Abstract
Viral infection is a major cause of ill health in wild chimpanzees (Pan troglodytes), but most evidence to date has come from conspicuous disease outbreaks with high morbidity and mortality. To examine the relationship between viral infection and ill health during periods not associated with disease outbreaks, we conducted a longitudinal study of wild eastern chimpanzees (P. t. schweinfurthii) in the Kanyawara and Ngogo communities of Kibale National Park, Uganda. We collected standardized, observational health data for four years and then used metagenomics to characterize gastrointestinal viromes (i.e., all viruses recovered from fecal samples) in individual chimpanzees before and during episodes of clinical disease. We restricted our analyses to viruses thought to infect mammals or primarily associated with mammals, discarding viruses associated with non-mammalian hosts. We found 18 viruses (nine of which were previously identified in this population) from at least five viral families. Viral richness (number of viruses per sample) did not vary by health status. By contrast, total viral load (normalized proportion of sequences mapping to viruses) was significantly higher in ill individuals compared to healthy individuals. Furthermore, when ill, Kanyawara chimpanzees exhibited higher viral loads than Ngogo chimpanzees, and males, but not females, exhibited higher infection rates with certain viruses and higher total viral loads as they aged. Post-hoc analyses, including the use of a machine-learning classification method, indicated that one virus, salivirus (Picornaviridae), was the main contributor to health-related and community-level variation in viral loads. Another virus, chimpanzee stool-associated virus (chisavirus; unclassified Picornavirales), was associated with ill health at Ngogo but not at Kanyawara. Chisavirus, chimpanzee adenovirus (Adenoviridae), and bufavirus (Parvoviridae) were also associated with increased age in males. Associations with sex and age are consistent with the hypothesis that non-lethal viral infections cumulatively reflect or contribute to senescence in long-lived species such as chimpanzees.
Keywords: Health, disease, virus, metagenomics, conservation, chimpanzee
Graphical Abstract

Introduction
Viruses are among the most important causes of ill health and mortality in wild chimpanzees and represent a grave threat to conservation (Dunay et al., 2018; Glasser et al., 2021; Köndgen et al., 2008; Leendertz et al., 2006; Melin et al., 2020; Williams et al., 2008). Respiratory diseases caused by viral infections, including those of coronaviruses, metapneumoviruses, and rhinoviruses, have been reported in chimpanzee populations across equatorial Africa (Kaur et al., 2008; Köndgen et al., 2008; Köndgen et al., 2010; Negrey et al., 2019; Patrono et al., 2018; Scully et al., 2018). However, most information about viral infection in wild chimpanzees comes from conspicuous disease outbreaks with high morbidity and mortality (Formenty et al., 1999; Köndgen et al., 2008; Negrey et al., 2019; Scully et al., 2018). Although virulence may, in some cases, augment viral transmission (Alizon & Michalakis, 2015), avirulent or minimally virulent pathogens can persist in populations due to greater long-term transmission success (Méthot, 2012). Therefore, many less virulent pathogens may have important but protracted health implications, perhaps acting cumulatively over time to affect health, as suggested for humans (Pawelec et al., 2010). Alternatively, viruses may respond to declines due to ill health by proliferating, offering a potentially useful biomarker of declining physiological or immunological function.
Viruses and other microparasites of low virulence can have important effects at both the individual and population levels (Anderson & May, 1979). Effects can be both direct (e.g., cellular damage) and indirect (e.g., limiting energy available for non-immunological processes). Furthermore, viruses can either cause ill health directly, or viral infection and shedding can result from ill health. In humans, several inflammatory bowel diseases are associated with expansion of the gastrointestinal virome (Norman et al., 2015), and in rhesus macaques (Macaca mulatta), pathogenic progression of simian immunodeficiency virus (SIV) infection entails expansion of the gut virome, including co-infection with potentially pathogenic agents such as adenovirus (Handley et al., 2016; Handley et al., 2012). A study of wild gorillas (Gorilla gorilla) reported that the gut virome differed between individuals with and without SIV infection; SIV-infected individuals not only shed greater quantities of viruses in the families Herpesviridae and Reoviridae but were the only individuals in the sample to shed viruses in the family Adenoviridae (D’arc et al., 2018).
In the present study, we used metagenomics to characterize gastrointestinal viromes (i.e., viral communities characterized in fecal samples) of wild eastern chimpanzees (P. t. schweinfurthii) from the Kanyawara and Ngogo communities of Kibale National Park, Uganda. The communities are separated by approximately 10 km within an estimated population of 1500-2000 individuals (Plumptre & Cox, 2006). For the last 27 to 34 years, both communities have grown in size and exhibit high individual survival, with life expectancy higher at Ngogo than at Kanyawara (Muller & Wrangham, 2014; Wood et al., 2017). Using methods previously applied to this chimpanzee population (Negrey et al., 2020), we investigated links between gastrointestinal viruses and general health. Our goal was to identify which viruses, if any, were associated with observed episodes of ill health in this population and may act as biomarkers of illness. We sampled individuals both before and during episodes of illness to assess within-individual changes in viral infection associated with ill health.
Materials and methods
Ethical approval
The noninvasive procedures used in this study were approved by the Uganda Wildlife Authority, the Uganda National Council for Science and Technology, and by the Institutional Animal Care and Use Committees (IACUCs) of Harvard University (protocol number 96-03), Tufts University (protocol number M2019-83), and the University of New Mexico (protocol number 18-200739-MC). The study was formally exempt from review by the IACUCs of the University of Michigan and Boston University. All procedures complied with the American Society of Primatologists Ethical Principles for the Treatment of Non-Human Primates.
Study sites, subjects, and sample collection
We conducted health monitoring and collected corresponding fecal samples in the Kanyawara and Ngogo chimpanzee communities of Kibale National Park, Uganda, from July 2015 to May 2019. On 1 January 2016, Kanyawara and Ngogo contained 49 and 194 individual chimpanzees, respectively. Research personnel collected daily health observations from all individually identified chimpanzees in the study communities. Observers noted the presence or absence of clinical signs and provided detailed descriptions when signs were severe. We converted these data into a categorical health scoring system: (1) no signs, (2) coughing or sneezing, (3) diarrhea or otherwise abnormal feces, (4) an unhealed wound, (5) skin abnormality or swelling, and (6) mobility deficits (e.g. reduced mobility / limping). Whenever a chimpanzee experienced one or more signs (categories 2-6), that individual was classified as ill.
As previously described (Negrey et al., 2020), we collected fecal samples on a quarterly basis and homogenized them at a 1:1 ratio with RNAlater buffer (Thermo Fisher Scientific, Waltham, MA, USA). We stored samples in an ice-filled thermos until returning to camp, at which point we transferred samples to freezers at −20 °C. We then transported samples on ice to the USA. To examine changes in viral infection corresponding to periods of ill health, our analyses only included chimpanzees that were classified as ill during the study period. For each episode of illness, we selected one pair of samples from the individual in the quarter prior to the illness event, and a second sample from the individual during the quarter in which they were ill. In three cases, fecal samples were not collected in the quarters immediately preceding observed illness, so we therefore used samples collected two or three quarters prior to observed illness (Table S1).
Viral identification and sequencing
We analyzed 40 samples from 19 individuals ranging in age from 2 to an estimated 54 years at the time of illness (Kanyawara Males = 4; Kanyawara Females = 3; Ngogo Males = 9; Ngogo Females = 3), collected during quarters when they exhibited clinical signs of illness outside of disease outbreaks (Table S1). We sampled one individual male at Ngogo for two different episodes of illness that occurred 2.1 years apart. Following previously described protocols (Goldberg et al., 2017; Goldberg et al., 2018; Goldberg et al., 2019; Negrey et al., 2020; Sibley et al., 2016; Toohey-Kurth et al., 2017), we used metagenomic methods to identify viruses in chimpanzee feces and to estimate their loads. In brief, we added 200 ul of feces (preserved in RNAlater) to 800 ul of Hanks’ Balanced Salt Solution and then homogenized the solution by bead beating. We treated the homogenate with nucleases to reduce unencapsidated nucleic acids (Allander et al., 2001) and performed subsequent extractions with the Qiagen QIamp MinElute Virus Spin Kit (Qiagen, Hilden, Germany). We synthesized cDNA from the extracted RNA, purified cDNA using Agencourt AmpureXP beads (Beckman Coulter, Brea, CA, USA), and prepared libraries using the Illumina Nextera XT kit (Illumina, San Diego, CA, USA). We sequenced libraries on an Illumina MiSeq instrument using 2x150 cycle paired-end chemistry. As previously described (Bennett et al., 2020; Toohey-Kurth et al., 2017), we mitigated “index hopping” or “bleed over” between samples by performing additional library cleanups to remove free adapters, storing libraries at −20°C, and sequencing libraries on a MiSeq instrument, which minimizes index hopping by using a nonpatterned flow cell and bridge amplification clustering (Bentley et al., 2008). We also ran blank samples and examined all data post-sequencing for evidence of cross-contamination with unique sequences.
To complement the methods above, we also analyzed samples by multiplex RT-PCR (NxTAG Respiratory Pathogen Panel, Luminex, Austin, TX, USA). The panel detects 20 human respiratory pathogens, including several, such as human respiratory syncytial virus, human metapneumovirus, and rhinovirus, that have caused respiratory disease in wild apes (Grützmacher et al., 2016; Negrey et al., 2019; Scully et al., 2018). Our prior studies of respiratory disease in these chimpanzee communities benefitted from this highly sensitive and specific assay (Negrey et al., 2019; Scully et al., 2018); see Scully et al. (2018) for further description of this method.
Bioinformatics
To identify and reconstruct viral sequences, we used CLC Genomics Workbench (CLC bio, Aarhus, Denmark). We trimmed short and low-quality sequences, removed reads that mapped to chimpanzee DNA and known contaminants, and assembled the remaining reads de novo. We used BLAST algorithms to compare the resulting contiguous sequences (contigs) to viral sequences in the GenBank sequence database at both the nucleotide and amino acid level (Altschul et al., 1990; Gish & States, 1993). For the purposes of this analysis, we retained only contigs that matched viruses with mammalian hosts or were primarily associated with mammals, under the assumption that contigs matching viruses of plants, insects and similar taxa were associated with dietary items. For all downstream analyses, we used replicase genes when possible; exceptions included viruses with genomes consisting of single open reading frames and those for which only the capsid-encoding gene was available. Because we used comparable genomic segments for each virus, we avoided classifying separate segments of the same genome as different viruses.
Following common methods for metagenomics analyses of viruses from fecal and sewage samples (Hjelmsø et al., 2017; Siqueira et al., 2018; Tisza & Buck, 2021), we measured viral loads by mapping reads from each sample to viral contigs and calculating the proportion of total reads that mapped to each identified virus, from which we also calculated the total load of all viruses detected in the sample. We subsequently normalized this proportion to 1 million reads and adjusted for target sequence length (kilobases). The resulting measure of viral load, viral reads per million per kilobase of target (vRPM/kb), has been validated by real-time quantitative polymerase chain reaction (Huang et al., 2019; Toohey-Kurth et al., 2017) and has proven useful for quantifying viral loads in wild chimpanzees (Negrey et al., 2020).
To infer viral phylogenetic relationships, we first used TranslatorX (Abascal et al., 2010) to align newly generated viral sequences with those of related viruses retrieved from GenBank, removing poorly aligned regions via the Gblocks algorithm (Castresana, 2000). We then used PhyML v3.0 (Lefort et al., 2017) to generate maximum likelihood phylogenetic trees with 1,000 bootstrap replicates and used FigTree v1.4.4 (Rambaut, 2018) to display the resulting trees.
Inferential statistics
We used the modified Wald method (Agresti & Coull, 1998) to calculate viral prevalence by health status, sex, and study community. We then conducted linear mixed model (LMM) analyses of the association of clinical status and covariates with viral richness (the number of distinct viruses per sample) and total viral load (vRPM/kb for all viruses) in R v4.0.3 (R Core Team, 2021). We used the “lmer” function in package lmerTest v3.1.3 (Kuznetsova et al., 2017) as modified from lme4 (Bates et al., 2015) to run LMMs with Gaussian error structures and fitted with restricted maximum likelihood. We ran two LMMs in which the response variables were viral richness and total viral load, respectively. The fixed effects included the individual’s health status, study community, sex, and age at time of sample collection. Chimpanzee age was determined as previously described (Negrey et al., 2020) and was included as a continuous variable. We also included interactions among health status with age, sex, and study community, as well as interaction between age with sex, based on prior findings from this population (Negrey et al., 2020). To mitigate type 1 and type 2 error, we removed interaction terms for which the p value was greater than or equal to alpha (0.05). Finally, we included subject ID as a random variable to control for multiple sampling of individuals. We calculated p values using the Satterthwaite method (Luke, 2017). After detecting that model residuals deviated moderately from normality (see Supporting Information), we ran both models again as robust linear mixed models (RLMMs) using the “rlmer” function in package robustlmm (Koller, 2016). We provide results for both the original LMMs and the accompanying RLMMs.
To determine which individual viruses contributed to patterns identified in the LMMs, we applied random forest classification using the “randomForest” function in R package randomForest (Liaw & Wiener, 2002). The random forest algorithm is a machine learning tool that generates a large number (i.e., from dozens to thousands) of decision trees via random, independent sampling of a dataset, and that can be used to predict or classify a given set of values (Breiman, 2001). Each decision tree is trained on a subset of the original dataset. We generated a series of random forests in which we analyzed response variables that were significant by mixed modeling (e.g., total viral load, community, age). For all random forests, predictor variables were viral loads (vRPM/kb) for each individual virus. Importance values of each virus for predicting the given response variable were calculated as the mean decrease in node impurity. As node impurity decreases, the forest becomes more effective at predicting and/or classifying samples (Segal & Xiao, 2011). For continuous and categorical response variables, node impurity was calculated using the residual sum of squares and Gini index, respectively (Liaw & Wiener, 2002).
R script and data are available on Figshare (Negrey et al., 2021).
Results
Virus detection
We selected and analyzed 20 episodes of ill health in 19 chimpanzees, all but one of whom were sexually mature (Table S1). Clinical signs included coughing and/or sneezing (19 episodes) and diarrhea (two episodes). One individual exhibited respiratory signs and diarrhea concurrently. No cases of skin abnormalities, wounding, or limping were included in our analyses. Episodes of ill health were acute: None of the observed episodes resulted in mortality, and at other times, each chimpanzee was apparently healthy.
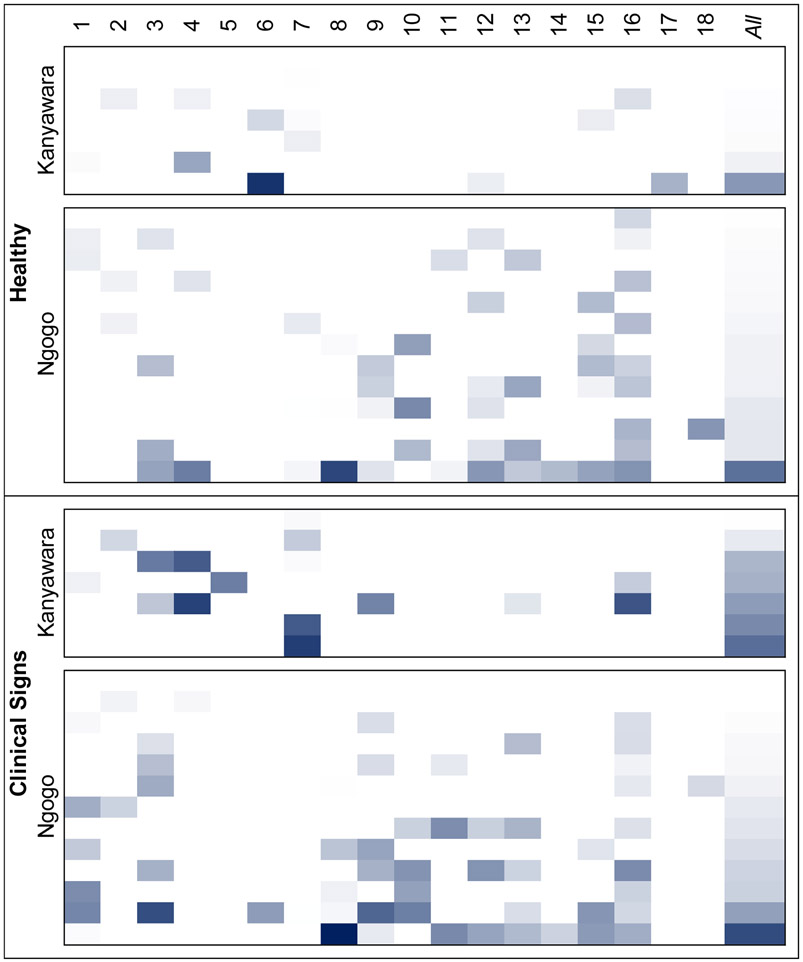
In fecal samples collected before and proximate to episodes of ill health, we found 18 viruses which are either thought to infect mammals or are primarily associated with mammals (Table 1), including 9 viruses previously identified in fecal samples from Kibale chimpanzees. Contigs derived from metagenomic analyses mostly corresponded to replication associated genes; only contigs for bufavirus, chisavirus, and salivirus represented most of the genome. Similarity of amino acid sequences to known viruses ranged from 46.3% to 99.8%. We did not detect any viruses in 2 of the 40 fecal samples. Viral prevalence varied considerably (Table S2), ranging from 2.5% for unclassified chimpanzee ssDNA virus 1 (95% CI: 0.0%, 14.0%) to 52.5% for eastern chimpanzee associated porprismacovirus 9 (95% CI: 37.5%, 67.1%). Eleven of the 18 viruses were present at both Ngogo and Kanyawara. Chimpanzee stool-associated RNA virus (chisavirus), three porprismacoviruses, and an unclassified single-stranded DNA (ssDNA) virus were detected only in Ngogo samples, whereas an enterovirus and a second unclassified ssDNA virus were found only in samples from Kanyawara. We found no evidence of “index hopping: or “bleed over” in read data from blanks or chimpanzee fecal samples.
Table 1.
Viruses detected in 40 fecal samples from wild chimpanzees in the Kanyawara and Ngogo communities of Kibale National Park, Uganda.
| Virus | Closest match (accession number1) | Family | Host (Country, Year) | Genome | Length (nt)2 | % Identity3 | Accession number4 | |
|---|---|---|---|---|---|---|---|---|
| 1 | chimpanzee adenovirus | chimpanzee adenovirus Y25 (YP_006272954) | Adenoviridae | Chimpanzee (−, 1969) | dsDNA | 2403 | 99.75 | MW876510 |
| 2 | chimpanzee bufavirus | human bufavirus (AOR39545) | Parvoviridae | Human (Tunisia, 2013) | ssDNA | 1500 | 81.40 | MT076200 |
| 3 | chimpanzee circovirus 1 | dromedary stool-associated circular ssDNA virus (AIY31253) | Circoviridae | Camel (United Arab Emirates, 2013) | ssDNA | 1134 | 60.65 | MW876514 |
| 4 | chimpanzee circovirus 2 | circovirus sp. (QBA83760) | Circoviridae | Pig (China, 2017) | ssDNA | 1179 | 46.31 | MW876515 |
| 5 | chimpanzee enterovirus | enterovirus B112 (AJA74399) | Picornaviridae | Chimpanzee (Gabon, 2009) | ssRNA | 6150 | 92.78 | MW876511 |
| 6 | chimpanzee picobirna-like virus | kumba picobirna-like virus (QAA77647) | Unclassified Picornavirales | Human (Cameroon, 2014) | RNA | 1188 | 93.43 | MT076202 |
| 7 | chimpanzee salivirus | salivirus FHB (YP_009067077) | Picornaviridae | Human (China, 2011) | ssRNA | 7125 | 98.27 | MW876512 |
| 8 | chisavirus (chimpanzee stool-associated RNA virus) | husavirus (AWU65954) | Unclassified Picornavirales | Human (Venezuela, 2015) | ssRNA | 8379 | 62.95 | MW876513 |
| 9 | eastern chimpanzee associated porprismacovirus 1 | macaca mulatta feces associated virus 4 (APG55823) | Smacoviridae | Rhesus macaque (USA, 2014) | ssDNA | 777 | 64.00 | MT076205 |
| 10 | eastern chimpanzee associated porprismacovirus 2 | porcine associated porprismacovirus (QBP37091) | Smacoviridae | Pig (Vietnam, 2013) | ssDNA | 735 | 71.97 | MT076206 |
| 11 | eastern chimpanzee associated porprismacovirus 3 | porcine associated porprismacovirus 8 (YP_009054991) | Smacoviridae | Pig (USA, 2011) | ssDNA | 786 | 55.91 | MT076207 |
| 12 | eastern chimpanzee associated porprismacovirus 4 | chimpanzee stool associated circular ssDNA virus (ADB24816) | Smacoviridae | Chimpanzee (Tanzania, 2004) | ssDNA | 816 | 99.26 | MT076208 |
| 13 | eastern chimpanzee associated porprismacovirus 6 | Macaca mulatta feces associated virus 7 (APG55812) | Smacoviridae | Rhesus macaque (USA, 2014) | ssDNA | 780 | 68.34 | MT076210 |
| 14 | eastern chimpanzee associated porprismacovirus 7 | chimpanzee associated porprismacovirus 2 (YP_009508863) | Smacoviridae | Chimpanzee (Tanzania, 2004) | ssDNA | 586 | 92.27 | MW876516 |
| 15 | eastern chimpanzee associated porprismacovirus 8 | chicken smacovirus mg4_964 (QIR82267) | Smacoviridae | Chicken (USA, 2017) | ssDNA | 786 | 77.27 | MW876517 |
| 16 | eastern chimpanzee associated porprismacovirus 9 | chicken smacovirus mg4_964 (QIR82267) | Smacoviridae | Chicken (USA, 2017) | ssDNA | 771 | 68.98 | MW876518 |
| 17 | unclassified chimpanzee ssDNA virus 1 | unidentified circular ssDNA virus (APG55818) | Unclassified | Macaque (USA, 2014) | ssDNA | 639 | 96.70 | MW876519 |
| 18 | unclassified chimpanzee ssDNA virus 2 | uncultured virus (AUM61717) | Unclassified | Wastewater (USA, 2015) | ssDNA | 1020 | 58.31 | MW876520 |
Accession number of closest match in GenBank
Length of the nucleotide sequence used to analyze phylogenetic relationships and viral load
% amino acid similarity of the new virus to its closest match in GenBank
GenBank accession number of viral sequence from this study
Targeted multiplex RT-PCR assays identified no viruses other than those identified using metagenomics. This assay identified adenovirus in 19 samples from 13 chimpanzees, compared to 10 samples identified using metagenomics. Concordance between the metagenomic and RT-PCR assay results identified 31 agreements (77.50%) and 9 disagreements (22.5%), yielding a Cohen’s kappa value of 0.538 ± SE 0.120 (95% CI: 0.303, 0.774).
Virus shedding and health status
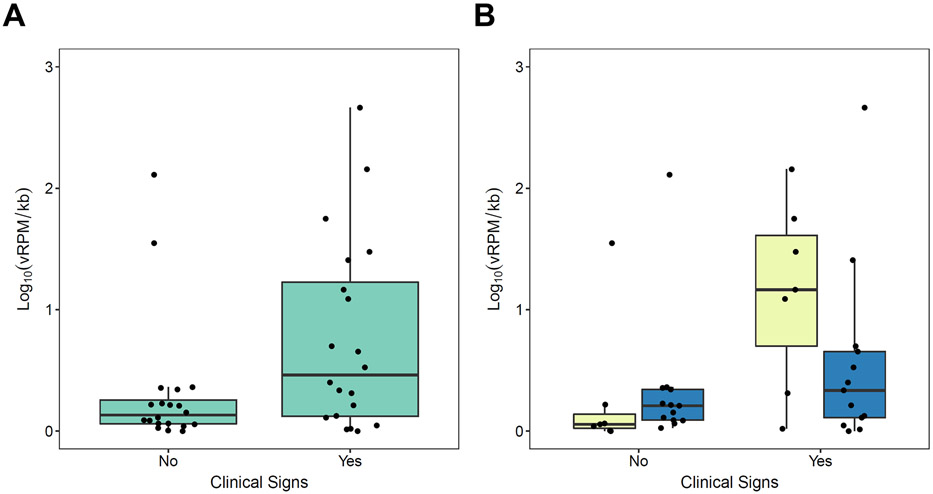
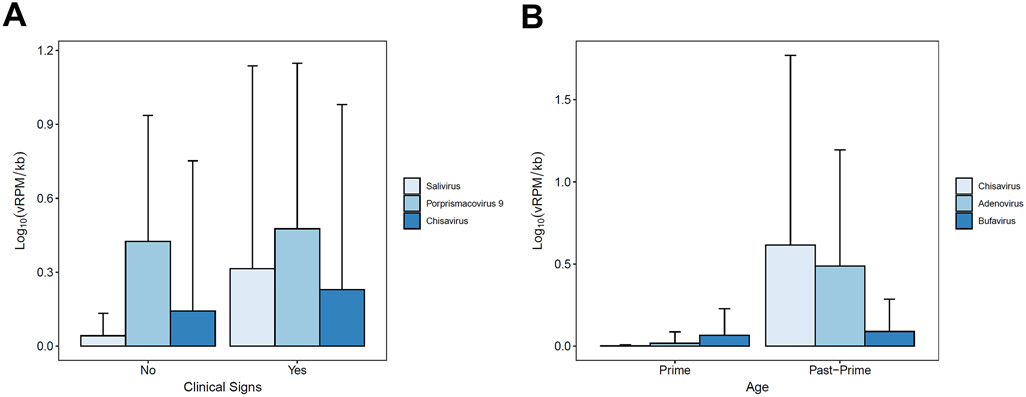
Virus prevalence varied by health status, sex, and study community (Table 2). Mixed models of viral richness showed no statistically significant variation by health status, age, sex, or community (Table S3). However, mixed models of viral load revealed substantial variation by these same predictors (Table 2; Fig. 1). Notably, overall viral loads were 2.4 times higher in chimpanzees exhibiting clinical signs than in healthy chimpanzees (Table 2; Fig. 2a). Healthy and ill chimpanzees exhibited mean total viral loads of 0.314 (SD ± 0.538) and 0.758 (SD ± 0.783) Log10(vRPM/kb), respectively. Random forest classification identified three viruses as most important for predicting total viral load in ill chimpanzees regardless of subject age, sex, or community, and these were (in descending order of importance): salivirus, porprismacovirus 9, and chisavirus (Table S4; Fig. 3a). Post-hoc modeling indicated the associations of salivirus and porprismacovirus 9 with total viral load were not driven by differences in prevalence: ill health did not predict overall prevalence of salivirus (β=−0.077, SE=0.994, p=0.938; Table S5a) or porprismacovirus 9 (β=0.272, SE=0.733, p=0.710; Table S5b). However, the overall prevalence of chisavirus was positively associated with ill health (β=27.4, SE=11.4 p=0.002; Table S5c).
Table 2.
Results of (a) a linear mixed model and (b) a robust linear mixed model assessing variation in total viral load. The marginal and conditional R2 for the linear mixed model were 0.37 and 0.69, respectively. Bold font denotes predictors for which p < 0.05.
| (a) Linear Mixed Model | ||||||
|---|---|---|---|---|---|---|
| Predictor | β | SE | 95% CI | DF | t | p |
| Intercept | 0.405 | 0.283 | ||||
| Health Status 1 | 0.857 | 0.224 | [0.406, 1.327] | 19.521 | 3.821 | 0.001 |
| Age | −0.295 | 0.248 | [−0.793, 0.192] | 14.856 | −1.191 | 0.252 |
| Sex2 | −0.024 | 0.265 | [−515,0.475] | 14.698 | −0.090 | 0.930 |
| Community3 | −0.052 | 0.345 | [−0.773,0.621] | 20.313 | -0.151 | 0.882 |
| Age x Sex | 0.681 | 0.263 | [0.161, 1.214] | 14.699 | 2.591 | 0.021 |
| Health Status x Community | −0.643 | 0.278 | [−1.232, −0.086] | 19.513 | −2.310 | 0.032 |
| (b) Robust Linear Mixed Model | ||||||
| Predictor | β | SE | - | - | t | p |
| Intercept | 0.359 | 0.178 | - | - | ||
| Health Status | 0.803 | 0.214 | - | - | 3.756 | 0.001 |
| Age | −0.327 | 0.136 | - | - | −2.404 | 0.030 |
| Sex | −0.132 | 0.144 | - | - | −0.916 | 0.374 |
| Community | 0.002 | 0.218 | - | - | 0.010 | 0.992 |
| Age x Sex | 0.539 | 0.143 | - | - | 3.769 | 0.002 |
| Health Status x Community | −0.619 | 0.265 | - | - | −2.335 | 0.030 |
The reference category is “healthy”.
The reference category is “female”.
The reference category is “Kanyawara”.
Figure 1.
Heatmap of viral loads for all 19 chimpanzees, grouped by community and health status. Values range from 0 (lightest) to 3.3 Log10(vRPM/kb) (darkest). Viruses are numbered as per Table 1.
Figure 2.
Chimpanzee gastrointestinal total viral load as a function of clinical signs (a) in all individuals and (b) by study community. In 2b, light and dark boxes represent Kanyawara and Ngogo, respectively. Thick horizontal bars represent medians, and the upper and lower bounds of each box represent the 75th and 25th percentiles, respectively.
Figure 3.
Mean viral loads for (a) all chimpanzees by health status and (b) male chimpanzees by age. “Prime” and “past-prime” refer to individuals aged <30 years and ≥30 years, respectively. Vertical bars indicate standard deviations.
The interaction of viral load and community was significant (Table 2; Fig. 2b), with higher total viral load in ill chimpanzees in Kanyawara than in Ngogo. Ill chimpanzees at Kanyawara and Ngogo exhibited mean total viral loads of 1.138 and 0.554 Log10(vRPM/kb), respectively. To determine if this relationship was affected by the time between observed illness and sample collection, we ran a robust multiple linear model (“rlm” function in R package “MASS”) on samples collected during quarters with observed clinical signs, with study community and days from health observation to sample collection as the two predictor variables. The relationship between community and total viral load remained significant (β=−0.937, SE=0.321, F=8.42, p=0.010); however, there was no relationship between the number of days between sample collection and total viral load (β=−0.187, SE=0.157, F=1.53, p=0.232). Random forest classification indicated that salivirus most strongly differentiated between ill chimpanzees at Kanyawara and Ngogo (Table S4). Post-hoc generalized mixed models controlling for subject sex indicated that, among ill chimpanzees, salivirus was more prevalent at Kanyawara than Ngogo (β=−0.705, SE=0.159, p<0.001; Table S5d), and that among Kanyawara chimpanzees, salivirus was more prevalent among ill individuals than healthy ones (β=15.2, SE=8.39, p=0.011; Table S5e). Similarly, chisavirus was only found in males at Ngogo (Table S2) and was more prevalent among ill males than healthy males (β=27.4, SE=11.2, p=0.002; Table S5f).
The age by sex interaction was also significant (Table 2), indicating that males, but not females, exhibited higher total viral loads as they aged. Random forest classification indicated that chisavirus, adenovirus, and bufavirus were most important for differentiating males by age (Table S4; Fig. 3b). Post-hoc generalized linear modeling indicated that male age was positively related to the prevalence of chisavirus (β=3.95, SE=2.85, p=0.007; Table S5g) and adenovirus (β=1.24, SE=0.565, p=0.012; Table S5h). Conversely, bufavirus prevalence decreased with age, but not significantly (β=−0.130, SE=0.470, p=0.781; Table S5i).
Discussion
We observed changes in the fecal viromes of clinically ill chimpanzees compared to when they were previously healthy. Specifically, we observed a 2.4-fold increase in total viral load when chimpanzees became ill. Of the viruses identified, salivirus was the strongest contributor to this trend. Salivirus was first described in humans suffering from gastroenteritis (Li et al., 2009) and has since been detected in sewage and water samples around the world (Adineh et al., 2019; Badru et al., 2018). Although members of family Picornaviridae cause a wide variety of illnesses in humans (Tuthill et al., 2010), the relationship between salivirus and gastroenteritis has been inconsistent, with some studies reporting a causal relationship (Itta et al., 2016; Shan et al., 2010) and others not (Aldabbagh et al., 2015; Yu et al., 2015). Interestingly, salivirus was also detected in a postmortem nasopharyngeal swab from a child suffering from a severe adenovirus co-infection (Pei et al., 2016), suggesting that salivirus infection may not be limited to the gastrointestinal tract. Moreover, in a previous study of this chimpanzee population, we identified salivirus in a higher proportion of aged males than in younger males (Negrey et al., 2020). In Kibale chimpanzees, salivirus may therefore be a cause of ill health, a biomarker of diminished immune function, or both.
Porprismacovirus 9 was also a strong contributor to total viral load, although prevalence and load did not differ greatly between healthy and ill chimpanzees. Porprismacoviruses (family: Smacoviridae) are small, circular, single-stranded DNA viruses found in the feces of primates and pigs (Varsani & Krupovic, 2018). Relatively little is known about porprimsmacoviruses, and any relationships between smacoviruses and vertebrate disease remain unclear. For instance, smacoviruses were detected in samples from a relatively large proportion of unsolved outbreaks of diarrhea, including 28% of outbreaks in the US and 67% in France (Ng et al., 2015), and from as many as 19% of diarrheal samples from Peruvian children (Phan et al., 2016). However, smacoviruses have also been routinely detected in the absence of illness or immune suppression in a number of vertebrates (Varsani & Krupovic, 2018). Recent evidence even suggests that archaea, rather than animals, act as hosts for smacoviruses (Díez-Villaseñor & Rodriguez-Valera, 2019). Given their ubiquity, uncertainty surrounding their life cycles, and comparable shedding by healthy and ill chimpanzees, porprismacoviruses are unlikely to cause ill health in chimpanzees. However, we cannot discount the possibility that porprismacoviruses have long-term and subtle clinical consequences.
Chimpanzee stool-associated RNA virus (chisavirus) was the third most important component of total viral load in ill chimpanzees, and its prevalence was predictive of ill health among Ngogo chimpanzees. Chisavirus is closely related to a cluster of unclassified members of Picornavirales infecting a taxonomically broad range of hosts; these viruses include husaviruses in humans, posaviruses in pigs, and fisaviruses in fish (Aoki et al., 2019; Oude Munnink et al., 2017). Often observed at low prevalence, husavirus has not displayed any consistent relationship to ill health in humans (Mohammad et al., 2020; Oude Munnink et al., 2017). However, genomic (Shan et al., 2011) and epidemiological (Siqueira et al., 2018) evidence suggests that husaviruses and their relatives could be shed by gastrointestinal nematodes. Chisavirus presence and load may therefore indirectly reflect burdens of nematode infection.
We also detected chimpanzee adenoviruses at high prevalence (14.3 and 35.0% in healthy and ill individuals, respectively). A prior study suggested that adenovirus was associated with a respiratory disease outbreak in chimpanzees in the Mahale Mountains, Tanzania (Tong et al., 2010). However, other studies indicate that adenoviruses are widespread and largely apathogenic in nonhuman primates (Hoppe et al., 2015; Negrey et al., 2019; Scully et al., 2018; Seimon et al., 2015; Wevers et al., 2011). In our study, adenovirus prevalence and load were not associated with ill health. However, adenovirus prevalence and load were associated with increasing age in male chimpanzees. These findings mirror a pattern we previously observed for salivirus, chisavirus, and a porprismacovirus, and suggest an association with immunosenescence (Negrey et al., 2020). In humans, some adenoviruses cause disease in elderly individuals (Kandel et al., 2015; Loeb, 2019), although less commonly than other frank pathogens such as rhinoviruses and influenza viruses (Chasqueira et al., 2018).
Notably, ill chimpanzees at Kanyawara exhibited greater total viral loads than did those at Ngogo. This accords with a recent study of female chimpanzees in the same population showing that Kanyawara chimpanzees shed more gastrointestinal parasites than those at Ngogo (Phillips et al., 2020). The Ngogo and Kanyawara territories exhibit differences in resource quality that may drive health differences. Notably, dietary quality impacts immune function (Scrimshaw & SanGiovanni, 1997), such that diminished or compromised nutrition may reduce resistance to viral infection or shedding. A 2009 study indicated that Ngogo chimpanzees exhibit higher noninvasive measures of energy balance than do Kanyawara chimpanzees (Emery Thompson et al., 2009), reflecting the greater availability of ripe fruit at Ngogo (Potts et al., 2011). Differences in energetic status may therefore manifest as differences in physiological function and tolerance to viruses at Ngogo than at Kanyawara through such processes as immune responsiveness. Community-level differences may also arise from other factors, such as ecological or behavioral differences in exposure to viruses. For example, the Kanyawara territory abuts the park boundary but the Ngogo territory does not, such that the Kanyawara chimpanzees interact more frequently with anthropogenically altered environments than do the Ngogo chimpanzees (Mackenzie et al., 2011). Differences in exposure to humans and their environments may also affect exposure to viruses of humans and domestic animals. For instance, the prevalence of salivirus was highest in samples from ill chimpanzees at Kanyawara, where excursions into surrounding villages occur more frequently than at Ngogo. Resolving the transmission dynamics of poorly known viruses such as salivirus would require more detailed epidemiological studies and accurate estimates of viral evolutionary rates. Regardless of sources and origins, however, viral systems may be particularly suited to examining host physiological/immunological function, including the influence of external factors such as ecology and sociality, given that viruses are obligate intracellular molecular parasites (Rivers, 1927).
In wild apes, certain anthroponoses are highly virulent and sometimes lethal (Grützmacher et al., 2016; Köndgen et al., 2008; Leendertz et al., 2006; Negrey et al., 2019; Palacios et al., 2011; Patrono et al., 2018; Scully et al., 2018). Our results show that wild chimpanzees also experience nonfatal infections of presumably low virulence, some of which increase in occurrence or load with ill health. In Kanyawara, respiratory disease was the most important cause of mortality between 1987 and 2017 (Emery Thompson et al., 2018). For example, a single epidemic of human rhinovirus C killed 8.9% of the community in 2013 (Scully et al., 2018). However, 53.2% of mortality during the same period remained unexplained (Emery Thompson et al., 2018). At Ngogo, respiratory disease caused by metapneumovirus killed 12.2% of the community in 2017 (Negrey et al., 2019). However, approximately 60% of deaths from 1995 to 2016 were from unknown causes (Wood et al., 2017). Infections of mild virulence during non-epidemic periods may account for some of the unexplained mortality in Kanyawara, Ngogo, and other wild chimpanzee communities, especially given that mortality rates are particularly high among younger and older individuals (Muller & Wrangham, 2014; Wood et al., 2017). If, as our data show, low-virulence viral infections are endemic and recurring within communities and affect individuals more as they senesce—especially males, as suggested in our present results and previously published work (Negrey et al., 2020)—such infections could cumulatively impact the chimpanzee aging process (i.e. “wear and tear”). Indeed, this idea has underlain key evolutionary theories of senescence. Even though the precise mechanisms whereby low-virulence infections cause cumulative health effects remain poorly understood (McHugh & Gil, 2018), aging seems to result from the accumulation of senescent cells (Childs et al., 2015). Cellular senescence, in turn, is caused by such factors as oxidative stress (Liguori et al., 2018) and telomere shortening (Oeseburg et al., 2010). Viral infections are known to contribute to such processes (Beck et al., 2000; Camini et al., 2017; Dowd et al., 2017; Gong et al., 2001; van de Berg et al., 2010).
We acknowledge that our inferences are based on small sample sizes. This limitation is inherent to studies of long-lived primates such as chimpanzees. We also note that the metagenomic methods used, although shown to be as sensitive as quantitative RT-PCR for detecting viruses in serum (Toohey-Kurth et al., 2017), may behave differently for other sample types such as feces. Such differences may account for lower detection rates of adenovirus using metagenomic methods than using RT-PCR. Because of these limitations, however, we suspect that including more chimpanzee communities over longer time periods (ideally, over the entire life course) and employing specific PCR assays for viruses of interest would likely increase the strength of the trends we have documented, and perhaps bring other trends to light.
We emphasize that the viruses associated with bouts of ill health we have identified here may not be causes of disease themselves but rather reflective of declines in health caused by other factors. Such patterns have been observed in humans. For example, anelloviruses, which are generally benign in humans (Kaczorowska & van der Hoek, 2020), are associated with compromised immune function and ill health. Anelloviruses flourish in transplant recipients (Young et al., 2015), AIDS patients (Shibayama et al., 2001), and children with acute encephalitis or meningoencephalitis (Eibach et al., 2019). We also note that, despite strong trends with viral load, we did not detect associations between viral richness and ill health or any demographic variables. This finding reinforces the notion that viruses within populations respond differently (and perhaps independently) to host-related factors (e.g., Kapusinszky et al. (2017)).
Conclusions
In summary, we observed that fecal viromes of wild chimpanzees change predictably as chimpanzees experience episodes of ill health. We found that viral loads increase during episodes of ill health and that the magnitude of this change varied by study community, suggesting ecological effects on host responses to viral exposure and infection. Furthermore, given that the best predictors of ill health were viruses not previously linked to disease outcomes (e.g., salivirus), our results emphasize the importance of broad-spectrum diagnostics for identifying causes or biomarkers of ill health. Identification and investigation of such little-known viruses may prove valuable, with implications for both the study of virology and health monitoring of free-ranging animals.
Supplementary Material
Research highlights.
Some viruses of wild primates are relatively benign but may be useful as biomarkers of physiology and health.
We used metagenomic methods to characterize viral communities in feces of wild chimpanzees in two long-term study communities (Kanyawara and Ngogo) in Kibale National Park, Uganda.
Chimpanzees exhibited higher viral loads (normalized abundances of viral genetic sequences) when they were ill than when those same individuals were previously healthy.
When ill, Kanyawara chimpanzees exhibited higher viral loads than Ngogo chimpanzees, and males, but not females, exhibited higher viral loads as they aged.
Acknowledgements
We are grateful to the Uganda Wildlife Authority, Uganda National Council for Science and Technology, and Makerere University Biological Field Station for granting research approval. We thank the field assistants of the Kibale and Ngogo Chimpanzee Projects for help with sample collection. For additional advice and technical assistance, we thank Samuel Angedakin, Christopher Dunn, and Taylor Weary.
Funding
This work was supported by National Institutes of Health Award R01AG049395 through the National Institute for Aging and the Office of Research on Women’s Health. Further support was provided by the National Science Foundation [Awards BCS-1355014, BCS-1613392, BCS-1613393, DGE-1256260, and NCS-FO 1926653], National Geographic Society [Awards 9742-15 and 9824-15], Leakey Foundation, Wenner-Gren Foundation, Nacey-Maggioncalda Foundation, Boston University, University of Michigan, University of New Mexico, and University of Wisconsin-Madison.
Footnotes
Conflicts of interest
The authors declare they have no conflicts of interest.
Data availability:
Viral nucleotide sequences are available on GenBank under accession numbers MT076199 to MT076210 and MW876510 to MW876520. Additional data and R code are available on Figshare (https://figshare.com/s/d7b44bc82c4086cf3824).
References
- Abascal F, Zardoya R, & Telford MJ (2010). TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic acids research, 38(Web Server Issue), W7–W13. 10.1093/nar/gkq291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adineh M, Ghaderi M, & Mousavi-Nasab SD (2019). Occurrence of salivirus in sewage and river water samples in Karaj, Iran. Food and Environmental Virology, 11(2), 193–197. 10.1007/s12560-019-09377-1 [DOI] [PubMed] [Google Scholar]
- Agresti A, & Coull BA (1998). Approximate is better than “exact” for interval estimation of binomial proportions. The American Statistician, 52(2), 119–126. 10.1080/00031305.1998.10480550 [DOI] [Google Scholar]
- Aldabbagh S, Eckerle I, Müller A, Delwart EL, & Eis-Hübinger AM (2015). Salivirus type 1 and type 2 in patients with acute gastroenteritis, Germany. Journal of Clinical Virology, 72, 16–19. 10.1016/j.jcv.2015.08.013 [DOI] [PubMed] [Google Scholar]
- Alizon S, & Michalakis Y (2015). Adaptive virulence evolution: the good old fitness-based approach. Trends in Ecology and Evolution, 30(5), 248–254. 10.1016/j.tree.2015.02.009 [DOI] [PubMed] [Google Scholar]
- Allander T, Emerson SU, Engle RE, Purcell RH, & Bukh J (2001). A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proceedings of the National Academy of Sciences USA, 98(20), 11609–11614. 10.1073/pnas.211424698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, & Lipman DJ (1990). Basic local alignment search tool. Journal of Molecular Biology, 215(3), 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Anderson RM, & May RM (1979). Population biology of infectious diseases: Part I. Nature, 280(5721), 361–367. 10.1038/280361a0 [DOI] [PubMed] [Google Scholar]
- Aoki H, Sunaga F, Ochiai H, Masuda T, Ito M, Akagami M, Naoi Y, Sano K, Katayama Y, Omatsu T, Oba M, Sakaguchi S, Furuya T, Ouchi Y, Shirai J, Mizutani T, Oka T, & Nagai M (2019). Phylogenetic analysis of novel posaviruses detected in feces of Japanese pigs with posaviruses and posa-like viruses of vertebrates and invertebrates. Archives of Virology, 164(8), 2147–2151. 10.1007/s00705-019-04289-8 [DOI] [PubMed] [Google Scholar]
- Badru S, Khamrin P, Kumthip K, Yodmeeklin A, Surajinda S, Supadej K, Sirilert S, Malasao R, Okitsu S, Ushijima H, & Maneekarn N (2018). Molecular detection and genetic characterization of Salivirus in environmental water in Thailand. Infection, Genetics and Evolution, 65, 352–356. 10.1016/j.meegid.2018.08.023 [DOI] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, & Walker S (2015). Fitting linear mixed-effects models using lme4. Journal of Statistical Software, 67(1), 48. 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Beck MA, Handy J, & Levander OA (2000). The role of oxidative stress in viral infections. Annals of the New York Academy of Sciences, 917(1), 906–912. 10.1111/j.1749-6632.2000.tb05456.x [DOI] [PubMed] [Google Scholar]
- Bennett AJ, Paskey AC, Ebinger A, Pfaff F, Priemer G, Höper D, Breithaupt A, Heuser E, Ulrich RG, Kuhn JH, Bishop-Lilly KA, Beer M, & Goldberg TL (2020). Relatives of rubella virus in diverse mammals. Nature, 586(7829), 424–428. 10.1038/s41586-020-2812-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, … Smith AJ (2008). Accurate whole human genome sequencing using reversible terminator chemistry. Nature, 456(7218), 53–59. 10.1038/nature07517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L (2001). Random forests. Machine Learning, 45(1), 5–32. 10.1023/A:1010933404324 [DOI] [Google Scholar]
- Camini FC, da Silva Caetano CC, Almeida LT, & de Brito Magalhães CL (2017). Implications of oxidative stress on viral pathogenesis. Archives of Virology, 162(4), 907–917. 10.1007/s00705-016-3187-y [DOI] [PubMed] [Google Scholar]
- Castresana J (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution, 17(4), 540–552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Chasqueira M-J, Paixão P, Rodrigues M-L, Piedade C, Caires I, Palmeiro T, Botelho M-A, Santos M, Curran M, Guiomar R, Pechirra P, Costa I, Papoila A, Alves M, & Neuparth N (2018). Respiratory infections in elderly people: viral role in a resident population of elderly care centers in Lisbon, winter 2013–2014. International Journal of Infectious Diseases, 69, 1–7. 10.1016/j.ijid.2018.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs BG, Durik M, Baker DJ, & van Deursen JM (2015). Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat Med, 21(12), 1424–1435. 10.1038/nm.4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’arc M, Furtado C, Siqueira JD, Seuánez HN, Ayouba A, Peeters M, & Soares MA (2018). Assessment of the gorilla gut virome in association with natural simian immunodeficiency virus infection. Retrovirology, 15(1), 19. 10.1186/s12977-018-0402-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díez-Villaseñor C, & Rodriguez-Valera F (2019). CRISPR analysis suggests that small circular single-stranded DNA smacoviruses infect Archaea instead of humans. Nature Communications, 10(1), 294. 10.1038/s41467-018-08167-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd JB, Bosch JA, Steptoe A, Jayabalasingham B, Lin J, Yolken R, & Aiello AE (2017). Persistent herpesvirus infections and telomere attrition over 3 years in the Whitehall II cohort. Journal of Infectious Diseases, 216(5), 565–572. 10.1093/infdis/jix255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunay E, Apakupakul K, Leard S, Palmer JL, & Deem SL (2018). Pathogen transmission from humans to great apes is a growing threat to primate conservation. EcoHealth, 15, 148–162. 10.1007/s10393-017-1306-1 [DOI] [PubMed] [Google Scholar]
- Eibach D, Hogan B, Sarpong N, Winter D, Struck NS, Adu-Sarkodie Y, Owusu-Dabo E, Schmidt-Chanasit J, May J, & Cadar D (2019). Viral metagenomics revealed novel betatorquevirus species in pediatric inpatients with encephalitis/meningoencephalitis from Ghana. Scientific Reports, 9(1), 2360. 10.1038/s41598-019-38975-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery Thompson M, Machanda ZP, Scully EJ, Enigk DK, Otali E, Muller MN, Goldberg TL, Chapman CA, & Wrangham RW (2018). Risk factors for respiratory illness in a community of wild chimpanzees (Pan troglodytes schweinfurthii). Royal Society Open Science, 5(9), 180840. 10.1098/rsos.180840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery Thompson M, Muller MN, Wrangham RW, Lwanga JS, & Potts KB (2009). Urinary C-peptide tracks seasonal and individual variation in energy balance in wild chimpanzees. Hormones and Behavior, 55(2), 299–305. 10.1016/j.yhbeh.2008.11.005 [DOI] [PubMed] [Google Scholar]
- Formenty P, Boesch C, Wyers M, Steiner C, Donati F, Dind F, Walker F, & Le Guenno B (1999). Ebola virus outbreak among wild chimpanzees living in a rain forest of Côte d'Ivoire. Journal of Infectious Diseases, 179(1), S120–S126. 10.1086/514296 [DOI] [PubMed] [Google Scholar]
- Gish W, & States DJ (1993). Identification of protein coding regions by database similarity search. Nature Genetics, 3(3), 266–272. 10.1038/ng0393-266 [DOI] [PubMed] [Google Scholar]
- Glasser DB, Goldberg TL, Guma N, Balyesiima G, Agaba H, Gessa SJ, & Rothman JM (2021). Opportunities for respiratory disease transmission from people to chimpanzees at an East African tourism site. American Journal of Primatology, 83(2), e23228. 10.1002/ajp.23228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg TL, Bennett AJ, Kityo R, Kuhn JH, & Chapman CA (2017). Kanyawara virus: A novel rhabdovirus infecting newly discovered nycteribiid bat flies infesting previously unknown pteropodid bats in Uganda. Scientific Reports, 7(1), 5287–5287. 10.1038/s41598-017-05236-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg TL, Clyde VL, Gendron-Fitzpatrick A, Sibley SD, & Wallace R (2018). Severe neurologic disease and chick mortality in crested screamers (Chauna torquata) infected with a novel Gyrovirus. Virology, 520, 111–115. 10.1016/j.virol.2018.05.014 [DOI] [PubMed] [Google Scholar]
- Goldberg TL, Sibley SD, Pinkerton ME, Dunn CD, Long LJ, White LC, & Strom SM (2019). Multidecade mortality and a homolog of Hepatitis C virus in bald eagles (Haliaeetus leucocephalus), the national bird of the USA. Scientific Reports, 9(1), 14953. 10.1038/s41598-019-50580-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong G, Waris G, Tanveer R, & Siddiqui A (2001). Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-κB. Proceedings of the National Academy of Sciences of the United States of America, 98(17), 9599–9604. 10.1073/pnas.171311298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grützmacher KS, Köndgen S, Keil V, Todd A, Feistner A, Herbinger I, Petrzelkova K, Fuh T, Leendertz SA, Calvignac-Spencer S, & Leendertz FH (2016). Codetection of respiratory syncytial virus in habituated wild western lowland gorillas and humans during a respiratory disease outbreak. EcoHealth, 13(3), 499–510. 10.1007/s10393-016-1144-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley SA, Desai C, Zhao G, Droit L, Monaco CL, Schroeder AC, Nkolola JP, Norman ME, Miller AD, Wang D, Barouch DH, & Virgin HW (2016). SIV infection-mediated changes in gastrointestinal bacterial microbiome and virome are associated with immunodeficiency and prevented by vaccination. Cell Host and Microbe, 19(3), 323–335. 10.1016/j.chom.2016.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, Abbink P, Maxfield LF, Kambal A, Duan E, Stanley K, Kramer J, Macri SC, Permar SR, Schmitz JE, Mansfield K, Brenchley JM, Veazey RS, Stappenbeck TS, … Virgin HW (2012). Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell, 151(2), 253–266. 10.1016/j.cell.2012.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmsø MH, Hellmér M, Fernandez-Cassi X, Timoneda N, Lukjancenko O, Seidel M, Elsässer D, Aarestrup FM, Löfström C, Bofill-Mas S, Abril JF, Girones R, & Schultz AC (2017). Evaluation of methods for the concentration and extraction of viruses from sewage in the context of metagenomic sequencing. PLoS One, 12(1), e0170199. 10.1371/journal.pone.0170199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe E, Pauly M, Gillespie TR, Akoua-Koffi C, Hohmann G, Fruth B, Karhemere S, Madinda NF, Mugisha L, Muyembe J-J, Todd A, Petrzelkova KJ, Gray M, Robbins M, Bergl RA, Wittig RM, Zuberbühler K, Boesch C, Schubert G, … Calvignac-Spencer S (2015). Multiple cross-species transmission events of human adenoviruses (HAdV) during hominine evolution. Molecular Biology and Evolution, 32(8), 2072–2084. 10.1093/molbev/msv090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Jennison A, Whiley D, McMahon J, Hewitson G, Graham R, De Jong A, & Warrilow D (2019). Illumina sequencing of clinical samples for virus detection in a public health laboratory. Scientific Reports, 9(1), 5409. 10.1038/s41598-019-41830-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itta KC, Patil T, Kalal S, Ghargi KV, & Roy S (2016). Salivirus in children with diarrhoea, western India. International Journal of Infectious Diseases, 52, 14–15. 10.1016/j.ijid.2016.09.015 [DOI] [PubMed] [Google Scholar]
- Kaczorowska J, & van der Hoek L (2020). Human anelloviruses: diverse, omnipresent and commensal members of the virome. FEMS Microbiology Reviews, 44(3), 305–313. 10.1093/femsre/fuaa007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel R, Srinivasan A, D'Agata EMC, Lu X, Erdman D, & Jhung M (2015). Outbreak of adenovirus type 4 infection in a long-term care facility for the ederly. Infection Control and Hospital Epidemiology, 31(7), 755–757. 10.1086/653612 [DOI] [PubMed] [Google Scholar]
- Kapusinszky B, Ardeshir A, Mulvaney U, Deng X, & Delwart E (2017). Case-control comparison of enteric viromes in captive rhesus macaques with acute or idiopathic chronic diarrhea. Journal of Virology, 91(18), e00952–00917. 10.1128/JVI.00952-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur T, Singh J, Tong S, Humphrey C, Clevenger D, Tan W, Szekely B, Wang Y, Li Y, Alex Muse E, Kiyono M, Hanamura S, Inoue E, Nakamura M, Huffman MA, Jiang B, & Nishida T (2008). Descriptive epidemiology of fatal respiratory outbreaks and detection of a human-related metapneumovirus in wild chimpanzees (Pan troglodytes) at Mahale Mountains National Park, Western Tanzania. American Journal of Primatology, 70(8), 755–765. 10.1002/ajp.20565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller M (2016). robustlmm: An R package for robust estimation of linear mixed-effects models. Journal of Statistical Software, 75(6), 1–24. 10.18637/jss.v075.i06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köndgen S, Kühl H, N'Goran PK, Walsh PD, Schenk S, Ernst N, Biek R, Formenty P, Mätz-Rensing K, Schweiger B, Junglen S, Ellerbrok H, Nitsche A, Briese T, Lipkin WI, Pauli G, Boesch C, & Leendertz FH (2008). Pandemic human viruses cause decline of endangered great apes. Current Biology, 18(4), 260–264. 10.1016/j.cub.2008.01.012 [DOI] [PubMed] [Google Scholar]
- Köndgen S, Schenk S, Pauli G, Boesch C, & Leendertz FH (2010). Noninvasive monitoring of respiratory viruses in wild chimpanzees. EcoHealth, 7(3), 332–341. [DOI] [PubMed] [Google Scholar]
- Kuznetsova A, Brockhoff PB, & Christensen RHB (2017). lmerTest package: Tests in linear mixed effects models. Journal of Statistical Software, 82(13), 1–26. 10.18637/jss.v082.i13 [DOI] [Google Scholar]
- Leendertz FH, Pauli G, Maetz-Rensing K, Boardman W, Nunn C, Ellerbrok H, Jensen SA, Junglen S, & Christophe B (2006). Pathogens as drivers of population declines: the importance of systematic monitoring in great apes and other threatened mammals. Biological Conservation, 131(2), 325–337. 10.1016/j.biocon.2006.05.002 [DOI] [Google Scholar]
- Lefort V, Longueville J-E, & Gascuel O (2017). SMS: Smart model selection in PhyML. Molecular Biology and Evolution, 34(9), 2422–2424. 10.1093/molbev/msx149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Victoria J, Kapoor A, Blinkova O, Wang C, Babrzadeh F, Mason CJ, Pandey P, Triki H, Bahri O, Oderinde BS, Baba MM, Bukbuk DN, Besser JM, Bartkus JM, & Delwart EL (2009). A novel picornavirus associated with gastroenteritis. Journal of Virology, 83(22), 12002–12006. 10.1128/JVI.01241-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw A, & Wiener M (2002). Classification and regression by randomForest. R News, 2(3), 18–22. [Google Scholar]
- Liguori I, Russo G, Curcio F, Bulli G, Aran L, Della-Morte D, Gargiulo G, Testa G, Cacciatore F, Bonaduce D, & Abete P (2018). Oxidative stress, aging, and diseases. Clinical interventions in aging, 13, 757–772. 10.2147/CIA.S158513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb M (2019). Immunosenescence and respiratory infections among nursing home residents. In Fulop T, Franceschi C, Hirokawa K, & Pawelec G (Eds.), Handbook of Immunosenescence: Basic Understanding and Clinical Implications (pp. 1789–1797). Springer. 10.1007/978-3-319-99375-1_130 [DOI] [Google Scholar]
- Luke SG (2017). Evaluating significance in linear mixed-effects models in R. Behavior Research Methods, 49(4), 1494–1502. 10.3758/s13428-016-0809-y [DOI] [PubMed] [Google Scholar]
- Mackenzie CA, Chapman CA, & Sengupta R (2011). Spatial patterns of illegal resource extraction in Kibale National Park, Uganda. Environmental Conservation, 39(1), 38–50. 10.1017/S0376892911000282 [DOI] [Google Scholar]
- McHugh D, & Gil J (2018). Senescence and aging: causes, consequences, and therapeutic avenues. Journal of Cell Biology, 217(1), 65–77. 10.1083/jcb.201708092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melin AD, Janiak MC, Marrone F, Arora PS, & Higham JP (2020). Comparative ACE2 variation and primate COVID-19 risk. Communications Biology, 3(1), 641. 10.1038/s42003-020-01370-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méthot P-O (2012). Why do parasites harm their host? On the origin and legacy of Theobald Smith's "Law of Declining Virulence" — 1900-1980. History and Philosophy of the Life Sciences, 34(4), 561–601. http://www.jstor.org/stable/43831447 [PubMed] [Google Scholar]
- Mohammad HA, Madi NM, & Al-Nakib W (2020). Analysis of viral diversity in stool samples from infants and children with acute gastroenteritis in Kuwait using Metagenomics approach. Virology Journal, 17(1), 10. 10.1186/s12985-020-1287-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller MN, & Wrangham RW (2014). Mortality rates among Kanyawara chimpanzees. Journal of Human Evolution, 66, 107–114. 10.1016/j.jhevol.2013.10.004 [DOI] [PubMed] [Google Scholar]
- Negrey JD, Emery Thompson M, Langergraber KE, Machanda ZP, Mitani JC, Muller MN, Otali E, Owens LA, Wrangham RW, & Goldberg TL (2020). Demography, life history trade-offs, and the gastrointestinal virome of wild chimpanzees. Philosophical Transactions of the Royal Society B, 375(1811), 20190613. 10.1098/rstb.2019.0613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrey JD, Mitani JC, Wrangham RW, Otali E, Reddy RB, Pappas TE, Grindle KA, Gern JE, Machanda ZP, Muller MN, Langergraber KE, Thompson ME, & Goldberg TL (2021). Data and R script: Viruses associated with ill health in wild chimpanzees. 10.6084/m9.figshare.14879934.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrey JD, Reddy RB, Scully EJ, Phillips-Garcia S, Owens LA, Langergraber KE, Mitani JC, Emery Thompson M, Wrangham RW, Muller MN, Otali E, Machanda Z, Hyeroba D, Grindle KA, Pappas TE, Palmenberg AC, Gern JE, & Goldberg TL (2019). Simultaneous outbreaks of respiratory disease in wild chimpanzees caused by distinct viruses of human origin. Emerging Microbes and Infections, 8(1), 139–149. 10.1080/22221751.2018.1563456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TFF, Zhang W, Sachsenröder J, Kondov NO, da Costa AC, Vega E, Holtz LR, Wu G, Wang D, Stine CO, Antonio M, Mulvaney US, Muench MO, Deng X, Ambert-Balay K, Pothier P, Vinjé J, & Delwart E (2015). A diverse group of small circular ssDNA viral genomes in human and non-human primate stools. Virus evolution, 1(1), vev017. 10.1093/ve/vev017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman Jason M., Handley Scott A., Baldridge Megan T., Droit L, Liu Catherine Y., Keller Brian C., Kambal A, Monaco Cynthia L., Zhao G, Fleshner P, Stappenbeck Thaddeus S., McGovern Dermot P. B., Keshavarzian A, Mutlu Ece A., Sauk J, Gevers D, Xavier Ramnik J., Wang D, Parkes M, & Virgin Herbert W. (2015). Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell, 160(3), 447–460. 10.1016/j.cell.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeseburg H, de Boer RA, van Gilst WH, & van der Harst P (2010). Telomere biology in healthy aging and disease. Pflügers Archiv 459(2), 259–268. 10.1007/s00424-009-0728-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oude Munnink BB, Phan MVT, Consortium V, Simmonds P, Koopmans MPG, Kellam P, van der Hoek L, & Cotten M (2017). Characterization of posa and posa-like virus genomes in fecal samples from humans, pigs, rats, and bats collected from a single location in Vietnam. Virus evolution, 3(2), vex022–vex022. 10.1093/ve/vex022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios G, Lowenstine LJ, Cranfield MR, Gilardi KVK, Spelman L, Lukasik-Braum M, Kinani J-F, Mudakikwa A, Nyirakaragire E, Bussetti AV, Savji N, Hutchison S, Egholm M, & Lipkin WI (2011). Human metapneumovirus infection in wild mountain gorillas, Rwanda. Emerging Infectious Diseases, 17(4), 711–713. 10.3201/eid1704.100883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrono LV, Samuni L, Corman VM, Nourifar L, Röthemeier C, Wittig RM, Drosten C, Calvignac-Spencer S, & Leendertz FH (2018). Human coronavirus OC43 outbreak in wild chimpanzees, Côte d´Ivoire, 2016. Emerging Microbes and Infections, 7(1), 1–4. 10.1038/s41426-018-0121-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelec G, Larbi A, & Derhovanessian E (2010). Senescence of the human immune system. Journal of Comparative Pathology, 142, S39–S44. 10.1016/j.jcpa.2009.09.005 [DOI] [PubMed] [Google Scholar]
- Pei N, Zhang J, Ma J, Li L, Li M, Li J, Sun Y, Ji J, Jiang H, Hou Y, Xu F, Lu H, Zhang R, Wei X, Xu X, & Deng J (2016). First report of human salivirus/klassevirus in respiratory specimens of a child with fatal adenovirus infection. Virus Genes, 52(5), 620–624. 10.1007/s11262-016-1361-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan TG, da Costa AC, del Valle Mendoza J, Bucardo-Rivera F, Nordgren J, O’Ryan M, Deng X, & Delwart E (2016). The fecal virome of South and Central American children with diarrhea includes small circular DNA viral genomes of unknown origin. Archives of Virology, 161(4), 959–966. 10.1007/s00705-016-2756-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips SR, Goldberg TL, Muller MN, Machanda ZP, Otali E, Friant S, Carag J, Langergraber KE, Mitani JC, Wroblewski EE, Wrangham RW, & Emery Thompson M (2020). Faecal parasites increase with age but not reproductive effort in wild female chimpanzees. Philosophical Transactions of the Royal Society B, 375(1811), 20190614. 10.1098/rstb.2019.0614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumptre AJ, & Cox D (2006). Counting primates for conservation: primate surveys in Uganda. Primates, 47(1), 65–73. 10.1007/s10329-005-0146-8 [DOI] [PubMed] [Google Scholar]
- Potts KB, Watts DP, & Wrangham RW (2011). Comparative feeding ecology of two communities of chimpanzees (Pan troglodytes) in Kibale National Park, Uganda. International Journal of Primatology, 32(3), 669–690. 10.1007/s10764-011-9494-y [DOI] [Google Scholar]
- R Core Team. (2021). R: A language and environment for statistical computing. In R Foundation for Statistical Computing. https://www.R-project.org [Google Scholar]
- Rambaut A (2018). FigTree, version 1.4.4. In http://tree.bio.ed.ac.uk/software/figtree/ [Google Scholar]
- Rivers TM (1927). Filterable viruses: A critical review. Journal of bacteriology, 14(4), 217–258. https://pubmed.ncbi.nlm.nih.gov/16559270 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC374955/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrimshaw NS, & SanGiovanni JP (1997). Synergism of nutrition, infection, and immunity: an overview. American Journal of Clinical Nutrition, 66(2), 464S–477S. 10.1093/ajcn/66.2.464S [DOI] [PubMed] [Google Scholar]
- Scully EJ, Basnet S, Wrangham RW, Muller MN, Otali E, Hyeroba D, Grindle KA, Pappas TE, Emery Thompson M, Machanda Z, Watters KE, Palmenberg AC, Gern JE, & Goldberg TL (2018). Lethal respiratory disease associated with human rhinovirus C in wild chimpanzees, Uganda, 2013. Emerging Infectious Diseases, 24(2), 267–274. 10.3201/eid2402.170778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M, & Xiao Y (2011). Multivariate random forests. WIREs Data Mining and Knowledge Discovery, 1(1), 80–87. 10.1002/widm.12 [DOI] [Google Scholar]
- Seimon TA, Olson SH, Lee KJ, Rosen G, Ondzie A, Cameron K, Reed P, Anthony SJ, Joly DO, Karesh WB, McAloose D, & Lipkin WI (2015). Adenovirus and herpesvirus diversity in free-ranging great apes in the Sangha region of the Republic Of Congo. PLoS One, 10(3), e0118543–e0118543. 10.1371/journal.pone.0118543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan T, Li L, Simmonds P, Wang C, Moeser A, & Delwart E (2011). The fecal virome of pigs on a high-density farm. Journal of Virology, 85(22), 11697–11708. 10.1128/JVI.05217-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan T, Wang C, Cui L, Yu Y, Delwart E, Zhao W, Zhu C, Lan D, Dai X, & Hua X (2010). Picornavirus salivirus/klassevirus in children with diarrhea, China. Emerging Infectious Diseases, 16(8), 1303–1305. 10.3201/eid1608.100087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibayama T, Masuda G, Ajisawa A, Takahashi M, Nishizawa T, Tsuda F, & Okamoto H (2001). Inverse relationship between the titre of TT virus DNA and the CD4 cell count in patients infected with HIV. AIDS (London, England), 15(5), 563–570. https://journals.lww.com/aidsonline/Fulltext/2001/03300/Inverse_relationship_between_the_titre_of_TT_virus.4.aspx [DOI] [PubMed] [Google Scholar]
- Sibley SD, Finley MA, Baker BB, Puzach C, Armién AG, Giehtbrock D, & Goldberg TL (2016). Novel reovirus associated with epidemic mortality in wild largemouth bass (Micropterus salmoides). Journal of General Virology, 97(10), 2482–2487. 10.1099/jgv.0.000568 [DOI] [PubMed] [Google Scholar]
- Siqueira JD, Dominguez-Bello MG, Contreras M, Lander O, Caballero-Arias H, Xutao D, Noya-Alarcon O, & Delwart E (2018). Complex virome in feces from Amerindian children in isolated Amazonian villages. Nature Communications, 9(1), 4270. 10.1038/s41467-018-06502-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisza MJ, & Buck CB (2021). A catalog of tens of thousands of viruses from human metagenomes reveals hidden associations with chronic diseases. Proceedings of the National Academy of Sciences, 118(23), e2023202118. 10.1073/pnas.2023202118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Singh J, Ruone S, Humphrey C, Yip CCY, Lau SKP, Anderson LJ, & Kaur T (2010). Identification of adenoviruses in fecal specimens from wild chimpanzees (Pan trogylodytes schweinfurthii) in western Tanzania. American Journal of Tropical Medicine and Hygiene, 82(5), 967–970. 10.4269/ajtmh.2010.09-0668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toohey-Kurth K, Sibley SD, & Goldberg TL (2017). Metagenomic assessment of adventitious viruses in commercial bovine sera. Biologicals, 47, 64–68. 10.1016/j.biologicals.2016.10.009 [DOI] [PubMed] [Google Scholar]
- Tuthill TJ, Groppelli E, Hogle JM, & Rowlands DJ (2010). Picornaviruses. In Johnson JE (Ed.), Cell Entry by Non-Enveloped Viruses (Vol. 343, pp. 43–89). Springer. 10.1007/82_2010_37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Berg PJEJ, Griffiths SJ, Yong S-L, Macaulay R, Bemelman FJ, Jackson S, Henson SM, ten Berge IJM, Akbar AN, & van Lier RAW (2010). Cytomegalovirus infection reduces telomere length of the circulating T cell pool. Journal of Immunology, 184(7), 3417–3423. 10.4049/jimmunol.0903442 [DOI] [PubMed] [Google Scholar]
- Varsani A, & Krupovic M (2018). Smacoviridae: a new family of animal-associated single-stranded DNA viruses. Archives of Virology, 163(7), 2005–2015. 10.1007/s00705-018-3820-z [DOI] [PubMed] [Google Scholar]
- Wevers D, Metzger S, Babweteera F, Bieberbach M, Boesch C, Cameron K, Couacy-Hymann E, Cranfield M, Gray M, Harris LA, Head J, Jeffery K, Knauf S, Lankester F, Leendertz SA, Lonsdorf E, Mugisha L, Nitsche A, Reed P, … Ehlers B (2011). Novel adenoviruses in wild primates: a high level of genetic diversity and evidence of zoonotic transmissions. Journal of Virology, 85(20), 10774–10784. 10.1128/jvi.00810-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JM, Lonsdorf EV, Wilson ML, Schumacher-Stankey J, Goodall J, & Pusey AE (2008). Causes of death in the Kasekela chimpanzees of Gombe National Park, Tanzania. American Journal of Primatology, 70(8), 766–777. 10.1002/ajp.20573 [DOI] [PubMed] [Google Scholar]
- Wood BM, Watts DP, Mitani JC, & Langergraber KE (2017). Favorable ecological circumstances promote life expectancy in chimpanzees similar to that of human hunter-gatherers. Journal of Human Evolution, 105, 41–56. 10.1016/j.jhevol.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Chehoud C, Bittinger K, Bailey A, Diamond JM, Cantu E, Haas AR, Abbas A, Frye L, Christie JD, Bushman FD, & Collman RG (2015). Viral metagenomics reveal blooms of anelloviruses in the respiratory tract of lung transplant recipients. American Journal of Transplantation, 15(1), 200–209. 10.1111/ajt.13031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J-M, Ao Y-Y, Liu N, Li L-L, & Duan Z-J (2015). Salivirus in children and its association with childhood acute gastroenteritis: a paired case-control study. PLoS One, 10(7), e0130977–e0130977. 10.1371/journal.pone.0130977 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Viral nucleotide sequences are available on GenBank under accession numbers MT076199 to MT076210 and MW876510 to MW876520. Additional data and R code are available on Figshare (https://figshare.com/s/d7b44bc82c4086cf3824).