Abstract
Confocal imaging has shown that CELLULOSE SYNTHASE (CESA) particles move through the plasma membrane as they synthesize cellulose. However, the resolution limit of confocal microscopy circumscribes what can be discovered about these tiny biosynthetic machines. Here, we applied Structured Illumination Microscopy (SIM), which improves resolution two-fold over confocal or widefield imaging, to explore the dynamic behaviors of CESA particles in living plant cells. SIM imaging reveals that Arabidopsis thaliana CESA particles are more than twice as dense in the plasma membrane as previously estimated, helping explain the dense arrangement of cellulose observed in new wall layers. CESA particles tracked by SIM display minimal variation in velocity, suggesting coordinated control of CESA catalytic activity within single complexes and that CESA complexes might move steadily in tandem to generate larger cellulose fibrils or bundles. SIM data also reveal that CESA particles vary in their overlaps with microtubule tracks and can complete U-turns without changing speed. CESA track patterns can vary widely between neighboring cells of similar shape, implying that cellulose patterning is not the sole determinant of cellular growth anisotropy. Together, these findings highlight SIM as a powerful tool to advance CESA imaging beyond the resolution limit of conventional light microscopy.
Super-resolution imaging improves detection and tracking of cellulose synthase particles in living cells, revealing undiscovered behaviors.
Introduction
Cellulose, the most abundant biopolymer on Earth, is composed of hydrogen-bonded chains of β-1,4-linked glucose and is produced in plants by Cellulose Synthase Complexes (CSCs; Turner and Kumar, 2018). Genetic, biochemical, structural, and electron microscopy data indicate that each CSC comprises 18 CELLULOSE SYNTHASE (CESA) enzymes (Desprez et al., 2007; Persson et al., 2007; Gonneau et al., 2014; Hill et al., 2014; Nixon et al., 2016). CESAs are integral membrane proteins that consume uridine diphosphate-glucose (UDP-glucose) to form β-1,4 glycosidic bonds between glucose monomers in growing glucan chains. The arrangement of CESA proteins as a hexamer of trimers forming a rosette-shaped complex ∼25 nm in diameter is thought to facilitate the production of partially crystalline, 18-chain cellulose microfibrils (Nixon et al., 2016; Purushotham et al., 2020). However, many questions remain concerning CSC behavior in living plant cells, including whether CESAs act coordinately within individual CSCs and how CSCs work together with the secretory machinery to assemble cell walls composed of layers of optimally organized cellulose and intervening matrix polymers (Anderson and Kieber, 2020).
For the last 15 years, confocal microscopy has been used to examine the density, trafficking, movements, and cytoskeletal interactions of fluorescently tagged CESA particles in living plants (Duncombe et al., 2020; Allen et al., 2021). However, the resolution limit of light microscopy (D = λ/2ΝΑ, where D is the diffraction limit, λ is the wavelength, and NA is the numerical aperture of the objective, or ∼200 nm in most cases for optical microscopy) is much larger than both the size of individual CSCs and their typical spacing in the plasma membrane (Reiss et al., 1984). This limitation prevents a clear determination of whether individual CESA particles, observed by light microscopy as fluorescent spots moving within the plasma membrane in a defined speed range, correspond to single or multiple CSCs (Chen et al., 2014). Freeze-fracture transmission electron microscopy data show CSCs in tight clusters in some cell types (Reiss et al., 1984; Herth, 1985), and confocal data rarely show fluorescently tagged CESA particles joining or splitting except immediately after delivery to the cell surface (Gutierrez et al., 2009), raising the possibility that CSCs might move coordinately in closely spaced groups that cannot be resolved by confocal imaging and work in tandem to generate cellulose microfibrils with 18 chains and/or aligned bundles of cellulose. Additionally, CESA particle density in living cells is reported from confocal studies to be ∼1 particle µm-2, which is well below the density of cellulose microfibrils observed in single wall layers by atomic force microscopy and field emission scanning electron microscopy (Paredez et al., 2006; Bashline et al., 2013; Xiao et al., 2016; Zhang et al., 2016; Zheng et al., 2017). This discrepancy raises questions about how widely spaced CSCs would be able to generate dense, partially aligned layers of cellulose during wall assembly.
The motility of plasma membrane-localized CESA particles is thought to be driven by the catalytic activity of CESA proteins and/or the crystallization of cellulose microfibrils (Diotallevi and Mulder, 2007). However, CESA particle speeds, as observed by confocal microscopy, vary widely across experimental conditions and even within individual cells (Paredez et al., 2006; Chen et al., 2010; Gu et al., 2010; Li et al., 2016; Xiao et al., 2016; Watanabe et al., 2018), calling into question how CESA particle movements might be mechanistically linked to cellulose polymerization. For example, the speed of an individual CSC might be modulated by substrate availability, catalytic coordination between individual CESAs in the complex, CESA modification status, interactions with other CSCs, microtubule guidance, membrane fluidity, the structure of the existing wall, or some combination of these (Turner and Kumar, 2018).
The factors that influence the directional movements of CESA particles in the plasma membrane are also incompletely defined. CESA particle trajectories are thought to be dominantly determined by cortical microtubules, with the patterning of previously deposited cellulose also exerting influence (Paredez et al., 2006; Chan and Coen, 2020). CESA particles have also been observed leaving microtubule tracks or switching from one track to another (Chan and Coen, 2020). Our knowledge of CSC–microtubule interactions and their precise physical positions is also blurred by the diffraction limit (Anderson, 2018): for instance, due to the difficulty of distinguishing microtubule bundles from single microtubules by light microscopy, it is unclear whether two CSCs moving in opposite directions along a single microtubule can pass each other without colliding. Despite advances in our understanding of the structures and catalytic mechanisms of CESAs and CSCs (Purushotham et al., 2020; Qiao et al., 2021; Zhang et al., 2021), many questions about CESA behaviors in vivo remain unanswered due to the resolution limit of light microscopy. These gaps in understanding can be bridged in part by super-resolution microscopy, which improves the resolution of light microscopy beyond the diffraction limit.
Understanding the nanoscale behaviors of CSCs would also help answer questions concerning the relationships between cellulose patterning and cell and tissue morphogenesis in plants. The high tensile strength of cellulose and its typical orientation perpendicular to the cellular growth axis, in combination with the observation that cells lose growth anisotropy in cellulose-deficient plants, implies that cellulose organization determines the direction and extent of growth anisotropy in plant cells (Baskin, 2005). However, accumulating evidence has called this model into question: microtubule orientation—and presumably cellulose deposition patterns—can change dynamically in cells undergoing anisotropic growth (Chan et al., 2010); loss of wall integrity sensing can partially rescue growth anisotropy without rescuing cellulose content (Hématy et al., 2007); and defects in pectin, another wall polymer, can disrupt growth anisotropy in elongating cells (Peaucelle et al., 2015). The relationship between CESA particle movements and growth anisotropy is still shrouded in mystery, but analyses of CESA particle movements in a wider variety of cell and tissue types might provide additional clarity.
In the past decade, super-resolution microscopy has become more widely available, allowing researchers to address new questions about the molecular dynamics and cellular processes that occur in living organisms, including plants (Fitzgibbon et al., 2010; Komis et al., 2015; Shaw et al., 2019). A form of super-resolution microscopy called Structured Illumination Microscopy (SIM) (Gustafsson, 2000) improves upon the resolution of conventional confocal microscopy by about two-fold, distinguishing objects spaced as little as ∼100 nm apart for SIM versus ∼200 nm for confocal microscopy, and can therefore help address questions about the in vivo behaviors of CESAs through improved detection and tracking of CESA particles. SIM also has the potential to help align insights gained from live-cell imaging with those from imaging modalities, such as atomic force and electron microscopy, that reveal ultrastructural details of CSCs, microtubules, and cell wall organization but cannot do so in living cells.
Here, we report the use of SIM to image and quantitatively analyze CESA particles in growing cells of Arabidopsis (Arabidopsis thaliana). SIM detects over twice as many CESA particles in these cells as confocal microscopy does, and time-lapse experiments performed with SIM show that individual CESA particles move steadily at a speed of ∼250 nm/min. Some CESA particles perform “U-turns” as they leave one microtubule and join another. Imaging CESA particle movements in cotyledon petioles, an elongating tissue, reveals widely varying patterns of CESA trajectories, but little variation in CESA particle speeds or cell shape.
Results
SIM can be used to detect the localization and behaviors of proteins in living plants
Super-resolution microscopy transcends the diffraction limit of light to provide improved imaging resolution (Figure 1). SIM (Gustafsson, 2000) accomplishes this by overlaying a grating pattern on the illumination beam, then translating and rotating that grating pattern to collect a series of images (Figure 1, A–C). Combining these images computationally produces moiré patterns that contain higher-order information about the fluorescence arising from individual molecules. Using a Fourier transform, the raw data can be reconstructed to generate a single image with roughly two-fold better resolution than confocal microscopy (compare Figure 1, D–F and G–I).
Figure 1.
CESA particle density is higher in SIM images. A, Four-sided slide chamber used for SIM imaging. Yellow rectangles are tape, light blue is half-strength MS medium, and the blue rectangle represents the coverslip. B, Cotyledon petiole cells that were imaged in the epidermal layer of 5-day-old light-grown cotyledons. C, Fifteen raw images were taken with SIM, with each column’s grating pattern represented above the corresponding five images. D, GFP–CESA3 image taken by confocal microscopy and scaled to fit the size of a SIM image. E, Zoomed-in region of (D). The green line traces a GFP–CESA3 track used to make a plot profile of CESA particles. F, Imaris automated particle tracking of (E), green circles are tracked particles. G, GFP–CESA3 image taken with near-TIRF microscopy and scaled to fit the size of a SIM image. H, Zoomed-in region of (G). The blue line traces a GFP–CESA3 track used to make a plot profile of CESA particles. I, Imaris automated particle tracking of (H), blue circles are tracked particles. J, GFP–CESA3 image taken by SIM. K, Zoomed-in region of (J). The purple line traces a GFP–CESA3 track used to make a plot profile of CESA particles. L, Imaris particle tracking of (K). The purple circles are tracked particles. M, Plot profiles from (E), (H), and (K). Black arrowheads mark peaks that were counted as particles. N, Number of particles along a track for confocal, near-TIRF, and SIM, n ≥ 15 cells, black horizontal lines indicate mean, different letters indicate significance, P < 0.0001, one-way ANOVA and Mann–Whitney test. O, Particle density determined by Imaris, n ≥ 15 cells, black horizontal lines indicate mean, different letters indicate significance, P < 0.0001, one-way ANOVA and Mann–Whitney test. Scale bar = 2 µm.
SIM can be used in living samples with the same fluorophores as confocal microscopy, making it a strong candidate for improved imaging of widely studied markers such as the green fluorescent protein (GFP)–CESA3 fusion between CESA3 and the GFP (Desprez et al., 2007). However, we determined that Arabidopsis seedlings must be pressed closely to the coverslip to ensure that CESA particles are in focus and overlaid with the grating pattern. When using a double-sided tape chamber to mount seedlings (Figure 1A), dark-grown hypocotyls, which have been widely used for CESA particle imaging (Paredez et al., 2006), are not pressed against the coverslip firmly enough to consistently place CESA particles in focus. For this reason, all SIM imaging in this study was performed in epidermal cells at the midvein of the cotyledon petiole of 5-day-old light-grown seedlings expressing GFP–CESA3 (Figure 1B). Cells in this region were close to the cover slip and flat enough to obtain consistent images of CESA particles in the plasma membrane. Additionally, tape placed on four sides of the seedling (Figure 1A) minimized drift during image collection. With these mounting procedures, SIM can be used to collect super-resolution imaging data of CESA particles and allows for the capture of time-lapse movies of up to 20 min at imaging intervals as small as 5 s. Reconstruction of super-resolution images for relatively slow-moving CESA particles in the plasma membrane was effective (Figure 1G), but images for fast-moving intracellular compartments or structures, such as Golgi or GFP–CESA-containing vesicles undergoing cytoplasmic streaming, led to streaking artifacts in reconstructed images (Movie 1). Although we did observe photobleaching over time, GFP proved bright and photostable enough to image GFP–CESA3 particles over long periods despite the requirement to acquire 15 images at each time point for 3D SIM.
SIM imaging detects a higher density of CESA particles than confocal imaging
Determining whether one fluorescent CESA particle corresponds to a single CSC has been impossible using conventional light microscopy including confocal, widefield, and near-total internal reflection fluorescence (TIRF) imaging (Bashline et al., 2013; Chen et al., 2014; Sanchez-Rodriguez et al., 2017; Chan and Coen, 2020), especially since CESA particles tend to cluster along microtubule-associated tracks at the plasma membrane (Paredez et al., 2006). To test whether individual CESA particles can be more effectively detected and resolved by SIM in these densely packed regions, we compared particle spacing along individual linear tracks to confocal data. We also collected widefield, near-TIRF images using the SIM microscope to control for differences in the cameras and objectives on the confocal and SIM microscopes used in this study. We traced lines along linear CESA tracks in average projections of time-lapse datasets and overlaid them onto single frames from the same datasets to measure fluorescence intensity profiles (Figure 1, D, E, G, H, J, K, and M). We then counted fluorescent peaks from these profiles: we obtained an average of 1.10 ± 0.17 (standard deviation [sd]) particles µm−1 for confocal tracks, 0.98 ± 0.16 particles µm−1 for near-TIRF tracks, and 2.38 ± 0.34 CESA particles µm−1 from SIM tracks (Figure 1N). We, therefore, detected more than twice as many CESA particles from SIM data than from confocal or near-TIRF data.
To determine whether this trend holds on a cellular scale, we also compared CESA particle density in petiole cells among confocal, near-TIRF, and SIM data. We outlined cells from still frames and cropped them in ImageJ (Schneider et al., 2012) to measure cell area, before counting particles using an automated algorithm in Imaris (Bitplane). Average particle density in SIM images (2.04 ± 0.38 particles µm−2, sd) was more than twice that of confocal images (0.89 ± 0.27 particles µm−2) or near-TIRF images (0.50 ± 0.12 particles µm−2) (Figure 1, F, I, L, and O), suggesting that CESA particles detected by confocal or near-TIRF imaging can often consist of at least two unresolved CESA particles and therefore multiple CSCs. To help determine whether the differences in CESA particle detection between imaging techniques are due to differences in either resolution or signal-to-noise, we estimated signal-to-noise ratio for confocal, near-TIRF, and SIM images. Confocal, near-TIRF, and SIM images had signal-to-noise ratios of 3.98, 8.51, and 3.05, respectively, for GFP–CESA3 (n = 10 images for each imaging method). This result suggests that improvements in particle detection from SIM images do not arise from improved signal-to-noise or a unique optical setup, but rather from improved resolution. Together, these data demonstrate the quantitative advantage provided by the improved resolution of SIM over confocal or near-TIRF microscopy for detecting and quantifying CESA particles in vivo.
SIM allows for improved tracking of CESA particle motility
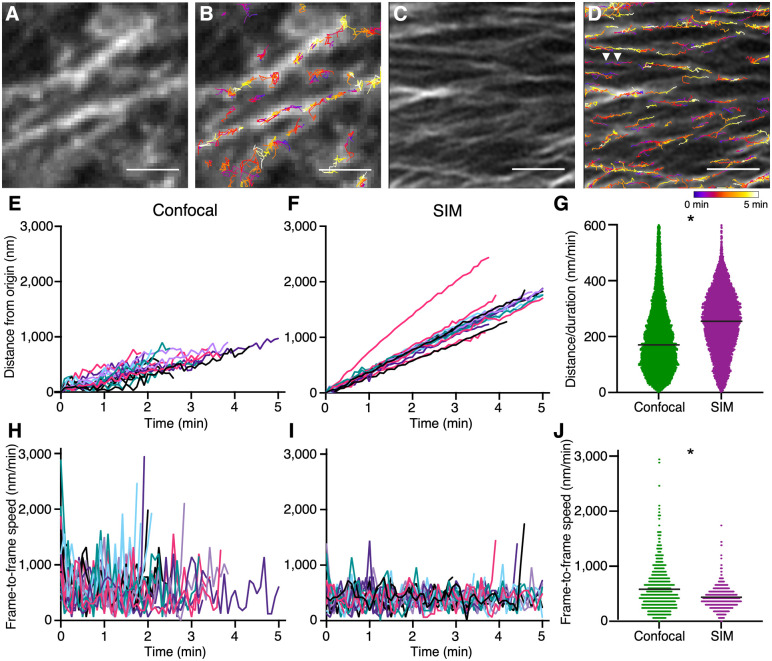
The increased CESA particle density observed in SIM images (Figure 1, N and O) suggests that single particles tracked in confocal time-lapse recordings might often consist of two particles moving in close proximity. Differences in the actual positions and/or speeds of closely spaced, unresolved CSCs that are tracked as single particles in kymograph or automated tracking analyses would result in errors in particle tracking, resulting in artifactually erratic particle trajectories and instantaneous velocities. CESA particles imaged using SIM at 5-s intervals over 5-min periods (Movie 1) typically moved along roughly linear trajectories that did not split (Figure 2D;Movie 1). Particles moving along a given track often moved in the same direction at very similar velocities (Figure 2D; Movie 1); quantifying particle directionality using kymographs showed that 69 out of 110 particles from 10 tracks moved in the predominant particle direction for a given track, a distribution that was nonrandom (P < 0.05, chi-squared test).
Figure 2.
SIM allows for more precise CESA particle tracking. A, Average projection of GFP–CESA3 from a confocal time-lapse movie. B, Imaris particle tracks overlaying (A), see Movie 1. The color of the tracks indicates the time point in the movie of the particle along its track, as seen in the heatmap below (D). C, Average projection of GFP–CESA3 from a SIM time-lapse movie. D, Imaris particle tracks overlaying (C), see Movie 1; white arrowheads indicate two particles moving in tandem. The color of the tracks indicates the time point in the movie of the particle along its track as seen in the heatmap below (D). E, Twenty confocal particle positions plotted as particle distance from particle origin, n ≥ 3 seedlings. Line colors are arbitrary and used solely to identify one particle track from another in (E), (F), (H), and (I). (F) Twenty SIM particle positions, plotted as particle distance from particle origin, n ≥ 3 seedlings. G, Particle speeds calculated as distance over duration, n = 12 cells from 12 different seedlings, black bars indicate mean, P < 0.0001, Student’s t test. H, Frame-to-frame speed calculated for 20 confocal particles, n ≥ 3 seedlings. I, Frame-to-frame speed calculated for 20 SIM particles, n ≥ 3 seedlings. J, Plot of instantaneous speed from every time point of particles in (H) and (I), black horizontal lines indicate mean, P < 0.0001, Student’s t test. Scale bar = 2 µm.
To compare precision in particle tracking between SIM and confocal imaging, we used identical procedures to track CESA particles in SIM, near-TIRF, and confocal time-lapse data using Imaris, but with a reduced estimated particle size in SIM images to match the smaller apparent size of the particles as detected by this technique (Figure 1, F and L). CESA particles tracked from SIM data did not deviate extensively from their trajectories, whereas CESA particles detected from confocal data meandered (Figure 2). We calculated particle positions for 20 particles each from SIM and confocal datasets that we confirmed visually to be single particles and with tracking information over at least 75 s. Plotting the distance traveled by each particle from its initial position over time revealed larger deviations from the primary trajectory for confocal particles than for SIM particles, suggesting that improved resolution allows for more accurate tracking of CESA particle movements (Figure 2, E and F). Calculating average speeds for larger sets of particles confirmed that the steeper slopes in the SIM particle plots in Figure 2F reflect a faster average particle speed of 250 ± 108 (sd) nm/min compared to an estimated speed of 194 ± 121 (sd) nm/min for confocal data (Figure 2G). Calculating frame-to-frame speeds for the same 20 particles from each dataset likewise showed a stark difference in average particle speeds (411 ± 184 [sd] nm/min for SIM versus 582 ± 395 [sd] for confocal; Figure 2, H–J), with confocal data showing higher average frame-to-frame speeds, likely due to imprecisions in particle tracking that exaggerated the distance each particle traveled between frames. The distribution of frame-to-frame particle speeds was also tighter for SIM data (Figure 2J). Both particle positions and speeds in SIM data showed less deviation from the linear, constant motility hypothesized for CESAs.
In time-lapse data collected from cells expressing GFP–CESA3 and the microtubule marker mCherry-TUA5, CESA particles tended to move along the sides of microtubules rather than directly on top of them (Movie 2). Using plot profiles to quantify the distance from the estimated center of a microtubule to the estimated center of a CESA particle moving along its track, we assigned particles to one of three categories: (1) those on top of microtubules (both edges of the CESA, defined as the position of one end of the extent of the Full Width at Half Max, are within the microtubule); (2) those overlapping microtubules (one edge of the CESA lies between the edges of the microtubule); and (3) those separated from microtubules (no CESA edges within the microtubule edges) (Figure 3F). Most particles did not sit directly on top of microtubules but localized slightly to one side. We also observed that the position of CESA particles moving along a microtubule track relative to that microtubule changed over time, often switching between the above three categories over the course of the time-lapse (Figure 3F). Many additional particles appeared to move in parallel with a microtubule but were separated (category three) for the duration of the movie. It is unclear whether this last group of CESA particles was tethered loosely to the microtubule, following the patterning of previously deposited cellulose, or were remnants from a parallel microtubule track that had recently depolymerized and was therefore absent from the dataset.
Figure 3.
A subset of CESA particles make U-turns. A, Average projection of a time-lapse movie of GFP–CESA3 and mCherry-TUB6 containing a particle that performs a U-turn (see Movie 2). The cyan arrowhead marks the start of the U-turn, the white arrowhead marks the end of the U-turn. B, Imaris tracks overlaid on (A). Color bar indicates the time point in the movie of the particle along its track. C, Model depicting microtubule polarity and stability influence on CESA particle U-turn behavior. D, Model depicting previously deposited cellulose as an influence on CESA particle U-turn behavior. E, Model depicting an obstruction in the plasma membrane as a driver for particle U-turn behavior. F, Colocalization analysis showing the number of particles moving along tracks that were on top of the microtubule, partially overlapping the microtubule, or separated from the microtubule. Particles are separated based on the distance between their measured fluorescent peak from a plot profile to the fluorescent peak of the microtubule. Black horizontal lines indicate mean. G, Duration of all 20 U-turn particles from the start of the U-turn to the return to linear movement. Black horizontal line indicates mean. Scale bar = 1 µm.
A subset of CESA particles makes U-turns
CESA particles that leave a microtubule-associated track, often due to depolymerization of the microtubule, can often continue to move along the same trajectory (Paredez et al., 2006). We observed CESA particles exhibiting these behaviors, but a subset of other particles followed a curving trajectory after leaving stable or unstable microtubules and eventually completed a turn greater than 90 degrees (Figure 3, A–E). These “U-turns” are uncommon but were observed multiple times across multiple experiments: from a total of 92 movies of either 20-min or 5-min duration, we observed U-turns in 17 movies. For the 20-min movies, 11 out of 24 showed examples of U-turns, while only 6 out of the 68 5-min movies contained U-turns. Within the movies that contained U-turns, U-turn particles made up only 0.01% of the tracked particles. From the initial deviation from a straight trajectory to the completion of a curve, U-turns lasted from 160 to 480 s (Figure 3G). Movies having a 5-min total duration would not be expected to include U-turns that initiated later in the movie due to the duration required to complete the U-turn, explaining the apparent underrepresentation of U-turn frequency in these shorter movies.
U-turns almost always coincided with at least one microtubule, although those microtubules displayed a full range of behaviors including depolymerization, lateral translation, and remaining stable (Movie 2). U-turn particles did not appear to be positioned on any specific region of the microtubule and could be found centered on a microtubule or along either edge. We did not observe stable microtubules in association with every U-turn, with one out of 20 U-turns having no stable microtubules and 10 out of 20 having at least one unstable microtubule, suggesting that microtubules are not required for U-turn behavior. Most often, multiple microtubules were involved in a U-turn, often in alignment with the initial trajectory and the final trajectory, and one or more of these microtubules were stable. In the U-turn displayed in Figure 3, A and B (Movie 2), a third microtubule appeared to “sweep” the CESA particle off the first microtubule and guide it toward the second microtubule in one of the few examples involving three microtubules. Overall, we observed particles performing U-turns between two parallel microtubules (9 out of 20), between acutely intersecting microtubules (9 out of 20), away from a microtubule into an area lacking microtubules (1 out of 20), and onto a microtubule from an area lacking microtubules (1 out of 20).
To determine whether CESA particle movement demonstrated a directional bias based on microtubule polarity that might influence this U-turn behavior, we analyzed 100 particles moving along newly polymerized microtubules. Newly polymerized microtubules are likely single microtubules, so CESA particle direction toward the plus or minus end of the microtubule can be determined by observing microtubule polymerization behavior. Of the 100 particles, 61 moved toward the fast-growing plus end, which represents a significant deviation from a random distribution, based on a Chi-squared analysis (P = 0.0278).
Most U-turns involved a single CESA particle, but a few cases involved multiple particles following the same U-turn trajectory. We scored multiple-particle U-turns as single U-turn occurrences and their data were taken as one data point, since all particles followed the same track at roughly the same speed. U-turns had an average radius of curvature of 0.29 µm and a range of 0.10 µm to 0.76 µm. Analyzing particle speeds using kymographs and automated particle tracking software (Imaris) showed that CESA particle speeds remain consistent throughout the U-turn, with variability in frame-to-frame speed matching that of particles moving along straighter tracks, suggesting that CESA particles do not speed up or slow down substantially when leaving or joining microtubule-aligned trajectories.
CESA track patterns vary in cotyledon petiole cells
The cotyledon petioles from which we collected SIM data displayed a diverse array of GFP–CESA3 densities, patterns, and cell shapes. The middle third of the petiole midvein (Figure 1B) contains long, rectangular cells of a similar size and shape. Cellular patterns of CESA particle trajectories varied widely in this tissue, even between neighboring cells (Figure 4). Unlike dark-grown hypocotyls where neighboring cells have similar CESA particle patterning and anisotropy (Barnes and Anderson, 2018), neighboring petiole cells often displayed transverse and longitudinal patterning, and yielded varying anisotropy scores, as calculated by the ImageJ plugin FibrilTool (Figure 4E; Boudaoud et al., 2014). Larger cells toward the hypocotyl or central region of the petiole also occasionally displayed several pattern types in a single cell, with transverse tracks morphing into longitudinal tracks. We assigned track patterns from 61 cells into one of four categories: longitudinal (<30 degrees relative to the growth axis of the cell); diagonal (30–60 degrees); transverse (>60 degrees); and multidirectional (Figure 4, A–D). We quantified the primary CESA trajectory angle relative to the cell growth axis for each cell using FibrilTool, which revealed the longitudinal category as the most common pattern (35 cells), followed by diagonal (15 cells), and lastly transverse (11 cells). We included multidirectionally patterned cells in the dataset, although they could not be scored visually for track angle, but were sorted into the other three categories based on the net track angle, as calculated using FibrilTool.
Figure 4.
Varied CESA trajectory patterns in epidermal petiole cells do not affect CESA speed. A–D, Representative images of the four most common track patterns seen in the epidermal cells of the cotyledon petiole. Scale bar = 2 µm. E, Depiction of the variety of track patterns seen in neighboring cells of the cotyledon petiole. F, Comparison of the average particle speeds of the 20 cells with the most transversely oriented and most longitudinally oriented CESA particle tracks. Black horizontal lines indicate mean. P = 0.093, Student’s t test. G, Comparison of the average particle speeds of the 20 cells with the highest and lowest anisotropy scores for CESA particle tracks, as determined by FibrilTool. Black bars indicate mean. P = 0.437, Student’s t test.
The difference in CESA patterns displayed in cells of similar size and shape in the epidermis of the petiole was unlike that of etiolated hypocotyls, which display similar track patterns in neighboring cells of the same size and shape. Since the above data came from a single developmental time frame, we imaged the plasma membrane marker, LOW TEMPERATURE-INDUCED PROTEIN 6B (LTI6B)-GFP, a fusion between LTI6B and GFP, in 3-, 5-, and 7-day-old petiole epidermal cells to observe cell shape over time. We measured the cell length to width ratios for each day: they were 3.82 on Day 3, 5.85 on Day 5, and 9.15 on Day 7 (n ≥ 38 cells from four seedlings for each time point). Because cell files contained the same number of cells over time, the increasing length-to-width ratios suggest anisotropic growth of the epidermal petiole cells between 3 and 7 days after sowing.
Although the average CESA particle speed across all of the cells imaged by SIM (249 ± 40 [sd] nm/min) was comparable to speeds obtained with confocal data for etiolated hypocotyls (Paredez et al., 2006; Chen et al., 2010; Gu et al., 2010; Xiao et al., 2016), we wondered whether the different trajectory patterns might lead to variability in CESA speed. Using FibrilTool to separate cells by both average trajectory angle relative to the growth axis and anisotropy score, we compared average CESA particle speeds from each cell based on track pattern and anisotropy score (Figure 4, F and G). Accordingly, we compared the top 20 scoring cells in each category to their opposite (transverse versus longitudinal, high anisotropy versus low anisotropy), but these comparisons showed no significant difference in CESA particle speeds between cells with different trajectory patterns or anisotropy scores. These data indicate that CESA particle speed is independent from trajectory patterning in the cell.
Discussion
Despite being very small, CESA complexes produce gigatons of cellulose in growing plants every year, but how these amazing molecular factories behave at the nanoscale in living cells to accomplish this feat has remained obscure. To begin to connect emerging information regarding the atomic structure and biochemical activity of plant CESAs (Purushotham et al., 2020; Qiao et al., 2021; Zhang et al., 2021) to their behaviors in living cells, we used super-resolution microscopy to reveal new details about how CESAs help construct the plant cell wall.
SIM data indicate that CESA particles as imaged by confocal microscopy can contain more than one CSC
Using confocal or near-TIRF microscopy, CESA particles show a density of ∼1 particle µm−2 (Xiao et al., 2016; Chan and Coen, 2020), but cellulose at the innermost layer of the cell wall appears to be much denser, as observed by atomic force microscopy and field emission scanning electron microscopy; these images represent a “fossil” image of recent cellulose deposition, whereas a single CESA image provides only a snapshot (Zhang et al., 2016; Zheng et al., 2017). The higher density of GFP–CESA3 in SIM images (Figure 1, N and O) suggests that many CESA particles in confocal images in fact contain two or more CSCs. This observation aligns more closely with freeze-fracture electron microscopy data estimating a CSC density in the base of caulonemal tip cells of bonfire moss (Funaria hygrometrica) of 3–4 particles µm−2 (Reiss et al., 1984). With a resolution limit of 100 nm, it is still possible for more than one CSC to exist in a CESA particle from a SIM image, but particle splitting and merging events are rarely seen, suggesting that particles in close proximity might be moving together to form larger or bundled cellulose microfibrils, as is seen at larger scales during secondary wall synthesis (Li et al., 2016). It is also possible that closely positioned CSCs influence each other’s behavior by linking catalytic rates to physical effects of the production and/or crystallization of aligned microfibrils, or by the inhibition of “overtaking” maneuvers along microtubule tracks.
Although 3D SIM shows improvements in axial resolution over confocal microscopy (roughly 269 nm axial resolution compared to 500 nm axial resolution, respectively; Gustafsson, 2000), we suspect that the lower estimated signal-to-noise ratio for SIM in comparison to that of near-TIRF comes from the speckling artifact of reconstruction that is especially prominent in the out-of-focus regions used for background measurements, as well as the fact that near-TIRF limits the illumination volume to near the coverslip.
Improved particle tracking shows that individual CESA particle speeds are relatively steady, but that particle speeds within a cell vary widely
Although CESA particle speeds in SIM images fall within the range of speeds published from confocal data (Paredez et al., 2006; Chen et al., 2010; Gu et al., 2010; Xiao et al., 2016; Liu et al., 2017), automated particle tracking improvements can more accurately reflect the motility of CESA particles and reduce the variability in speed averages previously seen in the field. The erratic particle trajectories seen in confocal data (Figure 2) might help explain why the average of frame-to-frame speeds for tracked CESA particles in confocal images is higher than in SIM images. CESA particles in confocal images that actually contain more than one CSC may be tracked inaccurately, as the tracked centroid shifts between complexes, resulting in higher frame-to-frame speeds. There is less discrepancy in SIM particle data between average frame-to-frame speed and distance over duration measurements, suggesting that the true speed of CESA particles is closer to the linear estimation given by the distance over duration calculation than confocal data suggest. By resolving these separate particles, SIM allows for more precise tracking of the movements of CESA particles, which will be especially useful in studying CESA–microtubule interactions. Because fast-moving objects are not captured by SIM due to the ∼5 s required to acquire 15 raw images at each time point, SIM also likely selects against small CESA compartments (SmaCCs)/microtubule-associated CESA compartments (MASCs), providing data enriched in well-resolved, slow-moving, plasma membrane-localized CESA particles; using a cutoff of 600 nm/min to roughly separate membrane-localized CESA particles from SmaCCs/MASCs, we determined that only 0.02% of tracked particles move faster than 600 nm/min in SIM time-lapse datasets, whereas 2.23% of tracked particles moved faster than 600 nm/min in confocal datasets. Observations of SIM data suggest a bias in particle movement along microtubules toward the plus end. Whether this bias is due to microtubule polarity in bundles, track orientation within the cell, or CSC coordination remains unknown. We also observed particles moving along the sides of microtubule bundles, which begs the question of whether there is variability in the strength of the connection between CSCs and microtubules. Particles moving along the sides of microtubule tracks might have a weaker connection and therefore be more likely to switch directions at intersections between cortical microtubules. Microtubules can form bundles and CESA particles are closely spaced along microtubule tracks; therefore, improvements in detection, localization, and tracking that exceed current algorithms such as Imaris, which can sometimes momentarily lose particles, promise to provide new insights into how CESA particle movements are influenced by microtubules and the conditions under which CESA particles leave, join, or switch microtubule tracks.
Although SIM improved CESA particle tracking (Figure 2, A–I), we still observed a wide range of CESA particle speeds within a single cell, although the speeds of individual particles remained remarkably constant and interparticle speed variation was far lower for particles moving along the same track. These data imply that fluctuation in local substrate availability is not a limiting factor for CSC motility and that the catalytic activity of individual CESA subunits might be coordinated to maintain the correct polymerization and crystallization of cellulose microfibrils (Harris et al., 2012; Fujita et al., 2013). Regional differences in microtubule organization and/or wall patterning across the cell surface might underlie the large variations in speed observed between distant CESA particles. The distribution of CESA particle speeds is more symmetrical in SIM data than in confocal data (Figure 2G), with more particles moving at faster speeds in SIM data, although the overall range of speeds is similar between the two forms of microscopy. A closer examination of where faster- and slower-moving particles reside within the cell, or how CESA particle speeds might vary over the course of the particle’s residence time at the plasma membrane, might provide additional clues concerning how CSC motility is regulated.
CSC particle trajectories are likely influenced by multiple factors
It is unclear whether the U-turns observed in the petiole are carefully regulated to produce a specific result in the cell wall, or if they are unregulated accidents of CSC behavior that have little effect on wall structure or function. Although U-turns appear to be a recurring behavior in CESA movies, the variability in circumstances surrounding each U-turn suggests a complicated mechanism for regulating CESA trajectory. U-turns can occur with or without microtubules, but do seem to be influenced by them when they are present, suggesting that several mechanisms are involved and that these mechanisms have a hierarchy for exerting control over CSC behavior.
Based on our observations, we propose three different hypotheses for the existence of the U-turn behavior. In cases involving microtubules, the slight bias in direction of movement along microtubules toward the plus end may encourage U-turn behavior (Figure 3C). Interestingly, our data differ from a previous report demonstrating a directional bias toward the minus end (Chan and Coen, 2020). A slight bias might explain the change in direction of particles displaying U-turn behavior, as well as the low frequency of this behavior. In many cases, other particles begin along the same trajectory as U-turn particles but continue on a linear path, so U-turn behavior does not appear to be the dominant pathway. Another potential regulator for particle behavior is previously deposited cellulose, which may be influencing particle trajectories independently from microtubules (Figure 3D). The existence of a curve in the newly deposited cellulose layer might cause the freshly extruded cellulose to follow the curve, pulling the CSC off its current linear track (on or off a microtubule) and into a curved trajectory. In cases with multiple particles following the same U-turn, this cellulose curve may be well established and therefore able to pull more particles away from their linear tracks. It is also possible that CESA particle trajectory is influenced by the patterned deposition of matrix polysaccharides, an especially intriguing direction for future SIM studies given the fact that both CESA particle behavior and cellulose organization are aberrant in mutants lacking matrix components such as xyloglucan or pectin (Xiao et al., 2016; Du et al., 2020). Lastly, U-turns may be a reaction to an obstruction in the plasma membrane that knocks the particle off its course (Figure 3E). This possibility might be the least likely, given the consistency of U-turn particle speeds, but resolution limits may obscure details of the movements of CESA particles. One or more of these mechanisms may be involved in each U-turn, suggesting a complex system of CSC trajectory regulation to create each layer of cellulose.
CESA track patterns vary widely in cells having similar shapes and degrees of growth anisotropy
The variety in CESA track patterns displayed in similarly shaped cells of the cotyledon petiole does not support the hypothesis that cellulose is deposited perpendicularly to the axis of growth. Not only did track patterns vary from cell to cell, but they also varied within a cell, resulting in arched and curved CESA tracks as they morphed from transverse to longitudinal patterning. Despite this diversity, these cells all had a uniformly thin, rectangular shape. Although cells closer to the cotyledon might eventually adopt rounded or complex shapes as pavement cells, cells near the hypocotyl that should remain long and rectangular also displayed large variations in CESA track patterns. Imaging of LTI6B-GFP revealed that these rectangular cells grow anisotropically and maintain a rectangular shape from 3 to 7 days after sowing. CESA tracks aligned closely with microtubules in all cotyledon petiole cells, so their patterning is likely still influenced by cortical microtubule organization (Paredez et al., 2006; Chan et al., 2010; Li et al., 2015).
Super-resolution imaging is helping to bridge the gap between dynamic in vivo imaging of CESAs and emerging structural data that tell us about CSC composition and molecular geometry; the broadening availability of super-resolution techniques, including SIM, PALM/STORM, or AIRYSCAN, will continue to improve our understanding of how the world’s most abundant biopolymer is produced and to open new avenues of research for designing renewable biomaterials.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana lines GFP–CESA3, mCherry-TUA5 (Gutierrez et al., 2009), and GFP–CESA3 cesa3je5 (Desprez et al., 2007) were used for all imaging experiments. Seeds were surface sterilized in 30% (v/v) bleach 0.1% (w/v) SDS (Fisher) for 20 min, with vortexing every 10 min. Seeds were then washed 4 times in sterile water before stratification in 0.15% agar at 4°C. Seedlings were grown on half-strength Murashige and Skoog (MS) medium (Caisson Laboratories) adjusted to pH 5.6 with 0.6 g 2-(N-morpholino)ethanesulfonic acid (MES; Research Organics), solidified with 0.8% (w/v) agar- (Research Organics), and supplemented with 1% (w/v) sucrose. Seeds were sown along two rows to allow adequate spacing. The plates were placed upright in a Percival CU36L5C8 growth chamber with Sylvania Octron Eco 4100K bulbs at 22°C under continuous light at 800 PPFD for 5 days before imaging. LTI6B-GFP (Cutler et al., 2000) seedlings were grown identically for 3–7 days before imaging.
GFP–CESA3 and mCherry-TUA5 imaging
SIM (3D SIM) and near-TIRF imaging were performed on a Nikon N-SIM/N-STORM microscope using a 100× 1.49 NA oil-immersion objective and an Andor back-thinned iXON 897 EMCCD camera (512 × 512 pixels). Confocal microscopy was performed on a Zeiss Observer SD spinning disk confocal microscope with a 100× 1.40 NA oil-immersion objective and a Photometrics QuantEM 512SC camera (512 × 512 pixels). It should be noted that the SIM-reconstructed images are 1,024 × 1,024 pixels, but that this difference in pixel count between the cameras does not improve the optical resolution of the microscope. Seedlings grown for 5 days were mounted on glass slides surrounded on all sides for SIM and two sides for confocal microscopy with double-sided tape (3M Permanent), immersed in 60 µL half-strength liquid MS medium, and covered with a 24 × 40 mm #1.5 coverslip (Corning). Dual-channel time-lapses were collected by imaging the 488-nm laser channel (500–545 nm filter for SIM and near-TIRF, 500–550 nm filter for confocal) and the 561-nm laser channel (570–640 nm filter for SIM, 581–653 nm filter for confocal) sequentially at each time point to collect imaging data on GFP–CESA3 and mCherry-TUA5. Images were all collected in the epidermal layer of the midvein region of the petiole of cotyledons. Confocal data were collected with 90% laser power, 200-ms exposure, and 300 EM gain. Near-TIRF data were collected with 0.3% laser power, 80-ms exposure, and 100 EM gain. SIM data were collected with 80% laser power, 150-ms exposure, and 100 EM gain. No frame averaging was used. While near-TIRF data were collected using the same objective and camera as SIM, we used the 1× module and the STORM driver instead of the SIM module to perform TIRF experiments without SIM. This 1× versus SIM module accounts for the difference in size of the imaging regions between the two modalities. The TIRF angle adjustment was set to 3,884.2. Both cameras were set to 16-bit depth, with each pixel being 16 × 16 μm. Raw SIM data were reconstructed in NIS Elements using a Fast Fourier Transform to optimize results. Settings for the reconstruction process were adjusted for each image, but variations were small. Illumination Modulation Contrast was set between 0.50 and 1.00. High-resolution noise suppression was always 0.10. Out of Focus Blur Suppression was set between 0.01 and 0.02.
Image analysis
Images were analyzed using ImageJ (Schneider et al., 2012) and Imaris version 7.4.1. No post-processing was performed other than the use of the ImageJ plugin StackReg to correct for minor sample drift (Thévenaz et al., 1998). Movies were cropped around cells to remove the cell outlines that are out of focus and would otherwise lead to falsely tracked particles along these edges in Imaris. For Imaris, estimated particle diameter was set empirically to 0.25 µm for confocal images, 0.4 µm for near-TIRF, and 0.15 µm for SIM images; connected components were used as the tracking algorithm, and the minimum track duration was set to 60 s. Tracked CESA particles that were traveling in Golgi or SmaCCs/MASCs or moving faster than 600 nm/min were manually removed from the tracking dataset. The ImageJ Plugin FibrilTool (Boudaoud et al., 2014) was used to determine the primary orientation and anisotropy score of CESA trajectories relative to the growth axis of the cell. The speeds of the 20 highest and lowest scores for anisotropy were compared using a Student’s t test. The speeds of the 20 most longitudinal and most transverse cells were compared similarly. Kymograph analysis for particle directionality along microtubules was performed using Kymobutler (Jakobs et al., 2019). Ten kymographs were collected along microtubule-associated CESA particle tracks from 10 seedlings. The frequency of particles moving in the dominant direction of CESA particle movement was compared to the expected value of 50% by chi-squared analysis.
Density and particle spacing
To determine density, the perimeter of a cell was traced using the polygon tool in ImageJ to collect the area (Schneider et al., 2012). The region was cropped and run through Imaris to determine particle count within that cell area to quantify particle density. Imaris settings were identical to all other analyses done for SIM movies. Particle spacing was determined by tracing an average projection of a CESA track using the segmented line tool in ImageJ. After overlaying the line onto the original time-lapse, a Plot Profile was taken and the peaks counted to determine the average number of particles per micron along a CESA track.
Signal-to-noise estimates
Ten images each from at least nine seedlings from confocal, near-TIRF, and SIM data were thresholded in ImageJ to select for GFP–CESA3 signal (Schneider et al., 2012). Average signal intensity was calculated and compared to the intensity from a selected region in the same image lacking GFP–CESA3 signal to estimate the signal-to-noise ratio.
Colocalization of CESA particles and microtubules
Plot profiles were drawn perpendicularly to microtubule tracks over 50 microtubule-associated CESA particles. Each particle was observed to move along the track to determine association. Profiles were plotted from each channel to get fluorescent peaks for the CESA particle and the microtubule. The distance between the peak of the CESA particle signal and peak of the microtubule signal was calculated for each particle. Particles were split into three categories: on top, overlapping, and separate, based on the relative positions of their outer edges. Outer edges of microtubules and CESA particles were defined as the boundaries of the Full Width at Half Max of the fluorescent peak. CESA particles in the “on top” category had both edges inside the edges of the microtubule; particles that were “overlapping” had one edge within the microtubule boundary; separate particles had no edges within the microtubule boundary. All particles came from two separate tracks each from five different seedlings with ten particles per seedling.
Microtubule polarity analysis
Microtubules that could be observed polymerizing were screened for CESA particle movement along the new segment of microtubule. A total of 100 CESA particles observed on newly polymerized microtubules were scored as moving toward the plus end or toward the minus end of microtubules.
Quantification of cell geometry in 3- to 7-day-old seedlings
LTI6B-GFP seedlings grown in the light for 3, 5, or 7 days were imaged in the same region as for GFP–CESA3 particles to quantify the cell length-to-width ratio in developing cotyledon petioles. Lengths and widths of cells close to the coverslip in the epidermal layer were measured for four different seedlings each day. At least seven cells were measured per seedling, and the average length-to-width ratio was calculated for Days 3, 5, and 7.
Statistical analysis
All data are from a minimum of three biological replicates. With the exception of the sets of 20 individually tracked particles from Figure 2, all data are from a minimum of three experimental replicates. Statistical significance was determined using a Mann–Whitney test, analysis of variance (ANOVA), or two-tailed unpaired t test. Deviation from a random distribution was determined using chi-squared analysis.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession number: CESA3 (At5g05170), LTI6B (At3g05890), and TUA5 (At5g19780).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Data Set S1. Summary of statistical analyses.
Supplementary Material
Acknowledgments
The authors thank members of the Anderson Lab for helpful comments on the manuscript and Missy Hazen for technical support. Super-resolution microscopy was performed in the Huck Microscopy Facility.
Funding
This work was supported as part of The Center for Lignocellulose Structure and Formation, an Energy Frontier Research Center funded by the US Department of Energy, Office of Science, Basic Energy Sciences under Award #DE-SC0001090. The Nikon N-SIM/N-STORM microscope was purchased with support from NSF MRI-1625473.
Conflict of interest statement. None declared.
S.G.D. and C.T.A. designed experiments; S.G.D. performed experiments; S.G.D. and S.G.C. analyzed data; S.G.D. and C.T.A. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Charles T. Anderson (cta3@psu.edu).
References
- Allen H, Wei D, Gu Y, Li S (2021) A historical perspective on the regulation of cellulose biosynthesis. Carbohydr Polymers 252: 117022. [DOI] [PubMed] [Google Scholar]
- Anderson CT (2018) Finding order in a bustling construction zone: quantitative imaging and analysis of cell wall assembly in plants. Curr Opin Plant Biol 46: 62–67 [DOI] [PubMed] [Google Scholar]
- Anderson CT, Kieber JJ (2020) Dynamic construction, perception, and remodeling of plant cell walls. Ann Rev Plant Biol 71: 39–69 [DOI] [PubMed] [Google Scholar]
- Barnes WJ, Anderson CT (2018) Cytosolic invertases contribute to cellulose biosynthesis and influence carbon partitioning in seedlings of Arabidopsis thaliana. Plant J Cell Mol Biol 94: 956–974 [DOI] [PubMed] [Google Scholar]
- Bashline L, Li S, Anderson CT, Lei L, Gu Y (2013) The endocytosis of cellulose synthase in Arabidopsis is dependent on μ2, a clathrin-mediated endocytosis adaptin. Plant Physiol 163: 150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin TI (2005) Anisotropic expansion of the plant cell wall. Ann Rev Cell Dev Biol 21: 203–222 [DOI] [PubMed] [Google Scholar]
- Boudaoud A, Burian A, Borowska-Wykręt D, Uyttewaal M, Wrzalik R, Kwiatkowska D, Hamant O (2014) FibrilTool, an ImageJ plug-in to quantify fibrillar structures in raw microscopy images. Nat Protocol 9: 457–463 [DOI] [PubMed] [Google Scholar]
- Chan J, Coen E (2020) Interaction between autonomous and microtubule guidance systems controls cellulose synthase trajectories. Curr Biol 30: 941–947 e942 [DOI] [PubMed] [Google Scholar]
- Chan J, Crowell E, Eder M, Calder G, Bunnewell S, Findlay K, Vernhettes S, Höfte H, Lloyd C (2010) The rotation of cellulose synthase trajectories is microtubule dependent and influences the texture of epidermal cell walls in Arabidopsis hypocotyls. J Cell Sci 123: 3490–3495 [DOI] [PubMed] [Google Scholar]
- Chen S, Ehrhardt DW, Somerville CR (2010) Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proc Natl Acad Sci USA 107: 17188–17193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Deffenbaugh NC, Anderson CT, Hancock WO (2014) Molecular counting by photobleaching in protein complexes with many subunits: best practices and application to the cellulose synthesis complex. Mol Biol Cell 25: 3630–3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR (2000) Random GFP∷cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97: 3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Hofte H, Gonneau M, Vernhettes S (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diotallevi F., Mulder B. (2007) The cellulose synthase complex: a polymerization driven supramolecular motor. Biophys J 92: 2666–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Kirui A, Huang S, Wang L, Barnes WJ, Kiemle S, Zheng Y, Rui Y, Ruan M, Qi S, et al. (2020) Mutations in the pectin methyltransferase QUASIMODO2 influence cellulose biosynthesis and wall integrity in Arabidopsis thaliana. Plant Cell 32: 3576–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncombe SG, Barnes WJ, Anderson CT (2020) Imaging the delivery and behavior of cellulose synthases in Arabidopsis thaliana using confocal microscopy. Methods Cell Biol 160: 201–213 [DOI] [PubMed] [Google Scholar]
- Fitzgibbon J, Bell K, King E, Oparka K (2010) Super-resolution imaging of plasmodesmata using three-dimensional structured illumination microscopy. Plant Physiol 153: 1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Himmelspach R, Ward J, Whittington A, Hasenbein N, Liu C, Truong TT, Galway ME, Mansfield SD, Hocart CH, et al. (2013) The anisotropy1 D604N mutation in the Arabidopsis cellulose synthase1 catalytic domain reduces cell wall crystallinity and the velocity of cellulose synthase complexes. Plant Physiol 162: 74–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonneau M, Desprez T, Guillot A, Vernhettes S, Hofte H (2014) Catalytic subunit stoichiometry within the cellulose synthase complex. Plant Physiol 166: 1709–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Kaplinsky N, Bringmann M, Cobb A, Carroll A, Sampathkumar A, Baskin TI, Persson S, Somerville CR (2010) Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc Natl Acad Sci USA 107: 12866–12871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson MGL (2000) Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc 198: 82–87 [DOI] [PubMed] [Google Scholar]
- Gutierrez R, Lindeboom JJ, Paredez AR, Emons AM, Ehrhardt DW (2009) Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol 11: 797–806 [DOI] [PubMed] [Google Scholar]
- Harris DM, Corbin K, Wang T, Gutierrez R, Bertolo AL, Petti C, Smilgies DM, Estevez JM, Bonetta D, Urbanowicz BR, et al. (2012) Cellulose microfibril crystallinity is reduced by mutating C-terminal transmembrane region residues CESA1A903V and CESA3T942I of cellulose synthase. Proc Natl Acad Sci USA 109: 4098–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hématy K, Sado PE, Van Tuinen A, Rochange S, Desnos T, Balzergue S, Pelletier S, Renou JP, Höfte H (2007) A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr Biol 17: 922–931 [DOI] [PubMed] [Google Scholar]
- Herth W (1985) Plasma-membrane rosettes involved in localized wall thickening during xylem vessel formation of Lepidium sativum L. Planta 164: 12–21 [DOI] [PubMed] [Google Scholar]
- Hill JL Jr., Hammudi MB, Tien M (2014) The Arabidopsis cellulose synthase complex: a proposed hexamer of CESA trimers in an equimolar stoichiometry. Plant Cell 26: 4834–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobs MAH, Dimitracopoulos A, Franze K (2019) KymoButler, a deep learning software for automated kymograph analysis. eLife 8: e42288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komis G, Mistrik M, Šamajová O, Ovečka M, Bartek J, Šamaj J (2015) Superresolution live imaging of plant cells using structured illumination microscopy. Nat Protocol 10: 1248–1263 [DOI] [PubMed] [Google Scholar]
- Li S, Lei L, Yingling YG, Gu Y (2015) Microtubules and cellulose biosynthesis: the emergence of new players. Curr Opin Plant Biol 28: 76–82 [DOI] [PubMed] [Google Scholar]
- Li S, Bashline L, Zheng Y, Xin X, Huang S, Kong Z, Kim SH, Cosgrove DJ, Gu Y (2016) Cellulose synthase complexes act in a concerted fashion to synthesize highly aggregated cellulose in secondary cell walls of plants. Proc Natl Acad Sci USA 113: 11348–11353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Zehfroosh N, Hancock BL, Hines K, Fang W, Kilfoil M, Learned-Miller E, Sanguinet KA, Goldner LS, Baskin TI (2017) Imaging cellulose synthase motility during primary cell wall synthesis in the grass Brachypodium distachyon. Sci Rep 7: 15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon BT, Mansouri K, Singh A, Du J, Davis JK, Lee JG, Slabaugh E, Vandavasi VG, O’Neill H, Roberts EM, et al. (2016) Comparative structural and computational analysis supports eighteen cellulose synthases in the plant cellulose synthesis complex. Sci Rep 6: 28696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW (2006) Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Peaucelle A, Wightman R, Hofte H (2015) The control of growth symmetry breaking in the Arabidopsis hypocotyl. Curr Biol 25: 1746–1752 [DOI] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR (2007) Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci USA 104: 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushotham P, Ho R, Zimmer J (2020) Architecture of a catalytically active homotrimeric plant cellulose synthase complex. Science 369: 1089–1094 [DOI] [PubMed] [Google Scholar]
- Qiao Z, Lampugnani ER, Yan XF, Khan GA, Saw WG, Hannah P, Qian F, Calabria J, Miao Y, Grüber G, et al. (2021) Structure of Arabidopsis CESA3 catalytic domain with its substrate UDP-glucose provides insight into the mechanism of cellulose synthesis. Proc Natl Acad Sci USA 118: e2024015118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss HD, Schnepf E, Herth W (1984) The plasma membrane of the Funaria caulonema tip cell: morphology and distribution of particle rosettes, and the kinetics of cellulose synthesis. Planta 160: 428–435 [DOI] [PubMed] [Google Scholar]
- Sanchez-Rodriguez C, Ketelaar K, Schneider R, Villalobos JA, Somerville CR, Persson S, Wallace IS (2017) BRASSINOSTEROID INSENSITIVE2 negatively regulates cellulose synthesis in Arabidopsis by phosphorylating cellulose synthase 1. Proc Natl Acad Sci USA 114: 3533–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SL, Thoms D, Powers J (2019) Structured illumination approaches for super-resolution in plant cells. Microscopy 68: 37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenaz P, Ruttimann UE, Unser M (1998) A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process 7: 27–41 [DOI] [PubMed] [Google Scholar]
- Turner S, Kumar M (2018) Cellulose synthase complex organization and cellulose microfibril structure. Philos Trans A Math Phys Eng Sci 376: 20170048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Schneider R, Barkwill S, Gonzales-Vigil E, Hill JL, Samuels AL, Persson S, Mansfield SD (2018) Cellulose synthase complexes display distinct dynamic behaviors during xylem transdifferentiation. Proc Natl Acad Sci USA 115: E6366–E6374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Zhang T, Zheng Y, Cosgrove DJ, Anderson CT (2016) Xyloglucan deficiency disrupts microtubule stability and cellulose biosynthesis in Arabidopsis, altering cell growth and morphogenesis. Plant Physiol 170: 234–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zheng Y, Cosgrove DJ (2016) Spatial organization of cellulose microfibrils and matrix polysaccharides in primary plant cell walls as imaged by multichannel atomic force microscopy. Plant J Cell Mol Biol 85: 179–192 [DOI] [PubMed] [Google Scholar]
- Zhang X, Xue Y, Guan Z, Zhou C, Nie Y, Men S, Wang Q, Shen C, Zhang D, Jin S, et al. (2021) Structural insights into homotrimeric assembly of cellulose synthase CesA7 from Gossypium hirsutum. Plant Biotechnol J 19: 1579–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Cosgrove DJ, Ning G (2017) High-resolution field emission scanning electron microscopy (FESEM) imaging of cellulose microfibril organization in plant primary cell walls. Microsc Microanal 23: 1048–1054 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.