with black shade. The areas in the interior green boxes and the red solid line indicate the C1-C6 and CAPE peptide, respectively.
2.3. Conserved Motifs of the CsPR-1 Protein Structure
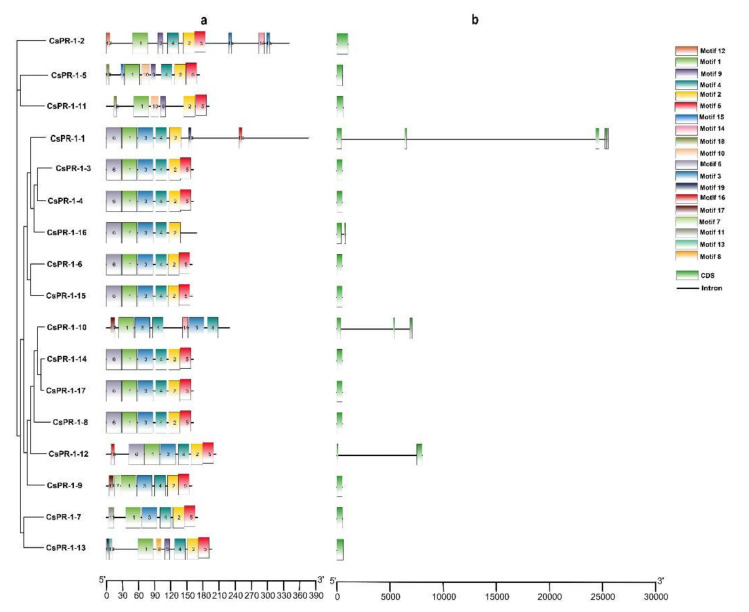
To explore the structural diversity of the CsPR-1 genes in tea plants, 20 conserved motifs in the CsPR-1 gene family were predicted by multiple-expectation maximization for motif elicitation (MEME) (Table S2). All CsPR-1 proteins contained motifs 1 and 2. Motif 3 was present in all CsPR-1 proteins, except in CsPR-1-2, -5, -11, and -13. Motif 4 was present in all CsPR-1 proteins, except in CsPR-1-11. Motif 5 was present in all CsPR-1 proteins, except in CsPR-1-1, -10, and -16. Motifs 7, 8, and 12 were present, along with CsPR-1-9, -13, and -2, respectively (Figure 2a). Exon-intron diagrams of the CsPR-1 genes were further generated based on their genome sequences and corresponding coding sequences. Most of the tea plant CsPR-1 genes did not contain introns, except for CsPR-1-1, -10, -12, and-16, which contained between one and three introns (Figure 2b). A visualization of the conserved motif and gene structure clearly shows that most members of the tea plant CsPR-1 gene family have similar structures.
Figure 2.
Phylogenetic clustering, motif compositions, and gene structures of the CsPR-1 family members in tea plants. (a) The motifs of each CsPR-1, as well as 20 different motifs, are represented by different colored boxes. (b) Exon-intron structural analyses of CsPR-1.
2.4. Functional Interaction Networks of CsPR-1 Proteins
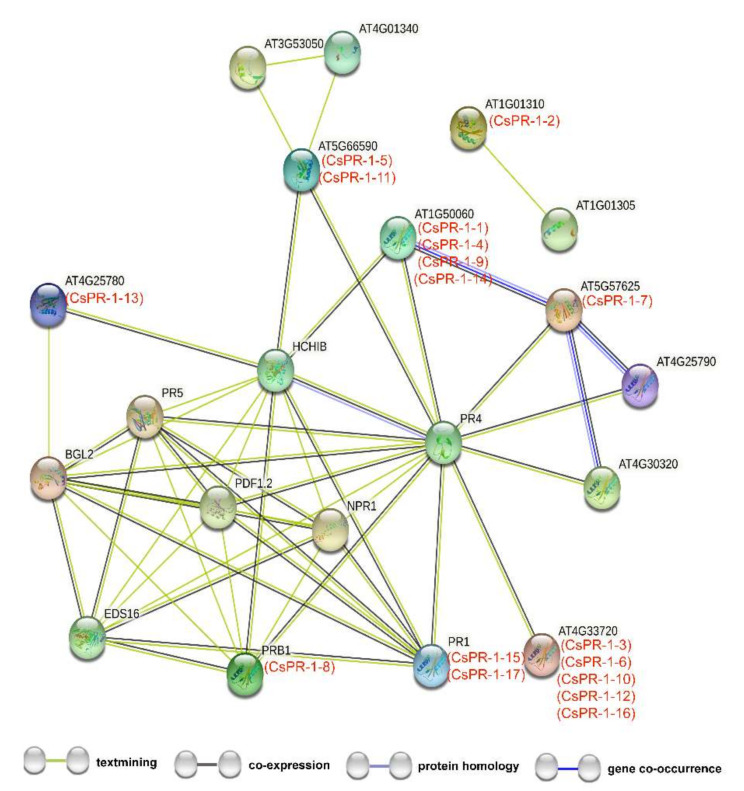
Functional interaction networks of the CsPR-1 proteins were constructed using STRING software on the basis of the homologous proteins in Arabidopsis (Table S3). The results show that the 17 CsPR-1 proteins participated in the interaction network, indicating a universal and complex interaction of CsPR-1 proteins (Figure 3). Most CsPR-1 proteins were involved in the SA-signaling pathway or JA/ET-signaling pathway by interacting with NPR1, EDS16, BGL2, PR4, and HCHIB. CsPR-1-12 was independent.
Figure 3.
Functional interaction networks of the CsPR-1 protein in the tea plant according to orthologs in Arabidopsis are shown in red and black.
2.5. Analysis of Cis Elements of the CsPR-1 Genes
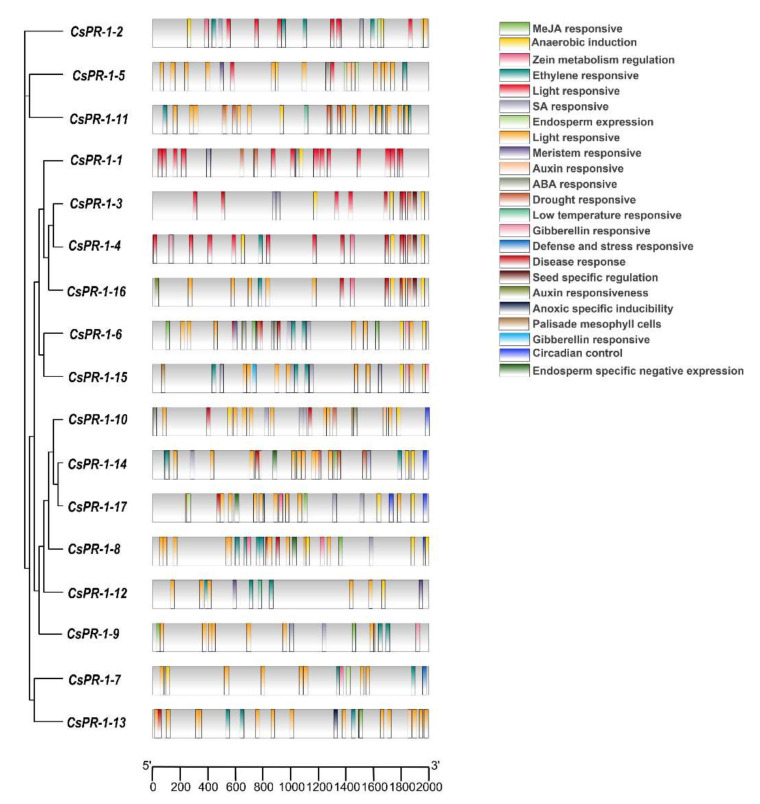
The cis element in the CsPR-1 promoters were analyzed using Plant CARE. The results show many hormone-response elements, biological stress-response elements, and abiotic stress-response elements in the CsPR-1 promoters (Figure 4). Various response-hormone elements, such as SA (TCA-element, as-1), Me-JA (TGACG-motif, CGTCA-motif), ABA (ABRE), GA (P-box, GARE-motif), ET (ERE), and IAA (TGA-element, AuxRR-core) were present. These hormones, especially SA and JA, are closely related to the resistance. SA and JA cis elements were present in the promoters of CsPR-1-1, -2, -3, -4, -5, -6, -7, -8, -9, -10, -13, -14, -15, and -17 genes, and JA cis elements were present in CsPR-1-2, -5, -6, -8, -9, -10, -13, -14, -15, and -17. Flavonoid biosynthesis (MBSI) and W-box elements were also found in the deduced CsPR-1 promoter, closely related to the response to fungal stress. In addition, cis -acting elements, in response to light (i.e., I-box, Box 4, G-box, GT1-motif, Box II, TCT motif, and L-box), low temperature (LTR), drought (MBS), anaerobic stress (ARE), and meristem (CAT-box) also exist in the promoter region of CsPR-1 family genes.
Figure 4.
Schematic model of the cis elements of the CsPR-1 promoter regions generated by TBtool software.
2.6. Predicted Secondary and 3D Structures of CsPR-1 Proteins
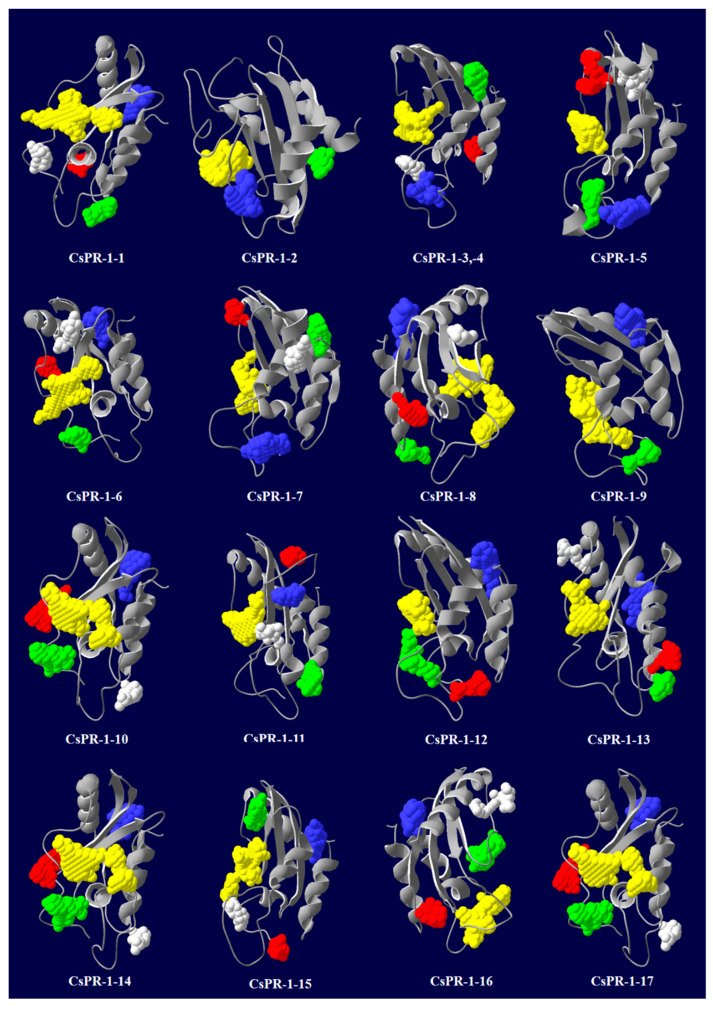
Secondary structures were analyzed using the SOPMA online server. The results show that α-helices, extended strands, beta turns, and random coils were found in the ranges of 23.82–39.88%, 8.24–22.45%, 1.73–7.05%, and 39.74–50.29%, respectively (Table 2). The 3D structures of the CsPR-1 proteins were predicted using the Phyre2 online server. The Ramachandran plot representing the residues in core, allowed, and generous regions exceeded 95%, revealing the quality and reliability of the protein structures (Table 2). The predicted channel structures of CsPR-1 proteins ranged from 2 to 9, and the overall percentages of disordered regions were between 6.47% and 61.76% (Table 2). The 3D structures of the CsPR-1 proteins were conserved, except for that of CsPR-1-2. The 3D structures and predicted pockets are shown in Figure 5.
Table 2.
Structural analyses of the CsPR-1 proteins.
| Proteins | α-Helices (%) | Extended Strand (%) | Beta Turn (%) | Random Coil (%) | Ramachandran Plot (%) | Number of Channels | Disordered Regions (%) |
|---|---|---|---|---|---|---|---|
| CsPR-1-1 | 38.65 | 13.50 | 4.29 | 43.56 | 98.3 | 2 | 23.93 |
| CsPR-1-2 | 23.82 | 8.24 | 4.12 | 63.82 | 99.2 | 2 | 61.76 |
| CsPR-1-3 | 37.04 | 14.20 | 6.17 | 42.59 | 98.3 | 4 | 24.07 |
| CsPR-1-4 | 37.04 | 14.20 | 6.17 | 42.59 | 98.3 | 4 | 24.07 |
| CsPR-1-5 | 30.64 | 17.34 | 1.73 | 50.29 | 96.0 | 3 | 20.23 |
| CsPR-1-6 | 29.38 | 20.62 | 4.38 | 45.62 | 98.3 | 3 | 23.75 |
| CsPR-1-7 | 29.41 | 17.65 | 4.71 | 48.24 | 97.4 | 3 | 6.47 |
| CsPR-1-8 | 32.72 | 16.05 | 2.47 | 48.77 | 99.1 | 2 | 17.28 |
| CsPR-1-9 | 33.33 | 15.09 | 3.77 | 47.80 | 99.1 | 3 | 18.24 |
| CsPR-1-10 | 36.54 | 16.67 | 7.05 | 39.74 | 99.1 | 3 | 16.03 |
| CsPR-1-11 | 39.27 | 15.18 | 4.71 | 40.84 | 96.7 | 5 | 9.42 |
| CsPR-1-12 | 32.21 | 17.45 | 6.04 | 44.30 | 99.1 | 3 | 22.15 |
| CsPR-1-13 | 28.57 | 22.45 | 3.06 | 45.92 | 96.7 | 9 | 16.33 |
| CsPR-1-14 | 33.95 | 15.43 | 4.32 | 46.30 | 99.1 | 3 | 17.28 |
| CsPR-1-15 | 38.12 | 16.25 | 4.38 | 41.25 | 98.3 | 3 | 27.50 |
| CsPR-1-16 | 39.88 | 11.31 | 4.76 | 44.05 | 97.5 | 6 | 32.74 |
| CsPR-1-17 | 37.04 | 16.05 | 4.32 | 42.59 | 99.1 | 3 | 18.52 |
Figure 5.
Predicted 3D structure of the CsPR-1 proteins using the Phyre2 server and pockets using the Pasaccca server. The top five predicted pockets are indicated as yellow, blue, green, red, and grey, respectively.
2.7. Histomorphological Observation of Tea Leaves
Healthy tea leaves had a complete cell structure, including cuticle, upper epidermis, palisade tissue, sponge tissue, down epidermis, and cuticle, and the glycogen and neutral mucus in the palisade tissue cells were red (Figure 6a,e). However, the cell structure of leaves invaded by tea blister blight disease was infected to varying degrees. When the pathogen of tea blister blight disease invaded in the early stage, an obvious hyphal invasion was observed in the gap of the tea plant in down-epidermal cells, but the down-epidermal cells were still intact (Figure 6b,f). In the middle stage of pathogen invasion, the hyphae proliferated rapidly in the gap between tea cells. Some down-epidermal cells of the tea leaves were broken by hyphae, and a small amount of basidiospores broke through the stratum corneum in clusters (Figure 6c,g). In the late stage of pathogen invasion, the hyphae occupied and destroyed most of the sponge-tissue cells, and a large number of basidiospores grew outward in clusters, while the down-epidermal cells of the tea leaves were almost invisible (Figure 6d,h).
Figure 6.
Observation of the stages and PAS staining of tea leaves. Healthy leaves (a,e) and tea leaves infected with tea blister blight at the different stages of infection: early stage (b,f), middle stage (c,g), and late stage (d,h). The orange arrow represents club-shaped hyphae structures of Exobasidium vexans. Up-EP: up-epidermis; Down-EP: down-epidermis; PT: palisade tissue; ST: spongy tissue. Bar = 100 μm.
2.8. Expression Patterns of CsPR-1 Genes
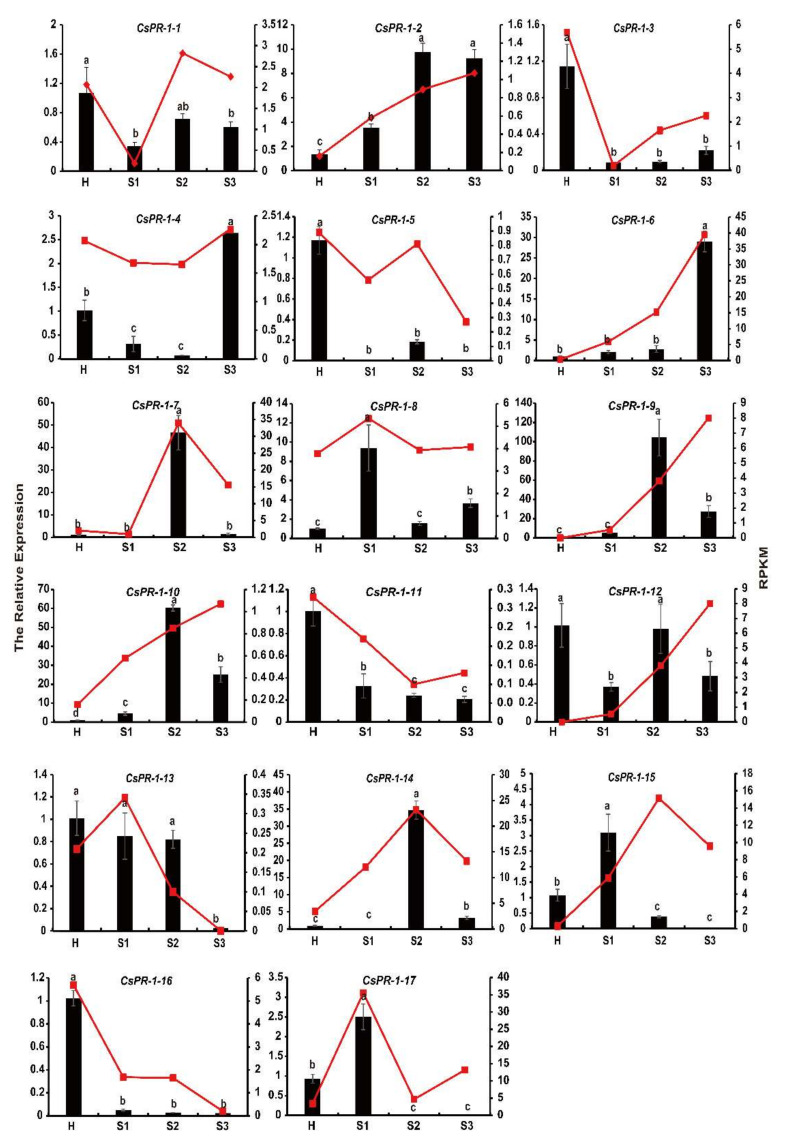
To further investigate the response of CsPR-1 genes to blister blight disease and to verify the accuracy of the RNA-seq data, the expression of 17 CsPR-1 genes in uninoculated (healthy) and inoculated (early, middle, and late stages) E. vexans was measured by qRT-PCR. The results show that the change trend of the expression pattern of most CsPR-1 genes after E. vexans invasion in tea plants was basically similar to that of transcriptome sequencing (Figure 7). A strong response of the CsPR-1 genes was observed, in which 10 CsPR-1 genes (CsPR-1-2, -4, -6, -7, -8, -9, -10, -14, -15, and -17) were obviously upregulated (corresponding to 6.86, 2.59, 28.78, 40.37, 9.36, 97.59, 25.84, 36.85, 2.87, and 2.67 folds), whereas 7 CsPR-1 genes (CsPR-1-1, -3, -5, -11, -12, -13, and -16) were significantly downregulated (corresponding to 0.32, 0.07, 0.15, 0.20, 0.36, 0.02, and 0.02 folds). The expression levels of these genes showed significant differences across the different disease stages. CsPR-1-2, -7, -8, -9, -10, -14, -15, and -17 expression was significantly increased during the early and middle stages, while the expression of CsPR-1-4 and -6 was upregulated in the late stage.
Figure 7.
Comparison of the transcript expression results from the RNA-seq and qRT-PCR analysis of the CsPR-1 family genes under tea blister-blight disease stress. The relative expression of genes validated using qRT-PCR (bar chart, left ordinate) and the fragments per kilobase of transcript per million mapped reads obtained from transcriptome sequencing (line diagram, right ordinate).
3. Discussion
PR-1 is a kind of PR protein induced by the host against a pathogen and encoded by multigene families in plants [8]. Seventeen members of the PR-1 gene family from the tea genome were identified in the current study, consistent with similar investigations in other plant species. Comprehensive analyses of the gene structures, conserved motifs, cis elements, protein-interaction networks, and expression patterns were performed, and the results could provide a scientific basis for future functional research.
The PR-1 family is highly conserved and exists in all plants studied. This family has been confirmed to possess an antifungal function. According to pI values, PR-1 proteins can be broadly classified as either acidic or basic proteins [12]. These two protein types have different inhibitory activities against fungal pathogens. In tomato plants, basic PR-1c and PR-1g exhibit greater antifungal activity against Phytophthora infestans than acidic PR-1a and PR-1b [32]. Overexpression of TMV-inducible basic pepper PR-1 gene in tobacco leaves could significantly enhance the resistance to heavy-metal stresses and pathogens [33]. In the current study, most basic CsPR-1 proteins were found to be more rapidly and strongly expressed than acidic CsPR-1 proteins in tea plants during interaction with E. vexans. This finding is similar to those reported by previous studies. The basic CsPR-1-2, -4, -7, -8, -9, -14, and -17 genes were obviously upregulated, and most of these genes responded rapidly during the early or middle stages of infection. These characteristics indicate that these genes may play an important role in the early pathogenesis of tea blister blight disease. Acidic CsPR-1-6, -10, and -15 were also upregulated. The expression trends of CsPR-1 genes were diverse, indicating a complex defense mechanism to tea blister blight in tea plants.
Notably, all deduced amino-acid sequences of CsPR-1 genes, except for those of CsPR-1-11 and-12, contained an SP at the N terminal. An SP is an RNA region encoding hydrophobic amino-acid sequences, which, in most cases, is a transient extension to the amino terminus of the protein and is responsible for guiding proteins into subcellular organelles with different membrane structures [34]. Previous studies on Triticum aestivum and Vitis showed that all PR-1 proteins contain an SP at the N terminal, which could secrete PR-1 proteins into the extracellular environment [14,24]. Two PnPR-1 proteins without SPs were also found in Piper nigrum, which is consistent with the present results [13]. Most CsPR-1 proteins containing an SP at the N terminal might be guided into special subcellular organelles for their biological functions. The CTE is supposed to be functionally similar to the carboxyl-terminal propeptide encoded by PR-2 and PR-3 genes, which is known to serve as a vacuolar targeting signal [16,35]. The CTE domain has been found in both dicot and monocot species, such as tobacco PR-1b and tomato PR-1a1 [36,37]. In this study, only CsPR-1-2 was found to contain the CTE domain, which may play an important role in guiding CsPR-1 proteins into vacuolar targeting. The 3D structures of CsPR-1 proteins are also conserved, except for those of CsPR-1-2. The structural variation was significantly connected with the CTE domain of the CsPR-1-2 gene and resulted in different function of PR-1 proteins in tea plants. Disordered regions contain flexibility-intrinsic conformation to bind multiple partners in transient interactions and are closely related to transcriptional regulation, signaling cascade, cell-cycle control, and chaperone activity [38]. The presence of disordered regions in CsPR-1-2 was up to 61.76%, which might be related to the metabolic roles of these proteins in cellular regulation. Further characterization may provide a theoretical basis for the exploration of the structural and functional diversity of CsPR-1 proteins in tea plants.
Previous studies have shown that the multigene family was formed by duplication and mutation of an ancestral gene [39]. The survival of duplicate genes made the regulatory and coding regions tend to differentiate, leading to differences in the structure and function of genes in the same family [39]. In the current study, the most closely related CsPR-1 genes in the same group were found to share a similar conserved motif and gene structures, indicating that analysis of the motif and gene structures was contributed to understanding of the evolutionary relationships of CsPR-1 family genes. Furthermore, introns are less conserved compared with exons. Intron density tended to decline during evolution in some genes that need to rapidly activate in response to stress, such as heat-shock proteins [40]. Using molecular characterization and sequence analysis, investigators in a previous study found that PR-1 genes lack introns [41]. In the current investigation, a majority of CsPR-1 genes were found to have no introns or only between one and three introns, which was conducive to the transcriptional regulation of these genes under stress. CsPR-1-7, -9, and -14 without introns in the same group responded more rapidly to tea blister blight disease stress than CsPR-1-1, which contains three introns.
Although substantial evidence has demonstrated that PR-1 acts as a defense protein in plant-pathogen interactions, the biological functions of PR-1 proteins remain obscure. A previous study showed that SA and JA can induce the expression of up- and downregulated PR genes, respectively [42]. PR-1 gene in wheat was strongly induced after ABA and SA treatments [43]. Similarly, under cold treatment and exogenous application of SA, the expression of PR-1 genes in Arabidopsis was significantly upregulated [44]. In the interaction networks of CsPR-1, many key genes engaged in activating and mediating diverse defense responses were involved. AT5G66590 (CsPR-1-5 and -11), AT5G57625 (CsPR-1-7), and AT4G33720 (CsPR-1-3, -6, -10, and -12) interacted with PR4, which was highly related to the defense of grapevine against downy mildew resistance and related to the JA/ET-signaling pathway [45]. HCHIB was basic endochitinase B, related to the JA/ET-mediated signaling pathway during systemic acquired resistance, which is an enzyme known for antifungal activity and close interaction with AT1G50060 (CsPR-1-1, -4, -9, and -14) and AT4G25780 (CsPR-1-13) [40]. CsPR-1-15 and -17 were involved in the defense of SA response. NPR1 was found to positively modulate SA signaling in plants and to interact with PRB1 (CsPR-1-8) [46]. CsPR-1 proteins were found to be involved in diverse disease resistance and to play a connecting role in the complex response network of different signal pathways.
4. Materials and Methods
4.1. Plant Materials
Tea plants (Camellia sinensis cv. Fuding dabai) were grown in the gardens of Northwest A&F University Tea Experimental Station (Xixiang, Shaanxi, China, 32°57′43″ N, 107°40′12″ E). Tea leaves (from the third leaf from the top of the plant) with typical tea blister-blight symptoms were sampled within 4 h. Each tea leaf only had one blister. The severity was classified into three grades on the basis of the course of the disease, as previously reported: early stage (S1, the formation of yellow, transparent patches with a diameter of 2–4 mm), middle stage (S2, the formation of features blisters), and late stage (S3, the formation of necrotic spots) [47]. Healthy tea leaves (H) free of any infestation at the same leaf positions were used as control. All samples were immediately frozen in liquid nitrogen and stored at −80 °C for periodic acid-Schiff (PAS) staining, RNA-seq, and real-time PCR (qRT-PCR) with three biological replicates.
4.2. Database Mining and Identification of CsPR-1 Genes
Published Arabidopsis PR-1 sequences were used as queries for BLASTP searches against the Tea Plant Genome Database (http://tpia.teaplant.org/index.html, accessed on 16 September 2021) [48]. All output genes were then verified using Pfam (http://pfam.xfam.org/family/PF00188, accessed on 16 September 2021). SMARAT (http://smart.embl-heidelberg.de/, accessed on 16 September 2021) and CCD (https://www.ncbi.nlm.nih.gov/cdd/, accessed on 16 September 2021) were used to confirm all the putative CsPR-1 genes. The chromosomal localization information of all CsPR-1 genes was obtained by local blast of TPIA (http://tpia.teaplant.org/index.html, accessed on 17 September 2021). pI and molecular weight (MW) were predicted using ProtParam (http://web.expasy.org/protpara-m/, accessed on 17 September 2021). The online program WoLF PSORT (https://wolfpsort.hgc.jp/, accessed on 17 September 2021) and SignalP-5.0 (http://www.cbs.dtu.dk/services/SignalP/, accessed on 17 September 2021) servers were used to predict the subcellar locations and signal peptides of the PR-1 proteins. Alignment and visualization of the PR-1 domain were performed through DNAMAN 7.0 software.
4.3. Analysis of the Conserved Motifs, Gene Structures, and Protein Functional Networks
The conserved motifs were analyzed with the MEME platform (http://meme-suite.org/tools/meme, v4.9.0, accessed on 20 September 2021). The number of motifs was set to 20, and other parameters were set to default values [49]. The exon-intron structures were retrieved from the gene-annotation file (http://www.plantkingd omgdb.com/tea_tree/data/gff3/, accessed on 20 September 2021), and the diagrams were drawn by using TBtools v1.098661 software [50]. The functional-interaction networks of the PR-1 proteins in tea plants were analyzed based on the STRING protein-interaction database (http://string-db.org/, accessed on 21 September 2021).
4.4. Analysis of the Cis Elements of the CsPR-1 Gene Promoters
The CsPR-1 promoter sequences (2 kb upstream the start codon) were retrieved from the TPIA (http://tpia.teaplant.org/index.html, accessed on 23 September 2021). PlantCARE online software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 23 September 2021) was used to search and analyze the cis elements in the extraction-promoter sequences. These elements were visualized using TBtool software [50].
4.5. Structural Analysis of CsPR-1 Proteins
Secondary structures were predicted with the SOPMA server (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html, accessed on 7 January 2022). Tertiary protein structures were predicted with the Phyre2 server (http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index, accessed on 7 January 2022). Disordered regions were predicted with the PONDR server (http://www.pondr.com/, accessed on 7 January 2022). The BetaCavityWeb server (http://voronoi.hanyang.ac.kr/betacavityweb, accessed on 7 January 2022) was used to search and analyze the channel structure. Web POCASA (http://g6altair.sci.hokudai.ac.jp/g6/service/pocasa/, accessed on 9 January 2022) was used to predict pocket structures.
4.6. PAS Staining
The samples were fixed with formaldehyde/acetic acid/ethanol fixative (containing 50% ethanol, 5% acetic acid, and 10% formaldehyde in H2O), dehydrated, transparented, waxed, and embedded. The paraffin sections were then routinely dewaxed in water and put into periodic acid solution for 10–15 min. Then, the slides were stained with PAS for 10 min. After being washed with water and dehydrated, the slices were covered with paraffin for further observation using the Olympus BX51 microscope (Olympus, Tokyo, Japan).
4.7. RNA Extraction and Quantitative RT-PCR Analysis
Total RNA was extracted using the Plant RNA Kit (Omega, Norcross, GA, USA). cDNA was synthesized by using the 5 × All-In-One RT MasterMix Kit (ABM, Richmond, BC, Canada) according to the manufacturer’s protocol, and the cDNA was diluted to 200 ng/μL for subsequent analysis. Then, qRT-PCR was performed using ChamQ SYBR qPCR Master Mix (Vazyme, Nanjing, China) on an iQ5 real-time PCR platform (Bio-Rad, Hercules, CA, USA) with the following PCR parameters: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s. Three independent biological replicates were performed, and the qPCR of each replicate was performed in triplicate. Relative transcript abundances of the PR-1 genes were calculated via the 2–ΔΔCT method [51]. All primers were designed using Primer5 software, and primer sequences are listed in Table S4.
5. Conclusions
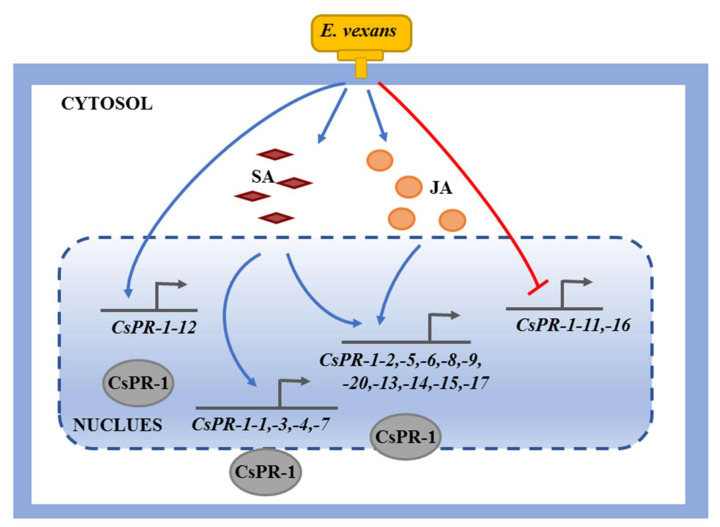
In the present study, 17 CsPR-1 genes were identified in tea plants, and bioinformatics and expression-profile analyses were performed to determine their potential functions (Figure 8). The CsPR-1 genes were found to be actively involved in the response to tea blister blight stress, and these processes are closely related to the signal transduction pathways involving TCA, NPR1, EDS16, BGL2, PR4, and HCHIB. The results provide new insights into the response to tea blister blight stress and also contribute to an important basis for subsequent functional studies investigating CsPR-1 in tea plants.
Figure 8.
Proposed model of CsPR-1-modulated blister blight in tea plants. The blue arrow indicates the upregulation of genes; the red arrow indicates the downregulation of genes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23031292/s1.
Author Contributions
Conceptualization, Q.Z. and S.L.; methodology, Q.Z. and Y.Z.; software, Q.Z. and Y.Z.; validation, Q.Z., N.G. and S.L.; formal analysis, N.G.; investigation, Q.Z.; resources, Y.Y. and S.L.; data curation, Q.Z., N.G. and Y.Z.; writing—original draft preparation, Q.Z.; writing—review and editing, S.L.; visualization, Q.Z.; supervision, Y.Y. and S.L.; project administration, S.L.; funding acquisition, Y.Y. and S.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a special fund for University-Supported Extension Model (grant number XTG2021-04), China Agriculture Research System of MOF and MARA (grant number CARS-19), and Natural Science Foundation of Shaanxi Province (grant number 2021JQ-135).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available within the article and Supplementary Material.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Takken F., Tameling W. To Nibble at Plant Resistance Proteins. Science. 2009;324:744–746. doi: 10.1126/science.1171666. [DOI] [PubMed] [Google Scholar]
- 2.Bigeard J., Colcombet J., Hirt H. Signaling Mechanisms in Pattern-Triggered Immunity (PTI) Mol. Plant. 2015;19:521–539. doi: 10.1016/j.molp.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 3.Tena G. PTI and ETI are One. Nat. Plants. 2021;7:1527. doi: 10.1038/s41477-021-01057-y. [DOI] [PubMed] [Google Scholar]
- 4.Silva M.S., Arraes F.B.M., Campos D.A., Grossi-de-Sa M., Fernandez D., Cândido E.S., Cardoso M.H., Franco O.L., Grossi-de-Sa M.F. Review: Potential Biotechnological Assets Related to Plant Immunity Modulation Applicable in Engineering Disease-Resistant Crops. Plant Sci. 2018;270:72–84. doi: 10.1016/j.plantsci.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Hamamouch N., Chunying L.I., Seo P.J., Park C.M. Expression of Arabidopsis Pathogenesis-Related Genes During Nematode Infection. Mol. Plant Pathol. 2011;12:355–364. doi: 10.1111/j.1364-3703.2010.00675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sung Y.C., Outram M.A., Breen S., Wang C., Dagvadorj B., Winterberg B., Kobe B., Williams S.J., Solomon P.S. PR1-Mediated Defence via C-Terminal Peptide Release is Targeted by a Fungal Pathogen Effector. New Phytol. 2021;229:3467–3480. doi: 10.1111/nph.17128. [DOI] [PubMed] [Google Scholar]
- 7.Datta K., Tu J., Oliva N., Ona I., Velazhahan R., Mew T.W., Muthukrishnan S., Datta S.K. Enhanced Resistance to Sheath Blight by Constitutive Expression of Infection-Related Rice Chitinase in Transgenic Elite Indica Rice Cultivars. Plant Sci. 2001;160:405–414. doi: 10.1016/S0168-9452(00)00413-1. [DOI] [PubMed] [Google Scholar]
- 8.Van Loon L.C., Rep M., Pieterse C.M.J. Significance of Inducible Defense-Related Proteins in Infected Plants. Annu. Rev. Phytopathol. 2006;44:135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- 9.Gibbs G.M., Kim R., O’Bryan M.K. The CAP Superfamily: Cysteine-Rich Secretory Proteins, Antigen 5, and Pathogenesis-Related 1 Proteins-Roles in Reproduction, Cancer, and Immune Defense. Endocr. Rev. 2008;7:865–897. doi: 10.1210/er.2008-0032. [DOI] [PubMed] [Google Scholar]
- 10.Sheng J., Olrichs N.K., Gadella B.M., Kaloyanova D.V., Helms J.B. Regulation of Functional Protein Aggregation by Multiple Factors: Implications for the Amyloidogenic Behavior of the CAP Superfamily Proteins. Int. J. Mol. Sci. 2020;21:6530. doi: 10.3390/ijms21186530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y.L., Lee C.Y., Cheng K.T., Chang W.H., Huang R.N., Nam H.G., Chen Y.R. Quantitative Peptidomics Study Reveals that a Wound-Induced Peptide from PR-1 Regulates Immune Signaling in Tomato. Plant Cell. 2014;26:4135–4148. doi: 10.1105/tpc.114.131185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiraga J., Mackiewicz P., Mackiewicz D., Kowalczuk M., Biecek P., Polak N., Smolarczyk K., Dudek M.R., Cebrat S. The Relationships Between theIsoelectric Point and: Length of Proteins, Taxonomy and Ecology of Organisms. BMC Genomics. 2007;8:163. doi: 10.1186/1471-2164-8-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kattupalli D., Sreenivasan A., Soniya E.V. A Genome-Wide Analysis of Pathogenesis-Related Protein-1 (PR-1) Genes from Piper nigrum Reveals Its Critical Role during Phytophthora capsici Infection. Genes. 2021;12:1007. doi: 10.3390/genes12071007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z.T., Dhekney S.A., Gray D.J. PR-1 Gene Family of Grapevine: A Uniquely Duplicated PR-1 Gene from a Vitis Interspecific Hybrid Confers High Level Resistance to Bacterial Disease in Transgenic Tobacco. Plant Cell Rep. 2011;30:1–11. doi: 10.1007/s00299-010-0934-5. [DOI] [PubMed] [Google Scholar]
- 15.Kiba A., Nishihara M., Nakatsuka T., Yamamure S. Pathogenesis-Related Protein 1 Homologue is an Antifungal Protein in Wasabia japonica Leaves and Confers Resistance to Botrytis cinerea in Transgenic Tobacco. Plant Biotechnol. 2007;24:247–253. doi: 10.5511/plantbiotechnology.24.247. [DOI] [Google Scholar]
- 16.Breen S., Williams S.J., Outram M., Kobe B., Solomon P.S. Emerging Insights into the Functions of Pathogenesis-Related Protein 1. Trends Plant Sci. 2017;22:871–879. doi: 10.1016/j.tplants.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 17.Baruah P.M., Krishnatreya D.B., Bordoloi K.S., Gill S.S., Agarwalaa N. Genome Wide Identification and Characterization of Abiotic Stress Responsive lncRnas in Capsicum annuum. Plant Physiol. Bioch. 2021;162:221–236. doi: 10.1016/j.plaphy.2021.02.031. [DOI] [PubMed] [Google Scholar]
- 18.Jiang M., Chen H., Liu J., Du Q., Lu S., Liu C. Genome-Wide Identification and Functional Characterization of Natural Antisense Transcripts in Salvia miltiorrhiza. Sci. Rep. 2021;11:4769. doi: 10.1038/s41598-021-83520-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kesawat M.S., Kherawat B.S., Singh A., Dey P., Kabi M., Debnath D., Saha D., Khandual A., Rout S., Manorama, et al. Genome-Wide Identification and Characterization of the Brassinazole-Resistant (BZR) Gene Family and Its Expression in the Various Developmental Stage and Stress Conditions in Wheat (Triticum aestivum L.) Int. J. Mol. Sci. 2021;22:8743. doi: 10.3390/ijms22168743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar M., Kherawat B.S., Dey P., Saha D., Singh A., Bhatia S.K., Ghodake G.S., Kadam A.A., Kim H.-U., Manorama, et al. Genome-Wide Identification and Characterization of PIN-FORMED (PIN) Gene Family Reveals Role in Developmental and Various Stress Conditions in Triticum aestivum L. Int. J. Mol. Sci. 2021;22:7396. doi: 10.3390/ijms22147396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tong T., Fang Y., Zhang Z., Zheng J., Zhang X., Li J., Niu C., Xue D., Zhang X. Genome-Wide Identification and Expression Pattern Analysis of the KCS Gene Family in Barley. Plant Growth Regul. 2021;93:89–103. doi: 10.1007/s10725-020-00668-3. [DOI] [Google Scholar]
- 22.Wang M., Chen B., Zhou W., Xie L., Wang L., Zhang Y., Zhang Q. Genome-Wide Identification and Expression Analysis of the AT-Hook Motif Nuclear Localized Gene Family in Soybean. BMC Genomics. 2021;22:361. doi: 10.1186/s12864-021-07687-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akbudak M.A., Yildiz S., Filiz E. Pathogenesis Related Protein-1 (PR-1) Genes in Tomato (Solanum lycopersicum L.): Bioinformatics Analyses and Expression Profiles in Response to Drought Stress. Genomics. 2020;112:4089–4099. doi: 10.1016/j.ygeno.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Lu S., Friesen T.L., Faris J.D. Molecular Characterization and Genomic Mapping of the Pathogenesis-Related protein 1 (PR-1) Gene Family in Hexaploid Wheat (Triticum aestivum L.) Mol. Gen. Genet. 2011;285:485–503. doi: 10.1007/s00438-011-0618-z. [DOI] [PubMed] [Google Scholar]
- 25.Almeida S.F., Venancio T.M. Pathogenesis-Related Protein 1 (PR-1) Genes in Soybean: Genome-Wide Identification, Structural Analysis and Expression Profiling under Multiple Biotic and Abiotic Stresses. Gene. 2021;809:146013. doi: 10.1016/j.gene.2021.146013. [DOI] [PubMed] [Google Scholar]
- 26.Higdon J.V., Frei B. Tea Catechins and Polyphenols: Health Effects, Metabolism, and Antioxidant Functions. Crit. Rev. Food Sci. 2003;43:89–143. doi: 10.1080/10408690390826464. [DOI] [PubMed] [Google Scholar]
- 27.Nagao H. Effect of Aqueous Vitamin B on the Growth of Blister Blight Pathogen, Exobasidium vexans. Songklanakarin J. Sci. Technol. 2012;34:601–606. [Google Scholar]
- 28.Punyasiri P.A., Abeysinghe S.B., Kumar V. Preformed and Induced Chemical Resistance of Tea Leaf against Exobasidium vexans Infection. J. Chem. Ecol. 2005;31:1315–1324. doi: 10.1007/s10886-005-5288-z. [DOI] [PubMed] [Google Scholar]
- 29.Ponmurugan P., Gnanamangai B.M., Manjukarunambika K. Architectural Effect of Different Tea Clones on the Development of Blister Blight Disease. J. Appl. Bot. Food Qual. 2019;92:7–14. [Google Scholar]
- 30.Jayaswall K., Mahajan P., Singh G., Parmar R., Seth R., Raina A., Swarnkar M.K., Singh A.K., Shankar R., Sharma R.K. Transcriptome Analysis Reveals Candidate Genes Involved in Blister Blight Defense in Tea (Camellia sinensis (L) Kuntze) Sci. Rep. 2016;6:30412. doi: 10.1038/srep30412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandra S., Chakraborty N., Panda K., Acharya K. Chitosan-Induced Immunity in Camellia sinensis (L.) O. Kuntze against Blister Blight Disease is Mediated by Nitric-Oxide. Plant Physiol. Biochem. 2017;115:298–307. doi: 10.1016/j.plaphy.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 32.Niderman T. Pathogenesis-Related PR-1 Proteins are Antifungal-Isolation and Characterization of 3 14-Kilodalton Proteins of Tomato and of a Basic PR-1 of Tobacco with Inhibitory Activity against Phytophthora -Infestans. Plant Physiol. 1995;108:17–27. doi: 10.1104/pp.108.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarowar S., Kim Y.J., Kim E.N., Kim K.D., Hwang B.K., Islam R., Shin J.S. Overexpression of a Pepper Basic Pathogenesis-Related Protein 1 Gene in Tobacco Plants Enhances Resistance to Heavy Metal and Pathogen Stresses. Plant Cell Rep. 2005;24:216–224. doi: 10.1007/s00299-005-0928-x. [DOI] [PubMed] [Google Scholar]
- 34.Xie S., Wang Y., Wei W., Lin Y., Yin W., Luo C. Development of Novel Methods for Functional Evaluation of the Signal Peptide of Secreted Protein. Physiol. Mol. Plant P. 2019;106:182–186. doi: 10.1016/j.pmpp.2019.02.002. [DOI] [Google Scholar]
- 35.Lu S., Faris J.D., Sherwood R., Friesen T.L., Edwards M.C. A Dimeric PR-1-Type Pathogenesis-Related Protein Interacts with ToxA and Potentially Mediates ToxA-Induced Necrosis in Sensitive Wheat. Mol. Plant Pathol. 2014;15:650–663. doi: 10.1111/mpp.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfitzner U.M., Goodman H.M. Isolation and Characterization of cDNA Clones Encoding Pathogenesis-Related Proteins from Tobacco Mosaic Virus Infected Tobacco Plants. Nucleic Acids Res. 1987;11:4449–4465. doi: 10.1093/nar/15.11.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tornero P., Conejero V., Vera P. A Gene Encoding a Novel Isoform of the PR-1 Protein Family from Tomato is Induced upon Viroid Infection. Mol. Gen. Genet. 1994;243:47–53. doi: 10.1007/BF00283875. [DOI] [PubMed] [Google Scholar]
- 38.Pazos F., Pietrosemoli N., García-Martín J.A., Solano R. Protein Intrinsic Disorder in Plants. Front. Plant Sci. 2013;4:00363. doi: 10.3389/fpls.2013.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He X., Zhang J. Rapid Subfunctionalization Accompanied by Prolonged and Substantial Neofunctionalization in Duplicate Gene Evolution. Genetics. 2005;169:1157–1164. doi: 10.1534/genetics.104.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J., Tong G., Wan S., Zhang Y., Yang J., Yu Y., Wang W. Genome-Wide Identification, Classification and Expression Analysis of the HSP Gene Superfamily in Tea Plant (Camellia sinensis) Int. J. Mol. Sci. 2018;19:2633. doi: 10.3390/ijms19092633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Q., Xue Q. Computational Identification of Novel PR-1-Type Genes in Oryza sativa. J. Genet. 2006;85:193–198. doi: 10.1007/BF02935330. [DOI] [PubMed] [Google Scholar]
- 42.Ali S., Mir Z.A., Tyagi A., Bhat J.A., Chandrashekar N., Papolu P.K., Rawat S., Grover A. Identification and Comparative Analysis of Brassica juncea Pathogenesis-Related Genes in Response to Hormonal, Biotic and Abiotic Stresses. Acta Physiol. Plant. 2017;39:268. doi: 10.1007/s11738-017-2565-8. [DOI] [Google Scholar]
- 43.Gao L., Wang S., Li X.Y. Expression and Functional Analysis of a Pathogenesis-Related Protein 1 Gene, TcLr19-PR1, Involved in Wheat Resistance against Leaf Rust Fungus. Plant Mol. Biol. Rep. 2015;33:797–805. doi: 10.1007/s11105-014-0790-5. [DOI] [Google Scholar]
- 44.Seo P.J., Lee A.K., Xiang F., Park C.M. Molecular and Functional Profiling of Arabidopsis Pathogenesis-Related Genes: Insights into Their Roles in Salt Response of Seed Germination. Plant Cell Physiol. 2008;49:334–344. doi: 10.1093/pcp/pcn011. [DOI] [PubMed] [Google Scholar]
- 45.Jiang W., Wu J., Zhang Y., Yin L., Lu J. Isolation of a WRKY30 Gene from Muscadinia rotundifolia (Michx) and Validation of Its Function Under Biotic and Abiotic Stresses. Protoplasma. 2015;252:1361–1374. doi: 10.1007/s00709-015-0769-6. [DOI] [PubMed] [Google Scholar]
- 46.Zhang D., Zhu Z., Gao J., Zhou X., Zhu S., Wang X.Y., Wang X.L., Ren G.D., Kuai B.K. The NPR1-WRKY46-WRKY6 Signaling Cascade Mediates Probenazole/Salicylic Acid-Elicited Leaf Senescence in Arabidopsis thaliana. J. Integr. Plant Biol. 2021;63:924–936. doi: 10.1111/jipb.13044. [DOI] [PubMed] [Google Scholar]
- 47.Xia E., Li F., Tong W., Li P., Wu Q., Zhao H., Ge H.R., Li R., Li Y., Zhang Z. Tea Plant Information Archive: A Comprehensive Genomics and Bioinformatics Platform for Tea Plant. Plant Biotechnol. J. 2019;17:1938–1953. doi: 10.1111/pbi.13111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bailey T.L., Williams N., Misleh C., Li W.W. MEME: Discovering and Analyzing DNA and Protein Sequence Motifs. Nucl. Acids Res. 2006;34:369–373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmittgen T.D., Livak K.J. Analyzing Real-Time PCR Data by the Comparative C-T Method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 50.Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 51.Livak K.J., Schmittgen T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available within the article and Supplementary Material.