Abstract
Background
Pantothenate kinase (PANK) is the first and rate-controlling enzymatic step in the only pathway for cellular coenzyme A (CoA) biosynthesis. PANK-associated neurodegeneration (PKAN), formerly known as Hallervorden–Spatz disease, is a rare, life-threatening neurologic disorder that affects the CNS and arises from mutations in the human PANK2 gene. Pantazines, a class of small molecules containing the pantazine moiety, yield promising therapeutic effects in an animal model of brain CoA deficiency. A reliable technique to identify the neurometabolic effects of PANK dysfunction and to monitor therapeutic responses is needed.
Methods
We applied 1H magnetic resonance spectroscopy as a noninvasive technique to evaluate the therapeutic effects of the newly developed Pantazine BBP-671.
Results
1H MRS reliably quantified changes in cerebral metabolites, including glutamate/glutamine, lactate, and N-acetyl aspartate in a neuronal Pank1 and Pank2 double-knockout (SynCre+ Pank1,2 dKO) mouse model of brain CoA deficiency. The neuronal SynCre+ Pank1,2 dKO mice had distinct decreases in Glx/tCr, NAA/tCr, and lactate/tCr ratios compared to the wildtype matched control mice that increased in response to BBP-671 treatment.
Conclusions
BBP-671 treatment completely restored glutamate/glutamine levels in the brains of the mouse model, suggesting that these metabolites are promising clinically translatable biomarkers for future therapeutic trials.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-022-03304-y.
Keywords: Pantothenate kinase, Coenzyme A, Neurodegeneration, Pantothenate kinase-associated neurodegeneration, 1H magnetic resonance spectroscopy, Metabolites, Therapeutics
Background
Pantothenate kinase (PanK) is the first and rate-controlling step in the only pathway for coenzyme A (CoA) biosynthesis [1]. CoA is essential for hundreds of metabolic reactions including the tricarboxylic acid (TCA) cycle, fatty acid oxidation and synthesis, amino acid metabolism, and neurotransmitter synthesis [1, 2]. A rare, life-threatening neurological disorder known as pantothenate kinase-associated neurodegeneration (PKAN) arises from mutations in the human PANK2 gene leading to a prominent extrapyramidal movement disorder and a characteristic deposition of iron in the basal ganglia [3]. CoA deficiency is thought to be the cause of the movement disorder and neurodegeneration in PKAN [4], but probes are not available to monitor CoA levels in the brain. Figure 1a depicts the effects of loss of CoA synthesis due to disruption of murine Pank1 and Pank2 genes in neurons. Pyruvate produced from glycolysis requires CoA to form acetyl-CoA, a substrate for the mitochondrial TCA cycle, and CoA limitation can disrupt TCA cycling [5] which, in turn, affects glutamate metabolism thereby causing cell death [6, 7] (Fig. 1a). Recent reviews discuss the various molecular consequences of CoA insufficiency in preclinical PKAN models [8, 9]. There are no disease-modifying treatments for PKAN, and therapeutics are desperately needed. A new potential PKAN therapeutic approach uses Pantazine small molecules that activate the PanK isoforms and thereby stimulate CoA production [10]. Pantazine PZ-2891 was previously reported to restore cerebral CoA levels in the SynCre+ Pank1,2 neuronal dKO mice [10]. In this study, we used proton magnetic resonance spectroscopy (1H MRS) to evaluate the brain metabolic derangements that are associated with neuronal CoA deficiency in this mouse model, with and without treatment with a newly developed Pantazine, BBP-671. 1H MRS is particularly advantageous for this study because it permits the noninvasive examination of the most abundant cerebral metabolites in vivo [11]. This Pantazine allosterically activates the alternate PanK3 isoform that is expressed in murine neurons and a proposed mechanism for the neurometabolic effect of CoA restoration by BBP-671 is shown in Fig. 1b.
Fig. 1.
a Schematic description of CoA loss due to Pank1,2 deletion in neurons leading to decreased TCA cycling and lower neuronal glutamate and NAA. b Schematic decription of CoA recovery after treated with BBP-671 therapeutic
1H MRS is a powerful and non-invasive technique which can provide information about neurometabolites in vivo [11, 12]. 1H MRS can identify biomarkers and be applied to evaluate clinical neurodegeneration and therapeutic strategies [13, 14]. In the limited clinical studies demonstrating the application of 1H MRS in studying PKAN, a reduced N-acetyl aspartate (NAA) level was commonly observed [15–18]. This is the first report using 1H MRS for monitoring PKAN therapeutics in a mouse model of the disease. In this study, we treated SynCre+Pank1,2 neuronal dKO mice with BBP-671 and used 1H MRS to identify the prominent neurochemical metabolites and evaluate treatment response (Fig. 2). We found that the cerebral metabolite levels, including glutamate + glutamine (Glx), NAA and lactate, were altered in the mid-brain of the animal model. Based on our observations, we show that Glx levels may be an indicator of restored CoA levels in the response to BBP-671 treatment.
Fig. 2.
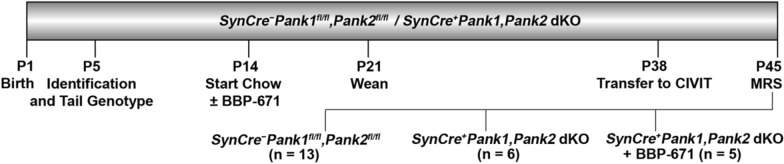
Timeline of events extended from birth at postnatal day 1 (P1) through P45 when animals were anesthetized and midbrains were evaluated by magnetic resonance spectroscopy (MRS). Individual animals were identified and tail genotyping was performed at P5. Animals were assigned to three groups: SynCre+Pank1,2 dKO ± BBP-671 and SynCre−Pank1fl/fl,Pank2fl/fl matched wildtype controls, and were housed with control nursing dams at P14. Animals were maintained on the purified chow diet ± 75 ppm BBP-671 starting at P14 through P45, and weaned at P21. Mice were transferred to the Center for In Vivo Imaging and Therapeutics (CIVIT) at approximately P38 and MRS was performed at approximately P45. Numbers of mice in each group are indicated in parentheses
Methods
Animal care and use
All procedures were reviewed and approved by the St. Jude Children’s Research Hospital Institutional Animal Care and Use Committee. The SynCre+Pank1,Pank2 neuronal dKO mice were generated as described in Sharma et al. [10]. The mice were maintained at room temperature, humidity 50 ± 10% and a 14/10-h light/dark cycle. Animals were maintained on a purified chow diet (Envigo TD.170542) ± 75 ppm BBP-671. Mice were put on study on a rolling basis as they emerged from the breeding program and were randomized into the treatment arms at postnatal day 14 (P14). Water was supplied ad libitum. SynCre+Pank1,2 neuronal dKO (n = 5) received BBP-671 fortified chow for approximately 31 days. The steady state levels of BBP-671 in plasma (1 mM) from mice on 75 ppm BBP-671 chow were determined 2–4 h after cessation of the dark cycle by mass spectrometry analyses as described previously [10] and found to be consistent within the treatment group. SynCre+Pank1,2 neuronal dKO (n = 6) and SynCre−Pank1fl/fl,Pank2fl/fl wild type controls (n = 13) received matched chow without BBP-671 for the same length of time. MRS was performed on mice at an age of 6–7 weeks (P42–P49). In a separate study, the SynCre+Pank1,Pank2 neuronal dKO mice treated with BBP-671 had improved lifespan, weight gain and locomotion similar to previous results using PZ-2891 [10]. Neuron-specific deletion of Pank1 and Pank2 was confirmed by PCR genotyping of brain and liver, and BBP-671 levels in brains were confirmed postmortem as described previously [10].
1H magnetic resonance spectroscopy in mice
MRI/MRS studies were performed on a Bruker Clinscan 7T magnetic resonance imaging (MRI) scanner (Bruker BioSpin MRI GmbH, Ettlingen, Germany). Mice were anesthetized using isoflurane mixed with oxygen (1–2%) and the respiration rate was monitored. The total scan time was 21 min. MRI was acquired with a mouse brain surface receive coil positioned over the mouse head and placed inside a 72 mm transmit/receive coil. After the localizer, a T2-weighted turbo spin echo sequence was performed in the coronal (TR/TE = 2290/41 ms, matrix size = 192 × 256, slice thickness 0.5 mm, number of slices = 14) and axial (TR/TE = 3841/50 ms, matrix size = 192 × 144, slice thickness 0.4 mm, number of slices = 42) orientations. The T2-weighted scans were used to position a 3.5 × 4.5 × 2.0 mm3 voxel for spectroscopy in the midbrain to cover thalamus and hippocampus. A 1H MR spectrum was generated with that voxel using a PRESS sequence (repetition time/echo time = 3000/11 ms, averages = 128, data length = 2048, spectral width = 2900 Hz). Voxel positioning is displayed in Additional file 1: Fig. S2.
1H MRS data processing
Metabolite to tCr ratios measured by in vivo 1H MRS were quantified using LCModel software (v.6.3), a widely applied MRS analysis tool that employs a least-squares-based prior-knowledge fitting program [19]. LCModel applied a 7 T spin echo (TE = 11 ms) basis set incorporating the following resonances: alanine (Ala), aspartic acid (Asp), creatine (Cre), phosphocreatine, g-amino butyric acid (GABA), glucose, glutamine (Gln), glutamate (Glu), glycerophosphocholine, phosphocholine, glutathione, myo-inositol (m-Ins), N-acetyl aspartate (NAA), NAA + Glu, sycllo-inositol and taurine, with lipid resonances at 0.9, 1.3 and 2.0 ppm and macromolecule resonances at 0.9, 1.2, 1.4, 1.7 and 2.0 ppm. The unsuppressed water signal was not acquired, therefore metabolite concentrations are reported relative to total creatine (tCr) as applied by others [20–23].
Statistics
Mean and standard deviation were calculated for MRS parameters in each mouse group. Wilcoxon Rank sum tests were used to test whether MRS parameters are different between wild type, SynCre+Pank1,Pank2 dKO or SynCre+Pank1,Pank2 dKO + BBP-671 groups. Bonferroni correction was done for the nine comparisons, and thus a p-value < 0.05 was deemed to be statistically significant. All the analyses were done using ‘R’ 4.0.2.
Results
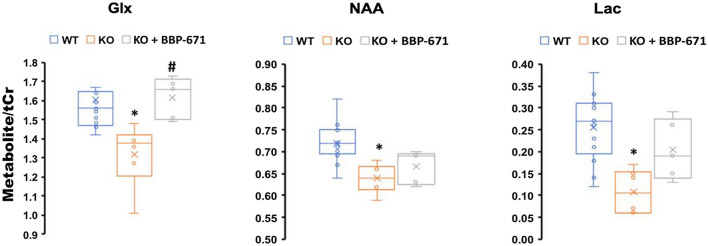
The SynCre+Pank1,2 neuronal dKO mice show clear physiological symptoms of CoA deficiency such as reduced growth rate and impaired locomotor activity [10]. Figure 3 represents the 1H MRS spectra acquired from the midbrain of representative mice in each group. There is a clear reduction in the Glx/tCr, NAA/tCr and lactate/tCr ratios in the midbrains of the SynCre+Pank1,2 neuronal dKO mice compared to WT (Wildtype). Glx is the summed group of Glu and Gln, where Glu is the most abundant excitatory neurotransmitter in brain and Gln is the main precursor for Glu [13, 24]. Reduction of Glx points toward impaired excitatory neuronal metabolism in the dKO mice and Glx fully recovered following treatment with BBP-671 (Fig. 4, Additional file 1: Table S1). Reduction of NAA indicated loss of neuronal integrity and function, and the NAA/tCr ratio trended toward improvement but was not significant following BBP-671 treatment. Reduction of NAA in the globus pallidus is consistently reported in patients with PKAN [15–18]. The lactate/tCr ratio was lower in the dKO mice representing the net balance between lactate production and consumption [13]. Reduced cerebral glycolysis and/or enhanced lactate oxidation to pyruvate to maintain the redox state in the neurons is suggested from the data. The lactate/tCr ratio trended toward improvement with treatment but without statistical significance. The metabolic recovery following therapy, together with a strong signal strength in the 1H MRS points to Glx as a promising biomarker. NAA and lactate are also candidate markers but require further study. While no changes were observed, additional metabolites—inositol, choline, and taurine—are shown in Additional file 1: Fig. S1.
Fig. 3.
Representative 1H MR spectra from three groups a Wild Type, b Neural Pank1,2 dKO, and c Neural Pank1,2 dKO mice treated with BBP-671
Fig. 4.
Metabolite to total creatine ratios of glutamate + glutamine (Glx), N-acetyl aspartate (NAA) and lactate (Lac); *p < 0.05 (WT vs KO); #p < 0.05 (KO vs. KO + BBP-671). The box in the box and whisker plot represents the first and third quartiles. The line within the box represents the median while the ‘x’ within the box represents the mean
Discussion
The treatment response to BBP-671 was associated with recovery of cerebral Glx. BBP-671 treatment activates the PANK3 isoform to increase CoA production that, in turn, has a direct role on TCA cycle metabolism (Fig. 1) [5] and hence Glx production. In contrast to our findings, a study performed on 3 human PKAN patients reported an increase of Glx in the white matter [25]. However, the clinical study was performed in patients with chronic disease progression and potential involvement of multiple brain cell types while this preclinical study was performed in a mouse model with rapidly progressing disease that is specific to neurons. Although neurons have been identified as a focal target of disease in PKAN [26], the role of glial cells in disease progression, for example, is not understood. A clinical MRS study reported elevated myo-inositol (m-Ins) levels in white matter due to gliosis and glial proliferation in patients with PKAN [16]. However, we did not find any significant changes in the m-Ins levels in the SynCre+Pank1,2 neuronal dKO mice.
The SynCre+Pank1,2 neuronal dKO mouse model represents only selected molecular effects of PKAN, and the study has limitations. Alternative mouse models were not appropriate for evaluation of cerebral MRS either due to the lack of disease-related movement dysfunction and normal lifespan [27] or early postnatal death [4, 28]. Analysis of human PKAN brains identified gene expression signatures that indicated neurons as a focal target of disease [26] and development of the SynCre+Pank1,2 neuronal dKO mouse model resulted in a longer-lived animal with consistent and measurable movement dysfunction [13] that was tractable for MRS. Previous studies reported white matter pathology in the patients of PKAN [29]. The previous 1H MRS studies in human patients were often performed in white matter [18], which is not easily detected in the mouse model since the mouse brain consists of less than 12% white matter [30], as compared to 43% white matter in humans [31]. Mouse models are recognized as not effective for studying white matter pathologies [32]. In addition, there was no observable iron accumulation in the model, in contrast with the iron accumulation often observed in the basal ganglia of PKAN patients. This study may be limited to detection of metabolic changes in the gray matter in the Pank1,2 neuronal dKO mice.
PKAN is a life-threatening neurological disorder that can be caused by a variety of PANK2 mutations that affect protein expression, enzyme activity and/or stability [33, 34], resulting in loss or reduction of kinase activity [33]. PKAN diagnosis is based on characteristics of the movement dysfunction, exon sequencing and MRI evidence for iron accumulation in the basal ganglia [35] with a characteristic “eye-of-the-tiger” pattern in T2-weighted images [25, 36]. Our study showed no qualitative differences among the three groups in the T2-weighted images. There are currently no approved clinical therapies for this genetic disorder [37]. Progress with newly developing PKAN therapeutics has been made by evaluating mouse models with deactivated Pank genes in which brain CoA biosynthesis has been disrupted [9, 10, 38]. One of the current challenges is to reliably identify molecular events associated with PKAN dysfunction in addition to monitoring therapeutic responses [39]. Thus, a technique which can quantitatively, non-invasively, and reproducibly analyze neurometabolism is needed and has potential value.
Conclusion
In summary, we have successfully applied 1H MRS to investigate neuronal chemical alterations in a mouse model of brain CoA deficiency, thought to be an underlying cause of PKAN. The effects of a potential PKAN therapeutic were evaluated by comparing the cerebral metabolic derangements among three mouse groups, where reductions of important metabolites (Glx, NAA, and Lactate) were observed in the model of brain CoA deficiency and recoveries of the same metabolites were found following treatment with a newly developed Pantazine, BBP-671. The most promising biomarker for this potential PKAN therapeutic was the recovery of the Glx/tCr ratio. This study shows that 1H MRS can be a powerful tool in evaluating therapeutics for metabolic responses of neurological disorders.
Supplementary Information
Additional file 1: Table S1. Mean values and standard deviations for Fig. 4. Table S2. The p values for Fig. 4. Figure S1. Metabolite to total creatine ratio for m-inositol, total choline, and taurine. KO, untreated Pank1/2 neuronal dKO mice; KO + BBP-671, BBP-671–treated Pank1/2 neuronal dKO mice; WT, wild-type. Figure S2. Voxel Positioning for Fig. 2 shown in a wild-type mouse. The viewpoints are A) Horizontal; B) Sagittal; C) Coronal.
Acknowledgements
We would like to acknowledge our colleagues in the Center for In Vivo Imaging and Therapeutics (CIVIT) of our institute for the MRI/MRS scanning of the mice. Graphpad Prism, LCModel, and Syngo.via were used in the making of figures. Excellent technical support was provided by Lois Richmond and Karen Miller.
Authors’ contributions
SJ, COR and PB conceived the study. JS, CS and WA performed in vivo MRI/MRS experiments. YL, JS, ZC, SW and PB performed and reviewed MRI/MRS data analysis. PB and SJ oversaw and guided experiments and analysis. YL performed the statistical analysis. YL, ZC and PB wrote the manuscript, and all authors reviewed and provided comments on the manuscript. All authors read and approved the final manuscript.
Funding
The study was funded by NIGMS Grant GM034496 (COR), CoA Therapeutics, Inc. (SJ), and the American Lebanese Syrian Associated Charities at St. Jude Children’s Research Hospital.
Declarations
Ethics approval and consent to participate
This study was limited to animals, and the statement mentioning the study protocol bein approved by an institutional ethics committee is already present in the Methods section.
Consent for publication
Not applicable.
Competing interests
SJ is a member of the Scientific Advisory Board of CoA Therapeutics, Inc. SJ, COR and CS are co-inventors of the patent #US2021/0061788. Other authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Leonardi R, Zhang Y-M, Rock CO, Jackowski S. Coenzyme A: back in action. Prog Lipid Res. 2005;44:125–153. doi: 10.1016/j.plipres.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Srinivasan B, Sibon OC. Coenzyme A, more than ‘just’ a metabolic cofactor. Biochem Soc Trans. 2014;42:1075–1079. doi: 10.1042/BST20140125. [DOI] [PubMed] [Google Scholar]
- 3.Zhou B, Westaway SK, Levinson B, Johnson MA, Gitschier J, Hayflick SJ. A novel pantothenate kinase gene (PANK2) is defective in Hallervorden-Spatz syndrome. Nat Genet. 2001;28:345–349. doi: 10.1038/ng572. [DOI] [PubMed] [Google Scholar]
- 4.Subramanian C, Yao J, Frank MW, Rock CO, Jackowski S. A pantothenate kinase-deficient mouse model reveals a gene expression program associated with brain coenzyme a reduction. Biochim Biophys Acta. 2020;1866:165663. doi: 10.1016/j.bbadis.2020.165663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subramanian C, Frank MW, Tangallapally R, Yun MK, Edwards A, White SW, Lee RE, Rock CO, Jackowski S. Pantothenate kinase activation relieves coenzyme A sequestration and improves mitochondrial function in mice with propionic acidemia. Sci Transl Med. 2021;13(611):eabf5965. doi: 10.1126/scitranslmed.abf5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martínez-Reyes I, Chandel NS. Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun. 2020;11:1–11. doi: 10.1038/s41467-019-13668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veyrat-Durebex C, Corcia P, Piver E, Devos D, Dangoumau A, Gouel F, Vourc’h, P., Emond, P., Laumonnier, F., and Nadal-Desbarats, L. Disruption of TCA cycle and glutamate metabolism identified by metabolomics in an in vitro model of amyotrophic lateral sclerosis. Mol Neurobiol. 2016;53:6910–6924. doi: 10.1007/s12035-015-9567-6. [DOI] [PubMed] [Google Scholar]
- 8.Mignani L, Gnutti B, Zizioli D, Finazzi D. Coenzyme A biochemistry: from neurodevelopment to neurodegeneration. Brain Sci. 2021;11:1031. doi: 10.3390/brainsci11081031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thakur N, Klopstock T, Jackowski S, Kuscer E, Tricta F, Videnovic A, Jinnah HA. Rational design of novel therapies for pantothenate kinase-associated neurodegeneration. Mov Disord. 2021;36(9):2005–2016. doi: 10.1002/mds.28642. [DOI] [PubMed] [Google Scholar]
- 10.Sharma LK, Subramanian C, Yun M-K, Frank MW, White SW, Rock CO, Lee RE, Jackowski S. A therapeutic approach to pantothenate kinase associated neurodegeneration. Nat Commun. 2018;9:1–15. doi: 10.1038/s41467-017-02088-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duarte JM, Lei H, Mlynárik V, Gruetter R. The neurochemical profile quantified by in vivo 1H NMR spectroscopy. Neuroimage. 2012;61:342–362. doi: 10.1016/j.neuroimage.2011.12.038. [DOI] [PubMed] [Google Scholar]
- 12.Preul MC, Caramanos Z, Collins DL, Villemure J-G, Leblanc R, Olivier A, Pokrupa R, Arnold DL. Accurate, noninvasive diagnosis of human brain tumors by using proton magnetic resonance spectroscopy. Nat Med. 1996;2:323–325. doi: 10.1038/nm0396-323. [DOI] [PubMed] [Google Scholar]
- 13.Choi JK, Dedeoglu A, Jenkins BG. Application of MRS to mouse models of neurodegenerative illness. NMR Biomed. 2007;20:216–237. doi: 10.1002/nbm.1145. [DOI] [PubMed] [Google Scholar]
- 14.Yang S, Salmeron BJ, Ross TJ, Xi Z-X, Stein EA, Yang Y. Lower glutamate levels in rostral anterior cingulate of chronic cocaine users—a 1H-MRS study using TE-averaged PRESS at 3 T with an optimized quantification strategy. Psychiatry Re Neuroimaging. 2009;174:171–176. doi: 10.1016/j.pscychresns.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Habibi AH, Razmeh S, Aryani O, Rohani M, Taghavian L, Alizadeh E, Moradian Kokhedan K, Zaribafian M. A novel homozygous variation in the PANK2 gene in two Persian siblings with atypical pantothenate kinase associated neurodegeneration. Neurol Int. 2019;11:9–11. doi: 10.4081/ni.2019.7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitis O, Tekgul H, Erdemir G, Polat M, Serdaroglu G, Tosun A, Coker M, Gokben S. Identification of axonal involvement in Hallervorden-Spatz disease with magnetic resonance spectroscopy. J Neuroradiol. 2006;33:129–132. doi: 10.1016/S0150-9861(06)77244-3. [DOI] [PubMed] [Google Scholar]
- 17.Parashari UC, Aga P, Parihar A, Singh R, Joshi V. Case report: MR spectroscopy in pantothenate kinase-2 associated neurodegeneration. Indian J Radiol Imaging. 2010;20:188. doi: 10.4103/0971-3026.69353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sener RN. Pantothenate kinase-associated neurodegeneration: MR imaging, proton MR spectroscopy, and diffusion MR imaging findings. Am J Neuroradiol. 2003;24:1690–1693. [PMC free article] [PubMed] [Google Scholar]
- 19.Provencher SW. Automatic quantitation of localized in vivo 1H spectra with LCModel. NMR Biomed. 2001;14:260–264. doi: 10.1002/nbm.698. [DOI] [PubMed] [Google Scholar]
- 20.Ackl N, Ising M, Schreiber YA, Atiya M, Sonntag A, Auer DP. Hippocampal metabolic abnormalities in mild cognitive impairment and Alzheimer's disease. Neurosci Lett. 2005;384:23–28. doi: 10.1016/j.neulet.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 21.Frederick BD, Lyoo IK, Satlin A, Ahn KH, Kim MJ, Yurgelun-Todd DA, Cohen BM, Renshaw PF. In vivo proton magnetic resonance spectroscopy of the temporal lobe in Alzheimer's disease. Prog Neuropsychopharmacol Biol Psychiatry. 2004;28:1313–1322. doi: 10.1016/j.pnpbp.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 22.Oberg J, Spenger C, Wang FH, Andersson A, Westman E, Skoglund P, Sunnemark D, Norinder U, Klason T, Wahlund LO, Lindberg M. Age related changes in brain metabolites observed by 1H MRS in APP/PS1 mice. Neurobiol Aging. 2008;29:1423–1433. doi: 10.1016/j.neurobiolaging.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Kickler N, Lacombe E, Chassain C, Durif F, Krainik A, Farion R, Provent P, Segebarth C, Rémy C, Savasta M. Assessment of metabolic changes in the striatum of a rat model of parkinsonism: an in vivo (1)H MRS study. NMR Biomed. 2009;22:207–212. doi: 10.1002/nbm.1305. [DOI] [PubMed] [Google Scholar]
- 24.Ramadan S, Lin A, Stanwell P. Glutamate and glutamine: a review of in vivo MRS in the human brain. NMR Biomed. 2013;26:1630–1646. doi: 10.1002/nbm.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hájek M, Adamovičová M, Herynek V, Škoch A, Jírů F, Křepelová A, Dezortová M. MR relaxometry and 1H MR spectroscopy for the determination of iron and metabolite concentrations in PKAN patients. Eur Radiol. 2005;15:1060–1068. doi: 10.1007/s00330-004-2553-4. [DOI] [PubMed] [Google Scholar]
- 26.Bettencourt C, Forabosco P, Wiethoff S, Heidari M, Johnstone DM, Botía JA, Collingwood JF, Hardy J, Milward EA, Ryten M, Houlden H, (UKBEC), U. B. E. C Gene co-expression networks shed light into diseases of brain iron accumulation. Neurobiol Dis. 2016;87:59–68. doi: 10.1016/j.nbd.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuo YM, Duncan JL, Westaway SK, Yang H, Nune G, Xu EY, Hayflick SJ, Gitschier J. Deficiency of pantothenate kinase 2 (Pank2) in mice leads to retinal degeneration and azoospermia. Hum Mol Genet. 2005;14(1):49–57. doi: 10.1093/hmg/ddi005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia M, Leonardi R, Zhang YM, Rehg JE, Jackowski S. Germline deletion of pantothenate kinases 1 and 2 reveals the key roles for CoA in postnatal metabolism. PLoS ONE. 2012;7(7):e40871. doi: 10.1371/journal.pone.0040871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoeter P, Roa-Sanchez P, Speckter H, Perez-Then E, Foerster B, Vilchez C, Oviedo J, Rodriguez-Raecke R. Changes of cerebral white matter in patients suffering from pantothenate kinase-associated neurodegeneration (PKAN): a diffusion tensor imaging (DTI) study. Parkinsonism Relat Disord. 2015;21:577–581. doi: 10.1016/j.parkreldis.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 30.Dorr A, Lerch JP, Spring S, Kabani N, Henkelman RM. High resolution three-dimensional brain atlas using an average magnetic resonance image of 40 adult C57Bl/6J mice. Neuroimage. 2008;42:60–69. doi: 10.1016/j.neuroimage.2008.03.037. [DOI] [PubMed] [Google Scholar]
- 31.Harris GJ, Barta PE, Peng LW, Lee S, Brettschneider PD, Shah A, Henderer JD, Schlaepfer TE, Pearlson GD. MR volume segmentation of gray matter and white matter using manual thresholding: dependence on image brightness. AJNR Am J Neuroradiol. 1994;15:225–230. [PMC free article] [PubMed] [Google Scholar]
- 32.Woltjer RL, Reese LC, Richardson BE, Tran H, Green S, Pham T, Chalupsky M, Gabriel I, Light T, Sanford L. Pallidal neuronal apolipoprotein E in pantothenate kinase-associated neurodegeneration recapitulates ischemic injury to the globus pallidus. Mol Genet Metab. 2015;116:289–297. doi: 10.1016/j.ymgme.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y-M, Rock CO, Jackowski S. Feedback regulation of murine pantothenate kinase 3 by coenzyme A and coenzyme A thioesters. J Biol Chem. 2005;280:32594–32601. doi: 10.1074/jbc.M506275200. [DOI] [PubMed] [Google Scholar]
- 34.Hong BS, Senisterra G, Rabeh WM, Vedadi M, Leonardi R, Zhang Y-M, Rock CO, Jackowski S, Park H-W. Crystal structures of human pantothenate kinases: insights into allosteric regulation and mutations linked to a neurodegeneration disorder. J Biol Chem. 2007;282:27984–27993. doi: 10.1074/jbc.M701915200. [DOI] [PubMed] [Google Scholar]
- 35.Hogarth P, Kurian MA, Gregory A, Csányi B, Zagustin T, Kmiec T, Wood P, Klucken A, Scalise N, Sofia F. Consensus clinical management guideline for pantothenate kinase-associated neurodegeneration (PKAN) Mol Genet Metab. 2017;120:278–287. doi: 10.1016/j.ymgme.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Schneider SA, Hardy J, Bhatia K. Iron accumulation in syndromes of neurodegeneration with brain iron accumulation 1 and 2: causative or consequential? J Neurol Neurosurg Psychiatry. 2009;80:589–590. doi: 10.1136/jnnp.2008.169953. [DOI] [PubMed] [Google Scholar]
- 37.Razmeh S, Habibi AH, Orooji M, Alizadeh E, Moradiankokhdan K, Razmeh B. Pantothenate kinase-associated neurodegeneration: clinical aspects, diagnosis and treatments. Neurol Int. 2018;10:32–34. doi: 10.4081/ni.2018.7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeong SY, Hogarth P, Placzek A, Gregory AM, Fox R, Zhen D, Hamada J, van der Zwaag M, Lambrechts R, Jin H, Nilsen A, Cobb J, Pham T, Gray N, Ralle M, Duffy M, Schwanemann L, Rai P, Freed A, Wakeman K, Woltjer RL, Sibon OC, Hayflick SJ. 4′-Phosphopantetheine corrects CoA, iron, and dopamine metabolic defects in mammalian models of PKAN. EMBO Mol Med. 2019;11:e10489. doi: 10.15252/emmm.201910489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackowski S. Proposed therapies for pantothenate-kinase-associated neurodegeneration. J Exp Neurosci. 2019;13:1179069519851118. doi: 10.1177/1179069519851118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Mean values and standard deviations for Fig. 4. Table S2. The p values for Fig. 4. Figure S1. Metabolite to total creatine ratio for m-inositol, total choline, and taurine. KO, untreated Pank1/2 neuronal dKO mice; KO + BBP-671, BBP-671–treated Pank1/2 neuronal dKO mice; WT, wild-type. Figure S2. Voxel Positioning for Fig. 2 shown in a wild-type mouse. The viewpoints are A) Horizontal; B) Sagittal; C) Coronal.