Summary
The nicotinic acetylcholine receptor, a pentameric ligand-gated ion channel, converts the free energy of binding of the neurotransmitter acetylcholine into opening of its central pore. Here we present the first high-resolution structure of the receptor type found in muscle-endplate membrane and in the muscle-derived electric tissues of fish. The native receptor was purified from Torpedo electric tissue and functionally reconstituted in lipids optimal for cryo-electron microscopy. The receptor was stabilized in a closed state by the binding of α-bungarotoxin. The structure reveals the binding of a toxin molecule at each of two subunit interfaces in a manner that would block the binding of acetylcholine. It also reveals a closed gate in the ion-conducting pore, formed by hydrophobic amino-acid side-chains, located ~60 Å from the toxin binding sites. The structure provides a framework for understanding gating in ligand-gated channels and how mutations in the acetylcholine receptor cause congenital myasthenic syndromes.
Keywords: Acetylcholine receptor, Cys-loop receptor, nicotinic receptor, ligand-gated ion channel, ion channel structure, Torpedo, membrane protein, cryo-EM, α-bungarotoxin, toxin
eTOC blurb
Rahman et al. report the high-resolution single-particle cryo-EM structure of a native muscle-type nicotinic acetylcholine receptor from the Torpedo electric ray, in complex with α-bungarotoxin from the banded krait. The structure was obtained in a lipidic environment shown to support channel function, and reveals a closed, hydrophobic ion channel gate.
Introduction
The nicotinic acetylcholine receptor (AChR) at the neuromuscular junction has been a prototype in disciplines spanning neuroscience, pharmacology, biochemistry and biophysics for more than a century. Langley located a nicotinic “receptive substance” on the muscle surface based on his application of nicotine to denervated striated muscle (Langley, 1905). The first kinetic models for ion channel function arose from studies of the AChR (Colquhoun and Hawkes, 1977; Katz and Thesleff, 1957); it was the first ligand-gated ion channel to be identified biochemically as a protein with a specific subunit composition (Changeux and Edelstein, 2005; Karlin, 2002); it was the target for the development of patch-clamp electrophysiology (Sakmann and Neher, 1984); it was the first channel to be imaged by electron microscopy (Cartaud et al., 1973); and its were the first ion channel genes to be cloned (Claudio et al., 1983; Devillers-Thiery et al., 1983; Noda et al., 1983).
Natural products have played pivotal roles in uncovering principles of nicotinic receptor structure and function and in making it the paradigm (Changeux and Edelstein, 2005). The discovery that electrogenic organs of Electrophorus eels and Torpedo rays contain extraordinary amounts of acetylcholinesterase (Nachmansohn et al., 1941) suggested that they might provide the raw material for defining how a chemical signal is transformed into an electrical one at cell membranes. α-Bungarotoxin, a snake venom peptide, blocks the effect of acetylcholine at the nicotinic receptor (Chang and Lee, 1963). It provided the high-affinity reagent for the first isolation and characterization of an ion channel, the receptor from the fish electric organ (Changeux et al., 1970; Karlin et al., 1971; Miledi et al., 1971). With a source of abundant receptor protein and a handle for assaying it came rapid progress, controversy and resolution over topics covering the number of subunits in the receptor, their threading through the membrane, their ordering around the ion channel ring, and the locations of acetylcholine sites and the nature of the transmembrane channel gate (Karlin, 2002).
The foundational efforts to visualize the 3D architecture of the nicotinic receptor were carried out initially by Stroud (Klymkowsky and Stroud, 1979) and in greater detail by Unwin and colleagues, who applied electron microscopy to tubular arrays of the receptor from Torpedo (Unwin, 2013). X-ray crystallographic studies of a soluble receptor-like domain (Brejc et al., 2001) and more recent x-ray and cryo-EM structural studies of intact members of the larger pentameric channel superfamily (Cys-loop receptors) provided higher resolution reference points for the AChR (Nemecz et al., 2016). Recent structural studies of subtypes of neuronal nicotinic receptors further refined our understanding of this important neurotransmitter-gated ion channel family (Gharpure et al., 2019; Morales-Perez et al., 2016; Walsh et al., 2018). Until now, there has been no high-resolution structure of the muscle-type AChR. Here we present α-bungarotoxin-bound structure of the Torpedo AChR at 2.7 Å resolution, which provides insight at the atomic-scale of mechanisms of agonist and antagonist binding, coupling of binding to gating, cation permeation, and the pathophysiology of myasthenic syndromes.
Results and Discussion
Biochemistry and functional validation
The native nicotinic acetylcholine receptor was purified from the electric organ of Tetronarce californica (formerly Torpedo californica) after extraction in detergent (Figure 1A, STAR methods). The choice of detergent and added lipids are known to affect the ability to functionally reconstitute the channel after purification (Baenziger and daCosta, 2013; Chak and Karlin, 1992). We settled on an approach of receptor extraction in Triton-X100 detergent, followed by on-column exchange into n-dodecylmaltoside (DDM). We synthesized a new affinity reagent based upon a stable acetylcholine pharmacophore for rapid purification and detergent exchange, as described in STAR methods, and Supplementary Items. Receptors purified in this manner were then reconstituted into soybean lipid liposomes for functional characterization. We performed proteoliposome patch-clamp electrophysiology experiments and found that the agonist carbachol stimulated channel activity and the antagonist α-bungarotoxin blocked this activity (Figure 1B, C, S1A). We concluded that receptors extracted in Triton, purified in DDM and exchanged into soy lipids maintained ligand-gated ion channel activity, consistent with earlier studies (Epstein and Racker, 1978; Lindstrom et al., 1980; McNamee et al., 1975; Nelson et al., 1980).
Figure 1. Biochemistry, functional reconstitution and overall architecture of the Torpedo nicotinic acetylcholine receptor in complex with α-bungarotoxin.
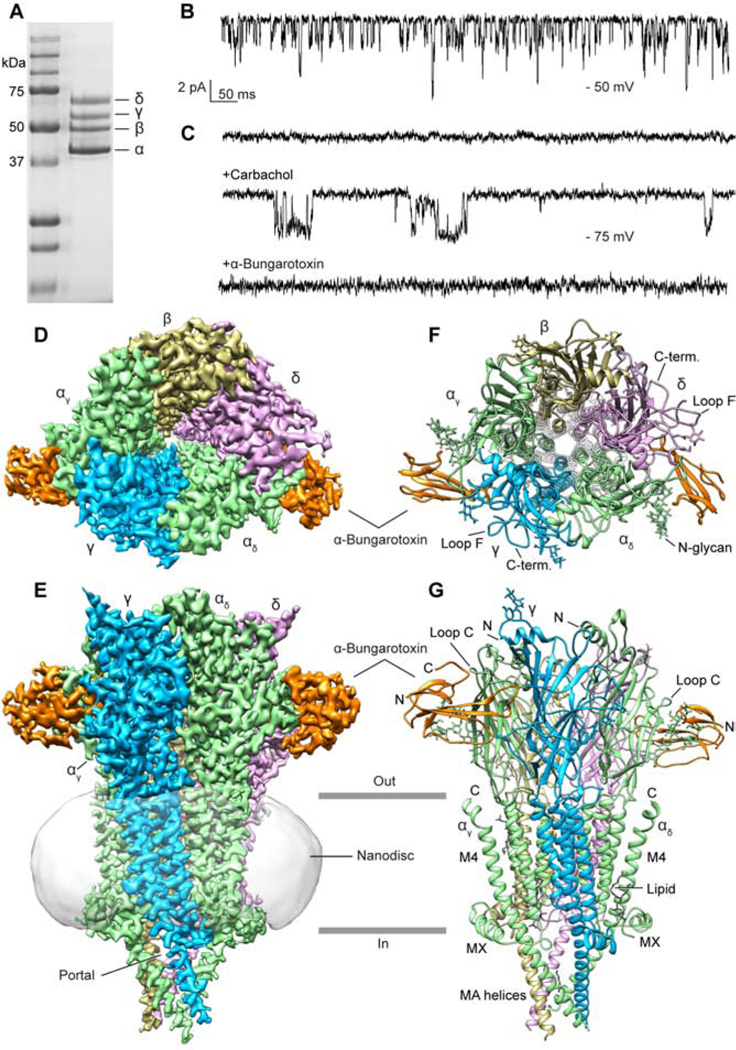
(A) SDS-PAGE of affinity chromatography purified receptor. (B) Liposome patch clamp recording from receptor purified as for EM, in the same lipids used in structural analysis and in the presence of carbachol. (C) Three sections from one continuous recording of an excised patch after liposome reconstitution. Upper trace is apo receptor. Middle trace is after adding carbachol dropwise to the bath solution. Lower trace is after adding α-bungarotoxin dropwise to the bath solution. (D, E) Top and side views of cryo-EM density map. (F, G) Top and side views of atomic model.
Torpedo nicotinic receptors in the electric organ present as dimers of pentamers, linked by a disulfide bond in the extracellular C-termini of δ subunits (Hamilton et al., 1979). Our purification strategy yielded mainly dimeric receptors. We reasoned that structural analysis of receptor monomers would be more straightforward than that of dimers, which we found to adopt different relative monomer orientations (Figure S1B). Accordingly, we reduced the dimers to monomeric receptors (Figure S1C), an approach that preserves channel function (Anholt et al., 1980). Guided by our electrophysiology data, we reconstituted monomeric receptors into soy lipid-saposin nanodiscs (Frauenfeld et al., 2016). The reconstituted receptors were further purified by size-exclusion chromatography to remove empty nanodiscs and reducing reagent.
Single particle cryo-electron microscopy (cryo-EM) was used to obtain the native Torpedo nicotinic acetylcholine receptor structure in complex with α-bungarotoxin. This 8 kDa polypeptide toxin stabilized the structure, facilitated particle alignment and elucidated the mechanism of toxin inhibition. Processing of the cryo-EM dataset yielded an overall map resolution of 2.7 Å, which is to date the highest resolution among reported cryo-EM structures of eukaryotic Cys-loop receptors (Figure 1D–G, Figure S1D–H, S2 and SI Table 1). Only the conformationally disordered linker in the intracellular domain that has been unresolved in all related Cys-loop receptor structures could not be modelled. We defined regions that have no structural precedent in previously solved structures in the Cys-loop receptor superfamily, including insertions in the extracellular domain (ECD), and C-terminal extensions that reach the ACh-binding site. Map quality was also sufficient to model waters, N-linked glycosylation in the ECD, a presumptive palmitoylation site and multiple lipids in the membrane domain.
Overall architecture
The Torpedo AChR – α-bungarotoxin complex forms a T-shaped structure, with the homologous subunits arranged α-γ-α-δ-β (Karlin et al., 1983) counterclockwise around the pseudo-five-fold symmetry axis, as viewed from outside of a cell (Figure 1F, G). Two toxin molecules extend as handles parallel to the membrane from the α-δ and α-γ interfaces in the ECD. Each subunit comprises a large N-terminal ECD with a short N-terminal helix and ten β-strands, a transmembrane domain (TMD) comprising four α-helices (M1-M4), and an intracellular domain (ICD) made up of an amphipathic post-M3 helix (MX), a disordered linker, and an MA helix that precedes and is continuous with M4. We call the unique α subunits αδ and αγ based on their neighbors. From the top (extracellular) view, the δ and γ subunits are readily distinguishable from other subunits due to their extended F Loops and C-termini in the ECD (Figure 1F, S3). Glycosylation sites are present in all subunits. A notably large N-linked glycan emanates from αN141 in the eponymous disulfide-cross-linked Cys-loop (C128-C142) toward the ACh-binding-site loop C (containing C192-C193). Except for the α-δ interface, several extra densities were observed at the TMD interface of each subunit pair, which were modeled speculatively as phosphatidylcholine, the most abundant lipid in the sample. A well-ordered density was also observed connected to C451 in the γ-subunit M4 helix, which was previously identified as a palmitoylation site important for channel function (Li et al., 1990); thus we modelled a thioester-linked palmitate lipid at this position. The TMD α-helices from each subunit assemble into a bundle familiar from earlier EM structures of the Torpedo receptor and from other structures in the superfamily, with the M2 helix lining the ion permeation pathway (Leonard et al., 1988), and M4 being most peripheral (Nemecz et al., 2016). The transmembrane pore is tightly closed and impermeable to hydrated ion flux, consistent with the antagonist activity of the bound toxin. The ICD forms a blunt point at its intracellular end, with lateral portals at each subunit interface poised to facilitate ion permeation parallel to the membrane.
Toxin interaction and mechanism
α-Neurotoxins poison the neuromuscular junction by antagonizing nicotinic receptors on striated muscle through direct competition with acetylcholine, which can result in paralysis, respiratory failure and death (Lester, 1972). α-Neurotoxins from the venom of kraits and cobras share an architecture comprising a core with either 4 or 5 disulfide bonds from which extend three pairs of β strands (“fingers”), the middle finger of which typically contains an arginine that is inserted in the ACh binding site. As a group, these are known as the 3-fingered toxins (Tsetlin, 1999). The most potent of these, α-bungarotoxin, is 74 residues long, contains 5 disulfide bonds, and binds to the Torpedo acetylcholine receptor practically irreversibly (Wang and Schmidt, 1980). Early work localized acetylcholine and α-toxin binding sites in nicotinic receptors to α-γ and α-δ subunit interfaces (Karlin, 2002). Mutagenesis, affinity labeling and structural studies uncovered determinants of toxin-receptor binding (Harel et al., 2001; Sine, 2012), and a preliminary 3D framework was provided by high-resolution crystal structures of α-bungarotoxin or cobratoxin bound to the ECD of α1 (Dellisanti et al., 2007) or α−9 (Zouridakis et al., 2014) single subunits and to homopentameric receptor-like ECDs (Bourne et al., 2005; Huang et al., 2013).
The Torpedo receptor cryo-EM structure reveals α-bungarotoxin molecules wedged deeply into the α-γ and α-δ subunit interfaces (Figure 2A,B). Roughly 25% of the total surface area of each α-bungarotoxin molecule (~1100 Å2) is buried by the receptor. Two structural elements each from the principal (α subunit) and complementary (γ or δ subunit) interface sides converge to stabilize toxin binding. From the principal (α) subunit, the toxin forms extensive contacts with both Loop C and a branched N-glycan extending from the Cys-loop. This N-glycan site is conserved in all Torpedo and human muscle subunits and stacks against loop C as well as Finger I of the toxin. On the complementary side of the interface, loop F makes prominent interactions with the toxin. Loop F is poorly conserved in sequence and length among Cys-loop superfamily members (Figure SI1,2). In γ and δ, it contains an insertion that forms a bowl-shaped scaffold around the base, side and top of Finger II of the toxin while making limited interactions with Finger III. This extended Loop F conformation is buttressed by long, well-ordered C-termini from the same subunits, an architectural feature likely conserved in the human muscle subunits (Figure S4, SI1,2). These C-termini have been absent or disordered in all previous Cys-loop receptor structures.
Figure 2. Receptor - toxin interactions.
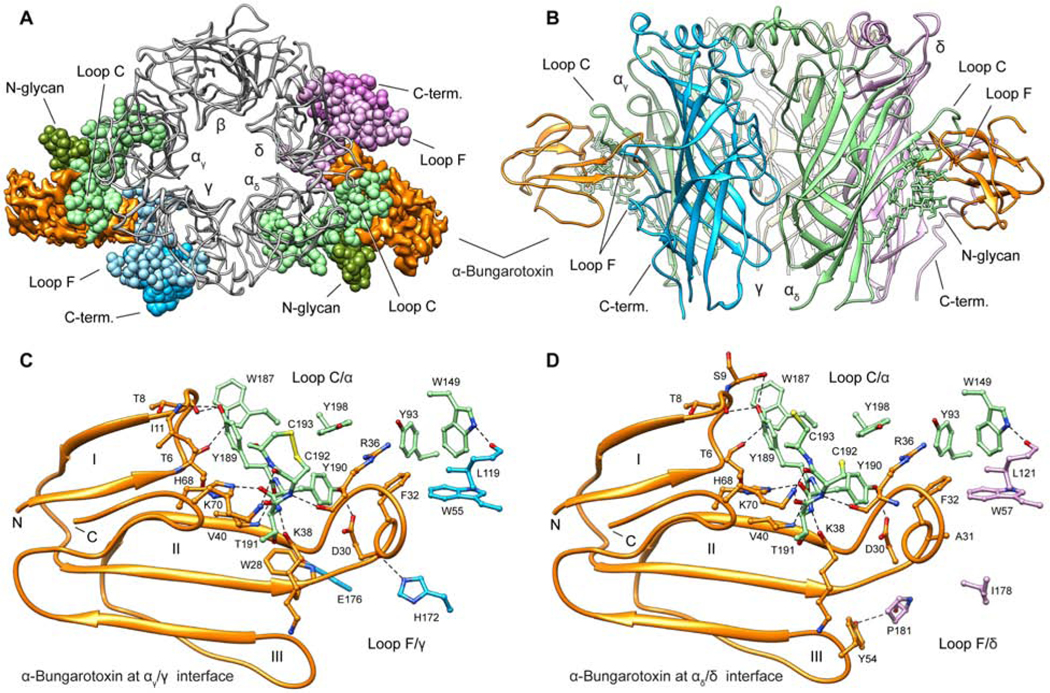
(A, B) Top and side views of receptor ECD – toxin complex showing the α-bungarotoxin bound to αγ/γ and αδ/δ interfaces; Loop C, N-glycan, Loop F and C-terminus residues are showing as sphere in top view. (C, D) Interacting residues are shown as sticks colored by subunit and α-bungarotoxin as orange ribbon. α subunits are in green, γ subunit in blue and δ subunit in violet. Dashed lines indicate electrostatic interactions. Roman numerals indicate the three fingers of α-bungarotoxin.
Finger II from the toxin reaches under loop C, propping it open, and penetrates deeply into the ACh binding pocket (Figure 2A,B). Its R36 guanidinium group positions in the aromatic box that constitutes the ACh binding site, in a manner that would directly compete with acetylcholine binding (Figure 2C,D). This basic side chain contacts principal subunit aromatics in the orthosteric site (αY93, Y190, Y198), and stacks in a cation-π sandwich between αY198 from loop C and F32 from its own Finger II. F32 packs against αW149 and γW55 or δW57, which are important for ACh binding. The net result of these interactions is steric interference with neurotransmitter binding and stabilization of the interface with loop C in a splayed-open conformation. This observation is consistent with the emerging consensus that agonists stabilize a more compact ECD conformation with loop C packed down tightly, which translates to opening of the ion channel gate. Antagonists, being generally larger than agonists, prevent this capping of loop C and overall compaction of the ECD (Karlin, 1969).
There are important differences between the interaction of toxin with the ACh binding site in our structure and those in previous structures of soluble pentameric complexes (Bourne et al., 2005; Huang et al., 2013) or of single subunit complexes (Dellisanti et al., 2007; Zouridakis et al., 2014). The binding interface in the single-subunit-toxin complex is incompatible with a pentameric subunit complex with complementary subunits (Figure S5). The soluble homopentameric ECD crystal structures lack the interactions with the Cys-loop glycan. Loop C, due to an insertion conserved in Torpedo and muscle α1 subunits but absent in the soluble ECD, adopts a different backbone conformation. The γ and δ loop F insertions we observe contacting the toxin are absent in the homomeric structures. These latter soluble models might better represent binding of the toxin to the α7 nicotinic receptor, a homopentameric neuronal subtype sensitive to 3-finger toxins. All structures show a cation-π sandwich corresponding to αY198 – Bgtx_R36 – Bgtx_F32, which undoubtedly is central to α-neurotoxin binding to the nicotinic ACh receptor not only in Torpedo and striated muscle but also to the neuronal α7 nicotinic receptor.
The Torpedo receptor structure provides a blueprint for understanding α-toxin selectivity between receptor subtypes and between different species. Firstly, mammalian muscle receptors undergo a developmental switch in subunit expression giving rise to embryonic/neonatal receptors with γ subunits and adult receptors with ε in place of γ (Mishina et al., 1986; Takai et al., 1985; Witzemann et al., 1987). While α-γ and α-δ interfaces have similar α-bungarotoxin sensitivities, α-ε has a lower affinity for homologous 3-finger α-toxins (Osaka et al., 1999). Loop F is the only region displaying sequence divergence in toxin-contacting residues among the three complementary subunits, highlighting the importance of loop F in toxin recognition. Secondly, the α4β2 neuronal nicotinic receptor, the most abundant subtype in the brain, is insensitive to α-bungarotoxin (Sine et al., 2019). Mutating two residues (K189Y, I196P) on the α4 subunit converts α4β2 into an α-bungarotoxin-sensitive receptor (Sine et al., 2019). In the Torpedo receptor, Y189 (K189 in α4), directly interacts with α-bungarotoxin, spanning both Finger I (T8, S9 and I11) and Finger II (V40). Lysine in position 189 would sterically clash with the toxin, making the sensitivity from this substitution easy to rationalize. P197 (equivalent to I196 in α4) on Loop C packs against Y189, stabilizing it in a toxin-binding-competent conformation.
Further insight into selectivity arises from several species that have evolved insensitivity to snake α-neurotoxins. Mongooses and cobras have the substitution of W187N that introduces a glycosylation site on loop C. Honey badgers, hedgehogs and pigs are resistant to α-bungarotoxin due to substitution in the same position by arginine (Drabeck et al., 2015; Sine, 2012). In the Torpedo – toxin structure, W187 is sandwiched between the Cys-loop glycan and Finger I on the toxin (Figure S4), an interaction missing from the α1 subunit ECD model due to the overall rotation of the toxin (Figure S5C). Substitution of W187 with arginine or a glycosylated asparagine would sterically interfere with toxin binding. Taken together, the new high-resolution structural information complements and extends decades of functional and structural analysis to explain the exquisite sensitivity to three-fingered neurotoxins at the muscle nicotinic receptor.
Permeation pathway and conformational state
Binding of the neurotransmitter acetylcholine to its receptor promotes a conformational transition from a resting state with a closed ion channel to an activated receptor with an open ion channel. In the sustained presence of agonist, most ligand-gated ion channels desensitize, adopting an agonist-bound, closed pore conformation, a phenomenon first described in the muscle AChR (Katz and Thesleff, 1957). Competitive antagonists like α-bungarotoxin arrest the channel in a non-conducting state resembling the resting state (Moore and McCarthy, 1995). Decades of analysis on the Cys-loop receptor superfamily suggest that the resting and desensitization gates are distinct (Gielen et al., 2015; Nemecz et al., 2016; Wilson and Karlin, 2001), with a predominant view that the desensitization gate is near the cytosolic end of the channel, and the resting gate is near the midpoint of the pore (Gielen and Corringer, 2018). High-resolution structural information is available for two neuronal nicotinic receptors in presumed desensitized states (Gharpure et al., 2019; Morales-Perez et al., 2016; Walsh et al., 2018) but there was until now no high-resolution information for a resting-state-like nicotinic receptor.
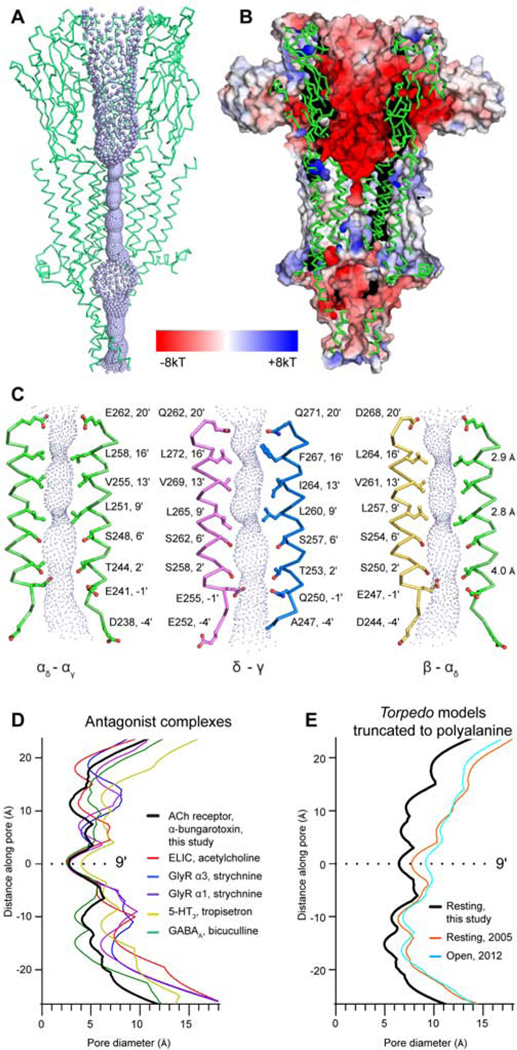
The ion permeation pathway in the Torpedo receptor – toxin complex comprises a wide extracellular vestibule, a tightly closed transmembrane channel, and an intracellular domain with polar side portals and an occluded axial pathway (Figure 3, 4). The electrostatic potential of the extracellular vestibule is strongly electronegative, which can serve to concentrate permeant cations (Hansen et al., 2008) (Figure 3B). M2 helices from each of the five subunits define the ion channel, where residues at the 2′, 6′, 9′, 13′, 16′ and 20′ positions face the lumen (Figure 3C). The M2 helices are oriented normal to the membrane plane and give rise to two prominent constriction points, one in the midpoint of the pore where the 9′Leu side chains orient toward the central axis (dmin = 2.8 Å), and one near the extracellular mouth of the pore, at the 16′ position (dmin = 2.9 Å). Both of these constrictions are hydrophobic and would be expected to exclude permeation of water and hydrated cations. Comparisons with other Cys-loop receptor structures in antagonist-bound states reveal conservation of the 9′Leu hydrophobic gate and variable degrees of constriction in the 16′ region (Basak et al., 2019; Du et al., 2015; Huang et al., 2015; Masiulis et al., 2019; Pan et al., 2012; Polovinkin et al., 2018) (Figure 3D, S6). The 9′Leu gate, formed by side chains oriented toward the pore axis, is generally conserved in apo receptor structures as well (Althoff et al., 2014; Basak et al., 2018b; Sauguet et al., 2014). Comparisons with the closely-related 5-HT3 receptor structures reveal differences, namely that constriction at the extracellular end of the pore is not observed in apo (Basak et al., 2018b) or antagonist-bound (Basak et al., 2019; Polovinkin et al., 2018) structures. A variable constriction is observed at the base of the pore in the 5-HT3 receptors, with a caveat that these −1′ constrictions are generated by glutamate side chains likely not resolvable at the resolutions of those datasets (4.3 (Polovinkin et al., 2018) and 4.5 Å (Basak et al., 2018b)). The Torpedo receptor pore is widely open at its intracellular end, as the −1′ side chains have rotated away from the central axis, where they provide negatively charged caps to the positively charged N-termini of the M2 helices in the neighboring subunits. This pore conformation is strikingly different from presumed desensitized-state structures of agonist-bound neuronal nicotinic receptors, where the pore is funnel-shaped tapering to a gate formed by side chains of −1′ residues (Gharpure et al., 2019; Morales-Perez et al., 2016; Walsh et al., 2018), but is consistent with the notion of distinct ion gates in these two states. Taken together, comparison of presumed resting-state pore conformations reveals substantial variability between receptor types, with the strictly conserved feature of a gate defined by the side chains at the hydrophobic 9′ position.
Figure 3. Permeation pathway.
(A) Axial ion permeation pathway with ribbon representation from three subunits (αγ-γ-αδ). Violet spheres represent solvent accessible surface. (B) Permeation pathway cutaway colored by electrostatic potential showing the transition from electronegative to uncharged at the gate and electropositive after the gate. (C) TMD pore profile from perspective of three pairs of opposing M2 α-helices. Minimal diameters are indicated at 3 positions on the right. (D) Pore diameter comparison of antagonist-bound Cys-loop receptors (ELIC, 3RQW; GlyR α3, 5CFB; GlyR α1, 3JAD; 5-HT3, 6HIS; GABAA, 6HUK). (E) Pore diameter comparison of toxin-bound Torpedo receptor with apo (PDB ID: 2BG9) and open state (PDB ID: 4AQ9) with side chains truncated. Structures are aligned with y=0 at the 9′ position.
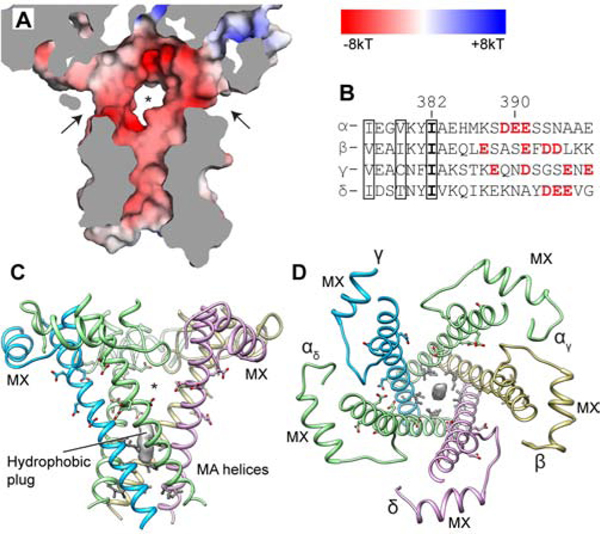
Figure 4. Permeation pathways in the ICD.
(A) ICD surface cutaway colored by electrostatic potential; arrows and asterisk indicate lateral portals for hydrated ion passage. (B) MA helix sequence alignment with charged portal-lining residues in red; boxed residues line the axial pathway and contact hydrophobic plug. (C, D) Side and top views of the ICD. Hydrophobic plug density is shown as gray surface. Negatively charged residues are shown as sticks; hydrophobic residues are in grey.
Earlier structures of the acetylcholine receptor from T. marmorata tubular crystals (Unwin, 2005) provide potential reference points for the α-toxin-bound receptor to understand gating. We confirm that these early models contain register discrepancies (Figure S7) in the transmembrane domain (Mnatsakanyan and Jansen, 2013). In order to compare pore conformations and to avoid ambiguity in the side chain positions, we truncated side chains to the alanine equivalent for these comparisons (Figure 3E, S7D). Notably, the pores of the earlier Torpedo receptor models (apo, 2BG9 (Unwin, 2005); open, 4AQ9 (Unwin and Fujiyoshi, 2012)) closely resemble each other and are substantially wider than in the toxin-bound structure (Figure 3E, S7 D,F). Systematic comparisons reveal that the toxin-bound structure is more similar to recent higher-resolution Cys-loop receptor structures (Figure S7–S9, SI Videos S1–4). Docking of the toxin bound model into earlier Torpedo receptor map suggests that these inconsistencies stem from magnification variances in processing of the earlier data combined with subunit registration differences (Figure S8), and further mechanistic inferences drawn from comparisons with the earlier Torpedo models were not attempted.
After passing the TMD pore, hydrated ions must pass through the ICD to the cytoplasm. Unwin and colleagues first proposed permeation through lateral portals (Figure 4A) in the ICD (Miyazawa et al., 1999), a concept supported by structure-function studies on the 5-HT3 and nicotinic receptors (Basak et al., 2018a; Gharpure et al., 2019; Kelley et al., 2003; Stokes et al., 2015). We observed a ‘hydrophobic plug’ of EM density in the lower axial ICD pathway, surrounded by hydrophobic residues including a conserved Ile residue (Figure 4 B–D). A recent structural and simulation analysis of the α3β4 nicotinic receptor suggested occupancy of this site with lipid promotes ICD stability (Gharpure et al., 2019). The portals are framed by the MX α-helix, post M3-loop and MA α-helices. All five portals are rich in polar and acidic resides that provide a favorable environment for hydrated cations to permeate. They are large enough, (8–12 Å between nearest atoms) to allow for hydrated sodium flux even in a presumed resting state, distinct from what was proposed for the 5-HT3 receptor (Basak et al., 2018a). Thus, as hydrated cations traverse the membrane through the nicotinic receptor, they enter the extracellular vestibule facilitated by the negative electrostatic surface potential and then encounter a hydrophobic constriction in the closed-pore state in the upper half of the transmembrane pore. Upon activation by neurotransmitter, this gate opens and becomes hydrated (Gupta et al., 2017), allowing passage through the pore and selectivity for cations based principally on the ring of negative charges at the −1′ position (Cymes and Grosman, 2016; Imoto et al., 1988; Wilson et al., 2000). Hydrated cations then exit through the portals rather than down the hydrophobic axial pathway. Both the extracellular vestibule and the intracellular portals provide determinants outside of the pore for fine-tuning of conductance and selectivity among cations.
Transduction mechanism, disease mutations and lipid sensitivity
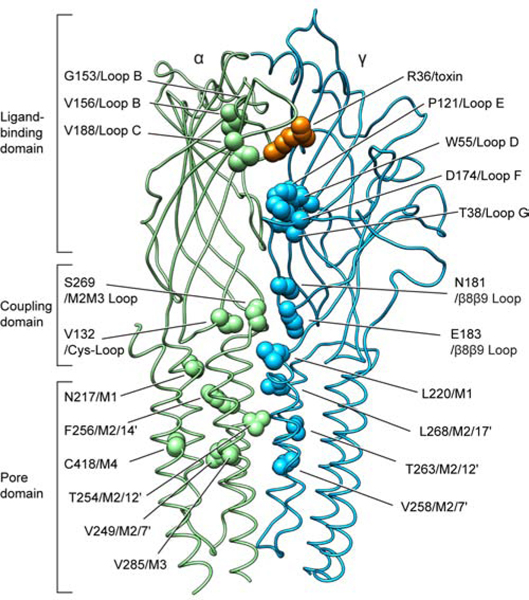
Mapping mutations that cause channel pathologies in the context of the new structural information allows for inferences into how agonist binding is coupled to channel gating. Many mutations in the muscle nicotinic receptor cause congenital muscle weakness (Engel et al., 2015). The mutations cluster into three principal zones mapped onto the Torpedo receptor structure (Figure 5 and SI Table 2). Ligand binding domain mutations that decrease sensitivity to acetylcholine, like εW55R (an important component of the aromatic box), result in attenuated channel activity dubbed the fast channel phenotype. By contrast, “slow-channel” mutants delay channel closing and result in larger integrated endplate currents. Mutations in the ligand binding domain that increase affinity for ACh, like αG153S and αV156M, are not found in the aromatic box where neurotransmitter binds but are positioned to exert indirect stabilization on the ACh-binding site.
Figure 5. Congenital myasthenic syndrome mutations in the receptor.
Amino acids identified in human fast- or slow-channel syndromes are shown as spheres mapped onto the Torpedo structure. Point mutations in the human muscle nicotinic receptor causing myasthenia are found predominantly on α1 and ε subunits. The majority of the residues at these positions are conserved in the Torpedo receptor α and γ subunits, respectively (α1/α: 80% and ε/γ: 56% amino acid sequence identity).
The interface between the ECD and TMD is the coupling zone through which agonist binding energy is relayed to the opening of the resting gate in the pore. Here the Cys-loop of the principal subunit and the β8β9 loop of the complementary subunit ECDs converge on the M2M3 loop of the principal subunit and the M1 helix of the complementary subunit TMDs (Figure 5). The new structural information sheds light on how mutations in all four of these coupling subdomains result in myasthenic syndromes. As representative examples, substitution of αV132L in the Cys-loop would destabilize a direct contact point with the M2M3 loop. γE183K on the β8β9 loop flips the side chain charge and would destabilize a local network of electrostatic interactions. Both of these mutants cause fast-channel syndromes consistent with a diminished coupling efficiency. As expected, the M2 pore-lining α-helix harbors many disease-related mutants. Interestingly, the toxin-bound presumed resting state structure reveals that M2 mutants in the α, γ and δ subunits do not face into the channel lumen but rather interact with the same or adjacent subunits in the TMD. These mutations, like αV249F and αT254I, would be expected to have indirect effects on the channel conformation by distorting local TMD structure and affecting gating transitions. The β subunit is an exception, with the conservative mutations in the pore-lining positions L9′M and V13′M, which cause slow-channel pathology.
A site removed from these three regional clusters is the αC418W point mutant in the peripheral M4 helix, which causes a gain of function and slow-channel syndrome (Domville and Baenziger, 2018). This cysteine sulfhydryl orients toward the core of the α subunit TMD helical bundle, in contrast to how it was modeled in the earlier Torpedo receptor structures (Unwin, 2005; Unwin and Fujiyoshi, 2012). Our structure shows αC418 interacting with residues in M1, consistent with mutagenesis studies (Domville and Baenziger, 2018). The M4 helix has been a focus in lipid sensitivity of the receptor due to its comparatively exposed position in the helical bundle (Henault et al., 2015). We observed density consistent with a bound lipid at the base of each α subunit M4, extending to the nearby αC418 residue; mutations in M4 that affect its interactions with the rest of the helical bundle may affect the lipid sensitivity of the receptor through this newly identified lipid site. Adjacent to this M4-interacting lipid, at the same γ-α interface, we observed density consistent with palmitate covalently linked to γC451, a finding supported by mutagenesis showing the importance of this residue in folding and trafficking of mature receptors (Li et al., 1990).
Conclusions
The muscle-type ACh receptor is the most extensively studied member of the Cys-loop receptor superfamily of ligand-gated ion channels. As such, structural information for this receptor forms an essential foundation for a comprehensive mechanistic understanding of channel function. The Torpedo receptor structure reveals the mechanism for α-toxin inhibition through direct competition at the neurotransmitter-binding site and stabilization of the transmembrane pore in a closed conformation. The transmembrane gates at L9′ and L/F16′ are the only obstructions for hydrated ion permeation, with the lateral portals in the intracellular domain remaining open. The quality of the underlying cryo-EM data in this study provides high confidence in atomic-level details underlying ligand recognition, ion permeation and how mutations linked to myasthenic syndromes cause pathologies in channel function. Comparisons with other resting-state structures in the superfamily reveal that fine details in structure-function, including positions of the channel obstruction in the resting state, are surprisingly variable among receptor subtypes. This structural diversity makes the pursuit of additional conformational states of the muscle-type receptor essential to achieving a complete structural and functional understanding of this archetypal ligand-gated ion channel.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding authors, Michael H. B. Stowell (stowellm@colorado.edu) and/or Ryan E. Hibbs (ryan.hibbs@utsouthwestern.edu).
Materials Availability
All unique reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
Cryo-EM maps and atomic model coordinates have been deposited in the EMDB and RCSB respectively (EMD-20928 and PDB ID 6UWZ).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Native source
The nicotinic receptor was purified from Tetronarce californica electric tissue (EastCoast Bio).
METHODS DETAILS
Receptor purification
Tissue frozen in liquid nitrogen (50 g) was thawed in 150 mL of buffer A (400 mM NaCl, 20 mM NaH2PO4, pH 7.4) with 75 mg of NEM (N-ethylmaleimide, Sigma) at room temperature. Subsequent steps were carried out at 4 °C or on ice. The tissue was homogenized for 2 min at top speed using a Kinematica Polytron Homogenizer (GmbH) and insoluble material was separated by centrifugation at 3,220 g for 15 min. The supernatant was filtered through cheesecloth (American Fiber & Finishing Inc.) in a beaker on ice. One complete protease inhibitor mini tablet (Sigma) was added and the suspension was gently stirred for 5 min. The membrane pellet was collected by centrifugation at 30,000 rpm in a Ti45 rotor for 30 min at 4 °C. Membranes were re-suspended in buffer B (80 mM NaCl, 20 mM Tris pH 11.0, 1 mM EDTA, 20% sucrose) using a Dounce homogenizer and were placed on ice for 30 min. The pellets were collected by centrifugation at 30,000 rpm in the Ti45 rotor for 30 min at 4 °C. The pellets were then washed 3 times with buffer C (80 mM NaCl, 20 mM NaH2PO4, pH 7.4) and stored at −80 °C.
The choice of detergent and added lipids are known to affect the ability to functionally reconstitute the Torpedo channel after purification (Baenziger and daCosta, 2013; Chak and Karlin, 1992). Historically, cholate was found to be effective in membrane extraction of receptors that could be functionally reconstituted, however this protein preparation aggregated after purification (Lindstrom et al., 1980). In contrast, Triton X-100 could be used in extraction and purification of receptors with superior biochemical behavior. While this Triton-based preparation resulted in channels that bound ligands with high affinity, ion channel activity could not be recovered after reconstitution into a membrane (Chak and Karlin, 1992; Lindstrom et al., 1980). Exchanging the receptor from Triton into cholate allowed for functional reconstitution, suggesting that the problem with Triton was at the reconstitution step and not in its use in extraction (McNamee et al., 1975). We observed the same profound aggregation with receptor extracted and purified in cholate as described in the literature. We thus took an approach of extracting in Triton and exchanging to DDM, which preserved both good biochemical behavior and the ability to reconstitute the functional receptor. The detailed procedure is as follows. Membrane pellets were thawed and re-suspended in 25x volume per weight of buffer C using a Dounce homogenizer. PMSF was added to a final concentration of 1 mM and Triton X-100 was added to a final concentration of 1.5% (v/v) and stirred for 1 hr. The supernatant was collected after centrifugation at 30,000 rpm in a Ti45 rotor for 30 min at 4 °C and was diluted 2X with buffer C. The affinity reagent ATM (2-[(4-aminobutanoyl)amino]-N,N,N-trimethylethanaminium) was synthesized and incorporated into NHS Sepharose resin (GE life sciences). The supernatant was mixed with 5 mL packed ATM resin while nutating for 1 hour at 4 °C. The resin was washed with buffer D (80 mM NaCl, 20 mM Tris pH 7.4, 1 mM EDTA, 1 mM n-dodecyl-β-D-maltoside, DDM, Anatrace) and eluted with buffer E (80 mM NaCl, 20mM Tris 7.4, 1mM EDTA, 1 mM DDM, 50 mM carbachol). For samples destined for cryo-EM grids, the eluted sample was treated with 50 mM βME (β-mercaptoethanol, Sigma) to produce monomeric receptors. The affinity chromatography elution samples were analyzed by size-exclusion chromatography monitoring intrinsic tryptophan fluorescence and concentrated to an A280 of ~7 with 100 kDa cutoff Amicon Ultra concentrators (Millipore).
Synthesis of the affinity reagent, ATM
The ATM (2-[(4-aminobutanoyl)amino]-N,N,N-trimethylethanaminium) affinity reagent was designed and synthesized in an effort to improve upon prior affinity reagent studies (Chak and Karlin, 1992; Sheng et al., 2010; Tierney et al., 2004).All reactions were carried out under an argon or nitrogen atmosphere with dry solvents using anhydrous conditions unless otherwise stated. 1H NMR (400 MHz) and 13C NMR (101 MHz) were recorded in C6D6 (internal standard: 7.15 ppm, 1H; 128.26 ppm, 13C), in THF-d4 (internal standard: 3.58 ppm, 1H; 67.57 ppm, 13C), in CDCl3 (internal standard: 7.26 ppm, 1H; 77.00 ppm, 13C), in MeCN-d3 (internal standard: 1.94 ppm, 1H; 118.3 ppm, 13C), in DMSO-d6 (internal standard: 2.50 ppm, 1H; 39.52 ppm, 13C), in MeOD-d3 (internal standard: 3.31 ppm, 1H; 49.15 ppm, 13C), on a Bruker DRX-400MHz spectrometer. Chemical shifts (δ) were reported as parts per million (ppm) and the following abbreviations were used to identify the multiplicities: s = singlet, d = doublet, t = triplet, q = quartet, sept. = septet, m = multiplet, b = broad and all combinations thereof can be explained by their integral parts. Column chromatography was carried out employing EMD (Merck) Geduran Silica Gel 60 (40–63 μm) with the indicated solvent mixtures. Infrared spectra were recorded using a Perkin Elmer Spectrum 100-FTIR spectrometer. Melting points were determined using a Thomas-Hoover capillary melting apparatus and are uncorrected. All chemicals were obtained from commercial sources and used as received unless otherwise noted within the context of use. The following chemicals were obtained from various commercial sources with varying purities and in all cases were used as received: 2-pyrrolidone (Chem-Impex, 99.3%), di-tert-butyl decarbonate (Boc2O, Oakwood Chemical, 99%), 4-dimethylaminopyridine (DMAP, Chem-Impex, 99%), N,N-dimethylethylenediamine (Aldrich, 98%), iodomethane (MeI, Aldrich, stabilized with copper, 99%), hydrogen chloride solution 4.0 M in dioxane (Oakwood Chemical). All deuterated solvents utilized in this study (C6D6, THF-d4, CDCl3, MeCN-d3, DMSO-d6, and MeOD-d3) were obtained from Cambridge Isotope Laboratories, Inc. and were used as received. The overview of the ATM synthesis and representative NMR spectra are included in Figure SI 3.

Synthesis of compound 1a: To a 500 mL round-bottomed flask equipped with a magnetic stir bar was added 5.00 grams (4.46 mL, 58.8 mmol, 1.00 equiv) of 2-pyrrolidinone and this was diluted with 120 mL (0.50 M) of reagent grade acetonitrile (MeCN). To this stirring solution was added 718 mgs (5.88 mmol, 0.10 equiv, 10.0 mol%) of DMAP followed by 15.4 grams (70.6 mmol, 1.20 equiv) of di-tert-butyl decarbonate (Boc2O) which was added in a single portion. The stirring yellow solution was capped with a 24/40 polyethylene stopper and stirred at room temperature overnight. After this period the volatiles were removed under reduced pressure and the residue was re-diluted with EtOAc (~150 mL), extracted with water (~150 mL, 2X), then brine (~150 mL, 1X), dried over Na2SO4, filtered, and concentrated to afford a crude yellow residue. This crude residue was purified by column chromatography (30% → 50% → 60% EtOAc/hexanes), concentration of fractions containing the desired material (Rf = 0.32, 50% EtOAc/hexanes, visualized by KMnO4) afforded 8.22 g (76%) of the title compound 1a as a yellow oil which was used in the next step with no further purifications.
Compound 1a:
yellow oil; 76% yield; Rf = 0.32 (TLC conditions: 50% EtOAc/hexanes, visualized by KMnO4); 1H NMR (300 MHz, CDCl3) δ = 3.74 (t, J = 7.1 Hz, 2H), 2.51 (t, J = 8.1 Hz, 2H), 1.99 (p, J = 7.8 Hz, 2H), 1.52 (bs, 9H); 13C NMR (70 MHz, CDCl3) δ = 174.39, 150.35, 82.85, 46.57, 33.06, 28.13, 17.51.
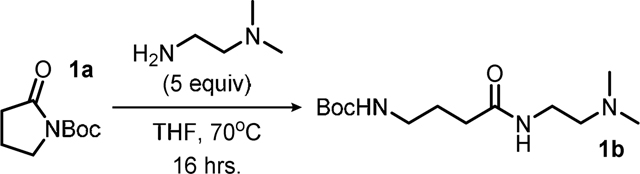
Synthesis of compound 1b: To a 250 mL round-bottomed flask equipped with a magnetic stir bar was added 5.00 grams (27.0 mmol, 1.00 equiv) of compound 1a and this was diluted with 27.0 mL (1.00 M) of reagent grade tetrahydrofuran (THF). To this stirring solution was added 14.8 mL (11.9 grams, 135.1 mmol, 5.00 equiv) of N,N-dimethylethylenediamine via syringe in a single portion, the flask was then equipped with a reflux condenser, placed into an equilibrated 70oC oil bath, and refluxed overnight. After this period the reaction was cooled to room temperature and the volatiles were removed under reduced pressure to afford 7.38 grams (quantitative) of the title compound 1b as an orange oil which was used in the next step with no further purifications.
Compound 1b:
orange oil; quantitative yield; Rf = 0.13 (TLC conditions: 10% MeOH/DCM, visualized by KMnO4); 1H NMR (300 MHz, CDCl3) δ = 6.25 (bs, 1H), 4.89 (bs, 1H), 3.31 (q, J = 5.9 Hz, 2H), 3.14 (q, J = 6.4 Hz, 2H), 2.40 (t, J = 6.1 Hz, 2H), 2.23 – 2.18 (m, 8H), 1.80 (p, J = 6.9 Hz, 2H), 1.42 (s, 9H); 13C NMR (70 MHz, CDCl3) δ = 172.68, 156.37, 79.26, 57.94, 45.25, 40.07, 36.94, 33.86, 28.54, 26.16.
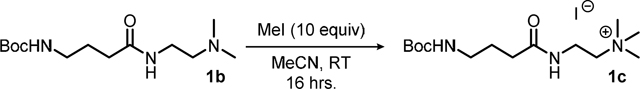
Synthesis of compound 1c: To a 100 mL round-bottomed flask equipped with a magnetic stir bar was added 7.38 grams (27.0 mmol, 1.00 equiv) of compound 1b and this was diluted with 30.0 mL (1.00 M) of reagent grade acetonitrile (MeCN). To this stirring solution was added 16.8 mL (38.3 grams, 270 mmol, 10.0 equiv) of iodomethane (MeI) and the yellow/orange solution was stirred overnight at room temperature. After this period the volatiles were removed under reduced pressure to afford a viscous yellow residue that was diluted with ~150 mL of diethylether (Et2O) and stirred rapidly overnight at room temperature. After this period the material was noted to have solidified and the precipitate was filtered, washed with additional portions of Et2O (~25.0 mL, 2X), and the filtrate was discarded. The remaining hydroscopic solid was dissolved in ~150 mL of DCM, transferred to a tared 500 mL round-bottomed flask, and the volatiles were removed under reduced pressure to afford 11.2 grams (quantitative) of the title compound 1c as a hygroscopic orange foam which was used in the next step with no further purifications.
Compound 1c:
hygroscopic orange foam; quantitative yield; Rf = n/a; 1H NMR (300 MHz, CDCl3) δ = 7.92 (t, J = 5.6 Hz, 1H), 4.99 (bs, 1H), 3.87 – 3.75 (m, 4H), 3.45 (s, 9H), 3.12 (bs, 2H), 2.33 (t, J = 7.2 Hz, 2H), 1.81 (p, J = 7.14 Hz, 2H), 1.42 (s, 9H); 13C NMR (70 MHz, CDCl3) δ = 174.00, 156.36, 79.34, 65.69, 40.09, 34.24, 33.57, 28.62, 25.99.
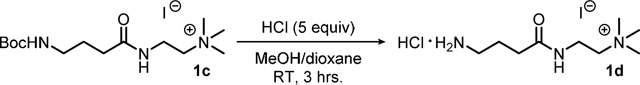
Synthesis of compound 1d: To a 250 mL round-bottomed flask equipped with a magnetic stir bar was added 10.5 grams (25.3 mmol, 1.00 equiv) of compound 1c (due to the hygroscopic nature of this material, it was found to best concentrate a methanol solution of compound 1c in a previously tared flask) and this was diluted with 25.0 mL (1.00 M) of reagent grade methanol (MeOH). To this stirring solution was added 31.6 mL (126 mmol, 5.00 equiv) of HCl (4.00 M in dioxane) in a single portion and the homogenous orange reaction was allowed to stir at room temperature with the flask left open to air to allow for evolving gas to escape. After ~3 hours an off-white solid was noted to have precipitated, the precipitate was filtered, washed with several portions of EtOAc (3X, ~25.0 mL), and allowed to dry under vacuum to afford 4.80 grams (54%) of the title compound 1d as an off-white solid which was used directly in affinity column preparation procedure with no further purifications.
Compound 1d:
off-white solid; 54% yield; Rf = n/a; 1H NMR (300 MHz, MeOD-d3) δ = 3.68 (t, J = 7.19 Hz, 2H), 3.51 (t, J = 6.46 Hz, 2H), 3.21 (bs, 9H), 2.99 (t, J = 7.77 Hz, 2H), 2.42 (t, J = 7.12 Hz, 2H), 1.95 (p, J = 7.40 Hz, 2H; 13C NMR (70 MHz, MeOD-d3) δ = 174.99, 65.77, 54.03, 40.36, 34.67, 33.49, 24.17.
Generation of affinity resin
NHS Sepharose 25 mL (GE Life Sciences) was gently vacuum filtered using a Steriflip 50 mL disposable vacuum filtration system (0.22 μm Millipore Express) and washed with 200 mL of cold MilliQ water, ensuring that the slurry did not dry out. The slurry was then transferred to a 250 mL centrifuge bottle with freshly-prepared 2.2 mM of ATM dissolved in 75 mL of 100 mM HEPES, pH 8.0 and nutating for 2 hrs at 4 °C. The slurry was gently vacuum filtered and washed with 400 mL of cold MilliQ water. The slurry was then re-suspended in 20% ethanol and stored at 4 °C where it was found to be stable for > 3 months.
Liposome patch-clamp electrophysiology
For proteoliposome patch-clamp experiments, receptors were extracted and affinity-purified as for cryo-EM leading up to nanodisc reconstitution, as described above, and further purified by using size-exclusion chromatography (Superose 6 Increase 10/300 GL column, GE Healthcare) in SEC buffer (80 mM NaCl, 20mM Tris pH 7.4, 1 mM DDM). For the case of experiments involving apo receptor, elution from affinity resin was accomplished by addition of 20 mM Tris pH 7.4, 1.6 M NaCl (rather than carbachol), 1 mM EDTA and 1 mM DDM. Soy polar lipids in chloroform (Avanti) were dried in a test tube under a stream of argon while rotating the tube to make a homogeneous thin lipid film. The lipid film was further dried under vacuum for 2 hours, then resuspended to 10 mg/mL with 20 mM Tris pH 7.4, 80 mM NaCl (TBS). To make uniform lipid vesicles, the lipid resuspension was sonicated for 15 min. Purified receptor (4 μg) was added into lipid vesicles in a protein to lipid mass ratio of 1:100 to 1:1000 (w/w). The mixture was rotated at room temperature for 1 hour to allow the protein to incorporate into lipid vesicles. Detergent was removed by incubating with Bio-Beads SM-2 (Bio-Rad) and the resultant liposomes were collected by ultracentrifugation, 4 °C, 30 min at 186,000 g. The pellet was resuspended in 6 μL TBS buffer. 2 μL of the suspension was spotted on a glass coverslip, and then desiccated overnight under vacuum at 4°C. Desiccated liposomes were rehydrated with 5 μL of buffer (320 mM sucrose, 10 mM KCl, and 2 mM MgCl2, 5 mM Hepes pH 8.0) for at least 2 hours at 4°C, and then used for patch-clamp recording. Torpedo receptor channel activity was examined in excised liposome patches. Data were acquired at +50, −50 and −75 mV at a sampling rate of 50 kHz with a 10 kHz filter, using an Axopatch 200b amplifier and pClamp10 software. The bath solution contained (in mM): 200 KCl, 40 MgCl2, 2 CaCl2, and 5 Hepes pH 7.3. Pipettes were filled with the same bath solution and initial pipette resistances ranged from 4–8 MΩ. After a stable baseline was observed, agonist (nicotine or carbachol) in bath solution was added to the bath to achieve a final concentration within a range of 15–60 μM. To block the channel activity, α-bungarotoxin was added in a similar manner to 2.5–30 μM. For experiments in Fig. 1B, n=8 patches and in Fig. 1C, n=2 patches.
Nanodisc reconstitution for cryo-EM
The Saposin A expression plasmid was provided by Salipro Biotech and the reconstitution protocol was modified from (Gharpure et al., 2019). The receptor was mixed with saposin and soy polar lipid extract (Avanti) in a molar ratio of 1:27:138. The detergent (DDM) was removed using Bio-Beads SM2 (Bio-Rad). The reconstituted receptor was polished using size-exclusion chromatography (Superose 6 Increase 10/300 GL column, GE Healthcare) in TBS to obtain a homogenous sample and remove empty nanodiscs and βME.
Cryo-EM sample preparation and data collection
The SEC-purified, reconstituted receptor was mixed with α-bungarotoxin (Vincent et al., 1998) (Tocris) to a 3–4 fold molar excess of receptor sites, and the complex was concentrated to an A280 of ~6.4. This sample was supplemented with 1 mM fluorinated Fos-Choline-8 (Anatrace) immediately prior to freezing to induce random orientations in the grid holes. Protein sample (3 μL) was applied to glow-discharged gold R1.2/1.3 300 mesh holey carbon grids (Quantifoil) and immediately blotted for 4 s at 100% humidity and 4°C before being plunge-frozen into liquid ethane cooled by liquid nitrogen using a Vitrobot Mark IV (FEI). Samples were screened on the Talos Arctica at UT Southwestern and the final dataset was collected on a 300 kV Titan Krios microscope (FEI) at the PNCC at OHSU (Portland, OR) equipped with a post-GIF K3 with 10 eV energy filter slit.
Cryo-EM data processing
The cryo-EM data were processed using the general workflow in RELION 3.0 (Zivanov et al., 2018). 7,426 movie stacks were gain normalized, 2x Fourier binned (pixel size 0.648 Å/pix), aligned, dose weighted, and summed with MotionCor2 (Zheng et al., 2017). Defocus value estimation and contrast transfer function correction were done with GCTF (Zhang, 2016). Approximately 150 particles were manually picked and subjected to 2D classification to generate 2D references. The 2D references were then used for auto-picking from a few micrographs to obtain improved 2D references for auto-picking. Auto-picking yielded 1,126,590 particles, from which 418,479 particles were selected by 2D classification. These particles were then used to generate a de novo initial model for 3D classification, which subsequently yielded 355,616 “good” particles. The best 3D class was low-pass filtered to 40 Å and used as an initial model for the 3D refinement followed by CTF refinement. Particles were then polished and used for another round of 3D classification. The 3D classes resulted in two distinct groups based on the presence vs. absence of ICD density. Refinement using particles from 3D classes with strong ICD signal (127,482 particles) resulted in a final map with an overall FSC = 0.143 resolution of 2.69 Å.
Model building, refinement and validation
The initial structural model was generated by Swissmodel (Schwede et al., 2003) using the α3β4 nicotinic receptor coordinates (PDB ID: 6PV7 (Gharpure et al., 2019)) as a template and α-bungarotoxin (SI Video 5) was built starting from PDB ID: 2QC1 (Dellisanti et al., 2007). Iterative cycles of manual building in Coot (Emsley et al., 2010) with global real space refinement in Phenix (Afonine et al., 2018) resulted in good stereochemistry and model-map correlation assessed by Molprobity and Phenix validation tools (Table SI1). The density map quality allowed modeling of 2032 out of the 2333 expected residues. The map for the C-terminal end of the MX helix in the γ subunit was ambiguous and this part of the model was not built. Noteworthy, the map resolution was sufficient to clearly observe that the C192-C193 disulfide bond is broken in the αδ subunit but not in the αγ subunit (Figure 2, Figure S2D). The most probable cause is radiation damage (Hattne et al., 2018) or utilization of the βME during sample preparation; no changes in backbone conformation or other interactions were observed as a consequence of the broken bond (as assessed by comparison of the two α subunits). In α subunits, the C-termini extending out of the membrane were poorly resolved and side chain positions were built with less confidence than in the rest of the model. Sequences were retrieved from the UniProtKB database (The UniProt, 2017) and sequence alignments were made using Clustal Omega (Madeira et al., 2019). Interface areas were analyzed by PDBePISA server (Krissinel and Henrick, 2007). The pore diameter was calculated by HOLE (Smart et al., 1996). The map and structural figures were generated using UCSF Chimera (Pettersen et al., 2004) and Pymol (Schrodinger, LLC) including the APBS electrostatics plugin (Baker et al., 2001).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| Electroplaque tissue from Torpedo californica | EastCoast Bio | Cat#DZ800 |
| Chemicals, Peptides | ||
| 2-[(4-aminobutanoyl)amino]-N,N,N-trimethylethanaminium (ATM) | This paper | N/A |
| N-ethylmaleimide (NEM) | Sigma | Cat#E3876 |
| cOmplete Mini | Sigma | Cat#11836153001 |
| Ethylenediaminetetraacetic acid (EDTA) | Sigma | Cat#E9884 |
| Sucrose | Sigma | Cat#S0389 |
| Triton X-100 | Acros | Cat#327371000 |
| n-Dodecyl-β-D-Maltopyranoside (DDM) | Anatrace | Cat#D310 |
| 2-Mercaptoethanol (βME) | Sigma | Cat#M6250 |
| Soy extract polar lipids | Avanti | Cat#541602 |
| Fos-choline-8, fluorinated | Anatrace | Cat#F300F |
| α-Bungarotoxin | Tocris | Cat#2133 |
| Critical Commercial Assays | ||
| NHS-activated Sepharose 4 Fast Flow | GE Healthcare | Cat#17–0906-01 |
| Superose 6 Increase 10/300 GL | GE Healthcare | Cat#29–0915-96 |
| Deposited Data | ||
| Coordinates of Torpedo nicotinic receptor in complex with α-bungarotoxin | This paper | PDB: 6UWZ |
| Cryo-EM map of Torpedo nicotinic receptor in complex with α-bungarotoxin | This paper | EMDB: EMD-20928 |
| Software and Algorithms | ||
| Serial EM | Mastronarde, 2005 | http://bio3d.colorado.edu/SerialEM/ |
| MotionCor2 | Zheng et al., 2017 | https://emcore.ucsf.edu/cryoem-software |
| GCTF | Zhang, 2016 | https://www.mrc-lmb.cam.ac.uk/kzhang/Gctf/ |
| Relion 3.0 | Zivanov et al., 2018 | https://www3.mrc-lmb.cam.ac.uk/relion/ |
| Swissmodel | Schwede et al., 2003 | https://www.swissmodel.expasy.org/ |
| UCSF Chimera | Petterson et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| Coot | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Phenix | Afonine et al., 2018 | https://www.phenix-online.org/ |
| Pymol | Schrodinger, LLC | http://www.pymol.org/ |
| HOLE | Smart et al., 1996 | http://www.holeprogram.org/ |
| pClamp 10 | Molecular Devices | https://www.moleculardevices.com/products/axon-patch-clamp-system/acquisition-and-analysis-software/pclamp-software-suite |
| Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Others | ||
| Quantifoil R1.2/1.3 Au 300 mesh | Electron Microscopy Sciences | Cat#Q3100AR1.3 |
Highlights.
High-resolution structure of a native muscle-type nicotinic acetylcholine receptor
Previously unresolved structural elements contribute to neurotoxin binding
Channel is stabilized in a closed conformation by α-bungarotoxin from snake venom
Transduction mechanism and myasthenic disease mutations
Acknowledgements
Single-particle cryo-EM grids were screened at the University of Texas Southwestern Medical Center Cryo-Electron Microscopy Facility, which is supported by the CPRIT Core Facility Support Award RP170644. We thank Craig Yoshioka for cryo-EM data collection at the PNCC under user proposal 50839 and Guipeun Kang for assistance with lipid reconstitution experiments. A portion of this research was supported by NIH grant U24GM129547 and performed at the PNCC at OHSU and accessed through EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research. Initial samples from the affinity column were screened at University of Colorado Boulder EM Services Core Facility. This work was supported by grants from the NIH (DA037492, DA042072, and NS095899 to R.E.H. and AG061829 to M.H.B.S) and the MCDB Neurodegenerative Disease Fund to M.H.B.S.
Footnotes
Competing interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, and Adams PD (2018). Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althoff T, Hibbs RE, Banerjee S, and Gouaux E. (2014). X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors. Nature 512, 333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anholt R, Lindstrom J, and Montal M. (1980). Functional equivalence of monomeric and dimeric forms of purified acetylcholine receptors from Torpedo californica in reconstituted lipid vesicles. European journal of biochemistry 109, 481–487. [DOI] [PubMed] [Google Scholar]
- Baenziger JE, and daCosta CJB (2013). Molecular mechanisms of acetylcholine receptor-lipid interactions: from model membranes to human biology. Biophys Rev 5, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, and McCammon JA (2001). Electrostatics of nanosystems: application to microtubules and the ribosome. Proceedings of the National Academy of Sciences of the United States of America 98, 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak S, Gicheru Y, Kapoor A, Mayer ML, Filizola M, and Chakrapani S. (2019). Molecular mechanism of setron-mediated inhibition of full-length 5-HT3A receptor. Nat Commun 10, 3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak S, Gicheru Y, Rao S, Sansom MSP, and Chakrapani S. (2018a). Cryo-EM reveals two distinct serotonin-bound conformations of full-length 5-HT3A receptor. Nature 563, 270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak S, Gicheru Y, Samanta A, Molugu SK, Huang W, Fuente M, Hughes T, Taylor DJ, Nieman MT, Moiseenkova-Bell V, et al. (2018b). Cryo-EM structure of 5-HT3A receptor in its resting conformation. Nature communications 9, 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne Y, Talley TT, Hansen SB, Taylor P, and Marchot P. (2005). Crystal structure of a Cbtx-AChBP complex reveals essential interactions between snake alpha-neurotoxins and nicotinic receptors. The EMBO journal 24, 1512–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, and Sixma TK (2001). Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature 411, 269–276. [DOI] [PubMed] [Google Scholar]
- Cartaud J, Benedetti EL, Cohen JB, Meunier JC, and Changeux JP (1973). Presence of a lattice structure in membrane fragments rich in nicotinic receptor protein from the electric organ of Torpedo marmorata. FEBS letters 33, 109–113. [DOI] [PubMed] [Google Scholar]
- Chak A, and Karlin A. (1992). Purification and reconstitution of nicotinic acetylcholine receptor. Methods in enzymology 207, 546–555. [DOI] [PubMed] [Google Scholar]
- Chang CC, and Lee CY (1963). Isolation of Neurotoxins from the Venom of Bungarus Multicinctus and Their Modes of Neuromuscular Blocking Action. Arch Int Pharmacodyn Ther 144, 241–257. [PubMed] [Google Scholar]
- Changeux J-P, and Edelstein SJ (2005). Nicotinic acetylcholine receptors : from molecular biology to cognition, 1st edn (New York: Odile Jacob; ). [Google Scholar]
- Changeux JP, Kasai M, and Lee CY (1970). Use of a snake venom toxin to characterize the cholinergic receptor protein. Proceedings of the National Academy of Sciences of the United States of America 67, 1241–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudio T, Ballivet M, Patrick J, and Heinemann S. (1983). Nucleotide and deduced amino acid sequences of Torpedo californica acetylcholine receptor gamma subunit. Proceedings of the National Academy of Sciences of the United States of America 80, 1111–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun D, and Hawkes AG (1977). Relaxation and fluctuations of membrane currents that flow through drug-operated channels. Proc R Soc Lond B Biol Sci 199, 231–262. [DOI] [PubMed] [Google Scholar]
- Cymes GD, and Grosman C. (2016). Identifying the elusive link between amino acid sequence and charge selectivity in pentameric ligand-gated ion channels. Proceedings of the National Academy of Sciences of the United States of America 113, E7106–E7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellisanti CD, Yao Y, Stroud JC, Wang ZZ, and Chen L. (2007). Crystal structure of the extracellular domain of nAChR alpha1 bound to alpha-bungarotoxin at 1.94 A resolution. Nature neuroscience 10, 953–962. [DOI] [PubMed] [Google Scholar]
- Devillers-Thiery A, Giraudat J, Bentaboulet M, and Changeux JP (1983). Complete mRNA coding sequence of the acetylcholine binding alpha-subunit of Torpedo marmorata acetylcholine receptor: a model for the transmembrane organization of the polypeptide chain. Proceedings of the National Academy of Sciences of the United States of America 80, 2067–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domville JA, and Baenziger JE (2018). An allosteric link connecting the lipid-protein interface to the gating of the nicotinic acetylcholine receptor. Sci Rep 8, 3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabeck DH, Dean AM, and Jansa SA (2015). Why the honey badger don’t care: Convergent evolution of venom-targeted nicotinic acetylcholine receptors in mammals that survive venomous snake bites. Toxicon : official journal of the International Society on Toxinology 99, 68–72. [DOI] [PubMed] [Google Scholar]
- Du J, Lu W, Wu S, Cheng Y, and Gouaux E. (2015). Glycine receptor mechanism elucidated by electron cryo-microscopy. Nature 526, 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K. (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel AG, Shen XM, Selcen D, and Sine SM (2015). Congenital myasthenic syndromes: pathogenesis, diagnosis, and treatment. Lancet Neurol 14, 420–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein M, and Racker E. (1978). Reconstitution of carbamylcholine-dependent sodium ion flux and desensitization of the acetylcholine receptor from Torpedo californica. The Journal of biological chemistry 253, 6660–6662. [PubMed] [Google Scholar]
- Frauenfeld J, Loving R, Armache JP, Sonnen AF, Guettou F, Moberg P, Zhu L, Jegerschold C, Flayhan A, Briggs JA, et al. (2016). A saposin-lipoprotein nanoparticle system for membrane proteins. Nature methods 13, 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharpure A, Teng J, Zhuang Y, Noviello CM, Walsh RM Jr., Cabuco R, Howard RJ, Zaveri NT, Lindahl E, and Hibbs RE (2019). Agonist Selectivity and Ion Permeation in the alpha3beta4 Ganglionic Nicotinic Receptor. Neuron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen M, and Corringer PJ (2018). The dual-gate model for pentameric ligand-gated ion channels activation and desensitization. J Physiol 596, 1873–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen M, Thomas P, and Smart TG (2015). The desensitization gate of inhibitory Cys-loop receptors. Nature communications 6, 6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Chakraborty S, Vij R, and Auerbach A. (2017). A mechanism for acetylcholine receptor gating based on structure, coupling, phi, and flip. The Journal of general physiology 149, 85–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton SL, McLaughlin M, and Karlin A. (1979). Formation of disulfide-linked oligomers of acetylcholine receptor in membrane from torpedo electric tissue. Biochemistry 18, 155–163. [DOI] [PubMed] [Google Scholar]
- Hansen SB, Wang HL, Taylor P, and Sine SM (2008). An ion selectivity filter in the extracellular domain of Cys-loop receptors reveals determinants for ion conductance. The Journal of biological chemistry 283, 36066–36070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel M, Kasher R, Nicolas A, Guss JM, Balass M, Fridkin M, Smit AB, Brejc K, Sixma TK, Katchalski-Katzir E, et al. (2001). The binding site of acetylcholine receptor as visualized in the X-Ray structure of a complex between alpha-bungarotoxin and a mimotope peptide. Neuron 32, 265–275. [DOI] [PubMed] [Google Scholar]
- Hattne J, Shi D, Glynn C, Zee CT, Gallagher-Jones M, Martynowycz MW, Rodriguez JA, and Gonen T. (2018). Analysis of Global and Site-Specific Radiation Damage in Cryo-EM. Structure 26, 759–766 e754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henault CM, Sun J, Therien JP, daCosta CJ, Carswell CL, Labriola JM, Juranka PF, and Baenziger JE (2015). The role of the M4 lipid-sensor in the folding, trafficking, and allosteric modulation of nicotinic acetylcholine receptors. Neuropharmacology 96, 157–168. [DOI] [PubMed] [Google Scholar]
- Huang S, Li SX, Bren N, Cheng K, Gomoto R, Chen L, and Sine SM (2013). Complex between alpha-bungarotoxin and an alpha7 nicotinic receptor ligand-binding domain chimaera. The Biochemical journal 454, 303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Chen H, Michelsen K, Schneider S, and Shaffer PL (2015). Crystal structure of human glycine receptor-alpha3 bound to antagonist strychnine. Nature 526, 277–280. [DOI] [PubMed] [Google Scholar]
- Imoto K, Busch C, Sakmann B, Mishina M, Konno T, Nakai J, Bujo H, Mori Y, Fukuda K, and Numa S. (1988). Rings of negatively charged amino acids determine the acetylcholine receptor channel conductance. Nature 335, 645–648. [DOI] [PubMed] [Google Scholar]
- Karlin A. (1969). Chemical modification of the active site of the acetylcholine receptor. The Journal of general physiology 54, 245–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin A. (2002). Emerging structure of the nicotinic acetylcholine receptors. Nature reviews Neuroscience 3, 102–114. [DOI] [PubMed] [Google Scholar]
- Karlin A, Holtzman E, Yodh N, Lobel P, Wall J, and Hainfeld J. (1983). The arrangement of the subunits of the acetylcholine receptor of Torpedo californica. The Journal of biological chemistry 258, 6678–6681. [PubMed] [Google Scholar]
- Karlin A, Prives J, Deal W, and Winnik M. (1971). Affinity labeling of the acetylcholine receptor in the electroplax. Journal of molecular biology 61, 175–188. [DOI] [PubMed] [Google Scholar]
- Katz B, and Thesleff S. (1957). A study of the desensitization produced by acetylcholine at the motor end-plate. J Physiol 138, 63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley SP, Dunlop JI, Kirkness EF, Lambert JJ, and Peters JA (2003). A cytoplasmic region determines single-channel conductance in 5-HT3 receptors. Nature 424, 321–324. [DOI] [PubMed] [Google Scholar]
- Klymkowsky MW, and Stroud RM (1979). Immunospecific identification and three-dimensional structure of a membrane-bound acetylcholine receptor from Torpedo californica. Journal of molecular biology 128, 319–334. [DOI] [PubMed] [Google Scholar]
- Krissinel E, and Henrick K. (2007). Inference of macromolecular assemblies from crystalline state. Journal of molecular biology 372, 774–797. [DOI] [PubMed] [Google Scholar]
- Langley JN (1905). On the reaction of cells and of nerve-endings to certain poisons, chiefly as regards the reaction of striated muscle to nicotine and to curari. J Physiol 33, 374–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard RJ, Labarca CG, Charnet P, Davidson N, and Lester HA (1988). Evidence that the M2 membrane-spanning region lines the ion channel pore of the nicotinic receptor. Science 242, 1578–1581. [DOI] [PubMed] [Google Scholar]
- Lester HA (1972). Blockade of acetylcholine receptors by cobra toxin: electrophysiological studies. Mol Pharmacol 8, 623–631. [PubMed] [Google Scholar]
- Li L, Schuchard M, Palma A, Pradier L, and McNamee MG (1990). Functional role of the cysteine 451 thiol group in the M4 helix of the gamma subunit of Torpedo californica acetylcholine receptor. Biochemistry 29, 5428–5436. [DOI] [PubMed] [Google Scholar]
- Lindstrom J, Anholt R, Einarson B, Engel A, Osame M, and Montal M. (1980). Purification of acetylcholine receptors, reconstitution into lipid vesicles, and study of agonist-induced cation channel regulation. The Journal of biological chemistry 255, 8340–8350. [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47, W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiulis S, Desai R, Uchanski T, Serna Martin I, Laverty D, Karia D, Malinauskas T, Zivanov J, Pardon E, Kotecha A, et al. (2019). GABAA receptor signalling mechanisms revealed by structural pharmacology. Nature 565, 454–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamee MG, Weill CL, and Karlin A. (1975). Purification of acetylcholine receptor from Torpedo californica and its incorporation into phospholipid vesicles. Annals of the New York Academy of Sciences 264, 175–182. [DOI] [PubMed] [Google Scholar]
- Miledi R, Molinoff P, and Potter LT (1971). Isolation of the cholinergic receptor protein of Torpedo electric tissue. Nature 229, 554–557. [DOI] [PubMed] [Google Scholar]
- Mishina M, Takai T, Imoto K, Noda M, Takahashi T, Numa S, Methfessel C, and Sakmann B. (1986). Molecular distinction between fetal and adult forms of muscle acetylcholine receptor. Nature 321, 406–411. [DOI] [PubMed] [Google Scholar]
- Miyazawa A, Fujiyoshi Y, Stowell M, and Unwin N. (1999). Nicotinic acetylcholine receptor at 4.6 A resolution: transverse tunnels in the channel wall. Journal of molecular biology 288, 765–786. [DOI] [PubMed] [Google Scholar]
- Mnatsakanyan N, and Jansen M. (2013). Experimental determination of the vertical alignment between the second and third transmembrane segments of muscle nicotinic acetylcholine receptors. J Neurochem 125, 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MA, and McCarthy MP (1995). Snake venom toxins, unlike smaller antagonists, appear to stabilize a resting state conformation of the nicotinic acetylcholine receptor. Biochim Biophys Acta 1235, 336–342. [DOI] [PubMed] [Google Scholar]
- Morales-Perez CL, Noviello CM, and Hibbs RE (2016). X-ray structure of the human alpha4beta2 nicotinic receptor. Nature 538, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachmansohn D, Coates CW, and Cox RT (1941). Electric Potential and Activity of Choline Esterase in the Electric Organ of Electrophorus Electricus (Linnaeus). The Journal of general physiology 25, 75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson N, Anholt R, Lindstrom J, and Montal M. (1980). Reconstitution of purified acetylcholine receptors with functional ion channels in planar lipid bilayers. Proceedings of the National Academy of Sciences of the United States of America 77, 3057–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemecz A, Prevost MS, Menny A, and Corringer PJ (2016). Emerging Molecular Mechanisms of Signal Transduction in Pentameric Ligand-Gated Ion Channels. Neuron 90, 452–470. [DOI] [PubMed] [Google Scholar]
- Noda M, Takahashi H, Tanabe T, Toyosato M, Kikyotani S, Furutani Y, Hirose T, Takashima H, Inayama S, Miyata T, et al. (1983). Structural homology of Torpedo californica acetylcholine receptor subunits. Nature 302, 528–532. [DOI] [PubMed] [Google Scholar]
- Osaka H, Malany S, Kanter JR, Sine SM, and Taylor P. (1999). Subunit interface selectivity of the alpha-neurotoxins for the nicotinic acetylcholine receptor. The Journal of biological chemistry 274, 9581–9586. [DOI] [PubMed] [Google Scholar]
- Pan J, Chen Q, Willenbring D, Yoshida K, Tillman T, Kashlan OB, Cohen A, Kong XP, Xu Y, and Tang P. (2012). Structure of the pentameric ligand-gated ion channel ELIC cocrystallized with its competitive antagonist acetylcholine. Nature communications 3, 714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Polovinkin L, Hassaine G, Perot J, Neumann E, Jensen AA, Lefebvre SN, Corringer PJ, Neyton J, Chipot C, Dehez F, et al. (2018). Conformational transitions of the serotonin 5-HT3 receptor. Nature 563, 275–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakmann B, and Neher E. (1984). Patch clamp techniques for studying ionic channels in excitable membranes. Annu Rev Physiol 46, 455–472. [DOI] [PubMed] [Google Scholar]
- Sauguet L, Shahsavar A, Poitevin F, Huon C, Menny A, Nemecz A, Haouz A, Changeux JP, Corringer PJ, and Delarue M. (2014). Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation. Proceedings of the National Academy of Sciences of the United States of America 111, 966–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, and Peitsch MC (2003). SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res 31, 3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng JR, Grimme S, Bhattacharya P, Stowell MH, Artinger M, Prabahakar BS, and Meriggioli MN (2010). In vivo adsorption of autoantibodies in myasthenia gravis using Nanodisc-incorporated acetylcholine receptor. Exp Neurol 225, 320–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sine SM (2012). End-plate acetylcholine receptor: structure, mechanism, pharmacology, and disease. Physiological reviews 92, 1189–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sine SM, Strikwerda JR, and Mazzaferro S. (2019). Structural basis for alpha-bungarotoxin insensitivity of neuronal nicotinic acetylcholine receptors. Neuropharmacology, 107660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart OS, Neduvelil JG, Wang X, Wallace BA, and Sansom MS (1996). HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph 14, 354–360, 376. [DOI] [PubMed] [Google Scholar]
- Stokes C, Treinin M, and Papke RL (2015). Looking below the surface of nicotinic acetylcholine receptors. Trends Pharmacol Sci 36, 514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai T, Noda M, Mishina M, Shimizu S, Furutani Y, Kayano T, Ikeda T, Kubo T, Takahashi H, Takahashi T, et al. (1985). Cloning, sequencing and expression of cDNA for a novel subunit of acetylcholine receptor from calf muscle. Nature 315, 761–764. [DOI] [PubMed] [Google Scholar]
- The UniProt C. (2017). UniProt: the universal protein knowledgebase. Nucleic Acids Res 45, D158D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney ML, Osborn KE, Milburn PJ, Stowell MH, and Howitt SM (2004). Phylogenetic conservation of disulfide-linked, dimeric acetylcholine receptor pentamers in southern ocean electric rays. The Journal of experimental biology 207, 3581–3590. [DOI] [PubMed] [Google Scholar]
- Tsetlin V. (1999). Snake venom alpha-neurotoxins and other ‘three-finger’ proteins. European journal of biochemistry 264, 281–286. [DOI] [PubMed] [Google Scholar]
- Unwin N. (2005). Refined structure of the nicotinic acetylcholine receptor at 4A resolution. Journal of molecular biology 346, 967–989. [DOI] [PubMed] [Google Scholar]
- Unwin N. (2013). Nicotinic acetylcholine receptor and the structural basis of neuromuscular transmission: insights from Torpedo postsynaptic membranes. Quarterly reviews of biophysics 46, 283–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwin N, and Fujiyoshi Y. (2012). Gating movement of acetylcholine receptor caught by plunge-freezing. Journal of molecular biology 422, 617–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent A, Jacobson L, and Curran L. (1998). Alpha-Bungarotoxin binding to human muscle acetylcholine receptor: measurement of affinity, delineation of AChR subunit residues crucial to binding, and protection of AChR function by synthetic peptides. Neurochemistry international 32, 427–433. [DOI] [PubMed] [Google Scholar]
- Walsh RM Jr., Roh SH, Gharpure A, Morales-Perez CL, Teng J, and Hibbs RE (2018). Structural principles of distinct assemblies of the human alpha4beta2 nicotinic receptor. Nature 557, 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GK, and Schmidt J. (1980). Primary structure and binding properties of iodinated derivatives of alpha-bungarotoxin. The Journal of biological chemistry 255, 11156–11162. [PubMed] [Google Scholar]
- Wilson G, and Karlin A. (2001). Acetylcholine receptor channel structure in the resting, open, and desensitized states probed with the substituted-cysteine-accessibility method. Proceedings of the National Academy of Sciences of the United States of America 98, 1241–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GG, Pascual JM, Brooijmans N, Murray D, and Karlin A. (2000). The intrinsic electrostatic potential and the intermediate ring of charge in the acetylcholine receptor channel. The Journal of general physiology 115, 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzemann V, Barg B, Nishikawa Y, Sakmann B, and Numa S. (1987). Differential regulation of muscle acetylcholine receptor gamma- and epsilon-subunit mRNAs. FEBS letters 223, 104–112. [DOI] [PubMed] [Google Scholar]
- Zhang K. (2016). Gctf: Real-time CTF determination and correction. Journal of structural biology 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouridakis M, Giastas P, Zarkadas E, Chroni-Tzartou D, Bregestovski P, and Tzartos SJ (2014). Crystal structures of free and antagonist-bound states of human alpha9 nicotinic receptor extracellular domain. Nature structural & molecular biology 21, 976–980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM maps and atomic model coordinates have been deposited in the EMDB and RCSB respectively (EMD-20928 and PDB ID 6UWZ).