Summary
To date, the direct causative mechanism of SARS-CoV-2-induced endotheliitis remains unclear. Here, we report that human ECs barely express surface ACE2, and ECs express less intracellular ACE2 than non-ECs of the lungs. We ectopically expressed ACE2 in hESC-ECs to model SARS-CoV-2 infection. ACE2-deficient ECs are resistant to the infection but are more activated than ACE2-expressing ones. The virus directly induces endothelial activation by increasing monocyte adhesion, NO production, and enhanced phosphorylation of p38 mitogen-associated protein kinase (MAPK), NF-κB, and eNOS in ACE2-expressing and -deficient ECs. ACE2-deficient ECs respond to SARS-CoV-2 through TLR4 as treatment with its antagonist inhibits p38 MAPK/NF-κB/ interleukin-1β (IL-1β) activation after viral exposure. Genome-wide, single-cell RNA-seq analyses further confirm activation of the TLR4/MAPK14/RELA/IL-1β axis in circulating ECs of mild and severe COVID-19 patients. Circulating ECs could serve as biomarkers for indicating patients with endotheliitis. Together, our findings support a direct role for SARS-CoV-2 in mediating endothelial inflammation in an ACE2-dependent or -independent manner.
Keywords: SARS-CoV-2, COVID-19, scRNA-seq, endothelial inflammation, monocyte adhesion, ACE2, TLR4, p38 MAPK, NF-κB, IL-1β
Highlights
-
•
The majority of adult and fetal ECs rarely express surface ACE2
-
•
ACE2 is dispensable for SARS-CoV-2-mediated endothelial activation
-
•
SARS-CoV-2 directly induces endothelial inflammation through TLR4 activation
-
•
ScRNA-seq reveals TLR4 pathway activation in circulating ECs of COVID-19 patients
In this article, Lui, Poon, and colleagues show that human ECs rarely express surface ACE2. Using human pluripotent stem cell-derived ECs that model SARS-CoV-2 infection, the team further reveals that ACE2 is indispensable for SARS-CoV-2 infection but is dispensable for activation of these cells. ACE2 protects rather than exacerbates endothelial inflammation and dysfunction resulted from SARS-CoV-2-induced TLR4 activation.
Introduction
Although primarily affecting the lungs, the SARS-CoV-2 virus is thought to cause vascular dysfunction as it has been well acknowledged that its entry receptor, ACE 2 (ACE2) (Hoffmann et al., 2020; Monteil et al., 2020), is ubiquitously expressed in the vasculature (Hamming et al., 2004). Detectable viral load is also found in the heart, liver, kidneys, and brain, which are highly vascularized organs (Puelles et al., 2020). Besides, mounting evidence identifies endotheliitis in rhesus macaques after SARS-CoV-2 infection (Aid et al., 2020) and in patients with fatal COVID-19 (Ackermann et al., 2020; Varga et al., 2020). There is also an unexplained association of the fatal systemic vascular disease, now termed as multisystem inflammatory syndrome in children, in pediatric patients with COVID-19 (Belhadjer et al., 2020). Furthermore, the vascular endothelium is responsible for recruitment of immune cells during inflammation, which are key drivers of cytokine release syndrome or cytokine storm, widespread intravascular coagulopathy, and complement-induced thromboembolism that are life-threatening complications leading to acute respiratory distress syndrome, systemic inflammation, and multi-organ failure in severe cases (for review, see Evans et al., 2020; Perico et al., 2020). These characteristic hyperinflammatory and procoagulatory features further suggest a critical role of the endothelium in the underlying pathogenesis of severe COVID-19.
During the systemic inflammatory response, including cytokine dysregulation possibly in fatal cases of COVID-19, the endothelium is directly exposed to pro-inflammatory cytokines that initiate the transcriptional programs to induce expression of chemokines and adhesion molecules, such as VCAM-1, to direct the migration, proliferation, and activation of immune cells (Chousterman et al., 2017). During the amplification cascade of inflammation, endothelial cells (ECs) might also synthesize pro-inflammatory cytokines including interleukin-1β (IL-1β) and IL-6 (Chousterman et al., 2017) that have been reported as features of cytokine storms in COVID-19 (Chen et al., 2020; Huang et al., 2020). In fact, our previous work has also demonstrated that immune cells and pro-inflammatory cytokines can directly cause EC inflammation, dysfunction, and apoptosis (Leung et al., 2018b; Liang et al., 2020). Therefore, systemic inflammation in the severe cases as a result of SARS-CoV-2-mediated cytokine dysregulation could further worsen endothelial barrier function and increase vascular permeability leading to sepsis and organ damage. Nevertheless, whether endotheliitis and endothelial dysfunction observed in severe COVID-19 patients are direct consequences of SARS-CoV-2 infection or secondary to systemic inflammation or both remain uninvestigated. A better understanding of the direct effects of SARS-CoV-2 infection on endothelial biology is, therefore, urgently required.
It is noteworthy that ACE2 has a specialized function in the endothelium. It is a carboxypeptidase that cleaves angiotensin (Ang)-II to Ang (1-7). In the classical renin-angiotensin-aldosterone system, the ACE/Ang-II/angiotensin type 1 receptor signaling has been implicated in vascular inflammation, vasoconstriction, and hypertension, leading to organ injury; whereas the ACE2/Ang (1-7)/Mas-1 receptor signaling protects tissues through vasodilation and anti-inflammatory responses (for review, see Zhang et al., 2020a). It has been previously reported that genetic knockout of ACE2 results in accumulation of Ang-II and cardiac injury (Yamamoto et al., 2006). In fact, internalization of both the virus and ACE2 has been observed in COVID-19 patients (South et al., 2020); whether ACE2 is required for SARS-CoV-2-mediated endothelial inflammation and dysfunction remains elusive. The molecular mechanisms underlying the pathogenic effects of SARS-CoV-2 in human ECs remain largely uninvestigated.
In this study, we re-assess the distribution of the surface receptor ACE2 in human ECs of both adult and fetal vascularized organs including the heart, lung, liver, umbilical cord, and placenta derived from the Chinese population, as well as that of common experimental human EC lines, such as umbilical vein ECs (HUVEC), lung microvascular ECs (HMVEC-L), and embryonic stem cell-derived ECs (hESC-ECs). Importantly, we demonstrate that the majority of human ECs do not express surface ACE2 and are resistant to SARS-CoV-2 infection; however, ectopic expression of ACE2 in hESC-ECs confers viral entry, replication, and infectivity. We also show that SARS-CoV-2 is capable of inducing endothelial inflammation without host cell entry through activation of the Toll-like receptor (TLR) 4/p38 mitogen-associated protein kinase (MAPK)/NF-κB signaling pathway. ACE2-independent endothelial activation is abolished by a TLR4 inhibitor or dexamethasone. Genome-wide, single-cell RNA sequencing (scRNA-seq) analyses further demonstrate activation of the TLR4/MAK14/RELA/IL1B signaling axis in circulating ECs collected from COVID-19 patients with mild or severe symptoms compared with that of the control cohort. Our results provide new clinically relevant insights into understanding the pathogenesis underlying SARS-CoV-2-induced vascular disease.
Results
Surface ACE2 is expressed by few human arterial and capillary ECs
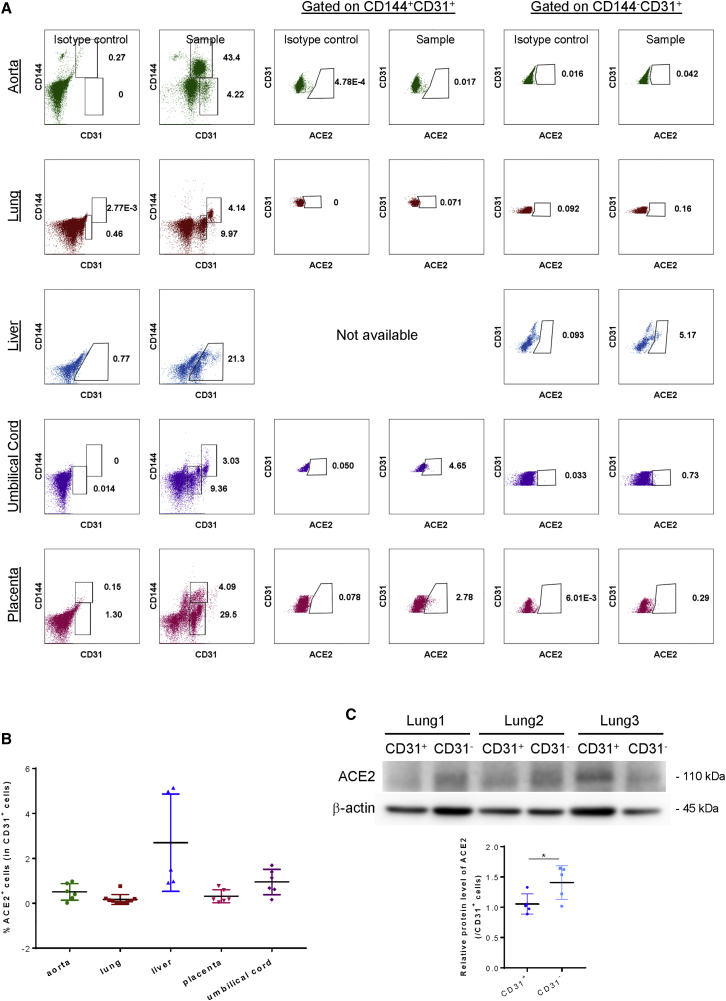
We first re-assessed whether all human ECs express ACE2 on cell surface by collecting human adult and fetal arteries and vascularized tissues (Table S1), respectively, for flow cytometric analyses at a single-cell resolution. The aorta, lung, and liver tissues were derived from adult patients, whereas the fetal umbilical cord and placenta were collected from pregnant women after delivery. We analyzed ACE2 expression in two distinct subsets of human ECs, including CD144+CD31+ cells that were observed in all except the liver samples, and CD144−CD31+ cells that were found in all samples (Figure 1A). Our results showed that the majority of CD144+CD31+ and CD144−CD31+ cells did not express surface ACE2. Further quantification revealed that ACE2 was only expressed by 0.53% ± 0.41%, 0.07% ± 0.05%, 2.70% ± 2.17%, 0.34% ± 0.32%, and 1.00% ± 0.61% of total CD31+ cells, including all EC subsets of the aorta (n = 5), lung (n = 10), liver (n = 5), umbilical cord (n = 5), and placenta (n = 5), respectively (Figure 1B). To further verify whether ACE2 is expressed in human lung ECs, we performed western blotting using an anti-ACE2 antibody of the same clone as the one used for flow cytometry. Antibody specificity was confirmed by western blotting targeting a recombinant human ACE2 protein (Figure S1). Our results showed that ACE2 expression was detected in human CD31+ lung ECs, albeit at a significantly lower level than that of CD31− cells of the lungs (Figure 1C). Altogether, our findings suggest that human ECs of large arteries and capillaries of adult and fetal tissues rarely express surface ACE2, respectively, but intracellular ACE2 expression was detected in human lung ECs.
Figure 1.
Surface ACE2 is expressed by few human arterial and capillary ECs
(A) Flow cytometric analysis showing distribution of ACE2 on CD144+CD31+ and CD144−CD31+ EC subsets derived from human aorta (n = 5), lungs (n = 10), liver (n = 5), umbilical cord (n = 5), and placenta (n = 5). (B) Quantification of (A) showing %ACE2+ cells in total CD31+ ECs, respectively, derived from these adult and fetal organs. (C) Western blotting analyses showing significantly reduced ACE2 expression in CD31+ than CD31− lung cells (n = 5). Data are presented as mean ± SD. Differences were determined by one-way ANOVA and Tukey’s HSD post hoc test. ∗p < 0.05
ACE2 expression is required for SARS-CoV-2 infection in human ECs
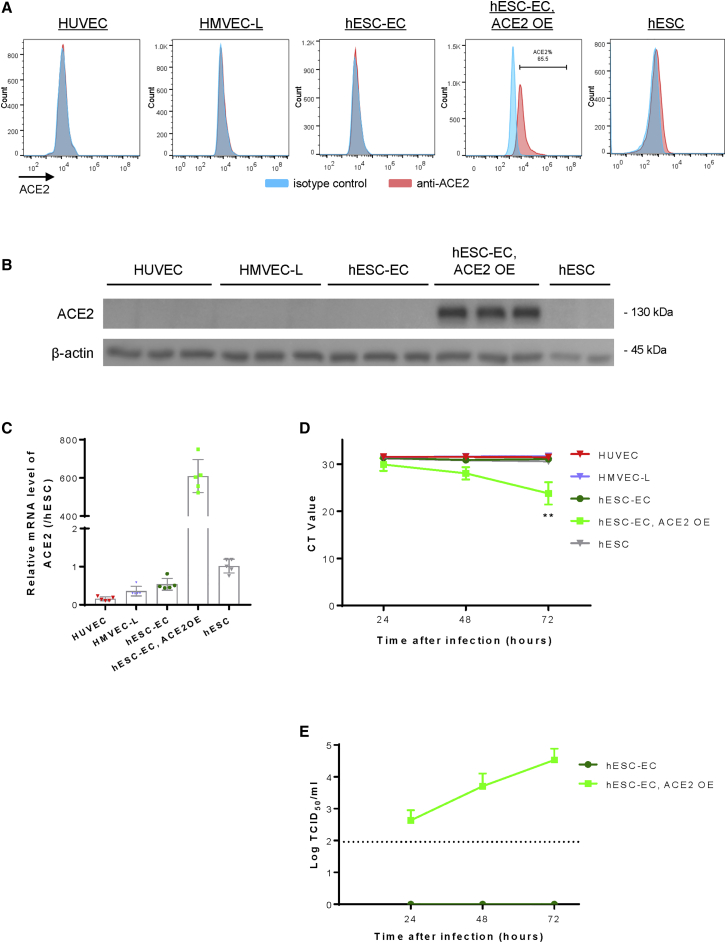
To model SARS-CoV-2 infection in human ECs in vitro, we examined the expression levels of ACE2 in common experimental cell lines. We used non-ECs such as hESCs as a negative control, and ACE2-expressing hESC-ECs (hESC-ECs, ACE2 OE) after lentiviral vector-mediated overexpression as a positive control. Compared with the controls, flow cytometry (Figure 2A), western blotting (Figure 2B), and qRT-PCR (Figure 2C), respectively, showed that HUVEC, HMVEC-L, and hESC-ECs did not naturally express surface or intracellular ACE2. With reference to the previous work in which hESC-derived blood vessels were used for ACE2-targeted drug screening without ectopic ACE2 expression (Monteil et al., 2020), our results highlighted the caution in experimental design and data interpretation as hESC-ECs derived from our protocol did not express ACE2.
Figure 2.
ACE2 expression is required for SARS-CoV-2 infection in human ECs
(A) Flow cytometry, (B) western blotting, and (C) qRT-PCR analyses respectively, showing a lack of cell surface and cellular expression of ACE2 in HUVEC, HMVEC-L, hESC-ECs, or hESCs compared with that of hESC-ECs after lentiviral vector-mediated ACE2 expression (ACE2 OE). (D) qRT-PCR analysis showing the lack of nucleocapsid (N) gene expression at 24, 48, and 72 h after inoculation with SARS-CoV-2 in ACE2-deficient HUVEC, HMVEC-L, hESC-ECs, or hESCs compared with that of hESC-ECs with ACE2 OE. (E) Median tissue culture infectious dose (TCID50) analysis showing a time-dependent increase in the infectivity of SARS-CoV-2 in ACE2-expressing but not -deficient hESC-ECs. (C–E) Data are presented as mean ± SD, n = 5 in (C) and n = 3 in (D and E), differences were determined by one-way ANOVA and Tukey’s HSD post hoc test, ∗∗p < 0.01.
Given that all human EC cell lines under investigation did not express ACE2, we asked whether SARS-CoV-2 is able to infect them. If infection is possible, ACE2 may not be the only entry receptor for SARS-CoV-2 in human ECs. We respectively inoculated HUVEC, HMVEC-L, hESC-ECs, ACE2-expressing hESC-ECs, and hESCs with SARS-CoV-2 at a multiplicity of infection (MOI) of 0.1 for 1 h followed by refreshment with culture medium. The cells were allowed to grow and the culture supernatants were collected at 24, 48, and 72 h after inoculation for viral quantification by qRT-PCR and by median tissue culture infectious dose assay, respectively. qRT-PCR data showed that viral RNA transcription of nucleocapsid (N) gene of SARS-CoV-2 was detected in ACE2-expressing hESC-ECs but not in ACE2-deficient ones after viral exposure (Figure 2D). Moreover, increased viral titers were detected in culture supernatants of ACE2-expressing but not ACE2-deficient hESC-ECs (Figure 2E). Taken together, our data revealed that ACE2 was required to confer replication and infectivity of SARS-CoV-2 in human ECs. Ectopic expression of ACE2 in hESC-ECs was, therefore, employed to model SARS-CoV-2-mediated endothelial dysfunction for more in-depth mechanistic studies below.
SARS-CoV-2 can directly activate human ECs without pro-inflammatory stimulus
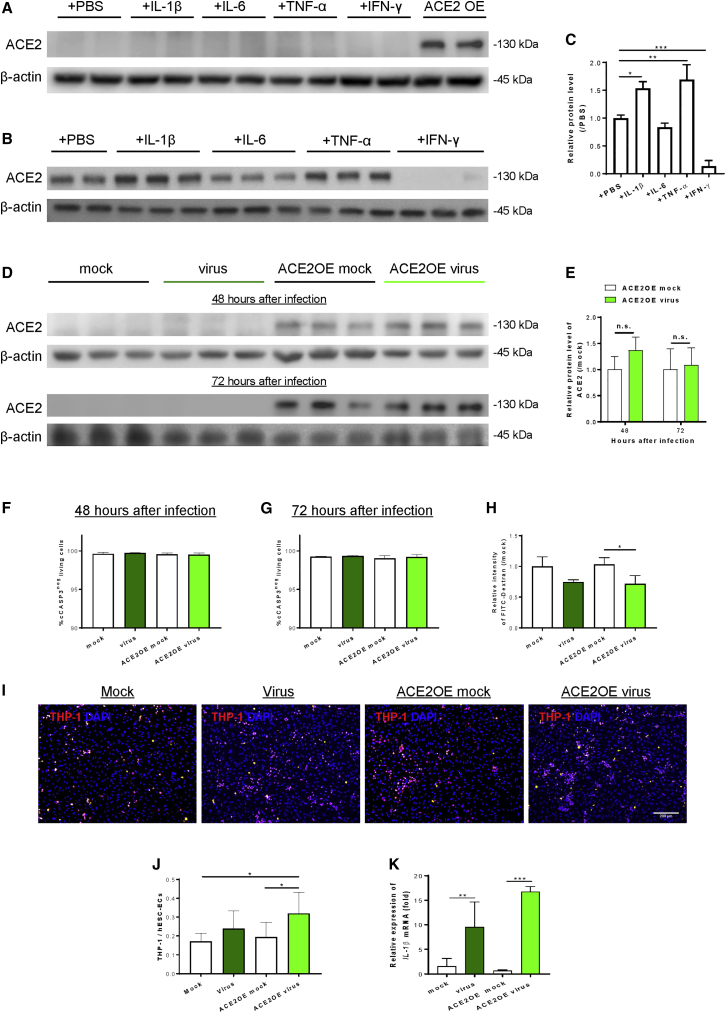
Next, we determined whether inflammation regulates ACE2 expression in hESC-ECs by western blotting. Treatment with pro-inflammatory cytokines including IL-1β, IL-6, tumor necrosis factor alpha (TNF-α), or interferon gamma (IFN-γ) for 48 h did not induce ACE2 expression in ACE2-deficient ECs (Figure 3A). On the other hand, treatment with IL-1β and TNF-α for 48 h significantly increased, whereas IFN-γ significantly reduced, endothelial ACE2 expression in ACE2-expressing ECs (Figures 3B and 3C). Moreover, SARS-CoV-2 did not induce ACE2 expression in ACE2-deficient ECs; nor did it promote its expression in ACE2-expressing ECs compared with the mock infection controls at 48 and 72 h after inoculation, respectively (Figures 3D and 3E). To examine whether the virus induces apoptosis of hESC-ECs during our study period, we performed immunostaining for the apoptotic marker cleaved caspase-3 (cCASP3, Figure S2) at 48 and 72 h after viral inoculation. Our data revealed that SARS-CoV-2 did not promote apoptosis of ACE2-deficient and -expressing ECs at 48 h (Figure 3F) or 72 h (Figure 3G) after inoculation, the duration of this study.
Figure 3.
SARS-CoV-2 can directly activate human ECs without pro-inflammatory stimulus
Western blotting analysis showing (A) a lack of pro-inflammatory cytokine-mediated ACE2 expression in hESC-ECs; or (B) pro-inflammatory cytokine-mediated ACE2 expression in hESC-ECs after ectopic expression of ACE2 at 48 h after treatments. (C) Quantification of (B) showing significantly increased ACE2 expression in IL-1β or TNF-α; but significantly reduced expression in IFN-γ-treated ACE2-expressing hESC-ECs. The relative protein expression level of each sample was compared with its mock infection control with a value 1. (D) Western blotting analysis showing SARS-CoV-2-mediated ACE2 expression in ACE2-deficient and -expressing hESC-ECs at 48 and 72 h after inoculation, respectively. (E) Quantification of (D) showing that SARS-CoV-2 did not induce ACE2 expression in ACE2-deficient hESC-ECs; nor did it promote its expression in ACE2-expressing hESC-ECs compared with the mock infection controls, respectively. (F and G) Immunostaining for cleaved caspase-3 (cCASP3) showing a lack of SARS-CoV-2-mediated apoptosis of ACE2-deficient and -expressing hESC-ECs at (F) 48 h and (G) 72 h after inoculation, respectively. (H) FITC-labeled 70 kDa dextran-based permeability assays showing a lack of SARS-CoV-2-mediated endothelial extravasation in ACE2-deficient and -expressing hESC-ECs at 72 h after inoculation. (I and J) Monocyte adhesion assays showing significantly increased numbers of adhered fluorescence-labeled human monocytes, THP-1 (red), to ACE2-expressing hESC-ECs compared with the mock infection controls at 72 h after inoculation. DAPI (blue) indicates cell nuclei. Scale bar, 200 μm. (K) qRT-PCR assays showing significantly increased gene expression levels of (K) IL1β in ACE2-deficient and -expressing hESC-ECs compared with their respective controls at 48 h after inoculation. (C, E–H, J, K) Data are presented as mean ± SD, n = 6–8, differences were determined by one- or two-way (if more than one factor) ANOVA and Tukey’s HSD post hoc test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Endothelial dysfunction is commonly characterized by increased permeability as a result of the loss of integrity and/or increased immune cell adhesion, including circulating monocytes through induction of adhesion molecules and cytokines. We asked whether SARS-CoV-2 can directly induce endothelial dysfunction at 72 h after inoculation as more significant viral replication and infection was observed at this time point (Figures 2C and 2D). We used FITC-labeled 70-kDa dextran to examine EC junctional alterations in transwell assays. At 72 h after viral inoculation, hESC-ECs at the upper chambers were supplemented with FITC-dextran for 30 min and the aggregated FITC at the lower compartment was measured thereafter. Surprisingly, significantly reduced levels of aggregated FITC indicated reduced apical to basolateral extravasation in ACE2-expressing ECs after viral inoculation compared with mock-infected cells (Figure 3H), suggesting that SARS-CoV-2 did not impair endothelial barrier integrity after cell entry. On the other hand, increased numbers of human monocyte THP-1 adhered to ACE2-deficient ECs that were even more significant to ACE2-expressing ones after viral inoculation compared with their respective mock controls (Figures 3I and 3J). Furthermore, we performed qRT-PCR to examine the gene expression levels of pro-inflammatory cytokines in hESC-ECs after viral inoculation. Significantly elevated levels of IL1B (Figure 3K) were detected in both ACE2-deficient and -expressing ECs, respectively, at 48 h after viral inoculation compared with their respective mock infection controls. Altogether, our data revealed that SARS-CoV-2 directly induced endothelial activation and monocyte adhesion in a microenvironment without pre-existing inflammation.
SARS-CoV-2 activates human ECs through p38 MAPK/NF-κB with or without ACE2
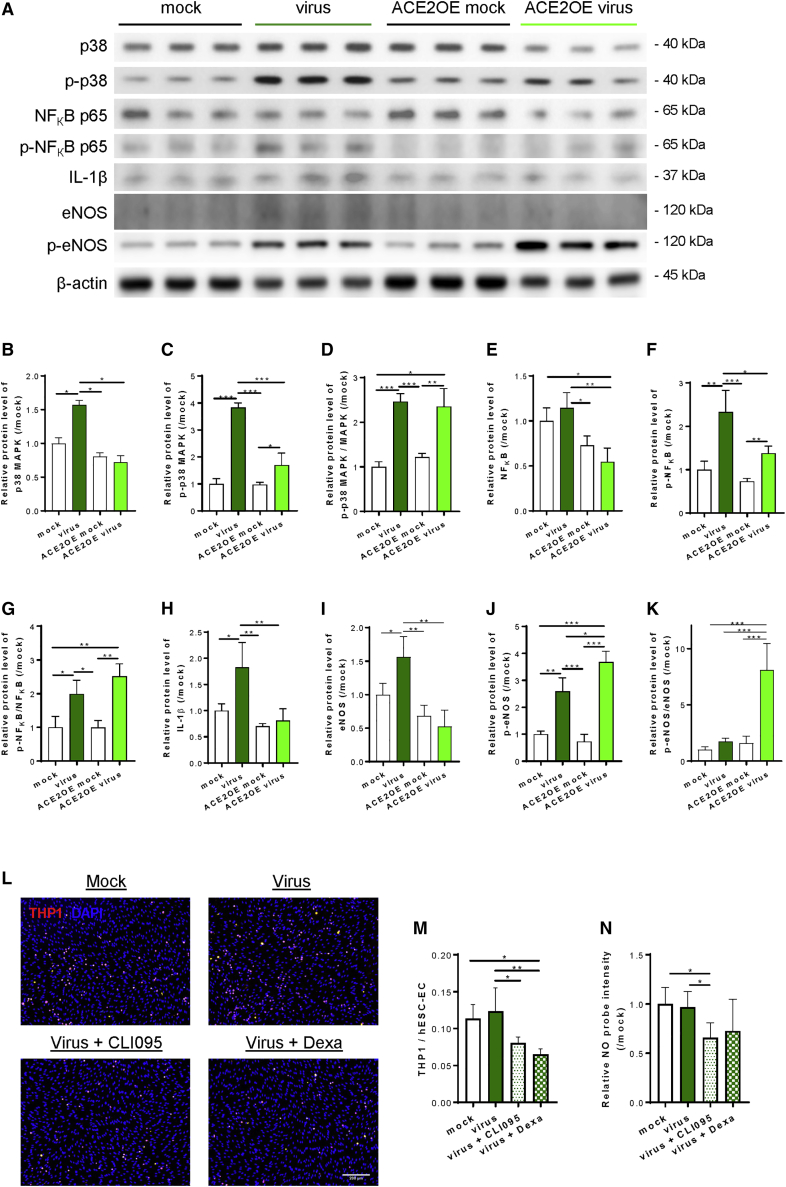
In fact, we did not expect to find upregulated IL1B expression in ACE2-deficient ECs after viral inoculation given that they were resistant to SARS-CoV-2 infection. To better understand the underlying molecular mechanisms by which SARS-CoV-2 mediated human endothelial activation and inflammation after viral exposure, we performed western blotting to determine the expression and phosphorylation levels of p38 MAPK, nuclear factor κB (NF-κB) p65, IL-1β, and endothelial nitric oxide synthase (eNOS), which are associated with endothelial inflammation and function at 72 h after viral exposure in both ACE2-deficient and -expressing hESC-ECs compared with their respective mock infection controls (Figure 4A). Expression levels of p38 MAPK were significantly upregulated in ACE2-deficient ECs (Figure 4B); its phosphorylation levels at Thr180/Tyr182 were also significantly elevated in both types of ECs (Figure 4C); and phosphorylated/total p38 MAPK protein levels were significantly increased in both types of ECs after viral exposure (Figure 4D). In addition, expression levels of NF-κB p65 were not significantly different after viral exposure compared with mock controls in both types of ECs (Figure 4E); but its phosphorylation levels at Ser536 (Figure 4F) and, thereby, phosphorylated/total protein levels (Figure 4G), were significantly increased after viral exposure in both types of ECs. Moreover, expression levels of IL-1β were also significantly upregulated in ACE2-deficient ECs after viral exposure (Figure 4H). Furthermore, expression levels of eNOS were significantly increased in ACE2-deficient ECs (Figure 4I); its phosphorylation levels at Ser632 were significantly upregulated in both types of ECs (Figure 4J); and phosphorylated/total protein levels were significantly elevated in ACE2-expressing ECs after viral exposure (Figure 4K).
Figure 4.
SARS-CoV-2 activates human ECs through p38 MAPK/NF-κB with and without ACE2
(A) Western blotting analysis at 72 h after viral exposure showing that p38 MAPK, NF-κB, and eNOS pathways were significantly elevated in both ACE2-deficient and -expressing hESC-ECs, respectively, compared with their respective mock infection controls; but the degrees of activation of p38 MAPK and NF-κB pathways were significantly attenuated by endothelial ACE2 expression. (B) p38 MAPK, (C) p-p38 MAPK at Thr180/Tyr182, (D) p-p38 MAPK/MAPK, (E) NF-κB, (F) p-NF-κB at Ser536, (G) p-NF-κB/NF-κB, (H) IL-1β, (I) eNOS, (J) p-eNOS at Ser632, and (K) p-eNOS/eNOS. Data are presented as mean ± SD, n = 6, the relative protein levels were compared with the mock infection controls (value = 1) of each time point, differences were determined by one-way ANOVA and Tukey’s HSD post hoc test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. (L and M) Monocyte adhesion assays showing increased numbers of adhered fluorescence-labeled THP-1 (red) to hESC-ECs at 72 h after inoculation compared with mock infection controls that were significantly reduced by CLI095 or dexamethasone. DAPI (blue) indicates cell nuclei. Scale bar, 200 μm. Data are presented as mean ± SD, n = 8, differences were determined by one-way ANOVA and Tukey’s HSD post hoc test, ∗p < 0.05, ∗∗p < 0.01. (N) NO production assays showing significantly reduced NO probes detected in hESC-ECs treated with virus and CLI095 compared with the virus alone group or mock infection control group. Data are presented as mean ± SD, n = 4, the relative probe intensities were compared with that of the mock infection controls (value = 1), differences were determined by one-way ANOVA and Tukey’s HSD post hoc test, ∗p < 0.05.
More intriguingly, our data also revealed that endothelial ACE2 appeared to mitigate SARS-CoV-2-mediated p38 MAPK/NF-κB activation in human ECs as demonstrated by significantly reduced expression (Figures 4B and 4E) and phosphorylation (Figures 4C and 4F) of p38 MAPK and NF-κB, respectively, after viral exposure in ACE2-expressing than that of ACE2-deficient ECs. Likewise, expression levels of IL-1β (Figure 4H) and eNOS (Figure 4I) were also significantly reduced in ACE2-expressing than -deficient ECs after viral exposure. Therefore, our findings revealed that SARS-CoV-2 directly activated the pro-inflammatory signaling pathways in both ACE2-deficient and -expressing human ECs through p38 MAPK, NF-κB p65, and eNOS activation, as well as increased IL-1β production. Nonetheless, total protein expression levels of p38 MAPK, NF-κB p65, IL-1β, and eNOS were not significantly elevated in human ECs with ACE2 after SARS-CoV-2 infection, suggesting its potential protective role against SARS-CoV-2 infection.
To functionally validate whether the observed molecular activation of the p38 MAPK/NF-κB pathway would lead to inflammation of hESC-ECs, we performed monocyte adhesion and NO production assays at 72 h after viral exposure. Although the majority of human ECs did not express ACE2, we speculated that they were still able to recognize SARS-CoV-2 directly through other cell surface receptors even without cell entry. Indeed, IL-1β is a downstream effector of TLR4 signaling in ECs (Dayang et al., 2019; Zeuke et al., 2002). As a member of the innate immune system, ECs also express pattern-recognition receptors, pathogen-associated molecular patterns, including TLR4. Therefore, we also treated hESC-ECs with a TLR4 inhibitor CLI095 or dexamethasone that has been reported to block TLR4-mediated endothelial inflammation (Goodwin et al., 2013), and examined if the virus-induced endothelial activation was mediated through TLR4. Our data revealed that the virus moderately increased the numbers of human monocytes THP-1 adhered to hESC-ECs that were significantly reduced after treatment with CLI095 or dexamethasone (Figures 4L and 4M). Nevertheless, increased NO production was not noticed in hESC-ECs after viral exposure compared with mock infection controls but that was significantly reduced after treatment with CLI095 (Figure 4N), indicating that TLR4 regulated NO production in hESC-ECs. Taken together, SARS-CoV-2 activated hESC-ECs through p38 MAPK/NF-κB with or without ACE2; however, its effects on monocyte adhesion and NO production were not significantly demonstrated in hESC-ECs.
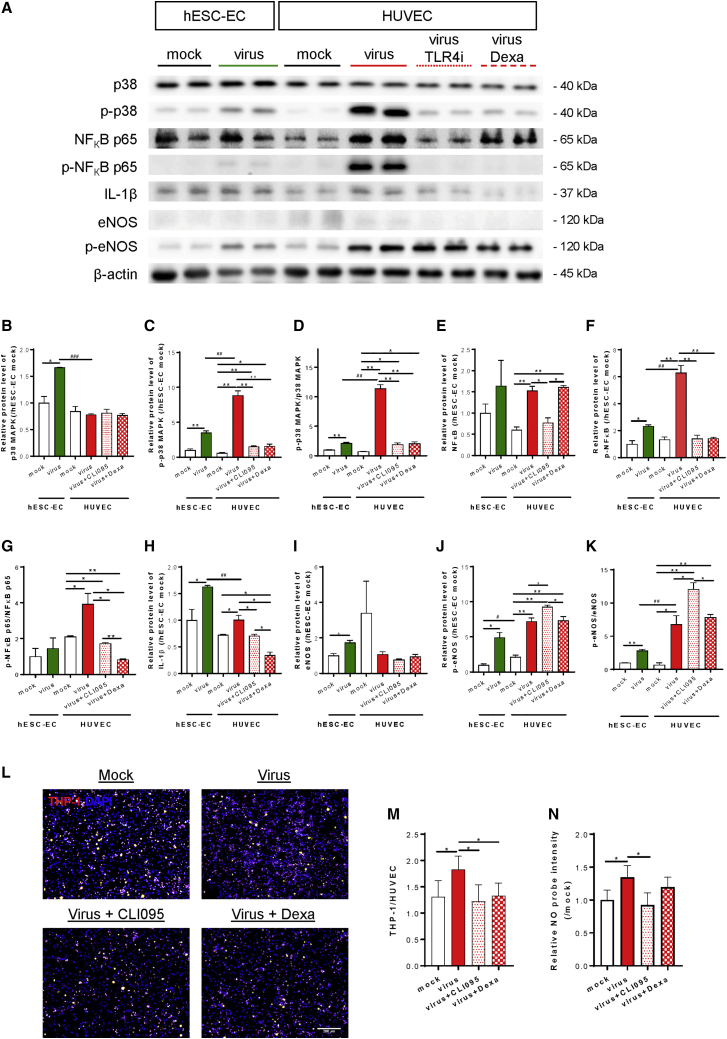
ACE2-deficient human ECs can recognize and be activated by SARS-CoV-2 through TLR4
Since hESC derivatives have been generally acknowledged as less mature than their human counterparts, we, therefore, employed HUVEC as an additional cell model to examine whether SARS-CoV-2 could induce inflammatory activation of human ECs without ACE2. We performed western blotting to compare the degree of TLR4 pathway activation in ACE2-deficient hESC-ECs and HUVEC at 72 h after viral exposure (Figure 5A). The levels of p38 MAPK expression (Figure 5B), phosphorylation at Thr180/Tyr182 (Figure 5C), and phosphorylated/total protein levels (Figure 5D); NF-κB p65 expression (Figure 5E), phosphorylation at Ser536 (Figure 5F), and phosphorylated/total protein levels (Figure 5G); IL-1β expression (Figure 5H); as well as eNOS expression (Figure 5I), phosphorylation at Ser632 (Figure 5J), and phosphorylated/total protein levels (Figure 5K) appeared to be more significantly elevated in hESC-ECs and HUVEC after viral exposure compared with their respective mock infection controls. Importantly, expression levels and activities of p38 MAPK/NF-κB/eNOS were more significantly elevated in HUVEC than that of hESC-ECs after viral exposure. Furthermore, CLI095 or dexamethasone significantly reduced SARS-CoV-2-mediated expression/activation levels of p38 MAPK (Figures 5B–5D), NF-κB p65 (Figures 5E–5G), and IL-1β (Figure 5H), suggesting that the virus directly induced endothelial inflammation via the TLR4/p38 MAPK/NF-κB signaling axis. Nevertheless, neither CLI095 nor dexamethasone mitigated the virus-induced eNOS activation in HUVEC (Figures 5I–5K).
Figure 5.
Human ECs can recognize and be activated by SARS-CoV-2 through TLR4
(A–K) (A) Western blotting analysis at 72 h after viral exposure showing that p38 MAPK, NF-κB, and eNOS pathways were significantly increased in ACE2-deficient hESC-ECs and HUVEC compared with their respective mock infection controls; and activation of p38 MAPK and NF-κB pathways were aborted by TLR blockade through CLI095 or dexamethasone in HUVEC. (B) p38 MAPK, (C) p-p38 MAPK at Thr180/Tyr182, (D) p-p38 MAPK/MAPK, (E) NF-κB, (F) p-NF-κB at Ser536, (G) p-NF-κB/NF-κB, (H) IL-1β, (I) eNOS, (J) p-eNOS at Ser632, and (K) p-eNOS/eNOS. (B–K) Data are presented as mean ± SD, n = 3–6, the relative protein levels were compared with hESC-EC mock infection controls (value = 1), differences were determined by two-way ANOVA and Tukey’s HSD post hoc test, ∗p < 0.05, ∗∗p < 0.01 compared within the same cell type; or #p < 0.05, ##p < 0.01, ###p < 0.001 compared with the same treatment conditions of another cell type. (L and M) Monocyte adhesion assays showing significantly increased numbers of adhered fluorescence-labeled THP-1 (red) to HUVEC at 72 h after inoculation compared with mock infection controls that were significantly blocked by CLI095 or dexamethasone. DAPI (blue) indicates cell nuclei. Scale bar, 200 μm. Data are presented as mean ± SD, n = 8, differences were determined by one-way ANOVA and Tukey’s HSD post hoc test, ∗p< 0.05. (N) NO production assays showing significantly increased NO probes detected in HUVEC at 72 h after inoculation compared with mock infection controls that were significantly aborted by CLI095. Data are presented as mean ± SD, n = 4, the relative probe intensities were compared with that of mock infection controls (value = 1), differences were determined by one-way ANOVA and Tukey’s HSD post hoc test, ∗p < 0.05.
Next, we examined whether the observed molecular activation of TLR4/p38 MAPK/NF-κB would lead to inflammation of HUVEC through monocyte adhesion and NO production assays at 72 h after viral exposure. In contrast to hESC-ECs (Figure 4L–N), SARS-CoV-2 directly and significantly increased the numbers of adhered THP-1 (Figures 5L and 5M) as well as NO production (Figure 5N) in HUVEC after viral inoculation compared with mock infection controls. More importantly, the virus-induced monocyte adhesion was aborted by CLI095 or dexamethasone (Figure 5M); and the virus-mediated NO production was also blocked by CLI095 (Figure 5N). Taken together, SARS-CoV-2 directly promoted inflammatory activation, monocyte adhesion, and NO production of HUVEC through TLR4 activation.
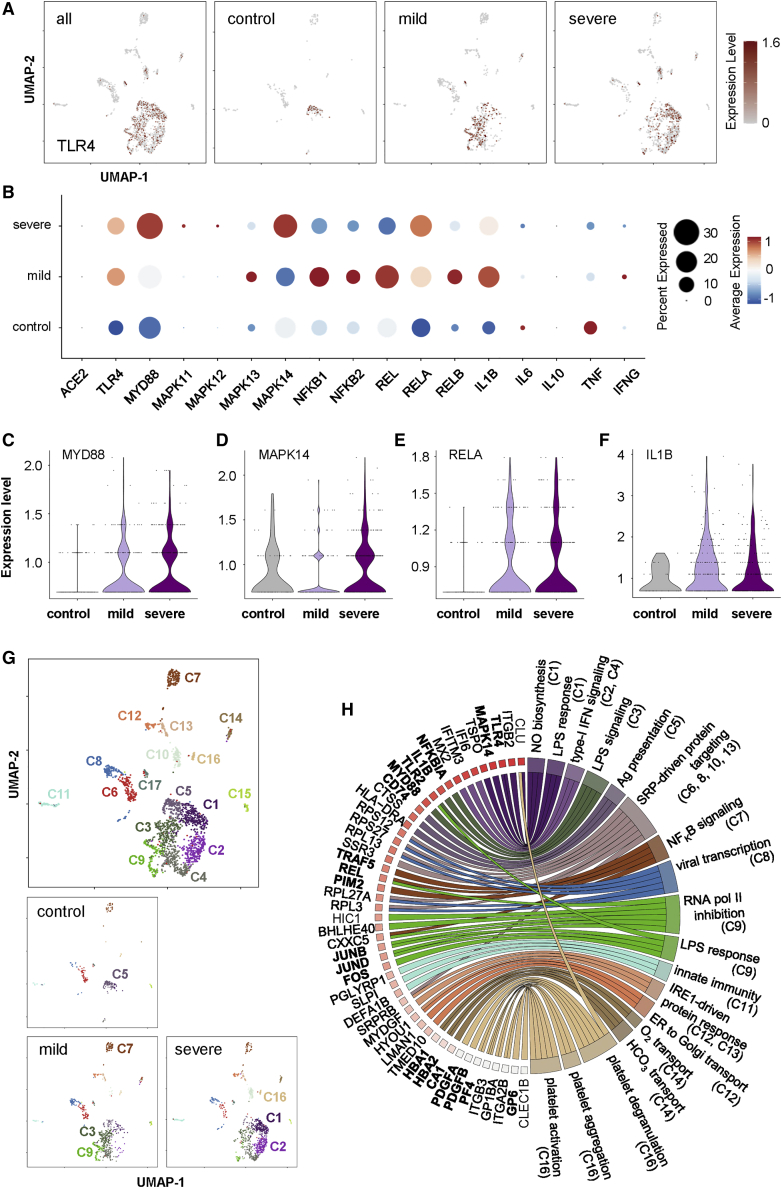
Genome-wide transcriptomic profiling reveals activation of the TLR4/p38 MAPK/NF-κB/IL1B axis in circulating ECs of COVID-19 patients
To date, circulating ECs are rare cells released from blood vessels during vascular damage, remodeling, or dysfunction, which have been regarded as non-invasive biomarkers of cardiovascular disease (Smadja et al., 2009). Essentially, they have also been demonstrated as biomarkers of endothelial injury in severe COVID-19 patients (Guervilly et al., 2020). To validate whether our in vitro findings are recapitulated in vivo after SARS-CoV-2 exposure to human ECs, we analyzed a droplet-based scRNA-seq dataset of 139,848 peripheral blood mononuclear cells (PBMCs) derived from a sex- and age-matched cohort containing 50 samples of 8 mild, 9 severe COVID-19 patients, and 14 samples of 13 controls (Schulte-Schrepping et al., 2020). We further analyzed the data using a total of 2,236 CD31+CD45− circulating ECs within the PBMC population after filtering. Expression analysis of TLR4 projected on Uniform Manifold Approximation and Projection (UMAP) (Figure 6A) and dot plots (Figure 6B) demonstrated increased numbers of TLR4-expressing circulating ECs in mild and severe COVID-19 patients compared with the controls. Importantly, the majority of circulating ECs of COVID-19 patients or controls demonstrated no or low expression levels of ACE2 (Figure 6B), further supporting our data in Figure 1. Moreover, increased numbers of circulating ECs with increased expressing levels of genes downstream of TLR4, including MYD88; MAPK11, MAPK12, MAPK13, and MAPK14 encoding p38 MAPK; NF-κB1, NFκB2, REL, RELA, and RELB encoding NF-κB; and IL1B were observed in both mild and severe COVID-19 patients compared with controls (Figure 6B). Of note, genes encoding IFN-α and IFN-β were not detected in circulating ECs of all groups (data not shown), and expression levels of IL-6, IL-10, TNF-α, and IFN-γ were not significantly increased in COVID-19 patients compared with the controls (Figure 6B), suggesting that endothelial expression of these cytokines might not be specific to COVID-19 pathogenesis. Furthermore, increased expression levels of MYD88 (Figure 6C), MAPK14 (Figure 6D), and RELA (Figure 6E) encoding NF-κB p65, and IL1B (Figure 6F), were found in circulating ECs of mild and severe COVID-19 patients compared with that of the controls, supporting inflammatory activation of the TLR4/p38 MAPK/NF-κB/IL1B signaling axis after endothelial exposure to SARS-CoV-2.
Figure 6.
Single-cell transcriptomic profiling reveals inflammatory activation of the TLR4/p38 MAPK/NF-κB/IL1B axis in human circulating ECs of COVID-19 patients
(A) Visualization of scaled expression of TLR4 in CD31+CD45− circulating ECs (2,236 out of 139,848 PBMCs) using UMAP; 50 samples from 8 mild, 9 severe COVID-19 patients, and 13 controls recruited in the same cohort of a previous study (Schulte-Schrepping et al., 2020). (B) Dot plot representation of scaled expression of ACE2, marker genes of TLR4/p38 MAPK/NF-κB and pro-inflammatory cytokines expressed in circulating ECs of all samples. (C–F) Violin plots showing expression of (C) MYD88, (D) MAPK14, (E) RELA, and (F) IL1B in circulating ECs of all samples. (G) UMAP visualization of 17 clusters of circulating ECs identified in all samples, cells are colored according to the identified clusters. (H) Circular gene ontology (GO) plot representing a selection of one to three pathways of each cluster determined by GO associated to genes detected as significantly upregulated (p < 0.05) comparing expression levels of that gene with the rest of clusters, differences were determined by Seurat’s implementation of the Wilcoxon rank-sum test.
To delineate the heterogeneity of circulating ECs during COVID-19 progression, we further performed unsupervised analyses that did not rely on known subset markers as described previously (Leung et al., 2018a; Li et al., 2019, 2020a, 2020b; Liang et al., 2020). UMAP and high-resolution cell-type classification further identified 17 distinct clusters of circulating ECs of COVID-19 patients and controls (Figure 6G, C1–C17). We then extracted the information regarding significantly and differentially expressed genes of each cluster by comparing gene expression levels of the cluster with that of the rest of clusters, and performed gene ontology (GO) functional annotations as demonstrated by both upregulated (Table S2) and downregulated (Table S3) pathway analyses (Figure S3). Circular GOplot showed some functional association with these clusters (Figure 6H). In particular, circulating ECs of mild COVID patients were distinguished by enrichment of C3, C7, and C9; of which, C3 was distinguished by increased lipopolysaccharide (LPS)-mediated signaling pathway (Table S2, GO:0031663) and reduced viral transcription (Table S3, GO:0019083); C7 was characterized by positive regulation of IκB kinase/NF-κB signaling (Table S2, GO:0043123) and reduced platelet degranulation (Table S3, GO:0002576); and C9 was described by increased negative regulation of transcription from RNA polymerase II promoter (Table S2, GO:0000122), enhanced response to LPS (Table S2, GO:0032496) and reduced viral transcription (Table S3, GO:0019083). Moreover, circulating ECs of severe patients were distinguished by enrichment of C1, C2, and C16; of which, C1 was marked by positive regulation of NO biosynthesis (Table S2, GO:0045429), increased cellular response to LPS (Table S2, GO:0071222) and reduced viral transcription (Table S3, GO:0019083); C2 was characterized by increased type-I IFN signaling pathway Table S2, GO:0060337) and reduced viral transcription (Table S3, GO:0019083); and C16 was described by increased platelet degranulation (Table S2, GO:0002576), aggregation (Table S2, GO:0070527), and activation (Table S2, GO:0030168). Furthermore, circulating ECs of control individuals were characterized by enrichment of C5 (Figure 6G) with increased immune response (Table S2, GO:0006955) and reduced viral transcription (Table S3, GO:0019083). Therefore, circulating ECs of mild and severe COVID-19 patients also demonstrated increased LPS-mediated signaling that is operated via the TLR4/NF-κB signaling axis.
Taken together, our genome-wide transcriptomic analyses further supported our in vitro findings that the TLR4/p38 MAPK/NF-κB/IL1B signaling axis was activated in human ECs after exposure to SARS-CoV-2. Moreover, human ECs could be the major contributors of IL1B during COVID-19 progression. Circulating ECs could also serve as biomarkers for indicating severe COVID-19 patients who demonstrated increased endothelial expression of genes related to NO biosynthesis, platelet degranulation, aggregation, and activation.
Discussion
In the field of cardiovascular studies, COVID-19 has been regarded as an endothelial disorder (Libby and Luscher, 2020). SARS-CoV-2 was detected in both adult (Varga et al., 2020) and pediatric (Colmenero et al., 2020) endothelial tissues. It has been widely acknowledged that ACE2 is ubiquitously expressed in the vasculature, including small and large arteries and veins of all human organs studied (Hamming et al., 2004). A recent study has also demonstrated that ECs derived from human pluripotent stem cells (hPSCs) express ACE2 (Yang et al., 2020). However, another report showed the lack of ACE2 expression and replicative infection by SARS-CoV-2 in human ECs through analyzing the RNA-seq experiments on ENCODE datasets (McCracken et al., 2021). Moreover, it has been documented that the endothelium of human lungs was not infected by SARS-CoV-2 in ex vivo cultures (Hui et al., 2020). These controversies prompt us to re-assess ACE2 expression in human adult and fetal ECs lining large vessels, such as the aorta and umbilical cord, as well as capillaries in various vascularized organs, including the lungs, liver, and placenta. Our studies demonstrated that the majority of adult and fetal human ECs rarely expressed ACE2 on cell surfaces, although intracellular ACE2 was detected in primary lung ECs. Experimental EC lines, including hESC-ECs, also did not express ACE2, as demonstrated by flow cytometry, western blotting, and qRT-PCR. Our results highlighted the need of ectopic expression of ACE2 in hPSC-derived ECs for modeling SARS-CoV-2-mediated endothelial dysfunction. Given that endothelial ACE2 was barely expressed on the cell surface, at least at the physiological state, how SARS-CoV-2 infects the endothelium remains to be investigated. Nevertheless, ECs also express other cell surface receptors including neuropilin-1, scavenger receptor B type 1 (SR-B1), and CD147, which have been reported to assist SARS-CoV-2 cell entry (for review, see Ma et al., 2021). Therefore, we asked whether ACE2-deficient ECs can still be infected by SARS-CoV-2. We demonstrated that ACE2 expression was indispensable for SARS-CoV-2 entry, replication, and infectivity in human ECs as ACE2-deficient ECs were not infected by the virus.
Since the majority of human ECs did not have surface ACE2 expression, we then investigated whether endotheliitis observed in COVID-19 patients is a direct consequence of SARS-CoV-2 infection. Essentially, activation of the vascular endothelium and disruption of endothelial barrier function contribute to production of pro-inflammatory factors and vascular leakage, respectively, which are early hallmarks of vascular disease and thrombosis in SARS-CoV-2-infected rhesus macaques (Aid et al., 2020). We showed that SARS-CoV-2 did not induce apoptosis or impair vascular integrity directly, but promoted endothelial activation as demonstrated by significantly increased monocyte adhesion to human ECs accompanied by significantly enhanced expression and phosphorylation of p38 MAPK and NF-κB, as well as elevated production of the COVID-19 cytokine IL-1β. Moreover, in contrast to the current beliefs (Nagele et al., 2020), SARS-CoV-2 did not impair but directly promoted eNOS activity and NO production in human ECs. In fact, it has been demonstrated that eNOS was activated in inflamed ECs (Lowry et al., 2013). In addition to blood vessels, NO also acts on other cells, such as monocytes and macrophages, as exogenous NO can be pro-inflammatory by inducing NF-κB activation and IL-6 production in macrophages (Connelly et al., 2001).
Interestingly, features of endothelial inflammation were observed in both ACE2-expressing and -deficient ECs after exposure to SARS-CoV-2. We, therefore, asked whether the virus can activate human ECs through TLR4 without host cell entry. In addition to their role in the circulatory system, ECs are also innate immune cells that elicit antiviral immunity after recognizing RNA viruses through TLRs. Among the known TLRs, TLR4 is expressed on the cell surface and is involved in immune response against other RNA viruses, such as influenza (Shirey et al., 2013). Our findings indicated that the expression/activation levels of p38 MAPK/NF-κB/IL-1β were significantly enhanced in ACE2-deficient human ECs in addition to ACE2-expressing ones after viral exposure. Treatment with a TLR4 inhibitor, CLI095, or dexamethasone, aborted the virus-induced monocyte adhesion and NO production in HUVEC accompanied by attenuated activation of p38 MAPK and NF-κB, and reduced IL-1β production. Interestingly, dexamethasone has been demonstrated to lower the 28-day mortality among patients hospitalized with COVID-19 receiving respiratory support (Group et al., 2021), providing clinical benefit that could be explained by our data, as dexamethasone also blocked TLR4/p38 MAPK/NF-κB activation in the endothelium. Altogether, our findings suggest that human ECs were not protected from SARS-CoV-2-mediated activation by being ACE2-deficient as they still recognized and responded to the virus through TLR4 activation. TLR4 blockade via specific inhibitors, such as CLI095, could also minimize side effects from general immunosuppression as a result of dexamethasone treatment.
We also examined whether endothelial ACE2 expression is regulated by cytokines, and investigated its potential role in SARS-CoV-2-mediated endothelial pathologies. Pro-inflammatory cytokines, including IL-1β, IL-6, TNF-α, or IFN-γ did not induce ACE2 expression in ACE2-deficient hESC-ECs; however, IL-1β and TNF-α significantly upregulated its protein levels in ACE2-expressing hESC-ECs. Since these cytokines are downstream effectors of TLR4, there could also be a crosstalk between the ACE2 and TLR4 signaling pathways in ACE2-expressing ECs. Nevertheless, the virus did not alter the expression levels of ACE2 in ACE2-expressing hESC-ECs. Moreover, ectopic or upregulated expression of endothelial ACE2 did not further aggravate endothelial dysfunction as there was no significant increase in the degree of vascular permeability, monocyte adhesion, expression levels of p38 MAPK, NF-κB, IL-1β, and eNOS in ACE2-expressing and -deficient hESC-ECs compared with their respective mock infection controls after viral exposure. These data might suggest that endothelial ACE2 could protect human ECs from TLR4-mediated inflammatory responses. Indeed, clinical evidence shows that the use of angiotensin-converting enzyme inhibitors or Ang-II receptor blockers that potentially increase ACE2 expression in COVID-19 patients with hypertension is associated with reduced risk of all-cause mortality compared with nonusers (Zhang et al., 2020b), further supporting our findings that ACE2 could be protective against SARS-CoV-2 infection at least in human ECs.
One limitation of this study is the lack of animal experimentations. Nevertheless, no mouse model recapitulates all aspects of COVID-19 in humans, especially the unusual features including vascular disease and hyperinflammatory syndromes observed in adults and children (Munoz-Fontela et al., 2020). In this study, we also provided in vivo data by analyzing genome-wide, scRNA-seq data of CD31+CD45− circulating ECs from a sex- and age-matched cohort containing 50 samples of 8 mild, 9 severe COVID-19 patients, and 13 controls using PBMC data collected from a previous study. We demonstrated increased numbers of TLR4-expressing cells as well as increased TLR4 expression levels in circulating ECs of mild and severe COVID-19 patients compared with that of the controls. Moreover, the expression levels of its downstream targets, such as MYD88, MAPK14, and RELA encoding NF-κB p65, and IL1B, were also increased in circulating ECs of mild and severe COVID-19 patients compared with controls, supporting the inflammatory activation of the TLR4/p38 MAPK/NF-κB/IL1B signaling axis after endothelial exposure to SARS-CoV-2. UMAP and high-resolution cell-type classification further demonstrated the heterogeneity of circulating ECs during COVID-19 progression, and identified 17 distinct clusters in these patients. Circulating ECs of mild patients were distinguished by enrichment of C3, C7, and C9; severe patients were marked by C1, C2, and C16; whereas the controls were characterized by C5. Of which, C3, C7, and C1 demonstrated upregulated responses to the TLR4 ligand, LPS, and increased IκB kinase/NF-κB signaling. Furthermore, our analyses might also provide some features of circulating ECs of severe patients, such as increased gene expression related to NO biosynthesis, platelet aggregation, and activation, potentially increasing the risks of thrombosis, suggesting that circulating ECs could serve as biomarkers for indicating severe COVID-19 patients.
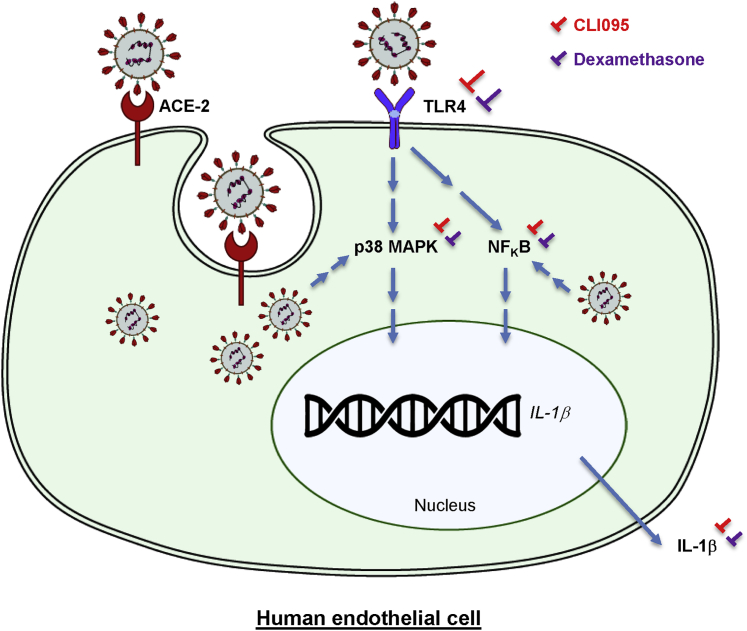
Taken together, our findings suggest that most human ECs could be resistant to SARS-CoV-2 infection by virtue of the lack of surface ACE2 expression. However, they were still susceptible to SARS-CoV-2-mediated inflammatory activation and dysfunction through endothelial TLR4 recognition. Therefore, in addition to ACE2, which was rarely expressed on the human EC surface, targeting endothelial TLR4 signaling could be necessary to combat SARS-CoV-2-induced endothelial inflammation and dysfunction (Figure 7).
Figure 7.
A figure summarizing the mechanisms by which SARS-CoV-2 activates the human endothelium
SARS-CoV-2 can enter human ECs via ACE2, and can also be recognized by human ECs through TLR4 without cell entry. Thereafter, SARS-CoV-2 directly activates endothelial inflammation through p38 MAPK/NF-κB activation to promote production of the COVID-19 cytokine IL-1β. Pharmacological treatment with a specific TLR4 inhibitor CLI095 or dexamethasone aborts SARS-CoV-2-mediated inflammatory activation and dysfunction of human ECs.
Experimental procedures
For details of this section, please also refer to the supplemental experimental procedures.
Human patients
Studies were approved by the Chinese University of Hong Kong (CUHK)-Hospital Authority (NTEC) Joint Clinical Research Ethics Committee. All procedures were performed in accordance with the Declaration of Helsinki; and informed consent was obtained from all patients. The aorta was derived from adult patients with aortic dissection. Normal lung and liver tissues surrounding tumors were excised from adult patients with malignancies. Fetal umbilical cord and placenta were collected from healthy pregnant women after delivery.
SARS-CoV-2 infection
The SARS-CoV-2 virus (BetaCoV/Hong Kong/VM20001061/2020) used in this study originated from a confirmed COVID-19 patient in Hong Kong (Hui et al., 2020). The indicated types of human ECs were seeded in 12-well plates and incubated with culture medium (mock infection control) or culture medium containing SARS-CoV-2 at an MOI of 0.1 for 1 h at 37°C. Virus inoculum was then replaced by fresh culture medium, and incubated at 37°C throughout the experiments. Infection was performed in triplicate. At the indicated time points, culture supernatants were collected for viral quantification, and cells were harvested for subsequent analysis. All the infectious work was conducted in a biosafety level 3 (BSL-3) facility at Li Ka Shing Faculty of Medicine, The University of Hong Kong.
scRNA-seq analysis and functional annotations
The scRNA-seq datasets were acquired from a published source with the accession number EGAS00001004571 (Schulte-Schrepping et al., 2020). We focused on the second cohort of 139,848 PBMCs obtained from 50 samples of 8 mild, 9 severe, and 13 controls, and analyzed using R package Seurat for data scaling, transformation, dimensionality reduction, clustering, visualization, and differentially expressed gene analyses (Butler et al., 2018). In brief, these samples were originally annotated as mild (WHO2-4) or severe (5-7) COVID-19 disease according to the WHO ordinal scale. Data were scaled, transformed and variable genes were identified using the SCTransform() function. Linear regression was performed to remove unwanted variation due to cellular complexity (number of genes per cell, number of UMIs per cell) or cell quality (%mitochondrial reads, %rRNA reads). CD31+CD45− circulating ECs were further filtered for downstream analyses based on the expression of endothelial and haematopoietic marker genes, CD31 and CD45, respectively, in each cell. Principal-component (PC) analysis was performed using variable genes, and the first 50 PCs were used to perform UMAP to map the datasets into two dimensions, and to construct a shared nearest-neighbor graph, which was used to cluster the datasets using a graph-based algorithm. Cluster identity was determined by finding differentially expressed genes of each cluster using Seurat’s implementation of the Wilcoxon rank-sum test, followed by annotations using DAVID. Circular GOplot was used to show the representative GO terms that were most significantly upregulated in each cluster. Dot plots, scatterplots, and violin plots of selected genes were shown by using the DotPlot(), FeaturePlot(), and VlnPlot() functions of Seurat.
Statistical analysis
The data are expressed as arithmetic mean ± SD of biological replicates performed independently under the same conditions three times. Statistical analysis was performed using the unpaired Student’s t test with data from two groups, while data from more than two groups were performed using an ANOVA followed by Tukey’s method for multiple comparisons. Significance was accepted when p < 0.05.
Author contributions
Z.M. performed most of the experiments with help from X.L., R.L.Y.F., K.Y.Y., Z.F., and P.Y. R.L.Y.F., A.W.H.C., and M.Y.A.L. performed all cell culture experiments containing live SARS-CoV-2. C.S.H.N., R.W.H.L., R.H.L.W., K.K.N., and C.C.W. recruited patients and collected human tissues. Z.M., X.L., K.Y.Y., X.Y.T., L.L.M., and K.O.L. analyzed and interpreted the data. Y.H., X.Y.T. and L.L.M.P. contributed reagents and critical comments throughout the study. L.L.M. and K.O.L. supervised the research. K.O.L. conceived the study, engaged in the overall direction and planning, and wrote the manuscript.
Conflict of interests
The authors declare no competing interests.
Acknowledgments
We thank Dr. Martin Chi Wai Chan (Department of Microbiology, CUHK) for all the discussion in the beginning of the study, and Dr. Michael Chi Wai Chan (Division of Public Health Laboratory Sciences, HKU) for contributing an anti-ACE2 antibody. This work was supported by the National Natural Science Foundation of China (81922077, 82070494); the Research Grants Council of Hong Kong (14100021, 14108420, C4026-17WF, M-402-20, T11-705/21-N); the University Grants Committee Research Matching Grant Scheme (2019, 2020, 2021); the Health and Medical Research Council (CID-HKU2); the Croucher Foundation Innovation Award; the Faculty Innovation Award; the Young Researcher Award; the Research Committee Funding, Direct Grants (2020.053), and postgraduate studentship (Z.M., X.L., Z.F., P.Y., and M.P.C.) from CUHK.
Published: February 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.01.015.
Contributor Information
Leo L.M. Poon, Email: llmpoon@hku.hk.
Kathy O. Lui, Email: kathyolui@cuhk.edu.hk.
Supplemental information
.
A total of 2,236 CD31+CD45- circulating ECs out of 139,848 PBMCs collected from 50 samples containing 8 mild, 9 severe COVID-19 patients, and 13 controls recruited in the same cohort of a previous study (Schulte-Schrepping et al., 2020). A selection of the top 3 pathways in terms of biological processes of each cluster determined by gene ontology (GO) associated to genes detected as significantly upregulated (p < 0.05) in scRNA-seq comparing expression of that gene with the rest of the clusters, differences were determined by Seurat’s implementation of the Wilcoxon rank-sum test.
A total of 2,236 CD31+CD45– circulating ECs out of 139,848 PBMCs collected from 50 samples containing 8 mild, 9 severe COVID-19 patients, and 13 controls recruited in the same cohort of a previous study (Schulte-Schrepping et al., 2020). A selection of the top 3 pathways in terms of biological processes of each cluster determined by gene ontology (GO) associated to genes detected as significantly downregulated (p < 0.05) in scRNA-seq comparing expression of that gene with the rest of the clusters, differences were determined by Seurat’s implementation of the Wilcoxon rank-sum test.
References
- Ackermann M., Verleden S.E., Kuehnel M., Haverich A., Welte T., Laenger F., Vanstapel A., Werlein C., Stark H., Tzankov A., et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in COVID-19. N. Engl. J. Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aid M., Busman-Sahay K., Vidal S.J., Maliga Z., Bondoc S., Starke C., Terry M., Jacobson C.A., Wrijil L., Ducat S., et al. Vascular disease and thrombosis in SARS-CoV-2-infected rhesus macaques. Cell. 2020;183:1354–1366.e13. doi: 10.1016/j.cell.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belhadjer Z., Meot M., Bajolle F., Khraiche D., Legendre A., Abakka S., Auriau J., Grimaud M., Oualha M., Beghetti M., et al. Acute heart failure in multisystem inflammatory syndrome in children (MIS-C) in the context of global SARS-CoV-2 pandemic. Circulation. 2020 doi: 10.1161/CIRCULATIONAHA.120.048360. [DOI] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhao B., Qu Y., Chen Y., Xiong J., Feng Y., Men D., Huang Q., Liu Y., Yang B., et al. Detectable serum severe acute respiratory syndrome coronavirus 2 viral load (RNAemia) is closely correlated with drastically elevated interleukin 6 level in critically ill patients with coronavirus disease 2019. Clin. Infect Dis. 2020;71:1937–1942. doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chousterman B.G., Swirski F.K., Weber G.F. Cytokine storm and sepsis disease pathogenesis. Semin. Immunopathol. 2017;39:517–528. doi: 10.1007/s00281-017-0639-8. [DOI] [PubMed] [Google Scholar]
- Colmenero I., Santonja C., Alonso-Riano M., Noguera-Morel L., Hernandez-Martin A., Andina D., Wiesner T., Rodriguez-Peralto J.L., Requena L., Torrelo A. SARS-CoV-2 endothelial infection causes COVID-19 chilblains: histopathological, immunohistochemical and ultrastructural study of seven paediatric cases. Br. J. Dermatol. 2020;183:729–737. doi: 10.1111/bjd.19327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly L., Palacios-Callender M., Ameixa C., Moncada S., Hobbs A.J. Biphasic regulation of NF-kappa B activity underlies the pro- and anti-inflammatory actions of nitric oxide. J. Immunol. 2001;166:3873–3881. doi: 10.4049/jimmunol.166.6.3873. [DOI] [PubMed] [Google Scholar]
- Dayang E.Z., Plantinga J., Ter Ellen B., van Meurs M., Molema G., Moser J. Identification of LPS-activated endothelial subpopulations with distinct inflammatory phenotypes and regulatory signaling mechanisms. Front Immunol. 2019;10:1169. doi: 10.3389/fimmu.2019.01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P.C., Ed Rainger G., Mason J.C., Guzik T.J., Osto E., Stamataki Z., Neil D., Hoefer I.E., Fragiadaki M., Waltenberger J., et al. Endothelial dysfunction in COVID-19: a position paper of the ESC working group for atherosclerosis and vascular biology, and the ESC Council of basic cardiovascular science. Cardiovasc. Res. 2020 doi: 10.1093/cvr/cvaa230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin J.E., Feng Y., Velazquez H., Sessa W.C. Endothelial glucocorticoid receptor is required for protection against sepsis. Proc. Natl. Acad. Sci. U S A. 2013;110:306–311. doi: 10.1073/pnas.1210200110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Group R.C., Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., Linsell L., Staplin N., Brightling C., Ustianowski A., et al. Dexamethasone in hospitalized patients with COVID-19. N. Engl. J. Med. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guervilly C., Burtey S., Sabatier F., Cauchois R., Lano G., Abdili E., Daviet F., Arnaud L., Brunet P., Hraiech S., et al. Circulating endothelial cells as a marker of endothelial injury in severe COVID-19. J. Infect Dis. 2020;222:1789–1793. doi: 10.1093/infdis/jiaa528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui K.P.Y., Cheung M.C., Perera R., Ng K.C., Bui C.H.T., Ho J.C.W., Ng M.M.T., Kuok D.I.T., Shih K.C., Tsao S.W., et al. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir. Med. 2020;8:687–695. doi: 10.1016/S2213-2600(20)30193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung C.S., Yang K.Y., Li X., Chan V.W., Ku M., Waldmann H., Hori S., Tsang J.C.H., Lo Y.M.D., Lui K.O. Single-cell transcriptomics reveal that PD-1 mediates immune tolerance by regulating proliferation of regulatory T cells. Genome Med. 2018;10:71. doi: 10.1186/s13073-018-0581-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung O.M., Li J., Li X., Chan V.W., Yang K.Y., Ku M., Ji L., Sun H., Waldmann H., Tian X.Y., et al. Regulatory T cells promote apelin-mediated sprouting angiogenesis in type 2 diabetes. Cell Rep. 2018;24:1610–1626. doi: 10.1016/j.celrep.2018.07.019. [DOI] [PubMed] [Google Scholar]
- Li J., Liang C., Yang K.Y., Huang X., Han M.Y., Li X., Chan V.W., Chan K.S., Liu D., Huang Z.P., et al. Specific ablation of CD4(+) T-cells promotes heart regeneration in juvenile mice. Theranostics. 2020;10:8018–8035. doi: 10.7150/thno.42943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yang K.Y., Tam R.C.Y., Chan V.W., Lan H.Y., Hori S., Zhou B., Lui K.O. Regulatory T-cells regulate neonatal heart regeneration by potentiating cardiomyocyte proliferation in a paracrine manner. Theranostics. 2019;9:4324–4341. doi: 10.7150/thno.32734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Yang K.Y., Chan V.W., Leung K.T., Zhang X.B., Wong A.S., Chong C.C.N., Wang C.C., Ku M., Lui K.O. Single-cell RNA-seq reveals that CD9 is a negative marker of glucose-responsive pancreatic beta-like cells derived from human pluripotent stem cells. Stem Cell Rep. 2020;15:1111–1126. doi: 10.1016/j.stemcr.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Yang K.Y., Chan V.W., Li X., Fung T.H.W., Wu Y., Tian X.Y., Huang Y., Qin L., Lau J.Y.W., Lui K.O. CD8(+) T-cell plasticity regulates vascular regeneration in type-2 diabetes. Theranostics. 2020;10:4217–4232. doi: 10.7150/thno.40663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby P., Luscher T. COVID-19 is, in the end, an endothelial disease. Eur. Heart J. 2020;41:3038–3044. doi: 10.1093/eurheartj/ehaa623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry J.L., Brovkovych V., Zhang Y., Skidgel R.A. Endothelial nitric-oxide synthase activation generates an inducible nitric-oxide synthase-like output of nitric oxide in inflamed endothelium. J. Biol. Chem. 2013;288:4174–4193. doi: 10.1074/jbc.M112.436022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Yang K.Y., Huang Y., Lui K.O. Endothelial contribution to COVID-19: an update on mechanisms and therapeutic implications. J. Mol. Cell Cardiol. 2021;164:69–82. doi: 10.1016/j.yjmcc.2021.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken I.R., Saginc G., He L., Huseynov A., Daniels A., Fletcher S., Peghaire C., Kalna V., Andaloussi-Mae M., Muhl L., et al. Lack of evidence of angiotensin-converting enzyme 2 expression and replicative infection by SARS-CoV-2 in human endothelial cells. Circulation. 2021;143:865–868. doi: 10.1161/CIRCULATIONAHA.120.052824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil V., Kwon H., Prado P., Hagelkruys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado Del Pozo C., Prosper F., et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913 e907. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Fontela C., Dowling W.E., Funnell S.G.P., Gsell P.S., Riveros-Balta A.X., Albrecht R.A., Andersen H., Baric R.S., Carroll M.W., Cavaleri M., et al. Animal models for COVID-19. Nature. 2020;586:509–515. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagele M.P., Haubner B., Tanner F.C., Ruschitzka F., Flammer A.J. Endothelial dysfunction in COVID-19: current findings and therapeutic implications. Atherosclerosis. 2020;314:58–62. doi: 10.1016/j.atherosclerosis.2020.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perico L., Benigni A., Casiraghi F., Ng L.F.P., Renia L., Remuzzi G. Immunity, endothelial injury and complement-induced coagulopathy in COVID-19. Nat. Rev. Nephrol. 2020 doi: 10.1038/s41581-020-00357-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles V.G., Lutgehetmann M., Lindenmeyer M.T., Sperhake J.P., Wong M.N., Allweiss L., Chilla S., Heinemann A., Wanner N., Liu S., et al. Multiorgan and renal tropism of SARS-CoV-2. N. Engl. J. Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Schrepping J., Reusch N., Paclik D., Bassler K., Schlickeiser S., Zhang B., Kramer B., Krammer T., Brumhard S., Bonaguro L., et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440 e1423. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirey K.A., Lai W., Scott A.J., Lipsky M., Mistry P., Pletneva L.M., Karp C.L., McAlees J., Gioannini T.L., Weiss J., et al. The TLR4 antagonist Eritoran protects mice from lethal influenza infection. Nature. 2013;497:498–502. doi: 10.1038/nature12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smadja D.M., Gaussem P., Mauge L., Israel-Biet D., Dignat-George F., Peyrard S., Agnoletti G., Vouhe P.R., Bonnet D., Levy M. Circulating endothelial cells: a new candidate biomarker of irreversible pulmonary hypertension secondary to congenital heart disease. Circulation. 2009;119:374–381. doi: 10.1161/CIRCULATIONAHA.108.808246. [DOI] [PubMed] [Google Scholar]
- South A.M., Diz D.I., Chappell M.C. COVID-19, ACE2, and the cardiovascular consequences. Am. J. Physiol. Heart Circ. Physiol. 2020;318:H1084–H1090. doi: 10.1152/ajpheart.00217.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga Z., Flammer A.J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A.S., Mehra M.R., Schuepbach R.A., Ruschitzka F., Moch H. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K., Ohishi M., Katsuya T., Ito N., Ikushima M., Kaibe M., Tatara Y., Shiota A., Sugano S., Takeda S., et al. Deletion of angiotensin-converting enzyme 2 accelerates pressure overload-induced cardiac dysfunction by increasing local angiotensin II. Hypertension. 2006;47:718–726. doi: 10.1161/01.HYP.0000205833.89478.5b. [DOI] [PubMed] [Google Scholar]
- Yang L., Han Y., Nilsson-Payant B.E., Gupta V., Wang P., Duan X., Tang X., Zhu J., Zhao Z., Jaffre F., et al. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27:125–136.e7. doi: 10.1016/j.stem.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeuke S., Ulmer A.J., Kusumoto S., Katus H.A., Heine H. TLR4-mediated inflammatory activation of human coronary artery endothelial cells by LPS. Cardiovasc. Res. 2002;56:126–134. doi: 10.1016/s0008-6363(02)00512-6. [DOI] [PubMed] [Google Scholar]
- Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Zhu L., Cai J., Lei F., Qin J.J., Xie J., Liu Y.M., Zhao Y.C., Huang X., Lin L., et al. Association of inpatient use of angiotensin-converting enzyme inhibitors and angiotensin II receptor blockers with mortality among patients with hypertension hospitalized with COVID-19. Circ. Res. 2020;126:1671–1681. doi: 10.1161/CIRCRESAHA.120.317134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
.
A total of 2,236 CD31+CD45- circulating ECs out of 139,848 PBMCs collected from 50 samples containing 8 mild, 9 severe COVID-19 patients, and 13 controls recruited in the same cohort of a previous study (Schulte-Schrepping et al., 2020). A selection of the top 3 pathways in terms of biological processes of each cluster determined by gene ontology (GO) associated to genes detected as significantly upregulated (p < 0.05) in scRNA-seq comparing expression of that gene with the rest of the clusters, differences were determined by Seurat’s implementation of the Wilcoxon rank-sum test.
A total of 2,236 CD31+CD45– circulating ECs out of 139,848 PBMCs collected from 50 samples containing 8 mild, 9 severe COVID-19 patients, and 13 controls recruited in the same cohort of a previous study (Schulte-Schrepping et al., 2020). A selection of the top 3 pathways in terms of biological processes of each cluster determined by gene ontology (GO) associated to genes detected as significantly downregulated (p < 0.05) in scRNA-seq comparing expression of that gene with the rest of the clusters, differences were determined by Seurat’s implementation of the Wilcoxon rank-sum test.