Abstract
Treatment of staphylococcal infections is difficult due to multidrug resistance with their persister forms posing an added threat of recalcitrant infections. Antibiotic combinations are widely studied as an alternative strategy to combat them; therefore, they merit further investigation into their effect on the number of persister cells. In the present study, the fractional inhibitory concentrations of antibiotic combinations ciprofloxacin-daptomycin, ciprofloxacin-vancomycin, daptomycin-tobramycin, and tobramycin-vancomycin (checkerboard assay) were determined against two previously studied clinical (S48 and J6) and one standard (NCIM 5021) isolate of Staphylococcus aureus. They showed synergistic effects with a 2 to 256-fold reduction in MICs. All combinations also resulted in inhibition and disruption of biofilms in a concentration-dependent manner. All antibiotic combinations, except ciprofloxacin-daptomycin, showed total biofilm inhibition at 100X MICs. Similarly, antibiotic combination at 100X MIC showed 77–97% disruption of preformed biofilms. Time-kill assays performed at a 100X MIC combination against stationary-phase cells showed a two to six log10 reduction in CFU followed by a plateau indicating the presence of persisters. Significant differences were observed in persister cell fraction remaining after treatment with antibiotic combinations compared to monotherapies (p < 0.05) and therefore merit further investigation in clinical use for treatment against persisters.
Keywords: Drug combination, Drug tolerance, Biofilm, Staphylococcus aureus
1. Introduction
The advent of multidrug-resistant bacteria has resulted from indiscriminate use of antibiotics and exposure to infections in hospitals ever since the discovery of penicillin [1,2]. The overall number of antibiotics effective against resistant pathogens is rapidly declining [2]. Newer antibiotics have been introduced into clinical practice at a prolonged rate [2,3]. While the search for novel antibiotics and strategies to combat resistant bacteria is ongoing, persister cells, known to cause recalcitrant infections [4], pose a new challenge in infection treatment [5]. Since persister cells are metabolically dormant, it is difficult to corrupt their function and kill them [6]. However, strategies to do so are underway [7]. One strategy is a cyclic application of antibiotics, in that each treatment will kill normal cells leaving persisters, and the removal will cause them to revert to their normal state [8]. Wood has described three primary anti-persister therapies that include the direct killing of persisters, resuscitation of persisters so as to sensitize them for conventional antibiotic treatment, and interfering with or reducing the formation of persister cells [9]. This last approach entails combining conventional antibiotics with different mechanisms of action. Combination therapy has already been beneficial in severe infections caused by multiple pathogens [10] as the combination of antibiotics exerts a synergistic effect more significantly than individual antibiotics [2,10]. However, its use in eliminating persister cells has not much been explored. While other highly advanced strategies are being developed to deal with persistent infections, a combination of drugs already in use provides a quick and easy way to implement a much effective regimen [11], an approach that warrants investigation as an anti-persister strategy.
We have previously shown that biofilms of clinical isolates of Staphylococcus aureus show a higher tolerance to antibiotics targeting various cellular processes than stationary-phase planktonic cells, possibly due to an increased number of persister cells [12]. In this study, we explore the effect of a combination of antibiotics that individually target different processes in the cell and have not yet been reported in combination against persister cells, on inhibition and disruption of biofilms and the number of persister cells of these clinical isolates of S. aureus.
2. Methodology
2.1. Isolates used for the study
Clinical isolates of S. aureus, S48, and J6, studied previously [12], were explored further. The standard isolate NCIM 5021 (ATCC 25923) was also tested with the clinical isolates.
2.2. Antibiotics used for the study
Ciprofloxacin (Cipla Ltd., India), daptomycin (Glenmark Pharmaceuticals Ltd., India), tobramycin sulfate (Sun Pharmaceutical Industries Ltd, India), and vancomycin hydrochloride (Chandra Bhagat Pharma Pvt. Ltd.) were used for all experiments.
2.3. Determination of fractional inhibitory concentration (FIC) and FIC Index (FICI)
The FIC and FICI of the four antibiotic combinations, namely ciprofloxacin-daptomycin, ciprofloxacin-vancomycin, daptomycin-tobramycin, and tobramycin-vancomycin, were determined using the checkerboard assay [13]. Briefly, 16–18 h old cultures (grown in Luria Bertani broth at 37 °C) were used to inoculate fresh Mueller Hinton broth to obtain a ∼105 CFU/mL density. A 100 μL of such pre-inoculated medium was distributed in flat-bottom 96-well microtitre plates (NEST®, USA). Antibiotic A and antibiotic B were added in increasing concentration (Table 1) in columns and rows, respectively. The plates were incubated at 37 °C for 24 h under static conditions, after which wells were observed for growth and O. D. recorded at 630 nm. For the first clear well in each row of the microtiter plate containing all antimicrobial agents, the fractional inhibitory concentration (FIC) was calculated according to the formula given below.
where MIC is the minimum inhibitory concentration.
Table 1.
Range of antibiotics tested in combination for FICI determination.
| Isolate | Antibiotic concentration (μg/mL) |
|||||||
|---|---|---|---|---|---|---|---|---|
| C + D |
C + V |
D + T |
T + V |
|||||
| C | D | C | V | D | T | T | V | |
| S48 | 0.007–4 | 0.063–8 | 0.007–4 | 0.063–8 | 0.015–8 | 0.007–1 | 0.002–1 | 0.063–8 |
| J6 | 0.039–1 | 0.031–2 | 0.039–1 | 0.015–1 | 0.008–2 | 0.008–0.5 | 0.002–0.5 | 0.039–1 |
| NCIM 5021 | 0.008–2 | 0.031–2 | 0.008–2 | 0.031–2 | 0.008–2 | 0.001–0.062 | 0.001–0.062 | 0.031–2 |
*C + D-ciprofloxacin + daptomycin; C + V- ciprofloxacin + vancomycin; D + T-daptomycin + tobramycin; T + V- tobramycin + vancomycin.
2.4. Inhibition assay of biofilm
Biofilm inhibition was carried out according to the protocol set by Kamble and Pardesi [12]. Briefly, 16–18 h old cultures (grown in Luria Bertani broth at 37 °C) were used to inoculate fresh Mueller Hinton broth to obtain a ∼105 CFU/mL density. A 100 μL of such pre-inoculated medium was distributed in flat-bottom 96-well microtitre plates (NEST®, USA). Antibiotic combinations C + D, C + V, D + T, and T + V, were added to the wells at 1X, 10X, and 100X of their individual MIC. No antibiotic was added to the control wells. The plates were incubated at 37 °C for 24 h under static conditions. Planktonic cells were removed, and the wells were gently washed with phosphate-buffered saline (PBS). Biofilms were stained with 100 μL of 0.4% (w/v) crystal violet and incubated for 15 min at room temperature. The wells were then washed with PBS to remove excess stain and air-dried. Bound crystal violet was released by adding 100 μL of 33% (v/v) acetic acid, and absorbance was read at 630 nm. The experiment was conducted three times in triplicates.
2.5. Disruption assay of preformed biofilm
Biofilm disruption was carried out as set by Kamble and Pardesi [12]. 18 h old culture was added to wells of a flat-bottom 96-well microtitre plate (NEST®, USA) containing 100 μL LB broth to a final 105 CFU/mL density. The plates were incubated at 37 °C for 24 h to allow biofilm formation. Then, media containing planktonic cells was removed from the wells, and biofilms were washed with PBS. Antibiotic combinations were added to the wells at 1X, 10X, and 100X MIC in combination. Untreated biofilms were used as a control. The plates were incubated further at 37 °C for 24 h. After treatment, the antibiotic was removed, and the wells were washed with PBS. As mentioned above, crystal violet staining was performed and absorbance was measured at 630 nm. The experiment was conducted three times in triplicates.
2.6. Calculation and statistical analysis
Percent biofilm inhibition and disruption was estimated by using the following formula:
where A is the absorbance at 630 nm. One-way ANOVA followed by Tukey Post Hoc test was carried out to evaluate significant differences between A630nm values obtained on treatment with various antibiotic concentrations and treatment with various antibiotics between the two isolates. Differences were considered statistically significant at p < 0.05.
2.7. Time-kill assay of stationary-phase cells using antibiotic combinations
The antibiotic combinations were tested for their ability to clear stationary-phase cells of S. aureus S48 and J6 in a time-dependent manner. LB broth tubes were inoculated with a single colony of the test isolate and incubated at 37 °C for 18 h till it reached the stationary phase of growth (109 CFU/ml). To each of these tubes, antibiotics were added individually as well as in combination at a 100X MIC. Untreated cultures were used as a control. Hundred microliter samples were drawn at 0 h, 3 h, 6 h, 9 h, and then at 24 h, 48h and 72 h. The samples were washed in saline to remove the antibiotics and serially diluted. Ten microliters of each dilution were spot inoculated on LB agar plates and incubated at 37 °C for 24 h. Colonies were counted to enumerate survivors. The experiment was conducted in triplicates two times. The CFU/mL was plotted against time to obtain the time-kill curve.
3. Results
3.1. Antibiotic combinations show synergistic activity against S. aureus
Antibacterial activity of the four combinations of antibiotics, namely, (C + D), (C + V), (D + T), and (T + V), was tested against the two clinical isolates, S. aureus S48 and J6, as well as the standard strain NCIM 5021 using the checkerboard assay. Of the four combinations tested, few showed synergistic activity resulting in lowered MICs, albeit in varying degrees (Table 2).
Table 2.
MIC, FICI, and effect of antibiotics in combination.
| Isolate | MIC alone (μg/mL) |
Effect of antibiotics used in combination |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | D | T | V | C + D |
C + V |
D + T |
T + V |
|||||||||
| MIC (μg/mL) |
FICI | MIC (μg/mL) |
FICI | MIC (10) |
FICI | MIC (μg/mL) |
FICI | |||||||||
| C | D | C | V | D | T | T | V | |||||||||
| S48 | 2 | 4 | 0.5 | 4 | 0.125 | 0.25 | 0.062 | 0.125 | 0.5 | 0.19 | 0.016 | 0.125 | 0.25 | 0.125 | 1 | 0.5 |
| J6 | 1 | 2 | 0.5 | 1 | >1 | >2 | NDa | >1 | >1 | ND | 0.125 | 0.062 | 0.19 | >0.5 | >1 | ND |
| NCIM 5021 |
2 | 2 | <0.062 | 2 | 0.062 | 0.5 | 0.28 | 0.125 | 0.5 | 0.31 | 0.016 | 0.031 | 0.26 | >0.062 | >2 | ND |
ND-Not determined. Result is interpreted as- FICI ≤0.5 = synergy; FICI >0.5 to 1 = partial synergy, FICI 1 to 2 = additive, FICI 2 to 4 = indifferent and FICI >4 = antagonism. C + D-ciprofloxacin + daptomycin; C + V- ciprofloxacin + vancomycin; D + T-daptomycin + tobramycin; T + V- tobramycin + vancomycin.
Antibiotics in all four combinations showed synergy in the case of S48. The MICs in combination reduced 2 to 256-fold compared to the individual antibiotic used alone. In the case of NCIM 5021, three of the four combinations, namely, C + D, C + V, and D + T, showed synergistic activity as their MIC in combination reduced 4 to 32-fold compared to the individual antibiotic used alone. The only combination that brought about a synergistic effect on J6 was daptomycin and tobramycin, lowering the MIC of each antibiotic to 8-fold and 2-fold, respectively. A combination of ciprofloxacin and daptomycin, ciprofloxacin and vancomycin, and tobramycin and vancomycin had an indifferent effect on J6. Combinations showing synergy were used for further experiments.
3.2. Antibiotic combinations show inhibition and disruption of biofilm at 100X MIC
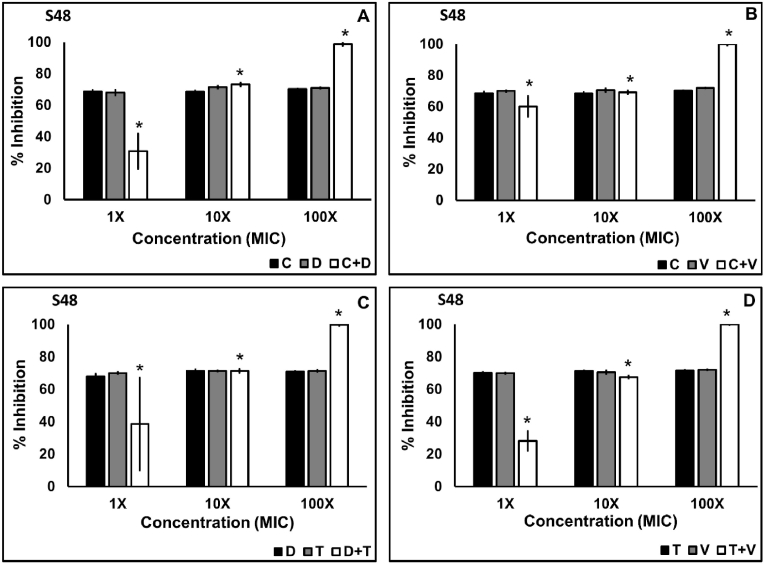
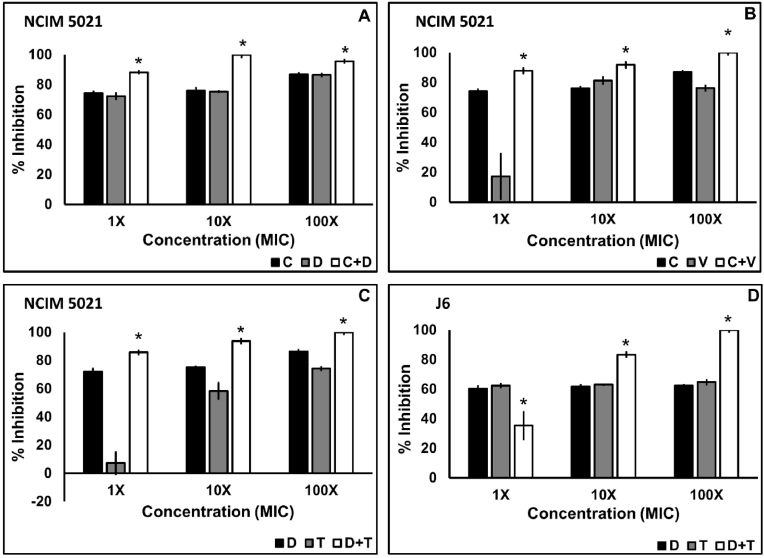
Inhibition and disruption of biofilm of the clinical isolates S48 and J6 and the standard culture NCIM 5021 was examined using the four combinations C + D, C + V, D + T, and T + V. The four combinations of antibiotics tested against S48 showed significant inhibition (p < 0.05) of biofilm at concentrations 10X and 100X MIC in combination according to the Tukey post hoc analysis (Fig. 1). However, inhibition in S48 biofilm brought about by the combination of antibiotics at 1X MIC was significantly lower than antibiotics applied alone. In the case of NCIM and J6, the combination of the antibiotics had a significantly higher (p < 0.05) effect than antibiotics alone (Fig. 2). At 1X MIC in combination, C + V showed maximum inhibition (60.11%) of S48 biofilm. Combinations C + D (88.17%), C + V (87.3%), and D + T (85.82%) at 1X MIC were more effective in inhibiting biofilm by NCIM 5021. Nearly 100% inhibition in S48 was observed at 100X MIC in combination using all four combinations, while the same was achieved at 10X MIC against NCIM 5021. All combinations showed significant biofilm inhibition (p < 0.05) in a concentration-dependent manner in the case of S48 and J6 but not NCIM 5021.
Fig. 1.
Biofilm inhibition of S. aureus S48 using antibiotic combinations ciprofloxacin plus daptomycin (A), ciprofloxacin plus vancomycin (B), daptomycin plus tobramycin (c), and tobramycin plus vancomycin (D) applied at 1X, 10X, and 100X MIC in combination. The data shown are the mean values of percent inhibition (n = 3); the error bars indicate standard deviation. (C-ciprofloxacin, D-daptomycin, T-tobramycin, V-vancomycin).
Fig. 2.
Biofilm inhibition of S. aureus NCIM 5021 and S. aureus J6 using antibiotic combinations. Ciprofloxacin plus daptomycin against NCIM (A), ciprofloxacin plus vancomycin against NCIM (B), daptomycin plus tobramycin against NCIM (C), and daptomycin plus tobramycin against J6 (D) applied at 1X, 10X, and 100X MIC in combination. The data shown are the mean values of percent inhibition (n = 3); the error bars indicate standard deviation. (C-ciprofloxacin, D-daptomycin, T-tobramycin, V-vancomycin).
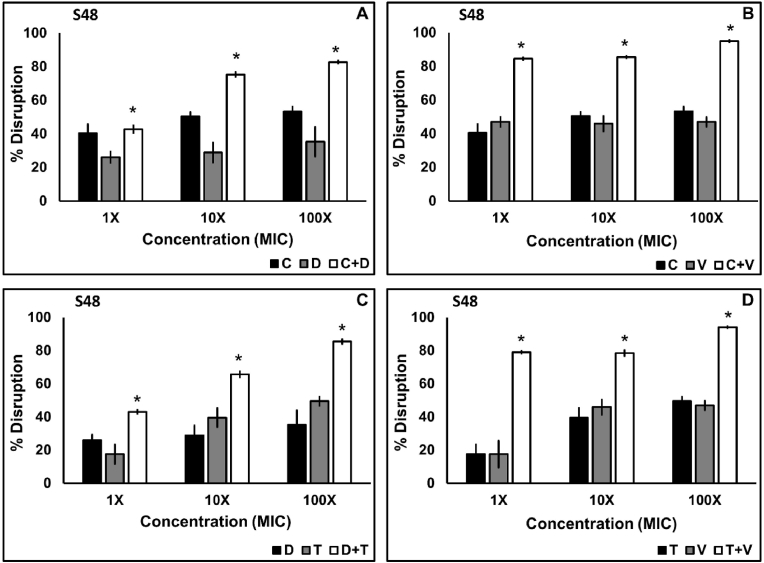
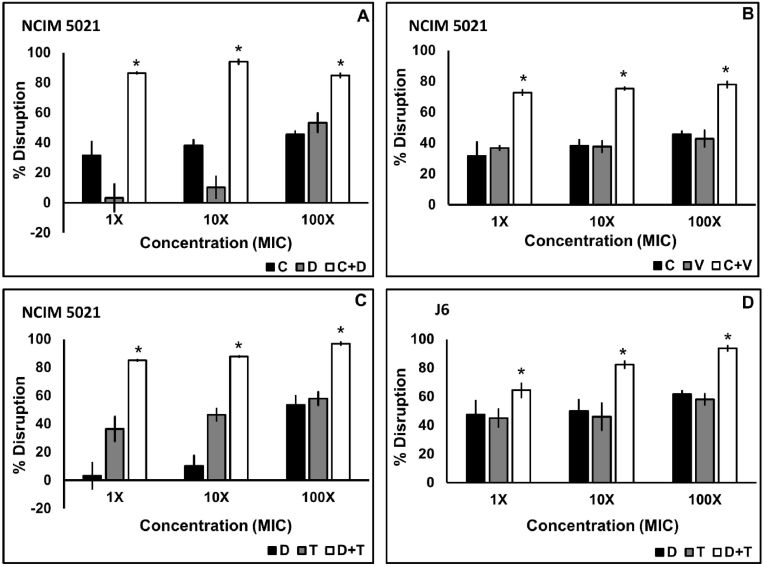
Biofilm disruption of the clinical and standard isolates using the four antibiotic combinations was concentration-dependent which ranged from 42.66 to 84.55% disruption at a 1X MIC, 65.67–85.5% disruption at a 10X MIC, and 82.73–94.92% at a 100X MIC in combination case of S48 (Fig. 3), 64.44%–93.67% in case of J6 (Fig. 4D) and 72.73%–86.33% at 1X MIC, 75.3%–93.93% at 10X MIC and 77.96%–96.88% at a 100X MIC in combination in case of NCIM 5021 (Fig. 4A–C). Tukey post hoc analysis revealed that each combination disrupted biofilm more significantly (p < 0.05) than their constituent antibiotics applied alone. 1X MIC in combination showed significantly high (p < 0.05) biofilm disruption of all three isolates. C + V and T + V showed the highest disruption at a 100X MIC in the case of S48. Even though there was considerable disruption using combination of antibiotics, no combination showed 100% inhibition.
Fig. 3.
Biofilm disruption of S. aureus S48 using antibiotic combinations. Twenty-four-hour old biofilms treated with ciprofloxacin plus daptomycin (A), ciprofloxacin plus vancomycin (B), daptomycin plus tobramycin (C), and tobramycin plus vancomycin (D) were applied at 1X, 10X, and 100X MIC in combination for 24 h. The data shown are the mean values of percent disruption (n = 3); the error bars indicate standard deviation. (C-ciprofloxacin, D-daptomycin, T-tobramycin, V-vancomycin).
Fig. 4.
Biofilm disruption of S. aureus NCIM 5021 and S. aureus J6 using antibiotic combinations. Twenty-four-hour-old biofilms were treated with a combination of antibiotics for 24 h. Ciprofloxacin plus daptomycin against NCIM 5021 (A), ciprofloxacin plus vancomycin against NCIM 5021 (B), daptomycin plus tobramycin against NCIM 5021 (C), and daptomycin plus tobramycin against J6 (D) applied at 1X, 10X, and 100X MIC in combination. The data shown are the mean values of percent disruption (n = 3); the error bars indicate standard deviation. (C-ciprofloxacin, D-daptomycin, T-tobramycin, V-vancomycin).
3.3. Treatment with antibiotic combinations results in a lesser number of persister cells
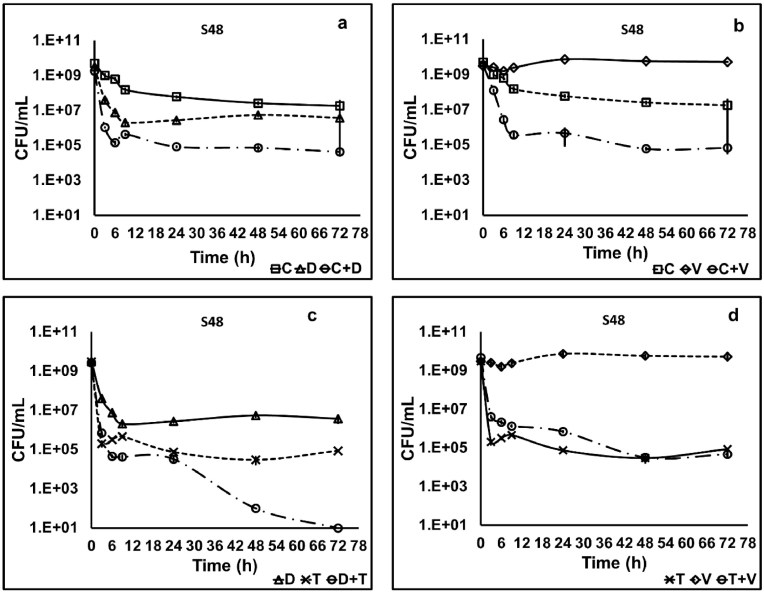
Antibiotic combinations (at a 100X MIC in combination) were tested for their ability to eradicate stationary-phase cells of the clinical isolates of S. aureus S48 and J6, and the standard isolate NCIM 5021 over 72 h. The time-kill curves obtained on treatment with combinations were compared with those obtained with individual antibiotics. Treatment of S48 with combinations C + D and C + V showed a steady decline in stationary-phase culture over 24 h (Fig. 5 a and b), resulting in a five log10 reduction leaving 104 CFU/mL. Comparison of surviving population after 24 h treatment with C + D and ciprofloxacin showed a lowering in the number of persisters by five log10, but that with daptomycin showed only one log10 reduction. The combination C + V showed a lower number of survivors than each ciprofloxacin and vancomycin used singly. Treatment with combinations D + T and T + V for 24 h showed a pattern similar to their constituent antibiotics alone (Fig. 5 c and d). The bulk of the population was killed rapidly within 3 h, resulting in a five log10 reduction in the CFU.
Fig. 5.
Time-kill curve of stationary-phase cells of S. aureus S48 showing the presence of a persister population against a 100X MIC of ciprofloxacin plus daptomycin (C + D) (a), ciprofloxacin plus vancomycin (C + V) (b), daptomycin plus tobramycin (D + T) (c) and tobramycin plus vancomycin (T + V) (d). The combination C + V showed a marked decline in persister cells compared to individual antibiotics. The data shown are the mean values of CFU/mL (n = 3); the error bars indicate standard deviation. (C-ciprofloxacin, D-daptomycin, T-tobramycin, V-vancomycin).
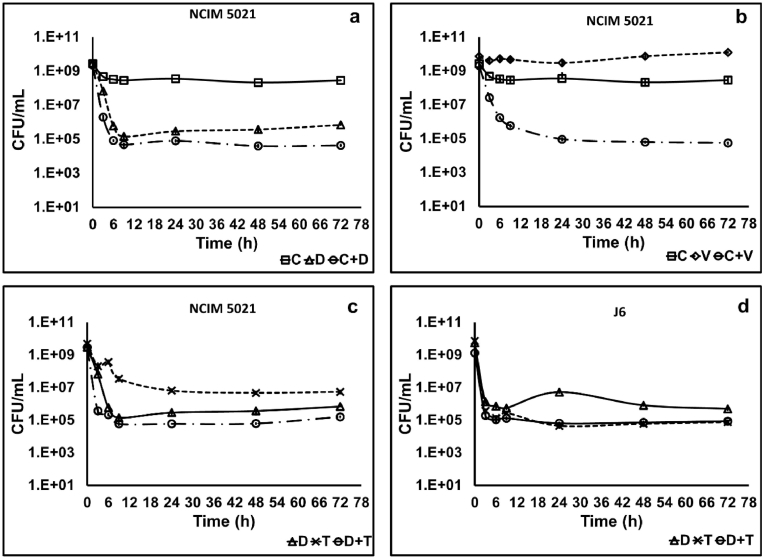
Treatment of NCIM 5021 with the two combinations C + D and D + T resulted in a similar killing pattern in stationary-phase culture over 24 h (four log10 reduction) as that of daptomycin alone (Fig. 6 a and d). Combination C + V showed a considerable decrease of five log10 in the number of survivors compared to ciprofloxacin and vancomycin alone (Fig. 6 b). Combination D + T did not affect the pattern of isolate J6 compared to daptomycin and tobramycin revealing persister cells (Fig. 6 d).
Fig. 6.
Time-kill curve of stationary-phase cells of S. aureus NCIM 5021and J6 using a 100X MIC of ciprofloxacin plus daptomycin against NCIM 5021 (a), ciprofloxacin plus vancomycin against NCIM 5021 (b), daptomycin plus tobramycin against NCIM 5021 (c), and daptomycin plus tobramycin against J6 (d). The data shown are the mean values of CFU/mL (n = 3); the error bars indicate standard deviation. (C-ciprofloxacin, D-daptomycin, T-tobramycin, V-vancomycin).
However, prolonged treatment with any of the combinations till 72 h did not reduce the number of survivors further, except in the case of D + T against S48 (Fig. 5 c).
4. Discussion
Staphylococcal persister cells pose a problem in treating recurrent infections due to their tolerance to antibiotics. It has also been shown that the otherwise sensitive planktonic cells show enhanced resistance to antibiotics in their biofilm form [14]. Alternative therapy appears to be the best approach against antibiotic resistance and tolerance. A combination of two or more antibiotics with each other and that of antibiotics with other agents such as antimicrobial peptides, phages, and nanoparticles are promising candidates for potential therapeutic application against persister cells [2,15]. In this regard, combination therapy as an alternative treatment strategy was examined by testing the ability to inhibit and disrupt biofilms and kill stationary-phase cells over 24 h.
In our previous study, antibiotics ciprofloxacin, daptomycin, tobramycin, and vancomycin showed the presence of persisters in stationary-phase cells of S. aureus isolates S48, J6, and NCIM 5021 [12]. To screen the antibiotic combinations that could reduce the number of persisters generated by these isolates, they were tested for synergy, indifference, or antagonism. Each isolate showed a different response to the combination of the antibiotics. These combinations were chosen such that one antibiotic targets the cell structure i. e., daptomycin and vancomycin, and the other targets an essential process like DNA replication or protein synthesis i. e. ciprofloxacin or tobramycin, respectively. The presumed synergism in these combinations is attributed to the fact that the antibiotic acting on the cell wall allows enhanced penetration of the other antibiotic, thereby killing the cell with better efficiency [16]. Of these, ciprofloxacin plus vancomycin and daptomycin plus tobramycin are reported to have synergistic activity, a promising strategy in clearing S. aureus infections [17,18].
The antibiotic combinations were also tested for their ability to inhibit biofilm formation and disrupt preformed biofilm by the clinical isolates at 1X, 10X, and 100X MIC in combination. These combinations have not previously been studied for said abilities to the best of our knowledge. Local antibiotic concentration in biofilms is affected due to several factors, which in turn decides the fate of the pathogen protected within. Monotherapies using antibiotics or other antimicrobials have shown limited biofilm inhibition and disruption activities compared to their combinations [12,19]. This phenomenon warrants antibiotic combination treatment [20,21]. Comparison with treatment using individual antibiotics showed significantly higher disruption (by 10–50%) at 10X and 100X MIC. Antibiotics like daptomycin and vancomycin also have shown promising results in disruption of biofilm of S. aureus in combination with other antibiotics like ceftaroline [22,23], rifampicin [24], and fosfomycin [25]. Biofilm disruption is attributed to the interaction of the antibiotics with the biofilm matrix and, therefore, their diffusion through it [26]. However, it is difficult to discern how a combination of antibiotics enhances biofilm disruption.
The combinations were tested for their anti-persister ability at a 100X MIC in combination against stationary-phase planktonic cultures of S. aureus S48, J6, and NCIM 5021. 100% killing of the isolates was not seen after treatment with any of the combinations tested. However, ciprofloxacin plus vancomycin showed the most pronounced reduction in the number of persisters. There are not many studies investigating the potential of ciprofloxacin with vancomycin in killing S. aureus. However, the remaining combinations showed no significant change in the number of persisters in the stationary-phase planktonic cells compared to the individual antibiotics. It is noteworthy that despite exhibiting a similar killing pattern as that shown by antibiotics alone, concentrations applied in combination to determine the number of persisters was much lower (100X of concentrations mentioned in Table 2). Therefore, treatment with higher concentrations in combination could show complete killing. Previous studies using a combination of antibiotics like daptomycin plus tobramycin [16] and daptomycin plus gentamicin [18] have demonstrated absolute killing of stationary-phase planktonic cells of S. aureus; however, the concentration of antibiotics applied in these studies was far greater as compared to our experiments and are not clinically achievable.
Peak serum levels define the concentration of drugs available in a particular compartment or test area of the body after administration of a dose available to exert its action [27]. In order to evaluate their potential applicability in the clinic, the peak serum concentration of each antibiotic was compared with its concentration applied in combination with other antibiotics to a stationary-phase planktonic culture of S. aureus. MIC of the four antibiotics used in combination ranged from 0.031 to 1 μg/mL. Therefore, the concentration of the antibiotics at 100X applied in combinations to determine the number of persisters ranged from 3.1 to 100 μg/mL. The peak serum concentration of ciprofloxacin 2 h after an intravenous dose is found to range between 0.5 and 7.27 μg/mL [28]. The concentration of ciprofloxacin in this study applied in combination with daptomycin was 12.5 μg/mL (S. aureus S48) and 6.2 μg/mL (S. aureus NCIM 5021), while that with vancomycin was 12.5 μg/mL (S. aureus S48 and NCIM 5021). These concentrations are higher than those achievable in serum. The peak serum concentrations of daptomycin and vancomycin are reported to be 95–165 μg/mL [29] and 40–60 μg/mL [30], respectively. In combination with ciprofloxacin, the concentration of daptomycin applied in this study was 6.2 μg/mL (S. aureus NCIM 5021) and 25 μg/mL (S. aureus S48). These concentrations are found to be well below the achievable range. In combination with tobramycin, the concentration of daptomycin applied was 1.6 μg/mL (S. aureus S48 and NCIM 5021) and 12.5 μg/mL (S. aureus J6) while that of vancomycin was 100 μg/mL (S. aureus S48). The concentration of daptomycin is thus achievable; however, that of vancomycin is not. Also, the peak serum concentration of tobramycin is reported as 20–35 μg/mL [31]. The tobramycin concentration applied in combination with daptomycin was 12.5 μg/mL (S. aureus S48), 6.2 μg/mL (S. aureus J6) and 3.1 μg/mL (S. aureus NCIM 5021) and that with vancomycin was 12.5 μg/mL (S. aureus S48). These concentrations of tobramycin are clinically achievable. Therefore, of the four combinations tested, ciprofloxacin plus vancomycin can be tested to examine its effectiveness in clearing infections in vivo since the concentration of each of these antibiotics are clinically achievable. Daptomycin plus tobramycin still showed presence of persister cells even at 100X MIC in combination against J6 and NCIM 5021. However, it can be tested at higher concentrations for eradication of stationary-phase planktonic culture of S. aureus which is still below the peak serum concentration of both antibiotics. More number of isolates need to be tested using these combinations at higher concentrations to conclusively establish them for potential use in therapy.
Overall, there was a reduction in the survivors of stationary phase population when treated against combinations as compared to the individual antibiotics over a period of 72 h. The combination D + T showed complete eradication of S. aureus S48 which signifies the potential of this combination in further exploration for its use against other isolates possibly in clinical settings.
Several studies have successfully shown the effectiveness of targeting persister cells by various antimicrobials [5,[32], [33], [34], [35], [36], [37]]. Since the mechanism of persistence is unclear most of these studies are conducted in a trial-and-error fashion. Schmidt et al. modified tobramycin by conjugating to it a 12 amino acid peptide [38]. The resultant compound named ‘Pentobra’ effectively killed S. aureus persisters. Other such compounds that have been shown to bring down the number of S. aureus persisters effectively include ADEP4 [39], Tosufloxacin (a fluoroquinolone) [40], and retinoids (CD437 and CD1530) [36]. The first-ever report of the effect of nanoparticles on persister cells showed that bimetal nanoparticles of CuO/ZnO were effective in controlling persister and biofilm cells of S. aureus, among other MDR bacteria [41]. However, this approach increases the rate of resistance occurring in the population. Therefore, strategies must be developed such that all types of cells in a given population are targeted without risk of inducing resistance. Acetic acid has been tested in this regard with promising results [42]. It is less likely that a population of cells will develop resistance against a combination of two or more antibiotics [10]. Therefore, antibiotic combinations prove to be promising to eliminate S. aureus persisters and could be studied for synergistic killing at higher concentrations along with eukaryotic cell cytotoxicity studies.
CRediT authorship contribution statement
Ekta Kamble: Conceptualization, Methodology, Investigation, Formal analysis, Writing – original draft. Purva Sanghvi: Investigation, Writing – original draft. Karishma Pardesi: Writing – review & editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by Department of Science and Technology, UPE-Phase II program (UGC-262-A-2) and DST-PURSE (GOI-A-670) program. Ekta Kamble is the recipient of Senior Research Fellowship granted by Council of Scientific and Industrial Research, India.
Contributor Information
Ekta Kamble, Email: ekta.kamble90@gmail.com.
Purva Sanghvi, Email: purvas97@gmail.com.
Karishma Pardesi, Email: karishmapardesi@gmail.com, karishma@unipune.ac.in.
References
- 1.Geneva S. World Health Organization; Geneva Switzerland: 2014. Antimicrobial resistance: global report on surveillance. [Google Scholar]
- 2.Mulani M.S., Kamble E.E., Kumkar S.N., Tawre M.S., Pardesi K.R. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: a review. Front Microbiol. 2019;10:539. doi: 10.3389/fmicb.2019.00539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ventola C.L. The antibiotic resistance crisis: causes and threats. PC Tech J. 2015;40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis K. Persister cells and the paradox of chronic infections. Microbe. 2010;5:429–437. doi: 10.1146/annurev.micro.112408.134306. [DOI] [Google Scholar]
- 5.Lewis K. Springer International Publishing; 2019. Persister cells and infectious disease. [DOI] [Google Scholar]
- 6.Keren I., Shah D., Spoering A., Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol. 2004;186:8172–8180. doi: 10.1128/JB.186.24.8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khan F., Pham D.T.N., Tabassum N., Oloketuyi S.F., Kim Y.M. Treatment strategies targeting persister cell formation in bacterial pathogens. Crit Rev Microbiol. 2020;46:665–688. doi: 10.1080/1040841X.2020.1822278. [DOI] [PubMed] [Google Scholar]
- 8.Fauvart M., de Groote V.N., Michiels J. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J Med Microbiol. 2011;60:699–709. doi: 10.1099/jmm.0.030932-0. [DOI] [PubMed] [Google Scholar]
- 9.Wood T.K. Combatting bacterial persister cells; Combatting bacterial persister cells. Biotechnol Bioeng. 2016;113:476–483. doi: 10.1002/bit.25721/abstract. [DOI] [PubMed] [Google Scholar]
- 10.Vazquez-grande G., Kumar A. Optimizing antimicrobial therapy of sepsis and septic shock. Focus on Antibiotic Combination Therapy. 2015;1:154–166. doi: 10.1055/s-0034-1398742. [DOI] [PubMed] [Google Scholar]
- 11.Broussou D.C., Lacroix M.Z., Toutain P.-L., Woehrlé F., El Garch F., Bousquet-Melou A., et al. Differential activity of the combination of vancomycin and amikacin on planktonic vs. Biofilm-growing Staphylococcus aureus bacteria in a hollow fiber infection model. Front Microbiol. 2018;9:572. doi: 10.3389/fmicb.2018.00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamble E., Pardesi K. Antibiotic tolerance in biofilm and stationary-phase planktonic cells of Staphylococcus aureus. Microb Drug Resist. 2020 doi: 10.1089/mdr.2019.0425. [DOI] [PubMed] [Google Scholar]
- 13.Doern C.D. When does 2 plus 2 equal 5? A review of antimicrobial synergy testing downloaded from. 4124. JcmAsmOrg J Clin Microbiol. 2014;52:4124–4128. doi: 10.1128/JCM.01121-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaidhani S.V., Raskar A.V., Poddar S., Gosavi S., Sahu P.K., Pardesi K.R., et al. Time-dependent enhanced resistance against antibiotics & metal salts by planktonic & biofilm form of Acinetobacter haemolyticus MMC 8 clinical isolate. Indian J Med Res. 2014;140:665–671. [PMC free article] [PubMed] [Google Scholar]
- 15.Chopade B., Ghosh, Patil, Ahire, Kitture, Jabgunde, et al. Synthesis of silver nanoparticles using Dioscorea bulbifera tuber extract and evaluation of its synergistic potential in combination with antimicrobial agents. Int J Nanomed. 2012;7:483. doi: 10.2147/ijn.s24793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lechner S., Lewis K., Bertram R. Staphylococcus aureus persisters tolerant to bactericidal antibiotics. J Mol Microbiol Biotechnol. 2012;22:235–244. doi: 10.1159/000342449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Çokça F., Arman D., Altay G. In vitro activity of vancomycin combined with rifampin, amikacin, ciprofloxacin or imipenem against methicillin-resistant and methicillin-susceptible Staphylococcus aureus. Clin Microbiol Infect. 1998;4:657–659. doi: 10.1111/j.1469-0691.1998.tb00349.x. [DOI] [PubMed] [Google Scholar]
- 18.Baltch A.L., Ritz W.J., Bopp L.H., Michelsen P., Smith R.P. Activities of daptomycin and comparative antimicrobials, singly and in combination, against extracellular and intracellular Staphylococcus aureus and its stable small-colony variant in human monocyte-derived macrophages and in broth. Antimicrob Agents Chemother. 2008;52:1829–1833. doi: 10.1128/AAC.01480-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pardesi A., Pable A., Bhagat D.S.S. Applications of metal nanoparticles to combat biofilm-forming ESKAPE pathogens. Recent Adv. Biotechnol. 2019:1–38. [Google Scholar]
- 20.Wu H., Moser C., Wang H.Z., Høiby N., Song Z.J. Strategies for combating bacterial biofilm infections. Int J Oral Sci. 2015;7:1–7. doi: 10.1038/ijos.2014.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ciofu O., Rojo-Molinero E., Macià M.D., Oliver A. Antibiotic treatment of biofilm infections. APMIS. 2017;125:304–319. doi: 10.1111/apm.12673. [DOI] [PubMed] [Google Scholar]
- 22.Barber K.E., Smith J.R., Ireland C.E., Boles B.R., Rose W.E., Rybaka M.J. Evaluation of ceftaroline alone and in combination against biofilm-producing methicillin-resistant Staphylococcus aureus with reduced susceptibility to daptomycin and vancomycin in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob Agents Chemother. 2015;59:4497–4503. doi: 10.1128/AAC.00386-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujimura S., Sato T., Hayakawa S., Kawamura M., Furukawa E., Watanabe A. Antimicrobial efficacy of combined clarithromycin plus daptomycin against biofilms-formed methicillin-resistant Staphylococcus aureus on titanium medical devices. J Infect Chemother. 2015;21:756–759. doi: 10.1016/j.jiac.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Jorgensen S.C.J., Rybak M.J. Meropenem and vaborbactam: stepping up the battle against carbapenem-resistant enterobacteriaceae. Pharmacotherapy. 2018;38:444–461. doi: 10.1002/phar.2092. [DOI] [PubMed] [Google Scholar]
- 25.Shi J., Mao N.-F., Wang L., Zhang H.-B., Chen Q., Liu H., et al. Efficacy of combined vancomycin and fosfomycin against methicillin-resistant Staphylococcus aureus in biofilms in vivo. PLoS One. 2014;9 doi: 10.1371/journal.pone.0113133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cirioni O., Mocchegiani F., Ghiselli R., Silvestri C., Gabrielli E., Marchionni E., et al. Daptomycin and rifampin alone and in combination prevent vascular graft biofilm formation and emergence of antibiotic resistance in a subcutaneous rat pouch model of staphylococcal infection. Eur J Vasc Endovasc Surg. 2010;40:817–822. doi: 10.1016/j.ejvs.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 27.Duffull S. Vol. 73. 2012. (Basic pharmacokinetics and pharmacodynamics, an integrated textbook and computer simulations). [DOI] [Google Scholar]
- 28.D'Espine M., Bellido F., Pechère J.C., Auckenthaler R., Rohner P., Lew D., et al. Serum levels of ciprofloxacin after single oral doses in patients with septicemia. Eur J Clin Microbiol Infect Dis. 1989;8:1019–1023. doi: 10.1007/BF01975162. [DOI] [PubMed] [Google Scholar]
- 29.Benvenuto M., Benziger D.P., Yankelev S., Vigliani G. Pharmacokinetics and tolerability of daptomycin at doses up to 12 milligrams per kilogram of body weight once daily in healthy volunteers. Antimicrob Agents Chemother. 2006;50:3245. doi: 10.1128/AAC.00247-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chapter 5. Vancomycin | Applied Clinical Pharmacokinetics, 2e | AccessPharmacy | McGraw Hill Medical n.d. https://accesspharmacy.mhmedical.com/content.aspx?sectionid=40843073&bookid=510 (accessed December 2, 2021).
- 31.VandenBussche H.L., Homnick D.N. Evaluation of serum concentrations achieved with an empiric once-daily tobramycin dosage regimen in children and adults with cystic fibrosis. J Pediatr Pharmacol Ther JPPT. 2012;17:67. doi: 10.5863/1551-6776-17.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asghari B., Sadeghi H.R., Mazaherylaghab H. Combatting Bacterial Persister cell infections by auranofin? Biomed Pharmacother. 2017;96:1565–1566. doi: 10.1016/j.biopha.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 33.Bojer M.S., Lindemose S., Vestergaard M., Ingmer H. Quorum sensing-regulated phenol-soluble modulins limit persister cell populations in Staphylococcus aureus. Front Microbiol. 2018;9:1–12. doi: 10.3389/fmicb.2018.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Breij A., Riool M., Cordfunke R.A., Malanovic N., de Boer L., Koning R.I., et al. The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci Transl Med. 2018;10:eaan4044. doi: 10.1126/scitranslmed.aan4044. [DOI] [PubMed] [Google Scholar]
- 35.Defraine V., Fauvart M., Michiels J. Fighting bacterial persistence: current and emerging anti-persister strategies and therapeutics. Drug Resist Updates. 2018;38:12–26. doi: 10.1016/j.drup.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 36.Kim W., Zhu W., Hendricks G.L., Van Tyne D., Steele A.D., Keohane C.E., et al. A new class of synthetic retinoid antibiotics effective against bacterial persisters. Nature. 2018;556:103–107. doi: 10.1038/nature26157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim W., Hendricks G.L., Tori K., Fuchs B.B., Mylonakis E. Strategies against methicillin-resistant Staphylococcus aureus persisters. Future Med Chem. 2018;10:779–794. doi: 10.4155/fmc-2017-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt N.W., Deshayes S., Hawker S., Blacker A., Kasko A.M., Wong G.C.L. Engineering persister-specific antibiotics with synergistic antimicrobial functions. ACS Nano. 2014;8:8786–8793. doi: 10.1021/nn502201a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conlon B.P., Nakayasu E.S., Fleck L.E., Lafleur M.D., Isabella V.M., Coleman K., et al. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature. 2013;503:365–370. doi: 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niu H., Cui P., Yee R., Shi W., Zhang S., Feng J., et al. A clinical drug library screen identifies tosufloxacin as being highly active against Staphylococcus aureus persisters. Antibiotics. 2015;4:329–336. doi: 10.3390/antibiotics4030329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manoharan R.K., Mahalingam S., Gangadaran P., Ahn Y.H. Antibacterial and photocatalytic activities of 5-nitroindole capped bimetal nanoparticles against multidrug resistant bacteria. Colloids Surf B Biointerfaces. 2020;188 doi: 10.1016/j.colsurfb.2020.110825. [DOI] [PubMed] [Google Scholar]
- 42.Tawre M.S., Kamble E.E., Kumkar S.N., Mulani M.S., Pardesi K.R. Antibiofilm and antipersister activity of acetic acid against extensively drug resistant Pseudomonas aeruginosa PAW1. PLoS One. 2021;16 doi: 10.1371/journal.pone.0246020. [DOI] [PMC free article] [PubMed] [Google Scholar]