Abstract
Seed size critically affects grain yield of crops and hence represents a key breeding target. The development of embryo-nourishing endosperm is a key driver of seed expansion. We here report unexpected dual roles of the transcription factor EIN3 in regulating seed size. These EIN3 functions have remained largely undiscovered because they oppose each other. Capitalizing on the analysis of multiple ethylene biosynthesis mutants, we demonstrate that EIN3 represses endosperm and seed development in a pathway regulated by ethylene. We, in addition, provide evidence that EIN3-mediated synergid nucleus disintegration promotes endosperm expansion. Interestingly, synergid nucleus disintegration is not affected in various ethylene biosynthesis mutants, suggesting that this promoting function of EIN3 is independent of ethylene. Whereas the growth-inhibitory ethylene-dependent EIN3 action appears to be encoded by sporophytic tissue, the growth-promoting role of EIN3 is induced by fertilization, revealing a generation conflict that converges toward the key signaling component EIN3.
Key words: seed size, EIN3, ethylene biosynthesis, fertilization, generation conflict, synergid disintegration
Seed size can have a critical influence on grain yield. We found that the transcription factor EIN3 has two opposing functions during seed development: a seed growth-promoting role and an ethylene-dependent seed growth-inhibiting role. The two functions are exerted by different tissues, suggesting an EIN3-mediated generation conflict that modulates seed development.
Introduction
Seeds are a key food source for humans and animals and an important basis for biofuel production, making them an important target of breeding programs. In flowering plants, the main components of the seed, the embryo and the embryo-nourishing endosperm, result from fertilization of an egg and an adjoining central cell. The two sperm necessary for this so-called double fertilization are delivered by a single pollen tube, which finds its way to the female gametes due to a sophisticated guidance system (Johnson et al., 2019; Hater et al., 2020; Hafidh and Honys, 2021). Short-range pollen tube attraction is mediated by two egg–cell-adjoining synergid cells, which secrete cysteine-rich peptides (Higashiyama et al., 2001; Márton et al., 2005; Okuda et al., 2009; Takeuchi and Higashiyama, 2012; Meng et al., 2019; Zhong et al., 2019) and mediate the discharge of two sperm from the pollen tubes arriving in the female gametophyte (Huck et al., 2003; Rotman et al., 2003; Amien et al., 2010). Pollen tube arrival is accompanied by programmed cell death of the first synergid. The disintegration of the second synergid and concomitant termination of pollen tube attraction require gamete fusion. This is evidenced by the work of Beale et al. and Kasahara et al., who have shown that incomplete fertilization or the delivery of gamete-fusion-defective sperm suppresses disintegration of the second synergid, resulting in the attraction of supernumerary pollen tubes (Beale et al., 2012; Kasahara et al., 2012).
Synergid disintegration and the establishment of a pollen tube block is a multiphasic process. It involves (i) fertilization-induced cleavage of LURE1 by the egg-secreted endopeptidases ECS1 and ECS2 (Yu et al., 2021), (ii) dilution of LURE by fusion of the synergid with the central cell in a process that requires central cell fertilization (Maruyama et al., 2015), and (iii) synergid nucleus disintegration (Völz et al., 2013; Maruyama et al., 2015). The last step is regulated by fertilization-independent seed–Polycomb Repressive Complex 2 as well as the transcription factors EIN3 and EIL1, which are components of the ethylene response pathway (Maruyama et al., 2013, 2015; Völz et al., 2013). In plants defective for any of these factors, the nucleus of the second synergid remains intact. We, in addition, have shown that synergid-derived nuclei initiate endosperm marker gene expression after fertilization and take on the cell-cycle regime of the endosperm in plants defective for EIN3 (Völz et al., 2013). Consequently, EIN3 prevents the formation of a maternal, haploid synergid-derived endosperm fraction and internuclear heterogeneity. Interploidy crosses have previously suggested that changes in the paternal-to-maternal genome ratio within endosperm nuclei have important implications for seed development, with a relative increase in paternal gene copies promoting seed development, whereas an increase in maternal copies results in smaller seeds (Scott et al., 1998). When analyzing the developmental implications of the internuclear heterogeneity, we found that EIN3 has dual and opposing roles during seed development: while EIN3 in the sporophytic tissue represses seed development, EIN3 signaling activated by fertilization promotes it through the selective degeneration of a synergid nucleus. Our results thus uncover an EIN3-mediated generation conflict that modulates seed development.
Results and discussion
Ploidy differences in the seed can be detected in the zygote stage
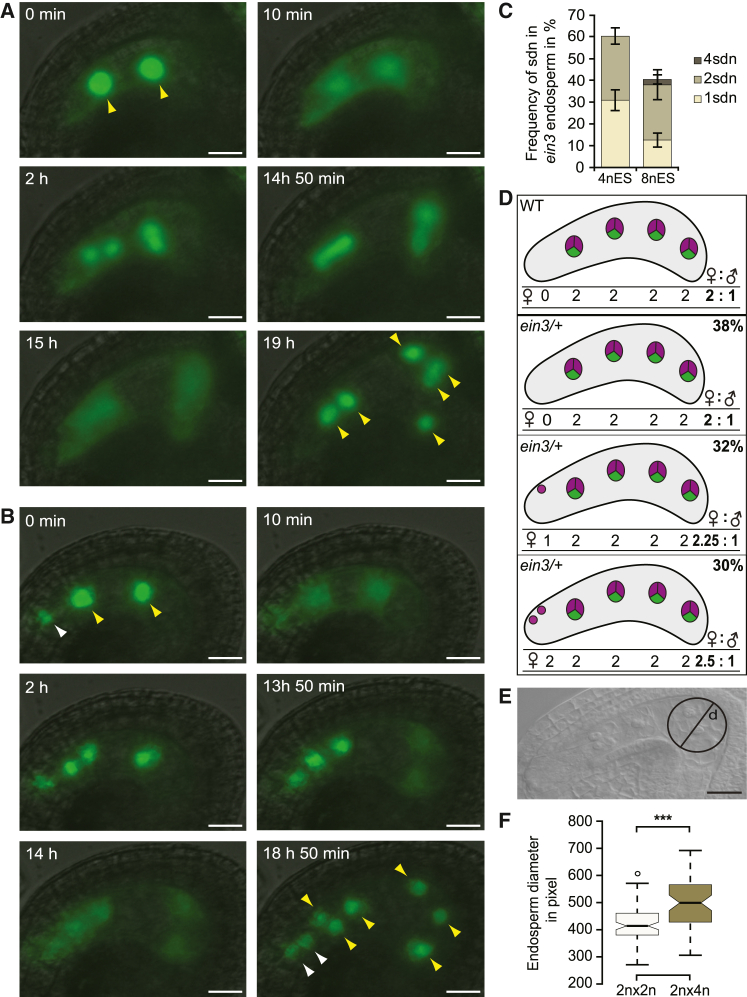
We have previously shown that the synergid-derived nucleus (sdn) in ein3eil1 mutants adopts the fate and division regime of the endosperm, which we also detected in single ein3 mutant (Figures 1A, 1B, and Supplemental Figures 1, and 2). In addition, the sdn incorporates a paternally introduced molecular marker, indicating an internuclear transfer of molecules from the biparental endosperm to the asexual, maternal nuclei (Völz et al., 2013) (Supplemental Figure 1A–1C). To investigate the developmental implications of the resulting parental heterogeneity on endosperm development, we aimed to analyze early seed development at the four-nucleate stage, when the difference in the parental architecture and the concomitant formal shift in maternal-to-paternal genome ratio between ein3 and wild-type endosperm is particularly pronounced (Figure 1C and 1D). In a first step, we asked whether young wild-type seeds at the four-nucleate endosperm stage are already susceptible to changes in maternal-to-paternal ratio. We therefore performed an interploidy cross of diploid wild-type plants with a tetraploid wild-type pollen donor and determined the dorsoventral endosperm diameter in the four-nucleate stage (Figure 1E).
Figure 1.
Ploidy differences in the seed can be detected at the zygote stage.
(A and B) Live-cell imaging of young ein3/− × wild-type seeds without (A) or with (B) sdn. The combinatorial multicolor marker FGR 7.0 confers green fluorescence to sdn and endosperm nuclei. Endosperm nuclei, yellow arrowheads; sdn, white arrowheads. See also Supplemental Videos 1 and 2.
(C) Frequency of one, two, or four sdn in ein3/− × wild-type seeds at the four- (4nES) and eight-nucleate endosperm stage (8nES) (n = 136/166 for 4nES/8nES respectively).
(D) Schematic representation indicating the changes in maternal- (magenta) to-paternal (green) genome ratio in the four-nucleate endosperm of a plant recovered from an ein3/- x wild-type cross containing no sdn, one sdn, or two sdn (n = 338).
(E) Wild-type seed in the four-nucleate endosperm stage. Dorsoventral endosperm diameter was determined by superimposing a circle inside the endosperm at its widest point using ImageJ.
(F) Dorsoventral endosperm diameter of interploidy crosses in wild-type seeds at the four-nucleate endosperm stage (n (2n × 2n) = 84; n (2n × 4n) = 58). Data are the mean ± SEM. Scale bar, 20 μm. Two-tailed Student’s t-test: ∗∗∗p < 0.001.
Figure 2.
Manipulation of EIN3 signaling suggests an inhibitory role of synergid-derived nuclei in endosperm expansion.
(A) Representative cleared whole mounts of ein3/− × wild-type seeds at the four-nucleate endosperm stage containing no sdn (left), one sdn (middle), or two sdn (right) (n = 338). Endosperm nuclei, black arrowheads; sdn, white arrowheads; zygote, asterisk.
(B) Deviation of dorsoventral endosperm diameter of ein3/− × wild-type seeds segregating either one or two sdn at the four-nucleate endosperm stage from the seeds segregating no sdn (n = 130/107/101 for no sdn/1sdn/2sdn, respectively). Endosperm diameter of seeds with either one or two sdn is shown relative to endosperm diameter of seeds without sdn.
(C) Time interval between nuclear disintegration of the two-nucleate and the four-nucleate endosperm stage (n = 42/36 for no sdn/sdn).
(D) Dorsoventral endosperm diameter of ein3-c1/− wild-type seeds segregating no, one, or two sdn at the four-nucleate endosperm stage (n = 45/70/207 for no sdn/1sdn/2sdn, respectively). Endosperm diameter of seeds with either one or two sdn is shown relative to endosperm diameter of seeds without sdn. See also Supplemental Figure 1D–1F.
(E) Dorsoventral endosperm diameter of pDD2::EIN3_SRDX × wild-type young seeds at the four-nucleate endosperm stage. L1, L2, and L3 indicate independent pDD2::EIN3_SRDX lines (n (L1) = 81/104; n (L2) = 73/16; n (L3) = 85/49 for no sdn/sdn, respectively). Data indicate the mean ± SEM. Scale bar, 20 μm. Two-tailed Student’s t-test: ∗p < 0.05; ∗∗∗p < 0.001.
We detected a significantly increased endosperm diameter in seeds having inherited twice as many paternal copies (2n × 4n) (Figure 1F). While we cannot exclude the possibility that increased pollen tube content of 4n plants contributed to this effect (Kasahara et al., 2016; Zhong et al., 2017), these data suggest that even young seeds in the zygotic stage are responsive to ploidy changes.
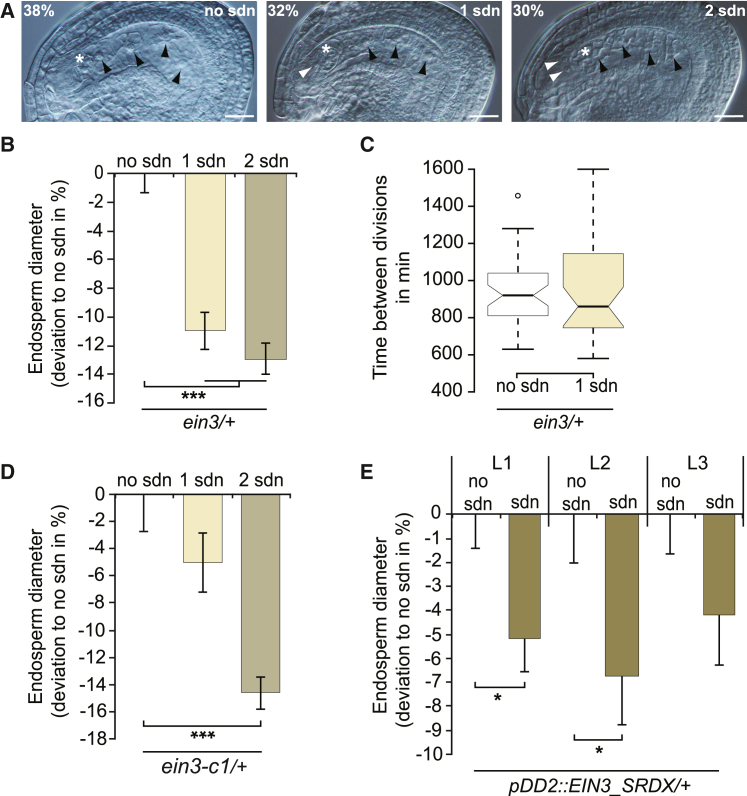
Manipulation of EIN3 signaling suggests an inhibitory role of synergid-derived nuclei in endosperm expansion
In contrast to interploidy crosses, which shift the ploidy of both endosperm and embryo, the overall maternal-to-paternal ratio in ein3 mutants is affected only in the endosperm. In addition, ein3 mutants exhibit a parental internuclear heterogeneity that contrasts with the homogeneous parental ratio characteristic of endosperm nuclei resulting from wild-type or interploidy crosses. To understand whether this idiosyncratic endosperm composition affects endosperm development, we measured dorsoventral endosperm diameter of ein3 seeds in the four-nucleate stage. We capitalized on our previous finding that the defect of synergid nucleus inheritance is not fully penetrant in ein3 mutants (Figures 1C and 2A) (Völz et al., 2013), i.e., we were able to compare seeds from the same flowers that either had or had not inherited sdn. We found that endosperm expansion was significantly reduced in ein3 seeds containing both biparental endosperm and sdn compared with ein3 seeds with biparental endosperm only (Figure 2B). While this result suggests that sdn negatively affect endosperm expansion, we could not rule out the possibility that development in sdn-segregating ovules was retarded. In fact, interploidy crosses performed by Scott et al. showed that increased maternal copies correlated with delayed mitotic progression and premature cellularization (Scott et al., 1998). To test this hypothesis, we assessed endosperm size dynamics on the basis of live-cell imaging. We introduced a combinatorial multicolor marker, FGR 7.0, into ein3 plants. FGR 7.0 confers fluorescence to synergids, zygotes, and endosperm (Völz et al., 2013). In addition, we established a protocol for visualization of nuclear dynamics in early endosperm. This allowed us to trace fertilized ovules over a period of 24 h, during which the endosperm underwent up to three mitotic divisions (Figure 1A and 1B; Supplemental Videos 1 and 2). To determine whether early seed development is slowed down in sdn-containing seeds, we used the decondensation of nuclei as a molecular timer. The durations of nuclear disintegration between the two-nucleate endosperm stage and the four-nucleate endosperm stage were comparable, independent of the segregation of sdn (Figure 2C). Together, these results indicate that the size differences are not an artifact introduced by retarded development, but that instead the segregation of sdn correlates with reduced early endosperm expansion.
Pictures were taken every 10 min. The last frame before nuclear decondensation in the two-nucleate endosperm stage was set to 0. Frame numbers are indicated in the video.
Pictures were taken every 10 min. The last frame before nuclear decondensation in the two-nucleate endosperm stage was set to 0. Frame numbers are indicated in the video.
To test whether this effect was indeed causally linked to the ein3 locus, we generated a CRISPR-induced EIN3 allele (ein3-c1). This allele contains a frameshift insertion at the position of the 495th base, resulting in a premature stop codon after 165 amino acids (Supplemental Figure 1D and 1E). The ein3-c1 allele exhibits a stronger phenotype than the ein3 allele with respect to the frequency of sdn-segregating seeds (Supplemental Figure 1F), whereas the overall effect of sdn on endosperm size was comparable (Figure 2D).
The correlation between sdn inheritance and endosperm size is compatible with two conceptually different scenarios: either endosperm expansion and the degeneration of the synergid nucleus are different effects of EIN3-dependent processes operating in the female gametophyte, or the persistence of the maternal haploid synergid nuclei is causally linked with reduced endosperm expansion. To discriminate between the two scenarios, we confined the EIN3-dependent defect to synergids only making use of the SRDX repressor motif (Hiratsu et al., 2003). We have previously shown that expression of the dominant negative EIN3_SRDX fusion under the control of the synergid-specific pDD2 promoter phenocopies the synergid nuclear disintegration defect of ein3 mutants (Völz et al., 2013). When analyzing the endosperm diameter of different pDD2::EIN3_SRDX transgenic lines, we observed substantial phenotypic variations. However, as a common denominator, we found that endosperm size is reduced in the presence of sdn compared with seeds without sdn (Figure 2E). As a control, we expressed the construct after fertilization only in the endosperm using the AtrBohD promoter (Völz et al., 2013). This approach did not affect synergid disintegration (n = 136/146/203 for wild type/L1/L2), nor did we observe reduced seed size (Supplemental Figure 1G), suggesting that the endosperm does not contribute to the effect in sdn-containing pDD2::EIN3_SRDX.
Together, our results indicate that internuclear heterogeneity caused by sdn and reduced endosperm size are causally linked. This also implies that fertilization-dependent synergid nuclei disintegration mediated by EIN3 promotes endosperm expansion. This finding is also in line with and in support of the parental conflict theory, which holds that both parents have different interests in the allocation of resources to a single seed of the same mother plant (Haig and Westoby, 1989).
Sporophytic EIN3-dependent signaling represses endosperm and seed expansion
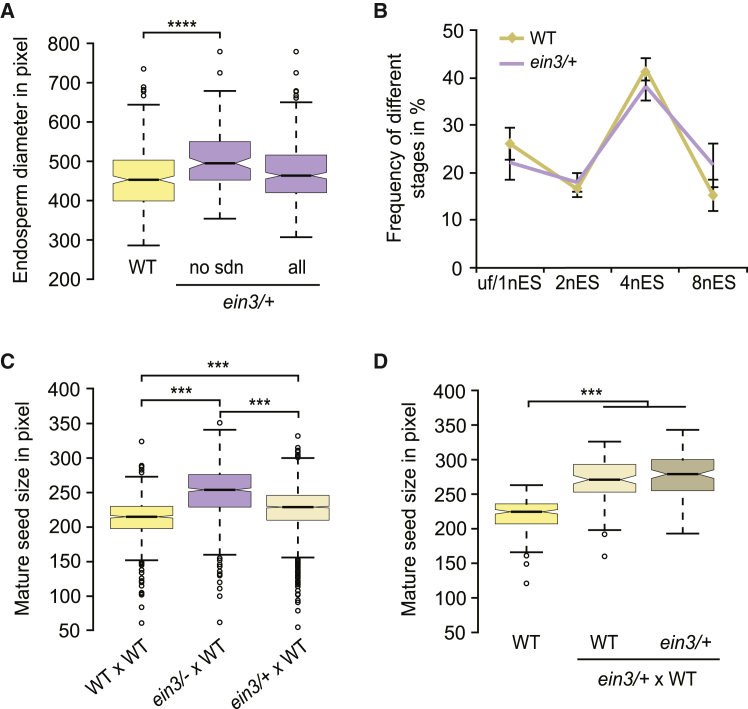
While young sdn-segregating ein3 seeds have a reduced endosperm diameter, previous results have reported an increased mature seed size of ein3 mutants resulting from inhibitory effects of EIN3 on embryo development (Meng et al., 2018). We similarly observed an effect on seed size in both ein3 and ein3-c1 alleles (Supplemental Figure 2A–2C). Notably, this effect was evident only when we introduced the mutation through the female (Supplemental Figure 2B). We next asked whether and to what extent a growth-inhibiting effect of EIN3 is observable at an early developmental stage. In fact, we detected an EIN3-dependent growth-inhibiting effect also in early developmental stages, where it is masked by the opposing sdn-dependent effect (Figure 3A). Combining the all, sdn-segregating, and no sdn-segregating ein3 categories yields an endosperm diameter comparable to that of wild type, while the endosperm diameter in ein3 seeds without sdn is significantly bigger compared with wild type (Figure 3A). Since there is no significant developmental shift between wild type and ein3 (Figure 3B), we can exclude that size deviations are caused by different developmental stages, which was further substantiated by live-cell imaging and by analyzing the ein3-c1 allele (Supplemental Figure 2D–2F).
Figure 3.
Sporophytic EIN3-dependent signaling represses endosperm and seed expansion.
(A) Dorsoventral endosperm diameter of wild-type (WT) × WT and ein3/− × WT seeds in the four-nucleate endosperm stage. “All” indicates ein3/+ seeds of ein3/− × WT crosses with and without sdn (n = 289/130/338 for WT seeds/ ein3/+ seeds with no sdn/ all of ein3/+ seeds).
(B) Frequency of WT × WT and ein3/− × WT seeds in different developmental stages 24 h after pollination: unfertilized and freshly fertilized seeds in the one-nucleate endosperm stage (uf/1nES) and two-, four-, and eight-nucleate endosperm staged seeds (2nES, 4nES, and 8nES, respectively) (n = 537/570 for WT/ein3/+).
(C) Mature seed size (n = 689/770/2079 for WT × WT seeds/ein3/− × WT seeds/ein3/+ × WT seeds, respectively).
(D) Independent experiment showing size of WT × WT and ein3/+ × WT mature seeds. Seeds were assigned according to the genotype of their progeny (n = 198/109/130 for WT/segregating WT/segregating ein3/+, respectively). Two-tailed Student’s t-test: ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001. See also Supplemental Figure 2.
We next asked whether the repressive effect of EIN3 originated from the gametophyte or the sporophyte. To discriminate between sporophytic and gametophytic function we compared seeds from wild-type, ein3 homozygous, or ein3 heterozygous plants pollinated with wild type. While ein3 heterozygous seeds were smaller than ein3 homozygous seeds, they were still significantly bigger than wild-type seeds (Figure 3C). This result is compatible with two different scenarios: either EIN3 has a dose-dependent sporophytic effect, which would affect the size of all seeds, or the intermediate seed size results from a mixed filial generation containing smaller wild type segregating and bigger ein3/+ segregating seeds. To distinguish between the two scenarios, we followed individual seeds to the seedling stage, where we genotyped them. Our results revealed that all seeds from ein3 heterozygous plants are significantly bigger than wild type but similar in size independent of the genotype of their embryo (Figure 3D). This result indicates that the EIN3 growth-inhibiting effect is attributable to the sporophytic tissue.
Together, these data indicate that EIN3 has opposing and spatially distinct roles in seed expansion: the EIN3 growth-inhibiting function is regulated by the sporophytic tissue, and this effect is counteracted in early endosperm stages by a growth-promoting EIN3 function, which is mediated by disintegration of the non-receptive synergid nucleus after fertilization. In mature seeds, the effect of sdn appears to become dominated by the growth-inhibiting effect of EIN3, potentially due to the fact that the endosperm is degraded during seed development.
Ethylene reduction affects the growth-inhibitory but not the growth-promoting function of EIN3
EIN3 is stabilized in the presence of ethylene (Guo and Ecker, 2003; Potuschak et al., 2003). In addition to its fundamental role in many developmental processes and stress responses, the plant hormone has a regulatory role in cell division, cell expansion, and growth (Abeles et al., 1992; Dubois et al., 2018). In light of the opposing effects of EIN3, we next asked whether the growth-promoting and the inhibitory roles of EIN3 equally respond to ethylene.
Ethylene biosynthesis is initiated by the production of S-adenosylmethionine from methionine by S-adenosylmethionine synthase (SAM), which is followed by two additional steps: first, S-adenosyl-L-methionine is converted to 1-aminocyclopropane-1-carboxylic acid (ACC) by aminocyclopropane-1-carboxylic acid synthase (ACS), and second, ACC is converted to ethylene by aminocyclopropane-1-carboxylic acid oxidase (ACO) (Adams and Yang, 1979; Pattyn et al., 2020).
To trace ethylene biosynthesis in time and space, we generated transcriptional and translational reporter lines and analyzed 15 members of the SAM, ACS, and ACO enzyme families involved in ethylene biosynthesis. Twenty-four hours after fertilization, when the endosperm effect of ein3 was already evident, we detected SAM1 and SAM2 in the sporophyte, endosperm, and zygote; SAM3 in the endosperm and zygote; pACS2 in the endosperm only; pACS6 and pACO2 in the sporophyte only; and pACO5 in the endosperm of young seeds (Supplemental Figure 3A–3C). This result is also supported by a recent study revealing that ethylene production as well as mRNA abundance of some ACS genes gradually increases during seed development (Sun et al., 2020).
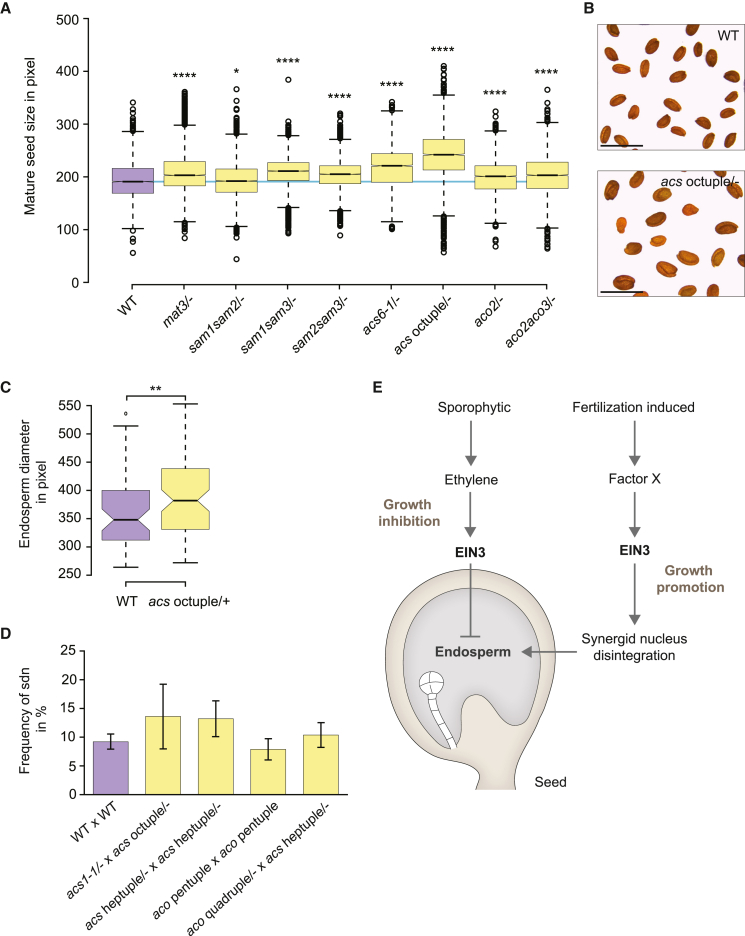
We next capitalized on ethylene biosynthesis mutants to address the functional relevance of ethylene production during seed development. It was previously reported that the level of ethylene production in Arabidopsis is directly regulated by ACSs (Tsuchisaka et al., 2009) and SAMs (Mao et al., 2015). When analyzing dry seed size of various mutants and mutant combinations targeting SAM, ACS, and/or ACO genes, we detected an increase in seed size compared with wild type (Figure 4A). The effect was particularly pronounced in acs octuple seeds in which ethylene production has previously been shown to be strongly reduced (Tsuchisaka et al., 2009) (Figure 4A and 4B). Except for acs octuple mutants, which show 49% non-developing/sterile ovules in mature siliques (n = 910), all mutants show fertile siliques, indicating that the larger seed size is not due to additional resources freed up by the formation of fewer seeds (Supplemental Figure 4A). On closer inspection of acs octuple mutants, we found that a substantial fraction exhibited integument abnormalities and early defects in female gametophyte development (Supplemental Figure 4D), which potentially contribute to a previously described defect of acs octuple mutants in pollen tube attraction (Mou et al., 2020). In addition, we observed an increased dorsoventral endosperm diameter in the four-nucleate stage, indicating that seed size deviation initiates early in acs seed development (Figure 4C).
Figure 4.
Ethylene reduction affects the growth-inhibitory but not the growth-promoting function of EIN3.
(A) Dry seed size of different ethylene biosynthetic mutants and wild type (WT) after selfing (n = 6508/5322/4751/4736/5390/2911/3453/4966/5597 for WT/mat3/−/sam1sam2/−/sam1sam3/−/sam2sam3/−/acs6-1/−/acs octuple/−/aco2/−/aco2aco3/−). The area of each seed collected from 15 individual plants sown at three different times was measured in ImageJ. A boxplot was generated based on individual seed area calculated in pixels. The blue line shows the WT median.
(B) Dry seeds of WT and acs octuple/− plants. Scale bars, 1 mm.
(C) Dorsoventral endosperm diameter of WT × WT and acs octuple/− × WT seeds in the four-nucleate endosperm stage. Seeds showing abnormal development at the micropylar end (see also Supplemental Figure 4B and 4C) are excluded (n = 53/79 for WT/acs octuple/−).
(D) Frequency of sdn in the seeds in the four- and eight-nucleate endosperm stages (n = 531, 125, 227, 418, 289 for WT × WT, acs1-1/− × acs octuple/−, acs heptuple/− × acs heptuple/−, aco pentuple × aco pentuple, aco quadruple/− × acs heptuple/−). Two-tailed Student’s t-test did not detect significant difference between mutants and WT for sdn frequency.
(E) Schematic model of dual and opposing roles of EIN3 during seed growth. Two-tailed Student’s t-test between WT and mutants: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001. See also Supplemental Figure 4.
We next asked whether synergid degeneration was also susceptible to a reduction of ethylene content. To characterize synergid disintegration in young acs octuple seeds, we analyzed cleared whole mounts 1 day after pollination. Interestingly, only 6% (n = 105) of the analyzed acs octuple seeds showed sdn in the four-nucleate endosperm stage, which is similar to the 4% observed in wild type (n = 53). Similar results were obtained when analyzing various combinations of acs and aco mutant lines, suggesting that synergid disintegration requires no or only a small amount of ethylene (Figure 4D). These results are consistent with recent work by Li et al., which shows that synergid disintegration is not affected in plants fully depleted of ACO function (Li et al., 2021). Our data suggest that synergid degeneration and the concomitant EIN3 growth-promoting function are not or are less responsive to ethylene, indicating that there might be a factor X activating EIN3 signaling after fertilization. Candidates include jasmonic acid (Zhu et al., 2011), salicylic acid (He et al., 2017), or salt (Peng et al., 2014), which have previously been implicated in the regulation of EIN3.
In conclusion, our findings unravel an unexpected dual role of EIN3 during early seed development, a function that is masked in wild-type plants because the effects oppose each other. Intriguingly, the effects are exerted by different tissues representing different generations: while sporophytic tissue represses endosperm and seed expansion in an EIN3-dependent manner, fertilization-triggered synergid disintegration promotes endosperm expansion, thereby ensuring the biparental origin of all endosperm nuclei. Our results, in addition, suggest that the dual and conflicting processes exerted by the two generations differ with respect to ethylene responsiveness: while the depletion of key ethylene biosynthesis genes affects seed expansion, synergid disintegration is not affected, suggesting that the latter process is either fully ethylene independent or sensitive to small traces of ethylene (Figure 4E). Given that ethylene integrates various external and internal stresses, it will be an attractive challenge for the future to determine whether and to what extent endosperm development is amenable to adaptation and how the different functions of EIN3 are regulated.
Methods
Plant materials and growth conditions
Seeds of Arabidopsis thaliana were sown and stratified at 4°C for 2 days. Stratified seeds were transferred to a Conviron MTPS growth chamber for germination and further growth under long-day conditions (16 h light/8 h dark) at 23°C. Plants were later transferred to 18°C after bolting.
The following plant lines are in the Ler background: ein3-1 (referred to as ein3 in this study), ein3-1eil1-2 (referred to as ein3eil1 in this study), ein3eil1 with pMEA::NLS_tdTomato or pRPS5a::NLS_GFP; pDD2::EIN3_SRDX (Völz et al., 2013). The ein3-1eil1-2 double mutant was kindly provided by Richard D. Vierstra. ein3-1 was crossed out with Ler wild type. For live-cell imaging, FGR 7.0 lines in the Ler and ein3 background were used (Völz et al., 2013). For interploidy assay, 2n Ler and 4n Ler kindly provided by Prof. Dr. Tobias Würschum were used.
Ethylene biosynthetic mutants were obtained from the European Arabidopsis Stock Center (NASC) (Nottingham, UK): sam1 (N573599), sam2 (N676306), sam3 (N552289), mat3 (N519375), acs6-1 (N16569), acs heptuple (N16650), acs octuple (N16651), aco1 (N682904), aco2 (N527311), aco3 (N582132), aco4 (N514965), and aco5-2 (N411335). sam1sam2, sam1sam3, sam2sam3, and aco2aco3, aco quadruple (aco1/−aco2/−aco3/−aco4/−), and aco pentuple (aco1/−aco2/−aco3/−aco4/−aco5-2/+) were generated by crossings of the respective single mutants. Col-0 was used as a control.
Generation of ein3-CRISPR line (ein3-c1)
All constructs as well as cloning procedures were described previously by Fauser et al. (2014). The protospacer used as a recognition site for the Cas9 nuclease was localized in the exon at position 478–498 of the coding sequence (Supplemental Table 1) and was followed by an AGG protospacer-adjacent motif. The thereby induced mutation has a 1 nt insertion between positions 494 and 495 of the coding sequence and was named ein3-c1 (Supplemental Figure 1D). This insertion disrupts an NlaIII restriction site important for genotyping and induces a frameshift resulting in a premature stop codon after 165 of 628 amino acids. The ein3-c1 mutant was outcrossed several times to remove the CAS9 gene and to reduce off-target mutations. The homozygous mutant was then used for further analyses. Ler and ein3 were used as controls.
Molecular cloning
To generate pSAMX::gSAMX_tdTomato_tNOS plasmids, the promoter and genomic loci of SAM1 (AT1G02500), SAM2 (AT4G01850), and SAM3 (AT3G17390) were amplified from the Arabidopsis Col-0 DNA library by the respective primers listed in Supplemental Table 1. The promoters digested with AscI and PacI and the genomic fragments digested with PacI and AvrII were subcloned into DR13 plasmid (pAt5g40260::NLS_tdTomato_tNOS) (Völz et al., 2013) followed by exchanging pAt5g40260 with pSAMX and NLS with gSAMX. In addition, to generate pACSX::NLS_GUS_tNOS plasmids, the promoters of ACS2 (AT1G01480), ACS4 (AT2G22810), ACS5 (AT5G65800), ACS6 (AT4G11280), ACS7 (AT4G26200), ACS8 (AT4G37770), ACS9 (AT3G49700), and ACS11 (AT4G08040) were amplified from the Arabidopsis Ler DNA library by the respective primers listed in Supplemental Table 1. The promoters pACS2, pACS6, and pACS11 were digested with AscI and PacI, whereas the promoters pACS4, pACS5, pACS7, and pACS9 were digested with AscI and PvuI, and the promoter pACS8 was digested with AscI and XhoI. Afterward, they were subcloned into pLIS::NLS_GUS_tNOS (Groß-Hardt et al., 2007), followed by exchanging pLIS with pACSX. Last, to generate pACOX::NLS_tdTomato_tNOS plasmids, the promoters of ACO1 (AT2G19590), ACO2 (AT1G62380), ACO3 (AT1G12010), and ACO5 (AT1G77330) were amplified from the Arabidopsis Ler DNA library by the respective primers listed in Supplemental Table 1. The promoters digested with AscI and PacI were subcloned into DR13 plasmid followed by exchanging pAt5g40260 with pACOX. All plasmids were then transformed into Col-0 plants by floral dip as previously described (Zhang et al., 2006).
PCR-based genotyping
Genotyping primers are listed in Supplemental Table 2.
Histology and microscopy
For the analysis of early seed development, the oldest closed flower bud of a given inflorescence was emasculated. One day after emasculation, the flowers were pollinated with wild-type pollen and harvested 24 h later.
For whole-mount clearings flowers were vacuum infiltrated in an ethanol:acetic acid solution (9:1) for 30 min, kept at 4°C overnight, washed for 1 h each with 80% and 70% ethanol, and mounted in chloral hydrate:glycerol:water solution (8:2:1; w:v:v). Cytochemical staining of GUS activity was performed on samples as described previously (Vielle-Calzada et al., 2000). GUS-stained samples as well as cleared whole mounts were then visualized under a Zeiss Axioscope (Zeiss, Oberkochen, Germany) and images were captured by a Canon PowerShot G10 camera. Fluorescence signals were detected by a Leica DMI6000B microscope (Leica Microsystems, Wetzlar, Germany).
Live-cell imaging
Flowers were used 20 h after pollination to perform live-cell imaging. Pistils were harvested and the two septa were separated by an apical–basal incision alongside the transmitting tract by using an insulin syringe (BD MicroFine). The septum halves with the attached ovules were transferred to an ovule medium modified after Palanivelu et al. (2003): 1 mM MgSO4, 4 mM CaCl2, 0.01% H2BO3, 3% PEG 4000, 14.5% sucrose (pH 5.9 adjusted with KOH), and 1.5% NuSieve GTG agarose (Lonza Bioscience). Subsequently the ovules were covered with 200 μl halocarbon oil 700 (Sigma-Aldrich). Live-cell imaging was performed using a Leica DMI6000B microscope (Leica Microsystems, Wetzlar, Germany) equipped with LAS AF version 2.2.1. Ovules in the two-endosperm stage were selected by using the mark and find function. The images were taken every 10 min over a period of 24 h. Four-nucleate endosperm duration was determined by using the time points when the fluorescence signal was still nuclear localized, shortly before nuclear division at the two- and four-nucleate endosperm stages.
Seed size measurement
Mature seeds were harvested and dried. Dry seeds were scanned by a CanoScan 9000F Mark II in black/white mode with transmitting light and 1200 dpi resolution. Seed area measurement was performed as described previously by using ImageJ (Herridge et al., 2011).
Data analysis
Datasets were analyzed using Microsoft Excel 2007. Bar charts were created in Microsoft Excel and modified in Adobe Illustrator. Boxplot graphs were generated with BoxPlotR (Spitzer et al., 2014) and modified in Adobe Illustrator. Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range (IQR) from the 25th and 75th percentiles, and outliers are represented by dots. Crosses indicate the mean. The notches are defined as ±1.58 ∗ IQR/sqrt(n) and represent the 95% confidence interval for each median. Statistical analyses were performed in the Analysis ToolPak of Microsoft Excel 2007.
Funding
We gratefully acknowledge financial support from the European Research Council to R.G. (ERC Consolidator Grant "bi-BLOCK" ID 646644, ERC Proof of Concept Grant "TriVolve" ID 957547).
Author contributions
Conceptualization, J.H., I.E.S., R.V., and R.G.; methodology, J.H., I.E.S., and R.G.; investigation, J.H., I.E.S., T.H., D.V., Y.M., R.V., S.G., and T.N; visualization, J.H., I.E.S., Y.M., and R.G.; writing – original draft, J.H., I.E.S., and R.G.; writing – review & editing, J.H., I.E.S., and R.G.; resources and funding acquisition, R.G.
Acknowledgments
We thank Prof. Dr. Holger Puchta (Karlsruhe Institute of Technology, Germany) for providing the pEN-Chimera and pDe-Cas9 plasmids and Prof. Dr. Tobias Würschum (The University of Hohenheim) for providing 4n Ler seeds. We thank members of the R.G. laboratory for comments on the manuscript. No conflict of interest is declared.
Published: November 27, 2021
Footnotes
Published by the Molecular Plant Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, SIBS, CAS.
Supplemental information is available at Molecular Plant Online.
Supplemental information
References
- Abeles F.B., Morgan P.W., Saltveit M.E. Ethylene in Plant Biology. 2nd edn. Academic Press; New York: 1992. pp. 297–398. [DOI] [Google Scholar]
- Adams D., Yang S. Ethylene biosynthesis: identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc. Natl. Acad. Sci. U S A. 1979;76:170–174. doi: 10.1073/pnas.76.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amien S., Kliwer I., Márton M.L., Debener T., Geiger D., Becker D., Dresselhaus T. Defensin-like ZmES4 mediates pollen tube burst in maize via opening of the potassium channel KZM1. PLoS Biol. 2010;8:e1000388. doi: 10.1371/journal.pbio.1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale K.M., Leydon A.R., Johnson M.A. Gamete fusion is required to block multiple pollen tubes from entering an Arabidopsis ovule. Curr. Biol. 2012;22:1090–1094. doi: 10.1016/j.cub.2012.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M., Van den Broeck L., Inzé D. The pivotal role of ethylene in plant growth. Trends Plant Sci. 2018;23:311–323. doi: 10.1016/j.tplants.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauser F., Schiml S., Puchta H. Both CRISPR/C as-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 2014;79:348–359. doi: 10.1111/tpj.12554. [DOI] [PubMed] [Google Scholar]
- Groß-Hardt R., Kägi C., Baumann N., Moore J.M., Baskar R., Gagliano W.B., Jürgens G., Grossniklaus U. LACHESIS restricts gametic cell fate in the female gametophyte of Arabidopsis. PLoS Biol. 2007;5:e47. doi: 10.1371/journal.pbio.0050047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Ecker J.R. Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell. 2003;115:667–677. doi: 10.1016/s0092-8674(03)00969-3. [DOI] [PubMed] [Google Scholar]
- Hafidh S., Honys D. Reproduction multitasking: the male gametophyte. Annu. Rev. Plant Biol. 2021;72:581–614. doi: 10.1146/annurev-arplant-080620-021907. [DOI] [PubMed] [Google Scholar]
- Haig D., Westoby M. Parent-specific gene expression and the triploid endosperm. Am. Nat. 1989;134:147–155. [Google Scholar]
- Hater F., Nakel T., Groß-Hardt R. Reproductive multitasking: the female gametophyte. Annu. Rev. Plant Biol. 2020;71:517–546. doi: 10.1146/annurev-arplant-081519-035943. [DOI] [PubMed] [Google Scholar]
- He X., Jiang J., Wang C.Q., Dehesh K. ORA59 and EIN3 interaction couples jasmonate-ethylene synergistic action to antagonistic salicylic acid regulation of PDF expression. J. Integr. Plant Biol. 2017;59:275–287. doi: 10.1111/jipb.12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herridge R.P., Day R.C., Baldwin S., Macknight R.C. Rapid analysis of seed size in Arabidopsis for mutant and QTL discovery. Plant Methods. 2011;7:3. doi: 10.1186/1746-4811-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama T., Yabe S., Sasaki N., Nishimura Y., Miyagishima S.-Y., Kuroiwa H., Kuroiwa T. Pollen tube attraction by the synergid cell. Science. 2001;293:1480–1483. doi: 10.1126/science.1062429. [DOI] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003;34:733–739. doi: 10.1046/j.1365-313x.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- Huck N., Moore J.M., Federer M., Grossniklaus U. The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development. 2003;130:2149–2159. doi: 10.1242/dev.00458. [DOI] [PubMed] [Google Scholar]
- Johnson M.A., Harper J.F., Palanivelu R. A fruitful journey: pollen tube navigation from germination to fertilization. Annu. Rev. Plant Biol. 2019;70:809–837. doi: 10.1146/annurev-arplant-050718-100133. [DOI] [PubMed] [Google Scholar]
- Kasahara R.D., Maruyama D., Hamamura Y., Sakakibara T., Twell D., Higashiyama T. Fertilization recovery after defective sperm cell release in Arabidopsis. Curr. Biol. 2012;22:1084–1089. doi: 10.1016/j.cub.2012.03.069. [DOI] [PubMed] [Google Scholar]
- Kasahara R.D., Notaguchi M., Nagahara S., Suzuki T., Susaki D., Honma Y., Maruyama D., Higashiyama T. Pollen tube contents initiate ovule enlargement and enhance seed coat development without fertilization. Sci. Adv. 2016;2:e1600554. doi: 10.1126/sciadv.1600554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li Q., Lyu M., Wang Z., Song Z., Zhong S., Gu H., Dong J., Dresselhaus T., Zhong S. Lack of ethylene does not affect reproductive success and synergid cell death in Arabidopsis. Mol. Plant. 2021 doi: 10.1016/j.molp.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao D., Yu F., Li J., Van de Poel B., Tan D., Li J., Liu Y., Li X., Dong M., Chen L. FERONIA receptor kinase interacts with S-adenosylmethionine synthetase and suppresses S-adenosylmethionine production and ethylene biosynthesis in Arabidopsis. Plant Cell Environ. 2015;38:2566–2574. doi: 10.1111/pce.12570. [DOI] [PubMed] [Google Scholar]
- Márton M.L., Cordts S., Broadhvest J., Dresselhaus T. Micropylar pollen tube guidance by egg apparatus 1 of maize. Science. 2005;307:573–576. doi: 10.1126/science.1104954. [DOI] [PubMed] [Google Scholar]
- Maruyama D., Hamamura Y., Takeuchi H., Susaki D., Nishimaki M., Kurihara D., Kasahara Ryushiro D., Higashiyama T. Independent control by each female gamete prevents the attraction of multiple pollen tubes. Dev. Cell. 2013;25:317–323. doi: 10.1016/j.devcel.2013.03.013. [DOI] [PubMed] [Google Scholar]
- Maruyama D., Völz R., Takeuchi H., Mori T., Igawa T., Kurihara D., Kawashima T., Ueda M., Ito M., Umeda M. Rapid elimination of the persistent synergid through a cell fusion mechanism. Cell. 2015;161:907–918. doi: 10.1016/j.cell.2015.03.018. [DOI] [PubMed] [Google Scholar]
- Meng J.G., Zhang M.X., Yang W.C., Li H.J. TICKET attracts pollen tubes and mediates reproductive isolation between relative species in Brassicaceae. Sci. China Life Sci. 2019;62:1413–1419. doi: 10.1007/s11427-019-9833-3. [DOI] [PubMed] [Google Scholar]
- Meng L.-S., Xu M.-K., Wan W., Wang J.-Y. Integration of environmental and developmental (or metabolic) control of seed mass by sugar and ethylene metabolisms in Arabidopsis. J. Agric. Food Chem. 2018;66:3477–3488. doi: 10.1021/acs.jafc.7b05992. [DOI] [PubMed] [Google Scholar]
- Mou W., Kao Y.-T., Michard E., Simon A.A., Li D., Wudick M.M., Lizzio M.A., Feijó J.A., Chang C. Ethylene-independent signaling by the ethylene precursor ACC in Arabidopsis ovular pollen tube attraction. Nat. Commun. 2020;11:1–11. doi: 10.1038/s41467-020-17819-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda S., Tsutsui H., Shiina K., Sprunck S., Takeuchi H., Yui R., Kasahara R.D., Hamamura Y., Mizukami A., Susaki D. Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature. 2009;458:357–361. doi: 10.1038/nature07882. [DOI] [PubMed] [Google Scholar]
- Palanivelu R., Brass L., Edlund A.F., Preuss D. Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell. 2003;114:47–59. doi: 10.1016/s0092-8674(03)00479-3. [DOI] [PubMed] [Google Scholar]
- Pattyn J., Vaughan-Hirsch J., Van de Poel B. The regulation of ethylene biosynthesis: a complex multilevel control circuitry. New Phytol. 2020;229:770–782. doi: 10.1111/nph.16873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Li Z., Wen X., Li W., Shi H., Yang L., Zhu H., Guo H. Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PLoS Genet. 2014;10:e1004664. doi: 10.1371/journal.pgen.1004664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T., Lechner E., Parmentier Y., Yanagisawa S., Grava S., Koncz C., Genschik P. EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell. 2003;115:679–689. doi: 10.1016/s0092-8674(03)00968-1. [DOI] [PubMed] [Google Scholar]
- Rotman N., Rozier F., Boavida L., Dumas C., Berger F., Faure J.-E. Female control of male gamete delivery during fertilization in Arabidopsis thaliana. Curr. Biol. 2003;13:432–436. doi: 10.1016/s0960-9822(03)00093-9. [DOI] [PubMed] [Google Scholar]
- Scott R.J., Spielman M., Bailey J., Dickinson H.G. Parent-of-origin effects on seed development in Arabidopsis thaliana. Development. 1998;125:3329–3341. doi: 10.1242/dev.125.17.3329. [DOI] [PubMed] [Google Scholar]
- Spitzer M., Wildenhain J., Rappsilber J., Tyers M. BoxPlotR: a web tool for generation of box plots. Nat Methods. 2014;11:121. doi: 10.1038/nmeth.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Li J.Q., Yan J.Y., Yuan J.J., Li G.X., Wu Y.R., Xu J.M., Huang R.F., Harberd N.P., Ding Z.J. Ethylene promotes seed iron storage during Arabidopsis seed maturation via ERF95 transcription factor. J. Integr. Plant. Biol. 2020;62:1193–1212. doi: 10.1111/jipb.12986. [DOI] [PubMed] [Google Scholar]
- Takeuchi H., Higashiyama T. A species-specific cluster of defensin-like genes encodes diffusible pollen tube attractants in Arabidopsis. PLoS Biol. 2012;10:e1001449. doi: 10.1371/journal.pbio.1001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka A., Yu G., Jin H., Alonso J.M., Ecker J.R., Zhang X., Gao S., Theologis A. A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics. 2009;183:979–1003. doi: 10.1534/genetics.109.107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calzada J.-P., Baskar R., Grossniklaus U. Delayed activation of the paternal genome during seed development. Nature. 2000;404:91–94. doi: 10.1038/35003595. [DOI] [PubMed] [Google Scholar]
- Völz R., Heydlauff J., Ripper D., von Lyncker L., Groß-Hardt R. Ethylene signaling is required for synergid degeneration and the establishment of a pollen tube block. Dev. Cell. 2013;25:310–316. doi: 10.1016/j.devcel.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Yu X., Zhang X., Zhao P., Peng X., Chen H., Bleckmann A., Bazhenova A., Shi C., Dresselhaus T., Sun M.-x. Fertilized egg cells secrete endopeptidases to avoid polytubey. Nature. 2021;592:433–437. doi: 10.1038/s41586-021-03387-5. [DOI] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.-S., Niu Q.-W., Chua N.-H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006;1:641. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- Zhong S., Zhang J., Qu L.-J. The signals to trigger the initiation of ovule enlargement are from the pollen tubes: the direct evidence. J. Integr. Plant Biol. 2017;59:600–603. doi: 10.1111/jipb.12577. [DOI] [PubMed] [Google Scholar]
- Zhong S., Liu M., Wang Z., Huang Q., Hou S., Xu Y.-C., Ge Z., Song Z., Huang J., Qiu X., et al. Cysteine-rich peptides promote interspecific genetic isolation in Arabidopsis. Science. 2019;364:eaau9564. doi: 10.1126/science.aau9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., An F., Feng Y., Li P., Xue L., Mu A., Jiang Z., Kim J.-M., To T.K., Li W., et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2011;108:12539–12544. doi: 10.1073/pnas.1103959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pictures were taken every 10 min. The last frame before nuclear decondensation in the two-nucleate endosperm stage was set to 0. Frame numbers are indicated in the video.
Pictures were taken every 10 min. The last frame before nuclear decondensation in the two-nucleate endosperm stage was set to 0. Frame numbers are indicated in the video.