Abstract
Carotenoid cleavage dioxygenases (CCDs) catalyzes the cleavage of various carotenoids into smaller apocarotenoids which are essential for plant growth and development and response to abiotic stresses. CCD family is divided into two subfamilies: 9-cis epoxycarotenoid dioxygenases (NCED) family and CCD family. A better knowledge of carotenoid biosynthesis and degradation could be useful for regulating carotenoid contents. Here, 23 CCD genes were identified from the Populus trichocarpa genome, and their characterizations and expression profiling were validated. The PtCCD members were divided into PtCCD and PtNCED subfamilies. The PtCCD family contained the PtCCD1, 4, 7, and 8 classes. The PtCCDs clustered in the same clade shared similar intron/exon structures and motif compositions and distributions. In addition, the tandem and segmental duplications resulted in the PtCCD gene expansion based on the collinearity analysis. An additional integrated collinearity analysis among poplar, Arabidopsis, rice, and willow revealed the gene pairs between poplar and willow more than that between poplar and rice. Identifying tissue-special expression patterns indicated that PtCCD genes display different expression patterns in leaves, stems, and roots. Abscisic acid (ABA) treatment and abiotic stress suggested that many PtCCD genes are responsive to osmotic stress regarding the comprehensive regulation networks. The genome-wide identification of PtCCD genes may provide the foundation for further exploring the putative regulation mechanism on osmotic stress and benefit poplar molecular breeding.
Keywords: carotenoid, CCD, NCED, ABA, poplar
1. Introduction
Terpenoids consist of various primary and secondary metabolites that function in all living organisms. Carotenoids and their apocarotenoids, considered C40 isoprenoids, participate in multiple essential biological functions in plants and animals [1,2]. For example, xanthophylls and violaxanthin are the critical components of plant light-harvesting protein complexes [3], and carotenoids in chloroplasts are involved in light absorption, electron transfer, and removal of triplet oxygen and superoxide anion in plant photosynthesis [4,5]. In addition, lycopene and β-carotene are important pigments that affect the color of plant flowers and fruits, and their contents are associated with fruit colors and qualities [6,7]. In addition, zeaxanthin aldehyde, a carotenoid derivative, can be catalyzed into abscisic acid (ABA). ABA, also known as a stress hormone, plays a vital role in regulating various physiological and developmental processes. It can not only act as a growth inhibitor to intervene in biological processes such as plant flowering [8], fruit maturation [9], and seed dormancy [10], but also directly responds to drought, salt, and low temperature through the ABA signal transduction pathway including mediation of stress-resistant genes expression [11], reducing transpiration [12,13], and inducing stomatal closure [14,15].
Carotenoid cleavage dioxygenases (CCDs) are mainly responsible for the oxidative cleavage of carotenoids in higher plants, which results in the biosynthesis of biologically smaller apocarotenoids [5]. CCDs are also a kind of non-heme iron dioxygenase, which contain the RPE65 (retinal segment epithelial membrane protein) domain responsible for binding Fe2+ [16,17]. In higher plants, the carotenoid cleavage dioxygenase (CCD) and 9-cis carotenoid cleavage dioxygenase (NCED) have been identified as a subfamily of CCDs based on the epoxy structure of substrate [18,19]. NCEDs can catalyze the cleavage of 11, 12 double bond of violaxanthin (C40) or neoxanthin (C40) to form the xanthoxin (C15), and this catalytic reaction carried out by NCEDs is considered as the rate-limiting step in ABA biosynthesis [20,21]. A variety of studies have been focused on the regulation mechanism of NCEDs in resistance to stress conditions. In Arabidopsis, NCED members were classified into NCED2, 3, 5, 6, and 9 according to phylogeny and gene function [18]. The transcript levels of AtNCED3 could be induced by drought stress, and AtNCED3 controls the level of endogenous ABA. Overexpression of AtNCED3 could improve drought tolerance of Arabidopsis by reducing leaf transpiration rates [22]. AtNCED6 and AtNCED9 were associated with developmental control of ABA biosynthesis in seeds. In addition, both AtNCED2, AtNCED3, and AtNCED5 have essential roles in endogenous ABA accumulation [23]. In rice, OsNCED members have also been classified into OsNCED1, OsNCED2, OsNCED1, OsNCED4, and OsNCED5, and the divergent expression of OsNCED members in tissues resulted in different functions [24]. OsNCED1 and OsNCED3 were involved in the drought resistance, and OsNCED2 was associated with the delay of seed germination [25]. Overexpression of OsNCED5 and OsNCED3 in Arabidopsis increased the tolerance to drought stress and delayed seed dormancy and changed plant size and leaf morphology [26,27]. CsNCED genes from Crocus sativus had a closer relationship with ABA accumulation under the drought, salt, and lower temperature [28]. These results suggested that NCEDs control ABA biosynthesis and then affect the ABA-mediated signal transduction pathway involved in plant stress responses.
In contrast, CCD enzymes do not have the specific cleavage sites of substrates. Some CCDs can cleave carotenoid or apocarotenoid substrates, while others recognize specific carotenoid or apocarotenoid substrates [29]. CCD subfamily of Arabidopsis was divided into CCD1, CCD4, CCD7, and CCD8 [17]. AtCCD1 and AtCCD4 mainly cleaved 9, 10 double bonds leading to catalyze the C40 carotenoids and C27 terpenoids to form the small molecule volatile substances, such as C13 β-ionone and C14 geranyl acetone geranyl acetone, which suggested that both AtCCD1 and AtCCD4 have an important influence on the formation of plant aroma [30,31]. AtCCD7 catalyzed β-carotene to form β-ionone and C27 10′-apo-β-carotenal, and AtCCD8 or AtCCD1 could further cleave C27 10′-apo-β-carotenal to generate C18 3′-apo-β-carotenal or β-ionone and one apo-10,10′-carotendial, respectively [32,33]. The observations illustrated that AtCCD7 and AtCCD8 could oxidize β-carotene to produce the strigolactone (SL) precursor related to the important biological processes such as branch formation, lateral root formation, seed germination, and response to drought and salt stresses [34]. The composition of CCD4 members is complex, and only one CCD4 member has been identified in Arabidopsis, while many CCD4 members have been identified in tomato, grape, and saffron [35,36,37]. The evidences on CCD4 members show that CCD4 members may be involved in forming a flower, peel, and pulp color. The β-cryptoxanthin and zeaxanthin, thought as the substrate of citrus CCD4b1, are cleaved at the 7, 8 double bonds to form unique C30 carotenoid associated with the color of orange peel [38]. In addition, the expression level of Glycine max CCDs has been significantly changed under salt, drought, low temperature, and high-temperature stresses, indicating that soybean CCDs are involved in its abiotic stress response process [39]. Brassica oleracea CCD1 and CCD4 are responsive to drought and salt stresses [40]. The expressions of Malus domestica CCDs are significantly affected under salt and drought stress, indicating that MdCCD members are involved in response to the abiotic stresses [41]. Given the above evidence, CCD members have various kinds of biological functions and participant in regulating plant growth and development, abiotic stresses, and color formation. However, so far, there is no genome-wide identification of CCD family genes in poplar.
Studies on genome-wide identification of gene families have focused on their characterizations and functions and provided valuable methods for analyzing gene networks or biological functions. For example, analysis of pin-formed (PIN) gene family in wheat displayed PIN may be involved in various developmental processes and biotic and abiotic stress conditions [42]. Identification of β-ketoacyl CoA synthetase (KCS) in barley exhibited barley KCS members function in regulating physiological and biochemical processes and participant in drought stress [43]. In addition, genome-wide identification and characterization of lncRNAs in Capsicum annuum showed that lncRNAs interaction with miRNAs takes part in different abiotic stress by regulating transcription factors (TFs), including tryptophan arginine lysine tyrosine (WRKY), myeloblastosis (MYB), basic leucine zipper domain (bZIP), and so on [44]. In addition, genome-wide analysis of the wheat brassinazole-resistant (BZR) gene family exhibited that BZRs play crucial roles in plant developmental processes and are associated with diverse biotic and abiotic stresses [45]. Genome-wide identification of Salvia miltiorrhiza antisense transcripts (NATs) considered as a class of long noncoding RNAs documented that NATs interaction with sense transcripts (STs) potentially plays significant regulatory roles in the biosynthesis of bioactive compounds [46]. Genome-wide investigation of the soybean AT-hook motif nuclear localized (AHL) gene family proved that AHLs mainly react to mediating stress responses [47].
Moreover, the identification of tobacco CCD family revealed that tobacco CCD genes have essential roles in response to different hormones, including ABA, methyl jasmonate (MeJA), indoleacetic acid (IAA), salicylic acid (SA), and abiotic stresses [48]. The above genome-wide identification provides an essential theoretical basis for understanding family gene physiological and biochemical functions in different species. Poplar and willow mainly distributed in the cold zone to the temperate zone of the northern hemisphere belong to Salicaceae Mirb [49,50]. Poplar is also a model plant to study the molecular mechanism of growth and development, material properties, stress responses, and other vital traits. In addition to being used as an energy tree for industrial production, poplar can also protect the soil structure, prevent and control soil erosion, and have a significant ecological value. In recent years, with the destruction of the ecological environment and the drastic change of the global climate, the inevitable natural disasters such as drought, salt, and freezing have seriously restricted the natural growth and development of poplar and have caused a decline in poplar production capacity and seriously damaged the balance of the local ecological environment. Considering the physiological functions of CCD members under the various stresses, we identify CCD family members of Populus trichocarpa at the whole genome level. We systematically analyze molecular evolution, gene structure, cis-acting elements, and conserved motifs of PtCCD family members. In addition, we evaluate the transcript levels of PtCCDs in various tissues and identify the expression patterns of PtCCDs under abiotic stress. The above results will lay a foundation for further elucidating the biological functions of PtCCD family members.
2. Results
2.1. Characterization of the CCD Family Members in Poplar
The 30 candidate PtCCD members were obtained from the genome database of P. trichocarpa (Phytozome https://phytozome-next.jgi.doe.gov/pz/portal.html (accessed on 28 July 2021)) based on sequence alignment with Arabidopsis CCD members (Table S1). The complete RPE65 domain was used as the standard for screening the above PtCCD members and four members (Potri.006G238500, Potri.006G239301, Potri.018G043201, Potri.018G044100) displayed no dominant RPE65 domain were eliminated from the PtCCD members. In addition, Potri.T074400 and Potri.T167700 remained on scaffolds, which were not spliced with the poplar chromosome, so Potri.T074400 and Potri.T167700 were abandoned for analysis. Finally, 23 PtCCD members possessing complete RPE65 domain were retrieved from the genome of poplar (Table 1). The PtCCD members were named based on their orthologous relationship with Arabidopsis and rice CCD family members (Table 1). Sequence alignment illustrated significant differences in PtCCD sequences, but the four histidines (His) sites are highly conserved (Figure S1).
Table 1.
The poplar, Arabidopsis, and rice CCD accession numbers and gene names.
| Arabidopsis thaliana | Oryza sativa | Populus trichocarpa | |||
|---|---|---|---|---|---|
| Accessions | Gene name | Accessions | Gene name | Accessions | Gene name |
| AT3G63520 | AtCCD1 | LOC_Os12g44310 | OsCCD1 | Potri.001G265400 | PtCCD1a |
| Potri.001G265600 | PtCCD1b | ||||
| Potri.001G265900 | PtCCD1c | ||||
| Potri.009G060500 | PtCCD1d | ||||
| AT4G19170 | AtCCD4 | LOC_Os02g47510 | OsCCD4a | Potri.004G190700 | PtCCD4a |
| Potri.005G069100 | PtCCD4b | ||||
| Potri.009G151900 | PtCCD4c | ||||
| LOC_Os12g24800 | OsCCD4b | Potri.009G152200 | PtCCD4d | ||
| Potri.009G152300 | PtCCD4e | ||||
| Potri.019G093400 | PtCCD4f | ||||
| AT2G44990 | AtCCD7 | LOC_Os04g46470 | OsCCD7 | Potri.014G056800 | PtCCD7 |
| AT4G32810 | AtCCD8 | LOC_Os01g54270 | OsCCD8a | Potri.006G239200 | PtCCD8a |
| Potri.006G239400 | PtCCD8b | ||||
| LOC_Os09g15240 | OsCCD8b | Potri.018G042650 | PtCCD8c | ||
| Potri.018G042900 | PtCCD8d | ||||
| LOC_Os01g38580 | OsCCD8c | Potri.018G043000 | PtCCD8e | ||
| Potri.018G043100 | PtCCD8f | ||||
| LOC_Os08g28240 | OsCCD8d | Potri.018G043400 | PtCCD8g | ||
| Potri.018G043500 | PtCCD8h | ||||
| AT4G18350 | AtNCED2 | LOC_Os12g42280 | OsNCED2 | Potri.011G084100 | PtNCED2 |
| AT3G14440 | AtNCED3 | LOC_Os07g05940 | OsNCED3 | Potri.001G393800 | PtNCED3a |
| Potri.011G112400 | PtNCED3b | ||||
| AT1G30100 | AtNCED5 | ||||
| AT3G24220 | AtNCED6 | Potri.003G176300 | PtNCED6 | ||
| AT1G78390 | AtNCED9 | LOC_Os03g44380 | OsNCED9 | ||
The open reading from (ORF) lengths ranged from 1221 to 1842, except for the incomplete sequence information on Potri.004G190700, Potri.009G152300, Potri.018G042900, and Potri.018G043400. The deduced amino acids of PtCCDs varied from 406 to 613, and the molecular weights (MWs) ranged from 46.28–69.07 KD. A high proportion of PtCCDs theoretical PI was less than 7, indicating that a large portion of them belongs to acidic protein, except PtCCD4a, 4c, 4d, 4e, PtCCD7, and PtNCED2. In addition, the grand average of hydropathicities of PtCCDs was less than 1, which illustrated that PtCCDs are hydrophilic and non-transmembrane proteins. The analysis of instability index showed that PtCCD4c, 4d, 4e, 4f, PtCCD8d, 8e 8f, 8g, 8h, and PtNCED2, 3a, and 3b belong to unstable proteins, while others belong to stable proteins. The signal peptide prediction showed that only PtCCD8a has a classical secretion signal peptide, and the cleavage site is located in positions 16 and 17. Other PtCCD proteins do not have signal peptides. In addition, subcellular localization prediction showed PtCCDs are distributed in chloroplast and cytoplasm.
2.2. Phylogenetic Analysis of PtCCD Family
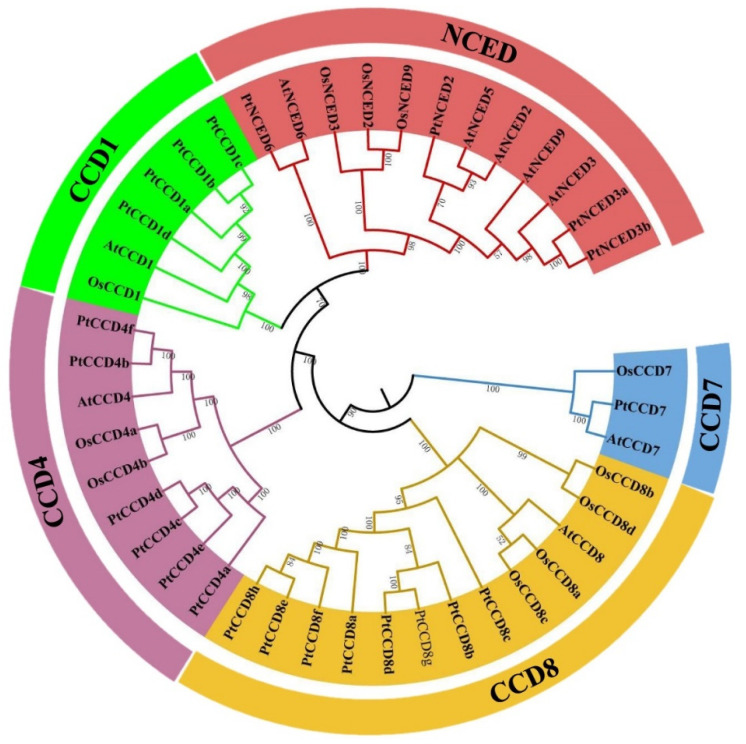
To better understand the evolutionary relationships among CCD members, 9 Arabidopsis, 11 rice, and 23 poplar CCD members were chosen to construct a phylogenetic tree. According to the evolutionary relationship of Arabidopsis CCDs and rice CCDs, PtCCD family members were divided into two families and named PtCCD subfamily (PtCCD1, PtCCD4, PtCCD7, and PtCCD8 classes) and PtNCED subfamily (Figure 1). Based on clustering analysis, four CCD members were clustered into the CCD1 class, whereas only one AtCCD1 or OsCCD1 was fell into the CCD1 class. In addition, 6 PtCCD4, 1 AtCCD4, and 2 OsCCD4 members were classified into CCD4 class. In addition, the CCD8 class contained eight members in poplar, four in rice, and only one in Arabidopsis, suggesting that CCD8 members were probably expanded only within some species.
Figure 1.
Phylogenetic tree displaying the evolutionary relationships among poplar, Arabidopsis, and rice CCDs. The neighbor-joining (NJ) method tree with 1000 bootstrap replicates was applied to draw a phylogenetic tree with the MEGA7 software. The CCD from poplar, Arabidopsis, and rice were involved in CCD and NCED families, and the CCD family were clustered into CCD1, 4, 7, and 8 classes.
Moreover, NCED5 and NCED9 classes were not found in poplar, but two members are identified as PtNCED3 class. Furthermore, the phylogenetic tree revealed that CCD4 and CCD8 classes have a closer evolutionary relationship, and CCD1 class and NCED subfamily were clustered together, indicating that the NCED type may be evolved from the CCD1 class. The CCD1, CCD4, CCD7, CCD8, and NCED among Arabidopsis, rice, and poplar were assembled in the evolutionary clade, suggesting that the plant CCD gene family is relatively more conservative among different species, and CCD evolution is later than that of herbs and woody plants, monocotyledons, and dicotyledons.
2.3. Gene Structures and Conserved Motifs of CCD Members
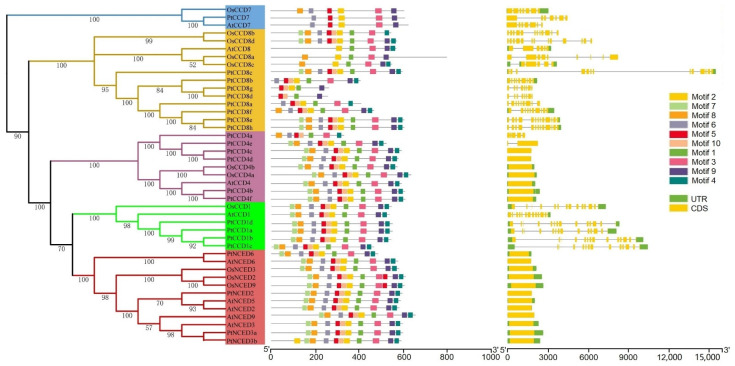
To better understand the relationship among the phylogenetic evolution, gene structures, and the conserved motifs of CCD family members, the exon/intron structures and distributions of conserved motifs together with the phylogenetic tree of the CCD family were analyzed. As expected, the CCD members possessed similar exons/introns were clustered into the same clade. All PtNCED members had no intron, and similar gene structures were identified in PtCCD4 (except PtCCD4a and PtCCD4e). Furthermore, the PtCCD1, 7, and 8 had many introns and relatively comprehensive exon/intron structures compared with PtNCED and PtCCD4 (Figure S2). Obviously, in addition to PtCCD gene structures, the AtCCD and OsCCD genes shared similar exon/intron structures (Figure 2).
Figure 2.
The poplar, Arabidopsis, and rice CCD gene structures and conserved motifs. Exons and introns were represented using bars and rectangles, respectively. The conserved motifs were displayed using colorful rectangles.
To explore the features of CCD member motifs, ten distinct motifs were chosen to investigate using the MEME tool. Interestingly, motif distributions of CCDs were similar in the same clade. The CCD1, CCD4, and NCED proteins contained motifs 1–10, suggesting that motifs 1–10 might be involved in their standard functions. In addition, the motif compositions of CCD8 members were divergent, which illustrated that CCD8 members may participate in various physiological processes and could be responsible for different functions. In addition, the number of motifs in CCD7s was lower than that in CCD1, CCD4, and NCED members, which illustrated that CCD7s are speculated to be lacking some specific biological functions compared with CCD1, CCD4, and NCED members (Figure 2). In conclusion, these observations suggested different conserved motifs and gene structures in CCD classes, which further supported the CCD family phylogenetic clustering.
2.4. Expansion and Contraction of PtCCD Genes
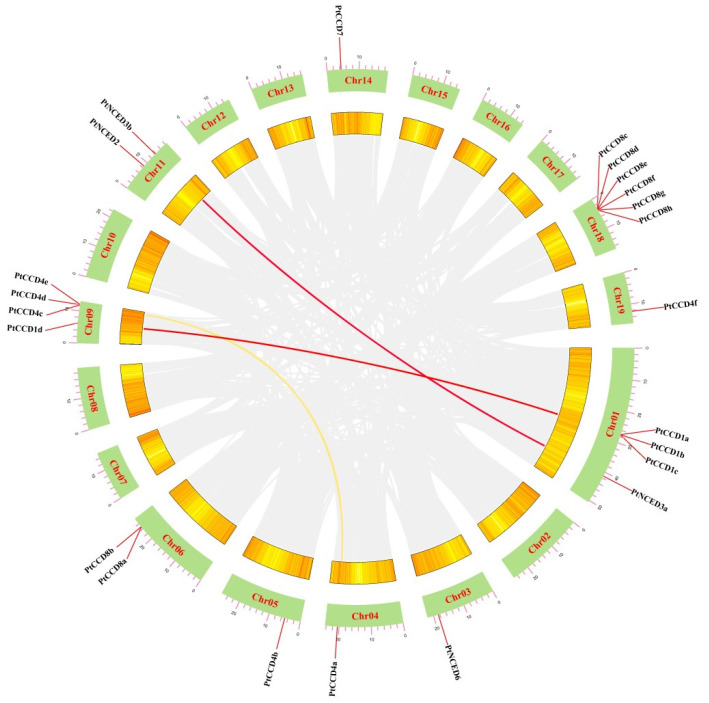
The chromosomal distributions of PtCCDs were determined and visualized based on the TBtools and poplar genome annotation information. In general, 23 PtCCD members were unevenly anchored to Chr01, Chr03–06, Chr09, Chr11, Chr14, Chr18, and Chr19 (Supplemental Figure S3). Chr01, as the largest chromosome, contained 4 PtCCD genes, and 6 PtCCD genes were mapped to Chr08, while Chr03–05, both 14 and 19, contained only one PtCCD gene, respectively. The method of gene family formation usually contains tandem, segmental, and whole-genome duplications. Gene duplication analysis might provide helpful information for CCD family gene formation. Here, tandem duplication and segmental duplication are evaluated by the multiple collinearity scan toolkit (MCScanX) method. In general, the tandem duplication origin from homologous genes located on the same chromosome. The gene pairs, including PtCCD4d and PtCCD4e, and PtCCD8e and PtCCD8f thought as tandem duplication events were identified in the PtCCD gene family. Meanwhile, the PtCCD4d and PtCCD4e, and PtCCD8e and PtCCD8f shared similar gene structures and motif distributions. In addition, homologous genes on the different chromosomes were probably the result of segmental duplication. Collinearity analysis of the PtCCDs showed that PtCCD1a and PtCCD1d, PtCCD4a and PtCCD4c, and PtNCED3a and PtNCED3b have collinearity relationships, respectively (Figure 3). The PtCCD1a and PtCCD1d, or PtNCED3a and PtNCED3b share 91.65% or 91.67% similarity, respectively, which illustrated that gene pairs are formed by segmental duplication. The above observations indicated that expansion of PtCCD genes might probably be original from both tandem duplication and segmental duplication. Moreover, PtCCD8c, 8d, 8e, 8f, 8g, and 8h are located on chromosome 18, PtCCD1a, 1b, and 1c located on chromosome 1 PtCCD4c, 4d, and 4e located on chromosome 9 formed various gene clusters. Furthermore, to clarify the role of selection pressure in the evolution of the PtCCD genes, TBtools were used to analyze the Ks, Ka, and Ka/Ks of homologous PtCCD genes. The results suggested that Ka/Ks values in 2 pairs of tandem duplication and 3 pairs of genome duplication are less than 1, indicating that the PtCCD family has undergone strong purification selection during the evolution process.
Figure 3.
The collinearity analysis of PtCCDs. Chromosomes 01–19 were indicated using green rectangles. The homologous PtCCD genes in the poplar genome were displayed using red and yellow curves. The poplar gene with collinearity relationship was represented using gray curves.
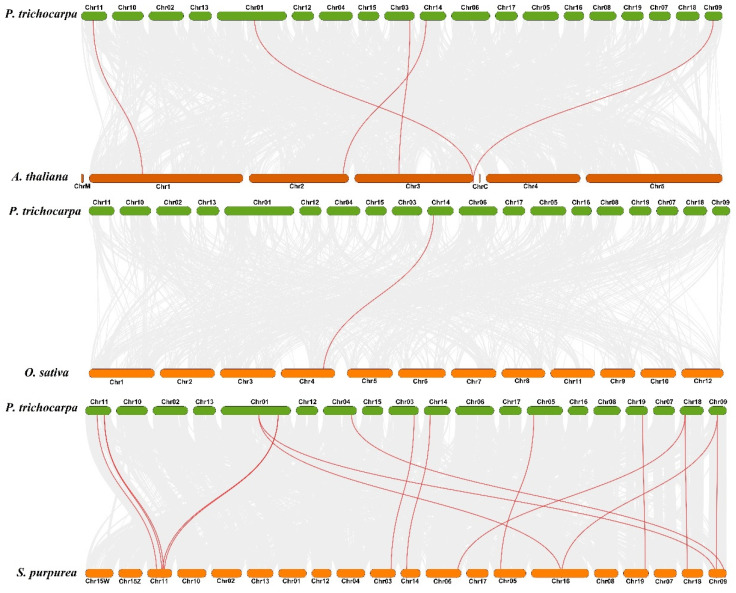
To explore the orthologous relationship among poplar, Arabidopsis, rice, and willow, the analysis of collinearity relationship was performed using TBtools with MCScanX. The result showed that 5 gene pairs are found between poplar and Arabidopsis, 1 gene pair is found between poplar and rice, and 16 gene pairs are found in poplar and willow, respectively (Figure 4). In the evolutionary relationship of species, poplar and willow were classified to Salicaceae Mirb, and poplar and Arabidopsis were classified to dicotyledonous plants. The collinearity relationship among poplar, Arabidopsis, rice, and willow suggested that poplar has a closer evolutionary relationship with willow.
Figure 4.
Analysis of collinearity among poplar, Arabidopsis, rice, and willow CCD genes. All putative orthologous genes were represented using gray curves, and the orthologous CCD genes among the genomes of poplar, Arabidopsis, rice, and willow were displayed using red curves.
2.5. Three-Dimensional (3D) Structures and Cis-Acting Elements Analysis
The results of 3D structure prediction showed that CCD proteins consist of coils, strands, and helixes (Supplemental Figure S3). The coils and strands occupied most CCD structures, while helixes only accounted for a small part of CCD structures. The same class, including CCD1, 4, and 7 and NCED, was visualized to possess the relatively higher similarity, and the divergences lay mainly in the coils and helixes. The 3D structures of CCD8 members were relatively comprehensive and various. The 3D structures of PtCCD8c, PtCCD8d, and PtCCD8g were dominantly divided into two parts marked A and B (Supplemental Figure S4), and the two parts of 3D structures were connected by coils or helixes.
In comparison, the other CCD8 members have no two significant parts of 3D structures. Based on the comparative analysis of CCD8 structures, nearly all 3D structures of CCD8 members could be highly merged in structure A, suggesting that structure A in CCD8 members may be relatively conservative compared to structure B. In addition, the CCD1 members had similar 3D structures with NCED members, which might explain the closer evolutionary relationship between CCD1 and NCED members in a phylogenetic tree.
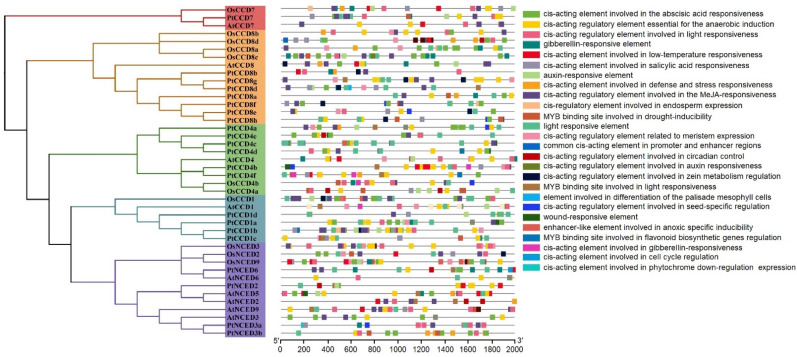
To understand the putative biological functions of CCD members, PlantCARE was used to analyze the cis-acting elements in the promoters of CCDs. The results showed that the CCD promoters contain various cis-acting elements, divided into two categories. One is abiotic stress response elements, such as anaerobic inducible element (ARE), stress response element (STRE), MYB drought inducible binding site (MBS), DREB/CBF transcription factor recognition site (DRE), disease resistance, and stress-inducing elements (TC-rich repeats) and low-temperature response elements (LTR). The other category is plant hormone response elements, such as auxin response elements (TGA-element), salicylic acid (SA) inducing elements (TCA-element), methyl jasmonate (MeJA) response element (CGTCA-motif), and ABA response element (ABRE). In addition, the number of light-responsive elements, anaerobic inducing elements, and ABA-responsive elements occupied a large part of cis-acting elements in the PtCCD and AtCCD promoters, and OsCCD promoters contained large numbers of light-responsive elements, MeJA-responsiveness elements, and ABA-responsive elements. Both of PtCCD promoters, PtCCD1b, PtCCD4d, PtCCD7, and PtCCD8f had a large proportion of MeJA-responsive elements in their promoters, and PtCCD4b and PtNCED2 contained relatively some low-temperature responsive elements. In addition, ABA-responsive elements were widely distributed in PtCCD4e, PtNCED3b, PtCCD1a, PtNCED3a, and PtCCD4e promoters, while PtCCD8a, PtCCD8b, and PtCCD8e contained fewer cis-acting elements (Supplemental Figure S5). For AtCCD promoters, ABA-responsive elements were widely present in AtNCED5, AtNCED9, and AtNCED3 promoters. MYB binding sites involved in drought-inducibility existed in AtNCED9, AtCCD4, AtCCD8, AtNCED3, and AtNCED6. For analysis of OsCCD promoters, OsCCD8c, OsCCD8a, OsNCED9, OsNCED3, and OsCCD8d contained large numbers of ABA-responsive elements. OsCCD8c, OsCCD7, OsNCED3, OsCCD8b, OsCCD4b, OsNCED2, and OsCCD1 contained the certain number of MeJA-responsive elements. MYB binding sites involved in drought-inducibility were predicted in OsCCD4a, OsCCD4b, OsCCD8c, OsNCED2, and OsNCED9 promoters. Overall, the analysis of cis-acting elements in CCD promotes illustrated that CCDs transcript levels may be regulated by light, hormone, and abiotic stress, and CCDs were speculated to be involved in diverse stress resistances (Figure 5).
Figure 5.
Analysis of poplar and Arabidopsis CCD gene promoter cis-elements. Left panel, the phylogenetic tree constructed by NJ method. Right panel, the kinds of CCD promoter cis-elements displayed using colorful rectangles.
To analyze the influence of CCDs evolution on their promoter elements, CCD cis-acting elements and CCDs phylogenetic tree were compared. The results showed a minor difference in PtCCDs cis-acting elements in the same clade. The compositions and distributions of PtCCDs cis-acting elements in different evolutionary clades have significant differences. In addition, the other species’ CCD cis-acting elements in the same clades showed substantial differences, suggesting that orthologous CCDs within the different species may exist the functional differentiation. All above evidence indicated that the categories of CCD cis-acting elements are different with CCD evolutionary relationship.
2.6. Interaction Networks of Protein-Protein Assays
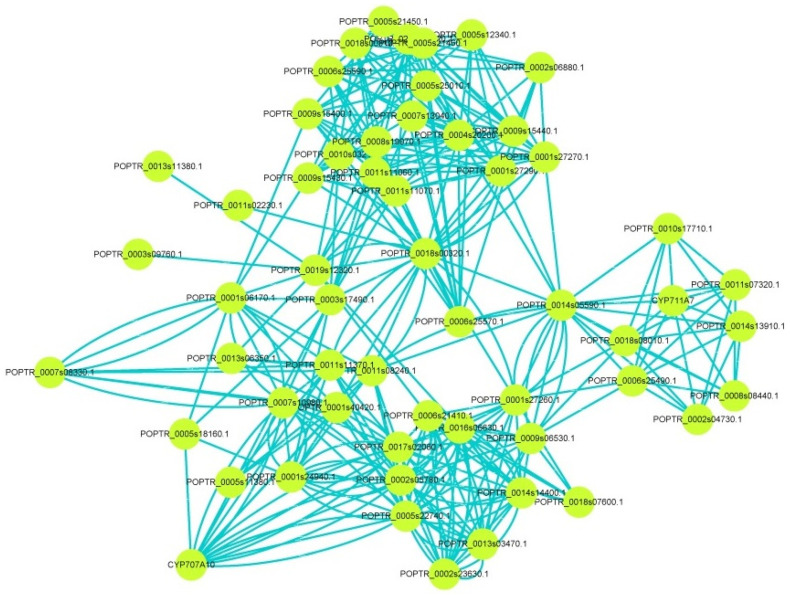
Interaction network analysis can find the relationship of protein–protein. These proteins may be mutually regulated, tightly related in function, or members involved in the same signaling pathway or physiological process. Here, the String database (https://string-db.org/ (accessed on 28 July 2021)) and Cytoscape software were used to identify the interaction network. As shown in Supplemental Figure S6, AtCCD7 was located at the core position in interaction network of AtCCD family members. In addition, the interaction network in PtCCD family members was more sophisticated than that in AtCCD family members. In interaction network of PtCCD family members, PtCCD8g, PtCCD7, and PtCCD4a were predicted to interact together and were in the core position. The above results predicted that PtCCD8g, PtCCD7, and PtCCD4a interaction with other PtCCDs functions together and participants in the same signal transduction and biological process.
Moreover, to further discover the functions of CCDs, interaction assays were performed to construct the relationship between CCD members and other proteins. The result showed that CCDs might interact with cytochrome P (CYP), phytoene synthase (PSY), abscisic acid insensitive (ABI), gibberellic oxidase (GAox), thaumatin-like protein (TCP), and so on, implying CCDs might exert functions by interacting with other genes (Figure 6 and Supplemental Figure S7). The above results predicted that PtCCDs might play an essential role in ABA and GA biosynthesis or ABA and GA signal transduction. Overall, interaction networks could provide crucial references for identifying the regulation mechanism of PtCCDs.
Figure 6.
The interaction network for PtCCD members in poplar. The String database (https://string-db.org/ (accessed on 28 July 2021)) was used to predict the interaction relationship between PtCCD members and other proteins, and Cytoscape software was applied to visualize the interaction network.
2.7. Expression Patterns of PtCCDs in Different Tissues
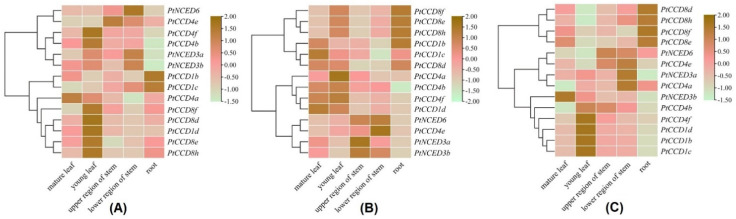
To gain insight into expression profiling of PtCCDs in different tissues, the qRT-PCR was used to illustrate gene expression levels in mature and young leaves, the upper and lower region of stems, and roots. There was a dominant gene expression divergence over different tissues and distinct expression patterns within poplar varieties (Figure 7). For tissue-specific expression patterns of PtCCDs in P. trichocarpa, the PtCCD1b and 1c were highly expressed in roots. The relatively higher expression accumulations of PtNCED3a, 3b, and 6 were presented in a lower region of stems. The mRNA transcript levels of PtCCD4e were abundant in the upper region of stems. The higher expression levels of PtCCD1d, 4b, 4f, 8d, 8e, 8f, and 8h were shown in young leaves (Figure 7A). In addition, for expression profiling of PtCCDs in ‘Nanlin 895’ (P. deltoides × P. euramericana), PtCCD1b, 8f, 8e, and 8h showed higher abundances in roots, the higher expression levels of PtNCED6 and PtCCD4e were accumulated in the lower region of stems, PtCCD4a, 4b, and 4f showed higher mRNA expression in young leaves, and PtCCD1c and 1d showed higher expression levels in the mature leaves (Figure 7B). In addition, for expression patterns of PtCCDs in ‘Shanxinyang’ (P. davidiana × P. bolleana Loucne), 4 (PtCCD8d, 8e, 8f, and 8h) were illustrated to be highly expressed in roots, 6 (PtCCD1b, 1c, 1d, 4b, and 4f) shared the highest expression in young leaves, and PtNCED3a and PtCCD4a and 4e were highly expressed in the lower region of stems (Figure 7C).
Figure 7.
The qRT-PCR analysis of PtCCDs expression patterns in different tissues of poplar varieties including P. trichocarpa (A), ‘Nanlin 895’ (B), and ‘Shanxinyang’ (C). The gene expression levels in mature and young leaves, upper and lower regions of stems and roots were identified using qRT-PCR. Three independent experiments were performed. The data are normalized to poplar Ptactin (XM-006370951). PtCCD transcript levels were normalized to that in mature leaves; color scale, log2-fold change.
To explore the putative relationship among the PtCCD genes, the clustering analysis was performed based on the PtCCD expression patterns. It is distinct that PtCCD genes in P. trichocarpa, ‘Nanlin 895’, and ‘Shanxinyang’ can be clustered into different clades, respectively. For example, the PtCCD8d, 8e, 8f, 8h, and 1d clustered into the same clade possessed relatively lower expression levels in young leaves, while the PtCCD1b and 1c clustered into the same clade showed the higher accumulations in roots. In addition, clustering analysis in ‘Nanlin 895’ indicated that PtCCD8e, 8f, and 8h clustered into the same clade shared similar tissue-specific expression patterns. In addition, PtCCD genes transcript patterns in ‘Shanxinyang’ were divided into three clusters. Cluster 1, 2, and 3 displayed higher expression levels in roots, stems, and leaves, respectively. All above observations revealed that PtCCD genes showed tissue-specific expression patterns in poplar, and diverse expression patterns of PtCCDs are presented at poplar varieties. The results could illustrate that PtCCD genes may be involved in various physiological processes, and the function of PtCCD genes possibly experience evolution in response to different environments.
2.8. PtCCD Expression in Response to Abiotic Stress
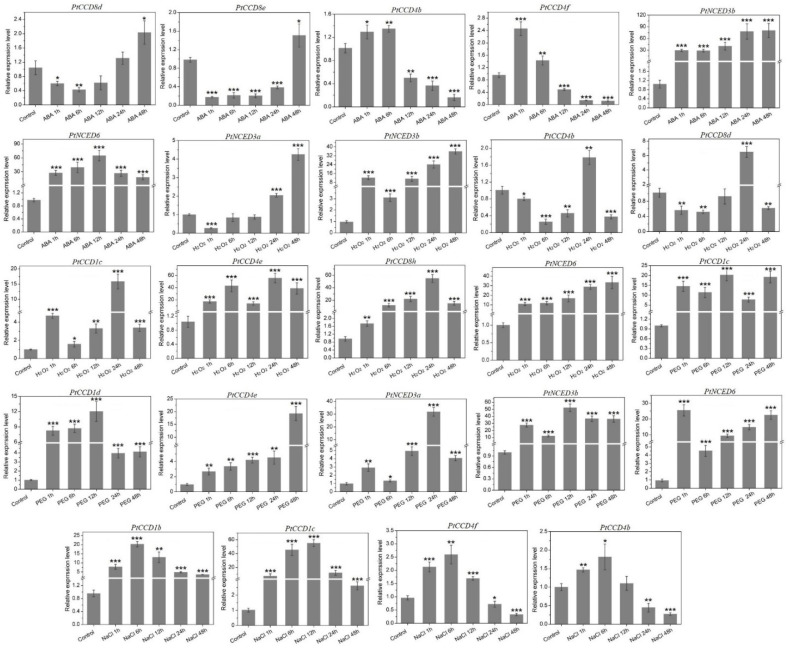
To illustrate the expression profiling of PtCCD genes under abiotic stress, time-course changes were analyzed according to the qRT-PCR. Figure 8 and Supplemental Figure S8 shows PtCCD gene expression levels were distinctly affected by abiotic stress, and there are connections and differences in the expression pattern of each PtCCD member. Under the ABA treatment, the expression of PtCCD8d and 8e were decreased during the early ABA treatment and recovered at 12 and 24 h, respectively; PtCCD4b and 4f expression was up-regulated with 6h and maintained low expression level during 12–48 h; and other PtCCD genes showed relatively higher expression levels within the ABA treatment period, especially PtNCED3b and 6. Under the H2O2 treatment, the expression abundances of PtNCED3a and 3b and PtCCD4b and 8d were downregulated during 0–12 h, and the higher expression level was found at 24 h; PtCCD1c, 4e, 8h, and PtNCED6 were upregulated and displayed the higher abundances after H2O2 stress, but difference existed in reaching the highest expression level. Under the PEG6000 treatment, the expression levels of PtCCD4b and 4f were increased firstly and then kept lower expression levels, while the transcript levels of PtCCD8d, 8e, and 8f displayed the negative results. Expression accumulations of PtCCD1c, 1d, and 4e and PtNCED3a, 3b, and 6 were dominantly induced during the period of PEG6000 treatment. Under the NaCl treatment, the higher accumulations of PtCCD1b and 1c were illustrated during 0–48 h; the expression levels of PtCCD4f and 4b were significantly promoted in early-stage and were dominantly decreased in the later stage. In addition, it is distinct noting that some PtCCD genes clustered into the same clade had relatively similar expression profiling. For example, PtNCED3a and 3b had similar mRNA transcript profiling under the H2O2 treatment. In addition, the expression levels of PtCCD8d, 8e, and 8f were generally downregulated in the early stage of PEG6000 treatment and then increased (Supplemental Figure S8). The above results showed that poplar CCD genes might be involved in response to abiotic and hormone stresses, and different PtCCD members had divergent response patterns.
Figure 8.
The qRT-PCR analysis of the PtCCD expression levels induced by abiotic treatments including ABA, H2O2, PEG6000, and NaCl. The vertical bars are representative of SD. Asterisk represents significant difference (t-test, * p < 0.05, ** p < 0.01, and *** p < 0.001). Three independent experiments were performed. The data are normalized to poplar Ptactin (XM-006370951). PtCCD transcript levels were normalized to that in untreated leaves (control).
3. Discussion
The CCD family containing the RPE65 domain participates in the carotenoid metabolic pathway. Almost all eukaryotes contain the CCD family, especially various CCD genes in plants, from yeast to human beings. For example, 9, 11, 9, and 19 CCD genes were identified in Arabidopsis [17], rice [35], tomato [51], and grape [36], respectively. In this study, 23 PtCCD genes were identified from poplar genome, and the compositions of PtCCD1, PtCCD4, PtCCD8, and PtNCED classes differed with other species. For example, each Arabidopsis CCD class contained one CCD gene. Rice contained above fourth CCD classes, and CCD8 class contained 8 OsCCD genes. While the PtCCD1, 4, and 8 classes contained 4, 6, and 8 PtCCD genes, respectively. Compared with Arabidopsis CCD classes, the number of poplar CCD genes was significantly increased, suggesting that poplar CCD genes might experience gene expansion in the process of evolution. In addition, Arabidopsis NCED subfamily contained AtNCED2, 3, 5, 6, and 9, while the poplar NCED subfamily only contained NCED3 and 6. Although the number of PtNCED genes was lower than the number of AtNCED genes, the NCED subfamily was closely associated with the CCD1 class, and PtCCD1 genes were dominantly more than AtCCD1 genes. It has been speculated that parts of PtCCD1 members may make up for the lack of PtNCED function. PtCCD family members are unevenly distributed on 10 poplar chromosomes, of which chromosome 18 contained the relatively more PtCCD genes, and only one PtCCD gene was identified to be distributed on chromosomes 03, 04, and 05. Moreover, 5 gene pairs of 23 PtCCD genes had a homologous evolutionary relationship, including tandem and segmental replication. Among them, PtCCD4d and PtCCD4e, and PtCCD8e and PtCCD8f originated from gene tandem replication events, while PtCCD1a and PtCCD1d, PtCCD4a, and PtCCD4c, and PtNCED3a and PtNCED3b originated from segmental replication events, which suggested that gene replication events may result in expansion of PtCCD gene family. The Ka/Ks ratios of 5 PtCCD pairs were far lower than 1, indicating that they have undergone strong purification selection in the process of evolution. In addition, the harmful non-synonymous substitution disappeared in the evolutionary process, indicating that gene duplication is the main reason for PtCCD gene expansion.
In Arabidopsis, AtCCD7 and AtCCD8 could catalyze β-carotene to form caprolactone, the precursor of strigolactone (SL) reported being involved in plant growth and development [52]. In addition, the CsCCD7 and CsCCD8 in saffron were identified to affect the synthesis of SL and control bud sprouting [53]. Similarly, CCD7 and CCD8 in kiwifruit [54], tomato [55], and rice [56] were involved in the regulation of cell senescence, root growth, branch, tiller, and flower organ morphogenesis. In this study, PtCCD7 and PtCCD8 groups showed a closer evolutionary relationship, and gene structures and motif compositions were usually similar in the same clade. Due to the conservation of CCD genes, CCD genes with similar functions are often clustered in the same clade, which provided an essential basis for studying the role of PtCCD7 and PtCCD8 classes. NCEDs were rate-limiting enzymes of ABA biosynthesis, and ABA as a crucial signal molecule participated in plant growth and development and stress response [57,58]. NCED could be associated with the contents of endogenous ABA and play an essential role in stress tolerance. In Arabidopsis, AtNCED3 expression was induced by drought stress and participated in response to drought treatment by regulating leaf transpiration rate and controlling the level of endogenous ABA [22]. Also, AtNCED6 and AtNCED9 were involved in ABA biosynthesis during seed development [59]. CsNCED from Crocus sativus was closely associated with the content of endogenous ABA under salt, low temperature, and drought stresses [28]. In the present study, PtNCED2, 3, and 6 had similar motif compositions and gene structures with AtNCED2, 3, and 6, respectively, which suggested that PtNCED2, 3, and 6 may be involved in accumulations of endogenous ABA and response to abiotic stress. Compared with the PtCCD subfamily, a part of PtNCED clades was absent from the PtNCED subfamily, which indicated that functions of some PtNCEDs are replaced in the process of poplar evolution. The cleavage of carotenoids catalyzed by CCD4 was related to the coloring of pulp and flower organs. Overexpression of Arabidopsis AtCCD4 in rice decreased contents of β-carotene and lutein and improved β-violone accumulation [60]. The loss function of CCD4 led to changes in the color of fruit and flower organs. For example, loss-of-function of CCD4 resulted in the change of azalea petal color from yellow to white [61] and the change of Eustoma grandiflorum petal color from light yellow to white [62]. The previous studies showed differences in the cleavage sites of carotenoids and substrates catalyzed by CCD4. In general, CCD4 cleaved carotenoids at 9′–10′ double bonds, while the cleavage position of CcCCD4b1 in citrus was 7′–8′ double bond, and the product was β-citraurin, β-citaurinene as the unique C30 carotenoids [63]. VvCCD4a from grape could catalyze red lycopene to form 6-methyl-5-heptene-2-one. In this study, 6 PtCCD4s were identified from poplar genome, and PtCCD4b and 4f had the closer relationship with AtCCD4. All the observations indicated that PtCCD4b and 4f might be involved in poplar pigment formation, and the activity of other PtCCD4 members may be complex. It should be comprehensively analyzed in further studies.
To identify the putative functions of PtCCDs associated with poplar growth and development, the transcript profiling of PtCCDs in young and mature leaves, upper and lower region of stems, and roots of poplar varieties were analyzed using qRT-PCR. The divergences in the PtCCD expression patterns were observed in poplar varieties, and even the same PtCCD was identified to have different expression levels in different poplar varieties. Those results indicated that the same PtCCD might be involved in various physiological processes in diverse poplar varieties. The Arabidopsis and petunia CCD1 genes were highly expressed in all tested tissues [17,64], while PtCCD1 genes had high expression levels in leaves of poplar varieties. PtCCD1 expression patterns were inconsistent with its homologs Arabidopsis and petunia CCD1 expression patterns, implying that PtCCD1 genes participate in different physiological processes from Arabidopsis and petunia CCD1 genes. Although the PtCCD1 class and PtNCED subfamily had the closer evolutionary relationship, the higher expression levels of PtNCED3a and PtNCED6 were found in stems, and PtCCD1 members highly expressed in leaves of poplar varieties, suggesting that PtNCED family and PtCCD1 class may have divergent functions in stems and leaves. In addition, PtCCD7 gene was orthologous with AtCCD7 gene, and previous studies showed CCD7 has a distinct expression accumulation in roots [65,66]. Interestingly, in this study, the relatively lower expression levels of PtCCD7 were identified in all tested tissues, which suggested that PtCCD7 in poplar may have different functions with A. thaliana CCD7 regarding root development and metabolite synthesis. However, the precise role and related regulation mechanism of PtCCD7 need to be confirmed in future studies. In addition, PtCCD8 members had been confirmed to have the higher expression levels in P. trichocarpa young leaves, while they were identified to have the expression abundances in ‘Shanxinyang’ and ‘Nanlin 895’ roots. These results suggested that PtCCD8 members have divergent functions in the leaves and roots development of poplar varieties.
Since little knowledge focuses on the functions of the poplar CCD genes in regulating abiotic stress responses, the PtCCDs expression patterns in response to abiotic stress were illustrated. Previous studies have pointed out that the CCDs play an essential role in ABA synthesis and ABA signal transduction in response to diverse pressures [34,40]. This study identified various kinds of cis-elements involved in hormone-responsive and stress-related elements in the PtCCD promoters. PtCCDs mRNA levels in leaves induced by abiotic stress were investigated using qRT-PCR. PtNCED3 and 6 expression abundances improved under the ABA treatment, similar to AtNCED gene accumulations caused by ABA treatment [67]. Although the distinct divergence was identified in the tissue-special expression of PtCCD1 class, the PtCCD1 members expression levels were also significantly up-regulated by ABA treatment. Compared with the previous conclusion that soybean CCD7 and CCD8 gene dominantly respond to ABA treatment [39], the PtCCD8d and 8e expression levels were dominantly down-regulated at the early stage of ABA treatment, and PtCCD8f and 8h expression levels were accumulated at the early stage of ABA treatment. Accordingly, PtCCD1 members expression was significantly improved after ABA treatment, and even PtCCD1a expression abundance underwent a 56-fold increase after a 48 h ABA treatment. These results suggested that PtCCDs may play an essential role in response to ABA treatment. The precise regulation mechanism of plant stress responses by the ABA signal complex network might need to be explored in further study. Osmotic stress could change the plant physiological processes and affect plant growth and development by decreasing the photosynthetic rate and transpiration [68]. Also, under osmotic stress, the level of membrane lipid peroxidation was increased significantly, which destroyed the plant cell membrane and affected cell integrity [69,70]. Generally, osmotic stress significantly increased the contents of superoxide anion and endogenous H2O2 in the plant. Excessive accumulation of superoxide anion would cause irreversible damage to the cell membrane and seriously inhibit the progress of photosynthesis [71]. To explore the putative physiological changes caused by osmotic stress, the expression patterns of PtCCDs under the NaCl, PEG6000, and H2O2 were analyzed. The expression levels of PtNCEDs were considerably upregulated under osmotic stress, and differences only existed in expression trends with time-course. Those results were consistent with the higher expression of Brassica rapa NCED under the osmotic stress [40].
In addition, apple MdCCD8a expression levels were significantly improved under salt and drought treatments, and the soybean CCD8 expression was decreased during drought and salt stresses. PtCCD8s showed diverse transcript levels under the NaCl and PEG6000 treatments. For example, PtCCD8f expression experienced an 11-fold increase in response to a 48-h PEG6000 treatment, while the peak of PtCCD8f expression was identified in 12 h under the H2O2 treatment. In addition, under the PEG6000 treatment, PtCCD4b and 4f expression levels were up-regulated from 1 to 6 h, but there was distinct down-regulation of their expression levels from 12 to 48 h. However, under the H2O2 treatment, PtCCD4b expression level was significantly higher than in control, except its expression at 24 h. It was noteworthy that PtCCDs have different response abilities under ABA treatment and osmotic stress, indicating that PtCCDs play vital roles in abiotic stress. This study lays the foundation for identifying the biological functions of PtCCDs and helps to find stress-resistant gene resources.
4. Materials and Methods
4.1. Identification and Classification of Poplar CCD Genes
The RPE65 (PF03055) was achieved from the Pfam database (http://pfam.xfam.org/ (accessed on 28 July 2021)). The sequence information and innovation of poplar, Arabidopsis, rice, and willow were downloaded from the Phytozome database (https://phytozome-next.jgi.doe.gov/ (accessed on 28 July 2021)). The RPE65 is considered as a query to search the putative CCD members from the poplar genome. In addition, the Arabidopsis AtCCDs as a query to search the PtCCD members. Then, the SMART database and NCBI Conserved Domain Search online were applied further to verify the conserved domains in putative PtCCD members. Based on the homologous relationship with AtCCDs, the PtCCD members were named CCD1s, 4s, 7, 8s, and NCEDs.
4.2. Evolutionary Relationship and CCD Sequence Analysis
To illustrate the characterizations and putative functions of PtCCDs, multiple sequence alignment was performed using ClustalX2 software. In addition, according to the neighbor-joining (NJ) method, a phylogenetic tree on PtCCDs, AtCCDs, and OsCCDs evolutionary relationship was constructed. In addition, the poplar, Arabidopsis, rice annotation, and whole-genome information were applied to build the CCD gene structures. The MEME was used to identify the CCD motif compositions and distributions. Finally, all those generated files related to CCD gene structures and motifs were visualized using TBtools software [72,73].
4.3. Chromosomal Localization and Collinearity Analysis of PtCCDs
According to the PtCCDs annotation, the 23 PtCCD genes were mapped onto poplar chromosomes. The TBtools with MCScanX was applied to analyze tandem and segmental duplication events of PtCCDs, and TBtools with synteny visualization was used to visualize the collinearity relationship. In addition, the TBtools with a simple Ka/Ks calculator was applied to calculate Ka/Ks values between gene pairs. In addition, MCScanX was also used to identify the gene pairs with collinearity relationship among poplar, Arabidopsis, rice, and willow. Similarly, TBtools was applied to visualize syntenic blocks of orthologous genes.
4.4. Analyses of 3D Structures and Cis-Elements
The SWISS-MODEL (https://swissmodel.expasy.org/ (accessed on 28 July 2021)) was used to predict the structures of PtCCDs, and the α-helix, random coil, and strand PtCCDs were represented using Chimera software. The 2000 bp sequences upstream of the translation start sites of CCD genes were obtained from poplar, Arabidopsis, and rice genome database, and Plant CARE online tool was applied to predict cis-elements of CCD genes.
4.5. Plant Treatments and qRT-PCR Analysis
The leaves, stems, and roots harvested from the P. trichocarpa, ‘Shanxinyang’ (P. davidiana × P. bolleana Loucne), and ‘Nanlin 895’ (P. deltoides × P. euramericana) were used for tissue-specific gene expression analysis. Additionally, the leaves of ‘Nanlin 895’ grown on MS medium were treated with 2 mM of H2O2, 10% PEG6000, 200 mM of NaCl, and 200 μM of ABA, and leaves were collected at 0 (untreated leaves served as a control), 1, 6, 12, 24, and 48 h after each treatment. The abiotic stress treatments were performed with three replicates, with three poplars per replicate. Leaves harvested from treated and untreated poplars were stored at −80 °C.
Total RNA was extracted from various tissues and treated and untreated leaves using an RNA extraction kit (Takara, Japan). The reverse transcriptase (Takara, Japan) was used to synthesize first-strand cDNA of poplar. The UltraSYBR Green I Mixture (CWBIO, China) with 10 μL of Green I Mixture in a 20-μL reaction volume was applied to identify the PtCCD gene expression patterns. The Primer3.0 online tool was used to design the primers used in qRT–PCR. The qRT–PCR procedure was as follows: 95 °C for 10 min; 40 cycles of 94 °C for 10 s, 60 °C for 30 s, and 72 °C for 30 s. The relative PtCCD expression levels were identified according to the 2−ΔΔCT method, with the Ptactin (XM-006370951), considered the internal control.
5. Conclusions
In this study, the characterizations and putative functions of 23 poplar CCD members were identified. These genes were divided into PtCCD1, PtCCD4, PtCCD7, PtCCD8, and PtNCED classes based on the molecular phylogenetic tree. The PtCCD gene structures and conserved motifs were also embodied in the evolutionary relationship. The 5 PtCCD pairs of homologous genes were identified in poplar genome, and 5 or 16 CCD pairs of orthologous genes were illustrated between poplar and Arabidopsis or willow, respectively. In addition, various kinds of stress-responsive cis-elements were identified in the promoters of PtCCDs, indicating that PtCCDs have a hand in comprehensive stress resistances. In addition, PtCCDs exhibited tissue-special expression patterns in poplar, implying they might be involved in divergent tissue and organ developments. Many PtCCDs expression levels were affected by ABA treatment and osmotic stress, suggesting that they may participate in ABA signal transduction or play an essential role in response to osmotic stress. The putative regulation mechanism of PtCCDs in response to abiotic and biotic stresses in poplar was predicted.
On one hand, abiotic and biotic stresses induce higher abundances of PtCCDs in poplar. PtNCEDs encode key enzymes for biosynthesis of ABA, ultimately causing the producing ABA signal transduction, resulting in abiotic and biotic stress resistance. On the other hand, other CCDs can catalyze carotenoids to form apocarotenoid substrates in response to stresses (Supplemental Figure S8). These will be beneficial for exploring PtCCD functions and potential regulatory mechanisms and lay the basis for illustrating corresponding gene networks involved in osmotic stress.
Acknowledgments
We gratefully thank Sheng Zhu (Nanjing Forestry University) for providing poplar varieties.
Abbreviations
Carotenoid cleavage dioxygenase: CCD; 9-cis epoxycarotenoid dioxygenases: NCED; abscisic acid: ABA; multiple collinearity scan toolkit: MCScanX; anaerobic inducible element: ARE; stress response element: STRE; MYB drought inducible binding site: MBS; DREB/CBF transcription factor recognition site: DRE; low-temperature response elements: LTR; auxin response elements: TGA-element; salicylic acid: SA; methyl jasmonate: MeJA; ABA response element: ABRE; cytochrome P: CYP; phytoene synthase: PSY; abscisic acid insensitive: ABI; gibberellic oxidase: GAox; thaumatin-like protein: TCP; strigolactone: SL; neighbor-joining: NJ.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms23031418/s1.
Author Contributions
J.Z. designed and funded experiments. H.W. wrote the first draft of the manuscript. J.Z. and A.M. revised the manuscript. H.W., G.L., Y.L., S.L., C.Y., Y.C., and F.Z. experimented. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by China Forestry Science and Technology Innovation and Promotion Project (2021TG03), Nantong University Scientific Research Start-up Project for Introducing Talents (135421609106), and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chen K., Li G.J., Bressan R.A., Song C.P., Zhu J.K., Zhao Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020;62:25–54. doi: 10.1111/jipb.12899. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez-Concepcion M., Avalos J., Bonet M.L., Boronat A., Gomez-Gomez L., Hornero-Mendez D., Limon M.C., Meléndez-Martínez A.J., Olmedilla-Alonso B., Palou A., et al. A global perspective on carotenoids: Metabolism, biotechnology, and benefits for nutrition and health. Prog. Lipid Res. 2018;70:62–93. doi: 10.1016/j.plipres.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Woitsch S., Romer S. Expression of xanthophyll biosynthetic genes during light-dependent chloroplast differentiation. Plant Physiol. 2003;132:1508–1517. doi: 10.1104/pp.102.019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartley G.E., Scolnik P.A. Plant carotenoids: Pigments for photoprotection, visual attraction, and human health. Plant Cell. 1995;7:1027. doi: 10.1105/tpc.7.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouvier F., Isner J.C., Dogbo O., Camara B. Oxidative tailoring of carotenoids: A prospect towards novel functions in plants. Trends Plant Sci. 2005;10:187–194. doi: 10.1016/j.tplants.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Ilahy R., Siddiqui M.W., Tlili I., Montefusco A., Piro G., Hdider C., Lenucci M.S. When color really matters: Horticultural performance and functional quality of high-lycopene tomatoes. Crit. Rev. Plant Sci. 2018;37:15–53. doi: 10.1080/07352689.2018.1465631. [DOI] [Google Scholar]
- 7.Wang C., Qiao A., Fang X., Sun L., Gao P., Davis A.R., Liu S., Luan F. Fine mapping of lycopene content and flesh color related gene and development of molecular marker-assisted selection for flesh color in watermelon (Citrullus lanatus) Front. Plant Sci. 2019;10:1240. doi: 10.3389/fpls.2019.01240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trivellini A., Ferrante A., Vernieri P., Mensuali-Sodi A., Serra G. Effects of promoters and inhibitors of ethylene and aba on flower senescence of Hibiscus rosa-sinensis L. J. Plant Growth Regul. 2011;30:175–184. doi: 10.1007/s00344-010-9181-9. [DOI] [Google Scholar]
- 9.Chernys J.T., Zeevaart J.A. Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiol. 2000;124:343–354. doi: 10.1104/pp.124.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shu K., Liu X.D., Xie Q., He Z.H. Two faces of one seed: Hormonal regulation of dormancy and germination. Mol. Plant. 2016;9:34–45. doi: 10.1016/j.molp.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 11.Seo M., Koshiba T. Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 2002;7:41–48. doi: 10.1016/S1360-1385(01)02187-2. [DOI] [PubMed] [Google Scholar]
- 12.Kuromori T., Sugimoto E., Shinozaki K. Arabidopsis mutants of AtABCG22, an ABC transporter gene, increase water transpiration and drought susceptibility. Plant J. 2011;67:885–894. doi: 10.1111/j.1365-313X.2011.04641.x. [DOI] [PubMed] [Google Scholar]
- 13.McAdam S.A., Brodribb T.J., Banks J.A., Hedrich R., Atallah N.M., Cai C., Geringer M.A., Lind C., Nichols D.S., Stachowski K., et al. Abscisic acid controlled sex before transpiration in vascular plants. Proc. Natl. Acad. Sci. USA. 2016;113:12862–12867. doi: 10.1073/pnas.1606614113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Islam M.M., Ye W., Matsushima D., Munemasa S., Okuma E., Nakamura Y., Biswas S., Mano J.I., Murata Y. Reactive carbonyl species mediate ABA signaling in guard cells. Plant Cell Physiol. 2016;57:2552–2563. doi: 10.1093/pcp/pcw166. [DOI] [PubMed] [Google Scholar]
- 15.Li X., Zhao J., Sun Y., Li Y. Arabidopsis thaliana CRK41 negatively regulates salt tolerance via H2O2 and ABA cross-linked networks. Environ. Exp. Bot. 2020;179:104210. [Google Scholar]
- 16.Kloer D.P., Ruch S., Al-Babili S., Beyer P., Schulz G.E. The structure of a retinal-forming carotenoid oxygenase. Science. 2005;308:267–269. doi: 10.1126/science.1108965. [DOI] [PubMed] [Google Scholar]
- 17.Auldridge M.E., Block A., Vogel J.T., Dabney-Smith C., Mila I., Bouzayen M., Magallanes-Lundback M., DellaPenna D., McCarty D.R., Klee H.J. Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 2006;45:982–993. doi: 10.1111/j.1365-313X.2006.02666.x. [DOI] [PubMed] [Google Scholar]
- 18.Tan B.C., Joseph L.M., Deng W.T., Liu L., Li Q.B., Cline K., McCarty D.R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003;35:44–56. doi: 10.1046/j.1365-313X.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- 19.Wang P., Lu S., Zhang X., Hyden B., Qin L., Liu L., Bai Y., Han Y., Wen Z., Xu J., et al. Double NCED isozymes control ABA biosynthesis for ripening and senescent regulation in peach fruits. Plant Sci. 2021;304:110739. doi: 10.1016/j.plantsci.2020.110739. [DOI] [PubMed] [Google Scholar]
- 20.Tan B.C., Schwartz S.H., Zeevaart J.A., McCarty D.R. Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. USA. 1997;94:12235–12240. doi: 10.1073/pnas.94.22.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwartz S.H., Tan B.C., Gage D.A., Zeevaart J.A., McCarty D.R. Specific oxidative cleavage of carotenoids by VP14 of maize. Science. 1997;276:1872–1874. doi: 10.1126/science.276.5320.1872. [DOI] [PubMed] [Google Scholar]
- 22.Iuchi S., Kobayashi M., Taji T., Naramoto M., Seki M., Kato T., Tabata S., Kakubari Y., Yamaguchi-Shinozaki K., Shinozaki K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001;27:325–333. doi: 10.1046/j.1365-313x.2001.01096.x. [DOI] [PubMed] [Google Scholar]
- 23.Fan J., Hill L., Crooks C., Doerner P., Lamb C. Abscisic acid has a key role in modulating diverse plant-pathogen interactions. Plant Physiol. 2009;150:1750–1761. doi: 10.1104/pp.109.137943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Changan S.S., Ali K., Kumar V., Garg N.K., Tyagi A. Abscisic acid biosynthesis under water stress: Anomalous behavior of the 9-cis-epoxycarotenoid dioxygenase1 (NCED1) gene in rice. Biol. Plantarum. 2018;62:663–670. doi: 10.1007/s10535-018-0807-2. [DOI] [Google Scholar]
- 25.Song S., Dai X., Zhang W.H. A rice F-box gene, Os Fbx352, is involved in glucose-delayed seed germination in rice. J. Exp. Bot. 2012;63:5559–5568. doi: 10.1093/jxb/ers206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hwang S.G., Chen H.C., Huang W.Y., Chu Y.C., Shii C.T., Cheng W.H. Ectopic expression of rice OsNCED3 in Arabidopsis increases ABA level and alters leaf morphology. Plant Sci. 2010;178:12–22. doi: 10.1016/j.plantsci.2009.09.014. [DOI] [Google Scholar]
- 27.Huang Y., Jiao Y., Xie N., Guo Y., Zhang F., Xiang Z., Wang R., Wang F., Gao Q., Tian L., et al. OsNCED5, a 9-cis-epoxycarotenoid dioxygenase gene, regulates salt and water stress tolerance and leaf senescence in rice. Plant Sci. 2019;287:110188. doi: 10.1016/j.plantsci.2019.110188. [DOI] [PubMed] [Google Scholar]
- 28.Ahrazem O., Rubio-Moraga A., Trapero A., Gómez-Gómez L. Developmental and stress regulation of gene expression for a 9-cis-epoxycarotenoid dioxygenase, CstNCED, isolated from Crocus sativus stigmas. J. Exp. Bot. 2012;63:681–694. doi: 10.1093/jxb/err293. [DOI] [PubMed] [Google Scholar]
- 29.Huang F.C., Molnár P., Schwab W. Cloning and functional characterization of carotenoid cleavage dioxygenase 4 genes. J. Exp. Bot. 2009;60:3011–3022. doi: 10.1093/jxb/erp137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baldermann S., Kato M., Kurosawa M., Kurobayashi Y., Fujita A., Fleischmann P., Watanabe N. Functional characterization of a carotenoid cleavage dioxygenase 1 and its relation to the carotenoid accumulation and volatile emission during the floral development of Osmanthus fragrans Lour. J. Exp. Bot. 2010;61:2967–2977. doi: 10.1093/jxb/erq123. [DOI] [PubMed] [Google Scholar]
- 31.Yahyaa M., Bar E., Dubey N.K., Meir A., Davidovich-Rikanati R., Hirschberg J., Aly R., Tholl D., Simon P.W., Tadmor Y., et al. Formation of norisoprenoid flavor compounds in carrot (Daucus carota L.) roots: Characterization of a cyclic-specific carotenoid cleavage dioxygenase 1 gene. J. Agric. food Chem. 2013;61:12244–12252. doi: 10.1021/jf404085k. [DOI] [PubMed] [Google Scholar]
- 32.Ilg A., Beyer P., Al-Babili S. Characterization of the rice carotenoid cleavage dioxygenase 1 reveals a novel route for geranial biosynthesis. FEBS J. 2009;276:736–747. doi: 10.1111/j.1742-4658.2008.06820.x. [DOI] [PubMed] [Google Scholar]
- 33.Schwartz S.H., Qin X., Loewen M.C. The biochemical characterization of two carotenoid cleavage enzymes from Arabidopsis indicates that a carotenoid-derived compound inhibits lateral branching. J. Biol. Chem. 2004;279:46940–46945. doi: 10.1074/jbc.M409004200. [DOI] [PubMed] [Google Scholar]
- 34.Alder A., Jamil M., Marzorati M., Bruno M., Vermathen M., Bigler P., Ghisla S., Bouwmeester H., Beyer P., Al-Babili S. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science. 2012;335:1348–1351. doi: 10.1126/science.1218094. [DOI] [PubMed] [Google Scholar]
- 35.Vallabhaneni R., Bradbury L.M., Wurtzel E.T. The carotenoid dioxygenase gene family in maize, sorghum, and rice. Arch. Biochem. Biophys. 2010;504:104–111. doi: 10.1016/j.abb.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lashbrooke J.G., Young P.R., Dockrall S.J., Vasanth K., Vivier M.A. Functional characterisation of three members of the Vitis vinifera L. carotenoid cleavage dioxygenase gene family. BMC Plant Biol. 2013;13:156. doi: 10.1186/1471-2229-13-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohmiya A., Kishimoto S., Aida R., Yoshioka S., Sumitomo K. Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol. 2006;142:1193–1201. doi: 10.1104/pp.106.087130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodrigo M.J., Alquézar B., Alós E., Medina V., Carmona L., Bruno M., Al-Babili S., Zacarías L. A novel carotenoid cleavage activity involved in the biosynthesis of Citrus fruit-specific apocarotenoid pigments. J. Exp. Bot. 2013;64:4461–4478. doi: 10.1093/jxb/ert260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang R.K., Wang C.E., Fei Y.Y., Gai J.Y., Zhao T.J. Genome-wide identification and transcription analysis of soybean carotenoid oxygenase genes during abiotic stress treatments. Mol. Biol. Rep. 2013;40:4737–4745. doi: 10.1007/s11033-013-2570-y. [DOI] [PubMed] [Google Scholar]
- 40.Kim Y., Hwang I., Jung H.J., Park J.I., Kang J.G., Nou I.S. Genome-wide classification and abiotic stress-responsive expression profiling of carotenoid oxygenase genes in Brassica rapa and Brassica oleracea. J. Plant Growth Regul. 2016;35:202–214. doi: 10.1007/s00344-015-9520-y. [DOI] [Google Scholar]
- 41.Chen H., Zuo X., Shao H., Fan S., Ma J., Zhang D., Zhao C., Yan X., Liu X., Han M. Genome-wide analysis of carotenoid cleavage oxygenase genes and their responses to various phytohormones and abiotic stresses in apple (Malus domestica) Plant Physiol. Bioch. 2018;123:81–93. doi: 10.1016/j.plaphy.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 42.Kumar M., Kherawat B.S., Dey P., Saha D., Singh A., Bhatia S.K., Ghodake G.S., Kadam A.A., Kim H.U., Chung S.M., et al. Genome-Wide Identification and Characterization of PIN-FORMED (PIN) Gene Family Reveals Role in Developmental and Various Stress Conditions in Triticum aestivum L. Int. J. Mol. Sci. 2021;22:7396. doi: 10.3390/ijms22147396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tong T., Fang Y.X., Zhang Z., Zheng J., Zhang X., Li J., Niu C., Xue D., Zhang X. Genome-wide identification and expression pattern analysis of the KCS gene family in barley. Plant Growth Regul. 2021;93:89–103. doi: 10.1007/s10725-020-00668-3. [DOI] [Google Scholar]
- 44.Baruah P.M., Krishnatreya D.B., Bordoloi K.S., Gill S.S., Agarwala N. Genome wide identification and characterization of abiotic stress responsive lncRNAs in Capsicum annuum. Plant Physiol. Biochem. 2021;162:221–236. doi: 10.1016/j.plaphy.2021.02.031. [DOI] [PubMed] [Google Scholar]
- 45.Kesawat M.S., Kherawat B.S., Singh A., Dey P., Kabi M., Debnath D., Saha D., Khandual A., Rout S., Ali A., et al. Genome-wide identification and characterization of the brassinazole-resistant (BZR) gene family and its expression in the various developmental stage and stress conditions in wheat (Triticum aestivum L.) Int. J. Mol. Sci. 2021;22:8743. doi: 10.3390/ijms22168743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang M., Chen H., Liu J., Du Q., Lu S., Liu C. Genome-wide identification and functional characterization of natural antisense transcripts in Salvia miltiorrhiza. Sci. Rep. 2021;11:4769. doi: 10.1038/s41598-021-83520-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang M., Chen B., Zhou W., Xie L., Wang L., Zhang Y., Zhang Q. Genome-wide identification and expression analysis of the AT-hook Motif Nuclear Localized gene family in soybean. BMC Genom. 2021;22:361. doi: 10.1186/s12864-021-07687-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Q., Li Q., Li P., Zhang S., Liu C., Jin J., Cao P., Yang Y. Carotenoid cleavage dioxygenases: Identification, expression, and evolutionary analysis of this gene family in tobacco. Int. J. Mol. Sci. 2019;20:5796. doi: 10.3390/ijms20225796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tuskan G.A., Difazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 50.Zhang J., Yuan H., Li Y., Chen Y., Liu G., Ye M., Yu C., Lian B., Zhong F., Jiang Y., et al. Genome sequencing and phylogenetic analysis of allotetraploid Salix matsudana Koidz. Hortic. Res. 2020;7:201. doi: 10.1038/s41438-020-00424-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei Y., Wan H., Wu Z., Wang R., Ruan M., Ye Q., Li Z., Zhou G., Yao Z., Yang Y. A comprehensive analysis of carotenoid cleavage dioxygenases genes in Solanum lycopersicum. Plant Mol. Biol. Rep. 2016;34:512–523. doi: 10.1007/s11105-015-0943-1. [DOI] [Google Scholar]
- 52.Bruno M., Vermathen M., Alder A., Wüst F., Schaub P., van der Steen R., Beyer P., Ghisla S., Al-Babili S. Insights into the formation of carlactone from in-depth analysis of the CCD 8-catalyzed reactions. FEBS Lett. 2017;591:792–800. doi: 10.1002/1873-3468.12593. [DOI] [PubMed] [Google Scholar]
- 53.Rubio-Moraga A., Ahrazem O., Pérez-Clemente R.M., Gómez-Cadenas A., Yoneyama K., López-Ráez J.A., Molina R.V., Gómez-Gómez L. Apical dominance in saffron and the involvement of the branching enzymes CCD7 and CCD8 in the control of bud sprouting. BMC Plant Biol. 2014;14:171. doi: 10.1186/1471-2229-14-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ledger S.E., Janssen B.J., Karunairetnam S., Wang T., Snowden K.C. Modified CAROTENOID CLEAVAGE DIOXYGENASE8 expression correlates with altered branching in kiwifruit (Actinidia chinensis) New Phytol. 2010;188:803–813. doi: 10.1111/j.1469-8137.2010.03394.x. [DOI] [PubMed] [Google Scholar]
- 55.Vogel J.T., Walter M.H., Giavalisco P., Lytovchenko A., Kohlen W., Charnikhova T., Simkin A.J., Goulet C., Strack D., Bouwmeester H.J., et al. SlCCD7 controls strigolactone biosynthesis, shoot branching and mycorrhiza-induced apocarotenoid formation in tomato. Plant J. 2010;61:300–311. doi: 10.1111/j.1365-313X.2009.04056.x. [DOI] [PubMed] [Google Scholar]
- 56.Kulkarni K.P., Vishwakarma C., Sahoo S.P., Lima J.M., Nath M., Dokku P., Gacche R.N., Mohapatra T., Robin S., Sarla N., et al. A substitution mutation in OsCCD7 cosegregates with dwarf and increased tillering phenotype in rice. J. Genet. 2014;93:389–401. doi: 10.1007/s12041-014-0389-5. [DOI] [PubMed] [Google Scholar]
- 57.Pei X., Wang X., Fu G., Chen B., Nazir M.F., Pan Z., He S., Du X. Identification and functional analysis of 9-cis-epoxy carotenoid dioxygenase (NCED) homologs in G. hirsutum. Int. J. Biol. Macromol. 2021;182:298–310. doi: 10.1016/j.ijbiomac.2021.03.154. [DOI] [PubMed] [Google Scholar]
- 58.Zhang J., Zhang P., Huo X., Gao Y., Chen Y., Song Z., Wang F., Zhang J. Comparative phenotypic and transcriptomic analysis reveals key responses of upland cotton to salinity stress during postgermination. Front. Plant Sci. 2021;12:606. doi: 10.3389/fpls.2021.639104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lefebvre V., North H., Frey A., Sotta B., Seo M., Okamoto M., Nambara E., Marion-Poll A. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 2006;45:309–319. doi: 10.1111/j.1365-313X.2005.02622.x. [DOI] [PubMed] [Google Scholar]
- 60.Song M.H., Lim S.H., Kim J.K., Jung E.S., John K.M., You M.K., Ahn S.N., Lee C.H., Ha S.H. In planta cleavage of carotenoids by Arabidopsis carotenoid cleavage dioxygenase 4 in transgenic rice plants. Plant Biotechnol. Rep. 2016;10:291–300. doi: 10.1007/s11816-016-0405-8. [DOI] [Google Scholar]
- 61.Ureshino K., Nakayama M., Miyajima I. Contribution made by the carotenoid cleavage dioxygenase 4 gene to yellow colour fade in azalea petals. Euphytica. 2016;207:401–417. doi: 10.1007/s10681-015-1557-2. [DOI] [Google Scholar]
- 62.Liu H., Kishimoto S., Yamamizo C., Fukuta N., Ohmiya A. Carotenoid accumulations and carotenogenic gene expressions in the petals of Eustoma grandiflorum. Plant Breed. 2013;132:417–422. doi: 10.1111/pbr.12043. [DOI] [Google Scholar]
- 63.Zheng X., Zhu K., Sun Q., Zhang W., Wang X., Cao H., Tan M., Xie Z., Zeng Y., Ye J., et al. Natural variation in CCD4 promoter underpins species-specific evolution of red coloration in citrus peel. Mol. Plant. 2019;12:1294–1307. doi: 10.1016/j.molp.2019.04.014. [DOI] [PubMed] [Google Scholar]
- 64.Simkin A.J., Underwood B.A., Auldridge M., Loucas H.M., Shibuya K., Schmelz E., Clark D.G., Klee H.J. Circadian regulation of the PhCCD1 carotenoid cleavage dioxygenase controls emission of β-ionone, a fragrance volatile of petunia flowers. Plant Physiol. 2004;136:3504–3514. doi: 10.1104/pp.104.049718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Booker J., Sieberer T., Wright W., Williamson L., Willett B., Stirnberg P., Turnbull C., Srinivasan M., Goddard P., Leyser O. MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev. Cell. 2005;8:443–449. doi: 10.1016/j.devcel.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 66.Delaux P.M., Xie X., Timme R.E., Puech-Pages V., Dunand C., Lecompte E., Delwiche C.F., Yoneyama K., Bécard G., Séjalon-Delmas N. Origin of strigolactones in the green lineage. New Phytol. 2012;195:857–871. doi: 10.1111/j.1469-8137.2012.04209.x. [DOI] [PubMed] [Google Scholar]
- 67.Iuchi S., Kobayashi M., Yamaguchi-Shinozaki K., Shinozaki K. A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol. 2000;123:553–562. doi: 10.1104/pp.123.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lawlor D.W., Cornic G. Photosynthetic carbon assimilation and associated metabolism in relation to water deficits in higher plants. Plant Cell Environ. 2002;25:275–294. doi: 10.1046/j.0016-8025.2001.00814.x. [DOI] [PubMed] [Google Scholar]
- 69.Candan N., Tarhan L. Tolerance or sensitivity responses of Mentha pulegium to osmotic and waterlogging stress in terms of antioxidant defense systems and membrane lipid peroxidation. Environ. Exp. Bot. 2012;75:83–88. doi: 10.1016/j.envexpbot.2011.08.014. [DOI] [Google Scholar]
- 70.Zhang C., Shi S. Physiological and proteomic responses of contrasting alfalfa (Medicago sativa L.) varieties to PEG-induced osmotic stress. Front. Plant Sci. 2018;9:242. doi: 10.3389/fpls.2018.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Q., Lv L.R., Teng Y.J., Si L.B., Ma T., Yang Y.L. Apoplastic hydrogen peroxide and superoxide anion exhibited different regulatory functions in salt-induced oxidative stress in wheat leaves. Biol. Plantarum. 2018;62:750–762. doi: 10.1007/s10535-018-0808-1. [DOI] [Google Scholar]
- 72.Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 73.Zhang J., zheng Shi S., Jiang Y., Zhong F., Liu G., Yu C., Lian B., Chen Y. Genome-wide investigation of the AP2/ERF superfamily and their expression under salt stress in Chinese willow (Salix matsudana) PeerJ. 2021;9:e11076. doi: 10.7717/peerj.11076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.