Abstract
A 1.6-kb DNA fragment isolated from a Campylobacter concisus genomic library gave C. concisus-specific restriction fragment length patterns when it was used as a probe in hybridization studies. All of the strains tested, including type strains and clinical isolates, contained a 0.5-kb HindIII fragment that hybridized to the probe. DNA sequencing of the 1.6-kb fragment identified three open reading frames (ORFs). One of the ORFs encodes the carboxy terminus of GyrB, and the translational products of ORF2 and ORF3 showed similarity to hypothetical proteins, previously identified in Campylobacter jejuni. DNA-DNA hybridization studies with a fragment internal to ORF3 showed that this sequence was responsible for the signal observed with the 0.5-kb HindIII fragment. A rapid PCR assay was developed and evaluated. Primers that annealed to the extremities of the 1.6-kb fragment were used to obtain an amplicon of the correct size from both reference and clinical strains of C. concisus.
Traditionally Campylobacter concisus has been recovered exclusively from and considered part of the bacterial flora of the human oral cavity (22). This species has been isolated predominantly from gingival crevices associated with the onset of periodontal disease (15, 22, 24). Subsequently, strains from outside the oral cavity have been isolated; however, these were initially misidentified or not classified (25).
Recent studies have increasingly reported on the isolation of C. concisus from the stools of patients with diarrhea (6, 12, 27). This has largely been attributed to an improvement in culture techniques, where the adoption of the membrane filter technique has improved isolation rates (13). Following the introduction of the Cape Town protocol (13), which utilizes the membrane filter technique in combination with a hydrogen-enriched microaerobic environment, the prevalence of C. concisus has increased to 31% of the total Campylobacter isolates from children at the Red Cross War Memorial Children's Hospital, Cape Town, South Africa. Of all the Campylobacter spp., only Campylobacter jejuni subsp. jejuni is more frequently isolated from children at this hospital. During a reevaluation of conventional isolation methods, similar results were obtained in a study in Denmark (6).
In Cape Town, South Africa, C. concisus displays clinical and seasonal characteristics similar to those of C. jejuni, an established gastrointestinal pathogen (A. J. Lastovica and M. E. Engel, Abstr. 100th Gen. Meet. Am. Soc. Microbiol. 2000, abstr. D250, 2000). Monthly isolation frequencies showed that both C. concisus and C. jejuni were notably higher in the 3-month period towards the end of summer (February to April). Clinical symptoms of diarrhea, such as severity and stool consistency, were similar to those associated with C. jejuni; however, C. concisus was predominantly isolated from children 1 year old or older, whereas C. jejuni was mostly found in children younger than 12 months. In addition, C. concisus was more likely to be associated with children of mixed descent than with indigenous black children. This observation could not be explained by age, gender differences, genetic susceptibility, eating habits or geographical location (Lastovica and Engel, Abstr. 100th Gen. Meet. Am. Soc. Microbiol.). Although some studies have suggested that C. concisus is not a primary pathogen (6, 27), the role of C. concisus and its association with diarrhea have yet to be clearly defined.
An understanding of the epidemiology of C. concisus has been hampered by the lack of a suitable identification tool. Like other members of the genus, C. concisus is fastidious and is characterized by low biochemical activity, properties that make the identification of C. concisus problematic, using conventional phenotypic techniques (7, 10, 17). DNA-DNA hybridization, immunotyping and whole-cell protein electrophoresis have been used to identify C. concisus (11, 23, 25). As these techniques are time-consuming and require considerable expertise, they are not suitable for large epidemiological studies. A limited number of molecular approaches for the identification of C. concisus have been described (3, 14). A PCR of the 23S ribosomal DNA (rDNA) (3) did not consistently identify C. concisus (6), probably because of the genotypic heterogeneity within the species (3, 25). A similar approach is based on the restriction profile of 16S rDNA PCR products (14). During the course of the study described in this report, a fragment of C. concisus genomic DNA was cloned and characterized. DNA-DNA hybridization studies and PCR assays indicate that this fragment could be used to identify C. concisus.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Reference Campylobacter strains used in this study included C. concisus NCTC 11485, C. concisus NCTC 11684, C. concisus CCUG 13144, C. concisus CCUG 19995, C. curvus NCTC 11649, C. rectus NCTC 11489, C. sputorum bv. fecalis NCTC 11415, C. coli CCUG 11283, C. lari NCTC 11352, C. upsaliensis NCTC 12183, C. helveticus NCTC 12470, C. jejuni subsp. doylei NCTC 11487, C. jejuni subsp. jejuni NCTC 11168, C. mucosalis NCTC 11000, and C. fetus subsp. fetus NCTC 10842. Bacteroides ureolyticus NCTC 10941 was also used. Local clinical isolates of C. concisus (n = 104) included in this study were collected during the period 1992 to 1999. One of the strains was a dental isolate from an adult; the remainder were obtained from children at the Red Cross War Memorial Children's Hospital. S. L. W. On (Veterinary Research Laboratory, Copenhagen, Denmark) kindly provided two Danish clinical isolates (SSI 63936 and SSI 11286) from children with diarrhea. Escherichia coli LK111 (30) was used as a recipient in transformation studies. The vectors, pEco251 (provided by M. Zabeau, Plant Genetics Systems N. V., Ghent, Belgium) and pUC19 (16) were used in cloning experiments.
Preparation and analysis of DNA.
Genomic DNA was extracted using guanidinium thiocyanate (19). Plasmid DNA was isolated using either the alkaline lysis method (8) or a Nucleobond kit (Macherey-Nagel). DNA was digested with various restriction enzymes, and the fragments were separated in horizontal gels of 0.8 to 1.2% (wt/vol) agarose dissolved in 0.4 M Tris-acetate and 0.01 M EDTA. Gels were stained with ethidium bromide, and DNA was visualized by UV transillumination.
Ligations.
Ligations were carried out as described (20), in 20-μl reaction mixtures containing 1× T4 DNA ligase buffer and 10 U of T4 DNA ligase (Boehringer Mannheim). Typically, an approximate ratio of 3 to 1 molar concentrations of insert and vector were used in ligations, and the reaction mixtures were incubated at 16°C for 4 to 16 h.
Transformation studies.
Competent E. coli LKIII cells were prepared as described (4). Ligations were added to competent cells, and transformants were selected on 2× YT agar containing appropriate antibiotics.
PCR assays.
Based on the DNA sequence (GenBank accession number AY026942) of the 1.6-kb BglII-XbaI fragment (Fig. 1), three pairs of oligonucleotide primers were used to amplify sequences internal to the three open reading frames (ORFs) identified. pcisus1 (5′-GAGCTTGTGGTAAAGA-3′) and pcisus2 (5′-GCGTGGTCTTGGATATAG-3′) were used to amplify ORF1. pcisus3 (5′-TTGACGTAGAGAGTGACCTTG-3′) and pcisus4 (5′-ATCAGAGCTGATCTCGATAAC-3′) were used to amplify ORF2, and pcisus5 (5′-AGCAGCATCTATATCACGTT-3′) and pcisus6 (5′-CCCGTTTGATAGGCGATAG-3′) were used to amplify ORF3. PCRs were carried out with a Perkin-Elmer thermocycler (Gene Amp 2400) in a final volume of 50 μl containing 50 ng of campylobacter DNA, a 50 pM concentration of each primer, a 2.5 mM concentration of each deoxynucleoside triphosphate, and 2.5 U of Taq DNA polymerase (TaKaRa Biochemicals) in the prescribed buffer. Initial denaturation (94°C for 120 s) was followed by 30 cycles of amplification. Each cycle consisted of 94°C for 60 s, 45°C for 60 s, and 72°C for 120 s. A final extension step (72°C for 10 min) completed the amplification. Aliquots of PCR products were separated by electrophoresis.
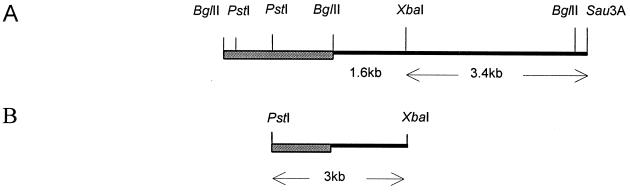
FIG. 1.
Restriction map of DNA fragment from C. concisus. (A) The shaded block represents pEco251, and the bold line (5.0 kb) represents the 5.0-kb fragment from C. concisus cloned in the vector, generating pB3C. (B) Shown is the 3.0-kb fragment that was cloned into pUC19, generating pNeo.
The primers pcisus1 and pcisus6 were used in PCR assays for the identification of C. concisus. PCR was carried out in a final volume of 50 μl and contained 50 ng of genomic DNA, a 2.5 nM concentration of each deoxynucleoside triphosphate, a 50 pM concentration of each primer, and 2.5 U of Taq DNA polymerase (TaKaRa) in the prescribed buffer. Amplification was carried out with a Perkin-Elmer (Gene Amp 2400) thermocycler, using the parameters described above.
Preparation of probes.
DNA of interest was eluted from agarose gels, purified (21), and labeled using an ECL labeling and detection kit (Amersham International).
DNA-DNA hybridization.
Probes were hybridized to genomic DNA restricted with HindIII, separated by agarose gel electrophoresis, and transferred to Hybond-N+ membranes (Amersham International). Prehybridization, hybridization, and the posthybridization washes were carried out according to the protocol supplied with the ECL gene detection system (Amersham International).
DNA sequencing.
DNA for sequencing was cloned into pUC19. Sequencing data were generated by automated laser fluorescence (Pharmacia Biotech AB, Uppsala, Sweden) in the Department of Molecular Biology, University of Cape Town. Internal oligonucleotide primers were used where necessary to ensure that both strands were sequenced. DNA sequences were analyzed with DNAMAN (version 4.0; Lynnon BioSoft). BLAST (1) was used to search databases in GenBank for nucleic acid and deduced amino acid sequence similarity with existing sequences. Protein alignments were carried out by the fast alignment method (28), using the Blosum matrix with a k-tuple value of 2 and a gap penalty of 4.
Construction of a DNA library.
A DNA library of C. concisus NCTC 11485 was prepared (2). Genomic DNA from C. concisus NCTC 11684 was digested partially with Sau3AI (Boehringer Mannheim). Size fractionation was carried out in a sucrose gradient; the 4- to 5-kb DNA fragments were pooled, recovered by ethanol precipitation, cloned into the BglII site of pEco251, and introduced into competent E. coli LKIII cells.
RESULTS
Identification of C. concisus DNA sequence for identification to species level.
To select a recombinant clone that could be used to identify C. concisus, the pEco251 library was screened using DNA-DNA hybridization studies. DNA inserts were restricted from pEco251 with BglII, labeled, and hybridized to HindIII-digested DNA from C. jejuni, C. mucosalis, C. concisus type strains, and clinical isolates from Cape Town (404-96 and 406-96) and Denmark (SSI 11286 and SS1 63936). One of the inserts (5.0 kb), from a recombinant plasmid, designated pB3C, hybridized to four bands (3.0, 1.2, 0.5, and 0.3 kb) in C. concisus NCTC 11485 and to two bands (1.2 and 0.5 kb) in each of the four clinical isolates (data not shown). In addition, signals were obtained with fragments in C. mucosalis and C. jejuni, but the banding patterns were entirely different from those observed in C. concisus. The presence of common bands, which hybridized to the probe in the C. concisus strains, combined with the fact that these bands were not present in C. jejuni and C. mucosalis, suggested that they could be used as identification markers.
To refine the probe further, a limited restriction map of pB3C containing the C. concisus insert was determined. Restriction digestion showed that the insert (5.0 kb) contains an internal BglII site 0.4 kb from one of the Sau3AI sites (Fig. 1). Digestion of the insert with BglII and XbaI generated two fragments of 1.6 and 3.0 kb.
When the 3.0-kb BglII-XbaI fragment was used to probe the DNA, a strong hybridization signal was obtained with a 3.0-kb band, and weak signals were obtained with 0.5- and 0.3-kb bands in C. concisus NCTC 11485 (data not shown). Hybridization signals, albeit weak, were also obtained with 1.2-kb bands in the local and Danish clinical isolates. No signals were obtained with DNA from the C. jejuni and C. mucosalis type strains.
Probing with the 1.6-kb BglII-XbaI fragment resulted in signals with bands of 1.2, 0.5, and 0.3 kb in C. concisus NCTC 11485 (data not shown). In addition, a signal was obtained with a 1.2-kb fragment in the Danish and local clinical isolates; a weak hybridization signal was observed with a 0.5-kb band in one of each of the local and Danish clinical isolates. No hybridization signal was obtained with DNA from C. jejuni, and in C. mucosalis, barely visible signals were detected with fragments (2.7 and 2.2 kb) that had hybridized to the 5.0-kb probe. The fact that similar hybridization profiles were observed in the C. concisus type strain and in the Danish and local clinical isolates suggested that the 1.6-kb sequence and its organization in the genome are conserved in C. concisus.
The specificity of the 1.6-kb fragment was investigated, using hybridization assays and an extended panel of Campylobacter type strains and B. ureolyticus, previously considered genotypically similar to campylobacter (26). An expected hybridization banding pattern (1.2, 0.5, and 0.3 kb) was obtained with C. concisus NCTC 11485 (data not shown). In addition, strong signals were obtained with DNA from 2 of the 11 Campylobacter isolates: bands of 2.2 and 1.7 kb in C. curvus and C. sputorum bv. fecalis, respectively, hybridized to the probe. A weak signal was obtained with a 2.6-kb fragment in C. mucosalis, but signals were not detected with DNA from C. rectus, C. coli, C. jejuni subsp. jejuni, C. lari, C. upsaliensis, C. helveticus, C. fetus subsp. fetus, C. jejuni subsp. doylei, and B. ureolyticus. It is important that when the membrane was hybridized with a 16S rDNA (5) probe, signals were obtained with DNA from these species, indicating that DNA had been transferred to the membrane.
Based on the above results it was concluded that probing with the 1.6-kb BglII-XbaI fragment generated a species-specific pattern with DNA from C. concisus. To test this further, DNA from 62 clinical isolates, including the dental isolate, and two Swedish reference strains (CCUG 13144 and CCUG 19995), suggested by pulsed-field gel electrophoresis to represent genetically diverse groups (data not shown), was probed with the 1.6-kb fragment. Analysis of the hybridization profiles showed a limited number of restriction fragment length polymorphisms (RFLPs) among the isolates. Five profiles were identified (Table 1). Common to all the profiles was a 0.5-kb fragment that hybridized to the probe, suggesting a genetic marker for the identification of C. concisus.
TABLE 1.
Hybridization profiles
| Profile group | Size (kb) of DNA fragments which hybridized to the 1.6-kb probe | No. of isolates |
|---|---|---|
| I | 1.2, 0.5, 0.3 | 12 |
| II | 1.6, 0.5, 0.3 | 9 |
| III | 0.8, 0.5 | 6 |
| IV | 1.2, 0.5 | 36 |
| V | 1.6, 1.2, 0.3 | 1 |
As a first step to gaining insight into the 0.5-kb fragment, which hybridized to the probe, the DNA sequence of the 1.6-kb BglII-XbaI fragment was determined. To facilitate sequencing, a 3.0-kb PstI-XbaI fragment (Fig. 1), which contained a portion of pEcoR251 (1.375-kb PstI-BglII fragment) and the 1.6-kb BglII-XbaI fragment from C. concisus, was subcloned into the PstI-XbaI site of pUC19 and introduced into E. coli LKIII. The recombinant plasmid was designated pNeo.
Sequence analysis of the 1.6-kb fragment from C. concisus in pNeo.
The DNA sequence of the 1.6-kb fragment was determined on both strands (GenBank accession number AY026942). Three major ORFs are contained on the sense strand, while no significant ORFs were identified on the other strand.
Analysis of the sequence upstream of ORF1 did not identify any bacterial transcription signals. This suggested that ORF1 extends upstream of the XbaI site and that the 5′ end is contained in the 3.4-kb XbaI-Sau3A fragment (Fig. 1). A comparison of the deduced amino acid sequence of this ORF with protein databases in GenBank showed 76 and 58% similarity to the carboxy termini of GyrB from C. jejuni and Helicobacter pylori, respectively.
ORF2 and ORF3 are preceded by ribosome-binding sites upstream of the ATG transcription initiation sites. The sequences (AAGGA) are identical to the C. jejuni consensus sequence (29) but shorter than that proposed previously for C. jejuni (9). Upstream of ORF2 is the sequence TTTAAAAGGT, which shows similarity to the −35 consensus sequence (TTTAAGTNNTT) for C. jejuni promoters (29). Based on the observation that the spacing between the −35 and −10 regions of C. jejuni promoters varies from 15 to 19 bp (29), two appropriately spaced putative −10 regions were identified. TTGACCT and CCTAGTT, which are separated from the putative −35 sequence by 14 and 18 bp, respectively, show similarity to the −10 consensus sequence, TATAATT, for C. jejuni promoters (29). Two regions, −35 (TTTAAACA) and −10 (TAAGCTA), separated by 18 bp precede ORF3. The −16 sequence (TTTTTTTG) (29) was not identified in either of the putative promoters.
The translational product of ORF2 showed 70% similarity to a conserved hypothetical protein of 127 amino acids in C. jejuni (CDS Cj 1724c), while that of ORF3 showed 70% similarity with the NH2-terminal of a conserved hypothetical protein (408 amino acids) in C. jejuni (CDS Cj 0015c).
DNA-DNA hybridization studies with sequences internal to ORF1, ORF2, and ORF3 as probes.
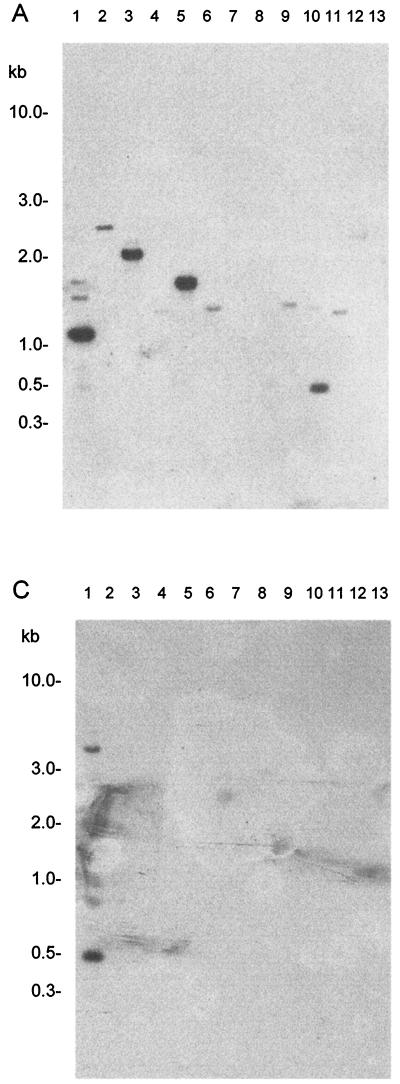
Internal portions of each ORF were amplified, labeled, and hybridized to DNA from Campylobacter spp. When the fragment internal to ORF1 (gyrB) was used to probe the DNA, a strong signal was obtained with the 1.2-kb band in C. concisus (Fig. 2A, lane 1). In addition, signals were obtained with a 2.2-kb fragment in C. curvus (Fig. 2A, lane 3), a 1.7-kb fragment in C. sputorum bv. fecalis (Fig. 2A, lane 5) and a 0.5-kb fragment in C. helveticus (Fig. 2A, lane 10). Weak hybridization signals were obtained with DNA fragments in C mucosalis, C. rectus, C. coli, C. upsaliensis, B. ureolyticus, and C. jejuni subsp. doylei (Fig. 2A, lanes 2, 4, 6, 9, 11, and 12). No signals were obtained with the DNA from C. jejuni subsp. jejuni, C. lari, and C. fetus subsp. fetus.
FIG. 2.
Genomic DNA from Campylobacter type strains digested with HindIII and hybridized to internal fragments of ORF1, ORF2, and ORF3. Autoradiographs of DNA probed with a 0.35-kb fragment internal to ORF1 (A), a 0.338-kb fragment internal to ORF2 (B), and a 0.441-kb fragment internal to ORF3 (C) are shown. Lanes: 1, C. concisus NCTC 11485; 2, C. mucosalis NCTC 11000; 3, C. curvus NCTC 11649; 4, C. rectus NCTC 11489; 5, C. sputorum bv. fecalis NCTC 11415; 6, C. coli CCUG 11283; 7, C. jejuni NCTC 11168; 8, C. lari NCTC 11352; 9, C. upsaliensis NCTC 12183; 10, C. helveticus NCTC 12470; 11, B. ureolyticus NCTC 10941; 12, C. jejuni subsp. doylei NCTC 10847; 13, C. fetus subsp. fetus NCTC 10842.
Similar studies using the probe for ORF2 resulted in strong signals with fragments of 1.2 and 0.3 kb in C. concisus (Fig. 2B, lane 1). That signals were obtained with two HindIII fragments was not unexpected, since this restriction site is present in ORF2. A strong signal was also obtained with a 2.2-kb fragment in C. curvus (Fig. 2B, lane 3). Weak signals were obtained with DNA from C. rectus, C sputorum bv. fecalis, and B. ureolyticus (Fig. 2B, lanes 4, 5, and 11).
Probing with the PCR product of ORF3 resulted in a strong signal with the 500-bp fragment in C. concisus (Fig. 2C, lane 1). A weak signal was obtained with a fragment of 5 kb in this strain (probably undigested DNA). Notwithstanding the background on the autoradiograph, there is a suggestion of a hybridization signal with a fragment of 2.6 kb in C. mucosalis (Fig. 2C, lane 2). No signals were obtained with DNA from any of the other Campylobacter spp. In another hybridization experiment, the internal fragment from ORF3 was used to probe DNA from clinical isolates representing three of the RFLP profile groups (II, III, and IV), previously identified (Table 1). A strong signal was obtained with a 500-bp fragment in each of the C. concisus isolates, further evidence that this sequence is common to and conserved in C. concisus.
PCR assay for the identification of C. concisus.
As hybridization studies are time consuming and not the preferred approach in a diagnostic laboratory, a PCR assay based on the DNA sequence of the 1.6-kb fragment was developed and evaluated. Using pcisus1 and pcisus6, which anneal to DNA sequences in ORF1 and ORF3, respectively, a product of the expected size (1.5 kb) was obtained from C. concisus NCTC 11485 (Fig. 3). No PCR products were obtained from C. jejuni, C. lari, C. upsaliensis, C. helveticus, B. ureolyticus, C. sputorum bv. fecalis, C. curvus, C. mucosalis, C. rectus and super-family member Helicobacter pylori. Amplicons were not obtained with DNA from unrelated bacteria Acinetobacter baumannii, Mycobacterium bovis BCG, E. coli, and Salmonella sp. It is noteworthy that, although hybridization signals were obtained with the DNA from C. curvus and C. sputorum bv. fecalis when it was probed with the 1.6-kb fragment, no PCR products were obtained from these species. It must also be noted that in a PCR assay using universal primers for 16S rDNA (5), a product of the appropriate size was obtained from all of the species tested, indicating the suitability of the DNA for PCR and the absence of inhibitors in the reaction mixture.
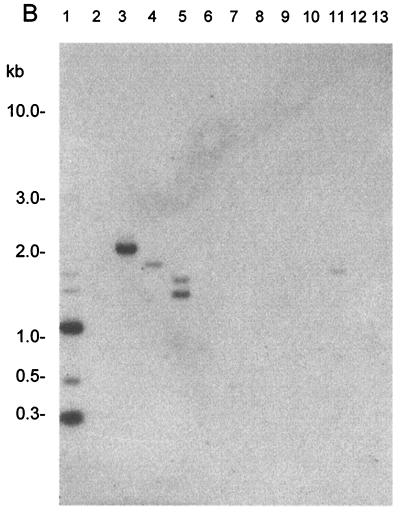
FIG. 3.
PCR analysis of DNA. (A) Lanes: 1, molecular weight marker (Promega, Madison, Wis.); 2, negative control (no DNA); 3, C. concisus NCTC 11485; 4, C. jejuni NCTC 11168; 5, C. lari NCTC 11352; 6, C. upsaliensis NCTC 2183; 7, C. helveticus NCTC 12470; 8, B. ureolyticus NCTC 10941. (B) Lanes: 1, molecular weight marker (Promega); 2, no DNA; 3, C. concisus; 4, C. sputorum bv. fecalis NCTC 11415; 5, C. curvus NCTC 116490; 6, C. mucosalis NCTC 11000; 7, C. rectus NCTC 11489; 8, Helicobacter pylori; 9, Acinetobacter baumannii; 10, Mycobacterium BCG; 11, Escherichia coli; 12, Salmonella sp.
Primers were used to carry out PCR assays of 91 clinical isolates, including the 49 clinical isolates used in the hybridization studies. A single product of the expected size was obtained from 88 of the isolates. Three products, of which one was the correct size, were obtained from one isolate. No amplicons were obtained from two of the isolates. As these two strains had not been previously characterized by hybridization studies, HindIII-digested DNA from the two PCR-negative isolates was probed with the 1.6-kb BglII-XbaI fragment. The hybridization profiles obtained were not consistent with those for C. concisus isolates (data not shown). In addition, when these strains were investigated using the 16S rDNA method (14), the resulting DdeI restriction profiles of the amplicons were different from the Campylobacter profiles described, including that of C. concisus. Taken together, these data suggested that the two strains are not C. concisus.
DISCUSSION
Hybridization studies and PCR assays carried out in this study indicate that a 1.6-kb BglII-XbaI fragment, which was cloned from C. concisus NCTC 11485, could be used to identify this species. When this fragment was used to probe DNA from Campylobacter spp., a C. concisus-specific HindIII hybridization profile was generated. Strong hybridization signals were obtained with the DNA from only two other species, C. sputorum bv. fecalis and C. curvus, suggesting that these species are more closely related to C. concisus than are the other species, a finding that is supported by phylogenetic rRNA studies (26). A weak signal was obtained with the DNA from C. mucosalis, but signals were not obtained with DNA from C. rectus, C. coli, C. jejuni, C. lari, C. upsaliensis, C. helveticus, B. ureolyticus, C. jejuni subsp. doylei, and C. fetus subsp. fetus.
Five RFLP profiles were observed in the C. concisus strains (n = 106) probed with the 1.6-kb fragment. All of the strains contained a 0.5-kb HindIII fragment that hybridized to the probe. The profiles obtained were consistent in both local and Danish clinical isolates. In addition, profiles indicative of C. concisus were observed in the two Swedish reference strains (CCUG 13144 and CCUG 19995), belonging to two separate DNA-DNA hybridization homology groups (25). These data suggest that the observed hybridization pattern is not confined to strains from a specific geographic region. Moreover, the stability of the profile is suggested by the fact that the strains were collected over a period of 7 years (1992 to 1999).
An analysis of the DNA sequencing data of the 1.6-kb fragment identified three ORFs. ORF1 contains the 3′ end of gyrB. It is assumed that the 5′ end of this gene is contained in the 3.4-kb XbaI-Sau3A portion of the 5.0-kb insert in pB3C (Fig. 1). As gyrB is contiguous with gyrA in C. jejuni it is probable that the latter gene is also contained in the 3.4-kb XbaI-Sau3A portion. The strong signals obtained with DNA from C. curvus, C. sputorum bv. fecalis, and C. helveticus when probed with a portion of ORF1 (Fig. 2) suggest that the 3′ ends of the gyrB genes in these species are closely related to their counterpart in C. concisus.
The fragment internal to ORF2 hybridized to DNA from C. concisus and C. curvus. As ORF2 contains a HindIII site, two fragments (1.2 and 0.3 kb) in C. concisus hybridized to the probe. On the other hand, only one band (2.2 kb) in the DNA from C. curvus hybridized to the probe. Since ORF1 also hybridized to a fragment of 2.2 kb in this strain, it is likely that as in C. concisus, ORF1 and ORF2 are contiguous in C. curvus.
Hybridization studies using a portion of ORF3 showed that this sequence was responsible for the signal obtained with the 0.5-kb HindIII fragment in all of the C. concisus strains investigated. The translational product of this ORF showed 70% similarity with the amino terminus of a hypothetical protein (408 amino acids) in C. jejuni (CDS Cj 0015c)
A rapid PCR assay for the identification of C. concisus was evaluated, using primers that annealed to the extremities of the 1.6-kb fragment. The assay was specific for C. concisus, and PCR products were not obtained from any of the other Campylobacter spp. tested. Amplicons were obtained from local (87 of 89) and Danish (n = 2) clinical isolates. Importantly, amplicons were also obtained from the two Swedish reference strains shown to be genetically diverse (25). It was not necessary to obtain a restriction enzyme profile of the amplicons, making the method rapid and suitable for epidemiological studies. It was not the purpose of this study to evaluate the use of the PCR assay to detect C. concisus directly from stools. In other studies, PCR assays have not always detected Campylobacter spp. in stools, suggesting the presence of inhibitors in the specimen (18).
The two clinical isolates from which PCR products were not obtained were investigated further. Hybridization studies, using the 1.6-kb fragment as a probe, supported the results of the PCR assay (data not shown). Using the 16S rDNA method (14), the resulting DdeI restriction profiles of the amplicons obtained from these two strains were different from the Campylobacter profiles described, including that of C. concisus. These data suggest that the two strains might have been misidentified as C. concisus and underscore the necessity for an unequivocal identification tool.
ACKNOWLEDGMENTS
We thank Harold Zappe for his help in the preparation of the C. concisus DNA library.
M. I. Matsheka and this study were funded by the Rockefeller and Carnegie Foundations through the University Science, Humanities, and Engineering Partnerships in Africa (USHEPiA) program. M. I. Matsheka thanks the University of Botswana for financial assistance received during the course of his studies. A. J. Lastovica thanks the South African Medical Research Council for financial assistance.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1989. [Google Scholar]
- 3.Bastyns K, Chapelle S, Vandamme P, Goosens H, De Wachter R. Specific detection of Campylobacter concisus by PCR amplification of 23S rDNA areas. Mol Cell Probes. 1995;9:247–250. doi: 10.1016/s0890-8508(95)90114-0. [DOI] [PubMed] [Google Scholar]
- 4.Dagert M, Ehrlich S D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979;6:23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- 5.Edwards U, Rogall T, Blocker H, Emde E M, Bottger C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engberg J, On S L W, Harrington C S, Gerner-Smidt P. Prevalence of Campylobacter, Arcobacter, Helicobacter, and Sutterella spp. in human fecal samples as estimated by a reevaluation of isolation methods for campylobacters. J Clin Microbiol. 2000;38:286–291. doi: 10.1128/jcm.38.1.286-291.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Figura N, Guglielmetti P, Zanchi A, Partini N, Armellini D, Bayeli P F, Bugnoli M, Verdiani S. Two cases of Campylobacter mucosalis enteritis in children. J Clin Microbiol. 1993;31:727–728. doi: 10.1128/jcm.31.3.727-728.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ish-Horowicz D, Burke J F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981;9:2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim N W, Gutell R R, Chan V L. Complete sequences and organization of the rRNA operon from Campylobacter jejuni TGH9011 (ATCC43431) Gene. 1995;164:101–106. doi: 10.1016/0378-1119(95)00471-h. [DOI] [PubMed] [Google Scholar]
- 10.Lastovica A, Le Roux E, Warren R, Klump H. Clinical isolates of Campylobacter mucosalis. J Clin Microbiol. 1993;31:2835–2836. doi: 10.1128/jcm.31.10.2835-2836.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lastovica A J, Le Roux E, Warren R, Klump H. Additional data on clinical isolates of Campylobacter mucosalis. J Clin Microbiol. 1994;32:2338–2339. doi: 10.1128/jcm.32.9.2338-2339.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lauwers S, Devreker T, Van Etterijck R, Breynaert J, Van Zeebroeck A, Smekens L, Kersters K, Vandamme P. Isolation of Campylobacter concisus from human faeces. Microb Ecol Health Dis. 1991;4(Suppl.):91. [Google Scholar]
- 13.Le Roux E, Lastovica A J. The Cape Town protocol: how to isolate the most campylobacters for your dollar, pound, franc, yen, etc. In: Lastovica A J, Newell D G, Lastovica E E, editors. Campylobacter, Helicobacter and related organisms. Proceedings of the 9th International Workshop. Cape Town, South Africa: Institute of Child Health, University of Cape Town; 1998. pp. 30–33. [Google Scholar]
- 14.Marshall S M, Melito P L, Woodward D L, Johnson W M, Rodgers F G, Mulvey M R. Rapid identification of Campylobacter, Arcobacter, and Helicobacter isolates by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene. J Clin Microbiol. 1999;37:4158–4160. doi: 10.1128/jcm.37.12.4158-4160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore W E, Holdeman L V, Cato E P, Smibert R M, Burmeister J A, Palcanis K G, Ranney R R. Comparative bacteriology of juvenile periodontitis. Infect Immun. 1985;48:507–519. doi: 10.1128/iai.48.2.507-519.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Norrander J, Kempe T, Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 17.On S L W. Confirmation of human Campylobacter concisus isolates misidentified as Campylobacter mucosalis and suggestions for improved differentiation between the species. J Clin Microbiol. 1994;32:2305–2306. doi: 10.1128/jcm.32.9.2305-2306.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oyofo B A, Thornton S A, Burr D H, Trust T J, Pavlovskis O R, Guerry P. Specific detection of Campylobacter jejuni and Campylobacter coli by using polymerase chain reaction. J Clin Microbiol. 1992;30:2613–2619. doi: 10.1128/jcm.30.10.2613-2619.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pitcher P G N, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidinium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 21.Seth A. A new method for linker ligation gene. Anal Technol. 1984;1:99–103. [Google Scholar]
- 22.Tanner A C, Dzink J L, Ebersole J L, Socransky S S. Wolinella recta, Campylobacter concisus, Bacteroides gracilis, and Eikenella corrodens from periodontal lesions. J Periodontal Res. 1987;22:327–330. doi: 10.1111/j.1600-0765.1987.tb01593.x. [DOI] [PubMed] [Google Scholar]
- 23.Tanner A C R. Characterization of Wolinella spp., Campylobacter concisus, Bacteroides gracilis, and Eikenella corrodens by polyacrylamide gel electrophoresis. J Clin Microbiol. 1986;24:562–565. doi: 10.1128/jcm.24.4.562-565.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanner A C R, Badger S, Lai C-H, Listgarten M A, Visconti R A, Socransky S S. Wolinella gen. nov., Wolinella succinogenes (Vibrio succinogens Wolin et al.) comb. nov., and description of Bacteroides gracilis sp. nov., Wolinella recta sp. nov., Campylobacter concisus sp nov., and Eikenella corrodens from humans with periodontal disease. Int J Syst Bacteriol. 1981;31:432–445. [Google Scholar]
- 25.Vandamme P, Falsen E, Pot B, Hoste B, Kersters K, De Ley J. Identification of EF group 22 campylobacters from gastroenteritis cases as Campylobacter concisus. J Clin Microbiol. 1989;27:1775–1781. doi: 10.1128/jcm.27.8.1775-1781.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vandamme P, Daneshvar M I, Dewhirst F E, Paster B J, Kersters K, Goossens H, Moss C W. Chemotaxonomic analyses of Bacteroides gracilis and Bacteroides ureolyticus and reclassification of B. gracilis as Campylobacter gracilis comb. nov. Int J Syst Bacteriol. 1995;45:145–152. doi: 10.1099/00207713-45-1-145. [DOI] [PubMed] [Google Scholar]
- 27.Van Etterijck R, Breynaert J, Revets H, Devreker T, Vandenplas Y, Vandamme P, Lauwers S. Isolation of Campylobacter concisus from feces of children with and without diarrhea. J Clin Microbiol. 1996;34:2304–2306. doi: 10.1128/jcm.34.9.2304-2306.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wibur W J, Lipman D J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci USA. 1983;80:727–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wosten M M, Boeve M, Koot M G, van Nuene A C, van der Zeijst B A. Identification of Campylobacter jejuni promoter sequences. J Bacteriol. 1998;180:594–599. doi: 10.1128/jb.180.3.594-599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zabeau M, Stanley K K. Enhanced expression of cro-beta-galactosidase fusion proteins under the control of the PR promoter of bacteriophage lambda. EMBO J. 1982;1:1217–1224. doi: 10.1002/j.1460-2075.1982.tb00016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]