Abstract
Background:
Diverse toxicants and mixtures that affect hormone responsive cells [endocrine disrupting chemicals (EDCs)] are highly pervasive in the environment and are directly linked to human disease. They often target the nuclear receptor family of transcription factors modulating their levels and activity. Many high-throughput assays have been developed to query such toxicants; however, single-cell analysis of EDC effects on endogenous receptors has been missing, in part due to the lack of quality control metrics to reproducibly measure cell-to-cell variability in responses.
Objective:
We began by developing single-cell imaging and informatic workflows to query whether the single cell distribution of the estrogen (ER), used as a model system, can be used to measure effects of EDCs in a sensitive and reproducible manner.
Methods:
We used high-throughput microscopy, coupled with image analytics to measure changes in single cell ER nuclear levels on treatment with toxicants, over a large number of biological and technical replicates.
Results:
We developed a two-tiered quality control pipeline for single cell analysis and tested it against a large set of biological replicates, and toxicants from the EPA and Agency for Toxic Substances and Disease Registry lists. We also identified a subset of potentially novel EDCs that were active only on the endogenous ER level and activity as measured by single molecule RNA fluorescence in situ hybridization (RNA FISH).
Discussion:
We demonstrated that the distribution of ER levels per cell, and the changes upon chemical challenges were remarkably stable features; and importantly, these features could be used for quality control and identification of endocrine disruptor toxicants with high sensitivity. When coupled with orthogonal assays, ER single cell distribution is a valuable resource for high-throughput screening of environmental toxicants. https://doi.org/10.1289/EHP9297
Introduction
The human population is constantly exposed to an increasing number of chemicals and complex mixtures that pose significant health risks. A subset of these is referred to as endocrine-disrupting chemicals (EDCs), because they interfere with actions of natural hormones altering central physiological mechanisms, ultimately causing disease (De Coster and van Larebeke 2012; Hall and Greco 2019; Kabir et al. 2015). One of the many potential EDC targets is the estrogen (ER), a transcription factor that regulates, among other physiological processes, female reproductive biology and is involved in several diseases, including obesity and breast cancer (Gibson and Saunders 2014; Heldring et al. 2007). The ER mechanistic pathway has been extensively studied in terms of EDC action, with large efforts spearheaded by the U.S. EPA and National Toxicology Program (NTP) ToxCast and Tox21 programs, using a battery of in vitro and in vivo assays (Huang et al. 2014; Judson et al. 2015; Mansouri et al. 2016; Richard et al. 2016; Rotroff et al. 2014; Stossi et al. 2014; Szafran et al. 2017). ToxCast efforts led to the important demonstration that 18 in vitro ER assays can be used as a substitute for expensive and less reliable animal models to predict in vivo EDC potential (Judson et al. 2015, 2017). The majority of high-throughput assays used for EDC screening measure end points based on a specific part of the ER pathway, usually in engineered systems (i.e., ER binding, dimerization, DNA binding, gene transcription changes, etc.). These approaches pay minimal, if any, attention to the impact on endogenous ER, where interrelations between multiple pathways occur, or attention to individual cell responses within the population. Cell-to-cell variability (here defined as phenotypic heterogeneity) in protein levels, phenotypes, responses to stimuli, gene transcription, and epigenetics is a common trait from bacteria to humans (Ackermann 2015; Bintu et al. 2016; Perlman et al. 2004; Raj and van Oudenaarden 2008; Schreiber et al. 2016; Singh et al. 2010), has been observed for many years but has rarely been exploited in high-throughput assays (Gough et al. 2016, 2017, 2014) or studies of environmental toxicants. Phenotypic heterogeneity is thought to serve as a “rheostat” for evolutionary processes and adaptive responses to stimuli (e.g., drugs, lack of nutrients, etc.) of a genetically homogeneous population (Ackermann 2015; Rubin 1990).
Although phenotypic heterogeneity is an important biological phenomenon, only a handful studies attempted to use phenotypic heterogeneity to quantify compound responses or discuss pipelines for quality control (QC) of such experiments. In excellent work from the Schurdak and Taylor groups (Gough et al. 2016, 2017, 2014), some phenotypic indexes have been proposed for both quality control and measurement of responses. These indexes inform on changes in cellular responses, presence of subpopulations, and response in only a subset of cells.
In this study, using the ER pathway as a test case, we sought to evaluate the reproducibility of phenotypic heterogeneity across multiple experiments (biological and technical replicates) with variations in sample preparation, acquisition, and analysis. The choice of ER nuclear levels is in line with the new consensus statement on key characteristics of EDCs [KC3, which alters hormone receptor expression (La Merrill et al. 2020)]. We used endogenous, nuclear levels in individual breast cancer cells as a model for phenotypic heterogeneity analysis. , the other estrogen receptor gene, is not expressed in the cell line used; hence, mention of ER in this study is always in reference to . The usefulness of this approach is that ER levels are modulated by ligands in different ways. For example, the natural agonist (E2) reduces ER levels through a negative feedback loop; the selective estrogen receptor modulator (SERM), 4-hydroxytamoxifen (4OHT) stabilizes ER; and the selective estrogen receptor down-regulator (SERD), Fulvestrant (ICI), immobilizes and ultimately degrades ER (Marsaud et al. 2003). The use of such control compounds allowed us to measure differences and reproducibility of ER single cell distributions upon treatments and across a large number of biological replicates. These controls served as basis to develop a two-tiered QC pipeline of ER distributions that identifies “bad,” i.e., nonreproducible, experiments. We further tested this pipeline by performing dose–response experiments with: a) a set of known EDCs (EPA45 from Judson et al. 2015) to determine the accuracy of this analysis method in comparison with the 18 assays in ToxCast; and; b) a set of 42 toxicants from the Agency for Toxic Substances and Disease Registry (ATSDR) list that were selected as environmentally relevant by the Texas A&M Superfund group (Chen 2020), and we identified a number of new toxicants that directly and indirectly affected ER levels and activity. We envision that similar approaches will greatly aid researchers venturing in single cell analysis of environmental toxicant actions.
Methods
Cell Culture
MCF-7 cells were obtained from BCM Cell Culture Core, which routinely validates their identity by genotyping, or directly from American Type Culture Collection (ATCC); all culture have constantly tested mycoplasma negative as determined by DAPI (Sigma-Aldrich) staining, which is evaluated by looking for extranuclear DAPI signal with a objective on a Cytiva DVLive epifluorescence deconvolution microscope. MCF-7 were routinely maintained in phenol-red free DMEM with the following additions, 10% FBS (Gemini), PenStrep, glutamine, and sodium pyruvate. Three days prior to experiments, cells were plated in media (DMEM) containing 5% charcoal-dextran stripped and dialyzed FBS. For 384 multiwell plates, 3,000 cells/well were plated; for other vessels it was proportionally scaled based on the well surface area (96-well plate: 10,000 cells/well, coverslip: 60,000/well).
Quality Control Experimental Design
We performed experiments over 3 y of time, using different cell passages and thaws (we always used up to eight passages for each round of thawing; all the thaws were derived from an initial cell culture stock that was frozen down in multiple vials), media and two-serum batches, personnel (as listed in Table 1), vessels (from coverslips to 384 multiwell plates), instruments for acquisition (manual vs. automated as listed in Table 1), analysis software (CellProfiler versions 2.0 and 3.0; https://cellprofiler.org/) [Carpenter et al. 2006 and mIA–myImageAnalysis (Szafran and Mancini 2014)], primary antibodies [from different sources with different epitopes, described below in the “IF and single molecule RNA FISH (smFISH).” section], secondary antibodies with two different fluorophores (Alexa488 or Alexa647), and number of cells acquired [indicated in Table 1 for dimethylsulfoxide (DMSO) samples as an example]. We specifically kept the magnification and numerical aperture constant () to ensure capture of a larger number of cells per run reducing the overall time of acquisition (Table 1) (example images in Figure 1A). From each experimental campaign ( biological replicates, with some experiments having “technical” replicate plates within the same biological replicate), we extracted single cell, nuclear ER mean intensity values, which represent an easy readout that is immediately apparent and biologically relevant, in comparison with other features obtainable from image analytics (e.g., texture and morphological features); however, in principle, the proposed framework is applicable to any feature and assay of interest.
Table 1.
Summary of experiments used for quality control pipeline establishment and testing.
| Experiment # | Instrument | Analysis software | Operator | # Cells DMSO |
|---|---|---|---|---|
| EXP01 | IC200 | mIA | RD | 11,691 |
| EXP02 | IC200 | CellProfiler | FS | 11,997 |
| EXP03 | DV | CellProfiler | FS | 2,274 |
| EXP04a | DV | CellProfiler | FS | 1,222 |
| EXP05a | DV | CellProfiler | RD | 588 |
| EXP06 | IC200 | CellProfiler | FS | 8,528 |
| EXP07 | IC200 | CellProfiler | RD | 7,202 |
| EXP09 | IC200 | CellProfiler | RD | 7,764 |
| EXP10 | IC200 | CellProfiler | RD | 7,795 |
| EXP11a | IC200 | mIA | FS | 4,527 |
| EXP12 | IC200 | mIA | FS | 4,492 |
| EXP13 | IC200 | CellProfiler | HJ | 9,744 |
| EXP14 | DV | CellProfiler | FS | 1,128 |
| EXP15a | DV | CellProfiler | HJ | 785 |
| EXP16 | IC200 | CellProfiler | HJ | 30,974 |
| EXP17 | IC200 | CellProfiler | HJ | 14,990 |
| EXP18 | IC200 | mIA | HJ | 30,055 |
| EXP19 | IC200 | CellProfiler | HJ | 2,068 |
| EXP20 | IC200 | CellProfiler | FS | 25,728 |
| EXP21a | IC200 | mIA | FS | 6,053 |
| EXP22 | IC200 | mIA | RM | 37,374 |
| EXP24 | IC200 | mIA | FS | 30,489 |
| EXP25 | IC200 | mIA | RM | 103,845 |
| EXP26_plate1 | IC200 | mIA | RM | 66,568 |
| EXP26_plate2 | IC200 | mIA | RM | 52,534 |
| EXP27_plate1 | IC200 | mIA | RM | 80,618 |
| EXP27_plate2 | IC200 | mIA | RM | 65,046 |
| EXP28 | IC200 | mIA | RM | 38,087 |
| EXP29 | IC200 | mIA | RM | 45,177 |
| EXP30 | IC200 | mIA | RM | 51,161 |
| EXP31_plate1 | IC200 | mIA | RM | 31,251 |
| EXP31_plate2 | IC200 | mIA | RM | 35,553 |
| EXP32_plate1 | IC200 | mIA | RM | 85,953 |
| EXP32_plate2 | IC200 | mIA | RM | 86,939 |
| EXP33_plate1 | IC200 | mIA | RM | 46,578 |
| EXP33_plate2 | IC200 | mIA | RM | 37,113 |
Note: IC200 refers to the Vala Sciences IC-200 high-throughput epifluorescence microscope with a Nikon PlanApo NA objective and a sCMOS camera; DV refers to Cytiva DVLive epifluorescence image restoration microscope, using an Olympus PlanApo NA objective and a sCMOS camera. # cells in DMSO indicates the number of objects counted by image analysis in vehicle (DMSO) treated wells in each experiment. DMSO, dimethylsulfoxide; QC, quality control.
Experiments that failed the QC analysis (Figure 1).
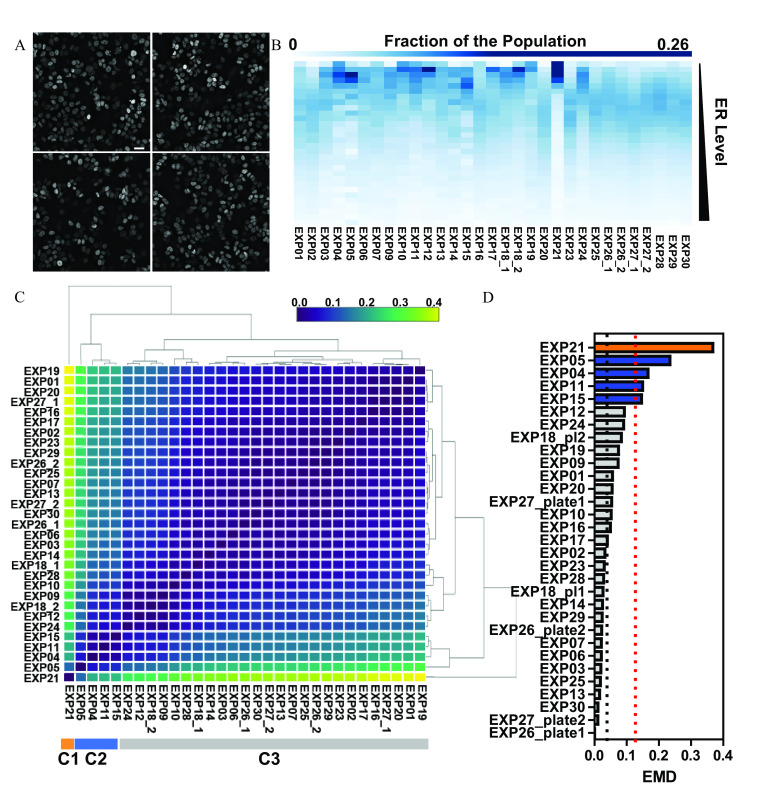
Figure 1.
Quality control pipeline for ER distribution under control conditions. (A) Random fields of view of ER-immunolabeled, untreated MCF-7 breast cancer cells automatically imaged at , shown as maximum intensity projection after image deconvolution. Scale bar: . (B) Heat map representation of the binned distribution of single cell ER levels across the indicated independent biological and technical replicates, with lower ER levels at the top and higher ER levels at the bottom of the heat map. (C) The distance between the ER distributions in two experiments was measured by earth mover’s distance (EMD), and the pairwise EMD distances were submitted to hierarchical clustering. The three main clusters “C1,” “C2,” and “C3” are indicated at the bottom of the heat map. (D) EMD distances calculated from the median ER distribution across all experiments in the main cluster from panel C. Experiments that deviated standard deviations from the mean of the main cluster EMDs are identified according to the cluster map in panel C. The dotted line above represents three standard deviations from the mean of the main cluster (dotted line above approximately 0.05 EMD). Summary data for Figure 1B and D are found in Excel Tables S1 and S2, respectively. Note: EMD, earth mover’s distance; ER, estrogen .
Chemicals
All chemicals were provided to us directly from the U.S. EPA (Dr. Keith Houck) or through the Texas A&M Superfund program (Dr. Ivan Rusyn and Dr. Weihsueh Chiu), which acquired them and provided aliquots arrayed in 96 well plates. A list of the chemicals and their CAS numbers has been added as Table 2. The chemicals’ highest tested concentration was from where the indicated dilutions (either half logs or logs) were made, as indicated in the figures (range was 10 pM–, except for Figure 2E and 3A, where the lower concentration was 10 fM and 10 pM, respectively). All treatments were conducted for 24 h before immunofluorescence (IF) or smRNA FISH. In each experiment, the following controls were included: vehicle (DMSO, 1:1,000), (E2, ), 4-hydroxytamoxifen (4-OHT, ), and Fulvestrant (ICI 182,780 ). Every chemical/concentration had a minimum of 4 wells/384 well plate (technical replicates), per biological replicate. Every ER IF experiment with chemical treatment was repeated a minimum of four times.
Table 2.
List of chemicals used.
| Name of chemical | CAS# |
|---|---|
| 17alpha-estradiol | 57-91-0 |
| 17alpha-ethinylestradiol | 57-63-6 |
| 17beta-estradiol | 50-28-2 |
| 17-methyltestosterone | 58-18-4 |
| 4-(1,1,3,3-Tetramethylbutyl)phenol | 140-66-9 |
| 4-Cumylphenol | 599-64-4 |
| 4-Hydroxytamoxifen | 68392-35-8 |
| 4-Nonylphenol | 104-40-5 |
| 5alpha-Dihydrotestosterone | 521-18-6 |
| Apigenin | 520-36-5 |
| Atrazine | 1912-24-9 |
| Benzyl butyl phthalate | 85-68-7 |
| Bisphenol A | 80-05-7 |
| Bisphenol B | 77-40-7 |
| Chrysin | 480-40-0 |
| Corticosterone | 50-22-6 |
| Cycloheximide | 66-81-9 |
| Daidzein | 486-66-8 |
| Di(2-ethylhexyl) phthalate | 117-81-7 |
| Dibutyl phthalate | 84-74-2 |
| Dicofol | 115-32-2 |
| Diethylstilbestrol | 56-53-1 |
| Estrone | 53-16-7 |
| Ethylparaben | 120-47-8 |
| Fenarimol | 60168-88-9 |
| Flutamide | 13311-84-7 |
| Genistein | 446-72-0 |
| Haloperidol | 52-86-8 |
| Hydroxyflutamide | 52806-53-8 |
| Kaempferol | 520-18-3 |
| Kepone | 143-50-0 |
| Ketoconazole | 65277-42-1 |
| Linuron | 330-55-2 |
| meso-Hexestrol | 84-16-2 |
| Methoxychlor | 72-43-5 |
| o,p′-DDT | 789-02-6 |
| p,p′-DDE | 72-55-9 |
| Phenobarbital sodium | 57-30-7 |
| Procymidone | 32809-16-8 |
| Progesterone | 57-83-0 |
| Raloxifene hydrochloride | 82640-04-8 |
| Reserpine | 50-55-5 |
| Spironolactone | 52-01-7 |
| Tamoxifen | 10540-29-1 |
| Tamoxifen citrate | 54965-24-1 |
| Benzo(a)anthracene | 56-55-3 |
| Naphthalene | 91-20-3 |
| Fluoranthene | 206-44-0 |
| DDT, p,p′- | 50-29-3 |
| Dieldrin | 60-57-1 |
| Aldrin | 309-00-2 |
| Heptachlor | 76-44-8 |
| Hexachlorocyclohexane, gamma- | 58-89-9 |
| Disulfoton | 298-04-4 |
| Endrin | 72-20-8 |
| Diazinon | 333-41-5 |
| Heptachlor Epoxide | 1024-57-3 |
| Pentachlorophenol | 87-86-5 |
| Di-n-butyl phthalate | 84-74-2 |
| Chlorpyrifos | 2921-88-2 |
| 2,4,6-Trichlorophenol | 88-06-2 |
| Ethion | 563-12-2 |
| Azinphos-Methyl | 86-50-0 |
| 2,4,5-Trichlorophenol | 95-95-4 |
| Parathion | 56-38-2 |
| Benzo(b)fluoranthene | 205-99-2 |
| Trifluralin | 1582-09-8 |
| Acenaphthene | 83-32-9 |
| DDD, p,p′- | 72-54-8 |
| Benzidine | 92-87-5 |
| Endosulfan | 115-29-7 |
| Methoxychlor | 72-43-5 |
| 2,4-Dinitrophenol | 51-28-5 |
| 2,4-Dinitrotoluene | 121-14-2 |
| Dicofol | 115-32-2 |
| Cresol, para- | 106-44-5 |
| DDT, o,p′- | 789-02-6 |
| 4,6-Dinitro-O-Cresol | 534-52-1 |
| 1,2,3-Trichlorobenzene | 87-61-6 |
| Lead (Nitrate) | 10099-74-8 |
| Cadmium (Chloride) | 10108-64-2 |
| Zinc (Chloride) | 7646-85-7 |
| Mercuric chloride | 7487-94-7 |
| Potassium chromate | 7789-00-6 |
| Cobalt (Chloride) | 7646-79-9 |
| Nickel (Chloride) | 7718-54-9 |
Figure 2.
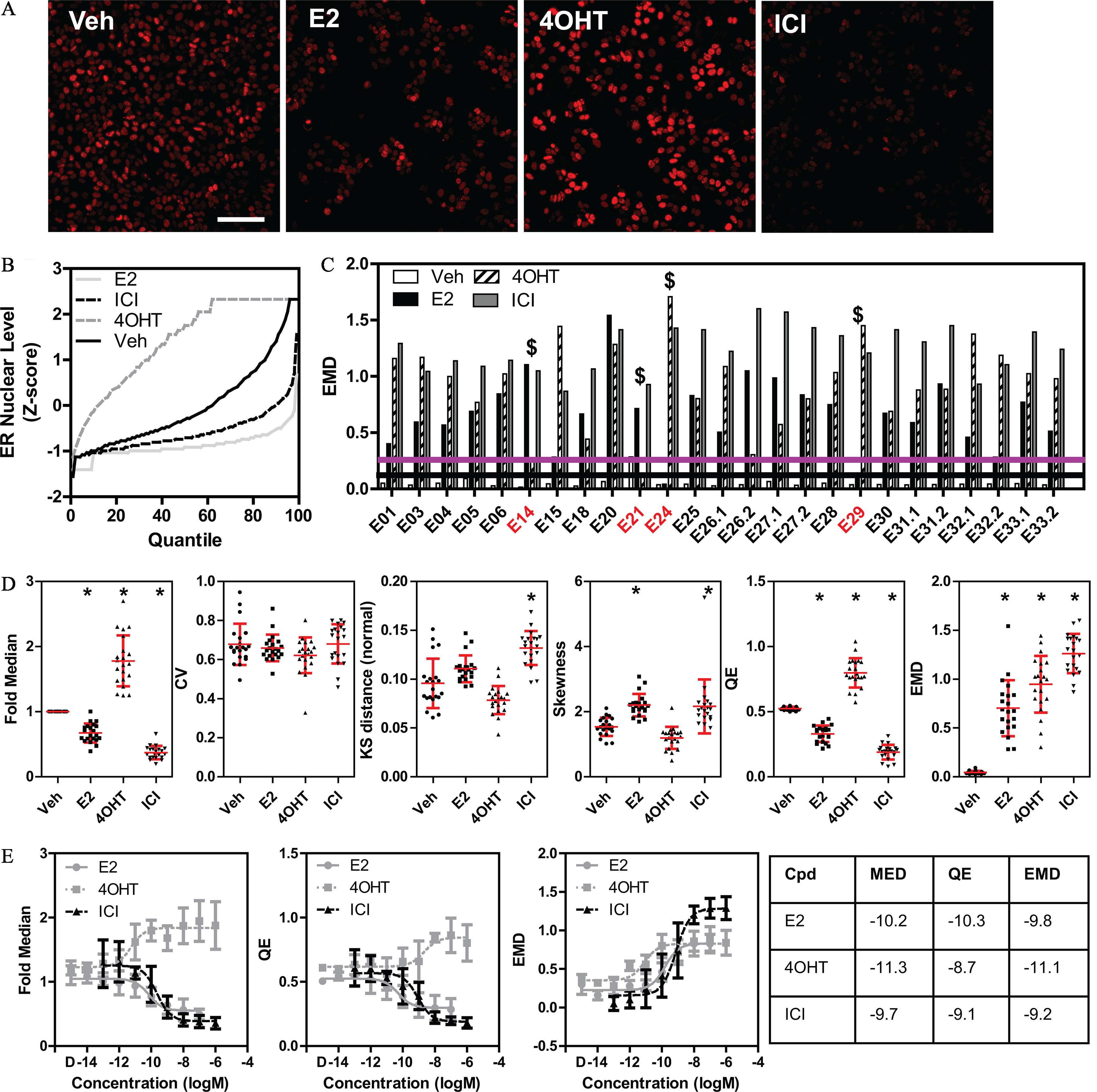
Quality control for experiments measuring changes in ER distribution after ligand treatment. (A) Representative random images of ER immunolabeling of MCF-7 cells treated for 24 h with DMSO (vehicle), (E2, ), 4-hydroxytamoxifen (4OHT, ), or Fulvestrant (ICI, ). Scale bar: . (B) Quantile plot curves of a representative experiment showing the effects of the indicated compounds on changing the ER distribution in comparison with vehicle (DMSO). ER levels are normalized by z-score based on the DMSO median/MAD. (C) EMD measure from the reference distribution showing the distances of the three control compounds across multiple independent replicate experiments. indicates experiments that failed to pass the QC test for ligand effects, also highlighted by red experiment names. The upper (purple) horizontal line represents three standard deviations from the vehicle (black, lower horizontal line). (D) Effects of the control ER ligands on several measured phenotypic heterogeneity indexes (CV, KS, and QE) across multiple experiments (every point in the graphs represents a single experiment). * using nonparametric ANOVA using vehicle as comparator. Mean and standard deviation are indicated. (E) Eight-point dose–response curves (mean and standard deviation of experiments, concentration is in logM) for the indicated descriptors in MCF-7 cells after 24 h of treatment with the control ligands and DMSO control (D). The table shows the (activity concentration 50%) values in molarity. Summary data for Figure 2B,C,D,E are found in Excel Tables S3-S6. Note: ANOVA, analysis of variance; Cpd, compound; CV, coefficient of variation; DMSO, dimethylsulfoxide; EMD, earth mover’s distance; ER, estrogen ; MAD, median absolute deviation; KS, Kolmogorov-Smirnov; MED, fold median of DMSO control; QE, quadratic entropy.
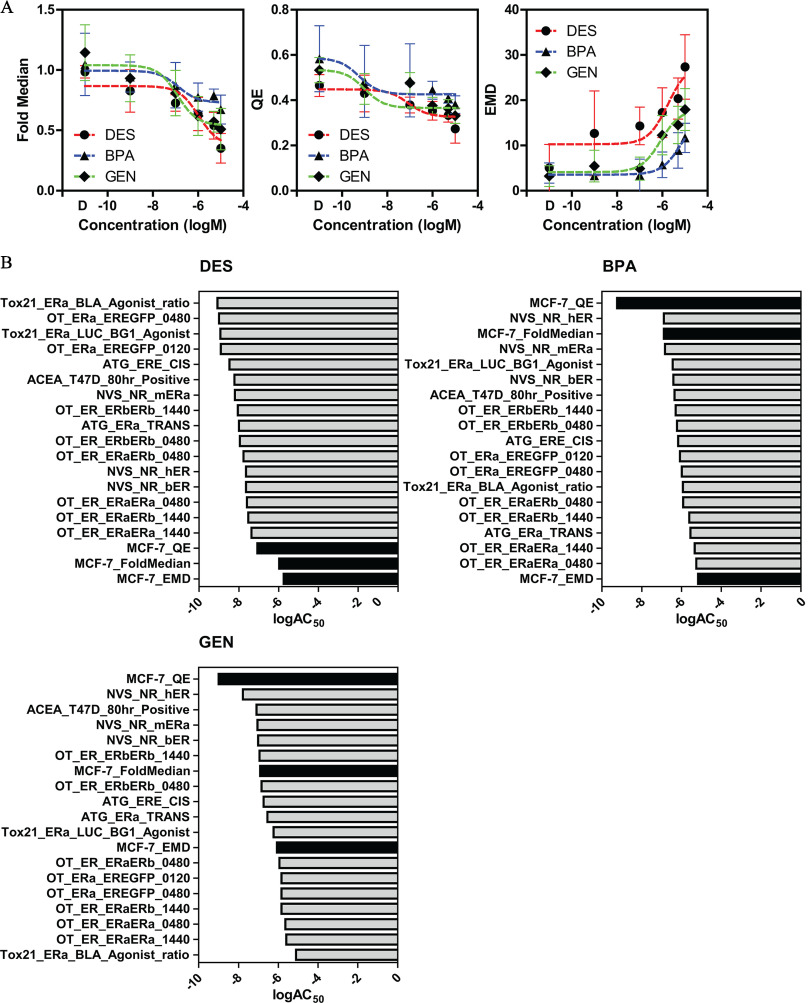
Figure 3.
Single cell analysis of three established estrogenic compounds. (A) MCF-7 cells treated for 24 h with six concentrations of diethylstilbestrol (DES, circles), bisphenol A (BPA, triangles), or genistein (GEN, diamonds) in comparison with DMSO control (D). Changes in single cell ER levels are represented by three descriptors: fold median of DMSO control, QE, and EMD. (B) The (activity concentration 50%) values of the three descriptors in panel A are ranked together with the ER assays present in ToxCast. Black columns indicate phenotypic index matrixes. Summary data are found in Excel Tables S7 and S8. Note: ER, estrogen ; EMD, earth mover’s distance; QE, quadratic entropy.
IF and single molecule RNA FISH (smFISH).
IF experiments were completed as previously described (Stossi et al. 2016) after cell seeding in different vessels to test whether vessel choice would impinge on the results (i.e., Greiner BioOne 384 glass-bottom plates, Aurora 384 optical plastic-bottom plates, poly D-lysine–coated glass coverslips in a 24-well plate). Briefly, cells were fixed in 4% EM-grade formaldehyde in phosphate-buffered saline (PBS) and permeabilized with 0.5% Triton X-100 for 30 min. Cells were incubated at room temperature in Blotto for 1 h, and then specific antibodies (1:1,000 dilution) were added overnight at prior to washing and 30 min of secondary antibody (AlexaFluor conjugates; Molecular Probes) at room temperature, followed by DAPI (Sigma-Aldrich, for 5 min at room temperature) staining. The primary mouse monoclonal antibody (mAb, clone 127) was generated in house (see below and Figure S9); the rabbit anti-ER (Millipore 04-820) and rat mAb (H-222, Santa Cruz Biotechnology) were obtained commercially. The three antibodies were used in a triple IF experiment (results are shown in Figure S1E) using the same protocol described above. Pairwise correlations between these antibodies were published in Stossi et al. 2020a.
GREB1 smFISH was performed as described in detailed protocols on poly D-lysine–coated coverslips (Mistry et al. 2020). Briefly, after fixation of cells as described for IF, cells were permeabilized overnight at in 70% ethanol, prior to a quick wash (10% formamide in 2X SSC buffer) and overnight hybridization ( in humidity chamber) with custom GREB1 intron- and exon-specific probe sets (LGC Biosearch Technologies, 1:300 dilution in LGC Biosearch Technologies hybridization buffer plus 10% formamide, probe locations are indicated in Excel Table S17). Next day, coverslips were washed twice, 15 min at , with wash buffer (10% formamide in 2X SSC buffer), followed by 5 min of DAPI staining in 2X SSC buffer. Coverslips were then mounted with Vectashield antifade media (Vector Labs) and imaged within 48 h.
Generation and Validation of the Clone 127 Antibody
In collaboration with mAbVista (Houston, Texas, USA) and LGC Biosearch Technologies (Novato, California, USA), a protein expression construct was designed based on information from the National Center for Biotechnology Information (NCBI) protein database (Gene ID: 2099, Ensembl:ENSG00000091831) and literature, containing a region of human with conserved domain structure and predicted to have high protein solubility and antigenicity. The resulting construct containing the N-terminal domain of human (aa 1-180) was expressed as a 6x His tag C-terminus fusion and purified by nickel affinity resins plus Hi-TRAP ion exchange by EZBiolab (Carmel, Indiana, USA). The purified antigen was used to immunize four 6-wk-old BALB/c female mice (The Jackson Laboratory). After three rounds of immunization [week protein in of PBS mixed with of Complete Freund’s Adjuvant, subcutaneous injection; week protein in of PBS mixed with of Incomplete Freund’s Adjuvant (IFA), intraperitoneal injection; week protein in of PBS mixed with of IFA, subcutaneous injection], serum isolated from tail bleeds was evaluated for antibody titers using enzyme-linked immunosorbent assay (ELISA) and IF methods by mAbVista. Indirect ELISA was performed using -coated ( in carbonate buffer [ , , (pH 9.6)] polystyrene 96-well microtiter plates, HRP-conjugated secondary antibodies (Abcam), and plate-based luminometer measuring relative light units (RLU) at . IF labeling performed as described elsewhere in the “Methods” section using the GFP-ER:PRL-HeLa cell line that stably expresses with an N-terminal green fluorescent protein (GFP) fusion. (Ashcroft et al. 2011). The mouse with the highest antibody titer was selected for final boost ( protein in of PBS, intraperitoneal injection) 3 d prior to sacrifice and spleen harvested into RPMI 1640 culture media (Gibco) supplemented with 10% FBS (VWR) and penicillin ()/streptomycin () (Gibco). Splenocytes were isolated by transferring the harvested spleen and media into a petri dish, dissecting, and manually teasing cells from the spleen capsule using sterile techniques. After washing, cells were transferred to RPMI 1640 culture media without FBS. Splenocytes were fused with mouse myeloma SP2/O cells [ATCC, maintained in DMEM cell culture media (Gibco) supplemented with 10% FBS] using the using the ClonaCell-HY kit (Stemcell Technologies) following the manufacturer’s protocol. Fused cells were plated (day 1) into ClonaCell methylcellulose semisolid media as a single cell suspension, and hybridoma selection was begun with ClonaCell Medium D, a DMEM-based media containing hypoxanthine-aminopterin-thymidine (HAT). On day 14, monoclonal colonies were isolated using an automated colony picking system (K Biosystems). Beginning on day 17, hybridoma supernatants were screened for the presence of antibodies using ELISA and IF assays as described above for antibody titers. IF labeling was quantified using mIA image analysis software and the amount/specificity of binding to the expressed and absence of background signal in GFP negative cells (Figure S9A–E).
Clone 127 was expanded by Baylor College of Medicine’s Monoclonal Antibody Advanced Technology Core by growth in IMDM growth media (Gibco) supplemented with 15% FBS (VWR). Purified antibody was obtained from supernatant media using protein G purification and ammonium sulfate precipitation.
Cell lysates were obtained from MCF-7 cells using ice-cold NP-40 lysis buffer, combined with LDS Sample Buffer (Novex) and boiled for 5 min. Samples were then loaded onto 8%–12% acrylamide gels and run at 200 V at for 90 min. Gels were then transferred onto Immobilon transfer membranes at for 2 h at 80 V. Nonspecific antibody binding was blocked with 5% milk in TBS-T. Blots were then probed using clone 127 supernatant (1:4 dilution) or clone 60C (Millipore, 04-820, 1:2,000 dilution). Proteins were visualized using antimouse (Abcam, ab205719; 1:5,000 dilution) and anti-rabbit (Abcam, ab205718; 1:50,000 dilution) secondary antibodies conjugated to HRP and a chemiluminescence detection system (Figure S9F).
Imaging
High-quality/high-resolution imaging for smFISH was performed on a Cytiva DVLive epifluorescence image restoration microscope using an Olympus PlanApo NA objective and a sCMOS camera. Z stacks () covering the whole nucleus () were acquired before applying a conservative restorative algorithm for quantitative image deconvolution.
High-throughput imaging for the QC analysis and follow-up experiments was performed on two different instruments: a) a Cytiva DVLive epifluorescence image restoration microscope using an Olympus PlanApo NA objective and a sCMOS camera; and, b) a Vala Sciences IC-200 high-throughputhigh-throughput epifluorescence microscope using a Nikon PlanApo NA objective and a sCMOS camera. Z stacks ( steps) covering the whole nucleus () were acquired for all instruments. For the DVLive, images were deconvolved using a conservative restorative algorithm. Max intensity projections were generated and used for image analysis.
Image Analysis
Automated image analyses for the high-throughput microscopy experiments was performed with previously published methods (Mistry et al. 2020; Stossi et al. 2020b), either via the open-source platform CellProfiler (Carpenter et al. 2006) and/or our custom PipelinePilot (Biovia)-based software mIA [myImageAnalysis (Szafran and Mancini 2014)]. Briefly, 16-bit grayscale images were background subtracted; DAPI signal was smoothed by a median filter; nuclei were segmented using the DAPI channel via a global minimum cross-entropy algorithm and filtered by size and intensity to eliminate poorly segmented nuclei (“cell clumps”), debris, dead and mitotic cells. Touching nuclei were separated using a watershed algorithm. Mean pixel intensity per nucleus for the ER channel was then extracted and used for downstream analysis.
Single Cell Analysis Quality Control Pipeline Development
The pipeline for quality control of single cell analysis is as follows. Single cell values were processed in the following manner: First, data were trimmed at the fourth and 96th percentiles to reduce the impact of outliers on the distribution range and shape; then, data was normalized using the median/median absolute deviation (MAD) values. This is an important step because absolute mean intensity values are highly variable between runs, due to intrinsic variability of experimental conditions (i.e., vessels, instrument light source, antibodies, etc.). For each experiment, we used the median and MAD of the DMSO vehicle control to normalize each compound including vehicle itself. We obtained the quantile values (first to 99th) for each compound.
For the second tier QC pipeline, after data normalization, we applied dynamic time warping (DTW) to warp the quantile curve of the compound to the quantile curve of the vehicle to mimic the relation between vehicle and each compound. We used the DTW to align the quantile curves for the compound and the vehicle and called this alignment function . DTW allowed us to approximately align two signals, or time series together, by finding a time-warping function that transforms, or warps, time (Sakoe and Chiba 1978). Next, we used DTW to obtain alignment function between the vehicle quantile curve and the quantile curve for the standard normal distribution. The composition of these two functions provided a closely aligned quantile curve with respect to the standard normal distribution and maintaining the relation between the compound and the vehicle. The original DTW algorithm used dynamic programming to compute a time-warping path that minimized misalignments in the time-warped signals while satisfying monotonicity, boundary, and continuity constraints.
Suppose we are given two time-series and of lengths and , respectively. and are termed as query and reference, respectively. Assuming that there is a nonnegative, local dissimilarity function f defined between any pair of elements and :
for all and . The goal is to find the warping curve , : with and such that the average accumulated distortion between the warped time series X and Y is minimized. This deformation brings these two time series as close as possible to each other. We used the implementation of the algorithm provided by the DTW package in R (Giorgino 2009) with a slight modification of converting the warping curve to a function by using rounded median value of the indices , corresponding to a fixed between and
As a last step, we calculated earth mover’s distance (EMD) for each control compound in comparison with vehicle-treated cells and the reference distribution described in Figure 1. EMD was calculated using the R package emdist. EMD was chosen because it is a cross-bin distance function and is very robust in comparison with other commonly used metrics, such as Kullback-Leibler divergence, Kolmogorov-Smirnov distance, and others. We used the Euclidean dissimilarity along with complete linkage to obtain the hierarchical clustering of the EMD (Rubner et al. 2000) distances (hclust function in R).
The phenotypic heterogeneity indexes used were the same as those described in Gough et al. 2016, 2017, 2014. In brief, the coefficient of variation (CV) was calculated as the standard deviation (SD) divided by the mean; the Kolmogorov-Smirnov distance (KS) was defined as maximum distance between the cumulative distribution functions (CDFs) of the experimental data and the reference distribution; skewness was a measure of the symmetry of a distribution; and quadratic entropy (QE) measured the entropy of the system by subdividing the distribution in equally separated bins.
The full QC analysis pipeline is available at https://github.com/pankajmath/SingleCellQC.
Simulations
To estimate the minimum number of experiments needed to generate a reference distribution that is close to the reference curve obtained from all the experiments, we mimicked the situation where we consider that we have experiments out of 31 available to begin with. For a fixed value of , we randomly picked combinations of experiments, where is the minimum of 100,000 and , total number of possible combinations of experiments. Next, we computed the functional median quantile curves for these combinations and obtained the EMD distances from the reference curve.
For estimating the minimum number of cells required to estimate the reference curve, we started with all the cells in the reference experiment (EXP26_1). For each fixed k between 1 and 665, we randomly sampled cells out of approximately 66,500 total cells in the reference experiment. We normalized these ER level values for the cells using median/MAD, obtained the corresponding warped quantile curve, and computed the EMD from the reference quantile curve. We repeated this process 1,000 times for each k.
Data Representation and Statistical Analysis
All graphs and analyses were generated in Python (version 3.7.10, Anaconda Individual Edition) (Figures 1C; 2B,C,D,E; 3A; 4A; 5A), R (version 3.6.0, R Development Core Team) (Figure 1C, 2B), Orange (Version 3.0, Orange Developing Team) (Figures 1B, 4A, 5A) (Demsar et al. 2013) and Prism (version 5.0, GraphPad) (Figures 1D, 2B, 2C, 2D, 2E, 3A, 3B, 4B, 4C, 4D, 5B, 5C, 6B). ToxCast (version 3.1) data was downloaded from https://www.epa.gov/chemical-research/exploring-toxcast-data, and the reported (Activity Concentration 50) was used for analysis based on the data from Judson et al. 2015. Nonparametric analysis of variance (ANOVA) using vehicle as comparator was used when indicated in the figure legends, and groups were considered significant at .
Figure 4.
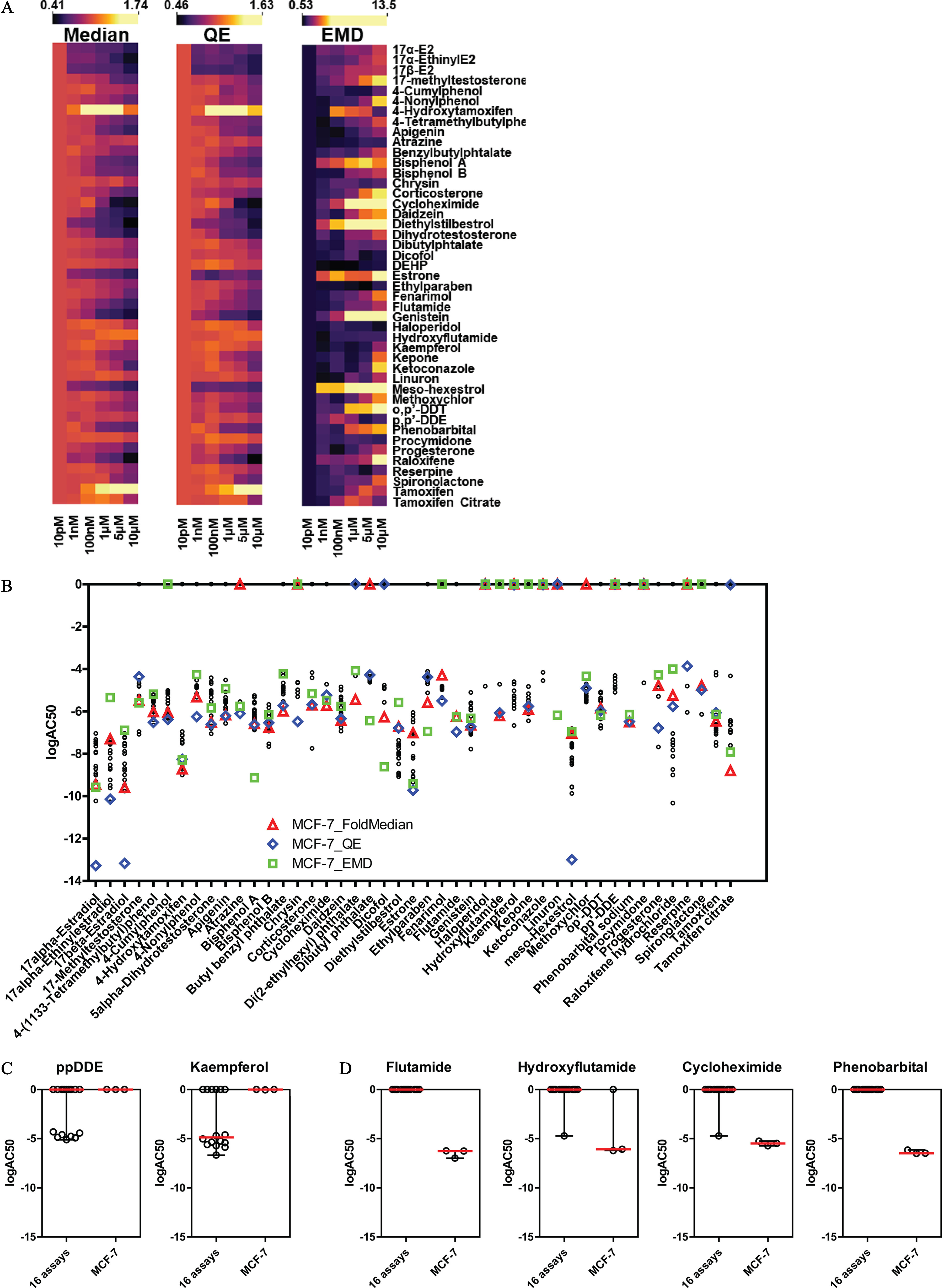
Evaluation of the EPA45 control set for estrogenic activity by single cell ER analysis. (A) Heat maps for the three indicated descriptors (fold median of DMSO control, QE, and EMD, independent biological replicates) after treatment of MCF-7 cells for 24 h with the EPA45 set of compounds, here shown in alphabetical order. (B) (activity concentration 50%) comparison between ER descriptors (fold median, triangles; QE, diamonds; EMD, squares) in MCF-7 and ToxCast assays (black dots). (C–D) False negative (C) and false positive (D) chemicals shown by . Summary data are found in Excel Tables S9-S12. Note: DMSO, Dimethylsulfoxide; ER, estrogen ; EMD, earth mover’s distance; QE, quadratic entropy.
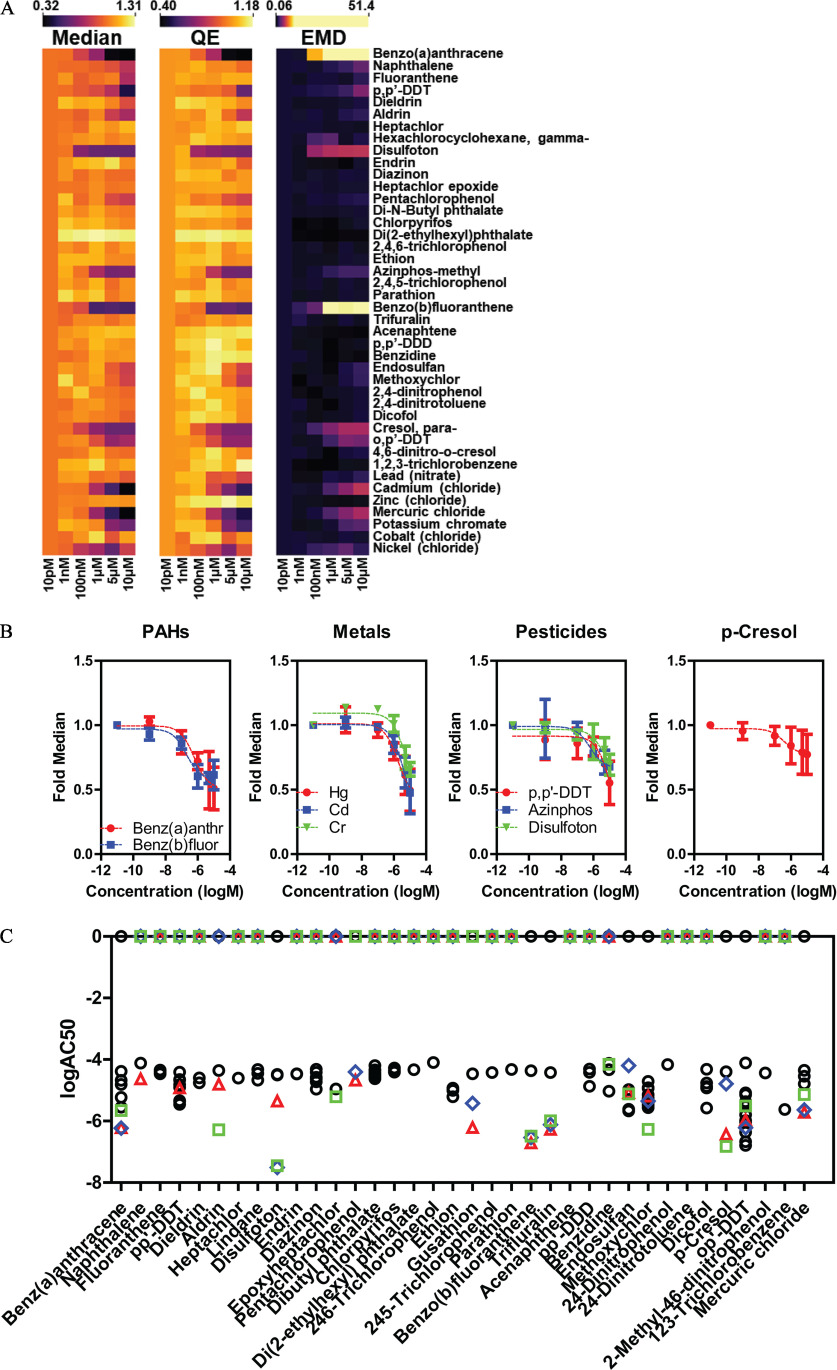
Figure 5.
Evaluation of 42 chemicals from the ATSDR list for estrogenic activity by single cell ER analysis. (A) Heat maps for the three indicated descriptors (fold median of DMSO control, QE, and EMD, independent biological replicates) after treatment of MCF-7 cells for 24 h with the ATSDR 42 set of compounds. (B) Fold median ER dose response data for three classes of chemicals, plus para-cresol, that where hits from the ATSDR 42 set. (C) (activity concentration 50%) comparison between ER descriptors (fold median, triangles; QE, diamonds; EMD, squares) and ToxCast assays (black dots) for the ATSDR 42 chemicals. Summary data are found in Excel Tables S13-S15. Note: ATSDR, Agency for Toxic Substances and Disease Registry; ER, estrogen ; EMD, earth mover’s distance; PAH, polycyclic aromatic hydrocarbons; QE, quadratic entropy.
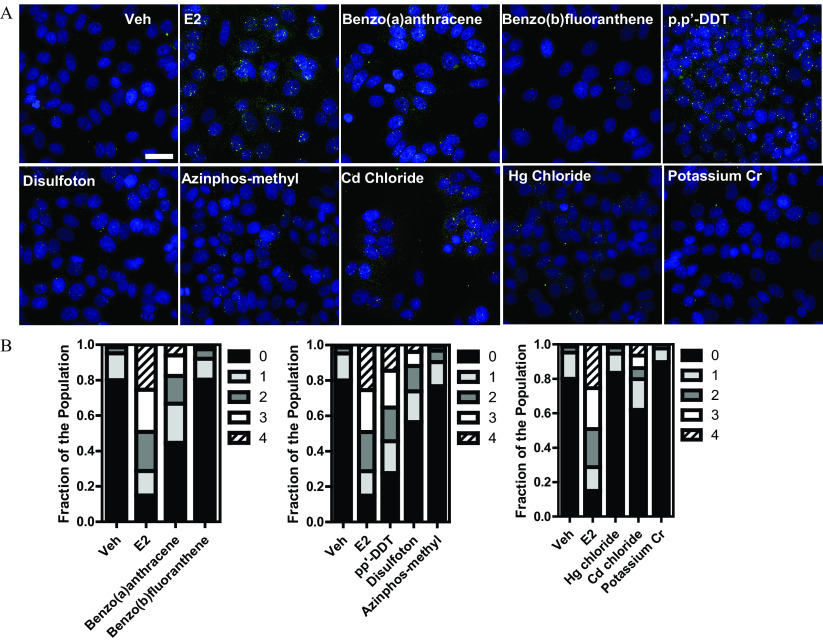
Figure 6.
GREB1 single molecule FISH (smFISH) identifies ATSDR 42 hits as potential activators of ER signaling. (A) MCF-7 cells were treated for 24 h with the indicated compounds (E2, ; metals, ; others, ) and subjected to smFISH using specific probes for GREB1 exons (green) and introns (red). NA deconvolved and max projected images are shown. Scale bar: . (B) Stacked bar graphs representing, with each part of the bar the fraction of cells in the population with the corresponding number of GREB1 active alleles (0–4). Results are from three independent evaluations with cells/treatment. Summary data found in Table S16. Note: E2, ; ER, estrogen . FISH, fluorescence in situ hybridization.
Results
Development of a Quality Control Pipeline for ER Phenotypic Heterogeneity
We used ER IF in breast cancer cells as a case study for reproducibility and quality control of single cell distributions and to examine responses of the system to environmental toxicants. MCF-7 is an established model where cell-to-cell variation in ER levels was observed more than two decades ago, yet largely remained unaddressed (Palmari et al. 1997; Resnicoff et al. 1987). Figure 1A shows ER immunolabeling of MCF-7 cells in random fields of a 384-well plate; imaged automatically on a high-throughput microscope under DMSO control treatment. It is visually evident that, in each field, there was cell-to-cell variation in ER nuclear levels that can be easily quantified by automated image analysis routines. We first addressed whether the single cell ER distribution was affected under basal conditions by experimental variation and then established a QC metric to qualify each experiment. We randomized several parameters, described in the “Methods” section, to partially mimic various laboratories performing the same experiment around the world, with likely access to resources different from our own. After data normalization, we calculated a frequency distribution plot by dividing the data into 30 bins, a number that was chosen based on the square root of the smallest number of data points acquired across the experiments. Figure 1B shows the frequency plots of ER single cell distributions as a heat map. For each experiment, the quantile curve was warped to that of the standard normal distribution. Because we do not know the ground truth (if it exists) of the distribution of ER levels in a cell population, we calculated pairwise EMD between the warped quantile curves for all the experiments and then performed unsupervised hierarchical clustering, followed by plotting them as a pairwise distance heat map (Figure 1C).
We identified an outlier experiment (EXP21, on the far left, cluster “C1”), a small (four experiments, cluster “C2”), and a large cluster (“C3”). The identification of these three groups of experiments was corroborated by performing principal component analysis (Figure S1A).
We defined a reference distribution for ER levels that allows a distance calculation from it for each new experiment as a QC metric. We considered the largest cluster as the one with the higher likelihood of containing the ground truth distribution, identified the functional median distribution of those experiments, and selected it as reference distribution (EXP26_1). We calculated EMD from such a reference distribution, and the mean and SD of the EMDs for all the experiments in the large cluster. We considered experiments with SDs away from the mean as failing to pass this first QC tier (Figure 1D), which is a common threshold used in high-throughput screening.
The results obtained across independent runs were very encouraging because 84% (26/31; Figure 1D) of the experiments had similar distribution of ER levels, allowing EMD to be used as a new QC metric. We next ran a simulation analysis to calculate how many initial experiments would be needed to generate a reference distribution for establishment of a new single cell-based assay. Figure S1B shows the mean EMD distance from the reference curve (EXP26_1), and it suggests that a minimum of five to six experiments would have been sufficient for establishing a reference curve for this specific assay and end point. We repeated a similar simulation to determine the minimum number of cells per experiment that need to be acquired to construct a reference distribution that is close to the ground truth. There were about 66,500 cells in the reference experiment (EXP26_1). Figure S1C suggests that a minimum of 1,200 to 1,500 cells would have been enough to closely approximate the reference curve. We also measured EMD across 28 replicate wells of untreated cells in the same plate (Figure S1D). Well-to-well variation was clearly identified, suggesting that this QC pipeline would be usable in identifying outlier wells due to procedural or other errors. We also measured EMD of a single sample using simultaneous triple immunolabeling with three different primary antibodies (Figure S1E), which showed that all three could be used because all fell in between QC parameters.
QC Pipeline to Assess Effects of Estrogenic Chemicals
Endogenous ER is a good model to study the effect of compounds because its protein levels are modulated differentially upon ligand binding. Specifically, as shown by immunolabeling in Figure 2A and quantified by quantile–quantile curves in Figure 2B, we used three control compounds at saturating concentrations, that, as far as we have tested, cover the spectrum and magnitude of responses in the MCF-7 model system: E2 (lower ER levels after 24 h), 4-hydroxytamoxifen (4-OHT) (higher ER levels), and Fulvestrant (ICI, lowest ER levels). The fact that ER levels could be modulated by ligands prompted us to develop a second tier QC pipeline, described in the “Methods” section, for qualifying experiments where the cells reproducibly responded to the three control compounds. For a compound to be considered significantly different from vehicle, we chose a cutoff value larger than three SDs from vehicle samples to pass QC (as determined in Figure 1D). By this criterion (upper line in Figure 2C), four experiments did not pass the second tier QC (EXP14, EXP21, EXP24, and EXP29), because one or more control compounds failed to respond as expected.
From the experiments that passed both QC tiers, we extracted a few of the possible indexes that could be used to quantify changes in ER levels upon ligand treatment. Specifically (Figure 2D), we measured the median intensity of ER levels (fold median of DMSO control), the dispersion of single cell data (CV), the normality of the single cell ER levels distribution (KS), the asymmetry of such distribution (skewness), the “ecological diversity” (QE), and the EMD. Of these measurements, apart from the expected fold median, only QE and EMD showed significant differences across all compound treatments and experiments (by nonparametric ANOVA), with EMD having the largest dynamic range (fold difference between maximum and minimum values; 32.4 average, range 43.6–11.8, CV 0.26), although it did not discriminate between 4OHT vs. E2 and ICI because it was always a positive value, measuring a distance from vehicle control. The dynamic range of the EMD was higher than both fold median (4.9 average, 6.4–3.3 range, CV 0.13) and QE (4.6 average, range 8.5–3, CV 0.32). All three parameters provided useful information on compound behavior either in magnitude, direction, or distribution of the response, and as such we kept all three for subsequent analyses.
To test the sensitivity of the single cell ER level assay, we performed a nine-point dose response (1 fM to ) of the three reference compounds (Figure 2E). The table in Figure 2E shows the values for three indexes (median ER level, QE, and EMD).
We first selected three additional estrogenic compounds of diverse chemistry, the synthetic estrogen diethylstilbestrol (DES), the plasticizer bisphenol A (BPA), and the phytoestrogen isoflavone genistein (GEN). We performed a six-point dose response (10 pM to ) for the three compounds and calculated the three indexes described above (Figure 3A), which allowed us to estimate and rank their activity concentration, 50% () to compare the results of the MCF-7 assay with the ToxCast database (Figure 3B). Interestingly, the ranking of the endogenous ER level assays was vastly different between compounds. In the case of DES, the three MCF-7 indexes ranked the worst (i.e., less sensitive), which might be a “false negative” result, because the indexes value at 1 pM were already different than vehicle-treated samples. For BPA, the three indexes showed a very different trend, EMD was again the worst; however, QE and fold median ranked first and third for sensitivity; GEN had a similar profile as BPA, with QE showing the highest sensitivity.
Analysis of the EPA45 Set of Control Compounds
We treated MCF-7 cells with a six-point dose response (10 pM to ) of three different batches of 45 compounds provided directly by the EPA (Judson et al. 2015), using multiple biological replicates. The multipoint dose response was designed to cover for each compound across a wide concentration range based on (Judson et al. 2015). In Figure 4A the dose–response data for all the compounds is shown as a heatmap for the three indexes, with the lowest dose (10 pM) set to 1. It is important to note that these dose–response experiments were repeated a minimum of four independent times to investigate reproducibility and variability of the responses and indexes with heat maps representing the average results, with full results shown in Figures S2–S4 and Excel Table S19. Moreover, we obtained three different batches of the 45 chemicals from the U.S. EPA, allowing us to check for batch-to-batch variations (Figure S5 and Excel Table S20), which proved that the results were fairly consistent, with only five chemicals being less-reproducible across batches. However, four of these nonreproducible or less reproducible chemicals were “inactive” (less than 20% change in ER levels), whereas one, hydroxyflutamide, showed some activity (see below) and would require additional confirmation using a different source of the chemical. We compared the values for the three phenotypic indexes with those obtained from 16 ToxCast assays run in a similar agonist mode (i.e., compounds alone). Figure 4B depicts the values for all 16 assays (black dots) and for the MCF-7 data obtained in this study (represented as colored shapes: triangle, square, and diamond). Overall, the MCF-7 data matched well with the ToxCast data, highlighting the fact that monitoring changes in endogenous ER levels is a relevant quantitative assay. Figure 4C–D shows compounds that had activity uniquely in the MCF-7 assay or that they have been missed in comparison with the 16 assays in Figure 4B.
Testing of Environmentally Relevant Chemicals (ATSDR42)
We then tested 42 chemicals (Table 2) selected (Chen 2020) from a list of maintained by the ATSDR, hazards frequently found at Superfund sites around the United States, that cover several pollutant classes: polycyclic aromatic hydrocarbons (PAHs, ), inorganic metals (), phthalates (), pesticides (), and other industrial chemicals ().
As shown in Figure 5A, we performed a six-point dose response for the ATSDR 42 compounds under same treatment conditions as those of the EPA45 set. From the three heat maps, this set of pollutants was considerably less active, with only 9 compounds reducing ER median levels by 30% or more: 2 PAHs [benzo(a)anthracene and benzo(b)fluoranthene], three heavy metals (mercury, chromium, and cadmium), three pesticides (p,p′-DDT, azinphos-methyl, and disulfoton); and an industrial intermediate (para-cresol). The dose–response curves for these chemicals, grouped by class, for the ER median level, are shown in Figure 5B. The data for all the chemicals in DRC for the three indexes is in Figures S6–S8. We compared the MCF-7 data for 36 out of 42 of the compounds that have corresponding results in ToxCast. Figure 5C shows the values for the ToxCast (circles) and MCF-7 (diamonds, squares, and triangles) assays. Interestingly, few chemicals were active mostly in MCF-7 cells, whereas they were not active at all, or with high log in ToxCast [i.e., disulfoton, benzo(a)anthracene and benzo(b)fluoranthene], and whereas others were equally active in both sets (i.e., methoxychlor, o,p′-DDT). Only a handful had activity exclusively in ToxCast assay, usually at concentrations at or higher than the highest tested concentration (i.e., ethion, dicofol).
Although measuring ER levels is certainly valuable, it is not necessarily linked to activity of the receptor, because its levels can be modulated similarly by agonists and antagonists, as we have shown above (i.e., E2 vs. ICI). For this reason, orthogonal assays are needed to ascertain the character of newly identified toxicants. We performed single molecule RNA FISH (smFISH) using the prototypic ER target gene GREB1, which is a good indicator of ER activity (Nwachukwu et al. 2016; Stossi et al. 2020a).
We treated MCF-7 for 24 h with E2 and several of the ATSDR compounds that are identified in Figure 5 at their highest non toxic concentration () and measured, as a proxy for transcriptional activity, the number of active GREB1 alleles per cell. Figure 6A shows representative images of GREB1 active alleles highlighted by the use of exon-specific and intron-specific fluorescent oligonucleotides (Mistry et al. 2020; Stossi et al. 2020a). In Figure 6B, results are represented as stacked bar graphs illustrating the fraction of the population of cells (four independent biological replicates, ) having 0–4 active GREB1 alleles per nucleus. From this analysis, coupled with statistical confirmation by two-sample Kolmogorov-Smirnov test, benzo(a)anthracene and p,p′-DDT were clearly the most active, followed by disulfoton, azinphos-methyl and cadmium, whereas benzo(b)fluoranthene, mercury, and chromium showed no GREB1 activation. This, or other, orthogonal analysis could thus help discriminate activities of each new chemical identified as potentially active through the pipeline measuring ER levels.
Discussion
ER can be bound by many ligands possessing diverse chemical structures, ranging from endogenous and synthetic steroids, to flavones, phenols, lignans, stilbenes, and many others, of both man-made and natural origin (Dahlman-Wright et al. 2006). Most of these chemicals end up in the environment thus positively and negatively influencing the ER pathway in humans and other fauna, often leading to disease (De Coster and van Larebeke 2012; Hall and Greco 2019; Kabir et al. 2015). One of the more interesting aspects of ER is that, upon interacting with its ligand binding pocket, each chemical structure elicits a unique conformation as a means to modulate its biological responses (Nwachukwu et al. 2017, 2016). Traditionally, the way to test estrogenic chemicals has been by either virtual screening sans biological context or engineered assays (i.e., ligand binding, reporter genes etc.). Although these approaches have proven to be effective in classifying chemicals and predicting in vivo uterotrophic data, there has been a paucity of studies looking at effects of chemicals on the endogenous receptor and especially at the single cell level.
Single cell analytics have been increasing in popularity because they can facilitate discovery of outlier effects or rare subpopulations and overall describe more accurately a biological response of a population to a stimulus vs. the use of bulk cellular assays. However, there have been only a few efforts to use single cell end points in high-throughput screening, environmental analysis and/or quality control metrics to establish robustness of single cell biological responses (Gough et al. 2016, 2014). In this study, we set out to elaborate on these concepts by defining a novel pipeline for QC of single cell features extracted by image analysis across technical and biological replicates. We then established a second level of QC for studies that are based on measuring response to chemicals; and finally, we tested the utility of such approach as an orthogonal way to quantify potential endocrine disruptor activity of environmentally relevant chemicals.
In the process of establishing our QC pipeline over several years and across independent biological replicates, we learned interesting lessons regarding ligand-dependent changes in the single cell distribution of ER. We found that ER phenotypic heterogeneity was a reasonably stable metric across the experimental conditions tested. This finding was exemplified by the fact that only one experiment had a completely different ER distribution (EXP_21), due to unknown reasons. Overall, this finding is suggestive of the fact that ER distribution was not only quantifiable, but also can be used to assess the quality of an experiment. The biological ramifications of this observation are still unclear because we have not yet found a chemical perturbation that would change the distribution of ER into a more uniform one (i.e., all cells with similar amounts of receptor) and the ramifications will be pursued in the future. One possible explanation may be that the population used in the study behaves similarly to bacterial colonies, because phenotypic heterogeneity is a hallmark supporting their adaptability to the environment and evolution. A second observation is that the cellular population also responded to ligands in a reproducible manner that could be quantified similarly by several metrics describing various aspects of the ER distribution (e.g., median, diversity, distance from vehicle). However, few experiments failed to pass our second tier QC; the reasons for this could be human error (i.e., researcher forgot to add one of the compounds to the wells), other experimental mistakes during processing (probably EXP21 falls into this category because it failed both QC tiers), or lack of cell responsiveness (Figure 2C), but the findings highlight the fact that controlling an experiment only by the distribution of ER under basal conditions is not enough to qualify an experiment as good or bad. Because no clear error pattern emerged (i.e., multiple ligands failing in the same experiment or always the same ligand failing, operator etc.), it is unlikely that the cause of our QC failure resides in compounds degradation, systematic errors, or complete lack of cell responsiveness.
When we examined the sensitivity of our assay in comparison with results in ToxCast on a set of well-established control compounds [EPA45 (Judson et al. 2015)], we found that, overall, the MCF-7 assay using endogenous ER was at least comparable. For example, the value for E2, in this assay, was 90 pM (SD of 59 pM), whereas the one across the 16 ER specific agonistic assays present in the ToxCast library (Judson et al. 2015) was ( SD). Interestingly, if we rank the metrics for endogenous ER (fold median of DMSO control, QE, and EMD) and then compare them to 16 ER agonist assays in ToxCast, at least for the compounds defined as strong/moderate agonists (Judson et al. 2015), the ER quadratic entropy ranked first between all assays. Although the fold median and EMD values were in the middle for weak and very weak agonists, the ER indexes performed even better, with QE ranking first and fold median third. This finding indicates that endogenous assays can be as sensitive as other engineered assays while being contextual. One caveat in these results was that, for very potent compounds (i.e., DES), a lower starting concentration should be tested; otherwise, the estimated will not be precise. As with every high-throughput assay, false positive and false negative results are expected; in the endogenous ER assay the false negatives were few, mostly linked to compounds with in the high micromolar range, which was close or above the upper limit of our testing concentrations. In terms of false positives, the ER endogenous assay is certainly prone to them; however, they can also be quite informative because they will often involve indirect ways to affect ER levels without directly binding to the receptor. Classical examples would be inhibitors of central molecular mechanisms (protein translation and degradation, gene transcription), crosstalk with other transcription factors (notably other steroid receptors and AhR), and modulators of intracellular/epigenetic signaling pathways [for example, histone deacetylase inhibitors (Stossi et al. 2020a)]. In the current study, eight chemicals that had low/minimal effect in ToxCast did show activity in MCF-7 cells (Figure 4C–D). Of the eight, cycloheximide is easily explainable as a product of assay interference because it is a protein translation inhibitor and as such will reduce ER levels through an ER independent pathway. Two steroids, corticosterone and progesterone, were also positive in few ToxCast assays (as were dicofol and atrazine), and this can be explained by very low affinity receptor binding, and/or cross-talk between pathways. The results for the antiandrogens flutamide and hydroxyflutamide are more puzzling because they had opposite effects on ER levels and only appeared active in the MCF-7 assay; because there is no evidence in the literature of flutamide and hydroxyflutamide binding to the ER, further studies will be needed to validate compound chemistry and their activities; phenobarbital is a similar case, even though a recent study (Xu et al. 2019) found it active in a transcriptional reporter assay. Only two chemicals (kaempferol and p,p′-DDE) appeared as false negatives; however, they have low activity in 10 and 7 individual ToxCast assays, out of 16, respectively. For kaempferol, the average in ToxCast was , whereas for DDE the average was . Because the upper limit of the tested dose response in MCF-7 was , this is the likely reason for missing DDE as a potential active chemical. Looking carefully at the data for kaempferol, we did see a small effect on fold median () at the highest tested concentration (Figure S2). Another limitation of this assay is that changes in ER level do not always directly relate to receptor activity and/or its being in an agonist or antagonist conformation, even though it was quite encouraging to see that in comparison with the 16 ToxCast agonist assays, we missed only two compounds that were classified as very weak (Judson et al. 2015). With the demonstration of testing 42 chemicals from the ATSDR list (Chen 2020), the ER assay was also capable of identifying potential new or less-established EDCs that we validated for transcriptional activity by single molecule RNA FISH. In a general sense, the largest limitation for this assay is that it would miss compounds that fail to alter ER levels but still affect its transcription activity. The number of compounds that exhibit this behavior in the chemical space is currently unknown. To move toward single cell methods reporting ER levels and transcriptional activity, we are currently validating a larger-scale IF/FISH screening platform to simultaneously measure ER levels and activation of its prototypical target gene, GREB1 (Mistry et al. 2020; Nwachukwu et al. 2016; Stossi et al. 2020a). As a proof of principle, we can already show that this method is effective because we recently published the results of the first small screen (Stossi et al. 2020a). In conclusion, this study highlights the efforts to quantify changes in endogenous receptors at the single cell level should be considered as an important complement to current assays.
Supplementary Material
Acknowledgments
M.A.M, F.S., and M.G.M. are funded in part by National Institute of Environmental Health Sciences (P42ES027704). M.A.M., F.S., H.J., R.M.M., A.U.R., and P.S. are supported via the Cancer Prevention and Research Institute of Texas (CPRIT)-funded GCC Center for Advanced Microscopy and Image Informatics (RP170719). P.S. and M.A.M. are also supported by the CPRIT-funded Combinatorial Drug Discovery Program (RP200668). M.A.M. is also partially funded by the GCC Center for Precision Environmental Health (P30ES030285). Imaging was supported by the Integrated Microscopy Core at Baylor College of Medicine with funding from the National Institutes of Health (NIH) (DK56338 and CA125123), CPRIT (RP150578), the Dan L. Duncan Comprehensive Cancer Center. Antibody preparation was supported by the Antibody-Base Proteomics Core at Baylor College of Medicine with funding from NIH (CA125123) and CPRIT (RP170005).
F.S., P.S.K., A.U.R., and M.A.M. conceptualized this study. F.S., R.M.M., H.L.J., R.D.D., and M.G.M. performed experiments or created reagents. F.S., P.S., R.M.M., A.U.R., H.L.J. provided data analysis, and F.S., P.S.K., and M.A.M. wrote the manuscript.
References
- Ackermann M. 2015. A functional perspective on phenotypic heterogeneity in microorganisms. Nat Rev Microbiol 13(8):497–508, PMID: , 10.1038/nrmicro3491. [DOI] [PubMed] [Google Scholar]
- Ashcroft FJ, Newberg JY, Jones ED, Mikic I, Mancini MA. 2011. High content imaging-based assay to classify estrogen receptor-α ligands based on defined mechanistic outcomes. Gene 477(1–2):42–52, PMID: , 10.1016/j.gene.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu L, Yong J, Antebi YE, McCue K, Kazuki Y, Uno N, et al. . 2016. Dynamics of epigenetic regulation at the single-cell level. Science 351(6274):720–724, PMID: , 10.1126/science.aab2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, et al. . 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7(10):R100, PMID: , 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Liu Y, Wright FA, Chiu WA, Rusyn I. 2020. Rapid hazard characterization of environmental chemicals using a compendium of human cell lines from different organs. ALTEX 37(4):623–638, PMID: 32521033, 10.14573/altex.2002291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlman-Wright K, Cavailles V, Fuqua SA, Jordan VC, Katzenellenbogen JA, Korach KS, et al. . 2006. International Union of Pharmacology. LXIV. Estrogen receptors. Pharmacol Rev 58(4):773–781, PMID: , 10.1124/pr.58.4.8. [DOI] [PubMed] [Google Scholar]
- De Coster S, van Larebeke N. 2012. Endocrine-disrupting chemicals: associated disorders and mechanisms of action. J Environ Public Health 2012:713696, PMID: , 10.1155/2012/713696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demsar J, Curk T, Erjavec A, Gorup C, Hocevar T, Milutinovic M, et al. . 2013. Orange: data mining toolbox in Python. J Mach Learn Res 14:2349–2353. [Google Scholar]
- Gibson DA, Saunders PTK. 2014. Endocrine disruption of oestrogen action and female reproductive tract cancers. Endocr Relat Cancer 21(2):T13–T31, PMID: , 10.1530/ERC-13-0342. [DOI] [PubMed] [Google Scholar]
- Giorgino T. 2009. Computing and visualizing dynamic time warping alignments in R: the DTW package. J Stat Software 31:1–24, 10.18637/jss.v031.i07. [DOI] [Google Scholar]
- Gough AH, Chen N, Shun TY, Lezon TR, Boltz RC, Reese CE, et al. . 2014. Identifying and quantifying heterogeneity in high content analysis: application of heterogeneity indices to drug discovery. PLoS One 9(7):e102678, PMID: , 10.1371/journal.pone.0102678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough A, Shun TY, Lansing Taylor D, Schurdak M. 2016. A metric and workflow for quality control in the analysis of heterogeneity in phenotypic profiles and screens. Methods 96:12–26, PMID: , 10.1016/j.ymeth.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough A, Stern AM, Maier J, Lezon T, Shun TY, Chennubhotla C, et al. . 2017. Biologically relevant heterogeneity: metrics and practical insights. SLAS Discov 22(3):213–237, PMID: , 10.1177/2472555216682725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JM, Greco CW. 2019. Perturbation of nuclear hormone receptors by endocrine disrupting chemicals: mechanisms and pathological consequences of exposure. Cells 9(1):13, PMID: , 10.3390/cells9010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, et al. . 2007. Estrogen receptors: how do they signal and what are their targets. Physiol Rev 87(3):905–931, PMID: , 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- Huang R, Sakamuru S, Martin MT, Reif DM, Judson RS, Houck KA, et al. . 2014. Profiling of the Tox21 10K compound library for agonists and antagonists of the estrogen receptor alpha signaling pathway. Sci Rep 4:5664, PMID: , 10.1038/srep05664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RS, Houck KA, Watt ED, Thomas RS. 2017. On selecting a minimal set of in vitro assays to reliably determine estrogen agonist activity. Regul Toxicol Pharmacol 91:39–49, PMID: , 10.1016/j.yrtph.2017.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RS, Magpantay FM, Chickarmane V, Haskell C, Tania N, Taylor J, et al. . 2015. Integrated model of chemical perturbations of a biological pathway using 18 in vitro high-throughput screening assays for the estrogen receptor. Toxicol Sci 148(1):137–154, PMID: , 10.1093/toxsci/kfv168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabir ER, Rahman MS, Rahman I. 2015. A review on endocrine disruptors and their possible impacts on human health. Environ Toxicol Pharmacol 40(1):241–258, PMID: , 10.1016/j.etap.2015.06.009. [DOI] [PubMed] [Google Scholar]
- La Merrill MA, Vandenberg LN, Smith MT, Goodson W, Browne P, Patisaul HB, et al. . 2020. Consensus on the key characteristics of endocrine-disrupting chemicals as a basis for hazard identification. Nat Rev Endocrinol 16(1):45–57, PMID: , 10.1038/s41574-019-0273-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri K, Abdelaziz A, Rybacka A, Roncaglioni A, Tropsha A, Varnek A, et al. . 2016. CERAPP: Collaborative Estrogen Receptor Activity Prediction Project. Environ Health Perspect 124(7):1023–1033, PMID: , 10.1289/ehp.1510267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsaud V, Gougelet A, Maillard S, Renoir JM. 2003. Various phosphorylation pathways, depending on agonist and antagonist binding to endogenous estrogen receptor α (ERα), differentially affect ERα extractability, Proteasome-Mediated stability, and transcriptional activity in human breast cancer cells. Mol Endocrinol 17(10):2013–2027, PMID: , 10.1210/me.2002-0269. [DOI] [PubMed] [Google Scholar]
- Mistry RM, Singh PK, Mancini MG, Stossi F, Mancini MA. 2020. Single cell analysis of transcriptionally active alleles by single molecule fish. J Vis Exp 2020:1–15, 10.3791/61680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwachukwu JC, Srinivasan S, Bruno NE, Nowak J, Wright NJ, Minutolo F, et al. . 2017. Systems structural biology analysis of ligand effects on ERα predicts cellular response to environmental estrogens and anti-hormone therapies. Cell Chem Biol 24(1):35–45, PMID: , 10.1016/j.chembiol.2016.11.014. [DOI] [PubMed] [Google Scholar]
- Nwachukwu JC, Srinivasan S, Zheng Y, Wang S, Min J, Dong C, et al. . 2016. Predictive features of ligand-specific signaling through the estrogen receptor. Mol Syst Biol 12(4):864, PMID: , 10.15252/msb.20156701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmari J, Dussert C, Berthois Y, Penel C, Martin PM. 1997. Distribution of estrogen receptor heterogeneity in growing MCF-7 cells measured by quantitative microscopy. Cytometry 27(1):26–35, PMID: . [PubMed] [Google Scholar]
- Perlman ZE, Slack MD, Feng Y, Mitchison TJ, Wu LF, Altschuler SJ. 2004. Multidimensional drug profiling by automated microscopy. Science 306(5699):1194–1198, PMID: , 10.1126/science.1100709. [DOI] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A. 2008. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135(2):216–226, PMID: , 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnicoff M, Medrano EE, Podhajcer OL, Bravo AI, Bover L, Mordoh J. 1987. Subpopulations of MCF7 cells separated by Percoll gradient centrifugation: a model to analyze the heterogeneity of human breast cancer. Proc Natl Acad Sci USA 84(20):7295–7299, PMID: , 10.1073/pnas.84.20.7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard AM, Judson RS, Houck KA, Grulke CM, Volarath P, Thillainadarajah I, et al. . 2016. ToxCast chemical landscape: paving the road to 21st century toxicology. Chem Res Toxicol 29(8):1225–1251, PMID: , 10.1021/acs.chemrestox.6b00135. [DOI] [PubMed] [Google Scholar]
- Rotroff DM, Martin MT, Dix DJ, Filer DL, Houck KA, Knudsen TB, et al. . 2014. Predictive endocrine testing in the 21st century using in vitro assays of estrogen receptor signaling responses. Environ Sci Technol 48(15):8706–8716, PMID: , 10.1021/es502676e. [DOI] [PubMed] [Google Scholar]
- Rubin H. 1990. On the nature of enduring modifications induced in cells and organisms. Am J Physiol 258(2 pt 1):L19–L24, PMID: , 10.1152/ajplung.1990.258.2.L19. [DOI] [PubMed] [Google Scholar]
- Rubner Y, Tomasi C, Guibas LJ. 2000. The earth mover’s distance as a metric for image retrieval. Int J Comput Vis 40(2):99–121, 10.1023/A:1026543900054. [DOI] [Google Scholar]
- Sakoe H, Chiba S. 1978. Dynamic programming algorithm optimization for spoken word recognition. IEEE Trans Acoust 26(1):43–49, 10.1109/TASSP.1978.1163055. [DOI] [Google Scholar]
- Schreiber F, Littmann S, Lavik G, Escrig S, Meibom A, Kuypers MMM, et al. . 2016. Phenotypic heterogeneity driven by nutrient limitation promotes growth in fluctuating environments. Nat Microbiol 1(6):16055, PMID: , 10.1038/nmicrobiol.2016.55. [DOI] [PubMed] [Google Scholar]
- Singh DK, Ku C-J, Wichaidit C, Steininger RJ, Wu LF, Altschuler SJ. 2010. Patterns of basal signaling heterogeneity can distinguish cellular populations with different drug sensitivities. Mol Syst Biol 6:369, PMID: , 10.1038/msb.2010.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stossi F, Bolt MJ, Ashcroft FJ, Lamerdin JE, Melnick JS, Powell RT, et al. . 2014. Defining estrogenic mechanisms of bisphenol A analogs through high throughput microscopy-based contextual assays. Chem Biol 21(6):743–753, PMID: , 10.1016/j.chembiol.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stossi F, Dandekar RD, Bolt MJ, Newberg JY, Mancini MAMG, Kaushik AK, et al. . 2016. High throughput microscopy identifies bisphenol AP, a bisphenol A analog, as a novel AR down-regulator. Oncotarget 7(13):16962–16974, PMID: , 10.18632/oncotarget.7655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stossi F, Dandekar RD, Mancini MG, Gu G, Fuqua SAW, Nardone A, et al. . 2020a. Estrogen-induced transcription at individual alleles is independent of receptor level and active conformation but can be modulated by coactivators activity. Nucleic Acids Res 48(4):1800–1810, PMID: , 10.1093/nar/gkz1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stossi F, Mistry RM, Singh PK, Johnson HL, Mancini MG, Szafran AT, et al. . 2020b. Single-cell distribution analysis of AR levels by high-throughput microscopy in cell models: application for testing endocrine-disrupting chemicals. SLAS Discov 25(7):684–694, PMID: , 10.1177/2472555220934420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafran AT, Mancini MA. 2014. The myImageAnalysis project: a web-based application for high-content screening. Assay Drug Dev Technol 12(1):87–99, PMID: , 10.1089/adt.2013.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafran AT, Stossi F, Mancini MG, Walker CL, Mancini MA. 2017. Characterizing properties of non-estrogenic substituted bisphenol analogs using high throughput microscopy and image analysis. PLoS One 12(7):e0180141, PMID: , 10.1371/journal.pone.0180141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Kirkpatrick A, Toperzer J, Ripp S, Close D. 2019. Improving estrogenic compound screening efficiency by using self-modulating, continuously bioluminescent human cell bioreporters expressing a synthetic luciferase. Toxicol Sci 168(2):551–560, PMID: , 10.1093/toxsci/kfz004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.