Abstract
Alagille Syndrome (ALGS) is a congenital disorder caused by mutations in the Notch ligand gene, JAGGED1, leading to neonatal loss of intrahepatic duct (IHD) cells and cholestasis. Cholestasis can resolve in certain ALGS patients, suggesting regeneration of IHD cells. However, the mechanisms driving IHD cell regeneration following Jagged loss remains unclear. Here, we show that cholestasis due to developmental loss of IHD cells can be consistently phenocopied in zebrafish with compound jagged1b and jagged2b mutations or knockdown. Leveraging the transience of jagged knockdown in juvenile zebrafish, we find that resumption of Jagged expression leads to robust regeneration of IHD cells via a Notch-dependent mechanism. Combining multiple lineage tracing strategies with whole liver 3D-imaging, we demonstrate that the extrahepatic duct (EHD) is the primary source of multipotent progenitors that contribute to the regeneration, but not to the development, of IHD cells. Hepatocyte-to-IHD cell transdifferentiation is possible, but rarely detected. Progenitors in the EHD proliferate and migrate into the liver with Notch signaling loss and differentiate into IHD cells if Notch signaling increases. Tissue-specific mosaic analysis with an inducible dominant-negative Fgf receptor suggests that Fgf signaling from the surrounding mesenchymal cells maintains this extrahepatic niche by directly preventing premature differentiation and allocation of EHD progenitors to the liver. Indeed, transcriptional profiling and functional analysis of adult mouse EHD organoids uncover their distinct differentiation and proliferative potential relative to IHD organoids.
Conclusion:
Our data show that IHD cells regenerate upon resumption of Jag/Notch signaling, from multipotent progenitors originating from an Fgf-dependent extrahepatic stem cell niche. We posit that if Jagged/Notch signaling is augmented, via normal stochastic variation, gene therapy, or a Notch agonist, regeneration of IHD cells in ALGS patients may be enhanced.
Introduction:
The liver is known for its robust regenerative capacity in vertebrates as diverse as mammals and fish. Perhaps analogous with the intestine, both of these organs require extensive regenerative potential to recover the large number of cells lost to their inherent functions. The regenerative abilities of these endodermal organs may be supported by their having multiple genetic mechanisms and cellular sources to replace lost or damaged cells. The particular mechanisms employed for regeneration appear to depend on the cause, severity, and persistence of the cell loss, whether it is due to normal turnover or to chemical, mechanical, or genetic damage(1-3). Therefore, investigations of regenerative mechanisms using damage models that more closely mimic specific human conditions will be most pertinent(4, 5).
To study a medically relevant liver damage condition, we genetically model the developmental intrahepatic duct (IHD) cell paucity and cholestasis associated with Alagille Syndrome (ALGS) using the zebrafish. The mammalian and zebrafish hepatopancreatic organ system, which includes the liver, pancreas, gallbladder, and particularly the extrahepatopancreatic ducts (EHPDs) that join these organs together with the intestine, are highly conserved in topology (Fig. S1). Together with conserved developmental genetics of this organ system, the zebrafish is a reliable vertebrate model for human liver and pancreas diseases(6, 7).
ALGS is an autosomal dominant disorder with a prevalence estimated at 1 in 40,000 births(8) and a 24% survival rate by 19 years of age for those with cholestasis (9, 10). Cholestasis in ALGS is caused by developmental loss of duct cells within the liver, leading to poor bile transport. Accumulation of waste metabolites such as bilirubin results in progressive liver damage, and ultimately liver failure, requiring organ replacement. Therefore, understanding whether and how intrahepatic cholangiocytes can be regenerated in an ALGS model may provide new therapeutic insights.
ALGS is associated with mutations primarily in JAGGED1 (JAG1, ~97%), and NOTCH2 (N2, ~2%)(11-14). By knocking down both jagged1b and jagged2b in zebrafish, we generated a robust ALGS genetic model with highly penetrant liver duct cell loss and regeneration. This genetic disease model provides a unique opportunity to investigate the cellular and molecular mechanisms driving cholangiocyte regeneration following neonatal loss resulting from Jagged insufficiency. Combined with novel lineage tracing strategies and functional mosaic analysis, we uncovered the EHD to be a regenerative stem cell niche that is directly regulated by Fgf signaling. Furthermore, we find that adult mouse EHD cells have a stem cell-like transcriptional profile and heightened proliferative potential relative to IHD cells. These insights into the mechanisms regulating liver cholangiocyte regeneration suggest that a mild increase in Notch signaling or a mild decrease in Fgf signaling may be potential therapeutic avenues for stimulating cholangiocyte regeneration.
Materials and Methods
Animal care and zebrafish strains:
Adult zebrafish and embryos were cared for and maintained under standard conditions. All research activity involving zebrafish was reviewed and approved by SBP Medical Discovery Institute Institutional Animal Care and Use Committee. The following mutants and transgenic (Tg) lines were used: jag1bb1105; jag2bhu3425; fgf10atbvbo, Tg(krt18a:eGFP)p315; Tg(kdrl:mCherry)uto2; Tg(T2KTp1bglob:hmgb1-mCherry)jh11, abbreviated as tp1:mCherry; Tg(Tp1bglob:eGFP)um14, abbreviated as tp1:GFP; Tg(ptf1a:GFP)jh1; Tg(fabp10:DsRed)gz15; TgBAC(hnf1ba:GFP)sid03 (generated here); Tg(sox17:GFP)s870 (9), TgBAC(ptf1a:CreERt2)mk201; Tg(fabp10a:CreERt2)pt602; Tg(UBB:loxP-CFP-STOP-Terminator-loxP-hmgb1-mCherry)jh66; Tg(sox17:CreERt2;cmlc2:DsRed)sid01; Tg(hsp70:loxP-3xSTOP-loxP-dnFGFR1-EGFP; cryaa:DsRed)s02. For references and details regarding to the transgenic zebrafish generation, see Supporting Material.
Morpholino injection.
Morpholinos for jag1b (5′-CTGAACTCCGTCGCAGAATCATGCC-3′) and jag2b (5′-TCCTGATACAATTCCACATGCCGCC-3′ (15) (Gene Tools) were combined and injected into the yolk of host embryos at the one cell stage in 0.2M KCl and phenol red. Morpholinos were delivered in 2nl at a final concentration of 0.25mM (jag1b) and 1mM (jag2b).
Immunofluorescence (IF) staining, Fluorescent in Situ Hybridization (FISH) and Imaging.
Whole-mount fluorescent in situ hybridization was performed as previously described using the fgf10 probe(16). Whole mount IF staining was performed as previously described (16). Antibodies used include rabbit anti-Prox1a (1:100, GTX128354, GeneTex), goat anti-Hnf4a (1:50, sc6556, Santa Cruz), mouse anti-Alcama (1:20, zn-8, DSHB) and mouse anti-Islet1/2 (1:20, 39.4D5, DSHB), rabbit anti-pan-Cadherin (1:5000; Sigma), mouse anti-Anxa4 (1:100, ab71286, aka 2F11, Abcam)(15), rabbit anti-Bhmt (1:200, ab96415, Abcam), mouse anti-Mdr1(1:200, sc-71557, Santa Cruz), chicken anti-GFP (1:300, GFP1010, Aves Labs), goat anti-mCherry (1:500, LS-C204207, LSBio), rabbit anti-Cytokeratin7 (1:200, ab181598, Abcam), mouse anti-Sox9 (1:200, ab76997, Abcam). Imaging was performed on a Zeiss LSM710 running Zen 2010 (Black) and images were processed with Adobe Photoshop.
Lineage tracing and cell counting.
For CreERt2 induction, embryos were incubated in 20μM 4-Hydroxytamoxifen (4-OHT, T176, Sigma) for 16 hours for sox17:CreERt2 starting at 8 hpf, and for 24 hrs for fabp10:CreERt2and ptf1a:CreERt2 starting at 24 hpf. After 4-OHT treatment, embryos were washed with egg water with 1% DMSO 3 times before placing into fresh egg water. These 4-OHT exposure strategies were applied to both Tg(UBB:loxP-CFP-STOP-Terminator-loxP-hmgb1-mCherry)jlh and Tg(hsp70:loxP-STOP-loxP-dnFGFR1-EGFP)sid02 responder lines. To reveal and quantify lineage traced cells, embryos were co-stained with anti-mCherry antibody and DAPI. mCherry/DAPI positive nuclei within whole livers and EHDs were manually counted from confocal z-stack images captured at a focal depth of 1.5μm. Control and experimental samples were scored blinded when possible.
Additional methods are available in the Supporting Material.
Results:
Intrahepatic duct agenesis due to loss of jag1b/2b is reversible via a Notch-dependent mechanism
Using jag1b and jag2b (jag1b/2b) mutant zebrafish, we were able to reduce Jag/Notch signaling and phenocopy ALGS biliary paucity and agenesis(15). We show here that compound homozygous jag1b/2b mutants (jag1b−/−; jag2b−/−) fail to initially specify cholangiocytes (IHD cells) in the developing liver bud at 72 hours post fertilization (hpf), resulting in a liver comprised entirely of hepatocytes that continues to grow (72 hpf to 9 dpf, Fig. 1A). Without the IHD network, bile (assessed with a vital, metabolized fluorescent lipid analog BODIPY-FL C5:0) failed to be transported to and through the extrahepatic duct (EHD; aka common hepatic duct), and into the gallbladder (GB; Fig. 1B). Consequently, the trapped BODIPY accumulates throughout the liver and forms large lipid droplets. These lipid droplets are located at the foci of rosette structures formed by hepatocytes surrounding a densely packed group of Mdr1+ canaliculi (Fig. 1B)(17-19). Further, senescence-associated beta-galactosidase (SA β-gal) staining reveals cellular senescence throughout much of the liver of jag1b/2b mutants (Fig.1B), suggesting extensive liver damage. These observations suggest severe cholestasis in jag1b/2b mutants (Fig. 1B-D)(17-19). Nearly all double mutants die by 9 dpf, without any detectable IHD cells (Fig. 1A-E).
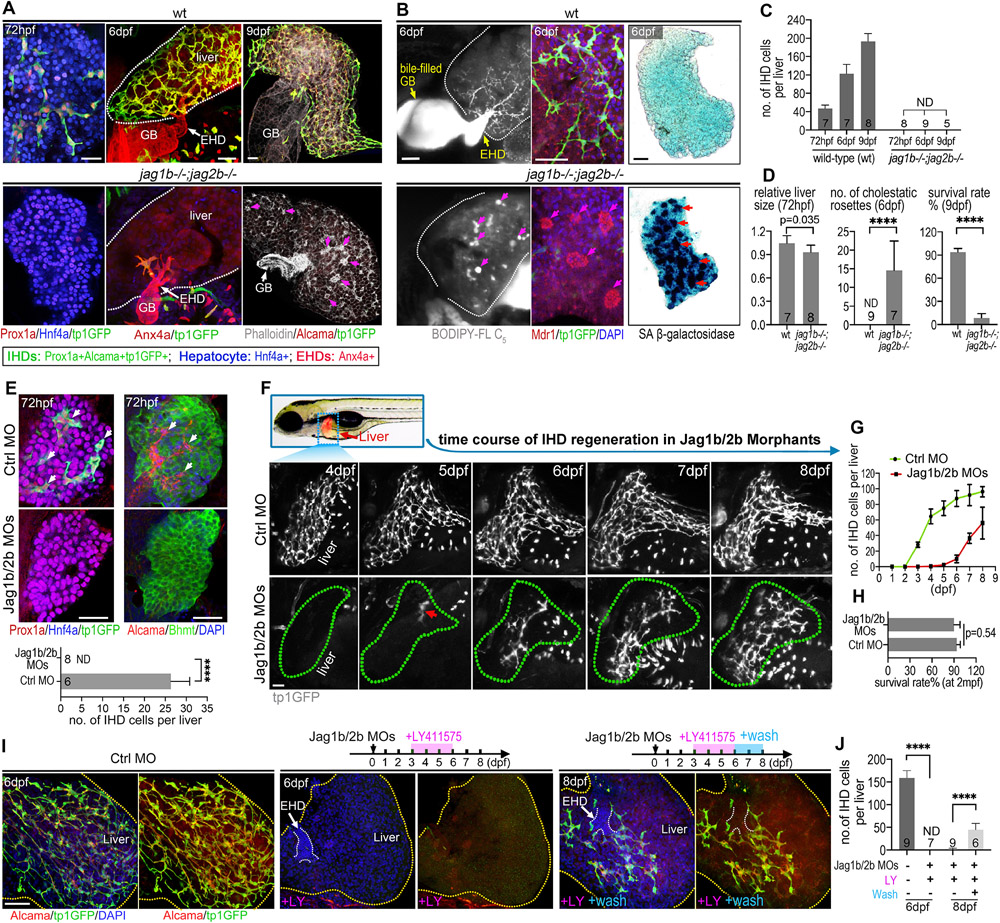
Figure 1. Developmental loss of intrahepatic duct cells regenerates upon resumption of Jagged/Notch signaling.
(A) Confocal images of zebrafish livers at embryonic (72 hpf) and juvenile (6 dpf and 9 dpf) stages in wild-type (wt) with IHD cells and jag1b/2b mutant (jag1b−/−;2b−/−) without IHD cells. IHD cells (tp1:GFP+/Prox1+/Alcama+/Anx4a+); hepatocytes (Hnf4a+/Prox1a+). F-actin (phalloidin+) staining reveals cholestatic rosettes (magenta arrows) in mutants. (B) BODIPY vital labeling shows ductal bile flow in wt but appears as pooled droplets (magenta arrows) in mutants. White dotted lines indicate liver margin; GB: gallbladder; EHD: extrahepatic duct; Mdr1 staining shows bile canaliculi distributed along the IHD in wt but densely aggregated at the center of cholestatic rosettes (magenta arrows) in mutants; SA β-gal staining shows cellular senescence (red arrows) in mutants but not in wt. (C) Number of IHD cells per liver in wt and jag1b/2b mutants. Animal numbers are indicated in graph. (D) Relative liver size, number of cholestatic rosettes, and survival rates of wt and jag1b/2b mutants. Survival rates from 4 independent experiments (n=80 wt; n=98 mutants). (E) IHD cells (arrows) found in developing livers from control are absent in Jagged1b/2b morphants at 72 hpf. IHD cells (Alcama+/tp1GFP+/Prox1a+), hepatocytes (Hnf4a+/Bhmt+). n is noted in graph. (F) (top) Dotted blue rectangle outlines area with liver (fabp10a:DsRed+) imaged in live zebrafish. (bottom) Live time course of the same liver from 4-8 dpf with IHD cells labeled (tp1:GFP+) in wt control and Jagged1b/2b morphant. Red arrow indicates initial regenerated IHD cells. Green dotted lines indicate liver margin. (G) Number of IHD cells in control (n=4) and Jag1b/2b morphant (n=5) livers from time course experiments. (H) Survival rates at 2 months post fertilization (mpf) (n=80 wt; n=79 morphants). (L) IHD cells (Alcama+/tp1:GFP+) in control and Jag1b/2b morphants with LY411575 treatment at 6 dpf and LY411575 wash out at 8 dpf. (M) Quantification of IHD cell number in L at 6 dpf and 8 dpf. Scale bars, 50μm. ND: not detected.
To determine whether restoring endogenous Jagged/Notch signaling following its loss during liver development will lead to IHD cell regeneration, both Jag1b and Jag2b were transiently knocked down. Antisense morpholinos (MOs) targeting jag1b and jag2b mRNA were microinjected into the zebrafish embryos, leading to a phenocopy of the IHD cell agenesis(20) observed in jag1b/2b mutants (72 hpf panels, Fig. 1A, E). As with wild-type at 72 hpf, livers in jag1b/2b mutants and morphants (animals with MO knockdown) continued to grow, indicating that liver development was not delayed in the absence of IHD cells (Fig. 1A, F). However, in contrast to mutants, Jag1b/2b morphants consistently start to show regeneration of the IHD cells at 4 dpf (Fig. 1F,G). The regenerated canonical Notch-active (tp1:GFP+) cells express multiple IHD cell markers (krt18a:GFP+/Pan-Cdh+/Alcama+) and are organized into a ductal network pattern, indicative of the cholangiocyte lineage (Fig. S2A,B). Further, live labeling with BODIPY and antibody staining for efflux transporter Mdr1 show that bile can be secreted from hepatocytes into the regenerated IHD and transported to the GB (Fig. S2C,E), demonstrating functional, regenerated IHDs.
We hypothesize that as the cellular concentration of the MOs decreases with the expansion of the liver, inhibition of Jag1b/2b translation also decreases, allowing endogenous Jag/Notch signaling to resume and stimulate IHD cell regeneration. To test this hypothesis, Jag1b/2b morphants were treated with the gamma-secretase inhibitor (GSI) LY411575 to prevent resumption of Jag/Notch signaling before IHD cell regeneration initiates (Fig. 1I). As expected, GSI treated morphants fail to regenerate IHD cells, even by 8 dpf (Fig. 1I,J). Furthermore, removal of GSI at later time points allows regeneration to proceed (Fig. 1I,J). These findings support a Notch-dependent regenerative mechanism.
The extrahepatic duct contributes multipotent progenitors to intrahepatic duct regeneration
To further investigate the regenerative mechanisms following resumption of Jagged/Notch signaling, we sought to determine the source of the regenerated IHD cells. Upon examination of the liver from the earliest stage of IHD regeneration, we found that the initial regenerated cholangiocytes did not appear from within the liver, as observed during normal liver development (50 hpf, Fig. S3A). Instead, they emerged from the proximal region of the liver, where the EHD is connected (Fig. 2A-C and Fig. S3B). As regeneration proceeds, regenerated IHD cells progressively repopulate the liver in a proximal to distal manner (Fig. S3C). Because the initial regenerated cholangiocytes are directly or closely associated with the EHD (Fig. 2A-C), we hypothesized that these cells arose from the EHD. However, testing this hypothesis in vivo via lineage tracing requires a Cre reporter line that can label cells in the EHD but not the liver. Such line is currently not available.
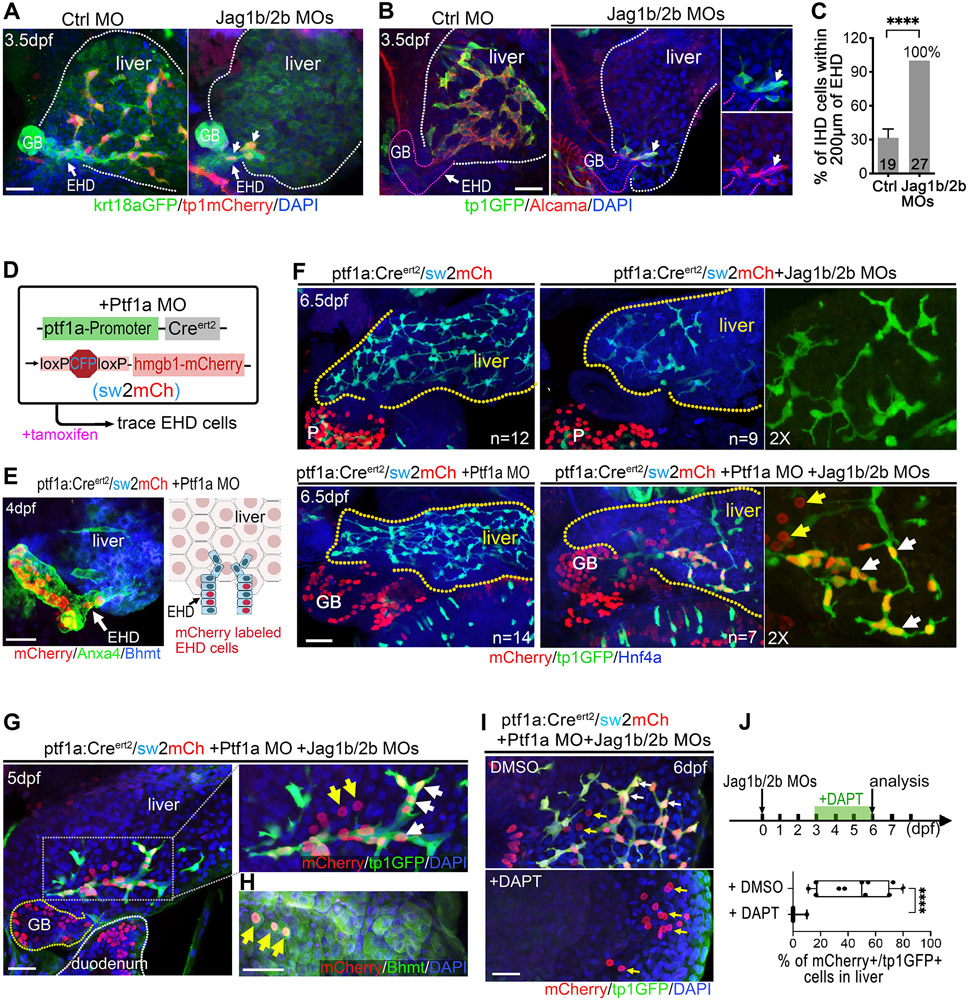
Figure 2. The extrahepatic duct contributes multipotent progenitors to intrahepatic duct regeneration.
(A, B) Confocal images of the EHD and proximal liver at 3.5 dpf in control and Jag1b/2b morphants. Earliest regenerated IHD cells (white arrows; krt18a:GFP+/tp1:dsRed+/ Alcama+/tp1:GFP+) are associated witht the EHD. (C) Percentage of IHD cells located within 200μm of the EHD at 3.5 dpf. Number of animals examined indicated in graph. (D) Lineage tracing strategy to label EHD cells using embryos with ptf1a:CreERt2, sw2mCh, and Ptf1a MO, dosed with tamoxifen at 24-48 hpf. (E) (left) The EHD and proximal liver from the experiment noted in D at 4 dpf show lineage labeled cells (mCherry+) in the EHD (~10% of EHD cells labeled, n=7) and GB (high Anxa4+), but not in the liver (Bhmt+ hepatocytes) as depicted (right). (F) Transgenic ptf1a:CreERt2/sw2mCh/tp1:GFP livers at 6 dpf with and without Ptf1a and Jag1b/2b MOs. mCherry+/ tp1:GFP+ IHD cells (white arrows) in the liver of Jag1b/2b morphants with Ptf1a MO. tp1:GFP−/mCherry+ cells (yellow arrows) in the same liver are larger and rounded, indicative of hepatocytes. P: pancreas; GB: gallbladder; Yellow dotted lines indicate liver margin. mCherry+/ tp1:GFP+ IHD cells were detected in ~13% (n=7/55) of livers in Jag1b/2b morphants with Ptf1a MO, but not detected in the livers of control zebrafish with only Ptf1a MO (n=0/14 number of livers analyzed. Total numbers of regenerated IHD cells are not significantly different (p=0.46) between livers with (n=7) and without (n=8) Ptf1a MO. (G, H) Lineage traced EHD cells in the Jag1b/2b morphant livers on 5 dpf, expressing tp1:GFP (white arrows) or hepatocyte marker Bhmt (H; yellow arrows). (I, J) Livers of Jag1b/2b morphants with EHD cells traced treated with DMSO (control, n=11) or DAPT (10μM; n=6), showing traced cells (mCherry+) in the liver can be tp1:GFP+ (white arrows) in control and only tp1:GFP− (yellow arrows) in GSI treated. Scale bars, 50μm.
Fortunately, previous work with the pancreatic gene ptf1a revealed that, in ptf1a loss-of-function zebrafish and mice, ptf1a:GFP reporter expression expands into the EHPD, including the common bile duct and a subset of cells in the EHD, but not the liver(21, 22). Leveraging this insight, we crossed a zebrafish ptf1a:CreERt2 reporter line(23) to the ubiquitously active Cre responder line, Tg(UBB:loxP-CFP-STOP-Terminator-loxP-hmgb1-mCherry)(15) (hereafter sw2mCh), and knocked down Ptf1a(22) (Fig. 2D). With this strategy, a small subpopulation of EHD cells, but not IHD cells, can be permanently labeled with mCherry expression (Fig. 2E). However, in Jag1b/2b morphants, we found that both EHD and regenerated IHD cells were lineage labeled (Fig. 2F). These results suggest that the EHD does contribute cells to IHD cell regeneration, but not to IHD cell development. Consistently, examination of the earliest IHD cells during normal development reveal that they arise within the liver and are not directly associated with the EHD, as observed with regenerated IHD cells. These findings provide the first in vivo evidence of EHD cells as a source of IHD regeneration. Interestingly, sporadic hepatocytes near the regenerated IHD cells were also lineage labeled (Fig. 2F,G), demonstrating that the EHD-derived cells can be multipotent, having the potential to give rise to either cholangiocytes or hepatocytes in the liver, depending on the availability of Jagged/Notch signaling (Fig. 2I,J).
Regenerated IHD cells arise almost exclusively from EHD progenitors, with minimal contribution from transdifferentiated hepatocytes
To assess whether EHD cells are the primary source of IHD regeneration, additional lineage tracing strategies were employed. First, we generated and utilized a sox17:CreERt2 line to specifically label endoderm cells(24, 25) (Fig. S4). We found that sox17:CreERt2 can efficiently label EHDs and hepatocytes at 3 dpf, when IHD cells have yet to regenerate (Fig. 3A,B). By 6.5 dpf, regenerated IHD cells are consistently lineage labeled, suggesting that the regenerated IHD cells are derived from either EHD cells or hepatocytes (Fig. 3B). Next, using fabp10a:CreERt2, we observed efficient and specific lineage labeling of nearly all hepatocytes, which comprise the entire Jag1b/2b morphant liver at 3 dpf (Fig. 3C,D and Fig. S5). However, by 6.5 dpf, we found no lineage labeling of regenerated IHD cells (Fig. 3D), ruling out hepatocytes as a significant cellular source. With lineage tracing results from these two Cre lines, it can be deduced that nearly all the regenerated IHD cells do indeed arise from the EHD, consistent with the ptf1a:CreERt2 lineage tracing studies described above.
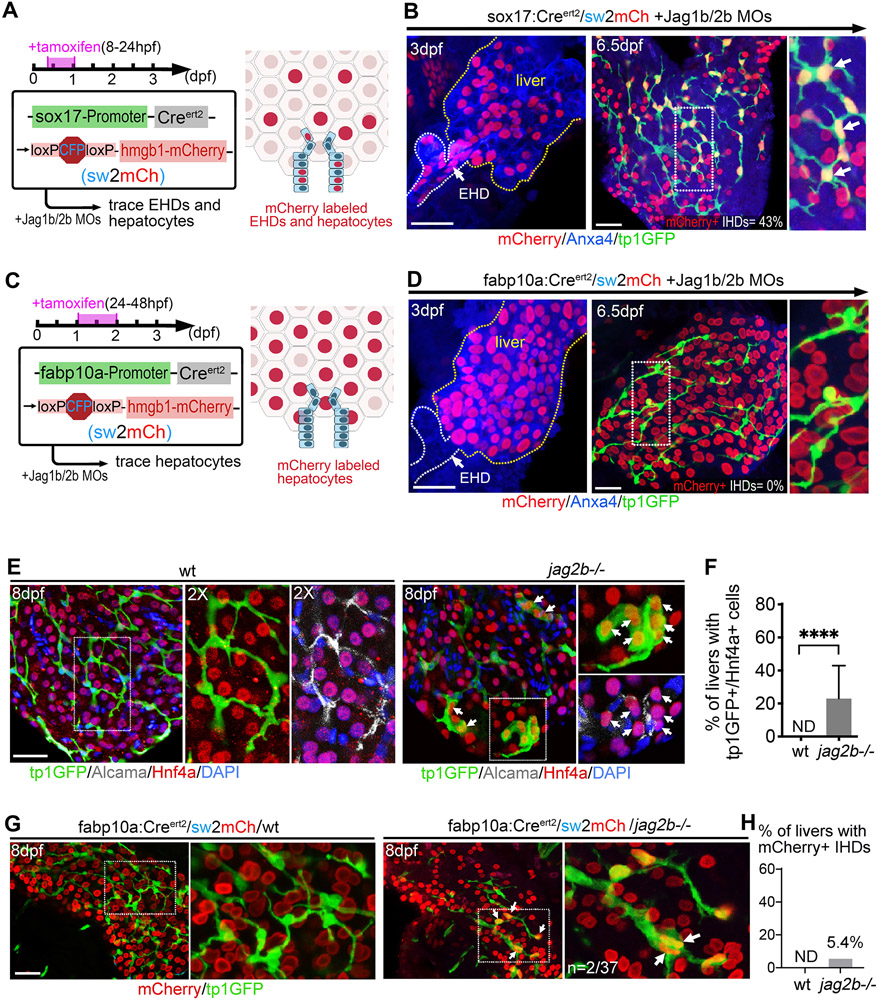
Figure 3. IHD cells regenerate primarily from EHD cells in Jag1b/2b morphants but hepatocytes transdifferentiation is possible in jag2b−/− mutants.
(A) (Left) Lineage tracing strategy to label both EHD cells and hepatocytes using sox17:CreERt2 with sw2mCh in Jag1b/2b morphants, dosed with tamoxifen at 8-24 hpf, (Right) as depicted. (B) Jag1b/2b morphant livers at 3 dpf (n=8) showing effective mCherry+ labeling of EHD cells (~27%) and hepatocytes (~46%) prior to the appearance of regenerated tp1:GFP+ IHD cells, and at 6.5 dpf (n=18) showing mCherry+ labeling of regenerated IHD cells (white arrows; percentage of IHD cells labeled indicated). Area in dotted box is magnified in right panel. (C) (Left) Lineage tracing strategy to genetically label hepatocytes using fabp10a:CreERt2 with sw2mCh in Jag1b/2b morphants, dosed with tamoxifen at 24-48 hpf, (Right) as depicted. (D) Jag1b/2b morphant livers at 3 dpf (n=22) showing extensive mCherry+ labeling of hepatocytes (~88%) prior to the appearance of regenerated tp1:GFP+ IHD cells, and at 6.5 dpf (n=120) showing no mCherry+ labeling of regenerated IHD cells. (E, F) (E) Wt and jag2b−/− mutant livers at 8 dpf, showing mutually exclusive expression of hepatocyte marker Hnf4a and IHD markers tp1:GFP/Alcama in wt liver, whereas tp1:GFP+/Hnf4a+ double positive cells (white arrows) are found in jag2b−/− liver (n=5/21)(F), boxed areas are magnified. (G, H) (G) Wt control and jag2b−/− mutant livers at 8 dpf with hepatocytes lineage traced using fabp10a:CreERt2, showing that all lineage labeled (mCherry+) hepatocytes in wt control livers lack tp1:GFP expression, whereas in jag2b−/− mutant livers, double positive tp1:GFP+/mCherry+ cells (white arrows) with cholangiocyte-like cell shape are found at low frequency (n=2/37) (H). Scale bars, 50μm.
Our extensive hepatocyte lineage tracing studies in Jag1b/2b morphants did not indicate a hepatocyte-to-IHD cell transdifferentiation mechanism of regeneration as was observed in another genetic model of developmental IHD cell agenesis and regeneration(26). However, that genetic damage model was more severe, with permanent hepatic loss of both RBPJ and HNF6, factors required for all canonical Notch signaling(27) and for EHD development(28), respectively. To test whether a more persistent loss of Jagged/Notch signaling in zebrafish could also lead to hepatocyte-to-IHD cell transdifferentiation, jag2b mutant homozygotes were examined. At the distal region of the liver, which is often devoid of regenerated IHD cells in jag2b−/− mutants at 6 dpf, we sporactically find mis-coexpression of Hnf4a and tp1:GFP, markers are normally mutually exclusive due to their distinct labeling of hepatocytes and IHD cells in wild-type livers (Fig. 3E,F and Fig. S6). These Hnf4a/tp1:GFP co-expressing cells express little to no duct marker Alcama. Yet they can also lack the typical round shape of a hepatocyte, instead exhibiting cellular extensions more characteristic of IHD cells. These molecular and morphological features suggest that the Hnf4a/tp1:GFP co-expressing cells in jag2b−/− mutant livers are hepatocytes in the process of transdifferentiating into IHD cells. To test this hypothesis, hepatocytes in jag2b−/− mutants were lineage traced using fabp10a:CreERt2 and examined at 8 dpf. In jag2b−/− mutants, tp1:GFP expression in hepatocyte-lineage-labeled cells can be found, albeit infrequently (Fig. 3G,H). These cells also display IHD cell morphology and are integrated within the IHD network (arrows in Fig. 3G). Together, these findings suggest that hepatocytes-to-IHD cell transdifferentiation is possible with permanent jag2b loss. Collectively, our findings suggest that most, if not all, IHD cells regenerate from the EHD upon the resumption of Jagged/Notch signaling, but transdifferentiation from hepatocytes is possible when loss of Notch signaling is more persistent.
EHD cells proliferate with Notch signaling loss and then differentiate into IHD cells with its resumption
We next investigated the proliferative mechanism mediating IHD repopulation. To label proliferation during the early stages of IHD regeneration in the Jag1b/2b morphants, EdU incorporation was assessed at 4-5 dpf (Fig. 4A). At this initial stage of regeneration, a strong proliferative response is observed in the EHD (Fig. 4B-D). Despite high EHD cell proliferation at this stage, the size and structure of the EHD showed little expansion (Fig. 4B), suggesting a loss of cells from the EHD, consistent with our lineage tracing results (Fig. 2). To determine the cell proliferation pattern during this regenerative process, a short 3-hour EdU pulse was applied at defined time points from 3 dpf to 7 dpf, when the repopulation of the IHD network is mostly complete (Fig. 4E). Consistent with an EHD source of regenerative cells, a strong proliferative response in the EHD was observed early, at 3 dpf, before the appearance of the earliest regenerating IHD cells (Fig. 4E). Assessment of EdU incorporation at subsequent stages reveals that although the EHD is highly proliferative prior to and during the initial stages of IHD regeneration (3-5 dpf), cell division decreases in the EHD as it increases in the newly regenerated IHD cells that have entered the liver (Fig. 4E,F). These results demonstrate that EHD cells initially proliferate to supply progenitors to the liver. Once in the liver, these progenitors differentiate into IHD cells, continue to robustly proliferate, and expand into in the distal liver (Fig. 4G). This pattern of EHD cell proliferation during IHD cell regeneration resembles that of quiescent stem cells from other tissues such as the intestine(29) and can explain how a small number of cells derived from EHD can self-renew and repopulate the entire IHD system (Fig. 3).
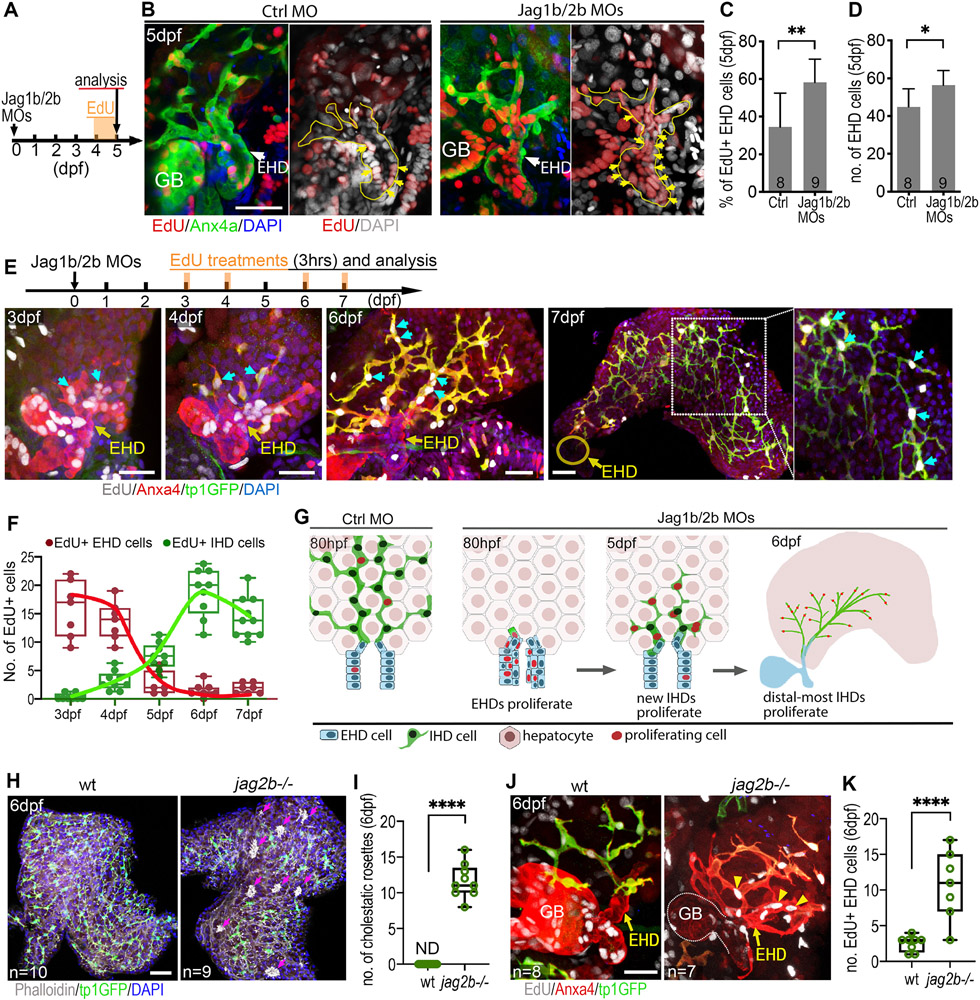
Figure 4. Extrahepatic duct cells contribute proliferative progenitors to the regenerating liver.
(A-D) (A) EdU incorporation experiment to assess cell proliferation from 4 to 5 dpf. (B) EdU+ EHD cells (yellow arrows) in the EHDs of control and Jag1b/2b morphants at 5 dpf. EHD margin outlined with yellow dotted lines. (C) Percentage of EdU+ EHD cells and (D) total EHD cell numbers in control and Jag1b/2b morphants at 5 dpf. Number of animals examined indicated. (E, F) (E, top) EdU treatment was pulsed in Jag1b/2b morphants for 3hrs at indicated time points. (E, bottom) EdU+ EHD cells (cyan arrows) in the EHDs and regenerated IHDs of Jag1b/2b morphants at 3 dpf (n=7), 4 dpf (n=7), 6 dpf (n=9), and 7 dpf (n=10). (F) Number of EdU+ cells in the EHDs or IHDs of Jag1b/2b morphants at indicated stages. (G) Model summarizing proliferation pattern during normal IHD development versus IHD regeneration in Jag1b/2b morphants. During development, proliferation is low in the EHD and scattered in the IHDs, whereas during regeneration, EHD cells initially proliferate to supply progenitors to the liver. These progenitors subsequently differentiate into IHD cells in the liver and continue to proliferate, more robustly in the distal-most IHD cells. (H, I) Liver in wt and jag2b−/− mutants at 6 dpf showing hepatic cholestatic rosettes (magenta arrows) revealed by phalloidin-dense staining (white clusters) in areas of the mutant liver devoid of tp1:GFP+ IHD cells, but not in wt, (I) as quantified. (J, K) EdU+ cells in the EHD of wt and jag2b−/− mutants at 6 dpf (following an 8hr EdU treatment) showing increased cell proliferation in an expanded mutant EHD (yellow arrowheads), (K) as quantified (n= # of livers). Scale bars, 50μm.
Compared to Jag1b/2b knock down animals, IHD regeneration is nominal in jag2b−/− mutants, which have fewer IHD cells and many more cholestatic rosettes (Fig. 4H,I), leading us to question whether this difference could result from compromised EHD proliferation in mutants. Unexpectedly, we found the EHD is still highly proliferative in jag2b−/− mutants, even up to 6 dpf (Fig. 4J,K). Moreover, the EHD appears structurally expanded into the liver (Fig. 4J). This finding, together with our finding of robust EHD cell proliferation in morphants at 3 dpf (Fig. 4E), well before the initial appearance of Notch active tp1:GFP+ IHD cells, suggests that the resumption of Jagged/Notch signaling is unlikely to trigger the proliferative response in the EHD. To test whether it is the loss of Notch signaling itself that can lead to EHD proliferation, we blocked Notch signaling in wild-type animals with a GSI. Treated with LY411575, we found a dramatic increase in EHD cell proliferation, suggesting that the loss of Notch signaling that can lead to increased EHD cell proliferation (Fig. S7).
Adult mouse EHD cells are distinct from IHD cells in their transcriptional profile and proliferative potential
The contiguous topology of the EHD and IHD, as well as their common biliary gene expression and ductal morphologies, suggest similar tissue identities. However, it remains unclear if these cholangiocyte populations have more substantial intrinsic and functional differences. To investigate potential differences between these cholangiocyte populations, we compared the transcriptional profiles of EHD and IHD cells by dissecting and isolating EpCAM+ biliary epithelial cells from the EHDs and livers of 3-month-old mice (Figs. 5A and S8A). After confirmation by expression of the ductal marker CK7 (Krt7) (Fig. S8B), these freshly isolated EpCAM+ ductal cell populations, as well as EpCAM– liver cells, were directly processed for bulk RNA-seq.
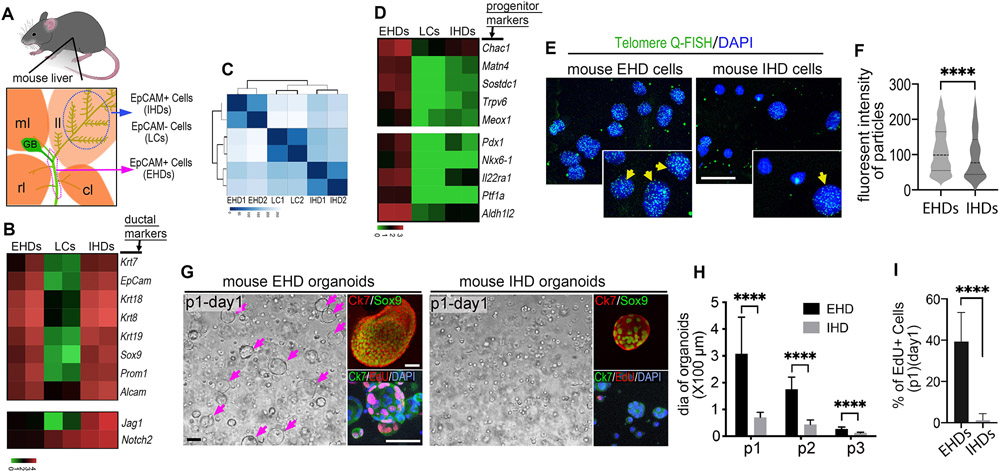
Figure 5. Adult mouse extrahepatic duct cells have distinctly high stem/progenitor cell gene expression and proliferative potential.
(A) Illustration depicting region of extrahepatic ducts (magenta dotted line) and the left lobe (ll; blue dotted line) of the mouse liver that was dissected from 8 weeks old C57BL/6 mice. EpCAM+ EHD cells (EHDs), EpCAM+ IHD cells (IHDs), and EpCAM– liver cells (LCs) were isolated by MACS using (EpCAM) microbeads (see supplementary Figure 5). Right lobe (rl), median lobe (ml), caudate lobe (cl) and gallbladder (GB). (B) Heatmap showing the biliary marker genes shared between EpCAM+ EHDs and IHDs, but not with EpCAM– LCs (log10 scaled bar). (C) Heatmap clustered by the Euclidean distances using unsupervised clustering analysis based on all significant differentially expressed genes among primary EHDs, IHDs, and LCs, showing greater transcriptome difference between EpCAM+ EHDs and IHDs than with EpCAM– LCs. (D) Heatmap showing markedly higher expression of stem/progenitor/quiescence implicated genes (see supplementary methods) in EHDs relative to IHDs and LCs (log10 scaled bar). (E) Telomere Q-FISH showing consistently stronger signals (green; yellow arrows) in nuclei (DAPI) of freshly isolated EHD versus IHD cells. (F) Quantification of fluorescent intensity in telomere labeled particles in EHD and IHD cells. (G) Bright-field and fluorescent microscopy of first passage (P1) organoids from the EHDs and IHDs showing some larger EHD-derived organoids (magenta arrows). Sox9 and Ck7 expression confirms biliary duct cell identity. More EdU+ cells in EHD-derived organoids following a 2hr EdU exposure. (H) Average diameter (dia) of organoids from EHDs and IHDs in first 3 passages. (I) Ratio of EdU+ cells in the organoids (passage1) from EHDs and IHDs. Scale bars, 20μm (E), 100μm (G).
As expected, EpCAM+ cells from the EHD and liver both express established cholangiocyte markers including Krt7, Krt18, Krt19, Alcam, and Sox9 (Fig. 5B). Using unsupervised clustering analysis, we further compared the transcriptome of the three populations and were surprised to find that EHD and IHD cells are less similar to each other than they are to the EpCAM– liver cells (Fig. 5C). However, the differences between these biliary populations are consistent with our previous findings using zebrafish, where we demonstrated that canonical Notch signaling is absent from EHD cells and active in the liver, where it is required for induction of IHD cell versus hepatocyte lineage identity(15). Given that Notch signaling is known for this evolutionarily conserved role in driving binary decisions for distinct cell identities(30), we examined Jag1 and Notch2 expression in these two EpCam+ mouse biliary populations. Consistent with zebrafish, we do find higher expression of these Notch ligand and receptor transcripts in mouse IHD cells compared to EHD cells (Fig. 5B), suggesting differential Notch regulation of their distinct cell identities. Further, consistent with the previous finding that pancreatic endocrine cells, including beta cells, can spontaneously arise in EHD of adult mouse(31), significant GO/KEGG enrichment categories relevant to pancreatic secretion and insulin secretion are found with EHD cells (Fig. S9). Together, these results confirm the identities of the population of EHD and IHD cells isolated.
Moreover, examination of progenitor and stem cell genes implicated in quiescence and maintenance reveals markedly higher expression of Chac1, Matn4, Sostdc1, Trpv6, and Meox1 in EHD cells compared to IHD cells (Fig. 5D). Moreover, specific foregut organ progenitor markers, including pancreatic (Pdx1, Ptf1a, Nkx6.1 and Il22ra1), and hepatic (Aldh1l2) progenitor genes are also more highly expressed in EHD cells (Fig. 5D), consistent with EHD cells having greater transcriptional heterogeneity and lineage potential. Accordingly, human EHD cells can differentiate into functional hepatic and pancreatic lineages in culture and transplants(32-34).
Given that telomere length is associated with proliferative potential and liver regeneration(35), we assessed telomere length using Q-FISH and found higher signals in EHD cells compared to IHD cells (Fig. 5E,F). To assess whether the higher telomere signals in EHD cells correlate with greater proliferative potential, we generated and examined organoids derived(36) from EpCAM+ cells from the EHDs and livers (IHD cells). Both EHD and IHD organoids express ductal cell markers, Sox9 and Krt7, consistent with their transcriptional profiles (Fig. 5B,G). However, a subset of EHD derived organoids appear consistently larger in size at each passage (Fig. 5G,H). Consistently, a short 2-hour EdU incorporation assay revealed extensive proliferation in the larger EHD organoids but not in IHD organoids (Fig. 5G,I). These results suggest that a subset of adult mouse EHD cells have a uniquely high proliferative capacity, consistent with their distinct progenitor signatures. Together with our in vivo proliferation and lineage tracing findings using zebrafish, these data support that the EHD contains a conserved source of distinctly quiescent, multipotent progenitors that can contribute significantly to liver regeneration.
Fgf signaling directly maintains progenitors in the EHD niche by preventing their premature differentiation and allocation into the liver
Our discovery of quiescent progenitors residing in the EHD led us to investigate the genetic mechanisms regulating this potential stem cell niche. fgf10a expression was previously reported in mesenchymal cells surrounding the entire developing EHPDs, alluding to a possible role in maintaining this niche(16). Upon closer examination at a later stage, we find that fgf10a expression continues to be enriched in mesenchymal cells adjacent to the EHD, but is sparse in the liver (Fig. 6A,B) as similarly reported in mice(37). In fgf10a−/− mutant zebrafish, the EHD can exhibit ectopic Hnf4a and Prox1a expression, suggesting hepatic lineage identities, and can also appear structurally incorporated into the liver (Fig. 6C, Fig. S10). These results lead us to hypothesize that Fgf signaling maintains multipotent progenitors in the EHD by preventing their premature differentiation into hepatic lineages. The incomplete penetrance of the EHD defects observed in fgf10a mutants suggests redundancy with other Fgf ligands, leading us to block Fgf signaling more effectively using the chemical inhibitor SU5402(38, 39). The use of this robust Fgf receptor tyrosine kinase inhibitor also allows for temporally controlled repression of Fgf signaling. Treatment of wild-type zebrafish with SU5402 at 4-6 dpf, after the EHD has fully developed, led to ectopic coexpression of IHD markers tp1:GFP and Alcama in the EHD (Fig. 6D), suggesting that Fgf signaling continues to be required to prevent differentiation of EHD cells into IHD cells. This finding is consistent with the fgf10a mutant results and adds further support to our transcriptional profiling and lineage tracing studies demonstrating the in vivo multipotency of EHD cells. Together, these loss-of-function studies suggest that Fgf signaling regulates the EHD niche by maintaining multipotent progenitor cells in the EHD in an undifferentiated state (Fig. 6E).
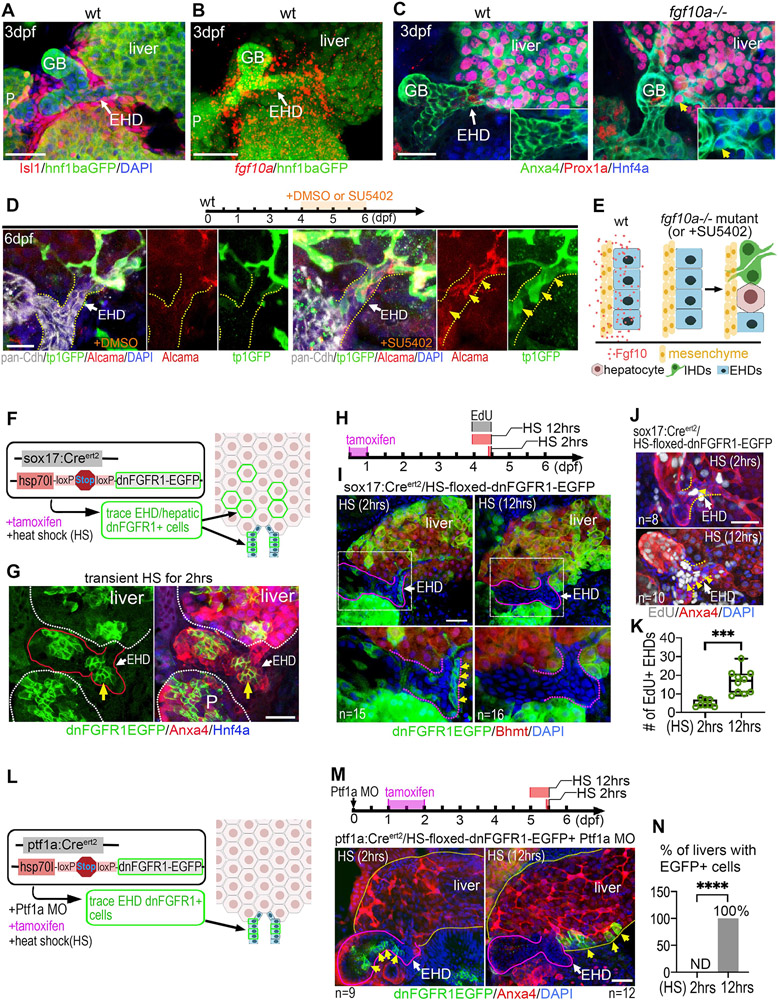
Figure 6. Fgf signaling directly maintains progenitors in the EHD niche by preventing their premature differentiation and allocation into the liver.
(A) Dense mesenchymal cells (Isl1+) surrounding the EHD of wt zebrafish at 3 dpf (n=9). hnf1ba:EGFP+ foregut endoderm showing the EHD and the liver, pancreas (P), and gallbladder (GB). (B) Fluorescent in situ hybridization (FISH) showing high fgf10a expression surrounding the EHD of wt zebrafish at 3 dpf (n=5). (C) EHD (Anxa4+) at 3 dpf showing ectopic co-expression of Prox1a and Hnf4a (inset; yellow arrow) in fgf10a−/− mutants but not in wt (n=16). (D) (top) Timeline of DMSO control and FGFR inhibitor SU5402 dosing experiment. (bottom) pan-Cdh+ EHD (yellow dotted lines) in wt at 6 dpf treated with DMSO (left) or SU5402 (right; 2μM) at 4-6 dpf showing ectopically expressed IHD markers (tp1:GFP and Alcama; yellow arrows) (n=7) in the EHD after SU5402 treatment. (E) Model depicting Fgf10a ligand signaling from mesenchymal cells directly to adjacent EHD cells to inhibit differentiation into liver cell fates. (F) Lineage tracing strategy to label clones of endoderm cells with inhibited Fgf receptor signaling using sox17:CreERt2 and heat-shock inducible Cre responder hs:switch2dnFGFR. (G) dnFGFR1-EGFP+ clones of cells appear in the EHD (yellow arrow), liver, and pancreas (P) at 4 dpf following tamoxifen treatment and a 2hr heat-shock. EHD and GB are outlined in red. (H) Tamoxifen (8-24 hpf) and EdU (4-4.5 dpf) treatments, with heat-shock for either 2 hours (control) or 12 hours (4-4.5 dpf) to induce dnFGFR1-EGFP expression. (I) dnFGFR1-EGFP+ clones after 2 or 12hr heat shock with EHD region (boxed) magnified below and outlined with purple dotted lines. dnFGFR1-EGFP+ cells (green membrane; yellow arrows) were found in all EHDs (n=15/15) following a 2hr heat shock, but not detected in the EHD (n=0/16) following a 12hr heat shock. (J) EdU+ cells in EHD (outlined with yellow dotted lines) with either 2 or 12hr heat shock, showing increased EdU+ EHD cells after a prolonged heat shock. 12hr EdU treatment. (K) Number of EdU+ EHD cells following 2hr (n=8) and 12hr (n=12) heat shock induction of dnFGFR1-EGFP. (L) Lineage tracing strategy to label clones of cells with inhibited Fgf receptor signaling in the EHD but not in the liver, using ptf1a:CreERt2 , Ptf1a MO, and hs:switch2dnFGFR as depicted. (M) (Top) Tamoxifen exposure at 24-48 hpf followed by heat shock induction of dnFGFR1-EGFP at 5 dpf for 2hrs (control) or 12hrs. (bottom) dnFGFR1-EGFP+ clones (green; yellow arrows) are found in the EHD (outlined with white dotted lines) after a 2hr heat shock (n=9), but located in the liver after a 12hr heat shock (n=12). (N) Percentage of livers with dnFGFR1-EGFP+ cells following a 2hr (n=9) and 12hr (n=12) heat-shock. Scale bars, 50μm.
To determine whether Fgf directly regulates EHD cells, we conditionally inhibited Fgf receptor (Fgfr) signal transduction in EHD cells. Specifically, a dominant-negative form of Fgfr1 (dnFGFR1a-EGFP)(40) was expressed in a mosaic pattern in the EHD using an endoderm specific inducible system. To achieve spatiotemporally controlled expression, we generated a transgenic line with a heat-shock inducible promoter driving a floxed STOP cassette upstream of dnFGFR1a-GFP (hsp70:3xSTOPFloxed-dnFGFR1a-EGFP, hereafter hs:switch2dnFGFR). This system allows us to induce (in a temporally controlled manner via heat-shock) repression of Fgfr signal transduction within Cre-induced mosaic clones of cells which can be tracked by their membrane EGFP expression. This line was first crossed to sox17:CreERt2 to spatially restrict dnFGFR1a-EGFP+ clones to the endoderm, which includes the entire hepatopancreatic system, but not the surrounding mesoderm-derived mesenchymal cells. With this mosaic loss-of-function approach, we generated and followed the behavior of clones of dnFGFR1-EGFP+ cells in the EHD and liver (Fig. 6F,G). We find that dnFGFR1-EGFP+ cells can initially be detected in the EHD after a brief 2-hour heat-shock, but are lost from the EHD after a continuous, 12-hour cyclic heat-shock (Fig. 6H,I), suggesting that Fgf signaling is required to cell-autonomously maintain cells in the EHD. EdU incorporation studies reveal increased EHD proliferation with the long-term, 12-hour heat-shock (Fig. 6J,K), suggesting that the remaining EHD cells with normal Fgf signaling activity can self-renew.
To specifically inhibit Fgf signaling in lineage traced cells in the EHD but not in the liver, the ptf1a:CreERt2/ptf1a MO approach (Fig. 2D) was combined with hs:switch2dnFGFR (Fig. 6L). Following long-term 12-hour heat-shock induction of dnFGFR1a-EGFP expression, we again find that the EHD becomes devoid of dnFGFR1a-EGFP+ cells (Fig. 6M), confirming results from the analogous experiments above with sox17:CreERt2 (Fig. 6I). Importantly, we find dnFGFR1a-EGFP+ cells localized within the liver, demonstrating that EHD cells with compromised Fgfr signaling can be allocated into the liver (Fig. 6M,N). These findings suggest that Fgf signals directly to the EHD cells to regulate their retention. It is unclear whether dnFGFR1a-EGFP+ cells actively vacate the EHD or are displaced by dividing cells via a mechanism previously described as neutral competition for stem cell niche space(29).
Because repression of Fgf signaling can be genetically induced to cause allocation of EHD progenitors to the liver (Fig. 6), we predicted that a controlled pharmacological inhibition of Fgf signaling could be leveraged to stimulate the EHD to contribute more progenitors to the liver and enhance IHD regeneration. To test this prediction, a short 8-hour pulse of SU5402 was applied to transgenic animals (ptf1a:CREert2/sw2mCh) to lineage trace EHD cells. Indeed, we found lineage labeled EHD cells localized to the liver after transient SU5402 treatment (Fig. 7A). Moreover, in Jag1b/2b morphants with the same transient inhibition of FGF signaling, a significant increase in the number of regenerating IHD cells was observed (Fig. 7B,C). These findings suggest that a measured pharmacological inhibition of Fgf signaling can coax progenitors from the EHD into the liver, to accelerate IHD regeneration. Collectively, we propose a model whereby that Fgf signals directly to the EHD cells from the adjacent mesenchyme to maintain a stem cell niche by regulating differentiation and allocation of progenitors that can self-renew (Fig. 7D).
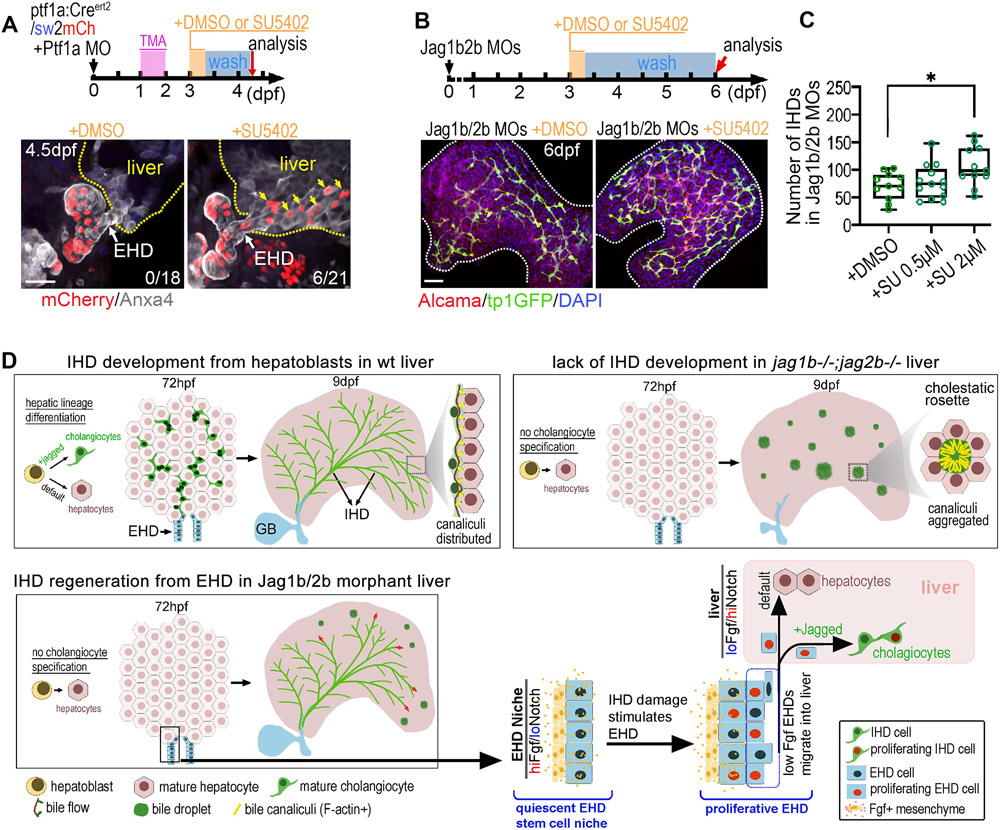
Figure 7. Chemical inhibition of Fgf signaling stimulates allocation of EHD cells into liver and accelerates IHD cell regeneration.
(A) (top) EHD lineage tracing approach using ptf1a:CreERt2, sw2mCh, and Ptf1a MO, with transient exposure to DMSO or SU5402 (2μM) for 8hrs at 3 dpf and analyzed at 4.5 dpf. (bottom) Lineage traced EHD cells (mCherry+) are restricted to EHD in DMSO treated controls (n=0/18), but appear in liver (yellow arrows) following SU5402 treatment (n=6/21). (B) (top) Jag1b/2b morphants treated with DMSO or SU5402 for 8hrs at 3 dpf and analyzed at 6 dpf. (bottom) Regenerating IHDs (tp1:GFP+/Alcama+) in whole liver of Jag1b/2b morphants at 6 dpf after DMSO or SU5402 treatment. (C) Number of regenerated IHD cells in Jag1b/2b morphants with DMSO or SU5402 treatment (8hrs; 0.5 μM and 2μM). (D) Working models of IHD development in wt zebrafish, IHD development failure in jag1b/2b mutants, and EHD-mediated regeneration of IHD cells in Jag1b/2b morphants. Scale bars, 50μm.
Discussion:
Structural birth defects are generally thought to be irreversible. For example, even with transient embryonic exposure to teratogenic chemicals, certain neonatal malformations, such as forelimb loss due to thalidomide(41), can persist. Despite the regenerative abilities of zebrafish, this permanence of developmental defects has also been observed in this vertebrate model following transient knockdown of genes required for lineages such as the pancreas(42) and forelimbs(43). Therefore, it was surprising to find that the complete developmental loss of IHD cells in zebrafish is reversible. Perhaps organs with ‘professional’ stem cells(29) that reside outside the damaged tissue area and are thus spared from insults, are more resilient to severe, chronic, and even developmental tissue loss.
We did not find a significant contribution of liver facultative stem cells to IHD cell regeneration due to transient Jagged loss. However, when Jag2b loss is permanent, we did observe, albeit infrequently, hepatocyte-to-IHD cell transdifferentiation. This mechanism of regeneration was previously reported in mice(26) with permanent loss of genes required for canonical Notch signaling(27) and EHD development(28). The combined loss of these of two factors may have precluded the regenerative mechanism uncovered here, which is both Notch and EHD dependent.
Given the extensive conservation between the zebrafish and mammalian EHPDs, a homologous stem cell niche likely also exists in mammals. Particularly, the conserved developmental expression of Fgf10 in the mesenchyme surrounding the EHD was reported to persist into later stages in mice(37) and therefore may continue to endow the adult mammalian EHD with a quiescent, multipotent progenitor pool that can contribute to liver homeostasis and regeneration. Because Fgf signaling also regulates differentiation, proliferation, and migration to maintain intestinal stem cells in adult mice(44), this function of Fgf may be a conserved niche regulatory mechanism.
The zebrafish regenerative EHD progenitor cells identified here may prove to be homologous to the mammalian EHD peribiliary cells, which were demonstrated to have the potential to expand and differentiate into liver cells in explant studies(32). Our comparison of adult mouse EHD and IHD cell transcriptional profiles revealed that these populations of biliary cells are more different from each other than they are from other liver cells, suggesting that the EHD is not merely an extension of the IHD. The EHD cells express more progenitor and quiescence markers, have longer telomeres, and have greater proliferative capacity than IHD cells, consistent with other established stem cells. Moreover, our studies lend critical in vivo data to support the debated model(29, 45) suggesting that cells in EHD are a potential source of stem cells for liver regeneration(33, 34). If the mammalian EHD cells are indeed a significant source of liver stem cells, it may be that exhaustion of these cells, or their inability to contribute to the liver, ultimately leads to liver regeneration failure in disease and aging. It will be important to examine this tissue to better understand liver disease states and to develop new diagnostic and therapeutic approaches.
Our studies demonstrating that a measured inhibition of Notch or Fgf signaling can be leveraged to increase EHD cell proliferation or contribution to the liver, respectively, suggest potential new therapeutic avenues for maintaining a healthy stem cell population or for stimulating an alternative progenitor source for liver regeneration. Indeed, loss of either signaling pathway was previously shown to also trigger proliferation of stem/progenitor cells in other tissues, implicating their conserved role in maintaining quiescence(46, 47). Intriguingly, Fgfr inhibitors are undergoing clinical trials for treating bile duct cancers, which are often associated with hyperactive Fgf signaling(48, 49). Based on our findings showing that chemical inhibition of Fgf signaling can also stimulate EHD cell differentiation and allocation to the liver, these clinical trials drugs may potentially be repurposed for liver regenerative therapies.
However, with regards to ALGS, where loss of Jagged/Notch signaling and IHD cells are developmental, increasing Notch activity may be therapeutic to enhance IHD regeneration. As presented here, upon developmental loss of Jagged/Notch signaling in jagged mutants, knockdowns, and Notch inhibitor-treated zebrafish, EHD cells proliferate and contribute robustly to the liver, but fail to differentiate into IHD cells due to insufficient Notch signaling (see models in Figs. 3G and 7D). We postulate that with the expanded number of cells in the EHD niche, certain progenitors become displaced and are localized farther from the mesenchyme, consequently receiving less Fgf ligands. The decreased Fgf signaling in these EHD cells would then lead to their allocation to the liver. When the Jagged knockdown or Notch inhibitor is later attenuated, Notch signaling increases, and these cells originating from the EHD can then differentiate into IHD cells, contributing to regeneration.
In ALGS patients, postnatal fluctuations in cholestasis severity is often observed, sometimes resolving(50), suggesting that IHD cells can also regenerate in humans. However, the mechanism driving this regenerative process is unknown. The partial and variable penetrance of pathologies among family members with the same heterozygous JAG1 mutation and even between identical twins(51-53), suggests that Notch is teetering on insufficiency in ALGS patients. Together with the Notch-depend mechanism of IHD cell regeneration uncovered here using a Jagged loss-of-function ALGS model, we posit that stochastic changes in Notch signaling can result in sufficiently elevated levels in certain ALGS patients to stimulate IHD cell regeneration and ameliorate cholestasis. For these reasons, we propose that a mild augmentation of Notch signaling will be sufficient to nudge this pathway to sufficiency and enhance IHD cell regeneration in ALGS. A Notch agonist will therefore likely be therapeutic for this disorder. Considering the well-established functions of Notch in other tissues in promoting cell segregation and self-organization(54, 55), it is reasonable that Notch active cells emerging from the EHD were able to migrate throughout the entire liver and assemble into a complex, integrated, and functional ductal network. Given this ability of Notch active cells to self-pattern, it will be intriguing to explore whether rescuing Notch signaling in other tissues will restore their remodeling potential and thereby help to resolve additional ALGS pathologies.
Supplementary Material
Acknowledgements:
We thank the zebrafish research community for the numerous reagents shared and members of our laboratory for critical discussions. This work was supported by funds from the W. M. Keck Foundation (2017-01), and the National Institutes of Health (DP2DK098092, U01DK105541, and R01DK124583) to P.D.S.D., the Larry L. Hillblom Foundation Fellowship (#2019-D-013-FEL) to C.Z., Diabetes Research Connection (Project #08) to J.J.L, and The National Natural Science Foundation of China (81773097, 81872068) to D.C.
List of abbreviations:
- ALGS
Alagille Syndrome
- Fgf
fibroblast growth factor
- EHD
extrahepatic duct
- IHD
intrahepatic duct
Footnotes
Competing interests: The authors declare no competing financial interests.
References:
- 1.Alison MR. The many ways to mend your liver: A critical appraisal. Int J Exp Pathol 2018;99:106–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bankaitis ED, Ha A, Kuo CJ, Magness ST. Reserve Stem Cells in Intestinal Homeostasis and Injury. Gastroenterology 2018;155:1348–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ko S, Russell JO, Molina LM, Monga SP. Liver Progenitors and Adult Cell Plasticity in Hepatic Injury and Repair: Knowns and Unknowns. Annu Rev Pathol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forbes SJ, Newsome PN. Liver regeneration - mechanisms and models to clinical application. Nat Rev Gastroenterol Hepatol 2016;13:473–485. [DOI] [PubMed] [Google Scholar]
- 5.Raven A, Lu WY, Man TY, Ferreira-Gonzalez S, O'Duibhir E, Dwyer BJ, Thomson JP, et al. Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature 2017;547:350–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goessling W, Sadler KC. Zebrafish: an important tool for liver disease research. Gastroenterology 2015;149:1361–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prince VE, Anderson RM, Dalgin G. Zebrafish Pancreas Development and Regeneration: Fishing for Diabetes Therapies. Curr Top Dev Biol 2017;124:235–276. [DOI] [PubMed] [Google Scholar]
- 8.Leonard LD, Chao G, Baker A, Loomes K, Spinner NB. Clinical utility gene card for: Alagille Syndrome (ALGS). Eur J Hum Genet 2014;22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamath BM, Ye W, Goodrich NP, Loomes KM, Romero R, Heubi JE, Leung DH, et al. Outcomes of Childhood Cholestasis in Alagille Syndrome: Results of a Multicenter Observational Study. Hepatol Commun 2020;4:387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emerick KM, Rand EB, Goldmuntz E, Krantz ID, Spinner NB, Piccoli DA. Features of Alagille syndrome in 92 patients: frequency and relation to prognosis. Hepatology 1999;29:822–829. [DOI] [PubMed] [Google Scholar]
- 11.Li L, Krantz ID, Deng Y, Genin A, Banta AB, Collins CC, Qi M, et al. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet 1997;16:243–251. [DOI] [PubMed] [Google Scholar]
- 12.McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, Krantz ID, Piccoli DA, Spinner NB. NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am J Hum Genet 2006;79:169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oda T, Elkahloun AG, Pike BL, Okajima K, Krantz ID, Genin A, Piccoli DA, et al. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet 1997;16:235–242. [DOI] [PubMed] [Google Scholar]
- 14.Turnpenny PD, Ellard S. Alagille syndrome: pathogenesis, diagnosis and management. Eur J Hum Genet 2012;20:251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang D, Gates KP, Barske L, Wang G, Lancman JJ, Zeng XI, Groff M, et al. Endoderm Jagged induces liver and pancreas duct lineage in zebrafish. Nat Commun 2017;8:769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong PD, Munson CA, Norton W, Crosnier C, Pan X, Gong Z, Neumann CJ, et al. Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nat Genet 2007;39:397–402. [DOI] [PubMed] [Google Scholar]
- 17.Ellis JL, Bove KE, Schuetz EG, Leino D, Valencia CA, Schuetz JD, Miethke A, et al. Zebrafish abcb11b mutant reveals strategies to restore bile excretion impaired by bile salt export pump deficiency. Hepatology 2018;67:1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagore N, Howe S, Boxer L, Scheuer PJ. Liver cell rosettes: structural differences in cholestasis and hepatitis. Liver 1989;9:43–51. [DOI] [PubMed] [Google Scholar]
- 19.Song JY, Van Noorden CJ, Frederiks WM. Rearrangement of hepatocellular F-actin precedes the formation of rosette-like structures in parenchyma of cholestatic rat liver. Hepatology 1998;27:765–771. [DOI] [PubMed] [Google Scholar]
- 20.Lorent K, Yeo SY, Oda T, Chandrasekharappa S, Chitnis A, Matthews RP, Pack M. Inhibition of Jagged-mediated Notch signaling disrupts zebrafish biliary development and generates multi-organ defects compatible with an Alagille syndrome phenocopy. Development 2004;131:5753–5766. [DOI] [PubMed] [Google Scholar]
- 21.Burlison JS, Long Q, Fujitani Y, Wright CV, Magnuson MA. Pdx-1 and Ptf1a concurrently determine fate specification of pancreatic multipotent progenitor cells. Dev Biol 2008;316:74–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong PD, Provost E, Leach SD, Stainier DY. Graded levels of Ptf1a differentially regulate endocrine and exocrine fates in the developing pancreas. Genes Dev 2008;22:1445–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang YJ, Park JT, Parsons MJ, Leach SD. Fate mapping of ptf1a-expressing cells during pancreatic organogenesis and regeneration in zebrafish. Dev Dyn 2015;244:724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fabian P, Tseng KC, Smeeton J, Lancman JJ, Dong PDS, Cerny R, Crump JG. Lineage analysis reveals an endodermal contribution to the vertebrate pituitary. Science 2020;370:463–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hockman D, Burns AJ, Schlosser G, Gates KP, Jevans B, Mongera A, Fisher S, et al. Evolution of the hypoxia-sensitive cells involved in amniote respiratory reflexes. Elife 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaub JR, Huppert KA, Kurial SNT, Hsu BY, Cast AE, Donnelly B, Karns RA, et al. De novo formation of the biliary system by TGFbeta-mediated hepatocyte transdifferentiation. Nature 2018;557:247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 2009;137:216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clotman F, Lannoy VJ, Reber M, Cereghini S, Cassiman D, Jacquemin P, Roskams T, et al. The onecut transcription factor HNF6 is required for normal development of the biliary tract. Development 2002;129:1819–1828. [DOI] [PubMed] [Google Scholar]
- 29.Post Y, Clevers H. Defining Adult Stem Cell Function at Its Simplest: The Ability to Replace Lost Cells through Mitosis. Cell Stem Cell 2019;25:174–183. [DOI] [PubMed] [Google Scholar]
- 30.Jeliazkova P, Jors S, Lee M, Zimber-Strobl U, Ferrer J, Schmid RM, Siveke JT, et al. Canonical Notch2 signaling determines biliary cell fates of embryonic hepatoblasts and adult hepatocytes independent of Hes1. Hepatology 2013;57:2469–2479. [DOI] [PubMed] [Google Scholar]
- 31.Dutton JR, Chillingworth NL, Eberhard D, Brannon CR, Hornsey MA, Tosh D, Slack JM. Beta cells occur naturally in extrahepatic bile ducts of mice. J Cell Sci 2007;120:239–245. [DOI] [PubMed] [Google Scholar]
- 32.Cardinale V, Wang Y, Carpino G, Cui CB, Gatto M, Rossi M, Berloco PB, et al. Multipotent stem/progenitor cells in human biliary tree give rise to hepatocytes, cholangiocytes, and pancreatic islets. Hepatology 2011;54:2159–2172. [DOI] [PubMed] [Google Scholar]
- 33.de Jong IEM, Matton APM, van Praagh JB, van Haaften WT, Wiersema-Buist J, van Wijk LA, Oosterhuis D, et al. Peribiliary Glands Are Key in Regeneration of the Human Biliary Epithelium After Severe Bile Duct Injury. Hepatology 2019;69:1719–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Overi D, Carpino G, Cardinale V, Franchitto A, Safarikia S, Onori P, Alvaro D, et al. Contribution of Resident Stem Cells to Liver and Biliary Tree Regeneration in Human Diseases. Int J Mol Sci 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin S, Nascimento EM, Gajera CR, Chen L, Neuhofer P, Garbuzov A, Wang S, et al. Distributed hepatocytes expressing telomerase repopulate the liver in homeostasis and injury. Nature 2018;556:244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shiota J, Zaki NHM, Merchant JL, Samuelson LC, Razumilava N. Generation of Organoids from Mouse Extrahepatic Bile Ducts. J Vis Exp 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berg T, Rountree CB, Lee L, Estrada J, Sala FG, Choe A, Veltmaat JM, et al. Fibroblast growth factor 10 is critical for liver growth during embryogenesis and controls hepatoblast survival via beta-catenin activation. Hepatology 2007;46:1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manfroid I, Delporte F, Baudhuin A, Motte P, Neumann CJ, Voz ML, Martial JA, et al. Reciprocal endoderm-mesoderm interactions mediated by fgf24 and fgf10 govern pancreas development. Development 2007;134:4011–4021. [DOI] [PubMed] [Google Scholar]
- 39.Mohammadi M, McMahon G, Sun L, Tang C, Hirth P, Yeh BK, Hubbard SR, et al. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science 1997;276:955–960. [DOI] [PubMed] [Google Scholar]
- 40.Lee Y, Grill S, Sanchez A, Murphy-Ryan M, Poss KD. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development 2005;132:5173–5183. [DOI] [PubMed] [Google Scholar]
- 41.Ito T, Ando H, Suzuki T, Ogura T, Hotta K, Imamura Y, Yamaguchi Y, et al. Identification of a primary target of thalidomide teratogenicity. Science 2010;327:1345–1350. [DOI] [PubMed] [Google Scholar]
- 42.Lin JW, Biankin AV, Horb ME, Ghosh B, Prasad NB, Yee NS, Pack MA, et al. Differential requirement for ptf1a in endocrine and exocrine lineages of developing zebrafish pancreas. Dev Biol 2004;274:491–503. [DOI] [PubMed] [Google Scholar]
- 43.Draper BW, Stock DW, Kimmel CB. Zebrafish fgf24 functions with fgf8 to promote posterior mesodermal development. Development 2003;130:4639–4654. [DOI] [PubMed] [Google Scholar]
- 44.Danopoulos S, Schlieve CR, Grikscheit TC, Al Alam D. Fibroblast Growth Factors in the Gastrointestinal Tract: Twists and Turns. Dev Dyn 2017;246:344–352. [DOI] [PubMed] [Google Scholar]
- 45.Gilgenkrantz H, Collin de l'Hortet A. Understanding Liver Regeneration: From Mechanisms to Regenerative Medicine. Am J Pathol 2018;188:1316–1327. [DOI] [PubMed] [Google Scholar]
- 46.Hsu YC, Fuchs E. A family business: stem cell progeny join the niche to regulate homeostasis. Nat Rev Mol Cell Biol 2012;13:103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lush ME, Diaz DC, Koenecke N, Baek S, Boldt H, St Peter MK, Gaitan-Escudero T, et al. scRNA-Seq reveals distinct stem cell populations that drive hair cell regeneration after loss of Fgf and Notch signaling. Elife 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dabney RS, Khalife M, Shahid K, Phan AT. Molecular pathways and targeted therapy in cholangiocarcinoma. Clin Adv Hematol Oncol 2019;17:630–637. [PubMed] [Google Scholar]
- 49.Wang J, Xing X, Li Q, Zhang G, Wang T, Pan H, Li D. Targeting the FGFR signaling pathway in cholangiocarcinoma: promise or delusion? Ther Adv Med Oncol 2020;12:1758835920940948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mouzaki M, Bass LM, Sokol RJ, Piccoli DA, Quammie C, Loomes KM, Heubi JE, et al. Early life predictive markers of liver disease outcome in an International, Multicentre Cohort of children with Alagille syndrome. Liver Int 2016;36:755–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Izumi K, Hayashi D, Grochowski CM, Kubota N, Nishi E, Arakawa M, Hiroma T, et al. Discordant clinical phenotype in monozygotic twins with Alagille syndrome: Possible influence of non-genetic factors. Am J Med Genet A 2016;170A:471–475. [DOI] [PubMed] [Google Scholar]
- 52.Kamath BM, Krantz ID, Spinner NB, Heubi JE, Piccoli DA. Monozygotic twins with a severe form of Alagille syndrome and phenotypic discordance. Am J Med Genet 2002;112:194–197. [DOI] [PubMed] [Google Scholar]
- 53.Ziesenitz VC, Loukanov T, Glaser C, Gorenflo M. Variable expression of Alagille syndrome in a family with a new JAG1 gene mutation. Cardiol Young 2016;26:164–167. [DOI] [PubMed] [Google Scholar]
- 54.Binshtok U, Sprinzak D. Modeling the Notch Response. Adv Exp Med Biol 2018;1066:79–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bocci F, Onuchic JN, Jolly MK. Understanding the Principles of Pattern Formation Driven by Notch Signaling by Integrating Experiments and Theoretical Models. Front Physiol 2020;11:929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.