Abstract
SARS-CoV-2 nucleocapsid protein-based COVID-19 diagnosis is a promising alternative to the high-priced, time-consuming, and labor-intensive RT-PCR tests. Here, we developed a rapid, dip-type, wash-free plasmonic fiber optic absorbance biosensor (P-FAB) strategy for the point-of-care detection of SARS-CoV-2 N-protein, expressed abundantly during the infection. P-FAB involves a sandwich assay with plasmonic labels on the surface of a U-bent fiber optic sensor probe with a high evanescent wave absorbance (EWA) sensitivity. The SARS-CoV-2 N-protein is quantified in terms of the change in the intensity of the light propagating through the U-bent sensor probe coupled to a green LED and a photodetector. Firstly, the optical fiber material (silica vs. polymeric optical fiber), was evaluated to realize a sensitive sensor platform. The optimal size of AuNP labels (20, 40, and 60 nm) to achieve high sensitivity and a lower limit of detection (LoD) was investigated. Following the P-FAB strategy, fused silica/glass optical fiber (GOF) U-bent senor probe and citrate-capped AuNP labels (size ~40 nm) gave rise to an LoD down to ~2.5 ng/mL within 10 mins of read-out time. Further, studies on development and validation of a point of care (PoC) read-out device, and preclinical studies are in progress.
Keywords: Biosensors, medical diagnosis, nanoparticles, nanophotonics, optical fiber sensor, COVID-19, SARS-CoV-2, N-protein, plasmonic fiberoptic absorbance biosensor
I. Introduction
The coronavirus disease-2019 (COVID-19), a highly transmissible respiratory viral infection caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has caused a public health emergency worldwide since the first case reported from Wuhan, China, in December 2019 [1]. WHO recommends mass screening to identify infected people (with or without symptoms), tracking of infected individuals, and tracing their contacts to effectively reduce and break the spread of the viral infection. Several methods are being developed to diagnose the COVID-19 either directly by detecting the viral RNA or antigens or indirectly by measuring the antibodies against the virus in the infected host [2]. The reverse transcription-polymerase chain reaction (RT-PCR) based nucleic acid detection is widely used to detect SARS-CoV-2 infection, although it is labor-intensive and requires numerous reagents and specialized infrastructure, and time-consuming. In addition, the RT-PCR assays amplify specific target loci, and the sample without the target locus will be reported negative. It is found that the deletion, insertion or recombination, and interchange are recurrent in coronaviruses [3], [4]. Hence, even a single nucleotide polymorphism at the probe binding site or primer could result as negative irrespective of the active viral infection. As an alternative, several rapid diagnostic tests are being developed to enable mass screening, and some of them have received conditional approval under emergency use authorization (EUA) [2].
During the initial stages of the COVID-19 pandemic, lateral flow assay (LFA) based serological (serum antibodies against the SARS-CoV-2 antigens) tests have been deployed in some countries as an alternative to RT-PCR, at least in the resource-poor settings, including India, due to the lack of PCR equipment and reagents. This alternative was tried with an anticipation to quickly trace the affected individuals and control the spread, mainly for emergency use [5]. However, antibody-based tests have failed to serve the purpose as it can take 2-3 weeks for the host to produce viral-specific antibodies, and false-positive due to interfering antibodies present in the serum because of prior COVID-like illness.
Among the SARS-CoV-2 proteins, the spike glycoprotein [6], [7] and the nucleocapsid protein (N-protein) in particular have gained attention as disease-specific antigens. SARS-CoV-2 N-protein, a structural protein that binds to the viral genome, is the preferred biomarker over the S-protein that facilitates viral attachment to the host cells and is known to be mutagenic [8]. Also, the S-protein of SARS-CoV-2 is 92% identical to that of SARS-CoV and therefore, may cause interference towards the specific diagnosis of COVID-19 [9]. Recently, ELISA and Simoa immunoassays based diagnostics are reported for the detection of the SARS-CoV-2 antigens, especially N-protein. However, these methods involve a multistep time-consuming (3-4 hr of incubation time) process, which needs to be performed in a laboratory setup and hence limiting their use, and thus not suitable for large-scale testing. Several other rapid detection strategies are being developed targeting the viral N-protein in biofluids, such as serum and saliva [8], [10]–[13]. Fabiani et. al., developed an electrochemical immunosensor that could detect recombinant SARS-CoV-2 N-protein in saliva at a concentration as low as 8 ng/mL [14]. Shao et. al., developed a sensitive field effect transistor-based sensor which could detect the recombinant SARS-CoV-2 N-protein down to 0.016 fg/mL in buffer solutions [15]. On the other hand, several lateral flow assay (LFA) based kits were developed, which are known to be simple and affordable for detecting the N-protein down to 0.65 ng/mL [8], [16], [17]. While the specificity of the LFA is acceptable, their utility is limited due to poor sensitivity (< 85%) with reference to RT-PCR [10]. Therefore, alternate point-of-care (PoC) diagnostic technologies with high sensitivity and the potential of quantifying N-protein concentrations in a saliva/swab sample are in high demand.
Recently, our research group has demonstrated a plasmonic fiberoptic absorbance biosensor (P-FAB) strategy for the detection of a urinary tuberculosis biomarker, i.e. mannose-lipoarabinomannan (manLAM), at sub-femtomolar concentration [18]. P-FAB works on the principle of the realization of a sandwich immunoassay with gold nanoparticle (AuNP) labels on a U-bent multimode fiber optic probe surface. The high evanescent wave absorbance sensitivity of the probes enables the detection of a small number of plasmonic labels, known to possess a high optical extinction coefficient. The light passing through the U-bent region of the probe is absorbed by the AuNP labels in proportion to the concentration of the analytes present in a sample. Given that the probes are made of a multimode fiber, these optical intensity measurements can easily be carried out with the help of a portable and inexpensive set-up consisting of a pair of a green light emitting diode (LED), a photodetector, and bare fiber adaptors [19]. Here, we report the development of a P-FAB for the detection of SARS CoV-2 N-protein. This study investigates the influence of optical fiber material, in particular the plastic optical fiber (POF) vis-à-vis the glass optical fiber (GOF), the physisorption vis-à-vis covalent immobilization of antibodies, and the AuNP label size towards the realization of COVID-19 diagnostic assay.
II. Methodology
A. Materials
Super Eska™ plastic optical fibers (POF) of core diameter
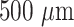 (SK 20) were procured from Mitsubishi Rayon Co., Ltd., Japan. Fused silica optical fibers (GOF) of core diameter
(SK 20) were procured from Mitsubishi Rayon Co., Ltd., Japan. Fused silica optical fibers (GOF) of core diameter
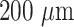 were obtained from CeramOptec GmbH. The bioreagents including mouse anti-SARS-CoV-2 Nucleocapsid protein monoclonal antibody IgG1 (capture antibody), and mouse anti-SARS-CoV-2 Nucleocapsid protein monoclonal antibody IgG2 (detector antibody) were purchased from Meridian Life Sciences, Inc., USA. SARS-CoV-2 N-protein was received from GenScript, USA. Gold(III) chloride (HAuCl4) Phosphate-buffered saline (PBS) tablets, (3-aminopropyl) trimethoxysilane (APTMS), ethyl(dimethylaminopropyl) carbodiimide (EDC), N-hydroxysuccinimide (NHS), poly (ethylene glycol) methyl ether thiol (6000 Da), hexamethylene diamine (HMDA), bovine serum albumin (BSA), sodium chloride, potassium chloride, boric acid, sodium tetraborate and sodium potassium monobasic were procured from Sigma-Aldrich (Merck, India). Gold nanoparticles of varying sizes with optical density (OD = 1) were obtained from BBI Solutions, UK. Sulphuric acid (98%), aqueous glutaraldehyde solution (25%), hydrogen peroxide (30%) were procured from Fisher Chemical (Fisher Scientific, USA). All chemicals used were of analytical grade.
were obtained from CeramOptec GmbH. The bioreagents including mouse anti-SARS-CoV-2 Nucleocapsid protein monoclonal antibody IgG1 (capture antibody), and mouse anti-SARS-CoV-2 Nucleocapsid protein monoclonal antibody IgG2 (detector antibody) were purchased from Meridian Life Sciences, Inc., USA. SARS-CoV-2 N-protein was received from GenScript, USA. Gold(III) chloride (HAuCl4) Phosphate-buffered saline (PBS) tablets, (3-aminopropyl) trimethoxysilane (APTMS), ethyl(dimethylaminopropyl) carbodiimide (EDC), N-hydroxysuccinimide (NHS), poly (ethylene glycol) methyl ether thiol (6000 Da), hexamethylene diamine (HMDA), bovine serum albumin (BSA), sodium chloride, potassium chloride, boric acid, sodium tetraborate and sodium potassium monobasic were procured from Sigma-Aldrich (Merck, India). Gold nanoparticles of varying sizes with optical density (OD = 1) were obtained from BBI Solutions, UK. Sulphuric acid (98%), aqueous glutaraldehyde solution (25%), hydrogen peroxide (30%) were procured from Fisher Chemical (Fisher Scientific, USA). All chemicals used were of analytical grade.
B. Fabrication of U-Bent Fiber Optic Probes
1). Plastic Optical Fiber (POF) Probes:
The plastic optical fiber consists of polymethylmethacrylate (PMMA) core and fluorinated polymer cladding with refractive indices of 1.49 and 1.41, respectively. The POF of 0.5 mm core diameter was bent into a U-shape with a bend ratio (bend diameter/core diameter) of 3 using glass capillary based thermal treatment method as reported earlier from our group [20]. Briefly, a 22 cm long POF is bent manually to bring the two ends close to each other and pushed into a glass capillary having an optimal inner diameter (Fig. S1A). Subsequently, the glass capillary loaded with POF was placed in a hot air oven and heated at 100°C for 10 min in order to obtain a permanent deformation into a U-shape. Thereafter, the U-bent region of the probe was dipped in ethyl acetate for 2 min to remove the cladding over a length of 5 mm as shown in Fig. S1A. Then, the decladded sensing portion of the probe was wiped with a lint-free tissue to remove the flakes over the bent portion and cleaned thoroughly with DI water.
2). Fused Silica Optical Fiber (GOF) Probes:
The GOF consists of a fused silica core surrounded by a silica clad, and a polymer buffer layer. U-bent GOF probes with optimal bend diameter were made using a customized CO2 laser bending machine (Fig. S1B). Briefly, a 20 cm long piece of GOF with a
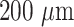 core diameter was subjected to the bending process with the help of a built-in-house fiber bending machine (Fig. S2), which is equipped with a CO2 laser and motors capable of performing buffer ablation followed by bending of a straight portion of the fiber with the desired bend diameter. Thereafter, the bent fiber probes were sonicated and wiped with acetone to remove the black char and debris due to the ablation process. The U-bent region of the fiber probes was dipped into 40% HF solution for 5 minutes to remove the fused silica clad. (Note: The 5 minutes of etching time was optimized by microscopic examination of the diameter of fiber probes every few min of etching. The fiber diameter was measured after every minute until it reduced to
core diameter was subjected to the bending process with the help of a built-in-house fiber bending machine (Fig. S2), which is equipped with a CO2 laser and motors capable of performing buffer ablation followed by bending of a straight portion of the fiber with the desired bend diameter. Thereafter, the bent fiber probes were sonicated and wiped with acetone to remove the black char and debris due to the ablation process. The U-bent region of the fiber probes was dipped into 40% HF solution for 5 minutes to remove the fused silica clad. (Note: The 5 minutes of etching time was optimized by microscopic examination of the diameter of fiber probes every few min of etching. The fiber diameter was measured after every minute until it reduced to
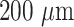 or lower to ensure complete etching of fluorinated silica clad layer). Then, the decladded probes were washed with DI water and sonicated in acetone for 2 to 5 mins.
or lower to ensure complete etching of fluorinated silica clad layer). Then, the decladded probes were washed with DI water and sonicated in acetone for 2 to 5 mins.
C. Functionalization of U-Bent Sensor Probes
The U-bent sensor probes were treated with suitable chemical agents to generate a functional surface for the immobilization of capture antibody molecules (anti-SARS CoV-2 N-protein IgG1).
1). POF Sensor Probes:
The POF sensor probes were modified using three different chemical treatment strategies, including (i) acid hydrolysis, (ii) acid-hydrolysis followed by EDC/NHC treatment, and (iii) acid hydrolysis followed by HMDA treatment. Briefly, in acid hydrolysis, the U-bent POF sensor probes were treated with 1 M H2SO4 for 1 hr followed by incubation in methanol:HCl (1:1) for 1 hr at room temperature (RT) to generate hydroxyl (-OH) groups on the surface. Later, the acid treated sensor probes were directly utilized for the immobilization of capture antibodies. In the second strategy, the sensor probes were incubated in methanol:HCl (1:1) for 1 hr at RT (anticipating generation of carboxyl groups) followed by a freshly prepared mixture of EDC (100 mM) and NHS (200 mM) (prepared in 0.1 M MES buffer in 0.9% NaCl, pH 6) for another 1 hr [21]. Then, they were heated in a hot air oven at 60°C for 20 mins and subsequently washed in MES buffer. In the third strategy, i.e. acid hydrolysis followed by HMDA treatment [20], the sensor probes were dipped in 1 M H2SO4 for 5 mins at RT to generate hydroxyl groups on the sensor probe surface. Then, the probes were incubated in 10% v/v HMDA (in 100 mM borate buffer) for 2 h and washed in borate buffer before condensation at 60°C for 15 mins. Further, the aminated sensor probes were incubated in 2.5% glutaraldehyde for 30 mins at RT to generate aldehyde functional groups. for covalent immobilization of capture antibodies.
2). GOF Sensor Probes:
The U-bent silica fiber probes were cleaned by sonication in acetone (15 min, 1000 Watt, 28 kHz). The cleaned U-bent sensing region of the fiber probes was further cleaned with piranha solution (20 min, 60°C) to oxidize and remove any organic contamination as well as generate hydroxyl groups on the sensor surface. Thereafter, the fiber probes were washed with DI water and followed by heat treatment for 1 hr at 115°C for dehydration of the sensor surface. For amino-silanization, the fiber probes were dipped in a 1% solution of APTMS in a 5:2 (v/v) mixture of ethanol and acetic acid (5 min). This was followed by three-times washing of the fiber probes with ethanol and sonication (15 min) and hot air drying (100°C, 1 h). Then, the silanized sensor probes were incubated into 1% glutaraldehyde solution for 30 mins to generate the aldehyde functional groups for covalent immobilization of the capture antibodies.
D. Capture Antibody Immobilization
The functionalized POF and GOF sensor probes were immobilized with anti-SARS CoV-2 N-protein IgG1, referred as capture antibody. Briefly, the functionalized U-bent fiber optic sensor probes were incubated in
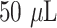 of
of
 /mL of capture antibody solution overnight at 4°C. Then, the antibody immobilized sensor probes were washed thrice in PBST (PBS, 0.01% Tween-20, pH 7.4) and dipped in
/mL of capture antibody solution overnight at 4°C. Then, the antibody immobilized sensor probes were washed thrice in PBST (PBS, 0.01% Tween-20, pH 7.4) and dipped in
 of 5 mg/mL of BSA solution for 15 – 20 mins in order to block the free functional groups (-OH and -CHO) on the sensor probe surface.
of 5 mg/mL of BSA solution for 15 – 20 mins in order to block the free functional groups (-OH and -CHO) on the sensor probe surface.
E. Synthesis of AuNPs and Bioconjugation
The gold nanoparticles were synthesized by citrate-mediated reduction of gold chloride [22]. Briefly, 1 mL of 12.7 mM HAuCl4 was added to 38 mL of DI water and heated to boil and followed by the addition of an aqueous solution of trisodium citrate dihydrate (0.349 mM, 1 mL) (citrate to gold molar ratio = 1.1). The heating was continued until the solution color turns pale pink, indicating the formation of AuNPs. Thereafter, the solution was allowed to cool to RT and then stored at 4°C. The colloidal solution of AuNP was characterized using UV-visible spectroscopy and transmission electron microscopy (TEM) for its optical absorption characteristics and their size and shape distribution, respectively. UV-vis spectroscopic analysis showed a strong absorption peak at 530 nm. TEM analysis revealed the presence of asymmetric AuNPs (elliptical) of size ~40 nm. The number of nanoparticles present in the colloidal solution was calculated as per the method described in section S1 in supporting information.
The plasmonic AuNP labels were prepared by utilizing the affinity of amine and thiol groups on the detection antibodies towards the gold nanoparticles. Commercial AuNPs of sizes 20, 40, and 60 nm (BBI solutions, UK) were also used as labels. For each nanoparticle label, the amount of antibodies required to saturate the AuNP surface was estimated by calculating the surface area of the nanoparticles and the foot-print area of the IgG antibody [23]. The optimal concentration of detector antibodies required to obtain stable conjugates was determined by the NaCl salt test. Briefly,
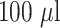 of AuNP solution (OD 1) was adjusted to pH 8.5 and then mixed with anti-SARS CoV-2 N-protein IgG2 antibody of varying quantity (0.125, 0.25, and
of AuNP solution (OD 1) was adjusted to pH 8.5 and then mixed with anti-SARS CoV-2 N-protein IgG2 antibody of varying quantity (0.125, 0.25, and
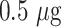 ). The reaction mixture was incubated for 15 mins at RT and followed by the addition of
). The reaction mixture was incubated for 15 mins at RT and followed by the addition of
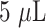 of 10% NaCl solution. After establishing the optimal antibody concentration for each AuNP size distribution, a larger volume (1 mL) of stable AuNP-antibody bioconjugates were prepared as described below. Briefly, an optimum quantity of the anti-SARS CoV-2 N-protein IgG 2 solution was added to the respective AuNP size distributions (1 mL, ~1 OD, pH 8.5) and incubated for 15 mins at RT. Thereafter,
of 10% NaCl solution. After establishing the optimal antibody concentration for each AuNP size distribution, a larger volume (1 mL) of stable AuNP-antibody bioconjugates were prepared as described below. Briefly, an optimum quantity of the anti-SARS CoV-2 N-protein IgG 2 solution was added to the respective AuNP size distributions (1 mL, ~1 OD, pH 8.5) and incubated for 15 mins at RT. Thereafter,
 of
of
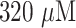 of SH-PEG was added and incubated for 15 mins. Then, the reaction mixture was centrifuged at a suitable RPM depending on the size of the AuNP (11000 rpm for 20 nm size; 8500 rpm for 40 nm, 60 nm and citrate-capped 40 nm) for 20 mins at 4°C to remove any unbound and loosely bound antibodies. The clear supernatant was discarded and the AuNP labels were resuspended in
of SH-PEG was added and incubated for 15 mins. Then, the reaction mixture was centrifuged at a suitable RPM depending on the size of the AuNP (11000 rpm for 20 nm size; 8500 rpm for 40 nm, 60 nm and citrate-capped 40 nm) for 20 mins at 4°C to remove any unbound and loosely bound antibodies. The clear supernatant was discarded and the AuNP labels were resuspended in
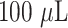 of resuspension buffer containing BSA, sodium salts, sucrose, trehalose, Tween-20, and sodium azide (Voxtur Bio Pvt., Ltd., Mumbai) to obtain
of resuspension buffer containing BSA, sodium salts, sucrose, trehalose, Tween-20, and sodium azide (Voxtur Bio Pvt., Ltd., Mumbai) to obtain
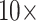 concentration of AuNP conjugates.
concentration of AuNP conjugates.
F. Optical Absorbance Measurements
The optical set-up for P-FAB involves a pair of simple LED and a photodetector. The capture antibody immobilized sensor probe was coupled to a green LED light source (525 nm wavelength, built-in-house) and a photodetector (S150C, Thorlabs Inc., USA) at either side using bare fiber adaptors. The intensity of the light propagating through (at a particular wavelength of choice, 530 nm) the sensor probe was monitored in real-time and recorded in a PC through PM 100 console (Thorlabs Inc., USA). Any change in the optical intensity arising due to the formation of the immunocomplex on the sensor probe surface was recorded. Later, an absorbance response was derived from the temporal sensor response by taking the logarithmic ratio of initial intensity and final intensity, mainly to appreciate small changes in the optical intensity.
G. Plasmonic Fiberoptic Absorbance Biosensor (P-FAB) Strategy
The P-FAB strategy involves the realization of a plasmonic sandwich immunoassay using AuNP labels on a U-bent fiber optic sensor probe, as depicted in Fig. 1. Firstly, the sample solution containing the analyte molecules was homogeneously mixed with the plasmonic AuNP label reagent, each of
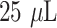 volume. The sample-reagent mixture was kept undisturbed for 10 mins at RT to facilitate the capturing of analyte molecules by the plasmonic labels leading to the formation of AuNP-IgG2-SARS CoV-2 N-protein complex. After the incubation time, the sensor probe connected to the optical setup is dipped into the sample-reagent mixture containing the AuNP-IgG2-SARS CoV-2 N-protein complex. A sandwich immunocomplex (AuNP-IgG2-SARS CoV-2 N-protein-IgG1 complex) is formed on the fiber probe surface. The drop in optical intensity as a result of the formation of the immunocomplex to the sensor probe was monitored in real-time and recorded using the optical set-up. After establishing the proof-of-concept to detect SARS-CoV-2 N-protein, the specificity of the antibodies and the selectivity test were done using SARS-CoV-2 S-protein at varying concentrations. In addition, the non-specific binding (NSB) was evaluated using the sensor probe immobilized with goat anti-human immunoglobin G (GaHIgG) as capture antibody.
volume. The sample-reagent mixture was kept undisturbed for 10 mins at RT to facilitate the capturing of analyte molecules by the plasmonic labels leading to the formation of AuNP-IgG2-SARS CoV-2 N-protein complex. After the incubation time, the sensor probe connected to the optical setup is dipped into the sample-reagent mixture containing the AuNP-IgG2-SARS CoV-2 N-protein complex. A sandwich immunocomplex (AuNP-IgG2-SARS CoV-2 N-protein-IgG1 complex) is formed on the fiber probe surface. The drop in optical intensity as a result of the formation of the immunocomplex to the sensor probe was monitored in real-time and recorded using the optical set-up. After establishing the proof-of-concept to detect SARS-CoV-2 N-protein, the specificity of the antibodies and the selectivity test were done using SARS-CoV-2 S-protein at varying concentrations. In addition, the non-specific binding (NSB) was evaluated using the sensor probe immobilized with goat anti-human immunoglobin G (GaHIgG) as capture antibody.
Fig. 1.
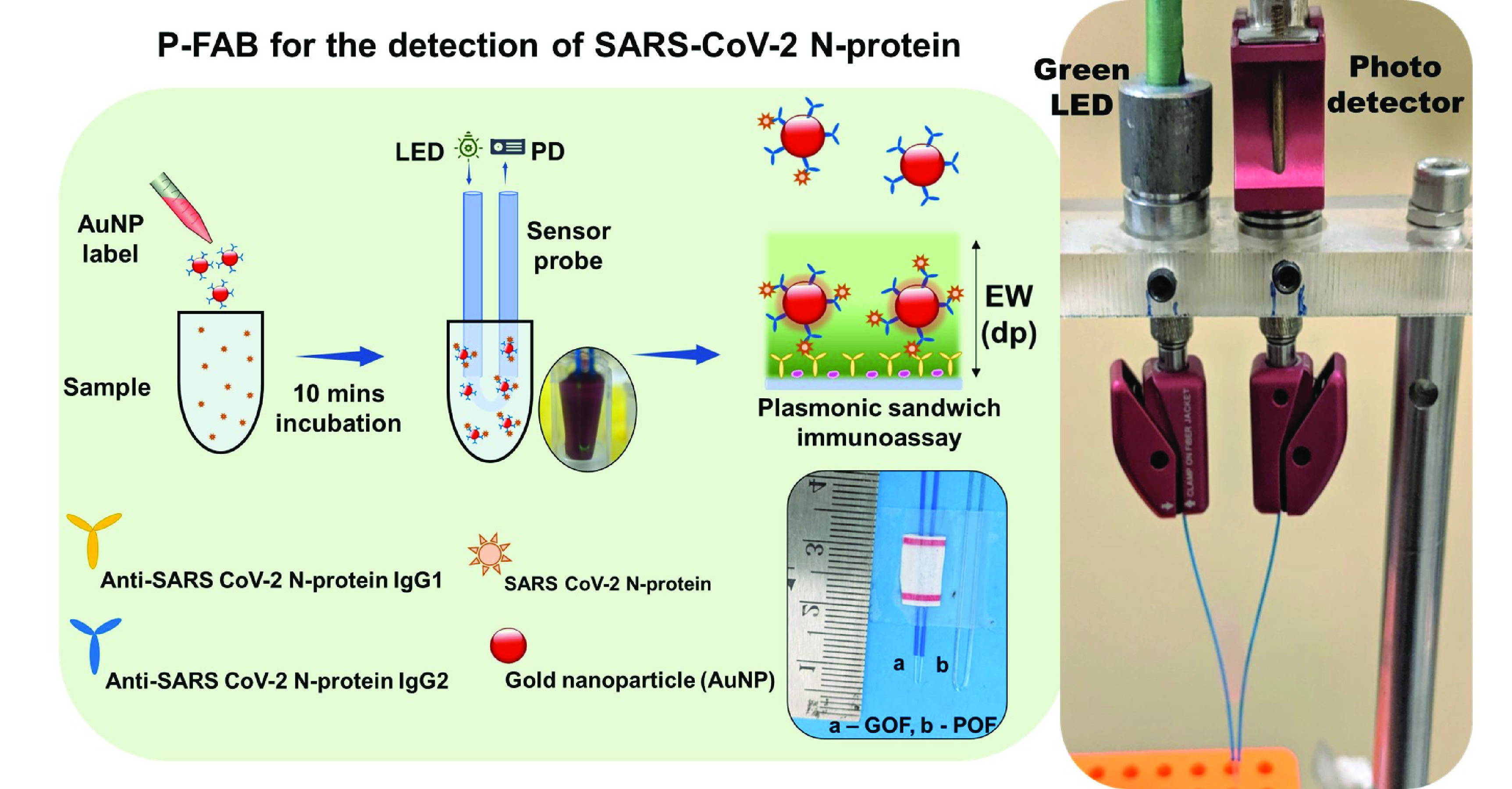
Schematic representation of plasmonic fiberoptic absorbance biosensor (P-FAB) strategy for the detection of SARS-CoV-2 N-protein, photographic image oof GOF and POF sensor probes, and the LED-PD based experimental set-up used for the optical absorbance measurement.
III. Results and Discussion
A. Realization of P-FAB for SARS-CoV-2 N-Protein Detection
P-FAB relies on the use of U-bent fiber optic probes either POF or GOF. Given the recent attention gained by POFs as an alternative to the GOFs. This study investigated the potential of U-bent POF probes as a possible candidate in addition to the conventional GOF with an established surface modification technique.
1). U-Bent POF Probes and Surface Modification Strategies:
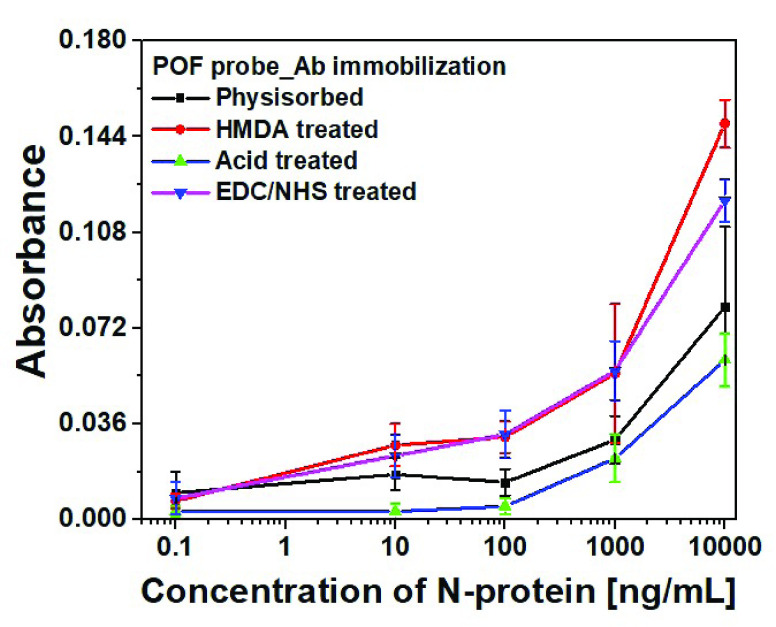
In order to realize U-bent POF probe-based P-FAB for SARS-CoV-2 N-protein detection, optimum conditions for antibody immobilization on POF probe surface were investigated. Capture antibodies were immobilized on U-bent POF probes by following four different methods including (i) simple physisorption of antibodies without any surface pre-treatment, (ii) direct immobilization over the H2SO4 acid-treated POF probes, (iii) EDC/NHS treated POF probes possessing amine-reactive NHS-ester surface groups and (iv) covalent immobilization over HMDA/glutaraldehyde treated POF probes. The sandwich assay was performed for each of the above-mentioned biofunctionalized POF probes.
The P-FAB absorbance response for different concentrations (0.1, 10, 100, and 10000 ng/mL) of SARS-CoV-2 N-protein spiked in PBS buffer (pH 7.4) was recorded as shown in Fig. 2 and S3. Fig. S3 shows the temporal response over a duration of 10 mins from the U-bent POF probes obtained from physisorption and HMDA based covalent immobilization of antibodies. The dose-response curves for each of the modification methods were compared in Fig. 2. We observed that physisorption and acid treatment derived biofunctionalized probes resulted in a lower response even at higher analyte concentrations, i.e. 1000 ng/mL and 10000 ng/mL. In addition, for both approaches, the standard deviation (n = 3) was found to be relatively higher in comparison to the covalent immobilization strategies involving HMDA/glutaraldehyde or EDC/NHS based antibody binding. The sensitivities of the HMDA and EDC/NHS-based strategies, within the range of 0.1 to 100 ng/mL, were found to be 0.0087
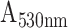 /(log ng/mL) and 0.0079
/(log ng/mL) and 0.0079
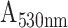 /(log ng/mL) with R-square values 0.98 and 0.98, respectively. Multiple effects such as steric hindrance, lower surface coverage of antibodies, and random orientation of capture antibodies are known to contribute to the high standard deviation in the sensor response. The relatively poor performance for the antibody immobilization by means of physisorption and a high standard deviation may be attributed to the lack of sufficient number of surface functional groups on the PMMA due to its chemical inertness [24], [25]. To obtain the maximum possible sensitivity, HMDA based covalent antibody immobilization was chosen in the subsequent studies.
/(log ng/mL) with R-square values 0.98 and 0.98, respectively. Multiple effects such as steric hindrance, lower surface coverage of antibodies, and random orientation of capture antibodies are known to contribute to the high standard deviation in the sensor response. The relatively poor performance for the antibody immobilization by means of physisorption and a high standard deviation may be attributed to the lack of sufficient number of surface functional groups on the PMMA due to its chemical inertness [24], [25]. To obtain the maximum possible sensitivity, HMDA based covalent antibody immobilization was chosen in the subsequent studies.
Fig. 2.
Dose-response curve obtained from POF sensor probes with capture antibody (Anti-SARS-CoV-2 N-protein IgG1) immobilized by means of physisorption, Acid-treatment, HMDA, and EDC/NHS based covalent binding methodology using a green LED-photodetector setup due to binding of immunocomplex with AuNP labels (40 nm,
 ) conjugated with anti-SARS CoV-2 -N-protein IgG2 (n = 3).
) conjugated with anti-SARS CoV-2 -N-protein IgG2 (n = 3).
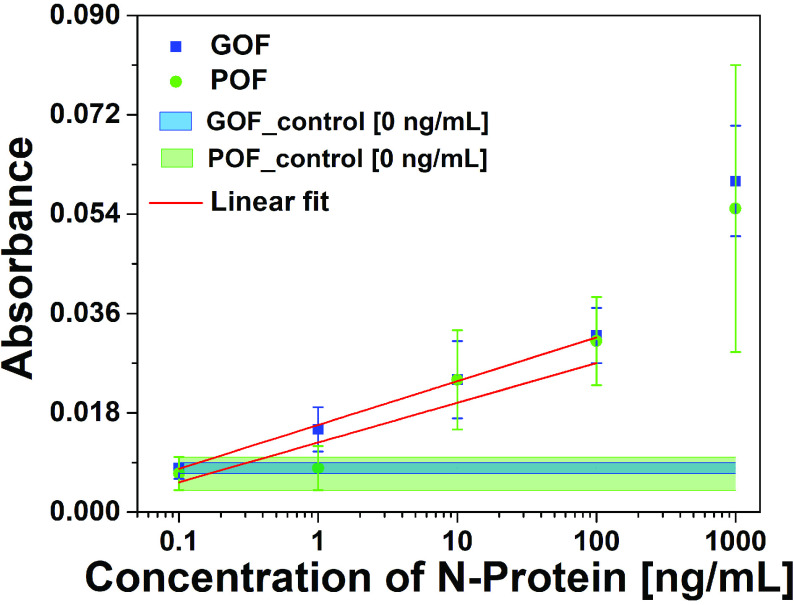
2). Comparison of P-FAB Response From POF and GOF Probes:
The absorbance response of HMDA functionalized POF and GOF sensor probes with covalent immobilization of antibodies were compared in order to identify the sensor probe with higher sensitivity to realize the detection of SARS-CoV-2 N-protein. The sensitivities of the POF and GOF sensor probes within the concentration range of interest between 0.1 ng/mL and100 ng/mL were found to be 0.0072
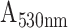 /(log ng/mL) and 0.0079
/(log ng/mL) and 0.0079
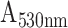 /(log ng/mL) with an R-square value of 0.80 and 0.99, respectively (Fig. 3 and S4).
/(log ng/mL) with an R-square value of 0.80 and 0.99, respectively (Fig. 3 and S4).
Fig. 3.
Absorbance response obtained for a range of SARS-CoV-2 N-protein concentrations from POF and GOF sensor probes chemisorbed with anti-SARS-CoV-2 -N-protein IgG1 with the help of a green LED-photodetector setup due to binding of immunocomplex with AuNP labels (40 nm,
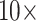 ) conjugated with anti-SARS CoV-2 -N-protein IgG2 (n = 3). The width of the control band represents the standard deviation of respective POF and GOF sensor probes (n = 3).
) conjugated with anti-SARS CoV-2 -N-protein IgG2 (n = 3). The width of the control band represents the standard deviation of respective POF and GOF sensor probes (n = 3).
The detection limits of POF and GOF probes were calculated using
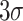 /S (where
/S (where
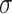 and S refer to the standard deviation in the control response and the sensitivity, respectively) give rise to 17.78 ng/mL and 5.62 ng/mL (Fig. S4). The GOF sensor response was found to be repeatable and with a lesser standard deviation. Also, the POF probes were not able to differentiate between 0.1 ng/mL and 1 ng/mL. Hence, GOF sensor probes were identified as the optimum probe for the P-FAB strategy to realize SARS-CoV-2-N-protein detection. The improved sensitivity of the GOF sensor probes in comparison to POF sensor probes is in agreement with the previous study on the detection of Mycobacterium tuberculosis lipoarabinomannan (M.TB LAM) in urinary samples [18].
and S refer to the standard deviation in the control response and the sensitivity, respectively) give rise to 17.78 ng/mL and 5.62 ng/mL (Fig. S4). The GOF sensor response was found to be repeatable and with a lesser standard deviation. Also, the POF probes were not able to differentiate between 0.1 ng/mL and 1 ng/mL. Hence, GOF sensor probes were identified as the optimum probe for the P-FAB strategy to realize SARS-CoV-2-N-protein detection. The improved sensitivity of the GOF sensor probes in comparison to POF sensor probes is in agreement with the previous study on the detection of Mycobacterium tuberculosis lipoarabinomannan (M.TB LAM) in urinary samples [18].
B. The Optimal Size of AuNP Labels for P-FAB
The sensitivity of the P-FAB-based sensing strategy originates from the unique ability of the U-bent optical fiber probes to measure the absorbance due to the plasmonic AuNP labels binding at the sensing region. The influence of AuNP label size on the P-FAB response is multi-faceted. The parameters that influence the P-FAB response include (i) the optical extinction property of the AuNP labels, which enables the ultra-low analyte detection limits, is highly dependent on their size and shape [18], [23], (ii) the number of active detector antibodies bound to the AuNP labels and the accessibility of their binding sites for the analyte molecules and (iii) the concentration of the AuNP labels (bioconjugates) that determines the availability of a sufficient number of bioconjugates towards the efficient capturing of analytes and the diffusion of the immunocomplex in the solution phase towards the probe surface. Here, AuNP labels of four different distributions were highly uniform and narrow size range around 20, 40, and 60 nm (procured from BBI solutions, UK) as well as asymmetric AuNP with a wider distribution (citrate-capped AuNP labels) prepared-in-house were investigated.
1). Theoretical Estimate for Antibody Conjugation to AuNP
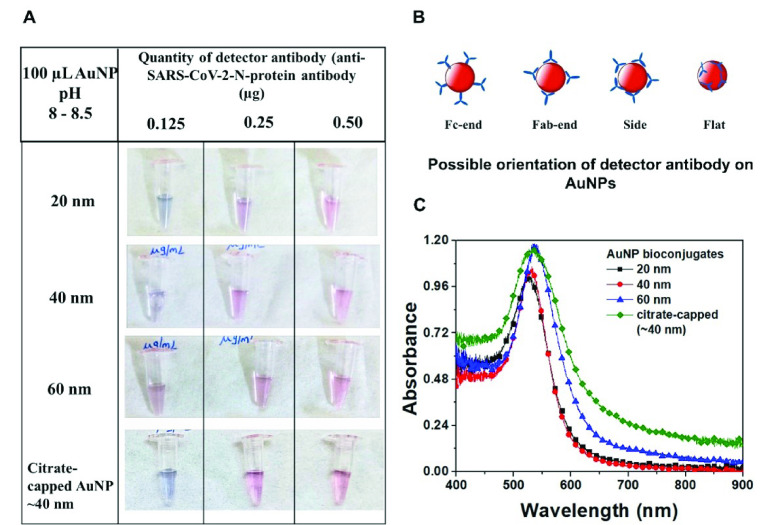
Evaluation of an optimum size of AuNP as plasmonic labels for the P-FAB with the highest sensitivity requires preparation of stable nanobioconjugates. Given that the optical density at the peak plasmonic wavelength of the AuNP of different sizes equal to unity is taken as reference, the optimum concentration of detector antibody (anti-SARS-CoV-2 N-protein IgG2) required to obtain stable conjugates with saturated surface coverage was theoretically estimated (Table S1). In general, the antibodies may be bound to the AuNP surface with one of the possible orientations, including Fc-end, Fab-end, side-on or flat-on (Fig 4B), depending on the pH used during the conjugation process [26]. The antibodies with Fc-end orientation offer a greater number of accessible binding sites. Subsequently, the antibody concentration required to achieve the saturated surface coverage for the chosen AuNP size distributions was estimated based on the surface area of nanoparticles and the footprint area of detector antibodies as 91 nm2.
Fig. 4.
(A) Photographic images of AuNP conjugates of varying sizes subjected to NaCl stability test. Stable conjugates remain pink, while the AuNP labels with insufficient surface coverage agglomerates and turns violet or colorless. (B) Schematic representation showing the possible orientation of IgG on the AuNP surface. (C) Absorbance spectra of AuNP labels for varying sizes obtained using USB 4000 spectrometer. A broader full-width half maxima of the citrate-capped AuNPs indicates a 30-40% wider size distribution in comparison to 40 nm AuNP.
Experimental investigations were carried out to verify the optimum antibody concentration required for the conjugation of the chosen AuNP size distributions. Fig. 4A shows the photographic images of
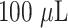 of AuNPs conjugated with 0.125, 0.25,
of AuNPs conjugated with 0.125, 0.25,
 of detector antibody and subsequently subjected to 10% NaCl solution. The AuNP conjugates with incomplete surface coverage tend to agglomerate, while the conjugates with complete or saturated surface coverage remain stable. Theoretically estimated minimum antibody concentration values correlate well with the experimental observations, where 20 nm, 40 nm, and citrate-capped 40 nm AuNPs require more antibodies (
of detector antibody and subsequently subjected to 10% NaCl solution. The AuNP conjugates with incomplete surface coverage tend to agglomerate, while the conjugates with complete or saturated surface coverage remain stable. Theoretically estimated minimum antibody concentration values correlate well with the experimental observations, where 20 nm, 40 nm, and citrate-capped 40 nm AuNPs require more antibodies (
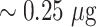 , containing
, containing
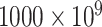 nos.) while 60 nm requires relatively less concentration (
nos.) while 60 nm requires relatively less concentration (
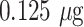 containing
containing
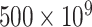 antibodies) for saturated surface coverage. The absorbance spectra of these bioconjugates of different AuNP sizes are compared with that of the respective bare AuNP (Fig. S5). No significant plateau around 600 nm was observed, confirming the absence of AuNP agglomeration and hence suggesting good stability at the saturated surface coverage of antibodies (Fig. 4C).
antibodies) for saturated surface coverage. The absorbance spectra of these bioconjugates of different AuNP sizes are compared with that of the respective bare AuNP (Fig. S5). No significant plateau around 600 nm was observed, confirming the absence of AuNP agglomeration and hence suggesting good stability at the saturated surface coverage of antibodies (Fig. 4C).
2). Optimum AuNP Label Size for N-Protein Detection:
The P-FAB response to various concentrations of SARS-CoV-2 N-protein for each of the four AuNP distributions was obtained Fig. S6. The representative temporal absorbance response curves for citrate-capped asymmetric AuNP prepared in-house are shown in Fig. 5A. Fig. 5B depicts the dose-response curves obtained for each of the AuNP size distributions. We observe that AuNP 20 nm labels gave rise to relatively a poor sensitivity towards the analyte detection at all concentration levels in comparison to the 40 and 60 nm AuNP labels. This could be attributed to the lower extinction coefficient of 20 nm size AuNP, which is the sum of absorption coefficient and scattering coefficient. These results corroborated with our previous study on IgG analyte detection [18].
Fig. 5.
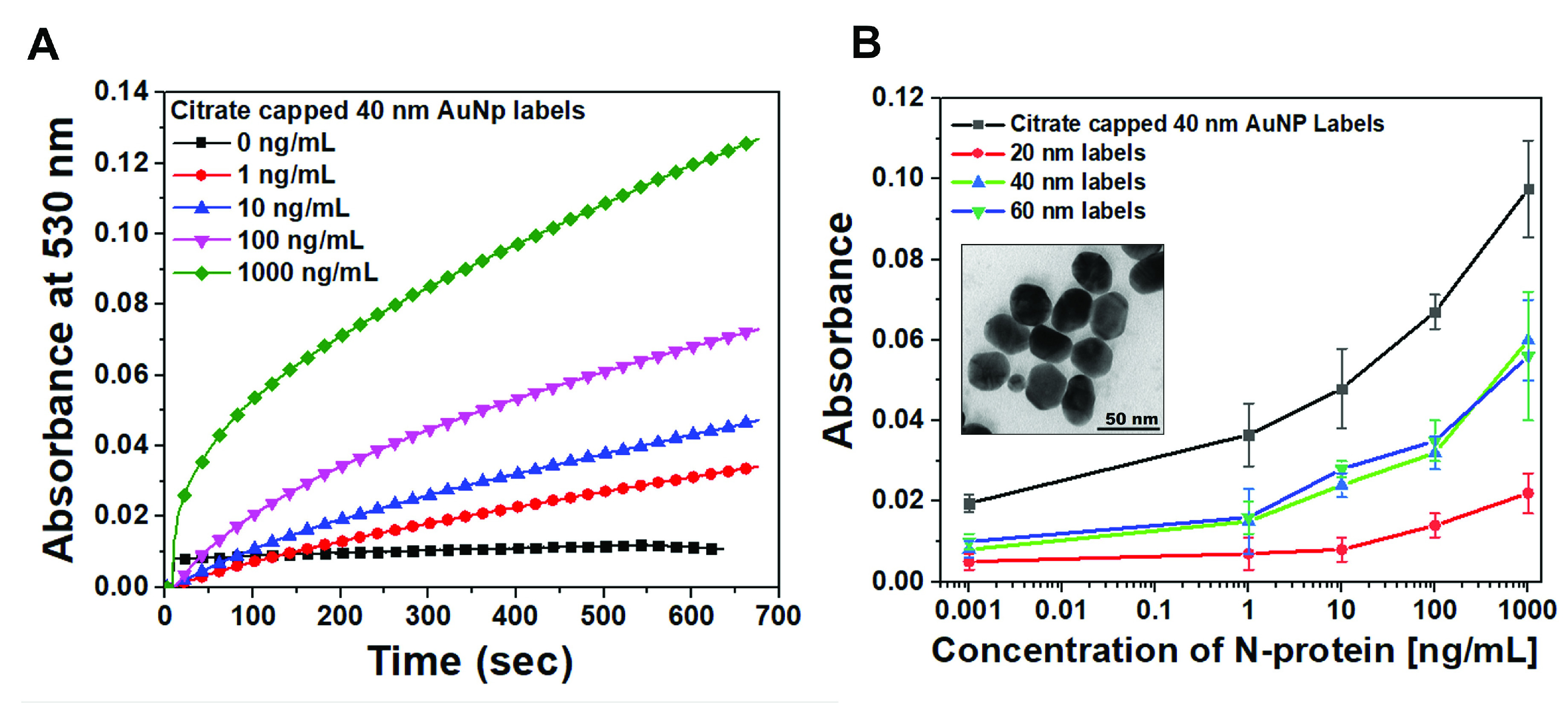
(A) Representative temporal absorbance response and (B) dose-response curves obtained for a range of SARS-CoV-2 N-protein concentrations from GOF sensor probes chemisorbed with anti-SARS-CoV-2-N-protein IgG1 with the help of the optical experimental setup due to binding of immunocomplex with AuNP labels varying sizes and
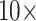 concentration conjugated with anti-SARS CoV-2 -N-protein IgG2 (n = 3). Inset: TEM images of citrate reduced AuNPs prepared in-house.
concentration conjugated with anti-SARS CoV-2 -N-protein IgG2 (n = 3). Inset: TEM images of citrate reduced AuNPs prepared in-house.
Interestingly, the citrate-capped 40 nm AuNP labels show approximately 2-fold higher sensitivity in comparison to the size-controlled distributions in the range of 40 and 60 nm size AuNP. The improved sensitivity could be attributed to the asymmetric spherical shape of AuNP, which facilitates more accessibility of binding sites of detector antibodies by reducing the steric hindrance and a higher extinction coefficient. A detailed understanding of the phenomenon requires a more elaborate investigation, which is outside the scope of this study. Overall, the sensitivity of the GOF sensor probes for SARS-CoV-2 N-protein detection using the citrate-capped labels within the range of interest between 0.1 ng/mL and 100 ng/mL was found to be 0.0151
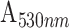 /(log ng/mL) with an R-square value of 0.99. The detection limit calculated using
/(log ng/mL) with an R-square value of 0.99. The detection limit calculated using
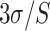 gives rise to ~2.5 ng/mL (Fig. S7). Furthermore, P-FAB was realized using a standard reference sample for N-protein as a simulated analyte, provided by the Indian Council of Medical Research (ICMR) to validate antigen-based diagnostic kits (Courtesy: Voxtur Bio Ltd, Mumbai, India). Under the optimum conditions using U-bent GOF probes and lab-prepared citrate-capped 40 nm AuNP labels, the P-FAB response for various dilutions of N-protein reference sample down to
gives rise to ~2.5 ng/mL (Fig. S7). Furthermore, P-FAB was realized using a standard reference sample for N-protein as a simulated analyte, provided by the Indian Council of Medical Research (ICMR) to validate antigen-based diagnostic kits (Courtesy: Voxtur Bio Ltd, Mumbai, India). Under the optimum conditions using U-bent GOF probes and lab-prepared citrate-capped 40 nm AuNP labels, the P-FAB response for various dilutions of N-protein reference sample down to
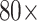 (in PBS) was obtained as shown in Fig. S8.
(in PBS) was obtained as shown in Fig. S8.
Table I showcases the summary of the relevant biosensing schemes reported in the literature to detect SARS-CoV-2 N-protein in buffer or serum matrix and the respective LoD values. The LoD of P-FAB was found to be comparable to the LFA-based system [8], [16], [17]. However, the LOD was slightly inferior to some of the electrochemical and FET based sensing strategies [15], [27], [28].
TABLE I. Biosensors Strategies Reported for SARS-CoV-2 N Protein Detection.
| Transducer Technique | Sample matrix | Limit of detection | Ref |
|---|---|---|---|
| LFA* | Buffer | 0.65 ng/mL (Genemedi) 3.03 ng/mL (GenScript) |
[8] |
| Electrochemical# | Whole serum 5× diluted serum | 0.23 ng/mL 0.1 ng/mL | [11] |
| Electrochemical* | Buffer Saliva |
4 ng/mL 8 ng/mL |
[14] |
| Field effect Transistor* | Buffer | 0.016 fg/mL | [15] |
| LFA* | Buffer | 2 ng/mL | [16] |
| LFA# | Buffer | 2.2 ng/mL | [17] |
| Electrochemical# | Buffer | ~0.7 pg/mL | [28] |
| Electrochemical# | Buffer | 8.33 pg/mL | [27] |
| Microfiber* | Buffer Fetal bovine serum | ~3 fg/mL 50 ng/mL | [29] |
| P-FAB* | PBS Buffer | 2.48 ng/mL | Present study |
Recombinant SARS-CoV-2 N-protein,
SARS-CoV-2 N-protein
It is interesting to note that the sensitivity and limit of detection values obtained here are inferior to our previous reports, where P-FAB is demonstrated to detect analytes down to sub-femtomolar concentration [30]. This could be attributed to the analyte molecular weight and the binding affinities of the antibodies used in this study [31]–[33]. It may also be noted that the results obtained in this study utilize a recombinant N-protein, which may not precisely corroborate with the native N-protein of SARS-CoV-2 [8]. Hence, we anticipate a better sensitivity using N-protein derived from SARS-CoV-2 and clinical samples, which is in progress. Given the advantages of P-FAB including ease in scalability for mass production, quantitative measurement, and simplicity in use it could be a viable alternative for lateral flow assay kits.
C. Antibody Selectivity Study
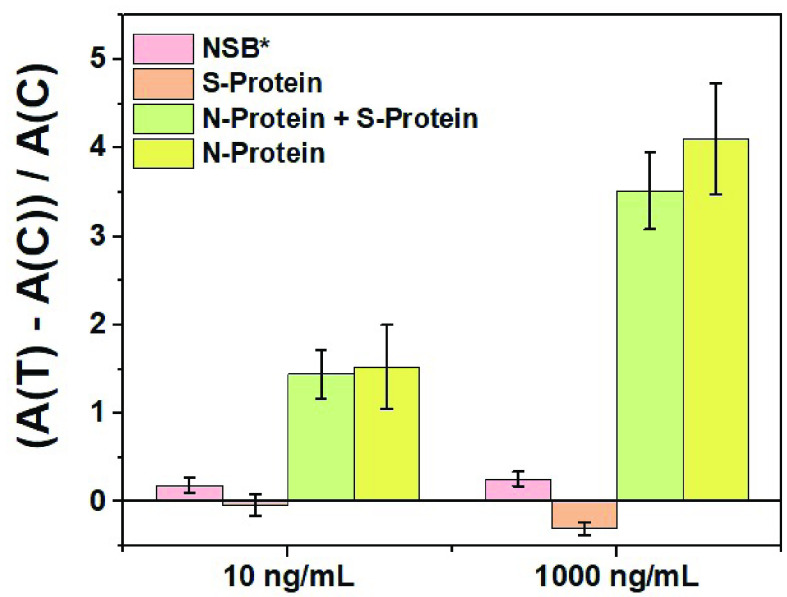
In continuation to the optimization and realization of the P-FAB strategy to detect SARS-CoV-2 N-protein, the antibody specificity and selectivity studies were carried out using SARS-CoV-2 S-protein as a negative control (Fig. 6 and S9). Firstly, to evaluate the non-specific binding (NSB) of anti-SARS-CoV-2 N-protein IgG2 conjugated AuNP labels to the sensor surface, the assay was performed using GOF sensor probes immobilized with GaHIgG antibodies, which are non-specific for the SARS CoV-2 N-protein (Fig. S9). In the absence of a specific analyte, the absorbance response was found to be negligible and consistent even with sensor probes immobilized with anti-SARS CoV-2 N-protein IgG1 (control = 0 ng/mL). To appreciate the selectivity and specificity of the sensor probe, the absorbance response obtained from each probe is normalized by taking the ratio of the difference in the absorbance value between the test probe and control probe (0 ng/mL) to the average response obtained from the control (0 ng/mL). The normalized sensor response obtained from GOF sensor probe immobilized with anti-SARS-CoV-2 N-protein IgG1 and anti-SARS-CoV-2 N-protein IgG2 as AuNP labels using SARS-CoV-2 S-protein as analyte (Fig. S9) shows no or negligible change in the absorbance even at high concentration (1000 ng/mL) of SARS-CoV-2 S-protein. This suggests the high selectivity of the capture and detector antibodies towards the SARS CoV-2 N-protein (Fig. 6). Further, the GOF sensor probes were subjected to a sample containing an equal amount of SARS-CoV-2 N-protein (specific analyte) and S-protein (non-specific analyte) to investigate any potential interference during the assay. The normalized results show a small drop of about 14% in the response due to the specific analyte binding even in the presence of interfering S-protein at a concentration as high as 1000 ng/mL (Fig. 6) in comparison to the SARS-CoV-2 N-protein alone. This could be due to limited diffusion in the presence of high protein concentration. However, such high analyte concentrations may fall outside the range of clinical relevance.
Fig. 6.
Normalized absorbance response obtained from U-bent GOF sensor probes immobilized with anti-SARS CoV-2 IgG1 and * goat anti-HIgG due to specific and non-specific binding (NSB) of anti-SARS CoV-2 N-protein IgG2-AuNP labels with at varying concentrations (n = 3), respectively. A (C) refers to the average absorbance response obtained from the control experiments for NSB, while A (T) refers to the absorbance response obtained from each of the analyte concentrations (specific or non-specific).
IV. Conclusion
P-FAB strategy for the detection of SARS-CoV-2 N-protein is optimized with respect to the type of optical fiber probe, and AuNP size to obtain an optimal sensitivity. The combination of fused silica sensor probes along with AuNP labels of irregular spheres (citrate-capped 40 nm AuNPs) and non-uniform distribution was found to be optimal to have maximum sensitivity and detection limits. A proof-of-concept for the detection of SARS-CoV-2 N-protein using the P-FAB strategy is realized with a detection limit of about ~2.5 ng/mL spiked in PBS buffer within 10 mins of the read-out time. Further studies are in progress to improve the sensitivity of the SARS-CoV-2 N-protein detection through analysing the binding affinity of the antibodies. In addition, development of a portable readout device, and its validation is in progress. These preliminary results established that the P-FAB strategy could be used to realize an affordable and rapid diagnostic device for mass-screening of COVID-19.
Supplementary Materials
Acknowledgment
The authors thank M/s Voxtur Bio Private Ltd., Mumbai, India for sparing N-protein reference sample.
Conflict of Interest
There are no conflict of interests to declare.
Biographies

M. Divagar received the B.Sc. degree in biotechnology from SRM University in 2014 and the M.Sc. degree in nanoscience and nanotechnology from the University of Madras, Chennai, in 2016. He is pursuing the Ph.D. degree in fiberoptic biosensors with the Department of Science and Technology (DST), IIT Madras, under the Innovation in Science Pursuit for Inspired Research (INSPIRE) Research Fellowship. He is the author of ten research articles and one invention. His research interests include fiber-optics, biosensors, spectroscopy, medical diagnostics, infectious diseases, nanomaterials, and nanostructures. He was a recipient of the Science Academies Summer Research Fellowship Award (2015), Newton-Bhabha Ph.D. Placement Award (2020–2021), and the Gandhian Young Technological Innovation Award (2020).

R. Gayathri received the B.Sc. degree in biotechnology from SRM Institute of Science and Technology, Chennai, India, in 2017, and the M.Sc. degree in biotechnology from D. G. Vaishnav College, University of Madras, Chennai, in 2019. She worked as a Project Assistant of Synthetic Biology at Anna University, Chennai, from 2019 to 2020. Later, she was a Project Associate with the Biosensor Laboratory, Department of Applied Mechanics, IIT Madras, Chennai, where she worked in medical diagnosis and biosensors. Her research interests include immunodiagnostics, diseases, and therapeutics.

Rahiel Rasool received the B.Tech. degree in biomedical engineering from SRM Institute of Science and Technology, Chennai, India, in 2018, and the M.Tech. degree in nanoscience and technology from Pondicherry Central University, Puducherry, India, in 2020. He is currently working as a Project Associate with the Biosensors Laboratory, Department of Applied Mechanics, IIT Madras. His research interests are fiber optic biosensors, immunodiagnostics, biomaterials, microfluidics, and nanomaterials.

J. Kuzhandai Shamlee received the B.Tech. degree in biotechnology from P. S. R. Engineering College, Sivakasi, India, in 2015, and the M.Tech. degree in biotechnology from P. S. G. College of Technology, Coimbatore, India, in 2017. She is pursuing the Ph.D. degree in fiberoptic biosensors with IIT Madras, Chennai. In 2017, she was a Research Scholar with the Biosensors Laboratory, Department of Applied Mechanics, IIT Madras, where she is an HTRA Research Fellow. Her research interests include bacterial pathogens sensing, disease biomarker detection, and nanomaterials-based sensor development.

Himanshu Bhatia received the M.D. degree from the University of Pune, the M.S. degree from the University of Texas, and the M.B.A. degree from the University of Rochester. He worked as a Research Scientist at Albert Einstein College of Medicine, University of Pittsburgh, and Roswell Park Cancer Institute. He is currently the CEO of RICOVR Healthcare Inc., which is focused on commercializing fiber-optic biosensors for various biomarkers.

Jitendra Satija received the B.Pharm. degree from Rajasthan University, India, in 2005, the M.Pharm. degree from Dr. H. S. Gour University, Sagar, India, in 2008, on pharmaceutics, and the Ph.D. degree from IIT Bombay, India, in 2013, on optical biosensors involving the development of dendrimer-based labeled and label-free plasmonic immunoassays. He is currently an Associate Professor at the Centre for Nanobiotechnology (CNBT), Vellore Institute of Technology (VIT), Vellore, India. His research interests include plasmonic biosensors, nanomaterials in theranostics, fiber-optic biosensors, and sensor matrices design.

V. V. R. Sai received the Ph.D. degree in biomedical engineering from the Indian Institute of Technology, Bombay, in 2009, for the work done on development of two novel fiber optic biosensing technologies for detection of biomolecules and pathogens. He spent two years of postdoctoral fellowship at the University of Idaho, USA, working on development of SERS-based DNA biosensor, receptor mediated detection of explosives, and application of nanomaterials for development of drug delivery systems for anti-sense and anti-gene oligonucleotide-based therapeutics. In August 2011, he came back to India and joined as an Assistant Professor of Biomedical Engineering with the Department of Applied Mechanics, IIT Madras, where he established biosensors laboratory to focus on development of affordable and indigenous technologies for clinical diagnostics, water, food, and environmental monitoring. Since July 2017, he has been working as an Associate Professor. He has authored 31 peer-reviewed research articles, more than 25 conference presentations/proceedings, and filed one U.S. and six Indian patents. He was a recipient of prestigious the Young Engineer Award from Indian National Academy of Engineering (INAE) and the Travel Award from Epson Research Foundation, Japan, in 2015 and 2016, respectively. Currently, two postdoctoral scholars and five research scholars are working with him towards Ph.D. and M.S. degrees.
Funding Statement
This work was supported in part by the COVID-19 Special Grants from the Science and Engineering Research Board (SERB) under Grant CVD/2020/000747 and in part by the Indo-U.S. Science and Technology Forum under Grant USISTEF/ COVID-II/132/2020. The work of M. Divagar was supported by the Innovation in Science Pursuit for Inspired Research (INSPIRE) Ph.D. Fellowship from the Department of Science and Technology, Ministry of Science and Technology, Government of India.
Contributor Information
Himanshu Bhatia, Email: himanshu.bhatia@ricovr.com.
Jitendra Satija, Email: jsatija11@gmail.com.
V. V. R. Sai, Email: vvrsai@iitm.ac.in.
References
- [1].COVID-19 Weekly Epidemiological Update, Coronavirus Dis. Situat. Rep., World Health Org., Geneva, Switzerland, Apr. 2021, pp. 1–3. [Google Scholar]
- [2].Giri B., Pandey S., Shrestha R., Pokharel K., Ligler F. S., and Neupane B. B., “Review of analytical performance of COVID-19 detection methods,” Anal. Bioanal. Chem., vol. 413, no. 1, pp. 35–48, Jan. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cui J., Li F., and Shi Z.-L., “Origin and evolution of pathogenic coronaviruses,” Nature Rev. Microbiol., vol. 17, no. 3, pp. 181–192, Mar. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Benvenuto D., Giovanetti M., Ciccozzi A., Spoto S., Angeletti S., and Ciccozzi M., “The 2019-new coronavirus epidemic: Evidence for virus evolution,” J. Med. Virol., vol. 92, no. 4, pp. 455–459, Apr. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].To K. K.-W.et al. , “Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study,” Lancet Infectious Diseases, vol. 20, no. 5, pp. 565–574, May 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cennamo N.et al. , “Proof of concept for a quick and highly sensitive on-site detection of SARS-CoV-2 by plasmonic optical fibers and molecularly imprinted polymers,” Sensors, vol. 21, no. 5, pp. 1–17, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lee S.-L., Kim J., Choi S., Han J., Seo G., and Lee Y. W., “Fiber-optic label-free biosensor for SARS-CoV-2 spike protein detection using biofunctionalized long-period fiber grating,” Talanta, Aug. 2021, Art. no. 122801. [DOI] [PMC free article] [PubMed]
- [8].Grant B. D.et al. , “SARS-CoV-2 coronavirus nucleocapsid antigen-detecting half-strip lateral flow assay toward the development of point of care tests using commercially available reagents,” Anal. Chem., vol. 92, no. 16, pp. 11305–11309, Aug. 2020. [DOI] [PubMed] [Google Scholar]
- [9].Wang N.et al. , “Serological evidence of bat SARS-related coronavirus infection in humans, China,” Virologica Sinica, vol. 33, no. 1, pp. 104–107, Feb. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Huggett J. F., Moran-Gilad J., and Lee J. E., “COVID-19 new diagnostics development: Novel detection methods for SARS-CoV-2 infection and considerations for their translation to routine use,” Current Opinion Pulmonary Med., vol. 27, no. 3, pp. 155–162, 2021. [DOI] [PubMed] [Google Scholar]
- [11].Li J. and Lillehoj P. B., “Microfluidic magneto immunosensor for rapid, high sensitivity measurements of SARS-CoV-2 nucleocapsid protein in serum,” ACS Sensors, vol. 6, no. 3, pp. 1270–1278, Mar. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sadighbayan D. and Ghafar-Zadeh E., “Portable sensing devices for detection of COVID-19: A review,” IEEE Sensors J., vol. 21, no. 9, pp. 10219–10230, May 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tayyab M., Sami M. A., Raji H., Mushnoori S., and Javanmard M., “Potential microfluidic devices for COVID-19 antibody detection at point-of-care (POC): A review,” IEEE Sensors J., vol. 21, no. 4, pp. 4007–4017, Feb. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Fabiani L.et al. , “Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: A reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva,” Biosensors Bioelectron., vol. 171, Jan. 2021, Art. no. 112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Shao W., Shurin M. R., Wheeler S. E., He X., and Star A., “Rapid detection of SARS-CoV-2 antigens using high-purity semiconducting single-walled carbon nanotube-based field-effect transistors,” ACS Appl. Mater. Interfaces, vol. 13, no. 8, pp. 10321–10327, Mar. 2021. [DOI] [PubMed] [Google Scholar]
- [16].Kim H.-Y.et al. , “Development of a SARS-CoV-2-specific biosensor for antigen detection using scFv-fc fusion proteins,” Biosensors Bioelectron., vol. 175, Mar. 2021, Art. no. 112868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Guo J.et al. , “5G-enabled ultra-sensitive fluorescence sensor for proactive prognosis of COVID-19,” Biosensors Bioelectron., vol. 181, Jun. 2021, Art. no. 113160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].M D., Bandaru R., Janakiraman V., and Sai V. V. R., “A plasmonic fiberoptic absorbance biosensor for mannose-capped lipoarabinomannan based tuberculosis diagnosis,” Biosensors Bioelectron., vol. 167, Nov. 2020, Art. no. 112488. [DOI] [PubMed] [Google Scholar]
- [19].Esposito F., Srivastava A., Sansone L., Giordano M., Campopiano S., and Iadicicco A., “Label-free biosensors based on long period fiber gratings: A review,” IEEE Sensors J., vol. 21, no. 11, pp. 12692–12705, Jun. 2021. [Google Scholar]
- [20].Gowri A. and Sai V. V. R., “Development of LSPR based U-bent plastic optical fiber sensors,” Sens. Actuators B, Chem., vol. 230, pp. 536–543, Jul. 2016. [Google Scholar]
- [21].Kling A., Dincer C., Armbrecht L., Horak J., Kieninger J., and Urban G., “Electrochemical microfluidic platform for simultaneous multi-analyte detection,” Procedia Eng., vol. 120, pp. 916–919, Jan. 2015. [Google Scholar]
- [22].Turkevich J., Stevenson P. C., and Hillier J., “A study of the nucleation and growth processes in the synthesis of colloidal gold,” Discuss. Faraday Soc., vol. 11, p. 55, May 1951. [Google Scholar]
- [23].Divagar M. and Sai V. V. R., “Fiber optic plasmonic sandwich immunosensor: Influence of AuNP label size and concentration,” in Proc. IEEE SENSORS, Oct. 2018, pp. 1–4. [Google Scholar]
- [24].Taheri R. A., Rezayan A. H., Rahimi F., Mohammadnejad J., and Kamali M., “Comparison of antibody immobilization strategies in detection of Vibrio cholerae by surface plasmon resonance,” Biointerphases, vol. 11, no. 4, Dec. 2016, Art. no. 041006. [DOI] [PubMed] [Google Scholar]
- [25].Vijayendran R. A. and Leckband D. E., “A quantitative assessment of heterogeneity for surface-immobilized proteins,” Anal. Chem., vol. 73, no. 3, pp. 471–480, Feb. 2001. [DOI] [PubMed] [Google Scholar]
- [26].Ruiz G., Tripathi K., Okyem S., and Driskell J. D., “PH impacts the orientation of antibody adsorbed onto gold nanoparticles,” Bioconjugate Chem., vol. 30, no. 4, pp. 1182–1191, Apr. 2019. [DOI] [PubMed] [Google Scholar]
- [27].Tian J., Liang Z., Hu O., He Q., Sun D., and Chen Z., “An electrochemical dual-aptamer biosensor based on metal-organic frameworks MIL-53 decorated with AuPt nanoparticles and enzymes for detection of COVID-19 nucleocapsid protein,” Electrochimica Acta, vol. 387, Aug. 2021, Art. no. 138553. [Google Scholar]
- [28].Raziq A., Kidakova A., Boroznjak R., Reut J., Öpik A., and Syritski V., “Development of a portable MIP-based electrochemical sensor for detection of SARS-CoV-2 antigen,” Biosensors Bioelectron., vol. 178, Apr. 2021, Art. no. 113029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jia H.et al. , “A graphene oxide coated tapered microfiber acting as a super-sensor for rapid detection of SARS-CoV-2,” Lab a Chip, vol. 21, no. 12, pp. 2398–2406, 2021. [DOI] [PubMed] [Google Scholar]
- [30].Ramakrishna B. and Sai V. V. R., “Evanescent wave absorbance based U-bent fiber probe for immunobiosensor with gold nanoparticle labels,” Sens. Actuators B, Chem., vol. 226, pp. 184–190, Apr. 2016. [Google Scholar]
- [31].Sztefko K., “Interferences in immunoassay,” Clin Biochem Rev, vol. 25, no. 105, pp. 477–480, 2004. [PMC free article] [PubMed] [Google Scholar]
- [32].Darwish I. A., “Immunoassay methods and their applications in pharmaceutical analysis: Basic methodology and recent advances,” Int. J. Biomed. Sci., vol. 2, no. 3, pp. 35–217, 2006. [PMC free article] [PubMed] [Google Scholar]
- [33].Wang D.et al. , “Rapid lateral flow immunoassay for the fluorescence detection of SARS-CoV-2 RNA,” Nature Biomed. Eng., vol. 4, no. 12, pp. 1150–1158, Dec. 2020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.