ABSTRACT
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection can induce mild to life-threatening symptoms. Especially individuals over 60 years of age or with underlying comorbidities, including heart or lung disease and diabetes, or immunocompromised patients are at a higher risk. Fatal multiorgan damage in coronavirus disease 2019 (COVID-19) patients can be attributed to an interleukin-6 (IL-6)-dominated cytokine storm. Consequently, IL-6 receptor (IL-6R) monoclonal antibody treatment for severe COVID-19 cases has been approved for therapy. High concentrations of soluble IL-6R (sIL-6R) were found in COVID-19 intensive care unit patients, suggesting the involvement of IL-6 trans-signaling in disease pathology. Here, in analogy to bispecific antibodies (bsAbs), we developed the first bispecific IL-6 trans-signaling inhibitor, c19s130Fc, which blocks viral infection and IL-6 trans-signaling. c19s130Fc is a designer protein of the IL-6 trans-signaling inhibitor cs130 fused to a single-domain nanobody directed against the receptor binding domain (RBD) of the SARS-CoV-2 spike protein. c19s130Fc binds with high affinity to IL-6:sIL-6R complexes as well as the spike protein of SARS-CoV-2, as shown by surface plasmon resonance. Using cell-based assays, we demonstrate that c19s130Fc blocks IL-6 trans-signaling-induced proliferation and STAT3 phosphorylation in Ba/F3-gp130 cells as well as SARS-CoV-2 infection and STAT3 phosphorylation in Vero cells. Taken together, c19s130Fc represents a new class of bispecific inhibitors consisting of a soluble cytokine receptor fused to antiviral nanobodies and principally demonstrates the multifunctionalization of trans-signaling inhibitors.
IMPORTANCE The availability of effective SARS-CoV-2 vaccines is a large step forward in managing the pandemic situation. In addition, therapeutic options, e.g., monoclonal antibodies to prevent viral cell entry and anti-inflammatory therapies, including glucocorticoid treatment, are currently developed or in clinical use to treat already infected patients. Here, we report a novel dual-specificity inhibitor to simultaneously target SARS-CoV-2 infection and virus-induced hyperinflammation. This was achieved by fusing an inhibitor of viral cell entry with a molecule blocking IL-6, a key mediator of SARS-CoV-2-induced hyperinflammation. Through this dual action, this molecule may have the potential to efficiently ameliorate symptoms of COVID-19 in infected individuals.
KEYWORDS: IL-6, SARS-CoV-2, sgp130, trans-signaling
INTRODUCTION
Since its emergence in 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spread globally, and as of 5 May 2021, about 200 million infections were recorded (https://coronavirus.jhu.edu/), threatening to overwhelm health care systems in many countries. Coronavirus disease 2019 (COVID-19) resulting from SARS-CoV-2 infection leads to a broad variety of outcomes ranging from very mild cases to life-threatening respiratory failure, shock, or multiorgan failure (1). Even though SARS-CoV-2 infection in many cases leads to no or only mild symptoms, millions of hospitalizations and mortalities are associated with COVID-19 worldwide (https://coronavirus.jhu.edu/). Mortality rates vary considerably among studies (2); however, a clear age dependence was observed with regard to the development of severe COVID-19 (3). Severe COVID-19 causes hyperinflammatory syndrome culminating in respiratory dysfunction and multiorgan damage (4). SARS-CoV-2-induced hyperinflammatory syndrome is often compared to cytokine-induced conditions known from other diseases, including sepsis (1), acute respiratory distress syndrome (1), and chimeric antigen receptor (CAR) T cell-induced cytokine release syndrome (CRS) (5). Interleukin-6 (IL-6) and soluble IL-6 receptor (sIL-6R) were identified among the key players in COVID-19-induced cytokine release syndrome (6–12). In classic signaling, IL-6 initially binds to the membrane-bound IL-6R followed by full receptor complex formation with the signal-transducing receptor chain gp130. In trans-signaling, complexes of IL-6 and sIL-6R bind to cell membrane-bound gp130 (13). During inflammation, membrane-bound IL-6R can be proteolytically cleaved into sIL-6R by a disintegrin and metalloprotease (ADAM) proteases, mainly ADAM10 and -17 (14–16). Consequently, serum levels of IL-6 and sIL-6R concomitantly rise under inflammatory conditions (17–22). Of note, whereas IL-6 classic signaling is considered beneficial, IL-6 trans-signaling has been shown to be the mostly detrimental driving force of ongoing inflammatory reactions (13, 23), including autoimmune disease, sepsis (24), cytokine release syndrome (24), and COVID-19 (25). Hence, antibody (Ab) IL-6R inhibitors are of great interest for the treatment of the COVID-19-induced hyperinflammatory syndrome (12, 26). In 2017, the IL-6R antibody tocilizumab was approved for the treatment of the CAR T cell-induced cytokine storm (5). Tocilizumab and sarilumab bind to soluble and membrane-bound IL-6R (27, 28), whereas siltuximab binds to IL-6 (29). However, all antibodies prevent the binding of IL-6 to IL-6R and inhibit classic and trans-signaling equally well (30). Consistently, the IL-6R antibodies tocilizumab and sarilumab demonstrated beneficial effects on survival rates in severe COVID-19 cases in preclinical and clinical studies (9–11, 31). However, IL-6 is required to control viral infection (32); hence, global blockade of IL-6 signaling, e.g., through tocilizumab, is associated with an increased risk of airway infections (33, 34). This might be detrimental for the treatment of COVID-19 with IL-6 inhibitors due to the potential increase in viral replication following IL-6 blockade. Whereas antibodies did not differentiate between classic and trans-signaling, soluble forms of gp130 (sgp130) are selective binders of IL-6:sIL-6R complexes, thereby interfering with only IL-6 trans-signaling (35). Therefore, inhibition of IL-6 trans-signaling by sgp130 molecules might offer an attractive alternative inhibitory pathway for cytokine release syndrome during severe SARS-CoV-2 infections. In mice, sgp130 prevents death caused by cecal ligation puncture-induced septic shock syndrome (36), and bacterial infections are better controllable after selective inhibition of IL-6 trans-signaling than after inhibition of both IL-6 classic and trans-signaling by monoclonal IL-6 antibodies (37, 38). However, the combination of IL-6 blockade with agents reducing uncontrolled viral infection might be more beneficial in the treatment of severe COVID-19 cases. The spike protein of SARS-CoV-2 binds to human ACE2 (hACE2) on the cell surface to facilitate viral cell entry (39, 40). Hence, preventive strategies as well as most efforts on the development of therapeutic antibodies focus on the inhibition of the interaction of the receptor binding domain (RBD) of the spike protein (S-RBD) with ACE2 (41, 42). There are more than 50 monoclonal antibodies against SARS-CoV-2 in various developmental stages (43), many of which are directed against the spike protein (43–45). Among them, the dimeric single-domain nanobody VHH72 was shown to efficiently block viral cell entry, whereas monomeric VHH72 was less effective (44). Here, we functionally combined our recently developed chimeric miniaturized sgp130 variant cs130 that selectively targets IL-6 trans-signaling (46) with the single-domain antibody VHH72. We demonstrate that this dimeric hybrid soluble gp130/VHH72-nanobody fusion protein, c19s130Fc, simultaneously blocks IL-6 trans-signaling and SARS-CoV-2 infection.
RESULTS
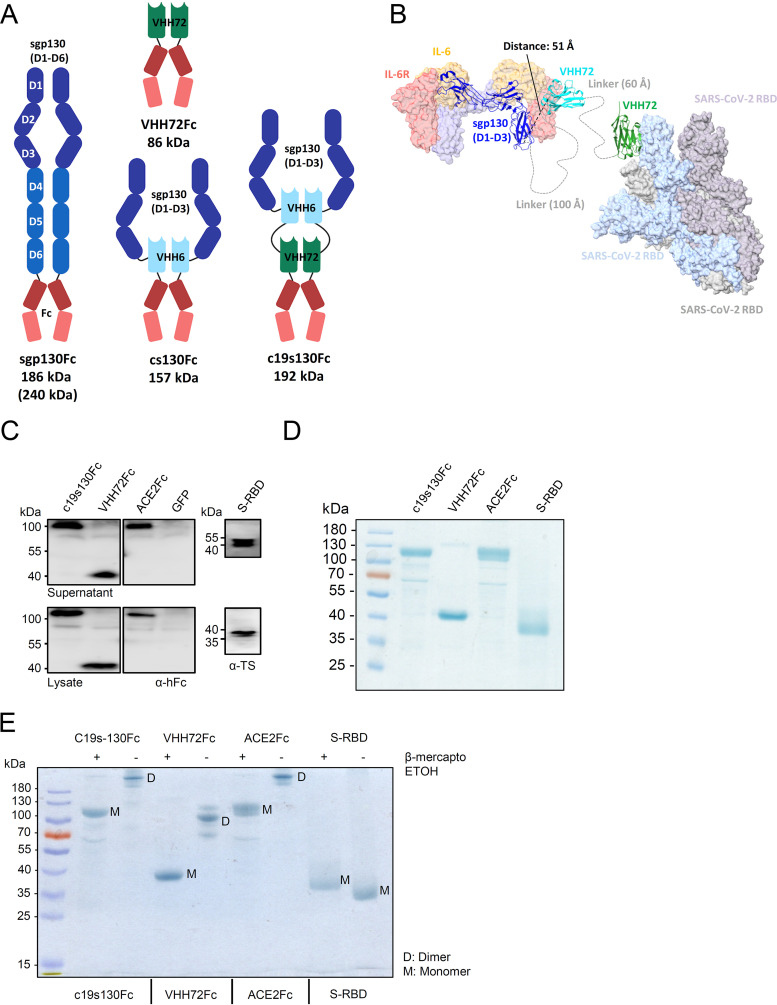
Modular architecture of the bispecific inhibitor c19s130Fc.
Several variants of soluble gp130 were described to selectively inhibit IL-6 trans-signaling. sgp130Fc consists of all six extracellular domains (ECDs) of gp130 fused to the Fc part of an IgG antibody. The dimerization of sgp130 increased the affinity for IL-6:sIL-6R complexes by a factor of 10 compared to monomeric sgp130 (35). Interestingly, only the first three extracellular domains of gp130 are needed for cytokine binding; however, sgp130 variants consisting of only these three domains (sgp130RAPS, sgp130-ELP, and sgp130E10) showed markedly reduced binding affinities for IL-6:sIL-6R complexes compared to sgp130 (47). The reason for this observation is not clear. Recently, we generated the miniaturized high-affinity sgp130 variant cs130, which consists of the first three extracellular cytokine binding domains, D1 to D3, of sgp130 (sgp130D1–D3) fused to the nanobody VHH6, which showed sgp130Fc-like binding affinities and inhibitory capacity (46). VHH6 specifically binds to IL-6:sIL-6R complexes, without inhibitory capacity (22, 46, 48). In particular, the monomeric fusion of VHH6 to sgp130D1–D3 resulted in an equally potent but an approximately three-times-smaller IL-6 trans-signaling inhibitor than sgp130Fc. Like IL-6, IL-11 signals via soluble and membrane-bound IL-11R and homodimeric gp130, and sgp130Fc inhibits IL-6 and IL-11 trans-signaling with comparable efficacy. Due to the incorporation of VHH6, the miniaturized sgp130 variant cs130Fc demonstrated increased specificity for IL-6 trans-signaling with diminished effects on IL-11 trans-signaling. As a consequence of the smaller size and modular architecture of cs130, we wondered if this design enables further upgrading into bispecificity with the binding of IL-6:sIL-6R complexes and a second IL-6-connected process/protein. Due to the involvement of IL-6 pathology in severe COVID-19 cases, we chose the binding of S-RBD to inhibit SARS-CoV-2 infection. To minimize the size of the resulting bispecific inhibitor, c19s130Fc, we chose to fuse the SARS-CoV-2 S-RBD nanobody VHH72 (44) to cs130 connected via a flexible linker sequence, T(GGGGS)2GGGGTG (Fig. 1A). Molecular modeling illustrated the possible complex formation of IL-6:sIL-6R/c19s130Fc/S-RBD protein complexes (Fig. 1B). Domains 1 to 3 of sgp130 and VHH72 trap and inactivate the IL-6:sIL-6R complex, while fusion to VHH72 via a long flexible linker allows the simultaneous blockade of S-RBD. c19s130Fc plus the control proteins VHH72 fused to IgG Fc (VHH72Fc) and cs130Fc were readily expressed and secreted in HEK293T cells (Fig. 1C) and were subsequently produced and purified from the supernatants of Expi293 cells. Following affinity purification, the proteins were >90% pure, as demonstrated by SDS-PAGE analysis and subsequent Coomassie staining (Fig. 1D). The disulfide-mediated dimerization of all proteins was assessed by nonreducing SDS-PAGE. In this analysis, all Fc-fused proteins demonstrated a shift to a higher molecular weight in the absence of a reducing agent (Fig. 1E), confirming disulfide-mediated dimerization. We did not produce monomeric c19s130 because it was shown previously that VHH72 inhibited cellular virus entry only in the dimeric form (44).
FIG 1.
Expression and purification of c19s130Fc, VHH72Fc, ACE2-Fc, and S-RBD. (A) Schematic overview of recombinant proteins utilized in this study. (B) Molecular modeling illustrating complex formation of IL-6:sIL-6R/c19s130Fc/spike protein complexes. The structure of the IL-6 signaling complex (PDB accession number 1P9M) was superpositioned with the structure of VHH6 bound to a complex of IL-6:sIL-6R (PDB accession number 5FUC). In addition, the structure of VHH72 bound to S-RBD (PDB accession number 6WAQ) was superpositioned onto the structure of trimeric S-RBD in an open conformation (PDB accession number 7CAC). Components of the c19s130Fc protein are depicted in a ribbon representation, and the IL-6 signal complex and S-RBD are depicted in a surface representation using ChimeraX. (C) Western blotting of supernatants and lysates of HEK293T cells expressing c19s130Fc, VHH72Fc, ACE2-Fc, and S-RBD. GFP, green fluorescent protein. (D) SDS-PAGE analysis of purified c19s130Fc, VHH72Fc, ACE2-Fc, and S-RBD followed by Coomassie staining. (E) SDS-PAGE analysis of c19s130Fc, VHH72Fc, ACE2-Fc, and S-RBD in the presence (+) or absence (−) of β-mercaptoethanol. ETOH, ethanol.
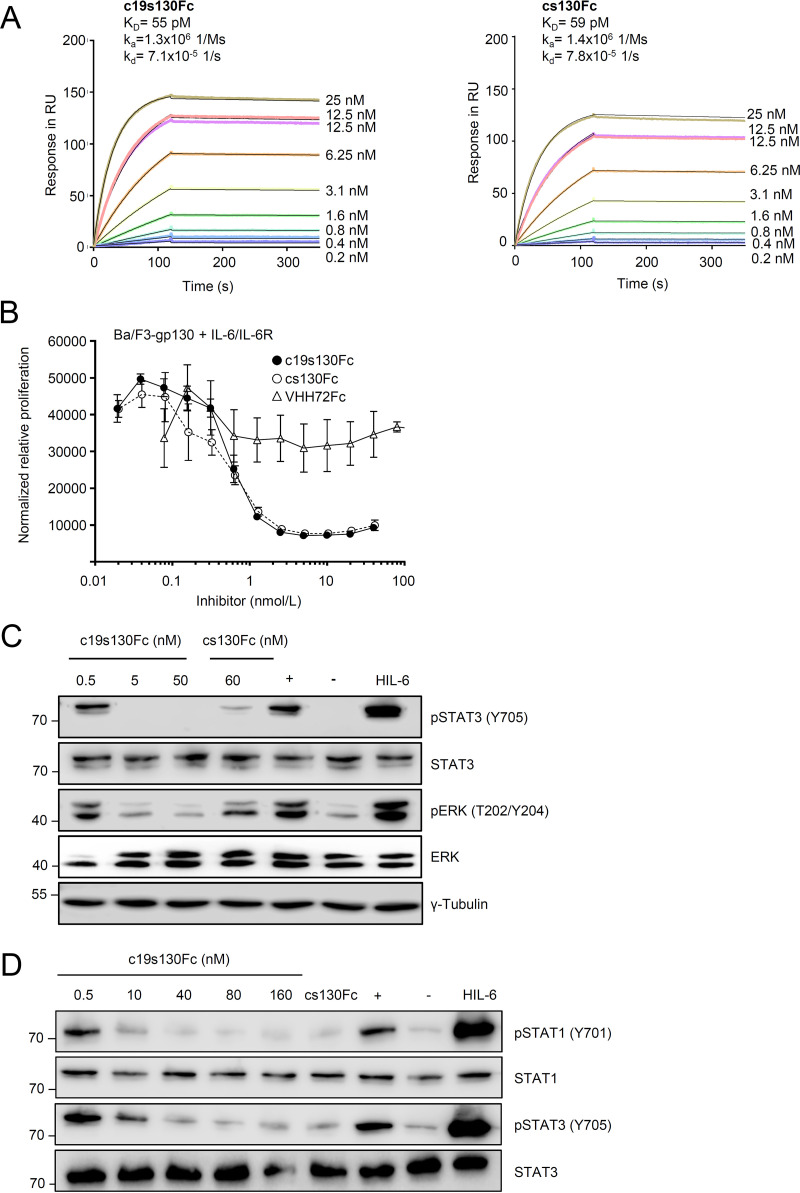
c19s130Fc efficiently inhibits IL-6 trans-signaling.
First, we determined and compared the affinities of c19s130Fc, cs130Fc, and VHH72Fc for hyper-IL-6 (HIL-6) in surface plasmon resonance (SPR) experiments. The trans-signaling designer cytokine HIL-6 is a fusion protein composed of IL-6 and the sIL-6R connected via a flexible peptide linker (49). Both c19s130Fc and cs130Fc displayed very high and almost identical affinities of 55 and 59 pM, respectively, for hyper-IL-6 (Fig. 2A). The kinetic analysis of the interaction revealed the formation of a very stable complex characterized by a very low koff rate of 7.1 × 10−5 1/s. Next, we analyzed the inhibitory potential of c19s130Fc toward IL-6 trans-signaling. To this end, Ba/F3 cells stably transduced with gp130 (Ba/F3-gp130) were stimulated with 100 ng/mL IL-6 and 200 ng/mL sIL-6R, which induced STAT3/extracellular signal-regulated kinase (ERK) phosphorylation-dependent cellular proliferation. This cell-based assay served as a surrogate model for the induction of IL-6 trans-signaling. We observed concentration-dependent inhibition of IL-6 trans-signaling through c19s130Fc and cs130Fc, while no effect was found for VHH72Fc (Fig. 2B). IC50 (50% inhibitory concentration) values of 1 ± 0.3 nM and 0.6 ± 0.2 nM were determined for c19s130Fc and cs130Fc, respectively. In an orthogonal experimental setup, IL-6:sIL-6R-stimulated Ba/F3-gp130 cells and Vero cells were utilized to examine the effect of c19s130Fc on STAT3 phosphorylation. In line with the proliferation data, c19s130Fc and cs130Fc inhibited STAT3 and ERK phosphorylation at concentrations above 5 nM in Ba/F3-gp130 cells (Fig. 2C). In Vero cells, STAT1 and STAT3 phosphorylations were blocked at concentrations above 10 nM (Fig. 2D). Taken together, c19s130Fc and cs130Fc but not VHH72 are highly potent inhibitors of IL-6 trans-signaling, as shown in biophysical and cell-based assays.
FIG 2.
c19s130Fc blocks IL-6 trans-signaling. (A) SPR analysis of HIL-6 binding to c19s130Fc. c19s130Fc was immobilized on a protein A chip, and increasing concentrations of HIL-6 were injected. Sensorgrams in response units (RU) over time are depicted as colored lines, and global fit data are displayed as black lines. KD, equilibrium dissociation constant; kd, dissociation constant. (B) Ba/F3-gp130 cells were stimulated with 100 ng/mL IL-6 and 200 ng/mL sIL-6R in the presence of increasing c19s130Fc, cs130Fc, or VHH72Fc concentrations. At 72 h poststimulation, cellular proliferation was detected using CellTiter-Blue. Assay results are representative of data from three independent experiments. (C) Western blot analysis of Ba/F3-gp130 cells stimulated for 30 min with 8 nM IL-6 and 1 nM sIL-6R in the presence of the indicated concentrations of c19s130Fc and cs130Fc. Prior to stimulation, IL-6, sIL-6R, and inhibitors were incubated separately for 30 min. Western blots were stained for pSTAT3, STAT3, pERK, and ERK. Western blots are representative of results from three independent experiments. Controls for unstimulated cells (−), cells in the absence of c19s130Fc (+), and stimulation with HIL-6 are included. (D) Western blot analysis of Vero cells stimulated for 30 min with 400 ng/mL IL-6 and 200 ng/mL sIL-6R in the presence of the indicated concentrations of c19s130Fc and cs130Fc. Prior to stimulation, IL-6, sIL-6R, and inhibitors were incubated separately for 30 min. Western blots were stained for pSTAT3, STAT3, pSTAT1, and STAT1. Western blots are representative of results from three independent experiments. Controls for unstimulated cells (−), cells in the absence of c19s130Fc (+), and stimulation with HIL-6 are included.
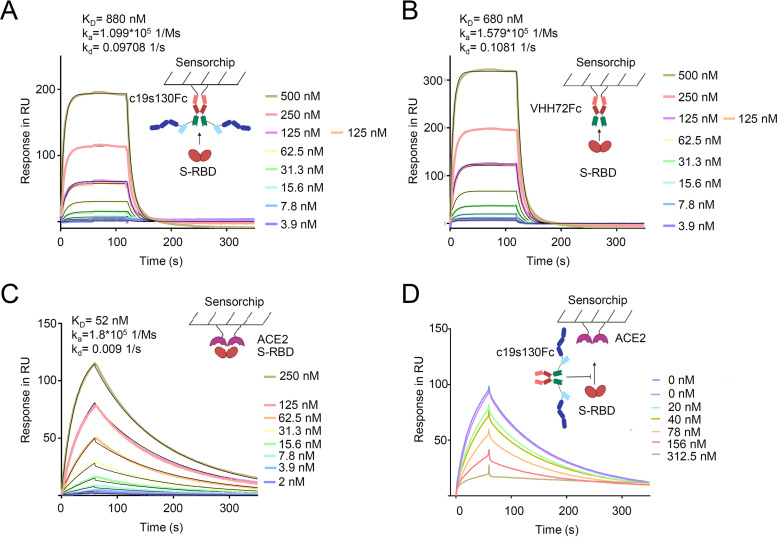
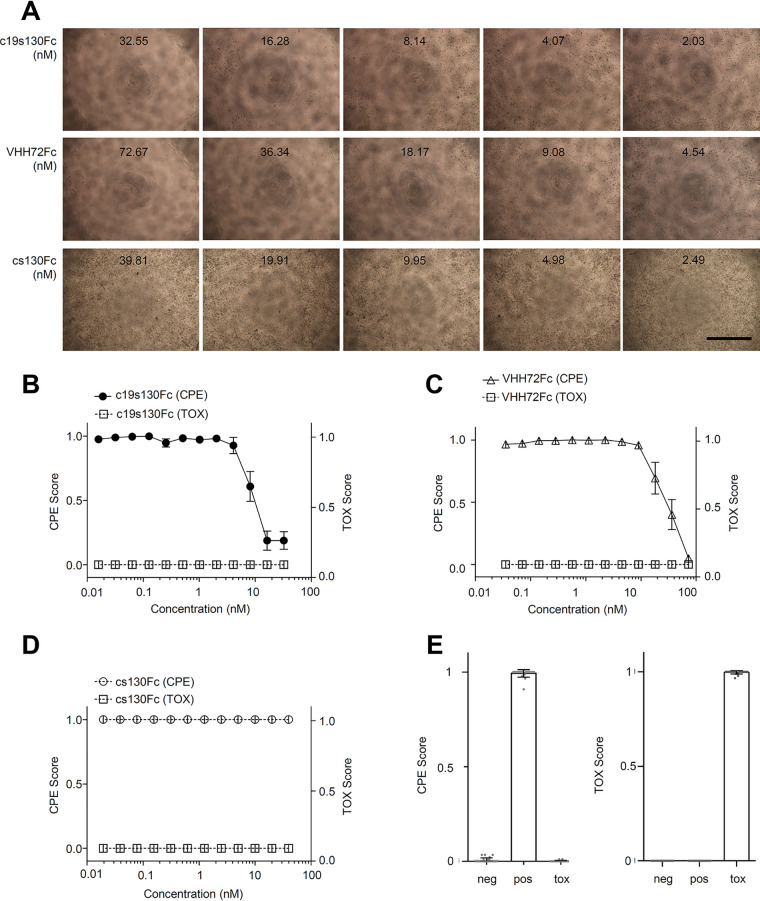
c19s130Fc binds to SARS-CoV-2 S-RBD and prevents viral entry.
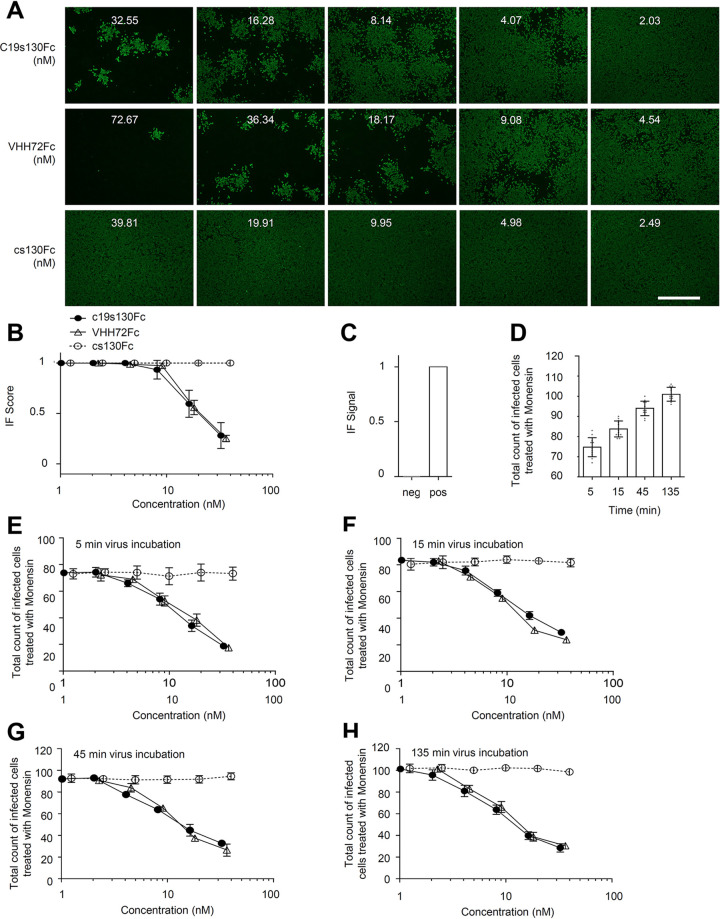
To determine the activity of the SARS-CoV-2-neutralizing entity in c19s130Fc, we first determined the binding kinetics with S-RBD. We determined the VHH72Fc binding affinity for S-RBD to be 680 nM using SPR (Fig. 3A), which deviates from the previously described affinity of 39 nM (44). Both setups contained monomeric S-RBD as an analyte, but the weaker binding in this study can primarily be attributed to a lower association rate constant (1.6 × 105 1/Ms [1 per molar times second]) and a higher dissociation rate constant (0.1 1/s). This can be explained by differences in the S-RBD protein compositions used. The previously described affinity of 39 nM was measured toward S-RBD and S-RBD subdomain 1 (S-RBD-SD1), in contrast to S-RBD, which was utilized in our study. This may lead to an altered dissociation rate constant. For c19s130Fc, highly comparable S-RBD binding kinetics with an affinity of 880 nM were found. Hence, VHH72 fully retains its activity in c19s130Fc. We further analyzed the affinity of purified S-RBD for immobilized ACE2-Fc via SPR (Fig. 3B). An affinity of 52 nM was detected, which is in good agreement with the previously described affinity of 44 nM (40). In competition assays with immobilized ACE2-Fc, S-RBD, and increasing concentrations of c19s130Fc, an inhibitor-dependent reduction of the binding of S-RBD to ACE2-Fc was found (Fig. 3C and D). A concentration of 78 nM c19s130Fc resulted in a reduction of ACE2:S-RBD by approximately 40%, as apparent by a reduction of the maximal binding response from 98.96 response units (RU) to 60.28 RU. This suggested that c19s130Fc binding to SARS-CoV-2 S-RBD prevents spike protein binding to ACE2 and, hence, neutralizes this key interaction required for viral cell entry. Next, we analyzed the effect of c19s130Fc on SARS-CoV-2-mediated cytopathic effects (CPE) on Vero cells. Vero cells can be efficiently infected with SARS-CoV-2 (50, 51) and serve as a model system for viral infection. Following incubation with SARS-CoV-2, a reduction of virus-induced CPE was found in the presence of c19s130Fc and VHH72Fc but not cs130Fc (Fig. 4A to E). IC50 values of 8.1 ± 0.8 nM and 32.3 ± 18.6 nM were determined for c19s130Fc and VHH72Fc, respectively (Fig. 4B and C). As for SPR-based affinity assays, IC50 values for VHH72Fc reported previously by Wrapp et al. (2.5 nM) differed slightly in pseudovirus neutralization assays (44). In addition to CPE assays, we investigated the ability of c19s130Fc to prevent infection of Vero cells by SARS-CoV-2. Virally infected Vero cells were visualized via immunofluorescence (IF) using anti-SARS-CoV-2 nucleocapsid antibodies (51). Vero cells treated with c19s130Fc and VHH72Fc showed reduced SARS-CoV-2 cell entry with comparable IC50 values of 15.1 ± 3.7 nM and 20.7 ± 1.6 nM, respectively (Fig. 5A to C). No effect was observed for cs130Fc. Next, we investigated the time-dependent effect of the inhibitors on viral entry during early phases of infection. Vero cells were incubated with SARS-CoV-2 for 5, 15, 45, or 135 min, and virus infection and uptake were then stopped by the addition of monensin. Total viral entry increased from 5 min to 135 min of incubation (Fig. 5D). Both c19s130Fc and VHH72Fc reduced SARS-CoV-2 uptake with very comparable IC50 values of 14.2 ± 5.4 nM, 11.8 ± 3.6 nM, 9.1 ± 1.4 nM, and 8.2 ± 1.4 nM for c19s130Fc and 16.2 ± 4.3 nM, 9.8 ± 1.2 nM, 11.9 ± 0.8 nM, and 10.7 ± 3.5 nM for VHH72Fc (Fig. 5E to H). These IC50 values are very comparable to the IC50 values determined for viral uptake after 48 h. Hence, the inhibitory proteins seem to be stable and maintain activity for at least 48 h in a cell culture setting.
FIG 3.
c19s130Fc binds to S-RBD and blocks its binding to ACE2. (A) SPR analysis of c19s130Fc binding to S-RBD. c19s130Fc was captured on a protein A chip, and increasing concentrations of S-RBD were injected. Sensorgrams in response units (RU) over time are depicted as colored lines, and global fit data are displayed as black lines. (B) SPR analysis of VHH72Fc binding to S-RBD. VHH72Fc was captured on a protein A chip, and increasing concentrations of S-RBD were injected. Sensorgrams in response units over time are depicted as colored lines, and global fit data are displayed as black lines. (C) SPR analysis of ACE2 binding to S-RBD. ACE2 was immobilized on a CM5 chip, and increasing concentrations of S-RBD were injected. Sensorgrams in response units over time are depicted as colored lines, and global fit data are displayed as black lines. (D) SPR analysis of ACE2 binding to S-RBD in the presence of c19s130Fc. ACE2 was immobilized on a CM5 chip, and 125 nM S-RBD was injected in the presence of increasing concentrations of c19s130Fc (colored lines).
FIG 4.
c19s130Fc limits SARS-CoV-2-mediated CPE on Vero cells. Vero cells were treated with the indicated concentrations of c19s130Fc, VHH72Fc, and cs130Fc and infected afterward with SARS-CoV-2 at an MOI of 0.03. (A) Bright-field images were taken on day 3 postinfection (one representative out of 4 is shown) (bar = 1 mm). (B to D) CPE scores and TOX scores of c19s130Fc (B), VHH72Fc (C), and cs130Fc (D) were determined from bright-field images from panel A by CPETOXnet and are shown in a concentration-dependent manner (n = 4). (E) CPE scores (left) and TOX scores (right) of the negative, positive, and toxic controls calculated by CPETOXnet on images 3 days after infection at an MOI of 0.03 or with staurosporine (5 μM)-treated cells.
FIG 5.
c19s130Fc inhibits SARS-CoV-2 infection of Vero cells. Vero cells were treated with the indicated concentrations of c19s130Fc, VHH72Fc, and cs130Fc; infected afterward with SARS-CoV-2 at an MOI of 0.03; and stained with SARS-CoV-2 nucleocapsid antibodies 2 days after infection. (A) Representative fluorescence images (n = 4; bar = 1 mm). (B) IF scores were determined from fluorescence images from panel A using IFnet and are shown in a concentration-dependent manner (n = 4). (C) IF signals of the negative- and positive-control images calculated by IFnet (n = 24). (D to H) An entry assay was performed by adding monensin to the cells at different time points after infection. (D) Positive control (n = 12). (E to H) Results of stopping infection after 5 min (E), 15 min (F), 45 min (G), and 135 min (H), in a concentration-dependent manner (n = 4).
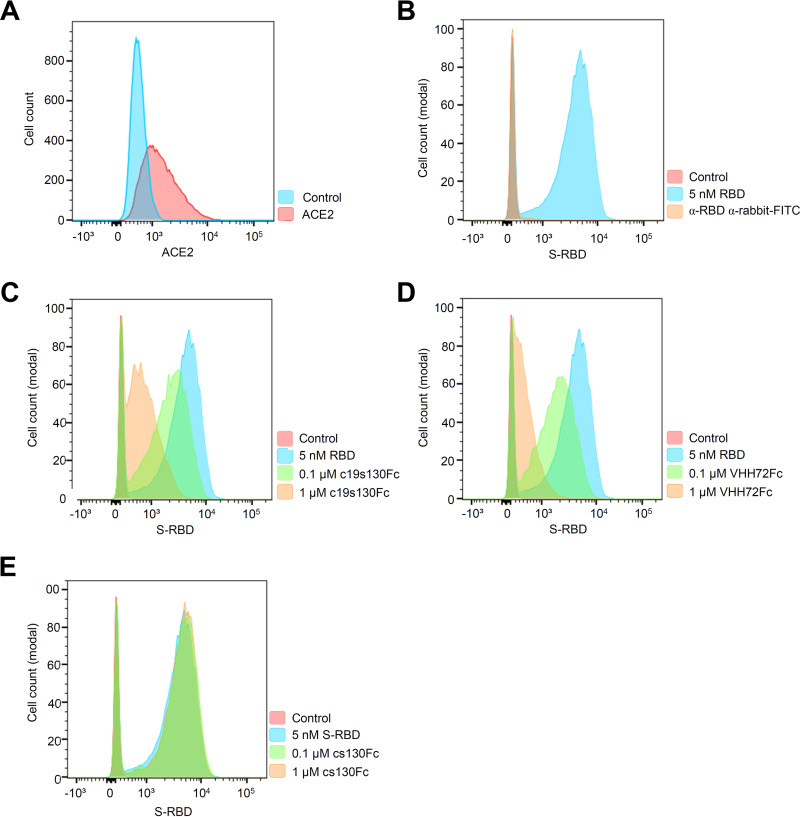
To further confirm the effect of c19s130Fc on SARS-CoV-2 cell entry, we stably expressed ACE2(1–615) on Ba/F3 cells (Ba/F3-ACE2). ACE2 was detected on the cell surface by flow cytometry (Fig. 6A). Following incubation of Ba/F3-ACE2 cells with S-RBD, surface binding of S-RBD was detected by flow cytometry (Fig. 6B). In the presence of c19s130Fc and VHH72Fc but not cs130Fc, a concentration-dependent reduction of surface-attached S-RBD was observed (Fig. 6C to E), indicating that c19s130Fc and VHH72Fc prevent the binding of the SARS-CoV-2 spike protein to ACE2.
FIG 6.
c19s130Fc prevents S-RBD binding to overexpressed ACE2 in Ba/F3 cells. (A) Flow cytometric analysis of cell surface expression of hACE2-gp130 (red population) in Ba/F3-gp130 cells detected by hACE2 antibody. The blue population indicates Ba/F3-gp130 cells incubated without hACE antibody (control). (B) Flow cytometric analysis of S-RBD binding to Ba/F3-ACE2 cells. Following incubation with S-RBD, S-RBD binding was detected (blue area) using an anti-spike S1 antibody (Sino Biological). Ba/F3-gp130-hACE2-gp130 cells without treatment served as controls (red area). (C to E) S-RBD binding in the presence of increasing concentrations of c19s130Fc, VHH72Fc, or cs130Fc. A total of 20,000 events were recorded, and the cell count was normalized. Histograms are representative of results from 3 independent experiments.
In summary, our data showed that c19s130Fc efficiently neutralized SARS-CoV-2 binding to ACE2 and blocked viral cell entry and infection.
DISCUSSION
Here, we define a new class of bispecific biomolecules based on a soluble cytokine receptor fused to two nanobodies, which together inhibit IL-6 trans-signaling and SARS-CoV-2 infection. Therapeutic bispecificity was initially described for bispecific antibodies (bsAbs), which bind two independent epitopes on the same or different antigens (52). Early bsAbs were selective T cell engagers with a binding site for a tumor-associated antigen and CD3 from T cells. Currently, more than 100 anticancer bsAbs are in clinical development (53). Apart from targeting tumor cells, bsAbs can interfere with pathogen infection and ligand/receptor activity and mediate intracellular drug delivery, cis- and trans-activation, or serum half-life extension. With our development of c19s130Fc, we expand the class of bispecific binders by the first example of a bispecific soluble cytokine receptor. c19s130Fc has a complex architecture and consists of the first three extracellular domains, D1 to D3, of gp130, which facilitates binding to IL-6:sIL-6R complexes. Since we and others have previously shown that for unknown reasons, these domains alone facilitate only low-affinity binding (47), we have fused sgp130D1–D3 to the nonneutralizing but IL-6:sIL-6R-selective nanobody VHH6 and converted sgp130D1–D3 into the high-affinity IL-6 trans-signaling inhibitor cs130 (46). Of note and in contrast to sgp130, fusion to VHH6 also prevents the inhibition of IL-11 trans-signaling (46). Bispecificity toward SARS-CoV-2 was achieved by fusion to VHH72, an antagonistic nanobody directed against the RBD of the spike protein (44). However, the antagonistic activity of VHH72 unfolded only after dimerization; therefore, VHH72 was directly placed between the IL-6 trans-signaling inhibitor and the Fc part of an IgG antibody, which served as a dimerizer and aided in the purification of c19s130Fc. In the future, bifunctional inhibition might also be achieved using nanobodies against SARS-CoV-2 that do not depend on dimerization (54), thereby further reducing the size of the bifunctional trans-signaling/SARS-CoV-2 inhibitor.
IL-6 initiates signal transduction via three different modes, termed classic, trans-, and cluster signaling (13, 22, 55, 56). Remarkably, these different signaling modes are associated with different physiological responses. While classic signaling is attributed mainly to regenerative functions and the induction of acute-phase responses, IL-6 trans-signaling is associated with chronic inflammatory processes (25), and IL-6 cluster signaling is required for the generation of pathogenic TH17 cells and thereby is also involved in detrimental inflammatory processes (55). IL-6 is a predictor of severity in COVID-19 patients (57–59), and the IL-6R antibodies sarilumab and tocilizumab, which block all types of IL-6 signaling, increase the survival rates of severe COVID-19 patients and have been approved for therapy (9–11, 31). On the other hand, sgp130 and variants thereof are potent IL-6 trans-signaling inhibitors without affecting classic signaling (35). They also inhibit trans-presentation (cluster signaling), albeit results are heterogeneous, and blocking might be context dependent (22, 55). Still, therapeutic targeting of IL-6 largely relies on the two IL-6R antibodies, while siltuximab directed against IL-6 has been approved for Castleman’s disease (60) and is currently being tested for COVID-19 patients (ClinicalTrials.gov identifier NCT04329650). The IL-6 trans-signaling inhibitor sgp130Fc (olamkicept) has recently shown remarkable results in phase II clinical studies for Crohn’s disease and ulcerative colitis (UC) (EudraCT identifier 2016-000205-36; ClinicalTrials.gov identifier NCT03235752), and phase III trials are in preparation (61).
Several lines of evidence point to a crucial role of IL-6 trans-signaling and not classic signaling in the development of hyperinflammatory states in severe COVID-19 (6–12), including multiorgan damage and respiratory failure (4). High levels of soluble IL-6R were found in COVID-19 intensive care unit patients, which was released by the increased activity of ADAM17 (26). IL-6 trans-signaling is also required for liver regeneration (62), and recent evidence indicates a key role of excessive IL-6 trans-signaling during SARS-CoV-2-induced liver damage (63–65). Mechanistically, SARS-CoV-2-induced IL-6 trans-signaling increased procoagulants like factor VIII and Von Willebrand factor (vWF), proinflammatory factors, platelet attachment to liver sinusoidal endothelial cells, and hepatocyte fibrinogen expression and induces endotheliopathy and subsequent liver damage (64). This view is further supported by the finding that sgp130Fc blocks SARS-CoV-2-induced increases of blood clotting and liver injury factors (64). Thereby, one motivation to generate the bispecific c19s130Fc was based on the notion that selective inhibition of trans-signaling would be superior to the simultaneous blocking of classic and trans-signaling by IL-6R antibodies in severe COVID-19 cases.
Mechanistically, the application of tocilizumab and sarilumab results in the binding of the antibody to sIL-6R but also to cells expressing membrane-bound IL-6R, including hepatocytes and immune cells. The life cycle of SARS-CoV-2 starts with the binding of the viral spike protein to host ACE2, cleavage of the spike protein, membrane fusion, and RNA injection. Virus-cell fusion is dependent on the cleavage of the spike protein by TMPRSS2 at the plasma membrane or by cathepsin L in endosomes (66). An imaginable bifunctional antibody for IL-6R and SARS-CoV-2 could be risky because it might bypass natural virus infection via a redirection of antibody–SARS-CoV-2 complexes to IL-6R-expressing cells. In this scenario, the spike protein in large IL-6R (target cell)–antibody–spike protein (SARS-CoV-2) complexes might still be processed by TMPRSS2 or cathepsin L to enable unintended drug-induced virus infection. Here, the virus-targeting antibody part would prevent the binding of the spike protein to ACE2 but not the binding and infection of IL-6R-expressing cells. To bypass this possibility, we opt for the trans-signaling inhibitor c19s130Fc. After binding to c19s130Fc, IL-6:sIL-6R complexes are still soluble, and unintended viral entries via membrane-bound IL-6R targeting are impossible and were not seen in our experiments using c19s130Fc during SARS-CoV-2 infection of Vero cells.
Taken together, c19s130Fc demonstrates the first example of the multifunctionalization of a trans-signaling inhibitor and thereby represents the first member of a new class of bispecific inhibitor molecules consisting of a soluble cytokine receptor fused to antiviral nanobodies.
Antiviral antibodies are effective only very early in COVID-19 disease progression (67). In later stages of severe COVID-19, patients develop a hyperinflammatory state, which is associated with multiorgan damage and respiratory failure (4). In this stage of the disease, antiviral antibodies seem to no longer be effective. Immunomodulatory treatments, including IL-6 blockade, were described to increase survival rates and decrease the need for mechanical oxygenation in later stages of the disease (9, 31, 58, 68, 69). Combined treatment using glucocorticoids and IL-6 blockade increased the observed positive effects (31). However, evidence suggests that immunomodulatory treatment, including glucocorticoid and anti-IL-6 treatments, while reducing hyperinflammation, may delay viral clearance (32, 70–72). Hence, immunomodulatory treatments may be a double-edged sword in the treatment of COVID-19 and may benefit from simultaneous antiviral treatment (73–75). In fact, there is evidence for the beneficial effects of combined antiviral and immunomodulatory treatments for SARS-CoV-2, hepatitis B, and influenza infections (76–80).
Here, we describe a bispecific molecule that incorporates antiviral and immunomodulatory entities. Such a bifunctional molecule may reduce viral infection in early-stage infections as well as reduce hyperinflammation observed in late-stage infections. In addition, bispecific inhibitors like c19s130Fc may also be useful during the transition state from early to late stages of COVID-19.
MATERIALS AND METHODS
Cloning of c19s130Fc variants.
The cDNAs encoding the single-domain antibody VHH72 (44), residues 1 to 615 of ACE2 (GenBank accession number Q9BYF1), and residues 319 to 591 of the SARS-CoV-2 RBD (GenBank accession number P0DTC2) were synthesized by Biocat (Heidelberg, Germany). VHH72 was subcloned via AgeI and NotI into the plasmid pcDNA3.1-Fc (Invitrogen) coding for an N-terminal signal peptide, a Myc tag (EQKLISEEDL), and a C-terminal human IgG1 Fc tag, thereby generating expression plasmid pcDNA3.1-VHH72Fc. c19s130Fc was cloned from pcDNA3.1-cs130Fc (46) by the insertion of a cDNA coding for VHH6-(GGGGS)3-VHH72 via XhoI and NotI. ACE2 was amplified by PCR using forward primer 5′-AGTCCTTAAGCCACCATGTCAAGCTCTTCCTGGC-3′ and reverse primer 5′-TGCGTATGCGGCCGCGTCTGCATATGGACTCCAG-3′ and subcloned into pcDNA3.1-Fc via AflII and NotI to generate pcDNA3.1-ACE2-Fc. The pcDNA3.1-RBD-TwinStrep expression vector was cloned via HindIII and NotI out of the cDNA encoding residues 319 to 591 of the SARS-CoV-2 RBD to extend the sequence with a TwinStrep tag (WSHPQFEK) connected with a (GGGS)3 linker. HIL-6 was subcloned via HindIII and NotI into the pcDNA3.1 vector containing an N-terminal signal peptide and a C-terminal TwinStrep tag, thereby generating pcDNA3.1-HIL-6-TS (twin Strep-tag) for expression. An analogous cloning strategy was used to generate the expression vector for IL-6–TS.
Cells and reagents.
The generation of Ba/F3-gp130 cells was described previously (81). For seeding and cultivation of Vero cells, cells were first washed with phosphate-buffered saline (PBS) and then incubated in the presence of a trypsin-EDTA solution (catalog number 4261.0110; Genaxxon Bioscience) until cells were detached. Cell lines were grown in Dulbecco’s modified Eagle’s medium (DMEM) high-glucose culture medium (Gibco, Life Technologies, Darmstadt, Germany) supplemented with 10% fetal bovine serum (Gibco, Life Technologies), 60 mg/L penicillin, and 100 mg/L streptomycin (Genaxxon Bioscience GmbH, Ulm, Germany) at 37°C with 5% CO2. The proliferation of Ba/F3-gp130 cells was maintained in the presence of HIL-6 (49). Ba/F3-gp130 cells were retrovirally transduced with a pMOWS expression plasmid coding for hACE2-gp130 composed of coding sequences for the ACE2 signal peptide (amino acids [aa] 1 to 17) followed by ACE2 (aa 18 to 615) and human gp130 (GenBank accession number P40189) comprising amino acids T607 to Q908, representing 13 aa of the extracellular domain (ECD), the complete transmembrane domain (TMD), and the intracellular domain (ICD) of the receptor. Selection of transduced Ba/F3-gp130 cells was performed with puromycin (1.5 μg/mL) (Carl Roth, Karlsruhe, Germany) for at least 2 weeks. Afterward, the generated Ba/F3-gp130 cell line was analyzed for receptor cell surface expression via flow cytometry. sIL-6R was obtained from Conaris Research Institute AG (Kiel, Germany). cs130Fc was produced and purified as described previously (46, 62). Expi-293F cells (Thermo Fisher Scientific) were cultured in 30 mL Expi293 expression medium without antibiotics until they reached a density of 3 × 106 to 5 × 106 cells/mL in a 37°C incubator with 8% CO2 on an orbital shaker at 125 rpm. The Expi293-F cells were cultured in a shaker flask until they reached a density of 3 × 106 to 5 × 106 cells/mL. Antibodies directed against STAT3 phosphorylated at Tyr705 (clone D3A7) and STAT3 (clone 124H6) were obtained from Cell Signaling Technology (Frankfurt, Germany). StrepMAP-Classic-HRP (horseradish peroxidase) (1:20,000) (catalog number 2-1509-001) was obtained from IBA GmbH (Göttingen, Germany). Peroxidase-conjugated secondary Abs (catalog numbers 31432, 31462, and 31423) were obtained from Pierce (Thermo Fisher Scientific, Waltham, MA, USA). Antibodies directed against phosphorylated ERK (pERK) (catalog number 4370) and ERK (catalog number 4695) were obtained from Cell Signaling Technology.
Proliferation assays.
Ba/F3-gp130 cells were washed, and 5,000 cells were cultured for 3 days in a final volume of 100 μL in the presence of cytokines and inhibitors. The CellTiter-Blue reagent was used to determine cellular viability by recording the fluorescence (excitation 560 nm and emission 590 nm) using an Infinite M200 Pro plate reader (Tecan, Crailsheim, Germany) immediately after the addition of 20 μL of the reagent per well (time point zero) and up to 120 min thereafter.
Cytokine stimulation of cells and lysate preparation.
A total of 106 Ba/F3-gp130 cells/mL were washed and starved in serum-free medium for 5 h. Vero cells were seeded at a density of 8 × 105 cells per 60-mm dish 24 h prior to stimulation and also washed five times with PBS before starvation in serum-free DMEM for at least 5 h. Prior to stimulation, cytokines and inhibitors were preincubated at room temperature for 30 min. Subsequently, cells were stimulated with the indicated cytokines and inhibitor combinations for 30 min, harvested by centrifugation at 4°C for 5 min at 500 × g, frozen, and lysed. The protein concentration of the cell lysates was determined by the bicinchoninic acid (BCA) protein assay (Pierce, Thermo Scientific). Analysis of STAT3 activation was performed by Western blotting of 25 to 75 μg of total protein from total cell lysates and subsequent detection steps using the anti-pSTAT3 (Tyr705) (1:1,000) and anti-STAT3 (1:1,000) antibodies described above.
Western blotting.
Proteins were separated by SDS-PAGE and transferred onto nitrocellulose membranes for 60 min (20 V, 1.0 Å). Membranes were blocked and probed with the indicated primary antibodies. After washing, the membranes were incubated with secondary peroxidase-conjugated antibodies or fluorescence-labeled secondary antibodies (1:2,500 dilution). Immobilon Western reagents (Millipore Corporation, Billerica, MA, USA) and the ChemoCam imager (Intas Science Imaging Instruments GmbH, Göttingen, Germany) or the Odyssey Fc imaging system (Li-Cor Biosciences, Bad Homburg, Germany) were used for signal detection. Control STAT3 blots were produced on separate membranes.
Expression and purification of VHH72Fc, c19s130Fc, the RBD, and ACE2Fc.
Mammalian expression plasmids encoding VHH72Fc, c19s130Fc, the SARS-CoV-2 RBD, and ACE2-Fc were transfected into Expi-293F cells using ExpiFectamine. After reaching 4.5 × 106 to 5.5 × 106 cells/mL, the cells were diluted to a final density of 3 × 106 cells/mL in 30 mL Expi293 expression medium for transfection. Thirty micrograms of the plasmid expression vectors was used for transfection. Thereafter, the culture was harvested by centrifugation at 450 × g at 4°C for 10 min, followed by centrifugation of the resulting supernatant at 4,000 × g at 4°C for 20 min. The supernatant from the second centrifugation step was filtered (0.45-μm filter, catalog number P667.1; Carl Roth) and purified by affinity chromatography. Constructs containing an Fc tag (c19s130Fc, VHH72Fc, and ACE2-Fc) were purified using protein A resin (1 mL) (HiTrap MabSelect PrismA) at a flow rate of 1 mL/min. The column was then washed with 30 column volumes of PBS. Proteins were eluted at pH 3.2 to 3.5 using 50 mM citric acid buffer. Fractions containing the protein peak were pooled, and the pH was adjusted to pH 7 with 1 M Tris. Constructs containing a C-terminal TwinStrep tag (SARS-CoV-2 RBD) were purified using Strep-Tactin resin (catalog number 2-5025-001; IBA) according to the manufacturer’s instructions. Proteins were buffer exchanged to PBS using illustra NAP25 columns (GE Healthcare Life Sciences, Munich, Germany). The protein concentration was determined by measuring the absorbance at 280 nm, and samples were flash-frozen in liquid nitrogen. A total of 2.5 μg of protein was loaded per lane and separated by SDS-PAGE under reducing (106 mM β-mercaptoethanol at 95°C for 10 min) and nonreducing (without β-mercaptoethanol and heating) conditions. The gel was stained with Coomassie staining solution (80% ethanol, 20% acetic acid, 4% Coomassie brilliant blue R250) for 1 h and destained overnight in a destaining solution (20% ethanol, 10% acetic acid).
Surface plasmon resonance.
For surface plasmon resonance experiments, the Biacore X100 instrument (GE Healthcare Life Sciences) was used. VHH72Fc or c19s130Fc was captured on a single flow cell of a protein A sensorchip at a level of ∼300 or 650 response units (RU), respectively, per cycle. Three samples containing only running buffer were injected over both the ligand and reference flow cells, followed by S-RBD serially diluted from 500 to 3.9 nM, with a replicate of the 125 nM concentration. The analyte S-RBD was injected at a flow rate of 30 μL/min for 120 s, and dissociation was measured for 300 s. ACE2 was immobilized in 10 mM acetate buffer (pH 4.5) by amine coupling on a CM5 chip (2,500 RU). After immobilization, S-RBD was injected at a flow rate of 30 μL/min at increasing concentrations (2 to 250 nM). Association was monitored in periods of 60 s, and dissociation was measured for 600 s. Immobilized ACE2 was regenerated with 2 M NaCl to remove bound S-RBD in multiple cycle measurements. ACE2 was immobilized on a CM5 chip, and 125 nM S-RBD was injected in the presence of increasing concentrations of c19s130Fc. Experiments were carried out at 25°C in PBS (pH 7.4), composed of 137 mM NaCl, 2.7 mM KCl, 12 mM HPO42− and H2PO4−, and 0.05% (vol/vol) surfactant P20 (GE Healthcare). The resulting data were reference subtracted and fit to a 1:1 binding model using Biacore X100 Evaluation software V2.0.1.
Viruses.
SARS-CoV-2 was used as described previously (51) (sequence accession number EPI_ISL_425126; https://www.gisaid.org/). A SARS-CoV-2 stock was obtained in Vero cells by infection at a multiplicity of infection (MOI) of 0.001. After 72 h, the supernatant was collected and stored at −80°C until use.
SARS-CoV-2 infection of Vero cells.
Vero cells were cultured as previously described (51). Vero cells were cultured in DMEM with the addition of 10% fetal calf serum (FCS), minimal essential amino acids, and penicillin/streptomycin at 37°C with 5% CO2. A total of 3 × 104 cells were seeded per well in a 96-well plate 1 day before infection. On the next day, the medium was changed to cell culture medium containing different dilutions of c19s130Fc, VHH72Fc, and cs130Fc dissolved in PBS and staurosporine (5 μM) as a toxic control. Moreover, 6 or 12 serial 2-fold dilutions were used. The cells were infected with SARS-CoV-2 20 min later at an MOI of 0.03. For the entry assay, monensin was added to the wells 5, 15, 45, and 135 min after infection to stop the further entry of SARS-CoV-2 into the cells. An overlay composed of DMEM with 1% methylcellulose was added at 2 h postinfection. The IC50 was measured using GraphPad Prism.
Immunofluorescence.
Two days after infection, the supernatant was discarded, and 4% formalin was added for 30 min. Hanks’ buffer containing Triton X-100 was applied to the cells for 20 min followed by 10% FCS in PBS for 1 h to block unspecific binding sites. The cells were stained with a SARS-CoV-2 nucleocapsid antibody (2019 novel coronavirus [nCoV]) (Sino Biology Inc., Eschborn, Germany) for 1 h. Following washing, fluorescein isothiocyanate (FITC)-conjugated AffiniPure goat anti-rabbit IgG(H+L) (Jackson Immuno Research, Cambridgeshire, UK) was added for 1 h. The cells were washed again and analyzed with a Nikon Eclipse TS100 fluorescence microscope. Pictures were taken with NIS-Elements F4.30.01 software. The images were quantified using deep transfer learning as previously described (51). ResNet18 was retrained to classify SARS-CoV-2-infected cell cultures. For recognizing CPE and toxic effects (TOX), we used CPETOXnet with three different classifications (CPE, TOX, and no CPE). For the quantification of SARS-CoV-2 immunofluorescence pictures, we used IFnet with two different classifications (IF signal and no signal) (51).
Cell surface detection via flow cytometry.
hACE2-gp130 cell surface expression of stably transfected Ba/F3-gp130 cells was detected by specific antibodies. A total of 5 × 105 cells were washed in fluorescence-activated cell sorter (FACS) buffer (PBS, 1% bovine serum albumin [BSA]) and then incubated in 50 μL containing the indicated primary antibody (anti-hACE2, catalog number AF933; Bio-Techne) (1:80) for 1 h at room temperature. Cells were washed and resuspended in 50 μL containing the secondary antibody (Northern Light 495-conjugated Fab anti-goat IgG, catalog number NL003; Bio-Techne) (1:100) and incubated for 1 h at room temperature. Cells were washed and resuspended in 500 μL of FACS buffer. A total of 20,000 cells were recorded and analyzed by flow cytometry (BD FACSCanto II flow cytometer using FACSDiva software; BD Biosciences). Data analysis was conducted using FlowJo version 10 (Tree Star Inc., USA).
Flow cytometry inhibition experiments were conducted with 5 × 105 Ba/F3-gp130-hACE2-gp130 cells in the presence of 5 nM S-RBD, inhibitory proteins, and the primary antibody (antispike, catalog number 40150-R007; Sino Biological) (1:50) for 1 h at room temperature. Cells were washed and resuspended in 50 μL containing a secondary antibody directed against antispike-IgG (anti-rabbit–Alexa Fluor 488; Cell Signaling Technology) (1:250) for 1 h at room temperature. Afterward, cells were treated as described above.
Molecular modeling.
For molecular modeling, the structure of the IL-6 signaling complex (PDB accession number 1P9M) was superpositioned with the structure of VHH6 bound to a complex of IL-6:sIL-6R (PDB accession number 5FUC). In addition, the structure of VHH72 bound to S-RBD (PDB accession number 6WAQ) was superpositioned onto the structure of trimeric S-RBD in an open conformation (PDB accession number 7CAC). Components of the c19s130Fc protein are depicted in a ribbon representation, and the IL-6 signal complex and S-RBD are depicted in a surface representation using ChimeraX (82).
Statistical analyses.
IC50 values were calculated by nonlinear regression analysis in GraphPad Prism 6.1 (version 6.1 for Windows; GraphPad Software, La Jolla, CA, USA) from 3 individual experiments. The data are presented as means ± standard deviations (SD).
Data availability.
All data needed to evaluate the conclusions in the paper are present in the paper.
ACKNOWLEDGMENTS
This study was supported by the DFG (SFB974 and RTG1949), the Jürgen Manchot Graduate School (MOI), and the Stiftung für Altersforschung, Düsseldorf.
J.E. and J.W. conducted most of the experiments. J.E. and J.M.M. performed the SPR experiments. H.T.W. supported protein purification and SPR experiments. J.W. and P.A.L. planned and performed virus infection experiments, E.M. and R.S. supported viral entry assays. All authors helped write the paper. J.S., J.M.M., and P.A.L. designed the study, analyzed the data, and wrote the paper.
J.E., J.W., P.A.L., J.S., and J.M.M. have applied for a patent covering c19s130Fc (EP 21189174.2). J.M.M. acts as a consultant for Ferring Pharmaceuticals.
Contributor Information
Jürgen Scheller, Email: jscheller@uni-duesseldorf.de.
Jens M. Moll, Email: jens.moll@uni-duesseldorf.de.
Tom Gallagher, Loyola University—Chicago.
REFERENCES
- 1.Li H, Liu L, Zhang D, Xu J, Dai H, Tang N, Su X, Cao B. 2020. SARS-CoV-2 and viral sepsis: observations and hypotheses. Lancet 395:1517–1520. 10.1016/S0140-6736(20)30920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wunsch H. 2020. Mechanical ventilation in COVID-19: interpreting the current epidemiology. Am J Respir Crit Care Med 202:1–4. 10.1164/rccm.202004-1385ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levin AT, Hanage WP, Owusu-Boaitey N, Cochran KB, Walsh SP, Meyerowitz-Katz G. 2020. Assessing the age specificity of infection fatality rates for COVID-19: systematic review, meta-analysis, and public policy implications. Eur J Epidemiol 35:1123–1138. 10.1007/s10654-020-00698-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryce C, Grimes Z, Pujadas E, Ahuja S, Beasley MB, Albrecht R, Hernandez T, Stock A, Zhao Z, AlRasheed MR, Chen J, Li L, Wang D, Corben A, Haines GK, III, Westra WH, Umphlett M, Gordon RE, Reidy J, Petersen B, Salem F, Fiel MI, El Jamal SM, Tsankova NM, Houldsworth J, Mussa Z, Veremis B, Sordillo E, Gitman MR, Nowak M, Brody R, Harpaz N, Merad M, Gnjatic S, Liu W-C, Schotsaert M, Miorin L, Aydillo Gomez TA, Ramos-Lopez I, Garcia-Sastre A, Donnelly R, Seigler P, Keys C, Cameron J, Moultrie I, Washington K-L, Treatman J, Sebra R, Jhang J, Firpo A, et al. 2021. Pathophysiology of SARS-CoV-2: the Mount Sinai COVID-19 autopsy experience. Mod Pathol 34:1456–1467. 10.1038/s41379-021-00793-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.England JT, Abdulla A, Biggs CM, Lee AYY, Hay KA, Hoiland RL, Wellington CL, Sekhon M, Jamal S, Shojania K, Chen LYC. 2021. Weathering the COVID-19 storm: lessons from hematologic cytokine syndromes. Blood Rev 45:100707. 10.1016/j.blre.2020.100707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Z-S, Shu T, Kang L, Wu D, Zhou X, Liao B-W, Sun X-L, Zhou X, Wang Y-Y. 2020. Temporal profiling of plasma cytokines, chemokines and growth factors from mild, severe and fatal COVID-19 patients. Signal Transduct Target Ther 5:100. 10.1038/s41392-020-0211-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, Péré H, Charbit B, Bondet V, Chenevier-Gobeaux C, Breillat P, Carlier N, Gauzit R, Morbieu C, Pène F, Marin N, Roche N, Szwebel T-A, Merkling SH, Treluyer J-M, Veyer D, Mouthon L, Blanc C, Tharaux P-L, Rozenberg F, Fischer A, Duffy D, Rieux-Laucat F, Kernéis S, Terrier B. 2020. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 369:718–724. 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506. 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guaraldi G, Meschiari M, Cozzi-Lepri A, Milic J, Tonelli R, Menozzi M, Franceschini E, Cuomo G, Orlando G, Borghi V, Santoro A, Di Gaetano M, Puzzolante C, Carli F, Bedini A, Corradi L, Fantini R, Castaniere I, Tabbì L, Girardis M, Tedeschi S, Giannella M, Bartoletti M, Pascale R, Dolci G, Brugioni L, Pietrangelo A, Cossarizza A, Pea F, Clini E, Salvarani C, Massari M, Viale PL, Mussini C. 2020. Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lancet Rheumatol 2:e474–e484. 10.1016/S2665-9913(20)30173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jordan SC, Zakowski P, Tran HP, Smith EA, Gaultier C, Marks G, Zabner R, Lowenstein H, Oft J, Bluen B, Le C, Shane R, Ammerman N, Vo A, Chen P, Kumar S, Toyoda M, Ge S, Huang E. 2020. Compassionate use of tocilizumab for treatment of SARS-CoV-2 pneumonia. Clin Infect Dis 71:3168–3173. 10.1093/cid/ciaa812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meleveedu KS, Miskovsky J, Meharg J, Abdelrahman A, Tandon R, Moody AE, Dasilva P, Masse G, LaPorte J, Saied Calvino A, Allen G, El-Bizri R, Roberts T, Armenio V, Katz SC. 2020. Tocilizumab for severe COVID-19 related illness—a community academic medical center experience. Cytokine X 2:100035. 10.1016/j.cytox.2020.100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen LYC, Biggs CM, Jamal S, Stukas S, Wellington CL, Sekhon MS. 2021. Soluble interleukin-6 receptor in the COVID-19 cytokine storm syndrome. Cell Rep Med 2:100269. 10.1016/j.xcrm.2021.100269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scheller J, Garbers C, Rose-John S. 2014. Interleukin-6: from basic biology to selective blockade of pro-inflammatory activities. Semin Immunol 26:2–12. 10.1016/j.smim.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Matthews V, Schuster B, Schütze S, Bussmeyer I, Ludwig A, Hundhausen C, Sadowski T, Saftig P, Hartmann D, Kallen K-J, Rose-John S. 2003. Cellular cholesterol depletion triggers shedding of the human interleukin-6 receptor by ADAM10 and ADAM17 (TACE). J Biol Chem 278:38829–38839. 10.1074/jbc.M210584200. [DOI] [PubMed] [Google Scholar]
- 15.Althoff K, Reddy P, Voltz N, Rose-John S, Mullberg J. 2000. Shedding of interleukin-6 receptor and tumor necrosis factor alpha. Contribution of the stalk sequence to the cleavage pattern of transmembrane proteins. Eur J Biochem 267:2624–2631. 10.1046/j.1432-1327.2000.01278.x. [DOI] [PubMed] [Google Scholar]
- 16.Müllberg J, Schooltink H, Stoyan T, Günther M, Graeve L, Buse G, Mackiewicz A, Heinrich PC, Rose-John S. 1993. The soluble interleukin-6 receptor is generated by shedding. Eur J Immunol 23:473–480. 10.1002/eji.1830230226. [DOI] [PubMed] [Google Scholar]
- 17.Jones SA. 2005. Directing transition from innate to acquired immunity: defining a role for IL-6. J Immunol 175:3463–3468. 10.4049/jimmunol.175.6.3463. [DOI] [PubMed] [Google Scholar]
- 18.Gaillard J, Pugniere M, Tresca J, Mani J, Klein B, Brochier J. 1999. Interleukin-6 receptor signaling. II. Bio-availability of interleukin-6 in serum. Eur Cytokine Netw 10:337–344. [PubMed] [Google Scholar]
- 19.Mitsuyama K, Toyonaga A, Sasaki E, Ishida O, Ikeda H, Tsuruta O, Harada K, Tateishi H, Nishiyama T, Tanikawa K. 1995. Soluble interleukin-6 receptors in inflammatory bowel disease: relation to circulating interleukin-6. Gut 36:45–49. 10.1136/gut.36.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montero-Julian FA. 2001. The soluble IL-6 receptors: serum levels and biological function. Cell Mol Biol (Noisy-le-grand) 47:583–597. [PubMed] [Google Scholar]
- 21.Akira S, Taga T, Kishimoto T. 1993. Interleukin-6 in biology and medicine. Adv Immunol 54:1–78. 10.1016/s0065-2776(08)60532-5. [DOI] [PubMed] [Google Scholar]
- 22.Baran P, Hansen S, Waetzig GH, Akbarzadeh M, Lamertz L, Huber HJ, Ahmadian MR, Moll JM, Scheller J. 2018. The balance of interleukin (IL)-6, IL-6.soluble IL-6 receptor (sIL-6R), and IL-6.sIL-6R.sgp130 complexes allows simultaneous classic and trans-signaling. J Biol Chem 293:6762–6775. 10.1074/jbc.RA117.001163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schumacher N, Rose-John S. 2019. ADAM17 activity and IL-6 trans-signaling in inflammation and cancer. Cancers (Basel) 11:1736. 10.3390/cancers11111736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka T, Narazaki M, Kishimoto T. 2016. Immunotherapeutic implications of IL-6 blockade for cytokine storm. Immunotherapy 8:959–970. 10.2217/imt-2016-0020. [DOI] [PubMed] [Google Scholar]
- 25.Rose-John S. 2012. IL-6 trans-signaling via the soluble IL-6 receptor: importance for the pro-inflammatory activities of IL-6. Int J Biol Sci 8:1237–1247. 10.7150/ijbs.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patra T, Meyer K, Geerling L, Isbell TS, Hoft DF, Brien J, Pinto AK, Ray RB, Ray R. 2020. SARS-CoV-2 spike protein promotes IL-6 trans-signaling by activation of angiotensin II receptor signaling in epithelial cells. PLoS Pathog 16:e1009128. 10.1371/journal.ppat.1009128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishimoto N, Kishimoto T. 2008. Humanized antihuman IL-6 receptor antibody, tocilizumab. Handb Exp Pharmacol 2008:151–160. 10.1007/978-3-540-73259-4_7. [DOI] [PubMed] [Google Scholar]
- 28.Boyce EG, Rogan EL, Vyas D, Prasad N, Mai Y. 2018. Sarilumab: review of a second IL-6 receptor antagonist indicated for the treatment of rheumatoid arthritis. Ann Pharmacother 52:780–791. 10.1177/1060028018761599. [DOI] [PubMed] [Google Scholar]
- 29.Rossi J-F, Négrier S, James ND, Kocak I, Hawkins R, Davis H, Prabhakar U, Qin X, Mulders P, Berns B. 2010. A phase I/II study of siltuximab (CNTO 328), an anti-interleukin-6 monoclonal antibody, in metastatic renal cell cancer. Br J Cancer 103:1154–1162. 10.1038/sj.bjc.6605872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang S, Tanaka T, Narazaki M, Kishimoto T. 2019. Targeting interleukin-6 signaling in clinic. Immunity 50:1007–1023. 10.1016/j.immuni.2019.03.026. [DOI] [PubMed] [Google Scholar]
- 31.Gordon AC, Angus DC, Derde LPG. 2021. Interleukin-6 receptor antagonists in critically ill patients with Covid-19. N Engl J Med 385:1147–1149. 10.1056/NEJMc2108482. [DOI] [PubMed] [Google Scholar]
- 32.Kopf M, Baumann H, Freer G, Freudenberg M, Lamers M, Kishimoto T, Zinkernagel R, Bluethmann H, Köhler G. 1994. Impaired immune and acute-phase responses in interleukin-6-deficient mice. Nature 368:339–342. 10.1038/368339a0. [DOI] [PubMed] [Google Scholar]
- 33.Geng Z, Yu Y, Hu S, Dong L, Ye C. 2019. Tocilizumab and the risk of respiratory adverse events in patients with rheumatoid arthritis: a systematic review and meta-analysis of randomised controlled trials. Clin Exp Rheumatol 37:318–323. [PubMed] [Google Scholar]
- 34.Pawar A, Desai RJ, Solomon DH, Santiago Ortiz AJ, Gale S, Bao M, Sarsour K, Schneeweiss S, Kim SC. 2019. Risk of serious infections in tocilizumab versus other biologic drugs in patients with rheumatoid arthritis: a multidatabase cohort study. Ann Rheum Dis 78:456–464. 10.1136/annrheumdis-2018-214367. [DOI] [PubMed] [Google Scholar]
- 35.Jostock T, Müllberg J, Ozbek S, Atreya R, Blinn G, Voltz N, Fischer M, Neurath MF, Rose-John S. 2001. Soluble gp130 is the natural inhibitor of soluble interleukin-6 receptor transsignaling responses. Eur J Biochem 268:160–167. 10.1046/j.1432-1327.2001.01867.x. [DOI] [PubMed] [Google Scholar]
- 36.Barkhausen T, Tschernig T, Rosenstiel P, van Griensven M, Vonberg R-P, Dorsch M, Mueller-Heine A, Chalaris A, Scheller J, Rose-John S, Seegert D, Krettek C, Waetzig GH. 2011. Selective blockade of interleukin-6 trans-signaling improves survival in a murine polymicrobial sepsis model. Crit Care Med 39:1407–1413. 10.1097/CCM.0b013e318211ff56. [DOI] [PubMed] [Google Scholar]
- 37.Hoge J, Yan I, Jänner N, Schumacher V, Chalaris A, Steinmetz OM, Engel DR, Scheller J, Rose-John S, Mittrücker H-W. 2013. IL-6 controls the innate immune response against Listeria monocytogenes via classical IL-6 signaling. J Immunol 190:703–711. 10.4049/jimmunol.1201044. [DOI] [PubMed] [Google Scholar]
- 38.Sodenkamp J, Waetzig GH, Scheller J, Seegert D, Grötzinger J, Rose-John S, Ehlers S, Hölscher C. 2012. Therapeutic targeting of interleukin-6 trans-signaling does not affect the outcome of experimental tuberculosis. Immunobiology 217:996–1004. 10.1016/j.imbio.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 39.Li F, Li W, Farzan M, Harrison SC. 2005. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309:1864–1868. 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 40.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. 2020. Structural basis of receptor recognition by SARS-CoV-2. Nature 581:221–224. 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kyriakidis NC, Lopez-Cortes A, Gonzalez EV, Grimaldos AB, Prado EO. 2021. SARS-CoV-2 vaccines strategies: a comprehensive review of phase 3 candidates. NPJ Vaccines 6:28. 10.1038/s41541-021-00292-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhattacharjee A, Saha M, Halder A, Debnath A, Mukherjee O. 2021. Therapeutics and vaccines: strengthening our fight against the global pandemic COVID-19. Curr Microbiol 78:435–448. 10.1007/s00284-020-02310-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deb P, Molla MMA, Saif-Ur-Rahman KM. 2021. An update to monoclonal antibody as therapeutic option against COVID-19. Biosaf Health 3:87–91. 10.1016/j.bsheal.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wrapp D, De Vlieger D, Corbett KS, Torres GM, Wang N, Van Breedam W, Roose K, van Schie L, VIB-CMB COVID-19 Response Team, Hoffmann M, Pöhlmann S, Graham BS, Callewaert N, Schepens B, Saelens X, McLellan JS. 2020. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell 181:1004–1015.e15. 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu Q, Zhang Z, Li H, Zhong K, Zhao Q, Wang Z, Wu Z, Yang D, Sun S, Yang N, Zheng M, Chen Q, Long C, Guo W, Yang H, Nie C, Tong A. 2021. Development of multivalent nanobodies blocking SARS-CoV-2 infection by targeting RBD of spike protein. J Nanobiotechnology 19:33. 10.1186/s12951-021-00768-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heise D, Derrac Soria A, Hansen S, Dambietz C, Akbarzadeh M, Berg AF, Waetzig GH, Jones SA, Dvorsky R, Ahmadian MR, Scheller J, Moll JM. 2021. Selective inhibition of IL-6 trans-signaling by a miniaturized, optimized chimeric soluble gp130 inhibits TH17 cell expansion. Sci Signal 14:eabc3480. 10.1126/scisignal.abc3480. [DOI] [PubMed] [Google Scholar]
- 47.Sommer J, Garbers C, Wolf J, Trad A, Moll JM, Sack M, Fischer R, Grötzinger J, Waetzig GH, Floss DM, Scheller J. 2014. Alternative intronic polyadenylation generates the interleukin-6 trans-signaling inhibitor sgp130-E10. J Biol Chem 289:22140–22150. 10.1074/jbc.M114.560938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams R, Burnley RJ, Valenzano CR, Qureshi O, Doyle C, Lumb S, Del Carmen Lopez M, Griffin R, McMillan D, Taylor RD, Meier C, Mori P, Griffin LM, Wernery U, Kinne J, Rapecki S, Baker TS, Lawson ADG, Wright M, Ettorre A. 2017. Discovery of a junctional epitope antibody that stabilizes IL-6 and gp80 protein:protein interaction and modulates its downstream signaling. Sci Rep 7:37716. 10.1038/srep37716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fischer M, Goldschmitt J, Peschel C, Brakenhoff JP, Kallen KJ, Wollmer A, Grötzinger J, Rose-John S. 1997. I. A bioactive designer cytokine for human hematopoietic progenitor cell expansion. Nat Biotechnol 15:142–145. 10.1038/nbt0297-142. [DOI] [PubMed] [Google Scholar]
- 50.Rosa RB, Dantas WM, do Nascimento JCF, da Silva MV, de Oliveira RN, Pena LJ. 2021. In vitro and in vivo models for studying SARS-CoV-2, the etiological agent responsible for COVID-19 pandemic. Viruses 13:379. 10.3390/v13030379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Werner J, Kronberg RM, Stachura P, Ostermann PN, Müller L, Schaal H, Bhatia S, Kather JN, Borkhardt A, Pandyra AA, Lang KS, Lang PA. 2021. Deep transfer learning approach for automatic recognition of drug toxicity and inhibition of SARS-CoV-2. Viruses 13:610. 10.3390/v13040610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brinkmann U, Kontermann R. 2021. Bispecific antibodies. Science 372:916–917. 10.1126/science.abg1209. [DOI] [PubMed] [Google Scholar]
- 53.Labrijn A, Janmaat M, Reichert J, Parren P. 2019. Bispecific antibodies: a mechanistic review of the pipeline. Nat Rev Drug Discov 18:585–608. 10.1038/s41573-019-0028-1. [DOI] [PubMed] [Google Scholar]
- 54.Güttler T, Aksu M, Dickmanns A, Stegmann KM, Gregor K, Rees R, Taxer W, Rymarenko O, Schünemann J, Dienemann C, Gunkel P, Mussil B, Krull J, Teichmann U, Groß U, Cordes VC, Dobbelstein M, Görlich D. 2021. Neutralization of SARS-CoV-2 by highly potent, hyperthermostable, and mutation-tolerant nanobodies. EMBO J 40:e107985. 10.15252/embj.2021107985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heink S, Yogev N, Garbers C, Herwerth M, Aly L, Gasperi C, Husterer V, Croxford AL, Möller-Hackbarth K, Bartsch HS, Sotlar K, Krebs S, Regen T, Blum H, Hemmer B, Misgeld T, Wunderlich TF, Hidalgo J, Oukka M, Rose-John S, Schmidt-Supprian M, Waisman A, Korn T. 2017. trans-presentation of IL-6 by dendritic cells is required for the priming of pathogenic TH17 cells. Nat Immunol 18:74–85. 10.1038/ni.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lamertz L, Rummel F, Polz R, Baran P, Hansen S, Waetzig GH, Moll JM, Floss DM, Scheller J. 2018. Soluble gp130 prevents interleukin-6 and interleukin-11 cluster signaling but not intracellular autocrine responses. Sci Signal 11:eaar7388. 10.1126/scisignal.aar7388. [DOI] [PubMed] [Google Scholar]
- 57.Dhar SK, K V, Damodar S, Gujar S, Das M. 2021. IL-6 and IL-10 as predictors of disease severity in COVID-19 patients: results from meta-analysis and regression. Heliyon 7:e06155. 10.1016/j.heliyon.2021.e06155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Galván-Román JM, Rodríguez-García SC, Roy-Vallejo E, Marcos-Jiménez A, Sánchez-Alonso S, Fernández-Díaz C, Alcaraz-Serna A, Mateu-Albero T, Rodríguez-Cortes P, Sánchez-Cerrillo I, Esparcia L, Martínez-Fleta P, López-Sanz C, Gabrie L, del Campo Guerola L, Suárez-Fernández C, Ancochea J, Canabal A, Albert P, Rodríguez-Serrano DA, Aguilar JM, del Arco C, de Los Santos I, García-Fraile L, de la Cámara R, Serra JM, Ramírez E, Alonso T, Landete P, Soriano JB, Martín-Gayo E, Fraile Torres A, Zurita Cruz ND, García-Vicuña R, Cardeñoso L, Sánchez-Madrid F, Alfranca A, Muñoz-Calleja C, González-Álvaro I, REINMUN-COVID Group. 2021. IL-6 serum levels predict severity and response to tocilizumab in COVID-19: an observational study. J Allergy Clin Immunol 147:72–80.e8. 10.1016/j.jaci.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sabaka P, Koščálová A, Straka I, Hodosy J, Lipták R, Kmotorková B, Kachlíková M, Kušnírová A. 2021. Role of interleukin 6 as a predictive factor for a severe course of Covid-19: retrospective data analysis of patients from a long-term care facility during Covid-19 outbreak. BMC Infect Dis 21:308. 10.1186/s12879-021-05945-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choy E, De Benedetti F, Takeuchi T, Hashizume M, John M, Kishimoto T. 2020. Translating IL-6 biology into effective treatments. Nat Rev Rheumatol 16:335–345. 10.1038/s41584-020-0419-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schreiber S, Aden K, Bernardes JP, Conrad C, Tran F, Höper H, Volk V, Mishra N, Blase JI, Nikolaus S, Bethge J, Kühbacher T, Röcken C, Chen M, Cottingham I, Petri N, Rasmussen BB, Lokau J, Lenk L, Garbers C, Feuerhake F, Rose-John S, Waetzig GH, Rosenstiel P. 2021. Therapeutic interleukin 6 trans-signaling inhibition by olamkicept (sgp130Fc) in patients with active inflammatory bowel disease. Gastroenterology 160:2354–2366. 10.1053/j.gastro.2021.02.062. [DOI] [PubMed] [Google Scholar]
- 62.Fazel Modares N, Polz R, Haghighi F, Lamertz L, Behnke K, Zhuang Y, Kordes C, Häussinger D, Sorg UR, Pfeffer K, Floss DM, Moll JM, Piekorz RP, Ahmadian MR, Lang PA, Scheller J. 2019. IL-6 trans-signaling controls liver regeneration after partial hepatectomy. Hepatology 70:2075–2091. 10.1002/hep.30774. [DOI] [PubMed] [Google Scholar]
- 63.Effenberger M, Grander C, Grabherr F, Griesmacher A, Ploner T, Hartig F, Bellmann-Weiler R, Joannidis M, Zoller H, Weiss G, Adolph TE, Tilg H. 2021. Systemic inflammation as fuel for acute liver injury in COVID-19. Dig Liver Dis 53:158–165. 10.1016/j.dld.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McConnell MJ, Kawaguchi N, Kondo R, Sonzogni A, Licini L, Valle C, Bonaffini PA, Sironi S, Alessio MG, Previtali G, Seghezzi M, Zhang X, Lee AI, Pine AB, Chun HJ, Zhang X, Fernandez-Hernando C, Qing H, Wang A, Price C, Sun Z, Utsumi T, Hwa J, Strazzabosco M, Iwakiri Y. 2021. Liver injury in COVID-19 and IL-6 trans-signaling-induced endotheliopathy. J Hepatol 75:647–658. 10.1016/j.jhep.2021.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kang S, Tanaka T, Inoue H, Ono C, Hashimoto S, Kioi Y, Matsumoto H, Matsuura H, Matsubara T, Shimizu K, Ogura H, Matsuura Y, Kishimoto T. 2020. IL-6 trans-signaling induces plasminogen activator inhibitor-1 from vascular endothelial cells in cytokine release syndrome. Proc Natl Acad Sci USA 117:22351–22356. 10.1073/pnas.2010229117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scudellari M. 2021. How the coronavirus infects cells—and why Delta is so dangerous. Nature 595:640–644. 10.1038/d41586-021-02039-y. [DOI] [PubMed] [Google Scholar]
- 67.Siemieniuk RA, Bartoszko JJ, Díaz Martinez JP, Kum E, Qasim A, Zeraatkar D, Izcovich A, Mangala S, Ge L, Han MA, Agoritsas T, Arnold D, Ávila C, Chu DK, Couban R, Cusano E, Darzi AJ, Devji T, Foroutan F, Ghadimi M, Khamis A, Lamontagne F, Loeb M, Miroshnychenko A, Motaghi S, Murthy S, Mustafa RA, Rada G, Rochwerg B, Switzer C, Vandvik PO, Vernooij RW, Wang Y, Yao L, Guyatt GH, Brignardello-Petersen R. 2021. Antibody and cellular therapies for treatment of covid-19: a living systematic review and network meta-analysis. BMJ 374:n2231. 10.1136/bmj.n2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gupta S, Wang W, Hayek SS, Chan L, Mathews KS, Melamed ML, Brenner SK, Leonberg-Yoo A, Schenck EJ, Radbel J, Reiser J, Bansal A, Srivastava A, Zhou Y, Finkel D, Green A, Mallappallil M, Faugno AJ, Zhang J, Velez JCQ, Shaefi S, Parikh CR, Charytan DM, Athavale AM, Friedman AN, Redfern RE, Short SAP, Correa S, Pokharel KK, Admon AJ, Donnelly JP, Gershengorn HB, Douin DJ, Semler MW, Hernán MA, Leaf DE, STOP-COVID Investigators. 2021. Association between early treatment with tocilizumab and mortality among critically ill patients with COVID-19. JAMA Intern Med 181:41–51. 10.1001/jamainternmed.2020.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hashimoto S, Yoshizaki K, Uno K, Kitajima H, Arai T, Tamura Y, Morishita H, Matsuoka H, Han Y, Minamoto S, Hirashima T, Yamada T, Kashiwa Y, Kameda M, Yamaguchi S, Tsuchihashi Y, Iwahashi M, Nakayama E, Shioda T, Nagai T, Tanaka T. 2021. Prompt reduction in CRP, IL-6, IFN-gamma, IP-10, and MCP-1 and a relatively low basal ratio of ferritin/CRP is possibly associated with the efficacy of tocilizumab monotherapy in severely to critically ill patients with COVID-19. Front Med (Lausanne) 8:734838. 10.3389/fmed.2021.734838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li J, Liao X, Zhou Y, Wang L, Yang H, Zhang W, Zhang Z, Kang Y. 2021. Association between glucocorticoids treatment and viral clearance delay in patients with COVID-19: a systematic review and meta-analysis. BMC Infect Dis 21:1063. 10.1186/s12879-021-06548-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Russell CD, Millar JE, Baillie JK. 2020. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet 395:473–475. 10.1016/S0140-6736(20)30317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Velazquez-Salinas L, Verdugo-Rodriguez A, Rodriguez LL, Borca MV. 2019. The role of interleukin 6 during viral infections. Front Microbiol 10:1057. 10.3389/fmicb.2019.01057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Akinosoglou K, Velissaris D, Ziazias D, Davoulos C, Tousis A, Tsiotsios K, Kalogeropoulou C, Spyridonidis A, Marangos M, Fligkou F, Gogos C. 2021. Remdesivir and tocilizumab: mix or match. J Med Virol 93:56–58. 10.1002/jmv.26117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feuillet V, Canard B, Trautmann A. 2021. Combining antivirals and immunomodulators to fight COVID-19. Trends Immunol 42:31–44. 10.1016/j.it.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu Q, Zhou YH, Yang ZQ. 2016. The cytokine storm of severe influenza and development of immunomodulatory therapy. Cell Mol Immunol 13:3–10. 10.1038/cmi.2015.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bockmann JH, Dandri M, Luth S, Pannicke N, Lohse AW. 2015. Combined glucocorticoid and antiviral therapy of hepatitis B virus-related liver failure. World J Gastroenterol 21:2214–2219. 10.3748/wjg.v21.i7.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ye Z-W, Yuan S, Chan JF-W, Zhang AJ, Yu C-Y, Ong CP, Yang D, Chan CC-Y, Tang K, Cao J, Poon VK-M, Chan CC-S, Cai J-P, Chu H, Yuen K-Y, Jin D-Y. 2021. Beneficial effect of combinational methylprednisolone and remdesivir in hamster model of SARS-CoV-2 infection. Emerg Microbes Infect 10:291–304. 10.1080/22221751.2021.1885998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Korolowicz KE, Suresh M, Li B, Huang X, Yon C, Leng X, Kallakury BV, Tucker RD, Menne S. 2021. Treatment with the immunomodulator AIC649 in combination with entecavir produces antiviral efficacy in the woodchuck model of chronic hepatitis B. Viruses 13:648. 10.3390/v13040648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zheng B-J, Chan K-W, Lin Y-P, Zhao G-Y, Chan C, Zhang H-J, Chen H-L, Wong SSY, Lau SKP, Woo PCY, Chan K-H, Jin D-Y, Yuen K-Y. 2008. Delayed antiviral plus immunomodulator treatment still reduces mortality in mice infected by high inoculum of influenza A/H5N1 virus. Proc Natl Acad Sci USA 105:8091–8096. 10.1073/pnas.0711942105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moseley CE, Webster RG, Aldridge JR. 2010. Peroxisome proliferator-activated receptor and AMP-activated protein kinase agonists protect against lethal influenza virus challenge in mice. Influenza Other Respir Viruses 4:307–311. 10.1111/j.1750-2659.2010.00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chalaris A, Rabe B, Paliga K, Lange H, Laskay T, Fielding CA, Jones SA, Rose-John S, Scheller J. 2007. Apoptosis is a natural stimulus of IL6R shedding and contributes to the proinflammatory trans-signaling function of neutrophils. Blood 110:1748–1755. 10.1182/blood-2007-01-067918. [DOI] [PubMed] [Google Scholar]
- 82.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE. 2021. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci 30:70–82. 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper.