Abstract
INTRODUCTION:
APOEε4 confers less risk for Alzheimer disease (AD) in carriers with African local genomic ancestry (ALA) than APOEε4 carriers with European local ancestry (ELA). Cell type specific transcriptional variation between the two local ancestries (LAs) could contribute to this disease risk differences.
METHODS:
Single-nucleus RNA sequencing was performed on frozen frontal cortex of homozygous APOEε4 AD patients: seven with ELA, four with ALA.
RESULTS:
60,908 nuclei were sequenced. Within the LA region (chr19:44–46Mb), APOEε4 was the gene most differentially expressed, with ELA carriers having significantly more expression (overall p < 1.8E−317) in 24 of 32 cell clusters. The transcriptome of one astrocyte cluster, with high APOEε4 expression and specific to ELA, is suggestive of A1 reactive astrocytes.
DISCUSSION:
AD patients with ELA expressed significantly greater levels of APOE than ALA APOEε4 carriers. These differences in APOEε4 expression could contribute to the reduced risk for AD seen in African APOEε4 carriers.
Keywords: APOE, Single-nucleus RNA-seq, Local Ancestry, Alzheimer Disease
1. Introduction
Alzheimer disease (AD) is the most common form of dementia among older adults, affecting an estimated fifty million individuals worldwide [1]. The APOEε4 allele is the strongest common genetic risk factor for AD [2]. However, the risk for AD conveyed by the APOEε4 allele varies with ancestral background. The homozygous APOEε4 genotype presents a stronger AD risk for Non-Hispanic Whites (NHW) (Odds Ratio (OR) ~15) [3–6] than for African Americans (AA) (OR~8) and Africans (OR~3) [3–9]. The lower AD risk in African and AA APOEε4 carriers is not due to different environments between the populations or different genetic structure across the AA and NHW genomes. Rather, the protective factor lowering the risk for AD from APOEε4 is associated with a genetic difference in the local genomic ancestry region (LA) surrounding APOEε4 [10,11]. That is, if you inherited your APOEε4 allele and its African local ancestry (ALA) from your African ancestor, you have the African APOEε4 risk for AD. If you inherited your APOEε4 allele and its surrounding European local ancestry (ELA) from your European ancestor, you have the European risk for AD associated with APOEε4.
There are no consistent differences observed in the APOEε4 amino acid coding sequence between AA and NHW. Thus, the most likely source of the relative protective effect for APOEε4 found in the ALA are non-coding variant(s). We hypothesized that these regulatory elements could affect the expression of genes mediating the risk difference. Thus, unlike previous gene expression studies comparing expression differences between unaffected individuals and those affected with AD, in this study we are specifically focused on identifying expression differences between the ALA and ELA surrounding the APOEε4 region. To do this we obtained brain tissue from both AA and NHW Alzheimer patients, all homozygous carriers of the APOEε4 genotype but differing in the ancestry of their surrounding LA (African or European).
The cellular heterogeneity of the human brain makes bulk transcriptome studies difficult to interpret for any specific cell, gene or genotype. Single-nucleus RNA-sequencing (snRNA-seq) provides a method to dissect this tissue complexity, allowing unbiased characterization and quantitative expression profiles from tens of thousands of individual cells from archived frozen brain. The large number of cells that can be examined increases the statistical power of the analyses.
We demonstrate here that in the dorsal lateral frontal cortex (Brodmann area 9), AA homozygous APOEε4 carriers with surrounding ALA expressed significantly less APOE than NHW homozygous APOEε4 carriers with ELA. Identifying the underlying mechanism of the APOEε4 risk differences between NHW and AA could provide insight into potential therapeutic interventions to reduce the risk posed by APOEε4 for AD.
2. Material and Methods
2.1. Sample Source
All patients presented clinically with a progressive dementia consistent with AD, had a confirmed diagnosis of AD upon neuropathological examination and were APOEε4 homozygotes. To identify African ancestry patients, the National Alzheimer Coordinating Center (NACC) was screened for patients who self-identified as AA and were APOEε4 carriers. Autopsy material for the NACC-selected AA samples was obtained from the Alzheimer Disease Research Centers (ADRC) at Emory University, Northwestern University, and the John P. Hussman Institute for Human Genomics (HIHG). APOEε4 expression was not measured prior to sample selection and thus was not a criterion in selecting tissue. All samples were acquired with informed consent for research use and approved by the institutional review board of each center.
2.2. Sample Genomic Characterization
Global and APOE local ancestry (LA) were assessed using genome-wide genotyping data as previously described [10]. Briefly, we phased the genotyping data with the SHAPEIT tool ver. 2 [12] using 1000 Genomes Phase 3 reference panel [13]with default settings. We then used the RFMix algorithm [14] using the Human Genome Diversity Project (HGDP) European and African data as a reference panel to label each admixture block using the RFMix estimates. We defined the primary LA region of interest as that within 1Mb on either side of APOE (chr19:44–46Mb), broad enough to include potential enhancers, primary topological associated domains, and other regulatory factors, while narrow enough to ensure contiguous LA blocks [10].
We selected individuals homozygous for ELA and ALA haplotypes. Whole Genome Sequencing (WGS) was performed at either the Center for Genome Technology at the John P. Hussman Institute for Human Genomics or The American Genome Center at Uniformed Services University of the Health Sciences (USUHS) using standard Illumina protocols and GATK Best Practices analysis recommendations [15,16]. All individuals used in the study were confirmed homozygous for the APOEε4 allele by Sanger and WGS and screened for pathogenic variants in known AD genes.
2.3. Nuclei isolation
Nuclei were isolated from ~100mg of frozen tissue from Brodmann area 9 at the HIHG using the Nuclei Isolation Kit: Nuclei EZ Prep (Sigma, #NUC101). All tissues were homogenized in ice-cold EZ Lysis buffer. Pelleted nuclei (500× g, 5 min and 4 °C) were washed in ice-cold EZ Lysis buffer, and Nuclei Suspension Buffer (NSB; consisting of 1X PBS, 1% BSA and 0.2 U/μl RNase inhibitor (NxGEN #97065–224). Isolated nuclei were resuspended in NSB, filtered through a 70μm and 40μm cell strainer and the pellet re-suspended in 2% BSA in PBS. Homogenates (2 mL) were layered onto a 1.8M sucrose cushion and ultra-centrifuged at 24,400 rpm at 4°C for 2 hours (Beckman Coulter Optimal centrifuge #L90K). The nuclear pellet was re-suspended in 2% BSA in 1X PBS. After resuspension, nuclei were re-filtered through a 40μm cell strainer and an aliquot was trypan-blue stained for visual quality assessment and counted using the Countess Automated Cell Counter (Thermo Fisher).
2.4. Single nucleus RNA sequencing
Single nucleus sequencing was performed in the Center for Genome Technology at the HIHG. Nuclei at a concentration of 1200 nuclei/μl were loaded on the 10X Genomics Chromium platform to isolate ~7,000 nuclei per sample and create individually barcoded Gel bead-in-Emulsions (GEMs) which were processed using the Chromium Single Cell 3’ Reagent Version 3 Kit. Sequencing libraries were evaluated for quality on the Agilent Tape Station (Agilent Technologies, Palo Alto, CA, USA), and quantified using a Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA) and qPCR before sequencing on the Illumina NovaSeq 6000 targeting 100,000 reads per cell with sequencing parameters: Read1, 28 cycles; Index1, 8 cycles; Read2, 98 cycles.
2.5. Single nucleus RNA Quality Control and Analysis
Reads were processed and analyzed as described in detail in the Supplementary Methods. Briefly, 10X Genomics CellRanger v3.0.2 software was used for primary bioinformatics with alignment to a customized GRCh38 reference genome that includes both intronic and exonic regions during gene quantification and further processing with the Seurat 3.1 pipeline [17]. We removed poor quality nuclei and potential doublets by excluding nuclei outside the 5th and 95th percentile of the number of genes and the number of unique molecular identifiers (UMIs) and nuclei with greater than 10% mitochondrial reads.
The first 100 principal components were calculated for each sample and plotted in an elbow plot (Supplementary Figure 1) to demonstrate that data integration considering 50 principal components captured most of the variability across the samples. Data integration was performed to identify shared cell states across different samples following a recently published Seurat protocol [18]. Similar nuclei were clustered using a final resolution of 0.4 in Seurat resulting in 32 distinct clusters. Differentially expressed genes (DEG) between ancestral groups within each cluster were identified using the MAST test which employs a generalized linear model framework using cell detection rate within replicates and across groups as a co-variate [19]. The MAST test has low error and false discovery rates in comparison with other single nuclei differential expression methods [20]. This was followed by pathway analysis performed using the gene enrichment analysis tools METASCAPE [21] and Enrichr [22,23].
2.5. Data Availability
Raw sequencing data (FASTQ) and CellRanger pipeline outputs (barcode, feature, and matrix files) are available for each sample by email request to the corresponding author.
3. Results
3.1. Brain Samples
Forty-five AA samples were identified in the NACC database, but only 14 AA samples were available at the time of the study, from the Alzheimer Disease Research Centers (ADRC) of Northwestern (four) and Emory University (ten). LA was assessed and identified three AA samples homozygous for ELA and three heterozygous for ELA and ALA, excluding them for the study. Two additional samples were eliminated due to the presence of other identified neurologic abnormalities (e.g. global head injury, glioblastoma) that could affect the analysis. This left four samples (three females, one male) that were homozygous for APOEε4 and ALA. The African global ancestry of these four samples ranged from 85 to 92%, with the remaining admixture European.
Initial RNA single nuclei seq analysis used four NHW AD samples (three females, one male) obtained from the HIHG Brain Bank, with samples collected prior to 2007 at Duke University. In order to eliminate the possibility of differences in processing between centers affecting APOEε4 expression, three additional homozygous APOEε4, homozygous ELA samples were obtained from the Emory ADC, added into the total analysis, and separately compared only with the ALA samples from that same site. Samples were chosen based on tissue availability, sex match, neuropathology (had no other identified neurologic abnormalities that would affect expression), homozygous APOEε4 genotype and ELA. Global ancestry for the NHW samples was >96% European. Comparison of the overall linkage disequilibrium structure between ALA and ELA was similar, with slightly smaller and more numerous LD blocks in ALA as would be expected (Supplementary Figure 2).
All donors included in the study were clinically diagnosed with AD using standard cognitive testing (Clinical Dementia Rating (CDR) or Mini-Mental State Exam (MMSE)), and met the neuropathological criteria of the National Institute on Aging-Alzheimer’s Association for the diagnosis of AD. We assessed brain pH levels (see Supplementary Methods) and an overview of the results is given on Table 1, with no statistically significant difference between the groups (p = 0.44). Description of the samples is shown in Table 1 and in Supplementary Table 1. Whole genome sequencing revealed absence of mutations in any known Mendelian genes for AD (PSEN1, PSEN2, APP, and MAPT) as well as absence of known risk rare variants in ABCA7, TREM2 and SORL1 in all samples.
Table 1.
Demographic characteristics of the samples.
| Sample | Center | Sex | AOD | APOE genotype | Local Ancestry | CDR Score | Freezer time (years) | PMI (hours) | BRAAK Score | ABC Score | pH |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Emory | Male | 86 | 4,4 | AF/AF | 3 | 3 | 7 | IV | A2B2C3 | 6.1 |
| 2 | Emory | Female | 82 | 4,4 | AF/AF | 3 | 10 | NA | V | A3B3C3 | 6.3 |
| 3 | NW | Female | 85 | 4,4 | AF/AF | 3 | 5 | 27 | V | A3B3C3 | 6.8 |
| 4 | Emory | Female | 80 | 4,4 | AF/AF | 2 | 5 | 14 | VI | A3B3C3 | 6.3 |
| 5 | Duke/UM | Male | 75 | 4,4 | EU/EU | 3 | 15 | 2 | IV | A2B2C3 | 6.5 |
| 6 | Duke/UM | Female | 76 | 4,4 | EU/EU | 3 | 15 | 6 | V | A3B3C3 | 6.4 |
| 7 | Duke/UM | Female | 72 | 4,4 | EU/EU | 3 | 15 | 12 | IV | A2B2C3 | 6.1 |
| 8 | Duke/UM | Female | 70 | 4,4 | EU/EU | 3 | 15 | 4 | VI | A3B3C3 | 6.4 |
| 9 | Emory | Female | 72 | 4,4 | EU/EU | 3 | 1 | 6 | VI | A3B3C3 | 6.3 |
| 10 | Emory | Female | 83 | 4,4 | EU/EU | NA | 19 | 12 | VI | A3B3C3 | 6.2 |
| 11 | Emory | Female | 76 | 4,4 | EU/EU | 3 | 12 | 3.5 | VI | A3B3C3 | 6.3 |
AOD: Age of Death, PMI: Post-Mortem Interval, UM: University of Miami, NW: Northwestern University, NA: Not Available
3.2. Single nucleus RNA-seq clusters
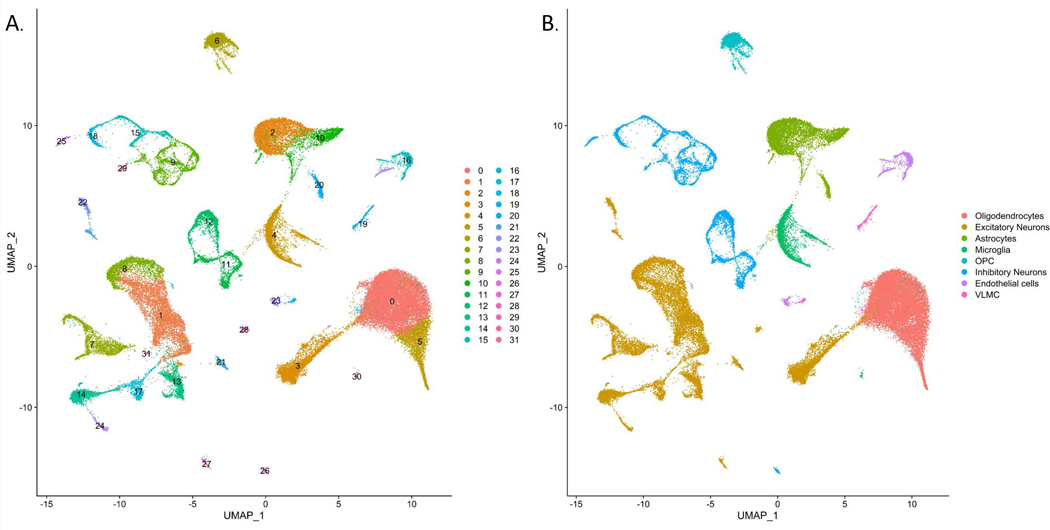
After quality control, we obtained data from a total of 60,908 total nuclei (2,805 – 7,774 nuclei per sample), sequenced at a median depth of ~126,000 reads per cell with on average ~1,900 genes/nucleus (Table 2). There was no significant difference between ELA and ALA in terms of percentage of aligned and unaligned reads (86 ± 8% vs 90% ± 4% of reads mapped to the hg38 genome in ALA and ELA, respectively). A UMAP plot for the integrated eleven samples was performed at a resolution of 0.4, resulting in 32 distinct clusters (Figure 1A). We identified thirteen excitatory neuron clusters (comprising ~33% of the total cells), eight inhibitory neuron clusters (~13% of total cells), two oligodendrocyte clusters (~29% of total cells), three astrocyte (~11% of total cells), two microglia clusters (~6% of total cells), one oligodendrocyte precursor cell (OPC) cluster (~5% of total cells), two endothelial cell cluster (~2% of total cells), and one vascular leptomeningeal cell (VLMC) cluster (~1% of total cells). (Figure 1B and Supplementary Table 2). A heatmap of the top ten marker genes defining each of the 32 clusters is shown in Supplementary Figure 3.
Table 2.
Summary of Single Nuclei RNA sequencing results.
| Sample | Raw # of cells | # of cells post-QC | Median # of reads/nucleus | Median # genes/nucleus |
|---|---|---|---|---|
| 1 | 10,160 | 3,587 | 169,439 | 2,593 |
| 2 | 6,660 | 5,592 | 126,054 | 2,677 |
| 3 | 5,984 | 4,993 | 180,736 | 1,331 |
| 4 | 3,233 | 2,805 | 322,773 | 1,388 |
| 5 | 8,853 | 7,774 | 86,528 | 1,253 |
| 6 | 10,492 | 7,765 | 96,055 | 3,332 |
| 7 | 5,151 | 4,353 | 204,838 | 1,735 |
| 8 | 7,100 | 6,001 | 126,487 | 1,759 |
| 9 | 12,472 | 6,752 | 78,945 | 1,820 |
| 10 | 11,566 | 5,618 | 86,167 | 1,900 |
| 11 | 18,596 | 5,668 | 47,890 | 1,117 |
QC: quality control
Figure 1.
Visualization of single nuclei clusters from 60,908 nuclei integrated across 11 samples. A) UMAP dimensionality reduction plot of using a resolution of 0.4 resulting in 32 unique clusters. B) UMAP dimensionality reduction plot with clusters colored by cell type.
3.3. Comparison of clusters between ancestries
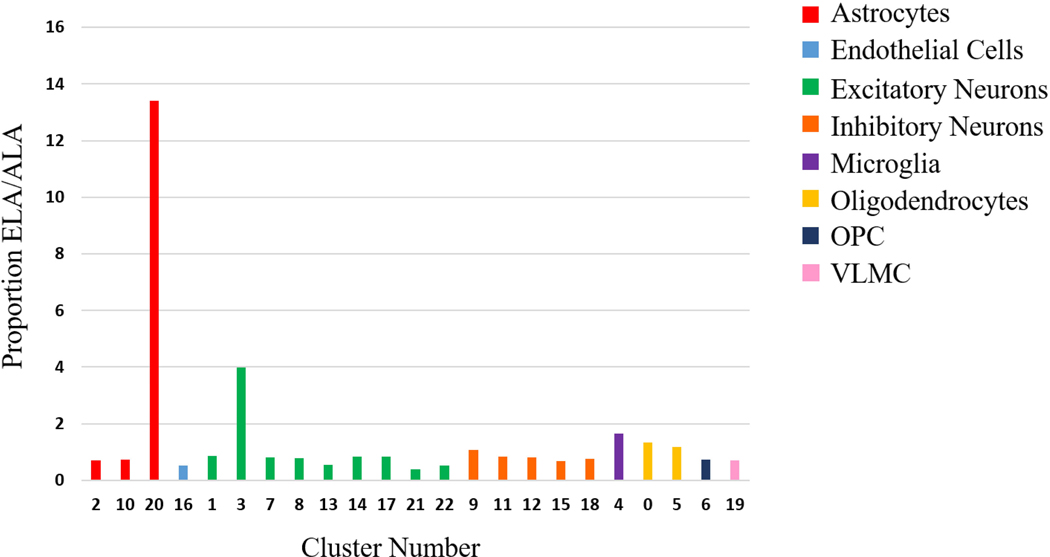
The cell count and percentage of the total cells found in each cluster, separated by ancestries, is shown in Figure 2 and Supplementary Table 2. The proportion of cells per cluster between the four cases with ALA and the seven cases with ELA was similar for all clusters with greater than 500 cells, with the exception of clusters 3, 20 and 21. Cluster 21 (excitatory neurons) was more than 2-fold greater in the ALA cluster and excitatory neuronal cluster 3 was more than 2-fold greater in ELA. Cluster 20’s proportion of the total cell number contributed by ELA was 13 times greater than the proportion of the total cells represented by cluster 20 from the ALA samples (Figure 2), but was derived from primarily two ELA samples (5 and 6).
Figure 2.
ELA/ALA cell proportions by clusters. The proportion of cells in ELA / proportion of cells in ALA plotted in a bar graph. Bars are grouped and colored by cell type of the cluster.
3.4. APOE expression
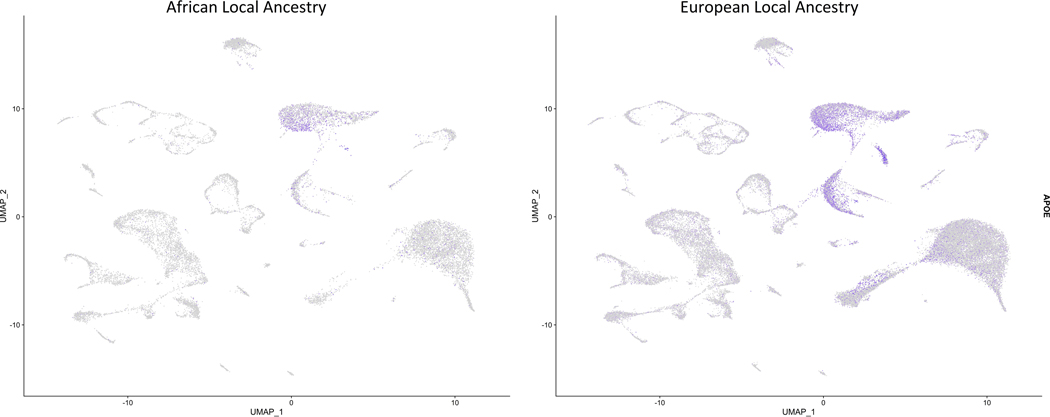
We compared APOEε4 expression between ALA and ELA clusters, as shown in Figure 3. APOEε4 expression was significantly increased overall in ELA versus ALA samples (adjusted p-value < 1.8E−317, FC = 1.56). APOEε4 demonstrated significant differential expression (DE) in 24 of the 32 clusters with all fold changes in the same direction (Table 3). The biggest fold change in APOEε4 expression was observed in a microglia cluster (cluster 4), in which ALA cells express 2.2-fold less APOEε4 than ELA cells.
Figure 3.
Visualization of APOE expression in nuclei from AA local ancestry and EU local ancestry. Cells are overlaid with gene expression information with expression depicted from gray (low) to purple (high).
Table 3.
APOE differential expression analysis.
| Cluster | Cell Type | Fold Change | adj-p-value |
|---|---|---|---|
| 2 | Astrocytes | −1.49 | 6.76E-93 |
| 16 | Endothelial cells | −1.64 | 1.22E-11 |
| 23 | −1.53 | 5.31E-07 | |
| 1 | Excitatory Neurons | −1.23 | 1.70E-162 |
| 13 | −1.22 | 5.33E-23 | |
| 14 | −1.22 | 3.21E-33 | |
| 17 | −1.21 | 1.92E-19 | |
| 21 | −1.21 | 2.67E-19 | |
| 22 | −1.21 | 8.54E-12 | |
| 3 | −1.79 | 1.95E-13 | |
| 7 | −1.21 | 2.53E-59 | |
| 8 | −1.26 | 2.50E-74 | |
| 11 | Inhibitory Neurons | −1.36 | 2.14E-39 |
| 12 | −1.30 | 3.69E-27 | |
| 15 | −1.47 | 1.10E-25 | |
| 18 | −1.32 | 2.40E-17 | |
| 26 | −1.20 | 0.00050 | |
| 29 | −1.36 | 0.02576 | |
| 9 | −1.34 | 5.73E-39 | |
| 4 | Microglia | −2.16 | 2.53E-76 |
| 0 | Oligodendrocytes | −1.30 | 1.18E-128 |
| 5 | −1.31 | 7.31E-29 | |
| 6 | OPC | −1.25 | 4.98E-25 |
| 19 | VLMC | −1.28 | 0.03288 |
To ensure that the differential expression for APOEε4 between the ALA and ELA was not secondary to differences in processing between ADRCs, we performed a second analysis using three ELA samples and three ALA samples, all from the Emory ADRC. Using the same methods as for the combined analysis, we identified a similar cluster pattern when integrating just these six samples (Supplementary Figure 4). Importantly, in these samples from the same ADRC, the expression of APOEε4 was still significantly higher in the ELA (p < 3.5E−318, FC = 1.42) (Supplementary Figure 5). Thus, the difference in APOEε4 expression is not due differences in processing of tissue between sites.
3.5. Characterization of astrocyte cluster 20
Cluster 20 was unusual in that the cluster had the highest proportion of cells expressing APOEε4 (74.9%) and APOEε4 expression was significantly higher in cluster 20 when compared to all other clusters (p = <1.39E−319, FC = 2.76). Examination of this cell cluster revealed that the vast majority of these cells (78%) originate from two of the Duke samples (5 and 6), and not the other ELA samples.
Examination of the transcriptional signature of cluster 20 revealed a strong enrichment for known astrocyte-specific markers (Supplementary File 1). To further characterize cluster 20, we compared the transcriptome of this cluster with the other astrocyte clusters (2 and 10). Analysis of the identified DEG indicates that cluster 20 preferentially expressed markers of reactive astrocytes [24–26], with significantly higher levels of GFAP (p= 2.88E-192, FC= 2.11), VIM (p= 9.33E-267, FC = 2.09), and HSPB1 (p= 2.39E-173, FC= 1.90) compared to other astrocyte clusters. Interestingly, compared to the other astrocyte clusters, cluster 20 also overexpressed IFITM3 (p = 1.08E-215, FC = 1.74), OLFM1 (p= 8.17E-204, FC= 1.90), B2M (p= 3.01E-160, FC= 1.56), TAPBP (p= 2.04E-168, FC= 1.35), CHI3L1 (p= 2.51E-43, FC= 1.19) which are specifically upregulated by neuroinflammation in A1-type reactive astrocytes in mice [24].
3.5. Other Local Ancestry and Known AD genes
We identified 116 genes (GENCODE version 32) that lie within the LA region. Of those 116, only three had significantly DE in any cluster: APOE, ERCC1, and MARK4. If we expand the analyzed region to two Mb on either side of the APOE locus, CALM3, PNMA8A, PLAUR, , and HIF3A were differentially expressed genes (Table 4).
Table 4.
Differentially expressed genes in the 2Mb local ancestry region.
| Cluster | Cell Type | Gene | Fold Change | adj-p-value |
|---|---|---|---|---|
| 2 | Astrocytes | CALM3 | −1.30 | 1.97E-108 |
| 2 | APOE | −1.50 | 6.76E-93 | |
| 2 | MARK4 | 1.22 | 2.94E-66 | |
| 2 | HIF3A | −1.20 | 3.50E-62 | |
| 16 | Endothelial cells | APOE | −1.64 | 1.22E-11 |
| 23 | APOE | −1.54 | 5.31E-07 | |
| 1 | Excitatory Neurons | APOE | −1.24 | 1.70E-162 |
| 1 | CALM3 | −1.27 | 4.66E-134 | |
| 3 | APOE | −1.80 | 1.95E-13 | |
| 7 | APOE | −1.22 | 2.53E-59 | |
| 7 | CALM3 | −1.31 | 1.70E-45 | |
| 8 | CALM3 | −1.34 | 1.70E-94 | |
| 8 | APOE | −1.27 | 2.50E-74 | |
| 13 | APOE | −1.23 | 5.33E-23 | |
| 13 | CALM3 | −1.32 | 1.85E-12 | |
| 14 | CALM3 | −1.51 | 8.38E-61 | |
| 14 | APOE | −1.23 | 3.21E-33 | |
| 17 | PNMA8A | −1.25 | 1.30E-19 | |
| 17 | APOE | −1.22 | 1.92E-19 | |
| 17 | CALM3 | −1.38 | 1.80E-18 | |
| 21 | APOE | −1.21 | 2.67E-19 | |
| 21 | CALM3 | −1.20 | 0.00683 | |
| 22 | APOE | −1.21 | 8.54E-12 | |
| 22 | CALM3 | −1.22 | 5.85E-05 | |
| 24 | CALM3 | −1.38 | 6.90E-11 | |
| 27 | CALM3 | −1.27 | 0.00283 | |
| 9 | Inhibitory Neurons | APOE | −1.34 | 5.73E-39 |
| 9 | CALM3 | −1.26 | 4.06E-38 | |
| 11 | APOE | −1.36 | 2.14E-39 | |
| 11 | CALM3 | −1.24 | 1.45E-16 | |
| 11 | ERCC1 | 1.25 | 4.00E-11 | |
| 12 | APOE | −1.31 | 3.69E-27 | |
| 12 | ERCC1 | 1.35 | 7.34E-19 | |
| 12 | CALM3 | −1.20 | 5.33E-15 | |
| 15 | CALM3 | −1.37 | 3.40E-27 | |
| 15 | APOE | −1.48 | 1.10E-25 | |
| 15 | PNMA8A | −1.19 | 3.88E-19 | |
| 18 | APOE | −1.33 | 2.40E-17 | |
| 18 | ERCC1 | 1.31 | 2.78E-09 | |
| 26 | MARK4 | 1.21 | 6.38E-05 | |
| 26 | CALM3 | −1.26 | 0.00018 | |
| 26 | APOE | −1.20 | 0.00049 | |
| 29 | CALM3 | −1.38 | 0.01493 | |
| 29 | APOE | −1.37 | 0.02575 | |
| 4 | Microglia | APOE | −2.16 | 2.53E-76 |
| 4 | CALM3 | −1.22 | 2.48E-17 | |
| 4 | PLAUR | 1.24 | 6.99E-13 | |
| 0 | Oligodendrocytes | APOE | −1.31 | 1.18E-128 |
| 0 | CALM3 | −1.30 | 3.33E-105 | |
| 5 | APOE | −1.31 | 7.31E-29 | |
| 5 | CALM3 | −1.29 | 2.71E-19 | |
| 6 | OPC | HIF3A | −1.25 | 4.31E-36 |
| 6 | CALM3 | −1.25 | 3.38E-33 | |
| 6 | APOE | −1.26 | 4.98E-25 | |
| 19 | VLMC | CALM3 | −1.45 | 0.00652 |
| 19 | APOE | −1.28 | 0.03287 |
We also evaluated DE of AD candidate genes, including Mendelian AD genes (APP, PS1, PS2, and MAPT), genes suggested by GWAS studies in both ethnic groups [27–29], and genes with rare variants recently reported to be associated with AD in the AA population. We observed DE between NHW and AA in five reported AD genes including, APOE, CLU, BIN1, SORL1, and SPI1. Interestingly, we also found significant DE in three recently suggested novel African American AD genes [29]: RBFOX1, ALCAM, and GPC6 (Table 5).
Table 5.
Alzheimer disease candidate genes differentially expressed. Negative fold changes = less expression in ALA.
| Gene | # of Clusters with DE | Average fold change | Average adjusted p-value |
|---|---|---|---|
| Genome-wide-significant loci in Non-Hispanic Whites [27] | |||
| CLU | 28 | −1.67 | 6.55E-05 |
| APOE | 24 | −1.37 | 2.46E-03 |
| SORL1 | 3 | −1.27 | 1.98E-04 |
| BIN1 | 2 | −1.22 | 5.94E-06 |
| SPI1 | 1 | 1.26 | 2.36E-15 |
| Novel loci in African American Meta-Analysis [29] | |||
| RBFOX1 | 7 | 1.28 | 2.21E-04 |
| ALCAM | 5 | −1.26 | 7.30E-03 |
| GPC6 | 7 | 1.44 | 5.41E-04 |
3.6. AA versus NHW transcriptome
While this study was designed to evaluate the differences in ALA versus ELA, it also provides one of the first looks at comparing snRNA-seq data between AA and NHW AD patients. The top ten significant DEG between AA and NHW across all clusters are shown in Supplementary Table 3. The DEG of the full set of clusters is shown in Supplementary File 2.
3.7. Cell type specific pathway analysis
To better understand ancestry-specific changes in gene expression, we performed an unbiased analysis of enriched pathways. We pooled the DEG from all clusters of a given cell-type to achieve a more complete cell-type transcriptome representation (Supplementary File 3). The DEG in the astrocytic lineages were enriched in GO and KEGG pathways containing the term “stress”, suggestive of differential cellular stress responses, and ontologies like “synaptic signaling” and “cellular responses to external stimuli”. Further, enriched analysis for terms “Alzheimer Disease”, “Cognition” and “Aging” were statistically significant. Pathways enriched in DEG in neurons (both, in inhibitory and excitatory) include neuronal development, synaptic transmission and response to toxicity. DEG in oligodendrocytes are enriched in pathways related to synaptic transmission and apoptosis. Microglia DEG were enriched in immune-related pathways. DEG in endothelial cells are enriched in pathways related to responses to stress and angiogenesis.
4. Discussion
We hypothesized that the DE of genes lying in the LA region between ALA and ELA could provide insight to the locus or gene(s) contributing to the risk difference between African and European LA carriers of APOEε4. We found that the gene with the greatest DE in the LA region was APOE. No other gene in the LA region displayed the amount or extent of DE that was seen for APOE. Carriers of the APOEε4 allele on the ELA surrounding the locus produced significantly more APOEε4 than did carriers of the APOEε4 allele on the ALA. Importantly, the expression of APOEε4 was low in all four ALA tested and higher in all seven ELA, supporting the biological importance of this DE in AD risk between the two groups. Further, the DE of APOE was found to be in the same direction in all the clusters in which it is differentially expressed.
Rajabli et al [10] used a region of 1Mb on either side of APOE as the area of LA. This should include any regulatory factors interacting with APOE and the strongest reported topologically associated domains [30], while providing the most contiguous LA. In this region, two other genes, ERCC1 and MARK4 demonstrated DE, but not to the extent seen in APOEε4 (Table 4). The gene ERCC excision Repair 1, Endonuclease Non-Catalytic Subunit (ERCC1) was the next most extensive DE gene in the LA area, with increased expression in the ALA relative to the ELA in the four clusters of inhibitory neurons. ERCC1 functions in DNA repair and interestingly has been associated with human longevity along with APOE [31]. DNA repair has been proposed to be a contributing pathway to AD pathogenesis as well [32]. As the LA region is not a fixed region in individuals, we also extended our analysis to genes lying within 2Mb upstream or downstream of APOE. Only CALM3, a subunit of calmodulin, had significant DE in multiple clusters, again with less expression in the ALA than ELA. Calmodulin and calcium homeostasis have been implicated in AD [33–35], though CALM3 itself was not. CALM1, on chromosome 10, is also DE and lower in AA, suggesting the DE of CALM3 is not likely due to local ancestry effects.
Our comparison of UMAP clusters between ALA and ELA revealed an obvious difference in the cell proportion in cluster 20, which was identified as potentially an A1 reactive astrocyte cluster. Reactive astrocytes have been categorized into two primary groups, A1 and A2. A1 reactive astrocytes, activated by neuroinflammatory conditions, are proposed to be “toxic” astrocytes, resulting in destructive actions towards neuronal synapses [36]. A2 astrocytes are activated by hypoxic or ischemic conditions and are postulated to help protect neuronal cells [24]. While there is heterogeneity in gene expression reported for these reactive astrocytes [26], elevated expression of GFAP, IFITM3,B2M and OLFM1 tags them as A1 astrocytes [24], all of which were significantly overexpressed in the ELA cluster 20 relative to the other astrocyte clusters. A1 reactive astrocytes have been previously reported to be increased in AD versus control brains [37] and are known to increase in the brain with increasing age [26]. However, mouse models have shown that the high expression of APOEε4 can stimulate the formation of A1 reactive astrocytes [38] and cluster 20 cells expressed a high proportion and expression level of APOEε4. Thus, whether the finding that two ELA and no ALA contributed to cluster 20 just represents the biological heterogeneity of AD or is secondary to the higher APOEε4 expression in ELA cells is not clear and requires further studies.
The mechanism leading to this difference in APOEε4 expression is currently not known. Sequence differences between the two LA leading to differences in methylation or enhancer/repressor activity seem most likely [39,40]. Whether these differences apply to APOEε3 homozygotes with different local ancestries is not known. We have recently shown that APOEε3 homozygotes with the Very Long repeat length of the TOMM40 523’ polyT repeat on the ELA background have less risk for AD than those APOEε3 homozygotes with smaller repeats [41]. This repeat is likely tagging a sequence difference in its haplotype, as the same effect is not seen in ALA homozygous APOEε3 carriers. Therefore, while we would expect an expression difference between APOEε3 carriers on different LAs, this may not be the case.
There is also a difference in age-of-onset between the two groups, though both are well within the normal age-of-onset for AD. As APOEε4 in NHW drives and earlier age-of-onset, this is perhaps not unexpected. This seems unlikely to have contributed to any differences between the two groups. First, age-of-onset for AD is at best an estimation, usually in retrospect. Second, it is now well accepted that AD starts pathologically years before its clinical manifestation, with amyloid deposition in APOEε4 homozygotes seen to rise in their 30s [42]. Thus, comparison of the clinical onset of the disease is not necessarily reflective of pathological differences, nor the more important duration of the course of pathological disease. Pathologically there was no significant difference in BRAAK staging or pH of the sample tissues between groups.
Recently, a SNP in the promoter of APOE has been suggested to be related to the risk difference for AD between NHW and Koreans (rs405509) [9]. However, rs405509 is not significantly different in allele frequency between AA and NHW LAs on the APOEε4 haplotype (Rajabli, personal communication). Thus, it does not appear to explain the risk difference between AA and NHW. Little is also known about the similarity of open reading frames or topologically associated domains between the two ancestries in the brain, which could affect expression differences as well.
We chose to compare clinically affected individuals in this study to minimize neuropathic heterogeneity of the disease and because the number of control African-American APOEε4 carriers with autopsies is very few, if any. There is no reason to expect that expression differences we observe here are not present preclinically in homozygous APOEε4 carriers, but confirmation of this will await the inclusion of many more autopsy samples from diverse ancestries.
One of the challenges in this study was identifying AA brains that were homozygous for APOEε4 and ALA The number of AA brain samples currently available for study is very limited relative to NHW samples. Further, as an admixed population, many AA samples are heterozygous for ELA and ALA or even homozygous for ELA ancestry surrounding APOEε4. In general, there is a great need for increased genomic and autopsy studies in diverse ancestries outside of NHW. Indeed, this is a primary research foci of the Alzheimer Disease Research Sequencing Project (ADSP) of the National Institutes of Aging (https://www.niagads.org/adsp/content/home).
Finally, there has been discussion by many authors debating whether increasing or decreasing APOEε4 production would be a therapeutic approach [43]. The data presented here, along with antisense oligonucleotides studies in mouse models [44], would support the premise that decreasing APOEε4 production could lead to a positive therapeutic effect, reducing the overall risk for AD in NHW APOEε4 carriers.
Supplementary Material
Elbow plot of percent variation explained (y-axis) by number of principal components (x-axis) indicating that beyond 50 PCs in each individual sample little additional variation is explained.
Linkage disequilibrium structure across the 2Mb local ancestry region using 1000 Genomes data for the A) CEU population (ELA) and B) YRI population (ALA). Structure is based on D’ calculation in Haploview [45]. The yellow star represents the location of the APOE gene.
UMAP dimensionality reduction plot visualization of single nuclei clusters from 30,002 nuclei integrated across three ELA and three ALA samples from Emory ADRC.
Visualization of APOE expression in nuclei from three ELA and three ALA samples from Emory ADRC. Cells are overlaid with gene expression information with expression depicted from gray (low) to purple (high).
Single cell Allen Brain Map heatmap for the top 10 marker genes for each cluster (one per page) used to identify cell types (https://celltypes.brain-map.org/rnaseq/human/cortex).
List of all differentially expressed genes between AA and NHW ancestry within each cluster. Avg_logFC is the log base 2 fold change. P_val_adj is the MAST FDR corrected p-value.
Pathway enrichment analysis in KEGG and GO Ontology Biological Process for the differentially expressed genes from all clusters of a given cell-type. Each tab represents a different cell type.
Heatmap of top 10 marker genes defining each cell type. Each column represents a cell and each row a gene. High expression is red and lower expression is blue.
Acknowledgements
This study was supported by the National Institute of Aging [grant numbers R01-AG059018, R01-AG059018, U01-AG052410, and U01-AG057659), the Alzheimer Disease Center (ADC) networks (NIA) [grant number AG054074], the BrightFocus Foundation and Alzheimer Association [grant number A2018425S]. The work was also funded by the National Institutes of Health [grant number P50-AG0256878 and P30-AG013854] from Emory and from Northwestern, respectively.
Genomic and data analysis was provided by the Center for Genome Technology (CGT) from the John P. Hussman Institute for Human Genomics (HIHG) from the University of Miami Miller School of Medicine. We thank Dr. Holly Cukier for helpful discussions of the data; Mr. Benjamin Goldstein for data analysis assistance; and the numerous participants, researchers, and staff from many studies who collected and contributed to the data.
Footnotes
Declarations of interest: none
REFERENCES
- [1].World Health Organization, Dementia. https://www.who.int/news-room/fact-sheets/detail/dementia (Published on September 19, 2019) [Accessed on: January 23, 2019] 2019. [Google Scholar]
- [2].Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA, Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 1993;261:921–923. [DOI] [PubMed] [Google Scholar]
- [3].Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM, Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA 1997;278:1349–1356. [PubMed] [Google Scholar]
- [4].Tang MX, Stern Y, Marder K, Bell K, Gurland B, Lantigua R, Andrews H, Feng L, Tycko B, Mayeux R, The APOE-epsilon4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. JAMA 1998;279:751–755. [DOI] [PubMed] [Google Scholar]
- [5].Tang MX, Cross P, Andrews H, Jacobs DM, Small S, Bell K, Merchant C, Lantigua R, Costa R, Stern Y, Mayeux R, Incidence of AD in African-Americans, Caribbean Hispanics, and Caucasians in northern Manhattan. Neurology 2001;56:49–56. [DOI] [PubMed] [Google Scholar]
- [6].Tang MX, Maestre G, Tsai WY, Liu XH, Feng L, Chung WY, Chun M, Schofield P, Stern Y, Tycko B, Mayeux R, Relative risk of Alzheimer disease and age-at-onset distributions, based on APOE genotypes among elderly African Americans, Caucasians, and Hispanics in New York City. Am J Hum Genet 1996;58:574–584. [PMC free article] [PubMed] [Google Scholar]
- [7].Sahota A, Yang M, Gao S, Hui SL, Baiyewu O, Gureje O, Oluwole S, Ogunniyi A, Hall KS, Hendrie HC, Apolipoprotein E-associated risk for Alzheimer’s disease in the African-American population is genotype dependent. Ann Neurol 1997;42:659–661. [DOI] [PubMed] [Google Scholar]
- [8].Hendrie HC, Murrell J, Baiyewu O, Lane KA, Purnell C, Ogunniyi A, Unverzagt FW, Hall K, Callahan CM, Saykin AJ, Gureje O, Hake A, Foroud T, Gao S, APOE epsilon4 and the risk for Alzheimer disease and cognitive decline in African Americans and Yoruba. Int Psychogeriatr 2014;26:977–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Graff-Radford NR, Green RC, Go RC, Hutton ML, Edeki T, Bachman D, Adamson JL, Griffith P, Willis FB, Williams M, Association between apolipoprotein E genotype and Alzheimer disease in African American subjects. Arch Neurol 2002;59:594–600. [DOI] [PubMed] [Google Scholar]
- [10].Rajabli F, Feliciano BE, Celis K, Hamilton-Nelson KL, Whitehead PL, Adams LD, Bussies PL, Manrique CP, Rodriguez A, Rodriguez V, Starks T, Byfield GE, Sierra Lopez CB, McCauley JL, Acosta H, Chinea A, Kunkle BW, Reitz C, Farrer LA, Schellenberg GD, Vardarajan BN, Vance JM, Cuccaro ML, Martin ER, Haines JL, Byrd GS, Beecham GW, Pericak-Vance MA, Ancestral origin of ApoE epsilon4 Alzheimer disease risk in Puerto Rican and African American populations. PLoS Genet 2018;14:e1007791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Blue EE, Horimoto ARVR, Mukherjee S, Wijsman EM, Thornton TA, Local ancestry at APOE modifies Alzheimer’s disease risk in Caribbean Hispanics. Alzheimers Dement 2019;15:1524–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Delaneau O, Marchini J, 1000 Genomes Project Consortium, 1000 Genomes Project Consortium, Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat Commun 2014;5:3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR, A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Maples BK, Gravel S, Kenny EE, Bustamante CD, RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am J Hum Genet 2013;93:278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ, A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ, A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Butler A, Hoffman P, Smibert P, Papalexi E, Satija R, Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R, Comprehensive Integration of Single-Cell Data. Cell 2019;177:1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, Linsley PS, Gottardo R, MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 2015;16:278–015-0844–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Soneson C, Robinson MD, Bias, robustness and scalability in single-cell differential expression analysis. Nat Methods 2018;15:255–261. [DOI] [PubMed] [Google Scholar]
- [21].Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK, Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A, Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44:W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A, Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 2013;14:128-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zamanian JL, Xu L, Foo LC, Nouri N, Zhou L, Giffard RG, Barres BA, Genomic analysis of reactive astrogliosis. J Neurosci 2012;32:6391–6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Teh DBL, Prasad A, Jiang W, Ariffin MZ, Khanna S, Belorkar A, Wong L, Liu X, All AH, Transcriptome Analysis Reveals Neuroprotective aspects of Human Reactive Astrocytes induced by Interleukin 1beta. Sci Rep 2017;7:13988-017-13174-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Clarke LE, Liddelow SA, Chakraborty C, Munch AE, Heiman M, Barres BA, Normal aging induces A1-like astrocyte reactivity. Proc Natl Acad Sci U S A 2018;115:E1896–E1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, Boland A, Vronskaya M, van der Lee SJ, Amlie-Wolf A, Bellenguez C, Frizatti A, Chouraki V, Martin ER, Sleegers K, Badarinarayan N, Jakobsdottir J, Hamilton-Nelson KL, Moreno-Grau S, Olaso R, Raybould R, Chen Y, Kuzma AB, Hiltunen M, Morgan T, Ahmad S, Vardarajan BN, Epelbaum J, Hoffmann P, Boada M, Beecham GW, Garnier JG, Harold D, Fitzpatrick AL, Valladares O, Moutet ML, Gerrish A, Smith AV, Qu L, Bacq D, Denning N, Jian X, Zhao Y, Del Zompo M, Fox NC, Choi SH, Mateo I, Hughes JT, Adams HH, Malamon J, Sanchez-Garcia F, Patel Y, Brody JA, Dombroski BA, Naranjo MCD, Daniilidou M, Eiriksdottir G, Mukherjee S, Wallon D, Uphill J, Aspelund T, Cantwell LB, Garzia F, Galimberti D, Hofer E, Butkiewicz M, Fin B, Scarpini E, Sarnowski C, Bush WS, Meslage S, Kornhuber J, White CC, Song Y, Barber RC, Engelborghs S, Sordon S, Voijnovic D, Adams PM, Vandenberghe R, Mayhaus M, Cupples LA, Albert MS, De Deyn PP, Gu W, Himali JJ, Beekly D, Squassina A, Hartmann AM, Orellana A, Blacker D, Rodriguez-Rodriguez E, Lovestone S, Garcia ME, Doody RS, Munoz-Fernadez C, Sussams R, Lin H, Fairchild TJ, Benito YA, Holmes C, Karamujic-Comic H, Frosch MP, Thonberg H, Maier W, Roshchupkin G, Ghetti B, Giedraitis V, Kawalia A, Li S, Huebinger RM, Kilander L, Moebus S, Hernandez I, Kamboh MI, Brundin R, Turton J, Yang Q, Katz MJ, Concari L, Lord J, Beiser AS, Keene CD, Helisalmi S, Kloszewska I, Kukull WA, Koivisto AM, Lynch A, Tarraga L, Larson EB, Haapasalo A, Lawlor B, Mosley TH, Lipton RB, Solfrizzi V, Gill M, Longstreth WT Jr, Montine TJ, Frisardi V, Diez-Fairen M, Rivadeneira F, Petersen RC, Deramecourt V, Alvarez I, Salani F, Ciaramella A, Boerwinkle E, Reiman EM, Fievet N, Rotter JI, Reisch JS, Hanon O, Cupidi C, Andre Uitterlinden AG, Royall DR, Dufouil C, Maletta RG, de Rojas I, Sano M, Brice A, Cecchetti R, George-Hyslop PS, Ritchie K, Tsolaki M, Tsuang DW, Dubois B, Craig D, Wu CK, Soininen H, Avramidou D, Albin RL, Fratiglioni L, Germanou A, Apostolova LG, Keller L, Koutroumani M, Arnold SE, Panza F, Gkatzima O, Asthana S, Hannequin D, Whitehead P, Atwood CS, Caffarra P, Hampel H, Quintela I, Carracedo A, Lannfelt L, Rubinsztein DC, Barnes LL, Pasquier F, Frolich L, Barral S, McGuinness B, Beach TG, Johnston JA, Becker JT, Passmore P, Bigio EH, Schott JM, Bird TD, Warren JD, Boeve BF, Lupton MK, Bowen JD, Proitsi P, Boxer A, Powell JF, Burke JR, Kauwe JSK, Burns JM, Mancuso M, Buxbaum JD, Bonuccelli U, Cairns NJ, McQuillin A, Cao C, Livingston G, Carlson CS, Bass NJ, Carlsson CM, Hardy J, Carney RM, Bras J, Carrasquillo MM, Guerreiro R, Allen M, Chui HC, Fisher E, Masullo C, Crocco EA, DeCarli C, Bisceglio G, Dick M, Ma L, Duara R, Graff-Radford NR, Evans DA, Hodges A, Faber KM, Scherer M, Fallon KB, Riemenschneider M, Fardo DW, Heun R, Farlow MR, Kolsch H, Ferris S, Leber M, Foroud TM, Heuser I, Galasko DR, Giegling I, Gearing M, Hull M, Geschwind DH, Gilbert JR, Morris J, Green RC, Mayo K, Growdon JH, Feulner T, Hamilton RL, Harrell LE, Drichel D, Honig LS, Cushion TD, Huentelman MJ, Hollingworth P, Hulette CM, Hyman BT, Marshall R, Jarvik GP, Meggy A, Abner E, Menzies GE, Jin LW, Leonenko G, Real LM, Jun GR, Baldwin CT, Grozeva D, Karydas A, Russo G, Kaye JA, Kim R, Jessen F, Kowall NW, Vellas B, Kramer JH, Vardy E, LaFerla FM, Jockel KH, Lah JJ, Dichgans M, Leverenz JB, Mann D, Levey AI, Pickering-Brown S, Lieberman AP, Klopp N, Lunetta KL, Wichmann HE, Lyketsos CG, Morgan K, Marson DC, Brown K, Martiniuk F, Medway C, Mash DC, Nothen MM, Masliah E, Hooper NM, McCormick WC, Daniele A, McCurry SM, Bayer A, McDavid AN, Gallacher J, McKee AC, van den Bussche H, Mesulam M, Brayne C, Miller BL, Riedel-Heller S, Miller CA, Miller JW, Al-Chalabi A, Morris JC, Shaw CE, Myers AJ, Wiltfang J, O’Bryant S, Olichney JM, Alvarez V, Parisi JE, Singleton AB, Paulson HL, Collinge J, Perry WR, Mead S, Peskind E, Cribbs DH, Rossor M, Pierce A, Ryan NS, Poon WW, Nacmias B, Potter H, Sorbi S, Quinn JF, Sacchinelli E, Raj A, Spalletta G, Raskind M, Caltagirone C, Bossu P, Orfei MD, Reisberg B, Clarke R, Reitz C, Smith AD, Ringman JM, Warden D, Roberson ED, Wilcock G, Rogaeva E, Bruni AC, Rosen HJ, Gallo M, Rosenberg RN, Ben-Shlomo Y, Sager MA, Mecocci P, Saykin AJ, Pastor P, Cuccaro ML, Vance JM, Schneider JA, Schneider LS, Slifer S, Seeley WW, Smith AG, Sonnen JA, Spina S, Stern RA, Swerdlow RH, Tang M, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Van Eldik LJ, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Wilhelmsen KC, Williamson J, Wingo TS, Woltjer RL, Wright CB, Yu CE, Yu L, Saba Y, Pilotto A, Bullido MJ, Peters O, Crane PK, Bennett D, Bosco P, Coto E, Boccardi V, De Jager PL, Lleo A, Warner N, Lopez OL, Ingelsson M, Deloukas P, Cruchaga C, Graff C, Gwilliam R, Fornage M, Goate AM, Sanchez-Juan P, Kehoe PG, Amin N, Ertekin-Taner N, Berr C, Debette S, Love S, Launer LJ, Younkin SG, Dartigues JF, Corcoran C, Ikram MA, Dickson DW, Nicolas G, Campion D, Tschanz J, Schmidt H, Hakonarson H, Clarimon J, Munger R, Schmidt R, Farrer LA, Van Broeckhoven C, C O’Donovan M, DeStefano AL, Jones L, Haines JL, Deleuze JF, Owen MJ, Gudnason V, Mayeux R, Escott-Price V, Psaty BM, Ramirez A, Wang LS, Ruiz A, van Duijn CM, Holmans PA, Seshadri S, Williams J, Amouyel P, Schellenberg GD, Lambert JC, Pericak-Vance MA, Alzheimer Disease Genetics Consortium (ADGC), European Alzheimer’s Disease Initiative (EADI), Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium (CHARGE), Genetic and Environmental Risk in AD/Defining Genetic, Polygenic and Environmental Risk for Alzheimer’s Disease Consortium (GERAD/PERADES), Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet 2019;51:414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Reitz C, Jun G, Naj A, Rajbhandary R, Vardarajan BN, Wang L, Valladares O, Lin C, Larson EB, Graff-Radford NR, Variants in the ATP-binding cassette transporter (ABCA7), apolipoprotein E ϵ4, and the risk of late-onset Alzheimer disease in African Americans. JAMA 2013;309:1483–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kunkle BW, Schmidt M, Klein HU, Naj AC, Hamilton-Nelson KL, Larson EB, Evans DA, De Jager PL, Crane PK, Buxbaum JD, Ertekin-Taner N, Barnes LL, Fallin MD, Manly JJ, Go RCP, Obisesan TO, Kamboh MI, Bennett DA, Hall KS, Goate AM, Foroud TM, Martin ER, Wang LS, Byrd GS, Farrer LA, Haines JL, Schellenberg GD, Mayeux R, Pericak-Vance MA, Reitz C, Alzheimer’s Disease Genetics Consortium (ADGC),Graff-Radford NR, Martinez I, Ayodele T, Logue MW, Cantwell LB, Jean-Francois M, Kuzma AB, Adams LD, Vance JM, Cuccaro ML, Chung J, Mez J, Lunetta KL, Jun GR, Lopez OL, Hendrie HC, Reiman EM, Kowall NW, Leverenz JB, Small SA, Levey AI, Golde TE, Saykin AJ, Starks TD, Albert MS, Hyman BT, Petersen RC, Sano M, Wisniewski T, Vassar R, Kaye JA, Henderson VW, DeCarli C, LaFerla FM, Brewer JB, Miller BL, Swerdlow RH, Van Eldik LJ, Paulson HL, Trojanowski JQ, Chui HC, Rosenberg RN, Craft S, Grabowski TJ, Asthana S, Morris JC, Strittmatter SM, Kukull WA, Novel Alzheimer Disease Risk Loci and Pathways in African American Individuals Using the African Genome Resources Panel: A Meta-analysis. JAMA Neurol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kikuchi M, Hara N, Hasegawa M, Miyashita A, Kuwano R, Ikeuchi T, Nakaya A, Enhancer variants associated with Alzheimer’s disease affect gene expression via chromatin looping. BMC Med Genomics 2019;12:128-019-0574-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Szymczak S, Dose J, Torres GG, Heinsen FA, Venkatesh G, Datlinger P, Nygaard M, Mengel-From J, Flachsbart F, Klapper W, Christensen K, Lieb W, Schreiber S, Hasler R, Bock C, Franke A, Nebel A, DNA methylation QTL analysis identifies new regulators of human longevity. Hum Mol Genet 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jensen HLB, Lillenes MS, Rabano A, Gunther CC, Riaz T, Kalayou ST, Ulstein ID, Bohmer T, Tonjum T, Expression of nucleotide excision repair in Alzheimer’s disease is higher in brain tissue than in blood. Neurosci Lett 2018;672:53–58. [DOI] [PubMed] [Google Scholar]
- [33].O’Day DH, Eshak K, Myre MA, Calmodulin Binding Proteins and Alzheimer’s Disease. J Alzheimers Dis 2015;46:553–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].O’Day DH, Myre MA, Calmodulin-binding domains in Alzheimer’s disease proteins: extending the calcium hypothesis. Biochem Biophys Res Commun 2004;320:1051–1054. [DOI] [PubMed] [Google Scholar]
- [35].Ibarreta D, Tao J, Parrilla R, Ayuso MS, Mutation analysis of chromosome 19 calmodulin (CALM3) gene in Alzheimer’s disease patients. Neurosci Lett 1997;229:157–160. [DOI] [PubMed] [Google Scholar]
- [36].Liddelow SA, Barres BA, Reactive Astrocytes: Production, Function, and Therapeutic Potential. Immunity 2017;46:957–967. [DOI] [PubMed] [Google Scholar]
- [37].Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, Bennett ML, Munch AE, Chung WS, Peterson TC, Wilton DK, Frouin A, Napier BA, Panicker N, Kumar M, Buckwalter MS, Rowitch DH, Dawson VL, Dawson TM, Stevens B, Barres BA, Neurotoxic reactive astrocytes are induced by activated microglia. Nature 2017;541:481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Shi Y, Yamada K, Liddelow SA, Smith ST, Zhao L, Luo W, Tsai RM, Spina S, Grinberg LT, Rojas JC, Gallardo G, Wang K, Roh J, Robinson G, Finn MB, Jiang H, Sullivan PM, Baufeld C, Wood MW, Sutphen C, McCue L, Xiong C, Del-Aguila JL, Morris JC, Cruchaga C, Alzheimer’s Disease Neuroimaging Initiative, Fagan AM, Miller BL, Boxer AL, Seeley WW, Butovsky O, Barres BA, Paul SM, Holtzman DM, ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature 2017;549:523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bray NJ, Jehu L, Moskvina V, Buxbaum JD, Dracheva S, Haroutunian V, Williams J, Buckland PR, Owen MJ, O’Donovan MC, Allelic expression of APOE in human brain: effects of epsilon status and promoter haplotypes. Hum Mol Genet 2004;13:2885–2892. [DOI] [PubMed] [Google Scholar]
- [40].Liu J, Zhao W, Ware EB, Turner ST, Mosley TH, Smith JA, DNA methylation in the APOE genomic region is associated with cognitive function in African Americans. BMC Med Genomics 2018;11:43-018-0363-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bussies PL, Rajabli F, Griswold A, Dorfsman DA, Whitehead P, Adams LD, Mena PR, Cuccaro M, Haines JL, Byrd GS, Beecham GW, Pericak-Vance MA, Young JI, Vance JM, Use of local genetic ancestry to assess TOMM40–523’ and risk for Alzheimer disease. Neurol Genet 2020;6:e404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Jack CR Jr, Holtzman DM, Biomarker modeling of Alzheimer’s disease. Neuron 2013;80:1347–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Belloy ME, Napolioni V, Greicius MD, A Quarter Century of APOE and Alzheimer’s Disease: Progress to Date and the Path Forward. Neuron 2019;101:820–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Huynh TV, Liao F, Francis CM, Robinson GO, Serrano JR, Jiang H, Roh J, Finn MB, Sullivan PM, Esparza TJ, Stewart FR, Mahan TE, Ulrich JD, Cole T, Holtzman DM, Age-Dependent Effects of apoE Reduction Using Antisense Oligonucleotides in a Model of beta-amyloidosis. Neuron 2017;96:1013–1023.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Barrett JC, Fry B, Maller J, Daly MJ, Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Elbow plot of percent variation explained (y-axis) by number of principal components (x-axis) indicating that beyond 50 PCs in each individual sample little additional variation is explained.
Linkage disequilibrium structure across the 2Mb local ancestry region using 1000 Genomes data for the A) CEU population (ELA) and B) YRI population (ALA). Structure is based on D’ calculation in Haploview [45]. The yellow star represents the location of the APOE gene.
UMAP dimensionality reduction plot visualization of single nuclei clusters from 30,002 nuclei integrated across three ELA and three ALA samples from Emory ADRC.
Visualization of APOE expression in nuclei from three ELA and three ALA samples from Emory ADRC. Cells are overlaid with gene expression information with expression depicted from gray (low) to purple (high).
Single cell Allen Brain Map heatmap for the top 10 marker genes for each cluster (one per page) used to identify cell types (https://celltypes.brain-map.org/rnaseq/human/cortex).
List of all differentially expressed genes between AA and NHW ancestry within each cluster. Avg_logFC is the log base 2 fold change. P_val_adj is the MAST FDR corrected p-value.
Pathway enrichment analysis in KEGG and GO Ontology Biological Process for the differentially expressed genes from all clusters of a given cell-type. Each tab represents a different cell type.
Heatmap of top 10 marker genes defining each cell type. Each column represents a cell and each row a gene. High expression is red and lower expression is blue.
Data Availability Statement
Raw sequencing data (FASTQ) and CellRanger pipeline outputs (barcode, feature, and matrix files) are available for each sample by email request to the corresponding author.