Abstract
Heparan sulfate 3-O-sulfotransferase (3-OST) transfers a sulfo group to the 3-OH position of a glucosamine saccharide unit to form 3-O-sulfated heparan sulfate. 3-O-sulfation is known to be critically important for bestowing anticoagulant activity and other biological functions of heparan sulfate. Here, we report two ternary crystal structures of 3-OST-5 with PAP (3’-phosphoadenosine 5’-phosphate) and two octasaccharide substrates. We also used 3-OST-5 to synthesize six 3-O-sulfated 8-mers. Results from the structural analysis of the six 3-O-sulfated 8-mers revealed the substrate specificity of 3-OST-5. The enzyme prefers to sulfate a 6-O-sulfo glucosamine saccharide that is surrounded by glucuronic acid over a 6-O-sulfo glucosamine saccharide that is surrounded by 2-O-sulfated iduronic acid. 3-OST-5 modified 8-mers display a broad range of anti-factor Xa activity, depending on the structure of the 8-mer. We also discovered that the substrate specificity of 3-OST-5 is not governed solely by the side chains from amino acid residues in the active site. The conformational flexibility of the 2-O-sulfated iduronic acid in the saccharide substrates also contributes to the substrate specificity. These findings advance our understanding for how to control the biosynthesis of 3-O-sulfated heparan sulfate with desired biological activities.
Keywords: Sulfotransferase, heparan sulfate biosynthesis, chemoenzymatic synthesis, oligosaccharide, protein/substrate complex
Heparan sulfates (HS) are sulfated polysaccharides that are widely present on the surface of mammalian cells and in the extracellular matrix. HS plays a wide range of biological functions, including regulating embryonic development, modulating inflammatory responses, and regulating blood coagulation 1. Heparin, a highly sulfated form of HS, is a commonly used drug to treat blood clotting disorders 2. Additionally, many viruses utilize cell surface HS on host cells as a receptor to establish infection. Notably, HS is reportedly involved in the infection of SARS-Cov-2 3. HS consists of repeating disaccharide units of either glucuronic acid (GlcA) or iduronic acid (IdoA) linked to a glucosamine (GlcN) saccharide and each saccharide unit can be sulfated. Common sulfations in HS are found at the N-position and 6-OH positions of GlcN saccharide units as well as the 2-OH position of IdoA saccharide unit. Sulfations of the 3-OH position of GlcN and 2-OH position of GlcA also exist in HS, but are exceedingly rare. The sulfation patterns, the position of both GlcA and IdoA saccharides, and the length of the saccharide chain contribute to the binding affinity of proteins and determine HS’s selectivity, regulating biological functions 4. A central goal in HS-related biological research is to identify the link between the sulfation patterns and biological functions.
A group of specialized enzymes, including glycosyltransferase, C5-epimerase, and a series of sulfotransferases, participate in the biosynthesis of HS. The concerted actions of these enzymes control the saccharide sequences and quantity of HS. The synthesis of the backbone, composed of a repeating disaccharide sequence of N-acetyl GlcN (GlcNAc) and GlcA is accomplished by glycosyltransferases referred as HS polymerases. The backbone can be further modified by N-deacetylase-N-sulfotransferase, converting a GlcNAc to a N-sulfo GlcN (GlcNS) unit. Subsequently, C5-epimerase converts the adjacent GlcA to an IdoA unit. The installation of additional sulfo groups is achieved by a 2-O-sulfotransferase, 6-O-sulfotransferases (three isoforms), and 3-O-sulfotransferases (3-OSTs, with seven isoforms). These enzymes act on the backbone in a step-wise manner, generating an extremely diverse set of heparin and HS molecules with diverse length and modification patterns. Because the process is not template-driven, product structures vary substantially and are primarily regulated by the enzyme tissue expression pattern and individual enzyme specificity 5. Recently, the HS biosynthetic enzymes have been used to synthesize structurally homogeneous HS oligosaccharides. High regioselectivity displayed by HS biosynthetic enzymes has eliminated the need for protection and deprotection steps used in chemical synthesis. As a result, enzyme-based synthesis has dramatically increased the synthesis efficiency, making it a promising alternative approach to synthesize structurally homogeneous oligosaccharides6, 7. The availability of structurally defined oligosaccharides accelerates efforts to study the relationship between the structure and function of HS and the development of HS-based therapeutics 8, 9. One example is the synthesis of anticoagulant dodecasaccharides to replace animal sourced low-molecular weight heparin 7, 9. Synthetic heparin reduces side effects and improves the safety of the supply chain for heparin and low-molecular weight heparin drugs. The 3-OST isoform 5 (3-OST-5) is a key enzyme used in the process of preparing the anticoagulant dodecasaccharides 7, 9.
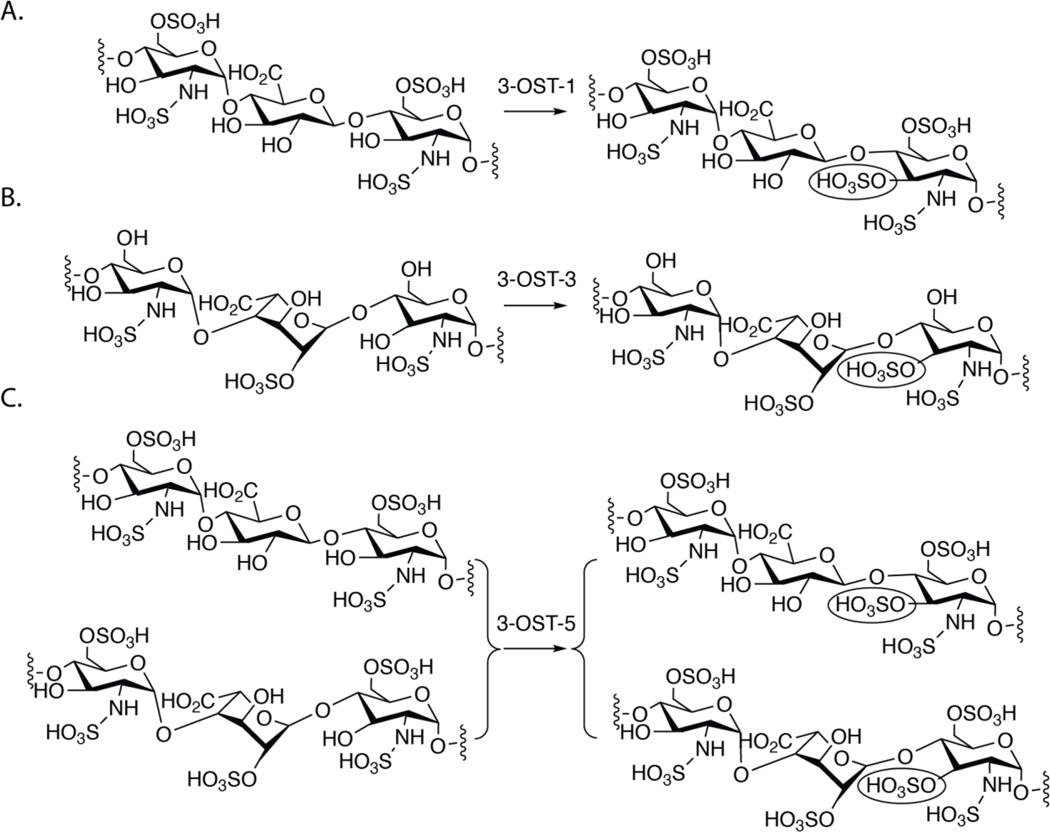
3-OST transfers a sulfo group to the 3-OH position of GlcNS. Although it is a rare modification in HS, the 3-O-sulfation is closely related to the biological functions of HS. Intriguingly, despite the scarcity of 3-O-sulfation in HS, 3-OSTs represent the largest family of HS sulfotransferase, with seven isoforms in the human genome 10. The isoforms recognize a saccharide sequence around the modification site, generating 3-O-sulfated HS products with distinct biological functions (Fig 1). Based on the current dogma in the field, 3-OST-1 sulfates a GlcNS6S saccharide that is linked to a GlcA on the non-reducing end, conferring anticoagulant activity to HS and heparin. However, 3-OST-3 sulfates a GlcNS that has no 6-O-sulfaion and is linked to an IdoA2S saccharide at the non-reducing end of the acceptor GlcNS. HS modified by 3-OST-3 is an entry receptor for herpes simplex virus-1 (HSV-1) 11. 3-OST-5 is an enzyme that appears to sulfate a GlcNS6S saccharide that is linked to either an IdoA2S or a GlcA on the non-reducing end. In a recent study, the substrate specificity of 3-OST-4 was explored, showing that it also sulfates a GlcNS saccharide that has no 6-O-sulfation 12. 3-O-sulfated HS have been reported to regulate axon guidance and growth of neurons, control the progenitor cell expansion for salivary gland development, and were found in high quantities in human granulosa cells, suggesting an importance in ovulation 13–15. Additionally, different 3-OST isoforms have been implicated in diseases including Alzheimer’s disease and cancers 16–18. The unique substrate specificity of 3-OST isoforms and the potential biological functions of HS modified by different 3-OSTs have attracted interest in understanding the mechanism used by different isoforms to distinguish among saccharide sequences.
Fig 1. The substrate specificity of different 3-OST isoforms.
A) shows the substrate specificity of 3-OST-1. B) Substrate specificity of 3-OST-3. The IdoA2S saccharide unit near the acceptor site is in 2S0 skew boat confirmation based on the crystal structure 21. C) Substrate specificity of 3-OST-5. The 3-O-sulfation site is circled.
In this manuscript, we used structurally homogeneous 8-mer substrates to investigate the substrate specificity of 3-OST-5. The crystal structures of the ternary complexes of 3-OST-5 with 3’-phosphoadenosine-5’-phosphate (PAP) and two different octasaccharide substrates reveal the key amino acid residues involved in interactions with the saccharide substrates at the resolution of 2.90 Å and 2.75 Å, respectively. Results from the structural analysis of 3-OST-5 modified 8-mers indicate that 3-OST-5 prefers to sulfate a GlcNS6S saccharide that is surrounded by glucuronic acid over one which is surrounded by 2-O-sulfated iduronic acid. 3-OST-5 can generate 8-mers both with and without anticoagulant activity, depending on the saccharide sequences around the 3-O-sulfation site. Compared with previously published structures of 3-OST-1 and 3-OST-3 19–21, we discovered that 3-OST-5 has a very similar structure in the active site and overall structure to 3-OST-1 and 3-OST-3. We also determined that the substrate specificity of 3-OST-5 is not only governed by the side chains from amino acid residues in the active site, but also by uronic acids flanking both sides of the acceptor. These findings advance our understanding of the regulatory mechanisms controlling the biosynthesis of 3-O-sulfated HS with desired biological activities. The results also expand the capability of synthesizing 3-O-sulfated oligosaccharides using the chemoenzymatic approach.
Results
3-OST-5 requires 6-O-sulfation to act on a substrate
Six octasaccharides (8-mers) were synthesized using the chemoenzymatic approach to evaluate the substrate specificity of 3-OST-5 (Suppl Fig S1A). The purities of 8-mers were confirmed by high performance liquid chromatography (HPLC) and molecular masses were verified by electrospray ionization mass spectrometry (ESI-MS) (Suppl Fig S2-S7). The substrates used varied in 6-O-sulfation and IdoA2S content. 3-OST-5 was assessed for activity against all substrates using a 35S-labeled sulfo donor, [35S]PAPS (3’-phosphoadenosine 5’-phosphosulfate). The results clearly show that the enzyme was more active toward octasaccharides containing 6-O-sulfation (8-mer-1, −2 and −3), than those without (8-mer-1a, −2a, and −3a) (Suppl Fig S1B). Additionally, 3-OST-5 had the highest activity toward the GlcA-only substrate (8-mer-1) and the lowest activity toward the substrate containing two IdoA2S residues (8-mer-3). However, there were multiple potential sites for sulfation by 3-OST-5 on each 8-mer substrate. Using radioactivity alone limited our ability to locate the 3-O-sulfation site. To further assess sites of 3-OST-5 modification, non-radioactive studies were employed.
Determination of the 3-O-sulfation sites on 8-mers after 3-OST-5 modification
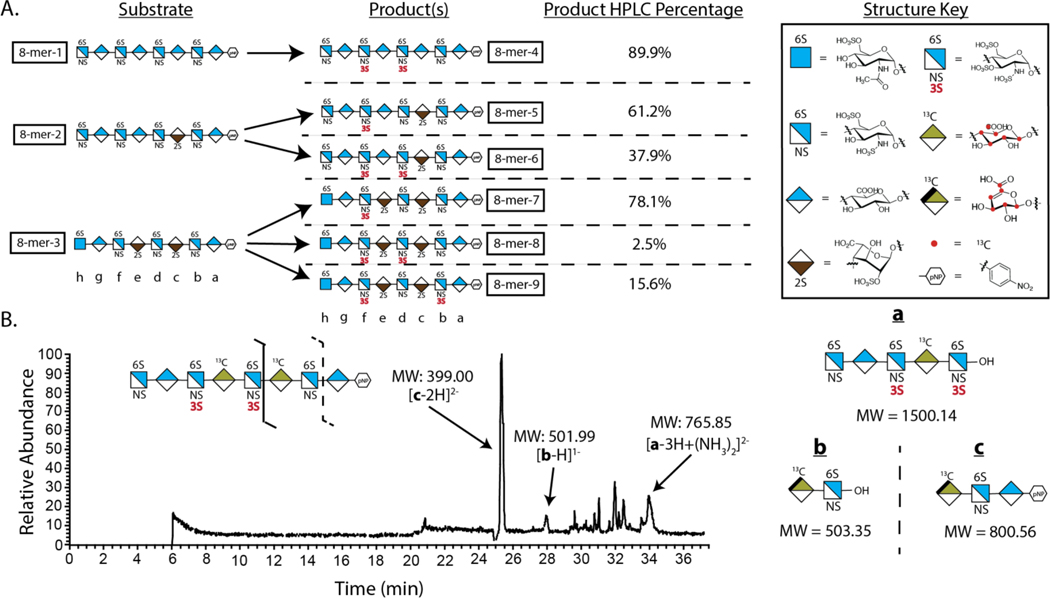
To determine the preferred sites for 3-OST-5 sulfation, we incubated 3-OST-5 enzyme with 8-mer-1, −2 and −3 along with PAPS to yield more than six 3-O-sulfated 8-mer products in 0.5–10 mg quantities (Fig 2A). The 3-O-sulfated 8-mers were then subjected to structural determination for the number and location of 3-O-sulfo groups. Two subgroups of 3-O-sulfated 8-mers were obtained based on molecular weight measurements (Suppl Table S1). One group contained mono-3-O-sulfated 8-mers, i.e. 8-mer-5 and 8-mer −7. Another group contained di-3-O-sulfated 8-mers, including 8-mer-4, 8-mer-6, 8-mer-8 and 8-mer-9. Structural analyses of the 3-O-sulfated 8-mer products, including 8-mer-4, −5, −6 and −9, were completed using an LC-MS/MS-based disaccharide/oligosaccharide analysis 7 (Fig 3, Suppl Figs S10 and S11 to S20). An example to prove the sites of 3-O-sulfation in 8-mer-4 is shown in Fig 2B. Characterization of 8-mer-4 was uniquely challenging using unlabeled 8-mer-1 substrate because it contained three repeated disaccharides of -GlcNS6S-GlcA-, with only two of the repeats carrying a 3-O-sulfo group. To differentiate the repeating disaccharide unit, a site-specific 13C-labeled 8-mer-1 was synthesized with both saccharides c and e in 8-mer-1 occupied by 13C-labeled GlcA residues (Suppl Fig S8). The corresponding 13C-labeled 8-mer-4 was generated by incubating 3-OST-5 and the 8-mer-1 (Suppl Fig S9). Digestion of the 13C-labeled 8-mer-4 by heparin lyases resulted in three fragments, a (pentasaccharide), b (disaccharide) and c (trisaccharide), based on the substrate specificity of heparin lyases 22, 23. All three fragments were found from the LC-MS/MS analysis, confirming the locations of 3-O-sulfations on saccharides d and f (Fig 2B). The 3-O-sulfation positions on 8-mer-4 were further confirmed by 2-D NMR (Suppl Figs S11 and S12). The structures of 8-mer-7 and −8 were verified by co-elution with 8-mer standards synthesized in previous studies 7, 24 (Suppl Figs S18 and S19).
Fig 2. Determination of the substrate specificity of 3-OST-5 using 8-mer substrates.
A) Sulfation reaction carried out by 3-OST-5 using 8-mer-1, 8-mer-2 and 8-mer-3 substrates. The structures of the 8-mers are presented in shorthand symbols. The key for the shorthand symbols is presented (right). Red dots in the structures represent sites with 13C-carbon. B) Structural analysis of 13C-labeled 8-mer-4 using the LC-MS/MS method. The schematic digestion of 8-mer by heparin lyases is presented (top). The digested fragments (a-c, diagrammed on right) were then assessed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Spectrum analysis allowed for matching of the product to a particular structure based on observed mass peaks.
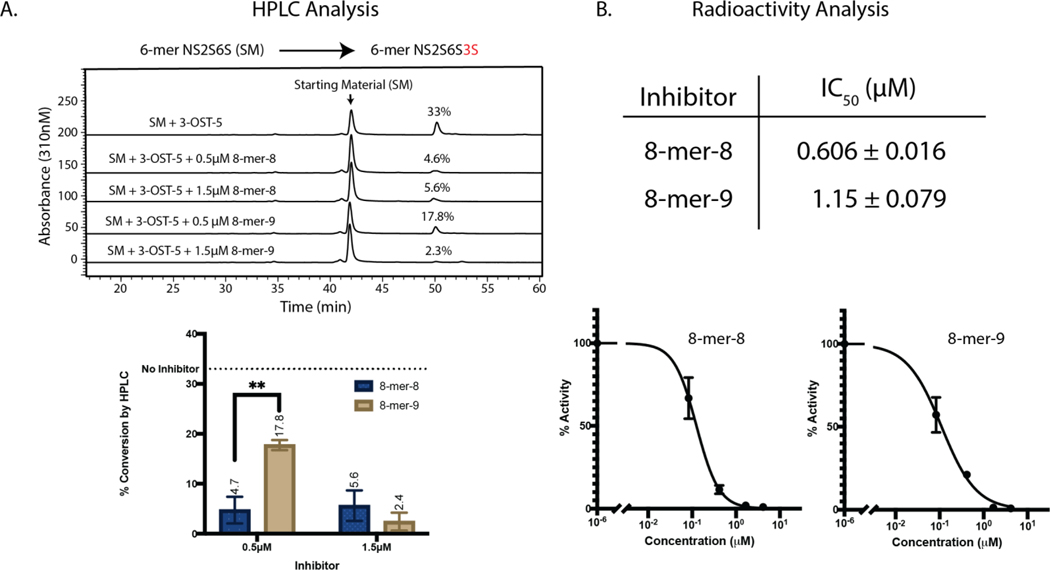
Fig 3. 3-OST-5 is inhibited by 3-O-sulfated 8-mer.
A) HPLC chromatograms of NS2S6S 6-mer modified by 3-OST-3 with or without spiked 8-mer-8 or 8-mer-9. Here, the NS2S6S 6-mer substrate has a saccharide sequence of GlcNS6S-GlcA-GlcNS6S-IdoA2S-GlcNS6S-GlcA-pNP. After 3-OST-5 modification, the 6-mer was transformed to NS2S6S3S 6-mer with a sequence of GlcNS6S-GlcA-GlcNS6S3S-IdoA2S-GlcNS6S-GlcA-pNP. B) IC50 values for 8-mer-8 and −9 were then determined in an 35S radioactivity assay. Inhibition curves from 8-mer-8 and −9 are presented. The NS2S6S 6-mer was also used as a substrate in the assay.
Knowing the structures of 3-O-sulfated 8-mer products, we observed trends in substrate preference for 3-OST-5 (Fig 2A). First, in the absence of IdoA2S residues (8-mer-1), 3-OST-5 was highly efficient at installing multiple 3-O-sulfations, generating the di-3-O-sulfated product 8-mer-4 (89.9%) (Fig 2A). The enzyme was less efficient at forming di-3-O-sulfated products with 8-mer-2 and −3 substrates, which contain one and two IdoA2S saccharides, respectively. Using 8-mer-2 substrate, 3-OST-5 generated 8-mer-5, a mono-3-O-sulfated product, as the major product (61.2%); and 8-mer-6, a di-3-O-sulfated product, was a minor product (37.9%) (Fig 2A). Additionally, when given the choice of modifying saccharide f or saccharide d in 8-mer-2, the enzyme chose to sulfate saccharide f to form trisaccharide domain of -GlcA-GlcNS3S6S-GlcA-. The data suggest that 3-OST-5 preferred the glucosamine site surrounded by GlcA saccharides, not by IdoA2S units (Fig 2A). Lastly, when a substrate contained two IdoA2S saccharides (8-mer-3), 3-OST-5 was able to install a second 3-O-sulfation on either saccharide b or saccharide d. However, the enzyme preferred to introduce the second 3-O-sulfo group on saccharide b to generate 8-mer-9, where the two 3-O-sulfated glucosamine saccharides are gapped (Fig 2A). It should be noted that incubation of the reaction mixtures with additional enzyme would lead to increased product formation. However, the identity of products formed as well as the modification site preference (i.e. 8-mer-9 site over 8-mer-8 site) remain consistent.
Product inhibition of 3-OST-5 by 8-mer-8 and 8-mer-9
We next attempted to explain why 3-OST-5 displayed a preference for producing 8-mer-9 over 8-mer-8. One hypothesis was that the 3-OST-5 enzyme is more susceptible to product inhibition by 8-mer-8 than that by 8-mer-9. The structural difference between 8-mer-8 and 8-mer-9 is the spacing of the 3-O-sulfo groups. In 8-mer-8, the two 3-O-sulfo groups are located on two consecutive glucosamine saccharides whereas in 8-mer-9, the two 3-O-sulfo groups are located on two glucosamine saccharides that are gapped by a glucosamine. To test the assertion of product inhibition, 3-OST-5 products 8-mer-8 and 8-mer-9 were first spiked into enzymatic reactions at 0.5 μM and 1.5 μM (Fig 3). A 6-mer was utilized as the substrate for these reactions to avoid generation of additional 8-mer product inhibitor. The products were analyzed by HPLC. Compared to the control, the presence of 8-mer-8 (at the concentration of 0.5 μM) reduced product generation 7.0-fold, while a 1.9-fold reduction was observed by 8-mer-9 at the same concentration (Fig 3A). The data suggest that 8-mer-8 displays stronger inhibition of the activity of 3-OST-5. To further assess the effect of product inhibition on 3-OST-5 activity, IC50 values for 8-mers 8 and 9 were determined using a radioactivity assay and the same 6-mer substrate used in the HPLC assay. The data from the inhibition study using 35S radioactivity were consistent with the HPLC results, with 8-mer-8 as a more potent inhibitor than 8-mer-9 (Fig 3B). The difference in the degree of product inhibition from 8-mer-8 and 8-mer-9 was small, but detectable by both radioactive and non-radioactive assays. The data suggest that product inhibition could serve as a contributing factor for 3-OST-5 to preferentially generate 8-mer-9, rather than 8-mer-8.
Crystal structures of 3-OST-5/PAP/8-mer-1 and 3-OST-5/PAP/8-mer-2
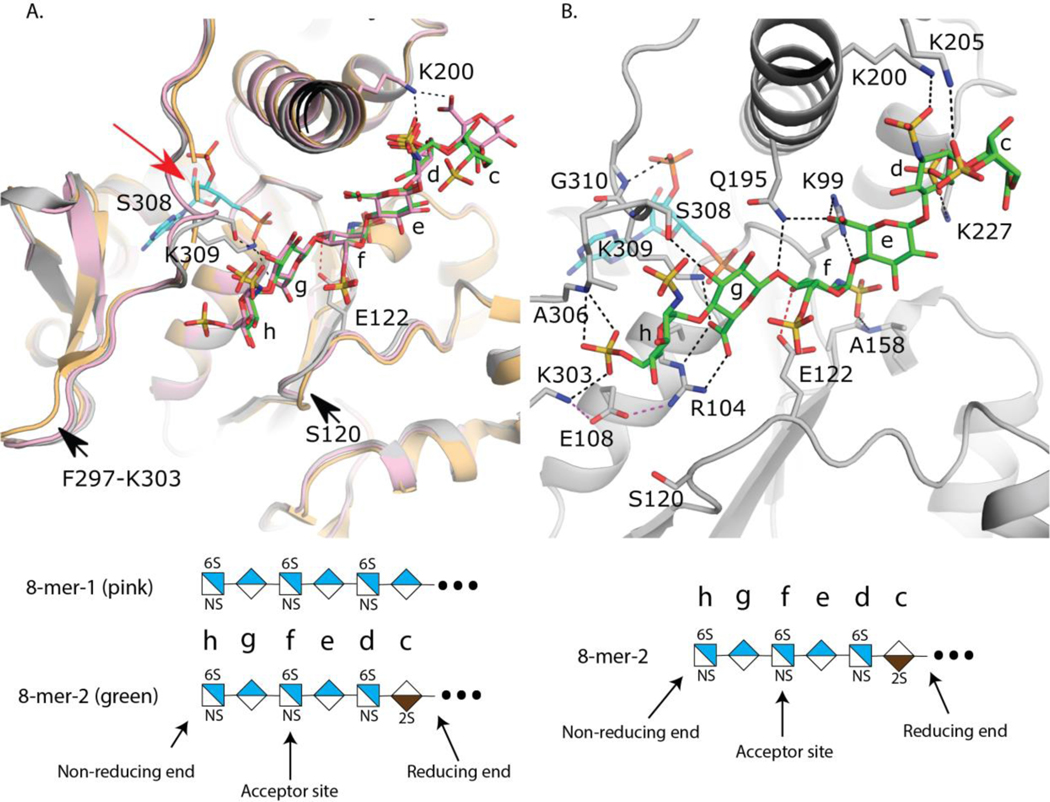
Diffraction quality ternary complex crystals were obtained utilizing a thioredoxin fusion of 3-OST-5 in the presence of PAP and either 8-mer-1 or 8-mer-2. The structures of the complexes with 8-mer-1 and 8-mer-2 were refined to 2.9 and 2.75 Å respectively (Table 1). A superposition of the 3-OST-5 structures shows a high degree of similarity (RMSD 0.333 Å over 260 Cα atoms) (Fig 4A). The addition of the N-terminal thioredoxin crystallization tag has negligible impact on the overall structure of 3-OST-5 as 3-OST-5 superimposes with an RMSD of 0.597 Å over 258 Cα atoms with the previous crystal structure of 3-OST-5 solved only in the presence of PAP without saccharide substrate and without a thioredoxin tag (Fig 4A) 25. The binding of substrates 8-mer-1 and 8-mer-2 to the substrate channel appears to induce small movements in two loops as well as small variations in sidechain rotomers (Fig 4A). The loop (Ala119-Phe125) containing the proposed catalytic base Glu122 has moved into the cleft upon binding to saccharide substrate with the largest shift of up to 2 Å at Ser120. Another loop (Phe297-Lys303) found at the non-reducing end of the binding cleft also appears to have moved into the cleft up to ~1.5–2 Å. Four residues on a loop overlying the PAP site that were disordered in the absence of 8-mer substrates (Gly307-Gly310) become ordered upon binding substrate, forming interactions with both 8-mers via the side chain of Ser308, and with the 3’-phosphate of PAP through the backbone amide of Gly310 (Fig 4A and 4B). Lys309, conserved in 3-OST-1 and 3-OST-3, forms interactions with both the substrate and the 5’-phosphate of PAP and is in position to interact with the sulfo group of PAPS (Fig 4A). Mutations of the equivalent lysine in 3-OST-1 to alanine yielded 17% of wt activity while abolishing activity for 3-OST-3 suggesting an important role in substrate binding and potentially catalysis 20, 26.
Table 1:
Data collection and refinement statistics
| Trx-3OST5 (I299E)/PAP/8mer-1a,b | Trx-3OST5 (I299E)/PAP/8mer-2a,b | |
|---|---|---|
| PDB ID code | 7SCD | 7SCE |
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 55.84, 58.82, 153.11 | 54.42, 59.00, 151.05 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50.00–2.90 (2.95–2.90)c | 50–2.75 (2.80–2.75) |
| Rsym (%) | 11.7 (54.7) | 16.3 (67.1) |
| I / σI | 5.46 (0.86) | 7.46 (1.54) |
| Completeness (%) | 90.1 (81.3) | 99.1 (98.6) |
| Redundancy | 3.2 (3.1) | 4.9 (4.6) |
| Refinement | ||
| Resolution (Å) | 40.50 (2.90)d | 40.00–2.75 |
| No. reflections | 9311 | 13196 |
| Rwork / Rfree (%) | 20.82/25.24 | 18.79/24.60 |
| No. atoms | ||
| Protein | 2872 | 2883 |
| PAP | 27 | 27 |
| Oligosaccharide | 94 | 97 |
| Water | 8 | 53 |
| B-factors | ||
| Protein | 50.87 | 30.72 |
| PAP | 29.05 | 17.50 |
| Oligosaccharide | 68.86 | 55.02 |
| Water | 18.71 | 23.95 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.006 | 0.004 |
| Bond angles (°) | 1.138 | 0.761 |
A single crystal was used to collect each data set
These crystals were collected on the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source at Argonne National Laboratory.
Values in parentheses are for highest-resolution shell.
The data for the 8mer-1 ternary complex was severely anisotropic and were therefore processed with ellipsoidal truncation by the Diffraction Anisotropy Server (http://www.services.mbi.ucla.edu/anisoscale).
Fig 4. Crystal structures of 3-OST-5.
A) Superposition of 3-OST-5 with substrates 8-mer-1 (pink, protein light pink), 8-mer-2 (green, protein gray), and no substrate bound (protein light orange, PDBidcode: 3bd9). All structures contain PAP co-factor. PAP from the 8-mer-2 structure is shown (cyan). Loops that change positions upon binding substrate are indicated with an arrow. Missing loop from the apo structure is represented by a dashed loop and denoted with a red arrow. B) Interactions between substrate 8-mer-2 (green) and 3-OST-5. Probable hydrogen bonds between 8-mer-2 and 3-OST-5 are shown in dashed black lines. Interaction between Glu122 and the acceptor 3-OH group is shown as a dashed red line and Glc108 interactions are displayed as magenta dashed lines. Saccharides are labeled c through h with shorthand structures of 8-mer-1 and −2 presented below each panel.
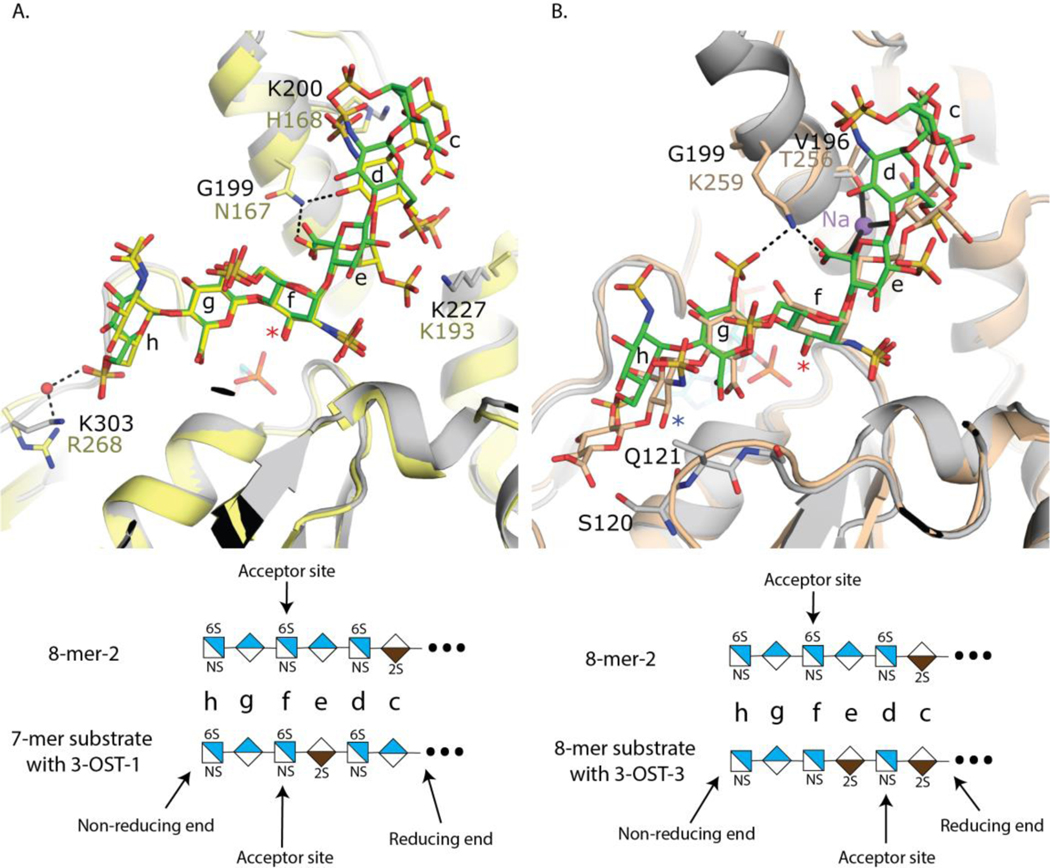
Despite the presence of an IdoA2S in the 8-mer-2 substrate, binding of the oligosaccharide within the cleft is nearly identical between the two 8-mers (Fig 4A). In the case of 8-mer-1 there is clear density in the channel for five saccharides (-GlcNS6S-GlcA-GlcNS6S-GlcA-GlcNS6S-) with a mostly disordered GlcA on the reducing end of the visible density (Suppl Fig S21A). Due to the repeating disaccharide nature of 8-mer-1, the exact register of the oligosaccharides is unknown as it is possible that additional disordered saccharides could be present on either end of the modeled hexasaccharide. Interestingly, 8-mer-2 binds in the identical conformation as 8-mer-1, with six ordered saccharides (Figs 4A, 4B, and Suppl Fig S21B). As both uronic acids present from the non-reducing end are clearly GlcA, it can be concluded that the terminal non-reducing end GlcNS6S (saccharide h) is bound in the pocket and the IdoA2S (saccharide c) is the last ordered residue on the reducing end. Due to the high similarity in binding and the better quality of data (higher resolution and not as anisotropic) for the 8-mer-2 structure, the analysis will focus on this structure. In the 8-mer-2 substrate, the IdoA2S is modeled in the 1C4 chair conformation, while all other saccharides are found in the expected 4C1 conformation (Fig 4B). The non-reducing end GlcNS6S (saccharide h) is found extended between residues Ala306 and Ser120 at the “top” of the cleft and “bottom” of the cleft, respectively. These residues have been speculated to participate in specificity of the 3-OST isoforms by regulating the trajectories of the substrates through the cleft, based on the sugar conformation of the saccharide at position g 25. In addition, the 6-O-sulfo group forms interactions with the sidechain from Lys303 and a backbone hydrogen bond with the amide nitrogen from Ala306. GlcA saccharide g is involved in a bidentate interaction between the carboxylate and the guanidinium group of Arg104 that lies beneath saccharide h in the cleft. The 3-OH of saccharide g forms a hydrogen bond with Ser308, helping to order this previously disordered loop. The acceptor GlcNS6S (saccharide f) is found at the active site with the 3-OH acceptor within hydrogen bonding distance to the proposed catalytic base, Glu122, and 5.5 Å from the phosphorous atom of the 5’ phosphate of the leaving group product PAP, consistent with a catalytically relevant position, based on the proposed in-line transfer catalytic mechanism 20, 27. Also contributing to binding is the NS moiety that forms a hydrogen bond with a backbone amide from Ala158. There are fewer interactions with the substrate on the reducing side of saccharide f. GlcA (saccharide e) forms interactions with Gln195 and Arg99 via interactions with the carboxylate while the NS group of GlcNS6S at position d forms an interaction with Arg200 and a possible interaction between the 6S group and Lys227. IdoA2S (saccharide c) has the weakest density of all the saccharides with only a single interaction between the 2-O-sulfo group and Lys205. In the case of 8-mer-1, the GlcA at position c forms a potential interaction with Lys200 via its carboxylate group (Fig 4A).
Structurally guided mutations of 3-OST-5 resulted in changes to substrate specificity
Amino acid residues near the substrate binding cleft were selected for site-directed mutagenesis to evaluate their roles in regulating substrate selectivity. The mutants were generated and screened against three oligosaccharides, including 8-mer-1, −2, and −3, in an HPLC assay (Suppl Table S2). While most mutants showed a modest effect on activity (25%−117% wt activity), the E108R mutant displayed a 95% decrease in enzymatic activity against all three 8-mer substrates. This suggests that the negatively charged amino acid side chain from Glu108 residue plays an essential role for substrate recognition/binding, potentially by helping to organize the non-reducing end of the binding pocket through hydrogen bonding With Arg104 and Lys303 (Fig 4b). The mutation of A306H converts the alanine at this position to a histidine that is found in 3-OST-1. This mutation was previously shown to give 3-OST-5 more 3-OST-1 like activity on heterogeneous substrates and was suggested to function as a substrate “gate” although the composition of the substrate was not analyzed 25. To better understand how this mutation might affect specificity within the active site, we analyzed the products produced using the three substrates 8-mer-1, −2 and −3. The A306H mutant showed different degrees of change in products produced from those three 8-mer substrates (Suppl Fig S22). No difference between the A306H mutant and wild type enzyme was observed when 8-mer-1 was used as a substrate. A small difference was observed in generating 8-mer-6 when 8-mer-2 was the substrate. An obvious decrease in generating di-3-O-sulfated products (or 8-mer-8 and 8-mer-9) was observed when 8-mer-3 was the substrate. The A306H mutant, unlike wild type enzyme, only produced 8-mer-7, a mono-3-O-sulfated product (Suppl Fig S22). The lack of 8-mer-8 and 8-mer-9 products for A306H mutant raise the possibility that the A306 residue confers the ability to generate the disaccharide domain of -IdoA2S-GlcNS3S6S- saccharide. This is consistent with the postulated role of the histidine at the “gate” in 3-OST-1 in selecting for -GlcA-GlcNS6S- containing substrates 25.
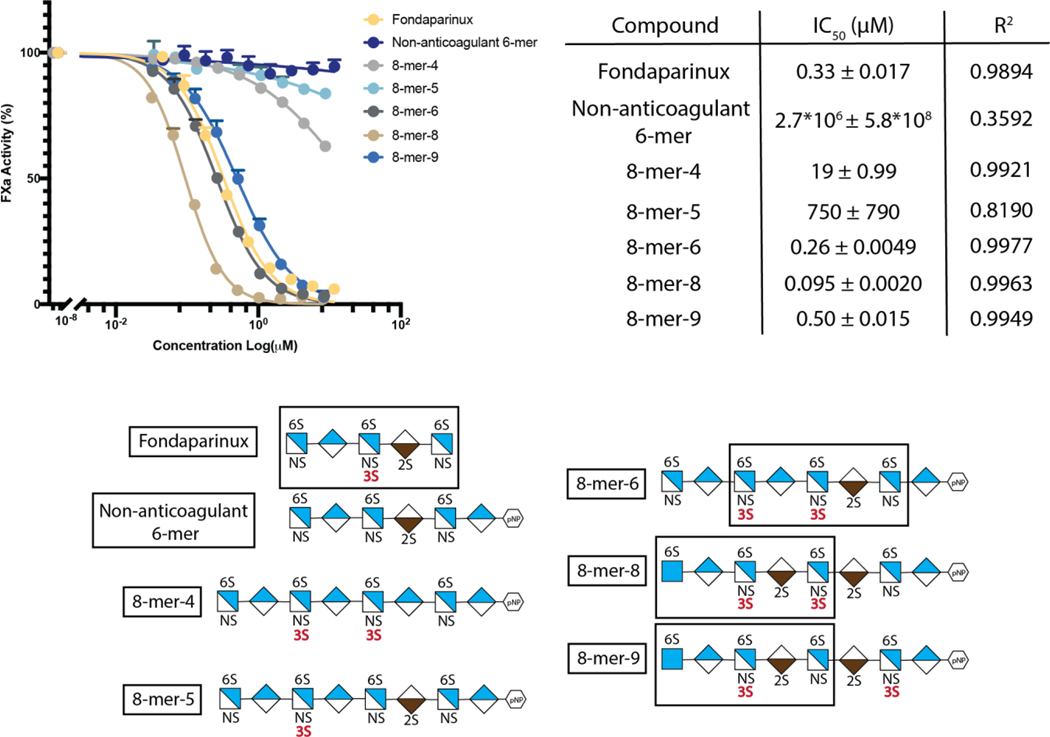
3-OST-5 generates 8-mer products with varying degrees of the anti-FXa activity
To assess the biological implications of 3-OST-5 modified HS, 8-mer products were analyzed for anticoagulant activity, as measured by anti-factor Xa (FXa) assay (Fig 5). We discovered that 8-mer products display a spectrum of anticoagulant activity, from very weak to very strong, depending on the saccharide sequences. The IC50 value of 8-mer-8 was ∼5-fold lower than that of 8-mer-6 and −9, suggesting that 8-mer-8 has the strongest anticoagulant activity. Comparing the saccharide sequences of 8-mer-6, 8-mer-8 and 8-mer-9, a difference in the relative location of the two 3-O-sulfated glucosamine (GlcNS3S6S) saccharides exists. In 8-mer-8, the two GlcNS3S6S saccharides are separated by an IdoA2S; whereas in 8-mer-6, the two GlcNS3S6S saccharides are separate by a GlcA. In 8-mer-9, the two GlcNS3S6S residues are separated by three sugar units consisting of two IdoA2S saccharides and one GlcNS6S saccharide. It should be noted that 8-mer-6, −8 and −9 all contain the antithrombin-binding pentasaccharide sequence, GlcNS6S (GlcNAc6S)-GlcA-GlcNS3S6S-IdoA2S-GlcNS6S- (boxed pentasaccharide sequence in Fig 5). The relationship between the pentasaccharide sequence and anticoagulant activity was further supported from the analysis of 8-mer-4 and −5, since each had no to weak anti-FXa activity. The 8-mer-4 does not contain an IdoA2S, which is a required saccharide unit for displaying anticoagulant activity 28. 8-mer-5 lacks anticoagulant activity because a GlcA saccharide, rather than an IdoA2S saccharide, is located on the reducing end of the GlcNS3S6S. It is known that an IdoA2S or an IdoA saccharide is required to be present at the reducing end of the GlcNS3S6S saccharide, in the context of -GlcNS3S6S-IdoA2S- or -GlcNS3S6S-IdoA-, to display anticoagulant activity 29.
Fig 5. 3-OST-5 products have varying anti-FXa activity.
8-mer products of 3-OST-5 were assessed for anti-FXa activity in a standard chromogenic assay. Fondaparinux and a non-anticoagulant 6-mer lacking 3-O-sulfation were used as positive and negative controls respectively. The antithrombin (AT) pentasaccharide binding sequence in 8-mers 6, 8, and 9 is boxed. These three 8-mers display strong anti-FXa activity. 8-mers 4 and 5 had minimal anti-FXa activity, consistent with their lack of the AT-binding pentasaccharide. Shorthand structures for 8-mers and non-anticoagulant 6-mer used in the analysis are shown at the bottom of the figure.
Discussion
The 3-OSTs are a unique family of HS biosynthetic enzymes. These enzymes transfer a sulfo group to synthesize 3-O-sulfated HS polysaccharides associated with widely variable biological functions 30. The substrate selectivity among different isoforms of 3-OSTs can be distinguished experimentally 31. Understanding the substrate recognition mechanism offers insights on how the biosynthesis of HS is regulated within cells. Recently, chemoenzymatic synthesis of structurally homogeneous HS oligosaccharides has been developed 2. In-depth understanding of the substrate specificity of new 3-OST isoforms permits the use of these new enzymes for the synthesis of 3-O-sulfated oligosaccharides for biological studies and the development of HS-based therapeutics 32. In this manuscript, we employed a series of HS octasaccharides to carry out the crystal structural analysis of 3-OST-5. For the first time, we solved the structures of 3-OST-5 in complex with oligosaccharide substrates. Comparing the crystal structures of 3-OST isoforms 1, 3 and 5 with substrate bound 19, 21, we found that the active sites are very similar, although subtle structural differences exist. To our surprise, we discovered that the presence of IdoA2S reduces the reactivity of 3-OST-5 for modification. Results from extensive structural comparison and homology analysis among 3-OST-1, 3-OST-3 and 3-OST-5 suggest that multiple factors contribute to the substrate specificity.
3-OST-5 and 3-OST-1 both require substrates carrying 6-O-sulfation, based on the enzyme activity measurements. The requirement for 6-O-sulfated substrate is supported by the crystal structures, which reveal interactions of the 6-O-sulfo groups on saccharide h with Lys303 and possibly saccharide d of the 8-mer-2 substrate with Lys227 (Fig 4B). The GlcNS6S at saccharide h superimposes well with that of 3-OST-1 substrate (Fig 6A). The 3-OST-1 mutant R268A reduced the activity to 7% of wild type enzyme, supporting its role in 6-O-sulfo group binding 19. While the positions of the 6-O-sulfo group at saccharide d for the 3-OST-1 and 3-OST-5 substrates differ in crystal structures, Lys193 of 3-OST-1, equivalent to Lys 227 of 3-OST-5 is mostly disordered and does not appear to participate in substrate binding. Interestingly, there are no interactions with the 6-O-sulfo group of the acceptor GlcNS6S saccharide f for either 3-OST-1 or 3-OST-5. The observation suggests that both 3-OST-5 and 3-OST-1 might be able to react with oligosaccharides that are partially 6-O-sulfated. Indeed, we demonstrated that 3-OST-1 sulfates partially 6-O-sulfated oligosaccharide substrates 33.
Fig 6. Superposition of 3-OST-1 and −3 onto 3-OST-5 with 8-mer-2 present.
A) Superposition of crystal structures of 3-OST-1 with heptasaccharide bound (yellow, protein light yellow; PDBidcode 3uan) onto 3-OST-5 with 8-mer-2 bound (protein in gray, 8-mer-2 in green). B) Superposition of ternary complex crystal structures of 3-OST-3 with PAP (cyan) and octasaccharide substrate (3-OST-3 in wheat for both protein and substrate PDB code 6xl8 3-OST-5 protein in gray and 8-mer-2 in green). The acceptor 3OH is marked with a red asterisk while the 3-OH on 3-OST-3 saccharide h is marked with a blue asterisk. Saccharides are labeled c-h and the Na ion bound to 3-OST-3 is shown as a purple sphere with chelating interactions shown in solid black lines. Select hydrogen bonds between 3-OST-1 and 3-OST-3 with their substrates are shown in black dashed lines. Shorthand structures for the oligosaccharide substrate in each crystal structure are presented below each panel.
3-OST-5 enzyme displays preferences to the saccharides around the acceptor site with the following order: 1) flanking GlcA on both sides of the acceptor 2) GlcA on non-reducing side and IdoA2S on reducing side 3) IdoA2S on non-reducing side and GlcA on reducing side and 4) least preferred flanking IdoA2S on both sides of acceptor. The crystal structures also provide insight into the uronic acid saccharide preference of GlcA for both saccharides flanking the acceptor GlcNS6S. Despite being given a substrate (8-mer-2) that would allow for 3-OST-5 to engage in three different ways, 3-OST-5 crystallized with GlcA flanking the acceptor GlcNS6S saccharide at saccharides g and e (Fig 4B). This is consistent with a preference for flanking GlcA saccharides to the acceptor GlcNS6S. Superpositions of the crystal structure of 3-OST-5 with 8-mer-2 bound with 3-OST-3 with the preferred two IdoA2S saccharides flanking the acceptor glucosamine reveals the acceptor glucosamines superimpose well and are in identical conformations (Fig 6B).
The conformational flexibility of the IdoA2S saccharide could play a role in substrate recognition. In 3-OST-3, both IdoA2S saccharides are in the skew boat ( 2S0 ) conformation 21. This places the carboxylate on the IdoA2S at saccharide e within 3.7 Å of the 2-O-sulfo group on the IdoA2S at saccharide g. This accumulation of negative charge is partially alleviated by Lys259 for 3-OST-3, which hydrogen bonds with both groups (Fig 6B) 31. Gly199 sits at the equivalent site in 3-OST-5 and is unable to form these interactions, resulting in a potential charge repulsion if IdoA2S was present at saccharide g. This would also place the 2-O-sulfo group within 4 Å of the N-sulfo group on the glucosamine at saccharide h. Finally, the 2S0 conformation results in a different position of the non-reducing end of the saccharide, which alters the trajectory of the substrate through the space between Ala306 and Ser120 in 3-OST-5, known as the gate structure 25. This could affect the protein interactions with the 6-O-sulfo and the N-sulfo on saccharide h. While the substrates for both 3-OST-1 and 3-OST-3 have IdoA2S on the reducing side of the acceptor, they are in different conformations. Based on crystal structures, 3-OST-3 interacts with the IdoA2S in the 2S0 conformation via an interaction with a bridging sodium ion that also interacts with Thr256 (Fig 6B). 3-OST-1 and 3-OST-5 contain a Val at this position (Val164 and Val196, respectively) that would preclude coordinating a sodium ion at this position. The IdoA2S on the reducing side of the acceptor for 3-OST-1 is in the 1C4 conformations (Fig 6A). Asn167 of 3-OST-1 (equivalent to Gly199 in 3-OST-5), forms interactions with both the carboxylate of the 2-O-sulfo group of IdoA2S at saccharide e and the 3-OH position of the GlcNS6S at saccharide d for 3-OST-1 (Fig 6A). This may help maximize the binding potential of a 1C4 conformation at this position. In addition, a His168 in 3-OST-1 is situated near the 2-O-sulfo from saccharide d. A H168A mutation reduced the activity of 3-OST-1 by 50% suggesting it contributes to substrate binding 19. In 3-OST-5, the equivalent residue Lys200 can extend further away from the binding pocket to interact with the N-sulfo group of the glucosamine at saccharide d due to the 4C1 conformation of the GlcA at saccharide e, supporting a preference for a GlcA at that saccharide. Thus, while binding of an IdoA2S on the non-reducing side could introduce charge repulsion in addition to disrupting interactions with the substrate, placement of an IdoA2S on the reducing side could simply reduce favorable interactions. The preference of 3-OST-5 to produce more of 8-mer-9 than 8-mer-8, in addition to product inhibition, could be due to two possible structurally based reasons; 1) having two flanking IdoA2S has the deleterious additive effects of the individual IdoA2Ss being present and/or 2) A 3-O-Sulfo group at saccharide h when there is a IdoA2S at saccharide e would result in clashes with residues Ser120 and Gln121, based on the position of the substrate orientation in 3-OST-3 (Fig 6B).
The results of these studies provide a starting point for understanding the specificity of 3-OST-5 utilizing smaller, 8-mer oligosaccharides. While the biological substrates of the enzyme are known to be much longer polysaccharides, current oligosaccharide sequencing methods limit our ability to conduct specificity studies using these longer structures. Additional studies of the sequence structure around modification sites on polysaccharides is of interest but would require more sophisticated sequencing methods. Such studies could reveal whether the observed specificity reported here is also observed in vivo.
An interesting finding from our study is that 3-OST-5 generates both anticoagulant and non-anticoagulant oligosaccharides, opening a new avenue for developing HS-based therapeutics. These oligosaccharides are all highly sulfated and show the same levels of sulfo groups per disaccharide unit. It was widely expected that fully sulfated HS oligosaccharides carrying 3-O-sulfation would display anticoagulant activity. The results from our studies suggest that 3-OST-5 can generate non-anticoagulant 3-O-sulfated 8-mers, i.e. 8-mer-4 and 8-mer-5. The synthesis of anticoagulant and non-anticoagulant 8-mers can be achieved by controlling the number and position of IdoA2S saccharide in the substrates subjected to 3-OST-5 modification. These non-anticoagulant 3-O-sulfated oligosaccharides could be used to target proteins in alternate pathways, such as inflammation, without the risk of bleeding side-effects.
Supplementary Material
Acknowledgements
We thank G. Mueller and P. Tumbale for critical reading of the manuscript. This work was supported in part by Division of Intramural Research of the National Institute of Environmental Health Sciences, National Institutes of Health Grants 1ZIA ES102645 (to L.C.P.) and by National Institutes of Health grants HL094463, HL144970, GM128484, GM134738, and HL139197. This work was also supported by GlycoMIP, a National Science Foundation Materials Innovation Platform funded through Cooperative Agreement DMR-1933525. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences [W-31-109-Eng-38].
YX and Jian Liu are founders of Glycan Therapeutics and have equity. VP is an employee of Glycan Therapeutics and has the equity of Glycan Therapeutics. TP is an employee of Glycan Therapeutics. Dr. Jian Liu’s lab at UNC has received a gift from Glycan Therapeutics to support research in glycoscience.
Footnotes
Competing interest
RW, AMK, ZW, ES, Jine Li, JMK and LCP declare no competing interest.
Supporting Information
Experimental procedures, supplementary tables, and supplementary figures included.
References
- 1.Bishop J; Schuksz M; Esko JD, Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 2007, 446, 1030–1037. [DOI] [PubMed] [Google Scholar]
- 2.Liu J; Linhardt RJ, Chemoenzymatic synthesis of heparan sulfate and heparin. Nat Prod Rep 2014, 31, 1676–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clausen TM; Sandoval DR; Spliid CB; Pihl J; Perrett HR; Painter CD; Narayanan A; Majowicz SA; Kwong EM; McVicar RN; Thacker BE; Glass CA; Yang Z; Torres JL; Golden GJ; Bartels PL; Porell RN; Garretson AF; Laubach L; Feldman J; Yin X; Pu Y; Hauser BM; Caradonna TM; Kellman BP; Martino C; M Gordts PLSM; Chanda SK; Schmidt AG; Godula K; Leibel SL; Jose J; Corbett KD; Ward AB; Carlin AF; Esko JD, SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell 2020, 183, 1043–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gama C; Tully SE; Sotogaku N; Clark PM; Rawat M; Vaidehi N; Goddard WA; Nishi A; Hsieh-Wilson LC, Sulfation patterns of glycosaminoglycans encode molecular recognition and activity. Nat Chem Biol 2006, 2, 467–473. [DOI] [PubMed] [Google Scholar]
- 5.Esko JD; Lindahl U, Molecular diversity of heparan sulfate. J. Clin. Invest 2001, 108, 169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Y; Masuko S; Takieddin M; Xu H; Liu R; Jing J; Mousa SA; Linhardt RJ; Liu J, Chemoenzymatic synthesis of homogeneous ultra-low molecular weight heparin. Science 2011, 334, 498–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Y; Cai C; Chandarajoti K; Hsieh P; Lin Y; Pham TQ; Sparkenbaugh EM; Sheng J; Key NS; Pawlinski RL; Harris EN; Linhardt RJ; Liu J, Homogeneous and reversible low-molecular weight heparins with reversible anticoagulant activity. Nat Chem Biol 2014, 10, 248–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z; Arnold K; Dhurandhare VM; Xu Y; Liu J, Investigation of the biological functions of heparan sulfate using a chemoenzymatic synthetic approach. RSC Chem Biol 2021, 2, 702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Y; Chandarajoti K; Zhang X; Pagadala V; Dou W; Hoppensteadt DM; Sparkenbaugh E; Cooley B; Daily S; Key NS; Severynse-Stevens D; Fareed J; Linhardt RJ; pawlinski R; Liu J, Synthetic oligosaccharides can replace animal-sourced low-molecular weight heparins. Sci. Transl. Med 2017, 9, eaan5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shworak NW; Liu J; Petros LM; Zhang L; Kobayashi M; Copeland NG; Jenkins NA; Rosenberg RD, Diversity of the extensive heparan sulfate D-glucosaminyl 3-O-sulfotransferase (3-OST) multigene family. J. Biol. Chem 1999, 274, 5170–5184. [DOI] [PubMed] [Google Scholar]
- 11.Shukla D; Liu J; Blaiklock P; Shworak NW; Bai X; Esko JD; Cohen GH; Eisenberg RJ; Rosenberg RD; Spear PG, A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 1999, 99, 13–22. [DOI] [PubMed] [Google Scholar]
- 12.Li J; Su G; Xu Y; Arnold K; Pagadala V; Wang C; Liu J, Synthesis of 3- O-Sulfated Heparan Sulfate Oligosaccharides Using 3- O-Sulfotransferase Isoform 4. ACS Chem Biol 2021, doi: 10.1021/acschembio.1c00474. [DOI] [PMC free article] [PubMed]
- 13.Thacker BE; Seamen E; Lawrence R; Parker MW; Xu Y; Liu J; Vander KCW; Esko JD, Expanding the 3-O-Sulfate Proteome--Enhanced Binding of Neuropilin-1 to 3-O-Sulfated Heparan Sulfate Modulates Its Activity. ACS Chem. Biol 2016, 11, 971–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel VN; Lombaert IMA; Cowherd SN; Shworak N; Xu Y; Liu J; Hoffman MP, 3-O-sulfated heparan sulfate controls epithelial Kit+FGFR2b+ progenitor expansion via rapid autocrine regulation of 3-O-sulfotransferases. Developmental cell 2014, 29, 662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Agostini AI; Dong JC; de Vantery Arrighi C; Ramus MA; Dentand-Quadri I; Thalmann S; Ventura P; Ibecheole V; Monge F; Fischer AM; HajMohammadi S; Shworak NW; Zhang L; Zhang Z; Linhardt RJ, Human follicular fluid heparan sulfate contains abundant 3-O-sulfated chains with anticoagulant activity. J Biol Chem 2008, 283 (42), 28115–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denys A; Allain F, The Emerging Roles of Heparan Sulfate 3- O-Sulfotransferases in Cancer. Front Oncol 2019, 9, 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sepulveda-Diaz JE; Alavi Naini SM; Huynh MB; Ouidja MO; Yanicostas C; Chantepie S; Villares J; Lamari F; Jospin E; van Kuppevelt TH; Mensah-Nyagan AG; Raisman-Vozari R; Soussi-Yanicostas N; Papy-Garcia D, HS3ST2 expression is critical for the abnormal phosphorylation of tau in Alzheimer’s disease-related tau pathology. Brain 2015, 138, 1339–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao J; Zhu Y; Song X; Xiao Y; Su G; Liu X; Wang Z; Xu Y; Liu J; Eliezer D; Ramlall TE; Lippens G; Gibson J; Zhang F; Linhardt RJ; Wang L; Wang C, 3-O-sulfation of heparan sulfate enhances tau interaction and cellular uptake. Angew. Chem. Int. Ed 2020, 59, 1818–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moon AF; Xu Y; Woody S; Krahn JM; Linhardt RJ; Liu J; Pedersen LC, Dissecting substrate recognition mode of heparan sulfate 3-O-sulfotransferases for the biosynthesis of anticoagulant heparin. Proc. Natl. Acad. Sci. USA 2012, 109, 5256–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon A; Edavettal SC; Krahn JX; Munoz EM; Negishi M; Linhardt RJ; Liu J; Pedersen LC, Structural analysis of the sulfotransferase (3-OST-3) involved in the biosynthesis of an entry receptor of herpes simplex virus 1. J. Biol. Chem 2004, 279, 45185–45193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wander R; Kaminski AM; Xu Y; Pagadala V; Krahn JM; Pham TQ; Liu J; Pedersen LC, Deciphering the substrate recognition mechanisms of the heparan sulfate 3-O-sulfotransferase-3 RSC Chem Biol 2021, 2, 1239–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhurandhare VM; Pagadala V; Ferreira A; Muynck LD; Liu J, Synthesis of 3-O-sulfated disaccharide and tetrasaccharide standards for compositional analysis of heparan sulfate Biochemistry 2020, 59, 3186–3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venkataraman G; Shriver Z; Raman R; Sasisekharan R, Sequencing complex polysaccharides. Science 1999, 286, 537–542. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z; Hsieh P-H; Xu Y; Thieker D; Chai EJE; Xie S; Cooley B; Woods RJ; Chi L; Liu J, Synthesis of 3-O-sulfated oligosaccharides to understand the relationship between structures and functions of heparan sulfate. J. Am. Chem. Soc 2017, 139, 5249–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu D; Moon A; Song D; Pedersen LC; Liu J, Engineering sulfotransferases to modify heparan sulfate. Nat Chem Biol 2008, 4, 200–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edavettal SC; Lee KA; Negishi M; Linhardt RJ; Liu J; Pedersen LC, Crystal structure and mutational analysis of heparan sulfate 3-O-sulfotransferase isoform 1. J. Biol. Chem 2004, 279, 25789–25797. [DOI] [PubMed] [Google Scholar]
- 27.Kakuta Y; Li L; Pedersen LC; Pedersen LG; Negishi M, Heparan sulphate N-sulphotransferase activity: reaction mechanism and substrate recognition. Biochem. Soc. Trans 2003, 31 (pt2), 331–334. [DOI] [PubMed] [Google Scholar]
- 28.Das S; Mallet J; J. E.; Driguez P; Duchaussoy P; Sizun P; Herault J; Herbert J; M. P.; P. S., Synthesis of conformationally locked L-iduronic acid derivatives: direct evidence for a critical role of the skew-boat 2S0 conformer in the activation of antithrombin by heparin. Chemistry 2001, 7, 4821–4834. [DOI] [PubMed] [Google Scholar]
- 29.Hsieh P-H; Thieker DF; Guerrini M; Woods RJ; Liu J, Uncovering the relationship between sulphation patterns and conformation of iduronic acid in heparan sulphate. Sci Rep 2016, 6, 29602; doi: 10.1038/srep29602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thacker B; Xu D; Lawrence R; Esko J, Heparan sulfate 3-O-sulfation: a rare modification in search of a function. Matrix Biol 2014, 35, 60–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J; Moon AF; Sheng J; Pedersen LC, Understanding the substrate specificity of the heparan sulfate sulfotransferases by a combined synthetic crystallographic approach. Curr Opin Struct Biol 2012, 22, 550–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnold K; Liao Y-E; Liu J, Potential use of anti-inflammatory synthetic heparan sulfate to attenuate liver damage Biomedicines 2021, 8, 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yi L; Xu Y; Kaminski AM; Chang X; Pagadala V; Horton M; Su G; Wang Z; Lu G; Conley P; Zhang Z; Pedersen LC; Liu J, Using enigneered 6-O-sulfotransferase to improve the synthesis of anticoagulant heparin. Org Biomol Chem 2020, 18, 8094–8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.