Abstract
Transfer of influenza A viruses from animal hosts to man may lead to the emergence of new human pandemic strains. The early detection and identification of such events are therefore paramount in the surveillance of influenza viruses. To detect and partially characterize influenza A viruses from different animal species, a combined reverse transcription (RT)-PCR heteroduplex mobility assay (HMA) was designed. This M gene RT-PCR was shown to be sensitive and specific for the detection of human, avian, and swine influenza A viruses. PCR amplicons from human, avian, and swine viruses of 15 different subtypes, with between 1.9 and 21.4% nucleotide divergence, were differentiated by HMA. Sequencing of the amplicons showed that the heteroduplex mobility patterns correlated with the sequence divergence between test and reference DNA. The application of the RT-PCR HMA method for rapid screening of samples was assessed with a reference panel of viruses of human, avian, and swine origin. The avian H9N2 virus A/HongKong/1073/99, which crossed the species barrier to humans, was screened against the reference panel. It was found to be most closely related to the avian A/Quail/HongKong/G1/97 H9N2 reference PCR product. Sequence analysis showed a nucleotide divergence of 1.1% between the A/Quail/HongKong/G1/97 and A/HongKong/1073/99 amplicons. From the results of our work, we consider the RT-PCR HMA method described to offer a rapid and sensitive means for screening for novel or unusual influenza viruses.
Influenza viruses are enveloped, negative-sense RNA viruses, which are classified into types A, B, and C. Influenza A viruses are further classified into subtypes on the basis of the antigenic properties of their two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA). To date, 15 HA and 9 NA subtypes have been identified. All influenza A virus subtypes have been found in aquatic and domestic birds, but only a few subtypes have been recovered from mammals and humans. In contrast, the natural host for influenza B and C viruses is man; influenza B has also been found to infect seals (28), and influenza C has been isolated from pigs (21).
The influenza A virus genome consists of eight single-stranded RNA segments, which code for a minimum of 10 gene products. The segmented nature of the genome allows reassortment of genes when different strains infect one host. Genetic reassortment, resulting in the generation of novel antigenic variants in humans, is known as antigenic shift. Several such antigenic shifts have occurred. For example, the catastrophic influenza pandemic of 1918 to 1920 apparently followed the introduction of an avian-like H1N1 virus into humans (33). Also, influenza viruses responsible for both the 1957 and 1968 human pandemics were generated by genetic reassortment between human and avian viruses (23, 35).
Since the first report of interspecies transmission of swine viruses to humans in 1974 (37), there have been sporadic isolations of them from humans, most notably from soldiers infected during an epidemic at Fort Dix in the United States, in 1976 (41). Subsequently, influenza A H3N2 avian-human reassortant viruses from pigs were shown to be responsible for two cases of influenza in young children in The Netherlands in 1993 (10). These reassortants possessed avian-like genes encoding the internal proteins, and they possessed human-like HA and NA genes.
Transmission of wholly avian viruses directly to humans, without passing through an intermediate host such as the pig, was thought to be restricted by human cell receptor specificity. However, infection of humans with avian influenza A viruses has now been documented. First, an avian influenza A H5N1 virus was transmitted from poultry to humans in 1997 (8). All eight gene segments of the virus were avian in origin (40), and the virus was highly pathogenic in poultry and killed 6 of the 18 people infected (39, 45). Second, early in 1999, the isolation of avian H9N2 viruses from five patients with influenza-like illness in China was reported (20); and in March 1999 in Hong Kong, influenza A H9N2 viruses (A/HongKong/1073/99 and A/HongKong/1074/99) were isolated from two children with self-limiting upper-respiratory infections (1, 30). All eight genes of the viruses from the two children in Hong Kong were of avian origin, the six genes encoding the internal components of these viruses being similar to those of the 1997 H5N1 human and avian isolates (26).
Although these interspecies transmission events are uncommon, they highlight the requirement for early identification of the organism responsible for outbreaks of respiratory infection. Rapid detection and characterization of influenza viruses are essential for comparison of new variants with recently circulating strains and with vaccine strains. For this purpose, influenza viruses isolated in diagnostic laboratories are routinely sent to reference laboratories for antigenic and genetic analysis. Identification of influenza viruses usually involves growth of virus in tissue culture or embryonated hens' eggs, prior to typing and subtyping by hemagglutination inhibition (HI) assays. However, this is time consuming and does not necessarily identify the species of origin of the virus. For this reason, we have developed a reverse transcription (RT)-PCR assay based on the matrix (M) gene, coupled with heteroduplex mobility assays (HMA) on the PCR product. The procedure from purification of viral nucleic acid to identification by HMA takes 24 to 36 h and can be performed directly with material in clinical specimens. The RT-PCR HMA described here will assist early recognition of interspecies transmission between animal and human hosts.
MATERIALS AND METHODS
Virus isolates.
Influenza viruses were grown in the allantoic cavities of embryonated hen's eggs. Virus-containing allantoic fluid was harvested and stored at −70°C until required. The viruses used in this study are listed in Table 1. The viruses A/Sw/Taiwan/7310/70, A/Sw/OMS/2899/82, A/Sw/OMS/3633/84, A/Sw/Scotland/410440/94, A/Sw/HongKong/168/93, A/HK/1774/99, and A/HK/1073/99 were kindly provided by Yipu Lin and Alan Hay, National Institute for Medical Research, Mill Hill, United Kingdom. The isolate A/Duck/Singapore-Q/F119-3/97 was kindly provided by the Director of Primary Production, Veterinary Laboratory Branch, Central Veterinary Laboratory, Singapore.
TABLE 1.
Influenza A viruses used in this study
| Virus | Abbreviation | Influenza A subtype | Virus host speciesa | M gene host speciesb |
|---|---|---|---|---|
| A/Bayern/7/95 | Bay/95 | H1N1 | H | H |
| A/Wuhan/371/95 | Wuh/371/95 | H1N1 | H | H |
| A/Sydney/5/97 | Syd/97 | H3N2 | H | H |
| A/Wuhan/359/95 | Wuh/359/95 | H3N2 | H | H |
| A/HongKong/1073/99 | HK/1073/99 | H9N2 | H | Av |
| A/HongKong/1774/99 | HK/1774/99 | H3N2 | H | Sw |
| A/Swine/Taiwan/7310/70 | Sw/Tw/70 | H3N2 | Sw | H |
| A/Swine/OMS/2899/82 | Sw/OMS/82 | H1N1 | Sw | Sw |
| A/Swine/OMS/3633/84 | Sw/OMS/84 | H3N2 | Sw | Sw |
| A/Swine/HongKong/168/93 | Sw/HK/93 | H1N1 | Sw | Av |
| A/Swine/Scotland/410440/94 | Sw/Scot/94 | H1N2 | Sw | Sw |
| A/Duck/Germany/1215/73 | Dk/Ger/73 | H2N3 | Av | Av |
| A/Duck/Ukraine/1/63 | Dk/Ukr/63 | H3N8 | Av | Av |
| A/Duck/Singapore-Q/F119-3/97 | Dk/Sing/97 | H5N3 | Av | Av |
| A/Duck/Potsdam/1402-6/86 | Dk/Pots/86 | H5N2 | Av | Av |
| A/Shearwater/Australia/1/72 | Shearw/Aus/72 | H6N5 | Av | Av |
| A/Conure/England/1234/94 | Con/Eng/94 | H7N1 | Av | Av |
| A/AfricanStarling/Q-England/983/79 | AfrStarl/Eng/79 | H7N1 | Av | Av |
| A/Turkey/Ontario/6118/68 | Ty/Ont/68 | H8N4 | Av | Av |
| A/Turkey/Wisconsin/1/66 | Ty/Wis/66 | H9N2 | Av | Av |
| A/Quail/HongKong/G1/97 | Qa/HK/G1/97 | H9N2 | Av | Av |
| A/Chicken/Germany/N/49 | Ck/Ger/49 | H10N1 | Av | Av |
| A/Turkey/Weybridge/PD250/79 | Ty/Wey/79 | H11N1 | Av | Av |
| A/Duck/Alberta/60/76 | Dk/Alb/76 | H12N5 | Av | Av |
| A/Gull/Maryland/704/77 | Gull/MD/77 | H13N6 | Av | Av |
Host of origin of the virus. H, human; Av, avian; Sw, swine.
Host of origin of the M gene segment.
Virus typing.
Influenza viruses were typed using ferret antisera in HI tests as described previously (7). All HI tests were carried out using eight HA U of virus and 0.5% (vol/vol) turkey red blood cells. All ferret sera were treated with receptor-destroying enzyme.
Nucleic acid extraction and cDNA synthesis.
Viral RNA was extracted from a 150-μl volume of sample by a guanidinium thiocyanate-silica binding method as described previously (4). Viral RNA was eluted in 30 μl of nuclease-free water (Promega, Southampton, England). RT of RNA to cDNA was performed by addition of 22.2 μl of RNA to 17.8 μl of RT mix containing 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 7.5 mM MgCl2 (Gibco BRL, Paisley, Scotland), 3 mM (each) deoxynucleoside triphosphate (Sigma-Aldrich, Dorset, England), 25 ng of random primer (pdN6) (Amersham Pharmacia Biotech, Little Chalfont, England), 1.6 U of RNasin (Promega), and 200 U of Moloney murine leukemia virus reverse transcriptase (Gibco BRL). The reaction mixture was incubated at 20°C for 10 min and then at 37°C for 45 min. Samples were then heated to 100°C for 5 min and quenched on ice.
Nested PCR.
Primer sequences were deduced following analysis of conserved regions of the M gene of influenza A viruses. The properties of the primers were analyzed with Oligo primer analysis software (version 6.0; National Biosciences Inc.). The primer pairs selected for use in the M gene nested RT-PCR are shown in Table 2. The outer primer, AMP71F, had previously been designed for use in a nested PCR to detect human influenza A viruses (47). Amplification using each primer pair was performed under a range of MgCl2, salt, and pH conditions (PCR Optimisation Kit II; Sigma-Aldrich). For the primary PCR, 20 μl of cDNA was added to 80 μl of PCR mix containing 8 μl of 10× PCR buffer (200 mM Tris-HCl [pH 8.4], 500 mM KCl [Gibco BRL]), 2.4 μl of 50 mM MgCl2, 1 μl (each) of outer primer at 5 pmol/μl (AMP71F; AMP831R), 67.3 μl of nuclease-free water, and 0.3 μl of Taq DNA polymerase (Gibco BRL). Optimal annealing temperatures for the primer pairs and cycling conditions were determined using a MasterCycler Gradient thermal cycler (Eppendorf, Cambridge, England). Primary amplification was performed by 1 cycle at 94°C for 2 min, followed by 30 cycles of denaturation at 94°C for 1 min, and combined annealing and extension at 68°C for 1.5 min. The primary products amplified from egg-grown viruses were diluted 1 in 10,000 prior to secondary amplification. An aliquot of the primary product (2 μl) was then transferred to 48 μl of secondary PCR mix containing the following: 5 μl of 10× PCR buffer (200 mM Tris-HCl [pH 8.4], 500 mM KCl [Gibco BRL]), 1 μl of deoxynucleoside triphosphate mix, 1.5 μl of 50 mM MgCl2, 3 μl (each) of inner primer at 25 pmol/μl (AMP227F; AMP622R), 34.2 μl of nuclease-free water, and 0.3 μl of Taq DNA polymerase (Gibco BRL). The samples were incubated at 94°C for 1 min and then subjected to 35 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min. Amplicons (413 bp) were visualized by ethidium bromide staining following electrophoresis on 2% agarose gels (Seakem GTG; Flowgen FMC Bioproducts, Lichfield, England).
TABLE 2.
Properties of primers used for influenza A virus M gene RT-PCR
| Primer | Sequence (5′–3′) | Gene position | Tma (°C) | Optimal annealing tempb (°C) | Maximum annealing temp (°C) | Amplicon size (bp) |
|---|---|---|---|---|---|---|
| AMP71F | CCGTCAGGCCCCCTCAAAGC | M1 | 78.8 | 56.3 | 72.0 | 780 |
| AMP831R | AGGCGATCAAGAATCCACAA | M2 | 69.1 | |||
| AMP227F | GTGCCCAGTGAGCGAGGAC | M1 | 73.0 | 55.3 | 69.8 | 413 |
| AMP622R | ATCTCCATGGCCTCTGCT | M1 | 66.8 |
Tm, melting temperature of an oligonucleotide complex calculated by the nearest-neighbor method.
The annealing temperature that gives the highest product yield when no false priming and primer dimerization occur (calculated by Oligo version 6.0 software).
Determination of RT-PCR assay sensitivity. (i) Titration of virus infectivity.
Infectivity assays were performed using previously described methods (38), with minor modifications. Ten-fold dilutions (10−1 to 10−12) of the viruses Syd/97 (H3N2), Dk/Sing/97 (H5N3), and HK/1073/99 (H9N2) were prepared in viral transport medium (VTM). The incubation time was modified to 72 h in a CO2 incubator at 37°C. Madin-Darby canine kidney cells were fixed with 5% glutaraldehyde, and plaques were visualized by staining with 5% vol/vol carbol fuchsin solution.
(ii) RT-PCR.
Freshly prepared aliquots (100 μl) of each virus dilution in VTM were subjected to nucleic acid extraction and RT-PCR as described above.
Heteroduplex mobility assays.
HMA analysis was adapted from previously described methods for the study of human immunodeficiency virus type 1 quasispecies (12). For HMA, 3 μl of sample PCR product was mixed with 3 μl of reference PCR product and 0.3 μl of HMA buffer containing 1 M NaCl, 100 mM Tris (pH 7.8), and 20 mM EDTA. Samples were then denatured at 95°C for 2 min, rapidly cooled to 4°C, and then placed on ice for 10 min. Gel loading buffer (1.3 μl) was then added (6×; Tris-boric acid-EDTA [TBE], glycerol, 1% bromophenol blue, 1% xylene cyanol). Samples were electrophoresed on 8% nondenaturing polyacrylamide gels in 1× TBE (NOVEX TBE gels) (Invitrogen, Groningen, The Netherlands) for 50 min at 200 V. Homoduplexes and heteroduplexes were visualized following staining of the gel with SYBR Green II nucleic acid stain (Invitrogen).
Sequencing and phylogenetic analysis.
Nucleotide sequencing of M gene PCR amplicons was performed using the Dye Deoxy terminator method (PE Applied Biosystems, Warrington, England) (17). The M gene PCR products were sequenced using the PCR inner primer pair AMP227F and AMP622R, at 3.2 pmol/reaction. Neighbour-joining phylogenetic analysis (34) was performed following the alignment of nucleotide sequences using the program Megalign 4.0 (DNASTAR Inc, Madison, Wis.).
RESULTS
M gene RT-PCR—sensitivity and specificity.
The sensitivity of detection of influenza A virus by the M gene RT-PCR was determined using influenza A viruses of three subtypes, Syd/97 (H3N2), Dk/Sing/97 (H5N3), and HK/1073/99 (H9N2). Dilution series of these freshly harvested viruses were prepared in VTM. From each dilution, nucleic acid was extracted for cDNA synthesis and PCR. An equivalent volume was taken from each dilution for infectivity assays, which were performed on the same day. In this manner, the end point of detection of infectious virus could be directly compared to the end point of detection of viral RNA by RT-PCR. The M gene nested RT-PCR detected ≤10 PFU of influenza A H3N2, H5N3, or H9N2 virus/ml (data not shown). This is comparable to the sensitivity reported for detection of influenza A or B viruses by nested RT-PCR assays, using primers specific for influenza A or B virus HA genes (18).
The RT-PCR was tested for its ability to detect influenza A viruses from different animal species and for its specificity for influenza A viruses. A product of the expected size (413 bp) was obtained when influenza A viruses of avian, human, or swine origin (Table 1) were tested (Fig. 1). These results show that M gene sequences of influenza A viruses from all animal species tested can be amplified with the primers listed in Table 2. No detectable PCR products were observed when influenza B viruses were tested (data not shown), indicating the specificity of the PCR for influenza A virus sequences.
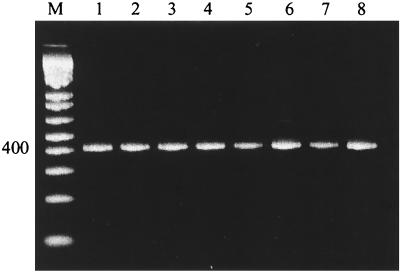
FIG. 1.
Detection of influenza A viruses from different animal species, by M gene RT-PCR. Influenza A viruses with M genes of human, swine, and avian origin (Table 1) were assayed by RT-PCR with primers specific for the M gene of all influenza A viruses. Amplicons (413 bp) were analyzed by electrophoresis on a 2% agarose gel stained with ethidium bromide. Lanes: 1, Wuh/359/95 (human); 2, Wuh/371/95 (human); 3, Sw/Tw/70 (human); 4, Sw/OMS/82 (swine); 5, Sw/OMS/84 (swine); 6, Dk/Ger/73 (avian); 7, Dk/Ukr/63 (avian); 8, Shearw/Aus/72 (avian). M, DNA molecular weight markers.
Characterization of M gene amplicons by HMA and sequence confirmation.
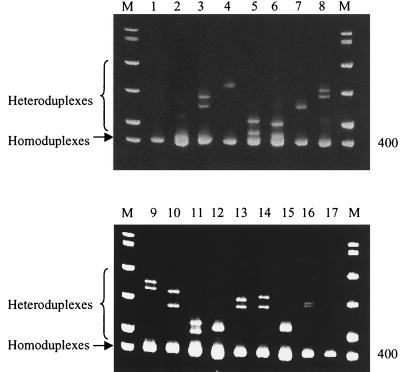
After amplification of the M gene by RT-PCR, the host species of origin of the PCR product was investigated by HMA. This was achieved by heteroduplex formation between amplicons of a reference human influenza A virus (Bay/95) and amplicons of test viruses of 15 different influenza A virus subtypes of either human, swine, or avian origin (Table 1). The results are shown in Fig. 2. Heteroduplexes were observed between amplicons of the reference strain Bay/95 and each test virus PCR product (lanes 2 to 16), reflecting the sequence variation between the reference virus and test virus DNA. No heteroduplexes were observed in lanes that contained reference PCR product only (lanes 1 and 17). The smallest reduction in heteroduplex mobility was observed where the reference virus (Bay/95) PCR product was mixed with the amplicon of the virus Wuh/371/95 (lane 2). Both Bay/7/95 and Wuh/371/95 viruses are human influenza A virus H1N1 strains. Sequence analysis indicated that the M gene amplicons of Bay/95 and Wuh/371/95 differ by 1.9%. (Table 3). Therefore, amplicons with <2% variation in nucleotide sequence could be differentiated by this HMA. No improvement in band resolution was seen when 10%, 20%, or gradient polyacrylamide gels were used (data not shown). The greatest retardation in mobility of heteroduplexes was seen in lanes where the reference amplicon from Bay/95 was mixed with PCR products from viruses that possess M genes of swine or avian origin (lanes 3, 4, and 7 to 16). The nucleotide divergence between the PCR amplicons from these swine and avian viruses and the reference human influenza strain Bay/95 was shown by sequence analysis to be between 14.5 and 21.4% (Table 3). Intermediate heteroduplex mobility patterns were observed in mixes containing PCR amplicons from the reference human H1N1 virus (Bay/95) and test human H3N2 viruses, Syd/97 and Wuh/359/95 (lanes 5 and 6, respectively). Sequence analysis revealed a nucleotide divergence of 6.7 to 7.9% between the reference human H1N1 virus and test human H3N2 virus M gene amplicons (Table 3).
FIG. 2.
Heteroduplex formation between influenza A viruses from different host species with a human H1N1 reference strain, Bay/95. Amplicons from viruses assayed by the M gene RT-PCR were mixed with the reference virus amplicon, heat denatured, cooled, and then analyzed by electrophoresis on 8% TBE polyacrylamide gels. Homoduplexes and heteroduplexes were visualized by staining with SYBR Green II. Lanes show Bay/95 mixed with H2O (lane 1), Wuh/371/95 (lane 2), Sw/OMS/82 (lane 3), Dk/Ger/73 (lane 4), Wuh/359/95 (lane 5), Syd/97 (lane 6), Dk/Ukr/63 (lane 7), Dk/Sing/97 (lane 8), Shearw/Aus/72 (lane 9), AfrStarl/Eng/79 (lane 10), Ty/Ont/68 (lane 11), Ty/Wis/66 (lane 12), Ck/Ger/49 (lane 13), Ty/Wey/79 (lane 14), Dk/Alb/76 (lane 15), Gull/MD/77 (lane 16), and Bay/95 (lane 17). M, DNA molecular weight markers.
TABLE 3.
Nucleotide divergence between M gene PCR amplicons
| M gene lineage | % Divergence in sequencea
|
|||
|---|---|---|---|---|
| Human
|
European swine (avian-like) | Avian | ||
| H1 | H3 | |||
| Human (H1) | 1.9 | 5.2–7.9 | 18.1–21.3 | 12.8–21.7 |
| Human (H3) | 2.2–4.0 | 16.2–21.0 | 10.8–19.9 | |
| European swine (avian-like) | 1.4–6.1 | 4.9–16.9 | ||
| Avian | 0.0–15.8 | |||
Viruses in Table 1 were analyzed by the M gene RT-PCR. Amplicons were sequenced, and divergence between sequences was determined using Megalign version 4.0 software. Except where a single value is given, values are ranges.
Evaluation of an HMA reference panel for detection of interspecies transmission.
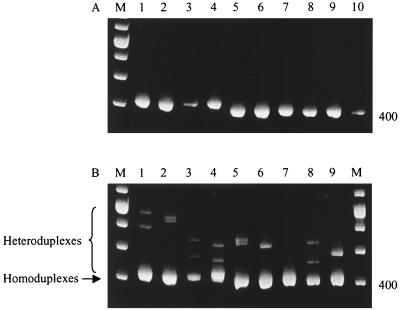
A panel of nine reference influenza A viruses with M genes representative of circulating human, swine, and avian lineages was constructed. RNA was extracted from these and a test virus, HK/1073/99, and subjected to RT-PCR. Amplicons of the expected size (413 bp) were visualized by agarose gel electrophoresis (Fig 3A). An aliquot of each reference amplicon was used for HMA with the test virus (HK/1073/99) PCR product. The results are shown in Fig. 3B. The greatest reduction in heteroduplex mobility was seen when the HK/1073/99 amplicon was mixed with human reference virus amplicons (lanes 1 and 2), indicating the large sequence distance of the M gene of the avian virus from human M gene sequences. The heteroduplexes formed when the HK/1073/99 virus PCR product was mixed with reference amplicons derived from swine and avian viruses showed various mobility patterns (lanes 3 to 9). The greatest similarity was with the PCR amplicon from the avian H9N2 virus Qa/HK/GI/97 (lane 7). No heteroduplexes were observed when the Qa/HK/G1/97 and HK/1073/99 amplicons were mixed, indicating that the divergence between these amplicons was less than the sensitivity of the HMA. The HMA results were confirmed by direct sequencing of the M gene PCR amplicons of the nine reference viruses and the test virus. Sequence analysis revealed that the nucleotide divergence between the Qa/HK/G1/97 and HK/1073/99 amplicons was 1.1%.
FIG. 3.
Characterization of the virus A/HK/1073/99 by HMA. (A) A panel of nine reference viruses from different host species and the test virus, A/HK/1073/99, were assayed by the M gene RT-PCR. Amplicons of the expected size (413 bp) were visualized on 2% agarose gels stained with ethidium bromide. Lanes: 1, Syd/97; 2, Bay/95; 3, HK/1774/99; 4, Sw/Scot/94; 5, Ty/Wis/66; 6, Dk/Alb/76; 7, Qa/HK/G1/97; 8, Dk/Sing/97; 9, Sw/HK/93; 10, HK/1073/99. (B) Lanes 1 to 9: amplicons of the reference viruses as in panel A and A/HK/1073/99 were mixed and subjected to HMA. Homoduplexes and heteroduplexes were analyzed by polyacrylamide gel electrophoresis on an 8% TBE gel. M, DNA molecular weight markers.
DISCUSSION
Although transmission to humans of influenza viruses of swine or avian origin is infrequent, such events can result in high mortality and have the potential for the generation of pandemic viruses. The early detection and characterization of newly emerging influenza variants are one of the aims of the World Health Organization global influenza surveillance network. Hence, there is a requirement for rapid tests that are able to detect and characterize novel or unusual viruses, which may have originated from an animal reservoir. Rapid methods, such as immunofluorescence and enzyme immunoassays, have been applied to the detection of influenza viruses in clinical material. These assays have been reported to have variable sensitivity and specificities (46), and it is unclear whether the reagents involved are equally capable of detecting different animal subtypes of influenza virus. The value of PCR assays for the detection and surveillance of influenza viruses in clinical material has been clearly demonstrated (6, 18, 32, 36). Amplification assays for detecting and subtyping influenza A viruses (2, 16, 18, 31, 44) and for differentiating influenza A, B, and C viruses (9) have been described. However, these assays have been designed specifically for the diagnosis and surveillance of human influenza viruses, and there is an additional need for rapid tests to detect viruses originating from nonhuman hosts. Recently a single-tube RT-PCR based on the M gene for the detection of influenza A viruses from multiple species (19) has been described.
In this study, we designed a combined PCR-HMA method to identify and partially characterize influenza A viruses from different species. The M gene was chosen as the target, since evidence suggests that the M1 open reading frame is highly conserved among influenza A viruses from different host species (22, 25). The M gene RT-PCR was shown to be sensitive and specific for the detection of influenza A viruses of 15 different subtypes, of human, avian, and swine origin.
Postamplification methods for rapidly analyzing sequence variation in PCR amplicons include restriction fragment length polymorphism (RFLP) analysis and HMAs. RFLP assays of M gene PCR products have been described for the subtyping of human influenza A viruses (29) and for the differentiation of the six internal genes, including the M gene of human H1N1 and H3N2 and avian H5N1 viruses (11). A possible drawback of the RFLP technique is that mutations in the nucleotide sequence of the gene of interest may lead to the loss or generation of a restriction site. The high mutability of RNA genomes, such as influenza viral genes, increases the possibility of this occurring. Since heteroduplex analysis has been shown to be ideally suited to the differentiation of rapidly evolving RNA viruses (14, 15, 24, 27, 43, 48, 49), and because no suitable restriction sites that differentiated human, avian, and swine viruses were identified in the M gene 413-bp amplicon, HMA was chosen for characterization of the M gene amplicon. HMA is based on the observation that sequence variation between amplicons from a test virus and that of the reference strain generates mismatches, and therefore heteroduplexes, following denaturation and reannealing. The heteroduplexes migrate more slowly than the homoduplex of the same size, and the reduction in mobility is proportional to the divergence between the two sequences in the mixture. The degree of variation required for good discrimination of heteroduplexes has been shown to be within the range of 5 and 25% (13). The degree of divergence between human, swine, and avian M gene amplicons is within this range, allowing detectable heteroduplexes to be formed (Table 3). The heteroduplex mobility shifts observed correlated with the sequence divergence between reference and test DNA, with as little as 1.1 to 1.9% nucleotide divergence being detectable. This is comparable with previous reports of the detection of nucleotide divergence of 1.4 to 4% (5, 14, 24, 42). In this work, the best discrimination by HMA was seen when the primary PCR products were diluted prior to secondary amplification. It is envisaged that this will not be necessary when directly testing clinical samples, which contain lower virus titers than egg- or cell-grown material.
The suitability of our RT-PCR HMA method for screening samples in an outbreak was assessed by challenging a test virus with a panel of reference viruses. The avian influenza A H9N2 virus strain, HK/1073/99, was chosen as the test virus, since this avian virus was isolated from a 4-year-old child in Hong Kong (1, 30). There was an excellent correlation between the HMA result and nucleotide divergence, as determined by direct sequencing and phylogenetic analysis of the reference and test amplicons. By HMA, the HK/1073/99 M gene PCR amplicon was most closely related to the Qa/HK/G1/97 reference PCR product. This correlates with the previously reported 1% nucleotide divergence between the Qa/HK/G1/97 and HK/1073/99 M genes (26).
In conclusion, the results of our work show that the combined M gene RT-PCR HMA is a powerful tool for the identification and genetic characterization of influenza viruses. HMA is a simple and sensitive means for screening the M genes of influenza virus isolates and takes only 24 to 36 h to perform. Although the RT-PCR HMA method was not used to analyze respiratory samples in this study, it is envisaged that the method could be applied to test viruses directly in clinical specimens, obviating the need to grow the virus first in cell culture or eggs. The sensitivity of the M gene nested RT-PCR described is comparable with sensitivities for nested RT-PCR assays which use primers specific for influenza A and B virus HA genes, to detect influenza A and B viruses directly in clinical specimens (18, 32, 38). Novel or unusual influenza viruses identified by HMA can be more extensively characterized by direct sequencing and HI typing. The HMA reference virus panel we constructed will need to be updated to reflect viruses known to be circulating in different animal species. Finally, the combined RT-PCR heteroduplex technique could also be applied to the identification of the other internal genes of influenza viruses.
ACKNOWLEDGMENTS
We thank Catherine Thompson and Katrina Sleeman for performing the virus infectivity assays; Jon Green, Chris Gallimore, and Katrina Barlow for their technical advice; and Ian Brown for strain nomenclature information.
REFERENCES
- 1.Anonymous. Influenza. Wkly Epidemiol Rec. 1999;74:111. [Google Scholar]
- 2.Atmar R L, Baxter B D, Dominguez E A, Taber L H. Comparison of reverse transcription-PCR with tissue culture and other rapid diagnostic assays for detection of type A influenza virus. J Clin Microbiol. 1996;34:2604–2606. doi: 10.1128/jcm.34.10.2604-2606.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barlow K L, Green J, Clewley J P. Viral genome characterisation by the heteroduplex mobility and heteroduplex tracking assays. Rev Med Virol. 2000;10:321–335. doi: 10.1002/1099-1654(200009/10)10:5<321::aid-rmv288>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 4.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van-Dillon P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calvo P L, Kansopon J, Sra K, Quan S, DiNello R, Guaschino R, Calabrese G, Danielle F, Brunetto M R, Bonino F, Massaro A L, Polito A, Houghton M, Weiner A J. Hepatitis C virus heteroduplex tracking assay for genotype determination reveals diverging genotype 2 isolates in Italian hemodialysis patients. J Clin Microbiol. 1998;36:227–233. doi: 10.1128/jcm.36.1.227-233.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carman W F, Wallace L A, Walker J, McIntyre S, Noone A, Christie P, Millar J, Douglas J D. Rapid virological surveillance of community influenza infection in general practice. Br Med J. 2000;321:736–737. doi: 10.1136/bmj.321.7263.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakraverty P. Antigenic relationships between influenza B viruses. Bull W H O. 1971;45:755–766. [PMC free article] [PubMed] [Google Scholar]
- 8.Claas E C, Osterhaus A D, van Beek R, De Jong J, Rimmelzwaan G, Senne D, Krauss S, Shortridge K, Webster R G. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 9.Claas E C, Sprenger M J W, Kleter G E M, van Beek R, Quint W G V, Masurel N. Type-specific identification of influenza viruses A, B and C by the polymerase chain reaction. J Virol Methods. 1992;39:1–13. doi: 10.1016/0166-0934(92)90120-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claas E C J, Kawaoka Y, de Jong J C, Masurel N, Webster R G. Infection of children with avian-human reassortant influenza virus from pigs in Europe. Virology. 1994;204:453–457. doi: 10.1006/viro.1994.1553. [DOI] [PubMed] [Google Scholar]
- 11.Cooper L A, Subbarao K. A simple restriction fragment length polymorphism-based strategy that can distinguish the internal genes of human H1N1, H3N2, and H5N1 influenza A viruses. J Clin Microbiol. 2000;38:2579–2583. doi: 10.1128/jcm.38.7.2579-2583.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delwart E L, Gordon C J. Tracking changes in HIV-1 envelope quasispecies using DNA heteroduplex analysis. Methods. 1997;12:348–354. doi: 10.1006/meth.1997.0489. [DOI] [PubMed] [Google Scholar]
- 13.Delwart E L, Herring B, Rodrigo A G, Mullins J I. Genetic subtyping of human immunodeficiency virus using a heteroduplex mobility assay. PCR Methods Appl. 1995;4:S202–S216. doi: 10.1101/gr.4.5.s202. [DOI] [PubMed] [Google Scholar]
- 14.Delwart E L, Sheppard H W, Walker B D, Goudsmit J, Mullins J I. Human immunodeficiency virus type I evolution in vivo tracked by DNA heteroduplex mobility assays. J Virol. 1994;68:6672–6683. doi: 10.1128/jvi.68.10.6672-6683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rubsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay. Analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 16.Donofrio J C, Coonrod J D, Davidson J N, Betts R F. Detection of influenza A and B in respiratory secretions with the polymerase chain reaction. PCR Methods Appl. 1992;1:263–268. doi: 10.1101/gr.1.4.263. [DOI] [PubMed] [Google Scholar]
- 17.Ellis J S, Chakraverty P, Clewley J P. Genetic and antigenic variation in the haemagglutinin of recently circulating human influenza A (H3N2) viruses in the United Kingdom. Arch Virol. 1995;140:1889–1904. doi: 10.1007/BF01322680. [DOI] [PubMed] [Google Scholar]
- 18.Ellis J S, Fleming D M, Zambon M C. Multiplex reverse transcription-PCR for surveillance of influenza A and B viruses in England and Wales in 1995 and 1996. J Clin Microbiol. 1997;35:2076–2082. doi: 10.1128/jcm.35.8.2076-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fouchier R A, Bestebroer T M, Herfst S, Van Der Kemp L, Rimmelzwaan G F, Osterhaus A D. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. J Clin Microbiol. 2000;38:4096–4101. doi: 10.1128/jcm.38.11.4096-4101.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo Y, Li J, Cheng X, Wang M, Zhou Y, Li X H, Cai F, Miao H L, Zhang H, Guo F. Discovery of men infected by avian influenza A(H9N2) virus. Chin J Exp Clin Virol. 1999;13:105–108. [PubMed] [Google Scholar]
- 21.Guo Y J, Jin F G, Wang P, Wang M, Zhu J M. Isolation of influenza C virus from pigs and experimental infection of pigs with influenza C virus. J Gen Virol. 1983;64:177–182. doi: 10.1099/0022-1317-64-1-177. [DOI] [PubMed] [Google Scholar]
- 22.Ito T, Gorman O T, Kawaoka Y, Bean W J, Webster R G. Evolutionary analysis of the influenza A virus M gene with comparison of the M1 and M2 proteins. J Virol. 1991;65:5491–5498. doi: 10.1128/jvi.65.10.5491-5498.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawaoka Y, Krauss S, Webster R G. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989;63:4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kreis S, Whistler T. Rapid identification of measles virus strains by the heteroduplex mobility assay. Virus Res. 1997;47:197–203. doi: 10.1016/s0168-1702(96)01413-x. [DOI] [PubMed] [Google Scholar]
- 25.Lamb R A, Lai C-J. Conservation of influenza virus membrane protein (M1) amino acid sequence and open reading frame of RNA segment 7 encoding a second protein (M2) in H1N1 and H3N2 strains. Virology. 1981;112:746–751. doi: 10.1016/0042-6822(81)90319-6. [DOI] [PubMed] [Google Scholar]
- 26.Lin Y P, Shaw M, Gregory V, Cameron K, Lim K, Klimov A, Subbarao K, Guan Y, Krauss S, Shortridge K, Webster R, Cox N, Hay A. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc Natl Acad Sci USA. 2000;97:9654–9658. doi: 10.1073/pnas.160270697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mattick K L, Green J, Punia P, Belda F J, Gallimore C I, Brown D W. The heteroduplex mobility assay (HMA) as a pre-sequencing screen for Norwalk-like viruses. J Virol Methods. 2000;87:161–169. doi: 10.1016/s0166-0934(00)00170-1. [DOI] [PubMed] [Google Scholar]
- 28.Osterhaus A D, Rimelzwaan G F, Martina B E, Bestebroer T M, Fouchier R A. Influenza B in seals. Science. 2000;288:1051–1053. doi: 10.1126/science.288.5468.1051. [DOI] [PubMed] [Google Scholar]
- 29.Park K Y, Lee M G, Ryu J C. Evolutionary stasis of M1 gene of human influenza A viruses and the possibility of their subtyping by restriction analysis of M1 gene polymerase chain reaction product. Acta Virol. 1997;41:231–239. [PubMed] [Google Scholar]
- 30.Peiris M, Yuen K Y, Leung C W, Chan K H, Ip P L, Lai R W, Orr W K, Shortridge K F. Human infection with influenza H9N2. Lancet. 1999;354:916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 31.Pisareva M, Bechtereva T, Plyusnin A, Dobretsova A, Kisselev O. PCR-amplification of influenza A virus specific sequences. Arch Virol. 1992;125:313–318. doi: 10.1007/BF01309648. [DOI] [PubMed] [Google Scholar]
- 32.Rebelo-de-Andrade H, Zambon M C. Different diagnostic methods for detection of influenza epidemics. Epidemiol Infect. 2000;124:515–522. doi: 10.1017/s0950268899003751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reid A H, Fanning T G, Janczewski T A, Taubenberger J K. Characterization of the 1918 “Spanish” influenza virus neuraminidase gene. Proc Natl Acad Sci USA. 2000;97:6785–6790. doi: 10.1073/pnas.100140097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saito N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 35.Scholtissek C, Rohde W, Von Hoyningen V, Rott R. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology. 1978;87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- 36.Schweiger B, Zadow I, Heckler R, Timm H, Pauli G. Application of a fluorogenic PCR assay for typing and subtyping of influenza viruses in respiratory samples. J Clin Microbiol. 2000;38:1552–1558. doi: 10.1128/jcm.38.4.1552-1558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith T F, Burgert E O, Dowdle W R, Noble G R, Campbell R J, Van Scoy R E. Isolation of swine influenza virus from autopsy lung tissue of man. N Engl J Med. 1976;294:708–710. doi: 10.1056/NEJM197603252941308. [DOI] [PubMed] [Google Scholar]
- 38.Stockton J, Ellis J S, Saville M, Clewley J P, Zambon M C. Multiplex PCR for typing and subtyping influenza and respiratory syncytial viruses. J Clin Microbiol. 1998;36:2990–2995. doi: 10.1128/jcm.36.10.2990-2995.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suarez D L, Perdue M L, Cox N, Rowe T, Bender C, Huang J, Swayne D E. Comparisons of highly virulent H5N1 influenza A viruses isolated from humans and chickens from Hong Kong. J Virol. 1998;72:6678–6688. doi: 10.1128/jvi.72.8.6678-6688.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 41.Top F H, Jr, Russell P K. Swine influenza A at Fort Dix, New Jersey (January-February 1976). IV. Summary and speculation. J Infect Dis. 1977;136:S376–S380. doi: 10.1093/infdis/136.supplement_3.s376. [DOI] [PubMed] [Google Scholar]
- 42.White P A, Li Z, Rawlinson W D. Sequence diversity in the 5′UTR region of GB virus C/hepatitis G virus assessed using sequencing, heteroduplex mobility assays and single-strand conformation polymorphism. J Virol Methods. 1999;83:91–101. doi: 10.1016/s0166-0934(99)00110-x. [DOI] [PubMed] [Google Scholar]
- 43.Wilson J J, Polyak S J, Day T D, Gretch D R. Characterization of simple and complex hepatitis C virus quasispecies by heteroduplex gel shift analysis: correlation with nucleotide sequencing. J Gen Virol. 1995;76:1763–1771. doi: 10.1099/0022-1317-76-7-1763. [DOI] [PubMed] [Google Scholar]
- 44.Wright K E, Wilson G A R, Novosad D, Dimock C, Tan D, Weber J M. Typing and subtyping of influenza viruses in clinical samples by PCR. J Clin Microbiol. 1995;33:1180–1184. doi: 10.1128/jcm.33.5.1180-1184.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuen K Y, Chan P K, Peiris M, Tsang D N, Que T L, Shortridge K F, Cheung P T, To W K, Ho E T, Sung R, Cheng A F. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet. 1998;351:467–471. doi: 10.1016/s0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]
- 46.Zambon M. Laboratory diagnosis and techniques. In: Nicholson K G, Webster R G, Hay A J, editors. Textbook of influenza. Oxford, United Kingdom: Blackwell Science Ltd.; 1998. pp. 291–316. [Google Scholar]
- 47.Zhang W, Evans D H. Detection and identification of human influenza viruses by the polymerase chain reaction. J Virol Methods. 1991;33:165–189. doi: 10.1016/0166-0934(91)90017-t. [DOI] [PubMed] [Google Scholar]
- 48.Zou S. A practical approach to genetic screening for influenza virus variants. J Clin Microbiol. 1997;35:2623–2627. doi: 10.1128/jcm.35.10.2623-2627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zou S, Stansfield C, Bridge J. Identification of new influenza B virus variants by multiplex reverse transcription-PCR and the heteroduplex mobility assay. J Clin Microbiol. 1998;26:1544–1548. doi: 10.1128/jcm.36.6.1544-1548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]