Abstract
Energy conservation in microorganisms is classically categorized into respiration and fermentation; however, recent work shows some species can use mixed or alternative bioenergetic strategies. We explored the use of extracellular electron transfer for energy conservation in diverse lactic acid bacteria (LAB), microorganisms that mainly rely on fermentative metabolism and are important in food fermentations. The LAB Lactiplantibacillus plantarum uses extracellular electron transfer to increase its NAD+/NADH ratio, generate more ATP through substrate-level phosphorylation, and accumulate biomass more rapidly. This novel, hybrid metabolism is dependent on a type-II NADH dehydrogenase (Ndh2) and conditionally requires a flavin-binding extracellular lipoprotein (PplA) under laboratory conditions. It confers increased fermentation product yield, metabolic flux, and environmental acidification in laboratory media and during kale juice fermentation. The discovery of a single pathway that simultaneously blends features of fermentation and respiration in a primarily fermentative microorganism expands our knowledge of energy conservation and provides immediate biotechnology applications.
Research organism: Other
eLife digest
Bacteria produce the energy they need to live through two processes, respiration and fermentation. While respiration is often more energetically efficient, many bacteria rely on fermentation as their sole means of energy production. Respiration normally depends on the presence of small soluble molecules, such as oxygen, that can diffuse inside the cell, but some bacteria can use metals or other insoluble compounds found outside the cell to perform ‘extracellular electron transfer’.
Lactic acid bacteria are a large group of bacteria that have several industrial uses and live in many natural environments. These bacteria survive using fermentation, but they also carry a group of genes needed for extracellular electron transfer. It is unclear whether they use these genes for respiration or if they have a different purpose.
Tejedor-Sanz, Stevens et al. used a lactic acid bacterium called Lactiplantibacillus plantarum to study whether and how this group of bacteria use extracellular electron transfer. Analysis of L. plantarum and its effect on its surroundings showed that these bacteria use a hybrid process to produce energy: the cells use aspects of extracellular respiration to increase the yield and efficiency of fermentation. Combining these two approaches may allow L. plantarum to adapt to different environments and grow faster, allowing it to compete against other species.
Tejedor-Sanz, Stevens et al. provide new information on a widespread group of bacteria that are often used in food production and industry. The next step will be to understand how the hybrid system is controlled and how it varies among species. Understanding this process could result in new biotechnologies and foods that are healthier, produce less waste, or have different tastes and textures.
Introduction
The ways in which microorganisms extract energy to maintain cellular functions are directly linked to their environment, including the availability of nutrients and cooperative or antagonistic interactions with other organisms (Haruta and Kanno, 2015). Microorganisms must also maintain redox homeostasis by responding to oxidative and reductive changes inside and outside the cell (Sporer et al., 2017). Ultimately, microorganisms that can effectively generate cellular energy while also managing redox requirements will maintain higher growth and survival rates, and therefore exhibit greater ecological fitness.
All organisms possess mechanisms to conserve energy, that is, to convert light or chemical energy into cellular energy in the form of ATP (Russell and Cook, 1995). During respiration, microorganisms rely on either oxygen (aerobic respiration) or other exogenous substrates (anaerobic respiration) as terminal electron acceptors. Some microorganisms, most notably Geobacter spp., can anaerobically respire using electron acceptors outside the cell, such as iron (III) oxides or an electrode (Renslow et al., 2013; Richter et al., 2012). This process is called extracellular electron transfer (EET). Regardless of the identity of the electron acceptor, ATP synthesis during respiration occurs via oxidative phosphorylation (Kim and Gadd, 2019). In oxidative phosphorylation, electrons from electron carriers are transported by an electron transport chain, which creates a proton motive force (PMF) for ATP generation. Under anaerobic conditions, some cells can also conserve energy using fermentation. In fermentation, microorganisms use internally supplied electron acceptors, and ATP is generated mainly through substrate-level phosphorylation (Kim and Gadd, 2019). In substrate-level phosphorylation, ATP is generated in the cytoplasm by transfer of phosphate from metabolic intermediates to ADP (Kim and Gadd, 2019).
Lactic acid bacteria (LAB) are a diverse group of aerotolerant, saccharolytic microorganisms in the Firmicutes phylum which mainly use fermentation for energy conservation. LAB are essential for many food fermentations, including fermented milk and meats, fruits and vegetables, and grains (Tamang et al., 2020). Strains of LAB are also used for industrial chemical production (Sauer et al., 2017) and as probiotics to benefit human and animal health (Vinderola et al., 2019). LAB are generally grouped by their differences in hexose metabolism (Salvetti et al., 2013). Some species perform homofermentation, reducing pyruvate to lactate as the sole metabolic end-product from glycolysis. Other LAB perform heterofermentation, producing lactate along with ethanol, acetate, and CO2 by the phosphoketolase pathway. However, for redox balancing, homofermentative LAB can also shift to a mixed acid fermentation and heterofermentative LAB use alternative electron acceptors, like fructose or citrate (Hansen, 2018). Although some LAB can respire in the presence of heme and menaquinone, those bacteria are unable to synthesize heme and many are also auxotrophic for menaquinone (Pedersen et al., 2012). Even those species capable of respiration still use fermentation metabolism as the primary mechanism to conserve energy (Pedersen et al., 2012). Therefore, LAB growth rates and cell yields are constrained by access to electron acceptors used to maintain intracellular redox balance during substrate-level phosphorylation.
The bioenergetics of anaerobic bacteria have been tightly linked to oxidative phosphorylation for anaerobic respiration and substrate-level phosphorylation for fermentation. However, experimental evidence shows a concurrent use of oxidative phosphorylation and substrate-level phosphorylation. For instance, some yeasts perform respiro-fermentation to enhance ATP production (Pfeiffer and Morley, 2014). Another example is the electron bifurcating mechanism used by some fermentative microorganisms such as Clostridium spp. (Herrmann et al., 2008; Li et al., 2008). Through that energy conservation strategy, cells can generate extra ATP through oxidative phosphorylation (Buckel and Thauer, 2013; Müller et al., 2018). Along with other examples that are not fully understood (Hunt et al., 2010,Kracke et al., 2018), these observations suggest metabolisms that combine aspects of fermentation and respiration may exist.
We recently discovered that Listeria monocytogenes, a facultative anaerobic pathogen known to rely on respiratory metabolism, uses EET to reduce Fe3+ or an anode through a flavin-based extracellular electron transfer pathway (Light et al., 2018). Use of this pathway allowed L. monocytogenes to maintain intracellular redox balance via NADH oxidation. This capacity was associated with the presence of a gene locus, called a flavin-based EET (FLEET) locus, that was identified in many Gram-positive species in the Firmicutes phylum, including LAB. Studies in individual species of LAB such as Lactococcus lactis (Freguia et al., 2009; Masuda et al., 2010), Enterococcus faecalis (Hederstedt et al., 2020; Keogh et al., 2018), and Lactiplantibacillus pentosus (Vilas Boas et al., 2015) show that they can perform EET with an anode endogenously, that is without addition of molecules foreign to their native niches. These observations are quite surprising because endogenous EET has been mainly associated with respiratory organisms, even though some of these organisms also possess fermentative-type metabolism (Glasser et al., 2014). Those observations also raise the question of whether the FLEET locus is functional in LAB and what, if any, role it plays in energy conservation and metabolism.
Here, we explored EET across LAB and studied the implications of this trait at a metabolic and energetic level in Lactiplantibacillus plantarum, a homofermentative LAB capable of mixed acid fermentation and which can respire in the presence of exogenous heme and menaquinone. L. plantarum is of particular interest as it is a generalist LAB species found in insect, animal, and human digestive tracts and is essential for the production of many fermented foods (Behera et al., 2018; Duar et al., 2017). These findings have significance for the understanding of energy conservation strategies in primarily fermentative microorganisms and on lactic acid fermentations in food biotechnology.
Results
L. plantarum reduces extracellular electron acceptors
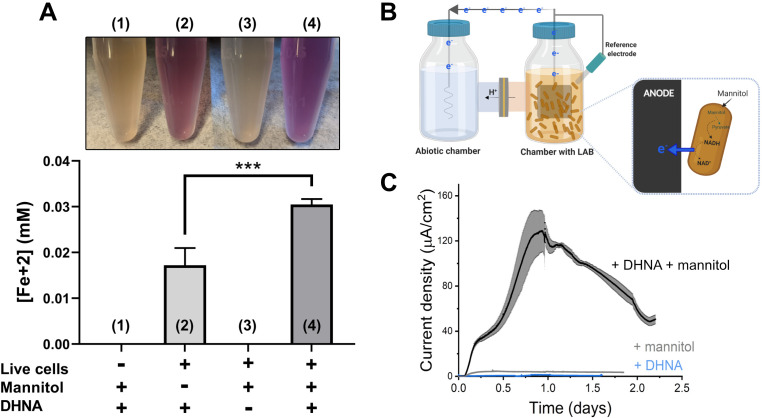
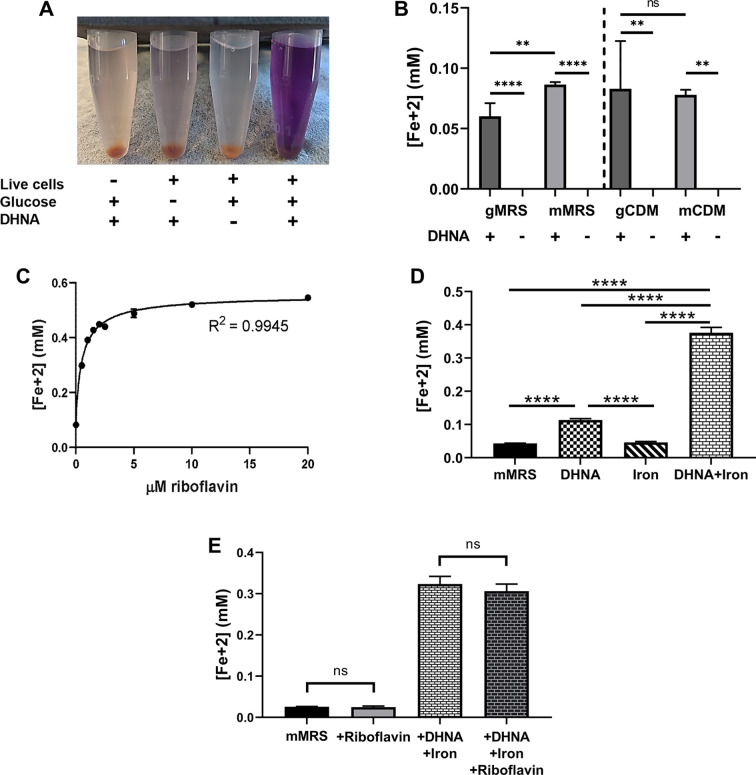
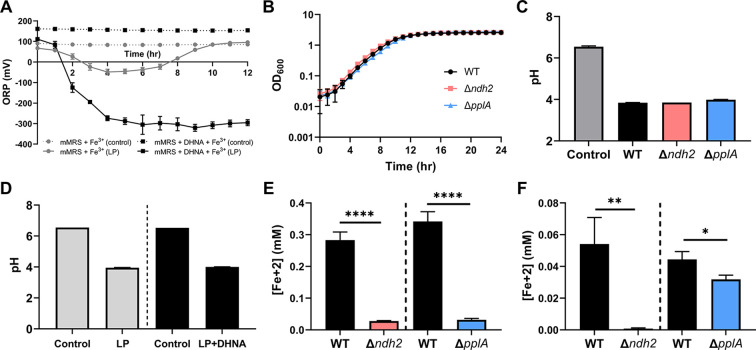
To determine whether L. plantarum can reduce extracellular electron acceptors, we first measured its ability to reduce insoluble ferrihydrite (iron (III) oxyhydroxide). Incubation of the model strain L. plantarum NCIMB8826 in the presence of ferrihydrite showed that this strain reduces Fe3+ to Fe2+ (Figure 1A and Figure 1—figure supplement 1A). Viable cells are required for iron reduction and this activity is dependent on the presence of exogenous quinone (DHNA, 1,4-dihydroxy-2-naphthoic acid) (Figure 1A and Figure 1—figure supplement 1A-B). The requirement for DHNA was hypothesized because DHNA is a precursor of demethylmenaquinone (DMK), a membrane electron shuttle utilized by L. monocytogenes for EET (Light et al., 2018), and L. plantarum lacks a complete DHNA biosynthetic pathway (Brooijmans et al., 2009a). For full activity, an electron donor (such as mannitol or glucose) was required to be present (Figure 1A and Figure 1—figure supplement 1A). Like L. monocytogenes (Light et al., 2018), the addition of riboflavin during the iron reduction assay also increased Fe3+ reduction in a dose-dependent manner (Figure 1—figure supplement 1C). Thus, L. plantarum reduces insoluble iron in a manner similar to L. monocytogenes.
Figure 1. L. plantarum can reduce both Fe3+ and an anode through EET.
(A) Reduction of Fe3+ (ferrihydrite) to Fe2+ by L. plantarum NCIMB8826 after growth in mMRS. The assays were performed in PBS supplemented with 20 µg/mL DHNA and/or 55 mM mannitol. Fe2+ was detected colorimetrically using 2 mM ferrozine. For L. plantarum inactivation, cells were incubated at 85℃ in PBS for 30 min prior to the assay. Significant differences were determined by one-way ANOVA with Tukey’s post-hoc test (n = 3), *** p < 0.001. (B) Two-chambered electrochemical cell setup for measuring current generated by L. plantarum. (C) Current density production over time by L. plantarum in CDM supplemented with 20 µg/mL DHNA and/or 110 mM mannitol. The anode was polarized at +0.2VAg/AgCl. The avg ± stdev of three biological replicates is shown. See also Figure 1—figure supplement 1 and Figure 1—figure supplement 2 and related data in Figure 1—source data 1.
Figure 1—figure supplement 1. Iron reduction by L. plantarum is dependent upon DHNA, carbon source, and riboflavin.
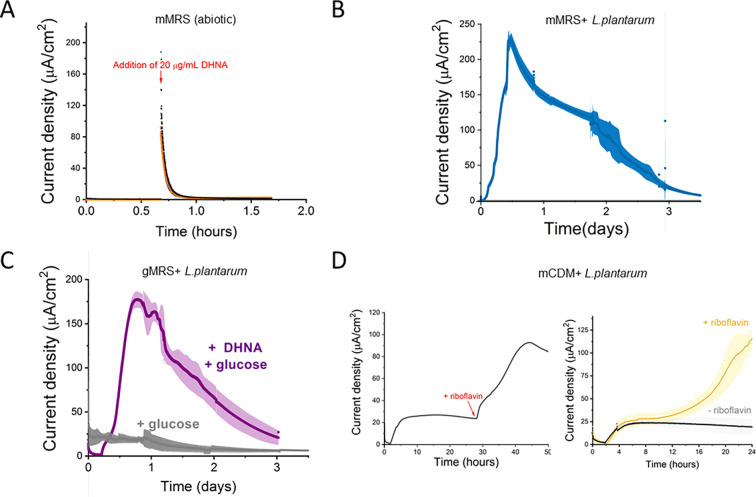
Figure 1—figure supplement 2. Current production by L. plantarum is a biotic process dependent on DHNA, carbon source, and riboflavin.
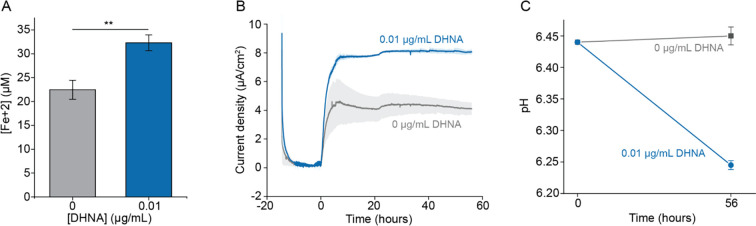
Figure 1—figure supplement 3. A sub-physiological concentration of DHNA stimulates EET in L. plantarum.
Next, we investigated whether the ability of L. plantarum to reduce insoluble iron was altered by growth media. L. plantarum was able to reduce iron after growth in either complete (MRS) medium or chemically defined medium (CDM) (Figure 1—figure supplement 1B). Iron reduction was greater when mannitol, a sugar alcohol, rather than glucose, was provided as the sole carbon source in MRS (Figure 1—figure supplement 1B). However, reduction was highest when L. plantarum was incubated in mannitol-containing MRS (mMRS) with both DHNA and ferric ammonium citrate present (Figure 1—figure supplement 1D). The addition of riboflavin to the growth medium did not further increase iron reduction by L. plantarum (Figure 1—figure supplement 1E), potentially because riboflavin is already present in high quantities in MRS, a medium containing yeast extract (Tomé, 2021). Thus, L. plantarum was grown in mMRS supplemented with DHNA and iron before ferrihydrite reduction assays in all subsequent experiments.
L. plantarum EET activity was confirmed in a bioelectrochemical reactor by quantifying electron output as current (Figure 1B). L. plantarum reduced a carbon electrode (anode) polarized to +200 mVAg/AgCl in the presence of both DHNA and an electron donor (mannitol) (Figure 1C). No current was observed in the absence of L. plantarum (Figure 1—figure supplement 2A), indicating that current production stems from a biological process. L. plantarum produced a maximum current of 129 ± 19 µA/cm2 in mCDM (Figure 1C) and 225 ± 9 µA/cm2 in mMRS (Figure 1—figure supplement 2B). Under EET conditions in mCDM, the L. plantarum biomass was 2.7 mg (dry cell mass). Assuming 50% of the dry cell mass was protein, the specific electron transfer rate was 57 µmol electrons/mg- protein/hr and the current production was 1.5 mA/mg-protein. This value is lower than that reported for Geobacter sulfurrenducens (4–8 mA/mg-protein) (Marsili et al., 2010; Rose and Regan, 2015), the model species for direct EET, and higher than that of Shewanella oneidensis (0.67 mA/mg-protein) (Marsili et al., 2008), the model species for mediated EET. It should be noted that these species, unlike L. plantarum, can synthesize riboflavin and quinones and do not require the addition of either for EET activity. Similar to our iron reduction experiments, EET to an anode occurred with different electron donors and growth media (Figure 1—figure supplement 2B-C), and current increased after supplementation of riboflavin when it was omitted from the growth medium (Figure 1—figure supplement 2D). Because of these differences, CDM was amended with mannitol and riboflavin in subsequent experiments.
DHNA is found in concentrations of 0.089–0.44 μg/mL in commercial fermented beverages (Eom et al., 2012), and under laboratory conditions, microbes can synthesize and secrete DHNA leading to concentrations of 0.37–48 μg/mL (Isawa et al., 2002; Furuichi et al., 2006; Kang et al., 2015). To test whether EET in L. plantarum is relevant under these physiological concentrations, we probed whether L. plantarum can perform EET with a sub-physiological DHNA concentration of 0.01 μg/mL. Indeed, L. plantarum can reduce iron and produce significant current density (Figure 1—figure supplement 3A-B), although the magnitude of iron reduction and current was smaller than what was observed with 20 μg/mL. These results show that the concentrations of DHNA found in niches of L. plantarum can support EET and suggest the magnitude of EET will depend on the DHNA concentration.
Iron reduction by LAB is associated with the presence of ndh2 and pplA
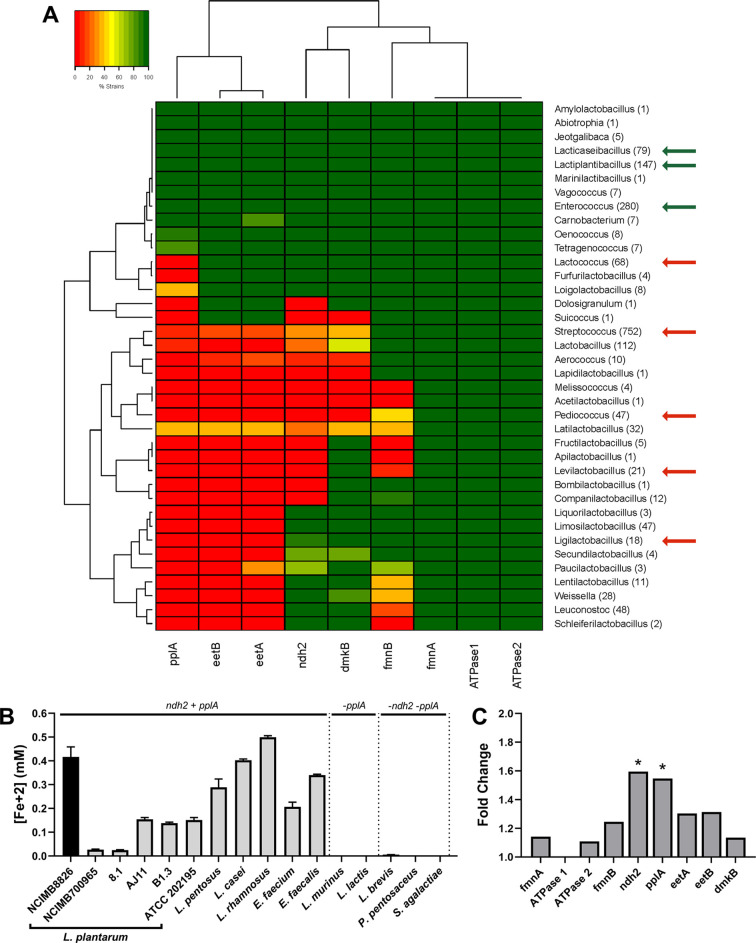
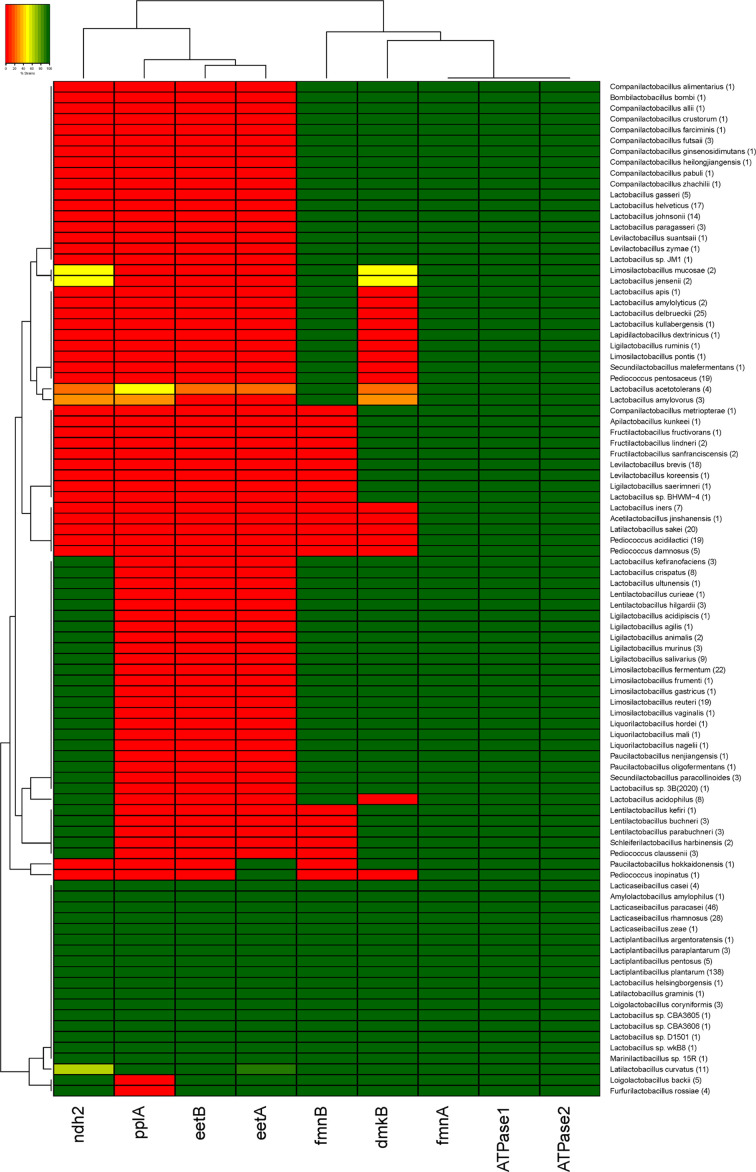
Because iron reduction by L. monocytogenes requires the genes in a 8.5 kb gene locus encoding a flavin-based EET (FLEET) pathway (Light et al., 2018), we looked for the presence of these genes in 1,788 LAB genomes deposited in NCBI. Homology searches identified the complete FLEET locus in 11 out of 38 genera including diverse LAB such as Enterococcus and Lacticaseibacillus (Figure 2A). The other LAB genera either lack multiple FLEET pathway genes or, as was observed for all 68 strains of Lactococcus, contain all genes except for pplA, which is predicted to encode an extracellular flavin-binding reductase. Among the lactobacilli, genomes of 19 out of 94 species contain the entire FLEET system (Figure 2—figure supplement 1). The lactobacilli species with the entire FLEET locus are homofermentative and are distributed between different phylogenetic groups (Zheng et al., 2020). These data show that the FLEET locus is conserved across LAB genera besides L. plantarum, including other homofermentative LAB species known to colonize host and food environments.
Figure 2. The FLEET genes ndh2 and pplA are associated with iron reduction by LAB.
(A) Heatmap showing the genera in the Lactobacillales order containing FLEET genes. Homology searches were conducted using tBLASTx for 1788 complete LAB genomes in NCBI (downloaded 02/25/2021) against the L. plantarum NCIMB8826 FLEET locus. A match was considered positive with a Bit-score >50 and an E-value of <10–3. Arrows designate genera tested for iron reduction activity; green = EET-active with Fe3+, red = EET-inactive with Fe3+. (B) Reduction of ferrihydrite in PBS with 20 μg/mL DHNA and 55 mM mannitol after growth in mMRS supplemented with 20 μg/mL DHNA and 1.25 mM ferric ammonium citrate. The avg ± stdev of three biological replicates per strain is shown. (C) Relative expression of NCIMB8826 FLEET locus genes in mMRS with 20 μg/mL DHNA and 1.25 mM ferric ammonium citrate compared to growth in mMRS. Significant differences in expression were determined by the Wald test (n = 3) with a Log2 (fold change) > 0.5 and an FDR-adjusted p-value of <0.05. See also Figure 2—figure supplement 1 and Figure 2—figure supplement 2 and related data in Figure 2—source data 1.
Figure 2—figure supplement 1. Conservation of FLEET locus genes among lactobacilli.
Figure 2—figure supplement 2. ndh2 and pplA are required for iron reduction through EET.
To determine whether LAB FLEET gene presence was associated with EET activity, a diverse collection of LAB strains were examined for their capacity to reduce ferrihydrite. The assay showed that isolates of L. plantarum, Lactiplantibacillus pentosus, Lacticaseibacillus rhamnosus, Lacticaseibacillus casei, Enterococcus faecium, and Enterococcus faecalis are capable of Fe3+ reduction (Figure 2B). The genomes of those species also contain a complete FLEET locus (Figure 2A and Figure 2—figure supplement 1). Conversely, strains of Lactococcus lactis, Ligilactobacillus murinus, Levilactobacillus brevis, Pediococcus pentosaceus, and Streptococcus agalactiae showed little to no iron reduction activity (Figure 2B). The presence of FLEET-associated genes varied between those species, but only strains of species found to contain both ndh2, a predicted membrane-bound, type-II NADH dehydrogenase, and pplA were able to reduce iron under the conditions tested.
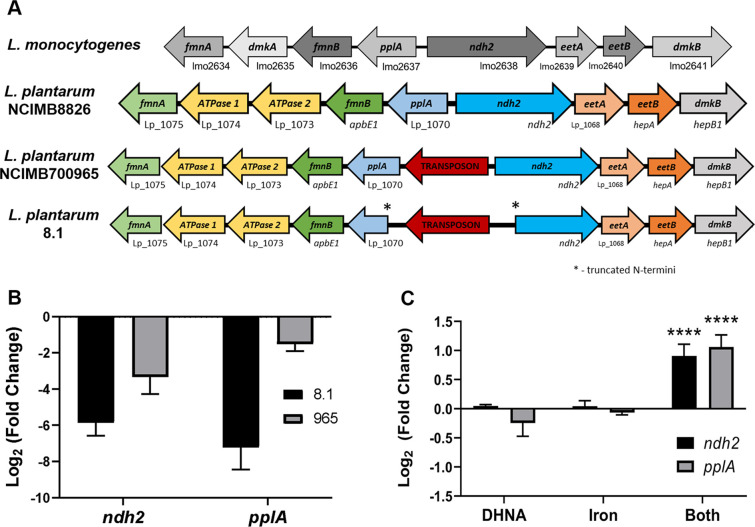
L. plantarum NCIMB8826 exhibited the highest EET activity resulting in at least 2.5-fold greater Fe3+ reduction than the other L. plantarum strains (Figure 2B). Remarkably, however, the L. plantarum NCIMB8826 genome and the genomes of 138 other L. plantarum strains queried all harbored a complete FLEET locus including ndh2 and pplA (Figure 2—figure supplement 1 and Figure 2—figure supplement 2A). Among those strains tested for the capacity to reduce Fe3+, L. plantarum NCIMB700965 and 8.1 could not reduce Fe3+ but possessed all genes in the FLEET pathway. Closer examination of both strains by aligning their FLEET loci with NCIMB8826 revealed unique IS30-family transposons in the intergenic promoter regions spanning ndh2 and pplA (Figure 2—figure supplement 2A). These genes were minimally expressed in L. plantarum NCIMB700965 and 8.1 in comparison to NCIMB8826 (Figure 2—figure supplement 2B). ndh2 and pplA were also the only two genes in the FLEET gene locus that were induced when L. plantarum NCIMB8826 was incubated in mMRS supplemented with DHNA and iron (Figure 2C and Figure 1—figure supplement 1D). Both ndh2 and pplA were induced (~1.6 fold, p < 0.05) in MRS containing mannitol, DHNA, and ferric ammonium citrate (Figure 2C), but were not upregulated when either DHNA or ferric ammonium citrate were omitted from the culture medium (Figure 2—figure supplement 2C). Taken together, these data show that widespread iron reduction in LAB is tightly associated with the presence and upregulation of ndh2 and pplA, suggesting they are required for EET.
Ndh2 is required and PplA is conditionally required for L. plantarum EET
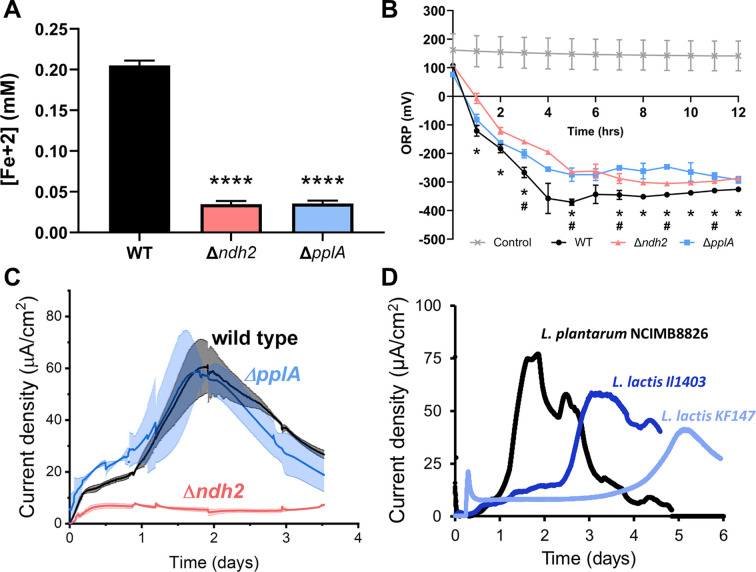
In order to confirm the necessity of ndh2 and pplA for EET in L. plantarum, we constructed ndh2 and ppA deletion mutants of L. plantarum NCIMB8826. Both mutants were significantly impaired in their capacities to reduce ferrihydrite compared with the wild-type strain (Figure 3A). The ndh2 and pplA deletion mutants also had different effects on the oxidation-reduction potential (ORP) of mMRS. ORP is defined as the ratio of all oxidative to reductive components in an environment (Killeen et al., 2018) and is an important environmental condition which influences the outcome of LAB fermentations such as flavor development in cheese (Morandi et al., 2016) and the growth of spoilage microorganisms (Olsen and Pérez-Díaz, 2009). Expectedly for the L. plantarum EET phenotype, significant reductions in mMRS ORP only occurred during L. plantarum growth when DHNA was included in the culture medium (Figure 3—figure supplement 1A). Although ORP declined for all three strains in a manner consistent with other ORP-reducing enzymatic activities (for example the reduction of oxygen by NADH oxidase) (Tachon et al., 2010), wild-type L. plantarum resulted in greater reductions in ORP compared to either mutant in mMRS, and these differences were significant at most time points measured over a 12 hr period (Figure 3B). The effects on ORP occurred in the absence of changes in growth rates, cell yields, and medium pH (Figure 3—figure supplement 1A-D). The ΔmVmax was reached during mid-exponential phase (approximately 5 hr) (Figure 3—figure supplement 1B), and at that time, wild-type L. plantarum cells but not the Δndh2 or ΔpplA strains were active in the ferrihydrite reduction assay (Figure 3—figure supplement 1E). This difference in ferrihydrite reduction activity similarly persisted in stationary phase cells (12 hr) (Figure 3—figure supplement 1F). These observations show that ndh2 and pplA contribute to the capacity of L. plantarum to reduce iron and have relevance to the ORP-dependent activities occurring during food fermentations (van Dijk et al., 2000).
Figure 3. L. plantarum requires ndh2 and conditionally pplA for EET.
(A) Reduction of Fe3+ (ferrihydrite) to Fe2+ with wild-type L. plantarum or EET deletion mutants in the presence of 20 μg/mL DHNA and 55 mM mannitol after growth in mMRS supplemented with 20 μg/mL DHNA and 1.25 mM ferric ammonium citrate. Significant differences determined by one-way ANOVA with Dunnett’s post-hoc test, **** p < 0.0001. (B) Redox potential of mMRS supplemented with 20 μg/mL DHNA and 1.25 mM ferric ammonium citrate after inoculation with wild-type L. plantarum or EET deletion mutants. Significant ORP differences between the wild-type and mutant strains determined by two-way RM ANOVA with Tukey’s post-hoc test, * p < 0.05 (WT vs. Δndh2); # p < 0.05 (WT vs. ΔpplA). (C) Current density generated by wild-type L. plantarum and deletion mutants in mCDM supplemented with 20 μg/mL DHNA. The avg ± stdev is shown. (D) Current density generated by L. plantarum and two L. lactis strains lacking pplA in mCDM. For L. plantarum, the mCDM was supplemented with 20 μg/mL DHNA. The data correspond to the average of two (D) or three (A to C) biological replicates per strain. See also Figure 3—figure supplement 1 and Figure 3—figure supplement 2 and related data in Figure 3—source data 1.
Figure 3—figure supplement 1. Impact of ndh2 and pplA deletion on growth, iron reduction, current density, and metabolites production.
Figure 3—figure supplement 2. Impact of ndh2 and pplA deletion on maximum current density, pH and metabolites production.
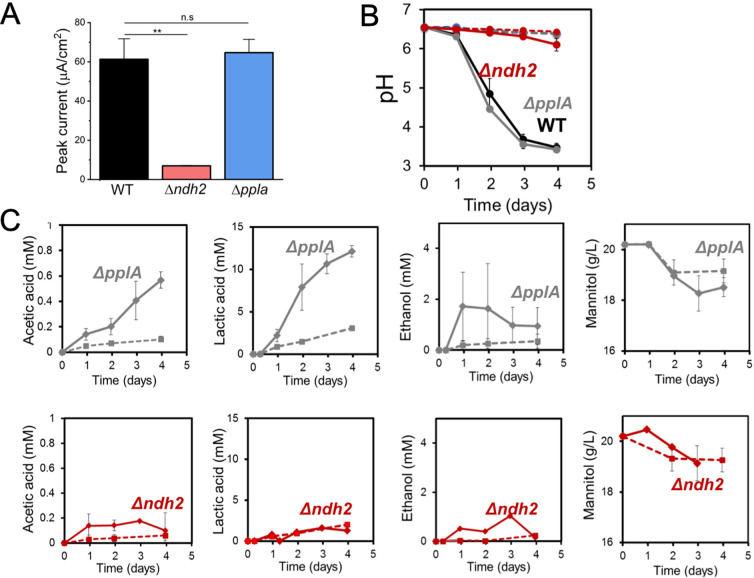
Use of an anode as an external electron acceptor instead of ferrihydrite showed a similar, but not identical genetic dependency. L. plantarum Δndh2 produced a significantly lower current density (Figure 3C) and a lower peak current (Figure 3—figure supplement 2A). Surprisingly, L. plantarum ΔpplA was able to produce the same amount of current as the wild-type strain, suggesting that the lipoprotein PplA is not essential and might not be involved in anode reduction through EET. This observation led us to investigate the anodic-EET ability of other LAB species lacking pplA like Lactococcus lactis (Figure 3D). DHNA was not provided to these strains because they can synthesize demethylmenaquinone and other quinones (Rezaïki et al., 2008). Both L. lactis strain IL1403 and strain KF147 were capable of current generation, confirming that PplA is not essential for LAB to produce current. This is consistent with the finding that other extracellular reductases besides PplA are responsible for EET activity in Gram-positive bacteria (Light et al., 2019). Taken together these results show that EET activity is dependent upon the presence of the putative FLEET locus, and specifically ndh2 and conditionally pplA.
L. plantarum increases energy conservation and balances intracellular redox state when performing EET
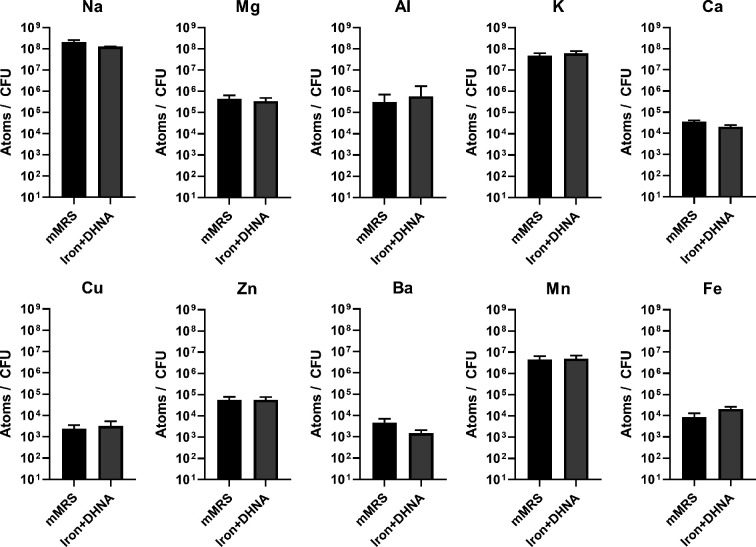
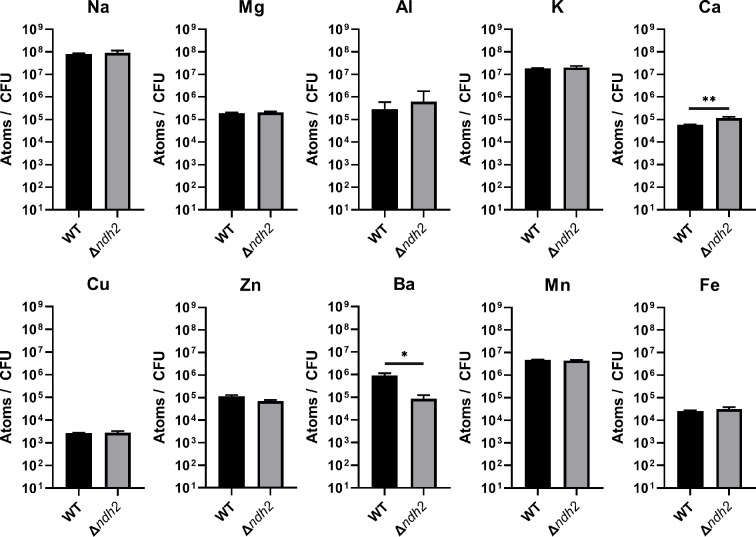
Building from studies in E. faecalis (Keogh et al., 2018), it has been suggested that EET improves growth by either enabling iron to be acquired as a macronutrient or by enhancing respiration (Jeuken et al., 2020). It is worth noting that several studies have shown that L. plantarum does not require iron to grow (Elli et al., 2000; Weinberg, 1997). To test whether EET allowed increased iron acquisition by L. plantarum, we measured intracellular iron by Inductively Coupled Plasma-Mass Spectrometry (ICP-MS). There was no significant difference in intracellular iron concentrations between L. plantarum growth in mMRS supplemented with DHNA and iron compared to growth in mMRS alone (Figure 4—figure supplement 1). Moreover, deletion of ndh2 did not significantly change the amount of intracellular iron (Figure 4—figure supplement 1). ICP-MS showed that other redox-active metals used for EET, such as manganese and copper (Kouzuma et al., 2012; Fan et al., 2018) were also not affected (Figure 4—figure supplements 1 and 2). In contrast to studies in E. faecalis in which iron supplementation leads to intracellular accumulation of this metal (Keogh et al., 2018), these data show that L. plantarum does not use EET to increase its acquisition of iron or other redox-active metals, suggesting EET may instead play a role in energy conservation.
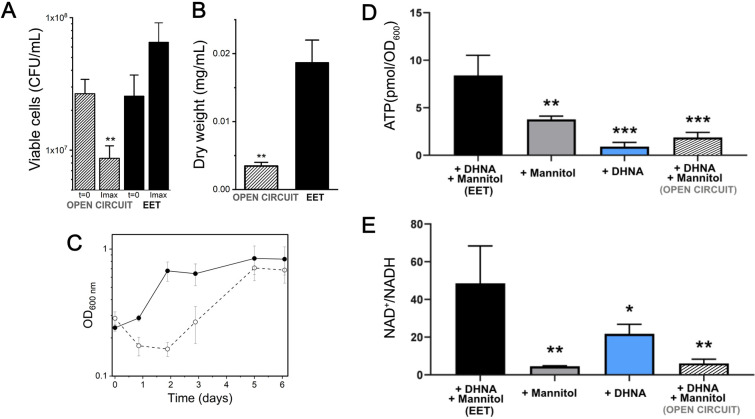
We next sought to understand if EET impacts energy conservation in L. plantarum by comparing its growth and ATP levels in the presence of a polarized anode. The highest current density (i.e. greatest EET activity) produced by L. plantarum in mannitol CDM typically occurred within 24 hr after inoculation into the bioreactor (Figure 1C). At this point, there was an approximately 4-fold higher dry cell weight and 2-fold higher numbers of viable cells compared to L. plantarum incubated in open circuit (OC) conditions (Figure 4A–B) Current density declined from its maximum value when L. plantarum cells performing EET were in exponential growth (Figures 1C and 4C). By comparison, growth was not observed until two days later under OC conditions (Figure 4C). During peak current production, intracellular ATP levels were significantly higher (4.5-fold) under EET compared to OC conditions (Figure 4D and Table 1). These results strongly suggest faster ATP accumulation under EET conditions allowed L. plantarum to exit lag phase more rapidly. ATP levels were also greater in L. plantarum when in the presence of both mannitol and DHNA, compared to either mannitol or DHNA separately (Figure 4D). Thus, EET allows L. plantarum to initiate growth and accumulate ATP more rapidly, indicating that EET significantly increases energy conservation in L. plantarum.
Figure 4. Growth, ATP, and redox balance of L. plantarum changes when an anode is provided as an extracellular electron acceptor.
These measurements and the current density plot shown in Figure 1C are from the same experiment. (A) Viable cells and (B) dry weight at the point of maximum current density under current circulating conditions (EET) and at open circuit conditions (OC) at the same time point. (C) Change in cell numbers measured by OD600 over time in the bioreactors under EET (continuous line) and OC conditions (dotted lines). (D) ATP production per OD600 unit and (E) NAD+/NADH ratios at the point of maximum current density. The bioreactors were shaken vigorously to dislodge cells before sampling. The avg ± stdev of three biological replicates is shown. Significant differences were determined by one-way ANOVA with (A and B) Dunn-Sidak post-hoc test (n = 3) and (D and E) Dunnett’s post-hoc test (n = 3), * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001. See also Figure 1 panel C and Figure 4—figure supplement 3 and related data in Figure 4—source data 1.
Figure 4—figure supplement 1. Intracellular metal concentrations in L. plantarum are not affected by EET-conducive growth conditions.
Figure 4—figure supplement 2. Redox-active metal concentrations in L. plantarum are not affected by the presence of ndh2.
Figure 4—figure supplement 3. Use of Fe3+ as an electron acceptor allows L. plantarum to regenerate NAD+.
Table 1. Bioenergetic balances suggest energy conservation under EET conditions occurs via substrate-level phosphorylation.
The reactors contained 20 µg/mL of DHNA and mannitol as the electron donor. Balances were calculated with data obtained by day four from Figure 5. See also Supplementary file 1. SLP stands for substrate-level phosphorylation. Yfermentation refers to the total fermentation products obtained (see Supplementary file 1) per mol of sugar consumed. Ymannitol is the ATP produced from the total fermentation products per mol of sugar consumed, and YATP is the dry weight measured per mol of ATP produced from fermentation products.
| NADH consumed* | Calculated ATP†(from metabolites) | Biomass yield | Yfermentation | Ymannitol | YATP | |
|---|---|---|---|---|---|---|
| Units | mM | mM | g-dw/mol-mannitol | mmol product/mmol-mannitol | mol ATP/mol mannitol | g dw/mol ATP |
| EET | 6.44 ± 0.48 via anode16.69 ± 2.72 via SLP | 16.6 ± 1.5 | 4.85 ± 0.33 | 1.53 ± 0.13 | 1.59 ± 0.13 | 3.09 ± 0.36 |
| OC | 5.51 ± 0.97 via SLP | 5.7 ± 0.6 | 7.21 ± 1.41 | 0.87 ± 0.09 | 0.89 ± 0.09 | 8.06 ± 0.86 |
Calculated based on production of 3 mol of NADH produced per mol of mannitol, 1 mol of NAD+ per lactate, 2 mol of NAD+ per ethanol, 2 mol of NAD+ mol per succinate produced and 0.5 mmol of NAD+ per mol of electrons harvested on the anode.
Calculated based on production of 1 mol of ATP per lactate, 2 mol per acetate, 1 mol per ethanol, and 3 mol per succinate produced.
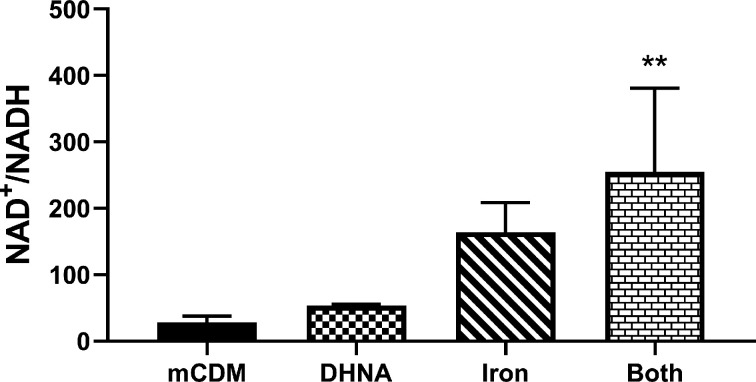
Because fermentation, anaerobic respiration, and aerobic respiration are each associated with a different NAD+/NADH ratio, energy conservation is linked to intracellular redox homeostasis (Holm et al., 2010). Therefore, we probed redox homeostasis in L. plantarum under EET conditions by measuring intracellular NAD+/NADH at the point of maximum current density (Figure 4E). L. plantarum showed an 8-fold higher NAD+/NADH ratio under EET conditions compared to OC (Figure 4E). This result was not limited to the presence of a polarized anode as L. plantarum also contained a significantly higher NAD+/NADH ratio when Fe3+ was available as a terminal electron acceptor (Figure 4—figure supplement 3). These NAD+/NADH ratios are more similar to those found for in E. coli performing aerobic respiration (de Graef et al., 1999) or G. sulfurreducens performing anaerobic respiration than in LAB performing fermentation (Guo et al., 2017). Taken together, our data indicate that EET is involved in energy conservation, and the intracellular redox balance during EET mimics a respiratory rather than a fermentative process.
EET increases fermentative metabolism through substrate-level phosphorylation and reduction in extracellular pH
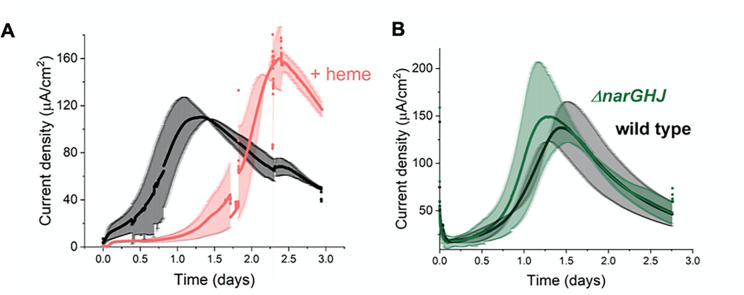
Metal-reducing bacteria use EET in anaerobic respiration (Richter et al., 2012; Shi et al., 2007). Ndh2 is considered an anaerobic respiratory protein, and L. plantarum can perform anaerobic respiration with exogenous menaquinone and heme using an electron transport chain (Brooijmans et al., 2009b). This led us to hypothesize that those electron transport proteins could also be involved for EET to conserve energy as part of anaerobic respiration. To test this hypothesis, we examined whether any of the known electron transfer proteins needed for PMF generation in aerobic and anaerobic respiration are required for L. plantarum EET. Neither the addition of heme to restore bd-type cytochrome (cydABCD) used in aerobic respiration, nor deletion of the respiratory nitrate reductase (ΔnarGHJI) significantly altered current production (Figure 5—figure supplement 1A-B). Because Ndh2 is a type-II NADH dehydrogenase which does not contribute to a proton gradient (Lin et al., 2008; Nakatani et al., 2020), these observations show that while EET does involve a respiratory protein, it does not involve any of the known PMF-generating electron transfer proteins in L. plantarum.
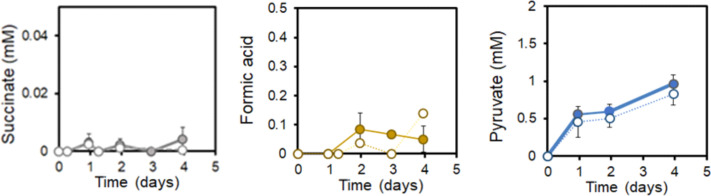
Respiration is also associated with the tricarboxylic acid (TCA) cycle. L. plantarum, like other LAB, does not possess an oxidative branch of the TCA cycle and only contains a reductive branch (Tsuji et al., 2013). To probe whether the reductive branch was active during EET, we also examined production of succinate, the terminal end-product of the reductive branch. EET did not increase the succinate concentration (Figure 5—figure supplement 2). Moreover, we did not detect any intermediates of the reductive branch of the TCA cycle, that is oxalacetate, malate, or fumarate. This indicates that EET did not cause additional metabolic flux through its TCA cycle. Thus, none of the known metabolic pathways or electron transport proteins associated with anaerobic respiration, besides Ndh2, are required for EET. These results suggest increased energy conservation during EET in L. plantarum is not through canonical anaerobic respiration.
An alternative hypothesis is that increased energy conservation under EET conditions is driven by changes in fermentation. L. plantarum uses glycolysis to convert mannitol to two molecules of pyruvate which are then converted mainly to lactate or ethanol via NADH-consuming steps, or acetate via an ATP-generating reaction using substrate-level phosphorylation (Dirar and Collins, 1972). Thus, shifting toward production of acetate from to lactate or ethanol production can increase ATP yield during fermentation. Additionally, NADH can be re-generated by oxidizing pyruvate to yield 2,3-butanediol, using acetoin as an intermediate. Fermentation in L. plantarum also decreases the pH of the surrounding media.
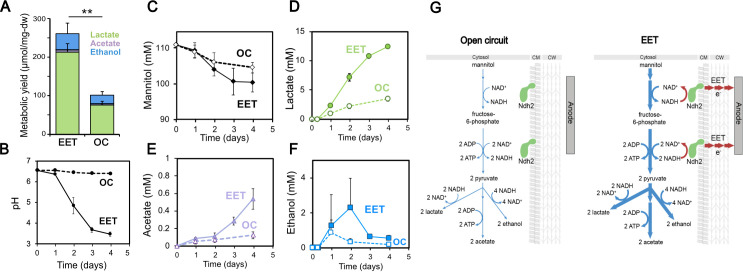
To probe changes in fermentation, we measured the concentrations of mannitol, acetate, lactate, ethanol, acetoin, 2,3-butanediol, formate, and pyruvate and the pH in L. plantarum cultures during OC and EET conditions. After four days, we accounted for ~80% and ~ 55% of the total carbon under EET and OC conditions (for all metabolite concentrations see Supplementary file 1), giving us a quantitative view of metabolism under EET conditions. Surprisingly, under EET conditions, the distribution of major end-fermentation products (acetate, lactate, and ethanol) did not change, but their yield per cell was 2.6-fold higher compared to OC conditions (Figure 5A). While we did not detect acetoin or 2,3-butanediol, formate was found at trace levels, and pyruvate was found at similar, low levels under EET and OC conditions (Figure 5—figure supplement 2). After accounting for mannitol consumption, we observed that EET allowed cells to produce ~1.75 x more fermentation products per each mol of mannitol utilized (Yfermentation, Table 1). The culture medium pH was also significantly lower than under OC (Figure 5B), a result which may indicate that EET conferred higher levels of acid stress on L. plantarum, and therefore, reductions in cell viability, despite EET leading to higher cell numbers overall (as measured by dry cell weight) (Figure 4A–B). A similar acidification of the medium was observed for ΔpplA, but not for Δndh2, when an anode was present as electron acceptor, indicating that ndh2-dependent EET is needed to decrease the pH (Figure 3—figure supplement 2B). When much lower, sub-physiological levels of DHNA were supplied (0.01 μg/mL), a smaller but significant decrease in the pH of the medium was also observed (Figure 1—figure supplement 3C). Overall, these results show that EET allows L. plantarum to ferment to ~1.75 x greater extent and to acidify the medium to a greater extent as well.
Figure 5. Fermentation fluxes are increased when an anode is provided as an extracellular electron acceptor.
Results are from the same set of experiments as the current density plot shown in Figure 3C. (A) Metabolic yields of L. plantarum end-fermentation products under open circuit conditions (OC) and current circulating conditions (EET) in mCDM supplemented with 20 μg/mL DHNA. (B) pH measurements and (C) mannitol, (D) lactate, (E) acetate, and (F) ethanol concentrations over time under OC and EET conditions. (G) Schematic of proposed model for NADH regeneration during fermentation of mannitol in the presence of an anode as electron sink for L. plantarum. The avg ± stdev of three biological replicates is shown. Significant differences were determined by one-way ANOVA with Dunn-Sidak post-hoc (n = 3), ** p < 0.01. See also Figure 5—figure supplement 1 and Figure 5—figure supplement 2 and related data in Figure 5—source data 1.
Figure 5—figure supplement 1. EET by L. plantarum is not dependent on aerobic or anaerobic respiration components.
Figure 5—figure supplement 2. EET by L. plantarum does not involve TCA cycle metabolites.
We also observed that EET led to higher cellular metabolic fluxes, that is, higher changes in metabolites per cell per unit time. Although the final OD600nm and dry cell weight were not significantly different (Supplementary file 1), L. plantarum utilized mannitol and produced acetate and lactate more rapidly under EET than OC conditions (Figure 5C–F). Cells performing EET were ~2 fold faster at consuming mannitol (Figure 5C) between days 1 and 3. Mannitol consumption increased between day 1 and day 2, approximately when the cells transitioned to higher current density (Figure 3C), suggesting that increased EET drove that increased consumption. The overall rates of acetate and lactate production also increased 3.4 and 3.6 times (Figure 5D–E), respectively. Measurements of metabolites produced by ΔpplA and Δndh2 strains confirmed that, like for current production to an anode, the EET-associated increased metabolic flux in L. plantarum requires the presence of Ndh2, but not PplA (Figure 3—figure supplement 2C). Overall, these data indicate that ndh2-dependent EET increases both the flux and final yield of fermentation in L. plantarum.
Because the production of acetate yields ATP, these results also suggested that the increase in ATP generation under EET conditions may be due to substrate-level phosphorylation. To probe whether EET-associated increase in fermentative flux could account for the changes in ATP generation, we calculated fermentation balances (Table 1). Our measurements account for 80% of the carbon under EET conditions (see Supplementary file 1), leaving a maximum of ~20% systematic uncertainty in these calculations. The concentrations of fermentation products detected (Supplementary file 1) were used to estimate the total ATP in the presence and absence of EET. The estimated ATP was 3-fold higher under EET conditions than OC conditions (Table 1), a result that is consistent with the ~2.5 fold higher accumulation of ATP measured at maximum current density (Figure 4D). Overall, this quantitative analysis shows that the vast majority of the increased energy conservation under EET conditions can be accounted for by an increase in fermentation yield and substrate-level phosphorylation.
EET shifts how L. plantarum uses electron acceptors and converts ATP into biomass
Thus far, our results provided an unusual picture of the energy metabolism of L. plantarum under EET conditions; while EET significantly shifted the intracellular redox state to a more respiratory-like balance, its increased ATP yield was mainly accounted for by an increased fermentative yield. Another major difference in fermentation and anaerobic respiration is the use of the endogenous versus exogenous electron acceptors. To more deeply understand how L. plantarum uses organic molecules and the anode as electron acceptors when performing EET, the electron balances under EET and OC conditions were calculated (Table 1 and Supplementary file 1). We estimated the NADH produced using two different methods (see Supplementary file 1 for methodology) and the NADH re-oxidized through the reduction of the anode (measured as current) and via substrate-level phosphorylation. This allowed us to obtain a global balance of the NAD+/NADH ratio. Under OC conditions between 35% and 66% of the NADH produced from the oxidation of mannitol to pyruvate (a range is given using the two methods used) was re-oxidized to NAD+ (5.5 mM NADH consumed, Table 1), qualitatively agreeing with the low NAD+/NADH ratios measured (Figure 4E). In contrast, electron balance calculations showed that between 77% and 96% of the NADH produced under EET conditions was re-oxidized (17 mM NADH consumed, Table 1), a result that is consistent with the significantly higher NAD+/NADH ratios measured (Figure 4E). Interestingly, these calculations estimate that 55–69% of the total NADH generated was oxidized through fermentation, while 21–28% of the NADH was oxidized using the electrode as a terminal electron acceptor (Table 1). Thus, L. plantarum growing under EET conditions achieves a more oxidized intracellular redox balance by more completely fermenting mannitol to lactate and ethanol and by using the electrode as a terminal electron acceptor (Figure 5G). These observations reinforce that the energy metabolism of L. plantarum under EET conditions utilizes elements of both fermentation and anaerobic respiration.
In rapidly dividing cells, energy conservation, a catabolic process, is associated with growth, an anabolic process. However, catabolism need not be coupled with anabolism (Russell and Cook, 1995). To determine how catabolic and anabolism are connected under EET conditions, the ATP requirements to grow biomass (YATP) were estimated using the calculated ATP and the measured dry weight. Under OC conditions, the YATP obtained (8.06 ± 0.8 g dw/mol ATP) for L. plantarum was similar to that observed previously (10.9 g-dw/mol ATP) (Dirar and Collins, 1972). Hence, without EET, the ATP generated from fermentation was converted into biomass nearly at the expected efficiency. In contrast, a significantly lower YATP was reached for L. plantarum performing EET (3.07 ± 0.35 g dw/mol ATP) (Table 1). This observation indicates that under EET conditions, either more ATP is required to produce biomass or more ATP is utilized by other functions such as for PMF-generation and intracellular pH regulation (Russell and Cook, 1995). EET conditions also resulted in 79% more ATP per mol of fermented mannitol (Ymannitol). Consequently, molar biomass yields (g-dw/mol-mannitol) under EET conditions were significantly lower (Table 1), in agreement with previous observations in respiratory electroactive species (Esteve-Núñez et al., 2005). These calculations show that when L. plantarum performs EET, anabolism and catabolism processes are differently coupled than under OC conditions. ATP is produced more efficiently, but this it is less efficiently utilized to make biomass. Overall, these results show an intriguing pattern of coupling between anabolism and catabolism, indicative of a novel energy metabolism in L. plantarum during EET.
EET is active in vegetable fermentations
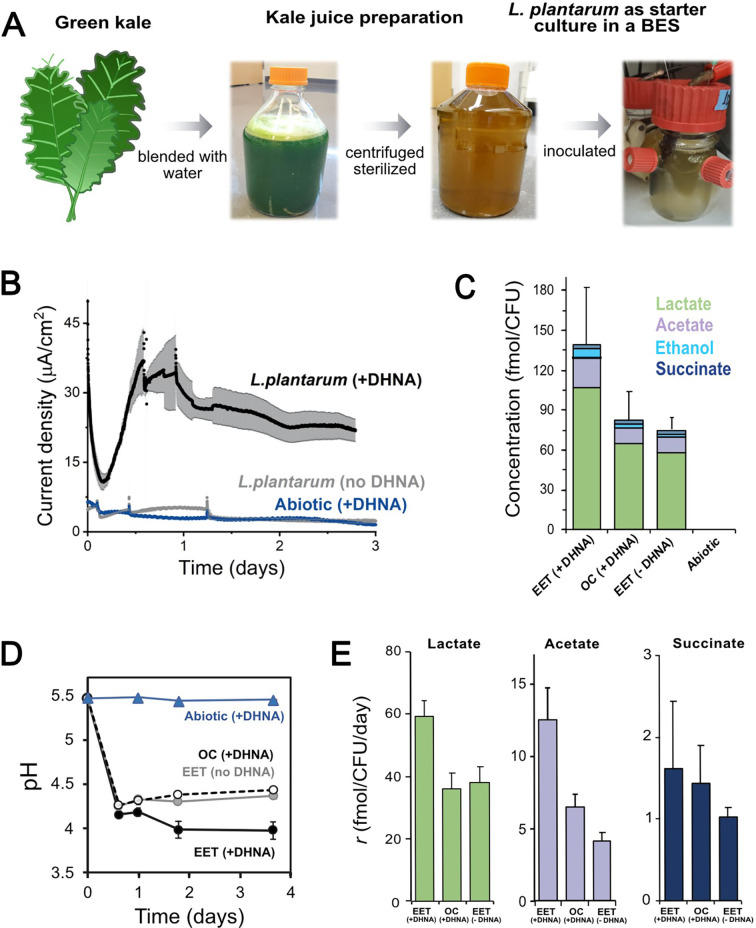
Our results inspired us to explore whether EET could occur in a physiological niche of LAB such as fermented foods. LAB are necessary for the making of many fermented fruit and vegetable foods and the properties of those foods depend on the metabolic diversity of the LAB strains present (Gänzle, 2015). Plant tissues also contain a much wider variety of carbon substrates and potential electron acceptors than the CDM used in our prior experiments. To study the physiological and biotechnological relevance of EET in food fermentations, kale juice was fermented using L. plantarum as a starter culture (Figure 6A). The fermentation of kale juice was measured under EET conditions (a polarized anode with or without DHNA), and an OC control (a nonpolarized anode with DHNA) was used to separate the role of DHNA and electron flow to the anode on the fermentation process. An additional bioreactor without cells, but with DHNA, was operated to identify any possible electrochemical-driven conversion of substrates. When L. plantarum was added to the prepared kale juice, approximately 10-fold more current was generated during EET conditions with DHNA (EET+ DHNA) as compared to abiotic and biological non-EET promoting conditions (no DHNA) (Figure 6B). This current was comparable to the current generated in laboratory medium (Figure 3C), indicating that robust EET by LAB is possible in the complex physiological conditions of a food fermentation.
Figure 6. EET in a kale juice increases the production of fermentation end products.
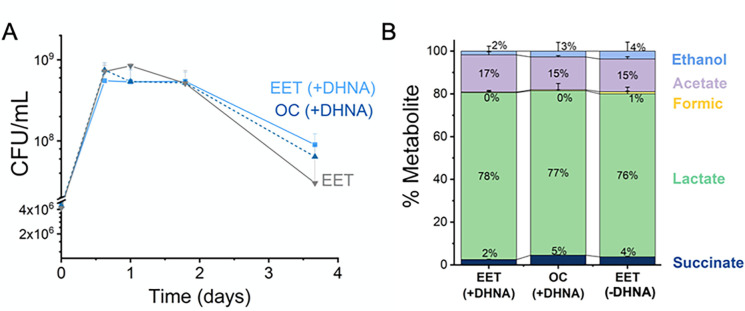
(A) Preparation of kale juice medium used for fermentation in bioelectrochemical reactors. (B) Current density production measured from kale juice medium over time in the presence of L. plantarum and 20 μg/mL DHNA, no DHNA, or under abiotic conditions with addition of 20 μg/mL DHNA. The anode polarization was maintained at 0.2 VAg/AgCl. (C) Normalized total quantities of the metabolites detected per cell (CFUmax used for calculations). (D) pH measurements over time under different conditions tested on a second set of kale juice fermentations performed under the same conditions. (E) Production rate per viable cell, r, of lactate, acetate, and succinate. The avg ± stdev of three biological replicates is shown. See also Figure 6—figure supplement 1 and related data in Figure 6—source data 1.
Figure 6—figure supplement 1. EET does not impact cell viability and distribution of metabolites in a kale fermentation.
We next investigated the impact of EET on L. plantarum growth and metabolism in the kale juice fermentation. Significant changes in the pH and fermentation products were detected under EET conditions (Figure 6C–D). These differences occurred in the absence of significant changes in viable cell numbers (Figure 6—figure supplement 1A) at the time points measured. As previously observed using laboratory culture media, an approximately 2-fold greater accumulation of total end-fermentation products per cell was obtained when cells interacted with an anode in the presence of DHNA (Figure 6C). In the kale juice fermentation, EET+ DHNA conditions enhanced both lactate and acetate production per cell without changing the distribution of metabolites (Figure 6E and Figure 6—figure supplement 1B). Thus, when DHNA was provided, EET enhanced the overall yield of fermentation end-products and their production rates per cell, mimicking our observations in laboratory medium (Figure 5C–D). EET also led to a significantly higher acidification of the kale juice compared to OC conditions, and the presence of DHNA dramatically enhanced this pH drop (Figure 6D). In general, when no DHNA was supplied but an anode was present as an electron acceptor, the fermentation process was very similar to OC conditions. This means in kale juice, a source of quinones is essential to support L. plantarum EET activity. Overall, these results show that EET under physiological conditions impacts cellular metabolism in L. plantarum by increasing metabolic flux which ultimately can affect the flavor profile of fermented foods (Chen et al., 2017).
Discussion
Increases in fermentation and energy conservation from EET have important bioenergetic implications for the mainly fermentative LAB. We showed that L. plantarum and other diverse LAB species perform EET if riboflavin and quinones are present. L. plantarum EET activity requires an NADH dehydrogenase (Ndh2) and conditionally requires an extracellular, flavin-binding reductase (PplA). EET in L. plantarum generates a high NAD+/NADH ratio, increases fermentation yield and flux, shortens lag phase, and increases ATP production. Thus, EET in L. plantarum is a hybrid energy metabolism that contains metabolic features of fermentation, redox features of anaerobic respiration, and predominately uses substrate-level phosphorylation to conserve energy. This pathway is active in L. plantarum with physiologically relevant DHNA concentrations and in a food fermentation and results in an increased metabolic flux and acidification rate.
The combined EET fermentation hybrid metabolism is distinct from anaerobic respiration, fermentation, and other energy conservation strategies
When performing EET, the metabolism of L. plantarum, a primarily fermentative bacterial species, is fundamentally different from EET-driven, anaerobic respiration of metal-reducing bacteria. Although aspects of EET in L. plantarum and metal-reducing Geobacter spp. are similar, such as the upregulation of NADH dehydrogenase, the reduction rate of extracellular electron acceptors, and the high NAD+/NADH ratio, other aspects of energy metabolism during EET in these two organisms are starkly different (see comparison in Supplementary file 2). Geobacter spp. direct their metabolic flux through the TCA cycle, rely almost exclusively on extracellular electron acceptors to regenerate NADH, and produce ATP exclusively through oxidative phosphorylation. In contrast, L. plantarum regenerates a substantial fraction of its NADH by directing metabolic flux through fermentative pathways. Additionally, oxidative phosphorylation is not a major mechanism of energy conservation in L. plantarum during EET, as supported by three lines of evidence: the marginal metabolic flux through the reduced branch of TCA cycle, no involvement of known PMF-generating proteins, and that increased ATP levels can be accounted for by increased substrate-level phosphorylation. While additional data are required to eliminate the possibility that oxidative phosphorylation is occurring in L. plantarum during EET, we can qualitatively state that substrate-level phosphorylation is the major mechanism for ATP generation.
Comparing EET and respiratory metabolism in LAB also reveals substantial differences in these metabolisms (Supplementary file 2). While both metabolisms require quinones, respiration also requires exogenous heme. Our findings and similar findings in E. faecalis (Pankratova et al., 2018) confirm that heme is not required for EET. Moreover, EET also differs from respiration in LAB because it occurs at the start of or prior to exponential phase growth, does not change the final cell density, and increases fermentation with no effect on the resultant proportions of lactate, acetate, and ethanol (Duwat et al., 2001). Thus, EET in LAB diverges from respiration in metal-reducing bacteria or LAB in its metabolic pattern and energetic consequences. In addition, while EET provokes a shift in fermentative metabolism in other bacteria upon the addition of artificial mediators (Vassilev et al., 2021; Emde and Schink, 1990), L. plantarum EET is active upon the presence of a mediator present in a complex food system.
This EET mechanism is also a novel energy conservation strategy compared to known fermentative metabolisms in LAB (comparison in Supplementary file 2). L. plantarum and other LAB, reduce alternative intracellular electron acceptors like citrate, fructose, and phenolic acids, resulting in increased intracellular NAD+/NADH ratios (Hansen, 2018). This metabolic activity is especially important for heterofermentative LAB in order to synthesize additional ATP through acetate kinase (Gänzle, 2015). Unlike these examples, however, the reduction of extracellular Fe3+ or an anode by EET requires a respiratory protein (Ndh2) and the shuttling of electrons outside of the cell. In addition, the reduction of the oxygen and organic compounds for cofactor regeneration by LAB leads to a metabolic shift toward acetate production and altered metabolic end-product ratios (Gänzle, 2015), which does not occur during EET. These differences show how the hybrid metabolism under EET conditions is distinct from other pathways that alleviate reduced intracellular conditions in LAB.
Previous studies have reported a simultaneous use of fermentation and electron transport elements, such as in respiro-fermentation in Saccharomyces cerevisiae (Blom et al., 2000). However, respiro-fermentation produces ATP and maintains intracellular redox balance through substrate-level phosphorylation and/or oxidative phosphorylation using separate pathways. Our data strongly suggests a single pathway is responsible for both ATP generation and intracellular redox balance. This hybrid fermentation mode is also different from the electron bifurcating mechanism, in which the extra ATP generation is driven by the creation of a H+ or Na+ potential from the oxidation of a ferredoxin (Buckel and Thauer, 2013). Unlike in this example, EET in LAB does not involve PMF creating elements and EET drives ATP generation through substrate-level phosphorylation. Another poorly understood example of the use of substrate-level phosphorylation and electron transport chains to balance intracellular redox state is found in the non-fermentative bacterium S. oneidensis. Although this species is a respiratory bacterium, it relies predominately on substrate-level phosphorylation to grow anaerobically with the exogenous electron acceptor fumarate (Hunt et al., 2010). In this scenario, it is unclear if there are changes in intracellular redox state or metabolism in this species.
In contrast to and expanding upon these studies, our work elucidates a qualitatively and quantitatively different blending of fermentation and respiration. L. plantarum EET-associated metabolism contains features of both fermentation (e.g. substrate-level phosphorylation, high fermentation product yields) and respiratory metabolisms (e.g. NAD+/NADH ratios, NADH dehydrogenase required) (Supplementary file 2). Quantitatively, this hybrid metabolism leads to an overall ~1.75x-more efficient and ~1.75x-faster energy conservation (increased Ymannitol, mannitol flux), but an overall ~1.5 fold weaker coupling between anabolism and catabolism (lower YATP). Additionally, the increased NAD+/NADH ratio arises from using an ~2:1 ratio of endogenous to extracellular electron acceptors. Thus, to our knowledge, the hybrid strategy that L. plantarum uses to generate ATP performing EET constitutes a novel mode of energy conservation in a primarily fermentative microorganism.
Different mechanisms of EET appear to be widespread in LAB
Based on our observations and others, we propose that EET is widespread in LAB and occurs by different mechanisms. Besides L. plantarum, we showed that L. lactis is able to generate current despite lacking pplA. Current generation by L. lactis was observed previously, found to be riboflavin dependent, and resulted in a small metabolic shift (yet to be defined) in which the flux through NADH-oxidizing pathways was reduced and ATP generating pathways were increased (Freguia et al., 2009; Masuda et al., 2010). L. lactis can also perform EET by reduction of tetrazolium violet and this activity depends on the presence of both quinones and an NADH dehydrogenase (NoxAB) (Tachon et al., 2009). E. faecalis is another LAB that performs EET, and similar to L. plantarum, it requires quinones (Pankratova et al., 2018) and a type-II NADH dehydrogenase (Ndh3) (Hederstedt et al., 2020) for Fe3+ reduction. In contrast to this mechanistic similarity, E. faecalis performs EET using matrix-associated iron resulting in both increased final cell biomass and intracellular iron (Keogh et al., 2018). Moreover, unlike L. plantarum and L. lactis, the presence of PplA is not necessary for anode reduction or Fe3+ reduction (Hederstedt et al., 2020). The conditional need for PplA in EET may be explained by the different prior growth conditions used and/or related to the existence of different mechanisms and proteins depending on the redox potential of the extracellular electron acceptor. Other flavin-binding, extracellular reductases amongst Gram-positive organisms, such as FrdA (acting on fumarate) have been identified in L. monocytogenes and UrdA (acting on urocanate) in Enterococcus rivorum (Light et al., 2019). Thus, there may exist a yet unidentified extracellular reductase in L. plantarum and L. lactis required for anode reduction. Thus, our findings elucidate a new pattern of metabolic changes associated with EET. It seems likely that these many mechanisms reflect the ability of EET to alleviate constraints of intracellular redox balance in fermentative metabolism across LAB.
EET has important implications for ecology and biotechnology of LAB
Conservation of the FLEET locus among different LAB species supports the premise that this hybrid fermentation with EET provides an important metabolic strategy for these bacteria in their natural habitats. LAB with a complete FLEET locus are homofermentative, thus underscoring the distinct ways homofermentative and heterofermentative LAB have evolved for energy conservation (Salvetti et al., 2013). L. plantarum and other LAB with FLEET systems such as L. casei are genetically and metabolically diverse and grow in a variety of nutrient rich environments including dairy and plant foods and the digestive tract (Cai et al., 2009; Martino et al., 2016; Siezen et al., 2010a). Those environments also are rich sources of sources of quinones, flavins, and extracellular electron acceptors such as iron (Cataldi et al., 2003; Fenn et al., 2017; Kim, 2017; Roughead and McCormick, 1990; Walther et al., 2013). Increased organic acid production and environmental acidification by LAB with this hybrid metabolism would provide an effective mechanism to inhibit competing microorganisms and confer a competitive advantage for growth. The increased ATP relative to biomass generation observed during growth on mannitol might also give sufficient readiness for using this energy later on to outcompete neighboring organisms (Russell and Cook, 1995). These effects of EET may be particularly important on plant tissues and intestinal environments, wherein LAB tend to be present in low numbers. Besides our observation that L. plantarum performs EET in kale juice, the FLEET pathway is important for intestinal colonization by both L. monocytogenes (Light et al., 2018) and E. faecalis (Lam et al., 2019), and L. plantarum FLEET genes including ndh2 and pplA were highly induced in the small intestine of rhesus macaques (Golomb et al., 2016).
The hybrid fermentation metabolism of LAB also has technological relevance. For many LAB food fermentations, acidification of the food matrix is required to prevent the growth of undesired microorganisms and result in a more consistent and reproducible product (Marco et al., 2021). Starter cultures are frequently selected based on their capacity for rapid growth and acid production (Bintsis, 2018). In the presence of an anode, exposure of L. plantarum to EET conditions during kale juice fermentation increased the acidification rate. Thus, this shows that EET metabolism is active in complex nutritive environments such as kale leaf tissues that contain other potential electron acceptors besides the anode and diverse electron donors (glucose, fructose, sucrose) (Thavarajah et al., 2016). This example also shows how electro-fermentation, the technological process by which fermentation is modulated using electrodes, can be used to control food fermentations (Moscoviz et al., 2016; Schievano et al., 2016; Vassilev et al., 2021). Because L. plantarum also increased fermentation flux when the electrode was available as an electron sink, higher quantities of organic acid flavor compounds were formed. Therefore, by the manipulation of extracellular redox potential, food electro-fermentations may be used to control microbial growth. This would allow the creation of new or altered sensory profiles in fermented foods, such as through altered organic acid production and metabolism or synthesis of other compounds that alter food flavors, aromas, and textures.
Final perspective
We expect that our study will improve the current understanding of energy conservation in primarily fermentative microorganisms and contribute to establishing the ecological relevance of EET in lactic acid bacteria. This work will ultimately allow the use of EET to electronically modulate the flavor and textural profiles of fermented foods and expand the use of lactic acid bacteria in bioelectronics, biomedicine, and bioenergy (Moscoviz et al., 2016). The identification of the precise components and full bioenergetics involved in L. plantarum EET will be key to unravel physiological and ecological questions and to develop other biotechnological applications.
Materials and methods
Key resources table.
| Reagent type (species) or resource |
Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Lactiplantibacillus plantarum) | NCIMB8826 | Dandekar, 2019 | Strain information listed in Supplementary file 3 | |
| Strain, strain background (Lactiplantibacillus plantarum) | NCIMB8826-R | Yin et al., 2018 | Rifampicin-resistant mutant of NCIMB8826 | Strain information listed in Supplementary file 3 |
| Strain, strain background (Lactiplantibacillus plantarum) | MLES100 | This study | Deletion mutant of NCIMB8826 lacking ndh2 | Plasmid information listed in Supplementary file 3 |
| Strain, strain background (Lactiplantibacillus plantarum) | MLES101 | This study | Deletion mutant of NCIMB8826 lacking pplA | Plasmid information listed in Supplementary file 3 |
| Strain, strain background (Lactiplantibacillus plantarum) | MLEY100 | This study | Deletion mutant of NCIMB8826 lacking narGHIJ | Plasmid information listed in Supplementary file 3 |
| Strain, strain background (Lactiplantibacillus plantarum) | B1.3 | Yin et al., 2018 | Strain information listed in Supplementary file 3 | |
| Strain, strain background (Lactiplantibacillus plantarum) | AJJ11 | Yu et al., 2021 | Strain information listed in Supplementary file 3 | |
| Strain, strain background (Lactiplantibacillus plantarum) | 8.1 | Yu et al., 2021 | Strain information listed in Supplementary file 3 | |
| Strain, strain background (Lactiplantibacillus plantarum) | ATCC 202195 | Wright et al., 2020 | Strain information listed in Supplementary file 3 | |
| Strain, strain background (Lactiplantibacillus plantarum) | NCIMB700965 | Heeney and Marco, 2019 | Strain information listed in Supplementary file 3 | |
| Strain, strain background (Lactiplantibacillus pentosus) | BGM48 | Golomb et al., 2013 | Strain information listed in Supplementary file 3 | |
| Strain, strain background (Lactiplantibacillus casei) | BL23 | Mazé et al., 2010 | Strain information listed in Supplementary file 3 | |
| Strain, strain background (Levilactobacillus brevis) | ATCC 367 | Makarova et al., 2006 | Strain information listed in Supplementary file 3 | |
| Strain, strain background (Lactococcus lactis) | KF147 | Siezen et al., 2010b | Strain information listed in Supplementary file 3 | |
| Strain, strain background (Lactococcus lactis) | IL1403 | Bolotin et al., 2001 | Strain information listed in Supplementary file 3 | |
| Strain, strain background (Lactiplantibacillus Rhamnosus) | GG | Kankainen et al., 2009 | Strain information listed in Supplementary file 3 | |
| Strain, strain background (Lactiplantibacillus murinus) | ASF361 | Wannemuehler et al., 2014 | Strain information listed in Supplementary file 3 | |
| Strain, strain background (Enterococcus faecalis) | ATCC 29212 | Minogue et al., 2014 | Strain information listed in Supplementary file 3 | |
| Strain, strain background (Enterococcus faecalis) | ATCC 8459 | Kopit et al., 2014 | Strain information listed in Supplementary file 3 | |
| Strain, strain background (Pediococcus pentosaceus) | ATCC 25745 | Makarova et al., 2006 | Strain information listed in Supplementary file 3 | |
| Strain, strain background (Streptococcus agalactiae) | ATCC 27956 | McDonald and McDonald, 1976 | Strain information listed in Supplementary file 3 | |
| Strain, strain background(Escherichia coli) | DH5α | Taylor et al., 1993 | fhuA2 lac(del)U169 phoA glnV44 Φ80’ lacZ(del) M15 gyrA96 recA1 relA1 endA1 thi−one hsdR17, amplification of cloning vector |
Strains and culture conditions
All strains and plasmids used in this study are listed in Supplementary file 3. Standard laboratory culture medium was used for routine growth of bacteria as follows: Lactiplantibacillus spp., Lacticaseibacillus spp., Levilactobacillus brevis, Ligilactobacillus murinus, and Pediococcus pentosaceus, MRS (BD, Franklin Lakes, NJ, USA); Lactococcus lactis and Streptococcus agalactiae, M17 (BD) with 2% w/v glucose; Enterococcus faecalis, and Enterococcus faecium, BHI (BD); and Escherichia coli, LB (Teknova, Hollister, CA, USA). Bacterial strains were incubated without shaking except for E. coli (250 RPM) and at either 30 or 37 °C. Where indicated, strains were grown in filter-sterilized MRS (De MAN et al., 1960) lacking beef extract with either 110 mM glucose [gMRS] or 110 mM mannitol [mMRS], or a chemically defined minimal medium (Supplementary file 4) with 125 mM glucose [gCDM] or 125 mM mannitol [mCDM] for 18 hr (Aumiller et al., 2021). Riboflavin (1 mg/L) was routinely added to the CDM. When indicated, culture medium was supplemented with 20 μg/mL of the quinone 1,4-dihydroxy-2-naphthoic acid (DHNA) (Alfa Aesar, Haverhill, MA, USA), 1.25 mM ferric ammonium citrate (C6H8FeNO7) (1.25 mM) (VWR, Radnor, PA, USA), riboflavin (Sigma-Aldrich, St. Louis, MO, USA), or 5 μg/mL erythromycin (VWR).
DNA sequence analysis
The FLEET gene locus of L. plantarum NCIMB8826 was identified using NCBI basic local alignment search tool (BLAST) (McGinnis and Madden, 2004) using the L. monocytogenes 10403S FLEET genes (lmo2634 to lmo2641) as a reference. L. plantarum genes were annotated based on predicted functions within the FLEET pathwa (Light et al., 2018). FLEET locus genes were identified in other LAB by examining 1,788 complete Lactobacillales genomes available at NCBI (downloaded 02/25/2021). A local BLAST (ver 2.10.1) database containing these genomes was queried using tBLASTx with NCIMB8826 FLEET genes a reference. A gene was considered to be present in the Lactobacillales strain genome if the Bit-score was >50 and the E-value was <10–3 (Pearson, 2013). Heatmaps showing the percentage of strains in Lactobacillales genera and the Lactobacillus-genus complex (Zheng et al., 2020) identified to contain individual FLEET genes were visualized using the R-studio package ggplot2 (Wickham, 2011) with clustering done through UPGMA. The FLEET loci of L. plantarum strain 8.1 and NCIMB700965 were aligned to the NCIMB8826 genome in MegAlign Pro (DNAstar Inc, Madison, WI, USA).
Insoluble iron reduction assays
Cells were collected by centrifugation at 10,000 g for 3 min, washed twice in phosphate-buffered saline (PBS), pH 7.2 (http://cshprotocols.cshlp.org), and adjusted to an optical density (OD) at 600 nm (OD600nm) of 2 in the presence of 2.2 mM ferrihydrite (Schwertmann and Fischer, 1973; Stookey, 2002) and 2 mM ferrozine (Sigma-Aldrich). Where indicated, 55 mM glucose or mannitol, 20 μg/mL DHNA, and riboflavin were added. After 3 hr incubation at 30 °C, the cells were collected by centrifugation at 10,000 g for 5 min and the absorbance of the supernatant was measured at 562 nm with a Synergy 2 spectrophotometer (BioTek, Winooski, VT, USA). Quantities of ferrihydrite reduced were determined using a standard curve containing a 2-fold range of FeSO4 (Sigma-Aldrich) (0.25 mM to 0.016 mM) and 2 mM ferrozine. The FeSO4 was dissolved in 10 mM cysteine-HCl (RPI, Mount Prospect, IL, USA) to prevent environmental re-oxidation of Fe2+ to Fe3+ in the standard curve. For testing iron reduction activity of cells with a DHNA concentration of 0.01 μg/mL in the medium, iron(III) oxide nanoparticles < 50 nm (Sigma-Aldrich) were used as insoluble iron form (Figure 1—figure supplement 3).
L. plantarum mutant construction
L. plantarum NCIMB8826 ndh2, pplA, and narGHIJ deletion mutants were constructed by double-crossover homologous recombination with the suicide plasmid pRV300 (Leloup et al., 1997). For mutant construction, upstream and downstream flanking regions of these genes were amplified using the A/B and C/D primers, respectively, listed in Supplementary file 5. Splicing-by-overlap extension (SOEing) PCR was used to combine PCR products as previously described (Heckman and Pease, 2007). PCR products were digested with restriction enzymes EcoRI, SacI, SacII, or SalI (New England Biolabs, Ipswich, MA, USA) for plasmid ligation and transformation into E. coli DH5α. The resulting plasmids were then introduced to L. plantarum NCIMB8826 by electroporation. Erythromycin-resistant mutants were selected and confirmed for plasmid integration by PCR (see Supplementary file 5 for primer sequences). Subsequently, deletion mutants were identified by a loss of resistance to erythromycin, PCR (see Supplementary file 5 for primer sequences) confirmation, and DNA sequencing (http://dnaseq.ucdavis.edu).
Bioelectrochemical reactors (BES) construction, operation, and electrochemical techniques
L. plantarum NCIMB8826 strains were grown overnight (approximately 16–18 hr) from glycerol stocks in MRS. Cells were harvested by centrifugation (5200 g, 12 min, 4 °C) and washed twice in PBS. When L. plantarum wild-type EET activity versus the Δndh2 (MLES100) and ΔpplA (MLES101) deletion mutants was compared, cells were grown as described and the number of cells was normalized across the three strains prior to inoculation in the BES. The bioreactors consisted of double-chamber electrochemical cells (Adams & Chittenden, Berkeley, CA) (Figure 1B) with a cation exchange membrane (CMI-7000, Membranes International, Ringwood, NJ) that separated them. A three-electrode configuration was used consisting of an Ag/AgCl sat KCl reference electrode (BASI, IN, USA), a titanium wire counter electrode, and a 6.35-mm-thick graphite felt working electrode (anode) of 4 × 4 cm (Alfa Aesar, MA, USA) with a piece of Ti wire threaded from bottom to top as a current collector and connection to the potentiostat. We used a Bio-Logic Science Instruments (TN, USA) potentiostat model VSP-300 for performing the electrochemical measurements (chronoamperometry). The bioreactors were sterilized by filling them with ddH2O and autoclaving at 121 °C for 30 min. The water was then removed and replaced with 150 mL of filter sterilized mMRS or mCDM media for the working electrode chamber, and 150 mL of M9 medium (6.78 g/L Na₂HPO₄, 3 g/L KH2PO4, 0.5 g/L NaCl, 1 g/L NH4Cl) (BD) for the counter electrode chamber. Both media of the working electrode chamber were supplemented with 20 μg/mL DHNA or 0.01 μg/mL diluted 1:1 in DMSO:ddH2O where appropriate. To test the role of bd-cytochrome, heme was added in a final concentration of 10 μg/mL (diluted 1:1 in DMSO: ddH2O). The medium in the working electrode chamber was continuously mixed with a magnetic stir bar and N2 gas was purged to maintain anaerobic conditions for the course of the experiment. The applied potential to the working electrode was of +0.2 V versus Ag/AgCl (sat. KCl) (BASI, IN, USA). Reactors run under OC conditions were similarly assembled but kept at open circuit and used as control for non-current circulating conditions. Once the current stabilized, the electrochemical cells were inoculated to a final OD600 of 0.12–0.15 with the cell suspensions prepared in PBS. Current densities are reported as a function of the geometric surface area of the electrode (16 cm2). The bioreactors were sampled by taking samples under sterilized conditions at different time points for subsequent analysis. The samples for organic acids analyses were centrifuged (15,228 g, 7 min) and the supernatant was separated for High-Performance Liquid Chromatography (HPLC) assessments. Samples for ATP and NAD+/NADH analyses were flash frozen in a dry ice/ethanol bath.
Metabolite analysis
Organic acids, ethanol, and sugar concentrations were measured by HPLC (Agilent, 1260 Infinity), using a standard analytical system (Shimadzu, Kyoto, Japan) equipped with an Aminex Organic Acid Analysis column (Bio-Rad, HPX-87H 300 × 7.8 mm) heated at 60 °C. The eluent was 5 mM of sulfuric acid, used at a flow rate of 0.6 mL min–1. We used a refractive index detector 1260 Infinity II RID. A five-point calibration curve based on peak area was generated and used to calculate concentrations in the unknown samples. The following standards were included in the HPLC measurements: acetate, formate, pyruvate, malate, lactate, succinate, oxalacetate, fumarate, ethanol, acetoin, butanediol, mannitol, and glucose. No gaseous products were measured.
BES biomass growth determination
Bioreactors were shaken to remove the cells attached to the working electrode and afterwards sampled to measure viable cells (colony forming units [CFUs]) and total biomass (dry weight). Samples for CFU enumeration were collected under sterile conditions at the time of inoculation and at the time of approximately maximum current density. Samples were serially diluted (1:1000 to 1:1000000) in sterile PBS and plated on MRS for CFUs enumeration after overnight incubation at 30 °C. Dry weight was determined using a 25 mL sample collected at approximately maximum current density. Cells were harvested by centrifugation (5250 g, 12 min, 4 °C) and washed twice in 50 mL ddH2O. Afterwards cells were resuspended in 1 mL of ddH2O and transferred to microfuge tubes (previously weighted). Cells were harvested by centrifugation (5250 g, 12 min, 4 °C), and the tubes were then transferred to an evaporator to remove humidity. The microfuge tubes were then cooled in a desiccator for 30 min and the weight of each tube was measured to determine cell weight. The difference between the weight of each tube with the pellet and before containing it allowed us to determine the dry weight/mL.
RNA-seq library construction and transcriptome analysis
L. plantarum NCIMB8826 was grown in triplicate to exponential phase (OD600 1.0) at 37 °C in mMRS with or without the supplementation of 20 μg/mL DHNA and 1.25 mM ferric ammonium citrate. Cells were collected by centrifugation at 10,000 g for 3 min at 4 °C, flash frozen in liquid N2 and stored at –80 °C prior to RNA extraction as previously described (Golomb et al., 2016). Briefly, frozen cell pellets were resuspended in cold acidic phenol:chloroform:isoamyl alcohol (pH 4.5) [125:24:1] (Invitrogen, Carlsbad, CA, USA) before transferring to 2 mL screw cap tubes containing buffer (200 mM NaCl, 20 mM EDTA), 20% SDS, and 300 mg 0.1 mm zirconia/silica beads. RNA was extracted by mechanical lysis with an MP Fastprep bead beater (MP Biomedicals, Santa Ana, CA, USA) at 6.5 m/s for 1 min. The tubes were centrifuged at 20,000 g at 4 °C for 3 min and the upper aqueous phase was transferred to a new tube. The aqueous phase was extracted twice with chloroform:isoamyl alcohol [24:1] (Fisher Scientific, Waltham, MA, USA), The aqueous phase was then transferred to a new tube for RNA ethanol precipitation (Green and Sambrook, 2020). RNA was then quantified on a Nanodrop 2000c (ThermoFisher), followed by double DNAse digestion with the Turbo DNA-free Kit (Invitrogen) according to the manufacturer’s protocols. The quality of the remaining RNA was checked using a Bioanalyzer RNA 6000 Nano Kit (Agilent Technologies, Santa Clara, CA, USA) (all RIN values > 9) and then quantified with the Qubit 2.0 RNA HS Assay (Life Technologies, Carlsbad, CA, USA). For reverse-transcription PCR (RT-PCR), 800 ng RNA was converted to cDNA with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocols. Quantitative RT-PCR was performed on a 7,500 Fast Real-Time PCR System (Applied Biosystems) using the PowerUp SYBR Green Master Mix (ThermoFisher) and RT-PCR primers listed in Supplementary file 5. The 2-ΔΔCt method was used for relative transcript quantification using rpoB as a control (Livak and Schmittgen, 2001).
For sequencing, ribosomal-RNA (rRNA) was depleted from 4 μg RNA using the RiboMinus Eukaryote Kit v2 with specific probes for prokaryotic rRNA (ThermoFisher) following the manufacturer’s instructions. RNA was then fragmented to approximately 200 bp, converted to cDNA, and barcoded using the NEBnext Ultra-directional RNA Library Kit for Illumina (New England Biolabs, Ipswitch, MA, USA) with NEBnext Multiplex Oligos for Illumina (Primer Set 1) (New England Biolabs) following the manufacturer’s protocols. cDNA libraries containing pooled barcoded samples was run across two lanes of a HiSeq400 (Illumina, San Diego, CA, USA) on two separate runs for 150 bp paired-end reads (http://dnatech.genomecenter.ucdavis.edu/). An average of 36,468,428 raw paired-end reads per sample was collected (Supplementary file 6). The DNA sequences were quality filtered for each of the 12 samples by first visualizing with FastQC (ver. 0.11.8) (Andrews, 2010) to check for appropriate trimming lengths, followed by quality filtering with Trimmomatic (ver. 0.39) (Bolger et al., 2014). Remaining reads then were aligned to the NCIMB8826 chromosome and plasmids using Bowtie2 (ver. 2.3.5) in the [-sensitive] mode (Langmead and Salzberg, 2012). The resulting ‘.sam’ files containing aligned reads from Bowtie2 were converted to ‘.bam’ files with Samtools (ver 1.9) (Li et al., 2009) before counting aligned reads with FeatureCounts in the [-- stranded = reverse] mode (ver. 1.6.4) (Liao et al., 2014). Reads aligning to noncoding sequences (e.g. rRNA, tRNA, trRNA, etc.) were excluded for subsequent analyses. Differential gene expression based on culture condition was determined with DESeq2 (Love et al., 2014) using the Wald test in the R-studio shiny app DEBrowser (ver 1.14.2) (Kucukural et al., 2019). Differential expression was considered significant with a False-discovery-rate (FDR)-adjusted p-value < 0.05 and a Log2 (fold-change) >0.5. Clusters of Orthologous Groups (COGs) were assigned to genes based on matches from the eggNOG (ver. 5.0) database (Huerta-Cepas et al., 2019).
Redox probe assays
Hamilton oxidation-reduction potential (ORP) probes (Hamilton Company, Reno, NV, USA) were inserted into air-tight Pyrex (Corning Inc, Corning, NY, USA) bottles containing mMRS supplemented with 20 μg/mL DHNA and/or 1.25 mM ferric ammonium citrate and incubated in a water bath at 37 °C. A custom cap for the Pyrex bottles was 3D printed with polylactic acid filament (2.85 mm diameter) such that the ORP probe threads into the cap and an o-ring seal can be used to provide an air-tight seal between the probe and the cap. The ORP was allowed to equilibrate over 40 min before L. plantarum NCIMB8826, Δndh2 (MLES100), or ΔpplA (MLES101) were inoculated at an OD600 of 0.10. Two uninoculated controls were used to measure baseline ORP over time. The ORP data was collected via Modbus TCP/IP protocol (Stride Modbus Gateway, AutomationDirect, Cumming, GA, USA) into a database (OSIsoft, San Leandro, CA, USA) and analyzed in MATLAB (Mathworks, Nantick, MA, USA). pH was measured using a Mettler Toledo SevenEasy pH meter (Mettler Toledo, Columbus, OH, USA). Cells were collected at either 24 hr or at the greatest ORP difference between the wild-type and mutant strains (ΔmVmax) by centrifugation at 10,000 g for 3 min and used for ferrihydrite reduction analyses.
ATP and NAD+/NADH quantification
Frozen cell pellets were suspended in PBS and lysed by mechanical agitation in a FastPrep 24 (MP Biomedicals) at 6.5 m/s for 1 min. The cell lysates were then centrifuged at 20,000 g for 3 min at 4 °C. ATP and NAD+ and NADH in the supernatants were then quantified with the Molecular Probes ATP Quantification Kit (ThermoFisher) and the Promega NAD/NADH-Glo Kit (Promega, Madison, WI, USA), respectively according to the manufacturers’ instructions.
Inductively coupled plasma-mass spectrometry (ICP-MS)
L. plantarum was inoculated in mMRS with or without 20 μg/mL DHNA and 1.25 mM ferric ammonium citrate at an OD600 of 0.10 for 3.5 hr. Cells were then collected by centrifugation at 10,000 x g for 3 min and washed twice in PBS to remove cell-surface-associated metals. Viable cell numbers were enumerated by plating serial dilutions on MRS laboratory culture medium and the resulting cell materials were digested by incubating at 95 °C for 45 min in a 60% concentrated trace metal grade HNO3, allowed to cool, then diluted with MilliQ water to a final concentration of 6% HNO3. The contents were quantified with internal standards with an Agilent 7,500Ce ICP-MS (Agilent Technologies, Palo Alto, CA) for simultaneous determination of select metals (Na, Mg, Al, K, Ca, Cu, Zn, Ba, Mn, Fe) at the UC Davis Interdisciplinary Center for Plasma Mass Spectrometry (http://icpms.ucdavis.edu/).
Kale juice fermentation assay
Green organic kale purchased from a market (Whole Foods) was washed with tap water and air dried for 1 hr as previously recommended (Kim, 2017). A total of 385 g of the leaves and stems were shredded with an electric food processor in 1 L ddH20. The kale juice was then diluted with 0.35 L ddH2O and autoclaved (121 °C, 15 min). The juice was then centrifuged under sterile conditions at 8000 rpm for 20 min and the supernatant was collected. A rifampicin-resistant variant of L. plantarum NCIMB8826-R (Tachon et al., 2014) (grown for 19 hr in MRS medium at 37 °C, 50 μg Rif/mL) was inoculated to an estimated final OD of approximately 0.05, and DHNA (20 μg/mL) was added where appropriate. Cells were collected and washed as previously described for the bioelectrochemical assays in mCDM. The anodic chambers of bioreactors assembled as previously described (anode of 4.3*6 cm) were filled with 125 mL of the inoculated kale juice and incubated at 30 °C purged with N2. After 1 hr, the anodes were polarized to 0.2 V versus Ag/AgCl (sat. KCl) (EET conditions) or kept at open circuit (OC, no EET). Viable cells were measured by plating 10-fold serial dilutions in MRS agar plates with 50 μg/mL of Rif.
Calculations
The total electrons harvested on the anode were estimated by integrating the area (charge) under the chronoamperometric curve (current response (A) over time (s)), which was corrected by subtracting the current baseline obtained before L. plantarum was added to the system. This obtained charge was then converted to mol of electrons using the Faraday constant (96,485.3 A*s/mol electrons).
Data accession numbers
L. plantarum RNA-seq data are available in the NCBI Sequence Read Archive (SRA) under BioProject accession no. PRJNA717240. A list of the completed Lactobacillales genomes used in the DNA sequence analysis is available in the Harvard Dataverse repository at https://doi.org/107910/DVN/IHKI0C.
Acknowledgements
This work was supported by the National Science Foundation grant #1650042, Office of Naval Research grant 0001418IP00037 (CMAF), and the USDA National Institute of Agriculture Multi-State Project (W4122). Work at the Molecular Foundry was supported by the Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. James Nelson was supported by the Rodgers University fellowship in Electrical and Computer Engineering.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Caroline M Ajo-Franklin, Email: cajo-franklin@rice.edu.
Maria L Marco, Email: mmarco@ucdavis.edu.
Gisela Storz, National Institute of Child Health and Human Development, United States.
Gisela Storz, National Institute of Child Health and Human Development, United States.
Funding Information
This paper was supported by the following grants:
National Science Foundation 1650042 to Eric T Stevens.
Office of Naval Research 0001418IP00037 to Caroline M Ajo-Franklin.
U.S. Department of Energy DE-AC02-05CH11231 to Sara Tejedor-Sanz, Eric T Stevens, Caroline M Ajo-Franklin, Maria L Marco.
U.S. Department of Agriculture W4122 to Maria L Marco.
Cancer Prevention and Research Institute of Texas RR190063 to Caroline M Ajo-Franklin, Siliang Li.
Additional information
Competing interests
No competing interests declared.
No competing interests declared.
Author contributions
Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review and editing.
Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review and editing.
Formal analysis, Investigation, Validation, Visualization, Writing – review and editing.
Data curation, Investigation, Software, Writing – review and editing.
Data curation, Investigation, Writing – review and editing.
Data curation, Methodology, Resources, Software, Supervision, Writing – review and editing.
Conceptualization, Writing – review and editing.
Conceptualization, Funding acquisition, Methodology, Supervision, Writing – original draft, Writing – review and editing.
Conceptualization, Funding acquisition, Methodology, Supervision, Writing – original draft, Writing – review and editing.
Additional files
Data availability
L. plantarum RNA-seq data are available in the NCBI Sequence Read Archive (SRA) under BioProject accession no. PRJNA717240. A list of the completed Lactobacillales genomes used in the DNA sequence analysis is available in the Harvard Dataverse repository at https://doi.org/10.7910/DVN/IHKI0C All other data generated or analysed during this study are included in the manuscript and supporting files.
The following dataset was generated:
Stevens E. 2021. Lactiplantibacillus plantarum transcriptome under extracellular electron transfer (EET)-conducive conditions. NCBI BioProject. PRJNA717240
References
- Andrews S. FastQC: a quality control tool for high throughput sequence data. v3Babraham Bioinformatics. 2010 http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- Aumiller K, Stevens E, Scheffler R, Güvener ZT, Tung E, Grimaldo AB, Carlson HK, Deutschbauer AM, Taga ME, Marco ML, Ludington WB. A Chemically-Defined Growth Medium to Support Lactobacillus – Acetobacter Community Analysis. bioRxiv. 2021 doi: 10.1101/2021.05.12.443930. [DOI] [PMC free article] [PubMed]
- Behera SS, Ray RC, Zdolec N. Lactobacillus plantarum with functional properties: an approach to increase safety and shelf-life of fermented foods. BioMed Research International. 2018;2018:9361614. doi: 10.1155/2018/9361614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintsis T. Lactic acid bacteria as starter cultures: an update in their metabolism and genetics. AIMS Microbiol. 2018;4:665–684. doi: 10.3934/microbiol.2018.4.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom J, De Mattos MJT, Grivell LA. Redirection of the respiro-fermentative flux distribution in Saccharomyces cerevisiae by overexpression of the transcription factor Hap4p. Applied and Environmental Microbiology. 2000;66:1970–1973. doi: 10.1128/aem.66.5.1970-1973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A, Wincker P, Mauger S, Jaillon O, Malarme K, Weissenbach J, Ehrlich SD, Sorokin A. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Research. 2001;11:731–753. doi: 10.1101/gr.gr-1697r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooijmans RJW, de Vos WM, Hugenholtz J. Lactobacillus plantarum WCFS1 electron transport chains. Applied and Environmental Microbiology. 2009a;75:3580–3585. doi: 10.1128/AEM.00147-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooijmans R, Smit B, Santos F, van Riel J, de Vos WM, Hugenholtz J. Heme and menaquinone induced electron transport in lactic acid bacteria. Microbial Cell Factories. 2009b;8:28. doi: 10.1186/1475-2859-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckel W, Thauer RK. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2013;1827:94–113. doi: 10.1016/j.bbabio.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Cai H, Thompson R, Budinich MF, Broadbent JR, Steele JL. Genome sequence and comparative genome analysis of Lactobacillus casei: insights into their niche-associated evolution. Genome Biology and Evolution. 2009;1:239–257. doi: 10.1093/gbe/evp019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldi TRI, Nardiello D, Carrara V, Ciriello R, De Benedetto GE. Assessment of riboflavin and flavin content in common food samples by capillary electrophoresis with laser-induced fluorescence detection. Food Chemistry. 2003;82:309–314. doi: 10.1016/S0308-8146(02)00567-8. [DOI] [Google Scholar]
- Chen C, Zhao S, Hao G, Yu H, Tian H, Zhao G. Role of lactic acid bacteria on the yogurt flavour: A review. International Journal of Food Properties. 2017;20:S316–S330. doi: 10.1080/10942912.2017.1295988. [DOI] [Google Scholar]
- Dandekar S. Lactiplantibacillus Plantarum Strain:NCIMB8826 (ID 527135)-BioProject. bioRxiv. 2019 doi: 10.1101/2021.05.26.445846. [DOI]
- de Graef MR, Alexeeva S, Snoep JL, Teixeira de Mattos MJ. The steady-state internal redox state (NADH/NAD) reflects the external redox state and is correlated with catabolic adaptation in Escherichia coli. Journal of Bacteriology. 1999;181:2351–2357. doi: 10.1128/JB.181.8.2351-2357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De MAN JC, Rogosa M, Sharpe ME. A MEDIUM FOR THE CULTIVATION OF LACTOBACILLI. Journal of Applied Bacteriology. 1960;23:130–135. doi: 10.1111/j.1365-2672.1960.tb00188.x. [DOI] [Google Scholar]
- Dirar H, Collins EB. End-products, fermentation balances and molar growth yields of homofermentative lactobacilli. Journal of General Microbiology. 1972;73:233–238. doi: 10.1099/00221287-73-2-233. [DOI] [PubMed] [Google Scholar]
- Duar RM, Lin XB, Zheng J, Martino ME, Grenier T, Pérez-Muñoz ME, Leulier F, Gänzle M, Walter J. Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiology Reviews. 2017;41:S27–S48. doi: 10.1093/femsre/fux030. [DOI] [PubMed] [Google Scholar]
- Duwat P, Sourice S, Cesselin B, Lamberet G, Vido K, Gaudu P, Le Loir Y, Violet F, Loubière P, Gruss A. Respiration capacity of the fermenting bacterium Lactococcus lactis and its positive effects on growth and survival. Journal of Bacteriology. 2001;183:4509–4516. doi: 10.1128/JB.183.15.4509-4516.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elli M, Zink R, Rytz A, Reniero R, Morelli L. Iron requirement of Lactobacillus spp. in completely chemically defined growth media. Journal of Applied Microbiology. 2000;88:695–703. doi: 10.1046/j.1365-2672.2000.01013.x. [DOI] [PubMed] [Google Scholar]
- Emde R, Schink B. Enhanced propionate formation by Propionibacterium freudenreichii subsp. freudenreichii in a three-electrode amperometric culture system. Applied and Environmental Microbiology. 1990;56:2771–2776. doi: 10.1128/aem.56.9.2771-2776.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom JE, Kwon SC, Moon GS. Detection of 1,4-dihydroxy-2-naphthoic Acid from commercial makgeolli products. Preventive Nutrition and Food Science. 2012;17:83–86. doi: 10.3746/pnf.2012.17.1.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve-Núñez A, Rothermich M, Sharma M, Lovley D. Growth of Geobacter sulfurreducens under nutrient-limiting conditions in continuous culture. Environmental Microbiology. 2005;7:641–648. doi: 10.1111/j.1462-2920.2005.00731.x. [DOI] [PubMed] [Google Scholar]
- Fan G, Dundas CM, Graham AJ, Lynd NA, Keitz BK. Shewanella oneidensis as a living electrode for controlled radical polymerization. PNAS. 2018;115:4559–4564. doi: 10.1073/pnas.1800869115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenn K, Strandwitz P, Stewart EJ, Dimise E, Rubin S, Gurubacharya S, Clardy J, Lewis K. Quinones are growth factors for the human gut microbiota. Microbiome. 2017;5:161. doi: 10.1186/s40168-017-0380-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freguia S, Masuda M, Tsujimura S, Kano K. Lactococcus lactis catalyses electricity generation at microbial fuel cell anodes via excretion of a soluble quinone. Bioelectrochemistry. 2009;76:14–18. doi: 10.1016/j.bioelechem.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Furuichi K, Hojo K, Katakura Y, Ninomiya K, Shioya S. Aerobic culture of Propionibacterium freudenreichii ET-3 can increase production ratio of 1,4-dihydroxy-2-naphthoic acid to menaquinone. Journal of Bioscience and Bioengineering. 2006;101:464–470. doi: 10.1263/jbb.101.464. [DOI] [PubMed] [Google Scholar]
- Gänzle MG. Lactic metabolism revisited: metabolism of lactic acid bacteria in food fermentations and food spoilage. Current Opinion in Food Science. 2015;2:106–117. doi: 10.1016/j.cofs.2015.03.001. [DOI] [Google Scholar]
- Glasser NR, Kern SE, Newman DK. Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton-motive force. Molecular Microbiology. 2014;92:399–412. doi: 10.1111/mmi.12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golomb BL, Morales V, Jung A, Yau B, Boundy-Mills KL, Marco ML. Effects of pectinolytic yeast on the microbial composition and spoilage of olive fermentations. Food Microbiology. 2013;33:97–106. doi: 10.1016/j.fm.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Golomb BL, Hirao LA, Dandekar S, Marco ML. Gene expression of Lactobacillus plantarum and the commensal microbiota in the ileum of healthy and early SIV-infected rhesus macaques. Scientific Reports. 2016;6:24723. doi: 10.1038/srep24723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MR, Sambrook J. Precipitation of RNA with Ethanol. Cold Spring Harbor Protocols. 2020;2020:101717. doi: 10.1101/pdb.prot101717. [DOI] [PubMed] [Google Scholar]
- Guo Y, Tian X, Huang R, Tao X, Shah NP, Wei H, Wan C. A physiological comparative study of acid tolerance of Lactobacillus plantarum ZDY 2013 and L. plantarum ATCC 8014 at membrane and cytoplasm levels. Annals of Microbiology. 2017;67:669–677. doi: 10.1007/s13213-017-1295-x. [DOI] [Google Scholar]
- Hansen EB. Redox reactions in food fermentations. Current Opinion in Food Science. 2018;19:98–103. doi: 10.1016/j.cofs.2018.03.004. [DOI] [Google Scholar]
- Haruta S, Kanno N. Survivability of microbes in natural environments and their ecological impacts. Microbes and Environments. 2015;30:123–125. doi: 10.1264/jsme2.ME3002rh. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman KL, Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nature Protocols. 2007;2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- Hederstedt L, Gorton L, Pankratova G. Two Routes for Extracellular Electron Transfer in Enterococcus faecalis. Journal of Bacteriology. 2020;202:e00725-19. doi: 10.1128/JB.00725-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeney DD, Marco ML. Complete Genome Sequence of the Plantaricin-Sensitive Strain Lactobacillus plantarum NCIMB 700965. Microbiology Resource Announcements. 2019;8:e01724-18. doi: 10.1128/MRA.01724-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann G, Jayamani E, Mai G, Buckel W. Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. Journal of Bacteriology. 2008;190:784–791. doi: 10.1128/JB.01422-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm AK, Blank LM, Oldiges M, Schmid A, Solem C, Jensen PR, Vemuri GN. Metabolic and transcriptional response to cofactor perturbations in Escherichia coli. The Journal of Biological Chemistry. 2010;285:17498–17506. doi: 10.1074/jbc.M109.095570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, von Mering C, Bork P. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Research. 2019;47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt KA, Flynn JM, Naranjo B, Shikhare ID, Gralnick JA. Substrate-level phosphorylation is the primary source of energy conservation during anaerobic respiration of Shewanella oneidensis strain MR-1. Journal of Bacteriology. 2010;192:3345–3351. doi: 10.1128/JB.00090-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isawa K, Hojo K, Yoda N, Kamiyama T, Makino S, Saito M, Sugano H, Mizoguchi C, Kurama S, Shibasaki M, Endo N, Sato Y. Isolation and identification of a new bifidogenic growth stimulator produced by Propionibacterium freudenreichii ET-3. Bioscience, Biotechnology, and Biochemistry. 2002;66:679–681. doi: 10.1271/bbb.66.679. [DOI] [PubMed] [Google Scholar]
- Jeuken LJC, Hards K, Nakatani Y, Mullineaux CW. Extracellular Electron Transfer: Respiratory or Nutrient Homeostasis? Journal of Bacteriology. 2020;202:4. doi: 10.1128/JB.00029-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JE, Kim TJ, Moon GS. A Novel Lactobacillus casei LP1 Producing 1,4-Dihydroxy-2-Naphthoic Acid, a Bifidogenic Growth Stimulator. Preventive Nutrition and Food Science. 2015;20:78–81. doi: 10.3746/pnf.2015.20.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankainen M, Paulin L, Tynkkynen S, von Ossowski I, Reunanen J, Partanen P, Satokari R, Vesterlund S, Hendrickx APA, Lebeer S, De Keersmaecker SCJ, Vanderleyden J, Hamalainen T, Laukkanen S, Salovuori N, Ritari J, Alatalo E, Korpela R, Mattila-Sandholm T, Lassig A, Hatakka K, Kinnunen KT, Karjalainen H, Saxelin M, Laakso K, Surakka A, Palva A, Salusjarvi T, Auvinen P, de Vos WM. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human- mucus binding protein. PNAS. 2009;106:17193–17198. doi: 10.1073/pnas.0908876106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh D, Lam LN, Doyle LE, Matysik A, Pavagadhi S, Umashankar S, Low PM, Dale JL, Song Y, Ng SP, Boothroyd CB, Dunny GM, Swarup S, Williams RBH, Marsili E, Kline KA, Hancock LE, Hultgren SJ. Extracellular Electron Transfer Powers Enterococcus faecalis Biofilm Metabolism. mBio. 2018;9:00626-17. doi: 10.1128/mBio.00626-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killeen DJ, Boulton R, Knoesen A. Advanced Monitoring and Control of Redox Potential in Wine Fermentation. American Journal of Enology and Viticulture. 2018;69:394–399. doi: 10.5344/ajev.2018.17063. [DOI] [Google Scholar]
- Kim SY. Production of Fermented Kale Juices with Lactobacillus Strains and Nutritional Composition. Preventive Nutrition and Food Science. 2017;22:231–236. doi: 10.3746/pnf.2017.22.3.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BH, Gadd GM. Prokaryotic Metabolism and Physiology. Cambridge University Press; 2019. [DOI] [Google Scholar]
- Kopit LM, Kim EB, Siezen RJ, Harris LJ, Marco ML. Safety of the surrogate microorganism Enterococcus faecium NRRL B-2354 for use in thermal process validation. Applied and Environmental Microbiology. 2014;80:1899–1909. doi: 10.1128/AEM.03859-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzuma A, Hashimoto K, Watanabe K. Roles of siderophore in manganese-oxide reduction by Shewanella oneidensis MR-1. FEMS Microbiology Letters. 2012;326:91–98. doi: 10.1111/j.1574-6968.2011.02444.x. [DOI] [PubMed] [Google Scholar]
- Kracke F, Lai B, Yu S, Krömer JO. Balancing cellular redox metabolism in microbial electrosynthesis and electro fermentation - A chance for metabolic engineering. Metabolic Engineering. 2018;45:109–120. doi: 10.1016/j.ymben.2017.12.003. [DOI] [PubMed] [Google Scholar]
- Kucukural A, Yukselen O, Ozata DM, Moore MJ, Garber M. DEBrowser: interactive differential expression analysis and visualization tool for count data. BMC Genomics. 2019;20:6. doi: 10.1186/s12864-018-5362-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam LN, Wong JJ, Matysik A, Paxman JJ, Chong KKL, Low PM, Chua ZS, Heras B, Marsili E, Kline KA. Sortase-Assembled Pili Promote Extracellular Electron Transfer and Iron Acquisition in Enterococcus Faecalis Biofilm. Cold Spring Harbor Laboratory; 2019. [DOI] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leloup L, Ehrlich SD, Zagorec M, Morel-Deville F. Single-crossover integration in the Lactobacillus sake chromosome and insertional inactivation of the ptsI and lacL genes. Applied and Environmental Microbiology. 1997;63:2117–2123. doi: 10.1128/aem.63.6.2117-2123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Hinderberger J, Seedorf H, Zhang J, Buckel W, Thauer RK. Coupled ferredoxin and crotonyl coenzyme a (coA) reduction with NADH catalyzed by the butyryl-coA dehydrogenase/Etf complex from Clostridium kluyveri. Journal of Bacteriology. 2008;190:843–850. doi: 10.1128/JB.01417-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics (Oxford, England) 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Light SH, Su L, Rivera-Lugo R, Cornejo JA, Louie A, Iavarone AT, Ajo-Franklin CM, Portnoy DA. A flavin-based extracellular electron transfer mechanism in diverse Gram-positive bacteria. Nature. 2018;562:140–144. doi: 10.1038/s41586-018-0498-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light SH, Méheust R, Ferrell JL, Cho J, Deng D, Agostoni M, Iavarone AT, Banfield JF, D’Orazio SEF, Portnoy DA. Extracellular electron transfer powers flavinylated extracellular reductases in Gram-positive bacteria. PNAS. 2019;116:26892–26899. doi: 10.1073/pnas.1915678116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SS, Kerscher S, Saleh A, Brandt U, Gross U, Bohne W. The Toxoplasma gondii type-II NADH dehydrogenase TgNDH2-I is inhibited by 1-hydroxy-2-alkyl-4(1H)quinolones. Biochimica et Biophysica Acta. 2008;1777:1455–1462. doi: 10.1016/j.bbabio.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, Shakhova V, Grigoriev I, Lou Y, Rohksar D, Lucas S, Huang K, Goodstein DM, Hawkins T, Plengvidhya V, Welker D, Hughes J, Goh Y, Benson A, Baldwin K, Lee JH, Díaz-Muñiz I, Dosti B, Smeianov V, Wechter W, Barabote R, Lorca G, Altermann E, Barrangou R, Ganesan B, Xie Y, Rawsthorne H, Tamir D. Comparative genomics of the lactic acid bacteria. PNAS. 2006;103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco ML, Sanders ME, Gänzle M, Arrieta MC, Cotter PD, De Vuyst L, Hill C, Holzapfel W, Lebeer S, Merenstein D, Reid G, Wolfe BE, Hutkins R. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on fermented foods. Nature Reviews Gastroenterology & Hepatology. 2021;18:196–208. doi: 10.1038/s41575-020-00390-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsili E, Baron DB, Shikhare ID, Coursolle D, Gralnick JA, Bond DR. Shewanella secretes flavins that mediate extracellular electron transfer. PNAS. 2008;105:3968–3973. doi: 10.1073/pnas.0710525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsili E, Sun J, Bond DR. Voltammetry and Growth Physiology of Geobacter sulfurreducens Biofilms as a Function of Growth Stage and Imposed Electrode Potential. Electroanalysis. 2010;22:865–874. doi: 10.1002/elan.200800007. [DOI] [Google Scholar]
- Martino ME, Bayjanov JR, Caffrey BE, Wels M, Joncour P, Hughes S, Gillet B, Kleerebezem M, van Hijum SAFT, Leulier F. Nomadic lifestyle of Lactobacillus plantarum revealed by comparative genomics of 54 strains isolated from different habitats. Environmental Microbiology. 2016;18:4974–4989. doi: 10.1111/1462-2920.13455. [DOI] [PubMed] [Google Scholar]
- Masuda M, Freguia S, Wang YF, Tsujimura S, Kano K. Flavins contained in yeast extract are exploited for anodic electron transfer by Lactococcus lactis. Bioelectrochemistry. 2010;78:173–175. doi: 10.1016/j.bioelechem.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Mazé A, Boël G, Zúñiga M, Bourand A, Loux V, Yebra MJ, Monedero V, Correia K, Jacques N, Beaufils S, Poncet S, Joyet P, Milohanic E, Casarégola S, Auffray Y, Pérez-Martínez G, Gibrat JF, Zagorec M, Francke C, Hartke A, Deutscher J. Complete genome sequence of the probiotic Lactobacillus casei strain BL23. Journal of Bacteriology. 2010;192:2647–2648. doi: 10.1128/JB.00076-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald TJ, McDonald JS. Streptococci isolated from bovine intramammary infections. American Journal of Veterinary Research. 1976;37:377–381. [PubMed] [Google Scholar]
- McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Research. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minogue TD, Daligault HE, Davenport KW, Broomall SM, Bruce DC, Chain PS, Coyne SR, Chertkov O, Freitas T, Gibbons HS, Jaissle J, Koroleva GI, Ladner JT, Palacios GF, Rosenzweig CN, Xu Y, Johnson SL. Complete Genome Assembly of Enterococcus faecalis 29212, a Laboratory Reference Strain. Genome Announcements. 2014;2:00968-14. doi: 10.1128/genomeA.00968-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morandi S, Silvetti T, Tamburini A, Brasca M. Changes in oxidation-reduction potential during milk fermentation by wild lactic acid bacteria. The Journal of Dairy Research. 2016;83:387–394. doi: 10.1017/S0022029916000339. [DOI] [PubMed] [Google Scholar]
- Moscoviz R, Toledo-Alarcón J, Trably E, Bernet N. Electro-fermentation: how to drive fermentation using electrochemical systems. Trends in Biotechnology. 2016;34:856–865. doi: 10.1016/j.tibtech.2016.04.009. [DOI] [PubMed] [Google Scholar]
- Müller V, Chowdhury NP, Basen M. Electron bifurcation: a long-hidden energy-coupling mechanism. Annual Review of Microbiology. 2018;72:331–353. doi: 10.1146/annurev-micro-090816-093440. [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Shimaki Y, Dutta D, Muench SP, Ireton K, Cook GM, Jeuken LJC. Unprecedented properties of phenothiazines unraveled by a NDH-2 bioelectrochemical assay platform. Journal of the American Chemical Society. 2020;142:1311–1320. doi: 10.1021/jacs.9b10254. [DOI] [PubMed] [Google Scholar]
- Olsen MJ, Pérez-Díaz IM. Influence of Microbial Growth on the Redox Potential of Fermented Cucumbers. Journal of Food Science. 2009;74:M149–M153. doi: 10.1111/j.1750-3841.2009.01121.x. [DOI] [PubMed] [Google Scholar]
- Pankratova G, Leech D, Gorton L, Hederstedt L. Extracellular electron transfer by the gram-positive bacterium Enterococcus faecalis. Biochemistry. 2018;57:4597–4603. doi: 10.1021/acs.biochem.8b00600. [DOI] [PubMed] [Google Scholar]
- Pearson WR. An Introduction to Sequence Similarity (“Homology”) Searching. Current Protocols in Bioinformatics. 2013;42:1. doi: 10.1002/0471250953.bi0301s42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen MB, Gaudu P, Lechardeur D, Petit MA, Gruss A. Aerobic respiration metabolism in lactic acid bacteria and uses in biotechnology. Annual Review of Food Science and Technology. 2012;3:37–58. doi: 10.1146/annurev-food-022811-101255. [DOI] [PubMed] [Google Scholar]
- Pfeiffer T, Morley A. An evolutionary perspective on the Crabtree effect. Frontiers in Molecular Biosciences. 2014;1:17. doi: 10.3389/fmolb.2014.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renslow RS, Babauta JT, Majors PD, Beyenal H. DIFFUSION IN BIOFILMS RESPIRING ON ELECTRODES. Energy & Environmental Science. 2013;6:595–607. doi: 10.1039/C2EE23394K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaïki L, Lamberet G, Derré A, Gruss A, Gaudu P. Lactococcus lactis produces short-chain quinones that cross-feed Group B Streptococcus to activate respiration growth. Molecular Microbiology. 2008;67:947–957. doi: 10.1111/j.1365-2958.2007.06083.x. [DOI] [PubMed] [Google Scholar]
- Richter K, Schicklberger M, Gescher J. Dissimilatory reduction of extracellular electron acceptors in anaerobic respiration. Applied and Environmental Microbiology. 2012;78:913–921. doi: 10.1128/AEM.06803-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose ND, Regan JM. Changes in phosphorylation of adenosine phosphate and redox state of nicotinamide-adenine dinucleotide (phosphate) in Geobacter sulfurreducens in response to electron acceptor and anode potential variation. Bioelectrochemistry. 2015;106:213–220. doi: 10.1016/j.bioelechem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Roughead ZK, McCormick DB. Qualitative and quantitative assessment of flavins in cow’s milk. The Journal of Nutrition. 1990;120:382–388. doi: 10.1093/jn/120.4.382. [DOI] [PubMed] [Google Scholar]
- Russell JB, Cook GM. Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol Rev. 1995;59:48–62. doi: 10.1128/mr.59.1.48-62.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvetti E, Fondi M, Fani R, Torriani S, Felis GE. Evolution of lactic acid bacteria in the order Lactobacillales as depicted by analysis of glycolysis and pentose phosphate pathways. Systematic and Applied Microbiology. 2013;36:291–305. doi: 10.1016/j.syapm.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Sauer M, Russmayer H, Grabherr R, Peterbauer CK, Marx H. The efficient clade: lactic acid bacteria for industrial chemical production. Trends in Biotechnology. 2017;35:756–769. doi: 10.1016/j.tibtech.2017.05.002. [DOI] [PubMed] [Google Scholar]
- Schievano A, Pepé Sciarria T, Vanbroekhoven K, De Wever H, Puig S, Andersen SJ, Rabaey K, Pant D. Electro-fermentation - merging electrochemistry with fermentation in industrial applications. Trends in Biotechnology. 2016;34:866–878. doi: 10.1016/j.tibtech.2016.04.007. [DOI] [PubMed] [Google Scholar]
- Schwertmann U, Fischer WR. Natural “amorphous” ferric hydroxide. Geoderma. 1973;10:237–247. doi: 10.1016/0016-7061(73)90066-9. [DOI] [Google Scholar]
- Shi L, Squier TC, Zachara JM, Fredrickson JK. Respiration of metal (hydr)oxides by Shewanella and Geobacter: a key role for multihaem c-type cytochromes. Molecular Microbiology. 2007;65:12–20. doi: 10.1111/j.1365-2958.2007.05783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen RJ, Bayjanov J, Renckens B, Wels M, van Hijum S, Molenaar D, van Hylckama Vlieg JET. Complete genome sequence of Lactococcus lactis subsp. lactis KF147, a plant-associated lactic acid bacterium. Journal of Bacteriology. 2010a;192:2649–2650. doi: 10.1128/JB.00276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen RJ, Tzeneva VA, Castioni A, Wels M, Phan HTK, Rademaker JLW, Starrenburg MJC, Kleerebezem M, Molenaar D, van Hylckama Vlieg JET. Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environmental Microbiology. 2010b;12:758–773. doi: 10.1111/j.1462-2920.2009.02119.x. [DOI] [PubMed] [Google Scholar]
- Sporer AJ, Kahl LJ, Price-Whelan A, Dietrich LEP. Redox-Based Regulation of Bacterial Development and Behavior. Annual Review of Biochemistry. 2017;86:777–797. doi: 10.1146/annurev-biochem-061516-044453. [DOI] [PubMed] [Google Scholar]
- Stookey LL. Ferrozine---a new spectrophotometric reagent for iron. Analytical Chemistry. 2002;42:779–781. doi: 10.1021/ac60289a016. [DOI] [Google Scholar]
- Tachon S, Michelon D, Chambellon E, Cantonnet M, Mezange C, Henno L, Cachon R, Yvon M. Experimental conditions affect the site of tetrazolium violet reduction in the electron transport chain of Lactococcus lactis. Microbiology. 2009;155:2941–2948. doi: 10.1099/mic.0.029678-0. [DOI] [PubMed] [Google Scholar]
- Tachon S, Brandsma JB, Yvon M. NoxE NADH oxidase and the electron transport chain are responsible for the ability of Lactococcus lactis to decrease the redox potential of milk. Applied and Environmental Microbiology. 2010;76:1311–1319. doi: 10.1128/AEM.02120-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachon S, Lee B, Marco ML. Diet alters probiotic Lactobacillus persistence and function in the intestine. Environmental Microbiology. 2014;16:2915–2926. doi: 10.1111/1462-2920.12297. [DOI] [PubMed] [Google Scholar]
- Tamang JP, Cotter PD, Endo A, Han NS, Kort R, Liu SQ, Mayo B, Westerik N, Hutkins R. Fermented foods in a global age: East meets West. Comprehensive Reviews in Food Science and Food Safety. 2020;19:184–217. doi: 10.1111/1541-4337.12520. [DOI] [PubMed] [Google Scholar]
- Taylor RG, Walker DC, McInnes RR. E. coli host strains significantly affect the quality of small scale plasmid DNA preparations used for sequencing. Nucleic Acids Research. 1993;21:1677–1678. doi: 10.1093/nar/21.7.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thavarajah D, Thavarajah P, Abare A, Basnagala S, Lacher C, Smith P, Combs GF. Mineral micronutrient and prebiotic carbohydrate profiles of USA-grown kale (Brassica oleracea L. var. acephala) Journal of Food Composition and Analysis. 2016;52:9–15. doi: 10.1016/j.jfca.2016.07.003. [DOI] [Google Scholar]
- Tomé D. Yeast Extracts: Nutritional and Flavoring Food Ingredients. ACS Food Science & Technology. 2021;1:487–494. doi: 10.1021/acsfoodscitech.0c00131. [DOI] [Google Scholar]
- Tsuji A, Okada S, Hols P, Satoh E. Metabolic engineering of Lactobacillus plantarum for succinic acid production through activation of the reductive branch of the tricarboxylic acid cycle. Enzyme and Microbial Technology. 2013;53:97–103. doi: 10.1016/j.enzmictec.2013.04.008. [DOI] [PubMed] [Google Scholar]
- van Dijk C, Ebbenhorst-Selles T, Ruisch H, Stolle-Smits T, Schijvens E, van Deelen W, Boeriu C. Product and redox potential analysis of sauerkraut fermentation. Journal of Agricultural and Food Chemistry. 2000;48:132–139. doi: 10.1021/jf990720t. [DOI] [PubMed] [Google Scholar]
- Vassilev I, Averesch NJH, Ledezma P, Kokko M. Anodic electro-fermentation: Empowering anaerobic production processes via anodic respiration. Biotechnology Advances. 2021;48:107728. doi: 10.1016/j.biotechadv.2021.107728. [DOI] [PubMed] [Google Scholar]
- Vilas Boas J, Oliveira VB, Marcon LRC, Pinto DP, Simões M, Pinto AMFR. Effect of operating and design parameters on the performance of a microbial fuel cell with Lactobacillus pentosus. Biochemical Engineering Journal. 2015;104:34–40. doi: 10.1016/j.bej.2015.05.009. [DOI] [Google Scholar]
- Vinderola G, Ouwehand A, Salminen S, von Wright A. Lactic Acid Bacteria: Microbiological and Functional Aspects. CRC Press; 2019. [Google Scholar]
- Walther B, Karl JP, Booth SL, Boyaval P. Menaquinones, Bacteria, and the Food Supply: The Relevance of Dairy and Fermented Food Products to Vitamin K Requirements. Advances in Nutrition. 2013;4:463–473. doi: 10.3945/an.113.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wannemuehler MJ, Overstreet AM, Ward DV, Phillips GJ. Draft genome sequences of the altered schaedler flora, a defined bacterial community from gnotobiotic mice. Genome Announcements. 2014;2:e00287-14. doi: 10.1128/genomeA.00287-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg ED. The Lactobacillus anomaly: total iron abstinence. Perspectives in Biology and Medicine. 1997;40:578–583. doi: 10.1353/pbm.1997.0072. [DOI] [PubMed] [Google Scholar]
- Wickham H. ggplot2. Wiley Interdisciplinary Reviews. 2011;3:180–185. doi: 10.1002/wics.147. [DOI] [Google Scholar]
- Wright ME, Yu AO, Marco ML, Panigrahi P. Genome Sequence of Lactiplantibacillus plantarum ATCC 202195, a Probiotic Strain That Reduces Sepsis and Other Infections during Early Infancy. Microbiology Resource Announcements. 2020;9:e00741-20. doi: 10.1128/MRA.00741-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Heeney DD, Srisengfa YT, Chen S-Y, Slupsky CM, Marco ML. Sucrose metabolism alters Lactobacillus plantarum survival and interactions with the microbiota in the digestive tract. FEMS Microbiology Ecology. 2018;94 doi: 10.1093/femsec/fiy084. [DOI] [PubMed] [Google Scholar]
- Yu AO, Goldman EA, Brooks JT, Golomb BL, Yim IS, Gotcheva V, Angelov A, Kim EB, Marco ML. Strain diversity of plant-associated Lactiplantibacillus plantarum. Microbial Biotechnology. 2021;14:1990–2008. doi: 10.1111/1751-7915.13871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Wittouck S, Salvetti E, Franz C, Harris HMB, Mattarelli P, O’Toole PW, Pot B, Vandamme P, Walter J, Watanabe K, Wuyts S, Felis GE, Gänzle MG, Lebeer S. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. International Journal of Systematic and Evolutionary Microbiology. 2020;70:2782–2858. doi: 10.1099/ijsem.0.004107. [DOI] [PubMed] [Google Scholar]