Abstract
Uterine corpus endometrial carcinoma (UCEC) is a malignant disease globally, and there is no unified prognostic signature at present. In our study, two clusters were identified. Cluster 1 showed better prognosis and higher infiltration level, such as tumor microenvironment (TME), tumor mutation burden (TMB), and immune checkpoint genes expression. Gene set enrichment analysis (GSEA) indicated that some tumor-related pathways and immune-associated pathways were exposed. What is more, six pyroptosis-related long noncoding RNAs (lncRNAs) (PRLs) were applied to establish a prognostic signature through multiple Cox regression analysis. In both training and testing sets, patients with higher risk score had poorer survival than patients with low risk. The area under the curve (AUC) of receiver operating characteristic (ROC) curves performed that the survival probability was better in people with lower risk score. Mechanism analysis revealed that high risk score was correlated with reduced immune infiltration and T cells exhaustion, matching the definition of an “immune-desert” phenotype. Patients with lower risk score were characterized by higher immune checkpoint gene expression and TMB and have a sensitive response to immunotherapy and chemotherapy compared with patients with high risk score. The signature has accurate prediction ability of UCEC and is a promising therapeutic target to improve the effect of immunotherapy.
Keywords: pyroptosis, lncRNAs, UCEC, prognostic signature, tumor microenvironment, TME, immunotherapy
Graphical abstract
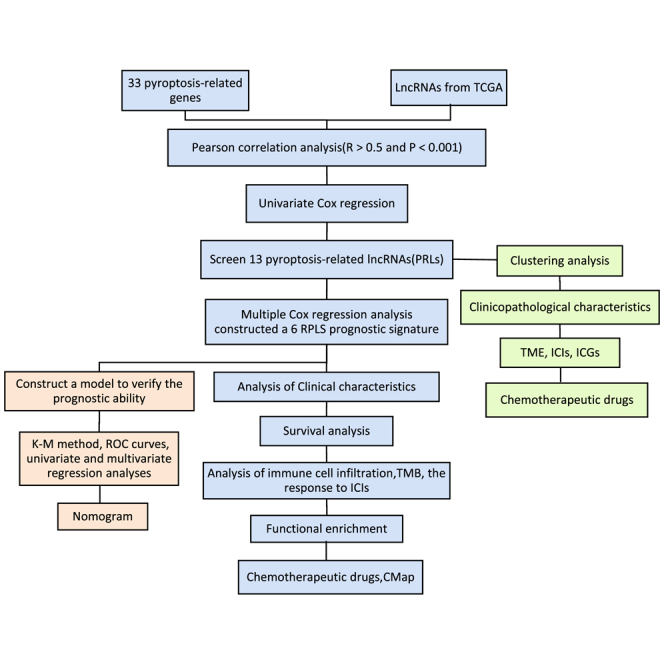
This research constructed and verified a prognostic signature on the basis of pyroptosis-related lncRNAs. The model can accurately predict the prognosis of UCEC patients and is a promising therapeutic target to improve the effect of immunotherapy. It contributes to the development of immunotherapy in patients with UCEC.
Introduction
Uterine corpus endometrial carcinoma (UCEC) is a gynecological malignant disease with high mortality,1 which came in fourth after breast cancer, rectum, and lung cancer.2 The incidence rate and mortality of UCEC are continuing to rise. In 2020, there were 417,367 new cases and 97,370 deaths.3 There are two pathological types of UCEC, one is estrogen-dependent endometrioid adenocarcinomas (EACs) and the other is estrogen-independent serous adenocarcinomas (SACs).4,5 Most patients have a good prognosis in early stage, while the 5-year survival rate is 95%.5 With the progress of the disease course, once metastasis occurs, the survival rate will also decrease significantly.6,7 At present, UCEC patients are often treated by surgery, radiotherapy, brachytherapy, and chemotherapy.8 However, these methods have poor therapeutic effect on advanced patients, and traditional risk assessment methods are not enough to explain the progress and prognostics of UCEC.9,10 Therefore, we urgently need to establish a predictive model to provide a new target for UCEC immunotherapy and improve the accuracy of prognosis prediction of UCEC.
Cell death (CD) can not only maintain the stability of internal environment but also an important tumor inhibition process.11 Resistance to CD is a characteristic of tumors.12,13 Pyroptosis is a programmed CD triggered by some inflammatory bodies,14,15 which will lead to cell swelling, plasma membrane dissolution, and chromatin rupture, accompanied by a series of inflammation and immune reaction.16,17 For one thing, as a kind of pro-inflammatory death, pyroptosis may lead to a microenvironment conducive to the progress of tumors. For another, it can prevent the occurrence and progression of cancer, although the mechanisms are not clear.16,18
Pyroptosis can affect tumor proliferation, invasion, and metastasis and has become a new study hotspot in tumor study in recent years.14,19 Previous research has shown that pyroptosis has close relationships with atherosclerosis and diabetic nephropathy20,21 and is also closely correlated with the occurrence and development of various cancers.22, 23, 24 Pyroptosis, as a programmed cell death, can be regulated by some noncoding RNAs (ncRNAs).14,25 In addition, pyroptosis inhibitors can also eliminate pyroptosis and long ncRNA (lncRNA) overexpression and inhibit the progression of tumor.26
lncRNA is an important regulator of gene expression, which widely affects the progression of cancer, cardiovascular disease, and diabetes.27 lncRNAs can participate in tumor growth and metastasis process through transcriptional and post-transcriptional mechanisms28 and are able to affect the expression of carcinogenic transcription factors.29,30 In the immune system, lncRNAs have different expression levels in different environments, stages, and cell types that contribute to coordinate some aspects of immune function. So far, hundreds of lncRNAs have been verified differentially expressing in immune cells.31 Some recent articles have shown that lncRNAs can be applied as biomarkers or immunotherapy targets to predict and diagnose many different types of cancer, including UCEC.32, 33, 34, 35 At present, several UCEC-related lncRNAs have been found, such as MEG3, GAS5, SRA, and HOTAIR.36,37 However, there is no unified biomarker used for UCEC.38
In this study, we identified six lncRNAs that might become prognostic signatures of UCEC. A risk model based on pyroptosis-related lncRNAs (PRLs) was established and verified to have the ability to judge the prognosis of UCEC patients and offer guidance for their treatments. The flow diagram of this study is shown in Figure S1.
Results
Analysis of pyroptosis-related genes (PRGs)
To evaluate the expression patterns of PRGs, we used profiles achieved from the The Cancer Genome Atlas (TCGA) dataset to find out the differential expression of 33 PRGs between UCEC and normal tissues. A heatmap provided the level of the 33 genes expression in UCEC tissues and normal tissues (Figure S2A). The color of red represents higher expression levels, and green represents lower expression levels. Of the 33 PRGs, 25 have significantly different gene expression between tumor and normal tissues (p < 0.001; Figure S2B). In tumor samples, 13 genes (CASP3, CASP5, CASP6, CASP8, GPX4, GSDMB, GSDMC, GSDMD, IL18, NLRP7, NOD2, PYCARD, and TNF) had higher expression. However, ELANE, GSDME, NLRP1, NLRP3, NOD1, PJVK, SCAF11, and TIRAP had lower expression in tumor samples (Figure S2C). A protein-protein interaction (PPI) network was established to show the relationships between PRGs (Figure S2D). Figure 2E shows the genes with more nodes. Among them, CASP1 and PYCARD were most closely related to other genes that had 23 nodes. After that, we conducted Pearson correlation analysis on these 33 genes (Figure S2F). GPX4 has a strong negative correlation with SCAF11 (cor = −0.45) and CASP8 (cor = −0.44). CASP1 was obviously positively correlated with CASP4 (cor = 0.61) and CASP5 (cor = 0.61). The above results showed that there were certain correlations between pyroptosis-related genes in UCEC.
Figure 2.
Immunoassay of two clusters
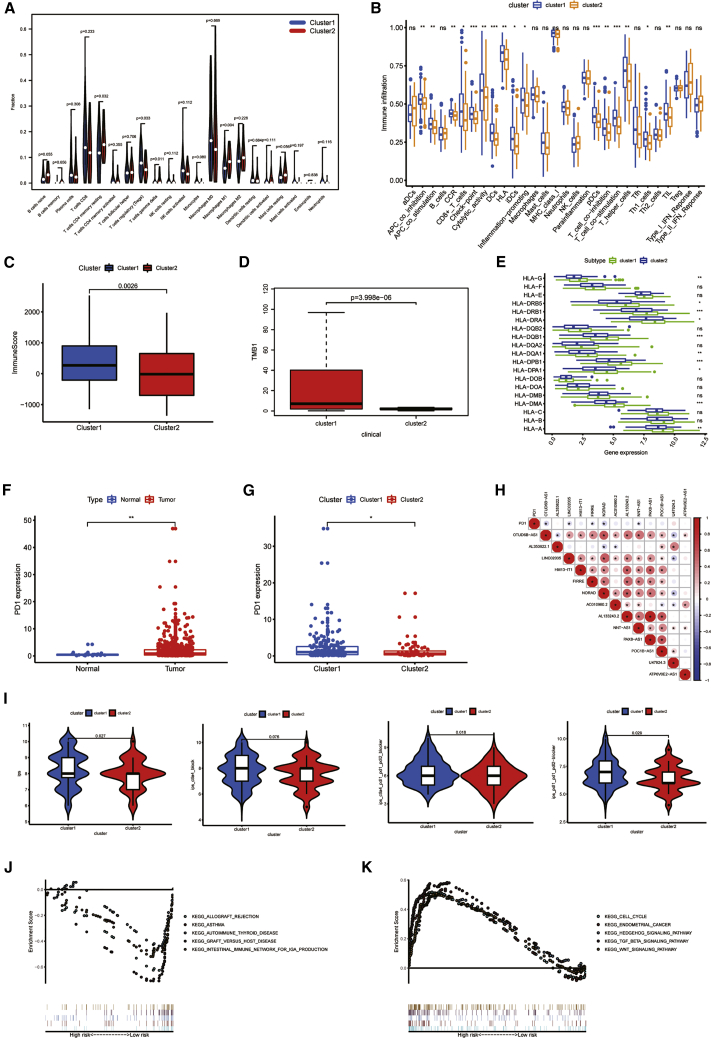
(A) The infiltrating levels of 22 immune cell types in cluster1 and cluster2 subtypes in the TCGA cohort. (B–E) Immune infiltration (B), immune score (C),TMB (D), and HLA expression (E) in two clusters are shown. (F) Up-regulation of PD1 expression in tumor tissue is shown. (G) Expression level of PD1 in cluster1 and cluster2 is shown. (H) The correlation between PD-L1 and PRLs is shown. (I) IPS analysis is shown. (J and K) GSEA was employed to predict the potential functions and pathways in cluster1 (J) and cluster2 (K). ∗P< 0.05 and ∗∗P< 0.01; ns, not significant.
Identification of PRLs
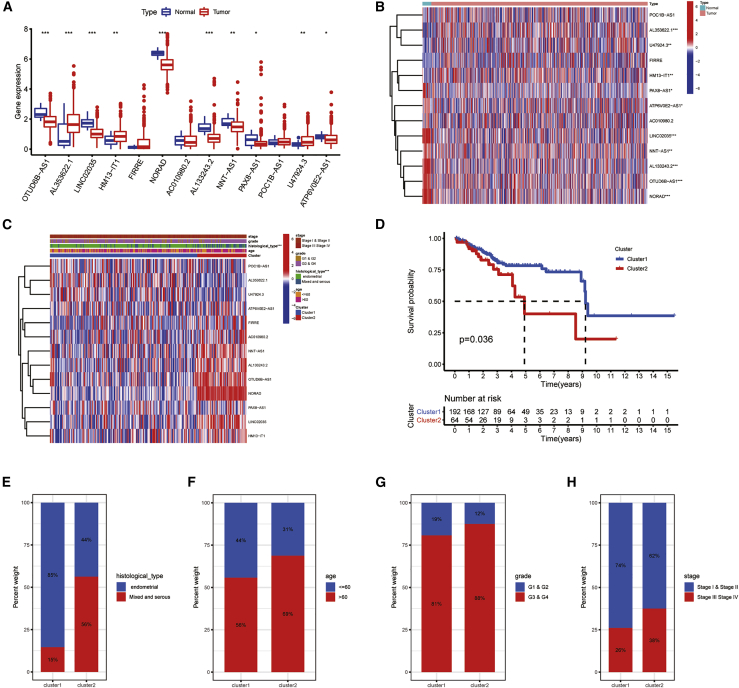
We screened a 384-lncRNA database that was closely related to PRGs from TCGA through Pearson correlation analysis. The selection criteria were correlation coefficient >0.5 and p < 0.001. After combining with clinical information, we achieved 511 samples. Samples of entire sets were assigned into training and testing sets for 1:1 ratio randomly. By performing univariate Cox regression analysis on the training set, we identified 13 PRLS (OTUD6B-AS1, AL353622.1, LINC02035, HM13-IT1, FIRRE, NORAD, AC010980.2, AL133243.2, NNT-AS1, PAX8-AS1, POC1B-AS1, U47924.3, and ATP6V0E2-AS1) that had potential prognosis value for UCEC (Table 1). A boxplot and heatmap of 13 PRLs expressed in normal and tumor tissues are shown in Figures 1A and 1B. Most of their expression was not the same. The abnormal expression of these PRLs means an important affect for the occurrence and progression of UCEC.
Table 1.
Univariate Cox regression analysis of pyroptosis-related lncRNAs in training set
| ID | HR | HR.95L | HR.95H | p value |
|---|---|---|---|---|
| OTUD6B-AS1 | 1.2494 | 1.0721 | 1.4561 | 0.0044 |
| AL353622.1 | 0.7589 | 0.6370 | 0.9041 | 0.0020 |
| LINC02035 | 1.5625 | 1.2069 | 2.0229 | 0.0007 |
| HM13-IT1 | 1.4665 | 1.1942 | 1.8009 | 0.0003 |
| FIRRE | 1.5388 | 1.2256 | 1.9321 | 0.0002 |
| NORAD | 1.0159 | 1.0039 | 1.0281 | 0.0095 |
| AC010980.2 | 1.5812 | 1.1270 | 2.2186 | 0.0080 |
| AL133243.2 | 1.8661 | 1.1751 | 2.9633 | 0.0082 |
| NNT-AS1 | 1.4395 | 1.1286 | 1.8359 | 0.0033 |
| PAX8-AS1 | 1.8934 | 1.2512 | 2.8653 | 0.0025 |
| POC1B-AS1 | 0.2332 | 0.0852 | 0.6386 | 0.0046 |
| U47924.3 | 0.3408 | 0.1603 | 0.7247 | 0.0052 |
| ATP6V0E2-AS1 | 1.6049 | 1.1671 | 2.2070 | 0.0036 |
HR, hazard rate
Figure 1.
Different clinicopathological features and survival probability of the two UCEC subtypes
(A) Boxplot of 13 PRLs’ expression in normal and tumor tissues. (B) Heatmap of 13 PRLs’ expression in normal and tumor tissues is shown. (C) The different expressions of PRLs and their clinicopathological features between the two clusters were shown by heatmap. (D) The OS rate of UCEC patients in the two groups was calculated by Kaplan-Meier curve. (E–H) The proportions of histological type (E), age (F), grade (G), and stage (H) between the two clusters were compared. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. ns, not significant.
Consistency clustering analysis
According to the proportion of similarity and fuzzy clustering measures on the basis of 13 PRL expression with prognostic value, it has the best clustering stability when k = 2 is determined. Figures S3A and S3B show the change of cumulative distribution function (CDF) of consensus cluster and area under the curve (AUC) from k = 2 to k = 9. 256 patients in the training set were assigned into two subtypes, cluster1 (n = 192) and cluster2 (n = 64), in terms of consensus matrix k = 2. A consensus cluster composed of 13 PRLs was constructed by using the “consumusclusterplus” package (Figure S2C).
Prognosis and clinical characteristics of the two clusters
The overall level of PRL expression was higher in cluster2 than in cluster1, especially the expression levels of OTUD6B-AS1 and NORAD (Figure 1C). The clinical characteristics between the two clusters were also compared. Samples in cluster2 were related to older, more advanced stage, grade, and historical type. At the same time, the survival probability of cluster2 was lower than that of cluster1 (p = 0.036; Figure 1D). A boxplot showed that patients in different clusters of UCEC have different clinical characteristics. As shown in Figures 1E–1H, the clinical symptoms of cluster2 patients were more serious. In cluster1, patients belonging to the essential histological type accounted for 85%, and 15% belonged to mixed and serous type, age ≤60 accounted for 44% and age >60 accounted for 56%, G1 and G2 accounted for 19% and G3 and G4 accounted for 81%, and stage I and stage II accounted for 74% and stage III and stage IV accounted for 26%. While in cluster2, patients belonging to endogenous histological type accounted for 44% and patients belonging to the mixed and serial type accounted for 56%, age ≤60 accounted for 31% and age >60 accounted for 69%, G1 and G2 accounted for 12% and G3 and G4 accounted for 88%, and stage I and stage II accounted for 62% and stage III and stage IV accounted for 38%.
Cluster2 had lower infiltration level of immune cell
To explore the immune infiltration level of different PRL expressions, we evaluated the fraction of 22 immune cells between the two clusters (Figure 2A). Regulatory T cells (Treg cells) showed a significantly lower fraction in cluster2. The immune information of checkpoint, dendritic cells (DCs), plasmacytoid DCs (pDCs), and T cell co-stimulation was higher in cluster1 (p < 0.001; Figure 2B). The immune score of cluster1 was also higher than in cluster2 (Figure 2C). The expression levels of some human leukocyte antigens (HLAs) were also different between the two subtypes. Figure 2D shows that the TMB of cluster1 was higher than cluster2. In particular, the expression levels of HLA-DRB1, HLA-DQB1, HLA-DPB1, and HLA-DMA in cluster1 were higher in cluster2 (Figure 2E). For the purpose of clarifying the correlations between immune checkpoint inhibitors (ICIs) and PRLs, we evaluated the expression discrepancy of programmed death-1 (PD1) and the association between PD1 and PRLs. PD1 expression was higher in UCEC tissue (p < 0.05; Figure 2F). Comparing the two clusters, the expression level of PD1 in cluster2 was down-regulated (p < 0.05; Figure 2G). Then, the association between PD1 and PRLs was further analyzed. In the TCGA cohort, the expression of PD1 had a negative correlation with the expression levels of OTUD6B-AS1, LINC02035, FIRRE, NORAD, and NNT-AS1 (Figure 2H). International Prognostic Score (IPS) analysis showed that IPS (p = 0.027), IPS_ ctla4_ pdl1_ pd1_ pdl2 (p = 0.018), and IPS_ pdl1_ pd1_ pdL2 (p = 0.029) had a higher level in cluster1, but not IPS_ ctla4 (p = 0.076; Figure 2I), which means cluster1 was associated with higher immunogenicity. In brief, in immune infiltration, proportion of T cells was lower in cluster2, which indicated that cluster2 might be a subtype of cold tumor. That means patients in cluster2 may have immune escape, and the efficacy of immunotherapy will be poor.
GSEA revealed the pathway that PRLs exposed
In order to explore the potential mechanism leading to the difference between the two groups, we conducted GSEA (Figures 2J and 2K). Enrichment scores of allograph rejection, asthma, autoimmune thyroid disease, graft versus host disease, and intrinsic immune network for immunoglobulin A (IgA) were evaluated both in cluster1 and cluster2. The results indicated that cluster1 had negative correlation with immune-related function, and cluster2 was positively related to immune functions. Therefore, the different prognoses of cluster1 and cluster2 may be due to the different immune statuses.
Analysis of drug sensitivity
Drug sensitivity analysis was carried out (Figures S4A–S4D). Patients in cluster1 were more sensitive to etoposide (p = 0.036), while patients in cluster2 were more sensitive to paclitaxel (p = 0.026). The sensitivity of patients in the two subtypes to cisplatin (p = 0.47) and doxorubicin (p = 0.14) were not significantly different.
Constructing and validating a risk signature based on PRLs
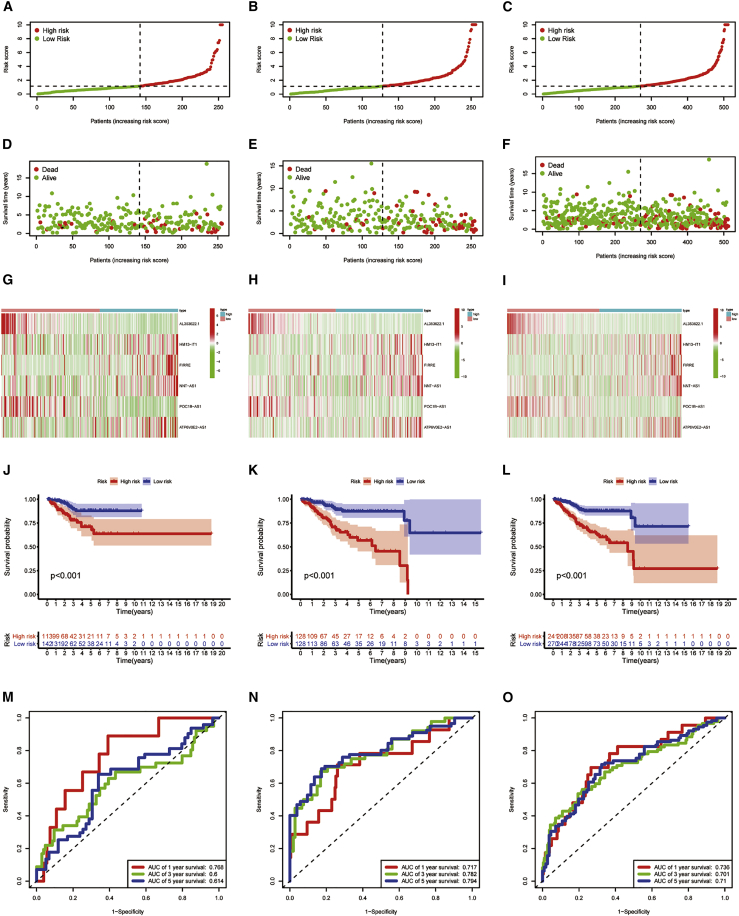
Multiple Cox regression analysis was then conducted on the training set, and six PRLs were verified to have independent prognostic value (Table 1). Among the six independent prognostic factors, HM13-IT1, FIRRE, NNT-AS1, and ATP6V0E2-AS1 were risk factors and AL353622.1 and POC1B-AS1 were protective factors. Then, we constructed a risk signature in the light of six PRLs. The formula is as follows: risk score = (0.2916 × HM13-IT1) − (0.2409 × AL353622.1) + (0.3454 × FIRRE) + (0.3754 × NNT-AS1) − (1.1937 × POC1B-AS1) + (0.3824 × ATP6V0E2-AS1) Table 2. After that, people were assigned into two risk groups in the light of median risk score (Figures 3A–3C). Subsequently, univariate and multivariate Cox regression analyses were applied to identify whether the model based on pyroptosis-related lncRNAs (PYLRM) can be thought as an independent prognostic factor. The results are shown in Table 3, indicating that the risk score can be considered as an important independent prognostic factor for UCEC. The distribution of risk score and survival time of different patient sets were further calculated, and the heatmap was used to test the prediction ability of PYLRM. The two risk groups had significant difference in risk score distribution, survival status, and survival time distribution (Figures 3D–3F). The results obtained from principal-component analysis (PCA) indicated that the two risk groups have different distributions (Figure S5). This means that the prognosis of UCEC patients can be judged by risk score. Heatmap showed that the expression levels of PRLs, such as HM13-IT1, FIRRE, NNT-AS1, and ATP6V0E2-AS1, were higher in patients with higher risk score, while the expression levels of PRLs, like AL353622.1 and POC1B-AS1, were up-regulated in patients with low risk (Figures 3G–3I). Kaplan-Meier survival analysis showed that the survival rate of patients with low risk was higher than that of patients with high risk (p < 0.01; Figures 3J–3L). As is shown in Figures 3M–3O, the PYLRM had great prediction sensitivity. It showed the same results in all 511 samples (Figures 3M–3O).
Table 2.
Multivariable Cox regression analysis of pyroptosis-related lncRNAs in the training set
| ID | Coef | HR | HR.95L | HR.95H | p value |
|---|---|---|---|---|---|
| AL353622.1 | −0.2409 | 0.7859 | 0.6548 | 0.9432 | 0.0097 |
| HM13-IT1 | 0.2916 | 1.3386 | 1.0957 | 1.6355 | 0.0043 |
| FIRRE | 0.3454 | 1.4126 | 1.1283 | 1.7685 | 0.0026 |
| NNT-AS1 | 0.3754 | 1.4555 | 1.1376 | 1.8622 | 0.0028 |
| POC1B-AS1 | −1.1937 | 0.3031 | 0.1066 | 0.8615 | 0.0251 |
| ATP6V0E2-AS1 | 0.3824 | 1.4658 | 1.0607 | 2.0257 | 0.0205 |
Figure 3.
Predictive value of risk model constructed by PRLs in different patients set
(A–C) Testing set (A), training set (B), and entire set (C) were divided into high- and low-risk groups according to the median risk score. (D–F) Distribution of survival time and survival status between high-risk group and low-risk group in testing set (D), training set (E), and entire set (F) is shown. (G–I) The heatmap of cluster analysis shows the expression levels of six PRLs in the testing set (G), training set (H), and entire set (I). (J–L) Kaplan-Meier survival curve of OS in low-risk group and high-risk group in testing set (J), training set (K), and entire set (L) is shown. (M–O) Prediction sensitivity in testing set (M), training set (N), and entire set (O) in 1, 3, and 5 years is shown.
Table 3.
Univariate and multivariate Cox regression analyses of the prognosis-related factors
| Variable | Univariable model |
Multivariable model |
||||||
|---|---|---|---|---|---|---|---|---|
| HR | HR.95L | HR.95H | p value | HR | HR.95L | HR.95H | p value | |
| Training set (n = 256) | ||||||||
| Age | 2.3114 | 1.2355 | 4.3241 | 0.0088 | 2.1003 | 1.0889 | 4.0511 | 0.0268 |
| Histological type | 2.7963 | 1.6241 | 4.8146 | 0.0002 | 1.4889 | 0.8113 | 2.7323 | 0.1988 |
| Grade | 4.4004 | 1.3628 | 14.2085 | 0.0132 | 2.3424 | 0.6819 | 8.0466 | 0.1764 |
| Stage | 3.9662 | 2.2870 | 6.8783 | 0.0000 | 2.7928 | 1.4988 | 5.2039 | 0.0012 |
| Risk score | 1.0614 | 1.0405 | 1.0827 | 0.0000 | 1.0489 | 1.0263 | 1.0720 | 0.0000 |
| Testing set (n = 255) | ||||||||
| Age | 1.4015 | 0.6890 | 2.8506 | 0.3515 | 1.2233 | 0.5728 | 2.6126 | 0.6026 |
| Histological type | 3.4650 | 1.7958 | 6.6859 | 0.0002 | 2.2780 | 1.1192 | 4.6365 | 0.0232 |
| Grade | 2.6125 | 0.8010 | 8.5213 | 0.1114 | 0.9889 | 0.2703 | 3.6183 | 0.9865 |
| Stage | 4.4158 | 2.2746 | 8.5726 | 0.0000 | 3.3562 | 1.6682 | 6.7520 | 0.0007 |
| Risk score | 1.0686 | 1.0250 | 1.1141 | 0.0018 | 1.0411 | 0.9912 | 1.0935 | 0.1083 |
| Entire set (n = 511) | ||||||||
| Age | 1.7782 | 1.1121 | 2.8432 | 0.0162 | 1.5435 | 0.9452 | 2.5204 | 0.0828 |
| Histological type | 3.0435 | 2.0032 | 4.6242 | 0.0000 | 1.7697 | 1.1192 | 2.7984 | 0.0146 |
| Grade | 3.3631 | 1.4671 | 7.7097 | 0.0042 | 1.5019 | 0.6199 | 3.6391 | 0.3677 |
| Stage | 4.1162 | 2.7000 | 6.2754 | 0.0000 | 3.0600 | 1.9218 | 4.8724 | 0.0000 |
| Risk score | 1.0616 | 1.0438 | 1.0797 | 0.0000 | 1.0481 | 1.0281 | 1.0684 | 0.0000 |
Different clinical features of patients with different risk
We calculated the differences of clinical features of UCEC between the groups and drew a heatmap for the entire set (Figure S6A). AL353622.1 and POC1B-AS1 had higher expression level in the low-risk group, while HM13-IT1, FIRRE, NNT-AS1, and ATP6V0E2-AS1 had higher expression in patients with high risk. At the same time, cases in cluster2 were mostly in a high-risk level. Patients with higher risk scores were related to older, more advanced stage, grade, and histological type. Immune score was also lower in patients with higher risk. In Figures S6B–S6G, a boxplot revealed the expression difference of clinical characteristics in the groups, and the discrepancy was significant (p < 0.05). There were differences in risk scores among the four UCEC immune subtypes (p < 0.05), except that there was no significant discrepancy in risk scores between C4 and other immune subtypes (Figure S6H). After that, the survival probability of the two groups were compared under different clinical characteristics (Figure S6I). Regardless of how old the patients were, survival probability was higher in patients with lower risk (p < 0.05). There was no statistical discrepancy of survival probability of patients with G1 and G2 between different risk groups, while in patients with G3 and G4, the survival probability of the low-risk group was significantly higher (p < 0.001). Survival probability of people with mixed and severe type was very low. However, the survival probability of patients that had lower risk score was higher in people with endometrial carcinoma (p < 0.001). Patients in different stages had significantly higher survival probability than people in the low-risk group in general (p < 0.05).
Construction and verification of nomogram
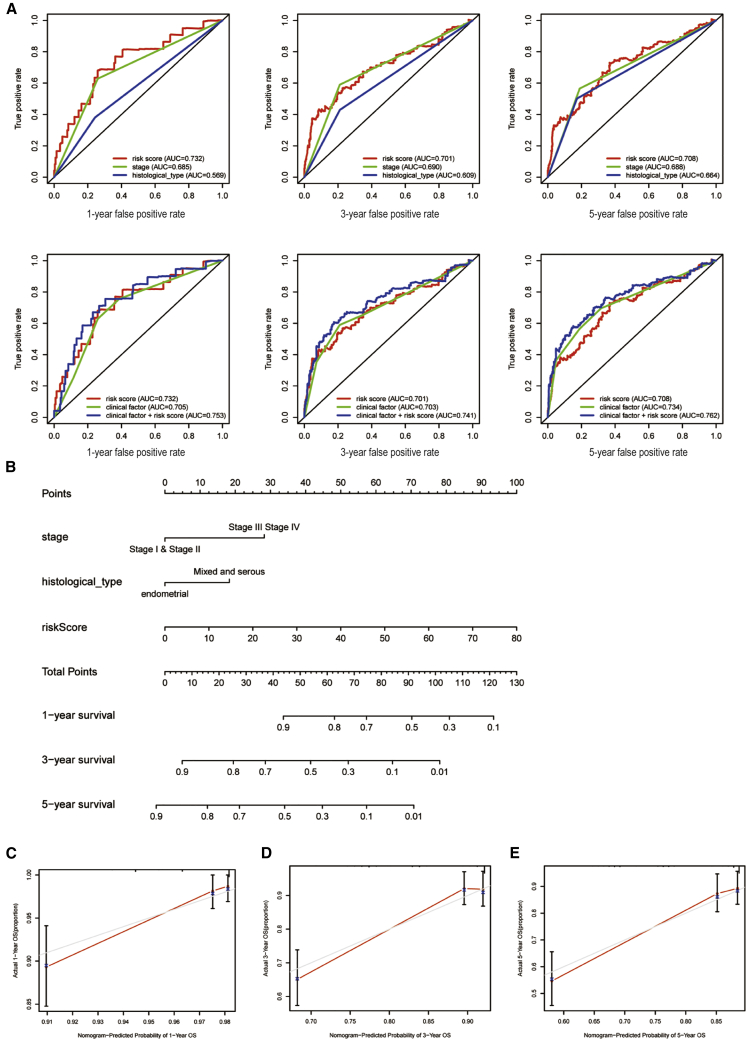
For the purpose of evaluating the prognostic accuracy of the PYLRM, we compared the true positive rates predicted by PYLRM, clinical factors, and the model combined with clinical factors. The analysis of receiver operating characteristic (ROC) curves for 1, 3, and 5 years is shown in Figure 4A. The AUCs of the PYLRM for the three different years were 0.732, 0.701, and 0.708, respectively, which were higher than that of single clinical characteristics. It is worth mentioning that it is better to combine PYLRM with clinical factors. From the results mentioned above, we inferred that the risk score evaluated by six PRLs can accurately forecast the prognosis of UCEC patients. According to the results of multivariate Cox regression in the entire cohort, a nomogram, including indicators such as stage, histological type, and risk score, was established to predict overall survival (OS) incidence rate in 1, 3, and 5 years. By comparing with clinical characteristics, the risk level of the PYLRM showed outstanding prediction value through nomogram (Figure 4B). The correlation diagram showed that the observed OS ratios in 1, 3, and 5 years are in good agreement with the predicted ratios (Figures 4C–4E).
Figure 4.
Construction and evaluation of a prognostic nomogram
(A) The area under the ROC curve (AUC) of risk score and clinical characteristics was determined according to the ROC curve. (B) The nomogram predicts the probability of the 1-, 3-, and 5-year OS. (C–E) The calibration plot of the nomogram predicts the probability of the 1- (C), 3- (D), and 5-year (E) OS.
Estimation of TME and response to ICI on the basis of the PRLs
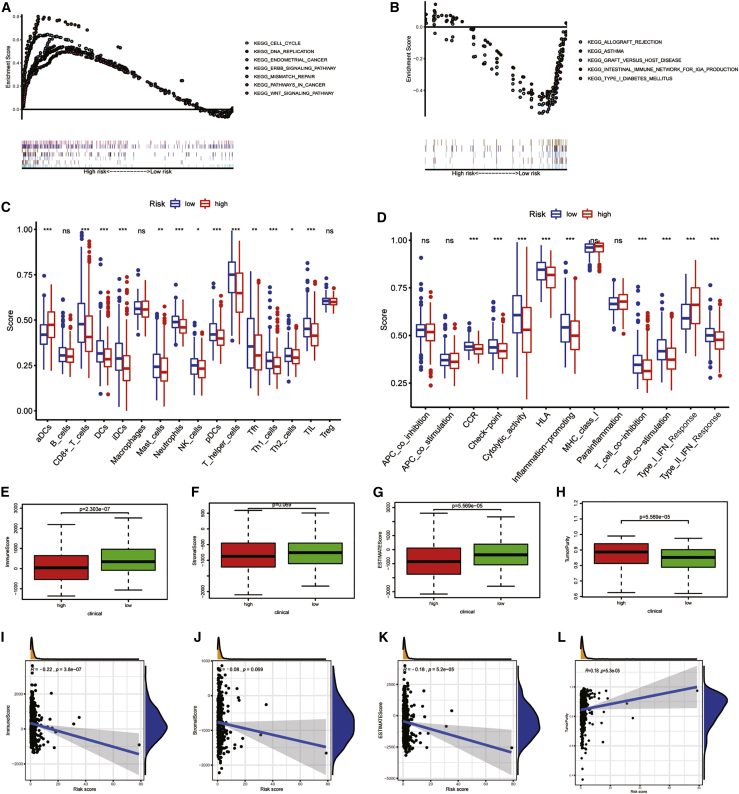
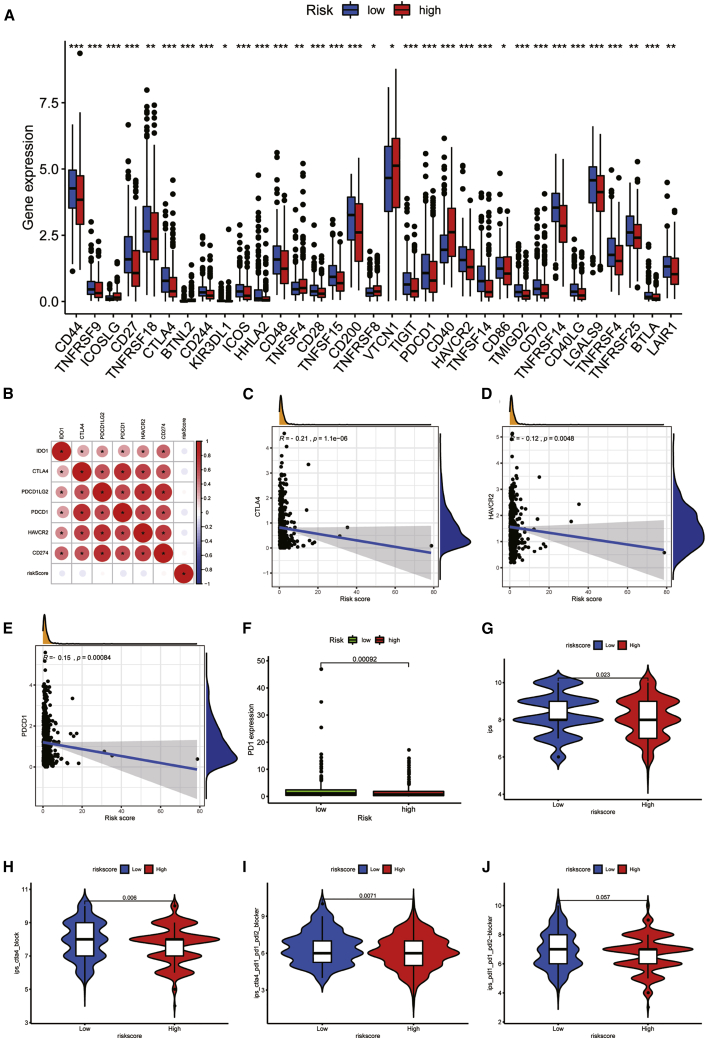
In this paragraph, we will further explore the tumor microenvironment (TME) of UCEC. In order to explore the potential mechanism leading to the difference between the two groups, we conducted GSEA. Tumor-related pathways, like cell cycle, DNA replication, endometrial cancer, ERBB signaling pathway, mismatch repair, pathways in cancer, and WNT signaling pathway, are mainly enriched in the high-risk groups (Figure 5A). However, immune-related pathways, such as allograft rejection, asthma, graft versus host disease, internal immune network for IgA production, and type 1 diabetes mellitus, are mainly enriched in low-risk groups (Figure 5B). This means that high-risk score was related to tumor-related pathways, and low-risk patients were associated with immune-related pathways. This may explain why high-risk patients have poor prognoses. Therefore, we used ESTIMATION and single sample gene set enrichment analysis (ssGSEA) analysis to verify the differences of immune status between groups. The study based on ssGSEA also confirmed that there were significant differences in immune cells and immune function between the two risk groups. The scores of immune cells and immune function in patient with higher risk score were generally lower than those with lower risk, except type I interferon (IFN) response scored higher in patients with higher risk score (Figures 5C and 5D). Immune cells and stromal cells are two important components of TME.39,40 We assessed immune cells (Figure 5E) and stromal cells (Figure 5F) in the two risk groups and added them together to get the estimated score (Figure 5G). The scores of the low-risk group were higher than the high-risk group, and there was significant discrepancy between the groups (p < 0.01). Higher estimated score means lower tumor purity, which is consistent with our results (Figure 5H): the tumor purity of low-risk patients was lower than patients with higher risk (p < 0.01). Figures 5I–5L showed the relationships between immune score, stromal score, estimate score, tumor purity, and the risk score. The immune score, stromal score, and estimate score were negatively related to the risk score, and tumor purity had a positive relationship with the risk score, which is consistent with the above results. Figure S7A shows the relative percent of 21 immune-infiltrating cells in two groups. Then, we analyzed the difference of the fraction of each immune cell between the groups (Figure S7B). Nine immune cells were different. The expressions of CD8 T cells (p = 0.014), Treg cells (p < 0.01), activated NK cells (p = 0.013), and monocytes (p = 0.02) in the high-risk group were lower. The expression of memory B cells (p = 0.013), M0 macrophages (p = 0.007), M1 macrophages (p = 0.031), M2 macrophages (p = 0.006), and activated DCs (p = 0.011) in the low-risk group were low. We further explored the relationships between 21 tumor-infiltrating cells and 6 PRLs (Figure S7C). We found that gamma delta T cells and naïve B cells had strong positive correlations with NNT-AS1, respectively (p < 0.001). Treg cells also had a positive correlation with POC1B-AS1 (p < 0.001). The negative relationship between neutrophils and ATP6V0E2-AS1 was the strongest (p < 0.001). After that, we assessed the relationships between the PYLRM risk score and immune-infiltrating cells. The risk score had positive relationships with memory B cells (R = 0.18; p = 0.0051), activated DCs (R = 0.22; p = 0.00059), M0 macrophages (R = 0.13; p = 0.045), and M1 macrophages (R = 0.17; p = 0.0087), while it had negative relationships with monocytes (R = −0.18; p = 0.005) and Treg cells (R = −0.23; p = 0.00035; Figure S7D), which showed that the level of T cell infiltration was lower in the high-risk group. These results suggested that the risk characteristics of PYLRM can distinguish the different characteristics of tumor immune cells. Then, we calculated the expression levels of immune checkpoints in the two groups (Figure 6A). Of the 47 immune checkpoints we analyzed, 33 had expression differences between the groups (p < 0.05). The relationship between immune checkpoint and risk score was evaluated. There was a significant positive relationship between immune checkpoints (Figure 6B). It was found that CTLA4 (R = −0.21; p < 0.01), HAVCR2 (R = −0.21; p = 0.0048), and PD1 (R = −0.15; p < 0.01) were negatively related to risk score (Figures 6C–6E). In addition, the level of PD1 expression was lower in high-risk groups (Figure 6F). The expression level of immune checkpoints of high-risk group was lower, which also confirmed that there may be T cell failure in patients with high risk. The results mentioned above suggest that immunosuppressants acting on immune checkpoints, such as PD1, can be applied to of UCEC patients for immunotherapy. IPS analysis showed that IPS (p = 0.023), IPS_ ctla4 (p = 0.006), and IPS_ ctla4_ pdl1_ pd1_ pdl2 (p = 0.0071) had a higher level in low-risk group, but not IPS _ pdl1_ pd1_ pdL2 (p = 0.057), which also showed that patients at low risk were related to higher immunogenicity (Figures 6G–6J). According to the above results, we can infer that the high-risk UCEC group belongs to the cold tumors subtype and may have poor response to immunotherapy.
Figure 5.
The results of GSEA, ssGSEA, and ESTIMATE analysis
(A and B) Gene set enrichment analysis based on Kyoto Encyclopedia of Genes and Genomes (KEGG) of low-risk group (A) and high-risk group (B). (C and D) The difference of immune cells (C) and immune functions between the two risk groups (D) is shown. (E–H) The difference of immune score (E), stromal score (F), ESTIMATE score (G), and tumor purity (H) in high- and low-risk groups is shown. (I–L) The relationships between immune score (I), stromal score (J), ESTIMATE score (K), and tumor purity (L) and risk score are shown. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. ns, not significant.
Figure 6.
The correlations between immune checkpoint and risk score
(A) The boxplot shows the expression of immune checkpoints in high- and low-risk groups. (B) The correlations between immune checkpoints are shown. (C–E) The relationships between CTLA4 (C), HAVCR2 (D), and PDCD1 (E) and the risk score are shown. (F) The expression difference of PD1 between high-and low-risk groups. (G–J) The differences of IPS (G), IPS_ ctla4 (H), IPS_ ctla4_ pdl1_ pd1_ pdl2 (I), and IPS _ pdl1_ pd1_ pdL2 (J) in patients with different risk are shown. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. ns, not significant.
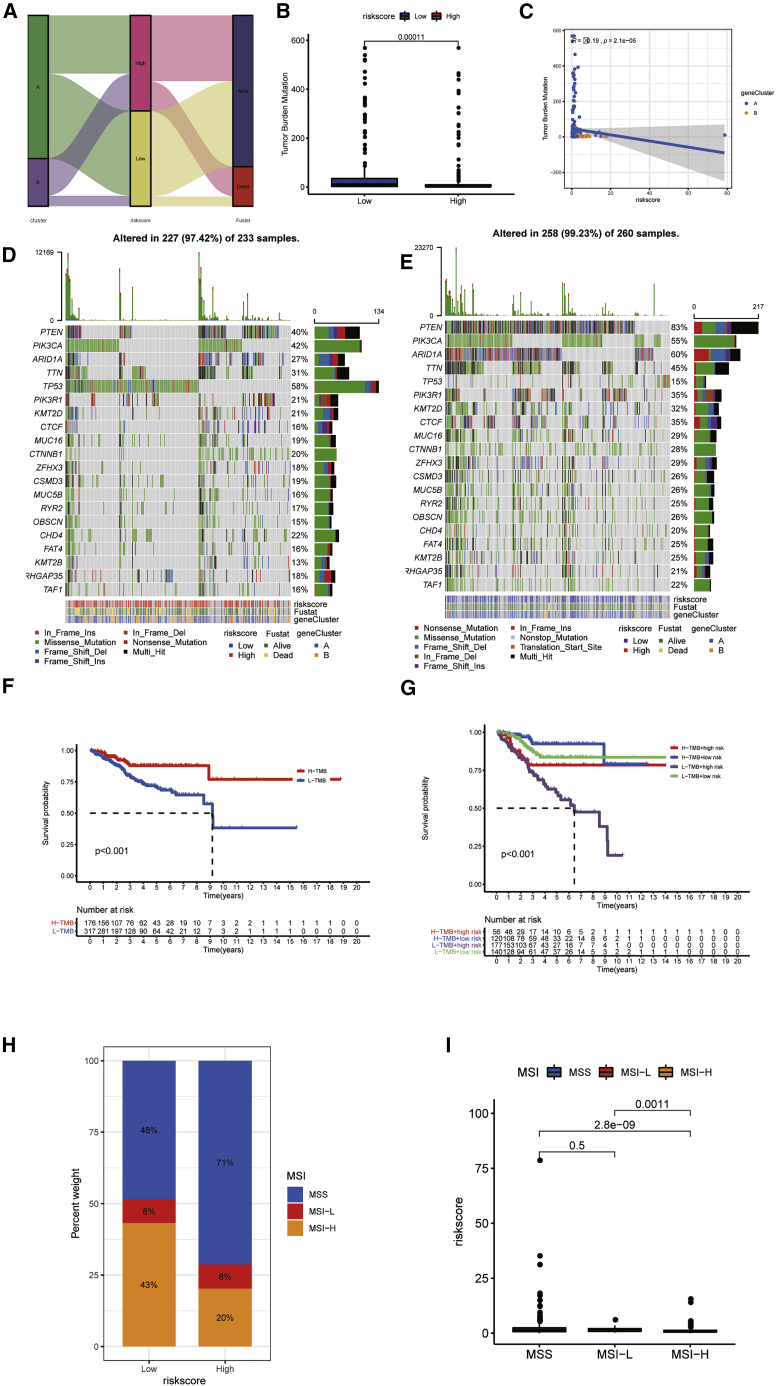
Prediction of immunotherapy effect TMB was employed to quantitatively evaluate the mutations taken by tumor cells, which is an effective molecular marker. Figure 7A showed the distribution of risk score and survival status of each sample. There were significant differences in TMBs between the two groups. TMB was higher in low-risk groups (Figure 7B). TMB was negatively related to the risk score (Figure 7C). Waterfall plot displayed mutation information of genes with high mutation frequency in high- (Figure 7D) and low-risk groups (Figure 7E). Survival probability of patients with high TMB was higher than patients with low TMB (p < 0.001; Figure 7F). The survival analysis of TMB combined with risk score indicated that the survival probability of patients with low TMB together with high risk was lower than other patients (Figure 7G). Microsatellite instability (MSI) is another tumor immune marker to reflect the effectiveness of immunotherapy. The proportion of three microsatellite states in two risk group is shown in Figure 7H. The proportion of MSI-high (MSI-H) in patients with higher risk score (20%) was lower than in patients with lower risk score (43%). The risk score of patients with MSI-H was lower than of patients with low MSI (p = 0.0011) and stable microsatellite (p < 0.001; Figure 7I). The stability of MSI in the low-risk group is poor, and the effect of immunotherapy may be better.
Figure 7.
The relationship between TMB and risk and survival
(A) The distribution of risk score and survival status of each sample. (B) The differences of TMB between high- and low-risk groups are shown. (C) Correlation between TMB and risk score is shown. (D and E) Waterfall plot displayed the mutation information of genes with high mutation frequency in high-risk group (D) and low-risk group (E). (F) Survival analysis of patients with different level of TMB is shown. (G) Survival analysis of patients with different TMBs combined with risk score is shown. (H) The percent weight of three microsatellite states in high- and low-risk groups is shown. (I) Differences in risk scores of microsatellites with different stabilities are shown.
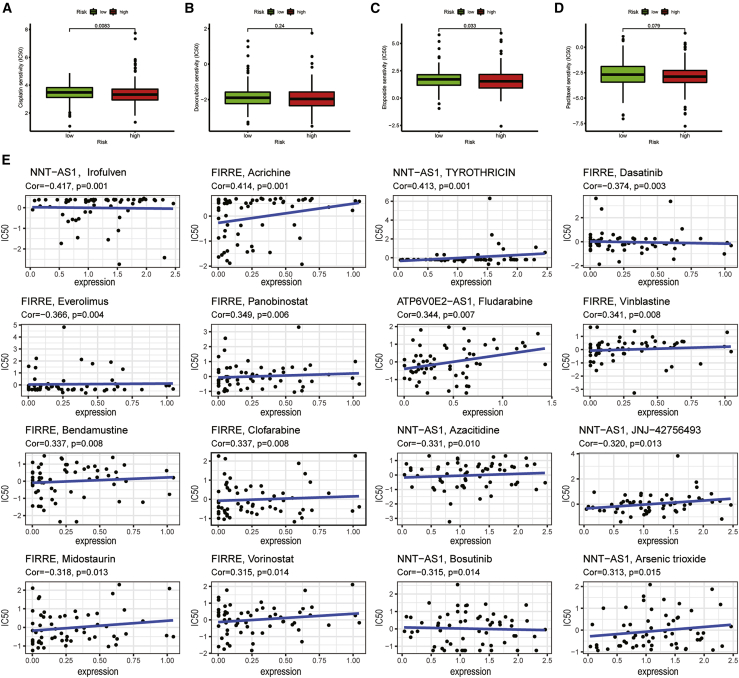
The sensitivity of chemotherapeutic drugs to UCEC
For the purpose of selecting more suitable chemotherapeutic drugs for UCEC patients, we evaluated the half-maximal inhibitory concentration (IC50) of four chemotherapeutic drugs in two risk groups and calculated the relationships between six PRLs and chemotherapeutic drugs. Paclitaxel (p = 0.079), cisplatin (p = 0.0083), etoposide (p = 0.033), and doxorubicin (p = 0.24) all had higher sensitivity in patients with lower risk score (Figures 8A–8D). It means that chemotherapy drugs may have better therapeutic effects in the low-risk group. Six PRLs were associated with the sensitivity of some chemotherapeutic drugs (p < 0.05; Figure 8E). For example, increased FIRRE expression was associated with increased resistance to midostaurin in UCEC patients, while the increase of FIRRE expression was related to the increased sensitivity of tumor cells to achine, vinblastine, bendamustine, clofarabine, and vorinostat.
Figure 8.
The relationships between six PRLs and drug sensitivity
(A–D) The differences of sensitivity of patients to cisplatin (A), doxorubicin (B), etoposide (C), and paclitaxel (D) in high- and low-risk groups. (E) Correlation between six PRLs and chemotherapeutic drugs is shown.
Finding potential drugs for UCEC
To clarify the new treatment scheme of UCEC, some potential therapeutic drugs for UCEC were screened from the connectivity map (CMAP) database. Functional enrichment analysis was employed to differential gene expression between the groups. In the Gene Ontology (GO) database, cilium organizing,collagen containing, and tubulin binding had the highest enrichments in biological process, cellular component, and molecular function, respectively (Figure S8A).The top ten related molecule drugs are listed in Table 4. Three small-molecule drugs (tacrolimus, aminoglutethimide, and emetine) with negative enrichment scores were selected to display the three-dimensional structure (Figures S8B–S8D). Due to lack of three-dimensional structure of tacrolimus, two-dimensional structure is shown instead.
Table 4.
Top ten results of CMAP analysis
| CMAP name | Mean | n | Enrichment | p value | Specificity | Percent non-null |
|---|---|---|---|---|---|---|
| Tomatidine | 0.363 | 4 | 0.816 | 0.00207 | 0 | 50 |
| Dirithromycin | 0.483 | 3 | 0.868 | 0.00427 | 0 | 66 |
| W-13 | −0.449 | 2 | −0.949 | 0.00561 | 0 | 100 |
| Glafenine | 0.357 | 4 | 0.737 | 0.00939 | 0.0355 | 50 |
| Ciclopirox | 0.477 | 4 | 0.711 | 0.01436 | 0.1159 | 75 |
| Tacrolimus | −0.353 | 3 | −0.785 | 0.02035 | 0.0645 | 66 |
| CAY-10397 | 0.492 | 3 | 0.784 | 0.02049 | 0.0305 | 66 |
| Capsaicin | 0.275 | 4 | 0.657 | 0.03177 | 0.0078 | 50 |
| Emetine | −0.32 | 4 | −0.602 | 0.06581 | 0.4588 | 50 |
| Aminoglutethimide | −0.535 | 3 | −0.659 | 0.07902 | 0.0595 | 66 |
Discussion
UCEC is a malignant gynecologic tumor.9,41 More and more evidence indicates that the uncommon expression of lncRNAs may be associated with the occurrence and development of many kinds of tumors.42, 43, 44, 45, 46, 47 Due to its high efficiency, high tissue specificity, and high stability, lncRNAs have the potential to become immunotherapeutic targets and biomarkers for UCEC diagnosis, prognosis, and treatment.48,49 Pyroptosis also has a close correlation with cancer progression.50, 51, 52 Inducing tumor cells apoptosis has been applied to eliminate malignant cells.53
In this study, a prognostic signature on the basis of PRLs was constructed and verified. PRLs may be an effective and reliable biomarker to forecast the prognosis of UCEC in the future. This is the first time analyzing the role of PRLs in UCEC.
The selection of 33 PRGs (AIM2, CASP1, CASP3, CASP4, CASP5, CASP6, CASP8, CASP9, ELANE, GPX4, GSDMA, GSDMB, GSDMC, GSDMD, GSDME, IL18, IL1B, IL6, NLRC4, NLRP1, NLRP2, NLRP3, NLRP6, NLRP7, NOD1, NOD2, PJVK, PLCG1, PRKACA, PYCARD, SCAF11, TIRAP, and TNF) refers to previous studies.54 The role of these genes in UCEC has not been studied before. Our study found that a large number of the 33 PRGs had different expressions between tumor and normal tissues, and there were interactions between the genes that may be used to judge the prognosis of UCEC.
The two clusters achieved from consensus clustering analysis had different prognosis, clinical features, and drug sensitivity of UCEC and had different level of PD-L1 expression, immune score, and immune-infiltrating cells. Cluster1 showed better prognosis, and the clinical symptoms of patients in cluster2 were more serious. The proportion of Treg cells was higher in cluster1, while the infiltration level of CD4 memory resetting T cells, gamma delta T cells, and M1 macrophages were higher in cluster2. It is considered that tumor-related macrophages can promote the invasion of UCEC tumor cells.55 The immune score assessed by estimation algorithm in cluster 1 was also higher than in cluster 2. Meanwhile, the immune infiltration level, TMB, PD1 expression level, HLA expression level, and IPS of cluster1 were higher, which means that people in cluster1 have higher immunogenicity. So it can be applied to forecast the immunotherapeutic effect of UCEC patients. Immune infiltration affects the survival rate of UCEC. Low TMB is related to low immune infiltration, which means poor immune response. TMB-related signals (GFAP, Edn3, CXCR3, plxna4, and SST) have been verified to be great predictors of the OS rate of UCEC.56 PD1 and PD-L1 inhibitors have important benefits for the immunotherapy of many types of recurrent or metastatic cancer,57, 58, 59 and UCEC showed high response to pembrolizumab, a PD1 immune checkpoint inhibitor.52,60,61 HLA alleles have been proven to stratify UCEC patients with high accuracy.62,63 The results indicated that cluster1 had negative correlation with immune-related functions, and cluster2 was positively correlated with immune-related functions. It may be the mechanism of different immunogenicity between the two groups. Then, we analyzed the drug sensitivity of the two clusters. Patients in cluster1 were more sensitive to etoposide, and cluster 2 patients were more sensitive to paclitaxel, which provided targeted guidance for patients with UCEC to choose therapeutic drugs.
After that, the PYLRM was constructed via multivariate Cox regression analyses. The survival status of people with low-risk score was better than high-risk persons in all three sets, indicating that the PYLRM has the potential to forecast the prognosis of UCEC patients. Among the six PRLs, the high expression of HM13-IT1, FIRRE, NNT-AS1, and ATP6V0E2-AS means that the patient has a high risk level and a poor prognosis. However, the high expression of AL353622.1 and POC1B-AS1 means that patients have a low risk level. Univariate and multivariate Cox regression analyses showed that the risk score could be an important independent prognostic factor for UCEC. Comparing the clinical characteristics of the two risk groups, it was found that there was significant discrepancy in age, grade, stage, historical type, and immune subtypes between the groups, and the prognosis of patients with different clinical characteristics is different, suggesting that the clinical characteristics also have the potential to forecast the prognosis of UCEC patients. However, the ROC curves suggested that the prognosis accuracy of risk score was much better. It is worth mentioning that the prediction of risk score combined with clinicopathological features was more accurate. Nomograph also confirms its excellent prediction ability.
Enrichment analysis based on GSEA suggested that the high-risk group was related to tumor-related pathways, while low-risk patients were closely related to immune-related pathways. This may explain why the high-risk group had poor prognosis. The results of ssGSEA and ESTIMATE showed that the scores of immune cells and immune function in patients with higher risk score were generally lower than people with lower risk score, which was in line with previous studies. There were statistical differences in the risk scores among the four UCEC immune subtypes, suggesting that immunotherapy may be effective for UCEC patients.
The infiltration of immune cells in UCEC was associated with the clinical prognosis.64 The expression level of tumor-infiltrating cells was different in the two risk groups. Memory B cells, M0 macrophages, M1 macrophages, M2 macrophages, and activated DCs had higher expression levels in patients with higher risk. The expression levels of CD8 cells, Treg cells, activated NK cells, and monocytes were higher in people with lower risk score, which can be used as a basis to select appropriate therapeutic targets and chemotherapeutic drugs. There were also some correlations between tumor-infiltrating cells and six PRLs. The risk score was positively related to memory B cells, activated DCs, M0 macrophages, and M1 macrophages, while it had negative relationships with monocytes and Treg cells. These results revealed that the risk characteristics of the PYLRM constructed by PRLs can distinguish the different factors of tumor immune cells in UCEC.
Immunotherapy has developed into an alternative or complementary therapeutic strategy to traditional radiotherapy and chemotherapy. CTLA-4, PD1, and PD-L1 antibodies showed great efficacy in tumor treatment especially.65 Previous literature also confirmed that advanced UCEC patients had great response to PD1 immunotherapy.60 In this study, the expression levels of immune checkpoints like CTLA4, HAVCR2, PDCD1, and PD1 significantly had negative correlations with risk score, so immunotherapy may be more effective in low-risk patients.
Cold tumor means that there are few infiltrating immune cells and a large proportion of immunosuppressive cells in the tumor, which means that the response to immunotherapy is weak.66 Our results suggested that patients with high risk and cluster2 belong to cold tumor subtype. The high-risk group and cluster2 are featured with immune infiltration reduction and T cells exhaustion, matching the definition of an “immune-desert” phenotype.67 This means that the immune surveillance functions of patients with higher risk score were weakened, which is conducive to immune escape, and the effect of immunotherapy will be poor.
IPS analysis indicated that the score of people with low risk was higher, which means that low-risk patients have higher immunogenicity. TMB is regarded as another important indicator of immunotherapy response. In most kinds of cancer, the higher the level of TMB, the longer the OS after immunotherapy;39,40,68 it was the same in UCEC. It was further proven that immunotherapy may be effective in UCEC patients. MSI is a phenomenon that arises in some tumors when the number of repeat units of specific microsatellite sites changed.69 MSI has been used in the diagnosis and treatment of patients with colorectal cancer. MSI status may affect the efficacy of ICIs by changing the TME of tumor. In colorectal cancer, patients with MSI-H benefit from ICIs treatment compared with MSS/low MSI (MSI-L) tumor patients.70 The stability of MSI in the low-risk group was poor, and the effect of immunotherapy was better, which was consistent with the previous results.
Paclitaxel, cisplatin, etoposide, and doxorubicin had higher sensitivity in the low-risk group. This means that the effect of chemotherapeutic drugs may have better therapeutic effect in the low-risk patients. Previous research has also verified that paclitaxel is a potential immunotherapeutic drug for UCEC.71
Due to immunotherapy and chemotherapy in patients with higher risk score being unsatisfactory, we consider that the combination of small molecular compounds may be effective. Through GO analysis, we enriched the differentially expressed genes in different risk groups. Through CMAP, we screened some potential small chemotherapeutic drugs for UCEC, such as tacrolimus, aminoglutethimide, and emetine. Aminoglutethimide can be used to treat Cushing's syndrome and breast cancer.72 Emetine can treat lung cancer in cooperation with cisplatin.73 The roles of aminoglutethimide and emetine in the treatment of UCEC still need further analysis.
Our study identified six UCEC prognostic markers associated with pyroptosis: HM13-IT1; FIRRE; NNT-AS1; ATP6V0E2-AS1; AL353622.1; and POC1B-AS1. Among these biomarkers, there is no research reporting the role of FIRRE and NNT-AS1 in UCEC. But NNT-AS1, the regulatory mechanism of microRNA (miRNA) sponge, may affect tumor cell proliferation, invasion, metastasis, and apoptosis74 and has been found to be involved in the progress of pneumonia.75 FIRRE can promote the growth of DLBCL cells.76 There are few reports that focus on HM13-IT1, POC1B-AS1, ATP6V0E2-AS1, and AL353622.1, but they have potential to be the prognostic biomarkers of UCEC.
Our study also has some deficiencies. First and foremost, insufficient raw data and small sample size for analysis may have some impact on the results. In future research, it is necessary to further improve the sample size, sequencing data, and clinical information of UCEC patients. In addition, our research is the result of bioinformatics analysis based on the dataset of a public database. It is necessary to further confirm the accuracy of the model through experiments and clinical studies.
Materials and methods
Data acquisition
Transcriptome and RNA sequencing (RNA-seq) profiles of UCEC were taken from the TCGA database: https://portal.gdc.cancer.gov/,77 and the transcriptome data files were “FPKM.” The following inclusion criteria were used: (1) removing all samples without clinical follow-up information and then (2) removing all samples with unknown survival time, <30 days, and no survival status. Finally, 511 patients with UCEC and the corresponding clinical information, such as age, grade, stage, and histological type, were enrolled for further study. All 511 patients with UCEC were randomly assigned into the training set (256 patients) and testing set (255 patients) at a 1:1 ratio via the caret package. Clinicopathology features of the samples are presented in Table 5 (p > 0.05; chi-square test).
Table 5.
The statistical data of clinical characteristics of the three sets
| Covariates | Type | Total | Test | Train | p value |
|---|---|---|---|---|---|
| Age | ≤60 | 199 (38.94%) | 94 (36.86%) | 105 (41.02%) | 0.3833 |
| Age | >60 | 312 (61.06%) | 161 (63.14%) | 151 (58.98%) | |
| Histological_type | endometrial | 384 (75.15%) | 192 (75.29%) | 192 (75%) | 1 |
| Histological_type | mixed and serous | 127 (24.85%) | 63 (24.71%) | 64 (25%) | |
| Grade | G1 and G2 | 91 (17.81%) | 46 (18.04%) | 45 (17.58%) | 0.9836 |
| Grade | G3 and G4 | 420 (82.19%) | 209 (81.96%) | 211 (82.42%) | |
| Stage | stage I and stage II | 370 (72.41%) | 188 (73.73%) | 182 (71.09%) | 0.571 |
| Stage | stage III and stage IV | 141 (27.59%) | 67 (26.27%) | 74 (28.91%) |
Identification of PRGs
Thirty-three PRGs were achieved from previous studies,16,54,78 including AIM2, CASP1, CASP3, CASP4, CASP5, CASP6, CASP8, CASP9, ELANE, GPX4, GSDMA, GSDMB, GSDMC, GSDMD, GSDME, IL18, IL1B, IL6, NLRC4, NLRP1, NLRP2, NLRP3, NLRP6, NLRP7, NOD1, NOD2, PJVK, PLCG1, PRKACA, PYCARD, SCAF11, TIRAP, and TNF. Profiles of 33 PRGs expression and clinicopathology characteristics of UCEC patients were achieved from the TCGA. “Limma” package was used to identify the different expressions of PRGs between UCEC and normal tissues. Heatmap showed the expression of 33 PRGs in tumor and normal tissues. Boxplot was used to show the differences of these genes between different tissues. In addition, volcano plot visualized the expression of these genes.
Constructing a PPI network
This study employed the interactive gene search tools STRING database: http://string-db.org/ to estimate the correlation between 33 PRGs, and the PPI network was constructed.79,80 Bar plot showed genes with more nodes and their quantity of nodes.
Screening PRLs
We identified 384 lncRNAs that had close correlation with PRGs from the TCGA database based on Pearson analysis. The standard used in this part was Pearson R > 0.5 and p < 0.001. Subsequently, univariate Cox regression analysis was conducted on the training set to screen 13 PRLs that had potential prognosis value of UCEC.81
Consensus clustering analysis
A consensus cluster consisting of 13 PRLs was constructed on the base of “ConsensusClusterPlus” package(K means cluster count).82 According to the similarity of PRLs’ expression level and the proportion of fuzzy clustering measurement, it was found that, when k = 2, the cluster had the best stability. Therefore, 256 patients with UCEC were assigned into two clusters: cluster1 (n = 192) and cluster2 (n = 64). After that, the differences of lncRNA expression, immunity, and clinical characteristics between the two clusters were compared.
Constructing and evaluating prognostic model
The model was constructed in the light of the training set; meanwhile, the testing set and the entire set were applied to test the predicted ability of the model. Multivariate Cox regression analysis was conducted in the training set and identified six PRLs.83 We named this model as “PYLRM.” The following formula was employed to evaluate the risk score: risk score = coef (lncRNA1) × expr (lncRNA1) + coef (lncRNA2) × expr (lncRNA2) + … + coef (lncRNAn) × expr (lncRNAn), where coef means the coefficients, coef (lncRNAn) indicates the coefficient of lncRNAs related to survival, and expr (lncRNAn) is the expression of lncRNAs.
Evaluation of predict ability of the PYLRM
In the light of the median risk score, all the samples were assigned into two groups. Kaplan-Meier was employed to calculate the OS.84,85 PCA was utilized to visualize the diversity of the groups.86 The distribution of clinical characteristics between two risk groups was visualized by “pheatmap” R package. Univariate and multivariate Cox regression analyses were applied to assess whether the risk score was an independent prognostic factor of UCEC patients when combined with other clinical characteristics.
Establishment and verity of a predictive nomogram
A nomogram was established in the light of risk score and clinicopathology factors that were applied to predict OS incidence rate in 1, 3, and 5 years. The modified curve calculated by Hosmer-Lemeshow test was utilized to show the consistency between the actual results and the predicted results.87 AUC and ROC curve were applied to evaluate the prognostic value of clinicopathological features.88 The AUC >0.6 showed that the PYLRM can accurately forecast the survival rate of UCEC patients.
GSEA and ssGSEA
For the purpose of clarifying the potential regulation mechanism causing the difference between the two clusters, we conducted GSEA. Profiles of gene sets in GSEA provide stable and interpretable measurement of biological function and pathways.89 ssGSEA was utilized to verify the differences of immune cells and immune function between the groups. ssGSEA worked at the single-sample level and is an extension of the GSEA method.90
Immune microenvironment assessment
Through CIBERSORT: http://cibersort.stanford.edu/, our study analyzed the composition and infiltration level of 21 infiltrating immune cells. CIBERSORT is an algorithm that can accurately evaluate the immune components of immune cells from complex gene expression profiles of tissues.91,92 After that, ESTIMATE was used to verify the discrepancy of immune microenvironment between different risk groups. The immune score, stromal score, estimate score, and tumor purity of each sample were evaluated by R “estimate” package. These scores reflect the proportion of immune and stromal components in TME. The relationships between immune score, stromal score, estimated score, tumor purity, and risk score were also evaluated by Pearson correlation coefficient method.
Mutational analysis
TMB indicates the number of somatic mutations per megabase genome sequence, which can be used to find patients who have more possibility to respond to ICIs.93 This study evaluated the difference of TMB between two risk groups and its relationship with risk score.
Immunotherapy response prediction
IPS range from 0 to 10 was evaluated in the light of the gene expression Z score. IPS level of the low-risk group was higher, which means patients with low risk have higher immunogenicity.94 The results were downloaded from the The Cancer Imaging Archive (TCIA) database.95 MSI refers to the change of allele size between tumor tissue and its corresponding normal tissue.96 MSI-H patients are more immunogenic than MSI-L and MSS patients. Meanwhile, the effect of immunotherapy for MSI-H patients will be better. The information about MSI was also downloaded from the TCIA database.
Analysis of drug sensitivity
By using R software package “PRROPHIC,” we evaluated IC50 of four chemotherapeutic drugs of two risk groups. The correlation between six PRLs and the sensitivity of some chemotherapeutic drugs was displayed by ggplot2.97 The NCI-60 database includes 60 different tumors cells from nine cancers evaluated by using CellMiner interface: https://discover.nci.nih.gov/cellminer.98,99 Pearson correlation analysis was conducted to explore the correlations between PRL expression and drug sensitivity.
Functional analysis
We used GO analysis to screen the differentially expressed genes (DEGs). This analysis utilized R package “clusterProfiler.” The threshold of GO analysis was decided by the q value, and p < 0.05 means that the functions were significantly enriched.
Exploration of potential chemotherapeutic drugs
CMAP was used to screen potential drugs for UCEC treatment.100 As we stated before, these small-molecule drugs were identified through 662 DEGs between two risk groups with | log2 fold change (FC) | > 1 and false discovery rate (FDR) < 0.05. The two-dimensional and three-dimensional structures of the drugs were taken from PubChem.101,102
Statistical analysis
R v.4.1.0 was used to do statistical tests. The differences of the two subgroups were calculated by Student’s t test and ANOVA. Kaplan-Meier analysis and log rank test were employed to calculate the discrepancy of OS between the two risk groups. The relationships between risk score and immune infiltration level were calculated by Pearson correlation test. p < 0.05 was defined to have statistical difference.
Availability of data and material
The data used to support the findings of this study are available from the corresponding author on reasonable request.
Acknowledgments
Not applicable.
Author contributions
J.B. and J.L. contributed to the conception of the study; L.C. contributed significantly to analysis and manuscript preparation; R.G. and S.N. performed the data analyses and wrote the manuscript; and S.Y. and F.S. helped perform the analysis with constructive discussions.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.01.018.
Supplemental information
References
- 1.Wang Y., Ren F., Song Z., Wang X., Ma X. Multiomics profile and prognostic gene signature of m6A regulators in uterine corpus endometrial carcinoma. J. Cancer. 2020;11:6390–6401. doi: 10.7150/jca.46386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 4.Bokhman J.V. Two pathogenetic types of endometrial carcinoma. Gynecol. Oncol. 1983;15:10–17. doi: 10.1016/0090-8258(83)90111-7. [DOI] [PubMed] [Google Scholar]
- 5.Colombo N., Creutzberg C., Amant F., Bosse T., Gonzalez-Martin A., Ledermann J., Marth C., Nout R., Querleu D., Mirza M.R., et al. ESMO-ESGO-ESTRO consensus conference on endometrial cancer: diagnosis, treatment and follow-up. Ann. Oncol. 2016;27:16–41. doi: 10.1097/IGC.0000000000000609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang X., Tang H., Chen T. Epidemiology of gynecologic cancers in China. J. Gynecol. Oncol. 2018;29:e7. doi: 10.3802/jgo.2018.29.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaudhry P., Asselin E. Resistance to chemotherapy and hormone therapy in endometrial cancer. Endocr. Relat. Cancer. 2009;16:363–380. doi: 10.1677/ERC-08-0266. [DOI] [PubMed] [Google Scholar]
- 8.Li B.L., Wan X.P. Prognostic significance of immune landscape in tumour microenvironment of endometrial cancer. J. Cell Mol. Med. 2020;24:7767–7777. doi: 10.1111/jcmm.15408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morice P., Leary A., Creutzberg C., Abu-Rustum N., Darai E. Endometrial cancer. Lancet. 2016;387:1094–1108. doi: 10.1016/S0140-6736(15)00130-0. [DOI] [PubMed] [Google Scholar]
- 10.Setiawan V.W., Yang H.P., Pike M.C., McCann S.E., Yu H., Xiang Y.B., Wolk A., Wentzensen N., Weiss N.S., Webb P.M., et al. Type I and II endometrial cancers: have they different risk factors? J. Clin. Oncol. 2013;31:2607–2618. doi: 10.1200/JCO.2012.48.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 12.Cerella C., Teiten M.H., Radogna F., Dicato M., Diederich M. From nature to bedside: pro-survival and cell death mechanisms as therapeutic targets in cancer treatment. Biotechnol. Adv. 2014;32:1111–1122. doi: 10.1016/j.biotechadv.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Crowley L.C., Marfell B.J., Scott A.P., Boughaba J.A., Chojnowski G., Christensen M.E., et al. Dead cert: measuring cell death. Cold Spring Harb. Protoc. 2016;2016 doi: 10.1101/pdb.top070318. [DOI] [PubMed] [Google Scholar]
- 14.Fang Y., Tian S., Pan Y., Li W., Wang Q., Tang Y., Yu T., Wu X., Shi Y., Ma P., et al. Pyroptosis: a new frontier in cancer. Biomed. Pharmacother. 2020;121:109595. doi: 10.1016/j.biopha.2019.109595. [DOI] [PubMed] [Google Scholar]
- 15.Cookson B.T., Brennan M.A. Pro-inflammatory programmed cell death. Trends Microbiol. 2001;9:113–114. doi: 10.1016/s0966-842x(00)01936-3. [DOI] [PubMed] [Google Scholar]
- 16.Xia X., Wang X., Cheng Z., Qin W., Lei L., Jiang J., Hu J. The role of pyroptosis in cancer: pro-cancer or pro-“host”? Cell Death Dis. 2019;10:650. doi: 10.1038/s41419-019-1883-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thi H.T.H., Hong S. Inflammasome as a therapeutic target for cancer prevention and treatment. J. Cancer Prev. 2017;22:62–73. doi: 10.15430/JCP.2017.22.2.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou C.B., Fang J.Y. The role of pyroptosis in gastrointestinal cancer and immune responses to intestinal microbial infection. Biochim. Biophys. Acta Rev. Cancer. 2019;1872:1–10. doi: 10.1016/j.bbcan.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Ruan J., Wang S., Wang J. Mechanism and regulation of pyroptosis-mediated in cancer cell death. Chem. Biol. Interact. 2020;323:109052. doi: 10.1016/j.cbi.2020.109052. [DOI] [PubMed] [Google Scholar]
- 20.Jia C., Chen H., Zhang J., Zhou K., Zhuge Y., Niu C., Qiu J., Rong X., Shi Z., Xiao J., et al. Role of pyroptosis in cardiovascular diseases. Int. Immunopharmacol. 2019;67:311–318. doi: 10.1016/j.intimp.2018.12.028. [DOI] [PubMed] [Google Scholar]
- 21.Pereira N.S., Queiroga T.B.D., Nunes D.F., Andrade C.M., Nascimento M.S.L., Do-Valle-Matta M.A., da Camara A.C.J., Galvao L., Guedes P.M.M., Chiari E. Innate immune receptors over expression correlate with chronic chagasic cardiomyopathy and digestive damage in patients. PLoS Negl. Trop. Dis. 2018;12 doi: 10.1371/journal.pntd.0006589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi J., Gao W., Shao F. Pyroptosis: gasdermin-mediated programmed necrotic cell death. Trends Biochem. Sci. 2017;42:245–254. doi: 10.1016/j.tibs.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Kovacs S.B., Miao E.A. Gasdermins: effectors of pyroptosis. Trends Cell Biol. 2017;27:673–684. doi: 10.1016/j.tcb.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia S., Hollingsworth L.R.t., Wu H. Mechanism and regulation of gasdermin-mediated cell death. Cold Spring Harb. Perspect. Biol. 2020;12:a036400. doi: 10.1101/cshperspect.a036400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J., Yao L., Zhang M., Jiang J., Yang M., Wang Y. Downregulation of LncRNA-XIST inhibited development of non-small cell lung cancer by activating miR-335/SOD2/ROS signal pathway mediated pyroptotic cell death. Aging. 2019;11:7830–7846. doi: 10.18632/aging.102291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren N., Jiang T., Wang C., Xie S., Xing Y., Piao D., Zhang T., Zhu Y. LncRNA ADAMTS9-AS2 inhibits gastric cancer (GC) development and sensitizes chemoresistant GC cells to cisplatin by regulating miR-223-3p/NLRP3 axis. Aging. 2020;12:11025–11041. doi: 10.18632/aging.103314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jonkhout N., Tran J., Smith M.A., Schonrock N., Mattick J.S., Novoa E.M. The RNA modification landscape in human disease. RNA. 2017;23:1754–1769. doi: 10.1261/rna.063503.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X., Li N. LncRNAs on guard. Int. Immunopharmacol. 2018;65:60–63. doi: 10.1016/j.intimp.2018.09.031. [DOI] [PubMed] [Google Scholar]
- 29.Zhou X., Liu S., Cai G., Kong L., Zhang T., Ren Y., Wu Y., Mei M., Zhang L., Wang X. Long non coding RNA MALAT1 promotes tumor growth and metastasis by inducing epithelial-mesenchymal transition in oral squamous cell carcinoma. Sci. Rep. 2015;5:15972. doi: 10.1038/srep15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tripathi V., Shen Z., Chakraborty A., Giri S., Freier S.M., Wu X., Zhang Y., Gorospe M., Prasanth S.G., Lal A., et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PloS Genet. 2013;9 doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atianand M.K., Fitzgerald K.A. Long non-coding RNAs and control of gene expression in the immune system. Trends Mol. Med. 2014;20:623–631. doi: 10.1016/j.molmed.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vallone C., Rigon G., Gulia C., Baffa A., Votino R., Morosetti G., Zaami S., Briganti V., Catania F., Gaffi M., et al. Non-Coding RNAs and endometrial cancer. Genes. 2018;9:187. doi: 10.3390/genes9040187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen B.J., Byrne F.L., Takenaka K., Modesitt S.C., Olzomer E.M., Mills J.D., Farrell R., Hoehn K.L., Janitz M. Transcriptome landscape of long intergenic non-coding RNAs in endometrial cancer. Gynecol. Oncol. 2017;147:654–662. doi: 10.1016/j.ygyno.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 34.Diaz G., Melis M., Tice A., Kleiner D.E., Mishra L., Zamboni F., Farci P. Identification of microRNAs specifically expressed in hepatitis C virus-associated hepatocellular carcinoma. Int. J. Cancer. 2013;133:816–824. doi: 10.1002/ijc.28075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fatima R., Akhade V.S., Pal D., Rao S.M. Long noncoding RNAs in development and cancer: potential biomarkers and therapeutic targets. Mol. Cell Ther. 2015;3:5. doi: 10.1186/s40591-015-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smolle M.A., Bullock M.D., Ling H., Pichler M., Haybaeck J. Long non-coding RNAs in endometrial carcinoma. Int. J. Mol. Sci. 2015;16:26463–26472. doi: 10.3390/ijms161125962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo Q., Qian Z., Yan D., Li L., Huang L. LncRNA-MEG3 inhibits cell proliferation of endometrial carcinoma by repressing Notch signaling. Biomed. Pharmacother. 2016;82:589–594. doi: 10.1016/j.biopha.2016.02.049. [DOI] [PubMed] [Google Scholar]
- 38.Hutt S., Tailor A., Ellis P., Michael A., Butler-Manuel S., Chatterjee J. The role of biomarkers in endometrial cancer and hyperplasia: a literature review. Acta Oncol. 2019;58:342–352. doi: 10.1080/0284186X.2018.1540886. [DOI] [PubMed] [Google Scholar]
- 39.Samstein R.M., Lee C.H., Shoushtari A.N., Hellmann M.D., Shen R., Janjigian Y.Y., Barron D.A., Zehir A., Jordan E.J., Omuro A., et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019;51:202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yarchoan M., Hopkins A., Jaffee E.M. Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med. 2017;377:2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 42.Dong H.X., Wang R., Jin X.Y., Zeng J., Pan J. LncRNA DGCR5 promotes lung adenocarcinoma (LUAD) progression via inhibiting I-mir-22-3p. J. Cell Physiol. 2018;233:4126–4136. doi: 10.1002/jcp.26215. [DOI] [PubMed] [Google Scholar]
- 43.Zhuang C., Ma Q., Zhuang C., Ye J., Zhang F., Gui Y. LncRNA GClnc1 promotes proliferation and invasion of bladder cancer through activation of MYC. FASEB J. 2019;33:11045–11059. doi: 10.1096/fj.201900078RR. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X.Z., Mao H.L., Zhang S.J., Sun L., Zhang W.J., Chen Q.Z., Wang L., Liu H.C. lncRNA PCAT18 inhibits proliferation, migration and invasion of gastric cancer cells through miR-135b suppression to promote CLDN11 expression. Life Sci. 2020;249:117478. doi: 10.1016/j.lfs.2020.117478. [DOI] [PubMed] [Google Scholar]
- 45.Martens-Uzunova E.S., Bottcher R., Croce C.M., Jenster G., Visakorpi T., Calin G.A. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur. Urol. 2014;65:1140–1151. doi: 10.1016/j.eururo.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 46.Terracciano D., Ferro M., Terreri S., Lucarelli G., D'Elia C., Musi G., de Cobelli O., Mirone V., Cimmino A. Urinary long noncoding RNAs in nonmuscle-invasive bladder cancer: new architects in cancer prognostic biomarkers. Transl. Res. 2017;184:108–117. doi: 10.1016/j.trsl.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 47.He Y., Hu H., Wang Y., Yuan H., Lu Z., Wu P., Liu D., Tian L., Yin J., Jiang K., et al. ALKBH5 inhibits pancreatic cancer motility by decreasing long non-coding RNA KCNK15-AS1 methylation. Cell Physiol. Biochem. 2018;48:838–846. doi: 10.1159/000491915. [DOI] [PubMed] [Google Scholar]
- 48.Rao A., Rajkumar T., Mani S. Perspectives of long non-coding RNAs in cancer. Mol. Biol. Rep. 2017;44:203–218. doi: 10.1007/s11033-017-4103-6. [DOI] [PubMed] [Google Scholar]
- 49.Malik B., Feng F.Y. Long noncoding RNAs in prostate cancer: overview and clinical implications. Asian J. Androl. 2016;18:568–574. doi: 10.4103/1008-682X.177123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu M., Wang Y., Yang D., Gong Y., Rao F., Liu R., Danna Y., Li J., Fan J., Chen J., et al. A PLK1 kinase inhibitor enhances the chemosensitivity of cisplatin by inducing pyroptosis in oesophageal squamous cell carcinoma. EBioMedicine. 2019;41:244–255. doi: 10.1016/j.ebiom.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruiz-Patino A., Barron F., Cardona A.F., Corrales L., Mas L., Martin C., Zatarain-Barron Z.L., Recondo G., Ricaurte L., Rojas L., et al. Antibiotics impair immune checkpoint inhibitor effectiveness in Hispanic patients with non-small cell lung cancer (AB-CLICaP) Thorac. Cancer. 2020;11:2552–2560. doi: 10.1111/1759-7714.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pezuk J.A. Pyroptosis in combinatorial treatment to improve cancer patie’ts' outcome, is that what we want? EBioMedicine. 2019;41:17–18. doi: 10.1016/j.ebiom.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fulda S. Targeting apoptosis for anticancer therapy. Semin. Cancer Biol. 2015;31:84–88. doi: 10.1016/j.semcancer.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 54.Ye Y., Dai Q., Qi H. A novel defined pyroptosis-related gene signature for predicting the prognosis of ovarian cancer. Cell Death Discov. 2021;7:71. doi: 10.1038/s41420-021-00451-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jing X., Peng J., Dou Y., Sun J., Ma C., Wang Q., Zhang L., Luo X., Kong B., Zhang Y., et al. Macrophage ERalpha promoted invasion of endometrial cancer cell by mTOR/KIF5B-mediated epithelial to mesenchymal transition. Immunol. Cell Biol. 2019;97:563–576. doi: 10.1111/imcb.12245. [DOI] [PubMed] [Google Scholar]
- 56.Zhou H., Chen L., Lei Y., Li T., Li H., Cheng X. Integrated analysis of tumor mutation burden and immune infiltrates in endometrial cancer. Curr. Probl. Cancer. 2021;45:100660. doi: 10.1016/j.currproblcancer.2020.100660. [DOI] [PubMed] [Google Scholar]
- 57.Makker V., Rasco D., Vogelzang N.J., Brose M.S., Cohn A.L., Mier J., Di Simone C., Hyman D.M., Stepan D.E., Dutcus C.E., et al. Lenvatinib plus pembrolizumab in patients with advanced endometrial cancer: an interim analysis of a multicentre, open-label, single-arm, phase 2 trial. Lancet Oncol. 2019;20:711–718. doi: 10.1016/S1470-2045(19)30020-8. [DOI] [PubMed] [Google Scholar]
- 58.Brahmer J., Reckamp K.L., Baas P., Crino L., Eberhardt W.E., Poddubskaya E., Antonia S., Pluzanski A., Vokes E.E., Holgado E., et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N. Engl. J. Med. 2015;373:123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weber J.S., D'Angelo S.P., Minor D., Hodi F.S., Gutzmer R., Neyns B., Hoeller C., Khushalani N.I., Miller W.H., Jr., Lao C.D., et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037):andomizedsed, controlled, open-label, phase 3 trial. Lancet Oncol. 2015;16:375–384. doi: 10.1016/S1470-2045(15)70076-8. [DOI] [PubMed] [Google Scholar]
- 60.Ott P.A., Bang Y.J., Berton-Rigaud D., Elez E., Pishvaian M.J., Rugo H.S., Puzanov I., Mehnert J.M., Aung K.L., Lopez J., et al. Safety and antitumor activity of pembrolizumab in advanced programmed death ligand 1-positive endometrial cancer: results from the KEYNOTE-028 study. J. Clin. Oncol. 2017;35:2535–2541. doi: 10.1200/JCO.2017.72.5952. [DOI] [PubMed] [Google Scholar]
- 61.Lee V., Murphy A., Le D.T., Diaz L.A., Jr. Mismatch repair deficiency and response to immune checkpoint blockade. Oncologist. 2016;21:1200–1211. doi: 10.1634/theoncologist.2016-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Callahan B.M., Tong W.L., Blanck G. T cell receptor-beta J usage, in combination with particular HLA class II alleles, correlates with better cancer survival rates. Immunol. Res. 2018;66:219–223. doi: 10.1007/s12026-018-8990-y. [DOI] [PubMed] [Google Scholar]
- 63.Clark K.R., Tong W.L., Callahan B.M., Yavorski J.M., Tu Y.N., Blanck G. TRB-J1 usage, in combination with the HLA-A∗01:01 allele, represents an apparent survival advantage for uterine corpus endometrial carcinoma: comparisons with microscopic assessments of lymphocyte infiltrates. Int. J. Immunogenet. 2019;46:31–37. doi: 10.1111/iji.12409. [DOI] [PubMed] [Google Scholar]
- 64.Zou R., Zheng M., Tan M., Xu H., Luan N., Zhu L. Decreased PTGDS expression predicting poor survival of endometrial cancer by integrating weighted gene Co-expression network analysis and immunohistochemical validation. Cancer Manag. Res. 2020;12:5057–5075. doi: 10.2147/CMAR.S255753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hugo W., Zaretsky J.M., Sun L., Song C., Moreno B.H., Hu-Lieskovan S., Berent-Maoz B., Pang J., Chmielowski B., Cherry G., et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2017;168:542. doi: 10.1016/j.cell.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 66.Nair S.S., Weil R., Dovey Z., Davis A., Tewari A.K. The tumor microenvironment and immunotherapy in prostate and bladder cancer. Urol. Clin. North Am. 2020;47:e17–e54. doi: 10.1016/j.ucl.2020.10.005. [DOI] [PubMed] [Google Scholar]
- 67.Zhang B., Wu Q., Li B., Wang D., Wang L., Zhou Y.L. m(6)A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol. Cancer. 2020;19:53. doi: 10.1186/s12943-020-01170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goodman A.M., Kato S., Bazhenova L., Patel S.P., Frampton G.M., Miller V., Stephens P.J., Daniels G.A., Kurzrock R. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol. Cancer Ther. 2017;16:2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tomlinson I., Halford S., Aaltonen L., Hawkins N., Ward R. Does MSI-low exist? J. Pathol. 2002;197:6–13. doi: 10.1002/path.1071. [DOI] [PubMed] [Google Scholar]
- 70.Lin A., Zhang J., Luo P. Crosstalk between the MSI status and tumor microenvironment in colorectal cancer. Front. Immunol. 2020;11:2039. doi: 10.3389/fimmu.2020.02039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaur D., Arora C., Raghava G.P.S. Prognostic biomarker-based identification of drugs for managing the treatment of endometrial cancer. Mol. Diagn. Ther. 2021;25:629–646. doi: 10.1007/s40291-021-00539-1. [DOI] [PubMed] [Google Scholar]
- 72.Santen R.J., Misbin R.I. Aminoglutethimide: review of pharmacology and clinical use. Pharmacotherapy. 1981;1:95–120. doi: 10.1002/j.1875-9114.1981.tb03557.x. [DOI] [PubMed] [Google Scholar]
- 73.Wu T.H., Chang S.Y., Shih Y.L., Huang T.W., Chang H., Lin Y.W. Emetine synergizes with cisplatin to enhance anti-cancer efficacy against lung cancer cells. Int. J. Mol. Sci. 2019;20:5914. doi: 10.3390/ijms20235914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou C., Duan S. The role of long non-coding RNA NNT-AS1 in neoplastic disease. Cancers. 2020;12:3086. doi: 10.3390/cancers12113086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang J.W., Luo X.Y., Li Z.H., Lang B.P. LncRNA NNT-AS1 regulates the progression of lung cancer through the NNT-AS1/miR-3666/E2F2 axis. Eur. Rev. Med. Pharmacol. Sci. 2020;24:238–248. doi: 10.26355/eurrev_202001_19916. [DOI] [PubMed] [Google Scholar]
- 76.Shi X., Cui Z., Liu X., Wu S., Wu Y., Fang F., Zhao H. LncRNA FIRRE is activated by MYC and promotes the development of diffuse large B-cell lymphoma via Wnt/beta-catenin signaling pathway. Biochem. Biophys. Res. Commun. 2019;510:594–600. doi: 10.1016/j.bbrc.2019.01.105. [DOI] [PubMed] [Google Scholar]
- 77.Tomczak K., Czerwinska P., Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Karki R., Kanneganti T.D. Diverging inflammasome signals in tumorigenesis and potential targeting. Nat. Rev. Cancer. 2019;19:197–214. doi: 10.1038/s41568-019-0123-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li T., Fu J., Zeng Z., Cohen D., Li J., Chen Q., Li B., Liu X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lian H., Han Y.P., Zhang Y.C., Zhao Y., Yan S., Li Q.F., Wang B.C., Wang J.J., Meng W., Yang J., et al. Integrative analysis of gene expression and DNA methylation through one-class logistic regression machine learning identifies stemness features in medulloblastoma. Mol. Oncol. 2019;13:2227–2245. doi: 10.1002/1878-0261.12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tao C., Huang K., Shi J., Hu Q., Li K., Zhu X. Genomics and prognosis analysis of epithelial-mesenchymal transition in glioma. Front. Oncol. 2020;10:183. doi: 10.3389/fonc.2020.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Su J., Miao L.F., Ye X.H., Cui M.S., He X.F. Development of prognostic signature and nomogram for patients with breast cancer. Medicine. 2019;98 doi: 10.1097/MD.0000000000014617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ranstam J., Cook J.A. Kaplan-Meier curve. Br. J. Surg. 2017;104:442. doi: 10.1002/bjs.10238. [DOI] [PubMed] [Google Scholar]
- 85.Barakat A., Mittal A., Ricketts D., Rogers B.A. Understanding survival analysis: actuarial life tables and the Kaplan-Meier plot. Br. J. Hosp. Med. 2019;80:642–646. doi: 10.12968/hmed.2019.80.11.642. [DOI] [PubMed] [Google Scholar]
- 86.Martin-Clemente R., Zarzoso V. On the link between L1-PCA and ICA. IEEE Trans. Pattern Anal. Mach. Intell. 2017;39:515–528. doi: 10.1109/TPAMI.2016.2557797. [DOI] [PubMed] [Google Scholar]
- 87.Hoshino N., Hida K., Sakai Y., Osada S., Idani H., Sato T., Takii Y., Bando H., Shiomi A., Saito N. Nomogram for predicting anastomotic leakage after low anterior resection for rectal cancer. Int. J. Colorectal Dis. 2018;33:411–418. doi: 10.1007/s00384-018-2970-5. [DOI] [PubMed] [Google Scholar]
- 88.Hoo Z.H., Candlish J., Teare D. What is an ROC curve? Emerg. Med. J. 2017;34:357–359. doi: 10.1136/emermed-2017-206735. [DOI] [PubMed] [Google Scholar]
- 89.Powers R.K., Goodspeed A., Pielke-Lombardo H., Tan A.C., Costello J.C. GSEA-InContext: identifying novel and common patterns in expression experiments. Bioinformatics. 2018;34:i555–i564. doi: 10.1093/bioinformatics/bty271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., Alizadeh A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen B., Khodadoust M.S., Liu C.L., Newman A.M., Alizadeh A.A. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol. Biol. 2018;1711:243–259. doi: 10.1007/978-1-4939-7493-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Merino D.M., McShane L.M., Fabrizio D., Funari V., Chen S.J., White J.R., Wenz P., Baden J., Barrett J.C., Chaudhary R., et al. Establishing guidelines to harmonize tumor mutational burden (TMB): in silico assessment of variation in TMB quantification across diagnostic platforms: phase I of the Friends of Cancer Research TMB Harmonization Project. J. Immunother. Cancer. 2020;8 doi: 10.1136/jitc-2019-000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Malhotra N. Induced pluripotent stem (iPS) cells in dentistry: a review. Int. J. Stem Cells. 2016;9:176–185. doi: 10.15283/ijsc16029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Prior F.W., Clark K., Commean P., Freymann J., Jaffe C., Kirby J., Moore S., Smith K., Tarbox L., Vendt B., et al. TCIA: an information resource to enable open science. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2013;2013:1282–1285. doi: 10.1109/EMBC.2013.6609742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sahin I.H., Akce M., Alese O., Shaib W., Lesinski G.B., El-Rayes B., Wu C. Immune checkpoint inhibitors for the treatment of MSI-H/MMR-D colorectal cancer and a perspective on resistance mechanisms. Br. J. Cancer. 2019;121:809–818. doi: 10.1038/s41416-019-0599-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu S., Xie X., Lei H., Zou B., Xie L. Identification of key circRNAs/lncRNAs/miRNAs/mRNAs and pathways in preeclampsia using bioinformatics analysis. Med. Sci. Monit. 2019;25:1679–1693. doi: 10.12659/MSM.912801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shankavaram U.T., Varma S., Kane D., Sunshine M., Chary K.K., Reinhold W.C., Pommier Y., Weinstein J.N. CellMiner: a relational database and query tool for the NCI-60 cancer cell lines. BMC Genomics. 2009;10:277. doi: 10.1186/1471-2164-10-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reinhold W.C., Sunshine M., Liu H., Varma S., Kohn K.W., Morris J., Doroshow J., Pommier Y. CellMiner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 2012;72:3499–3511. doi: 10.1158/0008-5472.CAN-12-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lamb J., Crawford E.D., Peck D., Modell J.W., Blat I.C., Wrobel M.J., Lerner J., Brunet J.P., Subramanian A., Ross K.N., et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 101.Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B., et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021;49:D1388–D1395. doi: 10.1093/nar/gkaa971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li H., Shi X., Jiang H., Kang J., Yu M., Li Q., Yu K., Chen Z., Pan H., Chen W. CMap analysis identifies atractyloside as a potential drug candidate for type 2 diabetes based on integration of metabolomics and transcriptomics. J. Cell Mol. Med. 2020;24:7417–7426. doi: 10.1111/jcmm.15357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author on reasonable request.