ABSTRACT
Hand-foot-and-mouth disease (HFMD) is a highly contagious viral disease commonly associated to Enteroviruses (EV). During 2018, Brazil faced massive HFMD outbreaks spread across the country. This study aimed to characterize the EV responsible for the HFMD outbreak that occurred in Paraiba State, Brazilian Northeastern region, in 2018, followed by a phylogenetic analysis to detail information on its genetic diversity. A total of 49 serum samples (one from each patient) collected from children ≤ 15 years old, clinically diagnosed with HFMD were tested for EV using conventional RT-PCR and RT-qPCR. EV infection was confirmed in 71.4% (35/49) of samples. The mean and median ages were 1.83 years and one year old, respectively. Twenty-two EV-positive samples were successfully sequenced and classified as EV-A species; 13 samples were also identified with the CV-A6 genotype. The phylogenetic analysis (VP1 region) of three samples revealed that the detected CV-A6 strains belonged to sub-lineage D3. The CV-A6 strains detected here clustered with strains from South America, Europe and West Asia strains that were also involved in HFMD cases during the 2017-2018 seasons, in addition to the previously detected Brazilian CV-A6 strains from 2012 to 2017, suggesting a global co-circulation of a set of different CV-A6 strains introduced in the country at different times. The growing circulation of the emerging CV-A6 associated with HFMD, together with the detection of more severe cases worldwide, suggests the need for a more intense surveillance system of HFMD in Brazil. In addition, this investigation was performed exclusively on serum samples, and the analysis of whole blood samples should be considered and could have shown advantages when employed in the diagnosis of enteroviral HFMD outbreaks.
KEYWORDS: Hand foot and mouth disease, Enterovirus, Coxsackievirus, Surveillance
INTRODUCTION
Hand-foot-and-mouth disease (HFMD) is a highly contagious acute viral disease that commonly affects children < 5 years old. HFMD is characterized by a mild febrile illness with multiple oral ulcers, and vesiculo-papular eruptions on the hands, feet, mouth and buttocks, which normally undergo spontaneous involution without major complications 1 .
The most commonly associated viruses leading to HFMD infections are enteroviruses (EV), non‐enveloped single‐stranded RNA viruses of the genus Enterovirus (family Picornaviridae; realm Riboviria) 1 . EV genome is composed of one open reading frame encoding four structural viral proteins (VP1 to VP4 ) and seven nonstructural proteins (2A-2C and 3A-3D). VP1 is the main coding region for neutralizing epitopes containing virulence determinants, being used for serotyping and molecular epidemiology studies. Seven EVs species have been associated with human disease: EV-A to -D and rhinovirus A to C 1,2 . EV-A is responsible for more than 90% of HFMD cases worldwide 3,4 . Enterovirus A 71 (EV-A71), coxsackievirus A6 (CVA6) and coxsackievirus A16 (CVA16) are the leading pathogens associated with HFMD outbreaks globally 2-4 . Although EVs have long been recognized as the most common etiological agents of HFMD1 1 , they have been poorly investigated in Brazil 5-7 . HFMD is not a notifiable disease in Brazil, and as a consequence, associated epidemiological data are scarce probably due to the lack of a continuous and systematic laboratory surveillance 6 . On the other hand, HFMD outbreaks must be notified 8 . During 2018, Brazil faced HFMD outbreaks spread across the country 9,10 , but the EV type (or types) involved in those outbreaks are still unknown.
The aim of the present study was to identify the EV genotype implicated in Brazilian HFMD outbreaks that occurred in Paraiba State, Northeastern region, during 2018. The phylogenetic analysis of VP1 sequences was also performed to obtain more detailed information on the genetic relationships with previously reported strains and to identify the lineage implicated in the outbreaks of 2018. In addition, this investigation was performed exclusively on serum samples, and the analysis of whole blood samples should be considered and could have shown advantages when employed in the diagnosis of enteroviral HFMD outbreaks.
MATERIAL AND METHODS
Study population
From February 26th to March 11th 2018, a total of 49 children (≤ 15 years old) sought the outpatient clinic of Arlinda Marques Children’s Hospital with suspected HFMD, were included in this study. Arlinda Marques Children’s Hospital is the largest pediatric hospital of the Joao Pessoa city, the capital of Paraiba State. Paraiba is the third most densely populated State of the Brazilian Northeastern region. It is a tourist and industrial hub, and the city of Joao Pessoa is the sea-bordered State capital, with an estimated population of 817,511 inhabitants 11 .
The definitive HFMD diagnosis was performed at the Arlinda Marques Children’s Hospital based on clinical and epidemiological criteria (Table 1). The HFMD cases were defined based on the presence of oral ulcers, mainly on the tongue and oral mucosa, as well as on the hard and the soft palate, accompanied by typical vesicular eruptions most commonly observed on the extensor surfaces of the hands, feet, buttocks and/or knees 1 . Basic demographic data were collected including age and gender, the date of the onset of disease, the date of blood sampling and clinical manifestations (cough, diarrhea, vomiting, fever and skin lesions). At the time of clinical onset, all 49 children were considered immunocompetent and were defined as mild HFMD, since no neurological and cardiopulmonary complications such as aseptic meningitis, encephalitis, encephalomyelitis, acute flaccid paralysis, as well as pulmonary hemorrhage or cardiorespiratory failure were observed 4,12 . Some patients presented with respiratory or gastrointestinal symptoms and both findings are recognized as part of EV infections 13 . The HFMD cases were reported to the Epidemiological Surveillance Center of Paraiba State (CIEVS/PB), which initiated an investigation in order to identify the EV genotype associated with the HFMD cases. A total of 49 serum samples (one from each patient) collected during the HFMD acute phase were initially sent to the Public Health Laboratory of Paraiba (LACEN-PB) and stored at -20 °C. Oropharyngeal swabs were not available. Frozen serum specimens were then sent to the Institute of Tropical Medicine, University of Sao Paulo (IMT-USP) in order to confirm the EV infection and identify the involved genotype.
Table 1. Clinical data from the 49 EV-positive patients presenting with HFMD infections, Paraiba State, Northeastern region, Brazil, 2018.
| ID | Gender | Age | Signs and symptoms | Other findings | RT-qPCR qualitative result | Ct | Nested RT-PCR result | Species | EV genotyping by 5’UTR | Accession Number | EV genotyping by VP1 | Accession Number |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2B | Female | 1 year | Hand, foot and mouth lesions, fever | Body blisters | - | - | - | - | - | - | - | - |
| 3B | Male | 2 years | Hand, foot and mouth lesions | - | - | - | + | - | - | - | - | - |
| 4B | Male | 1 year | Hand and foot lesions | Buttocks lesions | + | 36.5 | + | EV-A | CV-A6* | MZ457015 | - | - |
| 5B | Male | 1 year | Trunk and limbs lesions, fever | - | - | - | + | EV-A | CV-A6* | MZ464040 | - | - |
| 6B | Female | 2 years | Hand, foot and mouth lesions | Body blisters | - | - | - | - | - | - | - | - |
| 7B | Female | 8 months | Hand, foot and mouth lesions | - | - | - | + | - | - | - | - | - |
| 8B | Male | 3 months | Hand, foot and mouth lesions | - | - | - | + | EV-A | CV-A6* | MZ464041 | - | - |
| 9B | Female | 10 months | Hand, foot and mouth lesions | - | + | 34.9 | - | - | - | - | - | - |
| 10B | Female | 1 year | Trunk and mouth lesions, fever | - | + | 30.4 | + | EV-A | Not assigned | MZ464042 | - | - |
| 11B | Male | 1 year | Hand, foot and mouth lesions, fever | Body blisters | - | - | - | - | - | - | - | - |
| 12B | Male | 1 year | Hand, foot and mouth lesions, fever | - | - | - | + | EV-A | CV-A6* | MZ464043 | - | - |
| 13B | Male | 1 year | Hand, foot and mouth lesions, fever | Seizure, blisters on the face and buttocks | + | 36.5 | + | EV-A | Not assigned | MZ464044 | - | - |
| 14B | Female | 5 years | Hand, foot and mouth lesions, fever | Buttocks lesions | + | 34.6 | + | EV-A | CV-A6* | MZ464045 | - | - |
| 15B | Male | 2 years | Hand, foot and mouth lesions, fever | Buttocks lesions, vomiting, body blisters | - | - | + | - | - | - | - | - |
| 16B | Male | 1 year | Hand, foot and mouth lesions | - | + | 37.8 | + | EV-A | Not assigned | MZ464046 | - | - |
| 17B | Male | 2 years | Hand, foot and mouth lesions, fever | Body blisters, diarrhea | + | 36.6 | + | EV-A | Not assigned | MZ464047 | - | - |
| 18B | Male | 1 year | Hand, foot and mouth lesions, fever | Body blisters | + | 34.1 | + | EV-A | Not assigned | MZ464048 | - | - |
| 19B | Male | 1 year | Hand, foot and mouth lesions, fever | Body blisters, buttocks and elbow lesions | + | 31.9 | + | EV-A | Not assigned | MZ464049 | - | - |
| 21B | Male | 4 months | Hand, foot and mouth lesions | Body blisters | + | 33.2 | + | EV-A | Not assigned | MZ464050 | - | - |
| 22B | Female | 1 year | Hand, foot and mouth lesions, fever | Body blisters | + | 31.5 | + | EV-A | Not assigned | MZ464051 | - | - |
| 23B | Male | 15 years | Hand and mouth lesions, fever | Body blisters | - | - | - | - | - | - | - | - |
| 24B | Male | 10 years | Hand, foot and mouth lesions | Body blisters | - | - | - | - | - | - | - | - |
| 25B | Female | 1 year | Hand, foot and mouth lesions, fever | - | + | 34.5 | + | EV-A | CV-A6* | MZ464052 | - | - |
| 26B | Male | 1 year | Hand, foot and mouth lesions | - | - | - | - | - | - | - | - | - |
| 27B | Male | 9 months | Hand, foot and mouth lesions, fever | Buttocks lesions | + | 29.8 | - | - | - | - | - | - |
| 28B | Male | 1 year | Hand, foot and mouth lesions, fever | - | + | 31.4 | - | - | - | - | - | - |
| 29B | Male | 8 months | Trunk and mouth lesions, fever | - | + | 33.8 | + | EV-A | CV-A6* | MZ464053 | - | - |
| 30B | Male | 1 year | Trunk and limbs lesions | Body blisters | - | - | + | EV-A | Not assigned | MZ476967 | - | - |
| 31B | Male | 1 year | Hand, foot and mouth lesions, fever | Body blisters | + | 30.7 | + | EV-A | CV-A6* | MZ476968 | - | - |
| 32B | Male | 1 year | Fever | Body blisters | - | - | - | - | - | - | - | - |
| 33B | Female | 2 years | Hand, foot and mouth lesions, fever | Perianal lesions | - | - | - | - | - | - | - | - |
| 34B | Male | 1 year | Hand, foot and mouth lesions, fever | - | - | - | - | - | - | - | - | - |
| 35B | Female | 1 year | Hand, foot and mouth lesions, fever | - | - | - | - | - | - | - | - | - |
| 36B | Male | 8 months | Hand, foot and mouth lesions, fever | Body blisters | + | 33.1 | + | EV-A | CV-A6* | MZ476969 | - | - |
| 37B | Male | 1 year | Upper and lower limbs lesions, fever | Buttocks lesions | - | - | - | - | - | - | - | - |
| 38B | Male | 1 year | Trunk and mouth lesions, fever | Vomiting | + | 26.0 | + | EV-A | Not assigned | MZ476970 | CV-A6 | MZ779206 |
| 39B | Female | 8 years | Trunk and limbs lesions | - | - | - | + | - | - | - | - | - |
| 40B | Male | 1 year | Hand, foot and mouth lesions | - | - | - | - | - | - | - | - | - |
| 41B | Female | 1 year | Trunk and mouth lesions, fever | - | + | 31.5 | + | - | - | - | CV-A6 | MZ779207 |
| 42B | Male | 1 year | Trunk and limbs lesions | Vomiting, diarrhea | - | - | + | - | - | - | - | - |
| 43B | Male | 1 year | Hand and foot lesions | Head lesions, coughing | - | - | + | - | - | - | - | - |
| 44B | Female | 3 years | Hand lesions, fever | Coughing | - | - | + | - | - | - | - | - |
| 45B | Female | 3 years | Trunk and limbs lesions | - | - | - | + | - | - | - | - | - |
| 46B | Male | 5 months | Trunk and limbs lesions | - | + | 33.4 | + | - | - | - | - | - |
| 47B | Male | 4 years | Trunk and limbs lesions | - | + | 35.4 | + | EV-A | Not assigned | MZ476971 | Not assigned | - |
| 48B | Female | 8 months | Trunk and limbs lesions | - | + | 28.0 | + | EV-A | Not assigned | MZ476972 | CV-A6 | MZ779208 |
| 49B | Male | 10 months | Trunk and limbs lesions | - | - | - | + | EV-A | CV-A6* | MZ476973 | - | - |
| 50B | Female | 1 year | Mouth lesions, fever | Enlarged ganglia | - | - | - | - | - | - | - | - |
According to the data obtained by the Enterovirus Genotyping Tool.
EV detection
Viral RNA was extracted from serum samples using the PureLink RNA Mini kit (Invitrogen, Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. EV was screened by both real-time RT-PCR (RT-qPCR) 14 and conventional semi-nested RT-PCR targeting the 5’UTR region 15 . As a control for PCR inhibitors and to monitor RNA extraction efficiency, each sample was tested by qRT-PCR for the presence of the human ribonuclease (RNase) P gene following standards protocols 16 . Conventional semi-nested RT-PCR assays were monitored for false-positive or false-negative results using reactions controls containing a positive control, a negative control and sterile water.
Amplification of partial VP1
All EV-positive samples,obtained by RT-qPCR and/or conventional semi-nested RT-PCR, were submitted to VP1 amplification. Reverse transcription was carried out with random hexamer primers, and the region encoding the VP1 region of EV was used in a semi-nested PCR following the protocol described by Iturriza‐Gómara et al. 17 . The primers designed to EV genogroups A, B and C amplified fragments ranging from 355 to 414 base pairs (bp) 17 .
Sequencing and genotyping
PCR amplicons were sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) with the same primers used in the semi‐nested PCRs targeting the 5’ UTR and/or the VP1 regions 15,17 . Dye‐labeled products were sequenced in an ABI 3130 DNA Analyzer (Applied Biosystems). Sequences were edited with the Sequencher 4.7 (Gene Codes Corporation, Ann Harbor, MI). The Enterovirus Genotyping Tool (National Institute for Public Health and the Environment – RMVI) 18 was used to assign the genotype of the studied EV strains.
Phylogenetic analysis
The phylogenetic analysis was carried out using VP1 partial coding-region sequences (310 bp) in order to infer the lineages and phylogenetic relationships among relevant strains. The VP1 region sequences generated here and a set of prototype EV sequences available in GenBank were aligned using the CLUSTAL W algorithm in the BioEdit Sequence Alignment Editor software, version 7.0.5.2 (Ibis Therapeutics, Carlsbad, CA, USA). The selection of the best model considered the likelihood ratio test (LRT) implemented in the jModeltest software, version 2.1.10 (University of Vigo, Vigo, Spain), and the Generalized time reversible (GTR) model using gamma distribution with invariants sites (G + I) was applied. A maximum likelihood tree was constructed using the FastTree 2.1 (Lawrence Berkeley National Laboratory, Berkeley, CA, USA). To obtain reproducible data and provide a major tree clustering reliability pattern, the statistical support of branches was evaluated by the approximate likelihood ratio test (aLRT).
Accession numbers
The sequences of the present study were deposited in GenBank under the accession numbers: MZ457015, MZ464040-MZ464053 and MZ476967-MZ476973 for the 5´UTR region and MZ779206-MZ779208 for the VP1 region.
RESULTS
EV infections were laboratory confirmed in 71.4% of samples (35/49). RT-qPCR and conventional semi-nested RT-PCR were positive for EV in serum samples of 22 (44.9%) and 32 (65.3%) patients, respectively. Both methods resulted positive in 19 (38.8%) patients. The day of blood sampling ranged from the first day after the onset of symptoms to the sixth days, most samples were obtained on the first day (42.8%; 21/49). The percentage of confirmed viremia was the highest on the first day of illness (40%; 14/35) and viremia was lower in patients sampled after day 3 (14.3%; 5/35). Twenty-six of the cases presenting with viremia were detected within the first three days of disease (74.3%; 26/35).
Table 1 summarizes the clinical findings of HFMD patients. The age of the 49 patients ranged from four months to 15 years old, being 35 (71.4%) infants under the age of one year old, 11 (22.4%) were toddlers between 2-5 years old and three (6.1%) children were ≥ 6 years old. No significant difference in the frequency of EV infection was observed when children were sub-divided into these three age groups. The mean and median ages were 1.83 years and one year old, respectively. A total of 32 (65.3%) samples were from males, and 17 (34.7%) were from females (χ 2 =3.6234, p=0.056). Thirty-three (67.3%) patients showed common clinical signs and symptoms of HFDM, with fever and vesicles, mostly on hands, feet and oral mucosa. Eight cases (16.3 %) exhibited vesicular eruptions on the buttocks and perianal area. Twenty-two patients (44.9%) showed severe skin manifestations all over the body, and one had seizures associated with the severity of manifestations. There were no reported cases of meningitis, encephalitis or fatalities.
A 450 bp fragment of the 5’UTR region was sequenced to classify the detected EVs into species for all 32 conventional RT-PCR-positive samples. Twenty-two of the 32 (68.75%) positive samples could be successfully sequenced. All 22 sequenced samples were genotyped as members of EV-A. Ten of them were genotyped as CV-A6 according to the data obtained by the Enterovirus Genotyping Tool, and in twelve strains the genotype could not be assigned. CV-A6 could also be obtained for three extra samples by direct VP1 sequencing of RNA from serum samples, confirming the genotype previously obtained by the 5’UTR sequencing (Table 1).
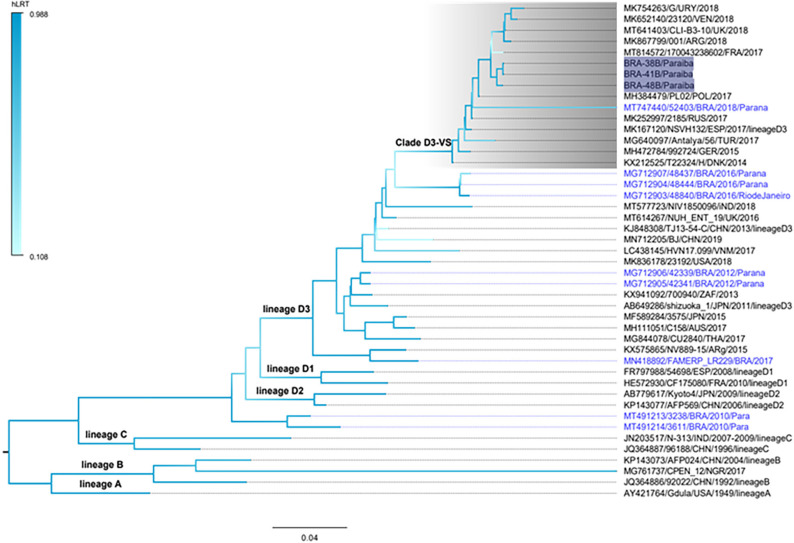
To investigate the genetic relationships among contemporaneous CV-A6 strains, the three Brazilian VP1 partial sequences (nucleotides at positions 2588-2888; 310 bp comprising 102 codons) were characterized here (BRA-38B, BRA-41B and BRA-48B strains) and compared to 42 homologous sequences from worldwide countries detected between 1949 and 2019, including nine CV-A6 sequences previously isolated in Brazil. The CV-A6 lineages (A-D) were determined as formerly proposed 19 . The three Brazilian CV-A6 samples detected in the city of Joao Pessoa were classified as belonging to clade D, sub-lineage D3. In addition, CV-A6 strains BRA-38B, BRA-41B and BRA-48B grouped together with strain 52403 (MT74440) detected in Brazil in 2018, but were separated from the other Brazilian CV-A6 strains sub-lineage D3 that had circulated earlier in the country, from 2012 to 2017 6,13 (Figure 1).
Figure 1. Maximum Likelihood phylogenetic tree of partial VP1 nucleotide sequences (∼310bp) generated with the FastTree software of coxsackievirus A6 (CV-A6) strains detected in patients presenting with HFMD in Paraiba State, Northeastern region, Brazil (highlighted in shaded dark blue). References of CV-A6 were obtained from the GenBank database. Forty-two homologous sequences were obtained from worldwide countries detected between 1949 and 2019, including nine CV-A6 sequences previously isolated in Brazil (highlighted in light blue color). The tree was rooted by using the prototype sequence Gdula (Accession number AY421764). The different CV-A6 lineages (A-D) and sub-lineages (D1-D3) are indicated in the branches. Clade D3-VS (named here) is highlighted in shaded gray. The branch support was achieved by the approximate likelihood ratio test (LRT) and is shown in a color scale on the left side. The tree was rooted in the middle and the scale is indicated at the bottom. Accession number, isolates, countries, and year of description of each strain are indicated.
The phylogenetic tree has also shown that the Brazilian CV-A6 strains appears to belong to a distinct phylloclade (herein designated D3-VS clade) and this finding was highly supported by the statistical test (hLRT=0.941). Clade D3-VS includes strains that were simultaneous circulating in Latin America countries, including Argentina, Uruguay and Venezuela 3,20 , as well as in European countries, such as France and the United Kingdom, in addition to West Asian countries like Turkey (Figure 1). All strains within this clade, with the exception of MK754263/G/URY/2018, share the same amino acid substitutions at positions I671V and A702S. In order to confirm that the clade D3-VS was not an artifact of convergent amino acid substitutions, a novel tree was inferred excluding codons 671 and 702. The resulting tree displayed the same topology of the original one, maintaining an elevated statistical support (hLRT=0.935) (data not shown).
Comparison of CV-A6 BRA-38B, BRA-41B and BRA-48B strains showed nucleotide similarities varying from 99.3% to 100%, and 83.2 to 83.5% when compared to the CV-A6 prototype (CV-A6/Gdula/USA/1949; GenBank accession AY421764).
DISCUSSION
During 2018, major HFMD outbreaks occurred throughout Brazil 9,10 . We tested 49 samples collected at the Pediatric Complex Arlinda Marques, Paraiba State, Brazilian Northeastern region, during the outbreak period, and CV-A6 was identified. Approximately three-fourth of the CV-A6 patients had viremia detected during the acute phase of disease. Cheng et al. 21 in a multi-center cohort study conducted in Taiwan showed that viremia associated with EV-71 occurred more frequently in children of one year of age in HFMD patients, agreeing with the results found in the present study. Cheng et al. 21 also demonstrated that after the third day of disease onset, the proportion of patients with viremia decreased to around one-fourth and no viremia was detected after the seventh day of disease onset, even if patients had persistent clinical symptoms, indicating that the resolution of EV viremia occur within seven days. Our study found a similar trend.
It is well known that RT-qPCR provides rapid diagnosis and increases the sensitivity for EV detection 22 ; however, our results have shown that conventional semi-nested RT-PCR was more sensitive than RT-qPCR. In fact, this is not an entirely surprise, as conventional PCR systems have often shown to be more sensitive than real-time PCR systems in the detection of different pathogens, including viruses 23,24 . Both methods targeted the EV 5’UTR region, but the sensitivity of the conventional PCR could be increased by adding a second round of amplification. On the other hand, conventional PCR methods are more prone to errors, including the lack of amplification products due to higher detection limits depending on the amount of cDNA per reaction 25 . Traditional detection of amplified DNA relies upon electrophoresis of nucleic acids in the presence of ethidium bromide (or similar) and visual analysis of the resulting bands in an ultraviolet light transillumination device 26 . Consequently, some fragments are not clearly detectable on agarose gels since they are visualized as faint bands or are not seen when the template cDNA concentrations was insufficient 25 , including the three EV samples (9B, 27B and 28B) recognized exclusively by RT-qPCR even when other samples with higher Ct values (i.e., samples 4B, 13B, 16B and 17B) were positive by both methods.
CV-A6 caused a massive HFMD outbreak in Finland in 2008, and then spread to other European countries and to North America 27-30 . Most of the largest CV-A6 HFMD- associated outbreaks have been reported in Asian Pacific countries, and CV-A6 has replaced EV-71 and CA-16 as the main enterovirus causing HFMD in the region since 2009 31-34 . In Latin America, CV-A6 associated with HFMD cases began to circulate between 2011 and 2012, with reports from Cuba 35 and Brazil 6 , reaching Peru in 2016 36 . Since 2018, CV-A6 has been more frequently detected in HFMD outbreaks, as well as in sporadic cases in the region 3,5,7,37 . The present study reinforces the importance of the emergence of CV-A6 causing HFMD in Latin America. CV-A6 is additionally characterized by an atypical HFMD clinical presentation with lesions in other anatomical areas, such as the trunk, limbs and buttocks 34 , supporting the clinical data observed here.
EV typing is crucial for assessing the enteroviral biodiversity associated with HFMD infections and also to improve our understanding on their potential public health burden. A limitation of the present report was the failure to genotype all EV‐positive samples. Simultaneous detection and identification of all EV genotypes in clinical samples is very challenging due to its high genetic diversity 20 . EV genotyping mainly depends on Sanger sequencing targeting the VP1 region 38 which clearly impaired our capacity to identify all EV‐positive samples.
Brazilian CV-A6 strains detected here are genetically linked to the strains concurrently circulating in Latin America and European countries 3,37 . Brazilian CV-A6 strains BRA-38B, BRA-41B and BRA-48B are also distantly related to those CV-A6 strains that circulated earlier in Brazil, from 2012 to 2017 6,13 . These data suggest that the Brazilian CV-A6 strains detected in 2018 in Paraiba State and the CV-A6-associated HFMD strains recently reported in Latin America countries, may have a common origin and did not seem to have emerged through the evolution of local CV-A6 strains that circulated previously in Brazil, therefore suggesting a global co-circulation of a set of different CV-A6 strains introduced in the country at different times. Moreover, the sub-lineage D3 became the main CV-A6 sub-lineage since 2008 associated not only to HFMD outbreaks 6,7,34 , but also to meningitis 13 and severe acute respiratory illness 19 . The sub-lineage D3 may have a stronger ability of transmission and it has been suggested that this sub-lineage can be responsible for the growing circulation of CV-A6 worldwide 39 .
Of outstanding significance in the present study was the detection and molecular characterization of EV strains in serum samples. It is well known that the detection of EV exclusively in one serum sample must be carefully interpreted. Single serum samples alone are not suitable for diagnosing EV-associated HFMD. In general, for the EV-associated HFMD diagnosis, it is recommended that oropharyngeal swabs or stool samples are accompanied by paired blood samples collected in the acute-phase and in the convalescent-phase of the disease. The diagnosis of EV is only confirmed when serological tests (neutralization test) present with a significant increase in the antibody titers (three times or more) in the paired serum samples, and these antibodies are raised to the same EV serotype isolated from feces or oropharyngeal swabs 40 . During the HFMD outbreak in Paraiba State reported here, only the acute-phase serum samples were collected, therefore they were the only samples available for the EV diagnosis. The lack of oropharyngeal swabs impaired the testing of EV in both samples in order to evaluate if blood EV PCR testing in patients with suspected HFMD is additionally favored when compared with PCR testing performed on oropharyngeal specimen. Despite this limitation, we have shown that serum samples collected in the acute phase of HFMD are suitable for the diagnosis of EV-associated HFMD by rapid molecular methods, especially during outbreaks emergencies.
CONCLUSION
In conclusion, our findings showed that CV-A6 was involved in the Brazilian HFMD outbreaks that occurred during 2018. The increasing circulation of the emerging CV-A6 associated with HFMD, along with the detection of more severe cases worldwide, suggest the need for a more intense surveillance system of HFMD in Brazil. Our data also support the use of the RT-qPCR method as a powerful tool for a rapid EV diagnosis in blood samples during HFMD outbreaks. The early and timely EV detection in serum samples could positively affect the patients’ management by reducing unnecessary antibiotic and antiviral therapies. In addition, the serum EV genotyping has the potential to assist health care planners to improve the health outcomes in EV-associated HFMD disease.
ACKNOWLEDGMENTS
The authors thank the Pediatric Complex Arlinda Marques staff for their assistance in the samples’ collection; the CGLAB/DAEVS/SVS-MS and the MP Biomedicals Inc. for the donation of reagents.
Footnotes
Ethical approval
A previous Ethics Committee approval was granted by the Faculdade de Medicina of the Universidade de Sao Paulo(CAAE Nº 53153916.7.0000.0065). Since the patients’ informed consents were entered in the study database in a way that the identification of an individual patient’s identification cannot be assessed, a novel informed consent was not needed.
FUNDING
This study was supported by the Sao Paulo Research Foundation (FAPESP) through the grant Nº 2017/00021-9 to ACC, 2019/21706-5 to VSM, 2020/02469-0 to YFVPS, 2020/11182-6 to RSM and 2020/14786-0 to AL. LSA is supported by Coordination for the Improvement of Higher Education Personnel (CAPES). EL is supported by Brazilian National Council for Scientific and Technological Development (CNPq), grant Nº 302677/2019‐4.
References
- 1.Esposito S, Principi N. Hand, foot and mouth disease: current knowledge on clinical manifestations, epidemiology, aetiology and prevention. Eur J Clin Microbiol Infect Dis. 2018;37:391–398. doi: 10.1007/s10096-018-3206-x. [DOI] [PubMed] [Google Scholar]
- 2.Yong W, Qiao M, Shi L, Wang X, Wang Y, Du X, et al. Genetic characteristics of coxsackievirus A16 associated with hand, foot, and mouth disease in Nanjing, China. J Infect Dev Ctries. 2016;10:168–175. doi: 10.3855/jidc.6876. [DOI] [PubMed] [Google Scholar]
- 3.Lizasoain A, Piegas S, Victoria M, Silva EE, Colina R. Hand-foot-and-mouth disease in uruguay: Coxsackievirus A6 identified as causative of an outbreak in a rural childcare center. J Med Virol. 2020;92:167–173. doi: 10.1002/jmv.25590. [DOI] [PubMed] [Google Scholar]
- 4.Xie J, Yang XH, Hu SQ, Zhan WL, Zhang CB, Liu H, et al. Co-circulation of coxsackieviruses A-6, A-10, and A-16 causes hand, foot, and mouth disease in Guangzhou city, China. BMC Infect Dis. 2020;20:271–271. doi: 10.1186/s12879-020-04992-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Justino MC, Mesquita DS, Souza MF, Farias FP, Alves JC, Ferreira JL, et al. Atypical hand-foot-mouth disease in Belém, Amazon region, northern Brazil, with detection of coxsackievirus A6. J Clin Virol. 2020;126:104307–104307. doi: 10.1016/j.jcv.2020.104307. [DOI] [PubMed] [Google Scholar]
- 6.Sousa IP, Jr, Burlandy FM, Costa EV, Tavares FN, Silva EE. Enteroviruses associated with hand, foot, and mouth disease in Brazil. J Infect. 2018;77:448–454. doi: 10.1016/j.jinf.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 7.Sousa IP, Jr, Giamberardino HI, Raboni SM, Debur MC, Oliveira ML, et al. Simultaneous enterovirus EV-D68 and CVA6 infections causing acute respiratory distress syndrome and hand, foot and mouth disease. Virol J. 2021;18:88–88. doi: 10.1186/s12985-021-01560-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brasil. Ministério da Saúde. Gabinete do Ministro Portaria de consolidação Nº 5, de 28 de setembro de 2017. Consolidação das normas sobre as ações e os serviços de saúde do Sistema Único de Saúde. [cited 2022 Jan 24]. Available from: http://portalsinan.saude.gov.br/images/documentos/Legislacoes/Portaria_Consolidacao_5_28_SETEMBRO_2017.pdf .
- 9.Mortari N, Yu AL, Liphaus B, Ferreira PM, Guinoza JA, Silva MR, et al. Doença mão-pé-boca: vigilância de surtos no estado de São Paulo, 2018. Braz J Infect Dis. 2018;22(Suppl 1):26–26. [Google Scholar]
- 10.Minas Gerais. Secretaria de Estado do Saúde SES-MG alerta sobre a síndrome mão-pé-boca. [cited 2022 Jan 24]. Available from: https://www.saude.mg.gov.br/cidadao/banco-de-noticias/story/10408-ses-mg-alerta-sobre-a-sindrome-mao-pe-boca .
- 11.Instituto Brasileiro de Geografia e Estatística Brasil/Paraíba/João Pessoa. [cited 2022 Jan 24]. Available from: https://cidades.ibge.gov.br/brasil/pb/joao-pessoa/panorama .
- 12.Yang X, Li Y, Zhang C, Zhan W, Xie J, Hu S, et al. Clinical features and phylogenetic analysis of severe hand-foot-and-mouth disease caused by Coxsackievirus A6. Infect Genet Evol. 2020;77:104054–104054. doi: 10.1016/j.meegid.2019.104054. [DOI] [PubMed] [Google Scholar]
- 13.Rocha LC, Estofolete CF, Milhim BH, Augusto MT, Zini N, Silva GC, et al. Enteric viruses circulating in undiagnosed central nervous system infections at tertiary hospital in São José do Rio Preto, São Paulo, Brazil. J Med Virol. 2021;93:3539–3548. doi: 10.1002/jmv.26216. [DOI] [PubMed] [Google Scholar]
- 14.Zhang S, Wang J, Yan Q, He S, Zhou W, Ge S, et al. A one-step, triplex, real-time RT-PCR assay for the simultaneous detection of enterovirus 71, coxsackie A16 and pan-enterovirus in a single tube. PLoS One. 2014;9:e102724. doi: 10.1371/journal.pone.0102724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayukekbong J, Kabayiza JC, Lindh M, Nkuo-Akenji T, Tah F, Bergström T, et al. Shift of Enterovirus species among children in Cameroon: identification of a new enterovirus, EV-A119. J Clin Virol. 2013;58:227–232. doi: 10.1016/j.jcv.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Emery SL, Erdman DD, Bowen MD, Newton BR, Winchell JM, Meyer RF, et al. Real-time reverse transcription-polymerase chain reaction assay for SARS-associated coronavirus. Emerg Infect Dis. 2004;10:311–316. doi: 10.3201/eid1002.030759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iturriza-Gómara M, Megson B, Gray J. Molecular detection and characterization of human enteroviruses directly from clinical samples using RT-PCR and DNA sequencing. J Med Virol. 2006;78:243–253. doi: 10.1002/jmv.20533. [DOI] [PubMed] [Google Scholar]
- 18.Kroneman A, Vennema H, Deforche K, Avoort HV, Peñaranda S, Oberste MS, et al. An automated genotyping tool for enteroviruses and noroviruses. J Clin Virol. 2011;51:121–125. doi: 10.1016/j.jcv.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 19.Hellferscee O, Tempia S, Walaza S, Variava E, Dawood H, Wolter N, et al. Enterovirus genotypes among patients with severe acute respiratory illness, influenza-like illness, and asymptomatic individuals in South Africa, 2012-2014. J Med Virol. 2017;89:1759–1767. doi: 10.1002/jmv.24869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oberste MS, Maher K, Kilpatrick DR, Pallansch MA. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol. 1999;73:1941–1948. doi: 10.1128/jvi.73.3.1941-1948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng HY, Huang YC, Yen TY, Hsia SH, Hsieh YC, Li CC, et al. The correlation between the presence of viremia and clinical severity in patients with enterovirus 71 infection: a multi-center cohort study. BMC Infect Dis. 2014;14:417–417. doi: 10.1186/1471-2334-14-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen XX, Qiu FZ, Zhao HL, Yang MJ, Hong L, Xu ST, et al. A novel and highly sensitive real-time nested RT-PCR assay in a single closed tube for detection of enterovirus. Diagn Microbiol Infect Dis. 2018;90:181–185. doi: 10.1016/j.diagmicrobio.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 23.Cattoli G, Drago A, Maniero S, Toffan A, Bertoli E, Fassina S, et al. Comparison of three rapid detection systems for type A influenza virus on tracheal swabs of experimentally and naturally infected birds. Avian Pathol. 2004;33:432–437. doi: 10.1080/03079450410001724058. [DOI] [PubMed] [Google Scholar]
- 24.Hierl T, Reischl U, Lang P, Hebart H, Stark M, Kyme P, et al. Preliminary evaluation of one conventional nested and two real-time PCR assays for the detection of Toxoplasma gondii in immunocompromised patients. J Med Microbiol. 2004;53:629–632. doi: 10.1099/jmm.0.45566-0. [DOI] [PubMed] [Google Scholar]
- 25.Zimmermann C, Hinrichs-Berger J, Moltmann E, Buchenauer H. Nested PCR (polymerase chain reaction) for detection of Xanthomonas fragariae in symptomless strawberry plants. J Plant Dis Prot. 2004;111:39–51. [Google Scholar]
- 26.Kidd IM, Clark DA, Emery VC. A non-radioisotopic quantitative competitive polymerase chain reaction method: application in measurement of human herpesvirus 7 load. J Virol Methods. 2000;87:177–181. doi: 10.1016/s0166-0934(00)00164-6. [DOI] [PubMed] [Google Scholar]
- 27.Osterback R, Vuorinen T, Linna M, Susi P, Hyypiä T, Waris M. Coxsackievirus A6 and hand, foot, and mouth disease, Finland. Emerg Infect Dis. 2009;15:1485–1488. doi: 10.3201/eid1509.090438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bracho MA, González-Candelas F, Valero A, Córdoba J, Salazar A. Enterovirus co-infections and onychomadesis after hand, foot, and mouth disease, Spain, 2008. Emerg Infect Dis. 2011;17:2223–2231. doi: 10.3201/eid1712.110395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirand A, Henquell C, Archimbaud C, Ughetto S, Antona D, Bailly JL, et al. Outbreak of hand, foot and mouth disease/herpangina associated with coxsackievirus A6 and A10 infections in 2010, France: a large citywide, prospective observational study. Clin Microbiol Infect. 2012;18:E110–E118. doi: 10.1111/j.1469-0691.2012.03789.x. [DOI] [PubMed] [Google Scholar]
- 30.Kimmis BD, Downing C, Tyring S. Hand-foot-and-mouth disease caused by coxsackievirus A6 on the rise. Cutis. 2018;102:353–356. [PubMed] [Google Scholar]
- 31.Han JF, Xu S, Zhang Y, Zhu SY, Wu DL, Yang XD, et al. Hand, foot, and mouth disease outbreak caused by coxsackievirus A6, China, 2013. J Infect. 2014;69:303–305. doi: 10.1016/j.jinf.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 32.Wu Y, Yeo A, Phoon MC, Tan EL, Poh CL, Quak SH, et al. The largest outbreak of hand; foot and mouth disease in Singapore in 2008: the role of enterovirus 71 and coxsackievirus A strains. Int J Infect Dis. 2010;14:e1076–e1081. doi: 10.1016/j.ijid.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 33.Puenpa J, Chieochansin T, Linsuwanon P, Korkong S, Thongkomplew S, Vichaiwattana P, et al. Hand, foot, and mouth disease caused by coxsackievirus A6, Thailand, 2012. Emerg Infect Dis. 2013;19:641–643. doi: 10.3201/eid1904.121666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujimoto T, Iizuka S, Enomoto M, Abe K, Yamashita K, Hanaoka N, et al. Hand, foot, and mouth disease caused by coxsackievirus A6, Japan, 2011. Emerg Infect Dis. 2012;18:337–339. doi: 10.3201/eid1802.111147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fonseca MC, Sarmiento L, Resik S, Martínez Y, Hung LH, Morier L, et al. Coxsackievirus A6 and enterovirus 71 causing hand, foot and mouth disease in Cuba, 2011-2013. Arch Virol. 2014;159:2451–2455. doi: 10.1007/s00705-014-2071-x. [DOI] [PubMed] [Google Scholar]
- 36.Rodríguez-Zúñiga MJ, Vértiz-Gárate K, Cortéz-Franco F, Qujiano-Gomero E. Enfermedad de mano, pie y boca en un hospital del Callao, 2016. Rev Peru Med Exp Salud Publica. 2017;34:132–138. doi: 10.17843/rpmesp.2017.341.2710. [DOI] [PubMed] [Google Scholar]
- 37.Sapia EY, Maroni C, Groisman C, Kromer H, Lihue Rojo G, Dastugue M, et al. Genotipificación del virus de la enfermedad mano-pie-boca atípica en un hospital pediátrico de la Ciudad de Buenos Aires, Argentina. Arch Argent Pediatr. 2020;118:e199–e203. doi: 10.5546/aap.2020.e199. [DOI] [PubMed] [Google Scholar]
- 38.Grädel C, Terrazos Miani MA, Barbani MT, Leib SL, Suter‐Riniker F, Ramette A. Rapid and cost‐efficient enterovirus genotyping from clinical samples using flonge flow cells. Genes (Basel) 2019;10:659–659. doi: 10.3390/genes10090659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song Y, Zhang Y, Ji T, Gu X, Yang Q, Zhu S, et al. Persistent circulation of Coxsackievirus A6 of genotype D3 in mainland of China between 2008 and 2015. Sci Rep. 2017;7:5491–5491. doi: 10.1038/s41598-017-05618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harvala H, Broberg E, Benschop K, Berginc N, Ladhani S, Susi P, Christiansen C, et al. Recommendations for enterovirus diagnostics and characterisation within and beyond Europe. J Clin Virol. 2018;101:11–17. doi: 10.1016/j.jcv.2018.01.008. [DOI] [PubMed] [Google Scholar]