Abstract
Multiplex PCR amplification followed by either agarose gel electrophoresis (PCR-AGE) or microchip electrophoresis (PCR-ME) was used to test a total of 120 fungal strains. The internal transcribed spacer 1 (ITS1) and ITS2 regions and the 5.8S ribosomal DNA (rDNA) region of the fungi were amplified by using universal primers ITS1 and ITS4. The ITS2 region was simultaneously amplified by using universal primers ITS3 and ITS4. Since Trichosporon asahi and T. asteroides showed similar lengths for two amplicons, 29 different gel patterns were demonstrated for 30 yeast species tested on the basis of differences in the lengths of one or two amplicons. Of 75 yeast isolates from clinical materials, 5 isolates (6.8%) which were incompletely identified or not identified by the phenotypic method were identified with our PCR-based method (2 isolates as Candida guilliermondii, 2 as C. krusei, and 1 as C. zeylanoides). No differences in discriminating power or sensitivity were observed between the PCR-AGE method and the PCR-ME method. These methods, prospectively applied to 24 yeast-positive blood culture bottles (16 patients), resulted in the correct detection of 24 yeast strains. In conclusion, multiplex PCR followed by electrophoresis seems to be a promising tool for the rapid identification of common and uncommon yeast strains from culture colonies and from yeast-positive blood culture bottles (5.5 h for the PCR-AGE method and 3 h for the PCR-ME method).
The virulence of Candida isolates differs according to the species, with Candida albicans being most virulent, followed by C. tropicalis (23); thus, the prompt and accurate detection and identification of yeast species are very important. Detection and identification have been made even more important because of the emergence of candidemia caused by innately fluconazole-resistant non-C. albicans Candida species (12). Various commercial systems that can identify these pathogens within 4 to 72 h have been developed (2). Although correct identification of clinically relevant yeast strains can be achieved with these systems, incomplete or incorrect identification may occur when certain new and emerging yeast strains are tested (2). Moreover, culture-based phenotypic identification of Candida species from clinical materials requires at least 1 day to obtain a pure culture.
PCR-based methods for the rapid detection and identification of Candida species have also been described. These include restriction fragment length polymorphism analysis (3), PCR-restriction fragment length polymorphism analysis (7, 11), PCR with species-specific probes (14, 15), and random amplification of polymorphic DNA analysis (16). These methods have already shown great promise in the field of species identification and strain delineation. However, none of these methods has yet been used in the clinical laboratory.
PCR with fungus-specific primers, targeting the conserved sequences of 5.8S and 28S ribosomal DNAs (rDNAs) as well as those of 18S and 28S rDNAs, results in the respective amplification of the species-specific internal transcribed spacer (ITS) 2 (ITS2) region and of the ITS1 and ITS2 regions, which vary in amplicon length and sequence according to species (4, 6). Recently, Turenne et al. reported that differences in the sizes of the ITS2 regions of fungi were useful for the rapid identification of clinically important fungi (20). However, they found only a three-nucleotide band difference when comparing the lengths of C. albicans and C. krusei ITS2 amplicons. We used the variability in the lengths of both ITS1 and ITS2 regions to achieve the specific identification of fungal strains, and we determined the lengths of PCR fragments by using both agarose gel electrophoresis (AGE) and microchip electrophoresis (ME).
MATERIALS AND METHODS
Organisms.
The 45 strains used in this study as control strains are listed in Table 1. These included 40 strains obtained from the Institute of Fermentation (IFO), Osaka, Japan; 4 strains obtained from the Japan Collection of Microorganisms (JCM), Institute of Physical and Chemical Research, Wako, Japan; and 1 strain (given the prefix K) which has been identified and maintained in our laboratory.
TABLE 1.
Summary of sizes of PCR-amplified fragments by species
| Organism | Strain(s) | Fragment size, in bp, found by PCR with the following primer pairsa:
|
|
|---|---|---|---|
| ITS1-ITS4 | ITS3-ITS4 | ||
| Candida albicans | IFO 1398, IFO 1974, IFO 1061 | 529–535 (532) | 335–342 (339) |
| Candida catenulata | IFO 0745 | 391–393 (392) | 262–266 (264) |
| Candida famata | IFO 0664, IFO 856 | 632–634 (633) | 369–370 (370) |
| Candida glabrata | IFO 0005, IFO 0622 | 869–879 (874) | 412–418 (415) |
| Candida guilliermondii | IFO 10279, IFO 1972, IFO 0566 | 598–607 (603) | 375–380 (378) |
| Candida holmii | IFO 1629 | 717–717 (717) | 431–432 (432) |
| Candida inconspicua | IFO 0621 | 447–449 (448) | 298–299 (299) |
| Candida intermedia | IFO 0761 | 384–388 (386) | 250–256 (253) |
| Candida kefyr | IFO 0586, IFO 10287, IFO 1065 | 720–723 (722) | 422–427 (425) |
| Candida krusei | IFO 0584 | 496–503 (500) | 332–338 (335) |
| Candida lambica | IFO 10289 | 432–434 (434) | 301–304 (303) |
| Candida lipolytica | IFO 1549, IFO 1548 | 358–359 (359) | 233–239 (236) |
| Candida lusitaniae | IFO 10059, IFO 10058 | 373–378 (375) | 245–254 (250) |
| Candida norvegensis | IFO 1020 | 480–483 (482) | 314–315 (315) |
| Candida parapsilosis | IFO 1396 | 511–521 (516) | 308–314 (311) |
| Candida rugosa | IFO 1364 | 416–419 (418) | 268–269 (269) |
| Candida saitoana | IFO 0768 | 611–614 (613) | 365–368 (367) |
| Candida tropicalis | IFO 1400, IFO 10241 | 517–524 (521) | 326–331 (329) |
| Candida utilis | IFO 10707 | 553–555 (554) | 354–357 (356) |
| Candida zoylanoides | IFO 10326 | 620–622 (621) | 367–369 (368) |
| Cryptococcus neoformans | K615 | 547–555 (550) | 370–375 (372) |
| Cryptococcus humicolus | IFO 10251 | 520–525 (522) | 335–339 (337) |
| Dipodascus capitatus | IFO 1197 | 454–457 (456) | 296–297 (297) |
| Pichia anomala | IFO 10213 | 614–615 (615) | 373–376 (375) |
| Pichia chambardii | IFO 1274 | 629–630 (630) | 380–382 (381) |
| Saccharomyces cerevisiae | IFO 0565 | 835–839 (837) | 414–417 (416) |
| Stephanoascus ciferrii | IFO 1854, JCM 9551 | 571–572 (572) | 354–356 (355) |
| Trichosporon asahii | JCM 2466 | 535–537 (536) | 352–356 (354) |
| Trichosporon asteroides | JCM 10016 | 537–540 (539) | 354–357 (356) |
| Trichosporon cutaneum | JCM 1462 | 521–523 (522) | 348–349 (349) |
| Aspergillus flavus | IFO 5839 | 581–585 (583) | 340–349 (345) |
| Aspergillus fumigatus | IFO 7839 | 580–588 (584) | 337–347 (342) |
| Aspergillus niger | IFO 4414 | 586–590 (588) | 342–350 (346) |
Range (mean) from three separate experiments.
Seventy-five clinical isolates were obtained from patients at the Kanazawa University Hospital. Presumptive identification of these isolates was made by means of colony color on CHROMagar Candida medium, and the isolates were definitively identified with an API 20C kit (bioMerieux-Vitek, Hazelwood, Mo.). When necessary, their abilities to produce germ tubes and chlamydospores and to grow at 42°C were tested. These isolates comprised C. albicans (n = 23), C. tropicalis (n = 12), C. parapsilosis (n = 6), C. guilliermondii (n = 9), C. glabrata (n = 8), C. krusei (n = 6), Cryptococcus neoformans (n = 6), and incompletely identified or unidentified strains (n = 5).
DNA extraction.
The Zymolyase-based method used to extract DNA has been described previously (3). In brief, yeast isolates were grown on CHROMagar Candida medium for 24 to 48 h at 35°C. Single colonies were inoculated into 1 ml of Tris-EDTA (TE) buffer at a McFarland standard of 0.5. Aspergillus strains were grown on Sabouraud dextrose agar for 3 days, and the conidia were inoculated into TE buffer. After centrifugation (5,000 × g, 3 min) in an Eppendorf microcentrifuge, the sediments were resuspended in 0.1 ml of 1 M sorbitol–1 M EDTA (pH 7.5) containing 5 μl of β-mercaptoethanol and 1 to 2 mg of Zymolyase; the mixture was incubated for 15 min at 37°C. For filamentous fungi, 1 to 2 mg of lysing enzymes (Sigma Chemical Co., St. Louis, Mo.) was added to the tube, and the mixture was incubated for a further 15 min at 37°C. Finally, 5 μl of proteinase K solution (20 mg/ml) was added to the tube, and the mixture was incubated at 55°C for 10 min and in a boiling water bath for 5 min.
Blood cultures.
A total of 4 to 8 ml of blood drawn from each patient was inoculated into aerobic and anaerobic blood culture bottles (Organon Teknika Corp., Durham, N.C.). All bottles were incubated in a BACTEC 9240 automated system. When microbial growth was detected by the system, microscopic examinations using Gram staining and subcultures were performed. Yeast-positive bottles were immediately processed for DNA extraction, and a prospective identification of the blood isolate was made on the basis of results obtained with the commercial system and PCR analysis. Ten randomely selected samples from patients with bacteremia were also tested as negative controls.
Processing of blood samples for PCR.
About 0.2 ml of blood culture fluid from each bottle was inoculated into a tube containing 1 ml of 0.17% sodium dodecyl sulfate, and this mixture was diluted with 8 to 10 ml of sterile distilled water. After centrifugation (3,000×g, 10 min), the pellets were placed in 1 ml of TE buffer and processed with the same method as that described above. Purified DNA was obtained with a MagExtract DNA extraction kit (Toyobo Co. Ltd., Osaka, Japan); about 1 h was required to disrupt and isolate DNA from positive blood culture bottles.
PCR procedure.
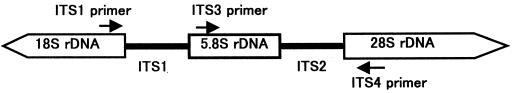
Primers ITS1 (5′-TCCGTAGGTGAACCTGCG-3′), ITS3 (5′-GCATCGATGAAGAACGCAGC-3′), and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′), targeting the conserved regions of 18S, 5.8S, and 28S rDNAs (22), respectively, were used for amplification. The ITS1-ITS4 primer pair was used to amplify the intervening 5.8S rDNA and the adjacent ITS1 and ITS2 regions, and the ITS3-ITS4 primer pair was used to amplify a large portion of the 5.8S rDNA and the adjacent ITS2 region (Fig. 1). PCR amplification was performed with a volume of 50 μl. Two microliters of each sample was added to the PCR master mixture, which consisted of 5 μl of 10× PCR buffer, 4 μl of a deoxynucleoside triphosphate mixture (0.1 mM each dNTP), 0.8 μl of each primer (40 pmol of each primer), and 0.4 μl (2.0 U) of ExTaq DNA polymerase (Takara Biomedicals, Osaka, Japan), with the remaining volume consisting of distilled water. Amplification consisted of an initial denaturation at 94°C for 4 min; 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min; and a final extension at 72°C for 4 min; a GeneAmp PCR system 9600 thermal cycler (Perkin-Elmer Corp., Emeryville, Calif.) was used. Negative control reactions without any template DNA were carried out simultaneously.
FIG. 1.
Schematic representation of the fungal ribosomal genes containing the primer target areas used in this study.
Electrophoresis.
Gel electrophoresis with 1.5% agarose gels was conducted with 1× TBE buffer (0.1 M Tris, 0.09 M boric acid, 1 mM EDTA) at 4.8 V/cm for 2 h. A 100-bp DNA ladder (Promega Corp., Madison, Wis.) was run concurrently with amplicons for sizing of the bands. Gels were stained with ethidium bromide-TBE solution for 20 min and then photographed. The computer program Diversity Database (PDI Inc., New York, N.Y.) was used to calculate molecular sizes. Fragment sizes of control strains were measured three times for each strain on different gels. In addition to AGE, PCR products were analyzed with an ME instrument (model SV1100; Hitachi Electronics Engineering Co. Ltd., Tokyo, Japan). The microchip had three sample wells, and wells were connected with a cross- channel 0.1 mm wide, 0.03 mm deep, and 45 mm long. After the channel was filled with 0.6% hydroxypropyl methylcellulose containing ethidium bromide, a mixture of 9 μl of PCR products and 1 μl of loading buffer containing the internal standards (100 and 800 bp) was loaded into one of the sample wells of the microchip; the program was run at 300 V for 1 min (injection time) and at 750 V for 3 min (separation time). Accurate fragment size analysis based on the electrophoretic mobility of the sample relative to the internal standards was achieved by using DNA size analysis software.
RESULTS
To determine the sensitivity of the PCR followed by electrophoresis, 0.5-ml aliquots of TE buffer were seeded with 0, 101, 102, 103, 104, or 105 C. albicans blastoconidia. The sensitivity of PCR-AGE and PCR-ME was 103 cells per 0.5 ml of buffer, or 20 cells per 2 μl of sample.
For the isolation of yeast DNA from culture colonies, 40 min was required; in comparison, 1 h is required for DNA isolation from yeast-positive blood culture bottles. For the PCR-AGE method, 2 h was required for PCR amplification, 2 h was required for AGE (4 min for ME), and 30 min was required for staining and fragment analysis. Therefore, the times from yeast-positive blood culture bottles to genus or species identification by our PCR-based method were about 5.5 h for the PCR-AGE method and about 3 h for the PCR-ME method.
Amplification of all fungi tested with ITS1 and ITS4 primers yielded fragments 350 to 880 bp long, while ITS3 and ITS4 primers yielded fragments 233 to 432 bp long. The sizes of the PCR products amplified with the former were nearly equal for some species: C. parapsilosis and C. tropicalis, about 520 bp; C. utilis and C. neoformans, about 554 bp; and C. saitoana and Pichia anomala, about 614 bp. However, these organisms showed species-specific differences in the sizes of the PCR products amplified with ITS3 and ITS4 primers. In contrast, the sizes of the PCR products amplified with ITS3 and ITS4 primers were identical for C. albicans, C. krusei, and C. humicolus and for C. famata, C. neoformans, and P. anomala. The PCR products amplified with ITS3 and ITS4 primers were about 356 bp long for C. utilis and Stephanoascus ciferrii. However, these organisms showed species-specific differences in the sizes of the PCR products amplified with ITS1 and ITS4 primers. On the other hand, the lengths of two amplicons of Trichosporon asahi and T. asteroides were almost equal. Three Aspergillus species tested also demonstrated fragments of equal or nearly equal lengths. Therefore, 30 genus- or species-specific patterns were recognized among the 33 fungal species on the basis of differences in the lengths of one or two fragments of amplicons (Table 1). No intraspecies variability was detected.
Seventy-five isolates identified with routine phenotypic identification methods were correctly identified by the species-specific patterns shown in Table 1. The results of electrophoresis of PCR products from eight Candida species are shown in Fig. 2. ME analysis of 120 fungal strains showed that the lengths of two amplicon were in agreement with the results obtained with AGE analysis. The results obtained with the ME method for three Candida species are shown in Fig. 3. The standard deviations (SDs) of calculated molecular sizes ranged from 1.20 to 3.36 with the AGE method and from 1.97 to 4.61 with the ME method (Table 2). Fragments having molecular sizes with more than 2 SDs were considered different, a value of about <7 bp in the fragment size range of 300 to 500 bp. The documented resolution of the ME instrument provided by the manufacturer was specified as 10 bp. We concluded that the AGE and ME methods could delineate strains which showed differences of 10 bp or more in the mean lengths of either one or two PCR products.
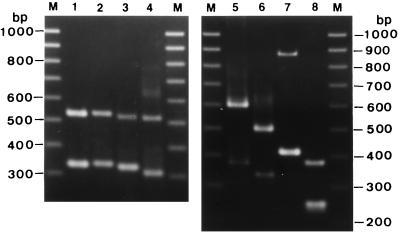
FIG. 2.
Gels showing amplification by PCR and length variations in the ITS regions of medically important Candida isolates. Lane 1, C. albicans; lane 2, C. dubliniensis; lane 3, C. tropicalis; lane 4, C. parapsilosis; lane 5, C. guilliermondii; lane 6, C. krusei; lane 7, C. glabrata; lane 8, C. lusitaniae; lanes M, 100-bp DNA ladder.
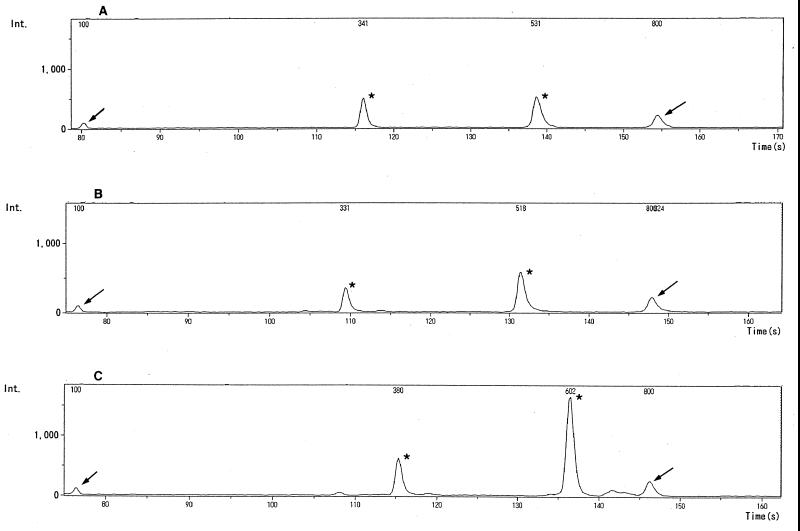
FIG. 3.
ME patterns of C. albicans IFO 0745 (A), C. tropicalis IFO 1400 (B), and C. guilliermondii IFO 0566 (C). Arrows indicate DNA size markers of 100 bp (left) and 800 bp (right), and asterisks indicate PCR products (A, 341 and 531 bp; B, 329 and 518 bp; and C, 380 and 602 bp). Int., intensity.
TABLE 2.
PCR fragment lengths of medically important Candida species
| Organism (no. of strains) | Base pair determination (mean ± SD) of PCR fragments by:
|
|
|---|---|---|
| AGE | ME | |
| C. albicans (36) | 337 ± 2.77 | 338 ± 3.04 |
| 533 ± 2.73 | 531 ± 3.09 | |
| C. tropicalis (14) | 327 ± 264 | 328 ± 2.79 |
| 521 ± 2.33 | 520 ± 3.09 | |
| C. parapsilosis (8) | 309 ± 1.20 | 309 ± 2.79 |
| 519 ± 3.31 | 518 ± 3.21 | |
| C. guilliermondii (13) | 376 ± 1.85 | 377 ± 2.89 |
| 602 ± 2.64 | 601 ± 3.02 | |
| C. krusei (7) | 333 ± 2.96 | 335 ± 3.02 |
| 499 ± 3.01 | 499 ± 3.12 | |
| C. glabrata (11) | 414 ± 2.54 | 416 ± 1.97 |
| 874 ± 3.36 | 873 ± 4.61 | |
Five strains could not be identified with the API 20C kit, and the code numbers of these strains were 6000004 (T. capitatum, C. krusei, and C. lipolytica) for K10320, 6556377 (C. famata and C. guilliermondii) for K7058, 6000104 (C. lipolytica and C. krusei) for K447, 6116333 (no code) for K10585, and 6402224 (no code) for K5701. The PCR-based method identified strains K10320 and K447 as C. krusei, strains K7058 and K10585 as C. guilliermondii, and strain K5701 as C. zeylanoides.
Twenty-four blood culture specimens obtained from 16 patients were positive for yeasts, as identified by primary Gram stain. These blood isolates were identified as C. albicans (12 patients, 14 specimens), C. parapsilosis (1 patient, 1 specimen), C. guilliermondii (1 patient, 4 specimens), C. glabrata (1 patient, 2 specimens), and C. neoformans (1 patient, 3 specimens) with the conventional phenotypic identification method. The results of this identification method were in agreement with the results of the PCR-based identification method.
DISCUSSION
The prompt and accurate identification of yeasts is becoming increasingly important because new antifungal agents with different activities against various species are being developed and because Candida species differ in their virulence characteristics (5, 23). Although yeast identification kits such as the RapID yeast system, API 20C kit, and ID 32C allow for simple identification of yeast isolates, the ability to correctly identify isolates at the species level differs significantly among the commercial systems (21). For the API 20C kit, 5 to 10% incomplete identification (i.e., supplemental tests were required for correct identification) has been reported for C. albicans, C. tropicalis, and C. glabrata strains (21). In our study, five Candida strains incompletely identified or not identified with the API 20C kit were correctly identified with the PCR-based method described here. Thanos et al. (19) have reported that three Candida isolates incompletely identified with the ID 32C system were correctly identified by PCR fingerprints (one each of C. albicans, C. guilliermondii, and C. parapsilosis).
Various molecular approaches have been used for the detection of fungi from clinical samples. Targets for the detection of fungi at the genus or species level have included 18S rDNA (9), mitochondrial DNA (4), intergenic spacer regions (13), and ITS regions (7). The sizes of the 18S, 5.8S, and 26S rRNA genes are essentially identical in all species, while the lengths of the ITS regions depend on the species. The ITS region is located between the 18S and 26S rRNA genes and is subdivided into the ITS1 region, between the 18S and 5.8S rRNA genes, and the ITS2 region, between the 5.8S and 26S rRNA genes. Interspecies differences in fragment sizes or sequences of the ITS1-5.8S rDNA-ITS2 regions (6, 7), ITS1 region (10), and ITS2 region (8, 20) have been used to detect and identify fungi. We utilized the differences in the lengths of both ITS1 and ITS2 regions to achieve specific identification of pathogenic fungi from culture colonies.
In this study, most yeast strains were correctly identified at the species level by means of gel electrophoresis, but C. albicans and C. dubliniensis could not be differentiated because of their nearly identical amplicon sizes (data not shown). C. dubliniensis is a recently reported species of chlamydospore- and germ tube-positive yeast which has been isolated from the oral cavities of human immunodeficiency virus-infected individuals. Despite the many phenotypic similarities between C. dubliniensis and C. albicans, pronounced genetic differences between the two species have been determined by DNA fingerprinting, karyotype analysis, and DNA sequence analysis of rDNA (18). However, no significant differences in fragment sizes were observed with our PCR-based method. Therefore, we routinely plate yeast isolates on CHROMagar Candida medium, and colonies which have turned dark green after 48 h of incubation are tested for their ability to grow at 42°C on Sabouraud agar medium. Isolates which fail to grow or which grow poorly at 42°C are tentatively identified as C. dubliniensis. On the other hand, five incompletely identified or unidentified strains were identified as C. guilliermondii (two strains), C. krusei (two strains), and C. zeylanoides (one strain) based on the characteristic lengths of two amplicons. The multiplex PCR-based method described here and used by us may thus be effective for the preliminary identification of clinically uncommon or emerging yeast isolates.
Sequences of the ITS1 and ITS2 regions of Trichosporon species have been reported by Sugita et al. (17). They found that the ITS1 and ITS2 regions of T. asahi and T. asteroides are about 298 bp long and that those of T. cutaneum and T. mucoides are about 285 or 286 bp long. In addition, the lengths of the PCR products, including the complete ITS1-5.8S rDNA-ITS2 regions and portions of the 18S (30 bp) and 28S (59 bp) rRNA genes, were identified as 595 bp for Aspergillus flavus, 596 to 598 bp for A. fumigatus, 599 bp for A. niger, and 609 to 613 bp for A. terreus (6). These previously reported results, which are similar to ours, indicate that differences in the lengths of the ITS regions of Trichosporon spp. and Aspergillus spp. are of little value for species identification.
As for rapid sizing of PCR products, Turenne et al. (20) used a capillary electrophoresis system which needs less than 30 min for analysis. Recently, electrophoretic analysis with a microchip was developed (1). To determine the lengths of PCR products more rapidly, we used an ME instrument. This instrument can detect PCR products within 4 min, whereas AGE used in our study needed 2 h to determine the lengths of amplicons. The former has enabled us to generate a profile of the expected amplicon sizes for a broad range of yeasts, thus allowing for their rapid identification. The advantages of using such an instrument include speed and the elimination of slab gels and staining steps as well as of species-specific probes or DNA sequencing. Furthermore, minimal manual labor is required, thus making the fragment detection system accurate and cost-effective. To the best of our knowledge, this is the first study in which the ME analysis system was used in a clinical microbiology laboratory for determining the lengths of PCR products.
Because our PCR-based method with universal primers amplified all kinds of genomic DNA from fungal strains, its use may be limited to culture colonies on agar plates. The major limitation of our PCR-based method for the direct detection of fungi from clinical samples is mixed flora. Mixed candidemia, which was not observed during our study, has a reported frequency of 3.3% (14 of 427 patients) (12). In our preliminary study, electrophoresis of PCR products obtained from a mixture of C. albicans and C. glabrata cultures showed four fragments, indicating mixed flora. Therefore, when four or more fragments are detected with the PCR-AGE or PCR-ME method, the presence of mixed flora should be taken into consideration and the identification of fungi may be difficult.
In summary, we have developed a method for the comprehensive identification of clinically relevant fungal isolates which is sensitive, rapid, and specific for all yeast organisms tested. Using this PCR-ME analysis system, we are now processing yeast-positive blood culture bottles as well as identifying problematic isolates of yeasts in routine laboratory work.
REFERENCES
- 1.Cheng J, Waters L C, Fortina P, Hvichia G, Jacobson S C, Ramsey J M, Kricka L, Wilding P. Degenerate oligonucleotide primed-polymerase chain reaction and capillary electrophoretic analysis of human DNA on microchip-based devices. Anal Biochem. 1998;257:101–106. doi: 10.1006/abio.1997.2531. [DOI] [PubMed] [Google Scholar]
- 2.Espinel-Ingroff A, Stockman L, Roberts G, Pincus D, Pollack J, Marler J. Comparison of RapID Yeast Plus system with API 20C system for identification of common, new, and emerging yeast pathogens. J Clin Microbiol. 1998;36:883–886. doi: 10.1128/jcm.36.4.883-886.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujita S, Hashimoto T. DNA fingerprinting patters of Candida species using HinfI endonuclease. Int J Syst Evol Microbiol. 2000;50:1381–1389. doi: 10.1099/00207713-50-3-1381. [DOI] [PubMed] [Google Scholar]
- 4.Gardes M, White T J, Fortin J A, Bruns T D, Taylor J W. Identification of indigenous and introduced symbiotic fungi in ectomycorrhizae by amplification of nuclear and mitochondrial ribosomal DNA. Can J Bot. 1991;69:180–190. [Google Scholar]
- 5.Hazen K C. New and emerging yeast pathogens. Clin Microbiol Rev. 1995;8:462–478. doi: 10.1128/cmr.8.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henry T, Iwen P C, Hinrichs S H. Identification of Aspergillus species using internal transcribed spacer regions 1 and 2. J Clin Microbiol. 2000;38:1510–1515. doi: 10.1128/jcm.38.4.1510-1515.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson C J, Barton R C, Evans E G V. Species identification and strain differentiation of dermatophyte fungi by analysis of ribosomal DNA intergenic spacer regions. J Clin Microbiol. 1999;37:931–936. doi: 10.1128/jcm.37.4.931-936.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lott T J, Burns B M, Zancope-Oliveira R, Elie C M, Reiss E. Sequence analysis of the internal transcribed spacer 2 (ITS2) from yeast species within the genus Candida. Curr Microbiol. 1998;36:63–69. doi: 10.1007/s002849900280. [DOI] [PubMed] [Google Scholar]
- 9.Makimura K, Murayama S, Yamaguchi H. Detection of a wide range of medically important fungi by the polymerase chain reaction. J Med Microbiol. 1994;40:358–364. doi: 10.1099/00222615-40-5-358. [DOI] [PubMed] [Google Scholar]
- 10.Makimura K, Tamura Y, Mochizuki T, Hasegawa A, Tajiri Y, Hanazawa R, Uchida K, Saito H, Yamaguchi H. Phylogenetic classification and species identification of dermatophyte strains based on DNA sequences of nuclear ribosomal internal transcribed spacer 1 regions. J Clin Microbiol. 1999;37:920–924. doi: 10.1128/jcm.37.4.920-924.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morace G, Sanguinetti M, Posteraro B, Cascio G L, Fadda G. Identification of various medically important Candida species in clinical specimens by PCR-restriction enzyme analysis. J Clin Microbiol. 1997;35:667–672. doi: 10.1128/jcm.35.3.667-672.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen M H, Peacock J E, Morris A J, Tanner D C, Nguyen M L, Snydman D R, Wabener M M, Rinaldi M G, Yu V L. The changing face of candidemia: emergence of non-Candida albicans species and antifungal resistance. Am J Med. 1996;100:617–623. doi: 10.1016/s0002-9343(95)00010-0. [DOI] [PubMed] [Google Scholar]
- 13.Radford S A, Johnson E M, Leeming J P, Millar M R, Cornish JM, Foot A B M, Warnock D W. Molecular epidemiology study of Aspergillus fumigatus in a bone marrow transplantation unit by PCR amplification of ribosomal intergenic sequences. J Clin Microbiol. 1998;36:1294–1299. doi: 10.1128/jcm.36.5.1294-1299.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin J H, Nolte F S, Morrison C J. Rapid identification of Candida species in blood cultures by a clinically useful PCR method. J Clin Microbiol. 1997;35:1454–1459. doi: 10.1128/jcm.35.6.1454-1459.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin J H, Nolte F S, Holloway B P, Morrison C J. Rapid identification of up to three Candida species in a single reaction tube by a 5′ exonuclease assay using fluorescent DNA probes. J Clin Microbiol. 1999;37:165–170. doi: 10.1128/jcm.37.1.165-170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stefan P, Vazquez J A, Boikov D, Xu C, Sobel J D, Akins R A. Identification of Candida species by randomly amplified polymorphic DNA fingerprinting of colony lysates. J Clin Microbiol. 1997;35:2031–2039. doi: 10.1128/jcm.35.8.2031-2039.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugita T, Nishikawa A, Ikeda R, Shinoda T. Identification of medically relevant Trichosporon species based on sequences of internal transcribed spacer regions and construction of a database for Trichosporon identification. J Clin Microbiol. 1999;37:1985–1993. doi: 10.1128/jcm.37.6.1985-1993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sullivan D, Coleman D. Candida dubliniensis:: characterstics and identification. J Clin Microbiol. 1998;36:329–334. doi: 10.1128/jcm.36.2.329-334.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thanos M, Schönian G, Meyer W, Schweynoch C, Gräser Y, Mitchell T G, Presber W, Tietz H-J. Rapid identification of Candida species by DNA fingerprinting with PCR. J Clin Microbiol. 1996;34:615–621. doi: 10.1128/jcm.34.3.615-621.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turenne C Y, Sanche S E, Hoban D J, Karlowsky J A, Kabani A M. Rapid identification of fungi by using the internal transcribed spacer 2 genetic region and an automated fluorescent capillary electrophoresis system. J Clin Microbiol. 1999;37:1846–1851. doi: 10.1128/jcm.37.6.1846-1851.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wadlin J K, Hanko G, Stewart R, Pape J, Nachamkin I. Comparison of three commercial systems for identification of yeasts commonly isolated in the clinical microbiology laboratory. J Clin Microbiol. 1999;37:1967–1970. doi: 10.1128/jcm.37.6.1967-1970.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White T J, Burns T, Lee S, Taylor J. Amplification and sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. A guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 315–322. [Google Scholar]
- 23.Wingard J R. Importance of Candida species other than C. albicans as pathogens in oncology. Clin Infect Dis. 1995;20:115–125. doi: 10.1093/clinids/20.1.115. [DOI] [PubMed] [Google Scholar]