Abstract
The fungal pathogen Candida albicans is surrounded by a cell wall that is the target of caspofungin and other echinocandin antifungals. Candida albicans can grow in several morphological forms, notably budding yeast and hyphae. Yeast and hyphal forms differ in cell wall composition, leading us to hypothesize that there may be distinct genes required for yeast and hyphal responses to caspofungin. Mutants in 27 genes reported previously to be caspofungin hypersensitive under yeast growth conditions were all caspofungin hypersensitive under hyphal growth conditions as well. However, a screen of mutants defective in transcription factor genes revealed that Cup9 is required for normal caspofungin tolerance under hyphal and not yeast growth conditions. In a hyphal-defective efg1Δ/Δ background, Cup9 is still required for normal caspofungin tolerance. This result argues that Cup9 function is related to growth conditions rather than cell morphology. RNA-seq conducted under hyphal growth conditions indicated that 361 genes were up-regulated and 145 genes were down-regulated in response to caspofungin treatment. Both classes of caspofungin-responsive genes were enriched for cell wall-related proteins, as expected for a response to disruption of cell wall integrity and biosynthesis. The cup9Δ/Δ mutant, treated with caspofungin, had reduced RNA levels of 40 caspofungin up-regulated genes, and had increased RNA levels of 8 caspofungin down-regulated genes, an indication that Cup9 has a narrow rather than global role in the cell wall integrity response. Five Cup9-activated surface-protein genes have roles in cell wall integrity, based on mutant analysis published previously (PGA31 and IFF11) or shown here (ORF19.3499, ORF19.851, or PGA28), and therefore may explain the hypersensitivity of the cup9Δ/Δmutant to caspofungin. Our findings define Cup9 as a new determinant of caspofungin susceptibility.
Keywords: Candida, cell wall integrity, regulation
Introduction
The fungus Candida albicans causes diverse infections with considerable morbidity and mortality. Invasive infections cause 10,000 deaths per year in the United States, and an estimated 400,000 deaths per year worldwide (Brown et al. 2012). The organism remains a threat for many reasons, including the limited selection and scope of antifungal drugs, the occurrence of drug resistance, the ability of the organism to grow as biofilm, and the diverse virulence determinants that enable it to infect almost any tissue (Mayer et al. 2013; Robbins et al. 2016).
This organism can grow in several morphological forms, notably yeast and hyphae (Sudbery 2011). Yeast-form cells, also called blastospores, are ovoid cells that grow by budding. Hyphae are filaments that have contiguous cell compartments separated by septa and grow by tip extension. Hyphae are closely tied to pathogenesis (Mayer et al. 2013) because (i) hyphae are prominent in infected tissue; (ii) hyphal growth is induced in vitro by infection-relevant conditions, such as 37°C temperature, neutral pH, and presence of serum; (iii) many genes that are highly induced in hyphae are required for pathogenicity; and (iv) all C. albicans mutant strains that are defective in hyphal formation in vivo are also defective in pathogenesis. Although there are other types of pathogenesis-defective mutants (Mayer et al. 2013), the program of hyphal formation is widely considered a major determinant of C. albicans pathogenicity during invasive infection.
C. albicans is surrounded by a cell wall that is the target of echinocandin antifungals, such as caspofungin, as well as several other therapeutic strategies under development (Robbins et al. 2016). The organism's response to stress caused by cell wall inhibitors enables it to survive echinocandin treatment (Ernst and Pla 2011). Defects in known cell wall stress response regulators, such as conserved Cell Wall Integrity MAP Kinase pathway components (Dichtl et al. 2016), cause C. albicans to become hypersensitive to echinocandins.
The transcriptional circuitry that governs the cell wall stress response has been characterized in considerable detail (Heredia et al. 2020a). Three key transcription factors—Cas5, Rlm1, and Sko1—were identified as insertion mutations that caused hypersensitivity to caspofungin. Cas5 has the broadest role among them in that it controls the largest number of genes, including over half of caspofungin-responsive genes (Bruno et al. 2006; Xie et al. 2017; Heredia et al. 2020a). Cas5 is also required for normal cell division and numerous stress responses (Xie et al. 2017; Heredia et al. 2020a). Sko1 and Rlm1 control fewer caspofungin-responsive genes, and are interconnected in that Rlm1 is required for induction of Sko1 RNA by caspofungin (Bruno et al. 2006; Rauceo et al. 2008; Heredia et al. 2020a, 2020b).
The yeast and hyphal growth forms differ considerably in cell wall constituents (Heilmann et al. 2011; Sudbery 2011; Hall 2015). For example, the highly expressed yeast-specific gene YWP1 encodes a cell wall protein; the highly expressed hyphal-specific genes ALS3, HWP1, and HYR1 encode cell wall proteins. On this basis, we suspected that the cell wall stress response may differ between yeast and hyphae. To date, the response to cell wall stress has been characterized primarily in yeast-form C. albicans cells (see, for example, Xie et al. 2017; Heredia et al. 2020a, 2020b). In this study, we characterize the cell wall stress response under hyphal growth conditions. Our findings provide evidence for an environmentally contingent response, though not a hyphal-specific response, that is mediated in part by the transcription factor Cup9.
Materials and methods
Media
Strains were grown in YPD (2% Bacto peptone, 2% dextrose, and 1% yeast extract), YPD with 25 mM HEPES or in RPMI-1640 with L-glutamine and 25 mM HEPES, without sodium bicarbonate (R4130 SIGMA). YPD with 25 mM HEPES and RPMI-1640 medium was adjusted to pH7.4 using NaOH. Transformants were selected on synthetic medium (2% dextrose, 1.7% Difco yeast nitrogen base with ammonium sulfate and auxotrophic supplements) or on YPD + 400 μg/ml nourseothricin [clonNAT, WERNER BioAgents] for nourseothricin-resistant isolates.
The medium for the mutant screen was RPMI+serum+caspofungin, buffered with NaHCO3 to prevent pH changes from a 5% CO2 atmosphere. One bottle of RPMI powder (R4130-10X*1L WITH BUFFER; Sigma) was mixed well with 20 g of Bacto-Agar and 876 ml of deionized water, then autoclaved with a stir bar in place. These components were added after autoclaving with mixing: 100 ml of Fetal Bovine Serum (S11150H; Atlanta Biologicals), 24 ml of filter sterilized 1 M Sodium Bicarbonate solution, and 0.120 ml of 250 µg/ml caspofungin stock solution. Plates were poured and allowed to solidify and dry before use.
Strains
All C. albicans strains used in this study are listed in Supplementary Table S1, and primer sequences are listed in Supplementary Table S2.
The C. albicans cup9 deletion mutant (mutant no. 991; orf19.6514) was obtained from the Noble homozygous deletion library (Homann et al. 2009) at the Fungal Genetics Stock Center. For complementation of the cup9Δ/Δ mutant, PCR primers were designed to amplify genomic DNA of strain SC5314 from 3 kb upstream to 0.5 kb downstream of the open reading frame (ORF). These primers (called CUP9_for and CUP9_rev) have 40 b flanking sequences with homology to pSN105+NruI. pSN105+NruI is a derivative of pSN105 (Noble et al. 2010) with a NruI site inserted right next to the LEU2 cassette. To construct a complementing plasmid containing the CUP9 gene, the resulting PCR product was co-transformed into S. cerevisiae with Nru1-digested pSN105+NruI. The resulting plasmid DNA was digested with HpaI and integrated at the native CUP9 locus (2.4 kb upstream of the ORF). pSN105 and pSN105+NruI digested with PmeI were transformed into WT (SN250) and the mutant strain respectively to generate prototrophic, marker-matched strains.
The C. albicans orf19.3499, orf19.851, and pga28 deletion mutants were constructed in the SN250 strain background through the transient CRISPR-Cas9 system (Min et al. 2016). The gene deletion construct was synthesized using the C.d. ARG4 plasmid pSN69 (Noble and Johnson 2005). The sgRNA cassette, the CaCAS9 expression cassette and the deletion construct were co-transformed into strain SN250 (Noble and Johnson 2005). PCR assays for each wild-type allele were accomplished with the “Gene Name for” and “Gene Name rev” primers indicated in Supplementary Table S2.
The orf19.851Δ/Δ pga28Δ/Δ orf19.3499Δ/Δ triple mutant was constructed using the transient CRISPR-Cas9 system (Min et al. 2016) in strain SN152. The deletion markers were C.d. ARG4 from pSN69 for ORF19.851, C.d. HIS1 from pSN52 for PGA28, and C.m. LEU2 from pSN40 for ORF19.3499 (Noble and Johnson 2005). The triple mutant was constructed sequentially by co-transforming a deletion marker construct, the CaCAS9 expression cassette and the appropriate sgRNA for the gene being targeted. Transformants were selected on drop out medium and screened by colony PCR for homozygous deletions of the targeted gene. The order of deletion was first orf19.851::ARG4, then pga28::HIS1, and finally orf19.3499::LEU2.
The efg1Δ/Δ and cup9Δ/Δ efg1Δ/Δ deletion mutants were constructed using the transient CRISPR-Cas9 system (Min et al. 2016). The NAT marker from pNAT (Min et al. 2016) was used as the deletion marker for EFG1. The sgRNA cassette (targeting EFG1), the CaCAS9 expression cassette, and the NAT deletion construct were co-transformed into strain SN250 (CUP9) and YI192 (cup9Δ/Δ) and transformants were selected on YPD + nourseothricin. NAT resistant transformants were checked by colony PCR for homozygous deletion strains.
The efg1Δ/Δ and cup9Δ/Δ efg1Δ/Δ strains that had EFG1 reconstituted were constructed using the transient CRISPR-Cas9 system (Min et al. 2016). An EFG1 cassette was amplified from wild type (SC5314) genomic DNA and co-transformed with an ARG4 cassette (also amplified from wild type genomic DNA), the CaCAS9 expression cassette and 2 sgRNA cassettes: one for targeting efg1Δ::NAT and one for targeting the arg4::dpl200 junction found in the SN250 strain background. Transformants were selected for growth on CSM-arg medium and then screened for nourseothricin sensitivity. Arg+ NATS strains were checked by colony PCR for the absence of arg4::dpl200 sequences and absence of NAT sequences to identify homozygous ARG/ARG EFG1/EFG1 strains. To distinguish EFG1/EFG1 reconstituted strains from EFG1/EFG1 wild-type strains we designate the reconstituted genotype “efg1(Δ/Δ)+/+.”
Spot dilution assay
Strains were grown overnight in 5 ml YPD medium at 30°C. Cell density was measured at OD600 for each strain, which were then diluted to an OD600 of 3.0 in H2O. Fivefold dilutions were made of the OD600 3.0 stock and these were plated on the indicated media. Plates were incubated at 30°C or 37°C (as indicated in figure legends) for 2–4 days.
Microscopy
Cells from an overnight 5 ml YPD 30°C culture were washed with 5 ml H2O and re-suspended again in 5 ml H2O. The cell suspension was diluted to an OD600 = 20 in H2O and finally diluted 1:100 into 5 ml RPMI medium. Cells were grown at 30°C with rotating for 6 hours and visualized with a Zeiss Axio Observer Z.1 fluorescence microscope and a 20x DIC objective.
RNA collection for RNA sequencing
For RNA sequencing, wild type (SC5314) and cup9Δ/Δ (YI192) cells from an overnight 5 ml YPD 30°C culture were washed with 5 ml H2O and re-suspended again in 5 ml H2O. The cell suspension was diluted to an OD600 = 20 in H2O and finally diluted 1:100 into 25 ml RPMI medium. Cells were grown at 37°C with 200 rpm shaking for 2 hours. Caspofungin was then added to a final concentration of 27.5 ng/ml, and the control cultures received an equal volume of water. Cells were collected by filtration 30 minutes after drug administration. RNA extractions were performed using Qiagen RNeasy mini kit (Cat#74104) with modifications as described previously (Xu et al. 2015).
RNA sequencing
RNA-seq libraries (nonstrand-specific and paired end) were prepared with the TruSeq RNA Sample Prep kit (Illumina). The total RNA samples were subject to poly(A) enrichment as part of the TruSeq protocol. In total, 150 nt of sequence was determined from both ends of each cDNA fragment using the HiSeq platform (Illumina). Sequencing reads were aligned to the C. albicans reference (Assembly A21) using TopHat2 (Kim et al. 2013). The alignment files from TopHat2 were used to generate read counts for each gene and a statistical analysis of differential gene expression was performed using the DEseq package from Bioconductor (Anders and Huber 2010). Each experimental group consisted of two biological replicates.
Data availability
Raw sequencing reads (RNA-seq) from this study have been submitted to the NCBI sequence read archive (SRA) under BioProject accession number PRJNA693694. The data are also available upon request. All other data are within the manuscript and its Supplementary materials files. Supplementary material is available at figshare: https://doi.org/10.25386/genetics.14568864.
Results
Identification of mutants hypersensitive to caspofungin under hyphal growth conditions
We hypothesized that some cell wall integrity regulators may affect caspofungin susceptibility in only yeast or hyphae. To explore this idea we sought to assay caspofungin susceptibility under hyphal inducing conditions. In pilot studies, we found that caspofungin susceptibility was difficult to assay in RPMI medium at 37°C, a typical hyphal growth condition. Therefore, we conducted assays in RPMI medium at 30°C, a condition in which hyphal growth of wild-type strain CW542 occurs (Supplementary Figure S1).
As an initial test of this hypothesis, we assayed caspofungin susceptibility of 27 previously identified cell wall integrity regulatory mutants in RPMI medium at 30°C. Susceptibility to cell wall inhibitors had been tested previously for these strains mainly under yeast growth conditions (Bruno et al. 2006; Rauceo et al. 2008; Blankenship et al. 2010; Roman et al. 2015; Heredia et al. 2020b). All 27 mutants were hypersensitive to caspofungin in RPMI medium at 30°C as well (Supplementary Figure S2). These results indicate that all of these regulators govern cell wall integrity under these growth conditions.
As a second test of this hypothesis, we screened 165 transcription factor deletion mutants (Homann et al. 2009) for sensitivity to caspofungin under hyphal growth conditions. Each strain was tested by a spot-dilution assay on RPMI + 10% serum + 30 ng/ml caspofungin in a 5% CO2 atmosphere at 37°C. Mutants that presented a caspofungin-hypersensitive phenotype included cas5Δ/Δ, cup9Δ/Δ, swi4Δ/Δ, mig1Δ/Δ, zcf14Δ/Δ, bas1Δ/Δ, sef1Δ/Δ, fgr15Δ/Δ, dal81Δ/Δ, orf19.2961Δ/Δ, and czf1Δ/Δ. Cell wall integrity defects have been reported previously for the mutants cas5Δ/Δ (Bruno et al. 2006; Xie et al. 2017), mig1Δ/Δ (Lagree et al. 2020), czf1Δ/Δ (Mottola et al. 2021), and swi4Δ/Δ (Xie et al. 2017). Therefore, this screen identified several new mutants that are hypersensitive to caspofungin.
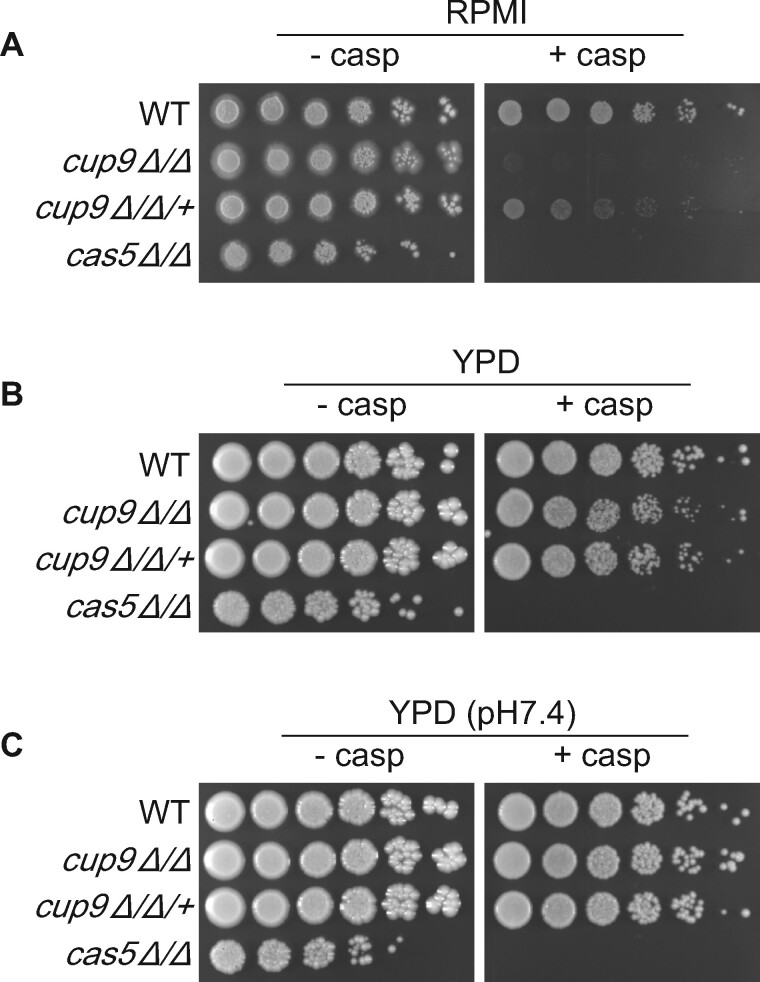
We focused on Cup9 in the present study because of the strength of the mutant phenotype (Figure 1A) and early indications that the mutant might cause a cell wall integrity defect only in hyphae (presented below). Cup9 has been characterized previously as a negative regulator of hyphal formation (Homann et al. 2009; Guan et al. 2013; Lu et al. 2014; Meir et al. 2018), but to our knowledge has not been tied previously to cell wall integrity. A complemented strain, made by introducing a wild-type copy of CUP9 into the mutant, showed greatly improved growth in the presence of caspofungin (Figure 1A, cup9Δ/Δ/+). Complementation was evident in RPMI medium at 30°C (Figure 1A) or 37°C (Supplementary Figure S3). We confirmed that the cup9Δ/Δ mutant grows as hyphae in RPMI (Supplementary Figure S1). The complementation test verified that the CUP9 deletion causes caspofungin hypersensitivity under hyphal growth conditions.
Figure 1.
Effect of growth conditions on cup9Δ/Δ mutant caspofungin hypersensitivity. The wild-type strain (CW542), cas5Δ/Δ caspofungin hypersensitive control (VIC1147), cup9Δ/Δ mutant (YI192), and cup9Δ/Δ/+ complemented strain (YI243) were serially diluted onto RPMI (A), YPD (B), or YPD buffered at pH7.4 (C) with or without 125 ng/ml caspofungin. Cells were incubated for 3 days at 30°C.
The cup9Δ/Δ mutant showed a comparably mild caspofungin-hypersensitive phenotype in YPD medium at 30°C, a yeast growth condition (Figure 1B). RPMI and YPD differ in pH, a possible cause of differential drug sensitivity. However, in YPD buffered at pH 7.4 (the pH of RPMI), the cup9Δ/Δ mutant remained only mildly caspofungin-hypersensitive (Figure 1C). These results show that the role of Cup9 in cell wall integrity is contingent upon growth conditions.
Caspofungin hypersensitivity of a yeast-form cup9Δ/Δ mutant
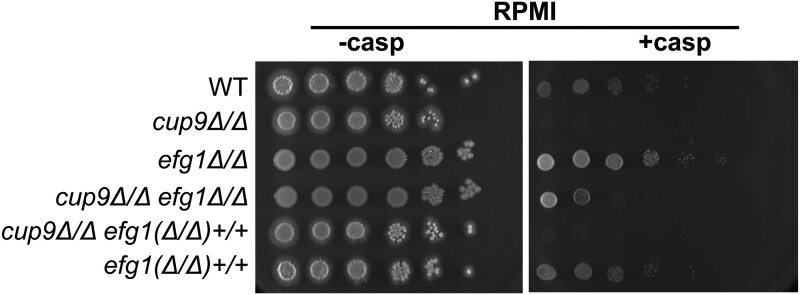
If Cup9 governs caspofungin sensitivity most strongly in the hyphal growth form, then a mutation that blocks hyphal growth may suppress the cup9Δ/Δ phenotype. We tested this prediction with a mutation in EFG1, which specifies a transcriptional regulator of hyphal development. An efg1Δ/Δ mutant is defective in hyphal-specific gene expression and hyphal morphogenesis (Lo et al. 1997; Stoldt et al. 1997; Braun and Johnson 2000; Sudbery 2011). We created a cup9Δ/Δ efg1Δ/Δ double deletion mutant and its EFG1/EFG1 reconstituted strain. Loss of EFG1 suppressed caspofungin hypersensitivity of the cup9Δ/Δ mutant (Figure 2, cup9Δ/Δ efg1Δ/Δ). Reconstitution of EFG1 restored caspofungin hypersensitivity [Figure 2, cup9Δ/Δ efg1(Δ/Δ)+/+]. This result is consistent with the model that Cup9 is required for cell wall integrity in hyphal cells but not in yeast-form cells. However, we also tested caspofungin sensitivity of an efg1Δ/Δ mutant made in a CUP9+/+ background on RPMI medium (Figure 2). The efg1Δ/Δ mutant (Figure 2, efg1Δ/Δ) showed less sensitivity than the cup9Δ/Δ efg1Δ/Δ mutant, and introduction of wild-type EFG1 alleles into the mutant increased caspofungin sensitivity [Figure 2, efg1(Δ/Δ)+/+]. Because the loss of CUP9 increases caspofungin sensitivity in an efg1Δ/Δ background, the role of Cup9 in cell wall integrity is not hyphal-specific; rather it is environmentally contingent.
Figure 2.
Effect of an efg1Δ/Δ mutation on cup9Δ/Δ mutant caspofungin hypersensitivity. The wild-type strain (CW1757), cup9Δ/Δ mutant (YI192), efg1Δ/Δ mutant (CW1792), cup9Δ/Δ efg1Δ/Δ mutant (CW1796), cup9Δ/Δ efg1(Δ/Δ)+/+ reconstituted strain (CW1785), and efg1(Δ/Δ)+/+ reconstituted strain (CW1779) were serially diluted onto RPMI with or without 125 ng/ml caspofungin. Cells were incubated for 4 days at 30°C.
Transcriptional response to caspofungin during hyphal growth
To determine the gene expression response to caspofungin in hyphal growth conditions, we conducted an RNA-seq comparison of the wild-type strain SC5314 with or without caspofungin treatment in RPMI at 37°C. We chose this temperature rather than 30°C because 37°C is very widely used to promote hyphal growth. In the wild type, 361 genes were up-regulated at least twofold and 145 genes were down regulated at least twofold after caspofungin treatment (Supplementary Table S3). Both up- and down-regulated genes were enriched significantly for GO descriptors related to the cell surface, as expected if the response alters the cell wall.
Genes regulated by Cup9 were identified through comparison of RNA-seq data of the cup9Δ/Δ mutant and the wild type, both treated with caspofungin. Among 361 genes that were up-regulated in the wild type, 40 genes were expressed at lower levels in the cup9Δ/Δ mutant (Table 1). This gene set showed some enrichment for cell surface-related functions. Among 145 genes that were down-regulated in the wild type, 8 genes were expressed at higher levels in the cup9Δ/Δ mutant (Table 1). These genes were enriched for zinc homeostasis functions. An additional 65 genes showed altered expression in the cup9Δ/Δ mutant compared to wild type, both treated with caspofungin (Supplementary Table S3). This gene set was not enriched significantly for any Gene Ontology terms. Overall, the narrow gene expression impact of a CUP9 defect suggests that Cup9 is not a global regulator of cell wall integrity.
Table 1.
Genes that respond in parallel to caspofungin and Cup9
| ORF ID | Gene symbol | Fold change Wt +caspo/Wt—caspo | Fold change cup9 +caspo/Wt +caspo | Signal peptide |
|---|---|---|---|---|
| orf19.2034 | — | >60 | 0.06 | — |
| orf19.3499 | — | 54.85 | 0.17 | x |
| orf19.6486 | LDG3 | 48.32 | 0.15 | x |
| orf19.937 | — | 24.55 | 0.28 | x |
| orf19.5302 | PGA31 | 23.66 | 0.41 | x |
| orf19.5303 | PGA30 | 18.41 | 0.33 | x |
| orf19.1539 | — | 17.30 | 6.26 | — |
| orf19.36.1 | — | 17.16 | 0.25 | — |
| orf19.1171 | — | 14.92 | 0.43 | x |
| orf19.5399 | IFF11 | 13.92 | 0.37 | x |
| orf19.5874 | — | 13.16 | 0.11 | x |
| orf19.4082 | DDR48 | 12.36 | 0.48 | — |
| orf19.1524 | SPR3 | 9.82 | 0.31 | — |
| orf19.2160 | NAG4 | 8.74 | 0.40 | — |
| orf19.6475 | — | 7.45 | 0.31 | x |
| orf19.4688 | DAG7 | 7.42 | 0.32 | x |
| orf19.7170 | — | 7.28 | 0.35 | x |
| orf19.2252 | — | 6.27 | 0.12 | — |
| orf19.2809 | CTN3 | 5.48 | 0.50 | — |
| orf19.1048 | IFD6 | 5.14 | 0.37 | — |
| orf19.2593 | BIO2 | 4.92 | 0.38 | — |
| orf19.3643 | — | 4.50 | 0.33 | — |
| orf19.1440.1 | — | 4.26 | 0.41 | — |
| orf19.5975 | — | 3.71 | 0.36 | — |
| orf19.4255 | ECM331 | 2.94 | 0.42 | x |
| orf19.938 | — | 2.89 | 0.43 | — |
| orf19.4934 | OP4 | 2.76 | 0.44 | x |
| orf19.1440.2 | — | 2.60 | 0.43 | — |
| orf19.24 | RTA2 | 2.57 | 0.34 | — |
| orf19.3710 | YHB5 | 2.48 | 0.46 | — |
| orf19.3375 | — | 2.43 | 0.38 | x |
| orf19.555 | — | 2.41 | 0.16 | x |
| orf19.1868 | RNR22 | 2.39 | 0.49 | — |
| orf19.6837 | FMA1 | 2.35 | 0.44 | — |
| orf19.4936.1 | — | 2.28 | 0.47 | — |
| orf19.851 | — | 2.17 | 0.24 | x |
| orf19.5542 | SAP6 | 2.09 | 0.22 | x |
| orf19.17 | SCP1 | 2.08 | 0.43 | — |
| orf19.5144 | PGA28 | 2.03 | 0.22 | x |
| orf19.7557 | FGR46 | 2.02 | 0.44 | x |
| orf19.3794 | ZAP1/CSR1 | 0.48 | 2.85 | — |
| orf19.1930 | CFL5 | 0.45 | 3.20 | x |
| orf19.5141 | — | 0.35 | 4.27 | x |
| orf19.3895 | CHT2 | 0.24 | 8.74 | x |
| orf19.6142 | — | 0.21 | 7.08 | — |
| orf19.3112 | ZRT1 | 0.17 | 2.44 | x |
| orf19.3111 | PRA1 | 0.16 | 2.83 | x |
| orf19.2038 | — | 0.15 | 3.61 | — |
Genes that are caspofungin-induced and Cup9-activated or caspofungin-repressed and Cup9-repressed are listed. Complete differentially expressed gene analysis is in Supplementary Table S3. Genes whose products have a predicted signal peptide are indicated.
Cup9-regulated genes related to cell wall integrity
Among caspofungin-induced Cup9-regulated genes, 26 encode possible secreted proteins or cell wall proteins based on presence of a signal peptide sequence (Table 1). Two genes in this group, PGA31 and IFF11, have been shown to be required for cell wall integrity (Bates et al. 2007; Plaine et al. 2008). We hypothesized that reduced expression of other potential secreted/cell wall protein genes may also contribute to the sensitivity of the cup9Δ/Δ mutant to caspofungin.
To see if Cup9-regulated secreted/cell wall protein genes may contribute to cell wall integrity, we tested three less characterized cell surface protein genes ORF19.3499, ORF19.851, and PGA28. Deletion mutants orf19.3499Δ/Δ, orf19.851Δ/Δ, and pga28Δ/Δ were slightly sensitive to caspofungin (Figure 3). No synergy was evident in a triple deletion mutant (Figure 3). These results suggest that ORF19.3499, ORF19.851, and PGA28 have a role in cell wall integrity. The relatively mild mutant phenotypes may indicate that the gene products have a minor role, or that their functions overlap with those of other Cup9-regulated genes.
Figure 3.
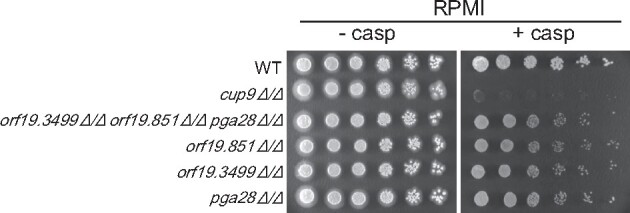
Roles of Cup9-regulated genes in cell wall integrity. Comparisons of deletion mutants were conducted. The wild-type strain (CW542), orf19.3499Δ/Δ orf19.851Δ/Δ pga28Δ/Δ triple mutant (CW1697), orf19.851Δ/Δ mutant (YI459), orf19.3499Δ/Δ mutant (YI463), pga28Δ/Δ mutant (YI464), and cup9Δ/Δ mutant (YI192), were serially diluted onto RPMI solid medium with or without 125 ng/ml caspofungin. Cells were incubated for 3 days at 30°C.
Discussion
We have characterized a new regulator of the C. albicans cell wall stress response, Cup9. The rationale that led us to Cup9 was that distinct regulatory functions may be required in hyphal cells as compared to yeast cells, given that their cell wall protein constituents are different. However, our results instead argue that Cup9 is required for cell wall integrity under hyphal growth conditions, but that Cup9 has a functional role in cell wall integrity even in yeast form cells. Therefore, it is environmental conditions rather than cell type that determines the need for Cup9 in cell wall integrity.
Cup9 has been characterized previously as a negative regulator of hyphal formation. For example, a cup9Δ/Δ mutant displays increased filamentation in various in vitro growth assays (Homann et al. 2009; Guan et al. 2013). Mechanistic insight into Cup9 function has come from analysis of its roles in farnesol inhibition of hyphal formation (Lu et al. 2014) and oral mucosal colonization (Meir et al. 2018). As a mediator of hyphal inhibition by farnesol, a quorum-sensing molecule, Cup9 functions as a repressor of SOK1 RNA accumulation; Sok1 in turn promotes degradation of the hyphal repressor Nrg1. Farnesol prevents Cup9 protein degradation (Lu et al. 2014). Therefore, farnesol promotes SOK1 repression, resulting in increased Nrg1 accumulation. The role of Cup9 in oral colonization is at least in part tied to its impact on hyphal formation. The cup9Δ/Δ mutant displays much more prominent hyphae in infected tongue samples than the wild-type strain (Meir et al. 2018). RNA-seq comparison of cup9Δ/Δ and wild-type cells grown on a semisolid surface at 37°C shows that Cup9 represses expression of some key hyphal-associated genes, including HWP1 and ECE1 (Meir et al. 2018). Thus, the previously known biological roles of Cup9 are tied to its negative regulation of hyphal formation.
Three observations argue that the impact of Cup9 on the cell wall stress response is independent of its regulation of hyphal formation. First, caspofungin hypersensitivity of the cup9Δ/Δ mutant is evident under hyphal-inducing conditions, when wild-type, mutant, and complemented strains all grow as hyphae. Second, in a hyphal-defective efg1Δ/Δ mutant background, a cup9Δ/Δ mutation causes increased caspofungin sensitivity. Third, little increased expression of hyphal-associated genes, except for a twofold increase in SOD5 RNA levels, is evident in the cup9Δ/Δ mutant under the hyphal-inducing conditions we used for RNA-seq analysis. These observations, together with previous work, suggest that Cup9 has two functions: it represses hyphal formation, and it promotes a response to cell wall damage.
Environmental conditions govern the functional activity of Cup9. This idea is supported by our observation that the cup9Δ/Δ mutant’s caspofungin hypersensitivity depends upon the growth medium. The idea is further supported by comparison of gene expression data for cup9Δ/Δ mutants. In the farnesol study, Lu et al. (2014) showed that a cup9Δ/Δ mutant has elevated SOK1 expression when grown in YPD+farnesol liquid medium at 37°C. In the oral colonization study, Meir et al. (2018) showed that a cup9Δ/Δ mutant has elevated HWP1 and ECE1 expression when grown on Todd–Hewitt solid medium at 37°C, but did not detect significantly elevated SOK1 expression. In our present study, the cup9Δ/Δ mutant did not cause increased SOK1, HWP1, or ECE1 expression in RPMI+caspofungin liquid medium at 37°C. In fact, only 10 genes display parallel regulation by Cup9 in our study and the analysis of Meier et al. (Meir et al. 2018): WOR1 and DAG7 are down-regulated in the mutant; SOD5, PGA26, PRA1, GIT1, orf19.4450.1, orf19.6586, MNN1, and CHT2 are up-regulated in the mutant. The observations above suggest that environmental conditions govern the gene set that is under Cup9 control. This relationship between environmental conditions and Cup9-responsive genes may explain why the cup9Δ/Δ mutant has environmentally contingent phenotypes.
How may the environment influence Cup9 activity? We note that Cup9 has a helix-turn-helix or homeodomain DNA binding domain, and such proteins often have binding partners that modify their binding specificity (Burglin and Affolter 2016). A simple model is that Cup9 has a few different environmentally responsive binding partners, and each directs it to act on a different gene set.
How does Cup9 promote cell wall integrity? We propose that several Cup9-responsive genes contribute to this process. Prior studies have shown that mutation of PGA31 or IFF11 confer hypersensitivity to cell wall inhibitors (Bates et al. 2007; Plaine et al. 2008). Both of these genes are induced by caspofungin and are expressed at reduced levels in a cup9Δ/Δ mutant. Three additional genes studied here—ORF19.3499, ORF19.851, and PGA28—share these regulatory features and mutant phenotypes, though mutation of any one or all three of those genes causes fairly mild caspofungin hypersensitivity. These observations argue that Cup9 promotes cell wall integrity by activating expression of several genes that encode cell wall- or surface-localized proteins, each of which contributes to overall cell wall integrity.
Acknowledgments
The authors thank Minju Kim, Liping Xiong, Anupam Sharma, and Yinhe Mao for comments on the manuscript, and Manning Huang, Katie Lagree, Kyunghun Min, Wenjie Xu, and Fred Lanni for many helpful discussions. They are grateful to Tatyana Aleynikov for outstanding technical support.
Funding
This work was supported by National Institutes of Health (NIH)/National Institutes of Allergy and Infectious Diseases (NIAID) grant 1R21AI137464 to APM. Website https://www.niaid.nih.gov/ The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflicts of interest
None declared.
Contributor Information
Yuichi Ichikawa, Department of Biological Sciences, Carnegie Mellon University, Pittsburgh, PA 15213, USA; Division of Cancer Biology, The Cancer Institute of JFCR, Koto-ku, Tokyo 135-8550, Japan.
Vincent M Bruno, Department of Microbiology and Immunology and Institute of Genome Sciences, University of Maryland School of Medicine, Baltimore, MD 21201, USA.
Carol A Woolford, Department of Biological Sciences, Carnegie Mellon University, Pittsburgh, PA 15213, USA.
Hannah Kim, Department of Biological Sciences, Carnegie Mellon University, Pittsburgh, PA 15213, USA; Institute for Genomics and Evolutionary Medicine, Temple University, Philadelphia, PA 19122, USA.
Eunsoo Do, Department of Microbiology, University of Georgia, Athens, GA 30602, USA.
Grace C Brewer, Department of Microbiology, University of Georgia, Athens, GA 30602, USA.
Aaron P Mitchell, Department of Biological Sciences, Carnegie Mellon University, Pittsburgh, PA 15213, USA; Department of Microbiology, University of Georgia, Athens, GA 30602, USA.
Literature cited
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S, de la Rosa JM, MacCallum DM, Brown AJ, Gow NA, et al. 2007. Candida albicans Iff11, a secreted protein required for cell wall structure and virulence. Infect Immun. 75:2922–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship JR, Fanning S, Hamaker JJ, Mitchell AP. 2010. An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 6:e1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BR, Johnson AD. 2000. TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics. 155:57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, et al. 2012. Hidden killers: human fungal infections. Sci Transl Med. 4:165rv113. [DOI] [PubMed] [Google Scholar]
- Bruno VM, Kalachikov S, Subaran R, Nobile CJ, Kyratsous C, et al. 2006. Control of the C. albicans cell wall damage response by transcriptional regulator Cas5. PLoS Pathog. 2:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burglin TR, Affolter M. 2016. Homeodomain proteins: an update. Chromosoma. 125:497–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl K, Samantaray S, Wagener J. 2016. Cell wall integrity signalling in human pathogenic fungi. Cell Microbiol. 18:1228–1238. [DOI] [PubMed] [Google Scholar]
- Ernst JF, Pla J. 2011. Signaling the glycoshield: maintenance of the Candida albicans cell wall. Int J Med Microbiol. 301:378–383. [DOI] [PubMed] [Google Scholar]
- Guan G, Xie J, Tao L, Nobile CJ, Sun Y, et al. 2013. Bcr1 plays a central role in the regulation of opaque cell filamentation in Candida albicans. Mol Microbiol. 89:732–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall RA. 2015. Dressed to impress: impact of environmental adaptation on the Candida albicans cell wall. Mol Microbiol. 97:7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann CJ, Sorgo AG, Siliakus AR, Dekker HL, Brul S, et al. 2011. Hyphal induction in the human fungal pathogen Candida albicans reveals a characteristic wall protein profile. Microbiology (Reading). 157:2297–2307. [DOI] [PubMed] [Google Scholar]
- Heredia MY, Gunasekaran D, Ikeh MAC, Nobile CJ, Rauceo JM. 2020a. Transcriptional regulation of the caspofungin-induced cell wall damage response in Candida albicans. Curr Genet. 66:1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heredia MY, Ikeh MAC, Gunasekaran D, Conrad KA, Filimonava S, et al. 2020b. An expanded cell wall damage signaling network is comprised of the transcription factors Rlm1 and Sko1 in Candida albicans. PLoS Genet. 16:e1008908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann OR, Dea J, Noble SM, Johnson AD. 2009. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 5:e1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, et al. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagree K, Woolford CA, Huang MY, May G, McManus CJ, et al. 2020. Roles of Candida albicans Mig1 and Mig2 in glucose repression, pathogenicity traits, and SNF1 essentiality. PLoS Genet. 16:e1008582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, et al. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell. 90:939–949. [DOI] [PubMed] [Google Scholar]
- Lu Y, Su C, Unoje O, Liu H. 2014. Quorum sensing controls hyphal initiation in Candida albicans through Ubr1-mediated protein degradation. Proc Natl Acad Sci USA. 111:1975–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer FL, Wilson D, Hube B. 2013. Candida albicans pathogenicity mechanisms. Virulence. 4:119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meir J, Hartmann E, Eckstein MT, Guiducci E, Kirchner F, et al. 2018. Identification of Candida albicans regulatory genes governing mucosal infection. Cell Microbiol. 20:e12841. [DOI] [PubMed] [Google Scholar]
- Min K, Ichikawa Y, Woolford CA, Mitchell AP. 2016. Candida albicans gene deletion with a transient CRISPR-Cas9 system. mSphere. 1:e00130-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottola A, Ramírez-Zavala B, Hünniger K, Kurzai O, Morschhäuser J. 2021. The zinc cluster transcription factor Czf1 regulates cell wall architecture and integrity in Candida albicans. Mol Microbiol. doi: 10.1111/mmi.14727 [DOI] [PubMed] [Google Scholar]
- Noble SM, French S, Kohn LA, Chen V, Johnson AD. 2010. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 42:590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, Johnson AD. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell. 4:298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaine A, Walker L, Da Costa G, Mora-Montes HM, McKinnon A, et al. 2008. Functional analysis of Candida albicans GPI-anchored proteins: roles in cell wall integrity and caspofungin sensitivity. Fungal Genet Biol. 45:1404–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauceo JM, Blankenship JR, Fanning S, Hamaker JJ, Deneault JS, et al. 2008. Regulation of the Candida albicans cell wall damage response by transcription factor Sko1 and PAS kinase Psk1. Mol Biol Cell. 19:2741–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins N, Wright GD, Cowen LE. 2016. Antifungal drugs: the current armamentarium and development of new agents. Microbiol Spectr. 4.doi:10.1128/microbiolspec.FUNK-0002-2016. [DOI] [PubMed] [Google Scholar]
- Roman E, Alonso-Monge R, Miranda A, Pla J. 2015. The Mkk2 MAPKK regulates cell wall biogenesis in cooperation with the Cek1-pathway in Candida albicans. PLoS One. 10:e0133476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoldt VR, Sonneborn A, Leuker CE, Ernst JF. 1997. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16:1982–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery PE. 2011. Growth of Candida albicans hyphae. Nat Rev Microbiol. 9:737–748. [DOI] [PubMed] [Google Scholar]
- Xie JL, Qin L, Miao Z, Grys BT, Diaz JC, et al. 2017. The Candida albicans transcription factor Cas5 couples stress responses, drug resistance and cell cycle regulation. Nat Commun. 8:499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Solis NV, Ehrlich RL, Woolford CA, Filler SG, et al. 2015. Activation and alliance of regulatory pathways in C. albicans during mammalian infection. PLoS Biol. 13:e1002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw sequencing reads (RNA-seq) from this study have been submitted to the NCBI sequence read archive (SRA) under BioProject accession number PRJNA693694. The data are also available upon request. All other data are within the manuscript and its Supplementary materials files. Supplementary material is available at figshare: https://doi.org/10.25386/genetics.14568864.