Abstract
In this study we seek to determine the efficacy of using automated mapping methods to reduce the manual mapping burden of laboratory data to LOINC(r) on a nationwide electronic health record derived oncology specific dataset. We developed novel encoding methodologies to vectorize free text lab data, and evaluated logistic regression, random forest, and knn machine learning classifiers. All machine learning models did significantly better than deterministic baseline algorithms. The best classifiers were random forest and were able to predict the correct LOINC code 94.5% of the time. Ensemble classifiers further increased accuracy, with the best ensemble classifier predicting the same code 80.5% of the time with an accuracy of 99%. We conclude that by using an automated laboratory mapping model we can both reduce manual mapping time, and increase quality of mappings, suggesting automated mapping is a viable tool in a real-world oncology dataset.
Background and Significance
Health data collected in the course of routine clinical care (real-world data [RWD]) are becoming a valuable part of the clinical research armamentarium, complementing and/or supplementing traditional prospective studies, and providing insights on aspects such as patterns of care or treatment effectiveness in populations underrepresented in clinical trials.1, 2 Electronic health records (EHRs) have emerged as a key oncology RWD source, with the potential to generate highly granular longitudinal data.3 The original purpose of EHRs however, is not research but patient care, administration, and reimbursement. Therefore, extraction of research-grade information from the EHR may become a multi-step process that requires optimization and quality controls.4
The utilization of EHR-derived data for research purposes involves extraction of structured and coded data, as well as unstructured data (narrative free text entered by users at the point of care). One domain in particular that requires significant pre-processing is that of laboratory (lab) data.5, 6 Laboratory data is crucial in describing the longitudinal patient journey and thus high quality laboratory data is essential in many areas of oncological research. For example, laboratory data is necessary to understand the efficacy of a patient's regimen or the performance of novel treatments.7 However inconsistent use of standards by laboratories and free text documentation leads to highly variable lab results data.6, 8 The Logical Observation Identifiers Names and Codes (LOINC(r))9 is a vocabulary standard used to identify and unify lab data under a common data model, but is often inappropriately or inconsistently used in clinical day-to-day settings.10, 11 Due to this, a manual mapping process where specialists assign laboratory data to LOINC codes is required before the data can be used in research datasets.12
This manual mapping process, however has some shortcomings:
-
1.
Ingestion of standardized and non-standardized lab data from multiple vendors, free text data entry at practice sites, and abbreviations, typos, and practice-specific documentation norms lead to a potentially infinite set of source terms.6, 8 In our study we have found that it takes an experienced clinical terminologist between 6-8 hours to map 1000 terms manually, meaning harmonizing large amounts of laboratory data to LOINC codes is an arduous process.
-
2.
As with any manual process, there is inevitable error in the mapping process, in spite of having mapping guidelines. As any error affects the accuracy of data that will be used for research and analysis, multiple levels of mapping review are required, further increasing the manual workload.
-
3.
Any free text terms that come in from source data not previously harmonized are required to be mapped before they are eligible for entry into research datasets. Depending on volume, harmonization can be a lengthy process, reducing the data recency of datasets.
Based on the above considerations, finding an automated approach to lower the burden and increase the accuracy of the mapping process has direct implications on the quality of real-world research datasets.6, 12-14 As a single mapping can affect many rows in a database, high accuracy mappings are of top importance when examining automated approaches.
Previous studies have attempted to automate LOINC mapping in a number of different scenarios. One study attempted to use a local high quality corpus and was able to achieve a best case accuracy of 79%.15 A second study that relied on using lexical methods achieved similar accuracy.16 A more recent study trained a machine learning classifier on a large national EHR database with noisy LOINC labels, and was able to achieve relatively high accuracy.17 The best classifiers in this study predicted the correct LOINC code in 85% of the unlabeled data and 96% of the labeled data by test frequency.17 More recently in a study focused on mapping COVID-19 labs to LOINC codes, a rules based algorithm was shown to have accuracy of 97.4%, prompting further investigation into deterministic rules based algorithms.18 Other studies have been successful at higher level grouping of laboratory data into categorical values, but did not focus on the standardization of individual lab records.19
To the best of our knowledge, there are no studies that have attempted to use automated LOINC mapping on a real-world dataset actively being used for clinical research, or on an oncology-specific dataset. These properties lead us to develop custom encoding methodologies, with the goal of high accuracy and high precision automated LOINC mappings. We used these novel encoding methodologies to process free text laboratory data for the use in supervised machine learning classifiers. We evaluated the ability for these classifiers to individually predict LOINC codes, as well as the ability for groups of classifiers to jointly make predictions through ensemble learning. Any reduction in manual mapping time or manual quality assessment work that an automated system can provide to clinical terminologists (without compromising accuracy) is our overall measure of success.
Methods
Index Of Lab Data
This study used the nationwide, longitudinal Flatiron Health electronic health record (EHR)-derived de-identified database. During the study period, the de-identified data originated from approximately 280 US cancer clinics (~800 sites of care). Flatiron Health creates EHR-derived research datasets comprising de-identified patient-level structured and unstructured data, curated via technology-enabled abstraction.20, 21 Flatiron Health has a harmonization process whereby research relevant clinical and administrative data are mapped by clinical terminologists to their appropriate terminology standards. All free text laboratory data undergoes this process, getting assigned LOINC codes. Once data is harmonized, it is re-used to determine standard codes for any current and future free text data.
Flatiron Health lab data is manually harmonized using term, unit, and panel information, with each distinct combination corresponding to a different row that each requires harmonization. As all three fields can potentially be free text fields in EHRs, thousands of new combinations require mapping each month.
The "term" field of the lab data contains the name of the lab result. This can vary from a fully spelled out name to a local acronym. An example of the term field is "white blood cell count", which also might appear as "white blood cell", "wbc", "wite bld cell", or any other abbreviation with or without typos.
The "unit" field aptly contains information about the unit of measurement. Examples of a few possible unit values can be seen in Table 1. As shown in Table 1, rows that have the same term value but differing unit values can result in different target LOINC codes. Lab results can also be expressed in equivalent units, for example the units mg/mL, g/dL and ug/mL, all measure mass/volume, but create separate rows for mapping.
Table 1:
Example Source Term, Unit, and Panel information, and the associated LOINC mapping.
| Source Term | Source Unit | Source Panel | LOINC Label |
| wbc | 10x3/ul | cbc | 26464-8 |
| white blood cell | 10x3/ul | cbc | 26464-8 |
| anc | 10x3/ul | cbc | 26499-4 |
| creat | mg/dl | urine, 24 hour | 20624-3 |
| creat | g/24 h | urine, 24 hour | 2162-6 |
| Albumin, URINE | mg/dL | ALBUMIN URINE 24HR | 1754-1 |
| Albumin, Urine | ug/mL | Microalbumin, Random Urine | 1754-1 |
The "panel" field represents a group of lab tests/results that are ordered and reported together. There are several bases for grouping individual labs together. For example, by the medical condition they are intended to help diagnose (cardiac risk panel), by the specimen type (complete blood count, CBC), by the tests most frequently requested by users (comprehensive chemistry profile), by the methodology employed in the test (viral panel by polymerase chain reaction), or by the types of components included (urine drug screen). Since the specimen for the lab result is not available as a distinct data point, we use the panel name to determine the system. In the Flatiron Health dataset the panel field contains the concatenation of all the panels that were ordered, adding variance and further increasing the amount of data that requires harmonization.
Datasets
There are three catalogs we used in our experiments for automated lab mapping harmonization:
-
1.
A source data catalog
-
2.
A target LOINC data catalog
-
3.
A mapping catalog containing associations between the source and target catalogs
The source data catalog contains all of the unique term, unit, and panel combinations that we have ever seen in our data, as well as an associated source ID. This table contains no LOINC code information. The "target" LOINC catalog contains all of the different LOINC codes that we might map the rows in the source data catalog to. Our harmonization team takes in the source catalog as input and manually determines which target code each row in the source catalog should map to.
Once a row from the source data catalog has been assigned an appropriate LOINC, it is added to the mapping catalog. Thus, this catalog contains the source ID and the associated target ID (LOINC) in a unique mapping. Despite the full LOINC catalog containing approximately 95,000 distinct LOINCs, because the source dataset is oncology specific we see only a small percentage of all possible LOINCs in our dataset. If the harmonization team determines that the information in the source row is insufficient to determine an accurate mapping, a label of "EXCLUDED" is attached and no target code is present. This provides an additional challenge for an automated system, as this label has a wide range of source data mapped to it, making it challenging to accurately predict.
There are different levels of LOINC interoperability, depending on the differences between two LOINC codes. Previous work has consolidated LOINC codes using these levels of interoperability17, but since the Flatiron Health dataset requires the most granular version of LOINCs, we do not do consolidation. While this makes automated LOINC prediction more challenging, it is required to keep the dataset as precise as possible.
Deterministic Automated LOINC Prediction
Before using learning-based automated LOINC prediction algorithms, we first set baselines using two computation-based algorithms. The two algorithms, maximal target string matching and maximal source string matching, are described below.
Maximal Target String Match
The first baseline algorithm we used compared source rows directly to the target LOINC catalog. We compared using Levenshtein distance, which is a string metric for measuring the difference between two character sequences.22 Specifically we used the Levenshtein ratio, which is a number between 0 and 1 representing how similar two strings are, with 0 being completely different and 1 being entirely the same. For each row in the source catalog, we compared the source term and source unit fields directly to the target LOINC catalog. We then combined the comparisons via a weighted sum of 4 term and 1 unit. This set of weights was evaluated against six other combinations (5/5 term and 0/5 unit, 4/5 term and 1/5 unit,…, 0/5 term and 5/5 unit) on a random sample of 100,000 rows from the source term catalog and had the highest performance of the six. Since the panel field contains a combination of all panels that were ordered, it has no directly comparable value in the LOINC catalog and was therefore excluded. For each row, the LOINC code that was most similar via the weighted sum of term and unit Levenshtein ratios was the one assigned.
Maximal Source String Match
The second baseline algorithm attempts to find the source row in the training data that is most similar to the test row being evaluated. Similar to the Maximal Target String Match algorithm, it uses Levenshtein ratio to measure similarity. To compare two rows, the Levenshtein ratio of the term, unit, and panel fields are calculated and combined via a weighted sum of 3/5 term, 1/5 unit, and 1/5 panel. We again tried multiple weighted combinations of fifths, and this one had the highest performance of the combinations we evaluated. Computing the Levenshtein ratio is a costly operation, and so comparing each row in the training data to every row in the source data is prohibitively expensive. Due to this limitation, we instead first pre-compute the five most common source terms for each LOINC, with ties being broken by random selection. We then compare each row in the test data to all of the pre-chosen source rows, and choose the LOINC that has a source row with the greatest similarity.
Feature Encodings
As the source data is composed of free text fields, we must first encode the free text to make it possible to pass as an input to a supervised machine learning model. Since lab data free text fields have an unlimited set of potential values, and are often short abbreviations, we found that traditional string encoding methods that rely on a finite corpus to encode such as One-Hot tokenization, TF-IDF, and Word2Vec perform poorly.23-25 With this in mind, we developed two new encoding methods specifically for this domain.
Levenshtein Distance Encoding
As mentioned above, Levenshtein distance is a string metric for measuring the difference between two character sequences.22 To encode each row, we compared the rows source term and source unit to every target description and target units in the LOINC target catalog, for each distinct LOINC in the training data. The weighted sum of the Levenshtein ratio for term and unit between the source catalog and each row in the LOINC catalog is captured in a vector. To determine the optimal set of weights to use, we tried every tenth fold combination (1/10 term, 9/10 unit,.., 9/10 term and 1/10 unit) and settled on 8/10 term and 2/10 unit. The vector of weighted Levenshtein ratios, which has a column for each LOINC representing how similar the source row is to that target LOINC, is the encoded row. Similar to the Maximal Target String Match, we exclude using panel as it has no directly comparable value in the LOINC catalog to compare to with Levenshtein distance.
Frequency Tokenization Encoding
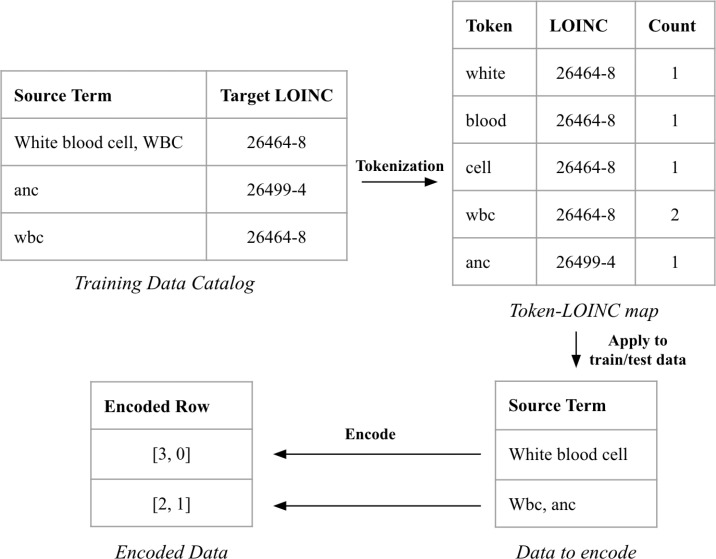
In a similar manner to TF-IDF, we created an approach to encode based on the frequency of distinct tokens in the source data. For each row in the training data set, we first clean the rows by removing any characters that are not alpha-numeric and replacing them with whitespace. This process is done for each of the term, unit, and panel fields. We then split the cleaned row on whitespace, creating a set of tokens for each field. These tokens are then mapped to the target LOINC, creating a token-LOINC map that details which source data tokens are associated with each LOINC. If tokens appear multiple times mapping to the same target LOINC, that count will be recorded in the token-LOINC map. This token-LOINC map can then be used to encode future rows.
To encode the term field, we first create tokens in the same manner as described above. These tokens will be used to create an encoded vector of length L, where L is the total number of LOINCs in the token-LOINC map. In this vector, each LOINC has a corresponding index, initialized at zero. For each LOINC that a token maps to, the corresponding index in the vector will be incremented by the count in the token-LOINC map. See Figure 1 for an example of this encoding. This process happens for each of the term, unit, and panel fields, resulting in a concatenated encoded vector of length 3*L.
Figure 1:
Tokenization Encoding process. Training data gets processed and tokenized, forming the token-LOINC map. This map is then used to encode both the training and test data, leading to vectorized rows. The encoded row has an index for each LOINC that exists in the training data catalog (here only 2 LOINCs), and the value at each index represents how many tokens overlap with training data tokens mapped to that LOINC. The first row gets encoded as [3, 0] since there are 3 tokens in the source term that match a token for the LOINC 26464-8, and 0 tokens that match the LOINC 26499-4.
Supervised Machine Learning Classification Models
For both of the feature encoding methods, we evaluate performance using Logistic Regression (L2 penalized)26, Random Forest27, and K Nearest Neighbors (KNN)28 classifiers. Model training and analyses were conducted using scikit-learn in Python 3.7.4.29
We used GridSearch to optimize key hyperparameters for each model.29 For logistic regression we focused on the number of iterations, and the inverse regularization parameter (C), and settled on values of 500 and 5 respectively. For random forest we focused on the number of estimators, and settled on 500. For KNN we focused on the number of nearest neighbors, and settled on 5.
Additionally, we evaluated the use of PCA to reduce the size of the input vectors and thereby decrease learning time. Using PCA with 95% variance we were able to decrease the total learning time by 50%. However, the increased efficiency came at the cost of a few percentage points of accuracy. As high accuracy is our top priority, the results in this paper do not use PCA or other compression schemes on the input vectors.
Ensemble Learning
Ensemble learning is the practice of using multiple learning algorithms together and then classifying new points based on a weighted vote of their individual predictions.30, 31 We explore the use of ensemble learning to increase accuracy of predictions in cases where multiple models predict the same LOINC. Our ensemble models are assessed based on the percent of the test set with prediction overlap, and the accuracy of the predictions on the overlap. The percent of the test set with overlap is simply the number of rows in the test set where all models make the same prediction, divided by the size of the test set. The accuracy is then measured in the standard way on the subset: by taking the predictions that all models made and comparing with the labeled data. A high performing ensemble model will have both a high percentage of test set overlap, and high accuracy on the overlap.32
It is important to note that adding additional models to an ensemble with these assessment criteria will not necessarily increase the ensemble's performance. For example, because we define prediction overlap to be only predictions where all models make the same prediction, adding a poorly performing model may significantly reduce prediction overlap, decreasing the subset the ensemble is able to make a prediction on. Moreover, a poorly performing model may only overlap on incorrect predictions, significantly decreasing the ensemble's accuracy. With this in mind, we evaluate various ensembles to attempt to find the set of models that most successfully complement each other.
Data Size and Model Performance
Models were trained using a subset of 450,000 randomly sampled and unique rows from the most recent two years of the labeled data corpus. We decided to use a large sample instead of training on the full dataset to increase investigative agility while maintaining high usability. Amongst the rows sampled, there are 482 distinct LOINCs that have been mapped to, including "EXCLUDED". Models were validated using an 80/20 split for training and testing respectively, and results are the average of 5 randomly subsampled splits. Models were assessed based on accuracy, F-1 score, and precision. Since models are multiclass classifiers, F-1 score and precision are measured as weighted averages of the F-1 score and precision across all classes. While accuracy is of the highest concern for prediction usability, F-1 score is important to assess whether accuracy is overfitting due to class imbalance and thereby simply predicting the most frequent LOINCs.
Results
Deterministic Algorithm Performance
To establish a baseline we first built and tested the deterministic prediction algorithms. These algorithms do not take advantage of larger data sets to learn, so modifying the size of the dataset only affects the variance. With this in mind, we averaged predictions from 5 trials run using 10,000 randomly sampled rows. The performance of these algorithms is in Table 2. For Maximal Target String Match, accuracy is low, but still much higher than random chance, implying that there is some inherent similarity between the source data and the target data. Given the level of abbreviation in the source data, this result is expected. Comparing to other source data instead of directly to target LOINC data in the Maximal Source String Match algorithm, performed significantly better. When solely basing future predictions on the most similar previously seen terms, we saw an accuracy of just below 50%. Incorporating unit and panel information as well, we saw a small increase, implying that there is value in using more than just the lab name represented in the term field when making predictions.
Table 2:
Deterministic (non-learning) algorithms performance for predicting LOINC, run on 10,000 LOINCs.
| Algorithm Name | Accuracy (%) |
| Maximal Target String Match (Term + Unit) | 16.0 |
| Maximal Source String Match (Term) | 48.4 |
| Maximal Source String Match (Term + Unit + Panel) | 52.0 |
Cross Validated Supervised Model Performance
After determining a baseline, we moved on to assessing the performance of different learning based models using our two different encoding schemes. The results from the Levenshtein Distance Encoding method are in Table 3. We can see that we have significantly outperformed the baseline, and that while the random forest classifier had the highest accuracy, all models have comparable performance at greater than 93% accuracy. KNN has marginally worse performance using this encoding scheme, but it is worth noting that training time for the KNN classification model is on average less than 50% that of logistic regression or random forest. We also see that the weighted F1 score and weighted precision have similar values to that of accuracy, implying that our models are not overfitting due to class imbalance.
Table 3:
Model performance using Levenshtein Distance Encoding trained using 80/20 split on dataset of size 450,000 (360,000 training, 90,000 testing). Top 10% and bottom 90% are measures of target LOINC frequency in testing dataset.
| Accuracy (%) | F1 Score (Weighted) | Precision (Weighted) | Top 10% LOINC Weighted Precision n=74866 | Bottom 90% LOINC Weighted Precision n=15134 | |
| Logistic Regression | 93.1 | 0.929 | 0.928 | 0.941 | 0.874 |
| Random Forest | 94.0 | 0.940 | 0.938 | 0.9489 | 0.899 |
| KNN | 93.0 | 0.931 | 0.928 | 0.946 | 0.854 |
As we are focused on any reduction in manual work, it is also important to see if there exist particular subsets that our models can reproducibly predict with higher success. One such subset we examined is the top 10% of LOINCs by labeled rows, a subset which on average accounts for over 84% of the test set. Unsurprisingly our models have a higher weighted precision on LOINCs in the top 10%, as these LOINCs have significantly more training data. However they still do relatively well on LOINCs that have less training data, once again suggesting that overfitting is not occuring.
Examining the results from the Frequency Tokenization Encoding in Table 4, we see that only the random forest classifier is able to outperform any model using the Levenshtein distance encoding. We also see that in this encoding method, random forest outperforms both logistic regression and KNN by a significant margin. Interestingly, when using the deterministic baseline algorithms, comparison to the LOINC catalog did significantly worse than comparison to the source data directly. Contrastly, when using supervised learning the Levenshtein encoding which encodes by comparing directly to the target LOINC catalog on average outperforms token encoding which encodes by comparing to previously seen source data. We also see slightly more variance between accuracy and weighted F1 Score as well as between the top 10% and bottom 90% of LOINCs, implying that there might be more class imbalance overfitting happening.
Table 4:
Model performance using Frequency Tokenization Encoding trained using 80/20 split on dataset of size 450,000 (360,000 training, 90,000 testing). Top 10% and bottom 90% are measures of target LOINC frequency in testing dataset.
| Accuracy (%) | F1 Score (Weighted) | Precision (Weighted) | Top 10% LOINC Weighted Precision n=74866 | Bottom 90% LOINC Weighted Precision n=15134 | |
| Logistic Regression | 87.3 | 0.859 | 0.864 | 0.896 | 0.6854 |
| Random Forest | 94.5 | 0.943 | 0.943 | 0.957 | 0.874 |
| KNN | 88.95 | 0.877 | 0.878 | 0.911 | 0.714 |
Ensemble Learning Performance
While supervised learning models did significantly better than baseline models, accuracy still has room for improvement. To increase usability, we attempted to use ensemble learning to see if we can achieve a higher accuracy for a subset of the predicted data. Results for different groupings of trained models are in Table 5. These results are extremely promising, with the combination of all trained models achieving an extremely high accuracy of 99% on the 80.5% of predictions that all models agreed upon. While using this method doesn't allow for full automation as it is only applicable to a subset of predictions, it has the potential to significantly reduce the work of manual harmonization and quality analysis.
Table 5:
Ensemble learning performance for various combinations of models and encoding methods.
| Prediction overlap raw count (out of 90000) | Percentage of Test Set (%) | Accuracy on prediction overlap (%) | |
| LR + KNN (Levi) | 86430 | 96.0 | 95.2 |
| LR + RF (Levi) | 87580 | 97.3 | 94.9 |
| KNN + RF (Levi) | 87211 | 96.9 | 95.1 |
| LR + KNN (Tokens) | 78469 | 87.2 | 94.7 |
| LR + RF (Tokens) | 79174 | 88.0 | 97.1 |
| KNN + RF (Tokens) | 81643 | 90.7 | 96.3 |
| LR Levi + Tokens | 80003 | 88.9 | 95.9 |
| RF Levi + Tokens | 83526 | 92.8 | 97.9 |
| KNN Levi + Tokens | 79103 | 87.9 | 97.0 |
| LR + RF + KNN (Levi) | 85774 | 95.3 | 95.6 |
| LR + RF + KNN (Tokens) | 75423 | 83.8 | 97.7 |
| LR + RF + KNN (Levi + Tokens) | 72477 | 80.5 | 99.0 |
Abbreviations: LR, logistic regression classifier. RF, random forest classifier. KNN, K-nearest neighbors classifier. Levi, Levenshtein distance encoding. Tokens, Frequency Tokenization Encoding.
In these experiments we observe that the combination of models across encoding methods generally leads to better performance when compared to the combination of models within an encoding method. One potential reason for this is that a feature of the encoding method itself leads different models to make similar predictions. This would also explain the high prediction overlap of the models trained with the Levenshtein distance encoding. As the different encoding methods emphasize unique aspects of the source data, the combination of the encoding methods allows the models to escape issues inherent to the encoding, generating a more robust prediction. We also observe that within encoding methods, increasing the number of models causes a drop in the overlap percentage, but results in a higher accuracy in the overlap.
Discussion
This study has shown that automated machine learning methods can be successful in mapping laboratory data to LOINC codes in a real-world oncology dataset. Overall our best performing model had an accuracy of 94.5% on the full hold-out dataset, and our ensemble method had an accuracy of 99.0% on 80.5% of the hold-out dataset. Additionally we have shown that it's possible to make predictions to LOINCs with the same level of specificity as clinical terminologists, including marking rows with insufficient information as EXCLUDED. Success in mapping to EXCLUDED is of particular value, as having source data without enough information to map accurately is a tenant of a real world dataset, and provides an additional challenge for machine learning models.
We examined cases where our model's predictions were incorrect and found that in the majority of cases, the incorrect prediction was extremely similar to the actual label. For example, in one instance the model predicted LOINC 20570-8 which has target term "Hematocrit Bld VFr Pt Qn", and the labeled LOINC was 4544-3 which has target term "Hematocrit Bld VFr Pt Qn Automated count". While these are very similar, because we want to maintain the highest degree of accuracy possible it still counts against the model's performance. This highlights the importance of having both correct and consistent mappings in our labeled dataset. While we don't have a measure of the accuracy of the manual mappings that we used to create our labels, in future work we plan to use the incorrect model predictions as a starting point to review the accuracy of existing labeled rows.
Our results are comparable in accuracy to the best reported results from prior studies involving automated laboratory data mapping.15-17 The high accuracy and precision of the ensemble learning allows for supplemental use of automated mapping methods alongside clinical terminologists in a range of potential applications. Aside from directly predicting new mappings, this method can also be used as an extra level of quality analysis for maps done manually. Additionally it can be used to verify existing maps, especially those done many years in the past, to identify potentially incorrect mappings to be reexamined. In summary this study has shown that using automated mapping methods has potential to not only reduce manual harmonization time by clinical terminologists, but also to provide an immediate quality improvement to real-world datasets.
The novelties and areas of strength in this study include (a) assessing the place of automated laboratory mapping methods in a real-world dataset, (b) demonstrating the efficacy of such methods on an oncology-specific dataset, (c) implementation of two novel laboratory free text encoding schemes, and (d) high prediction accuracy using ensemble learning. In future work, we would like to explore other encoding methods, as well as attempt to use multiple encoding methods for different fields within the same model. Furthermore, we would like to explore the extensibility of these techniques to other domains, such as free text medication administration data.
This study has a few notable limitations. Firstly, while the methods in this study are reproducible at no cost, the labeled dataset of oncology laboratory data to LOINC codes used in this study is not publicly available. However, there may be opportunity in the future to open source some of these mappings for the larger research community. A second limitation is that while these encoding and models performed well on oncology data, there may be oncology-specific aspects about laboratory data that would cause these methods to perform poorly in other medical domains. A limitation with our encoding methodology is that the encoded vectors created are very wide, which can lead to long model train times and reduced development speed. Encoding with Levenshtein ratios further increases this issue, as comparing strings is an expensive operation. Another limitation is that because machine learning classifiers can only predict LOINCs that they have been trained on, any new additions to the LOINC catalog or changes in mapping policy would be impossible for the classifier to predict. Whenever this happens all models would need to be retrained on additional applicable training data. Lastly, our primary focus is reducing the burden of clinical terminologists and we are therefore satisfied with high accuracy on a subset of data. Other applications however might require full automation, for which our study has comparably high accuracy, but decreases the applicableness of our ensemble learning methods.
Conclusion
As the use of real-world data continues to grow, automated methods that allow for accurate aggregation and harmonization of data from EHRs become increasingly important. Mapping free text laboratory data to LOINC is important before the data can effectively be leveraged for use in research, however the manual mapping process is extremely time intensive. This study has shown that through the use of automated methods, we can significantly lower this burden. Free text medical data is challenging to encode for existing NLP methods, and so we developed specific encoding methodologies to effectively capture and encode the information required to map laboratory data. We demonstrated that with these encodings and the use of ensemble learning, training an automated laboratory data classifier with high accuracy is not only possible, but can provide immediate value to the creation of a high-quality real-world oncology dataset.
Figures & Table
References
- [1].Chaudhry B, Wang J, Wu S, Maglione M, Mojica W, Roth E, et al. Systematic review: impact of health information technology on quality, efficiency, and costs of medical care. Annals of internal medicine. 2006;144(10):742–752. doi: 10.7326/0003-4819-144-10-200605160-00125. [DOI] [PubMed] [Google Scholar]
- [2].Murdoch TB, Detsky AS. The inevitable application of big data to health care. Jama. 2013;309(13):1351–1352. doi: 10.1001/jama.2013.393. [DOI] [PubMed] [Google Scholar]
- [3].Khozin S, Blumenthal GM, Pazdur R. Real-world data for clinical evidence generation in oncology. JNCI: Journal of the National Cancer Institute. 2017;109(11):djx187. doi: 10.1093/jnci/djx187. [DOI] [PubMed] [Google Scholar]
- [4].Hersh WR. Adding value to the electronic health record through secondary use of data for quality assurance, research, and surveillance. Clin Pharmacol Ther. 2007;81:126–128. [PubMed] [Google Scholar]
- [5].McDonald CJ, Huff SM, Suico JG, Hill G, Leavelle D, Aller R, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clinical chemistry. 2003;49(4):624–633. doi: 10.1373/49.4.624. [DOI] [PubMed] [Google Scholar]
- [6].Baorto DM, Cimino JJ, Parvin CA, Kahn MG. Combining laboratory data sets from multiple institutions using the logical observation identifier names and codes (LOINC) International journal of medical informatics. 1998;51(1):29–37. doi: 10.1016/s1386-5056(98)00089-6. [DOI] [PubMed] [Google Scholar]
- [7].Duong CD, Loh JY. Laboratory monitoring in oncology. Journal of Oncology Pharmacy Practice. 2006;12(4):223–236. doi: 10.1177/1078155206072982. [DOI] [PubMed] [Google Scholar]
- [8].Kim H, El-Kareh R, Goel A, Vineet F, Chapman WW. An approach to improve LOINC mapping through augmentation of local test names. Journal of biomedical informatics. 2012;45(4):651–657. doi: 10.1016/j.jbi.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Loinc® Indianapolis, IN: Regenstrief Institute, Inc. Logical Observation Identifiers Names and Codes (LOINC®). http://www.loinc.org. Accessed March 1, 2021.
- [10].Lin MC, Vreeman DJ, McDonald CJ, Huff SM. AMIA Annual Symposium Proceedings. vol. 2010. American Medical Informatics Association; 2010. Correctness of voluntary LOINC mapping for laboratory tests in three large institutions; p. 447. [PMC free article] [PubMed] [Google Scholar]
- [11].Khan AN, Griffith SP, Moore C, Russell D, Rosario AC, Jr, Bertolli J. Standardizing laboratory data by mapping to LOINC. Journal of the American Medical Informatics Association. 2006;13(3):353–355. doi: 10.1197/jamia.M1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Barda AJ, Ruiz VM, Gigliotti T, Tsui F. An argument for reporting data standardization procedures in multi-site predictive modeling: case study on the impact of LOINC standardization on model performance. JAMIA open. 2019;2(1):197–204. doi: 10.1093/jamiaopen/ooy063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lin MC, Vreeman DJ, Huff SM. AMIA Annual Symposium Proceedings. vol. 2011. American Medical Informatics Association; 2011. Investigating the semantic interoperability of laboratory data exchanged using LOINC codes in three large institutions; p. 805. [PMC free article] [PubMed] [Google Scholar]
- [14].Lau LM, Johnson K, Monson K, Lam SH, Huff SM. Proceedings of the AMIA Symposium. American Medical Informatics Association; 2000. A method for the automated mapping of laboratory results to LOINC; p. 472. [PMC free article] [PubMed] [Google Scholar]
- [15].Fidahussein M, Vreeman DJ. A corpus-based approach for automated LOINC mapping. Journal of the American Medical Informatics Association. 2014;21(1):64–72. doi: 10.1136/amiajnl-2012-001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sun JY, Sun Y. A system for automated lexical mapping. Journal of the American Medical Informatics Association. 2006;13(3):334–343. doi: 10.1197/jamia.M1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Parr SK, Shotwell MS, Jeffery AD, Lasko TA, Matheny ME. Automated mapping of laboratory tests to LOINC codes using noisy labels in a national electronic health record system database. Journal of the American Medical Informatics Association. 2018;25(10):1292–1300. doi: 10.1093/jamia/ocy110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dong X, Li J, Soysal E, Bian J, DuVall SL, Hanchrow E, et al. COVID-19 TestNorm: A tool to normalize COVID-19 testing names to LOINC codes. Journal of the American Medical Informatics Association. 2020;27(9):1437–1442. doi: 10.1093/jamia/ocaa145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kim M, Shin SY, Kang M, Yi BK, Chang DK. Developing a standardization algorithm for categorical laboratory tests for clinical big data research: retrospective study. JMIR medical informatics. 2019;7(3):e14083. doi: 10.2196/14083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ma X, Long L, Moon S, Adamson BJ, Baxi SS. Comparison of population characteristics in real-world clinical oncology databases in the US: Flatiron Health, SEER, and NPCR. medRxiv. 2020 [Google Scholar]
- [21].Birnbaum B, Nussbaum N, Seidl-Rathkopf K, Agrawal M, Estevez M, Estola E, et al. Model-assisted cohort selection with bias analysis for generating large-scale cohorts from the EHR for oncology research. arXiv preprint arXiv:200109765. 2020 [Google Scholar]
- [22].Levenshtein VI. Soviet physics doklady. vol. 10. Soviet Union; 1966. Binary codes capable of correcting deletions, insertions, and reversals; pp. 707–710. [Google Scholar]
- [23].Liu Y, Ge T, Mathews KS, Ji H, McGuinness DL. Exploiting task-oriented resources to learn word embeddings for clinical abbreviation expansion. arXiv preprint arXiv:180404225. 2018 [Google Scholar]
- [24].Wu Y, Denny JC, Rosenbloom ST, Miller RA, Giuse DA, Xu H. AMIA annual symposium proceedings. vol. 2012. American Medical Informatics Association; 2012. A comparative study of current clinical natural language processing systems on handling abbreviations in discharge summaries; p. 997. [PMC free article] [PubMed] [Google Scholar]
- [25].Moon S, McInnes B, Melton GB. Challenges and practical approaches with word sense disambiguation of acronyms and abbreviations in the clinical domain. Healthcare informatics research. 2015;21(1):35. doi: 10.4258/hir.2015.21.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hoerl AE, Kennard RW. Ridge regression: Biased estimation for nonorthogonal problems. Technometrics. 1970;12(1):55–67. [Google Scholar]
- [27].Segal MR. Machine learning benchmarks and random forest regression. 2004 [Google Scholar]
- [28].Zhang ML, Zhou ZH. ML-KNN: A lazy learning approach to multi-label learning. Pattern recognition. 2007;40(7):2038–2048. [Google Scholar]
- [29].Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: Machine learning in Python. the Journal of machine Learning research. 2011;12:2825–2830. [Google Scholar]
- [30].Dietterich TG. Ensemble methods in machine learning. In: International workshop on multiple classifier systems. Springer; 2000. pp. 1–15. [Google Scholar]
- [31].Zhang C, Ma Y. Ensemble machine learning: methods and applications. Springer; 2012. [Google Scholar]
- [32].Dietterich TG, et al. Ensemble learning. The handbook of brain theory and neural networks. 2002;2:110–125. [Google Scholar]