Abstract
The molecular evolution processes underlying the acquisition of the placenta in eutherian ancestors are not fully understood. Mouse NCK-interacting kinase (NIK)-related kinase (NRK) is expressed highly in the placenta and plays a role in preventing placental hyperplasia. Here, we show the molecular evolution of NRK, which confers its function for inhibiting placental cell proliferation. Comparative genome analysis identified NRK orthologs across vertebrates, which share the kinase and citron homology (CNH) domains. Evolutionary analysis revealed that NRK underwent extensive amino acid substitutions in the ancestor of placental mammals and has been since conserved. Biochemical analysis of mouse NRK revealed that the CNH domain binds to phospholipids, and a region in NRK binds to and inhibits casein kinase-2 (CK2), which we named the CK2-inhibitory region (CIR). Cell culture experiments suggest the following: 1) Mouse NRK is localized at the plasma membrane via the CNH domain, where the CIR inhibits CK2. 2) This mitigates CK2-dependent phosphorylation and inhibition of PTEN and 3) leads to the inhibition of AKT signaling and cell proliferation. Nrk deficiency increased phosphorylation levels of PTEN and AKT in mouse placenta, supporting our hypothesis. Unlike mouse NRK, chicken NRK did not bind to phospholipids and CK2, decrease phosphorylation of AKT, or inhibit cell proliferation. Both the CNH domain and CIR have evolved under purifying selection in placental mammals. Taken together, our study suggests that placental mammals acquired the phospholipid-binding CNH domain and CIR in NRK for regulating the CK2–PTEN–AKT pathway and placental cell proliferation.
Keywords: NRK, molecular evolution, placental development, CK2, PTEN, AKT
Introduction
The placenta is the defining organ of eutherians (placental mammals). The eutherian placenta is evolutionarily novel, is derived from the chorioallantoic membrane, and varies in morphology across species. It mediates the exchange of nutrients and gas between the mother and the fetus, regulates pregnancy as an endocrine tissue, and plays a role in suppressing the mother’s immune system (Imakawa and Nakagawa 2017). The development of the eutherian placenta is regulated by various extracellular or intracellular factors (Woods et al. 2018; Knofler et al. 2019). However, the molecular mechanisms by which they regulate placental development are not fully understood. Elucidating these molecular mechanisms will provide a basis for developing diagnostic and therapeutic methods for pregnancy complications, including fetal growth retardation. Moreover, it is also not fully understood how molecular evolution underlay the acquisition of the placenta in eutherian ancestors. Eutherian ancestors seem to have acquired several novel genes that code proteins involved in placental development, such as those derived from retrotransposons (Ono et al. 2006; Rawn and Cross 2008; Bolze et al. 2017; Imakawa and Nakagawa 2017). This has led to the increasing importance of research into the molecular evolution of proteins that regulate placental development.
The NCK-interacting kinase (NIK)-related kinase (NRK) is involved in mouse placental development. The mature mouse placenta comprises three histologically distinct layers: the labyrinth, spongiotrophoblast, and maternal decidual layers (Rossant and Cross 2001). Mouse Nrk is highly expressed in spongiotrophoblasts during late pregnancy, when the placenta weight reaches a plateau (Denda et al. 2011). Loss of Nrk leads to placental hyperplasia due to the hyperproliferation of the spongiotrophoblasts and difficulty during delivery (Denda et al. 2011). These observations suggest that mouse NRK plays a role in preventing hyperplasia of the placenta. Spongiotrophoblasts produce several hormones (e.g., placental lactogens) and regulate nutrient distribution between the fetus and placenta (Tunster et al. 2016). Mouse NRK may regulate the endocrine and nutritional environment in pregnancy by controlling spongiotrophoblast proliferation.
NRK is a member of the germinal center kinase (GCK) family subgroup IV. This kinase family exhibits an N-terminal kinase domain, an uncharacterized middle region, and a C-terminal citron homology (CNH) domain (Delpire 2009). In Caenorhabditiselegans and flies, this family comprises only one member, namely, MIG-15 and MSN, respectively. They function as MAP kinase kinase kinase kinase (MAP4K), which is similar to the prototype Ste20 in yeast, and are involved in morphogenic events, including cell migration (Chapman et al. 2008; Plutoni et al. 2019). In mice and humans, the family comprises four kinases, namely, NIK, TRAF2- and NCK-interacting protein kinase (TNIK), misshapen-like kinase 1 (MINK1), and NRK (Delpire 2009). Experiments with mice and human cells have revealed that only NRK suppresses cell proliferation (Yu et al. 2014; Daulat et al. 2016; Masuda et al. 2016; Morioka et al. 2017; Naito et al. 2020). However, the evolutionary origin and molecular determinants of eutherian NRK function remain poorly understood.
Recently, we and the other research groups observed the ability of mouse NRK in suppressing AKT signaling, one of the major signaling pathways involved in the regulation of proliferation (Morioka et al. 2017; Naito et al. 2020). Phosphatidylinositol-3 kinase (PI3K), an upstream factor of the AKT signaling, is activated in response to growth factor stimuli and converts phosphatidylinositol 4,5-diphosphate (PIP2) to phosphatidylinositol 3,4,5-triphosphate (PIP3). AKT then recognizes PIP3 and localizes at the plasma membrane, where threonine 308 (T308) and serine 473 (S473) in AKT are phosphorylated by PDK1 and mTORC2, respectively. This induces the activation of AKT kinase, which phosphorylates various substrate proteins and ultimately promotes cell proliferation (Risso et al. 2015). Mouse NRK suppresses AKT phosphorylation (Morioka et al. 2017; Naito et al. 2020). However, the underlying mechanism remains elusive.
AKT signaling is regulated by various pathways, including the CK2–PTEN–AKT pathway. CK2 is a tetrameric protein complex consisting of two molecules of the catalytic subunit (α or α′ subunit) and two molecules of the regulatory subunit (β subunit). The CK2 complex may contain identical (two α or two α′) or nonidentical (a pair of α and α′) catalytic subunits. CK2α and α′ exhibit overlapped functions. CK2 is ubiquitously expressed, is observed in various cellular compartments, and phosphorylates many substrate proteins (Litchfield 2003). PTEN, a lipid phosphatase that dephosphorylates the third phosphate group of PIP3, is one of the substrate proteins of CK2. CK2 phosphorylates multiple serine/threonine residues (S370, S380, T382, T383, and S385) near the C-terminus of PTEN. Unphosphorylated-PTEN demonstrates high phosphatase activity and is localized at the plasma membrane, thereby converting PIP3 to PIP2. In contrast, phosphorylated PTEN shows reduced phosphatase activity and is localized in the cytosol and the nucleus. Through these mechanisms, CK2 inhibits PTEN and increases PIP3 levels at the plasma membrane, thereby enhancing AKT signaling (Vazquez et al. 2000; Torres and Pulido 2001; Rahdar et al. 2009). However, the types of physiological situations in which AKT signaling is regulated via the CK2–PTEN–AKT pathway remains unclear.
This study aimed to reveal molecular evolution of the placental protein NRK, which confers its function for inhibiting placental cell proliferation.
Results
Nrk Orthologs Were Identified Across Vertebrates
Our synteny analysis showed that the Nrk gene was widely present in vertebrates, including eutherians, marsupials, birds, reptiles, amphibians, and fish (fig. 1A; supplementary table 1, Supplementary Material online). We also found that the genes of other GCK IV family members (Nik, Tnik, and Mink1) were widely present in mammals, birds, reptiles, amphibians, and fish (supplementary fig. S1 and table 2, Supplementary Material online). Notably, we did not find the Mink1 gene in chicken and turtle genomes. These results indicate that each gene of the GCK IV family members was present in the common ancestor of vertebrates.
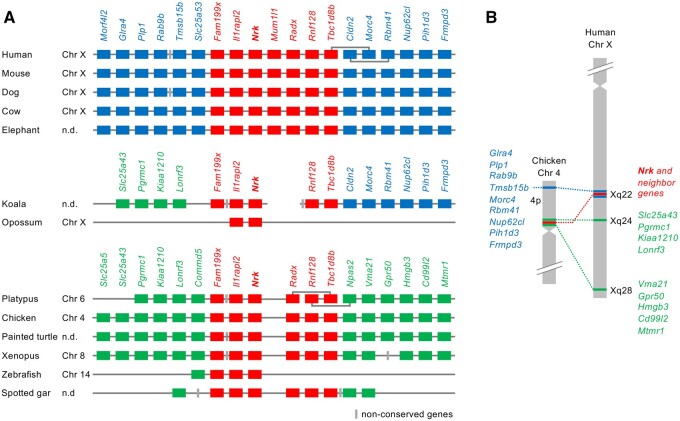
Fig. 1.
Synteny analysis of the Nrk gene. (A) Synteny conservation of the region surrounding the Nrk gene. Red, blue, and green boxes indicate syntenic genes across vertebrates, eutherians, and those observed only in platypus and nonmammals, respectively. Gray boxes indicate nonconserved genes. (B) Loci of Nrk and syntenic genes in chicken and human chromosomes.
Genomic Region Containing Nrk Was Relocated During Early Mammalian Evolution
We compared gene loci and synteny of vertebrate Nrk orthologs (fig. 1A). Human, mouse, dog, and cow Nrk genes were located on the X chromosome. In platypus and chicken, Nrk genes were located on chromosomes 6 and 4, which contain regions orthologous to the eutherian X chromosome (Veyrunes et al. 2008). Synteny analysis of Nrk and the neighboring genes showed three different groups of syntenic genes. Red-colored genes (Nrk and some neighboring genes) retain their synteny from Xenopus to humans. In eutherians, blue-colored genes upstream and downstream of red genes were syntenic. However, in platypus and nonmammals, a different group of genes (green genes) showed synteny. Incomplete koala genome data showed that the genomic region on one side of the Nrk gene was orthologous to the regions of platypus and nonmammals, and the genomic region on the other side was orthologous to certain regions of eutherians. Hence, the genomic region in the koala was maintained in a transitional state. Opossum Nrk was adjacent to Il1rapl2, but no other Nrk synteny genes found in mouse or chicken were identified in the opossum. This indicated that a small genomic region containing Nrk and Il1rapl2 was translocated in the ancestor of the opossum. The human Xq is highly homologous to the 20-Mb region of chicken 4p (Ross et al. 2005). We compared the loci of Nrk and neighboring genes in chicken and human chromosomes (fig. 1B). In chicken, the red and green genes were located near the centromere of 4p, and the blue genes were located near the end of 4p. In humans, the red and blue genes were located at Xq22. The green genes on the left side of Nrk in figure 1A were located at Xq24, whereas the green genes on the right side were located at Xq28.
Nrk Gene Underwent Rapid Molecular Evolution During Early Mammalian Evolution
Next, we identified coding sequences of vertebrate Nrk genes in the orthologous loci (supplementary table 1, Supplementary Material online) based on several public data of validated mRNA sequences, gene predictions, and RNA-seq assemblies (see Materials and Methods). We compared the exon structures of vertebrate Nrk orthologs, and classified them into three types (eutherian, marsupial, and platypus and nonmammalian; supplementary fig. S2A, Supplementary Material online). The human Nrk gene consisted of 29 exons, all of which were coding exons. Almost all eutherian Nrk genes demonstrated orthologs for the aforementioned 29 exons. The therian exon 13 was longer than the orthologous exon of platypus and nonmammals. The orthologous sequences of the eutherian exons 4, 12, and 25 were not found in noneutherians. The eutherian exons 18 and 19 were longer than those of the noneutherians. These changes led to the insertions of amino acid sequences that were found only in the eutherian NRK protein. We searched the origin of the inserted sequences in the transposable element database, but no similar element was found.
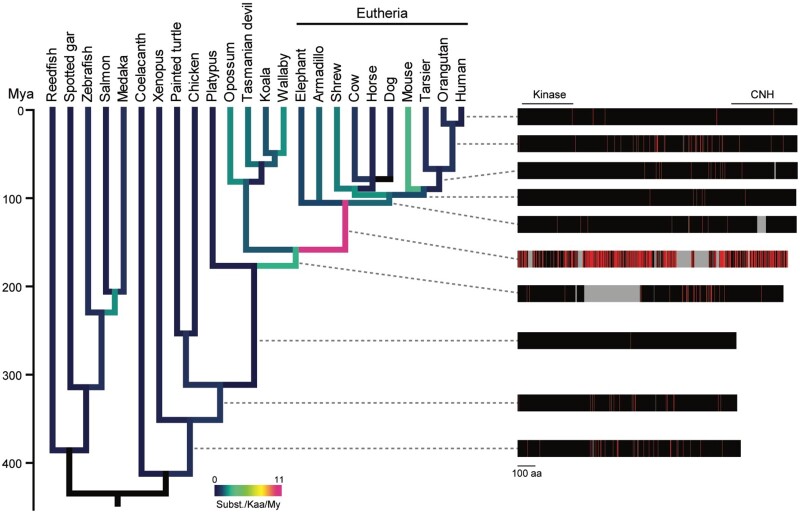
Using the Nrk coding sequences, we estimated the evolutionary rate of the amino acid sequences based on the divergence times of the species in TimeTree (Kumar et al. 2017; fig. 2, left panel). We also analyzed the distribution of amino acid changes in the NRK sequences occurring in each branch from the tetrapod ancestor to humans (fig. 2, right panel; black, no change; gray, insertion; red, substitution). The amino acid evolutionary rate in the eutherian ancestor was 10.8 × 10−9 substitutions/site/year, much higher than those in the therian ancestor (2.6 × 10−9 substitutions/site/year) and the mammalian ancestor (0.012 × 10−9 substitutions/site/year). The eutherian-specific high evolutionary rate was not observed in a control group that comprised of an amino acid sequence collection of 831 orthologous genes in vertebrates reported previously (Takezaki and Nishihara 2016; supplementary fig. S2B, Supplementary Material online), suggesting that the high rate in NRK did not result from a eutherian-specific genome-wide substitution rate. The results demonstrated an accelerated evolutionary rate of NRK in the eutherian ancestor, approximately 100–180 million years ago (Mya). During this period, the NRK sequence was lengthened owing to certain insertions as shown in the exon structure analysis. The protein sequence underwent many substitutions in several regions, even including the kinase and CNH domains. Indeed, the adaptive branch-site random effects likelihood (aBSREL) analysis revealed that the CNH domain experienced positive selection in the eutherian ancestor with ω = 1.91 for 73% of the amino acid sites (P < 0.01 with Bonferroni correction). The evolutionary rates after the most recent common ancestor of eutherians were low (1.2 × 10−9 substitutions/site/year on average of the terminal branch; fig. 2), suggesting the sequence and functional conservation of Nrk among eutherians.
Fig. 2.
Phylogenetic analysis of the amino acid sequences of vertebrate NRK. Amino acid substitutions of NRK along with the vertebrate phylogeny. The amino acid substitutions in each branch were estimated based on the reconstructed ancestral sequences of NRK proteins. The divergence times were retrieved from TimeTree. The colors of the branches indicate the average number of substitutions per 1,000 amino acid sequences over a million years. The right panel shows the changes in amino acid lengths and sequences of ancestral proteins of human NRK. The positions of the kinase and CNH domains of the human NRK are indicated. Sites of amino acid substitutions, unchanged sites, and insertions in each branch are shown in red, black, and gray, respectively. My, million years.
We found that vertebrate NRK protein shares a similar domain architecture: the N-terminal kinase domain and the C-terminal CNH domain. The neighbor-joining (NJ) trees for the amino acid sequences of these domains suggest that both domains of eutherians were distinct from those of noneutherians (supplementary fig. S2C and D, Supplementary Material online), consistent with the result of the evolutionary rate analysis.
Nrk Gene Became to Be Expressed Preferentially in the Placenta During Early Mammalian Evolution
Using public databases, we compared the tissue expression patterns of the mouse Nrk (mNrk) gene and the other GCK IV family members (Nik, Tnik, and Mink1). Mouse Nrk was restrictedly expressed in the placenta, whereas other genes were ubiquitously expressed (supplementary fig. S3A, Supplementary Material online). We also compared the tissue expression pattern of Nrk orthologs in different species. The data were limited, but those of human, sheep, chicken, and Xenopus were available. Human and sheep Nrk genes were restrictedly expressed in the placenta. Chicken and Xenopus Nrk genes were highly expressed in other tissues (supplementary fig. S3B, Supplementary Material online). These results suggest a eutherian-specific pattern of Nrk gene expression.
Mouse NRK Suppressed Cell Proliferation and AKT Signaling, for Which the Middle Region of NRK and the CNH Domain Are Required
Next, we analyzed the molecular function of mouse NRK (mNRK) and compared it with the functions of NRK orthologs and the other GCK IV family members. To the best of our knowledge, no monotypic cell line is known in which endogenous NRK protein can be detected. Using the HEK293 Flp-In T-REx cell system, we generated cells that expressed mNRK or green fluorescence protein (GFP) in a doxycycline (dox)-dependent manner (Naito et al. 2020) and examined two representative proliferative pathways: the AKT and MAPK pathways. Compared with the control cells, dox-treated cells showed decreased cell numbers (fig. 3A) and inhibited AKT phosphorylation without affecting MAPK phosphorylation (fig. 3B). Immunostaining analysis confirmed that mNRK expression inhibited AKT phosphorylation (fig. 3C; supplementary fig. S4A, Supplementary Material online). mNRK expression did not induce cleavage of PARP1 (supplementary fig. S4B, Supplementary Material online), indicating that it did not mediate cellular apoptosis. We then examined the effects of the other GCK IV family members (mouse NIK, TNIK, and MINK1) and chicken NRK (cNRK) on AKT phosphorylation. The expression of these proteins did not decrease AKT phosphorylation (fig. 3D). The protein levels were varied, probably owing to differences in protein stabilities under our experimental conditions. These results demonstrate the unique inhibitory function of mNRK in mediating AKT signaling.
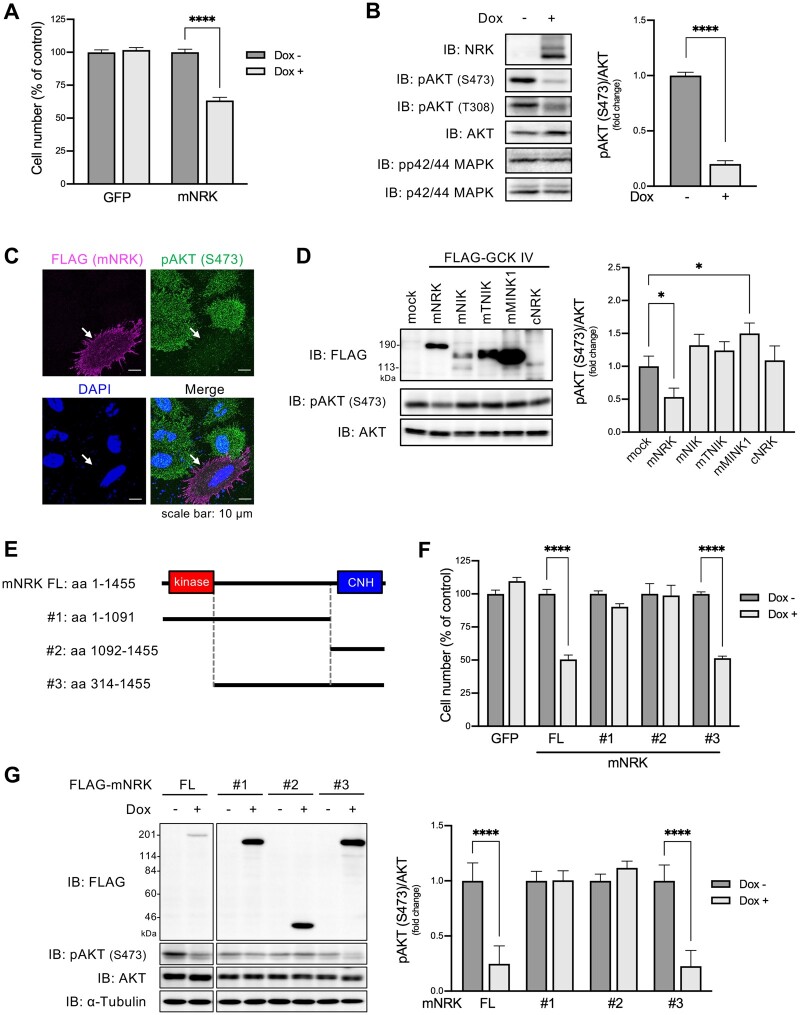
Fig. 3.
Effects of mNRK on cell proliferation and AKT signaling and the corresponding responsible regions of mNRK. (A) Effects of mNRK on cell proliferation. We generated Flp-In T-REx 293 cell lines that expressed mNRK and GFP in doxycycline (dox)-dependent manner. The cells were treated with dox (1 μg/ml) for 2 days and then subjected to MTT assay. The graph shows the mean ± SD of three independent experiments. ****P ≤ 0.0001 (unpaired two-tailed Student’s t-test). (B) Effects of mNRK on AKT signaling analyzed by immnoblotting. Cells prepared by a method similar to that shown in (A) were subjected to immunoblotting. Densitometric analyses were performed, and phosphorylation levels of AKT (S473) were normalized to protein levels of AKT. The graph shows the mean ± SD of three independent experiments. ****P ≤ 0.0001 (unpaired two-tailed Student’s t-test). (C) Effects of mNRK on AKT signaling analyzed by immnostaining. HeLa cells expressing FLAG-tagged NRK were stimulated with EGF (100 ng/ml) for 10 min and subjected to cell staining with anti-FLAG antibody (magenta), anti-phospho AKT (S473) antibody (green), and DAPI (blue). Arrows indicate NRK-positive cells. Scale bar, 10 µm. (D) Effects of the other GCK IV family members (mNIK, mTNIK, and mMINK1) and cNRK on AKT signaling. HEK293 cells were transfected with plasmids encoding mNRK, mNIK, mTNIK, mMINK1, and cNRK. The lysates were subjected to immunoblotting, followed by densitometric analyses. The graph shows the mean ± SD of three independent experiments. *P ≤ 0.05 (one-way analysis of variance [ANOVA], followed by Dunnett’s multiple comparisons test). (E) Schematic structures of the truncated forms of mNRK used in (F) and (G). (F) Effects of the truncated forms of mNRK on cell proliferation. We generated Flp-In T-REx 293 cell lines that expressed the truncates in a dox-dependent manner. The cells were treated with dox (1 μg/ml) for 2 days and then subjected to the MTT assay. (G) Effects of the truncated forms of mNRK on AKT signaling. Cells treated by a method similar to that shown in (F) were subjected to immunoblotting, followed by densitometric analyses. The graphs show the mean ± SD of three independent experiments. ****P ≤ 0.0001 (unpaired two-tailed Student’s t-test).
We investigated molecular mechanisms underlying the inhibition of cell proliferation by mNRK. To identify the region(s) required for mediating the antiproliferation effect of mNRK, we developed cells that expressed deletion mutants of mNRK in a dox-dependent manner (fig. 3E). The expression of full-length mNRK and mutant #3 decreased cell numbers to a similar extent, whereas mutants #1 and #2 exerted a minor or no effect (fig. 3F). Full-length mNRK and mutant #3 suppressed AKT phosphorylation, whereas mutants #1 and #2 did not (fig. 3G). These results suggest that the middle region and CNH domain are required for the antiproliferation effect of mNRK.
NRK CNH Domain Acquired Lipid-Binding Ability During Mammalian Evolution, Thereby Recruiting NRK to the Plasma Membrane
We then attempted to elucidate the function of the mNRK CNH domain. As mNRK localizes at the plasma membrane independent of the kinase activity (Nakano et al. 2003), we speculated that the CNH domain may be important for mNRK localization. The CNH domain of mNRK showed ∼40% amino acid similarity to the sequences of the other GCK IV family members and cNRK (fig. 4A). We examined the cellular localization of these full-length proteins and CNH domains. Although mNRK is consistently localized at the plasma membrane, mNIK, mTNIK, mMINK1, and cNRK were primarily present in the cytosol (fig. 4B, upper panels). The CNH domain of mNRK was preferentially localized at the plasma membrane, but the CNH domain of NIK, TNIK, MINK1, and cNRK showed different localization (fig. 4B, lower panels). These results suggest a distinct function of the mNRK CNH domain.
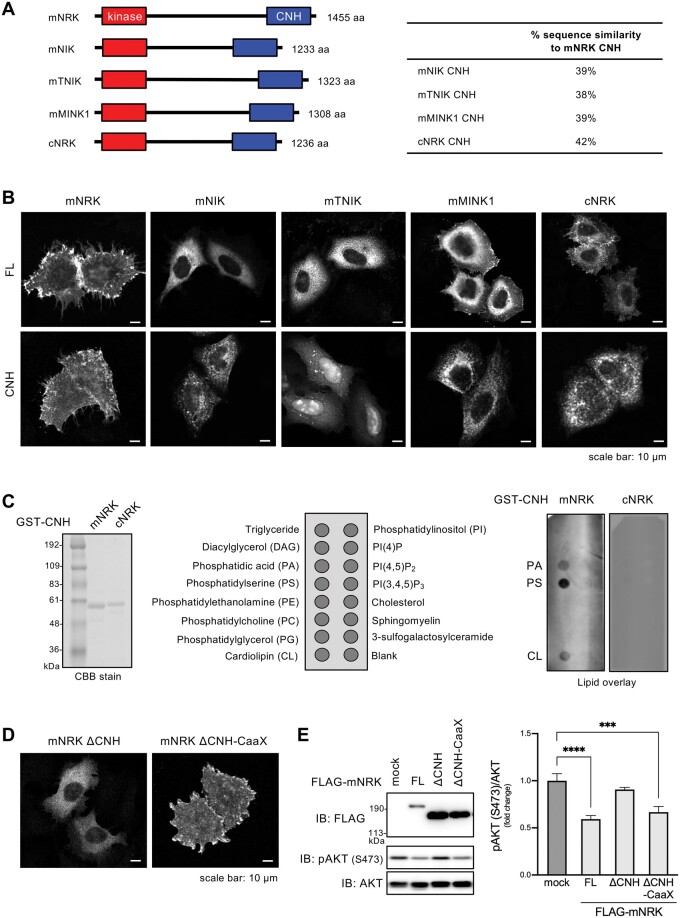
Fig. 4.
Function of the CNH domain of mNRK and related proteins. (A) Domain architectures of mNRK and related proteins, and sequence similarities of their CNH domains. (B) Intracellular localization of full length (FL) proteins of mNRK, mNIK, mTNIK, mMINK1, and cNRK and their CNH domains. HeLa cells expressing indicated proteins were subjected to immunostaining with anti-FLAG antibody. Scale bar, 10 µm. (C) Profiles of phospholipids that were bound to the CNH domain of mNRK. Left panel, CBB staining of the CNH domain of mNRK and cNRK. Middle panel, phospholipids spotted on the lipid strips. Right panel, lipid overlay assay of the CNH domain of mNRK and cNRK. The data shown are representative of two independent experiments. (D) Effects of the deletion of the CNH domain and the fusion of CaaX motif to mNRK on mNRK localization. HeLa cells expressing indicated proteins were subjected to immunostaining. Scale bar, 10 µm. (E) Their effects of on AKT signaling. HEK293 cells expressing indicated proteins were subjected to immunoblotting, followed by densitometric analyses. The graph shows the mean ± SD of three independent experiments. ***P ≤ 0.001; ****P ≤ 0.0001 (one-way analysis of variance [ANOVA], followed by Dunnett’s multiple comparisons test).
Lipid-protein overlay assay showed that the mNRK CNH domain bound strongly to phosphatidylserine and moderately to phosphatidic acid and cardiolipin (fig. 4C). The cNRK CNH domain showed no detectable binding to any phospholipid (fig. 4C). These results demonstrate that the mNRK CNH domain is a phospholipid-binding domain, and the NRK CNH domain acquired lipid-binding ability during mammalian evolution.
We further examined whether the CNH domain is necessary for mNRK plasma membrane localization and whether mNRK plasma membrane localization is important for inhibition of AKT signaling. The mNRK mutant lacking the CNH domain (ΔCNH) was present in the cytosol and did not suppress AKT phosphorylation (fig. 4D and E, ΔCNH). The fusion of K-Ras prenylation motif (CaaX motif) to ΔCNH recovered the plasma membrane localization and decreased AKT phosphorylation (fig. 4D and E, ΔCNH-CaaX), indicating that the deficiency of the CNH domain was compensated by the fusion of a short plasma membrane localization motif. These results indicate that the CNH domain recruited mNRK to the plasma membrane, thereby enabling mNRK to suppress AKT signaling.
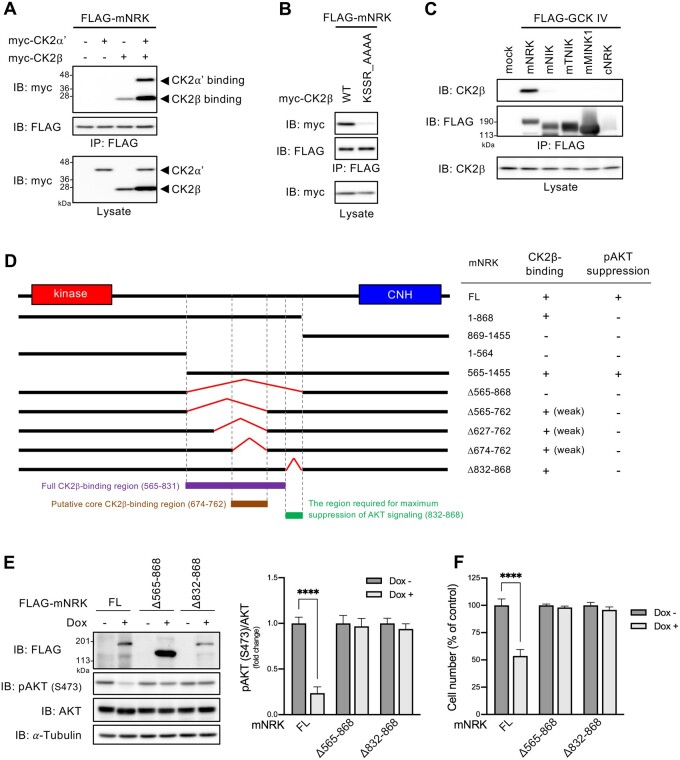
NRK Acquired CK2-Binding Ability During Mammalian Evolution, and the Region of mNRK Comprising Amino Acids 565–868 Was Required for Binding to CK2β and Suppression of AKT Signaling
We previously identified mNRK-interacting proteins including a CK2 catalytic subunit (CKα′) and a CK2 regulatory subunit (CK2β; Naito et al. 2020). To further elucidate the mechanisms underlying the antiproliferating effect of mNRK, we focused on CK2 because it can modulate AKT signaling (Litchfield 2003). We performed coimmunoprecipitation and immunoblotting analysis using cells overexpressing mNRK, CK2α′, and/or CK2β. CK2α′ was coimmunoprecipitated with mNRK only in the presence of exogenous CK2β. CK2β was coimmunoprecipitated with mNRK in the presence and absence of exogenous CK2α′ (fig. 5A). These results suggest that mNRK interacted with the CK2 complex by directly binding to CK2β. Next, we attempted to determine the mNRK-binding site in CK2β. The KSSR motif is one of the protein interaction motifs known in CK2β (Cao et al. 2014). The mutation in the motif (substitution of AAAA for KSSR) drastically reduced binding to mNRK (fig. 5B), indicating that it is responsible for the binding.
Fig. 5.
Ability of mNRK and related proteins to bind to CK2, and the relationship between mNRK binding to CK2 and its effect on AKT signaling. (A) Binding of mNRK to CK2β. HEK293T cells expressing indicated proteins were subjected to immunoprecipitation and immunoblotting. (B) mNRK-binding site in CK2β. The experimental method was similar to that used for (A). (C) No binding to CK2 was detected with mNIK, mTNIK, mMINK1, and cNRK. The experimental method was similar to that used for (A). (D) Binding of mNRK truncated mutant to CK2 and its effects on AKT signaling. Left panel, schematic structure of mNRK truncated mutants. We mapped the full CK2β-binding region (aa 565–831, red), the putative core CK2β-binding region (aa 674–762, brown), and the region not necessary for binding to CK2β but required for the suppression of AKT signaling (aa 832–868, green). Right panel, summary of the binding of mNRK mutants to CK2β and their effects on AKT phosphorylation (see supplementary fig. S5A and B, Supplementary Material online). (E) Effects of the deletion of the regions comprising aa 565–868 and aa 832–868 of mNRK on AKT signaling. We generated Flp-In T-REx 293 cell lines that expressed mNRKΔ565–868 and mNRKΔ832–868 in a dox-dependent manner. The cells were treated with dox (1 μg/ml) for 2 days. The cells were subjected to immunoblotting, followed by densitometric analyses. (F) Effects of the deletion of the regions comprising aa 565–868 and aa 832–868 of mNRK on cell proliferation. Cells treated by a method similar to that shown in (E) were subjected to MTT assay. The graphs show the mean ± SD of three independent experiments. ****P ≤ 0.0001 (unpaired two-tailed Student’s t-test).
We then examined the binding ability of the other GCK IV family members and cNRK to CK2β. These proteins did not bind to CK2β (fig. 5C), suggesting that NRK acquired CK2-binding ability during mammalian evolution.
To determine the CK2β-binding site in mNRK, we constructed several mNRK deletion mutants and performed coimmunoprecipitation analyses. The summary of the results is shown in figure 5D. The regions comprising amino acids (aa) 1–868 of mNRK (mNRK1–868) and aa 565–1,455 (mNRK565–1,455) were bound to CK2β, but the regions of aa in mNRK869–1,455, mNRK1–564, and the mutant lacking aa 565–868 (mNRKΔ565–868) did not demonstrate any binding (supplementary fig. S5A, left panel, Supplementary Material online). Hence, aa 565–868 are required for the binding activity. As described later, aa 565–762 and aa 832–868 were well conserved among eutherians. Thus, we further constructed mNRKΔ565–762, mNRKΔ627–762, mNRKΔ674–762, and mNRKΔ832–868 mutants. The mNRKΔ565–762, mNRKΔ627–762, and mNRKΔ674–762 bound weakly to CK2β (supplementary fig. S5A, center panel, Supplementary Material online), suggesting that the region comprising aa 674–762 was a core binding region; however, the other region weakly contributed to the binding. The mNRKΔ832–868 was bound to CK2β to an extent similar to that observed with full-length mNRK (supplementary fig. S5A, right panel, Supplementary Material online), suggesting that the region comprising aa 832–868 is not required for the binding.
Next, we investigated the effects of these mNRK deletion mutants on AKT phosphorylation; a summary is shown in figure 5D. The expression of mNRK565–1,455 decreased AKT phosphorylation; however, mNRK1–868, mNRK869–1,455, mNRK1–564, and mNRKΔ565–868 expressions did not reduce the phosphorylation (supplementary fig. S5B, left panel, Supplementary Material online). It should be noted that the expression level of mNRK869–1,455 was much lower than others under this experimental condition. These results confirm that the CNH domain and the middle region comprising aa 565–868 were required for AKT suppression. The mNRKΔ565–762, mNRKΔ627–762, and mNRKΔ674–762 did not decrease AKT phosphorylation (supplementary fig. S5B, right panel, Supplementary Material online), suggesting that aa 674–762, a region that is crucial for binding to CK2β, was required for AKT suppression. Surprisingly, mNRKΔ832–868 did not decrease AKT phosphorylation (supplementary fig. S5B, right panel, Supplementary Material online), suggesting that the region comprising aa 832–868 was required for AKT suppression despite not contributing toward binding to CK2β.
We then generated cells with dox-induced expression of mNRKΔ565–868 and mNRKΔ832–868. As expected, the expression of these mutants did not decrease AKT phosphorylation (fig. 5E) and cell proliferation (fig. 5F).
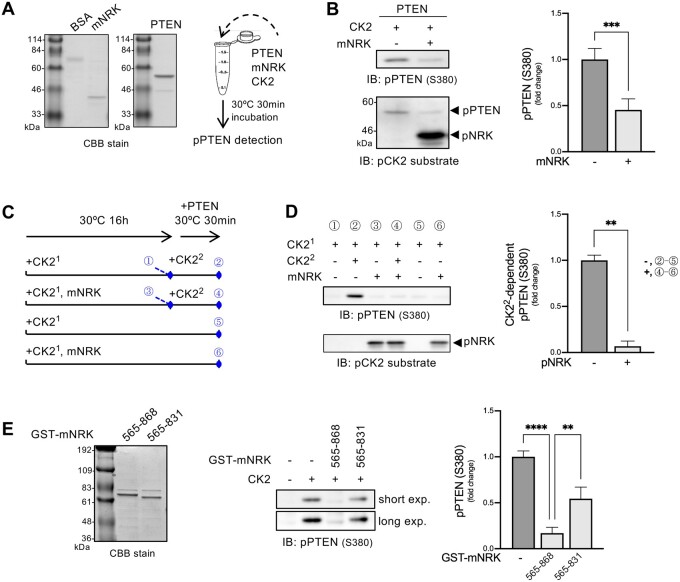
Region Comprising Amino Acids 565–868 of mNRK Inhibited CK2 Activity In Vitro
Next, we investigated whether mNRK565–868 inhibited CK2 activity in vitro (fig. 6A). A CK2 substrate protein PTEN190–403 (mutant lacking N-terminal phosphatase domain) was phosphorylated by a commercial CK2 complex in the presence or absence of mNRK565–868 (fig. 6A). The phosphorylation levels were measured by immunoblotting with phospho-PTEN (S380) antibody and phospho-CK2 substrate antibody. The result showed that mNRK565–868 inhibited CK2 activity (fig. 6B). We named this region the CK2-inhibitory region (CIR).
Fig. 6.
Effects of the middle region of mNRK on CK2 kinase activity. (A) CBB staining of purified mNRK565–868 and GST-PTEN190–403 and schematic representation of in vitro kinase assay in (B). (B) Inhibition of CK2 activity by mNRK565–868. CK2, GST-PTEN190–403, and mNRK565–868 were mixed in a kinase assay buffer containing ATP and incubated at 30 °C for 30 min. The samples were then subjected to immunoblotting, followed by densitometric analyses. The graph shows the mean ± SD of four independent experiments. ***P ≤ 0.001 (unpaired two-tailed Student’s t-test). (C) Schematic representation of the in vitro kinase assay and the sampling time of each sample (#1–6) shown in (D). (D) mNRK565–868 inhibition of CK2 activity via a noncompetitive mechanism. mNRK565–868 was phosphorylated by preincubation with CK2 (denoted as CK21) in a kinase assay buffer containing ATP at 30 °C for 16 h. The samples were then mixed with CK2 (denoted as CK22), GST-PTEN190–403, and ATP and incubated at 30 °C for 30 min, followed by immunoblotting and densitometric analyses. We subtracted the value of sample #5 from sample #2 and sample #6 from sample #4, marking them as those with phosphorylation of PTEN by CK22 in the absence or presence of phospho-NRK in the graph, respectively. The graph shows the mean ± SD of four independent experiments. **P ≤ 0.01 (unpaired two-tailed Student’s t-test). (E) Importance of the region comprising aa 832–868 for maximum inhibition of CK2 activity by mNRK565–831. CK2, GST-PTEN190–403, and GST-mNRK (mNRK565–868 or mNRK565–831) were mixed in a kinase assay buffer containing ATP and incubated at 30 °C for 30 min. The samples were then subjected to immunoblotting, followed by densitometric analyses. The graph shows the mean ± SD of three independent experiments. **P ≤ 0.01; ****P ≤ 0.0001 (one-way analysis of variance [ANOVA], followed by Tukey’s multiple comparisons test).
We also observed CK2-dependent phosphorylation of mNRK565–868 (fig. 6B). This raised the possibility that the CIR may compete with PTEN as a substrate for CK2. To test this hypothesis, we prepared phospho-mNRK565–868 by preincubation with CK2 (denoted as CK21) and examined its effect on the activity of CK2 that was added later (denoted as CK22; fig. 6C). The results show that PTEN190–403 was strongly phosphorylated by CK22 in the absence of phospho-mNRK565–868 (fig. 6D, #1 vs #2). However, it was phosphorylated to a small extent by CK22 in the presence of phospho-mNRK565–868 (#3 vs #4). CK21 added at the start of the 16 h preincubation showed little activity in the subsequent 30-min incubation (#5 and #6), probably because CK21 might have lost its activity during long-term preincubation. We also found similar phosphorylation levels of mNRK565–868 in samples #3, #4, and #6 (fig. 6D, bottom panel), confirming that almost all mNRK565–868 was phosphorylated by CK21. Densitometric analyses showed that phospho-mNRK565–868 inhibited CK22-dependent phosphorylation of PTEN190–403 (fig. 6D, right panel). These data suggest that the CIR inhibited CK2 activity via a noncompetitive mechanism.
In figure 5, we show that the region comprising aa 832–868 of mNRK did not contribute toward binding to CK2β but was essential for AKT suppression. We examined the effects of GST-tagged mNRK565–868 and mNRK565–831 (lacking the region comprising aa 832–868) on CK2 activity. The results show that mNRK565–868 strongly decreased CK2-dependent phosphorylation of PTEN190–403, whereas mNRK565–831 only demonstrated a minor reduction in phosphorylation (fig. 6E). These results indicate that the region of mNRK comprising aa 832–868 plays an important role in the inhibition of CK2 activity.
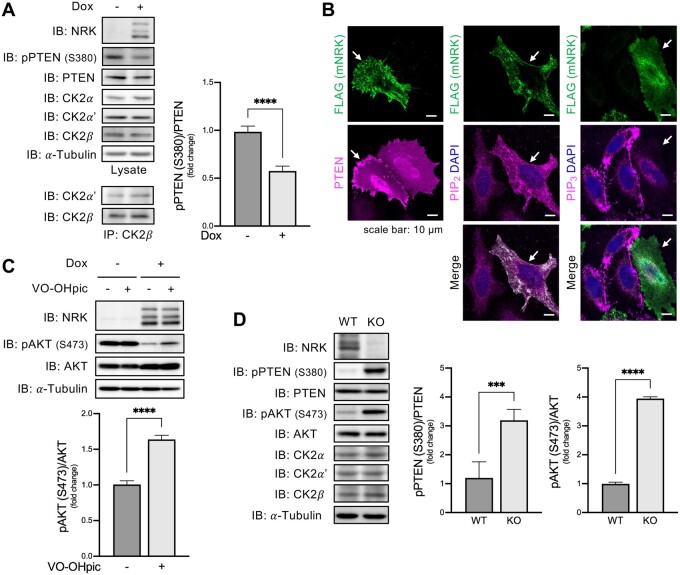
mNRK Suppressed AKT Signaling by Modulating the CK2–PTEN–AKT Pathway
We then examined whether mNRK inhibited CK2 activity in cells. Dox-induced mNRK expression reduced PTEN S380 phosphorylation (fig. 7A). The mNRK expression did not affect the protein levels of each CK2 subunit and the interaction of CK2α′ with CK2β (fig. 7A). These results indicate that mNRK inhibited cellular CK2 activity without affecting the formation of the CK2 complex.
Fig. 7.
Effects of mNRK on the CK2–PTEN–AKT pathway. (A) Effects of mNRK on PTEN phosphorylation in cultured cells. Flp-In T-REx 293 cells with dox-inducible expression of mNRK were treated with dox (1 μg/ml) for 2 days. The cells were subjected to immunoblotting, followed by densitometric analyses. The graph shows the mean ± SD of four independent experiments. ****P ≤ 0.0001 (unpaired two-tailed Student’s t-test). (B) Effects of mNRK on PTEN localization and levels of PIP2 and PIP3. Left panel, HeLa cells expressing myc-mNRK and FLAG-PTEN were subjected to cell staining with anti-myc antibody (green) and anti-FLAG antibody (magenta). Middle and right panels, cells expressing myc-mNRK were subjected to staining with anti-myc antibody (green), anti-PIP2 or PIP3 antibodies (magenta), and DAPI (blue). Arrows indicate NRK-positive cells. Scale bars, 10 μm. (C) Restoration of AKT phosphorylation by PTEN inhibitor in mNRK-expressing cells. Flp-In T-REx 293 cells with dox-inducible expression of mNRK were treated with dox (1 μg/ml) for 2 days and treated with a PTEN inhibitor VO-OHpic (1 μM) for the last 24 h. The cells were subjected to immunoblotting, followed by densitometric analyses. The graph shows the mean ± SD of four independent experiments. ****P ≤ 0.0001 (unpaired two-tailed Student’s t-test). (D) Effects of mNRK on PTEN phosphorylation in mouse placentas. Placenta tissues were lysed at 18.5 dpc and subjected to immunoblotting with indicated antibodies, then densitometric analyses. The graph shows the mean ± SD (WT, n = 5; KO, n = 5). ***P ≤ 0.001, ****P ≤ 0.0001 (unpaired two-tailed Student’s t-test).
We examined the effect of mNRK expression on PTEN localization. PTEN was detected in the cytosol and nucleus in mNRK-negative cells. A part of PTEN was also detected at the plasma membrane in mNRK-positive cells (fig. 7B; supplementary fig. S6, Supplementary Material online). We observed higher levels of PIP2 and lower levels of PIP3 in the mNRK-positive cells compared with the mNRK-negative cells (fig. 7B; supplementary fig. S6, Supplementary Material online). These observations suggest that mNRK inhibited CK2-mediated phosphorylation of PTEN, thereby mediating the localization of PTEN on the plasma membrane and PIP3 dephosphorylation.
Next, we examined whether mNRK-induced AKT suppression was mediated by PTEN. We used cells with or without dox-inducible expression of mNRK and treated them with PTEN inhibitor VO-OHpic. The PTEN inhibitor showed minor effects on AKT phosphorylation in cells without mNRK expression. Dox-induced mNRK expression decreased AKT phosphorylation; PTEN inhibitor restored it (fig. 7C). These results suggest that PTEN is mostly inactive in cells without mNRK expression and that NRK activates PTEN, thereby leading to AKT suppression.
Next, we examined the role of mNRK in the regulation of the CK2–PTEN–AKT pathway under in vivo conditions using NRK knockout (KO) mice. Consistent with earlier reports, KO placentas did not express mNRK and were larger than WT placentas. KO placentas showed increased phosphorylation levels of PTEN S380 as well as AKT S473 compared with WT placentas (fig. 7D). These results suggest that mNRK inhibits phosphorylation of PTEN by CK2 and activates PTEN, leading to the suppression of AKT signaling in the placenta.
Functional Regions in Eutherian NRK Underwent Negative Selection
Given that mNRK plays an important role in regulating placental development, we speculated that its functional regions would be affected by negative selection in eutherians. To evaluate selective pressure during Nrk evolution, we compared coding sequences of 15 eutherian Nrk orthologs and calculated the ratio of nonsynonymous to synonymous substitution rates (dN/dS). The kinase and CNH domains showed low dN/dS values (0.14 and 0.20 on average, respectively; dN/dS < 1; fig. 8A), suggesting that they are under strong purifying selection. The amino acid sequence alignment of the CNH domain in vertebrate NRK orthologs shows a similarity in eutherian sequences and dissimilarity among eutherian and other sequences (fig. 8B), consistent with the dN/dS and evolutionary rate analysis (fig. 2).
Fig. 8.
Evolutionary pressure on mNRK functional regions. (A) Negative selection of eutherian Nrk sequences. The graph shows the ratio of nonsynonymous per synonymous substitution (dN/dS) estimated using 15 eutherian Nrk sequences; dN/dS<1 predicts negative selection. Functional domains and regions are indicated: 565–831, full CK2β-binding region; 674–762, putative core CK2β-binding region; 832–868, the region required for the maximum inhibition of CK2 activity. (B) Conservation of amino acid sequences of NRK CNH domains among eutherians. We selected eutherians (human, mouse, cow, and elephant) and nonmammals (chicken and Xenopus), aligned their NRK CNH domain sequences, and colored them according to the ClustalX color scheme. (C) Conservation of amino acid sequences of NRK CK2-binding regions among eutherians. We selected 15 eutherians, aligned their regions orthologous to the mNRK CIR, and colored them according to the ClustalX color scheme.
The CIR showed low dN/dS values (fig. 8A). We performed amino acid sequence alignment of the CIR. The sequences of noneutherian NRK could not be aligned owing to their much lower similarity to mNRK. Two functional regions were examined; a region (aa 674–762; dN/dS = 0.38) important for binding to CK2β, and a region (aa 832–868; dN/dS = 0.23) not required for binding to CK2β but necessary for maximum inhibition of the CK2 activity. As expected, these two regions appear to be well conserved among eutherians (fig. 8C).
Discussion
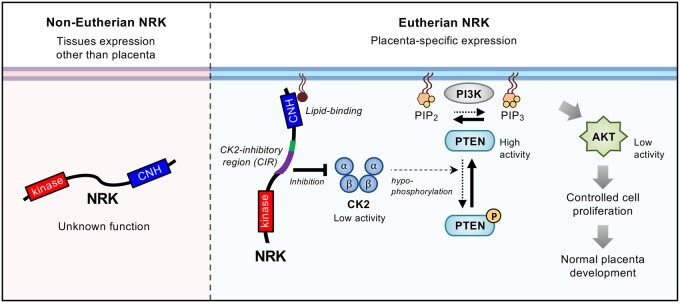
This study showed that the eutherian Nrk gene underwent multiscale evolution in terms of genomic location, exon structure, sequence of the protein-coding region, and gene expression regulation. NRK underwent extensive amino acid substitutions in the ancestor of placental mammals and has been conserved since then. Through this, eutherian NRK seems to have acquired the lipid-binding CNH domain and the CIR. We also found that mNRK inhibits placental cell proliferation by modulating the CK2–PTEN–AKT pathway, and this function can be attributed to this molecular evolution (fig. 9, working hypothesis).
Fig. 9.
Model of molecular mechanisms underlying inhibition of placental cell proliferation by eutherian NRK and its molecular evolution to acquire this function. NRK is highly expressed in the placenta and underwent rapid molecular evolution during early mammalian evolution. The changes in amino acid sequences in the CNH domain and the middle region conferred plasma membrane localization and CK2 inhibition activities, respectively. Eutherian NRK is localized at the plasma membrane and inhibited CK2, thereby mitigating CK2-dependent inhibition of PTEN, leading to the suppression of AKT signaling and cell proliferation. Through these mechanisms, eutherian NRK played an essential role in normal placental development by preventing hyperplasia of the placenta.
Evolutionary History of Nrk Leading to an X-Linked Placenta-Specific Gene
Mouse and human Nrk genes are located on Xq and are descended from proto-X chromosome orthologous to chicken 4p. A previous report indicated that approximately 30 subregions with sequence homology were observed upon comparing human Xq with chicken 4p. However, their positions and directions differed, implying many recombination events during the evolution of their common ancestor (Ross et al. 2005). Our result suggests that the genomic region containing Nrk was relocated owing to multiple genomic recombinations during the early evolution of mammals.
Consistent with a theory that selection favors an increase in the frequency of genes with sexually antagonistic traits on the X chromosome (Rice 1984), the mouse X chromosome is enriched for genes preferentially expressed in the placenta (Khil et al. 2004). Our study added eutherian Nrk to the list of X-linked placenta-specific genes. Recently, Liu et al. screened the eutherian X chromosome to detect the regions with accelerated evolution in the ancestral lineage of eutherians and found that the region with the highest accelerated evolution is located in the coding regions of Nrk (Liu et al. 2021). Consistent with this, our study revealed that Nrk underwent accelerated evolution in terms of exon gain/loss, sequence insertion events in the eutherian and therian ancestors, and rapid sequence substitutions in the eutherian ancestor. Accelerated evolution has been frequently observed in duplicated genes such as those in teleost fish (Brunet et al. 2006). However, we found no paralogous gene of Nrk duplicated in or before the eutherian ancestor, suggesting that the eutherian-specific rapid evolution of Nrk was not caused by gene duplication. Although the expression of Nrk genes in noneutherians and the function of their coding proteins are largely unclear, Nrk seems to have been co-opted to generate eutherian placenta. We speculate that Nrk became a placental-specific gene in the eutherian or therian ancestor and was subsequently subjected to strong positive evolutionary pressure to optimize its protein function in the placenta. After optimization, it was subjected to negative evolutionary pressure to maintain the function (fig. 8A).
Many proteins are specifically expressed in the placenta and contribute to placental function and regulation (Rossant and Cross 2001; Rawn and Cross 2008; Woods et al. 2018). They can be divided into two groups, namely, those found only in a few mammalian species and those widely shared among mammals. NRK is included in the latter group. Most proteins in the former group may enable diversity among species in the placental morphology and mother–fetus interactions. In contrast, proteins in the latter group are thought to be involved in the fundamental development, function, and regulation of the placenta. The evolutionary history of the latter group can be further classified into several patterns. The first pattern involves proteins encoded by genes that were newly acquired during early mammalian evolution. For example, a placenta-specific secreted protein PLAC1 is not found in marsupials or monotremes but is found in eutherians (Devor 2014). A retrotransposon-derived PEG10 is found only in placental mammals (Suzuki et al. 2007). The next pattern involves those present in many taxons of vertebrates, whereas they have come to be expressed in the placenta during early mammalian evolution, without changing their primary protein function. For example, HAND1, a transcription factor found in vertebrates, regulates gene expression in neurons in chickens (Howard et al. 1999). It is also expressed in the placenta in mice and thought to regulate placental gene expression as transcriptional factor (Riley et al. 1998). NRK does not fit into these patterns; it was present in mammalian ancestors but acquired new functional domains/regions during early mammalian evolution, changing its molecular functions to regulate placental development. To our knowledge, no placenta-specific protein other than NRK is known with such an evolutionary history.
Unique Role of Eutherian NRK in Regulating AKT Signaling Among GCK IV Kinases
We also found that other members of GCK IV kinases (mNIK, mTNIK, mMINK1) and cNRK were not localized at the plasma membrane and did not interact with CK2 or inhibited AKT signaling under our experimental conditions. NIK is reported to enhance the invasion of some cancer cells (Wright et al. 2003). TNIK potentiates WNT and AKT signaling to promote tumorigenesis (Yu et al. 2014; Masuda et al. 2016). MINK1 interacts with mTORC2 to enhance AKT signaling and promote cancer cell metastasis (Daulat et al. 2016). Although the function of noneutherian NRK remains unclear, these facts demonstrate that, among all GCK IV kinases, only eutherian NRK functions as a negative regulator of AKT signaling.
Roles of the Kinase Domain, CNH Domain, and CIR of mNRK
This study shows distinct functions of the kinase domain, CNH domain, and CIR of mNRK. The kinase domain of eutherian NRK underwent negative selection (fig. 8A), indicating a crucial function mediated by this domain. mNRK phosphorylates cofilin, thereby enhancing actin polymerization (Nakano et al. 2003). mNRK can also activate the JNK pathway in a kinase activity-dependent manner (Nakano et al. 2000; Kakinuma et al. 2005). However, in this study, the kinase domain was not required for mNRK-induced inhibition of AKT signaling or cell proliferation. We speculate that NRK may inhibit AKT signaling, possibly not via mechanisms involving actin polymerization or JNK activation.
The CNH domain of mNRK can bind to phospholipids and facilitates the recruitment of mNRK to a site near the plasma membrane. The sequence of the CNH domain showed similarity among eutherians and dissimilarity between eutherians and noneutherians, indicating that the CNH domain acquired its lipid-binding ability during mammalian evolution. The localization of mNRK on the plasma membrane was required to inhibit AKT signaling. It was unclear whether the lipid-binding activity of mNRK was sufficient for its localization on the plasma membrane. Further study is required to investigate the detailed mechanisms. Researchers have reported several phospholipid-binding domains, most of which are well conserved among eukaryotes (Lemmon 2008). To our knowledge, this is the first report of the mammalian-specific intracellular phospholipid-binding domain. The CNH domain of NRK may be useful to study the molecular evolution of the phospholipid-binding domain.
The CIR, the region comprising aa 565–868 of mNRK, was bound to the CK2β regulatory subunit and inhibited the kinase activity of the CK2 complex via a noncompetitive mechanism. In the CIR, the region comprising aa 565–831 is responsible for binding to CK2β. CK2β is thought to recruit substrates and/or regulators of the CK2 complex (Bibby and Litchfield 2005). The region comprising aa 565–831 weakly inhibited the CK2 activity in vitro (fig. 6E), indicating that this region may impair the ability of CK2β to recruit substrates to the CK2α/α′ catalytic subunit. We also found that the region comprising aa 832–868 was not required for binding to CK2β; however, it was required for the maximal inhibition of the CK2 activity (figs. 5D and 6E). The region comprising aa 832–868 has putative phosphorylation sites for CK2, but how this region is involved in the inhibition remains unclear. CK2 is known to be inhibited by several proteins (Homma et al. 2002; Llorens et al. 2003), including the intrinsically disordered protein Nopp140. Nopp140 primarily binds to CK2β and is phosphorylated by the CK2 complex (Li et al. 1997). The phosphorylated sequence in Nopp140 binds to the active site of CK2α, thereby inhibiting the kinase activity (Lee et al. 2013). These studies suggest that Nopp140 may have different regions that interact with CK2α and CK2β, and the two regions work in concert to inhibit CK2 effectively. We speculate that the CIR also inhibits the CK2 complex possibly by interacting both with CK2α/α′ and CK2β. The region comprising aa 832–868 may weakly interact with the CK2α/α′ catalytic subunit and inhibit the kinase activity, although this interaction would be too weak to be detected in our experimental conditions. This interaction may be subsisted by the strong interaction between the region comprising aa 565–831 and CK2β. Further analyses are required to elucidate how the region comprising aa 832–868 is involved in the inhibition. The sequences of the CIR were observed to be well conserved in eutherians, but no homologous sequence was found in noneutherians. This indicates that the CIR was acquired during mammalian evolution. Similar to Nopp140, a large part of the CIR is predicted to be intrinsically disordered (Naito et al. 2020). In general, the disordered regions of proteins appear to evolve rapidly owing to relaxed purifying selection (Brown et al. 2011). We speculate that the disordered regions in the middle of NRK underwent rapid evolution in the ancestor of placental mammals and became the CIR.
Molecular Mechanism Underlying the Inhibition of AKT Signaling by mNRK
We observed that mNRK overexpression in cultured cells inhibited the PTEN phosphorylation activity of CK2. Based on previous studies, mNRK must mitigate CK2-dependent negative regulation of PTEN (Vazquez et al. 2000; Torres and Pulido 2001; Rahdar et al. 2009). Consistently, mNRK overexpression decreased PIP3 levels accompanied by an increase in PIP2 levels, leading to the suppression of AKT signaling and cell proliferation. In addition to phosphorylating PTEN, CK2 can directly phosphorylate AKT S129 and thereby enhance AKT kinase activity (Risso et al. 2015). CK2 may also inhibit phosphatases targeting phospho-T308 and phospho-S473 of AKT, leading to the enhancement of AKT kinase activity (Trotman et al. 2006; Chatterjee et al. 2013). Therefore, it cannot be denied that additional mechanisms are involved in the attenuation of AKT signaling by mNRK.
Mechanism for the Regulation of Placental Development
We found that Nrk deficiency increased PTEN as well as AKT phosphorylation in mouse placenta. CK2, PTEN, and AKT have been implicated in the regulation of placental development. CK2 is expressed in the placenta from early pregnancy, and is required for cell proliferation, migration, and differentiation (Abi Nahed et al. 2020). PTEN+/− mice showed an enlarged spongiotrophoblast layer and approximately 20% increase in placental weight (Church et al. 2012), similar to NRK KO mice (Denda et al. 2011), albeit with a milder phenotype. Phosphatase of regenerating liver (PRL)-2 is a protein responsible for PTEN degradation, and PRL-2 KO mice showed a small spongiotrophoblast layer (Dong et al. 2012). AKT1 deficiency reduced the weight of mouse placenta (Yang et al. 2003), and an AKT inhibitor inhibited placental cell proliferation (Morioka et al. 2017). Based on the evidence, we believe that NRK prevents hyperplasia of the placenta by modulating the CK2–PTEN–AKT pathway. So far, the types of physiological situations in which AKT signaling is regulated via the CK2–PTEN–AKT pathway remain unclear. Given that Nrk expression is pronounced in the placenta during late pregnancy, our study suggests that tissue-specific expression of mNRK is the first mechanism to regulate AKT signaling via the CK2–PTEN–AKT pathway.
Limitations of This Study and Future Directions
1) The data on tissue expression patterns of the Nrk gene in vertebrates were limited. In order to better understand the involvement of Nrk in placental evolution, it is important to investigate their expression in tissues that supply nutrients and oxygen to the fetus, such as the primitive placenta in marsupials, the chorioallantoic membrane in reptiles and birds, and placenta-like structures in some fish. 2) It is also important to identify the regulatory mechanism underlying placenta-specific expression of eutherian Nrk gene. Multiple recombination events in the genomic region containing the Nrk gene occurred during early mammalian evolution. This might have led to the interchange of transcriptional regulatory elements between genes, resulting in placenta-specific expression of the Nrk gene. 3) We investigated the molecular mechanism by which mNRK suppresses AKT signaling using HEK293-derived cells. However, in vivo, mNRK is highly expressed in placental spongiotrophoblasts. As spongiotrophoblasts specifically express some proteins that regulate AKT signaling (Takao et al. 2012), the molecular mechanism may be more complicated in these cells. 4) The organization of eutherian placentas is diverse, and it is unclear whether nonmouse placenta possesses cells similar to mouse spongiotrophoblasts. Recent single-cell RNA-seq analysis showed that human NRK is expressed in placental syncytiotrophoblasts and cytotrophoblasts (Vento-Tormo et al. 2018). The well-conserved amino acid sequences of the functional domains/regions of eutherian NRK imply their conserved molecular functions. It is important to investigate how NRK functions in the placenta of species other than mice.
Conclusions
This is the first report suggesting that NRK prevents overproliferation of placental cells by modulating the CK2–PTEN–AKT pathway. Reduced NRK levels, the loss of NRK function due to mutations in the CNH domain or the CIR, or abnormalities in the downstream pathway of NRK may lead to pregnancy complications. We also provided evidence for the unique and rapid molecular evolution of NRK, during which it acquired the antiproliferation function. We believe that NRK evolution facilitated the proper control of placental development in mammals.
Materials and Methods
Synteny Analysis
We evaluated genomic data using Ensembl (https://www.ensembl.org, last accessed January 21, 2022) and the University of California, Santa Cruz (UCSC) genome browser (http://genome.ucsc.edu, last accessed January 21, 2022). We organized vertebrate Nrk and the neighboring genes and identified syntenic genes by referring to the neighboring genes located around Nrk in the mouse and chicken genome. We also identified syntenic genes around vertebrate Nik, Tnik, and Mink1 genes by referring to the neighboring genes located around Nik, Tnik, and Mink1 genes in the human genome. Nonsyntenic genes smaller than 10 kb in genomic size were omitted as such genes may have been mapped by mistake in gene prediction.
Exon Structure Analysis
We retrieved the validated coding sequences of the human and mouse Nrk from the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov, last accessed January 21, 2022). The therian Nrk coding regions were identified from the genome sequences in supplementary table 1, Supplementary Material online by referring to the human and mouse exon structures. RNA-seq data were obtained from NCBI SRA for platypus (SRX081892 and SRX122683), chicken (ERX697668 and ERX2593575), and zebrafish (DRX045951 and DRX045952); each species data were assembled using Bridger (Chang et al. 2015). The Nrk coding regions in the other vertebrates were identified from the genome sequences in supplementary table 1, Supplementary Material online by referring to the above RNA-seq assemblies and the gene prediction data in NCBI (platypus: XM_029067556.1, Xenopus: XM_031891524.1, spotted gar: XM_006632839.2, and reedfish: XM_028815688.1) and Ensembl (painted turtle: ENSCPBT00000034663.1, coelacanth: ENSLACT00000019211.1, and zebrafish: ENSDARG00000098680). We searched the origin of the inserted sequences in the transposable element collection (Repbase repeat library ver. 20181026) using RepeatMasker (http://www.repeatmasker.org, last accessed January 21, 2022).
Protein Evolutionary Rate Analysis
The NRK amino acid sequences from 24 species were aligned for each exon using MAFFT version 7 (Katoh and Standley 2013) with the linsi option followed by partial adjustments with MUSCLE implemented in MEGA (Tamura et al. 2021). The JTT + F + G substitution model was selected as the best evolutionary model for the NRK sequences based on the Bayesian information criterion using ModelFinder (Kalyaanamoorthy et al. 2017). We reconstructed the ancestral amino acid sequences by using FastML (Ashkenazy et al. 2012) with the JTT + G model and ML-based indel reconstruction under the assumption of known species tree in TimeTree (accessed on August 5, 2020; Kumar et al. 2017). The number of amino acid substitutions was counted for each branch by comparing the estimated ancestral sequences of the two corresponding nodes, and the substitution rates were calculated based on the divergence times available in TimeTree. The estimated indel information was used only for visualization and ignored in the substitution rate calculations. As a control of the evolutionary rate comparison, we used an amino acid sequence collection of 831 orthologous genes in vertebrates reported previously (Takezaki and Nishihara 2016). The substitution rates were calculated for each of the 831 genes using the same method described above. For the comparison, the rates of the mammalian and the therian stem lineages of NRK were merged because the corresponding platypus sequence in the 831 gene collection was not available. Positive selection was tested for the CNH domain corresponding to the exons 21–29, excluding the eutherian-specific exon 25 of Nrk. The aBSREL method was used in the selection analysis through the Datamonkey 2.0 webserver (Smith et al. 2015; Weaver et al. 2018). NJ tree for the amino acid sequences of the NRK domains was estimated under the JTT + F + G model with the pairwise deletion option.
Gene Expression Analysis
We collected gene expression data of mouse Nik, Tnik, Mink1, Nrk, and human NRK in various tissues analyzed by the cap analysis of gene expression in RIKEN FANTOM5 project from the public database RefEx (Reference Expression data set; http://refex.dbcls.jp, last accessed January 21, 2022; Ono et al. 2017) and Expression Atlas (https://www.ebi.ac.uk/gxa/home, last accessed January 21, 2022). We also collected gene expression data of sheep (Jiang et al. 2014), chicken (Merkin et al. 2012), and Xenopus Nrk (Barbosa-Morais et al. 2012) in various tissues from the Expression Atlas. The data were then visualized using an online tool, Heatmapper (http://heatmapper.ca/expression/, last accessed January 21, 2022).
dN/dS Calculation
The ratio of nonsynonymous to synonymous substitution (dN/dS) was calculated to estimate the selective constraint of the Nrk sequences in 15 eutherian mammals (human, orangutan, tarsier, mouse, squirrel, dog, cat, horse, microbat, minke whale, sheep, cow, shrew, elephant, and tenrec) under the species tree topology. This calculation was performed based on the Bayes Empirical Bayes method with the M8 model by using CODEML in the Phylogenetic Analysis by Maximum Likelihood (PAML) 4.9j package (Yang 2007).
Antibodies, Reagents, and Plasmids
The details of antibodies, reagents, and plasmids were described in Supplementary Material online.
Cell Culture and Transfection
HEK293 cells that express mNRK in a dox-dependent manner were developed using Flp-In T-REx 293 cells (Invitrogen), according to the manufacturer’s instructions. They were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin, 7.5 μg/ml blasticidin (Wako), and 16 μg/ml hygromycin (Invitrogen). HEK293, HEK293T, and HeLa cells were cultured in DMEM supplemented with 10% FBS, penicillin, and streptomycin. DNA transfection was performed using polyethylenimine (PEI, linear; molecular weight, 25,000; Polyscience, Warrington, PA).
MTT Assay
HEK293 cells expressing NRK in a dox-dependent manner were seeded in collagen-coated 96-well plates at a density of 2 × 103/well and cultured for 24 h. The cells were treated with dox (1 μg/ml) and incubated for 48 h. A total of 10 µl of cell count reagent SF (#07553-15, Nacalai Tesque) was added, and cells were incubated for an additional 2 h. The absorbance was then measured at 450 nm using microplate readers (Varioskan LUX, Thermo Fisher Scientific).
Immunoprecipitation and Immunoblotting
Cells were lysed with ice-cold lysis buffer (20 mM Tris–HCl, pH 7.4, 100 mM NaCl, 50 mM NaF, 0.5% Nonidet P-40, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 µg/ml aprotinin, 1 µg/ml leupeptin, and 1 µg/ml pepstatin A). For the detection of phosphorylation signals, 1 mM Na3VO4 was added to the lysis buffer. The lysates were then centrifuged at 15,000 rpm for 15 min. The supernatants were subjected to immunoprecipitation and immunoblotting according to the standard protocols. Immunoblotting signals were detected using the ECL prime western blotting detection reagents (GE Healthcare) and the ImageQuant LAS 4000 mini-imager (GE Healthcare). Densitometric analyses were performed using the ImageJ program (version 1.52).
Immunostaining
Immunostaining analyses were performed according to standard procedures. Briefly, HeLa cells were cultured on glass coverslips and transfected with PEI. After 48 h of transfection, cells were fixed with 4% paraformaldehyde (PFA) in phosphate buffer saline (PBS) for 10 min at 20–25 °C, permeabilized with 0.2% Triton X-100 in PBS, and blocked with 5% FBS in PBS. The samples were then incubated with primary antibodies for 16 h at 4 °C, followed by secondary antibodies for 1 h at 20–25 °C. DAPI (1 μg/ml, Nacalai Tesque) was added at the end for nuclear staining before mounting. As an exception, immunostaining of PIP2 and PIP3 was performed according to the protocol provided by Echelon Biosciences. Briefly, the transfected HeLa cells were fixed with 4% PFA and permeabilized with 0.01% digitonin for 20 min each at 20–25 °C. After blocking with 10% goat serum in TBS for 30 min at 37 °C, cells were then incubated with anti-PIP2 or anti-PIP2 antibodies followed by secondary antibodies for 1 h each at 37 °C. DAPI was added at the end for nuclear staining before mounting. Fluorescence images were acquired using a laser-scanning confocal microscope (LSM 780, Carl Zeiss, Oberkochen, Germany).
Lipid-Protein Overlay Assay
GST-tagged CNH domains of mNRK and cNRK (GST–CNH) were expressed in Escherichiacoli strain Rosetta (DE3)pLysS and purified using Glutathione-Sepharose 4B beads (GE Healthcare). Membrane lipid strips (Echelon Biosciences) were blocked with blocking buffer containing PBS, 3% bovine serum albumin (BSA), and 0.1% Tween 20 for 1 h at 20–25 °C. Membranes were then incubated with 3 µg/ml of GST–CNH in blocking buffer for 1 h at 20–25 °C followed by washing thrice for 10 min each in a washing buffer containing PBS and 0.1% Tween 20. Subsequently, the membranes were incubated with HRP-conjugated antibody (Cytiva) for 1 h at 20–25 °C, washed thrice for 10 min each with washing buffer, and subjected to chemiluminescence detection using the ECL prime reagents and the ImageQuant LAS 4000 mini-imager.
In Vitro CK2 Kinase Assay
A purified CK2 complex was purchased (#P6010S, New England BioLabs, Beverly, MA). GST-mNRK565–868, GST-mNRK565–831, and GST-PTEN190–403 fusion proteins were expressed in the E. coli strain Rosetta (DE3) pLysS, adsorbed to Glutathione-Sepharose 4B beads, and eluted with reduced glutathione. Nontagged mNRK565–868 was eluted with PreScission Protease (Cytiva) according to the manufacturer’s recommended protocol. SDS-PAGE (sodium dodecyl sulphate–polyacrylamide gel electrophoresis) and CBB staining confirmed the purity of proteins used in this study. For the in vitro kinase assay, CK2 (20 units) and the substrate GST-PTEN190–403 (1 µg) were mixed with recombinant mNRK proteins (0.5 µg) or BSA (0.25 µg) in a kinase assay buffer (20 mM Tris–HCl, pH 7.5, 5 mM MgCl2, 150 mM KCl, 0.5 mM DTT, 0.3 µM ATP) and incubated at 30 °C for 30 min. Reactions were stopped by the addition of SDS-PAGE sample buffer and boiling for 5 min. Samples were subjected to SDS-PAGE and immunoblotting.
To test whether mNRK565–868 competes with PTEN as a substrate for CK2, mNRK565–868 (0.25 µg) or BSA (0.25 µg) was mixed with CK2 (10 units; denoted as CK21) in the kinase assay buffer and preincubated at 30 °C for 16 h to prepare phosphorylated mNRK565–868 and the negative control. In some samples, the reactions were stopped just after preincubation. Certain samples were mixed with CK2 (10 units; denoted as CK22), GST-PTEN190–403 (1 µg), and ATP (0.3 µM) and incubated at 30 °C for 30 min. The remaining samples were processed similarly except for the absence of CK22. The reactions were stopped, and the samples were analyzed as described above.
Animal Experiment
Female heterozygous mice (X−X+) were mated with male null mice (X−Y). The placentas at 18.5 dpc were dissected, genotyped, and divided into two groups (WT, placentas of male fetuses [X+Y] and female fetuses [X−X+]; KO, placentas of male fetuses [X−Y], and female fetuses [X−X−]). Note that the paternally derived X chromosome is preferentially inactivated in the extraembryonic tissues of mice (Takagi and Sasaki 1975). Placentas were dissected at 18.5 days postcoitum (dpc) from pregnant female mice and cut into small pieces. Placental proteins were then lysed and sonicated in lysis buffer containing 20 mM Tris–HCl pH 7.4, 100 mM NaCl, 50 mM NaF, 1% Nonidet P-40, 0.1% sodium dodecyl sulfate, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 µg/ml aprotinin, 1 µg/ml leupeptin, and 1 µg/ml pepstatin A. The lysates were subjected to immunoblotting. All animal experiments were approved and conducted in compliance with the Institutional Animal Care and Research Advisory Committee of the Tokyo Institute of Technology guidelines.
Statistical Analysis
The number of independent experiments is indicated in figure legends. All data are presented as mean ± standard deviation (SD) of independent experiments. Graphs and statistical analyses were processed using GraphPad Prism 9. Asterisks on figures indicate values that were statistically different (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We would like to thank the Biomaterials Analysis Division, Tokyo Institute of Technology, for DNA sequencing analysis, confocal microscopy analysis, and mice breeding. We thank Dr Hiroshi Iwasaki and Dr Hitoshi Nakatogawa (Tokyo Institute of Technology) for helpful discussions. We thank Dr Mikiko Tanaka for the chicken embryo DNA library. We thank Dr Yuka Madoka for help with the plasmid preparation. This work was supported by the Sasakawa Scientific Research Grant from the Japan Science Society (to A.E., grant number: 2018-4012) and the Takeda Science Foundation (to T.F.). We thank Editage (www.editage.com, last accessed January 21, 2022) for English language editing.
Authors Contributions
T.F. and M.K. conceived and supervised the study; B.L., S.N., A.E., and T.F. designed experiments, performed experiments and analyzed data; B.L., S.N., H.N., E.W., and A.K. performed in silico genomic analyses; S.N., B.L., A.K., M.K., and T.F. wrote the manuscript; A.E., H.N., and K.D. revised the manuscript.
Data Availability
The data underlying this article are available in the article and in its online supplementary material.
References
- Abi Nahed R, Reynaud D, Lemaitre N, Lartigue S, Roelants C, Vaiman D, Benharouga M, Cochet C, Filhol O, Alfaidy N.. 2020. Protein kinase CK2 contributes to placental development: physiological and pathological implications. J Mol Med (Berl). 98(1):123–133. [DOI] [PubMed] [Google Scholar]
- Ashkenazy H, Penn O, Doron-Faigenboim A, Cohen O, Cannarozzi G, Zomer O, Pupko T.. 2012. FastML: a web server for probabilistic reconstruction of ancestral sequences. Nucleic Acids Res. 40(Web Server issue):W580–W584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al. 2012. The evolutionary landscape of alternative splicing in vertebrate species. Science 338(6114):1587–1593. [DOI] [PubMed] [Google Scholar]
- Bibby AC, Litchfield DW.. 2005. The multiple personalities of the regulatory subunit of protein kinase CK2: CK2 dependent and CK2 independent roles reveal a secret identity for CK2beta. Int J Biol Sci. 1(2):67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolze PA, Mommert M, Mallet F.. 2017. Contribution of syncytins and other endogenous retroviral envelopes to human placenta pathologies. Prog Mol Biol Transl Sci. 145:111–162. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Johnson AK, Dunker AK, Daughdrill GW.. 2011. Evolution and disorder. Curr Opin Struct Biol. 21(3):441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet FG, Roest Crollius H, Paris M, Aury JM, Gibert P, Jaillon O, Laudet V, Robinson-Rechavi M.. 2006. Gene loss and evolutionary rates following whole-genome duplication in teleost fishes. Mol Biol Evol. 23(9):1808–1816. [DOI] [PubMed] [Google Scholar]
- Cao JY, Shire K, Landry C, Gish GD, Pawson T, Frappier L.. 2014. Identification of a novel protein interaction motif in the regulatory subunit of casein kinase 2. Mol Cell Biol. 34(2):246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z, Li G, Liu J, Zhang Y, Ashby C, Liu D, Cramer CL, Huang X.. 2015. Bridger: a new framework for de novo transcriptome assembly using RNA-seq data. Genome Biol. 16:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JO, Li H, Lundquist EA.. 2008. The MIG-15 NIK kinase acts cell-autonomously in neuroblast polarization and migration in C. elegans. Dev Biol. 324(2):245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Chatterjee U, Ghosh MK.. 2013. Activation of protein kinase CK2 attenuates FOXO3a functioning in a PML-dependent manner: implications in human prostate cancer. Cell Death Dis. 4:e543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church DN, Phillips BR, Stuckey DJ, Barnes DJ, Buffa FM, Manek S, Clarke K, Harris AL, Carter EJ, Hassan AB.. 2012. Igf2 ligand dependency of Pten(+/-) developmental and tumour phenotypes in the mouse. Oncogene 31(31):3635–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daulat AM, Bertucci F, Audebert S, Serge A, Finetti P, Josselin E, Castellano R, Birnbaum D, Angers S, Borg JP.. 2016. PRICKLE1 contributes to cancer cell dissemination through its interaction with mTORC2. Dev Cell. 37(4):311–325. [DOI] [PubMed] [Google Scholar]
- Delpire E. 2009. The mammalian family of sterile 20p-like protein kinases. Pflugers Arch. 458(5):953–967. [DOI] [PubMed] [Google Scholar]
- Denda K, Nakao-Wakabayashi K, Okamoto N, Kitamura N, Ryu JY, Tagawa Y, Ichisaka T, Yamanaka S, Komada M.. 2011. Nrk, an X-linked protein kinase in the germinal center kinase family, is required for placental development and fetoplacental induction of labor. J Biol Chem. 286(33):28802–28810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devor EJ. 2014. Placenta-specific protein 1 is conserved throughout the Placentalia under purifying selection. ScientificWorldJournal 2014:537356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Zhang L, Zhang S, Bai Y, Chen H, Sun X, Yong W, Li W, Colvin SC, Rhodes SJ, et al. 2012. Phosphatase of regenerating liver 2 (PRL2) is essential for placental development by down-regulating PTEN (phosphatase and tensin homologue deleted on chromosome 10) and activating Akt protein. J Biol Chem. 287(38):32172–32179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma MK, Li D, Krebs EG, Yuasa Y, Homma Y.. 2002. Association and regulation of casein kinase 2 activity by adenomatous polyposis coli protein. Proc Natl Acad Sci U S A. 99(9):5959–5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard M, Foster DN, Cserjesi P.. 1999. Expression of HAND gene products may be sufficient for the differentiation of avian neural crest-derived cells into catecholaminergic neurons in culture. Dev Biol. 215(1):62–77. [DOI] [PubMed] [Google Scholar]
- Imakawa K, Nakagawa S.. 2017. The phylogeny of placental evolution through dynamic integrations of retrotransposons. Prog Mol Biol Transl Sci. 145:89–109. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Xie M, Chen W, Talbot R, Maddox JF, Faraut T, Wu C, Muzny DM, Li Y, Zhang W, et al. 2014. The sheep genome illuminates biology of the rumen and lipid metabolism. Science 344(6188):1168–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakinuma H, Inomata H, Kitamura N.. 2005. Enhanced JNK activation by NESK without kinase activity upon caspase-mediated cleavage during apoptosis. Cell Signal 17(11):1439–1448. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD.. 2004. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat Genet. 36(6):642–646. [DOI] [PubMed] [Google Scholar]
- Knofler M, Haider S, Saleh L, Pollheimer J, Gamage T, James J.. 2019. Human placenta and trophoblast development: key molecular mechanisms and model systems. Cell Mol Life Sci. 76(18):3479–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB.. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 34(7):1812–1819. [DOI] [PubMed] [Google Scholar]
- Lee WK, Son SH, Jin BS, Na JH, Kim SY, Kim KH, Kim EE, Yu YG, Lee HH.. 2013. Structural and functional insights into the regulation mechanism of CK2 by IP6 and the intrinsically disordered protein Nopp140. Proc Natl Acad Sci U S A. 110(48):19360–19365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. 2008. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 9(2):99–111. [DOI] [PubMed] [Google Scholar]
- Li D, Meier UT, Dobrowolska G, Krebs EG.. 1997. Specific interaction between casein kinase 2 and the nucleolar protein Nopp140. J Biol Chem. 272(6):3773–3779. [DOI] [PubMed] [Google Scholar]
- Litchfield DW. 2003. Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem J. 369(Pt 1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Zhang C, Wang Y, Dai G, Liu SQ, Wang W, Pan YH, Ding J, Li H.. 2021. New exon and accelerated evolution of placental gene Nrk occurred in the ancestral lineage of placental mammals. Placenta 114:14–21. [DOI] [PubMed] [Google Scholar]
- Llorens F, Roher N, Miro FA, Sarno S, Ruiz FX, Meggio F, Plana M, Pinna LA, Itarte E.. 2003. Eukaryotic translation-initiation factor eIF2beta binds to protein kinase CK2: effects on CK2alpha activity. Biochem J. 375(Pt 3):623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda M, Uno Y, Ohbayashi N, Ohata H, Mimata A, Kukimoto-Niino M, Moriyama H, Kashimoto S, Inoue T, Goto N, et al. 2016. TNIK inhibition abrogates colorectal cancer stemness. Nat Commun. 7:12586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB.. 2012. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science 338(6114):1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morioka Y, Nam JM, Ohashi T.. 2017. Nik-related kinase regulates trophoblast proliferation and placental development by modulating AKT phosphorylation. PLoS One 12(2):e0171503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito S, Fukushima T, Endo A, Denda K, Komada M.. 2020. Nik-related kinase is targeted for proteasomal degradation by the chaperone-dependent ubiquitin ligase CHIP. FEBS Lett. 594(11):1778–1786. [DOI] [PubMed] [Google Scholar]
- Nakano K, Kanai-Azuma M, Kanai Y, Moriyama K, Yazaki K, Hayashi Y, Kitamura N.. 2003. Cofilin phosphorylation and actin polymerization by NRK/NESK, a member of the germinal center kinase family. Exp Cell Res. 287(2):219–227. [DOI] [PubMed] [Google Scholar]
- Nakano K, Yamauchi J, Nakagawa K, Itoh H, Kitamura N.. 2000. NESK, a member of the germinal center kinase family that activates the c-Jun N-terminal kinase pathway and is expressed during the late stages of embryogenesis. J Biol Chem. 275(27):20533–20539. [DOI] [PubMed] [Google Scholar]
- Ono H, Ogasawara O, Okubo K, Bono H.. 2017. RefEx, a reference gene expression dataset as a web tool for the functional analysis of genes. Sci Data 4:170105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono R, Nakamura K, Inoue K, Naruse M, Usami T, Wakisaka-Saito N, Hino T, Suzuki-Migishima R, Ogonuki N, Miki H, et al. 2006. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat Genet. 38(1):101–106. [DOI] [PubMed] [Google Scholar]
- Plutoni C, Keil S, Zeledon C, Delsin LEA, Decelle B, Roux PP, Carreno S, Emery G.. 2019. Misshapen coordinates protrusion restriction and actomyosin contractility during collective cell migration. Nat Commun. 10(1):3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahdar M, Inoue T, Meyer T, Zhang J, Vazquez F, Devreotes PN.. 2009. A phosphorylation-dependent intramolecular interaction regulates the membrane association and activity of the tumor suppressor PTEN. Proc Natl Acad Sci U S A. 106(2):480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawn SM, Cross JC.. 2008. The evolution, regulation, and function of placenta-specific genes. Annu Rev Cell Dev Biol. 24:159–181. [DOI] [PubMed] [Google Scholar]
- Rice WR. 1984. Sex chromosomes and the evolution of sexual dimorphism. Evolution 38(4):735–742. [DOI] [PubMed] [Google Scholar]
- Riley P, Anson-Cartwright L, Cross JC.. 1998. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat Genet. 18(3):271–275. [DOI] [PubMed] [Google Scholar]
- Risso G, Blaustein M, Pozzi B, Mammi P, Srebrow A.. 2015. Akt/PKB: one kinase, many modifications. Biochem J. 468(2):203–214. [DOI] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, et al. 2005. The DNA sequence of the human X chromosome. Nature 434(7031):325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J, Cross JC.. 2001. Placental development: lessons from mouse mutants. Nat Rev Genet. 2(7):538–548. [DOI] [PubMed] [Google Scholar]
- Smith MD, Wertheim JO, Weaver S, Murrell B, Scheffler K, Kosakovsky Pond SL.. 2015. Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol Biol Evol. 32(5):1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Ono R, Narita T, Pask AJ, Shaw G, Wang C, Kohda T, Alsop AE, Marshall Graves JA, Kohara Y, et al. 2007. Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet. 3(4):e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi N, Sasaki M.. 1975. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 256(5519):640–642. [DOI] [PubMed] [Google Scholar]
- Takao T, Asanoma K, Tsunematsu R, Kato K, Wake N.. 2012. The maternally expressed gene Tssc3 regulates the expression of MASH2 transcription factor in mouse trophoblast stem cells through the AKT-Sp1 signaling pathway. J Biol Chem. 287(51):42685–42694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takezaki N, Nishihara H.. 2016. Resolving the phylogenetic position of coelacanth: the closest relative is not always the most appropriate outgroup. Genome Biol Evol. 8(4):1208–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Kumar S.. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J, Pulido R.. 2001. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. J Biol Chem. 276(2):993–998. [DOI] [PubMed] [Google Scholar]
- Trotman LC, Alimonti A, Scaglioni PP, Koutcher JA, Cordon-Cardo C, Pandolfi PP.. 2006. Identification of a tumour suppressor network opposing nuclear Akt function. Nature 441(7092):523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunster SJ, Creeth HDJ, John RM.. 2016. The imprinted Phlda2 gene modulates a major endocrine compartment of the placenta to regulate placental demands for maternal resources. Dev Biol. 409(1):251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Ramaswamy S, Nakamura N, Sellers WR.. 2000. Phosphorylation of the PTEN tail regulates protein stability and function. Mol Cell Biol. 20(14):5010–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, Park JE, Stephenson E, Polanski K, Goncalves A, et al. 2018. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563(7731):347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrunes F, Waters PD, Miethke P, Rens W, McMillan D, Alsop AE, Grutzner F, Deakin JE, Whittington CM, Schatzkamer K, et al. 2008. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 18(6):965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S, Shank SD, Spielman SJ, Li M, Muse SV, Kosakovsky Pond SL.. 2018. Datamonkey 2.0: a modern web application for characterizing selective and other evolutionary processes. Mol Biol Evol. 35(3):773–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods L, Perez-Garcia V, Hemberger M.. 2018. Regulation of placental development and its impact on fetal growth-new insights from mouse models. Front Endocrinol (Lausanne) 9:570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JH, Wang X, Manning G, LaMere BJ, Le P, Zhu S, Khatry D, Flanagan PM, Buckley SD, Whyte DB, et al. 2003. The STE20 kinase HGK is broadly expressed in human tumor cells and can modulate cellular transformation, invasion, and adhesion. Mol Cell Biol. 23(6):2068–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang ZZ, Tschopp O, Hemmings-Mieszczak M, Feng J, Brodbeck D, Perentes E, Hemmings BA.. 2003. Protein kinase B alpha/Akt1 regulates placental development and fetal growth. J Biol Chem. 278(34):32124–32131. [DOI] [PubMed] [Google Scholar]
- Yu DH, Zhang X, Wang H, Zhang L, Chen H, Hu M, Dong Z, Zhu G, Qian Z, Fan J, et al. 2014. The essential role of TNIK gene amplification in gastric cancer growth. Oncogenesis 3:e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.