Abstract
Species-specific polymorphisms in the noncoding internal transcribed spacer 2 (ITS2) region of the rRNA operon provide accurate identification of clinically significant yeasts. In this study, we tested the hypothesis that ITS1 noncoding regions contain diagnostically useful alleles. The length of ITS1 region PCR products amplified from 40 species (106 clinical strains, 5 reference strains, and 30 type strains) was rapidly determined with single-base precision by automated capillary electrophoresis. Polymorphisms in the PCR product length permitted 19 species to be distinguished by ITS1 alone, compared with 16 species distinguished by using only ITS2. However, combination of both ITS alleles permitted identification of 30 species (98% of clinical isolates). The remaining 10 species with PCR products of similar sizes contained unique ITS alleles distinguishable by restriction enzyme analysis. DNA sequence analysis of amplified ITS1 region DNA from 79 isolates revealed species-specific ITS1 alleles for each of the 40 pathogenic species examined. This provided identification of unusual clinical isolates, and 53 diagnostic ITS1 sequences were deposited in GenBank. Phylogenetic analyses based on ITS sequences showed a similar overall topology to 26S rRNA gene-based trees. However, different species with identical 26S sequences contained distinct ITS alleles that provided species identification with strong statistical support. Together, these data indicate that the analysis of ITS polymorphisms can reliably identify 40 species of clinically significant yeasts and that the capacity for identifying potentially new pathogenic species by using this database holds significant promise.
The complexity of opportunistic fungal infections is increasing as more patients are adversely affected by an expanding diversity of yeasts. Invasive procedures or immunosuppression increases the risk of fungemia (24, 34), and drug-resistant strains have emerged (9, 21). Although Candida albicans is still the predominate agent of nosocomial infection, serious infections caused by other yeasts have increased in frequency (8, 11, 29, 34). For example, species of Cryptococcus other than Cryptococcus neoformans, previously considered to be nonpathogenic saprophytes, have been reported to cause cryptococcosis (28). Adequate treatment of these infections depends on early detection and accurate identification of the pathogens. However, conventional identification by evaluation of morphological and physiological characteristics can be laborious, sometimes leads to incorrect classification and identification (2, 3, 13), and can be impeded by database limitations (4, 6).
To develop a rapid molecular method for identifying yeasts, we recently analyzed the length and sequence polymorphisms in the DNA of noncoding internal transcribed spacer region 2 (ITS2) of the rRNA operon (2). ITS2 region polymorphisms permitted accurate identification of over 400 clinical strains representing 34 species of pathogenic yeasts. ITS2 region PCR product length determined by automated capillary electrophoresis permitted single-base-pair precision, with between-run standard deviations (SD) of ≤0.5 base, and 92% of the clinical strains examined were rapidly and correctly identified by using only the unique length of their PCR products. The remaining 8% could be identified by either restriction endonuclease or DNA sequence analysis of their ITS2 region PCR products.
In this study, we investigated the length and sequence polymorphisms in the ITS1 region and determined their diagnostic and phylogenetic utility for identification of medically important yeasts. These DNA sequences contained unique alleles for all 40 pathogenic species examined, and ITS1 alleles displayed greater interspecies variability than ITS2 region sequences. Thirty species, comprising 98% of the clinical isolates, were easily distinguished by simply determining the length of the ITS1 and ITS2 region PCR products. The remaining 10 species could be differentiated by either restriction endonuclease or DNA sequence analysis of ITS PCR products. The sequence diversity among ITS1 alleles was useful for distinguishing closely related species, particularly those that contained identical DNA sequences in the D1/D2 variable domain of the 26S rRNA gene. Thus, our ITS sequence data provide a reliable means with which to rapidly identify known yeast pathogens, will facilitate the discovery of potentially novel pathogens, and establish a foundation for further expansion of an ITS sequence database of medically important fungi.
MATERIALS AND METHODS
Yeast isolates.
We examined 106 clinical strains and five reference strains (Tables 1 and 2) previously characterized genetically, morphologically, and biochemically in detail (2). These strains were from our clinical laboratory at the University of Washington Medical Center. Twenty-three type strains from the American Type Culture Collection (ATCC) and 7 type strains from the Centraalbureau voor Schimmelcultures (CBS) were also included in this study (Table 2). These 141 strains represented 40 different species of pathogenic yeasts. Nitrate assimilation, which distinguishes Pichia fabianii from Pichia veronae, was performed on a slant of nitrate agar (1.4 g of potassium nitrate, 1.6 g of yeast carbon base from Difco, 0.12 g of bromthymol blue, 16 g of Noble agar, 1 liter of distilled water [pH 5.9 to 6.0]) incubated at 30°C for about 1 week until the agar turned from yellow or yellow-green (negative) to blue (positive). Cryptococcus albidus and Candida albicans were used as positive and negative controls, respectively (17).
TABLE 1.
Lengths of ITS1 and ITS2 region PCR products from clinical isolates and type strains
| Speciesa (no. of isolates) | ITS1 PCR product lengthb (mean no. of bp) | SDc (bp) | Range of product lengths (bp)
|
ITS2 PCR product lengthd (mean no. of bp) | |
|---|---|---|---|---|---|
| Minimum | Maximum | ||||
| Yarrowia lipolytica (2) | 134.43 | 0.29 | 134.10 | 134.60 | 236.66 |
| Candida rugosaT (1) | 142.91 | NAe | NA | 270.99 | |
| Candida lusitaniae (4) | 143.87 | 0.35 | 143.54 | 144.44 | 251.14 |
| Candida intermedia (1) | 148.48 | NA | NA | 255.10 | |
| Pichia ohmeriT (1) | 154.01 | NA | NA | 268.84 | |
| Candida lambica (1) | 156.39 | NA | NA | 302.18 | |
| Candida pararugosa (1)f | 161.34 | NA | NA | 269.57 | |
| Candida krusei (5) | 181.44 | 0.64 | 180.39 | 182.27 | 344.61 |
| Cryptococcus laurentiiT (1) | 190.84 | NA | NA | 362.83 | |
| Trichosporon jiroveciiT (1) | 192.33 | NA | NA | 349.89 | |
| Trichosporon cutaneumT (1) | 193.72 | NA | NA | 350.00 | |
| Trichosporon mucoides (1) | 194.25 | NA | NA | 349.30 | |
| Cryptococcus neoformans (5) | 197.93 | 0.26 | 197.59 | 198.47 | 370.76 |
| Cryptococcus humicolusT (1) | 197.94 | NA | NA | 353.17 | |
| Trichosporon inkin (1) | 197.97 | NA | NA | 353.27 | |
| Trichosporon asahii (1) | 200.00 | NA | NA | 355.87 | |
| Cryptococcus uniguttulatus (1) | 208.00 | NA | NA | 428.73 | |
| Candida utilisT (1) | 213.92 | NA | NA | 362.34 | |
| Candida dubliniensis (7)g | 214.06 | 0.46 | 213.40 | 214.81 | 342.33 |
| Candida albicans (8) | 214.35 | 0.41 | 214.00 | 214.99 | 339.34 |
| Candida tropicalis (11) | 214.78 | 0.45 | 213.95 | 215.73 | 327.42 |
| Candida freyschussiiT (1) | 223.46 | NA | NA | 347.27 | |
| Candida parapsilosis (4) | 225.43 | 0.11 | 225.30 | 225.58 | 309.93 |
| Sporobolomyces salmonicolorT (1) | 227.45 | NA | NA | 400.19 | |
| Rhodotorula rubra (7) | 228.36 | 0.33 | 228.41 | 229.41 | 400.46 |
| Rhodotorula glutinisT (1) | 229.92 | NA | NA | 390.93 | |
| Cryptococcus albidusT (1) | 231.03 | NA | NA | 403.67 | |
| Cryptococcus liquefaciens (1)h | 231.39 | NA | NA | 403.45 | |
| Cryptococcus diffluens (1)h | 231.41 | NA | NA | 403.75 | |
| Candida guilliermondii (5) | 243.75 | 0.21 | 243.51 | 244.11 | 374.61 |
| Hansenula anomala (3) | 257.35 | 0.12 | 257.21 | 257.53 | 372.58 |
| Pichia veronaeT (1) | 261.44 | NA | NA | 365.95 | |
| Pichia fabianii (1)f | 265.45 | NA | NA | 364.78 | |
| Candida zeylanoides (2) | 267.72 | 0.29 | 267.27 | 268.03 | 371.65 |
| Candida famata (2) | 273.52 | 0.21 | 273.32 | 273.73 | 376.21 |
| Endomyces fibuliger (2) | 291.13 | 1.14 | 289.56 | 292.25 | 374.98 |
| Pichia farinosa (2) | 303.19 | 0.00 | 303.18 | 303.19 | 379.18 |
| Candida kefyr (3) | 305.45 | 0.15 | 305.31 | 305.60 | 427.88 |
| Saccharomyces cerevisiae (6) | 436.52 | 2.09 | 434.75 | 440.03 | 416.35 |
| Candida glabrata (17) | 475.24 | 0.29 | 474.78 | 475.68 | 413.51 |
Identification of species by biochemical, morphological, and genotypic assessment. The number of isolates analyzed is indicated in parentheses. Type strains are designated by a superscript T.
PCR product sizes determined by capillary electrophoresis as described in Materials and Methods.
SD are calculated for species with two or more strains.
ITS2 PCR product lengths are from our previous study (2).
NA, nonapplicable.
Candida pararugosa and Pichia fabianii were designated as isolate UWFP-348 and UWFP-345, respectively, in a previous study (2).
Three strains obtained from the University of Texas Health Science Center at San Antonio (2).
C. liquefaciens UWFP-357 and C. diffluens UWFP-359 were both misidentified as Cryptococcus albidus by the VITEK and API systems (2).
TABLE 2.
Length and sequence polymorphisms of ITS1 region DNAs from clinical and type strains
| Species | Strain(s)a | ITS1 PCR product lengthb (bp) | Sequenced PCR productc (bp) | % Identity with type strains | GenBank accession no. | Source or reference |
|---|---|---|---|---|---|---|
| Candida albicans | ATCC 11006 | 215.09 | 219 | AF336831d | TSk | |
| UWFP-60 | 214.17 | 218 | 99.5 | AF336832d | TS | |
| ATCC 28516 | NTl | 218 | 99.5 | AF217609 | ||
| Candida dubliniensis | UWFP-92 | 214.06 | 218 | AF336833 | TS | |
| CD36 | NT | 218 | 100e | AJ249485 | ||
| Candida famata | CBS1795T | 273.63 | 278 | AF336834 | TS | |
| UWFP-352 | 273.48 | 278 | 100 | AF336834 | TS | |
| CBS767 | NT | 278 | 100 | AF210327 | ||
| Candida freyschussii | ATCC 18737T | 223.46 | 231 | AF336835 | TS | |
| Candida glabrata | ATCC 2001T | 475.50 | 482 | AF336836 | TS | |
| UWFP-115, -118 | 475.24 | 482 | 99.8 | AF336837 | TS | |
| UWFP-192 | 475.31 | 482 | 99.8 | AF336838 | TS | |
| IFO 0622 | NT | 452 | 100 | AB032177 | ||
| Candida guilliermondii | ATCC 6260T | 243.08 | 248 | AF336839 | TS | |
| UWFP-195 | 243.73 | 248 | 100 | AF336839 | TS | |
| Taxon:4929 | NT | 249 | 99.6 | L47110 | 33 | |
| Candida intermedia | ATCC 14439T | 147.42 | 151 | AF336840 | TS | |
| UWFP-347 | 148.48 | 151 | 100 | AF336840 | TS | |
| Candida kefyr | UWFP-208 | 305.60 | 309 | AF336841 | TS | |
| JCM1630 | NT | 279 | 99.7f | AB011519 | 22 | |
| Candida krusei | ATCC 6258T | 180.41 | 182 | AF411417 | TS | |
| Candida krusei sequevar 1 | UWFP-210, -211 | 181.87 | 183 | 98.4e | AF336842 | TS |
| ATCC 6258T | NT | 182 | 100 | AF246989 | ||
| Candida lambica | ATCC 24750T | 156.42 | 158 | AF336843 | TS | |
| UWFP-346 | 156.39 | 158 | 100 | AF336843 | TS | |
| Candida lusitaniae | ATCC 34449T | 144.34 | 148 | AF336844 | TS | |
| UWFP-233, -234 | 144.07 | 148 | 100 | AF336844 | TS | |
| ATCC 34449T | NT | 148 | 98.0 | AF009215 | ||
| Candida parapsilosis | UWFP-254, -285 | 225.44 | 229 | AF336846 | TS | |
| ATCC 22019 | NT | 229 | 100e | U10987 | 16 | |
| Candida pararugosa | ATCC 38774T | 161.37 | 164 | AF411418 | TS | |
| UWFP-348 | 161.34 | 164 | 99.4 | AF335925d | TS | |
| Candida pelliculosa (Hansenula anomala) | CBS605T | 257.34 | 262 | AF335926 | TS | |
| UWFP-396 | 257.36 | 262 | 100 | AF335926 | TS | |
| FY-102 | NT | 262 | 100 | AF270936 | 19 | |
| Candida rugosa | ATCC 10571T | 142.91 | 145 | AF335927 | TS | |
| Candida tropicalis | ATCC 750T | 214.83 | 218 | AF335928 | TS | |
| UWFP-313, -321, -324 | 214.94 | 218 | 100 | AF335928 | TS | |
| Taxon:5482 | NT | 218 | 99.5 | L47112 | 33 | |
| Candida utilis | ATCC 22023T | 213.92 | 220 | AF335929 | TS | |
| Candida zeylanoides | ATCC 7351T | 267.81 | 272 | AF335930 | TS | |
| UWFP-349 | 267.64 | 272 | 100 | AF335930 | TS | |
| Cryptococcus albidus | ATCC 10666T | 231.03 | 234 | AF335931g | TS | |
| CBS142T | NT | 234 | 100 | AF145321 | 26 | |
| Cryptococcus diffluens | UWFP-359 | 231.41 | 234 | AF335932 | TS | |
| CBS160T | NT | 234 | 100e | AF145330 | 26 | |
| Cryptococcus humicolus | ATCC 14438T | 197.94 | 200 | AF335933 | TS | |
| UWFP-362 | 197.91 | 200 | 99.5 | AF335934 | TS | |
| JCM1457 | NT | 170 | 100h | AB035572 | 29 | |
| Cryptococcus laurentii | ATCC 18803T | 190.84 | 193 | AF335935 | TS | |
| JCM9066 | NT | 163 | 99.4h | AB035043 | 28 | |
| Cryptococcus liquefaciens | UWFP-357 | 231.39 | 234 | AF336845g | TS | |
| Cryptococcus neoformans | ATCC 32045T | 198.20 | 201 | AF335936 | TS | |
| UWFP-360 | 197.74 | 201 | 99.0 | AF335937 | TS | |
| Taxon:37769 | NT | 201 | 99.0 | AF196312 | ||
| Cryptococcus uniguttulatus | CBS1730T | 207.71 | 212 | AF335938 | TS | |
| UWFP-364 | 208.00 | 212 | 100 | AF335938 | TS | |
| JCM3685 | NT | 212 | 100 | AB032692 | 31 | |
| Endomyces fibuliger | CBS329.83T | 290.92 | 296 | AF335939 | TS | |
| UWFP-397 | 290.62 | 295 | 99.7 | AF335940 | TS | |
| UWFP-398 | 291.64 | 297 | 99.7 | AF335941 | TS | |
| 8014 | NT | 296 | 100 | U10409 | ||
| Pichia fabianii | CBS5640T | 265.62 | 270 | AF335942d | TS | |
| UWFP-345 | 265.45 | 270 | 100 | AF335942 | TS | |
| Pichia farinosa | CBS185T | 304.46 | 307 | AF335944 | TS | |
| Pichia farinosa sequevar 1 | UWFP-389, -390 | 303.19 | 307 | 98.4 | AF335945 | TS |
| Pichia ohmeri | ATCC 46053T | 154.01 | 156 | AF335946 | TS | |
| Pichia ohmeri sequevar 1 | UWFP-388 | 155.48 | 158 | 88.9 | AF335947g | TS |
| Pichia veronae | CBS6591T | 261.44 | 265 | AF335943d | TS | |
| Rhodotorula glutinis | ATCC 32765T | 229.92 | 232 | AF335948 | TS | |
| JCM8208 | NT | 232 | 98.3 | AB026018 | ||
| Rhodotorula rubra | ATCC 32763T | 229.53 | 232 | AF335949 | TS | |
| UWFP-370, -371, -374 | 228.80 | 232 | 100i | AF335950d | TS | |
| UWFP-373, -380 | 228.61 | 232 | 99.1i | AF335951d | TS | |
| SY-93 | NT | 232 | 100 | AB026003 | ||
| Saccharomyces cerevisiae | UWFP-382, -384 | 435.09 | 441 | 99.8fj | AF335952 | TS |
| UWFP-383 | 439.90 | 446 | 98.7fj | AF335953 | TS | |
| UWFP-387 | 437.11 | 443 | 99.3fj | AF335954 | TS | |
| CBS4903 | NT | 424g | Z95940 | |||
| Sporobolomyces salmonicolor | ATCC 36400T | 227.45 | 231 | AF335955 | TS | |
| JCM1841 | NT | 231 | 98.7 | AB030341 | 30 | |
| Trichosporon asahii | UWFP-391 | 200.00 | 203 | AF335956 | TS | |
| CBS2530 | NT | 173 | 100f | AB018014 | 27 | |
| Trichosporon cutaneum | ATCC 28592T | 193.72 | 196 | AF335957 | TS | |
| CBS2466T | NT | 166 | 100h | AB018020 | 27 | |
| Trichosporon inkin | ATCC 18020T | 198.47 | 201 | AF335958 | TS | |
| UWFP-399 | 197.97 | 201 | 100 | AF335958 | TS | |
| CBS5585T | NT | 170 | 99.4h | AB018024 | 27 | |
| Trichosporon jirovecii | ATCC 34499T | 192.33 | 195 | AF335959 | TS | |
| CBS6950 | NT | 165 | 100h | AB018026 | 27 | |
| Trichosporon mucoides | UWFP-363, -366, -367 | 194.37 | 197 | AF335960d | TS | |
| M9478 | NT | 167 | 100f | AB018031 | 27 | |
| Yarrowia lipolytica | ATCC 18942T | 133.97 | 139 | AF335961 | TS | |
| Y. lipolytica sequevar 1 | UWFP-400, -401 | 134.48 | 139 | 97.8 | AF335962 | TS |
ATCC, American Type Culture Collection; UWFP, University of Washington Fungal Project; CD, Cardiff Dental; CBS, Centraalbureau voor Schimmelcultures; IFO, Institute of Fermentation; JCM, Japan Collection of Microorganisms; SY, deep sea yeast; M, Meiji Pharmaceutical; type strains are labeled with superscript T.
PCR product length determined with capillary electrophoresis as described in Materials and Methods.
Exact number of nucleotides determined by direct sequencing of the PCR products.
In addition to ITS1 data listed, new ITS2 sequence data include C. albicans ATCC 11006 (AF335963). C. albicans UWFP-60 (AF335964), C. pararugosa UWFP-348 (AF335965), P. fabianii CBS5640T (AF335967), P. veronae CBS6591T (AF335966), R. rubra UWFP-370 (AF335969), R. rubra UWFP-373 (AF335968), and T. mucoides UWFP-366 (AF335970).
Complete sequence compared to ITS1 region DNA sequences from clinical strains in this study.
Partial sequence compared to ITS1 region DNA sequences from clinical strains in this study.
UWFP-357 and -359 were misidentified as Cryptococcus albidus by the VITEK system (2). Their ITS2 sequences were identical and submitted as C. albidus under accession no. AF219002.
Partial sequence compared to ITS1 region DNA sequences from type strains in this study.
UWFP-373 and -380 had identical ITS2 sequences, and they were 99.3% similar to the R. rubra type strain. UWFP-370 had 99.5% sequence similarity to the R. rubra type strain.
ITS1 sequence similarities between UWFP-383 and -387, UWFP-382 and -383, and UWFP-382 and -387 were 98.4, 98.9, and 99.5%, respectively.
TS, this study.
NT, not tested.
A hierarchical approach to building a cross-validated data set was employed when selecting isolates for molecular analyses. This was based on our observations that (i) congenic strains identified according to their genetic, biochemical, and morphological characters generally show >99% sequence similarity at ITS2 (2) and ITS1 (Table 2); (ii) capillary electrophoresis permits single-base-pair precision in determining the length of PCR products, with SD of ≤0.5 base between runs; and (iii) the apparent PCR product length strongly correlates (R2 = 0.9992) with the actual number of nucleotides enumerated by direct sequencing (2). Type and reference strains were characterized by directly sequencing ITS1 region DNA. ITS1 PCR product length polymorphisms were determined for randomly selected representatives of each of 40 species (Table 1): for 18 species, multiple isolates were characterized, and for 22 species, only single isolates were characterized for PCR product length. All length polymorphisms (Table 1) were subsequently confirmed by direct sequence analysis of, in most cases, more than one representative isolate of each species (Table 2). Among the 22 singly listed isolates in Table 1, 7 isolates represent type strains added to this study for completeness and for which no clinical isolates were available, 7 isolates showed >99% sequence similarity with their respective type strains, 7 isolates showed >99% sequence similarity with another sequenced clinical isolate, and 1 clinical isolate, Cryptococcus liquefaciens, was the only representative of that species characterized by analyzing DNA sequences from ITS1 (Table 2) and ITS2 (Table 3) and the D1/D2 variable domain of the 26S rRNA gene (Table 4).
TABLE 3.
Species with similar ITS region PCR product lengths are distinguishable by species-specific ITS1 and ITS2 DNA sequences or restriction fragments of the ITS1-ITS2-region
| Species | Length (bp)
|
% Sequence similarity
|
Restriction fragment size(s)b (bp)
|
|||
|---|---|---|---|---|---|---|
| ITS1 | ITS2a | ITS1 | ITS2a | BsaHI | HincII | |
| T. jiroveciiT (A) | 192.33 | 349.89 | A | A | 527 | 49/478 |
| T. cutaneumT (B) | 193.72 | 350.00 | 98.0, B | 97.7, B | 529 | 53/74/102 |
| T. mucoides (C) | 194.25 | 349.30 | 97.0, 97.5 | 99.4, 97.2 | 528 | 528 |
| C. humicolusT | 197.94 | 353.17 | 84.6 | 85.2 | 535 | 535 |
| T. inkin | 197.97 | 353.27 | 350/189 | 539 | ||
| C. dubliniensis | 214.06 | 342.33c | 93.2 | 92.1 | 457/84 | 541 |
| C. albicans | 215.09 | 338.63c | 536 | 536 | ||
| S. salmonicolorT | 227.45 | 400.19 | 80.6 | 83.7 | 612 | 612 |
| R. rubraT | 229.53 | 400.00 | 41/575 | 616 | ||
| C. albidusT (A) | 231.03 | 403.67 | A | A | 621 | 621 |
| C. liquefaciens (B) | 231.39 | 403.45 | 96.2, B | 98.0, B | 621 | 363/258 |
| C. diffluens (C) | 231.41 | 403.75 | 96.6, 99.6 | 98.0, 100 | 621 | 363/258 |
TABLE 4.
Comparing three diagnostic loci for identifying unusual isolates
| Group and strain(s)a | Identity by biochemical testsb | Actual identity by genetic analysesc | Sequence similarity among clinical and type strains (%)d
|
Accession no. from this study for 26S | ||
|---|---|---|---|---|---|---|
| ITS1 | ITS2 | 26S | ||||
| Group 1 | ||||||
| UWFP-208 | C. kefyr | C. kefyr | 99.7 | 98.4 | 100 | AF335978 |
| Group 2 | ||||||
| ATCC 6258T | C. krusei | C. krusei | ||||
| UWFP-210, -211 | C. krusei | C. krusei | 98.4 | 100 | 100 | AF335979 |
| Group 3 | ||||||
| ATCC 10571T | C. rugosa | C. rugosa (A) | A | A | A | |
| UWFP-348 | C. rugosa | C. pararugosa (B) | 69.5, B | 62.8, B | 63.7, B | AF335972 |
| ATCC 38774T | C. pararugosa | C. pararugosa (C) | 68.9, 99.4 | 61.1, 100 | 63.7, 100 | AF421856 |
| Group 4 | ||||||
| ATCC 10666T | C. albidus | C. albidus (A) | A | A | A | AF335982 |
| UWFP-359 | C. albidus | C. diffluens (B) | 96.6, B | 98.0, B | 97.0, B | AF335981 |
| UWFP-357 | C. albidus | C. liquefaciens (C) | 96.2, 99.6 | 98.0, 100 | 97.3, 99.5 | AF335980 |
| Group 5 | ||||||
| ATCC 32045T | C. neoformans | C. neoformans | 100 | AF335984 | ||
| UWFP-360 | C. neoformans | C. neoformans | 99.0 | 99.0 | 99.5 | AF335983 |
| Group 6 | ||||||
| CBS185T | P. farinosa | P. farinosa | 100 | AF335974 | ||
| UWFP-389, -390 | P. farinosa | P. farinosa sequevar 1 | 98.4 | 100 | 99.7 | AF335973 |
| Group 7 | ||||||
| ATCC 46053T | P. ohmeri | P. ohmeri | 100 | AF335976 | ||
| UWFP-388 | P. ohmeri | P. ohmeri sequevar 1 | 88.9 | 99.6 | 100 | AF335975 |
| Group 8 | ||||||
| ATCC 32765T | R. glutinis | R. glutinis (A) | A | A | A | AF335985 |
| ATCC 32763T | R. rubra | R. rubra (B) | 90, B | 91, B | 94.3, B | AF335986 |
| UWFP-370 | R. glutinis | R. rubra (C) | 90, 100, C | 90, 99, C | 94.3, 100, C | AF335986 |
| UWFP-373, -380 | R. glutinis | R. rubra (D) | 90, 99, 99 | 91, 99, 100 | 94.3, 99.8, 99.8 | AF335987 |
| Group 9 | ||||||
| ATCC 18942T | Y. lipolytica | Y. lipolytica | 100 | AF335977 | ||
| UWFP-400, -401 | Y. lipolytica | Y. lipolytica sequevar 1 | 97.8 | 100 | 100 | AF335977 |
| Group 10 | ||||||
| UWFP-345 | Unknowne | P. fabianii (A) | A | A | A | AF335971 |
| CBS5640T | P. fabianii | P. fabianii (B) | 100, B | 100, B | 100, B | |
| CBS6591T | P. veronae | P. veronae (C) | 90.1, 90.1 | 94.1, 94.1 | 100, 100 | |
| Group 11 | ||||||
| UWFP-363, -366, -367 | Unknowne | T. mucoides | 100 | 100 | 99.7 | AF335988 |
Type strains are designated by a superscript T.
API and VITEK systems, supplemented by conventional methods, were used to perform biochemical tests (2).
Identification confirmed by ≥99% 26S rDNA gene sequence similarity with type strains and published sequences. Published sequences used for similarity comparison are given in parentheses as follows: C. kefyr (no. U94924T), C. krusei (no. U76347T), C. pararugosa (no. U62306T), C. albidus (no. AF075474T), C. diffluens (no. AF075502T), C. liquefaciens (no. AF181515T), C. neoformans (no. AF075484T), P. farinosa (no. U45739T), P. ohmeri (no. U45702T), R. glutinis (no. AF070430), R. rubra (no. AF189960T), Y. lipolytica (no. U40080T), P. fabianii (no. U73573T), P. veronae (no. U73576T), and T. mucoides (no. AF075515T). Sequences from this study are identical with published 26S sequences except as follows: there was 99.8% similarity to published sequences for strains of C. pararugosa (UWFP-348), R. glutinis (ATCC 32765T), and R. rubra (UWFP-373 and -380); 99.7% similarity for strains of P. farinosa (UWFP-389 and -390) and T. mucoides (UWFP-363, -366, and -367); and 99.5% similarity for C. neoformans (UWFP-360).
ITS1 and ITS2 sequence similarities were compared among clinical and type strains from our study, except for C. kefyr, C. krusei, and T. mucoides, which were compared to published sequences: C. kefyr (JCM1630 for ITS1 [AB011519], taxon 4911 for ITS2 [L47107], NRRL Y-8281T for 26S [U94924]), C. krusei (NRRL Y-5396T for 26S [U76347]), and T. mucoides (M9478 for ITS1 and ITS2 [AB018031], CBS7625T for 26S [AF075515]).
Clinical strains UWFP-345, -363, -366, and -367 were unidentifiable by routine automated biochemical methods. Strain UWFP-345 has 100% 26S sequence identity with P. fabianii and P. veronae. However, ITS sequence comparison clearly identified this strain as P. fabianii, which was concordant with its positive nitrate assimilation test. Strains UWFP-363, -366, and -367 were identified by their 100% ITS2 sequence identity with T. mucoides in the ITS2 study (2).
PCR and DNA sequencing.
DNA was extracted from yeasts after 48 h of growth on Sabouraud's agar and PCR amplified as previously described (2). ITS1 region DNA was amplified with 900 nM primer ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′; GIBCO BRL, Grand Island, N.Y.), 300 nM primer ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′) (18), and the following thermocycler parameters: 95°C for 6 min, followed by 25 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, followed by one final extension at 72°C for 10 min. The same parameters were used to amplify the ITS1 and ITS2 regions simultaneously as a single product by using 300 nM ITS1 and ITS4 primers (5′-TCCTCCGCTTATTGATATGC-3′) (18). Similarly, 600 nM (each) NL-1 (5′-GCATATCAATAAGCGGAGGAAAAG-3′) and NL-4 (5′-GGTCCGTGTTTCAAGACGG-3′) (14) was used to amplify the D1/D2 variable domain of the 26S ribosomal DNA (rDNA) gene: denaturation at 94°C for 5 min was followed by 30 cycles of amplification at 94°C for 1 min, 60°C for 2 min, and 72°C for 2 min, followed by one final extension at 72°C for 7 min. Determination of the length of ITS1 PCR products by automated capillary electrophoresis and DNA sequence analysis of both the forward and reverse strands of PCR products (listed in Table 2) were performed as previously described (2).
Sequence similarity and phylogenetic analyses.
DNA sequences were assembled, edited, and subjected to phylogenetic analyses as described previously (2). Sequence similarities were expressed as the percentage of nucleotide differences determined by pairwise sequence comparisons. Saccharomyces cerevisiae was used as the outgroup instead of Pneumocystis carinii (2): the ITS1 sequence of S. cerevisiae is distant from all the other sequences (except Candida glabrata) and thus forms a natural outgroup. The branching order of the neighbor-joining (23) dendrograms (Fig. 1) was evaluated with 1,000 bootstrap analyses by using the SEQBOOT program in the PHYLIP software package (version 3.573) (J. Felsenstein, Department of Genetics, University of Washington, Seattle; http://evolution.genetics.washington.edu/phylip.html).
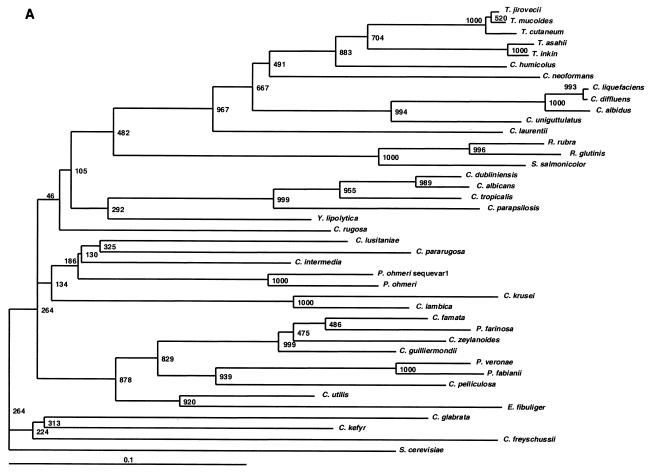
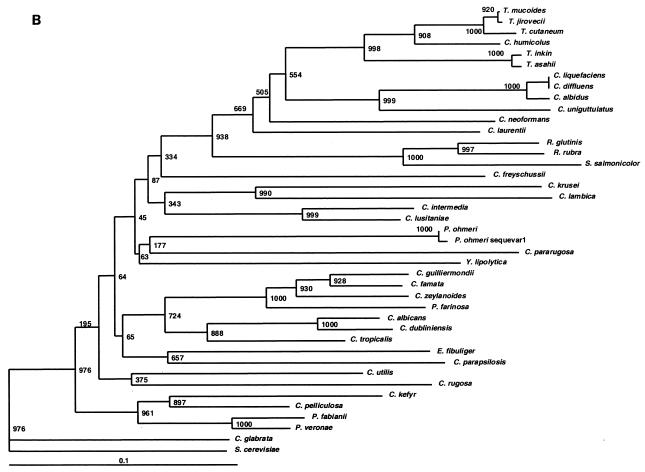
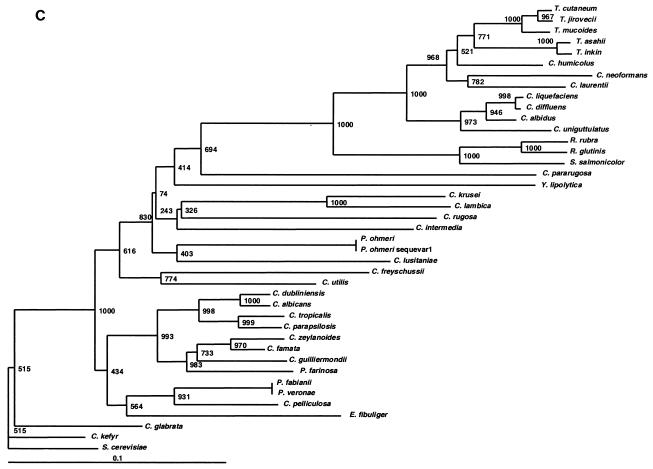
FIG. 1.
(A) ITS1 sequence-based phylogenetic tree of 41 clinically significant yeast species. A consensus neighbor-joining dendrogram with 1,000 bootstrap replicates was based on 419 aligned positions of complete ITS1 sequences with adjacent partial sequences of 18S and 5.8S rRNA genes. GenBank accession numbers of sequences generated in this study are presented in Table 2. Numbers at the nodes indicate the bootstrap values. Lower bars indicate relative genetic distance. (B) ITS2 sequence-based phylogenetic tree of 42 clinically significant yeast species. Consensus neighbor-joining dendrogram with 1,000 bootstrap replicates was based on 432 aligned positions of complete ITS2 sequences with adjacent partial sequences of 5.8S and 26S rRNA genes. The sequences of C. albicans (no. L28817) and C. parapsilosis (no. U10988), as well as our sequences used for tree building, were previously published (2). Accession numbers of additional ITS2 sequences generated in this study are shown in Table 2 (footnote d). (C) 26S sequence-based phylogenetic tree of 41 clinically significant yeast species. Consensus neighbor-joining dendrogram with 1,000 bootstrap replicates was based on 565 aligned positions of the D1/D2 region of the 26S rRNA genes. Sequences of the following organisms (accession numbers in parentheses) were retrieved from GenBank: C. albicans (no. U45776), C. dubliniensis (no. U57685), C. famata (no. U45808), C. freyschussii (no. AF017242), C. glabrata (no. U44808), C. guilliermondii (no. U45709), C. intermedia (no. U44809), C. kefyr (no. U94924), C. krusei (no. U76347), C. lambica (no. AF020435), C. lusitaniae (no. U44817), C. parapsilosis (no. U45754), C. pararugosa (no. U62306), C. pelliculosa (no. U74592), C. rugosa (no. U45727), C. tropicalis (no. AB034689), C. utilis (no. U73570), C. zeylanoides (no. U45832), C. diffluens (no. AF075502), C. humicolus (no. AF189836), C. laurentii (no. AF075469), C. liquefaciens (no. AF181515), C. uniguttulatus (no. AF075468), E. fibuliger (no. U40088), P. fabianii (no. U73573), P. veronae (no. U73576), S. cerevisiae (no. U44806), S. salmonicolor (no. AF070439), T. asahii (no. AF105393), T. cutaneum (no. AF075483), T. inkin (no. AF105396), T. jirovecii (no. AF105398), and T. mucoides (no. AF075515). Sequences of the following organisms (accession numbers in parentheses) are from this study: C. albidus (no. AF335982), C. neoformans (no. AF335984), P. farinosa (no. AF335974), P. ohmeri (no. AF335976), P. ohmeri sequevar 1 (no. AF335975), R. glutinis (no. AF335985), R. rubra (no. AF335986), and Y. lipolytica (no. AF335977).
RESULTS
To determine if DNA polymorphisms in the ITS1 region contained diagnostically useful information, we analyzed single PCR products that had been amplified from 141 strains of yeasts, including 30 type and 5 reference strains. Each product contained the 3′ end of the 18S rDNA gene, the entire ITS1 region, and the 5′ end of the 5.8S rDNA gene. The PCR product length was analyzed for each strain by capillary electrophoresis under denaturing conditions (Table 1), a method that rapidly provides single-base-pair precision, low run-to-run variation, and an excellent correlation (R2 = 0.9992) with the actual number of nucleotides enumerated by direct sequencing (2). To confirm the specificity of length polymorphisms and identify species-specific ITS1 alleles, the forward and reverse strands of ITS1 region PCR products from 79 strains were directly sequenced (Table 2).
ITS1 sequence polymorphisms.
ITS1 PCR product lengths determined by capillary electrophoresis ranged from 134.43 bp (Yarrowia lipolytica) to 475.24 bp (C. glabrata) with intraspecies strain variation generally less than 0.5 bp, except for Candida krusei (SD = 0.64 bp), Endomyces fibuliger (SD = 1.14 bp), and S. cerevisiae (SD = 2.09 bp) (Table 1). Similar intraspecies strain variation was observed among ITS2 alleles for Candida dubliniensis, Candida tropicalis, and S. cerevisiae (2), but the distribution of ITS2 region PCR product lengths among all tested species was more limited and ranged from 236.66 to 428.73 bp (Table 1). Of 40 species examined, 19 could be identified based solely on the apparent length of their ITS1 PCR products (species listed individually in Table 1).
Direct DNA sequence analysis of the ITS1 region PCR products (Table 2) confirmed a close correlation of the apparent ITS1 PCR product lengths measured by capillary electrophoresis with the actual product length (2) and revealed species-specific ITS1 DNA sequences for all 40 pathogenic species examined. Intraspecies sequence variability was detected among clinical strains and between clinical and type strains (Table 2). Nucleotide insertions or deletions in the ITS1 region were observed between type and clinical strains of C. albicans and E. fibuliger, as well as among clinical strains of E. fibuliger and S. cerevisiae. For these species, this variation accounts for the SD of ≥0.5 bp observed for the PCR product lengths measured by capillary electrophoresis (Table 1). Similar nucleotide insertions and deletions were found previously in the ITS2 regions of type and clinical strains of C. glabrata and C. tropicalis, as well as among clinical strains of C. tropicalis and S. cerevisiae (2). Conspecific strains examined among a total of 79 isolates in this study, including 30 type strains and 5 reference strains, generally displayed ≥99% sequence similarity (Table 2). Together with the 53 new species-specific diagnostic alleles identified (Table 2), these data provide two important measures of the diagnostic utility of ITS1 sequences for identifying medically important yeasts.
Complementary ITS1 and ITS2 polymorphisms.
Twenty-one species in Table 1 comprise five groups with nearly identical ITS1 PCR product lengths, yet 11 of these species can be identified if their ITS2 PCR product lengths are also determined: Candida rugosa, Candida lusitaniae; Pichia ohmeri, Candida lambica, Cryptococcus laurentii, Cryptococcus neoformans, Candida utilis, C. dubliniensis, C. albicans, C. tropicalis, and Rhodotorula glutinis. Notably, C. rugosa, P. ohmeri, C. neoformans, C. laurentii, and C. utilis could only be identified by using a combination of ITS1 and -2 length polymorphisms (Table 1) (2). That is, neither ITS PCR product length alone permitted unambiguous identification. ITS2 length polymorphisms alone identified 16 species of yeasts (2) compared with 19 identified by using ITS1 alone (Table 1). However, the combination of ITS1 and ITS2 length polymorphisms accurately identified 30 species of yeasts (Tables 1 and 2). The isolation rate of these 30 species in our clinical laboratory, which serves a spectrum of patients ranging from primary to tertiary care and provides reference laboratory services for five Western states, indicates that >98% of clinical isolates from our laboratory could be identified by ITS length polymorphisms.
The remaining 10 species (Table 1) included four groups that could not be distinguished by ITS region PCR product lengths (Table 3). These isolates could be readily and accurately identified by DNA sequence analysis of either ITS allele (Table 3) or restriction endonuclease analysis of the PCR product containing both ITS1 and ITS2 (Table 3). Two exceptions were Cryptococcus liquefaciens and Cryptococcus diffluens. These closely related species (7) contained identical ITS2 alleles (Table 3) (2), provided indistinguishable BsaHI and HincII restriction analyses from their ITS1 and -2 PCR products, and displayed phenotypic characteristics of Cryptococcus albidus by routine automated biochemical methods and therefore were only distinguishable by the DNA sequences of their ITS1 alleles (Table 3).
Identification of unusual isolates.
In a previous study (2), we observed >98% concordance between biochemical and ITS2 genotypic identifications validated with over 400 clinical isolates. However, four clinical isolates could not be readily identified by automated biochemical systems (UWFP-345, -363, -366, and -367). Six additional clinical isolates (UWFP-348, -357, -359, -370, -373, and -380) had questionable designations: their identity predicted by biochemical characteristics was clearly not concordant with their ITS2 genotype. The identity of these 10 isolates was established in Table 4, which includes analysis of the hypervariable D1/D2 region of the 26S rRNA gene (5, 13). These data confirm the utility of ITS sequences for correctly identifying C. rugosa and C. pararugosa (Table 4, group 3), C. albidus, C. diffluens, and C. liquefaciens (group 4), R. glutinis and R. rubra (group 8), P. fabianii and P. veronae (group 10), and T. mucoides (group 11). Notably, P. veronae and P. fabianii have identical D1/D2 hypervariable sequences and could only be identified by using ITS alleles (Table 4, group 10).
Table 4 also contains analyses of isolates with ITS1 (Table 2) or ITS2 (2) alleles that showed ≤99% sequence similarity between type and clinical strains or among clinical strains. Interestingly, the newly designated C. diffluens and C. liquefaciens (7) strains share 99.6, 100, and 99.5% sequence similarity at ITS1, ITS2, and the 26S rRNA hypervariable loci, respectively (Table 4, group 4). Although >99% similarity is generally typical of conspecific strains (2, 5, 13, 27), Table 4 provides several examples where this may not be the case: Candida kefyr (row 1), C. krusei (row 2), P. farinosa (group 6), P. ohmeri (group 7), and Y. lipolytica (group 9). Because species delineation now encompasses the integration of both physiological and genotypic characteristics, we use the term “sequevar” to distinguish clinical isolates with the following characteristics: one diagnostic allele <99% similar to the type strain while biochemical and two (or more) other diagnostic loci (defined as ≥99% similar) concur for the designation of a particular species—e.g., P. farinosa ITS1 sequevar 1, P. ohmeri ITS1 sequevar 1, and Y. lipolytica ITS1 sequevar 1 (Table 4, groups 6, 7, and 9). It is possible that further investigation(s) will confirm these as new species, considering the similarity of the three diagnostic loci examined for C. diffluens and C. liquefaciens (Table 4). Furthermore, ITS2 sequevars, such as for C. keyfr (Table 4, row 1), may also exist. Together, these data indicate that ITS analyses are useful for distinguishing closely related clinical isolates.
Phylogenetic analysis.
In addition to discriminating among closely related species, we sought to determine if ITS1 alleles could accurately indicate the relationships among distantly related taxa. Phylogenetic trees were constructed with ITS1, ITS2, and 26S sequences (Fig. 1A, B, and C, respectively). Dendrogram topologies derived from the three sets of sequences were highly similar. Similar to the 26S tree, high bootstrap values were observed at the deep branches separating the heterogeneous ascomycetous yeasts and the different clades of the basidiomycetous yeasts. Within each clade, the relationships among species of monophyletic taxa with statistically well-supported peripheral branches were generally concordant between all three markers. Moreover, closely related species with identical 26S sequences could be separated by ITS sequences with strong bootstrap support (e.g., P. veronae and P. fabiani) and species with identical ITS2 sequences could be separated by examining their ITS1 alleles (e.g., C. diffluens and C. liquefaciens). We therefore conclude that ITS loci could be used for taxonomic placement and to infer phylogenetic relationships of previously undescribed species and novel pathogens isolated in the clinical laboratory.
DISCUSSION
DNA-based methods have been used successfully to characterize pathogenic yeasts and provide evidence of novel species (1, 2, 5, 7, 10, 12, 14, 25, 27–29, 31). We have established an ITS database that provides accurate identification of at least 40 species of medically important yeasts. Nineteen of these can be rapidly identified by capillary electrophoresis to determine ITS1 PCR product lengths alone, compared with 16 identified by using ITS2 alone (Table 1). The use of both alleles identified 30 species (Table 1), including 4 species that could only be unambiguously designated by using both ITS alleles (see Results). Our diagnostic library of ITS length polymorphisms is validated with over 400 clinical strains (Tables 1 and 2) (2), and together, the ITS1 and -2 length polymorphisms identify >98% of the isolates received by our clinical laboratory.
The direct sequence analysis of 79 isolates confirmed the accuracy of estimating PCR product lengths by capillary electrophoresis (2) and provided 53 new diagnostic ITS1 alleles from a well-characterized group of clinical isolates with concordant biochemical analysis-, ITS1-, and ITS2-based identifications (Table 2). ITS1 alleles showed species specificity and permitted 40 species of pathogenic yeasts to be unambiguously identified by restriction endonuclease or DNA sequence analysis of ITS1 region PCR products (Tables 2 and 3). In combination with sequences from ITS2 (55 alleles total from reference 2 and Table 2) and the D1/D2 hypervariable region of the 26S rRNA gene, unusual clinical isolates were also readily identified (Table 4). Together with the ITS1 alleles, the additional ITS2 and D1/D2 region sequences from this study provide a total of 67 new diagnostic alleles (Table 2, footnote d, and Table 4).
Data from three diagnostic loci, the D1/D2 hypervariable region of the 26S rRNA gene, ITS1, and ITS2, allowed ambiguous biochemical and genotypic designations to be clarified (Table 4). In this respect, we found ITS1 to be most useful, because it distinguished among closely related isolates with identical sequences at either the 26S rRNA gene or ITS2 loci. Similarly, others have found ITS sequences particularly useful for distinguishing among closely related yeasts (5, 12, 14, 27, 28, 31). Furthermore, our data indicating that ATCC 11066 (originally designated Candida stellatoidea) is C. albicans (Table 2) agrees with other molecular data (20) that C. stellatoidea does not merit species status (15, 32). Although we observed intraspecies ITS allelic variation, as has been reported previously (2, 27), conspecific strains generally demonstrated ≥99% sequence similarity (Table 2). This result is in agreement with studies including other loci and large numbers of diverse isolates (2, 5, 12, 14, 27, 28). However, the possibility exists for exceptions to this benchmark (Table 4), because the converse argument implies that ≤99% sequence similarity connotes different species: for example, Cryptococcus diffluens and C. liquefaciens demonstrate >99% similarity at three diagnostic loci (Table 4 [see also Results and reference 7]). To facilitate accurate reporting of genetic data and to maintain the integrity of public databases for the purposes of identifying clinically significant yeasts, we use the term “sequevar” to indicate isolates that differ at one locus (≤99% sequence similarity) but share two (or more) loci (≥99% similarity) concordantly with biochemical and morphological indicators for a particular species designation. Clearly, additional investigation will be necessary to accurately determine the species relationships among taxa containing such isolates.
The data in Table 4 indicate the utility of ITS sequences for distinguishing among closely related yeasts, and we also determined the relevance of ITS sequences for establishing the relationships among distantly related taxa. The phylogenetic relationships observed in the ITS1 and ITS2 trees were highly concordant with those inferred from 26S rDNA sequences (Fig. 1). ITS2 and ITS1 were better than 26S at resolving the taxonomic position of Cryptococcus humicolus. In agreement with our data, this species has been previously shown to belong to the Trichosporon clade by analysis of 18S rDNA and ITS sequences (29), as well as 26S rDNA (10). In the ascomycetous group, Pichia species were polyphyletic in all three dendrograms. These results are concordant with the data of Kurtzman et al., who also showed that Pichia species are polyphyletic and widely distributed among the ascomycetous yeasts based on 26S rDNA phylogenies (13). Thus, our data confirm that ITS1 and ITS2 regions are not only phylogenetically informative, as shown by other investigators (5, 28, 29, 31), but are also useful in distinguishing closely as well as distantly related taxa (Fig. 1). By extrapolation, an expanded ITS sequence database could be used for taxonomic placement and to infer phylogenetic relationships of previously undescribed species and novel pathogens. Ready access to these species-specific DNA sequences, amplified with universal primers, predicts that array-based hybridization schemes will become useful diagnostic tools in the clinical microbiology laboratory.
REFERENCES
- 1.Baleiras Couto M M, Eijsma B, Hofstra H, in't Veld J H, van der Vossen J M B M. Evaluation of molecular typing techniques to assign genetic diversity among Saccharomyces cerevisiae strains. Appl Environ Microbiol. 1996;62:41–46. doi: 10.1128/aem.62.1.41-46.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y C, Eisner J D, Kattar M M, Rassoulian-Barrett S L, LaFe K, Yarfitz S L, Limaye A P, Cookson B T. Identification of medically important yeasts using PCR-based detection of DNA sequence polymorphisms in the internal transcribed spacer 2 region of the rRNA genes. J Clin Microbiol. 2000;38:2302–2310. doi: 10.1128/jcm.38.6.2302-2310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dooley D P, Beckius M L, Jeffrey B S. Misidentification of clinical yeast isolates by using the updated Vitek Yeast Biochemical Card. J Clin Microbiol. 1994;32:2889–2892. doi: 10.1128/jcm.32.12.2889-2892.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.el-Zaatari M, Pasarell L, McGinnis M R, Buckner J, Land G A, Salkin I F. Evaluation of the updated Vitek yeast identification data base. J Clin Microbiol. 1990;28:1938–1941. doi: 10.1128/jcm.28.9.1938-1941.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fell J W, Boekhout T, Fonseca A, Scorzetti G, Statzell-Tallman A. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int J Syst Evol Microbiol. 2000;50:1351–1371. doi: 10.1099/00207713-50-3-1351. [DOI] [PubMed] [Google Scholar]
- 6.Fenn J P, Segal H, Barland B, Denton D, Whisenant J, Chun H, Christofferson K, Hamilton L, Carroll K. Comparison of updated Vitek Yeast Biochemical Card and API 20C yeast identification systems. J Clin Microbiol. 1994;32:1184–1187. doi: 10.1128/jcm.32.5.1184-1187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fonseca A, Scortzetti G, Fell J W. Diversity in the yeast Cryptococcus albidus and related species as revealed by ribosomal DNA sequence analysis. Can J Microbiol. 2000;46:7–27. [PubMed] [Google Scholar]
- 8.Gilfillan G D, Sullivan D J, Haynes K, Parkinson T, Coleman D C, Gow N A. Candida dubliniensis: phylogeny and putative virulence factors. Microbiology. 1998;144:829–838. doi: 10.1099/00221287-144-4-829. [DOI] [PubMed] [Google Scholar]
- 9.Gleason T G, May A K, Caparelli D, Farr B M, Sawyer R G. Emerging evidence of selection of fluconazole-tolerant fungi in surgical intensive care units. Arch Surg. 1997;132:1197–1201. doi: 10.1001/archsurg.1997.01430350047008. [DOI] [PubMed] [Google Scholar]
- 10.Gueho E, Improvisi L, Christen R, de Hoog G S. Phylogenetic relationships of Cryptococcus neoformans and some related basidiomycetous yeasts determined from partial large subunit rRNA sequences. Antonie Leeuwenhoek. 1993;63:175–189. doi: 10.1007/BF00872392. [DOI] [PubMed] [Google Scholar]
- 11.Irobi J, Schoofs A, Goossens H. Genetic identification of Candida species in HIV-positive patients using the polymerase chain reaction and restriction fragment length polymorphism analysis of its DNA. Mol Cell Probes. 1999;14:401–406. doi: 10.1006/mcpr.1999.0266. [DOI] [PubMed] [Google Scholar]
- 12.Kurtzman C P. Four new yeasts in the Pichia anomala clade. Int J Syst Evol Microbiol. 2000;50:395–404. doi: 10.1099/00207713-50-1-395. [DOI] [PubMed] [Google Scholar]
- 13.Kurtzman C P, Robnett C J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Leeuwenhoek. 1998;73:331–371. doi: 10.1023/a:1001761008817. [DOI] [PubMed] [Google Scholar]
- 14.Kurtzman C P, Robnett C J. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol. 1997;35:1216–1223. doi: 10.1128/jcm.35.5.1216-1223.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon-Chung K J, Hicks J B, Lipke P N. Evidence that Candida stellatoidea type II is a mutant of Candida albicans that does not express sucrose-inhibitable α-glucosidase. Infect Immun. 1990;58:2804–2808. doi: 10.1128/iai.58.9.2804-2808.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin D, Wu L C, Rinaldi M G, Lehmann P F. Three distinct genotypes within Candida parapsilosis from clinical sources. J Clin Microbiol. 1995;33:1815–1821. doi: 10.1128/jcm.33.7.1815-1821.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lodder J, editor. The yeasts: a taxonomic study. 2nd ed. Amsterdam, The Netherlands: North-Holland Publishing Company; 1970. [Google Scholar]
- 18.Lott T J, Kuykendall R J, Reiss E. Nucleotide sequence analysis of the 5.8S rDNA and adjacent ITS2 region of Candida albicans and related species. Yeast. 1993;9:1199–1206. doi: 10.1002/yea.320091106. [DOI] [PubMed] [Google Scholar]
- 19.Masih E I, Alie I, Paul B. Can the grey mould disease of the grape-vine be controlled by yeast? FEMS Microbiol Lett. 2000;189:233–237. doi: 10.1111/j.1574-6968.2000.tb09236.x. [DOI] [PubMed] [Google Scholar]
- 20.McCullough M J, Clemons K V, Stevens D A. Molecular and phenotypic characterization of genotypic Candida albicans subgroups and comparison with Candida dubliniensis and Candida stellatoidea. J Clin Microbiol. 1999;37:417–421. doi: 10.1128/jcm.37.2.417-421.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metzgar D, van Belkum A, Field D, Haubrich R, Wills C. Random amplification of polymorphic DNA and microsatellite genotyping of pre- and posttreatment isolates of Candida spp. from human immunodeficiency virus-infected patients on different fluconazole regimens. J Clin Microbiol. 1998;36:2308–2313. doi: 10.1128/jcm.36.8.2308-2313.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagahama T, Hamamoto M, Nakase T, Horikoshi K. Kluyveromyces nonfermentans sp. nov., a new yeast species isolated from the deep sea. Int J Syst Bacteriol. 1999;49:1899–1905. doi: 10.1099/00207713-49-4-1899. [DOI] [PubMed] [Google Scholar]
- 23.Oda Y, Yabuki M, Tonomura K, Fukunaga M. A phylogenetic analysis of Saccharomyces species by the sequence of 18S–28S rRNA spacer regions. Yeast. 1997;13:1243–1250. doi: 10.1002/(SICI)1097-0061(199710)13:13<1243::AID-YEA173>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 24.Pfaller M A, Messer S A, Houston A, Rangel-Frausto M S, Wiblin T, Blumberg H M, Edwards J E, Jarvis W, Martin M A, Neu H C, Saiman L, Patterson J E, Dibb J C, Roldan C M, Rinaldi M G, Wenzel R P. National epidemiology of mycoses survey: a multicenter study of strain variation and antifungal susceptibility among isolates of Candida species. Diagn Microbiol Infect Dis. 1998;31:289–296. doi: 10.1016/s0732-8893(97)00245-9. [DOI] [PubMed] [Google Scholar]
- 25.Reiss E, Tanaka K, Bruker G, Chazalet V, Coleman D, Debeaupuis J P, Hanazawa R, Latge J P, Lortholary J, Makimura K, Morrison C J, Murayama S Y, Naoe S, Paris S, Sarfati J, Shibuya K, Sullivan D, Uchida K, Yamaguchi H. Molecular diagnosis and epidemiology of fungal infections. Med Mycol. 1998;36(Suppl. 1):249–257. [PubMed] [Google Scholar]
- 26.Scorzetti G, Petrescu I, Yarrow D, Fell J W. Cryptococcus adeliensis sp. nov., a xylanase producing basidiomycetous yeast from Antarctica. Antonie Leeuwenhoek. 2000;77:153–157. doi: 10.1023/a:1002124504936. [DOI] [PubMed] [Google Scholar]
- 27.Sugita T, Nishikawa A, Ikeda R, Shinoda T. Identification of medically relevant Trichosporon species based on sequences of internal transcribed spacer regions and construction of a database for Trichosporon identification. J Clin Microbiol. 1999;37:1985–1993. doi: 10.1128/jcm.37.6.1985-1993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugita T, Takashima M, Ikeda R, Nakase T, Shinoda T. Intraspecies diversity of Cryptococcus laurentii as revealed by sequences of internal transcribed spacer regions and 28S rRNA gene and taxonomic position of C. laurentii clinical isolates. J Clin Microbiol. 2000;38:1468–1471. doi: 10.1128/jcm.38.4.1468-1471.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugita T, Takashima M, Ikeda R, Nakase T, Shinoda T. Phylogenetic and taxonomic heterogeneity of Cryptococcus humicolus by analysis of the sequenes of the internal transcribed spacer regions and 18S rDNA, and the phylogenetic relationships of C. humicolus, C. curvatus, and the genus Trichosporon. Microbiol Immunol. 2000;44:455–461. doi: 10.1111/j.1348-0421.2000.tb02520.x. [DOI] [PubMed] [Google Scholar]
- 30.Takashima M, Nakase T. Four new species of the genus Sporobolomyces isolated from leaves in Thailand. Mycoscience. 2000;41:357–369. [Google Scholar]
- 31.Takashima M, Nakase T. Molecular phylogeny of the genus Cryptococcus and related species based on the sequences of 18S rDNA and internal transcribed spacer regions. Microbiol Cult Coll. 1999;15:35–47. [Google Scholar]
- 32.Wickes B L, Golin J E, Kwon-Chung K J. Chromosomal rearrangement in Candida stellatoidea results in a positive effect on phenotype. Infect Immun. 1991;59:1762–1771. doi: 10.1128/iai.59.5.1762-1771.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams D W, Wilson M J, Lewis M A O, Potts A J C. Identification of Candida species in formalin fixed, paraffin wax embedded oral mucosa by sequencing of ribosomal DNA. Mol Pathol. 1996;49:M23–M28. doi: 10.1136/mp.49.1.m23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright W L, Wenzel R P. Nosocomial Candida. Epidemiology, transmission, and prevention. Infect Dis Clin N Am. 1997;11:411–425. doi: 10.1016/s0891-5520(05)70363-9. [DOI] [PubMed] [Google Scholar]