Abstract
Trafficking of intracellular cargo is essential to cellular function and can be defective in pathological states including cancer and neurodegeneration. Tools to quantify intracellular traffic are thus necessary for understanding this fundamental cellular process, studying disease mechanisms, and testing the effects of therapeutic pharmaceuticals. In this article we introduce an algorithm called QuoVadoPro that autonomously quantifies the movement of fluorescently tagged intracellular cargo. QuoVadoPro infers the extent of intracellular motility based on the variance of pixel illumination in a series of time-lapse images. The algorithm is an unconventional approach to the automatic measurement of intracellular traffic and is suitable for quantifying movements of intracellular cargo under diverse experimental paradigms. QuoVadoPro is particularly useful to measure intracellular cargo movement in non-neuronal cells, where cargo trafficking occurs as short movements in mixed directions. The algorithm can be applied to images with low temporal or spatial resolutions and to intracellular cargo with varying shapes or sizes, like mitochondria or endoplasmic reticulum: situations in which conventional methods such as kymography and particle tracking cannot be applied. In this article we present a stepwise protocol for using the QuoVadoPro software, illustrate its methodology with common examples, discuss critical parameters for reliable data analysis, and demonstrate its use with a previously published example.
Keywords: autonomous software, intracellular trafficking, motility quantification, pixel occupancy
INTRODUCTION
Intracellular transport of proteins and organelles is vital for maintaining cellular organization and physiology. As local metabolic needs transiently arise within a cell or as environmental cues change, the rapid trafficking of intracellular cargoes enables a swift reorganization of the cell’s proteins and organelles to adapt it to the new conditions. In eukaryotes, the rapid movement and reorganization of intracellular cargoes is powered by molecular motors such as kinesin, dynein, and myosin, which carry the cargo on a network of microtubules and actin filaments spanning the entire cytosol (Hartman & Spudich, 2012; Hirokawa, 1998; Miki, Okada, & Hirokawa, 2005; Pilling, Horiuchi, Lively, & Saxton, 2006; Reck-Peterson, Redwine, Vale, & Carter, 2018; Schnapp & Reese, 1989; Titus, 2018; Verhey, Kaul, & Soppina, 2011).
Multiple factors affect the rates and directions of intracellular traffic. The motors involved and the principles that govern their movement differ from cargo to cargo and are further shaped by cellular health, intracellular location, and signaling mechanisms. Changes in intracellular trafficking can influence pathologies such as cancer (Caino et al., 2016; Goldenring, 2013; Parachoniak & Park, 2012) and neurodegeneration (Baloh, Schmidt, Pestronk, & Milbrandt, 2007; Collard, Cote, & Julien, 1995; Gunawardena & Goldstein, 2001; Hafezparast et al., 2003; Williamson & Cleveland, 1999). Studying the rates of intracellular traffic in both normal and disease states is thus vital to understanding an essential cell biological process, elucidating disease mechanisms and revealing potential points of therapeutic intervention (Misgeld & Schwarz, 2017; Schwarz, 2013; Shlevkov etal., 2019; Vale, 2003).
We have developed two algorithms, called Kymolyzer (Basu et al., 2020) and QuoVadoPro, to quantify intracellular movement. Kymolyzer is semi-autonomous and utilizes kymography to analyze intracellular trafficking. Kymolyzer is useful to measure the speeds, directionality, and flux of intracellular cargo in cells where intracellular movement occurs along a specified path, such as that along the aligned microtubules found in neuronal axons and dendrites. These conditions, however, are not met in most non-neuronal cell lines or in neuronal cell bodies. In those places, the intracellular cargoes move in short processive runs on microtubules and actin filaments that have mixed orientations. To analyze such movements characterized by short runs in heterogeneous directions, we have developed QuoVadoPro. QuoVadoPro is fully autonomous and uses the variance in pixel occupancy in a series of time-lapse images as an indicator of the extent of movement of a fluorescently tagged intracellular cargo.
In this article, we describe QuoVadoPro. QuoVadoPro is useful to measure intracellular traffic in most cell types and is especially effective in quantifying movement in cells whose microtubules are not in parallel arrays and thus are not readily analyzable by kymography. Traditionally, automated object detection and tracking has been used to quantify intracellular trafficking in such cell lines (Chenouard et al., 2014; Jaqaman et al., 2008; Lee et al., 2019; Wasim & Treanor, 2018). However, object-tracking algorithms are often computationally intensive, are not easily adaptable to tracking different organelles, and require images of high temporal and spatial resolutions. Most significantly, object tracking is not reliable in tracking the movement of reticular or partially reticular structures such as mitochondria or endoplasmic reticulum (ER). Manually tracking the movement of individual mitochondria in every frame of a movie (Caino et al., 2016; Caino et al., 2017) has overcome some of the challenges of automated detection and tracking. These approaches, however, are extremely low throughput and may ignore mitochondria in a fused reticulum.
QuoVadoPro takes a nontraditional approach that does not employ conventional object detection and tracking. Through QuoVadoPro, we infer the motility along an x-y plane by analyzing the variance in pixel illumination over time. QuoVadoPro has several advantages over conventional object tracking: it is not influenced by the heterogeneity of sizes, shapes, or reticular structure of objects to be tracked; it can infer motility from images with low temporal and spatial resolutions and from images with high object densities; it is computationally light; and it can be used to monitor the movement of different types of cargo with minimal user-driven changes to input parameters. QuoVadoPro is packaged as an ImageJ-based macro set that can be installed and used through a graphical user interface. It can accept most microscopy file formats and outputs motility scores and two-dimensional (2D) motility heatmaps for visual representation.
QuoVadoPro, AN AUTONOMOUS TOOL TO MEASURE INTRACELLULAR DYNAMICS USING TEMPORAL VARIANCE
QuoVadoPro is a new method to quantify intracellular movement. In this algorithm, we use the temporal variance in pixel illumination by fluorescently tagged intracellular structures to infer on motility within a cell. It provides a quick, fully autonomous, and computationally light way to measure intracellular movements.
Time-lapse images of fluorescently tagged intracellular objects with high motility display a high pixel variance over time. This is because the pixels have a high probability of being transiently occupied by moving objects. On the other hand, time-lapse images of less motile intracellular objects have pixels that either are constantly occupied by the objects or are empty, and thereby have low variance in intensity over time. Thus, although not a direct measure of motility, temporal variance in pixel illumination serves as a proxy for motility. Based on this principle, QuoVadoPro can quantify intracellular traffic under diverse experimental and imaging conditions, even in samples that are otherwise poorly analyzed by conventional methods such as kymography or automated object tracking. Further, by normalizing the variance in pixel illumination with the sum of pixel illumination during a time lapse, QuoVadoPro distinguishes processive movements from short discontinuous back-and-forth movements (also referred to as jitters or jiggle) and weights the former more heavily.
The algorithm works as a set of three macros that can be installed in Fiji (a distribution of ImageJ that is bundled with commonly used plugins) and can be run sequentially through a user-friendly graphical interface. In the first step, the user selects an image series to be analyzed (representing a time-lapse sequence of fluorescently tagged intracellular objects). During analysis, the user can choose to have parts of the image analyzed separately (for example, if a single image has two cells that need to be quantified separately) or to have the image analyzed as a whole. QuoVadoPro then extracts the part of the image (if indicated) or uses the whole image to create a binary mask of the fluorescently tagged objects in all the frames of the time lapse. In the second step, the algorithm analyzes the variance in the binary image stack created in step 1 and creates a 2D heatmap of movement. A motility value is then derived from such a heatmap for quantification. The optional third step can be used to automatically collate data from multiple such quantifications through a reiterative loop.
QuoVadoPro uses the contrast of the fluorescently tagged objects over their local backgrounds to convert them into binary masks. The algorithm can thus work robustly irrespective of object shape, size, and absolute fluorescence. In addition, QuoVadoPro does not rely on tracking movements of individual objects from one frame to another and can thus work on images irrespective of frame-to-frame object displacements (governed by temporal and spatial resolutions and movement speeds of the intracellular objects). These features render QuoVadoPro a versatile software to analyze intracellular traffic with minimal user supervision.
For those starting out with QuoVadoPro, a sample file is provided. The file contains a time-lapse image stack of a Cos-7 cell expressing a fluorescent tag that marks the mitochondria (mito-DsRed, Addgene 55838) and a cytosolic GFP (meGFP-N1, Addgene 54767) that helps visualize the cell boundary. To use this sample file, the user should download it into a separate folder on their computer before processing the file through QuoVadoPro. The cell depicted in the sample file is used as an example in the protocol to illustrate the steps and features of QuoVadoPro in several figures referred to within the protocol. A video made from the cell shown in the sample file is also provided as supplementary material (Movie S1). The video depicts moving mitochondria in the Cos-7 cell along with the corresponding motility heatmap.
Necessary Resources
Hardware
Workstation with internet access
Software
Please read Instructions_Readme file and download the folder
“QuoVadoPro_<date>.zip” from https://github.com/ThomasSchwarzLab/QuoVadoPro
Sample File
https://drive.google.com/file/d/1yJZzhlsAGEFrIBoIS0iB6nUDzguzjQcu/view?usp=sharing
- Installation instructions:
- Download and install Fiji from https://imagej.net/Fiji/Downloads.
- Download the zipped folder named QuoVadoPro_<date>.zip from https://github.com/ThomasSchwarzLab/QuoVadoPro, and extract it into a separate folder anywhere easily accessible on the computer (this folder can be deleted after installation).
- From Fiji, go to the following tab: Plugins > Macros > run. In the file selection window that opens up, select the file named installer_QuoVadoPro_.ijm within the downloaded and unzipped folder, and run it.
- Restart Fiji to complete macro installation. Following the restart, a new tab should appear as follows: Plugins > Macros > QuoVadoPro.
- If images have drift, the optional installation of the Fiji registration plugin Stack-Reg (https://imagej.net/StackReg) may be needed. To install this plugin, go to the following tab in Fiji: Help > Update. Then click on the “manage update sites” button in the update window, select the “BIG-EPFL” update site from the list of update sites and click the “Close” button to go back to the update window. In the update window, press the “Apply changes” button. Then restart Fiji.
-
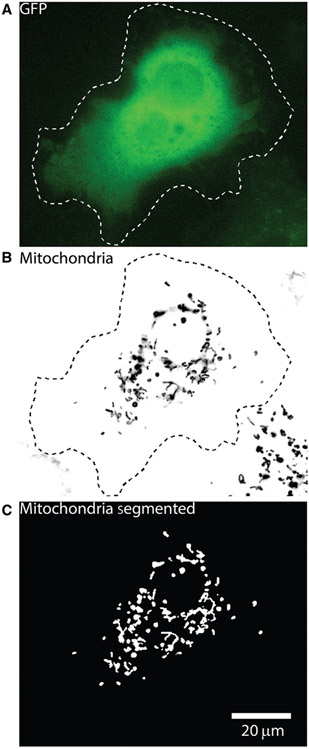
Step 1: Segmentation of objects by local-intensity-based thresholding. As the first step of QuoVadoPro, the user selects a file containing images representing a time-lapse sequence of fluorescently tagged intracellular objects. The user then has the choice to outline a specific region of interest (ROI) in the image or use the whole image for analysis. All fluorescently tagged objects in the whole image or defined ROI are converted to binary masks. This conversion (known as thresholding or segmenting) ensures that the quantification done by QuoVadoPro is independent of absolute intensities. To create the binary masks, the fluorescent objects are thresholded based on their signal over the local background (Fig. 1). At this step, the user has the option of using multiple tools to remove the background from the images while creating the binary masks of the objects.
Instructions for running step 1:- To run step 1 of QuoVadoPro, go to the following tab in Fiji: Plugins>Macros>QuoVadoPro>Step1_preprocess_Cells_IntensitybasedSegmention. A file selection window will open to select the file containing the movie (time lapse) to be analyzed.
Figure 1.
Example of a Cos-7 cell with fluorescently tagged mitochondria, illustrating the object segmentation carried out during step 1 of QuoVadoPro. A Cos-7 cell transfected with GFP to facilitate determining the outline of the cell (A) and with a red fluorescent marker of the mitochondrial matrix, mitoDsRed (B), is shown. The cell was imaged at 3.3 Hz for 3 min. Mitochondria at each time frame are segmented into binary masks by local intensity-based thresholding. During this step, the user has the option to select a region of interest (ROI), such as the cell outline (dotted line). All objects outside the ROI are removed. The images in the time-lapse sequence are then corrected for local background. Following this, the mitochondria are segmented using a predefined intensity threshold (C). Due to local background correction, all mitochondria are faithfully segmented as binary objects, irrespective of their individual intensities.
QuoVadoPro uses the bioformats reader in Fiji to open image files. Most microscopy image file formats are supported by this reader. In the case of an unsupported file format, the user should save the file as a multipage tiff stack before opening it in step 1.
The user can choose to run step 1 on an already opened file. However, we recommend opening the file using the file selection window that is triggered at step 1a. This ensures that QuoVadoPro can read the metadata and file location correctly.
b. After selecting the movie file, the user is asked to draw a coarse outline marking the cell periphery (ROI). The outline should include all the organelles to be tracked (Fig. 1A and B). If the movie has multiple channels or z planes, the macro will ask the user to select one channel and one z plane. If no selection or ROI is indicated, the macro will proceed to analyze the full frame of the currently active z plane and channel. If the user wants to analyze multiple z planes, they can choose to either run QuoVadoPro on each individual z plane or use a single projection for all the z planes.
QuoVadoPro analyzes the movement in one 2D plane (z plane). Given the thickness of the cell imaged and the mode of imaging, the user may choose to supply one z plane or a projection of all the z planes for analysis. To analyze a projection of all the z slices, the user must perform the z projection and save the time lapse as a multi-image tiff stack before running step 1 of QuoVadoPro. Typically, most cell lines that are used to monitor intracellular trafficking (such as Cos-7 or U2OS) have a relatively thin cytoplasm, most of which can be captured in one optical section using an epifluorescence microscope, and therefore do not require a z projection. However, certain cell lines have thicker cytoplasm (for example, dividing HeLa cells), in which case multiple confocal optical sections are needed to capture the cytoplasm, and thus may need to be projected after acquisition and before analysis.
- The image or the ROI (if selected in step 1b) is then subjected to a local background subtraction to correct for local variations in background and illumination. The resulting image is then adjusted to a standardized intensity scale (16-bit). This processing standardizes images of different bit depths (8-bit, 16-bit, and 32-bit). The user is then asked to supply a threshold intensity value for segmenting the objects in the movie, along with additional optional inputs to ensure that the binary masks do not incorporate noise artifacts. The following values are asked for:
-
Gaussian smoothing radius for background removal: Local background correction for each frame is done by subtracting a background image from the original image. The background image is generated by smoothing the original image through a Gaussian disk of a defined radius. When the smoothed background image is subtracted from the original image, diffuse background fluorescence is eliminated while the sharp signals in the original image are retained. Each pixel in the resulting image is also normalized for its local background.The “Gaussian smoothing radius” thus dictates the strength of the background removal. A lower Gaussian smoothing radius results in a strong background correction, whereas a higher radius reduces the strength of the background correction. By default, the radius is set to 5 pixels.The Gaussian smoothing radius necessary for optimal background correction depends on the signal-to-noise ratio (SNR) of the image. The user should decide on a value empirically. A value that is set too high (low background correction) will cause large, diffuse background objects to be included in the binary mask. Conversely, a Gaussian radius value that is set too low (high background correction) will cause over-amplification of object edges and thus may result in artifacts in the binary mask. To prevent the over-amplification of object edges, the “Gaussian smoothing radius” should be set to a value that is at minimum equal to the average cross-sectional distance of the fluorescently tagged objects.
- ii. Threshold level: This value is the threshold intensity used for segmenting the fluorescent objects after local background subtraction and standardization of the image intensity to a 16-bit range. The user can decide on a threshold anywhere from 0 to 65,535 (maximum intensity in a 16-bit image). The default threshold is set to 5000, and the working value that needs to be set is dictated by the SNR of the image.
- For images with high SNR, a strong background subtraction can be applied. In such cases, the background intensities will be greatly reduced (brought close to 0) and the threshold for segmentation can thus be set to a low value (just above the background).
- If the image is highly noisy (or grainy), the Gaussian radius for background removal should be set to a high value (i.e., weak background subtraction) to prevent the amplification of noise pixels. In this case, the background intensities will be equalized throughout the image, but low levels of noise may remain. The threshold value should thus be set relatively high, enough to remove any remaining background while retaining the fluorescent objects.
-
Time smoothing: In images with a high degree of noise, choosing a threshold that completely removes all background pixels may not be possible even after local background subtraction. In such cases, the user can choose to select a threshold that captures most of the fluorescent objects, even if it retains some background pixels. To further remove the background, the user can then utilize the time-smoothing feature in QuoVadoPro. This feature removes any pixels that were not constantly illuminated in the defined set of frames. The time-smoothing feature works on the principle that noise pixels will not be detected at the same position in consecutive frames, whereas the position of fluorescent objects will not change as fast.This value can be kept as 1 (no time smoothing) for images with high SNR and increased if noise is detected after segmentation. To use the time-smoothing feature (i.e., for any value > 1), the images must have a high temporal resolution (i.e., the frame-to-frame displacement of each object should be much lower than the size of the object). This ensures the fluorescently tagged objects illuminate overlapping sets of pixels in consecutive frames, whereas noise pixels do not.
-
It is important to note that both local background subtraction and time smoothing in QuoVadoPro correct for noise pixels that appear randomly due to camera noise or indistinct background fluorescence. These features, however, will not eliminate background caused by unintended fluorescent objects (for example, autofluorescent cytosolic granules), if visible.
Minimum particle size: In addition to time smoothing, QuoVadoPro gives the user another option to remove noise pixels by filtering the images for a minimum particle size. The minimum particle size allows the user to define a lower area limit for all objects detected in the binary mask, such that any object occupying an area less than this limit is removed. By default, this value is set to 1 pixel. The user can choose to increase it if small clusters of pixels are seen to be illuminated in the binary mask due to noise. The minimum size filter should be used in conditions where the objects to be masked have a substantially larger area than noise clusters.
As discussed later, it is important that the binary masks created have minimal noise (pixels that do not represent fluorescent objects). To ensure optimal noise removal, the user should run step 1 multiple times and identify the optimal parameters for background removal and thresholding with every new imaging condition. Once the parameters have been decided, the user should not change them between samples of the same experiment.
d. Post segmentation, the macro will ask the user to fine-tune the ROI to be analyzed (made in step 1b). If no specific ROI was selected in step 1b (i.e., the whole frame was considered), the user should click “OK” and move on to the next step. However, if the user selected an ROI in step 1b, the edge of the selection may have been erroneously amplified during background subtraction and segmented within the binary mask. This is especially true if there is high background fluorescence (for example, autofluorescence from growth medium), and it can introduce artifacts in the quantifications. In this case, the user should amend the selection (ROI) at this step to remove any edge artifacts, if present.
- e. At the end of step 1, the macro will save the binarized image (Fig. 1C) and the thresholding information in a folder created at the same location as the image, with the same name as the image along with a time stamp.
-
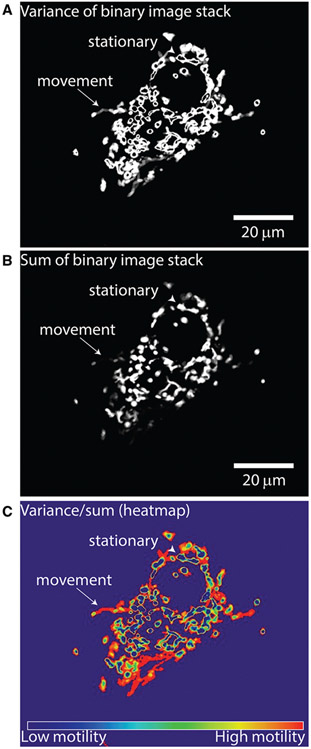
Step 2: Construction of motility heatmap and quantification. In step 2 of QuoVadoPro, the binarized video frames (from step 1) are used to generate a motility heatmap, depicting the movement of fluorescent objects. During this step, the variance in illumination over time and the total time of illumination (obtained by summation of intensities over time) is calculated for each binary pixel. The final motility heatmap is constructed as the ratio of an image representing the variance in pixel illumination (Fig. 2A) to an image representing the total time (sum) of pixel illumination (Fig. 2B). Parts of the image containing moving objects display high variance and low sum and thereby appear as hot spots on the heatmap. Conversely, parts of the image containing stationary objects have less variation in pixel residence and thereby have low variance and high sum, and appear as cold spots on the heat map (Fig. 2C). Pixels that were never occupied by an object have a sum of 0 and are excluded from the analysis. They are assigned a value of NaN (not a number). For representation purposes, they appear as cold pixels on the heatmap.Normalizing the variance in pixel illumination to the total time of pixel illumination (sum) also allows QuoVadoPro to give more weight to processive runs than to back-and-forth jitters (see Guidelines for Understanding Results for more details).The motility heatmap is used to derive a motility score for the whole image. To calculate the motility score, the intensities of the heatmap pixels are summed and normalized to the object area (derived from the number of pixels occupied by the binary mask in the first frame). The normalization for the total object area factors out differences in number or size of objects between images.The user should note that although the motility score is normalized for the number and area of objects, QuoVadoPro should not be utilized to analyze samples with highly varying object numbers (see Commentary for more details).Instructions for running step 2:
-
a. To run step 2 of QuoVadoPro, go to the following tab in Fiji: Plugins>Macros>QuoVadoPro>Step2_variance_Calculation. A folder-selection window will appear in which the user can select the folder containing the binarized image stack created in step 1. This step can also be run directly on the binarized image that is already open at the end of step 1.
b. After the user selects the folder, the macro will automatically find and open the binary image stack and ask the user for the number of substacks to split the time-lapse movie into. By default, this value is kept as 1, whereby the macro will process all the images in the movie as a single stack. In the case of long movies or movies in which the experiment may have caused motility to change over time, the user may want to break up the movie into substacks to gain information on how movement changed during the course of the movie.
- c. During processing, the macro will create the variance image and divide it by the sum image as described before. The final image is represented as a 32-bit heatmap (Fig. 2C). As this step is finished, the quantification is shown in a text window and is automatically saved as a text file in the same folder as the binary image. The macro then asks the user to set a display range that is used to convert the 32-bit heatmap into RGB images. It is important that all the images be set to the same display range within one experiment. The original variance image and the RGB heatmap are stored in the same image folder.
-
4. Step 3: Data collation. At this step, the user can interactively consolidate the quantifications generated for different cells into one table.Instructions for running step 3:To run step 3 of QuoVadoPro, go to the following tab in Fiji: Plugins > Macros >QuoVadoPro>Step3_CollateDataFiles. A folder selection window will appear. In that window, the user should select the parent folder containing the subfolders with the quantification text files. The macro will automatically go through the subfolders and reiteratively assemble all the data into one table, which the user can then save through a dialogue box.
-
Figure 2.
Demonstration of step 2 of QuoVadoPro for the Cos-7 cell whose segmentation was illustrated in Figure 1. Following the binarization of images, in step 2, QuoVadoPro calculates the variance (A) and the sum (B) of each binary pixel over all time frames. The final motility heatmap (C) is represented as the ratio of variance over sum of intensity of each pixel. Pixels that contain a moving object, for example the mitochondrion that has moved toward the edge of the cell (arrow), display low sum and high variance. These pixels will therefore be represented as hot points on the heatmap. Pixels that contain a stationary object will have a low variance but a high sum, and will therefore appear as cold points on the heatmap (arrowhead).
GUIDELINES FOR UNDERSTANDING RESULTS
Explanation of Motility Scores Quantified by QuoVadoPro
QuoVadoPro infers the motility of fluorescent objects based on the variation in pixel illumination. To quantify movement from a time-lapse of fluorescently tagged objects, as the first step, QuoVadoPro converts the fluorescence signal into binary masks (Fig. 1). At the second step, the algorithm uses the binarized images to generate a motility heatmap. Each pixel in the heatmap corresponds to the ratio of the temporal variance to the temporal sum of that pixel across all the binarized images of the time-lapse (Fig. 2). To generate a motility score, the pixel intensities of the heatmap are summed and normalized to the area of the masked objects.
The final motility scores as calculated from binary time-lapse images can also be represented by the formula:
where variancetime represents the variance of pixel intensities over time, sumtime represents the sum of pixel intensities over time, and npixelssegmentedfirstframe represents the number of pixels that were segmented in the first frame (indicating the total area occupied by the fluorescent objects). Pixels with sumtime = 0 are assigned NaN (not a number) values and are not considered in the calculations. For representation, these are shown as cold spots on the heatmap.
The motility scores generated by QuoVadoPro are saved as a text file in which the following quantification parameters are reported:
a. Motility score: This value represents the final quantified motility score as detailed above. The reported motility score is on an arbitrary scale (i.e., does not have a unit).
b. Number of slices: This is the number of time points that were present in the time lapse.
c. Number of substacks considered: During the analysis of an image stack representing a time lapse, QuoVadoPro can split the image stack into multiple substacks (if requested by the user during step 2). The “Number of substacks considered” value represents the number of substacks the original image stack was split into. If the user has chosen not to split the image stack into smaller subsets, this value will be stated as 1. Splitting the time lapse into smaller segments is often useful (especially for long movies), to get information on how movement may have changed during the image acquisition. Once the number of substacks to be considered is set at step 2, it should not be changed between samples of the same experiment.
d. Thresholding parameters: The values represented under this tab are the values that the user inputs in step 1 of QuoVadoPro—the strength of background subtraction, the threshold value used for image segmentation and the noise removal parameters.
e. Total pixel intensity and area occupied in heatmaps of each substack: The sum of intensities of all pixels from the heatmaps constructed from each substack and the area occupied by the objects in each substack are shown under this tab. To calculate the motility score for each substack, the sum of intensities of the heatmap pixels is normalized to the object area.
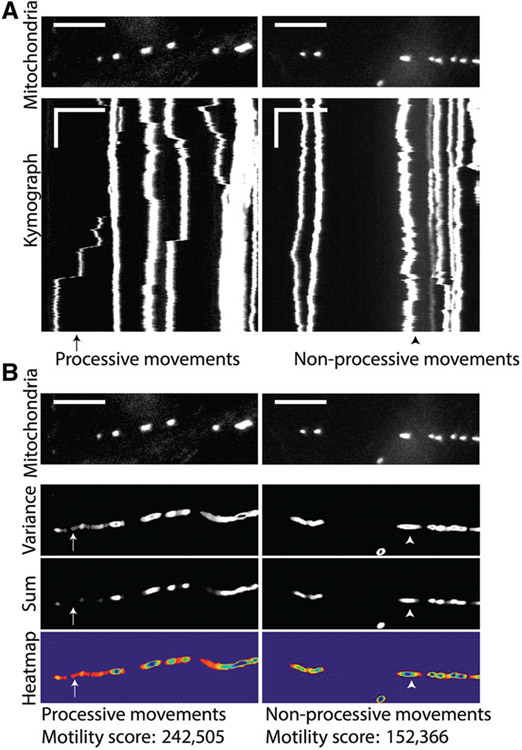
QuoVadoPro Gives More Weight to Processive Movements than to Back and Forth Jitters
Normalizing the temporal variance to the temporal sum of each binary pixel to construct the motility heatmaps allows QuoVadoPro to give more weight to processive movements than to short back-and-forth jitters. With processive movement, an object transiently illuminates several sets of pixels, each of which thus has a high variance and low sum and appears as a hot spot on the image (Fig. 3A and B, left panels). When an object moves back and forth (i.e., displays non-processive movement), it illuminates the same set of pixels repeatedly (Fig. 3A and B, right panels). Each pixel illuminated by an object moving back and forth shows a high sum and a lower variance than pixels depicting processive movements, and thus creates a less intense spot on the heatmap.
Figure 3.
Example of neuronal axons with fluorescently tagged mitochondria analyzed by QuoVadoPro, demonstrating that processive movements are weighted more heavily than back-and-forth movements. Two axon segments expressing fluorescent tags on mitochondria (mitoDsRed) are shown (A, top panels) along with their kymographs (A, bottom panels). These segments were analyzed by QuoVadoPro, and their respective variance images, sum images, and heatmaps along with final motility scores are depicted (B). One of the axons has processively moving mitochondria (an example is marked by the arrow), whereas the other contains mitochondria that are largely jiggling back and forth (an example is marked by the arrowhead). When analyzed over time, pixels containing processive movements show high variance and low sum (arrow, panel B). Binarized pixels depicting back-and-forth movements have a high variance and a high sum (arrowhead, panel B). The motility heatmaps are constructed as a ratio of variance over sum. Thus, processive movements create more hot points on the motility heatmaps compared to non-processive movements. Heatmaps that contain processively moving objects thus have a higher motility score than heatmaps of non-processive movements. Horizontal scale bars depict 15 μm and vertical scale bars indicate 30 s.
COMMENTARY
QuoVadoPro takes a unique approach to automatically measure intracellular movements. The algorithm uses the variation in pixel illumination in time-lapse images of fluorescently tagged intracellular objects as a proxy parameter that can reveal the extent of intracellular movements. This unique approach makes QuoVadoPro suitable for analyzing intracellular traffic in a variety of experimental systems, even where traditional methods of measuring intracellular motility, such as kymography and object tracking, cannot be applied.
Kymography depends on the ability to identify discrete intracellular objects and track them along a well-defined path. As such, it works well to follow organelles or protein complexes in axons, dendrites, filopodia, and fungal hyphae (Alieva et al., 2019; Bielska et al., 2014; Kerber et al., 2009; Klinman & Holzbaur, 2016; Maday, Twelvetrees, Moughamian, & Holzbaur, 2014).
Although kymography is restricted to measuring movement along a defined path, QuoVadoPro can measure intracellular movements irrespective of the directions of movements. It is thus applicable to measuring motility in more complex systems where microtubules and actin filaments (the paths of intracellular traffic) have mixed orientations. Alternative manual or automated solutions that track objects from frame to frame have been employed to quantify intracellular traffic in cells where intracellular cargo trafficking primarily occurs as short runs in heterogeneous directions (Chenouard et al., 2014; Dokka, Park, Jansen, DeAngelis, & Angelaki, 2019; Jaqaman et al., 2008; Wasim & Treanor, 2018).
Manual object tracking approaches are low throughput and subjective to user bias. Automatic object tracking approaches, although capable of high-throughput data analysis, have limited fidelity, which depends on the spatial and temporal resolutions and the heterogeneity of sizes and shapes of the objects tracked. QuoVadoPro is automated, fast, and unbiased, and may be the method of choice when the right conditions for automated object tracking do not prevail. For example, QuoVadoPro can be used to measure for example in quantifying the movement of complex reticular structures like mitochondria and the ER, where individual organelles may vary from submicron to >100 μm in length (Bosch & Calvo, 2019; Costantini & Snapp, 2013; Friedman, Webster, Mastronarde, Verhey, & Voeltz, 2010; Koopman, Visch, Smeitink, & Willems, 2006; Leonard et al., 2015). This heterogeneity can prevent object-tracking algorithms from faithfully detecting and therefore tracking the movements of the network. Since QuoVadoPro not depend on object detection, it can infer the movement of such cellular structures irrespective of size and shape. In addition, unlike tracking algorithms, QuoVadoPro does not need to track objects from frame to frame to quantify motility. It can thus operate on data sets with low temporal and spatial resolutions and high object densities: conditions under which individual cellular objects cannot be reliably assigned in every frame of a movie, thus preventing analysis by object-tracking algorithms.
Unlike kymography or object-tracking algorithms, however, QuoVadoPro is not intended to extract in-depth parameters describing different features of the motility of individual intracellular objects. For that analysis, the previous methods are preferable if they can be accurately applied. Instead, the QuoVadoPro algorithm is designed to be a fast and automatic method to obtain a proxy readout for total motility within a cell. The algorithm is versatile, is computationally light, and requires minimal user input and supervision. It should be stressed, however, that the variance algorithm is only a proxy for motility. Drastic differences in the shapes of organelles or cells between experimental samples can also alter the output, even if these differences do not represent the movement of intracellular cargo.
Critical Parameters
To ensure that a change in the variance of pixel occupancy faithfully reflects a change in intracellular motility, the following parameters should be carefully noted.
• Rate of image acquisition: As QuoVadoPro analyzes the variance of pixel illumination over the entire span of a time-lapse sequence, the total number of frames in the time lapse (as governed by the image acquisition rate) has a significant impact on the final motility score. Thus, when comparing two samples, variation in frame rates should be kept to a minimum.
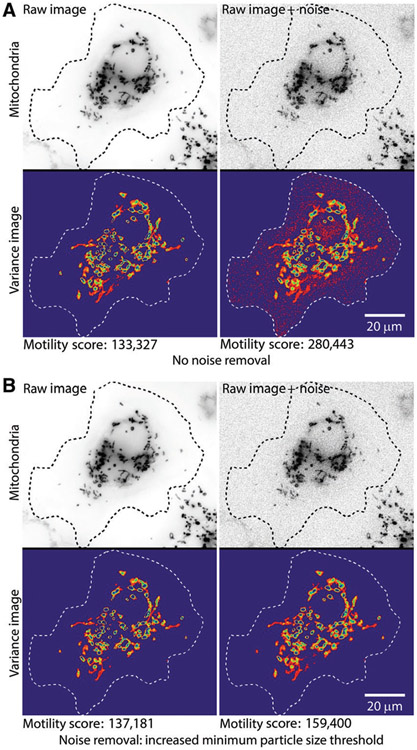
• Noise removal from images: In step 1, QuoVadoPro generates a stack of binary masks from a time-lapse movie of fluorescent objects. The binarization of fluorescence allows the algorithm to omit differences in absolute fluorescence during quantification. To make the binary masks, QuoVadoPro uses local contrast to segment the fluorescently tagged objects. Accurate quantification depends on reliable elimination of noise pixels. Thus, images with high SNRs are preferred. The erroneous detection of noise pixels in certain frames can influence the variance (Fig. 4A). The user can choose from multiple tools described in step 1, such as smoothing in time and defining a lower limit of object area, to eliminate noise pixels (Fig. 4B).
Figure 4.
Impact of pixel noise and demonstration of noise removal during step 1 of QuoVadoPro. To demonstrate the effect of pixel noise on quantification by QuoVadoPro, artificial noise is introduced in the sample cell with fluorescent mitochondria (previously shown in Figures 1 and 2). When the noise in the images is not filtered out, the noise pixels are retained in the 2D motility heatmap and significantly affect the motility score (A). However, when the noise pixels are filtered out, the impact of noise is greatly reduced or eliminated (B). QuoVadoPro gives the user multiple tools to remove image noise during its operation in step 1. In this example, one such tool is used, wherein a minimum size threshold can be declared for all discrete objects in the image. Any object below the minimum size threshold is eliminated. Because most image noise is in the form of isolated pixels, the user can choose a minimum size threshold of a few pixels (typically <3 pixels), to remove most of the noise.
• Number of objects: The edges of objects may be scored slightly differently from frame to frame, even for stationary objects. This results in an increased variance at object edges. Though the algorithm weights processive movement more heavily than this edge jitter and normalizes for total object area, the signal due to edge jitter can still contribute to the calculated variance. Because edge jitter scales with the number of discrete objects in an image, the total number of discrete objects has an impact on the variance. Thus, QuoVadoPro should be used to analyze experimental samples with similar numbers of discrete objects. For example, QuoVadoPro can be used to analyze mitochondrial movement only in cases where the samples do not have large differences in mitochondrial fission or fusion.
• Cell shape: Quantification through QuoVadoPro is done on a single plane or on 2D projections of 3D images. Thus, the algorithm is best used to samples that do not exhibit large changes in cell shape, as that may influence the distribution of objects in the plane or projection analyzed.
Anticipated Results
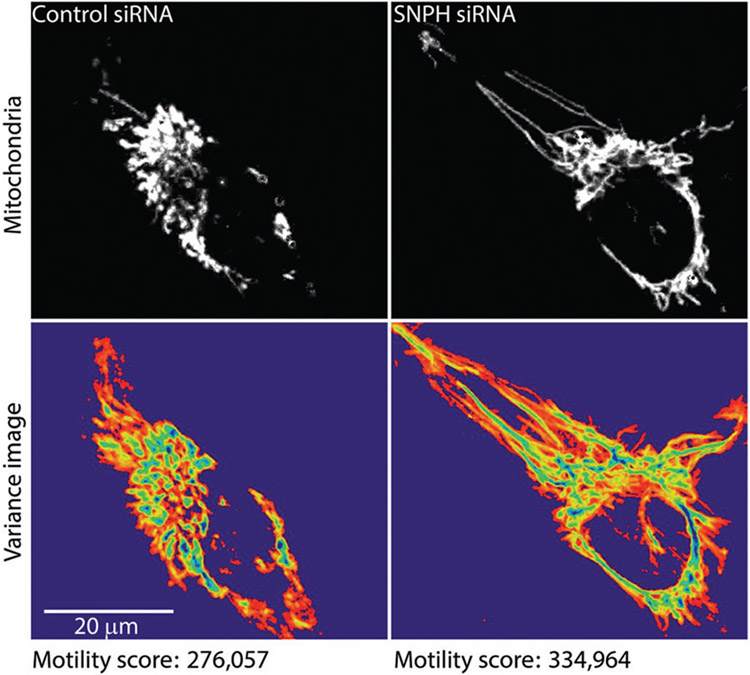
As a demonstration of data analysis using QuoVadoPro, we used the algorithm to analyze previously published data of mitochondrial movement in LN229 cells (Caino et al., 2016). The authors in this study show that mitochondrial movement in tumor cells is essential to promote cell invasion and membrane dynamics. They study mitochondrial movement in the presence and absence of an anchoring protein called syntaphilin (SNPH). The authors manually track individual mitochondria in these cells to demonstrate that knocking down SNPH increases mitochondrial motility. We have reanalyzed the published video of the LN229 cells using QuoVadoPro (Fig. 5). Our observations indicate a 20% increase in mitochondrial motility upon knockdown of SNPH, similar to the authors’ observation.
Figure 5.
Demonstration of mitochondrial motility analysis by QuoVadoPro on previously published example. To demonstrate the utility and robustness of QuoVadoPro, the algorithm was used to analyze a previously published video (Caino et al., 2016) of mitochondrial movement in LN229 cells. The mitochondria are shown (top panels) along with their heatmaps (bottom panels). In concordance with the data previously obtained by manual tracking (Caino et al., 2016), the analysis by QuoVadoPro demonstrates a 20% increase in mitochondrial motility upon knockdown of an anchoring protein, syntaphilin (SNPH).
Supplementary Material
Acknowledgments
We thank the members of the T.L. Schwarz laboratory for fruitful discussions, especially Dr. Jill Falk for critical reading of the manuscript and for her inputs. We also thank Dr. Gergana Shipkovenska for her help with editing of the manuscript. We thank S. Vasquez for help with hippocampal dissections from embryonic rats; we are grateful to the Harvard NeuroDiscovery Center’s Enhanced Neuroimaging Core (supported by NINDS P30 Core Center grant no. NS072030) and the Cellular Imaging Core at Boston Children’s Hospital (supported by NIH grant no. U54 HD090255) for support with imaging. This research was generously supported by the National Institutes of Health grant R01 GM069808 (NIGMS) to T.L.S.
Literature Cited
- Alieva NO, Efremov AK, Hu S, Oh D, Chen Z, Natarajan M, … Bershadsky AD (2019). Myosin IIA and formin dependent mechanosensitivity of filopodia adhesion. Nature Communications, 10(1), 3593. doi: 10.1038/s41467-019-10964-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloh RH, Schmidt RE, Pestronk A, & Milbrandt J (2007). Altered axonal mitochondrial transport in the pathogenesis of Charcot-Marie-Tooth disease from mitofusin 2 mutations. Journal of Neuroscience, 27(2), 422–430. doi: 10.1523/JNEUROSCI.4798-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu H, Ding L, Pekkurnaz G, Cronin M, & Schwarz TL (2020). Kymolyzer, a semi-autonomous kymography tool to analyze intracellular motility. Current Protocols in Cell Biology, 87, e107. doi: 10.1002/cpcb.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielska E, Higuchi Y, Schuster M, Steinberg N, Kilaru S, Talbot NJ, & Steinberg G (2014). Long-distance endosome trafficking drives fungal effector production during plant infection. Nature Communications, 5, 5097. doi: 10.1038/ncomms6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch A, & Calvo M (2019). Automated quantitative analysis of mitochondrial morphology. Methods in Molecular Biology, 2040, 99–115. doi: 10.1007/978-1-4939-9686-5_6. [DOI] [PubMed] [Google Scholar]
- Caino MC, Seo JH, Aguinaldo A, Wait E, Bryant KG, Kossenkov AV, … Altieri DC (2016). A neuronal network of mitochondrial dynamics regulates metastasis. Nature Communications, 7, 13730. doi: 10.1038/ncomms13730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caino MC, Seo JH, Wang Y, Rivadeneira DB, Gabrilovich DI, Kim ET, … Altieri DC (2017). Syntaphilin controls a mitochondrial rheostat for proliferation-motility decisions in cancer. Journal of Clinical Investigation, 127(10), 3755–3769. doi: 10.1172/JCI93172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenouard N, Smal I, de Chaumont F, Maska M, Sbalzarini IF, Gong Y, … Meijering E (2014). Objective comparison of particle tracking methods. Nature Methods, 11(3), 281–289. doi: 10.1038/nmeth.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collard JF, Cote F, & Julien JP (1995). Defective axonal transport in a transgenic mouse model of amyotrophic lateral sclerosis. Nature, 375(6526), 61–64. doi: 10.1038/375061a0. [DOI] [PubMed] [Google Scholar]
- Costantini L, & Snapp E (2013). Probing endoplasmic reticulum dynamics using fluorescence imaging and photobleaching techniques. Current Protocols in Cell Biology, 60, 21.7.1–21.7.29. doi: 10.1002/0471143030.cb2107s60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokka K, Park H, Jansen M, DeAngelis GC, & Angelaki DE (2019). Causal inference accounts for heading perception in the presence of object motion. Proceedings of the National Academy of Sciences of the United States of America, 116(18), 9060–9065. doi: 10.1073/pnas.1820373116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Webster BM, Mastronarde DN, Verhey KJ, & Voeltz GK (2010). ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. Journal of Cell Biology, 190(3), 363–375. doi: 10.1083/jcb.200911024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenring JR (2013). A central role for vesicle trafficking in epithelial neoplasia: Intracellular highways to carcinogenesis. Nature Reviews Cancer, 13(11), 813–820. doi: 10.1038/nrc3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardena S, & Goldstein LS (2001). Disruption of axonal transport and neuronal viability by amyloid precursor protein mutations in Drosophila. Neuron, 32(3), 389–401. doi: 10.1016/s0896-6273(01)00496-2. [DOI] [PubMed] [Google Scholar]
- Hafezparast M, Klocke R, Ruhrberg C, Marquardt A, Ahmad-Annuar A, Bowen S, … Fisher EM (2003). Mutations in dynein link motor neuron degeneration to defects in retrograde transport. Science, 300(5620), 808–812. doi: 10.1126/science.1083129. [DOI] [PubMed] [Google Scholar]
- Hartman MA, & Spudich JA (2012). The myosin superfamily at a glance. Journal of Cell Science, 125(Pt 7), 1627–1632. doi: 10.1242/jcs.094300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N (1998). Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science, 279(5350), 519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, & Danuser G (2008). Robust single-particle tracking in live-cell time-lapse sequences. Nature Methods, 5(8), 695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerber ML, Jacobs DT, Campagnola L, Dunn BD, Yin T, Sousa AD, … Cheney RE (2009). A novel form of motility in filopodia revealed by imaging myosin-X at the singlemolecule level. Current Biology, 19(11), 967–973. doi: 10.1016/j.cub.2009.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinman E, & Holzbaur EL (2016). Comparative analysis of axonal transport markers in primary mammalian neurons. Methods in Cell Biology, 131, 409–424. doi: 10.1016/bs.mcb.2015.06.011. [DOI] [PubMed] [Google Scholar]
- Koopman WJ, Visch HJ, Smeitink JA, & Willems PH (2006). Simultaneous quantitative measurement and automated analysis of mitochondrial morphology, mass, potential, and motility in living human skin fibroblasts. Cytometry Part A, 69(1), 1–12. doi: 10.1002/cyto.a.20198. [DOI] [PubMed] [Google Scholar]
- Lee Y, Phelps C, Huang T, Mostofian B, Wu L, Zhang Y, … Nan X (2019). High-throughput, single-particle tracking reveals nested membrane domains that dictate KRasG12D diffusion and trafficking. Elife, 8, e46393. doi: 10.7554/eLife.46393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard AP, Cameron RB, Speiser JL, Wolf BJ, Peterson YK, Schnellmann RG, … Rohrer B (2015). Quantitative analysis of mitochondrial morphology and membrane potential in living cells using high-content imaging, machine learning, and morphological binning. Biochimica et Biophysica Acta, 1853(2), 348–360. doi: 10.1016/j.bbamcr.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S, Twelvetrees AE, Moughamian AJ, & Holzbaur EL (2014). Axonal transport: Cargo-specific mechanisms of motility and regulation. Neuron, 84(2), 292–309. doi: 10.1016/j.neuron.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Okada Y, & Hirokawa N (2005). Analysis of the kinesin superfamily: Insights into structure and function. Trends in Cell Biology, 15(9), 467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Misgeld T, & Schwarz TL (2017). Mitostasis in neurons: Maintaining mitochondria in an extended cellular architecture. Neuron, 96(3), 651–666. doi: 10.1016/j.neuron.2017.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parachoniak CA, & Park M (2012). Dynamics of receptor trafficking in tumorigenicity. Trends in Cell Biology, 22(5), 231–240. doi: 10.1016/j.tcb.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Pilling AD, Horiuchi D, Lively CM, & Saxton WM (2006). Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Molecular Biology of the Cell, 17(4), 2057–2068. doi: 10.1091/mbc.e05-06-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck-Peterson SL, Redwine WB, Vale RD, & Carter AP (2018). The cytoplasmic dynein transport machinery and its many cargoes. Nature Reviews Molecular Cell Biology, 19(6), 382–398. doi: 10.1038/s41580-018-0004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp BJ, & Reese TS (1989). Dynein is the motor for retrograde axonal transport of organelles. Proceedings of the National Academy of Sciences of the United States of America, 86(5), 1548–1552. doi: 10.1073/pnas.86.5.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz TL (2013). Mitochondrial trafficking in neurons. Cold Spring Harbor Perspectives in Biology, 5(6), a011304. doi: 10.1101/cshperspect.a011304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlevkov E, Basu H, Bray MA, Sun Z, Wei W, Apaydin K, … Schwarz TL (2019). A high-content screen identifies TPP1 and Aurora B as regulators of axonal mitochondrial transport. Cell Reports, 28(12), 3224–3237. e3225. doi: 10.1016/j.celrep.2019.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus MA (2018). Myosin-driven intracellular transport. Cold Spring Harbor Perspectives in Biology, 10(3), a021972. doi: 10.1101/cshperspect.a021972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD (2003). The molecular motor toolbox for intracellular transport. Cell, 112(4), 467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- Verhey KJ, Kaul N, & Soppina V (2011). Kinesin assembly and movement in cells. Annual Review of Biophysics, 40, 267–288. doi: 10.1146/annurev-biophys-042910-155310. [DOI] [PubMed] [Google Scholar]
- Wasim L, & Treanor B (2018). Single-particle tracking of cell surface proteins. Methods in Molecular Biology, 1707, 183–192. doi: 10.1007/978-1-4939-7474-0_13. [DOI] [PubMed] [Google Scholar]
- Williamson TL, & Cleveland DW (1999). Slowing of axonal transport is a very early event in the toxicity of ALS-linked SOD1 mutants to motor neurons. Nature Neuroscience, 2(1), 50–56. doi: 10.1038/4553. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.