Abstract
Sperm storage tubules (SST) are specialized invaginations of the oviductal epithelium that permit avian species to store spermatozoa for extended periods of time, without compromising sperm fertilization capacity. The molecular and physiological mechanisms behind sperm storage tubule differentiation, sperm protection, and regression remain largely unknown, but most likely have potential implications for substantially improving hen fertility, sperm storage, and semen cryopreservation in commercial poultry species. RNA sequencing was performed on sperm storage tubules isolated from the epithelium of the uterovaginal junction (UVJ) from hens at d 1, 7, 30, 60, and 90 postinsemination (n = 4 per timepoint). Read mapping and differential expression analysis were performed using CLC Genomics Workbench. A total of 2,340 differentially expressed genes were subjected to pathway analysis through Ingenuity Pathway Analysis (IPA). Through functional annotation of differentially expressed genes during early, peak, and late egg production, novel insights regarding the role of innate and acquired immune response to sperm, lipid synthesis and transfer, steroid hormone signalling, cytoskeletal reorganization, and regulation of ion homeostasis in SST were obtained. Additionally, potential pathways were identified that could be involved with suppressing sperm motility while sperm reside within the SST. Upstream analysis identified potential regulatory roles for 18 upstream regulators that could modulate sperm storage tubule function, including suppression of sperm motility. Understanding sperm storage tubule function throughout the laying cycle, especially with regards to sperm preservation may allow for the development of industry-based protocols for semen storage and cryopreservation that mimic the sperm preservation capabilities of SST and improve fertility.
Key words: sperm storage tubule, transcriptome, artificial insemination, sperm motility, sperm storage
INTRODUCTION
Avian species possess the ability to store sperm within the female reproductive tract for extended periods of time within specialized invaginations of the oviductal epithelium. Sperm storage occurs in blind-ended tubular invaginations termed sperm storage tubules (SST), which are primarily located in the uterovaginal junction (UVJ), but are also found to a lesser extent in the infundibulum (Matsuzaki and Sasanami, 2017). Microscopic examination revealed that SST differentiate from the oviductal epithelium prior to sexual maturation and consist of a simple columnar epithelium characterized by a supranuclear vacuole (Bakst, 1987). The duration of successful sperm storage in the SST varies among avian species and correlates with both the number and depth of SST present in the female reproductive tract. For example, in the chicken hen, roughly 5,000 small, short SST are found in the UVJ and the duration of sperm storage is 21 d; while, in the turkey, roughly 30,000 longer, larger SST are found in the UVJ with a much longer sperm storage duration of 70 d (Birkhead and Moller, 1992; Bakst et al., 2010). Additionally, within a given species, the number of SST has been correlated with fertility rates, where increased SST counts were correlated with increased numbers of sperm penetrating the vitelline membrane (Brillard and Bakst, 1990).
Upon artificial insemination or natural mating, only 1% of sperm enter SST, while the remaining sperm are expelled from the oviduct or targeted for degradation. It has been noted that SST likely function as gatekeepers for sperm quality, preventing selection of non-motile sperm, possibly by requiring a certain velocity threshold for sperm entry (Mendonca et al., 2019). Moreover, there is additional evidence that SST also limit entrance for damaged sperm, particularly regarding alterations to surface proteins or the glycocalyx of sperm (Peláez and Long, 2008; Peláez et al., 2011). Sperm entrance is also impacted by timing within the ovulatory cycle, with SST filling capacity reduced when insemination occurs near oviposition (Birkhead et al., 1996). Additionally, researchers have noted that SST does not fill at the same rate, potentially indicating alternative factors involved in sperm uptake into SST (McIntyre and Christensen, 1983). Multifactorial components of regulation for sperm attraction to and sperm quality assessment by the SST appear to determine the proportion of sperm that enter SST, though the governing mechanisms are still undefined.
Within the SST, sperm arrange in a tight bundle with sperm heads parallel and oriented toward the distal end of the SST. During the storage process, sperm are relatively quiescent, with slowed metabolic processes and motility, though sperm are observed to exhibit a slow synchronous movement during storage (Bakst, 1992). Though metabolism is reduced, sperm still require metabolic inputs, such as fatty acids or lipids, as an energy source to perform basic cell functions (Douard et al., 2000). Furthermore, lipid containing microvillus blebs have been observed on the apical tips of SST, and have been hypothesized to supply sperm with metabolic substrates and/or alter the surface composition of the sperm membranes to induce decreased motility (Bakst and Bauchan, 2015). The mechanisms surrounding the reduction in motility while sperm reside in SST are not well defined, but lactic acid and the associated reduction in pH has been implicated as a major source for the reduced sperm motility (Matsuzaki et al., 2015). Certain ions, such as calcium and zinc, have been shown to impact sperm motility (Holm et al., 2000). Despite some key aspects related to reduce motility of sperm having been identified, other key components of fertility and survival including regulators of reduced metabolism, sperm orientation, and sperm deactivation and subsequent reactivation are still poorly understood.
Similar to sperm entrance and storage within the SST, the systematic cues and molecular mechanisms that lead to sperm release from the SST largely remain an enigma. Initially, it was suggested that sperm release was regulated mechanically, either 1) with egg development and progression through the oviduct exerting pressure on the SST, or 2) by slow, constant sperm release (Van Krey et al., 1967; Burke and Ogasawara, 1969). However, additional work has shown that sperm release is differentially regulated throughout the ovulatory cycle, with the highest rate of release close to ovulation (Hemmings et al., 2015). There is also evidence for neuronal control of sperm release, as neurons and small ganglia terminate on or run parallel to the SST. This combined with the dense, F-actin rich microfilament network within the SST epithelium, could potentially regulate sperm release through contractile mechanisms (Freedman et al., 2001). More recently, progesterone has been shown to act as a sperm-releasing factor in SST, indicating hormonal regulation of sperm release (Ito et al., 2011). Thus, while some of the individual and collective roles of the several elements in the regulation of sperm release have been explored, many roles remain unclear and require further research to determine the mechanisms involved.
Currently, the commercial poultry industry faces extremely limited options for in vitro storage of semen, including cryopreservation. The fertility rates of hypothermically stored turkey sperm are especially poor, with fertility rates falling below economically acceptable levels after just 6 h of in vitro storage at 4°C (Long and Conn, 2012). In stark contrast, the natural in vivo semen storage of the turkey hen maintains viable sperm capable of fertilizing eggs for up to 10 wk after a single insemination (McCartney, 1951); however, the molecular mechanisms responsible for this prolonged storage are poorly characterized. The current working hypothesis is that in vitro stored sperm are missing key nutrients, substrates, or external cues that are provided by the hens’ SST during in vivo storage, resulting in loss of sperm function during in vitro storage and, ultimately, low fertility rates after insemination of in vitro stored semen. The objective of this study was to examine SST transcriptome changes from the day of post-lay insemination throughout the duration of fertility to gain a better understanding of the genomic dynamics involved in sperm entrance into, maintenance within, and release from SST.
MATERIALS AND METHODS
Bird Husbandry and Insemination
All protocols used in this study were approved by the Institutional Animal Care and Use Committee of the Beltsville Animal Research Center, United States Department of Agriculture. Feed and water were provided ad libitum to NRC standards to both hens and toms. Commercial line hens and toms were used for this study (Hendrix Genetics, Kitchener, Ontario, Canada). Hens were housed individually in cages. Hens were photo-stimulated at 27 wk of age, by increasing the light exposure from 6L:18D to 14L:10D to initiate ovarian development and egg production. Per industry standards, hens were artificially inseminated twice prior to the onset of egg production, a practice which ensures that SST are filled with sperm before the first egg is laid approximately 14 d after photostimulation. These 2 pre-lay inseminations occurred 8 and 11 d after photostimulation. A single, post-lay insemination was conducted 14 d after photo-stimulation; this is defined as D0 (hen age was 29 wk at D0) and is the reference for the experimental timepoints (D1, D7, D30, D60, D90). Average egg production during the experimental period was as follows: 0.81 eggs for wk 1, 2.92 eggs for wk 2, 4.75 eggs for wk 3, 4.58 eggs for wk 4, 5.13 eggs for wk 5, 5.00 eggs for wk 6, 4.88 eggs for wk 7, 5.00 eggs for wk 8, 5.00 eggs for wk 9, 5.25 eggs for wk 10, 4.75 eggs for wk 11, and 4.50 eggs for wk 12. Artificial insemination (AI) was performed using 50 μL of pooled semen extended 1:1 with room temperature Beltsville Turkey Semen Extender (Continental, Delavan, WI) with a dose of 2.5 × 108 sperm/hen. All inseminations were performed with a ratio of one tom to 4 hens. Toms were kept in floor pens in groups of 8 to 10. Toms were photostimulated at 28 wk of age, by increasing light exposure from 12L:12D to 14L:10D cycle to initiate testicular development and semen production. The abdominal massage semen collection method (Burrows and Quinn, 1935) was used to pre-milk toms at 30 wk of age and screen for semen quality at 31 and 32 wk of age. Ejaculate collected at 31 and 32 wk of age was evaluated for visual abnormalities (ex. presence of blood), sperm viability, and sperm mobility using previously published methods (Long and Kulkarni, 2004). Toms with abnormal ejaculate parameters and/or low sperm mobility were not used in the semen pool. Toms were 33 to 34 wk of age when used in the study. The same pool of toms was used for all artificial inseminations.
UVJ Tissue and SST Isolation
A total of 20 hens were euthanized via injection of pentobarbital sodium and phenyotoin sodium (390:50 mg) at D1, D7, D30, D60, and D90 following the post-lay AI (D0), with four replicates per timepoint (n = 4). Immediately, the UVJ tissue was dissected from the reproductive tract as previously described (Bakst, 1992). Briefly, the UVJ was dissected from the vagina and the shell gland, and the remaining connective tissue was removed from the UVJ prior to cutting longitudinally with to expose the mucosal region containing the SST. This region was examined microscopically to determine specific location of SST. Selected regions were divided into multiple segments that were individually embedded in optimal cutting temperature compound (Tissue Tek, Leica Microsystems, Buffalo Grove, IL) in 25 × 20 × 5 mm cryomolds and snap frozen using liquid nitrogen and stored at −80°C. The length of time from euthanasia to snap freezing was less than 20 min.
Flash frozen segments were sectioned at a thickness of 10 μm using a Leica CM1860 Cryostat and placed onto PEN Membrane 2 μm slides (Leica Microsystems). Slides were treated with RNAse Away (Thermo Fisher Scientific, Waltham, MA), rinsed with RNA free water (Thermo Fisher Scientific, Waltham, MA), and irradiated to ensure no contamination, and all reagents and plastics used were RNA/DNA free. After sectioning, slides were fixed in fresh acetone (Sigma) at −20°C for 30 s, then air dried for 30 s. Two rinses of 200 μL of 1X Dulbecco's Phosphate Buffered Saline (DPBS) (Thermo Fisher Scientific) were done, followed by 30 s of staining using 197 μL Nuclear Fast Red (Sigma-Aldrich, Inc., St. Louis, MO) and 3 μL RNase Inhibitor (Thermo Fisher Scientific). A second round of rinses using 200 μL of 1X DPBS were done, followed by dehydration in ethanol (Sigma-Aldrich, Inc.) for 30 s each at 70%, 95%, and twice with 100%. Slides were then air dried for 3 min and immediately sectioned via laser capture microdissection. SST were individually identified and isolated using the Leica LMD7 (Leica Microsystems) at 10X magnification. Sections were dropped via gravity into a tube containing 25 μL of 2-Mercaptoethanol (Bio-Rad, Hercules, CA) and RLT Buffer (Qiagen, Valencia, CA) at a concentration of 1:100. After laser sectioning was completed, tubes were centrifuged for 10 s, vortexed for 30 s, then stored at −80°C until RNA extraction. An average of 342.35 SST was isolated from each slide with an average surface area of 1,469,543.9 μm.
RNA Isolation, cDNA Library Construction, and Sequencing
Total RNA was extracted from isolated SST using RNeasy Plus Micro Kit (Qiagen), including on-column deoxyribonuclease digestion. RNA quality was assessed using Tapestation RNA HS Assay (Agilent, Santa Clara, CA) and quantified via Qubit RNA HS assay (Thermo Fisher Scientific). Average RNA integrity number (RIN) value was 7.75. Libraries were constructing using SMARTer Stranded RNA-Seq Kit (Takara Bio Inc., San Jose, CA) using standard manufacture recommendations. Quantity of the libraries was measured by KAPA SYBR FAST qPCR and quality via Tapestation RNA HS Assay (Agilent). Equimolar pooling of libraries was done on basis of qPCR values. Sample were sequenced using 150 bp paired-end sequencing (2 × 150 bp) on an Illumina HiSeq platform and run according to standard Illumina protocols. Images generated by sequencers were converted into nucleotide sequences by the base calling pipeline using RTA 1.18.64.0 and were stored in FASTQ format.
Bioinformatic Analysis of Sequencing Data
Sequencing files for each sample were submitted to the NIH Short Read Archive (accession numbers SAMN21925817- SAMN21925836) (https://www.ncbi.nlm.nih.gov/sra). The program CLC Genomics Workbench 20.0 (QiagenA; https://digitalinsights.qiagen.com) was used to perform the bioinformatic analysis of the sequenced data. To assess quality of both raw and trimmed sequencing reads, FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used (Andrews, 2010). Raw reads were trimmed for quality (Phred quality score <20). Additionally, poly-A and adapter sequencing trimming was performed using the trimming tool of CLC Genomics Workbench platform. Trimmed reads were mapped to the most recent Meleagris gallopavo reference genome (Turkey_5.1; NCBI annotation release 103; https://www.ncbi.nlm.nih.gov/assembly/GCF_000146605.3). Mapped reads were analyzed through principle component analysis and heat map analysis using CLC Genomics Workbench tools. Heat map analysis was generated through hierarchal clustering using Euclidean distance metric and complete linkage clustering method. Differential expression analysis was performed through pairwise comparisons between D1 and D7, D7 and D30, D30 and D60, and D60 and D90 timepoints. The gene set test tool of CLC Genomics Workbench was used to determine enriched gene ontology (GO) biological processes for DEGs common across multiple comparisons. Species specific gene ontology annotations were used for analysis of enriched biological processes. These sequential comparisons provided a linear timepoint analysis from the onset to cessation of egg lay.
Ingenuity Pathway Analysis (IPA; Qiagen; https://www.qiagenbioinformatics.com/products/ingenuity- pathway-analysis) was used to perform functional annotation of the obtained differential expressed genes (DEGs) (Krämer et al., 2014). Only DEGs with a q-value less than 0.05, an absolute fold change greater than 1.5, and a FPKM value greater than 1 were subjected to function annotation analysis (Liu and Di, 2020). The core analysis and comparison analysis functions of IPA were used to examine individual timepoints, as well as compare and contrast all five timepoints, respectively. Fold change data resulting from differential gene expression analysis was used as input for IPA analysis. For the comparison analysis, only upregulated genes for each individual timepoint were analyzed. For analysis of canonical pathways, biological functions, and upstream regulators an absolute z-score greater than 2 and P-value less than 0.05 were considered to be significantly activated or inhibited. For regulator effects in IPA, only effects with a positive consistency score, an absolute z-score greater than 2, and a P-value less than 0.05 were analyzed. For networks in IPA, only networks with scores greater than 35 were considered.
Confirmation of RNAseq Gene Expression Results
Quantified RNA was subjected to reverse transcription (RT) using SuperScript III and random hexamer primers (Thermo Fisher Scientific, Waltham, MA). To control for genomic DNA contamination, a reaction without reverse transcriptase was also performed on pooled RNA from all experimental samples. Expression profiles of randomly selected 10 DEGs were determined using RT-qPCR. Primers (Integrated DNA Technologies, Skokie, IL) used in the PCR reactions were designed using NCBI primer BLAST Software (NCBI, Bethesda, MD) with the following characteristics: 1) a target area located in the 3’ region of the transcript, 2) a location spanning an intron, 3) a melting temperature (Tm) of 58 to 60°C, 4) a GC content (GC%) of 40 to 60%, 5) a length of 18 to 30 nucleotides, and 6) a product length of 90 to 250 nucleotides (Table 1). PCR reactions (15 μL) contained 1 μL of cDNA, 0.6 μL of each primer (final concentration in PCR reaction of 0.4 μM), 5.3 μL of nuclease free water, and 7.5 μL of 2X iTaq Universal SYBR Green Supermix (Bio-Rad). All reactions were carried out using CFX Connect Real-Time PCR System (Bio-Rad). The PCR cycling conditions were as follows: initial denaturation at 95°C for 30 s followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. Dissociation curve analysis and gel electrophoresis were conducted to ensure that a single PCR product of appropriate size was amplified in each reaction and was absent from RT and PCR water controls. Additionally, primer pairs were validated through amplification efficiencies and sequencing of resulting PCR products as described previously (Brady et al., 2019). Relative mRNA levels were calculated using the 2−DDCT method using averaged expression of TUBB3, UBB, and BTUBB as housekeeping genes (Livak and Schmittgen, 2001). Data are presented relative to average D1 mRNA levels for each gene examined. A one-way ANOVA using SAS software (SAS Institute, Cary, NC) was used to analyze normalized, log2 transformed data. The least squares means were compared using the PDIFF statement.
Table 1.
Primer sequences used for confirmation of gene expression results obtained from RNA sequencing.
| Gene | Forward | Reverse |
|---|---|---|
| SCD | CAATGCCACCTGGCTAGTGA | GGTGGAGTAGTCGTAGGGGA |
| SPARC | AAGTGCACCTTGGAGGGAAC | GCGCTTCTCATTCTCGTGGA |
| SERPINB5 | AAGCTACGTTTTGCCTGGGT | TCAGGGGTGAGTGCCTTTTC |
| CA4 | ACTATCGGTACGAGGGCTCC | GCACCGGACCAGTACACATA |
| ELAPOR2 | CCTGCGATGGATGCACTTTC | ACCAGCTCCAACCTTTAGCC |
| OSGIN1 | TACCCTGAGCAGAGGGGAAT | ATCTTCCTGTGCGTGGTTGG |
| KCNMB1 | CAGCCTCAGGACAAGAGGTC | GTGAAGAGGAGGCCTTTGGG |
| SPINT4 | ATTTCTGCACGGTCACTCCC | CGCGTTGTAGAAGAAGCGGAT |
| CYGB | GGAGAACCTCAACGACCCAG | CACGTGGGTGTAGATGAGGG |
| GAREM1 | CGCTCTCCTAGCCCTACTCT | CTGGCCTTCCGTAGTTCCTG |
| UBB | TCAAGCAAGATGCACAGCAC | TTTCAACATACAGATCAGCAG |
| TUBB3 | CAGTTTTGGGAGGTGATCAGCGA | CCCGCTCTGACCGAAAATGA |
RESULTS
A total of 1.6 billion reads were obtained across the 20 samples sequenced, with an average read count of 82.2 million reads per sample. On average, 85.2% of reads mapped in pairs, 2.1% of reads mapped in broken pairs, and 12.7% of reads did not map to the turkey genome (Supplementary File 1). Of the mapped fragments, a total of 71.6% mapped to gene regions, while 28.4% mapped to intergenic regions (Supplementary File 1). No significant differences were identified between timepoint comparisons related to read or fragment mapping (P > 0.05). Principle component analysis showed sample clustering by timepoint, with the largest degree of variation seen in the D1 and D90 samples (Supplementary File 2). Clustering by timepoint was also seen through heat map analysis of the mapped reads across samples (Supplementary File 2). RNA sequencing gene expression results were confirmed using quantitative PCR methods, with similar mRNA expression profiles for the 10 genes examined (Supplementary File 3). In addition, true fertility means and SEM were calculated for each timepoint group on a weekly basis for the duration of the experimental period (Supplementary File 4).
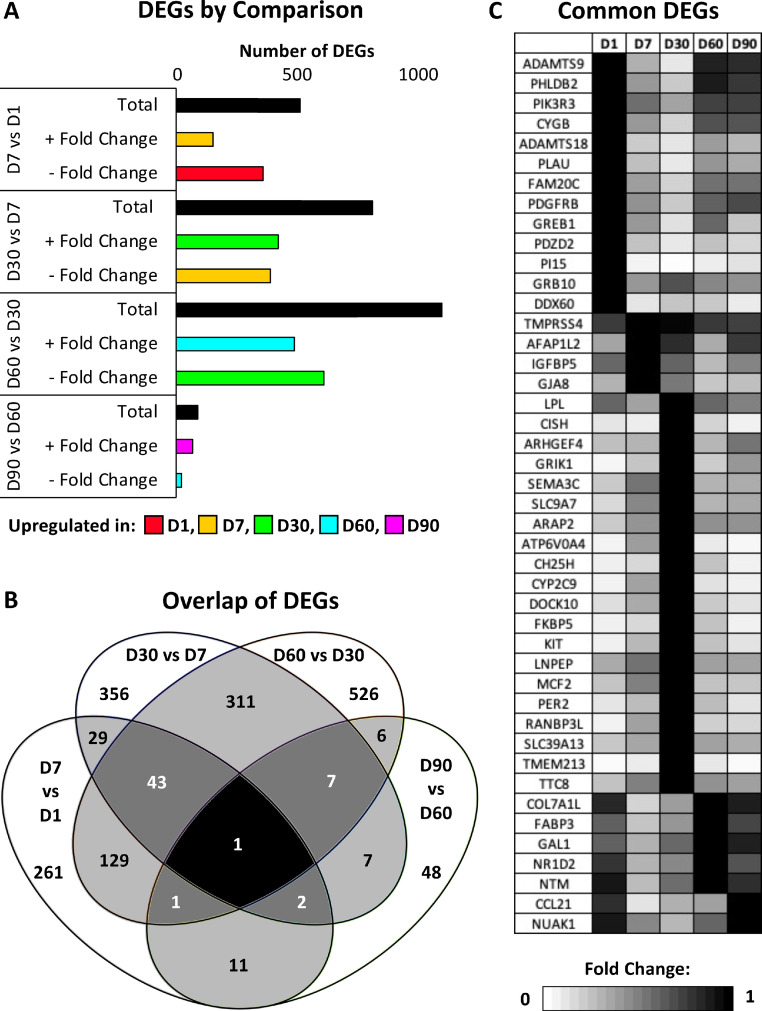
A total of 2,340 DEGs were identified across the four timepoint comparisons (Figure 1A). The largest number of DEGs was identified in the D30 and D60 comparison while the smallest number of DEGs was identified in the D60 and D90 comparison. A significant degree of overlap occurred when DEGs were examined across timepoint comparisons, with 54 DEGs appearing in 3 or more of the timepoint comparisons (Figure 1B). Several of the common DEGs in multiple timepoint comparisons exhibited patterned expression in relation to timepoint (Figure 1C). Enriched biological processes for the common DEGs included chemotaxis, chemical homeostasis, transmembrane receptor protein tyrosine kinase signaling pathway, response to external stimulus, and regulation of phosphoprotein phosphatase activity (Table 2).
Figure 1.
Differentially expressed genes (DEGs) characterization. (A) Total number of DEGs for each comparison, with the number of DEGs upregulated in each timepoint (FPKM > 1, |fold change| >1.5, q < 0.05). (B) Venn diagram showing the number of common DEGs among each timepoint comparison (FPKM > 1, |fold change| >1.5, q < 0.05). White text indicates DEGs common to at least three timepoint comparisons (C) Heat map showing the gene name and expression profile across D1, D7, D30, D60, and D90 timepoints for each annotated DEG common to at least three timepoint comparisons. Fold change is presented relative to the timepoint with the highest level of expression.
Table 2.
Enriched biological processes associated with common DEGs.
| Term | Count | P value | Genes | Fold |
|---|---|---|---|---|
| Enzyme linked receptor protein signaling pathway | 5 | 0.012 | PDGFRB, AFAP1L2, KIT, GRB10, LNPEP | 5.012 |
| Chemotaxis | 4 | 0.014 | PDGFRB, TTC8, SEMA3C, KIT | 7.175 |
| Chemical homeostasis | 5 | 0.017 | SLC9A7, LPL, SLC39A13, ATP6V0A4, NR1D2 | 4.656 |
| Transmembrane receptor protein tyrosine kinase signaling pathway | 4 | 0.018 | PDGFRB, AFAP1L2, KIT, GRB10 | 6.713 |
| Response to external stimulus | 6 | 0.044 | PDGFRB, TTC8, SEMA3C, PLAU, KIT, NR1D2 | 2.825 |
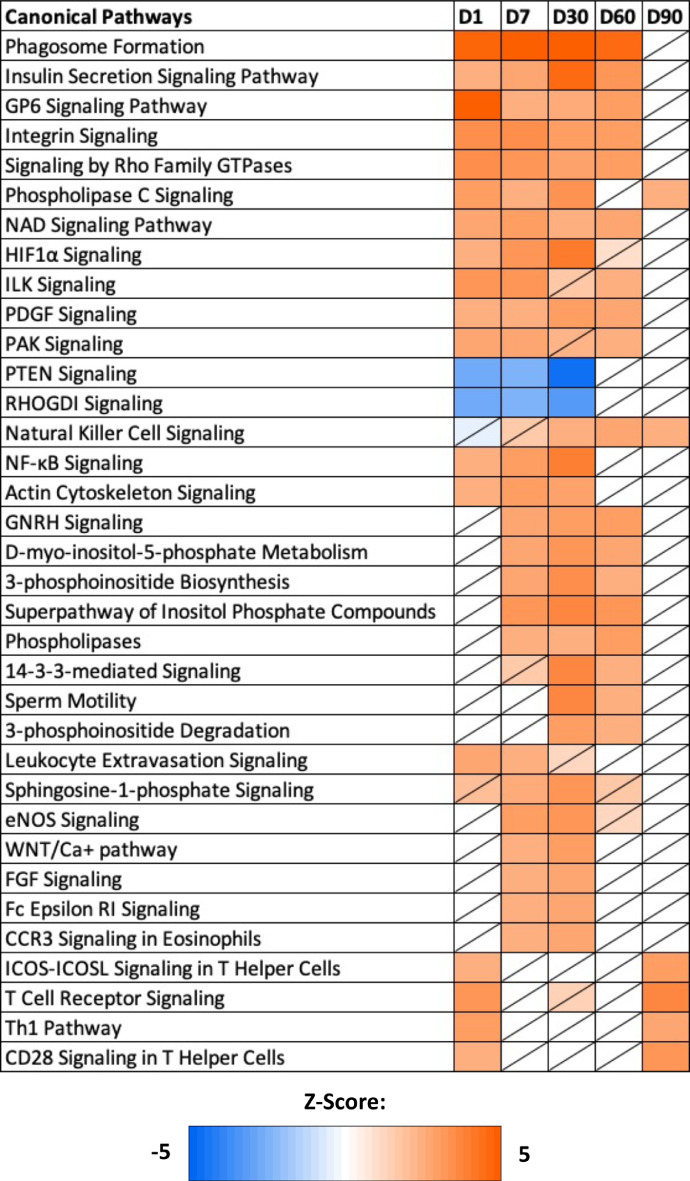
Common canonical pathways were established from experimental timepoint comparisons based on gene expression profiles (Figure 2). Predicted activation of several cellular-based immune response pathways were seen at D1 and D90 timepoints, with T cell receptor signaling, signaling in T helper cells, and Th1 pathway activation. Proinflammatory related pathways, including NF-xB signaling and leukocyte extravasation signaling, were only predicted to be activated during early timepoints, while natural killer cell signaling, an innate immune pathway, was only predicted to be activated in the later timepoints. Multiple canonical pathways related to phagosome formation and cytoskeletal reorganization, collagen and extracellular signaling, energy metabolism, and chemotaxis were predicted to be activated in the 4 of the 5 timepoints. In the early timepoints, D1, D7, and D30, actin cytoskeleton signaling was predicted to be activated while negative cell growth and development regulators including PTEN signaling and RHOGDI signaling were predicted to be inhibited. Pathways related to gonadotrophin-releasing hormone (GnRH) signaling and lipid and carbohydrate metabolism showed predicted activation at D7, D30, and D60 timepoints, with sperm motility predicted to be activated in D30 and D60 timepoints. Lastly, D7 and D30 timepoints exhibited predicted activation of pathways associated with generation of nitric oxide, WNT/Ca+ signaling, and eosinophil function.
Figure 2.
Common canonical pathways across timepoints. The common canonical pathways appearing in two or more timepoint comparisons, obtained via the comparison analysis tool of Ingenuity Pathway Analysis (Qiagen, Valencia, CA). Z-score is denoted by color, with orange representing activation and blue representing inhibition. Nonsignificant z-scores are denoted with a slash through the cell (|z-score| > 2, P-value < 0.05).
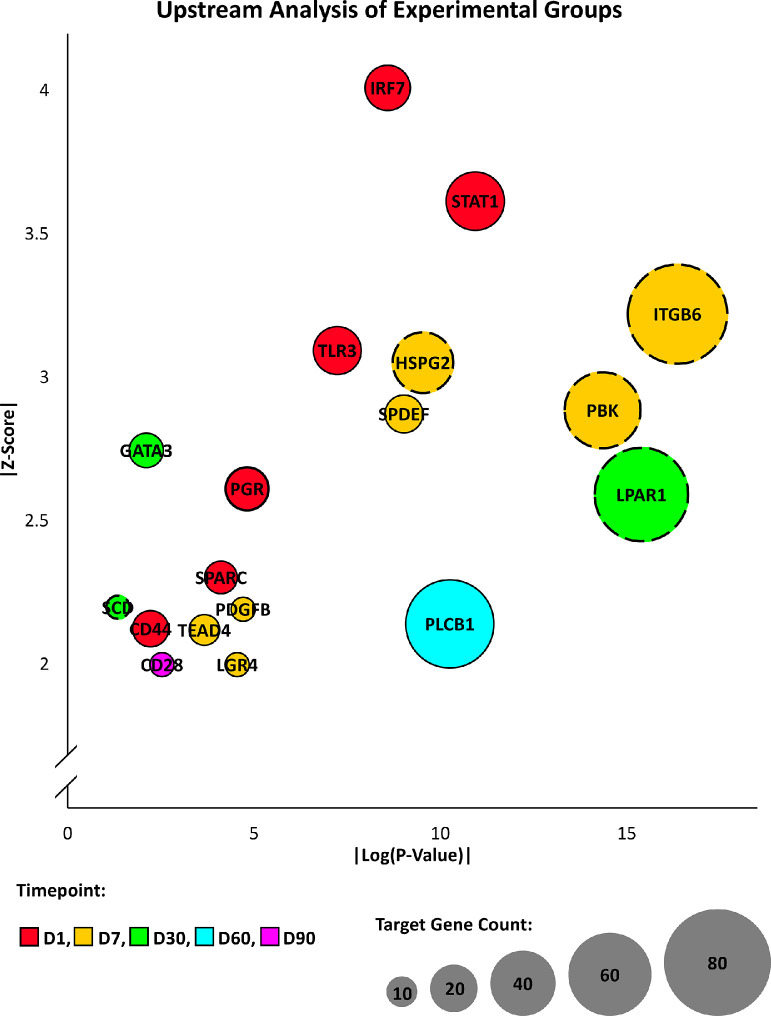
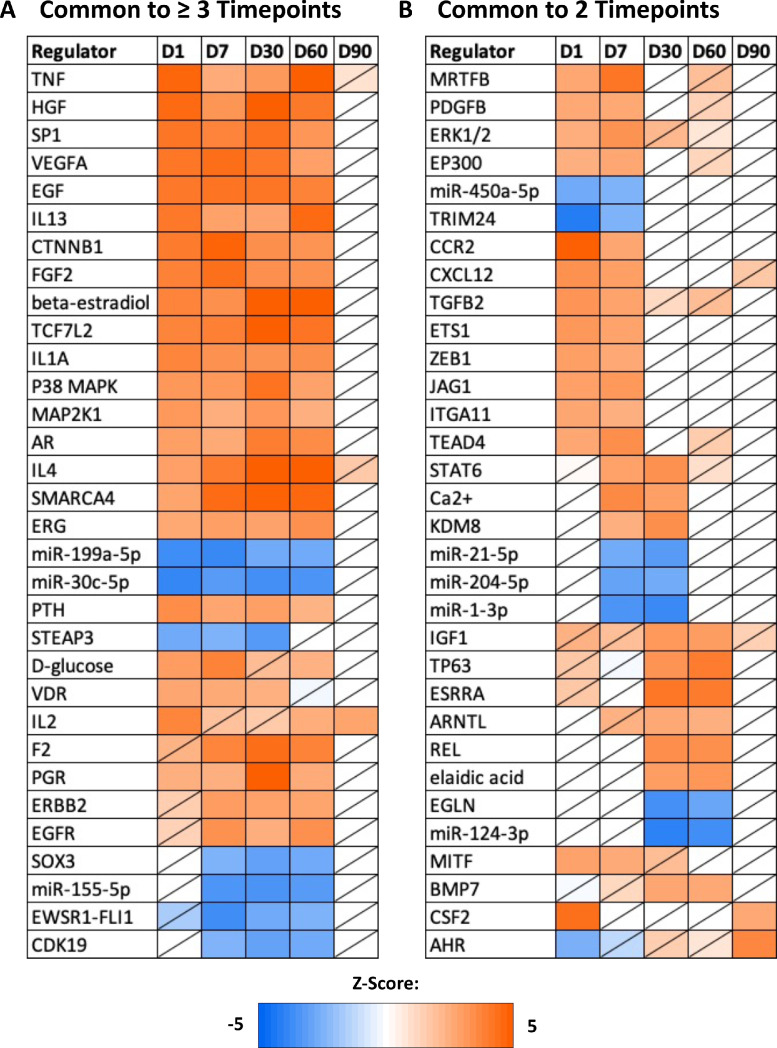
Upstream analysis across multiple timepoint comparisons identified 18 regulators with significant predicted activation or inhibition that also showed differential gene expression across the experimental timepoints (Figure 3). Interferon regulatory factor 7 (IRF7), signal transducer and activator of transcription 1 (STAT1), toll-like receptor 3 (TLR3), CD44 molecule (CD44), secreted protein acidic and cysteine rich (SPARC), and the progesterone receptor (PGR) were predicted to be activated D1 samples, and showed increased expression levels in D1 samples in comparison to D7 samples. The D7 samples showed downregulation and predicted inhibition of heparan sulfate proteoglycan 2 (HSPG2), PDZ binding kinase (PBK), and integrin subunit beta 6 (ITGB6) compared to D1 samples. In D7 samples, upregulation and predicted activation of platelet-derived growth factor subunit B (PDGFB), TEA domain transcription factor 4 (TEAD4), leucine rich repeat containing G protein-coupled receptor 4 (LGR4), and SAM pointed domain containing ETS transcription factor (SPDEF) when compared to D30 samples. By comparison, the D30 samples exhibited predicted activation and upregulation of GATA binding protein 3 (GATA3) compared to D7 as well as predicted inhibition and downregulation of stearoyl-CoA desaturase (SCD) and lysophosphatidic acid receptor 1 (LPAR1). Lastly, D60 samples displayed predicted activation and upregulation of phospholipase C beta 1 (PLCB1) compared to D30 samples, and D90 samples displayed predicted activation and upregulation of CD28 molecule (CD28) compared to D60 samples. In addition to the predicted upstream regulators that also showed gene expression differences across timepoints, an evaluation of upstream analysis results from each experimental comparison revealed 31 significant upstream regulators common to three or more timepoints and 32 significant upstream regulators common to two timepoints (Figure 4).
Figure 3.
Upstream analysis by timepoint. Predicted upstream regulators for each experimental timepoint from the core analysis tool of Ingenuity Pathway Analysis (Qiagen, Valencia, CA) are presented. Only upstream regulators with significant predicted activity (|z-score| > 2, P < 0.05) and that exhibit differential expression in the given timepoint comparison (|fold change| > 1.5, FPKM > 1, q-value < 0.05) are represented. The z-score described the degree of activity, the circle size pertains to the number of target genes that appear in the dataset, and the circle outline indicates if the upstream regulator is predicted to be activated (solid) or inhibited (dashed) in the given timepoint.
Figure 4.
Common upstream regulators. The common upstream regulators appearing in two or more timepoint comparisons, resulting from the comparison analysis tool of Ingenuity Pathway Analysis (Qiagen, Valencia, CA). Z-score is denoted by color, with orange representing activation and blue representing inhibition. Nonsignificant z-scores are denoted with a slash through the cell (|z-score| > 2, P-value < 0.05).
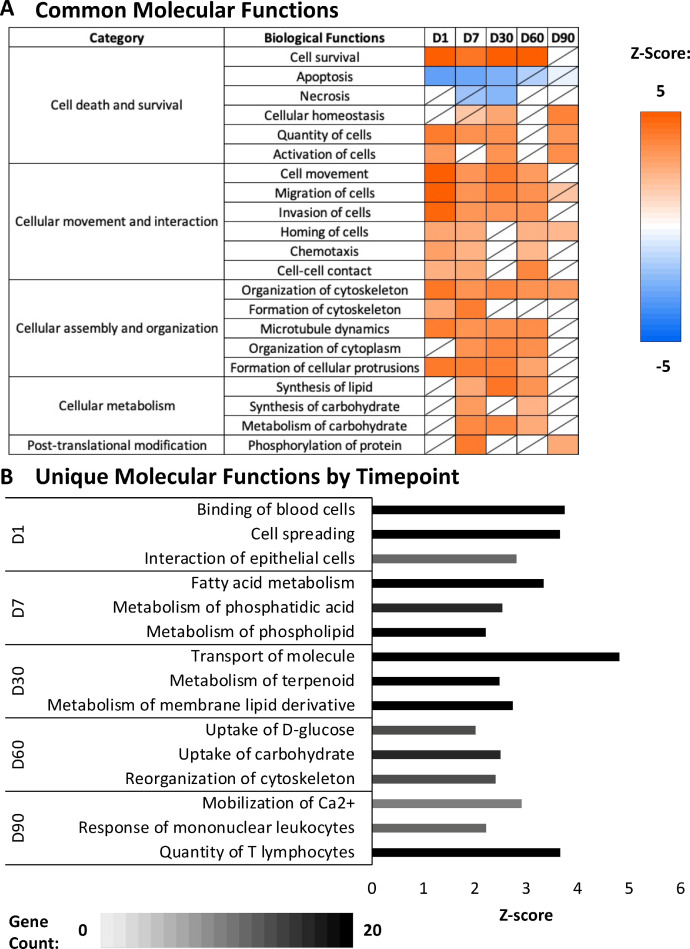
Examination of activated molecular functions revealed that functions related to cell death and survival, cellular movement and interaction, cellular assembly and organization, cellular metabolism, and post-translational modifications were common across multiple timepoints (Figure 5A). Cell death and survival had contrasting predicted statuses with cell survival functions activated while cell death functions inhibited. Cellular movement functions predominately displayed decreased predicted activation status across increased timepoints, with the exception of homing of cells, chemotaxis, and cell-cell contact functions, which only showed activation at D1, D7, and D60 timepoints. Cellular assembly and organization functions mostly exhibited decreased predicted activation with increased timepoints, however, the formation of cytoskeleton was only predicted to be activated at D1 and D7 timepoints. The predicted activation of cell metabolism functions was only seen at D7, D30, and D60, while post-translational modification functions displayed significant activation only at D7 and D90.
Figure 5.
Common and unique molecular functions. (A) The common molecular functions appearing in two or more timepoint comparisons, resulting from the comparison analysis tool of Ingenuity Pathway Analysis (Qiagen, Valencia, CA). Z-score is denoted by color, with orange representing activation and blue representing inhibition. Nonsignificant z-scores are denoted with a slash through the cell (|z-score| > 2, P-value < 0.05). (B) The molecular functions unique to each timepoint that were produced from the core analysis tool of Ingenuity Pathway Analysis (Qiagen, Valencia, CA). Z-score is denoted on the x-axis (|z-score| > 2, P-value < 0.05). Gene count is indicated by bar color.
In addition to molecular functions common to multiple timepoints, the enrichment of molecular functions unique to each timepoint were noted (Figure 5B). Gene expression of SST isolated at D1 showed enrichment for binding of blood cells, cell spreading, and interaction of epithelial cells. The D7 SST gene levels demonstrated enrichment for molecular functions associated with fatty acid metabolism, metabolism of phosphatidic acid, and metabolism of phospholipid. Gene expression profiles from D30 SST showed enriched molecular functions related to molecule transport, metabolism of terpenoid, and metabolism of membrane lipid derivatives. In D60, the SST gene expression was consistent with enrichment for D-glucose uptake, carbohydrate uptake, and cytoskeleton reorganization. Finally, gene expression of SST isolated at D90 displayed enrichment for mobilization of Ca2+, mononuclear leukocyte response, and T lymphocyte quantity.
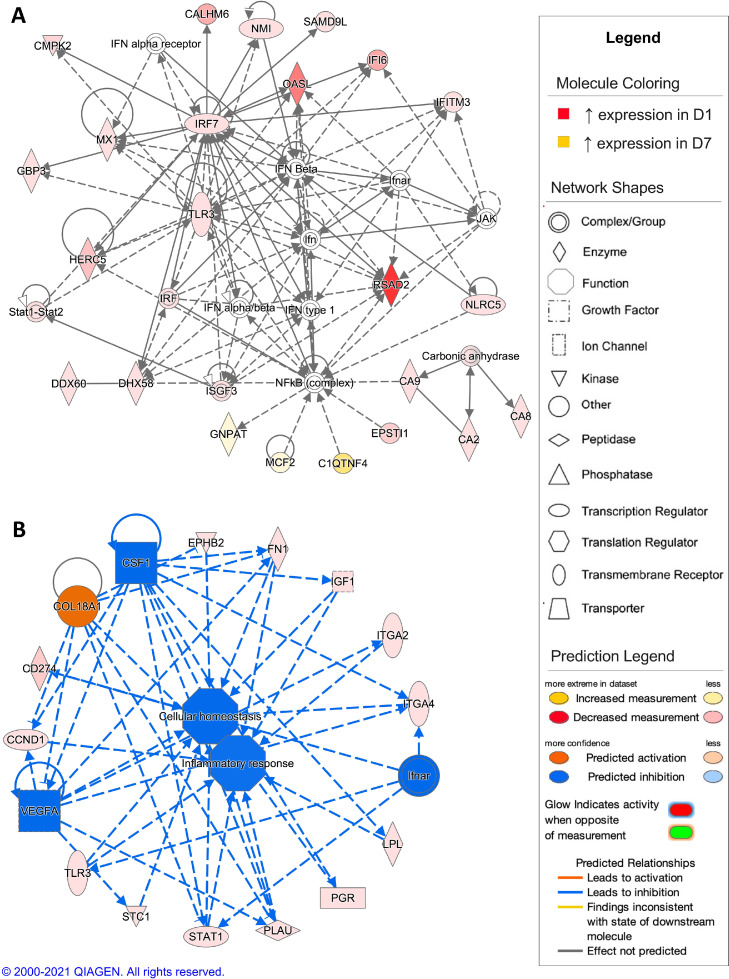
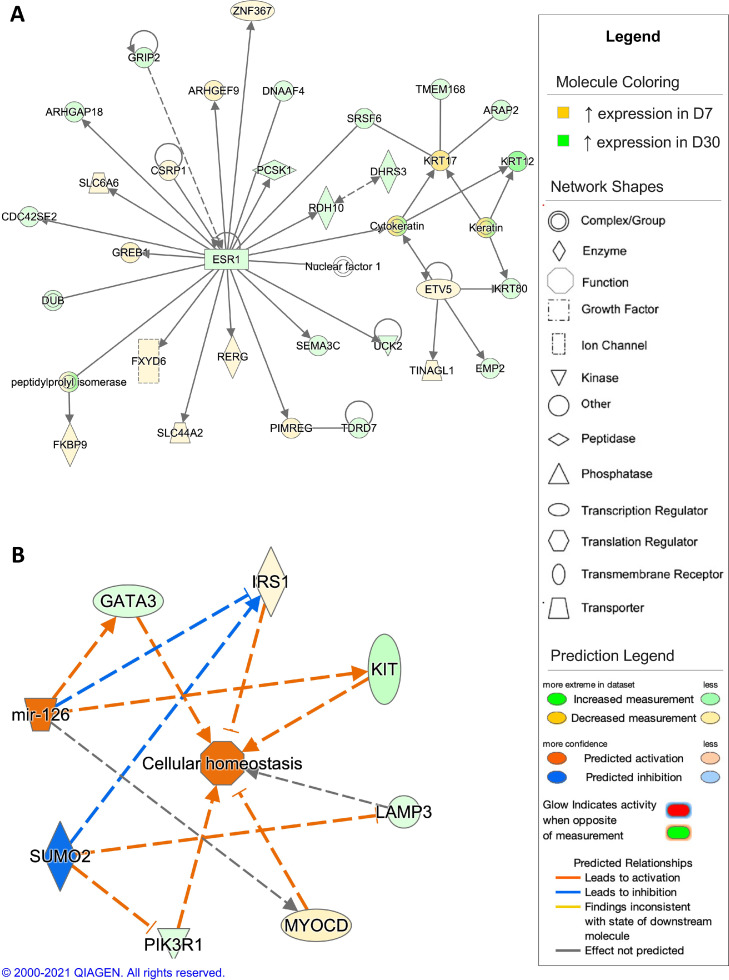
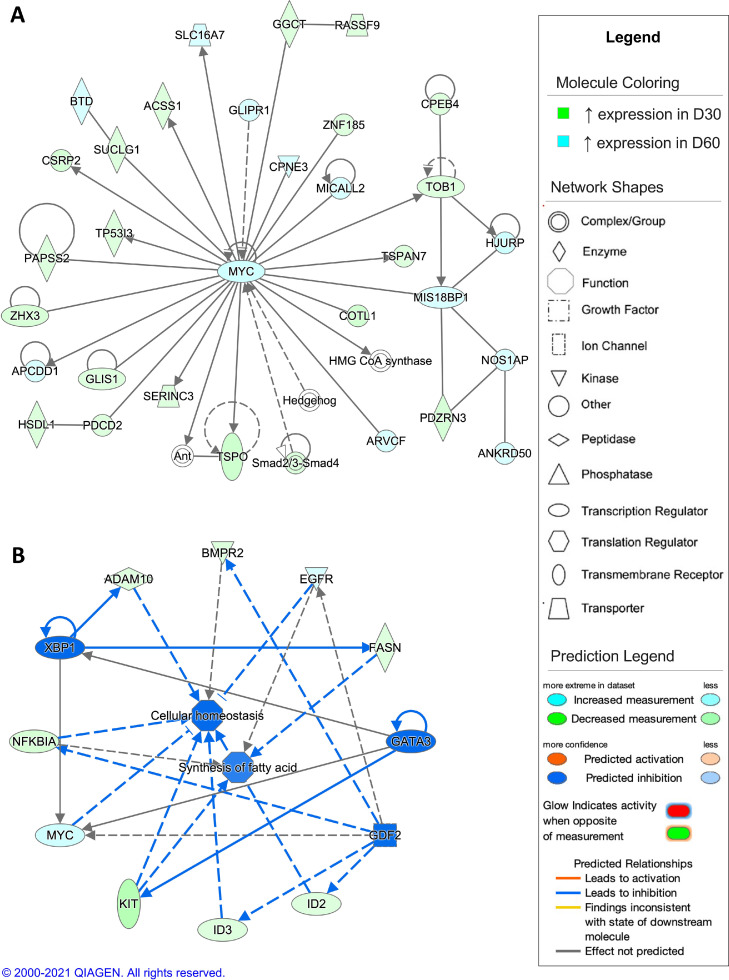
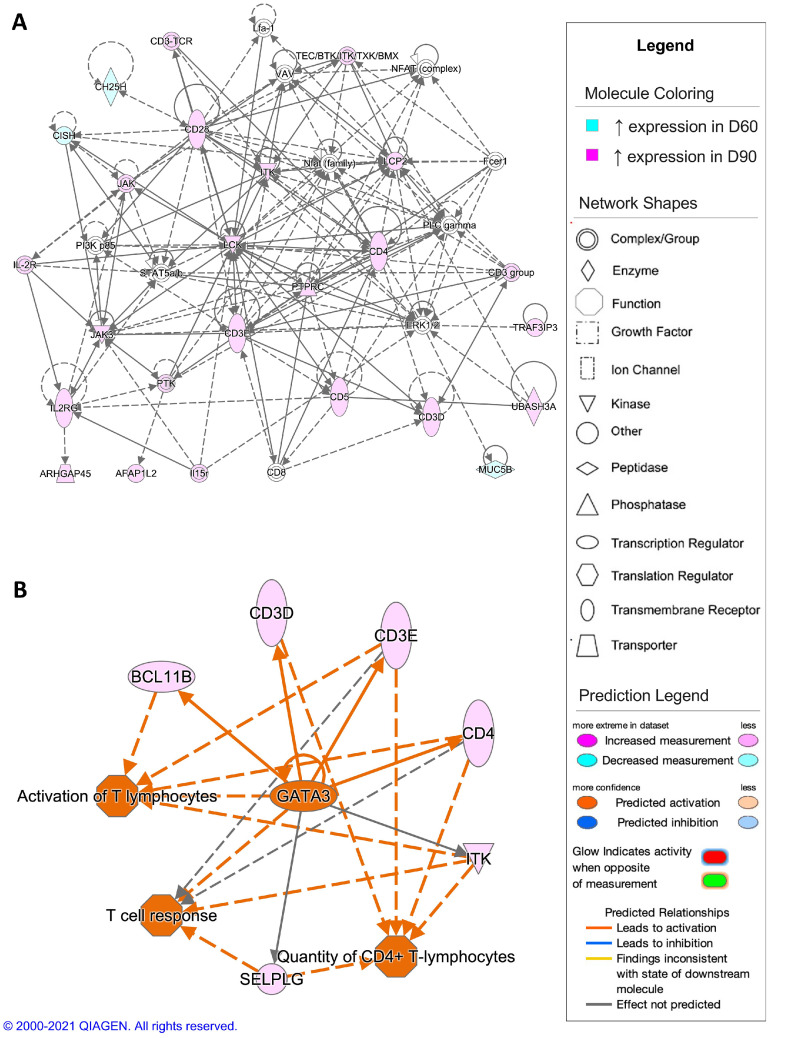
The top scoring network resulting from analysis of DEGs from the comparison of D1 and D7 timepoints showed gene expression associated with innate immune responses (Figure 6A). When comparing the D7 timepoint to the D1 timepoint, the top scoring regulatory network was predicted activation of collagen type XVIII alpha 1 chain (COL18A1) and inhibition of vascular endothelial growth factor A (VEGFA), colony stimulating factor 1 (CSF1), and interferon alpha and beta receptor (IFNAR), leading to an inhibition of cellular homeostasis and inflammatory responses (Figure 6B). For the comparison of the D7 and D30 timepoints, differential gene expression in the top scoring network was associated with regulation of cell morphology (Figure 7A). The regulatory network with highest consistency score for this comparison was predicted activation of microRNA-126 (mir-126) and inhibition of small ubiquitin like modifier 2 (SUMO2), resulting in activation of cellular homeostasis in the D30 timepoint compared to the D7 timepoint (Figure 7B). In the D30 and D60 timepoint comparison, the top scoring network showed differential gene expression associated with carbohydrate metabolism (Figure 8A). In comparison, the top scoring regulatory network predicted inhibition of x-box binding protein 1 (XBP1), GATA3, and growth differentiation factor 2 (GDF2), promoting inhibition of cellular homeostasis and fatty acid synthesis in the D60 timepoint compared to the D30 timepoint (Figure 8B). In the comparison of the D60 and D90 timepoints, differential gene expression in the top scoring network was associated with regulation of T lymphocytes (Figure 9A), with the top scoring regulatory network predicted activation of GATA3, causing activation of T lymphocytes and the T cell response in the D90 timepoint compared to the D60 timepoint (Figure 9B).
Figure 6.
Network analysis of sperm storage tubules (SST) isolated at D1 and D7. The core analysis tool from Ingenuity Pathway Analysis (Qiagen, Valencia, CA) was used to biologically interpret differentially expressed genes (DEGs). Copyright permission from Qiagen has been obtained for use of the images presented. (A) The top network generated through pathway analysis of DEGs between D1 and D7 timepoints (network score = 36, top function = inflammatory response) (FPKM > 1, q-value < 0.05, |fold change| > 1.5). (B) The top regulator effects network generated through pathway analysis of DEGs between D1 and D7 (consistency score = 15.841) (FPKM > 1, q-value < 0.05, |fold change| > 1.5).
Figure 7.
Network analysis of sperm storage tubules (SST) isolated at D7 and D30. The core analysis tool from Ingenuity Pathway Analysis (Qiagen, Valencia, CA) was used to biologically interpret differentially expressed genes (DEGs). Copyright permission from Qiagen has been obtained for use of the images presented. (A) The top network generated through pathway analysis of DEGs between D7 and D30 timepoints (network score = 48, top function = cell morphology) (FPKM > 1, q-value < 0.05, |fold change| > 1.5). (B) The top regulator effects network generated through pathway analysis of DEGs between D7 and D30 (consistency score = 2.041) (FPKM > 1, q-value < 0.05, |fold change| > 1.5).
Figure 8.
Network analysis of sperm storage tubules (SST) isolated at D30 and D60. The core analysis tool from Ingenuity Pathway Analysis (Qiagen, Valencia, CA) was used to biologically interpret differentially expressed genes (DEGs). Copyright permission from Qiagen has been obtained for use of the images presented. (A) The top network generated through pathway analysis of DEGs between D30 and D60 timepoints (network score = 46, top function = carbohydrate metabolism) (FPKM > 1, q-value < 0.05, |fold change| > 1.5). (B) The top regulator effects network generated through pathway analysis of DEGs between D30 and D60 (consistency score = 3.207) (FPKM > 1, q-value < 0.05, |fold change| > 1.5).
Figure 9.
Network analysis of sperm storage tubules (SST) isolated at D60 and D90. The core analysis tool from Ingenuity Pathway Analysis (Qiagen, Valencia, CA) was used to biologically interpret differentially expressed genes (DEGs). Copyright permission from Qiagen has been obtained for use of the images presented. (A) The top network generated through pathway analysis of DEGs between D60 and D90 timepoints (network score = 40, top function = cell-mediated immune response) (FPKM > 1, q-value < 0.05, |fold change| > 1.5). (B) The top regulator effects network generated through pathway analysis of DEGs between D60 and D90 (consistency score = 5.307) (FPKM > 1, q-value < 0.05, |fold change| > 1.5).
DISCUSSION
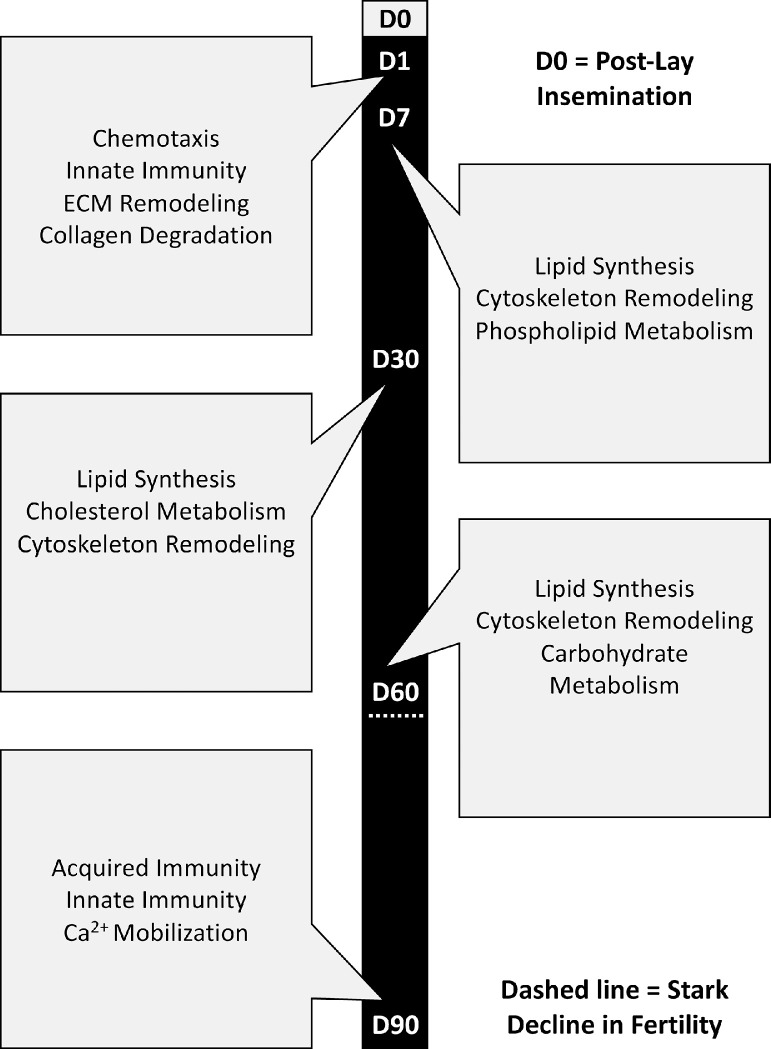
This study was the first to examine SST-specific transcriptome changes of inseminated turkey hens throughout the duration of fertility. The results from this study confirm existing findings and present new potential mechanisms related to the biology of extended sperm survival in SST. The experiment timepoints corresponded with early (D1, D7), peak (D30, D60), and late (D90) egg production. With a single post-lay insemination, turkey hens can produce fertile eggs for at least 10 wk, although the percentage of fertile eggs steadily declines without the constant refilling of SST that normally occurs with the industry standard practice of weekly inseminations. Our study was designed to evaluate the changes in gene expression over time with the natural decline in fertility that occurs when AI occurs only at the onset of egg production. During early egg production (D1 and D7 timepoints) and maximal SST capacity pathways related to chemotaxis, polarity, and innate immune responses showed the most significant upregulation. During peak egg production (D30 and D60 timepoints) with presumably high numbers of sperm residing within the SST, pathways related to carbohydrate and lipid metabolism, steroid hormone signaling, ion balance, and cytoskeletal reorganization exhibited the most significant upregulation. Notably, several pathways related to inhibition of sperm motility were also found to be significantly upregulated during peak egg production. During late egg production (D90 timepoint) when SST likely had more sperm exiting the SST than were being stored, increased progesterone signaling, calcium ion concentrations, and neural contraction were noted to be significantly upregulated. Pathways associated with adaptive immunity, appear to be activated at the end of egg production, perhaps as a mechanism to clear remaining sperm and remodel SST tissue. It is also possible that specific pathways identified in this study are also related to cycling of sperm entrance, maintenance and egress from the SST. For example, changes in polarity and chemokines may direct or orient sperm correctly upon entry, while maintenance of sperm in the SST may supported by presence of critical lipids or carbohydrates. Mobilization of calcium ions or hormone signaling may assist egress of a greater number of sperm in a natural effort to maintain fertility as the hen approaches the end of egg production.
Artificial insemination induced several innate and acquired immune pathways within SST cells, indicating the SST does initiate an immune response to sperm. The immune response may help to aid in the targeting of nonoptimal sperm and serve as a sperm selection mechanism. In particular, the predicted activation of CCR3 signaling in eosinophils and natural killer cell signaling, at both early and late timepoints, respectively, indicate presence of an innate immune response within SST. Previous studies have not identified any innate immunity pathways that potentially serve to regulate sperm entrance into SST. Several genes associated with innate immunity pathways displayed upregulation in D1 compared to D7, including N-myc and STAT interactor (NMI), which is released by activated macrophages, and 2′-5′-oligoadenylate synthetase like (OASL), which is induced by type I interferons (Zhu et al., 1999). Moreover, genes classically associated with interferon signaling, such as interferon regulatory factor 7 (IRF7), toll-like receptor 3 (TLR3), and signal transducer and activator of transcription 1 (STAT1), were also upregulated at D1 compared to D7. In addition, interferon regulatory factor 7 (IRF7) and radical S-adenosyl methionine domain containing 2 (RSAD2) showed increased expression at D1 compared to D7, with RSAD2 responsible for dendritic cell maturation via IRF7 mediated signaling (Jang et al., 2018). Lastly, calcium homeostasis modulator family member 6 (CALHM6) and toll-like receptor 3 (TLR3) exhibited increased expression at D1 compared to D7, with TLR3 previously shown to induce the immune modulation properties of CALHM6 (Malik et al., 2017).
Adaptive immune pathways, specifically those associated with T cell activation and function, exhibited predicted activation in both the D1 and D90 timepoints. The 2 pre-lay inseminations prior to the onset of egg production, typically done in commercial practice, may have induced the antigen-presenting capabilities of T lymphocytes to target post-lay insemination sperm, similar to documented changes in T cells UVJ population following repeated artificial inseminations (Das et al., 2005). This is an interesting concept that could relate to the suboptimal fertility rates often observed in commercial production during the first full week of egg production. In the aforementioned study, increased T lymphocytes were associated with decreased fertility. Genes classically associated with T lymphocyte differentiation and function, such as cluster of differentiation 3D, 3E, 4, 5, and 28 (CD3D, CD3E, CD4, CD5, and CD28) showed increased expression levels at D90 when compared with D60, indicating upregulation of adaptive immune pathways at the D90 timepoint. In addition to classical T lymphocyte genes, signaling by NF-xB, a pathway that controls immune related transcription and leukocyte extravasation signaling, which allows for leukocyte movement, both support innate and adaptive immune responses and were predicted to be activated in early timepoints. Lastly, GATA3, a well-known regulator of both innate and adaptive immune responses, was identified as a predicted upstream regulator in all experimental comparisons and could serve to coordinate the multiple types of innate and adaptive immune responses seen in SST during the duration of fertility (Tindemans et al., 2014).
A unique finding from this study is the enrichment of genes related to chemotaxis as chemotaxis factors have not been previously examined in SST. The enrichment of chemotaxis associated genes could potentially represent a signaling response to a diverse array of immune cells to target suboptimal sperm or signaling to the sperm themselves to direct sperm into and keep sperm within the SST. Platelet-derived growth factor subunit B (PDGFB) is a key gene involved in chemotaxis and was identified as a predicted and activated upstream regulator in the D1 and D7 timepoints. Furthermore, the associated receptor, platelet-derived growth factor receptor beta (PDGFRB) was a common DEG across multiple comparisons, with the highest expression in D1 samples. PDGFRB is part of family receptor tyrosine kinases, which are involved in signal transduction, and has been implicated as the main chemotactic factor driving the migration of the precursors of spermatogonial stem cells in mammals (Basciani et al., 2008). PDGFRB could potentially play a role in aiding sperm to reach the SST and thereafter maintaining sperm within the SST through chemotactic mechanisms.
Maintenance of viable sperm within SST requires sperm immobilization and available metabolic substrates for sperm consumption and long-term survival. Previous studies have noted that sperm motility can be downregulated by increased lactic acid and by the presence of zinc ions (Matsuzaki et al., 2015; Holm et al., 2000). In this study, upregulation of several members of the carbonic anhydrase family, a group of zinc metalloenzymes that function to decrease pH levels, was seen in D1 samples compared to D7 samples. Carbonic anhydrases have been previously localized to SST tissue in chickens and turkeys and shown to impact pH regulation in SST (Holm et al., 1996; Holm and Ridderstråle, 1998). Other genes including associated with pH regulation including ATPase H+ transporting V0 subunit A4 (ATP6V0A4) and solute carrier family 9 member A (SLC9A7), a sodium and potassium/proton antiporter, as well as gap junction protein alpha 8 (GJA8), which is a component of a pH dependent gap junction channel, were differentially expressed in multiple experimental timepoint comparisons. ATP6V0A4 was also found to be upregulated in the UVJ of high fertility laying hens in comparison to low fertility chicken hens, indicating a potential role in SST function (Yang et al., 2021a).
Transforming growth factor (TGFB) signaling is also implicated in reducing sperm motility, with UVJ TGFB expression increasing in inseminated chicken hens (Das et al., 2006a; Yoshimura, 2006). In this study, transforming growth factor beta receptor 2 (TGFB2) was predicted to be an activated upstream regulator in D1 and D7 timepoints, potentially reducing the motility of sperm that have newly entered into the SST. Moreover, growth differentiation factor 2 (GDF2), which encodes a secreted ligand of the TGF-beta superfamily of proteins, appeared as a top upstream regulator in the D30 and D60 comparison, with a predicted inhibition in D60 samples compared to D30 samples. D60 is relatively close to the duration of fertility in turkey hens, and inhibition of TGF-beta genes at that timepoint may terminate the previously established sperm protection pathways and initiate immune related pathways for sperm clearance. Additionally, predicted inhibition of PTEN signaling and RHOGDI signaling, which are negative cell growth and development regulators, are both required for proper sperm motility in the epididymis and oviduct, in mammals, and were seen at D1, D7, and D30, potentially indicating a role for these two pathways in the reduced sperm motility associated with sperm storage in SST (Xu et al., 2014; Ducummon and Berger, 2006).
Slowed sperm metabolism is hypothesized to occur as a result of sperm storage within SST, though sufficient metabolic activity is required for sperm cell homeostasis and survival. In vitro, zinc has been shown to be a metabolic inhibitor in sperm by reducing sperm oxygen uptake without compromising fertilization ability (Bakst, 1985). Research also noted that zinc concentrations in the UVJ were increased compared to other oviductal segments (Bakst and Richards, 1985). We have characterized expression of solute carrier family 39 member 13 (SLC39A13) for the first time in avian reproductive tissue. Moreover, SLC39A13, a ZIP transporter that controls cellular zinc homeostasis, was identified as a common DEG across multiple comparisons, with peak expression at the D30 timepoint. Upregulation of zinc transport at the D30 timepoint may serve to slow the greater metabolic activity of sperm at that timepoint. Sperm cell homeostasis also includes using metabolic inputs for production of membrane components, such as ether phospholipids, for cell maintenance. Interestingly, glyceronephosphate O-acyltransferase (GNPAT), which plays a key role in ether phospholipid formation, exhibited upregulation at D7 compared to D1 (Parks and Lynch, 1992). It is possible that SST may produce important cell maintenance requirements for resident sperm to keep alive during periods of reduced metabolism throughout storage.
Though sperm metabolism is quiescent during in vivo storage, it is hypothesized that sperm metabolize fatty acids derived from SST cells to perform basic cell functions. Microvillus blebs, which contain lipid material, are shed from SST cells and are observed on the apical tips of SST interacting with resident sperm. It is plausible that the microvillus blebs found in the SST represent a lipid delivery system for sperm basic cellular metabolism (Bakst and Bauchan, 2015). MYC proto-oncogene, BHLH transcription factor (MYC), which regulates anabolic and catabolic aspects of lipid metabolism, was central to the differential gene expression seen between the D30 and D60 timepoints, with these timepoints corresponding to predicted activation of lipid metabolism (Edmunds et al., 2014). A common DEG found across multiple timepoint was (LPL) gene encodes the enzyme lipoprotein lipase, which metabolizes fat into triglycerides. In chickens, LPL expression has been documented in UVJ tissue and was found to be downregulated 24 h after AI (Huang et al., 2016). Additionally, phospholipase activity, which hydrolyzes phospholipids into fatty acids, was upregulated D7, D30, and D60 timepoints and could correspond to the generation of lipid material for packing in microvillus blebs. Previous work has identified high phospholipase activity in turkey oviductal fluid (Douard et al., 2004), and in this study, phospholipase C beta 1 (PLCB1) was identified as a predicted activated upstream regulator, with highest expression levels in D60. Additionally, PLCB1 has been previously shown to be upregulated in UVJ isolated from laying hens with longer durations of fertility (Adetula et al., 2018). Phospholipid metabolism appears to be initiated at the D7 timepoint, peaks at the D30 timepoint, and continues to a lesser extent through the D60 timepoint. Another common DEG across multiple timepoints was (NR1D2), which encodes a protein that may be involved with lipid and carbohydrate metabolism. Interestingly, carbohydrate metabolism is also upregulated in the D7, D30, and D60 timepoints and this could indicate a shift in metabolic inputs for fatty acid synthesis or could serve to restock energy reserves depleted during exocytosis of microvillus blebs. While sperm have key metabolic inputs for survival, excessive fatty acid synthesis has been shown to have negative consequences on sperm storage capabilities, indicating the requirement for structured regulation of both microvillus bleb formation and utilization of fatty acids and phospholipase (Wen et al., 2020).
The formation of microvillus blebs requires the reorganization of cytoskeletal elements and remodeling of extracellular matrix components to allow for exocytosis. Cytoskeletal organization was predicted to be activated across timepoints and several key genes involved in cytoskeletal signaling and remodeling, such as actin filament associated protein 1 like 2 (AFAP1L2) and arfGAP with RhoGAP domain, ankyrin repeat and pH domain 2 (ARAP2), were common DEGs among the experimental comparisons, with increased expression during timepoints with upregulated lipid synthesis. Moreover, CDC42 small effector 2 (CDC42SE2) displayed increased expression at D30 compared to D7, and acts downstream of the key regulator of actin dynamics, CDC42 (Momboisse et al., 2009). Increased gene expression associated with cytoskeletal formation and organization was also noted in UVJ tissue isolated from younger layer hens with higher fertility than in older hens with diminished fertility (Yang et al., 2021a). In addition to cytoskeletal related genes, enrichment for genes related to collagen components of the extracellular matrix, extracellular matrix degradation, and integrin signaling, which links extracellular matrix and cytoskeletal elements, were also found in the current dataset. ADAM metallopeptidase with thrombospondin type 1 motif 9 and 18 (ADAMTS9 and ADAMTS18), regulators of extracellular matrix structure, were common across all timepoints, with the highest expression at D1 and D60/D90 timepoints when SST remodeling is occurring. Additionally, collagen degradation enzymes matrix metalloproteinases 1 and 3 (MMP1 and MMP3) exhibited increased at the earlier timepoints, potentially contributing to SST formation. The enrichment of genes related to extracellular matrix remodeling and associated receptor signaling has also been documented in transcriptome analysis of chicken SST (Yang et al., 2020; Yang et al., 2021b). Cytoskeletal elements and the F-actin rich areas of SST, combined with neuronal components including small ganglia and neurons within the tissue surrounding SST, have also been hypothesized to regulate SST contraction for sperm release (Freedman et al., 2001). Several genes associated with neurite outgrowth (neurotrimin, NTM), axonal guidance (galectin-1, GAL1; semaphorin 3C, SEMA3C), and formation of the postsynaptic apparatus (pleckstrin homology like domain family B member 2, PHLDB2) were common DEGs across experimental comparisons, supporting a regulatory role for neuronal functional components in normal SST operation.
Steroid hormones also play a role in oviduct maintenance and sperm release from SST. PGR showed differential expression during early egg production, with increased expression in the D1 timepoint compared to the D7 timepoint, as PGR was identified as a predicted activated upstream regulator in all 4 experimental contrasts. This finding is consistent with previous reports that indicate PGR expression within SST tissue and progesterone regulation play a part in sperm release (Foye-Jackson et al., 2011). In addition to progesterone, estradiol was identified as an activated potential upstream regulator in the D1, D7, D30, and D60 timepoints and estrogen receptor 1 (ESR1) exhibited upregulation in the D30 timepoint. Expression of both PGR and ESR1 was found to increase significantly in the UVJ during sexual maturation and SST differentiation in quail (Khillare et al., 2018). Estradiol has also been previously implicated in oviduct formation and maintenance but the specific regulatory role of estradiol in SST formation and function and warrants further investigation (Das et al., 2006b,Ruh and Spelsberg, 1983). In particular, the estradiol regulation of cytoskeletal maintenance is of interest. Interestingly, androgen receptor (AR) was also identified with predicted activation at D1, D7, D30, and D60, with high testosterone levels in the seminal plasma, signifying that the presence of sperm may impact SST gene expression. Previous studies have shown that testosterone rich ejaculates from roosters influenced embryo quality following artificial insemination, indicating that ejaculate steroid hormone concentrations persist through sperm storage and release (Lelono et al., 2019). Increased activity of steroid hormones coupled with predicted activation of GnRH signaling at D7, D30, and D60, indicates that the hypothalamo-pituitary-gonadal axis may play a larger role in SST function than initially thought, with potential roles in SST formation and maintenance in addition to sperm release.
From this study, potential individual roles for and coordination of 1) innate and adaptive immunity in sperm selection, 2) chemotaxis in sperm homing to SST invaginations, 3) pH regulation, TGF beta signaling, PTEN signaling, and RHOGDI signaling in the downregulation of sperm mobility within SST, 4) zinc regulation in the downregulation of sperm metabolism, 5) apical bled derived lipid metabolic inputs in sperm metabolism, 6) steroid hormones in SST function, and 7) combined cytoskeletal and neural elements in SST contraction were identified (Figure 10). However, future research is needed to more precisely experimentally validate the analysis of transcriptome data, especially in regards to the role of previously mentioned pathways in the sperm entrance, maintenance, and exit of SST tissue. The interplay between the genomically identified molecular pathways and the interactions between sperm and SST during storage are complex, and for this reason, future studies will be required to more completely determine the how regulation of sperm and SST function are related, as well as gauge the significance of these interactions in order to optimize turkey hen insemination and in vitro semen storage protocols.
Figure 10.
Schematic drawing of key sperm storage tubule features by timepoint.
DISCLOSURES
The authors listed do not have any conflicts of interest to report.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2022.101704.
Appendix. Supplementary materials
Supplemental File 4. True fertility data for the sample group timepoints over the 13-week experimental period. Weekly true fertility mean and SEM values for each timepoint group.
Supplemental File 5. Differential expression data for experimental comparisons. Differential expression output from CLC Genomics Workbench for each experimental timepoint comparisons.
REFERENCES
- Adetula A.A., Gu L., Nwafor C.C., Du X., Zhao S., Li S. Transcriptome sequencing reveals key potential long non-coding RNAs related to duration of fertility trait in the uterovaginal junction of egg-laying hens. Sci. Rep. 2018;8:1–12. doi: 10.1038/s41598-018-31301-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Accessed October 2020. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Bakst M.R. Zinc reduces turkey sperm oxygen uptake in vitro. Poult. Sci. 1985;64:564–566. doi: 10.3382/ps.0640564. [DOI] [PubMed] [Google Scholar]
- Bakst M.R. Anatomical basis of sperm-storage in the avian oviduct. Scanning Microsc. 1987;1:1257–1266. [PubMed] [Google Scholar]
- Bakst M.R. Observations on the turkey oviductal sperm-storage tubule using differential interference contrast microscopy. J. Repro. Fert. 1992;95:877–883. doi: 10.1530/jrf.0.0950877. [DOI] [PubMed] [Google Scholar]
- Bakst M.R., Bauchan G. Apical blebs on sperm storage tubule epithelial cell microvilli: their release and interaction with resident sperm in the turkey hen oviduct. Theriogenology. 2015;83:1438–1444. doi: 10.1016/j.theriogenology.2015.01.016. [DOI] [PubMed] [Google Scholar]
- Bakst M.R., Richards M.P. Concentrations of selected cations in turkey serum and oviductal mucosae. Poult. Sci. 1985;64:555–563. doi: 10.3382/ps.0640555. [DOI] [PubMed] [Google Scholar]
- Bakst M.R., Donoghue A.M., Yoho D.E., Moyle J.R., Whipple S.M., Camp M.J., Liu G.Q., Bramwell R.K. Comparisons of sperm storage tubule distribution and number in 4 strains of mature broiler breeders and in turkey hens before and after the onset of photostimulation. Poult. Sci. 2010;89:986–992. doi: 10.3382/ps.2009-00481. [DOI] [PubMed] [Google Scholar]
- Basciani S., De Luca G., Dolci S., Brama M., Arizzi M., Mariani S., Rosano G., Spera G., Gnessi L. Platelet-derived growth factor receptor beta-subtype regulates proliferation and migration of gonocytes. Endocrinology. 2008;149:6226–6235. doi: 10.1210/en.2008-0349. [DOI] [PubMed] [Google Scholar]
- Birkhead T.R., Cunningham E.J.A., Cheng K.M. The insemination window provides a distorted view of sperm competition in birds. Proc. Royal Soc. B P Roy Soc. B-Biol. Sci. 1996;263:1187–1192. [Google Scholar]
- Birkhead T.R., Moller A.P. Numbers and size of sperm storage tubules and the duration of sperm storage in birds: a comparative study. Biological Journal of the Linnean Society. 1992;45(4):363–372. [Google Scholar]
- Brady K., Porter T.E., Liu H.C., Long J.A. Characterization of gene expression in the hypothalamo-pituitary-gonadal axis during the preovulatory surge in the turkey hen. Poult. Sci. 2019;98:7041–7049. doi: 10.3382/ps/pez437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brillard J.P., Bakst M.R. Quantification of spermatozoa in the sperm-storage tubules of turkey hens and the relation to sperm numbers in the perivitelline layer of eggs. Biol. Reprod. 1990;43:271–275. doi: 10.1095/biolreprod43.2.271. [DOI] [PubMed] [Google Scholar]
- Burke W.H., Ogasawara F.X. Presence of spermatozoa in uterovaginal fluids of the hen at various stages of the ovulatory cycle. Poult. Sci. 1969;48:408–413. doi: 10.3382/ps.0480408. [DOI] [PubMed] [Google Scholar]
- Burrows W.H., Quinn J.P. A Method of Obtaining Spermatozoa from the Domestic Fowl. Poultry Science. 1935;251(253) [Google Scholar]
- Das S.C., Isobe N., Nishibori M., Yoshimura Y. Expression of transforming growth factor-β isoforms and their receptors in utero-vaginal junction of hen oviduct in presence or absence of resident sperm with reference to sperm storage. Reproduction. 2006;132:781–790. doi: 10.1530/rep.1.01177. [DOI] [PubMed] [Google Scholar]
- Das S.C., Nagasaka N., Yoshimura Y. Changes in the localization of antigen presenting cells and T cells in the utero-vaginal junction after repeated artificial insemination in laying hens. J. Reprod. Dev. 2005;51:683–687. doi: 10.1262/jrd.17027. [DOI] [PubMed] [Google Scholar]
- Das S.C., Nagasaka N., Yoshimura Y. Changes in the expression of estrogen receptor mRNA in the utero-vaginal junction containing sperm storage tubules in laying hens after repeated artificial insemination. Theriogenology. 2006;65:893–900. doi: 10.1016/j.theriogenology.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Douard V., Gassama-Diagne A., Hermier D., Blesbois E. Activity of phospholipases A and lysophospholipase in turkey semen and oviducal fluid. Poult. Sci. 2004;83:1385–1393. doi: 10.1093/ps/83.8.1385. [DOI] [PubMed] [Google Scholar]
- Douard V., Hermier D., Blesbois E. Changes in turkey semen lipids during liquid in vitro storage. Biol. Reprod. 2000;63:1450–1456. doi: 10.1095/biolreprod63.5.1450. [DOI] [PubMed] [Google Scholar]
- Ducummon C.C., Berger T. Localization of the Rho GTPases and some Rho effector proteins in the sperm of several mammalian species. Zygote. 2006;14:249–257. doi: 10.1017/S0967199406003790. [DOI] [PubMed] [Google Scholar]
- Foye-Jackson O.T., Long J.A., Bakst M.R., Blomberg L.A., Akuffo V.G., Silva M.V., Guthrie H.D., McMurtry J.P. Oviductal expression of avidin, avidin-related protein-2, and progesterone receptor in turkey hens in relation to sperm storage: effects of oviduct tissue type, sperm presence, and turkey line. Poult. Sci. 2011;90:1539–1547. doi: 10.3382/ps.2010-01159. [DOI] [PubMed] [Google Scholar]
- Freedman S.L., Akuffo V.G., Bakst M.R. Evidence for the innervation of sperm storage tubules in the oviduct of the turkey (Meleagris gallopavo) Reproduction. 2001;121:809–814. [PubMed] [Google Scholar]
- Hemmings N., Birkhead T.R., Brillard J.P., Froment P., Briere S. Timing associated with oviductal sperm storage and release after artificial insemination in domestic hens. Theriogenology. 2015;83:1174–1178. doi: 10.1016/j.theriogenology.2014.12.022. e1. [DOI] [PubMed] [Google Scholar]
- Holm L., Ridderstråle Y. Localization of carbonic anhydrase in the sperm-storing regions of the turkey and quail oviduct. Histochem. J. 1998;30:481–488. doi: 10.1023/a:1003247504288. [DOI] [PubMed] [Google Scholar]
- Holm L., Ekwall H., Wishart G.J., Ridderstråle Y. Localization of calcium and zinc in the sperm storage tubules of chicken, quail and turkey using X-ray microanalysis. J. Reprod. Fert. 2000;118:331–336. [PubMed] [Google Scholar]
- Holm L., Ridderstråle Y., Knutsson P.G. Localisation of carbonic anhydrase in the sperm storing regions of the domestic hen oviduct. Acta. Anat. 1996;156:253–260. doi: 10.1159/000147853. [DOI] [PubMed] [Google Scholar]
- Huang A., Isobe N., Obitsu T., Yoshimura Y. Expression of lipases and lipid receptors in sperm storage tubules and possible role of fatty acids in sperm survival in the hen oviduct. Theriogenology. 2016;85:1334–1342. doi: 10.1016/j.theriogenology.2015.12.020. [DOI] [PubMed] [Google Scholar]
- Ito T., Yoshizaki N., Tokumoto T., Ono H., Yoshimura T., Tsukada A., Kansaku N., Sasanami T. Progesterone is a sperm-releasing factor from the sperm-storage tubules in birds. Endocrinology. 2011;152:3952–3962. doi: 10.1210/en.2011-0237. [DOI] [PubMed] [Google Scholar]
- Jang J.S., Lee J.H., Jung N.C., Choi S.Y., Park S.Y., Yoo J.Y., Song J.Y., Seo H.G., Lee H.S., Lim D.S. Rsad2 is necessary for mouse dendritic cell maturation via the IRF7-mediated signaling pathway. Cell Death Dis. 2018;9:823. doi: 10.1038/s41419-018-0889-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khillare G.S., Sastry K., Agrawal R., Saxena R., Mohan J., Singh R.P. Expression of gonadotropin and sex steroid hormone receptor mRNA in the utero-vaginal junction containing sperm storage tubules of oviduct during sexual maturation in Japanese quail. Gen. Comp. Endocr. 2018;259:141–146. doi: 10.1016/j.ygcen.2017.11.015. [DOI] [PubMed] [Google Scholar]
- Krämer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelono A., Riedstra B., Groothuis T. Ejaculate testosterone levels affect maternal investment in red junglefowl (Gallus gallus gallus) Sci. Rep. 2019;9:12126–12135. doi: 10.1038/s41598-019-48563-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds L.R., Sharma L., Kang A., Lu J., Vockley J., Basu S., Uppala R., Goetzman E.S., Beck M.E., Scott D., Prochownik E.V. c-Myc programs fatty acid metabolism and dictates acetyl-CoA abundance and fate. J. Bio. Chem. 2014;289:25382–25392. doi: 10.1074/jbc.M114.580662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. H. and Y. P. Di. 2020. Analysis of RNA sequencing data using CLC genomics workbench. In: Keohavong P., Singh K., Gao W. (eds) Molecular Toxicology Protocols. Methods in Molecular Biology, vol 2102. Humana, New York, NY. [DOI] [PubMed]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Long J.A., Conn T.L. Use of phosphatidylcholine to improve the function of turkey semen stored at 4°C for 24 hours. Poult. Sci. 2012;91:1990–1996. doi: 10.3382/ps.2011-02028. [DOI] [PubMed] [Google Scholar]
- Long J.A., Kulkarni G. An effective method for improving the fertility of glycerol-exposed poultry semen. Poult. Sci. 2004;83:1594–1601. doi: 10.1093/ps/83.9.1594. [DOI] [PubMed] [Google Scholar]
- Malik U., Javed A., Ali A., Asghar K. Structural and functional annotation of human FAM26F: a multifaceted protein having a critical role in the immune system. Gene. 2017;597:66–75. doi: 10.1016/j.gene.2016.10.029. [DOI] [PubMed] [Google Scholar]
- Matsuzaki M., Mizushima S., Hiyama G., Hirohashi N., Shiba K., Inaba K., Suzuki T., Dohra H., Ohnishi T., Sato Y., Kohsaka T., Ichikawa Y., Atsumi Y., Yoshimura T., Sasanami T. Lactic acid is a sperm motility inactivation factor in the sperm storage tubules. Sci. Rep. 2015;1:17643–17655. doi: 10.1038/srep17643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M., Sasanami T. Sperm storage in the female reproductive tract: a conserved reproductive strategy for better fertilization success. Adv. Exp. Med. Biol. 2017;1001:173–186. doi: 10.1007/978-981-10-3975-1_11. [DOI] [PubMed] [Google Scholar]
- McCartney M.G. The physiology of reproduction in turkeys: 2. Degree and duration of fertility and hatchability in broody and non-broody pullets. Poult. Sci. 1951;30:663–667. [Google Scholar]
- McIntyre D.R., Christensen V.L. Filling rates of the uterovaginal sperm storage glands in the turkey. Poult. Sci. 1983;62:1652–1656. doi: 10.3382/ps.0621652. [DOI] [PubMed] [Google Scholar]
- Mendonca T., Cadby A.J., Hemmings N. Sperm gatekeeping: 3D imaging reveals a constricted entrance to zebra finch sperm storage tubules. Biophys. J. 2019;117:2180–2187. doi: 10.1016/j.bpj.2019.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momboisse F., Ory S., Calco V., Malacombe M., Bader M.F., Gasman S. Calcium-regulated exocytosis in neuroendocrine cells: intersectin-1L stimulates actin polymerization and exocytosis by activating Cdc42. Ann. N. Y. Acad. Sci. 2009;1152:209–214. doi: 10.1111/j.1749-6632.2008.03998.x. [DOI] [PubMed] [Google Scholar]
- Parks J.E., Lynch D.V. Lipid composition and thermotropic phase behavior of boar, bull, stallion, and rooster sperm membranes. Cryobiol. 1992;29:255–266. doi: 10.1016/0011-2240(92)90024-v. [DOI] [PubMed] [Google Scholar]
- Peláez J., Long J.A. Characterizing the glycocalyx of poultry spermatozoa: II. In vitro storage of Turkey semen and mobility phenotype affects the carbohydrate component of sperm membrane glycoconjugates. J. Androl. 2008;29:L431–L439. doi: 10.2164/jandrol.107.004259. [DOI] [PubMed] [Google Scholar]
- Peláez J., Bongalhardo D.C., Long J.A. Characterizing the glycocalyx of poultry spermatozoa: III. Semen cryopreservation methods alter the carbohydrate component of rooster sperm membrane glycoconjugates. Poult. Sci. 2011;90:435–443. doi: 10.3382/ps.2010-00998. [DOI] [PubMed] [Google Scholar]
- Ruh T.S., Spelsberg T.C. Acceptor sites for the oestrogen receptor in hen oviduct chromatin. Biochem. J. 1983;210:905–912. doi: 10.1042/bj2100905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindemans I., Serafini N., Di Santo J.P., Hendriks R.W. GATA-3 function in innate and adaptive immunity. Immunity. 2014;41:191–206. doi: 10.1016/j.immuni.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Van Krey H.P., Ogasawara F.X., Pangborn J. Light and electron microscopic studies of possible sperm gland emptying mechanisms. Poult. Sci. 1967;46:69–78. doi: 10.3382/ps.0460069. [DOI] [PubMed] [Google Scholar]
- Wen C., Mai C., Wang B., Li J., Sun C., Yang N. Detrimental effects of excessive fatty acid secretion on female sperm storage in chickens. J. Anim. Sci. Biotechnol. 2020;11:26–36. doi: 10.1186/s40104-020-0432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Washington A.M., Hinton B.T. PTEN signaling through RAF1 proto-oncogene serine/threonine kinase (RAF1)/ERK in the epididymis is essential for male fertility. Proc. Natl. Acad. Sci. U. S. A. 2014;111:18643–18648. doi: 10.1073/pnas.1413186112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Li S., Zhao Q., Chu J., Zhou B., Fan S., Shi F., Wei X., Hu X., Zheng X., Liu Z., Zhou X., Tao Y., Li S., Mou C. Transcriptomic and metabolomic insights into the variety of sperm storage in oviduct of egg layers. Poult. Sci. 2021;100:101087–101097. doi: 10.1016/j.psj.2021.101087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Li S., Mo C., Zhou B., Fan S., Shi F., Wei X., Zhao Q., Yang G., Li S., Mou C. Transcriptome analysis and identification of age-associated fertility decreased genes in hen uterovaginal junction. Poult. Sci. 2021;100:100892–100902. doi: 10.1016/j.psj.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Zheng X., Mo C., Li S., Liu Z., Yang G., Zhao Q., Li S., Mou C. Transcriptome analysis and identification of genes associated with chicken sperm storage duration. Poultry science. 2020;99:1199–1208. doi: 10.1016/j.psj.2019.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura Y. Expression of transforming growth factor-beta isoforms and their receptors in utero-vaginal junction of hen oviduct in presence or absence of resident sperm with reference to sperm storage. Reproduction (Cambridge, England) 2006;132:781–790. doi: 10.1530/rep.1.01177. [DOI] [PubMed] [Google Scholar]
- Zhu M., John S., Berg M., Leonard W.J. Functional association of Nmi with Stat5 and Stat1 in IL-2- and IFN gamma-mediated signaling. Cell. 1999;96:121–130. doi: 10.1016/s0092-8674(00)80965-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental File 4. True fertility data for the sample group timepoints over the 13-week experimental period. Weekly true fertility mean and SEM values for each timepoint group.
Supplemental File 5. Differential expression data for experimental comparisons. Differential expression output from CLC Genomics Workbench for each experimental timepoint comparisons.