Abstract
Hepatocellular carcinoma (HCC) is a common malignant tumor with high mortality and poor prognosis in the world. The low rate of early diagnosis, as well as the high risk of postoperative metastasis and recurrence, led to the poor clinical prognosis of HCC patients. Currently, it mainly depends on serum markers, imaging examination, and tissue biopsy to diagnose and determine the recurrence and metastasis of HCC after treatments. Nevertheless, the accuracy and sensitivity of serum markers and imaging for early HCC diagnosis are suboptimal. Tissue biopsy, containing limited tissue samples, is insufficient to reveal comprehensive tumor biology information and is inappropriate to monitor dynamic tumor progression due to its invasiveness. Thus, low invasive diagnostic methods and novel biomarkers with high sensitivity and reliability must be found to improve HCC detection and prediction. As a non-invasive, dynamic, and repeatable detection method, “liquid biopsy”, has attracted much attention to early diagnosis and monitoring of treatment response, which promotes the progress of precision medicine. This review summarizes the clinical applications of liquid biopsy in HCC, including circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), and exosome in early diagnosis, prognostic evaluation, disease monitoring, and guiding personalized treatment.
Keywords: clinical application, liquid biopsy, circulating tumor cells, circulating tumor DNA, exosome, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the main pathological type of primary malignant tumor of the liver, ranking as the sixth common cancer and the fourth leading cause of cancer-related death in the world (1). It mainly develops in the background of cirrhosis, resulting from hepatitis B and C virus infection, excessive drinking, or non-alcoholic fatty liver disease (2). There will be more than 1 million people to die in HCC by 2030 from the prediction of World Health Organization (WHO) (3). Surgical intervention including surgical resection and liver transplantation is the primary therapy to obtain satisfactory long-term results for HCC patients. However, due to the insidious onset and rapid progression of HCC, most patients have already reached the advanced stage of HCC during the first diagnosis and lost the opportunity to access surgical treatment (4). Currently, it mainly depends on serum markers, like alpha-fetoprotein (AFP), imaging examination as well as tissue biopsy to diagnose and determine recurrence and metastasis of HCC after treatments. Although clinical practice guidelines recommend that high-risk individuals undergo ultrasound (US) and serum AFP monitoring every 6 months (5), the sensitivity of this method for the detection of early-stage HCC is only 63% (6). Besides, elevated AFP may also be detected in some other diseases, for example, cirrhosis, hepatitis, intrahepatic cholangiocarcinoma, and metastatic colon cancer (7). AFP is no longer recommended as a part of the diagnostic evaluation by the latest American Association for the Study of Liver Disease (AASLD) guidelines (5). Therefore, low invasive diagnostic methods and novel biomarkers with high sensitivity and reliability must be found to detect the early-stage HCC and monitor the tumor recurrence.
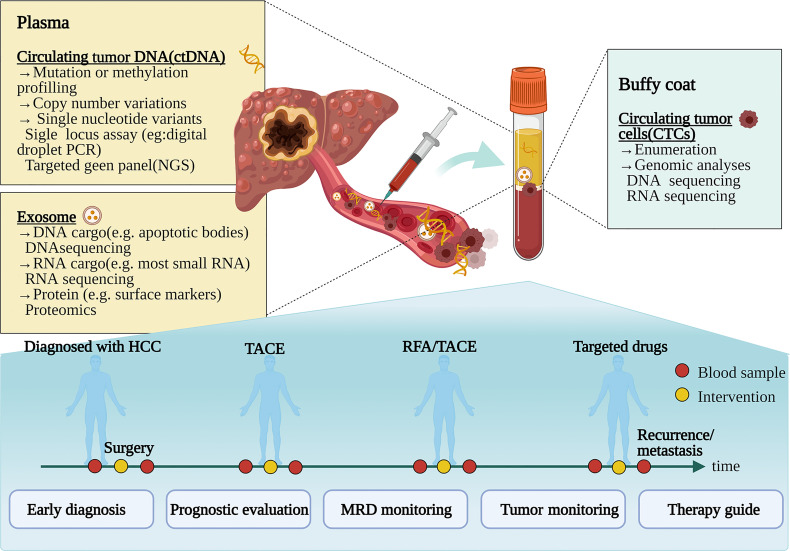
Liquid biopsy is a noninvasive, dynamic, and repeatable approach, which has emerged and shown significant prospects for HCC. Liquid biopsy obtains tumor-related information by collecting samples of body fluids such as blood and detecting circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), and exosomes (8, 9) ( Figure 1 ). Liquid biopsy has shown promising clinical application in several tumors, for instance, colorectal cancer (10), prostate cancer (11), lung cancer, and breast cancer (12). The increased studies of liquid biopsy in HCC have also emerged in recent years. Owing to HCC heterogeneity, only depend on a single biopsy might not be sufficient to reveal comprehensive tumor biology. Meanwhile, tissue biopsy is also inappropriate to monitor dynamic tumor progression as a routine practice due to its invasiveness. Fortunately, liquid biopsy can overcome these disadvantages to provide a real-time sample for the disease in a non-invasive and convenient way. As novel biomarkers, CTCs, ctDNA, and exosomes have made excellent progress in liquid biopsy of HCC (13–15). Analysis of specific gene mutations in ctDNA will help to better select treatment options and deal with drug resistance (14). This review summarizes the clinical applications of liquid biopsy in HCC, including CTCs, ctDNA, and exosomes in early diagnosis, prognostic evaluation, disease monitoring, and guiding personalized treatment.
Figure 1.
Clinical application pattern of liquid biopsy in patients with hepatocellular carcinoma. Tumor composition analyses, such as circulating tumor cells, circulating tumor DNA, and exosomes, are released by tumors to the bloodstream. During the various treatments such as surgery, transcatheter arterial chemoembolization (TACE), radiofrequency ablation (RFA), and targeting molecular treatment, liquid biopsy can be used for diagnosis, prognosis, and progress monitoring of HCC patients. NGS, next-generation sequencing; MRD, minimal/molecular residual disease. Figure 1 created in BioRender (https://biorender.com/).
Circulating Tumor Cells (CTCS)
Thomas R. Ashworth first found CTCs from the peripheral blood in the 1860s (16). CTC is released into the blood circulation from primitive or metastatic tumor cells (17). Tumor cells migrate into the bloodstream becoming CTCs by secreting matrix metalloproteinase to break the basement membrane (18). Then they can invade different parts of the body through blood circulation, thus being vividly described as “seeds” of the tumors (14). A crowd of tumor cells is released into circulation every day. However, less than 0.01% of CTCs could eventually survive and lead to a fatal metastasis (19). The half-life of CTCs is 1-2.4h, most CTCs introduced into the circulation are eliminated by shear stress, immune attack, and anoikis (20, 21). At present, the CTC defined by the CellSearch™ system is an accepted standard: CTC is a kind of epithelial cell with an intact nucleus, which is positive for EpCAM and/or cytokeratin 8, 18, and 19, but do not express CD45 (13). Meanwhile, CTCs gain the mesenchymal features by downregulating epithelial cell adhesion molecule EpCAM expression (22), thus enhancing the ability to enter the lymph vascular system (23), this process is called the epithelial-to-mesenchymal transition (EMT). In the process of tumor metastasis, EMT is triggered as a consequence of the interaction between various cellular signaling pathways, like Notch, Wnt, PDGF, TGF-β, Akt, and NF-κB (24). There are three subtypes of CTC in the process of EMT, including epithelial CTCs, mesenchymal CTCs, and mixed CTCs, which have both epithelial and mesenchymal CTCs (25). Also, CTCs may aggregate to form CTC clusters with or without leukocytes, platelets fibroblasts, or endothelial cells and move together in the bloodstream. CTC clusters are rare compared to individual CTC, but they tend to have increased survival ability and metastatic potential (26). Because of its short half-life and limited detection methods, the importance of CTC clusters may be underestimated.
Several detection and isolation techniques of CTCs have been produced, which are mainly divided into biological and physical methods. CTCs can be separated from other nucleated cells or normal epithelial cells by utilizing their distinct physical properties, and molecular characteristics, such as density, measurement, electric charge, deformation ability, and transfer capacity, as well as biological parameters, including cell surface markers, or combining both characteristics (13). The widely used technologies for CTCs detection are fluorescence in situ hybridization (FISH), reverse transcriptase-polymerase chain reaction (RT-PCR), immunofluorescence (IF), next-generation sequencing (NGS), and microfluidic-based techniques (14). CellSearch™ system that targets EpCAM to quantify CTCs remains the first and only clinically verified technology for enrichening and counting CTCs approved by the Food and Drug Administration (FDA). However, there is only a small portion of the HCC patients with EpCAM positive, and this method may underestimate the number of CTCs due to the existence of EMT. Hence, it is imperative to explore novel technologies with high accuracy to improve the capture rate of CTCs and expand the clinical application.
Clinical Applications of CTCS IN HCC
CTC has a strong prognostic effect, especially after resection, it also shows potential in monitoring the progress of HCC and guiding treatment (14). Decades of CTCs research have made great progress in the clinical application of HCC ( Table 1 ).
Table 1.
Clinical applications of CTC in HCC.
| Region | Patient | Method | CTC markers | Function of the marker | Positive rate | Ref. |
|---|---|---|---|---|---|---|
| Prognostic evaluation | ||||||
| China | 123 | CellSearch™ | EpCAM | Epithelial marker | 67% | (27) |
| China | 49 | negative enrichment qRT-PCR | EpCAM, CD4+CD25+Foxp3Treg cells | EpCAM: epithelial marker; Treg cells: immune escape | 35% | (28) |
| United Kingdom | 69 | ImageStream | AFP, EpCAM, GPC3, DNA- PK | AFP, GPC3: related biomarkers of HCC; DNA- PK: candidate biomarker for treatment stratification in HCC; EpCAM: epithelial biomarker | 65% | (29) |
| China | 47 | flow cytometry qRT-PCR | MAGE-3, Survivin, CEA | MAGE-3, Survivin: metastasis-associated markers; CEA:carcinoembryonic antigen | Not applicable | (30) |
| China | 139 | CellSearch™ | EpCAM | Epithelial marker | 44%(Pre) 54%(post) | (31) |
| China | 112 | CanPatrol™ | EpCAM, CK, Vimentin, Twist | EpCAM, CK: epithelial marker; Vimentin, Twist: mesenchymal marker | 90% | (32) |
| Germany | 57 | CellSearch™ | EpCAM | Epithelial marker | 16% | (33) |
| China | 89 | CellSearch™ | EpCAM | Epithelial marker | 56% | (34) |
| United States | 20 | CellSearch™ | EpCAM | Epithelial marker | 40% | (35) |
| China | 42 | CTC-Chip | EpCAM | Epithelial marker | 60% | (36) |
| China | 195 | CanPatrol™ | CK, EpCAM, Twist, Cadherin, Vimentin, AKT2 | EpCAM, Cadherin,CK: epithelial marker; Twist, Vimentin, Snail, AKT2: mesenchymal marker; | 95% | (37) |
| Germany | 59 | CellSearch™ | EpCAM | Epithelial marker | 31% | (38) |
| China | 299 | negative enrichment qRT-PCR | EpCAMmRNA+ | Epithelial marker | 43% | (39) |
| China | 73 | CellSearchTM qRT-PCR | EpCAM, E-cadherin, N-cadherin, Vimentin, Snail, Slug | EpCAM, E-cadherin: epithelial marker; N-cadherin, Vimentin, Snail, Slug: mesenchymal marker | 68%(PV) 45%(PA) 81%(HV) 40%(IHIVC) 59%(PoV) | (40) |
| China | 14 | SE-Ifish | Aneuploid chromosome 8 | genomic instability | 8% (EpCAM+ CTSC) 86% (EpCAM- CTC) | (41) |
| Korea | 105 | Tapered slit filter, immunofluorescence | CK, CD45 | CK: epithelial marker; CD45: leukocyte marker | 24%(ΔCTC>0) | (42) |
| China | 137 | CellSearch™ | EpCAM | Epithelial marker | 34% | (43) |
| China | 214 | CanPatrol™ RNA-ISH | CD45, EpCAM, DAPI CK8/18/19, vimentin/twist, | EpCAM, CK: epithelial marker; Twist, Vimentin, mesenchymal marker; CD45: leukocyte marker; DAPI: nuclei marker | 42% | (44) |
| Monitoring and guide therapy | ||||||
| China | 136 | CanPatrol™ | CD45, EpCAM, CK8/18/19, vimentin, twist | EpCAM, CK: epithelial marker; Twist, Vimentin, mesenchymal marker; CD45: leukocyte marker | 92% | (45) |
| China | 30 | PowerMag negative selection system | EpCAM, Hoechst, CD45, | EpCAM: epithelial marker; CD45: leukocyte marker; Hoechst: nuclei marker | 100% | (46) |
| China | 109 | immunofluorescence staining | pERK | Sorafenib-related targets | 93% | (47) |
| United States | 6 | IFC scRNA-seq | ASGPR1, pan-CK, GPC3, EPCAM, CD45 | EpCAM, pan-CK: epithelial marker; CD45: leukocyte marker; GPC3: related biomarkers of HCC; ASGPR1: expressed in hepatocytes | 67% | (48) |
qRT-PCR, quantitative real-time polymerase chain reaction; PV, peripheral vein; PA, peripheral artery; HV, hepatic veins; IHIVC, infrahepatic inferior vena cava; PoV, portal vein; IFC, Imaging Flow Cytometry; scRNA-seq, Single-cell RNA sequencing.
Prognostic Evaluation
Current data do not provide clear evidence to support CTCs as an early-stage HCC diagnostic tool (14, 49). But as for prognosis, a great deal of research supports its prospect in predicting therapeutic outcome and monitoring disease progression, particularly after resection (3). Sun et al. (27) applied CellSearch™, for the first time, to capture and analyze EpCAM+ CTCs in 123 patients before the resection of HCC and 1 month thereafter. They found that EpCAM+ CTCs≥2 per 7.5 ml of blood were proved to be the strongest prognosticator, especially in the subgroup of patients whose AFP level was lower than that of 400ng/ml. Additional similar studies proved that the expression of EpCAM+ CTCs in HCC patients showed a significant positive correlation to the serum AFP level (35), BCLC stage (38) and was associated with vascular invasion (40), disease progression (35), higher recurrence rate (33), and shorter disease-free survival (DFS) and overall survival (OS) (31). Prospective research on 135 HCC patients demonstrated that surgical resection of tumors reduced the quantity of CTC. They suggest that the postoperative CTC continued to be at a higher level (≥5), which has a better predictive validity for prognosis than AFP>400cmg/L and tumor diameter>5cm (43).
However, EpCAM was positive in less than 20% of HCC cases (50), and some of them also develop into EMT. This will reduce the detection rate of EpCAM+CTCs and limit its clinical application. The Canpatrol™ CTC analysis platform was developed for EMT, which uses microfiltration and various markers to characterize CTCs. This technology not only uses epithelial markers (EpCAM, CK8/9/19) but also adds mesenchymal markers (Vimentin and Twist) (51). Qi et al. (32) used the CanPatrol™ CTC-enrichment technique in 112 HCC patients with a positive rate of more than 90%, even for early-stage diseases. Before resection, the number of CTCs≥16 and the percentage of mesenchymal CTCs≥2% were greatly correlated with intrahepatic recurrence and distant lung metastasis. Some negative enrichment methods and combinations of different markers or technologies of CTCs have also been established to optimize the platform (29, 46, 52).
Except for the number of preoperative CTCs, the forms of CTCs also influenced the prognosis of HCC patients. Mesenchymal CTCs have more invasion and metastatic potential. Bai et al. (53) showed that high expression of CXCR4 protein in mixed CTCs was more common, which might be associated with the progression and metastasis. Another study found that the proportion of mixed and mesenchymal CTCs was also a prognostic indicator of HCC (37). In a prospective study, Sun et al (40). examined CTCs at five important sites of blood vessels in HCC patients, and showed that the presence of multi-vascular CTC cluster could prognosticate recurrence and metastasis. The CTC clusters are also been reported in some studies. Gkountela et al. (54) showed that the specific hypermethylation of the binding sites for transcription factors related to stemness and proliferation in the CTC cluster promoted metastasis. The CTC-associated white blood cell clusters of peripheral blood in patients with HCC were also related to DFS and OS (44).
Tumor Monitoring And Guiding Personalized Therapy
CTCs may serve as an index for long-term monitoring of HCC, and its dynamic changes reflected the therapeutic response treated with locoregional therapies, such as transcatheter arterial chemoembolization (TACE), radiotherapy, and radiofrequency ablation (55–57). Rau et al. (46) used a negative selection method for the enrichment of CTCs and found that the analyzes of CTCs at a different time during the treatment is helpful to dynamically monitor the progression of HCC patients, especially those without elevated serum AFP levels. Fan et al. (58) investigated a new technique which was combined flow cytometry in vivo with orthotopic tumor models and showed that the number of CTCs decreased significantly after resection and early metastases were also reduced. The tumor size and the number of distant metastases of HCC were in accord with the dynamic change of CTCs. Moreover, Zhang et al. (59) used microfluidic chip technology to obtain CTCs and discovered that the number of spheroids formed by CTCs substantially reduced after treated with sorafenib or oxaliplatin. This reflects the potential of CTCs in the analysis of sensitivity and drug resistance of chemotherapeutic drugs.
Identifying CTCs that express specific tumor markers and drug targets enable doctors to better guide personalized therapy. In 2016, Li et al. (47) indicated that CTCs can replace tumor tissue to characterize the expression of pERK/pAkt. The pERK+/pAkt- CTCs were most sensitive to sorafenib, which is helpful to choose the appropriate treatment scheme. In recent years, immunotherapy is a research hotspot, and immune checkpoint inhibitors, including Programmed cell death protein-1 (PD-1) and programmed death-ligand 1 (PD-L1), are promising in the treatment of advanced HCC (60). Winograd et al. (61) firstly evaluated PD-L1+ CTCs in HCC. They reported that PD-L1+ CTCs could serve as a predictor of immunotherapy. Because 3 of all 6 patients who received anti-PD1 therapy showed responses to treatment and had PD-L1+ CTCs (61). In 2020, the same team conducted a larger, prospective cohort to enumerate/phenotype CTCs. They showed that PD-L1+ CTCs mainly present in advanced HCC, and related to beneficial therapeutic responses of patients with HCC who received anti-PD-1 therapy (62). The expression of PD-L1 varies with immune statuses, treatment response, and disease progression. Precise assessment of PD-L1 expression in CTCs could be used to evaluate and monitor the immune status of tumor cells in real-time.
Circulating Tumor DNA (ctDNA)
When it comes to ctDNA, it is inevitable to mention circulating cell-free DNA(cfDNA), that derives from lymphocytes or dying benign host cells (63). It is generally believed that ctDNA is the fragmented DNA shedding from necrotic and apoptotic tumor cells, occupying only a small portion of total cfDNA (14, 64). The mechanism of how ctDNA is released into the bloodstream is still unclear. It might be associated with apoptosis and necrosis (65), and exposure to intermittent hypoxia is likely to promote ctDNA fall off into the circulation (66). At present, there are still some difficulties in separating ctDNA from cfDNA using the existing technology. Methylation of ctDNA and cfDNA is research hotspots (14). DNA methylation is involved in the epigenetic regulation of gene expression and often leads to gene silencing. The ctDNA has the molecular characteristics of methylation changes, and it has been found that the tumor DNA methylation profiles of HCC are highly correlated with the paired plasma ctDNA (67). Methylation changes in ctDNA usually occur early in carcinogenesis (14), therefore, the detection of methylation genes in ctDNA has a certain clinical potential in HCC (67–69). Moreover, ctDNA comprises a complete tumor genome, including variants derived from a plurality of independent tumors, so ctDNA has a greater advantage in overcoming tumor heterogeneity than single tissue biopsy.
Since only 10 ng cfDNA can be extracted per milliliter of blood (70), the detection method of ctDNA should be highly sensitive and specific. Various methods can be selected according to different detection purposes. The ctDNA carries tumor-specific information in terms of genetic or epigenetic alterations, like methylation changes, single-nucleotide variants (SNVs), and copy number variations (CNVs). There are two types of analytical techniques based on ctDNA: quantitative detection (measure the number of ctDNA) and qualitative detection (detect tumor-specific genetic aberration). The length of ctDNA fragments is less than 167 base pairs, approximately the size of 1 nucleosome (71). Moreover, the half-life of ctDNA is short, usually, no more than 2 hours, which can relatively accurately reflect the real-time change of the tumor burden during cancer therapy. The ctDNA carries the same genetic mutation as the primitive tumor cell, thus the qualitative and quantitative analysis of ctDNA is mainly based on detecting the aberrations in cfDNA. Digital PCR (dPCR) and NGS are two popular methods nowadays techniques to detect ctDNA.
Clinical Application OF ctDNA IN HCC
The ctDNA contains genomes derived from multiple independent tumors (72), thus, detecting ctDNA is expected to overcome temporal and spatial heterogeneity of tumor tissues. We summarized the clinical application of ctDNA in patients with HCC ( Table 2 ).
Table 2.
Clinical applications of ctDNA in HCC.
| Region | Patient | Target site | Methods | Function of the gene | Ref. |
|---|---|---|---|---|---|
| Early detection and prognosis | |||||
| China | 37 HCC 33 healthy | DBX2, THY1 | Targeted bisulfite sequencing | Hypermethylation of DBX2, THY1 may result in HCC development | (73) |
| Hong Kong | 26 HCC 32 healthy | Hypomethylation, CNAs | Massively parallel bisulfite sequencing | / | (74) |
| United States | 66 HCC 43 benign chronic liver diseases | INK4A | Pyrosequencing and MSP | Promoter hypermethylation of INK4A leads to loss of p16 expression | (75) |
| China | 121 HCC 37 chronic hepatitis B 31 healthy | MT1M, MT1G promoter | MSP | Methylation of MT1M and MT1G promoters is associated with vascular invasion or metastasis | (76) |
| China | 100 HCC 29 healthy | HOXA9 | MSP, bisulfite sequencing, and Q-MSP | Hypermethylation of HOXA9 may be present in precancerous lesion during carcinogenesis | (77) |
| China | 1098 HCC 835 healthy | BMPR1A, PSD, ARHGAP25, KLF3, PLAC8, ATXN1, Chr 6:170, Chr 6:3, ATAD2, Chr 8:20 | Targeted bisulfite sequencing | / | (67) |
| Taiwan | 180 HCC | APC, COX2, RASSF1A miRNA | qMSP | Hypermethylation of RASSF1A suggests the early stage of HCC. Hypermethylation of APC and COX2 is associated with liver carcinogenesis | (78) |
| Taiwan | 237 HCC 257 controls | TBX2 | Pyrosequencing assay, Real-time PCR | Hypermethylation of TBX2 is associated with increased HCC risk | (79) |
| France and Germany | 98 HCC 191 cirrhosis | SEPT9 | MSP | Hypermethylation of SEPT9 is associated with liver carcinogenesis | (68) |
| United States | 116 HCC 81cirrhosis 98 healthy | HOXA1, EMX1, AK055957, ECE1, PFKP, CLEC11A | qMSP | / | (69) |
| China | 1204 HCC 392 CH/cirrhosis 958 healthy | 5hmC modifications | 5hmC-Seal technique | Serve as ideal markers for specific gene/locus activation in chromatin | (80) |
| China | 508 HCC 2250 cirrhosis 476 healthy | 5-hmc, NF, 5′end motif, fragmentation | NGS | / | (81) |
| ctDNA mutation | |||||
| United States | 66 HCC 35 cirrhosis 41HCV-related chronic hepatitis | hTERT | real-time PCR | The amount of hTERT gene in plasma served as serves as a surrogate of cfDNA | (82) |
| China | 48 HCC | TP53, CTNNB1, TERT | Droplet digital PCR | Mutation of TP53 and CTNNB1 suggests the occurrence of HCC; TERT promoter mutation is an early event in liver carcinogenesis; | (83) |
| China | 41 HCC | TERT, CTNNB1, TP53 | MiSeq sequencing | Mutation of TP53 and CTNNB1 suggests the occurrence of HCC; TERT promoter mutation is an early event in liver carcinogenesis; | (84) |
| China | 65 HCC 70 non- HCC 331 at risk patients | TP53, CTNNB1, AXIN1, the TERT promoter, HBV insertion site, AFP, DCP | HCCscreen | AXIN1 mutation is associated with HCC | (85) |
| China | 384 HCC | SCNA | WGS | / | (86) |
| Monitoring and guide therapy | |||||
| China | 34 HCC | SNVs, CNVs | Target sequencing Whole exome sequencing | / | (87) |
| United States | 14 HCC | TP53, CTNNB1, PTEN, CDKN2A, ARID1A, MET; CDK6, EGFR, MYC, BRAF, RAF1, FGFR1, CCNE1, PIK3CA, ERBB2/HER2 | NGS | / | (88) |
| United States | 26 HCC | TP53, CTNNB1, ARID1A | NGS | Mutations of TP53, CTNNB1 and ARID1A are associated with treatment response | (89) |
Q-MSP, Quantitative methylation-specific PCR; MSP, methylation-specific PCR; qMSP, real-time quantitative methylation-specific PCR; WGS, whole-genome sequencing; NGS, next-generation sequencing; SNVs, single nucleotide variants; CNVs, copy-number variants; SCNA, somatic copy number aberration.
Early Diagnosis And Prognostic Evaluation
Epigenetic changes induced by DNA methylation and DNA methylation are involved in the process of tumor occurrence and development (90). The methylation pattern of ctDNA has tremendous potential in the early diagnosis of HCC patients. Changes in methylation of multiple genes in plasma/serum, such as p15, p16, GSTP1, INK4A, RASSF1A, and so on, have been confirmed in many studies to distinguish HCC from controls. The methylation characteristics of tumor DNA in HCC are highly correlated with paired plasma ctDNA (67), so some biomarkers of DNA methylation in HCC may also be used in ctDNA. Kotoh et al. (91) developed a methylated SEPT9 assay in HCC, with 63.2% sensitivity and 90.0% specificity. They pointed out that combined diagnosis with AFP can improve the diagnosis rate of early-stage HCC. Yan et al. (92) proposed the HCC index, which was a combined diagnostic model of cfDNA, age, and AFP. The HCC index was more accurate in HCC diagnosis than cfDNA or AFP, alone. Currently, Chen et al. (81) conducted large-scale, multi-center research and constructed a diagnostic model based on HIFI (5-Hydroxymethylcytosine/motIf/Fragmentation/nucleosome footprInt) method, playing a key role in differentiating HCC from non-HCC. Both the test set and the verification set had a sensitivity and specificity of more than 95%.
In addition to the role of diagnosis, ctDNA methylation can also be served as a prognostic indicator. The study of Kotoh et al. also showed that the copy number of methylated SEPT9 was related to BCLC stage, macrovascular invasion, tumor number, and size (91). Li et al. (93) indicated that IGFBP7 promoter methylation was significantly related to OS and early tumor recurrence after hepatectomy. In a large-scale study involving 1098 HCC patients, a diagnostic prediction model was constructed by screening 10 overlapping markers of cfDNA methylation using Random Forest and Least Absolute Shrinkage and Selection Operator (LASSO) methods. The diagnostic sensitivity and specificity of this model were 85.7% and 94.3%, respectively (67). Kisiel et al. (69) identified 6 best-methylated DNA markers (MDMs) in HCC, that include ECE1, HOXA1, CLEC11A, AK055957, PFKP and EMX1, and combined Phase I Pilot, and Phase II Clinical Validation. In the diagnosis of HCC, the area-under-the-receiver-operating-curve (AUC) of this 6-marker MDM panel was 0.96, the sensitivity was 95%, and the specificity was 92%. Moreover, elevated levels of cfDNA are a positive correlation with poor prognosis. The existence and quantity of ctDNA are usually determined by detecting the mutation of cfDNA. Some hot mutants, such as TP53, CTNNB1, and TERT, are usually selected to detect ctDNA aberrations (14). Ren N et al. (94) found that allelic imbalance at D8S258 in circulating plasma DNA was also related to the poor prognosis of HCC. A recent study focused on SNVs of ctDNA and showed that the existence of MLH1 SNV, coupled with elevated ctDNA levels, can predict poor OS of HCC patients (95). In short, the different mutants and epigenetic modifications in ctDNA have significance in early diagnosis and prognostic outcomes of HCC.
Tumor Monitoring and Guiding Personalized Therapy
Considering that liver biopsy is invasive and unnecessary in advanced HCC, ctDNA can be a reliable biomarker for dynamically monitoring tumor progression and assessing the treatment efficacy. These include identifying new mutations that drive acquired drug resistance and capturing heterogeneity between tumors. According to the study of Park et al. (96), the high level of cfDNA after radiotherapy was related to the poor outcome of treatment. Thus, ctDNA can be used as an indicator to evaluate the curative effect after radiotherapy. Recently, Zhao et al. (97) prospectively enrolled 42 patients with unresectable liver cancer. They found that TP53 mutation was related to disease progression and interventional treatment was more effective in patients without TP53 mutation. The follow-up study showed that plasma levels of CNVs and SNVs in ctDNA dynamically correlated with patients’ tumor burden in HCC (80). They decreased after surgery and increased in cases with tumor recurrence, showing that ctDNA was a feasible biomarker to monitor treatment response (80). Cai et al. (87) collected the information of postoperative ctDNA and protein biomarkers which included AFP, AFP-L3, and des-gamma-carboxy prothrombin (DCP), then evaluated the results with corresponding MRI scan images during follow-up. They confirmed that both SNVs and CNVs of ctDNA could dynamically monitor the tumor load of HCC. The combination of ctDNA and DCP could improve the detection rate of minimal/molecular residual disease (MRD) in patients undergoing hepatectomy (87).
On other hand, the detection of ctDNA mutations can guide the choice of treatment. Ikeda et al. (98) used digital ctDNA sequencing to evaluate the mutant associated with wild-type allele fraction in 14 advanced HCC patients. The level of DCP and AFP decreased with the treatment of palbociclib (CDK4/6 inhibitor) and celecoxib (COX-2/Wnt inhibitor) after two months in a patient with CDKN2A-inactivating and CTNNB1-activating mutation. And AFP declined by 63% in another patient treated with sirolimus (mechanistic target of rapamycin inhibitor) and cabozantinib (MET inhibitor). This patient had PTEN-inactivating and MET-activating mutations of ctDNA (98). Compared with tissue biopsies, cfDNA identified clinically related drug resistance changes more frequently in a prospective cohort study of patients with gastrointestinal tumors. In 78% of cases, cfDNA could detect drug resistance gene mutations that were not found in matched tumor biopsies (99). A recent study reported that the sequential mutation profiling of ctDNA can be used for molecular detection of drug resistance in HCC (100). The gene mutation of PI3K/MTOR pathway was associated with the worse PFS in HCC patients receiving tyrosine kinase inhibitors but was not associated with immunosuppression therapy (100). It shows that monitoring the mutation of the drug target gene or drug resistance gene in advanced patients can better guide the choice of personalized treatment. All in all, genomic profiles of patients with HCC can be obtained from ctDNA, which can guide the treatment to some extent.
Extracellular Vesicles (EVs): Exosomes
Exosomes were thought to originate from mature sheep reticulocytes (101). Exosome belongs to extracellular vesicles (EVs), which is a nano-sized phospholipid bilayer membrane vesicle and responsible for intercell communication (102). It is secreted by living cells and formed by the separation of intracellular poly vesicles with cell membranes in the process of being released out of the cell (103). The component transported in exosomes contains several molecular biomarkers including proteins, RNA, DNA, which range in size from 50 to 140 nm (88, 104). Currently studies suggest that the cargos carried by exosomes associated with HCC, including non-coding RNAs(ncRNAs), messenger RNAs (mRNAs), and proteins, can serve as potential biomarkers for the clinical application of HCC (105). They transferred to target cells by exosomes, affecting drug resistance, tumor angiogenesis and metastasis (106–108). Exosomes show significant superiority in liquid biopsy. Exosomal substances are hard to be degraded due to the protection of phospholipid bilayer membrane (15). Its high biological stability improves the clinical applicability of exosomes, which can not only cut down the cost of short-term sample preservation but also reduce the challenges of transportation (109). Moreover, exosomes carry biological information from parental cells, so they are more typically than cfDNA (110). Exosomes are bound up with the growth and metastasis of HCC (108), tumor angiogenesis (111), and immune regulation (112).
The capture and enrichment of exosomes require the identification of their markers, such as heat shock protein 70 (HSP 70), CD9, CD63, CD81, and ALIX (113). There are some common technologies used for exosomes isolation according to their composition or physical properties, such as ultracentrifugation (UC), transmission electron microscopy (TEM), magnetic associated cell sorting (MACS), filtration, polymer-based precipitation, nanoparticle tracking analysis, fluorescence, colorimetric ELISA assays, and size exclusion chromatography (SEC). However, the eventual clinical utility of exosomes is still in its preliminary stages and needs more validation.
Clinical Application OF Exosome IN HCC
Exosomes are related to the establishment of the tumor microenvironment and participate in the occurrence, development of HCC (114). Substantial researches have demonstrated that exosomes play a part in clinical applications of HCC over the years ( Table 3 ).
Table 3.
Clinical applications of exosomes in HCC.
| Region | Patients | Method | Target | Function of the Cargo | Reference |
|---|---|---|---|---|---|
| Korea | 20 HCC 20 CH 20 cirrhosis | Ultracentrifugation | miR-18a, miR-101, miR-106b, miR-122, miR-195, miR-221, miR-222, miR-224 | / | (115) |
| Korea | 84 HCC 26 CH 32 cirrhosis 26 healthy | ExoQuick™ Exosome Precipitation Solution | The panel based on miR-4661-5p | Immune escape | (116, 117) |
| Korea | 90 HCC 60 CLD 28 healthy | Ultracentrifugation | miR-10b-5p, miR-215-5p | Invasion and metastasis of HCC | (118) |
| Spain | 29 HCC 32 healthy 43 CCA 30 PSC | Filtration, Ultracentrifugation, | FIBG | unknow | (119) |
| China | 74 HCC 26 Cirrhosis 34 CHB 72 healthy | ExoQuick™ Exosome Precipitation Solution | lncRNAs X-inactive-specific transcript | Regulate proliferation and metastasis of HCC | (120) |
| China | 115 HCC 156 CHB 85 LC 120 healthy | Total Exosome Isolation Kit | ENSG00000258332.1, LINC00635, AFP | Regulate proliferation and metastasis of HCC | (121) |
| China | 29 HCC 37 healthy and benign hepatomas | ExoQuick™ Exosome Precipitation Solution | SMAD3 | Promoted cell adhesion | (122) |
| China | 50 HCC 40 cirrhosis | Ultracentrifugation, filtration, and precipitation | A panel combining miR-122, miR-148a, and AFP | Inhibit proliferation and multidrug resistance | (123) |
| Korea | 79 HCC | ExoQuick™ Exosome Precipitation Solution | miRNA-21 lncRNA-ATB | Regulate proliferation, invasion and metastasis of HCC | (124) |
| China | 82 HCC 47 healthy | ExoQuick-TC exosome precipitation solution | circPTGR1 | Promote metastasis of HCC | (125) |
| China | 240 HCC | ExoQuick™ Exosome Precipitation Solution | circUHRF1 | Immune escape | (126) |
| China | 71 HCC 40 HD | Ultracentrifugation | A panel combining circ_0004001, circ_0004123, and circ_0075792 | Regulate the proliferation, migration, and invasion of HCC | (127) |
| Korea | 32 HCC 28CH 35 cirrhosis | ExoQuick™ Exosome Precipitation Solution | LINC00853 | unknow | (128) |
| China | 122 HCC 43 cirrhosis | Ribo™ Exosome Isolation Reagent | Lnc85 | Inhibit proliferation and migration of HCC | (129) |
| India | 38 HCC 35 CH 25 cirrhosis | ExoEnrich™ instant exosome isolation kit and immunoaffinity capture (anti-ASGR2) | A panel combining miR-10b-5p, miR-221-3p, miR-223-3p, and miR-21-5p | / | (130) |
CCA, Cholangiocarcinoma; PSC, primary sclerosing cholangitis; CHB, chronic hepatitis B; LC, liver cirrhosis.
Early Diagnosis and Prognostic Evaluation
Exosomes contain bioactive compounds, such as RNA, DNA, proteins, and cholesterol (15), some of which can be expressed uniquely by tumor cells. MicroRNAs (miRNAs), lncRNA and circular RNAs (circRNA) have been described in many reports as noninvasive biomarker for better prediction and prognosis of HCC progression. The miRNAs, a class of conserved RNAs (usually 22-25 nucleotides), can inhibit the translation of mRNA or promote the degradation of mRNA by combining with their response element on the 3-untranslated region of the target mRNA (107). Exosomes can actively secrete miRNAs to regulate the progress of HCC (15, 131). Sohn et al (115) found that serum exosomal miR-222, miR-221, and miR-18a in patients with chronic hepatitis or cirrhosis were significantly lower than that in HCC patients. Wang et al. (132) showed that the serum exosomal miR-21 also elevated in HCC patients, which could be used to distinguish HCC from people and chronic hepatitis B patients. Ghosh et al. (130) found that the combination of four miRNAs (miR-221-3p, miR-223-3p, miR-10b-5p and miR-21-5p) showed good diagnostic ability in patients with low expression of AFP. Currently, Cho et al (116) showed that serum exo-miR-10b-5p had great potential for early diagnosis of HCC with a sensitivity of 90.7%, specificity of 75.0%, and the AUC was 0.934. The same team developed a panel that included exo-miR-4746-5p and exo-miR-4661-5p for the early diagnosis of HCC. The sensitivity, specificity, and AUC were 81.8%, 91.7%, and 0.947 respectively (118). Along with the miRNA, lncRNA and cirRNA in exosomes have also shown potential in the early detection of HCC. The expression of LINC00853, lnc85, ENSG00000248932.1, ENST00000440688.1 and ENST00000457302.2, have shown promise for the tumorigenesis prediction (128, 129, 133). Xu et al. (121) suggested that AFP combined with serum exosomal LINC00635 and ENSG00000258332.1 could discriminate HCC from chronic hepatitis B, gaining an AUC of 0.894. By comparing the level of exosomal Trna-derived small RNA (tsRNA) between the healthy and patients with liver cancer, Zhu et al. (134)found a significant increase of tsRNAs in plasma exosomes of liver cancer patients, which provided new insight into the HCC diagnostic potential of the exosome. Currently, a study suggested that the combination of three circRNAs, including circ_0004001, circ_0004123 and circ_0075792, has been served as a valuable diagnostic biomarker for HCC (127).
In addition to ncRNA, proteins are also used as biomarkers of HCC. Compared with miRNA, the study of protein in exosomes is a relatively less explored direction to detect HCC. According to a study that the proteomes of HCC, cirrhosis and healthy people have different compositions (135). Arbelaiz et al. (119) suggested that the differentially expressed exosomal proteins might be a kind of potential biomarkers for differential diagnosis. Mass spectrometry was used to detect the proteomes of exosomes in patients with HCC, primary sclerosing cholangitis, and cholangiocarcinoma. Some of these proteins including LG3BP and FIBG demonstrated superior diagnostic ability to AFP. The AUC of LG3BP for identifying HCC patients from the healthy was 0.904, and the AUC of FIBG for distinguishing intrahepatic cholangiocarcinoma from HCC was 0.894 (119). Interestingly, LG3BP and PIGR can promote the transformation, invasion, and proliferation of tumor cells, which are associated with poor prognosis (136). Fu et al. (122) found that patients with advanced HCC had higher levels of Smad3 in exosomes and suggested that combined AFP and detection of exosomes containing SMAD3 can improve the diagnosis of HCC.
Exosomes also have great potential in terms of the prognosis of HCC. Lee et al. (124) demonstrated that circulating exosomal miRNA-21 and lncRNA-ATB were associated with the T stage, the TNM stage, and portal vein thrombosis. The HCC patients who had the level of exosomal miRNA-21≥0.09 and lncRNA-ATB ≥0.0016 tend to have significantly lower OS and PFS (P<0.05). The high level of LINC00635 and ENSG00000258332.1 in HCC was related to lymph node metastasis, TNM stage, and OS (121). Some studies showed that the lower levels of exosomal miR-125b (137) and miR-638 (138) predicted poor prognosis of HCC patients. The abundance of exosomes containing SMAD3 was negatively correlated with DFS in postoperative patients of HCC (122). Moreover, circUHRF1 might be associated with resistance to anti-PD1 immunotherapy in HCC patients (126). Luo et al. (139) measured the level of exosomal circular RNA (circAKT3) from 124 patients with HCC and 100 healthy controls and found that patients with high exosomal circAKT3 tend to have a higher risk of recurrence and death.
Conclusion and Perspective
With the continuous enrichment of molecular tumor information and the frequent breakthroughs of molecular technology, precision oncology has revolutionized the field of medicine. Noninvasive liquid biopsy has competitive advantages for accurate diagnosis and individualized management (140). The detection and analysis of CTCs, ctDNA, and exosomes provide a promising strategy for early diagnosis, prognostic evaluation, the guidance of treatment, and monitoring of MRD and recurrence ( Figure 1 ). DNA methylation is a widely used epigenetic biomarker, and the analysis of ctDNA methylation has the potential of early diagnosis of high-risk patients with HCC. This may soon become an alternative method for long-term monitoring of HCC, and be a supplement to the existing clinical detection methods. The molecular characterization of ctDNA and CTCs are, at least in part, dependent on tumor burden, so they may be more useful in intermediate or advanced settings in prognosis or predicting treatment response. In general, the increase of CTCs level after treatment indicates tumor recurrence and reduced survival. Further studies of CTC and ctDNA will better understand the emergence of resistance to sorafenib or TACE. It also provides new insights into the development of more personalized diagnosis and treatment programs for HCC. The substances in exosomes, especially miRNAs, provide a new direction for improving the early diagnosis of HCC. Thus, liquid biopsies can provide researchers with more detailed and personalized information from cancer diagnosis to tumor monitoring by collecting samples continuously. However, there still stands challenges in the way that translates liquid biopsy from bench to bedside. The clinical application of liquid biopsy requires accurate biomarkers and standardized detection methods. In summary, liquid biopsy seems a convenient, noninvasive, and potential method for HCC.
Author Contributions
J-CY wrote the draft and prepared the tables and figures. J-JH collected the literature. WL designed the tables and figures. Y-XL and J-ZL revised it critically. D-WY designed this review and revised the manuscript. All authors contribute to the article and approved the submitted version.
Funding
This research was supported by National Natural Science Foundation of China (No. 81873732). This study was funded by Shanxi Province136 Revitalization Medical Project Construction Fund.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2020. CA Cancer J Clin (2020) 70:7–30. doi: 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2. Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A Global View of Hepatocellular Carcinoma: Trends, Risk, Prevention and Management. Nat Rev Gastroenterol Hepatol (2019) 16:589–604. doi: 10.1038/s41575-019-0186-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. von Felden J, Garcia-Lezana T, Schulze K, Losic B, Villanueva A. Liquid Biopsy in the Clinical Management of Hepatocellular Carcinoma. Gut (2020) 69:2025–34. doi: 10.1136/gutjnl-2019-320282 [DOI] [PubMed] [Google Scholar]
- 4. European Association for the Study of the Liver . Electronic Address E E E and L European Association for the Study of the, EASL Clinical Practice Guidelines: Management of Hepatocellular Carcinoma. J Hepatol (2018) 69:182–236. doi: 10.1016/j.jhep.2018.03.019 [DOI] [PubMed] [Google Scholar]
- 5. Bruix J, Sherman M, D American Association for the Study of Liver . Management of Hepatocellular Carcinoma: An Update. Hepatology (2011) 53:1020–2. doi: 10.1002/hep.24199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tzartzeva K, Obi J, Rich NE, Parikh ND, Marrero JA, Yopp A, et al. Surveillance Imaging and Alpha Fetoprotein for Early Detection of Hepatocellular Carcinoma in Patients With Cirrhosis: A Meta-Analysis. Gastroenterology (2018) 154:1706–18.e1701. doi: 10.1053/j.gastro.2018.01.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zamcheck N, Pusztaszeri G. CEA, AFP and Other Potential Tumor Markers. CA Cancer J Clin (1975) 25:204–14. doi: 10.3322/canjclin.25.4.204 [DOI] [PubMed] [Google Scholar]
- 8. Mann J, Reeves HL, Feldstein AE. Liquid Biopsy for Liver Diseases. Gut (2018) 67:2204–12. doi: 10.1136/gutjnl-2017-315846 [DOI] [PubMed] [Google Scholar]
- 9. Zhang Q, Rong Y, Yi K, Huang L, Chen M, Wang F. Circulating Tumor Cells in Hepatocellular Carcinoma: Single-Cell Based Analysis, Preclinical Models, and Clinical Applications. Theranostics (2020) 10:12060–71. doi: 10.7150/thno.48918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Issa IA, Noureddine M. Colorectal Cancer Screening: An Updated Review of the Available Options. World J Gastroenterol (2017) 23:5086–96. doi: 10.3748/wjg.v23.i28.5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morrison GJ, Goldkorn A. Development and Application of Liquid Biopsies in Metastatic Prostate Cancer. Curr Oncol Rep (2018) 20:35. doi: 10.1007/s11912-018-0683-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hench IB, Hench J, Tolnay M. Liquid Biopsy in Clinical Management of Breast, Lung, and Colorectal Cancer. Front Med (Lausanne) (2018) 5:9. doi: 10.3389/fmed.2018.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ahn JC, Teng PC, Chen PJ, Posadas E, Tseng HR, Lu SC, et al. Detection of Circulating Tumor Cells and Their Implications as a Novel Biomarker for Diagnosis, Prognostication, and Therapeutic Monitoring in Hepatocellular Carcinoma. Hepatology (2020) 73:422–36. doi: 10.1002/hep.31165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ye Q, Ling S, Zheng S, Xu X. Liquid Biopsy in Hepatocellular Carcinoma: Circulating Tumor Cells and Circulating Tumor DNA. Mol Cancer (2019) 18:114. doi: 10.1186/s12943-019-1043-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li X, Li C, Zhang L, Wu M, Cao K, Jiang F, et al. The Significance of Exosomes in the Development and Treatment of Hepatocellular Carcinoma. Mol Cancer (2020) 19:1. doi: 10.1186/s12943-019-1085-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li J, Han X, Yu X, Xu Z, Yang G, Liu B, et al. Clinical Applications of Liquid Biopsy as Prognostic and Predictive Biomarkers in Hepatocellular Carcinoma: Circulating Tumor Cells and Circulating Tumor DNA. J Exp Clin Cancer Res (2018) 37:213. doi: 10.1186/s13046-018-0893-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pantel K, Speicher MR. The Biology of Circulating Tumor Cells. Oncogene (2016) 35:1216–24. doi: 10.1038/onc.2015.192 [DOI] [PubMed] [Google Scholar]
- 18. Kleiner DE, Stetler-Stevenson WG. Matrix Metalloproteinases and Metastasis. Cancer Chemother Pharmacol (1999) 43(Suppl):S42–51. doi: 10.1007/s002800051097 [DOI] [PubMed] [Google Scholar]
- 19. Miller MC, Doyle GV, Terstappen LW. Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients With Metastatic Breast Colorectal and Prostate Cancer. J Oncol (2010) 2010:617421. doi: 10.1155/2010/617421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Massague J, Obenauf AC. Metastatic Colonization by Circulating Tumour Cells. Nature (2016) 529:298–306. doi: 10.1038/nature17038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mohme M, Riethdorf S, Pantel K. Circulating and Disseminated Tumour Cells - Mechanisms of Immune Surveillance and Escape. Nat Rev Clin Oncol (2017) 14:155–67. doi: 10.1038/nrclinonc.2016.144 [DOI] [PubMed] [Google Scholar]
- 22. Thompson EW, Haviv I. The Social Aspects of EMT-MET Plasticity. Nat Med (2011) 17:1048–9. doi: 10.1038/nm.2437 [DOI] [PubMed] [Google Scholar]
- 23. Liu X, Li J, Cadilha BL, Markota A, Voigt C, Huang Z, et al. Epithelial-Type Systemic Breast Carcinoma Cells With a Restricted Mesenchymal Transition are a Major Source of Metastasis. Sci Adv (2019) 5:eaav4275. doi: 10.1126/sciadv.aav4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pantel K, Alix-Panabieres C. Circulating Tumour Cells in Cancer Patients: Challenges and Perspectives. Trends Mol Med (2010) 16:398–406. doi: 10.1016/j.molmed.2010.07.001 [DOI] [PubMed] [Google Scholar]
- 25. Li YM, Xu SC, Li J, Han KQ, Pi HF, Zheng L, et al. Epithelial-Mesenchymal Transition Markers Expressed in Circulating Tumor Cells in Hepatocellular Carcinoma Patients With Different Stages of Disease. Cell Death Dis (2013) 4:e831. doi: 10.1038/cddis.2013.347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cheung KJ, Ewald AJ. A Collective Route to Metastasis: Seeding by Tumor Cell Clusters. Science (2016) 352:167–9. doi: 10.1126/science.aaf6546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun YF, Xu Y, Yang XR, Guo W, Zhang X, Qiu SJ, et al. Circulating Stem Cell-Like Epithelial Cell Adhesion Molecule-Positive Tumor Cells Indicate Poor Prognosis of Hepatocellular Carcinoma After Curative Resection. Hepatology (2013) 57:1458–68. doi: 10.1002/hep.26151 [DOI] [PubMed] [Google Scholar]
- 28. Zhou Y, Wang B, Wu J, Zhang C, Zhou Y, Yang X, et al. Association of Preoperative EpCAM Circulating Tumor Cells and Peripheral Treg Cell Levels With Early Recurrence of Hepatocellular Carcinoma Following Radical Hepatic Resection. BMC Cancer (2016) 16:506. doi: 10.1186/s12885-016-2526-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ogle LF, Orr JG, Willoughby CE, Hutton C, McPherson S, Plummer R, et al. Imagestream Detection and Characterisation of Circulating Tumour Cells - A Liquid Biopsy for Hepatocellular Carcinoma? J Hepatol (2016) 65:305–13. doi: 10.1016/j.jhep.2016.04.014 [DOI] [PubMed] [Google Scholar]
- 30. Shi J, Li Y, Liang S, Zeng J, Liu G, Mu F, et al. Circulating Tumour Cells as Biomarkers for Evaluating Cryosurgery on Unresectable Hepatocellular Carcinoma. Oncol Rep (2016) 36:1845–51. doi: 10.3892/or.2016.5050 [DOI] [PubMed] [Google Scholar]
- 31. Yu JJ, Xiao W, Dong SL, Liang HF, Zhang ZW, Zhang BX, et al. Effect of Surgical Liver Resection on Circulating Tumor Cells in Patients With Hepatocellular Carcinoma. BMC Cancer (2018) 18:835. doi: 10.1186/s12885-018-4744-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qi LN, Xiang BD, Wu FX, Ye JZ, Zhong JH, Wang YY, et al. Circulating Tumor Cells Undergoing EMT Provide a Metric for Diagnosis and Prognosis of Patients With Hepatocellular Carcinoma. Cancer Res (2018) 78:4731–44. doi: 10.1158/0008-5472.CAN-17-2459 [DOI] [PubMed] [Google Scholar]
- 33. von Felden J, Schulze K, Krech T, Ewald F, Nashan B, Pantel K, et al. Circulating Tumor Cells as Liquid Biomarker for High HCC Recurrence Risk After Curative Liver Resection. Oncotarget (2017) 8:89978–87. doi: 10.18632/oncotarget.21208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shen J, Wang WS, Zhu XL, Ni CF. High Epithelial Cell Adhesion Molecule-Positive Circulating Tumor Cell Count Predicts Poor Survival of Patients With Unresectable Hepatocellular Carcinoma Treated With Transcatheter Arterial Chemoembolization. J Vasc Interv Radiol (2018) 29:1678–84. doi: 10.1016/j.jvir.2018.07.030 [DOI] [PubMed] [Google Scholar]
- 35. Kelley RK, Magbanua MJ, Butler TM, Collisson EA, Hwang J, Sidiropoulos N, et al. Circulating Tumor Cells in Hepatocellular Carcinoma: A Pilot Study of Detection, Enumeration, and Next-Generation Sequencing in Cases and Controls. BMC Cancer (2015) 15:206. doi: 10.1186/s12885-015-1195-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang S, Zhang C, Wang G, Cheng B, Wang Y, Chen F, et al. Aptamer-Mediated Transparent-Biocompatible Nanostructured Surfaces for Hepotocellular Circulating Tumor Cells Enrichment. Theranostics (2016) 6:1877–86. doi: 10.7150/thno.15284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen J, Cao SW, Cai Z, Zheng L, Wang Q. Epithelial-Mesenchymal Transition Phenotypes of Circulating Tumor Cells Correlate With the Clinical Stages and Cancer Metastasis in Hepatocellular Carcinoma Patients. Cancer Biomark (2017) 20:487–98. doi: 10.3233/CBM-170315 [DOI] [PubMed] [Google Scholar]
- 38. Schulze K, Gasch C, Staufer K, Nashan B, Lohse AW, Pantel K, et al. Presence of EpCAM-Positive Circulating Tumor Cells as Biomarker for Systemic Disease Strongly Correlates to Survival in Patients With Hepatocellular Carcinoma. Int J Cancer (2013) 133:2165–71. doi: 10.1002/ijc.28230 [DOI] [PubMed] [Google Scholar]
- 39. Guo W, Yang XR, Sun YF, Shen MN, Ma XL, Wu J, et al. Clinical Significance of EpCAM mRNA-Positive Circulating Tumor Cells in Hepatocellular Carcinoma by an Optimized Negative Enrichment and qRT-PCR-Based Platform. Clin Cancer Res (2014) 20:4794–805. doi: 10.1158/1078-0432.CCR-14-0251 [DOI] [PubMed] [Google Scholar]
- 40. Sun YF, Guo W, Xu Y, Shi YH, Gong ZJ, Ji Y, et al. Circulating Tumor Cells From Different Vascular Sites Exhibit Spatial Heterogeneity in Epithelial and Mesenchymal Composition and Distinct Clinical Significance in Hepatocellular Carcinoma. Clin Cancer Res (2018) 24:547–59. doi: 10.1158/1078-0432.CCR-17-1063 [DOI] [PubMed] [Google Scholar]
- 41. Wang L, Li Y, Xu J, Zhang A, Wang X, Tang R, et al. Quantified Postsurgical Small Cell Size CTCs and EpCAM(+) Circulating Tumor Stem Cells With Cytogenetic Abnormalities in Hepatocellular Carcinoma Patients Determine Cancer Relapse. Cancer Lett (2018) 412:99–107. doi: 10.1016/j.canlet.2017.10.004 [DOI] [PubMed] [Google Scholar]
- 42. Ha Y, Kim TH, Shim JE, Yoon S, Jun MJ, Cho YH, et al. Circulating Tumor Cells are Associated With Poor Outcomes in Early-Stage Hepatocellular Carcinoma: A Prospective Study. Hepatol Int (2019) 13:726–35. doi: 10.1007/s12072-019-09994-9 [DOI] [PubMed] [Google Scholar]
- 43. Zhou J, Zhang Z, Zhou H, Leng C, Hou B, Zhou C, et al. Preoperative Circulating Tumor Cells to Predict Microvascular Invasion and Dynamical Detection Indicate the Prognosis of Hepatocellular Carcinoma. BMC Cancer (2020) 20:1047. doi: 10.1186/s12885-020-07488-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Luo Q, Wang C, Peng B, Pu X, Cai L, Liao H, et al. Circulating Tumor-Cell-Associated White Blood Cell Clusters in Peripheral Blood Indicate Poor Prognosis in Patients With Hepatocellular Carcinoma. Front Oncol (2020) 10:1758. doi: 10.3389/fonc.2020.01758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Qi LN, Ma L, Chen YY, Chen ZS, Zhong JH, Gong WF, et al. Outcomes of Anatomical Versus non-Anatomical Resection for Hepatocellular Carcinoma According to Circulating Tumour-Cell Status. Ann Med (2020) 52:21–31. doi: 10.1080/07853890.2019.1709655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rau K-M, Liu C-T, Hsiao Y-C, Hsiao K-Y, Wang T-M, Hung W-S, et al. Sequential Circulating Tumor Cell Counts in Patients With Locally Advanced or Metastatic Hepatocellular Carcinoma: Monitoring the Treatment Response. J Clin Med (2020) 9:188. doi: 10.3390/jcm9010188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li J, Shi L, Zhang X, Sun B, Yang Y, Ge N, et al. pERK/Pakt Phenotyping in Circulating Tumor Cells as a Biomarker for Sorafenib Efficacy in Patients With Advanced Hepatocellular Carcinoma. Oncotarget (2016) 7:2646–59. doi: 10.18632/oncotarget.6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. D’Avola D, Villacorta-Martin C, Martins-Filho SN, Craig A, Labgaa I, von Felden J, et al. High-Density Single Cell mRNA Sequencing to Characterize Circulating Tumor Cells in Hepatocellular Carcinoma. Sci Rep (2018) 8:11570. doi: 10.1038/s41598-018-30047-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sun C, Liao W, Deng Z, Li E, Feng Q, Lei J, et al. The Diagnostic Value of Assays for Circulating Tumor Cells in Hepatocellular Carcinoma: A Meta-Analysis. Med (Baltimore) (2017) 96:e7513. doi: 10.1097/MD.0000000000007513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Went PT, Lugli A, Meier S, Bundi M, Mirlacher M, Sauter G, et al. Frequent EpCam Protein Expression in Human Carcinomas. Hum Pathol (2004) 35:122–8. doi: 10.1016/j.humpath.2003.08.026 [DOI] [PubMed] [Google Scholar]
- 51. Wang Z, Luo L, Cheng Y, He G, Peng B, Gao Y, et al. Correlation Between Postoperative Early Recurrence of Hepatocellular Carcinoma and Mesenchymal Circulating Tumor Cells in Peripheral Blood. J Gastrointest Surg (2018) 22:633–9. doi: 10.1007/s11605-017-3619-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hamaoka M, Kobayashi T, Tanaka Y, Mashima H, Ohdan H. Clinical Significance of Glypican-3-Positive Circulating Tumor Cells of Hepatocellular Carcinoma Patients: A Prospective Study. PloS One (2019) 14.e0217586. doi: 10.1371/journal.pone.0217586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bai T, Mai R, Ye J, Chen J, Qi L, Tang J, et al. Circulating Tumor Cells and CXCR4 in the Prognosis of Hepatocellular Carcinoma. Trans Cancer Res (2020) 9:1384–94. doi: 10.21037/tcr.2020.01.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gkountela S, Castro-Giner F, Szczerba BM, Vetter M, Landin J, Scherrer R, et al. Circulating Tumor Cell Clustering Shapes DNA Methylation to Enable Metastasis Seeding. Cell (2019) 176:98–112.e114. doi: 10.1016/j.cell.2018.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu X, Yang C, Yu H, Cao F, Shan Y, Zhao W. The Predictive Values of Serum Dickkopf-1 and Circulating Tumor Cells in Evaluating the Efficacy of Transcatheter Arterial Chemoembolization Treatment on Hepatocellular Carcinoma. Med (Baltimore) (2019) 98:e16579. doi: 10.1097/MD.0000000000016579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li Y, Huang N, Wang C, Ma H, Zhou M, Lin L, et al. Impact of Liver Tumor Percutaneous Radiofrequency Ablation on Circulating Tumor Cells. Oncol Lett (2018) 16:2839–50. doi: 10.3892/ol.2018.9019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Guo W, Sun YF, Shen MN, Ma XL, Wu J, Zhang CY, et al. Circulating Tumor Cells With Stem-Like Phenotypes for Diagnosis, Prognosis, and Therapeutic Response Evaluation in Hepatocellular Carcinoma. Clin Cancer Res (2018) 24:2203–13. doi: 10.1158/1078-0432.CCR-17-1753 [DOI] [PubMed] [Google Scholar]
- 58. Fan ZC, Yan J, Liu GD, Tan XY, Weng XF, Wu WZ, et al. Real-Time Monitoring of Rare Circulating Hepatocellular Carcinoma Cells in an Orthotopic Model by In Vivo Flow Cytometry Assesses Resection on Metastasis. Cancer Res (2012) 72:2683–91. doi: 10.1158/0008-5472.CAN-11-3733 [DOI] [PubMed] [Google Scholar]
- 59. Zhang Y, Zhang X, Zhang J, Sun B, Zheng L, Li J, et al. Microfluidic Chip for Isolation of Viable Circulating Tumor Cells of Hepatocellular Carcinoma for Their Culture and Drug Sensitivity Assay. Cancer Biol Ther (2016) 17:1177–87. doi: 10.1080/15384047.2016.1235665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xu F, Jin T, Zhu Y, Dai C. Immune Checkpoint Therapy in Liver Cancer. J Exp Clin Cancer Res (2018) 37:110. doi: 10.1186/s13046-018-0777-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Winograd P, Hou S, Court CM, Sadeghi S, Finn RS, DiPardo B, et al. Evaluation of Hepatocellular Carcinoma Circulating Tumor Cells Expressing Programmed Death-Ligand 1. Hpb (2018) 20:S2–3. doi: 10.1016/j.hpb.2018.02.004 [DOI] [Google Scholar]
- 62. Winograd P, Hou S, Court CM, Lee YT, Chen PJ, Zhu Y, et al. Hepatocellular Carcinoma-Circulating Tumor Cells Expressing PD-L1 Are Prognostic and Potentially Associated With Response to Checkpoint Inhibitors. Hepatol Commun (2020) 4:1527–40. doi: 10.1002/hep4.1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. van der Vaart M, Pretorius PJ. The Origin of Circulating Free DNA. Clin Chem (2007) 53:2215. doi: 10.1373/clinchem.2007.092734 [DOI] [PubMed] [Google Scholar]
- 64. Alix-Panabieres C, Schwarzenbach H, Pantel K. Circulating Tumor Cells and Circulating Tumor DNA. Annu Rev Med (2012) 63:199–215. doi: 10.1146/annurev-med-062310-094219 [DOI] [PubMed] [Google Scholar]
- 65. Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M. Origins, Structures, and Functions of Circulating DNA in Oncology. Cancer Metastasis Rev (2016) 35:347–76. doi: 10.1007/s10555-016-9629-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cortese R, Almendros I, Wang Y, Gozal D. Tumor Circulating DNA Profiling in Xenografted Mice Exposed to Intermittent Hypoxia. Oncotarget (2015) 6:556–69. doi: 10.18632/oncotarget.2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Xu RH, Wei W, Krawczyk M, Wang W, Luo H, Flagg K, et al. Circulating Tumour DNA Methylation Markers for Diagnosis and Prognosis of Hepatocellular Carcinoma. Nat Mater (2017) 16:1155–61. doi: 10.1038/nmat4997 [DOI] [PubMed] [Google Scholar]
- 68. Oussalah A, Rischer S, Bensenane M, Conroy G, Filhine-Tresarrieu P, Debard R, et al. Plasma Msept9: A Novel Circulating Cell-Free DNA-Based Epigenetic Biomarker to Diagnose Hepatocellular Carcinoma. EBioMedicine (2018) 30:138–47. doi: 10.1016/j.ebiom.2018.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kisiel JB, Dukek BA, R VSRK, Ghoz HM, Yab TC, Berger CK, et al. Hepatocellular Carcinoma Detection by Plasma Methylated DNA: Discovery, Phase I Pilot, and Phase II Clinical Validation. Hepatology (2019) 69:1180–92. doi: 10.1002/hep.30244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yong E. Cancer Biomarkers: Written in Blood. Nature (2014) 511:524–6. doi: 10.1038/511524a [DOI] [PubMed] [Google Scholar]
- 71. Mouliere F, Chandrananda D, Piskorz AM, Moore EK, Morris J, Ahlborn LB, et al. Enhanced Detection of Circulating Tumor DNA by Fragment Size Analysis. Sci Transl Med (2018) 10. doi: 10.1126/scitranslmed.aat4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bergman Y, Cedar H. DNA Methylation Dynamics in Health and Disease. Nat Struct Mol Biol (2013) 20:274–81. doi: 10.1038/nsmb.2518 [DOI] [PubMed] [Google Scholar]
- 73. Zhang P, Wen X, Gu F, Deng X, Li J, Dong J, et al. Methylation Profiling of Serum DNA From Hepatocellular Carcinoma Patients Using an Infinium Human Methylation 450 BeadChip. Hepatol Int (2013) 7:893–900. doi: 10.1007/s12072-013-9437-0 [DOI] [PubMed] [Google Scholar]
- 74. Chan KC, Jiang P, Chan CW, Sun K, Wong J, Hui EP, et al. Noninvasive Detection of Cancer-Associated Genome-Wide Hypomethylation and Copy Number Aberrations by Plasma DNA Bisulfite Sequencing. Proc Natl Acad Sci USA (2013) 110:18761–8. doi: 10.1073/pnas.1313995110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Huang G, Krocker JD, Kirk JL, Merwat SN, Ju H, Soloway RD, et al. Evaluation of INK4A Promoter Methylation Using Pyrosequencing and Circulating Cell-Free DNA From Patients With Hepatocellular Carcinoma. Clin Chem Lab Med (2014) 52:899–909. doi: 10.1515/cclm-2013-0885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ji XF, Fan YC, Gao S, Yang Y, Zhang JJ, Wang K. MT1M and MT1G Promoter Methylation as Biomarkers for Hepatocellular Carcinoma. World J Gastroenterol (2014) 20:4723–9. doi: 10.3748/wjg.v20.i16.4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kuo CC, Lin CY, Shih YL, Hsieh CB, Lin PY, Guan SB, et al. Frequent Methylation of HOXA9 Gene in Tumor Tissues and Plasma Samples From Human Hepatocellular Carcinomas. Clin Chem Lab Med (2014) 52:1235–45. doi: 10.1515/cclm-2013-0780 [DOI] [PubMed] [Google Scholar]
- 78. Lu CY, Chen SY, Peng HL, Kan PY, Chang WC, Yen CJ. Cell-Free Methylation Markers With Diagnostic and Prognostic Potential in Hepatocellular Carcinoma. Oncotarget (2017) 8:6406–18. doi: 10.18632/oncotarget.14115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wu HC, Yang HI, Wang Q, Chen CJ, Santella RM. Plasma DNA Methylation Marker and Hepatocellular Carcinoma Risk Prediction Model for the General Population. Carcinogenesis (2017) 38:1021–8. doi: 10.1093/carcin/bgx078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cai J, Chen L, Zhang Z, Zhang X, Lu X, Liu W, et al. Genome-Wide Mapping of 5-Hydroxymethylcytosines in Circulating Cell-Free DNA as a non-Invasive Approach for Early Detection of Hepatocellular Carcinoma. Gut (2019) 68:2195–205. doi: 10.1136/gutjnl-2019-318882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chen L, Abou-Alfa GK, Zheng B, Liu JF, Bai J, Du LT, et al. Genome-Scale Profiling of Circulating Cell-Free DNA Signatures for Early Detection of Hepatocellular Carcinoma in Cirrhotic Patients. Cell Res (2021) 31:589–92. doi: 10.1038/s41422-020-00457-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Piciocchi M, Cardin R, Vitale A, Vanin V, Giacomin A, Pozzan C, et al. Circulating Free DNA in the Progression of Liver Damage to Hepatocellular Carcinoma. Hepatol Int (2013) 7:1050–7. doi: 10.1007/s12072-013-9481-9 [DOI] [PubMed] [Google Scholar]
- 83. Huang A, Zhang X, Zhou SL, Cao Y, Huang XW, Fan J, et al. Detecting Circulating Tumor DNA in Hepatocellular Carcinoma Patients Using Droplet Digital PCR Is Feasible and Reflects Intratumoral Heterogeneity. J Cancer (2016) 7:1907–14. doi: 10.7150/jca.15823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Liao W, Yang H, Xu H, Wang Y, Ge P, Ren J, et al. Noninvasive Detection of Tumor-Associated Mutations From Circulating Cell-Free DNA in Hepatocellular Carcinoma Patients by Targeted Deep Sequencing. Oncotarget (2016) 7:40481–90. doi: 10.18632/oncotarget.9629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Qu C, Wang Y, Wang P, Chen K, Wang M, Zeng H, et al. Detection of Early-Stage Hepatocellular Carcinoma in Asymptomatic HBsAg-Seropositive Individuals by Liquid Biopsy. Proc Natl Acad Sci USA (2019) 116:6308–12. doi: 10.1073/pnas.1819799116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tao K, Bian Z, Zhang Q, Guo X, Yin C, Wang Y, et al. Machine Learning-Based Genome-Wide Interrogation of Somatic Copy Number Aberrations in Circulating Tumor DNA for Early Detection of Hepatocellular Carcinoma. EBioMedicine (2020) 56:102811. doi: 10.1016/j.ebiom.2020.102811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Cai Z, Chen G, Zeng Y, Dong X, Li Z, Huang Y, et al. Comprehensive Liquid Profiling of Circulating Tumor DNA and Protein Biomarkers in Long-Term Follow-Up Patients With Hepatocellular Carcinoma. Clin Cancer Res (2019) 25:5284–94. doi: 10.1158/1078-0432.CCR-18-3477 [DOI] [PubMed] [Google Scholar]
- 88. Wang Y, Balaji V, Kaniyappan S, Kruger L, Irsen S, Tepper K, et al. The Release and Trans-Synaptic Transmission of Tau via Exosomes. Mol Neurodegener (2017) 12:5. doi: 10.1186/s13024-016-0143-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ikeda S, Lim JS, Kurzrock R. Analysis of Tissue and Circulating Tumor DNA by Next-Generation Sequencing of Hepatocellular Carcinoma: Implications for Targeted Therapeutics. Mol Cancer Ther (2018) 17:1114–22. doi: 10.1158/1535-7163.MCT-17-0604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Liu A, Wu Q, Peng D, Ares I, Anadon A, Lopez-Torres B, et al. A Novel Strategy for the Diagnosis, Prognosis, Treatment, and Chemoresistance of Hepatocellular Carcinoma: DNA Methylation. Med Res Rev (2020) 40:1973–2018. doi: 10.1002/med.21696 [DOI] [PubMed] [Google Scholar]
- 91. Kotoh Y, Suehiro Y, Saeki I, Hoshida T, Maeda M, Iwamoto T, et al. Novel Liquid Biopsy Test Based on a Sensitive Methylated SEPT9 Assay for Diagnosing Hepatocellular Carcinoma. Hepatol Commun (2020) 4:461–70. doi: 10.1002/hep4.1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yan L, Chen Y, Zhou J, Zhao H, Zhang H, Wang G. Diagnostic Value of Circulating Cell-Free DNA Levels for Hepatocellular Carcinoma. Int J Infect Dis (2018) 67:92–7. doi: 10.1016/j.ijid.2017.12.002 [DOI] [PubMed] [Google Scholar]
- 93. Li F, Qiao CY, Gao S, Fan YC, Chen LY, Wang K. Circulating Cell-Free DNA of Methylated Insulin-Like Growth Factor-Binding Protein 7 Predicts a Poor Prognosis in Hepatitis B Virus-Associated Hepatocellular Carcinoma After Hepatectomy. Free Radic Res (2018) 52:455–64. doi: 10.1080/10715762.2018.1443448 [DOI] [PubMed] [Google Scholar]
- 94. Ren N, Qin LX, Tu H, Liu YK, Zhang BH, Tang ZY. The Prognostic Value of Circulating Plasma DNA Level and Its Allelic Imbalance on Chromosome 8p in Patients With Hepatocellular Carcinoma. J Cancer Res Clin Oncol (2006) 132:399–407. doi: 10.1007/s00432-005-0049-5 [DOI] [PubMed] [Google Scholar]
- 95. Kim SS, Eun JW, Choi JH, Woo HG, Cho HJ, Ahn HR, et al. MLH1 Single-Nucleotide Variant in Circulating Tumor DNA Predicts Overall Survival of Patients With Hepatocellular Carcinoma. Sci Rep (2020) 10:17862. doi: 10.1038/s41598-020-74494-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Park S, Lee EJ, Rim CH, Seong J. Plasma Cell-Free DNA as a Predictive Marker After Radiotherapy for Hepatocellular Carcinoma. Yonsei Med J (2018) 59:470–9. doi: 10.3349/ymj.2018.59.4.470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zhao W, Qiu L, Liu H, Xu Y, Zhan M, Zhang W, et al. Circulating Tumor DNA as a Potential Prognostic and Predictive Biomarker During Interventional Therapy of Unresectable Primary Liver Cancer. J Gastrointest Oncol (2020) 11:1065–77. doi: 10.21037/jgo-20-409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ikeda S, Tsigelny IF, Skjevik AA, Kono Y, Mendler M, Kuo A, et al. Next-Generation Sequencing of Circulating Tumor DNA Reveals Frequent Alterations in Advanced Hepatocellular Carcinoma. Oncologist (2018) 23:586–93. doi: 10.1634/theoncologist.2017-0479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Parikh AR, Leshchiner I, Elagina L, Goyal L, Levovitz C, Siravegna G, et al. Liquid Versus Tissue Biopsy for Detecting Acquired Resistance and Tumor Heterogeneity in Gastrointestinal Cancers. Nat Med (2019) 25:1415–21. doi: 10.1038/s41591-019-0561-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. von Felden J, Craig AJ, Garcia-Lezana T, Labgaa I, Haber PK, D’Avola D, et al. Mutations in Circulating Tumor DNA Predict Primary Resistance to Systemic Therapies in Advanced Hepatocellular Carcinoma. Oncogene (2021) 40:140–51. doi: 10.1038/s41388-020-01519-1 [DOI] [PubMed] [Google Scholar]
- 101. Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle Formation During Reticulocyte Maturation. Association of Plasma Membrane Activities With Released Vesicles (Exosomes). J Biol Chem (1987) 262:9412–20. doi: 10.1016/S0021-9258(18)48095-7 [DOI] [PubMed] [Google Scholar]
- 102. Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, et al. Biological Properties of Extracellular Vesicles and Their Physiological Functions. J Extracell Vesicles (2015) 4.27066. doi: 10.3402/jev.v4.27066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Raab-Traub N, Dittmer DP. Viral Effects on the Content and Function of Extracellular Vesicles. Nat Rev Microbiol (2017) 15:559–72. doi: 10.1038/nrmicro.2017.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, et al. Proteomic Comparison Defines Novel Markers to Characterize Heterogeneous Populations of Extracellular Vesicle Subtypes. Proc Natl Acad Sci USA (2016) 113:E968–77. doi: 10.1073/pnas.1521230113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wang M, Wang Y, Ye F, Yu K, Wei W, Liu M, et al. Exosome Encapsulated ncRNAs in the Development of HCC: Potential Circulatory Biomarkers and Clinical Therapeutic Targets. Am J Cancer Res (2021) 11:3794–812. [PMC free article] [PubMed] [Google Scholar]
- 106. Wang B, Zhang Y, Ye M, Wu J, Ma L, Chen H. Cisplatin-Resistant MDA-MB-231 Cell-Derived Exosomes Increase the Resistance of Recipient Cells in an Exosomal miR-423-5p-Dependent Manner. Curr Drug Metab (2019) 20:804–14. doi: 10.2174/1389200220666190819151946 [DOI] [PubMed] [Google Scholar]
- 107. Pan JH, Zhou H, Zhao XX, Ding H, Li W, Qin L, et al. Role of Exosomes and Exosomal microRNAs in Hepatocellular Carcinoma: Potential in Diagnosis and Antitumour Treatments (Review). Int J Mol Med (2018) 41:1809–16. doi: 10.3892/ijmm.2018.3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zhou Y, Ren H, Dai B, Li J, Shang L, Huang J, et al. Hepatocellular Carcinoma-Derived Exosomal miRNA-21 Contributes to Tumor Progression by Converting Hepatocyte Stellate Cells to Cancer-Associated Fibroblasts. J Exp Clin Cancer Res (2018) 37:324. doi: 10.1186/s13046-018-0965-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zhou B, Xu K, Zheng X, Chen T, Wang J, Song Y, et al. Application of Exosomes as Liquid Biopsy in Clinical Diagnosis. Signal Transduct Target Ther (2020) 5:144. doi: 10.1038/s41392-020-00258-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Cai X, Janku F, Zhan Q, Fan JB. Accessing Genetic Information With Liquid Biopsies. Trends Genet (2015) 31:564–75. doi: 10.1016/j.tig.2015.06.001 [DOI] [PubMed] [Google Scholar]
- 111. Lin XJ, Fang JH, Yang XJ, Zhang C, Yuan Y, Zheng L, et al. Hepatocellular Carcinoma Cell-Secreted Exosomal MicroRNA-210 Promotes Angiogenesis In Vitro and In Vivo . Mol Ther Nucleic Acids (2018) 11:243–52. doi: 10.1016/j.omtn.2018.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Rao Q, Zuo B, Lu Z, Gao X, You A, Wu C, et al. Tumor-Derived Exosomes Elicit Tumor Suppression in Murine Hepatocellular Carcinoma Models and Humans In Vitro. Hepatology (2016) 64:456–72. doi: 10.1002/hep.28549 [DOI] [PubMed] [Google Scholar]
- 113. Kalluri R, LeBleu VS. The Biology, Function, and Biomedical Applications of Exosomes. Science (2020) 367. doi: 10.1126/science.aau6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wang H, Lu Z, Zhao X. Tumorigenesis, Diagnosis, and Therapeutic Potential of Exosomes in Liver Cancer. J Hematol Oncol (2019) 12:133. doi: 10.1186/s13045-019-0806-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Sohn W, Kim J, Kang SH, Yang SR, Cho JY, Cho HC, et al. Serum Exosomal microRNAs as Novel Biomarkers for Hepatocellular Carcinoma. Exp Mol Med (2015) 47:e184. doi: 10.1038/emm.2015.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Cho HJ, Baek GO, Seo CW, Ahn HR, Sung S, Son JA, et al. Exosomal microRNA-4661-5p-Based Serum Panel as a Potential Diagnostic Biomarker for Early-Stage Hepatocellular Carcinoma. Cancer Med (2020) 9:5459–72. doi: 10.1002/cam4.3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Ma F, Liu X, Li D, Wang P, Li N, Lu L, et al. MicroRNA-466l Upregulates IL-10 Expression in TLR-Triggered Macrophages by Antagonizing RNA-Binding Protein Tristetraprolin-Mediated IL-10 mRNA Degradation. J Immunol (2010) 184:6053–9. doi: 10.4049/jimmunol.0902308 [DOI] [PubMed] [Google Scholar]
- 118. Cho HJ, Eun JW, Baek GO, Seo CW, Ahn HR, Kim SS, et al. Serum Exosomal MicroRNA, miR-10b-5p, as a Potential Diagnostic Biomarker for Early-Stage Hepatocellular Carcinoma. J Clin Med (2020) 9:281. doi: 10.3390/jcm9010281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Arbelaiz A, Azkargorta M, Krawczyk M, Santos-Laso A, Lapitz A, Perugorria MJ, et al. Serum Extracellular Vesicles Contain Protein Biomarkers for Primary Sclerosing Cholangitis and Cholangiocarcinoma. Hepatology (2017) 66:1125–43. doi: 10.1002/hep.29291 [DOI] [PubMed] [Google Scholar]
- 120. Ma X, Yuan T, Yang C, Wang Z, Zang Y, Wu L, et al. X-Inactive-Specific Transcript of Peripheral Blood Cells is Regulated by Exosomal Jpx and Acts as a Biomarker for Female Patients With Hepatocellular Carcinoma. Ther Adv Med Oncol (2017) 9:665–77. doi: 10.1177/1758834017731052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Xu H, Chen Y, Dong X, Wang X. Serum Exosomal Long Noncoding RNAs ENSG00000258332.1 and LINC00635 for the Diagnosis and Prognosis of Hepatocellular Carcinoma. Cancer Epidemiol Biomarkers Prev (2018) 27:710–6. doi: 10.1158/1055-9965.EPI-17-0770 [DOI] [PubMed] [Google Scholar]
- 122. Fu Q, Zhang Q, Lou Y, Yang J, Nie G, Chen Q, et al. Primary Tumor-Derived Exosomes Facilitate Metastasis by Regulating Adhesion of Circulating Tumor Cells via SMAD3 in Liver Cancer. Oncogene (2018) 37:6105–18. doi: 10.1038/s41388-018-0391-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Wang Y, Zhang C, Zhang P, Guo G, Jiang T, Zhao X, et al. Serum Exosomal microRNAs Combined With Alpha-Fetoprotein as Diagnostic Markers of Hepatocellular Carcinoma. Cancer Med (2018) 7:1670–9. doi: 10.1002/cam4.1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Lee YR, Kim G, Tak WY, Jang SY, Kweon YO, Park JG, et al. Circulating Exosomal Noncoding RNAs as Prognostic Biomarkers in Human Hepatocellular Carcinoma. Int J Cancer (2019) 144:1444–52. doi: 10.1002/ijc.31931 [DOI] [PubMed] [Google Scholar]
- 125. Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo J, et al. Three Isoforms of Exosomal Circptgr1 Promote Hepatocellular Carcinoma Metastasis via the Mir449a-MET Pathway. EBioMedicine (2019) 40:432–45. doi: 10.1016/j.ebiom.2018.12.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, et al. Cancer Cell-Derived Exosomal Circuhrf1 Induces Natural Killer Cell Exhaustion and may Cause Resistance to Anti-PD1 Therapy in Hepatocellular Carcinoma. Mol Cancer (2020) 19:110. doi: 10.1186/s12943-020-01222-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Sun XH, Wang YT, Li GF, Zhang N, Fan L. Serum-Derived three-circRNA Signature as a Diagnostic Biomarker for Hepatocellular Carcinoma. Cancer Cell Int (2020) 20:226. doi: 10.1186/s12935-020-01302-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Kim SS, Baek GO, Ahn HR, Sung S, Seo CW, Cho HJ, et al. Serum Small Extracellular Vesicle-Derived LINC00853 as a Novel Diagnostic Marker for Early Hepatocellular Carcinoma. Mol Oncol (2020) 14:2646–59. doi: 10.1002/1878-0261.12745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Huang X, Sun L, Wen S, Deng D, Wan F, He X, et al. RNA Sequencing of Plasma Exosomes Revealed Novel Functional Long Noncoding RNAs in Hepatocellular Carcinoma. Cancer Sci (2020) 111:3338–49. doi: 10.1111/cas.14516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Ghosh S, Bhowmik S, Majumdar S, Goswami A, Chakraborty J, Gupta S, et al. The Exosome Encapsulated microRNAs as Circulating Diagnostic Marker for Hepatocellular Carcinoma With Low Alpha-Fetoprotein. Int J Cancer (2020) 147:2934–47. doi: 10.1002/ijc.33111 [DOI] [PubMed] [Google Scholar]
- 131. Zhang YC, Xu Z, Zhang TF, Wang YL. Circulating microRNAs as Diagnostic and Prognostic Tools for Hepatocellular Carcinoma. World J Gastroenterol (2015) 21:9853–62. doi: 10.3748/wjg.v21.i34.9853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Wang H, Hou L, Li A, Duan Y, Gao H, Song X. Expression of Serum Exosomal microRNA-21 in Human Hepatocellular Carcinoma. BioMed Res Int (2014) 2014:864894. doi: 10.1155/2014/864894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Lu Y, Duan Y, Xu Q, Zhang L, Chen W, Qu Z, et al. Circulating Exosome-Derived Bona Fide Long non-Coding RNAs Predicting the Occurrence and Metastasis of Hepatocellular Carcinoma. J Cell Mol Med (2020) 24:1311–8. doi: 10.1111/jcmm.14783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Zhu L, Li J, Gong Y, Wu Q, Tan S, Sun D, et al. Exosomal tRNA-Derived Small RNA as a Promising Biomarker for Cancer Diagnosis. Mol Cancer (2019) 18:74. doi: 10.1186/s12943-019-1000-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Taleb RSZ, Moez P, Younan D, Eisenacher M, Tenbusch M, Sitek B, et al. Quantitative Proteome Analysis of Plasma Microparticles for the Characterization of HCV-Induced Hepatic Cirrhosis and Hepatocellular Carcinoma. Proteomics Clin Appl (2017) 11. doi: 10.1002/prca.201700014 [DOI] [PubMed] [Google Scholar]
- 136. Jiang SS, Weng DS, Wang QJ, Pan K, Zhang YJ, Li YQ, et al. Galectin-3 Is Associated With a Poor Prognosis in Primary Hepatocellular Carcinoma. J Transl Med (2014) 12:273. doi: 10.1186/s12967-014-0273-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Liu W, Hu J, Zhou K, Chen F, Wang Z, Liao B, et al. Serum Exosomal miR-125b Is a Novel Prognostic Marker for Hepatocellular Carcinoma. Onco Targets Ther (2017) 10:3843–51. doi: 10.2147/OTT.S140062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Shi M, Jiang Y, Yang L, Yan S, Wang YG, Lu XJ. Decreased Levels of Serum Exosomal miR-638 Predict Poor Prognosis in Hepatocellular Carcinoma. J Cell Biochem (2018) 119:4711–6. doi: 10.1002/jcb.26650 [DOI] [PubMed] [Google Scholar]
- 139. Luo Y, Liu F, Gui R. High Expression of Circulating Exosomal Circakt3 is Associated With Higher Recurrence in HCC Patients Undergoing Surgical Treatment. Surg Oncol (2020) 33:276–81. doi: 10.1016/j.suronc.2020.04.021 [DOI] [PubMed] [Google Scholar]
- 140. Macias M, Alegre E, Diaz-Lagares A, Patino A, Perez-Gracia JL, Sanmamed M, et al. Liquid Biopsy: From Basic Research to Clinical Practice. Adv Clin Chem (2018) 83:73–119. doi: 10.1016/bs.acc.2017.10.003 [DOI] [PubMed] [Google Scholar]