Abstract
Within primates, the great apes are outliers both in terms of body size and lifespan, since they include the largest and longest-lived species in the order. Yet, the molecular bases underlying such features are poorly understood. Here, we leveraged an integrated approach to investigate multiple sources of molecular variation across primates, focusing on over 10,000 genes, including approximately 1,500 previously associated with lifespan, and additional approximately 9,000 for which an association with longevity has never been suggested. We analyzed dN/dS rates, positive selection, gene expression (RNA-seq), and gene regulation (ChIP-seq). By analyzing the correlation between dN/dS, maximum lifespan, and body mass, we identified 276 genes whose rate of evolution positively correlates with maximum lifespan in primates. Further, we identified five genes, important for tumor suppression, adaptive immunity, metastasis, and inflammation, under positive selection exclusively in the great ape lineage. RNA-seq data, generated from the liver of six species representing all the primate lineages, revealed that 8% of approximately 1,500 genes previously associated with longevity are differentially expressed in apes relative to other primates. Importantly, by integrating RNA-seq with ChIP-seq for H3K27ac (which marks active enhancers), we show that the differentially expressed longevity genes are significantly more likely than expected to be located near a novel “ape-specific” enhancer. Moreover, these particular ape-specific enhancers are enriched for young transposable elements, and specifically SINE–Vntr–Alus. In summary, we demonstrate that multiple evolutionary forces have contributed to the evolution of lifespan and body size in primates.
Keywords: ageing, cancer, evolutionary genomics, cis-regulatory evolution, natural selections, apes, transposable elements, cell senescence
Introduction
Uncovering the molecular and evolutionary bases of ageing in the tree of life is important to increase our understanding of the natural mechanisms of disease resistance (Galis and Metz 2003; Caulin and Maley 2011; Ganten and Nesse 2012; Nesse et al. 2012; Thomas et al. 2013; Petralia et al. 2014; Abegglen et al. 2015; Harris et al. 2017; Ciccarelli and DeGregori 2020). The evolution of lifespan in animals has been shaped by selective pressures arising from different environments, ecological niches, habitats, and diets (Healy et al. 2014; Tollis et al. 2017; Kacprzyk et al. 2021). Ageing and senescence are the result of gradual declines in biological functions, which lead to increased vulnerability, disease predisposition, and ultimately death (López-Otín et al. 2013). With the exception of a few species that show no signs of ageing (e.g., some jellyfishes and hydras; Petralia et al. 2014), most animals undergo this process. Nevertheless, maximum lifespan (MLS) is a highly variable trait across the tree of life, suggesting that long-lived species may have adopted several evolutionary strategies which may have favored longevity by reducing disease susceptibility (Gorbunova et al. 2014; Kacprzyk et al. 2021; Yu et al. 2021). Yet, most of these strategies remain unknown.
One of the main consequences of ageing is the increased risk of developing cancer. Cancer has been reported in almost all multicellular organisms (Aktipis et al. 2013; Albuquerque et al. 2018), and it is often associated with somatic genetic mutations that inactivate tumor suppressor genes or activate oncogenes. These mutations ultimately affect cell metabolism, promoting uncontrolled cell division (Stratton et al. 2009; Tomasetti and Vogelstein 2015). Statistically, animal species with larger body sizes and longer lifespans accumulate more cell divisions during their lives and are therefore expected to accumulate more deleterious genetic mutations. Nonetheless, large and long-lived species overall do not develop cancer at higher frequency than smaller species (Nunney 2018; Seluanov et al. 2018). This scientific paradox was first noted by Richard Peto (the Peto’s Paradox; Peto et al. 1977; Nunney 1999).
Specific anticancer mechanisms have been discovered in long-lived species. These mechanisms include early contact inhibition in naked mole rats (Seluanov et al. 2009; Tian et al. 2013), positive selection in DNA repair and inflammatory genes in the giant tortoises from Galapagos (Quesada et al. 2019), and immunity pathways, telomere maintenance, and cellular senescence (CS) genes in bats (Zhang et al. 2013; Foley et al. 2018; Tian et al. 2018; Wilkinson and Adams 2019). Particularly, CS—a process in which otherwise replicating cells reach the maximum number of divisions and cease proliferating—is another important mechanism of cancer suppression; previous studies indicate that genes regulating the senescent phenotype are strongly conserved among vertebrates compared with other protein-coding genes (Avelar et al. 2020). Despite the importance of senescence processes in inhibiting cancer, CS itself is detrimental for health; senescent cells produce pro-inflammatory cytokines that can paradoxically promote cancer (Rao and Jackson 2016). A previous study found that senolytics—drugs that specifically target and eliminate senescent cells—can enhance life- and health-span in old mice (Xu et al. 2018). On the other hand, in larger mammals such as elephants and whales, molecular substitutions in CS genes and increases in the copy number of important tumor suppressor genes have contributed to significantly reduce the risk of developing cancer (Abegglen et al. 2015; Caulin et al. 2015; Keane et al. 2015; Tollis et al. 2019). Specific changes in the regulation and expression of specific genes have also been associated with ageing (de Magalhães et al. 2009; Donlon et al. 2017; Morris et al. 2019; Chatsirisupachai et al. 2021). Yet, the extent of the contribution of cis-regulatory evolution in ageing is still poorly understood.
In primates, MLS and body mass (BM) are correlated, despite being highly variable across species, with great apes (gorillas, orangutans, humans, and chimpanzees) representing outliers (Finch and Austad 2012). The BM ranges from approximately 5 kg in the gray mouse lemur (Microcebus murinus) to approximately 140 kg in the gorilla (Gorilla gorilla). Similarly, the MLS varies from 13 years in the calabar angwantibo (Arctocebus calabarensis) to 55–122 years in great apes (AnAge database: https://genomics.senescence.info/ (accessed 5th November 2021); Tacutu et al. 2018). Outside great apes, just a few primate species can reach 50 years (e.g., the capuchin monkeys; Muntané et al. 2018; Orkin et al. 2021).
Consistent with Peto’s Paradox, great apes develop cancer at lower rates than other primates (Fowler et al. 1980; Lowenstine 1986; Cho et al. 2007; Lowenstine et al. 2016). Humans represent an exception, possibly as a consequence of their life style (Finch 2010; Hochberg and Noble 2017; Albuquerque et al. 2018). In fact, in some human populations cancer rates are approximately 25% (Ferlay et al. 2015), and some cancer types seem to be unique to our species (e.g., prostate cancer and lung cancer). Several studies have recently investigated the link between longevity, BM and disease resistance in several mammalian species (Tollis et al. 2017; Muntané et al. 2018; Boddy et al. 2020; Huang et al. 2021; Kacprzyk et al. 2021). On the other hand, the mechanisms of ageing in great apes are largely uncharacterized, and several questions remain unanswered.
Here, we aimed at investigating the relative contribution of different sources of molecular variation (molecular evolution in coding genes, positive selection, cis-regulatory, and gene expression evolution) to the evolution of longevity in great apes. We interrogated 19 mammalian genomes, focusing on approximately 10,500 genes. We evaluated: 1) potential correlations between MLS, BM, and coding gene evolution (nonsynonymous/synonymous mutations); 2) positive selection signal in the great-ape ancestor; 3) species-specific gene expression (GE) patterns (RNA-seq) on a list of approximately 1,500 genes previously associated with MLS; 4) species-specific gene-regulation patterns (ChIP-seq for H3 lysine 27 acetylation [H3K27ac], which marks active cis-regulatory elements).
We identified 276 genes whose rate of evolution positively correlates with MLS. Importantly, 25/276 genes were previously associated with longevity based on scientific literature. These genes are also enriched for diverse processes such as body size, immunity, central nervous system, and developmental pathways. Further, we report five genes, related with tumor suppression, adaptive immunity, metastasis, and inflammation, under positive selection exclusively in the great ape lineage. In addition, RNA-seq and ChIP-seq data, generated from the liver of six species representing all the primate lineages (Trizzino et al. 2017), revealed that approximately 8% of the longevity genes investigated in this study are differentially expressed in apes relative to other primates, and that these lineage-specific GE patterns are associated with the rise of novel ape-specific enhancers, most of which derived from the insertion of young transposable elements (TEs).
Results
Ape-Specific Patterns in the Evolution of Lifespan
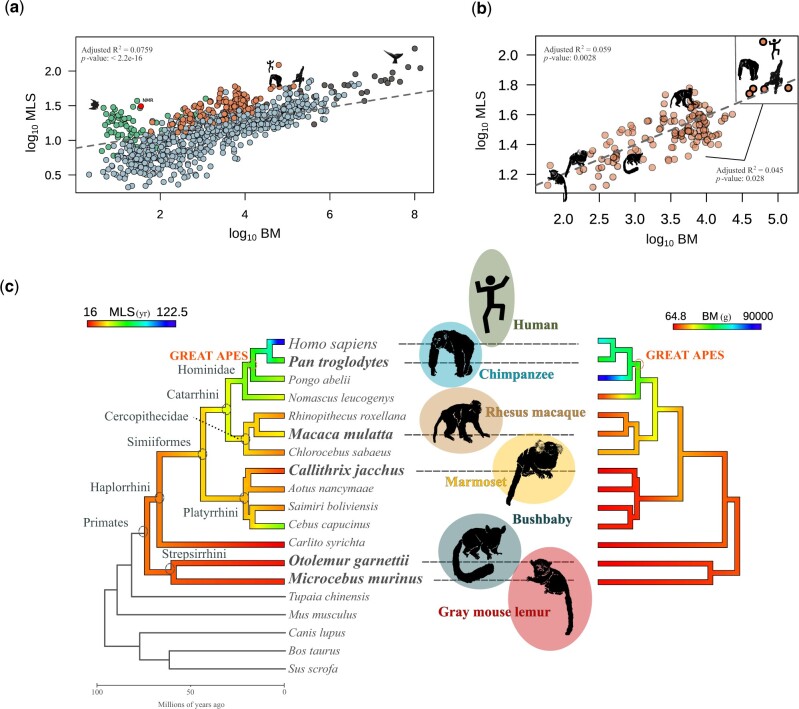
The evolution of lifespan across mammals has a large degree of variability. Some lineages are characterized by species with unexpected longevity and disease resistance (e.g., naked mole rat, bats, and whales, fig. 1a) (Tollis et al. 2017). In primates, the great apes include the species with the largest body size, and are also the most long-lived species in the entire order (fig. 1b and c). Given the high diversity of phenotypes across primates, and considering that closely related species could have evolved with similar traits due to the effect of ancestry, we used phylogenetic generalized least squares (PGLS) models (Orme et al. 2012; Revell 2012; Pennell et al. 2014; Symonds and Blomberg 2014) to evaluate if BM and MLS evolved independently in great apes. The PGLS is a phylogenetic comparative method that estimates the covariance among traits, MLS and BM in this case, taking into account the effect of the phylogenetic signal across the tree. The estimation of the PGLS regression allows to establish the association between those traits controlled by the statistical nonindependence effect of the shared evolutionary history. We found that the allometric expectations are maintained across primates, with a positive correlation between BM and MLS (fig. 1b). Our model revealed that the BM predicts approximately 6% of the variation in longevity across primate species (adjusted R2: 0.05904, 131 degrees of freedom, P = 0.0028). Nonetheless, the relation between MLS and BM in great apes is significantly different from the other primates (adjusted R2: 0.04568, degrees of freedom, P = 0.02888). This suggests that lineage-specific molecular evolution may have shaped lifespan and BM in great apes.
Fig. 1.
The evolution of BM and MLS across primates. (a) PGLS models correlating MLS ∼ BM across mammals. The highlighted species are from the primate order (orange) and other lineages that have been previously associated with cancer resistance and extreme longevity: bats (green), naked mole rat (NMR, red), cetaceans (dark gray). (b) PGLS correlating MLS ∼ BM across primates. The dashed lines represent a positive correlation between log10-transformed BM and MLS. The continuous line displays the correlation between great apes and other primates. (c) Phylogenomic design. The molecular evolution analysis included 19 mammalian species. The six species highlighted are representatives from the main primate lineages (Catarrhini, Platyrrhini, and Strepsirrhini). For these six species, RNA-seq and ChIP-seq data were publicly available (Trizzino et al. 2017). The colors in the phylogenetic tree reflect the values of MLS and BM in each primate branch respectively, from lowest (red) to highest (blue).
The Rate of Evolution (dN/dS) in 276 Genes Positively Correlates with the MLS across Primates
The ratio between synonymous (dS) and nonsynonymous substitutions (dN) is considered a reliable measure of natural selection in the evolution of phenotypic diversity. In line with this, several studies have reported a positive relationship between dN/dS and either BM or MLS (Romiguier et al. 2010; Weber et al. 2014; Figuet et al. 2016). Nonetheless, the contribution of individual genes to the evolution of body size and lifespan remains largely unexplored, especially in primates. Even if MLS and BM are positively correlated in most mammalian lineages, their molecular evolution is likely largely independent, as it has been previously reported for example in rodents (Seluanov et al. 2018). To investigate the molecular evolution of longevity and BM in primates, we focused on a set of 10,516 coding genes that have a six-way ortholog in six species (human, chimpanzee, rhesus macaque, marmoset, bushbaby, mouse lemur; supplementary table S1.2—file S1, Supplementary Material online) representative of the three main primate lineages and for which we have also analyzed publicly available next generation sequencing data (see below).
We correlated the rates of evolution (dN/dS) with the two life history traits (BM and MLS) together and independently (supplementary table S1.3–S1.5, Supplementary Material online). As a first step, from the list of 10,516 genes, we removed 678 genes with ω > 2 due to the possible overestimation of ω in the branch, possibly as a consequence of saturation or miscalculation of synonymous (dS) or nonsynonymous (dN) substitutions. The final PGLS models were thus performed on a total of 9,838 genes.
Next, we used the variance of the residuals from the first PGLS model (MLS ∼ BM), in order to account for the expected covariance structure between the two variables and evaluate the contribution of each gene (dN/dS) to the evolution of MLS and BM simultaneously and independently.
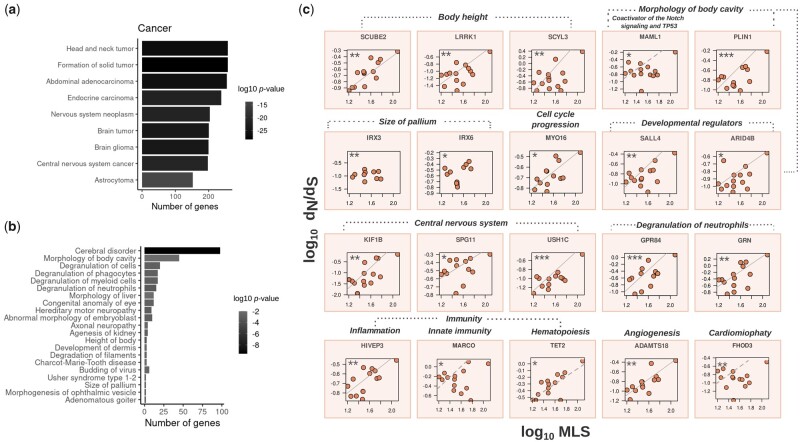
In the first PGLS model, we focused exclusively on MLS. With this analysis, we identified 276 genes whose rate of evolution (dN/dS) positively correlates with longevity across primates (FDR < 0.1, table S1—supplementary table S1.5, Supplementary Material online). Gene enrichment analysis on the 276 genes revealed an overrepresentation for cancer, inflammatory response, development, body size, immune system, and nervous system-related pathways (fig. 2a and b; supplementary table S1.6, Supplementary Material online). Importantly, 25 out of the 276 genes were previously associated with longevity (Zhao et al. 2013; Liu et al. 2017; Avelar et al. 2020), and this number was lower than expected by chance (one-tailed Fisher’s exact test: P value: 1.7e-19). This indicates that a large number of genes that were so far never associated with longevity may play an important role for this life trait in great apes.
Fig. 2.
Examples of genes whose evolution correlated with the evolution of MLS. (a, b) Ingenuity Pathway Analysis for 276 genes whose rates of evolution are positively correlated with the MLS. Panels (a) and (b), respectively, show enriched categories for cancer and developmental pathways. (c) Independent PGLS models were performed in order to evaluate the independent contribution of each gene in the evolution of MLS across primates (PGLS dN/dS ∼ MLS). The dN/dS values were obtained from the branch model in PAML. The genes with a FDR <0.1 were considered statistically significant.
The 276 genes include several tumor suppressor, oncogenes, and senescence genes (e.g., SALL4, ARID4B, and several others). SALL4 and ARID4B are important developmental regulators (Hirsch et al. 2015; Buttgereit et al. 2016; Wu et al. 2019; Bon-Baret et al. 2021; Guven and Cizmecioglu 2021) with roles in chromatin remodeling (Kim et al. 2017), human development (SALL4;Hirsch et al. 2015; Buttgereit et al. 2016; Wu et al. 2019; Bon-Baret et al. 2021; Guven and Cizmecioglu 2021), and leukemia (Wu et al. 2008).
Interestingly, several of the genes whose rate of evolution positively correlates with MLS were found to be enriched in immunity-related pathways. Particularly, GPR84, HIVEP3, and MARCO are associated with inflammation and immunity-related functions (Hicar et al. 2001; Areschoug and Gordon 2009; Gaidarov et al. 2018; Recio et al. 2018; Krovi et al. 2020), and could have positively contributed to diseases protection in primates. Similarly, TET2 is an important hematopoietic and ageing regulator (Moran-Crusio et al. 2011; Fuster et al. 2017). Other important genes that could have positively contributed to the evolution of longevity in primates are related with cell cycle regulation. For example, ADAMTS18 has a role in cell division (Wei et al. 2014) and angiogenesis (Mushimiyimana et al. 2021), whereas MAML1, is a coactivator in the notch signaling (Wu et al. 2000; Shen et al. 2006), and it also regulates the important tumor suppressor gene TP53 (Zhao et al. 2007).
Additionally, we found enrichment for pathways related to organismal development, such as body height (e.g., SCUBE2, LRRK1, and SCYL3) and the size of the pallium (IRX3, IRX6). It is worth mentioning that, due to pleiotropic effects, these genes could have been associated with the evolution of important traits in primates (e.g., BM) and at the same time could have contributed indirectly to lifespan regulation and ageing. For example, SCUBE2 expression is reduced in endometrial cancers (Skrzypczak et al. 2013). This gene has also been linked to metabolism, hedgehog signaling (Kawakami et al. 2005; Tsai et al. 2009), body mass index (Cousminer 2020), and limb/bone development (Xavier and Cobourne 2011). LRRK1 has been implicated with short stature in humans (van Duyvenvoorde et al. 2014) and with the regulation of bone mass (Iida et al. 2016). Likewise, the paralogs IRX3 and IRX6 are transcription factors active during brain development (Anselme et al. 2007), and have been related to obesity (Zou et al. 2017; de Araújo and Velloso 2020) and inflammation (Yao et al. 2021). These genes regulate the size of the pallium, which is an important brain structure that covers the upper surface of the brain (Medina and Abellán 2009). This structure has been implicated with the evolution of complex cognitive capabilities in primates (Doty 2005; Passingham and Wise 2012; Herculano-Houzel 2017; Smaers et al. 2017).
KIF1B is an axonal precursor which encodes for an important motor protein that transports mitochondria (Aulchenko et al. 2008). In humans, KIF1B mutations lead to autoimmune and neurological diseases, including multiple sclerosis (Aulchenko et al. 2008) and Charcot-Marie-Tooth (Zhao et al. 2001). KIF1B-knockout mice do not survive after birth due to atrophies in the nervous system, resembling what is observed in Charcot-Marie-Tooth patients (Zhao et al. 2001). MYO16 is important for cell cycle regulation, DNA replication stress (Cameron et al. 2013), and variants in this gene have been found in patients with schizophrenia (Rodriguez-Murillo et al. 2014). Finally, USH1C and USH2A are biomarkers for Usher syndrome, an inheritable disorder that causes hearing loss and blindness (Reiners et al. 2005; Sun et al. 2018).
In summary, we found that, across primates, the dN/dS ratio positively correlates with MLS in a total of 276 genes (FDR < 0.1). These genes are involved in multiple biological functions directly associated with ageing regulation and with species/organism development and diversification.
On the other hand, when we performed the PGLS models for body size alone and for body size and MLS combined, not a single gene passed the chosen FDR threshold.
Longevity Genes under Positive Selection in the Ancestor of Great Apes Are Involved in Tumor Suppression, Adaptive Immunity, Metastasis, and Inflammation
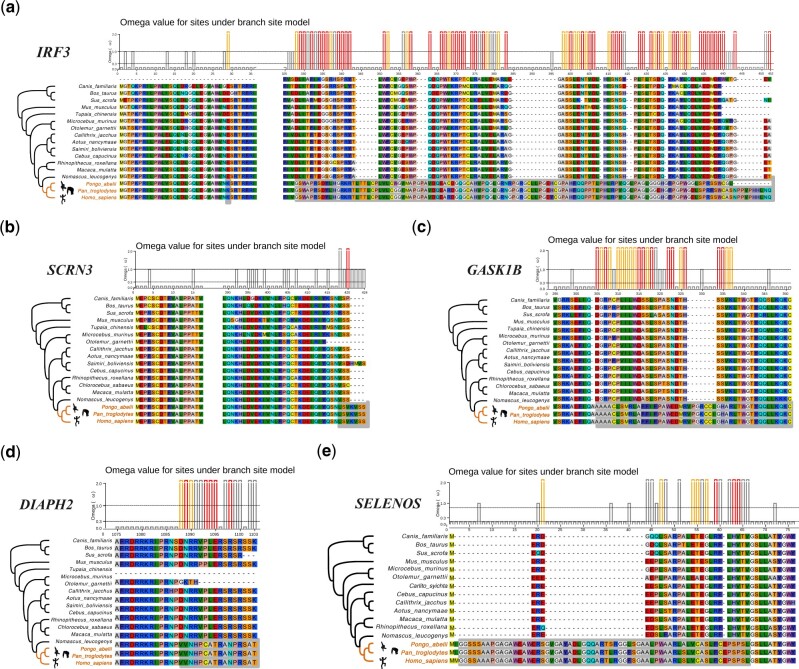
The integration of an evolutionary perspective in the study of genes related to human health could reveal the discovery of novel molecular variants that underlie important biological questions, such as how to live longer and healthier lives, or how to be more resistant to diseases. To this purpose, we studied the signal of positive selection in the evolution of 10,516 genes in the ancestors of great apes, which is the lineage including the species with largest body sizes and longest lifespan across primates. We specifically focused on genes under positive selection exclusively in the branch leading to the great ape ancestor, and not in any other primate lineage. We identified five such genes: IRF3, SCRN3, DIAPH2, GASK1B, and SELENO (fig. 3a–e and supplementary tables S1.7 and S1.8, Supplementary Material online). Importantly, IRF3 is considered a tumor suppressor gene (Yanai et al. 2018; Tian et al. 2020), it is a precursor of CS (Frisch and MacFawn 2020) and mediates an antiviral mechanism regulated via apoptosis (Chattopadhyay et al. 2016). Moreover, this gene is involved in the type I interferon signaling pathway (Li et al. 2011), which represents a key process in adaptive immune responses (Wang and Fish 2019; Schwanke et al. 2020). Interestingly, only a specific isoform of this gene (IRF3-CL) was found under positive selection. This isoform is characterized by a unique C-terminal insertion of 125 amino acids (fig. 3a), likely originated from an alternative splicing event in the great ape ancestor. Recent studies have demonstrated that this isoform is involved in the self-regulation of the IRF3 gene, acting as a negative regulator (Li et al. 2011).
Fig. 3.
Genes under positive selection exclusively in the great ape ancestor (a–g): IRF3-CL, SCRN3, GASK1B, DIAPH2, and SELENOS. The color gradients are used to display the degree and typology of amino acid change. The bars indicate the positive sites under the Bayes Empirical Bayes (BEB) analysis with a foreground lineages Prob(ω > 1): *** (red), ** (orange), and * (gray).
SCRN3 has been linked with survival of breast cancer patients, however its function remains poorly understood (Liu et al. 2019). DIAPH2 and GASK1B (also known as FAM198B) have been associated with cancer metastasis (Kawamata et al. 2003; Hsu et al. 2018). Particularly, DIAPH2 is involved in laryngeal squamous cell carcinoma, a type of cancer with low survival rate (Kostrzewska-Poczekaj et al. 2019). Finally, SELENO (also known as SEPS1) encodes for a protein that induces inflammatory responses as a result of cellular stress (Curran et al. 2005) and diabetes (Li et al. 2019).
Longevity Genes with Human-Specific Expression Patterns
We reasoned that different strategies may have contributed to the evolution of longevity genes in great apes. On the one hand, positive selection may lead to important changes in the amino acid composition, and thus in the structure and function of the encoded protein, as well as in the sequence of noncoding regulatory regions. On the other hand, the coding sequence, and hence the structure of the protein, may remain unaltered across species but the expression level of the gene may change. Changes in GE would be reflected in the amount of translated protein, which could have a significant impact on cell metabolism. Our data so far demonstrated that a number of longevity genes are under positive selection in great apes. We surmised that changes in the expression of longevity genes may provide an additional contribution to the differences in lifespan and size between the great apes and the other primates. To test this hypothesis, we leveraged a publicly available RNA-seq data set generated from the liver of the six primate species analyzed in the present study (humans, chimpanzee, rhesus macaque, marmoset, mouse lemur, and bushbaby; Trizzino et al. 2017). The liver is particularly relevant for this study because most genes associated with longevity and BM are highly expressed in the liver, and also because liver GE displays high variation across species, likely reflecting adaptation to different diets and environments, in spite of a conserved core function (Trizzino et al. 2017; Berthelot et al. 2018). Consistent with this premise, the liver has been employed in studies that investigated biomarkers of aging (Lee et al. 2012; Bochkis et al. 2014; White et al. 2015).
To study the evolution of ageing-related liver GE and regulation across primates, we first aimed at compiling a list of genes associated with CS, ageing, longevity, and related functions. We started from Cell Senescence (CS). To this purpose, and especially with the goal to make this list accessible, we first compiled a Build 2 of the CellAge data set by means of a scientific literature search of gene manipulation experiments in primary, immortalized, or cancer human cell lines that caused cells to induce or inhibit CS. The novel CellAge build comprises 915 distinct CS genes, of which 169 affect replicative CS, 198 affect stress-induced CS, 245 are related to oncogene-induced CS, and 380 have uncharacterized function. Of the 915 total genes, 383 genes induce CS (∼ 41.86%), 508 inhibit it (∼ 55.52%), and 24 genes have unclear effects, both inducing and inhibiting CS depending on experimental conditions (∼ 2.62%). The genes in the data set are also classified according to the experimental context used to determine these associations (supplementary table S2, Supplementary Material online).
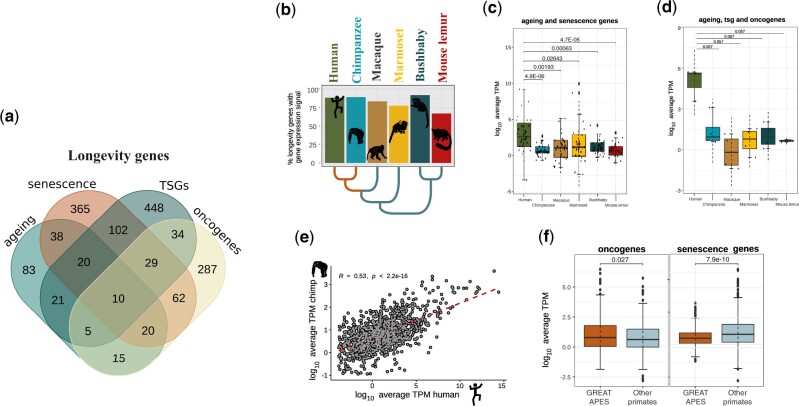
Next, we added to the list a number of genes previously associated with tumor suppression, oncogenic function, and ageing, based on literature searches and public databases (supplementary table S2, Supplementary Material online). Overall, the cumulative list of genes included 2,268 genes associated with either CS, tumor suppression, oncogenic function, or ageing (hereafter: longevity genes; supplementary table S1.9, Supplementary Material online). Of these 2,268 longevity genes, 1,539 have a six-way ortholog in the six primate species for which there were available RNA-seq and ChIP-seq data (Trizzino et al. 2017; supplementary table S1.10, Supplementary Material online). These 1,539 longevity genes were hence used for the downstream analysis.
As a first step, we examined the expression patterns of all the 1,539 genes, and found that genes involved in both ageing and senescence are significantly more expressed in humans relative to all the other primates grouped together (Wilcoxon rank-sum test, P < 0.001; fig. 4a). Similarly, genes that are involved in both ageing and tumor suppression are more expressed in humans than in other primates grouped together, although the P value was only marginally significant, possibly as a consequence of the small sample size (N = 3; Wilcoxon rank-sum test, P = 0.057; fig. 4b).
Fig. 4.
Evolution of longevity GE in the primate liver. (a) Venn diagram representing the total number of orthologous longevity genes (1,539) shared by at least one species in each of main primate lineages, broken down based on the different categories (tumor suppressors, oncogenes, CS and ageing genes). The genes were divided into categories: ageing, senescence-related genes, tumor suppressor genes (TSGs), and oncogenes. (b) Expression levels for the 1,539 genes in the liver were comparable across primates. (c) Expression levels for ageing-related genes and senescence genes. (d) Expression levels for ageing-related genes, tumor suppressors, and oncogenes. In both groups (c, d), humans display higher expression compared with other primates (Wilcoxon’s rank-sum test). (e) The expression of the 1,539 longevity genes in the liver is positively correlated between humans and chimpanzee (Spearman’s correlation coefficient, R = 0.53, P < 0.001). (f) Oncogenes are significantly more expressed in great apes compared with other primates. Senescence genes are significantly less expressed in great apes relative to the other primates (Wilcoxon’s rank-sum test, P < 0.05).
Longevity Genes with Ape-Specific Expression Patterns
Next, we compared the expression of the longevity genes between apes and other primates. Overall, apes have a higher number of longevity genes expressed in the liver relative to other primates (fig. 4c; Spearman’s correlation coefficient R = 0.53, P = 2.2×10−16), and the expression level of the longevity genes is strongly correlated between humans and chimpanzees (fig. 4d). We decomposed the longevity genes across the different categories (fig. 4e), and observed that the oncogenes are significantly more expressed in apes relative to other primates grouped together (Wilcoxon rank-sum test, P = 0.027), whereas the senescence genes are significantly less expressed in apes (Wilcoxon rank-sum test, P = 7.9×10−10). This pattern could reflect the findings from recent studies that reported that senescence genes are beneficial in the younger ages, during which they increase reproductive fitness, whereas they might have a negative impact later in life (Campisi 2003; Blagosklonny 2010; Di Micco et al. 2021). Accordingly, GE meta-analysis across human tissues showed that cancer genes promote longevity, whereas senescence genes showed antilongevity patterns (Chatsirisupachai et al. 2019).
Ape-Specific Enhancers Drive the Differential Expression of Longevity Genes in Apes
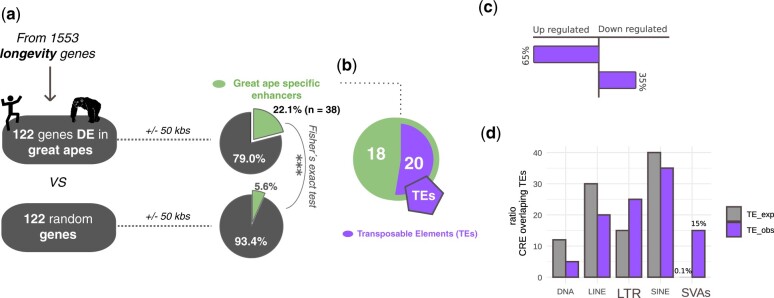
We performed a differential GE analysis, comparing the expression of the longevity genes in apes (human, chimpanzee) versus the other four primate species grouped together (rhesus macaque, marmoset, bushbaby, mouse lemur). Overall, 122/1,539 (∼7.9%) longevity genes were found to be differentially expressed between apes and the other primates grouped together (DESeq2 FDR < 0.05; supplementary table S3, Supplementary Material online). Of those, 61 genes were upregulated and 61 downregulated. We investigated if cis-regulatory evolution could underlie the changes in expression of the 122 genes identified as differentially expressed in apes compared with other primates. To this purpose, we leveraged H3K27ac ChIP-seq data generated in the same study from the same liver samples of the same individuals (Trizzino et al. 2017). Namely, this specific histone modification marks active cis-regulatory elements (enhancers and promoters). Importantly, in the original study, Trizzino et al. (2017) identified a set of ape-specific enhancers. We thus scanned the surrounding regions of the 122 differentially expressed genes found in the apes versus other primates comparison (fig. 5a). We focused on the 50 kb up- or downstream of the transcription start site (TSS) of the 122 genes, and observed that 27 of the 122 differentially expressed genes (22.1%) have an ape-specific enhancer in the 50 kb surrounding the TSS (38 total enhancers, median distance from TSS = 19.8 kb; fig. 5a and supplementary table S3, Supplementary Material online). To test if this number is higher than expected by chance, we performed a permutation test (1,000 permutations), by randomly extracting (for 1,000 times) 122 genes from a list including all the annotated human genes (Ensembl), and found that, on average, only 4/122 random genes (3.3%) were located within 50 kb from an ape-specific enhancer. Together, these data indicate that the longevity genes identified as differentially expressed in apes versus other primates are approximately six times more likely than expected to be found near an ape-specific enhancer (22.1% vs. 3.3%; permutation test P < 2.2×10−16; fig. 5a).
Fig. 5.
Ape-specific enhancers are enriched near differentially expressed longevity genes. (a) 27/122 differentially expressed (DE) longevity genes (22.1%) are located within 50 kb of an ape-specific enhancer (a total of 38 ape-specific enhancers for 27 DE genes). In comparison, only 4/122 (3.3%) randomly selected genes (1,000 permutations) are located within 50 kb of an ape-specific enhancer (permutation test P < 2.2×10−16). (b) 20/38 ape-specific enhancers (52%) located within 50 kb of the DE longevity genes overlap annotated TEs. (c) Of the DE longevity genes found near TE-derived ape-specific enhancers, 65% were identified as upregulated, 35% as downregulated (Fisher’s exact test P = 0.0425). (e) LTR and SVA transposons are overrepresented in the sequence of the ape-specific enhancers located within 50 kb of a DE longevity gene (Fisher’s exact test P < 2.2×10−16 for SVAs; P < 0.0001 for LTRs).
Ape-Specific Enhancers Associated with Differentially Expressed Longevity Genes Are Derived from Recent TE Insertions
Overall, a total of 38 ape-specific enhancers were found in the 50 kb surrounding 27 of the 122 differentially expressed longevity genes (supplementary table S1.11, Supplementary Material online). Since multiple studies have demonstrated that TEs are an important source of gene regulatory novelty in the primate gene regulation (Jacques et al. 2013; Chuong et al. 2016; Trizzino et al. 2017, 2018), we tested for potential association between the 38 ape-specific enhancers and TE insertions. We found that 20/38 (∼52.6%) of ape-specific enhancers are derived from TE insertions (fig. 5b). This was not higher than expected by chance, considering that TEs represent approximately 50% of the human genome. Nonetheless, TE-derived ape-specific enhancers were significantly more associated with upregulated (65%) than to downregulated (35%) longevity genes (fig. 5c, Fisher’s exact test P = 0.0425).
Finally, we investigated if specific TE families were overrepresented in the TE-derived ape-specific enhancers associated with differentially expressed longevity genes (fig. 5d). Notably, 15% of those were derived from SINE–Vntr–Alu (SVA) insertions. SVAs are the youngest TE family, with most copies being either ape- or human-specific, and represent only 0.1% of all human (and 0.3% of chimpanzee’s) annotated TEs. Overall, our data suggest that SVAs are significantly overrepresented in our set of ape-specific enhancers associated with differentially expressed longevity genes (expected: 0.1%—observed: 15%; Fisher’s exact test P < 2.2×10−16). These findings are consistent with recent studies which demonstrated that SVA insertions contribute to human gene-regulatory networks (Trizzino et al. 2017, 2018; Pontis et al. 2019). Similarly, the LTR transposons were also overrepresented in our set of ape-specific enhancers (Fisher’s exact test P < 0.0001). These findings are also consistent with recent literature on the contribution of LTRs to human gene regulation (Wang et al. 2007; Cohen et al. 2009; Sundaram et al. 2014; Chuong et al. 2016; Janousek et al. 2016; Fuentes 2018).
In summary, our analysis revealed that lineage-specific enhancers may have contributed to the evolution of the expression of longevity genes in great apes and that young transposon insertions may have had a significant role in this process.
Discussion
Natural selection and gene regulatory evolution likely underlay the evolution of most phenotypic traits. Here, we specifically focused on the evolution of lifespan and BM in great apes. This primate lineage includes species which evolved with a lifespan and BM significantly different from the other primates. We carried out comparative genomic analyses, examining different sources of molecular variation, including both coding genes and noncoding genomic regions (i.e., cis-regulatory elements: enhancers, and promoters). Cis-regulatory evolution plays an essential role in phenotypic diversification (Trizzino et al. 2017; Berthelot et al. 2018; Farré et al. 2019; Feigin et al. 2019; Sundaram and Wysocka 2020; Marand et al. 2021), and can lead to evolutionary innovations in disease resistance across species (Gorbunova et al. 2007; MacRae et al. 2015; Tollis et al. 2020).
Our comparative genomic analysis revealed 276 genes whose rate of evolution positively correlates with maximum life-span. Importantly, 25 of these genes were previously associated with longevity. These 276 genes encompass a high variation of phenotypes, including immunity, inflammation, CS, and organismal development (e.g., body height and body mass index). Among those genes, we also identified several associated with neurodevelopment and brain functions, implicated with the regulation of memory and emotions, all of which are peculiar great ape features. This finding is consistent with several studies that have proposed a link between increased life span and increased cognitive functions (Ghirlanda et al. 2014; Barton and Venditti 2017; Orkin et al. 2021). Since the increase in longevity elicits a greater risk to develop cancer, the rise and fixation of new molecular variants (nonsynonymous substitutions) could have positively contributed to improve the functionality of the immune system and thus be beneficial for the evolution of the most recent primate lineage. Similar outcomes have been recently reported by a study on mammals focused on copy number variation in tumor suppressor genes (Sulak et al. 2016; Tollis et al. 2020). Consistent with these lines of evidence, previous studies have found that the rate of protein evolution in anticancer and DNA damage response genes is accelerated in long-lived mammals (Li and de Magalhães 2013; Tollis et al. 2019; Tejada-Martinez et al. 2021).
Importantly, we also identified approximately 700 genes with an increased evolutionary constraint in relation with the MLS across primates (i.e., rate of evolution which negatively correlated with lifespan). From those, approximately 10% were in some longevity-associated categories, including tumor suppression or oncogenesis. Interestingly, a recent genomic comparative study across mammals has shown that genes involved in cancer regulation are under higher evolutionary pressure in long-lived species (Kowalczyk et al. 2020). Small changes in those genes with increased evolutionary constraint could affect the fitness and as a result slower rates of evolution are expected. Similar patterns have been found in genes involved in cell cycle regulation and pro-senescence processes (Chatsirisupachai et al. 2019). Nevertheless, our investigation predominantly focused on genes for which the evolutionary rates positively correlated with the maximum life-span. The increase in evolutionary rates can be related to a decrease in evolutionary constraint. In this scenario, nonsynonymous changes could be accumulated in genes that do not affect the phenotype (Kowalczyk et al. 2020). On the other hand, a positive correlation due to the accumulation of nonsynonymous mutations could point out signs of molecular innovations that could have arisen in the evolution of longevity-associated traits. A greater lifespan correlates with a higher likelihood of being exposed to pathogens and diseases. Consequently, the immune system of long-lived species is expected to be under particularly strong pressure to sustain the prolonged arm-race with the pathogens (Quesada et al. 2019; Singh et al. 2019). Consistent with this premise, we found a significant number of immune, inflammation, and senescence genes that are positively correlated with longevity in primates. This points toward an evolutionary strategy in which disease resistance and lifespan evolved together in primates, and particularly in apes, as it has been reported for other long-lived species (Harris et al. 2017; Muntané et al. 2018; Vazquez et al. 2018).
In the great ape ancestor, we identified five genes under positive selection involved in tumor suppression, adaptive immunity, metastasis, and inflammation. Characterizing their molecular evolution could help us to better understand the rise of genetic disorders in humans, such as several types of cancer, schizophrenia, and cognitive disorders. Different molecular strategies may have contributed to the evolution of longevity genes across primates. Positive selection can lead to changes in the structure and function of a protein through amino acid substitutions, without affecting GE, and thus the amount of protein produced by a specific cell type. In parallel, the expression level of genes important for longevity and BM could be affected by evolutionary changes in the associated gene regulatory network. For example, a novel, species-specific enhancer could influence the expression of the associated gene, ultimately leading to higher levels of protein production without any modification to the structure of the protein itself. Here, using RNA-seq, we have investigated how the expression of the longevity genes varies across primates, using the liver as a proof of principle. We demonstrated that oncogenes are significantly more expressed, and senescence genes less expressed, in great apes relative to other primates, suggesting a tradeoff between living longer (which leads to increased likelihood to produce offspring) and disease susceptibility later in life (Campisi 2003; Blagosklonny 2010; Rodríguez et al. 2017; Di Micco et al. 2021). Interestingly, the “ageing and senescence genes” seemingly exhibited increased expression only in humans and not in the chimpanzee. Although this may seem controversial, several comparative GE studies (including the original paper where these data were generated; Trizzino et al. 2017; Berthelot et al. 2018) have indicated that there are major transcriptomic differences between the two ape species. Future studies will be necessary to assess functional genomic differences in ageing-related genes between humans and chimpanzees.
Overall, we report that approximately 8% of the longevity genes tested in this work were differentially expressed in the liver of great apes relative to higher primates, and we demonstrate that lineage-specific (i.e., ape-specific) enhancers have significantly contributed to this process. In fact, longevity genes differentially expressed between great apes and other primates are approximately six times more likely than expected to be located near an ape-specific enhancer. It is important to take into account that these changes in expression levels may have occurred due to other selective pressures unrelated to longevity, and therefore these results should be interpreted with caution.
Consistent with recent studies that suggested an important contribution of young TEs in the evolution of primate gene regulation (Jacques et al. 2013; Trizzino et al. 2017; Pontis et al. 2019), we demonstrate that SVA transposons are significantly overrepresented in the DNA sequence of ape-specific enhancers located near differentially expressed longevity genes. The SVAs are the youngest family of TEs, and include ape- and/or human-specific copies.
In summary, our work has shed light on the evolution of thousands of longevity genes in great apes, highlighting a significant contribution to this process for both positive selection on coding genes, as well as evolutionary changes in the noncoding regulatory regions.
Materials and Methods
Relationship between BM and Longevity across Primates
To determine if the BM contributes to the variation in longevity independently in primates relative to other mammals, and to assess ape-specific patterns, we examined the evolution of the life history traits MLS and adult BM across 932 mammals. We fit a linear regression through the PGLS (Orme et al. 2012), while controlling for potential phylogenetic signals. Two independent regression models were implemented: the first one across primates, and in the second model, the regression was performed between great apes versus other primates (log10 MLS ∼ log10 BM×great_apes). The phylogenetic tree used was derived from (Uyeda et al., 2017) and the MLS and BM data were gathered from AnAge database (Tacutu et al. 2018).
Homology Inference
We inferred homology relationships among the 2,268 longevity genes and the 19 mammalian species included in our study using the program OMA standalone v.2.3.1 (Roth et al. 2008; Altenhoff et al. 2019). We inferred the OMA groups (OG), containing the sets of orthologous genes, for which we performed natural selection analyses. The amino acid sequences were aligned using the L-INS-i algorithm from MAFFT v.7 (Katoh and Standley 2013) and the codon alignments using the function pxaa2cdn in phyx (Brown et al. 2017). Finally, to reduce the chance of false positives given for low-quality alignment regions, we used the codon.clean.msa algorithm of the rphast package (Hubisz et al. 2011) with the associated longevity gene from the OG as reference sequence.
Evolution of Longevity Genes and Life History Traits
To evaluate the possible coevolution between the longevity genes and the life history traits MLS and BM in primates, the rate of evolution (ω = dN/dS) was calculated. The ω ratio per gene was estimated for each tip in the 14 primates species using the branch model (Yang 2007) as is implemented in ETE-toolkit with the ete-evol function (Huerta-Cepas et al. 2016). To calculate the ω ratios, the treeshrew, dog, cow, and pig were used as outgroups. Three independent PGLS models were performed per OG: 1) between the ω and the MLS; 2) between the ω and the BM; and 3) between the ω and the PGLS residuals between the MLS ∼ BM. Previous to the analysis, all variables were transformed to log10. The genes with a dN/dS > 2 were removed from the analysis.
To test whether longevity genes were positively correlated with the life history traits more than expected by chance, we performed a one-tailed Fisher’s exact test using the GeneOverlap R package (Shen and Sinai 2021). The conserved orthologs among the six ape species was used as the overlap background.
Positive Selection Analysis
To evaluate the role of natural selection in the evolution of longevity genes in the ancestor of great apes, we used codon-based models through a maximum likelihood approach using the program PAML v4.9 (Yang 2007), as is implemented in ETE-toolkit with the ete-evol function (Huerta-Cepas et al. 2016). We calculated the branch-site model in order to estimate changes in the ω value of individual sites. We compared the null model, where the ω value in the foreground branch was set to 1, with the model in which the ω value was estimated from the data (Zhang et al. 2005). The comparisons between models were made using likelihood ratio tests (LRT) and the P values from the LRT were corrected with FDR (Benjamini and Hochberg 1995).
In order to discover the genes under positive selection exclusive to the branch leading to the great ape ancestor, we performed additional branch-site models across 22 independent branches of the tree: 14, one for each primate species, and eight for stem branches leading to: Hominidae, Catherine, Platyrrhini, Cercopithecidae, Simiiformes, Haplorrini, Strepsirhini, and the Primates ancestor. This analysis was applied for the genes that were initially found under positive selection in the great ape ancestor.
Genomic Sampling, Longevity Associated Genes and Build 2 of CellAge Database of CS Genes
To study the evolution of longevity in primates, we carried out a phylogenetic design that included genomes from 19 mammalian species. Our taxonomic sampling included three species from the family Hominidae (Homo sapiens—human, Pan troglodytes—chimpanzee, and Pongo abelii—sumatran orangutan), one representant from Hylobatidae (Nomascus leucogenys—white-cheeked gibbon), three Cercopithecidae (Macaca mulatta—Rhesus macaque, Rhinopithecus roxellana—Golden snub-nosed monkey, and Chlorocebus sabaeus—vervet), four Platyrrhini (Callithrix jacchus—white tufted ear marmoset, Aotus nancymaae—night monkey, Saimiri boliviensis—bolivian squirrel monkey, and Cebus imitator—white-faced capuchin), two Strepsirrhine (Otolemur garnettii—bushbaby and Microcebus murinus—Gray mouse lemur), and finally five other mammalian species outside of primates (Tupaia chinensis—Chinese tree shrew, Mus musculus—common mice, Canis lupus familiaris—dog, Bos taurus—cow, and Sus scrofa—pig).
The coding sequences of each species were downloaded from Ensembl v.96 and NCBI databases (supplementary table S1, Supplementary Material online). To remove low-quality records, sequences were clustered using CD-HIT-est v.4.6 (Fu et al. 2012) with a sequence identity threshold of 90% and an alignment coverage control of 80%. The longest open reading frame was kept using TransDecoder LongOrfs and TransDecoder-predicted in TransDecoder v3.0.1 (https://github.com/TransDecoder/TransDecoder/ (accessed 5th November 2021)).
The longevity-associated coding genes, 2,268 in total, were gathered in four different categories: ageing genes from GenAge Database (build 20—307 protein-coding genes, https://genomics.senescence.info/genes/index.html); TSGs from the Tumor Suppressor gene database (TSGene 2.0—1,018 protein-coding genes, https://bioinfo.uth.edu/TSGene/ (accessed 5th November 2021); Zhao et al. 2013); oncogenes from the Oncogene database (698 protein-coding genes, http://ongene.bioinfo-minzhao.org/ (accessed 5th November 2021); Liu et al. 2017); and finally, we compiled build 2 of the CellAge Database of Cell Senescence Genes with an additional 641 genes, complementary to the 274 genes previously reported; https://genomics.senescence.info/cells/ (accessed 5th November 2021), (Avelar et al. 2020).
Build 2 of CellAge was compiled in the same way as build 1. A robust scientific literature search was performed for relevant papers before curation and annotation; genes were appended to the database if they met the following criteria:
Only gene manipulation experiments were used to identify the role of the genes in CS to ensure objectivity in the selection process.
The genetic manipulation caused cells to induce or inhibit the CS process in wet lab experiments. CS was detected by growth arrest, increased SA-β-galactosidase activity, SA-heterochromatin foci, a decrease in BrdU incorporation, changes in morphology, and/or specific GE signatures such as p21 and p16.
The experiments were performed in primary, immortalized, or cancer human cell lines.
The resulting list comprised 915 genes that in human cell lines can induce or inhibit the CS process (supplementary table S2, Supplementary Material online). The full CellAge database, including all annotations regarding cell types and cell lines is available at https://genomics.senescence.info/cells/ (accessed 5th November 2021).
RNA-Seq Analysis
To evaluate the contribution of the gene regulation to the evolution of longevity genes in great apes, we took advantage of recently published RNAseq and Chip-seq data for histone H3 lysine 27 acetylation (H3K27ac) from liver of six species representatives of the main groups of primates (human, chimpanzee, rhesus macaque, marmoset, bushbaby, and gray mouse lemur; Trizzino et al. 2017). As described in the original study (PMID: 28855262), the liver samples were collected from young adults (of comparable age) in all species, always including both males and females. The livers were healthy, and the samples were always immediately preserved in RNA-later and immediately flash-frozen in liquid nitrogen. QC was performed on all extracted RNAs, and only samples with RIN >8 were used (see Materials and Methods section of the original study).
The list of the genes differentially expressed between Apes and other primates was downloaded from the Supplementary Material online of the same paper (Trizzino et al. 2017). To generate box-plots comparing GE across species, we leveraged the transcripts per million (TPM), also available from the Supplementary Materials of Trizzino et al. (2017). The TPMs were quantile normalized using R version 3.6.3 (R Studio Team 2020).
Enhancer Evolution Analysis
From the publicly available Chip-seq data (Trizzino et al. 2017), we downloaded the list and coordinates all the enhancers previously identified as “ape-specific” by Trizzino and collaborators. We then selected all the ape-specific enhancers overlapping a region encompassing ±50 kb from the TSS of each differentially expressed longevity gene using BEDTools v2.29.2 (Quinlan and Hall 2010). Based on this association, we then calculated how many longevity genes were both differentially expressed in the ape versus other primates comparison AND also associated with an ape-specific enhancer. To evaluate if this number was higher than expected by chance, we selected 122 random genes in the human genome and examined how many of them had an ape-specific enhancer within 50 kb of the TSS and assessed statistical significance by means of Fisher’s exact test. Finally, using BEDTools, we looked for overlap between TEs and ape-specific enhancers associated with genes differentially expressed in ape versus primate comparison. The list and coordinates of the TEs were downloaded from the UCSC Genome Browser (RepeatMasker; hg38 assembly).
Enrichment Analysis
To gain insight into the particular functions of the longevity genes of interest, the enrichment analysis was performed using Ingenuity Pathway Analysis (IPA; Yu et al. 2016) and WebGestaltR v0.4.4 (Liao et al. 2019). We tested for significant pathway associations using the hypergeometric test for Over-Representation Analysis (ORA; Khatri et al. 2012). We selected the categories of gene ontology, biological processes, molecular pathways, diseases OMIM, and human phenotype and we considered overrepresented categories to be those with a significance level above that of an FDR of 0.01 after correction with the Benjamini–Hochberg multiple test. An independent enrichment analysis was made using as a background the protein-coding genes and relative to the 2,268 longevity genes.
Libraries in R Associated with the Data Treatment, Statistical Analysis, and Graphics
Bioconductor, msa, ape, geiger, nlme, pythools, caper, ggpubr, gridExtra, tidyverse, grid, corrplot, ggplot2, ggpubr, Hmisc, gmodels, car, DescTools, qqplotr, dplyr, GeneOverlap, calecopal.
Data Availability
The full CellAge database, including all annotations regarding cell types and cell lines is available at https://genomics.senescence.info/cells/.
Supplementary Material
Acknowledgments
This study was funded by NIH R35 GM138344-01, awarded to M.T. AnAge, GenAge, and CellAge are supported by funding from the Biotechnology and Biological Sciences Research Council (BB/R014949/1) to J.P.M. The authors are grateful to Alejandra Tejada-Martinez for the graphical support.
Author Contributions
D.T.-M. and M.T. conceived the study and performed the genomic analyses. R.A.A., J.P.M., B.Z., and G.N. compiled the Build 2 of the CellAge database. I.L. contributed to the Cellage website development. D.T.-M., M.T., and R.A.A. wrote the manuscript. M.T. and D.T.-M. directed the research. All authors read and approved the manuscript.
References
- Abegglen LM, Caulin AF, Chan A, Lee K, Robinson R, Campbell MS, Kiso WK, Schmitt DL, Waddell PJ, Bhaskara S, et al. 2015. Potential mechanisms for cancer resistance in elephants and comparative cellular response to DNA damage in humans. JAMA 314(17):1850–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktipis CA, Boddy AM, Gatenby RA, Brown JS, Maley CC.. 2013. Life history trade-offs in cancer evolution. Nat Rev Cancer. 13(12):883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque TAF, Drummond do Val L, Doherty A, de Magalhães JP.. 2018. From humans to hydra: patterns of cancer across the tree of life. Biol Rev Camb Philos Soc. 93(3):1715–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenhoff AM, Levy J, Zarowiecki M, Tomiczek B, Warwick Vesztrocy A, Dalquen DA, Müller S, Telford MJ, Glover NM, Dylus D, et al. 2019. OMA standalone: orthology inference among public and custom genomes and transcriptomes. Genome Res. 29(7):1152–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anselme I, Laclef C, Lanaud M, Rüther U, Schneider-Maunoury S.. 2007. Defects in brain patterning and head morphogenesis in the mouse mutant fused toes. Dev Biol. 304(1):208–220. [DOI] [PubMed] [Google Scholar]
- Areschoug T, Gordon S.. 2009. Scavenger receptors: role in innate immunity and microbial pathogenesis. Cell Microbiol. 11(8):1160–1169. [DOI] [PubMed] [Google Scholar]
- Aulchenko YS, Hoppenbrouwers IA, Ramagopalan SV, Broer L, Jafari N, Hillert J, Link J, Lundström W, Greiner E, Dessa Sadovnick A, et al. 2008. Genetic variation in the KIF1B locus influences susceptibility to multiple sclerosis. Nat Genet. 40(12):1402–1403. [DOI] [PubMed] [Google Scholar]
- Avelar RA, Ortega JG, Tacutu R, Tyler EJ, Bennett D, Binetti P, Budovsky A, Chatsirisupachai K, Johnson E, Murray A, et al. 2020. A multidimensional systems biology analysis of cellular senescence in aging and disease. Genome Biol. 21(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton RA, Venditti C.. 2017. Rapid evolution of the cerebellum in humans and other great apes. Curr Biol. 27(8):1249–1250. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 57(1):289–300. [Google Scholar]
- Berthelot C, Villar D, Horvath JE, Odom DT, Flicek P.. 2018. Complexity and conservation of regulatory landscapes underlie evolutionary resilience of mammalian gene expression. Nat Ecol Evol. 2(1):152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny MV. 2010. Revisiting the antagonistic pleiotropy theory of aging: TOR-driven program and quasi-program. Cell Cycle. 9(16):3151–3156. [DOI] [PubMed] [Google Scholar]
- Bochkis IM, Przybylski D, Chen J, Regev A.. 2014. Changes in nucleosome occupancy associated with metabolic alterations in aged mammalian liver. Cell Rep. 9(3):996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy AM, Abegglen LM, Pessier AP, Aktipis A, Schiffman JD, Maley CC, Witte C.. 2020. Lifetime cancer prevalence and life history traits in mammals. Evol Med Public Health. 2020(1):187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bon-Baret V, Chignon A, Boulanger M-C, Li Z, Argaud D, Arsenault BJ, Thériault S, Bossé Y, Mathieu P.. 2021. System genetics including causal inference identify immune targets for coronary artery disease and the lifespan. Circ Genom Precis Med. 14(2):e003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JW, Walker JF, Smith SA.. 2017. Phyx: phylogenetic tools for Unix. Bioinformatics 33(12):1886–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttgereit A, Lelios I, Yu X, Vrohlings M, Krakoski NR, Gautier EL, Nishinakamura R, Becher B, Greter M.. 2016. Sall1 is a transcriptional regulator defining microglia identity and function. Nat Immunol. 17(12):1397–1406. [DOI] [PubMed] [Google Scholar]
- Cameron RS, Liu C, Pihkala JPS.. 2013. Myosin 16 levels fluctuate during the cell cycle and are downregulated in response to DNA replication stress. Cytoskeleton (Hoboken) 70(6):328–348. [DOI] [PubMed] [Google Scholar]
- Campisi J. 2003. Cellular senescence and apoptosis: how cellular responses might influence aging phenotypes. Exp Gerontol. 38(1–2):5–11. [DOI] [PubMed] [Google Scholar]
- Caulin AF, Graham TA, Wang L-S, Maley CC.. 2015. Solutions to Peto’s paradox revealed by mathematical modelling and cross-species cancer gene analysis. Philos Trans R Soc Lond B Biol Sci. 370(1673):20140222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulin AF, Maley CC.. 2011. Peto’s Paradox: evolution’s prescription for cancer prevention. Trends Ecol Evol. 26(4):175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatsirisupachai K, Lesluyes T, Paraoan L, Van Loo P, de Magalhães JP.. 2021. An integrative analysis of the age-associated multi-omic landscape across cancers. Nat Commun. 12(1):2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatsirisupachai K, Palmer D, Ferreira S, de Magalhães JP.. 2019. A human tissue-specific transcriptomic analysis reveals a complex relationship between aging, cancer, and cellular senescence. Aging Cell 18(6):e13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Kuzmanovic T, Zhang Y, Wetzel JL, Sen GC.. 2016. Ubiquitination of the transcription factor IRF-3 activates RIPA, the apoptotic pathway that protects mice from viral pathogenesis. Immunity 44(5):1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H-S, Kim Y-S, Choi C, Lee J-H, Kurkure NV, Subramanian M, Park N-Y.. 2007. Neuroendocrine tumor in the lung of a captive black spider monkey (Ateles paniscus). J Vet Med Sci. 69(7):783–785. [DOI] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C.. 2016. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351(6277):1083–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli FD, DeGregori J.. 2020. Approaching cancer evolution from different angles. iScience 23(11):101661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen CJ, Lock WM, Mager DL.. 2009. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene 448(2):105–114. [DOI] [PubMed] [Google Scholar]
- Cousminer DL. 2020. Pubertal timing and body mass: genes involved. Curr Opin Endocr Metab Res. 14:117–126. [Google Scholar]
- Curran JE, Jowett JBM, Elliott KS, Gao Y, Gluschenko K, Wang J, Abel Azim DM, Cai G, Mahaney MC, Comuzzie AG, et al. 2005. Genetic variation in selenoprotein S influences inflammatory response. Nat Genet. 37(11):1234–1241. [DOI] [PubMed] [Google Scholar]
- de Araújo TM, Velloso LA.. 2020. Hypothalamic IRX3: a new player in the development of obesity. Trends Endocrinol Metab. 31(5):368–377. [DOI] [PubMed] [Google Scholar]
- de Magalhães JP, Curado J, Church GM.. 2009. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics 25(7):875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Krizhanovsky V, Baker D, d’Adda di Fagagna F.. 2021. Cellular senescence in ageing: from mechanisms to therapeutic opportunities. Nat Rev Mol Cell Biol. 22(2):75–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlon TA, Morris BJ, Chen R, Masaki KH, Allsopp RC, Willcox DC, Elliott A, Willcox BJ.. 2017. FOXO3 longevity interactome on chromosome 6. Aging Cell 16(5):1016–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doty RW. 2005. Principles of brain evolution. Sunderland (MA: ): Sinauer Associates. [Google Scholar]
- Farré M, Kim J, Proskuryakova AA, Zhang Y, Kulemzina AI, Li Q, Zhou Y, Xiong Y, Johnson JL, Perelman PL, et al. 2019. Evolution of gene regulation in ruminants differs between evolutionary breakpoint regions and homologous synteny blocks. Genome Res. 29(4):576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigin CY, Newton AH, Pask AJ.. 2019. Widespread cis-regulatory convergence between the extinct Tasmanian tiger and gray wolf. Genome Res. 29(10):1648–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F.. 2015. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 136(5):E359–E386. [DOI] [PubMed] [Google Scholar]
- Figuet E, Nabholz B, Bonneau M, Mas Carrio E, Nadachowska-Brzyska K, Ellegren H, Galtier N.. 2016. Life history traits, protein evolution, and the nearly neutral theory in amniotes. Mol Biol Evol. 33(6):1517–1527. [DOI] [PubMed] [Google Scholar]
- Finch CE. 2010. Evolution of the human lifespan and diseases of aging: roles of infection, inflammation, and nutrition. Proc Natl Acad Sci U S A. 107(Suppl 1):1718–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch CE, Austad SN.. 2012. Primate aging in the mammalian scheme: the puzzle of extreme variation in brain aging. Age (Dordr). 34(5):1075–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley NM, Hughes GM, Huang Z, Clarke M, Jebb D, Whelan CV, Petit EJ, Touzalin F, Farcy O, Jones G, et al. 2018. Growing old, yet staying young: the role of telomeres in bats’ exceptional longevity. Sci Adv. 4(2):eaao0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler ME, Benirschke K, Garner DM, Jones TC.. 1980. Pathology of laboratory animals. J Z Anim Med. 11(4):132. [Google Scholar]
- Frisch SM, MacFawn IP.. 2020. Type I interferons and related pathways in cell senescence. Aging Cell 19(10):e13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W.. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28(23):3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes A. 2018. How humans and apes are different, and why it matters. J Anthropol Res. 74(2):151–167. [Google Scholar]
- Fuster JJ, Maclauchlan S, Zuriaga MA, Polackal MN, Walsh K.. 2017. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis in hyperlipidemic mice. Atherosclerosis 263:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidarov I, Anthony T, Gatlin J, Chen X, Mills D, Solomon M, Han S, Semple G, Unett DJ.. 2018. Embelin and its derivatives unravel the signaling, proinflammatory and antiatherogenic properties of GPR84 receptor. Pharmacol Res. 131:185–198. [DOI] [PubMed] [Google Scholar]
- Galis F, Metz JAJ.. 2003. Anti-cancer selection as a source of developmental and evolutionary constraints. Bioessays 25(11):1035–1039. [DOI] [PubMed] [Google Scholar]
- Ganten D, Nesse R.. 2012. The evolution of evolutionary molecular medicine. J Mol Med (Berl). 90(5):467–470. [DOI] [PubMed] [Google Scholar]
- Ghirlanda S, Enquist M, Lind J.. 2014. Coevolution of intelligence, behavioral repertoire, and lifespan. Theor Popul Biol. 91:44–49. [DOI] [PubMed] [Google Scholar]
- Gorbunova V, Seluanov A, Mao Z, Hine C.. 2007. Changes in DNA repair during aging. Nucleic Acids Res. 35(22):7466–7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V, Seluanov A, Zhang Z, Gladyshev VN, Vijg J.. 2014. Comparative genetics of longevity and cancer: insights from long-lived rodents. Nat Rev Genet. 15(8):531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guven G, Cizmecioglu NT.. 2021. Arid4b alters cell cycle and cell death dynamics during mouse embryonic stem cell differentiation. Turk J Biol. 45:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris VK, Schiffman JD, Boddy AM.. 2017. Chapter 7 – evolution of cancer defense mechanisms across species. In: Ujvari B, Roche B, Thomas F, editors. Ecology and evolution of cancer. Cambridge, MA, USA: Academic Press, Elsevier. p. 99–110. [Google Scholar]
- Healy K, Guillerme T, Finlay S, Kane A, Kelly SBA, McClean D, Kelly DJ, Donohue I, Jackson AL, Cooper N.. 2014. Ecology and mode-of-life explain lifespan variation in birds and mammals. Proc Biol Sci. 281(1784):20140298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S. 2017. Numbers of neurons as biological correlates of cognitive capability. Curr Opin Behav Sci. 16:1–7. [Google Scholar]
- Hicar MD, Liu Y, Allen CE, Wu L-C.. 2001. Structure of the human zinc finger protein HIVEP3: molecular cloning, expression, exon–intron structure, and comparison with paralogous genes HIVEP1 and HIVEP2. Genomics 71(1):89–100. [DOI] [PubMed] [Google Scholar]
- Hirsch S, El-Achkar T, Robbins L, Basta J, Heitmeier M, Nishinakamura R, Rauchman M.. 2015. A mouse model of Townes-Brocks syndrome expressing a truncated mutant Sall1 protein is protected from acute kidney injury. Am J Physiol Renal Physiol. 309(10):F852–F863. [DOI] [PubMed] [Google Scholar]
- Hochberg ME, Noble RJ.. 2017. A framework for how environment contributes to cancer risk. Ecol Lett. 20(2):117–134. [DOI] [PubMed] [Google Scholar]
- Hsu C-Y, Chang G-C, Chen Y-J, Hsu Y-C, Hsiao Y-J, Su K-Y, Chen H-Y, Lin C-Y, Chen J-S, Chen Y-J, et al. 2018. FAM198B is associated with prolonged survival and inhibits metastasis in lung adenocarcinoma via blockage of ERK-mediated MMP-1 expression. Clin Cancer Res. 24(4):916–926. [DOI] [PubMed] [Google Scholar]
- Huang X, Sun D, Wu T, Liu X, Xu S, Yang G.. 2021. Genomic insights into body size evolution in carnivora support Peto’s paradox. BMC Genomics 22(1):429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz MJ, Pollard KS, Siepel A.. 2011. PHAST and RPHAST: phylogenetic analysis with space/time models. Brief Bioinform. 12(1):41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Serra F, Bork P.. 2016. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol. 33(6):1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida A, Xing W, Docx MKF, Nakashima T, Wang Z, Kimizuka M, Van Hul W, Rating D, Spranger J, Ohashi H, et al. 2016. Identification of biallelic LRRK1 mutations in osteosclerotic metaphyseal dysplasia and evidence for locus heterogeneity. J Med Genet. 53(8):568–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques P-É, Jeyakani J, Bourque G.. 2013. The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet. 9(5):e1003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janousek V, Laukaitis CM, Yanchukov A, Karn RC.. 2016. The role of retrotransposons in gene family expansions in the human and mouse genomes. Genome Biol Evol. 8(9):2632–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacprzyk J, Locatelli AG, Hughes GM, Huang Z, Clarke M, Gorbunova V, Sacchi C, Stewart GS, Teeling EC.. 2021. Evolution of mammalian longevity: age-related increase in autophagy in bats compared to other mammals. Aging (Albany NY). 13(6):7998–8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami A, Nojima Y, Toyoda A, Takahoko M, Satoh M, Tanaka H, Wada H, Masai I, Terasaki H, Sakaki Y, et al. 2005. The zebrafish-secreted matrix protein you/scube2 is implicated in long-range regulation of hedgehog signaling. Curr Biol. 15(5):480–488. [DOI] [PubMed] [Google Scholar]
- Kawamata H, Furihata T, Omotehara F, Sakai T, Horiuchi H, Shinagawa Y, Imura J, Ohkura Y, Tachibana M, Kubota K, et al. 2003. Identification of genes differentially expressed in a newly isolated human metastasizing esophageal cancer cell line, T.Tn-AT1, by cDNA microarray. Cancer Sci. 94(8):699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane M, Semeiks J, Webb AE, Li YI, Quesada V, Craig T, Madsen LB, van Dam S, Brawand D, Marques PI, et al. 2015. Insights into the evolution of longevity from the bowhead whale genome. Cell Rep. 10(1):112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri P, Sirota M, Butte AJ.. 2012. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol. 8(2):e1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Xu S, Xiong L, Yu L, Fu X, Xu Y.. 2017. SALL4 promotes glycolysis and chromatin remodeling via modulating HP1α-Glut1 pathway. Oncogene 36(46):6472–6479. [DOI] [PubMed] [Google Scholar]
- Kostrzewska-Poczekaj M, Byzia E, Soloch N, Jarmuz-Szymczak M, Janiszewska J, Kowal E, Paczkowska J, Kiwerska K, Wierzbicka M, Bartochowska A, et al. 2019. DIAPH2 alterations increase cellular motility and may contribute to the metastatic potential of laryngeal squamous cell carcinoma. Carcinogenesis 40(10):1251–1259. [DOI] [PubMed] [Google Scholar]
- Kowalczyk A, Partha R, Clark NL, Chikina M.. 2020. Pan-mammalian analysis of molecular constraints underlying extended lifespan. Elife 9:e51089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krovi SH, Zhang J, Michaels-Foster MJ, Brunetti T, Loh L, Scott-Browne J, Gapin L.. 2020. Thymic iNKT single cell analyses unmask the common developmental program of mouse innate T cells. Nat Commun. 11:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Ward WO, Ren H, Vallanat B, Darlington GJ, Han E-S, Laguna JC, DeFord JH, Papaconstantinou J, Selman C, et al. 2012. Meta-analysis of gene expression in the mouse liver reveals biomarkers associated with inflammation increased early during aging. Mech Ageing Dev. 133(7):467–478. [DOI] [PubMed] [Google Scholar]
- Li C, Ma L, Chen X.. 2011. Interferon regulatory factor 3-CL, an isoform of IRF3, antagonizes activity of IRF3. Cell Mol Immunol. 8(1):67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Mao A, Fu X, She Y, Wei X.. 2019. Correlation between SEPS1 gene polymorphism and type 2 diabetes mellitus: a preliminary study. J Clin Lab Anal. 33(8):e22967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, de Magalhães JP.. 2013. Accelerated protein evolution analysis reveals genes and pathways associated with the evolution of mammalian longevity. Age (Dordr). 35(2):301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B.. 2019. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47(W1):W199–W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhou S, Wang J, Zhang Q, Yang S, Feng J, Xu B, Zhong S.. 2019. Identification of genes associated with survival of breast cancer patients. Breast Cancer. 26(3):317–325. [DOI] [PubMed] [Google Scholar]
- Liu Y, Sun J, Zhao M.. 2017. ONGene: a literature-based database for human oncogenes. J Genet Genomics. 44(2):119–121. [DOI] [PubMed] [Google Scholar]
- López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G.. 2013. The hallmarks of aging. Cell 153(6):1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenstine LJ. 1986. Neoplasms and proliferative disorders in nonhuman primates. In: Benirschke K. (eds) Primates. Proceedings in Life Sciences. New York: Springer. p. 781–814. DOI: 10.1007/978-1-4612-4918-4_53. [DOI] [Google Scholar]
- Lowenstine LJ, McManamon R, Terio KA.. 2016. Comparative pathology of aging great apes: bonobos, chimpanzees, gorillas, and orangutans. Vet Pathol. 53(2):250–276. [DOI] [PubMed] [Google Scholar]
- MacRae SL, Zhang Q, Lemetre C, Seim I, Calder RB, Hoeijmakers J, Suh Y, Gladyshev VN, Seluanov A, Gorbunova V, et al. 2015. Comparative analysis of genome maintenance genes in naked mole rat, mouse, and human. Aging Cell 14(2):288–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marand AP, Chen Z, Gallavotti A, Schmitz RJ.. 2021. A cis-regulatory atlas in maize at single-cell resolution. Cell 184(11):3041–3055.e21. [DOI] [PubMed] [Google Scholar]
- Medina L, Abellán A.. 2009. Development and evolution of the pallium. Semin Cell Dev Biol. 20(6):698–711. [DOI] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. 2011. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 20(1):11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris BJ, Willcox BJ, Donlon TA.. 2019. Genetic and epigenetic regulation of human aging and longevity. Biochim Biophys Acta Mol Basis Dis. 1865(7):1718–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntané G, Farré X, Rodríguez JA, Pegueroles C, Hughes DA, de Magalhães JP, Gabaldón T, Navarro A.. 2018. Biological processes modulating longevity across primates: a phylogenetic genome-phenome analysis. Mol Biol Evol. 35(8):1990–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushimiyimana I, Niskanen H, Beter M, Laakkonen JP, Kaikkonen MU, Ylä-Herttuala S, Laham-Karam N.. 2021. Characterization of a functional endothelial super-enhancer that regulates ADAMTS18 and angiogenesis. Nucleic Acids Res. 49(14):8078–8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesse RM, Ganten D, Gregory TR, Omenn GS.. 2012. Evolutionary molecular medicine. J Mol Med (Berl). 90(5):509–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunney L. 1999. Lineage selection and the evolution of multistage carcinogenesis. Proc Biol Sci. 266(1418):493–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunney L. 2018. Size matters: height, cell number and a person’s risk of cancer. Proc Biol Sci. 285(1889):20181743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin JD, Montague MJ, Tejada-Martinez D, de Manuel M, Del Campo J, Cheves HS, Di Fiore A, Fontsere C, Hodgson JA, Janiak MC, et al. 2021. The genomics of ecological flexibility, large brains, and long lives in capuchin monkeys revealed with fecalFACS. Proc Natl Acad Sci U S A. 118(7):e2010632118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orme D, Freckleton R, Thomas G, Petzoldt T, Fritz S, Isaac N, Pearse W, et al. 2012. Caper: comparative analyses of phylogenetics and evolution in R. Proc R Soc B 2:458. [Google Scholar]
- Passingham RE, Wise SP.. 2012. The neurobiology of the prefrontal cortex: anatomy, evolution, and the origin of insight. Oxford: OUP. [Google Scholar]
- Pennell MW, Eastman JM, Slater GJ, Brown JW, Uyeda JC, FitzJohn RG, Alfaro ME, Harmon LJ.. 2014. geiger v2.0: an expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics 30(15):2216–2218. [DOI] [PubMed] [Google Scholar]
- Peto R, Epidemiology MM, Tests STM.. 1977. In: Hiatt HH, Watson JD, Winsten JA, editors. The origins of human cancer. New York: Cold Spring Harbor Laboratory. p. 1473–1481.
- Petralia RS, Mattson MP, Yao PJ.. 2014. Aging and longevity in the simplest animals and the quest for immortality. Ageing Res Rev. 16:66–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontis J, Planet E, Offner S, Turelli P, Duc J, Coudray A, Theunissen TW, Jaenisch R, Trono D.. 2019. Hominoid-specific transposable elements and KZFPs facilitate human embryonic genome activation and control transcription in naive human ESCs. Cell Stem Cell. 24(5):724–735.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Freitas-Rodríguez S, Miller J, Pérez-Silva JG, Jiang Z-F, Tapia W, Santiago-Fernández O, Campos-Iglesias D, Kuderna LFK, Quinzin M, et al. 2019. Giant tortoise genomes provide insights into longevity and age-related disease. Nat Ecol Evol. 3(1):87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SG, Jackson JG.. 2016. SASP: tumor suppressor or promoter? Yes! Trends Cancer Res. 2(11):676–687. [DOI] [PubMed] [Google Scholar]
- Recio C, Lucy D, Purvis GSD, Iveson P, Zeboudj L, Iqbal AJ, Lin D, O’Callaghan C, Davison L, Griesbach E, et al. 2018. Activation of the immune-metabolic receptor GPR84 enhances inflammation and phagocytosis in macrophages. Front Immunol. 9:1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiners J, van Wijk E, Märker T, Zimmermann U, Jürgens K, te Brinke H, Overlack N, Roepman R, Knipper M, Kremer H, et al. 2005. Scaffold protein harmonin (USH1C) provides molecular links between Usher syndrome type 1 and type 2. Hum Mol Genet. 14(24):3933–3943. [DOI] [PubMed] [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 3(2):217–223. [Google Scholar]
- Rodríguez JA, Marigorta UM, Hughes DA, Spataro N, Bosch E, Navarro A.. 2017. Antagonistic pleiotropy and mutation accumulation influence human senescence and disease. Nat Ecol Evol. 1(3):55. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Murillo L, Xu B, Roos JL, Abecasis GR, Gogos JA, Karayiorgou M.. 2014. Fine mapping on chromosome 13q32–34 and brain expression analysis implicates MYO16 in schizophrenia. Neuropsychopharmacology 39(4):934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romiguier J, Ranwez V, Douzery EJP, Galtier N.. 2010. Contrasting GC-content dynamics across 33 mammalian genomes: relationship with life-history traits and chromosome sizes. Genome Res. 20(8):1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth ACJ, Gonnet GH, Dessimoz C.. 2008. Algorithm of OMA for large-scale orthology inference. BMC Bioinformatics 9:518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Studio Team (2020). R Studio: Integrated Development for R. R Studio, PBC, Boston, MA. http://www.rstudio.com/.
- Schwanke H, Stempel M, Brinkmann MM.. 2020. Of keeping and tipping the balance: host regulation and viral modulation of IRF3-dependent IFNB1 expression. Viruses 12(7):733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluanov A, Gladyshev VN, Vijg J, Gorbunova V.. 2018. Mechanisms of cancer resistance in long-lived mammals. Nat Rev Cancer. 18(7):433–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluanov A, Hine C, Azpurua J, Feigenson M, Bozzella M, Mao Z, Catania KC, Gorbunova V.. 2009. Hypersensitivity to contact inhibition provides a clue to cancer resistance of naked mole-rat. Proc Natl Acad Sci U S A. 106(46):19352–19357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, McElhinny AS, Cao Y, Gao P, Liu J, Bronson R, Griffin JD, Wu L.. 2006. The Notch coactivator, MAML1, functions as a novel coactivator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev. 20(6):675–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Sinai I.. 2021. GeneOverlap: test and visualize gene overlaps. R package version 1.28.0. Available from: http://shenlab-sinai.github.io/shenlab-sinai/ (accessed 5th November 2021).
- Singh PP, Demmitt BA, Nath RD, Brunet A.. 2019. The genetics of aging: a vertebrate perspective. Cell 177(1):200–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzypczak M, Lattrich C, Häring J, Schüler S, Ortmann O, Treeck O.. 2013. Expression of SCUBE2 gene declines in high grade endometrial cancer and associates with expression of steroid hormone receptors and tumor suppressor PTEN. Gynecol Endocrinol. 29(12):1031–1035. [DOI] [PubMed] [Google Scholar]
- Smaers JB, Gómez-Robles A, Parks AN, Sherwood CC.. 2017. Exceptional evolutionary expansion of prefrontal cortex in great apes and humans. Curr Biol. 27(10):1549. [DOI] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA.. 2009. The cancer genome. Nature 458(7239):719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulak M, Fong L, Mika K, Chigurupati S, Yon L, Mongan NP, Emes RD, Lynch VJ.. 2016. Correction: TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. Elife 5:e11994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Xu K, Ren Y, Xie Y, Zhang X, Tian L, Li Y.. 2018. Comprehensive molecular screening in Chinese Usher syndrome patients. Invest Ophthalmol Vis Sci. 59(3):1229–1237. [DOI] [PubMed] [Google Scholar]
- Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, Snyder MP, Wang T.. 2014. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 24(12):1963–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram V, Wysocka J.. 2020. Transposable elements as a potent source of diverse cis-regulatory sequences in mammalian genomes. Philos Trans R Soc Lond B Biol Sci. 375(1795):20190347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds MRE, Blomberg SP.. 2014. A primer on phylogenetic generalised least squares. In: Modern phylogenetic comparative methods and their application in evolutionary biology. p. 105–130. Available from: 10.1007/978-3-662-43550-2_5 10.1007/978-3-662-43550-2_5 (accessed 5th November 2021). [DOI] [Google Scholar]
- Tacutu R, Thornton D, Johnson E, Budovsky A, Barardo D, Craig T, Diana E, Lehmann G, Toren D, Wang J, et al. 2018. Human ageing genomic resources: new and updated databases. Nucleic Acids Res. 46(D1):D1083–D1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejada-Martinez D, de Magalhães JP, Opazo JC.. 2021. Positive selection and gene duplications in tumour suppressor genes reveal clues about how cetaceans resist cancer. Proc Biol Sci. 288(1945):20202592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas F, Fisher D, Fort P, Marie J-P, Daoust S, Roche B, Grunau C, Cosseau C, Mitta G, Baghdiguian S, et al. 2013. Applying ecological and evolutionary theory to cancer: a long and winding road. Evol Appl. 6(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Wang X, Sun J, Lin W, Chen L, Liu S, Wu X, Shi L, Xu P, Cai X, et al. 2020. IRF3 prevents colorectal tumorigenesis via inhibiting the nuclear translocation of β-catenin. Nat Commun. 11(1):5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Azpurua J, Hine C, Vaidya A, Myakishev-Rempel M, Ablaeva J, Mao Z, Nevo E, Gorbunova V, Seluanov A.. 2013. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature 499(7458):346–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Doerig K, Park R, Can Ran Qin A, Hwang C, Neary A, Gilbert M, Seluanov A, Gorbunova V.. 2018. Evolution of telomere maintenance and tumour suppressor mechanisms across mammals. Philos Trans R Soc Lond B Biol Sci. 373(1741):20160443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollis M, Robbins J, Webb AE, Kuderna LFK, Caulin AF, Garcia JD, Bèrubè M, Pourmand N, Marques-Bonet T, O’Connell MJ, et al. 2019. Return to the sea, get huge, beat cancer: an analysis of cetacean genomes including an assembly for the humpback whale (Megaptera novaeangliae). Mol Biol Evol. 36(8):1746–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollis M, Schiffman JD, Boddy AM.. 2017. Evolution of cancer suppression as revealed by mammalian comparative genomics. Curr Opin Genet Dev. 42:40–47. [DOI] [PubMed] [Google Scholar]
- Tollis M, Schneider-Utaka AK, Maley CC.. 2020. The evolution of human cancer gene duplications across mammals. Mol Biol Evol. 37(10):2875–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasetti C, Vogelstein B.. 2015. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 347(6217):78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trizzino M, Kapusta A, Brown CD.. 2018. Transposable elements generate regulatory novelty in a tissue-specific fashion. BMC Genomics 19(1):468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trizzino M, Park Y, Holsbach-Beltrame M, Aracena K, Mika K, Caliskan M, Perry GH, Lynch VJ, Brown CD.. 2017. Transposable elements are the primary source of novelty in primate gene regulation. Genome Res. 27(10):1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M-T, Cheng C-J, Lin Y-C, Chen C-C, Wu A-R, Wu M-T, Hsu C-C, Yang R-B.. 2009. Isolation and characterization of a secreted, cell-surface glycoprotein SCUBE2 from humans. Biochem J. 422(1):119–128. [DOI] [PubMed] [Google Scholar]
- Uyeda JC, , PennellMW, , MillerET, , MaiaR, , McClain CR.. 2017. The Evolution of Energetic Scaling across the Vertebrate Tree of Life. The American Naturalist. 190(2):185–199. [DOI] [PubMed] [Google Scholar]
- van Duyvenvoorde HA, Lui JC, Kant SG, Oostdijk W, Gijsbers ACJ, Hoffer MJV, Karperien M, Walenkamp MJE, Noordam C, Voorhoeve PG, et al. 2014. Copy number variants in patients with short stature. Eur J Hum Genet. 22(5):602–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez JM, Sulak M, Chigurupati S, Lynch VJ.. 2018. A zombie LIF gene in elephants is upregulated by TP53 to induce apoptosis in response to DNA damage. Cell Rep. 24(7):1765–1776. [DOI] [PubMed] [Google Scholar]
- Wang BX, Fish EN.. 2019. Global virus outbreaks: interferons as 1st responders. Semin Immunol. 43:101300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, Burgess SM, Brachmann RK, Haussler D.. 2007. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci U S A. 104(47):18613–18618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CC, Nabholz B, Romiguier J, Ellegren H.. 2014. Kr/Kc but not dN/dS correlates positively with body mass in birds, raising implications for inferring lineage-specific selection. Genome Biol. 15(12):542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Liu C-J, Li Z.. 2014. ADAMTS-18: a metalloproteinase with multiple functions. Front Biosci. 19:1456–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RR, Milholland B, MacRae SL, Lin M, Zheng D, Vijg J.. 2015. Comprehensive transcriptional landscape of aging mouse liver. BMC Genomics 16:899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson GS, Adams DM.. 2019. Recurrent evolution of extreme longevity in bats. Biol Lett. 15(4):20180860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Griffin JD.. 2000. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet. 26(4):484–489. [DOI] [PubMed] [Google Scholar]
- Wu M-Y, Eldin KW, Beaudet AL.. 2008. Identification of chromatin remodeling genes Arid4a and Arid4b as leukemia suppressor genes. J Natl Cancer Inst. 100(17):1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R-C, Young I-C, Chen Y-F, Chuang S-T, Toubaji A, Wu M-Y.. 2019. Identification of the PTEN-ARID4B-PI3K pathway reveals the dependency on ARID4B by PTEN-deficient prostate cancer. Nat Commun. 10(1):4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier GM, Cobourne MT.. 2011. Scube2 expression extends beyond the central nervous system during mouse development. J Mol Histol. 42(5):383–391. [DOI] [PubMed] [Google Scholar]
- Xu M, Pirtskhalava T, Farr JN, Weigand BM, Palmer AK, Weivoda MM, Inman CL, Ogrodnik MB, Hachfeld CM, Fraser DG, et al. 2018. Senolytics improve physical function and increase lifespan in old age. Nat Med. 24(8):1246–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai H, Chiba S, Hangai S, Kometani K, Inoue A, Kimura Y, Abe T, Kiyonari H, Nishio J, Taguchi-Atarashi N, et al. 2018. Revisiting the role of IRF3 in inflammation and immunity by conditional and specifically targeted gene ablation in mice. Proc Natl Acad Sci U S A. 115(20):5253–5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yao J, Wu D, Zhang C, Yan T, Zhao Y, Shen H, Xue K, Huang X, Wang Z, Qiu Y.. 2021. Macrophage IRX3 promotes diet-induced obesity and metabolic inflammation. Nat Immunol. 22(10):1268–1279. [DOI] [PubMed] [Google Scholar]
- Yu J, Gu X, Yi S.. 2016. Ingenuity pathway analysis of gene expression profiles in distal nerve stump following nerve injury: insights into Wallerian degeneration. Front Cell Neurosci. 10:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Seim I, Yin M, Tian R, Sun D, Ren W, Yang G, Xu S.. 2021. Comparative analyses of aging-related genes in long-lived mammals provide insights into natural longevity. Innovation (NY). 2(2):100108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Cowled C, Shi Z, Huang Z, Bishop-Lilly KA, Fang X, Wynne JW, Xiong Z, Baker ML, Zhao W, et al. 2013. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science 339(6118):456–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z.. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 22(12):2472–2479. [DOI] [PubMed] [Google Scholar]
- Zhao C, Takita J, Tanaka Y, Setou M, Nakagawa T, Takeda S, Yang HW, Terada S, Nakata T, Takei Y, et al. 2001. Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bβ. Cell 105(5):587–597. [DOI] [PubMed] [Google Scholar]
- Zhao M, Sun J, Zhao Z.. 2013. TSGene: a web resource for tumor suppressor genes. Nucleic Acids Res. 41(Database issue):D970–D976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Katzman RB, Delmolino LM, Bhat I, Zhang Y, Gurumurthy CB, Germaniuk-Kurowska A, Reddi HV, Solomon A, Zeng M-S, et al. 2007. The notch regulator MAML1 interacts with p53 and functions as a coactivator. J Biol Chem. 282(16):11969–11981. [DOI] [PubMed] [Google Scholar]
- Zou Y, Lu P, Shi J, Liu W, Yang M, Zhao S, Chen N, Chen M, Sun Y, Gao A, et al. 2017. IRX3 promotes the browning of white adipocytes and its rare variants are associated with human obesity risk. EBioMedicine 24:64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full CellAge database, including all annotations regarding cell types and cell lines is available at https://genomics.senescence.info/cells/.