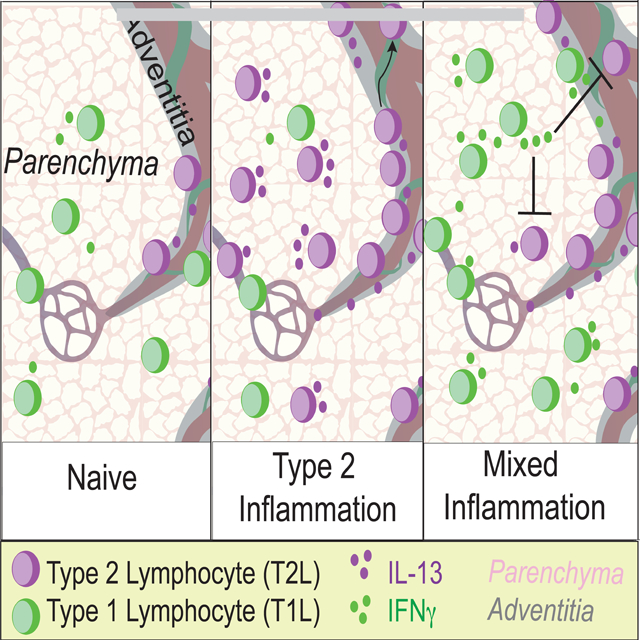
SUMMARY
Allergic immunity is orchestrated by group 2 innate lymphoid cells (ILC2s) and type 2 helper T (Th2) cells prominently arrayed at epithelial- and microbial-rich barriers. However, ILC2s and Th2 cells are also present in fibroblast-rich niches within the adventitial layer of larger vessels and similar boundary structures in sterile deep tissues, and it remains unclear if they undergo dynamic repositioning during immune perturbations. Here we used thick-section quantitative imaging to show that allergic inflammation drives invasion of lung and liver non-adventitial parenchyma by ILC2s and Th2 cells. However, during concurrent type 1 and type 2 mixed inflammation, IFNγ from broadly distributed type 1 lymphocytes directly blocked both ILC2 parenchymal trafficking and subsequent cell survival. ILC2 and Th2 cell confinement to adventitia limited mortality by the type 1 pathogen Listeria monocytogenes. Our results suggest that the topography of tissue lymphocyte subsets is tightly regulated to promote appropriately timed and balanced immunity.
eTOC BLURB:
Effector type 2 lymphocytes are involved in both physiologic and pathologic responses, yet their physical tissue-niches are poorly described. Here Cautivo and colleagues identify a secondary domain in tissue parenchyma for type 2 lymphocytes that is dominantly restricted by IFNγ-producing type 1 lymphocytes, a restriction necessary to mount a beneficial mixed type 1 - type 2 inflammatory response.
Graphical Abstract
INTRODUCTION
Type 2 allergic immunity limits excessive colonization by helminths and protozoa but can also drive asthma, atopic dermatitis, and allergy, with over 20% of the world’s population suffering from allergic pathology (Lambrecht and Hammad, 2017). Recent work supports non-canonical contributions of type 2 immune cells and cytokines in tissue development, physiology, and remodeling (Allen and Sutherland, 2014; Cautivo and Molofsky, 2016; Gieseck et al., 2018; Lloyd and Snelgrove, 2018; Nguyen et al., 2020; Vainchtein et al., 2018). Type 2 immune responses are directed by the cytokines Interleukin (IL)-4, IL-5, IL-9 and IL-13, which are abundantly produced by both adaptive CD4+ type 2 helper T (Th2) cells and group 2 innate lymphoid cells (ILC2s) (Fort et al., 2001; Hurst et al., 2002; Moro et al., 2010; Neill et al., 2010; Price et al., 2010; Ruterbusch et al., 2020). ILC2s lack T cell receptors and respond directly to local stress signals associated with tissue perturbation and damage (Colonna, 2018; Dahlgren and Molofsky, 2018). However, in non-lymphoid tissues ILC2s and Th2 cell subsets are transcriptionally similar, expressing both IL-5 and IL-13, responding directly to shared upstream tissue signals, and often displaying functional redundancy (Van Dyken et al., 2016; Endo et al., 2015; Hondowicz et al., 2016; Tibbitt et al., 2019).
As tissue lymphocytes, tissue ILC2 and Th2 cell function is tightly controlled by the integration of multiple local signals (Fan and Rudensky, 2016; Gasteiger et al., 2015), including the cytokines Interleukin-33 (IL-33) and thymic stromal lymphopoietin (TSLP) (Corren and Ziegler, 2019; Han et al., 2017a; Liew et al., 2016; Molofsky et al., 2015a). ILC2 and Th2 subsets, both marked by IL-5 expression, reside in adventitial regions of multiple deep tissues such as lung and liver that lack significant numbers of resident microbes (Dahlgren and Molofsky, 2019). Adventitial regions are the outermost, collagen-rich boundary layer of larger vessels, airways, and ducts, and are defined by adventitial fibroblasts, a stromal cell type that can produce both IL-33 and TSLP and function as a ‘niche’ cell for effector IL-5+ lymphocytes (i.e. ILC2s and Th2 cells), type-2-like regulatory T (Treg) cells, and other immune subsets (Buechler et al., 2021; Dahlgren and Molofsky, 2019; Dahlgren et al., 2019; Mahlakõiv et al., 2019; Puttur et al., 2019; Rana et al., 2019; Spallanzani et al., 2019). Allergic insults drive the expansion of IL-5+ lymphocytes and fibroblasts at adventitial niches but also leads to the accumulation of IL-5+ ILC2s and Th2 cells within non-adventitial ‘parenchymal’ sites (Dahlgren et al., 2019). This suggests the possibility of inflammation-driven de novo type 2 lymphocyte domains, but neither the signals that regulate lymphocyte accumulation at these areas nor the topographic significance is known (Cautivo et al., 2020; Dahlgren and Molofsky, 2019).
Many immune challenges that drive ILC2s and Th2 cells also engage other arms of lymphocytic immunity. The best studied is concurrent type 1 and type 2 mixed inflammation, where we and others previously identified IFNγ, the canonical type 1 cytokine, as a potent repressor of ILC2 activation, proliferation, and type 2 cytokine production (Duerr et al., 2016; Han et al., 2017b; Molofsky et al., 2015b; Moro et al., 2016; Stier et al., 2017). Reciprocal regulation of CD4+ Th1 and Th2 cells occurs both during T helper cell development in secondary lymphoid organs and in fully differentiated lymphocyte subsets in non-lymphoid tissues (Coyle et al., 1996; Gavett et al., 1995; Maggi et al., 1992; Marsland et al., 2004; Reinhold et al., 1993; Ruterbusch et al., 2020). However, the impact of topography on IFNγ-producing type 1 lymphocytes (T1Ls) restricting effector ILC2s and Th2 cells, particularly in non-lymphoid tissues, is not well described.
Here, we found that type 2 inflammation driven by helminth infection or IL-33 treatment promotes trafficking and accumulation of both ILC2s and IL-5+ Th2 cells in lung and liver parenchyma. IFNγ production by T1Ls that were distributed across both parenchymal and adventitial sites directly acted on ILC2s and IL-5+ Th2 cells, restricting their IL-13 production and parenchymal trafficking and survival. Using models of mixed type 1 - type 2 inflammation and infection, we found that IFNγ-mediated repression of IL-5+ ILC2 and Th2 cell parenchymal redistribution was required to limit morbidity and mortality associated with infection by the model type-1 immune-inducing bacteria, Listeria monocytogenes. Together, our work supports an emergent paradigm, whereby type 1 and type 2 lymphocyte topographic cross-regulation at discrete tissue microdomains is a critical determinant of beneficial and pathologic organ inflammation and repair.
RESULTS:
ILC2s and IL-5+ Th2 cells transiently expand in lung and liver parenchyma during type 2 inflammation.
To determine the distribution of ILC2s and Th2 cells across tissue adventitial niches and parenchymal sites (Fig. 1A), we used IL-5 lineage tracker mice (Fig. S1A; Il5dtomato-Cre; Rosa26fsf-RFP-Ai14) (Dahlgren et al., 2019; Molofsky et al., 2013, 2015b; Nussbaum et al., 2013) and performed confocal imaging and histocytometry from thick sections of cleared tissue (Gerner et al., 2012; Li et al., 2017; Tang et al., 2013). The IL-5 fate mapper mice were both sensitive and specific, with virtually all tissue ILC2s and rare T cells marked in naïve mice, whereas myeloid or non-hematopoietic cells remained unlabeled (Fig. S1B) (Cameron et al., 2019; Dahlgren et al., 2019; Molofsky et al., 2013, 2015b; Nussbaum et al., 2013). After infection with the helminth Nippostrongylus brasiliensis (Nb), ILC2s and a subset of tissue CD4+ Th2 cells were IL-5 lineage marked (Fig. S1B–C). IL-5+ ILC2s and Th2 cells have shared properties and partially redundant functions (Van Dyken et al., 2016; Endo et al., 2015; Hondowicz et al., 2016; Tibbitt et al., 2019), and as such we collectively refer to them as effector IL-5+ type 2 lymphocytes (IL-5+ T2Ls or IL-5+ lymphocytes), noting that their regulation and impact may diverge in certain settings (Schuijs and Halim, 2016). In resting lungs, IL-5+ lymphocytes (predominantly ILC2s) resided in adventitial niches around larger vessels and airways (Fig. 1B, Video S1) (Dahlgren et al., 2019). However, after activation of type 2 immunity with exogenous IL-33, which dominantly drives ILC2s, or infection with Nb helminths, which drives mixed Th2 cells and ILC2s, IL-5+ lymphocytes accumulated in both adventitial niches and de novo parenchymal sites, with 40% of cells now residing in the parenchyma in proximity to alveoli (Fig. 1C–F, Video S2,3). In the liver and perigonadal adipose tissue (GAT), type 2 challenges also resulted in IL-5+ ILC2 (IL-33 treatment) or IL-5+ Th2 cell and ILC2 (Nb infection) expansion and appearance within parenchymal areas in proximity to hepatocytes (Fig. 1G–I, Video S4) or adipocytes (not shown). After Nb infection, IL-5+ Th2 cells accounted for a majority of IL-5+ lymphocytes (~70%); however, both ILC2s and IL-5+ Th2 cells had similar topographic distributions across adventitial niches and parenchymal sites (Fig. S1C–D) (Dahlgren et al., 2019). Parenchymal areas are enriched with microvasculature; however, after Nb infection or IL-33 treatment, only a minority of IL-5+ lymphocytes transiently resided in the microvasculature of lung, as revealed by anti-CD45 labeling (Anderson et al., 2014) (Fig S1E–F). Liver IL-5+ ILC2s and Th2 cells were variably vascular accessible after type 2 challenge (ivCD45+, not shown), and consistent with the porous nature of hepatic sinus microvasculature. Together, these data demonstrate that ILC2s and IL-5+ Th2 cells accumulate within bona fide parenchymal interstitial sites in response to innate and adaptive type 2 inflammatory stimulants.
Figure 1: IL-5+ type 2 lymphocytes expand in lung and liver parenchyma during type 2 inflammation.
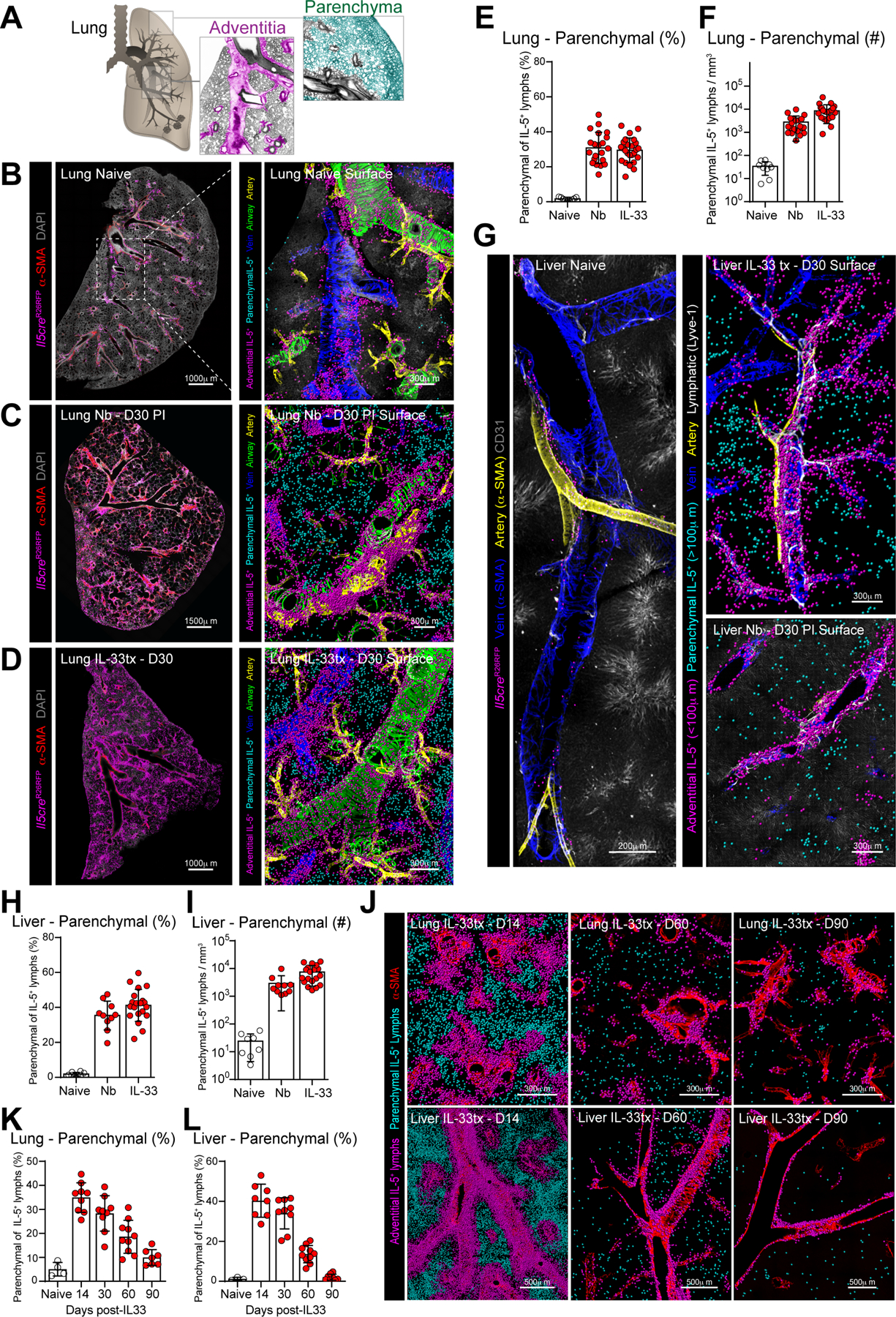
(A) Schematics of lung topography highlighting adventitial (pink) and parenchymal (teal) domains.
(B-D) Confocal imaging of lung thick-sections (left) or surface-rendered three-dimensional reconstruction (right) from IL-5+ T2L lineage tracker naïve mice, Nippostrongylus brasiliensis infected (Nb) at day 30 (D30) post-infection (PI), or IL-33 treated at D30 post treatment, as indicated. 3 independent experiments, total N=5–6 mice/group.
(E-F, H-I) Quantitative analysis of lung and liver thick-section confocal images as percent parenchymal IL-5+ T2Ls of total IL-5+ T2Ls (E and H) or parenchymal IL-5+ T2L numbers per tissue volume (F and I). Pooled from 3 independent experiments, total N=5–6 mice/group.
(G) Confocal imaging of liver thick-sections at resting, D30 PI with Nb and D30 post IL-33 treatment showing surface-rendered reconstruction, as described in B-D.
(J-L) Confocal imaging of lung and liver sections at indicated days post IL-33 treatment showing surface-rendered 3D reconstruction, as described in B-D and quantitative imaging analysis (K-L) of parenchymal IL-5+ T2L accumulation as a percent of total IL-5+ T2L. Images and quantification from 2 independent experiments, total N=4–5 mice per time point.
See also Figure S1.
Next, we examined the kinetics of IL-5+ type 2 lymphocyte parenchymal accumulation. As assessed by flow cytometry, IL-33 stimulated ILC2s remained elevated in the lungs, liver, and GAT, and only gradually declined over several months (Fig S1G–H, not shown). However, lung and liver thick-section imaging revealed the preferential loss of parenchymal ILC2s over this time frame (Fig 1J–L, Fig S1I–J). To assess the tissue-intrinsic determinants underlying the adventitial or parenchymal localization of IL-5+ lymphocytes, we transferred sort-purified IL-5-RFP+ ILC2s into naive hosts and examined their accumulation and localization in lungs (Fig S1K). We found that adoptively transferred IL-5+ ILC2s required GPCR-mediated signaling sensitive to pertussis toxin to accumulate within lung tissue, but not to expand in vitro or localize inside of lung vasculature (Fig S1L–N). In lymphocyte-deficient (Il7ra−/−) recipients, a majority of ILC2s resided in lung adventitial niches (~75%); in contrast, lymphocyte-replete littermates (Il7ra+/−) had reduced total lung ILC2 accumulation with relatively increased parenchymal occupancy (Fig S1L–P, ~50%). These data suggest that ILC2s preferentially sustain long-term accumulation at adventitial niches. Parenchymal regions may represent secondary sites engaged during acute to sub-acute type 2 inflammation and/or during times of adventitial niche occupancy, possibly associated with type 2 immune perturbations (Dahlgren et al., 2019).
IFNγ-producing type 1 lymphocytes restrict ILC2 and Th2 cell parenchymal expansion during mixed inflammation.
Next, we compared the topographic distribution of IL-5+ ILC2s and Th2 cells with IFNγ+ T1Ls. First, using the Ifng-YFP ‘Yeti’ reporter mouse (Fig 2A; IfngYFP-Yeti/+) (Reinhardt et al., 2015; Stetson et al., 2003), we found that Ifng-YFP+ T1Ls were diffusely distributed across adventitial and parenchymal domains in the liver, lung, and GAT (Fig 2B–E, Fig S2A, Video S5). Ifng-YFP+ cells included innate NKs and ILC1s as well as adaptive CD4+ T cells, CD8+ T cells, and CD4− CD8− CD3e+ unconventional T cells; tissue-resident (ivCD45−) T1Ls were predominantly comprised of T cells (CD3e+) and were distributed throughout tissue with some enrichment in adventitial regions (Fig S2B–F). Similar results were found using Tbet-zsGreen reporter mice (Zhu et al., 2012), labeling cells that express the Tbx21 transcription factor associated with competency for IFNγ production (Fig S2G–J). To drive type 1 immunity, we infected mice with Listeria monocytogenes (Lm), an intracellular bacteria that replicates in the liver and spleen and drives an IFNγ+ T1L response that is required for bacterial clearance (Chávez-Arroyo and Portnoy, 2020; Radoshevich and Cossart, 2018). Lm infection promoted a trend towards increased parenchymal liver T1Ls (Fig 2D–E, Video S5) with multiple liver granuloma in parenchymal regions that were strongly T1L-enriched (Fig S2K, Video S5) (Kang et al., 2008).
Figure 2: IFNγ-producing type 1 lymphocytes restrict IL-5+ type 2 lymphocyte parenchymal expansion during mixed inflammatory challenges.
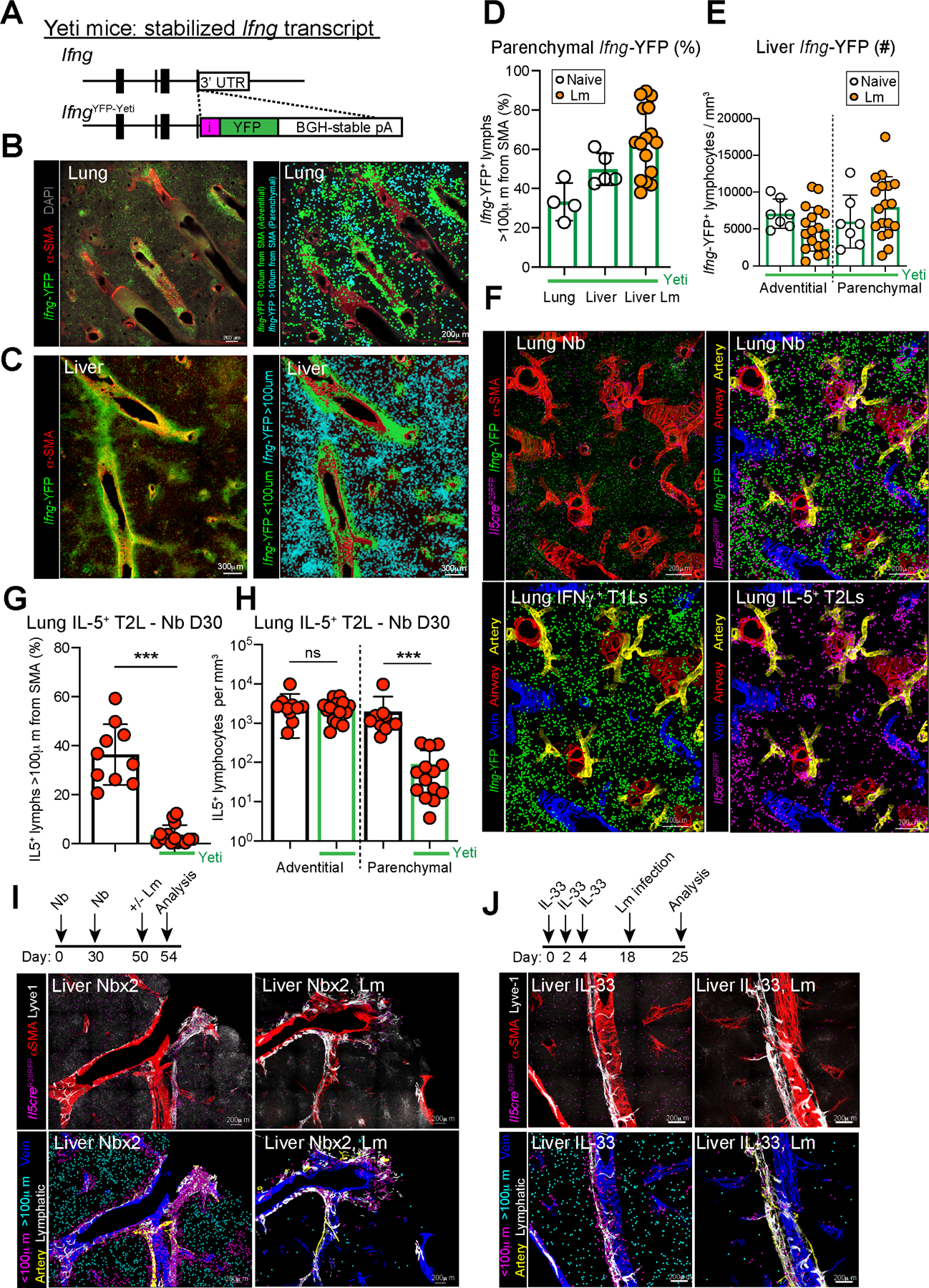
(A) Schematic of the IfngYFP Yeti reporter mouse.
(B and C) Confocal thick-section images from lung and liver of IfngYFP/+ mice with all Ifng-YFP+ cells (left, green) or with surfaces rendered (right) on adventitial cells (IFNγ+, green, <100μm from SMA) and parenchymal cells (IFNγ+, teal, >100μm from SMA). Images are representative of N=8 mice.
(D and E) Quantitative imaging analysis of (D) lung and liver sections as percent parenchymal IFNγ+ T1Ls of total T1Ls or (E) total IFNγ+ T1Ls per volume from liver of IfngYFP/+ mice at rest (open white dots) and D7 post Listeria monocytogenes (Lm) infection (orange dots). Pooled from 2 independent experiments, total N=4 mice.
(F) Confocal imaging and surfacing analysis of lung thick-sections of T1L-Yeti/T2L-lineage tracker mice infected with Nb and imaged at D30 PI. 2 independent experiments, total N=7 mice/group.
(G and H) Quantitative imaging analysis of lung as (G) percent parenchymal IL-5+ T2Ls and (H) IL-5+ T2Ls per volume in thick lung sections from T1L-Yeti/T2L-lineage tracker mice (bars outlined in green), or littermate T2L-lineage tracker only mice (bars outlined in black) at D30 post Nb infection. Pooled from 2 independent experiments, total N=7 mice/group.
(I and J) Confocal imaging of liver thick sections from IL-5 reporter mice (after the indicated challenges, showing native images and rendered surfaces of adventitial and parenchymal IL-5tdtomato+ T2Ls. 2 independent experiments, total N=6 mice/group.
Bar graphs indicate mean (±SD), unpaired Student’s t test. See also Figure S2.
Next, we asked how a mixed type 1 - type 2 inflammatory setting would impact IL-5+ ILC2 and Th2 cell localization. Heterozygous Yeti mice (IfngYFP/+) have one copy of a stabilized IFNγ transcript, leading to spontaneous low-level T1L activation (Reinhardt et al., 2015; Stetson et al., 2003) which is sufficient to partially restrict type 2 responses to Nb helminths, including the expansion of ILC2s and Th2 cells (Molofsky et al., 2015b). In Yeti+ strains, we found that Nb-driven IL-5+ lymphocyte parenchymal accumulation was profoundly restricted, with little impact on numbers of adventitial lymphocytes (Fig 2F–H, Video S6). Additionally, we used IL-5 lineage tracker mice pre-challenged with type 2 stimuli (Nb infection x2 or IL-33 treatment) and then infected with Lm to drive type 1 immunity. As in the mice with genetically enhanced IFNγ (Yeti+), we observed a similar restriction of IL-5+ lymphocytes that was predominant in parenchymal domains (Fig 2I–J). Thus, activated IFNγ-producing T1Ls preferentially restrict the parenchymal accumulation of IL-5+ lymphocytes during concurrent or serial challenges with mixed type 2 - type 1 inflammation in the lung and liver.
IFNγ directly restricts activated IL-5+ ILC2s and Th2 cells.
We hypothesized that IFNγ+ T1Ls restrict ILC2 and IL-5+ Th2 cell parenchymal accumulation and function directly via IFNγ signaling; however, IFNγ targets many cell types and acts in an autocrine manner to promote T1Ls and type 1 immunity (Castro et al., 2018; Schroder et al., 2004). To determine the direct impact of IFNγ signaling on IL-5+ lymphocytes, we generated mice with loss of IFNγR1 (Lee et al., 2013) from all IL-5+ T2Ls (Il5Cre/Cre; Ifngr1flox/flox, i.e. Il5creΔIfngr1; Fig 3A, Fig S3A). Of note, the Il5Cre ‘Red5’ allele disrupted the first exon of the Il5 gene; as two copies of Cre led to optimal deletion of Ifngr1 (not shown), these mice were IL-5-deficient as were their littermate controls. ILC2s and rare GAT IL-5+ Th2 cells expressed IL-5 in resting mice, resulting in both sensitive and specific loss of IFNγR1 (Fig 3B–C, Fig S3B). We found that naive ‘IL-5+ T2L IFNγ-blind’ mice (Il5creΔIfngr1) had normal ILC2 numbers and Il5-tdTomato fluorescence in the lung (Fig S3C) with a modest IFNγ-mediated repression of ILC2s in the GAT of aged mice (Fig S3D). We conclude that ILC2s in naïve mice are minimally repressed by IFNγ, at least in the organs examined and conditions of our UCSF specific pathogen free mouse colony.
Figure 3: IFNγ directly restricts activated IL-5+ type 2 lymphocytes.
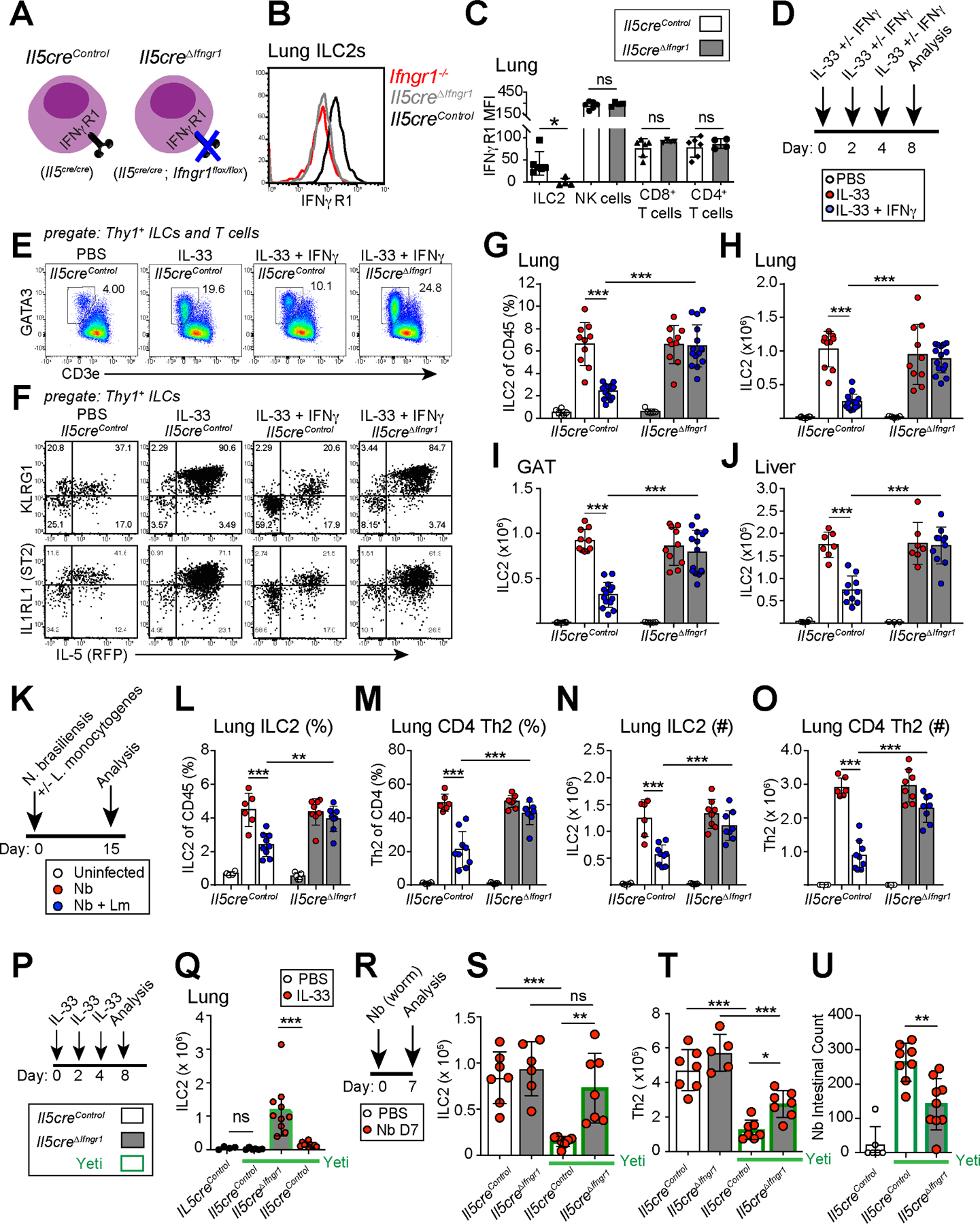
(A) Schematic of IL-5+ T2L IFNγ-blind conditional mouse strain (Il5creΔIfngr1) or littermate controls.
(B and C) Flow histograms and mean fluorescence intensity (MFI) of IFNγR1 expression on resting lung ILC2s and other lung lymphocytes in control (white bars) and IL-5+ T2L IFNγ-blind (grey bars) mice. 2 independent experiments, total N=6 mice/ group.
(D) Schematic showing IL33 +/− IFNγ administration schedule, relevant to E-J.
(E and F) Flow cytometry of ILC2s or total ILCs expression of KLRG1, IL1RL1 (ST2), or IL-5RFP in lungs at D8. 3 independent experiments, N≥7 mouse/group.
(G-J) Flow cytometry of ILC2 as (G) percent or (H) total number in lungs (I), gonadal adipose tissue (GAT), and (J) liver from the indicated mice treated with PBS (white dots), IL-33 in the absence (red dots) or presence (blue dots) of IFNγ. Pooled from 3 independent experiments, N≥7 mouse/group.
(K) Schematic showing co-infection with Nippostrongylus brasiliensis followed by inoculation with Listeria monocytogenes, relevant to L-O.
(L-O) Flow cytometry showing percent and numbers of ILC2 and Th2 cells at D15 PI. Pooled from 3 independent experiments, N≥6 mouse/group.
(P and Q) Schematic of IL-33 treatment and flow analysis of ILC2s in lungs on D8 post PBS control (white dots) or IL-33 treatment (red dots) from the indicated strains, with mice on an IfngYFP/+ Yeti background (bars outlined in green). Pooled from 2 independent experiments, N≥8 mouse/group.
(R-T) Schematic of Nb helminth infection with flow analysis of ILC2s and Th2 cells on D7 PI from the indicated mouse strains. Pooled from 2 independent experiments, N≥6 mouse/group.
(U) Intestinal Nb larvae at D7 PI. Pooled from 2 independent experiments, N≥6 mouse/group.
Bar graphs indicate mean (±SD), Two-Way ANOVA with Sidak post-test. See also Figure S3
Next, we used models of mixed type 1 - type 2 inflammation to test the cell-intrinsic impact of IFNγ on activated ILC2s and IL-5+ Th2 cells. First, we used a reductionist approach, co-administering the cytokines IFNγ and IL-33. Exogenous IFNγ directly restricted IL-33 driven ILC2 expansion, activation (KLRG1 expression), IL-5 production, and proliferation (Ki-67+) in multiple tissues; this effect was abrogated in littermate ‘IL-5+ T2L IFNγ-blind’ mice (Il5creΔIfngr1, Fig 3D–J, not shown). We did not observe mouse morbidity or gross tissue pathology with the IL-33 +/− IFNγ doses given. We next used an infection-driven model, co-infecting mice with Nb and Lm (Fig 3K–O) (Molofsky et al., 2015b). Tissue Th2 cells accumulated aberrantly in both Il5creΔIfngr1 and CD4creΔIfngr1 mice, suggesting IFNγ also acts directly to restrict helminth-driven IL-5-expressing Th2 cells (Fig S3E–F). IFNγ repressed ILC2s independently of T cells, since mice with loss of IFNγR1 in all T cells (Cd4Cre; Ifngr1flox/flox) or specifically in Tregs (Foxp3Cre; Ifngr1flox/flox) maintained IFNγ-mediated repression of ILC2s (Fig S3G). Thus, IFNγ induced during type 1 inflammation acts directly on effector IL-5+ ILC2s and Th2 cells to limit their IL-33- or helminth-driven expansion and activation.
In a third model, we challenged IL-5+ T2L IFNγ-blind mice carrying one Yeti allele (IfngYFP/+;Il5creΔIfngr1) with Nb and found similar results (Fig 3P–T, Fig S3I–J), as well as improved Nb intestinal worm clearance (Fig 3U) and consistent with our prior work (Molofsky et al., 2015b). On the other hand, IL-5+ T2L IFNγ-blind mice had mildly reduced IFNγYFP+ T cells (Fig S3K–P). Although IL-5 can restrict lung IFNγ+ NK cells (T1Ls) and tumor metastasis via activation of eosinophils (Schuijs et al., 2020), our use of IL-5 deficient mice suggested additional potential mechanisms of T2L to T1L feedback, which were not further examined here. Together, our findings showed that IFNγ, whether administered exogenously, generated during infection, or genetically driven, directly repressed activation and accumulation of IL-5+ ILC2 and Th2 cells and impaired Nb clearance.
IFNγ directly limits IL-5+ lymphocyte accumulation at parenchymal domains.
We next hoped to dissociate effects of IFNγ attenuating ILC2 early activation and proliferation from potential impacts on ILC2 parenchymal accumulation. We found that IL-33 treatment drove ILC2 proliferation in the first two weeks post-challenge, a period that correlated with elevated blood ILC2s and detectable blood IL-13 (Fig 4A–C) and similar to Nb infection (Huang et al., 2018; Ricardo-Gonzalez et al., 2020). Although blood ILC2s fell dramatically by 2–3 weeks post challenge, they remained detectably elevated up to several months after IL-33 challenge (Fig 4D). Given these results, we asked whether delaying IFNγ induction could still impact the accumulation of tissue ILC2s. After treating with IL-33, we waited several weeks until early ILC2 proliferation and activation declined, and then administered IFNγ. IFNγ again limited ILC2 accumulation in multiple tissues (Fig S4A–D). Similarly, two weeks after IL-33-mediated ILC2 expansion, Lm infection drove induction of IFNγ that restricted liver ILC2 accumulation and tissue IL-13 production (Fig 4E–G, Fig S4E–F), with similar albeit reduced impacts on systemic ILC2s in the lung and GAT (Fig S4G–H). Lm infection also enhanced the accumulation of liver T1Ls, although their total numbers and IFNγ production was not altered in IL-5+ T2L IFNγ-blind mice (Fig S4I–L). In contrast, mice with IFNγ-blind T2Ls had decreased accumulation of liver neutrophils (PMN) 2 days post Lm, whereas liver monocytes and blood neutrophil pools were unchanged (Fig S4M–O). Thus, ILC2s were restricted by exogenous or bacterially driven IFNγ even weeks after the cessation of IL-33-induced ILC2 activation and proliferation, leading to impaired local accumulation of liver neutrophils after Lm infection.
Figure 4: IFNγ directly limits IL-5+ type 2 lymphocyte accumulation at parenchymal domains.
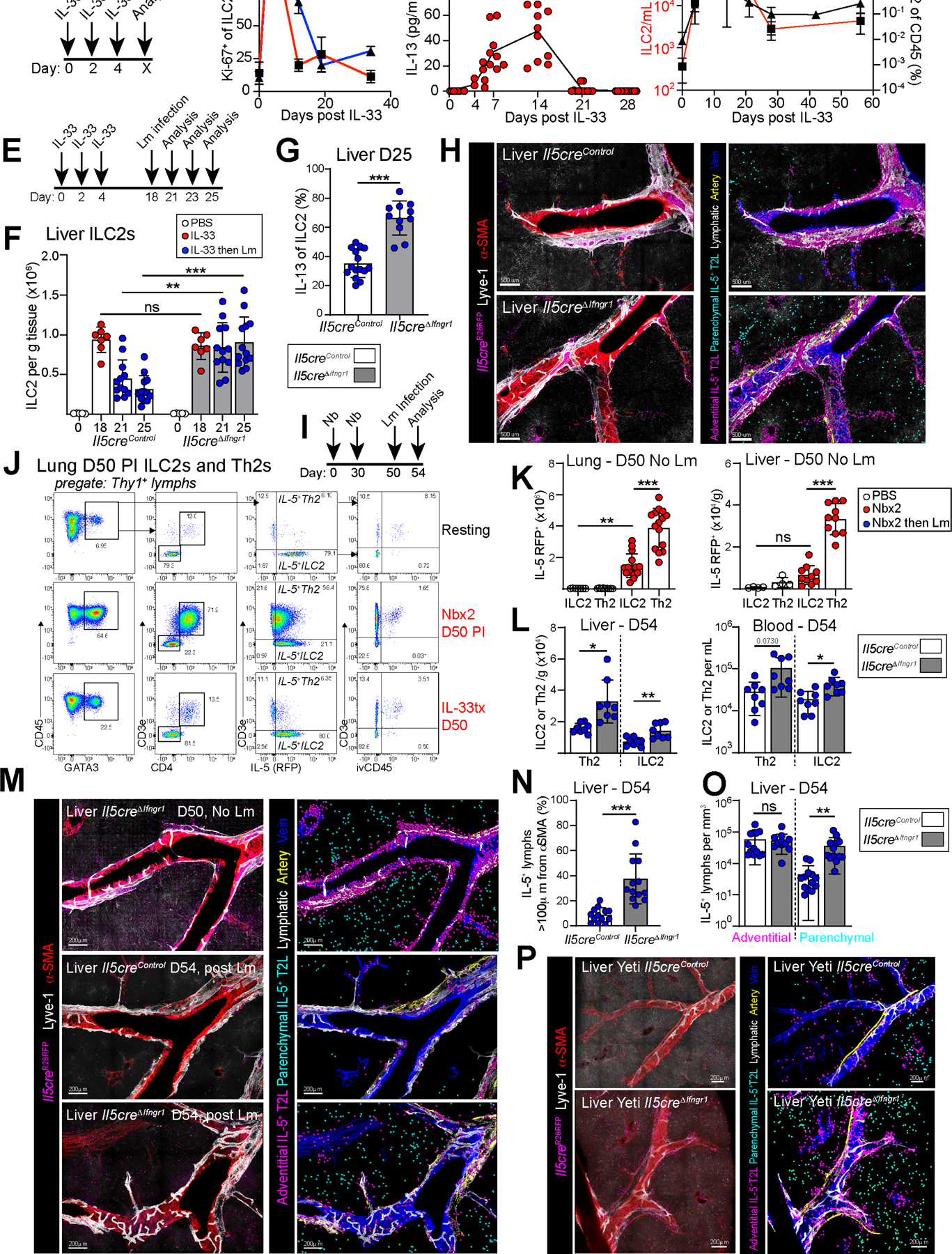
(A) Schematic of IL-33 treatment, relevant to B-D.
(B) Flow cytometry showing percent of Ki-67+ ILC2. Pooled from 3 independent experiments, N≥ 8 mice per time point
(C) Plasma IL-13 at the indicated time points. Pooled from 3 independent experiments, N≥ 6 mice per time point.
(D) Flow cytometry showing blood ILC2s per mL (red) or percentages (black). Pooled from 3 independent experiments with N≥ 8 mice per time point.
(E) Schematic of IL-33 treatment followed by infection with Lm on D18 post IL-33 treatment, relevant to F-H.
(F and G) Flow cytometry quantitation of (F) liver ILC2s and (G) percent IL-13+ of ILC2s elicited upon in vitro restimulation on D25. Red dots indicate IL-33 treatment only and blue dots indicate mice with IL-33 treatment, then Lm infection. Pooled from 3 independent experiments, with N≥ 7 mice per group.
(H) Confocal thick-section imaging, surface rendered and distance analysis for liver IL-5RFP+ T2Ls at D23 post initial IL-33 treatment, (D5 Lm PI).
(I) Schematic of Nb s.c. infections, followed by Lm i.v. infection. Relevant to J-O.
(J) Flow cytometry plots comparing IL-5+ ILC2 and IL-5+ Th2 cells after Nb infections or IL-33 treatment in IL-5+ T2L lineage tracker mice. 2 independent experiments, total N=6 mice/group.
(K) Flow cytometry of IL-5+ ILC2 and IL-5+ Th2 cell numbers at D50 post Nbx2 (no Lm infected mice). Pooled from 3 independent experiments, N≥ 8 mice/group.
(L) Flow cytometry quantitation of liver and blood ILC2s in T2L IFNγ-blind mice (grey bars) and controls, 4 days post Lm infection (D54).
(M-O) Confocal images of liver thick sections (M) and (N and O) quantitation of IL-5+ lymphocyte localization. Pooled from 2 independent experiments, total N=5–6 mice/group.
(P) Confocal imaging with distance analysis in livers from IfngYFP/+; Il5creΔIfngr1 mice and controls at D8 post IL-33 treatment. 2 independent experiments, total N≥4.
Bar graphs indicate mean (±SD), Two-Way ANOVA with Sidak post-test for F and unpaired t test for G, K, L, N, O. See also Figure S4
Thus far, our data suggested that type 1 inflammation confines IL-5+ lymphocytes to adventitial niches, with IFNγ directly limiting IL-5+ lymphocytes both during acute and post-acute phases of type 2 inflammation. Combining these observations, we next tested if IFNγ could directly restrict ILC2 and IL-5+ Th2 cell accumulation at parenchymal regions. We found that Lm-driven IFNγ directly restricted IL-33 driven parenchymal ILC2s, even when induced several weeks after type 2 stimulation (Fig 4H). Next, we used two rounds of Nb infection to preferentially expand systemic IL-5+ Th2 cells (Fig 4I–K). We found that subsequent Lm infection also drove robust liver Tbx21zsgreen+ T1L expansion, with more modest lung expansion of T1Ls (Fig S4P–W); liver Th2 cells and less prevalent ILC2s were fewer in number after Lm infection (Fig 4L), with particularly fewer IL-5+ T2Ls in liver parenchymal domains (Fig 4M–O) and near Lm-induced granulomas (Fig S4X). We found similar results in mice with genetic overexpression of IFNγ (IfngYFP/+, Fig 4P). Together, these data suggest that activated type 1 lymphocytes, present in parenchymal domains, limit IL-5+ ILC2 and Th2 cell parenchymal occupancy directly via IFNγ.
IFNγ restriction of ILC2s and IL-5+ Th2 cells limits the morbidity and mortality of Listeria infection during mixed inflammation.
Next we tested the functional consequences of IFNγ-blind ILC2s and IL-5+ Th2 cells during mixed inflammation. Without prior type 2 activation, mice with IFNγ-blind ILC2s did not display altered morbidity or mortality from Lm infection, consistent with low type 2 immune activation in naïve lab mice (Fig S3C–D). When ILC2s were first expanded by IL-33 and mice subsequently infected with Lm, control mice demonstrated mildly elevated bacterial counts in spleen and liver and increased mortality (Fig. 5A–C). However, mice with IFNγ-blind ILC2s had further increased liver and spleen bacterial counts, enlarged and necrotic liver granulomas, and increased overall mortality (Fig. 5D–F).
Figure 5: IFNγ directly acts on IL-5+ type 2 lymphocytes to restrict morbidity and mortality of Listeria infection during mixed inflammation.
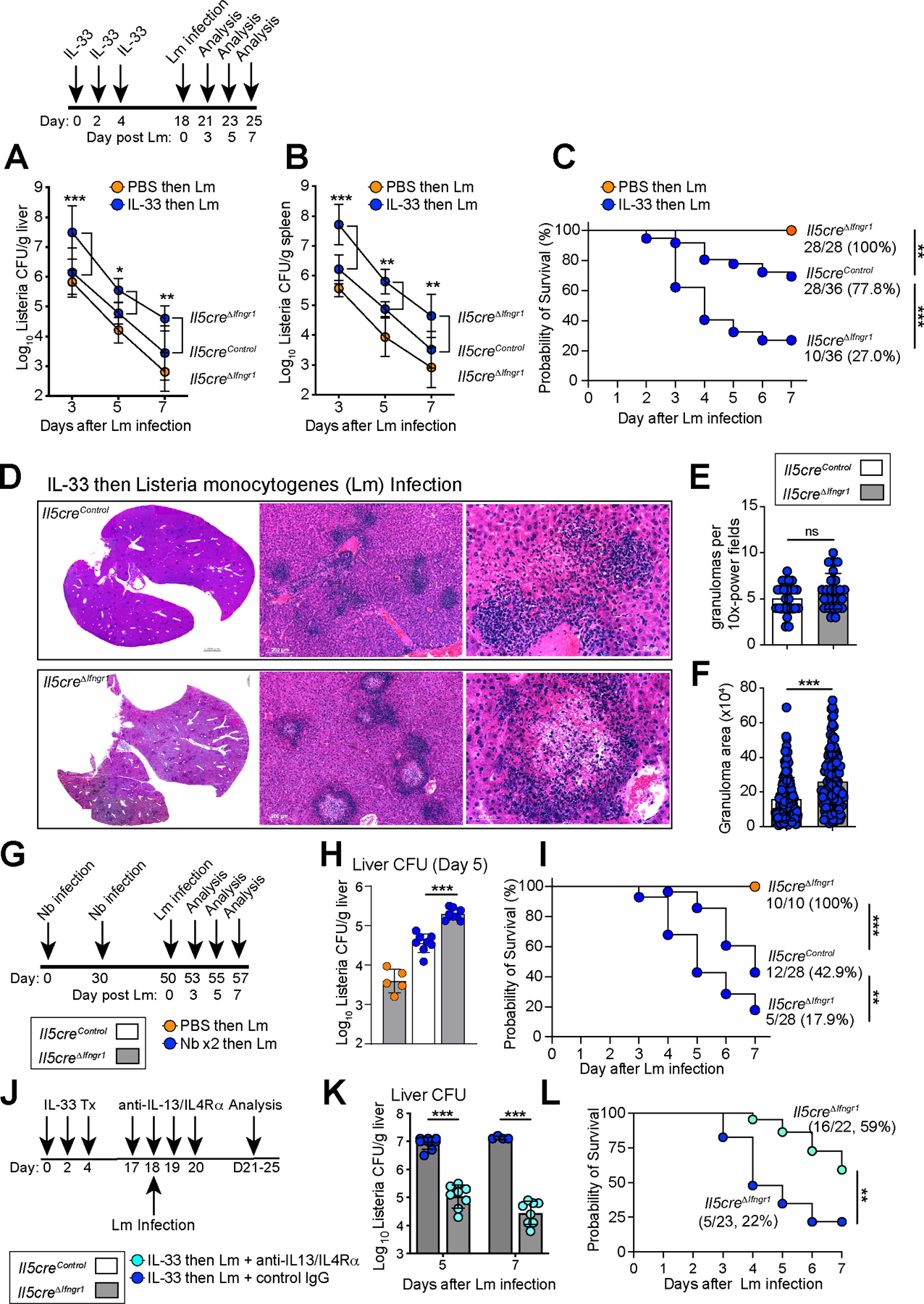
(A-B) Schematic of IL-33 treatment then infection with Lm (relevant to A-F) and colony forming units (CFUs) from (A) liver and (B) spleen. Pooled from 3 independent experiments, N≥8 mice.
(C) Survival curves after Lm infection. Pooled from 3 independent experiments with N=28 mice for Il5creΔIfngr1; PBS tx, N=36 for Il5creΔIfngr1; IL-33 tx and, N=36 for Il5creControl; IL-33 tx.
(D) H&E staining of 7μm paraffin liver sections at D7 post Lm. 3 independent experiments, N=6 mice/group.
(E and F) Granuloma numbers and size in 10X magnification fields. Pooled from 3 independent experiments, total N=6 mice/group.
(G-I) Schematic of Nb infections, followed by Lm infection. Analysis of (H) liver CFUs at D5 PI and (I) Survival curves. Orange dots indicate mice infected with Lm, blue dots indicate mice infected with 2 rounds of Nb, then Lm infection. Pooled from 3 independent experiments, total N≥10/group.
(J-L) Schematic of the treatment regimen with neutralizing antibodies (teal dots) i.v injected into Il5creΔIfngr1 mice pre-treated with IL-33, then infected with Lm. (K) liver CFUs at D5 and D7 PI and (L) Survival curves (N=22 mice with neutralizing Abs, N=23 mice with isotypes Abs). Pooled from 2 independent experiments.
Bar graphs indicate mean (±SD), two-way ANOVA followed by Dunnett test for A, B. Kaplan-Meier survival curves are compared using the log-rank (Mantel-Cox) analysis, for C, I, L. Students t test for E, F, H and K. See also Figure S5
As IL-33 treatment drives an ILC2-dominant response, we also tested our Th2 cell skewed helminth model and assessed Listeria-associated morbidity and mortality (Fig 5G, Nbx2, followed by Lm). In the liver and spleen, IL-5+ T2L IFNγ-blind mice had increased necrotic granuloma formation, bacterial loads, weight loss, and mortality, resembling mice that had been activated with IL-33 (Fig. 5H–I, S5A–D). To test if IFNγ-blind ILC2s and IL-5+ Th2 cells promote Lm bacterial-driven mortality via production of IL-13, we concurrently blocked IL-13 and its obligate receptor IL4Rα during Lm infection (Fig 5J). IL-13 blockade did not impact blood or liver ILC2 numbers, but did improve Lm clearance, weight loss, and overall survival (Fig 5K–L, Fig 5SE–I). Unlike IL-13, our data did not suggest a role for the cytokine IL-5, as heterozygous mice with preserved IL-5 and specific loss of IFNgR1 on IL-5+ T2Ls (Il5cre/+; Ifngr1flox/flox) phenocopied IL-5 deficient mice (Il5cre/cre; Ifngr1flox/flox, Fig S5J–M). Together our results suggested dual functions for IFNγ signaling during type 1 - type 2 mixed inflammation: (1) activating protective type 1 immunity (Lee et al., 2013) and (2) suppressing ILC2s and IL-5+ Th2 cell parenchymal accumulation and production of maladaptive IL-13, a signal that can limit beneficial neutrophil recruitment to Lm-associated granulomas (Haak-Frendscho et al., 1992; Woytschak et al., 2016).
ILC2 transcriptome induced by IL-33 and repressed by IFNγ includes cellular trafficking and cell death related programs.
Having uncovered a cell-intrinsic role for IFNγ signaling in ILC2s, we used RNA sequencing to assess how IFNγ signaling may mechanistically restrict ILC2s in vivo. We treated Il5creΔIfngr1 or littermate controls with IL-33 +/− IFNγ and two weeks later purified lung IL-5+ ILC2s for RNA sequencing (Fig 6A). We chose this delayed time point to examine how IFNγ might restrict not just early ILC2 activation, but also potential pathways associated with trafficking or survival at parenchymal sites. Principal component analysis demonstrated a shift in PC1 (68% variance) driven by IL-33 and directly repressed by IFNγ (Fig 6B). PC2 was driven in part by IFNγ-regulated genes that may have been impacted indirectly via IFNγ signaling in IL-5-negative populations (e.g., macrophages, fibroblasts), although we cannot exclude effects from incomplete Il5Cre activity (Fig S6I; Table S2). Analysis of differentially expressed genes (DEGs) revealed IL-33 induced genes (top; e.g. Il13, Klrg1, Mki67, Id2, Il1r2, Il17rb), or IL-33 repressed genes (bottom; e.g. Cd44, Cd69, Areg, Nr4a1, Tnfaip3, Klf4), with smaller subsets of IFNγ–induced (red; Cxcr3, Ly6a, H2-Aa) or repressed (blue) genes (Fig 6C; Table S1). Direct comparison across conditions highlighted the genes above with preservation of lung ILC2 lineage-defining genes (Gata3, Bcl11b, Il1rl1, Fig 6D–E). Ingenuity Pathways Analysis (IPA, Fig 6F) found expected IL-33-driven pathways associated with ILC2 proliferation and cytokine production (Molofsky et al., 2015b; Moro et al., 2016) that were repressed by IFNγ in control (blue) but not cells from ILC2 IFNγ-blind mice (Il5creΔIfngr1, red). We also identified IFNγ-repressed pathways associated with cell survival, movement, and cellular interactions (Fig 6F–G). ILC2 tissue retention-associated genes were repressed by IL-33, including Cd44 and Cd69, whereas multiple trafficking-associated genes were induced, including S1pr1, S1pr4, Ccr1, Ccr3, Ccr4, Ccr7, and several integrins (Fig. 6G). The sphingosine 1-phosphate receptors (S1PRs) were notable, as S1P signaling regulates ILC2 tissue-extrusion into blood during type 2 inflammation (Campbell et al., 2019; Dutton et al., 2019; Germain and Huang, 2019; Huang et al., 2018; Ricardo-Gonzalez et al., 2020). IFNγ also activated a number of cell death-related genes (Fig 6H, Fig S6H), some of which were previously identified in vitro (Moro et al., 2016), and suggesting IFNγ may also promote ILC2 death. ILC2s isolated from GAT displayed similar transcriptomics to lung ILC2s, although the magnitude of IFNγ-driven transcriptomic changes was greater (Fig S6A–H).
Figure 6: ILC2 lung transcriptome induced by IL-33 and repressed by IFNγ includes programs for cellular trafficking and regulation of cell death.
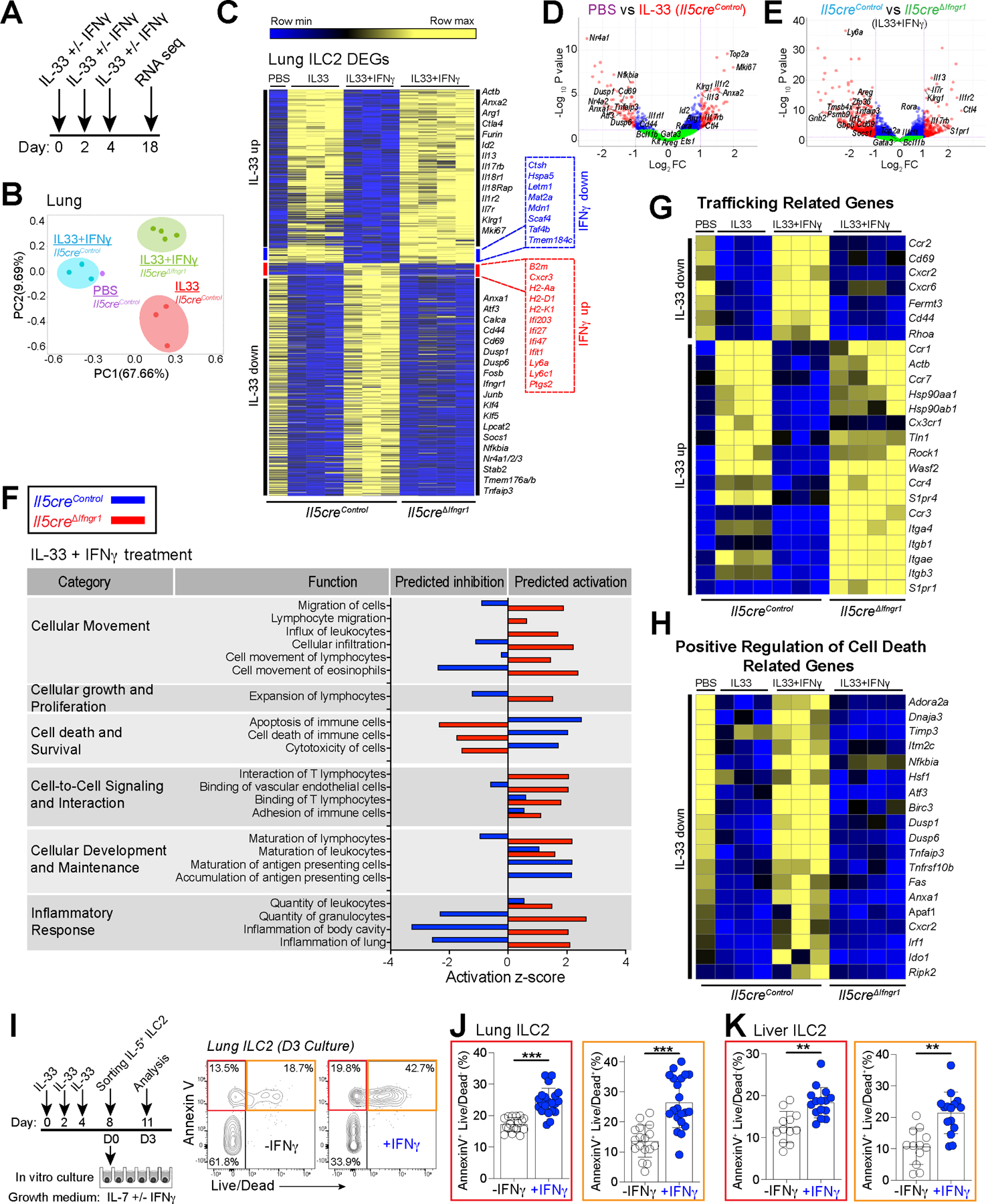
(A) Schematic of mouse treatment regimen and flow sorting of IL-5RFP+ ILC2s.
(B) Principal component analysis (PCA) plots.
(C) Heat map of differentially expressed genes (DEGs) in lung ILC2s highlighting genes for IL-33 induced (top), IFNγ-repressed (blue), IFNγ-activated (red), and IL-33 repressed (bottom)
(D and E) Volcano plots comparing (D) Il5control mice treated with PBS or IL-33 or (E) Il5creControl or Il5creΔIfngr1 mice both treated with IL-33 + IFNγ, with signature genes highlighted in red regions and borderline significance in blue.
(F) Pathway analysis (IPA) of mice treated with IL33 + IFNγ comparing Il5creControl versus Il5creΔIfngr1 mice.
(G-H) Heatmaps of differentially expressed trafficking-related and cell-death related genes highlighted after IPA analysis.
(I-K) Schematic of in vivo ILC2 expansion and in vitro cell culture conditions to evaluate IFNγ-mediated cell death. Percentages of annexin V versus live/dead cell subsets. 3 independent experiments.
Bar graphs indicate mean (±SD), unpaired Student’s t-test with welch’s correction. See also Figure S6.
IFNγ and Lm infection promote ILC2 cell death.
To test the relevance of these IFNγ-regulated pathways in ILC2s, we first examined whether IFNγ could directly induce ILC2 death. In vitro we found that IFNγ impaired the proliferation and cytokine production of activated ILC2s (not shown), as described (Molofsky et al., 2015b; Moro et al., 2016). However, when cultured with low-level survival signals (IL-7 alone), IFNγ increased lung and liver ILC2 cell death, as measured by Annexin V positivity and live-dead cell dye (Fig 6I–J). In vivo, Lm infection also induced rapid liver ILC2 cell death (TUNEL+), preferentially acting on parenchymal ILC2s including those in proximity to Lm granulomas enriched in IFNγ+ lymphocytes and TUNEL+ apoptotic cells (Fig 7A–D). In the lung, Lm infection also increased ILC2 cell death preferentially in parenchymal cells (Fig 7SA–D), although the magnitude of the impact was more modest and correlated with the less robust accumulation of lung T1Ls (Fig S5R-W). Together, we conclude that IFNγ can directly drive ILC2 cell death, with the degree of impact correlating with the accumulation of local T1Ls, and accounting for at least one mechanism by which ILC2 parenchymal occupancy is restricted during mixed inflammation.
Figure 7: S1P-mediated trafficking and IL-5+ lymphocyte cell death impact parenchymal accumulation and Listeria associated morbidity and mortality.
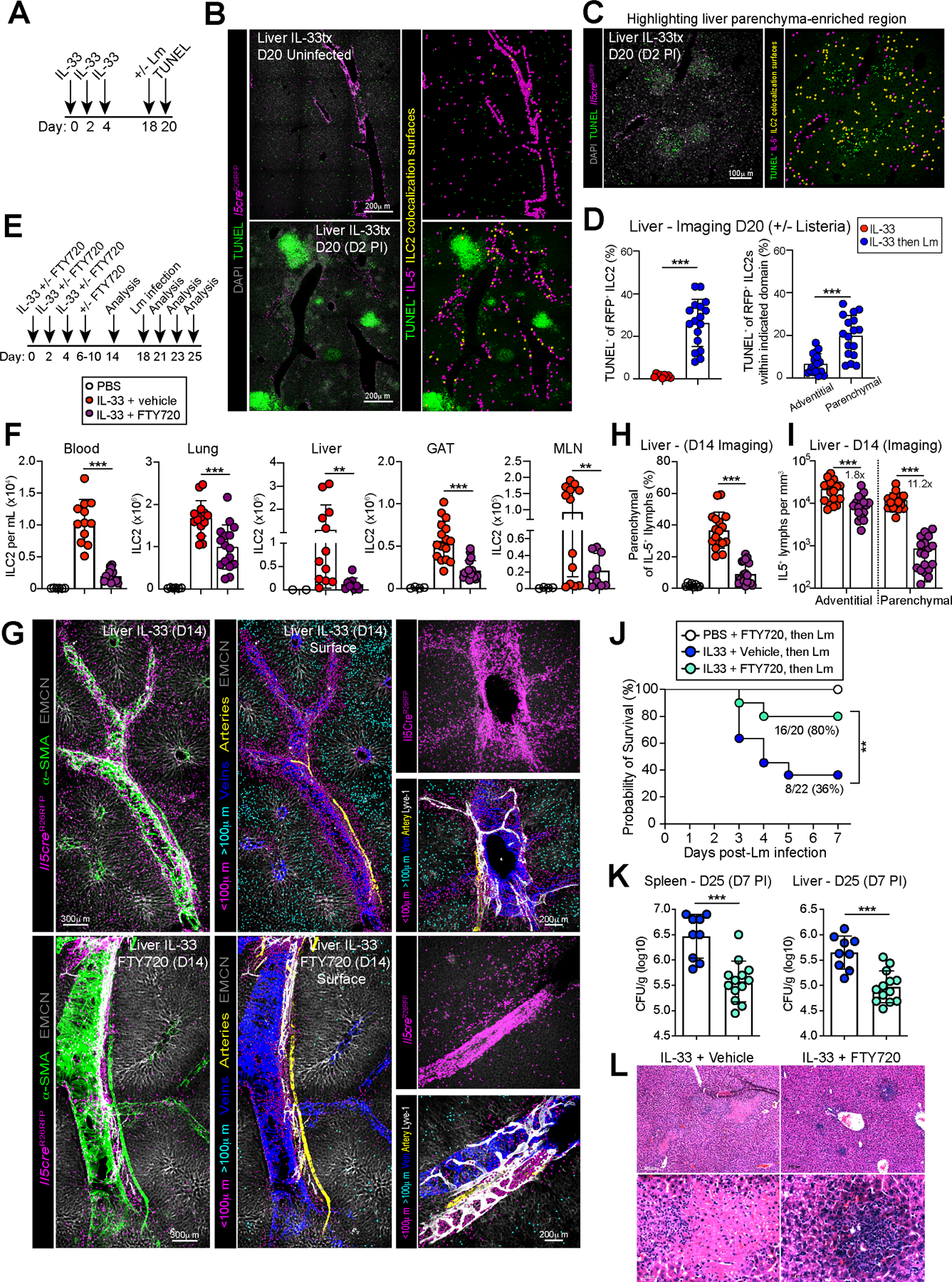
(A) Schematic of mouse treatment regimen for TUNEL cell death assay after Lm infection.
(B) Paraffin-embedded sections of livers from Lm infected IL-5 lineage tracker mice (D2 PI) assessed by TUNEL staining and surfacing analysis for TUNEL and IL-5 colocalization (yellow spots) at adventitial and (C) parenchymal domains with (D) imaging quantification. 3 independent experiments, N=7 mice/group.
(E) Schematic of mouse treatment with IL-33 ± FTY720 ± infection with or without Lm on D18. Relevant to F-L
(F) Flow cytometry of ILC2s in multiple tissues on D14. Pooled data from 3 independent experiments, total N≥12 mouse/group.
(G) Confocal thick-section imaging, surfaces rendered and distance analysis for adventitial and parenchymal IL-5+ T2Ls in livers at D14. 3 independent experiments, total N≥6 mouse/group.
(H and I) Imaging quantification as percent parenchymal IL-5+ T2Ls of total IL-5+ T2Ls or parenchymal IL-5+ T2L numbers per tissue volume. 3 independent experiments, total N≥6 mouse/group.
(J) Survival curves after Lm infection. Data pooled from 3 independent experiments, N=20 or n=22 mice for Il5creΔIfngr1 treated with or without FTY720, respectively.
(K) Lm CFUs on day 25 (D7 PI) from spleen and liver of Il5creΔIFNγR1 mice pre-treated with IL-33 with (teal dots) or without FTY720 (blue dots). Data pooled from three independent experiments, total N≥ 8 mice/ group.
(L) H&E staining of paraffin liver sections at D7 post Lm infection. 3 independent experiments, total N=6 mice/group.
Bar graphs indicate mean (±SD). Unpaired t test with Welch’s correction for F, H, I, K. Kaplan-Meier survival curves are compared using the log-rank (Mantel-Cox) analysis. See also Figure S7.
S1P-mediated trafficking is required for optimal ILC2 parenchymal accumulation and Listeria-associated morbidity and mortality.
We next examined the functional impacts of ILC2 trafficking via Sphingosine-1-phosphate (S1P) (Campbell et al., 2019; Dutton et al., 2019; Germain and Huang, 2019; Huang et al., 2018; Ricardo-Gonzalez et al., 2020). We hypothesized that IL-5+ lymphocytes, including ILC2s and possibly pre-existing tissue Th2 cells, reach parenchymal niches via migratory pathways, as suggested by our RNA sequencing. To test this idea, we first administered FTY720 (Fingolimod, a S1P functional antagonist) concurrent with IL-33 treatment, allowed the drug to clear, and assessed the impact on lung and liver ILC2 numbers and localization two weeks later (Fig 7E). FTY720 efficiently blocked ILC2 tissue egress into blood, with variably reduced ILC2s in multiple tissues (Fig 7F). FTY720 was particularly efficient at restricting the accumulation of parenchymal-localized ILC2s in the liver (Fig 7G–I, Video S7) and lung (Fig S7E–H). While adventitial ILC2 accumulation in the liver was mildly impaired (~1.8x), liver parenchymal ILC2s were profoundly restricted (~12x) (Fig 7I). These data suggest that robust IL-33 driven ILC2 expansion and parenchymal redistribution depends on S1P-dependent trafficking. To test the functional impact of this S1P-dependent ILC2 trafficking, we treated IL-5+ T2L IFNγ-blind mice with IL-33 ± FTY720, waited two weeks to allow for drug clearance, and infected with Lm) As seen previously, IL-33 treated mice had increased Lm-induced necrosis within granulomas, elevated bacterial burdens in spleen and liver, and impaired overall survival; in contrast, FTY720 co-treatment improved Lm-morbidity and mortality (Fig 7J–L). FTY720 did not directly impair ILC2 expansion or activation, as treatment with FTY720 one week after initial ILC2 activation did not significantly alter liver and lung numbers or localization, with blood ILC2s only modestly impaired (Fig S7I–K). We found that IL-33 driven ILC2 blood occupancy could also be partially blocked by specific antagonists to either S1PR4 (robust effect) or S1PR1 (modest effect), two receptors expressed by trafficking ILC2s (Huang et al., 2018) and confirmed in our RNAseq data (Fig S7L–O). Our experimental setup does not address whether similar trafficking occurs for tissue-resident IL-5+ Th2 cells, although FTY720 will also restrict recently generated lymphoid Th2 cells from reaching parenchymal niches. Together, our results suggest that ILC2 topographic expansion from adventitial niches into parenchymal sites may be a conserved mechanism to increase tissue type 2 immune influence, with IFNγ+ T1Ls dispersed across tissue domains and acting as a dominant and functionally critical brake to this niche expansion. IFNγ acted at multiple steps to restrict IL-5+ T2Ls to adventitial regions, including preferentially promoting parenchymal cell death and limiting S1P dependent cell trafficking.
DISCUSSION:
Inflammation impacts both tissue physiology and pathology, but the role of lymphocyte positioning on this critical balance has not been well defined. Our previous work describe effector IL-5+ lymphocyte niches within adventitial boundary regions in proximity to adventitial fibroblasts, a stromal cell subset that can both support and activate IL-5+ ILC2s and Th2 cells (Dahlgren and Molofsky, 2019; Dahlgren et al., 2019). Here, we showed that IL-5+ ILC2 and Th2 cells localized to de novo parenchymal domains after allergic inflammation, an event that required S1P-dependent trafficking and was critically and directly restricted by IFNγ and type 1 lymphocytes.
T1Ls produce IFNγ to restrain intracellular organisms and target cells with altered-self for elimination; in contrast, effector T2Ls produce IL-4, IL-5, and IL-13 to restrict extracellular helminths and protozoa (Colonna, 2018; Fan and Rudensky, 2016; Vivier et al., 2018). However, both infectious and sterile insults frequently engage type 1 and type 2 activation and crosstalk. First described for CD4+ T cells in lymphoid tissues, the presence of IFNγ reinforces Th1 cell differentiation while suppressing a Th2 cell fate (Maggi et al., 1992). However, cross-regulation of type 1 and type 2 immunity also occurs in non-lymphoid tissues; for example, infection with Influenza A virus or Mycobacterium, or exogenous dosing of IL-12, drives IFNγ+ T1Ls that restrict lung type 2 lymphocytes and downstream allergic responses (Erb et al., 1998; Gavett et al., 1995; Marsland et al., 2004). IFNγ also ameliorates established atopic dermatitis (Chang and Stevens, 2002; Reinhold et al., 1993) with skin NK cells and ILC2s acting to cross-regulate each other (Mack et al., 2020). ILC2s produce IL-5 to promote eosinophil accumulation and limit NK cell (T1L) function in models of lung cancer metastasis (Schuijs et al., 2020), suggesting reciprocal pathways also exist. Childhood allergic asthma is driven by ILC2s and Th2 cells but is also associated with severe respiratory viral infections that drive potent type 1 immunity (Lambrecht and Hammad, 2015, 2017; Martinez, 2009). This cross-regulation during mixed inflammatory states is also evident in metabolic derangements such as obesity, type 2 diabetes, and cardiovascular disease (Cautivo and Molofsky, 2016; Gisterå and Hansson, 2017; Wensveen et al., 2015), suggesting it is may be broadly applicable to tissue health and disease. As cytokine signals commonly act on neighboring cells, the localization of T1Ls and T2Ls could be an important factor in determining healthy versus pathologic mixed inflammatory settings. Indeed, our data suggested liver parenchymal T2Ls produce IL-13 that represses local neutrophil accumulation at forming granulomas (Kang et al., 2008). It will be interesting to determine the upstream regulatory signals and cellular targets of parenchymal IL-5+ T2Ls.
After allergic inflammation, parenchymal domains were short- to mid-term type 2 niches, with at least a portion of IL-5+ T2Ls ultimately maintaining long-term residence in adventitial niches. Although we detected few IL-5+ lymphocytes in parenchyma of naïve mice, it is possible that parenchymal domains are occupied in animals exposed to broader environmental and infectious challenges. Indeed, recent work has defined how commensal bacterial signals organize myeloid and lymphoid topography in the liver (Gola et al., 2021). As such, parenchymal sites may represent ‘overflow’ niches in specific-pathogen free mice but physiologic sites of IL-5+ lymphocyte occupancy in wild mammals. In contrast to IL-5+ lymphocytes, IFNγ+ T1Ls were broadly distributed in parenchymal sites and could produce elevated local concentrations of IFNγ to drive local ILC2 cell death. Additionally, adventitial niche fibroblasts or other cells may provide survival signals that allow adventitial ILC2s and Th2 cells to resist IFNγ-driven cell death, although the nature of these signals is unknown. In this scenario, adventitial regions may act as ‘safe zones’ to limit excessive and unnecessary parenchymal tissue damage by effector lymphocytes, a potential driver of tissue pathology. Future work will need to delineate how broadly this type 1 - type 2 spatial cross regulation impacts diverse mixed inflammatory challenges and whether specific T1L or T2L subsets may mediate unique inflammatory or reparative outcomes.
We envision at least two mechanisms by which IL-5+ T2Ls reached parenchymal sites: (1) activated ILC2s and Th2 cells directly move from tissue adventitial niches through interstitial spaces to parenchymal locations, or (2) tissue ILC2s and Th2 cells, or T2L-precursors (ILC2 precursors, naïve Th2 cells), move from circulation directly into tissue parenchyma. Arguing for contributions of the former, ILC2s have been originally described as tissue-resident lymphocytes that develop post-birth and proliferate in situ (Gasteiger et al., 2015; Moro et al., 2016). However, circulatory pools also contribute to tissue ILC2s, both at rest and particularly following type 2 inflammation (Gasteiger et al., 2015; Moro et al., 2016; Schneider et al., 2019). Circulatory ILC2s could originate from bone marrow ILC or ILC2 progenitors (Stier et al., 2018). However, after tissue allergic inflammation, tissue-resident ILC2s and T cells also reach the blood via retrograde trafficking, a pathway by which tissue-resident lymphocytes are activated by local inflammation to enter lymphatics and ultimately spill into blood, potentially acting as a mechanism to systemically distribute type 2 cells and signals (Campbell et al., 2019; Dutton et al., 2019; Huang et al., 2018; Miller et al., 2020; Ricardo-Gonzalez et al., 2020)(Beura et al., 2018; Klicznik et al., 2019; Stolley et al., 2020). During early type 2 inflammation, we found that parenchymal ILC2 accumulation was blunted with S1P blockade (FTY720), a pathway strongly implicated in lymphocyte retrograde trafficking, and suggesting this may be a potential dominant route by which ILC2s, and possibly subsets of tissue resident Th2 cells, reach tissue parenchyma.
The relevant molecular targets downstream of IFNγ signaling in ILC2s or Th2 cells remain obscure, although IFNγ likely acts via STAT1 (Duerr et al., 2016; Moro et al., 2016; Stier et al., 2017) to interfere with aspect(s) of ILC2 activation, survival, and trafficking pathways. Our RNAseq data highlighted several candidate negative repressors of NF-κB/MAP kinase pathways (e.g. Socs1, Nr4a1/2/3, Tnfaip3). These targets could mediate some of the direct impacts of IFNγ, including repression of S1PR1 and S1PR4 upregulation and subsequent trafficking program in adventitial IL-5+ lymphocytes, as well as promoting cell death in parenchymal lymphocytes. Together, our work suggests that a better understanding of tissue lymphocyte trafficking and topographic crosstalk may lead to improved therapeutics that impact diverse physiologic and pathologic inflammatory states.
Limitations of Study:
Our work does not resolve the trafficking route and the tissue source by which ILC2s or Th2s reached parenchymal niches during type 2 challenges. Critical future questions include delineating the possible contribution of local or circulatory ILC2s and ILC progenitors to parenchymal and adventitial ILC2 pools (Ghaedi et al., 2020; Zeis et al., 2020) and precisely defining the regulation of ILC2s versus Th2 cell subsets. While our studies focused on ILC2s, we were able to verify similar findings in IL-5+ Th2 cells, at least regarding topographic positioning and counter-regulation by IFNγ. However, as ILC2s and Th2 cells have unique developmental trajectories, responsiveness to antigen, and functional contributions under certain conditions (Schuijs and Halim, 2016; Sonnenberg and Hepworth, 2019), their localization and function may also diverge in certain inflammatory settings or time frames.
STAR METHODS:
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ari B. Molofsky (ari.molofsky@ucsf.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The accession number for the 3’Taq RNA-seq data reported in this paper is GSE190208. Accession code for ILC2s (Moro et al., 2016) can be found in the referenced study.
EXPERIMENTAL MODEL AND DETAILS:
Mice
Cytokine reporter mice used in these studies include Red5 (Il5-Tdtomato-Cre, Jackson 030926) for tracking IL-5-producing type 2 lymphocytes (Nussbaum et al., 2013). Yeti mice contain an IRES-eYFP cassette with a bovine growth hormone poly-A tail inserted into the 3’ UTR (Stetson et al., 2003), leading to stabilization of the IFNγ transcript. Yeti mice used in these studies were heterozygous to avoid inflammatory toxicity observed in mice with homozygous Yeti alleles (Reinhardt et al., 2015). T-bet (Tbx21)-zsGreen transgenic mice are previously described (Zhu et al., 2012), a kind gift of Jinfang Zhu, Lab of Immune System Biology, NIH. Where indicated, thick section imaging was performed in Red5 homozygous mice, or Red5 heterozygous mice crossed with R26-CAG-RFP mice (Ai14 line) containing a flox-stop-flox sequence upstream of a CAG-RFP-WPRE- cassette in the constitutively expressed ROSA26 (R26) locus (Jackson 007914), serving as an IL-5 lineage- or fate-mapping reporter IL-5 lineage tracker mice (Il5dtomato-Cre; Rosa26fsf-RFP-Ai14) (Dahlgren et al., 2019). Where indicated, Red5 homozygous mice were crossed with conditional IFNγ receptor 1 gene targeted mice (Ifngr1flox/flox) to selectively eliminate IFNγ signaling in IL-5 producing cells (Lee et al., 2013). Additional mice utilized include IFNγR1 deficient (Jackson 003288), CD4-Cre (Jackson 022071) or Foxp3Cre-YFP (Jackson 016959) crossed to Ifngr1flox/flox, IL7Rα deficient (Jackson 002295) and PDGFRα-H2B-eGFP (Jackson 007669). All mice were bred and maintained in specific-pathogen-free conditions at the animal facilities of UCSF and were used in accordance with institutional guidelines and under approved study protocols. All mouse strains were backcrossed on C57BL/6 for at least 10 generations. If not otherwise stated, all experiments were performed with 10–12 weeks old mice and both male and females were included.
Tissue preparation for flow cytometry
Single cells suspensions were prepared from tissues including blood, lung, liver, gonadal adipose tissue (GAT), spleen and mesenteric lymph nodes (MLN). Mice were euthanized with CO2. Immediately after, peripheral blood was collected through the posterior vena cava into heparin tubes, and mice were perfused by flushing the left ventricle with 10 mL 1X DPBS. For lung and liver, whole organs were excised, cut in small pieces with an automated tissue dissociator (GentleMacs; Miltenyi Biotec) and then digested in 1X Hanks’ Balanced Salt Solution (HBSS) with 0.2 mg/mL Liberase Tm (Roche, Cat# 5401127001), 25 μg/mL DNase 1 (Roche, Cat# 10104159001), 0.5% bovine serum albumin (BSA, Sigma-Aldrich, Cat# A2153) for 30 min at 37°C on a shaker. Samples were subsequently processed on the GentleMacs using the “lung2” program, followed by filtration through 70μm filters, washed, and subjected to red blood cell lysis (1X Pharm-Lyse lysing solution; BD Biosciences) before final suspension in FACS buffer (1X DPBS, 3% FCS, 0.05% NaN3). Liver samples were spun at 30 x g, 3min, 4C to remove hepatocytes then further separated using 40% Percoll-HBSS gradient (GE Healthcare #17–0891-01) and centrifugation (1400 x g, 20 min, room temperature, no brake). Perigonadal adipose tissue (GAT) was harvested, cut into small pieces, digested in 10mL of low-glucose DMEM containing 0.2 mg/mL Liberase Tm, 25μg/mL DNase, 0.2 M HEPES and 10 mg/mL BSA for 45 min at 37°C with gentle agitation, passed through 100um filters and centrifuged at 1000 x g for 10min. Red blood cells were lysed using 1X Pharm-Lyse and the remaining cell pellet were resuspended in FACS buffer. Spleen and MLN were prepared by mashing tissue through 70 μm filters without tissue digestion, followed by centrifugation and red cells lysis steps. Blood samples were centrifuged for 5 minutes at 1500 x g. The plasma was collected for further analysis while the remaining cell pack was resuspended in 1X Pharm-Lyse for 10–12 min at RT, followed by centrifugation and final suspension in FACS buffer. Cell counts were performed using a NucleoCounter (Chemometic). Samples were stained in 96-well V-bottom plates. Surface staining was performed at 4°C for 30 minutes in 50–100 μL staining volume. For experiment involving intra-cellular staining, cells were fixed and permeabilized using Foxp3 Transcription Factor Staining Buffer Set (eBioscience, cat# 00–5523-00). For cytokine production, 5×106 cells were plated in 96 well U-bottom plate and stimulated ex vivo with 1X Stimulation Cocktail (Tonbo Biosciences, cat# TNB-4975) in complete media (RPMI1640 supplemented with 1 mM nonessential amino acids, 2 mM glutamine, 1 mM sodium pyruvate, 100 U/mL penicillin/streptomycin and 50 μM β-mercaptoethanol) for 3 hours at 37C. For detection of vascular-associated lymphocytes, 3–5 min before euthanizing each mouse, 3μg of anti-mouse fluorophore-conjugated CD45 antibody diluted in 200 uL of 1X DPBS were intravenously injected via the tail vein (Anderson et al., 2014). All samples were acquired on a BD LSRFortessa X-20 or a BD FACSAria II for cell sorting. Live lymphocytes were gated based on their forward and side scatter followed by Zombie NIR fixable (Biolegend) or DAPI exclusion. ILC2s were identified as lineage negative (CD11b−, CD11c−, CD3−, CD4−, CD8−, CD19−, NK1.1−), CD45+, Thy1.2+ (CD90.2), and Gata3hi, IL1RL1+ (ST2), or KLRG1+, as indicated. Th2 cells were identified as CD45+ CD3ε+ CD4+ Gata3hi FoxP3−. In some cases, IL-5Cre-tdtomato; R26-RFP mice were used to identify ILC2 as Lineage−, CD90+ RFP+ and CD4+ Th2 cells as CD45+, CD3ε+, CD4+ RFP+. Myeloid populations were identified as CD45+, CD3−, CD19−, CD11b+, CD11c−, MHCII−, SiglecF−, Ly6G+ (Neutrophils) and Ly6G−Ly6C+ (monocytes). Data were analyzed using FlowJo software (TreeStar, USA) and compiled using Prism (Graphpad Software).
Flow cytometry Antibodies
For cell sorting and flow cytometry analysis, cell suspension were prepared as described above, incubated with fixable viability dye and subsequently stained with following fluorophore-conjugated monoclonal antibodies: anti-CD45 (30-F11, BD Biosciences), anti-CD45.2 (104, Biolegend), anti-CD90.2 (thy1.2) (53–2.1, Biolegend), anti-CD3 (17A2, Biolegend), anti-CD4 (Biolegend, RM4–5), anti-CD8 (53–6.7, Biolegend), anti-CD11b (M1/70; Biolegend), anti-CD11c (N418, Biolegend), anti-Ly6C (HK1.4, Biolegend), anti-Ly6G (1A8, BD Biosciences), anti-Siglec-F (E50–2440, BD Bioscience), anti-mouse MHC Class II (I-A/I-E) (M5/114.15.2, eBioscience), anti-NK1.1 (PK136, Biolegend), anti-CD19 (6D5, Biolegend), anti-T1/ST2 (DJ8, MD Biosciences), anti-KLRG1 (2F1, Biolegend), anti-FoxP3 (FJK-16S, eBiosciences), anti-Ki-67 (16A8, Biolegend), anti-Gata3 (TWAJ, eBioscience), anti-IL13 (eBio13A, eBioscience), anti-IFNgR1 (XMG1.2, BD Bioscience).
Imaging Antibodies
Primary antibodies used include Living Colors anti-DsRed Rabbit Polyclonal Pan Antibody (1:500; TaKaRa), Chicken Polyclonal anti-GFP (1:300, Aves labs), Rat Monoclonal anti-mouse CD31 (1:100, clone MEC 13.3, BD Pharmingen), Rat Monoclonal anti-Endomucine (1:200, clone V.7C7.1, Abcam), Rat Monoclonal anti mouse CD4 (1:100, clone RM4–5, Biolegend), Goat anti mouse VEGFR3 (1:300, clone Flt-4, R&D systems), Alexa Fluor 488 anti-aSMA monoclonal Antibody (1:200, clone 1A4, eBioscience), eFluor 660 anti-LYVE1 monoclonal Antibody (1:300, clone ALY7, eBioscience), Alexa Fluor 647 anti-CD4 (1:100, Clone RM4–5, Biolegend,), APC anti-CD45 (1:100, clone 30-F11, Biolegend). As necessary the following secondary antibodies were used at 1:400 dilution: Alexa Fluor 555 donkey anti-rabbit IgG (H+L) cross-adsorbed (ThermoFisher Scientific) and Alexa Fluor 647 donkey anti-rat IgG (H+L) cross-adsorbed (Abcam). CF™ 488A donkey anti-chicken IgY (H+L), cross-adsorbed (Sigma-Aldrich)
3D Tissue Preparation and Imaging
Animals were euthanized with CO2. Shortly thereafter, transcardial perfusion with pre-warmed 1X phosphate buffered saline with 0.2% heparin (1X DPBS/0.2% heparin) followed by 4% paraformaldehyde (PFA) (Thermo Scientific, catalog # 28906) was performed. Lungs were inflated with 37°C 1.5% low-melting point agarose (Lonza). Tissues (liver, lung and gonadal adipose tissue) were harvested and post-fixed in fresh 4% PFA overnight at 4°C. After wash with 1X DPBS, coronal sections of 200–500 μm sections were prepared using a vibratome (Leica VT1000S or Precisionary Compresstome VF-310–0Z). Samples were washed and incubated in permeabilization buffer (DPBS/0.2%Triton X-100/0.3M glycine) for 1 day at room temperature, then blocked in DPBS/0.2% TritonX-100/5% serum (from the same host species as the secondary antibody) at 4°C overnight. After, samples were washed in DPBS/0.2% Tween-20 once and incubated with primary antibodies diluted in DPBS/0.2% Tween-20/3% serum, room temperature until the next day. Next, samples were washed in DPBS/0.2% Tween-20 for 30 min, 3–4 times, then incubated with secondary antibodies diluted in DPBS/0.2%Tween-20/3% serum at room temperature for 6–8 h. Samples were washed in DPBS/0.2% Tween-20 for 1 day and then dehydrated in an ascending ethanol series (20, 30, 50, 70, 95, 100%), 10 min each step and then cleared by soaking in methyl salicylate. For T-bet-ZsGreen imaging, after secondary antibody staining step, samples were washed and incubated in RIMS (Refractive Index Matching Solution: 80% Histodenz in 1X PBS, 0.01% sodium azide, 0.1% Tween20) until transparent, then mounted in fresh RIMS solution and imaged.
For identification and localization of IL-5-tdTomato+ T2Ls to anatomical structures, lung tissues were stained with anti-tdTomato (1:500), anti-CD4 (1:100), anti-Lyve1 (1:200) and anti-αSMA (1:200) and in some cases, anti-CD31 (MEC13.3, 1:100, Biolegend) or anti-Endomucin. IFNγ–YFP+ Yeti tissue sections were stained with anti-GFP/YFP (1:300). All preparations were scanned using a Nikon A1R laser scanning confocal including 405, 488, 561, and 650 laser lines for excitation and imaging with 16X/0.8 or 25X1.1, NA Plan Apo long working distance water immersion objectives. Z steps were acquired every 2–6μm.
Image Analysis and quantification
z-stacks images were rendered in 3D dimensions and quantitatively analyzed using Bitplane Imaris v9.5 software package (Andor Technology PLC, Belfast, N. Ireland). Individual IL5+ and IFNγ+ lymphocytes were annotated using the Imaris spots function based on the fluorescent reporter signal and using the Ortho slicer function to visualize size, morphology, and nuclear staining (DAPI). 3D reconstructions of alpha smooth muscle actin (αSMA)-labeled structures were performed using Imaris surface function and based on the morphological characteristics of orientation, location, and SMA content to subdivide blood vessels types and airways (Dahlgren et al., 2019). Three dimensional distances between lymphocytes and SMA+ surfaces were calculated using the Imaris Distance Transform Matlab XTension and volumetric decile calculations were performed using a Matlab-based Imaris XTension. Adventitial lymphocytes were defined as spots localized <100μm from SMA+ smooth muscle surface and parenchymal lymphocytes as >100μm away from SMA+ surfaces.
Histology
Animals were euthanized by CO2 and then perfused with 1X DPBS/0.2% heparin followed by 4% PFA. Tissues were harvested, post fixed overnight in 4% PFA/DPBS with rotation, then stored in 70% ethanol prior to processing. For liver histology, embedding, sectioning and Hematoxylin and Eosin (H&E) staining were performed at UCSF Core Facility.
Infections
For helminth infections, 500 third-stage (L3) larvae of Nippostrongylus brasiliensis (Nb) were injected subcutaneously at the base of the tail as previously described (Molofsky et al., 2015b). Mice were sacrificed at the indicated time points and tissues were harvested and analyzed. For infections using wild-type Listeria monocytogenes strain 10403s, mice were injected intravenously via the tail veins with 1×104 colony-forming units (CFU) in 200 μL sterile 1X DPBS, or 2.5×104 CFU in Yeti recipients. Livers and spleens from infected mice were removed at different time points after infection, and Listeria CFU were calculated by plating dilutions of tissue homogenates on brain heart infusion (BHI) plates.
Cytokine and antibody in vivo treatments
For cytokine injections, recombinant mouse Interleukin-33 (R&D systems, Cat# 3626-ML-010/CF) was given as 500ng in 200 μL DPBS i.p every other day for three doses. Recombinant mouse IFNγ (R&D systems, Cat# 485-MI-100/CF) doses were administrated as 10μg in 200 μL DPBS. For antibody injections, mice were injected i.v with 50μg neutralizing mAb against IL-13 (IgG1, clone 8H8, InvivoGen) and 50ug purified NA/LE mAb against IL-4Rα mAB against ILR4a (IgG2a, clone mIL4R-M1, BD Biosciences) or corresponding isotypes controls, for 4 consecutive days following the scheme presented in figure 5J.
Drug Preparation and Administration.
FTY720 (Fingolimod, Cayman Chemicals, Cambridge, UK) was freshly prepared by dissolving 10mg/mL in absolute ethanol and then diluted in corn oil (1:20 ratio, vehicle solution). Mice were injected intraperitoneally with 5 mg/kg FTY720 in 100 μl vehicle solution. W146 (TOCRIS, Cat. 3602 Batch N.2), a selective S1PR1 antagonist, was first dissolved in 2 mM NaOH at 5 mg/mL and finally diluted in sterile water to a final concentration of 2 mg/mL and given at 10 mg/kg body weight. CYM50358 hydroxichloride (TOCRIS, Cat. 4679), a selective antagonist of S1PR4, was dissolved in sterile water at x 50 mg/mL and given at 10 mg/kg.
Cytokine measurement in plasma by ELISA
To measure concentrations of circulating IL-13, plasma from resting and IL33-treated mice were collected at different time points and measured by ELISA using mouse IL-13 antibody pair kit that detect native and recombinant mouse IL-13 with 0.2 pg/mL sensitivity (Abcam, Cat. ab221432). Amounts of IFNγ in blood were analyzed 2 days after Lm infection using 0.2ug/mL anti-IFNγ capture antibody (R4–6A2 Biolegend Cat. 505702) and 0.2ug/mL biotinylated anti-IFNγ detector antibody (XMG1.2 Biolegend Cat. 505804). Overall, plasma, positive controls, and recombinant standard proteins were diluted, added to ELISA plates (Corning, NY USA) pre-coated with capture antibody and incubated for 2 hours at RT or overnight at 4°C. Biotinylated detector antibody was then added and incubated at 37°C for 1 h followed by streptavidin-conjugated HRP (R&D Systems, MN USA) and visualized with slow kinetic-form TMB (Sigma Aldrich, MO USA). The reaction was stopped with 1N H2SO4 (Fisher Scientific, MA USA). Absorbance was measured at 450nm and corrected for absorbance at 540nm using a SpectraMax 5 spectrophotometer (Molecular Devices, CA USA). Measurements were performed in duplicate, and the results were averaged.
ILC2 cell sorting and 3’ Taq RNASeq
Approximately 10,000–20,000 ILC2s from lungs and adipose tissue were sorted into 100–200 ul of lysis/binding buffer (Qiagen) using a BD FACSAria II sorting system with a 100um nozzle size and 4 way-purity sort mode. ILC2s were sorted from IL-5Cre-tdtomato Red5 mice as live (DAPI−), lineage-negative (CD3, CD4, CD8, CD11b, CD11c, CD19, NK1.1, Ter119) CD45+ Thy1.2+ tdtomato+. Total RNA was isolated using QIAGEN RNeasy Plus Micro Kit according to the manufacturer’s instructions. RNA quality and quantitation were assessed on a Bioanalyzer. 3’-Tag RNAseq library preparation and sequencing was carried by the DNA Technologies and Expression Analysis Core at the UC Davis Genome Center, supported by NIH Shared Instrumentation Grant 1S10OD010786–01. Briefly, Gene expression profiling was carried out using a 3’ Tag-RNA-Seq protocol. Barcoded sequencing libraries were prepared using the QuantSeq FWD kit (Lexogen, Vienna, Austria) for multiplexed sequencing according to the recommendations of the manufacturer (Lexogen). The fragment size distribution of the libraries was verified via micro-capillary gel electrophoresis on a Bioanalyzer 2100 (Agilent, Santa Clara, CA). The libraries were quantified by fluorometry on a Qubit fluorometer (LifeTechnologies, Carlsbad, CA), and pooled in equimolar ratios. Up to forty-eight libraries per lane were sequenced on a HiSeq 4000 sequencer (Illumina, San Diego, CA).
To read raw sequence data and set quality checks we used FastQC v0.72. FASTQ files were trimmed with Trimmomatic v0.38.1 to remove low quality reads and any adapter. The reads were mapped with HISAT2 v2.1.0 to the mouse genome (mm10 / GRCm38). After mapping, all BAM files were used as input for HTSeq-count v0.91 to calculate transcript coverage. DESeq2 (v2.11.40) was used to find differentially expressed transcripts between samples for each sequencing depth. Differentially expressed genes (DEG) were kept if the adjusted p value was equal or below 0.05 and if a log2 fold-change of 1 or greater was observed. Data are deposited in the NCBI Gene Expression Omnibus (GEO) database (GSE190208). Volcano plots were build using EnhancedVolcano v1.8.0 and heatmaps (FDR 0.05) with GENE-E v3.0.215. In order to do pathway enrichment analysis, we use Ingenuity Pathways Analysis taking differential expressed genes with a FDR 0.05 as determined from the DESeq2 differential gene expression analysis above.
RNA preparation and qRT-PCR
Indicated populations were sorted into RLT Plus lysis buffer (Qiagen) and stored at −80 °C, then processed using Allprep DNA/RNA micro kit (Qiagen) per manufacturer’s protocol. For qPCR analyses, RNA was reverse transcribed using SuperScript III cDNA synthesis kit (ThermoFisher) and amplified using Power SYBR Green PCR master mix (ThermoFisher). Transcripts were normalized to Ppia (Cyclophilin A) expression and relative expression shown as 2-ΔCt. Primer sequences: Ppia, 5′ TGGAGAGCACCAAGACAGACA-3′, 5′- TGCCGGAGTCGACAATGAT-3′; Il4, 5′- ACAGGAGAAGGGACGCCAT-3′, 5′- GAAGCCCTACAGACGAGCTCA-3′; Il13, 5 GGAGCTGAGCAACATCACACA-3′, 5′- GGTCCTGTAGATGGCATTGCA-3′.
ILC2 adoptive transfers
In vivo expansion of ILC2s were performed in Il5dtomato-Cre/+; Rosa26fsf-RFP-Ai14 mice with 3 injections of 500ng IL-33 as described before. Lung ILC2s were sorted as viable, lineage negative RFP+ cells and collected in complete media. In some cases, cells were washed in 1X DPBS before i.v. transfer the same day. For Pertussis toxin (PTX) treatment, cells were cultured for 24 h in complete media supplemented with hIL-2 and mIL-7, +/− 100mg/mL PTX (Millipore sigma) before DPBS wash and transfer. 50,000–200,000 (ex-vivo) or 500,000 (in vitro expanded) IL-5+ ILC2s were transferred into wild type, Il7ra+/− controls or Il7ra−/− recipients. Recipient mice were euthanized 3- or 14-days post transfer and lungs were prepared for flow cytometry or imaging as described in separate sections.
Cell death assays.
For in vitro cell culture, IL-5tdtomato+ ILC2s were purified by FACS sorting as described before. After sorting, cells were seeded in 96-well U-bottom plates at a density of 3–5,000 cell/well in complete medium (RPMI1640 supplemented with 1 mM nonessential amino acids, 2 mM glutamine, 1 mM pyruvate, 100 U/mL penicillin/streptomycin and 50 μM β-mercaptoethanol) with 20 ng/mL rmIL-7 (R&D Systems Cat. 407-ML) and incubated at 37°C and 105% CO2. To evaluated IFNγ mediated cell death, cells were either left untreated or treated with IFNγ (20 ng/mL). After 72h, cells were washed with 1X annexin binding buffer and resuspended in cold binding buffer containing APC-conjugated AnnexinV and 1 μM SYTOX® Green stain, incubated at 37°C for 15 minutes and then immediately analyzed by flow cytometry. Viable cells were defined as those negative for both SYTOX and Annexin V, while early apoptotic cells were Annexin V+SYTOX-. Double-positive cells were late apoptotic or necrotic.
TUNEL: For detection of in situ cell death, DNA fragmentation in Il5dtomato-Cre; Rosa26fsf-RFP-Ai14 lymphocytes after listeria infection was detected by terminal dUTP nick-end labeling (TUNEL) method using DeadEnd Fluorometric TUNEL System (Promega, Madison, WI) according to the manufacturer’s protocol and co-stained with antibodies as described (Cautivo et al., 2016). In brief, 7um paraffin-embedded liver and lung sections were deparaffinized, rehydrated, washed and then permeabilized with 20μg/mL Proteinase K solution for 10 minutes. After, samples were treated with Equilibration buffer for 10 min and then incubated with TdT reaction mix for 60 minutes at 37°C in the dark. Samples were subsequently washed with DPBS, then blocked, stained with anti-tdTomato (1:300) overnight at 4°C, washed again and then incubated with Alexa Fluor 555 donkey anti-rabbit IgG for 1hr at room temperature. Finally, slides were mounted in ProLong Gold antifade reagent with DAPI (Molecular Probes Cat. P36935), covered and analyzed under a confocal microscope
QUANTIFICATION AND STATISTICAL ANALYSIS
All data were analyzed by comparison of means using unpaired two-tailed Student’s t-tests, or for multiple comparisons ANOVA with Holm-Sidak post hoc test (Prism, GraphPad Software, La Jolla, CA), with ns = not significant, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001. Figures display means ± standard deviation (SD) unless otherwise noted. When possible, results from independent experiments were pooled. All data points reflect individual biological mouse replicates (flow analysis) or individual tissue slices analyzed (confocal imaging). For imaging experiments, all analysis was performed at least three mice from at least two independent experiments, with two or more fields analyzed per mouse.
Supplementary Material
Table S1: Complete RNA-sequencing results with DEGs from lung and perigonadal adipose tissue (GAT) ILC2s from control and T2L IFNγ-blind mice treated +/− IL-33 +/− IFNγ, related to Figure 6 and Figure S6.
Table S2: List of lung ILC2 genes differentially regulated between IL-33 treated control mice and IL-33 + IFNγ treated “IL-5+ T2L IFNγ-blind mice, related to Figure 6 and Figure S6.
Video S1: IL-5+ T2L in naïve lung, thick-section confocal imaging, 3D. Related to Fig. 1
Video S2: IL-5+ T2L in lung one-month post Nippostrongylus brasiliensis (Nb) infection, thick-section confocal imaging, 3D. Related to Fig. 1
Video S3: IL-5+ T2L in lung one-month after IL-33 treatment, thick-section confocal imaging, 3D. Related to Fig. 1
Video S4: IL-5+ T2L in naïve and post IL33 treatment (D30) livers, thick-section confocal imaging, 3D. Related to Fig. 1
Video S5: IFNγ+ T1L in lung and liver from naïve IfngYFP/+ Yeti mice and after infection with Listeria monocytogenes. Thick-section confocal imaging, 3D. Related to Fig. 2.
Video S6: IL-5+ T2L in lung from IL-5 lineage tracker mice crossed to IfngYFP/+ Yeti mouse, one-month post Nippostrongylus brasiliensis (Nb) infection. Related to Fig. 2
Video S7: IL-5+ T2L in livers from mice treated with IL33 + FTY720, Thick-section confocal imaging, 3D. Related to Fig. 7.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse CD45 BUV395 (clone 30-F11) | BD Biosciences | Cat#565967; RRID: AB_2739420 |
| Anti-mouse CD45 APC (clone 30-F11) | Biolegend | Cat#103112; RRID: AB_312977 |
| Anti-mouse CD45.2 AF647 (clone 104) | Biolegend | Cat#109817; RRID: AB_492871 |
| Anti-mouse CD90.2 BV605 (Thy1.2) (clone 53-2.1) | Biolegend | Cat#140318; RRID: AB_2650924 |
| Anti-mouse CD3 PerCP Cy5.5 (clone 17A2) | Biolegend | Cat#100218, RRID: AB_1595492 |
| Anti-mouse CD3 AF488 (clone 17A2) | Biolegend | Cat#100210; RRID: AB_389301 |
| Anti-mouse CD3e APC (clone 17A2) | Biolegend | Cat#100236; RRID: AB_2561456 |
| Anti-mouse CD4 BV711 (clone RM4-5) | Biolegend | Cat#100557; RRID: AB_2562607 |
| Anti-mouse CD8 BV786 (clone 53-6.7) | Biolegend | Cat#100750; RRID: AB_2562610 |
| Anti-mouse CD11b BV650 (clone M1/70) | Biolegend | Cat#101259; RRID: AB_2566568 |
| Anti-mouse CD11b Pacific Blue (clone M1/70) | Biolegend | Cat#101224 RRID: AB_755986 |
| Anti-mouse CD11b FITC (clone M1/70) | Biolegend | Cat#101206; RRID: AB_312789) |
| Anti-mouse CD11c Pacific Blue (clone N418) | Biolegend | Cat#117321; RRID: AB_755987 |
| Anti-mouse CD11c PE-Cy7 (clone N418) | Biolegend | Cat#117318; RRID: AB_493568 |
| Anti-mouse Ly6C BV605 (clone HK1.4) | Biolegend | Cat#128035; RRID:AB_2562352 |
| Anti-mouse Ly6G PerCP Cy5.5 (clone 1A8) | BD Biosciences | Cat#560602; RRID:AB_1727563 |
| Anti-mouse Siglec-F PE (clone E50-2440) | BD Biosciences | Cat#552126; RRID: AB_394341 |
| Anti-mouse Siglec-F BV786 (clone E50-2440) | BD Biosciences | Cat#740956 RRID: AB_2740581 |
| Anti-mouse mouse MHC Class II (I-A/I-E) FITC (clone M5/114.15.2) | Thermo Fisher Scientific | Cat#11-5321-85; RRID: AB_465233 |
| Anti-mouse NK1.1 Pacific Blue (clone PK136) | Biolegend | Cat#108722; RRID: AB_2132712 |
| Anti-mouse NK1.1 PE-Cy7 (clone PK136) | Biolegend | Cat#108714; RRID: AB_389364 |
| Anti-mouse NK1.1 BV650 (clone PK136) | Biolegend | Cat#108736; RRID: AB_2563159 |
| Anti-mouse CD19 Pacific Blue (clone 6D5) | Biolegend | Cat#115523; RRID: AB_439718 |
| Anti-mouse T1/ST2 PE (clone DJ8) | MD Biosciences | Cat#101001PE |
| Anti-mouse KLRG1 PE-Cy7 (clone 2F1) | Biolegend | Cat#138416; RRID: AB_2561736 |
| Anti-mouse FoxP3 PE-Cy7 (clone FJK-16S) | Thermo Fisher Scientific | Cat#25-5773-82; RRID: AB_891552 |
| Anti-mouse FoxP3 AF488 (clone FJK-16S) | Thermo Fisher Scientific | Cat#53-5773-82; RRID: AB_763537 |
| Anti-mouse Ki-67 FITC (clone 16A8) | Biolegend | Cat#652410; RRID: AB_2562141 |
| Anti-mouse Ki-67 PE (clone 16A8) | Biolegend | Cat#652404; RRID: AB_2561525 |
| Anti-mouse Gata3 eF660 (clone TWAJ) | Thermo Fisher Scientific | Cat#50-9966-42; RRID: AB_10596663 |
| Anti-mouse Gata3 PE (clone TWAJ) | Thermo Fisher Scientific | Cat#12-9966-41; RRID: AB_1963601 |
| Anti-mouse IL13 PE (clone eBio13A) | Thermo Fisher Scientific | Cat#12-7133-82; RRID: AB_763559 |
| Anti-mIL-13-IG antibody (clone 8H8) | InvivoGen | Cat#mabg-mil13; RRID:AB_2722583 |
| Purified Rat Anti-Mouse CD124 (clone mIL4R-M1) | BD Biosciences | Cat#551853; RRID:AB_394274 |
| Living Colors anti-DsRed Rabbit Polyclonal Pan Antibody | TaKaRa | Cat#632496 |
| Chicken Polyclonal anti-GFP | Aves Labs | Cat#GFP-1020 |
| Rat Monoclonal anti-mouse CD31 (clone MEC 13.3) | BD Pharmigen | Cat#550274; RRID:AB_393571 |
| Rat Monoclonal anti mouse CD4 (clone RM4-5) | Biolegend | Cat#100506; RRID:AB_312709 |
| Goat anti mouse VEGFR3 (clone Flt-4) | R&D Systems | Cat#AF743 |
| Alexa Fluor 488 anti-aSMA monoclonal Antibody (clone 1A4) | eBioscience | Cat#53-9760-82; RRID:AB_2574461 |
| eFluor 660 anti-LYVE1 monoclonal Antibody (clone ALY7) | eBioscience | Cat#53-0443-82; RRID:AB_1633415 |
| Alexa Fluor 647 anti-CD4 (clone RM4-5) | Biolegend | Cat#100530; RRID:AB_389325 |
| Alexa Fluor 555 donkey anti-rabbit IgG (H+L) cross-adsorbed | Invitrogen | Cat#A31572 |
| Alexa Fluor 647 donkey anti-rat IgG (H+L) | Abcam | Cat#ab150155 |
| Rat Monoclonal anti-Endomucin (clone V.7C7.1) | Abcam | Cat#ab106100 |
| Anti-Chicken IgY (H+L), highly cross-adsorbed, CF™ 488A antibody | Sigma-Aldrich | Cat# SAB4600031 |
| FITC Rat Anti-Mouse IFN-γ (clone XMG1.2) | BD Pharmigen | Cat#562019; RRID:AB_395375 |
| Purified anti-mouse CD3 Antibody (clone 17A2) | Biolegend | Cat#100202; RRID:AB_312659 |
| Purified anti-mouse IFN-γ Antibody (clone R4-6A2) | Biolegend | Cat#505702; RRID:AB_315390 |
| Biotin anti-mouse IFN-γ Antibody (clone XMG1.2) | Biolegend | Cat#505804; RRID:AB_315398 |
| anti-mouse TER-119/Erythroid Cells Pacific Blue (clone TER-119) | Biolegend | Cat#116232; RRID:AB_2251160 |
| Bacterial and Virus Strains | ||
| Listeria monocytogenes strain 10403s | L. monocytogenes is maintained and grown in Ari Molofsky lab. | |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant mouse Interleukin-33 | R&D systems | Cat#3626-ML-010/CF |
| Recombinant mouse IFNg | R&D systems | Cat#485-MI-100/CF |
| FTY720 | Cayman Chemicals | Cat#10006292 |
| W146 | TOCRIS | Cat#3602 Batch N.2 |
| CYM50358 hydroxichloride | TOCRIS | Cat#4679 |
| 1X Hanks’ Balanced Salt Solution (1X HBSS), with calcium, with magnesium, no phenol red | Gibco | Cat#14025092 |
| 1X Dulbecco’s phosphate-buffered saline (1X DPBS), no calcium, no magnesium | Gibco | Cat#14190144 |
| RPMI 1640 Medium | Gibco | Cat#11875093 |
| MEM Non-Essential Amino Acids Solution (100X) | Gibco | Cat#11140050 |
| GlutaMAX™ Supplement (100X) | Thermo Scientific | Cat#35050038 |
| Sodium Pyruvate (100 mM) | Gibco | Cat#11360070 |
| Penicillin-Streptomycin (10,000 U/mL) | Gibco | Cat#15140122 |
| 2-Mercaptoethanol (50 mM) | Gibco | Cat#31350010 |
| Pierce™ 16% Formaldehyde (w/v), Methanol-free | Thermo Scientific Pierce | Cat#28906 |
| SeaPlaque™ Agarose | Lonza | Cat#50101 |
| HistoDenz | Sigma-Aldrich | Cat#D2158 |
| Bovine Serum Albumin (BSA) | Sigma-Aldrich | A9418 |
| HEPES (1 M) | Gibco | Cat#15630080 |
| DMEM, low glucose | Gibco | Cat#12320032 |
| Catalog number: | ||
| Liberase Tm | Roche | Cat#5401127001 |
| DNase I grade II, from bovine pancreas | Roche | Cat#10104159001 |
| Methyl salicylate ReagentPlus®, ≥99% (GC) | Sigma-Aldrich | Cat#M6752 |
| BD Difco™ Brain Heart Infusion Agar | BD Biosciences | Cat#241830 |
| Brain Heart Infusion Broth | Millipore | Cat#53286 |
| Corn Oil | Sigma-Aldrich | Cat#C8267 |
| BD Pharm Lyse™ lysing solution (10X) | BD Biosciences | Cat#555899 |
| Percoll | GE Healthcare | Cat#GE17-0891-01 |
| Recombinant Mouse IL-7 Protein | R&D systems | Cat#407-ML-200/CF |
| Fetal Bovine Serum (FBS) | Atlanta Biologicals | Cat#S11150 |
| Critical Commercial Assays | ||
| DeadEnd Fluorometric TUNEL System | Promega | Cat#G3250 |
| Dead Cell Apoptosis Kit with Annexin V Alexa Fluor™ 488 & Propidium Iodide (PI) | Invitrogen | Cat#V13241 |
| Allprep DNA/RNA micro kit | Qiagen | Cat#80284 |
| Zombie NIR™ Fixable Viability Kit | Biolegend | Cat#423105 |
| CountBright™ Absolute Counting Beads | Invitrogen | Cat#C36950 |
| eBioscience™ Foxp3 / Transcription Factor Staining Buffer Set | Invitrogen | Cat#00-5523-00 |
| Cell Stimulation Cocktail (500X) | Tonbo Biosciences | Cat#TNB-4975 |
| Mouse IL-13 Matched Antibody Pair Kit | Abcam | Cat#ab221432 |
| Power SYBR Green Master Mix | Applied Biosystems | Cat#4367659 |
| SuperScript™ III First-Strand Synthesis System | Invitrogen | Cat#18080051 |
| Deposited Data | ||
| 3’ Tag-RNA-Seq on sorted ILC2s. Raw and analyzed data | This paper | GSE190208 |
| RNA-Seq data on wild type sorted ILC2 post treated in vitro with IL33 or IL33+IFNg | Moro et al., 2016 | GSE73272 |
| Experimental Models: Organisms/Strains | ||
| Mouse: B6(C)-Il5tm1.1(icre)Lky/J (Red5) | Nussbaum et al., 2013 | RRID: IMSR_JAX:030926 |
| Mouse: B6.129S4-Ifngtm3.1Lky/J (YFP-Enhanced Transcript for IFN-γ or Yeti) | Stetson et al., 2003 | Richard Locksley Lab |
| Mouse: Tbet (Tbx21)-zsGreen | Zhu et al., 2012 | Jinfang Zhu Lab |
| Mouse: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J (Ai14) | Madisen et al., 2010 | RRID: IMSR_JAX:007914 |
| Mouse: B6(C)-Ifngr1tm1.1Rds/J (Ifngr1flox/flox) | Lee et al., 2013 | RRID: IMSR_JAX:025394 |
| Mouse: B6.129S7-Ifngr1tm1Agt/J (IFNgR1 deficient) | Huang et al., 1993 | RRID: IMSR_JAX:003288 |
| Mouse: B6.Cg-Tg(Cd4-cre)1Cwi/BfluJ (CD4-Cre) | Lee et al., 2001 | RRID: IMSR_JAX:022071 |
| Mouse: B6.129(Cg)-Foxp3tm4(YFP/icre)Ayr/J (Foxp3Cre-YFP) | Rubtsov et al., 2008 | RRID: IMSR_JAX:016959 |
| Mouse: B6.129S7-Il7rtm1/mx/J (IL7Ra deficient) | Peschon et al., 1994 | RRID: IMSR_JAX:002295 |
| Mouse: B6.129S4-Pdgfratm11(EGFP)Sor/J (PDGFRa-H2B-eGFP) | Hamilton et al., 2003 | RRID: IMSR_JAX:007669 |
| Oligonucleotides | ||
| Primer: Il4 Forward: 5’- ACAGGAGAAGGGACGCCAT-3’ | Elim biopharmaceuticals | N/A |
| Primer: Il4 Reverse: 5’- GAAGCCCTACAGACGAGCTCA-3’ | Elim biopharmaceuticals | N/A |
| Primer: Il13 Forward: 5’-GGAGCTGAGCAACATCACACA-3’ | Elim biopharmaceuticals | N/A |
| Primer: Il13 Reverse: 5’- GGTCCTGTAGATGGCATTGCA-3’ | Elim biopharmaceuticals | n/a |
| Software and Algorithms | ||
| GraphPad Prism (v9.1.2) | GraphPad Software https://www.graphpad.com | |
| FlowJo (v10.7.2) | https://www.flowjo.com | Tree Star, USA |
| Bitplane Imaris (v9.5) | https://imaris.oxinst.com | Andor Technology PLC, Belfast, N. Ireland |
| FastQC (v0.72) | ||
| Trimmomatic (v0.38.1) | ||
| HISAT2 (v2.1.0) | ||
| HTSeq-count (v0.9) | ||
| DESeq2 (v2.11.40) | ||
| EnhancedVolcano (v1.8.0) | ||
| Ingenuity Pathways Analysis | Qiagen | |
| Fiji | https://imagej.net/software/fiji/ | |
| Other | ||
| Nippostrongylus brasiliensis | Third-stage larvae (L3) of N. brasiliensis were produced and maintained in Richard Locksley lab. | |
HIGHLIGHTS:
3D imaging defines IL-5+ ILC2 and Th2 cell secondary niches in tissue parenchyma.
IFNγ directly restricts IL-5+ lymphocyte dispersion and parenchymal accumulation.
IFNγ acts by restricting parenchymal IL-5+ cell trafficking and promoting cell death.
Restricting parenchymal IL-5+ lymphocytes limits Listeria-associated mortality.
ACKNOWLEDGEMENTS
We thank the UCSF Parnassus Flow Core RRID:SCR_018206 and DRC Center Grant NIH P30 DK063720 and UCSF Biological Imaging and Development Core (BIDC) for instruments and services; Tien Peng, Anna V. Molofsky, Michael Rosenblum (comments and discussion), Jorge Ortiz-Carpena and Sepideh Farhat (technical assistance); KMC is supported the NHLBI (NIH R01HL142701 and R56HL142701). RML by NIH R01AI26918 and HHMI. ABM is supported by the NLHBI (NIH R01HL142701 and R56HL142701), NIDDK (K08DK101604), Larry L. Hillblom Foundation Grant, Nina Ireland Program for Lung Health, Sandler Asthma Basic Research Center (SABRE), and the UCSF Liver Center, RAP and PBBR grants.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ajendra J, Chenery AL, Parkinson JE, Chan BHK, Pearson S, Colombo SAP, Boon L, Grencis RK, Sutherland TE, and Allen JE (2020). IL-17A both initiates, via IFNγ suppression, and limits the pulmonary type-2 immune response to nematode infection. Mucosal Immunol. 13, 958–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JE, and Sutherland TE (2014). Host protective roles of type 2 immunity: Parasite killing and tissue repair, flip sides of the same coin. Semin. Immunol. 26, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KG, Mayer-Barber K, Sung H, Beura L, James BR, Taylor JJ, Qunaj L, Griffith TS, Vezys V, Barber DL, et al. (2014). Intravascular staining for discrimination of vascular and tissue leukocytes. Nat. Protoc. 9, 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura LK, Mitchell JS, Thompson EA, Schenkel JM, Mohammed J, Wijeyesinghe S, Fonseca R, Burbach BJ, Hickman HD, Vezys V, et al. (2018). Intravital mucosal imaging of CD8+ resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory article. Nat. Immunol. 19, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J, Cui L, Yu G, Yang X, Chen Y, and Wan X (2017). NK Cells Alleviate Lung Inflammation by Negatively Regulating Group 2 Innate Lymphoid Cells. J. Immunol. 198, 3336–3344. [DOI] [PubMed] [Google Scholar]
- Buechler MB, Pradhan RN, Krishnamurty AT, Cox C, Calviello AK, Wang AW, Yang YA, Tam L, Caothien R, Roose-girma M, et al. (2021). Cross-tissue organization of the fibroblast lineage. Nature. [DOI] [PubMed] [Google Scholar]
- Cameron GJM, Cautivo KM, Loering S, Jiang SH, Deshpande AV, Foster PS, McKenzie ANJ, Molofsky AB, Hansbro PM, and Starkey MR (2019). Group 2 Innate Lymphoid Cells Are Redundant in Experimental Renal Ischemia-Reperfusion Injury. Front. Immunol. 10, 826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell L, Hepworth MR, Whittingham-Dowd J, Thompson S, Bancroft AJ, Hayes KS, Shaw TN, Dickey BF, Flamar AL, Artis D, et al. (2019). ILC2s mediate systemic innate protection by priming mucus production at distal mucosal sites. J. Exp. Med. 216, 2714–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro F, Cardoso AP, Gonçalves RM, Serre K, and Oliveira MJ (2018). Interferon-gamma at the crossroads of tumor immune surveillance or evasion. Front. Immunol. 9, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cautivo KM, and Molofsky AB (2016). Regulation of metabolic health and adipose tissue function by group 2 innate lymphoid cells. Eur. J. Immunol. 46, 1315–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cautivo KM, Lizama CO, Tapia PJ, Agarwal AK, Garg A, Horton JD, and Cortés VA (2016). AGPAT2 is essential for postnatal development and maintenance of white and brown adipose tissue. Mol. Metab. 5, 491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cautivo KM, Steer CA, and Molofsky AB (2020). Immune outposts in the adventitia: One foot in sea and one on shore. Curr. Opin. Immunol. 64, 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TT, and Stevens SR (2002). Atopic dermatitis: the role of recombinant interferon-gamma therapy. Am. J. Clin. Dermatol. 3, 175–183. [DOI] [PubMed] [Google Scholar]
- Chávez-Arroyo A, and Portnoy DA (2020). Why is Listeria monocytogenes such a potent inducer of CD8+ T-cells? Cell. Microbiol. 22, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M (2018). Innate Lymphoid Cells: Diversity, Plasticity, and Unique Functions in Immunity. Immunity 48, 1104–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corren J, and Ziegler SF (2019). TSLP : from allergy to cancer. Nat. Immunol. 20, 1603–1609. [DOI] [PubMed] [Google Scholar]
- Coyle AJ, Tsuyuki S, Bertrand C, Huang S, Aguet M, Alkan SS, and Anderson GP (1996). Mice lacking the IFN-γ receptor have an impaired ability to resolve a lung eosinophilic inflammatory response associated with a prolonged capacity of T cells to exhibit a Th2 cytokine profile. J. Immunol. 156, 2680–265. [PubMed] [Google Scholar]
- Dahlgren MW, and Molofsky AB (2018). All along the watchtower: group 2 innate lymphoid cells in allergic responses. Curr Opin Immunol 54, 13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren MW, and Molofsky AB (2019). Adventitial Cuffs: Regional Hubs for Tissue Immunity. Trends Immunol. 40, 877–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren MW, Jones SW, Cautivo KM, Dubinin A, Ortiz-Carpena JF, Farhat S, Yu KS, Lee K, Wang C, Molofsky AVAB, et al. (2019). Adventitial Stromal Cells Define Group 2 Innate Lymphoid Cell Tissue Niches. Immunity 50, 707–722.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr CU, McCarthy CDA, Mindt BC, Rubio M, Meli AP, Pothlichet J, Eva MM, Gauchat J-F, Qureshi ST, Mazer BD, et al. (2016). Type I interferon restricts type 2 immunopathology through the regulation of group 2 innate lymphoid cells. Nat Immunol 17, 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton EE, Gajdasik DW, Willis C, Fiancette R, Bishop EL, Camelo A, Sleeman MA, Coccia M, Didierlaurent AM, Tomura M, et al. (2019). Peripheral lymph nodes contain migratory and resident innate lymphoid cell populations. Sci. Immunol. 4, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y, Hirahara K, Iinuma T, Shinoda K, Tumes DJ, Asou HK, Matsugae N, Obata-Ninomiya K, Yamamoto H, Motohashi S, et al. (2015). The interleukin-33-p38 kinase axis confers memory T helper 2 cell pathogenicity in the airway. Immunity 42, 294–308. [DOI] [PubMed] [Google Scholar]
- Erb KJ, Holloway JW, Sobeck A, Moll H, and Le-Gros G (1998). Infection of mice with mycobacterium bovis-bacillus calmette-guerin (BCG) suppresses allergen-induced airway eosinophilia. Pneumologie 52, 420–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, and Rudensky AY (2016). Hallmarks of Tissue-Resident Lymphocytes. Cell 164, 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustino LD, Griffith JW, Rahimi RA, Nepal K, Hamilos DL, Cho JL, Medoff BD, Moon JJ, Vignali DAA, and Luster AD (2020). Interleukin-33 activates regulatory T cells to suppress innate γδ T cell responses in the lung. Nat. Immunol. 21, 1371–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort MM, Cheung J, Yen D, Li J, Zurawski SM, Lo S, Menon S, Clifford T, Hunte B, Lesley R, et al. (2001). IL-25 induces IL-4, IL-5, and IL-13 and Th2-associated pathologies in vivo. Immunity 15, 985–995. [DOI] [PubMed] [Google Scholar]
- Gasteiger G, Fan X, Dikiy S, Lee SY, and Rudensky AY (2015). Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science (80-. ). 350, 981–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavett SH, O’Hearn DJ, Li X, Huang SK, Finkelman FD, and Wills-Karp M (1995). Interleukin 12 inhibits antigen-induced airway hyperresponsiveness, inflammation, and Th2 cytokine expression in mice. J. Exp. Med. 182, 1527–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain RN, and Huang Y (2019). ILC2s — resident lymphocytes pre-adapted to a specific tissue or migratory effectors that adapt to where they move? Curr. Opin. Immunol. 56, 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner MY, Kastenmuller W, Ifrim I, Kabat J, and Germain RN (2012). Histocytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity 37, 364–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaedi M, Shen ZY, Orangi M, Martinez-Gonzalez I, Wei L, Lu X, Das A, Heravi-Moussavi A, Marra MA, Bhandoola A, et al. (2020). Single-cell analysis of RORα tracer mouse lung reveals ILC progenitors and effector ILC2 subsets. J. Exp. Med. 217, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieseck RL, Wilson MS, and Wynn TA (2018). Type 2 immunity in tissue repair and fibrosis. Nat. Rev. Immunol. 18, 62–76. [DOI] [PubMed] [Google Scholar]
- Gisterå A, and Hansson GK (2017). The immunology of atherosclerosis. Nat. Rev. Nephrol. 13, 368–380. [DOI] [PubMed] [Google Scholar]
- Gola A, Dorrington MG, Speranza E, Sala C, Shih RM, Radtke AJ, Wong HS, Baptista AP, Hernandez JM, Castellani G, et al. (2021). Commensal-driven immune zonation of the liver promotes host defence. Nature 589, 131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo XJ, Dash P, Crawford JC, Allen EK, Zamora AE, Boyd DF, Duan S, Bajracharya R, Awad WA, Apiwattanakul N, et al. (2018). Lung γδ T Cells Mediate Protective Responses during Neonatal Influenza Infection that Are Associated with Type 2 Immunity. Immunity 49, 531–544.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak-Frendscho M, Brown JF, Iizawa Y, Wagner RD, and Czuprynski CJ (1992). Administration of anti-IL-4 monoclonal antibody 11B11 increases the resistance of mice to Listeria monocytogenes infection. J. Immunol. 148, 3978–3985. [PubMed] [Google Scholar]
- Han H, Roan F, and Ziegler SF (2017a). The atopic march: current insights into skin barrier dysfunction and epithelial cell-derived cytokines. Immunol. Rev. 278, 116–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Hong JY, Jaipalli S, Rajput C, Lei J, Hinde JL, Chen Q, Hershenson NM, Bentley JK, and Hershenson MB (2017b). IFN-γ Blocks Development of an Asthma Phenotype in Rhinovirus-Infected Baby Mice by Inhibiting Type 2 Innate Lymphoid Cells. Am. J. Respir. Cell Mol. Biol. 56, 242–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmers S, Schizas M, and Rudensky AY (2021). T reg cell-intrinsic requirements for ST2 signaling in health and neuroinflammation. J. Exp. Med. 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondowicz BD, An D, Schenkel JM, Kim KS, Steach HR, Krishnamurty AT, Keitany GJ, Garza EN, Fraser KA, Moon JJ, et al. (2016). Interleukin-2-Dependent Allergen-Specific Tissue-Resident Memory Cells Drive Asthma. Immunity 44, 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Mao K, Chen X, Sun MA, Kawabe T, Li W, Usher N, Zhu J, Urban JF, Paul WE, et al. (2018). S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science (80-. ). 359, 114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst SD, Muchamuel T, Gorman DM, Gilbert JM, Clifford T, Kwan S, Menon S, Seymour B, Jackson C, Kung TT, et al. (2002). New IL-17 Family Members Promote Th1 or Th2 Responses in the Lung: In Vivo Function of the Novel Cytokine IL-25. J. Immunol. 169, 443–453. [DOI] [PubMed] [Google Scholar]
- Kang SJ, Liang HE, Reizis B, and Locksley RM (2008). Regulation of Hierarchical Clustering and Activation of Innate Immune Cells by Dendritic Cells. Immunity 29, 819–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klicznik MM, Morawski PA, Höllbacher B, Varkhande SR, Motley SJ, Kuri-Cervantes L, Goodwin E, Rosenblum MD, Alice Long S, Brachtl G, et al. (2019). Human CD4+CD103+ cutaneous resident memory T cells are found in the circulation of healthy individuals. Sci. Immunol. 4, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlgruber AC, Gal-Oz ST, LaMarche NM, Shimazaki M, Duquette D, Nguyen HN, Mina AI, Paras T, Tavakkoli A, von Andrian U, et al. (2018). γδ T cells producing interleukin-17A regulate adipose regulatory T cell homeostasis and thermogenesis. Nat Immunol 19, 464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrecht BN, and Hammad H (2015). The immunology of asthma. Nat Immunol 16, 45–56. [DOI] [PubMed] [Google Scholar]
- Lambrecht BN, and Hammad H (2017). The immunology of the allergy epidemic and the hygiene hypothesis. Nat Immunol 18, 1076–1083. [DOI] [PubMed] [Google Scholar]
- Lee SH, Carrero JA, Uppaluri R, White JM, Archambault JM, Lai KS, Chan SR, Sheehan KCF, Unanue ER, and Schreiber RD (2013). Identifying the initiating events of anti-Listeria responses using mice with conditional loss of IFN-γ receptor subunit 1 (IFNGR1). J. Immunol. 191, 4223–4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Germain RN, and Gerner MY (2017). Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (Ce3D). Proc. Natl. Acad. Sci. U. S. A. 114, E7321–E7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew FY, Girard J-P, and Turnquist HR (2016). Interleukin-33 in health and disease. Nat. Rev. Immunol. 16, 676–689. [DOI] [PubMed] [Google Scholar]
- Liu Q, Dwyer GK, Zhao Y, Li H, Mathews LR, Chakka AB, Chandran UR, Demetris JA, Alcorn JF, Robinson KM, et al. (2019). IL-33–mediated IL-13 secretion by ST2+ Tregs controls inflammation after lung injury. JCI Insight 4, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd CM, and Snelgrove RJ (2018). Type 2 immunity: Expanding our view. Sci. Immunol. 3, 1–12. [DOI] [PubMed] [Google Scholar]
- Mack MR, Brestoff JR, Berrien-Elliott MM, Trier AM, Yang TLB, McCullen M, Collins PL, Niu H, Bodet ND, Wagner JA, et al. (2020). Blood natural killer cell deficiency reveals an immunotherapy strategy for atopic dermatitis. Sci. Transl. Med. 12, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi E, Parronchi P, Manetti R, Simonelli C, Piccinni MP, Rugiu FS, De Carli M, Ricci M, and Romagnani S (1992). Reciprocal regulatory effects of IFN-γ and IL-4 on the in vitro development of human Th1 and Th2 clones. J. Immunol. 148, 2142–2147. [PubMed] [Google Scholar]
- Mahlakõiv T, Flamar AL, Johnston LK, Moriyama S, Putzel GG, Bryce PJ, and Artis D (2019). Stromal cells maintain immune cell homeostasis in adipose tissue via production of interleukin-33. Sci. Immunol 4, eaax0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsland BJ, Harris NL, Camberis M, Kopf M, Hook SM, and Le Gros G (2004). Bystander suppression of allergic airway inflammation by lung resident memory CD8+ T cells. Proc. Natl. Acad. Sci. U. S. A. 101, 6116–6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez FD (2009). The origins of asthma and chronic obstructive pulmonary disease in early life. Proc. Am. Thorac. Soc. 6, 272–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MM, Patel PS, Bao K, Danhorn T, O’Connor BP, and Reinhardt RL (2020). BATF acts as an essential regulator of IL-25-responsive migratory ILC2 cell fate and function. Sci. Immunol. 5, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AB, Nussbaum JC, Liang H-E, Van Dyken SJ, Cheng LE, Mohapatra A, Chawla A, and Locksley RM (2013). Innate lymphoid type 2 cells sustain visceral adipose tissue eosinophils and alternatively activated macrophages. J. Exp. Med. 210, 535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AB, Savage AK, and Locksley RM (2015a). Interleukin-33 in Tissue Homeostasis, Injury, and Inflammation. Immunity 42, 1005–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AB, Van Gool F, Liang H-EE, Van Dyken SJ, Nussbaum JC, Lee J, Bluestone JA, Locksley RM, Dé Ric F, Gool V, et al. (2015b). Interleukin-33 And Interferon-gamma Counter-Regulate Group 2 Innate Lymphoid Cell Activation During Immune Perturbation. Immunity 43, 161–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, Kawamoto H, Furusawa J-I, Ohtani M, Fujii H, and Koyasu S (2010). Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature 463, 540–544. [DOI] [PubMed] [Google Scholar]
- Moro K, Kabata H, Tanabe M, Koga S, Takeno N, Mochizuki M, Fukunaga K, Asano K, Betsuyaku T, and Koyasu S (2016). Interferon and IL-27 antagonize the function of group 2 innate lymphoid cells and type 2 innate immune responses. Nat. Immunol. 17, 76–86. [DOI] [PubMed] [Google Scholar]
- Neill DR, Wong SH, Bellosi A, Flynn RJ, Daly M, Langford TKA, Bucks C, Kane CM, Fallon PG, Pannell R, et al. (2010). Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature 464, 1367–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen PT, Dorman LC, Pan S, Vainchtein ID, Han RT, Nakao-Inoue H, Taloma SE, Barron JJ, Molofsky AB, Kheirbek MA, et al. (2020). Microglial Remodeling of the Extracellular Matrix Promotes Synapse Plasticity. Cell 182, 388–403.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum JC, Van Dyken SJ, Von Moltke J, Cheng LE, Mohapatra A, Molofsky AB, Thornton EE, Krummel MF, Chawla A, Liang HE, et al. (2013). Type 2 innate lymphoid cells control eosinophil homeostasis. Nature 502, 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AE, Liang H-E, Sullivan BM, Reinhardt RL, Eisley CJ, Erle DJ, and Locksley RM (2010). Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc. Natl. Acad. Sci. 107, 11489–11494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttur F, Denney L, Gregory LG, Vuononvirta J, Oliver R, Entwistle LJ, Walker SA, Headley MB, McGhee EJ, Pease JE, et al. (2019). Pulmonary environmental cues drive group 2 innate lymphoid cell dynamics in mice and humans. Sci. Immunol. 4, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoshevich L, and Cossart P (2018). Listeria monocytogenes: Towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 16, 32–46. [DOI] [PubMed] [Google Scholar]
- Rana BMJ, Jou E, Barlow JL, Rodriguez-Rodriguez N, Walker JA, Knox C, Jolin HE, Hardman CS, Sivasubramaniam M, Szeto A, et al. (2019). A stromal cell niche sustains ILC2-mediated type-2 conditioning in adipose tissue. J. Exp. Med. 216, 1999–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt RL, Liang H-E, Bao K, Price AE, Mohrs M, Kelly BL, and Locksley RM (2015). A Novel Model for IFN-γ–Mediated Autoinflammatory Syndromes. J. Immunol. 194, 2358–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold U, Kukel S, Brzoska J, and Kreysel HW (1993). Systemic interferon gamma treatment in severe atopic dermatitis. J. Am. Acad. Dermatol. 29, 58–63. [DOI] [PubMed] [Google Scholar]
- Ricardo-Gonzalez RR, Schneider C, Liao C, Lee J, Liang HE, and Locksley RM (2020). Tissue-specific pathways extrude activated ILC2s to disseminate type 2 immunity. J. Exp. Med. 217, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruterbusch M, Pruner KB, Shehata L, and Pepper M (2020). In Vivo CD4 + T Cell Differentiation and Function: Revisiting the Th1/Th2 Paradigm. Annu. Rev. Immunol. 38, 705–725. [DOI] [PubMed] [Google Scholar]
- Schneider C, Lee J, Koga S, Ricardo-Gonzalez RR, Nussbaum JC, Smith LK, Villeda SA, Liang HE, and Locksley RM (2019). Tissue-Resident Group 2 Innate Lymphoid Cells Differentiate by Layered Ontogeny and In Situ Perinatal Priming. Immunity 50, 1425–1438.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K, Hertzog PJ, Ravasi T, and Hume DA (2004). Interferon-γ: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 75, 163–189. [DOI] [PubMed] [Google Scholar]
- Schuijs MJ, and Halim TYF (2016). Group 2 innate lymphocytes at the interface between innate and adaptive immunity. Ann. N. Y. Acad. Sci. 1417, 87–103. [DOI] [PubMed] [Google Scholar]
- Schuijs MJ, Png S, Richard AC, Tsyben A, Hamm G, Stockis J, Garcia C, Pinaud S, Nicholls A, Ros XR, et al. (2020). ILC2-driven innate immune checkpoint mechanism antagonizes NK cell antimetastatic function in the lung. Nat. Immunol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg GF, and Hepworth MR (2019). Functional interactions between innate lymphoid cells and adaptive immunity. Nat. Rev. Immunol. 19, 599–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spallanzani RG, Zemmour D, Xiao T, Jayewickreme T, Li C, Bryce PJ, Benoist C, and Mathis D (2019). Distinct immunocyte-promoting and adipocyte-generating stromal components coordinate adipose tissue immune and metabolic tenors. Sci. Immunol 4, eaaw3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann S, Schoedsack M, Heinrich F, Breda PC, Ochel A, Tiegs G, and Neumann K (2020). Hepatic ILC2 activity is regulated by liver inflammation-induced cytokines and effector CD4+ T cells. Sci. Rep. 10, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Mohrs M, Reinhardt RL, Baron JL, Wang Z-E, Gapin L, Kronenberg M, and Locksley RM (2003). Constitutive Cytokine mRNAs Mark Natural Killer (NK) and NK T Cells Poised for Rapid Effector Function. J. Exp. Med. 198, 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stier MT, Goleniewska K, Cephus JY, Newcomb DC, Sherrill TP, Boyd KL, Bloodworth MH, Moore ML, Chen K, Kolls JK, et al. (2017). STAT1 Represses Cytokine-Producing Group 2 and Group 3 Innate Lymphoid Cells during Viral Infection. J. Immunol. 199, 510–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stier MT, Zhang J, Goleniewska K, Cephus JY, Rusznak M, Wu L, Van Kaer L, Zhou B, Newcomb DC, and Peebles RS (2018). IL-33 promotes the egress of group 2 innate lymphoid cells from the bone marrow. J. Exp. Med. 215, 263–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolley JM, Johnston TS, Soerens AG, Beura LK, Rosato PC, Joag V, Wijeyesinghe SP, Langlois RA, Osum KC, Mitchell JS, et al. (2020). Retrograde migration supplies resident memory t cells to lung-draining ln after influenza infection. J. Exp. Med. 217, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, van Panhuys N, Kastenmüller W, and Germain RN (2013). The future of immunoimaging - Deeper, bigger, more precise, and definitively more colorful. Eur. J. Immunol. 43, 1407–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbitt CA, Stark JM, Martens L, Ma J, Mold JE, Deswarte K, Oliynyk G, Feng X, Lambrecht BN, De Bleser P, et al. (2019). Single-Cell RNA Sequencing of the T Helper Cell Response to House Dust Mites Defines a Distinct Gene Expression Signature in Airway Th2 Cells. Immunity 51, 169–184.e5. [DOI] [PubMed] [Google Scholar]
- Vainchtein ID, Chin G, Cho FS, Kelley KW, Miller JG, Chien EC, Liddelow SA, Nguyen PT, Nakao-Inoue H, Dorman LC, et al. (2018). Astrocyte-derived interleukin-33 promotes microglial synapse engulfment and neural circuit development. Science (80-. ). 359, 1269–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyken SJ, Nussbaum JC, Lee J, Molofsky AB, Liang HE, Pollack JL, Gate RE, Haliburton GE, Ye CJ, Marson A, et al. (2016). A tissue checkpoint regulates type 2 immunity. Nat. Immunol. 17, 1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie ANJ, Mebius RE, et al. (2018). Innate Lymphoid Cells: 10 Years On. Cell 174, 1054–1066. [DOI] [PubMed] [Google Scholar]
- Wensveen FM, Valentić S, Šestan M, Turk Wensveen T, and Polić B (2015). The “Big Bang” in obese fat: Events initiating obesity-induced adipose tissue inflammation. Eur. J. Immunol. 45, 2446–2456. [DOI] [PubMed] [Google Scholar]
- Woytschak J, Keller N, Krieg C, Impellizzieri D, Thompson RW, Wynn TA, Zinkernagel AS, and Boyman O (2016). Type 2 Interleukin-4 Receptor Signaling in Neutrophils Antagonizes Their Expansion and Migration during Infection and Inflammation. Immunity 45, 172–184. [DOI] [PubMed] [Google Scholar]
- Zeis P, Lian M, Fan X, Herman JS, Hernandez DC, Gentek R, Elias S, Symowski C, Knöpper K, Peltokangas N, et al. (2020). In Situ Maturation and Tissue Adaptation of Type 2 Innate Lymphoid Cell Progenitors. Immunity 53, 775–792.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Jankovic D, Oler AJ, Wei G, Sharma S, Hu G, Guo L, Yagi R, Yamane H, Punkosdy G, et al. (2012). The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity 37, 660–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Complete RNA-sequencing results with DEGs from lung and perigonadal adipose tissue (GAT) ILC2s from control and T2L IFNγ-blind mice treated +/− IL-33 +/− IFNγ, related to Figure 6 and Figure S6.
Table S2: List of lung ILC2 genes differentially regulated between IL-33 treated control mice and IL-33 + IFNγ treated “IL-5+ T2L IFNγ-blind mice, related to Figure 6 and Figure S6.
Video S1: IL-5+ T2L in naïve lung, thick-section confocal imaging, 3D. Related to Fig. 1
Video S2: IL-5+ T2L in lung one-month post Nippostrongylus brasiliensis (Nb) infection, thick-section confocal imaging, 3D. Related to Fig. 1
Video S3: IL-5+ T2L in lung one-month after IL-33 treatment, thick-section confocal imaging, 3D. Related to Fig. 1
Video S4: IL-5+ T2L in naïve and post IL33 treatment (D30) livers, thick-section confocal imaging, 3D. Related to Fig. 1
Video S5: IFNγ+ T1L in lung and liver from naïve IfngYFP/+ Yeti mice and after infection with Listeria monocytogenes. Thick-section confocal imaging, 3D. Related to Fig. 2.
Video S6: IL-5+ T2L in lung from IL-5 lineage tracker mice crossed to IfngYFP/+ Yeti mouse, one-month post Nippostrongylus brasiliensis (Nb) infection. Related to Fig. 2
Video S7: IL-5+ T2L in livers from mice treated with IL33 + FTY720, Thick-section confocal imaging, 3D. Related to Fig. 7.
Data Availability Statement
The accession number for the 3’Taq RNA-seq data reported in this paper is GSE190208. Accession code for ILC2s (Moro et al., 2016) can be found in the referenced study.


