Abstract
Proper embryonic and post-natal skeletal development require coordination of a myriad of complex molecular mechanisms. Disruption of these processes, through genetic mutation, contributes to variation in skeletal development. We developed a high-throughput ENU-induced saturation mutagenesis skeletal screening approach in mice to identify genes required for proper skeletal development. Here, we report initial results from live-animal X-ray and dual-energy X-ray absorptiometry (DXA) imaging of 27,607 G3 mice from 806 pedigrees, testing the effects of 32,198 coding/splicing mutations in 13,020 genes. 39.7% of all autosomal genes were severely damaged or destroyed by mutations tested twice or more in the homozygous state. Results from our study demonstrate the feasibility of in vivo mutagenesis to identify mouse models of skeletal disease. Furthermore, our study demonstrates how ENU mutagenesis provides opportunities to create and characterize putative hypomorphic mutations in developmentally essential genes. Finally, we present a viable mouse model and case report of recessive skeletal disease caused by mutations in FAM20B. Results from this study, including engineered mouse models, are made publicly available via the online Mutagenetix database.
Keywords: DXA, genetic animal models, Molecular pathways-development, Wnt/B-catenin/LRPs, Osteoblasts
INTRODUCTION
Development and maintenance of the skeleton is regulated by multiple cell types within the bone periosteum, growth plate, and bone marrow, as well as numerous other organ systems, such as the muscular(1), nervous(2), vascular(3), and endocrine(4) systems. In addition to environmental influences such as age, gender, ethnicity, and physical activity, genetic factors also contribute to variation in bone phenotypes, for example bone mineral density (BMD)(5).
Early positional cloning studies identified the genetic basis for numerous human Mendelian skeletal diseases(6). Subsequent microarray-based genome-wide association studies (GWAS) enabled identification of numerous genetic loci associated with variation in complex human phenotypes, such as BMD and fracture risk(7,8). However, GWAS often require subsequent fine-mapping approaches to resolve causal genes underlying association signals. There is an emerging potential for GWAS polygenic risk scores to explain more of the genetic variation in quantitative bone measures(9); however, the majority of the variation in bone traits remains unexplained, reflecting the limitations of GWAS, which focus on common variants that confer small to modest effect sizes. Genome sequencing has potential to identify significant associations with increasingly low-frequency or rare variants(10), but practical challenges of cost, demands for large-scale data storage, and the need to develop computationally efficient means to analyze these datasets persist.
In vivo forward-genetic screens provide an alternative approach to identify genes required for diverse developmental processes. Forward-genetic screens utilize model systems that are amenable to mutagenesis and that allow for high-throughput, accurate measurement of phenotypic variation, such as in Drosophila(11), zebrafish(12,13), and mice(14). To identify genes with non-redundant functions in skeletal development, we implemented a live-animal Dual-Energy X-ray Absorptiometry (DXA) and X-ray radiography phenotyping approach as part of a previously published large-scale forward-genetic mouse screen(15).
Here, we report selected examples demonstrating how our approach advances identification of pre-clinical mouse models of human skeletal disease and how ENU mutagenesis provides opportunities to characterize viable recessive alleles in essential genes.
MATERIALS AND METHODS
ENU mutagenesis, breeding, and genotyping
All animal protocols were approved by the IACUC of UT Southwestern Medical Center. Male C57BL/6J mice were purchased from the Jackson Laboratory, and male mice were mutagenized with ENU as previously described(16). Following mutagenesis, mice were bred as shown in Figure 1A. ENU-induced alleles were detected in G1 male mice by exome sequencing, and all non-synonymous ENU-induced alleles were genotyped in all G2- (female) and G3-generation mice using a targeted capture and sequencing approach, as previously described(15).
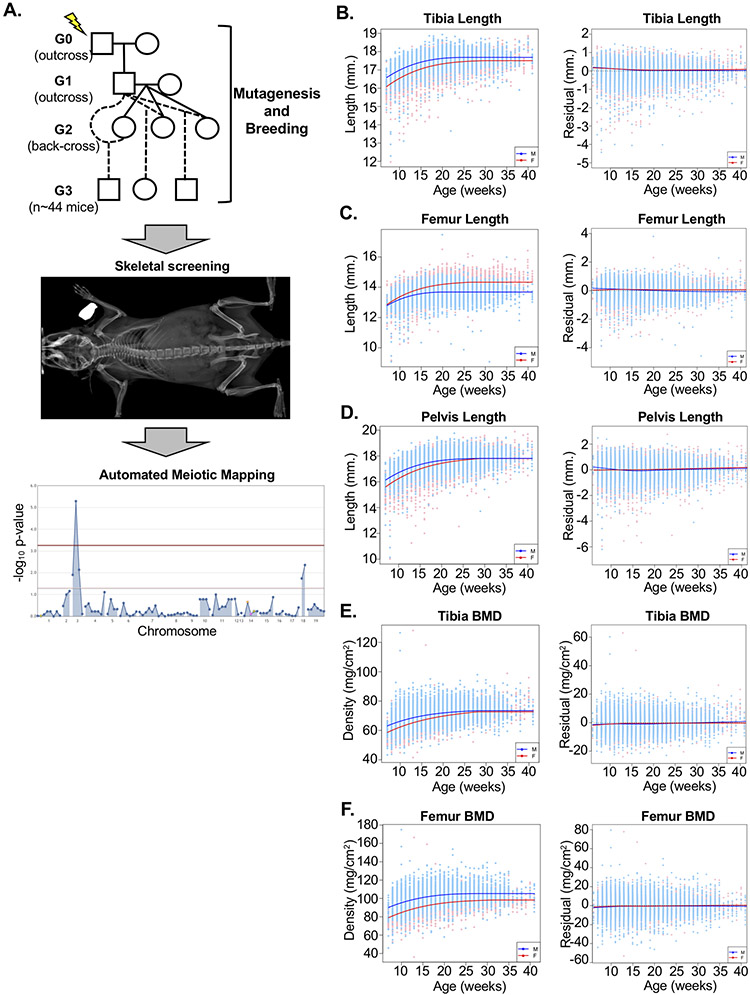
Figure 1.
ENU mutagenesis and skeletal screening strategy. (A) ENU-mutagenized G0 male mice are outcrossed with C57BL/6J females to generate male G1 mice, which are then outcrossed with C57BL/6J females. Resulting G2 female mice are back-crossed to their G1 sire, and all resulting G3 mice undergo DXA and X-ray imaging. Automated meiotic mapping identifies ENU-induced alleles segregating with phenotypic variation, and statistical significance is visualized by Manhattan plot. Alleles exceeding Bonferroni multiple-test correction (red line) are considered associated with phenovariance. (B-F) Left: Prior to statistical modeling, all phenotypes were significantly correlated with age and gender in both male (blue) and female (pink) mice. Numbers of mice used for modeling and percent of variation explained by age and gender are shown in Supplementary Table 2. Right: Residual differences between predicted and actual values among N~25,300 mice confirm statistical modeling (see Methods) successfully removes variation due to age and gender. Dashed horizontal line indicates no variation. Solid colored lines are smoothing lines (estimated by lowess) indicating the trends in residual measures against age. Individual male mice are shown in blue and female mice shown in pink, with male and female trend lines shown in blue and red, respectively.
Skeletal screening
Mice were anesthetized with 2% isoflurane via precision vaporizer and imaged with a Faxitron UltraFocusDXA instrument with continued anesthesia. Standard X-ray and DXA images were acquired for two mice simultaneously. Skeletal phenotypes were measured using Faxitron software. For each trait, the average of the measurements for each side was calculated and phenotypes uploaded to the Mutagenetix database for automated meiotic mapping. Additional details regarding automated meiotic mapping were previously reported(15).
Generation of validation mice using CRISPR
Super-ovulation of 3-week-old C57BL/6N females was performed by administering 5 International Units (IU) PMSG (pregnant mare serum gonadotropin) followed by 5 IU hCG 48hrs later, then females were mated with C57BL/6N stud males and embryos harvested next day (D0.5).
crRNA (Integrated DNA Technologies, Coralville, IA) was annealed to tracRNA (Integrated DNA Technologies, Coralville, IA) by heating to 95°C for 5 mins then ramping down to 25°C at 5°C/min intervals. The Ribonucleoprotein (RNP) complex was generated by annealing the crRNA/tracRNA complex with Cas9 endonuclease (Integrated DNA Technologies, Coralville, IA) for 10 minutes at room temperature.
For knock-in models, single stranded oligodeoxynucleotides (ssODN) were added. CRISPR reagents (50ng/ul Cas9, 50ng/ul sgRNA, and 50ng/ul ssODN) were injected into the cytoplasm of embryos. Alternatively, CRISPR reagents (final concentrations of 300ng/ul Cas9, 300ng/ul sgRNA, and 500ng/ul ssODN) were delivered via electroporation using a NEPA21 Super Electroporator (NEGPAGENE, Ichikawa, Japan) or Gene Pulser (Biorad, Hercules, CA). Embryos were implanted into pseudo-pregnant mothers at D0.5 and resulting pups screened by Sanger sequencing.
CRISPR crRNA and single-stranded donor template sequences are provided in Supplementary Table 1. All genotyping primers are available upon request or are available on the Mutagenetix website.
Micro-CT analysis
Proximal tibiae were imaged using a Skyscan 1072 (Bruker, Aartselaar, Belgium) set at 50kV/200μA and using a 0.5mm Al filter. Images were obtained at 8μm pixel size with a rotation step of 0.4° between each image. The 3D image stack was reconstructed using NRecon version 1.7.4.6 (Bruker), and trabecular parameters were measured using methods recommended by Bruker. Using a reference level at the growth plate, the trabecular region was then defined as 200 slices offset by 50 slices from the reference using CTan software (Bruker). Trabecular segmentation excluded cortical bone, and following thresholding, all samples were analyzed for trabecular bone parameters. Statistically significant differences were detected using 2-sided T-tests.
Analysis of DMP1 protein
Femurs were embedded in paraffin, sectioned, and analyzed by immunofluorescent staining using a DMP1 antibody (provided by Dr. Chunlin Qin at Texas A&M University, 1:400) and DAPI (for nuclei) as previously described(17). Images were processed using ImageJ software.
Western blot analysis for DMP1 secretion was performed as previously described(18). Briefly, MC3T3-E1 preosteoblast cells (ATCC, Manassas, VA) were cultured in α-minimum essential media (MEM) supplemented with 10% fetal bovine serum (FBS) in a humidified incubator with 5% CO2 at a temperature of 37 °C. Cells were seeded into 6-well plates at a density of 12x104 cells per well. On the next day, cells were transfected with 2 μg of a pCDNA3 empty vector or a construct expressing hemagglutinin (HA)-tagged DMP1 or DMP1L10P using X-tremeGENE 9 reagent (Roche, Indianapolis, IN) according to the manufacturer’s instructions. Total cell lysates and conditioned media were harvested 24 hours after transfection.
For Western-blotting, 5 μg of the total cell lysates as well as total protein extracted from 500 μl of conditioned medium by StrataClean resin (Agilent Technologies, Inc., Santa Clara, CA) were electrophoresed on a 4-15% gradient SDS-PAGE gel and transferred onto PVDF membrane (EMD Millipore). Membranes were blocked in 5% milk (LabScientific, Highlands, NJ) for 1 hour at room temperature and immunoblotted with mouse anti-HA monoclonal antibody (BioLegend, San Diego, CA; 1:1000) overnight at 4°C, followed by incubation with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Santa Cruz Biotechnology, Inc., Dallas, TX; 1:1000) for 2 hrs at room temperature. The immunostained protein bands were detected with ECL™ Chemiluminescent detection reagents (Pierce Biotechnology, Inc., Rockford, IL) and imaged using a CL-XPosure film (Pierce Biotechnology, Inc., Rockford, IL). Experiments were repeated three times.
Exome sequencing and analysis
The affected subject was enrolled in the “Diagnosis and Treatment of Patients with Inborn Errors of Metabolism and Other Genetic Disorders” protocol approved by the Institutional Review Board at the National Institutes of Health (NCT00369421, IRB#76HG0238, approved 01/18/2021). The subject’s guardians provided informed consent. Genomic DNA was obtained from lymphocytes, and exome sequencing was performed through a clinical laboratory using a proprietary approach. Reads were aligned to the human reference genome (GRCh37/hg19) and variants analyzed using the GeneDx XomeAnalyzer tool. Variants in FAM20B were reported by the clinical laboratory following validation by Sanger sequencing.
In silico analysis of FAM20 kinases
Normal Mode Analysis (NMA) and Elastic Network Contact Model (ENCoM) analyses were performed using DynaMut, with verification of results in standalone Bio3d and ENCoM implementations, using established methods(19-21). Data was visualized in R using ggplot2, and structure depictions and alignments were generated using PyMol(22-24).
Statistical analysis
The relationship between skeletal phenotypes and age and gender of mice were estimated using linear regression models, including polynomial (quadratic and cubic) terms for age. Since the growth patterns in bone length must be non-decreasing, peak growth was estimated from polynomial regression models (above) and the values of phenotypes were assumed to be constant beyond the age of maximal growth. Regression models also included gender-by-age interaction terms, allowing for a different prediction equation for male and female mice. Outliers (with residual values >3 SD from the mean) were excluded when estimating the final prediction equation. Residualized phenotypes (calculated as the observed minus predicted value) were used to test for association with ENU-induced mutations.
RESULTS
Design and implementation of the skeletal screen
A schematic of the skeletal phenotype screening pipeline is shown in Figure 1A. C57BL/6J male mice, designated G0, are treated with ENU and subsequently crossed to non-mutagenized C57BL/6J females. Each resulting G1 male carries transmitted ENU-induced mutations in the heterozygous state, which are detected by exome sequencing. G1 male mice are crossed to C57BL/6J females, and the resulting G2 female offspring are subsequently back-crossed to their G1 sire to produce G3 pups that are screened for skeletal phenotypes. Non-synonymous ENU-induced mutations (~60 per pedigree(15)) are genotyped in G2 females and in all G3 mice by targeted sequencing, yielding REF (homozygous for the reference allele), HET (heterozygous for the variant allele), and VAR (homozygous for the variant allele) genotypes for each variant locus with respect to the C57BL/6J reference genome (GRCm38.p6)(15). All breeding is performed on the C57BL/6J background to eliminate variation due to mouse strains or C57BL/6 sub-strain differences.
G3 mice and a small independent control cohort of non-mutagenized C57BL/6J mice undergo skeletal screening by live-animal DXA and standard X-ray radiography to measure tibia and femur BMD (measured by DXA) as well as tibia, femur, and pelvis length (measured by X-ray) phenotypes. Because the ages of G3 mice vary at the time of screening, we first evaluated the contribution of non-genetic factors, such as age and gender, to variation in skeletal phenotypes. Using a sub-set of G3 and C57BL/6J mice (n~11,650 mice for lengths; n~16,330 for BMD), we used linear regression models (see Methods) to estimate the normal physiologic relationship between skeletal phenotypes, age, and gender. Together, these factors accounted for 33-43% of variance in measured skeletal phenotypes (Supplementary Table 2). Regression models were used to estimate predicted phenotypic measures for each animal, and residual differences were calculated for each phenotype. These models were subsequently validated using a larger set of N~25,300 G3 and C57BL/6J mice, confirming that our approach successfully removed variation due to age and gender (Figure 1B-F). This approach approximates the use of human Z-scores to identify clinically significant differences compared to age- and sex-specific population averages.
To identify ENU-induced alleles associated with variation in age- and gender-adjusted skeletal phenotypes, automated meiotic mapping tests for association between genotype and phenotype residuals using dominant, recessive, and additive genetic models.
Novel mouse models of human mucopolysaccharidoses
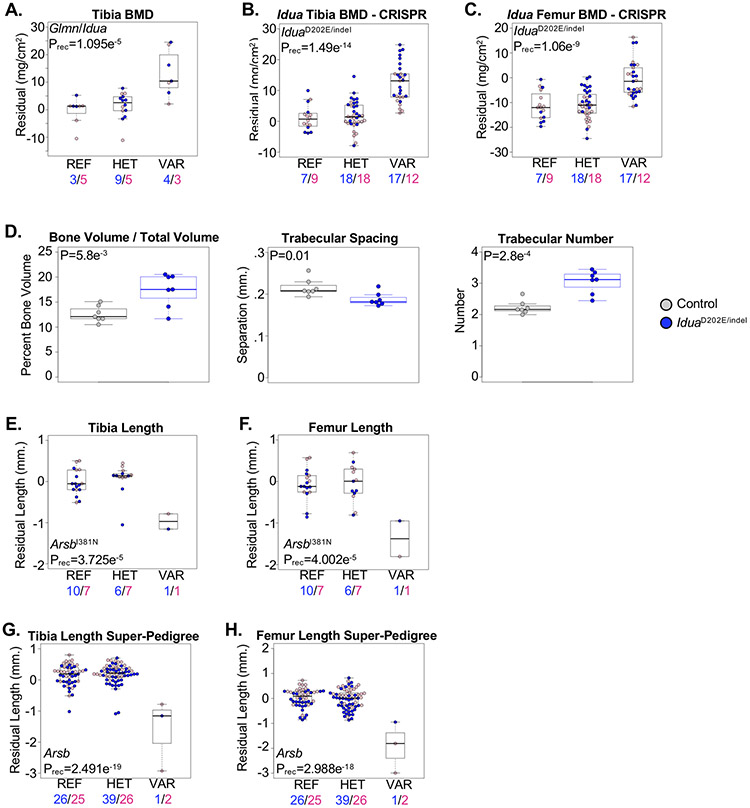
Random mutagenesis provides opportunities to discover novel ENU-induced mouse models of previously-reported human diseases. Here, we describe two novel mouse models developing skeletal phenotypes associated with variants in genes known to cause human mucopolysaccharidoses (MPS). First, we identified a locus co-segregating alleles in the Alpha-L-iduronidase (IduaD202E) and the FKBP-associated glomulin (GlmnS284P) genes associated with a recessive increase in BMD (Figure 2A). Recessive mutations in human IDUA cause MPS1 (Hurler syndrome), which presents with a variety of skeletal abnormalities(25). Idua knock-out mice develop a high bone mass phenotype(26); therefore, we selected Idua for validation by generating IduaD202E knock-in mice. IduaD202E knock-in mice were inter-crossed with mice harboring a predicted loss-of-function frameshift mutation (Iduaindel), and skeletal phenotypes were evaluated in 4-month-old male and female IduaD202E/indel compound heterozygous mice. Compound heterozygous IduaD202E/indel mice developed a high bone mass phenotype, and μCT analysis of the proximal tibia from age-matched male mice confirmed increased trabecular bone volume (BV/TV), increased trabeculae number (Tb.N.), and reduced trabeculae separation (Tb.S.) compared to control (Figure 2B-D). These results confirm the IduaD202E allele is loss-of-function and represents a novel mouse model of human MPS1. Furthermore, these results demonstrate that our residuals-based approach is capable of identifying significantly-associated phenotypes that are recapitulated using age- and gender-matched mice.
Figure 2.
Mouse models of human Mucopolysaccharidoses. (A) Mice homozygous for co-segregating alleles in the Glmn and Idua genes (VAR) developed a high bone mass phenotype compared to mice heterozygous for the alleles (HET) or homozygous for the reference allele (REF). Male and female mice are shown with blue and pink symbols, respectively, and the numbers of mice are shown below. (B,C) Compound heterozygous or homozygous mice harboring the IduaD202E and Idua p.S204Ffs*1 (Iduaindel) alleles (VAR) developed high bone mass evident in the (B) tibia and (C) femur compared to mice heterozygous for either allele (HET) or homozygous for the reference allele (REF). Male and female mice are shown with blue and pink symbols, respectively, and the numbers of mice are shown below. (D) μCT analysis of the proximal tibia of 5-month-old male mice revealed increased trabecular bone volume/tissue volume (BV/TV), decreased trabecular separation (Tb. S.), and increased trabecular number (Tb. N.) in compound heterozygous IduaD202E/indel mice (blue) compared to litter-matched control (Idua+/+, Idua+/D202E, and Idua+/indel pooled) mice (grey). Statistically significant differences were determined using 2-sided T-tests. (E,F) Mice homozygous for the ArsbI381N allele (VAR) developed shorter (E) tibia and (F) femur lengths compared to mice heterozygous for the allele (HET) or homozygous for the reference allele (REF). Male and female mice are shown with blue and pink symbols, respectively, and the numbers of mice are shown below. (G,H) Super-pedigree analysis combining two pedigrees segregating alleles in Arsb. Mice homozygous for the ENU alleles (VAR) developed reduced (G) tibia and (H) femur lengths compared to mice heterozygous for either allele (HET) and homozygous for the reference allele (REF). Male and female mice are shown with blue and pink symbols, respectively, and the numbers of mice are shown below. Data are median with interquartile range (IQR); whiskers extend up to 1.5x the IQR.
MPS6 (Maroteaux-Lamy syndrome), which presents with short stature and other skeletal abnormalities, is caused by recessive mutations in ARSB, encoding Arylsulfatase B(27,28). Similar to Idua knock-out mice, mice lacking Arsb develop skull and limb abnormalities(29). We identified the ArsbI381N allele significantly associated with a recessive reduction in tibia and femur lengths (Figure 2E,F). We surveyed additional Arsb alleles in the Mutagenetix database and identified a critical splice site mutation (Arsbsplice) predicted to result in a loss of function. However as only a single homozygous G3 mouse was identified in the pedigree, we used a “super-pedigree” approach to investigate the combined association of both the Arsbsplice and ArsbI381N alleles with variation in bone lengths. Indeed, combining results from both pedigrees strengthened the association of Arsb with reduced skeletal growth (Figure 2G,H).
Mouse models of congenital hypothyroidism causing growth restriction
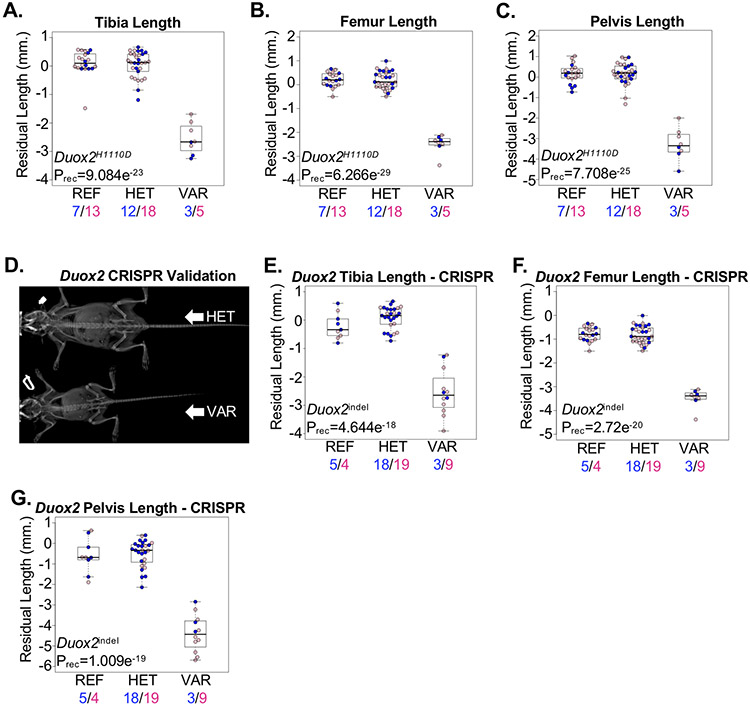
As illustrated by the identification of multiple MPS-associated genes, random mutagenesis is powered to independently identify phenotypes associated with alleles in different genes converging on the same molecular, biochemical, or developmental pathway. Thyroglobulin, encoded by the Tg gene, is an iodoglycoprotein expressed by the thyroid gland. Iodination of thyroglobulin requires the oxidation of iodide from hydrogen peroxide (H2O2) by thyroid peroxidase. Generation of H2O2 at the apical membrane of thyrocytes is dependent on the activity of Dual-function oxidases 1 and 2, encoded by the Duox1 and Duox2 genes, respectively(30). Recessive loss-of-function mutations in the human TG or DUOX2 genes cause recessive thyroid dyshormonogeneses 3 and 6, respectively(31,32). We identified a significant association of the TgI1352K allele with recessive dwarfism (Supplementary Figure 1A-C). The reduced growth observed in homozygous TgI1352K mice was similar to other Tg-mutant mouse lines reported by us and others(33,34). Similarly, we identified a significant association of the Duox2H1110D allele with recessive dwarfism (Figure 3A-C). To validate the association with Duox2, we engineered Duox2 knock-out mice. Consistent with results from G3 mice and a previously published mouse model harboring a spontaneous Duox2V674G mutation(35), knock-out of Duox2 resulted in recessive dwarfism (Figure 3D-G). Although the developmental manifestations of Duox2 and Tg mouse lines reported here are consistent with a congenital hypothyroidism in these mice, this was not directly tested. Furthermore, consistent with a known primary role for DUOX2 in the thyroglobulin biosynthesis pathway and a lack of any observable phenotype in Duox1 knockout mice(36,37), mice homozygous for the Duox1Y514X or Duox1Q196X nonsense alleles showed no significant differences in skeletal development (Supplementary Figure 2).
Figure 3.
Dwarfism caused by recessive mutations in Duox2. (A-C) Mice homozygous for the Duox2H1110D allele (VAR) developed reduced (A) tibia, (B) femur, and (C) pelvis lengths compared to mice heterozygous for the allele (HET) or mice homozygous for the reference allele (REF). Male and female mice are shown with blue and pink symbols, respectively, and the numbers of mice are shown below. (D) Representative radiograph of CRISPR-engineered mice heterozygous (HET) for a Duox2 p.G497Gfs*5 frameshift indel or homozygous (VAR) for a Duox2 p.F500Ifs*6 frameshift indel. (E-G) CRISPR-engineered mice homozygous (VAR) for the Duox2 frameshift indels developed shorter (E) tibiae, (F) femurs, and (G) pelvis bone lengths compared to heterozygous (HET) mice or mice homozygous for the reference allele (REF). Male and female mice are shown with blue and pink symbols, respectively, and the numbers of mice are shown below. Data are median with interquartile range (IQR); whiskers extend up to 1.5x the IQR.
Mouse models of altered bone mineralization
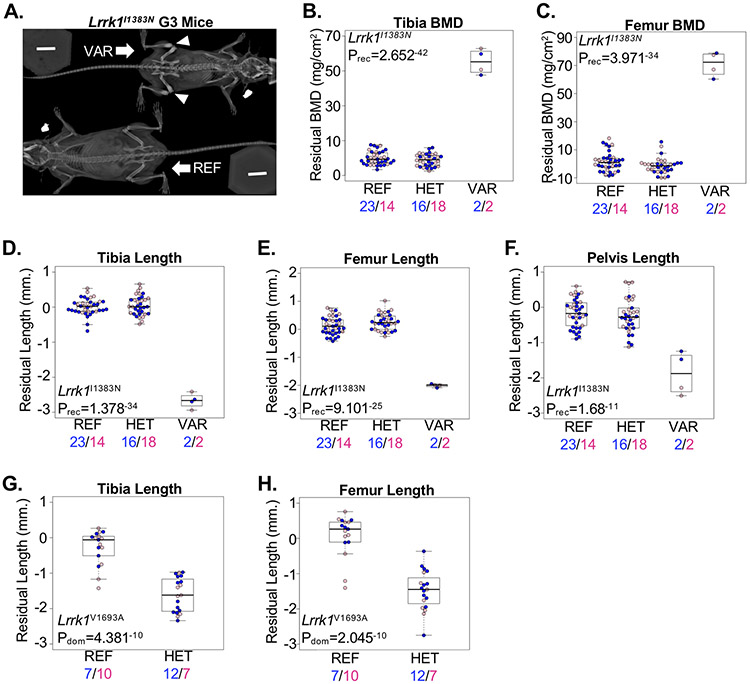
In addition to differences in longitudinal growth, our skeletal screen detects ENU-induced alleles associated with differences in bone mineralization. For example, recessive loss-of-function mutations in the LRRK1 gene, encoding the Leucine rich repeat kinase 1 protein, were recently identified in patients with osteosclerotic metaphyseal dysplasia(38,39). We detected two alleles (Lrrk1I1383N and Lrrk1V1693A) within the Lrrk1 gene significantly associated with variation in skeletal development. The Lrrk1I1383N allele was associated with a recessive increase in BMD and reduced bone lengths that were evident upon X-ray imaging (Figure 4A-F). Results from the Lrrk1I1383N allele are similar to homozygous knock-out (Lrrk1Δex16-19) mice that develop a high bone mass phenotype due to osteoclast dysfunction and impaired bone resorption(40). Interestingly, we also identified a second, more distal allele (Lrrk1V1693A) associated with a dominant reduction in longitudinal bone growth but not a BMD phenotype (Figure 4G,H). No homozygous mice were available for screening, and the Lrrk1V1693A allele was nominally associated (p=0.003) with a recessive lethality phenotype, though this was not significant after correcting for the number of ENU-induced alleles tested in the pedigree. In contrast to the Lrrk1Δex16-19 allele, mice homozygous for a distal C-terminal deletion allele (Lrrk1Δex24-29;Lrrk1tm1.1Mjff) presented with a pre-weaning lethality phenotype(41). Thus, we identified two independent alleles within Lrrk1 associated with different recessive and dominant skeletal phenotypes. These results suggest allelic heterogeneity within Lrrk1 that differentially affects longitudinal skeletal growth, bone mineralization/resorption, and possibly survival.
Figure 4.
Allelic heterogeneity in the BMD-associated Lrrk1 gene. (A) Radiograph of mice homozygous for the reference allele (REF) or homozygous for the Lrrk1I1383N allele (VAR) showing osteosclerotic bone at the distal femur and proximal tibia (arrowhead). (B,C) Mice homozygous for the Lrrk1I1383N allele (VAR) developed a high (B) tibia and (C) femur bone mass phenotype compared to mice heterozygous for the same allele (HET) or homozygous for the reference allele (REF). (D-F) Mice homozygous for the Lrrk1I1383N allele (VAR) developed reduced (D) tibia, (E) femur, and (F) pelvis lengths compared to mice heterozygous for the same allele (HET) or homozygous for the reference allele (REF). (G,H) Mice heterozygous for the Lrrk1V1693A allele (HET) developed shorter (G) tibia and (H) femur lengths compared to mice homozygous for the reference allele (REF); no mice homozygous for the variant allele were observed. Male and female mice are shown with blue and pink symbols, respectively, and the numbers of mice are shown below. Data are median with interquartile range (IQR); whiskers extend up to 1.5x the IQR.
Characterizing a series of Lrp5 alleles in vivo
The Low density lipoprotein receptor-related protein 5 (LRP5) functions as a co-receptor with LRP6 and frizzled to regulate canonical WNT signaling(42). In mice, activation and inhibition of WNT signaling through different genetic perturbations results in increased and decreased bone mass, respectively(43). Loss-of-function mutations in human LRP5 are associated with recessive low bone mass, while mutations affecting interactions between LRP5 and its inhibitors (i.e., gain-of-function) are associated with dominant high bone mass(44-46). Multiple mouse models have been engineered that demonstrate the high and low bone mass phenotypes associated with Lrp5 mutations(47-49).
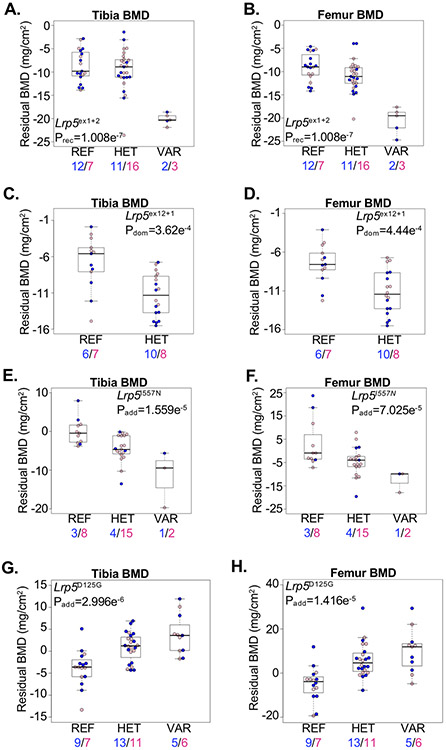
We tested whether an allelic series of ENU-induced alleles in mice could identify both gain- and loss-of-function mutations associated with opposite skeletal phenotypes. We identified multiple alleles in Lrp5 associated with variation in bone mass. Three alleles, including two predicted loss-of-function canonical splice site mutations (Lrp5ex1+2 and Lrp5ex12+1) and a missense mutation (Lrp5I557N) were associated with reduced bone density (Figure 5A-F). Interestingly, the Lrp5ex1+2 allele was associated with a recessive low bone mass phenotype and spontaneous tibia fracture (Prec=0.0001), while the Lrp5ex12+1 and Lrp5I557N alleles were associated with additive reductions in bone mass, with heterozygous mice developing an intermediate phenotype. No mice homozygous for the Lrp5ex12+1 allele were available for screening, and the allele was only nominally associated with lethality (p=0.03), suggesting too few mice were tested in the pedigree to observe homozygotes.
Figure 5.
Allelic series identifies pathogenic Lrp5 mutations in mice. (A,B) Mice homozygous for the Lrp5ex1+2 splice allele (VAR) developed reduced (A) tibia and (B) femur BMD compared to mice heterozygous for the allele (HET) or homozygous for the reference allele (REF). (C,D) Mice heterozygous for the Lrp5ex12+1 splice allele (HET) developed reduced (C) tibia and (D) femur BMD compared to mice homozygous for the reference allele (REF). (E,F) Mice homozygous for the Lrp5I557N allele developed reduced (E) tibia and (F) femur BMD compared to mice homozygous for the reference allele (REF). Mice heterozygous for the allele (HET) develop an intermediate reduction in BMD. (G,H) Mice homozygous for the Lrp5D125G allele (VAR) developed increased (G) tibia and (H) femur BMD compared to mice homozygous for the reference allele (REF). Mice heterozygous for the allele (HET) developed an intermediate reduction in BMD. Male and female mice are shown with blue and pink symbols, respectively, and the numbers of mice are shown below. Data are median with interquartile range (IQR); whiskers extend up to 1.5x the IQR.
In contrast, we identified the Lrp5D125G allele significantly associated with increased bone mass (Figure 5G,H). The Lrp5D125G allele is located within blade 2 of the first YWTD beta-propeller domain, altering the Aspartic acid of the second Y-W-T-D motif to Glycine. The additive high bone mass phenotype associated with the Lrp5D125G allele is consistent with a gain-of-function mechanism, likely through interfering with binding of LRP5 inhibitors DKK1, SOST, or MESD(50,51). This is further supported by the prior identification of the human LRP5G171V allele, also located within the first beta-propeller domain, causing a high bone mass phenotype that was subsequently verified in mice harboring the orthologous Lrp5G170V allele(46,48). These results demonstrate the feasibility of our ENU-induced approach to characterize an allelic series including both gain-of-function and loss-of-function mutations associated with opposing skeletal phenotypes in mice.
Secretion-deficient Dmp1L10P mice do not phenocopy Dmp1−/− mice
Mice lacking the Dentin matrix acidic phosphoprotein 1 gene (Dmp1) develop severe chondrodysplasia-like skeletal deformities, including severely shortened long bones, and, consistent with its role in osteocyte function, loss of Dmp1 results in a disorganized osteocyte-lacunocanalicular system and reduced bone mineralization(52). Both nuclear and extra-cellular (secreted) functions for DMP1 have been proposed. In support of this, DMP1 localizes to the nucleus in undifferentiated cultured pre-osteoblast progenitor cells, but it is quickly exported from the nucleus at the onset of osteogenic differentiation(53,54). To study this in vivo, nuclear- and secretion-specific forms of DMP1 were conditionally expressed in osteoblasts (using Col1a13.6kb-cre) of Dmp1−/− mice. Osteoblast-specific expression of nuclear DMP1 failed to rescue Dmp1−/− skeletal defects(55); however, expression of nuclear-specific DMP1 in osteochondroprogenitors was not tested.
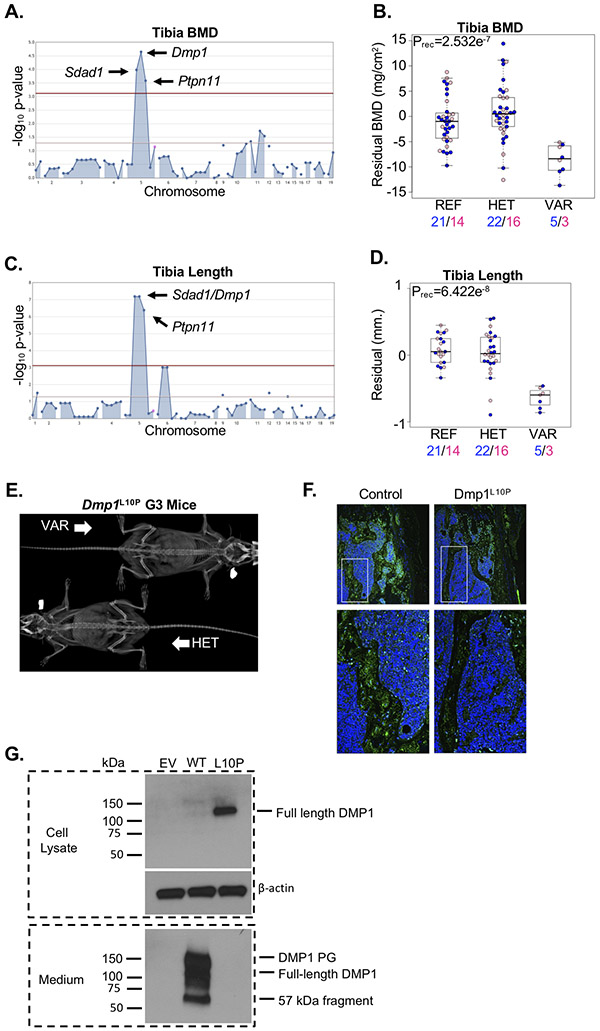
We detected a locus harboring linked alleles in Dmp1 (Dmp1L10P), the SDA1 domain containing 1 gene (Sdad1D144G), and the Protein tyrosine phosphatase non-receptor type 11 gene (Ptpn11H419L) significantly associated with reduced tibia BMD and slightly reduced tibia length (Figure 6A-D). Sdad1 knock-out mice are uncharacterized, and no human disease has previously been associated with SDAD1 mutations. Gain-of-function and loss-of-function mutations in PTPN11 cause the Noonan syndrome Ras-opathy and metachondromatosis, respectively(56). In mice, knock-out of Ptpn11 is embryonic lethal(57), and conditional post-natal deletion of Ptpn11 in chondrocytes (using Col2a1-creERT2) resulted in metachondromatoses(58) that were not evident in homozygous G3 mice (Figure 6E). Although contribution from the Ptpn11H419L or Sdad1D144G alleles cannot be definitively excluded, we selected the Dmp1L10P allele as the most compelling because it is located at a highly conserved residue within the DMP1 signal peptide and may possibly disrupt protein secretion. To test this, we evaluated DMP1 protein within the distal femur of a single mouse homozygous for the Dmp1L10P allele compared to a pedigree-matched control. Secretion of mutant DMP1 was markedly reduced in the homozygous Dmp1L10P mouse, localizing predominantly to the nucleus (Figure 6F). Moreover, wild-type Dmp1 and Dmp1L10P transcripts were transiently expressed in MC3T3-E1 cells, and DMP1 protein was evaluated in cell lysates and in the culture medium(18). Consistent with in vivo immunofluorescence localization, mutant DMP1L10P accumulated in cell lysates and was undetectable in the culture medium (Figure 6G). The phenotypic differences between published Dmp1−/− mice and homozygous Dmp1L10P mice described here suggest a model whereby DMP1 secreted by osteoblasts and osteocytes is required for maintenance of bone mineralization, while nuclear DMP1, possibly expressed in progenitor populations, is required for proper chondrogenesis and longitudinal skeletal growth.
Figure 6.
Loss of DMP1 secretion does not cause severe chondrodysplasia. (A) A single locus segregating alleles in Sdad1, Dmp1, and Ptpn11 were significantly associated with variation in tibia BMD. (B) Mice homozygous for the Dmp1L10P allele (VAR) developed reduced tibia BMD compared to mice heterozygous for the allele (HET) and mice homozygous for the reference allele (REF). Male and female mice are shown with blue and pink symbols, respectively, and the numbers of mice are shown below. Data are median with interquartile range (IQR); whiskers extend up to 1.5x the IQR. (C) A single locus segregating alleles in Sdad1, Dmp1, and Ptpn11 were significantly associated with variation in tibia length. (D) Mice homozygous for the Dmp1L10P allele (VAR) developed slightly reduced tibia length compared to mice heterozygous for the allele (HET) and mice homozygous for the reference allele (REF). Male and female mice are shown with blue and pink symbols, respectively, and the numbers of mice are shown below. Data are median with interquartile range (IQR); whiskers extend up to 1.5x the IQR. (E) Representative radiograph of a mouse homozygous (VAR) or heterozygous (HET) for the Dmp1L10P allele. (F) Immunofluorescence localization of DMP1 protein (green) in the distal femur of a mouse homozygous for the reference allele (Control) or homozygous for the Dmp1L10P allele. DMP1 protein is primarily restricted to the nucleus with little evidence of secretion in the homozygous Dmp1L10P mouse. (G) Western blot analysis of MC3T3-E1 cells transiently expression HA-tagged wild-type DMP1 (WT) or mutant DMP1L10P (L10P). Cell lysate and medium were analyzed separately to detect intracellular and secreted DMP1 protein, respectively. The DMP1 proteoglycan (DMP1 PG) and 57kDa fragment are evident in the culture medium. Beta-actin is shown as a loading control. Empty vector (EV) is shown as negative control.
ENU-induced skeletal phenotypes in essential genes
Multiple studies have shown that up to 34% of genes in the mouse genome may result in knock-out lethality that prohibits comprehensive phenotyping(59,60). One benefit of ENU mutagenesis is the potential to introduce putative hypomorphic alleles in developmentally essential genes, thereby providing opportunities to discover skeletal phenotypes associated with genes that otherwise remain unstudied or that are restricted to conditional approaches(60). As proof of concept, we sought examples of putative recessive hypomorphic ENU-induced alleles in essential genes previously associated with skeletal phenotypes.
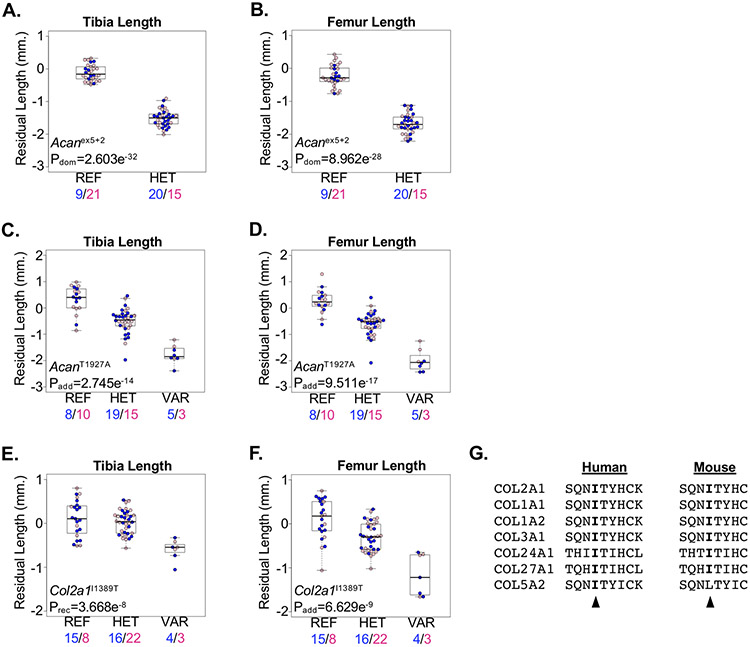
Aggrecan, encoded by the Acan gene, is a proteoglycan essential to the extracellular matrix of the skeleton(61). Mutations in the C-type lectin domain of human aggrecan have been associated with short stature and advanced bone age, while recessive mutations cause severe dysplasia and dwarfism(62,63). In mice, knock-out of the Acan gene results in lethality prior to weaning(59). Two spontaneous mouse lines harboring loss-of-function mutations in Acan develop recessive severe dwarfism with reduced lengths of long bones as well as other skeletal abnormalities, and both are homozygous lethal(64-67). More recently, the ENU-induced AcanA1946V allele, located within the C-type lectin domain, was associated with late-onset obesity and joint disease; however, no other skeletal abnormalities were noted(68). We identified significant association of the Acanex5+2 predicted loss-of-function allele with reduced bone lengths in heterozygous mice; no homozygous mice were observed (Figure 7A,B). We also identified the AcanT1927A allele, located within the C-type lectin domain, associated with an additive reduction in bone lengths, with heterozygous mice developing an intermediate phenotype (Figure 7C,D). The reduced long bone growth observed in mice homozygous for the AcanT1927A allele was similar to mice heterozygous for the Acanex5+2 loss-of-function allele, suggesting AcanT1927A is hypomorphic and viable in the homozygous state.
Figure 7.
ENU-alleles in essential genes cause skeletal phenotypes in mice. (A,B) Mice heterozygous for the Acanex5 allele (HET) developed reduced (A) tibia and (B) femur lengths compared to mice homozygous for the reference allele (REF). Male and female mice are shown with blue and pink symbols, respectively, and the numbers of mice are shown below. (C,D) Mice homozygous for the AcanT1927A allele (VAR) developed reduced (C) tibia and (D) femur lengths compared to mice homozygous for the reference allele (REF). Mice heterozygous for the AcanT1927A allele (HET) develop an intermediate phenotype. Male and female mice are shown with blue and pink symbols, respectively, and the numbers of mice are shown below. (E,F) Mice homozygous for the Col2a1I1389T allele (VAR) developed reduced (E) tibia and (F) femur lengths compared to mice homozygous for the reference allele (REF). Mice heterozygous for the Col2a1I1389T allele (HET) developed an intermediate femur length phenotype. Male and female mice are shown with blue and pink symbols, respectively, and the numbers of mice are shown below. Data are median with interquartile range (IQR); whiskers extend up to 1.5x the IQR. (G) Protein sequence alignment of mouse (left) and human (right) fibrillar collagens showing conservation of the human COL2A1 p.Ile1389 and mouse COL2A1I1389 residues (arrowhead), respectively.
In addition to aggrecan and other proteoglycans, the extracellular matrix is composed of collagen fibers that provide critical structural integrity to the skeleton. For example, mutations in human type 2 Collagen, encoded by the COL2A1 gene, cause a variety of skeletal chondrodysplasias(69). In mice, homozygous loss of Col2a1 resulted in multiple skeletal defects, including dwarfism, and perinatal lethality(70). We detected a significant recessive association of the Col2a1I1389T allele with reduced bone lengths (Figure 7E,F). The COL2A1I1389 amino acid is located within the C-terminal fibrillar collagen domain and is highly conserved among mouse and human fibrillar collagens (Figure 7G). The orthologous amino acid among all human fibril collagens is highly conserved, with only a single heterozygous individual identified within the gnomAD database (N~136,000) harboring a COL27A1 p.(Ile1763Thr) (rs754072876) mutation. Defining the mechanism through which the Col2a1I1389T allele results in shorter limbs in mice remains unclear and requires further study.
Viable mouse models of human proteoglycan synthesis disorders
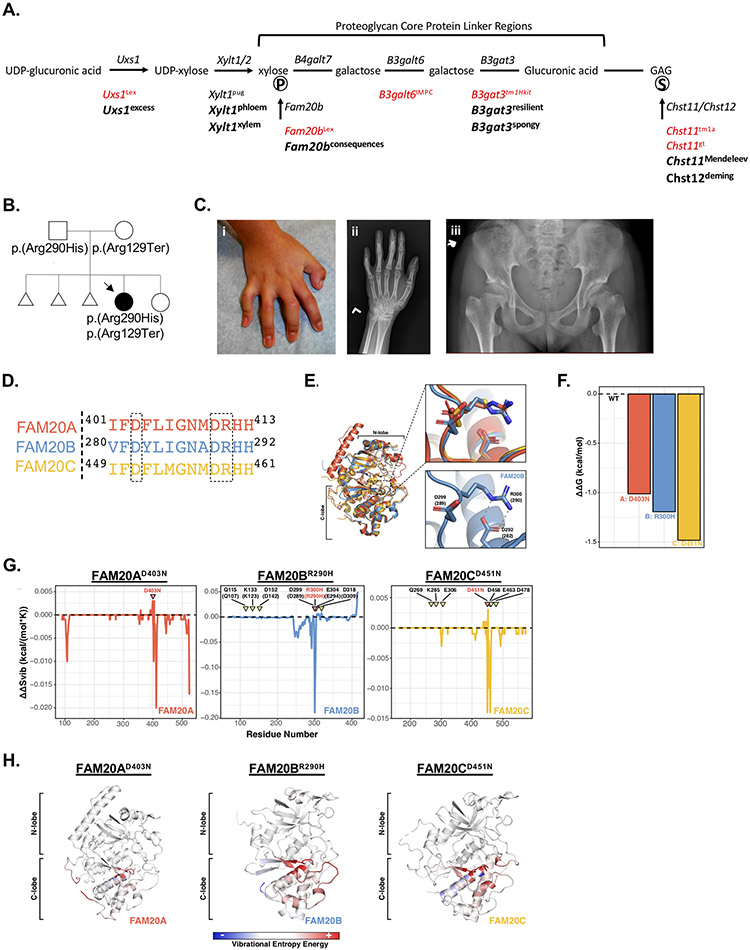
Proteoglycans (PGs) are complex macromolecules with varied distributions in the skeleton and cartilage that consist of a core protein, tetrasaccharide linker region, and covalently bound glycosaminoglycans (GAGs) that may undergo modification, such as sulfation(71). PGs regulate pathways required for proper endochondral ossification and chondrogenesis, and dysregulation of PG biosynthesis or degradation pathways results in various skeletal diseases(71,72). Both spontaneous and genetically-engineered mouse models of GAG defects have been characterized, many developing shortened limbs with varying severity(73-75); however, numerous genes required for PG biosynthesis are essential and display lethality when knocked out in mice (Figure 8A). Using our saturation mutagenesis approach, we queried potential hypomorphic alleles in these and other genes involved in PG biosynthesis that may be associated with skeletal differences. Indeed, we identified multiple viable alleles associated with recessive skeletal defects within essential genes required for PG linker synthesis, GAG sulfation, or GAG elongation (Figure 8A; Supplementary Figures 3-6). These results reproducibly demonstrate that genetic mutations in genes required for PG biosynthesis in mice result in reduced skeletal growth or long bone dysplasia resembling human patients with skeletal disorders caused by recessive mutations in these genes(76-80).
Figure 8.
Mouse model and case report of FAM20B-associated skeletal syndrome. (A) Schematic diagram showing genes required for synthesis of the proteoglycan tetrasaccharide linker region. Published lethal mouse alleles are shown in red and novel ENU alleles are bolded below. Phosphorylation and sulfation are shown with encircled “P” and “S”, respectively. (B) Pedigree demonstrating recessive inheritance of FAM20B mutations in a pediatric patient with skeletal disease. Compound heterozygous variants were confirmed in the patient. (C) The patient developed camptodactyly, as demonstrated from the (i) clinical photo and (ii) radiograph of the patient’s left hand. Pelvis radiograph (iii) demonstrates the patient’s prominent lesser trochanters bilaterally (monkey wrench appearance). (D) Protein sequence alignment of human FAM20A (red), FAM20B (blue), and FAM20C (yellow) surrounding the location of the FAM20B p.(Arg290His) mutation. (E) Superimposition of human FAM20A (red, PDB ID: 5yh3, chain A), Hydra magnipapillata Fam20b (blue, PDB ID:5xom, chain A), and human FAM20C (yellow, PDB ID:5yh3, chain C) protein structures. Upper panel: Alignment of residues surrounding the FAM20B p.(Arg290His) mutation shows conserved structure among FAM20A (red), Fam20b (blue), and FAM20C (yellow). Lower panel: Protein structure of the Hydra Fam20b showing the salt bridge (dashed lines) between residues. Orthologous human amino acid positions are shown in parentheses. (F) Predicted destabilization (ΔΔG (kcal/mol)) outcome from DynaMut analysis. Zero indicates no alteration from wild-type (WT) structure. Predictions were performed for disease-associated variants modeled onto human FAM20A (red, PDB ID: 5yh3, chain A), Hydra magnipapillata Fam20b (blue, PDB ID: 5xom, chain A), and human FAM20C (yellow, PDB ID: 5yh3, chain C). (G) Predicted ΔΔS (kcal/(mol*K)) per residue plotted along the length of the protein. Results are shown for human FAM20A (red, PDB ID: 5yh3, chain A), Hydra magnipapillata Fam20b (blue, PDB ID: 5xom, chain A), and human FAM20C (yellow, PDB ID: 5yh3, chain C). Negative values indicate greater disorder. Variant residues are indicated in red and by pink inverted triangles. Residues important to FAM20B and FAM20C kinase activity are shown in black and by yellow inverted triangles. FAM20B labels correspond to hydra residue numbering, with human FAM20B orthologues indicated in parentheses. (H) Representations of variation in the vibrational entropy energy of the variant protein indicated above compared to wild-type protein. Red indicates increased flexibility, blue indicates decreased flexibility, and white indicates no alteration compared to wild-type. N- and C-lobes are indicated to provide domain perspective.
Case report and mouse model of FAM20B-associated skeletal disease
The FAM20B gene encodes the FAM20B glycosaminoglycan xylosylkinase responsible for phosphorylating the initial xylose within the PG linker region (Figure 8A), which was shown to modulate GAG concentrations in vitro(81). In mice, loss of Fam20b (Fam20bLex) resulted in embryonic lethality(82). However, we identified the Fam20bW224R allele associated with reduced skeletal growth in homozygous mice (Figure 8A; Supplementary Figure 3). The reduced skeletal growth observed in mice homozygous for the Fam20bW224R allele was similar to other mouse models harboring variants in genes required for PG synthesis or sulfation (Supplementary Figures 4-6), suggesting the Fam20bW224R allele is likely hypomorphic.
We then sought patients with undiagnosed skeletal disease harboring putative recessive hypomorphic alleles in FAM20B. We identified a female patient born to non-consanguineous unaffected parents of Bolivian descent who presented with intrauterine growth retardation and, at birth (35 weeks), preaxial polydactyly with partial duplication of the left distal phalanx, bilateral finger contractures, and muscular ventricular septal defect that closed spontaneously by 6 months of age (Figure 8B). She developed glaucoma requiring surgery at 6 months of age. At 11 years of age, her weight and height were 24.9kg (−2.6 standard deviations (SD)) and 120.3cm (−3.5 SD). At 12 years of age, her physical examination revealed prominent eyes, retrognathia, high-arched palate, camptodactyly (Figure 8C), and pes planus with mild foot eversion bilaterally, but no joint laxity was elicited. Skeletal imaging showed subluxation of the proximal interphalangeal joints (Figure 8C), prominence of the lesser trochanters (Figure 8C), and large epiphysis of the distal phalanx of the thumb. Exome sequencing performed on DNA from the patient identified bi-allelic variants in FAM20B, including a rare predicted loss-of-function mutation (NM_014864.3:c.385C>T/p.(Arg129Ter), rs1222347878) and a rare nonsynonymous mutation (c.869G>A/p.(Arg290His), rs1183872117) located at an evolutionarily highly conserved residue (Supplementary Figure 7). Both mutations were computationally predicted to be damaging, and both variants were confirmed compound heterozygous in the patient (Figure 8B). No other recessive variants were identified in candidate genes potentially associated with the patient’s phenotype.
Recently, two siblings from a single family presented with a lethal skeletal syndrome characterized by a spectrum of skeletal abnormalities including short limb dwarfism that resembled Desbuquois dysplasia, and exome sequencing identified compound heterozygous loss-of-function alleles in FAM20B in one patient(83). To evaluate whether the FAM20B p.(Arg290His) allele in our index patient was potentially hypomorphic, we performed multiple computational analyses and compared results to the FAM20A p.(Asp403Asn) and FAM20C p.(Asp451Asn) alleles known to cause Amelogenesis Imperfecta 1G and Raine syndrome, respectively(84,85). All three alleles occur within an evolutionarily conserved structural element shared among FAM20 kinases (Figure 8D). Homologous structures suggest the FAM20B p.Arg290 residue forms a salt bridge with the FAM20B p.Asp282 residue, which is homologous to the disease-causing FAM20A p.Asp403 and FAM20C p.Asp451 residues and serves to properly align the active site architecture of FAM20 kinases(86) (Figure 8D,E). After modeling these missense alleles on the hydra (Hydra magnipapillata) Fam20 kinases(86), Normal Mode Analysis (NMA) predicts that introduction of each allele destabilizes the proteins, as indicated by alterations in Gibbs free energy of protein folding (ΔΔG) (Figure 8F). Simulation of protein dynamics utilizing an Elastic Network Contact Model (ENCoM) revealed alterations in vibrational entropy (ΔΔSvib) associated with each variant allele, with the greatest disruption occurring proximal to the variant residue and throughout the C-lobe (Figure 8G,H). These results, together with analysis of pathogenic variants in FAM20A and FAM20C, consistently suggest the FAM20B p.(Arg290His) variant destabilizes the protein, likely leading to the hypomorphic phenotypes observed in the index patient.
DISCUSSION
We present results from a saturation mutagenesis screen in mice to identify genes required for proper skeletal development. The integration of ENU mutagenesis, massively-parallel sequencing, and high-throughput live-animal imaging enables testing of a wide breadth of non-synonymous alleles for potential effects on skeletal development. Results from this screen provide opportunities to identify novel mouse models of known human skeletal disease and enable gene- and variant-level resolution of associated skeletal phenotypes that may potentially inform human GWAS-associated loci.
Due to the high-throughput design of our screen, individual G3 pedigrees undergoing skeletal screening consist of mice of different genders and ages. Therefore, we developed and validated statistical models to quantify phenotypic variation due to differences in age and gender. Using this approach, we estimated the amount of variation due to these factors and performed genetic mapping using residual phenotypic differences. It is possible, however, that other factors, such as body mass (i.e., fat mass and lean mass), contribute to variation in skeletal phenotypes among G3 mice, and continued screening of G3 mice will further improve the precision of our statistical models. Our approach is similar to the application of population-based Z-scores to detect clinical differences in human skeletal development. In support of our residuals-based approach, we present multiple alleles in loci previously implicated with essential roles in skeletal development and provide validation using novel CRISPR-engineered lines. Similarly, we demonstrate the comparability of results using our residuals-based approach in G3 mice with that of age- and gender-matched CRISPR-engineered mice (Figure 2).
We and others estimated that up to 34% of genes in the mouse genome are essential for survival(59,60). The ENU mutagenesis approach employed here is uniquely capable of generating and phenotypically screening adult mice harboring viable hypomorphic alleles in these essential genes that are otherwise unavailable using traditional knock-out approaches, for example Acan and Col2a1. The COL2A1I1389 residue is conserved among fibrillar collagens, suggesting the homologous Col3a1I1366T or Col27a1I1748T alleles may result in viable homozygous mice, potentially developing skeletal abnormalities, despite these genes being essential for survival(87,88). Moreover, we demonstrate that independent testing of multiple non-synonymous alleles within the same gene may identify allelic heterogeneity associated with different phenotypic traits, or potentially survival, as was observed for alleles within the Lrrk1 and Lrp5 genes.
One limitation of our forward-genetic screen is the inability to know, a priori, how many homozygous mice will be screened for each allele. Here, we report results of screening 24,931 mice harboring predicted damaging or loss-of-function alleles tested in at least 2 homozygous mice across 8,294 genes (39.7% of autosomal genes). We have previously shown that as few as two or three homozygous mice is sufficient to identify bona-fide loci associated with non-skeletal phenotypes(15,89,90). In addition to Arsb (Figure 2E,F) presented here, we identified other associated loci that either phenocopied known knock-out mouse models(91) (Supplementary Figure 8) or were validated using CRISPR-engineered mouse lines (Supplementary Figures 9-11). These results demonstrate the power of our approach to identify significantly-associated recessive loci from as few as two homozygous mice within a pedigree.
Finally, we anticipate phenotypic associations identified from ENU-mutagenized mice can be integrated with large-scale human sequencing studies, such as the TOPMed, All of Us, and Deciphering Developmental Disorders(92) studies, the Centers for Mendelian Genetics, and the Undiagnosed Diseases Network to provide evidence that a putative disease gene plays an essential role in skeletal development. We present the second independent case report of a patient with skeletal disease associated with recessive mutations in FAM20B. Our patient exhibited similar features as the siblings previously described(83), including prenatal and postnatal growth retardation, abnormalities in the femora, and preaxial digit involvement. Interestingly, the long survival observed in our patient may be attributed to the predicted hypomorphic effect of the missense allele in trans with a single loss-of-function mutation, while the previously-reported siblings inherited recessive loss-of-function mutations. Our patient also developed glaucoma, a known complication of Desbuquois dysplasia(93,94). Glaucoma and hyperopia, as well as short stature and pes planus, have been described in patients with other GAG synthesis disorders, particularly linkeropathies(95). Furthermore, we report the first viable mouse model of skeletal disease associated with recessive mutations in Fam20b.
Phenotypic alleles are made publicly available online via the Mutagenetix website (https://mutagenetix.utsouthwestern.edu/home.cfm), and mouse models, including ENU-induced alleles and CRISPR-engineered mice, are made publicly available through the Mutant Mouse Resource and Research Center (MMRRC). Results from our saturation mutagenesis skeletal screen serve as an important resource that will advance understanding of the molecular signaling required for proper skeletal development.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Beutler and Rios laboratories and the staff of the UTSW Transgenic Technology Center. This work was supported by Scottish Rite for Children (J.J.R.) and by National Institutes of Health grants R01AI125581 (B.B.) and U19AI100627 (B.B.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
COMPETING INTERESTS
The authors declare no competing interests
DATA AVAILABILITY
Results and mouse models are publicly available through the Mutagenetix website at https://mutagenetix.utsouthwestern.edu/home.cfm and through the Mutant Mouse Resource and Research Center (MMRRC), respectively.
REFERENCES
- 1.DiGirolamo DJ, Kiel DP, Esser KA. Bone and skeletal muscle: neighbors with close ties. J Bone Miner Res. Jul 2013;28(7):1509–18. Epub 2013/05/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bajayo A, Goshen I, Feldman S, Csernus V, Iverfeldt K, Shohami E, et al. Central IL-1 receptor signaling regulates bone growth and mass. Proc Natl Acad Sci U S A. Sep 6 2005;102(36):12956–61. Epub 2005/08/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stegen S, Carmeliet G. The skeletal vascular system - Breathing life into bone tissue. Bone. Oct 2018;115:50–8. Epub 2017/08/29. [DOI] [PubMed] [Google Scholar]
- 4.Wit JM, Camacho-Hubner C. Endocrine regulation of longitudinal bone growth. Endocr Dev. 2011;21:30–41. Epub 2011/08/26. [DOI] [PubMed] [Google Scholar]
- 5.Peacock M, Turner CH, Econs MJ, Foroud T. Genetics of osteoporosis. Endocr Rev. Jun 2002;23(3):303–26. Epub 2002/06/07. [DOI] [PubMed] [Google Scholar]
- 6.Warman ML, Cormier-Daire V, Hall C, Krakow D, Lachman R, LeMerrer M, et al. Nosology and classification of genetic skeletal disorders: 2010 revision. Am J Med Genet A. May 2011;155A(5):943–68. Epub 2011/03/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Estrada K, Styrkarsdottir U, Evangelou E, Hsu YH, Duncan EL, Ntzani EE, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. Apr 15 2012;44(5):491–501. Epub 2012/04/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kemp JP, Morris JA, Medina-Gomez C, Forgetta V, Warrington NM, Youlten SE, et al. Identification of 153 new loci associated with heel bone mineral density and functional involvement of GPC6 in osteoporosis. Nat Genet. Oct 2017;49(10):1468–75. Epub 2017/09/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SK. Identification of 613 new loci associated with heel bone mineral density and a polygenic risk score for bone mineral density, osteoporosis and fracture. PLoS One. 2018;13(7):e0200785. Epub 2018/07/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng HF, Forgetta V, Hsu YH, Estrada K, Rosello-Diez A, Leo PJ, et al. Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature. Oct 1 2015;526(7571):112–7. Epub 2015/09/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao FB, Brenman JE, Jan LY, Jan YN. Genes regulating dendritic outgrowth, branching, and routing in Drosophila. Genes Dev. Oct 1 1999;13(19):2549–61. Epub 1999/10/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. Dec 1996;123:37–46. Epub 1996/12/01. [DOI] [PubMed] [Google Scholar]
- 13.Gray RS, Gonzalez R, Ackerman SD, Minowa R, Griest JF, Bayrak MN, et al. Postembryonic screen for mutations affecting spine development in zebrafish. Dev Biol. Dec 5 2020;471:18–33. Epub 2020/12/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moresco EM, Li X, Beutler B. Going forward with genetics: recent technological advances and forward genetics in mice. Am J Pathol. May 2013;182(5):1462–73. Epub 2013/04/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang T, Zhan X, Bu CH, Lyon S, Pratt D, Hildebrand S, et al. Real-time resolution of point mutations that cause phenovariance in mice. Proc Natl Acad Sci U S A. Feb 3 2015;112(5):E440–9. Epub 2015/01/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgel P, Du X, Hoebe K, Beutler B. ENU mutagenesis in mice. Methods Mol Biol. 2008;415:1–16. Epub 2008/03/29. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Jing Y, Zhang R, Zhang Q, Wang J, Martin A, et al. Hypophosphatemic rickets accelerate chondrogenesis and cell trans-differentiation from TMJ chondrocytes into bone cells via a sharp increase in beta-catenin. Bone. Feb 2020;131:115151. Epub 2019/11/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang T, Meng T, Wang S, Qin C, Lu Y. The LPV Motif Is Essential for the Efficient Export of Secretory DMP1 From the Endoplasmic Reticulum. J Cell Physiol. Jul 2016;231(7):1468–75. Epub 2015/11/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodrigues CH, Pires DE, Ascher DB. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. Jul 2 2018;46(W1):W350–W5. Epub 2018/05/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frappier V, Najmanovich RJ. A coarse-grained elastic network atom contact model and its use in the simulation of protein dynamics and the prediction of the effect of mutations. PLoS Comput Biol. Apr 2014;10(4):e1003569. Epub 2014/04/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grant BJ, Skjaerven L, Yao XQ. The Bio3D packages for structural bioinformatics. Protein Sci. Jan 2021;30(1):20–30. Epub 2020/08/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Team RC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2017. [Google Scholar]
- 23.Wickham H ggplot2: Elegant Graphics for Data Analysis. New York: Springer; 2009. [Google Scholar]
- 24.Schrodinger LLC. The PyMOL Molecular Graphics System. Version 1.2r3pre ed2015. [Google Scholar]
- 25.Morishita K, Petty RE. Musculoskeletal manifestations of mucopolysaccharidoses. Rheumatology (Oxford). Dec 2011;50 Suppl 5:v19–25. Epub 2012/01/11. [DOI] [PubMed] [Google Scholar]
- 26.Kuehn SC, Koehne T, Cornils K, Markmann S, Riedel C, Pestka JM, et al. Impaired bone remodeling and its correction by combination therapy in a mouse model of mucopolysaccharidosis-I. Hum Mol Genet. Dec 15 2015;24(24):7075–86. Epub 2015/10/03. [DOI] [PubMed] [Google Scholar]
- 27.Black SH, Pelias MZ, Miller JB, Blitzer MG, Shapira E. Maroteaux-Lamy syndrome in a large consanguineous kindred: biochemical and immunological studies. Am J Med Genet. Oct 1986;25(2):273–9. Epub 1986/10/01. [DOI] [PubMed] [Google Scholar]
- 28.Azevedo AC, Schwartz IV, Kalakun L, Brustolin S, Burin MG, Beheregaray AP, et al. Clinical and biochemical study of 28 patients with mucopolysaccharidosis type VI. Clin Genet. Sep 2004;66(3):208–13. Epub 2004/08/25. [DOI] [PubMed] [Google Scholar]
- 29.Evers M, Saftig P, Schmidt P, Hafner A, McLoghlin DB, Schmahl W, et al. Targeted disruption of the arylsulfatase B gene results in mice resembling the phenotype of mucopolysaccharidosis VI. Proc Natl Acad Sci U S A. Aug 6 1996;93(16):8214–9. Epub 1996/08/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Citterio CE, Targovnik HM, Arvan P. The role of thyroglobulin in thyroid hormonogenesis. Nat Rev Endocrinol. Jun 2019;15(6):323–38. Epub 2019/03/20. [DOI] [PubMed] [Google Scholar]
- 31.Ieiri T, Cochaux P, Targovnik HM, Suzuki M, Shimoda S, Perret J, et al. A 3' splice site mutation in the thyroglobulin gene responsible for congenital goiter with hypothyroidism. J Clin Invest. Dec 1991;88(6):1901–5. Epub 1991/12/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreno JC, Bikker H, Kempers MJ, van Trotsenburg AS, Baas F, de Vijlder JJ, et al. Inactivating mutations in the gene for thyroid oxidase 2 (THOX2) and congenital hypothyroidism. N Engl J Med. Jul 11 2002;347(2):95–102. Epub 2002/07/12. [DOI] [PubMed] [Google Scholar]
- 33.Andrews TD, Whittle B, Field MA, Balakishnan B, Zhang Y, Shao Y, et al. Massively parallel sequencing of the mouse exome to accurately identify rare, induced mutations: an immediate source for thousands of new mouse models. Open Biol. May 2012;2(5):120061. Epub 2012/06/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim PS, Hossain SA, Park YN, Lee I, Yoo SE, Arvan P. A single amino acid change in the acetylcholinesterase-like domain of thyroglobulin causes congenital goiter with hypothyroidism in the cog/cog mouse: a model of human endoplasmic reticulum storage diseases. Proc Natl Acad Sci U S A. Aug 18 1998;95(17):9909–13. Epub 1998/08/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson KR, Marden CC, Ward-Bailey P, Gagnon LH, Bronson RT, Donahue LR. Congenital hypothyroidism, dwarfism, and hearing impairment caused by a missense mutation in the mouse dual oxidase 2 gene, Duox2. Mol Endocrinol. Jul 2007;21(7):1593–602. Epub 2007/04/19. [DOI] [PubMed] [Google Scholar]
- 36.Muzza M, Fugazzola L. Disorders of H2O2 generation. Best Pract Res Clin Endocrinol Metab. Mar 2017;31(2):225–40. Epub 2017/06/27. [DOI] [PubMed] [Google Scholar]
- 37.Donko A, Ruisanchez E, Orient A, Enyedi B, Kapui R, Peterfi Z, et al. Urothelial cells produce hydrogen peroxide through the activation of Duox1. Free Radic Biol Med. Dec 15 2010;49(12):2040–8. Epub 2010/12/15. [DOI] [PubMed] [Google Scholar]
- 38.Iida A, Xing W, Docx MK, Nakashima T, Wang Z, Kimizuka M, et al. Identification of biallelic LRRK1 mutations in osteosclerotic metaphyseal dysplasia and evidence for locus heterogeneity. J Med Genet. Aug 2016;53(8):568–74. Epub 2016/04/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miryounesi M, Nikfar A, Changi-Ashtiani M, Shahrooei M, Dinmohammadi H, Shahani T, et al. A novel homozygous LRRK1 stop gain mutation in a patient suspected with osteosclerotic metaphyseal dysplasia. Ann Hum Genet. Jan 2020;84(1):102–6. Epub 2019/10/02. [DOI] [PubMed] [Google Scholar]
- 40.Xing W, Liu J, Cheng S, Vogel P, Mohan S, Brommage R. Targeted disruption of leucine-rich repeat kinase 1 but not leucine-rich repeat kinase 2 in mice causes severe osteopetrosis. J Bone Miner Res. Sep 2013;28(9):1962–74. Epub 2013/03/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baptista MA, Dave KD, Sheth NP, De Silva SN, Carlson KM, Aziz YN, et al. A strategy for the generation, characterization and distribution of animal models by The Michael J. Fox Foundation for Parkinson's Research. Dis Model Mech. Nov 2013;6(6):1316–24. Epub 2013/09/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He X, Semenov M, Tamai K, Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: arrows point the way. Development. Apr 2004;131(8):1663–77. Epub 2004/04/16. [DOI] [PubMed] [Google Scholar]
- 43.Baron R, Kneissel M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat Med. Feb 2013;19(2):179–92. Epub 2013/02/08. [DOI] [PubMed] [Google Scholar]
- 44.Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. Nov 16 2001;107(4):513–23. Epub 2001/11/24. [DOI] [PubMed] [Google Scholar]
- 45.Little RD, Carulli JP, Del Mastro RG, Dupuis J, Osborne M, Folz C, et al. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am J Hum Genet. Jan 2002;70(1):11–9. Epub 2001/12/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, et al. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med. May 16 2002;346(20):1513–21. Epub 2002/05/17. [DOI] [PubMed] [Google Scholar]
- 47.Kato M, Patel MS, Levasseur R, Lobov I, Chang BH, Glass DA, 2nd, et al. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol. Apr 15 2002;157(2):303–14. Epub 2002/04/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cui Y, Niziolek PJ, MacDonald BT, Zylstra CR, Alenina N, Robinson DR, et al. Lrp5 functions in bone to regulate bone mass. Nat Med. Jun 2011;17(6):684–91. Epub 2011/05/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Babij P, Zhao W, Small C, Kharode Y, Yaworsky PJ, Bouxsein ML, et al. High bone mass in mice expressing a mutant LRP5 gene. J Bone Miner Res. Jun 2003;18(6):960–74. Epub 2003/06/24. [DOI] [PubMed] [Google Scholar]
- 50.Ai M, Holmen SL, Van Hul W, Williams BO, Warman ML. Reduced affinity to and inhibition by DKK1 form a common mechanism by which high bone mass-associated missense mutations in LRP5 affect canonical Wnt signaling. Mol Cell Biol. Jun 2005;25(12):4946–55. Epub 2005/06/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Wang Y, Li X, Zhang J, Mao J, Li Z, et al. The LRP5 high-bone-mass G171V mutation disrupts LRP5 interaction with Mesd. Mol Cell Biol. Jun 2004;24(11):4677–84. Epub 2004/05/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng JQ, Ward LM, Liu S, Lu Y, Xie Y, Yuan B, et al. Loss of DMP1 causes rickets and osteomalacia and identifies a role for osteocytes in mineral metabolism. Nat Genet. Nov 2006;38(11):1310–5. Epub 2006/10/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Narayanan K, Ramachandran A, Hao J, He G, Park KW, Cho M, et al. Dual functional roles of dentin matrix protein 1. Implications in biomineralization and gene transcription by activation of intracellular Ca2+ store. J Biol Chem. May 9 2003;278(19):17500–8. Epub 2003/03/05. [DOI] [PubMed] [Google Scholar]
- 54.Siyam A, Wang S, Qin C, Mues G, Stevens R, D'Souza RN, et al. Nuclear localization of DMP1 proteins suggests a role in intracellular signaling. Biochem Biophys Res Commun. Aug 3 2012;424(3):641–6. Epub 2012/07/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin S, Zhang Q, Cao Z, Lu Y, Zhang H, Yan K, et al. Constitutive nuclear expression of dentin matrix protein 1 fails to rescue the Dmp1-null phenotype. J Biol Chem. Aug 1 2014;289(31):21533–43. Epub 2014/06/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Digilio MC, Conti E, Sarkozy A, Mingarelli R, Dottorini T, Marino B, et al. Grouping of multiple-lentigines/LEOPARD and Noonan syndromes on the PTPN11 gene. Am J Hum Genet. Aug 2002;71(2):389–94. Epub 2002/06/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang W, Klaman LD, Chen B, Araki T, Harada H, Thomas SM, et al. An Shp2/SFK/Ras/Erk signaling pathway controls trophoblast stem cell survival. Dev Cell. Mar 2006;10(3):317–27. Epub 2006/03/07. [DOI] [PubMed] [Google Scholar]
- 58.Kim HK, Feng GS, Chen D, King PD, Kamiya N. Targeted disruption of Shp2 in chondrocytes leads to metachondromatosis with multiple cartilaginous protrusions. J Bone Miner Res. Mar 2014;29(3):761–9. Epub 2013/08/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, et al. High-throughput discovery of novel developmental phenotypes. Nature. Sep 22 2016;537(7621):508–14. Epub 2016/09/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang T, Bu CH, Hildebrand S, Jia G, Siggs OM, Lyon S, et al. Probability of phenotypically detectable protein damage by ENU-induced mutations in the Mutagenetix database. Nat Commun. Jan 30 2018;9(1):441. Epub 2018/02/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kiani C, Chen L, Wu YJ, Yee AJ, Yang BB. Structure and function of aggrecan. Cell Res. Mar 2002;12(1):19–32. Epub 2002/04/11. [DOI] [PubMed] [Google Scholar]
- 62.Tompson SW, Merriman B, Funari VA, Fresquet M, Lachman RS, Rimoin DL, et al. A recessive skeletal dysplasia, SEMD aggrecan type, results from a missense mutation affecting the C-type lectin domain of aggrecan. Am J Hum Genet. Jan 2009;84(1):72–9. Epub 2008/12/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stattin EL, Wiklund F, Lindblom K, Onnerfjord P, Jonsson BA, Tegner Y, et al. A missense mutation in the aggrecan C-type lectin domain disrupts extracellular matrix interactions and causes dominant familial osteochondritis dissecans. Am J Hum Genet. Feb 12 2010;86(2):126–37. Epub 2010/02/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bell L, Juriloff M, Harris MJ. A new mutation at the cmd locus in the mouse. J Hered. May-Jun 1986;77(3):205–6. Epub 1986/05/01. [DOI] [PubMed] [Google Scholar]
- 65.Krueger RC, Jr., Kurima K, Schwartz NB. Completion of the mouse aggrecan gene structure and identification of the defect in the cmd-Bc mouse as a near complete deletion of the murine aggrecan gene. Mamm Genome. Dec 1999;10(12):1119–25. Epub 1999/12/14. [DOI] [PubMed] [Google Scholar]
- 66.Rittenhouse E, Dunn LC, Cookingham J, Calo C, Spiegelman M, Dooher GB, et al. Cartilage matrix deficiency (cmd): a new autosomal recessive lethal mutation in the mouse. J Embryol Exp Morphol. Feb 1978;43:71–84. Epub 1978/02/01. [PubMed] [Google Scholar]
- 67.Watanabe H, Kimata K, Line S, Strong D, Gao LY, Kozak CA, et al. Mouse cartilage matrix deficiency (cmd) caused by a 7 bp deletion in the aggrecan gene. Nat Genet. Jun 1994;7(2):154–7. Epub 1994/06/01. [DOI] [PubMed] [Google Scholar]
- 68.Potter PK, Bowl MR, Jeyarajan P, Wisby L, Blease A, Goldsworthy ME, et al. Novel gene function revealed by mouse mutagenesis screens for models of age-related disease. Nat Commun. Aug 18 2016;7:12444. Epub 2016/08/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spranger J, Winterpacht A, Zabel B. The type II collagenopathies: a spectrum of chondrodysplasias. Eur J Pediatr. Feb 1994;153(2):56–65. Epub 1994/02/01. [DOI] [PubMed] [Google Scholar]
- 70.Li SW, Prockop DJ, Helminen H, Fassler R, Lapvetelainen T, Kiraly K, et al. Transgenic mice with targeted inactivation of the Col2 alpha 1 gene for collagen II develop a skeleton with membranous and periosteal bone but no endochondral bone. Genes Dev. Nov 15 1995;9(22):2821–30. Epub 1995/11/15. [DOI] [PubMed] [Google Scholar]
- 71.Paganini C, Costantini R, Superti-Furga A, Rossi A. Bone and connective tissue disorders caused by defects in glycosaminoglycan biosynthesis: a panoramic view. FEBS J. Aug 2019;286(15):3008–32. Epub 2019/07/10. [DOI] [PubMed] [Google Scholar]
- 72.Taylan F, Makitie O. Abnormal Proteoglycan Synthesis Due to Gene Defects Causes Skeletal Diseases with Overlapping Phenotypes. Horm Metab Res. Nov 2016;48(11):745–54. Epub 2016/11/22. [DOI] [PubMed] [Google Scholar]
- 73.Cardone M, Polito VA, Pepe S, Mann L, D'Azzo A, Auricchio A, et al. Correction of Hunter syndrome in the MPSII mouse model by AAV2/8-mediated gene delivery. Hum Mol Genet. Apr 1 2006;15(7):1225–36. Epub 2006/03/01. [DOI] [PubMed] [Google Scholar]
- 74.Ou L, Herzog T, Koniar BL, Gunther R, Whitley CB. High-dose enzyme replacement therapy in murine Hurler syndrome. Mol Genet Metab. Feb 2014;111(2):116–22. Epub 2013/10/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mizumoto S, Yamada S, Sugahara K. Human genetic disorders and knockout mice deficient in glycosaminoglycan. Biomed Res Int. 2014;2014:495764. Epub 2014/08/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bui C, Huber C, Tuysuz B, Alanay Y, Bole-Feysot C, Leroy JG, et al. XYLT1 mutations in Desbuquois dysplasia type 2. Am J Hum Genet. Mar 6 2014;94(3):405–14. Epub 2014/03/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schreml J, Durmaz B, Cogulu O, Keupp K, Beleggia F, Pohl E, et al. The missing "link": an autosomal recessive short stature syndrome caused by a hypofunctional XYLT1 mutation. Hum Genet. Jan 2014;133(1):29–39. Epub 2013/08/29. [DOI] [PubMed] [Google Scholar]
- 78.Munns CF, Fahiminiya S, Poudel N, Munteanu MC, Majewski J, Sillence DO, et al. Homozygosity for frameshift mutations in XYLT2 result in a spondylo-ocular syndrome with bone fragility, cataracts, and hearing defects. Am J Hum Genet. Jun 4 2015;96(6):971–8. Epub 2015/06/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Baasanjav S, Al-Gazali L, Hashiguchi T, Mizumoto S, Fischer B, Horn D, et al. Faulty initiation of proteoglycan synthesis causes cardiac and joint defects. Am J Hum Genet. Jul 15 2011;89(1):15–27. Epub 2011/07/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shabbir RMK, Nalbant G, Ahmad N, Malik S, Tolun A. Homozygous CHST11 mutation in chondrodysplasia, brachydactyly, overriding digits, clino-symphalangism and synpolydactyly. J Med Genet. Jul 2018;55(7):489–96. Epub 2018/03/09. [DOI] [PubMed] [Google Scholar]
- 81.Koike T, Izumikawa T, Tamura J, Kitagawa H. FAM20B is a kinase that phosphorylates xylose in the glycosaminoglycan-protein linkage region. Biochem J. Jun 26 2009;421(2):157–62. Epub 2009/05/29. [DOI] [PubMed] [Google Scholar]
- 82.Vogel P, Hansen GM, Read RW, Vance RB, Thiel M, Liu J, et al. Amelogenesis imperfecta and other biomineralization defects in Fam20a and Fam20c null mice. Vet Pathol. Nov 2012;49(6):998–1017. Epub 2012/06/27. [DOI] [PubMed] [Google Scholar]
- 83.Kuroda Y, Murakami H, Enomoto Y, Tsurusaki Y, Takahashi K, Mitsuzuka K, et al. A novel gene (FAM20B encoding glycosaminoglycan xylosylkinase) for neonatal short limb dysplasia resembling Desbuquois dysplasia. Clin Genet. Jun 2019;95(6):713–7. Epub 2019/03/09. [DOI] [PubMed] [Google Scholar]
- 84.Wang SK, Reid BM, Dugan SL, Roggenbuck JA, Read L, Aref P, et al. FAM20A mutations associated with enamel renal syndrome. J Dent Res. Jan 2014;93(1):42–8. Epub 2013/11/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mameli C, Zichichi G, Mahmood N, Elalaoui SC, Mirza A, Dharmaraj P, et al. Natural history of non-lethal Raine syndrome during childhood. Orphanet journal of rare diseases. Apr 16 2020;15(1):93. Epub 2020/04/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang H, Zhu Q, Cui J, Wang Y, Chen MJ, Guo X, et al. Structure and evolution of the Fam20 kinases. Nat Commun. Mar 23 2018;9(1):1218. Epub 2018/03/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Plumb DA, Ferrara L, Torbica T, Knowles L, Mironov A Jr., Kadler KE, et al. Collagen XXVII organises the pericellular matrix in the growth plate. PLoS One. 2011;6(12):e29422. Epub 2011/12/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu X, Wu H, Byrne M, Krane S, Jaenisch R. Type III collagen is crucial for collagen I fibrillogenesis and for normal cardiovascular development. Proc Natl Acad Sci U S A. Mar 4 1997;94(5):1852–6. Epub 1997/03/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Choi JH, Zhong X, Zhang Z, Su L, McAlpine W, Misawa T, et al. Essential cell-extrinsic requirement for PDIA6 in lymphoid and myeloid development. J Exp Med. Apr 6 2020;217(4). Epub 2020/01/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McAlpine W, Wang KW, Choi JH, San Miguel M, McAlpine SG, Russell J, et al. The class I myosin MYO1D binds to lipid and protects against colitis. Dis Model Mech. Sep 27 2018;11(9). Epub 2018/10/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Conover CA, Bale LK, Overgaard MT, Johnstone EW, Laursen UH, Fuchtbauer EM, et al. Metalloproteinase pregnancy-associated plasma protein A is a critical growth regulatory factor during fetal development. Development. Mar 2004;131(5):1187–94. Epub 2004/02/20. [DOI] [PubMed] [Google Scholar]
- 92.Deciphering Developmental Disorders S Large-scale discovery of novel genetic causes of developmental disorders. Nature. Mar 12 2015;519(7542):223–8. Epub 2014/12/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Faivre L, Cormier-Daire V, Young I, Bracq H, Finidori G, Padovani JP, et al. Long-term outcome in Desbuquois dysplasia: a follow-up in four adult patients. Am J Med Genet A. Jan 1 2004;124A(1):54–9. Epub 2003/12/18. [DOI] [PubMed] [Google Scholar]
- 94.Faden M, Al-Zahrani F, Arafah D, Alkuraya FS. Mutation of CANT1 causes Desbuquois dysplasia. Am J Med Genet A. May 2010;152A(5):1157–60. Epub 2010/04/29. [DOI] [PubMed] [Google Scholar]
- 95.Arunrut T, Sabbadini M, Jain M, Machol K, Scaglia F, Slavotinek A. Corneal clouding, cataract, and colobomas with a novel missense mutation in B4GALT7-a review of eye anomalies in the linkeropathy syndromes. Am J Med Genet A. Oct 2016;170(10):2711–8. Epub 2016/06/21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Results and mouse models are publicly available through the Mutagenetix website at https://mutagenetix.utsouthwestern.edu/home.cfm and through the Mutant Mouse Resource and Research Center (MMRRC), respectively.