Highlights
-
•
PDE4D is the predominant subtype of PDE4 in renal cancer cells.
-
•
CRISPR/Cas9-mediated knockout of PDE4D or inhibition by roflumilast reduced the proliferation and colony formation of Caki-1 ccRCC cells, and inhibited MAPK/ERK signaling in a CRAF-dependent manner.
-
•
PDE4D targeting enhanced the ability of sorafenib to stunt the survival of Caki-1 cells.
-
•
Knockout of PDE4D enhanced sorafenib-mediated induction of apoptosis in Caki-1 cells.
-
•
PDE4D targeting may restore negative feedback on MAPK/ERK signaling in tumors with CRAF-dependent ERK activation.
Keywords: Phosphodiesterase 4D, cAMP, Renal cell carcinoma, Tyrosine kinase inhibitor, MAPK/ERK pathway, CRAF
Abbreviations: cAMP, cyclic adenosine monophosphate; ccRCC, clear cell renal cell carcinoma; ERK, extracellular signal-regulated kinase; KO, knockout; MAPK, mitogen-activated protein kinase; PDE4, phosphodiesterase 4; PI3K, phosphatidylinositol 3-kinase; PKA, protein kinase A; sgRNA, single guide RNA; TKI, tyrosine kinase inhibitor
Abstract
Clear cell renal cell carcinoma (ccRCC) is the most lethal form of kidney cancer and effective treatment regimens are yet to be established. Tyrosine kinase inhibitors (TKI) have widely been used as ccRCC therapeutics, but their efficacy is limited due to accompanying resistance mechanisms. Previous studies have provided substantial evidence for crosstalk between cAMP and the MAPK/ERK signaling pathway. Low levels of intracellular cAMP have been found in several human malignancies and some data suggest that elevation of cAMP expression can be achieved by phosphodiesterase 4 (PDE4) inhibition, resulting in cell growth arrest and/or cell death. The effects of crosstalk between cAMP and the MAPK/ERK pathway on the development progression in ccRCR, however, remain to be fully understood. In this study, we sought to explore the involvement of PDE4 in ccRCC and to assess its potential as a target for therapeutic intervention. We demonstrated that PDE4D is the predominant subtype of PDE4 expressed in healthy and cancerous renal cell lines, particularly in metastatic Caki-1 cells. We generated a CRISPR/Cas9-mediated PDE4D-KO Caki-1 cell model and showed that PDE4D depletion reduced cell proliferation and recovered cAMP expression in these cells. PDE4D-KO and/or PDE4 inhibition with the FDA approved PDE4 inhibitor, roflumilast, also attenuated MAPK/ERK signaling in a CRAF-dependent manner. Most interestingly, we showed that PDE4D-KO enhanced the effectiveness of the TKI, sorafenib, to stunt cell survival. In conclusion, we provide preliminary evidence of PDE4 involvement in ccRCC and suggest a rationale for dual tyrosine kinase/PDE4D targeting in patients with CRAF-dependent MAPK activation.
Introduction
Clear cell renal cell carcinoma (ccRCC) is the most common and aggressive histological type of renal cell carcinoma, accounting for approximately 75% of all RCC cases and 85% of metastatic tumors. The overall five-year survival rate of ccRCC patients with metastasis is only 12% [1], [2], [3].
Dysregulation of receptor tyrosine kinase (RTK) signaling is a prominent feature of many cancer types including ccRCC and contributes to tumor development and progression. Overexpression of growth factors and their receptors constitutes one mechanism of RTK dysregulation and of its oncogenic effects. Aberrant signaling through RTKs, including vascular endothelial growth factor receptor (VEGFR) and platelet-derived growth factor receptor (PDGFR), has been described in ccRCC. Increased activation of such receptors enhances signaling through downstream pathways like PI3K/AKT and MAPK/ERK, and results in oncogenic effects like tumor cell proliferation, angiogenesis, invasion, and metastasis [4], [5], [6], [7]. Several receptor tyrosine kinase inhibitors (TKIs) have been developed in the past decade to address the adverse effects of dysregulated RTK signaling. These therapeutics primarily function as anti-angiogenic agents to inhibit endothelial cell growth and intratumoral vascular formation, but they also impede signaling involved in the proliferation and survival of tumor cells. TKIs like sorafenib have become the most successful class of therapeutics in the treatment of ccRCC [1,8]. Unfortunately, acquired resistance to TKIs can develop in the first year of treatment and limits their long-term efficacy [9]. Various mechanisms underlying tumor cell resistance to TKIs have been postulated; however, the precise mechanisms remain to be fully understood [8,10,11]. Although pathways like MAPK/ERK, which are activated directly downstream of surface receptors, may be the intended targets of TKIs, the effect of their inhibition transitively extends to other associated signaling mechanisms. In this way, the development of resistance can in part be thought of as an unintentional interference in additional signaling pathways that are secondary to those such as MAPK/ERK. Sorafenib, which targets intracellular signaling components of the MAPK/ERK pathway in addition to surface receptors [12], may be particularly susceptible to developing resistance due to its wider range of targets. As a consequence, tyrosine kinase inhibition can also result in transitive deregulation of negative feedback mechanisms intrinsic to TKI-targeted pathways and a loss of therapeutic efficacy. Thus, in order to compensate for such effects of RTK inhibition, TKIs should be combined with additional therapeutics targeting the components of inadvertently inhibited RTK-associated pathways. Such strategies may serve to counteract the development and persistence of resistance in cancer therapy.
Phosphodiesterases (PDEs) are categorized into 11 families in humans and function to hydrolyze the cyclic nucleotide secondary messenger(s) cAMP and/or cGMP. The PDE4 family of enzymes specifically hydrolyze cAMP and include variants categorized into 3 groups based on the expression of N-terminal upstream conserved region (UCR) domains (long form, short form, and super-short form). Interestingly, some residues involved in the regulation of PDE4 activity are located in the N-terminal UCR domains, which are only present in PDE4 long forms including PDE4D. It is well established that PDE4D can be activated by protein kinase A (PKA) and inhibited by ERK [13]. Some studies have shown that initial elevation of cAMP levels resulting from ERK-mediated PDE4D inhibition can lead to PKA activation and net activation of PDE4 enzymatic activity [14]. This type of counter feedback mechanism may explain the depletion of cAMP levels observed in various human tumors with enhanced ERK signaling. Moreover, cAMP-activated PKA can also exert regulatory effects on components of the MAPK/ERK pathway, namely CRAF and BRAF. Whereas PKA inhibits CRAF directly, it can also activate BRAF, albeit indirectly [15]. In the context of tyrosine kinase inhibition, the MAPK/ERK and cAMP/PKA pathways should be considered critically related because TKI-mediated inhibition of ERK may impede ERK-mediated PDE4D inhibition and promote PKA-mediated PDE4D activation, culminating in sustained cAMP depletion. As a result, cAMP-mediated PKA activation may be hindered, eliminating PKA-mediated negative feedback of the MAPK/ERK pathway, particularly in tumor cells with CRAF-dependent ERK activation. Given that PDEs directly regulate cAMP levels, targeting these enzymes presents a viable option for therapeutic intervention in conditions where cAMP expression is depleted. Several PDEs have already been targeted for treatment of a number of physiological conditions and PDE4 inhibitors have been approved for the treatment of inflammatory diseases like arthritis and chronic obstructive pulmonary disease [16,17]. Recent studies have also investigated the potential of PDE4 inhibitors as therapeutics for lung cancer, melanoma, and leukemia [16]. While some reports indicate that increased cAMP levels promote cell proliferation in multiple normal cells and some tumor cells [18,19], others have demonstrated that cAMP inhibits cell proliferation and contributes to apoptosis induction [20], [21], [22]. Elevation of cAMP levels by inhibition of PDE4 has also been found to result in growth arrest and cell death [21], [22], [23], [24], [25], [26], [27], [28]. Thus, the network of interactions between cAMP/PKA, MAPK/ERK, and PDE4 make this an interesting area in cancer research, particularly in ccRCC, where the expression profiles and involvement of PDE4 remain unknown. The present study aimed to answer these questions and to explore the potential of PDE4 targeting as a combination strategy to supplement TKIs like sorafenib in the treatment of ccRCC.
Materials and methods
Cell lines and reagents
All cell lines were obtained from the American Tissue Culture Collection (ATCC; Manassas, VA, USA). HEK 293T human embryonic kidney cells, Caki-1 metastatic ccRCC cells, and 786-O, 769-P non-metastatic ccRCC cells were maintained in RPMI 1640 (ATCC) with 10% FBS and 1% Pen/Strep. Human renal proximal tubule epithelial cells (RPTEC) were maintained in DMEM:F12 (1:1) (ATCC) supplemented according to the manufacturer's instructions. All cell lines were cultured at 37 ˚C and 5% CO2. Forskolin, roflumilast, and sorafenib were purchased from Cayman Chemical Company (Ann Arbor, MI, USA).
Vectors and transfection
CRISPR/Cas9 PDE4D-KO plasmid was purchased from GenScript (Piscataway, NJ, USA). The control lentiCRISPR v2 plasmid was purchased from Addgene (Watertown, MA, USA). The CRISPR/Cas9 PDE4D-KO plasmid encoding the Cas9 nuclease and a target-specific 20 nucleotide single guide RNA (sgRNA) was designed for maximum knockout efficiency. The PDE4D-specific sgRNA sequence was 5′-GATTGTGACTCCATTTGCTC-3′. CRISPR/Cas9 PDE4D-KO or control CRISPR/Cas9 plasmids were transfected into Caki-1 cells using Lipofectamine™ CRISPRMAX™ Cas9 Transfection Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Caki-1 cells were seeded in a 6-well plate and cultured for 24 h until the cells reached 60–70% confluence. The Lipofectamine™ CRISPRMAX™ Cas9 Transfection Reagent was mixed and added directly to the culture media. At 24 h post transfection, 3 mL of selection medium (complete medium containing 2 μg/mL of puromycin) was added. After 1 week, the selection medium was removed and cells were cultured in complete medium for 1 week. Cells were then seeded in a 96-well plate diluted to 1 cell per well. After another 2 weeks of culture, clones were directly sequenced and protein expression analyzed to confirm successful knockout.
qRT-PCR analysis
Total RNA was isolated using an RNA extraction kit (Qiagen, Hilden, Germany). RNA quantity and quality were assessed using the Nanodrop-ND-1000 instrument (Nanodrop Technologies, Wilmington, DE, USA). Single stranded complementary DNA was synthesized using a High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. Transcript levels were measured by qRT-PCR using TaqMan primer on a 7300 RT-PCR system (Applied Biosystems) and normalized to GAPDH.
Western blotting analyses
Cells were lysed in RIPA buffer (Invitrogen, Carlsbad, CA, USA) supplemented with a protease/phosphatase inhibitor cocktail. Cell lysates were resolved by SDS–polyacrylamide gel electrophoresis and transferred on PVDF membrane. Membranes were probed with the following primary antibodies: phospho-ERK1/2 (p-ERK), ERK1/2, phospho-BRAF (p-BRAF), BRAF, phospho-CRAF (p-CRAF), CRAF, PDE4B, β-actin (Cell Signaling Technology, Beverly, MA, USA); PDE4A (Santa Cruz Biotechnology, Dallas, TX, USA); PDE4D, phospho-AKT (p-AKT), AKT, (R&D System, Minneapolis, MN, USA). Protein expression was visualized using an enhanced chemiluminescence kit (Thermo Fisher Scientific, Lafayette, CO, USA) on a Syngene ImageQuant Imaging System (Frederick, MD, USA) and quantified using Image J software (NIH, Bethesda, MD, USA).
cAMP assays
Cells were seeded in a 96-well plate (5000 cells/well). Following overnight incubation at 37 ˚C and 5% CO2, cells were treated with roflumilast (0.1 μM) for 6 h and/or forskolin (20 μM) for 10 min. The concentration of cAMP was determined using a cAMP direct immunoassay kit (Millipore Sigma, Burlington, MA, USA) according to the manufacturer's instructions.
Cell viability assay
Cells were cultured in a 96-well plate and treated with the following concentrations: 0, 0.01, 0.1, 1, 10, and 100 μM. Dual tyrosine kinase/PDE4 inhibition was performed by addition of 0.1 μM roflumilast to each sorafenib concentration group. For combination treatment with roflumilast and sorafenib, a constant dose of 0.1 μM roflumilast was used at varying concentrations of sorafenib (0, 0.01, 0.1, 0.5, 1, 2, 4, 8, 10 and 100 μM). After 72 h of incubation, CellTiter-Glo luminescent cell viability assay reagent (Promega, Fitchburg, WI, USA) was added according to the manufacturer's instructions. Luminescence was detected using the Veritas Microplate Luminometer (Turner BioSystems, Inc., Sunnyvale, CA, USA). MATLAB software was used to calculate dose-response curves and IC50.
Cell cycle analysis
For quantification of cell cycle stage distribution, cells were seeded in a 6-well plate at a density of 3 × 105 cells/well. After 24 h, cells were harvested and analyzed using propidium Iodide (PI) staining solution (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Subsequently the samples were analyzed using a BD FACSCanto II instrument (BD Biosciences, Franklin Lakes, NJ, USA). A total of 10,000 events were acquired for each sample. Data were analyzed with FACSDiva software (BD Biosciences).
Apoptosis detection of Annexin V by flow cytometry
Cells were seeded in a 6-well plate at a density of 3 × 105 cells/well. After 24 h, cells were harvested and analyzed using the Annexin V-FITC Apoptosis Detection Kit (Cell Signaling Technology) according to the manufacturer's instructions. The samples were analyzed using a BD FACSCanto II instrument (BD Biosciences). A total of 10,000 events were acquired for each sample. Data were analyzed with FACSDiva software (BD Biosciences).
Clonogenic assay
To measure clonogenicity, 1 × 103 cells/well were seeded in a 6‑well plate, incubated overnight at 37°C and 5% CO2, then treated with DMSO, 1 μM sorafenib, and/or 0.1 roflumilast for 7 days at 37°C. Colonies were stained with 0.5% crystal violet solution in 25% methanol.
Soft agar assay
WT or PDE4D KO Caki-1 cells (5 × 103 cells/well) were mixed with 0.36% top agar (Sigma‑Aldrich; Saint Louis, MO, USA) and seeded on 0.72% bottom agar in a 12‑well plate to visualize their anchorage‑independent growth. Treatment conditions (DMSO, 1 μM sorafenib and/or 0.1 roflumilast) were maintained for 21 days at 37°C with 5% CO2. Colonies were stained with 0.005% crystal violet solution for 1 h at room temperature and counted.
Immunofluorescence staining
Cells were plated on a multi-chamber slide (MilliporeSigma) and treated with 0.1 µM sorafenib for 72 h. Immunofluorescence staining was performed as previously described. In brief, cells were fixed with 4% PFA for 15 min and permeabilized using 1% Triton X-100 for 15 min. Two percent normal sheep serum containing 5% BSA was used for blocking for 1 h. Primary antibodies (anti-Caspase 3, dilution of 1:100, Cell Signaling Technology; anti-FasL, dilution of 1:50, Abcam; anti-Bax, dilution of 1:50 Santacruz Biotechnology; anti-p53, dilution of 1:100, MilliporeSigma) were added and incubated overnight at 4 °C in 5x diluted blocking buffer containing 0.1% Tween 20. After three washes with PBST (0.1% Tween 20), Alexa Fluor 488 or Alexa Fluor 595 labeled secondary antibodies (Thermo Fisher Scientific) were added and incubated for 1 h at room temperature. Slides were washed and mounted using a medium containing DAPI. Images were captured on a Nikon Confocal Imaging system (Nikon C1 Eclipse, Nikon, Tokyo, Japan).
Apoptosis detection of Annexin V by immunofluorescence
Cells were plated on a multi-chamber slide (MilliporeSigma) and treated with 0.1 µM sorafenib for 72 h. Cells were incubated with Alexa488 labeled Annexin V reagent (Thermo Fisher Scientific) for 1 h. Cells were then fixed with 4% PFA, washed with PBST three times, and mounted with DAPI-containing medium. Images were captured on a Nikon Confocal Imaging system (Nikon C1 Eclipse, Nikon).
Statistical analysis
The experiments were performed for a minimum of three independent tests. Student's t-test and/or ANOVA were performed for statistical analyses. All values were expressed as mean ± SD and p ≤ 0.05 was considered statistically significant.
Results
PDE4D is the predominant subtype of PDE4 in ccRCC cell lines
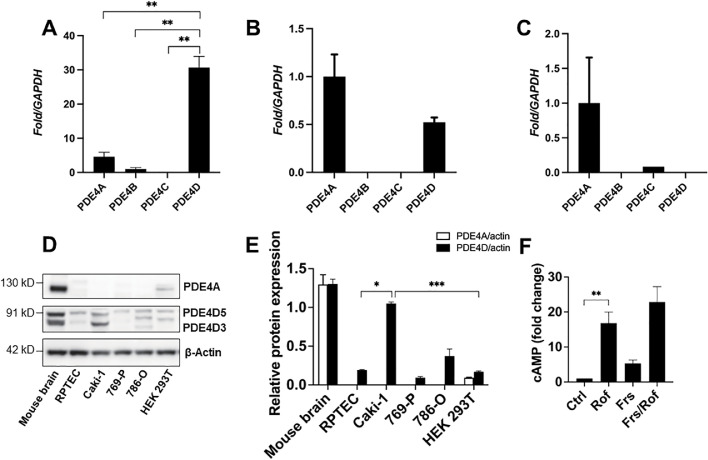
To gain insight into the involvement of PDE4 in ccRCC, we first analyzed expression levels of the PDE4 subfamily genes in several ccRCC cell lines. Our results indicated significantly higher expression of PDE4D in metastatic Caki-1 cells relative to all other PDE4 subtypes (Fig. 1A). Moreover, PDE4D gene expression was found to be approximately 60-fold lower in 769-P cells than in Caki-1 cells, and undetectable in 786-O cells (Fig. 1A–C). PDE4A, PDE4B, and PDE4C expression was found to be varying and minimal among the investigated ccRCC cell lines (Fig. 1A–C). Next, we analyzed protein expression of PDE4A and PDE4D in ccRCC and healthy renal cell lines. Our results revealed no visible expression of PDE4A in the ccRCC cell lines, and of our healthy renal cell lines, only HEK 293T cells showed PDE4A expression, albeit minimal. Interestingly, PDE4D protein expression was detected in all investigated cell lines; however, only Caki-1 cells exhibited markedly high expression relative to the healthy renal cell lines (Fig. 1D, E). Resultantly, Caki-1 cells were selected as the cell model of PDE4D-KO in subsequent experiments. Given that PDE4D functions specifically to hydrolyze cAMP and that depleted cAMP levels have been found in various other human tumors, we sought to determine if the observed overexpression of PDE4D in Caki-1 cells contributes to cAMP depletion. We measured intracellular cAMP expression under basal conditions and following PDE4 inhibition. Our results revealed severely depleted basal cAMP levels, which were recovered following cell treatment with the subtype-non-specific PDE4 inhibitor, roflumilast (0.1 μM). Furthermore, treatment of cells with the adenylate cyclase activator, forskolin (20 μM), also increased cAMP expression, though not nearly to the extent achieved by PDE4 inhibition with roflumilast. Combined treatment of Caki-1 cells with roflumilast and forskolin increased cAMP levels in an additive manner; however, this increase was not significantly greater than roflumilast treatment alone (Fig. 1F). Taken together, our results suggest that although dampened adenylate cyclase activity may lessen cAMP production in these cells, PDE4 is the predominant component responsible for the observed cAMP depletion.
Fig. 1.
PDE4D is the predominant subtype of PDE4 in ccRCC. (A) PDE4D gene expression was significantly higher than all other PDE4 subtypes in Caki-1 cells. (B, C) Gene expression of all investigated PDE4 subtypes was minimal in 769-P and 786-O cells, respectively. (D) PDE4A protein expression was absent in all investigated ccRCC cell lines and minimal in the HEK 293T healthy renal cell line. PDE4D was detected in all investigated cell lines and found to be significantly overexpressed in Caki-1 cells. (E) Quantification of protein expression analysis by Western blot. (F) Basal intracellular cAMP expression was depleted in Caki-1 cells and recovered by PDE4 inhibition with roflumilast (0.1 μM). Mouse brain tissues served as a positive control for PDE4A and PDE4D protein expression analysis. Ctrl, control; Rof, roflumilast; Frs, forskolin. Mean±SD was used, n = 3, * p ≤ 0.05, ** p<0.001, *** p < 0.0001.
PDE4D knockout reduces cell viability and inhibits ERK-mediated signaling in Caki-1 cells
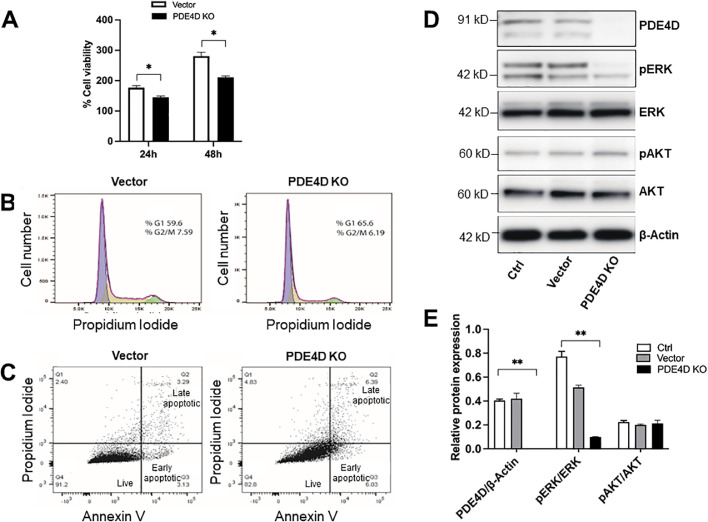
To gain insight into the therapeutic potential of PDE4D targeting in ccRCC, we developed a CRISPR/Cas9-mediated PDE4D-KO Caki-1 cell model (Supplemental Fig. 1A). Single cell sequencing confirmed successful deletion of two base pairs at the targeted site (Supplemental Fig. 1B). Western blot analysis additionally confirmed successful knockout of PDE4D in Caki-1 cells (clone no. 2; Supplemental Fig. 1C). Using this cell model, we assessed the effect of PDE4D-KO on cell proliferation by analyzing viability, cell cycle, and apoptosis. Our results showed significantly reduced viability of PDE4D-KO cells at 24 h post-recovery, and to an even greater extent, at 48 h (Fig. 2A). Moreover, PDE4D-KO also resulted in G1 cell cycle arrest and an increased proportion of apoptotic cells; however, these effects appeared to be moderate (Fig. 2B, C). A more comprehensive follow-up analysis of critical proteins involved in apoptosis confirmed no significant effect of PDE4D-KO alone on apoptosis induction in Caki-1 cells (Fig. 5A, C). Nonetheless, the significant reduction of cell viability in PDE4D-KO cells prompted us to analyze the canonical oncogenic pathways, PI3K/AKT and MAPK/ERK. Interestingly, our results indicated no effect of PDE4D-KO on AKT phosphorylation, but a significant reduction in ERK phosphorylation was observed (Fig. 2D, E). These data suggest that PDE4D targeting may attenuate cell proliferation by interfering with ERK activation and downstream oncogenic signaling.
Fig. 2.
CRISPR/Cas9-mediated PDE4D knockout reduced cell viability and inhibited ERK phosphorylation in Caki-1 cells. (A) Cell viability was significantly reduced in PDE4D-KO cells at 24 and 48 h post-recovery relative to vector cells. (B) Cell cycle analysis by flow cytometry showed that PDE4D-KO resulted in moderate G1 cell cycle arrest. (C) Annexin V apoptosis analysis by flow cytometry showed that PDE4D-KO resulted in a moderate increase in the proportion of apoptotic cells. (D) PDE4D-KO resulted in depletion of phosphorylated-ERK expression. (E) Quantification of protein expression analysis by Western blot. Ctrl, control; Vector, vector only. Mean±SD was used, n = 3, * p ≤ 0.05, ** p < 0.001.
Fig. 5.
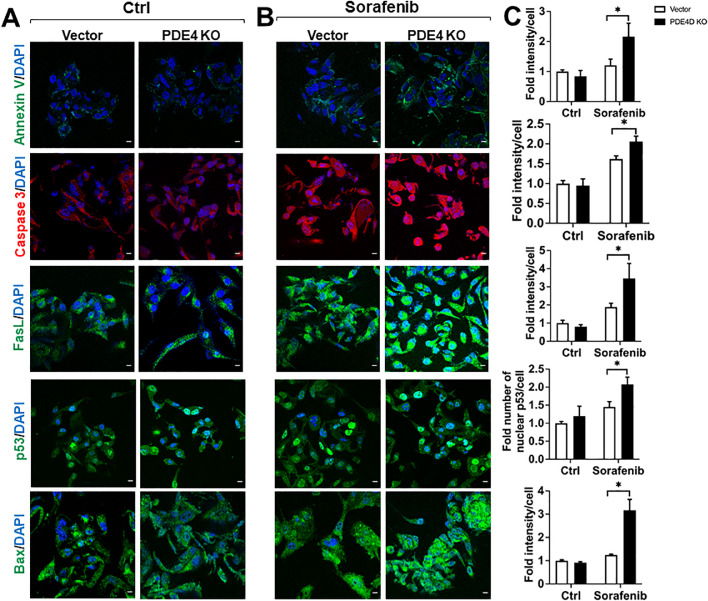
PDE4D enhanced sorafenib-mediated induction of apoptosis in Caki-1 cells. (A) Untreated vector cells exhibited no observable increase in apoptotic marker expression. (B) Expression of Annexin V, Caspase 3, FasL, p53, and Bax was significantly increased in PDE4D-KO cells. (C) Quantification of median fluorescence intensity per cell for each investigated marker. Ctr, control; Vector, vector only. Images obtained at 60x magnification, scale bar = 10 μm. Mean±SD was used, n = 3, * p ≤ 0.05.
PDE4D targeting by CRISPR/Cas9-mediated knockout or inhibition with roflumilast recovers intracellular cAMP levels and inhibits MAPK signaling in a CRAF-dependent manner
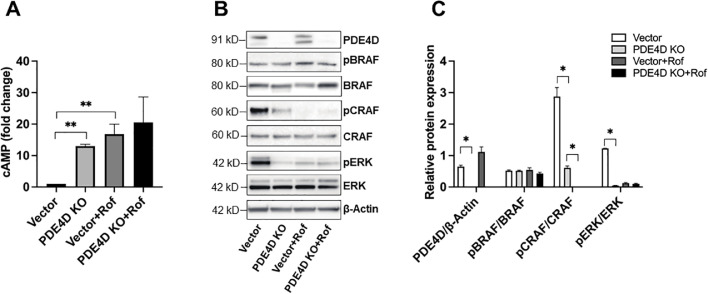
To further explore our findings concerning PDE4-associated cAMP depletion and ERK inhibition, we assessed the expression of cAMP and activation of the MAPK/ERK pathway upstream kinases, BRAF and CRAF, in the PDE4D-KO Caki-1 cell model. Our results showed significantly elevated intracellular cAMP levels in PDE4D-KO cells versus vector only (vector) cells (Fig. 3A). The recovery of cAMP expression observed here mimicked the result of subtype-non-specific PDE4 inhibition with roflumilast (Fig. 1F). Moreover, treatment of vector or PDE4D-KO cells with roflumilast (0.1 μM) resulted in a moderately additive result on elevation of cAMP; however, this increase was not significant relative to untreated PDE4D-KO cells (Fig. 3A). These data confirmed our speculation that PDE4D exerts a profound effect on regulating cAMP expression in the Caki-1 cell line. Given the well-described associations between cAMP and the MAPK/ERK signaling pathway, we analyzed the activation of BRAF and CRAF to gain insight into the observed attenuation of ERK activation in PDE4D-KO cells. Interestingly, whereas BRAF phosphorylation was unaltered in PDE4D-KO cells, phosphorylated-CRAF expression was almost completely depleted. Additionally, treatment of either vector or PDE4D-KO cells with roflumilast further depleted phosphorylated-CRAF expression (Fig. 3B, C). Taken together, our results suggest that although other PDE4 subtypes may contribute to cAMP regulation and promote activation of MAPK signaling, PDE4D predominates.
Fig. 3.
CRISPR/Cas9-mediated PDE4D knockout or PDE4 inhibition by roflumilast recovered intracellular cAMP expression and inhibited MAPK/ERK signaling in a CRAF-dependent manner. (A) Intracellular cAMP expression was elevated in PDE4D-KO and roflumilast-treated Caki-1 cells. (B) PDE4D knockout resulted in depletion of phosphorylated-CRAF and phosphorylated-ERK protein expression. Treatment of PDE4D-KO Caki-1 cells with roflumilast resulted in a slight additive effect. (C) Quantification for protein expression analysis by Western Blot. Roflumilast concentration was maintained at 0.1 μM. Vector, vector only; Rof, roflumilast. Mean±SD was used, n = 3, ** p < 0.001, * p < 0.01.
PDE4D targeting by CRISPR/Cas9-mediated knockout or inhibition by roflumilast enhances the effectiveness of sorafenib to stunt survival of Caki-1 cells in vitro
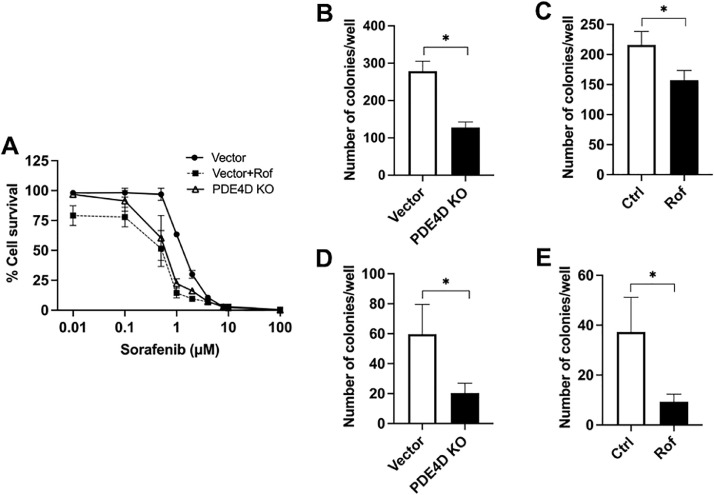
Next, we sought to explore the potential of PDE4 targeting as a combination strategy with the TKI, sorafenib. To do so, we conducted a dose-response viability assessment of PDE4D-KO Caki-1 cells and vector cells treated with the FDA approved PDE4 inhibitor, roflumilast (0.1 μM). Our results showed that targeting PDE4 or PDE4D either by small molecule inhibition or CRISPR/Cas9-mediated knockout resulted in an enhancement of the effectiveness of sorafenib to stunt cell survival compared to untreated vector cells (Fig. 4A). Moreover, the IC50 for sorafenib-treated cells was determined to be 1.33 μM, 0.60 μM, and 0.59 μM in vector, PDE4D-KO, and roflumilast-treated Caki-1 cells, respectively (data not shown). Thus, no significant additive enhancement of sorafenib efficacy could be attributed to subtype-non-specific PDE4 inhibition, suggesting that PDE4D is largely responsible. To further explore the effect of PDE4 targeting in combination with sorafenib treatment, we assessed the colony formation of PDE4D-KO and wild-type control cells treated with roflumilast. Our results revealed that both methods of PDE4 or PDE4D targeting enhanced the ability of sorafenib to reduce colony formation of Caki-1 cells (Fig. 4B, C). Together, these findings demonstrate that either selective small molecule inhibition of PDE4 or CRISPR/Cas9-mediated PDE4D depletion can enhance the sensitivity of Caki-1 cells to treatment with sorafenib.
Fig. 4.
CRISPR/Cas9-mediated PDE4D knockout or PDE4 inhibition by roflumilast enhanced the effectiveness of sorafenib to stunt cell survival and inhibit colony formation in Caki-1 cells. (A) Dose-response viability assessment of sorafenib. (B, C) Clonogenic assay showing reduced colony formation in PDE4D-KO and roflumilast-treated cells in response to sorafenib treatment. (D, E) Soft agar assay showing reduced colony formation in PDE4-KO and roflumilast-treated cells in response to sorafenib treatment. Concentrations of roflumilast and sorafenib were maintained at 0.1 μM and 1 μM, respectively. Vector, vector only; Rof, roflumilast; Ctrl, control. Mean±SD was used, n = 3, * p ≤ 0.05.
PDE4D knockout enhances sorafenib-mediated induction of apoptosis in Caki-1 cells
To follow up on our findings regarding apoptosis induction in PDE4D-KO cells, we conducted a comprehensive analysis of apoptotic proteins with and without sorafenib treatment. Using immunofluorescence staining, we assessed the expression of Annexin V, Caspase 3, Fas ligand, p53, and Bax. Our results showed that PDE4D-KO alone did not result in any significant upregulation of apoptotic protein expression (Fig. 5A, C). This confirmed our previous finding that PDE4D-KO did not result in a substantial increase in the proportion of apoptotic cells (Fig. 2C). However, we found that when treated with sorafenib, PDE4D-KO cells exhibited a significant increase in the expression of all analyzed apoptosis markers versus sorafenib-treated vector cells (Fig. 5B, C). Thus, we provide additional evidence demonstrating that targeting PDE4D can enhance the tumor-suppressive function of sorafenib in Caki-1 cells.
Discussion
The present study sought to provide preliminary insights into the expression and involvement of PDE4 in ccRCC. Initial analysis of PDE4 subfamily genes revealed varying expression in Caki-1, 769-P, and 786-O ccRCC cell lines. However, of the investigated cell lines, only the metastatic Caki-1 cells showed markedly high expression of PDE4, particularly of the PDE4D subtype. This suggests that PDE4 may be upregulated in advanced ccRCC. Given the observed upregulation of PDE4D expression in the Caki-1 cell line, we measured intracellular cAMP in wild-type cells and found its expression to be severely depleted. We also found that inhibition of PDE4 with the subtype-non-specific inhibitor, roflumilast, resulted in a recovery of cAMP expression. These findings were in accordance with other studies of human malignancies showing depleted cAMP expression that can be recovered with PDE4 inhibition [21], [22], [23], [24], [25], [26], [27], [28]. Furthermore, we showed that combined treatment with roflumilast and the activator of adenylate cyclase, forskolin, did not significantly enhance intracellular cAMP expression. This suggests that even though adenylate cyclase activity may contribute to cAMP depletion in Caki-1 cells, PDE4 and PDE4D in particular may be largely responsible. Resultantly, we generated a CRISPR/Cas9-mediated PDE4D-KO Caki-1 ccRCC cell model. Analysis of cAMP expression in PDE4D-KO cells revealed elevated cAMP levels and mimicked the result of roflumilast-treated wild-type Caki-1 cells. Moreover, treatment of PDE4D-KO cells with roflumilast did not significantly enhance cAMP expression, confirming our postulation that PDE4D is the predominant subtype of PDE4 responsible for cAMP depletion in Caki-1 cells.
To gain insight into the therapeutic potential of PDE4D targeting in ccRCC, we analyzed several cell proliferation parameters in PDE4D-KO Caki-1 cells. We found that cell viability was significantly lower in PDE4D-KO cells at 24 and 48 h post-recovery when compared to vector only cells. This finding was in agreement with previous studies showing that cAMP inhibits cell proliferation and contributes to apoptosis induction [20], [21], [22]. However, our results showed that cell cycle arrest and apoptosis were only moderately induced in PDE4D-KO cells. Nevertheless, to gain insight into the mechanism involved in the observed reduction of cell viability, we analyzed signaling through the canonical oncogenic pathways, PI3K/AKT and MAPK/ERK. Interestingly, we found that PDE4D-KO had no effect on AKT phosphorylation, but resulted in a significant reduction in ERK phosphorylation. We suggest that RAP1B, which, when activated by cAMP, inhibits AKT [29], is inefficiently expressed in Caki-1 cells . Lessened RAP1B expression may also account for the observation that BRAF phosphorylation was unaffected by PDE4 or PDE4D targeting, as will be discussed later on. Our future work will seek to elucidate the regulatory function of RAP1B in the crosstalk between cAMP/PKA and MAPK/ERK signaling. To further explore the finding that PDE4D-KO resulted in phosphorylated-ERK depletion, we assessed the activation of MAPK/ERK pathway upstream kinases, BRAF and CRAF. Interestingly, our results showed that PDE4D-KO had no effect on BRAF phosphorylation; however, phosphorylated-CRAF expression was found to be nearly completely depleted. It has been well documented that components of the cAMP/PKA and MAPK/ERK pathways exert feedback on one another [30], [31], [32]. PDE4, which specifically hydrolyzes cAMP, is an intermediary component in feedback regulation of these pathways. Previous studies have shown that cAMP-activated PKA can exert regulatory effects on components of the MAPK/ERK pathway, namely BRAF and CRAF. Whereas PKA activates BRAF indirectly through RAP1B [33,34], it can also directly inhibit CRAF [35,36]. Our finding that phosphorylated-CRAF was depleted in PDE4D-KO cells may be explained by direct inhibition of CRAF activation by PKA resulting from elevated cAMP expression. Taken together, our results suggest that PDE4D-KO attenuated cell proliferation by interfering with ERK activation; however, given the slight-to-moderate effects of PDE4D-KO on cell cycle and apoptosis, PDE4D targeting alone may be insufficient to suppress survival of these cells. Resultantly, we sought to analyze the additive therapeutic potential of dual tyrosine kinase/PDE4 targeting.
Dysregulation of RTK signaling is a prominent feature in ccRCC and results in enhanced oncogenic effects. RTKs mediate these effects through downstream pathways like PI3K/AKT and MAPK/ERK. TKIs like sorafenib have become a moderately successful class of therapeutics to combat dysregulated RTK signaling. Thus, we sought to determine if treating PDE4D-KO Caki-1 cells with sorafenib could enhance the effects of PDE4 targeting alone. We conducted a dose-response viability assessment to sorafenib on PDE4D-KO Caki-1 cells and vector cells treated with the FDA approved PDE4 inhibitor, roflumilast. Our results showed that targeting PDE4 either by CRISPR/Cas9-mediated knockout or small molecule inhibition resulted in an enhancement of the effectiveness of sorafenib to stunt cell survival compared to untreated vector cells. We likewise assessed the ability of sorafenib to inhibit colony formation in PDE4D-KO and roflumilast-treated wild-type control cells. Our results revealed that either method of PDE4 targeting enhanced the ability of sorafenib to reduce colony formation, again suggesting that PDE4D predominates among other PDE4 subtypes. Given these results, we followed up on our previous finding regarding apoptosis induction in PDE4D-KO Caki-1 cells. We conducted a comprehensive expression analysis of the apoptotic markers Annexin V, Caspase 3, Fas ligand, p53, and Bax in PDE4D-KO and vector cells with and without sorafenib treatment. Our results showed that PDE4D-KO alone did not result in significant upregulation of any of the analyzed apoptotic proteins. This confirmed our previous finding that PDE4D-KO did not result in a substantial increase in the proportion of apoptotic cells. However, our results showed that when treated with sorafenib, PDE4D-KO Caki-1 cells exhibited a significant increase in the expression of all analyzed apoptosis markers versus sorafenib-treated vector cells. Taken together, our results suggest that targeting PDE4D can enhance the anti-tumor function of sorafenib in Caki-1 cells. Although the mechanism by which this occurs remains to be elucidated, we postulate that it is critically related with crosstalk between the cAMP/PKA and MAPK/ERK pathways. Tumor cells exhibiting PDE4D overexpression and cAMP depletion may be incapable of PKA-mediated CRAF inhibition and this may contribute to the dysregulation of MAPK/ERK signaling frequently observed in ccRCC. Since decreased PDE enzymatic activity can elevate expression of cAMP, which directly activates PKA, targeting PDE4D may result in a recovery of negative feedback regulation on MAPK/ERK signaling. Thus, our finding that PDE4 or PDE4D targeting enhances the effectiveness of sorafenib may be a result of additive sorafenib-mediated MAPK/ERK inhibition and induction of negative feedback inhibition of CRAF. However, since PKA can also activate BRAF indirectly, the expression of intermediary components facilitating BRAF activation (e.g., RAP1B) must be analyzed when considering this mechanism for application in therapy so as not to inadvertently promote BRAF-mediated ERK activation. Our results showed no effect of PDE4D-KO on BRAF phosphorylation; therefore, we think that the intermediary components facilitating BRAF activation are inefficiently expressed in Caki-1 cells. Although our study focuses only on the expression, function, and potential of dual tyrosine kinase/PDE4 targeting in ccRCC, our findings may be of particular relevance in the context of tumor cell resistance to TKIs. It has previously been shown that initial elevation of cAMP levels resulting from ERK-mediated PDE4D inhibition can lead to PKA activation and net activation of PDE4 enzymatic activity [37]. In spite of the fact that TKIs like sorafenib directly target MAPK/ERK signaling and inhibit ERK, prevailing ERK activation may explain the depletion of cAMP levels as a result of the dominant effect of PKA on PDE4D activation. Thus, targeting PDE4D may eliminate the intermediary component in the crosstalk between cAMP/PKA and MAPK/ERK signaling. Our future studies will focus on the long-term efficacy of sorafenib in combination with therapeutics targeting PDE4 or PDE4D.
Data availability
All datasets generated for this study are included in the manuscript files. The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
CRediT authorship contribution statement
Minghua Cao: Conceptualization, Investigation, Writing – review & editing, Formal analysis, Writing – original draft. Karol Nawalaniec: Investigation, Writing – review & editing, Formal analysis, Writing – original draft, Visualization. Amrendra K. Ajay: Investigation, Writing – review & editing, Formal analysis. Yueming Luo: Investigation, Writing – review & editing. Romana Moench: Investigation, Writing – review & editing. Yanfei Jin: Writing – review & editing. Sheng Xiao: Writing – review & editing. Li-Li Hsiao: Conceptualization, Investigation, Writing – review & editing, Writing – original draft, Visualization. Ana Maria Waaga-Gasser: Conceptualization, Investigation, Writing – review & editing, Formal analysis, Writing – original draft, Visualization.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
This work was supported by the Sundry Fund for the Global Kidney Health Innovation Center. Minghua Cao was supported by grants from The Sixth People's Hospital of Nantong, Affiliated Nantong Hospital of Shanghai University (grant no. MS22019020). Amrendra K. Ajay was supported by The American Heart Association Career Development Award (grant no. 19CDA34480005).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2022.101377.
Contributor Information
Li-Li Hsiao, Email: lhsiao@bwh.harvard.edu.
Ana Maria Waaga-Gasser, Email: awaaga@bwh.harvard.edu.
Appendix. Supplementary materials
References
- 1.Atkins M.B., Tannir N.M. Current and emerging therapies for first-line treatment of metastatic clear cell renal cell carcinoma. Cancer Treat. Rev. 2018;70:127–137. doi: 10.1016/j.ctrv.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Inamura K. Renal cell tumors: understanding their molecular pathological epidemiology and the 2016 WHO classification. Int. J. Mol. Sci. 2017;18:2195–2210. doi: 10.3390/ijms18102195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gnarra J.R., Tory K., Weng Y., Schmidt L., Wei M.H., Li H., Latif F., Liu S., Chen F., Duh F.M., et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat. Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 5.Latif F., Tory K., Gnarra J., Yao M., Duh F.M., Orcutt M.L., Stackhouse T., Kuzmin I., Modi W., Geil L., et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 6.Mukhopadhyay D., Knebelmann B., Cohen H.T., Ananth S., Sukhatme V.P. The von Hippel-Lindau tumor suppressor gene product interacts with Sp1 to repress vascular endothelial growth factor promoter activity. Mol. Cell. Biol. 1997;17:5629–5639. doi: 10.1128/mcb.17.9.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu L., Tong R., Cochran D.M., Jain R.K. Blocking platelet-derived growth factor-D/platelet-derived growth factor receptor beta signaling inhibits human renal cell carcinoma progression in an orthotopic mouse model. Cancer Res. 2005;65:5711–5719. doi: 10.1158/0008-5472.CAN-04-4313. [DOI] [PubMed] [Google Scholar]
- 8.Gotink K.J., Verheul H.M. Anti-angiogenic tyrosine kinase inhibitors: what is their mechanism of action? Angiogenesis. 2010;13:1–14. doi: 10.1007/s10456-009-9160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bielecka Z.F., Czarnecka A.M., Solarek W., Kornakiewicz A., Szczylik C. Mechanisms of acquired resistance to tyrosine kinase inhibitors in clear - cell renal cell carcinoma (ccRCC) Curr. Signal Transduct. Ther. 2014;8:218–228. doi: 10.2174/1574362409666140206223014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khattak M., Larkin J. Sequential therapy with targeted agents in metastatic renal cell carcinoma: beyond second-line and overcoming drug resistance. World J. Urol. 2014;32:19–29. doi: 10.1007/s00345-012-1013-z. [DOI] [PubMed] [Google Scholar]
- 11.Buczek M., Escudier B., Bartnik E., Szczylik C., Czarnecka A. Resistance to tyrosine kinase inhibitors in clear cell renal cell carcinoma: from the patient's bed to molecular mechanisms. Biochim. Biophys. Acta. 2014;1845:31–41. doi: 10.1016/j.bbcan.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Wilhelm S.M., Adnane L., Newell P., Villanueva A., Llovet J.M., Lynch M. Preclinical overview of sorafenib, a multikinase inhibitor that targets both Raf and VEGF and PDGF receptor tyrosine kinase signaling. Mol. Cancer Ther. 2008;7:3129–3140. doi: 10.1158/1535-7163.MCT-08-0013. [DOI] [PubMed] [Google Scholar]
- 13.Omori K., Kotera J. Overview of PDEs and their regulation. Circ. Res. 2007;100:309–327. doi: 10.1161/01.RES.0000256354.95791.f1. [DOI] [PubMed] [Google Scholar]
- 14.Houslay M.D., Adams D.R. PDE4 cAMP phosphodiesterases: modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization. Biochem. J. 2003;370:1–18. doi: 10.1042/BJ20021698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houslay M.D., Kolch W. Cell-type specific integration of cross-talk between extracellular signal-regulated kinase and cAMP signaling. Mol. Pharmacol. 2000;58:659–668. [PubMed] [Google Scholar]
- 16.Zebda R., Paller A.S. Phosphodiesterase 4 inhibitors. J. Am. Acad. Dermatol. 2018;78:S43–S52. doi: 10.1016/j.jaad.2017.11.056. [DOI] [PubMed] [Google Scholar]
- 17.Li H., Zuo J., Tang W. Phosphodiesterase-4 inhibitors for the treatment of inflammatory diseases. Front. Pharmacol. 2018;9:1048–1069. doi: 10.3389/fphar.2018.01048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tirosh A., Jin D.X., De Marco L., Laitman Y., Friedman E. Activating genomic alterations in the Gs alpha gene (GNAS) in 274 694 tumors. Genes Chromosomes Cancer. 2020;59:503–516. doi: 10.1002/gcc.22854. [DOI] [PubMed] [Google Scholar]
- 19.Choudhary P., Gutteridge A., Impey E., Storer R.I., Owen R.M., Whiting P.J., Bictash M., Benn C.L. Targeting the cAMP and transforming growth factor-β pathway increases proliferation to promote Re-epithelialization of human stem cell-derived retinal pigment epithelium. Stem Cells Transl. Med. 2016;5:925–937. doi: 10.5966/sctm.2015-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stork P.J., Schmitt J.M. Crosstalk between cAMP and MAP kinase signaling in the regulation of cell proliferation. Trends Cell Biol. 2002;12:258–266. doi: 10.1016/s0962-8924(02)02294-8. [DOI] [PubMed] [Google Scholar]
- 21.Massimi M., Ragusa F., Cardarelli S., Giorgi M. Targeting cyclic AMP signalling in hepatocellular carcinoma. Cells. 2019;8:1511–1528. doi: 10.3390/cells8121511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reggi E., Diviani D. The role of A-kinase anchoring proteins in cancer development. Cell. Signal. 2017;40:143–155. doi: 10.1016/j.cellsig.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 23.Nam J., Kim D.U., Kim E., Kwak B., Ko M.J., Oh A.Y., Park B.J., Kim Y.W., Kim A., Sun H., Jung Y., Lee J.H., Shin H.J., Park I., Song D.K., Jeong J.Y., Lee Y.H., Kim S.W. Disruption of the Myc-PDE4B regulatory circuitry impairs B-cell lymphoma survival. Leukemia. 2019;33:2912–2923. doi: 10.1038/s41375-019-0492-y. [DOI] [PubMed] [Google Scholar]
- 24.Mishra R.R., Belder N., Ansari S.A., Kayhan M., Bal H., Raza U., Ersan P.G., Tokat Ü.M., Eyüpoğlu E., Saatci Ö., Jandaghi P., Wiemann S., Üner A., Cekic C., Riazalhosseini Y., Şahin Ö. Reactivation of cAMP pathway by PDE4D inhibition represents a novel druggable axis for overcoming tamoxifen resistance in ER-positive breast cancer. Clin. Cancer Res. 2018;24:1987–2001. doi: 10.1158/1078-0432.CCR-17-2776. [DOI] [PubMed] [Google Scholar]
- 25.Marquette A., André J., Bagot M., Bensussan A., Dumaz N. ERK and PDE4 cooperate to induce RAF isoform switching in melanoma. Nat. Struct. Mol. Biol. 2011;18:584–591. doi: 10.1038/nsmb.2022. [DOI] [PubMed] [Google Scholar]
- 26.Fukuda M., Williams K.W., Gautron L., Elmquist J.K. Induction of leptin resistance by activation of cAMP-Epac signaling. Cell Metab. 2011;13:331–339. doi: 10.1016/j.cmet.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan J., Mei F.C., Cheng H., Lao D.H., Hu Y., Wei J., Patrikeev I., Hao D., Stutz S.J., Dineley K.T., Motamedi M., Hommel J.D., Cunningham K.A., Chen J., Cheng X. Enhanced leptin sensitivity, reduced adiposity, and improved glucose homeostasis in mice lacking exchange protein directly activated by cyclic AMP isoform 1. Mol. Cell. Biol. 2013;33:918–926. doi: 10.1128/MCB.01227-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim D.U., Nam J., Cha M.D., Kim S.W. Inhibition of phosphodiesterase 4D decreases the malignant properties of DLD-1 colorectal cancer cells by repressing the AKT/mTOR/Myc signaling pathway. Oncol. Lett. 2019;17:3589–3598. doi: 10.3892/ol.2019.9996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lou L., Urbani J., Ribeiro-Neto F., Altschuler D.L. cAMP inhibition of Akt is mediated by activated and phosphorylated Rap1b. J. Biol. Chem. 2002;277:32799–32806. doi: 10.1074/jbc.M201491200. [DOI] [PubMed] [Google Scholar]
- 30.Hoffmann R., Baillie G.S., MacKenzie S.J., Yarwood S.J., Houslay M.D. The MAP kinase ERK2 inhibits the cyclic AMP-specific phosphodiesterase HSPDE4D3 by phosphorylating it at Ser579. EMBO J. 1999;18:893–903. doi: 10.1093/emboj/18.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim J., Pahlke G., Conti M. Activation of the cAMP-specific phosphodiesterase PDE4D3 by phosphorylation. Identification and function of an inhibitory domain. J. Biol. Chem. 1999;274:19677–19685. doi: 10.1074/jbc.274.28.19677. [DOI] [PubMed] [Google Scholar]
- 32.Liu H., Maurice D.H. Phosphorylation-mediated activation and translocation of the cAMP-specific phosphodiesterase PDE4D3 by cyclic cAMP-dependent protein kinase and mitogen-activated protein kinases. A potential mechanism allowing for the coordinated regulation of PDE4D activity and targeting. J. Biol. Chem. 1999;274:10557–10565. doi: 10.1074/jbc.274.15.10557. [DOI] [PubMed] [Google Scholar]
- 33.Vossler M.R., Yao H., York R.D., Pan M.G., Rim C.S., Stork P.J. cAMP activates MAP kinase and Elk-1 through a B-Raf- and Rap1-dependent pathway. Cell. 1997;89:73–82. doi: 10.1016/s0092-8674(00)80184-1. [DOI] [PubMed] [Google Scholar]
- 34.Ohtsuka T., Shimizu K., Yamamori B., Kuroda S., Takai Y. Activation of brain B-Raf protein kinase by Rap1B small GTP-binding protein. J. Biol. Chem. 1996;271:1258–1261. doi: 10.1074/jbc.271.3.1258. [DOI] [PubMed] [Google Scholar]
- 35.Haefner S., Adler H.S., Mischak H., Janosch P., Heidecker G., Wolfman A., Pippig S., Lohse M., Ueffing M., Kolch W. Mechanism of inhibition of Raf-1 by protein kinase A. Mol. Cell. Biol. 1994;14:6696–6703. doi: 10.1128/mcb.14.10.6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mischak H., Seitz T., Janosch P., Eulitz M., Steen H., Schellerer M., Philipp A., Kolch W. Negative regulation of Raf-1 by phosphorylation of serine 621. Mol. Cell. Biol. 1996;16:5409–5418. doi: 10.1128/mcb.16.10.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Houslay M.D., Adams D.R. PDE4 cAMP phosphodiesterases: modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization. Biochem. J. 2003;370:1–18. doi: 10.1042/BJ20021698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the manuscript files. The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.