Abstract
Numerous processes occur simultaneously within the cell for both normal function and in response to changes in the environment. The ability of cells to segregate biochemical reactions into separate compartments is essential for ensuring specificity and efficiency in cellular processes. The discovery of liquid-liquid phase separation as a mechanism of compartmentalization has revised our thinking regarding the intracellular organization of molecular pathways such as signal transduction. Here, we highlight recent studies that advance our understanding of how phase separation impacts the organization of biochemical processes, with a particular focus on the tools used to study the functional impact of phase separation. In addition, we offer some of our perspectives on the pathological consequences of dysregulated phase separation within biochemical pathways.
Compartmentalizing cellular processes
Our environment is ever changing and the cells in our body must dynamically adapt to these changes. A network of dynamic molecules is responsible for orchestrating the appropriate responses by decoding the input signals and passing along the cellular information. Mechanistically, each molecule relays the signaling information downstream by altering the next molecule in the pathway through a cascade of biochemical reactions, a process termed signal transduction. A pressing question in signal transduction research is one of specificity. Many of the cell’s critical processes are regulated by a few key signaling nodes; thus, how is specificity achieved? One solution, proposed nearly 40 years ago, lies in the idea of compartmentation [1,2]: More than being just a random collection of molecular interactions and biochemical reactions, intracellular signaling networks are organized into distinct compartments embedded in the framework of a living cell and intricately regulated in cellular space and time. Segregating the biochemical machinery into separate cellular domains through compartmentation allows cells to precisely control both the location and timing of biochemical activities being turned on and off, creating a sophisticated and dynamic biochemical activity architecture [3,4]. Such spatiotemporal regulation is essential for achieving specific control of cellular processes.
The typical intracellular compartments that often first come to mind are membrane-bound organelles, which enclose specific biochemical pathways within lipid membranes. In the past decade though [5], the discovery of liquid-liquid phase separation occurring in cells has shifted our understanding of how compartmentation can be achieved via membraneless compartments [6,7]. Liquid-liquid phase separation occurs when a given molecule that is uniformly dispersed in solution spontaneously de-mixes into two phases - a highly concentrated, condensed phase and a dilute phase - upon reaching a critical threshold concentration (Box 1 describes the molecular forces driving phase separation). Excitingly, there has been a recent acceleration in the discovery of phase-separated systems within cells, many of which are involved in signal transduction [8–14]. These phase-separated systems go by many names, including membraneless organelles [15], biomolecular condensates [16], phase-separated bodies [17], liquid droplets [6], granules [18], or assemblies [19], and these are used interchangeably in this review. Given the specialized, liquid-like properties of phase-separated condensates, it is possible that unique biochemical activities can arise by virtue of phase separating key components involved in signaling. Thus, this review will discuss how phase separation contributes to signaling activity architectures and explore the biochemical, physiological, and pathological consequences of such phase separation.
Box 1: Formation and regulation of phase-separated bodies
Phase-separated systems in biology are generally composed of “scaffolds” and “clients” [17]. Scaffold molecules drive phase separation, as they are necessary and sufficient for spontaneous droplet formation in vitro and in cells. Client molecules partition into scaffold-driven condensates and can influence the properties of the phase-separated system. Biomolecular phase separation is driven by networks of interactions between polymeric biomolecules, such as proteins and nucleic acids [20–23], and these multivalent interactions regulate the dynamics of phase separation by the scaffold molecule and determine which clients are sequestered into the condensate [16,24,25]. The stable multivalent interactions necessary for phase separation can be mediated by interactions between multiple folded domains or short linear motifs, such as Src homology domain 3 (SH3) modules or proline-rich motifs (PRMs), respectively [16,26]. In addition, phase separation can be driven by weak multivalent interactions from intrinsically disordered regions (IDRs) with multiple interaction motifs, a classic example being interactions between RNA and RNA-binding proteins [27,28]. Borrowing terminology from the polymer field, both folded domains and disordered interaction motifs can be viewed as “stickers” that mediate valency and are separated by “spacers” for flexibility [29].
While it is still hard to predict which proteins can phase separate, finding common features of scaffold proteins and mutagenesis experiments have unveiled several amino acid interactions that contribute to the phase separation of scaffold proteins [27,30–32]. Cation-TT interactions from positively charged and aromatic residues and cation-cation interactions are necessary and sufficient for phase separation of certain systems [27,33]. Electrostatic forces also regulate phase separation by altering interdomain connections [34,35], and this may explain why some post-translational modifications (PTMs) such as phosphorylation can regulate condensation [36,37]. In addition, IDRs are enriched with glycine (Gly), serine (Ser), and Gln (Gln) residues [27]. While the cation-TT and cation-cation interactions can be considered as stickers in determining the propensity for condensation, the spacer regions within IDRs influence the phase behavior of droplets. Gly residues enhance liquidity of condensates possibly due to Gly enhancing backbone flexibility or altering hydrophobicity, whereas Gln and Ser residues promote hardening [27].
Condensates shape the biochemical activity landscape
The concentration of biomolecules throughout the cell is rarely uniform, in part due to membrane-bound organelles. Phase-separated bodies can also cause inhomogeneous distribution of biomolecules by concentrating these molecules into membraneless compartments. If the biomolecules sequestered into condensates were components of a biochemical pathway, such as an enzyme and its effector/substrate, unique activity dynamics and thus functions may arise due to these compartments. Condensation may enhance molecular interactions/collisions, which in turn will affect the kinetics of biochemical activities within and also influence processes ocurring outside the droplet. Here, we present three examples that highlight the functional impact of phase separation. Along with these examples, we also highlight the innovative tools and methods developed and used to characterize the activity and function of biomolecular condensates.
Dynamic sequestration of key effector molecules in condensates
By sequestering clients, condensates can act as reservoirs. Our recent discovery that a regulatory subunit of the cyclic AMP (cAMP)-dependent protein kinase (PKA), Rlα, undergoes liquid-liquid phase separation and dynamically buffers cAMP demonstrates this principle [38]. Spatial compartmentation of the ubiquitous second messenger cAMP has been central to our understanding of cAMP signaling specificity for more than three decades [2]. However, plausible mechanisms of cAMP compartmentation have been lacking given that cAMP-degrading phosphodiesterases (PDEs) have modest catalytic properties [39,40] and that cAMP diffusion can be fast [41–43], thus raising the question of whether and how cAMP is compartmentalized. Recent studies address these questions by showing that most cAMP is not freely diffusible within cells and is instead dynamically sequestered in Rlα phase-separated bodies [38,40].
Specifically, we used a split-GFP tagging system [44,45] to label endogenously expressed Rlα and observed the formation of dynamic, liquid-like Rlα puncta in live cells, while we also found that purified Rlα formed liquid droplets in vitro [38]. The formation of phase-separated Rlα bodies was driven by multivalent interactions through its dimerization/docking domain and intrinsically disordered linker region and closely mirrored intracellular cAMP elevations, with acute activation of cAMP signaling rapidly inducing Rlα phase separation in cells. Interestingly, overexpressing fluorescent protein (FP)-tagged PKA catalytic subunit (PKAcat) together with Rlα revealed PKAcat co-phase separation into Rlα condensates [38]. These findings prompted us to explore the dynamics of cAMP and PKA signaling within Rlα bodies. To directly measure the cAMP and PKA dynamics inside native condensates in cells, we developed fluorescent sensors targeted to endogenous proteins (FluoSTEPs), a novel platform that combines genetically encoded biosensors with genome editing for straightforward monitoring of signaling activities at endogenously expressed proteins [38,46]. Like other FRET-based biosensors [47], FluoSTEPs utilize a molecular switch to sense a specific biochemical input and control the distance and orientation, and thus FRET, between a pair of FPs. Crucially, however, FluoSTEPs are split into two components: a small fragment of GFP tagged to a protein of interest (POI) via CRISPR/Cas technology, plus the remaining biosensor containing the other GFP fragment, RFP, and molecular switch (Figure 1) [38,46]. When both sensor components are expressed, the GFP fragments spontaneously reconstitute to yield an intact biosensor with a functional, fluorescent FRET donor localized to the POI. This scheme ensures that only the properly targeted biosensor will produce a FRET signal, allowing native signaling dynamics to be monitored around the POI without affecting its endogenous expression. FluoSTEPs are especially useful for investigating POIs that undergo phase separation, as they do not perturb endogenous stoichiometry and expression levels, which dictate droplet formation.
Figure 1. Fluorescent biosensors illuminate cAMP buffering and compartmentation by phase-separated PKA Rlα.

(Upper panel) Liquid-like condensates of the PKA regulatory subunit Rlα function like sponges that dynamically sequester cAMP. Zhang et al. were able to monitor this phenomenon directly in living cells using FluoSTEP-ICUE, a FRET-based cAMP indicator targeted to endogenously expressed Rlα [38]. FluoSTEP-ICUE uses the first 10 β-strands of GFP (GFP1–10) as a partial FRET donor, along with the 11th β-strand (GFP11) inserted into the Rlα genomic locus via CRISPR/Cas9. When co-expressed, these two fragments spontaneously reconstitute and yield an intact sensor fused to Rlα, enabling visualization of local cAMP dynamics both within and outside Rlα condensates. (Middle panel) cAMP sequestration by Rlα condensates is essential to reduce the levels of free cAMP in the cytosol and thus allow cAMP phosphodiesterases (PDEs) to function as cAMP sinks and control cAMP compartmentation. (Lower panel) Zhang et al. were able to measure this effect by fusing the cAMP indicator ICUE4 directly to the PDE4D2 catalytic subunit (PDE4D2cat).
To monitor cAMP levels near endogenously tagged Rlα, we generated a FluoSTEP version of our FRET-based cAMP sensor Indicator of cAMP using Epac (FluoSTEP-ICUE), in which a fragment of the cAMP effector Epac1 is sandwiched between a pair of FPs [48]. Binding of cAMP induces the Epac domain to adopt a more open, extended conformation that translates increases in cAMP levels to changes in FRET efficiency. Stimulating cells expressing FluoSTEP-ICUE tagged to endogenously expressed Rlα revealed a clear increase in cAMP levels detected in diffuse Rlα regions, whereas the puncta-localized sensor showed no FRET change. Further investigation suggested that the FluoSTEP-ICUE FRET response was already fully saturated inside existing Rlα puncta and that newly forming Rlα puncta showed larger increases in cAMP levels compared with diffuse Rlα regions (Figure 1), suggesting that Rlα condensates are cAMP-rich structures that actively sequester cAMP [38]. Strikingly, we observed a similar level of cAMP enrichment in vitro when we treated Rlα liquid droplets with a fluorescently labeled cAMP analogue, with 99% of labeled cAMP being sequestered into Rlα droplets [38].
A key feature of the current model of cAMP compartmentation is the ability of PDEs to function as “sinks” that locally deplete cAMP levels. While these PDE-mediated cAMP sinks can be observed experimentally [39,40], mathematical modeling suggests that PDE catalytic activity alone is insufficient to produce these compartments [39]. Given the strong enrichment of cAMP that we observed within Rlα phase-separated bodies, we hypothesized that this behavior limits cAMP levels outside Rlα bodies, thus enabling the formation of nanometer-sized PDE-mediated cAMP sinks [38–40]. We tested this hypothesis by using a cAMP compartmentation assay based on an established cAMP “nanoruler” design [39,40] in which a FRET-based cAMP biosensor is fused directly to a PDE catalytic domain to detect the presence of cAMP within the immediate vicinity of the PDE. Specifically, we fused our cAMP reporter ICUE [48] to the catalytic domain of PDE4 (Figure 1), predicting that cAMP levels around PDE4 should be low when Rlα phase separation is present and high when Rlα bodies are disrupted, which will be reflected in the PDE-tethered ICUE response. Indeed, when Rlα bodies were either pharmacologically or genetically disrupted, we observed that PDE4 could not keep pace with stimulated cAMP production and lost the ability to locally deplete cAMP to form sinks (Figure 1). Our results suggested that Rlα bodies act as a collective sponge, soaking up cAMP and thus restricting cAMP levels outside condensates to facilitate compartmentalized signaling. This study highlights the capabilities of biomolecular condensates to regulate signaling outside their borders.
Phase transitions mediate non-linear amplification of signaling cascades
Bringing together key components of the same pathway can augment pathway output. This has been seen in various signaling systems such as the Erk pathway, where the Kinase Suppressor of Ras acts as a scaffold in binding to Erk and its direct upstream effectors Raf and MEK [49]. Signaling condensates can also concentrate pathway components, potentially allowing for more diverse signaling dynamics such as non-linear signal amplification, as seen through the clustering behavior of Linker for activation of T-cells (LAT) [50].
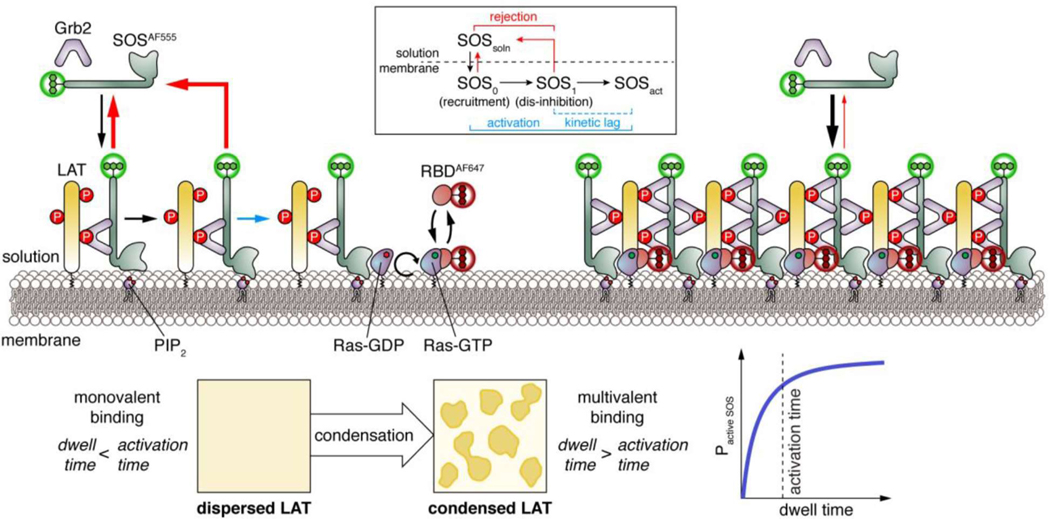
The transmembrane protein LAT is an intermediate component of the T-cell receptor (TCR) signaling cascade, connecting upstream TCR activation - such as through antigen presentation - to downstream effects including actin polymerization and Erk signaling, two processes necessary for proper T-cell responses. TCR activation triggers phosphorylation of the LAT cytoplasmic tail, which serves as a scaffold for assembly of the adaptor protein Grb2 and subsequent recruitment and activation of the Ras guanine exchange factor Son of sevenless homolog 1 (Sos1) to signal downstream to the Ras/Raf/MEK/Erk pathway [51]. Assembly of these components into submicron-scale plasma membrane clusters has been known for over a decade to be required for proper TCR signaling, yet the precise mechanistic and functional implications of LAT clustering were unclear [50]. In 2016, Su and colleagues showed using in vitro reconstitution that LAT signaling clusters arise through liquid-liquid phase separation driven by multivalent interactions between phosphorylated LAT (pLAT), Grb2, and Sos1 [50]. These liquid-like structures selectively recruit pathway activators and exclude repressors (e.g. phosphatases), yielding a distinct signaling compartment that greatly enhances TCR downstream signaling. Although this phase transition has important biological roles, the mechanisms by which LAT phase separation regulates TCR signaling are unclear. For example, LAT clustering increases Ras activation by 8-fold compared to no clustering, despite increasing Sos1 recruitment by only 2-fold [52]. Therefore, concentration increases alone are not sufficient to explain the non-linear amplification of downstream events.
In a recent study, Huang et al. combined biophysical modeling with single-molecule imaging on supported lipid membranes (SLMs) to understand the amplification of downstream TCR signaling by LAT condensation [52]. SLMs are simplified membrane models containing various user-inputted biomolecules such as lipids, carbohydrates, and proteins, whose concentration, stoichiometry, and membrane environment can be precisely controlled [53], making them useful for deciphering the biochemical influence of phase transitions. For their study, Huang et al. constructed SLMs containing pLAT, Grb2, Sos1, and GDP-loaded Ras, with recruitment of green-fluorophore-labeled Sos1 by Grb2 and red-fluorophore-labeled Ras binding domain (RBD) by GTP-bound (i.e. active) Ras serving as readouts for activation of this system (Figure 2). By performing single-particle tracking of Sos1 and RBD fluorescence via total internal reflection fluorescence microscopy, Huang et al. observed a temporal distribution of Sos1 recruitment that concomitantly led to different outcomes for RBD binding. Prolonged Sos1 recruitment (> 1 min) led to successful Ras activation and RBD translocation, whereas transient Sos recruitment (< 1 min) was not sufficient for Ras activation.
Figure 2. Recapitulating non-linear signal amplification by phase-separated LAT on supported lipid membranes.
Efficient activation of Ras by the guanine exchange factor Son of sevenless (SOS) is required to stimulate ERK signaling downstream of T-cell receptors. To investigate the role of membrane clustering by linker of activated T-cells (LAT), which recruits SOS to the membrane, in SOS activation, Huang et al. incubated synthetic membranes with phosphorylated LAT cytoplasmic tail (pLAT), the adaptor protein Grb2, and SOS labeled with AlexaFluor 555 (SOSAF555) and monitored SOS membrane recruitment, as well as Ras activation via recruitment of AlexaFluor 647-labeled Ras binding domain (RBDAF647) [52]. Combined with mathematical modeling, this approach revealed that SOS activation is subject to a kinetic proofreading step such that downstream Ras activation is largely restricted to pLAT phase-separated clusters. Specifically, full SOS activation required both pLAT-mediated recruitment (fast) and PIP2-mediated dis-inhibition (slow). In the absence of LLPS, monovalent binding yields very short SOS membrane dwell times, favoring dissociation before dis-inhibition can occur (rejection, red arrows). Conversely, LAT condensation promotes multivalent binding that dramatically extends SOS dwell times above the threshold required for activation.
These kinetic requirements results in delayed, switch-like Sos1 activation, which the authors recapitulated computationally by modeling Sos1 activation as a 2-step process: 1) Sos1 recruitment by Grb2 bound to pLAT, and 2) PIP2-facilitated release of Sos1 autoinhibition. Further investigation suggested that Sos1 deinhibition is rate limiting and acts as a kinetic-proofreading mechanism to filter out noise from transient Sos1 membrane recruitment [54,55]. Huang et al. further tested the role of LAT phase separation in their system by inputting the Sos1 proline-rich domain (PRD), which not only mediates Grb2 binding but also tunes LAT phase separation. PRD addition would be expected to decrease Sos1 activity through competition with full-length Sos1 if Sos1 sequestration in LAT clusters were purely stoichiometric. Instead, inputting the PRD increased Sos1 recruitment by 2-fold and Ras activation by 8-fold, an effect lost at low LAT densities, indicating a striking supra-stoichiometric enhancement caused by LAT phase separation. Interestingly, Sos1 membrane retention was increased 4-fold by LAT phase transitions [52], consistent with multivalent interactions greatly enhancing Sos1 membrane dwell times (Figure 2) [56]. Modeling the probability of Sos1 activation as a function of dwell time (Figure 2) further suggested that Sos1 activation exhibits switch-like behavior past a dwell-time threshold [52], whereby small increases in Sos1 dwell times, such as that achieved via co-phase separation into LAT condensates, promote sharp increases in Sos1 activation. Thus, Huang et al. were able to reveal a role for phase-separation in both confining and enhancing signaling activities.
Substrate channeling within liquid-like multi-enzyme assemblies
Controlled condensation of the various substrates and enzymes involved in a complex pathway can dictate the kinetics of biochemical activities, as seen in the previous examples. Moreover, localization of these complex structures with other cellular compartments that play a role in the same pathway can further fine-tune the flux of reactions by directing the flow of molecules between and within compartments. This process is exemplified by the formation of higher-order assemblies, called metabolons, containing enzymes responsible for catalyzing sequential steps in a metabolic pathway, which recently have been revealed to be essential for coordinating multi-step metabolic pathways [57].
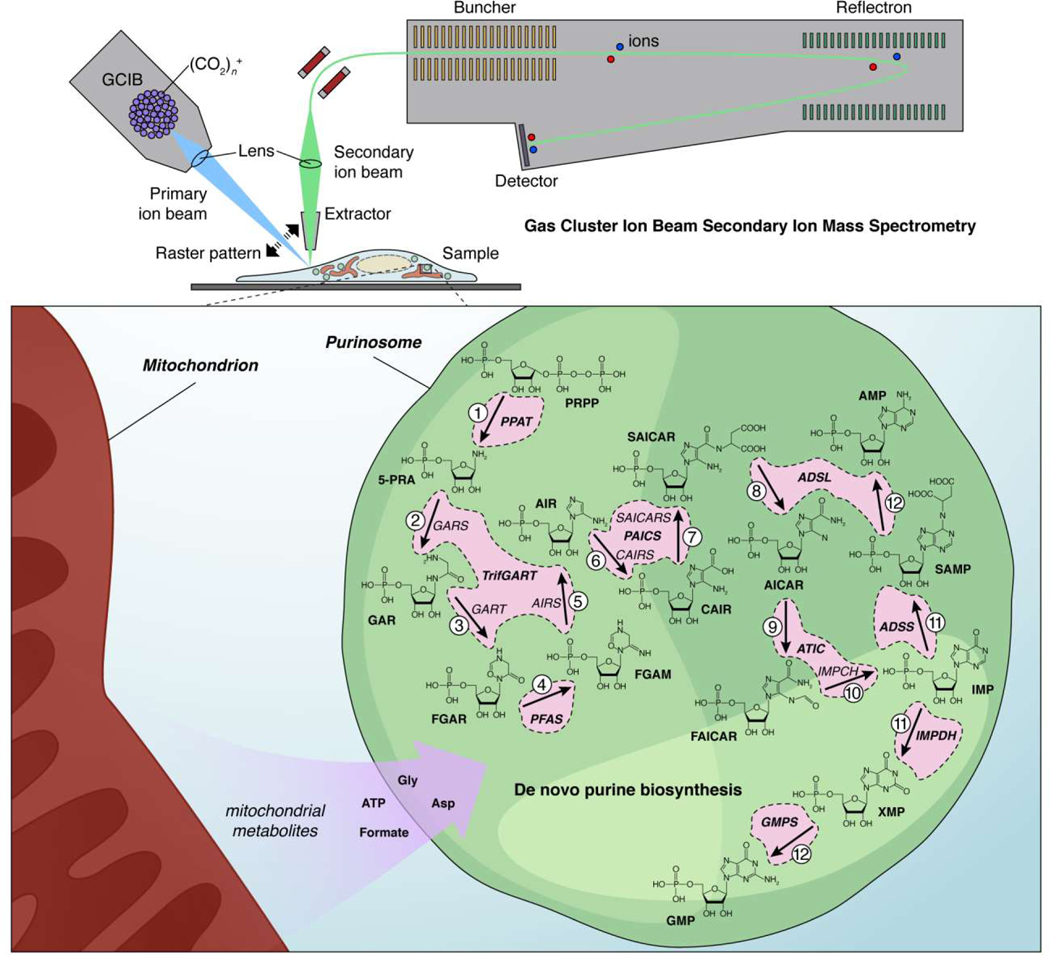
The existence of metabolons was first hypothesized over 30 years ago [58,59], and several examples have been identified in connection to various metabolic pathways [57,59,60]. Among the more famous examples is the purinosome, which contains the 6 enzymes that catalyze the 10-step de novo purine biosynthesis pathway in eukaryotes (Figure 3) [59,61]. Purinosomes rapidly and reversibly form under purine-depleted conditions [62], depend on multivalent interactions through enzyme oligomerization [63,64], and exhibit dynamic, liquid-like properties [57,65]. These behaviors are strongly suggestive of phase-separated condensates; however, additional studies are needed to determine whether LLPS indeed drives purinosome formation. These macromolecular granules are essential for dynamically regulating metabolic flux [66] by promoting substrate channeling, whereby clustering enhances transfer of metabolic intermediates between sequential enzymes [57]. Location is a key factor in this process: Purinosomes are often found near the mitochondrial surface [67], and these are thought to represent the enzymatically active pool of purinosomes [68]. Mitochondrial dysregulation can alter purinosome numbers in cells [67], highlighting a clear functional link, and indeed, mitochondrial metabolism generates several key factors required for de novo purine biosynthesis, such as Gly and formate (Figure 3) [59]. Thus, spatially linking purinosomes with mitochondria likely promotes substrate channeling by facilitating influx of these necessary materials.
Figure 3. Mass spectrometry imaging reveals substrate channeling by the purinosome, a liquid-like multi-enzyme assembly.
(Upper panel) In gas cluster ion beam secondary ion mass spectrometry (GCIB-SIMS), clusters containing many thousands of ionized gas molecules (e.g., [CO2]n+, where n>10,000) are a fired at high velocity (primary ion beam) to liberate molecular ions from the surface of a frozen sample (e.g., a cell). These molecular ions are then extracted and focused into a secondary ion beam that travels into the detector. The primary ion beam is focused into a small spot (e.g., 1 μm × 1 μm × 400 nm) and raster scanned at different depths across the sample to create a 3D “image” where each voxel corresponds to a mass spectrum. (Lower panel) Pareek et al. used this technology to directly visualize the substrate-channeling behavior of the purinosome, a multi-enzyme condensate that controls de novo purine biosynthesis, in HeLa cells [68]. Based on their studies, the authors were ultimately able to conclude that active purinosomes comprise all 9 enzymes responsible for catalyzing the 14-step synthesis of AMP and GMP from PRPP in a highly channeled process, insulated from the bulk cytosol, and that localization near mitochondria further allows purinosomes to effectively channel necessary cofactors, produced via mitochondrial metabolism, directly into the de novo purine biosynthesis pathway.
In a recent effort to test the hypothesis of spatiotemporal coordination of enzymatic activity by metabolons, Pareek and colleagues utilized mathematical modeling, isotopic labeling experiments, and mass spectrometry imaging (MSI) to investigate purinosome activity and directly probe metabolon function [68]. The authors cultured HeLa cells in purine-depleted media supplemented with [13C3,15N] Ser, such that mitochondrial conversion of Ser to Gly and formate would introduce isotopic labels into purine biosynthetic intermediates. They further modeled de novo purine biosynthesis as a completely diffusive process where each step occurs independently and all intermediates are fully equilibrated in the cytosol. Using this model, they then calculated the expected distribution of isotope-labeled species (isotopomers) that should form in the absence of purinosomes and compared these values with the actual distribution measured using high-resolution LC-MS of HeLa cell extracts. Remarkably, isotope-labeling experiments revealed that the actual isotopomer distribution, as well as the specific enrichment of 13C and 15N isotopes derived from formate and Gly, respectively, differed significantly from what would be expected in the absence of purinosomes and was instead consistent with highly channeled synthesis directed by an active metabolon [68]. Through further testing, the authors ultimately concluded that fully active purinosomes comprise all 9 enzymes responsible for catalyzing the 14-step synthesis of AMP and GMP near the mitochondrial surface, efficiently channeling mitochondrially derived substrates through de novo purine biosynthesis to increase pathway flux 7-fold.
Seeking to directly visualize metabolon function in situ, Pareek et al. then used gas cluster ion beam secondary ion mass spectrometry (GCIB-SIMS) to image purinosome biochemical activity within intact HeLa cells (Figure 3) [68]. GCIB-SIMS is an MSI approach that allows label-free 3D spatial profiling of chemical species within biological specimens [69,70]. In particular, this approach utilizes a primary ion beam composed of clusters of highly charged gas molecules accelerated to high velocity, enabling high-yield molecular desorption with relatively minimal chemical damage (e.g., molecular fragmentation) [69,70], with the resulting secondary ions extracted and analyzed via SIMS (Figure 3). Using a (CO2)n+ (n>10000) GCIB focused to a 1-μm spot, the authors scanned frozen, dehydrated HeLa cells to collect mass spectra corresponding to individual 1 μm × 1 μm × 400 nm voxels, close to the estimated spatial dimensions of individual purinosomes [59]. Isotopic labeling with [15N]Ser was used to promote enrichment of labeled metabolites within mitochondrially associated purinosomes, and the MS peak corresponding to the purine biosynthetic intermediate 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) was used as a specific reporter for these active metabolons. GCIB-SIMS imaging revealed a striking heterogeneity in the distribution of 15N-labeled AICAR, with specific voxels showing 300- to 1000fold AICAR enrichment versus that predicted for a uniform cytosolic distribution [68]. Further analysis showed that these AICAR hotspots were also enriched in labeled ATP, a pathway end product. These results highlight the utility of GCIB-SIMS to map out the molecular components of micron-sized compartments and provide evidence that purinosomes are active, substrate-channeling metabolons [68].
By selectively recruiting and localizing pathway components to functionally related compartments, these liquid-like multi-enzyme assemblies can thus coordinate various molecules and enzymes to organize complex, multi-step processes.
Concluding remarks and future perspectives
It is clear now that membraneless, liquid-like condensates play a major role in driving cellular compartmentation. Concentrating specific biomolecules into these phase-separated bodies allows for unique chemistries and reaction kinetics within these microdomains. When applied to biochemical machinery, condensation not only allows for unique pathway dynamics but also enables specific cellular processes that are critical, as their disruption leads to pathological consequences [8,38,71]. This shift in our paradigm of cellular compartmentation encourages us to think about new possibilities for spatiotemporal regulation of biochemical activity, which is fundamental to our basic understanding of how pathway specificity is achieved.
What happens, then, when phase separation behavior becomes dysfunctional? Emerging evidence suggests that protein aggregates found in various neurodegenerative diseases are phase-separated bodies [72] that become more gel-like during disease progression [35,73], thus providing new insights into decades-old questions regarding the origins of these pathogenic structures in patients [74,75]. Dysregulation of phase separation is also seen in other diseases such as cancer [7,76]. In the case of prostate cancer, mutations in the tumor suppressor and component of an E3 ubiquitin ligase, speckle-type POZ protein (SPOP) inhibit its interaction with substrates, thus blocking proper ubiquitination of various signaling cascade effectors, disrupting SPOP nuclear condensation and enhancing cell proliferation [77,78]. In breast cancer tissues, increased TAZ expression and more nuclear TAZ condensates compared to healthy breast tissue may serve as a potential biomarker [79]. In another example, the fusion of DnaJB1 exon 1 with the last 9 exons of PKAcat (DnaJB1-PKAcat) is exclusively detected in patients with fibrolamellar carcinoma (FLC) [80], a rare liver cancer that has little similarity with other liver cancers [81]. While it is clear that this chimeric fusion enzyme drives FLC, as it is sufficient to induce FLC-like tumors in mice [82], biochemical and structural studies show little difference between DnaJB1-PKAcat and wildtype PKAcat in terms of either activity or regulation [83–85]. Recent work has offered new insight into the oncogenic mechanisms of this fusion oncoprotein by revealing that DnaJB1-PKAcat disrupts Rlα phase separation by recruiting Hsp70 and disrupting PKAcat myristoylation [38]. Furthermore, DnaJB1-PKAcat expression induces loss of cAMP compartmentation, while the loss of Rlα phase separation alone is sufficient to promote increased cell proliferation and transformation, suggesting that Rlα phase separation has tumor-suppressive roles.
In this review, we have highlighted three modes by which phase separation can shape the biochemical landscape, but there are many other ways that condensation can play a role in signal transduction. Uncovering more phase-separating systems and dissecting their functional effects of via new tools and methods [28,86–88] will be crucial for advancing our general understanding of how these membraneless compartments help construct biochemical architectures (see Outstanding Questions). Furthermore, the functional specificity imparted by these non-membrane-bound compartments presents a pharmacologically attractive opportunity [89—91 ] to therapeutically target specific cellular processes. Indeed, recent efforts have sought pharmacological agents that can disrupt phase-separated bodies [92,93], with the ultimate goal of developing small molecules, peptides, or antibodies that can influence the behavior of select phase-separated bodies [94]. Advancing our understanding of phase separation and developing new biomolecular tools and methods to target these compartments will push the boundaries for therapeutics and drug delivery in treating diseases.
Outstanding Questions:
What are the guiding principles for determining what is included and excluded from phase-separated bodies?
How general is phase separation as an organizer for proper signal transduction?
Are there other disease-related mutations that can alter the behavior of phase-separated systems?
To characterize the functional impact of condensation, can tools be generated to induce, inhibit, or alter the material properties of specific phase-separated systems?
Is specific targeting of pathological phase separation a viable therapeutic avenue?
Acknowledgements
This work was supported by the National Institutes of Health (R35 CA197622, R01 DK073368, and R01 DE030497 to J.Z.) and the Air Force Office of Scientific Research (FA9500-18-1-0051 to J.Z.).
Footnotes
Declaration of Interest
The authors declare no conflicts of interest.
References
- 1.Buxton IL and Brunton LL (1983) Compartments of cyclic AMP and protein kinase in mammalian cardiomyocytes. J. Biol. Chem 258, 10233–10239 [PubMed] [Google Scholar]
- 2.Brunton LL et al. (1981) Functional compartmentation of cyclic AMP and protein kinase in heart. Adv. Cyclic Nucleotide Res 14, 391–397 [PubMed] [Google Scholar]
- 3.Mehta S. and Zhang J. (2021) Biochemical Activity Architectures Visualized-Using Genetically Encoded Fluorescent Biosensors to Map the Spatial Boundaries of Signaling Compartments. Acc. Chem. Res 54, 2409–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mehta S. and Zhang J. (2017) Illuminating the Cell’s Biochemical Activity Architecture. Biochemistry 56, 5210–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brangwynne CP et al. (2009) Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 [DOI] [PubMed] [Google Scholar]
- 6.Courchaine EM et al. (2016) Droplet organelles? EMBO J. 35, 1603–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hyman AA et al. (2014) Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol 30, 39–58 [DOI] [PubMed] [Google Scholar]
- 8.Chong PA and Forman-Kay JD (2016) Liquid-liquid phase separation in cellular signaling systems. Curr. Opin. Struct. Biol 41, 180–186 [DOI] [PubMed] [Google Scholar]
- 9.Case LB et al. (2019) Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science (80-.). 363, 1093–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tulpule A. et al. (2021) Kinase-mediated RAS signaling via membraneless cytoplasmic protein granules. Cell 184, 2649–2664.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou W. et al. (2021) cGAS phase separation inhibits TREX1-mediated DNA degradation and enhances cytosolic DNA sensing. Mol. Cell 81, 739–755.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu J. et al. (2020) GIT/PIX Condensates Are Modular and Ideal for Distinct Compartmentalized Cell Signaling. Mol. Cell 79, 782–796.e6 [DOI] [PubMed] [Google Scholar]
- 13.Gammons MV et al. (2016) Wnt Signalosome Assembly by DEP Domain Swapping of Dishevelled. Mol. Cell 64, 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z. et al. (2020) Par complex cluster formation mediated by phase separation. Nat. Commun 11,2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boeynaems S. et al. (2018) Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 28, 420–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banani SF et al. (2017) Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol 18, 285–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banani SF et al. (2016) Compositional Control of Phase-Separated Cellular Bodies. Cell 166, 651–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mateju D. et al. (2017) An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 36, 1669–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nair SJ et al. (2019) Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nat. Struct. Mol. Biol 26, 193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perdikari TM et al. (2020) SARS-CoV-2 nucleocapsid protein undergoes liquid-liquid phase separation stimulated by RNA and partitions into phases of human ribonucleoproteins. bioRxiv DOI: 10.1101/2020.06.09.141101 [DOI] [Google Scholar]
- 21.Markmiller S. et al. (2018) Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 172, 590–604.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheu-Gruttadauria J. and MacRae IJ (2018) Phase Transitions in the Assembly and Function of Human miRISC. Cell 173, 946–957.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiu Y-P et al. (2020) Liquid-liquid phase separation and extracellular multivalent interactions in the tale of galectin-3. Nat. Commun 11, 1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bracha D. et al. (2018) Mapping Local and Global Liquid Phase Behavior in Living Cells Using Photo-Oligomerizable Seeds. Cell 175, 1467–1480.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitrea DM and Kriwacki RW (2016) Phase separation in biology; functional organization of a higher order. Cell Commun. Signal 14, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li P. et al. (2012) Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J. et al. (2018) A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 174, 688–699.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin Y. et al. (2017) Spatiotemporal Control of Intracellular Phase Transitions Using Light-Activated optoDroplets. Cell 168, 159–171.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi J-M et al. (2020) Physical Principles Underlying the Complex Biology of Intracellular Phase Transitions. Annu. Rev. Biophys 49, 107–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Q. et al. (2020) LLPSDB: a database of proteins undergoing liquid-liquid phase separation in vitro. Nucleic Acids Res. 48, D320–D327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vernon RM and Forman-Kay JD (2019) First-generation predictors of biological protein phase separation. Curr. Opin. Struct. Biol 58, 88–96 [DOI] [PubMed] [Google Scholar]
- 32.Saar KL et al. (2021) Learning the molecular grammar of protein condensates from sequence determinants and embeddings. Proc. Natl. Acad. Sci 118, e2019053118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gallego LD et al. (2020) Phase separation directs ubiquitination of gene-body nucleosomes. Nature 579, 592–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nott TJ et al. (2015) Phase Transition of a Disordered Nuage Protein Generates Environmentally Responsive Membraneless Organelles. Mol. Cell 57, 936–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molliex A. et al. (2015) Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell 163, 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wegmann S. et al. (2018) Tau protein liquid-liquid phase separation can initiate tau aggregation. EMBO J. 37, 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rai AK et al. (2018) Kinase-controlled phase transition of membraneless organelles in mitosis. Nature 559, 211–216 [DOI] [PubMed] [Google Scholar]
- 38.Zhang JZ et al. (2020) Phase separation of a PKA regulatory subunit controls cAMP compartmentation and oncogenic signaling. Cell 182, 1531–1544.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lohse C. et al. (2017) Experimental and mathematical analysis of cAMP nanodomains. PLoS One 12, e0174856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bock A. et al. (2020) Optical mapping of cAMP signaling at the nanometer scale. Cell 182, 1519–1530.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen C. et al. (1999) Cyclic AMP diffusion coefficient in frog olfactory cilia. Biophys. J 76, 2861–2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nikolaev VO et al. (2004) Novel single chain cAMP sensors for receptor-induced signal propagation. J. Biol. Chem 279, 37215–37218 [DOI] [PubMed] [Google Scholar]
- 43.Bacskai BJ et al. (1993) Spatially resolved dynamics of cAMP and protein kinase A subunits in Aplysia sensory neurons. Science 260, 222–226 [DOI] [PubMed] [Google Scholar]
- 44.Leonetti MD et al. (2016) A scalable strategy for high-throughput GFP tagging of endogenous human proteins. Proc. Natl. Acad. Sci. U. S. A 113, E3501-E3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamiyama D. et al. (2016) Versatile protein tagging in cells with split fluorescent protein. Nat.Commun 7, 11046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tenner B. et al. (2021) FluoSTEPs: Fluorescent biosensors for monitoring compartmentalizedsignaling within endogenous microdomains. Sci. Adv 7, eabe4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greenwald EC et al. (2018) Genetically Encoded Fluorescent Biosensors Illuminate the Spatiotemporal Regulation of Signaling Networks. Chem. Rev 118, 11707—11794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DiPilato LM and Zhang J. (2009) The role of membrane microdomains in shaping beta2-adrenergic receptor-mediated cAMP dynamics. Mol. Biosyst 5, 832–837 [DOI] [PubMed] [Google Scholar]
- 49.Witzel F. et al. (2012) How scaffolds shape MAPK signaling: What we know and opportunities forsystems approaches. Front. Physiol 3 DEC, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su X. et al. (2016) Phase separation of signaling molecules promotes T cell receptor signaltransduction. Science 352, 595–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balagopalan L. et al. (2015) The linker for activation of T cells (LAT) signaling hub: from signaling complexes to microclusters. J. Biol. Chem 290, 26422–26429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang WYC et al. (2019) A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science (80-.). 363, 1098–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu C. and Groves JT (2010) Engineering supported membranes for cell biology. Med. Biol. Eng. Comput 48, 955–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee YK et al. (2017) Mechanism of SOS PR-domain autoinhibition revealed by single-moleculeassays on native protein from lysate. Nat. Commun 8, 15061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sondermann H. et al. (2004) Structural Analysis of Autoinhibition in the Ras Activator Son of Sevenless. Cell 119, 393–405 [DOI] [PubMed] [Google Scholar]
- 56.Huang WYC et al. (2016) Phosphotyrosine-mediated LAT assembly on membranes drives kinetic bifurcation in recruitment dynamics of the Ras activator SOS. Proc. Natl. Acad. Sci. U. S. A 113,8218–8223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sweetlove LJ and Fernie AR (2018) The role of dynamic enzyme assemblies and substrate channelling in metabolic regulation. Nat. Commun 9, 2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Srere PA (1987) Complexes of sequential metabolic enzymes. Annu. Rev. Biochem 56, 89–124 [DOI] [PubMed] [Google Scholar]
- 59.Pedley AM and Benkovic SJ (2017) A New View into the Regulation of Purine Metabolism: The Purinosome. Trends Biochem. Sci 42, 141–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmitt DL and An S. (2017) Spatial Organization of Metabolic Enzyme Complexes in Cells. Biochemistry 56, 3184–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao H. et al. (2013) The purinosome, a multi-protein complex involved in the de novo biosynthesis of purines in humans. Chem. Commun. (Camb) 49, 4444–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.An S. et al. (2008) Reversible Compartmentalization of de Novo Purine Biosynthetic Complexes in Living Cells. Science (80-. ). 320, 103–106 [DOI] [PubMed] [Google Scholar]
- 63.Pareek V. et al. (2021) Human de novo purine biosynthesis. Crit. Rev. Biochem. Mol. Biol 56, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doigneaux C. et al. (2020) Hypoxia drives the assembly of the multienzyme purinosome complex. J. Biol. Chem 295, 9551–9566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.An S. et al. (2019) Chapter One - Phase-separated condensates of metabolic complexes in living cells: Purinosome and glucosome. In Enzyme Activity in Single Cells 628 (Allbritton NL and Kovarik MLBT-M in E., eds), pp. 1–17, Academic Press; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kyoung M. et al. (2015) Dynamic architecture of the purinosome involved in human de novo purine biosynthesis. Biochemistry 54, 870–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.French JB et al. (2016) Spatial colocalization and functional link of purinosomes with mitochondria. Science (80-. ). 351, 733–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pareek V. et al. (2020) Metabolomics and mass spectrometry imaging reveal channeled de novo purine synthesis in cells. Science (80-. ). 368, 283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winograd N. (2018) Gas cluster ion beams for secondary ion mass spectrometry. Annu. Rev. Anal. Chem 11, 29–48 [DOI] [PubMed] [Google Scholar]
- 70.Tian H. et al. (2017) Gas Cluster Ion Beam Time-of-Flight Secondary Ion Mass Spectrometry High-Resolution Imaging of Cardiolipin Speciation in the Brain: Identification of Molecular Losses after Traumatic Injury. Anal. Chem 89, 4611–4619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cai D. et al. (2019) Phase separation of YAP reorganizes genome topology for long-term, YAP target gene expression. Nat. Cell Biol 21, 1578–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Elbaum-Garfinkle S. (2019) Matter over mind: Liquid phase separation and neurodegeneration. J. Biol. Chem 294, 7160–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peskett TR et al. (2018) A Liquid to Solid Phase Transition Underlying Pathological Huntingtin Exon1 Aggregation. Mol. Cell 70, 588–601.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Selkoe DJ and Hardy J. (2016) The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med 8, 595–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kametani F. and Hasegawa M. (2018) Reconsideration of Amyloid Hypothesis and Tau Hypothesis in Alzheimer’s Disease. Front. Neurosci 12, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alberti S. and Dormann D. (2019) Liquid-Liquid Phase Separation in Disease. Annu. Rev. Genet DOI: 10.1146/annurev-genet-112618-043527 [DOI] [PubMed] [Google Scholar]
- 77.Bouchard JJ et al. (2018) Cancer Mutations of the Tumor Suppressor SPOP Disrupt the Formation of Active, Phase-Separated Compartments. Mol. Cell 72, 19–36.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim MS et al. (2013) Mutational and expressional analyses of SPOP, a candidate tumor suppressor gene, in prostate, gastric and colorectal cancers. APMIS 121, 626–633 [DOI] [PubMed] [Google Scholar]
- 79.Lu Y. et al. (2020) Phase separation of TAZ compartmentalizes the transcription machinery to promote gene expression. Nat. Cell Biol 22, 453–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Honeyman JN et al. (2014) Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science 343, 1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maniaci V. et al. (2009) Fibrolamellar hepatocellular carcinoma: prolonged survival with multimodality therapy. Eur. J. Surg. Oncol 35, 617–621 [DOI] [PubMed] [Google Scholar]
- 82.Kastenhuber ER et al. (2017) DNAJB1-PRĈA fusion kinase interacts with (3-catenin and the liver regenerative response to drive fibrolamellar hepatocellular carcinoma. Proc. Natl. Acad. Sci. U. S. A 114, 13076–13084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cao B. et al. (2019) Structures of the PKA Rlα Holoenzyme with the FLHCC Driver J-PKAcα or Wild-Type PKAcα. Structure 27, 816–828.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Riggle KM et al. (2016) Enhanced cAMP-stimulated protein kinase A activity in human fibrolamellar hepatocellular carcinoma. Pediatr. Res 80, 110–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cheung J. et al. (2015) Structural insights into mis-regulation of protein kinase a in human tumors. Proc. Natl. Acad. Sci. U. S. A 112, 1374–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tang L. (2019) Optogenetic tools light up phase separation. Nat. Methods 16, 139. [DOI] [PubMed] [Google Scholar]
- 87.Mitrea DM et al. (2018) Methods for Physical Characterization of Phase-Separated Bodies and Membrane-less Organelles. J. Mol. Biol 430, 4773–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alberti S. et al. (2019) Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 176, 419–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Verdile V. et al. Aberrant Phase Transitions: Side Effects and Novel Therapeutic Strategies in Human Disease., Frontiers in Genetics, 10. (2019), 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sabari BR et al. (2018) Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Klein IA et al. (2020) Partitioning of cancer therapeutics in nuclear condensates. Science (80-.). 368, 1386–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wheeler RJ et al. (2019) Small molecules for modulating protein driven liquid-liquid phase separation in treating neurodegenerative disease. bioRxiv DOI: 10.1101/721001 [DOI] [Google Scholar]
- 93.Babinchak WM et al. (2020) Small molecules as potent biphasic modulators of protein liquid-liquid phase separation. Nat. Commun 11, 5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wheeler RJ (2020) Therapeutics—how to treat phase separation-associated diseases. Emerg. Top. Life Sci DOI: 10.1042/ETLS20190176 [DOI] [PMC free article] [PubMed] [Google Scholar]