Abstract
The SARS-CoV-2 virus, the pathogen causing COVID-19, has caused more than 200 million confirmed cases, resulting in more than 4.5 million deaths worldwide by the end of August, 2021. Upon detection of SARS-CoV-2 infection by pattern recognition receptors (PRRs), multiple signaling cascades are activated, which ultimately leads to innate immune response such as induction of type I and III interferons, as well as other antiviral genes that together restrict viral spread by suppressing different steps of the viral life cycle. Our understanding of the contribution of the innate immune system in recognizing and subsequently initiating a host response to an invasion of SARS-CoV-2 has been rapidly expanding from 2020. Simultaneously, SARS-CoV-2 has evolved multiple immune evasion strategies to escape from host immune surveillance for successful replication. In this review, we will address the current knowledge of innate immunity in the context of SARS-CoV-2 infection and highlight recent advances in the understanding of the mechanisms by which SARS-CoV-2 evades a host's innate defense system.
Keywords: SARS-CoV-2, innate immune response, signaling transduction, interferon, inflammation, antiviral targets
Graphical abstract
Abundant research of innate immune responses to SARS-CoV-2 infection has been carried out in the past 2 years. Here, Zhang et al. summarize the current progress of innate immune responses to SARS-CoV-2 infection and how SARS-CoV-2 evades innate immunity, and they discuss the potential of this crosstalk as a therapeutic target.
Introduction
Coronaviruses are enveloped, positive-sense, single-stranded RNA viruses in the Coronaviridae family, which have a broad spectrum of hosts such as humans, bats, camels,and avian species, including livestock and companion animals, posing a threat to public health.1 Coronaviruses are classified in the subfamily of Orthocoronavirinae, which is further divided into four genera, based on differences in protein sequences: α-coronavirus, β-coronavirus, γ-coronavirus, and δ-coronavirus.2 The α-coronaviruses and β-coronaviruses only infect mammals, whereas γ-coronaviruses and δ-coronaviruses primarily infect birds, though some of them can infect mammals.3 HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU1, SARS-CoV, MERS-CoV, and SARS-CoV-2 are the seven coronaviruses that have been identified to infect humans.4 Among them, SARS-CoV and MERS-CoV, which have emerged in the human population in 2002 and 2012, are highly pathogenic.5,6 Whereas human coronavirus (HCoV)-229E, HCoV-NL63, HCoV-OC43, or HCoV-HKU1 strains circulating in the human population cause only the common cold,7 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causal agent of COVID-19, is a novel β-coronavirus, which early appeared at the end of 2019 and has resulted in devastating deaths. The primary symptoms of COVID-19 are similar to those of SARS-CoV and MERS-CoV: fever, tiredness, dry cough, upper chest pain, sometimes diarrhea, and dyspnea.8 Unlike past coronavirus (CoV) infections, the rapid global dissemination, high transmission rate, longer incubation time, more asymptomatic infections, and disease severity of SARS-CoV-2 require in-depth knowledge regarding the viral immune evasion strategies.
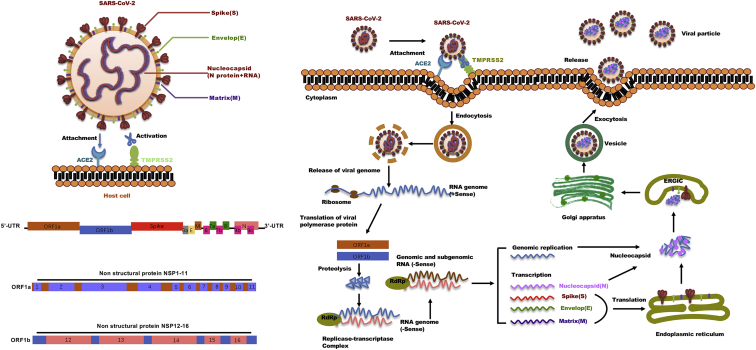
Like other human coronaviruses (SARS-CoV-2, MERS-CoV), SARS-CoV-2 also has a single-stranded, positive-sense RNA genome of around 30 kb in size.9 As shown in Figure 1, the viral nucleocapsid (N) proteins bundle the genome into a large ribonucleoprotein (RNP) complex, which is then enveloped by lipids and viral proteins S (spike), M (membrane), and E (envelope). The 5′ end of the genome has two large open reading frames (ORFs), ORF1a and ORF1b, encoding polypeptides pp1a and pp1b, which are produced into 16 nonstructural proteins (NSPs) involving every aspect of viral replication by viral proteases NSP3 and NSP5 that harbor a papain-like protease domain and a 3C-like protease domain, respectively.9 The 3′ end of the genome encodes structural proteins and the accessory proteins, of which ORF3a, ORF6, ORF7a, and ORF7b have been proven to be viral structural proteins involved in the formation of viral particles and ORF3b and ORF6 function as interferon antagonists. According to the current annotation on the basis of sequence similarity to other β-coronaviruses, SARS-CoV-2 includes predictions of six accessory proteins (3a, 6, 7a, 7b, 8, and 10).10,11 However, not all of these ORFs have been experimentally validated yet, and the exact number of accessory genes of SARS-CoV-2 is still a point of contention. Therefore, it is still unclear which accessory genes are actually expressed by this compact genome.
Figure 1.
The life cycle of SARS-CoV-2 in host cells
SARS-CoV-2 enters into the host cell through the binding of the Spike protein to the receptor ACE2, together with TMPRSS2, followed by multiple steps including release of the viral genome RNA, translation of the viral RNA, proteolytic cleavage of polyproteins (pp1ab and pp1a) by nsp3 (papain-like protease; PLpro) and nsp5 (main protease, Mpro), replication and translation by the RTC, packaging and assembly of new virions, and virion release from the cell through exocytosis. RdRP: RNA-dependent RNA polymerase; RTC: replication and transcription complex; ERGIC: ER-to-Golgi intermediate compartment. ACE2: angiotensin-converting enzyme 2; TMPRSS2: transmembrane serine protease 2. (Right) 5′ UTR: 5′ untranslated region; 3′ UTR: 3′ untranslated region; ORF1a: open reading frame 1a; ORF1b: open reading frame 1b; 3a: ORF3a; E: envelop; M: membrane protein; 6: ORF6; 7a: ORF7a; 7b: ORF7b; 8: ORF8; 9b: ORF9b; 9c: ORF9c; N: nucleocapsid protein.
The spike (S) glycoprotein, a large transmembrane protein that covers the surface of SARS-CoV-2, mediates the entry of the coronavirus into the host cell through binding the receptor angiotensin-converting enzyme 2 (ACE2) that is found in human cells in the lungs, heart, kidneys, and intestines.12,13 The S protein is demarcated into two subunits, S1 containing the receptor binding domain, which binds the ACE2 receptor on the host cell surface, and S2 carrying membrane fusion peptide, which mediates membrane fusion, respectively. Proteolytic cleavage of the S protein by the host transmembrane proteases such as serine 2 (TMPRSS2) at the S1/S2 site promotes virus entry into the cell by activating the S protein.14 Neuropilin-1 (NRP1), which is known to bind furin-cleaved substrates, is abundantly expressed in the respiratory and olfactory epithelium, with highest expression in endothelial and epithelial cells. In addition to ACE2 and TMPRSS2, NRP1 was identified as a cellular factor binding furin-cleaved S1 fragment of S protein, which facilitates the entry of SARS-CoV-2 into host cells, thus increasing its infectivity and contributing to its tropism.15,16 As shown in Figure 1, once the virus enters the cell, the viral genome is released into the host cell cytoplasm, hijacking the cell replication machinery and mounting a complex program of viral gene expression, which is highly regulated in space and time. Briefly, the viral genome is translated as a messenger RNA (mRNA) by the host cell machinery and generates the polyprotein ORF1a and ORF1b, which then is processed into some essential enzymes (Nsp1-16 complex) for RNA synthesis (Nsp12: RNA-dependent RNA polymerase, RdRp), proofreading (Nsp14), and capping (Nsp14-Nsp16 complex).17,18 Meanwhile, the viral genome also serves as a template for negative-strand RNA transcription. Subsequent assembly of replication and transcription complex (RTC) facilitates the synthesis of genomic RNA (gRNA) and subgenomic RNAs (sgRNAs) inside virus infection-induced double-membrane vesicles (DMVs) to replicate the viral genome and transcription of viral protein-encoding genes, respectively.19,20 The newly synthesize sgRNAs released from the DMV encode viral structural and accessory proteins. Finally, translated structural proteins translocate into the ER membranes and transit through the ER-to-Golgi intermediate compartment (ERGIC), where a newly generated gRNA is encapsidated with N proteins, enclosed by a viral envelope, and released from the infected cells by exocytosis.21,22
Since there are many functional similarities between SARS-CoV, SARS-CoV-2, and MERS-CoV, understanding the biology and pathology of coronavirus family members has greatly promoted the development of vaccines and therapeutics against SARS-CoV-2. Generally, therapeutic approaches for viral diseases can be divided into directly acting antivirals and host-directed therapies.23 Ideally, directly acting antivirals often only target the virus without impairing host processes, which may reduce unwanted adverse effects. Thus, plenty of compounds with antiviral activity have been identified by high-throughput screening and molecular target-based assays, which can be divided into several categories based on the specific targets of SARS-CoV-2 life cycle: (1) some compounds targeting enzymes or functional proteins (Mpro, Plpro, RdRp), preventing the virus RNA synthesis and replication;24, 25, 26, 27 (2) some compounds targeting structural proteins of virus (E, M, S), blocking virus binding to the receptors, or inhibiting the virus's self-assembly process;28, 29, 30 (3) some compounds acting on the host's specific receptors or enzymes (ACE2, TMPRSS2), blocking viral attachment and entry into the host's cells.31, 32, 33, 34 Although most of the identified compounds were performed by cell-based assays, several trials have been initiated to test some of them specifically targeting SARS-CoV-2.35
Although drugs that target viral proteins offer a substantial benefit, as they may be more specific against viruses and have fewer side effects on humans, drug resistance may develop rapidly after treatment, particularly in RNA viruses with high polymerase error rates where mutations happen frequently. By contrast, if host factors that benefit viral replication or help the virus escape antiviral innate immune are identified as drug targets, viral mutations will usually not affect the interaction of the drug with its host target. Thus, understanding the innate immune responses to SARS-CoV-2 and its immune evasion strategies will help us better understand pathogenesis and aid in the development of antiviral drugs as well as immunotherapies. Here, we will discuss how the innate immune system and critical molecules are involved in the antiviral defense against SARS-CoV-2, as well as how SARS-CoV-2 antagonizes different immune responses to achieve a successful infection and highlight the potential of some host targets for developing medicines against COVID-19.
Host innate immune response against SARS-CoV-2
Accumulating evidence indicates that SARS-CoV-2 infects the epithelial cells of the respiratory tract and lungs, as well as epithelial and nonepithelial cells of other organs that express the ACE2 receptor, which initiates an antiviral immune response upon detection of the virus (Figure 2).36,37 As an evolutionarily conserved system of cellular and chemical defenses, the innate immune system is crucial to pathogen detection and restriction as well as the subsequent activation of an adaptive immune response. Here, we outline several of the innate defense mechanisms directed against SARS-CoV-2 infections. Figure 3 summarizes the recognition of SARS-CoV-2 by PRRs.
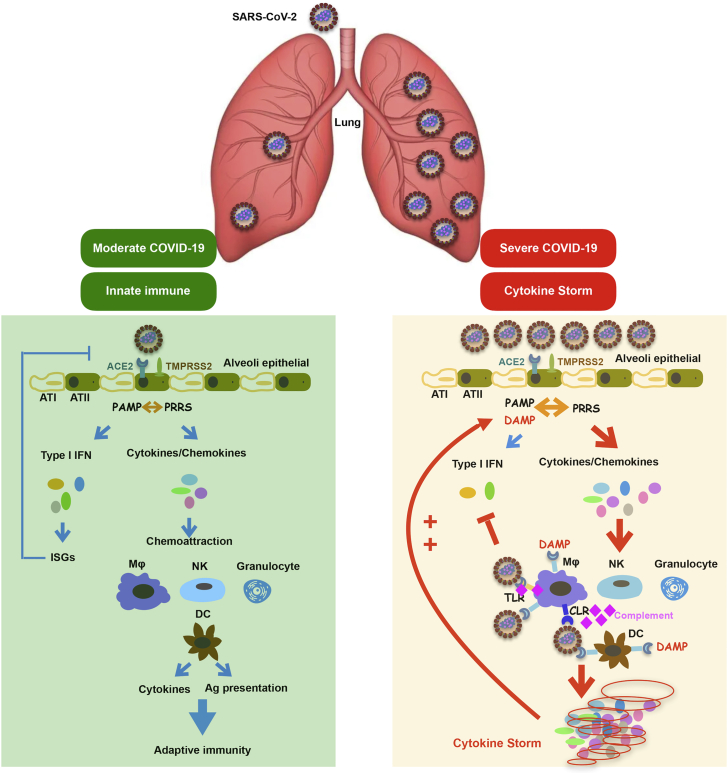
Figure 2.
The innate immune responses to SARS-CoV-2 infection
Infection with SARS-CoV-2 in airway epithelial cells triggers innate and adaptive immune responses. In severe COVID-19, a high virus load hyperactivates the innate immune system results in the production of high levels of inflammatory cytokines called a “cytokine storm.” AT I, alveolar type I cells; AT II: alveolar type II cells; MΦ, macrophage; DC, dendritic cells; NK, NK cells; Ag presentation, antigen presentation; DAMP, damage-associated molecular pattern; PAMP, pathogen-associated molecular pattern; PRRS, pattern recognition receptors; TLR, Toll-like receptors; CLR, C-type lectin receptors; ISG, interferon-stimulated gene.
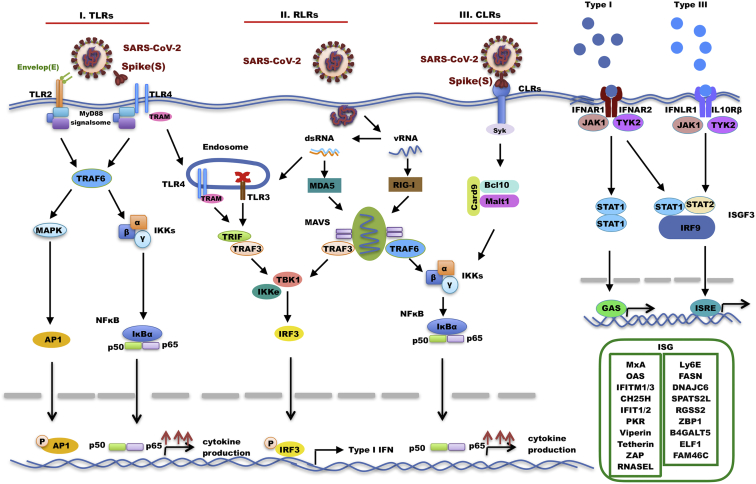
Figure 3.
PRRs-mediated recognition of SARS-CoV-2
SARS-CoV-2 is recognized by the innate immune system by members of distinct classes of PRRs (with their respective ligands indicated): Toll-like receptors (TLRs), retinoic acid-inducible gene-I (RIG-I) receptors, and C-type lectin receptors (CLRs). Upon recognition, signal transduction occurs through downstream transcription regulators called interferon regulatory factors (IRFs) to elicit interferons production. The secreted interferons interact with their receptors, which results in activation of JAK-Stat signaling pathway that governs the expression of various IFN-stimulated genes. TRAM, Toll receptor–associated molecule; TRAF6, TNF receptor associated factor 6; MAPK, mitogen-activated protein kinase; AP1, activator protein 1; IKKs, IκB kinase; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; TRIF, TIR domain-containing adaptor-inducing interferon-β; TRAF3, TNF receptor associated factor 3; IKKe, IkappaB kinase-epsilon; TBK1, TANK binding kinase 1; IRF3, interferon regulatory factor 3; Syk, spleen tyrosine kinase; Card9, caspase recruitment domain family member 9; Bcl10, B-cell lymphoma/leukemia 10; Malt1, mucosa-associated lymphoid tissue lymphoma translocation protein 1; IFNAR, interferon alpha/beta receptor; JAK1, tyrosine-protein kinase JAK1; Tyk2, non-receptor tyrosine-protein kinase 2; IFNLR1, interferon lambda receptor 1; IL10Rβ, interleukin 10 receptor subunit beta; Stat1, signal transducer and activator of transcription 1; Stat2, signal transducer and activator of transcription 2, IRF9: interferon regulatory factor 9; ISGF3, interferon-stimulated gene factor 3; GAS, gamma interferon activation site; ISRE, interferon-sensitive response element.
Sensing of SARS-CoV-2 infection by PRR receptors of the innate immune system
The innate immune system detects SARS-CoV-2 infection through the recognition of pathogen-associated molecular patterns (PAMPs) that are specifically present in the viral particle or generated during viral replication by PRRs that initiate antiviral signaling cascades, leading to the production of interferons (IFNs), cytokines, and chemokines.38, 39, 40 SARS-CoV-2 is recognized by the innate immune system by members of distinct classes of PRRs (with their respective ligands indicated): Toll-like receptors (TLRs), retinoic acid-inducible gene-I (RIG-I) receptors (RLRs), and C-type lectin receptors (CLRs).
TLR-mediated recognition of SARS-CoV-2
The cell surface innate immune sensor TLR2 forms heterodimers with TLR1 and TLR6 to detect diverse ligands from bacteria, fungi, parasites, and viruses, which is the initial step in a cascade of events leading to significant innate immune responses to pathogens.41,42 TLR2 has been reported to recognize multiple viral proteins, including the dUTPase of Epstein-Barr virus, the glycoprotein B of cytomegalovirus, and the capsid of hepatitis B virus.43, 44, 45 Proud et al. revealed that prophylactic intranasal administration of TLR2/6 agonist INNA-051 reduces SARS-CoV-2 transmission and provides protection against COVID-19 in a ferret model.46 Stimulation of TLR2 leads to activation of the innate immune response, suppression of excessive inflammation and tissue damage, as well as promotion of the integrity of local epithelial barrier function. However, the pathologic role of TLR2 in excessive inflammation in COVID-19 has also been revealed. Min Zheng et al. reported that TLR2 and its adaptor MyD88 are associated with the severity of COVID-19 and required for SARS-CoV-2-induced inflammatory responses during infection independent of viral entry through sensing the SARS-CoV-2 envelop protein as its ligand. Blocking TLR2 by oxPAPC also protects against SARS-CoV-2-induced lethality in vivo, indicating that TLR2 may contribute to the disease development of COVID-19.47
Among TLR genes, TLR3 was identified as an IFN-inducing dsRNA sensor with the ability to sense double-stranded RNA, a common intermediate of replication among many viruses, highlighting the TLR3 pathway as a “major sentinel” against viral infection.40,48 Several studies have shown that TLR3 via the TRIF pathway leads to a protective response in SARS-CoV infection.49 Although TLR3 knockout mice are more susceptible to SARS-CoV without an increase in mortality, TRIF, adaptor of TLR3, deficient mice are highly susceptible to SARS-CoV with a high risk of mortality.49 Q. Zhang et al. used a candidate gene approach and identified at least 3.5% of patients with life-threatening COVID-19 pneumonia who have mutations in genes involved in TLR3-dependent induction and amplification of type I IFNs.50 This discovery reveals essential roles for the double-stranded RNA sensor TLR3 in the control of SARS-CoV-2 infection.51
TLR4 is a remarkable PRR that recognizes multiple PAMPs from bacteria, viruses, and other pathogens.52 In addition, TLR4 also senses certain damage-associated molecular patterns (DAMPs) such as high-mobility group box 1 (HMGB1)53 and heat shock proteins (HSPs)54 released from dying or lytic cells during host tissue injury or viral infection. Like TLR2, TLR4 has also been reported to sense several viral proteins after infection, including the fusion protein of respiratory syncytial virus (RSV),55 the glycoprotein of EBOV,56 the glycoprotein of vesicular stomatitis virus (VSV G),57 and the dengue virus nonstructural protein 1 (NS1).58 Sohn et al. recently reported that the expression of TLR4 itself and its downstream signaling mediators including TRAF6, IRAK1, and TRIF in COVID-19 patients were significantly upregulated in peripheral blood mononuclear cells, compared with those in healthy controls, which indicated a correlation between increased TLR4 expression and its activation by a component of the SARS-CoV-2 virus, similar to that which occurs in bacterial sepsis.59 Choudhury et al. undertook an in silico study and found that the S glycoprotein of SARS-CoV-2 had the strongest protein-protein interaction with TLR4, compared with other TLRs.60 Yingchi Zhao et al. revealed that different coronaviruses (SARS-CoV-2, SARS-CoV, MHV-A59, and HCoV-229E) were able to activate TLR4 via their S proteins independent of ACE2 and TMPRSS2, thus inducing inflammatory IL-1β production.61 The studies add evidence to the possibility that the binding of S protein to TLR4 receptor plays a role in the hyperinflammation of COVID-19, thus implicating TLR4 as a therapeutic target for COVID-19. Interleukin-1β (IL-1β) is a proinflammatory cytokine that is central for host responses to infection.62 Typical IL-1β activation is mediated by two steps: priming and activation. In the priming step, upon activation by PAMPs or DAMPs, expression of canonical inflammasome-related components such as inactive NLRP3, pro-Caspase-1, pro-IL-1β, and pro-IL-18 are driven by NF-κB. In the activation step, assembly of NLRP3, ASC, and pro-caspase-1 into a complex mediates activation of caspase-1, which promotes the cleavage of pro-IL-1β and pro-IL-18 into their mature, biologically active forms.63 Tamara S. Rodrigues et al. found that the NLRP3 inflammasome is activated in response to SARS-CoV-2 infection and is active in COVID-19 patients, suggesting that inflammasomes participate in the pathophysiology of the disease.64 SARS-CoV-2 can encode several proteins, such as N protein, Spike, and open reading frame 3a to trigger NLRP3 inflammasome activation and induce hyperinflammation. Pan et al. identified the direct interaction between N protein and NLRP3 protein, which promotes the binding of NLRP3 with ASC, and facilitates NLRP3 inflammasome assembly.65 Huanzhou et al. showed a distinct mechanism by which ORF3 protein promotes NLRP3 inflammasome activation to induce hyperinflammation. The ORF3a triggers IL-1β expression via NF-κB, thus priming the inflammasome while also activating it via ASC-dependent and -independent modes, and it accelerates the inflammasome assembly by increasing the potassium efflux and releasing ROS generation.66
Toll-like receptor 7 (TLR7) is intracellular sensor located in endosomes that recognize single-stranded RNA. Mutations in the gene encoding this receptor have been found in men who develop severe COVID-19. Van der Made et al. analyzed primary immune cells isolated from the patients and family members and found that four young male patients with severe COVID-19 had rare putative loss-of-function variants of X-chromosomal TLR7 that were associated with impaired type I and II IFN responses.67 It has been shown that IRF3 is a major transcription factor for TLR3 and RIG-I to prime type I IFN production, whereas IRF7 is important for TLR7 and TLR9.68 Qian Zhang et al. identified genetic defects of IRF7 in patients with severe COVID-19 pneumonia. Plasmacytoid dendritic cells from IRF7-deficient patients produced no type I IFN on infection with SARS-CoV-2, and IRF7−/− fibroblasts were susceptible to SARS-CoV-2 infection in vitro.50 These preliminary findings reveal essential roles for TLR7- and IRF7-dependent signaling in the control of SARS-CoV-2 infection. Type I IFN administration may be of therapeutic benefit in selected patients, at least early in the course of SARS-CoV-2 infection.
RLR-mediated recognition of SARS-CoV-2
RLRs encompass three members, RIG-I, melanoma differentiation-associated protein 5 (MDA-5), and laboratory of genetics and physiology 2 (LGP2), which are critical cytosolic RNA sensors in the detection of RNA virus infection, leading the transcriptional induction of type I IFNs and other genes that collectively establish an antiviral host response.69 RIG-I has been implicated in the recognition of viral RNA that harbor an uncapped 5′-di/triphosphate end and a short, blunt-ended, double-stranded portion, two essential features facilitating discrimination from self-RNAs.70,71 Although the properties of MDA-5 physiological ligands have not been fully described yet, it is acknowledged that RIG-I and MDA-5 have different dsRNA length dependencies: RIG-I prefers short dsRNA, whereas MDA-5 selectively binds long dsRNA.72 Children have reduced SARS-CoV-2 infection rates and a substantially lower risk for disease development compared with adults. J. Loske et al. reported that children displayed higher basal expression of MDA-5 and RIG-I in upper airway epithelial cells, macrophages, and dendritic cells, resulting in stronger early innate antiviral responses to SARS-CoV-2 infection than in adults.73 The role of RIG-I in sensing SARS-CoV-2 viral RNA is controversial. Lucy et al. revealed that SARS-CoV-2 triggers a robust innate immune response in lung epithelial cells Calu-3 through the activation of cytoplasmic RNA sensors RIG-I and MDA-5.74 Takahisa et al. introduced SARS-CoV-2 viral RNA fragment into WT, RIG-I knockout, and MDA-5 knockout HEK293 cells, and they found that not only MDA-5 but also RIG-I recognize SARS-CoV-2 viral RNAs and subsequently induce type I IFN production.75 Nevertheless, several independent groups have also reported that MDA-5, not RIG-I, governs the innate immune response to SARS-CoV-2 in lung epithelial cells Calu-3.76, 77, 78 The technical differences and cell line origin differences may explain the disparity between these findings. Thus, more compelling experimental approaches, like RIG-I mutant mice or human lung organoids, should be used to determine if RIG-I is essential for type I IFN production in response to SARS-CoV-2. As an ISG (IFN-stimulated gene), RIG-I sufficiently restrains SARS-CoV-2 replication in human lung cells in an IFN-independent manner. Taisho et al. revealed that RIG-I, by recognizing the 3′ untranslated region of the SARS-CoV-2 RNA genome via the helicase domains, directly abrogates viral RNA-dependent RNA polymerase mediation of the first step of replication.79
CLR-mediated recognition of SARS-CoV-2
CLRs expressed by antigen-presenting cells, including macrophages and dendritic cells (DCs), are crucial for tailoring immune responses to pathogens by sensing carbohydrate-based PAMPs.80 A number of enveloped viruses interact with CLRs at the cell surface to facilitate the transfer toward their particular entry receptors, which trigger the fusion of viral and host membranes.81,82 CLRs, especially L-SIGN (or DC-SIGNR) or DC-SIGN, are well-known for their role in the entry of viruses like HIV, cytomegalovirus, dengue, Ebola, and Zika virus.83, 84, 85 Densely glycosylated S protein of SARS-CoV-2 potentially presents ligands for CLRs, which are known to bind specific glycans mostly in a Ca2+-dependent manner.86 Qiao Lu et al. identified several C-type lectins (DC-SIGN, L-SIGN, LSECtin, ASGR1, and CLEC10A) and Tweety family member 2 (TTYH2) as glycan-dependent binding partners of the SARS-CoV-2 S protein using a myeloid cell receptor-focused ectopic expression screen.87 Although these receptors do not support active replication of SARS-CoV-2, their engagement with the virus-induced robust proinflammatory responses in myeloid cells that correlated with COVID-19 severity. Meanwhile, another two independent groups also reported the involvement of LSECtin and DC-SIGN in recognition of S glycoprotein of SARS-CoV-2.82,88 Considering that recognition of S protein by CLRs leads to the release of pro-inflammatory cytokines like IL-6, which has been found to contribute to the cytokine storm seen in severe COVID-19, Qiao Lu et al. also generated a bispecific anti-S nanobody that not only blocked ACE2-mediated infection but also the myeloid receptor-mediated proinflammatory responses.87 Michel Thépaut et al. revealed that polyman26 (PM26), a multivalent glycomimetic mannoside tailored for optimal interaction with DC-SIGN, not only binds the lectin but also enhances its internalization, reducing the number of CLRs available for the virus to bind it and transfect susceptible cells.82 These studies point to the importance of developing CLRs antagonists to reduce the severity of infection by inhibiting their activity or expression, which could help reduce the severity of COVID-19.
Type I IFNs in host defense against SARS-CoV-2
Upon recognition of viral PAMPs by PRRs, signal transduction occurs through downstream transcription regulators called IFN regulatory factors (IRFs) to elicit IFN production.89 IFNs can be broadly characterized into three groups: IFN-I, type II (IFN-II), and type III (IFN-III). On the binding of type I IFNs to their receptor (IFNAR), multiple downstream signaling pathways can be activated, leading to the induction of a large number of ISGs to trigger a global antiviral state.90,91 Although the products of these ISGs exert broad-spectrum antiviral effector functions against viruses including influenza, SARS-CoV-1, and MERS-CoV, many of these are still not fully described in SARS-CoV-2 infection.92 Recent attempts have focused on identifying which ISGs are anti-SARS-CoV-2 and further describing their mechanisms of action. Given the hundreds of ISGs induced by IFN, it is reasonable to expect that every stage of the viral life cycle (entry, uncoating, transcription, translation, assembly, and egress) might be targeted for suppression.
Cholesterol 25-hydroxylase (CH25H) is an ISG that encodes an enzyme that synthesizes the oxysterol 25-hydroxycholesterol (25HC) from cholesterol.93 CH25H has been shown to have broad antiviral activity against a variety of enveloped viruses by inhibiting virus entry into the host cells.94,95 For SARS-CoV-2, three independent groups revealed the anti-SARS-CoV-2 activities of CH25H and its enzymatic product 25HC in vitro and in vivo. Mechanistically, 25HC activates the ER-localized enzyme ACAT, whose activity depletes accessible cholesterol on the plasma membrane. Furthermore, internalized 25HC accumulates in the late endosomes and potentially restricts SARS-CoV-2 S protein catalyzed membrane fusion via blockade of cholesterol export.96, 97, 98 In addition, lymphocyte antigen 6 complex, locus E (LY6E) was also shown to restrict multiple CoVs, including SARS-CoV, SARS-CoV-2, and MERS-CoV, entry into cells by interfering with S protein-mediated membrane fusion.99 Base on a gain-of-function screen of human ISGs, more ISGs (AXIN2, EPSTI1, GBP5, IFIH1, IFITM2, IFITM3, CLEC4D, UBD, ELF1, FAM46C, and REC8) were identified as inhibitors of S protein-mediated SARS-CoV-2 entry into cells.100
Following releasing of the viral genome into the host cell cytoplasm, nonstructural proteins are produced via the transcription and translation of the replicase gene from viral genomic RNA, which then assemble the RTC to facilitate viral RNA replication and protein synthesis.22 The overexpression screen identified multiple ISGs able to reduce SARS-CoV-2 mRNA levels, SARS-CoV-2 protein translation, or genome synthesis, including ZAP, IFIT1, IFIT3, IFIT5, SPATS2L, DNAJC6, RGSS2, and LOC152225, as well as ZBP1 and B4GALT5.100 In addition, BST-2, known as an inhibitor of the retrovirus, filovirus, arenavirus, and herpesvirus families, was also found to potentially restrict SARS-CoV-2 release.100 Through genome-wide CRISPR/Cas9 screen, DEAD box RNA helicase DDX42 and Death domain-associated protein 6 (DAXX) were identified as broad antiviral inhibitors against both retroviruses and RNA viruses including SARS-CoV-2.101,102
Innate immune evasion by SARS-CoV-2
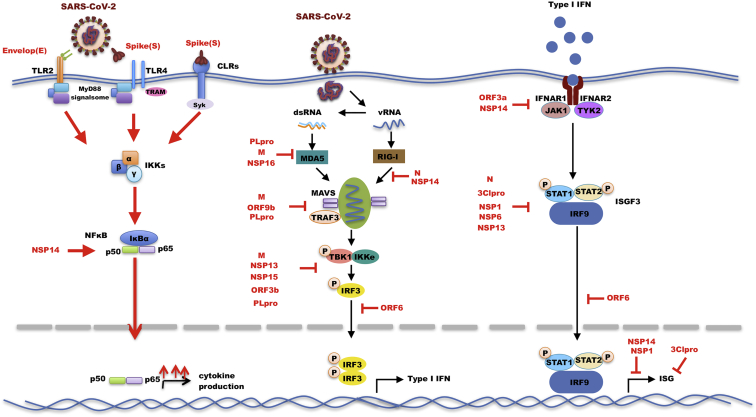
The immune imbalances in COVID-19 are characterized by poor type I IFNs production and an exacerbated release of proinflammatory cytokines, contributing to the severe manifestations of the disease.103 Several studies have uncovered the multitude of strategies employed by SARS-CoV-2 to limit the global cellular antiviral state, both directly and indirectly (Table 1). This is assumed to be mostly due to viral targeting of the IFN induction and signaling cascades at multiple levels, as described below (Figure 4).
Table 1.
Specific viral antagonism mechanisms at each stage of interferon signaling
| Working model | Viral protein | Targets | Mechanisms | References |
|---|---|---|---|---|
| Evasion of PRRs recognition of viral RNA | NSP10 | viral RNA | cofactor of NSP16 for RNA cap methylation | Wilamowski et al.104, Viswanathan et al105 |
| NSP14 | viral RNA | guanine-N7-methyltransferase activity that can mimic 5′cap structure on the viral RNA | Ogando et al.106 | |
| NSP16 | viral RNA | 2′-O-methyltransferase activity involved in viral RNA capping | Wilamowski et al.104, Viswanathan et al.105 | |
| NSP15 | viral RNA | processes viral RNA and limits the formation of dsRNA intermediates | Deng et al.107 | |
| Inhibition of PRRs-mediated signaling cascades | N | TRIM25 | interferes with the interaction of TRIM25 and RIG-I | Wang et al.108, Zhao et al.109 |
| STAT1/STAT2 | suppresses phosphorylation and nuclear translocation of STAT1 and STAT2 | Mu et al.110 | ||
| M | MAVS | impairs MAVS aggregation and its recruitment of downstream TRAF3, TBK1, and IRF3 | Fu et al.111, Zheng et al.112 | |
| TBK1 | interacts with TBK1, thus preventing the formation of the multiprotein complex | Fu et al.111, Zheng et al. 112 | ||
| ORF9b | TOM70 | suppresses type I interferon (IFN-I) responses through association with TOM70 | Jiang et al.113 | |
| ORF3b | IRF3 | prevents the translocation of IRF3 into the nucleus | Konno et al.114 | |
| NSP13 | TBK1 | interacts with TBK1 and inhibits TKB1/IRF3 activation | Guo et al.115, Lei et al.116 | |
| NSP15 | IRF3 | interacts with RNF41, an E3 ligase associated with activation of IRF3 | Gordon et al.117 | |
| ORF3a | STAT1 | suppresses STAT1 phosphorylation via upregulating SOCS1 | Wang et al.118 | |
| NSP6 | TBK1 | binds TBK1 to suppress IRF3 phosphorylation | Xia et al.119 | |
| ORF6 | IRF3 | binds importin karyopherin α 2 (KPNA2) to inhibit IRF3 nuclear translocation | Miorin et al.120 | |
| STAT1 | binds directly to the Nup98-Rae1 complex to block STAT nuclear import | Miorin et al.120 | ||
| Modulation of ubiquitination and deubiquitination | N | RIG-I | associates with TRIM25 and suppresses the ubiquitination of RIG-I | Wang et al.108 |
| M | TBK1 | induces TBK1 degradation via K48-linked ubiquitination | Sui et al.121 | |
| PLpro | ISG15 | cleavage of ISG15 from IRF3 and attenuates type I interferon responses | Shin et al.122 | |
| mediates de-ISGyaltion of MDA5 and inhibit IFN response | Liu et al.123 | |||
| ORF9b | NEMO | interrupts K63-linked ubiquitination of NEMO | Wu et al.124 | |
| Viral proteases-mediated cleavage | PLpro | IRF3 | direct cleavage of IRF3 by NSP3 results in reduced type I IFN response | Moustaqil et al.125 |
| 3Clpro | NEMO | 3Clpro cleaves NEMO to induces the death of human brain endothelial cells | Josephine Lampe et al.126 | |
| Host translation shutoff | NSP1 | 40S subunit | shuts down host protein translation and subsequently blocks host immune functions | Zhang et al.127 |
| NSP14 | Unknown | exerts translational inhibition to abolish the IFN-I-dependent induction of ISGs | Hsu et al.128 | |
| NSP10 | NSP14 | cofactor of NSP14 Exerts translational inhibition to abolish the IFN-I-dependent induction of ISGs | Hsu et al.128 |
Figure 4.
Innate immune evasion by SARS-CoV-2
Recognition by PRRs triggers a signaling cascade that culminates in the transcription and subsequent generation of interferons. SARS-CoV-2 has evolved to antagonize these pathways at virtually all stages, indicated by red blunt end arrows. Red solid arrows indicate hyperactivated signal cascade by SARS-CoV-2. Black solid arrows indicated pathway connection.
Evasion of PRRs recognition of viral RNA
The cytosolic RLRs receptors are the principal detector of pathogenic RNAs that recognize the 5′cap in order to distinguish the host mRNA from viral RNA, thereby preventing autoimmune pathologies.129 However, many viruses have evolved mechanisms for capping their genomes and/or transcripts with methylation at the N7 position of the capped guanine and the ribose 2′-O-position of the first nucleotide during replication, which help viral RNAs evade recognition by the host RNA sensors of the innate immune system.130 In SARS coronaviruses, the non-structural protein 16 (nsp16), in conjunction with nsp10, methylates the 5′-end of virally encoded mRNAs to mimic cellular mRNAs, thus protecting the virus from host innate immune restriction.104,105 In addition, the conserved uridine-specific endoribonuclease (NSP15), an integral component of the coronaviral RTC complex, processes viral RNA and limits the formation of dsRNA intermediates to escape the recognition of viral RNA sensors and subsequent IFN response.107
Inhibition of PRRs-mediated signaling cascades
The induction of the IFN system starts with cellular recognition of viral infection. Upon activation, the cellular viral RNA sensors launch a signaling cascade that results in transcriptional induction of IFN. These signaling events comprise many molecules of cellular adaptors, regulatory enzymes, and transcription factors, particularly the IRFs, many of which are targets for direct suppression by viral products that attach to them and impede their action.131 SARS-CoV-2 encodes a plethora of viral proteins to counteract the IFN induction at various stages. Wang et al. reported that NP can disrupt the interaction between MAVS and RIG-I, as well as the interaction between RIG-I and TRIM31, thus limiting the host IFN response to the virus.108 In addition, SARS-CoV-2 membrane glycoprotein M was also identified as a negative regulator of the innate immune response. The interaction between M and MAVS impaired MAVS aggregation and its recruitment of downstream TRAF3, TBK1, and IRF3, leading to attenuation of the innate antiviral response.111,112 ORF9b, an alternative ORF in the N protein, was reported to localize on mitochondria and thus suppress IFN response through association with adaptor protein TOM70.113
TBK1, an IKK-related serine/threonine kinase, is pivotal for the induction of antiviral type I IFN by TLR and RLR signaling pathways. Yi Zheng et al. revealed that M protein interacts with TBK1, thus preventing the formation of the multiprotein complex containing RIG-I, MAVS, TRAF3, and TBK1 and subsequently impeding the phosphorylation, nuclear translocation, and activation of IRF3.112 Furthermore, two independent groups identified NSP13 as a TBK1 interactor that downregulates IFN signaling by limiting TBK1 and IRF3 activation.115,116 Nonstructural protein 6 (nsp6) also binds TBK1 to suppress IRF3 phosphorylation.119 As a pivotal transcriptional factor driving IFN production, IRF3 is also targeted by SARS-CoV-2. ORF6 binds importin karyopherin α 2 (KPNA2) to inhibit IRF3 nuclear translocation, not IRF3 phosphorylation.120 Based on the IFN-β luciferase screening, Wenjing et al. reported that NSP12 attenuates IFN production by inhibiting IRF3 nuclear translocation.132 However, Aixin et al. found that, although NSP12 could suppress IFN-β promoter luciferase activity, it showed no inhibitory effect on IFN-β production or its downstream signaling.133
After binding to its receptor, IFN activates the Jak-STAT pathway, triggering STAT dimerization and nuclear translocation. Hongjie et al. identified two sets of viral proteins that antagonize type I IFN signaling through blocking signal transducer and activator of transcription (STAT1)/STAT2 phosphorylation or nuclear translocation.119 Remarkably, Orf6 localizes at the nuclear pore complex where it binds directly to the Nup98-Rae1 complex to block STAT nuclear import and antagonize IFN signaling.120 In addition, N protein was also reported to antagonize type I IFN signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2.110 The unique accessory protein of SARS-CoV-2, ORF8 forms intracellular aggregates and inhibits not only basal expression of several antiviral ISGs, including DHX58, ZBP1, MX1, and MX2, but also the induction of antiviral molecules by IFN in lung epithelial cells.134
Modulation of ubiquitination and deubiquitination
Ubiquitination, a crucial type of protein posttranslational modification catalyzed by the sequential action of ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin-ligating (E3) enzymes, plays a crucial role in regulating antiviral innate immune responses.135 To maintain proper protein homeostasis, all the major posttranslational modifications, including the ubiquitination process, can be reversed. The process of cleaving ubiquitin molecules from ubiquitin-conjugated protein substrates by deubiquitinating enzymes (DUBs) is called deubiquitination.136 Yinghua et al. found that low-dose N protein combined with TRIM25 could suppress the ubiquitination and activation of RIG-I.109 SARS-CoV-2 M could interact with MDA-5, TRAF3, IKKε, and TBK1, and induce TBK1 degradation via K48-linked ubiquitination.121 SARS-CoV ORF3a overexpression has been reported to induce the lysosome-mediated degradation of IFNAR1.137 For SARS-CoV-2, Nsp14 induces lysosomal degradation of IFNAR1, thereby preventing STAT transcription factor activation.138 Jing Wu et al. revealed that ORF9b targets the NEMO and interrupts its K63-linked polyubiquitination upon viral stimulation, thereby inhibiting the canonical NF-κB signaling and subsequent IFN production.124 A more general strategy by which SARS-CoV-2 interferes with ubiquitination involves viral PLpro protease that removes ubiquitin-like ISG15 protein and ubiquitin from host proteins to negatively impact the innate immune system. Although it shares 83% sequence identity with SARS-CoV-PLpro, SARS-CoV-2-PLpro exhibits different host substrate preferences; SARS-CoV-2-PLpro preferentially cleaves the ubiquitin-like ISG15, whereas SARS-CoV-PLpro predominantly targets ubiquitin chains.122 Donghyuk et al. reported that SARS-CoV2-PLpro contributes to the cleavage of ISG15 from IRF3 and attenuates type I IFN responses.122 In addition, ISG15 conjugation is essential for antiviral IFN responses mediated by the viral RNA sensor MDA-5. The ISG15-dependent activation of MDA-5 is also antagonized through direct de-ISGylation mediated by the PLpro of SARS-CoV-2.123
Viral proteases-mediated cleavage
Many viral proteases that are involved in the cleavage and processing of viral polyproteins, such as those found in positive-strand RNA viruses, have also been demonstrated to cleave factors essential for the IFN response.139 The genome of SARS-CoV-2 encodes two viral proteases, PLpro and 3Clpro, that are responsible for cleaving viral polyproteins during replication. For PLpro, IRF3 has been identified as its substrate. Direct cleavage of IRF3 by NSP3 would result in reduced type I IFN response during SARS-CoV-2 infections.125 As a main protease, 3CLpro is also highly conserved among coronaviruses. STAT2 and NEMO, the essential modulators of IFN signaling, are known to be cleaved by coronavirus main protease.140,141 The cleavage of STAT2 by 3Clpro has not been reproduced for SARS-CoV-2 yet. However, Josephine et al. reported that 3Clpro cleaves NEMO, the essential modulator of both NF-κB and IFN signaling, in human brain endothelial cells.126 Human Bre1/RNF20 (hBre1), the E3 ubiquitin ligase for histone 2B monoubiquitination, is necessary for ISGs transcription.142 Zhang et al. revealed that 3Clpro cleaves RNF20 to disrupt the RNF20/RNF40 complex, which promotes SARS-CoV-2 replication.143 Although the host proteolytic targets of 3Clpro are still anonymous, multifarious host proteins have been predicted as 3Clpro. Among these, TRIF, IRF2, STAT1/4, IFI6, RIP1/2, NEDD4, and SMURF2 are reported to be involved in the regulation of IFN signaling.144
Host translation shutoff by virus
Many viruses have evolved ways to shut off host gene expression by interacting with the host translational machinery and degrading host mRNA.145 This global inhibition of cellular protein synthesis serves to ensure maximal viral gene expression and to prevent the synthesis of antiviral genes.146 SARS-CoV-2 infection leads to the accelerated degradation of cytosolic cellular mRNA including innate immune genes. Nonstructural protein 1 (Nsp1) specifically shuts down host protein translation and subsequently blocks host immune functions through competing with RNA segment to bind the small ribosomal 40S subunit to stall mRNA translation at various stages during imitation and binding to the ribosome for endonucleolytic cleavage and degradation of host mRNAs.127,147 In addition, coronavirus NSP14 protein, known to have 3′–5′ exoribonuclease (ExoN) activity and guanine-N7-methyltransferase activity (N7-MTase), also has the translational inhibition activity.106 Jack Chun-Chieh et al. revealed that ExoN and N7-MTase activities enable NSP14 to exert translational inhibition to abolish the type I IFN-dependent induction of ISGs.128 Another nonstructural protein NSP10 also participates in host translational shutoff through forming the complex of NSP10-NSP14 that enhances translation inhibition executed by NSP14.128
Complement system in severe COVID-19
The complement system, also known as complement cascade, is a critical part of the innate immune system that enhances antibodies and phagocytic cells to clear pathogens and damaged cells from an organism, promote inflammation, and attack the pathogen's cell membrane.148 The complement system consists of more than 50 proteins in the plasma and on cell surfaces that exert their functions through highly ordered interactions.149 There are three major pathways through which the complement system can be activated: classical, lectin, and alternative.150 Multiple lines of evidence pointed to complement cascade hyperactivation being involved in the pathogenesis and disease severity of COVID-19. SARS-CoV-2 can directly activate the complement cascade through the lectin pathway, the alternative pathway, and classical pathway. Mechanistically, multiple lectin pathway recognition molecules of the complement system can bind to SARS-CoV-2 S and N proteins.151 S protein (S1 and S2), but not the N protein, directly activates the alternative pathway of complement on cell surfaces.152 In addition, N protein activates the lectin pathway via directly binding to the lectin pathway effector enzyme MASP-2.151,153 These studies suggest that SARS-CoV-2 infection involves the hyperactivation of multiple complement cascades. Currently, little is known about the role of the complement system in the elimination of coronavirus. However, its deleterious involvement in the severe COVID-19 is becoming clear. Gao et al. reported that N protein of SARS-CoV, MERS-CoV, and SARS-CoV-2 bound to MASP-2, resulting in aberrant complement activation and aggravated inflammatory lung injury. Either blocking the N protein-MASP-2 interaction or deleting MASP-2 can significantly attenuate N protein-induced complement hyperactivation and lung injury in vitro and in vivo.153 Holter et al. described the systemic complement activation in hospitalized COVID-19 patients and its association with the development of respiratory failure.154
SARS-CoV-2 activates NF-κB signaling to exert hyperinflammation
The transcription factor NF-κB is a crucial modulator of proinflammatory gene induction and functions in both innate and adaptive immune cells. Accumulating evidence suggests that the hyperactivation of NF-κB signaling, i.e., cytokine storm, has been implicated in the pathogenesis of COVID-19, which was supported by the fact that severity of COVID-19 is associated with an increased level of NF-κB-mediated inflammatory mediators including cytokines and chemokines such as interleukin IL-1β, IL-6, IL-8, MCP-1, MIP-1α, MIP-1β, and TNF-α in blood upon SARS-CoV-2 infection.8,155, 156, 157, 158 The relevance of TLR receptors to stimulate excessive inflammation in COVID-19 has been documented. Upon SARS-CoV-2, viral envelop and S protein could induce hyperactivation of NF-κB-mediated inflammation through directly binding to TLR2 and TLR4.47,61 In addition, S protein can also be sensed by various CLRs on the surface of immune cells, which not only helps SARS-CoV-2 get into cells but also exacerbates proinflammatory responses (Figure 2).86, 87, 88 For SARS-CoV-1, nucleocapsid (N), ORF3a, and ORF7a proteins were able to activate NF-κB and significantly enhanced IL-8 and IL-6 expression.159 For SARS-CoV-2, Chia-Ming et al. identified ORF3a, M, ORF7a, and N proteins of SARS-CoV-2 as NF-κB activators.160 Particularly, the ORF7a protein induced the NF-κB dictated proinflammatory cytokines including IL-1α, IL-1β, IL-6, IL-8, IL-10, and TNF-α, and different chemokines including CCL11, CCL17, CCL19, CCL20, CCL21, CCL22, CCL25, CCL26, CCL27, and CXCL9.160 Notably, SARS-CoV-2 has evolved some unique proteins with dual-role in NF-κB activation and the suppression of IFN signaling. ORF8, the unique protein encoded by SARS-CoV-2, has an inhibitory effect on the IFN pathway.134,161 On the other hand, Xiaoyuan et al. demonstrated that ORF8 could interact with IL-17 receptor and subsequently aggravate the secretion of pro-inflammatory factors by activating NF-κB signaling pathway.162 NSP14, well documented in inhibiting IFN signaling, was also reported to contribute to the viral activation of NF-κB signaling, which was characterized by the nuclear translocation of NF-κB p65 subunit and upregulation of IL-6 and IL-8.163 Mechanistically, NSP14 interacts with host inosine-5′-monophosphate dehydrogenase 2 (IMPDH2) protein, which is known to regulate NF-κB signaling.163 The imbalanced immune features of severe COVID-19 are defined as hyperactivation of NF-κB-mediated inflammation but a relatively lower level of antiviral IFNs. The dual-role of ORF8 and NSP14 in NF-κB activation and IFN signaling inhibition would contribute to the understanding of the imbalanced immune response of COVID-19 and provide drug targets for controlling COVID-19.
Conclusions and perspectives
SARS-CoV-2 emerged in the human population in 2019 and has become a serious global health threat that impacts all countries. During host-virus co-evolution, SARS-CoV-2 has evolved well to adapt to avoiding the host innate immune response and inhibiting the host antiviral IFN signaling. This adaption of SARS-CoV-2 depends on the interaction between the host innate immune system and the virus. By this interaction, SARS-CoV-2 escapes host antiviral machinery and hijacks host innate immunity and accomplishes the life cycle. Although a plethora of viral proteins encoded by SARS-CoV-2 have been identified as IFN antagonists,116,119 we are still far from clearly understanding the complex immune regulation of SARS-CoV-2, due to most of these viral IFN antagonist proteins being identified in the contexts of ectopic expression by transfection in cell lines. Thus, it is important to validate these findings using more comparative analyses and multiple experimental systems in the contexts of authentic viral infection in both cultured cells and animal models. In combination with reverse genetic systems of SARS-CoV-2, identification of viral proteins with antagonistic action against innate immune or IFN will facilitate the development of live-attenuated vaccine candidates.36,164,165
The traditional design and screening of antivirus drugs often target viral proteins that offer a substantial benefit, as identified compounds may be more specific against viruses and have fewer side effects on humans. However, as an RNA virus, SARS-CoV-2 has a high rate of mutation leading to the constant generation of many different strains with an altered viral evasion or resistant protein, which could render the therapy ineffective.166,167 Thus, developing novel antivirals targeting virus-host innate immune interaction offers a huge potential for alleviating the pathology of virus infections as well as assisting the immune system to clear viral infection. The strategies for designing antiviral drugs based on the knowledge of crosstalk between virus and host innate immunity can be divided into several categories: (1) targeting TLRs and CLRs as well as NF-κB to prevent hyperinflammation in COVID-19; (2) designing novel IFN agonists with comparable or better antiviral activities but with less toxicity than IFN itself; (3) targeting ISGs that produce natural antiviral products, like CH25 produces natural antiviral 25HC; (4) screening compounds that can target both viral protein and host innate immune. The increasing knowledge on the interaction between viruses and hosts has been and continues to guide the development of broad-spectrum antiviral drugs not only in the fight against SARS-CoV-2 but also in regulating the host immune system.
Currently, plenty of compounds have been screened to target SARS-CoV-2 proteases PLpro and 3Clpro, which not only inhibit viral polyprotein processing but also would rescue host antiviral IFN signaling.27,168, 169, 170, 171 Aside from PLpro and 3Clpro, multiple viral proteins are also involved in the dysregulation of antiviral IFN signaling. Thus, combinatory usage of multiple drugs targeting viral IFN antagonists might be a promising strategy for anti-SARS-CoV-2 therapy. Unlike PLpro and 3Clpro with enzymatic activities, other viral proteins are undruggable or difficult-to-target proteins that may demand more innovative thinking and novel therapeutic approaches. Proteolysis targeting chimeras (PROTAC) technology shows many advantages over small-molecule inhibitors, such as inducing the durative and fast depletion of target proteins, overcoming mutation-caused drug resistance, targeting “undruggable” and mutant proteins.172 Potent and selective PROTACs might be considered in the study of anti-SARS-CoV-2 therapy by degrading viral IFN antagonists. Particularly, we propose to use the small compounds library to screen small molecules targeting both viral protein and host factors to generate selective PROTACs with the potential to degrade viral protein and reactivate host antiviral innate immunity.
Elucidating how SARS-CoV-2 evades the innate immune response is critical for understanding the pathogenicity and developing innovative therapeutic approaches to limit the sequelae of viral infections. The discovery of critical immune evasion proteins paves the way for the development of more effective antivirals that target these viral proteins. In addition, the identification of pivotal cellular antiviral pathways against SARS-CoV-2 infection also facilitates the development of strategies targeting these pathways to overwhelm incoming viruses.
Acknowledgments
This study was supported by National Institute of Health (NIH) grants (AI069120, AI158154 and AI149718), the UCLA AIDS Institute and UCLA David Geffen School of Medicine—Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research Award Program.
Author contributions
The conception and design of the paper were done by S.Z. and G.C. The initial draft was completed by S.Z. and L.W. Editing and further drafts were done by S.Z. and G.C. All authors reviewed the final version of the paper.
Declaration of interests
The authors declare no conflict of interest.
References
- 1.Totura A.L., Bavari S. Broad-spectrum coronavirus antiviral drug discovery. Expert Opin. Drug Discov. 2019;14:397–412. doi: 10.1080/17460441.2019.1581171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 6.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 7.Woldemeskel B.A., Kwaa A.K., Garliss C.C., Laeyendecker O., Ray S.C., Blankson J.N. Healthy donor T cell responses to common cold coronaviruses and SARS-CoV-2. J. Clin. Invest. 2020;130:6631–6638. doi: 10.1172/JCI143120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rouchka E.C., Chariker J.H., Chung D. Variant analysis of 1,040 SARS-CoV-2 genomes. PLoS One. 2020;15 doi: 10.1371/journal.pone.0241535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bar-On Y.M., Flamholz A., Phillips R., Milo R. SARS-CoV-2 (COVID-19) by the numbers. Elife. 2020;9 doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 13.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daly J.L., Simonetti B., Klein K., Chen K.E., Williamson M.K., Antón-Plágaro C., Shoemark D.K., Simón-Gracia L., Bauer M., Hollandi R., et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu D.X., Fung T.S., Chong K.K., Shukla A., Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antivir. Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolff G., Melia C.E., Snijder E.J., Bárcena M. Double-membrane vesicles as platforms for viral replication. Trends Microbiol. 2020;28:1022–1033. doi: 10.1016/j.tim.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein S., Cortese M., Winter S.L., Wachsmuth-Melm M., Neufeldt C.J., Cerikan B., Stanifer M.L., Boulant S., Bartenschlager R., Chlanda P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020;11:5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartenian E., Nandakumar D., Lari A., Ly M., Tucker J.M., Glaunsinger B.A. The molecular virology of coronaviruses. J. Biol. Chem. 2020;295:12910–12934. doi: 10.1074/jbc.REV120.013930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brant A.C., Tian W., Majerciak V., Yang W., Zheng Z.M. SARS-CoV-2: from its discovery to genome structure, transcription, and replication. Cell Biosci. 2021;11:136. doi: 10.1186/s13578-021-00643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lou Z., Sun Y., Rao Z. Current progress in antiviral strategies. Trends Pharmacol. Sci. 2014;35:86–102. doi: 10.1016/j.tips.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coelho C., Gallo G., Campos C.B., Hardy L., Würtele M. Biochemical screening for SARS-CoV-2 main protease inhibitors. PLoS One. 2020;15 doi: 10.1371/journal.pone.0240079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo S., Xie H., Lei Y., Liu B., Zhang L., Xu Y., Zuo Z. Discovery of novel inhibitors against main protease (Mpro) of SARS-CoV-2 via virtual screening and biochemical evaluation. Bioorg. Chem. 2021;110:104767. doi: 10.1016/j.bioorg.2021.104767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Douangamath A., Fearon D., Gehrtz P., Krojer T., Lukacik P., Owen C.D., Resnick E., Strain-Damerell C., Aimon A., Ábrányi-Balogh P., et al. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat. Commun. 2020;11:5047. doi: 10.1038/s41467-020-18709-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao Y., Du X.Y., Duan Y.K., Pan X.Y., Sun Y.F., You T., Han L., Jin Z., Shang W., Yu J., et al. High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors. Protein Cell. 2021;12:877–888. doi: 10.1007/s13238-021-00836-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandala V.S., McKay M.J., Shcherbakov A.A., Dregni A.J., Kolocouris A., Hong M. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat. Struct. Mol. Biol. 2020;27:1202–1208. doi: 10.1038/s41594-020-00536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhowmik D., Nandi R., Jagadeesan R., Kumar N., Prakash A., Kumar D. Identification of potential inhibitors against SARS-CoV-2 by targeting proteins responsible for envelope formation and virion assembly using docking based virtual screening, and pharmacokinetics approaches. Infect. Genet. Evol. 2020;84:104451. doi: 10.1016/j.meegid.2020.104451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., Burgstaller-Muehlbacher S., De Jesus P.D., Teriete P., Hull M.V., et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J.S., Petitjean S.J.L., Koehler M., Zhang Q.R., Dumitru A.C., Chen W.Z., Derclaye S., Vincent S.P., Soumillon P., Alsteens D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2021;11:4541. doi: 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gopinath K., Jokinen E.M., Kurkinen S.T., Pentikäinen O.T. Screening of natural products targeting SARS-CoV-2-ACE2 receptor interface - a MixMD based HTVS pipeline. Front. Chem. 2020;8:589769. doi: 10.3389/fchem.2020.589769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu X., Shrimp J.H., Guo H., Xu M., Chen C.Z., Zhu W., Zakharov A.V., Jain S., Shinn P., Simeonov A., et al. Discovery of TMPRSS2 inhibitors from virtual screening as a potential treatment of COVID-19. Acs Pharmacol. Transl. 2021;4:1124–1135. doi: 10.1021/acsptsci.0c00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y.W., Lear T.B., Evankovich J.W., Larsen M.B., Lin B., Alfaras I., Kennerdell J., Salminen L., Camarco D., Lockwood K., et al. A high-throughput screen for TMPRSS2 expression identifies FDA-approved compounds that can limit SARS-CoV-2 entry. Nat. Commun. 2021;12:3907. doi: 10.1038/s41467-021-24156-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tu Y.F., Chien C.S., Yarmishyn A.A., Lin Y.Y., Luo Y.H., Lin Y.T., Lai W.Y., Yang D.M., Chou S.J., Yang Y.P., et al. A review of SARS-CoV-2 and the ongoing clinical trials. Int. J. Mol. Sci. 2020;21:2657. doi: 10.3390/ijms21072657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., Kato T., Lee R.E., Yount B.L., Mascenik T.M., et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawai T., Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 2009;21:317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brennan K., Bowie A.G. Activation of host pattern recognition receptors by viruses. Curr. Opin. Microbiol. 2010;13:503–507. doi: 10.1016/j.mib.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 40.Thompson M.R., Kaminski J.J., Kurt-Jones E.A., Fitzgerald K.A. Pattern recognition receptors and the innate immune response to viral infection. Viruses. 2011;3:920–940. doi: 10.3390/v3060920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin M.S., Kim S.E., Heo J.Y., Lee M.E., Kim H.M., Paik S.G., Lee H., Lee J.O. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 42.Farhat K., Riekenberg S., Heine H., Debarry J., Lang R., Mages J., Buwitt-Beckmann U., Röschmann K., Jung G., Wiesmüller K.H., Ulmer A.J. Heterodimerization of TLR2 with TLR1 or TLR6 expands the ligand spectrum but does not lead to differential signaling. J. Leukoc. Biol. 2008;83:692–701. doi: 10.1189/jlb.0807586. [DOI] [PubMed] [Google Scholar]
- 43.Ariza M.E., Glaser R., Kaumaya P.T., Jones C., Williams M.V. The EBV-encoded dUTPase activates NF-kappa B through the TLR2 and MyD88-dependent signaling pathway. J. Immunol. 2009;182:851–859. doi: 10.4049/jimmunol.182.2.851. [DOI] [PubMed] [Google Scholar]
- 44.Cai M., Li M., Wang K., Wang S., Lu Q., Yan J., Mossman K.L., Lin R., Zheng C. The herpes simplex virus 1-encoded envelope glycoprotein B activates NF-κB through the Toll-like receptor 2 and MyD88/TRAF6-dependent signaling pathway. PLoS One. 2013;8 doi: 10.1371/journal.pone.0054586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cooper A., Tal G., Lider O., Shaul Y. Cytokine induction by the hepatitis B virus capsid in macrophages is facilitated by membrane heparan sulfate and involves TLR2. J. Immunol. 2005;175:3165–3176. doi: 10.4049/jimmunol.175.5.3165. [DOI] [PubMed] [Google Scholar]
- 46.Proud P.C., Tsitoura D., Watson R.J., Chua B.Y., Aram M.J., Bewley K.R., Cavell B.E., Cobb R., Dowall S., Fotheringham S.A., et al. Prophylactic intranasal administration of a TLR2/6 agonist reduces upper respiratory tract viral shedding in a SARS-CoV-2 challenge ferret model. EBioMedicine. 2021;63:103153. doi: 10.1016/j.ebiom.2020.103153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng M., Karki R., Williams E.P., Yang D., Fitzpatrick E., Vogel P., Jonsson C.B., Kanneganti T.D. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 2021;22:829–838. doi: 10.1038/s41590-021-00937-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perales-Linares R., Navas-Martin S. Toll-like receptor 3 in viral pathogenesis: friend or foe? Immunology. 2013;140:153–167. doi: 10.1111/imm.12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Totura A.L., Whitmore A., Agnihothram S., Schäfer A., Katze M.G., Heise M.T., Baric R.S. Toll-like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus infection. mBio. 2015;6 doi: 10.1128/mBio.00638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lévy R., Bastard P., Lanternier F., Lecuit M., Zhang S.Y., Casanova J.L. IFN-α2a therapy in two patients with inborn errors of TLR3 and IRF3 infected with SARS-CoV-2. J. Clin. Immunol. 2021;41:26–27. doi: 10.1007/s10875-020-00933-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu Y.C., Yeh W.C., Ohashi P.S. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145–151. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 53.Tsung A., Sahai R., Tanaka H., Nakao A., Fink M.P., Lotze M.T., Yang H., Li J., Tracey K.J., Geller D.A., Billiar T.R. The nuclear factor HMGB1 mediates hepatic injury after murine liver ischemia-reperfusion. J. Exp. Med. 2005;201:1135–1143. doi: 10.1084/jem.20042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vabulas R.M., Ahmad-Nejad P., Ghose S., Kirschning C.J., Issels R.D., Wagner H. HSP70 as endogenous stimulus of the toll/interleukin-1 receptor signal pathway. J. Biol. Chem. 2002;277:15107–15112. doi: 10.1074/jbc.M111204200. [DOI] [PubMed] [Google Scholar]
- 55.Kurt-Jones E.A., Popova L., Kwinn L., Haynes L.M., Jones L.P., Tripp R.A., Walsh E.E., Freeman M.W., Golenbock D.T., Anderson L.J., Finberg R.W. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 2000;1:398–401. doi: 10.1038/80833. [DOI] [PubMed] [Google Scholar]
- 56.Lai C.Y., Strange D.P., Wong T.A.S., Lehrer A.T., Verma S. Ebola virus glycoprotein induces an innate immune response in vivo via TLR4. Front. Microbiol. 2017;8:1571. doi: 10.3389/fmicb.2017.01571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Georgel P., Jiang Z., Kunz S., Janssen E., Mols J., Hoebe K., Bahram S., Oldstone M.B., Beutler B. Vesicular stomatitis virus glycoprotein G activates a specific antiviral Toll-like receptor 4-dependent pathway. Virology. 2007;362:304–313. doi: 10.1016/j.virol.2006.12.032. [DOI] [PubMed] [Google Scholar]
- 58.Chao C.H., Wu W.C., Lai Y.C., Tsai P.J., Perng G.C., Lin Y.S., Yeh T.M. Dengue virus nonstructural protein 1 activates platelets via Toll-like receptor 4, leading to thrombocytopenia and hemorrhage. Plos Pathog. 2019;15:e1007625. doi: 10.1371/journal.ppat.1007625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sohn K.M., Lee S.G., Kim H.J., Cheon S., Jeong H., Lee J., Kim I.S., Silwal P., Kim Y.J., Paik S., et al. COVID-19 patients upregulate toll-like receptor 4-mediated inflammatory signaling that mimics bacterial sepsis. J. Korean Med. Sci. 2020;35:e343. doi: 10.3346/jkms.2020.35.e343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choudhury A., Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J. Med. Virol. 2020;92:2105–2113. doi: 10.1002/jmv.25987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao Y., Kuang M., Li J., Zhu L., Jia Z., Guo X., Hu Y., Kong J., Yin H., Wang X., You F. SARS-CoV-2 spike protein interacts with and activates TLR41. Cell Res. 2021;31:818–820. doi: 10.1038/s41422-021-00495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garlanda C., Dinarello C.A., Mantovani A. The interleukin-1 family: back to the future. Immunity. 2013;39:1003–1018. doi: 10.1016/j.immuni.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao N., Di B., Xu L.L. The NLRP3 inflammasome and COVID-19: activation, pathogenesis and therapeutic strategies. Cytokine Growth Factor. Rev. 2021;61:2–15. doi: 10.1016/j.cytogfr.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodrigues T.S., de Sá K.S.G., Ishimoto A.Y., Becerra A., Oliveira S., Almeida L., Gonçalves A.V., Perucello D.B., Andrade W.A., Castro R., et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J. Exp. Med. 2021;218:e20201707. doi: 10.1084/jem.20201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pan P., Shen M., Yu Z., Ge W., Chen K., Tian M., Xiao F., Wang Z., Wang J., Jia Y., et al. SARS-CoV-2 N protein promotes NLRP3 inflammasome activation to induce hyperinflammation. Nat. Commun. 2021;12:4664. doi: 10.1038/s41467-021-25015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu H., Chitre S.A., Akinyemi I.A., Loeb J.C., Lednicky J.A., McIntosh M.T., Bhaduri-McIntosh S. SARS-CoV-2 viroporin triggers the NLRP3 inflammatory pathway. bioRxiv. 2020 doi: 10.1101/2020.10.27.357731. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van der Made C.I., Simons A., Schuurs-Hoeijmakers J., van den Heuvel G., Mantere T., Kersten S., van Deuren R.C., Steehouwer M., van Reijmersdal S.V., Jaeger M., et al. Presence of genetic variants among young men with severe COVID-19. JAMA. 2020;324:663–673. doi: 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Honda K., Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 69.Loo Y.M., Gale M. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Y., Ludwig J., Schuberth C., Goldeck M., Schlee M., Li H., Juranek S., Sheng G., Micura R., Tuschl T., et al. Structural and functional insights into 5'-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat. Struct. Mol. Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu C., Ranjith-Kumar C.T., Hao L., Kao C.C., Li P. Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5' triphosphate. Nucleic Acids Res. 2011;39:1565–1575. doi: 10.1093/nar/gkq974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J., et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 73.Loske J., Rohmel J., Lukassen S., Stricker S., Magalhaes V.G., Liebig J., et al. Pre-activated antiviral innate immunity in the upper airways controls early SARS-CoV-2 infection in children. Nat. Biotechnol. 2021 doi: 10.1038/s41587-021-01037-9. [DOI] [PubMed] [Google Scholar]
- 74.Thorne L.G., Reuschl A.K., Zuliani-Alvarez L., Whelan M.V.X., Turner J., Noursadeghi M., Jolly C., Towers G.J. SARS-CoV-2 sensing by RIG-I and MDA5 links epithelial infection to macrophage inflammation. Embo J. 2021;40 doi: 10.15252/embj.2021107826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kouwaki T., Nishimura T., Wang G., Oshiumi H. RIG-I-Like receptor-mediated recognition of viral genomic RNA of severe acute respiratory syndrome coronavirus-2 and viral escape from the host innate immune responses. Front. Immunol. 2021;12:700926. doi: 10.3389/fimmu.2021.700926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang D.M., Geng T.T., Harrison A.G., Wang P.H. Differential roles of RIG-I like receptors in SARS-CoV-2 infection. Mil. Med. Res. 2021;8:49. doi: 10.1186/s40779-021-00340-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yin X., Riva L., Pu Y., Martin-Sancho L., Kanamune J., Yamamoto Y., Sakai K., Gotoh S., Miorin L., De Jesus P.D., et al. MDA5 governs the innate immune response to SARS-CoV-2 in lung epithelial cells. Cell Rep. 2021;34:108628. doi: 10.1016/j.celrep.2020.108628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rebendenne A., Valadao A.L.C., Tauziet M., Maarifi G., Bonaventure B., McKellar J., Planès R., Nisole S., Arnaud-Arnould M., Moncorgé O., et al. SARS-CoV-2 triggers an MDA-5-dependent interferon response which is unable to control replication in lung epithelial cells. J. Virol. 2021;95 doi: 10.1128/JVI.02415-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yamada T., Sato S., Sotoyama Y., Orba Y., Sawa H., Yamauchi H., Sasaki M., Takaoka A. RIG-I triggers a signaling-abortive anti-SARS-CoV-2 defense in human lung cells. Nat. Immunol. 2021;22:820–828. doi: 10.1038/s41590-021-00942-0. [DOI] [PubMed] [Google Scholar]
- 80.Geijtenbeek T.B., Gringhuis S.I. Signalling through C-type lectin receptors: shaping immune responses. Nat. Rev. Immunol. 2009;9:465–479. doi: 10.1038/nri2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moris A., Nobile C., Buseyne F., Porrot F., Abastado J.P., Schwartz O. DC-SIGN promotes exogenous MHC-I-restricted HIV-1 antigen presentation. Blood. 2004;103:2648–2654. doi: 10.1182/blood-2003-07-2532. [DOI] [PubMed] [Google Scholar]
- 82.Thépaut M., Luczkowiak J., Vivès C., Labiod N., Bally I., Lasala F., Grimoire Y., Fenel D., Sattin S., Thielens N., et al. DC/L-SIGN recognition of spike glycoprotein promotes SARS-CoV-2 trans-infection and can be inhibited by a glycomimetic antagonist. PLoS Pathog. 2021;17:e1009576. doi: 10.1371/journal.ppat.1009576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Alvarez C.P., Lasala F., Carrillo J., Muñiz O., Corbí A.L., Delgado R. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J. Virol. 2002;76:6841–6844. doi: 10.1128/JVI.76.13.6841-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Carbaugh D.L., Baric R.S., Lazear H.M. Envelope protein glycosylation mediates Zika virus pathogenesis. J. Virol. 2019;93 doi: 10.1128/JVI.00113-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Navarro-Sanchez E., Altmeyer R., Amara A., Schwartz O., Fieschi F., Virelizier J.L., Arenzana-Seisdedos F., Desprès P. Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. Embo Rep. 2003;4:723–728. doi: 10.1038/sj.embor.embor866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gao C., Zeng J., Jia N., Stavenhagen K., Matsumoto Y., Zhang H., Li J., Hume A.J., Mühlberger E., van Die I., et al. SARS-CoV-2 spike protein interacts with multiple innate immune receptors. bioRxiv. 2020 doi: 10.1101/2020.07.29.227462. Preprint at. [DOI] [Google Scholar]
- 87.Lu Q., Liu J., Zhao S., Gomez Castro M.F., Laurent-Rolle M., Dong J., Ran X., Damani-Yokota P., Tang H., Karakousi T., et al. SARS-CoV-2 exacerbates proinflammatory responses in myeloid cells through C-type lectin receptors and Tweety family member 2. Immunity. 2021;54:1304–e9. doi: 10.1016/j.immuni.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Amraei R., Yin W.Q., Napoleon M.A., Suder E.L., Berrigan J., Zhao Q., Olejnik J., Chandler K.B., Xia C., Feldman J., et al. CD209L/L-SIGN and CD209/DC-SIGN act as receptors for SARS-CoV-2. Acs Cent. Sci. 2021;7:1156–1165. doi: 10.1021/acscentsci.0c01537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Levy D.E., Marié I.J., Durbin J.E. Induction and function of type I and III interferon in response to viral infection. Curr. Opin. Virol. 2011;1:476–486. doi: 10.1016/j.coviro.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yan H., Krishnan K., Lim J.T., Contillo L.G., Krolewski J.J. Molecular characterization of an alpha interferon receptor 1 subunit (IFNaR1) domain required for TYK2 binding and signal transduction. Mol. Cell Biol. 1996;16:2074–2082. doi: 10.1128/mcb.16.5.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Barbieri G., Velazquez L., Scrobogna M., Fellous M., Pellegrini S. Activation of the protein tyrosine kinase tyk2 by interferon alpha/beta. Eur. J. Biochem. 1994;223:427–435. doi: 10.1111/j.1432-1033.1994.tb19010.x. [DOI] [PubMed] [Google Scholar]
- 92.Schoggins J.W., Rice C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu S.Y., Sanchez D.J., Aliyari R., Lu S., Cheng G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc. Natl. Acad. Sci. U S A. 2012;109:4239–4244. doi: 10.1073/pnas.1114981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li C., Deng Y.Q., Wang S., Ma F., Aliyari R., Huang X.Y., Zhang N.N., Watanabe M., Dong H.L., Liu P., et al. 25-Hydroxycholesterol protects host against Zika virus infection and its associated microcephaly in a mouse model. Immunity. 2017;46:446–456. doi: 10.1016/j.immuni.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu S.Y., Aliyari R., Chikere K., Li G., Marsden M.D., Smith J.K., Pernet O., Guo H., Nusbaum R., Zack J.A., et al. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity. 2013;38:92–105. doi: 10.1016/j.immuni.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zang R., Case J.B., Yutuc E., Ma X., Shen S., Gomez Castro M.F., Liu Z., Zeng Q., Zhao H., Son J., et al. Cholesterol 25-hydroxylase suppresses SARS-CoV-2 replication by blocking membrane fusion. Proc. Natl. Acad. Sci. U S A. 2020;117:32105–32113. doi: 10.1073/pnas.2012197117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang S., Li W., Hui H., Tiwari S.K., Zhang Q., Croker B.A., Rawlings S., Smith D., Carlin A.F., Rana T.M. Cholesterol 25-Hydroxylase inhibits SARS-CoV-2 and other coronaviruses by depleting membrane cholesterol. Embo J. 2020;39:e106057. doi: 10.15252/embj.2020106057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zu S., Deng Y.Q., Zhou C., Li J., Li L., Chen Q., Li X.F., Zhao H., Gold S., He J., et al. 25-Hydroxycholesterol is a potent SARS-CoV-2 inhibitor. Cell Res. 2020;30:1043–1045. doi: 10.1038/s41422-020-00398-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pfaender S., Mar K.B., Michailidis E., Kratzel A., Boys I.N., V'kovski P., Fan W., Kelly J.N., Hirt D., Ebert N., et al. LY6E impairs coronavirus fusion and confers immune control of viral disease. Nat. Microbiol. 2020;5:1330–1339. doi: 10.1038/s41564-020-0769-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Martin-Sancho L., Lewinski M.K., Pache L., Stoneham C.A., Yin X., Becker M.E., Pratt D., Churas C., Rosenthal S.B., Liu S., et al. Functional landscape of SARS-CoV-2 cellular restriction. Mol. Cell. 2021;81:2656–2668.e8. doi: 10.1016/j.molcel.2021.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bonaventure Boris R.A., Garcia de Gracia F., Tauziet M., McKellar J., chaves Valadão A.L., Courgnaud V., et al. A genome-wide CRISPR/Cas9 knock-out screen identifies the DEAD box RNA helicase DDX42 as a broad antiviral inhibitor. bioRxiv. 2020 doi: 10.1101/2020.10.28.359356. Preprint at. [DOI] [Google Scholar]
- 102.Alice Mac Kain G.M., Sophie-Marie A., Arhel N., Baidaliuk A., Vallet T., Tran Q.D., et al. Identification of DAXX as A restriction factor of SARS-CoV-2 through A CRISPR/cas9 screen. bioRxiv. 2021 doi: 10.1101/2021.05.06.442916. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wilamowski M., Sherrell D.A., Minasov G., Kim Y., Shuvalova L., Lavens A., Chard R., Maltseva N., Jedrzejczak R., Laven A., et al. 2 '-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography. P Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2100170118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Viswanathan T., Arya S., Chan S.H., Qi S., Dai N., Misra A., Park J.G., Oladunni F., Kovalskyy D., Hromas R.A., et al. Structural basis of RNA cap modification by SARS-CoV-2. Nat. Commun. 2020;11:3718. doi: 10.1038/s41467-020-17496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ogando N.S., Zevenhoven-Dobbe J.C., van der Meer Y., Bredenbeek P.J., Posthuma C.C., Snijder E.J. The enzymatic activity of the nsp14 exoribonuclease is critical for replication of MERS-CoV and SARS-CoV-2. J. Virol. 2020;94 doi: 10.1128/JVI.01246-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Deng X., Hackbart M., Mettelman R.C., O'Brien A., Mielech A.M., Yi G., Kao C.C., Baker S.C. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. U S A. 2017;114:E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang S., Dai T., Qin Z., Pan T., Chu F., Lou L., Zhang L., Yang B., Huang H., Lu H., Zhou F. Targeting liquid-liquid phase separation of SARS-CoV-2 nucleocapsid protein promotes innate antiviral immunity by elevating MAVS activity. Nat. Cell Biol. 2021;23:718–732. doi: 10.1038/s41556-021-00710-0. [DOI] [PubMed] [Google Scholar]
- 109.Zhao Y., Sui L., Wu P., Wang W., Wang Z., Yu Y., Hou Z., Tan G., Liu Q., Wang G. A dual-role of SARS-CoV-2 nucleocapsid protein in regulating innate immune response. Signal. Transduct Target. Ther. 2021;6:331. doi: 10.1038/s41392-021-00742-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mu J., Fang Y., Yang Q., Shu T., Wang A., Huang M., Jin L., Deng F., Qiu Y., Zhou X. SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2. Cell Discov. 2020;6:65. doi: 10.1038/s41421-020-00208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fu Y.Z., Wang S.Y., Zheng Z.Q., Yi Huang Y., Li W.W., Xu Z.S., Wang Y.Y. SARS-CoV-2 membrane glycoprotein M antagonizes the MAVS-mediated innate antiviral response. Cell Mol. Immunol. 2021;18:613–620. doi: 10.1038/s41423-020-00571-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zheng Y., Zhuang M.W., Han L.L., Zhang J., Nan M.L., Zhan P., Kang D., Liu X., Gao C., Wang P.H. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal. Transduct. Target. Ther. 2020;5:299. doi: 10.1038/s41392-020-00438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jiang H.W., Zhang H.N., Meng Q.F., Xie J., Li Y., Chen H., Zheng Y.X., Wang X.N., Qi H., Zhang J., et al. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell Mol. Immunol. 2020;17:998–1000. doi: 10.1038/s41423-020-0514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Konno Y., Kimura I., Uriu K., Fukushi M., Irie T., Koyanagi Y., Sauter D., Gifford R.J., USFQ-COVID19 Consortium. Nakagawa S., Sato K. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep. 2020;32:108185. doi: 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Guo G.J., Gao M., Gao X.C., Zhu B.B., Huang J.Z., Luo K.T., Zhang Y., Sun J., Deng M., Lou Z. SARS-CoV-2 non-structural protein 13 (nsp13) hijacks host deubiquitinase USP13 and counteracts host antiviral immune response. Signal. Transduct. Target. Ther. 2021;6 doi: 10.1038/s41392-021-00509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L., Ren L., et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang R., Yang X.F., Chang M.K., Xue Z.Y., Wang W.R., Bai L., Zhao S., Liu E. ORF3a protein of severe acute respiratory syndrome coronavirus 2 inhibits interferon-activated janus kinase/signal transducer and activator of transcription signaling via elevating suppressor of cytokine signaling 1. Front. Microbiol. 2021;12:752597. doi: 10.3389/fmicb.2021.752597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xia H., Cao Z., Xie X., Zhang X., Chen J.Y., Wang H., Menachery V.D., Rajsbaum R., Shi P.Y. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33:108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Miorin L., Kehrer T., Sanchez-Aparicio M.T., Zhang K., Cohen P., Patel R.S., Cupic A., Makio T., Mei M., Moreno E., et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. U S A. 2020;117:28344–28354. doi: 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sui L.Y., Zhao Y.H., Wang W.F., Wu P., Wang Z.D., Yu Y., Hou Z., Tan G., Liu Q. SARS-CoV-2 membrane protein inhibits type I interferon production through ubiquitin-mediated degradation of TBK1. Front. Immunol. 2021;12:662989. doi: 10.3389/fimmu.2021.662989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., Schulz L., Widera M., Mehdipour A.R., Tascher G., et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu G., Lee J.H., Parker Z.M., Acharya D., Chiang J.J., van Gent M., Riedl W., Davis-Gardner M.E., Wies E., Chiang C., Gack M.U. ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nat. Microbiol. 2021;6:467–478. doi: 10.1038/s41564-021-00884-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wu J., Shi Y.H., Pan X.Y., Wu S., Hou R.X., Zhang Y., Zhong T., Tang H., Wang L., Wo J., et al. SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Rep. 2021;34:108761. doi: 10.1016/j.celrep.2021.108761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Moustaqil M., Ollivier E., Chiu H.P., Van Tol S., Rudolffi-Soto P., Stevens C., Bhumkar A., Hunter D.J.B., Freiberg A.N., Jacques D., et al. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species. Emerg. Microbes Infec. 2021;10:178–195. doi: 10.1080/22221751.2020.1870414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Josephine Lampe J.W., Müller-Fielitz H., Müller K., Schuster R., Zhang L., Zille M., Krohn M., Özorhan Ü., Jiang Y., Neve V. The SARS-CoV-2 main protease Mpro causes microvascular brain pathology by cleaving NEMO in brain endothelial cells. Res. Square. 2020;24:1522–1533. doi: 10.1038/s41593-021-00926-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang K., Miorin L., Makio T., Dehghan I., Gao S., Xie Y., Zhong H., Esparza M., Kehrer T., Kumar A., et al. Nsp1 protein of SARS-CoV-2 disrupts the mRNA export machinery to inhibit host gene expression. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abe7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hsu J.C.C., Laurent-Rolle M., Pawlak J.B., Wilen C.B., Cresswell P. Translational shutdown and evasion of the innate immune response by SARS-CoV-2 NSP14 protein. P Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2101161118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Herbert K.M., Nag A. A tale of two RNAs during viral infection: how viruses antagonize mRNAs and small non-coding RNAs in the host cell. Viruses. 2016;8:154. doi: 10.3390/v8060154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zeng C., Wu A., Wang Y., Xu S., Tang Y., Jin X., Wang S., Qin L., Sun Y., Fan C., et al. Identification and characterization of a ribose 2'-O-methyltransferase encoded by the Ronivirus branch of Nidovirales. J. Virol. 2016;90:6675–6685. doi: 10.1128/JVI.00658-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rehwinkel J., Gack M.U. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020;20:537–551. doi: 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang W., Zhou Z., Xiao X., Tian Z., Dong X., Wang C., Li L., Ren L., Lei X., Xiang Z., Wang J. SARS-CoV-2 nsp12 attenuates type I interferon production by inhibiting IRF3 nuclear translocation. Cell Mol. Immunol. 2021;18:945–953. doi: 10.1038/s41423-020-00619-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li A., Zhao K., Zhang B., Hua R., Fang Y., Jiang W., Zhang J., Hui L., Zheng Y., Zhu C., et al. SARS-CoV-2 NSP12 protein is not an interferon-beta antagonist. J. Virol. 2021;95 doi: 10.1128/JVI.00747-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Geng H., Subramanian S., Wu L., Bu H.F., Wang X., Du C., De Plaen I.G., Tan X.D. SARS-CoV-2 ORF8 forms intracellular aggregates and inhibits IFNγ-induced antiviral gene expression in human lung epithelial cells. Front. Immunol. 2021;12:679482. doi: 10.3389/fimmu.2021.679482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hu H., Sun S.C. Ubiquitin signaling in immune responses. Cell Res. 2016;26:457–483. doi: 10.1038/cr.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sun S.C. Deubiquitylation and regulation of the immune response. Nat. Rev. Immunol. 2008;8:501–511. doi: 10.1038/nri2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Minakshi R., Padhan K., Rani M., Khan N., Ahmad F., Jameel S. The SARS coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS One. 2009;4 doi: 10.1371/journal.pone.0008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hayn M., Hirschenberger M., Koepke L., Nchioua R., Straub J.H., Klute S., Hunszinger V., Zech F., Prelli Bozzo C., Aftab W., et al. Systematic functional analysis of SARS-CoV-2 proteins uncovers viral innate immune antagonists and remaining vulnerabilities. Cell Rep. 2021;35:109126. doi: 10.1016/j.celrep.2021.109126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lei J., Hilgenfeld R. RNA-virus proteases counteracting host innate immunity. Febs Lett. 2017;591:3190–3210. doi: 10.1002/1873-3468.12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhu X., Wang D., Zhou J., Pan T., Chen J., Yang Y., et al. Porcine deltacoronavirus nsp5 antagonizes type I interferon signaling by cleaving STAT2. J. Virol. 2017;91 doi: 10.1128/JVI.00003-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang D., Fang L., Shi Y., Zhang H., Gao L., Peng G., Chen H., Li K., Xiao S. Porcine epidemic diarrhea virus 3C-like protease regulates its interferon antagonism by cleaving NEMO. J. Virol. 2016;90:2090–2101. doi: 10.1128/JVI.02514-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fonseca G.J., Thillainadesan G., Yousef A.F., Ablack J.N., Mossman K.L., Torchia J., Mymryk J.S. Adenovirus evasion of interferon-mediated innate immunity by direct antagonism of a cellular histone posttranslational modification. Cell Host Microbe. 2012;11:597–606. doi: 10.1016/j.chom.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 143.Zhang S., Wang J., Cheng G. Protease cleavage of RNF20 facilitates coronavirus replication via stabilization of SREBP1. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2107108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Prescott L. SARS-CoV-2 3CLpro whole human proteome cleavage prediction and enrichment/depletion analysis. bioRxiv. 2020 doi: 10.1101/2020.08.24.265645. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gaglia M.M., Covarrubias S., Wong W., Glaunsinger B.A. A common strategy for host RNA degradation by divergent viruses. J. Virol. 2012;86:9527–9530. doi: 10.1128/JVI.01230-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., Rice C.M. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Finkel Y., Gluck A., Nachshon A., Winkler R., Fisher T., Rozman B., Mizrahi O., Lubelsky Y., Zuckerman B., Slobodin B., et al. SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis. Nature. 2021;594:240–245. doi: 10.1038/s41586-021-03610-3. [DOI] [PubMed] [Google Scholar]
- 148.Lo M.W., Woodruff T.M. Complement: bridging the innate and adaptive immune systems in sterile inflammation. J. Leukoc. Biol. 2020;108:339–351. doi: 10.1002/JLB.3MIR0220-270R. [DOI] [PubMed] [Google Scholar]
- 149.Walport M.J. Complement. First of two parts. N. Engl. J. Med. 2001;344:1058–1066. doi: 10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- 150.Noris M., Remuzzi G. Overview of complement activation and regulation. Semin. Nephrol. 2013;33:479–492. doi: 10.1016/j.semnephrol.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ali Y.M., Ferrari M., Lynch N.J., Yaseen S., Dudler T., Gragerov S., Demopulos G., Heeney J.L., Schwaeble W.J. Lectin pathway mediates complement activation by SARS-CoV-2 proteins. Front. Immunol. 2021;12:714511. doi: 10.3389/fimmu.2021.714511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yu J., Yuan X., Chen H., Chaturvedi S., Braunstein E.M., Brodsky R.A. Direct activation of the alternative complement pathway by SARS-CoV-2 spike proteins is blocked by factor D inhibition. Blood. 2020;136:2080–2089. doi: 10.1182/blood.2020008248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ting Gao M.H., Zhang X., Li H., Zhu L., Liu H., Dong Q., Zhang Z., Wang Z., Hu Y., Fu Y., et al. Highly pathogenic coronavirus N protein aggravates lung injury by MASP-2-mediated complement over-activation. medRxiv. 2020 doi: 10.1101/2020.03.29.20041962. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Holter J.C., Pischke S.E., de Boer E., Lind A., Jenum S., Holten A.R., Tonby K., Barratt-Due A., Sokolova M., Schjalm C., et al. Systemic complement activation is associated with respiratory failure in COVID-19 hospitalized patients. Proc. Natl. Acad. Sci. U S A. 2020;117:25018–25025. doi: 10.1073/pnas.2010540117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Liu Y., Yang Y., Zhang C., Huang F., Wang F., Yuan J., Wang Z., Li J., Li J., Feng C., et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci. China Life Sci. 2020;63:364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kanzawa N., Nishigaki K., Hayashi T., Ishii Y., Furukawa S., Niiro A., Yasui F., Kohara M., Morita K., Matsushima K., et al. Augmentation of chemokine production by severe acute respiratory syndrome coronavirus 3a/X1 and 7a/X4 proteins through NF-kappaB activation. Febs Lett. 2006;580:6807–6812. doi: 10.1016/j.febslet.2006.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Su C.M., Wang L., Yoo D. Activation of NF-κB and induction of proinflammatory cytokine expressions mediated by ORF7a protein of SARS-CoV-2. Sci. Rep. 2021;11:13464. doi: 10.1038/s41598-021-92941-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Li J.Y., Liao C.H., Wang Q., Tan Y.J., Luo R., Qiu Y., Ge X.Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286:198074. doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lin X., Fu B., Yin S., Li Z., Liu H., Zhang H., Xing N., Wang Y., Xue W., Xiong Y., et al. ORF8 contributes to cytokine storm during SARS-CoV-2 infection by activating IL-17 pathway. iScience. 2021;24:102293. doi: 10.1016/j.isci.2021.102293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Li T., Kenney A.D., Liu H., Fiches G.N., Zhou D., Biswas A., Que J., Santosa N., Yount J.S., Zhu J. SARS-CoV-2 Nsp14 activates NF-kappaB signaling and induces IL-8 upregulation. bioRxiv. 2021 doi: 10.1101/2021.05.26.445787. Preprint at. [DOI] [Google Scholar]
- 164.Amarilla A.A., Sng J.D.J., Parry R., Deerain J.M., Potter J.R., Setoh Y.X., Rawle D.J., Le T.T., Modhiran N., Wang X., et al. A versatile reverse genetics platform for SARS-CoV-2 and other positive-strand RNA viruses. Nat. Commun. 2021;12:3431. doi: 10.1038/s41467-021-23779-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Xie X., Lokugamage K.G., Zhang X., Vu M.N., Muruato A.E., Menachery V.D., Shi P.Y. Engineering SARS-CoV-2 using a reverse genetic system. Nat. Protoc. 2021;16:1761–1784. doi: 10.1038/s41596-021-00491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.De Maio N., Walker C.R., Turakhia Y., Lanfear R., Corbett-Detig R., Goldman N. Mutation rates and selection on synonymous mutations in SARS-CoV-2. Genome Biol. Evol. 2021;13 doi: 10.1093/gbe/evab087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Van Egeren D., Novokhodko A., Stoddard M., Tran U., Zetter B., Rogers M., Pentelute B.L., Carlson J.M., Hixon M., Joseph-McCarthy D., Chakravarty A. Risk of rapid evolutionary escape from biomedical interventions targeting SARS-CoV-2 spike protein. Plos One. 2021;16:e0250780. doi: 10.1371/journal.pone.0250780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Klemm T., Ebert G., Calleja D.J., Allison C.C., Richardson L.W., Bernardini J.P., Lu B.G., Kuchel N.W., Grohmann C., Shibata Y., et al. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. Embo J. 2020;39:e106275. doi: 10.15252/embj.2020106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Smith E., Davis-Gardner M.E., Garcia-Ordonez R.D., Nguyen T.T., Hull M., Chen E., Baillargeon P., Scampavia L., Strutzenberg T., Griffin P.R., et al. High-throughput screening for drugs that inhibit papain-like protease in SARS-CoV-2. Slas Discov. 2020;25:1152–1161. doi: 10.1177/2472555220963667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Drayman N., J K.A., Azizi S.-A., Froggatt H.M., Tan K., Maltseva N.I., Chen S., Vlad N., Steve D. Drug repurposing screen identifies masitinib as a 3CLpro inhibitor that blocks replication of SARS-CoV-2 in vitro. bioRxiv. 2020 doi: 10.1101/2020.08.31.274639. Preprint at. [DOI] [Google Scholar]
- 171.Mody V., Ho J., Wills S., Mawri A., Lawson L., Ebert M.C.C.J.C., Fortin G.M., Rayalam S., Taval S. Identification of 3-chymotrypsin like protease (3CLPro) inhibitors as potential anti-SARS-CoV-2 agents. Commun. Biol. 2021;4:93. doi: 10.1038/s42003-020-01577-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pettersson M., Crews C.M. PROteolysis TArgeting Chimeras (PROTACs) - past, present and future. Drug Discov. Today Technol. 2019;31:15–27. doi: 10.1016/j.ddtec.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]