Abstract
Protein Kinase C isoenzymes (PKCs) are the key mediators of the phosphoinositide signaling pathway, which involves regulated hydrolysis of phosphatidylinositol(4,5)-bisphosphate to diacylglycerol (DAG) and inositol-1,4,5-trisphosphate. Dysregulation of PKCs is implicated in many human diseases making this class of enzymes an important therapeutic target. Specifically, the DAG-sensing cysteine-rich conserved homology-1 (C1) domains of PKCs have emerged as promising targets for pharmaceutical modulation. Despite significant progress, the rational design of the C1 modulators remains challenging due to difficulties associated with structure determination of the C1-ligand complexes. Given the dearth of experimental structural data, computationally derived models have been instrumental in providing atomistic insight into the interactions of the C1 domains with PKC agonists. In this review, we provide an overview of the in silico approaches for seven classes of C1 modulators and outline promising future directions.
Keywords: Protein Kinase C, Phosphoinositide signaling, diacylglycerol, C1 domains, PKC agonists, in silico
1. Introduction
Lipid membranes are essential structural barriers that provide a highly dynamic scaffold for signaling events and maintain homeostasis by confining cellular milieu (Dowhan et al., 2008; Fernandis and Wenk, 2007; van Meer et al., 2008). In signal transduction pathways, catalytic protein complexes are actively recruited to membrane regions that are enriched in signaling lipids (Groves and Kuriyan, 2010; Kholodenko et al., 2000; Kusumi et al., 2012). The allosteric activation of PKC family of serine/threonine kinases in the phosphoinositide pathway aptly exemplifies this process (Newton, 2009). PKCs are involved in regulation of vital cellular processes including survival, proliferation, motility, and apoptosis (Nishizuka, 1986; Rosse et al., 2010). The kinase function of PKC is activated by the transient membrane recruitment of the enzyme. This process is driven by the PKC interactions with signaling lipids, such as diacylglycerol (DAG) (Oancea and Meyer, 1998; Takai et al., 1979), Phosphatidylserine (PtdSer) (Johnson et al., 2000), Phosphatidyl inositols (PIs) (Evans et al., 2006; Nakanishi et al., 1993), and Phosphatidic acid (PA) (Limatola et al., 1994).
The involvement of PKC isoenzymes in cancer development and progression is supported by a wealth of experimental data. The loss/gain of function PKC mutations, as well as aberrant expression and activation profiles have been observed in cancer cell models (Black and Black, 2020; Leitges, 2020; Rahimova et al., 2020). The characteristic cellular changes associated with tumor initiation, promotion, and metastasis can often be correlated to the PKC involvement (Rahimova et al., 2020). PKCs have been traditionally viewed as promoting kinases, in large part due to their activation in response to binding tumor-promoting phorbol esters. However, in recent years, this perspective has been transformed into a more expansive view of PKCs acting as both, tumor promoter and suppressors, depending on the context (Antal et al., 2015; Black and Black, 2020; Newton and Brognard, 2017). Apart from cancer, the dysregulation of PKC isoenzymes is implicated in cardiac failure (Ferreira et al., 2011), diabetes (Das Evcimen and King, 2007; Nishikawa et al., 2000), autoimmune (Zanin-Zhorov et al., 2011), and neurodegenerative disorders (Garrido et al., 2002; Gordon et al., 2016). Currently, the corrective modulation of PKC function is considered a promising therapeutic approach for the treatment of these diseases (Hardman et al., 2020; Mochly-Rosen et al., 2012; Talman et al., 2016).
The development of pharmacological modulators of PKC has proven challenging mainly due to isoenzyme-specific differences in the subcellular localization, numerous physiological outcomes resulting from these differences, and the contrasting up/down-regulatory response seen in disease (Majumder et al., 2000; Mochly-Rosen et al., 2012; Newton, 2018; Rybin et al., 2004; Wang, Q.J. et al., 1999). There are several comprehensive reviews that discuss these challenges, along with the current repertoire of available PKC modulator classes and their application in PKC-linked diseases (Cooke et al., 2017; Das and Rahman, 2014; Igumenova, 2015; Irie et al., 2012; Mochly-Rosen et al., 2012; Nakagawa, 2012; Newton, 2018). The existing modulators target either the N- terminal regulatory or C-terminal catalytic regions (Figure 1A). These regions are responsible for the PKC membrane-binding and kinase functions, respectively (Steinberg, 2008). Agents targeting the catalytic ATP-binding site suffer from poor selectivity and off-target effects due to ubiquitous occurrence of serine/threonine kinases (Karaman et al., 2008; Soltoff, 2007). The regulatory region varies significantly among the PKCs and thereby provides more opportunities for the development of isoenzyme-selective agents.
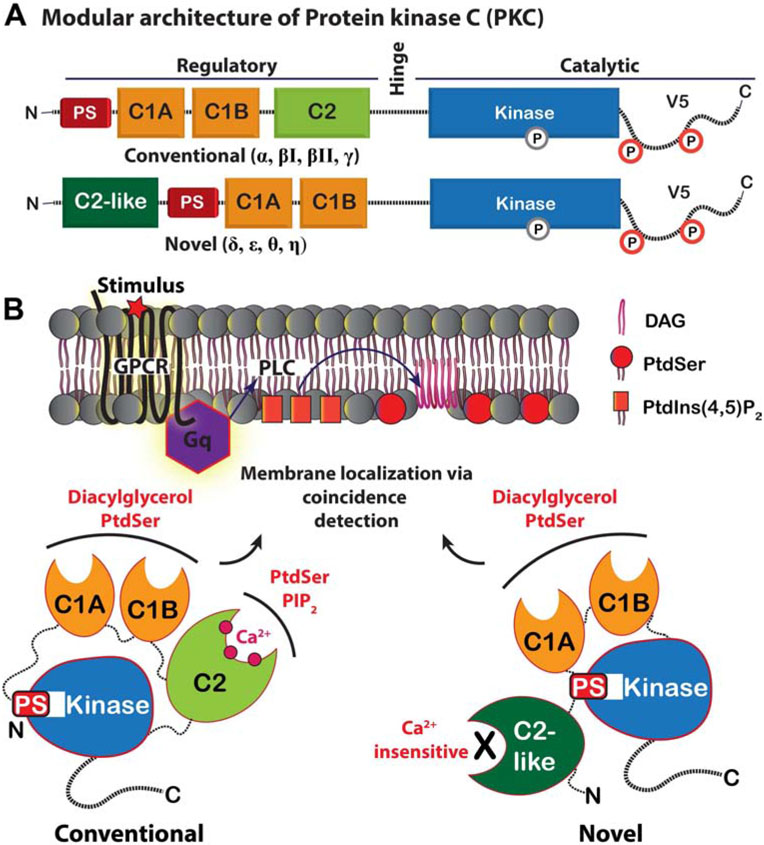
Figure 1. Modular architecture and second-messenger preferences of conventional and novel PKC isoenzymes.
(A) The N-terminal regulatory region of PKCs comprises the pseudosubstrate (PS) region, tandem C1 domains, and the C2 domain. (B) The membrane translocation of PKC in response to DAG and PtdSer is achieved via the combined action of C1 and C2 (conventional isoenzymes) or C1 alone (novel isoenzymes). The autoinhibition of the kinase domain by the PS region is relieved allosterically as a result of membrane recruitment.
The differences in the regulatory region substructure and second messenger preferences define three classes of PKC isoenzymes (Steinberg, 2008). The conventional (cPKC: α, βI, βII, γ) and novel (nPKC: δ, ε, θ, η) isoenzymes are activated by their recruitment to anionic membranes that contain diacylglycerol (DAG) (Gallegos and Newton, 2008). The atypical ζ, λ/ι isoenzymes are DAG-independent and will not be discussed here. The modular arrangement of specialized lipid-binding domains within the regulatory region is responsible for the isoenzyme-specific detection of membrane signals (Coussens et al., 1986; Ono et al., 1988). In cPKCs, the coincidence detection of DAG and anionic phospholipids, PtdSer and PtdIns(4,5)P2, results from the combined action of tandem conserved homology 1 (C1) domains and a Ca2+-dependent conserved homology 2 (C2) domain, respectively (Figure 1B) (Newton, 1995). In contrast, nPKCs sense DAG and PtdSer through the action of their C1 domains, as their consensus C2-like domains are not Ca2+-dependent membrane-targeting modules (Figure 1B) (Sossin and Schwartz, 1993).
The DAG-sensing C1 domains of PKC have emerged as promising drug targets (Blumberg et al., 2008; Boije af Gennas et al., 2011). C1 domains exhibit isoenzyme-specific variations in their dynamic properties and DAG affinities (Dries et al., 2007; Giorgione et al., 2006; Stewart et al., 2014; Stewart and Igumenova, 2017; Stewart, M.D. et al., 2011). In addition, the C1 domains are promiscuous and bind structurally diverse natural products such as phorbol esters, macrocyclic lactones, teleocidins, and aplysiatoxins (Mochly-Rosen et al., 2012; Nakagawa, 2012). Using the pharmacophores of these compounds and the endogenous activator DAG as templates, a number of synthetic modulators have been developed and tested (Beans et al., 2013; Cooke et al., 2018; DeChristopher et al., 2012; Nakagawa, 2012). Apart from their general DAG-mimicking function, some of these modulators induce opposing PKC-mediated cellular responses. One example is the tumor-promoting effect of phorbol esters and the antineoplastic effect of bryostatins (Choi et al., 2006; Szallasi et al., 1994).
This account summarizes the progress that has been made over the recent years in the development of C1 domain modulators, and the use of in silico methods to predict their binding modes. We also include cases where atomistic molecular dynamics simulations were used to obtain structural insight into the interactions of C1 domains with membranes and membrane–embedded agonists. These developments have the potential to guide future design of PKC agonists/antagonists for therapeutic and research applications.
2. C1 domains and their ligands
C1 domains were originally identified within PKC isoenzymes as modules of ~50 amino acids with the characteristic HX12-CX2-CXn-CX2-CX4-HX2-CX7-C sequence motif, where the cysteine (C), and histidine (H) residues are conserved (Figure 2A) (Ono et al., 1988; Parker et al., 1986). These domains are responsible for mediating the kinase activation by endogenous second messenger DAG and naturally occurring lipophilic diterpenoids called phorbol esters (Konig et al., 1985; Ono et al., 1989). The potent activation response induced by phorbol esters, unlike that of DAG, promotes tumor formation (Fournier and Murray, 1987; Nishizuka, 1984). This tumorigenic response induced directly by the C1 ligand provided the first piece of evidence that PKC regulation plays an important role in carcinogenesis. In conventional and novel PKC isoenzymes, the C1 domains occur in a tandem arrangement and are designated as C1A and C1B (Figure 1A). Despite high sequence homology, C1A and C1B domains have differential affinities to DAG and phorbol esters (Ananthanarayanan et al., 2003; Bogi et al., 1998; Giorgione et al., 2003; Stahelin et al., 2004). In addition to PKC, the C1 domains are found in other DAG effector proteins such as Protein Kinase D, RasGRPs, Chimaerins, DAG kinases, and UNC-13s (Colon-Gonzalez and Kazanietz, 2006; Hurley et al., 1997).
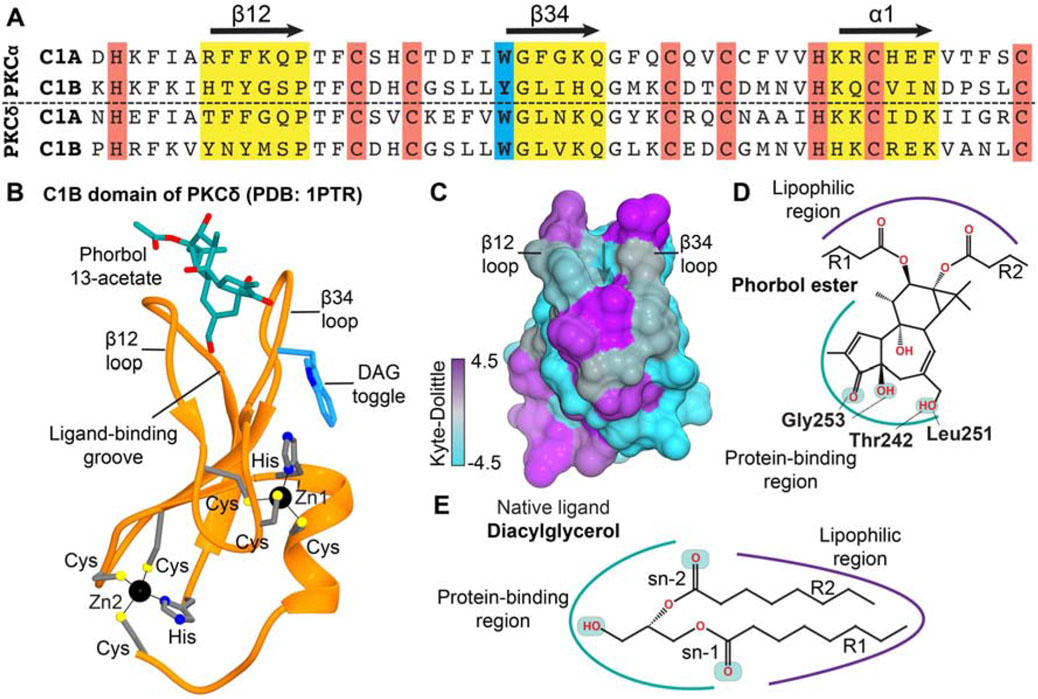
Figure 2. Structural features of a representative C1 domain and its ligand-binding site.
(A) Sequence alignment of the C1 domains from conventional PKCα and novel PKCδ isoenzymes showing: the agonist binding loops, β12 and β34, and the a1 helix (yellow); Zn2+-coordinating residues (red); and the DAG “toggling” position in C1B domains (blue). (B) Crystal structure of PKCδ C1B domain complexed to the hydrophilic phorbol ester, phorbol-13 acetate (1ptr) (Zhang et al., 1995). (C) Surface representation of apo C1Bδ color-coded according to the Kyte-Dolittle hydrophobicity scale; positive (purple) and negative (cyan) values correspond to regions of high and low hydrophobicity, respectively. (D) The H-bonding interactions of phorbol group with the C1Bδ residues observed in the crystal structure of the complex (1ptr). (E) Amphiphilic nature of the endogenous PKC agonist DAG.
The first structures of the C1 domains were determined in 1994 and 1995 using solution NMR spectroscopy (C1B from PKCα, C1Bα) and X-ray crystallography (C1B from PKCδ, C1Bδ), respectively (Hommel et al., 1994; Zhang et al., 1995). The three-dimensional fold made of two β sheets and an α helix is classified as a “treble-clef” Zn2+ finger (Grishin, 2001) (Figure 2B). Each of the two structural Zn2+ ions is coordinated by three conserved Cys and one His residues. The crystal structure of C1Bδ complexed to a partial agonist, phorbol 13-acetate, revealed that the agonist-binding region is located between the two loops, β12 and β34 (Figure 2B) (Zhang et al., 1995). The loops are flexible on the microsecond timescale, as demonstrated using the relaxation-dispersion NMR experiments carried out on the C1Bα and C1Bδ domains (Stewart and Igumenova, 2017; Stewart, M.D. et al., 2011). The concentric arrangement of hydrophobic and charged residues around the loop region gives the domain an amphiphilic character. Specifically, the “belt” of charged residues surrounding the loop region was hypothesized to be involved in electrostatic interactions with anionic phospholipids, such as PtdSer (Bittova et al., 2001; Johnson et al., 2000; Zhang et al., 1995). The aromatic residue, Trp or Tyr, at the N-terminal hinge of the β34 loop (Figure 2B) facilitates the agonist-independent membrane partitioning of the C1B domains (Stewart et al., 2014), and tunes their affinity to diacylglycerol (Dries et al., 2007; Giorgione et al., 2006; Stewart et al., 2014; Stewart and Igumenova, 2017; Stewart, M.D. et al., 2011). Preferential partitioning of Trp vs Tyr into the lipid headgroup region and changes in loop dynamics are the factors proposed to contribute to high-affinity interactions of the C1B Trp variants with DAG-containing membranes (Stewart and Igumenova, 2017; Stewart, M.D. et al., 2011). The significance of other loop residues in binding to the phorbol-ester was established by a series of mutagenesis studies (Kazanietz et al., 1995; Rahman et al., 2013).
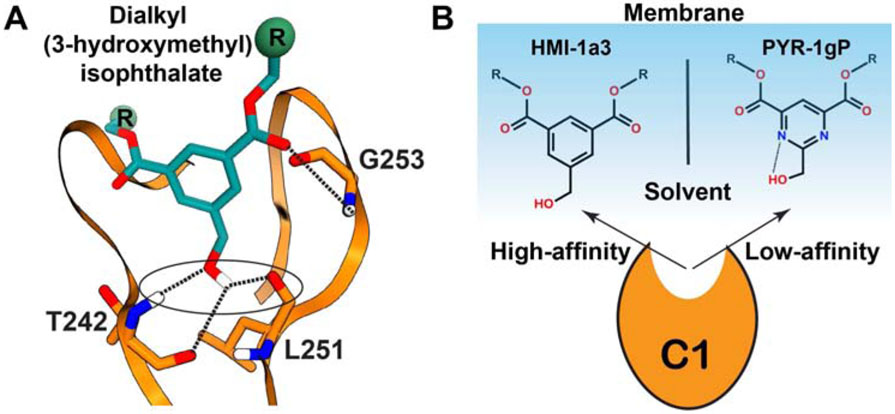
The amphiphilicity and conformational plasticity of the ligand-binding groove enables C1 to interact with membrane–embedded lipophilic ligands. The ligands themselves contain both, polar and hydrophobic chemical moieties (Figure 3). For example, the polar groups of the phorbol-13 acetate engage in hydrogen-bonding interactions with the backbone atoms of the residues that line the binding grove: T242, L251, and G253 (Figure 2D) (Zhang et al., 1995). The phorbol rings and the 13-acetyl group forms hydrophobic contacts with the amino acids lining the sides of the groove. The less lipophilic Phorbol-13 acetate is considered a “partial activator”, due to its low efficacy in inducing PKC-mediated growth inhibitory effects in yeast when compared to lipophilic phorbol esters (Saraiva et al., 2004). The atomistic picture of how C1 domains are recruited to membranes and recognize/capture the membrane-embedded potent PKC activators remains poorly understood.
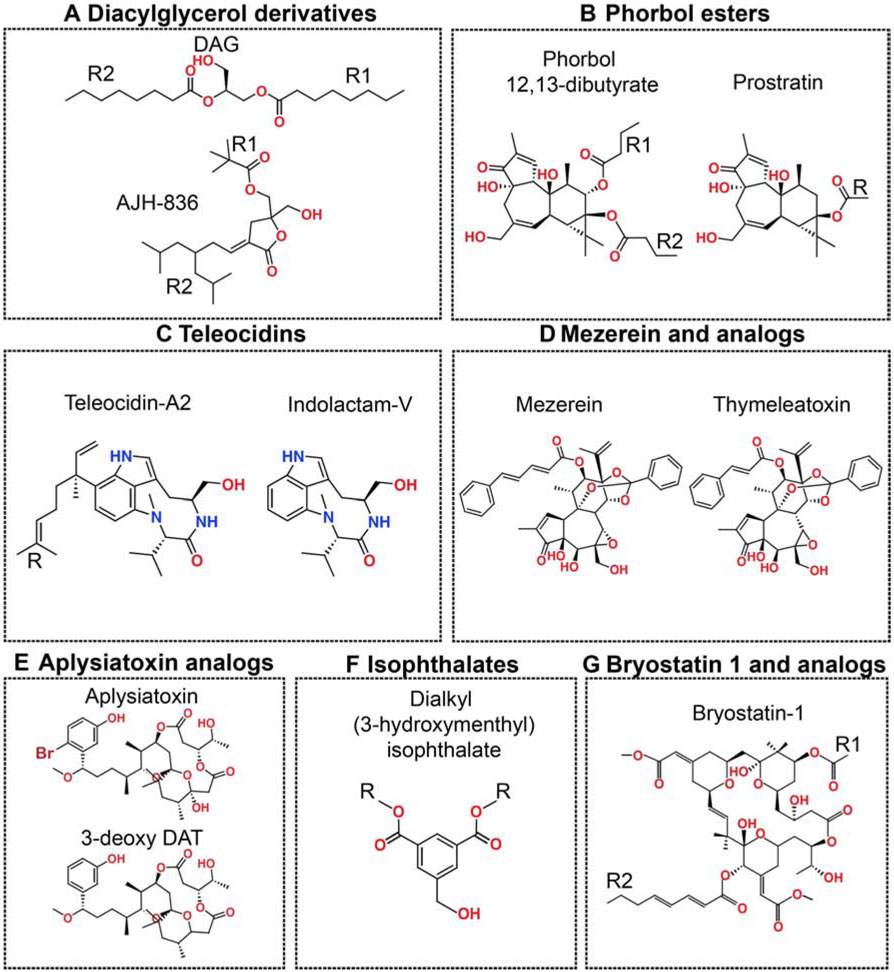
Figure 3. Chemical diversity of the C1 domain ligands.
The polar oxygen- and nitrogen-containing groups are colored red and blue, respectively. “R” represents variable lipophilic segments.
The PKC agonists that bind to the C1 domains will be referred to as C1 ligands or modulators in this article. The C1 ligands have considerable chemical diversity, and are usually grouped into classes according to their C1-interacting pharmacophores (Figure 3) (Das and Rahman, 2014). This chemical diversity combined with inter- and intra-isoenzyme differences between C1 domains suggests that the design of isoenzyme-specific modulatory C1 agents is feasible. The key requirement for tapping into the therapeutic potential of C1 domains is understanding the structural and dynamical basis of the C1-ligand-membrane interactions. However, structural biology of the C1 complexes remains challenging due to their poor solubility. While several apo C1 structures (only two determined using X-ray crystallography, the rest deposited as NMR ensembles) are available (Hommel et al., 1994; Rahman et al., 2013; Shanmugasundararaj et al., 2012; Xu et al., 1997; Zhang et al., 1995), there are no structures of C1 domains complexed to the endogenous activator DAG or ligands with therapeutic potential. Computational methods such as molecular docking and molecular dynamics simulations have been applied to gain atomistic insight and guide the design of potential modulators. These advances and their implications for future design of the C1 modulators are discussed in subsequent sections for seven major classes of the C1 ligands (Figure 3).
3. In silico studies of the C1-ligand interactions
3.1. Diacylglycerol and its derivatives
The diacylglycerol (DAG) levels inside the cellular membranes are stringently regulated due to the key function of this lipid as a modulator of membrane morphology, second messenger in signaling pathways, and the building block of other phospholipids (Eichmann and Lass, 2015). The isoenzyme-specific sub-cellular localization and activation of conventional and novel PKCs are intricately coupled to the membrane distribution of diacylglycerol (Carrasco and Merida, 2004; Gallegos and Newton, 2008; Melowic et al., 2007; Stahelin et al., 2004). It is well established that the recognition of membrane-embedded diacylglycerol by PKC C1 domains is stoichiometric (Konig et al., 1985), highly stereospecific towards sn-1,2 isomer (Rando and Young, 1984), and cannot be simply attributed to the DAG-induced membrane perturbations (Goni and Alonso, 1999; Rando and Young, 1984). The metabolic turnover of DAG by DAG-kinases, lipases, and acyl/phospho-transferases is one of the major routes of PKC downregulation (Eichmann and Lass, 2015).
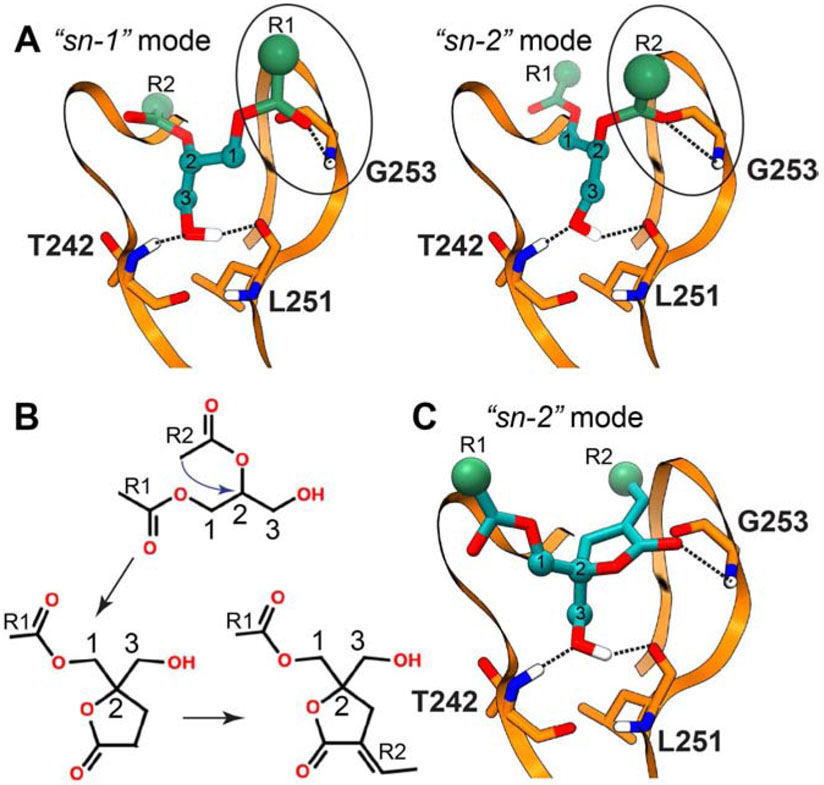
The hydrophobic nature of the C1-DAG complex and the dynamics of the DAG acyl chain moieties present challenges for conventional structural biology methods. In the absence of experimental structural data, molecular docking approaches have provided valuable insights into the putative binding modes. In the docked models of C1Bδ, the polar moieties of the DAG glycerol backbone interact with the same set of residues that were identified within the phorbol 13-acetate bound crystal structure: T242, L251, and G253 (Figure 4A) (Marquez and Blumberg, 2003). The two most energetically favorable DAG-binding orientations, referred to as “sn-1” and “sn-2”, differ in the identity of the carbonyl oxygen that forms a hydrogen bond with the amide hydrogen of G253 (Figure 4A). For several DAG derivatives, the “sn-1”orientation was consistently identified as having more favorable H-bonding energies (Marquez and Blumberg, 2003). The docking studies of C1Bδ and C1Bδ suggest that hydrophobic contacts are formed between the DAG aliphatic tails and the side-chains of the residues lining the agonist-binding pocket (Marquez and Blumberg, 2003; Sigano et al., 2003).
Figure 4. Schematic representation of the interaction modes of DAG and DAG lactones with C1Bδ obtained in docking studies.
The hydrogen bonding patterns obtained in computational docking studies cited in the text are shown with dashed lines. (A) The two distinct binding orientations of DAG defined as “sn-1” and “sn-2”. (B) The conceptual strategy of DAG lactone design developed by Blumberg and Marquez. (C) The DAG lactone “sn-2” binding orientation mode that was found to be more energetically favorable than “sn-1” in the docking studies (Marquez and Blumberg, 2003).
Additional insight into the C1-DAG interactions comes from the atomistic molecular dynamics simulations conducted with PKCα C1 domains (C1Aα and C1Bα) in the PtdCho:PtdSer membranes (Li et al., 2014). For both domains, a stable “sn-1” orientation of the bound DAG is reported. DAG binding to C1 decreased the distance between the tips of the loops that form the binding groove and attenuated its conformational plasticity. The bound DAG molecule formed a H-bonding network with analogous set of residues seen for the C1Bδ in docking studies. An additional side-chain H-bond was also observed between Q63 (C1Aα)/Q128 (C1Bα) and the sn-2 ester oxygen atom. Analysis of the hydrophobic contacts between the bound DAG and respective domains in this study show systematically greater occurrences for C1Aα compared to those with C1Bα. Further work is required to determine whether or not tandem C1 domains in other PKC isoenzymes show such differences.
The endogenous activator DAG served as a template for the development of DAG lactones by the laboratories of Blumberg and Marquez (Lee et al., 1993). The enclosed five-membered ring structure of these synthetic derivatives was designed to conformationally restrain the otherwise dynamic glycerol backbone of DAG (Figure 4B) (Lee, J. et al., 1996a). Given the retention of the identical H-bonding moieties, the substantially enhanced C1 affinities of DAG lactones over DAG can be attributed to the lower entropic penalty associated with a constrained pharmacophore (Nacro et al., 2000). Docking studies reported on DAG lactones show the presence of “sn-1/2” binding orientations with C1 domains (Figure 4C) (Eleonora et al., 2020; Kang et al., 2004; Marquez and Blumberg, 2003; Sigano et al., 2003). Unlike DAG, the “sn-2” orientation of DAG lactones showed more favorable H-bonding energies (Marquez and Blumberg, 2003). In either orientation, only one carbonyl oxygen is involved in the H-bonding interactions with C1. However, the isosteric removal of either the sn-1 vs sn-2 carbonyl functionalities in the DAG lactones revealed that both carbonyls are essential for high-affinity binding to PKCα. This was assessed by the relative abilities of these derivatives to displace the C1-bound phorbol ester, PDBu (Kang et al., 2003). Based on the experiments conducted on isolated C1Bδ with modified DAG lactones in the presence/absence of PtdSer membranes, it is proposed that the role of unbound carbonyl group of DAG lactones (i.e. “sn-1”) could be to interact with phospholipids (Benzaria et al., 1998; Kang et al., 2005).
The synthetic accessibility of DAG lactones enabled their modifications to include a variety of hydrophobic groups (Choi et al., 2003; Lee et al., 2001; Lee et al., 2004; Lee, J. et al., 1996a; Lee, J. et al., 1996b; Nomura et al., 2011; Ohashi et al., 2017), as a route to achieve isoenzyme-specificity (Ann et al., 2015; Cooke et al., 2018; Garcia-Bermejo et al., 2002; Pu et al., 2005). For instance, the synthetic DAG lactone AJH-836 (Figure 3A) preferentially binds, activates, and selectively down-regulates novel PKC isoenzymes δ and ε (Cooke et al., 2018). The structural basis of such specificity is still unclear. Studies of DAG lactones in cancer cell models attest to the therapeutic potential of this class of compounds (Cooke et al., 2019; Eleonora et al., 2020; Garcia-Bermejo et al., 2002). Further development of DAG-lactones as C1-targeting therapeutic agents will benefit from structural characterization of their complexes with C1 domains and modes of membrane interactions.
3.2. Phorbol esters
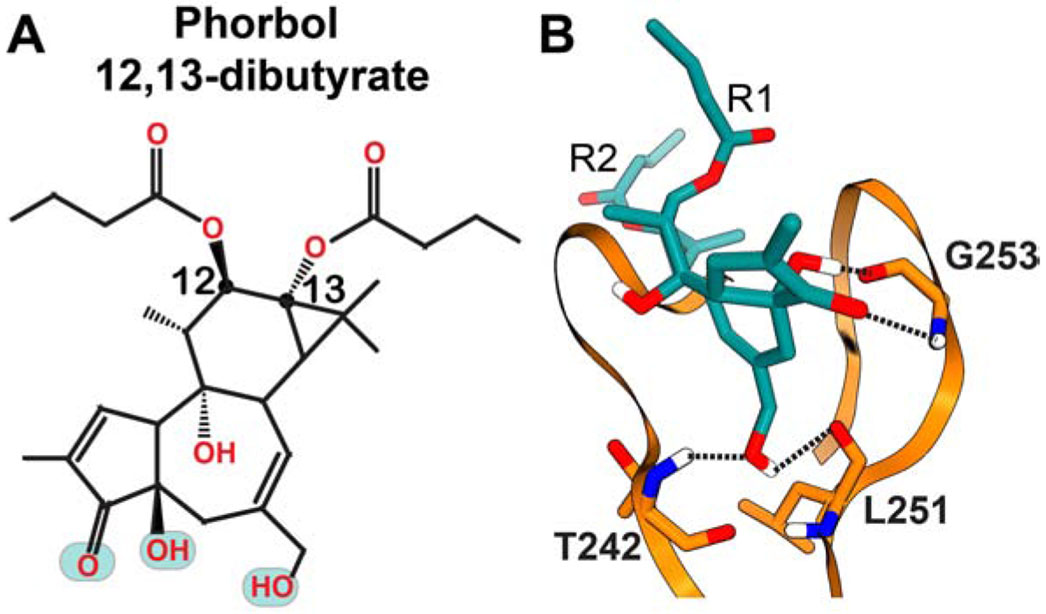
Derived first from the Euphorbiaceae family of plants, the diterpenoid phorbol esters (PEs) modulate the activity of conventional and novel PKC isoenzymes through direct interactions with the C1 domains (Hecker, 1967; Hecker and Schmidt, 1974; Ono et al., 1989). Unlike the transient activation of PKC caused by endogenous metabolic cycling of DAG, certain phorbol esters induce hyper-activation of PKC that is linked to tumor promotion (Ryves et al., 1991; Sharkey and Blumberg, 1986). One of the first identified examples is 12-O-tetradecanoylphorbol 13-acetate (TPA) that acts as a tumor-promoting agent in mouse models exposed to carcinogens (Slaga et al., 1980). Due to the availability of phorbol 13-acetate bound crystal structure of C1Bδ (Zhang et al., 1995), the C1 binding mode of the PE pharmacophore is known. The interaction mode between C1 and a potent membrane agonist, phorbol 12,13-dibutyrate (PDBu), was investigated using computational docking approaches (Figure 5) (Pak et al., 2001). Based on these studies, two factors were proposed to contribute to the C1-PDBu high-affinity interactions: (i) H-bonding interactions between the polar oxygenated moieties on the phorbol group and protein residues, and (ii) reduction of entropic penalty upon binding due to the conformationally constrained nature of the tigliane moiety.
Figure 5. (A) Chemical structure of PDBu and (B) schematic representation of its interactions with C1Bδ obtained in docking studies.
The polar groups implicated in the interactions with C1 are highlighted in cyan in the chemical structure. The hydrogen bonding patterns obtained in computational docking studies cited in the text are shown with dashed lines.
The lipophilicity of the phorbol esters can vary significantly depending upon the identity of the hydrophobic groups attached at the C12/13 positions of the tigliane skeleton (Figure 5A) (Wang et al., 2000). The PEs with relatively low lipophilicity induced disperse sub-cellular localization, and moderate non-tumorigenic activation of PKC (Szallasi et al., 1993; Wang et al., 2000). Compared to the tumor promoting phorbol esters TPA, or PDBu; the short-chain congeners such as phorbol 12,13-diacetate and C12-deoxyphorbol 13-acetate (Prostratin) are non-tumorigenic. Among those, Prostratin and its synthetic analogs are promising candidates for reactivation of the latent HIV-1 provirus expression (Beans et al., 2013; Korin et al., 2002). These distinct cellular responses might be connected to differential lipophilicities of phorbol esters and the resulting modulation of the C1 membrane partitioning (Ryckbosch et al., 2017).
The support for this idea comes from the results of atomistic molecular dynamics simulations involving a ternary C1-PE-membrane system (Li et al., 2014; Ryckbosch et al., 2017). In one of these studies, tumorigenic and therapeutic phorbol esters, PDBu and Prostratin, were compared with respect to their ability to drive the membrane partitioning of C1Bδ (Ryckbosch et al., 2017). The free energy landscape of membrane embedded C1Bδ-PDBu complex showed a single minimum attributed to a dominant, deeply membrane-embedded state. In contrast, the free energy landscape of the C1Bδ-Prostratin complex showed an additional, less-prominent free energy basin that was attributed to a shallow and tilted (with respect to the bilayer normal) membrane-bound state. This behavior likely originates from the reduced lipophilicity of Prostratin compared to PDBu, as the former lacks lipophilic moieties at the C12/13 positions (Figure 3B, 5A).
Introduction of new functionalities into PEs is limited exclusively to their C12/13 positions. Retaining the tigliane skeleton while altering the groups attached at these positions is a viable strategy to design novel modulators of variable lipophilicities (Nakagawa, 2012). This approach was applied to generate the PE derivatives with PKC inhibitory activities. For instance, the hybrid phorbol ester-PtdSer derivative (PEPS) significantly inhibited the PKC activation by PMA in vitro (Sodeoka et al., 1998). In another study, modification of the C12 ester group with the hydrophilic tetra-ethylene glycol chain produced the PE that failed to activate PKCα despite high binding affinity (Wada et al., 2002). Correlating the effects of these modifications to the membrane binding properties of the C1 domains is essential to understand the therapeutic value of PE derivatives as PKC modulators.
3.3. Teleocidins and their analogs
Teleocidins produced by actinomycetes and blue-green algae constitute the class of indole-alkaloids that activate PKCs through interactions with their C1 domains (Fujiki et al., 1981; Fujiki et al., 1984). Depending upon the nature of the monoterpenoid moieties at the indole ring, teleocidins are divided into sub-classes, namely teleocidin A (1-2) and B (1-4) (Irie et al., 1990). The least chemically complex member of this class is (−)-Indolactam V (IL-V) (Figure 6A), which consists of an indole attached to a nine-membered lactam ring (Endo et al., 1986). Unlike the potent tumorigenic members (Fujiki et al., 1981), IL-V is less effective as a tumor promoter, attributed to its reduced hydrophobicity (Nakagawa, 2012). The computer modelling and NMR spectroscopy revealed that the amide bond of the lactam ring undergoes a cis-trans isomerization between the ‘twist’ and ‘sofa’ forms (Endo et al., 1986; Ohno et al., 1993), with the former identified as the biologically active form (Endo et al., 1996; Ohno et al., 1993). Experiments conducted on the synthetic conformationally-restricted teleocidin analogs showed that certain C1 domains of nPKCs can bind both conformers, while those of cPKCs can bind only the ‘twist’ form with high potency (Masuda et al., 2002). The structural basis of C1 interactions with different members of this class and their conformers require further exploration.
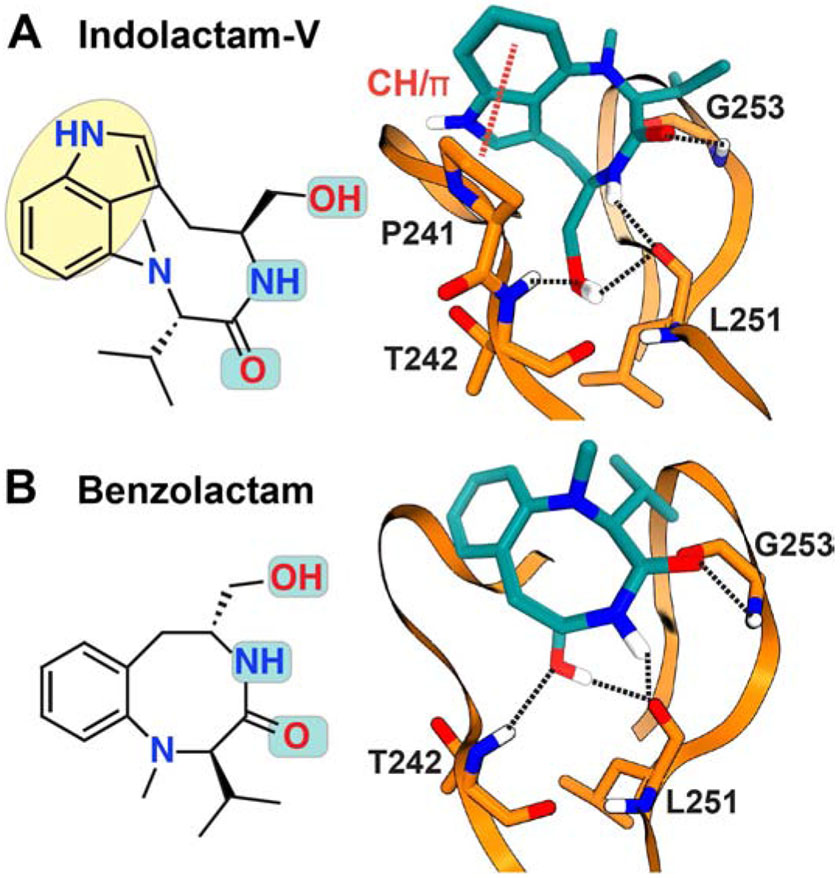
Figure 6. Chemical structures and C1Bδ interaction modes of (A) (−)-Indolactam V and (B) Benzolactam.
The polar groups implicated in interactions with C1 are highlighted in cyan in the chemical structures. The hydrogen bonding patterns (dashed lines) were obtained in computational docking studies cited in the text.
Computational docking studies of IL-V to C1Bδ were carried out to gain insight into the structural basis of the interactions (Figure 6A) (Endo et al., 1998; Nakagawa et al., 2005; Pak et al., 2001; Wang, S. et al., 1999). The obtained models show that similar to phorbol esters, the primary hydroxyl and the carbonyl group of the lactam form the H-bonds with the T242-L251 pair and G253, respectively. Also present in those models is the H-bond between the cis-amide of the ligand and the backbone carbonyl of the L251, which is unique to teleocidins. In addition to the H-bonding interactions, several hydrophobic contacts are formed between IL-V and hydrophobic residues of the β12 and β34 loops: M239, S240, P241, L250, and L254. The identity of the interacting residues was verified by mutagenesis (Pak et al., 2001; Wang, S. et al., 1999). An additional docking study revealed a previously uncharacterized CH/π interaction between the indole and a highly conserved Proline residue (P241 in case of C1Bδ) of the β12 loop (Nakagawa et al., 2005). The extensive nature of these predicted interactions compared to native agonist DAG is likely responsible for the high-affinity of teleocidins towards C1 domains.
The indolactam pharmacophore of teleocidins is amenable to synthetic modifications (Nakagawa, 2012). Benzolactams (BLs) are the synthetic analogs of IL-V that have the benzene ring attached to the lactam instead of the indole (Figure 6B) (Endo et al., 1996). Benzolactams exclusively adopt either ‘twist’ or the ‘sofa’ forms, depending upon whether the lactam ring is eight- or nine-membered (Endo et al., 1996; Ohno et al., 1993). The binding orientation of the eight-membered compound, benzolactam-V8 (BL-V8) with C1Bδ shows the H-bonding pattern identical to that of IL-V (Figure 6B) (Endo et al., 1998; Kozikowski et al., 1997). However, the CH/π interaction observed between IL-V and conserved Proline is not reported in the docked models of BL-V8. The weaker activation of PKC by BL-V8 compared to IL-V can potentially be attributed to the lack of this interaction (Nakagawa et al., 2005). The hydrophobic modifications to the benzene ring markedly improve the activity of BL-V8 over IL-V (Nakagawa, 2012). Computationally predicted models of the IL-V, BL-V8, and some of their modified analogs were instrumental for the synthetic development of this class of ligands (Endo et al., 1998; Kozikowski et al., 2009; Kozikowski et al., 2003; Kozikowski et al., 1997; Nakagawa et al., 2005; Pak et al., 2001; Wang, S. et al., 1999; Wei et al., 2002; Yanagita et al., 2008).
The activation of PKC elicited by IL-V and BL-V8 analogs makes them promising lead compounds for Alzheimer’s disease treatment and HIV latency reversal (Endo et al., 1994; Kozikowski et al., 2009; Kozikowski et al., 2003; Mach et al., 2006; Matsuda et al., 2019). PKC isoenzyme-specific design has proven to be feasible, in part due to the synthetic accessibility of these modulators. The IL-V modifications of the indole ring with alkyl, prenyl, or isopropyl groups (Nakagawa, 2012), as well as the indole to benzofuran substitution, remarkably changed the selectivity for novel vs conventional isoenzymes (Kozikowski et al., 1995; Nakagawa, 2012). Some of these modifications are predicted to alter or compensate for the CH/π interaction of the compounds with the conserved Proline residue of C1 β12 loop (Nakagawa, 2012). Among the benzolactams, 8-decynyl-BL-V8 is selective for conventional over novel PKCs (Kozikowski et al., 1997). In Alzheimer’s disease cell models, 8-decynyl-BL-V8 enhances the secretion of soluble amyloid precursor protein (sAPPα) produced by α-secretase (Ibarreta et al., 1999). In another study, the synthetic thioacetate derivatives of BL-V8 were designed to simultaneously activate PKC and inhibit histone deacetylase (HDAC) using a single scaffold to treat Alzheimer’s and alleviate oxidative stress (Kozikowski et al., 2009). Consistent with their predicted dual activity, these derivatives increased the sAPPα production and histone H4 acetylation in cell models. The versatile design potential of teleocidins makes them promising candidates for future development of pharmaceutical modulators.
3.4. Mezerein and analogs
Mezerein is a non-phorbol diterpene that was isolated from the plant Daphne mezereum as the main component exhibiting antileukemic activity in mice (Kupchan and Baxter, 1975). Mezerein and its analogs, such as thymeleatoxin (ThX) and daphnetoxin, have a daphnane-type skeleton that resembles the tigliane skeleton of the phorbol esters (Figure 7). Members of this class compete with phorbol esters and activate PKCs in phospholipid-dependent manner both in vitro and in vivo (Brooks et al., 1989; Kazanietz et al., 1993; Miyake et al., 1984; Roivainen and Messing, 1993; Ryves et al., 1991; Saraiva et al., 2001; Slaga et al., 1980). Mezerein and ThX are classified as second-stage tumor promoters, whose effect is fully manifested only after treatment by more potent tumorigenic agents such as TPA (Fürstenberger et al., 1981; Slaga et al., 1980). Both mezerein and ThX showed higher affinities for cPKCs than for the nPKCs in vitro (Ryves et al., 1991). However, it was subsequently revealed that in PC12 cells, ThX can cause membrane-translocation of both c/nPKCs (Roivainen and Messing, 1993). Contrary to the observations in vitro, ThX is highly potent in triggering the activation of novel isoenzymes PKC δ and ε in mesangial cells (Huwiler et al., 1994).
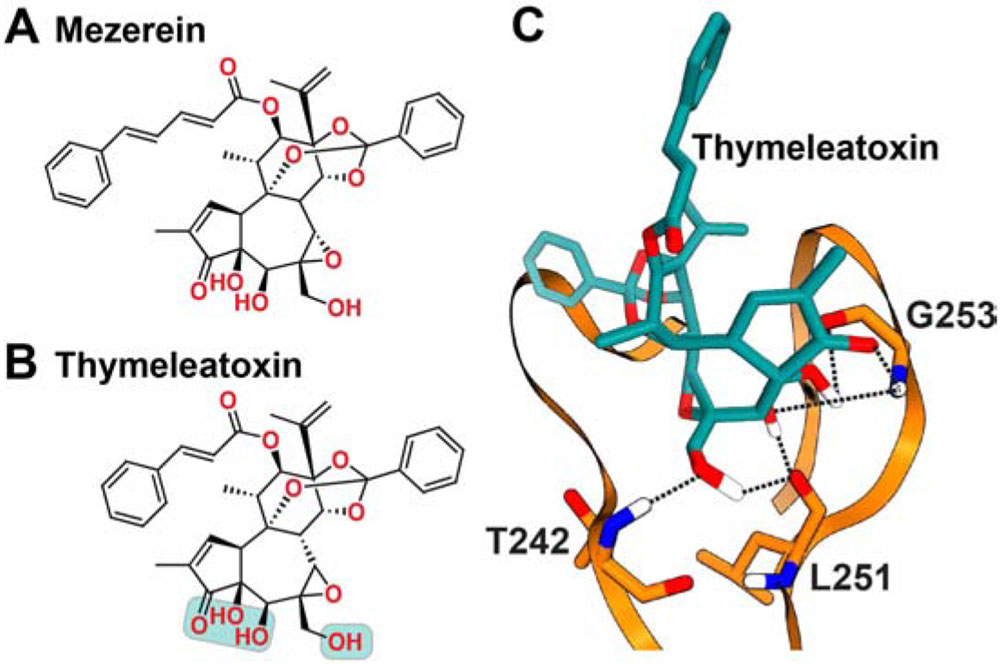
Figure 7. Chemical structures of (A) Mezerein and (B) Thymeleatoxin, along with the schematic representation of the C1Bδ- Thymeleatoxin interaction mode (C).
The polar groups of Thymeleatoxin implicated in interactions with C1 are highlighted in cyan. The hydrogen bonding patterns (dashed lines) were obtained in the computational docking study cited in the text.
The interactions of ThX with C1Bδ (Pak et al., 2001) and C1Bα (Caloca et al., 2001) were studied by molecular docking (Figure 7C). The structural model of the C1Bδ complex has six H-bonds that are formed between the three hydroxyl groups of the ligand and protein residues: T242, L251, and G253 (Pak et al., 2001). The ThX binding mode to C1Bα is identical (Caloca et al., 2001). Of note, the ThX complex shows 3 more H-bonds than in the DAG/DAG lactone complexes (Figure 4) and 2 more than the phorbol ester complexes (Figure 5). Similar to other ligands, these docking studies predict the existence of hydrophobic contacts between ThX and the conserved hydrophobic residues of C1 that line the binding groove: P241, L250, and L254 (C1Bδ numbering scheme).
Mezerein analogs or daphnane congeners in general have limited synthetic accessibility and natural supply (Wender et al., 2011b). Although a synthetic approach to produce fused tricyclic daphnane pharmacophore is reported (Stewart, C. et al., 2011), selective modifications to the core skeleton remain challenging. Key advances in the synthetic development of this class were reported by Wender’s group. Those include complete synthesis of resiniferatoxin (Wender et al., 1997), and a gateway strategy that produced two new congeners with nano- to sub-nanomolar PKC affinities (Wender et al., 2011b). Several daphnanes showed promising results in melanoma (Miyamae et al., 2009), lung cancer (Hong et al., 2010), and HIV-AIDS (Zhang et al., 2010) cell models. However, whether or not there is a direct correlation between observed therapeutic benefits and PKC activation remains unclear for most members of this class.
3.5. Aplysiatoxin derivatives
Halogenated aplysiatoxin (ATX) (Figure 8A), and its derivative debromo-aplysiatoxin (DAT) were first isolated from the sea hare Stylocheilus longicauda (Kato and Scheuer, 1974). Both compounds were shown to inhibit the PDBu binding in transformable mouse cell lines (Shimomura et al., 1983), and subsequently identified as potent PKC activators (Arcoleo and Weinstein, 1985). The potency of ATX and DAT with respect to PKC activation does not appear to directly correlate with a tumorigenic response. The tumor-promoting activity of ATX is comparable to that of phorbol esters (Horowitz et al., 1983; Suganuma et al., 1984), while tumor-promoting activity of DAT is significantly lower (Shimomura et al., 1983). These findings suggest that the differences in the aplysiatoxin framework, in this case reduced hydrophobicity of DAT lacking a bromine atom, can significantly alter the tumor-promoting activity (Nakagawa, 2012; Shimomura et al., 1983).
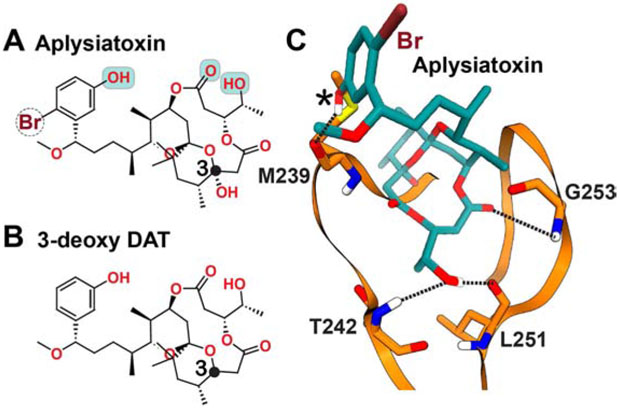
Figure 8. Chemical structures of (A) Aplysiatoxin and (B) 3-deoxy DAT, along with the schematic representation of the interaction mode of Aplysiatoxin with C1Bδ (C).
The polar groups of Aplysiatoxin implicated in interactions with C1 are highlighted in cyan. The hydrogen bonding patterns (dashed lines) were obtained in the computational docking and membrane MD simulation (additional H bond denoted by asterisk) studies cited in the text.
The C1Bδ-ATX interactions were characterized using molecular docking followed by the atomistic molecular dynamics simulations in the presence of PtdSer membrane (Ashida et al., 2016). The initial conformational search produced two energetically comparable conformers of ATX that were subsequently docked to C1Bδ. Valid binding modes were identified on the basis of their H-bonding networks. The structural model that showed H-bonds between ATX and the trio of C1Bδ residues, T242, L251, and G253, (Figure 8C) was selected for atomistic simulations with membrane. During the course of a 10 ns simulation of C1Bδ-ATX complex embedded in the bilayer, an additional H-bond was formed between the phenolic hydroxyl group of ATX (labeled with asterisk in Figure 8C) and the oxygen of the M239 carbonyl (Ashida et al., 2016). The MD results also showed that non-polar moieties of ATX and hydrophobic side-chains lining the C1Bδ binding groove are involved in direct interactions with lipids. A similar molecular dynamics-based approach was applied to the naphthalene-ring derivatives of DAT complexed to C1Bδ. These derivatives were designed to introduce CH/π interactions with the conserved Proline residue (P241 for C1Bδ) of the β12 loop, in order to increase the binding affinity (Kobayashi et al., 2020). The simultaneous formation of H-bonds and CH/π interactions with C1 was indeed observed in the simulations, prompting further optimizations to naphthalene ring placement in future studies.
The natural abundance and synthetic accessibility of ATX/DAT is limited (Nakagawa, 2012). Furthermore, ATX and DAT have a labile hydroxyl group at position 3 (Figure 8A) that is prone to elimination (Moore et al., 1984). A stable ATX derivative, the 3-deoxy DAT, was generated in an attempt to achieve stability without the loss of PKC activation potency (Figure 8B) (Nakamura et al., 1989). Given significantly lower tumor-promoting ability of DAT over ATX, modifications to the 3-deoxy DAT framework can potentially produce high-affinity PKC modulators with desired biological properties (Nakagawa, 2012; Nakagawa et al., 2009). A highly promising lead compound that emerged from this line of exploration was “Analog 1”. Analog 1 stimulated the translocation of PKCδ to nuclear membranes, caused PKCδ activation, and showed anti-tumorigenic activity in multiple cancer cell lines (Nakagawa et al., 2009). These findings highlight the therapeutic potential of the ATX analogs as anti-cancer agents that can be explored in future.
3.6. Isophthalate derivatives
The derivatives of dialkyl (3-hydroxymethyl) isophthalate (HMI) (Figure 9) are high-affinity synthetic C1 domain ligands that can be generated using a 4-step synthesis procedure (Boije af Gennas et al., 2009). Their potency as PKC modulators was demonstrated by the ability to displace radiolabeled PDBu from PKCα and PKCδ isoenzymes with the Ki values in the 200-900 nM range (Boije af Gennas et al., 2009; Talman et al., 2014). Isophthalates were shown to have anti-proliferative properties in HeLa cervical carcinoma and SH-SY5Y neuroblastoma cell lines (Talman et al., 2013; Talman et al., 2011).
Figure 9. Schematic representation of isophthalate interactions with C1Bδ.
(A) The hydrogen-bonding pattern (dotted lines) was obtained in the computational docking studies cited in the text. (B) Membrane-stabilized conformations of HMI-1a3 and PYR-1gP that have high- and low-affinity to C1Bδ, respectively.
Molecular docking approaches were used to gain insight into the interactions of HMIs with C1Bδ (Boije af Gennas et al., 2009; Provenzani et al., 2018). In these studies, four H-bonds between the HMI’s oxygen-containing moieties and the trio of C1Bδ residues: T242, L251, G253 were observed (Figure 9A). Similar to DAG and DAG lactones, only one of the two carbonyl functionalities is directly H-bonded to C1Bδ (Figure 9A). The other one is predicted to form a water-mediated H-bond with the carbonyl of M239 or with the lipid headgroups (Boije af Gennas et al., 2009). Additional docking studies showed identical H-bonding patterns for other HMI derivatives complexed to C1Bδ. These HMIs were derivatized with branched alkyl chains, trifluromethyl-attached benzene groups, or pyrimidine scaffold (Provenzani et al., 2018).
The role of membrane lipids in the formation of the C1Bδ-HMI complexes is most clearly illustrated by the differences between the behavior of two derivatives, HMI-1a3 and PYR-1gP that have phenyl and pyrimidine scaffolds, respectively (Figure 9B). In contrast to HMI-1a3, PYR-1gP is unable to displace radiolabeled PDBu from PKCα (Boije af Gennas et al., 2009; Provenzani et al., 2018), despite both ligands adopting identical poses when docked to C1Bδ (Lautala et al., 2020; Provenzani et al., 2018). Atomistic MD simulations in the presence of PtdSer membranes suggest a possible explanation (Lautala et al., 2020). The hydrophobic membrane environment facilitated the formation of the intra-molecular H-bond between 3-hydoxymethyl group and the pyrimidine of PYR-1gP (Figure 9B). The resulting sequestration of the hydroxyl group and the orientation of the PYR-1gP in the membrane preclude high-affinity interactions with C1 domains. These results demonstrate the significance of incorporating membranes in the modeling and MD simulations of the C1-ligand complexes.
3.7. Bryostatin-1 and analogs
The bryostatin family of 21 naturally occurring macrolides is one of the most promising classes of therapeutic PKC modulators (Wu et al., 2020; Yu et al., 2015). Bryostatin-1 (Bryo-1), the first member isolated from bryozoan Bugula neritina (Pettit et al., 1982), has been studied extensively for its synaptogenic (Ly et al., 2020), antineoplastic (Steube and Drexler, 1993; Wang et al., 2003), and anti-HIV latency (Bullen et al., 2014; Gutierrez et al., 2016) effects. The chemical structure of Bryo-1 is more complex than that of other C1 modulators. It comprises a macrolactone framework of 26 carbon atoms, 3 integrated tetrahydropyran rings (denoted by A, B, and C), and 11 chiral centers (Figure 10A). The pharmacological potential of Bryo-1 has been explored in more than 30 clinical trials (Blackhall et al., 2001; Farlow et al., 2019; Nelson et al., 2017; Nezhat et al., 2004; Pavlick et al., 2009; Varterasian et al., 2001), with the latest phase II trial planned this year (2020) for the treatment of Alzheimer’s disease (ClinicalTrials.gov identifier: NCT04538066).
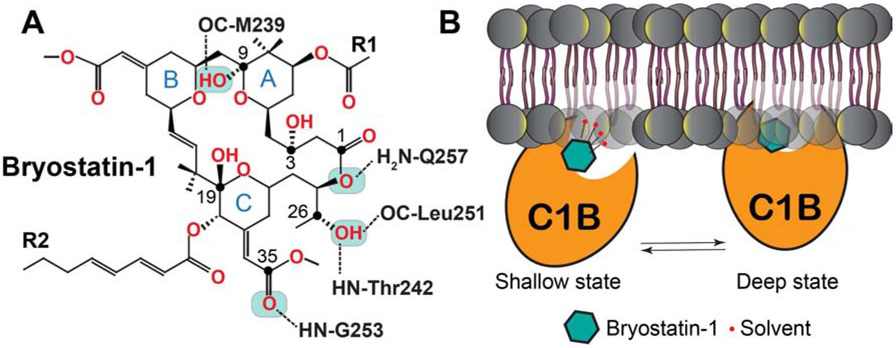
Figure 10. Interaction network (A) and schematic membrane positioning (B) of the C1Bδ-Bryo-1.
complex obtained using computational methods cited in the text. The “shallow” membrane-embedded state of the C1Bδ-Bryo-1 complex is stabilized by the interactions of Bryo-1 with the solvent molecules in the vicinity of the membrane headgroup region. The desolvation of the interaction interface results in the formation of the “deep” state, where Bryo-1 engages in direct interactions with lipids.
In the PKC context, the mechanisms involved in the unique pharmacological effects of Bryo-1 are not well understood. Bryo-1 causes membrane translocation of PKCs through direct interactions with their C1 domains (Kazanietz et al., 1994; Lee, H.W. et al., 1996). However, the resulting acute PKC activation does not elicit same set of cellular responses as tumorigenic phorbol esters (Sako et al., 1987). Furthermore, Bryo-1 shows protective effects by counteracting tumor promoters such as TPA both in vitro and in vivo (Hennings et al., 1987; Kraft et al., 1986; Sako et al., 1987; Steube and Drexler, 1993). The proposed models attribute these effects to selective down-regulation of certain PKC isoenzymes (Huwiler et al., 1994), and their patterns of membrane localization (Wang, Q.J. et al., 1999). Further work is needed to develop complete understanding of the Bryo-1 effects in the context of exceedingly complex cancer biology of PKC isoenzymes.
The interaction mode of Bryo-1 with C1Bδ was investigated by molecular docking (Keck et al., 2010; Kimura et al., 1999). Due to the complexity of the Bryo-1 chemical structure, the search for energetically stable conformers was carried out prior to docking. The most energetically favorable conformer resembled the crystal structure of Bryo-1 (Pettit et al., 1982), and when docked produced the lowest energy C1Bδ complex with five potential inter-molecular H-bonds (Figure 10A) (Keck et al., 2010; Kimura et al., 1999). The pattern of three H-bonds formed by the C26 hydroxyl group and the C35 carbonyl oxygen with the protein residues resembles that observed for C1Bδ complexed to the phorbol-13-acetate (Figure 2D) (Zhang et al., 1995). There are two additional H-bonds that involve the C9-OH (donor) and the carbonyl oxygen of M239 (acceptor) (Keck et al., 2010; Kimura et al., 1999), and a weak bond between the C1-adjacent ester oxygen (donor) and the side-chain NH of Q257 (acceptor) (Keck et al., 2010). Experimental chemical modification studies validated the essential role of the C26-OH group in the PKC-Bryo-1 interactions (Wender et al., 1998b). The C9-OH group however is expendable, since the affinity of the C9-deoxy Bryo-1 to PKCα is comparable to that of unmodified Bryo-1 (Keck et al., 2010). The intra-molecular H-bonds in the C1Bδ-complexed Bryo-1 are identical to those observed in the crystal structure of free Bryo-1 (Pettit et al., 1982). Both hydrophobic groups of Bryo-1 (denoted as R in Figure 10A), along with the tetrahydropyran rings A and B, appear to form an extensive capping surface over the C1Bδ binding groove (Keck et al., 2010; Kimura et al., 1999). The expansive nature of the amphiphilic surface created by the C1-complexed Bryo-1 suggests that the ligand-lipid interactions would contribute significantly to the stability of the ternary C1-Bryo-1-membrane complex.
Extensive atomistic molecular dynamics simulations conducted on the Bryo-1-C1Bδ complex in the presence of PtdSer membranes provided unprecedented insight into the membrane positioning of this complex (Ryckbosch et al., 2017). The pre-docked binary C1Bδ-Bryo-1 complex, initially placed in the vicinity of the PtdSer bilayer, partitioned into the membrane within ~200 ns. Analysis of the productions runs (400 μs aggregate) revealed the presence of two energetically favorable states of the complex. The states differed in their depth of membrane insertion and tilt angle (Figure 10B). In the “shallow” state, the oxygen-containing moieties of Bryo-1 form H-bonding interactions with the water molecules in the headgroup region of the membrane, stabilizing the complex. These solvent interactions are eliminated in the deeply embedded state, where the ternary complex is stabilized by interactions of the protein hydrophobic residues with the acyl chains of the lipid. In contrast, the tumor promoting phorbol ester, PDBu shows only a single, deeply embedded state. The ability of Bryo-1 to engage in solvent interactions while in the protein-complexed form could not have been predicted solely from docking studies. These observations open up a possibility that modulation of the depth of membrane partitioned C1-Bryo-1 complex could alter its interactions with other PKC regulatory proteins (Ryckbosch et al., 2017).
The atomistic MD simulations were subsequently combined with the solid-state REDOR (rotational-echo double-resonance) NMR experiments to gain insight into the conformational preferences of bryolog 1, a potent Bryo-1 analog (Yang et al., 2018). In the MD simulations, bryolog-1 adopted multiple low energy conformations when complexed to C1Bδ in PtdSer membranes. The existence of multiple conformers was corroborated by the NMR-derived heteronuclear 13C-19F distance distributions in isotopically labeled bryolog 1. The dynamic nature of the C1-agonist complexes in their membrane-bound states is apparent from these studies.
Initially, the low natural abundance of bryostatins presented significant obstacles towards realizing their full therapeutic potential. These limitations were overcome with the report of complete chemical synthesis of Bryo-1(Keck et al., 2011; Wender et al., 2017) that can now be produced in gram quantities via 29-steps (Wender et al., 2017). The synthetic routes to several other natural bryostatins have also been reported (Evans et al., 1999; Kageyama et al., 1990; Lu et al., 2011; Ohmori et al., 2000; Trost and Dong, 2008; Trost et al., 2020; Wender and Schrier, 2011; Zhang et al., 2018). These remarkable advancements present opportunities to design bryostatin derivatives for potential applications in the treatment of cancers, HIV-AIDS, and neurodegenerative diseases (Abramson et al., 2020; DeChristopher et al., 2012; Hardman et al., 2020; Wender et al., 2002; Wender et al., 2011a; Wender et al., 1998a; Wender et al., 2020). One of the examples is the computationally guided, function-oriented synthesis of bryologs reported by Wender’s group (Wender et al., 2020).
4. Concluding remarks and outlook
The current challenges in establishing the structure-activity relationships of C1 modulators stem from lack of experimental structural information, tissue-specific variations in the PKC expression profiles (Kim et al., 1999; Rybin and Steinberg, 1994), and potentially opposing roles of the isoenzymes in diseases (Chen et al., 2001). Despite these challenges, the results of cell biology, biophysical and in silico studies have successfully guided the development of C1 modulators that reached preclinical stages. In this account, we discussed seven classes of promising C1-targeted PKC agonists (Figure 3), with the primary focus on their binding modes predicted by computational docking methods. In addition, we highlighted the outcomes of atomistic molecular dynamics simulations that illustrate the importance of membranes in understanding the formation of the C1-ligand complexes. Below, we outline two additional avenues that could facilitate further development of therapeutic agents targeting the DAG-sensing function of PKC.
4.1. Characterization of ternary C1-ligand-membrane complexes
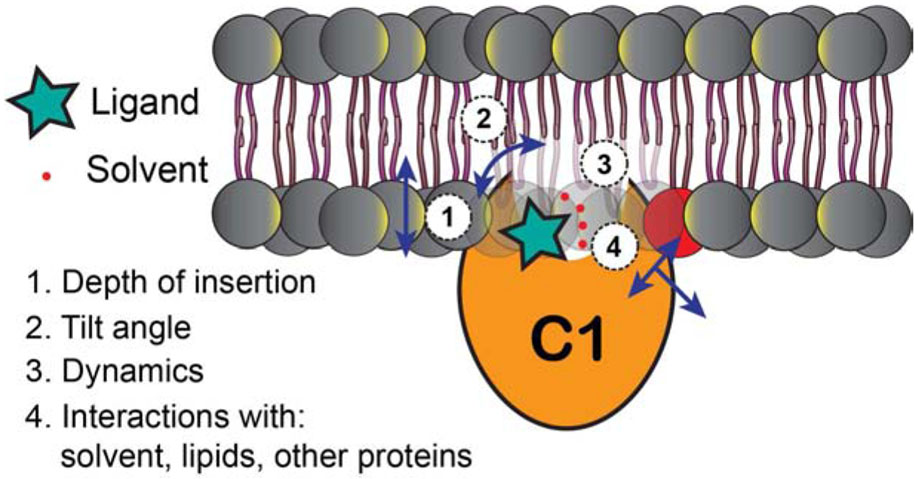
Due to their amphiphilicity (Figure 2C), the C1 domains can exist in solution state as well as partition conditionally into membranes (Zhang et al., 1995). The C1 ligands differ in their lipophilic properties. It is the combination of the C1 and specific ligand characteristics that determines the properties of the ternary C1-ligand-membrane complexes (Figure 11). Understanding these properties may provide insight into the structural and dynamical basis of differential functional responses elicited by the PKC agonists. Of equal significance would be to elucidate the mechanism of the ternary complex formation: the initial membrane recruitment of C1, recognition of the PKC agonists, and their subsequent capture.
Figure 11.
Parameters of the ternary C1-ligand-membrane complexes that are important for establishing structure-activity relationships for different PKC agonists.
Atomistic MD simulations of the C1-ligand-membrane systems show significant promise in addressing these structural and mechanistic questions. This is illustrated by the work on isophthalates (Lautala et al., 2020) and bryostatins (Ryckbosch et al., 2017; Yang et al., 2018) that was discussed in earlier sections. With respect to experimental approaches, we have demonstrated that solution NMR spectroscopy of C1 domains in membrane mimetics can be applied to probe their interactions with agonists in residue-specific manner (Stewart et al., 2014; Stewart, M.D. et al., 2011), quantify the dynamics of the ligand-binding groove (Stewart and Igumenova, 2017; Stewart, M.D. et al., 2011), and probe the depth of membrane insertion using paramagnetic lipids (Stewart et al., 2014). Characterization of the ternary C1-ligand-membrane complexes, using a combination of computational and experimental approaches, will inform the rational design of therapeutic agents targeting the C1 domains.
4.2. Exploiting the specificities of tandemly organized C1 domains
The tandem C1 domains of PKC isoenzymes are not equivalent with respect to their functional properties. For instance, the C1A and C1B domains of PKCδ show opposing affinities for DAG (C1A) and phorbol esters (C1B) (Stahelin et al., 2004), while those of PKCε lack such distinction (Stahelin et al., 2005). Similarly, based on in vitro experiments, the hydrophobic residues of the C1A, but not C1B are implicated in DAG dependent membrane binding, and activation of PKCα (Medkova and Cho, 1999). Moreover, when compared between isoenzymes, the C1 domains of PKCδ show two-orders of magnitude higher DAG affinity compared to those of PKCδ, compensating for the lack of Ca2+-sensitive C2 domains (Dries et al., 2007). These differences extend further to subcellular localization of the isolated C1 domains. The membrane-targeting studies in human neuroblastoma cells of GFP-tagged C1 domains show highly preferential localization of C1B, but not C1A (of PKCδ, ε, and η isoenzymes) to trans-Golgi network (Schultz et al., 2004), where DAG production can be stimulated (Kunkel and Newton, 2010). Sensitivity to anionic second messenger PtdIns(4,5)P2 has been attributed to C1B domain of PKCε in conjunction with the pseudosubstrate region (Shirai et al., 2007).
The determinants of differential second-messenger/lipid preferences of C1 domains are not completely understood. Mutagenesis studies have been instrumental in identifying the contributions of individual residues to the agonist/lipid specificities (Kazanietz et al., 1995; Pak et al., 2001; Szallasi et al., 1996; Wang et al., 1996). However, inherent dynamics of the C1 ligand-binding regions (Stewart and Igumenova, 2017; Stewart, M.D. et al., 2011) makes it challenging to completely understand the structural basis of these specificities. Computer simulations, in combination with structurally non-invasive methods for quantifying protein dynamics such as solution NMR spectroscopy, are uniquely suited for characterization of individual C1 domains and their conformational ensembles. The inclusion of lipid membranes with varying compositions into MD simulations will provide insight into the lipid preferences of the C1 domains and enable to mimic scenarios where PKC shows agonist-specific sub-cellular localization (Wang, Q.J. et al., 1999).
The differences between the C1A and C1B domains present opportunities for the design of isoenzyme-selective PKC modulators. To this end, the approach based on the design of bivalent C1 modulators shows promise (Blumberg et al., 2008). The dimeric phorbol ester (Giorgione et al., 2003), and benzolactam (Sridhar et al., 2003) derivatives have already been reported that can potentially target C1 domains as a tandem unit. The individual counterparts of these agents can be further tuned to achieve the desired specificity towards either C1A or C1B domain. It might also be feasible to alter the membrane interactions of individual C1 domains to “tune” the agonistic or antagonistic PKC response required for the corrective action in a particular disease state.
As a final remark, we would like to note that in the context of rational drug design, the computer-generated models cannot fully replace the experimental structures obtained by X-ray crystallography or NMR spectroscopy. So far, the C1-agonist complexes have eluded structure elucidation attempts due to their obligatory requirement for the membrane mimics. While the X-ray structure determination of large membrane proteins in lipid cubic phases (Landau and Rosenbusch, 1996) and bicelles (Ujwal and Bowie, 2011) has been successful, small and highly dynamic C1 domains that peripherally associate with membranes may not be amenable to this approach. Until methodological advances avail direct structural characterization of the C1-agonist complexes, the field will rely on experimentally validated computational models for the development of PKC modulators that target its DAG-sensing function.
Acknowledgements
We acknowledge the support from National Institutes of Health grant R01 GM108998, National Science Foundation grant CHE-1905116, and Welch Foundation grant A-1784, all awarded to Tatyana I. Igumenova.
Abbreviations
- PKC
Protein Kinase C
- DAG
Diacylglycerol
- C1
Cysteine-rich Conserved Homology-1
- PtdSer
Phosphatidylserine
- PIs
Phosphatidyl Inositols
- PA
Phosphatidic Acid
- cPKCs
Conventional PKCs
- nPKCs
Novel PKCs
- H-bond
Hydrogen bond
- PEs
Phorbol Esters
- TPA
12-O-Tetradecanoylphorbol 13-Acetate
- PDBu
Phorbol 12,13-Dibutyrate
- IL-V
(−)-Indolactam V
- BL
Benzolactam
- sAPPα
Soluble Amyloid Precursor Protein
- HDAC
Histone Deacetylase
- ThX
Thymeleatoxin
- ATX
Aplysiatoxin
- DAT
Debromo-Aplysiatoxin
- HMI
Dialkyl (3-hydroxymethyl) Isophthalate
- Bryo-1
Bryostatin-1
Footnotes
CRediT Author Statement
Sachin Katti: Conceptualization, Writing - original draft, Writing - review & editing, Visualization. Tatyana I. Igumenova: Conceptualization, Writing - original draft, Writing - review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abramson E, Hardman C, Shimizu A, Hwang S, Hester LD, Snyder SH, Wender PA, Kim PM, Kornberg MD, 2020. Designed PKC-targeting bryostatin analogs modulate innate immunity and neuroinflammation. bioRxiv, 2020.2009.2012.294918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthanarayanan B, Stahelin RV, Digman MA, Cho W, 2003. Activation mechanisms of conventional protein kinase C isoforms are determined by the ligand affinity and conformational flexibility of their C1 domains. J. Biol. Chem 278(47), 46886–46894. [DOI] [PubMed] [Google Scholar]
- Ann J, Yoon S, Baek J, Kim DH, Lewin NE, Hill CS, Blumberg PM, Lee J, 2015. Design and synthesis of protein kinase C epsilon selective diacylglycerol lactones (DAG-lactones). Eur. J. Med. Chem 90, 332–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antal CE, Hudson AM, Kang E, Zanca C, Wirth C, Stephenson NL, Trotter EW, Gallegos LL, Miller CJ, Furnari FB, Hunter T, Brognard J, Newton AC, 2015. Cancer-associated protein kinase C mutations reveal kinase's role as tumor suppressor. Cell 160(3), 489–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcoleo JP, Weinstein IB, 1985. Activation of protein kinase C by tumor promoting phorbol esters, teleocidin and aplysiatoxin in the absence of added calcium. Carcinogenesis 6(2), 213–217. [DOI] [PubMed] [Google Scholar]
- Ashida Y, Yanagita RC, Takahashi C, Kawanami Y, Irie K, 2016. Binding mode prediction of aplysiatoxin, a potent agonist of protein kinase C, through molecular simulation and structure-activity study on simplified analogs of the receptor-recognition domain. Bioorg. Med. Chem 24(18), 4218–4227. [DOI] [PubMed] [Google Scholar]
- Beans EJ, Fournogerakis D, Gauntlett C, Heumann LV, Kramer R, Marsden MD, Murray D, Chun T-W, Zack JA, Wender PA, 2013. Highly potent, synthetically accessible prostratin analogs induce latent HIV expression in vitro and ex vivo. Proc. Natl. Acad. Sci. U.S.A 110(29), 11698–11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzaria S, Bienfait B, Nacro K, Wang S, Lewin NE, Beheshti M, Blumberg PM, Marquez VE, 1998. Conformationally constrained analogues of diacylglycerol (DAG). 15. The indispensable role of the sn-1 and sn-2 carbonyls in the binding of DAG-lactones to protein kinase C (PK-C). Bioorg. Med. Chem. Lett 8(23), 3403–3408. [DOI] [PubMed] [Google Scholar]
- Bittova L, Stahelin RV, Cho W, 2001. Roles of ionic residues of the C1 domain in protein kinase C-alpha activation and the origin of phosphatidylserine specificity. J. Biol. Chem 276(6), 4218–4226. [DOI] [PubMed] [Google Scholar]
- Black AR, Black JD, 2020. The complexities of PKCalpha signaling in cancer. Adv Biol Regul 80, 100769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackhall FH, Ranson M, Radford JA, Hancock BW, Soukop M, McGown AT, Robbins A, Halbert G, Jayson GC, Cancer Research Campaign Phase, I.I.I.C., 2001. A phase II trial of bryostatin 1 in patients with non-Hodgkin's lymphoma. Br. J. Cancer 84(4), 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg PM, Kedei N, Lewin NE, Yang D, Czifra G, Pu Y, Peach ML, Marquez VE, 2008. Wealth of opportunity - the C1 domain as a target for drug development. Curr. Drug Targets 9(8), 641–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogi K, Lorenzo PS, Szallasi Z, Acs P, Wagner GS, Blumberg PM, 1998. Differential selectivity of ligands for the C1a and C1b phorbol ester binding domains of protein kinase Cdelta: possible correlation with tumor-promoting activity. Cancer Res. 58(7), 1423–1428. [PubMed] [Google Scholar]
- Boije af Gennas G, Talman V, Aitio O, Ekokoski E, Finel M, Tuominen RK, Yli-Kauhaluoma J, 2009. Design, synthesis, and biological activity of isophthalic acid derivatives targeted to the C1 domain of protein kinase C. J. Med. Chem 52(13), 3969–3981. [DOI] [PubMed] [Google Scholar]
- Boije af Gennas G, Talman V, Yli-Kauhaluoma J, Tuominen RK, Ekokoski E, 2011. Current status and future prospects of C1 domain ligands as drug candidates. Curr. Top. Med. Chem 11(11), 1370–1392. [DOI] [PubMed] [Google Scholar]
- Brooks G, Evans AT, Aitken A, Evans FJ, 1989. Tumour-promoting and hyperplastic effects of phorbol and daphnane esters in CD-1 mouse skin and a synergistic effect of calcium ionophore with the non-promoting activator of protein kinase C, sapintoxin A. Carcinogenesis 10(2), 283–288. [DOI] [PubMed] [Google Scholar]
- Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF, 2014. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat. Med 20(4), 425–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caloca MJ, Wang H, Delemos A, Wang S, Kazanietz MG, 2001. Phorbol esters and related analogs regulate the subcellular localization of beta 2-chimaerin, a non-protein kinase C phorbol ester receptor. J. Biol. Chem 276(21), 18303–18312. [DOI] [PubMed] [Google Scholar]
- Carrasco S, Merida I, 2004. Diacylglycerol-dependent binding recruits PKCtheta and RasGRP1 C1 domains to specific subcellular localizations in living T lymphocytes. Mol. Biol. Cell 15(6), 2932–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Hahn H, Wu G, Chen CH, Liron T, Schechtman D, Cavallaro G, Banci L, Guo Y, Bolli R, Dorn GW 2nd, Mochly-Rosen D, 2001. Opposing cardioprotective actions and parallel hypertrophic effects of delta PKC and epsilon PKC. Proc. Natl. Acad. Sci. U.S.A 98(20), 11114–11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Hyman T, Blumberg PM, 2006. Differential effect of bryostatin 1 and phorbol 12-myristate 13-acetate on HOP-92 cell proliferation is mediated by down-regulation of protein kinase Cdelta. Cancer Res. 66(14), 7261–7269. [DOI] [PubMed] [Google Scholar]
- Choi Y, Kang JH, Lewin NE, Blumberg PM, Lee J, Marquez VE, 2003. Conformationally constrained analogues of diacylglycerol. 19. Synthesis and protein kinase C binding affinity of diacylglycerol lactones bearing an N-hydroxylamide side chain. J. Med. Chem 46(13), 2790–2793. [DOI] [PubMed] [Google Scholar]
- Colon-Gonzalez F, Kazanietz MG, 2006. C1 domains exposed: from diacylglycerol binding to protein-protein interactions. Biochim. Biophys. Acta 1761(8), 827–837. [DOI] [PubMed] [Google Scholar]
- Cooke M, Casado-Medrano V, Ann J, Lee J, Blumberg PM, Abba MC, Kazanietz MG, 2019. Differential Regulation of Gene Expression in Lung Cancer Cells by Diacyglycerol-Lactones and a Phorbol Ester Via Selective Activation of Protein Kinase C Isozymes. Scientific Reports 9(1), 6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke M, Magimaidas A, Casado-Medrano V, Kazanietz MG, 2017. Protein kinase C in cancer: The top five unanswered questions. Mol. Carcinog 56(6), 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke M, Zhou X, Casado-Medrano V, Lopez-Haber C, Baker MJ, Garg R, Ann J, Lee J, Blumberg PM, Kazanietz MG, 2018. Characterization of AJH-836, a diacylglycerol-lactone with selectivity for novel PKC isozymes. J. Biol. Chem 293(22), 8330–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens L, Parker PJ, Rhee L, Yang-Feng TL, Chen E, Waterfield MD, Francke U, Ullrich A, 1986. Multiple, distinct forms of bovine and human protein kinase C suggest diversity in cellular signaling pathways. Science 233(4766), 859–866. [DOI] [PubMed] [Google Scholar]
- Das Evcimen N, King GL, 2007. The role of protein kinase C activation and the vascular complications of diabetes. Pharmacol. Res 55(6), 498–510. [DOI] [PubMed] [Google Scholar]
- Das J, Rahman GM, 2014. C1 domains: structure and ligand-binding properties. Chem. Rev 114(24), 12108–12131. [DOI] [PubMed] [Google Scholar]
- DeChristopher BA, Loy BA, Marsden MD, Schrier AJ, Zack JA, Wender PA, 2012. Designed, synthetically accessible bryostatin analogues potently induce activation of latent HIV reservoirs in vitro. Nature Chemistry 4(9), 705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowhan W, Bogdanov M, Mileykovskaya E, 2008. CHAPTER 1 - Functional roles of lipids in membranes, in: Vance DE, Vance JE (Eds.), Biochemistry of Lipids, Lipoproteins and Membranes (Fifth Edition). Elsevier, San Diego, pp. 1–37. [Google Scholar]
- Dries DR, Gallegos LL, Newton AC, 2007. A single residue in the C1 domain sensitizes novel protein kinase C isoforms to cellular diacylglycerol production. J. Biol. Chem 282(2), 826–830. [DOI] [PubMed] [Google Scholar]
- Eichmann TO, Lass A, 2015. DAG tales: the multiple faces of diacylglycerol--stereochemistry, metabolism, and signaling. Cell. Mol. Life Sci 72(20), 3931–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleonora E, Ana B, Mariana C, Antonella S, Larry VP, Megan P, Lucía G-D, Marcelo K, Maria Julieta C, 2020. Design, Synthesis and Characterization of Novel sn-1 Heterocyclic DAG lactones as PKCe Activators. ChemRxiv. Preprint. [Google Scholar]
- Endo Y, Ohno M, Hirano M, Fujiwara T, Sato A, Hinuma Y, Shudo K, 1994. TELEOCIDINS AND BENZOLACTAMS INHIBIT CELL KILLING BY HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 (HIV-1). Biol. Pharm. Bull 17(8), 1147–1149. [DOI] [PubMed] [Google Scholar]
- Endo Y, Ohno M, Hirano M, Itai A, Shudo K, 1996. Synthesis, Conformation, and Biological Activity of Teleocidin Mimics, Benzolactams. A Clarification of the Conformational Flexibility Problem in Structure–Activity Studies of Teleocidins. J. Am. Chem. Soc 118(8), 1841–1855. [Google Scholar]
- Endo Y, Shudo K, Itai A, Hasegawa M, Sakai S.-i., 1986. Synthesis and stereochemistry of indolactam-v, an active fragment of teleocidins. Structural requirements for tumor-promoting activity. Tetrahedron 42(21), 5905–5924. [Google Scholar]
- Endo Y, Takehana S, Ohno M, Driedger PE, Stabel S, Mizutani MY, Tomioka N, Itai A, Shudo K, 1998. Clarification of the binding mode of teleocidin and benzolactams to the Cys2 domain of protein kinase Cdelta by synthesis of hydrophobically modified, teleocidin-mimicking benzolactams and computational docking simulation. J. Med. Chem 41(9), 1476–1496. [DOI] [PubMed] [Google Scholar]
- Evans DA, Carter PH, Carreira EM, Charette AB, Prunet JA, Lautens M, 1999. Total Synthesis of Bryostatin 2. J. Am. Chem. Soc 121(33), 7540–7552. [DOI] [PubMed] [Google Scholar]
- Evans JH, Murray D, Leslie CC, Falke JJ, 2006. Specific translocation of protein kinase Calpha to the plasma membrane requires both Ca2+ and PIP2 recognition by its C2 domain. Mol. Biol. Cell 17(1), 56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farlow MR, Thompson RE, Wei L-J, Tuchman AJ, Grenier E, Crockford D, Wilke S, Benison J, Alkon DL, 2019. A Randomized, Double-Blind, Placebo-Controlled, Phase II Study Assessing Safety, Tolerability, and Efficacy of Bryostatin in the Treatment of Moderately Severe to Severe Alzheimer’s Disease. J. Alzheimer's Dis 67, 555–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandis AZ, Wenk MR, 2007. Membrane lipids as signaling molecules. Curr. Opin. Lipidol 18(2), 121–128. [DOI] [PubMed] [Google Scholar]
- Ferreira JC, Brum PC, Mochly-Rosen D, 2011. betaIIPKC and epsilonPKC isozymes as potential pharmacological targets in cardiac hypertrophy and heart failure. J. Mol. Cell. Cardiol 51(4), 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier A, Murray AW, 1987. Application of phorbol ester to mouse skin causes a rapid and sustained loss of protein kinase C. Nature 330(6150), 767–769. [DOI] [PubMed] [Google Scholar]
- Fujiki H, Mori M, Nakayasu M, Terada M, Sugimura T, Moore RE, 1981. Indole alkaloids: dihydroteleocidin B, teleocidin, and lyngbyatoxin A as members of a new class of tumor promoters. Proc. Natl. Acad. Sci. U.S.A 78(6), 3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki H, Suganuma M, Hakii H, Bartolini G, Moore RE, Takayama S, Sugimura T, 1984. A two-stage mouse skin carcinogenesis study of lyngbyatoxin A. J. Cancer Res. Clin. Oncol 108(1), 174–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürstenberger G, Berry DL, Sorg B, Marks F, 1981. Skin tumor promotion by phorbol esters is a two-stage process. Proc. Natl. Acad. Sci. U.S.A 78(12), 7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos LL, Newton AC, 2008. Spatiotemporal dynamics of lipid signaling: protein kinase C as a paradigm. IUBMB Life 60(12), 782–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bermejo ML, Leskow FC, Fujii T, Wang Q, Blumberg PM, Ohba M, Kuroki T, Han KC, Lee J, Marquez VE, Kazanietz MG, 2002. Diacylglycerol (DAG)-lactones, a new class of protein kinase C (PKC) agonists, induce apoptosis in LNCaP prostate cancer cells by selective activation of PKCalpha. J. Biol. Chem 277(1), 645–655. [DOI] [PubMed] [Google Scholar]
- Garrido JL, Godoy JA, Alvarez A, Bronfman M, Inestrosa NC, 2002. Protein kinase C inhibits amyloid beta peptide neurotoxicity by acting on members of the Wnt pathway. FASEB J. 16(14), 1982–1984. [DOI] [PubMed] [Google Scholar]
- Giorgione J, Hysell M, Harvey DF, Newton AC, 2003. Contribution of the C1A and C1B domains to the membrane interaction of protein kinase C. Biochemistry 42(38), 11194–11202. [DOI] [PubMed] [Google Scholar]
- Giorgione JR, Lin JH, McCammon JA, Newton AC, 2006. Increased membrane affinity of the C1 domain of protein kinase Cdelta compensates for the lack of involvement of its C2 domain in membrane recruitment. J. Biol. Chem 281(3), 1660–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goni FM, Alonso A, 1999. Structure and functional properties of diacylglycerols in membranes. Prog. Lipid Res 38(1), 1–48. [DOI] [PubMed] [Google Scholar]
- Gordon R, Singh N, Lawana V, Ghosh A, Harischandra DS, Jin H, Hogan C, Sarkar S, Rokad D, Panicker N, Anantharam V, Kanthasamy AG, Kanthasamy A, 2016. Protein kinase Cdelta upregulation in microglia drives neuroinflammatory responses and dopaminergic neurodegeneration in experimental models of Parkinson's disease. Neurobiol. Dis 93, 96–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishin NV, 2001. Treble clef finger--a functionally diverse zinc-binding structural motif. Nucleic Acids Res. 29(8), 1703–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves JT, Kuriyan J, 2010. Molecular mechanisms in signal transduction at the membrane. Nat. Struct. Mol. Biol 17(6), 659–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C, Serrano-Villar S, Madrid-Elena N, Perez-Elias MJ, Martin ME, Barbas C, Ruiperez J, Munoz E, Munoz-Fernandez MA, Castor T, Moreno S, 2016. Bryostatin-1 for latent virus reactivation in HIV-infected patients on antiretroviral therapy. AIDS 30(9), 1385–1392. [DOI] [PubMed] [Google Scholar]
- Hardman C, Ho S, Shimizu A, Luu-Nguyen Q, Sloane JL, Soliman MSA, Marsden MD, Zack JA, Wender PA, 2020. Synthesis and evaluation of designed PKC modulators for enhanced cancer immunotherapy. Nat. Commun 11(1), 1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker E, 1967. Phorbol esters from croton oil chemical nature and biological activities. The Science of Nature 54(11), 282–284. [DOI] [PubMed] [Google Scholar]
- Hecker E, Schmidt R, 1974. Phorbolesters — the Irritants and Cocarcinogens of Croton Tiglium L, in: Andersen NH, Brady SF, Harris CM, Harris TM, Hecker E, Hindley KB, McGregor DN, Marshall JA, Roberts JC, Schmidt R, Schrauzer GN, Swan GA, Tamm C, Wagner H, Winterfeldt E, Herz W, Grisebach H, Kirby GW (Eds.), Fortschritte der Chemie Organischer Naturstoffe / Progress in the Chemistry of Organic Natural Products. Springer Vienna, Vienna, pp. 377–467. [DOI] [PubMed] [Google Scholar]
- Hennings H, Blumberg PM, Pettit GR, Herald CL, Shores R, Yuspa SH, 1987. Bryostatin 1, an activator of protein kinase C, inhibits tumor promotion by phorbol esters in SENCAR mouse skin. Carcinogenesis 8(9), 1343–1346. [DOI] [PubMed] [Google Scholar]
- Hommel U, Zurini M, Luyten M, 1994. Solution structure of a cysteine rich domain of rat protein kinase C. Nat. Struct. Biol 1(6), 383–387. [DOI] [PubMed] [Google Scholar]
- Hong J, Nam J, Seo E-K, Lee S, 2010. Daphnane diterpene esters with anti-proliferative activities against human lung cancer cells from Daphne genkwa. Chem. Pharm. Bull. (Tokyo) 58 2, 234–237. [DOI] [PubMed] [Google Scholar]
- Horowitz AD, Fujiki H, Weinstein IB, Jeffrey A, Okin E, Moore RE, Sugimura T, 1983. Comparative effects of aplysiatoxin, debromoaplysiatoxin, and teleocidin on receptor binding and phospholipid metabolism. Cancer Res. 43(4), 1529–1535. [PubMed] [Google Scholar]
- Hurley JH, Newton AC, Parker PJ, Blumberg PM, Nishizuka Y, 1997. Taxonomy and function of C1 protein kinase C homology domains. Protein Sci. 6(2), 477–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huwiler A, Fabbro D, Pfeilschifter J, 1994. Comparison of different tumour promoters and bryostatin 1 on protein kinase C activation and down-regulation in rat renal mesangial cells. Biochem. Pharmacol 48(4), 689–700. [DOI] [PubMed] [Google Scholar]
- Ibarreta D, Duchén M, Ma D, Qiao L, Kozikowski AP, Etcheberrigaray R, 1999. Benzolactam (BL) enhances sAPP secretion in fibroblasts and in PC12 cells. Neuroreport 10(5). [DOI] [PubMed] [Google Scholar]
- Igumenova TI, 2015. Dynamics and Membrane Interactions of Protein Kinase C. Biochemistry 54(32), 4953–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie K, Kajiyama S.-i., Funaki A, Koshimizu K, Hayashi H, Arai M, 1990. Biosynthesis of indole alkaloid tumor promoters teleocidins (I) possible biosynthetic pathway of the monoterpenoid moieties of teleocidins. Tetrahedron 46(8), 2773–2788. [Google Scholar]
- Irie K, Yanagita RC, Nakagawa Y, 2012. Challenges to the development of bryostatin-type anticancer drugs based on the activation mechanism of protein kinase Cdelta. Med. Res. Rev 32(3), 518–535. [DOI] [PubMed] [Google Scholar]
- Johnson JE, Giorgione J, Newton AC, 2000. The C1 and C2 domains of protein kinase C are independent membrane targeting modules, with specificity for phosphatidylserine conferred by the C1 domain. Biochemistry 39(37), 11360–11369. [DOI] [PubMed] [Google Scholar]
- Kageyama M, Tamura T, Nantz MH, Roberts JC, Somfai P, Whritenour DC, Masamune S, 1990. Synthesis of bryostatin 7. J. Am. Chem. Soc 112(20), 7407–7408. [Google Scholar]
- Kang JH, Chung HE, Kim SY, Kim Y, Lee J, Lewin NE, Pearce LV, Blumberg PM, Marquez VE, 2003. Conformationally constrained analogues of diacylglycerol (DAG). Effect on protein kinase C (PK-C) binding by the isosteric replacement of sn-1 and sn-2 esters in DAG-lactones. Bioorg. Med. Chem 11(12), 2529–2539. [DOI] [PubMed] [Google Scholar]
- Kang JH, Kim SY, Lee J, Marquez VE, Lewin NE, Pearce LV, Blumberg PM, 2004. Macrocyclic diacylglycerol-bis-lactones as conformationally constrained analogues of diacylglycerol-lactones. Interactions with protein kinase C. J. Med. Chem 47(16), 4000–4007. [DOI] [PubMed] [Google Scholar]
- Kang JH, Peach ML, Pu Y, Lewin NE, Nicklaus MC, Blumberg PM, Marquez VE, 2005. Conformationally constrained analogues of diacylglycerol (DAG). 25. Exploration of the sn-1 and sn-2 carbonyl functionality reveals the essential role of the sn-1 carbonyl at the lipid interface in the binding of DAG-lactones to protein kinase C. J. Med. Chem 48(18), 5738–5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, Faraoni R, Floyd M, Hunt JP, Lockhart DJ, Milanov ZV, Morrison MJ, Pallares G, Patel HK, Pritchard S, Wodicka LM, Zarrinkar PP, 2008. A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol 26(1), 127–132. [DOI] [PubMed] [Google Scholar]
- Kato Y, Scheuer PJ, 1974. Aplysiatoxin and debromoaplysiatoxin, constituents of the marine mollusk Stylocheilus longicauda. J. Am. Chem. Soc 96(7), 2245–2246. [DOI] [PubMed] [Google Scholar]
- Kazanietz MG, Areces LB, Bahador A, Mischak H, Goodnight J, Mushinski JF, Blumberg PM, 1993. Characterization of ligand and substrate specificity for the calcium-dependent and calcium-independent protein kinase C isozymes. Mol. Pharmacol 44(2), 298. [PubMed] [Google Scholar]
- Kazanietz MG, Lewin NE, Gao F, Pettit GR, Blumberg PM, 1994. Binding of [26-3H]bryostatin 1 and analogs to calcium-dependent and calcium-independent protein kinase C isozymes. Mol. Pharmacol 46(2), 374–379. [PubMed] [Google Scholar]
- Kazanietz MG, Wang S, Milne GW, Lewin NE, Liu HL, Blumberg PM, 1995. Residues in the second cysteine-rich region of protein kinase C delta relevant to phorbol ester binding as revealed by site-directed mutagenesis. J. Biol. Chem 270(37), 21852–21859. [DOI] [PubMed] [Google Scholar]
- Keck GE, Poudel YB, Cummins TJ, Rudra A, Covel JA, 2011. Total Synthesis of Bryostatin 1. J. Am. Chem. Soc 133(4), 744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck GE, Poudel YB, Rudra A, Stephens JC, Kedei N, Lewin NE, Peach ML, Blumberg PM, 2010. Molecular modeling, total synthesis, and biological evaluations of C9-deoxy bryostatin 1. Angew. Chem. Int. Ed. Engl 49(27), 4580–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholodenko BN, Hoek JB, Westerhoff HV, 2000. Why cytoplasmic signalling proteins should be recruited to cell membranes. Trends Cell Biol. 10(5), 173–178. [DOI] [PubMed] [Google Scholar]
- Kim TT, Saunders T, Bieber E, Phillippe M, 1999. Tissue-specific protein kinase C isoform expression in rat uterine tissue. J. Soc. Gynecol. Investig 6(6), 293–300. [DOI] [PubMed] [Google Scholar]
- Kimura K, Mizutani MY, Tomioka N, Endo Y, Shudo K, Itai A, 1999. Docking Study of Bryostatins to Protein Kinase Cδ Cys2 Domain. Chem. Pharm. Bull. (Tokyo) 47(8), 1134–1137. [Google Scholar]
- Kobayashi T, Yanagita RC, Irie K, 2020. Synthesis and biological activities of simplified aplysiatoxin analogs focused on the CH/π interaction. Bioorganic Med. Chem. Lett 30(24), 127657. [DOI] [PubMed] [Google Scholar]
- Konig B, DiNitto PA, Blumberg PM, 1985. Stoichiometric binding of diacylglycerol to the phorbol ester receptor. J. Cell. Biochem 29(1), 37–44. [DOI] [PubMed] [Google Scholar]
- Korin YD, Brooks DG, Brown S, Korotzer A, Zack JA, 2002. Effects of Prostratin on T-Cell Activation and Human Immunodeficiency Virus Latency. J. Virol 76(16), 8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozikowski AP, Chen Y, Subhasish T, Lewin NE, Blumberg PM, Zhong Z, D'Annibale MA, Wang WL, Shen Y, Langley B, 2009. Searching for disease modifiers-PKC activation and HDAC inhibition - a dual drug approach to Alzheimer's disease that decreases Abeta production while blocking oxidative stress. ChemMedChem 4(7), 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozikowski AP, Ma D, Du L, Lewin NE, Blumberg PM, 1995. Effect of Alteration of the Heterocyclic Nucleus of Indolactam V on Its Isoform Selectivity for PKC. Palladium-Catalyzed Route to Benzofuran Analogs of ILV. J. Am. Chem. Soc 117(25), 6666–6672. [PubMed] [Google Scholar]
- Kozikowski AP, Nowak I, Petukhov PA, Etcheberrigaray R, Mohamed A, Tan M, Lewin N, Hennings H, Pearce LL, Blumberg PM, 2003. New amide-bearing benzolactam-based protein kinase C modulators induce enhanced secretion of the amyloid precursor protein metabolite sAPPalpha. J. Med. Chem 46(3), 364–373. [DOI] [PubMed] [Google Scholar]
- Kozikowski AP, Wang S, Ma D, Yao J, Ahmad S, Glazer RI, Bogi K, Acs P, Modarres S, Lewin NE, Blumberg PM, 1997. Modeling, Chemistry, and Biology of the Benzolactam Analogues of Indolactam V (ILV). 2. Identification of the Binding Site of the Benzolactams in the CRD2 Activator-Binding Domain of PKCδ and Discovery of an ILV Analogue of Improved Isozyme Selectivity. J. Med. Chem 40(9), 1316–1326. [DOI] [PubMed] [Google Scholar]
- Kraft AS, Smith JB, Berkow RL, 1986. Bryostatin, an activator of the calcium phospholipid-dependent protein kinase, blocks phorbol ester-induced differentiation of human promyelocytic leukemia cells HL-60. Proc. Natl. Acad. Sci. U.S.A 83(5), 1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel MT, Newton AC, 2010. Calcium transduces plasma membrane receptor signals to produce diacylglycerol at Golgi membranes. J. Biol. Chem 285(30), 22748–22752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupchan SM, Baxter RL, 1975. Mezerein: antileukemic principle isolated from Daphne mezereum L. Science 187(4177), 652. [DOI] [PubMed] [Google Scholar]
- Kusumi A, Fujiwara TK, Morone N, Yoshida KJ, Chadda R, Xie M, Kasai RS, Suzuki KGN, 2012. Membrane mechanisms for signal transduction: The coupling of the meso-scale raft domains to membrane-skeleton-induced compartments and dynamic protein complexes. Semin. Cell Dev. Biol 23(2), 126–144. [DOI] [PubMed] [Google Scholar]
- Landau EM, Rosenbusch JP, 1996. Lipidic cubic phases: A novel concept for the crystallization of membrane proteins. Proc. Natl. Acad. Sci. U.S.A 93(25), 14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautala S, Provenzani R, Koivuniemi A, Kulig W, Talman V, Róg T, Tuominen RK, Yli-Kauhaluoma J, Bunker A, 2020. Rigorous Computational Study Reveals What Docking Overlooks: Double Trouble from Membrane Association in Protein Kinase C Modulators. J. Chem. Inf. Model 60(11), 5624–5633. [DOI] [PubMed] [Google Scholar]
- Lee HW, Smith L, Pettit GR, Bingham Smith J, 1996. Dephosphorylation of activated protein kinase C contributes to downregulation by bryostatin. Am. J. Physiol 271(1 Pt 1), C304–311. [DOI] [PubMed] [Google Scholar]
- Lee J, Han KC, Kang JH, Pearce LL, Lewin NE, Yan S, Benzaria S, Nicklaus MC, Blumberg PM, Marquez VE, 2001. Conformationally constrained analogues of diacylglycerol. 18. The incorporation of a hydroxamate moiety into diacylglycerol-lactones reduces lipophilicity and helps discriminate between sn-1 and sn-2 binding modes to protein kinase C (PK-C). Implications for isozyme specificity. J. Med. Chem 44(25), 4309–4312. [DOI] [PubMed] [Google Scholar]
- Lee J, Kim SY, Kang JH, Acs G, Acs P, Blumberg PM, Marquez VE, 2004. Conformationally constrained diacylglycerol (DAG) analogs: 4-C-hydroxyethyl-5-O-acyl-2,3-dideoxy-D-glyceropentono-1,4-lactone analogs as protein kinase C (PKC) ligands. Eur. J. Med. Chem 39(1), 69–77. [DOI] [PubMed] [Google Scholar]
- Lee J, Lewin NE, Acs P, Blumberg PM, Marquez VE, 1996a. Conformationally constrained analogues of diacylglycerol. 13. Protein kinase C ligands based on templates derived from 2,3-dideoxy-L-erythro(threo)-hexono-1,4-lactone and 2-deoxyapiolactone. J. Med. Chem 39(25), 4912–4919. [DOI] [PubMed] [Google Scholar]
- Lee J, Marquez VE, Blumberg PM, Krausz KW, Kazanietz MG, 1993. Conformationally constrained analogues of diacylglycerol (DAG)--II. Differential interaction of delta-lactones and gamma-lactones with protein kinase C (PK-C). Bioorg. Med. Chem 1(2), 119–123. [DOI] [PubMed] [Google Scholar]
- Lee J, Sharma R, Wang S, Milne GW, Lewin NE, Szallasi Z, Blumberg PM, George C, Marquez VE, 1996b. Conformationally constrained analogues of diacylglycerol. 12. Ultrapotent protein kinase C ligands based on a chiral 4,4-disubstituted heptono-1,4-lactone template. J. Med. Chem 39(1), 36–45. [DOI] [PubMed] [Google Scholar]
- Leitges M, 2020. Investigations of mouse models during tumorigenesis revealed essential but distinct in vivo functions among the PKC family. Adv Biol Regul 78, 100756. [DOI] [PubMed] [Google Scholar]
- Li J, Ziemba BP, Falke JJ, Voth GA, 2014. Interactions of protein kinase C-alpha C1A and C1B domains with membranes: a combined computational and experimental study. J. Am. Chem. Soc 136(33), 11757–11766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limatola C, Schaap D, Moolenaar WH, van Blitterswijk WJ, 1994. Phosphatidic acid activation of protein kinase C-zeta overexpressed in COS cells: comparison with other protein kinase C isotypes and other acidic lipids. Biochem. J 304 (Pt 3), 1001–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Woo SK, Krische MJ, 2011. Total Synthesis of Bryostatin 7 via C–C Bond-Forming Hydrogenation. J. Am. Chem. Soc 133(35), 13876–13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly C, Shimizu AJ, Vargas MV, Duim WC, Wender PA, Olson DE, 2020. Bryostatin 1 Promotes Synaptogenesis and Reduces Dendritic Spine Density in Cortical Cultures through a PKC-Dependent Mechanism. ACS Chemical Neuroscience 11(11), 1545–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach UR, Lewin NE, Blumberg PM, Kozikowski AP, 2006. Synthesis and pharmacological evaluation of 8- and 9-substituted benzolactam-v8 derivatives as potent ligands for protein kinase C, a therapeutic target for Alzheimer's disease. ChemMedChem 1(3), 307–314. [DOI] [PubMed] [Google Scholar]
- Majumder PK, Pandey P, Sun X, Cheng K, Datta R, Saxena S, Kharbanda S, Kufe D, 2000. Mitochondrial translocation of protein kinase C delta in phorbol ester-induced cytochrome c release and apoptosis. J. Biol. Chem 275(29), 21793–21796. [DOI] [PubMed] [Google Scholar]
- Marquez VE, Blumberg PM, 2003. Synthetic diacylglycerols (DAG) and DAG-lactones as activators of protein kinase C (PK-C). Acc. Chem. Res 36(6), 434–443. [DOI] [PubMed] [Google Scholar]
- Masuda A, Irie K, Nakagawa Y, Ohigashi H, 2002. Binding selectivity of conformationally restricted analogues of (−)-indolactam-V to the C1 domains of protein kinase C isozymes. Biosci. Biotechnol. Biochem 66(7), 1615–1617. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Kobayakawa T, Tsuchiya K, Hattori SI, Nomura W, Gatanaga H, Yoshimura K, Oka S, Endo Y, Tamamura H, Mitsuya H, Maeda K, 2019. Benzolactam-related compounds promote apoptosis of HIV-infected human cells via protein kinase C-induced HIV latency reversal. J. Biol. Chem 294(1), 116–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medkova M, Cho W, 1999. Interplay of C1 and C2 domains of protein kinase C-alpha in its membrane binding and activation. J. Biol. Chem 274(28), 19852–19861. [DOI] [PubMed] [Google Scholar]
- Melowic HR, Stahelin RV, Blatner NR, Tian W, Hayashi K, Altman A, Cho W, 2007. Mechanism of diacylglycerol-induced membrane targeting and activation of protein kinase Ctheta. J. Biol. Chem 282(29), 21467–21476. [DOI] [PubMed] [Google Scholar]
- Miyake R, Tanaka Y, Tsuda T, Kaibuchi K, Kikkawa U, Nishizuka Y, 1984. Activation of protein kinase C by non-phorbol tumor promoter, mezerein. Biochem. Biophys. Res. Commun 121(2), 649–656. [DOI] [PubMed] [Google Scholar]
- Miyamae Y, Villareal MO, Abdrabbah MB, Isoda H, Shigemori H, 2009. Hirseins A and B, Daphnane Diterpenoids from Thymelaea hirsuta That Inhibit Melanogenesis in B16 Melanoma Cells. J. Nat. Prod 72(5), 938–941. [DOI] [PubMed] [Google Scholar]
- Mochly-Rosen D, Das K, Grimes KV, 2012. Protein kinase C, an elusive therapeutic target? Nat. Rev. Drug Discov 11(12), 937–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RE, Blackman AJ, Cheuk CE, Mynderse JS, Matsumoto GK, Clardy J, Woodard RW, Craig JC, 1984. Absolute stereochemistries of the aplysiatoxins and oscillatoxin A. J. Org. Chem 49(13), 2484–2489. [Google Scholar]
- Nacro K, Bienfait B, Lewin NE, Blumberg PM, Marquez VE, 2000. Diacylglycerols with lipophilically equivalent branched acyl chains display high affinity for protein kinase C (PK-C). A direct measure of the effect of constraining the glycerol backbone in DAG lactones. Bioorg. Med. Chem. Lett 10(7), 653–655. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, 2012. Artificial analogs of naturally occurring tumor promoters as biochemical tools and therapeutic leads. Biosci. Biotechnol. Biochem 76(7), 1262–1274. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Irie K, Yanagita RC, Ohigashi H, Tsuda K.-i., 2005. Indolactam-V Is Involved in the CH/π Interaction with Pro-11 of the PKCδ C1B Domain: Application for the Structural Optimization of the PKCδ Ligand. J. Am. Chem. Soc 127(16), 5746–5747. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Yanagita RC, Hamada N, Murakami A, Takahashi H, Saito N, Nagai H, Irie K, 2009. A simple analogue of tumor-promoting aplysiatoxin is an antineoplastic agent rather than a tumor promoter: development of a synthetically accessible protein kinase C activator with bryostatin-like activity. J. Am. Chem. Soc 131(22), 7573–7579. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Kishi Y, Pajares MA, Rando RR, 1989. Structural basis of protein kinase C activation by tumor promoters. Proc. Natl. Acad. Sci. U.S.A 86(24), 9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H, Brewer KA, Exton JH, 1993. Activation of the zeta isozyme of protein kinase C by phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem 268(1), 13–16. [PubMed] [Google Scholar]
- Nelson TJ, Sun MK, Lim C, Sen A, Khan T, Chirila FV, Alkon DL, 2017. Bryostatin Effects on Cognitive Function and PKCvarepsilon in Alzheimer's Disease Phase IIa and Expanded Access Trials. J. Alzheimer's Dis 58(2), 521–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton AC, 1995. Protein Kinase C: Seeing two domains. Curr. Biol 5(9), 973–976. [DOI] [PubMed] [Google Scholar]
- Newton AC, 2009. Lipid activation of protein kinases. J. Lipid Res 50 Suppl, S266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton AC, 2018. Protein kinase C as a tumor suppressor. Semin. Cancer Biol 48, 18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton AC, Brognard J, 2017. Reversing the Paradigm: Protein Kinase C as a Tumor Suppressor. Trends Pharmacol. Sci 38(5), 438–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezhat F, Wadler S, Muggia F, Mandeli J, Goldberg G, Rahaman J, Runowicz C, Murgo AJ, Gardner GJ, 2004. Phase II trial of the combination of bryostatin-1 and cisplatin in advanced or recurrent carcinoma of the cervix: a New York Gynecologic Oncology Group study. Gynecologic Oncology 93(1), 144–148. [DOI] [PubMed] [Google Scholar]
- Nishikawa T, Edelstein D, Brownlee M, 2000. The missing link: a single unifying mechanism for diabetic complications. Kidney Int. Suppl 77, S26–30. [DOI] [PubMed] [Google Scholar]
- Nishizuka Y, 1984. The role of protein kinase C in cell surface signal transduction and tumour promotion. Nature 308(5961), 693–698. [DOI] [PubMed] [Google Scholar]
- Nishizuka Y, 1986. Studies and perspectives of protein kinase C. Science 233(4761), 305–312. [DOI] [PubMed] [Google Scholar]
- Nomura W, Narumi T, Ohashi N, Serizawa Y, Lewin NE, Blumberg PM, Furuta T, Tamamura H, 2011. Synthetic caged DAG-lactones for photochemically controlled activation of protein kinase C. ChemBioChem 12(4), 535–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oancea E, Meyer T, 1998. Protein Kinase C as a Molecular Machine for Decoding Calcium and Diacylglycerol Signals. Cell 95(3), 307–318. [DOI] [PubMed] [Google Scholar]
- Ohashi N, Kobayashi R, Nomura W, Kobayakawa T, Czikora A, Herold BK, Lewin NE, Blumberg PM, Tamamura H, 2017. Synthesis and Evaluation of Dimeric Derivatives of Diacylglycerol-Lactones as Protein Kinase C Ligands. Bioconjug. Chem 28(8), 2135–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori K, Ogawa Y, Obitsu T, Ishikawa Y, Nishiyama S, Yamamura S, 2000. Total Synthesis of Bryostatin 3. Angew. Chem. Int. Ed 39(13), 2290–2294. [PubMed] [Google Scholar]
- Ohno M, Endo Y, Hirano M, Itai A, Shudo K, 1993. Designed molecules reproducing the two conformations of teleocidins. Tetrahedron Lett. 34(50), 8119–8122. [Google Scholar]
- Ono Y, Fujii T, Igarashi K, Kuno T, Tanaka C, Kikkawa U, Nishizuka Y, 1989. Phorbol ester binding to protein kinase C requires a cysteine-rich zinc-finger-like sequence. Proc. Natl. Acad. Sci. U.S.A 86(13), 4868–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y, Fujii T, Ogita K, Kikkawa U, Igarashi K, Nishizuka Y, 1988. The structure, expression, and properties of additional members of the protein kinase C family. J. Biol. Chem 263(14), 6927–6932. [PubMed] [Google Scholar]
- Pak Y, Enyedy IJ, Varady J, Kung JW, Lorenzo PS, Blumberg PM, Wang S, 2001. Structural basis of binding of high-affinity ligands to protein kinase C: prediction of the binding modes through a new molecular dynamics method and evaluation by site-directed mutagenesis. J. Med. Chem 44(11), 1690–1701. [DOI] [PubMed] [Google Scholar]
- Parker PJ, Coussens L, Totty N, Rhee L, Young S, Chen E, Stabel S, Waterfield MD, Ullrich A, 1986. The complete primary structure of protein kinase C--the major phorbol ester receptor. Science 233(4766), 853–859. [DOI] [PubMed] [Google Scholar]
- Pavlick AC, Wu J, Roberts J, Rosenthal MA, Hamilton A, Wadler S, Farrell K, Carr M, Fry D, Murgo AJ, Oratz R, Hochster H, Liebes L, Muggia F, 2009. Phase I study of bryostatin 1, a protein kinase C modulator, preceding cisplatin in patients with refractory non-hematologic tumors. Cancer Chemother. Pharmacol 64(4), 803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit GR, Herald CL, Doubek DL, Herald DL, Arnold E, Clardy J, 1982. Isolation and structure of bryostatin 1. J. Am. Chem. Soc 104(24), 6846–6848. [Google Scholar]
- Provenzani R, Tarvainen I, Brandoli G, Lempinen A, Artes S, Turku A, Jantti MH, Talman V, Yli-Kauhaluoma J, Tuominen RK, Boije Af Gennas G, 2018. Scaffold hopping from (5-hydroxymethyl) isophthalates to multisubstituted pyrimidines diminishes binding affinity to the C1 domain of protein kinase C. PLoS One 13(4), e0195668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Y, Perry NA, Yang D, Lewin NE, Kedei N, Braun DC, Choi SH, Blumberg PM, Garfield SH, Stone JC, Duan D, Marquez VE, 2005. A novel diacylglycerol-lactone shows marked selectivity in vitro among C1 domains of protein kinase C (PKC) isoforms alpha and delta as well as selectivity for RasGRP compared with PKCalpha. J. Biol. Chem 280(29), 27329–27338. [DOI] [PubMed] [Google Scholar]
- Rahimova N, Cooke M, Zhang S, Baker MJ, Kazanietz MG, 2020. The PKC universe keeps expanding: From cancer initiation to metastasis. Adv Biol Regul 78, 100755. [DOI] [PubMed] [Google Scholar]
- Rahman GM, Shanker S, Lewin NE, Kedei N, Hill CS, Prasad BV, Blumberg PM, Das J, 2013. Identification of the activator-binding residues in the second cysteine-rich regulatory domain of protein kinase Ctheta (PKCtheta). Biochem. J 451(1), 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando RR, Young N, 1984. The stereospecific activation of protein kinase C. Biochem. Biophys. Res. Commun 122(2), 818–823. [DOI] [PubMed] [Google Scholar]
- Roivainen R, Messing RO, 1993. The phorbol derivatives thymeleatoxin and 12-deoxyphorbol-13-O-phenylacetate-10-acetate cause translocation and down-regulation of multiple protein kinase C isozymes. FEBS Lett. 319(1–2), 31–34. [DOI] [PubMed] [Google Scholar]
- Rosse C, Linch M, Kermorgant S, Cameron AJM, Boeckeler K, Parker PJ, 2010. PKC and the control of localized signal dynamics. Nature Reviews Molecular Cell Biology 11(2), 103–112. [DOI] [PubMed] [Google Scholar]
- Rybin VO, Guo J, Sabri A, Elouardighi H, Schaefer E, Steinberg SF, 2004. Stimulus-specific differences in protein kinase C delta localization and activation mechanisms in cardiomyocytes. J. Biol. Chem 279(18), 19350–19361. [DOI] [PubMed] [Google Scholar]
- Rybin VO, Steinberg SF, 1994. Protein kinase C isoform expression and regulation in the developing rat heart. Circ. Res 74(2), 299–309. [DOI] [PubMed] [Google Scholar]
- Ryckbosch SM, Wender PA, Pande VS, 2017. Molecular dynamics simulations reveal ligand-controlled positioning of a peripheral protein complex in membranes. Nat. Commun 8(1), 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryves WJ, Evans AT, Olivier AR, Parker PJ, Evans FJ, 1991. Activation of the PKC-isotypes α, β1, γ, δ, and ε by phorbol esters of different biological activities. FEBS Lett. 288(1), 5–9. [DOI] [PubMed] [Google Scholar]
- Sako T, Yuspa SH, Herald CL, Pettit GR, Blumberg PM, 1987. Partial parallelism and partial blockade by bryostatin 1 of effects of phorbol ester tumor promoters on primary mouse epidermal cells. Cancer Res. 47(20), 5445–5450. [PubMed] [Google Scholar]
- Saraiva L, Fresco P, Pinto E, Goncalves J, 2004. Characterization of phorbol esters activity on individual mammalian protein kinase C isoforms, using the yeast phenotypic assay. Eur. J. Pharmacol 491(2–3), 101–110. [DOI] [PubMed] [Google Scholar]
- Saraiva L, Fresco P, Pinto E, Portugal H, Goncalves J, 2001. Differential activation by daphnetoxin and mezerein of PKC-isotypes alpha, beta I, delta and zeta. Planta Med. 67(9), 787–790. [DOI] [PubMed] [Google Scholar]
- Schultz A, Ling M, Larsson C, 2004. Identification of an amino acid residue in the protein kinase C C1b domain crucial for its localization to the Golgi network. J. Biol. Chem 279(30), 31750–31760. [DOI] [PubMed] [Google Scholar]
- Shanmugasundararaj S, Das J, Sandberg WS, Zhou X, Wang D, Messing RO, Bruzik KS, Stehle T, Miller KW, 2012. Structural and functional characterization of an anesthetic binding site in the second cysteine-rich domain of protein kinase Cdelta*. Biophys. J 103(11), 2331–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey NA, Blumberg PM, 1986. Comparison of the activity of phorbol 12-myristate 13-acetate and the diglyceride glycerol 1-myristate 2-acetate. Carcinogenesis 7(4), 677–679. [DOI] [PubMed] [Google Scholar]
- Shimomura K, Mullinix MG, Kakunaga T, Fujiki H, Sugimura T, 1983. Bromine residue at hydrophilic region influences biological activity of aplysiatoxin, a tumor promoter. Science 222(4629), 1242. [DOI] [PubMed] [Google Scholar]
- Shirai Y, Murakami T, Kuramasu M, Iijima L, Saito N, 2007. A novel PIP2 binding of epsilonPKC and its contribution to the neurite induction ability. J. Neurochem 102(5), 1635–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigano DM, Peach ML, Nacro K, Choi Y, Lewin NE, Nicklaus MC, Blumberg PM, Marquez VE, 2003. Differential binding modes of diacylglycerol (DAG) and DAG lactones to protein kinase C (PK-C). J. Med. Chem 46(9), 1571–1579. [DOI] [PubMed] [Google Scholar]
- Slaga TJ, Fischer SM, Nelson K, Gleason GL, 1980. Studies on the mechanism of skin tumor promotion: Evidence for several stages in promotion. Proc. Natl. Acad. Sci. U.S.A 77(6), 3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodeoka M, Arai MA, Adachi K, Uotsu K, Shibasaki M, 1998. Rational Design, Synthesis, and Evaluation of a New Type of PKC Inhibitor. J. Am. Chem. Soc 120(2), 457–458. [Google Scholar]
- Soltoff SP, 2007. Rottlerin: an inappropriate and ineffective inhibitor of PKCdelta. Trends Pharmacol. Sci 28(9), 453–458. [DOI] [PubMed] [Google Scholar]
- Sossin WS, Schwartz JH, 1993. Ca(2+)-independent protein kinase Cs contain an amino-terminal domain similar to the C2 consensus sequence. Trends Biochem. Sci 18(6), 207–208. [DOI] [PubMed] [Google Scholar]
- Sridhar J, Wei Z-L, Nowak I, Lewin NE, Ayres JA, Pearce LV, Blumberg PM, Kozikowski AP, 2003. New Bivalent PKC Ligands Linked by a Carbon Spacer: Enhancement in Binding Affinity. J. Med. Chem 46(19), 4196–4204. [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Digman MA, Medkova M, Ananthanarayanan B, Melowic HR, Rafter JD, Cho W, 2005. Diacylglycerol-induced membrane targeting and activation of protein kinase Cepsilon: mechanistic differences between protein kinases Cdelta and Cepsilon. J. Biol. Chem 280(20), 19784–19793. [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Digman MA, Medkova M, Ananthanarayanan B, Rafter JD, Melowic HR, Cho W, 2004. Mechanism of diacylglycerol-induced membrane targeting and activation of protein kinase Cdelta. J. Biol. Chem 279(28), 29501–29512. [DOI] [PubMed] [Google Scholar]
- Steinberg SF, 2008. Structural basis of protein kinase C isoform function. Physiol. Rev 88(4), 1341–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steube KG, Drexler HG, 1993. Differentiation and growth modulation of myeloid leukemia cells by the protein kinase C activating agent bryostatin-1. Leuk. Lymphoma 9(1-2), 141–148. [DOI] [PubMed] [Google Scholar]
- Stewart C, McDonald R, West FG, 2011. Expedient Route to the Tigliane-Daphnane Skeleton via Oxonium Ylide [1,2]-Shift. Org. Lett 13(4), 720–723. [DOI] [PubMed] [Google Scholar]
- Stewart MD, Cole TR, Igumenova TI, 2014. Interfacial partitioning of a loop hinge residue contributes to diacylglycerol affinity of conserved region 1 domains. J. Biol. Chem 289(40), 27653–27664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart MD, Igumenova TI, 2017. Toggling of Diacylglycerol Affinity Correlates with Conformational Plasticity in C1 Domains. Biochemistry 56(21), 2637–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart MD, Morgan B, Massi F, Igumenova TI, 2011. Probing the determinants of diacylglycerol binding affinity in the C1B domain of protein kinase Calpha. J. Mol. Biol 408(5), 949–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma M, Fujiki H, Tahira T, Cheuk C, Moore RE, Sugimura T, 1984. Estimation of tumor promoting activity and structure-function relationships of aplysiatoxins. Carcinogenesis 5(3), 315–318. [DOI] [PubMed] [Google Scholar]
- Szallasi Z, Bogi K, Gohari S, Biro T, Acs P, Blumberg PM, 1996. Non-equivalent roles for the first and second zinc fingers of protein kinase Cdelta. Effect of their mutation on phorbol ester-induced translocation in NIH 3T3 cells. J. Biol. Chem 271(31), 18299–18301. [DOI] [PubMed] [Google Scholar]
- Szallasi Z, Krsmanovic L, Blumberg PM, 1993. Nonpromoting 12-deoxyphorbol 13-esters inhibit phorbol 12-myristate 13-acetate induced tumor promotion in CD-1 mouse skin. Cancer Res. 53(11), 2507–2512. [PubMed] [Google Scholar]
- Szallasi Z, Smith CB, Pettit GR, Blumberg PM, 1994. Differential regulation of protein kinase C isozymes by bryostatin 1 and phorbol 12-myristate 13-acetate in NIH 3T3 fibroblasts. J. Biol. Chem 269(3), 2118–2124. [PubMed] [Google Scholar]
- Takai Y, Kishimoto A, Kikkawa U, Mori T, Nishizuka Y, 1979. Unsaturated diacylglycerol as a possible messenger for the activation of calcium-activated, phospholipid-dependent protein kinase system. Biochem. Biophys. Res. Commun 91(4), 1218–1224. [DOI] [PubMed] [Google Scholar]
- Talman V, Amadio M, Osera C, Sorvari S, Boije af Gennäs G, Yli-Kauhaluoma J, Rossi D, Govoni S, Collina S, Ekokoski E, Tuominen RK, Pascale A, 2013. The C1 domain-targeted isophthalate derivative HMI-1b11 promotes neurite outgrowth and GAP-43 expression through PKCα activation in SH-SY5Y cells. Pharmacol. Res 73, 44–54. [DOI] [PubMed] [Google Scholar]
- Talman V, Pascale A, Jantti M, Amadio M, Tuominen RK, 2016. Protein Kinase C Activation as a Potential Therapeutic Strategy in Alzheimer's Disease: Is there a Role for Embryonic Lethal Abnormal Vision-like Proteins? Basic Clin. Pharmacol. Toxicol 119(2), 149–160. [DOI] [PubMed] [Google Scholar]
- Talman V, Provenzani R, Boije af Gennas G, Tuominen RK, Yli-Kauhaluoma J, 2014. C1 domain-targeted isophthalates as protein kinase C modulators: structure-based design, structure-activity relationships and biological activities. Biochem. Soc. Trans 42(6), 1543–1549. [DOI] [PubMed] [Google Scholar]
- Talman V, Tuominen RK, Gennäs G.B.a., Yli-Kauhaluoma J, Ekokoski E, 2011. C1 Domain-Targeted Isophthalate Derivatives Induce Cell Elongation and Cell Cycle Arrest in HeLa Cells. PLoS One 6(5), e20053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trost BM, Dong G, 2008. Total synthesis of bryostatin 16 using atom-economical and chemoselective approaches. Nature 456(7221), 485–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trost BM, Wang Y, Buckl AK, Huang Z, Nguyen MH, Kuzmina O, 2020. Total synthesis of bryostatin 3. Science 368(6494), 1007. [DOI] [PubMed] [Google Scholar]
- Ujwal R, Bowie JU, 2011. Crystallizing membrane proteins using lipidic bicelles. Methods 55(4), 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW, 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol 9(2), 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varterasian ML, Pemberton PA, Hulburd K, Rodriguez DH, Murgo A, Al-Katib AM, 2001. Phase II Study of Bryostatin 1 in Patients with Relapsed Multiple Myeloma. Investigational New Drugs 19(3), 245–247. [DOI] [PubMed] [Google Scholar]
- Wada R, Suto Y, Kanai M, Shibasaki M, 2002. Dramatic Switching of Protein Kinase C Agonist/Antagonist Activity by Modifying the 12-Ester Side Chain of Phorbol Esters. J. Am. Chem. Soc 124(36), 10658–10659. [DOI] [PubMed] [Google Scholar]
- Wang QJ, Bhattacharyya D, Garfield S, Nacro K, Marquez VE, Blumberg PM, 1999. Differential localization of protein kinase C delta by phorbol esters and related compounds using a fusion protein with green fluorescent protein. J. Biol. Chem 274(52), 37233–37239. [DOI] [PubMed] [Google Scholar]
- Wang QJ, Fang TW, Fenick D, Garfield S, Bienfait B, Marquez VE, Blumberg PM, 2000. The lipophilicity of phorbol esters as a critical factor in determining the pattern of translocation of protein kinase C delta fused to green fluorescent protein. J. Biol. Chem 275(16), 12136–12146. [DOI] [PubMed] [Google Scholar]
- Wang S, Kazanietz MG, Blumberg PM, Marquez VE, Milne GW, 1996. Molecular modeling and site-directed mutagenesis studies of a phorbol ester-binding site in protein kinase C. J. Med. Chem 39(13), 2541–2553. [DOI] [PubMed] [Google Scholar]
- Wang S, Liu M, Lewin NE, Lorenzo PS, Bhattacharrya D, Qiao L, Kozikowski AP, Blumberg PM, 1999. Probing the binding of indolactam-V to protein kinase C through site-directed mutagenesis and computational docking simulations. J. Med. Chem 42(18), 3436–3446. [DOI] [PubMed] [Google Scholar]
- Wang S, Wang Z, Dent P, Grant S, 2003. Induction of tumor necrosis factor by bryostatin 1 is involved in synergistic interactions with paclitaxel in human myeloid leukemia cells. Blood 101(9), 3648–3657. [DOI] [PubMed] [Google Scholar]
- Wei ZL, Sakamuri S, Petukhov PA, George C, Lewin NE, Blumberg PM, Kozikowski AP, 2002. Synthesis and modeling study of (2S,5R,6R)- and (2S,5R,6S)-6-hydroxy- 8-(1-decynyl)-benzolactam-V8 as protein kinase C modulators. Org. Lett 4(13), 2169–2172. [DOI] [PubMed] [Google Scholar]
- Wender PA, Baryza JL, Bennett CE, Bi FC, Brenner SE, Clarke MO, Horan JC, Kan C, Lacôte E, Lippa B, Nell PG, Turner TM, 2002. The Practical Synthesis of a Novel and Highly Potent Analogue of Bryostatin. J. Am. Chem. Soc 124(46), 13648–13649. [DOI] [PubMed] [Google Scholar]
- Wender PA, Baryza JL, Brenner SE, DeChristopher BA, Loy BA, Schrier AJ, Verma VA, 2011a. Design, synthesis, and evaluation of potent bryostatin analogs that modulate PKC translocation selectivity. Proc. Natl. Acad. Sci. U.S.A 108(17), 6721–6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wender PA, Buschmann N, Cardin NB, Jones LR, Kan C, Kee J-M, Kowalski JA, Longcore KE, 2011b. Gateway synthesis of daphnane congeners and their protein kinase C affinities and cell-growth activities. Nature Chemistry 3(8), 615–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wender PA, De Brabander J, Harran PG, Jimenez J-M, Koehler MFT, Lippa B, Park C-M, Shiozaki M, 1998a. Synthesis of the First Members of a New Class of Biologically Active Bryostatin Analogues. J. Am. Chem. Soc 120(18), 4534–4535. [Google Scholar]
- Wender PA, DeBrabander J, Harran PG, Jimenez J-M, Koehler MFT, Lippa B, Park C-M, Siedenbiedel C, Pettit GR, 1998b. The design, computer modeling, solution structure, and biological evaluation of synthetic analogs of bryostatin 1. Proc. Natl. Acad. Sci. U.S.A 95(12), 6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wender PA, Hardman CT, Ho S, Jeffreys MS, Maclaren JK, Quiroz RV, Ryckbosch SM, Shimizu AJ, Sloane JL, Stevens MC, 2017. Scalable synthesis of bryostatin 1 and analogs, adjuvant leads against latent HIV. Science 358(6360), 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wender PA, Jesudason CD, Nakahira H, Tamura N, Tebbe AL, Ueno Y, 1997. The First Synthesis of a Daphnane Diterpene: The Enantiocontrolled Total Synthesis of (+)-Resiniferatoxin. J. Am. Chem. Soc 119(52), 12976–12977. [Google Scholar]
- Wender PA, Schrier AJ, 2011. Total Synthesis of Bryostatin 9. J. Am. Chem. Soc 133(24), 9228–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wender PA, Sloane JL, Luu-Nguyen QH, Ogawa Y, Shimizu AJ, Ryckbosch SM, Tyler JH, Hardman C, 2020. Function-Oriented Synthesis: Design, Synthesis, and Evaluation of Highly Simplified Bryostatin Analogues. J. Org. Chem 85 (23), 15116–15128. [DOI] [PubMed] [Google Scholar]
- Wu R, Chen H, Chang N, Xu Y, Jiao J, Zhang H, 2020. Unlocking the Drug Potential of the Bryostatin Family: Recent Advances in Product Synthesis and Biomedical Applications. Chemistry – A European Journal 26(6), 1166–1195. [DOI] [PubMed] [Google Scholar]
- Xu RX, Pawelczyk T, Xia TH, Brown SC, 1997. NMR structure of a protein kinase C-gamma phorbol-binding domain and study of protein-lipid micelle interactions. Biochemistry 36(35), 10709–10717. [DOI] [PubMed] [Google Scholar]
- Yanagita RC, Nakagawa Y, Yamanaka N, Kashiwagi K, Saito N, Irie K, 2008. Synthesis, Conformational Analysis, and Biological Evaluation of 1-Hexylindolactam-V10 as a Selective Activator for Novel Protein Kinase C Isozymes. J. Med. Chem 51(1), 46–56. [DOI] [PubMed] [Google Scholar]
- Yang H, Staveness D, Ryckbosch SM, Axtman AD, Loy BA, Barnes AB, Pande VS, Schaefer J, Wender PA, Cegelski L, 2018. REDOR NMR Reveals Multiple Conformers for a Protein Kinase C Ligand in a Membrane Environment. ACS Cent. Sci 4(1), 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H-B, Yang F, Li Y-Y, Gan J-H, Jiao W-H, Lin H-W, 2015. Cytotoxic Bryostatin Derivatives from the South China Sea Bryozoan Bugula neritina. J. Nat. Prod 78(5), 1169–1173. [DOI] [PubMed] [Google Scholar]
- Zanin-Zhorov A, Dustin ML, Blazar BR, 2011. PKC-θ function at the immunological synapse: prospects for therapeutic targeting. Trends Immunol. 32(8), 358–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Kazanietz MG, Blumberg PM, Hurley JH, 1995. Crystal structure of the cys2 activator-binding domain of protein kinase C delta in complex with phorbol ester. Cell 81(6), 917–924. [DOI] [PubMed] [Google Scholar]
- Zhang L, Luo R-H, Wang F, Dong Z-J, Yang L-M, Zheng Y-T, Liu J-K, 2010. Daphnane diterpenoids isolated from Trigonostemon thyrsoideum as HIV-1 antivirals. Phytochemistry 71(16), 1879–1883. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Guo Q, Sun X, Lu J, Cao Y, Pu Q, Chu Z, Gao L, Song Z, 2018. Total Synthesis of Bryostatin 8 Using an Organosilane-Based Strategy. Angew. Chem. Int. Ed 57(4), 942–946. [DOI] [PubMed] [Google Scholar]