Abstract
Selenicereus is a genus of perennial shrub from the family Cactaceae, and some of them play an important role in the food industry, pharmaceuticals, cosmetics and medicine. To date, there are few reports on Selenicereus plastomes, which limits our understanding of this genus. Here, we have reported the complete plastomes of four Selenicereus species (S. monacanthus, S. annthonyanus, S. grandifloras, and S. validus) and carried out a comprehensive comparative analysis. All four Selenicereus plastomes have a typical quartile structure. The plastome size ranged from 133,146 to 134,450 bp, and contained 104 unique genes, including 30 tRNA genes, 4 rRNA genes and 70 protein-coding genes. Comparative analysis showed that there were massive losses of ndh genes in Selenicereus. Besides, we observed the inverted repeat regions had undergone a dramatic expansion and formed a previously unreported small single copy/inverted repeat border in the intron region of the atpF gene. Furthermore, we identified 6 hypervariable regions (trnF-GAA-rbcL, ycf1, accD, clpP-trnS-GCU, clpP-trnT-CGU and rpl22-rps19) that could be used as potential DNA barcodes for the identification of Selenicereus species. Our study enriches the plastome in the family Cactaceae, and provides the basis for the reconstruction of phylogenetic relationships.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12298-021-01121-z.
Keywords: Plastome, Hylocereus, Selenicereus, Gene, Phylogenomics
Introduction
Hylocereus species are perennial herbs from the family Cactaceae. The species in this genus are native to Central America, and nearly 20 species of Hylocereus are recognized by most researchers and they can be found naturally occurring from Southern Mexico to throughout Central America and even Northern South America (Nunes et al. 2014). All Hylocereus species have varying edible fruits and are commercially developed in different ways. Although the white pitaya (H. undatus) is the primary species found in grocery stores and street markets, red fleshed dragon fruit has gained more popularity. The red fleshed pitaya (Selenicereus monacanthus (Lem.) D.R.Hunt), formerly known as H. lemairei, not only has an attractive red–purple appearance and unique taste, due to its rich content of high-value functional compounds(Zhuang et al. 2012), it is also widely used in pharmaceutical, cosmetic and medical applications. For example, the pulp of red fleshed pitaya is rich in β-carotene and anthocyanin, which can effectively prevent and treat some chronic diseases (especially cancer) (Bai and Zhang 2017; Guimaraes et al. 2017; Villalobos et al. 2012).
The specific definitions of Hylocereus and Selenicereus have always been controversial (Cálix de Dios 2009). Britton and Rose (1963) divided Selenicereus and Hylocereus into different genera from morphology. However, based on many plastids and nuclear DNA sequences, morphology and anatomical data, it was proved that the two genera were not separated, and Hylocereus was nested in Selenicereus (Arias et al. 2005; Gómez-Hinostrosa et al. 2014; Plume et al. 2013; Miguel Ángel et al. 2016). Different perspectives on classification standards and limited genomic information further complicated the taxonomic definition of this genus. Therefore, it is important to explore the phylogenetic relationship of the Selenicereus species based on genomics. There are few studies on the phylogenetic relationship between Hylocereus and Selenicereus based on the complete plastomes (Korotkova et al. 2017).
Organelle genome sequencing is essential for understanding the phylogenetic relationship between closely related species. (Ivanova et al. 2017). Chloroplast is an essential organelle in plants with a semi-autonomous genetic system. Its genome is called plastid genome or plastome (Palmer et al. 1985). Most plastomes in angiosperms are a typical quadripartite structure (Palmer 1985), consisting of two inverted repeats (IRa and IRb) and two single copy regions (LSC and SSC) (Yang et al. 2016), and the size of the plastome ranges from 72 to 220 kb (Pervaiz et al. 2015), including about 110–130 unique genes, many are involved in photosynthesis (Choi et al. 2015). Plastomes have been widely used in taxonomic and evolutionary studies (Daniell et al. 2016) due to their small size, simple structure and maternal inheritance (Maliga 2002; Palmer et al. 1988). Entire plastome and nuclear DNA clusters are important in distinguishing between closely related or recessive species (Krawczyk et al. 2018; Yang et al. 2013; Myszczyński et al. 2017). Besides, although the plastomes are generally conserved in terms of sequence differences and structural organization, some non-coding regions may experience an unexpectedly high frequency of nucleotide substitutions, and these hypervariable regions could be used as DNA barcodes for species identification (Dong et al. 2012).
In this study, we sequenced, assembled and annotated the plastomes of four Selenicereus species, including the red-fleshed pitaya (S. monacanthus, formerly classified as Hylocereus) and three traditional Selenicereus species (S. annthonyanus, S. grandifloras and S. validus). Our main tasks were as follows: (1) To provide four high-quality reference Selenicereus plastomes; (2) To analyze the structural characteristics and sequence divergence of the plastomes in Selenicereus; (3) To identify simple sequence repeats (SSRs) loci and repeat sequences for further studies on population genetic structure; (4) To understand the phylogenetic relationships of Selenicereus in Cactaceae based on the complete plastome sequences; and (5) To identify the hypervariable regions that could be used as DNA barcodes for commercial identification of pitaya varieties.
Materials and methods
Sampling, DNA extraction and sequencing
Fresh stems of the red-fleshed pitaya (S. monacanthus) were collected from Yulin, Guangxi, China (22°94' N, 110°49' E). The fresh stems of the other three analyzed Selenicereus species were collected from the local flower market of Beibei, Chongqing, China (29°81' N, 106°40' E). S. monacanthus, S. annthonyanus, S. grandifloras, and S. validus were identified by Professor Jie Yu based on morphological characteristics and related DNA barcoding. These species were cultivated for edible use or ornamental plants, and no permission is required to collect these samples. Our experimental work, including the collection of plant materials, complies with institutional, national or international guidelines. All the samples were deposited in the Herbarium of Southwest University, Chongqing, China (voucher code: YJ-swu002, YJ-swu027 ~ YJ-swu029). Total genomic DNA was extracted by using the CTAB method (Arseneau et al. 2017). The DNA library with an insert size of 350 bp was constructed using a NEBNext® library construction kit and sequenced by using the HiSeq Xten PE150 sequencing platform. Sequencing produced a total of 6.04–6.85 Gb of raw data per species. Clean data were obtained by using Trimmomatic (Bolger et al. 2014): by removing the low-quality sequences with more than 5% bases being “N”, and a quality value of Q < 19 accounted for more than 50% of the total base. The detailed sequencing data were shown in Table S6.
Genome assembling and annotation
The plastome assembly from the clean data was accomplished utilizing GetOrganelle (v1.7.3) with a default setting. The correctness of the assembly was confirmed by using Bowtie2 (v2. 0.1) (Langmead et al. 2009) to manually edit and map all the raw reads to the assembled genome sequence under the default settings. Detailed assembly information was shown in Table S1. The plastomes were initially annotated by using GeSeq (Tillich et al. 2017) with two reference genomes (Carnegiea gigantea, GenBank: NC_027618.1 and Lophocereus schottii, GenBank: NC_041727.1). Subsequently, the annotations with problems were manually edited by using Apollo (Misra and Harris 2005), and genome maps were drawn by OGDRAW (Greiner et al. 2019).
Repeats and SSR analysis
The GC content was determined by using the cusp program provided by EMBOSS (v6.3.1) (Rice et al. 2000). Simple Repeat Sequences (SSRs) were available through the online site MISA (https://webblast.ipk-gatersleben.de/misa/). Additionally, REPuter (https://bibiserv.cebitec.uni-bielefeld.de/reputer/) was used to calculate palindromic repeats, forward repeats, reverse repeats, and complementary repeats with the following settings: hamming distance of three and minimal repeat size of 30 bp (Kurtz et al. 2001).
Sequence divergence analysis
The BLASTn (Chen et al. 2015) program was used to search for the homologous sequences of ndh, rpoC1 and rpl2 genes among these plastomes, including: the four Selenicereus species, Opuntia quimilo (MN114084.1) and Portulaca oleracea (NC_036236.1). The parameters were as follows: -evalue 1e-5, -word_size 9, gapopen 5, gapextend 2, reward 2, penalty 3, and dust no. The BLASTn results were visualized on TBtools (Chen et al. 2020). Furthermore, the sequence similarity analysis results of the four plastomes we analyzed were obtained in shuffle-LAGAN mode by using the online site mVISTA (http://genome.lbl.gov/cgi-bin/VistaInput?num_seqs=4). With the help of PhyloSuite (v1.2.1), we extracted the orthologous genes of the four taxa and aligned the sequences by using the plugin MAFFT (v7.313) embedded in PhyloSuite. The percentage of variable sites was calculated based on the comparison of protein-coding genes by MEGA (v6.0) (Tamura et al. 2013). A sliding window with both window length and step size of 500 bp was set using DnaSP (v6.0) software to obtain nucleotide polymorphisms (Pi) of four plastomes. IRscope was used for visualizing the IR/SC boundaries (https://irscope.shinyapps.io/irapp/) and the adjacent genes.
Phylogenetic analysis
The data sources for phylogenetic analysis were shown in Table S2. A total of 56 orthologous genes among the analyzed plastomes were identified and extracted by using PhyloSuite (v1.2.1) (Zhang et al. 2020). The 56 shared plastid protein-coding genes includes atpA, atpB, atpE, atpF, atpH, atpI, ccsA, cemA, clpP, infA, matK, petA, petB, petD, petG, petL, petN, psaA, psaB, psaC, psaI, psaJ, psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbM, psbN, psbT, rbcL, rpl14, rpl16, rpl20, rpl22, rpl2, rpoA, rpoB, rpoC1, rpoC2, rps11, rps12, rps14, rps15, rps19, rps2, rps3, rps4, rps7, rps8 and ycf3. The corresponding nucleotide sequences were aligned by using MAFFT (v7.450) (Rozewicki et al. 2019) implemented in PhyloSuite. These aligned nucleotide sequences were concatenated, and used to construct the phylogenetic trees by using the maximum likelihood (ML) method implemented in RAxML (v8.2.4). The parameters were “raxmlHPC-PTHREADS-SSE3 -f a -N 1000 -m GTRGAMMA—× 551,314,260 -p 551,314,260”. The bootstrap analysis was performed with 1,000 replicates. Bayesian inferences (BI) analysis was performed in MrBayes (v3.2.6) using the Markov Chain Monte Carlo method with 200,000 generations and sampling trees every 100 generations. The first 20% of trees were discarded as burn-in with the remaining trees being used for generating a consensus tree.
Results
Overall organization and features of the four plastomes
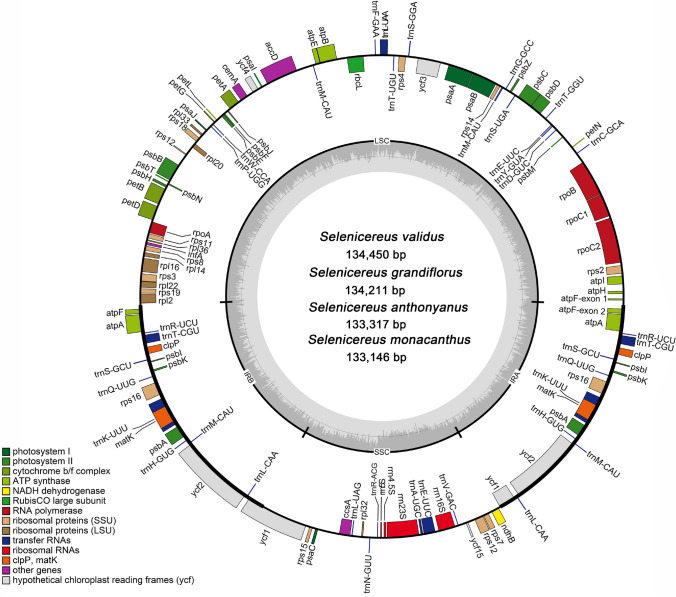
The plastome size of these four taxa ranged from 133,146 bp (S. monacanthus) to 134,450 bp (S. validus). They were typical quadripartite structure, consisting of a large single-copy region (LSC, 68,076—68,877 bp), a small single-copy region (SSC, 21,716—22,023 bp), and a pair of inverted repeat region (IRs, 21,674—21,775 bp). Figure 1 showed the plastid genome map. In addition to the differences in length, the GC content of these conserved plastomes also showed slight changes. According to the analysis, the GC content of the four plastome ranged from 36.29 to 36.43%, and the GC content in SSC region (39.39–39.69%) was significantly higher compared to LSC region (36.22%—36.36%) and IR region (34.83%—34.98%) (Table 1).
Fig. 1.
Plastid genome map of Selenicereus species and corresponding pictures of four plants. The thick line spacing in the inner circle represents a conservative quaternary structure, with LSC region, SSC region and a pair of IR region, and the dark gray area and light gray parts inside represent the ratio of GC and AT content, respectively
Table 1.
Plastome features of the four Selenicereus species
| Species | S. monacanthus | S. anthonyanus | S. grandiflorus | S. validus |
|---|---|---|---|---|
| Accession number | MW553055 | MW553068 | MW553069 | MW553070 |
| Length (bp) | ||||
| Total length | 133,146 | 133,317 | 134,211 | 134,450 |
| LSC | 68,076 | 68,203 | 68,839 | 68,877 |
| SSC | 21,716 | 21,766 | 22,014 | 22,023 |
| IR | 21,677 | 21,674 | 21,679 | 21,775 |
| GC content (%) | ||||
| Total GC content | 36.40 | 36.43 | 36.34 | 36.29 |
| LSC | 36.25 | 36.36 | 36.24 | 36.22 |
| SSC | 39.69 | 39.54 | 39.40 | 39.39 |
| IR | 34.98 | 34.98 | 34.95 | 34.83 |
| Gene numbers | ||||
| Total number of genes | 104 | 104 | 104 | 104 |
| tRNA | 30 | 30 | 30 | 30 |
| rRNA | 4 | 4 | 4 | 4 |
| Protein-coding | 70 | 70 | 70 | 70 |
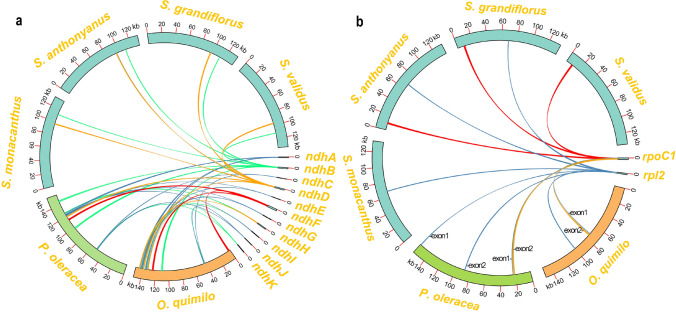
Like previous reports in cacti plastomes, the genome annotation results showed that the 11 ndh genes in the analyzed plastome were partially lost, including ndhA, ndhC, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, and ndhK. However, all these genes existed in the plastome of Opuntia Quimilo and Portulaca Oleracea (Köhler et al. 2020; Liu et al. 2018). We used all 11 ndh genes of O. Quimilo as query sequences to search for homologous sequences in the four Selenicereus plastomes based on the BLASTn program. The results confirmed the absence of most ndh genes (Fig. 2a). The second exon of ndhB gene was also lost, and only the first exon remained (Fig. S1). By contrast, only the ndhD gene was intact. Overall, the four plastomes were all composed of 104 unique genes, including 30 unique tRNA genes, 4 unique rRNA genes and 70 unique protein-coding genes. Moreover, we observed the loss of the first exon of clpP gene based on BLASTn search (Fig. S2), which might be pseudogenes similar to gene ndhB (Table 2).
Fig. 2.
Visualization of BLASTn results on TBtools. a Schematic diagram of significant loss of ndh gene in the four Selenicereus plastomes. b Schematic diagram of rpoC1 and rpl2 gene’s intron in the four Selenicereus plastomes
Table 2.
Gene composition in the plastomes of Selenicereus
| Category of genes | Group of genes | Name of genes |
|---|---|---|
| Ribosomal RNA | rRNA | rrn16S, rrn23S, rrn5S, rrn4.5S |
| Transfer RNA | tRNA | 30 unique trna genes |
| Photosynthesis | Subunits of ATP synthase | atpA (× 2), atpA, atpB, atpE, atpF* (× 2), atpH, atpI |
| Subunits of photosystem II | psbA (× 2), psbB, psbC, psbD, psbE, psbF, psbI (× 2), psbJ, psbK (× 2), psbM, psbN, psbT, psbZ | |
| Subunits of NADH-dehydrogenase | ndhBψ, ndhD | |
| Subunits of cytochrome b/f complex | petA, petB*, petD*, petG, petL, petN | |
| Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ | |
| Subunit of rubisco | rbcL | |
| Self-replication | Large subunit of ribosome | rpl14, rpl16*, rpl2, rpl20, rpl22, rpl32, rpl33, rpl36 |
| DNA dependent RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | |
| Small subunit of ribosome | rps11, rps12*, rps14, rps15, rps16, rps16*, rps18, rps19, rps2, rps3, rps4, rps7, rps8 | |
| Other genes | Subunit of Acetyl-CoA-carboxylase | accD |
| c-type cytochrom synthesis gene | ccsA | |
| Envelope membrane protein | cemA | |
| Protease | clpPψ (× 2) | |
| Translational initiation factor | infA | |
| Maturase | matK (× 2) | |
| Unknown | Conserves open reading frames | ycf1, ycf1ψ, ycf3**, ycf2 (× 2), ycf4 |
(× 2) indicates that the gene located in the IRs and thus had two complete copies, * and ** indicate that genes containing one/two introns. ‘ψ’ indicates that it is a pseudogene
Furthermore, we also clearly observed the loss of intron in two genes: rpl2 and rpoC1 (Fig. 2b). Due to the loss of some genes, exons and introns, the number of intron-containing functional genes in Selenicereus species' plastomes were significantly reduced. Except for the trans-splicing gene, rps12, there were only 5 protein-coding genes (petB, petD, rpl16, rps16 and atpF,) containing one intron, and only one gene (ycf3) containing two introns. Moreover, there were 5 tRNA genes containing one intron (trnL-UAA, trnT-CGU, trnK-UUU, trnA-UGC and trnE-UUC).
In the four Selenicereus plastomes, there were 10 protein-coding genes (atpF, atpA, clpP, psbI, psbK, rps16, matK, psbA, ycf2, ycf1) and 8 tRNA genes (trnR-UCU, trnT-CGU, trnS-GCU, trnQ-UUG, trnK-UUU, trnH-GUG, trnM-CAU and trnL-CAA) were observed located in IR regions, and they duplicated in the IR regions, so they existed as two copies. Among the protein-coding genes, two genes (ycf1 and atpF) are partially located in IR region, and all rRNA are located in the SSC region.
Repeat and SSR analysis
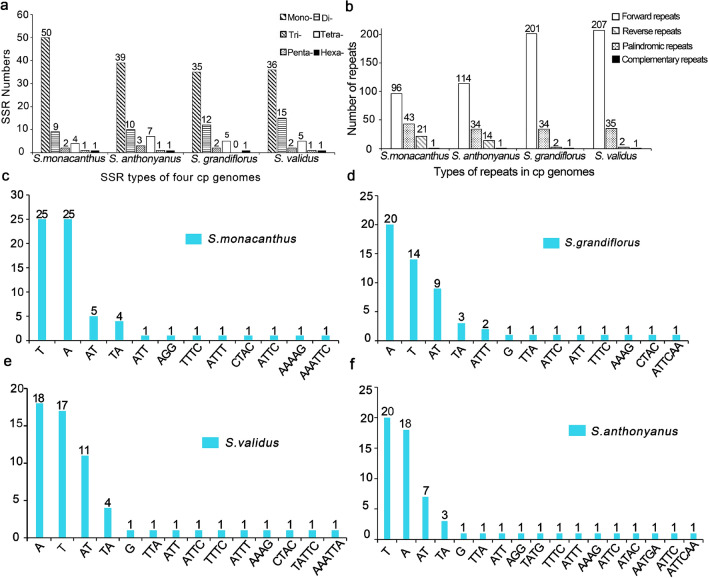
Microsatellites (also called simple repeat sequences, SSRs) are usually 6 bp tandem sequences in eukaryotic genomes (Lovin et al. 2009). Their high polymorphism and codominant inheritance make them become popular molecular markers (Morgante et al. 2002; Naranpanawa et al. 2020). They play an important role in the identification of species and the evaluation of evolutionary relationships (Guang et al. 2019). Among the four plastomes, S. monacanthus had the most significant number of SSR i.e. 67, followed by S. anthonyanus with 61 SSR and S. validus with 60 SSR, and finally, S. grandiflorus with 55 SSR. Most of these SSRs were homopolymers of A/T mononucleotide, and on average, they accounted for 64.60% of the total SSRs. Dinucleotides, tetranucleotides, and trinucleotides account for 18.93%, 8.64%, and 3.70% of the total SSR. Pentanucleotide and hexanucleotide repeats were rare in Selenicereus plastomes, accounting for 1.23% and 1.64% of all SSRs, respectively (Table S3 and Fig. 3).
Fig. 3.
Comparison of repeated sequences in the 4 Selenicereus plastomes. a Types and numbers of SSRs detected in the 4 plastomes. b Types and numbers of repeats detected in the 4 chloroplast genomes. c-f Types and numbers of SSR motifs detected in the 4 plastomes
We detected a large number of dispersed repeats in the four plastomes. A total of 807 dispersed repeats were identified, including 618 forward repeats (with length ranging from 30 to 415 bp), 146 palindromic repeats (30 to 415 bp), 39 reverse repeats (30 to 41 bp), and 4 complementary repeats of 30 bp in length (Table S4). Notably, the number of forward repeats in S. grandiflorus and S. validus was significantly compared to the other two taxa (Fig. 3b). The dispersed repeats not only serve as potential markers for rearrangement, but were also crucial for inducing mutations (Lopez et al. 2015).
Genomic divergence
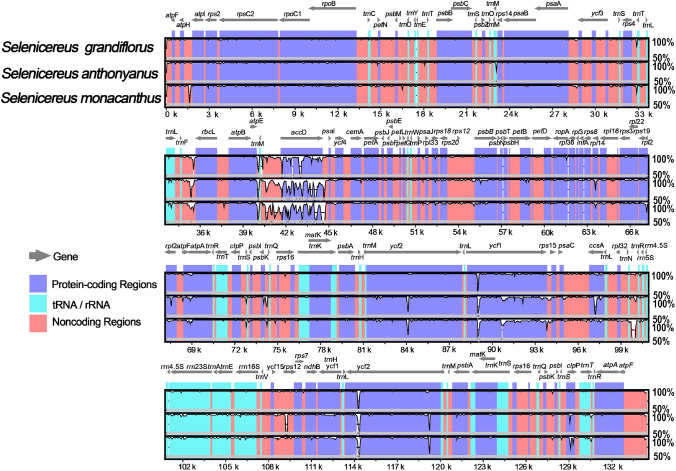
Sequence similarity analysis based on mVISTA (Frazer et al. 2004) was performed among the 4 plastomes, with the reference being the plastome of S. validus. We observed that the plastome sequences of the four species were quite conservative. In general, IR regions were more conserved than LSC and SSC regions, and the hypervariable regions were mainly observed in non-coding sequences. Nevertheless, several coding-regions showed significant differences in the sequences (Fig. 4), such as accD, clpP, ycf1 and ccsA; particularly, for gene accD, which showed a high-level of sequence divergence. In addition, there were significant differences among several non-coding regions: trnF-rbcL, trnM-accD and trnN-trnR.
Fig. 4.
Sequence similarity of 4 Selenicereus species by using S.validus as a reference sequence and visualized in mVISTA. Different color markers represent different areas, the pink regions are conserved noncoding sequences, the purple regions are protein-coding sequences, the light blue regions are tRNA or rRNA and the gray arrows are the gene and its direction. The percentage of identity ranges from 50 to 100%, shown on the Y-axis
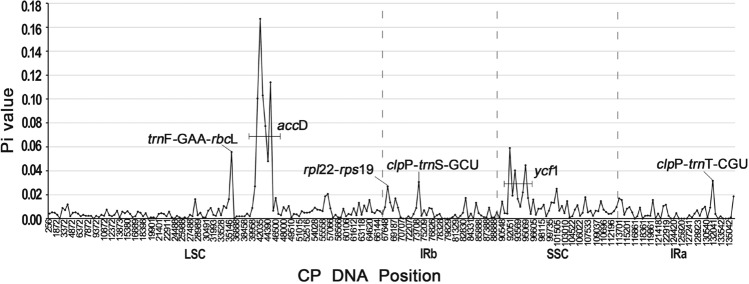
According to the results of DNA sequence polymorphism obtained by DnaSP (v6.0) (Rozas et al. 2017), we detected six hypervariable regions, trnF-GAA-rbcL (Pi = 0.05567), ycf1 (Pi = 0.059), clpP-trnS-GCU (Pi = 0.03067), clpP-trnT-CGU (Pi = 0.03167), rpl22-rps19 (Pi = 0.02067), and the highest Pi value of accD gene, including the intergenic region trnM-accD, with Pi value ranging from 0.00667 to 0.167 (Fig. 5). The maximum Pi value for six hypervariable regions is given in parentheses. The results were similar to those based on mVISTA, suggesting that these regions could be used as potential DNA barcodes.
Fig. 5.
The nucleotide diversity (Pi) of four Selenicereus plastomes (analyzed using DnaSP with a sliding window analysis (window length: 500 bp, step size: 500 bp)). The horizontal and vertical axes respectively represent the midpoint position of the window and the Pi value of each window. Pi values in one intergenic (trnF-GAA-rbcL, 0.05567) and two protein-coding genes (accD, 0.00667–0.167; ycf1, 0.004–0.059) were greater than 0.05
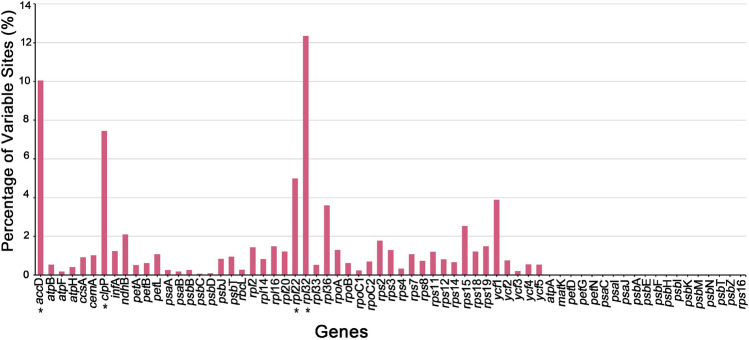
We analyzed 67 orthologous genes in the protein-coding regions of the four plastomes. In our study, a total of 19 genes (atpA, matK, petD, petG, petN, psaC, psaI, psaJ, psbA, psbE, psbF, psbH, psbI, psbK, psbM, psbN, psbT, psbZ, rps16) in the four species were completely conserved, and 27 genes had a mutation rate of less than 1.0%. However, we also observed that some protein-coding genes had a high-level of mutation (Table S5 and Fig. 6). For example, the mutation rates of 2 genes were more than 2%, and the mutation rates of 3 genes (rpl36, ycf1 and rpl22) were more than 3%. The highest mutation rates were observed in three genes: rpl32 (12.34%), accD (10.05%) and clpP (7.44%).
Fig. 6.
Percentage of variable sites for 67 shared plastidial genes of 4 Selenicereus species calculated by MEGA v6.0. The four genes with the highest mutation rate have been marked with an ‘*’ in the figure, and they are rpl32 (12.34%), accD (10.05%), clpP (7.44%) and rpl22 (5.00%), respectively
Contraction and expansion of inverted repeats
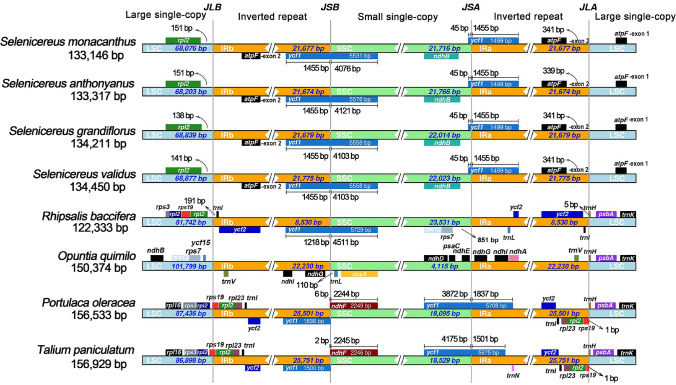
We analyzed the IR/SC boundaries and their adjacent genes in the four plastomes, and compared them to previously published related plastomes. The IR/SC border and the adjacent genes of Selenicereus plastomes were very similar in structural characteristics except for small differences in gene position. However, we observed that the IR lengths and IR boundaries of the four plastomes which have been newly reported here varied greatly from those previously reported in cacti and related species. The length of IR regions was observed as more than 20,000 bp in Opuntia quimilo and all other reported non-cactus species in the order Caryophyllales (Su et al. 2018). However, it was only 8530 bp in Rhipsalis baccifera, and less than 2000 bp in most cacti genera, such as Mammillaria, Carnegiea, and Lophocereus (Solórzano et al. 2019a; Oulo et al. 2020b). Here, in our four Selenicereus plastomes, the IR lengths ranged from 21,674 to 21,775 bp, indicating that the cacti had undergone a drastic expansion/contraction event in IR regions.
Furthermore, we also analyzed the IR boundaries of plastomes longer than 2000 bp in the IR region. As shown in Fig. 7, in two non-cactus species, the rps19 gene span the LSC/IRb border, and a rps19 pseudogene was duplicated in the IRa. The ycf1 gene span the SSC/IRa border, and an ycf1 pseudogene was duplicated in the IRb.
Fig. 7.
Comparision of borders among the LSC, SSC and IR region of 8 species. JLB, JSB, JSA and JLA represents the boundary between LSC/IRb, IRb/SSC, SSC/IRa and IRa/LSC
In O. quimilo, the two LSC/IR boundaries were ycf15-trnV and trnV-trnH, and the two SSC/IR boundaries were ndhG-trnL and ndhG-ndhE, respectively. By contrast, in R. baccifera, the two LSC/IR boundaries were rpl23-trnI and trnI-trnH, and the two SSC/IR boundaries were inside ycf1. Due to the dynamic changes of IRs, the IR boundaries were also changed in the four Selenicereus plastomes. Although the two SSC/IR boundaries were similar to R. baccifera, the second exon of atpF captured by the IR region, while the first exon of atpF was still located in the LSC region. Thus, a previously unreported LSC/IR boundary at the intron region of atpF was formed. This result suggested that the IR boundaries in cacti plastomes were extremely unstable compared with other Caryophyllales plastomes.
Phylogenetic analysis based on conserved protein-coding genes
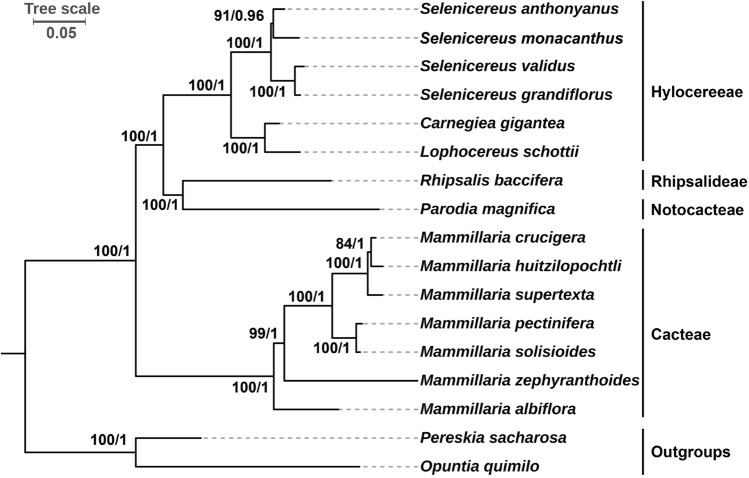
In this study, we constructed phylogenetic trees by using the 56 shared plastid genes as datasets. The tree reconstruction based on maximum likelihood (ML) method and Bayesian Inference (BI) method had a highly consistent topology. The stable topological structure and high bootstrap/posterior probability support values of each node indicated the reliability of phylogenetic tree (Fig. 8).
Fig. 8.
Phylogenetic relationships among 17 Cactaceae species. The 56 shared plastid protein coding genes (atpA, atpB, atpE, atpF, atpH, atpI, ccsA, cemA, clpP, infA, matK, petA, petB, petD, petG, petL, petN, psaA, psaB, psaC, psaI, psaJ, psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbM, psbN, psbT, rbcL, rpl14, rpl16, rpl20, rpl22, rpl2, rpoA, rpoB, rpoC1, rpoC2, rps11, rps12, rps14, rps15, rps19, rps2, rps3, rps4, rps7, rps8 and ycf3) were used as datasets to construct the phylogenetic trees by using the maximum likelihood (ML) method and Bayesian inference (BI) method. Pereskia sacharosa and Opuntia quimilo were used as outgroups. The scale number 0.05 indicates the length of the branch and the frequency of substitutions at 0.01 of the base at each site of the genome
The phylogenetic analysis involved 15 species of the subfamily Cereoideae and two outgroups (Pereskia sacharosa and O. quimilo). In our trees, the four Selenicereus species form a monophyletic clade supported by strong support values. The red-fleshed pitaya (S. monacanthus) was most closely related to S. anthonyanus compared to other two Selenicereus species.
Discussion
Changes in the content of plastomes: gene gain/loss and intron loss
In this study, we reported the complete plastomes of S. monacanthus, S. annthonyanus, S. grandiflorus, and S. Validus. According to the assembly results, the plastomes of these four taxa were typical quartile structure, with a pair of inverted repeats separated by a large single-copy region and a small single-copy region. Interestingly, we observed two phenomena in this study. Firstly, the phenomenon of massive losses of ndh genes in the plastome was observed, which was similar to the report by Sanderson et al. (2015), and only the ndhD gene was relatively complete. The ndh genes in plastids are important for the formation of the NADH dehydrogenase-like complex, which plays a role in the circulating electron flow (CEF) in the photosystem of most land plants. CEF is attributed to plant maintenance of effective photosynthesis, water stress and light protection (Burrows et al. 1998; Wang et al. 2006). Under favorable conditions, plants lacking NADH dehydrogenase-like complexes usually do not show significant growth and development defects (Horváth et al. 2000), most likely because there is a second pathway in CEF independent of plastid ndh genes. Secondly, we observed that compared with most species in the cactus family, the number of intron-containing genes in the plastomes of Selenicereus species was significantly reduced, such as O. quimilo plastome includes 16 intron-containing genes (Köhler et al. 2020). The main reason for this phenomenon is the losses of exons (ndhB and clpP) and introns (rpl2 and rpoC1). It was confirmed that the rpl2 intron was lost in the common ancestor of Caryophyllales (Downie et al. 1991). Plastid-encoded Plastid RNA polymerase (PEP) and nucleus-encoded Plastid RNA polymerase (NEP) are important in plastid gene transcription in higher plants during photosynthesis. The rpoC1 gene encode the DNA-directed RNA polymerase (PEP) subunit beta, and the lack of PEP activity leads to photosynthetic defects in plants, and there is no functional copy of rpoC1 outside the plastid that can complement the plastid rpo gene (Serino and Maliga 1998). Introns can effectively improve the expression level of genes under certain conditions and play an indispensable role in regulating gene expression (Yi et al. 2012). Whether the loss of rpoC1 intron has an effect on the photosynthesis of Selenicereus plants still needs further study. This loss has also been observed in other plastomes of subfamily Cereoideae (Oulo et al. 2020b; Sanderson et al. 2015; Solórzano et al. 2019b), and it probably is a feature unique to this clade.
SSRs and the repeats are crucial for the plastome rearrangement, and are widely used to detect population genetic diversity (Khan et al. 2019), as well as being considered as markers for DNA fingerprinting (Bodin et al. 2013). We analyzed the SSRs and repeat sequences in the four plastomes. First, the number of SSRs ranged from 55 to 67. Most SSRs were mononucleotide (A/T) polymers, accounting for 64.60% of all SSRs. This is one of the reasons for the low GC content in the plastome. Second, compared with SSRs, there were a lot of dispersed repeats in the four analyzed plastomes, and the length of forward/palindromic repeats was even more than 400 bp. The repeated sequences have previously been reported to have the potential to form secondary structures, they can be used to identify the recombination process (Kawata et al. 1997). In our study, these large numbers of short dispersed repeats most likely facilitated the plastome rearrangement. Unfortunately, our Illumina short reads have not been able to confirm this, and the long reads will be needed to confirm the presence of genomic recombination in the future.
The expansion of inverted repeats resulted in a rare boundary
The contraction and expansion of IRs are common in angiosperms (Zhu et al. 2016), which is also one of the factors affecting the length of plastome (Xue et al. 2019). According to the comparative analysis results, we observed that the length of IR regions in the four Selenicereus plastomes exceeded 20 kb. Although this phenomenon also exists in O. quimilo, the IR length of most reported genera in cacti such as Mammillaria, Carnegiea and Lophoereus were usually less than 2 kb (Solórzano et al. 2019a). Other studies have observed that the IR length of R. baccifera was only 8,530 bp (Oulo et al. 2020a). Apparently, cacti species have undergone dramatic expansion/contraction events in IR region. Besides, through the analysis of the IR boundaries, we noticed that the positions of each gene in the IR/SC border of the four Selenicereus plastomes were not significantly different. However, due to the expansion of IRs, some genes originally located in the LSC region were access to the IR region and formed a new IR boundary in the intron region of gene atpF that had not been reported before. In general, compared with other plastomes of Caryophyllales (Yao et al. 2019), the IRs of the cactus family are extremely unstable.
Hypervariable regions were identified based on plastome sequences
According to the results of sequence similarity analysis by mVISTA, the four Selenicereus plastomes were highly conserved, and there were few regions of difference. The hypervariable regions in plastomes were mainly identified in non-coding regions, which is consistent with the other plastomes in angiosperm (Gao et al. 2018; Zhang et al. 2016). Although there is little difference in plastomes as a whole, some hypervariable regions deserve our attention. Significant differences were observed in some protein-coding genes, such as clpP, ycf1, ccsA and accD, particular in gene accD, the mutation rates were even higher compared to the non-coding region. While in contrast, the gene with the greatest difference among the other plastomes usually was observed in gene ycf1. The differences in accD genes might be due to the presence of a large number of forward repeats in this region, which tend to mediate genome rearrangement. A large number of repeats in this region have been previously observed in passion fruit (Cauz-Santos et al. 2020), leading to the rearrangement of plastomes. Our results suggested that this region is also highly variable in cactus, and that they probably also contribute to genomic recombination in the genus Selenicereus. The gene accD and ycf1 both are indispensable for plant adaptation and leaf development (Kode et al. 2005; de Vries et al. 2015), and the high variability of nucleotide sequences of these two genes might be the result of environmental adaptation during evolution (Park et al. 2017; Thode and Lohmann 2019; de Vries et al. 2017). However, whether they cause physiological differences between Selenicereus and other cactus plants remains to be observed. On the whole, these hot-spots of mutations could be used as resources for system biology analysis and identification of DNA barcodes in plants. Our results provide a wealth of genetic information for the identification of species for the development of new DNA barcodes in Selenicereus (Dong et al. 2012).
Phylogenomic analysis revealed a close relationship among Selenicereus species
The phylogenetic relationship of the Cactaceae has long been a problem because of hybridization and complex evolutionary pattern of convergence in life forms and other traits (Korotkova et al. 2017). Plastid phylogenomics has provided new ideas and insights for the phylogenetic relationship of Cactaceae family and solved some taxonomic problems. In this study, we have constructed the high-resolution phylogenetic tree by using the 56 shared plastid genes as datasets. The results show undisputed monophyly of the 4 Selenicereus species. However, it is worth noting that S. monacanthus, once classified as Hylocereus (synonym: H. lemairei), is more closely related to S. anthonyanus, the traditional Selenicereus species. Our results support the previous studies, namely the two genera were not separated, and they have a very close phylogenetic relationship (Arias et al. 2005; Gómez-Hinostrosa et al. 2014; Plume et al. 2013; Miguel Ángel et al. 2016). However, considering the existence of interspecific or even intergeneric hybridization for Selenicereus plants (Tel-Zur et al. 2004), it is one-side to perform phylogenetic inferences about species with hybridization origin based on organelle genomes, as organelles are matrilineal inheritance (Liu et al. 2016). The combination of nuclear and organelle genes should be considered and used for phylogenetic inference in the future. In addition, future studies on Selenicereus can consider comprehensively exploring the delimits of unknown species, such as S. triangularis, S. murrillii and S. costaricensis, based on a wide range of molecular, morphological and ecological data.
Conclusion
In this study, we reported the complete plastomes of four Selenicereus species. The plastomes of these four species were similar to those of other angiosperms with typical quadripartite structure. In general, the genomic changes of the four plastomes were interesting: The large losses of ndh genes and the losses of introns/exons for several split genes (ndhB, rpoC1, clpP and rpl2). This implies that these changes of plastome in Selenicereus species is likely correlated with the adaption of arid climate. Furthermore, the IR region underwent a dramatic expansion and formed a previously unreported SC/IR border in the intron region of the atpF gene. These observations provide new insights into the plastome evolution associated with drought-tolerant plants and deepen our understanding of the genetics of Cactaceae plants.
Supplementary Information
Below is the link to the electronic supplementary material.
Table S1. Summary of the assembly information of the 4 Selenicereus species. Table S2. List of plastomes used for phylogenetic analysis. Table S3. Statistics on simple sequence repeats (SSRs) in the 4 plastomes. Table S4. Repeats (>= 30bp) identified in the four Selenicereus species. Table S5. Percentages of variable sites in 67 orthologous genes among the 4 Selenicereus species. Table S6. Summary of sequencing data quality. (PDF 390 kb)
Acknowledgements
The authors are grateful to the technical support provided by Novogene. This work was supported by the National Natural Science Foundation of China [31772260] and Chongqing Study Abroad Innovation Project [cx2019052]. The funders were not involved in the study design, data collection, and analysis, decision to publish, or manuscript preparation.
Abbreviations
- SSR
Simple sequence repeat
- IRs
Inverted repeats
- LSC
Large single-copy
- SSC
Small single-copy
- ML
Maximum-likelihood
- BI
Bayesian inference
- DnaSP
DNA Sequence Polymorphism
- CTAB
Cetyl trimethylammonium bromide
- NCBI
National Center for Biotechnology Information
- Pi
Nucleotide diversity/polymorphism
Author contribution
JY conceived the study and designed experiments; FH collected the samples and extracted DNA for sequencing by using the Illumina platform; YCX assembled and annotated the plastid genomes; SYZ and JLL carried out the comparative chloroplast analysis; QLQ drafted the manuscript. All authors have read and approved the final manuscript.
Data availability
The raw sequencing data generated in this study and the four plastome sequences were deposited in NCBI (https://www.ncbi.nlm.nih.gov/)with accession number: SAMN18357737, SAMN18357760, SAMN18357760, SAMN18357760, MW553055, MW553068, MW553069 and MW553070. All the samples are deposited at the Herbarium of Southwest University, Chongqing, China. All other data and material generated in this manuscript are available from the corresponding author upon reasonable request.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics declarations
The four collected Selenicereus species are widely cultivated in China as ornamental or edible fruits. Experimental researches do not include the genetic transformation, preserving the genetic background of the species used, and any other processes requiring ethics approval.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qiulin Qin and Jingling Li have contributed equally to this work.
References
- Arias S, Terrazas T, Arreola-Nava HJ, Vázquez-Sánchez M, Cameron KM. Phylogenetic relationships in Peniocereus (Cactaceae) inferred from plastid DNA sequence data. J Plant Res. 2005;118(5):317–328. doi: 10.1007/s10265-005-0225-3. [DOI] [PubMed] [Google Scholar]
- Arseneau JR, Steeves R, Laflamme M. Modified low-salt CTAB extraction of high-quality DNA from contaminant-rich tissues. Mol Ecol Resour. 2017;17(4):686–693. doi: 10.1111/1755-0998.12616. [DOI] [PubMed] [Google Scholar]
- Bai X, Zhang H. P41 Microwave-assisted extraction and HPLC analysis of polyphenols from pitaya peel and its inhibitory effect on human lung cancer cell line A549. Biochem Pharmacol. 2017;139:139–140. doi: 10.1016/j.bcp.2017.06.042. [DOI] [Google Scholar]
- Bodin SS, Kim JS, Kim J-H. Complete chloroplast genome of chionographis japonica (Willd.) maxim. (melanthiaceae): comparative genomics and evaluation of universal primers for liliales. Plant Mol Biol Rep. 2013;31(6):1407–1421. doi: 10.1007/s11105-013-0616-x. [DOI] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England) 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton NL, Rose JN (1963) The Cactaceae: descriptions and illustrations of plants of the cactus family, vol 3. Courier Corporation
- Burrows PA, Sazanov LA, Svab Z, Maliga P, Nixon PJ. Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. The EMBO J. 1998;17(4):868–876. doi: 10.1093/emboj/17.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cálix de Dios H. A new subspecies of Hylocereus undatus (Cactaceae) from Southeastern México. Haseltonia. 2009;11:11–17. doi: 10.2985/1070-0048(2005)11[11:ANSOHU]2.0.CO;2. [DOI] [Google Scholar]
- Cauz-Santos LA, da Costa ZP, Callot C, Cauet S, Zucchi MI, Bergès H, van den Berg C, Vieira MLC, A repertory of rearrangements and the loss of an inverted repeat region in passiflora chloroplast genomes. Genome Biol Evol. 2020;12(10):1841–1857. doi: 10.1093/gbe/evaa155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- Chen Y, Ye W, Zhang Y, Xu Y. High speed BLASTN: an accelerated MegaBLAST search tool. Nucl Acids Res. 2015;43(16):7762–8. doi: 10.1093/nar/gkv784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KS, Son OG, Park S. The chloroplast genome of elaeagnus macrophylla and trnh duplication event in elaeagnaceae. PLoS One. 2015;10(9):0138727. doi: 10.1371/journal.pone.0138727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Lin C-S, Yu M, Chang W-J. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17(1):134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, Archibald JM, Gould SB. The carboxy terminus of YCF1 contains a motif conserved throughout >500 myr of streptophyte evolution. Genome Biol Evol. 2017;9(2):473–479. doi: 10.1093/gbe/evx013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, Sousa FL, Bölter B, Soll J, Gould SB. YCF1: a green TIC? The Plant cell. 2015;27(7):1827–1833. doi: 10.1105/tpc.114.135541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W, Liu J, Yu J, Wang L, Zhou S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS One. 2012;7(4):e35071. doi: 10.1371/journal.pone.0035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie SR, Olmstead RG, Zurawski G, Soltis DE, Soltis PS, Watson JC, Palmer JD. Loss of the chloroplast DNA rpl2 intron demarcates six lineages of dicotyledons: Molecular and phylogenetic implications. Evolution. 1991;45:1245–1259. doi: 10.1111/j.1558-5646.1991.tb04390.x. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Zhang X, Meng H, Li J, Zhang D, Liu C. Comparative chloroplast genomes of Paris Sect Marmorata: insights into repeat regions and evolutionary implications. BMC Genom. 2018;19(Suppl 10):878. doi: 10.1186/s12864-018-5281-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Hinostrosa C, Hernández H, Terrazas T, Correa M. Studies on Mexican Cactaceae. V. Taxonomic notes on Selenicereus tricae. Brittonia. 2014;66:51–59. doi: 10.1007/s12228-013-9308-y. [DOI] [Google Scholar]
- Greiner S, Lehwark P, Bock R. OrganellarGenomeDRAW (OGDRAW) version 131: expanded toolkit for the graphical visualization of organellar genomes. Nucl Acids Res. 2019;47(W1):W59–w64. doi: 10.1093/nar/gkz238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang XM, Xia JQ, Lin JQ, Yu J, Wan QH, Fang SG. IDSSR: an efficient pipeline for identifying polymorphic microsatellites from a single genome sequence. Int J Mol Sci. 2019;20(14):3497. doi: 10.3390/ijms20143497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes DAB, DeCastro D, deOliveira FL, Nogueira EM, daSilva MAM, Teodoro AJ. Pitaya extracts induce growth inhibition and proapoptotic effects on human cell lines of breast cancer via downregulation of estrogen receptor gene expression. Oxid Med Cell Longev. 2017;2017:7865073. doi: 10.1155/2017/7865073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth EM, Peter SO, Joët T, Rumeau D, Cournac L, Horváth GV, Kavanagh TA, Schäfer C, Peltier G, Medgyesy P. Targeted inactivation of the plastid ndhB gene in tobacco results in an enhanced sensitivity of photosynthesis to moderate stomatal closure. Plant Physiol. 2000;123(4):1337–1350. doi: 10.1104/pp.123.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova Z, Sablok G, Daskalova E, Zahmanova G, Apostolova E, Yahubyan G, Baev V. Chloroplast genome analysis of resurrection tertiary relict haberlea rhodopensis highlights genes important for desiccation stress response. Front Plant Sci. 2017;8:204. doi: 10.3389/fpls.2017.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata M, Harada T, Shimamoto Y, Oono K, Takaiwa F. Short inverted repeats function as hotspots of intermolecular recombination giving rise to oligomers of deleted plastid DNAs (ptDNAs) Curr Genet. 1997;31(2):179–184. doi: 10.1007/s002940050193. [DOI] [PubMed] [Google Scholar]
- Khan A, Asaf S, Khan AL, Al-Harrasi A, Al-Sudairy O, AbdulKareem NM, Khan A, Shehzad T, Alsaady N, Al-Lawati A, Al-Rawahi A, Shinwari ZK. First complete chloroplast genomics and comparative phylogenetic analysis of Commiphora gileadensis and C foliacea: Myrrh producing trees. PLoS One. 2019;14(1):e0208511. doi: 10.1371/journal.pone.0208511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kode V, Mudd EA, Iamtham S, Day A. The tobacco plastid accD gene is essential and is required for leaf development. Plant J. 2005;44(2):237–244. doi: 10.1111/j.1365-313X.2005.02533.x. [DOI] [PubMed] [Google Scholar]
- Köhler M, Reginato M, Souza-Chies TT, Majure LC. Insights into chloroplast genome evolution across opuntioideae (cactaceae) reveals robust yet sometimes conflicting phylogenetic topologies. Front Plant Sci. 2020;11:729. doi: 10.3389/fpls.2020.00729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkova N, Borsch T, Arias S (2017) A phylogenetic framework for the Hylocereeae (Cactaceae) and implications for the circumscription of the genera. Phytotaxa 327:1–46. 10.11646/phytotaxa.327.1.1
- Krawczyk K, Nobis M, Myszczyński K, Klichowska E, Sawicki J. Plastid super-barcodes as a tool for species discrimination in feather grasses (Poaceae: Stipa) Sci Rep. 2018;8(1):1924. doi: 10.1038/s41598-018-20399-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29(22):4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang Z, Shao W, Ye Z, Zhang J. Phylogenetic and taxonomic status analyses of the abaso section from multiple nuclear genes and plastid fragments reveal new insights into the North America origin of populus (salicaceae) Front Plant Sci. 2016;7:2022. doi: 10.3389/fpls.2016.02022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yang H, Zhao J, Zhou B, Li T, Xiang B. The complete chloroplast genome sequence of the folk medicinal and vegetable plant purslane (Portulaca oleracea L) J Hortic Sci Biotechnol. 2018;93(4):356–365. doi: 10.1080/14620316.2017.1389308. [DOI] [Google Scholar]
- Lopez L, Barreiro R, Fischer M, Koch MA. Mining microsatellite markers from public expressed sequence tags databases for the study of threatened plants. BMC Genom. 2015;16:781. doi: 10.1186/s12864-015-2031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovin DD, Washington KO, deBruyn B, Hemme RR, Mori A, Epstein SR, Harker BW, Streit TG, Severson DW. Genome-based polymorphic microsatellite development and validation in the mosquito Aedes aegypti and application to population genetics in Haiti. BMC Genom. 2009;10:590. doi: 10.1186/1471-2164-10-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliga P. Engineering the plastid genome of higher plants. Curr Opin Plant Biol. 2002;5(2):164–172. doi: 10.1016/s1369-5266(02)00248-0. [DOI] [PubMed] [Google Scholar]
- Miguel Ángel C, Salvador A, Teresa T. Molecular phylogeny and taxonomy of the genus Disocactus (Cactaceae), based on the DNA sequences of six chloroplast markers. Willdenowia. 2016;46(1):145–164. doi: 10.3372/wi.46.46112. [DOI] [Google Scholar]
- Misra S, Harris N. Using Apollo to browse and edit genome annotations. Curr Protoc Bioinform. 2005;12(1):9.5.1–9.5.28. doi: 10.1002/0471250953.bi0905s12. [DOI] [PubMed] [Google Scholar]
- Morgante M, Hanafey M, Powell W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet. 2002;30(2):194–200. doi: 10.1038/ng822. [DOI] [PubMed] [Google Scholar]
- Myszczyński K, Bączkiewicz A, Buczkowska K, Ślipiko M, Szczecińska M, Sawicki J. The extraordinary variation of the organellar genomes of the Aneura pinguis revealed advanced cryptic speciation of the early land plants. Sci Rep. 2017;7(1):9804. doi: 10.1038/s41598-017-10434-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranpanawa DNU, Chandrasekara C, Bandaranayake PCG, Bandaranayake AU. Raw transcriptomics data to gene specific SSRs: a validated free bioinformatics workflow for biologists. Sci Rep. 2020;10(1):18236. doi: 10.1038/s41598-020-75270-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes E, Sousa A, Lucena C, Silva S, Lucena R, Alves CA, Alves R. Pitaia (Hylocereus sp.): Uma revisão para o Brasil. Gaia Scientia. 2014;8:90–98. [Google Scholar]
- Oulo MA, Yang J-X, Dong X, Wanga VO, Mkala EM, Munyao JN, Onjolo VO, Rono PC, Hu G-W, Wang Q-F. Complete chloroplast genome of rhipsalis baccifera, the only cactus with natural distribution in the old world: genome rearrangement, intron gain and loss, and implications for phylogenetic studies. Plants. 2020;9(8):979. doi: 10.3390/plants9080979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oulo MA, Yang JX, Dong X, Wanga VO, Mkala EM, Munyao JN, Onjolo VO, Rono PC, Hu GW, Wang QF. Complete chloroplast genome of rhipsalis baccifera, the only cactus with natural distribution in the old world: genome rearrangement, intron gain and loss, and implications for phylogenetic studies. Plants (Basel, Switzerland) 2020;9(8):979. doi: 10.3390/plants9080979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD. Comparative organization of chloroplast genomes. Ann Rev Genet. 1985;19:325–354. doi: 10.1146/annurev.ge.19.120185.001545. [DOI] [PubMed] [Google Scholar]
- Palmer JD, Jansen RK, Michaels HJ, Chase MW, Manhart JR. Chloroplast DNA variation and plant phylogeny. Ann Missouri Bot Garden. 1988;75(4):1180–1206. doi: 10.2307/2399279. [DOI] [Google Scholar]
- Palmer JD, Jorgensen RA, Thompson WF. Chloroplast DNA variation and evolution in pisum - patterns of change and phylogenetic analysis. Genetics. 1985;109(1):195–213. doi: 10.1093/genetics/109.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Ruhlman TA, Weng ML, Hajrah NH, Sabir JSM, Jansen RK. Contrasting patterns of nucleotide substitution rates provide insight into dynamic evolution of plastid and mitochondrial genomes of geranium. Genome Biol Evol. 2017;9(6):1766–1780. doi: 10.1093/gbe/evx124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervaiz T, Sun X, Zhang Y, Tao R, Zhang J, Fang J. Association between chloroplast and mitochondrial dna sequences in chinese prunus genotypes (prunus persica, prunus domestica, and prunus avium) BMC Plant Biol. 2015;15:4. doi: 10.1186/s12870-014-0402-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plume O, Straub S, Tel Zur N, Cisneros A, Schneider B, Doyle J. Testing a hypothesis of intergeneric allopolyploidy in vine cacti (cactaceae: hylocereeae) Syst Bot. 2013;38:737. doi: 10.1600/036364413X670421. [DOI] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34(12):3299–3302.10.1093/molbev/msx248 [DOI] [PubMed]
- Rozewicki J, Li S, Amada KM, Standley DM, Katoh K. MAFFT-DASH: integrated protein sequence and structural alignment. Nucl Acids Res. 2019;47(W1):W5–w10. doi: 10.1093/nar/gkz342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ, Copetti D, Búrquez A, Bustamante E, Charboneau JL, Eguiarte LE, Kumar S, Lee HO, Lee J, McMahon M, Steele K, Wing R, Yang TJ, Zwickl D, Wojciechowski MF. Exceptional reduction of the plastid genome of saguaro cactus (Carnegiea gigantea): Loss of the ndh gene suite and inverted repeat. Am J Bot. 2015;102(7):1115–1127. doi: 10.3732/ajb.1500184. [DOI] [PubMed] [Google Scholar]
- Serino G, Maliga P. RNA polymerase subunits encoded by the plastid rpo genes are not shared with the nucleus-encoded plastid enzyme. Plant Physiol. 1998;117(4):1165–1170. doi: 10.1104/pp.117.4.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solórzano S, Chincoya DA, Sanchez-Flores A, Estrada K, Díaz-Velásquez CE, González-Rodríguez A, Vaca-Paniagua F, Dávila P, Arias S. De novo assembly discovered novel structures in genome of plastids and revealed divergent inverted repeats in mammillaria (cactaceae, caryophyllales) Plants. 2019;8(10):392. doi: 10.3390/plants8100392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solórzano S, Chincoya DA, Sanchez-Flores A, Estrada K, Díaz-Velásquez CE, González-Rodríguez A, Vaca-Paniagua F, Dávila P, Arias S. De novo assembly discovered novel structures in genome of plastids and revealed divergent inverted repeats in mammillaria (cactaceae, caryophyllales) Plants (Basel, Switzerland) 2019;8(10):392. doi: 10.3390/plants8100392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su CK, Myounghai K, Byoungyoon L, Seonjoo P, Tzen-Yuh C. Complete chloroplast genome of Tetragonia tetragonioides: molecular phylogenetic relationships and evolution in Caryophyllales. PLoS One. 2018;13(6):e0199626. doi: 10.1371/journal.pone.0199626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 60. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tel-Zur N, Abbo S, Bar-Zvi D, Mizrahi Y. Genetic relationships among Hylocereus and Selenicereus vine cacti (Cactaceae): evidence from hybridization and cytological studies. Ann Bot. 2004;94(4):527–534. doi: 10.1093/aob/mch183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thode VA, Lohmann LG. Comparative chloroplast genomics at low taxonomic levels: a case study using amphilophium (bignonieae, bignoniaceae) Front Plant Sci. 2019;10:796. doi: 10.3389/fpls.2019.00796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45(W1):W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalobos M, Schweiggert R, Carle R, Esquivel P. Chemical characterization of Central American pitaya (Hylocereus sp.) seeds and seed oil. CyTA J Food. 2012;10:78–83. doi: 10.1080/19476337.2011.580063. [DOI] [Google Scholar]
- Wang P, Duan W, Takabayashi A, Endo T, Shikanai T, Ye JY, Mi H. Chloroplastic NAD(P)H dehydrogenase in tobacco leaves functions in alleviation of oxidative damage caused by temperature stress. Plant Physiol. 2006;141(2):465–474. doi: 10.1104/pp.105.070490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Shi T, Luo W, Ni X, Iqbal S, Ni Z, Huang X, Yao D, Shen Z, Gao Z. Comparative analysis of the complete chloroplast genome among Prunus mume, P armeniaca, and P salicina. Hortic Res. 2019;6:89. doi: 10.1038/s41438-019-0171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JB, Yang SX, Li HT, Yang J, Li DZ. Comparative chloroplast genomes of camellia species. PLoS One. 2013;8(8):e73053. doi: 10.1371/journal.pone.0073053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhou T, Duan D, Yang J, Feng L, Zhao G. Comparative analysis of the complete chloroplast genomes of five quercus species. Front Plant Sci. 2016;7:959. doi: 10.3389/fpls.2016.00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao G, Jin JJ, Li HT, Yang JB, Mandala VS, Croley M, Mostow R, Douglas NA, Chase MW, Christenhusz MJM, Soltis DE, Soltis PS, Smith SA, Brockington SF, Moore MJ, Yi TS, Li DZ. Plastid phylogenomic insights into the evolution of Caryophyllales. Mol Phylogenet Evol. 2019;134:74–86. doi: 10.1016/j.ympev.2018.12.023. [DOI] [PubMed] [Google Scholar]
- Yi DK, Lee HL, Sun BY, Chung MY, Kim KJ. The complete chloroplast DNA sequence of Eleutherococcus senticosus (Araliaceae); comparative evolutionary analyses with other three asterids. Mol Cells. 2012;33(5):497–508. doi: 10.1007/s10059-012-2281-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Gao F, Jakovlic I, Zou H, Zhang J, Li WX, Wang GT. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 2020;20(1):348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Du L, Liu A, Chen J, Wu L, Hu W, Zhang W, Kim K, Lee SC, Yang TJ, Wang Y. The complete chloroplast genome sequences of five epimedium species: lights into phylogenetic and taxonomic analyses. Front Plant Sci. 2016;7:306. doi: 10.3389/fpls.2016.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A, Guo W, Gupta S, Fan W, Mower JP. Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New Phytol. 2016;209(4):1747–1756. doi: 10.1111/nph.13743. [DOI] [PubMed] [Google Scholar]
- Zhuang Y, Zhang Y, Sun L. Characteristics of fibre-rich powder and antioxidant activity of pitaya (Hylocereus undatus) peels. Int J Food Sci Technol. 2012;47(6):1279–1285. doi: 10.1111/j.1365-2621.2012.02971.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Summary of the assembly information of the 4 Selenicereus species. Table S2. List of plastomes used for phylogenetic analysis. Table S3. Statistics on simple sequence repeats (SSRs) in the 4 plastomes. Table S4. Repeats (>= 30bp) identified in the four Selenicereus species. Table S5. Percentages of variable sites in 67 orthologous genes among the 4 Selenicereus species. Table S6. Summary of sequencing data quality. (PDF 390 kb)
Data Availability Statement
The raw sequencing data generated in this study and the four plastome sequences were deposited in NCBI (https://www.ncbi.nlm.nih.gov/)with accession number: SAMN18357737, SAMN18357760, SAMN18357760, SAMN18357760, MW553055, MW553068, MW553069 and MW553070. All the samples are deposited at the Herbarium of Southwest University, Chongqing, China. All other data and material generated in this manuscript are available from the corresponding author upon reasonable request.