Abstract
The PBAF complex, a member of SWI/SNF family of chromatin remodelers, plays an essential role in transcriptional regulation. We revealed a disease progression associated elevation of PHF10 subunit of PBAF in clinical melanoma samples. In melanoma cell lines, PHF10 interacts with MYC and facilitates the recruitment of PBAF complex to target gene promoters, therefore augmenting MYC transcriptional activation of genes involved in the cell cycle progression. Depletion of either PHF10 or MYC induced G1 accumulation and a senescence-like phenotype. Our data identify PHF10 as a pro-oncogenic mechanism and an essential novel link between chromatin remodeling and MYC-dependent gene transcription.
Keywords: chromatin remodeling, transcriptional activation, MYC, PHF10, SWI/SNF, PBAF, melanoma cells
INTRODUCTION
The SWI/SNF chromatin remodeling complex has multiple functions in eukaryotes1,2. SWI/SNF is a positive or negative regulator of gene transcription that interacts with different transcription factors3. Mammalian SWI/SNF complexes are comprised of two major subfamilies, BAF and PBAF, each containing common and variable subunits which control the functional specificity of the complex. In addition, a non-canonical BAF complex has been described2.
The exact mechanism of SWI/SNF function is not completely understood. Individual core subunits such as BRG1 or BRM ATPase, BAF47, BAF155 and BAF170 are directly involved in chromatin-remodeling activity4, whereas the function(s) of BAF57, BAF60, SS18 or BAF as well as PBAF specific subunits BAF250A/B BAF200, BAF180, BAF45C-D, and PHF10 (BAF45A) is largely limited to cell viability and development2. The specific subunits of PBAF (BAF200, BAF180, BRD7 and PHF10) or BAF (BAF250A/B) have been considered important for recruitment of the complexes to target genes through association with transcription regulators, RNA polymerase complex and chromatin5. Many subunits contain DNA-binding domains and domain(s) recognizing histone modifications1. Components of the SWI/SNF complex can physically interact with transcription factors3. Chromatin remodeling allows the promoter activation by the exogenous stimuli; one example is the NFκB pathway in which RelA interacts with DPF1,2,3b and PHF106. Due to the diverse subunit composition, the mammalian SWI/SNF complex is involved in transcriptional regulation of a variety of genes5.
Modulation of the chromatin state in oncogenes is a critical molecular mechanism of tumorigenesis. Individual SWI/SNF subunits play differential roles in tumor biology depending on cell type and protein partner(s). In malignant rhabdoid tumors, the bi-allelic inactivation of SMARCB1 (BAF47) leads to impairment of SWI/SNF complex and its inability to oppose the repression function of the Polycomb7,8. In synovial sarcoma, SS18 subunit of BAF is fused with SSX1/2 protein9,10. The SS18–SSX fusion incorporates into BAF complex thereby recruiting it to SOX2 and PAX6 oncogenic loci. Subsequently, derepression of these genes is mechanistically associated with malignant transformation11. In general, the role of SWI/SNF in regulation of tumorigenesis is determined by epigenetic changes that may lead to different consequences depending on a particular gene or genome region.
The transcription factor MYC is an established regulator of gene expression. In so doing MYC interacts with different transcriptional co-activators12; the major net effect is cell cycle progression. Furthermore, elevated levels of MYC protein have been found in 60-70% of all cancers including solid and hematopoietic malignancies13. It is plausible to hypothesize that pro-oncogenic effects of MYC are mediated by its physical and/or functional association with the chromatin remodeling machinery.
PHF10/BAF45a is a specific subunit of PBAF subfamily of SWI/SNF complexes in metazoan organisms9,14. This non-core subunit of PBAF is recruited to the complex at the final stage of its assembly15. PHF10 possesses the double C-terminal PHD (DPF) domain. A similar domain has been identified in other proteins; it is capable of interacting with acetylated residues in histone 3 (H3K14 and H3K9) and recruiting the complex to active chromatin16,17. PHF10 has been initially discovered in D. melanogaster where its homozygous loss-of-function mutation resulted in lethality whereas a weak mutation led to a reduced viability, an attenuated transcription of the yellow gene, and sterility18-20. Furthermore, PHF10 has been found to be essential for maintenance of proliferation of mouse neuroblasts21, hematopoietic precursors22 and transcriptional activation in myelogenesis23. Almost all mice with PHF10 knockout died at the late stage of embryogenesis; in the survived animals the defects of hematopoiesis were observed22.
The functional role of chromatin-remodeling complexes, SWI/SNF in particular, in MYC-dependent transcriptional regulation is poorly investigated and appears controversial24-26. In the present study we demonstrate that, in cultured melanoma cells, PHF10 physically interacts with MYC thereby facilitating the recruitment of PBAF complex to the regulatory regions of target genes. Furthermore, PHF10 is required for MYC-dependent transcriptional activation, cell cycle progression and attenuation of the senescence phenotype. Accordingly, we demonstrated that PHF10 is frequently overexpressed in human melanomas suggesting its pro-oncogenic function. Alone, PHF10 and MYC phenocopy each other whereas together these proteins cooperate in transcriptional activation of genes important for melanoma maintenance.
RESULTS
PHF10 transcription levels increase along with melanoma progression
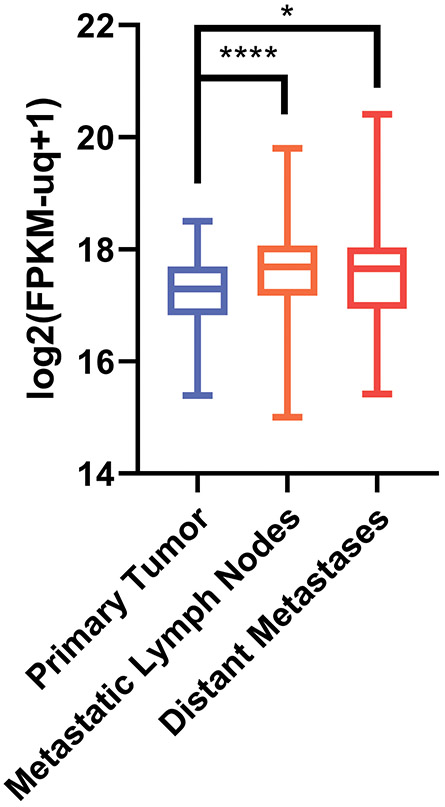
According to the analysis of TCGA database, PHF10 mRNA levels increased from primary melanoma foci to regional cutaneous metastases and further to the distant metastatic nodes, indicating a correlation of PHF10 expression with the disease progression (Fig. 1). Also, MYC protein levels increased along with progression from primary tumors to distant metastases27.
Figure 1. PHF10 mRNA expression (RSEM counts) in cutaneous melanoma.
In GDC TCGA Melanoma cohort (n=477) primary tumors exhibited a lower expression of PHF10 compared to metastatic lymph nodes and distant sites (Kruskal-Wallis test with post hoc Mann Whitney U test and Benjamini-Hochberg correction for multiple comparisons was used to assess the significance). TCGA cutaneous melanoma RNA-Seq dataset (https://cancergenome.nih.gov/) revealed a statistically significant increase of PHF10 transcripts in metastatic vs primary melanoma specimens (*p<0.01, ****p<0.00001). Y axis: log2 FPKM (fragments/kilobase of exon model/million reads mapped).
MYC-associated PBAF complexes in melanoma cell lines contain PHF10
To address potential interactions between PHF10 and MYC, we used melanoma cell extracts for immunoprecipitation with antibodies against the core (BRG1, BAF200) and specific (PHF10, BAF200, BAF180, BRD7) subunits of the PBAF complex. The immunoprecipitated material was probed with the antibodies against PHF10 and other PBAF subunits, as well as against MYC. Antibodies against PHF10 and other PBAF subunits efficiently co-precipitated the endogenous MYC from Sk-Mel28 and Sk-Mel29 melanoma cells (Fig. 2A, B). Consistently, the same antibodies co-precipitated the recombinant FLAG-MYC from A375 melanoma cells that stably expressed this construct (Fig. 2C).
Figure 2. MYC interacts with PBAF complex in melanoma cells.
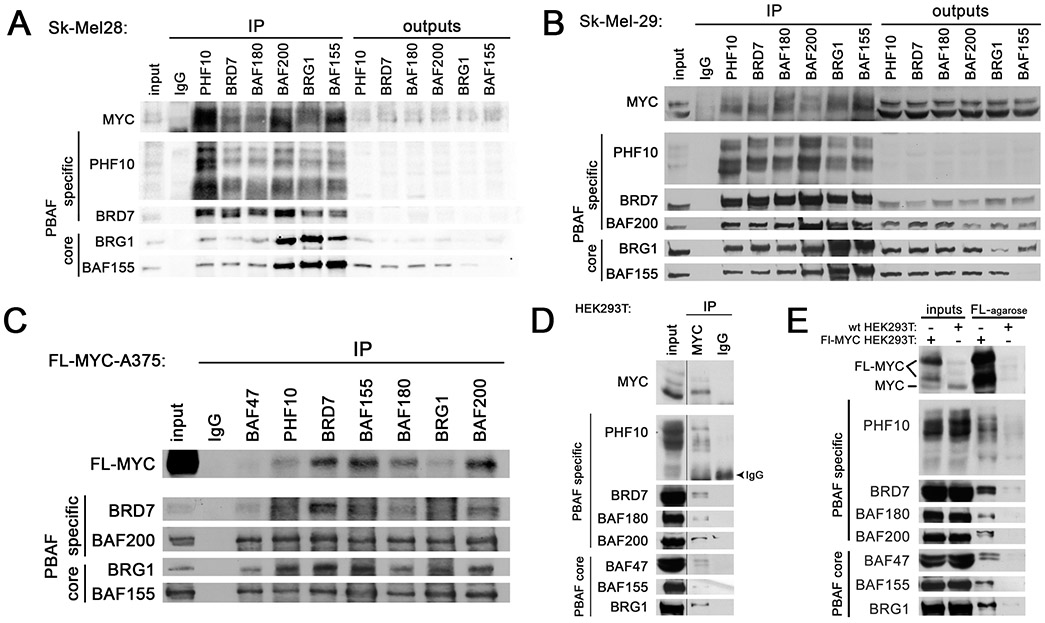
Immunoprecipitation of the endogenous MYC with antibodies against the core and specific PBAF subunits from Sk-Mel28 (A) and Sk-Mel29 (B) cell extracts. PHF10 is detectable as several bands because it has deltaDPF isoform and is highly phosphorylated14. (C) Immunoprecipitation of FLAG-tagged MYC with antibodies against the core and specific PBAF subunits from extracts of A375 cells stably transfected with N-terminal FL-MYC construct. (D) Immunoprecipitation of the core and specific PBAF subunits from HEK293T extracts with antibodies against the endogenous MYC. One percent of IP volume was loaded in MYC-IP. (E) precipitation on FLAG agarose of the core and specific PBAF subunits from extracts of the wild type HEK293T cell line and HEK293T transformed by FLAG-MYC. The output indicates the unbound cell extract.
In reciprocal experiments, we demonstrated that the endogenous or the ectopically expressed MYC was able to co-immunoprecipitate all tested PBAF subunits including PHF10 from the wild type HEK293T cells or HEK293T cells expressing FLAG-MYC (Fig. 2D, E and Fig. S1). These results demonstrate that PHF10 was present in PBAF-MYC complexes.
PBAF-associated PHF10 interacts with MYC
To test whether PHF10 interacts with MYC directly, we down-regulated the endogenous PHF10 using PHF10 siRNA-I and siRNA-II (Fig. S2) in A375 cells that expressed FLAG-tagged MYC (Fig. 3A). Depletion of PHF10 did not lead to disruption of PBAF complexes as confirmed in co-immunoprecipitation with subunit specific antibodies (Fig. 3B and Fig. S3). However, PHF10 knockdown substantially impaired the interaction between PBAF and MYC (Fig. 3C) as evidenced by the fact that, in PHF10 knockdown cells, the amounts of MYC co-precipitated with anti-BAF200 or anti-BAF150 antibodies were significantly smaller than the respective amounts in siControl treated cells.
Figure 3. Influence of PHF10 knockdown on MYC-PBAF interaction in A375 cells.
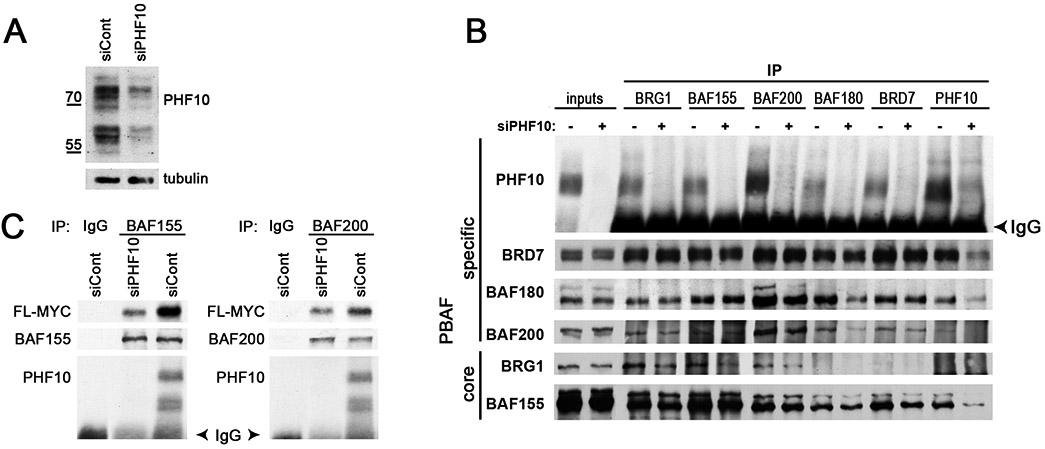
(A) Knockdown of PHF10 significantly decreases its intracellular abundance. (B) PHF10 knockdown does not disrupt PBAF complexes. Immunoprecipitation with antibodies against different PBAF subunits (indicated on top) from the wild type (siPHF10−) and PHF10 knockdown (siPHF10+) cells. (C) Co-immunoprecipitation of MYC with antibodies against BAF200 and BAF155 subunits or with control IgG from extracts of the wild type and PHF10 knockdown cells. Note that the knockdown significantly reduces the amounts of MYC co-precipitated with PBAF.
PHF10 interacts with MYC through MBI, MBII and CTD domains
We were interested in defining the specific regions of the MYC protein responsible for interactions with PHF10. HA-PHF10 were transiently co-expressed in HEK293T cells with the full-length FLAG-MYC or with FLAG-MYC deletion mutants (Fig. 4A) followed by immunoprecipitation with anti-HA antibodies and immunoblotting. In agreement with Fig. 2A-C, PHF10 efficiently co-immunoprecipitated the wild type MYC (Fig. 4B). On the contrary, deletion of MYC Box I and II domains (MBI and MBII, respectively) known to interact with transcriptional co-factors Mediator, TRRAP, p300/CBP, GCN5 and others12, as well as deletion of C-terminal domain bHLH-ZipDNA domain of MYC required for binding with MYC associated factor X (MAX), all affected PHF10-MYC interaction. Deletion of C-terminal DPF domain of HA-PHF10 (deltaDPF) did not alter its association with FLAG-MYC (Fig. 4B, compare upper and lower panels) suggesting that DPF is not involved in PHF10-MYC interaction.
Figure 4. MYC regions responsible for interaction with PHF10.
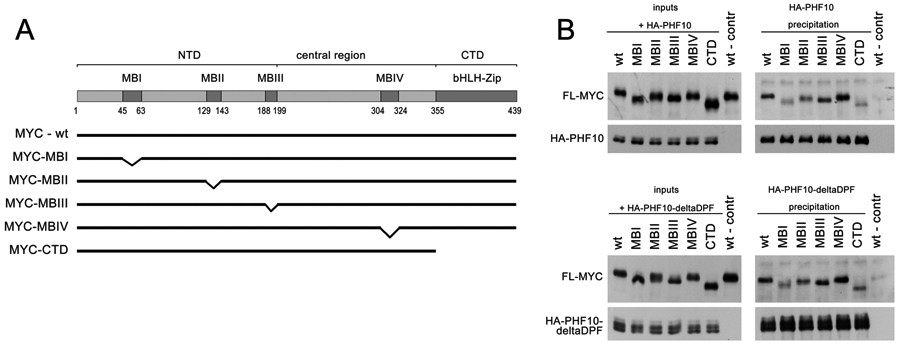
(A) Deletion mutants used in experiments. (B) Interaction of FLAG-MYC with full-length PHF10 and PHF10 lacking DPF domain (deltaDPF). The FLAG tagged full length MYC (wt) or different MYC mutants were overexpressed in HEK293T cells together with HA-tagged PHF10 (top panel) or PHF10 with deleted DPF domain (bottom panel). Shown are inputs (left) and co-immunoprecipitation with anti-HA antibodies (right).
PHF10 cooperates with MYC dependent transcription
To identify whether interaction with PHF10 affects MYC-dependent transcriptional activation, we performed luciferase-based reporter assays. The construct containing the luciferase gene under the promoter of MYC-responsive CDK4 gene was co-transfected with Control or PHF10 siRNAs into HEK293T cells followed by measurement of luciferase activity 48 h post-transfection. A significant decrease of the reporter activity was detected in cells treated with PHF10 siRNA compared with cells treated with Control siRNA, suggesting that PHF10 co-activates MYC-dependent transcription (Fig. 5).
Figure 5. PHF10 knockdown decreases MYC reporter transactivation in HEK293T cells.
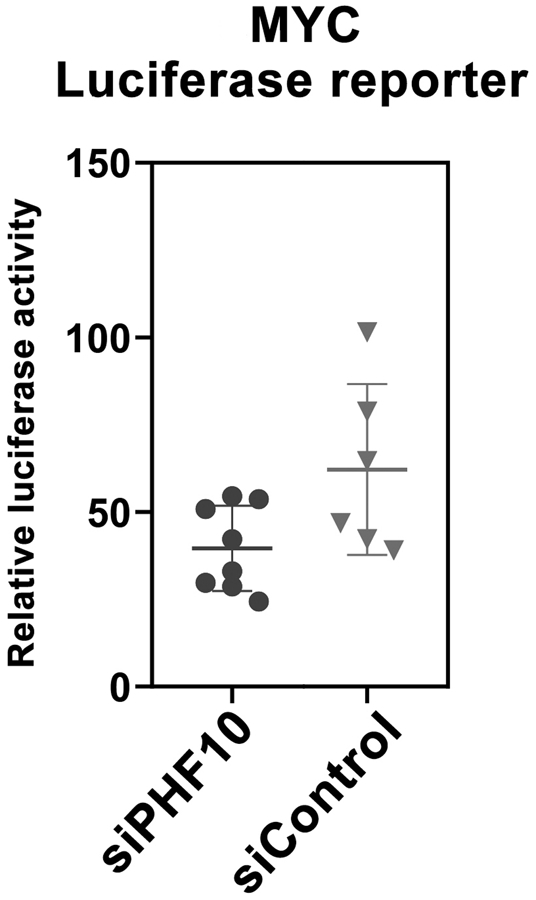
PHF10 was knocked down in HEK293T cells for 48 h. Then cells were transfected with FL-MYC (wt)-pcDNA and MYC-Luc-reporter plasmids. The Firefly luciferase signal was normalized to the Renilla signal. Values on graphs are mean ± SD from 3 independent experiments. Statistical analysis was performed using one-way ANOVA.
PHF10 and MYC regulate the expression of similar sets of genes
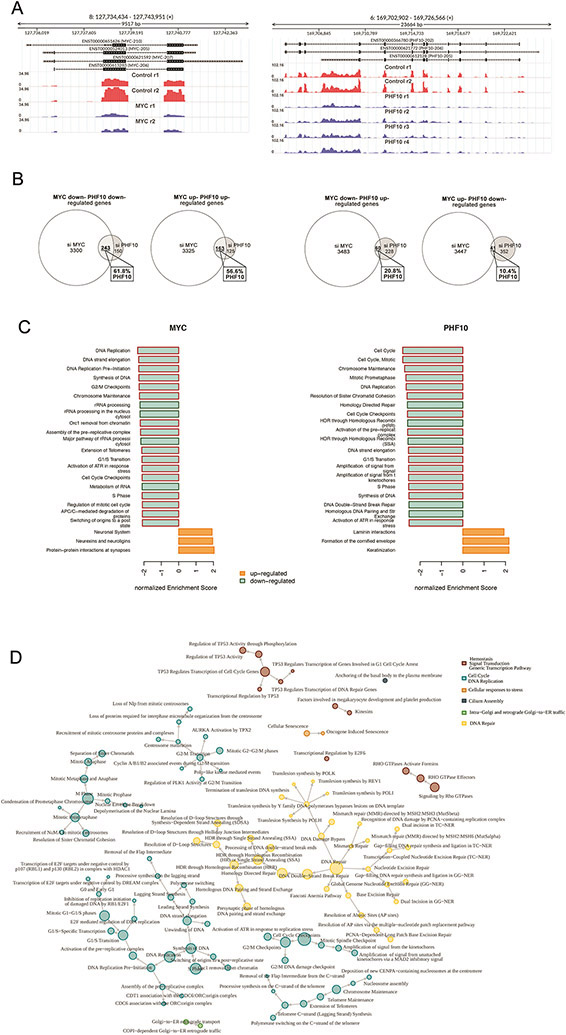
To evaluate the importance of PHF10 for transcriptional regulation by the endogenous MYC, we compared the effects caused by depletion of PHF10 or MYC on global gene expression. The RNASeq assay was performed in A375 melanoma cells transfected with control siRNA or PHF10 siRNA-I and siRNA-II or MYC siRNA-I and siRNA-II (Fig. 6A). The analysis of RNA-Seq data revealed that the majority of differentially expressed genes (DEGs) between control and PHF10-depleted cell populations overlapped significantly with DEGs between control and MYC-depleted cell populations ((FDR<0.05; Fig. 6B). In particular, 61.8% of all DEGs down-regulated by PHF10 knockdown overlapped with genes down-regulated by MYC knockdown. Consistently, 56% of genes up-regulated in PHF10 knockdown cells (compared to control cells) were also up-regulated by MYC knockdown. In contrast, only 20.8% of genes activated by PHF10 knockdown decreased upon MYC knockdown. As little as 10.4% of genes elevated by PHF10 knockdown were down-regulated in MYC knockdown cells (Fig. 6B). Thus, the patterns of genes activated or suppressed by either PHF10 or MYC were highly coincident, indicating that PHF10 cooperates with MYC in transcriptional regulation.
Figure 6. Depletion of PHF10 or MYC induces similar changes in global gene expression.
(A) MYC and PHF10 expression in A375 cells was down-regulated as verified by subsequent RNA-seq. Distribution of RNA-Seq reads over the MYC and the PHF10 genes are shown in red (controls) and blue (knockdown) replicas. Y axis: RPKM normalized expression. (B) Venn’s diagrams of genes down- and up-regulated upon depletion of MYC or PHF10 revealed by RNA-Seq. (C) Top-20 enriched Reactome pathways (sorted by normalized enrichment score) for MYC- and PHF10-dependent genes according to GSEA. Red frames indicate pathways related to Cell Cycle in the Reactome pathways hierarchy. (D) Pathway interaction network for Reactome pathways enriched in intersected MYC/PHF10 down-regulated genes according to ORA analysis. The network was constructed using the Reactome pathway hierarchy. Pathways were clustered based on information about the parent’s nodes in the hierarchy.
Furthermore, the gene set enrichment analysis (GSEA) using Reactome database identified the significantly enriched pathways in MYC and PHF10 knockdown cells (FDR<0.05) (Fig. 6C, Supplementary Tables). Among top 20 pathways enriched by MYC or PHF10 knockdown the majority is relevant to cell cycle progression. Two hundred forty three genes down-regulated by either PHF10 or MYC knockdown were passed to Overrepresentation analysis (ORA) using Reactome database. All enriched pathways were used to construct a network using Reactome pathways hierarchy relationship information and were grouped into clusters named after their parent node in the hierarchy. Most frequently, the genes common for both comparisons (PHF10 siRNA/Control siRNA and MYC siRNA/Control siRNA) belonged to the pathways that regulate the cell cycle, DNA replication, or DNA repair pathways (Fig. 6D) in agreement with the MYC functions in the cell. Likewise, PHF10 regulates the expression of genes that control the abovementioned processes.
Cooperation of PHF10 and MYC in transcriptional activation was further substantiated by global gene expression analysis following knockdown of MYC or PHF10. The presence of PHF10 and MYC on the MYC-binding sites close to the promoter was confirmed by chromatin immunoprecipitation (ChIP; Fig. 7A). The wild type A375 cells and PHF10 knockdown counterparts were cultured in a serum free medium followed by serum replenishment to activate MYC responses (Fig. 7B). As expected, in the wild type cells the serum significantly increased mRNA abundancies of all studied genes (Fig. 7B). However, in cells with PHF10 knockdown these genes were serum-stimulated to a substantially lower extent. These results confirm that PHF10 acts as a co-activator of MYC dependent transcription.
Figure 7. PHF10 cooperates with MYC dependent transcription in A375 melanoma cells.
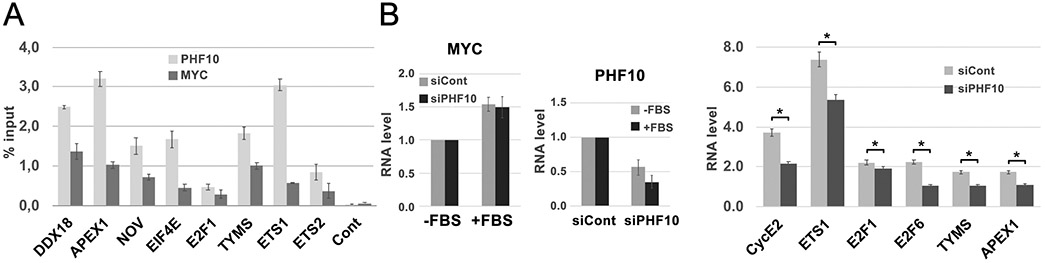
(A) Presence of PHF10 and MYC on the regulatory regions of MYC-dependent genes (ChIP analysis). Regions included the promoters and proximal (+/−150 bp) MYC binding cites. (B) Left: levels of PHF10 and MYC mRNAs in control (siCont) and PHF10 (siPHF10) knockdown cells after serum addition (+FBS). MYC mRNA level in serum starved cells (-FBS) was taken as 1. Middle: serum replenishment does not change PHF10 mRNA abundance after PHF10 down-regulation. PHF10 mRNA level in cells treated with siControl and serum starved was taken as 1. Right: serum replenishment activates MYC responsive genes in a PHF10 dependent manner Shown are the ratios of transcription of an individual gene after serum replenishment to the respective value in serum starved, siControl treated (grey bars) or PHF10 knockdown (black bars) cells (mean ± SD from three independent experiments). Statistically significant differences were marked as *(p < 0.05 for pairs of compared data (one-way ANOVA, Holm-Sidak's multiple comparison test).
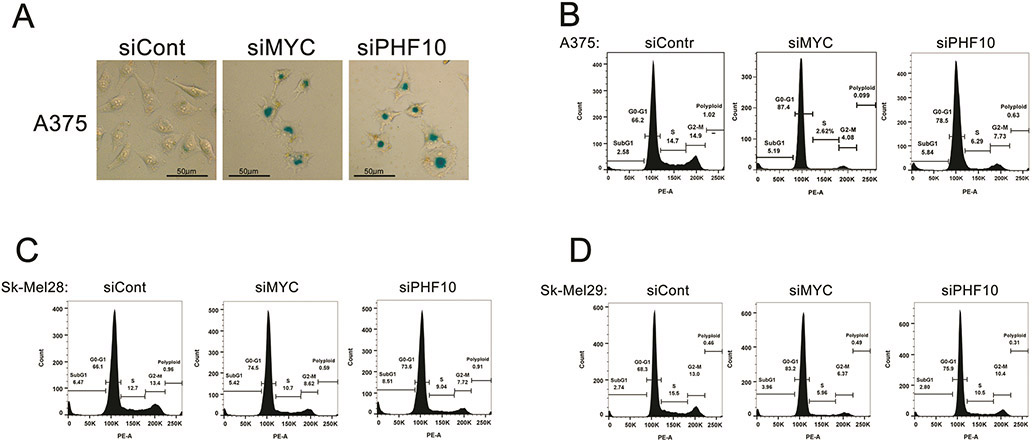
PHF10 depletion induces senescence and G1 arrest in melanoma cells
We have demonstrated that MYC depletion in melanoma cells causes senescence-like phenotype(s)27. Now we investigated whether PHF10 down-regulation can phenocopy MYC depletion. Indeed, PHF10 depletion in A375 cells was accompanied by changes in cell morphology and an increased activity of senescence-associated (SA) β-galactosidase (Fig. 8A). Consistently, MYC depletion caused similar phenotypes (Fig. 8A). Furthermore, knockdown of MYC or PHF10 in Sk-Mel-28, Sk-Mel-29 and A375 melanoma cells led to accumulation in G1 and a concomitant decrease of S and G2/M phases of cell cycle demonstrating similar role of MYC and PHF10 in cell cycle regulation (Fig. 8B). These phenotypic changes substantiate the role of PHF10 as a MYC partner.
Figure 8. Role of PHF10 and MYC knockdown in senescence and cell cycle distribution in melanoma cells.
(A) Knockdowns of PHF10 and MYC cause senescence of A375 cells as determined by SA-β-gal staining (blue). Smaller magnification shows more stained cells (Fig. S4). (B) Knockdown of PHF10 or MYC in A375, Sk-Mel-28 and Sk-Mel-29 cells leads to accumulation in G1 phase.
DISCUSSION
The MYC transcription factor plays a unique role in promoting cell cycle progression and neoplastic transformation. Multiple studies demonstrate that MYC recruits different chromatin-modifying co-factors and co-activators such as histone acetyltransferases, histone demethylases, Mediator and others12. The SWI/SNF chromatin remodeler has been shown to be necessary for MYC-mediated transactivation. The core subunit BAF47 (INI) interacts with MYC helix-loop-helix (bHLH) C-terminal domain, and this interaction helps to recruit the SWI/SNF complex24. BAF47 also opposes MYC function in transcriptional activation25,28. Yet, not much is known about the mechanism of cooperation of SWI/SNF with MYC in transcription.
Here we show that MYC interacts with PHF10, the specific subunit of PBAF family of SWI/SNF. PHF10 has been shown to be important for proliferation of normal and SV40 immortalized human fibroblasts29. However, the mechanism of cell growth maintenance by PHF10 is unclear. We found that PHF10 mediates MYC-PBAF interaction providing the association of PBAF with MYC regulated genes. Importantly, our data demonstrate that PHF10 is critical for activation of MYC regulated genes, as PHF10 depletion significantly decreases MYC dependent transcription. In line with these results the Drosophila SAYP, a homologue of PHF10, has been shown to be essential for recruitment of PBAP, a homologue of PBAF complex in the fly, to the target promoters20. SAYP can directly interact with transcription activators DHR3 or STAT and mediate activator dependent PBAP accumulation on the target genes30,31. Thus, it is conceivable that PHF10 may be recruited by MYC as well as by other transcriptional activators.
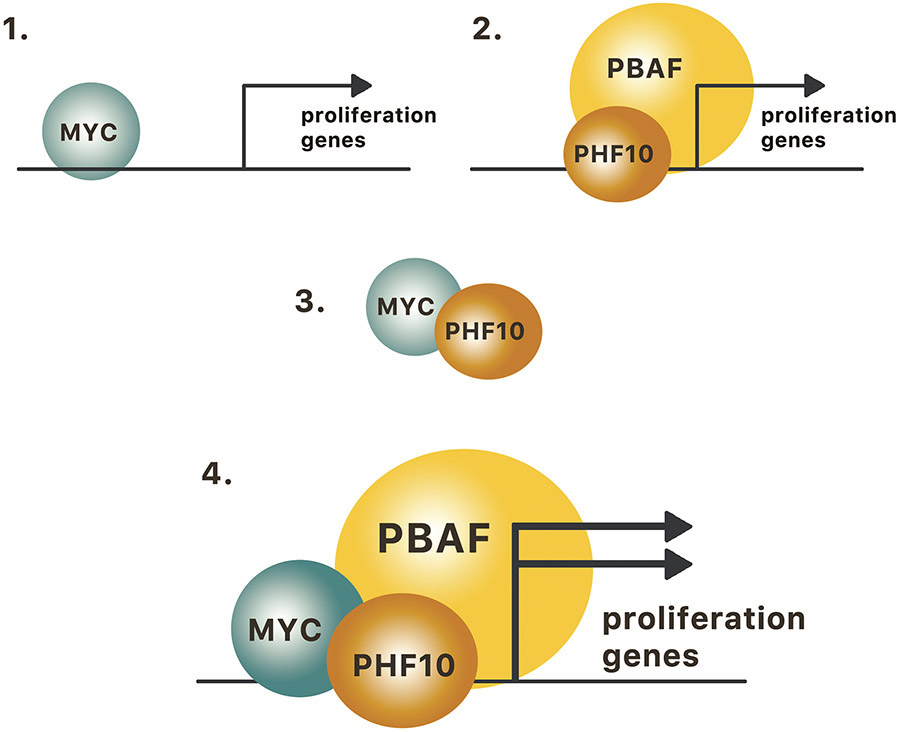
Our data demonstrate that PHF10 is a genome-wide co-activator of MYC driven transcription, a mechanism important for regulation of cell cycle progression. In line with MYC being a powerful activator of proliferation32 the analysis of the global transcriptome in melanoma cells with depleted PHF10 demonstrated that the majority of genes up-regulated by PHF10 are related to cell cycle progression and overlap with MYC activated genes. Similarly to MYC depletion, the depletion of PHF10 led to accumulation of cells in G1 phase concomitant with the decrease of S and G2/M phases and induction of senescence. The similarity between PHF10- and MYC-dependent phenotypes reflects the role of PHF10 as a MYC transcriptional co-activator whose common targets are the cell cycle regulated genes (Fig. 9). Genes whose expression changed upon depletion of PHF10 or MYC may be direct transcriptional targets; otherwise (or in addition to) these genes may be regulated indirectly by secondary mechanisms activated by PHF10/MYC cooperativity. Our findings of PHF10-MYC mediated regulation of cell cycle genes can provide new therapeutic opportunities.
Figure 9. MYC and PBAF cooperate in activation of genes involved in proliferation.
1. MYC regulates proliferation genes; 2. PHF10 in the context of PBAF regulates proliferation genes; 3. MYC interacts with PHF10; 4. MYC together with PHF10-containing PBAF complex cooperatively activate cell proliferation.
Next, we found that MBI, MBII, and bHLH domains of MYC protein are involved in interaction with PHF10. The conserved MBI and MBII domains participate in interaction with distinct transcriptional co-regulators and chromatin modifiers such as BRD4, p400, GCN5, and TRRAP, while bHLH domain interacts with MAX and BAF4712. The PHF10-MYC interactions thus follow a predictable pattern of interactions proven for other chromatin modifiers.
MYC is overexpressed in the course of progression of malignancies including melanoma27. MYC dependent suppression of oncogene-induced senescence is a prerequisite for transition from benign nevi to melanoma. The TCGA database demonstrated the correlation of PHF10 expression levels with melanoma progression (this study). Depletion of either MYC or PHF10 was associated with senescence-like phenotypes in melanoma cells, suggesting that PHF10 may facilitate MYC-driven melanoma progression.
SWI/SNF subunits are implicated in different types of cancers33-35. BAF47 (SMARCB1) inactivation by biallelic mutations has been detected in nearly 100% of rhabdoid tumors7,36,37 while Brg1/SMARCA4 inactivation in > 90% small cell ovarian carcinomas38. On the other hand, BRG1 is required to maintain proliferation and survival of acute myeloid leukemia cells through enhancer-mediated MYC regulation39. The PBAF subunit Brg1 can be strongly increased in melanoma cells while down-regulation of the BRG1 gene suppressed cell proliferation40. Recent study suggested that PHF10 is a potential tumor suppressor: PHF10 depletion down-regulated adhesion and migration in Mel202 and 92.1 uveal melanoma cell lines. Moreover, a homozygous PHF10 deletion was found in patients’ uveal melanoma cells41. These findings indicate that functions of SWI/SNF subunits are highly cell type- and context-specific33.
The role of SWI/SNF complex in MYC-dependent phenotypes is context-specific and therefore ambiguous. MYC has been shown to interact with integrase interactor 1 (INI1) protein, a core subunit of the SWI/SNF complex, and to recruit it to the promoters of MYC target genes24,42. Interaction with INI1 has been attributed to the transcriptional activation by MYC24. On the other hand, INI1 and MYC exert opposite functions on transactivation of individual MYC regulated genes important for the cell cycle progression25. Identification of PHF10 as a co-activator of MYC-dependent transcription in melanoma cells provides a novel mechanistic insight into the role of chromatin remodeling complexes in oncogenic functions of MYC.
METHODS
Reagents were purchased in Sigma-Aldrich unless specified otherwise.
Plasmids
For FL-MYC (wt)-pcDNA expressing vector, MYC cDNA was cloned into the FL-pcDNA plasmid. Mutants MBI (deletion of 45-63 aa), MBII (deletion of 129-143 aa), MBIII (deletion of 188-199 aa), MBIV (deletion of 304-324 aa) and DelCTD (deletion of 355-429 aa) were cloned using NEBuilder (NEB) and primers described in Table S1. To obtain A375 cells carrying FL-MYC inducible by doxycycline, FL-MYC was cloned from FL-MYC-pcDNA into pSLIK plasmid (Addgene #2573543). Lentiviral particles were produced using pCMV-VSV-G (Addgene #845444) and pCMV-dR8.2 (Addgene #845544) plasmids. The LucReporter plasmid contained CDK4 promoter with four MYC binding sites (Addgene #1656445). The RenillapRL plasmid (Promega) was used for signal normalization.
Antibodies
Purified polyclonal antibodies against PHF10, BRD7, BAF155, BAF200, BAF180, and BRG1 subunits were produced by us as described14,46. The M2 clone was used as anti-FLAG antibodies. MYC antibodies were from Cell Signaling (E5Q6W). HPR-conjugated anti-rabbit IgG and HPR-conjugated anti-mouse goat IgG were from DHGB.
Cell lines
Human melanoma Sk-Mel28, Sk-Mel29 and A375 cell lines as well as human embryo kidney HEK293T cells (all from American Type Culture Collection, Manassas, VA) were propagated in Dulbecco modified Eagle's medium (DMEM; PanEco, Russia) with 10% fetal bovine serum (HyClone, Logan, UT) supplemented with 2 mM L-glutamine (Merck) and penicillin/streptomycin at 37°C, 5% CO2 in a humidified atmosphere. All cell lines were routinely tested for Mycoplasm contamination by DAPI staining.
Transfection of HEK293T cells was performed with FL-MYC (wt)-pcDNA, FL-MYC (mut)-pcDNA or FL-MYC-pSLIK with pCMV-VSV-G and pCMV-dR8.2 plasmids. 10 μg of plasmids were mixed with 20 μl (20 μg) polyethylenimine (Polysciences) for transfection in 60 mm Petri dish; 30 μg plasmids and 60 μl polyethylenimine for 90 mm Petri dish. Mixtures were incubated for 15 min at room temperature and added to cells grown to 50% confluence. Cells were harvested 48 h post-transfection.
Genetic inactivation of PHF10 or MYC
Knockdown of PHF10 and MYC in A375, SK-Mel-28 and Sk-Mel-29 cells was performed with siRNA-I and siRNA-II added simultaneously. Sequences of PHF10-siRNA, MYC-siRNA and control siRNA as well as immunoblotting validation are listed in Supplementary (Fig. S2, Table S2). 0,5x106 cells were seeded per one 60 mm Petri dish. Metafecten Pro (Biontex; 10 μl) and siRNAs (total 200 pMol) were mixed with 0.5 ml Opti-MEM media (Gibco), then the mixtures were pooled, incubated for 15 min at room temperature and added to cells in the antibiotic free DMEM. Cells were incubated at 37°C, 5% CO2 for 48 h or 72 h and harvested.
Lentiviral transduction
To obtain FL-MYC-A375 subline stably expressing the doxycycline-inducible FL-MYC, FL-MYC-pSLIK was co-transfected into HEK293T cells together with VSV-G and pCMV-dR8.2 vectors (10 μg of each plasmid per 90 mm Petri dish). Supernatants with viral particles were collected at the 2nd day after transfection, filtered through 0.22 μm membrane, then 10 ml was added to A375 cells. Next day the media was replaced with the media containing 0.5 μg/ml G418. Selection continued for 10 days followed by immunostaining.
Luciferase reporter assays
PHF10 knockdown using siPHF10 was performed in A375 cells cultured on 60 mm Petri dishes for 48 h as described above. Control cells were treated with siControl. Then cells were plated into a 96-well plate and transfected with the mixture of plasmids containing FL-MYC (wt)-pcDNA (48.5%), MYC-Luc-reporter (48.5%) and Renilla luciferase (3%). Twenty four h later cells were harvested. The luciferase signals were measured using single tube assay kit (Biotium) on a Biotek Synergy 4 luminometer according to the manufacturer’s protocol. Statistical analysis was performed using one-way ANOVA. ANOVA assumptions were testes using Shapiro-Wilk test (normality in each group) and Levene's test (homogeneity of variance across groups). The data meet both assumptions.
Immunoprecipitation and immunoblotting
For immunoprecipitation, the harvested cells were lysed in the buffer containing 10 mM HEPES pH 7.9, 5 mM MgCl2, 0.5% NP-40, 0.45 M NaCl, 1 mM DTT, protease inhibitor cocktail (PIC; Roche), phosphatase inhibitor cocktails II and III (PhIC) supplemented with DNase I and RNAse I (both from Thermo Fisher Sci.) at 4°C for 15 min. Lysates were centrifuged at 10,000 rpm at 4°C for 10 min, the supernatant was diluted 4-fold with the same buffer without NaCl. Immunoprecipitation was started by adding 15 μl of antibody-saturated protein A-Sepharose beads or 15 μl HA-agarose to the cell extract (3 × 106 cells per round) followed by overnight incubation at 4°C on a rotary platform. Beads were washed twice with IP-500 buffer (25 mM Tris-HCl pH 7.9; 5 mM MgCl2; 10% glycerol; 500 mM NaCl; 0.1% NP-40) and once with IP-100 buffer (25 mM Tris-HCl pH 7.9; 5 mM MgCl2; 10% glycerol; 100 mM NaCl; 0.1% NP-40) supplemented with PIC and PhIC. Precipitated proteins were eluted with 4×Laemmli buffer (200 mM Tris-HCl pH 6.8; 4% SDS; 40% glycerol; 0.01% bromophenol blue; 100 mM DTT) and boiled for 10 min. For immunoblotting, cells were lysed in RIPA buffer (50 mM Tris-HCl pH 7.4; 1% NP-40; 0.5% Na deoxycholate; 0.1% SDS; 150 mM NaCl; 2 mM EDTA; PIC and PhIC) and centrifuged at 12,000 rpm, 4°C. Protein concentration was measured using Qubit protein assay kit (Thermo Fisher Sci.).
Chromatin immunoprecipitation
Cells (3 × 106) were cross-linked using 1% formaldehyde solution in PBS, treated with 2.5 M glycine and washed three times with cold PBS supplemented by PIC. Then cells were resuspended in a sonication buffer (50 mM HEPES-KOH pH 7.9; 140 mM NaCl; 1 mM EDTA; 1% Triton X-100; 0.1% Na deoxycholate; 0.1% SDS) with PIC. DNA was sheared to ~ 500 bp by sonication for 30 s with 30 s intervals, 10 cycles, and then centrifuged for 15 min at 12000 rpm at 4°C. Mab-select beads (GE Health) were pre-incubated with 1% BSA in PBS with 0.1% NP-40 at 4°C overnight, then 15 μl beads were added to the chromatin solution and anti-MYC (5 μl) or anti-PHF10 (15 μl) antibodies. After precipitation the beads were washed, DNA was eluted by Elution Buffer (SDS 1% and 100mM NaHCO3) and extracted by phenol-chloroform method. Sequences of primers for quantitative PCR are presented in Supplementary (Table S3). The MYC and PHF10 enrichments on promoters were calculated as the percentage of the input material. As a control of specificity of anti-MYC and anti-PHF10 antibodies the binding to the chromosome 3 intergenic region was used.
Gene expression analysis
RNA was isolated from 3 x 106 cells using TriReagent (MRC) according to the manufacturer's protocol. Two μg RNA was used for cDNA synthesis with oligo(dT) primer and MMLV reverse transcriptase (Thermo Fisher Sci.). PCR primers are listed in Supplementary (Table S4). Values were normalized to the RPLP0 housekeeping gene. At least three independent experiments were performed; values are mean ± SD. Statistical analysis was performed using a one-way ANOVA with Holm-Sidak's multiple comparison test, GraphPadPrism6 software. P values < 0.05 were considered significant. ANOVA assumptions were testes using Shapiro-Wilk test (normality in each group) and Levene's test (homogeneity of variance across groups). The data meet both assumptions.
Preparation of libraries and NGS
Total RNA from A375 cells after knockdown of MYC or PHF10 (isolated as described above) was treated with DNase I (Thermo Fisher Sci.) at 37°C for 15 min followed by phenol-chloroform extraction and 96% ethanol precipitation. For NGS library preparation, one μg of RNA and NEBNext Ultra II Directional RNA library prep kit for Illumina with NEBNext Poly(A) mRNA magnetic isolation module (NEB) were used according to the manufacturer’s protocol. Adaptor ligation was performed using NEBNext multiplex oligos for Illumina index primer set #1 (NEB). Quality of RNA and cDNA libraries was analyzed with Bioanalyzer RNA 6000 Nano Chip and Bioanalyzer DNA 1000 Chip kits (Agilent). RNA and DNA yields were assessed using Qubit assay kits (Thermo Fisher Sci.). Libraries were sequenced on an Illumina NovaSeq 6000 with single strand 100 bp length reads. Approximately 3x107 reads were obtained per one sample.
Analysis of RNA-Seq data
Raw data were the single end reads from two biological replicates for MYC knockdowns and four for PHF10 knockdowns. Data are available under the accession number GSE164726. Reads were preprocessed as follows: cutadapt software47 was used to remove the adapters and poly-A tails and trimming the low quality ends (quality threshold was set to 20); the reads shorter than 20 bp post-trimming were discarded. The processed reads were aligned to Homo sapiens genome (assembly GRCh38.p13) using the splice-aware STAR algorithm48. Gene read counts were calculated using STAR quantMode option (quantModeGeneCounts). The analysis was performed in R version 3.4.4. Differentially expressed genes were identified using edgeR package version 3.20.949 as follows. First, the read counts were normalized using TMM algorithm50 after discarding the reads with multiple mapping, those overlapping with more than one gene (ambiguous) and not overlapping with genes (noFeature). Genes differentially expressed between each group of samples and appropriate control were identified using the estimateDisp, glmFit and glmLRT functions with 0.05 FDR significance threshold. GSEA was applied to the set of differentially expressed genes identified for each sample group. ORA was used to obtain the pathways enriched in the set of intersecting genes down-regulated upon MYC and PHF10 depletion. ORA and GSEA were performed using WebGestalt toolkit51 and Reactome database52. Pathways with FDR<0.05 were considered as significantly enriched. Pathways interaction network was constructed using the hierarchy information from the Reactome database52. Pathways were clustered according to the parent nodes in the hierarchy. The coverage tracks were obtained using the deeptools53 bamCoverage function for the forward and reverse strand separately with RPKM normalization and 50 bp binning. Visualization of MYC and PHF10 down-regulation during knockdown was performed using svist4get software54.
Cell cycle analysis
A375, SK-Mel-28 and Sk-Mel-29 cells were transfected with siRNA as described above. After 48 h of incubation cells were washed with ice cold PBS, detached with 0.25 mM EDTA solution in PBS and lysed in a buffer containing 0.1% sodium citrate, 0.3% NP-40, 10 μg/ml RNAse A and 10 μg/ml propidium iodide for 30 min in the dark. Cells were analyzed on a BD FACS Canto II flow cytometer in the PerCP-Cy5 channel. Ten thousand events were collected per each sample. Data were analyzed using FlowJo software.
SA-β-gal staining
A375, SK-Mel-28 and Sk-Mel-29 cells were transfected with siRNA as described above. Forty eight h later cells were rinsed with PBS and fixed (15 min at room temperature) with the buffer containing 0.5% glutaraldehyde and 1 mM MgCl2 in PBS. After fixation cells were rinsed with PBS, stained with the solution containing 1 mM MgCl2, 3 mM K3Fe(CN)6, 3 mM K4Fe(CN)6, 0.02% NP-40, and 0.04% bromo-chloro-indolyl-galactopyranoside (X-gal) in saline pH 6.0 for 16 h at 37°C, washed with PBS and photographed under a light microscope.
Supplementary Material
Acknowledgment
We thank the Center for Precision Genome Editing and Genetic Technologies for Biomedicine (Institute of Gene Biology) supported by Ministry of Science and Higher Education of the Russian Federation (075-15-2019-1661) for providing the equipment.
Funding:
This study was supported by the Russian Foundation of Basic Research (grant #17-54-33031 (to S.G.G.) and NIH R21 CA220096 grant (to M.A.N).
Footnotes
Conflict of interest
The authors declare no competing financial interests.
References
- 1.Kadoch C and Crabtree GR Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci Adv. 197, 804–809 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfert A, Moreno N and Kerl K The BAF complex in development and disease. Epigenetics and Chromatin 12, 1–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centore RC, Sandoval GJ, Soares LMM, Kadoch C and Chan HM Mammalian SWI/SNF Chromatin Remodeling Complexes: Emerging Mechanisms and Therapeutic Strategies. Trends Genet. 36, 936–950 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Phelan ML, Sif S, Narlikar GJ and Kingston RE Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3, 247–253 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Trotter KW and Archer TK The BRG1 transcriptional coregulator. Nucl. Recept. Signal 6, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishizaka A, Mizutani T, Kobayashi K, Tando T, Sakurai K, Fujiwara T et al. Double plant homeodomain (PHD) finger proteins DPF3a and −3b are required as transcriptional co-activators in SWI/SNF complex-dependent activation of NF-κB RelA/p50 heterodimer. J. Biol. Chem 287, 11924–11933 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394, 203–206 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Roberts СW, Galusha SA, McMenamin ME, Fletcher CD, and Orkin SH Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. PNAS 5, 13796–13800 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Middeljans E, Wan X, Jansen PW, Sharma V, Stunnenberg HG, Logie C SS18 together with animal-specific factors defines human BAF-type SWI/SNF complexes. PLoS One 7, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crew AJ, Clark J, Fisher C, Gill S, Grimer R, Chand A et al. Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel-associated box in human synovial sarcoma. EMBO J. 14, 2333–2340 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet 45, 592–602 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poole CJ & van Riggelen J MYC—master regulator of the cancer epigenome and transcriptome. Genes (Basel). 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dang CV MYC on the path to cancer. Cell 149, 22–35 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brechalov AV, Georgieva SG & Soshnikova NV Mammalian cells contain two functionally distinct PBAF complexes incorporating different isoforms of PHF10 signature subunit. Cell Cycle 13, 1970–1979 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mashtalir N, D'Avino AR, Michel BC, Luo J, Pan J, Otto JE et al. Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes. Cell 175, 1272–1288.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng L, Zhang Q, Li S, Plotnikov AN, Walsh MJ, Zhou M et al. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature 466, 258–62 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soshnikova NV, Sheynov AA, Tatarskiy EV and Georgieva SG The DPF Domain As a Unique Structural Unit Participating in Transcriptional Activation, Cell Differentiation, and Malignant Transformation. 12, 115–123 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shidlovskii YV, Krasnov AN, Nikolenko JV, Lebedeva LA, Kopantseva M, Ermolaeva MA et al. A novel multidomain transcription coactivator SAYP can also repress transcription in heterochromatin. EMBO J. 24, 97–107 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chalkley GE, Moshkin YM, Langenberg K, Bezstarosti K, Blastyak A, Gyurkovics H et al. The Transcriptional Coactivator SAYP Is a Trithorax Group Signature Subunit of the PBAP Chromatin Remodeling Complex. Mol. Cell. Biol 28, 2920–2929 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vorobyeva NE, Soshnikova NV, Nikolenko JV, Kuzmina JL, Nabirochkina EN, Georgieva S et al. Transcription coactivator SAYP combines chromatin remodeler Brahma and transcription initiation factor TFIID into a single supercomplex. Proc. Natl. Acad. Sci. U. S. A 106, 11049–11054 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl B et al. An Essential Switch in Subunit Composition of a Chromatin Remodeling Complex during Neural Development. Neuron 55, 201–215 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krasteva V, Crabtree GR and Lessard JA The BAF45a/PHF10 subunit of SWI/SNF-like chromatin remodeling complexes is essential for hematopoietic stem cell maintenance. Exp. Hematol 48, 58–71.e15 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Viryasova GM, Tatarskiy VV Jr., Sheynov AA, Tatarskiy Eu. V., Sud'ina GF, Georgieva SG et al. PBAF lacking PHD domains maintains transcription in human neutrophils. Biochim. Biophys. Acta - Mol. Cell Res 1866, (2019). [DOI] [PubMed] [Google Scholar]
- 24.Cheng SW, Davies KP, Yung E, Beltran RJ, Yu J, Kalpana GV c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat. Genet 22, 102–105 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Stojanova A, Tu WB, Ponzielli R, Kotlyar M, Chan P, Boutros PC et al. MYC interaction with the tumor suppressive SWI/SNF complex member INI1 regulates transcription and cellular transformation. Cell Cycle 15, 1693–1705 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park J, Wood MA & Cole MD BAF53 Forms Distinct Nuclear Complexes and Functions as a Critical c-Myc-Interacting Nuclear Cofactor for Oncogenic Transformation. Mol. Cell. Biol 22, 1307–1316 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhuang D, Mannava S, Grachtchouk V, Tang W-H, Wawrzyniak JA, Berman AE et al. c-MYC overexpression is required for continuous suppression of oncogene-induced senescence in melanoma cells. Oncogene 27, 6623–6634 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sammak S, Allen MD, Hamdani N, Bycroft M & Zinzalla G The structure of INI1/hSNF5 RPT1 and its interactions with the c-MYC:MAX heterodimer provide insights into the interplay between MYC and the SWI/SNF chromatin remodeling complex. FEBS J. 285, 4165–4180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banga SS, Peng L, Dasgupta T, Palejwala V & Ozer HL PHF10 is required for cell proliferation in normal and SV40-immortalized human fibroblast cells. Cytogenet. Genome Res 126, 227–242 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panov VV, Kuzmina JL, Doronin SA, Kopantseva MR, Nabirochkina EN, Georgieva SG et al. Transcription co-activator SAYP mediates the action of STAT activator. Nucleic Acids Res. 40, 2445–2453 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vorobyeva NE, Nikolenko JV, Nabirochkina EN, Krasnov AN, Shidlovskii YV, Georgieva SG et al. SAYP and Brahma are important for ‘repressive’ and ‘transient’ Pol II pausing. Nucleic Acids Res. 40, 7319–7331 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.García-Gutiérrez L, Delgado MD & León J Myc oncogene contributions to release of cell cycle brakes. Genes (Basel). 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mittal P & Roberts CWM The SWI/SNF complex in cancer — biology, biomarkers and therapy. Nat. Rev. Clin. Oncol 17, 435–448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ribeiro-Silva C, Vermeulen W & Lans H SWI/SNF: Complex complexes in genome stability and cancer. DNA Repair (Amst). 77, 87–95 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Masliah-Planchon J, Bièche I, Guinebretière J-M, Bourdeaut F & Delattre O SWI/SNF Chromatin Remodeling and Human Malignancies. Annu. Rev. Pathol 145–171 (2014). doi: 10.1146/annurev-pathol-012414-040445 [DOI] [PubMed] [Google Scholar]
- 36.Biegel JA, Zhou JY, Rorke LB, Stenstrom LB, Wainwright LM, Fogelgren B et al. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 59, 74–79 (1999). [PubMed] [Google Scholar]
- 37.Sévenet N, Sheridan E, Amram D, Schneider P, Handgretinger R, Delattre O Constitutional mutations of the hSNF5/INI1 gene predispose to a variety of cancers. Am. J. Hum. Genet 65, 1342–1348 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu B & Shi H An in-depth look at small cell carcinoma of the ovary, hypercalcemic type (SCCOHT): Clinical implications from recent molecular findings. J. Cancer 10, 223–237 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buscarlet M, Krasteva V, Ho L, Simon C, Hebert J, Wilhelm B et al. Essential role of BRG, the ATPase subunit of BAF chromatin remodeling complexes, in leukemia maintenance. Blood 123, 1720–1728 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saladi SV and de la Serna IL ATP dependent chromatin remodeling enzymes in embryonic stem cells. Bone 6, 62–73 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anbunathan H, Verstraten R, Singh AD, Harbour WJ and Bowcock AM Integrative copy number analysis of uveal melanoma reveals novel candidate genes involved in tumorigenesis including a tumor suppressor role for PHF10/BAF45A. Clin. Cancer Res 25, 5156–5166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bagnasco L, Tortolina L, Biasotti B, Castagnino N, Ponassi R, Tomati V et al. Inhibition of a protein-protein interaction between INI1 and c-Myc by small peptidomimetic molecules inspired by Helix-1 of c-Myc: identification of a new target of potential antineoplastic interest. FASEB J. 21, 1256–1263 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Shin KJ, Wall EA, Zavzavadjian JR, Santat LA, Liu J, Hwang J et al. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc. Natl. Acad. Sci. U. S. A 103, 13759–13764 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. Rna 9, 493–501 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett JF, Obaya AJ et al. Identification of CDK4 as a target of c-MYC. Proc. Natl. Acad. Sci. U. S. A 97, 2229–2234 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brechalov AV, Valieva ME, Georgieva SG & Soshnikova NV PHF10 isoforms are phosphorylated in the PBAF mammalian chromatin remodeling complex. Mol. Biol 50, 278–283 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011). [Google Scholar]
- 48.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson MD, McCarthy DJ and Smyth GK edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson MD and Oshlack A A scaling normalization method for differential expression analysis of RNA-seq data. 11, 1–12 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liao Y, Wang J, Jaehnig EJ, Shi Z & Zhang B WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47, W199–W205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Croft D, Mundo AF, Haw R, Milacic M, Weiser, Wu G et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 42, 472–477 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramírez F, Dündar F, Diehl S, Grüning BA & Manke T DeepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, 187–191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Egorov AA, Sakharova EA, Anisimova AS, Dmitriev SE, Gladyshev VN, Kulakovskiy IV Svist4get: A simple visualization tool for genomic tracks from sequencing experiments. BMC Bioinformatics 20, 4–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.