Abstract
Simultaneous delivery of mRNA to multiple populations of antigen (Ag)–specific CD8+ T cells is challenging given the diversity of peptide epitopes and polymorphism of class I major histocompatibility complexes (MHCI). We developed Ag-presenting nanoparticles (APNs) for mRNA delivery using pMHCI molecules that were refolded with photocleavable peptides to allow rapid ligand exchange by UV light and site-specifically conjugated with a lipid tail for postinsertion into preformed mRNA lipid nanoparticles. Across different TCR transgenic mouse models (P14, OT-1, and Pmel), UV-exchanged APNs bound and transfected their cognate Ag-specific CD8+ T cells equivalent to APNs produced using conventionally refolded pMHCI molecules. In mice infected with PR8 influenza, multiplexed delivery of UV-exchanged APNs against three immunodominant epitopes led to ~50% transfection of a VHH mRNA reporter in cognate Ag-specific CD8+ T cells. Our data show that UV-mediated peptide exchange can be used to rapidly produce APNs for mRNA delivery to multiple populations of Ag-specific T cells in vivo.
Light-induced rapid production of antigen-presenting nanoparticles enables RNA delivery to multiple virus-specific T cell subsets.
INTRODUCTION
Antigen (Ag)–specific CD8+ T cells express T cell receptors (TCRs) that recognize processed peptide Ags bound to major histocompatibility complex class I (MHCI) molecules expressed on the cell surface. The TCR–peptide MHCI (pMHCI) interaction forms the basis for the exquisite specificity of CD8+ T cell recognition and their cytotoxic activity against target cells bearing cognate pMHCI Ags. This central mechanism has driven increasing interest in delivery approaches that can target and modulate T cells for immunotherapy. Recent studies include delivery of immunomodulatory molecules (e.g., transforming growth factor–β inhibitors) using nanoparticles decorated with antibodies against T cell surface markers, including CD3 and programmed cell death protein-1 (PD-1), to enhance effector functions within the tumor microenvironment (1–4). Programming endogenous CD3+ or CD8+ T cells with polymer/lipid nanoparticles (LNPs) loaded with nucleic acids [e.g., CD45 small interfering RNA (siRNA) and chimeric Ag receptor (CAR)–encoded DNA] has shown potential to silence target genes in T cells or for in situ manufacturing of CAR T cells (5–9). The ability to target Ag-specific T cells offers opportunities to selectively augment disease-relevant T cell subsets (e.g., viral or tumor Ag-specific T cells) in vivo while maintaining homeostasis and self-tolerance of the immune system (10). To target Ag-specific T cells in vivo, strategies include engineered human pMHCI [human leukocyte Ag (HLA)]–Fc fusion dimers to expand human papillomavirus (HPV)–specific CD8+ T cells against HPV-associated malignancies (11) or track virus-specific CD8+ T cells by immuno–positron emission tomography imaging (12), artificial Ag-presenting cells composed of pMHCI on nanoparticles or engineered red blood cells to activate Ag-specific T cells and enhance their effector function for cancer treatment (13–15), tumor-targeting antibodies to deliver viral peptides that are cleaved by tumor proteases and then loaded onto MHCI on the tumor cell surface to redirect virus-specific T cells against tumors (16, 17), and nanoparticles decorated with pMHCII molecules to reprogram autoantigen-reactive CD4+ T cells into disease-suppressing regulatory T cells (Treg) (18, 19). These studies highlight the broad applications of in vivo delivery to Ag-specific T cells.
Despite considerable interest, however, multiplexed delivery to distinct populations of Ag-specific T cells remains challenging owing to the complexity of the immune response. For example, more than 500 severe acute respiratory syndrome coronavirus 2 CD8+ T cell epitopes that are restricted across 26 HLA class I alleles have been described so far (20). Conventionally, pMHCI molecules are expressed by individual refolding reactions to assemble three components—an invariant light chain, a polymorphic heavy chain, and a peptide ligand—into the heterotrimeric structure of endogenous pMHCI molecules (21). This serial process precludes production of large pMHCI libraries (22) until the development of peptide exchange strategies mediated by ultraviolet (UV) light (21, 23), temperature (24), dipeptides (25), or chaperone proteins (26). With UV light–mediated peptide exchange, the heavy and light chains are refolded with a sacrificial peptide containing a photolabile group, such that upon photocleavage by UV light, the sacrificial peptide dissociates to allow an exchange peptide to bind to the MHCI presentation groove (21, 23). For a particular MHC allele, a single batch of UV-sensitive pMHCI molecules can be conventionally refolded and then used to produce hundreds of pMHCI molecules carrying different peptides in one step. For example, pMHCI tetramer libraries with >1000 peptide specificities have been described for the detection of neoAg-specific T cells (27).
Here, we developed Ag-presenting nanoparticles (APNs) synthesized using UV light–mediated ligand exchange for mRNA delivery to multiple influenza-specific CD8+ T cells (Fig. 1). Our approach increases the precision of T cell delivery compared to antibodies (e.g., CD3 and CD8), is rapidly scalable to different peptide epitopes, and, through mRNA delivery, will enable a range of applications from in situ manufacturing of T cell therapies to genome editing and regulation (28, 29). We used UV light–mediated ligand exchange to produce a panel of pMHCI molecules from a sacrificial pMHCI precursor that was site-specifically modified with a lipid tail. This allowed postinsertion after peptide exchange to preformed LNPs, which was formulated on the basis of a similar D-Lin-MC3-DMA-based composition as the first U.S. Food and Drug Administration–approved siRNA drug (ONPATTRO) (30), encapsulating a model mRNA reporter encoding a camelid single variable domain on a heavy chain (VHH) antibody (31). We found that APNs decorated with conventionally refolded or peptide-exchanged pMHCI molecules targeted and transfected Ag-specific CD8+ T cells in multiple TCR transgenic mouse models in vivo (P14, Pmel-1, and OT-1) regardless of the MHC allotype (H2-Db for P14 and Pmel and H2-Kb for OT-1). In a mouse model of recombinant influenza A viral infection (A/Puerto Rico/8/34 H1N1 modified with GP33 Ag, abbreviated as PR8-GP33), intravenous administration of a three-plex cocktail of peptide-exchanged APNs (NP366/Db, PA224/Db, and GP33/Db) resulted in the simultaneous transfection of the top three immunodominant PR8-GP33–specific T cell populations that was significantly more efficient compared to other major cell populations in the spleen and liver. Our data show that UV light–mediated peptide exchange allows for parallel production of APNs for functional mRNA delivery to multiple Ag-specific CD8+ T cell populations in vivo.
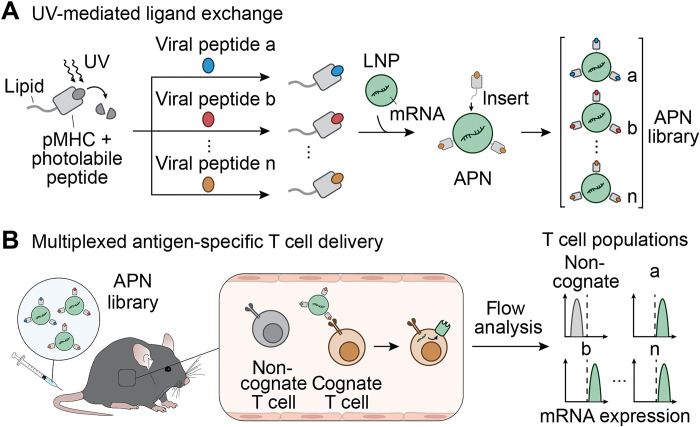
Fig. 1. Schematic of UV-mediated peptide exchange of MHCI APNs for in vivo multiplexed delivery to virus-specific T cells.
(A) We refold pMHC molecules with photolabile peptides and then conjugated them to a lipid tail to allow subsequent formulation with preformed lipid nanoparticles (NPs). The presence of UV light cleaves the photolabile peptide and induces replacement of the resulting empty MHC groove with a library of viral peptides. After the UV-mediated peptide ligand exchange, we functionalize pMHC molecules on the surface of preformed LNPs via postinsertion to form the APN library for multiplexed delivery to virus-specific T cells. (B) After intravenous injection into living mice, APNs selectively target cognate T cell populations and transfect them with model mRNA. We validate the mRNA expression using flow analysis.
RESULTS
APNs bind to Ag-specific T cells and induce internalization for mRNA transfection
The insertion of derivatives modified with lipids to preformed nanoparticles is a well-established approach (32) to decorate nanoparticles with ligands that are stabilized by hydrophobic interactions. For example, postinsertion is commonly used to PEGylate liposomes or LNPs using poly(ethylene glycol) (PEG) polymers derivatized with lipid tails (33). We therefore first sought to express recombinant pMHCI molecules with a site-specific handle for conjugation of a lipid such that the complex could serve as the starting point for peptide exchange before postinsertion to LNPs (Fig. 1A). We expressed and refolded the lymphocytic choriomeningitis virus (LCMV) Ag GP33/Db (KAVYNFATM/Db) with a C-terminal cysteine in the heavy chain to prevent disruption of native disulfide bonds (34) and retain proper pMHCI orientation for TCR recognition (35). These were then reacted with 1,2-distearoyl-sn-glycero-3-phosphorylethanolamine (DSPE)-PEG2000-maleimide to generate lipid-modified GP33/Db molecules. In parallel, we synthesized MC3-based LNPs encapsulating enhanced green fluorescent protein (eGFP) mRNA by microfluidic mixers that were characterized by an average diameter of 93.85 nm and a zeta potential of −30.20 ± 0.5 mV by dynamic light scattering (fig. S1, A and B). Post-insertion of lipid-modified GP33/Db pMHCI molecules did not appreciably increase LNP size nor alter the zeta potential (107.9 ± 7.34 nm and −22.73 ± 4.7 mV, respectively) (fig. S1, B and C). Successful postinsertion across various APN formulations was confirmed using bicinchoninic acid (BCA) protein quantification (table S1). We also assessed eGFP mRNA concentration using the RiboGreen assay and found that bare LNPs and APNs were comparable (80.20 ± 0.50% to 73.62 ± 0.58%, respectively) (fig. S1C).
We next tested whether GP33/Db APNs can selectively bind to their cognate CD8+ T cells isolated from TCR transgenic P14 mice whose CD8+ T cells express a TCR that specifically recognizes the LCMV GP33/Db Ag (Fig. 2A) (36). We found that GP33/Db APNs bound to ~97% of P14 CD8+ T cells, whereas noncognate GP100/Db (KVPRNQDWL/Db) APNs showed minimal staining (3.22%) (Fig. 2B). We further tested APN binding using H2-Kb restricted OVA/Kb (SIINFEKL/Kb) APNs and observed similar Ag-specific binding when coincubated with their cognate CD8+ T cells isolated from OT-1 transgenic mice compared to noncognate NS2/Kb (RTFSFQLI/Kb) APNs (Fig. 2C). The observed 10% cross-reactivity in NS2/Kb APNs to OT-1 CD8+ splenocytes could be due to the binding affinity of H2-Kb MHC to CD8 co-receptors on CD8+ T cells (37). We next investigated whether the binding of APNs to T cells would induce internalization by T cells given that pMHCI multimers are known to be rapidly taken up by T cells through TCR clustering and receptor-mediated endocytosis at physiological temperatures (38). To do this, we studied the fate of APNs after engaging cognate T cells at 37°C compared to 4°C, which is typically used for pMHCI multimer staining to minimize T cell activation and TCR internalization (39). We incubated DiIC18(5) solid (1,1’-dioctadecyl-3,3,3’,3’-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt) (DiD)-labeled OVA/Kb APNs with OT-1 CD8+ T cells at 4° and 37°C, followed by an acidic wash to strip uninternalized APNs bound to the TCRs on T cell surface. We found that DiD fluorescence decreased for cells incubated at 4°C, indicating that APNs remained on the cell surface before acid wash (Fig. 2, D and E). For cells treated at 37°C, however, we observed no change in fluorescence, suggesting efficient T cell internalization of APNs. By contrast, we found no binding and internalization of noncognate GP100/Db APNs by OT-1 CD8+ T cells under all conditions (Fig. 2E). To determine whether pMHCI-induced TCR internalization could result in functional mRNA delivery to T cells, we incubated P14 splenocytes with GP33/Db APNs loaded with eGFP mRNA. We observed a dose-dependent eGFP expression by APN-transfected CD8+ T cells [mean fluorescent intensity (MFI), 397 and 506 for 1- and 2-μg mRNA doses, respectively] in contrast to splenocytes treated with phosphate-buffered saline (PBS; MFI, 153) or free mRNA (MFI, 118) (Fig. 2F). Collectively, these results demonstrate that cognate APNs target, bind, and induce T cell uptake for functional mRNA delivery in vitro.
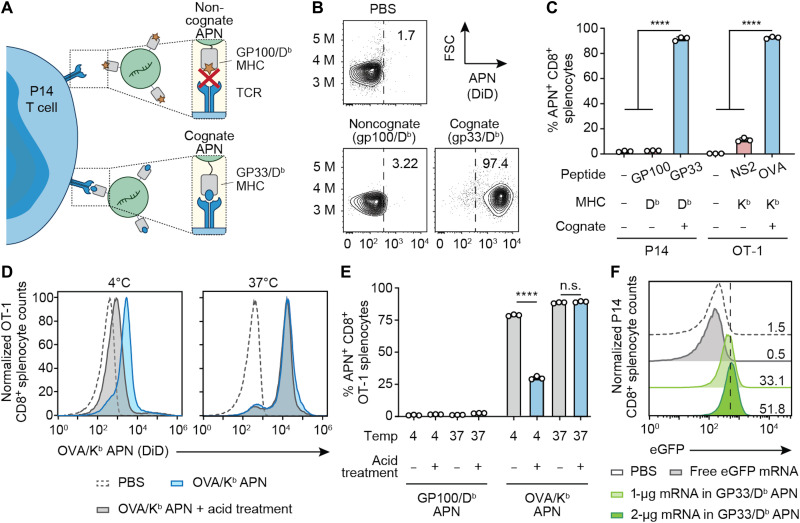
Fig. 2. APNs target Ag-specific T cells and induce cell uptake in vitro.
(A) Illustration of the interaction of P14 CD8+ T cells with its cognate APNs (GP33/Db), in contrast to the lack of binding to the noncognate control (GP100/Db APN). (B) Representative flow plots of noncognate APNs and cognate APNs binding to CD8+ T cells in splenocytes from P14 TCR transgenic mice. Frequencies depicted are based on gating on CD8+ cells. (C) Ag-specific binding of cognate and noncognate APNs to CD8+ T cells isolated from TCR transgenic P14 or OT-1 mice. ****P < 0.0001, one-way analysis of variance (ANOVA) and Tukey post-test and correction. All data are means ± SD; n = 3 biologically independent wells. (D and E) OT-1 CD8+ splenocytes stained with OVA/Kb APNs at 4° or 37°C and analyzed by flow cytometry before and after treatment with an acetate buffer to strip cell surface proteins. GP100/Db APNs served as a noncognate control. ****P < 0.0001 between OVA/Kb APN treatment with and without acid treatment at 4°C; n.s., not significant, where P = 0.61 between OVA/Kb APN with and without acid treatment at 37°C; two-way ANOVA and Sidak post-test and correction. All data are means ± SD; n = 3 biologically independent wells. (F) eGFP mRNA expression in P14 CD8+ T cells after coincubation with free-form eGFP mRNA or eGFP mRNA loaded in GP33/Db APNs for 24 hours.
APNs transfect Ag-specific T cells in TCR transgenic mice
We next quantified in vivo biodistribution and transfection efficiency of GP33/Db APNs in TCR transgenic P14 mice (Fig. 3A). We tested functional delivery to major organs using APNs loaded with firefly luciferase (Fluc) mRNA (Fig. 3, B and C) and found significantly higher luminescence in the spleens isolated from mice treated with GP33/Db APNs compared to noncognate GP100/Db APNs. Notably, no significant difference was observed in the other major organs between the two groups. To quantify delivery to T cells, we harvested P14 splenocytes 24 hours after infusion of DiD-labeled APNs encapsulating VHH mRNA and observed that cognate GP33/Db APNs targeted >95% of P14 CD8+ T cells, while GP100/Db APN controls resulted in <2% binding, as quantified by DiD fluorescence (Fig. 3D). To quantify functional delivery, we used mRNA encoding glycosylphosphatidylinositol (GPI) membrane–anchored VHH as a reporter gene, as it has been shown to achieve durable surface expression (>28 days) that can be detected by immunofluorescence staining with anti-VHH antibodies (25). Therefore, we stained for surface expression of VHH and found that GP33/Db APNs resulted in significantly higher transfection efficiency compared to its noncognate counterpart (~40% versus <2%, respectively; Fig. 3, E and F). Notably, we observed negligible transfection (<2%) of CD8− splenocytes in mice treated with either GP33/Db APNs or GP100/Db APNs. Combined with the results that we observed at the organ level by IVIS imaging (Fig. 3, B and C), our data suggested that the observed Fluc luminescence in the spleen was mainly from the transfection of cognate CD8+ splenocytes. We further confirmed our results in a different model using Pmel mice that have been engineered to express the cognate TCR against GP100/Db. Similar to our results in P14 mice, we observed ~30 to 40% in vivo transfection of Pmel CD8+ splenocytes treated with GP100/Db APNs compared to ~3% transfection with GP33/Db noncognate APNs (fig. S2). Together, these results demonstrate that APNs enable T cell targeting and functional mRNA delivery in an Ag-specific manner.
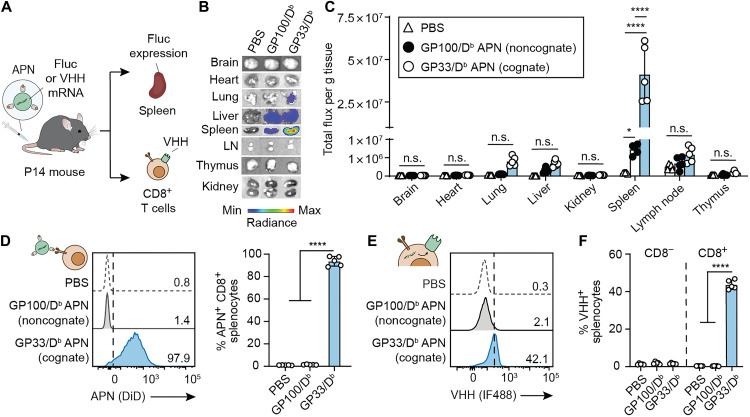
Fig. 3. APNs target and transfect Ag-specific T cells in TCR transgenic P14 mice.
(A) Intravenous injection of GP33/Db APNs to TCR transgenic P14 mice. mRNA encoding Fluc or camelid antibody VHH was loaded in APNs as a reporter. Major organs or splenocytes were harvested for IVIS imaging of Fluc expression or flow analysis of VHH protein expression on the CD8+ splenocytes. GP100/Db APNs were used as a noncognate control. (B) Representative bioluminescence images of various organs were recorded after 6 hours after APN injection to P14 mice. (C) Quantification data of bioluminescence images show in (B). (D) In vivo targeting of GP33/Db APNs to CD8+ splenocytes in P14 mice. *P = 0.0132 between PBS and GP100/Db APNs; ****P < 0.0001 between PBS and GP33/Db APNs and between GP100/Db APNs and GP33/Db APNs; one-way ANOVA and Tukey post-test and correction. (E) Representative flow plot showing in vivo APN-mediated transfection in P14 CD8+ splenocytes. (F) In vivo APN-mediated transfection in both P14 CD8− and CD8+ splenocytes. ****P < 0.0001 between PBS and GP33/Db APNs and between GP100/Db APNs and GP33/Db APNs; one-way ANOVA and Tukey post-test and correction. All data are means ± SD; n = 5 biologically independent mice.
APNs synthesized by UV-mediated ligand exchange transfect T cells equivalently to folded APNs
Sacrificial peptides that contain a photolabile amino acid and stabilize the MHCI complex during refolding have been previously developed for prevalent alleles including H2-Db and H2-Kb in mice (21, 40). We validated UV-mediated peptide exchange by comparing staining of P14 splenocytes using fluorescent GP33/Db tetramers where the pMHCI monomers were either produced by peptide exchange from ASNENJETM/Db (J represents photocleavable amino acid) or conventionally refolded. We first found that tetramers formed with UV-labile peptide present on H2-Db MHCI did not cause any nonspecific binding to CD8+ splenocytes isolated from P14 and Pmel mice (fig. S3). After UV-mediated peptide exchange into GP33 peptides, UV-exchanged tetramers showed comparable staining of CD8+ P14 splenocytes to folded GP33/Db tetramers (>95%), whereas noncognate GP100/Db tetramers produced by refolding or UV exchange resulted in minimal binding (fig. S4). We further tested UV-exchanged tetramers to detect endogenous immune responses where T cells have a broad range of Ag specificity and binding affinities to their cognate Ags. To do this, we used the well-characterized mouse model of influenza virus PR8 (A/PR/8/34) modified to express the LCMV GP33 Ag (PR8-GP33). Infection of mice with PR8-GP33 leads to CD8+ T cell responses against at least 16 PR8-derived peptide epitopes including NP366 (ASNENMETM/Db), PA224 (SSLENFRAYV/Db), PB1-703 (SSYRRPVGI/Kb), PB1-F2 (LSLRNPILV/Db), and NP55 (RLIQNSLTI/Db), as well as against GP33 (41–44), which served as a positive control epitope. We produced a panel of pMHCI tetramers against these six epitopes by peptide exchange (Fig. 4, A and B) (45) and validated Ag-specific splenic T cell responses 10 days after infection (Fig. 4C) (44, 46).
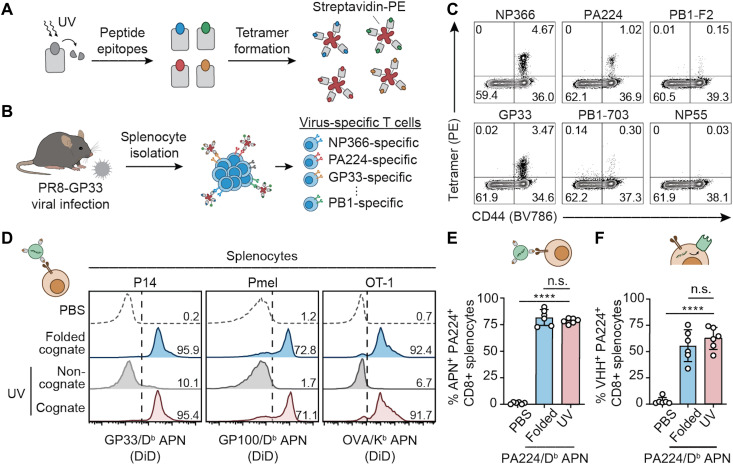
Fig. 4. UV-exchanged APNs transfect Ag-specific T cells equivalently to folded APNs.
(A) Light-triggered peptide exchange technology for high-throughput production of pMHC molecules with various peptide epitopes. pMHC molecules were folded with photolabile peptides that can be cleaved and exchanged with target peptides, followed by tetramer formation with streptavidin conjugated with R-phycoerythrin (PE). (B) Using the UV-exchanged tetramer library to stain virus-specific T cells in a mouse model of PR8-GP33 flu infection. (C) Flow cytometry validation of five epitopes showing diverse Ag specificity of T cell responses to PR8-GP33 flu infection. (D) Equivalent CD8+ splenocyte binding efficiency of UV-exchanged APNs to conventionally folded APNs in three TCR transgenic mouse models in vitro. Numbers indicate the percentage of APN+ cells of CD8+ cells. (E) In vivo targeting activity of UV-exchanged PA224/Db APNs and folded PA224/Db APNs to PA224-specific CD8+ T cells in a mouse model of PR8 infection. n.s., not significant, where P = 0.4548 between folded and UV-exchanged PA224/Db APNs; ****P < 0.0001 between PBS and UV-exchanged PA224/Db APNs; one-way analysis of variance (ANOVA) and Tukey post-test and correction. (F) In vivo transfection efficiency of UV-exchanged PA224/Db APNs and folded PA224/Db APNs to PA224-specific CD8+ T cells in a mouse model of PR8 infection. n.s., not significant, where P = 0.5191 between folded and UV-exchanged PA224/Db APNs; ****P < 0.0001 between PBS and UV-exchanged PA224/Db APNs; one-way ANOVA and Tukey post-test and correction. For (E) and (F), all data are means ± SD; n = 6 biologically independent mice.
We next sought to integrate UV exchange for APN production. To do this, we synthesized a panel of three UV-exchanged APNs inserted with GP33/Db, GP100/Db, and OVA/Kb pMHCI molecules to compare with APNs inserted with pMHCI molecules synthesized using the conventional refolding protocol. In splenocytes isolated from three strains of transgenic mice (P14, Pmel, and OT-1), we found that cognate UV-exchanged APNs bound to CD8+ T cells similar to the folded APNs (Fig. 4D). By contrast, we only observed minimal background staining from the noncognate control APNs. Last, we tested whether the UV-exchanged APNs can target and transfect virus-specific T cells in vivo using PR8-infected mice. To do this, we intravenously injected DiD-labeled PA224/Db APNs to PR8-infected mice (44, 46). At 24 hours after injection, we found that, consistent with the in vitro staining results (Fig. 4D), both folded and UV-exchanged PA224/Db APNs targeted ~80% of PA224-specific T cells (Fig. 4E) and resulted in comparable transfection efficiency of the model VHH mRNA (Fig. 4F).
Multiplexed T cell transfection using a mouse model of PR8 flu infection
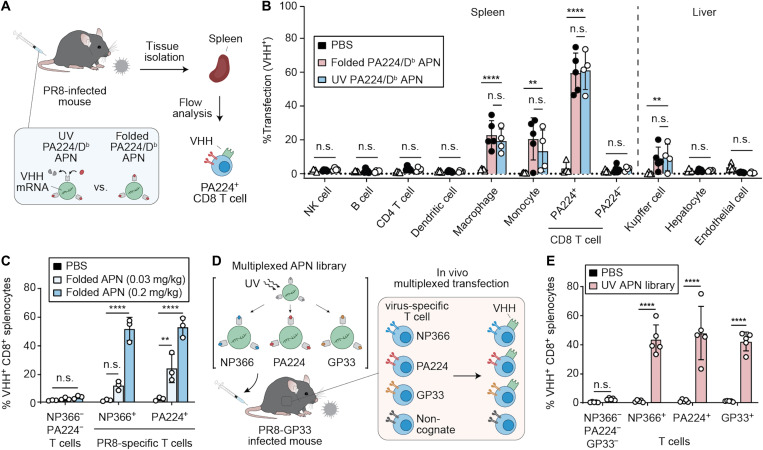
Antibodies against T cell surface markers, including CD3 and CD8, have been used to target polymeric nanoparticles to T cells in vivo irrespective of Ag specificity (47, 48). We therefore examined the ability of APNs to transfect virus-specific T cells compared to noncognate cell populations (Fig. 5A). We focused our analysis on liver and spleen, as these were the major organs that showed APN accumulation after intravenous administration (fig. S5). Flow cytometry analysis of the major cell types [natural killer (NK) cells, B cells, CD4 T cells, dendritic cells, macrophages, monocytes, PA224+ flu-specific CD8 T cells, noncognate PA224− CD8 T cells, Kupffer cells, hepatocytes, and endothelial cells] revealed that APNs preferentially transfected flu virus–specific T cells (PA224+ CD8 T cells, 59.46 ± 11.81%) compared to noncognate T cells (PA224− CD8 T cells, 2.63 ± 2.17%), which comprise a population diversity of approximately 106 to 108 (Fig. 5B) (49). As anticipated, transfection was also observed by the reticuloendothelial system, including monocytes and macrophages in the spleen and Kupffer cells in the liver, but at significantly lower transfection efficiency than that of PA224-specific CD8 T cells (****P < 0.0001). Compared to cohorts that received PBS, mice that were given folded or UV-exchanged PA224/Db APNs resulted in similar transfection levels across all cell populations studied, supporting their equivalency.
Fig. 5. APNs transfect multiplexed T cell subsets with significantly higher transfection efficiency than noncognate cell populations.
(A) Schematic of functional biodistribution study at cellular level comparing UV-exchanged PA224/Db APNs with folded PA224/Db APNs. (B) Transfection efficiency of PBS and PA224/Db APNs in the major cell populations of spleen and liver. **P = 0.0010 between PBS and UV-exchanged PA224/Db APNs in monocyte cell population; **P = 0.0025 between PBS and UV-exchanged PA224/Db APNs in Kupffer cells; ****P < 0.0001; two-way ANOVA and Dunnett post-test and correction. All data are means ± SD; n = 4 to 5 biologically independent mice. (C) Dose-dependent transfection in two immunodominant flu-specific T cells in PR8 model. Infected mice were treated with a mixture of folded NP366/Db APNs and PA224/Db APNs. n.s., not significant, where P = 0.0606 between PBS and folded APNs (0.03 mg/kg total mRNA dose) in NP366+ flu-specific T cell population; **P = 0.0015 between PBS and folded APNs (0.03 mg/kg) in PA224+ flu-specific T cell population (PA224+); ****P < 0.0001; two-way ANOVA and Sidak post-test and correction. All data are means ± SD; n = 3 biologically independent mice. (D) Schematic of multiplexed transfection study. (E) Multiplexed transfection study showing the UV-exchanged APN library transfect three virus-specific T cell populations simultaneously. ****P < 0.0001; two-way ANOVA and Sidak post-test and correction. All data are means ± SD; n = 5 biologically independent mice.
To demonstrate simultaneous transfection of distinct Ag-specific T cell populations in vivo, we administered a mixture of DiD-labeled, conventionally refolded NP366/Db and PA224/Db APNs to PR8-infected mice at an mRNA dose of 0.1 and 0.015 mg/kg for each APNs. We found that APNs specifically targeted NP366- and PA224-specific T cells in a dose-dependent manner (fig. S6), whereas no detectable DiD fluorescence was observed in NP366−PA224− T cells (49). This resulted in a dose-dependent transfection of NP366+ or PA224+ T cells with the model VHH mRNA (Fig. 5C) compared to minimal transfection (<5%) of NP366− and PA224− CD8+ T cells, which was consistent with our earlier observations (Fig. 5B). Last, to demonstrate multiplexed transfection with UV-exchanged APNs, we synthesized and pooled a three-plex panel of APNs presenting the top three immunodominant epitopes (NP366, PA224, and GP33) for PR8-GP33 (Fig. 5D). This APN library efficiently transfected the three selective clones of T cells with significantly higher expression of the model VHH protein than T cells in mice treated with PBS (Fig. 5E). Notably, the transfection efficiency across the three T cell clones were comparable with that of PA224/Db APNs administrated alone (Figs. 4F and 5B), suggesting that the APN-mediated multiplexed transfection did not compromise the transfection efficiency in each T cell population tested.
DISCUSSION
Ag-specific CD8+ T cells are key players in adaptive immunity, and their ability to directly kill target cells expressing cognate peptide Ags restricted to MHCI presentation is being harnessed for important applications in cell therapy, vaccines, and autoimmunity (11, 12, 16–19). Whereas previous work on delivery to T cells via antibodies against cell surface markers (CD3, CD8, etc.) shows great promise, these markers are expressed by all T cells. Moreover, Ag-specific T cell responses are polyclonal (50, 51); for instance, across five prevalent HLA-A alleles (HLA-A*01:01, HLA-A*02:01, HLA-A*03:01, HLA-A*11:01, and HLA-A*24:02), more than 110 flu-specific peptide epitopes have been identified for human influenza A virus (PR8) (52). Therefore, we developed APNs for multiplexed mRNA delivery to Ag-specific T cells using UV-mediated peptide exchange to expedite production of APNs against a panel of peptide epitopes. Our in vivo data using PR8-infected mice showed that APNs with UV-exchanged pMHCI molecules transfected PA224-specific T cells equivalently to APNs synthesized with conventionally folded pMHCI molecules. This allowed us to construct a three-plex APN library using UV-mediated peptide exchange to simultaneously transfect the top three immunodominant T cell populations (NP366, PA224, and GP33 specific) in a mouse model of PR8-GP33 flu infection. Notably, while the frequencies of the top three immunodominant T cells in the spleen range from 1 to 4% of the total CD8+ T cells, APNs achieved ~50% transfection efficiency with a model mRNA encoding membrane-anchored VHH. This targeting sensitivity has not been demonstrated before with antibody– and chemical composition–mediated targeting. Moreover, the in vivo transfection efficiency of APNs in T cells was comparatively higher than T cell delivery technologies previously reported, including a poly(beta-amino ester)-based polymeric nanoparticle system functionalized with anti-CD3 antibodies (~10 to 20%) (8) and a lipid-derived polymeric nanoparticle system (~1.5%) (53). However, note that the mRNA used in our study was different from the two studies, which used mRNA encoding CAR and Fluc, respectively.
The use of APNs has the potential to be expanded for more than three peptide epitopes and beyond the two MHC alleles (H-Db and H-Kb) demonstrated in this study. On the basis of prior studies using the UV exchange technology to generate pMHCI libraries with thousands of peptide epitopes, we expect that UV exchange would be sufficient to produce an APN library with 10 to 20 viral peptide epitopes per MHC allele, the scale that we anticipate in common viral infection settings (e.g., cytomegalovirus, Epstein-Barr virus, and flu) (52, 54, 55). Moreover, sacrificial UV-labile peptides have been developed for most prevalent HLA alleles, including HLA-A*01:01 (STAPGJLEY), HLA-A*02:01 (KILGFVFJV), and HLA-A*11:01 (RVFAJSFIK) (21, 56, 57). Therefore, APNs are amenable to other pMHCI molecules, including HLA expressed by human CD8+ T cells. The capability of APNs in transfecting multiple virus-specific T cell populations may be used to induce in vivo proliferation of virus-specific T cells to treat virus-mediated cancers. For instance, a fusion protein composed of dimerized pMHCI and interleukin-2 (IL-2) has been developed to expand HPV16 E711-20-specific CD8+ T cells to treat HPV-mediated cancers (11), and a recent study suggests that HPV-specific T cells recognizing peptide epitopes derived from HPV E2 and E5 proteins should also be considered to elicit maximal tumor-reactive CD8+ T cell responses against HPV-positive head and neck cancer (58).
Other than CD8+ T cells, it may be possible to expand the transfection capability of APNs to CD4+ T cells by generating APNs with pMHCII. Unlike the cytotoxicity effects triggered by the CD8+ TCR-pMHCI interaction, the interaction between CD4+ TCR and MHCII induces the differentiation and proliferation of CD4+ helper T cells and CD4+ Treg cells, which help CD8+ T cell responses and suppress pathogenic autoimmunity, respectively. Therefore, prior studies have developed pMHCII-functionalized nanoparticles to expand Ag-specific Tregs through pMHC-TCR interaction for the treatment of autoimmune diseases (18, 19). However, this is likely more challenging as binding of class II–bound peptides is less stable compared to that of class I–bound peptides (59), and the interaction between the pMHCII and CD4+ T cells is weaker than that between the class I counterpart and CD8+ T cells (60). Together, our data support the use of APNs for multiplexed mRNA delivery to virus-specific T cells, which can potentially be expanded to transfect broader Ag-specific T cell subsets.
MATERIALS AND METHODS
Animals
Six- to 8-week-old female mice were used at the outsets of all experiments. P14 [B6;D2-Tg(TcrLCMV)327Sdz/JDvsJ], Pmel [B6.Cg-Thy1a/Cy Tg(TcraTcrb)8Rest/J], and OT-1 [C57BL/6-Tg(TcraTcrb)1100Mjb/J] transgenic mice were bred in house using breeding pairs purchased from the Jackson Laboratory. C57BL/6 for PR8 viral infections were purchased from the Jackson Laboratory. All animal procedures were approved by Georgia Tech Institutional Animal Care and Use Committee (protocol numbers: Kwong-A100191, Kwong-A100193, and Santangelo-A100169D).
pMHCI refolding and purification
Peptides used for pMHC refolding were synthesized in house using the Liberty Blue Peptide Synthesizer (CEM) and validated using liquid chromatography–mass spectrometry (Agilent). To generate pMHC molecules for bioconjugation, codon-optimized gBlocks for H2-Db and H2-Kb β2m were purchased from Integrated DNA Technologies and cloned into pET3a vectors (Novagen). H2-Db and H2-Kb genes were engineered with a C-terminal cysteine by site-directed mutagenesis (New England Biolabs), and pMHC molecules were expressed and refolded as described previously (23).
Preparation and characterization
Lipids, including 1,2-distearoyl-sn-glycero-3-phosphocholine), cholesterol, 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glyco (DMG-PEG), DSPE-PEG (18:0 PEG2000 PE), and DSPE-PEG2000-maleimide, were purchased from Avanti Polar Lipids. Ionizable lipid D-Lin-MC3-DMA was purchased from MedKoo Biosciences Inc. Fluorescent, lipophilic carbocyanine dye DiD was purchased from Thermo Fisher Scientific. LNP was synthesized as described previously (61). Briefly, lipid mixture containing MC3, DSPC, cholesterol, DMG-PEG, DSPE-PEG (50:10:38:1.5:0.5 molar ratio), and DiD (1% molar ratio of lipid mix) in ethanol was combined with three volumes of mRNA in acetate buffer [10 mM, pH 4.2, 16:1 (w/w) lipid to mRNA] and injected into microfluidic mixing device NanoAssemblr (Precision NanoSystems) at a total flow rate of 12 ml/min (3:1 flow rate ratio aqueous buffer to ethanol). mRNA encoding eGFP, Fluc, and membrane-anchored VHH antibody were gifts from P.J.S. The resultant LNPs were diluted 40× in PBS and concentrated down using Amicon spin filter (10 kDa; Millipore).
To functionalize the synthesized LNPs with pMHC, pMHC was first coupled with DSPE-PEG-maleimide and decorated on LNPs via postinsertion (62, 63). Briefly, a lipid solution of DSPE-PEG and DSPE-PEG2000-maleimide at 4:1 molar ratio was dried under nitrogen and placed in a vacuum chamber for 2 hours to form a thin film. Lipids were rehydrated in PBS at 6.4 mg/ml in a 60°C water bath for 15 min and sonicated in an ultrasonic bath (Branson) for 5 min. Refolded pMHCI monomers with C-terminal cysteine were reduced with TCEP [1:3 pMHC to tris(2-carboxyethyl)phosphine hydrochloride (TCEP) molar ratio] at 37°C for 2 hours and mixed with the lipid mixture at room temperature (RT) overnight at 2:1 pMHC/maleimide molar ratio (64). Lipid-modified pMHCI molecules were incubated with LNPs at 1:50 maleimide/D-Lin-MC3-DMA molar ratio at RT for 6 hours to incorporate pMHCI onto LNPs. The resultant postinsertion mixture was placed in 1 MDa Float-A-Lyzer (Spectrum) and dialyzed against PBS for 16 hours.
The sizes of APNs in PBS were measured by dynamic light scattering with Malvern Nano ZS Zetasizer (Malvern). Final lipid concentration was quantified using a phospholipid assay kit (Sigma-Aldrich). The concentration of conjugated pMHCI was determined using a BCA assay kit (Sigma-Aldrich). The mRNA encapsulation efficiency was quantified by Quant-iT RiboGreen RNA assay (Life Technologies) as previously described (7). Briefly, 50 μl of diluted APNs was incubated with 50 μl of 2% Triton X-100 (Sigma-Aldrich) in TE buffer (10 mM tris-HCl and 20 mM EDTA) in a 96-well fluorescent plate (Costar, Corning) for 10 min at 37°C to permeabilize the particle. Then, 100 μl of 1% RiboGreen reagent in TE buffer was added into each well, and the fluorescence (excitation wavelength, 485 nm; emission wavelength, 528 nm) was measured using a plate reader (BioTek).
Primary T cell isolation and activation
Spleens isolated from P14, Pmel, or OT-1 TCR transgenic mice were dissociated in complete RPMI media [RPMI 1640 (Gibco) + 10% fetal bovine serum (FBS; Gibco) + 1% penicillin-streptomycin (Gibco)], and red blood cells were lysed using RBC lysis buffer (BioLegend). CD8+ T cells were isolated using a CD8a+ T cell isolation kit (Miltenyi Biotec). For T cell activation, isolated CD8 T cells were cultured in T cell media [complete RPMI media supplemented with 1× nonessential amino acids (Gibco) + 1 × 10−3 M sodium pyruvate (Gibco) + 0.05 × 10−3 M 2-mercaptoethanol (Sigma-Aldrich)] supplemented with soluble anti-mouse CD28 (5 μg/ml; BD Pharmingen) and rhIL-2 (30 U/ml; Roche) at 1 × 106 cells/ml in wells coated with anti-mouse CD3e (3 μg/ml; BD Pharmingen).
In vitro T cell binding and transfection by APNs
P14, Pmel, and OT-1 CD8+ T cells (1 × 106 cells per sample) were isolated and incubated with APNs (10 μg/ml) in fluorescence-activated cell sorting (FACS) buffer (1× Dulbecco’s PBS + 2% FBS + 1 mM EDTA + 25 mM HEPES) for 30 min at 37°C. Cells were washed three times with 1 ml of FACS buffer before analysis on a BD Accuri C6. For validation of in vitro transfection, P14 CD8+ T cells were activated for 24 hours as described above and resuspended in T cell media + rhIL-2 (30 U/ml; Roche) at 2 × 106 cell/ml. Cells (5 × 105) were coincubated with GP33/Db APN containing eGFP mRNA (1 μg) in 24-well plates at 37°C. After 4 hours, 700 μl of T cell media + rhIL-2 (30 U/ml; Roche) was added to each well. After an additional 48-hour incubation, cells were washed three times and stained against αCD8 monoclonal antibody (mAb; clone 53-6.7, BioLegend; table S2) at 4°C for 30 min. Cells underwent another two washes with FACS buffer before analysis on BD Accuri C6.
Quantification of APN internalization by acid wash
OT-1 CD8+ T cells were isolated as described above and incubated with OVA/Kb or GP33/Db APNs at 10 μg/ml and αCD8 mAb (clone 53-6.7, BioLegend; table S2) at 4° or 37°C for 30 min. Cells were washed with FACS buffer, and a portion of stained cells was analyzed on a BD Accuri C6. The remaining cells were incubated in an acid wash solution (0.5 M NaCl + 0.5 M acetic acid, pH 2.5) for 5 min to strip cell surface proteins as described previously before reanalysis on a BD Accuri C6 (64).
In vivo transfection at organ level using P14 TCR transgenic mice
P14 TCR transgenic mice were injected intravenously with GP33/Db or GP100/Db APNs loaded with mRNA encoding Fluc (0.1 mg/kg). Organs were harvested 6 hours after injection and incubated in PBS on ice before IVIS analysis. Organs were soaked in d-luciferin solution (2 mM luciferin) in PBS for 5 min. After 5-min incubation, bioluminescence images were collected with a Xenogen IVIS Spectrum Imaging System (Xenogen, Alameda, CA). The same type of organs was separated from other organs and imaged together (i.e., spleens from all treatment groups were imaged together).
In vivo T cell targeting and transfection in TCR transgenic mice
P14 or Pmel TCR transgenic mice were injected intravenously with GP33/Db or GP100/Db APNs loaded with GPI-anchored camelid VHH antibody mRNA (0.2 mg/kg). Splenocytes were harvested 24 hours later and stained against αCD8 mAb (clone 53-6.7, BioLegend), anti-camelid VHH antibody (clone 96A3F5, GenScript), and pMHC tetramers (streptavidin, 2 μg/ml) on ice for 30 min. The working concentrations of antibodies were listed in table S2. Epitope pMHC tetramers for staining were synthesized in house by mixing biotinylated pMHC with fluorescently labeled streptavidin at a 4:1 molar ratio (23). Cells were then fixed with IC fixation buffer (Thermo Fisher Scientific) for the flow analysis (LSRFortessa, BD). All flow data in this study were analyzed with FlowJo v.10 (Tree Star).
In vivo T cell targeting and transfection in PR8-infected mice
PR8 virus was a gift from P.J.S. PR8-GP33 was a gift from R.A. (Emory University) and E. J. Wherry (University of Pennsylvania). Six- to 8-week-old PR8-infected C57BL/6 mice were intranasally infected with either PR8 virus or PR8-GP33 recombinant virus, as specified in Results and figure captions. PR8-infected mice were injected intravenously with NP366/Db and PA224/Db APNs containing the GPI-anchored camelid VHH antibody mRNA (0.03 or 0.2 mg/kg) on day 10 after viral infection. Twenty-four hours after the injection, splenocytes were harvested as described above for immunofluorescent staining. Cells were stained against tetramers (NP366/Db, PA224/Db, 0.2 μg of streptavidin/staining sample), αCD8a mAb (clone 53-6.7, BD), αNK1.1 mAb (clone PK136, Tonbo), αB220 mAb (clone RA3-6B2, Tonbo), αCD4 mAb (clone RM4-2, Tonbo), and anti-camelid VHH antibody (clone 96A3F5, GenScript) on ice for 30 min (65–67). Antibodies were all used at 1:100 dilutions, and the specific working concentrations were listed in table S2. Cells were then fixed with IC fixation buffer (Thermo Fisher Scientific) for the flow analysis (LSRFortessa, BD).
Functional distribution of APNs at cellular level
Six- to 8-week-old PR8-infected C57BL/6 mice were injected intravenously with conventional or UV-exchanged PA224/Db APNs containing the GPI-anchored camelid VHH antibody mRNA (0.1 mg/kg) on day 10 after viral infection. Twenty-four hours after the injection, cells from spleen and liver were harvested as described above. Cells were stained against αCD8a mAb (clone 53-6.7, BD), αNK1.1 mAb (clone PK136, Tonbo), αB220 mAb (clone RA3-6B2, Tonbo), αCD31 mAb (clone PK136, Tonbo), αCD45 mAb (clone 30-F11, BioLegend), αCD4 mAb (clone RM4-2, Tonbo), αCD11b mAb (clone M1/70, BioLegend), αCD11c mAb (clone N418, BioLegend), αLy6c mAb (clone HK1.4, BioLegend), αF4/80 mAb (clone BM8, BioLegend), and anti-camelid VHH antibody (clone 96A3F5, GenScript) on ice for 30 min. Antibodies were all used at 1:100 dilutions, and the specific working concentrations were listed in table S2. Cells were then fixed with IC fixation buffer (Thermo Fisher Scientific) for the flow analysis (LSRFortessa, BD). Cells were identified by a combination of surface markers: macrophages (CD45+, CD11b+, CD11c−, and Ly6c−/low), dendritic cells (CD45+, CD11c+, and CD11b−), endothelial cells (CD45− and CD31+), monocytes (CD45+ CD11b+, CD11c−, and Ly6c+), B cells (CD45+ and B220+), CD4+ T cells (CD45+ and CD4+), CD8+ T cells (CD45+, CD8+, and NK1.1−), flu-specific CD8+ T cells (CD45+, CD8+, NK1.1−, and tet+), NK cells (CD45+ and NK1.1+), hepatocytes (CD31−, CD45−, and F4/80−), and Kupffer cells (CD31−, CD45+, and F4/80+).
Statistical analysis
Significant differences between control and treatment groups were determined by various statistical analyses. Student’s t test was used for two-group comparison. One-way analysis of variance (ANOVA) was used for multiple-group comparison. Two-way ANOVA was used when there were subgroups in each group. Data represent means ± SD in each figure and table as indicated. Statistical analyses were performed using GraphPad Prism 8.0.2 software (GraphPad Software) (*P < 0.05, **P < 0.005, ***P < 0.0005, and ****P < 0.0001).
Acknowledgments
Funding: This work was supported by Defense Advanced Research Projects Agency grant HR00111920008 (R.A., P.J.S., and G.A.K.), NIH grant DP2HD091793 (G.A.K.), NIH grant R01CA237210-01 (G.A.K.), NSF grant ECCS-1542174 (G.A.K.), The Burroughs Wellcome Fund, Career Award at the Scientific Interface (G.A.K.), NSF Graduate Research Fellowships Program DGE-1650044 (S.N.D.), NSF Integrative Graduate Education and Research Traineeship DGE-0965945 (S.N.D.), Alfred P. Sloan Foundation (L.G.), NIH GTBioMAT Training Grant 5T32EB006343 (L.G.), NSF Graduate Research Fellowships Program DGE-1451512 (L.G.), NSF Graduate Research Fellowships Program DGE-2039655 (A.D.S.T.), NIH Cell and Tissue Engineering (CTEng) Training Program 5T32GM8433-30 (A.D.S.T.), and Georgia Tech President’s Fellowship (A.Z.)
Author contributions: Conceptualization: F.-Y.S., S.N.D., and G.A.K. Methodology: F.-Y.S., Q.H.Z., S.N.D., L.G., S.S.B., A.Z., H.P., and G.A.K. Investigation: F.-Y.S., Q.H.Z., S.N.D., L.G., A.D.S.T., A.Z., and G.A.K. Supervision: R.A., P.J.S., and G.A.K. Writing (original draft): F.-Y.S., Q.H.Z., S.N.D., and H.P. Writing (review and editing): F.-Y.S. and G.A.K.
Competing interests: G.A.K. is cofounder of Glympse Bio and Port Therapeutics, and consults for Glympse Bio, Port Therapuetics, and Satellite Bio. F.-Y.S., S.N.D., L.G., P.J.S., and G.A.K. are listed as inventors on a patent application pertaining to the results of this paper. The patent applicant is the Georgia Tech Research Corporation. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Tables S1 and S2
REFERENCES AND NOTES
- 1.Zheng Y., Stephan M. T., Gai S. A., Abraham W., Shearer A., Irvine D. J., In vivo targeting of adoptively transferred T-cells with antibody- and cytokine-conjugated liposomes. J. Control. Release 172, 426–435 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmid D., Park C. G., Hartl C. A., Subedi N., Cartwright A. N., Puerto R. B., Zheng Y., Maiarana J., Freeman G. J., Wucherpfennig K. W., Irvine D. J., Goldberg M. S., T cell-targeting nanoparticles focus delivery of immunotherapy to improve antitumor immunity. Nat. Commun. 8, 1747 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng Y., Tang L., Mabardi L., Kumari S., Irvine D. J., Enhancing adoptive cell therapy of cancer through targeted delivery of small-molecule immunomodulators to internalizing or noninternalizing receptors. ACS Nano 11, 3089–3100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Y.-S. S., Moynihan K. D., Bekdemir A., Dichwalkar T. M., Noh M. M., Watson N., Melo M., Ingram J., Suh H., Ploegh H., Stellacci F. R., Irvine D. J., Targeting small molecule drugs to T cells with antibody-directed cell-penetrating gold nanoparticles. Biomater. Sci. 7, 113–124 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramishetti S., Kedmi R., Goldsmith M., Leonard F., Sprague A. G., Godin B., Gozin M., Cullis P. R., Dykxhoorn D. M., Peer D., Systemic gene silencing in primary T lymphocytes using targeted lipid nanoparticles. ACS Nano 9, 6706–6716 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Smith T. T., Stephan S. B., Moffett H. F., McKnight L. E., Ji W., Reiman D., Bonagofski E., Wohlfahrt M. E., Pillai S. P. S., Stephan M. T., In situ programming of leukaemia-specific T cells using synthetic DNA nanocarriers. Nat. Nanotechnol. 12, 813–820 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kedmi R., Veiga N., Ramishetti S., Goldsmith M., Rosenblum D., Dammes N., Hazan-Halevy I., Nahary L., Leviatan-Ben-Arye S., Harlev M., Behlke M., Benhar I., Lieberman J., Peer D., A modular platform for targeted RNAi therapeutics. Nat. Nanotechnol. 13, 214–219 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Parayath N. N., Stephan S. B., Koehne A. L., Nelson P. S., Stephan M. T., In vitro-transcribed antigen receptor mRNA nanocarriers for transient expression in circulating T cells in vivo. Nat. Commun. 11, 6080 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su F.-Y., Mac Q. D., Sivakumar A., Kwong G. A., Interfacing biomaterials with synthetic t cell immunity. Adv. Healthc. Mater. 10, 2100157 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S., Symonds A. L. J., Miao T., Sanderson I., Wang P., Modulation of antigen-specific T-cells as immune therapy for chronic infectious diseases and cancer. Front. Immunol. 5, 293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quayle S. N., Girgis N., Thapa D. R., Merazga Z., Kemp M. M., Histed A., Zhao F., Moreta M., Ruthardt P., Hulot S., Nelson A., Kraemer L. D., Beal D. R., Witt L., Ryabin J., Soriano J., Haydock M., Spaulding E., Ross J. F., Kiener P. A., Almo S., Chaparro R., Seidel R., Suri A., Cemerski S., Pienta K. J., Simcox M. E., CUE-101, a novel E7-pHLA-IL2-Fc fusion protein, enhances tumor antigen-specific T-cell activation for the treatment of HPV16-driven malignancies. Clin. Cancer Res. 26, 1953–1964 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Woodham A. W., Zeigler S. H., Zeyang E. L., Kolifrath S. C., Cheloha R. W., Rashidian M., Chaparro R. J., Seidel R. D., Garforth S. J., Dearling J. L., Mesyngier M., Duddempudi P. K., Packard A. B., Almo S. C., Ploegh H. L., In vivo detection of antigen-specific CD8+ T cells by immuno-positron emission tomography. Nat. Methods 17, 1025–1032 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L., Song S., Jin X., Wan X., Shahzad K. A., Pei W., Zhao C., Shen C., An artificial antigen-presenting cell delivering 11 immune molecules expands tumor antigen–specific CTLs in ex vivo and in vivo murine melanoma models. Cancer Immunol. Res. 7, 1188–1201 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Zhang X., Luo M., Dastagir S. R., Nixon M., Khamhoung A., Schmidt A., Lee A., Subbiah N., McLaughlin D. C., Moore C. L., Gribble M., Bayhi N., Amin V., Pepi R., Pawar S., Lyford T. J., Soman V., Mellen J., Carpenter C. L., Turka L. A., Wickham T. J., Chen T. F., Engineered red blood cells as an off-the-shelf allogeneic anti-tumor therapeutic. Nat. Commun. 12, 2637 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ugel S., Zoso A., de Santo C., Li Y., Marigo I., Zanovello P., Scarselli E., Cipriani B., Oelke M., Schneck J. P., Bronte V., In vivo administration of artificial antigen-presenting cells activates low-avidity T cells for treatment of cancer. Cancer Res. 69, 9376–9384 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Millar D. G., Ramjiawan R. R., Kawaguchi K., Gupta N., Chen J., Zhang S., Nojiri T., Ho W. W., Aoki S., Jung K., Chen I., Shi F., Heather J. M., Shigeta K., Morton L. T., Sepulveda S., Wan L., Joseph R., Minogue E., Khatri A., Bardia A., Ellisen L. W., Corcoran R. B., Hata A. N., Pai S. I., Jain R. K., Fukumura D., Duda D. G., Cobbold M., Antibody-mediated delivery of viral epitopes to tumors harnesses CMV-specific T cells for cancer therapy. Nat. Biotechnol. 38, 420–425 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sefrin J. P., Hillringhaus L., Mundigl O., Mann K., Ziegler-Landesberger D., Seul H., Tabares G., Knoblauch D., Leinenbach A., Friligou I., Dziadek S., Offringa R., Lifke V., Lifke A., Sensitization of tumors for attack by virus-specific CD8+ T-cells through antibody-mediated delivery of immunogenic T-cell epitopes. Front. Immunol. 10, 1962 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clemente-Casares X., Blanco J., Ambalavanan P., Yamanouchi J., Singha S., Fandos C., Tsai S., Wang J., Garabatos N., Izquierdo C., Agrawal S., Keough M. B., Yong V. W., James E., Moore A., Yang Y., Stratmann T., Serra P., Santamaria P., Expanding antigen-specific regulatory networks to treat autoimmunity. Nature 530, 434–440 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Singha S., Shao K., Yang Y., Clemente-Casares X., Solé P., Clemente A., Blanco J., Dai Q., Song F., Liu S. W., Yamanouchi J., Umeshappa C. S., Nanjundappa R. H., Detampel P., Amrein M., Fandos C., Tanguay R., Newbigging S., Serra P., Khadra A., Chan W. C. W., Santamaria P., Peptide–MHC-based nanomedicines for autoimmunity function as T-cell receptor microclustering devices. Nat. Nanotechnol. 12, 701–710 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Tarke A., Sidney J., Kidd C. K., Dan J. M., Ramirez S. I., Yu E. D., Mateus J., da Silva Antunes R., Moore E., Rubiro P., Methot N., Phillips E., Mallal S., Frazier A., Rawlings S. A., Greenbaum J. A., Peters B., Smith D. M., Crotty S., Weiskopf D., Grifoni A., Sette A., Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2, 100204 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toebes M., Coccoris M., Bins A., Rodenko B., Gomez R., Nieuwkoop N. J., van de Kasteele W., Rimmelzwaan G. F., Haanen J. B. A. G., Ovaa H., Schumacher T. N. M., Design and use of conditional MHC class I ligands. Nat. Med. 12, 246–251 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Gee M. H., Han A., Lofgren S. M., Beausang J. F., Mendoza J. L., Birnbaum M. E., Bethune M. T., Fischer S., Yang X., Gomez-Eerland R., Bingham D. B., Sibener L. V., Fernandes R. A., Velasco A., Baltimore D., Schumacher T. N., Khatri P., Quake S. R., Davis M. M., Garcia K. C., Antigen identification for orphan T cell receptors expressed on tumor-infiltrating lymphocytes. Cell 172, 549–563.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodenko B., Toebes M., Hadrup S. R., van Esch W. J. E., Molenaar A. M., Schumacher T. N. M., Ovaa H., Generation of peptide-MHC class I complexes through UV-mediated ligand exchange. Nat. Protoc. 1, 1120–1132 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Luimstra J. J., Garstka M. A., Roex M. C. J., Redeker A., Janssen G. M. C., van Veelen P. A., Arens R., Falkenburg J. H. F., Neefjes J., Ovaa H., A flexible MHC class I multimer loading system for large-scale detection of antigen-specific T cells. J. Exp. Med. 215, 1493–1504 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saini S. K., Schuster H., Ramnarayan V. R., Rammensee H. G., Stevanović S., Springer S., Dipeptides catalyze rapid peptide exchange on MHC class I molecules. Proc. Natl. Acad. Sci. U.S.A. 112, 202–207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Overall S. A., Toor J. S., Hao S., Yarmarkovich M., O’Rourke S. M., Morozov G. I., Nguyen S., Japp A. S., Gonzalez N., Moschidi D., Betts M. R., Maris J. M., Smibert P., Sgourakis N. G., High throughput pMHC-I tetramer library production using chaperone-mediated peptide exchange. Nat. Commun. 11, 1909 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bentzen A. K., Marquard A. M., Lyngaa R., Saini S. K., Ramskov S., Donia M., Such L., Furness A. J. S., McGranahan N., Rosenthal R., Straten P., Szallasi Z., Svane I. M., Swanton C., Quezada S. A., Jakobsen S. N., Eklund A. C., Hadrup S. R., Large-scale detection of antigen-specific T cells using peptide-MHC-I multimers labeled with DNA barcodes. Nat. Biotechnol. 34, 1037–1045 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Sahin U., Karikó K., Türeci Ö., mRNA-based therapeutics—Developing a new class of drugs. Nat. Rev. Drug Discov. 13, 759–780 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Atsavapranee E. S., Billingsley M. M., Mitchell M. J., Delivery technologies for T cell gene editing: Applications in cancer immunotherapy. EBioMedicine 67, 103354 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Multi-discipline review: Patisiran. Population PK and/or PD analyses (2018).
- 31.Tiwari P. M., Vanover D., Lindsay K. E., Bawage S. S., Kirschman J. L., Bhosle S., Lifland A. W., Zurla C., Santangelo P. J., Engineered mRNA-expressed antibodies prevent respiratory syncytial virus infection. Nat. Commun. 9, 3999 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sou K., Endo T., Takeoka S., Tsuchida E., Poly(ethylene glycol)-modification of the phospholipid vesicles by using the spontaneous incorporation of poly(ethylene glycol)-lipid into the vesicles. Bioconjug. Chem. 11, 372–379 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Wang Y., Miao L., Satterlee A., Huang L., Delivery of oligonucleotides with lipid nanoparticles. Adv. Drug Deliv. Rev. 87, 68–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agarwal P., Bertozzi C. R., Site-specific antibody–drug conjugates: The nexus of bioorthogonal chemistry, protein engineering, and drug development. Bioconjug. Chem. 26, 176–192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen M., Rusling J., Dixit C. K., Site-selective orientated immobilization of antibodies and conjugates for immunodiagnostics development. Methods 116, 95–111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puglielli M. T., Zajac A. J., van der Most R. G., Dzuris J. L., Sette A., Altman J. D., Ahmed R., In vivo selection of a lymphocytic choriomeningitis virus variant that affects recognition of the GP33-43 epitope by H-2Db but not H-2Kb. J. Virol. 75, 5099–5107 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moody A. M., Chui D., Reche P. A., Priatel J. J., Marth J. D., Reinherz E. L., Developmentally regulated glycosylation of the CD8alphabeta coreceptor stalk modulates ligand binding. Cell 107, 501–512 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Whelan J. A., Dunbar P. R., Price D. A., Purbhoo M. A., Lechner F., Ogg G. S., Griffiths G., Phillips R. E., Cerundolo V., Sewell A. K., Specificity of CTL interactions with peptide-MHC class I tetrameric complexes is temperature dependent. J. Immunol. 163, 4342–4348 (1999). [PubMed] [Google Scholar]
- 39.Altman J. D., Moss P. A. H., Goulder P. J. R., Barouch D. H., McHeyzer-Williams M. G., Bell J. I., McMichael A. J., Davis M. M., Phenotypic analysis of antigen-specific T lymphocytes. Science 274, 94–96 (1996). [DOI] [PubMed] [Google Scholar]
- 40.Grotenbreg G. M., Roan N. R., Guillen E., Meijers R., Wang J. H., Bell G. W., Starnbach M. N., Ploegh H. L., Discovery of CD8+ T cell epitopes in Chlamydia trachomatis infection through use of caged class I MHC tetramers. Proc. Natl. Acad. Sci. U.S.A. 105, 3831–3836 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mueller S. N., Langley W. A., Li G., García-Sastre A., Webby R. J., Ahmed R., Qualitatively different memory CD8+ T cells are generated after lymphocytic choriomeningitis virus and influenza virus infections. J. Immunol. 185, 2182–2190 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Pauken K. E., Godec J., Odorizzi P. M., Brown K. E., Yates K. B., Ngiow S. F., Burke K. P., Maleri S., Grande S. M., Francisco L. M., Ali M. A., Imam S., Freeman G. J., Haining W. N., Wherry E. J., Sharpe A. H., The PD-1 pathway regulates development and function of memory CD8 T cells following respiratory viral infection. Cell Rep. 31, 107827 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ichinohe T., Pang I. K., Kumamoto Y., Peaper D. R., Ho J. H., Murray T. S., Iwasaki A., Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. U.S.A. 108, 5354–5359 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laidlaw B. J., Decman V., Ali M. A. A., Abt M. C., Wolf A. I., Monticelli L. A., Mozdzanowska K., Angelosanto J. M., Artis D., Erikson J., Wherry E. J., Cooperativity between CD8+ T cells, non-neutralizing antibodies, and alveolar macrophages is important for heterosubtypic influenza virus immunity. PLOS Pathog. 9, e1003207 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mueller S. N., Langley W. A., Carnero E., García-Sastre A., Ahmed R., Immunization with live attenuated influenza viruses that express altered NS1 proteins results in potent and protective memory CD8+ T-cell responses. J. Virol. 84, 1847–1855 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu T., Guan J., Handel A., Tscharke D. C., Sidney J., Sette A., Wakim L. M., Sng X. Y. X., Thomas P. G., Croft N. P., Purcell A. W., la Gruta N. L., Quantification of epitope abundance reveals the effect of direct and cross-presentation on influenza CTL responses. Nat. Commun. 10, 2846 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenblum D., Joshi N., Tao W., Karp J. M., Peer D., Progress and challenges towards targeted delivery of cancer therapeutics. Nat. Commun. 9, 1410 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cevaal P. M., Ali A., Czuba-Wojnilowicz E., Symons J., Lewin S. R., Cortez-Jugo C., Caruso F., In vivo T cell-targeting nanoparticle drug delivery systems: Considerations for rational design. ACS Nano 15, 3736–3753 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Nikolich-Zugich J., Slifka M. K., Messaoudi I., The many important facets of T-cell repertoire diversity. Nat. Rev. Immunol. 4, 123–132 (2004). [DOI] [PubMed] [Google Scholar]
- 50.de Vos van Steenwijk P. J., Heusinkveld M., Ramwadhdoebe T. H., Löwik M. J., van der Hulst J. M., Goedemans R., Piersma S. J., Kenter G. G., van der Burg S. H., An unexpectedly large polyclonal repertoire of HPV-specific t cells is poised for action in patients with cervical cancer. Cancer Res. 70, 2707–2717 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Andersen R. S., Thrue C. A., Junker N., Lyngaa R., Donia M., Ellebæk E., Svane I. M., Schumacher T. N., thor Straten P., Hadrup S. R., Dissection of T-cell antigen specificity in human melanoma. Cancer Res. 72, 1642–1650 (2012). [DOI] [PubMed] [Google Scholar]
- 52.National Institute of Allergy and Infectious Diseases, Immune epitope database and analysis resource (2021).
- 53.McKinlay C. J., Benner N. L., Haabeth O. A., Waymouth R. M., Wender P. A., Enhanced mRNA delivery into lymphocytes enabled by lipid-varied libraries of charge-altering releasable transporters. Proc. Natl. Acad. Sci. U.S.A. 115, E5859–E5866 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Catalina M. D., Sullivan J. L., Bak K. R., Luzuriaga K., Differential evolution and stability of epitope-specific CD8(+) T cell responses in EBV infection. J. Immunol. 167, 4450–4457 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Elkington R., Walker S., Crough T., Menzies M., Tellam J., Bharadwaj M., Khanna R., Ex vivo profiling of CD8+-T-cell responses to human cytomegalovirus reveals broad and multispecific reactivities in healthy virus carriers. J. Virol. 77, 5226–5240 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bakker A. H., Hoppes R., Linnemann C., Toebes M., Rodenko B., Berkers C. R., Hadrup S. R., van Esch W. J. E., Heemskerk M. H. M., Ovaa H., Schumacher T. N. M., Conditional MHC class I ligands and peptide exchange technology for the human MHC gene products HLA-A1, -A3, -A11, and -B7. Proc. Natl. Acad. Sci. U.S.A. 105, 3825–3830 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chandran P. A., Heidu S., Zelba H., Schmid-Horch B., Rammensee H.-G., Pascolo S., Gouttefangeas C., A simple and rapid method for quality control of major histocompatibility complex-peptide monomers by flow cytometry. Front. Immunol. 8, 96 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eberhardt C. S., Kissick H. T., Patel M. R., Cardenas M. A., Prokhnevska N., Obeng R. C., Nasti T. H., Griffith C. C., Im S. J., Wang X., Shin D. M., Carrington M., Chen Z. G., Sidney J., Sette A., Saba N. F., Wieland A., Ahmed R., Functional HPV-specific PD-1+ stem-like CD8 T cells in head and neck cancer. Nature 597, 279–284 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao W., Sher X., Systematically benchmarking peptide-MHC binding predictors: From synthetic to naturally processed epitopes. PLoS Comput. Biol. 14, e1006457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sugata K., Matsunaga Y., Yamashita Y., Nakatsugawa M., Guo T., Halabelian L., Ohashi Y., Saso K., Rahman M. A., Anczurowski M., Wang C. H., Murata K., Saijo H., Kagoya Y., Ly D., Burt B. D., Butler M. O., Mak T. W., Hirano N., Affinity-matured HLA class II dimers for robust staining of antigen-specific CD4+ T cells. Nat. Biotechnol. 39, 958–967 (2021). [DOI] [PubMed] [Google Scholar]
- 61.Veiga N., Goldsmith M., Granot Y., Rosenblum D., Dammes N., Kedmi R., Ramishetti S., Peer D., Cell specific delivery of modified mRNA expressing therapeutic proteins to leukocytes. Nat. Commun. 9, 4493 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ishida T., Iden D. L., Allen T. M., A combinatorial approach to producing sterically stabilized (Stealth) immunoliposomal drugs. FEBS Lett. 460, 129–133 (1999). [DOI] [PubMed] [Google Scholar]
- 63.Lainé A. L., Gravier J., Henry M., Sancey L., Béjaud J., Pancani E., Wiber M., Texier I., Coll J. L., Benoit J. P., Passirani C., Conventional versus stealth lipid nanoparticles: Formulation and in vivo fate prediction through FRET monitoring. J. Control. Release 188, 1–8 (2014). [DOI] [PubMed] [Google Scholar]
- 64.Dahotre S. N., Romanov A. M., Su F.-Y., Kwong G. A., Synthetic antigen-presenting cells for adoptive T cell therapy. Adv. Ther. 4, 2100034 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dahotre S. N., Chang Y. M., Romanov A. M., Kwong G. A., DNA-barcoded pMHC tetramers for detection of single antigen-specific T cells by digital PCR. Anal. Chem. 91, 2695–2700 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dahotre S. N., Chang Y. M., Wieland A., Stammen S. R., Kwong G. A., Individually addressable and dynamic DNA gates for multiplexed cell sorting. Proc. Natl. Acad. Sci. U.S.A. 115, 4357–4362 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kwong G. A., Radu C. G., Hwang K., Shu C. J., Ma C., Koya R. C., Comin-Anduix B., Hadrup S. R., Bailey R. C., Witte O. N., Schumacher T. N., Ribas A., Heath J. R., Modular nucleic acid assembled p/mhc microarrays for multiplexed sorting of antigen-specific T cells. J. Am. Chem. Soc. 131, 9695–9703 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 and S2