Abstract
Organisms throughout the tree of life accumulate chemical resources, in particular forms or compartments, to secure their availability for future use. Here we review microbial storage and its ecological significance by assembling several rich but disconnected lines of research in microbiology, biogeochemistry, and the ecology of macroscopic organisms. Evidence is drawn from various systems, but we pay particular attention to soils, where microorganisms play crucial roles in global element cycles. An assembly of genus-level data demonstrates the likely prevalence of storage traits in soil. We provide a theoretical basis for microbial storage ecology by distinguishing a spectrum of storage strategies ranging from surplus storage (storage of abundant resources that are not immediately required) to reserve storage (storage of limited resources at the cost of other metabolic functions). This distinction highlights that microorganisms can invest in storage at times of surplus and under conditions of scarcity. We then align storage with trait-based microbial life-history strategies, leading to the hypothesis that ruderal species, which are adapted to disturbance, rely less on storage than microorganisms adapted to stress or high competition. We explore the implications of storage for soil biogeochemistry, microbial biomass, and element transformations and present a process-based model of intracellular carbon storage. Our model indicates that storage can mitigate against stoichiometric imbalances, thereby enhancing biomass growth and resource-use efficiency in the face of unbalanced resources. Given the central roles of microbes in biogeochemical cycles, we propose that microbial storage may be influential on macroscopic scales, from carbon cycling to ecosystem stability.
Subject terms: Soil microbiology, Biogeochemistry, Microbial ecology, Microbial ecology, Biogeochemistry
Introduction
Storage is a widespread trait in many organisms, familiar from everyday experience with animal fats or plant storage organs. We define storage as the accumulation of chemical resources in a particular form or compartment, in order to secure their availability for future use by the storing organism. This definition is applicable to storage traits across the domains of life, and broadly consistent with macroscopic ecology [1, 2]. Storage is conceptually distinguished from recycling, which degrades materials that were originally synthesized for other functions. Intracellular storage of carbon (C) and energy, as well as other nutrients, has long been documented among fungi and bacteria and is currently a subject of research for industrial applications [3]. However, despite its acknowledged importance for macroscopic life [1, 2, 4], the implications of storage by microorganisms have been largely overlooked in ecology.
Here, we assemble evidence from microbiology and biogeochemistry to assess the prevalence and importance of storage for microbial life. Various microbial ecosystems are considered but with a particular focus on soils, where microbes play critical roles in terrestrial nutrient availability, primary productivity, and global C fluxes [5]. We highlight the inherently dynamic nature of storage, with reference to observations from soil. We then introduce storage concepts from macroscopic ecology and use these to develop theories of storage for microbial ecology that are applicable across the breadth of microbial resource allocation and life-history strategies. The prevalence of storage among microorganisms carries implications for contemporary concepts of microbial growth, ecological stoichiometry, and element cycling in soils. We explore these implications theoretically and disentangle the dynamical implications with a numerical model of intracellular C storage. We highlight challenges for future work to test our predictions and integrate storage physiology into microbial ecology and soil biogeochemistry.
Microbial storage
Overview of microbial storage
Storage compounds are known throughout the microbial world (Table 1, with additional information in Supplementary Information 1). Storage has been widely recognized in aquatic phototrophs. Daily oscillations of triacylglyceride (TAG) storage in the North Pacific account for 23% of primary production by nanophytoplankton [6], and oscillations of polyhydroxyalkanoate (PHA) have been observed in photosynthetic microbial mats [7]. Cyanophycin was first identified in diazotrophic cyanobacteria [8], which use it to balance nitrogen (N) supply during daily cycles of photosynthesis and N fixation [9, 10]. Polyphosphate is well known in cyanobacteria [10, 11] and algae [12], and is an important pool in the marine P cycle [13] that influences microbial stoichiometry and affects global biogeochemistry through P sedimentation [14]. The ecological importance of storage by marine heterotrophs has received less attention, although storage might underlie the variability observed in the stoichiometry of aquatic heterotrophic biomass [15].
Table 1.
Overview of microbial macronutrient storage compounds with key references and characteristics.
| Storage compound | Occurrence | Structure | Comments |
|---|---|---|---|
|
Triacylglycerides (TAG) [139, 140] Hydrophobic lipids; as intracellular inclusions |
Widespread in bacteria and fungi, not in archaea |  |
High energy density, but can only be mobilized for energy under aerobic conditions |
|
Polyhydroxyalkanoates (PHA) [141, 142] High molecular-weight, hydrophobic lipid; as intracellular inclusions |
Bacteria and archaea, not in eukaryotes |  |
Intracellular PHA can comprise >80% of cell dry weight; can only be mobilized for energy under aerobic conditions |
|
Hydrophilic, high molecular-weight polymer of glucose; as intracellular granules |
Bacteria, fungi, animals, possibly archaea, not plants |  |
Polymer enables glucose storage without increasing osmotic pressure |
|
Nonreducing water-soluble glucose dimer |
Bacteria, archaea, fungi, plants, and invertebrates |  |
Plays roles in osmotic regulation and protection against desiccation |
|
Hydrophobic lipid; as intracellular inclusions |
Bacteria |  |
Also in eukaryotes, e.g., in hydrophobic leaf cuticles, but not as storage |
|
Storage of P and energy, as intracellular granules or in acidocalcisomes |
Ubiquitous, but extent of accumulation differs |  |
Multifunctional molecule also involved in pH buffering, heavy metal chelation, cell signaling, motility, and virulence |
|
Storage of N; as intracellular granules |
Cyanobacteria, some other bacteria | 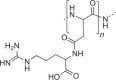 |
Up to 18% of cell dry mass of cyanobacteria and >40% in Acinetobacter calcoaceticus |
More details are provided in Supplementary Information 1.
Storage by heterotrophic bacteria has been most intensively studied in wastewater treatment, particularly enhanced biological phosphate removal (EBPR) systems [16]. These systems alternate between anaerobic and aerobic conditions. During the C-rich anaerobic phase, when growth is limited by low oxygen availability, some bacteria convert organic matter into intracellular PHA. The switch to aerobic conditions triggers these microbes to degrade their PHA stores to power growth as well as accumulating intracellular polyphosphate, which can then be separated from the water with the microbial biomass [17]. By alternating between anaerobic and aerobic conditions, these systems select for microorganisms that perform this cyclic storage, enabling the removal of P from the water [16]. Conditions during wastewater treatment are imposed by human design, but these systems nevertheless indicate that microbial storage can play important roles in ecosystem function.
Various storage forms have been described, besides the well-known macronutrient storage compounds (Table 1). Iron is sequestered in ferritin and bacterioferritin structures by bacteria [18] and likely also fungi [19]. Iron accumulation serves two purposes: detoxification of high intracellular iron concentrations, which present a dangerous oxidative risk, and a storage function since the iron can later be remobilized to avoid deficiency [20]. Acidocalcisomes, acidic organelles containing polyphosphate and metal cations, which are conserved between prokaryotes and eukaryotes, are implicated in the storage of Ca [21] and Mn [22]. New storage forms are still being discovered. Intracellular storage of crystalline guanine was recently reported as a eukaryotic functional analog of cyanophycin, which enables the marine dinoflagellate Amphidinium carterae to support multiple cycles of cell division without additional N supply [23]. It has been postulated that extracellular P storage may account for the as-yet unexplained prevalence of inositol phosphate stereoisomers in soil [24]. External storage has also been proposed in extracellular polymeric substances (EPS) [25], since EPS production is enhanced under conditions of high C availability [26, 27]. However, subsequent reuse of EPS by the producing organism has seldom been investigated. Reuse of soluble organic components of the EPS matrix was reported for a cyanobacterium [10] and modeling has suggested an EPS storage function, with a trade-off between maintaining EPS for protection against dehydration or degrading it as a source of C [28]. However, although production of both EPS and the PHA polyhydroxybutyrate (PHB) by Azotobacter beijerinckii were favored by C-excess, N-limited conditions, only PHB showed a subsequent decline after C depletion [29]. Ralstonia eutropha also produced more PHB and EPS with greater glucose supply, but whereas PHB content was suppressed by N supply, EPS showed the opposite relationship [27]. Many other functions are attributed to EPS [25], and more evidence is necessary to determine the extent of its storage role, and especially to distinguish this from recycling of EPS that was produced for other purposes. In any event, it is safe to predict that many microbial storage traits remain to be discovered.
Occurrence of storage in soil
Storage traits are known for many microbial taxa from diverse ecosystems, but here we focus on the evidence from soil, where storage has been relatively neglected in comparison to oceans and wastewater. Soils are among the most biodiverse ecosystems on Earth, with critical roles in terrestrial cycles of C and other nutrients [5]. Soil habitats present a challenging environment for microorganisms, in which the availability of nutrients, their element ratios, and physicochemical conditions vary across all temporal and spatial scales [30]. Such a habitat should offer numerous opportunities for organisms that can save resources to meet future needs. Occasional studies across more than four decades have accumulated evidence of microbial storage in soil, although sustained research has been lacking.
Storage compounds and their synthesis in soil
Soils have proven to be rich sources of organisms that produce TAG, PHB, or wax esters [31]. Out of 73 bacterial isolates from a temperate clay-loam soil, 23 were found to produce PHB [32]. Random selection of 60 isolates from each of two Chernozem soils yielded 20 and 28 PHB-producing strains, respectively [33]. Trehalose production has been demonstrated in bacterial and fungal isolates from soil [34, 35], and is an important sink of photosynthetic C in ectomycorrhizae [36]. Polyphosphate was produced by three of eight ascomycetes isolates from two Australian soils, accounting for between 10 and 30% of extractable cellular P [37]. Storage compounds have also been directly observed in soil organisms. Genet et al. identified glycogen granules in the ectomycorrhizal hyphae of Fagus sylvatica-Lactarius subdulcis symbiosis [38], and Frey et al. reported glycogen as well as probable polyphosphate granules in the Hartig net of Picea abies-Hebeloma crustuliniforme ectomyccorhizae [39]. Clearly, the physiological capacity for storage biosynthesis is present in soil communities.
Some microbial storage compounds have already been quantified in soils. PHB contents of 1–4 µg C g−1 soil have been reported for untreated soils [40, 41], with a tenfold increase observed after glucose addition [41]. In the soil literature, TAG content has generally been reported in terms of constituent neutral lipid fatty acids which imply total lipid contents of around 2–20 µg C g−1 soil [42, 43]. These values can be compared to typical extractable soil microbial biomass of a few hundred µg C g−1 [44]. As with PHB, large increases in TAG have been demonstrated in response to enhanced C availability, which were suppressed by simultaneous supply of N and P [45], indicating a strong link between element stoichiometry and storage. The responses to C supply suggest that storage may be an alternative C allocation strategy for microorganisms in hotspots of C availability, such as the rhizosphere, instead of the more widely recognized response of maximized growth rates [30]. Extracellular degradation of microbial storage compounds has only occasionally been studied in soil. The TAG triolein was 38% degraded over 23 weeks [46]. An immediate and sustained increase in soil respiration was observed after trehalose addition, comparable to that induced by glucose [47] and soil calorimetric and respiratory responses to glycogen addition were comparable to alanine [48], which is rapidly degraded [49]. PHA is degraded in soil [50], but the degradation rates of micro-scale PHB granules have not yet been quantified. More systematic investigation of degradation rates is needed to assess how microbial necromass contributes to total storage compound levels in soils.
Dynamics of storage in soil
Storage pools are dynamic by nature, and thus their importance can only be assessed with respect to a particular timeframe. Diurnal fluctuations in glycogen storage have been reported for Microcoleus from biological soil crusts [51], but most investigations in soil have examined longer timeframes. TAG accumulation in soil was induced by the addition of glucose, and TAGs still remained above control levels after 3 months [45]. This suggests that TAGs may have seasonally-relevant turnover times in soil. Soil trehalose and TAG contents are reportedly higher in summer than winter [52], which is consistent with metatranscriptomic evidence that carbohydrate storage compounds are catabolized during winter [53]. On the other hand, direct observation of glycogen granules in ectomycorrhizal fungi revealed accumulation in autumn [38]. It is certainly tempting to interpret these findings as resource storage for winter scarcity, but in situ storage turnover times remain to be demonstrated before this can be concluded. This is because the dynamics of storage compounds at the natural population or community level might either reflect long-term storage in individuals, or enduring environmental conditions that promote storage by successive generations. In the latter case, individual organisms would not survive into the following season to exploit their stored resources. The many changes that occur over a year could also confound the roles of storage and other functions. For example, seasonal differences in soil trehalose levels might reflect either C storage to balance changes in C supply, or trehalose accumulation as an osmoticant in response to changes in soil moisture [52]. These seasonal time-scales are far longer than the periods over which storage physiology has typically been investigated in the laboratory. A rare exception was the laboratory demonstration of PHB-enhanced survival in the diazotrophic soil bacterium Sinorhizobium meliloti during 528 days of C starvation [54].
Prevalence of storage among soil microbial taxa
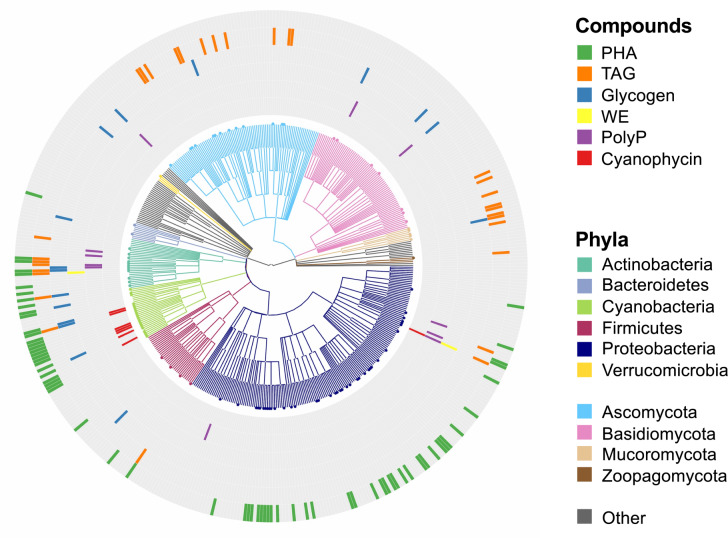
Depicting the soil microbial taxa that accumulate storage compounds on a tree of fungal and bacterial diversity reveals the potential significance of microbial storage in soil (Fig. 1). A comprehensive literature survey identified genera with storage traits as follows: (i) storage has been phenotypically demonstrated for at least one member of the genus as either: (a) the build-up of at least 5% of cell dry weight as a known storage compound, or (b) build-up of sufficient storage compounds for observation by light microscopy; and (ii) the genus has at least one member that occurs in soil. Literature was assembled by searching Web of Science using storage compound names and storage terms, supplemented by literature citing or cited by relevant studies from this search. This yielded a shortlist of 89 bacterial and 40 fungal genera that fulfilled criterion (i), based on 126 peer-reviewed journal articles. For each of these genera, a second search was performed for evidence of their occurrence in soil. The 106 genera fulfilling both criteria were depicted on a cladogram constructed using the NCBI taxonomy database [55] alongside a selection of representative bacterial and fungal taxa for context [56, 57]. Genera are detailed in Supplementary Fig. S2.1, with sources provided in Supplementary Table S2.1, S2.2.
Fig. 1. Known microbial storage by genera occurring in soil.
The cladogram presents a representative tree of fungal and bacterial diversity to highlight genera that meet the following criteria: (i) storage traits have been phenotypically demonstrated for at least one member of the genus as either the build-up of at least 5% of cell dry weight as a known storage compound, or build-up of storage compounds to a sufficient degree for observation by light microscopy; and (ii) the genus has at least one member that occurs in soil. Organisms with storage compounds are displayed in a standard NCBI taxonomic hierarchy together with representative microbial genera [56, 57]. Clades are colored at the phylum level. Color markers in the outer rings indicate which storage compounds are accumulated by the corresponding genus. PHA polyhydroxyalkanoate, TAG triacylglyceride, WE wax ester, PolyP polyphosphate. In the vast majority of cases, gray indicates a lack of data on storage traits for a particular genus. Genera and sources are detailed in Supplementary Information 2.
This analysis found that, for the vast majority of taxa, the presence or absence of storage compounds has not yet been investigated. The distribution of known storage compound producers therefore underestimates the true prevalence of storage. Nonetheless, it is clear that TAG is widespread, and neither restricted to eukaryotes nor to actinomycetes among the prokaryotes. On the other hand, PHA storage seems to be restricted to prokaryotes. Glycogen and polyphosphate storage occurs in diverse soil organisms, but, to date, only one soil genus outside the cyanobacteria is known to accumulate substantial cyanophycin (Acinetobacter [58]). Only one genus (Saccharomyces) met the criteria for trehalose [59], which casts doubt on the importance of trehalose as a C storage form among soil microorganisms. The literature on storage compound occurrence is strongly biased towards bioprospecting efforts, especially for PHA and TAG producers, so the relative occurrence of particular storage compounds in Fig. 1 should not be seen as a measure of their importance in nature. Caveats notwithstanding, however, one conclusion is clear: storage traits occur across diverse bacterial and fungal genera in soil.
Principles of storage from macroscopic ecology
The internal storage of resources has been extensively studied in plants and animals, yielding theoretical concepts that can help to interpret patterns of microbial storage. We therefore make a digression to introduce some useful principles of macroscopic storage ecology, before returning to microorganisms in the next section.
Modes of storage in resource allocation
Chapin et al. distinguished between different storage modes [2], which may be generalized as follows:
Surplus storage is the storage of resources that are available in excess of immediate requirements for reproduction or maintenance. Allocation of surplus resources to storage does not compete with other metabolic demands.
Reserve storage is the biosynthesis of storage compounds that diverts resources from other metabolic demands. There is therefore an opportunity cost of reserve storage in the form of reduced metabolic activity or reproduction in the short-term.
A sharp boundary between supply-independent reserve storage and supply-dependent surplus storage is not to be expected, and these are better viewed as extremes along a spectrum of storage strategies. Nonetheless, the conceptual distinction between storage modes is valuable for understanding resource allocation. The surplus storage concept predicts that storage is formed during periods of resource excess, with low opportunity cost [2]. For example, weight gain in humans results (in part) from prolonged energy intake in excess of metabolic needs [60]. However, surplus storage is not restricted to favorable conditions: trees often increase starch pools even when suffering from drought, because growth and respiration are more strongly suppressed than photosynthesis [61].
Reserve storage, in contrast, involves a trade-off against other physiological functions, and can be viewed as an investment in future reproduction [62]. Prioritizing storage might increase reproductive success in the future, as seen in biennial plants [63], or ensure survival of future scarcity, such as lipid storage by animals in summer to prepare for resource-poor winters [4]. The importance of reserve storage is underlined by observations that body fat of different individuals of the same animal species is often inversely related to their environmental food supply [1, 64], suggesting particular advantages under resource scarcity.
Degradation of functional biomass components can also support the energy or nutrient needs of the organism, as occurs during starvation [65]. Such recycling supplies future needs from internal resources and, at times, could be important for an organism’s resource budget. However, we do not include recycling as a mode of storage because in these cases storage was not the purpose of the original resource allocation. It is nevertheless important to recognize that storage compounds themselves may simultaneously serve other roles, such as metabolic water supply in desert animals [66] a function also hypothesized in microbes [67] or protection from cellular oxidative stress [68]. There is therefore not only a spectrum of storage strategies between surplus storage and reserve storage but also a spectrum of strategies between storage and recycling.
Advantages of storage
At the most general level, storage decouples the activity of an organism from the immediate supply of resources. Thus, storage is a widespread strategy to deal with fluctuations in resource availability by stockpiling during productive periods to support survival or sustained activity through unfavorable times [4]. Storage can also enable variable levels of activity (i.e., variable resource demand) under conditions of relatively stable resource supply. This can be an adaptation to variation in other environmental factors, for example to concentrate reproductive investment in periods of reduced predation risk [4], or can enable intense activity [69]. It can also serve as insurance against unpredictable environmental challenges, and modeling indicates that simultaneous allocation to storage as well as to reproduction and maintenance can be a successful strategy in unpredictable environments [70]. Even without environmental variability, resource-poor environments can necessitate storage in order to assemble the resources needed for reproduction [71]. On the other hand, when a resource is abundant, surplus storage can be a competitive strategy to restrict that resource’s availability to competitors, so as to reduce competition for other, more limited resources [72].
Net benefits of storage depend on the trade-off between advantages and costs (both direct and opportunity costs). Storage can carry various direct costs, such as the development and maintenance of storage structures, the additional energy required for motile organisms to move stores around [1], and enhanced risk of predation [64]. Opportunity costs are largely due to forgone growth or reproduction, which are minimal for surplus storage but characteristic of reserve storage [2], though periods of low reproductive value can represent low opportunity costs [73]. Of course, the future reproductive value of storage can only be realized by an organism that survives to remobilize the stored resource. Mortality risk therefore plays an important role in the trade-off between storage costs and advantages [62], particularly by causes that are not mitigated by storage, such as predation.
Toward microbial storage ecology
Storage modes among microbes
Numerous lines of evidence indicate that the storage mode concepts from macroecology (section “modes of storage in resource allocation”) are applicable in microbiology. As for plants and animals, microbial reproduction can be constrained by environmental conditions or resource limitations, so that another resource is available in excess of immediate needs. High levels of storage compounds have been widely observed in microbes under these conditions, consistent with surplus storage. PHA, TAG, and glycogen are accumulated by diverse microorganisms under C-rich, N-limited conditions [74–77]. Expression of the glycogen synthase gene GSY2 in Saccharomyces cerevisiae is precisely coordinated to the exhaustion of N in the media [78]. When exponential-phase S. cerevisiae was transferred to media lacking N, P, or S, growth was arrested before the glucose supply was exhausted, and glycogen and trehalose were accumulated [59]. Resupply of the missing nutrient restarted growth, confirming that C-supply was not limiting. These observations indicate that C storage under N limitation does not compete with growth in S. cerevisiae. Similarly, cyanobacteria have low levels of cyanophycin storage when rapidly growing, but accumulation of this N-storage compound is stimulated by N excess or by limitations of light, P or sulfur [11]. Microbial surplus storage has not only been observed in the laboratory but also applied in industrial settings, for example in the wastewater EBPR process (section “overview of microbial storage”), where oxygen limitation drives PHB accumulation [79], and the use of N-limiting conditions in the production of microbial TAG [80]. The surplus storage concept formalizes the longstanding interpretation of microbial storage as accumulation of surplus resources.
Aside from surplus storage, microbes also make use of reserve storage. Matin et al. reported that PHA accumulation was highest under more C-limited conditions, which they interpreted as a survival strategy of the oligotrophic Spirillum species they investigated [81]. Accumulation of C-rich compounds under C-limited conditions has been confirmed in other bacteria as well: PHA in Pseudomonas putida up to 26% of cell dry mass [82], up to 12% in Bacillus megaterium [83] and 21% of TAG in Rhodococcus opacus [84]. Polyphosphate accounted for up to 25% of cellular P in Trichodesmium sampled from P-poor waters [85]. These observations appear paradoxical if microbes are assumed to maximize short-term growth and store only surplus resources, but are understandable once the advantages of reserve storage are recognized. Similarly, the reserve storage mode can explain the expression of PHA synthesis genes by oligotrophic bacteria deep within the Earth’s crust [86]. Upregulation of storage synthesis in response to declining resources has been reported for glycogen and trehalose in S. cerevisiae [59], glycogen in Escherichia coli [87] and cyanophycin in Anabaena cylindrica [88], a strategy previously predicted from modeling [89]. This suggests a prioritization of storage over growth especially when resource depletion is imminent. Recognition of the reserve storage mode cautions against ruling out storage in times of scarcity. Rather, we suggest that microbial storage is expected when the future value of the resource greatly exceeds its immediate utility to the organism.
Microorganisms recycle functional biomolecules such as proteins, cell walls and cell membrane lipids. Storage compounds sometimes also serve other functions, so that storage and recycling define a continuum of strategies rather than strict categories. Trehalose is a case in point. Though often considered a form of microbial C storage [36, 53, 90], its protective functions, particularly against desiccation, are well documented [91]. Polyphosphate also plays multiple roles besides storage, including cellular pH buffering, heavy metal chelation, and involvement in metabolic regulation, amongst others [92].
Advantages of microbial storage
Microbial storage is expected to provide survival and reproductive advantages by decoupling activity from immediate resource supply. Storage mutants have enabled demonstrations of this principle through experimental starvation, in which the supply of an essential element is insufficient for maintaining metabolic activity or growth. E. coli uses glycogen stores to maintain metabolic activity following C-supply depletion, which enhances its growth under fluctuating nutritional supply relative to mutants that are unable to mobilize glycogen [93]. A S. cerevisiae mutant deficient in trehalose and glycogen synthesis experienced a rapid loss of viability when C-starved [94]. Similarly, a deletion in the gene for polyphosphate kinase, which catalyses polyphosphate synthesis, rendered Vibrio cholerae unable to maintain the cellular ATP concentrations needed for an effective stress response in low P medium [95].
The advantages of storage should be obtained through its degradation to access the stored resources. PHA degradation supported growth under C starvation in Sinorhizobium meliloti [96]. Mobilization of storage compounds during nutrient starvation has also been observed for polyphosphate in S. cerevisiae [97], trehalose in Cellulomonas [98] and PHA in Alcaligenes eutrophus [76]. However, straightforward advantages are not always evident, as for Alcanivorax borkumensis mutants with reduced TAG stores but unchanged survival over 26 days of C starvation [99]. This perhaps reflects the diversity of roles that storage compounds can play in starvation responses, quiescence and dormancy. Starvation studies with Pseudomonas and Streptococcus reported survival benefits of storage that outlasted the storage compounds themselves [100, 101], suggesting that storage can support the transition to a stable quiescent state, rather than simply powering ongoing metabolism [93]. On the other hand, in S. cerevisiae, glycogen and trehalose catabolism support reactivation from starvation when C availability increases again [94], and glycogen plays a similar role in cyanobacterial reactivation from dormancy following N resupply [102]. Hence storage can improve microbial survival of starvation by compensating for the shortage of external resources, or by supporting the transition into or out of starvation-adapted physiological states.
Microorganisms also incur costs when responding to non-starvation stressors. Some storage compounds, including PHA, polyphosphate and trehalose, participate in stress responses that do not involve mobilization of the stored resource [92, 103, 104]. These would not be considered storage advantages in the sense used here (section “Introduction”). However, Ayub et al. demonstrated that reducing equivalents from PHA degradation enhance the cold-shock survival of an Antarctic Pseudomonas species by sustaining the cell’s oxidative stress response [105]. Ruiz et al. showed that Pseudomonas oleovorans was substantially better at adapting to and surviving ethanol and heat stress than a mutant that was unable to degrade its PHA stores [101]. These examples suggest that microorganisms can benefit from the insurance function of storage when facing temporally variable stressors.
Even for an abundant resource, the accumulation of appropriate storage compounds will incur metabolic costs of building synthetic machinery and storage structures. There may be further indirect costs arising from altered cell morphology, motility costs and osmotic homeostasis. These trade-offs have not been rigorously quantified in microbes, but the prevalence of microbial storage in nature indicates that, in many cases, the advantages indeed outweigh the costs.
Microbial life-history strategies
So far we have largely discussed storage traits in terms of resource allocation and direct survival and reproductive benefits. The storage concepts presented here can also be seen in the context of microbial life-history strategies. Trait-based approaches to understanding life history propose that the enormous range of microbial traits can be simplified by recognizing correlations between traits, which arise as a result of unavoidable life-history trade-offs. Grime’s competitor-stress tolerator-ruderal (CSR) framework [106] provides one such approach that is suitable for microbial ecology [107, 108]. This is based on the principle that organisms face a compromise between competitive ability (C), resistance to stressful environments (S), and the ability to rapidly colonize niches released by disturbance (ruderal, R). This trait-based framework provides a helpful illustration of the interface between storage concepts and microbial life history.
In the CSR framework, “competitors” are organisms adapted to productive habitats with low external disturbance, and therefore face strong competition. Under these conditions, storage would allow an organism to: (i) deprive competing organisms of an abundant resource without having to use the resource immediately, thereby reducing competition for other, more limiting resources [72] (for example, surplus storage of C to reduce competition for P), and (ii) grow on stoichiometrically imbalanced resources by mobilizing storage compounds to provide the limiting elements, and thus exploit resources that are unavailable to non-storing competitors that lack the complementary nutrients required for growth (for example, by mobilizing polyphosphate stores to grow on an available C resource).
“Stress tolerators” are adapted to survive adverse abiotic or resource-poor conditions that limit productivity. Storage could support these strategies by (i) enabling sufficient resources to be assembled through reserve storage for short periods of high metabolic activity, such as reproduction; (ii) facilitating the transition into or out of resilient starvation states, and (iii) serving an insurance function by providing resources for effective stress responses.
“Ruderals” are adapted to elevated rates of biomass destruction caused by frequent disturbance. For unicellular organisms, a loss of biomass through disturbance equates to the death of individuals, and therefore a high-disturbance regime sensu Grime is equivalent to a habitat with high levels of externally-induced mortality. Most benefits of investing in storage are attained in the future, when the stored resources are used. Since higher external mortality increases the risk of death before stored resources can be remobilized, it can be expected that microorganisms following ruderal strategies make less use of storage than their competitor or stress-tolerator counterparts.
The diversity of storage chemistries (section “Overview of microbial storage”) and strategies suggests that storage contributes to the differentiation of resource use between taxa. Differences in resource-use strategy have profound implications for ecosystem structure and function. Coexistence theory predicts that diversification of strategies enables species to stably share a habitat [109], suggesting that microbial storage could contribute to the extraordinary biodiversity found in soil. Moreover, when organisms in the same habitat use storage to different degrees, and in pursuance of different resource strategies, the outcome will be a redistribution of resource demand through time. Differences in the timing and speed of microbial responses to environmental fluctuations, including resource-use patterns, have been found to underlie ecosystem resistance and resilience [110]. Storage may therefore have a stabilizing influence on microbial communities exposed to extreme events, which are predicted to occur with increasing frequency as a result of global change.
Implications for soil biogeochemistry
The notion of microbial biomass and the stoichiometry of its constituent elements will be influenced by the extent of storage at a given time. Assimilation of nutrients into biomass is a crucial step in biogeochemical transformations. For example, C use efficiency plays a decisive role in soil C balances [111]. The diversion of resources into storage and their later remobilization could affect how we conceptualize, measure and model microbial element fluxes.
Soil microbial biomass
Soil microbial biomass is a central pool in process-based biogeochemical models [112, 113]. Storage generally involves the incorporation of resources into intracellular structures, and thus contributes to biomass in the usual sense of the word. However, reconciling the concepts of storage and microbial biomass face methodological challenges, because many of the prevailing methods of biomass estimation in soil do not accurately reflect storage pools [41]. There is no physiological proportionality between storage compounds and proxy measures of microbial biomass, such as cell membrane phospholipids (measured in phospholipid fatty acid analysis—PLFA), substrate-induced respiration or DNA-based proxies. The chloroform fumigation-extraction method, widely used for biomass estimation in soil, involves aqueous extraction and therefore overlooks high molecular weight (e.g., PHB, glycogen) or highly hydrophobic (e.g., PHB, TAG) storage compounds. The alternative is targeted chemical analysis [41, 43, 114, 115], although protocols are lacking for some key storage compounds in soil (including glycogen and cyanophycin). Storage therefore represents a form of microbial biomass—and, by extension, biomass growth—that is overlooked both conceptually and by current analytical methods.
Ecological stoichiometry
A key implication of storage as an alternative mode of growth emerges in the elemental composition of microbial biomass. Growth of microbial biomass is commonly viewed as the proliferation of cells, requiring complementary nutrients so that the elemental stoichiometry of individual cells and of the total biomass remains within a narrow range [116]. While this may be the case when averaged across the community over the long-term, the accumulation of C-, N-, or P-rich storage compounds to substantial proportions of cell mass clearly has the potential to skew organismal stoichiometry. Storage therefore represents biomass that does not conform to the C:N:P ratios of the organism as a whole, enabling the incorporation of resources that would be considered unbalanced from a conventional stoichiometric perspective. Extending this from storage synthesis to its mobilization leads to the hypothesis that storage can correct for nutrient imbalances across time, much as fungal hyphae connecting contrasting soil patches can balance nutrient availability in space [117].
Carbon sequestration
The formation of new microbial biomass from fresh organic matter is increasingly viewed as the first step towards soil C sequestration, due to the major contribution of dead microbial biomass to long-lasting soil organic matter [118]. Storage synthesis at certain times may constitute large flows of C to biomass. However, storage compounds must be resource-dense and easily degradable to fulfill their function, and may be mobilized in response to stress before a cell dies. Degradation of storage compounds during lytic viral infection or digestion by a predator are yet to be investigated, but are presumably significant. We hypothesize that biodegradation of these compounds is more rapid than for other components of biomass, and C flows to storage are therefore less likely to become sequestered in soil. The same reasoning would suggest that storage compounds contribute less to the biological pump that sequesters C from the ocean surface into deep sediments [119]. If this expectation is correct, models of soil organic matter stabilization may need to account for the peculiarities of storage relative to other biomass constituents, in contrast to current approaches that neglect internal storage [120]. In addition, storage may have indirect effects on C and nutrient cycling by altering the efficiency of resource use, which we explore with the help of a dynamic process-based model.
Modeling of microbial storage dynamics
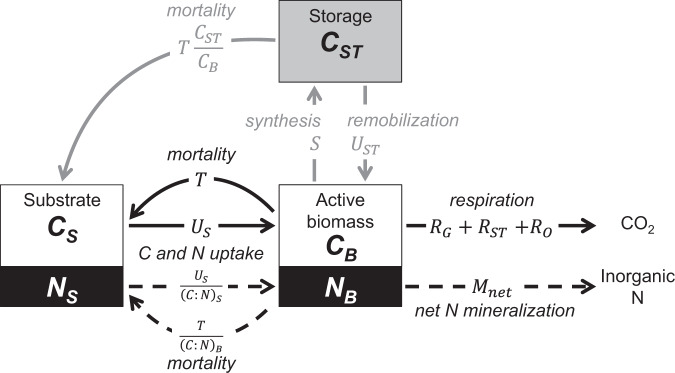
Foreseeing the outcomes of dynamic processes, such as storage, can be difficult without the help of dynamical modeling. Most soil biogeochemical models consider a single, homogenous biomass compartment without distinguishing storage compounds [112, 120]. We explore the implications of storage for microbial resource use by incorporating a C storage pool into a widely applied microbial growth model (Fig. 2) [121].
Fig. 2. Schematic of a dynamic microbial model that includes intracellular C storage.
C and N compartments are shown as white and black boxes, and C storage in gray (subscripts “S”, “B”, and “ST” refer to substrate, active microbial biomass, and storage); solid and dashed arrows indicate C and N rates, respectively (US substrate C uptake, S and UST, storage C synthesis and remobilization, RG, RST, and RO, respiration associated with growth on substrate, growth on storage, and overflow processes, Mnet net N mineralization, T microbial mortality).
Incorporating storage into a dynamic microbial model
A complete description of the model is provided in Supplementary Information 3 (including Table S3.1 and Fig. S3.1, S3.2), and here we only give a summary. The model tracks C flow from the available organic substrate (CS) to the “active microbial biomass” (CB) and storage (CST) via uptake rate US. Active microbial biomass comprises non-storage biomass components, which are involved in immediate physiological functions. This is in contrast to storage biomass, which is produced for future use. C taken up is primarily allocated to growth respiration (RG) and storage synthesis (S), and microbial mortality (T) returns C to the substrate compartment. Microbial storage is remobilized at a rate UST and is converted to active biomass after accounting for a respiration cost (RST). C dynamics are linked to N dynamics via stoichiometric ratios. N flow from the organic substrate (NS) to biomass (NB) is proportional to the C flow (according to the substrate C:N ratio, (C:N)S), and microbial biomass regulates C and N release, or C storage synthesis and remobilization, to maintain a constant C:N ratio (C:N)B for the active (but not total) microbial biomass. Under conditions of C limitation, excess N is released via net N mineralization (Mnet) (as in most soil C and N cycling models, [112]). When N from the substrate is insufficient for microbial requirements, N can be immobilized from inorganic sources (i.e., Mnet < 0), but immobilization is limited to a maximum rate to represent inorganic N availability. If this limit is reached, microorganisms become N limited and excess C is released via overflow respiration, RO (as in [121]).
Three modes of microbial C storage were explored as: (i) no storage; (ii) reserve storage; and (iii) surplus storage. For reserve storage, storage synthesis is modeled as a fraction of substrate uptake (e.g., [122]), with remobilization of storage in proportion to the storage pool. This mode allows microorganisms to store C when substrate C is abundant and use it during starvation, but storage is not reliant on a C surplus. Since C allocation to reserve storage is not regulated by substrate C:N ratio, excess N and C are released via RO or Mnet. With the surplus storage mode, excess C is stored when N is in limited supply (so RO = 0) and used later when C becomes limiting (so Mnet = 0 as long as CST > 0). To compare the effects of the different storage modes, a pulse of organic substrate was provided at the start of the numerical experiments, with substrate C:N ratio as an adjustable parameter.
Predictions for microbial biomass
When storage is included in a microbial model, both storage modes alleviate stoichiometric imbalances caused by a C-rich amendment by enabling microbes to store C until N becomes available. Storage therefore allows more microbial biomass to grow after a substrate addition (Fig. 3A) compared to the null model without storage, because less C and N are lost to overflow or mineralization processes. This is consistent with the expectation that storage confers advantages (section “Advantages of microbial storage”). The model predicts that storage allows communities to sustain growth of active biomass beyond the depletion of substrate. This has not been tested in complex soil communities, although it has been observed in pure culture [23].
Fig. 3. Modeled effects of storage on microbial processes.
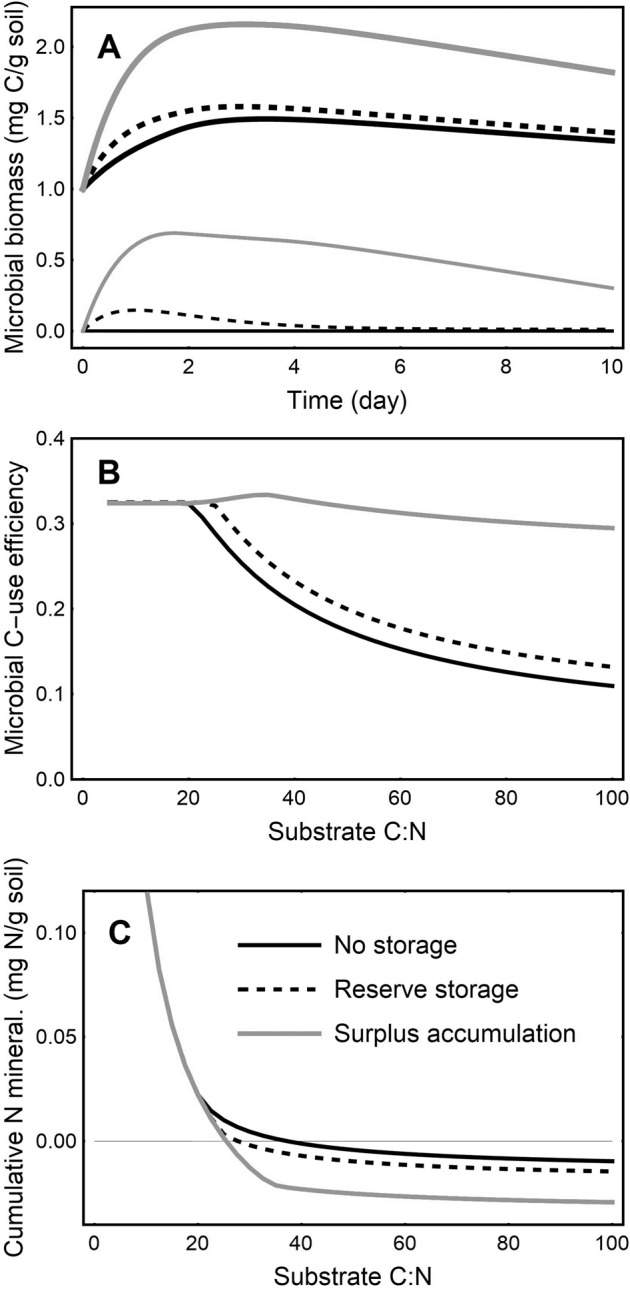
(A) Temporal changes of total microbial biomass C (including storage, CB + CST thick lines) and storage C (CST thin lines) for an initial substrate C:N ratio of 50; and (B) C-use efficiency (CUE) and (C) cumulative net N mineralization as a function of initial substrate C:N ratio, integrated over a 10-day period after substrate addition. CUE is calculated as active microbial biomass growth divided by the sum of biomass growth and respiration. Negative cumulative N mineralization indicates immobilization.
Predictions for ecological stoichiometry
After adding a C-rich substrate, modeled storage C is synthesized faster by microorganisms adopting the surplus storage mode, compared to those using only reserve storage. This suggests that surplus storage is a more effective buffer against stoichiometric imbalances. In contrast, as a consequence of model construction, storage utilization starts sooner after resources are added to soil in the reserve storage strategy, and only later—when N is no longer limiting—in the surplus storage mode.
Storage affects the element ratios of the total biomass, reaching C:N ratios of up to 15.8 and 9.8 for surplus storage and reserve storage, respectively, relative to the fixed C:N of 8.9 in the no-storage scenario. For comparison, microbial biomass C:N and C:P ratios in aquatic bacteria were found to vary several-fold when grown on a wide range of substrate C:P ratios [123], indicating that even larger stoichiometric shifts can occur. Increases in microbial C:P and decreases in C-use efficiency allowed those bacteria to remain C-limited, whereas only regulating C-use efficiency would have resulted in severe P limitation at substrate C:P > 1200 [123]. In that experiment, cells were collected from the water sample with a filter, so that—unlike in soil analyses—total C, P, and N contents were measured. This evidence and our model suggest that storage of C can buffer nutrient limitation and help to explain the plastic microbial C:N or C:P ratios found when total (not only non-storage) biomass is measured [124].
Terrestrial ecosystem models that have accounted for flexible microbial stoichiometry are based on empirical relations between biomass and substrate C:N:P ratios [125, 126]. In these models, stoichiometric flexibility is intended to represent shifts in microbial community structure, rather than short-term variations in storage. Because they neglect internal storage dynamics, these empirical approaches cannot capture short-term microbial responses to fluctuating resource quantity and quality, which may be decisive in determining element transformations in soil.
Predictions for soil carbon and nutrient transformations
The incorporation of C into new biomass as a proportion of total C taken up, termed C-use efficiency, is a central parameter in microbially-explicit models of the soil C cycle [120], because it reflects the retention of C in microbial biomass [111]. Microorganisms in our simulations that employ either surplus or reserve storage achieve higher C-use efficiency with respect to active (non-storage) biomass growth, and this is especially pronounced for surplus storage from substrates with high C:N ratios (Fig. 3B). This efficiency advantage persists even if all storage compounds are consumed, because the cycle of storage and remobilization reduces C loss. The strong storage effects on C-use efficiency in our pulse-response simulations contrast with the lack of effect from steady-state metabolic flux modeling [90], reflecting the importance of temporal dynamics in storage. It remains to be determined how storage affects long-term dynamics of soil organic matter. Lower C losses and higher C-use efficiency could promote soil C sequestration, if the increased biomass C is ultimately stabilized, but could also lower soil organic C stocks if the increased biomass promotes decomposition. If the first mechanism dominates, our model predicts that even if storage compounds are not directly stabilized in soil, they could indirectly enhance C sequestration by supporting more efficient formation of non-storage biomass.
When substrate becomes C-depleted and N-limitation ends, remobilization of stored C allows available N to be used for biomass production. This coupling of C and N cycles through storage means that even C-only storage reduces N mineralization. As a result, cumulative N mineralization at the end of the simulation is lower under any storage mode than without storage (Fig. 3C). Therefore, C storage is predicted to reduce losses of both C and N, a win-win strategy for microbes experiencing frequent fluctuations in resource supply.
These model predictions are not easily tested with existing soil datasets, which do not capture storage compounds. C-use efficiency has been shown to decrease with substrate C:N ratio in soil and litter [127, 128], but in other cases the change was small or positive, even when substrate C:N was increased substantially [129]. Our modeled responses suggest that storage may contribute to the wide range of C-use efficiencies observed in past studies [130].
Outlook
Microbial communities are instrumental in nutrient and energy flows through the biosphere [5]. Based on our taxonomic analysis and mathematical modeling of storage traits, we argue that microbial storage is widespread and may directly modulate these flows over the short-term as well as indirectly enhancing the efficiency of resource use over longer periods. At a global scale, ongoing efforts to incorporate microbial processes into Earth system models may benefit from considering the effect of storage on resource-use stoichiometry and efficiency, particularly with respect to C flows. Explicit modeling of storage might not be necessary over the long-term or under steady-state conditions, but the dynamic nature of storage suggests that system responses to perturbations may be sensitive to its buffering effects. Investigating these possibilities could yield new insights into how the Earth system will respond to climate disturbances, when temporal variability is taken into account [131].
Though we propose that microbial storage should be considered at larger temporal and spatial scales, considerable work is still required to achieve an adequate understanding of storage in microbial ecology. Three areas stand out in particular: our limited knowledge of storage trait occurrence; the conceptual and methodological challenges posed by storage compounds that serve multiple functions; and the necessity of considering storage in its dynamic context.
Assessment of storage traits
Our knowledge of storage in specific microorganisms is largely limited to culturable species, notwithstanding evidence from in situ sampling of fungi [38]. The culturability bias is exacerbated by uneven screening of microbes for storage compound synthesis. The search for general patterns of storage requires more representative screening with broader coverage of compounds and organisms. Moreover, investigation of storage in diverse environments will enhance our understanding of what drives the selection of storage traits and the magnitude of resource flows to and from storage. Culture-independent techniques such as fluorescence- or Raman-activated cell sorting and single-cell sequencing may prove valuable for coupling phenotypic observation of storage to genetic characterization of organisms under natural conditions [132, 133]. High-resolution confirmation of storage synthesis could be achieved through compound-specific staining combined with isotopic labeling and NanoSIMS [22]. Here we have avoided reliance on metagenomic evidence, since a genetically inferred synthetic capacity cannot guarantee that the organism utilizes that compound for storage functions. However, combining molecular genetics techniques with methods for characterizing storage traits holds great promise for studies of complex communities [134].
Mutants for genes involved in storage compound biosynthesis, regulation, or degradation have yielded new insights into the roles of storage compounds, but have been studied in the context of clonal populations rather than from an (eco)system or community-level perspective. Nonetheless, studies of isolates can be powerful when integrated with metabolomic analysis. A compelling example was the recent use of real-time metabolomics [135], involving the direct injection of living cells into a mass-spectrometer. This demonstrated that rapid glycogen mobilization provides a survival advantage to E. coli exposed to nutrient pulses, in comparison to a glycogen-storage mutant [93]. Integration of genetic and metabolomic approaches for studying storage in complex communities may draw on advances in environmental metabolomics, where emerging techniques of lipidomics may be particularly relevant for storage research [136].
Multifunctional storage
Large intracellular inclusions of known storage compounds are strong indicators of storage capabilities, but experimentally distinguishing storage from other functions is not straightforward. It was originally proposed that storage can be identified by its synthesis under conditions of resource surplus and its degradation under deficit [137], but the recognition of different modes of storage undermines such straightforward criteria. More sophisticated experimental manipulations will be needed to assess storage functions, which should at a minimum demonstrate that the advantage of the accumulated resource is obtained through its subsequent degradation. This sort of evidence would be further strengthened by evidence that the advantage is reduced when ample extracellular resources are available. Manipulations of resource supply have provided valuable insights through experimentally controlled feeding in animal studies and shading in plants. In microbiological research, strategies of resource manipulation can further benefit from storage-deficient mutants (section “Advantages of microbial storage”). Chemical inhibitors of storage metabolism have also proven useful in the past [77], and may enable less targeted manipulations within complex communities.
Storage dynamics
Microbial storage is by definition a dynamic process, so understanding storage functions will require consideration of the time dimension. Storage compound levels can only be properly interpreted in the context of the community’s past, which necessitates careful consideration of sampling and storage procedures. Storage dynamics present microbiologists and biogeochemists with numerous open questions, including what factors drive storage compound accumulation and degradation; what the turnover times of these compounds are and what influences these; and how past storage accumulation affects future patterns of nutrient uptake and allocation. If nutrient availability and elemental composition determine patterns of microbial storage, the amount of storage at a particular time relative to other cellular components might provide a useful indicator of microbial nutritional history [138], a possibility that is yet to be explored in soil. In other cases, changes in storage compound levels may better reflect ongoing ecological processes than the absolute levels.
There is a temporal mismatch between microbiological experiments conducted over periods of minutes to days and the longer-term view taken by soil studies (section “Dynamics of storage in soil”). This is indicative of the general challenge, not limited to storage ecology, to determine the appropriate temporal scale for elucidating dynamic processes. Theoretical analyses as in section “Modeling of microbial storage dynamics” could guide investigations by providing testable hypotheses on storage dynamics.
Concluding remarks
Current evidence shows that storage is: (i) a widespread trait among diverse microorganisms in soil that (ii) provides important advantages and plays fundamental roles in their life-history strategies, (iii) storage is not only associated with resource surplus, but regulated by physiological and environmental cues that may lead to storage even in times of scarcity, and finally, (iv) storage can—in certain contexts and timeframes—modulate microbial transformations of energy, C or other nutrients in soil. It is highly likely that in soils, as in other ecosystems, storage plays a crucial role in resource allocation and survival strategies. Closer consideration of storage will enrich our understanding of microbial lifestyles and their biogeochemical roles at micro- to global scales.
Supplementary information
Acknowledgements
Many thanks to Aliia Gilmullina, whose hospitality supported the initiation of this work, and to Emilia Hannula and Andreas Schweiger for their comments on early drafts. We are grateful for the critical input of three anonymous reviewers. KMJ acknowledges the Dutch Research Council (NWO) for funding of the Vital Soils project under the “Programma NWO Groen 2015” (ALWGR.2015.5) and the NWO Veni project VI.Veni.202.086. SLR acknowledges support from the National Science Foundation Graduate Research Fellowship (00039202) and a Graduate Research Opportunities Worldwide (GROW) fellowship supported by the NSF and NWO (040.15.054/6097). SM received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 101001608).
Competing interests
The authors declare no competing interests: KMJ conceived of the work, SM undertook modeling, all authors contributed to data collection and conceptual development, KMJ, SLR, and SM drafted the paper, and all authors were involved in paper revision and have approved the final version.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-01110-w.
References
- 1.Pond C. Storage. In: Townsend C, Calow P, editors. Physiological ecology. Oxford: Blackwell Scientific; 1981. p. 190–219.
- 2.Chapin FS, Schulze E, Mooney HA. The ecology and economics of storage in plants. Annu Rev Ecol Syst. 1990;21:423–47. [Google Scholar]
- 3.Moradali MF, Rehm BHA. Bacterial biopolymers: from pathogenesis to advanced materials. Nat Rev Microbiol. 2020;18:195–210. doi: 10.1038/s41579-019-0313-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varpe Ø. Life history adaptations to seasonality. Integr Comp Biol. 2017;57:943–60. doi: 10.1093/icb/icx123. [DOI] [PubMed] [Google Scholar]
- 5.Paul EA. Soil microbiology, ecology and biochemistry. 4th ed. Waltham, MA: Academic Press; 2015.
- 6.Becker KW, Collins JR, Collins BP, Groussman RD, White AE, Fredricks HF, et al. Daily changes in phytoplankton lipidomes reveal mechanisms of energy storage in the open ocean. Nat Commun. 2018;9:1–9. doi: 10.1038/s41467-018-07346-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rothermich MM, Guerrero R, Lenz RW, Goodwin S. Characterization, seasonal occurrence, and diel fluctuation of poly(hydroxyalkanoate) in photosynthetic microbial mats. Appl Environ Microbiol. 2000;66:13. doi: 10.1128/aem.66.10.4279-4291.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borzi A. Le comunicazioni intracellulari delle Nostochinee. Malpighia. 1887;1:28–74. [Google Scholar]
- 9.Sherman LA, Meunier P, Colón-López MS. Diurnal rhythms in metabolism: a day in the life of a unicellular, diazotrophic cyanobacterium. Photosynth Res. 1998;58:25–42. [Google Scholar]
- 10.Stuart RK, Mayali X, Boaro AA, Zemla A, Everroad RC, Nilson D, et al. Light regimes shape utilization of extracellular organic C and N in a cyanobacterial biofilm. mBio. 2016;7:e00650–16. doi: 10.1128/mBio.00650-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen MM. Cyanobacterial cell inclusions. Annu Rev Microbiol. 1984;38:1–25. doi: 10.1146/annurev.mi.38.100184.000245. [DOI] [PubMed] [Google Scholar]
- 12.Sanz-Luque E, Bhaya D, Grossman AR. Polyphosphate: a multifunctional metabolite in cyanobacteria and algae. Front Plant Sci. 2020;11:938. doi: 10.3389/fpls.2020.00938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin P, Lauro FM, Sarkar A, Goodkin N, Prakash S, Vinayachandran PN. Particulate polyphosphate and alkaline phosphatase activity across a latitudinal transect in the tropical Indian Ocean: polyphosphate in the tropical Indian Ocean. Limnol Oceanogr. 2018;63:1395–406. [Google Scholar]
- 14.Diaz J, Ingall E, Benitez-Nelson C, Paterson D, de Jonge MD, McNulty I, et al. Marine polyphosphate: a key player in geologic phosphorus sequestration. Science. 2008;320:652–5. doi: 10.1126/science.1151751. [DOI] [PubMed] [Google Scholar]
- 15.Godwin CM, Cotner JB. Aquatic heterotrophic bacteria have highly flexible phosphorus content and biomass stoichiometry. ISME J. 2015;9:2324–7. doi: 10.1038/ismej.2015.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oehmen A, Lemos P, Carvalho G, Yuan Z, Keller J, Blackall L, et al. Advances in enhanced biological phosphorus removal: From micro to macro scale. Water Res. 2007;41:2271–300. doi: 10.1016/j.watres.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 17.Dorofeev AG, Nikolaev YuA, Mardanov AV, Pimenov NV. Role of phosphate-accumulating bacteria in biological phosphorus removal from wastewater. Appl Biochem Microbiol. 2020;56:1–14. [Google Scholar]
- 18.Carrondo MA. Ferritins, iron uptake and storage from the bacterioferritin viewpoint. EMBO J. 2003;22:1959–68. doi: 10.1093/emboj/cdg215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Canessa P, Larrondo LF. Environmental responses and the control of iron homeostasis in fungal systems. Appl Microbiol Biotechnol. 2013;97:939–55. doi: 10.1007/s00253-012-4615-x. [DOI] [PubMed] [Google Scholar]
- 20.Harrison PM, Arosio P. The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta. 1996;1275:161–203. doi: 10.1016/0005-2728(96)00022-9. [DOI] [PubMed] [Google Scholar]
- 21.Docampo R, Moreno SNJ. Acidocalcisomes. Cell Calcium. 2011;50:113–9. doi: 10.1016/j.ceca.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsednee M, Castruita M, Salomé PA, Sharma A, Lewis BE, Schmollinger SR, et al. Manganese co-localizes with calcium and phosphorus in Chlamydomonas acidocalcisomes and is mobilized in manganese-deficient conditions. J Biol Chem. 2019;294:17626–41. doi: 10.1074/jbc.RA119.009130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mojzeš P, Gao L, Ismagulova T, Pilátová J, Moudříková Š, Gorelova O, et al. Guanine, a high-capacity and rapid-turnover nitrogen reserve in microalgal cells. Proc Natl Acad Sci USA. 2020;117:32722–30. [DOI] [PMC free article] [PubMed]
- 24.Turner BL. Inositol phosphates in soil: Amounts, forms and significance of the phosphorylated inositol stereoisomers. In: Turner BL, Richardson AE, Mullaney EJ, editors. Inositol phosphates: linking agriculture and the environment. 2007. Wallingford: CABI; 2007. p. 186–206.
- 25.Flemming H-C, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–33. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 26.Otero A, Vincenzini M. Nostoc (Cyanophyceae) goes nude: Extracellular polysaccharides serve as a sink for reducing power under unbalanced C/N metabolism. J Phycol. 2004;40:74–81. [Google Scholar]
- 27.Wang J, Yu H-Q. Biosynthesis of polyhydroxybutyrate (PHB) and extracellular polymeric substances (EPS) by Ralstonia eutropha ATCC 17699 in batch cultures. Appl Microbiol Biotechnol. 2007;75:871–8. doi: 10.1007/s00253-007-0870-7. [DOI] [PubMed] [Google Scholar]
- 28.Brangarí AC, Fernàndez-Garcia D, Sanchez-Vila X, Manzoni S. Ecological and soil hydraulic implications of microbial responses to stress—a modeling analysis. Adv Water Resour. 2018;116:178–94. [Google Scholar]
- 29.Pal S, Manna A, Paul AK. Production of poly(β-hydroxybutyric acid) and exopolysaccharide by Azotobacter beijerinckii WDN-01. World J Microbiol Biotechnol. 1999;15:11–6. [Google Scholar]
- 30.Kuzyakov Y, Blagodatskaya E. Microbial hotspots and hot moments in soil: Concept & review. Soil Biol Biochem. 2015;83:184–99. [Google Scholar]
- 31.Hauschild P, Röttig A, Madkour MH, Al-Ansari AM, Almakishah NH, Steinbüchel A. Lipid accumulation in prokaryotic microorganisms from arid habitats. Appl Microbiol Biotechnol. 2017;101:2203–16. doi: 10.1007/s00253-017-8149-0. [DOI] [PubMed] [Google Scholar]
- 32.Wang JG, Bakken LR. Screening of soil bacteria for poly-β-hydroxybutyric acid production and its role in the survival of starvation. Micro Ecol. 1998;35:94–101. doi: 10.1007/s002489900063. [DOI] [PubMed] [Google Scholar]
- 33.Hanzlíková A, Jandera A, Kunc F. Poly-3-hydroxybutyrate production and changes of bacterial community in the soil. Folia Microbiologica. 1985;30:58–64. [Google Scholar]
- 34.Iwahara S, Miki S. Production of α-α-trehalose by a bacterium isolated from soil. Agric Biol Chem. 1988;52:867–8. [Google Scholar]
- 35.Treseder KK, Lennon JT. Fungal traits that drive ecosystem dynamics on land. Microbiol Mol Biol Rev. 2015;79:243–62. doi: 10.1128/MMBR.00001-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.López MF, Männer P, Willmann A, Hampp R, Nehls U. Increased trehalose biosynthesis in Hartig net hyphae of ectomycorrhizas. N Phytol. 2007;174:389–98. doi: 10.1111/j.1469-8137.2007.01983.x. [DOI] [PubMed] [Google Scholar]
- 37.Bünemann EK, Smernik RJ, Doolette AL, Marschner P, Stonor R, Wakelin SA, et al. Forms of phosphorus in bacteria and fungi isolated from two Australian soils. Soil Biol Biochem. 2008;40:1908–15. [Google Scholar]
- 38.Genet P, Prevost A, Pargney JC. Seasonal variations of symbiotic ultrastructure and relationships of two natural ectomycorrhizae of beech (Fagus sylvatica/Lactarius blennius var. viridis and Fagus sylvatica/Lactarius subdulcis) Trees. 2000;14:465–74. [Google Scholar]
- 39.Frey B, Brunner I, Walther P, Scheidegger C, Zierold K. Element localization in ultrathin cryosections of high-pressure frozen ectomycorrhizal spruce roots. Plant Cell Environ. 1997;20:929–37. [Google Scholar]
- 40.Hanzlíkova A, Jandera A, Kunc F. Formation of poly-3-hydroxybutyrate by a soil microbial community during batch and heterocontinuous cultivation. Folia Microbiol. 1984;29:233–41. [Google Scholar]
- 41.Mason-Jones K, Banfield CC, Dippold MA. Compound‐specific 13C stable isotope probing confirms synthesis of polyhydroxybutyrate by soil bacteria. Rapid Commun Mass Spectrom. 2019;33:795–802. doi: 10.1002/rcm.8407. [DOI] [PubMed] [Google Scholar]
- 42.Hedlund K. Soil microbial community structure in relation to vegetation management on former agricultural land. Soil Biol Biochem. 2002;34:1299–307. [Google Scholar]
- 43.White PM, Potter TL, Strickland TC. Pressurized liquid extraction of soil microbial phospholipid and neutral lipid fatty acids. J Agric Food Chem. 2009;57:7171–7. doi: 10.1021/jf901257n. [DOI] [PubMed] [Google Scholar]
- 44.Xu X, Thornton PE, Post WM. A global analysis of soil microbial biomass carbon, nitrogen and phosphorus in terrestrial ecosystems: Global soil microbial biomass C, N and P. Glob Ecol Biogeogr. 2013;22:737–49. [Google Scholar]
- 45.Bååth E. The use of neutral lipid fatty acids to indicate the physiological conditions of soil fungi. Micro Ecol. 2003;45:373–83. doi: 10.1007/s00248-003-2002-y. [DOI] [PubMed] [Google Scholar]
- 46.Soliman AH, Radwan SS. Degradation of sterols, triacylglycerol, and phospholipids by soil microorganisms. Zbl Bakt II Abt. 1981;136:420–6. [Google Scholar]
- 47.Diakhaté S, Gueye M, Chevallier T, Diallo NH, Assigbetse K, Abadie J, et al. Soil microbial functional capacity and diversity in a millet-shrub intercropping system of semi-arid Senegal. J Arid Environ. 2016;129:71–9. [Google Scholar]
- 48.Bölscher T, Wadsö L, Börjesson G, Herrmann AM. Differences in substrate use efficiency: impacts of microbial community composition, land use management, and substrate complexity. Biol Fertil Soils. 2016;52:547–59. [Google Scholar]
- 49.Mason-Jones K, Schmücker N, Kuzyakov Y. Contrasting effects of organic and mineral nitrogen challenge the N-Mining Hypothesis for soil organic matter priming. Soil Biol Biochem. 2018;124:38–46. [Google Scholar]
- 50.Muhammadi S, Afzal M, Hameed S. Bacterial polyhydroxyalkanoates-eco-friendly next generation plastic: Production, biocompatibility, biodegradation, physical properties and applications. Green Chem Lett Rev. 2015;8:56–77. [Google Scholar]
- 51.Jose NA, Lau R, Swenson TL, Klitgord N, Garcia-Pichel F, Bowen BP, et al. Flux balance modeling to predict bacterial survival during pulsed-activity events. Biogeosciences. 2018;15:2219–29. [Google Scholar]
- 52.Medeiros PM, Fernandes MF, Dick RP, Simoneit BRT. Seasonal variations in sugar contents and microbial community in a ryegrass soil. Chemosphere. 2006;65:832–9. doi: 10.1016/j.chemosphere.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 53.Žifčáková L, Větrovský T, Lombard V, Henrissat B, Howe A, Baldrian P. Feed in summer, rest in winter: microbial carbon utilization in forest topsoil. Microbiome. 2017;5:1–12. doi: 10.1186/s40168-017-0340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ratcliff WC, Denison RF. Individual-level bet hedging in the bacterium Sinorhizobium meliloti. Curr Biol. 2010;20:1740–4. doi: 10.1016/j.cub.2010.08.036. [DOI] [PubMed] [Google Scholar]
- 55.Schoch CL, Ciufo S, Domrachev M, Hotton CL, Kannan S, Khovanskaya R, et al. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database. 2020;2020:baaa062. doi: 10.1093/database/baaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi J, Kim S-H. A genome tree of life for the fungi kingdom. Proc Natl Acad Sci USA. 2017;114:9391–6. doi: 10.1073/pnas.1711939114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jun S-R, Sims GE, Wu GA, Kim S-H. Whole-proteome phylogeny of prokaryotes by feature frequency profiles: An alignment-free method with optimal feature resolution. Proc Natl Acad Sci USA. 2010;107:133–8. doi: 10.1073/pnas.0913033107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elbahloul Y, Krehenbrink M, Reichelt R, Steinbuchel A. Physiological conditions conducive to high cyanophycin content in biomass of Acinetobacter calcoaceticus strain ADP1. Appl Environ Microbiol. 2005;71:858–66. doi: 10.1128/AEM.71.2.858-866.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lillie SH, Pringle JR. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol. 1980;143:1384–94. doi: 10.1128/jb.143.3.1384-1394.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hall KD, Guo J. Obesity energetics: Body weight regulation and the effects of diet composition. Gastroenterology. 2017;152:1718–27.e3. doi: 10.1053/j.gastro.2017.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sala A, Woodruff DR, Meinzer FC. Carbon dynamics in trees: feast or famine? Tree Physiol. 2012;32:764–75. doi: 10.1093/treephys/tpr143. [DOI] [PubMed] [Google Scholar]
- 62.Varpe Ø, Ejsmond MJ. Trade-offs between storage and survival affect diapause timing in capital breeders. Evol Ecol. 2018;32:623–41. [Google Scholar]
- 63.Heilmeier H, Freund M, Steinlein T, Schulze E-D, Monson RK. The influence of nitrogen availability on carbon and nitrogen storage in the biennial Cirsium vulgare (Savi) Ten. I. Storage capacity in relation to resource acquisition, allocation and recycling. Plant Cell Environ. 1994;17:1125–31. [Google Scholar]
- 64.Pond CM. Ecology of storage. In: Levin SA, editor. Encyclopedia of biodiversity, 2nd ed. Amsterdam: Academic Press; 2013. p. 23–38.
- 65.McCue MD. Starvation physiology: reviewing the different strategies animals use to survive a common challenge. Comp Biochem Physiol A Mol Integr Physiol. 2010;156:1–18. doi: 10.1016/j.cbpa.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 66.Donald J, Pannabecker TL. Osmoregulation in desert-adapted mammals. In: Hyndman KA, Pannabecker TL, editors. Sodium and water homeostasis. New York: Springer New York; 2015. p. 191–211.
- 67.Röttig A, Hauschild P, Madkour MH, Al-Ansari AM, Almakishah NH, Steinbüchel A. Analysis and optimization of triacylglycerol synthesis in novel oleaginous Rhodococcus and Streptomyces strains isolated from desert soil. J Biotechnol. 2016;225:48–56. doi: 10.1016/j.jbiotec.2016.03.040. [DOI] [PubMed] [Google Scholar]
- 68.Bailey AP, Koster G, Guillermier C, Hirst EMA, MacRae JI, Lechene CP, et al. Antioxidant role for lipid droplets in a stem cell niche of Drosophila. Cell. 2015;163:340–53. doi: 10.1016/j.cell.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jenni-Eiermann S, Jenni L. Fasting in birds: general patterns and the special case of endurance flight. In: McCue MD, editor. Comparative physiology of fasting, starvation, and food limitation. 2012. Berlin: Springer; 2012. p. 171–92.
- 70.Fischer B, Dieckmann U, Taborsky B. When to store energy in a stochastic environment. Evolution. 2011;65:1221–32. doi: 10.1111/j.1558-5646.2010.01198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bonnet X, Bradshaw D, Shine R. Capital versus income breeding: An ectothermic perspective. Oikos. 1998;83:333. [Google Scholar]
- 72.de Mazancourt C, Schwartz MW. Starve a competitor: evolution of luxury consumption as a competitive strategy. Theor Ecol. 2012;13:37–49. [Google Scholar]
- 73.Ejsmond MJ, Varpe Ø, Czarnoleski M, Kozłowski J. Seasonality in offspring value and trade-offs with growth explain capital breeding. Am Nat. 2015;186:E111–25. [Google Scholar]
- 74.Kourmentza C, Plácido J, Venetsaneas N, Burniol-Figols A, Varrone C, Gavala HN, et al. Recent advances and challenges towards sustainable polyhydroxyalkanoate (PHA) production. Bioengineering. 2017;4:55. doi: 10.3390/bioengineering4020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilson WA, Roach PJ, Montero M, Baroja-Fernández E, Muñoz FJ, Eydallin G, et al. Regulation of glycogen metabolism in yeast and bacteria. FEMS Microbiol Rev. 2010;34:952–85. doi: 10.1111/j.1574-6976.2010.00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doi Y, Kawaguchi Y, Koyama N, Nakamura S, Hiramitsu M, Yoshida Y, et al. Synthesis and degradation of polyhydroxyalkanoates in Alcaligenes eutrophus. FEMS Microbiol Lett. 1992;103:103–8. [Google Scholar]
- 77.Alvarez AHM, Kalscheuer R, Steinbüchel A. Accumulation of storage lipids in species of Rhodococcus and Nocardia and effect of inhibitors and polyethylene glycol. Lipid. 1997;99:239–46. [Google Scholar]
- 78.Parrou JL, Enjalbert B, Plourde L, Bauche A, Gonzalez B, François J. Dynamic responses of reserve carbohydrate metabolism under carbon and nitrogen limitations in Saccharomyces cerevisiae. Yeast. 1999;15:191–203. doi: 10.1002/(SICI)1097-0061(199902)15:3<191::AID-YEA358>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 79.Gebremariam SY, Beutel MW, Christian D, Hess TF. Research advances and challenges in the microbiology of enhanced biological phosphorus removal-A critical review. Water Environ Res. 2011;83:195–219. doi: 10.2175/106143010x12780288628534. [DOI] [PubMed] [Google Scholar]
- 80.Ratledge C. Fatty acid biosynthesis in microorganisms being used for single cell oil production. Biochimie. 2004;86:807–15. doi: 10.1016/j.biochi.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 81.Matin A, Veldhuis C, Stegeman V, Veenhuis M. Selective advantage of a Spirillum sp. in a carbon-limited environment. Accumulation of poly-β-hydroxybutyric acid and its role in starvation. J Gen Microbiol. 1979;112:349–55. doi: 10.1099/00221287-112-2-349. [DOI] [PubMed] [Google Scholar]
- 82.Poblete-Castro I, Escapa IF, Jäger C, Puchalka J, Chi Lam C, Schomburg D, et al. The metabolic response of P. putida KT2442 producing high levels of polyhydroxyalkanoate under single- and multiple-nutrient-limited growth: Highlights from a multi-level omics approach. Micro Cell Fact. 2012;11:34. doi: 10.1186/1475-2859-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wilkinson JF, Munro ALS. The influence of growth limiting conditions on the synthesis of possible carbon and energy storage polymers in Bacillus megaterium. In: Powell EO, Evans CGT, Strange RE, Tempest DW, editors. Microbial physiology and continuous culture, Proceedings of the Third International Symposium. Salisbury, United Kingdom: Her Majesty’s Stationery Office; 1967. p. 173–85.
- 84.Alvarez HM, Mayer F, Fabritius D, Steinbüchel A. Formation of intracytoplasmic lipid inclusions by Rhodococcus opacus strain PD630. Arch Microbiol. 1996;165:377–86. doi: 10.1007/s002030050341. [DOI] [PubMed] [Google Scholar]
- 85.Orchard ED, Benitez-Nelson CR, Pellechia PJ, Lomas MW, Dyhrman ST. Polyphosphate in Trichodesmium from the low-phosphorus Sargasso Sea. Limnol Oceanogr. 2010;55:2161–9. [Google Scholar]
- 86.Li J, Mara P, Schubotz F, Sylvan JB, Burgaud G, Klein F, et al. Recycling and metabolic flexibility dictate life in the lower oceanic crust. Nature. 2020;579:250–5. doi: 10.1038/s41586-020-2075-5. [DOI] [PubMed] [Google Scholar]
- 87.Preiss J, Romeo T. Molecular biology and regulatory aspects of glycogen biosynthesis in bacteria. Prog Nucleic Acid Res Mol Biol. 1994;47:299–329. doi: 10.1016/s0079-6603(08)60255-x. [DOI] [PubMed] [Google Scholar]
- 88.Mackerras AH, de Chazal NM, Smith GD. Transient accumulations of cyanophycin in Anabaena cylindrica and Synechocystis 6308. J Gen Microbiol. 1990;136:2057–65. [Google Scholar]
- 89.Parnas H, Cohen D. The optimal strategy for the metabolism of reserve materials in micro-organisms. J Theor Biol. 1976;56:19–55. doi: 10.1016/s0022-5193(76)80044-6. [DOI] [PubMed] [Google Scholar]
- 90.Dijkstra P, Salpas E, Fairbanks D, Miller EB, Hagerty SB, van Groenigen KJ, et al. High carbon use efficiency in soil microbial communities is related to balanced growth, not storage compound synthesis. Soil Biol Biochem. 2015;89:35–43. [Google Scholar]
- 91.Empadinhas N, da Costa MS. Osmoadaptation mechanisms in prokaryotes: distribution of compatible solutes. Int Microbiol. 2008;11:151–61. [PubMed] [Google Scholar]
- 92.Albi T, Serrano A. Inorganic polyphosphate in the microbial world. Emerging roles for a multifaceted biopolymer. World J Microbiol Biotechnol. 2016;32:27. doi: 10.1007/s11274-015-1983-2. [DOI] [PubMed] [Google Scholar]
- 93.Sekar K, Linker SM, Nguyen J, Grünhagen A, Stocker R, Sauer U. Bacterial glycogen provides short-term benefits in changing environments. Appl Environ Microbiol. 2020;86:e00049–20. doi: 10.1128/AEM.00049-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Silljé HH, Paalman JW, ter Schure EG, Olsthoorn SQ, Verkleij AJ, Boonstra J, et al. Function of trehalose and glycogen in cell cycle progression and cell viability in Saccharomyces cerevisiae. J Bacteriol. 1999;181:396–400. doi: 10.1128/jb.181.2.396-400.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jahid IK, Silva AJ, Benitez JA. Polyphosphate stores enhance the ability of Vibrio cholerae to overcome environmental stresses in a low-phosphate environment. Appl Environ Microbiol. 2006;72:7043–9. doi: 10.1128/AEM.00924-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ramírez-Trujillo JA, Dunn MF, Suárez-Rodríguez R, Hernández-Lucas I. The Sinorhizobium meliloti glyoxylate cycle enzyme isocitrate lyase (AceA) is required for the utilization of poly-β-hydroxybutyrate during carbon starvation. Ann Microbiol. 2016;66:921–4. [Google Scholar]
- 97.Vagabov VM, Trilisenko LV, Kulaev IS. Dependence of inorganic polyphosphate chain length on the orthophosphate content in the culture medium of the yeast Saccharomyces cerevisiae. Biochemistry. 2000;65:6. [PubMed] [Google Scholar]
- 98.Schimz K-L, Irrgang K, Overhoff B. Glycogen, a cytoplasmic reserve polysaccharide of Cellulomonas sp. (DSM20108): Its identification, carbon source-dependent accumulation, and degradation during starvation. FEMS Microbiol Lett. 1985;30:165–9. [Google Scholar]
- 99.Kalscheuer R, Stöveken T, Malkus U, Reichelt R, Golyshin PN, Sabirova JS, et al. Analysis of storage lipid accumulation in Alcanivorax borkumensis: Evidence for alternative triacylglycerol biosynthesis routes in bacteria. J Bacteriol. 2007;189:918–28. doi: 10.1128/JB.01292-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Busuioc M, Mackiewicz K, Buttaro BA, Piggot PJ. Role of intracellular polysaccharide in persistence of Streptococcus mutans. J Bacteriol. 2009;191:7315–22. doi: 10.1128/JB.00425-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ruiz JA, Lopez NI, Fernandez RO, Mendez BS. Polyhydroxyalkanoate degradation Is associated with nucleotide accumulation and enhances stress resistance and survival of Pseudomonas oleovorans in natural water microcosms. Appl Environ Microbiol. 2001;67:225–30. doi: 10.1128/AEM.67.1.225-230.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Klotz A, Georg J, Bučinská L, Watanabe S, Reimann V, Januszewski W, et al. Awakening of a dormant cyanobacterium from nitrogen chlorosis reveals a genetically determined program. Curr Biol. 2016;26:2862–72. doi: 10.1016/j.cub.2016.08.054. [DOI] [PubMed] [Google Scholar]
- 103.Elbein AD. New insights on trehalose: a multifunctional molecule. Glycobiology. 2003;13:17R–27R. doi: 10.1093/glycob/cwg047. [DOI] [PubMed] [Google Scholar]
- 104.Obruca S, Sedlacek P, Koller M. The underexplored role of diverse stress factors in microbial biopolymer synthesis. Bioresour Technol. 2021;326:124767. doi: 10.1016/j.biortech.2021.124767. [DOI] [PubMed] [Google Scholar]
- 105.Ayub ND, Tribelli PM, López NI. Polyhydroxyalkanoates are essential for maintenance of redox state in the Antarctic bacterium Pseudomonas sp. 14-3 during low temperature adaptation. Extremophiles. 2009;13:59–66. doi: 10.1007/s00792-008-0197-z. [DOI] [PubMed] [Google Scholar]
- 106.Grime JP. Evidence for the existence of three primary strategies in plants and its relevance to ecological and evolutionary theory. Am Nat. 1977;111:1169–94. [Google Scholar]
- 107.Ho A, Kerckhof F-M, Luke C, Reim A, Krause S, Boon N, et al. Conceptualizing functional traits and ecological characteristics of methane-oxidizing bacteria as life strategies: Functional traits of methane-oxidizing bacteria. Environ Microbiol Rep. 2013;5:335–45. doi: 10.1111/j.1758-2229.2012.00370.x. [DOI] [PubMed] [Google Scholar]
- 108.Santillan E, Seshan H, Constancias F, Wuertz S. Trait‐based life‐history strategies explain succession scenario for complex bacterial communities under varying disturbance. Environ Microbiol. 2019;21:3751–64. doi: 10.1111/1462-2920.14725. [DOI] [PubMed] [Google Scholar]
- 109.Chesson P. Mechanisms of maintenance of species diversity. Annu Rev Ecol Syst. 2000;31:343–66. [Google Scholar]
- 110.Loreau M, de Mazancourt C. Biodiversity and ecosystem stability: a synthesis of underlying mechanisms. Ecol Lett. 2013;16:106–15. doi: 10.1111/ele.12073. [DOI] [PubMed] [Google Scholar]
- 111.Geyer KM, Kyker-Snowman E, Grandy AS, Frey SD. Microbial carbon use efficiency: accounting for population, community, and ecosystem-scale controls over the fate of metabolized organic matter. Biogeochemistry. 2016;127:173–88. [Google Scholar]
- 112.Manzoni S, Porporato A. Soil carbon and nitrogen mineralization: Theory and models across scales. Soil Biol Biochem. 2009;41:1355–79. [Google Scholar]
- 113.Schultz P, Urban NR. Effects of bacterial dynamics on organic matter decomposition and nutrient release from sediments: a modeling study. Ecol Model. 2008;210:1–14. [Google Scholar]
- 114.Torres-Dorante LO, Claassen N, Steingrobe B, Olfs H-W. Polyphosphate determination in calcium acetate-lactate (CAL) extracts by an indirect colorimetric method. J Plant Nutr Soil Sci. 2004;167:701–3. [Google Scholar]
- 115.Micić V, Köster J, Kruge MA, Engelen B, Hofmann T. Bacterial wax esters in recent fluvial sediments. Org Geochem. 2015;89–90:44–55. [Google Scholar]
- 116.Mooshammer M, Wanek W, Zechmeister-Boltenstern S, Richter A. Stoichiometric imbalances between terrestrial decomposer communities and their resources: mechanisms and implications of microbial adaptations to their resources. Front Microbiol. 2014;5:1–10. doi: 10.3389/fmicb.2014.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Op De Beeck M, Troein C, Siregar S, Gentile L, Abbondanza G, Peterson C, et al. Regulation of fungal decomposition at single-cell level. ISME J. 2020;14:896–905. doi: 10.1038/s41396-019-0583-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liang C, Amelung W, Lehmann J, Kästner M. Quantitative assessment of microbial necromass contribution to soil organic matter. Glob Chang Biol. 2019;25:3578–90. doi: 10.1111/gcb.14781. [DOI] [PubMed] [Google Scholar]
- 119.Ducklow H, Steinberg D, Buesseler K. Upper ocean carbon export and the biological pump. Oceanography. 2001;14:50–8. [Google Scholar]
- 120.Wieder WR, Allison SD, Davidson EA, Georgiou K, Hararuk O, He Y, et al. Explicitly representing soil microbial processes in Earth system models: Soil microbes in Earth system models. Glob Biogeochem Cycles. 2015;29:1782–800. [Google Scholar]
- 121.Schimel J, Weintraub MN. The implications of exoenzyme activity on microbial carbon and nitrogen limitation in soil: a theoretical model. Soil Biol Biochem. 2003;35:549–63. [Google Scholar]
- 122.Ni B-J, Fang F, Rittmann BE, Yu H-Q. Modeling microbial products in activated sludge under feast-famine conditions. Environ Sci Technol. 2009;43:2489–97. doi: 10.1021/es8026693. [DOI] [PubMed] [Google Scholar]
- 123.Godwin CM, Cotner JB. Stoichiometric flexibility in diverse aquatic heterotrophic bacteria is coupled to differences in cellular phosphorus quotas. Front Microbiol. 2015;6:1–15. doi: 10.3389/fmicb.2015.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Camenzind T, Philipp Grenz K, Lehmann J, Rillig MC. Soil fungal mycelia have unexpectedly flexible stoichiometric C:N and C:P ratios. Ecol Lett. 2021;24:208–18. doi: 10.1111/ele.13632. [DOI] [PubMed] [Google Scholar]
- 125.Fatichi S, Manzoni S, Or D, Paschalis A. A mechanistic model of microbially mediated soil biogeochemical processes: a reality check. Glob Biogeochem Cycles. 2019;33:620–48. [Google Scholar]
- 126.Sistla SA, Rastetter EB, Schimel JP. Responses of a tundra system to warming using SCAMPS: a stoichiometrically coupled, acclimating microbe–plant–soil model. Ecol Monogr. 2014;84:151–70. [Google Scholar]
- 127.Lashermes G, Gainvors-Claisse A, Recous S, Bertrand I. Enzymatic strategies and carbon use efficiency of a litter-decomposing fungus grown on maize leaves, stems, and roots. Front Microbiol. 2016;7:1–14. doi: 10.3389/fmicb.2016.01315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lee ZM, Schmidt TM. Bacterial growth efficiency varies in soils under different land management practices. Soil Biol Biochem. 2014;69:282–90. [Google Scholar]
- 129.Camenzind T, Lehmann A, Ahland J, Rumpel S, Rillig MC. Trait‐based approaches reveal fungal adaptations to nutrient‐limiting conditions. Environ Microbiol. 2020;22:3548–60. doi: 10.1111/1462-2920.15132. [DOI] [PubMed] [Google Scholar]
- 130.Manzoni S, Čapek P, Mooshammer M, Lindahl BD, Richter A, Šantrůčková H. Optimal metabolic regulation along resource stoichiometry gradients. Ecol Lett. 2017;20:1182–91. doi: 10.1111/ele.12815. [DOI] [PubMed] [Google Scholar]
- 131.Tang J, Riley WJ. Weaker soil carbon–climate feedbacks resulting from microbial and abiotic interactions. Nat Clim Chang. 2015;5:5. [Google Scholar]
- 132.Lee KS, Pereira FC, Palatinszky M, Behrendt L, Alcolombri U, Berry D, et al. Optofluidic Raman-activated cell sorting for targeted genome retrieval or cultivation of microbial cells with specific functions. Nat Protoc. 2021;16:634–76. doi: 10.1038/s41596-020-00427-8. [DOI] [PubMed] [Google Scholar]
- 133.Günther S, Trutnau M, Kleinsteuber S, Hause G, Bley T, Röske I, et al. Dynamics of polyphosphate-accumulating bacteria in wastewater treatment plant microbial communities detected via DAPI (4′,6′-diamidino-2-phenylindole) and tetracycline labeling. Appl Environ Microbiol. 2009;75:2111–21. doi: 10.1128/AEM.01540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Singleton CM, Petriglieri F, Kristensen JM, Kirkegaard RH, Michaelsen TY, Andersen MH, et al. Connecting structure to function with the recovery of over 1000 high-quality metagenome-assembled genomes from activated sludge using long-read sequencing. Nat Commun. 2021;12:2009. doi: 10.1038/s41467-021-22203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Link H, Fuhrer T, Gerosa L, Zamboni N, Sauer U. Real-time metabolome profiling of the metabolic switch between starvation and growth. Nat Methods. 2015;12:1091–7. doi: 10.1038/nmeth.3584. [DOI] [PubMed] [Google Scholar]
- 136.Warren CR. Altitudinal transects reveal large differences in intact lipid composition among soils. Soil Res. 2021;59:644–59. [Google Scholar]
- 137.Wilkinson J. The problem of energy-storage compounds in bacteria. Exp Cell Res. 1959;7:111–30. [Google Scholar]
- 138.Nickels JS, King JD, White DC. Poly-β-hydroxybutyrate accumulation as a measure of unbalanced growth of the estuarine detrital microbiota. Appl Environ Microbiol. 1979;37:459–65. doi: 10.1128/aem.37.3.459-465.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Murphy DJ. The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma. 2012;249:541–85. doi: 10.1007/s00709-011-0329-7. [DOI] [PubMed] [Google Scholar]
- 140.Alvarez HM. Triacylglycerol and wax ester-accumulating machinery in prokaryotes. Biochimie. 2016;120:28–39. doi: 10.1016/j.biochi.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 141.Koller M, Maršálek L, de Sousa Dias MM, Braunegg G. Producing microbial polyhydroxyalkanoate (PHA) biopolyesters in a sustainable manner. N Biotechnol. 2017;37:24–38. doi: 10.1016/j.nbt.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 142.Obruca S, Sedlacek P, Slaninova E, Fritz I, Daffert C, Meixner K, et al. Novel unexpected functions of PHA granules. Appl Microbiol Biotechnol. 2020;104:4795–810. doi: 10.1007/s00253-020-10568-1. [DOI] [PubMed] [Google Scholar]
- 143.Roach PJ, Depaoli-Roach AA, Hurley TD, Tagliabracci VS. Glycogen and its metabolism: some new developments and old themes. Biochem J. 2012;441:763–87. doi: 10.1042/BJ20111416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang L, Wang M, Wise MJ, Liu Q, Yang T, Zhu Z, et al. Recent progress in the structure of glycogen serving as a durable energy reserve in bacteria. World J Microbiol Biotechnol. 2020;36:14. doi: 10.1007/s11274-019-2795-6. [DOI] [PubMed] [Google Scholar]
- 145.Ruhal R, Kataria R, Choudhury B. Trends in bacterial trehalose metabolism and significant nodes of metabolic pathway in the direction of trehalose accumulation: Trehalose metabolism in bacteria. Micro Biotechnol. 2013;6:493–502. doi: 10.1111/1751-7915.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kalscheuer R. Genetics of wax ester and triacylglycerol biosynthesis in bacteria. In: Timmis KN, editor. Handbook of hydrocarbon and lipid microbiology. Berlin, Heidelberg: Springer Berlin Heidelberg; 2010. p. 527–35.
- 147.Rao NN, Gómez-García MR, Kornberg A. Inorganic polyphosphate: essential for growth and survival. Annu Rev Biochem. 2009;78:605–47. doi: 10.1146/annurev.biochem.77.083007.093039. [DOI] [PubMed] [Google Scholar]
- 148.Denoncourt A, Downey M. Model systems for studying polyphosphate biology: a focus on microorganisms. Curr Genet. 2021;67:331–46. doi: 10.1007/s00294-020-01148-x. [DOI] [PubMed] [Google Scholar]
- 149.Füser G, Steinbüchel A. Analysis of genome sequences for genes of cyanophycin metabolism: Identifying putative cyanophycin metabolizing prokaryotes. Macromol Biosci. 2007;7:278–96. doi: 10.1002/mabi.200600207. [DOI] [PubMed] [Google Scholar]
- 150.Watzer B, Forchhammer K. Cyanophycin: a nitrogen-rich reserve polymer. In: Tiwari A, editor. Cyanobacteria. London: InTech; 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.