Significance
Multiprotein complexes in mammalian cells are thought to form by interactions between domains of mature, fully folded proteins. However, in some cases interprotein interaction is obstructed by “buried” or inaccessible binding domains. One such example is the interaction between EBP50 and ezrin, proteins linking the plasma membrane and cytoskeleton; self-association of domains in ezrin masks the site recognized by EBP50. Here, we show EBP50 overcomes this obstacle by cotranslationally binding to nascent ezrin’s otherwise masked domain emerging from the translating ribosome. Our study extends the function of mRNA translation beyond “simple” generation of linear peptide chains that fold into mature proteins for subsequent complex assembly; additionally, cotranslation can facilitate interactions with sterically inaccessible domains to form functional multiprotein complexes.
Keywords: cotranslational assembly, protein–protein interaction, ezrin, EBP50, mRNA translation
Abstract
Multiprotein assemblages are the intracellular workhorses of many physiological processes. Assembly of constituents into complexes can be driven by stochastic, domain-dependent, posttranslational events in which mature, folded proteins specifically interact. However, inaccessibility of interacting surfaces in mature proteins (e.g., due to “buried” domains) can obstruct complex formation. Mechanisms by which multiprotein complex constituents overcome topological impediments remain enigmatic. For example, the heterodimeric complex formed by EBP50 and ezrin must address this issue as the EBP50-interacting domain in ezrin is obstructed by a self-interaction that occupies the EBP50 binding site. Here, we show that the EBP50-ezrin complex is formed by a cotranslational mechanism in which the C terminus of mature, fully formed EBP50 binds the emerging, ribosome-bound N-terminal FERM domain of ezrin during EZR mRNA translation. Consistent with this observation, a C-terminal EBP50 peptide mimetic reduces the cotranslational interaction and abrogates EBP50-ezrin complex formation. Phosphorylation of EBP50 at Ser339 and Ser340 abrogates the cotranslational interaction and inhibits complex formation. In summary, we show that the function of eukaryotic mRNA translation extends beyond “simple” generation of a linear peptide chain that folds into a tertiary structure, potentially for subsequent complex assembly; importantly, translation can facilitate interactions with sterically inaccessible domains to form functional multiprotein complexes.
Protein complexes contribute to most cellular activities, including signal transduction, transport, catalysis, and structural integrity, among others. Accurate assembly of protein complexes, under appropriate conditions and in correct cellular locale, is essential, and often subject to stringent control. The formation of multiprotein complexes can be constitutive or stimulus-dependent, the latter generally depends on posttranslational modifications (1). In nonpolycistronic eukaryotes, in which genes encoding functionally related proteins are scattered among multiple chromosomes, these complexes are generally thought to be generated by stochastic, domain-specific interactions between fully formed, mature constituents: that is, by “posttranslational assembly” (2). However, assembly of certain constitutive complexes take advantage of a newly recognized “cotranslational assembly” mechanism in which a fully formed constituent interacts with the nascent peptide of a partner constituent as it emerges from the exit tunnel of the ribosome traversing its encoding mRNA (3–6). Reports of complex assembly in yeast revealed the process is widespread, with 9 of 12 and 12 of 31 complexes assembled cotranslationally (4, 6). Several mammalian nuclear complexes form cotranslationally, including TFIID (transcription factor IID), TREX2 (transcription and mRNA export 2), and the SAGA (Spt-Ada-Gcn5-acetyltransferase coactivator) complex (3). Cotranslational interaction offers multiple advantages, including protection of a free constituent that is susceptible to aggregation or degradation, or neutralization of a deleterious free constituent (3–7). Possibly, the emerging peptide offers target domains and conformations not accessible in the fully formed protein, facilitating complex formation between proteins in which one of more interacting domains is buried in the mature proteins. In addition, the cotranslational mechanism can direct formation of topologically unfavorable assemblies not readily formed from mature proteins. To our knowledge, examples of complex assembly that takes advantage of cotranslation to overcome these spatial obstacles have not been reported.
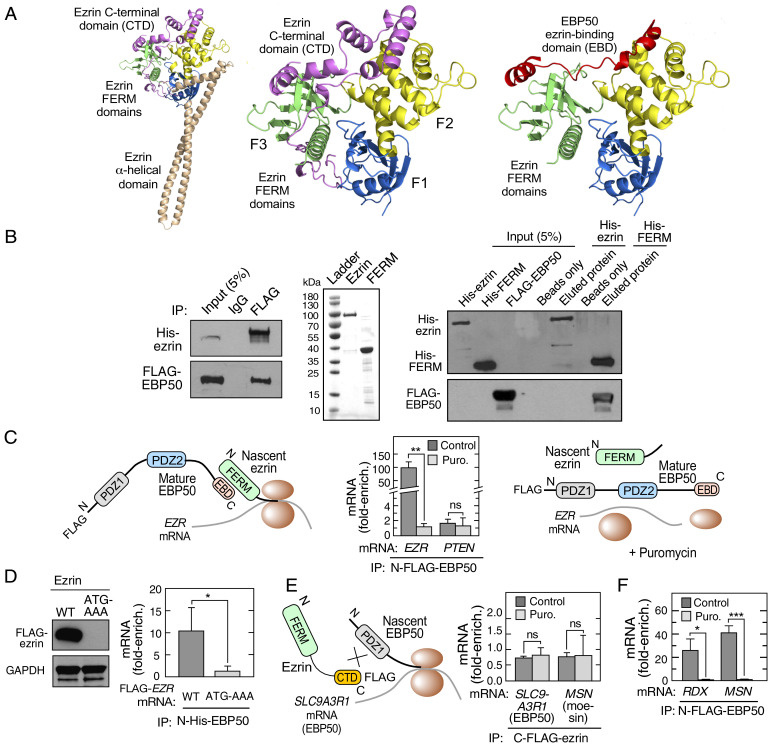
Ezrin is a member of the ezrin-radixin-moesin (ERM) family that links the plasma membrane to the underlying actin cytoskeleton (8). Ezrin consists of an N terminus, 296-amino-acid FERM (4.1 protein ERM) domain joined to a C-terminal domain (CTD) by an elongated helix-turn-helix as revealed in the structure of the full-length (FL), closed ezrin conformer based on the crystal structure of moesin (9) (Fig. 1 A, Left). Free cytoplasmic ezrin is generally present in a closed, “dormant” conformation, in which the CTD interacts with two of the three lobes of the tripartite FERM domain as determined by X-ray crystallography (10) (Fig. 1 A, Center). Ezrin colocalizes with EBP50 (ERM-binding phosphoprotein 50) at the cell periphery, for example, at the apical membrane of polarized epithelia, and their interaction confirmed by coimmunoprecipitation experiments (11). EBP50 is a member of the NHERF family of scaffold proteins, bearing two protein-binding PDZ domains, and an ezrin-binding domain (EBD) through which it binds ezrin and other ERM family members (12–14). Based on the moesin FERM-EBP50 C-terminal crystal structure (PDB ID code 1SGH), a homology model of the complex formed between the EBP50 C terminus and the ezrin FERM domain, indicates the C termini of EBP50 and ezrin occupy similar FERM domain regions (15) (Fig. 1 A, Right). The binding site is supported by in vitro studies showing the EBP50 EBD binds recombinant ezrin FERM domain (16). These observations suggest binding of EBP50 EBD to “closed-form” ezrin is obstructed by the CTD, consistent with the finding that EBP50 fails to bind FL ezrin in vitro (16, 17). These observations raise an enigma: namely, how does EBP50 overcome the steric obstacle presented by the ezrin CTD to form the EBP50-ezrin heterodimer in cells?
Fig. 1.
EBP50 interacts with ezrin cotranslationally. (A) Homology model of closed conformation of human ezrin (Left). Crystal structure showing interaction between C terminus of ezrin with its N-terminal FERM domain (PDB ID code 4RM9) (Center) (10). Homology model of the complex formed between the EBP50 C terminus and the ezrin FERM domain, based on the moesin FERM-EBP50 C-terminal crystal structure (PDB ID code 1SGH) (15) (Right). (B) EBP50 and ezrin interact in cells, but not in vitro. HEK293T cells were cotransfected with His-ezrin and FLAG-EBP50, and proteins detected following immunoprecipitation with anti-FLAG antibody (Left). Purification of His-tagged FL ezrin and ezrin FERM domain from prokaryotic expression system (Center). His-tagged FL ezrin and ezrin FERM domain were immobilized on Ni2+-NTA magnetic beads. The interaction with EBP50 was determined using lysates from HEK293T cells overexpressing FLAG-EBP50, followed by elution by heating in SDS-containing sample buffer (Right). (C) Cotranslational interaction between EBP50 and ezrin. Schematic of cotranslational interaction (Left). HEK293T cells were transfected with N-FLAG-EBP50, incubated with or without puromycin (Puro.), and lysates subjected to RIP with IgG or anti-FLAG antibodies followed by qRT-PCR with EZR and PTEN mRNA probes. mRNA was expressed as fold-enrichment compared to isotype-specific IgG RIP-qRT-PCR (Center). Schematic of absent cotranslational interaction in presence of puromycin (Right). Mean + SD, n = 3; **P < 0.01; ns, not significant. (D) Translation is required for EBP50-ezrin interaction. HEK293T cells were transfected with wild type (WT) or start codon mutant (ATG-AAA) FLAG-ezrin construct, and lysates probed with anti-FLAG and anti-GAPDH antibodies (Left). Cells were cotransfected with C-FLAG-ezrin constructs and N-His-EBP50 as in C, and lysates subjected to anti-His RIP-qRT-PCR. mRNA is expressed as fold-enrichment compared to isotype-specific IgG RIP-qRT-PCR (Right). Mean + SD, n = 3; *P < 0.05. (E) Cotranslational interaction of ezrin and EBP50 is unidirectional. Schematic of failed cotranslational interaction of ezrin and nascent EBP50 (Left). HEK293T cells were transfected with C-FLAG-ezrin and subjected to anti-FLAG RIP-qRT-PCR with probes for SLC9A3R1 and MSN mRNA (Right). mRNA is expressed as fold-enrichment compared to isotype-specific IgG RIP-qRT-PCR (Right). Mean + SD, n = 3. (F) Cotranslational interaction of EBP50 with ERM family members. HEK293T cells were transfected with N-FLAG-EBP50 and subjected to anti-FLAG RIP-qRT-PCR with probes for RDX and MSN mRNA. mRNA is expressed as fold-enrichment compared to isotype-specific IgG RIP-qRT-PCR. Mean + SD, n = 3; *P < 0.05, ***P < 0.001.
Here, we show that EBP50 and ezrin interact by a cotranslational mechanism in which mature EBP50 binds the nascent N terminus of ezrin during EZR mRNA translation. Our findings expand the known functions of cotranslational binding to enabling protein-protein interactions in situations in which a binding domain is inaccessible in the mature protein.
Results
Cotranslational Interaction of EBP50 and Ezrin.
To verify the interaction between EBP50 and ezrin in cells, N-terminal FLAG-tagged EBP50 cDNA and N-terminal His-tagged ezrin cDNA were coexpressed in HEK293T cells. Their intracellular interaction was confirmed by immunoprecipitation with anti-FLAG antibody followed by immunoblot (Fig. 1 B, Left). To evaluate the in vitro interaction, recombinant FL human N-terminal 6xHis-tagged ezrin and its FERM domain (amino acids 1 to 296) were purified from Escherichia coli (Fig. 1 B, Center) and separately immobilized on Ni2+-nitrilotriacetic acid (NTA) magnetic beads. Ezrin- and FERM domain-bound beads were incubated with lysates from HEK293T cells overexpressing N-FLAG-tagged EBP50 protein. Following washing and elution, robust binding was observed between EBP50 and the FERM domain, whereas FL ezrin failed to bind EBP50 (Fig. 1 B, Right). These experiments confirm the paradoxical results reported by others that EBP50 and ezrin interact within cells, but the FL proteins fail to interact in vitro (16, 17).
We considered the possibility that cotranslational interaction of EBP50 and ezrin contribute to the observed binding in cells. Due to the technical difficulty of detecting a protein-protein interaction involving a nascent, emerging peptide, cotranslational interactions are generally interrogated by a surrogate assay in which the interaction of a fully formed protein with the nascent partner protein associated with its encoding mRNA is determined by RNA immunoprecipitation (RIP) followed by qRT-PCR or RNA sequencing (6). We first investigated the binding of mature EBP50 to nascent EZR (gene encoding ezrin) mRNA (Fig. 1 C, Left). HEK293T cells were transfected with N-FLAG-tagged EBP50 and its interaction with EZR mRNA was determined by RIP with monoclonal anti-FLAG, or isotype-specific IgG control antibody, followed by qRT-PCR using EZR primers. An ∼100-fold enrichment of EZR mRNA compared to pulldown with control IgG was observed, consistent with cotranslational interaction (Fig. 1 C, Center). As an additional control, cells were treated with puromycin before RIP to disassemble the translational machinery, and release nascent peptide from the ribosome and mRNA (Fig. 1 C, Right). This control distinguishes cotranslational assembly from direct binding of a regulatory protein to target mRNA (e.g., for transcript-selective control of mRNA translation or stability). Puromycin completely disrupted the interaction of EBP50 with EZR mRNA, consistent with a cotranslational mechanism (Fig. 1 C, Center).
We queried the possible cotranslational interaction of EBP50 with PTEN that binds the N-terminal PDZ1 domain of EBP50, not the ezrin-binding C-terminus (18). No enrichment of PTEN mRNA in the FLAG-EBP50 pulldown was observed, indicating cotranslation-independent binding (Fig. 1 C, Center). As an additional confirmatory control, we evaluated the requirement for translation of EZR mRNA for cotranslational interaction. A mutation of the start codon of the mRNA encoding the target blocks translation-initiation, and prevents cotranslational interaction (6). N-His-tagged EBP50 was coexpressed with a C-FLAG-tagged EZR cDNA construct bearing an ATG-to-AAA mutation of the initiation codon. As expected, the wild-type construct generated protein, but the mutant construct failed to express FLAG-ezrin (Fig. 1 D, Left). Following RIP with anti-His antibody, qRT-PCR revealed enrichment of wild-type, but not the mutant, EZR mRNA (Fig. 1 D, Right), indicating that translation is essential for the interaction between EBP50 and ezrin. Together, these results reveal a cotranslational mechanism that directs formation of topologically unfavorable interaction between EBP50 and ezrin that is not readily formed with mature proteins. To our knowledge, this is a unique example of a cotranslational event that overcomes a spatial obstacle to complex assembly.
Specificity and Generality of EBP50–Ezrin Cotranslation.
Cotranslational interaction can be unidirectional or bidirectional in nature (3, 4). We investigated the potential bidirectionality of the cotranslational interaction: that is, the interaction of mature ezrin with nascent EBP50 (Fig. 1 E, Left). C-FLAG-tagged ezrin was expressed in HEK293T cells and subjected to RIP with anti-FLAG antibody. Bound SLC9A3R1 mRNA, the gene encoding EBP50, was determined by qRT-PCR, as well as MSN mRNA, which encodes moesin that can heterodimerize with ezrin (19). Enrichment of mRNA encoding EBP50 was not observed (Fig. 1 E, Right). This observation is consistent with the interaction of ezrin with the C terminus of EBP50, and thus the PDZ-bearing Nterminus of EBP50 emerging from the ribosome is not a preferred target of the FERM domain of ezrin. Likewise, MSN mRNA enrichment following RIP was not observed, indicating the interaction of ezrin and moesin, and likely other ERM pairs, is independent of cotranslation. These results show a unidirectional, cotranslational interaction of mature EBP50 with nascent, translating ezrin. To further investigate target generality, the cotranslational interaction of EBP50 with other ERM family binding partners, namely moesin and radixin, was determined. Transfection of N-FLAG-tagged EBP50, followed by anti-FLAG RIP-qRT-PCR with primers targeting RDX (encodes radixin) and MSN mRNA, revealed marked puromycin-inhibited interaction (Fig. 1F). To determine cell-type specificity of the cotranslational interaction between EBP50 and ezrin, we probed two tumor-derived cell lines (i.e., HCT116 and Jurkat). In both lines, RIP-qRT-PCR of endogenous EBP50 showed enrichment of EZR, but not PTEN, mRNA, revealing a generality of the cotranslational interaction (Figure S1).
The Ezrin N Terminus and EBP50 C Terminus Are Required for Cotranslational Interaction.
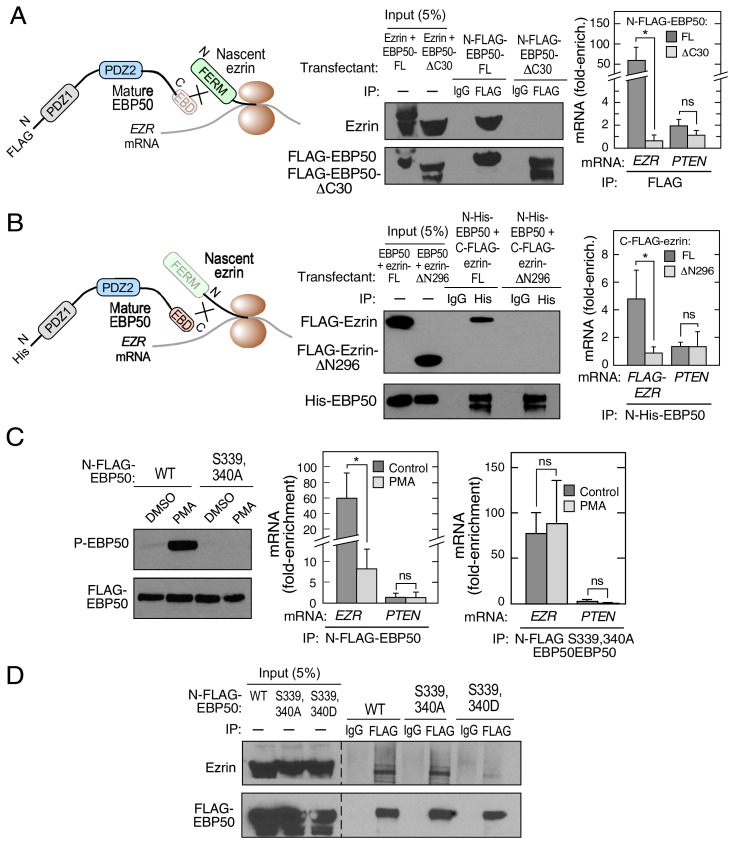
The N-terminal FERM domain of ezrin is a 296-aminoacid trimeric structure that forms heterotypic interactions with the C termini of multiple partners, including EBP50, CD44, ICAM-1, and ICAM-2 (11, 20, 21). The 30-amino-acid C terminus of EBP50 harbors the EBD, essential for binding the ezrin FERM domain (16). The role of these domains in the cotranslational interaction was investigated. N-terminal FLAG-tagged EBP50, with a 30-amino-acid C-terminal deletion (N-FLAG-EBP50-ΔC30), was constructed and interaction with nascent ezrin determined (Fig. 2 A, Left). Immunoprecipitation of N-FLAG-tagged FL EBP50 efficiently pulled down ezrin, whereas the ΔC30 deletion mutant failed to do so (Fig. 2 A, Center). Likewise, RIP of FL EBP50, but not EBP50-ΔC30, showed robust enrichment of EZR mRNA (Fig. 2 A, Right). To determine the role of the ezrin FERM domain in cotranslation, an ezrin construct with an N-terminal deletion of the 296-amino-acid FERM domain (ezrin-ΔN296) was constructed with a C-terminal-FLAG tag. N-terminal-His-tagged EBP50 was cotransfected with either C-FLAG-ezrin-FL or C-FLAG-ezrin-ΔN296 cDNA constructs (Fig. 2 B, Left). Coimmunoprecipitation showed deletion of the ezrin FERM domain abrogated the interaction with EBP50 (Fig. 2 B, Center). RIP with anti-His antibody of cells cotransfected with N-terminal His-tagged EBP50 and either C-FLAG-ezrin-FL or C-FLAG-ezrin-ΔN296 constructs, followed by qRT-PCR, revealed enrichment of FL, but not ΔN296, EZR mRNA (Fig. 2 B, Right). These results indicate the C-terminal EBD of EBP50 and the N-terminal FERM domain of ezrin are essential for cotranslational assembly of the heterodimer in cells.
Fig. 2.
Domains and posttranslational modification determine cotranslational interaction between ezrin and EBP50. (A) Schematic showing cotranslational requirement of EBP50 C-terminus (Left). HEK293T cells were transfected with either N-FLAG-EBP50 or N-FLAG-EBP50-ΔC30 and lysates subjected to immunoprecipitation with anti-FLAG and isotype-specific IgG antibodies (Center). HEK293T cells were transfected with same EBP50 constructs and subjected to anti-FLAG RIP- qRT-PCR with probes for EZR and PTEN mRNA. mRNA was expressed as fold-enrichment compared to isotype-specific IgG RIP-qRT-PCR. Mean + SD, n = 3; *P < 0.05; ns, not significant. (B) Schematic showing cotranslational requirement of ezrin FERM domain (Left). HEK293T cells were cotransfected with N-His-EBP50 and either C-FLAG-ezrin or C-FLAG-ezrin-ΔN296, and lysates subjected to immunoprecipitation with anti-His and isotype-specific IgG antibodies (Center). HEK293T cells were transfected with same EBP50 and ezrin constructs and subjected to anti-His RIP-qRT-PCR with probes for FLAG and PTEN mRNA. mRNA is expressed as fold-enrichment compared to isotype-specific IgG RIP-qRT-PCR. Mean + SD, n = 3; *P < 0.05. (C) Phosphorylation of EBP50 inhibits cotranslational interaction with ezrin. EBP50 deficient HEK293T cells were transfected with N-FLAG–tagged wild type (WT) or S339,440A phospho-deficient EBP50, and treated with PMA or DMSO as control. P-EBP50 and total EBP50 were determined by immunoblot (Left). HEK293T cells were transfected with N-FLAG-EBP50 and treated with PMA or DMSO. Lysates were subjected to anti-FLAG RIP-qRT-PCR with probes for EZR and PTEN mRNA. mRNA is expressed as fold-enrichment compared to isotype-specific IgG RIP-qRT-PCR (Center). HEK293T cells were transfected with N-FLAG-EBP50 S339,340A mutant and treated with PMA or DMSO as control. Lysates were subjected to anti-FLAG RIP-qRT-PCR with probes for EZR and PTEN mRNA. mRNA is expressed as fold-enrichment compared to isotype-specific IgG RIP-qRT-PCR (Right). Mean + SD, n = 3; *P < 0.05. (D) HEK293T cells were transfected with N-FLAG-tagged WT, S339,340A phospho-deficient, or S339,340D phospho-mimetic EBP50 constructs and subjected to immunoprecipitation with anti-FLAG and isotype-specific IgG antibodies (Right).
Posttranslational Modification of EBP50 Regulates Cotranslational Interaction with Ezrin.
EBP50 phosphorylation at multiple sites regulates conformation as well as interaction with partners. For example, protein kinase C (PKC)-mediated phosphorylation of canine EBP50 at Ser347 and Ser348 in the EBD (equivalent to Ser339 and Ser340 in human EBP50) reduces its affinity for ezrin, and induces EBP50 relocalization from the plasma membrane to cytoplasm (22). To explore the role of EBP50 phosphorylation in cotranslational complex assembly, HEK293T cells lacking EBP50 were generated by CRISPR for transfection with phosphorylation mutants. Wild-type and phospho-defective (S339,340A) N-FLAG-EBP50 was expressed, and cells treated with phorbol 12-myristate 13-acetate (PMA) to activate PKC. Immunoblot with anti-P-Ser-EBP50, which recognizes dual phosphorylated P-Ser339, P-Ser340 EBP50, showed marked induction of EBP phosphorylation in PMA-treated cells expressing wild-type EBP50, but not the phospho-defective S339,340A mutant (Fig. 2 C, Left). As shown by RIP-qRT-PCR, PMA markedly inhibited N-FLAG-EBP50 binding to EZR mRNA in HEK293T cells, indicating substantial phosphorylation-mediated inhibition of cotranslational interaction (Fig. 2 C, Center). However, PMA did not reduce binding of S339,340A mutant EBP50 to EZR mRNA in HEK293T cells, indicating the specificity of the effect of PMA (Fig. 2 C, Right). PMA stimulation of PKC induces phosphorylation of a plethora of proteins, including both EBP50 and ezrin. To confirm that phosphorylation of the C-terminal EBD of EBP50 is directly responsible for reducing cotranslational interaction, a FLAG-tagged EBP50 mutant was constructed bearing phosphorylation-mimetic S339,340D residues. Immunoprecipitation with anti-FLAG antibody, followed by immunoblot, showed the wild-type and S339,340A mutant exhibited comparable binding to ezrin, but the phospho-mimetic S339,340D EBP50 mutant showed markedly reduced interaction, even in the absence of PMA (Fig. 2D). These experiments represent a unique example of posttranslational modification in influencing the cotranslational mechanism, and consequent protein-protein interactions.
Interaction of Ezrin with EBP50 Maintains Ezrin Expression and Juxta-Membrane Localization.
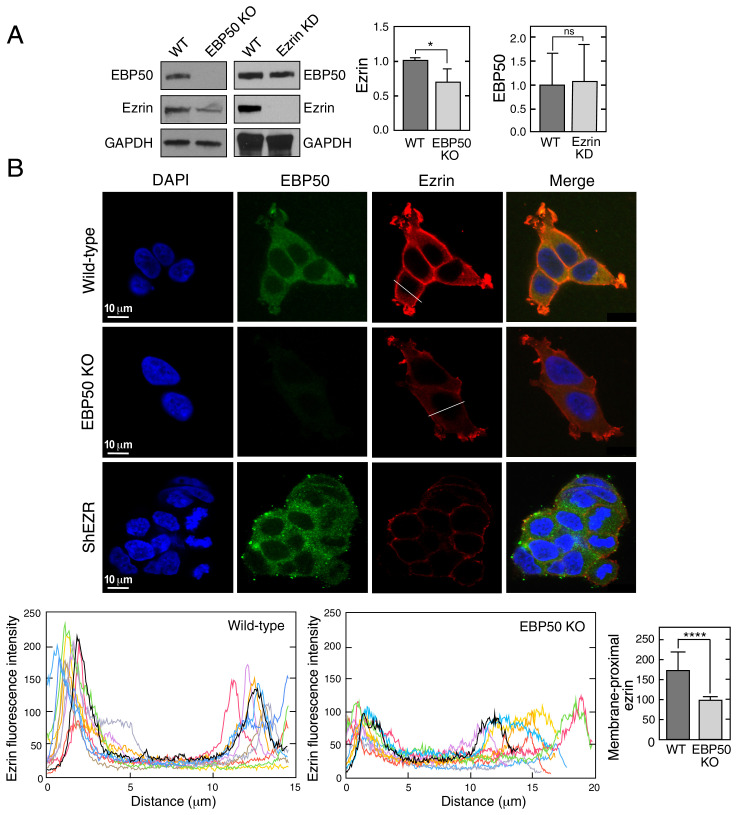
A principal advantage of cotranslational interaction is reduced degradation of one or both interacting partners. For example, TAF8, a TFIID subunit that cotranslates with TAF10, and Set1 that cotranslates with Swd1 and Swd3, are rapidly degraded in the absence of their cotranslation partners (3, 5). To determine the influence of EBP50 and ezrin on expression of its binding partner, we generated a shRNA-mediated stable knockdown of ezrin and a CRISPR-mediated knockout of EBP50, both in HEK293T cells. Immunoblot analysis showed near-complete inhibition of ezrin expression by knockdown had little effect on EBP50 expression; however, knockout of EBP50 moderately reduced ezrin expression (Fig. 3A). More impressive is the reduction of plasma membrane localization of ezrin in the EBP50 knockout cells as shown by immunofluorescence (Fig. 3 B, Upper); quantitative analysis of multiple cross-sections indicated a reduction of juxta-membrane localization of ezrin by about 50% (Fig. 3 B, Lower). Thus, interaction of EBP50 with ezrin contributes to both expression and plasma membrane accumulation of the latter.
Fig. 3.
EBP50 regulates ezrin expression and localization. (A) Influence of EBP50 on ezrin expression. Wild-type (WT), EBP50 knockout (KO), and ezrin knockdown (KD) HEK293T cells were subjected to immunoblot with anti-EBP50, anti-ezrin and anti-GAPDH antibodies (Left). Densitometry reported as mean + SD, n = 3; *P < 0.05; ns, not significant (Center, Right). (B) Immunofluorescence analysis of cells in A using anti-EBP50 and anti-ezrin antibodies (Upper). Line plots showing ezrin localization in WT and EBP50 knockout cells (n = 10 cells, Lower, Left two panels). Bar graph reports peak maximum fluorescence intensities at the membrane-proximal regions; mean + SD; n = 10 cells; ****P < 0.001 (Lower, Right).
EBP50 C-Terminal Peptide Mimetic Abrogates Cotranslational Interaction.
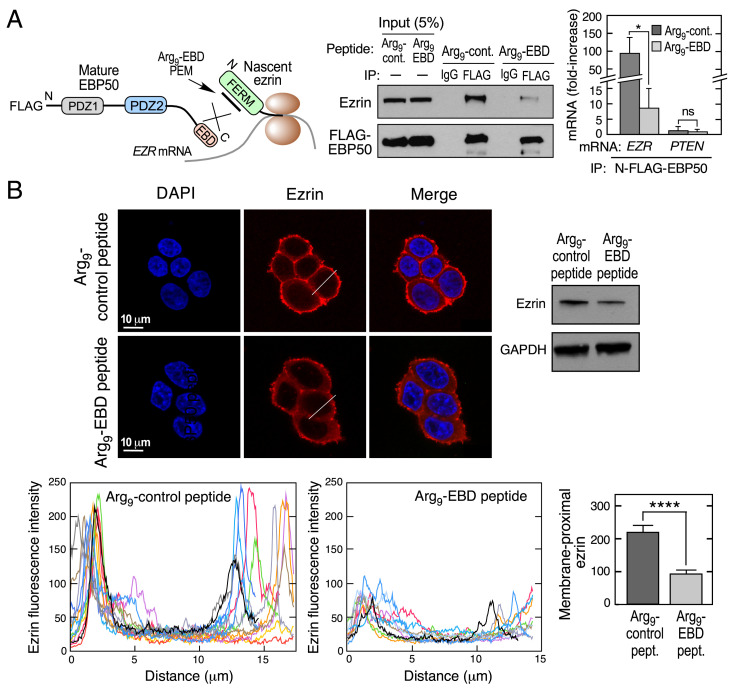
To determine the specific influence of cotranslation on ezrin expression and localization, we explored the activity of an inhibitory peptide. We investigated whether the 28-amino-acid Cterminus of the EBP50 EBD behaves as a peptide epitope mimetic (PEM) that can abrogate the cotranslational interaction of EBP50 with ezrin. This peptide was selected since it reduces the EBP50-ezrin interaction in T-cells (23). HEK293T cells were transfected with a construct expressing N-FLAG-EBP50, and incubated with the PEM bearing an upstream 9-arginine (Arg9) sequence to promote membrane permeation (Fig. 4 A, Left) and with a control, Arg9-bearing EBD peptide containing three-point mutations that does not bind ezrin (23). As shown by immunoprecipitation with anti-FLAG antibody, the EBD PEM markedly reduced the interaction between ezrin and EBP50 compared to control peptide (Fig. 4 A, Center). As shown by RIP followed by qRT-PCR, the EBD PEM reduced cotranslational interaction by about 10-fold compared to the control, indicating that an intervention that blocks cotranslation can interfere with complex formation (Fig. 4 A, Right). The functional consequence of the EBD PEM was investigated by its influence on ezrin expression and localization. Incubation of HEK293T cells with the Arg9-EBD PEM reduced ezrin expression as determined by confocal immunofluorescence and immunoblot (Fig. 4 B, Upper). Quantitative analysis of multiple cross-sections revealed markedly diminished juxta-membrane localization of ezrin following PEM treatment (Fig. 4 B, Lower).
Fig. 4.
Peptide mimetic of EBD abrogates cotranslational interaction of EBP50 with ezrin. (A) Schematic depicting inhibitory activity of EBD mimetic peptide (Left). HEK293T cells were transfected with N-FLAG- EBP50 and then treated with Arg9-EBD or control peptides, and subjected to immunoprecipitation with anti-FLAG and isotype-specific IgG antibodies (Center). HEK293T cells were transfected with same EBP50 constructs and lysates subjected to anti-FLAG RIP-qRT-PCR with probes targeting EZR or PTEN mRNA (Right). mRNA is expressed as fold-enrichment compared to isotype-specific IgG RIP-qRT-PCR. Mean + SD, n = 3; *P < 0.05; ns, not significant. (B) Influence of EBD peptide mimetic on ezrin expression and localization. Analysis of HEK293T cells were treated with Arg9-EBD or control peptides. Ezrin localization was detected by Immunofluorescence using anti-ezrin antibodies (Upper, Left), and expression determined by immunoblot (Upper, Right). Line plots showing ezrin localization in cells treated with Arg9-EBD or control peptides (n = 10 cells, Lower, Left two panels). Bar graph reports peak maximum fluorescence intensities at membrane-proximal regions; mean + SD; n = 10 cells; ****P < 0.001 (Lower, Right).
Discussion
Near-faultless assembly of protein complexes is essential for normal functioning of cellular pathways, including gene expression, intracellular signaling, and metabolism, among others. To achieve accurate complex formation, individual proteins and subcomplexes must overcome a host of obstacles: for example, discovery of binding partners within the crowded cellular milieu, avoidance of nonspecific interactions, maintenance of appropriate stoichiometry, as well as surmounting structural obstructions preventing domain interaction (16, 24–26). Cells have evolved multiple countermeasures, both transcriptional and posttranscriptional, to ensure accurate complex assembly (1, 27–31).
Ezrin exhibits multiple critical functions in diverse physiological and pathological processes (32–34). Many of these functions are defined by ezrin’s interaction with partner proteins, which in turn depend on the ezrin conformational state (35). A key ezrin partner is EBP50, which harbors a C-terminal EBD that binds the FERM domain of ezrin and other ERM family members, and a substantial amount of EBP50-ezrin complex is constitutively present in multiple cell types (11, 18). However, this heterotypic interaction is challenged by the self-association of ezrin’s FERM domain and CTD, also known as N- and C-ERMADs (ezrin-radixin-moesin-association domains), respectively (16). The self-association of ezrin in its closed conformation masks the binding site for the EBD of EBP50, which shares significant sequence and structural homology with the CTD of ezrin (16). Site-specific phosphorylation of EBP50 regulates its interaction with several proteins, including ezrin (22, 36). Similarly, PKC-mediated phosphorylation of ezrin Thr567 has been suggested to drive its transformation from a closed to open conformation, thereby increasing its association with EBP50 (35, 37). We show that PKC-mediated phosphorylation of two Ser residues in the EBP50 EBD reduces its affinity for ezrin, consistent with the report that EBP50 phosphorylation reduces binding to ezrin, but facilitates interaction of newly accessible PDZ domains with other target proteins (22). Thus, the opposing consequences of phosphorylation on EBP50-ezrin binding suggest alternative binding mechanisms. Our results show that cotranslational interaction between EBP50 and ezrin overcomes the domain-masking obstacle, directing a noncompetitive interaction between fully formed EBP50 with nascent, ribosome-bound ezrin. Moreover, EBP50 binds cotranslationally with other ERM family members, moesin and radixin, consistent with ∼85% sequence identity in their N-terminal FERM domains (38).
PEMs have emerged as therapeutic agents with potential to block pathological protein-protein interactions (39). The C-terminal PEM of EBP50 abrogated the cotranslational interaction between EBP50 and ezrin, and markedly reduced ezrin amount and plasma membrane localization. Thus, the PEM presents an attractive therapeutic approach to reduce potential pathological influences of EBP50-ezrin interactions, ezrin plasma membrane localization, and ezrin cellular levels. For example, ezrin interaction with P-glycoprotein-1 (Pgp, also known as multidrug resistance protein 1) at the plasma membrane of osteosarcoma cells enhances multiple drug resistance (40). Inhibition of the interaction by overexpression of an ezrin deletion mutant relocalizes Pgp to the cytoplasm restoring drug susceptibility. Also, plasma membrane-associated ezrin enhances the migration and invasive potential of cancer cells (41, 42). Finally, ezrin binding to membrane PIP2 precedes phosphorylation at T567, thereby unmasking its membrane- and actin-binding sites necessary for ezrin conformational activation (43), potentially contributing to tumor progression (44–46).
Based on these and other studies, ezrin has been proposed as a therapeutic target against cancer, and small-molecule inhibitors that block ezrin phosphorylation inhibit invasion by osteosarcoma cells (47, 48). Inhibition of cotranslational interaction of EBP50-ezrin (e.g., by the EBP50 PEM) represents an alternative therapeutic modality. The approach has certain advantages, including relative ease of delivery as an arginylated peptide, as well as high specificity. Also, because the approach alters ezrin intracellular localization and amount, but not its structure or function, homeostatic activities depending on unaltered ezrin are likely to remain intact. It is possible that the PEM abrogates posttranslational interaction of EBP50 with partially or completely open ezrin molecules. In this event, the PEM will effect ezrin expression and membrane-proximal localization indistinguishable from its effect on cotranslational interaction. Despite showing that the mature, FL proteins fail to interact in vitro, the inability to quantitatively determine the role of cotranslation as a determinant of total protein-protein interaction remains a technical limitation of the approach.
In summary, our results resolve a long-standing enigma concerning EBP50-ezrin complex formation. In specific, we show a critical role for cotranslational interaction between the two proteins. Complexes from yeast to mammals have recently been found to take advantage of this mechanism. Unique features of the EBP50-ezrin interaction include negative regulation by posttranslational modification, and application of the mechanism to direct a sterically unfavorable interaction. These features complement previously recognized advantages of cotranslational interaction, including ordered assembly of complex constituents and resistance to degradation.
Materials and Methods
Cell Culture, Reagents, Constructs, and Antibodies.
HEK293T, Jurkat, and HCT116 cells were purchased from ATCC. HEK293T cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin solution, in a humidified 5% CO2 chamber. HCT116 and Jurkat cells were similarly cultured in McCoy’s 5A and RPMI-1640 supplemented media, respectively. Ezrin-specific and control shRNAs, mouse anti-FLAG M2 antibody, phorbol 12-myristate 13-acetate (PMA), polybrene, bovine serum albumin and CelLytic buffer were purchased from Sigma-Aldrich. Rabbit anti-ezrin and anti-GAPDH antibodies were from Proteintech. Mouse anti-EBP50 antibody was from Novus Bio. Rabbit anti-mouse light chain-specific antibody was from Cell Signaling. Phospho-EBP50 antibody was a generous gift from T. S. Jou, National Taiwan University, Taiwan. Goat anti-mouse and anti-rabbit antibodies, and ECL and ECL prime reagents were purchased from GE Healthcare. Alexa-488 goat anti-mouse and Alexa-565 goat anti-rabbit secondary antibodies were from Invitrogen. Lipofectamine 2000 transfection reagent, isopropyl β-d-1-thiogalactopyranoside (IPTG), HisPur Ni2+-NTA magnetic beads, qPCR probes, Halt Protease Inhibitor mixture, RNase Out, DNaseI, glycogen, and One-step TaqMan reaction mixture were from ThermoFisher. LipoD293 transfection reagent was purchased from Signagen. Custom TaqMan probes against ezrin, and primers for mutants, were from Integrated DNA Technologies. Amicon Ultra4 concentrators were from Millipore-Sigma. Puromycin was obtained from Invivogen. DAPI-containing mounting media was from Vector Laboratories. cDNA expressing C-terminal FLAG-tagged ezrin was obtained from Genscript. N-FLAG-EBP50, N-His-EBP50, and N-His-ezrin cDNAs were from Sino Biological. BL-21 (DE3)-pLysS competent cells were from Novagen. DH5α cells and restriction enzymes were from New England Biolabs. Ni2+-NTA-coupled agarose beads were purchased from Qiagen and chambered culture slides from BD.
shRNA-Mediated Gene Knockdown and CRISPR-Mediated Gene Knockout Using Lentivirus.
Recombinant lenti-ezrin virus was generated as described previously (32). Briefly, 2.5 × 105 HEK293T cells were transduced with ezrin shRNA lentiviral particles in the presence of polybrene (2 μg/mL). After 3 d, the medium was replaced with fresh medium containing puromycin (1 μg/mL), and replaced with fresh medium containing puromycin every 3 d for 3 to 4 wk for selection of stably transfected ShEzrin cells, and the knockdown was validated by immunoblot. For generation of the EBP50 knockout cell line, single-guide (sg)RNAs designed using an online tool (https://www.benchling.com/) were cloned into lentiCRISPR v2-Nickase vector. Lentiviral vector containing the gRNA targeting EBP50 was cotransfected with packaging plasmids in HEK293T cells, and culture medium containing lentiviral particles concentrated as above. Next, 2.5 × 105 HEK293T cells were transduced with the virus, and single-cell clones isolated by fluorescence-activated cell sorting, and expanded in DMEM. EBP50 knockout cells were validated by mRNA expression and by immunoblot.
Ezrin cDNA Cloning and Protein Expression.
FL human ezrin coding sequence was amplified by PCR with appropriate primers bearing NdeI and XhoI restriction sites using Q5 polymerase, and cloned into pET28a with an N-terminal His-tag. The resulting plasmid, pHuEzrin, was transformed in E. coli DH5α, and validated by restriction analysis and sequencing. pHuEzrin was transformed in E. coli BL21 (DE3)-pLysS, and following IPTG induction ezrin expression was confirmed in the lysate soluble fraction. The freshly transformed colony was grown in Luria-Bertani (LB) broth (500 mL) supplemented with 50 μg/mL kanamycin at 37 °C. When the culture density reached A600 nm ∼0.5, ezrin was induced by IPTG (0.5 mM) for 4 h at 37 °C. Pelleted cells (6,000 rpm, 10 min) were washed in sodium chloride-tris-EDTA (STE) buffer (10 mM Tris⋅HCl, pH 8.0, 1 M NaCl, 1 mM EDTA, and 10% [vol/vol] glycerol), and resuspended in buffer A (20 mM Tris–HCl [pH 8.0], 100 mM NaCl, 10% glycerol, 5 mM 2-mercaptoethanol, 1 mM PMSF). The cells were lysed by sonication, and debris removed by centrifugation at 30,000 rpm for 1 h. Following resin equilibration with buffer A, the cell-free supernatant was affinity-purified on a 2-mL Ni2+-NTA-agarose column. The resin was washed with 50 mL of buffer A containing 10 mM imidazole, and bound protein eluted with a linear gradient of imidazole (50 to 400 mM) in buffer A. Ezrin-containing fractions were pooled and dialyzed against buffer B (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 20% glycerol). Ezrin amount was determined by dye-binding using bovine serum albumin as standard, and purified protein was stored at −80 °C.
The ezrin FERM domain (amino acids 1 to 296) was amplified with Q5 polymerase using pHuEzrin as a template and appropriate primers bearing NdeI and XhoI restriction sites, and cloned into pET28a vector with an N-terminal His-tag. The resulting plasmid pHuEzrin-FERM was validated by sequencing and transformed in E. coli BL21 (DE3)-pLysS. Following IPTG induction, the cells were grown at 18 °C and expression confirmed in the lysate soluble fraction. The ezrin-FERM domain protein was purified using the above-described procedure. Following purification, the ezrin-FERM domain was quantitated and stored as above.
Immunoblot Assay.
Cells were scraped and washed with ice-cold phosphate-buffered saline (PBS) and pelleted at 1,000 rpm, 5 min. The pellet was resuspended in CelLytic cell lysis reagent for 15 min at 4 °C in the presence of 1X protease inhibitor mixture, and debris removed by centrifugation at 15,000 rpm for 15 min at 4 °C. Samples were subjected to sodium dodecyl sulphate (SDS)/polyacrylamide gel electrophoresis (PAGE), and proteins transferred by electrophoresis to a polyvinylidene fluoride (PVDF) membrane for 45 min at 250 mA. The membrane was blocked with 5% dried nonfat milk in Tris-buffered saline containing 0.05% Tween 20 (TBST) for 1 h at room temperature, and then incubated overnight at 4 °C with target-specific antibodies. Following three washes with TBST, membranes were incubated with horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. The blots were washed and developed using ECL reagent according to the manufacturer’s protocol.
Immunoprecipitation and In Vitro Interaction Assays.
For immunoprecipitation assays, HEK293T cells were grown to ∼80% confluence and transfected with FLAG- or His-tagged protein-expressing constructs for 48 h. The cells were washed with ice-cold PBS, pelleted, and lysed in CelLytic buffer containing 1X protease inhibitors for 45 min, and centrifuged at 15,000 rpm for 15 min. Protein A/G beads were used for preclearing cell-free extracts for 1 h at 4 °C in an end-to-end rocker, and removed using a magnetic rack. For immunoprecipitations, IgG, anti-Flag, or anti-His antibodies were added with beads (20 μL) and mixed end-to-end at 4 °C overnight. The beads were washed four times in NT2 buffer (50 mM Tris⋅HCl, pH 7.4, 150 mM NaCl, 1 mM MgCl2, 0.05% Nonidet P-40), and the immunoprecipitated proteins extracted by boiling beads in SDS sample buffer for 5 min. Eluted proteins were subjected to SDS-PAGE followed by western blotting with appropriate antibodies.
For in vitro interaction assays, purified, His-tagged FL ezrin or ezrin FERM domain were immobilized on Ni2+-NTA magnetic beads which were then incubated with HEK293T cell lysates (prepared in CelLytic buffer as above) overexpressing FLAG-tagged EBP50 protein for 30 min at 4 °C. The beads were collected with a magnet and washed four times in ice-cold NT2 buffer. Bound proteins were eluted by boiling beads in SDS sample buffer for 5 min, and loaded onto SDS-PAGE gels.
Confocal Microscopy and Image Analysis.
Cells (5 × 103 cells per chamber) were seeded into two-chamber BD culture slides and allowed to adhere for 24 h. Cells were rinsed with ice-cold PBS and fixed with 4% paraformaldehyde for 10 min at room temperature, and permeabilized with 0.1% Triton X-100. The cells were incubated with anti-ezrin antibody (1:500) and anti-EBP50 antibody (1:100) overnight at 4 °C, washed with cold PBS containing 1% Tween 20 three times for 5 min, and incubated with Alexa-565-labeled anti-rabbit and Alexa-488-labeled anti-mouse secondary antibodies (1:500) respectively at room temperature for 1 h. After washing again as above, cells were mounted in Vectashield containing DAPI for staining nuclei. Images were acquired at 63×/1.40 NA using a Leica TCS-SP8-AOBS inverted confocal microscope (Leica Microsystems). Image-Pro Plus 10 (Media Cybernetics) was used for quantification of line profiles for 10 cells in each condition.
RNA Immunoprecipitation (RIP) and qRT-PCR.
HEK293T cells were grown to about ~80% confluence and transfected with constructs expressing FLAG- or His-tagged proteins. Briefly, 150-cm dishes were treated with puromycin (50 μg/mL) or water control, and incubated at 37 °C for 30 min. The cells were washed with ice-cold PBS, pelleted, and lysed in polysome lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM Hepes, pH 7.0, 0.5% Nonidet P-40, 1 mM DTT, 100 units/mL RNase Out, 1X protease inhibitor cocktail) for 45 min, and centrifuged at 15,000 rpm for 15 min. Cell-free extracts were precleared by incubation with protein A/G magnetic beads for 1 h at 4 °C in an end-to-end rocker, and beads removed with a magnetic rack. For immunoprecipitations, IgG, anti-Flag, anti-His, or anti-EBP50 antibodies (or IgG control) was added with beads (20 μL) and mixed end-to-end at 4 °C overnight. The beads were washed four times in NT2 buffer (50 mM Tris⋅HCl pH 7.4, 150 mM NaCl, 1 mM MgCl2, 0.05% Nonidet P-40) and resuspended in TRIzol for RNA extraction. RNA was subjected to qRT-PCR using TaqMan probes, and expression determined as fold-enrichment compared to IgG.
Cell Treatments.
For experiments requiring EBP50 phosphorylation, HEK293T cells were transfected with N-FLAG wild-type and S339,340A EBP50 constructs overnight. Cells were serum-deprived for 18 h then incubated with 1 μM PMA (or dimethyl sulfoxide [DMSO]) for 2 h, and collected. For peptide treatment, HEK293T cells were transfected with N-FLAG-EBP50 construct overnight, and cells incubated with EBD and control peptides (100 μM) with an Arg9 leader to facilitate cell uptake (23). Sequences were as follows: Arg9-EBD, RRRRRRRRRKERAHQKRSSKRAPQMDWSKKNELFSNL; Arg9-Control, RRRRRRRRRKERAHQKRSSKRAPQMDASKANELASNL. After 5 h the cells were collected for pulldown and RIP-qRT-PCR.
Molecular Modeling.
The model for FL ezrin was built by homology modeling using the crystal structure of moesin from Spodoptera frugiperda (PDB ID code 2I1K) (9). Peptide additions and deletions were built de novo with SWISS-MODELER (49). Homology model of the complex formed between the EBP50 C terminus and the ezrin FERM domain was based on the moesin-EBP50 C-terminal crystal structure (PDB ID code 1SGH) (15), and the FERM domain was docked to the EBP50 CTD using the program PatchDock (50).
Statistical Analysis.
The experiments were performed in triplicate unless mentioned otherwise. The data are expressed as the mean + SD. Statistical analysis was done by Student’s t-test using Prism 7.0 (GraphPad) software. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
Phospho-EBP50 antibody was a generous gift from Dr. T.S. Jou (National Taiwan University, Taiwan). This research was supported by National Institute of Diabetes and Digestive and Kidney Diseases R01 DK124203 and R01 DK123236 (to P.L.F.); National Institute on Aging R01 AG067146 (to P.L.F.); National Institute of Neurological Disorders and Stroke R01 NS124547 (to P.L.F. and V.G.); a Research Accelerator Program Grant from the Lerner Research Institute, Cleveland Clinic (to P.L.F.); and the VelaSano 6 Pilot Award (to P.L.F.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2115799119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Eisenberg A. R., et al. , Precise post-translational tuning occurs for most protein complex components during meiosis. Cell Rep. 25, 3603–3617.e2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luijsterburg M. S., et al. , Stochastic and reversible assembly of a multiprotein DNA repair complex ensures accurate target site recognition and efficient repair. J. Cell Biol. 189, 445–463 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamenova I., et al. , Co-translational assembly of mammalian nuclear multisubunit complexes. Nat. Commun. 10, 1740 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shiber A., et al. , Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature 561, 268–272 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halbach A., et al. , Cotranslational assembly of the yeast SET1C histone methyltransferase complex. EMBO J. 28, 2959–2970 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duncan C. D., Mata J., Widespread cotranslational formation of protein complexes. PLoS Genet. 7, e1002398 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarz A., Beck M., The benefits of cotranslational assembly: A structural perspective. Trends Cell Biol. 29, 791–803 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Fiévet B., Louvard D., Arpin M., ERM proteins in epithelial cell organization and functions. Biochim. Biophys. Acta 1773, 653–660 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Li Q., et al. , Self-masking in an intact ERM-merlin protein: An active role for the central alpha-helical domain. J. Mol. Biol. 365, 1446–1459 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phang J. M., et al. , Structural characterization suggests models for monomeric and dimeric forms of full-length ezrin. Biochem. J. 473, 2763–2782 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Morales F. C., Takahashi Y., Kreimann E. L., Georgescu M. M., Ezrin-radixin-moesin(ERM)-binding phosphoprotein 50 organizes ERM proteins at the apical membrane of polarized epithelia. Proc. Natl. Acad. Sci. U.S.A. 101, 17705–17710 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reczek D., Berryman M., Bretscher A., Identification of EBP50: A PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family. J. Cell Biol. 139, 169–179 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bretscher A., Chambers D., Nguyen R., Reczek D., ERM-Merlin and EBP50 protein families in plasma membrane organization and function. Annu. Rev. Cell Dev. Biol. 16, 113–143 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Garbett D., LaLonde D. P., Bretscher A., The scaffolding protein EBP50 regulates microvillar assembly in a phosphorylation-dependent manner. J. Cell Biol. 191, 397–413 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finnerty C. M., et al. , The EBP50-moesin interaction involves a binding site regulated by direct masking on the FERM domain. J. Cell Sci. 117, 1547–1552 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Reczek D., Bretscher A., The carboxyl-terminal region of EBP50 binds to a site in the amino-terminal domain of ezrin that is masked in the dormant molecule. J. Biol. Chem. 273, 18452–18458 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Bonilha V. L., Rodriguez-Boulan E., Polarity and developmental regulation of two PDZ proteins in the retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci. 42, 3274–3282 (2001). [PubMed] [Google Scholar]
- 18.Morales F. C., et al. , NHERF1/EBP50 head-to-tail intramolecular interaction masks association with PDZ domain ligands. Mol. Cell. Biol. 27, 2527–2537 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gary R., Bretscher A., Ezrin self-association involves binding of an N-terminal domain to a normally masked C-terminal domain that includes the F-actin binding site. Mol. Biol. Cell 6, 1061–1075 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heiska L., et al. , Association of ezrin with intercellular adhesion molecule-1 and -2 (ICAM-1 and ICAM-2). Regulation by phosphatidylinositol 4, 5-bisphosphate. J. Biol. Chem. 273, 21893–21900 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Chen X., et al. , Phosphatidylinositol 4,5-bisphosphate clusters the cell adhesion molecule CD44 and assembles a specific CD44-Ezrin heterocomplex, as revealed by small angle neutron scattering. J. Biol. Chem. 290, 6639–6652 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J. Y., Lin Y. Y., Jou T. S., Phosphorylation of EBP50 negatively regulates β-PIX-dependent Rac1 activity in anoikis. Cell Death Differ. 19, 1027–1037 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stokka A. J., Mosenden R., Ruppelt A., Lygren B., Taskén K., The adaptor protein EBP50 is important for localization of the protein kinase A-Ezrin complex in T-cells and the immunomodulating effect of cAMP. Biochem. J. 425, 381–388 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Johnson M. E., Hummer G., Nonspecific binding limits the number of proteins in a cell and shapes their interaction networks. Proc. Natl. Acad. Sci. U.S.A. 108, 603–608 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Launay G., Ceres N., Martin J., Non-interacting proteins may resemble interacting proteins: Prevalence and implications. Sci. Rep. 7, 40419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taggart J. C., Zauber H., Selbach M., Li G. W., McShane E., Keeping the proportions of protein complex components in check. Cell Syst. 10, 125–132 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taggart J. C., Li G. W., Production of protein-complex components is stoichiometric and lacks general feedback regulation in eukaryotes. Cell Syst. 7, 580–589.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wells J. N., Bergendahl L. T., Marsh J. A., Operon gene order is optimized for ordered protein complex assembly. Cell Rep. 14, 679–685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shieh Y. W., et al. , Operon structure and cotranslational subunit association direct protein assembly in bacteria. Science 350, 678–680 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Panasenko O. O., et al. , Co-translational assembly of proteasome subunits in NOT1-containing assemblysomes. Nat. Struct. Mol. Biol. 26, 110–120 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Mingle L. A., et al. , Localization of all seven messenger RNAs for the actin-polymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J. Cell Sci. 118, 2425–2433 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan K., Long B., Deshpande G. M., Fox P. L., Bidirectional tumor-promoting activities of macrophage ezrin. Int. J. Mol. Sci. 21, 7716 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaffer M. H., et al. , Ezrin and moesin function together to promote T cell activation. J. Immunol. 182, 1021–1032 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pore D., et al. , Cutting edge: Deletion of zzrin in B cells of Lyn-deficient mice downregulates lupus pathology. J. Immunol. 201, 1353–1358 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viswanatha R., Wayt J., Ohouo P. Y., Smolka M. B., Bretscher A., Interactome analysis reveals ezrin can adopt multiple conformational states. J. Biol. Chem. 288, 35437–35451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song G. J., et al. , Phosphorylation of ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) by Akt promotes stability and mitogenic function of S-phase kinase-associated protein-2 (Skp2). J. Biol. Chem. 290, 2879–2887 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren L., et al. , The actin-cytoskeleton linker protein ezrin is regulated during osteosarcoma metastasis by PKC. Oncogene 28, 792–802 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yonemura S., et al. , Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43, and ICAM-2. J. Cell Biol. 140, 885–895 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson J. A., Demarco S., Gombert F., Moehle K., Obrecht D., The design, structures and therapeutic potential of protein epitope mimetics. Drug Discov. Today 13, 944–951 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Brambilla D., et al. , P-glycoprotein binds to ezrin at amino acid residues 149-242 in the FERM domain and plays a key role in the multidrug resistance of human osteosarcoma. Int. J. Cancer 130, 2824–2834 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Antelmi E., et al. , ß1 Integrin binding phosphorylates ezrin at T567 to activate a lipid raft signalsome driving invadopodia activity and invasion. PLoS One 8, e75113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Endo K., et al. , Phosphorylated ezrin is associated with EBV latent membrane protein 1 in nasopharyngeal carcinoma and induces cell migration. Oncogene 28, 1725–1735 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fiévet B. T., et al. , Phosphoinositide binding and phosphorylation act sequentially in the activation mechanism of ezrin. J. Cell Biol. 164, 653–659 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen C., Ye C., Xia J., Zhou Y., Wu R., Ezrin T567 phosphorylation regulates migration and invasion of ectopic endometrial stromal cells by changing actin cytoskeleton. Life Sci. 254, 117681 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Zhan X. H., et al. , LOXL2 upregulates phosphorylation of ezrin to promote cytoskeletal reorganization and tumor cell invasion. Cancer Res. 79, 4951–4964 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Wang A., et al. , Tumor-associated macrophages promote Ezrin phosphorylation-mediated epithelial-mesenchymal transition in lung adenocarcinoma through FUT4/LeY up-regulation. Oncotarget 8, 28247–28259 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bulut G., et al. , Small molecule inhibitors of ezrin inhibit the invasive phenotype of osteosarcoma cells. Oncogene 31, 269–281 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song Y., et al. , Ezrin mediates invasion and metastasis in tumorigenesis: A review. Front. Cell Dev. Biol. 8, 588801 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waterhouse A., et al. , SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H. J., PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 33, W363–W367 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.