Summary
Germinal center T follicular helper (GCTfh) cells are defined by a Bcl6+CXCR5hiPD-1hi phenotype, but only a minor fraction of these reside in GCs. Here we examined whether GC-resident and -nonresident Tfh cells share a common physiology and function. Fluorescently-labeled, GC-resident Tfh cells in different mouse models were distinguished by low expression of CD90. CD90neg/lo GCTfh cells required antigen-specific, MHCII+ B cells to develop, and stopped proliferating soon after differentiation. In contrast, non-resident, CD90hi Tfh (GCTfh-like) cells developed normally in the absence of MHCII+ B cells and proliferated continuously during primary responses. The TCR repertoires of both Tfh subsets overlapped initially but later diverged in association with dendritic cell-dependent proliferation of CD90hi GCTfh-like cells, suggestive of TCR-dependency seen also during TCR-transgenic adoptive transfer experiments. Further, the transcriptomes of CD90neg/lo and CD90hi GCTfh-like cells were enriched in different functional pathways. Thus, GC-resident and non-resident Tfh cells have distinct developmental requirements and activities, implying distinct functions.
Keywords: Germinal centers, Tfh cells, humoral immunity
Graphical Abstract
eTOC blurb
T follicular helper (Tfh) cells within the germinal center (GC) arbitrate antibody affinity maturation. Yeh et al. utilize various models to distinguish GC resident Tfh cells, showing that previous phenotypic definition of GC Tfh cells include a large subset that does not enter GCs. These CD90hi Tfh cells have different developmental requirements and activities than the rarer GC-resident Tfh cells (CD90neg/lo), implying distinct functions.
Introduction
T follicular helper cells represent a differentiation lineage distinct from other CD4+ helper T cell types, with Bcl6 serving as the lineage-defining transcription factor (Johnston et al., 2009; Nurieva et al., 2009; Yu et al., 2009). The Tfh fate decision appears to be made prior to Bcl6 expression and may be determined by the initial strength of TCR signaling when encountering antigen-presenting dendritic cells (DCs) (Choi et al., 2013; Tubo et al., 2013). Whereas non-Tfh effector cells (e.g., Th1, Th2, Th17) predominantly emigrate to distal sites, Tfh cells largely remain in situ and play a crucial role in T-dependent B-cell responses (Crotty, 2014). Early Tfh cells express the chemokine receptor CXCR5, enabling them to migrate to the B-cell follicle border. There, they interact with activated B cells, undergo further maturation, and subsequently penetrate the follicle, where a subset of Tfh cells help initiate GCs (Ansel et al., 1999; Breitfeld et al., 2000; Haynes et al., 2007; Schaerli et al., 2000). In organized GCs, Tfh cells residing within the GC light zone (LZ) are necessary for the maintenance of the GC reaction. These GCTfh cells are thought to arbitrate affinity-driven competition among GC B cells and to influence GCB cell differentiation into plasma cells or memory B cells in a manner that reflects the quality of cognate interaction between GCTfh and GCB cells (Crotty, 2014; Wan et al., 2019). Whereas all Tfh cells share a defining Bcl6+CXCR5+ signature, not every Tfh cell enters or remains in GCs (Crotty, 2019; Shulman et al., 2013). Tfh cells that do not enter GCs are unlikely to provide direct selection signals to GCB cells; instead, these Tfh may have effector activities outside GCs, and may be a source of memory Tfh cells (Choi et al., 2013; Suan et al., 2015).
A reliable cellular marker to distinguish GC-resident from -nonresident Tfh cells would facilitate efforts to determine whether GC-resident and -nonresident Tfh share a common physiology and function. Higher expression of surface CXCR5 and PD-1 on GCTfh cells generally has been used to discriminate them from other Tfh populations (Crotty, 2011; Tubo et al., 2013; Yusuf et al., 2010). However, this distinction can be difficult, because efficient resolution of CXCR5hi cells requires special adjustments to standard staining methods (Meli and King, 2015; Pepper et al., 2011; Tubo et al., 2013; Yusuf et al., 2010). Moreover, even the strictest definition of GCTfh cells (CD4+FoxP3−Bcl6+CXCR5hiPD-1hi) is too broad, because such cells constitute ≥20% of GC lymphocytes in mice (Cho et al., 2018; Nance et al., 2015; Wu et al., 2015; Yu et al., 2009), in humans (Dan et al., 2019), and in non-human primates (Cirelli et al., 2019; Havenar-Daughton et al., 2019; Havenar-Daughton et al., 2016), whereas more accurate direct histologic analyses reveal that T cells account for only ~10% of GC cellularity (Kelsoe, 1996; Wittenbrink et al., 2011; Wollenberg et al., 2011). Thus, investigating the functions of GCTfh cells and GC-extrinsic Tfh cells would benefit from more precise identification of these subsets.
In the present study, we provide a precise definition for GC-resident Tfh cells in primary responses: CXCR5hiPD-1hiCD90neg/lo. These cells were S1pr2+, required antigen-specific MHCII+ B cells for development, and ceased proliferating after differentiation. In contrast, non-resident GCTfh-like cells were CXCR5hiPD-1hiCD90hiS1pr2−, arose in the absence of antigen presentation by B cells, and continued proliferating throughout the primary response. Strikingly, these two Tfh populations acquired distinct TCRβ repertoires and gene expression profiles, implying their functional heterogeneity and underscoring the importance of dissecting the roles of these discrete cell types in humoral responses.
Results
The CXCR5hiPD-1hi GCTfh phenotype comprises mostly GC non-resident Tfh cells
Flow cytometric analyses and direct histologic observations produce substantially different estimates of GCTfh cell abundance (Cho et al., 2018; Nance et al., 2015; Wittenbrink et al., 2011; Wollenberg et al., 2011; Wu et al., 2015; Yu et al., 2009). To confirm this discrepancy, we used standard phenotypic definitions for GCB cells [B220+Bcl6+Ki-67+ (Cirelli et al., 2019; Yeh et al., 2018)] and GCTfh cells [CD4+FoxP3−Bcl6+CXCR5hiPD-1hi (Choi et al., 2011; Pepper et al., 2011)] to enumerate by flow cytometry GCB and GCTfh cells in primary GCs elicited by NP-Ova+alum. CXCR5hiPD-1hi GCTfh-phenotype cells constituted ≈20% of GC lymphocytes at days 8 and 16 post-immunization (p.i.), rising to ≈33% at days 16 and 24 (Figs. 1A, 1B and S1A). In contrast, examination of the same LN tissues by immunofluorescence (IF) microscopy indicated GCTfh cells constituted ≈10% of GC lymphocytes from day 8 to 16, rising to ≈14% at day 24 (Figs. 1C, 1D and S1B)(Wittenbrink et al., 2011). From the literature and our own experience, we conclude that the Bcl6+CXCR5hiPD-1hi phenotype substantially overestimates the size of the true GCTfh cell compartment. GCTfh cells, defined by their physical location in GCs, are a minor subset of Bcl6+CXCR5hiPD-1hi Tfh cells.
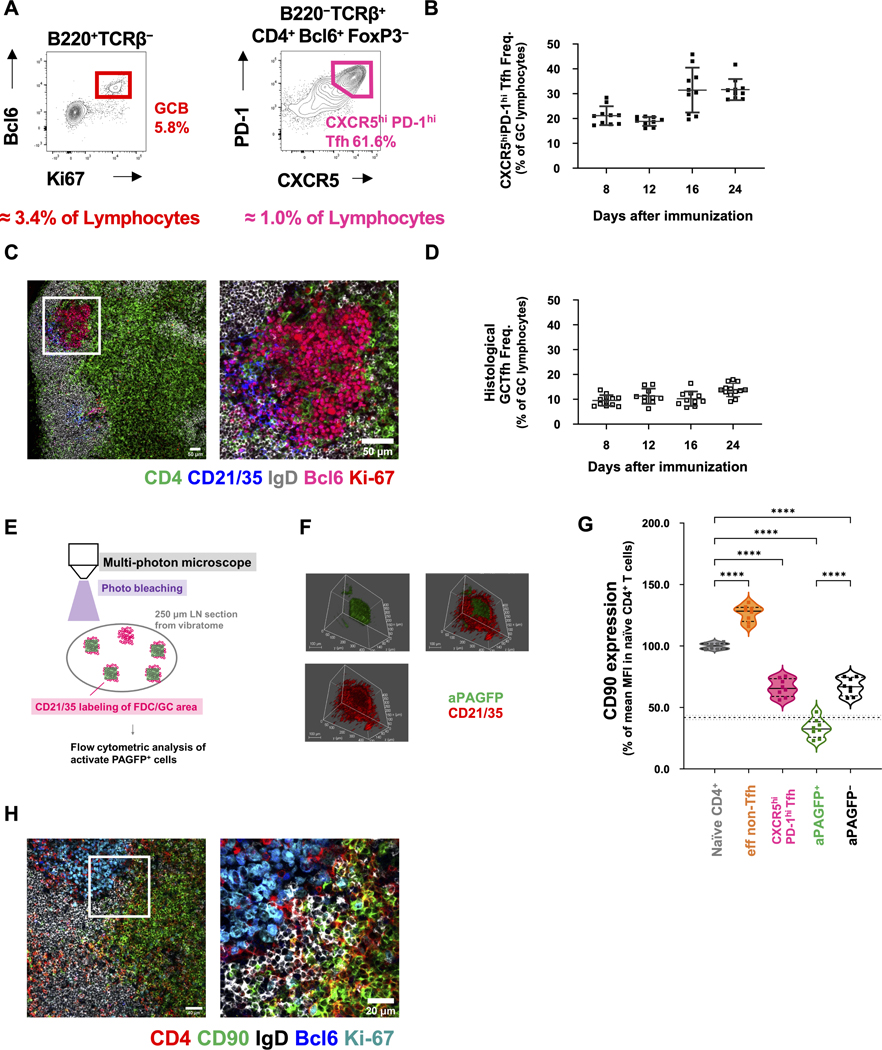
Figure 1. The CXCR5hiPD-1hi GCTfh cell phenotype comprises mostly GC non-resident Tfh cells.
(A-D) B6 mice were footpad-immunized with 20 μg of NP-Ova+alum. (A) Flow cytometry contour plots showing the frequencies of GC B cells and CXCR5hiPD-1hi Tfh cells among total pLN lymphocytes 8 days p.i. (B) Frequencies of CXCR5hiPD-1hi Tfh cells (as determined in A) in individual pLNs 8 to 24 days p.i. (n = 10 at each time point; mean ±S.D.). (C) Representative IF image of GCs, B cell follicles and adjacent T cell zone in pLNs 8 days p.i. The right panel depicts the white boxed area from the left panel, at higher magnification. CD4 (green), CD21/CD35 (blue), IgD (gray), Bcl6 (magenta), Ki-67 (red). Magnification: x100 (left), x200 (right), scale bars 50 μm. (D) Frequencies of GCTfh cells in individual GCs, as determined by IF analysis (n = 10–12 GCs at each time point; mean ± S.D.). (E-G) PAGFP Tg mice were footpad-immunized with 20 μg of NP-Ova+alum 11 days prior to analysis. (E) Diagram of the experimental design. (F) Representative 3D microscope image showing photo-activation of a region (activated PAGFP, green) within the FDC network (CD21/CD35, red). Original optical magnification: x25, scale bars 100 μm. (G) CD90 MFI of each cell population relative to naïve CD4+ T cells (n = 8; mean ± S.D.). The dashed horizontal line indicates the 10th percentile of CD90 expression in naïve CD4+ T cells (mean ± S.D.). (H) B6 mice were immunized as described in 1A for IF analysis 8 days p.i. Representative images show the GC, B cell follicle, T-B border, interfollicular region and nearby T cell zone in the draining pLN. The right panel depicts the white boxed area from the left panel, at higher magnification. CD4 (red), CD90 (green), IgD (gray), Bcl6 (blue), Ki-67 (cyan). Magnification: x200 (left); x630 (right), scale bars 40 μm (left) or 20 μm (right). Detailed flow cytometry gating strategies are shown in Fig. S1.
To determine a more precise phenotype of GCTfh cells, we turned to mice carrying a photoactivatable green fluorescent protein (PAGFP) transgene (Victora et al., 2010). We immunized PAGFP mice with NP-Ova+alum and 10 days later injected AF594-labeled CD21/CD35 mAb to identify the follicular dendritic cell (FDC) network (Fig. 1E). After another 16 hours, GCs and FDC networks were readily identified in vibratome sections of draining popliteal LNs (pLNs) (Fig. 1F). The central regions of AF594-labeled GCs were then activated using a multi-photon laser and recovered for analysis by flow cytometry. Cells containing activated PAGFP (aPAGFP) comprised both T and B cells within the central GC area (Figs. 1F and S1C). In this way, GCB cells were enriched ~8-fold in the aPAGFP+ B-cell fraction (75% vs 9%; Fig. S1C). aPAGFP+ T cells were comparably enriched for Bcl6+ cells: 78% of aPAGFP+ T cells were Bcl6+, compared to only 10% of aPAGFP− T cells (Fig. S1C). Almost all aPAGFP+Bcl6+ CD4 T cells were FoxP3− and exclusively exhibited the CXCR5hiPD-1hi phenotype (Fig. S1C).
In addition to the CXCR5 and PD-1 markers for Tfh cells, we previously noted that CD4 T cells in mouse GCs express little or no CD90 (Thy-1) (Zheng et al., 1996). Consequently, we determined levels of CD90 on aPAGFP+ GCTfh cells in comparison to resting naïve CD4 T cells, all CXCR5hiPD-1hi Tfh cells, and effector non-Tfh cells (eff non-Tfh) (Figs. S1A, S1C and S1D). Compared with resting naïve CD4 T cells, the photoactivated GC Tfh cells expressed reduced levels of CD90. The MFI of CD90 on aPAGFP+ GCTfh was lower than the 10th percentile of CD90 expression in resting naïve CD4 T cells (Fig. 1G). Loss of CD90 expression was an atypical property of antigen-activated CD4+ T cells, as activated eff non-Tfh cells, which expanded in parallel with GC Tfh populations (Figs. S1D), increased and maintained high CD90 expression throughout the primary response (Fig. 1G).
In B6 mice, analysis of the broadest Tfh compartment (CD4+Bcl6+FoxP3−) in day 12 LNs showed that CD90neg/lo Tfh cells were predominantly (>88%) CXCR5hiPD-1hi, whereas only 56% of those cells expressing normal levels of CD90 were CXCR5hiPD-1hi (Fig. S1E). Loss of CD90 by Tfh cells correlated with that fraction of GCTfh-phenotype cells resident in GCs, a conclusion confirmed by histologic inspection (Figs. 1H and S1F). Whereas CD4+CD90hi T cells were detected in paracortex, interfollicular regions and even in follicles of immunized LNs, CD4+ T cells in GCs expressed little or no CD90 (Figs. 1H and S1F).
Decreased expression of CD90 in S1pr2-marked GCTfh cells
In a separate approach to identifying GC-resident Tfh cells, we studied primary GC responses in S1pr2ERT2Cre-Rosa26lox-stop-lox-tdTomato (S1pr2-RFP) mice (Shinnakasu et al., 2016). Sphingosine-1-phosphate receptor (S1pr2), a G-protein-coupled receptor, is expressed by both GCB and GCTfh cells and promotes their anatomic retention (Green et al., 2011; Moriyama et al., 2014). To identify GC-resident Tfh cells (i.e., those retained by S1pr2) within the Bcl6+CXCR5hiPD-1hi population, we immunized S1pr2-RFP mice in the footpad with NP-Ova+alum, followed by daily doses of tamoxifen i.p. to induce Cre activity on days 5 to 7 (Fig. S2A). Histologic analysis of LN tissue from S1pr2-RFP mice indicated that Tfh cells expressing S1pr2 during the period of tamoxifen treatment were confined to GCs (Fig. 2A). Histologic analyses on days 10, 12, and 16 p.i. consistently showed ≈40% of GC-resident CD3+ T cells were RFP+. In contrast, <1% of CD3+ T cells outside of GCs were RFP+ over the same period (Figs. 2A, 2B). These observations confirmed S1pr2 is a stringent GCTfh cell marker. The constancy of RFP+ frequencies within GCs implies that GCTfh cell fate is fixed prior to day 10 p.i., with little or no migration to or from other anatomical sites. The near absence of RFP+ T cells in the B-cell follicles from days 10–16 p.i. is consistent with no GC-to-GC migration during primary responses (Suan et al., 2015).
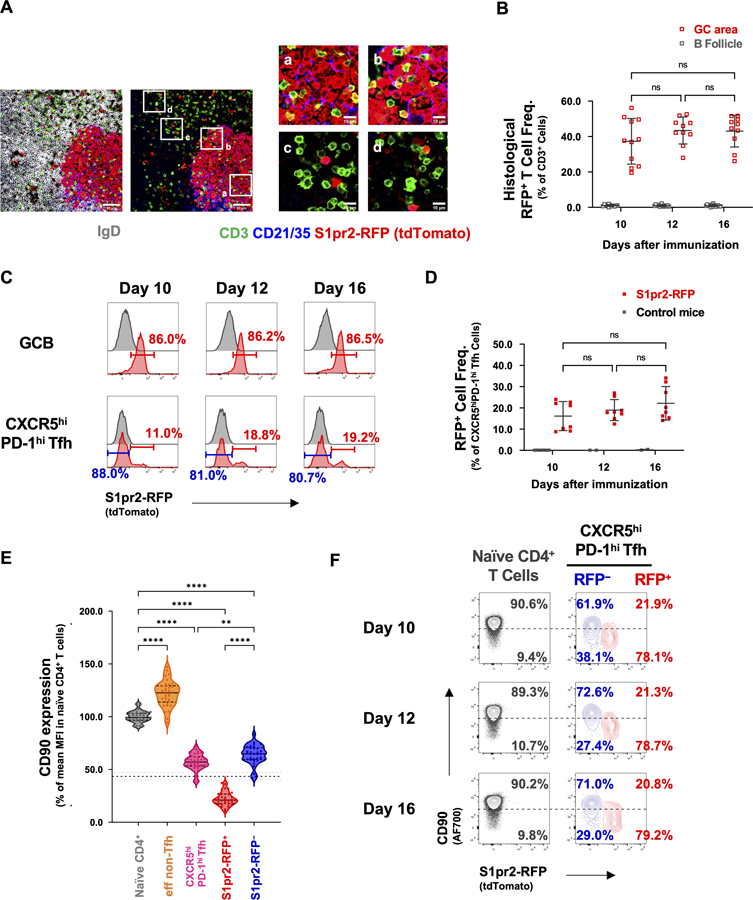
Figure 2. Decreased expression of CD90 in S1pr2-marked GCTfh cells.
S1pr2-RFP mice were footpad-immunized with 20 μg of NP-Ova+alum and treated with 5 mg tamoxifen (i.p.) at days 5–7 p.i. The draining pLNs were harvested at day 10, 12 and 16 p.i. (A) IF staining of pLNs from S1pr2-RFP mice 12 days p.i.; CD3 (green), CD21/CD35 (blue), IgD (gray), tdTomato (red). Magnification: x200, scale bars 50 μm (left) or 10 μm (right). IgD signal is omitted in the right panels to better visualize T cells in the B follicle. (B) Frequencies of RFP+ Tfh cells in individual GCs or B follicle areas, as determined by IF analysis (n = 10–14 sections from 3 animals at each time point; mean ± S.D.). (C) Representative flow histogram plots and (D) dot plots showing the frequencies of RFP+ Tfh cells (n = 8 at each time point; mean ± S.D.). (E) CD90 expression in each cell population relative to naïve CD4+ T cells (n = 34; mean ± S.D.). The dashed line indicates the 10th percentile of CD90 expression in naïve CD4+ T cells (mean ± S.D.). (F) Representative flow cytometry contour plots showing the frequencies of naïve (grey), RFP− Tfh (blue) and RFP+ GCTfh (red) cells above (CD90hi) or below (CD90neg/lo) the 10th percentile of CD90 expression in naïve CD4 T cells (dashed line). Each symbol represents an individual mouse. Data were pooled from ≥2 independent experiments. Statistical significance was measured using ordinary ANOVA followed by Tukey’s post-test (E; **P<0.01; **** P<0.0001).
More than 85% of GCB-phenotype cells were RFP+ on days 10–16 p.i.; in contrast, only 15–20% of Bcl6+CXCR5hiPD-1hi Tfh cells were marked by RFP (Figs. 2C–D and S2B). Mature follicular (MF) B and eff non-Tfh cells did not express RFP (Figs. S2C–D). Regardless of RFP expression, in S1pr2-RFP mice, Bcl6+CXCR5hiPD-1hi Tfh cells constituted ≈18% of all GC lymphocytes (GCB + Tfh cells), a frequency identical to that in B6 controls (Figs. 1B and S2E). However, RFP+ GCTfh cells constituted only 4% of GC cellularity (Fig. S2E). Taking the RFP-labeling efficiency into consideration, the size of the S1pr2-expressing GCTfh cell subset was consistent with the number of GC-resident Tfh cells observed directly by histology (Fig. 2B). Therefore, most Bcl6+CXCR5hiPD-1hi Tfh cells do not reside in GCs.
In S1pr2-RFP mice, RFP+ GCTfh-phenotype cells expressed substantially lower surface CD90 than did naïve CD4+ T cells, eff non-Tfh or RFP− GCTfh-phenotype cells (Fig. 2E). Indeed, RFP+ and RFP− CXCR5hiPD-1hi Tfh subsets could be reliably distinguished by their CD90 expression, with most of the former population expressing CD90 at levels below the 10th percentile of naïve CD4 T cells, while the great majority of the latter expressed CD90 at levels above the 10th percentile (Fig. 2E, 2F). Consequently, we hereafter used this 10th percentile cutoff to define the CD90neg/lo Tfh population in our experiments (Figs. 2E, 2F and S2F). We note that within the RFP− GCTfh compartment (Fig. 2C), ≈35% of cells were CD90neg/lo on days 10–16 p.i. (Figs. 2F and S2G), consistent with the inefficient RFP labeling in GC-resident Tfh cells (Fig. 2B). In contrast, >75% of RFP+ CXCR5hiPD-1hi Tfh cells were categorized as CD90neg/lo (Figs. 2F and S2H).
Primary GCTfh and GCTfh-like cell populations show different dynamics
GCTfh cells have long been considered to be non-dividing, as determined by the lack of Ki-67 expression in GC LZ T cells (Gulbranson-Judge et al., 1997), a key factor in the rationale that limiting numbers of GCTfh cells promotes stringent GCB cell selection (Meyer-Hermann et al., 2006; Wang et al., 2016). This notion was challenged recently by experiments demonstrating that phenotypically defined (CD4+CD62lowCD44hi Bcl6+CXCR5hiPD-1hi) Tfh cells proliferate throughout the GC response (Merkenschlager et al., 2021). To address the issue, we enumerated Ki-67+ GC cells in immunized S1pr2-RFP mice on days 10–16 p.i. Unsurprisingly, some 95% of RFP+ GCB cells were Ki-67+ (Fig. 3A and S3A–C). In contrast, RFP+ GCTfh cells were exclusively Ki-67−, but ~26% of RFP− GCTfh phenotype cells were Ki-67+ (Figs. 3A and S3A–D). Histological examination of the same LN tissues confirmed that Ki-67+ Tfh cells were common in interfollicular regions and B-cell follicles but virtually absent from GCs (Fig. 3B).
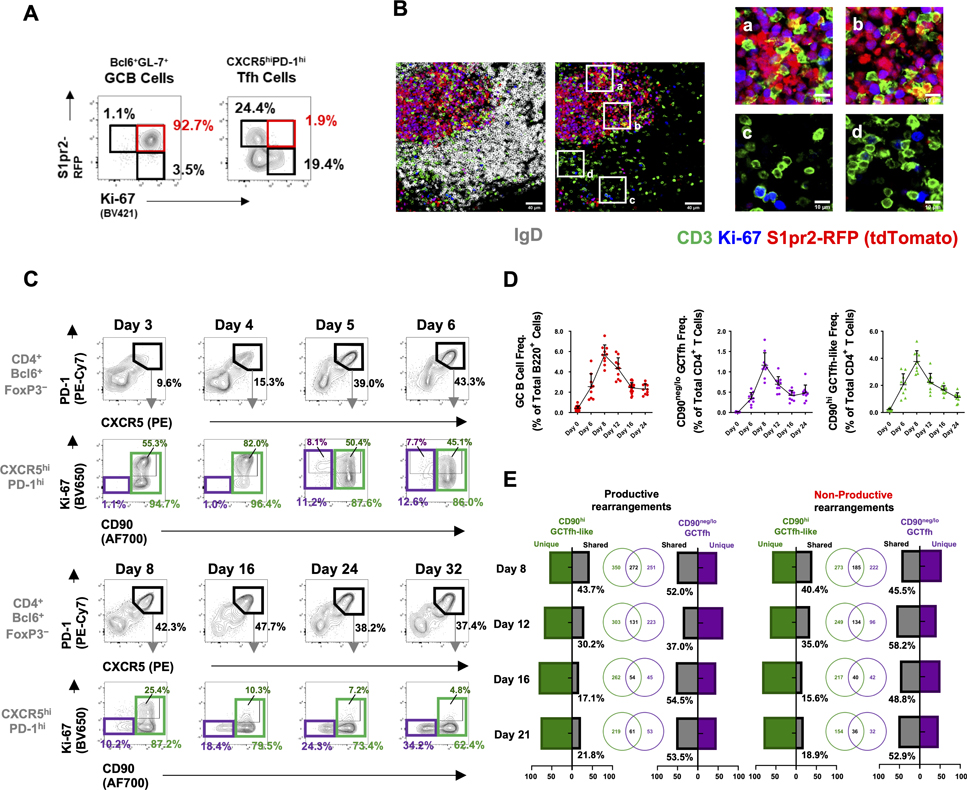
Figure 3. GCTfh and GCTfh-like cells follow different dynamics in primary responses.
(A-B) S1pr2-RFP mice were immunized and treated as described in Fig. S2A. (A) Representative flow cytometry contour plots show the frequencies of RFP+Ki-67+ GC B cells and RFP+Ki-67− Tfh cells. (B) IF analysis of pLNs from S1pr2-RFP mice 12 days p.i.; CD3 (green), IgD (gray), Ki-67 (blue), tdTomato (red). Magnification: x200, scale bars indicate 50 μm (left) or 10 μm (right). IgD signal is omitted in the right panels to better visualize T cells in the B follicle. (C-D) B6 mice were immunized as in Fig. 1A. and pLN cells were analyzed at indicated time points. (C) Flow cytometry plots indicating the frequencies of CXCR5hiPD-1hi Tfh cells among TCRβ+CD4+Bcl6+FoxP3− cells; and the frequencies of CD90neg/lo GCTfh (CD90neg/lo; purple), proliferating GCTfh-like (CD90hiKi-67+; dark green) or total CD90hi GCTfh-like (light green) populations among all CXCR5hiPD-1hi Tfh cells. (D) The population kinetics of GCB cells (left), CD90neg/lo GCTfh cells (center), and CD90hi GCTfh-like cells (right) after immunization (n = 10 at each time point; mean ± S.D.). (E) FoxP3EGFP mice were footpad-immunized with 20 μg of NP-Ova+alum and pLN cells were analyzed at indicated time points. CD90neg/lo GCTfh (purple) and CD90hi GCTfh-like cells (green) were sorted from the same pLN and were subjected to high-throughput TCRβ sequencing. Venn diagrams depict the numbers and bar charts the frequencies of unique and shared VDJ rearrangements. TCRβ sequencing data represent one of two independent experiments with similar results. Gating strategies are shown in Fig. S7A.
Our histological, photo-activation and S1pr2-driven labeling experiments demonstrated that even the most stringent previous definition for GCTfh cells (CD4+FoxP3−Bcl6+CXCR5hiPD-1hi) is too broad, since 50–65% of these cells were not GC residents. Therefore, most prior studies of “GCTfh” cells likely described properties of the larger, GC-nonresident Tfh cell population. To characterize potential functional differences between CD90neg/lo resident GCTfh and CD90hi “GCTfh-like” cells, we followed both populations in LNs after immunization. Cells with the standard CXCR5hiPD-1hi Tfh phenotype were identifiable as early as day 3 p.i. These cells expressed high levels of CD90, and ≈50% were Ki-67+, indicating proliferation (Fig. 3C). By day 4, before GC B cells could be identified, the CD90hi GCTfh-like population was >80% Ki-67+ and had expanded (Figs. 3C and S3E–F). On day 5, ≈10% of the proliferating GCTfh-like population reduced CD90 expression; this shift coincided with the appearance of Bcl6+Ki-67+ GCB cells and presumably represents the emergence of GC-resident Tfh cells (Figs. 3C and S3F). Later, as GCs organized into light- and dark zones (Liu et al., 1991), the CD90neg/lo GCTfh compartment became Ki-67− (Fig. 3C). However, the numbers and frequencies of CD90neg/lo GCTfh and CD90hi GCTfh-like cells both increased through day 8, implying continued recruitment of antigen-specific T cells into both compartments through the peak GCB cell response (Figs. 3D and S3G). Following the peak GC response, the numbers and frequencies of GCB, CD90neg/lo GCTfh and CD90hi GCTfh-like cells initially declined with similar kinetics (Figs. 3D, S3F–G), but after day 16, frequencies of GCB and CD90neg/lo GCTfh cells stabilized, whereas CD90hi GCTfh-like cell numbers continued to decrease (Figs. 3C and 3D). Consequently, over time, CD90neg/lo GCTfh cells increased their representation among the broader Bcl6+CXCR5hiPD-1hi population (Figs. 3C and S3H). Similar population dynamics of CD90neg/lo GCTfh and CD90hi GCTfh-like cells were elicited by multiple antigens, including B. anthracis protective antigen and influenza H1 hemagglutinin (Fig. S3H). Taken together, CD90neg/lo GCTfh cells are non-proliferative, but the proportion of this population among all Bcl6+CXCR5hiPD-1hi Tfh cells increases as the GC reaction wanes, suggesting local stability while CD90hi GCTfh-like cells migrate or die.
To determine whether CD90neg/lo GCTfh cells are unique to transient GCs elicited by primary immunization, we analyzed the composition of constitutive GCs in Peyer’s patches (PPs) of B6 mice (Figs. S3I–L). CD90neg/lo GCTfh cells were as abundant in PPs as in primary GCs elicited by NP-Ova (Fig. 3C and S3K); in PPs, CD90neg/lo GCTfh cells constituted ≈18% of the Bcl6+CXCR5hiPD-1hi Tfh cell population (Fig. S3M). Notably, CXCR5hiPD-1hi Tfh cells represented ≈25% of GC lymphocytes, whereas CD90neg/lo GCTfh cells constituted only ≈5% (Fig. S3N). Thus, like the GC responses elicited by primary immunization, chronic GC responses driven by dietary and environmental antigens support CD90neg/lo GCTfh cells.
To investigate the clonal origins of CD90neg/lo GCTfh and CD90hi GCTfh-like cells, we isolated both populations from a common LN and used high-throughput sequencing to recover the Tcrβ rearrangements present in each (Carlson et al., 2013). From pLNs at days 8, 12, 16 and 21 p.i., we obtained 6,179 and 8,977 Tcrβ VDJ templates from CD90neg/lo GCTfh and CD90hi GCTfh-like cells, respectively. Among these, 2,902 and 1,877 unique rearrangements were found in each compartment. At day 8, there was substantial overlap in the Tcrβ rearrangements expressed by CD90neg/lo GCTfh and CD90hi GCTfh-like cells, with ≈44% of all Tcrβ rearrangements from CD90hi GCTfh-like cells being shared with CD90neg/lo GCTfh cells. Thereafter, sharing diminished significantly, such that by day 21 only 22% of Tcrβ rearrangements were shared. Whereas about half of the Tcrβ rearrangements in CD90neg/lo GCTfh cells were always shared with CD90hi GCTfh-like cells over the course of the response, the converse was not true (Figs. 3E and S3O). Similar patterns of divergence were observed for both productive and non-productive Tcrβ rearrangements, implying discordant selection/expansion between the two populations (Figs. 3E).
Differentiation and maintenance of CD90neg/lo GCTfh cells requires antigen-specific, MHCII+ B cells
Although Tfh cell commitment occurs within the first few rounds of cell division after activation by antigen, Tfh cell differentiation is a process that includes stepwise priming by DCs and cognate interaction with B cells at the border of T- and B-cell zones (Choi et al., 2013; DiToro et al., 2018; Tubo et al., 2013). Given that CD90hi GCTfh-like and CD90neg/lo GCTfh cells extensively shared Tcrβ rearrangements early but not late after immunization (Figs. 3E and S3O), and that differentiation of the CD90neg/lo GCTfh cell subset coincided with the appearance of GCB cells, we hypothesized that CD90neg/lo GCTfh cell differentiation requires continued B-cell interaction whereas the CD90hi GCTfh-like population does not. To test this hypothesis, we generated experimental mixed BM chimeras in which all B cells (but only 20% of DCs) were MHCII-deficient; in isogenic control chimeras, B cells expressed MHCII normally (Fig. 4A). Finally, IgD+Igλ+ B1-8i NP-specific B cells (Sonoda et al., 1997) were transferred i.v. to experimental chimeras one day before immunization with NP-Ova or Ova to provide cohorts of MHCII-sufficient and antigen-specific (NP-Ova) or unspecific (Ova) B-cells (Fig. 4A).
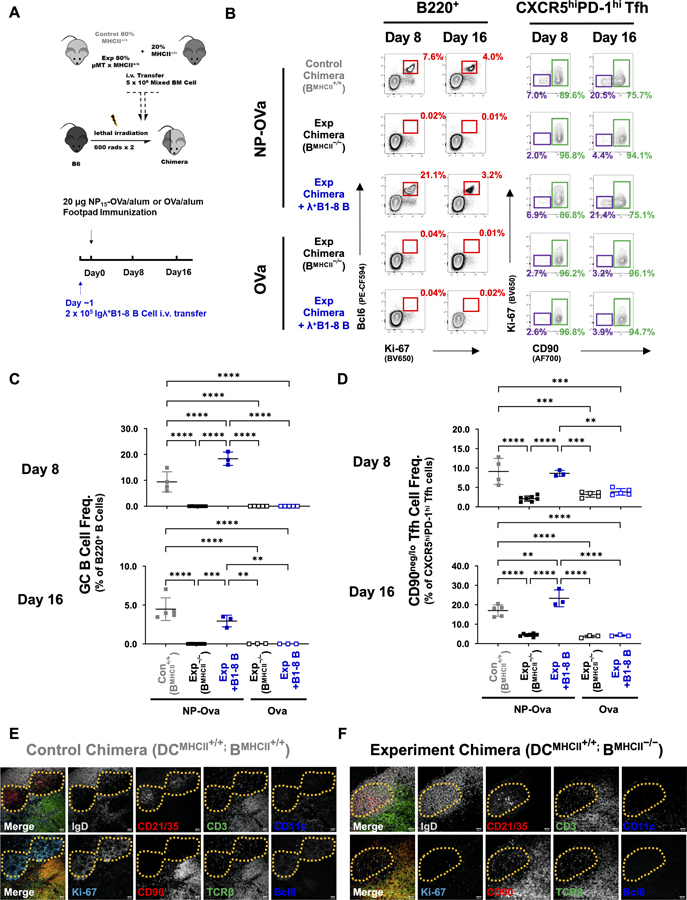
Figure 4. Differentiation and maintenance of CD90neg/lo GCTfh cells requires antigen-specific, MHCII+ B cells.
(A) Diagram of the experimental design. (B) Representative flow cytometry contour plots depicting the frequencies of B220+Bcl6+Ki-67+ GCB cells among B220+ cells (red; left panel) and of CD90neg/lo Ki-67− (CD90neg/lo GCTfh; purple) or CD90hi GCTfh-like (green) cells among all CXCR5hiPD-1hi Tfh cells in pLNs at days 8 and 16 p.i. (C-D) Frequencies of (C) GCB cells among total B220+ B cells and (D) CD90neg/lo GCTfh cells among CXCR5hiPD-1hi Tfh cells at indicated time points (n = 3–8 at each time point; mean ±S.D.). Each symbol represents an individual mouse; data were pooled from at least two independent experiments. Statistical significance was measured using ordinary ANOVA followed by Tukey’s post-test (**P<0.01; *** P<0.001). (E-F) Representative images showing IF analysis of serial sections from the LNs of (E) a control chimera (DCMHCII+/+; BMHCII+/+) and (F) an experimental chimera (DCMHCII+/+; BMHCII−/−). The far left panel in each series depicts the merged signals from the subsequent four panels. Top row: IgD (gray), CD21/35 (red), CD3 (green), CD11c (blue). Bottom row: Ki-67 (cyan), CD90 (red), TCRβ (green), Bcl6 (blue). Dashed yellow lines circumscribe the GC (E) or FDC network (F). Magnification: x200, scale bars 40 μm. Data represent one of two independent experiments with similar results.
After immunization, control chimeras generated potent GC responses, but in experimental chimeras with only MHCII-deficient B cells, no GCB cells were generated (Figs. 4B and 4C). Transfer of B1-8i B cells fully restored GCB responses in mice immunized with NP-Ova, but not with Ova alone (Figs. 4B and 4C). In concert with the presence or absence of GCB cells, we observed significant changes in the frequencies of CD90neg/lo GCTfh cells within the CXCR5hiPD-1hi Tfh compartment. In control chimeras, CD90neg/lo GCTfh cells represented 9%−17% of all CXCR5hiPD-1hi Tfh cells on days 8 and 16 p.i. Similar frequencies were observed in experimental chimeras given B1-8i cells and immunized with NP-Ova. In contrast, in both experimental chimeras or experimental chimeras given B1-8i cells and immunized with Ova, CD90neg/lo GCTfh cells constituted just 2–3% of CXCR5hiPD-1hi Tfh cells on days 8 and 16 (Figs. 4B, 4D and S4A–D). We note that Tfh cells in non-responding experimental chimeras expressed lower levels of PD-1 compared to controls (Fig. S4A). Importantly, the presence or absence of MHCII-sufficient B cells did not affect the frequencies of eff non-Tfh cells (Figs. S4B).
Histologic examination of pLNs from control and experimental chimeras (Figs. 4E–F) confirmed the flow cytometry results. Eight days after immunization, control chimeras generated Ki-67+Bcl6+ GCs within the CD21/CD35+ reticula of FDC networks in B-cell follicles (Fig. 4E). In contrast, whereas typical follicles and FDC networks were present in experimental chimeras with MHCII-deficient B cells, no GCs were generated (Fig. 4F). In control chimeras, CD90neg/lo GCTfh cells were observed in GCs while CD90hi GCTfh-like cells were present in the interfollicular regions and at the intersection of T-cell zones and follicles (Fig. 4E; compare to 1H). However, in experimental chimeras, CD3+CD90hi or TCRβ+CD90hi Tfh cells were uniformly distributed over the B-cell follicle (Fig. 4F). We conclude that antigen-specific B cells, presumably as cognate partners, are necessary for the differentiation of CD90neg/lo GCTfh cells, whereas CD90hi GCTfh-like cells readily develop when DCs are the exclusive antigen-presenting cell. Cognate T:B interaction provides a specific cue for CD90neg/lo GCTfh cell development.
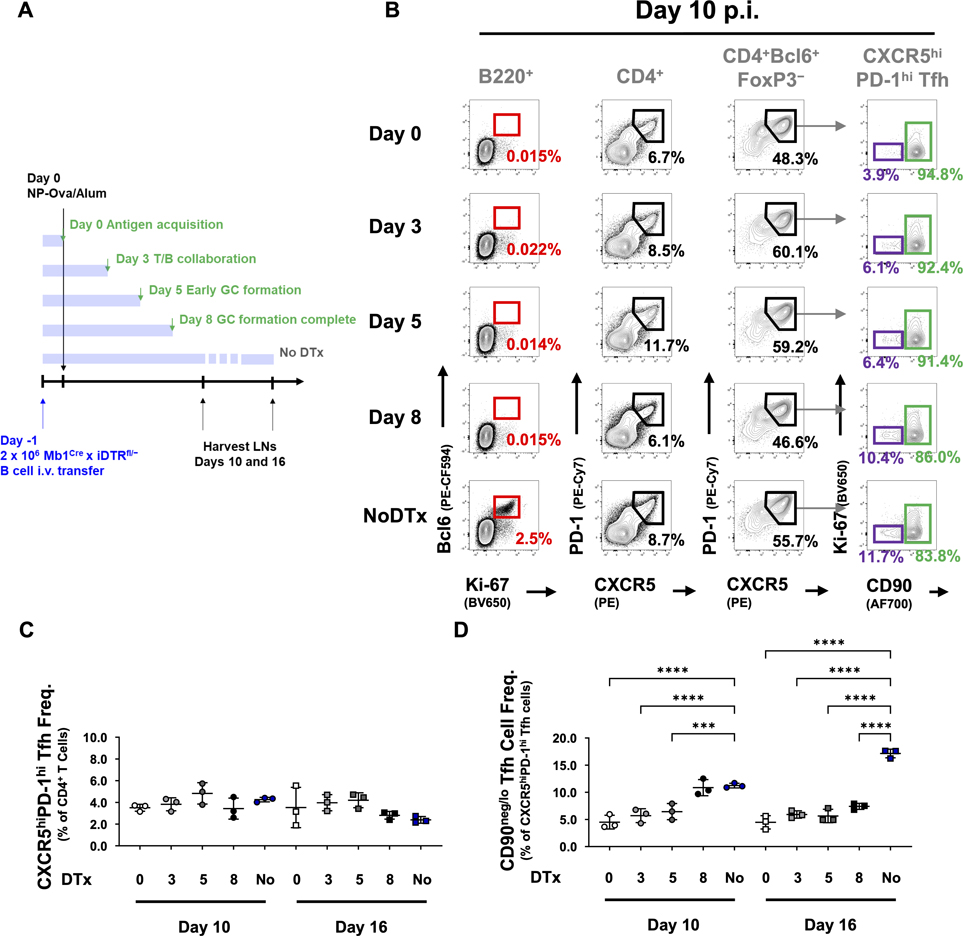
To define the window during which antigen-specific, MHCII-sufficient B cells confer the signal for CD90neg/lo GCTfh cell differentiation, we transferred Mb1CrexDTRLSL B cells into chimeric mice in which all other B cells (but not DCs) were MHCII−/− and thus incapable of antigen presentation (Fig. 4A). We then immunized the mice and administered diphtheria toxin (DTx) after various intervals to kill antigen-presenting B cells (Fig. 5A and S4E). DTx administration began either 1) on the day of immunization (d0) to block any cognate T:B interaction; 2) during early T/B collaboration (d3); 3) at the initiation of GC formation (d5), or 4) after GC organization was complete (d8) (Crotty, 2019). Once initiated, i.p. injections of DTx occurred every other day to maintain effective serum concentrations of DTx (Meredith et al., 2012). In all cohorts, GCB, CD90neg/lo GCTfh, and CD90hi GCTfh-like cells were enumerated at days 10 and 16 p.i. (Fig. 5A).
Figure 5. CD90neg/lo GCTfh cell differentiation requires B cells to present cognate pMHCII until GCs completely coalesce.
(A) Diagram of the experimental design. Chimeric mice were generated as in Fig. 4A. 2 × 106 MF B cells from Mb1CrexDTRLSL mice were i.v. transferred 1 day prior to immunization. DTx was i.p. injected starting at indicated time points. Draining pLNs were harvested at day 10 or 16 p.i. (B) Representative flow cytometry contour plots depicting the frequencies of B220+Bcl6+Ki-67+ GCB cells (red; left column), CD4+CXCR5hiPD-1hi Tfh cells (second column), CD4+FoxP3−Bcl6+CXCR5hiPD-1hi Tfh cells (third column) and CD90neg/lo Ki-67− (CD90neg/lo GCTfh; purple, fourth column) and CD90hi GCTfh-like (green, fourth column) populations in pLNs harvested from each group of chimeras at day 10 p.i. (C-D) Frequencies of (C) CXCR5hiPD-1hi Tfh cells among total CD4+ T cells and (D) CD90neg/lo GCTfh cells among CXCR5hiPD-1hi Tfh cells at indicated time points (n = 3 at each time point; mean ±S.D.). Each symbol represents an individual mouse; results were pooled from at least two independent experiments. Statistical significance was determined using ordinary ANOVA followed by Tukey’s post-test (***P<0.01; **** P<0.001).
DTx injection effectively depleted all transferred B cells, ending any capacity for humoral responses and abrogating GCs. Regardless of when DTx administration began, GCB cells were completely absent in all treated mice (Fig. S4F). However, B cell depletion at any time point did not affect the size of the total CXCR5hiPD-1hi Tfh cell population present on days 10 and 16 p.i. (Figs. 5B and 5C). In contrast, depleting GCB cells significantly reduced the size of the CD90neg/lo GCTfh compartment. DTx given in the early phases of GC response (day 0, 3, or 5) reduced CD90neg/lo GCTfh cell frequencies 2- to 3-fold on day 10 and 3- to 4-fold on day 16 (Fig. 5D). Treatment with DTx beginning at day 8, after GCs become fully organized, did not affect CD90neg/lo GCTfh cell frequencies on day 10, but on day 16, CD90neg/lo GCTfh cell frequencies fell to half that of controls (Figs. 5B, 5D and S4G–I). This loss after GC organization suggests that sustained cognate T:B interaction is necessary to maintain CD90neg/lo GCTfh populations. Notably, DTx-derived B cell depletion at any time did not affect the frequencies of eff non-Tfh cells (Fig. S4J).
Continued proliferation of CD90hi GCTfh-like cells is DC dependent
The dispensability of B cells for robust responses of CXCR5hiPD-1hiCD90hi GCTfh-like cells implies that conventional DCs (cDCs) alone are sufficient for the differentiation and maintenance of this T-cell population. To test this hypothesis, we generated Zbtb46-DTR BM chimeras to restrict DTR expression to cDCs (Meredith et al., 2012). We immunized these animals with NP-Ova+alum, and then depleted the cDC compartment by DTx injection on day 7 p.i., after GC organization was complete (Fig. S5A). On days 8, 12, and 16 p.i., we enumerated and characterized MHCIIhiCD11c+ migratory cDCs (mDCs) and MHCII+CD11chi LN-resident cDCs (rDCs), along with GCB, CD90neg/lo GCTfh, and CD90hi GCTfh-like cells (Fig. S5B–J).
Whereas DTx injection effectively reduced both mDC and rDC frequencies (Fig. S5B, S5E–F), the frequencies of GCB (Figs. S5C and S5G) and CD90neg/lo GCTfh cells (Figs. S5D and S5H) were unaffected. Frequencies of CD90hi GCTfh-like cells were also unchanged (Figs. S5D and S5I), but cDC ablation significantly reduced CD90hi GCTfh-like cell proliferation at days 12 and 16, as measured by lower frequencies of Ki-67+CD90hi cells (Figs. S5D and S5J). We conclude that Tfh cell proliferation in the CD90hi compartment is driven by cognate interaction with antigen-presenting cDCs; an observation that might explain the diverging TCRβ repertoires of CD90hi GCTfh-like and CD90lo GCTfh cell populations.
The influence of antigen receptors on CD90neg/lo GCTfh cell differentiation
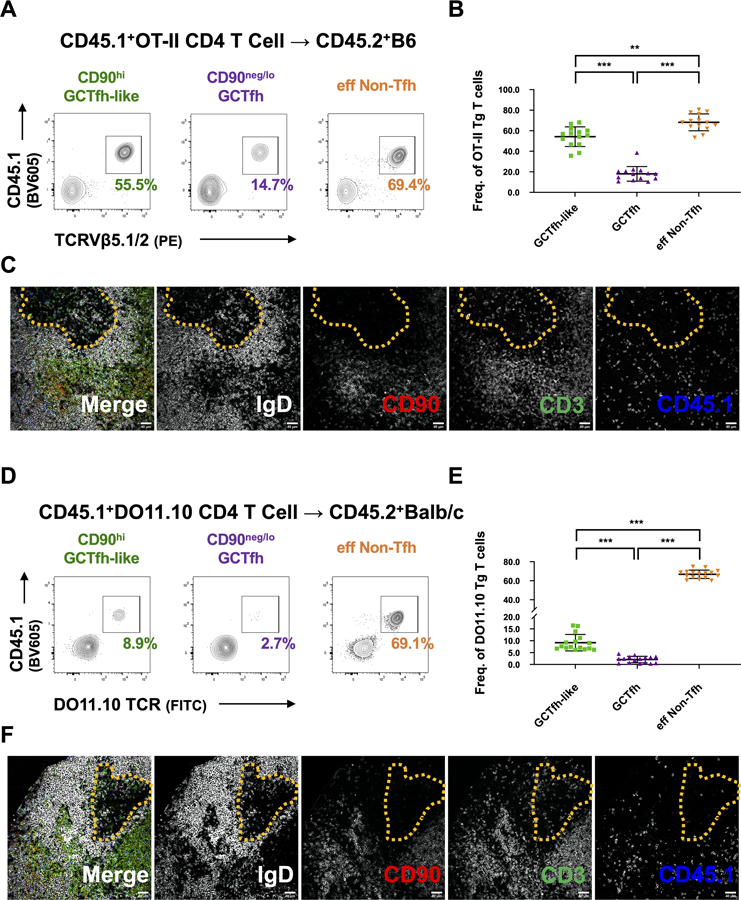
That the TCR repertoires of CD90neg/lo GCTfh and CD90hi GCTfh-like cells diverged over time (Fig. 3E) suggested to us that not all antigen-specific TCRs equally support GCTfh cell differentiation. To investigate the potential impact of individual TCRs on CD4+ T cell fates, we adoptively transferred CD4+ T cells from CD45.1+ OT-II or DO11.10 TCR transgenic mice into naïve CD45.2+ B6 or BALB/c hosts, respectively. The OT-II and DO11.10 TCRs are specific for the Ova323–339 peptide presented on I-Ab or I-Ad (Robertson et al., 2000). We then immunized recipient mice with NP-Ova and analyzed draining pLNs by flow cytometry on day 8 p.i. to determine the participation of OT-II or DO11.10 T cells in the CD90neg/lo GCTfh, CD90hi GCTfh-like, and eff non-Tfh cell compartments.
Transferred OT-II T cells were capable of differentiating into GCTfh, GCTfh-like and eff non-Tfh cells in the presence of endogenous competitors; however, the frequency of OT-II cells in each subset varied significantly (Figs. 6A and 6B). While the OT-II cells constituted 70% and 50% of eff non-Tfh and GCTfh-like subsets, respectively, only about 15% of GCTfh cells were OT-II T cells (Figs. 6A and 6B). Histology confirmed that most of the transferred OT-II T cells expressed CD90 and were located at the T-B border, in B cell follicles or in T cell zones (Fig. 6C). The propensity toward non-Tfh fates was even more extreme in DO11.10 transgenic T cells transferred to BALB/c recipients: DO11.10 T cells constituted 67% the eff non-Tfh subset, but only 10% and 2% of the GCTfh-like and GCTfh populations, respectively (Figs. 6D–F). We conclude that these commonly used transgenic TCRs are better suited for studying GCTfh-like and eff non-Tfh cell responses than for probing the physiology of GCTfh cells.
Figure 6. TCR influences the likelihood of CD90neg/lo GCTfh cell differentiation.
CD4+ T cells from (A-C) CD45.1+ OT-II or (D-F) CD45.1+ DO11.10 mice were adoptively transferred into B6 or Balb/c mice. Hosts were subsequently footpad-immunized with 20 μg of NP-Ova+alum. Draining pLNs were analyzed at day 8 p.i. (A) Representative flow cytometry contour plots and (B) summary graph depicting the frequencies of CD45.1+TCRVβ5+ transferred OT-II cells among CD90hi GCTfh-like, GCTfh and eff non-Tfh cells (n = 14; mean ±S.D.). (C) Representative IF images showing cryostat sections from draining pLNs. (D) Representative flow cytometry contour plots and (E) summary graph depicting the frequencies of CD45.1+DO11.10Tg+ transferred cells among CD90hi GCTfh-like, GCTfh and eff non-Tfh cells (n = 16; mean ±S.D.). (F) Representative IF images showing cryostat sections from draining pLNs. (B and E) Each symbol represents an individual mouse LN. Results were pooled from at least two independent experiments. Statistical significance was measured using ordinary ANOVA followed by Tukey’s post-test (**P<0.01; *** P<0.001). (C and F) The far left panel depicts the merged signals from the subsequent four panels. IgD (gray), CD90 (red), CD3 (green), CD45.1 (blue). Dashed yellow lines circumscribe the GC region. Magnification: x200, scale bars 40 μm.
B cells carrying low-affinity BCRs are fully capable of T-dependent immune responses (Dal Porto et al., 1998), but it is unclear how BCR affinity affects GCTfh cell development. To address this question, we used NP-conjugated human serum albumin (NP-HSA)+alum to immunize B6.H50Gμ transgenic mice, which express an IgM BCR that, with the λ1 light chain, binds the NP hapten with an association constant (Ka) of ≈1.2 × 105 M−1 (Dal Porto et al., 1998). We also immunized a second cohort of B6.H50Gμ mice that had received 1 × 105 NP-specific λ+ B cells from B1-8i mice (Ka = ≈1.0 × 106 M−1 (Dal Porto et al., 1998; Sonoda et al., 1997)) to determine whether higher BCR affinity affects GCTfh cell differentiation. As expected, immunized B6.H50Gμ mice that had received B1-8i B cells supported ≥5-fold larger GCB cell responses than immunized control B6.H50Gμ mice (Figs. S6A–D). Despite the significant increases in GCB cell responses, the frequencies of total CXCR5hiPD-1hi Tfh and CD90hi GCTfh-like cells were comparable in both cohorts (Figs. S6A and S6E). In contrast, CD90neg/lo GCTfh cell frequencies and numbers were significantly increased in B6.H50Gμ mice supplemented with B1-8i B cells (Fig. S6F). Thus, higher BCR affinity promotes CD90neg/lo GCTfh cell differentiation but has limited effect on CD90hi GCTfh-like cell populations.
Transcriptional profiling implies functional divergence of CD90neg/lo GCTfh and CD90hi GCTfh-like cells
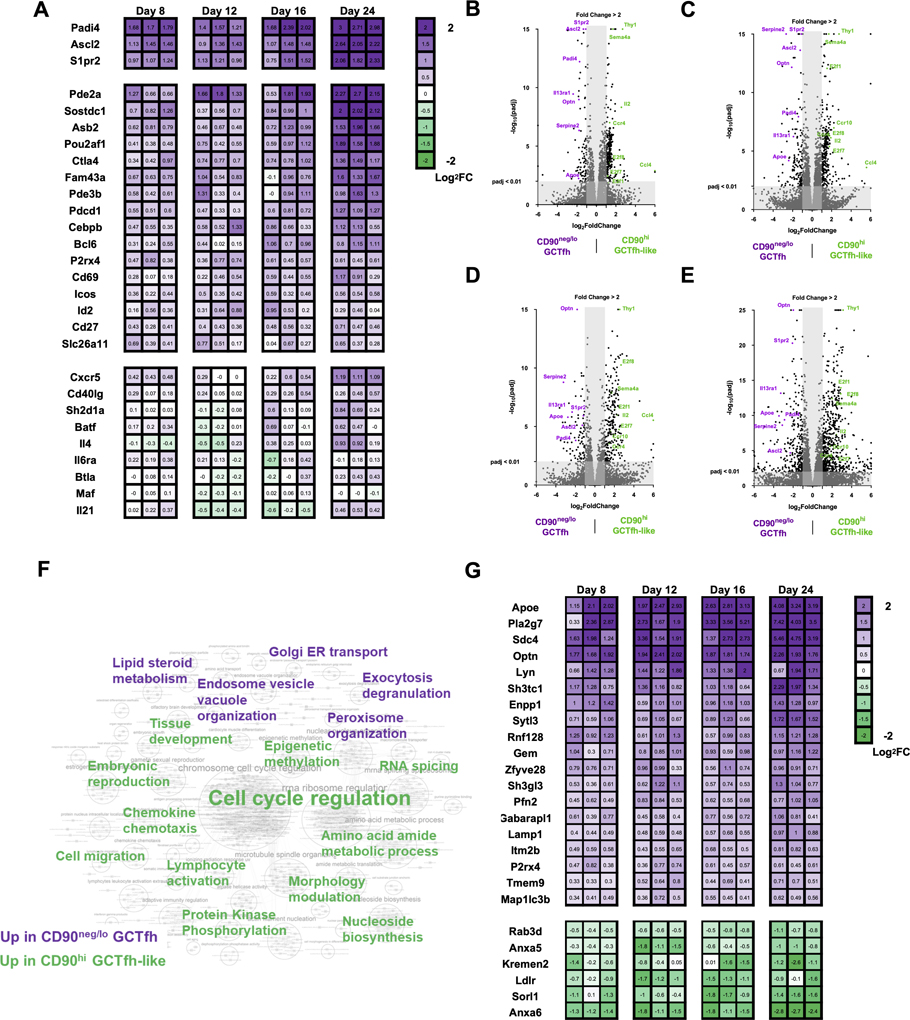
To identify functional differences between CD90neg/lo GCTfh and CD90hi GCTfh-like cells, we performed RNA sequencing (RNASeq) on both populations, together with eff non-Tfh and TFR cells isolated from the same LN of FoxP3EGFP mice (Haribhai et al., 2007) 8, 12, 16 and 24 days p.i. (Fig. S7A). Transcriptomes for each T-cell population were obtained by deep-sequencing cDNA libraries. Transcriptional profiles of CD90neg/lo GCTfh cells relative to CD90hi GCTfh-like, eff non-Tfh and TFR cells were compared by principal component analysis of each cohort and grouping d8 and d12 (Fig. S7B), or d16 and d24 samples (Fig. S7C). The transcriptome of each T-cell group was clearly distinct: eff non-Tfh and TFR cells displayed higher intra-subset variability than did the CD90neg/lo GCTfh and CD90hi GCTfh-like cohorts (Figs. S7B and S7C). Despite shared clonal origins for many CD90neg/lo GCTfh and CD90hi GCTfh-like cells (Fig. 3E), z-score normalized heatmap analysis revealed significant differences (P < 0.05 and ≥2-fold-change) in gene expression between these groups (Fig. S7D). Compared to CD90hi GCTfh-like cells, the transcriptome of CD90neg/lo GCTfh cells was significantly enriched for the Tfh-related genes Ascl2, Padi4 and S1pr2 (Liu et al., 2014; Moriyama et al., 2014; Wing et al., 2017) at every time point (Figs. 7A–E). Other Tfh-related genes, including Asb2, Bcl6, Cd27, Cd69, Cebpb, Ctla4, Fam43a, Icos, Id2, P2rx4, Pdcd1, Pde2a, Pde3b, Pou2af1, Slc26a11 and Sostdc1 (Choi et al., 2015; Wing et al., 2017), were significantly elevated (P < 0.05) in the CD90neg/lo GCTfh compartment but occasionally did not meet the ≥2-fold threshold at some time points (Fig. 7A). Still other Tfh-related genes, including Batf, Btla, Cd40lg, Cxcr5, Il4, Il6ra, Il21, Maf and Sh2d1a (Choi et al., 2015; Moriyama et al., 2014; Wing et al., 2017), were comparably expressed in both CD90neg/lo GCTfh and CD90hi GCTfh-like cells (Fig. 7A). We also identified a number of genes (Ccl4, Ccr4, Ccr10, E2f1, E2f7, E2f8, Il2 and Sema4a) that were significantly upregulated in CD90hi GCTfh-like cells compared to CD90neg/lo GCTfh cohorts (Fig. 7B–E). These genes are not known to be Tfh-related but are associated with T-cell activation, proliferation and migration (Attwooll et al., 2004; Lu et al., 2018; Stein and Nombela-Arrieta, 2005).
Figure 7. Transcriptional profiling implies functional divergence of CD90neg/lo GCTfh and CD90hi GCTfh-like cells.
FoxP3EGFP mice were footpad-immunized with 20 μg of NP-Ova+alum. Draining pLNs were harvested at days 8, 12, 16 and 24 p.i. CD90neg/lo GCTfh (purple) and CD90hi (green) GCTfh-like cells were sorted from the same pLN and were subjected to ultra-low RNA sequencing. Detailed gating strategies are shown in Fig. S7A. RNA was extracted and then subjected to library preparation and sequencing. Differential gene expression analysis was performed in R by DEseq2. (A) Heatmap graph showing the fold-change of Tfh-related gene expression in CD90neg/lo GCTfh over CD90hi GCTfh-like cells in each individual pLN. Numbers indicate the log2 fold-change in gene expression (n = 3 at each time point). (B-E) Volcano plots depicting genes up- or down-regulated with fold-change ≥ 2 and adjusted P value < 0.01 in CD90neg/lo GCTfh cells relative to CD90hi GCTfh-like cells at (B) day 8, (C) day 12, (D) day 16 or (E) day 24. (F) GSEA of RNASeq data. Significant gene sets with FDR < 0.1 or p < 0.01 were visualized with Cytoscape and Enrichment Map. The keyword graph represents the annotated results for clustered gene set comparisons. Purple keywords denote the physiological signature enriched in CD90neg/lo GCTfh cells and green words represent the physiological signature enriched in CD90hi GCTfh-like cells. A detailed graph and lists of gene set comparison information are shown in Fig. S7E and Table S1. (G) Heatmap graph showing the fold-change in expression of significantly (p<0.05) up- or down-regulated genes associated with endosomal vesicle organization and exocytosis/degranulation in CD90neg/lo GCTfh over CD90hi GCTfh-like cells. Numbers indicate the value of log2 fold-change in gene expression (n = 3 at each time point).
To infer functional differences from the distinctive transcriptomes of the CD90neg/lo GCTfh and CD90hi GCTfh-like populations, we performed gene set enrichment analysis (GSEA) against the Gene Ontology database (C5; MSigDB) to identify ontological pathways associated with the gene expression patterns of these two populations. Significant gene sets (FDR < 0.1 or P < 0.01) were visualized as interaction networks with Cytoscape and Enrichment Map (Figs. 7F, S7E and Table S1) (Merico et al., 2010). Compared with CD90hi GCTfh-like cells, the CD90neg/lo GCTfh subset was significantly enriched for gene expression profiles linked to endosome/vesicle organization, and exocytosis/degranulation (Figs. 7F, S7E and Table S1). Expression of genes associated with vesicle organization and/or exocytosis (Apoe, Enpp1, Gaparapl1, Gem, Itmb2b, Lamp1, Lyn, Map1lc3b, Optn, P2rx4, Pfn2, Pla2g7, Rnf128, Sdc4, Sh3gl3, Sh3tc1, Sytl3, Tmem9 and Zfyve28) was significantly elevated in CD90neg/lo GCTfh cells (Fig. 7G), whereas genes downregulated during vesicle organization and/or exocytosis (Anxa5, Anxa6, Kremen2, Ldlr, Rab3d and Sorl1) were suppressed (Fig. 7G). Notably, gene expression indicative of activated lipid and steroid metabolism (Table S1) was also significantly enriched in CD90neg/lo GCTfh cells.
In contrast, CD90hi GCTfh-like cells showed patterns of gene expression associated with cell division (Figs. 7F, S7E–F and Table S1), consistent with the more abundant numbers of Ki-67+ cells in this compartment (Figs. 3A and 3C). CD90hi GCTfh-like cells also expressed genes related to cell migration and chemotaxis (Ccl3, Ccl4, Ccr4, Ccr5, Ccr6, Ccr7, Ccr10, Cxcl13, Cxcr3, Dock4, Gpr18, Gpr183, Hmgb2, Selplg, Sell, Tbx21) (Moriyama et al., 2014) (Fig. S7G) and protein kinase phosphorylation (Acvrl1, Aurka, Aurkb, Bub1, Bub1b, Ccnb1, Ccnb2, Cdk1, Chek1, Cit, Clspn, Gsg2, Mastl, Melk, Nek2, Nek6, Pbk, Plk1, Thy1, Ttk) (Bolanos-Garcia and Blundell, 2011; den Hollander et al., 2010; Gong and Ferrell, 2010; O’Regan et al., 2007) (Fig. S7H). Genes generally downregulated during protein kinase phosphorylation, including Dapk2, Lyn, Matk, Nrpb2, Pnck and Spock2, were suppressed in CD90hi GCTfh-like cells (Fig. S7H). In addition, gene expression indicative of activated tissue development and amino acid metabolism was also significantly enriched in CD90hi GCTfh-like cells (Figs. 7F, S7E and Table S1). These results indicate distinctive physiologies for CD90neg/lo GCTfh and CD90hi GCTfh-like cells, which previously was obscured by the cellular diversity in the Bcl6+CXCR5hiPD-1hi Tfh population.
Discussion
High-affinity antibody and humoral memory arise in GCs elicited by immunization or infection. In GCs, specialized GCTfh cells appear to act as principal regulators of affinity maturation by selecting higher affinity GCB cells in the LZ to return to the DZ for additional rounds of mutation and proliferation (Allen et al., 2007; Gitlin et al., 2014). GCTfh cells also direct the output of plasmacyte and memory B cell progeny (Foy et al., 1994; Han et al., 1995; Ise et al., 2018; Randall et al., 1998; Takahashi et al., 1998). Understanding how GCTfh cells guide these humoral reactions is critical to understanding the strength, breadth, and persistence of antibody responses.
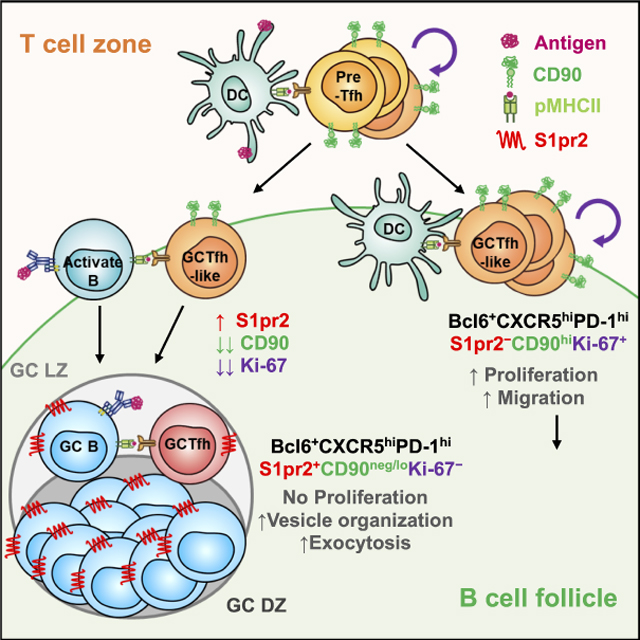
We showed by photoactivation and S1pr2-driven labeling that in primary responses GC-resident Tfh cells are in fact a small subset of the CD4+FoxP3−Bcl6+CXCR5hiPD-1hi population usually designated “GCTfh”; the former can be identified by reduced or absent expression of CD90. This observation is not novel (Harriman et al., 1990; Zheng et al., 1996), but largely has been neglected. Using CD90 expression to enrich GC-resident Tfh cells from the larger non-resident GCTfh-like cell population allowed the demonstration of distinctive physiologies for these T-cell subsets, which are otherwise phenotypically similar absent artificial genetic marking systems. Loss of CD90 on resident GCTfh cells was observed for multiple immunogens and in the chronic GCs of PPs. Given that TCR signaling in CD4+ cells is impaired in CD90-deficient mice or by CD90-blocking antibody (Beissert et al., 1998; Furlong et al., 2017), decreased CD90 on GCTfh cells may be a mechanism for increasing TCR triggering thresholds, perhaps to avoid exhaustion by repetitive interaction with GCB cells (Good-Jacobson et al., 2010). Alternatively, the CD90neg/lo phenotype may represent a novel or unrecognized specialization.
In primary humoral responses, activated Tfh cells leave the T-B border for follicles at day 3, shortly before antigen-activated B cells do the same (Kerfoot et al., 2011; Kitano et al., 2011). With B-cell migration, clusters of B cells can be identified at FDC networks, establishing nascent GCs (Kerfoot et al., 2011). In contrast, Tfh cell emigrants are not confined to these primitive GCs, but distribute throughout the follicle (Kerfoot et al., 2011). During this early phase of the response, we observed only CD90hi GCTfh-like cells. Reduction of CD90 was observed only after day 5, coincident with the organization of GC LZs and DZs, suggesting that CD90neg/lo GCTfh cells are a component of GC organization. Although peptide MHCII (pMHCII) presentation on B cells was necessary for the differentiation of CD90neg/lo GCTfh cells, it was not required for the generation of CD90hi GCTfh-like cells. This dichotomy explains how robust Tfh cell migration into follicles, but not FDC networks, can be driven solely by peptide-pulsed DCs (Xu et al., 2013). Together, these findings imply that commitment to the CD90neg/lo GCTfh cell fate is not fixed until DC-activated CD90hi GCTfh-like cells interact with antigen-presenting B cells, and perhaps GCB cells.
The origin of CD90neg/lo GCTfh cells is linked to the CD90hi GCTfh-like cell compartment: some 50% of all Tcrβ rearrangements from CD90neg/lo GCTfh cells were shared with the CD90hi GCTfh-like population. This sharing was stable over time but the reverse was not: Tcrβ rearrangements from CD90hi GCTfh-like cells diverged from those of the CD90neg/lo GCTfh subset as the response progressed. The most likely explanation for this asymmetric divergence is that as GC responses progress past day 5, input into the non-dividing CD90neg/lo GCTfh subset ends, whereas recruitment, activation and differentiation of CD90hi GCTfh-like cells persists. We surmise that early in the response, when antigen is abundant, DC-activated pre-Tfh cells become CD90hi GCTfh-like cells and in turn, may become CD90neg/lo GCTfh cells on cognate interaction with antigen-presenting B cells. Later, when most antigen is retained by DCs and FDCs (Baumjohann et al., 2013; Heesters et al., 2013), pre-Tfh cells can still be activated by DCs but have little chance of encountering activated B cells at the T:B border; consequently, their differentiation is limited to CD90hi GCTfh-like cells. Successful GCTfh cell development requires the antigen-specific T cell clone to experience serial activation and selection from both DCs and B cells. In the event that pMHCII complexes of DC and B cells differ, the divergence of TCR repertoires in the CD90neg/lo GCTfh and CD90hi GCTfh-like cell compartments might represent selection.
The concept of differing selection by distinct antigen-presenting cell types is consistent with our finding that neither OT-II nor DO11.10 T cells efficiently differentiated into CD90neg/lo GCTfh cells but were highly competent to generate eff non-Tfh and GCTfh-like cells in the presence of endogenous T-cell competitors. Both DO11.10 (Murphy et al., 1990) and OT-II (Barnden et al., 1998) transgenic mouse lines express TCRs recovered from T-cell hybridomas generated from CD4+ T cells selected for continued proliferation in vitro in response to irradiated, Ova-pulsed splenocytes (Barnden et al., 1998; White et al., 1983). Given that splenic B cells’ ability to present antigen and co-stimulatory signals is radiosensitive, it is likely that selection of Ova-specific blasts was driven by myeloid-derived antigen-presenting cells (Ashwell et al., 1988). We infer that the conditions were not optimal for selecting T-cell clones with high potential for GCTfh cell differentiation.
GCTfh cells provide survival and proliferation signals to promote proliferative expansion by higher-affinity GCB cells (Gitlin et al., 2014; Schwickert et al., 2011). This task does not require – and might even be impaired by – GCTfh cell proliferation (Crotty, 2014). Additionally, limiting GCTfh cell numbers may help prevent the dysregulated GC expansion observed in autoimmunity (Vinuesa et al., 2009); indeed, whereas Tfh cells can produce IL-2, they are resistant to IL-2-driven proliferation (Ballesteros-Tato et al., 2012; DiToro et al., 2018). A recent study, however, showed that CXCR5hiPD-1hi Tfh cells continue to divide during the GC reaction (Merkenschlager et al., 2021). Our work now demonstrates the proliferative CXCR5hiPD-1hi Tfh cells do not include CD90neg/lo GC-resident Tfh cells but rather the CD90hi GCTfh-like cell compartment that carries a transcriptomic signature of persistent cell activation. These CD90hi GCTfh-like cells are generated in the absence of B-cell antigen-presentation, but presumably interact with DCs (Baumjohann et al., 2011; Merkenschlager et al., 2021). Another study reported a quiescent Bcl6-Low Tfh population during GC responses that has some similarities to CD90neg/lo GCTfh cells (Kitano et al., 2011). However, unlike the Bcl6-Low Tfh cells, CD90neg/lo GC-resident Tfh cells did not upregulate Klf2, Il7r, Ccr7, or S1pr1 transcripts; in fact, CD90neg/lo GCTfh cells downregulated Klf2, Ccr7, S1pr1 and other migration-related genes.
Using CD90 expression to enrich GCTfh from GCTfh-like cells, we demonstrated distinctive physiologies between these phenotypically similar T-cell subsets. The population generally described as “GCTfh” cells is, in fact, a composite of subpopulations with dramatic transcriptomic differences with only a minority representing true GC-resident Tfh cells. Prior studies of GCTfh cells using the Bcl6+CXCR5hiPD-1hi phenotype would have encompassed both the dominant GCTfh-like and the less abundant CD90neg/lo GCTfh cells. Indeed, the GCTfh-cell gene signature identified by some of those studies (Choi et al., 2015; Liu et al., 2014; Wing et al., 2017) was highly evident in CD90neg/lo GCTfh cells but less so in GCTfh-like cells; this disparity increased over time as the GC response proceeded from d8 to d24. This observation agrees with previous findings that genes essential for Tfh cell function were expressed most abundantly in S1pr2hi GCTfh cells (Moriyama et al., 2014). Some Tfh-related genes were comparably expressed by CD90neg/lo GCTfh and CD90hi GCTfh-like cells, e.g., Batf, Btla, Cd40lg, Cxcr5, Il4, Il6ra, Il21, Maf and Sh2d1a (Choi et al., 2015; Wing et al., 2017). These are presumably important for initial Tfh cell development or common functions in B-cell follicles. Finally, the expression of genes related to cell migration and chemotaxis was significantly lower in CD90neg/lo GCTfh cells than in CD90hi GCTfh-like cells, a finding also noted in comparisons of S1pr2hi and S1pr2low Tfh cells (Moriyama et al., 2014); this difference likely reflects their anatomical segregation in and outside of GCs.
Cytokine production by GCTfh cells is limited (Dan et al., 2016), perhaps to focus helper activity to individual GCB cells to avoid bystander activity (Dan et al., 2016; Wan et al., 2019). Indeed, the essential functions of GCTfh are thought to be the repeated expression of membrane CD154 and delivery of neurotransmitters across the T:B-cell synapse (Papa et al., 2017; Wan et al., 2019). These findings are fully consistent with our RNASeq data showing that CD90neg/lo GCTfh cells are specialized for endosomal/vesicle organization and exocytosis/degranulation. Since cognate GCTfh cell interactions with GCB cells are brief, lasting ≤5 minutes on average (Shulman et al., 2014), a transcriptome enriched for exocytosis and vesicle transport is consistent with CD90neg/lo GCTfh cells’ being capable of efficient and individualized help to GCB cells via immune synapses (Papa and Vinuesa, 2018). Interestingly, transfer of CD154 across the immunological synapse by vesicles to antigen-presenting B cells occurs in vitro (Gardell and Parker, 2017), raising the possibility of T-cell-help “to go” for GCB cells (Dustin, 2017). That CD90neg/lo GCTfh cells are enriched for vesicle organization and exocytosis pathways is consistent with synapse-dependent help and provides in vivo evidence to support the “help to-go” hypothesis (Dustin, 2017). That GCTfh cells can transfer microRNA to GCB cells via extracellular vesicles at synapse formation also supports the potential role of GCTfh cell exosomes in GC development and antibody production (Fernandez-Messina et al., 2020).
GC-resident CD90neg/lo GCTfh cells are spatially, functionally and physiologically distinct from CD90hi GCTfh-like cells, despite sharing the Bcl6+CXCR5hiPD-1hi phenotype. Whereas these two subsets appear to share a common origin, the Tcrβ repertoire differences imply a distinct program of clonal activation and selection for these cohorts, perhaps as a consequence of fate determination driven by cognate interaction with B cells. Regardless of the exact mechanisms that drive this differentiation, identification of CD90neg/lo GCTfh cells has revealed a previously obscured transcriptional program for GC-resident Tfh cells that implies the delivery of individualized help to GCB cells by vesicle exocytosis. Furthermore, the role of residual CD90hi GCTfh-like cells outside the GCs is unclear. Additional investigation of these different Tfh cell subsets will likely provide novel insights into how T and B cells collaborate during humoral responses to protein antigens.
Limitations
Our study focuses only on murine Tfh cells participating in primary or chronic GC responses. By histology, all GC-resident Tfh cells downregulate CD90 but only 40% become RFP+ by S1pr2-driven Cre activity. With flow cytometry, all RFP+ Tfh cells reduced CD90 expression as did an equivalent population of RFP− Tfh; we assume these CD90neg/lo RFP− Tfh cells represent the RFP− Tfh cells observed histologically in the GC LZ. We cannot exclude the possibility of S1pr2-independent CD90neg/lo GC-resident Tfh cells. The Zbtb46-DTR model is useful for only short periods (≤8 d) of DC depletion, this limits the window for determining the role of cDCs in GCTfh and GCTfh-like cell differentiation.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Requests for further information, resources and reagents should be directed to the Lead Contact, Garnett Kelsoe (garnett.kelsoe@duke.edu).
Materials availability
All materials in this study are available from the lead contact upon reasonable request.
Data and code availability
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. All Tcrβ sequence data sets are available from Adaptive Biotechnologies immuneACCESS and are publicly available as of the date of publication. Direct links are listed in the key resources table. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-mouse CD3 AF488 (Clone 17A2) | BioLegend | Cat#100210; RRID: AB_389301 |
| anti-mouse CD3 AF647 (Clone 145-2C11) | BioLegend | Cat#100324; RRID: AB_492861 |
| anti-mouse CD4 Biotin (Clone GK1.5) | BioLegend | Cat#100404; RRID: AB_312689 |
| anti-mouse CD4 AF647 (Clone GK1.5) | BioLegend | Cat#100424; RRID: AB_389324 |
| anti-mouse CD4 AF647 (Clone RM4-5) | BioLegend | Cat#100533; RRID: AB_493372 |
| anti-mouse CD4 BV421 (Clone GK1.5) | BioLegend | Cat#100443; RRID: AB_2562557 |
| anti-mouse CD4 BV510 (Clone RM4-5) | BD Biosciences | Cat#563106; RRID: AB_2687550 |
| anti-mouse CD8 BV421 (Clone 53-6.7) | BD Biosciences | Cat#563898; RRID: AB_2738474 |
| anti-mouse CD11b PE-Cy7 (Clone M1/70) | BioLegend | Cat#101215; RRID: AB_312798 |
| anti-mouse CD11c Biotin (Clone N418) | BioLegend | Cat#117304; RRID: AB_313773 |
| anti-mouse CD11c AF647 (Clone N418) | BioLegend | Cat#117312; RRID: AB_389328 |
| anti-mouse CD19 APC-R700 (Clone 1D3) | BD Biosciences | Cat#565473; RRID: AB_2739253 |
| anti-mouse CD21/35 AF594 (Clone 7E9) | BioLegend | Cat#123426; RRID: AB_2632698 |
| anti-mouse CD21/35 AF647 (Clone 7E9) | BioLegend | Cat#123424; RRID: AB_2629578 |
| anti-mouse CD25 BV421 (Clone PC61) | BioLegend | Cat#102043; RRID: AB_2562611 |
| anti-mouse CD43 Biotin (Clone S7) | BD Biosciences | Cat#553269; RRID: AB_2255226 |
| anti-mouse/human CD44 FITC (Clone IM7) | BioLegend | Cat#103006; RRID: AB_312957 |
| anti-mouse/human CD44 PerCP-Cy5.5 (Clone IM7) | BioLegend | Cat#103006; RRID: AB_2076204 |
| anti-mouse/human CD44 PE-594 (Clone IM7) | BioLegend | Cat#103056; RRID: AB_2564044 |
| anti-mouse CD45.1 BV421 (Clone A20) | BioLegend | Cat#110732; RRID: AB_2562563 |
| anti-mouse CD45.2 FITC (Clone 104) | Thermo Fisher Scientific | Cat#11-0454-82; RRID: AB_465061 |
| anti-mouse CD62L PerCP-Cy5.5 (Clone MEL-14) | BioLegend | Cat#104430; RRID: AB_2187124 |
| anti-mouse CD62L PE (Clone MEL-14) | BioLegend | Cat#104408; RRID: AB_313095 |
| anti-mouse CD62L BV786 (Clone MEL-14) | BD Biosciences | Cat#564109; RRID: AB_2738598 |
| anti-mouse CD69 PE-Cy5 (Clone H1.2F3) | BioLegend | Cat#104510; RRID: AB_313113 |
| anti-mouse CD90.2 Biotin (Clone 30H12) | BioLegend | Cat#105304; RRID: AB_313175 |
| anti-mouse CD90.2 AF488 (Clone 30H12) | BioLegend | Cat#105316; RRID: AB_492886 |
| anti-mouse CD90.2 PE (Clone 30H12) | BioLegend | Cat#105307; RRID: AB_313178 |
| anti-mouse CD90.2 AF647 (Clone 30H12) | BioLegend | Cat#105318; RRID: AB_492888 |
| anti-mouse CD90.2 AF700 (Clone 30H12) | BioLegend | Cat#105320; RRID: AB_493725 |
| anti-mouse CD90.2 AF700 (Clone 53-2.1) | BioLegend | Cat#140324; RRID: AB_2566740 |
| anti-mouse CD90.2 BV421 (Clone 53-2.1) | BioLegend | Cat#140327; RRID: AB_2686992 |
| anti-mouse CD138 BV308 (Clone 281-2) | BD Biosciences | Cat#563147; RRID: AB_2721029 |
| anti-mouse CD185 (CXCR5) Biotin (Clone L138D7) | BioLegend | Cat#145510; RRID: AB_2562126 |
| anti-mouse CD185 (CXCR5) Biotin (Clone 2G8) | BD Biosciences | Cat#551960; RRID: AB_394301 |
| anti-mouse CD279 (PD-1) PE-Cy7 (Clone 29F.1A12) | BioLegend | Cat#135216; RRID: AB_10689635 |
| anti-mouse CD279 (PD-1) BV421 (Clone 29F.1A12) | BioLegend | Cat#135221; RRID: AB_2562568 |
| anti-mouse/human B220 APC/Fire750 (Clone RA3-6B2) | BioLegend | Cat#103259; RRID: AB_2572108 |
| anti-mouse/human B220 BV605 (Clone RA3-6B2) | BioLegend | Cat#103244; RRID: AB_2563312 |
| anti-mouse/human B220 BV785 (Clone RA3-6B2) | BioLegend | Cat#103246; RRID: AB_2563256 |
| anti-mouse/human Bcl-6 PE (Clone K112-91) | BD Biosciences | Cat#561522; RRID: AB_10717126 |
| anti-mouse/human Bcl-6 PE-CF594 (Clone K112-91) | BD Biosciences | Cat#562401; RRID: AB_11152084 |
| anti-mouse/human Bcl-6 AF647 (Clone K112-91) | BD Biosciences | Cat#561525; RRID: AB_10898007 |
| anti-mouse DO11.10 TCR FITC (Clone KJ1-26) | Thermo Fisher Scientific | Cat#11-5808-82; RRID: AB_465248 |
| anti-mouse F4/80 Biotin (Clone BM8) | BioLegend | Cat#123106; RRID: AB_893501 |
| anti-mouse F4/80 PE (Clone BM8) | BioLegend | Cat#123110; RRID: AB_893486 |
| anti-mouse FoxP3 AF488 (Clone FJK-16s) | Thermo Fisher Scientific | Cat#53-5773-82; RRID: AB_763537 |
| anti-mouse FoxP3 AF647 (Clone MF23) | BD Biosciences | Cat#560401; RRID: AB_1645201 |
| anti-GFP AF488 (Clone FM264G) | BioLegend | Cat#338008; RRID: AB_2563288 |
| anti-mouse GL7 FITC (Clone GL7) | BD Biosciences | Cat#553666; RRID: AB_394981 |
| anti-mouse GL7 PE (Clone GL7) | BD Biosciences | Cat#561530; RRID: AB_10715834 |
| anti-mouse Gr-1 PE (Clone RB6-8C5) | BioLegend | Cat#108404; RRID: AB_313369 |
| anti-human HB-EGF (Goat Polyclonal) | R&D Systems | Cat#BAF259; RRID: AB_2114598 |
| anti-mouse I-A/I-E (MHCII) AF647 (Clone M5/114.15.2) | BioLegend | Cat#107618; RRID: AB_493525 |
| anti-mouse I-A/I-E (MHCII) BV711 (Clone M5/114.15.2) | BD Biosciences | Cat#563414; RRID: AB_2738191 |
| anti-mouse I-Ab (MHCII) PE-CF594 (Clone AF6-120.1) | BD Biosciences | Cat#562824; RRID: AB_2737819 |
| anti-mouse IgD BV421 (Clone 11-26c.2a) | BioLegend | Cat#405725; RRID: AB_2562743 |
| anti-mouse IgD BV510 (Clone 11-26c.2a) | BioLegend | Cat#405723; RRID: AB_2562742 |
| anti-mouse Ig κ light chain BV421 (Clone 187.1) | BD Biosciences | Cat#562888; RRID: AB_2737867 |
| anti-mouse Ig λ1, λ2, & λ3 light chain FITC (Clone R26-46) | BD Biosciences | Cat#553434; RRID: AB_394854 |
| anti-mouse/human Ki-67 BV421 (Clone 11F6) | BioLegend | Cat#151208; RRID: AB_2629748 |
| anti-mouse/human Ki-67 BV421 (Clone B56) | BD Biosciences | Cat#562899; RRID: AB_2686897 |
| anti-mouse/human Ki-67 AF647 (Clone B56) | BD Biosciences | Cat#558615; RRID: AB_647130 |
| anti-mouse/human Ki-67 BV650 (Clone B56) | BD Biosciences | Cat#563757; RRID: AB_2688008 |
| anti-mouse Ly6C FITC (Clone AL21) | BD Biosciences | Cat#553104; RRID: AB_394628 |
| anti-RFP (Rabbit Polyclonal) | Rockland | Cat#600-401-379; RRID: AB_2209751 |
| anti-mouse TCRβ FITC (Clone H57-597) | BioLegend | Cat#109205; RRID: AB_313428 |
| anti-mouse TCRβ PE-Cy7 (Clone H57-597) | BioLegend | Cat#109221; RRID: AB_893627 |
| anti-mouse TCRβ BV711 (Clone H57-597) | BD Biosciences | Cat#563135; RRID: AB_2738023 |
| anti-mouse TCR Vβ5.1/5.2 PE (Clone MR9-4) | BD Biosciences | Cat#562086; RRID: AB_394698 |
| anti-mouse Ter-119 Biotin (Clone TER-119) | BioLegend | Cat#116204; RRID: AB_313705 |
| anti-Rabbit IgG (H+L) AF594 (Goat Polyclonal) | Thermo Fisher Scientific | Cat#A-11012; RRID: AB_ 2534079 |
| anti-mouse CD16/CD32 (Mouse BD Fc Block™) | BD Biosciences | Cat#553142; RRID: AB_394656 |
| Chemicals, peptides, and recombinant proteins | ||
| Acetone | Sigma-Aldrich | Cat#179124 |
| Alhydrogel® adjuvant 2% | InvivoGen | Cat#vac-alu-250 |
| Corn oil | Sigma-Aldrich | Cat#PHR2897 |
| Diphtheria Toxin from Corynebacterium diphtheriae | Sigma-Aldrich | Cat#D0564 |
| IgG from rat serum | Sigma-Aldrich | Cat#I4131 |
| Methanol | Sigma-Aldrich | Cat#34860 |
| NP10-HSA (Human Serum Albumin) | Biosearch Technologies | Cat#N-5059-10 |
| NP15-Ova (Ovalbumin) | Biosearch Technologies | Cat#N-5051-100 |
| Ovalbumin | Biosearch Technologies | Cat#O-1000-100 |
| recombinant PA (Protective antigen) | ||
| recombinant HA (A/Solomon Islands/3/2006) | S. Harrison | (Schmidt et al., 2015) |
| 16% Formaldehyde (w/v), Methanol-free | Thermo Fisher Scientific | Cat#28906 |
| Streptavidin AF488 | BioLegend | Cat#405235 |
| Streptavidin PE | BioLegend | Cat#405204 |
| Streptavidin APC | BioLegend | Cat#405207 |
| Streptavidin BV421 | BioLegend | Cat#405225 |
| Streptavidin BV650 | BioLegend | Cat#405232 |
| Sucrose | Sigma-Aldrich | Cat#S9378 |
| Tamoxifen | Sigma-Aldrich | Cat#T5648 |
| Tissue-Tek® O.C.T. Compound | Sakura Finetek USA | Cat#4583 |
| Critical commercial assays | ||
| Direct-zol RNA Miniprep | Zymo Research | Cat#R2051 |
| immunoSEQ mouse Tcrβ assay | Adaptive Biotechnologies | https://www.immunoseq.com/assays/ |
| KAPA HyperPlus Kit | Roche Molecular Systems | Cat#07962401001 |
| KAPA Pure Beads | Roche Molecular Systems | Cat#07983280001 |
| LIVE/DEAD™ Fixable Near-IR Dead Cell Stain Kit | Thermo Fisher Scientific | Cat#L34976 |
| Pan T Cell Isolation Kit II | Miltenyi Biotec | Cat#130-095-130 |
| Qubit dsDNA HS Assay Kit | Thermo Fisher Scientific | Cat#Q32851 |
| Qubit RNA IQ Assay | Thermo Fisher Scientific | Cat#Q33222 |
| RNA 6000 Pico kit | Agilent | Cat#5067-1513 |
| SMART-Seq® v4 Ultra Low Input RNA Kit | Takara Bio | Cat#634890 |
| Streptavidin MicroBeads | Miltenyi Biotec | Cat#130-048-101 |
| Transcription Factor Buffer Set | BD Biosciences | Cat#562574 |
| Deposited data | ||
| RNA-seq data | This paper | GSE147035 |
| Tcrβ sequence data | This paper |
https://clients.adaptivebiotech.com/pub/yeh-2021-immunity
DOI: https://dx.doi.org/10.21417/CY2021I |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J (B6) | The Jackson Laboratory | JAX: 000664 |
| Mouse: B6.129S2-H2dlAb1-Ea/J (MHCII−/−) | The Jackson Laboratory | JAX: 003584 |
| Mouse: B6.SJL-Ptprca Pepcb/BoyJ (B6.CD45.1) | The Jackson Laboratory | JAX: 002014 |
| Mouse: B6.Cg-Ptprca Tg(UBC-PA-GFP)1Mnz/J (PAGFP) | The Jackson Laboratory | JAX: 022486 |
| Mouse: B6.Cg-Foxp3tm2Tch/J (FoxP3EGFP) | The Jackson Laboratory | JAX: 006772 |
| Mouse: B6(Cg)-Zbtb46tm1(HBEGF)Mnz/J (zbtb46-DTR) | The Jackson Laboratory | JAX: 019506 |
| Mouse: B6.129S2-Ighmtm1Cgn/J (μMT) | The Jackson Laboratory | JAX: 002288 |
| Mouse: B6.C(Cg)-Cd79atm1(cre)Reth/EhobJ (Mb1Cre) | The Jackson Laboratory | JAX: 020505 |
| Mouse: B6.Cg-Tg(TcraTcrb)425Cbn/J (OT-II Tg) | The Jackson Laboratory | JAX: 004194 |
| Mouse: C57BL/6-Gt(ROSA)26Sortm1(HBEGF)Awai/J (DTRLSL) | The Jackson Laboratory | JAX: 007900 |
| Mouse: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | The Jackson Laboratory | JAX: 007914 |
| Mouse: Balb/cJ (Balb/c) | The Jackson Laboratory | JAX: 000651 |
| Mouse: C.Cg-Tg(DO11.10)10Dlo/J (DO11.10 Tg) | The Jackson Laboratory | JAX: 003303 |
| Mouse: CByJ.SJL(B6)-Ptprca/J (Balb/c.CD45.1) | The Jackson Laboratory | JAX: 006584 |
| Mouse: Tg(S1pr2-cre/ERT2)#Kuro (S1pr2ERT2Cre) | (Shinnakasu et al., 2016) | MGI: 6435090 |
| Mouse: Ightm2Cgn (B1-8i) | (Sonoda et al., 1997) | MGI:2388486 |
| Mouse: B6.H50Gμ | (Dal Porto et al., 2002) | N/A |
| Software and algorithms | ||
| BD FACSDiva Software | BD Biosciences | http://www.bdbiosciences.com/instruments/software/facsdiva/index.jsp RRID:SCR_001456 |
| Bioconductor | Huber et al., 2015 | http://www.bioconductor.org/ RRID:SCR_006442 |
| Cytoscape | National Institute of General Medical Sciences | http://cytoscape.org RRID:SCR_003032 |
| DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html RRID:SCR_015687 |
| EnrichmentMap | Merico et al., 2010 | http://baderlab.org/Software/EnrichmentMap RRID:SCR_016052 |
| Fiji (ImageJ) | National Institutes of Health | http://fiji.sc/ RRID: SCR_002285 |
| FlowJo | BD | https://www.flowjo.com/solutions/flowjo RRID:SCR_008520 |
| GraphPad Prism_V9 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ RRID: SCR_002798 |
| HTSeq tool | European Molecular Biology Laboratory | http://www-huber.embl.de/users/anders/HTSeq/ RRID:SCR_004473 |
| immunoSEQ ANALYZER | Adaptive Biotechnologies (Carlson et al., 2013) | https://clients.adaptivebiotech.com/ |
| STAR RNAseq alignment tool | Dobin et al., 2013 | http://code.google.com/p/rna-star/ RRID:SCR_016533 |
| Trim Galore toolkit | Martin, 2011 | http://www.bioinformatics.babraham.ac.uk/projects/trim_galore RRID:SCR_011847 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice and immunizations
B6, B6.CD45.1, MHCII−/−, PAGFP, FoxP3EGFP, Zbtb46-DTR, μMT, Mb1Cre, OT-II Tg, DTRLSL, DO11.10 Tg and Balb/c.CD45.1 mice were purchased from the Jackson Laboratory (see KEY RESOURCES TABLE). S1pr2ERT2Cre-Rosa26lox-stop-lox-tdTomato mice (Shinnakasu et al., 2016) were provided by T. Kurosaki at Osaka University, B1-8i mice (Sonoda et al., 1997) were provided by K. Rajewsky at MDC Berlin, and B6.H50Gμ mice originated in our laboratory (Dal Porto et al., 1998). All mice were maintained under specific pathogen-free, temperature- and humidity-controlled conditions at the Duke University Animal Care Facility and used in experiments at 8 to 12 weeks of age. Due to the limited availability of special strains and chimeric mice, no randomization was used. The investigator was not blinded to group allocation during the animal experiments. Sample size to ensure adequate statistical power was based on prior experience in the laboratory. Mice were footpad-immunized with 20 μg of NP15-Ova, NP10-HSA, Ova, rPA, or rHA (A/Solomon Islands/3/2006)(Schmidt et al., 2015) in Alhydrogel ® adjuvant 2% (1:1, v/v) in a final volume of 20 μL. Draining pLN samples were collect at indicated time points post-immunization. Deletion of the loxP-flanked STOP cassette in S1pr2ERT2Cre-Rosa26lox-stop-lox-tdTomato mice was induced by i.p. injection of 5 mg tamoxifen in corn oil once daily on day 5–7. Depletion of Mb1CrexDTRLSL B cells or Zbtb46-DTR cDCs in chimeric mice was induced by i.p. injection of 20 ng/gwt diphtheria toxin in PBS at indicated starting time points, followed by 4 ng/gwt injection every two days. All experimental procedures involving animals were approved by the Duke University Institutional Animal Care and Use Committee.
METHOD DETAILS
Antibodies and flow cytometry
For surface marker detection, cells were suspended in PBS containing 0.5% bovine serum albumin, 0.1% sodium azide and 1mM EDTA (FACS buffer). Samples were blocked with rat anti-mouse CD16/32 and rat IgG in FACS buffer for 30 minutes and stained with fluorochrome-conjugated antibodies at 4°C for 30–40 minutes (for antibody clones see KEY RESOURCES TABLE). LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit was used to exclude dead cells. For nuclear or intracellular staining, samples were fixed, permeabilized, and stained using BD Transcription Factor Buffer Set. Subsequently, cells were washed twice with FACS buffer, and the resuspended cells were then analyzed on LSRII or LSRFortessa cell analyzers (BD Biosciences). Fluorescence-activated cell sorting was performed with a FACSAria sorter (BD Biosciences). Data analysis was performed with FACSDiva and FlowJo software. Cell gating strategies are described in Figs. S1, S3A, S3I and S7A. Flow cytometry was performed in the Duke Human Vaccine Institute Flow Cytometry Facility (Durham, NC).
Mutiphoton imaging and photoactivation
PAGFP mice were footpad-immunized with 20 μg of NP-Ova+alum. AF594-conjugated anti-CD21/CD35 antibody (5 μg) was s.c. injected into the hock 16–24 hours prior to tissue harvest. Draining pLNs were harvested and immediately embedded in 4% low-melting-point agarose, followed by sectioning into 250 μm slices with a Leica VT1200S vibratome. LN slices were firmly attached to the bottom of cell culture dish filled with 1x HBSS. All imaging was performed on a Leica SP8 multi-photon DIVE microscope fitted with a 25X 1.05NA dipping objective and two tunable fSec Ti:Saph lasers (680–1080 nm and 680–1300 nm). Background GFP and FDC networks labeled with AF594-conjugated anti-CD21/CD35 antibody were visualized using λ = 940 nm and 1100 nm excitation, simultaneously. GC area were photoactivated using λ = 830 nm light and the photoactivated area was subsequently visualized with λ = 940 and 1100 nm excitation light. The photoactivated tissue sections were recovered and subjected to flow cytometric analysis.
Adoptive B cell transfer and mixed bone marrow chimeric mice
For short-term cell transfers, single-B-cell suspensions were harvested and processed from spleens of B1-8i or Mb1CrexDTRLSL mice. Splenocytes were first stained with a mixture of biotinylated-Abs (anti-CD4, anti-CD11c, anti-CD43, anti-CD90.2, anti-F4/80, anti-Gr-1 and anti-Ter119) and subsequently labeled with Streptavidin MicroBeads. B cells were then negatively purified using magnet-activated cell sorting in a CS column on a VarioMACS separator (Miltenyi Biotec). For B1-8i B cells, B cell-enriched samples were stained and sorted using flow cytometry to acquire B220+Igλ+IgD+ B1-8i B cells. Single-cell suspension containing indicated numbers of purified B cells in 200 μL PBS were i.v. transferred to individual recipient mice. To generate mixed BM chimeric mice, C57BL/6 mice were lethally irradiated with two doses of 600 rad X-ray 3 hours apart and then i.v. injected 5 × 106 mixed BM cells. The BM mixture were made with 80:20 ratio of BM cells harvested from B6 or μMT and MHCII−/− mice. Reconstituted mice were rested for 8 weeks before use in experiments. To generate zbtb46-DTR chimeric mice, C57BL/6 mice were lethally irradiated with two doses of 600 rad X-ray 3 hours apart and then i.v. injected with 5 × 106 of BM cells harvested from B6 or zbtb46-DTR mice. Reconstituted mice were rested for 8 weeks before use in experiments.
Adoptive T cell transfer
For transgenic T cell transfers, single-cell suspensions were harvested and processed from spleens of CD45.1+ OT-II or DO11.10 TCR transgenic mice. Splenocytes were first stained with a mixture of biotinylated-Abs (anti-CD8a, anti-CD11b, anti-CD11c, anti-CD19, anti-CD25, anti-CD45R (B220), anti-CD49b (DX5), anti-CD105, Anti-MHCII, anti-Ter-119, and anti-TCRγ/δ) and subsequently labeled with Streptavidin MicroBeads. CD4+ T cells were then negatively purified using magnet-activated cell sorting in a CS column on a VarioMACS separator (Miltenyi Biotec). T cell-enriched samples were stained and analyzed using flow cytometry to determine the purity and percentage of transgenic TCR-bearing populations. Single cell suspensions containing 2 × 106 of CD4+ T cells in 200 μL PBS were i.v. transferred to individual recipient CD45.2+ B6 or Balb/c mice. Reconstituted mice were rested overnight (16–24 hours) before immunization.
Immunofluorescence staining and microscope
Harvested pLN samples were embedded in Tissue-Tek OCT Compound and frozen at −80°C. Cryosectioning was performed on a Leica CM1850 Cryostat and fixed in cold acetone/methanol (1:1) at −20°C for 10 minutes. For S1pr2-RFP mice, the pLN samples were pre-fixed with 1% PFA overnight, followed by gradient sucrose dehydration. Tissue sections (5–10 μm-thick) were mounted on glass slides and rehydrated by soaking in wash solution (PBS containing 0.5% BSA and 0.1% Tween-20) at RT for 30 minutes. Samples were then blocked with rat anti-mouse CD16/CD32 and rat IgG for 15 min at room temperature. After washing, the samples were incubated with antibodies for CD3 (17A2 or 145-2C11), CD4 (GK1.5 or RM4-5), CD21/CD35 (7E9), CD90.2 (53-2.1 or 30-H12), Bcl6 (K112-91), Ki-67 (11F6), IgD (11-26c.2a) and anti-RFP Ab in a humid, dark chamber for 3 hours at RT or 4°C overnight (see KEY RESOURCES TABLE). After washing, the samples were then incubated with secondary or enhancing antibodies for 1 hour at RT. Images were acquired by confocal microscopy using a Zeiss LSM 780 confocal microscope. Image processing, including counting cells in GCs, was performed using ImageJ software (Fiji package).
DNA extraction and deep sequencing for Tcrβ repertoire analysis
The TCRβ repertoire of CD4+ T cells was analyzed using the immunoSEQ mouse Tcrβ assay (Adaptive Biotechnologies; (Carlson et al., 2013)). FoxP3EGFP mice were footpad-immunized with 20μg of NP-Ova+alum. pLNs were harvested at indicated time points. CD90neg/lo GCTfh and CD90hi GCTfh-like cells sorted from the same pLN were subjected to genomic DNA extraction using a phenol/chloroform method (Kuraoka et al., 2009). Isolated genomic DNA was sent to Adaptive Biotechnologies, which performed multiplex PCR amplification of all possible rearranged Tcrb genes from gDNA samples and high-throughput deep sequencing using Illumina HiSeq platform. The raw HiSeq sequence data were preprocessed to remove errors and to compress the data. Tcrβ sequences were characterized and analyzed with the Adaptive immunoSEQ Analyzer (Carlson et al., 2013).
RNA extraction, library preparation and sequencing
RNA was extracted from sorted cell populations using a Direct-zol RNA Kit. RNA quality and concentration were determined with a Qubit 4.0 fluorimeter (Thermo Fisher Scientific) with the RNA IQ Assay and a Bioanalyzer (Agilent) with Agilent RNA 6000 Pico Kit. Only samples with RIN > 8 were proceed to reverse transcription. cDNA was synthesized with the SMART-Seq® v4 Ultra Low Input RNA Kit following manufacturer’s recommendations. Adapters were used as priming sites for cDNA synthesis and downstream PCR to amplify the cDNA. Amplified cDNA was purified using KAPA Pure Beads, and the yield and quality were determined with a Qubit 4.0 fluorimeter using the dsDNA HS Assay Kit. The DNA library was constructed using a KAPA HyperPlus Kit, following the manufacturer’s recommendations. Sequencing was performed using a HiSeq 4000 system (Illumina) at the Duke University Center for Genomic and Computational Biology.
RNA Sequencing data analysis
RNAseq data were processed using the TrimGalore toolkit which employs Cutadapt (Martin, 2011) to trim low-quality bases and Illumina sequencing adapters from the 3’ end of the reads. Only reads that were 20 nucleotides or longer after trimming were retained for further analysis. Reads were mapped to the GRCm38v73 version of the mouse genome and transcriptome (Kersey et al., 2012) using the STAR RNAseq alignment tool (Dobin et al., 2013). Reads were retained for subsequent analysis if they mapped to a single genomic location. Gene counts were compiled using the HTSeq tool. Only genes that had at least 10 reads in any given library were used in subsequent analyses. Normalization and differential expression analysis was carried out using the DESeq2 (Love et al., 2014) Bioconductor (Huber et al., 2015) package with the R statistical programming environment. The false discovery rate was calculated to control for multiple hypothesis testing. Gene set enrichment analysis was performed to identify gene ontology terms and pathways associated with altered gene expression for each of the comparisons performed (Mootha et al., 2003). Network visualization of gene set enrichment was performed using Cytoscape Version 3.7.2 and the plugin “Enrichment Map” to build the network and the plugin “AutoAnnotate” to build clusters with visual annotations (Merico et al., 2010).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed using ordinary analysis of variance (ANOVA) followed by Tukey’s or Dunnett’s post-tests or two-way ANOVA followed by Sidak’s post-test. Differences were considered statistically significant at P values < 0.05 (*P<0.05; **P<0.01; ***P<0.001; ****P<0.0001). Details about statistical analyses are described in the figure legends. Data were visualized with GraphPad Prism V9.
Supplementary Material
Highlights.
Primary Bcl6+CXCR5hiPD-1hi Tfh cells encompass both GC-resident and non-resident cells
GC-resident Tfh cells are S1pr2+CD90neg/lo; non-resident, GCTfh-like are S1pr2−CD90hi
CD90neg/lo GCTfh and CD90hi GCTfh-like cells have distinct developmental requirements
GCTfh and GCTfh-like cells have distinct TCR repertoires and transcriptomic profiles
Acknowledgements:
We thank J. Garnett, D. Liao, X. Nie, A. Watanabe, X. Liang, W. Zhang and R. Qi for technical assistance. We thank Drs. M. Kuraoka, S. Song and R. Kotaki for helpful discussion and feedback on the manuscript. We thank Drs. R. Ji and C. Jiang for advice and vibratome use. We thank Dr. E.A. Moseman for advice on multi-photon microscopy. We thank Drs. B. Carlson, Y. Gao and L. Cameron of the Duke University Light Microscopy Core Facility for imaging equipment and consultation. We thank the Duke University School of Medicine for the use of the Sequencing and Genomic Technologies / Genomic Analysis and Bioinformatics Shared Resource. This work was supported by AI100645, the Duke Center for HIV/AIDS Vaccine Immunology-Immunogen Discovery (CHAVI-ID), and AI14437, the Duke Consortium for HIV/AIDS Vaccine Development (CHAVD), Division of AIDS, NIAID, NIH and by NIH grants AI089618 and AI117892.
Footnotes
Declaration of Interests
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen CD, Okada T, Tang HL, and Cyster JG (2007). Imaging of germinal center selection events during affinity maturation. Science 315, 528–531. [DOI] [PubMed] [Google Scholar]
- Ansel KM, McHeyzer-Williams LJ, Ngo VN, McHeyzer-Williams MG, and Cyster JG (1999). In vivo-activated CD4 T cells upregulate CXC chemokine receptor 5 and reprogram their response to lymphoid chemokines. J Exp Med 190, 1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwell JD, Jenkins MK, and Schwartz RH (1988). Effect of gamma radiation on resting B lymphocytes. II. Functional characterization of the antigen-presentation defect. J Immunol 141, 2536–2544. [PubMed] [Google Scholar]
- Attwooll C, Lazzerini Denchi E, and Helin K (2004). The E2F family: specific functions and overlapping interests. Embo J 23, 4709–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros-Tato A, Leon B, Graf BA, Moquin A, Adams PS, Lund FE, and Randall TD (2012). Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity 36, 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnden MJ, Allison J, Heath WR, and Carbone FR (1998). Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol 76, 34–40. [DOI] [PubMed] [Google Scholar]
- Baumjohann D, Okada T, and Ansel KM (2011). Cutting Edge: Distinct waves of BCL6 expression during T follicular helper cell development. J Immunol 187, 2089–2092. [DOI] [PubMed] [Google Scholar]
- Baumjohann D, Preite S, Reboldi A, Ronchi F, Ansel KM, Lanzavecchia A, and Sallusto F (2013). Persistent Antigen and Germinal Center B Cells Sustain T Follicular Helper Cell Responses and Phenotype. Immunity 38, 596–605. [DOI] [PubMed] [Google Scholar]
- Beissert S, He HT, Hueber AO, Lellouch AC, Metze D, Mehling A, Luger TA, Schwarz T, and Grabbe S (1998). Impaired cutaneous immune responses in Thy-1-deficient mice. J Immunol 161, 5296–5302. [PubMed] [Google Scholar]
- Bolanos-Garcia VM, and Blundell TL (2011). BUB1 and BUBR1: multifaceted kinases of the cell cycle. Trends Biochem Sci 36, 141–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitfeld D, Ohl L, Kremmer E, Ellwart J, Sallusto F, Lipp M, and Forster R (2000). Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J Exp Med 192, 1545–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CS, Emerson RO, Sherwood AM, Desmarais C, Chung MW, Parsons JM, Steen MS, LaMadrid-Herrmannsfeldt MA, Williamson DW, Livingston RJ, et al. (2013). Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun 4, 2680. [DOI] [PubMed] [Google Scholar]
- Cho S, Lee HM, Yu IS, Choi YS, Huang HY, Hashemifar SS, Lin LL, Chen MC, Afanasiev ND, Khan AA, et al. (2018). Differential cell-intrinsic regulations of germinal center B and T cells by miR-146a and miR-146b. Nat Commun 9, 2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Gullicksrud JA, Xing S, Zeng Z, Shan Q, Li F, Love PE, Peng W, Xue HH, and Crotty S (2015). LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat Immunol 16, 980–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Kageyama R, Eto D, Escobar TC, Johnston RJ, Monticelli L, Lao C, and Crotty S (2011). ICOS Receptor Instructs T Follicular Helper Cell versus Effector Cell Differentiation via Induction of the Transcriptional Repressor Bcl6. Immunity 34, 932–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Yang JA, Yusuf I, Johnston RJ, Greenbaum J, Peters B, and Crotty S (2013). Bcl6 expressing follicular helper CD4 T cells are fate committed early and have the capacity to form memory. J Immunol 190, 4014–4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli KM, Carnathan DG, Nogal B, Martin JT, Rodriguez OL, Upadhyay AA, Enemuo CA, Gebru EH, Choe Y, Viviano F, et al. (2019). Slow Delivery Immunization Enhances HIV Neutralizing Antibody and Germinal Center Responses via Modulation of Immunodominance. Cell 177, 1153–1171 e1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S (2011). Follicular helper CD4 T cells (TFH). Annu Rev Immunol 29, 621–663. [DOI] [PubMed] [Google Scholar]
- Crotty S (2014). T follicular helper cell differentiation, function, and roles in disease. Immunity 41, 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S (2019). T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity 50, 1132–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Porto JM, Haberman AM, Shlomchik MJ, and Kelsoe G (1998). Antigen drives very low affinity B cells to become plasmacytes and enter germinal centers. J Immunol 161, 5373–5381. [PubMed] [Google Scholar]
- Dan JM, Havenar-Daughton C, Kendric K, Al-Kolla R, Kaushik K, Rosales SL, Anderson EL, LaRock CN, Vijayanand P, Seumois G, et al. (2019). Recurrent group A Streptococcus tonsillitis is an immunosusceptibility disease involving antibody deficiency and aberrant TFH cells. Sci Transl Med 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan JM, Lindestam Arlehamn CS, Weiskopf D, da Silva Antunes R, Havenar-Daughton C, Reiss SM, Brigger M, Bothwell M, Sette A, and Crotty S (2016). A Cytokine-Independent Approach To Identify Antigen-Specific Human Germinal Center T Follicular Helper Cells and Rare Antigen-Specific CD4+ T Cells in Blood. J Immunol 197, 983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hollander J, Rimpi S, Doherty JR, Rudelius M, Buck A, Hoellein A, Kremer M, Graf N, Scheerer M, Hall MA, et al. (2010). Aurora kinases A and B are up-regulated by Myc and are essential for maintenance of the malignant state. Blood 116, 1498–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiToro D, Winstead CJ, Pham D, Witte S, Andargachew R, Singer JR, Wilson CG, Zindl CL, Luther RJ, Silberger DJ, et al. (2018). Differential IL-2 expression defines developmental fates of follicular versus nonfollicular helper T cells. Science 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin ML (2017). Help to go: T cells transfer CD40L to antigen-presenting B cells. Eur J Immunol 47, 31–34. [DOI] [PubMed] [Google Scholar]
- Fernandez-Messina L, Rodriguez-Galan A, de Yebenes VG, Gutierrez-Vazquez C, Tenreiro S, Seabra MC, Ramiro AR, and Sanchez-Madrid F (2020). Transfer of extracellular vesicle-microRNA controls germinal center reaction and antibody production. EMBO Rep 21, e48925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foy TM, Laman JD, Ledbetter JA, Aruffo A, Claassen E, and Noelle RJ (1994). gp39-CD40 interactions are essential for germinal center formation and the development of B cell memory. J Exp Med 180, 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong S, Power Coombs MR, and Hoskin DW (2017). Thy-1 stimulation of mouse T cells induces a delayed T cell receptor-like signal that results in Ca2+independent cytotoxicity. Mol Med Rep 16, 5683–5692. [DOI] [PubMed] [Google Scholar]
- Gardell JL, and Parker DC (2017). CD40L is transferred to antigen-presenting B cells during delivery of T-cell help. Eur J Immunol 47, 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin AD, Shulman Z, and Nussenzweig MC (2014). Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature 509, 637–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong D, and Ferrell JE Jr. (2010). The roles of cyclin A2, B1, and B2 in early and late mitotic events. Mol Biol Cell 21, 3149–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good-Jacobson KL, Szumilas CG, Chen L, Sharpe AH, Tomayko MM, and Shlomchik MJ (2010). PD-1 regulates germinal center B cell survival and the formation and affinity of long-lived plasma cells. Nat Immunol 11, 535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JA, Suzuki K, Cho B, Willison LD, Palmer D, Allen CD, Schmidt TH, Xu Y, Proia RL, Coughlin SR, and Cyster JG (2011). The sphingosine 1-phosphate receptor S1P(2) maintains the homeostasis of germinal center B cells and promotes niche confinement. Nat Immunol 12, 672–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulbranson-Judge A, Casamayor-Palleja M, and MacLennan IC (1997). Mutually dependent T and B cell responses in germinal centers. Ann N Y Acad Sci 815, 199–210. [DOI] [PubMed] [Google Scholar]
- Han S, Hathcock K, Zheng B, Kepler TB, Hodes R, and Kelsoe G (1995). Cellular interaction in germinal centers. Roles of CD40 ligand and B7-2 in established germinal centers. J Immunol 155, 556–567. [PubMed] [Google Scholar]
- Haribhai D, Lin W, Relland LM, Truong N, Williams CB, and Chatila TA (2007). Regulatory T cells dynamically control the primary immune response to foreign antigen. J Immunol 178, 2961–2972. [DOI] [PubMed] [Google Scholar]
- Harriman GR, Lycke NY, Elwood LJ, and Strober W (1990). T lymphocytes that express CD4 and the alpha beta-T cell receptor but lack Thy-1. Preferential localization in Peyer’s patches. J Immunol 145, 2406–2414. [PubMed] [Google Scholar]
- Havenar-Daughton C, Carnathan DG, Boopathy AV, Upadhyay AA, Murrell B, Reiss SM, Enemuo CA, Gebru EH, Choe Y, Dhadvai P, et al. (2019). Rapid Germinal Center and Antibody Responses in Non-human Primates after a Single Nanoparticle Vaccine Immunization. Cell Rep 29, 1756–1766 e1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C, Carnathan DG, Torrents de la Pena A, Pauthner M, Briney B, Reiss SM, Wood JS, Kaushik K, van Gils MJ, Rosales SL, et al. (2016). Direct Probing of Germinal Center Responses Reveals Immunological Features and Bottlenecks for Neutralizing Antibody Responses to HIV Env Trimer. Cell Rep 17, 2195–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes NM, Allen CD, Lesley R, Ansel KM, Killeen N, and Cyster JG (2007). Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J Immunol 179, 5099–5108. [DOI] [PubMed] [Google Scholar]
- Heesters BA, Chatterjee P, Kim YA, Gonzalez SF, Kuligowski MP, Kirchhausen T, and Carroll MC (2013). Endocytosis and recycling of immune complexes by follicular dendritic cells enhances B cell antigen binding and activation. Immunity 38, 1164–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al. (2015). Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12, 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ise W, Fujii K, Shiroguchi K, Ito A, Kometani K, Takeda K, Kawakami E, Yamashita K, Suzuki K, Okada T, and Kurosaki T (2018). T Follicular Helper Cell-Germinal Center B Cell Interaction Strength Regulates Entry into Plasma Cell or Recycling Germinal Center Cell Fate. Immunity 48, 702–715 e704. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, and Crotty S (2009). Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 325, 1006–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsoe G (1996). The germinal center: a crucible for lymphocyte selection. Semin Immunol 8, 179–184. [DOI] [PubMed] [Google Scholar]
- Kerfoot SM, Yaari G, Patel JR, Johnson KL, Gonzalez DG, Kleinstein SH, and Haberman AM (2011). Germinal center B cell and T follicular helper cell development initiates in the interfollicular zone. Immunity 34, 947–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey PJ, Staines DM, Lawson D, Kulesha E, Derwent P, Humphrey JC, Hughes DS, Keenan S, Kerhornou A, Koscielny G, et al. (2012). Ensembl Genomes: an integrative resource for genome-scale data from non-vertebrate species. Nucleic Acids Res 40, D91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano M, Moriyama S, Ando Y, Hikida M, Mori Y, Kurosaki T, and Okada T (2011). Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity 34, 961–972. [DOI] [PubMed] [Google Scholar]
- Kuraoka M, Liao D, Yang K, Allgood SD, Levesque MC, Kelsoe G, and Ueda Y (2009). Activation-induced cytidine deaminase expression and activity in the absence of germinal centers: insights into hyper-IgM syndrome. J Immunol 183, 3237–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Chen X, Zhong B, Wang A, Wang X, Chu F, Nurieva RI, Yan X, Chen P, van der Flier LG, et al. (2014). Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature 507, 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Zhang J, Lane PJ, Chan EY, and MacLennan IC (1991). Sites of specific B cell activation in primary and secondary responses to T cell-dependent and T cell-independent antigens [published erratum appears in Eur J Immunol 1992 Feb;22(2):615]. Eur J Immunol 21, 2951–2962. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu N, Li Y, Zhang Z, Xing J, Sun Y, Yao S, and Chen L (2018). Human Semaphorin-4A drives Th2 responses by binding to receptor ILT-4. Nat Commun 9, 742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt Removes Adapter Sequences From High-Throughput Sequencing Reads. EMBnet.journal 17, 10–12. [Google Scholar]
- Meli AP, and King IL (2015). Identification of mouse T follicular helper cells by flow cytometry. Methods Mol Biol 1291, 3–11. [DOI] [PubMed] [Google Scholar]
- Meredith MM, Liu K, Darrasse-Jeze G, Kamphorst AO, Schreiber HA, Guermonprez P, Idoyaga J, Cheong C, Yao KH, Niec RE, and Nussenzweig MC (2012). Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J Exp Med 209, 1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, Emili A, and Bader GD (2010). Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5, e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager J, Finkin S, Ramos V, Kraft J, Cipolla M, Nowosad CR, Hartweger H, Zhang W, Olinares PDB, Gazumyan A, et al. (2021). Dynamic regulation of TFH selection during the germinal centre reaction. Nature 591, 458–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Hermann ME, Maini PK, and Iber D (2006). An analysis of B cell selection mechanisms in germinal centers. Math Med Biol 23, 255–277. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. (2003). PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34, 267–273. [DOI] [PubMed] [Google Scholar]
- Moriyama S, Takahashi N, Green JA, Hori S, Kubo M, Cyster JG, and Okada T (2014). Sphingosine-1-phosphate receptor 2 is critical for follicular helper T cell retention in germinal centers. J Exp Med 211, 1297–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KM, Heimberger AB, and Loh DY (1990). Induction by antigen of intrathymic apoptosis of CD4+CD8+TCRlo thymocytes in vivo. Science 250, 1720–1723. [DOI] [PubMed] [Google Scholar]
- Nance JP, Belanger S, Johnston RJ, Takemori T, and Crotty S (2015). Cutting edge: T follicular helper cell differentiation is defective in the absence of Bcl6 BTB repressor domain function. J Immunol 194, 5599–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, and Dong C (2009). Bcl6 mediates the development of T follicular helper cells. Science 325, 1001–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Regan L, Blot J, and Fry AM (2007). Mitotic regulation by NIMA-related kinases. Cell Div 2, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa I, Saliba D, Ponzoni M, Bustamante S, Canete PF, Gonzalez-Figueroa P, McNamara HA, Valvo S, Grimbaldeston M, Sweet RA, et al. (2017). TFH-derived dopamine accelerates productive synapses in germinal centres. Nature 547, 318–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa I, and Vinuesa CG (2018). Synaptic Interactions in Germinal Centers. Frontiers in immunology 9, 1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper M, Pagan AJ, Igyarto BZ, Taylor JJ, and Jenkins MK (2011). Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity 35, 583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall TD, Heath AW, Santos-Argumedo L, Howard MC, Weissman IL, and Lund FE (1998). Arrest of B lymphocyte terminal differentiation by CD40 signaling: mechanism for lack of antibody-secreting cells in germinal centers. Immunity 8, 733–742. [DOI] [PubMed] [Google Scholar]
- Robertson JM, Jensen PE, and Evavold BD (2000). DO11.10 and OT-II T cells recognize a C-terminal ovalbumin 323–339 epitope. J Immunol 164, 4706–4712. [DOI] [PubMed] [Google Scholar]
- Schaerli P, Willimann K, Lang AB, Lipp M, Loetscher P, and Moser B (2000). CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J Exp Med 192, 1553–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AG, Therkelsen MD, Stewart S, Kepler TB, Liao HX, Moody MA, Haynes BF, and Harrison SC (2015). Viral receptor-binding site antibodies with diverse germline origins. Cell 161, 1026–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwickert TA, Victora GD, Fooksman DR, Kamphorst AO, Mugnier MR, Gitlin AD, Dustin ML, and Nussenzweig MC (2011). A dynamic T cell-limited checkpoint regulates affinity-dependent B cell entry into the germinal center. J Exp Med 208, 1243–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnakasu R, Inoue T, Kometani K, Moriyama S, Adachi Y, Nakayama M, Takahashi Y, Fukuyama H, Okada T, and Kurosaki T (2016). Regulated selection of germinal-center cells into the memory B cell compartment. Nat Immunol 17, 861–869. [DOI] [PubMed] [Google Scholar]
- Shulman Z, Gitlin AD, Targ S, Jankovic M, Pasqual G, Nussenzweig MC, and Victora GD (2013). T follicular helper cell dynamics in germinal centers. Science 341, 673–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman Z, Gitlin AD, Weinstein JS, Lainez B, Esplugues E, Flavell RA, Craft JE, and Nussenzweig MC (2014). Dynamic signaling by T follicular helper cells during germinal center B cell selection. Science 345, 1058–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E, Pewzner-Jung Y, Schwers S, Taki S, Jung S, Eilat D, and Rajewsky K (1997). B cell development under the condition of allelic inclusion. Immunity 6, 225–233. [DOI] [PubMed] [Google Scholar]
- Stein JV, and Nombela-Arrieta C (2005). Chemokine control of lymphocyte trafficking: a general overview. Immunology 116, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suan D, Nguyen A, Moran I, Bourne K, Hermes JR, Arshi M, Hampton HR, Tomura M, Miwa Y, Kelleher AD, et al. (2015). T follicular helper cells have distinct modes of migration and molecular signatures in naive and memory immune responses. Immunity 42, 704–718. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Dutta PR, Cerasoli DM, and Kelsoe G (1998). In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. V. Affinity maturation develops in two stages of clonal selection. Journal of Experimental Medicine 187, 885–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubo NJ, Pagan AJ, Taylor JJ, Nelson RW, Linehan JL, Ertelt JM, Huseby ES, Way SS, and Jenkins MK (2013). Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell 153, 785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, and Nussenzweig MC (2010). Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell 143, 592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuesa CG, Sanz I, and Cook MC (2009). Dysregulation of germinal centres in autoimmune disease. Nat Rev Immunol 9, 845–857. [DOI] [PubMed] [Google Scholar]
- Wan Z, Lin Y, Zhao Y, and Qi H (2019). TFH cells in bystander and cognate interactions with B cells. Immunol Rev 288, 28–36. [DOI] [PubMed] [Google Scholar]
- Wang P, Shih CM, Qi H, and Lan YH (2016). A Stochastic Model of the Germinal Center Integrating Local Antigen Competition, Individualistic T-B Interactions, and B Cell Receptor Signaling. J Immunol 197, 1169–1182. [DOI] [PubMed] [Google Scholar]
- White J, Haskins KM, Marrack P, and Kappler J (1983). Use of I region-restricted, antigen-specific T cell hybridomas to produce idiotypically specific anti-receptor antibodies. J Immunol 130, 1033–1037. [PubMed] [Google Scholar]
- Wing JB, Kitagawa Y, Locci M, Hume H, Tay C, Morita T, Kidani Y, Matsuda K, Inoue T, Kurosaki T, et al. (2017). A distinct subpopulation of CD25(−) T-follicular regulatory cells localizes in the germinal centers. Proc Natl Acad Sci U S A 114, E6400–E6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenbrink N, Klein A, Weiser AA, Schuchhardt J, and Or-Guil M (2011). Is there a typical germinal center? A large-scale immunohistological study on the cellular composition of germinal centers during the hapten-carrier-driven primary immune response in mice. J Immunol 187, 6185–6196. [DOI] [PubMed] [Google Scholar]
- Wollenberg I, Agua-Doce A, Hernandez A, Almeida C, Oliveira VG, Faro J, and Graca L (2011). Regulation of the germinal center reaction by Foxp3+ follicular regulatory T cells. J Immunol 187, 4553–4560. [DOI] [PubMed] [Google Scholar]
- Wu H, Xu LL, Teuscher P, Liu H, Kaplan MH, and Dent AL (2015). An Inhibitory Role for the Transcription Factor Stat3 in Controlling IL-4 and Bcl6 Expression in Follicular Helper T Cells. J Immunol 195, 2080–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Li X, Liu D, Li J, Zhang X, Chen X, Hou S, Peng L, Xu C, Liu W, et al. (2013). Follicular T-helper cell recruitment governed by bystander B cells and ICOS-driven motility. Nature 496, 523–527. [DOI] [PubMed] [Google Scholar]
- Yeh CH, Nojima T, Kuraoka M, and Kelsoe G (2018). Germinal center entry not selection of B cells is controlled by peptide-MHCII complex density. Nat Commun 9, 928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, et al. (2009). The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity 31, 457–468. [DOI] [PubMed] [Google Scholar]
- Yusuf I, Kageyama R, Monticelli L, Johnston RJ, Ditoro D, Hansen K, Barnett B, and Crotty S (2010). Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150). J Immunol 185, 190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Han S, and Kelsoe G (1996). T helper cells in murine germinal centers are antigen-specific emigrants that downregulate Thy-1. J Exp Med 184, 1083–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. All Tcrβ sequence data sets are available from Adaptive Biotechnologies immuneACCESS and are publicly available as of the date of publication. Direct links are listed in the key resources table. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-mouse CD3 AF488 (Clone 17A2) | BioLegend | Cat#100210; RRID: AB_389301 |
| anti-mouse CD3 AF647 (Clone 145-2C11) | BioLegend | Cat#100324; RRID: AB_492861 |
| anti-mouse CD4 Biotin (Clone GK1.5) | BioLegend | Cat#100404; RRID: AB_312689 |
| anti-mouse CD4 AF647 (Clone GK1.5) | BioLegend | Cat#100424; RRID: AB_389324 |
| anti-mouse CD4 AF647 (Clone RM4-5) | BioLegend | Cat#100533; RRID: AB_493372 |
| anti-mouse CD4 BV421 (Clone GK1.5) | BioLegend | Cat#100443; RRID: AB_2562557 |
| anti-mouse CD4 BV510 (Clone RM4-5) | BD Biosciences | Cat#563106; RRID: AB_2687550 |
| anti-mouse CD8 BV421 (Clone 53-6.7) | BD Biosciences | Cat#563898; RRID: AB_2738474 |
| anti-mouse CD11b PE-Cy7 (Clone M1/70) | BioLegend | Cat#101215; RRID: AB_312798 |
| anti-mouse CD11c Biotin (Clone N418) | BioLegend | Cat#117304; RRID: AB_313773 |
| anti-mouse CD11c AF647 (Clone N418) | BioLegend | Cat#117312; RRID: AB_389328 |
| anti-mouse CD19 APC-R700 (Clone 1D3) | BD Biosciences | Cat#565473; RRID: AB_2739253 |
| anti-mouse CD21/35 AF594 (Clone 7E9) | BioLegend | Cat#123426; RRID: AB_2632698 |
| anti-mouse CD21/35 AF647 (Clone 7E9) | BioLegend | Cat#123424; RRID: AB_2629578 |
| anti-mouse CD25 BV421 (Clone PC61) | BioLegend | Cat#102043; RRID: AB_2562611 |
| anti-mouse CD43 Biotin (Clone S7) | BD Biosciences | Cat#553269; RRID: AB_2255226 |
| anti-mouse/human CD44 FITC (Clone IM7) | BioLegend | Cat#103006; RRID: AB_312957 |
| anti-mouse/human CD44 PerCP-Cy5.5 (Clone IM7) | BioLegend | Cat#103006; RRID: AB_2076204 |
| anti-mouse/human CD44 PE-594 (Clone IM7) | BioLegend | Cat#103056; RRID: AB_2564044 |
| anti-mouse CD45.1 BV421 (Clone A20) | BioLegend | Cat#110732; RRID: AB_2562563 |
| anti-mouse CD45.2 FITC (Clone 104) | Thermo Fisher Scientific | Cat#11-0454-82; RRID: AB_465061 |
| anti-mouse CD62L PerCP-Cy5.5 (Clone MEL-14) | BioLegend | Cat#104430; RRID: AB_2187124 |
| anti-mouse CD62L PE (Clone MEL-14) | BioLegend | Cat#104408; RRID: AB_313095 |
| anti-mouse CD62L BV786 (Clone MEL-14) | BD Biosciences | Cat#564109; RRID: AB_2738598 |
| anti-mouse CD69 PE-Cy5 (Clone H1.2F3) | BioLegend | Cat#104510; RRID: AB_313113 |
| anti-mouse CD90.2 Biotin (Clone 30H12) | BioLegend | Cat#105304; RRID: AB_313175 |
| anti-mouse CD90.2 AF488 (Clone 30H12) | BioLegend | Cat#105316; RRID: AB_492886 |
| anti-mouse CD90.2 PE (Clone 30H12) | BioLegend | Cat#105307; RRID: AB_313178 |
| anti-mouse CD90.2 AF647 (Clone 30H12) | BioLegend | Cat#105318; RRID: AB_492888 |
| anti-mouse CD90.2 AF700 (Clone 30H12) | BioLegend | Cat#105320; RRID: AB_493725 |
| anti-mouse CD90.2 AF700 (Clone 53-2.1) | BioLegend | Cat#140324; RRID: AB_2566740 |
| anti-mouse CD90.2 BV421 (Clone 53-2.1) | BioLegend | Cat#140327; RRID: AB_2686992 |
| anti-mouse CD138 BV308 (Clone 281-2) | BD Biosciences | Cat#563147; RRID: AB_2721029 |
| anti-mouse CD185 (CXCR5) Biotin (Clone L138D7) | BioLegend | Cat#145510; RRID: AB_2562126 |
| anti-mouse CD185 (CXCR5) Biotin (Clone 2G8) | BD Biosciences | Cat#551960; RRID: AB_394301 |
| anti-mouse CD279 (PD-1) PE-Cy7 (Clone 29F.1A12) | BioLegend | Cat#135216; RRID: AB_10689635 |
| anti-mouse CD279 (PD-1) BV421 (Clone 29F.1A12) | BioLegend | Cat#135221; RRID: AB_2562568 |
| anti-mouse/human B220 APC/Fire750 (Clone RA3-6B2) | BioLegend | Cat#103259; RRID: AB_2572108 |
| anti-mouse/human B220 BV605 (Clone RA3-6B2) | BioLegend | Cat#103244; RRID: AB_2563312 |
| anti-mouse/human B220 BV785 (Clone RA3-6B2) | BioLegend | Cat#103246; RRID: AB_2563256 |
| anti-mouse/human Bcl-6 PE (Clone K112-91) | BD Biosciences | Cat#561522; RRID: AB_10717126 |
| anti-mouse/human Bcl-6 PE-CF594 (Clone K112-91) | BD Biosciences | Cat#562401; RRID: AB_11152084 |
| anti-mouse/human Bcl-6 AF647 (Clone K112-91) | BD Biosciences | Cat#561525; RRID: AB_10898007 |
| anti-mouse DO11.10 TCR FITC (Clone KJ1-26) | Thermo Fisher Scientific | Cat#11-5808-82; RRID: AB_465248 |
| anti-mouse F4/80 Biotin (Clone BM8) | BioLegend | Cat#123106; RRID: AB_893501 |
| anti-mouse F4/80 PE (Clone BM8) | BioLegend | Cat#123110; RRID: AB_893486 |
| anti-mouse FoxP3 AF488 (Clone FJK-16s) | Thermo Fisher Scientific | Cat#53-5773-82; RRID: AB_763537 |
| anti-mouse FoxP3 AF647 (Clone MF23) | BD Biosciences | Cat#560401; RRID: AB_1645201 |
| anti-GFP AF488 (Clone FM264G) | BioLegend | Cat#338008; RRID: AB_2563288 |
| anti-mouse GL7 FITC (Clone GL7) | BD Biosciences | Cat#553666; RRID: AB_394981 |
| anti-mouse GL7 PE (Clone GL7) | BD Biosciences | Cat#561530; RRID: AB_10715834 |
| anti-mouse Gr-1 PE (Clone RB6-8C5) | BioLegend | Cat#108404; RRID: AB_313369 |
| anti-human HB-EGF (Goat Polyclonal) | R&D Systems | Cat#BAF259; RRID: AB_2114598 |
| anti-mouse I-A/I-E (MHCII) AF647 (Clone M5/114.15.2) | BioLegend | Cat#107618; RRID: AB_493525 |
| anti-mouse I-A/I-E (MHCII) BV711 (Clone M5/114.15.2) | BD Biosciences | Cat#563414; RRID: AB_2738191 |
| anti-mouse I-Ab (MHCII) PE-CF594 (Clone AF6-120.1) | BD Biosciences | Cat#562824; RRID: AB_2737819 |
| anti-mouse IgD BV421 (Clone 11-26c.2a) | BioLegend | Cat#405725; RRID: AB_2562743 |
| anti-mouse IgD BV510 (Clone 11-26c.2a) | BioLegend | Cat#405723; RRID: AB_2562742 |
| anti-mouse Ig κ light chain BV421 (Clone 187.1) | BD Biosciences | Cat#562888; RRID: AB_2737867 |
| anti-mouse Ig λ1, λ2, & λ3 light chain FITC (Clone R26-46) | BD Biosciences | Cat#553434; RRID: AB_394854 |
| anti-mouse/human Ki-67 BV421 (Clone 11F6) | BioLegend | Cat#151208; RRID: AB_2629748 |
| anti-mouse/human Ki-67 BV421 (Clone B56) | BD Biosciences | Cat#562899; RRID: AB_2686897 |
| anti-mouse/human Ki-67 AF647 (Clone B56) | BD Biosciences | Cat#558615; RRID: AB_647130 |
| anti-mouse/human Ki-67 BV650 (Clone B56) | BD Biosciences | Cat#563757; RRID: AB_2688008 |
| anti-mouse Ly6C FITC (Clone AL21) | BD Biosciences | Cat#553104; RRID: AB_394628 |
| anti-RFP (Rabbit Polyclonal) | Rockland | Cat#600-401-379; RRID: AB_2209751 |
| anti-mouse TCRβ FITC (Clone H57-597) | BioLegend | Cat#109205; RRID: AB_313428 |
| anti-mouse TCRβ PE-Cy7 (Clone H57-597) | BioLegend | Cat#109221; RRID: AB_893627 |
| anti-mouse TCRβ BV711 (Clone H57-597) | BD Biosciences | Cat#563135; RRID: AB_2738023 |
| anti-mouse TCR Vβ5.1/5.2 PE (Clone MR9-4) | BD Biosciences | Cat#562086; RRID: AB_394698 |
| anti-mouse Ter-119 Biotin (Clone TER-119) | BioLegend | Cat#116204; RRID: AB_313705 |
| anti-Rabbit IgG (H+L) AF594 (Goat Polyclonal) | Thermo Fisher Scientific | Cat#A-11012; RRID: AB_ 2534079 |
| anti-mouse CD16/CD32 (Mouse BD Fc Block™) | BD Biosciences | Cat#553142; RRID: AB_394656 |
| Chemicals, peptides, and recombinant proteins | ||
| Acetone | Sigma-Aldrich | Cat#179124 |
| Alhydrogel® adjuvant 2% | InvivoGen | Cat#vac-alu-250 |
| Corn oil | Sigma-Aldrich | Cat#PHR2897 |
| Diphtheria Toxin from Corynebacterium diphtheriae | Sigma-Aldrich | Cat#D0564 |
| IgG from rat serum | Sigma-Aldrich | Cat#I4131 |
| Methanol | Sigma-Aldrich | Cat#34860 |
| NP10-HSA (Human Serum Albumin) | Biosearch Technologies | Cat#N-5059-10 |
| NP15-Ova (Ovalbumin) | Biosearch Technologies | Cat#N-5051-100 |
| Ovalbumin | Biosearch Technologies | Cat#O-1000-100 |
| recombinant PA (Protective antigen) | ||
| recombinant HA (A/Solomon Islands/3/2006) | S. Harrison | (Schmidt et al., 2015) |
| 16% Formaldehyde (w/v), Methanol-free | Thermo Fisher Scientific | Cat#28906 |
| Streptavidin AF488 | BioLegend | Cat#405235 |
| Streptavidin PE | BioLegend | Cat#405204 |
| Streptavidin APC | BioLegend | Cat#405207 |
| Streptavidin BV421 | BioLegend | Cat#405225 |
| Streptavidin BV650 | BioLegend | Cat#405232 |
| Sucrose | Sigma-Aldrich | Cat#S9378 |
| Tamoxifen | Sigma-Aldrich | Cat#T5648 |
| Tissue-Tek® O.C.T. Compound | Sakura Finetek USA | Cat#4583 |
| Critical commercial assays | ||
| Direct-zol RNA Miniprep | Zymo Research | Cat#R2051 |
| immunoSEQ mouse Tcrβ assay | Adaptive Biotechnologies | https://www.immunoseq.com/assays/ |
| KAPA HyperPlus Kit | Roche Molecular Systems | Cat#07962401001 |
| KAPA Pure Beads | Roche Molecular Systems | Cat#07983280001 |
| LIVE/DEAD™ Fixable Near-IR Dead Cell Stain Kit | Thermo Fisher Scientific | Cat#L34976 |
| Pan T Cell Isolation Kit II | Miltenyi Biotec | Cat#130-095-130 |
| Qubit dsDNA HS Assay Kit | Thermo Fisher Scientific | Cat#Q32851 |
| Qubit RNA IQ Assay | Thermo Fisher Scientific | Cat#Q33222 |
| RNA 6000 Pico kit | Agilent | Cat#5067-1513 |
| SMART-Seq® v4 Ultra Low Input RNA Kit | Takara Bio | Cat#634890 |
| Streptavidin MicroBeads | Miltenyi Biotec | Cat#130-048-101 |
| Transcription Factor Buffer Set | BD Biosciences | Cat#562574 |
| Deposited data | ||
| RNA-seq data | This paper | GSE147035 |
| Tcrβ sequence data | This paper |
https://clients.adaptivebiotech.com/pub/yeh-2021-immunity
DOI: https://dx.doi.org/10.21417/CY2021I |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J (B6) | The Jackson Laboratory | JAX: 000664 |
| Mouse: B6.129S2-H2dlAb1-Ea/J (MHCII−/−) | The Jackson Laboratory | JAX: 003584 |
| Mouse: B6.SJL-Ptprca Pepcb/BoyJ (B6.CD45.1) | The Jackson Laboratory | JAX: 002014 |
| Mouse: B6.Cg-Ptprca Tg(UBC-PA-GFP)1Mnz/J (PAGFP) | The Jackson Laboratory | JAX: 022486 |
| Mouse: B6.Cg-Foxp3tm2Tch/J (FoxP3EGFP) | The Jackson Laboratory | JAX: 006772 |
| Mouse: B6(Cg)-Zbtb46tm1(HBEGF)Mnz/J (zbtb46-DTR) | The Jackson Laboratory | JAX: 019506 |
| Mouse: B6.129S2-Ighmtm1Cgn/J (μMT) | The Jackson Laboratory | JAX: 002288 |
| Mouse: B6.C(Cg)-Cd79atm1(cre)Reth/EhobJ (Mb1Cre) | The Jackson Laboratory | JAX: 020505 |
| Mouse: B6.Cg-Tg(TcraTcrb)425Cbn/J (OT-II Tg) | The Jackson Laboratory | JAX: 004194 |
| Mouse: C57BL/6-Gt(ROSA)26Sortm1(HBEGF)Awai/J (DTRLSL) | The Jackson Laboratory | JAX: 007900 |
| Mouse: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | The Jackson Laboratory | JAX: 007914 |
| Mouse: Balb/cJ (Balb/c) | The Jackson Laboratory | JAX: 000651 |
| Mouse: C.Cg-Tg(DO11.10)10Dlo/J (DO11.10 Tg) | The Jackson Laboratory | JAX: 003303 |
| Mouse: CByJ.SJL(B6)-Ptprca/J (Balb/c.CD45.1) | The Jackson Laboratory | JAX: 006584 |
| Mouse: Tg(S1pr2-cre/ERT2)#Kuro (S1pr2ERT2Cre) | (Shinnakasu et al., 2016) | MGI: 6435090 |
| Mouse: Ightm2Cgn (B1-8i) | (Sonoda et al., 1997) | MGI:2388486 |
| Mouse: B6.H50Gμ | (Dal Porto et al., 2002) | N/A |
| Software and algorithms | ||
| BD FACSDiva Software | BD Biosciences | http://www.bdbiosciences.com/instruments/software/facsdiva/index.jsp RRID:SCR_001456 |
| Bioconductor | Huber et al., 2015 | http://www.bioconductor.org/ RRID:SCR_006442 |
| Cytoscape | National Institute of General Medical Sciences | http://cytoscape.org RRID:SCR_003032 |
| DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html RRID:SCR_015687 |
| EnrichmentMap | Merico et al., 2010 | http://baderlab.org/Software/EnrichmentMap RRID:SCR_016052 |
| Fiji (ImageJ) | National Institutes of Health | http://fiji.sc/ RRID: SCR_002285 |
| FlowJo | BD | https://www.flowjo.com/solutions/flowjo RRID:SCR_008520 |
| GraphPad Prism_V9 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ RRID: SCR_002798 |
| HTSeq tool | European Molecular Biology Laboratory | http://www-huber.embl.de/users/anders/HTSeq/ RRID:SCR_004473 |
| immunoSEQ ANALYZER | Adaptive Biotechnologies (Carlson et al., 2013) | https://clients.adaptivebiotech.com/ |
| STAR RNAseq alignment tool | Dobin et al., 2013 | http://code.google.com/p/rna-star/ RRID:SCR_016533 |
| Trim Galore toolkit | Martin, 2011 | http://www.bioinformatics.babraham.ac.uk/projects/trim_galore RRID:SCR_011847 |