Abstract
CTCF is crucial to the organization of mammalian genomes into loop structures. According to recent studies, the transcription apparatus is compartmentalized and concentrated at super-enhancers to form phase-separated condensates and drive the expression of cell-identity genes. However, it remains unclear whether and how transcriptional condensates are coupled to higher-order chromatin organization. Here, we show that CTCF is essential for RNA polymerase II (Pol II)-mediated chromatin interactions, which occur as hyperconnected spatial clusters at super-enhancers. We also demonstrate that CTCF clustering, unlike Pol II clustering, is independent of liquid-liquid phase-separation and resistant to perturbation of transcription. Interestingly, clusters of Pol II, BRD4, and MED1 were found to dissolve upon CTCF depletion, but were reinstated upon restoration of CTCF, suggesting a potent instructive function for CTCF in the formation of transcriptional condensates. Overall, we provide evidence suggesting that CTCF-mediated chromatin looping acts as an architectural prerequisite for the assembly of phase-separated transcriptional condensates.
INTRODUCTION
Organization of the mammalian genome into three-dimensional (3D) chromatin architecture is tightly linked with gene regulation (1–3). Topologically associated domains (TADs), the structural basis for chromatin organization at a scale of hundreds of kilobases or below, are characterized by preferential interactions among the chromatin located within them and have distinct boundaries that often form loops (4–9). CCCTC-binding factor (CTCF), a transcription factor with 11 zinc finger domains, is well known to demarcate TADs via binding to boundary elements and to promote the formation of chromatin loops in association with the cohesin complex that forms tripartite rings around chromatin (6,7,10–17). Recently, a loop extrusion model was proposed to explain a coordinating function for CTCF and cohesin complex in the establishment and maintenance of TADs and loop structures (18–26). According to this model, the cohesin complex associates with chromatin and translocates along the fiber in opposite directions, thereby extruding an intra-chromosomal chromatin loop, until it is stalled by the amino terminus of CTCF at boundary elements with convergently-oriented CTCF-binding sites.
Regulation of eukaryotic gene expression depends on the efficient assembly of transcription machinery, including sequence-specific transcription factors, various transcriptional co-activators, such as MED1 and BRD4, and RNA polymerase II, at specific genomic sites (27). Enhancers, which regulate transcription at the promoters of the genes that they control in response to intrinsic and external signals, are typically bound by multiple transcription factors in a cooperative manner (27–31). Interestingly, super-enhancers, clusters of enhancers that are occupied by an unusually high density of transcriptional machinery, have been found to control mammalian genes that play essential roles in cell-type-specific processes (32,33).
Liquid–liquid phase separation (LLPS) is a process by which a homogeneous liquid solution of macromolecules, such as proteins or nucleic acids, separates into two distinct phases: a dense and a dilute phase (34). The dense phase has liquid-like properties, including the rapid exchange of components and droplet coalescence (35). LLPS is now thought to underlie the formation of non-membrane-bound compartments, such as nucleoli, and to generate distinct microenvironments that can compartmentalize specific biochemical reactions (36,37). These membraneless organelles formed by LLPS are called biomolecular condensates and are composed of higher-order assemblies of biomolecules that engage in numerous weak, multivalent interactions (35,38). Many nuclear processes, including DNA replication, DNA damage repair, transcription and RNA processing, occur within biomolecular condensates wherein specific biomolecules required for each process interact at higher concentrations relative to other regions in the nucleus (34).
According to recent research, the assembly of the transcription machinery at super-enhancers occurs through LLPS, leading to the formation of transcriptional condensates (34,39–41). For instance, specific transcription factors, coactivator BRD4, subunits of the Mediator complex, and RNA polymerase II, contain intrinsically disordered, low-complexity domains that are thought to drive their phase separation. In support thereof, a set of experiments has shown that the long intrinsically disordered C-terminal domain of Pol II, where repeated amino acids (YSPTSPS) are phosphorylated during transcription, could be a key regulator of condensate formation, and others have demonstrated that RNA amounts produced during individual stages of gene transcription determine the fate of transcriptional condensates (42–47). Therefore, since phosphates and RNA are both electrically charge-rich entities, the formation and dissolution of transcriptional condensates may involve a transition between a hydrophobic, predominantly neutral state and an electrically repulsive state. Indeed, research has indicated that the formation and dissolution of transcriptional condensates in live cells are highly dynamic and show differential sensitivity to transcription inhibitors that perturb selective transcription processes (41,48–50). These results suggest that transcriptional condensates are not merely physical property-driven, but transcriptionally adjustable entities.
Key insights into the molecular mechanisms that drive the formation of transcriptional condensates in vitro and in living cells have emerged from recent studies. One critical step in the formation of condensates appears to be the creation of a dense network of interacting macromolecules (38,40). The valence of these interacting components, the affinity of a given interaction module for its ligand, and the reversibility of a binding reaction, among others, are suggested to be important determinants affecting condensate formation (51). Since transcriptional condensates incorporate both promoter-bound Pol II and enhancer-bound transcription coactivators, researchers have proposed that transcriptional condensates may function within the framework of 3D chromatin structures (40,52–57). However, there are few studies on whether and how transcriptional condensates are coupled to higher-order genome organization.
Thus, we sought to determine whether CTCF, a major component in TAD organization and chromatin looping, plays a role in the formation of transcriptional condensates. To address this possibility, we first generated an auxin-inducible degron system to acutely deplete CTCF protein in a human colorectal carcinoma cell line, HCT116. Genome-wide 3D chromatin structures were then analyzed at the level of TADs and Pol II-mediated chromatin interactions by in situ Hi-C and HiChIP, respectively. The organization and dynamics of CTCF clusters, as well as transcriptional condensates, containing Pol II, BRD4 and MED1, in live cells were examined via a combination of genetic engineering and super-resolution microscopy.
MATERIALS AND METHODS
Cell lines
The human colorectal cancer cell line HCT116, which constitutively expresses OsTIR1 under the control of the CMV promoter (HCT116 CMV-OsTIR1), was kindly provided by Dr Masato T. Kanemaki (58). HCT116 CMV-OsTIR1 cells and all CRISPR/Cas9 knock-in cell lines derived from them were cultured at 37°C in 5% CO2 in RPMI 1640 (HyClone) supplemented with 10% FBS (HyClone) and 100 U/ml penicillin and 100μg/ml streptomycin (HyClone). Cell lines were tested for Mycoplasma contamination using the e-Myco™ Mycoplasma PCR Detection kit (iNtRON).
Plasmid construction of the homology-directed repair (HDR) DNA template
The homology arm sequences flanking the stop codon of CTCF (250 bp each) were PCR amplified from HCT116 genomic DNA and cloned into LITMUS28 vector (New England Biolabs). The stop codon of CTCF was mutated to insert BamHI restriction enzyme site, and a silent mutation was introduced on the sgRNA target site to prevent re-cutting of the repair template by Cas9. The mAID-mClover cassette containing a neomycin (Neo) or hygromycin (Hygro) resistance marker was excised from pMK289 (mAID-mClover-NeoR) (Addgene #72827) or pMK290 (mAID-mClover-Hygro) (Addgene #72828), respectively, and cloned at the BamHI site between the homology arms to generate the CTCF-mAID-mClover-Neo and the CTCF-mAID-mClover-Hygro targeting vectors.
The sequence encoding Halo tag was amplified by PCR using pENTR4-HaloTag plasmid (Addgene #29644) as a template and inserted in place of the mClover sequence of the CTCF-mAID-mClover targeting vectors to generate the CTCF-mAID-Halo-Neo and the CTCF-mAID-Halo-Hygro targeting vectors.
Primers for the Dendra2 repair template with homology arm sequences for RPB1 (170 bp each from the ATG start codon) were designed (Integrated DNA Technologies). The sequence encoding Dendra2 (without the stop codon) was PCR-amplified with the primers from pDendra2-C plasmid (Clontech #632546). The repair template was cloned into pUC19 plasmid (Addgene #50005) to generate the Dendra2-RPB1 targeting vectors. Silent mutations were introduced on the sgRNA target sites to avoid Cas9 degradation of the repair templates.
The homology arm sequences for BRD4 and MED1 genes (250 bp each) were designed from the ATG start codon. The Dendra2 sequence was inserted between the homology arms without the stop codon of Dendra2 to fuse Dendra2 with BRD4 and MED1. Silent mutations were introduced on the sgRNA target sites to avoid Cas9 degradation of the repair templates. The full repair template sequences for BRD4 and MED1 were synthesized (Integrated DNA Technologies), PCR-amplified, and cloned into LITMUS28 vector (New England Biolabs) to generate the Dendra2-BRD4 and Dendra2-MED1 targeting vectors, respectively.
The homology arm sequences for CTCF (250 bp each) were excised from the CTCF-mAID-mClover-Neo targeting vector. The Dendra2 sequence with stop codon was amplified by PCR using the Dendra2-RPB1 targeting vector as a template, digested with BamHI, and inserted between the homology arms for the CTCF gene to generate CTCF-Dendra2 targeting vector.
Plasmid construction for Cas9 and single-guide RNA (sgRNA)
sgRNA targeting CTCF was cloned into pX330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene #42230) by annealing caccgTCAGCATGATGGACCGGTGA and aaacTCACCGGTCCATCATGCTGAc. sgRNA targeting RPB1 was cloned into pSpCas9(BB)-2A-Puro (PX459) V2.0 (Addgene #62988) by annealing caccgGGGCATGCGCTGTCCCCCGA and aaacTCGGGGGACAGCGCATGCCCc. sgRNA targeting BRD4 was cloned into pX330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene #42230) by annealing caccgTGGGATCACTAGCATGTCTG and aaacCAGACATGCTAGTGATCCCAc. sgRNA targeting MED1 was cloned into pX330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene #42230) by annealing caccgCTTCAGGATGAAAGCTCAGG and aaacCCTGAGCTTTCATCCTGAAGc.
CRISPR/Cas9-mediated genome editing
Genome-editing was performed as previously described (59). To generate the CTCF auxin-inducible degron system with mClover reporter, HCT116 CMV-OsTIR1 cells were grown in a 12-well plate before two repair templates containing neomycin and hygromycin resistance markers (CTCF-mAID-mClover-Neo and CTCF-mAID-mClover-Hygro targeting vectors, respectively) and a plasmid encoding Cas9 and sgRNA targeting CTCF were transfected using FuGene HD (Promega). One day after transfection, cells were removed and diluted in 10-cm dishes, followed by selection in the presence of 0.7mg/ml of G418 (Sigma) and 0.1mg/ml of hygromycin (Sigma). After 11–13 days, colonies were picked for further selection in a 96-well plate.
To generate the CTCF auxin-inducible degron system with the HaloTag reporter, HCT116 CMV-OsTIR1 cells were grown in a 12-well plate before two repair templates containing neomycin and hygromycin resistance markers (CTCF-mAID-Halo-Neo and CTCF-mAID-Halo-Hygro targeting vectors, respectively) and a plasmid encoding Cas9 and sgRNA targeting CTCF were transfected using FuGene HD (Promega). One day after transfection, cells were removed and diluted in 10-cm dishes, followed by selection in the presence of 0.7mg/ml of G418 (Sigma) and 0.1 mg/ml of hygromycin (Sigma). After 11–13 days, colonies were picked for further selection in a 96-well plate.
To introduce Dendra2 into endogenous CTCF, HCT116 CMV-OsTIR1 cells were grown in a six-well plate before a repair plasmid (CTCF-Dendra2 targeting vector) and a plasmid encoding Cas9 and sgRNA targeting CTCF were transfected using FuGene HD (Promega).
To introduce Dendra2 into endogenous RPB1, BRD4 or MED1, CTCF-mAID-Halo degron cells were grown in a six-well plate before a repair plasmid (Dendra2-RPB1, Dendra2-BRD4 or Dendra2-MED1 targeting vector, respectively) and a plasmid encoding Cas9 and sgRNA targeting each gene were transfected using FuGene HD (Promega). Two days after transfection, Dendra2-expressing cells were isolated by FACS sorting using FACSAria II and grown in a 96-well plate for further selection.
Genomic PCR for genotyping
All cells stably integrated with knock-in cassette were harvested for DNA extraction using DirectPCR DNA extraction reagent (Fiat international) according to the manufacturer's instruction. Specific primer sets recognizing the 5′ and 3′ knock-in boundaries were designed to test the cells for insertion of the mAID or Dendra2 cassette at the desired target (Supplementary Table S1). Target DNA for genotyping was amplified using MG Taq DNA Polymerase (MGmed) with initial denaturation at 95°C for 3 min, followed by 35 cycles of denaturation at 95°C for 10 s, annealing at 63°C for 30 s, extension at 72°C for 1 to 1.5 min, and a final elongation step at 72°C for 5 min. Stable clones that carried a homozygous insertion were selected for subsequent experiments.
Auxin-induced degradation
For induction of the auxin-inducible degron, indole-3-acetic acid (IAA, chemical analog of auxin) was added in the medium for 24 h at 500 μM from 1000× stock diluted in sterile water. In order to restore CTCF protein levels, auxin-treated cells were washed three times with fresh medium, followed by an additional culture for 24 h in regular medium.
Western blot
Cells lysate was prepared using T-PER Tissue Protein Extraction Reagent (Thermo Scientific) supplemented with Halt™ phosphatase and protease inhibitor cocktail (100X) (Thermo Scientific). Proteins were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto a polyvinylidene fluoride membrane (Millipore). After blocking with 5% skim milk, the membrane was incubated with primary antibodies, followed by incubation with horseradish peroxidase-conjugated secondary antibody. Target proteins were visualized using Super Signal West Pico Chemiluminescent Substrate and ImageQuant 800 (Amersham).
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
One million cells were subjected to RNA extraction using Hybrid-R Total RNA kits (GeneAll Biotechnology) according to the manufacturer's instruction. RNA was reverse transcribed using PrimeScript™ RT Master Mix (Takara Bio). The resulting cDNA was used for qRT-PCR using the QuantStudio3 real-time PCR system (Applied Biosystems) for monitoring the synthesis of double-stranded DNA during various PCR cycles using SYBR Green (QIAGEN). For each sample, duplicate test reactions were analyzed for the expression of the gene of interest, and results were normalized to GAPDH mRNA. Primer sequences are listed in Supplementary Table S1.
RNA sequencing
Strand-specific libraries were generated using the TruSeq PolyA Stranded mRNA sample preparation kit (Illumina) according to the manufacturer's protocol. Barcoded libraries were pooled and sequenced on the Illumina HiSeq 2500 platform, generating 100 bp paired-end reads. Three biological replicates were performed for each condition.
Chromatin immunoprecipitation sequencing (ChIP-seq)
ChIP-seq was performed as previously described (60). Briefly, chromatin samples prepared from appropriate numbers of fixed cells (2 × 105 for H3K27ac and 1 × 107 for CTCF, SMC1 and Pol II) were sonicated and subsequently immunoprecipitated with individual antibodies recognizing CTCF (Cell Signaling Technology), SMC1 (Bethyl lab), Phospho-Rpb1 CTD (Cell Signaling Technology), and H3K27ac (Abcam). Antibody-chromatin complexes were captured with protein A and G Dynabeads (Thermo Fisher Scientific) and washed with low salt wash buffer, high salt wash buffer, and LiCl wash buffer. Chromatin-antibody immobilized on magnetic beads were then subjected to tagmentation. Eluted DNA was purified using SPRI Ampure XP beads (Beckman Coulter) and amplified for 8–12 cycles using KAPA HiFi HotStart Ready mix (KAPA biosystems) and Nextera PCR primers (Illumina). Libraries were purified using dual (0.65–0.25×) SPRI Ampure XP beads (Beckman Coulter) and paired-end sequenced (100 bp) on the Illumina HiSeq2500 platform.
In situ Hi-C
In situ Hi-C was performed as described previously (7). In brief, two million cells were crosslinked with 1% formaldehyde (Thermo Fisher Scientific) for 10 min and subsequently quenched with 0.125M glycine (Thermo Fisher Scientific). Chromatin was digested using DpnII restriction enzyme (New England Biolabs), followed by biotin incorporation with Biotin-14-dATP (Jena bioscience). After de-crosslinking, ligated DNA was purified and sheared to 200–300 bp. DNA was purified with Phenol/Chloroform (Sigma) and quantified using the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific). 150 ng was used for capture with Dynabeads MyOne Streptavidin C-1 (Thermo Fisher Scientific), and an appropriate amount of Tn5 enzyme (Illumina) was added to captured DNA to generate sequencing libraries. Each library was paired-end sequenced (100 bp) on the Illumina NovaSeq6000 platform. Two biological replicates were performed in each condition.
HiChIP
HiChIP was performed as described previously (61). Briefly, twenty million cells were crosslinked with 1% formaldehyde (Thermo Fisher Scientific) for 10 min and subsequently quenched with 0.125 M glycine (Thermo Fisher Scientific). Chromatin was digested using DpnII restriction enzyme (New England Biolabs), followed by biotin incorporation with Biotin-14-dATP (Jena bioscience) in end-repair, ligation, and sonication. Sheared chromatin was then incubated with antibodies recognizing Phospho-Rpb1 CTD (Cell Signaling Technology) at 4°C overnight. Chromatin-antibody complexes were captured by Protein-A magnetic beads (Thermo Fisher Scientific), washed with low salt wash buffer, high salt wash buffer, and LiCl wash buffer, and eluted. DNA was purified with 1.8× SPRI Ampure XP beads (Beckman Coulter) and quantified using the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific). 50–150 ng was used for capture with Dynabeads MyOne Streptavidin C-1 (Thermo Fisher Scientific), and an appropriate amount of Tn5 enzyme (Illumina) was added to captured DNA to generate sequencing libraries. Each library was paired-end sequenced (100 bp) on the Illumina NovaSeq6000 platform. Two biological replicates were performed in each condition.
RNA-seq data processing
Paired-end sequencing reads were trimmed using Trim Galore with default parameters and mapped to the hg19 human reference genome using STAR with the parameters –sjdbOverhang 100 –chimSegMin 20 –twopassMode Basic (version 2.6.0a) (62). Gene expression was quantified using RSEM with the parameters –paired-end –estimate-rspd (version 1.2.31) (63). Differentially expressed genes were determined using DEseq2 (64), with an adjusted P-value <0.01 and a fold-change >2 (version 1.26.0).
ChIP-seq data processing
Fastq files were trimmed using Trim Galore (version 0.6.4) with default parameters to remove adapter sequences. Trimmed reads were mapped to the hg19 human reference genome using bwa with default parameters (version 0.7.17) (65). Low quality reads were filtered using SAMtools with parameters -q 30 -F 1804 -f 2 (version 1.9) (66). Reads mapped to mitochondrial chromosomes and marked as duplicates by Picard (version 2.18.23) were removed using SAMtools. For visualization and downstream analysis, bam files containing uniquely mapped reads were converted to bigwig files using deeptools with parameters –normalizeUsing CPM –binSize 1 (version 3.4.3) (67). Chip-seq peaks were called on each replicate individually using MACS2 with a q-value (false discovery rate) threshold of 0.01 (version 2.2.7.1) (68). The consensus peak list was obtained by retaining peaks that overlapped at least 1bp between biological replicates.
in situ Hi-C and HiChIP data analysis
Paired-end .fastq files from in situ Hi-C and HiChIP experiments were processed using HiC-Pro (version 2.11.4) (69). Default settings were used to align reads to the hg19 human genome, remove duplicate reads, assign reads to DpnII restriction fragments, filter for valid interactions, and generate binned interaction matrices. After confirmation of good reproducibility between biological replicates using 3DChromatin_ReplicateQC, the replicates data were merged for re-processing as combined results (Supplementary Figure S1, Tables S2 and S3) (70). The validated contact pairs were transformed to Juicer .hic files with hicpro2juicebox. To segregate A and B compartments, eigenvector values for each chromosome of each sample were generated from the HiC data using Juicer tools ‘eigenvector’ command with KR normalization at 100 kb resolution (version 1.22.01) (71). The Juicer .hic files were converted to .cool files using hic2cool with default parameters. Saddle plots were generated at 100 kb resolution using cooltools (version 0.3.2), and the strength of compartmentalization was defined as the ratio of (A–A + B–B)/(A–B + B–A) interactions. Insulation score was calculated as described previously (72), using an algorithm that aggregated the number of interactions that occurred across chromosome bins, which were then divided by the mean number of interactions for the whole chromosome and then logarithmized. To identify topological domain boundaries following an insulation square analysis (72), contact matrix files were generated from Hi-C data and utilized for calculation of insulation scores using matrix2insulation.pl with parameters -b 500000 -ids 200000 -im mean -bmoe 3 -nt 0.1. Intra-TAD DNA interactions, represented as TAD strengths, were determined using FAN-C with parameters –tad-strength (version 0.9.10) (73). Loop-calling for the Pol II HiChIP experiments was performed using FitHiChIP with 10 kb bin sizes, bias correction by coverage, false discovery rate <0.01, a minimum genomic distance of 20 kb, and a maximum genomic distance of 2 Mb (version 8.0) (74). To quantify the level of interactions genome-wide, we performed a pileup analysis using our Hi-C and HiChIP data. The values in each corner of the APA plots were determined using coolpup.py by calculating the mean value for five central squares (version 0.9.5) (75).
Definition of regulatory elements for annotating HiChIP loop anchors
A total of 57 820 unique gene transcripts was obtained from gene annotation files downloaded from GENCODE V19 (76). Promoters were defined as ±2 kb from the transcription start site of each annotated protein-coding gene. Enhancers were defined as regions with an H3K27ac peak as determined by ChIP-seq. H3K27ac peaks that overlapped a gene promoter were removed from this list. Super-enhancers were defined by applying the ROSE algorithm to H3K27ac peaks with the default stitching size of 12.5 kb (33). The presence of one or more promoter was considered a promoter HiChIP anchor. The absence of any promoter, but the presence of an enhancer constituted an enhancer HiChIP anchor. The presence of at least one CTCF ChIP-seq, but the absence of any promoter and enhancer was considered as a CTCF HiChIP anchor.
Virtual 4C plots
Contact matrices of 10 kb resolution were generated from .hic files using the Juicer tools ‘dump’ command with parameters set to ‘NONE.’ Bins containing the TSS of genes of interest or super-enhancers were determined as ‘viewpoints.’ Depth normalization was achieved by scaling counts by the total number of de-duplicated valid interactions in each experiment. To visualize interactions of surrounding regions to a viewpoint, the row of the viewpoint in the contact matrix with the columns within a selected range were plotted as a line with the R package ggplot2.
3D clique analysis
3D clique analysis was performed following the same procedure as reported previously (77). In brief, an undirected graph of Pol II-mediated chromatin interactions was constructed from Pol II HiChIP data, wherein each vertex was a loop anchor and each edge was a significant Pol II HiChIP loop. ‘3D cliques’ were defined by spectral clustering of Pol II-mediated chromatin interactions using the cluster_louvain function in the igraph R package with default parameters. 3D clique connectivity was defined as the number of edges connecting vertices within the clique. The connectivity of cliques was ranked in ascending order and plotted against the rank. The cutoff for ‘hyper-connected 3D cliques’ was set to the elbow of the curve, and a tangent line at the cutoff was shown. Cliques below the elbow point of the curve were defined as regular-connected 3D cliques.
Cell preparation for imaging
Cells were cultured on glass bottom dishes (Cellvis, D35-25-1.5-N) at 37ºC and 5% CO2 in phenol red-free DMEM (Gibco, 21063-029) supplemented with 10% FBS (Gibco, 12483–020, Canada origin, Qualified) and 100 U/ml penicillin and 100μg/ml streptomycin (Gibco, 15140-122). Before imaging, cell culture medium was exchanged with L-15 (Leibovitz's, Gibco, 21083-027) supplemented with 10% FBS after washing with PBS (Phosphate buffered saline, Gibco, 10010-023). For fixed cell imaging, growth medium was exchanged with 4% paraformaldehyde after washing with PBS. After 10-min incubation at room temperature, paraformaldehyde was removed and PBS was added to completely remove paraformaldehyde. PBS incubation for 5 min was repeated three times. Transcription inhibitors were added in growth media with 1 μM JQ1 (Sigma-Aldrich, SML0974), 125 nM triptolide (Sigma-Aldrich, T3652), 10 μM flavopiridol (Sigma-Aldrich, F3055) and 100 μM DRB (Sigma-Aldrich, D1916) diluted in dimethyl sulfoxide (Sigma-Aldrich, D8418).
Fluorescent tag ligands incubation for imaging
For HaloTag-labeled CTCF imaging, 100 nM JF646-HaloTag ligand (Janelia Fluor® HaloTag Ligands, Promega, GA1120) diluted in dimethyl sulfoxide was added to cell culture media and incubated for 10 min at 37ºC and 5% CO2 to induce binding to HaloTag (78). Non-bound JF646-HaloTag ligands were removed by 15-min incubation with cell culture media, repeated twice.
Highly inclined and laminated optical (HILO) microscopy
All super-resolution imaging data in the study were acquired from a custom-built microscope based on the Nikon microscope body Ti2e with HILO illumination. HILO illumination has an incident angle smaller than the critical angle of total reflection so that it is transmitted in the form of a thin sheet, allowing excitation in depth. Fluorescence in cells was excited with 405-, 488-, 531-, 640-nm lasers (CUBE, Coherent). Images were acquired using an EMCCD camera (Andor, iXon Ultra 888) and processed using NIS-Elements. Image analysis was performed using MATLAB R2020a and ImageJ scripts.
Super-resolution imaging
For PALM imaging of Dendra2, cells were illuminated with a 405-nm (for photoconversion of Dendra2) and 561-nm laser (for excitation of photoconverted Dendra2) (79,80). For each image, 6000 frames were acquired with a temporal resolution of 50 ms/frame, until most of the photoconverted Dendra2 signals were bleached in the nucleus. For dSTORM imaging of JF646-Halo-CTCF, cells were excited with a 640-nm laser for 600 frames with a temporal resolution of 50 ms/frame (81). In this study, lasers with a longer wavelength were illuminated prior to shorter ones to minimize photo-bleaching. Finally, collected images were analyzed and reconstructed into super-resolved images using MTT (82) and qSR (83).
Density-based spatial clustering of applications with noise (DBSCAN) analysis
We defined separated clusters on super-resolved images using the density-based spatial clustering of applications with noise (DBSCAN) algorithm (84). We investigated groups of each detected molecule based on single molecule localizations. Each group by DBSCAN was defined as a single cluster. For DBSCAN, we used two parameters, the minimum number of detections in a group (N) and the minimum distance between detections in a group (R). In the study, we used N = 10–30 (points) and R = 80–100 (nm) to determine clusters. Since expression levels of a protein of interest and shapes of nuclei are not consistent across cells, the optimum N, R values for grouping clusters are iteratively chosen from an inspection of each single cell image. Considering that the transcription condensate is formed through LLPS (39,41), the shape of a cluster we defined in the DBSCAN analysis is expected to be spherical by surface tension, which will be represented as an intensified normal Gaussian profile in the super-resolved color-code image. Thus, we compared a DBSCAN-clustering result with a matched super-resolved color code image and a set of parameters (N, R) were iteratively chosen such that most of intensified regions with normal Gaussian profiles are grouped as clusters in the DBSCAN result, as shown in Supplementary Figure S6.
Fluorescence recovery after photobleaching (FRAP)
FRAP experiments were performed using a spinning disk confocal microscope (NIKON CSU-W1) with digital micromirror devices. Images of a single confocal plane were taken with an exposure time of 200 ms. The bleaching spot was centered on a cluster, and images were taken for 1 min at 1-s intervals to assess fluorescence recovery in the cluster. To quantify FRAP recovery, we followed the approach described in a previous study (25). We measured integrated intensities of the bleached cluster over time and then corrected them by subtracting nearby nuclear background intensities in the same cell. The corrected data were normalized with the pre-bleached intensity of the cluster. To define a recovery fraction (A) and a mean recovery time (τ), we processed fitting with a single exponential model: I(t) = A · (1 – exp(– t/τ)).
Bleaching step analysis for CTCF clusters
To measure the number of CTCF molecules per CTCF cluster, we counted the number of steps during photobleaching of CTCF clusters. To define each fluorescence step from background noise, we used HaMMy software (85). We defined CTCF clusters as locally bright spots of 3 × 3 pixels from raw images of JF646-Halo-CTCF. To remove photo-bleaching effects in defining steps, we subtracted the intensity of the background from the selected CTCF clusters.
Analysis of distances between clusters
From DBSCAN with qSR software, we collected center positions of defined clusters per each nucleus. We measured center-to-center distances between all defined clusters in a single nucleus and determined the distances between the closest clusters: for example, to obtain the distance between neighboring CTCF and Pol II clusters, we matched each CTCF cluster to every Pol II cluster and measured the minimal distances between them.
RNA fluorescence in situ hybridization (FISH)
We designed customized RNA FISH probes using the Stellaris Probe Designer, targeting intron regions of genes of interest to visualize nascent transcription loci. Cells were cultured on glass bottom dishes and fixed with 4% paraformaldehyde. After washing twice with 1× PBS, 2 ml of 70% ethanol was added to the dishes for permeabilization at 4°C for 1 h. 20% Stellaris RNA FISH Wash Buffer A (Biosearch Technologies, SMF-WA1-60), 70% nuclease-free water (Ambion, AM9932), and 10% deionized formamide (Ambion, AM9344) were mixed to make wash buffer A, which was added to fixed cells after the aspiration of 70% ethanol at RT for 5 min. Then, wash buffer A was removed, and 100 μl of hybridization buffer (90% Stellaris RNA FISH Hybridization Buffer [Biosearch Technologies, MSF-HB1-10]) and 10% deionized formamide containing 1 μl of probes were added to the cells. Samples were maintained in a humidified chamber at 37°C overnight. After removal of hybridization buffer, 1 ml of wash buffer A was added and incubated in the dark at 37°C for 30 min. Then, wash buffer A was changed to 1 ml of wash buffer B (Biosearch Technologies, MSF-WB1-20) and incubated at RT for 5 min. Wash buffer B was changed to PBS for imaging.
Reagents
The reagents or resources used in this study were listed in the Supplementary Table S4.
RESULTS
Acute depletion of endogenous CTCF protein maintains compartment organization, but compromises global TAD insulation
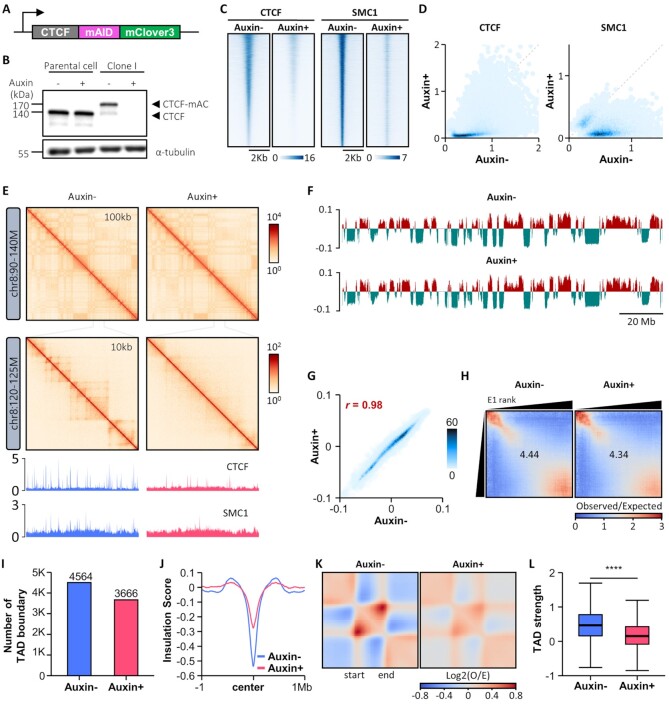
To investigate the roles of CTCF in 3D chromatin organization, a mAID-mClover3 cassette was introduced to the C-terminus of endogenous CTCF (both alleles) in HCT116 cells, which constitutively express a OsTIR1 transgene to allow for rapid and specific depletion of mAID-tagged protein via proteasome-dependent degradation upon auxin treatment (Figure 1A; Supplementary Figure S1A and B) (58). Immunoblotting and fluorescence-activated cell sorting (FACS) demonstrated efficient depletion of the CTCF-miniAID-mClover3 (CTCF-mAC) fusion protein, with little effect on apoptosis. Parental HCT116 cells exhibited no change in CTCF expression when treated with auxin for 24 h (Figure 1B; Supplementary Figure S1C and S1D). Two degron clones (I and M clones) showing almost complete CTCF depletion were selected and used as biological replicates. CTCF binding, as measured by ChIP-Seq, was lost or severely reduced at most of its binding sites in auxin-treated cells, further confirming the depletion of CTCF protein upon auxin treatment (left panels of Figure 1C and D; Supplementary Figure S1E). Genome-wide binding of SMC1, a subunit of the cohesin complex functionally associated with CTCF, was also strongly depleted at CTCF binding sites upon auxin treatment (right panels of Figure 1C and D; Supplementary Figure S1F).
Figure 1.
Acute depletion of endogenous CTCF protein maintains compartment organization, but compromises global TAD insulation. (A) The mAID-mClover3 cassette for auxin-inducible degron is tagged to the C-terminus of CTCF on both alleles in a HCT116 cell line wherein OsTIR1 is integrated at the AAVS1 locus. (B) Immunoblotting shows complete degradation of endogenous mAID-mClover3-tagged CTCF (CTCF-mAC) upon auxin treatment. (C and D) Heatmaps (C) and scatterplots (D) of ChIP-Seq signals called for CTCF (left) and SMC1 (right) show marked reductions in the global occupancies of both proteins upon auxin treatment. (E) Hi-C contact maps generated by HiCExplorer at 100- and 10-kb resolution (version 3.4.3) (113). ChIP-seq signal tracks for CTCF and SMC1 were aligned below the contact maps at 10-kb resolution. (F) Distributions of cis Eigenvector 1 values across the entirety of chromosome 8 with and without auxin treatment. (G) Scatterplot shows that CTCF depletion does not affect genome-wide cis Eigenvector 1 values. (H) Saddle plots of compartmentalization strength with and without auxin treatment. (I) Number of TAD boundaries obtained with Hi-C data. (J) Genome-wide averaged insulation scores plotted against distance around insulation center at WT TAD boundaries. (K) Heatmaps show the average observed/expected Hi-C interactions in the TAD regions. (L) Boxplot shows TAD strength with and without auxin treatment.
Additionally, we performed in situ Hi-C to explore how CTCF depletion affects higher-order chromatin organization in HCT116 cells. Given the high reproducibility of Hi-C data between biological replicates (Supplementary Figure S1H), we combined raw Hi-C data generated from both degron clones for subsequent analysis. Contact maps (Figure 1E), the first eigenvector values from principal component analysis (Figure 1F and G), and compartmentalization strength represented by saddle plots (Figure 1H) indicated that genomic segmentation of active and inactive chromosome domains into A and B compartments was mostly maintained after CTCF depletion.
Also, defining TADs using insulation scores, we found that fewer TAD boundaries were maintained after auxin treatment (Figure 1I) and that the insulation capacity at TAD boundaries was severely compromised by CTCF loss (Figure 1J). We also found that genome-wide intra-TAD chromatin interactions were diminished after CTCF depletion (Figure 1K and L). These data suggested that CTCF is dispensable in compartment organization, but essential for global TAD insulation in HCT116 cells, consistent with several other cellular models (11,86,87).
CTCF is essential for Pol II-mediated chromatin interactions
Next, we performed RNA-seq to profile gene expression in CTCF-depleted cells. Consistent with previous reports (11,86), the overall impact of CTCF depletion on transcript abundance was limited: differential RNA expression analysis revealed 262 deregulated genes (adjusted P-value < 0.01, fold-change > 2.0), 160 upregulated and 102 downregulated, upon CTCF depletion (Supplementary Figure S2A). Integration of these differentially expressed genes with CTCF ChIP-seq data revealed that 72% of the downregulated genes had CTCF bound to promoter prior to depletion, as opposed to <32% of the upregulated genes. These results indicated that direct binding of CTCF at promoters play a crucial role in the activation of a subset of CTCF-bound promoters (Supplementary Figure S2B), consistent with a previous report (11). Further, we examined whether CTCF depletion has any effect on active transcription events by quantifying occupancy of Pol II with C-terminal domain Ser5 phosphorylation, a conserved mark of Pol II initiating transcription (88). Pol II binding patterns, as measured by ChIP-seq, were highly similar between untreated and auxin-treated cells, indicating that CTCF loss does not affect Pol II binding and genome-wide transcription activities overall (Figure 2A and B; Supplementary Figure S1G).
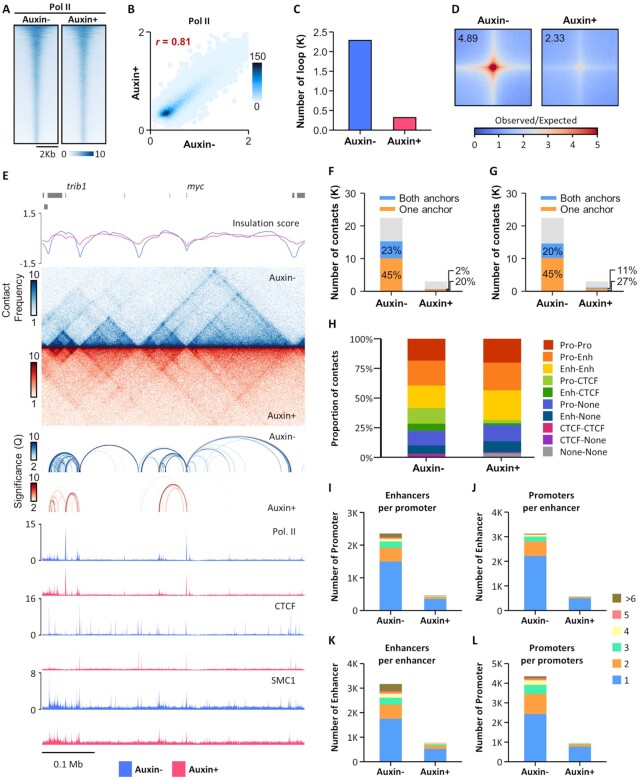
Figure 2.
Pol II-mediated chromatin interactions are greatly reduced by CTCF depletion. (A and B) Heatmaps (A) and scatterplots (B) of Pol II ChIP-Seq signals with or without auxin treatment. (C) Number of Pol II HiChIP loops with or without auxin treatment. (D) Aggregate peak analysis (APA) for Pol II HiChIP loops called from untreated cells, comparing Pol II HiChIP (top) or Hi-C (bottom) data generated from untreated (left) or auxin-treated (right) cells. (E) Snapshot of insulation score curves, Pol II HiChIP contact maps, Pol II HiChIP loops, and ChIP-seq signal tracks for Pol II, CTCF, and SMC1 with and without auxin treatment. (F and G) Percentage of overlap of CTCF (F) and SMC1 (G) ChIP-seq peaks with either one or both Pol II HiChIP loop anchors. (H) Distribution of regulatory elements at the anchors of Pol II HiChIP loops. (I) Number of enhancer-interacting promoters. (J) Number of promoter-interacting enhancers. (K) Number of promoter-interacting promoters. (L) Number of enhancer-interacting enhancers.
We then explored Pol II-mediated chromatin looping by performing Pol II HiChIP in HCT116 cells using antibodies against Pol II with C-terminal domain Ser5 phosphorylation. Statistically significant Pol II-based chromatin interactions were called using FitHiChIP at a resolution of 10 kb, with a false discovery rate of less than 10–2, a minimum genomic distance of 20 kb, and a maximum genomic distance of 2 Mb. CTCF depletion dramatically decreased the number of Pol II HiChIP loops present (22 842 in untreated and 3,192 in auxin-treated cells) (Figure 2C). Moreover, genome-wide loop aggregation analysis of Pol II HiChIP data revealed that Pol II-mediated loop strength was greatly reduced in auxin-treated cells (Figure 2D).
When examining local genomic regions, we noticed that most of the Pol II HiChIP loops were called within TADs and overlapped with ChIP-seq peaks for CTCF and SMC1 at their anchors (Figure 2E). Indeed, genome-wide analysis of Pol II loops showed that binding of CTCF and SMC1 was highly enriched overall for at least one of the loop anchors in untreated cells, but less so in auxin-treated cells (Figure 2F and G). We also analyzed the distribution of regulatory elements at loop anchors and found that >95% of Pol II loops had either promoters or enhancers present on at least one of the loop anchors (Figure 2H). Additionally, we discovered that individual promoters could interact with multiple enhancers acting in concert and that multiple promoters could be regulated by a single enhancer (Figure 2I and J). Finally, we noted several chromatin interactions between promoter pairs and that enhancers could interact with multiple enhancers (Figure 2K and L). These results suggested that CTCF is essential in the maintenance of Pol II-centric chromatin interactions, most of which may contribute to the regulatory functions of promoters and enhancers for gene expression.
Pol II-mediated chromatin interactions at super-enhancers typically occur as hyperconnected spatial clusters that require CTCF
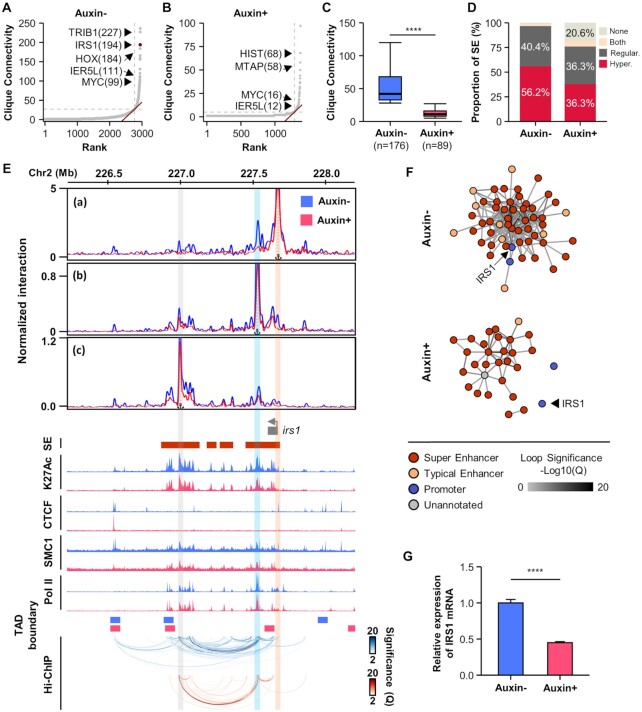
Given that a large fraction of Pol II loops were connected to promoter and enhancer pairs, we next searched for Pol II-mediated, highly interacting, enhancer and promoter spatial clusters, called 3D cliques, as previously reported (77). Showing asymmetric distribution, 176 and 89 cliques were identified as hyperconnected 3D cliques in untreated and auxin-treated cells, respectively (Figure 3A and B). Of note, the number of connections in each of the hyperconnected 3D cliques was significantly reduced by CTCF depletion (Figure 3C). In untreated cells, more than half of the super-enhancers (56%) contributed to the formation of hyperconnected 3D cliques; the remaining super-enhancers (40%) were associated with regular 3D cliques (Figures 3D; Supplementary Figure S3). In CTCF-depleted cells, however, markedly fewer super-enhancers (36%) were involved in the hyperconnected 3D cliques, and around 20% of them showed no Pol II-mediated spatial interactions (Figure 3D; Supplementary Figure S3).
Figure 3.
Pol II-mediated hyperconnected 3D cliques are disrupted by CTCF depletion. (A and B) 3D clique total connectivity based on Pol II HiChIP loops in untreated (A) or auxin-treated (B) cells. Hyperconnected 3D cliques are defined as those above the elbow of the total connectivity ranking. Examples of hyperconnected 3D cliques are marked and named with their representative genes. The number of interactions in each clique is provided in parenthesis. (C) Number of interactions for each hyperconnected 3D clique in untreated or auxin-treated cells. (D) Percentage of overlap of super-enhancers with hyperconnected 3D cliques. (E) Snapshots displaying virtual 4C plots, ChIP-seq signal tracks, and Pol II HiChIP loops (from top to bottom) at the IRS1 locus with and without auxin treatment. Virtual 4C plots show normalized Pol II HiChIP loop strength with the transcription start site (a), proximal super-enhancer (b), and distal super-enhancer (C) as viewpoints. The locations of super-enhancers are shown on top of H3K27ac ChIP-seq signal tracks. The locations of TAD boundaries are shown on top of Pol II HiChIP loops. Orange, sky, and gray vertical bars highlight the location of the viewpoints at the transcription start site, proximal super-enhancer, and distal super-enhancer, respectively. (F) 3D cliques in the IRS1 locus, wherein each edge represents a significant Pol II-mediated chromatin interaction. The color of each edge indicates loop strength (–log10(Q)). The color of each node represents promoter, typical enhancer, super-enhancer, or unannotated regions. (G) Decreased mRNA expression of the IRS1 by CTCF depletion was validated with qRT-PCR. Error bars show mean ± standard errors of the mean (s.e.m). **** P < 0.0001, unpaired two-tailed Student's t-tests.
One clique containing IRS1 was identified as a hyperconnected 3D clique, ranking among the top 3 most connected 3D cliques in untreated cells (Figure 3A). Examination of the IRS1 cliques showed that IRS1 promoter was connected to multiple super-enhancers via highly interacting Pol II loops in the presence of CTCF, while CTCF depletion led to the exclusion of IRS1 promoter from nearby super-enhancers, which still form hyperconnected 3D cliques at the same TAD, albeit with less connectivity (Figure 3E and F). Virtual 4C (v4C) analysis of the IRS1 locus, with the promoter, proximal super-enhancer, or distal super-enhancer as viewpoints, showed significant interactions between super-enhancers and the IRS1 promoter in the presence of CTCF, which were abrogated by CTCF depletion (Figure 3E). The disconnection of IRS1 promoter from super-enhancers could be the reason for the decreased mRNA expression of IRS1 in CTCF-depleted cells (Figure 3G). The functional significance of CTCF in Pol II-mediated enhancer-promoter interactions was also observed at MYC and IER5L loci, where the promoters were connected to super-enhancers via hyperconnected 3D cliques (Figure 3A and Supplementary Figure S4) and the expression of these genes were decreased due to abrogated interactions between promoters and super-enhancers (Supplementary Figure S4). Taken together, these data suggested that Pol II-centric chromatin interactions at super-enhancers typically occur as hyperconnected spatial clusters, the formation of which requires CTCF.
Dual-color super-resolution imaging captures CTCF and RNA polymerase II clusters in the nuclei of living cells
We next sought to characterize the organization and dynamics of CTCF and Pol II molecules within the nuclei of live cells with super-resolution microscopy. To do so, we generated a dual-labeled homozygous HCT116 cell line wherein a mAID-Halo cassette and a transgene encoding Dendra2 protein were introduced to the C-terminus of the CTCF gene and the N-terminus of the POLR2A gene encoding RPB1, respectively (Figure 4A; Supplementary Figure S5A–D) (41,50,89).
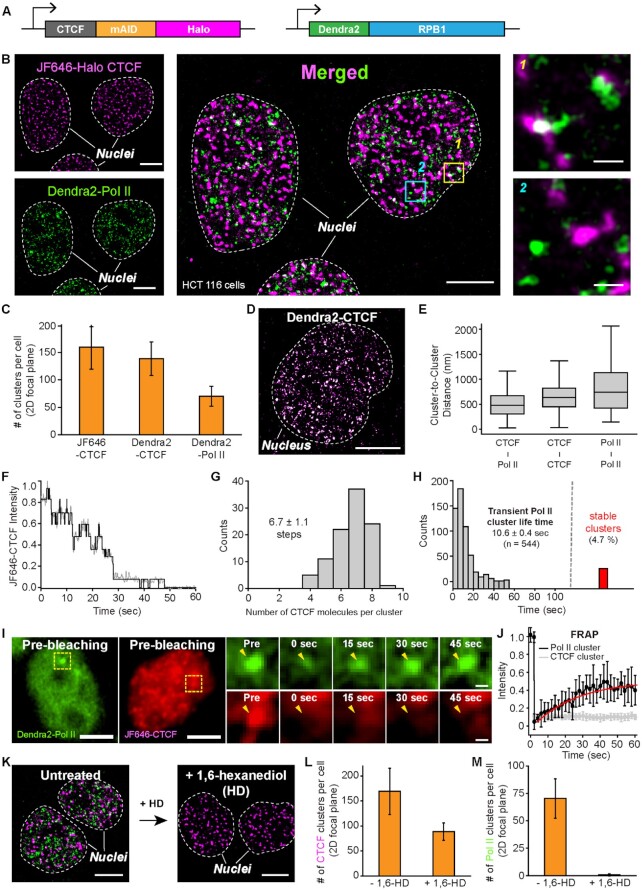
Figure 4.
Dual-color super-resolution imaging captures CTCF and Pol II clusters in a cell nucleus. (A) The mAID cassette, as well as HaloTag, are inserted at the C-terminus of CTCF on both alleles in a HCT116 cell line wherein Dendra2 is homozygously tagged to endogenous Pol II and the OsTIR1 gene is integrated at the AAVS1 locus. (B) Endogenous JF646-Halo-CTCF (left top, magenta) and Dendra2-Pol II (left bottom, green) form clusters in HCT116 nuclei. A super-resolved merged image shows both clusters in each nucleus (middle). Some CTCF and Pol II clusters were colocalized (right top), while others were not (right bottom). (C) Numbers of CTCF and Pol II clusters per cell per 2D focal plane for each cell line (from N = 20 cells for each). (D) A representative super-resolved image of Dendra2-CTCF clusters. (E) Center-to-center distance plots for nearby CTCF-Pol II, CTCF-CTCF, and Pol II-Pol II clusters, from N = 475, 1039 and 419 cluster pairs, respectively. (F) A representative intensity plot over time for photo-bleaching analysis to count numbers of CTCF molecules in a single CTCF cluster. (G) Distribution of CTCF molecules per cluster. N = 100 CTCF foci from nine cells were selected for the estimation. (H) Distribution of Pol II cluster life time (mean ± s.e.; n = number of clusters). 4.7% (red bar) of the clusters were temporally stable. (I) Representative images of FRAP experiments for CTCF and Pol II clusters. Yellow dotted boxes indicate specific photobleached regions. (J) Normalized fluorescence recovery for Dendra2-Pol II (black, N = 9 clusters) and JF646-CTCF (grey, N = 8 clusters). (K) Representative images for JF646-CTCF (magenta) and Dendra2-Pol II (green) before and after 1,6-hexanediol (1%, 10 min) treatment. (L and M) Number of CTCF and Pol II clusters per cell before (N = 20 cells) and after (N = 16 cells) 1,6-hexanediol treatment. Scale bars represent 5 μm for entire nuclei images and 500 nm for zoom-in images.
When Halo-tagged CTCF molecules were labeled with JF646-Halo-ligands for single-molecule localization-based super-resolution imaging with direct stochastic optical reconstruction microscopy (dSTORM), clusters of CTCF molecules were clearly observed in the living HCT116 cells (Figure 4B; left upper panel). High resolution imaging by photoactivated localization microscopy (PALM) with Dendra2, a photo-switchable fluorescent protein, also revealed the formation of Pol II clusters in the nuclei of living cells, which was previously demonstrated in other cells (Refs; U2OS, MEF, mESCs) (Figure 4B; left lower panel) (41,48–50). The average numbers of CTCF and Pol II clusters per cell per 2D-focal plane were 159.2 ± 39.8 and 70.3 ± 17.9, respectively (Figure 4C, Supplementary Figures S5E–G, S6).
In order to verify that the super-resolved JF646-CTCF clusters detected by dSTORM actually represent regions of clustered CTCF molecules, not just multiple counts resulting from long-lived JF646 dye, we generated a HCT116 cell line wherein a transgene encoding Dendra2 protein was homozygously introduced to the C-terminus of CTCF alleles (Figure 4D, Supplementary Figure S5H-S5J). Super-resolution imaging by PALM for Dendra2-labeled CTCF revealed a similar number of CTCF clusters as that for JF646-labeled CTCF (138.5 ± 30.6 and 159.2 ± 39.8 clusters per cell per 2D-focal plane, respectively) (Figure 4C), confirming that CTCF molecules indeed formed clusters.
We then performed dual-color super-resolution imaging to merge JF646 and Dendra2 images and found that several CTCF clusters colocalized with Pol II clusters, while some of them were separated from each other (right panel in Figure 4B; Supplementary Figure S7A). Separated clusters for CTCF and Pol II could be defined using density-based spatial clustering of applications with noise (DBSCAN) (Supplementary Figures S5E–G, S6) (84,90), and the average center-to-center distance between CTCF and Pol II clusters was estimated at 510.4 ± 11.9 nm, which was shorter than center-to-center distances between CTCF cluster pairs and Pol II cluster pairs (651.7 ± 8.5 and 836.2 ± 25.2 nm, respectively) (Figure 4E and Supplementary Figure S7B).
We then quantified the dynamics of the observed CTCF clusters by photo-bleaching analysis (Figure 4F) and found that most CTCF clusters (>82.8%) contained six to eight molecules, which is consistent with a previous report (Figure 4G) (91). It should be noted, however, that the average number of CTCF molecules per cluster (6.7 ± 1.1) could have been over-estimated, since background noise from JF646-CTCF in the nucleus made it difficult to detect CTCF-bound foci containing less than four molecules.
Previously, two distinct classes of Pol II clusters were observed in the nuclei of live mouse embryonic stem cells (mESCs): small, temporally transient clusters and large, but stable clusters (41). The small, temporal Pol II clusters were only detectable with single-molecule localization-based super-resolution imaging. In good agreement with this and other cells (Figure 4H), we were able to observe these two distinct classes of Pol II clusters in our HCT116 cell line, with an average lifetime of 10.6 ± 0.4 s for the transient clusters (Figure 4H; red bar represents counts for stable clusters). Meanwhile, transcriptional condensates are thought to be highly dynamic, with components freely exchanging within and with the surrounding nucleoplasm (39,41). To examine this, we conducted fluorescence recovery after photobleaching (FRAP), which is widely used to assess condensate fluidity and to estimate protein diffusion (38). Our FRAP analyses of Pol II clusters revealed very rapid dynamics and turnover of Pol II molecules with a characteristic recovery time of 25.3 ± 1.5 s (Figure 4I and J, Supplementary Figure S7C-S7F). Moreover, treatment of HCT116 cells with 1,6-hexanediol, commonly used to dissolve LLPS condensates by perturbing hydrophobic protein interactions, resulted in the complete dissolution of Pol II clusters (Figure 4K–M). In contrast, fluorescence signals from bleached CTCF clusters were not recovered (Figure 4I and J; Supplementary Figure S7C-S7F), and 1,6-hexanediol treatment showed a relatively modest effect on the number of CTCF clusters (Figure 4K–M). These results indicated that LLPS plays a crucial role in the Pol II clustering, but not in CTCF clustering.
Transcription inhibition, which disturbs Pol II clustering, slightly enhances CTCF clustering
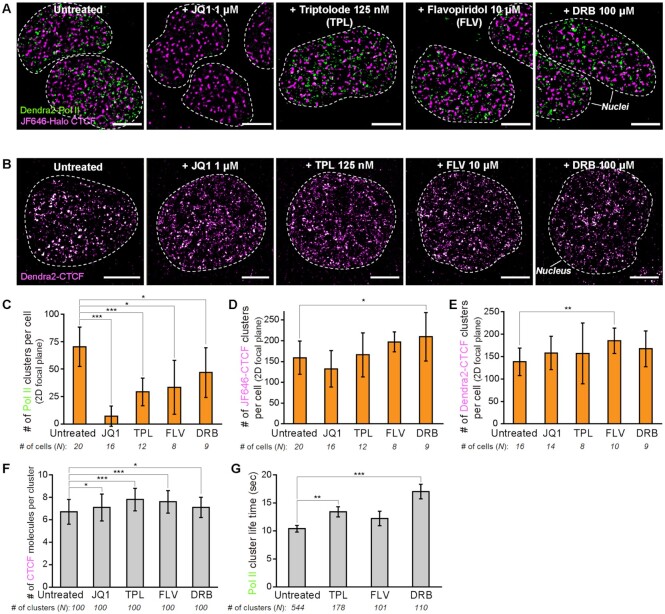
Given that transcription-related processes have been shown to be strongly associated with the behavior of RNA polymerase II complex and to play a role in shaping the spatial organization of the genome (56,92), we set out to explore whether transcription inhibition can affect the formation of Pol II and CTCF clusters (Figure 5A and B, Supplementary Figures S8). As expected, dual-color super-resolution imaging of CTCF-mAID-Halo/Dendra2-PolII cells revealed dramatic effects for transcription inhibitors on Pol II clustering. JQ1, which inhibits interactions between BRD4 and enhancers, almost completely dissolved Pol II clusters, and inhibition of transcription initiation (using triptolide) and transcription elongation (using DRB and flavopiridol) significantly decreased the formation of Pol II clusters (Figure 5C). In contrast, the numbers of CTCF clusters estimated in JF646-labeled cells (Figure 5D) and in Dendra2-tagged cells (Figure 5E) did not significantly change in the presence of JQ1 and slightly increased after treatment with each transcription inhibitor. These results suggested that transcription-related processes, such as initiation, elongation, and communication with enhancers, exert distinct and independent effects on the formation of Pol II and CTCF clusters.
Figure 5.
CTCF and Pol II clusters respond differently to transcription inhibition. (A) Representative super-resolved images of JF646-Halo-CTCF (magenta) and Dendra2-Pol II (green) clusters in cell nuclei before and after treatment with JQ1 (1 μM, 2 h), triptolide (TPL; 125 nM, 2 h), flavopiridol (FLV; 10 μM, 2 h) and DRB (100 μM, 2 h). (B) Representative super-resolved images of Dendra2-CTCF clusters before and after treatment with JQ1, TPL, FLV and DRB. (C–E) Numbers of (C) Dendra2-Pol II, (D) JF646-Halo-CTCF and (E) Dendra2-CTCF clusters per cell for each transcription inhibitor. Number of cells analyzed for estimation is indicated below each condition. (F) Numbers of CTCF molecules per cluster for each transcription inhibitor (from N = 100 clusters for each condition). (G) Life times of Pol II clusters for each transcription inhibitor. Number of analyzed clusters is indicated below each condition. Scale bars represent 5 μm. Error bars show mean ± standard errors of the mean (s.e.m.). *, **, *** indicate statistical significance at P < 0.10, P < 0.05 and P < 0.01, unpaired two-tailed Student's t-tests.
Accordingly, we wondered whether transcription inhibition would affect the dynamics of CTCF and Pol II clusters. While the numbers of CTCF molecules per cluster were not affected by JQ1, they were slightly increased by transcription inhibitors (Figure 5F). Moreover, the lifetime of Pol II clusters was markedly increased after treatment with transcription inhibitors (Figure 5G), which is in agreement with a recent report (41,48,50). We could not measure the lifetime of Pol II clusters under JQ1 treatment, since it was difficult to capture Pol II clusters in this condition.
Taken together, transcription inhibition appears to have distinct effects on the formation of CTCF and Pol II clusters, modestly affecting CTCF clustering, but perturbing Pol II clustering. Transcription inhibition exhibited similar effects on the stabilization of already formed CTCF and Pol II clusters, however.
CTCF is required for the formation of phase-separated transcriptional condensates
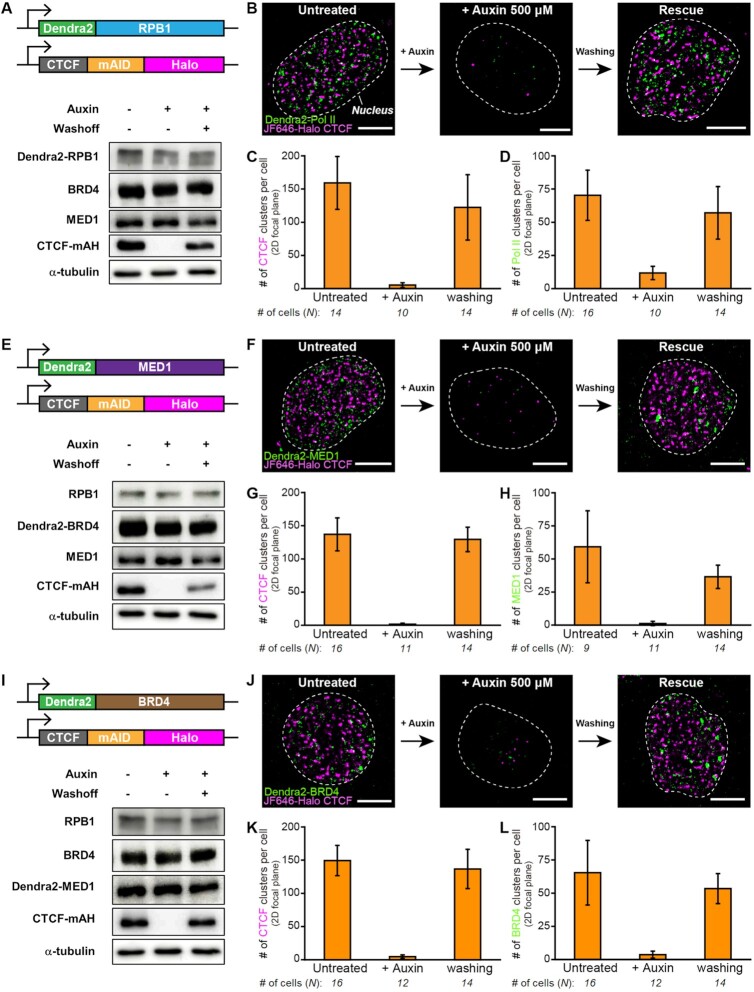
The C-terminal-domain of Pol II contains a series of YSPTSPS heptad repeats that are multiply-phosphorylated during the eukaryotic transcription cycle (88). For example, Ser5 phosphorylation is implicated in promoter clearance to shift from initiation to early elongation, while Ser2 phosphorylation occurs during productive elongation and the 3′-end processing of the transcript (88). Recent report suggested an RNA feedback model in which low levels of short RNAs produced during transcription initiation promote formation of transcriptional condensates, whereas high levels of longer RNAs produced during elongation can cause condensate dissolution (45). Given the HiChIP experiments using antibodies against Pol II with C-terminal domain Ser5 phosphorylation where hyperconnected spatial clusters of Pol II-centric chromatin interactions are severely attenuated by CTCF depletion, we examined whether the chromatin looping protein CTCF is required for the formation of Pol II clusters using super-resolution microscopy. As with CTCF protein tagged with the mAID-mClover3 cassette (Figure 1A and B), immunoblotting analysis of dual-labeled HCT116 cells showed that CTCF-mAID-Halo (CTCF-mAH) fusion protein was efficiently depleted in the presence of auxin, while washout of auxin readily rescued its expression (Figure 6A). The reversible expression of CTCF protein, which could be controlled by auxin treatment, was clearly reflected by the presence of CTCF clusters under fluorescence super-resolution microscopy: CTCF clusters were almost completely depleted by auxin treatment and efficiently restored after washout of auxin (Figure 6B and C, Supplementary Figures S9A). To our surprise, dual-color super-resolution imaging demonstrated that Pol II clusters almost completely disappeared when CTCF was depleted by auxin treatment for 24 h, but was restored to their original levels when CTCF expression was rescued after washout of auxin (Figure 6B and D, Supplementary Figure S9A). Further analysis at different time points showed that around half of all CTCF and Pol II clusters disappeared within as little as 20 min after auxin treatment, and nearly all disappeared after treatment with auxin for 6 and 12 h, respectively (Supplementary Figure S10A-S10C). Recovery of the Pol II cluster, like that of the CTCF cluster, occurred readily after washout of auxin and was almost completed by 6 h (Supplementary Figure S10D–F). The large degree of similarity between both clusters with respect to kinetics in depletion and recovery demonstrated a potent instructive function for CTCF in the formation of the Pol II cluster.
Figure 6.
Transcriptional condensates are dramatically disassembled and reassembled depending on CTCF degradation/restoration. (A, E, I) Dendra2 is homozygously labelled to endogenous (A) Pol II, (E) MED1 and (I) BRD4 wherein the mAID cassette and HaloTag are homozygously tagged to the C-terminus of CTCF. The OsTIR1 gene is integrated at the AAVS1 locus. Representative immunoblots show that the endogenous mAID/HaloTag CTCF (CTCF-mAH) is degraded completely upon auxin treatment and recovers after auxin removal (confirmed N = 3 times). (B, F, J) Representative dual-color super-resolved images of (B) Pol II, (F) MED1 and (J) BRD4 clusters, along with CTCF clusters, upon CTCF degradation and restoration. (C, G, K) Numbers of CTCF clusters per cell per 2D-focal plane for each cell line upon CTCF degradation and restoration. (D, H, L) Numbers of (D) Pol II, (H) MED1, and (L) BRD4 clusters per cell per 2D-focal plane upon CTCF degradation and restoration. Auxin (500 μM) was treated for 24 h for CTCF degradation and washed out for 24 h for CTCF restoration. Number of cells analyzed for estimation is indicated below each condition. Scale bars represent 5 μm.
We next tested whether the disappearance of Pol II clusters upon auxin treatment is simply due to decreased expression of Pol II protein complex, as in the case of CTCF clusters. However, immunoblotting analysis showed that Dendra2-tagged RPB1 protein levels were not affected by treatment or washout of auxin (Figure 6A). Moreover, expression of Dendra2-tagged RPB1 protein estimated by fluorescence signals was similar regardless of auxin treatment (Figure 6A; Supplementary Figure S10G and H). These results indicate that CTCF is crucial for the formation of Pol II clusters despite having little effect on Pol II protein levels.
The coactivator proteins MED1 and BRD4 are well known to form phase-separated transcriptional condensates along with Pol II to mediate multi-factor assembly for efficient transcription activation (39). In order to investigate whether CTCF has any effect on the formation of transcriptional condensates containing coactivator proteins, we generated additional dual-labeled homozygous HCT116 cell lines in which a mAID-Halo cassette was introduced to the C-terminus of CTCF and a transgene encoding Dendra2 protein was introduced to the N-terminus of MED1 and BRD4 genes (Supplementary Figure S11A–H). Depletion of endogenous CTCF protein upon auxin treatment and tagging of Dendra2 to MED1 and BRD4 proteins, respectively, were verified by immunoblotting (Figure 6E and I; Supplementary Figure S11D and H).
Consistent with previous reports in other cells (39,41), PALM analysis for Dendra2 clearly revealed the formation of MED1 and BRD4 clusters in the nuclei of living cells at similar quantities to that for Pol II clusters (Figure 6F, H, J and L). Furthermore, combined analysis of RNA FISH with super-resolution microscopy revealed that actively transcribed genes, exemplified by MYC, CYP24A1 and CDKN2AIPNL, which were selected based on RNA-seq analysis, were in close physical proximity to transcriptional condensates containing Pol II, MED1 and BRD4 (Supplementary Figure S12). Interestingly, both MED1 and BRD4 clusters, as for Pol II clusters, almost completely disappeared when CTCF was depleted by auxin treatment and returned when CTCF expression was rescued by washout of auxin (Figure 6F–H and J–L, Supplementary Figure S9B and C). These changes in cluster formation upon reversible CTCF depletion can be recapitulated with minimal cell division in which untreated cells were exposed to auxin for 3 h, after which auxin was withdrawn for 6 h (Supplementary Figure S13). The expression levels of MED1 and BRD4 were also not changed by CTCF depletion, which was confirmed by Western blot and fluorescence intensities of nuclear background (Figure 6E and I; Supplementary Figure S11I and J). All of these results indicated that the chromatin architectural protein CTCF plays a crucial instructive role in the formation of transcriptional condensates.
DISCUSSION
In mammalians cells, a typical TAD encompasses several transcription units and a large number of enhancers, and enhancer-promoter interactions are a hallmark of mammalian gene expression control (27). However, the molecular mechanisms of how regulatory signals are integrated and transmitted from enhancers to promoters has long been unresolved. The recently proposed loop-extrusion model, orchestrated by cohesin and CTCF, provides a compelling mechanism for regulatory elements to be in close physical proximity and for higher-order chromatin organization: while the action range of enhancers is restricted within CTCF-marked TAD boundaries, promoter choice by an enhancer can be determined by the binding of CTCF at promoters (93–102). In support of this, our analysis by combining a CTCF degron system and Pol II HiChIP revealed that loop connections among transcription regulatory elements are dramatically diminished upon depletion of CTCF. In addition, we demonstrated that CTCF is essential to form Pol II-dependent hyperconnected spatial clusters of chromatin interactions, which generally occurred at super-enhancers.
Meanwhile, researchers have also suggested that long-range enhancer–promoter interactions may be driven by an affinity between transcription factors bound to active regulatory elements (1,57,103–110). In addition, the intrinsically disordered regions of BRD4 and MED1, as well as Pol II, have been shown to form phase-separated condensates at super-enhancers that may help to stabilize the physical proximities between enhancers and promoters (39,40,52–57). However, treatment with JQ1, which is known to dissolve Pol II-containing phase condensates (39,41), did not disrupt enhancer-promoter interactions, but did reduce binding of BRD4 and Mediator at enhancers and strongly affected gene transcription (111). Other researchers have also demonstrated that depletion of Pol II and Mediator elicits little to no change in enhancer-promoter interactions (112) and that dissolution of phase condensate structures with 1,6-hexanediol does not perturb enhancer–promoter interactions, despite having a marked effect on global BRD4 and MED1 binding and gene expression (111). Altogether, these results suggest that looping structures between enhancers and promoters do not depend on high levels of transcriptional coactivators and Pol II that form phase-separated transcriptional condensates, but are likely maintained by the chromatin architectural proteins CTCF and cohesin.
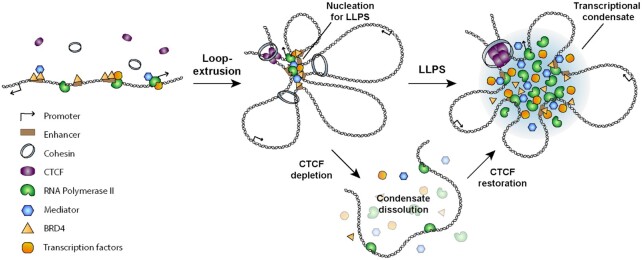
In contrast, however, our study demonstrated that CTCF-mediated chromatin looping functions as a critical structural determinant that promotes the assembly of transcriptional condensates. For transcriptional condensates to be formed at an active gene spot, interacting molecules are needed in excess of a critical concentration threshold in the initial nucleation process of phase separation. Given that enhancers and promoters contain large numbers of binding sites for transcription factors, hyperconnected spatial clusters of chromatin interactions, the formation of which depends on CTCF, can lead to the recruitment of enough transcription factors with which to exceed the threshold for condensate formation. Supporting this process, loop extrusion is thought to create spatial confinement that allows for local concentrations of protein complexes bound to intervening DNA sequences to increase. In light of these findings, we suspect that CTCF-demarcated loops can serve as local structural hubs for the accumulation of Pol II and other molecules associated with enhancers and promoters (Figure 7). Accordingly, sufficient enrichment of transcription machinery components within this pre-looped topology might drive LLPS to form transcriptional condensates, which then stabilize specific interactions between enhancers and promoters (Figure 7). In support of this hypothesis, our experimental evidence achieved through localization-based super-resolution microscopy revealed that transcriptional condensates composed of transcription coactivators and Pol II, which exhibits liquid properties, were completely dissolved upon CTCF degradation, but reestablished upon restoration of CTCF (Figure 7). It should also be noted that CTCF clustering, unlike Pol II clustering, is independent of LLPS and insensitive to the perturbation of transcription. Altogether, this implies that CTCF-mediated chromatin looping and formation of transcriptional condensates are distinct events, but associated in a structurally hierarchical manner.
Figure 7.
CTCF-mediated chromatin looping provides a topological framework for the formation of transcriptional condensates via LLPS. Chromatin looping mediated by loop-extrusion provides a spatial framework for the accumulation of transcriptional factors, coactivators, and RNA polymerase II. Sufficient enrichment of transcription machinery components within this pre-looped topology drives the formation of transcriptional condensates via LLPS.
Building on recent studies of the characteristics of transcriptional condensates, we provide an evidence showing that chromatin looping mediated by CTCF provides a spatial framework for the accumulation of transcription machinery and acts as a critical prerequisite for the assembly of transcriptional condensates.
DATA AVAILABILITY
All RNA-seq, ChIP-seq, in situ Hi-C and Pol II HiChIP data are available under the GEO under accession number GSE179545: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE179545.
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: H.K. and W.C. conceived the project and designed the experiments. M.K., C.Y., E.L. and J.J. performed genome editing to generate degron and reporter cell lines. B.Y., E.L. and J.J. generated RNA-seq, ChIP-seq, Hi-C and HiChIP data. Y.K., S.K., K.K. and W.J. analyzed RNA-seq, ChIP-seq, Hi-C and HiChIP data. R.L., H.S., B.M. and G.P. performed super-resolution fluorescence imaging and image data processing. R.L., M.K., Y.K., B.Y. and H.S. drafted the manuscript. H.K. and W.C. edited the manuscript and supervised the project.
Contributor Information
Ryanggeun Lee, Department of Biological Sciences, Korea Advanced Institute of Science and Technology (KAIST), 291 Daehak-ro, Yuseong-gu, Daejeon 34141, Korea; College of Natural Sciences, Korea Advanced Institute of Science and Technology (KAIST), 291 Daehak-ro, Yuseong-gu, Daejeon 34141, Korea.
Moo-Koo Kang, Department of Environmental Medical Biology, Institute of Tropical Medicine, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea; Brain Korea 21 PLUS Project for Medical Science, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea.
Yong-Jin Kim, Department of Environmental Medical Biology, Institute of Tropical Medicine, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea; Brain Korea 21 PLUS Project for Medical Science, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea.
Bobae Yang, Department of Environmental Medical Biology, Institute of Tropical Medicine, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea; Brain Korea 21 PLUS Project for Medical Science, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea.
Hwanyong Shim, Department of Biological Sciences, Korea Advanced Institute of Science and Technology (KAIST), 291 Daehak-ro, Yuseong-gu, Daejeon 34141, Korea.
Sugyung Kim, Department of Environmental Medical Biology, Institute of Tropical Medicine, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea; Brain Korea 21 PLUS Project for Medical Science, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea.
Kyungwoo Kim, Department of Environmental Medical Biology, Institute of Tropical Medicine, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea; Brain Korea 21 PLUS Project for Medical Science, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea.
Chul Min Yang, Department of Environmental Medical Biology, Institute of Tropical Medicine, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea.
Byeong-gyu Min, Department of Biological Sciences, Korea Advanced Institute of Science and Technology (KAIST), 291 Daehak-ro, Yuseong-gu, Daejeon 34141, Korea.
Woong-Jae Jung, Department of Environmental Medical Biology, Institute of Tropical Medicine, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea.
Eun-Chong Lee, Department of Environmental Medical Biology, Institute of Tropical Medicine, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea.
Jung-Sik Joo, Department of Environmental Medical Biology, Institute of Tropical Medicine, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea; Brain Korea 21 PLUS Project for Medical Science, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea.
Gunhee Park, Department of Biological Sciences, Korea Advanced Institute of Science and Technology (KAIST), 291 Daehak-ro, Yuseong-gu, Daejeon 34141, Korea.
Won-Ki Cho, Department of Biological Sciences, Korea Advanced Institute of Science and Technology (KAIST), 291 Daehak-ro, Yuseong-gu, Daejeon 34141, Korea; KI for Health Science and Technology (KIHST), Korea Advanced Institute of Science and Technology (KAIST), 291 Daehak-ro, Yuseong-gu, Daejeon 34141, Korea.
Hyoung-Pyo Kim, Department of Environmental Medical Biology, Institute of Tropical Medicine, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea; Brain Korea 21 PLUS Project for Medical Science, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea; Yonsei Genome Center, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea (NRF) grants funded by the Korean government (MSIP) [2017M3C9A5029978, 2018M3A9D3079290, 2020R1A2C2013258 to H.-P.K., 2020R1C1C1014599, 2020R1A4A3079755, 2019M3A9H1103711, 2019R1A6A1A10073887 to W.-K.C.]; Suh Kyungbae Foundation (to W.-K.C.). Funding for open access charge: National Research Foundation of Korea.
Conflict of interest statement. None declared.
REFERENCES
- 1.Oudelaar A.M., Higgs D.R.. The relationship between genome structure and function. Nat. Rev. Genet. 2021; 22:154–168. [DOI] [PubMed] [Google Scholar]
- 2.Schoenfelder S., Fraser P.. Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet. 2019; 20:437–455. [DOI] [PubMed] [Google Scholar]
- 3.Rowley M.J., Corces V.G.. Organizational principles of 3D genome architecture. Nat. Rev. Genet. 2018; 19:789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dekker J., Mirny L.. The 3D genome as moderator of chromosomal communication. Cell. 2016; 164:1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang L.H., Ghosh S., Noordermeer D.. TADs and their borders: free movement or building a wall?. J. Mol. Biol. 2020; 432:643–652. [DOI] [PubMed] [Google Scholar]
- 6.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B.. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012; 485:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S.et al.. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., Hoichman M., Parrinello H., Tanay A., Cavalli G.. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012; 148:458–472. [DOI] [PubMed] [Google Scholar]
- 9.Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J.et al.. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012; 485:381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rao S.S.P., Huang S.C., Glenn St Hilaire B., Engreitz J.M., Perez E.M., Kieffer-Kwon K.R., Sanborn A.L., Johnstone S.E., Bascom G.D., Bochkov I.D.et al.. Cohesin loss eliminates all loop domains. Cell. 2017; 171:305–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nora E.P., Goloborodko A., Valton A.L., Gibcus J.H., Uebersohn A., Abdennur N., Dekker J., Mirny L.A., Bruneau B.G.. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell. 2017; 169:930–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowen J.M., Young R.A.. SMC complexes link gene expression and genome architecture. Curr. Opin. Genet. Dev. 2014; 25:131–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haarhuis J.H.I., van der Weide R.H., Blomen V.A., Yanez-Cuna J.O., Amendola M., van Ruiten M.S., Krijger P.H.L., Teunissen H., Medema R.H., van Steensel B.et al.. The cohesin release factor WAPL restricts chromatin loop extension. Cell. 2017; 169:693–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Splinter E., Heath H., Kooren J., Palstra R.J., Klous P., Grosveld F., Galjart N., de Laat W.. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006; 20:2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou C., Zhao H., Tanimoto K., Dean A.. CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:20398–20403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parelho V., Hadjur S., Spivakov M., Leleu M., Sauer S., Gregson H.C., Jarmuz A., Canzonetta C., Webster Z., Nesterova T.et al.. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008; 132:422–433. [DOI] [PubMed] [Google Scholar]
- 17.Wendt K.S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T.et al.. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008; 451:796–801. [DOI] [PubMed] [Google Scholar]
- 18.Sanborn A.L., Rao S.S., Huang S.C., Durand N.C., Huntley M.H., Jewett A.I., Bochkov I.D., Chinnappan D., Cutkosky A., Li J.et al.. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E6456–E6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fudenberg G., Imakaev M., Lu C., Goloborodko A., Abdennur N., Mirny L.A.. Formation of chromosomal domains by loop extrusion. Cell Rep. 2016; 15:2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davidson I.F., Bauer B., Goetz D., Tang W., Wutz G., Peters J.M.. DNA loop extrusion by human cohesin. Science. 2019; 366:1338–1345. [DOI] [PubMed] [Google Scholar]
- 21.Kim Y., Shi Z., Zhang H., Finkelstein I.J., Yu H.. Human cohesin compacts DNA by loop extrusion. Science. 2019; 366:1345–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golfier S., Quail T., Kimura H., Brugues J.. Cohesin and condensin extrude DNA loops in a cell cycle-dependent manner. Elife. 2020; 9:e53885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ganji M., Shaltiel I.A., Bisht S., Kim E., Kalichava A., Haering C.H., Dekker C.. Real-time imaging of DNA loop extrusion by condensin. Science. 2018; 360:102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen A.S., Cattoglio C., Darzacq X., Tjian R.. Recent evidence that TADs and chromatin loops are dynamic structures. Nucleus. 2018; 9:20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen A.S., Pustova I., Cattoglio C., Tjian R., Darzacq X.. CTCF and cohesin regulate chromatin loop stability with distinct dynamics. Elife. 2017; 6:e25776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vos E.S.M., Valdes-Quezada C., Huang Y., Allahyar A., Verstegen M., Felder A.K., van der Vegt F., Uijttewaal E.C.H., Krijger P.H.L., de Laat W.. Interplay between CTCF boundaries and a super enhancer controls cohesin extrusion trajectories and gene expression. Mol. Cell. 2021; 81:3082–3095. [DOI] [PubMed] [Google Scholar]
- 27.Furlong E.E.M., Levine M.. Developmental enhancers and chromosome topology. Science. 2018; 361:1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bulger M., Groudine M.. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011; 144:327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malik S., Roeder R.G.. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 2010; 11:761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ong C.T., Corces V.G.. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 2011; 12:283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spitz F., Furlong E.E.. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012; 13:613–626. [DOI] [PubMed] [Google Scholar]
- 32.Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-Andre V., Sigova A.A., Hoke H.A., Young R.A.. Super-enhancers in the control of cell identity and disease. Cell. 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A.. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013; 153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabari B.R., Dall’Agnese A., Young R.A.. Biomolecular condensates in the nucleus. Trends Biochem. Sci. 2020; 45:961–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin Y., Brangwynne C.P.. Liquid phase condensation in cell physiology and disease. Science. 2017; 357:aaf4382. [DOI] [PubMed] [Google Scholar]
- 36.Stoeger T., Battich N., Pelkmans L.. Passive noise filtering by cellular compartmentalization. Cell. 2016; 164:1151–1161. [DOI] [PubMed] [Google Scholar]
- 37.Banani S.F., Lee H.O., Hyman A.A., Rosen M.K.. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017; 18:285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hyman A.A., Weber C.A., Julicher F.. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014; 30:39–58. [DOI] [PubMed] [Google Scholar]
- 39.Sabari B.R., Dall’Agnese A., Boija A., Klein I.A., Coffey E.L., Shrinivas K., Abraham B.J., Hannett N.M., Zamudio A.V., Manteiga J.C.et al.. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018; 361:aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hnisz D., Shrinivas K., Young R.A., Chakraborty A.K., Sharp P.A.. A phase separation model for transcriptional control. Cell. 2017; 169:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho W.K., Spille J.H., Hecht M., Lee C., Li C., Grube V., Cisse II.. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018; 361:412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boehning M., Dugast-Darzacq C., Rankovic M., Hansen A.S., Yu T., Marie-Nelly H., McSwiggen D.T., Kokic G., Dailey G.M., Cramer P.et al.. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 2018; 25:833–840. [DOI] [PubMed] [Google Scholar]
- 43.Chong S., Dugast-Darzacq C., Liu Z., Dong P., Dailey G.M., Cattoglio C., Heckert A., Banala S., Lavis L., Darzacq X.et al.. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science. 2018; 361:aar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu H., Yu D., Hansen A.S., Ganguly S., Liu R., Heckert A., Darzacq X., Zhou Q.. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature. 2018; 558:318–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henninger J.E., Oksuz O., Shrinivas K., Sagi I., LeRoy G., Zheng M.M., Andrews J.O., Zamudio A.V., Lazaris C., Hannett N.M.et al.. RNA-mediated feedback control of transcriptional condensates. Cell. 2021; 184:207–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nosella M.L., Forman-Kay J.D.. Phosphorylation-dependent regulation of messenger RNA transcription, processing and translation within biomolecular condensates. Curr. Opin. Cell Biol. 2021; 69:30–40. [DOI] [PubMed] [Google Scholar]
- 47.Guo Y.E., Manteiga J.C., Henninger J.E., Sabari B.R., Dall’Agnese A., Hannett N.M., Spille J.H., Afeyan L.K., Zamudio A.V., Shrinivas K.et al.. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature. 2019; 572:543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cho W.K., Jayanth N., English B.P., Inoue T., Andrews J.O., Conway W., Grimm J.B., Spille J.H., Lavis L.D., Lionnet T.et al.. RNA polymerase II cluster dynamics predict mRNA output in living cells. Elife. 2016; 5:e13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cisse II, Izeddin I., Causse S.Z., Boudarene L., Senecal A., Muresan L., Dugast-Darzacq C., Hajj B., Dahan M., Darzacq X.. Real-time dynamics of RNA polymerase II clustering in live human cells. Science. 2013; 341:664–667. [DOI] [PubMed] [Google Scholar]
- 50.Cho W.K., Jayanth N., Mullen S., Tan T.H., Jung Y.J., Cisse II.. Super-resolution imaging of fluorescently labeled, endogenous RNA Polymerase II in living cells with CRISPR/Cas9-mediated gene editing. Sci. Rep. 2016; 6:35949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi J.M., Holehouse A.S., Pappu R.V.. Physical principles underlying the complex biology of intracellular phase transitions. Annu. Rev. Biophys. 2020; 49:107–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamamoto T., Sakaue T., Schiessel H.. Slow chromatin dynamics enhances promoter accessibility to transcriptional condensates. Nucleic Acids Res. 2021; 49:5017–5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dean A., Larson D.R., Sartorelli V.. Enhancers, gene regulation, and genome organization. Genes Dev. 2021; 35:427–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heist T., Fukaya T., Levine M.. Large distances separate coregulated genes in living Drosophila embryos. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:15062–15067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boija A., Klein I.A., Sabari B.R., Dall’Agnese A., Coffey E.L., Zamudio A.V., Li C.H., Shrinivas K., Manteiga J.C., Hannett N.M.et al.. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 2018; 175:1842–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Steensel B., Furlong E.E.M.. The role of transcription in shaping the spatial organization of the genome. Nat. Rev. Mol. Cell Biol. 2019; 20:327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hua P., Badat M., Hanssen L.L.P., Hentges L.D., Crump N., Downes D.J., Jeziorska D.M., Oudelaar A.M., Schwessinger R., Taylor S.et al.. Defining genome architecture at base-pair resolution. Nature. 2021; 595:125–129. [DOI] [PubMed] [Google Scholar]
- 58.Natsume T., Kiyomitsu T., Saga Y., Kanemaki M.T.. Rapid protein depletion in human cells by auxin-inducible degron tagging with short homology donors. Cell Rep. 2016; 15:210–218. [DOI] [PubMed] [Google Scholar]
- 59.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmidl C., Rendeiro A.F., Sheffield N.C., Bock C.. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat. Methods. 2015; 12:963–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mumbach M.R., Rubin A.J., Flynn R.A., Dai C., Khavari P.A., Greenleaf W.J., Chang H.Y.. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods. 2016; 13:919–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li B., Dewey C.N.. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011; 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramirez F., Dundar F., Diehl S., Gruning B.A., Manke T.. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014; 42:W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Servant N., Varoquaux N., Lajoie B.R., Viara E., Chen C.J., Vert J.P., Heard E., Dekker J., Barillot E.. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015; 16:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yardimci G.G., Ozadam H., Sauria M.E.G., Ursu O., Yan K.K., Yang T., Chakraborty A., Kaul A., Lajoie B.R., Song F.et al.. Measuring the reproducibility and quality of Hi-C data. Genome Biol. 2019; 20:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Durand N.C., Shamim M.S., Machol I., Rao S.S., Huntley M.H., Lander E.S., Aiden E.L.. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 2016; 3:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Crane E., Bian Q., McCord R.P., Lajoie B.R., Wheeler B.S., Ralston E.J., Uzawa S., Dekker J., Meyer B.J.. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015; 523:240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kruse K., Hug C.B., Vaquerizas J.M.. FAN-C: a feature-rich framework for the analysis and visualisation of chromosome conformation capture data. Genome Biol. 2020; 21:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bhattacharyya S., Chandra V., Vijayanand P., Ay F.. Identification of significant chromatin contacts from HiChIP data by FitHiChIP. Nat. Commun. 2019; 10:4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Flyamer I.M., Illingworth R.S., Bickmore W.A.. Coolpup.py: versatile pile-up analysis of Hi-C data. Bioinformatics. 2020; 36:2980–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J.et al.. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019; 47:D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Petrovic J., Zhou Y., Fasolino M., Goldman N., Schwartz G.W., Mumbach M.R., Nguyen S.C., Rome K.S., Sela Y., Zapataro Z.et al.. Oncogenic notch promotes long-range regulatory interactions within hyperconnected 3D cliques. Mol. Cell. 2019; 73:1174–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grimm J.B., English B.P., Chen J., Slaughter J.P., Zhang Z., Revyakin A., Patel R., Macklin J.J., Normanno D., Singer R.H.et al.. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods. 2015; 12:244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chudakov D.M., Lukyanov S., Lukyanov K.A.. Tracking intracellular protein movements using photoswitchable fluorescent proteins PS-CFP2 and Dendra2. Nat. Protoc. 2007; 2:2024–2032. [DOI] [PubMed] [Google Scholar]
- 80.Betzig E., Patterson G.H., Sougrat R., Lindwasser O.W., Olenych S., Bonifacino J.S., Davidson M.W., Lippincott-Schwartz J., Hess H.F.. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006; 313:1642–1645. [DOI] [PubMed] [Google Scholar]
- 81.Rust M.J., Bates M., Zhuang X.. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods. 2006; 3:793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Serge A., Bertaux N., Rigneault H., Marguet D.. Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat. Methods. 2008; 5:687–694. [DOI] [PubMed] [Google Scholar]
- 83.Andrews J.O., Conway W., Cho W.K., Narayanan A., Spille J.H., Jayanth N., Inoue T., Mullen S., Thaler J., Cisse II.. qSR: a quantitative super-resolution analysis tool reveals the cell-cycle dependent organization of RNA Polymerase I in live human cells. Sci. Rep. 2018; 8:7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ester M., Kriegel H.P., Sander J., Xu X.. A density-based algorithm for discovering clusters in large spatial databases with noise. Proceedings of the Second International Conference on Knowledge Discovery and Data Mining 1996. 1996; IAAA Press; 226–231. [Google Scholar]
- 85.McKinney S.A., Joo C., Ha T.. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 2006; 91:1941–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hyle J., Zhang Y., Wright S., Xu B., Shao Y., Easton J., Tian L., Feng R., Xu P., Li C.. Acute depletion of CTCF directly affects MYC regulation through loss of enhancer-promoter looping. Nucleic Acids Res. 2019; 47:6699–6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stik G., Vidal E., Barrero M., Cuartero S., Vila-Casadesus M., Mendieta-Esteban J., Tian T.V., Choi J., Berenguer C., Abad A.et al.. CTCF is dispensable for immune cell transdifferentiation but facilitates an acute inflammatory response. Nat. Genet. 2020; 52:655–661. [DOI] [PubMed] [Google Scholar]
- 88.Phatnani H.P., Greenleaf A.L.. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006; 20:2922–2936. [DOI] [PubMed] [Google Scholar]
- 89.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A.et al.. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Narayanan A., Meriin A., Andrews J.O., Spille J.H., Sherman M.Y., Cisse II.. A first order phase transition mechanism underlies protein aggregation in mammalian cells. Elife. 2019; 8:e39695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gu B., Comerci C.J., McCarthy D.G., Saurabh S., Moerner W.E., Wysocka J.. Opposing effects of cohesin and transcription on CTCF organization revealed by super-resolution imaging. Mol. Cell. 2020; 80:699–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hilbert L., Sato Y., Kuznetsova K., Bianucci T., Kimura H., Julicher F., Honigmann A., Zaburdaev V., Vastenhouw N.L.. Transcription organizes euchromatin via microphase separation. Nat. Commun. 2021; 12:1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.de Wit E., Vos E.S., Holwerda S.J., Valdes-Quezada C., Verstegen M.J., Teunissen H., Splinter E., Wijchers P.J., Krijger P.H., de Laat W.. CTCF binding polarity determines chromatin looping. Mol. Cell. 2015; 60:676–684. [DOI] [PubMed] [Google Scholar]
- 94.Dowen J.M., Fan Z.P., Hnisz D., Ren G., Abraham B.J., Zhang L.N., Weintraub A.S., Schujiers J., Lee T.I., Zhao K.et al.. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014; 159:374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gomez-Marin C., Tena J.J., Acemel R.D., Lopez-Mayorga M., Naranjo S., de la Calle-Mustienes E., Maeso I., Beccari L., Aneas I., Vielmas E.et al.. Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:7542–7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guo Y., Xu Q., Canzio D., Shou J., Li J., Gorkin D.U., Jung I., Wu H., Zhai Y., Tang Y.et al.. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell. 2015; 162:900–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hanssen L.L.P., Kassouf M.T., Oudelaar A.M., Biggs D., Preece C., Downes D.J., Gosden M., Sharpe J.A., Sloane-Stanley J.A., Hughes J.R.et al.. Tissue-specific CTCF-cohesin-mediated chromatin architecture delimits enhancer interactions and function in vivo. Nat. Cell Biol. 2017; 19:952–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lupianez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., Horn D., Kayserili H., Opitz J.M., Laxova R.et al.. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015; 161:1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Narendra V., Rocha P.P., An D., Raviram R., Skok J.A., Mazzoni E.O., Reinberg D. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015; 347:1017–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Oh S., Shao J., Mitra J., Xiong F., D’Antonio M., Wang R., Garcia-Bassets I., Ma Q., Zhu X., Lee J.H.et al.. Enhancer release and retargeting activates disease-susceptibility genes. Nature. 2021; 595:735–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Paliou C., Guckelberger P., Schopflin R., Heinrich V., Esposito A., Chiariello A.M., Bianco S., Annunziatella C., Helmuth J., Haas S.et al.. Preformed chromatin topology assists transcriptional robustness of Shh during limb development. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:12390–12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Symmons O., Pan L., Remeseiro S., Aktas T., Klein F., Huber W., Spitz F.. The Shh topological domain facilitates the action of remote enhancers by reducing the effects of genomic distances. Dev. Cell. 2016; 39:529–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kagey M.H., Newman J.J., Bilodeau S., Zhan Y., Orlando D.A., van Berkum N.L., Ebmeier C.C., Goossens J., Rahl P.B., Levine S.S.et al.. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010; 467:430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Weintraub A.S., Li C.H., Zamudio A.V., Sigova A.A., Hannett N.M., Day D.S., Abraham B.J., Cohen M.A., Nabet B., Buckley D.L.et al.. YY1 is a structural regulator of enhancer-promoter loops. Cell. 2017; 171:1573–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim S., Shendure J.. Mechanisms of Interplay between Transcription Factors and the 3D Genome. Mol. Cell. 2019; 76:306–319. [DOI] [PubMed] [Google Scholar]
- 106.Krivega I., Dale R.K., Dean A.. Role of LDB1 in the transition from chromatin looping to transcription activation. Genes Dev. 2014; 28:1278–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Deng W., Lee J., Wang H., Miller J., Reik A., Gregory P.D., Dean A., Blobel G.A.. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012; 149:1233–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Drissen R., Palstra R.J., Gillemans N., Splinter E., Grosveld F., Philipsen S., de Laat W.. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 2004; 18:2485–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vakoc C.R., Letting D.L., Gheldof N., Sawado T., Bender M.A., Groudine M., Weiss M.J., Dekker J., Blobel G.A.. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol. Cell. 2005; 17:453–462. [DOI] [PubMed] [Google Scholar]
- 110.Tolhuis B., Palstra R.J., Splinter E., Grosveld F., de Laat W.. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 2002; 10:1453–1465. [DOI] [PubMed] [Google Scholar]
- 111.Crump N.T., Ballabio E., Godfrey L., Thorne R., Repapi E., Kerry J., Tapia M., Hua P., Lagerholm C., Filippakopoulos P.et al.. BET inhibition disrupts transcription but retains enhancer-promoter contact. Nat. Commun. 2021; 12:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.El Khattabi L., Zhao H., Kalchschmidt J., Young N., Jung S., Van Blerkom P., Kieffer-Kwon P., Kieffer-Kwon K.R., Park S., Wang X.et al.. A pliable mediator acts as a functional rather than an architectural bridge between promoters and enhancers. Cell. 2019; 178:1145–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ramirez F., Bhardwaj V., Arrigoni L., Lam K.C., Gruning B.A., Villaveces J., Habermann B., Akhtar A., Manke T.. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun. 2018; 9:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq, ChIP-seq, in situ Hi-C and Pol II HiChIP data are available under the GEO under accession number GSE179545: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE179545.