Abstract
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as a global threat to human health has highlighted the need for the development of novel therapies targeting current and emerging coronaviruses with pandemic potential. The coronavirus main protease (Mpro, also called 3CLpro) is a validated drug target against coronaviruses and has been heavily studied since the emergence of SARS-CoV-2 in late 2019. Here, we report the biophysical and enzymatic characterization of native Mpro, then characterize the steady-state kinetics of several commonly used FRET substrates, fluorogenic substrates, and six of the 11 reported SARS-CoV-2 polyprotein cleavage sequences. We then assessed the suitability of these substrates for high-throughput screening. Guided by our assessment of these substrates, we developed an improved 5-carboxyfluorescein-based FRET substrate, which is better suited for high-throughput screening and is less susceptible to interference and false positives than existing substrates. This study provides a useful framework for the design of coronavirus Mpro enzyme assays to facilitate the discovery and development of therapies targeting Mpro.
Keywords: high-throughput screening, FRET, viral protease, protease substrate, SARS-CoV-2, main protease (Mpro), 3CL protease (3CLpro), enzyme kinetics, coronavirus
Abbreviations: AMC, 7-amino-4-methylcoumarin; COVID-19, coronavirus disease 2019; DABCYL, 4-(4-dimethylaminophenylazo)benzoic acid; DLS, dynamic light scattering; DMSO, dimethyl sulfoxide; DNP, 2,4-dinitrophenyl; EDANS, 5-(2-aminoethylamino)-1-naphthalenesulfonic acid; FAM, 5-carboxyfluorescein; HCoV, human coronavirus; HTS, high-throughput screen; MCA, 7-methoxycoumarin-4-acetic acid; MERS-CoV, Middle East respiratory syndrome coronavirus; Mpro, main protease; nanoDSF, nano-differential scanning fluorimetry; PDB, Protein Data Bank; PLpro, papain-like protease; pp1a, polyprotein 1a; pp1ab, polyprotein 1ab; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2
Coronaviruses are a family of viruses commonly found in wildlife, companion animals, livestock, and humans. Human coronaviruses include HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1 continuously circulating in the population and mostly cause mild symptoms associated with the common cold. In contrast, severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and SARS-CoV-2 are highly pathogenic coronaviruses causing the SARS epidemic, MERS epidemic, and most recently, the coronavirus disease 2019 (COVID-19) pandemic (1). To date, there have been over 220 million reported cases of COVID-19 and 4.6 million reported deaths. Despite the development of efficacious vaccines against COVID-19, SARS-CoV-2 transmission continues at high levels, and case numbers continue to increase (2, 3). As a result, there is an urgent need for effective antiviral drugs targeting SARS-CoV-2 that can be used to treat COVID-19.
SARS-CoV-2 is an enveloped positive-stranded RNA virus, which infects cells of the upper and lower respiratory tract. Upon entry into the host cell, the viral RNA genome is translated into two polyproteins, pp1a and pp1ab. These polyproteins are proteolytically processed by two viral proteases, a papain-like protease (PLpro) and a chymotrypsin-like main protease (Mpro, also called 3CLpro), releasing 16 nonstructural proteins (1). Mpro inhibitors can effectively block SARS-CoV-2 replication in cell culture, demonstrating that Mpro is a valid drug target (4, 5, 6, 7, 8). SARS-CoV-2 Mpro has been shown to preferentially recognize the A-X-L-Q-(A/S) consensus sequence (where X is any amino acid), with cleavage occurring after the glutamine (9, 10). Interestingly, other coronaviruses including the related SARS-CoV and MERS-CoV share similar substrate specificity with SARS-CoV-2 Mpro, suggesting that inhibitors of SARS-CoV-2 Mpro could serve as broad-spectrum antiviral drugs against future epidemic- or pandemic-causing coronaviruses (4, 11).
Discovery of Mpro inhibitors has relied heavily on the use of high-throughput screens (HTS) using a FRET-based peptide substrate to monitor protease activity (5, 7, 12, 13, 14, 15, 16, 17). Fluorogenic substrates that cleave an aminocoumarin-based fluorophore attached to the carboxyl terminus of a peptide have also been used (14, 15, 18). A number of Mpro enzyme assays have been developed using different substrates, Mpro constructs, and buffer conditions (14, 15, 19, 20, 21, 22). As a result, there have been inconsistent findings regarding the identification of potential Mpro inhibitors (20, 21, 23). This has highlighted a clear need for an improved SARS-CoV-2 Mpro assay that delivers better performance and improved consistency.
Here, we provide the detailed biophysical and enzymatic characterization of SARS-CoV-2 Mpro with native N termini and C termini and assess the steady-state kinetic parameters of three commonly used SARS-CoV-2 Mpro fluorescent substrates (4, 7, 18). We measured the kinetic efficiency of six SARS-CoV2 Mpro polyprotein cleavage sequences to determine the optimal substrate amino acid sequence. Guided by these results, an improved 5-carboxyfluorescein (FAM)-based FRET substrate was developed that is better suited for HTS and is less susceptible to interference and false positives than previously reported substrates. This study provides a useful framework for the design of assays aimed at discovering and developing new coronavirus Mpro inhibitors.
Results
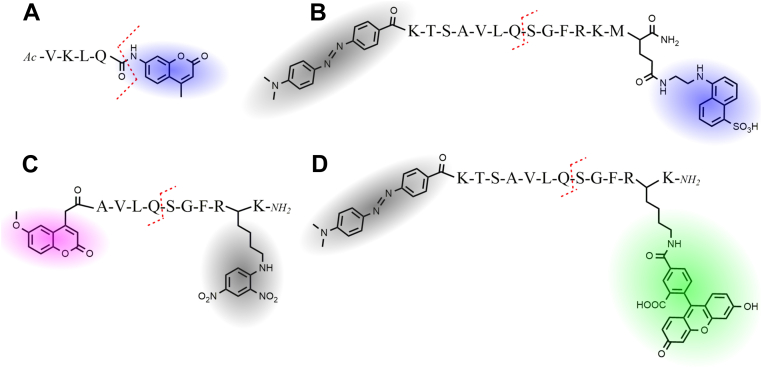
Both fluorogenic and FRET-based substrates were used in this work (Fig. 1). The previously reported VKLQ–7-amino-4-methylcoumarin (AMC) fluorogenic substrate consists of a short peptide with a cleavable AMC fluorophore at the P1′ position (Fig. 1A) (18). The FRET substrates consist of a fluorophore and quencher pair separated by a SARS-CoV-2 polyprotein cleavage sequence. The previously reported FRET nsp4–5-5-(2-aminoethylamino)-1-naphthalenesulfonic acid (EDANS) substrate uses an EDANS fluorophore and a 4-(4-dimethylaminophenylazo)benzoic acid quencher (Fig. 1B), whereas the nsp4–5-7-methoxycoumarin-4-acetic acid (MCA) substrate uses an MCA fluorophore with a 2,4-dinitrophenyl quencher (Fig. 1C) (4, 7). The FAM-based FRET substrate developed in this work uses a 4-(4-dimethylaminophenylazo)benzoic acid quencher (Fig. 1D). Table 1 lists the amino acid sequence, cleavage site location, fluorophore, and quencher used for each substrate tested in this work.
Figure 1.
Structure and names of fluorescent substrates used in this study.A, fluorogenic VKLQ–AMC substrate, with AMC fluorophore shown in blue. B, nsp4–5-EDANS FRET substrate uses an EDANS fluorophore shown in blue, and a 4-(4-dimethylaminophenylazo)benzoic acid (DABCYL) quencher shown in gray. C, nsp4–5-MCA FRET substrate uses an MCA fluorophore shown in purple with a 2,4-dinitrophenyl (DNP) quencher shown in gray. D, nsp4–5-FAM FRET substrate uses a FAM fluorophore shown in green with a DABCYL quencher shown in gray. The red dashed line indicates the cleavage site on the substrates. AMC, 7-amino-4-methylcoumarin; EDANS, 5-(2-aminoethylamino)-1-naphthalenesulfonic acid; FAM, 5-carboxyfluorescein; MCA, 7-methoxycoumarin-4-acetic acid.
Table 1.
Steady-state kinetic parameters for SARS-CoV-2 Mpro fluorescent substrates
| Substrate | Sequencea | kcat/Km (M−1 s−1) |
|---|---|---|
| AKLQ–AMC | Ac-VKLQ ↓ {AMC} | 18.5 ± 1.0 |
| 24.5 ± 5.0b | ||
| nsp4–5-MCA | {MCA}-AVLQ ↓ SGFR{K(DNP)}K-NH2 | 14,190 ± 420 |
| nsp4–5-EDANS | {DABCYL}-KTSAVLQ ↓ SGFRKM{E(EDANS)}-NH2 | 1960 ± 190 |
| nsp4–5-FAM | {DABCYL}-KTSAVLQ ↓ SGFR{K(FAM)}K-NH2 | 2448 ± 85 |
| nsp5–6-FAM | {DABCYL}-KSGVTFQ ↓ SAVK{K(FAM)}K-NH2 | 77.5 ± 4.2 |
| nsp6–7-FAM | {DABCYL}-KKVATVQ ↓ SKMS{K(FAM)}K-NH2 | 68 ± 10 |
| nsp8–9-FAM | {DABCYL}-KSAVKLQ ↓ NNEL{K(FAM)}K-NH2 | 6.01 ± 0.61 |
| nsp10–12-FAM | {DABCYL}-KREPMLQ ↓ SADA{K(FAM)}K-NH2 | 4.74 ± 0.48 |
| nsp14–15-FAM | {DABCYL}-KTFTRLQ ↓ SLEN{K(FAM)}K-NH2 | 38.0 ± 5.2 |
Text in brackets denotes fluorophore or quencher, ↓ denotes cleavage site.
Kinetic parameters determined from Michaelis–Menten plot. Each value reported as the mean ± 1 standard deviation, n = 3.
Biophysical characterization demonstrates excellent protein quality
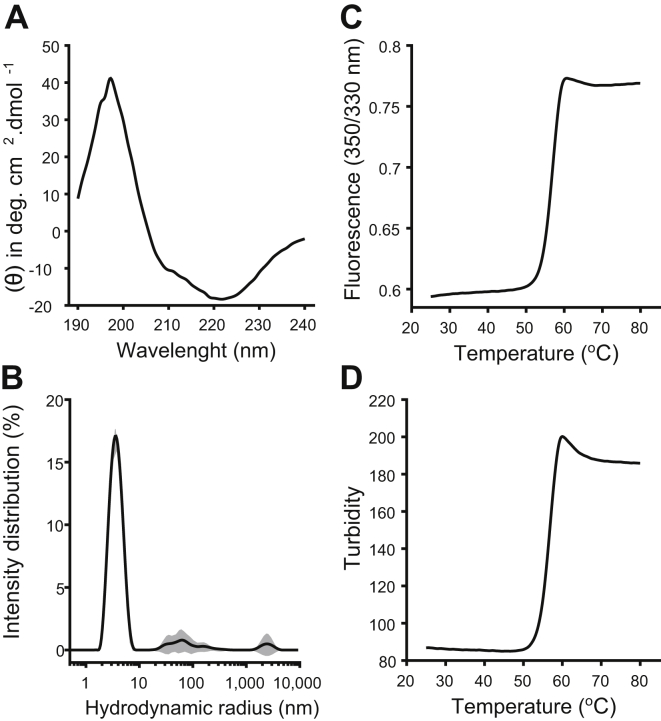
Following the method developed by Xue et al. (24), SARS-CoV-2 Mpro with native N and C termini was expressed and purified to apparent homogeneity (Fig. S1) for further biophysical and enzymatic characterization. Nano-differential scanning fluorimetry (nanoDSF), dynamic light scattering (DLS), and CD spectroscopy were used to establish the quality of Mpro used in this work. CD measurements confirmed that the Mpro secondary structure is in agreement with what is expected based on the crystal structure (Protein Data Bank [PDB] ID: 6Y2E) (Fig. 2A and Table S1). DLS was used to measure the hydrodynamic radius (RH) of Mpro and to measure the state of Mpro self-oligomerization (Fig. 2B). Using the size distribution analysis model, the major intensity peak had an RH of 3.76 ± 0.14 nm with a polydispersity index of 0.18 ± 0.03. The measured RH of 3.76 ± 0.14 nm agrees with the expected RH for the Mpro dimer based on the crystal structure (PDB ID: 6Y2E). The nanoDSF melt curve measured by the 350/330 nm fluorescence ratio showed a melting onset beginning at 51.1 °C and an inflection point or melting point (Tm) of 56.8 °C (Fig. 2C). The baseline turbidity measurement is stable from 20 °C until the onset in turbidity increased beginning at 47.9 °C, indicating Mpro was stable against aggregation until 3.2 °C before the onset of melting begins (Fig. 2D). The inflection point of the turbidity measurement was 56.8 °C corresponding to the Tm of Mpro. The measured Tm of 56.8 °C was slightly higher than previously reported values of between 48.5 and 54.2 °C (25, 26). Taken together, these results show Mpro is highly pure, properly folded, thermodynamically stable, and monodisperse in solution with very little aggregation or higher order oligomerization present.
Figure 2.
Biophysical characterization of SARS-CoV-2 Mpro.A, CD measurement showing secondary structure content of Mpro. B, DLS intensity distribution with major peak showing a hydrodynamic radius (RH) of 3.76 ± 0.14 nm corresponding to Mpro, black line shows mean intensity distribution with ±1 standard deviation shown in gray-shaded area, n = 10. C, 350/330 nm fluorescence ratio shows a melting onset at 51.1 °C and a Tm of 56.8 °C. D, turbidity shows an onset in aggregation beginning at 47.9 °C with an inflection point at 56.8 °C. DLS, dynamic light scattering; Mpro, main protease; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Optimization of assay buffer conditions
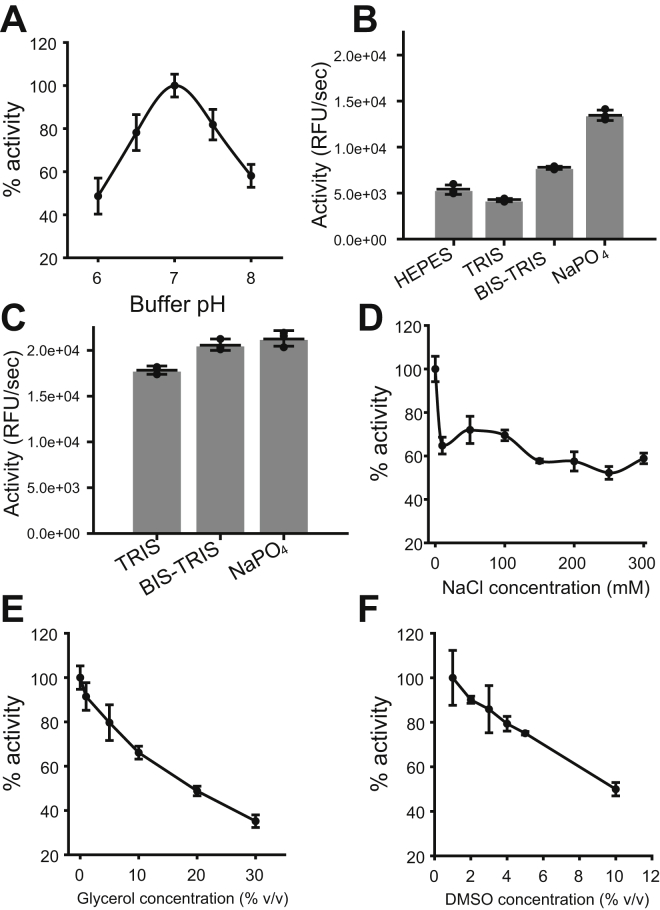
The effects of different buffer conditions on Mpro activity were assessed to determine optimal assay conditions. The AKLQ–AMC substrate was chosen for buffer optimization because it displayed good solubility up to a concentration of around 2.5 mM in all buffers tested. The optimal pH for Mpro was found to be pH 7.0 (Fig. 3A). Mpro had a strong preference for NaPO4 as a buffering agent, followed by Bis–Tris, Hepes, and Tris, when tested at 20 mM buffering agent, 150 mM NaCl, pH 7.0 (Fig. 3B); however, this preference was lessened when tested at 20 mM buffering agent (pH 7.0) without NaCl (Fig. 3C). The highest enzyme activity was achieved when NaCl was omitted from the buffer while adding between 10 and 300 mM NaCl decreases the enzyme activity roughly the same degree (Fig. 3D). Both glycerol and dimethyl sulfoxide (DMSO) were found to have a negative effect on enzyme activity (Fig. 3, E and F). Based on these results, the optimized buffer found in this study consists of 20 mM NaPO4 at pH 7.0. It was found that the FRET substrates used in this work showed better solubility in buffers of low ionic strength; therefore, 20 mM Bis–Tris (pH 7.0) was used instead of a NaPO4-based buffer. These optimized buffer conditions closely agree with other work showing that Mpro is most thermodynamically stable at pH 7.0 in the absence of NaCl (26).
Figure 3.
Assessing the effects of common buffer conditions on Mproinitial rate. All tests are done in 100 μl buffer containing 100 μM VKLQ–AMC substrate and 200 nM enzyme. A, pH optimization in 20 mM NaPO4 (pH 6.0–8.0) with 150 mM NaCl. B, preference of Mpro for Hepes, Tris, Bis–Tris, or NaPO4, in 20 mM buffering agent, pH 7.0, with 150 mM NaCl. C, preference of Mpro for Tris, Bis–Tris, and NaPO4 in 20 mM buffering agent, pH 7.0. D, effect of 0 to 300 mM NaCl in 20 mM NaPO4 (pH 7.0). E, effect of 0 to 30% v/v glycerol in 20 mM NaPO4 (pH 7.0) with 150 mM NaCl. F, effect of 1 to 10% v/v DMSO in 20 mM NaPO4 (pH 7.0) with 150 mM NaCl. Each measurement is reported as the mean with error bars showing ±1 standard deviation, n = 3. AMC, 7-amino-4-methylcoumarin; DMSO, dimethyl sulfoxide; Mpro, main protease.
Measuring the reaction progress curve for the complete hydrolysis of substrate can help assess and identify nonoptimal buffer conditions and loss of enzyme activity because of inactivation and inhibition. It is also helpful to verify that the measured initial rate corresponds to the true linear steady-state portion of the reaction progress curve (27). When hydrolyzing the FRET substrates, the Mpro began to lose activity after about 400 s of reaction time in 20 mM Bis–Tris (pH 7.0) (Fig. S2). Adding 2 mM EDTA and 2 mM DTT to the assay buffer prevented this inactivation for the nsp4–5-FAM and nsp4–5-EDANS substrates (Fig. S2, B and C) but not for the nsp4–5-MCA substrate (Fig. S2D), which was inactive in 20 mM Bis–Tris (pH 7.0) and 2 mM EDTA. As previously discussed, the FRET substrates showed reduced solubility in buffers with higher ionic strength. The inactivity of the nsp4–5-MCA substrate in the 20 mM Bis–Tris (pH 7.0) and 2 mM EDTA buffer may be caused by the increase in ionic strength of the buffer from the addition of 2 mM EDTA, reducing the solubility of the nsp4–5-MCA substrate. In contrast to the FRET substrates, Mpro did not fully lose activity when hydrolyzing the VKLQ–AMC substrate (Fig. S2A), and addition of EDTA and DTT to the reaction buffer had a minimal effect on the enzyme’s behavior. The linear initial rate portion of the reaction progress curve could be measured for at least the first 600 s of reaction time with the VKLQ–AMC substrate and about 200 s for the FRET substrates. This initial rate was unaffected by the addition of EDTA and DTT (Fig. S2).
Substrate steady-state kinetic parameters
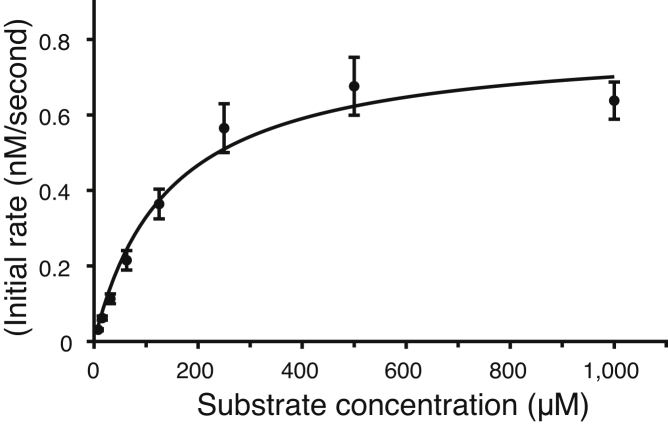
Measurements were performed with each substrate to determine their utility for characterizing Mpro. kcat/Km was measured at low substrate concentrations where the initial rate increased linearly with substrate concentration. The results show that the nsp4–5-MCA substrate had the highest kcat/Km, followed by the nsp4–5-FAM and nsp4–5-EDANS substrates, whereas the VKLQ–AMC substrate had the lowest kcat/Km value (Fig. S3, A–D and Table 1). The FRET substrates suffered from poor solubility and large inner filter effects when used at high concentrations needed to reach saturating substrate concentrations (Vmax). These are commonly reported problems for FRET substrates in general, including FRET substrates used for SARS-CoV-2 Mpro (18, 28, 29, 30). As a result, a full Michaelis–Menten plot that reaches saturating substrate concentrations could only be measured using the VKLQ–AMC substrate (Fig. 4 and Table 1).Mpro recognizes and cleaves 11 sites on the viral pp1a and pp1ab during viral replication. The nsp4–5 cleavage sequence at the N terminus of Mpro is the sequence commonly used in Mpro FRET substrates and is the sequence used in each of the nsp4–5-MCA, nsp4–5-EDANS, and nsp4–5-FAM substrates (Table 1). To test the kinetic efficiency of other SARS-CoV-2 polyprotein cleavage sequences, the kcat/Km of five additional FAM-based FRET substrates utilizing the nsp5–6, nsp6–7, nsp8–9, nsp10–12, and nsp14–15 cleavage sequences were tested and compared with the nsp4–5-FAM substrate (Table 1). The kcat/Km values for these substrates show that nsp4–5 has by far the highest kcat/Km value followed by nsp5–6, nsp6–7, and nsp14–15, whereas nsp8–9 and nsp10–12 cleavage sites have by far the lowest kcat/Km values (Fig. S3, E–I and Table 1). These results show that out of the substrates tested, the nsp4–5 sequence has the most desirable kinetic properties for use in enzyme assays.
Figure 4.
Michaelis–Menten plot for the VKLQ–AMC substrate. Values of 172 ± 28 μM, 0.842 ± 65 nM s−1; and 0.00421 ± 53 s−1 are measured for the Km, Vmax, and kcat, respectively. kcat/Km was found to be 24.5 ± 5.0 M−1 s−1. Each data point is the mean with error bars showing ±1 standard deviation, n = 3. AMC, 7-amino-4-methylcoumarin.
Characterizing the improved nsp4–5-FAM substrate
To assess the suitability of each of the three FRET substrates for HTS, the Z′-factor for each substrate was determined. The Z′-factor is a statistical parameter used to evaluate the quality of an HTS (31). The Z′-factor reflects two critical criteria that a good quality HTS must have. The first criterion is the signal dynamic range, which describes the difference in signal produced by a positive control and a negative control. When the assay signal dynamic range is large, the signal produced by an active compound can more confidently be distinguished from an inactive compound. The second criterion reflected in the Z′-factor is the variability or standard deviation of the signal produced by the positive and negative controls. When the positive and negative controls produce a consistent signal with low variability, the signal produced by an active compound can be more confidently differentiated from signal variability. To assess the Z′-factor for a FRET substrate, the mean and standard deviation of the initial rate measured for 16 positive and 16 negative controls was calculated. The signal dynamic range and Z′-factor were calculated as described in the Experimental procedures section. Baicalein is a noncovalent inhibitor of SARS-CoV-2 (32) and was used as a positive control; the negative control contained DMSO instead of baicalein. This assay was repeated in triplicate for each FRET substrate, with results reported in Table 2. Of the FRET substrates tested in this work, the nsp4–5-EDANS substrate produces the smallest standard deviation in signal for the positive and negative controls, followed by the nsp4–5-FAM and nsp4–5-MCA substrates. The nsp4–5-FAM substrate produced the largest signal dynamic range followed by the nsp4–5-MCA and nsp4–5-EDANS substrates (Table 2). The large signal dynamic range produced by the nsp4–5-FAM substrate is largely attributed to the much higher brightness of the FAM fluorophore in comparison to the MCA and EDANS fluorophores (Fig. S4). In this study, the nsp4–5-MCA and nsp4–5-EDANS substrates produced an Z′-factor of between 0.72 and 0.79, whereas the nsp4–5-FAM substrate produces a considerably higher Z′-factor of between 0.82 and 0.85 (Table 2).
Table 2.
HTS assay quality statistics for SARS-CoV-2 Mpro FRET substrates
| Substrate | Replicate | Signal mean (RFU/s)a |
Signal standard deviation (RFU/s)a |
Signal dynamic range (RFU/s) | Z′-factor | ||
|---|---|---|---|---|---|---|---|
| Positive control | Negative control | Positive control | Negative control | ||||
| nsp4–5-EDANS | 1 | 287 | 6182 | 141 | 315 | 5895 | 0.768 |
| 2 | 699 | 6638 | 132 | 382 | 5939 | 0.741 | |
| 3 | 551 | 6022 | 179 | 297 | 5471 | 0.739 | |
| nsp4–5-MCA | 1 | 1190 | 84,193 | 407 | 6521 | 83,003 | 0.750 |
| 2 | 2389 | 98,870 | 646 | 6115 | 96,482 | 0.790 | |
| 3 | 3023 | 91,495 | 580 | 7568 | 88,472 | 0.724 | |
| nsp4–5-FAM | 1 | 5004 | 162,872 | 575 | 8923 | 157,868 | 0.820 |
| 2 | 3418 | 140,138 | 471 | 6135 | 136,720 | 0.855 | |
| 3 | 4194 | 156,189 | 1086 | 7013 | 151,955 | 0.840 | |
Abbreviation: RFU, relative fluorescent unit.
Signal mean and standard deviation, n = 16.
Discussion
The SARS-CoV-2 Mpro is a validated drug target, and Mpro inhibitors have been shown to block viral replication in cell culture (4, 5, 6, 7, 8). In addition, Mpro inhibitors could have broad-spectrum antiviral activity against related coronaviruses because of the conserved features of the Mpro recognition sequence (4, 11). Fluorescent substrates are commonly used to study Mpro enzymatic activity, identify inhibitors through HTS, and test inhibitor efficacy. In this study, we perform a biophysical characterization of SARS-CoV-2 Mpro and assess the steady-state kinetic parameters of three commonly used substrates, as well as six polyprotein cleavage sequences. We then develop the improved nsp4–5-FAM substrate that is better suited for HTS when compared with commonly used FRET substrates, resulting from the higher brightness of the FAM fluorophore. In addition, the FAM fluorophore is less susceptible to interference and false positives because of the green-shifted absorption and emission spectra of the FAM fluorophore.
The protease used in this study was produced recombinantly in Escherichia coli following a previously described method (24). Mpro produced by this method has been successfully used for structural and enzymatic studies (4, 7, 8, 18). The primary advantage of this method is that it generates Mpro with native N and C termini, which are known to be structurally different, and more catalytically active than Mpro with non-native N or C termini (4, 24). In addition, conflicting Mpro enzymatic data have been published in the literature, which has been in part attributed to the use of different Mpro constructs with non-native termini (20, 21, 23). Work on the closely related SARS-CoV Mpro has recommended the standard adoption of native termini Mpro for enzymatic and structural studies (30). For these reasons, native SARS-CoV-2 Mpro was used for the biophysical and enzymatic work done in this study.
Characterization of substrate kinetic parameters is critical for understanding the behavior of both the substrate and enzyme and can also help guide the development of a properly optimized enzyme assay. kcat/Km is an informative and useful parameter that gives a measure of substrate specificity and is the apparent second-order rate constant for product formation. We found that the value of 14,190 ± 420 M−1 s−1 for the nsp4–5-MCA substrate was six to seven times higher than the value of 2448 ± 85 M−1 s−1 and 1960 ± 190 M−1 s−1 measured for the nsp4–5-FAM and nsp4–5-EDANS substrates, respectively, which is consistent with values reported in the literature (4, 7). The kcat/Km value of 18.5 ± 1.0 M−1 s−1 for the VKLQ–AMC substrate is far lower than the FRET substrates because of the shorter recognition sequence of the VKLQ–AMC substrate, which lacks the C-terminal residues to the cleavage site (18).
The VKLQ–AMC substrate was the only substrate that could be used at concentrations needed to reach Vmax. By measuring the Michaelis–Menten kinetics of the VKLQ–AMC substrate, we report a kcat/Km of 24.5 ± 5.0 M−1 s−1, which agrees with the value of 18.5 ± 1.0 M−1 s−1 obtained using low substrate concentrations. This demonstrates that the behavior of the VKLQ–AMC substrate is consistent at low and high concentrations; therefore, the VKLQ–AMC substrate is relatively unaffected by the inner filter effect. Others have also found that these fluorogenic substrates are better suited for use at high concentrations than FRET substrates (18). A chromogenic substrate very similar to the fluorogenic VKLQ–AMC substrate was more useful for catalytic mechanism studies of SARS-CoV Mpro than the nsp4–5-EDANS FRET substrate (33).
Of the Mpro polyprotein cleavage sequences tested, we found that the nsp4–5 cleavage sequence has by far the highest kcat/Km and therefore is best suited for use in enzyme assays. This result is consistent with measurements done on the SARS-CoV Mpro, which also show that the nsp4–5 cleavage sequence has the highest kcat/Km of the 11 polyprotein cleavage sequences (34). N-terminomics studies have identified the preferred cleavage sequence of SARS-CoV-2 Mpro to be A-X-L-Q↓(A/S) (9, 10). Of the cleavage sequences we tested, the nsp4–5 sequence is the only one that strictly represents this consensus sequence.
By characterizing the properties of the previously published nsp4–5-EDANS, nsp4–5-MCA, and VKLQ–AMC substrates, we recognized that an improved substrate for HTS could be developed. Because the kcat/Km of the VKLQ–AMC substrate is low, high concentrations of substrate and enzyme are needed to generate a measurable fluorescent signal. This makes the VKLQ–AMC substrate undesirable for HTS because of the larger amounts of enzyme and substrate that would be consumed. We also recognized that both the nsp4–5-MCA and nsp4–5-EDANS substrates use fluorophores with relatively low brightness and undesirable excitation and emission spectra that makes them susceptible to interference from assay compounds (35). To develop an improved FRET substrate for HTS, we chose to use a FAM fluorophore because of its higher brightness and spectral properties that are less prone to interference. In addition, FAM is an inexpensive and readily available fluorophore that can easily be incorporated into the peptide substrate by most custom peptide synthesis companies.
To assess whether our new nsp4–5-FAM substrate is better for HTS than existing nsp4–5-MCA and nsp4–5-EDANS substrates, we characterized the Z′-factor for these substrates. An ideal assay produces an Z′-factor of 1; however, an experimental assay could never achieve this value. An Z′-factor of greater than 0.5 is usually considered an excellent quality assay (31). We found that both the nsp4–5-MCA and nsp4–5-EDANS substrates produced an Z′-factor of 0.75, indicating our assay conditions are well optimized. However, because of the low brightness of the MCA and EDANS fluorophores, these substrates produce a low-signal dynamic range, which limits the Z′-factor. The most effective way to develop a substrate that is better suited for HTS is to use a brighter fluorophore. We found that by using the brighter FAM fluorophore, we were able to greatly increase the signal dynamic range of the assay and increase the Z′-factor of the assay to 0.84. This demonstrates that the improved nsp4–5-FAM substrate is better suited for HTS.
False positives are another common issue encountered in HTS assays and are especially problematic when screening large libraries of compounds (35). Fluorescent-based assays are especially susceptible to false positives caused by compounds that interfere with the measured fluorescent signal. In addition, the vast majority of compounds tested in HTS absorb and fluoresce at wavelengths in the ultraviolet and blue regions of the spectrum (<490 nm) (36). This makes the nsp4–5-MCA and nsp4–5-EDANS substrates (excitation/emission of 320/405 nm and 350/480 nm, respectively) especially susceptible to interference and false positives. In contrast, the nsp4–5-FAM substrate absorbs and emits green light (excitation/emission of 490/530 nm) and is therefore largely unaffected by this source of interference, reducing the potential for false positives (35).
The biophysical and enzymatic characterization of the native SARS-CoV-2 Mpro described in this work will serve as a valuable reference for future studies investigating the activity of SARS-CoV-2 Mpro. Using optimized assay conditions, we were able to compare properties of commonly used Mpro substrates and develop an improved nsp4–5-FAM substrate that is better suited for HTS. When compared with commonly used Mpro FRET substrates, this substrate generates a better-quality HTS assay because of the higher brightness of the FAM fluorophore and is less susceptible to interference from assay compounds because of its green-shifted absorbance and emission spectra. This substrate will thus serve as a valuable tool in the development and design of future HTS assays aimed at identifying and characterizing novel direct-acting antivirals targeting the SARS-CoV-2 Mpro.
Experimental procedures
Construct design, enzyme expression, and storage
Design of the expression vector followed previously reported methods (4, 24). The codon-optimized SARS-CoV-2 Mpro open reading frame was inserted at the BamHI and XhoI restriction sites of a PGEX-6p-1 expression vector. The Mpro open reading frame contained the N-terminal autocleavage site AVLQ↓SGFRK (↓ denotes cleavage site) and a modified version of the C-terminal autocleavage site VTFQ↓GP followed by a His6 tag. Autocleavage occurs during protein expression to produce a native N terminus. The modified C-terminal autocleavage site is not cleaved by Mpro but can be cleaved by human rhinovirus 3C protease to produce the native Mpro C terminus during protein purification. This SARS-CoV-2 Mpro expression vector was synthesized by GenScript.
The SARS-CoV-2 Mpro expression vector was transformed into E. coli strain BL21-Gold (DE3) (37). A single colony was used to inoculate a 50 ml culture of Miller's LB containing 100 μg/ml ampicillin overnight at 30 °C with shaking. About 10 ml of overnight culture was used to inoculate 600 ml of LB containing 100 μg/ml ampicillin. This culture was grown at 37 °C until an absorbance of around 0.6 at 600 nm was reached and then induced with 0.5 mM IPTG for 14 h at 20 °C. Cells were harvested at 4 °C by centrifugation at 4000g for 20 min. The cell pellet was resuspended in a minimal volume of 20 mM Tris (pH 8.0), 300 mM NaCl, and stored at −20 °C. Cells were thawed and then lysed by sonication on ice. Lysate was clarified at 4 °C by centrifugation at 48,000g for 20 min. The supernatant was passed onto a 5 ml HisTrap HP (Cytiva) equilibrated with buffer A (20 mM Tris [pH 8.0], 20 mM imidazole, 500 mM NaCl, and 1 mM DTT). The column was washed with 25 ml of buffer A, then Mpro was eluted with a linear gradient from 0 to 100% buffer B (20 mM Tris [pH 8.0], 500 mM NaCl, 500 mM NaCl, and 1 mM DTT) over 75 ml, and fractions of 4 ml were collected. Fractions were analyzed by SDS-PAGE, those containing Mpro were pooled and mixed with human rhinovirus 3C protease (Sigma–Aldrich) in a 40:1 ratio, and dialyzed into 50 mM Tris (pH 8.0), 150 mM NaCl, 1 mM EDTA, and 1 mM DTT overnight at room temperature. Next, the protein was exchanged into buffer C (50 mM Tris [pH 8.0] and 1 mM DTT) and concentrated to 10 mg/ml using a 10 kDa nominal molecular weight centrifugal filter. The protein was loaded onto a Mono Q 4.6/100 PE anion exchange column (Cytiva) pre-equilibrated with buffer C. The column was washed with 20 ml of buffer C, then Mpro was eluted with a linear gradient from 0 to 30% buffer D (50 mM Tris [pH 8.0], 500 mM NaCl, and 1 mM DTT) over 20 ml, and 0.5 ml fractions were collected. Fractions were analyzed by SDS-PAGE, those containing pure Mpro free of detectable contamination were pooled and buffer exchanged into 20 mM Tris (pH 8.0), 150 mM NaCl, 1 mM Tris(2-carboxyethyl)phosphine, and concentrated using a centrifugal filter.
Protein concentration was measured spectrophotometrically using an extinction coefficient of 32,890 M−1 cm−1 and a molecular weight of 33,796 Da, calculated by ProtParam (38). Mpro was stored at a concentration of 45.3 μM in 50% v/v ethylene glycol at −80 °C for long-term storage and −20 °C for short-term storage. There was no substantial loss of enzyme activity under these storage conditions.
nanoDSF and DLS
Thermodynamic stability and particle size distribution of Mpro were measured using a Prometheus Panta (NanoTemper Technologies GmbH). Mpro at a concentration of 1.1 mg/ml in 20 mM Tris (pH 8.0), 150 mM NaCl, 1 mM EDTA, 1 mM DTT was filtered through a 0.1 μm Ultrafree centrifugal filter (Merck Millipore). Standard Prometheus capillaries (PR-C002) were used. nanoDSF measurements were done from 25 to 80 °C with a temperature gradient of 1 °C/min, and intrinsic tryptophan fluorescence was measured at 350 and 330 nm. For DLS measurements, 10 acquisitions of 5000 ms each were done at a temperature of 20 °C with 100% DLS power. Buffer viscosity was calculated to be 1.02139 mPa•s using the buffer builder incorporated in the Panta Control software, version 1.2.1. The autocorrelation function was fit to a size distribution analysis model. Data analysis was done with the Panta analysis software, version 1.2.
CD spectroscopy
CD measurements of Mpro were performed at a concentration of 0.5 mg/ml in 10 mM Na2HPO4 (pH 8.0). CD measurements were taken with a Jasco J-810 (JASCO Corporation) spectropolarimeter at 20 °C in a 0.05 cm path length quartz cuvette. Raw data were converted to mean residue ellipticity, and secondary structure deconvolution was done using the CDSSTR algorithm and the SMP180 reference set on the DichroWeb server (39, 40). Experimental secondary structure fractions were compared to the protease crystal structure (PDB ID: 6Y2E) using PDBsum (41).
Fluorescent substrates
Amino acid sequences of the substrates used in this study can be found in Table 1 and Figure 1. The nsp4–5-MCA substrate was purchased from CanPeptide, Inc; all other substrates were purchased from GenScript. All substrates had a purity greater than 95% confirmed by HPLC and the molecular weight confirmed by mass spectrometry (testing done by supplier). The excitation and emission wavelengths used for each substrate are as follows: VKLQ–AMC 360/460 nm excitation/emission; nsp4–5-EDANS 350/480 nm excitation/emission; nsp4–5-MCA 320/405 nm excitation/emission; and FAM-based substrates 490/530 nm excitation/emission. All substrates came lyophilized as the trifluoroacetic acid salt; stock solutions were prepared in DMSO and stored protected from light at −20 °C.
Enzyme assay general methods
All measurements were taken on a SpectraMax iD5 microplate reader controlled by Softmax pro 7.1 software (Molecular Devices). All readings were done in a black 96-well flat-bottom polypropylene microplate (Greiner Bio-One; ref 655209). Readings were taken every 20 s for 600 s to measure initial reaction rates and up to 1.5 h to measure complete hydrolysis. Measurements were done at ambient temperature. Initial rates were fit to the linear portion of the reaction progress, usually the first 200 s corresponding to less than 10% substrate hydrolysis. Fluorescence units were converted to concentration using a standard curve generated using a fluorophore standard in 20 mM Bis–Tris (pH 7.0). MCA and EDANS were purchased from Fisher Scientific; FAM was purchased from Cayman Chemical Company; and AMC was purchased from Sigma–Aldrich. All parameter fittings by linear and nonlinear regression were done in QtiPlot (IonDev Software). All measurements were performed in triplicate, and final values are expressed as the mean ± 1 standard deviation of the three measurements.
Steady-state enzyme kinetics
kcat/Km measurements were done in 20 mM Bis–Tris (pH 7.0) with a well volume of 100 μl. For the nsp4–5-FAM, nsp4–5-EDANS, and nsp4–5-MCA substrates, 80 nM enzyme was used with substrate ranging from 15 to 1.3 μM for nsp4–5-FAM and nsp4–5-EDANS or 15 to 0.88 μM for nsp4–5-MCA. For the VKLQ–AMC, nsp5–6-FAM, and nsp6–7-FAM substrates, 200 nM enzyme was used, with substrate concentrations ranging from 25 to 0.78 μM for the VKLQ–AMC substrate and 25 to 1.46 μM for the nsp5–6-FAM and nsp6–7-FAM substrates. For the nsp8–9-FAM and nsp14–15-FAM substrates, 200 nM enzyme was used with substrate concentrations from 50 to 2.93 μM. For the nsp14–15-FAM substrate, the baseline rate of substrate hydrolysis in the absence of enzyme was subtracted from the rate of hydrolysis with enzyme. For the nsp10–12-FAM substrate, 400 nM enzyme was used with substrate concentrations ranging from 100 to 13.2 μM. Initial rate measured in relative fluorescent unit/second was converted to M/second. This rate was divided by the molar concentration of enzyme used in the assay, to give the rate of product formation per second per enzyme active site in units of second−1. A plot of this rate against molar substrate concentration was made; the slope of this plot gives the value of kcat/Km. To measure the full Michaelis–Menten plot for the VKLQ–AMC substrate, 100 μl of 200 nM enzyme and between 1000 and 7.8 μM substrate in 20 mM Bis–Tris (pH 7.0) was used. Initial rate in relative fluorescent unit/second was converted to M/second. A plot of reaction rate in M/second versus the molar substrate concentration was fit to the Michaelis–Menten equation to obtain values of Km and Vmax. To calculate kcat, Vmax was divided by the molar concentration of enzyme used in the assay. With these values of kcat and Km, the value of kcat/Km can be calculated independently from the method described previously.
HTS assessment
The Z′-factor was assessed by measuring enzyme activity of 16 positive and 16 negative controls and repeated in triplicate for each FRET substrate. Baicalein (CAS number: 491-67-8; Sigma–Aldrich), a noncovalent inhibitor of SARS-CoV-2 Mpro, was used as a positive control; the negative control contained DMSO instead of baicalein. The reaction contained 100 μl of 10 μM substrate, 40 nM enzyme, and either 50 μM baicalein or DMSO as the positive and negative controls, respectively. The final buffer composition was 20 mM Bis–Tris (pH 7.0) and 1.2% v/v DSMO. For each assay, the mean and standard deviation of the initial rate for positive and negative controls were calculated. The signal dynamic range was calculated according to the following, where and are the mean of the negative and positive controls, respectively.
The Z′-factor was calculated according to Zhang et al. (31), where and are the standard deviation of the positive and negative controls, respectively.
Data availability
All data are contained within the article and in the supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This research is funded by the Canadian Institutes of Health Research (grant number: 201610PJT-152935) and the Natural Sciences and Engineering Research Council of Canada (grant number: RGPIN-004954-2017).
Author contributions
S. L., F. H., B. A. B.-E., and J. S. conceptualization; S. L., F. H., B. A. B.-E., and J. S. methodology; S. L. and F. H. validation; S. L. and F. H. formal analysis; S. L. and F. H. investigation; S. L. writing–original draft; S. L., F. H., B. A. B.-E., and J. S. writing–review & editing; S. L. visualization; J. S. funding acquisition.
Funding and additional information
J. S. is a Tier-1 Canada Research Chair in Structural Biology and Biophysics.
Edited by Craig Cameron
Contributor Information
Scott Legare, Email: legares@myumanitoba.ca.
Jörg Stetefeld, Email: jorg.stetefeld@umanitoba.ca.
Supporting information
References
- 1.V'kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CDC COVID-19 Vaccine Breakthrough Case Investigations Team COVID-19 vaccine breakthrough infections reported to CDC — United States, January 1–April 30, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70:792–793. doi: 10.15585/mmwr.mm7021e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y., Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vuong W., Khan M.B., Fischer C., Arutyunova E., Lamer T., Shields J., Saffran H.A., McKay R.T., van Belkum M.J., Joyce M.A., Young H.S., Tyrrell D.L., Vederas J.C., Lemieux M.J. Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2020;11:4282. doi: 10.1038/s41467-020-18096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 8.Dai W., Zhang B., Jiang X.-M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pablos I., Machado Y., de Jesus H.C.R., Mohamud Y., Kappelhoff R., Lindskog C., Vlok M., Bell P.A., Butler G.S., Grin P.M., Cao Q.T., Nguyen J.P., Solis N., Abbina S., Rut W., et al. Mechanistic insights into COVID-19 by global analysis of the SARS-CoV-2 3CLpro substrate degradome. Cell Rep. 2021;37:109892. doi: 10.1016/j.celrep.2021.109892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koudelka T., Boger J., Henkel A., Schönherr R., Krantz S., Fuchs S., Rodríguez E., Redecke L., Tholey A. N-terminomics for the identification of in vitro substrates and cleavage site specificity of the SARS-CoV-2 main protease. Proteomics. 2021;21 doi: 10.1002/pmic.202000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L., Lin D., Kusov Y., Nian Y., Ma Q., Wang J., von Brunn A., Leyssen P., Lanko K., Neyts J., de Wilde A., Snijder E.J., Liu H., Hilgenfeld R. α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: Structure-based design, synthesis, and activity assessment. J. Med. Chem. 2020;63:4562–4578. doi: 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- 12.Coelho C., Gallo G., Campos C.B., Hardy L., Würtele M. Biochemical screening for SARS-CoV-2 main protease inhibitors. PLoS One. 2020;15 doi: 10.1371/journal.pone.0240079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jo S., Kim S., Kim D.Y., Kim M.-S., Shin D.H. Flavonoids with inhibitory activity against SARS-CoV-2 3CLpro. J. Enzyme Inhib. Med. Chem. 2020;35:1539–1544. doi: 10.1080/14756366.2020.1801672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breidenbach J., Lemke C., Pillaiyar T., Schäkel L., Hamwi G.A., Diett M., Gedschold R., Geiger N., Lopez V., Mirza S., Namasivayam V., Schiedel A.C., Sylvester K., Thimm D., Vielmuth C., et al. Targeting the main protease of SARS-CoV-2: From the establishment of high throughput screening to the design of tailored inhibitors. Angew. Chem. Int. Ed. Engl. 2021;60:10423–10429. doi: 10.1002/anie.202016961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker J.D., Uhrich R.L., Kraemer G.C., Love J.E., Kraemer B.C. A drug repurposing screen identifies hepatitis C antivirals as inhibitors of the SARS-CoV2 main protease. PLoS One. 2021;16 doi: 10.1371/journal.pone.0245962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuzikov M., Costanzi E., Reinshagen J., Esposito F., Vangeel L., Wolf M., Ellinger B., Claussen C., Geisslinger G., Corona A., Iaconis D., Talarico C., Manelfi C., Cannalire R., Rossetti G., et al. Identification of inhibitors of SARS-CoV-2 3CL-pro enzymatic activity using a small molecule in vitro repurposing screen. ACS Pharmacol. Transl. Sci. 2021;4:1096–1110. doi: 10.1021/acsptsci.0c00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu W., Xu M., Chen C.Z., Guo H., Shen M., Hu X., Shinn P., Klumpp-Thomas C., Michael S.G., Zheng W. Identification of SARS-CoV-2 3CL protease inhibitors by a quantitative high-throughput screening. ACS Pharmacol. Transl. Sci. 2020;3:1008–1016. doi: 10.1021/acsptsci.0c00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rut W., Groborz K., Zhang L., Sun X., Zmudzinski M., Pawlik B., Wang X., Jochmans D., Neyts J., Młynarski W., Hilgenfeld R., Drag M. SARS-CoV-2 Mpro inhibitors and activity-based probes for patient-sample imaging. Nat. Chem. Biol. 2021;17:222–228. doi: 10.1038/s41589-020-00689-z. [DOI] [PubMed] [Google Scholar]
- 19.Dražić T., Kühl N., Leuthold M.M., Behnam M.A.M., Klein C.D. Efficiency improvements and discovery of new substrates for a SARS-CoV-2 main protease FRET assay. SLAS Discov. 2021;26:1189–1199. doi: 10.1177/24725552211020681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z., Li X., Huang Y.-Y., Wu Y., Liu R., Zhou L., Lin Y., Wu D., Zhang L., Liu H., Xu X., Yu K., Zhang Y., Cui J., Zhan C.-G., et al. Identify potent SARS-CoV-2 main protease inhibitors via accelerated free energy perturbation-based virtual screening of existing drugs. Proc. Natl. Acad. Sci. U. S. A. 2020;117:27381–27387. doi: 10.1073/pnas.2010470117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma C., Wang J. Dipyridamole, chloroquine, montelukast sodium, candesartan, oxytetracycline, and atazanavir are not SARS-CoV-2 main protease inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2024420118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hattori S., Higashi-Kuwata N., Hayashi H., Allu S.R., Raghavaiah J., Bulut H., Das D., Anson B.J., Lendy E.K., Takamatsu Y., Takamune N., Kishimoto N., Murayama K., Hasegawa K., Li M., et al. A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2021;12:688. doi: 10.1038/s41467-021-20900-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z., Lin Y., Huang Y.-Y., Liu R., Zhan C.-G., Wang X., Luo H.-B. Reply to Ma and Wang: Reliability of various in vitro activity assays on SARS-CoV-2 main protease inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2024937118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue X., Yang H., Shen W., Zhao Q., Li J., Yang K., Chen C., Jin Y., Bartlam M., Rao Z. Production of authentic SARS-CoV Mpro with enhanced activity: Application as a novel tag-cleavage endopeptidase for protein overproduction. J. Mol. Biol. 2007;366:965–975. doi: 10.1016/j.jmb.2006.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abian O., Ortega-Alarcon D., Jimenez-Alesanco A., Ceballos-Laita L., Vega S., Reyburn H.T., Rizzuti B., Velazquez-Campoy A. Structural stability of SARS-CoV-2 3CLpro and identification of quercetin as an inhibitor by experimental screening. Int. J. Biol. Macromol. 2020;164:1693–1703. doi: 10.1016/j.ijbiomac.2020.07.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferreira J.C., Rabeh W.M. Biochemical and biophysical characterization of the main protease, 3-chymotrypsin-like protease (3CLpro) from the novel coronavirus SARS-CoV 2. Sci. Rep. 2020;10:22200. doi: 10.1038/s41598-020-79357-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Copeland R.A. 2nd Ed. John Wiley & Sons Inc; New York, NY: 2000. Enzymes: A Practical Introduction to Structure Mechanism and Data Analysis. [Google Scholar]
- 28.Ferreira J.C., Fadl S., Ilter M., Pekel H., Rezgui R., Sensoy O., Rabeh W.M. Dimethyl sulfoxide reduces the stability but enhances catalytic activity of the main SARS-CoV-2 protease 3CLpro. FASEB J. 2021;35 doi: 10.1096/fj.202100994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y., Kati W., Chen C.-M., Tripathi R., Molla A., Kohlbrenner W. Use of a fluorescence plate reader for measuring kinetic parameters with inner filter effect correction. Anal. Biochem. 1999;267:331–335. doi: 10.1006/abio.1998.3014. [DOI] [PubMed] [Google Scholar]
- 30.Grum-Tokars V., Ratia K., Begaye A., Baker S.C., Mesecar A.D. Evaluating the 3C-like protease activity of SARS-coronavirus: Recommendations for standardized assays for drug discovery. Virus Res. 2008;133:63–73. doi: 10.1016/j.virusres.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J.H., Chung T.D.Y., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 32.Su H., Yao S., Zhao W., Zhang Y., Liu J., Shao Q., Wang Q., Li M., Xie H., Shang W., Ke C., Feng L., Jiang X., Shen J., Xiao G., et al. Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease. Nat. Commun. 2021;12:3623. doi: 10.1038/s41467-021-23751-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solowiej J., Thomson J.A., Ryan K., Luo C., He M., Lou J., Murray B.W. Steady-state and pre-steady-state kinetic evaluation of severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro cysteine protease: Development of an ion-pair model for catalysis. Biochemistry. 2008;47:2617–2630. doi: 10.1021/bi702107v. [DOI] [PubMed] [Google Scholar]
- 34.Fan K., Wei P., Feng Q., Chen S., Huang C., Ma L., Lai B., Pei J., Liu Y., Chen J., Lai L. Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase. J. Biol. Chem. 2004;279:1637–1642. doi: 10.1074/jbc.M310875200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simeonov A., Jadhav A., Thomas C.J., Wang Y., Huang R., Southall N.T., Shinn P., Smith J., Austin C.P., Auld D.S., Inglese J. Fluorescence spectroscopic profiling of compound libraries. J. Med. Chem. 2008;51:2363–2371. doi: 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- 36.Simeonov A., Davis M.I. Eli Lilly & Company and the National Center for Advancing Translational Sciences; Bethesda, MD: 2004. Assay Guidance Manual. [PubMed] [Google Scholar]
- 37.Inoue H., Nojima H., Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 38.Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A. Humana Press; Totowa, NJ: 2005. The Proteomics Protocols Handbook. [Google Scholar]
- 39.Whitmore L., Wallace B.A. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 40.Abdul-Gader A., Miles A.J., Wallace B.A. A reference dataset for the analyses of membrane protein secondary structures and transmembrane residues using circular dichroism spectroscopy. Bioinformatics. 2011;27:1630–1636. doi: 10.1093/bioinformatics/btr234. [DOI] [PubMed] [Google Scholar]
- 41.Laskowski R.A., Jabłońska J., Pravda L., Vařeková R.S., Thornton J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018;27:129–134. doi: 10.1002/pro.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and in the supporting information.