SUMMARY
TH2 and innate lymphoid cells 2 (ILC2) can stimulate tumor growth by secreting pro-tumorigenic cytokines such as IL-4, IL-5 and IL-13. However, the mechanisms by which type 2 immune cells traffic to the tumor microenvironment (TME) are unknown. Here, we show that oncogenic KrasG12D increases IL-33 expression in pancreatic ductal adenocarcinoma (PDAC) cells, which recruits and activates TH2 and ILC2 cells. Correspondingly, cancer cell-specific deletion of IL-33 reduces TH2 and ILC2 recruitment and promotes tumor regression. Unexpectedly, IL-33 secretion is dependent on the intratumoral fungal mycobiome. Genetic deletion of IL-33 or anti-fungal treatment decreases TH2 and ILC2 infiltration and increases survival. Consistently, high IL-33 expression is observed in approximately 20% of human PDAC, and expression is mainly restricted to cancer cells. These data expand our knowledge of the mechanisms driving PDAC tumor progression and identify therapeutically targetable pathways involving intratumoral mycobiome-driven secretion of IL-33.
Keywords: Type 2 immune response, cytokines, innate lymphoid cells (ILC2), TH2, fungal mycobiome, Kras, PDAC
Graphical Abstract
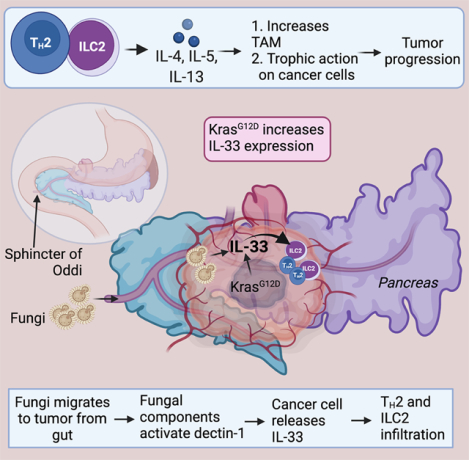
eTOC Blurb
Alam et al. discover that IL-33 is a downstream target of KrasG12D. Further, he reports that intratumor mycobiome activates a signaling pathway in cancer cells that facilitate IL-33 secretion. The secretion of IL-33 promotes type 2 immune response and accelerates pancreatic cancer progression.
INTRODUCTION
PDAC is associated with a distinctive tumor immune profile dominated by immune suppressor cells, such as tumor-associated macrophages (TAMs), T regulatory (Treg) cells, CD4+ TH2 cells and myeloid-derived suppressor cells (MDSCs) which act in concert to inhibit effector T cell activation, expansion and function, thereby contributing to suppression of anti-tumor T cell immunity and PDAC progression (Clark et al., 2007, Steele et al., 2013, Feig et al., 2012). In addition to its classical roles in tumor immunity, immune cells can contribute to additional cancer hallmarks including a unique metabolic mechanism by which CD4+ TH2 cells support glycolysis to fuel PDAC progression (Dey et al., 2020). Specifically, TH2 cells infiltrate the pancreas in the early stages of tumorigenesis and secrete type 2 cytokines [interleukin (IL-4) and IL-13)] that promote metabolic reprogramming and proliferation of cancer cells in murine KrasG12D-driven PDAC. Consistent with type 2 immune responses driving PDAC progression in mouse models, PDAC patients with predominantly TH2 (CD45+CD3+CD4+Gata3+)-polarized lymphoid cell infiltration exhibit reduced survival, compared to patients with a higher infiltration of TH1 (CD45+CD3+CD4+Tbet+) cells (De Monte et al., 2011). Moreover, the circulating levels of IL-4 negatively correlate with disease-free survival in PDAC patients (Piro et al., 2017).
Given the emerging importance of TH2 cells in PDAC (Dey et al., 2020), we sought to determine the molecular mechanisms driving their recruitment and expansion in the tumor microenvironment (TME). Our studies identified oncogenic KRASG12D-mediated upregulation of IL-33, which is a known potent activator of TH2, innate lymphoid cells 2 (ILC2), Tregs, and eosinophils (Liew et al., 2016). IL-33 is both a damage-associated molecular pattern (DAMP) and a cytokine that belongs to the IL-1 cytokine superfamily (Schmitz et al., 2005), which plays important roles in innate immunity, inflammation and tumor development (Wang et al., 2017, Fournie and Poupot, 2018, Li et al., 2019, Cui et al., 2018). IL-33 exerts its biological function by binding to its cognate receptor, suppression of tumorigenicity (ST)2 (also called IL1RL1) (Lingel et al., 2009), which interacts with its co-receptor, the IL-1 receptor accessory protein (IL1RAcP) (Lingel et al., 2009). Both receptors are expressed by innate and type 2 immune cells that include TH2 cells, ILC2s, Treg, eosinophils, basophils and mast cells (Rank et al., 2009) (Ali et al., 2007).
ILC2s are the most prominent target of IL-33 and stimulate the activation of ILC2 in response to several stimuli, such as allergens and parasites. ILC2s are primarily tissue-resident immunocytes that remain in close proximity to the epithelial cells, enabling ILC2 cells to respond to an immune insult within hours by producing cytokines such as IL-4, IL-13 and IL-5 which in turn activate additional players of the type 2 immune response (Van Gool et al., 2014, Vivier et al., 2018). Moreover, activation of ILC2 cells is independent of antigen presentation and uses ligand receptors often specific to the tissue where they reside.
Gut microbes can interact with the host and modulate disease pathogenesis and response to therapy (Gopalakrishnan et al., 2018, Matson et al., 2018). It is now well known that microbes can colonize the pancreas and play a role in PDAC tumorigenesis and progression (Riquelme et al., 2019, Aykut et al., 2019). Specifically, a recent study found that the mycobiome present in the gut lumen migrates to the pancreas via the sphincter of Oddi (Aykut et al., 2019). The translocation of endoluminal fungi to the pancreas allows the fungal population to increase by >3000-fold in PDAC compared to the normal pancreas.
In this study, oncogenic KrasG12D is shown to induce IL-33 expression and secretion by cancer cells through a pathway that depends on fungi within the TME and that genetic deletion of IL-33 or anti-fungal treatment each leads to robust PDAC tumor regression. This mechanism of cooperative interactions of intratumor fungi with cancer cells and priming type 2 immune responses to accelerate tumor progression identifies potential therapeutic strategies for PDAC.
RESULTS
Type 2 immune cell infiltration increases significantly in PDAC tumor microenvironment
The observations that CD4+Gata3+ TH2 cells infiltrate the TME and promote tumorigenesis via IL-4 and IL-13 (Venmar et al., 2015, Dey et al., 2020) prompted a more thorough characterization of the type 2 immunocytes present within the PDAC tumors of KPC (KrasG12D;Trp53R172H;Pdx-Cre) mice (Figure 1A). Flow cytometry (Figure S1A) revealed a dramatic expansion of TH2 (CD4+Gata3+CCR4+) cells within the CD4+ T cell population in the PDAC TME (72.1%) compared to the normal pancreas (8.33%) and spleen (0.4%) (Figure 1B–D, Figure S1B). This was accompanied by a significant increase in ILC2 cells (Lin−Sca1+ST2+) in the PDAC TME (74.2%) compared to the normal pancreas (7.96%), spleen (0.18%) and bone marrow (0.25%) (Figure 1E–G, Figure S1C). Specifically, the frequency of ILC2 cells was approximately 60% of Lin− cells in the PDAC (KPC model) compared to <10% in the normal pancreas and ~25% in PanIN (Figure 1H). Similarly, the frequency of ILC2 cells was approximately 45% of Lin− cells in the PDAC (iKPC model) compared to <10% in the normal pancreas (Figure S1D).
Figure 1: Type 2 immune cell infiltration increases significantly in PDAC tumor microenvironment.
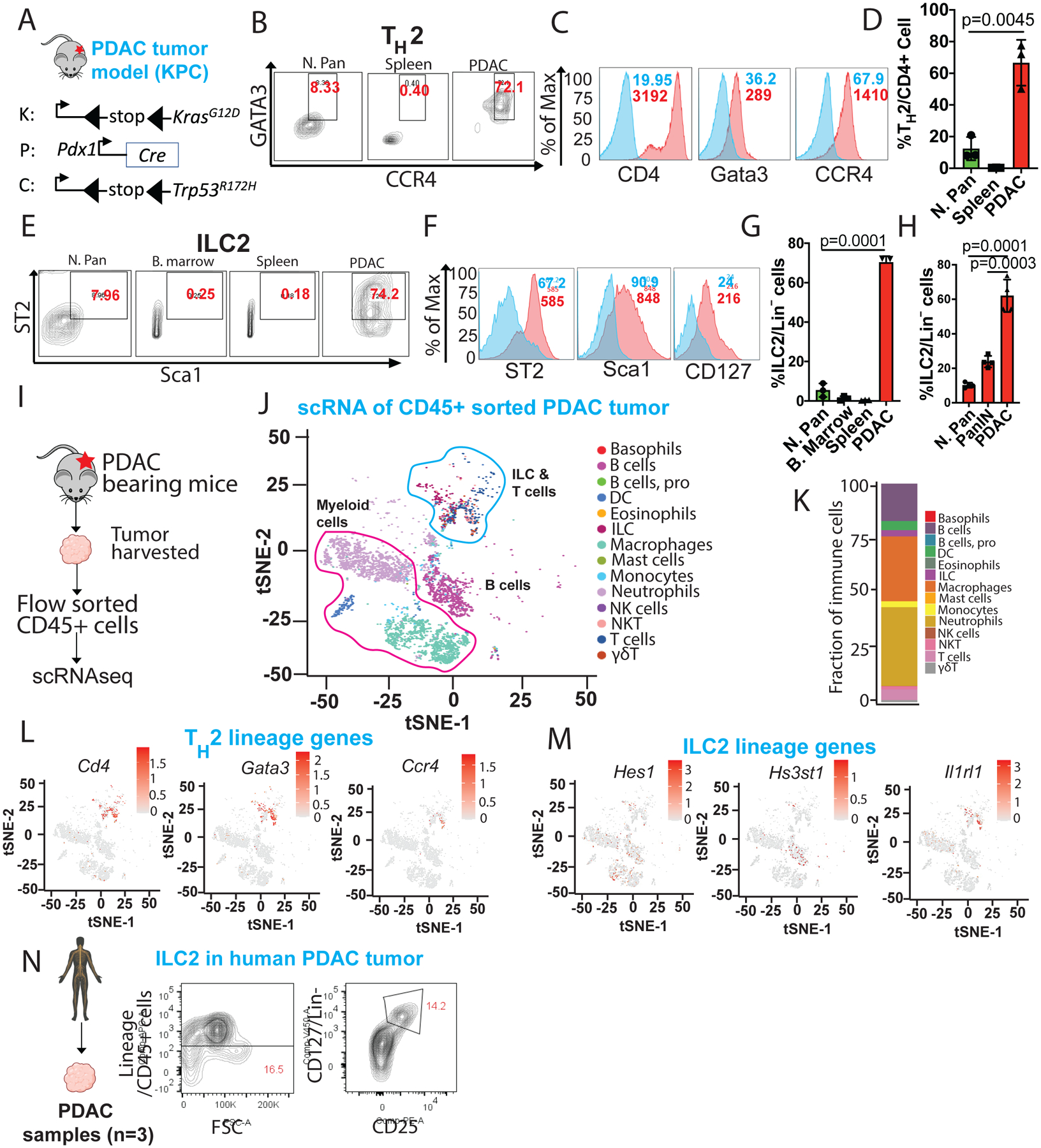
(A) Schematic diagram showing strategy for KPC (KrasG12D;p53R172H;pdx-Cre) PDAC mouse modeling.
(B) Flow cytometry gating strategy and frequency of TH2 cells out of total CD4+ T cells in normal pancreas, spleen and PDAC tumor.
(C) Representative flow cytometry histogram of TH2 cell phenotype stained with either isotype control (blue histogram) and CD4, Gata3 and CCR4 antibodies (red histogram).
(D) Frequency of TH2 cells out of total CD4+ T cells in normal pancreas, spleen and PDAC tumor (n=3).
(E) Gating strategy and frequency of ILC2 cells out of total lineage negative [Lin−] (CD3, Ly6G, Ly6C, CD11b, CD45R/B220 and TER-119) cells in normal pancreas, bone marrow, spleen and PDAC tumor (n=3).
(F) Representative flow cytometry histogram of ILC2s cell phenotype stained with either isotype control (blue histogram) and ST2, Sca-1 and CD127 antibodies (red histogram).
(G) Frequency of ILC2 cells out of total Lin− cells in normal pancreas, bone marrow, spleen and PDAC tumor (n=4).
(H) Frequency of ILC2 cells out of total Lin− cells in normal pancreas, PanIN and PDAC tumor (n=4).
(I) Schematic showing experimental strategy for single-cell RNA sequencing (scRNA-seq) from PDAC tumor-bearing mice. CD45+ cells were flow-sorted from PDAC tumor and 10,000 live CD45+ cells were used for scRNA-seq.
(J) t-SNE plot of immune cells showing 14 clusters belonging to 3 major groups in PDAC sample.
(K) Bar graph showing the proportion of major immune cell clusters in PDAC sample.
(L) t-SNE plots showing TH2 lineage genes (Cd4, Gata3 and Ccr4) expression in sub-cluster of immune cells. The color key bar represents gene expression level.
(M) t-SNE plots showing expression of ILC2 lineage genes (Hes1, Hs3st1 and Il1rl1) expression in sub-cluster of immune cells. The color key represents gene expression level.
(N) Gating and frequency of ILC2 out of total lineage negative (CD3, CD14, CD16, CD19, CD20, and CD56) cells in human PDAC tumor.
Data are presented as mean ±SD. p-values were calculated using the Student t-test. ns, no significance. Individual p-values are indicated in the figures.
See also Figure S1.
To solidify our flow-based immunophenotyping data, we used single-cell RNA-seq (scRNA-seq) analysis to identify the presence of the type 2 immunocytes in the PDAC TME. We sort-purified CD45+ cells and used the 10X Genomics platform for scRNA-seq of the immune populations in the PDAC tumor samples (Figure 1I–J). The majority of immune cell populations identified were myeloid cells (macrophages, monocytes, DCs and neutrophils: ~74.72%), followed by lymphoid populations (16.45%) (Figure 1K, Figure S1E). Using previously reported gene signatures for TH2 (Tibbitt et al., 2019) and ILC2 (Robinette et al., 2015), we found that the TH2 cluster was enriched for Cd4, Gata3 and Ccr4 genes (Figure 1L, Figure S1F) and ILC2 clusters were enriched for Hes1, Hs3st1 and Il1rl1 genes (Figure 1M, Figure S1G), which are bona fide markers of TH2 and ILC2 cells, respectively. Finally, we analyzed fresh human PDAC samples by flow cytometry and found that ILC2 cells accounted for 14.2% of the Lin− cells (Figure 1N). Overall, these results show that the murine and human PDAC TME contain abundant TH2 and ILC2s cells.
IL-33 is a downstream target of oncogenic KrasG12D
Type 2 immunocytes are detected in the early stages of PDAC tumorigenesis (Dey et al., 2020), prompting speculation that a chemotactic factor secreted by cancer cells may recruit and activate them. Given that KRAS mutation is an early genetic alteration and drives tumor initiation, we examined the KrasG12D-regulated transcript of multiple PDAC cell lines derived from the inducible model of PDAC (iKPC: LSL-tet-O-KrasG12D; LSL-Trp53+/−;p48-Cre;LSL-Rosa26-rtTA) (Figure 2A) (Ying et al., 2016). RNA-seq analysis of iKPC cell lines included KrasG12D ON (doxycycline ON), KrasG12D OFF 2 days and KrasG12D OFF 4 days. Among the top cytokine genes that are regulated by KrasG12D and enriched in the KRAS signaling network is the alarmin gene, Il33 (Figure 2B–D). Il33 expression is ~30-fold higher in KrasG12D ON compared to KrasG12D OFF (4 days) samples, as validated by quantitative real-time PCR (qRT-PCR) analysis (Figure 2E, Figure S2A-B).
Figure 2: IL-33 is a downstream target of oncogenic KrasG12D.

(A) Schematic showing doxycycline (Dox) inducible KrasG12D transgenic mouse model (iKPC) (top). Strategy to turn ON and OFF Kras signaling in cell lines followed by transcriptome analysis (bottom).
(B) Pathways enriched upon GSEA analysis of RNAseq dataset comparing Kras ON vs Kras OFF.
(C) GSEA analysis of RNAseq dataset showing enrichment of hallmark Kras signaling comparing Kras ON vs Kras OFF.
(D) Heatmap of genes enriched in Kras signaling pathway upon comparing Kras ON, OFF-2 and 4 days in 4 murine cell lines.
(E) qRT-PCR analysis of Il33 expression in the Kras ON, OFF-2 and 4 days (n=3).
(F) Western blot analysis of IL-33 (Lo=low and Hi=high exposure), pERK1/2 (P-p42/44) and ERK1/2 (p42/44) in Kras ON, OFF-1, 2 and 3 days in the murine cell line.
(G) Western blot analysis of IL-33 and pERK1/2 in Kras ON, OFF and re-ON in murine cell line.
(H) Western blot analysis of IL-33, pERK1/2 (P-p42/44) and pAkt-S307 upon treatment with MEK inhibitors (CI-1040 and Trametinib) in the murine cell line.
(I) Representative confocal images of IL-33 and α-smooth muscle actin (α-SMA) staining in mouse PDAC tumor. Magnification 63x. Scale bar 50 μm.
(J) Representative IHC images of IL-33 in human PDAC tumor. Inset (red box) showing 100x magnification. (n=121). Scale bar 25 μm.
(K) Statistical analysis of IL-33 staining of human PDAC TMA. The intensity of IHC staining was scored as negative (0), low (1), medium (2), and high (3) (left). Table showing statistical analysis of the IL-33 expression, between normal pancreas and tumor tissues (right). The N vs T comparison was done considering all staining (score 1–3) intensities. See also Figure S2.
Data are presented as mean ±SD. p-values were calculated using the Student t-test. ns, no significance. GSEA: Gene signature enrichment analysis. Individual p-values are indicated in the figures.
See also Figure S2.
The above findings were further validated by western blot analysis showing a loss of IL-33 expression upon abolishing KrasG12D signaling (Figure 2F–G). We further validated this finding using cell lines derived from the KPC model. Treatment with MEK inhibitors (CI-1040, Trametinib), a downstream target of KRAS signaling, resulted in complete inhibition of IL-33 expression in these cell lines (Figure 2H, Figures S2C). However, treatment with PI3K inhibitors (Buparlisib and GSK-690696) did not lead to a reduction in IL-33 expression, suggesting that IL-33 is induced via a KRAS-MEK-ERK signaling pathway independent of PI3K (Figures S2C).
We next evaluated IL-33 expression in the KPC mouse model and in human PDAC tissue. In the KPC model, IL-33 expression was mostly observed in the cancer cells with nominal expression observed in fibroblasts or immune cells (Figure 2I). Furthermore, these murine data align with the human tumor data showing that IL-33 expression was increased, with ~20% PDAC cases showing high and ~52% showing moderate IL-33 expression (Figure 2J and K). As with mice, IL-33 expression in human PDAC specimens was most prominent in cancer cells with minimal staining in the stromal compartment (Figure 2J, Figure S2D-E). The normal pancreas did not express detectable IL-33 (Figure 2J, Figure S2F). These data show that KrasG12D-MEK signaling induces IL-33 in PDAC cells.
IL-33 expression is required for the recruitment and activation of type 2 immunocytes
The TH2 and ILC2 cellular infiltration increases significantly during PDAC tumorigenesis. To test the requirement of IL-33 expression by cancer cells to recruit type 2 immunocytes, Il33 was depleted in cancer cells by lentivirus transduction of small hairpin (sh)RNA (Figure 3A–B). While shRNA-mediated depletion of Il33 had no effect on cell growth in vitro (Figure S3A), decreased Il33 levels in cancer cells resulted in reduced tumor burden and increased survival in a syngeneic orthotopic model of PDAC (Figure 3C–D, Figure S3B-C). Immunofluorescence of these tumors confirmed that PDAC cancer cells are the major source of IL-33 and that the IL-33 signal is diminished by shRNA-mediated depletion (Figure 3E). Moreover, malignant ascites, observed in ~20% of PDAC patients (Zervos et al., 2006), is also a feature of the PDAC mouse model. In tumor-bearing mice, Il33 depletion in cancer cells led to decreased IL-33 protein levels in the ascites fluid (Figure 3F).
Figure 3: IL-33 is required for recruitment of type 2 immunocytes.
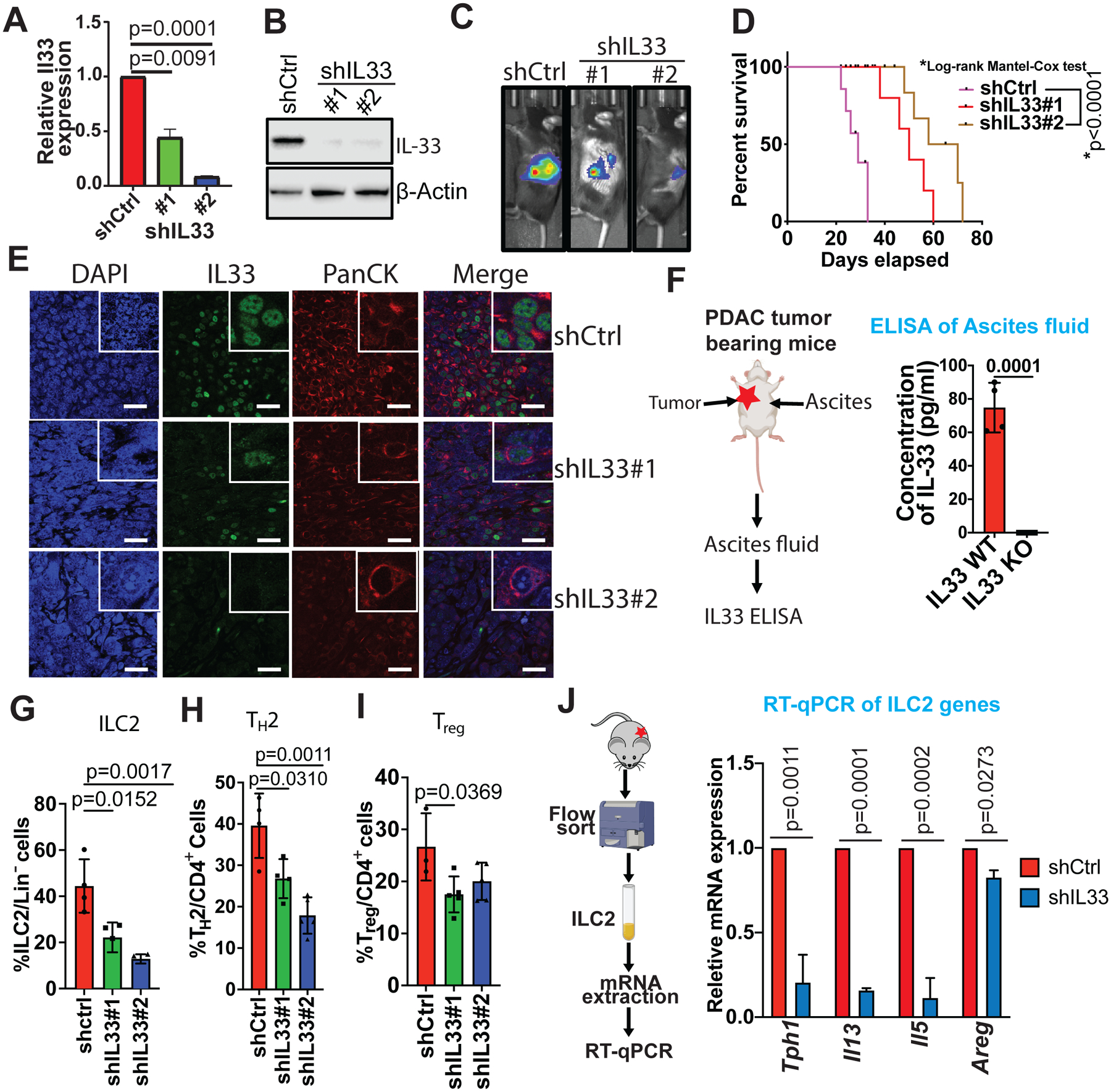
(A) Il33 gene expression was determined by qRT-PCR relative to β-actin in non-target (shCtrl) vs shIL33 (#1 and #2) stable murine cell line (n=3).
(B) Western blot analysis showing stable knockdown of Il33 in shIL33 (#1 and #2) murine cell lines. β-actin was used as a loading control.
(C) Intrapancreatic injection of shCtrl (n=23) and shIL33 (#1 [n=25] and #2 [n=24]) stable PDAC isogenic mouse cell lines. Representative bioluminescence images showing orthotopically transplanted PDAC tumors.
(D) Kaplan–Meier survival curves of mice orthotopically transplanted with shCtrl and shIL33 (#1 and #2) stable PDAC isogenic mouse cell line (n=10).
(E) Representative confocal images showing cancer cell-specific nuclear expression of IL-33 (green) in orthotopically transplanted shCtrl and shIL33 PDAC tumors. DAPI (blue) was used as nuclear marker and PanCK (red) was used as an epithelial cell marker. Magnification 63x. Scale bar 100 μm.
(F) Schematic showing a accumulation of ascites fluid on day 25–28 of orthotopically transplanted shCtrl and shIL33 PDAC tumor-bearing mice (left). IL-33 was quantified in ascites fluid using ELISA (n=3) (right).
(G) Frequency of ILC2s in orthotopically transplanted shCtrl and shIL33 PDAC tumors relative to total Lin− cells.
(H) Frequency of TH2 in orthotopically transplanted shCtrl and shIL33 PDAC tumors relative to total CD4+ cells.
(I) Frequency of Tregs in orthotopically transplanted shCtrl and shIL33 PDAC tumors relative to total CD4+ cells.
(J) Schematic showing flow sorting of ILC2 cells from orthotopically transplanted shCtrl and shIL33 PDAC tumors (left). qRT-PCR analysis was performed for ILC2 lineage signature genes Tph1, Il13, Il5, and Areg (right).
Data are presented as mean ±SD. p-values were calculated using the Student t-test. ns, no significance. Individual p-values are indicated in the figures.
See also Figure S3.
To establish the mechanistic link between IL-33 secretion and ILC2 trafficking, flow cytometry analysis of the TME showed that Il33 depletion reduced both TH2 and ILC2 infiltration (Figure 3G–H, Figure S3D-G). In addition to ILC2 and TH2 cells, Treg cells are known to express ST2 and respond to IL-33 signaling (Pastille et al., 2019). Accordingly, IL-33 depletion resulted in a small but significant reduction in Treg infiltration in the TME (Figure 3I). The above shRNA depletion findings mirrored those from CRISPR-Cas9 knockout (KO) of Il33 which showed inhibition of tumor growth and a strong reduction in TH2 infiltration (Figure S3H-K). In addition, IL-33 KO produced reductions in other IL-33 responding immunocytes, such as MDSCs and Tregs, however, no significant change was observed in CD8+ T cells, neutrophils, and B cell populations (Figure S3L-Q). Notably, in addition to the diminished infiltration of ILC2 cells in IL-33-deficient tumors, the activation marker required for ILC2 function, Tph1 (Flamar et al., 2020) and the effector cytokines, Il13, Il5 and Areg mRNA levels were also reduced, which suggests that the small population of resident ILC2 cells present within the PDAC TME are functionally inactive with decreased IL-33 (Figure 3J). Together, these data establish that cancer cell derived IL-33 recruits and activates type 2 immune cells into the PDAC TME.
Intratumor fungi facilitate the secretion of IL-33 from PDAC cells
Immunohistochemistry analysis of PDAC tumors revealed that IL-33 expression was predominantly nuclear in the preneoplastic stages (pancreatic intraepithelial neoplasia, (PanIN)1–3) compared to advanced adenocarcinoma, when its expression becomes wide-spread across the cytosol and extracellular milieu (6 vs. 24-week-old PDAC tumors) (Figure 4A). In contrast, immunofluorescence staining of IL-33 of PDAC cells in culture showed staining primarily in the nucleus (Figure 4B), which was further confirmed by subcellular fractionation studies (Figure 4C). These observations prompted us to consider the possibility that the secretion of IL-33 is regulated by pathways and factors beyond KrasG12D signaling that are operative in the TME (Figure S4A). The notion that IL-33 secretion requires an environmental stimulus is further fueled by the fact that IL-33 is a DAMP molecule, and its extracellular secretion is tightly regulated in normal cells to avoid unwanted immune responses (Bonilla et al., 2012, Miller, 2011).
Figure 4: Intratumor fungi facilitate the secretion of IL-33 from PDAC cells.

(A) Representative IHC images of IL-33 in the spleen, normal pancreas, PanIN (KC mice) and PDAC (KPC mice) of 6-, 12- and 24-weeks old mice. Scale bar 50 μm.
(B) Fluorescence images showing nuclear expression of IL-33 (green) in PDAC cell line. DAPI (blue) was used for nuclear staining. Magnification 40x, Scale 75μm.
(C) Subcellular fractionation of dox inducible murine PDAC cell line showing IL-33 expression in cytoplasm and nucleus. Lamin A/C and β-tubulin were used as nuclear and cytoplasmic loading control respectively.
(D) The gut and intrapancreatic (n=3, biologically independent samples) mycobiomes of PDAC tumor bearing mice were analyzed by 18S internal transcribed space (ITS) sequencing. The heatmap of relative abundance of the fungal genus (left) and family (right) in gut vs PDAC.
(E) Fluorescence in-situ hybridization (FISH) showing fungal population in normal pancreas vs PDAC. D223 fungal-specific probe was used to detect the fungal species in the normal pancreas.
(F) Schematic showing strategy for fungal extract treatment followed by biochemical assays to determine IL-33 expression in cells and conditioned media upon treatment with Alternaria alternata.
(G) Western blot analysis of IL-33 in PDAC cell line treated with vehicle, fungal extract (Alternaria alternata) for different time points (2, 3, 6, and 24 hrs) and shIL33 PDAC cell line. β-actin was used as a loading control.
(H) IL-33 was measured in conditioned media using ELISA in PDAC cell line treated with Alternaria alternata extract for different time points (2, 3 and 6 hrs), n=3.
(I) Schematic showing strategy for quantification of IL-5 in flow-sorted ILC2 cells cultured with PDAC cell conditioned media which was earlier treated with the fungal extract.
(J) IL-5 measured using ELISA produced from ILC2s upon treatment with fungal extract-treated cancer cell conditioned media.
Data are presented as mean ±SD. p-values were calculated using the Student t-test. ns, no significance. Individual p-values are indicated in the figures.
See also Figure S4.
Prior studies in a murine allergic model showed that IL-33 can be secreted by lung epithelial cells in the presence of fungus, thereby exacerbating allergic responses (Snelgrove et al., 2014). Importantly, recent studies reported the presence of intra-tumor micro- and myco-biome in PDAC, which supports tumor progression (Riquelme et al., 2019, Aykut et al., 2019). As a first step, 18S ribosomal RNA (rRNA) sequencing confirmed the presence of mycobiome in our PDAC mouse model (Figure 4D, Figure S4B). Fungal communities were detected in both tumor and gut samples of PDAC-bearing mice and were present in much higher abundance in tumors. Malassezia and Alternaria, were documented as the most abundant fungi in PDAC tumors. Fluorescence in situ hybridization (FISH) is used to probe for the presence of fungal DNA in the normal pancreas and PDAC tumor, confirmed higher fungal abundance in the PDAC tumor compared to the normal pancreas (Figure 4E).
Next, in vitro assays were performed to ascertain the direct role of fungus on IL-33 secretion. We used an array of fungi identified in our 18S rRNA sequencing, as well as those reported earlier in PDAC tumors (Aykut et al., 2019). There was a time-course dependent loss of IL-33 in PDAC cells treated with extracts of Alternaria alternata (Figure 4F–G, Figure S4C, E), as shown by western blot analysis. The secretion of IL-33 peaks at 3 hours post-fungal extract treatment and tapers off at 6 hours. The secretion completely ceases at about 24 hours post-treatment. Simultaneously, an ELISA detected IL-33 in the spent media, which coincides with the loss of IL-33 in cell lysates (Figure 4H, Figure S4D, F). Interestingly, a similar experiment conducted with Aspergillus and Candida spp. did not yield similar results, indicating that specific fungi species regulate IL-33 secretion in PDAC cells (Figure S4G-J). In a separate experiment, cancer cells treated with Alternaria alternata showed a time-course dependent loss of IL-33 as shown by immunofluorescence (Figure S4K).
To determine whether the IL-33 secreted into the spent media can activate ILC2 cells, we conducted a functional assay where we sort-purified ILC2s from mouse spleen and treated the ILC2s with the fungal-treated spent media from the PDAC cell line (Figure 4I). An ELISA showed robust IL-5 secretion by ILC2 cells (Figure 4J).
Intratumor fungi accelerate PDAC tumor growth
We next assessed the impact of depleting fungi on IL-33 secretion and the type 2 immune response in the PDAC TME. Gastrointestinal fungi in tumor-bearing mice were depleted by oral amphotericin B (anti-fungal) treatment (Figure 5A). Amphotericin B treatment or IL-33 depletion resulted in a significant decrease in tumor burden and increased survival (Figure 5B–D, Figure S5A-C) as well as reduced tumor infiltrating ILC2 (Figure 5E) and TH2 cells (Figure 5F).
Figure 5: Intratumor fungus accelerates PDAC tumor growth.

(A) Schematic showing fungal depletion strategy using amphotericin B (200 ug/dose) followed by orthotopic transplantation of PDAC cells and tumor progression studies.
(B) Representative MRI images with their relative volumes (n=5) on day 26, showing orthotopically transplanted shCtrl and shIL33 PDAC tumors with or without amphotericin B treatment. Red dotted circles define the tumor boundaries.
(C) Bar graph showing tumor volume of orthotopically transplanted shCtrl and shIL33 PDAC tumors (n=14) and shIL33 (#1 [n=9] and #2 [n=10]) with or without amphotericin B treatment.
(D) Kaplan–Meier survival curves of mice orthotopically transplanted with shCtrl and shIL33 PDAC tumors with or without amphotericin B treatment (n=10). Statistical analysis was done using the Gehan-Breslow Wilcoxon test.
(E) Frequency of ILC2 in orthotopically transplanted shCtrl and shIL33 PDAC tumors with or without amphotericin B treatment relative to total Lin− cells.
(F) Frequency of TH2 cells in orthotopically transplanted shCtrl and shIL33 PDAC tumors with or without amphotericin B treatment relative to total CD4+ cells.
(G) Schematic showing strategy for fungal transplantation followed by orthotopic transplantation of PDAC cells and tumor progression studies.
(H) Representative IHC (top) and immunofluorescence (middle) images of PDAC tumors showing IL33 expression in control and fungal transplanted mice by oral gavage (n=10), Magnification 40x. Scale bar 100 μm. FISH (bottom) showing fungal colonization of PDAC tumors in fungal transplanted mice by oral gavage. Scale bar 10 μm.
(I) 18S rRNA sequence showing fungal species in PDAC tumors of the Alternaria alternata and Malassezia globosa transplanted mice by oral gavage. Also, shown are the 18S rRNA sequencing in stool samples of the Alternaria alternata and Malassezia globosa transplanted mice by oral gavage. Positive control-Alternaria is a sample of pure Alternaria alternata culture extract.
(J) Bar Graph showing the wet weight of PDAC tumors of the control, amphotericin B and fungal transplanted mice (n=10).
(K) Frequency of ILC2/total Lin− cells in PDAC tumors of control, amphotericin B and fungal transplanted mice.
Data are presented as mean ±SD. p-values were calculated using the Student t-test. ns, no significance. Individual p-values are indicated in the figures.
See also Figure S5.
To further test the role of fungi in IL-33 secretion and recruitment of ILC2 and TH2 cells in the PDAC TME, we administered Malassezia globosa or Alternaria alternata by oral gavage to PDAC-bearing mice (Figure 5G). First, fungi resident in the mice were depleted by a course of amphotericin B (i.e., 5 doses of amphotericin B, 200 μg/day by oral gavage), followed by a maintenance dose of 0.5 μg/ml in the drinking water for 20 days. Following depletion of fungi, we administered either Malassezia globosa or Alternaria alternata (108 fungal spores per mouse) by oral gavage into the tumor-bearing mice. At 28 days post-fungal treatment, the tumors were harvested for analyses (Figure S5D). IHC and IF showed that fungal-depleted PDAC tumors exhibited an IL-33 signal restricted predominantly to the nucleus of cancer cells, whereas fungi-administered PDAC tumors showed extracellular expression of IL-33 (Figure 5H, top and middle). Intratumoral fungal depletion and repopulation were confirmed by fungal FISH analysis (Figure 5H, bottom). In addition, 18S rRNA sequencing confirmed increased presence of Alternaria and Malassezia spp. in the tumor and stool of the mouse receiving fungal transplantation (Figure 5I, Figure S5E-G). Notably, amphotericin B treatment significantly decreased tumor burden, whereas the administration of either Malassezia globosa or Alternaria alternata promoted tumor growth (Figure 5J). Similarly, Malassezia globosa or Alternaria alternata administration augmented the infiltration of ILC2 (Figure 5K) and TH2 (Figure S5H) cell populations within the TME. Moreover, Alternaria alternata administration decreased CD8+ T cells, and no significant changes were observed in total CD4+ and B cells (Figure S5I-K).
IL-33-mediated ILC2 activation is necessary for tumor progression
To establish a direct link between IL-33-mediated ILC2 activation and tumor progression, we orthotopically co-transplanted ILC2 cells with PDAC cells (Figure 6A). The co-transplantation of PDAC-derived ILC2 cells (donor) into a syngeneic host (recipient) led to a significant increase in tumor burden, while IL-33 KO in cancer cells resulted in significantly reduced tumor burden, which was unaltered with ILC2 co-transplantation (Figure 6B–E). A complementary experiment demonstrated that retro-orbital injection of ILC2 cells enabled their recruitment to the TME in response to IL-33-expressing PDAC cells. First, to ensure the proper transfer of the retro-orbitally injected ILC2 cells to the PDAC TME, we established a protocol in which we labelled ILC2 cells with a Vybrant-DiD dye. The DiD-labeled ILC2 cells were then injected retro-orbitally and 7 days post-injection, the PDAC tumors were collected and DiD-labelled ILC2 cells were measured by flow cytometry (Figure S6A). The assay confirmed the presence of DiD-labeled ILC2 cells in the PDAC tumor, as shown by the mean fluorescence intensity (MFI) levels of the ILC2 cells (Figure S6B-C). Thereafter, we transplanted ILC2 (1×105 cells) into either IL-33 WT or IL-33 KO PDAC-bearing mice. ILC2 transplantation in IL-33-WT mice led to a small increase in tumor growth, whereas IL-33 KO tumors showed minimal change in tumor growth (Figure S6D-F).
Figure 6: IL-33 mediated ILC2 activation is necessary for tumor progression.
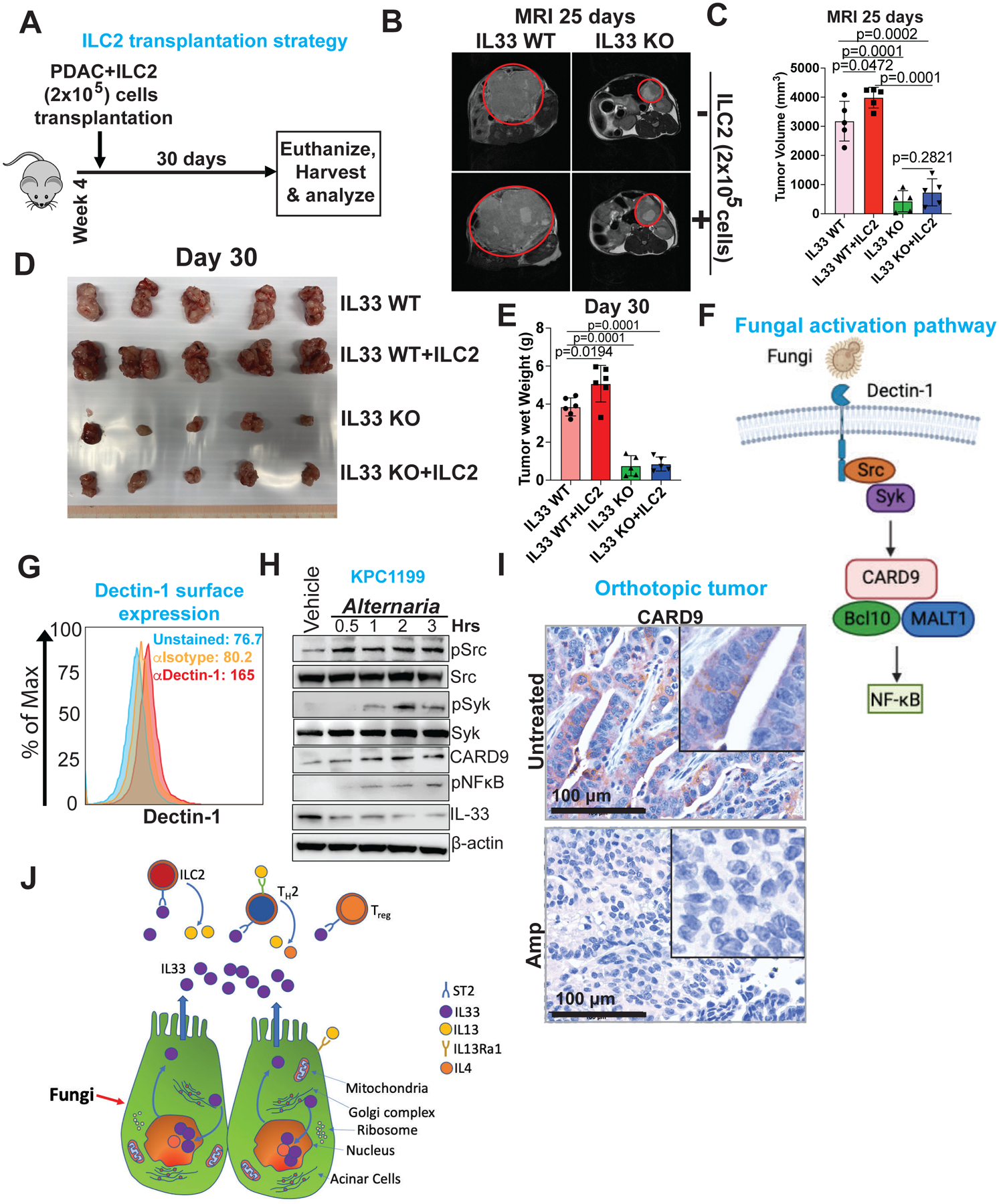
(A) Schematic showing strategy for orthotopic co-transplantation of PDAC and ILC2 cells.
(B) Representative MRI scans showing axial images of CRISPR-Cas9 knockout tumors (IL33 WT vs IL33 KO), with or without ILC2 co-transplantation.
(C) Bar graph showing tumor volume calculated by MRI image analysis (n=5–7).
(D) Picture showing gross PDAC tumor with or without ILC2 in IL33 WT vs IL33 KO mice (n=5).
(E) Bar graph showing PDAC tumor wet weight with or without ILC2 in IL33 WT vs IL33 KO mice (n=5–7).
(F) Schematic showing fungal activation pathway, where dectin-1 receptor ligates fungal components and induce Src-Syk-CARD9 signaling cascade.
(G) Histogram showing dectin-1 expression on PDAC cell line, analyzed by flow cytometry (n=3).
(H) Western blot showing expression of pSrc, Src, pSyk, Syk, CARD9, p-NFκB and IL-33 upon treatment with Alternaria alternata. β-actin was used as a loading control.
(I) Representative IHC image showing CARD9 expression in orthotopic transplanted tumor with or without antifungal treatment. Scale bar 100 μm.
(J) Working model showing fungus mediated secretion of IL-33 from PDAC tumor, attracting type 2 immune cells (ILC2, TH2, and Treg) thereby promoting tumor progression. Results are shown as mean ±SD. p-values were calculated using the Student t-test.
Individual p-values are indicated in the figures.
See also Figure S6.
Prior studies have demonstrated that fungal-derived lectin (e.g., β-glucans) are recognized by a pattern recognition receptor (PRR), dectin-1, expressed primarily by macrophages, neutrophils, and dendritic cells. These studies showed that the dectin-1 pathway is involved in the release of cytokines, such as IL-1β by macrophages (Rosas et al., 2008, Hernanz-Falcon et al., 2009). Thus, to understand the mechanisms governing fungal-mediated IL-33 secretion from PDAC cells, we conducted cell culture studies wherein PDAC cells were exposed to fungal extracts and analyzed for activation of the dectin-1 pathway (Figure 6F). We observed activation of the dectin-1-mediated Src-Syk-CARD9-NFkB pathway, which tracks with the secretion pattern of IL-33 from the PDAC cells (Figure 6G–H, Figure S6G). Of note, dectin-1 is not expressed in normal human pancreatic cells (Figure S6H-I). In addition, PDAC tumor-bearing mice treated with amphotericin B showed low expression of CARD9, compared to the untreated tumor-bearing counterparts (Figure 6I). Based on these findings, we conclude that intratumoral fungi or fungal products provokes IL-33 secretion by PDAC cells, which promotes type 2 immune responses and tumor progression (Figure 6J).
DISCUSSION
PDAC tumors are infiltrated by pro-tumorigenic immune cells that include TH2 and ILC2 cells, which via their cytokine networks, foster a pro-tumorigenic program that leads to PDAC progression [32–35]. However, it was unknown which factors mediate the recruitment and activation of TH2 and ILC2 cells. In this study, we establish that KrasG12D regulates the expression of a chemoattracting cytokine, IL-33, that recruits TH2 and ILC2 cells. The TH2 and ILC2 cells via their pro-tumorigenic cytokine production accelerate PDAC tumor progression. Specifically, we established IL-33 as a bona fide downstream target of KrasG12D and that IL-33 expression is significantly upregulated in PDAC patients. We also identified an unanticipated role for the intratumoral mycobiome in facilitating the type 2 immune response in the PDAC TME by stimulating the extracellular secretion of IL-33. This role of intratumor fungus is distinct from prior observation where fungi were shown to activate a complement system (Aykut et al., 2019) and might act in tandem to promote PDAC progression. These mechanistic insights governing the extracellular secretion of IL-33 suggest that IL-33-ILC2/TH2 axis is a potential target in PDAC. Accordingly, in a preclinical proof-of-concept study, genetic deficiency of IL-33 or an anti-fungal treatment decreased tumor burden and prolonged survival in mice.
Although the definitive molecular link between fungal components and IL-33 secretion remains to be defined, our study has revealed an important connection between the intratumoral mycobiome and the spatiotemporal secretion of IL-33 in the PDAC TME. We found that fungal derived components can activate a dectin-1-dependent pathway, reminiscent of similar fungal response pathways observed in macrophages and DCs. Specifically, fungi can activate a dectin-1-mediated Src-Syk-CARD9 pathway which, via downstream mechanisms, secretes IL-33. In addition to fungi, biochemical factors, such as reactive oxygen species (ROS) and oxidative stress (Taniguchi et al., 2020, Uchida et al., 2017), can promote the extracellular secretion of IL-33. These aforementioned factors might act in concert with the mycobiome as a stimulator and amplifier of IL-33 secretion. Notably, we and others have found that fungal components can be detected in the early stages of PDAC tumorigenesis, such as in PanIN (Aykut et al., 2019) when a large-scale oxidative stress has yet to be detected in the TME. This raises the question of whether the mycobiome is responsible for initiating the type 2 immune response in PDAC, which is further reinforced by factors such as oxidative stress and other yet to be discovered processes or vice versa.
Recent studies using metagenomic sequencing have demonstrated the presence of intratumoral fungi in PDAC (Aykut et al., 2019). A possible route for microbial migration from the duodenum to the pancreas is the retrograde transfer via the opening of the sphincter of Oddi, which controls the flow of digestive juices (bile and pancreatic) from the pancreas and gall bladder. We have validated this finding in our mouse model where we found that PDAC tumors are infiltrated mostly by fungal genera such as Alternaria and Malassezia. The question is how early in the PDAC tumorigenesis process does this retrograde transfer of fungi occur. The chain of events is crucial to understanding the mechanism of IL-33 secretion into the extracellular space, the recruitment and activation of ILC2 and TH2 cells and the secretion of pro-tumorigenic cytokines. In addition to the intratumor fungal microbiome, the role of the bacterial microbiome as a collaborator cannot be ruled out as studies have showed that gram-negative bacteria can stimulate IL-33 secretion in the nasopharynx (Hentschke et al., 2017). Recent studies have also demonstrated the presence of intratumor bacterial microbiome in PDAC models and consequent antibiotic treatment reduces tumor growth, increases M1-type macrophage abundance, promotes TH1 differentiation and stimulates CD8+ T cell activation (Riquelme et al., 2019).
Although IL-33 plays a predominant role in PDAC progression, as its deletion leads to significant tumor regression and increased survival, however, the role of IL-33 is context-dependent and sometimes have opposite effects in various cancer types. Moreover, IL-33 is involved in a myriad of functions depending on the spatial context of the protein. For example, the immune function of IL-33 described here is distinct from its cancer cell-intrinsic function that has been described recently, in which IL-33 mediates a pancreas tissue injury program in Kras mutant mice (Park et al., 2021) (Alonso-Curbelo et al., 2021). Importantly, IL-33 has been shown to cooperate with mutant Kras to initiate pancreatic neoplasia by a chromatin switch (Alonso-Curbelo et al., 2021).
Given the divergent function of ILC2s in different tumor types, and also the context-dependent role of ILC2 in tumor progression, further study is necessary to tease apart the role of ILC2 cells in PDAC and other cancers. For example, mice lacking the IL-33 receptor ST2 have slower tumor progression by increasing TH1 and NK cell activity, hinting towards a pro-tumorigenic role of an IL-33-ILC2 axis (Jovanovic et al., 2011, Ercolano et al., 2019). In contrast, a recent study found that ILC2 infiltration in PDAC cells positively correlate with long-time survival in patients (Moral et al., 2020). According to that study, the infiltrating ILC2 cells are anti-tumorigenic due to their ability to engage CD103+ DCs, but that PD-1 expression constrains the anti-tumor activity of ILC2 cells; thus, anti-PD-1 plus rIL-33 should provide clinical benefit. The stark contrast between this and our study can be explained at various levels. First, the study focused exclusively on the ILC2-CD103+ DC-CD8+ T cell axis, although the main effector cytokines of ILC2 cells are IL-4 and IL-13, which are potent inducers of M2-like macrophage polarization and are central players in immune suppression in the PDAC TME (Zhu et al., 2017). Second, our study and those of other investigators showed that oncogenic KRAS and/or tissue damage (Alonso-Curbelo et al., 2021, Park et al., 2021) already potently upregulates IL-33 expression in PDAC, raising uncertainty as to the utility of injecting rIL-33 as a treatment regimen for PDAC. Finally, the tumor models used in the study induced a robust CD8+ T cell response which contrasts to the poorly immunogenic PDAC models used in our study, suggesting that IL-33 may have different outcomes in immunogenic tumors that are capable of priming endogenous anti-tumor CD8+ T cells.
Finally, while anti-fungal treatment may appear to be an attractive therapeutic option for PDAC; our in vivo studies show that anti-fungal treatment might be insufficient to inhibit IL-33 secretion and suppress tumor growth. It is possible that fungal components as opposed to live fungus are sufficient for IL-33 secretion as demonstrated by our in vitro studies. Along these lines, it is worth noting that our attempts to culture live fungus from fresh tumor samples have been unsuccessful to date. This may relate to a lack of live fungus or phagocytic elimination of fungus by intratumoral macrophages which, in turn, release fungal components such as chitin, β-glucans, DNA/RNA fragments into the PDAC TME. Thus, the absence of live fungi would render anti-fungal treatment inconsequential in a prolonged treatment paradigm. Alternatively, it is possible that live fungi are present, but we are unable to culture the fungi because >75% of the microbiome are not culturable using conventional culturing techniques (Watterson et al., 2020) and efforts are being made to improve such culturing methodologies. Nevertheless, the fungal components are crucial for regulating an integral pro-tumorigenic cancer-immune cell axis which, if targeted, can yield potentially significant benefit to PDAC patients.
Finally, it is likely that ILC2 plays a variable role in the early and late stages of the tumorigenesis. Seen in this light, the effects of fungi in priming the IL-33-ILC2 axis may have different consequences based on the tumor type, location, and extent of tumor burden, and the involvement of fungi adds another layer of complexity to the IL-33-ILC2 axis.
STAR METHODS
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Prasenjit Dey (prasenjit.dey@roswellpark.org).
Materials availability
Upon request mouse cell lines (PJ-B6-4291, PJ-B6-4298 and PJ-B6-4271) generated in the lab will be made available.
Data and code availability
Processed single-cell RNA-seq and bulk RNA-seq data of this study can be obtained from Gene Expression Omnibus (GEO) with an accession number of GSE190103 and GSE190468, respectively. 18s rRNA seq data can be obtained from BioSample accessions: SAMN23523449, SAMN23523450, SAMN23523451, SAMN23523452, SAMN23523453, SAMN23523454, SAMN23523455, SAMN23523456.
All other data are available from the corresponding authors upon request.
Experimental model and subject details
Ethics statement and animal modeling
All animal procedures were approved by Institutional Animal Care and Use Committee (IACUC) at Roswell Park Comprehensive Cancer Center. All animals were maintained in pathogen-free conditions and cared for in accordance with the International Association for Assessment and Accreditation of Laboratory Animal Care policies and certification. All surgeries were performed with isoflurane anesthesia. Analgesic (Buprenorphine) was administered after surgery along with temperature-controlled post-surgical monitoring to minimize suffering. TetO_Lox-Stop-Lox-KrasG12D (tetO_KrasG12D), ROSA26-LSL-rtTA-IRES-GFP (R26_rtTA), Ptf1a-Cre, LSL-Trp53, LSL-KrasG12D, LSL-Trp53R172H and Pdx1-Cre strains were described previously (Ying et al., 2012, Olive et al., 2004, Hingorani et al., 2003). Mice were backcrossed to the C57BL/6 background for more than 8 generations to achieve a pure B6 mouse, and the purity and zygosity of iKPC mouse was validated by Charles River. Mice with spontaneous pancreatic tumors were euthanized at designated time points for tumor collection. Owing to the internal location of these tumors, we used signs of lethargy, reduced mobility, and morbidity, rather than maximal tumor size, as a protocol-enforced end point.
Human PDAC primary tumor samples
Fresh human PDAC samples were obtained from Roswell Park Comprehensive Cancer Center’s tissue Biobank. Human studies were approved by Roswell Park Comprehensive Cancer Center’s Institutional Review Board, and prior informed consent was obtained from all subjects and the tissues were distributed under IRB protocol STUDY00001407/BDR 135920. Fresh human PDAC samples were processed to obtain single cell suspension using a tissue dissociation cocktail consisting of Collagenase IV (Cat. No.: 07909, Stem Cell Technologies), Dispase (Cat. No.: 07923, Stem Cell Technologies) and DNase I (Cat. No.: 9003-98-9, Fisher Scientific) in a DMEM/F12 with 15 mM HEPES media. For flow cytometry analysis samples were further processed as described below in the section “Flow cytometry”. Fluorochrome-conjugated antibodies against CD45 (304014), Foxp3 (320124 and 320112), CD3 (317318), CD4 (317414), CD8 (344740), KLRG1 (138408), Lin− cocktail (348803), CD127 (351324), CD25 (302604), TruStain FcX (422302), Gata-3 (653812) were purchased from Biolegend and ST2 (FAB5231A) from R&D. To assess cell viability, cells were incubated with Zombie UV (423107, Biolegend), prior to FACS analysis. All samples were acquired with the BD LSR analyzer (Becton Dickinson) and analyzed with FlowJo software (Tree Star).
Human PDAC TMA samples were obtained from MD Anderson Cancer Center Biobank. Human studies were approved by MD Anderson’s Institutional Review Board (IRB), and prior informed consent was obtained from all subjects under IRB protocol LAB05-0854. The samples were stained using the standard IHC protocol. The antibodies used were IL-33 (R&D systems, AF3626) and secondary antibody (HRP-polymers, Biocare Medical). The stained samples were imaged using Aperio ScanScope XT scanner and Aperio ImageScope software was used for image visualization and analysis. Staining intensity of tissue sections was scored in a ‘blinded’ manner by a pathologist.
Mice and tumor models
For all experiments, C57BL/6J (Stock 000664) mice, aged 4–6 weeks were obtained from Jackson Laboratory unless otherwise mentioned. For orthotopic pancreas transplantation, mice were anaesthetized using isoflurane. A 2×2-mm portion of the left abdomen was shaved to facilitate transplantation. An incision was made in the left abdomen and the pancreas was gently exposed along with the spleen. Luciferase-expressing cells (RFP-Luc) were slowly injected into the tail of the pancreas using a Hamilton syringe. Twenty microliters of cells (5 × 105) mixed with 20 μl of Matrigel were injected. For the orthotopic model, animals were imaged (IVIS Spectrum, PerkinElmer) 2 days after surgery to assess successful implantation of the tumors. Only orthotopic tumors of similar luciferase intensity were used further for the study. For iKPC cell line, chow supplemented with Dox was started after transplantation. Furthermore, in early stages the progress of the tumor is monitored by MRI and/or IVIS Spectrum imaging and in the late stages of the tumor development by MRI alone. The upper limit for the tumor volume is set at 4000 mm3. However, owing to the internal location of the tumors, we used signs of lethargy, reduced mobility, and morbidity, rather than maximal tumor size, as a protocol-enforced endpoint. Typically, mice are sacrificed around day 28 or if moribund because of tumor burden, which is considered as an endpoint.
Method details
In vivo imaging
In vivo live imaging was performed at the Animal Imaging Facility at Roswell Park Comprehensive Cancer Center. Magnetic resonance imaging (MRI) was performed using a 4.7 Tesla, preclinical scanner using the ParaVision 3.0.2 platform and a 35 mm I.D. radiofrequency coil (Bruker Biospin, Billerica, MA). Mice were anesthetized with isoflurane. Temperature and respiration were regulated using an MR-compatible small animal monitoring system (Model 1250, SAII, Stony Brook, NY.) Following scout scans, tumor burden was determined using multislice, fast spin echo scans in the axial and coronal planes. Both sets of images used a TE/TR = 40/2500 ms, an echo train length =8, slice thickness = 1mm, acquisition matrix = 256×192 and 4 averages/NEX. Coronal images were acquired with a field of view (FOV) = 48×32mm, while axial images were acquired with a FOV = 32×32 mm. A subset of axial scans required a larger number of slices to capture tumor growth, which increased the repetition time to 3600 ms. Tumor volumes were calculated by manual segmentation and voxel summation using commercially available, medical image processing software (Analyze 10.0, AnalyzeDirect, Overland Park, KS).
For bioluminescent imaging, animals were anesthetized with isoflurane, injected intraperitoneally with 3 mg of D-Luciferin (Perkin Elmer) and imaged using IVIS Spectrum Imaging System (Perkin Elmer). The Living Image 4.7 software (Perkin Elmer) was used for analysis of the images post acquisition.
Transcriptomic profiling by RNA-seq and qRT-PCR
For bulk RNAseq, total RNA was isolated using QIAzol extraction (Cat. No.: 79306, Qiagen) followed by purification with the Qiagen RNAeasy kit as described previously (Dey et al., 2012). Libraries were generated using Illumina’s TruSeq RNA Library Prep Kit v2 and were sequenced using the Illumina HiSeq 2000 Sequencer with 76 nt single-end read. Raw read RNA-seq data were mapped to GRCm38-mm10 reference genome using STAR aligner (Dobin et al., 2013). HTSEQ-COUNT (Anders et al., 2015) was used to count raw reads in each gene for each sample. Differential expression analysis was performed with edgeR (Robinson et al., 2010) and pathway enrichment analysis was performed with GSEA software. For qRT-PCR, RNA samples were reverse transcribed into cDNA using the High-Capacity cDNA Reverse Transcript kit (Life Technologies). cDNA samples were subjected to qRT-PCR quantification in duplicates using Power SYBR Green PCR Master Mix (Life Technologies) according to the product guides on an Agilent Mx3005P or Applied Biosystems AB7500 Fast Real Time machine. The primer sequences used for qRT-PCR are the following: Il33 (Fwd 5’ TGAGACTCCGTTCTGGCCTC 3’, Rev 5’ CTCTTCATGCTTGGTACCCGAT 3’), Actb (Fwd 5’ GGCTGTATTCCCCTCCATCG 3’, Rev 5’ CCAGTTGGTAACAATGCCATGT 3’), Tph1 (Fwd 5’ CACGAGTGCAAGCCAAGGTTT 3’, Rev 5’ AGTTTCCAGCCCCGACATCAG 3’), Il13 (Fwd 5’ TGAGGAGCTGAGCAACATCACACA 3’, Rev 5’ TGCGGTTACAGAGGCCATGCAATA 3’), Il5 (Fwd 5’ TCACCGAGCTCTGTTGACAA 3’, Rev 5’ CCACACTTCTCTTTTTGGCG 3’), and Amphiregulin (Areg) (Fwd 5’ GGTCTTAGGCTCAGGACATTA 3’, Rev 5’ CGCTTATGGAAACCTCTC 3’) (Table S1).
Single cell RNA sequencing (scRNAseq)
For scRNAseq, live CD45+ cells were flow sorted by BD FACSAria. Single cell libraries were generated using the 10X Genomics Chromium Single Cell 3′ GEM, Library & Gel Bead Kit v3, (10x Genomics, Cat.: 1000075). Briefly, single cell suspensions were first assessed with Trypan Blue using Thermo Fisher Countess II FL Automated Cell Counter (ThermoFisher), to determine concentration, viability and the absence of clumps and debris that could interfere with single cell capture. The cells were then loaded into the Chromium Controller (10x Genomics) where they are partitioned into nanoliter-scale Gel Beads-in-emulsion. Reverse transcription was performed, and the resulting cDNA was amplified following the manufacturer’s instructions. The full-length amplified cDNA was used to generate gene expression libraries by enzymatic fragmentation, end-repair, a-tailing, adapter ligation, and PCR to add Illumina compatible sequencing adapters. The resulting libraries were evaluated on D1000 screentape using a TapeStation 4200 (Agilent Technologies) and quantitated using Kapa Biosystems qPCR quantitation kit for Illumina. Final libraries were then pooled, denatured, and diluted to 300pM with 1% PhiX control library added. The resulting pool was then loaded into the Illumina NovaSeq Reagent cartridge and sequenced on a NovaSeq 6000 following the manufacturer’s recommended protocol (Illumina Inc.).
scRNAseq data analysis:
FASTQ files were processed using 10x Genomics Cell Ranger 6.0.0 pipeline to align reads and generate feature-barcode matrices. The Cell Ranger output was further used for detailed data analysis. For the Chromium 10x Genomics libraries, the raw sequencing data were processed using Cell Ranger software to generate the fastq files and count matrices (Zheng et al., 2017). Then the filtered gene-barcode matrices which contain barcodes with the Unique Molecular Identifier (UMI) counts that passed the cell detection algorithm was used for further analysis. The downstream analyses were performed primarily using Seurat single cell data analysis R package (Hao et al., 2021). First, cells with unique feature counts over 5000 or less than 200, or cells that have >20% mitochondrial counts were filtered out from the analysis to remove dead cells. Additionally, a cell cycle score for S and G2/M phases based on the well-defined gene sets was assigned to each cell using CellCycleScoring function from Seurat. Then the normalized and scaled UMI counts were calculated using the SCTransform method and be regressed against the cell cycle scores. Dimension reductions including principal component analysis (PCA), UMAP and tSNE were performed for the highly variable genes. Data clustering were identified using the shared nearest neighbor (SNN)-based clustering on the first 30 principal components. SingleR package was utilized to identify the immune cell types using ImmGen reference dataset (Aran et al., 2019).
Flow cytometry
Single cells from tumor were isolated using the Mouse Tumor Dissociation kit (Cat.: 130-096-730, Miltenyi Biotec) based on manufacturer’s protocol. Cells from spleen were isolated by mincing with a 5-mL syringe plunger against a 70 μm cell strainer into a 50 ml falcon tube using Roswell Park Memorial Institute (RPMI) medium. The cells were depleted of erythrocytes by RBC lysis buffer (Biolegend, Cat.: 420302). Peripheral blood (100 μL) was drawn using retroorbital bleeding and depleted of erythrocytes by RBC lysis buffer. Next, tumor, spleen or blood cells were incubated with CD16/CD32 antibody (clone 2.4G2, BD Biosciences) to block FcγR binding for 10 minutes then with a antibody mix for 30 minutes at room temperature. Fluorochrome-conjugated antibodies against CD45 (103115), CD11b (101220), CD11b (101237), Gr-1 (108405), Ly-6C (128015), Lineage cocktail (133311), B220 (103247), Ki67 (350521), KLRG1 (138419), ICOS (313533), GR1 (108405), CD45RB (151607), EpCAM (118207), CD62L (104405), CCR3 (144509), CD90.2 (140303), CD25 (102027), CD11c (117307), CD44 (103023), CCR4 (131203), CD117 (105807), F4/80 (123109), CD86 (105016), CD127 (135015), ST2 (146609), Ly-6c (128015), CD3 (100235), Sca1(108111), Gata3 (653812), Ly-6G (127617), CD4 (100421), CD3 (100235), CD8 (100749) were purchased from BioLegend. To assess cell viability, cells were incubated with Zombie UV (423107, Biolegend), prior to FACS analysis. All samples were acquired with the BD LSR analyzer (Becton Dickinson) and analyzed with FlowJo software (Tree Star).
Immunoblotting and antibodies
Cell culture medium were removed, and the cells were washed twice in ice-cold phosphate-buffered saline (PBS). Cells are scraped and transferred to an Eppendorf tube, followed by centrifugation at 1700g for 5 min. The pelleted cells were incubated in radio immune precipitation assay (RIPA) buffer with proteinase and phosphatase inhibitors for 15 mins. Lysates were then collected and centrifuged at 12000 rpm for 15 min at 4°C. Protein concentration was measured using the DC Protein Assay Kit (Biorad, 5000111). SDS–PAGE and immunoblotting were performed as described previously in a pre-cast bis-Tris 4–20% gradient gels (Invitrogen) (Dey et al., 2017). The following antibodies were used: The following antibodies were used: IL-33 (R&D systems, AF3626); pAKT-S473 (CST, 9271); pERK1/2 (CST, 4370); CARD9 (CST, 2283s); Src (CST, 2108s); Phospho-Syk-Y525/526 (CST, 2710s); Phospho-Src-Y416 (CST, 6943s); Syk (CST, 2712s) and β-Actin (Sigma-Aldrich, A2228).
Immunohistochemistry and Immunofluorescence
Harvested tissues were immediately fixed in 10% formalin overnight followed by incubation in 70% ethanol for 48 hrs. Fixed tissues are then processed in tissue processor and embedded in paraffin. IHC was performed as described previously (Dey et al., 2014). Briefly, endogenous peroxidases were inactivated by 3% hydrogen peroxide. Non-specific signal was blocked using 5% BSA for 30 mins in 0.1% Tween 20. Tumor samples were stained with the following primary antibodies: CD45 (Abcam, ab10558), mouse IL-33 antibody (R&D systems, AF3626); human IL-33 antibody (R&D systems, AF3625); SMA (abcam, ab5694) and Pan-Keratin (CST, 4545S). After overnight incubation, the slides were washed and incubated with secondary antibody (HRP-polymers, Biocare Medical) for 30 min at room temperature. The slides were washed three times and stained with DAB substrate (ThermoFisher Scientific). The slides were then counterstained with haematoxylin and mounted with mounting medium. For immunofluorescence tissues were stained with primary antibodies overnight followed by staining with fluorescence labelled secondary antibodies at room temperature for 2 hrs. The nuclei is stained with DAPI. Immunofluorescence slides were imaged with Leica confocal Microscope and quantified with ImageJ.
Cell Culture and establishment of primary PDAC lines
Following primary mouse cell lines were established in the laboratory (PJ-B6-4291, PJ-B6-4298 and PJ-B6-4271). The following cell lines are a gift from Dr. Ronald DePinho (AK-B6, AK14838, AK192, HY19636, HY15559) as described previously (Aguirre et al., 2003) and Dr. Michael Feigin (KPC1199). All cell lines were routinely cultured in RPMI 1640 (Invitrogen) in 10% FBS (Invitrogen), 100 U/ml penicillin and 100 U/ml streptomycin. For inducible KPC derived cell lines, 1 μg/ml of doxycycline was directly added to the media. The cell lines were mycoplasma free, based on tests done monthly in the laboratory using Lonza’s MycoAlert Mycoplasma Detection Kit assays with confirmatory tests by PCR-based assays.
shRNA and CRISPR-Cas9 knockdown
shRNA knockdown was performed as described previously (Dey et al., 2017). We screened 3–5 hairpins targeting the gene of interest and found two independent sequences that reduced mRNA levels by >60%. The shRNA sequences were as follows: Il-33 5’ CCGGGCATCCAAGGAACTTCACTTTCTCGAGAAAGTGAAG TTCCTTGGATGCTTTTTTG 3’ (TRCN0000173352) and 5’ CCGGCCATAAGAAAG GAGACTAGTTCTCGAGAACTAGTCTCCTTTCTTATGGTTTTTTG 3’ (TRCN0000176387); 3’ (Table S1). A non-targeting shRNA (shCtrl) was used as a control. The shRNA-expressing pLKO.1 vector was introduced into cancer cell lines by lentiviral infection. Recombinant lentiviral particles were produced by transient transfection of 293T cells following a standard protocol. Briefly, 10 μg of the shRNA plasmid, 5 μg of psPAX2 and 2.5 μg of pMD2.G were transfected using Lipofectamine 3000 (Invitrogen) into 293T cells plated in a 100-mm dish. Viral supernatant was collected 72 h after transfection, centrifuged to remove any 293T cells and filtered (0.45 μm). For transduction, viral solutions were added to cell culture medium containing 4 μg/ml polybrene; 48 h after infection, cells were selected using 2 μg/ml puromycin and tested for gene depletion by qRT–PCR or immunoblotting. For CRISPR-Cas9 knockdown of Il33, sgRNAs were purchased from Synthego (Sanger CRISPR clones). The sgRNAs along with Cas9 protein (sigma) was transfected into PDAC cells and single cell clones were FACS sorted. The sgIL33 sequence used for Il33 DNA sequence targeting. 5’ AUAGUAGCGUAGCGUAGUAGCACC 3’ and 5’ AUCUCUUCCUAGAAUCCCG 3’ (Table S1). The clones were validated by western blot for deletion of IL-33.
Antifungal treatment and fungal challenge experiments
For anti-fungal treatment experiments, PDAC cell lines were treated with 0.1 μg/ul (final concentration) of fungal extract (Alternaria alternata, XPM1D3a.5, Candida albicans, XPLM73X1A2 and Aspergilus fumigatus, XPM3D3A4, Stallergenes Greer Lab) for different time points (2h, 3h, 6h and 24h). The fungal extract treated spent media from PDAC cell culture was used for IL-33 cytokine profiling by ELISA. The fungal extract treated PDAC cell extract was used for western blotting. All the experiments were performed in a biosafety cabinet following Roswell Park Cancer Center bio-safety guidelines. For gastrointestinal fungal depletion, C57BL/6 mice were treated with 200 μg amphotericin B per day by oral gavage for five consecutive days, followed by 0.5 μg/ml amphotericin B treatment in drinking water for 21 days. Control groups were gavage with 200 μl of PBS for 5 consecutive days. After completion of antifungal treatment, species specific fungal repopulation was done with Alternaria alternata (ATCC, 36376) and Malassezia globosa (MYA-4889). Fungi were administered (1 × 108 CFU/ml) by oral gavage. Seven days after fungal administration, PDAC cells were orthotopically transplanted.
Fungal fluorescence in-situ hybridization (FISH)
FISH was done on a 4 μm thick paraffin embedded pancreatic tissue sections. Sections were pretreated using a commercially available kit (Cytocell, Inc) according to manufacturer’s instructions. For Hybridization, D223 28S rRNA gene probe (Molecular probe, Thermo Fisher Scientific) (Table S1) labeled with the 6-FAM fluorophore (extinction wavelength, 488 nm; emission wavelength, 530 nm) was used to detect the fungal colonization within the mouse pancreatic tissues. Hybridization and post hybridization washes were conducted according to standard procedures (Alamri et al., 2017). Slides were visualized on an Olympus BX61 microscope.
18S rRNA sequencing and Mycobiome analysis
For 18s rRNA sequencing fresh stool and tumor tissues were used. Approximately 25 mg of tissue samples were first homogenized with using Navy RINO Lysis Kit in a Bullet Blender Homogenizer (Next Advance) for 5 minutes followed by overnight treatment with proteinase K (2.5 μg/ml; Thermo Fisher Scientific, Cat.: EO0491) at 55 °C. Subsequently, DNA extraction was performed using Qiagen DNeasy Powersoil Pro Kit as per manufacturer’s instructions (Qiagen, Cat.: 47014). Quantitative assessment of the purified DNA was then accomplished by using a Qubit Broad Range DNA kit (Thermo Fisher Scientific, Cat.: Q32853). The sequencing libraries were prepared using a two-step PCR method using the primer set ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′) (Table S1). The first PCR (25-cycle) uses 25 ng of DNA to amplify the target region, where the PCR primers have overhang adapter sequence necessary for the second PCR step. After purification, the amplicon from the first step is amplified with 8 cycles of PCR using the Nextera Index Kit (Illumina Inc.), which uses primers that target the overhang adaptor sequence added during the first round of PCR. The second round PCR adds one of 384 different combinations of indexed tags to each sample which allows pooling of libraries and multiplex sequencing. Prior to pooling, each individual sample’s amplified DNA is visualized on a Tapestation 4200 D1000 tape (Agilent Technologies) for expected amplicon size, purity and concentration. Validated libraries are pooled equal molar in a final concentration of 4 nM in Tris-HCI 10 mM, pH 8.5, before 2 × 300 cycle sequencing on a MiSeq (Illumina, Inc.).
Mycobiome analysis:
Paired-end fastq reads were demultiplexed, processed and analyzed using QIIME (v1.9.1) (Caporaso et al., 2010). Then, operational taxonomic units (OTUs) were assigned using QIIME’s uclust-based open-reference OTU-picking pipeline. OTUs with less than 0.001% assigned sequences were removed from each sample to avoid biased and inflated diversity estimates. ITS samples were processed following the QIIME pipeline steps using UNITE’s Fungi taxon (v8.4) (Koljalg et al., 2013) referenceannotation adapted for QIIME. Chimeras were removed before taxonomy assignments with vsearch (v2.15.0) (Rognes et al., 2016) using the UCHIME reference dataset (v7.2) available at the UNITE website (Abarenkov et al., 2010). Positive and negative control samples were checked for QC purposes.
Taxonomy assignments from all samples were then compiled in a raw-counts matrix. Raw counts were formatted, processed, and analyzed using phyloseq package (v1.28.0) (McMurdie and Holmes, 2013) [8] in R (v4.0.0). 16S data is summarized to OTUs at the genus level. Observed, Chao1, Shannon and Simpson’s Reciprocal diversity indices were estimated for alpha-diversity scores; mean estimates were obtained performing 100 bootstrapped rarefactions. Same analyses were performed for ITS, but at the species and genus levels. For Beta-diversity, Bray-Curtis dissimilarity score paired with classical multidimensional scaling was estimated and plotted using the vegan package (v2.5.6) (Oksanen, 2007).
Enzyme linked immunosorbent assay (ELISA)
ELISA was done using cell culture conditioned media from PDAC mouse cell lines and ascites fluid from PDAC tumor bearing mice. Cell culture media was concentrated using Amicon Ultra centrifugal filter units (Millipore, Z717185) before conducting ELISA. IL-33 ELISA was performed using LEGEND MAX™ Mouse IL-33 ELISA Kit (Biolegend) using manufacturer’s standard protocol. IL-5 ELISA was performed using Mouse IL-5 Quantikine ELISA Kit (R&D systems) using manufacturer’s standard protocol.
ILC2 enrichment and adoptive transfer of ILC2
Orthotopically transplanted PDAC tumors (donor) were minced, and single cell suspension was prepared using mouse tumor dissociation Kit (Milteny biotec). Ficoll column was used to separate the immune cells. ILC2 cells was purified using ILC2 enrichment kit (Stem cells Technology, Cat.: 19842) according to manufacturer’s instructions. Orthotopically transplanted PDAC tumor bearing mice (recipient) were used for adoptive transfer experiments. After 10 days of orthotopic pancreatic injections of PDAC cells, 1×105 ILC2 cells were adoptively transferred via retroorbital injection. Tumor progression was monitored using MRI imaging post transplantation and volume was measured. Mice were sacrificed between day 28–30 and tumor weight was measured. For localization of adoptively transferred ILC2 cells in the orthotopically transplanted mouse tumor, injected ILC2 cells was labelled with a lipid binding dye DiD’ solid (1,1’-Dioctadecyl-3,3,3’,3’-Tetramethylindodicarbocyanine, 4-Chlorobenzene sulfonate Salt (Invitrogen, D7757). After 7 days, PDAC tumor is harvested and DiD-labelled ILC2 cells were measured by flow cytometry using APC channel.
ILC2 Co-transplantation:
For co-transplantation experiment, PDAC cells (5×105) were co-transplanted with ILC2 cells (2×105). The ILC2 cells used for this experiment was isolated as described in ILC2 adaptive transfer experiment described above. PDAC cells and ILC2 cells were mixed with Matrigel (Corning, USA) in 1:1 ratio and orthotopically injected into mouse pancreas. Tumor progression was monitored using MRI imaging post transplantation and tumor size and volume was measured. Mice were sacrificed between day 28–30 and tumor weight was measured.
Quantification and statistical analysis
Statistical Analysis
GraphPad Prism software was used to conduct the statistical analysis of all data except for qRT-PCR data, where Microsoft excel was used. Data are presented as mean ± SD.
All quantitative results were assessed by unpaired Student’s t-test after confirming that the data met appropriate assumptions (normality and independent sampling). The Student t-test assumed two-tailed distributions to calculate statistical significance between groups. Unless otherwise indicated, for all in vitro experiments, three technical replicates were analyzed. Sample size estimation was done taking into consideration previous experience with animal strains, assay sensitivity and tissue collection methodology used. Animal survival impact was determined by the Kaplan–Meier analysis. p<0.05 was considered statistically significant; the p values are indicated in the figures.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| mIL-33 | R&D | AF3626 AB_884269 |
| hIL-33 | R&D | AF3625 AB_1151900 |
| beta-actin | Sigma | A2228 AB_476697 |
| CD4 | Abcam | ab133616 AB_2750883 |
| CD45 | Abcam | ab10558 AB_442810 |
| p-STAT6 | Cell signaling technology | 56554S AB_2799514 |
| pAKT-S473 | Cell signaling technology | 9271S AB_329825 |
| pERK-p44/42 | Cell signaling technology | 4370S AB_2315112 |
| pan-keratin | Cell signaling technology | 4545S AB_490860 |
| CARD9 | Cell signaling technology | 12283S AB_2797869 |
| Src | Cell signaling technology | 2108s AB_331137 |
| Phospho-Syk (Tyr525/526) | Cell signaling technology | 2710s AB_2197222 |
| Phospho-Src Family (Tyr416) | Cell signaling technology | 6943s AB_10013641 |
| Syk | Cell signaling technology | 2712s AB_2197223 |
| Phospho-NF-κB p65 (Ser536) | Cell signaling technology | 13346T AB_2798185 |
| Phospho-IRF-3 (Ser396) | Cell signaling technology | 29047T AB_2773013 |
| Phospho-IRF-7 (Ser477) | Cell signaling technology | 12390T AB_2797896 |
| Anti-rabbit IgG, HRP-linked | Cell signaling technology | 7074P2 AB_2099233 |
| Mouse on mouse horseradish peroxidase polymer | Biocare medical | MM510 |
| Rabbit on rodent horseradish peroxidase polymer | Biocare medical | RMR622 |
| Goat on rodent horseradish peroxidase polymer | Biocare medical | GHP516 |
| Goat probe | Biocare medical | GP626L |
| Donkey anti Goat IgG Alexa Flour Plus 488 | Invitrogen |
A32814 AB_2762838 |
| Donkey anti Rabbit Alexa Flour Plus 555 | Invitrogen | A32794 AB_2762834 |
| A32814 secondary antibody IgG goat | Biorad | 1721034 AB_11125144 |
| A32794 secondary antibody anti IgG rabbit | Biorad | 1706515 AB_11125142 |
| Zombi UV | Biolegend | 423107 |
| BV421 Lin− | Biolegend | 133311 AB_11203535 |
| BV510 B220 | Biolegend | 103247 AB_2561394 |
| BV605 CD11b | Biolegend | 101237 AB_11126744 |
| BV605 KLRG1 | Biolegend | 138419 AB_2563357 |
| BV785 ICOS | Biolegend | 313533 AB_2629728 |
| FITC GR1 | Biolegend | 108405 AB_313370 |
| FITC CD90.2 | Biolegend | 140303 AB_10642686 |
| PerCP CD25 | Biolegend | 102027 AB_893290 |
| PE CD11c | Biolegend | 117307 AB_313776 |
| PE CCR4 | Biolegend | 131203 AB_1236369 |
| PE F4/80 | Biolegend | 123109 AB_893498 |
| PE-Cy5 CD127 | Biolegend | 135015 AB_1937262 |
| PE Cy7 Ly6G | Biolegend | 127617 AB_1877262 |
| PE Cy7 CD4 | Biolegend | 100421 AB_312706 |
| PE Cy7 ST2 | Biolegend | 146609 AB_2728178 |
| APC Ly6C | Biolegend | 128015 AB_1732087 |
| APC CD3 | Biolegend | 100235 AB_2561455 |
| APC Sca1 | Biolegend | 108111 AB_313348 |
| AF647 eFlour 640 MHC II | Biolegend | 107617 AB_493526 |
| AF700 CD11b | Biolegend | 101220 AB_493546 |
| APC-Cy7 CD45 | Biolegend | 103115 AB_312980 |
| Brilliant Violet 421 Rat IgG2b | Biolegend | 400639 AB_10895758 |
| Ultra-LEAF Purified Rat IgG2b | Biolegend | 400644 AB_11149687 |
| PE/Cy7 Rat IgG1 | Biolegend | 400416 AB_326522 |
| APC Rat IgG2a | Biolegend | 400512 AB_2814702 |
| PE/Cyanine5 Rat IgG2a | Biolegend | 400510 |
| PerCP/Cyanine5.5 Mouse IgG2b | Biolegend | 400338 |
| Brilliant Violet 421 FOXP3 | Biolegend | 126419 AB_2565933 |
| Brilliant Violet 785 CD8a | Biolegend | 100749 AB_2562610 |
| Brilliant Violet 421(TM) anti-human FOXP3 | Biolegend | 320124 AB_2565972 |
| APC/Cyanine7 anti-human CD45 | Biolegend | 304014 AB_314402 |
| APC anti-human CD3 | Biolegend | 317318 AB_1937212 |
| PE/Cyanine7 anti-human CD4 | Biolegend | 317414 AB_571959 |
| Brilliant Violet 785(TM) anti-human CD8 | Biolegend | 344740 AB_2566202 |
| PE anti-mouse/human KLRG1 (MAFA) | Biolegend | 138408 AB_10574313 |
| APC anti-human Lineage Cocktail (CD3, CD14, CD16, CD19, CD20, CD56) | Biolegend | 348803 |
| PE/Cyanine5 anti-human CD127 (IL-7Ralpha) | Biolegend | 351324 AB_10915554 |
| FITC anti-human CD25 | Biolegend | 302604 AB_314273 |
| Human TruStain FcX™ | Biolegend | 422302 AB_2818986 |
| Alexa Fluor(R) 488 anti-human FOXP3 | Biolegend | 320112 AB_430883 |
| PerCP/Cyanine5.5 anti-GATA3 | Biolegend | 653812 AB_2563219 |
| Mouse Anti-Human CD4 | Becton Dickinson | 560345 AB_1645572 |
| CD3 | Becton Dickinson | 560176 AB_1645475 |
| CD8 | Becton Dickinson | 560347 AB_1645581 |
| CD127 (IL-7 Receptor a subunit) | Becton Dickinson | 557938 AB_2296056 |
| Human ST2/IL-33R APC | R&D | FAB5231A |
| Brilliant Violet 510™ anti-human Lineage Cocktail (CD3, CD14, CD16, CD19, CD20, CD56) | Biolegend | 348807 |
| PE anti-human CD25 | Biolegend | 302606 AB_314276 |
| Bacterial and Virus Strains | ||
| Alternaria alternata | ATCC | ATCC 36376 |
| Malassezia globosa | ATCC | MYA-4889 |
| Alternaria alternata extract | Stallergenes Greer Lab | XPM1D3a.5 |
| Candida albicans extract | Stallergenes Greer Lab | XPLM73X1A2 |
| Aspergilus fumigatus extract | Stallergenes Greer Lab | XPM3D3A4 |
| Biological Samples | ||
| Human fresh PDAC samples | Roswell Park Cancer Center | Project approval number MOD00008843/BDR 135920 |
| Human PDAC TMA | MD Anderson Cancer Center | Project approval number LAB05-0854 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DiD solid | Fisher | D7757 |
| PD184352 (CI-1040) | Selleck Chem | S1020 |
| Trametinib (GSK1120212) | Selleck Chem | S2673 |
| ECL select western blot detection | Fisher | 45-000-999 |
| Chitinase from S. griseus | Sigma-Aldrich | 2593-74-2 |
| xylenes | Fisher | 1330-20-7 |
| hematoxylin | Fisher | 517-28-2 |
| tween 20 | Biorad | 9005-64-5 |
| trizol | Sigma-Aldrich | 136426-54-5 |
| chloroform | Sigma-Aldrich | 67-66-3 |
| proteinase K | Thermo Fisher Scientific | EO0491 |
| blotting grade buffer | Biorad | 999999-99-4 |
| 20X MOPS buffer | Fisher | 1132-61-2 |
| sodium n-dodecyl sulfate | Fisher | 151-21-3 |
| Triton-x 100 | Fisher | 9002-93-1 |
| Tween 40 | Fisher | 9005-66-7 |
| ox-bile | Sigma-Aldrich | 8008-63-7 |
| peptone | Sigma-Aldrich | 91079-38-8 |
| tris buffered saline tween 20 (TBST) | Corning | 7732-18-5 |
| western blot stripping buffer | Fisher | 21059 |
| formaldehyde solution | Fisher | 50-00-0 |
| 10% neutral buffered formalin | Fisher | 7558-80-7 |
| glucose solution | Corning | 50-99-7 |
| penicillin-streptomycin 100x | Corning | 63936-85-6 |
| glutamax 100x | Corning | 333-93-7 |
| bambanker freezing media | Corning | NC9582225 |
| MEM non essential amino acids (NEAA) | Corning | 70-47-3 |
| sodium pyruvate | Corning | 113-24-6 |
| fetal bovine syrum (FBS) | Fisher | 9014-81-7 |
| 0.25% trypsin | Corning | 2594-14-1 |
| l-glutamate 100x | Corning | 56-86-0 |
| collagenase IV | stem cell technologies | 07909 |
| dispase | stem cell technologies | 07923 |
| ficoll-paque plus | Fisher | 17-1440-02 |
| amphotericin b | Fisher | 1397-89-3 |
| doxycycline | Sigma-Aldrich | 564-25-0 |
| DNase I | Fisher | 9003-98-9 |
| puromycin | Fisher | 58-58-2 |
| buparlisib (BKM120) | Selleck Chem | 944396-07-0 |
| RPMI | Corning | 10-040-CV |
| DMEM | Corning | 10-017-CV |
| IL-7 | R&D | 407-ML-005 |
| IL-33 | Peprotech | 210-33 |
| IL-2 | Peprotech | 212-12 |
| Qiazol | Qiagen | 79306 |
| Critical Commercial Assays | ||
| LEGEND MAX™ Mouse IL-33 ELISA Kit | Biolegend | 436407 |
| Mouse IL-5 Quantikine ELISA Kit | R&D | M5000 |
| Mouse IL-13 Culture Media Quantikine ELISA | R&D | M1300CB |
| Cell fractionation kit | Abcam | ab109719 |
| Lipofectamine 3000 | Invitrogen | L3000015 |
| NextSeq 500 High Output Kit | Illumina | 20024906 |
| Mouse Tumor Dissociation kit | Miltenyi Biotec | 130-096-730 |
| Qubit Broad Range DNA kit | Thermofisher | Q32853 |
| MycoAlert Mycoplasma Detection Kit | Lonza | LT07-218 |
| DNeasy Powersoil Pro Kit | Qiagen | 47014 |
| DC Protein Assay Kit | Biorad | 5000111 |
| Nextera Index Kit | Illumina | FC-131-1024 |
| iTaq Universal SYBR Green Supermix | BioRad | 1725121 |
| iScript™ cDNA Synthesis Kit | Biorad | 1708891 |
| EasySep™ Mouse ILC2 Enrichment Kit | Stem Cell Technologies | 19842 |
| Cas9 Protein | Sigma-Aldrich | CAS9PROT-50UG |
| Deposited Data | ||
| UNITE’s Fungi taxon (v8.4) | https://unite.ut.ee/repository.php | N/A |
| The Human Protein Atlas | https://www.proteinatlas.org/ | N/A |
| Oncomine | https://www.oncomine.org/resource/login.html | N/A |
| RNAseq of mouse PDAC cell lines | This paper | GEO: GSE190468 |
| scRNAseq of CD45 cells | This paper | GEO: GSE190103 |
| 18s rRNA sequencing | This paper | BioSample accessions: SAMN23523449, SAMN23523450, SAMN23523451, SAMN23523452, SAMN23523453, SAMN23523454, SAMN23523455, SAMN23523456 |
| Experimental Models: Cell Lines | ||
| AK-B6 | From Ron DePinho | PMID: 32046984 |
| KPC1199 | From Michael Feigin | N/A |
| PJ-B6-4298 | This study | N/A |
| PJ-B6-4291 | This study | N/A |
| PJ-B6-4271 | This study | N/A |
| AK138 | From Ron DePinho | PMID: 32046984 |
| AK192 | From Ron DePinho | PMID: 32046984 |
| HY15549 | From Ron DePinho | PMID: 32046984 |
| HY19636 | From Ron DePinho | PMID: 32046984 |
| AK14838 | From Ron DePinho | PMID: 32046984 |
| HEK 293T | ATCC | CRL-3216 |
| Experimental Models: Organisms/Strains | ||
| Mouse:C57BL/6J | The Jackson Laboratory | 000644 IMSR_JAX:006362 |
| Mouse:B6.129S4-Krastm4Tyj/J | The Jackson Laboratory | 008179 IMSR_JAX:008179 |
| Mouse:B6.FVB-Tg(Pdx1-cre)6Tuv/J | The Jackson Laboratory | 014647 IMSR_JAX:014647 |
| Mouse:B6.129P2-Trp53tm1Brn/J | The Jackson Laboratory | 008462 IMSR_JAX:008462 |
| Mouse:TetO_Lox-Stop-Lox-KrasG12D (tetO_KrasG12D) | From Ron DePinho | PMID: 22541435 |
| Mouse:ROSA26-LSL-rtTA-IRES-GFP (ROSA_rtTA) | From Ron DePinho | PMID: 22541435 |
| 129S-Trp53tm2Tyj/J | The Jackson Laboratory | 008652 IMSR_JAX:008652 |
| Mouse:p48-Cre | From Ron DePinho | PubMed:17301087 |
| Oligonucleotides | ||
| See Supplemental Table S1 | N/A | N/A |
| Recombinant DNA | ||
| pLKO.1 | Addgene | 10878 A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE Cell. 2006 Mar 24. 124(6):1283-98. |
| psPAX2 | Addgene | 12260 psPAX2 was a gift from Didier Trono (Addgene plasmid # 12260 ; http://n2t.net/addgene:12260 ; RRID:Addgene_12260) |
| pMD2.G | Addgene | 12259 pMD2.G was a gift from Didier Trono (Addgene plasmid # 12259 ; http://n2t.net/addgene:12259 ; RRID:Addgene_12259) |
| Software and Algorithms | ||
| FlowJo | FlowJo, LLC | N/A |
| GraphPad Prism 8.4.3 | GraphPad Software | N/A |
| Biorad CFX Manager | BioRad | 184500 |
| Leica DM3000 LED | Leica Microsystems | 11888844 |
| Stelaris 5 Confocal Microscope | Leica Microsystems | N/A |
| Li-Cor Odyssey Fc | Li-Cor Biosciences | 2800 |
| CFX Connect Real-Time PCR Detection System | BioRad | 1855201 |
| IVIS Spectrum Imaging System | Perkin Elmer | N/A |
| ParaVision 3.0.2 | Paravision | N/A |
| Living Image 4.7 | Perkin Elmer | N/A |
| Illumina HiSeq2000 | Illumina | N/A |
| Agilent 2200 Tapestation | Agilent | N/A |
| BD LSR | Becton Dickinson | N/A |
| Aperio ScanScope XT | Aperio | N/A |
| Aperio ImageScope | Aperio | N/A |
| Olympus BX61 | Olympus | N/A |
| Tapestation 4200 D1000 | Agilent | N/A |
| MiSeq | Illumina | N/A |
| R Studio | R Studio | N/A |
| Other | ||
HIGHLIGHTS.
Oncogenic KRASG12D upregulates IL-33 to promote type 2 immunity in PDAC
Intratumoral mycobiome enhances cancer cell secretion of IL-33
Secreted IL-33 recruits and activates TH2 and ILC2 cells in the PDAC TME
Genetic deletion of IL-33 or anti-fungal treatment causes PDAC tumor regression
ACKNOWLEDGEMENTS
We thank the Roswell Park core facilities supported by National Cancer Institute (NCI) grant P30CA016056, including Genomic, Bioinformatics and Biostatistics, Data Bank and repository, Pathology Network and Flow and Imaging Cytometry shared resources. In addition, this study was supported by 5R00CA218891-04 (P.D.); 1R01CA262822-01 (P.D.), The Roswell Park Alliance Foundation-62-2839-01 (P.D.); R01CA188900 (B.I.S.); R01CA172105 (S.I.A.), P01CA234212 (P.K.), NCI P01 CA117969 (R.A.D.); NCI R01 CA225955 (R.A.D.); the Burkhart III Distinguished University Chair in Cancer Research Endowment (R.A.D.). We thank Dr. Michael Feigin for sharing the KPC1199 cell line. We also thank Dr. Shyamananda Singh Mayengbam for his technical support during revision. Graphical abstract was created with BioRender.com.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
P.D. and A.A. have a patent pending on targeting IL-33 and mycobiome in cancer. U.S. Provisional Patent Application Serial No. 63/238,531. R.A.D. is a co-founder, advisor, and/or director of Tvardi Therapeutics, Asylia Therapeutics, Stellanova Therapeutics, Nirogy Therapeutics and Sporos Bioventures. Tvardi and Nirogy are developing STAT3 and MCT inhibitors, respectively. A.M. receives royalties from Cosmos Wisdom Biotechnology and Thrive Earlier Detection, an Exact Sciences Company. A.M. is also a consultant for Freenome and Tezcat Biotechnology. The other authors declare no competing interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science.
REFERENCES
- ABARENKOV K, HENRIK NILSSON R, LARSSON KH, ALEXANDER IJ, EBERHARDT U, ERLAND S, HOILAND K, KJOLLER R, LARSSON E, PENNANEN T, SEN R, TAYLOR AF, TEDERSOO L, URSING BM, VRALSTAD T, LIIMATAINEN K, PEINTNER U & KOLJALG U 2010. The UNITE database for molecular identification of fungi--recent updates and future perspectives. New Phytol, 186, 281–5. [DOI] [PubMed] [Google Scholar]
- AGUIRRE AJ, BARDEESY N, SINHA M, LOPEZ L, TUVESON DA, HORNER J, REDSTON MS & DEPINHO RA 2003. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev, 17, 3112–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALAMRI A, NAM JY & BLANCATO JK 2017. Fluorescence In Situ Hybridization of Cells, Chromosomes, and Formalin-Fixed Paraffin-Embedded Tissues. Methods Mol Biol, 1606, 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALI S, HUBER M, KOLLEWE C, BISCHOFF SC, FALK W & MARTIN MU 2007. IL-1 receptor accessory protein is essential for IL-33-induced activation of T lymphocytes and mast cells. Proc Natl Acad Sci U S A, 104, 18660–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALONSO-CURBELO D, HO YJ, BURDZIAK C, MAAG JLV, MORRIS JPT, CHANDWANI R, CHEN HA, TSANOV KM, BARRIGA FM, LUAN W, TASDEMIR N, LIVSHITS G, AZIZI E, CHUN J, WILKINSON JE, MAZUTIS L, LEACH SD, KOCHE R, PE’ER D & LOWE SW 2021. A gene-environment-induced epigenetic program initiates tumorigenesis. Nature, 590, 642–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ANDERS S, PYL PT & HUBER W 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics, 31, 166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ARAN D, LOONEY AP, LIU L, WU E, FONG V, HSU A, CHAK S, NAIKAWADI RP, WOLTERS PJ, ABATE AR, BUTTE AJ & BHATTACHARYA M 2019. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol, 20, 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AYKUT B, PUSHALKAR S, CHEN R, LI Q, ABENGOZAR R, KIM JI, SHADALOEY SA, WU D, PREISS P, VERMA N, GUO Y, SAXENA A, VARDHAN M, DISKIN B, WANG W, LEINWAND J, KURZ E, KOCHEN ROSSI JA, HUNDEYIN M, ZAMBRINIS C, LI X, SAXENA D & MILLER G 2019. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature, 574, 264–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BONILLA WV, FROHLICH A, SENN K, KALLERT S, FERNANDEZ M, JOHNSON S, KREUTZFELDT M, HEGAZY AN, SCHRICK C, FALLON PG, KLEMENZ R, NAKAE S, ADLER H, MERKLER D, LOHNING M & PINSCHEWER DD 2012. The alarmin interleukin-33 drives protective antiviral CD8(+) T cell responses. Science, 335, 984–9. [DOI] [PubMed] [Google Scholar]
- CAPORASO JG, KUCZYNSKI J, STOMBAUGH J, BITTINGER K, BUSHMAN FD, COSTELLO EK, FIERER N, PENA AG, GOODRICH JK, GORDON JI, HUTTLEY GA, KELLEY ST, KNIGHTS D, KOENIG JE, LEY RE, LOZUPONE CA, MCDONALD D, MUEGGE BD, PIRRUNG M, REEDER J, SEVINSKY JR, TURNBAUGH PJ, WALTERS WA, WIDMANN J, YATSUNENKO T, ZANEVELD J & KNIGHT R 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods, 7, 335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLARK CE, HINGORANI SR, MICK R, COMBS C, TUVESON DA & VONDERHEIDE RH 2007. Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res, 67, 9518–27. [DOI] [PubMed] [Google Scholar]
- CUI G, YUAN A, PANG Z, ZHENG W, LI Z & GOLL R 2018. Contribution of IL-33 to the Pathogenesis of Colorectal Cancer. Front Oncol, 8, 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE MONTE L, RENI M, TASSI E, CLAVENNA D, PAPA I, RECALDE H, BRAGA M, DI CARLO V, DOGLIONI C & PROTTI MP 2011. Intratumor T helper type 2 cell infiltrate correlates with cancer-associated fibroblast thymic stromal lymphopoietin production and reduced survival in pancreatic cancer. J Exp Med, 208, 469–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEY P, BADDOUR J, MULLER F, WU CC, WANG H, LIAO WT, LAN Z, CHEN A, GUTSCHNER T, KANG Y, FLEMING J, SATANI N, ZHAO D, ACHREJA A, YANG L, LEE J, CHANG E, GENOVESE G, VIALE A, YING H, DRAETTA G, MAITRA A, WANG YA, NAGRATH D & DEPINHO RA 2017. Genomic deletion of malic enzyme 2 confers collateral lethality in pancreatic cancer. Nature, 542, 119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEY P, JONSSON P, HARTMAN J, WILLIAMS C, STROM A & GUSTAFSSON JA 2012. Estrogen receptors beta1 and beta2 have opposing roles in regulating proliferation and bone metastasis genes in the prostate cancer cell line PC3. Mol Endocrinol, 26, 1991–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEY P, LI J, ZHANG J, CHAURASIYA S, STROM A, WANG H, LIAO WT, CAVALLARO F, DENZ P, BERNARD V, YEN EY, GENOVESE G, GULHATI P, LIU J, CHAKRAVARTI D, DENG P, ZHANG T, CARBONE F, CHANG Q, YING H, SHANG X, SPRING DJ, GHOSH B, PUTLURI N, MAITRA A, WANG YA & DEPINHO RA 2020. Oncogenic KRAS-Driven Metabolic Reprogramming in Pancreatic Cancer Cells Utilizes Cytokines from the Tumor Microenvironment. Cancer Discov, 10, 608–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEY P, STROM A & GUSTAFSSON JA 2014. Estrogen receptor beta upregulates FOXO3a and causes induction of apoptosis through PUMA in prostate cancer. Oncogene, 33, 4213–25. [DOI] [PubMed] [Google Scholar]
- DOBIN A, DAVIS CA, SCHLESINGER F, DRENKOW J, ZALESKI C, JHA S, BATUT P, CHAISSON M & GINGERAS TR 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ERCOLANO G, FALQUET M, VANONI G, TRABANELLI S & JANDUS C 2019. ILC2s: New Actors in Tumor Immunity. Front Immunol, 10, 2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FEIG C, GOPINATHAN A, NEESSE A, CHAN DS, COOK N & TUVESON DA 2012. The pancreas cancer microenvironment. Clin Cancer Res, 18, 4266–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FLAMAR AL, KLOSE CSN, MOELLER JB, MAHLAKOIV T, BESSMAN NJ, ZHANG W, MORIYAMA S, STOKIC-TRTICA V, RANKIN LC, PUTZEL GG, RODEWALD HR, HE Z, CHEN L, LIRA SA, KARSENTY G & ARTIS D 2020. Interleukin-33 Induces the Enzyme Tryptophan Hydroxylase 1 to Promote Inflammatory Group 2 Innate Lymphoid Cell-Mediated Immunity. Immunity, 52, 606–619 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FOURNIE JJ & POUPOT M 2018. The Pro-tumorigenic IL-33 Involved in Antitumor Immunity: A Yin and Yang Cytokine. Front Immunol, 9, 2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOPALAKRISHNAN V, SPENCER CN, NEZI L, REUBEN A, ANDREWS MC, KARPINETS TV, PRIETO PA, VICENTE D, HOFFMAN K, WEI SC, COGDILL AP, ZHAO L, HUDGENS CW, HUTCHINSON DS, MANZO T, PETACCIA DE MACEDO M, COTECHINI T, KUMAR T, CHEN WS, REDDY SM, SZCZEPANIAK SLOANE R, GALLOWAY-PENA J, JIANG H, CHEN PL, SHPALL EJ, REZVANI K, ALOUSI AM, CHEMALY RF, SHELBURNE S, VENCE LM, OKHUYSEN PC, JENSEN VB, SWENNES AG, MCALLISTER F, MARCELO RIQUELME SANCHEZ E, ZHANG Y, LE CHATELIER E, ZITVOGEL L, PONS N, AUSTIN-BRENEMAN JL, HAYDU LE, BURTON EM, GARDNER JM, SIRMANS E, HU J, LAZAR AJ, TSUJIKAWA T, DIAB A, TAWBI H, GLITZA IC, HWU WJ, PATEL SP, WOODMAN SE, AMARIA RN, DAVIES MA, GERSHENWALD JE, HWU P, LEE JE, ZHANG J, COUSSENS LM, COOPER ZA, FUTREAL PA, DANIEL CR, AJAMI NJ, PETROSINO JF, TETZLAFF MT, SHARMA P, ALLISON JP, JENQ RR & WARGO JA 2018. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science, 359, 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAO Y, HAO S, ANDERSEN-NISSEN E, MAUCK WM 3RD, ZHENG S, BUTLER A, LEE MJ, WILK AJ, DARBY C, ZAGER M, HOFFMAN P, STOECKIUS M, PAPALEXI E, MIMITOU EP, JAIN J, SRIVASTAVA A, STUART T, FLEMING LM, YEUNG B, ROGERS AJ, MCELRATH JM, BLISH CA, GOTTARDO R, SMIBERT P & SATIJA R 2021. Integrated analysis of multimodal single-cell data. Cell, 184, 3573–3587 e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HENTSCHKE I, GRASER A, MELICHAR VO, KIEFER A, ZIMMERMANN T, KROSS B, HAAG P, XEPAPADAKI P, PAPADOPOULOS NG, BOGDAN C & FINOTTO S 2017. IL-33/ST2 immune responses to respiratory bacteria in pediatric asthma. Sci Rep, 7, 43426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HERNANZ-FALCON P, JOFFRE O, WILLIAMS DL & REIS E SOUSA C 2009. Internalization of Dectin-1 terminates induction of inflammatory responses. Eur J Immunol, 39, 507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HINGORANI SR, PETRICOIN EF, MAITRA A, RAJAPAKSE V, KING C, JACOBETZ MA, ROSS S, CONRADS TP, VEENSTRA TD, HITT BA, KAWAGUCHI Y, JOHANN D, LIOTTA LA, CRAWFORD HC, PUTT ME, JACKS T, WRIGHT CV, HRUBAN RH, LOWY AM & TUVESON DA 2003. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell, 4, 437–50. [DOI] [PubMed] [Google Scholar]
- JOVANOVIC I, RADOSAVLJEVIC G, MITROVIC M, JURANIC VL, MCKENZIE AN, ARSENIJEVIC N, JONJIC S & LUKIC ML 2011. ST2 deletion enhances innate and acquired immunity to murine mammary carcinoma. Eur J Immunol, 41, 1902–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOLJALG U, NILSSON RH, ABARENKOV K, TEDERSOO L, TAYLOR AF, BAHRAM M, BATES ST, BRUNS TD, BENGTSSON-PALME J, CALLAGHAN TM, DOUGLAS B, DRENKHAN T, EBERHARDT U, DUENAS M, GREBENC T, GRIFFITH GW, HARTMANN M, KIRK PM, KOHOUT P, LARSSON E, LINDAHL BD, LUCKING R, MARTIN MP, MATHENY PB, NGUYEN NH, NISKANEN T, OJA J, PEAY KG, PEINTNER U, PETERSON M, POLDMAA K, SAAG L, SAAR I, SCHUSSLER A, SCOTT JA, SENES C, SMITH ME, SUIJA A, TAYLOR DL, TELLERIA MT, WEISS M & LARSSON KH 2013. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol, 22, 5271–7. [DOI] [PubMed] [Google Scholar]
- LI A, HERBST RH, CANNER D, SCHENKEL JM, SMITH OC, KIM JY, HILLMAN M, BHUTKAR A, CUOCO MS, RAPPAZZO CG, ROGERS P, DANG C, JERBY-ARNON L, ROZENBLATT-ROSEN O, CONG L, BIRNBAUM M, REGEV A & JACKS T 2019. IL-33 Signaling Alters Regulatory T Cell Diversity in Support of Tumor Development. Cell Rep, 29, 2998–3008 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIEW FY, GIRARD JP & TURNQUIST HR 2016. Interleukin-33 in health and disease. Nat Rev Immunol, 16, 676–689. [DOI] [PubMed] [Google Scholar]
- LINGEL A, WEISS TM, NIEBUHR M, PAN B, APPLETON BA, WIESMANN C, BAZAN JF & FAIRBROTHER WJ 2009. Structure of IL-33 and its interaction with the ST2 and IL-1RAcP receptors--insight into heterotrimeric IL-1 signaling complexes. Structure, 17, 1398–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATSON V, FESSLER J, BAO R, CHONGSUWAT T, ZHA Y, ALEGRE ML, LUKE JJ & GAJEWSKI TF 2018. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science, 359, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCMURDIE PJ & HOLMES S 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One, 8, e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MILLER AM 2011. Role of IL-33 in inflammation and disease. J Inflamm (Lond), 8, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORAL JA, LEUNG J, ROJAS LA, RUAN J, ZHAO J, SETHNA Z, RAMNARAIN A, GASMI B, GURURAJAN M, REDMOND D, ASKAN G, BHANOT U, ELYADA E, PARK Y, TUVESON DA, GONEN M, LEACH SD, WOLCHOK JD, DEMATTEO RP, MERGHOUB T & BALACHANDRAN VP 2020. ILC2s amplify PD-1 blockade by activating tissue-specific cancer immunity. Nature, 579, 130–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OKSANEN J, KINDT R, LEGENDRE P, O’HARA B, STEVENS MHH, OKSANEN MJ AND SUGGESTS MASS 2007. The vegan package. Community ecology package. 719. [Google Scholar]
- OLIVE KP, TUVESON DA, RUHE ZC, YIN B, WILLIS NA, BRONSON RT, CROWLEY D & JACKS T 2004. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell, 119, 847–60. [DOI] [PubMed] [Google Scholar]
- PARK JH, AMERI AH, DEMPSEY KE, CONRAD DN, KEM M, MINO-KENUDSON M & DEMEHRI S 2021. Nuclear IL-33/SMAD signaling axis promotes cancer development in chronic inflammation. EMBO J, 40, e106151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PASTILLE E, WASMER MH, ADAMCZYK A, VU VP, MAGER LF, PHUONG NNT, PALMIERI V, SIMILLION C, HANSEN W, KASPER S, SCHULER M, MUGGLI B, MCCOY KD, BUER J, ZLOBEC I, WESTENDORF AM & KREBS P 2019. The IL-33/ST2 pathway shapes the regulatory T cell phenotype to promote intestinal cancer. Mucosal Immunol, 12, 990–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PIRO G, SIMIONATO F, CARBONE C, FRIZZIERO M, MALLEO G, ZANINI S, CASOLINO R, SANTORO R, MINA MM, ZECCHETTO C, MERZ V, SCARPA A, BASSI C, TORTORA G & MELISI D 2017. A circulating TH2 cytokines profile predicts survival in patients with resectable pancreatic adenocarcinoma. Oncoimmunology, 6, e1322242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RANK MA, KOBAYASHI T, KOZAKI H, BARTEMES KR, SQUILLACE DL & KITA H 2009. IL-33-activated dendritic cells induce an atypical TH2-type response. J Allergy Clin Immunol, 123, 1047–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RIQUELME E, ZHANG Y, ZHANG L, MONTIEL M, ZOLTAN M, DONG W, QUESADA P, SAHIN I, CHANDRA V, SAN LUCAS A, SCHEET P, XU H, HANASH SM, FENG L, BURKS JK, DO KA, PETERSON CB, NEJMAN D, TZENG CD, KIM MP, SEARS CL, AJAMI N, PETROSINO J, WOOD LD, MAITRA A, STRAUSSMAN R, KATZ M, WHITE JR, JENQ R, WARGO J & MCALLISTER F 2019. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell, 178, 795–806 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBINETTE ML, FUCHS A, CORTEZ VS, LEE JS, WANG Y, DURUM SK, GILFILLAN S, COLONNA M & IMMUNOLOGICAL GENOME C 2015. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol, 16, 306–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBINSON MD, MCCARTHY DJ & SMYTH GK 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROGNES T, FLOURI T, NICHOLS B, QUINCE C & MAHE F 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ, 4, e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROSAS M, LIDDIARD K, KIMBERG M, FARO-TRINDADE I, MCDONALD JU, WILLIAMS DL, BROWN GD & TAYLOR PR 2008. The induction of inflammation by dectin-1 in vivo is dependent on myeloid cell programming and the progression of phagocytosis. J Immunol, 181, 3549–57. [DOI] [PubMed] [Google Scholar]
- SCHMITZ J, OWYANG A, OLDHAM E, SONG Y, MURPHY E, MCCLANAHAN TK, ZURAWSKI G, MOSHREFI M, QIN J, LI X, GORMAN DM, BAZAN JF & KASTELEIN RA 2005. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity, 23, 479–90. [DOI] [PubMed] [Google Scholar]
- SNELGROVE RJ, GREGORY LG, PEIRO T, AKTHAR S, CAMPBELL GA, WALKER SA & LLOYD CM 2014. Alternaria-derived serine protease activity drives IL-33-mediated asthma exacerbations. J Allergy Clin Immunol, 134, 583–592 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEELE CW, JAMIESON NB, EVANS TR, MCKAY CJ, SANSOM OJ, MORTON JP & CARTER CR 2013. Exploiting inflammation for therapeutic gain in pancreatic cancer. Br J Cancer, 108, 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TANIGUCHI S, ELHANCE A, VAN DUZER A, KUMAR S, LEITENBERGER JJ & OSHIMORI N 2020. Tumor-initiating cells establish an IL-33-TGF-beta niche signaling loop to promote cancer progression. Science, 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TIBBITT CA, STARK JM, MARTENS L, MA J, MOLD JE, DESWARTE K, OLIYNYK G, FENG X, LAMBRECHT BN, DE BLESER P, NYLEN S, HAMMAD H, ARSENIAN HENRIKSSON M, SAEYS Y & COQUET JM 2019. Single-Cell RNA Sequencing of the T Helper Cell Response to House Dust Mites Defines a Distinct Gene Expression Signature in Airway Th2 Cells. Immunity, 51, 169–184 e5. [DOI] [PubMed] [Google Scholar]
- UCHIDA M, ANDERSON EL, SQUILLACE DL, PATIL N, MANIAK PJ, IIJIMA K, KITA H & O’GRADY SM 2017. Oxidative stress serves as a key checkpoint for IL-33 release by airway epithelium. Allergy, 72, 1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UHLEN M, FAGERBERG L, HALLSTROM BM, LINDSKOG C, OKSVOLD P, MARDINOGLU A, SIVERTSSON A, KAMPF C, SJOSTEDT E, ASPLUND A, OLSSON I, EDLUND K, LUNDBERG E, NAVANI S, SZIGYARTO CA, ODEBERG J, DJUREINOVIC D, TAKANEN JO, HOBER S, ALM T, EDQVIST PH, BERLING H, TEGEL H, MULDER J, ROCKBERG J, NILSSON P, SCHWENK JM, HAMSTEN M, VON FEILITZEN K, FORSBERG M, PERSSON L, JOHANSSON F, ZWAHLEN M, VON HEIJNE G, NIELSEN J & PONTEN F 2015. Proteomics. Tissue-based map of the human proteome. Science, 347, 1260419. [DOI] [PubMed] [Google Scholar]
- UHLEN M, ZHANG C, LEE S, SJOSTEDT E, FAGERBERG L, BIDKHORI G, BENFEITAS R, ARIF M, LIU Z, EDFORS F, SANLI K, VON FEILITZEN K, OKSVOLD P, LUNDBERG E, HOBER S, NILSSON P, MATTSSON J, SCHWENK JM, BRUNNSTROM H, GLIMELIUS B, SJOBLOM T, EDQVIST PH, DJUREINOVIC D, MICKE P, LINDSKOG C, MARDINOGLU A & PONTEN F 2017. A pathology atlas of the human cancer transcriptome. Science, 357. [DOI] [PubMed] [Google Scholar]
- VAN GOOL F, MOLOFSKY AB, MORAR MM, ROSENZWAJG M, LIANG HE, KLATZMANN D, LOCKSLEY RM & BLUESTONE JA 2014. Interleukin-5-producing group 2 innate lymphoid cells control eosinophilia induced by interleukin-2 therapy. Blood, 124, 3572–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VENMAR KT, KIMMEL DW, CLIFFEL DE & FINGLETON B 2015. IL4 receptor alpha mediates enhanced glucose and glutamine metabolism to support breast cancer growth. Biochim Biophys Acta, 1853, 1219–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VIVIER E, ARTIS D, COLONNA M, DIEFENBACH A, DI SANTO JP, EBERL G, KOYASU S, LOCKSLEY RM, MCKENZIE ANJ, MEBIUS RE, POWRIE F & SPITS H 2018. Innate Lymphoid Cells: 10 Years On. Cell, 174, 1054–1066. [DOI] [PubMed] [Google Scholar]
- WANG K, SHAN S, YANG Z, GU X, WANG Y, WANG C & REN T 2017. IL-33 blockade suppresses tumor growth of human lung cancer through direct and indirect pathways in a preclinical model. Oncotarget, 8, 68571–68582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATTERSON WJ, TANYERI M, WATSON AR, CHAM CM, SHAN Y, CHANG EB, EREN AM & TAY S 2020. Droplet-based high-throughput cultivation for accurate screening of antibiotic resistant gut microbes. Elife, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YING H, DEY P, YAO W, KIMMELMAN AC, DRAETTA GF, MAITRA A & DEPINHO RA 2016. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev, 30, 355–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YING H, KIMMELMAN AC, LYSSIOTIS CA, HUA S, CHU GC, FLETCHERSANANIKONE E, LOCASALE JW, SON J, ZHANG H, COLOFF JL, YAN H, WANG W, CHEN S, VIALE A, ZHENG H, PAIK JH, LIM C, GUIMARAES AR, MARTIN ES, CHANG J, HEZEL AF, PERRY SR, HU J, GAN B, XIAO Y, ASARA JM, WEISSLEDER R, WANG YA, CHIN L, CANTLEY LC & DEPINHO RA 2012. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell, 149, 656–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZERVOS EE, OSBORNE D, BOE BA, LUZARDO G, GOLDIN SB & ROSEMURGY AS 2006. Prognostic significance of new onset ascites in patients with pancreatic cancer. World J Surg Oncol, 4, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHENG GX, TERRY JM, BELGRADER P, RYVKIN P, BENT ZW, WILSON R, ZIRALDO SB, WHEELER TD, MCDERMOTT GP, ZHU J, GREGORY MT, SHUGA J, MONTESCLAROS L, UNDERWOOD JG, MASQUELIER DA, NISHIMURA SY, SCHNALL-LEVIN M, WYATT PW, HINDSON CM, BHARADWAJ R, WONG A, NESS KD, BEPPU LW, DEEG HJ, MCFARLAND C, LOEB KR, VALENTE WJ, ERICSON NG, STEVENS EA, RADICH JP, MIKKELSEN TS, HINDSON BJ & BIELAS JH 2017. Massively parallel digital transcriptional profiling of single cells. Nat Commun, 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHU Y, HERNDON JM, SOJKA DK, KIM KW, KNOLHOFF BL, ZUO C, CULLINAN DR, LUO J, BEARDEN AR, LAVINE KJ, YOKOYAMA WM, HAWKINS WG, FIELDS RC, RANDOLPH GJ & DENARDO DG 2017. Tissue-Resident Macrophages in Pancreatic Ductal Adenocarcinoma Originate from Embryonic Hematopoiesis and Promote Tumor Progression. Immunity, 47, 323–338 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Processed single-cell RNA-seq and bulk RNA-seq data of this study can be obtained from Gene Expression Omnibus (GEO) with an accession number of GSE190103 and GSE190468, respectively. 18s rRNA seq data can be obtained from BioSample accessions: SAMN23523449, SAMN23523450, SAMN23523451, SAMN23523452, SAMN23523453, SAMN23523454, SAMN23523455, SAMN23523456.
All other data are available from the corresponding authors upon request.


